Insights into mechanisms coordinating membrane remodeling, local actin nucleation, and postsynaptic scaffolding during postsynapse formation are important for understanding vertebrate brain function. Gene knockout and RNAi in individual neurons reveal that the F-BAR protein syndapin I is a crucial postsynaptic coordinator in formation of excitatory synapses. Syndapin I deficiency caused significant reductions of synapse and dendritic spine densities. These syndapin I functions reflected direct, SH3 domain–mediated associations and functional interactions with ProSAP1/Shank2. They furthermore required F-BAR domain-mediated membrane binding. Ultra-high-resolution imaging of specifically membrane-associated, endogenous syndapin I at membranes of freeze-fractured neurons revealed that membrane-bound syndapin I preferentially occurred in spines and formed clusters at distinct postsynaptic membrane subareas. Postsynaptic syndapin I deficiency led to reduced frequencies of miniature excitatory postsynaptic currents, i.e., to defects in synaptic transmission phenocopying ProSAP1/Shank2 knockout, and impairments in proper synaptic ProSAP1/Shank2 distribution. Syndapin I–enriched membrane nanodomains thus seem to be important spatial cues and organizing platforms, shaping dendritic membrane areas into synaptic compartments.
Introduction
Brain development and function relies on remodeling of neuronal membranes. The postsynapses of glutamatergic neurons often protrude from dendrites as dendritic spines and contain neurotransmitter receptors and signaling components interconnected by scaffold proteins, such as ProSAP/Shanks, which additionally interact with F-actin binding proteins (Hering and Sheng, 2003; Qualmann et al., 2004). Receptor clustering and linkage to F-actin thus represent two prominent mechanisms in synaptogenesis (Tada and Sheng, 2006). ProSAP/Shank deficiencies led to reduced synapse or spine densities and seem related to autism spectrum disorders (Grabrucker et al., 2011; Peça et al., 2011; Berkel et al., 2012; Schmeisser et al., 2012). Yet, the mechanisms that shape dendritic spines during formation and help to coordinate membrane remodeling, local actin nucleation, and postsynaptic scaffold formation remain elusive.
Membrane shaping can be mediated by cytoskeletal forces and membrane-associated proteins. Syndapins (PACSINS) belong to the F-BAR subfamily of BAR domain proteins that are thought to shape membranes by scaffolding and/or partial insertion into one membrane leaflet (Qualmann et al., 2011). Syndapins have the potential to combine cytoskeletal and membrane shaping mechanisms. They interact with proteins promoting actin filament formation, interconnect SH3 domain binding partners via F-BAR domain–mediated self-association, and bind to membranes via their F-BAR domain (Qualmann et al., 1999; Itoh et al., 2005; Kessels and Qualmann, 2006; Dharmalingam et al., 2009; Schwintzer et al., 2011). The first loss-of-function analyses have indeed revealed a role for syndapin I in membrane shaping processes. Syndapin I knockout (KO) mice had defects in retrieval and shaping synaptic vesicles in presynapses (Koch et al., 2011). Syndapin I was furthermore found to be crucial for early neuromorphogenesis and for ciliogenesis (Dharmalingam et al., 2009; Schwintzer et al., 2011; Schüler et al., 2013).
Considering its molecular and functional properties, we addressed a potential role of syndapin I in postsynapse formation by gene KO and by RNAi at specifically postsynaptic sides of evaluated excitatory synapses. Our biochemical and functional studies demonstrated that syndapin I plays a crucial role in dendritic spine and synapse formation based on SH3 domain–mediated associations with ProSAP1/Shank2 and F-BAR domain–mediated membrane binding. Consistently, syndapin I RNAi led to impairments in synaptic activity similar to ProSAP1/Shank2 KO and to defects in ProSAP2/Shank1 organization. Imaging of specifically membrane-associated, endogenous syndapin I at ultra-high resolution revealed that it preferentially occurred in spines and formed clusters at membrane subareas of spines. Membrane-bound syndapin I nanodomains thereby can provide spatial cues and molecular organizing platforms during the formation of postsynapses.
Results
Neurons of syndapin I KO mice have reduced densities of dendritic spines
Syndapin I KO mice suffer from generalized seizures correlating with altered neuronal network activity (Koch et al., 2011). To evaluate whether defects in synaptic organization may contribute to this phenotype, we analyzed Golgi-stained CA1 hippocampal sections. The density of spines protruding from syndapin I KO neurons was decreased by ∼15% when compared with wild type (WT; Fig. 1, A and B).
Significant reduction of dendritic spine density in neurons of syndapin I KO mice. (A) Maximal intensity projections of inverted brightfield images of dendrites of Golgi-stained hippocampal CA1 neurons from adult WT (+/+) and syndapin I KO mouse (−/−) brain sections (top) and corresponding reconstruction and classification of dendritic spines with Imaris software showing stubby (red), thin (blue), and mushroom spines (green) as well as filopodia-like spines (magenta; bottom). Thin, white lines are Imaris evaluation grids. Bar, 4 µm. (B–F) Quantitative analyses of all types of spiny protrusions. **, P < 0.01; ***, P < 0.001. Data represent mean ± SEM (error bars).
Significant reduction of dendritic spine density in neurons of syndapin I KO mice. (A) Maximal intensity projections of inverted brightfield images of dendrites of Golgi-stained hippocampal CA1 neurons from adult WT (+/+) and syndapin I KO mouse (−/−) brain sections (top) and corresponding reconstruction and classification of dendritic spines with Imaris software showing stubby (red), thin (blue), and mushroom spines (green) as well as filopodia-like spines (magenta; bottom). Thin, white lines are Imaris evaluation grids. Bar, 4 µm. (B–F) Quantitative analyses of all types of spiny protrusions. **, P < 0.01; ***, P < 0.001. Data represent mean ± SEM (error bars).
More detailed analyses revealed that filopodia-like spine density was not changed (Fig. 1 C). Instead, the density of thin spines declined by 18%, and the densities of stubby and mushroom spines declined by 25% and 27%, respectively (Fig. 1, D–F).
Synapse formation requires postsynaptic SH3 domain–dependent syndapin I functions
In line with putative postsynaptic functions, syndapin I did not just localize presynaptically; a subpool of syndapin I was found in dendrites and colocalized with the postsynaptic marker PSD-95. Tracing the complete morphology of neurons with GFP confirmed presence of endogenous syndapin I in the dendritic/postsynaptic compartment. Also, Xpress–syndapin I clearly localized to both axonal/presynaptic and dendritic/postsynaptic compartments (Fig. S1, A–G).
To evaluate whether the loss of postsynaptic syndapin I specifically underlies the impaired formation of dendritic spines observed in syndapin I KO brains (Fig. 1), we used primary hippocampal cultures transfected with syndapin I RNAi plasmids (Dharmalingam et al., 2009). This allowed for analysis of the spines of syndapin I–depleted cells and thereby for evaluation of specifically postsynaptic effects of syndapin I depletion (Fig. 2, A–D). As observed in brains of syndapin I KO animals (−27%; Fig. 1 F), the density of mushroom spines was reduced in syndapin I–depleted neurons when compared with controls (−35%; Fig. 2, A and C).
Impaired spine and synapse formation upon syndapin I loss-of-function is caused by a loss of SH3 domain-dependent syndapin I functions in the postsynaptic compartment. (A and H) PM-mCherry signals of dendrites of neurons transfected as indicated at DIV 12 and fixed at DIV 14. Bars, 5 µm. (B–D) Quantitative analyses of general spine density (B) and of individual morphology groups (C and D) upon syndapin I RNAi. (E) Anti–PSD-95 (postsynaptic) and anti–synapsin 1 (presynaptic) immunolabeling along dendrites of transfected neurons. Bar, 5 µm. (F and G) Quantitation of PSD-95– (F) and synapsin 1–positive puncta (G) spatially overlapping with transfected neurons. (I–K) Quantitative analyses of general spine density (I) and of individual morphology groups (J and K) of syndapin I–depleted cells expressing Sdp IΔSH3 compared with pRNAT control cells transfected in parallel. **, P < 0.01; ***, P < 0.001. Data represent mean ± SEM (error bars).
Impaired spine and synapse formation upon syndapin I loss-of-function is caused by a loss of SH3 domain-dependent syndapin I functions in the postsynaptic compartment. (A and H) PM-mCherry signals of dendrites of neurons transfected as indicated at DIV 12 and fixed at DIV 14. Bars, 5 µm. (B–D) Quantitative analyses of general spine density (B) and of individual morphology groups (C and D) upon syndapin I RNAi. (E) Anti–PSD-95 (postsynaptic) and anti–synapsin 1 (presynaptic) immunolabeling along dendrites of transfected neurons. Bar, 5 µm. (F and G) Quantitation of PSD-95– (F) and synapsin 1–positive puncta (G) spatially overlapping with transfected neurons. (I–K) Quantitative analyses of general spine density (I) and of individual morphology groups (J and K) of syndapin I–depleted cells expressing Sdp IΔSH3 compared with pRNAT control cells transfected in parallel. **, P < 0.01; ***, P < 0.001. Data represent mean ± SEM (error bars).
Reintroduction of syndapin I rescued the syndapin I RNAi phenotype. Thus, the observed RNAi-induced impairments in dendritic spine formation are specifically due to loss of syndapin I in the same cell (Fig. 2, A–D).
In line with the impaired formation of mushroom spines, anti–PSD-95 immunolabeling showed that postsynaptic structures were less abundant at dendrites of syndapin I–depleted cells when compared with control cells (Fig. 2, E and F; absolute numbers, Fig. S2 A). Anti–synapsin 1 immunolabeling also showed that presynapses spatially overlapping with dendrites of transfected cells were less frequent (Fig. 2, E and G; and Fig. S2 B).
These observations reflected the fact that the majority of mushroom spines displayed anti–PSD-95 immunolabeling and were contacted by synapsin 1–marked presynapses, highlighting the crosstalk between pre- and postsynapses during neuronal network formation (Fig. S2, C–F). Both syndapin I RNAi phenotypes were specific, as shown by rescue experiments (Fig. 2, E–G). Consistently, syndapin I overexpression had opposite effects, i.e., increased spine densities, in particular of stubby and mushroom spines, whereas, similar to syndapin I KO (Fig. 1), the filopodia-containing group of dendritic protrusions was not affected (Fig. S2, G–K). In line with these data, syndapin I overexpression also increased synapse densities (Fig. S2, L–N).
Interestingly, expression of RNAi-insensitive syndapin IΔSH3 failed to rescue the syndapin I loss-of-function phenotypes, and both postsynapse and spine formation were strongly impaired when compared with corresponding controls (Fig. 2, E–K; and Fig. S2, O and P).
Identification of ProSAP1/Shank2 and ProSAP2/Shank3 as postsynaptically enriched syndapin I interaction partners
Our studies revealed that syndapin I plays an important role in formation of postsynapses, yet, its interaction partners in this specialized compartment were unknown. Yeast two-hybrid screening with syndapin I did not identify any established PSD-enriched SH3 domain interaction candidates (unpublished data). However, genome-wide in silico searches using a recently identified consensus for syndapin SH3 domain interactions (+++APPPP; Schwintzer et al., 2011) led to identification of two postsynaptic candidates, ProSAP1/Shank2 and ProSAP2/Shank3. Because ProSAP1/Shank2 (for simplicity, referred to as ProSAP1 throughout) was described as the isoform of the ProSAP/Shank family appearing at synapses first (Grabrucker et al., 2011), we mainly concentrated on ProSAP1. Coprecipitation analyses with immobilized GST–syndapin I indeed revealed interactions with GFP-ProSAP1 (Fig. 3 A). The same was observed for syndapin II and III (Fig. 3 A).
Identification of ProSAP1/Shank2 and ProSAP2/Shank3 as postsynaptically enriched Syndapin I interaction partners. (A) GST–syndapin I, II, and III specifically precipitate GFP-ProSAP1 expressed in HEK293 cells. (B) Coprecipitation analysis with GST–syndapin I and deletion mutants thereof. The SH3 domain is critical and sufficient for binding. A mutant SH3 domain (P434L; SH3*) did not bind. White lines indicate lanes omitted from blots (B and F). (C) Syndapin I SH3 precipitates GFP-ProSAP1 and GFP-ProSAP2 but not GFP-Shank1. (D) Alignment of +++APPPP motifs in ProSAP1 (NCBI Protein database accession no. NP_001004133), ProSAP2 (accession no. NP_067708), and Cobl (accession no. NP_766084; conserved amino acids are highlighted) and of corresponding residues in Shank1 (accession no. Q9WV48). (E) Scheme of rat ProSAP1b and deletion mutants used. Indicated are the N-terminal PDZ domain (medium grey), several proline-rich motifs (dark grey lines), and the C-terminal SAM (sterile alpha motif) domain (light grey). (F) GST–syndapin I precipitated GFP-ProSAP1 1–235 but none of the other ProSAP1 deletion mutants. (G) GFP fusion peptides encompassing the +++APPPP motifs of ProSAP1, ProSAP2, and Cobl associated with syndapin I SH3. (H and I) RKKAPPPPKR to GAGAAAAAAG mutation (amino acids 141–150 in ProSAP1; ProSAP1 1–235*) disrupted direct binding of ProSAP1 to syndapin I in both in vitro reconstitutions with purified proteins (H) and in coprecipitation analyses (I).
Identification of ProSAP1/Shank2 and ProSAP2/Shank3 as postsynaptically enriched Syndapin I interaction partners. (A) GST–syndapin I, II, and III specifically precipitate GFP-ProSAP1 expressed in HEK293 cells. (B) Coprecipitation analysis with GST–syndapin I and deletion mutants thereof. The SH3 domain is critical and sufficient for binding. A mutant SH3 domain (P434L; SH3*) did not bind. White lines indicate lanes omitted from blots (B and F). (C) Syndapin I SH3 precipitates GFP-ProSAP1 and GFP-ProSAP2 but not GFP-Shank1. (D) Alignment of +++APPPP motifs in ProSAP1 (NCBI Protein database accession no. NP_001004133), ProSAP2 (accession no. NP_067708), and Cobl (accession no. NP_766084; conserved amino acids are highlighted) and of corresponding residues in Shank1 (accession no. Q9WV48). (E) Scheme of rat ProSAP1b and deletion mutants used. Indicated are the N-terminal PDZ domain (medium grey), several proline-rich motifs (dark grey lines), and the C-terminal SAM (sterile alpha motif) domain (light grey). (F) GST–syndapin I precipitated GFP-ProSAP1 1–235 but none of the other ProSAP1 deletion mutants. (G) GFP fusion peptides encompassing the +++APPPP motifs of ProSAP1, ProSAP2, and Cobl associated with syndapin I SH3. (H and I) RKKAPPPPKR to GAGAAAAAAG mutation (amino acids 141–150 in ProSAP1; ProSAP1 1–235*) disrupted direct binding of ProSAP1 to syndapin I in both in vitro reconstitutions with purified proteins (H) and in coprecipitation analyses (I).
Further analyses showed that the syndapin I SH3 domain was both critical and sufficient for the interaction. P434L mutation (Sdp I SH3*) showed that complex formation was based on classical SH3 domain–PxxP interactions (Fig. 3 B). The syndapin SH3 domain interaction was restricted to ProSAP1 and ProSAP2 isoforms, whereas Shank1 (ProSAP3) did not bind (Fig. 3 C), as the respective site in Shank1 (aa 765–779) does not follow the +++APPPP consensus (Fig. 3 D).
ProSAP1 contains a plethora of proline-rich motifs. Yet, of all deletion mutants tested only ProSAP1 1–235 containing the residues RKKAPPPP (aa 141–148) associated with syndapin I (Fig. 3, E and F). To directly prove that the suggested RKKAPPPP motif is the syndapin binding site, we next tested the +++APPPP motifs found in ProSAP1 and ProSAP2. GFP fusions of the +++APPPP peptides were specifically precipitated by the syndapin I SH3 domain (Fig. 3 G).
In vitro reconstitutions with purified proteins demonstrated that the syndapin I interaction with the ProSAP1 N terminus was direct. HisTrx–syndapin I bound to GST-ProSAP1 1–235 but not to GST (Fig. 3 H). Mutation of the amino acids 141–150 of ProSAP1 (RKKAPPPPKR to GAGAAAAAAG; ProSAP1 1–235*; underlined characters indicate the conserved motif) strongly decreased the interaction with syndapin I in both in vitro reconstitutions and coprecipitation studies with lysates of HEK293 cells overexpressing GFP-ProSAP1 1–235* (Fig. 3, H and I).
Together, these observations experimentally proved that the RKKAPPPP motif of ProSAP1 suggested by our in silico searches is a direct binding site for the SH3 domain of syndapin I in vitro.
Syndapin I and ProSAP1 interact in vivo
ProSAP1 is part of the PSD, which hampers interaction studies due to indirect interactions occurring in the PSD scaffold. We circumvented this problem by first conducting coimmunoprecipitation experiments in nonneuronal cells. GFP-ProSAP1 was specifically coimmunoprecipitated with FLAG–syndapin I from HEK293 cell extracts (Fig. 4 A). In experiments done in a reciprocal manner, consistently, FLAG–syndapin I, specifically coimmunoprecipitated with anti-GFP antibodies immunoprecipitating GFP-ProSAP1 (Fig. 4 B).
Syndapin I interacts with ProSAP1 in vivo. (A) Specific coimmunoprecipitation of GFP-ProSAP1 with anti-FLAG antibodies immunoprecipitating FLAG–syndapin I. (B) Consistently, FLAG–syndapin I (arrowhead) was specifically coimmunoprecipitated with GFP-ProSAP1. (C) Immobilized GST–syndapin I SH3 specifically precipitated endogenous ProSAP1 from mouse brain cytosol (MBC). (D) Endogenous syndapin I was precipitated from rat brain preparations (RBC) with immobilized GST-ProSAP1 139–153 comprising the RKKAPPPP motif. (E–H) Syndapin I constitutively targeted to outer mitochondrial membranes recruited GFP-ProSAP1 (E) and GFP-ProSAP1 1–235 (F) in intact COS-7 cells, whereas Sdp IΔSH3 did not (G and H). Bars, 10 µm. (I) Syndapin I and ProSAP1 immunolabeling of brain sections from adult mice. Colocalization in synapses of mossy fibers with dendrites of pyramidal cells in the stratum lucidum in the hippocampus CA3 is shown. Blue signal in merge, DAPI. Insets, 2.5-fold enlargements of the boxed areas. Bars, 25 µm. (J) Immunolabeling of neurons transfected with Xpress–syndapin I at DIV 12 and stained for syndapin I, ProSAP1, and the dendritic marker MAP2 at DIV 14. Insets, 1.5-fold enlargements of boxed areas. Bar, 10 µm. (K) Endogenous syndapin I colocalized with ProSAP1 and synapsin 1 (DIV 21). Insets, twofold enlargements of boxed areas. Bars: (main panels) 5 µm; (insets) 2 µm.
Syndapin I interacts with ProSAP1 in vivo. (A) Specific coimmunoprecipitation of GFP-ProSAP1 with anti-FLAG antibodies immunoprecipitating FLAG–syndapin I. (B) Consistently, FLAG–syndapin I (arrowhead) was specifically coimmunoprecipitated with GFP-ProSAP1. (C) Immobilized GST–syndapin I SH3 specifically precipitated endogenous ProSAP1 from mouse brain cytosol (MBC). (D) Endogenous syndapin I was precipitated from rat brain preparations (RBC) with immobilized GST-ProSAP1 139–153 comprising the RKKAPPPP motif. (E–H) Syndapin I constitutively targeted to outer mitochondrial membranes recruited GFP-ProSAP1 (E) and GFP-ProSAP1 1–235 (F) in intact COS-7 cells, whereas Sdp IΔSH3 did not (G and H). Bars, 10 µm. (I) Syndapin I and ProSAP1 immunolabeling of brain sections from adult mice. Colocalization in synapses of mossy fibers with dendrites of pyramidal cells in the stratum lucidum in the hippocampus CA3 is shown. Blue signal in merge, DAPI. Insets, 2.5-fold enlargements of the boxed areas. Bars, 25 µm. (J) Immunolabeling of neurons transfected with Xpress–syndapin I at DIV 12 and stained for syndapin I, ProSAP1, and the dendritic marker MAP2 at DIV 14. Insets, 1.5-fold enlargements of boxed areas. Bar, 10 µm. (K) Endogenous syndapin I colocalized with ProSAP1 and synapsin 1 (DIV 21). Insets, twofold enlargements of boxed areas. Bars: (main panels) 5 µm; (insets) 2 µm.
Complex formation with endogenous ProSAP1 was addressed by coprecipitation from brain lysates. Endogenous ProSAP1 was specifically precipitated with the immobilized SH3 domain of syndapin I (Fig. 4 C). Consistently, endogenous syndapin I specifically associated with the ProSAP1-RKKAPPPP motif (Fig. 4 D).
The harsh extraction conditions for PSD proteins may lead to a postsolubilization artifact in interaction studies. We therefore reconstituted complex formation in intact cells by attaching syndapin I to a defined membrane compartment (mitochondria). Both GFP-ProSAP1 full-length and ProSAP1 1–235 were effectively accumulated at syndapin I–coated mitochondria membranes (Fig. 4, E and F).
Control experiments confirmed our in vitro finding that the ProSAP1 interaction was dependent on the SH3 domain of syndapin I. Mitochondrially targeted syndapin I lacking the SH3 domain (Mito-Sdp IΔSH3) neither accumulated GFP-ProSAP1 full-length nor 1–235 (Fig. 4, G and H). Collectively, syndapin I interacts with ProSAP1 in an SH3 domain–dependent manner in vitro as well as in cells.
Immunostaining of slices of murine brains show a good spatial overlap of syndapin I with ProSAP1 in, for example, the CA3 region of the hippocampus. Synapses of mossy fibers with pyramidal cell dendrites in the stratum lucidum were clearly immunopositive for both proteins (Fig. 4 I). Analyses of dissociated hippocampal neurons demonstrated that both Xpress-tagged and endogenous syndapin I colocalize with ProSAP1 in dendritic spines (Fig. 4, J and K).
Spine head enlargement caused by ProSAP1 overexpression depends on SH3–PxxP interactions with syndapin I
In the case that ProSAP1 functions in postsynaptic scaffolding critically involve ProSAP1–syndapin I complexes, disruptions of complex formation should suppress ProSAP1-induced spine head expansion. Indeed, cooverexpression of the syndapin I SH3 domain suppressed ProSAP1-mediated spine head expansion completely (Fig. 5, A and B).
ProSAP1-mediated functions in spine head enlargement rely on complex formation with syndapin I. (A–G) Absence of ProSAP1-mediated spine head enlargement upon coexpression of the syndapin I SH3 domain blocking the syndapin I binding site of ProSAP1 (A and B), upon use of ProSAP1* (C–E), and upon concomitant syndapin I RNAi (F and G), respectively. (A, D, and F) Representative images of neurons transfected as indicated (cotransfected with PM-mCherry for morphological analysis). (B, E, and G) Quantification of head width of mushroom spines. (C) Coprecipitation studies with immobilized syndapin I and Abp1 SH3 domains and GFP-ProSAP1 versus GFP-ProSAP1* showing specific disruption of syndapin I interaction. (H–J) Quantitative analysis of head width of mushroom spines. Neither syndapin I RNAi nor overexpression of syndapin I modulate head sizes of mushroom spines. (J) ProSAP RNAi causes a decrease in head width not seen upon syndapin I RNAi and not rescued by syndapin I coexpression. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Data represent mean ± SEM (error bars).
ProSAP1-mediated functions in spine head enlargement rely on complex formation with syndapin I. (A–G) Absence of ProSAP1-mediated spine head enlargement upon coexpression of the syndapin I SH3 domain blocking the syndapin I binding site of ProSAP1 (A and B), upon use of ProSAP1* (C–E), and upon concomitant syndapin I RNAi (F and G), respectively. (A, D, and F) Representative images of neurons transfected as indicated (cotransfected with PM-mCherry for morphological analysis). (B, E, and G) Quantification of head width of mushroom spines. (C) Coprecipitation studies with immobilized syndapin I and Abp1 SH3 domains and GFP-ProSAP1 versus GFP-ProSAP1* showing specific disruption of syndapin I interaction. (H–J) Quantitative analysis of head width of mushroom spines. Neither syndapin I RNAi nor overexpression of syndapin I modulate head sizes of mushroom spines. (J) ProSAP RNAi causes a decrease in head width not seen upon syndapin I RNAi and not rescued by syndapin I coexpression. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Data represent mean ± SEM (error bars).
To specifically address the involvement of the syndapin I binding site, we constructed a ProSAP1 full-length protein with that site mutated (ProSAP1*). Biochemical examinations confirmed that the ProSAP1* mutant was unable to bind to syndapin I. As ProSAP1* still associated with Abp1, another SH3 domain containing an interaction partner of ProSAP1 involved in spine and synapse formation (Qualmann et al., 2004; Haeckel et al., 2008), ProSAP1* was suitable to dissect ProSAP1/Abp1 from ProSAP1–syndapin I functions (Fig. 5 C). Expression of ProSAP1* in primary neurons failed to cause the ProSAP1-mediated head width increase; spine heads instead resembled those of controls (Fig. 5, D and E). Therefore, the syndapin I–binding motif of ProSAP1 is crucial for ProSAP1-mediated functions in spine head expansion.
The experiments described thus far clearly demonstrated the importance of SH3 domain–mediated protein complex formation with the RKKAPPPP motif of ProSAP1 in ProSAP1-mediated functions. Yet, experimental evidence that these observations explicitly reflect a crucial importance of syndapin I was still lacking. We therefore depleted syndapin I in ProSAP1-overexpressing primary neurons. These cells also failed to develop the ProSAP1-overexpression phenotype and resembled control neurons (Fig. 5, F and G). Thus, ProSAP1 functions in the formation of morphologically recognizable postsynapses critically depend on syndapin I.
Interestingly, neither syndapin I gain-of-function (Fig. 5 H) nor loss-of-function (Fig. 5 I) altered the size of established mushroom spine heads. Likewise, syndapin I overexpression did not modulate the decrease in head width induced upon ProSAP1 RNAi (Fig. 5 J). Thus, syndapin I does not modulate the size of established mushroom spine heads.
Syndapin I and ProSAP1 cooperate in spine and synapse formation
If syndapin I cooperates with ProSAP1 in spine and synapse formation, ProSAP1 loss-of-function should lead to somewhat similar phenotypes to those revealed for syndapin I deficiency. Indeed, in line with previous analyses of synapse densities upon ProSAP1 RNAi (Grabrucker et al., 2011; Berkel et al., 2012), we observed a significant reduction of both PSD-95–marked and synapsin 1–marked synapses in ProSAP1-depleted cells (Fig. 6, A–C; and Fig. S3, A and B).
ProSAP1 RNAi phenocopies syndapin I RNAi in synapse formation. (A) Examples of dendrites of neurons transfected as indicated and stained for PSD-95 or synapsin 1. Bar, 5 µm. (B and C) Quantitative evaluation of the relative density of PSD-95– (B) and synapsin 1–positive puncta (C) in contact with transfected neurons. *, P < 0.05; ***, P < 0.001. Data represent mean ± SEM (error bars).
ProSAP1 RNAi phenocopies syndapin I RNAi in synapse formation. (A) Examples of dendrites of neurons transfected as indicated and stained for PSD-95 or synapsin 1. Bar, 5 µm. (B and C) Quantitative evaluation of the relative density of PSD-95– (B) and synapsin 1–positive puncta (C) in contact with transfected neurons. *, P < 0.05; ***, P < 0.001. Data represent mean ± SEM (error bars).
With a deviation of about −20% from control, the ProSAP1 loss-of-function phenotype was relatively similar to the syndapin I loss-of-function phenotype (Fig. 2, E–G). Thus, in spine and synapse formation, syndapin I loss-of-function mirrors ProSAP1 loss-of-function, implicating a critical role of both proteins in this process.
Importantly, in line with syndapin working together closely with ProSAP1 in this cell biological process, syndapin I overexpression was able to modulate the ProSAP1 RNAi-induced reduction of synapse density. Syndapin I coexpression raised synapse densities toward those of control cells (Fig. 6, A–C; and Fig. S3, A and B).
It can be hypothesized that all the experimentation that interfered with syndapin I–ProSAP1 complex formation and ProSAP1 function in enlargement of established heads (Fig. 5, A–G) should also have some negative impact on spine formation when compared with ProSAP1 overexpression, which on its own under our experimental conditions did not lead to statistically significant increases in the densities of mushroom spines and postsynapses, respectively (Fig. S3, C–E). Indeed, coexpression of the syndapin I SH3 domain, expression of the syndapin I binding-deficient ProSAP1 mutant ProSAP1*, and coexpression of syndapin I RNAi also led consistently to decreased mushroom spine densities when compared with ProSAP1 overexpression (Fig. S3, F–H). Together these observations suggested an organizational role of syndapin I in spines that needed to be unraveled.
Syndapin I functions rely on F-BAR domain–mediated membrane association
We hypothesized that a role of syndapin I as an upstream organizer of ProSAP1 functions in postsynapse formation may involve plasma membrane (PM) targeting and spatial control of ProSAP1. To specifically address a putative requirement of the PM-related aspects of syndapin I function, we constructed a rat syndapin I mutant with the lipid-intercalating wedges of the F-BAR domain disrupted in accordance to human syndapin I (Wang et al., 2009). In vitro reconstitutions with liposomes confirmed that, in contrast to WT syndapin I, syndapin II122E/M123E did not float together with liposomes in density gradients (F2) but remained at the bottom of the gradient (F4–6; Fig. 7 A).
Wedge loop–mediated membrane association is crucial for Syndapin I’s role in synapse formation and links ProSAP1 to membranes. (A) Immunoblot analyses of gradient fractions of in vitro reconstitution of protein–membrane interactions. Syndapin I floats with liposomes (to F2), whereas syndapin II122E/M123E stays at the bottom (F4–6). (B–G) RNAi-insensitive syndapin II122E/M123E fails to rescue syndapin I RNAi phenotypes. (B) Representative dendrites of PM-mCherry–transfected and anti–PSD-95–stained and anti–synapsin 1–stained neurons, respectively. Bar, 5 µm. (C–E) Quantitative evaluations of general spine density (C) and of individual morphology groups (D and E). (F and G) Counts of postsynaptic (F) and presynaptic (G) puncta in contact with transfected neurons. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Data indicate mean ± SEM (error bars). (H) Immunoblot analyses of complexes of GST–syndapin I and GST–syndapin II122E/M123E, respectively, with GFP-ProSAP1 1–235 that were incubated with liposomes and subjected to density gradient fractionations. Syndapin I–ProSAP1 1–235 complexes float to fraction F2, whereas syndapin II122E/M123E/ProSAP1 1–235 complexes do not.
Wedge loop–mediated membrane association is crucial for Syndapin I’s role in synapse formation and links ProSAP1 to membranes. (A) Immunoblot analyses of gradient fractions of in vitro reconstitution of protein–membrane interactions. Syndapin I floats with liposomes (to F2), whereas syndapin II122E/M123E stays at the bottom (F4–6). (B–G) RNAi-insensitive syndapin II122E/M123E fails to rescue syndapin I RNAi phenotypes. (B) Representative dendrites of PM-mCherry–transfected and anti–PSD-95–stained and anti–synapsin 1–stained neurons, respectively. Bar, 5 µm. (C–E) Quantitative evaluations of general spine density (C) and of individual morphology groups (D and E). (F and G) Counts of postsynaptic (F) and presynaptic (G) puncta in contact with transfected neurons. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Data indicate mean ± SEM (error bars). (H) Immunoblot analyses of complexes of GST–syndapin I and GST–syndapin II122E/M123E, respectively, with GFP-ProSAP1 1–235 that were incubated with liposomes and subjected to density gradient fractionations. Syndapin I–ProSAP1 1–235 complexes float to fraction F2, whereas syndapin II122E/M123E/ProSAP1 1–235 complexes do not.
Further characterizations of syndapin II122E/M123E addressed the second established function of BAR domains: self-assembly (Kessels and Qualmann, 2006). Chemical cross-linking as well as heterologous coimmunoprecipitations revealed that self-association was not affected by I122E/M123E mutation (Fig. S4, A and B).
Functional analyses in neurons revealed that reexpression of an RNAi-insensitive version of syndapin II122E/M123E instead of WT syndapin I failed to rescue the syndapin I RNAi-mediated impairments of synapse formation and the defects in mushroom spine formation (Fig. 7, B–G). Instead, syndapin II122E/M123E coexpression even seemed to have an additional negative effect on mushroom spine formation. Thus, besides SH3 domain–mediated complex formation with ProSAP1, F-BAR domain–mediated membrane binding is absolutely crucial for the role of syndapin I in synapse formation.
The requirement of both SH3 and membrane interactions suggested that syndapin I may interconnect ProSAP1 with membranes. To experimentally address, this we formed protein complexes of GST–syndapin I and GFP-ProSAP1 expressed in HEK293 cells, and subjected them to liposome binding assays. ProSAP1 fusion proteins effectively floated with liposomes to fraction 2 when WT syndapin I was present. In contrast, syndapin II122E/M123E/ProSAP1 complexes remained at the bottom of the gradients (Fig. 7 H).
Syndapin I is enriched at membranes of dendritic spines
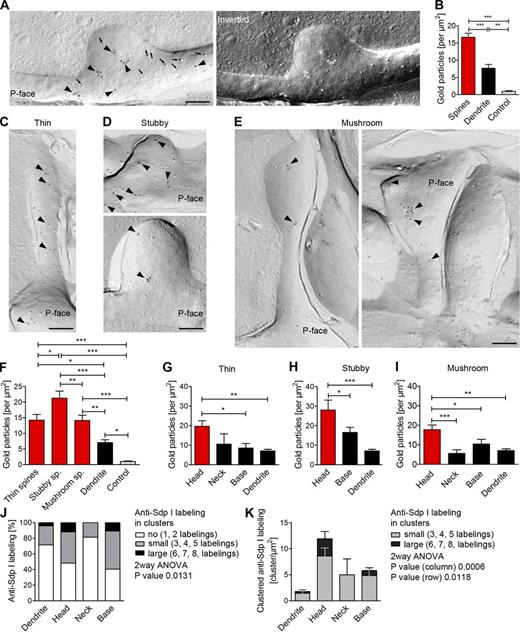
Syndapin I exists in a membrane-associated, larger cytosolic subpool (Qualmann et al., 1999). Our functional analyses clearly demonstrated that membrane association is a key aspect in syndapin I’s functions in synapse formation (Fig. 7). To be able to explicitly study membrane-associated, endogenous syndapin I at high resolution in membranes of neurons undergoing spine and synapse formation, we established a combination of freeze-fracturing and immunolabeling (Fujimoto, 1995; Severs and Robenek, 2008) for syndapin I detection. Subsequent transmission electron microscopy (TEM) provided high resolution and views of large membrane areas.
On sapphire discs, primary rat hippocampal neurons formed typical neuronal networks and a plethora of synapses (Fig. S5, A–D). Rapid-freezing preserved neuronal morphology to the extent that even fragile structures, such as neurites and dendritic spines, were maintained. Although filopodia-like protrusions were rare, morphologically intact thin, stubby, and mushroom spines were observable (Fig. S5 E).
As expected for freeze-fracturing, fractures usually predominantly separated the outer and the inner membrane leaflet. Whereas the E-face (outer leaflet of the PM facing the extracellular space) had the typical smooth appearance, P-faces (inner leaflet of the PM facing the cytosol) were easily identifiable based on the abundance of integral membrane protein complexes protruding into the cytoplasm (Fig. 8, A and C–E; and Fig. S5 E).
Visualization of membrane-associated syndapin I via immunolabeling of freeze-fractured replica of hippocampal neurons. (A) Electron micrograph of an anti–syndapin I immunogold labeled (arrowheads) replica of a dendrite section of rapid-frozen, freeze-fractured primary rat hippocampal neurons (DIV 16). Examples of intramembrane particles (arrows) identifying the P-face. Right, inverted image (light = C/Pt shadowing direction). (B) Quantitation of anti–syndapin I labeling densities in spine, dendrite, and control areas. (C–E) Electron micrographs of anti–syndapin I–labeled (arrowheads) freeze-fracture replica showing thin (C), stubby (D), and mushroom spines (E). Bars, 200 nm. (F) Labeling densities in the different spines. (G–I) Intra-spine distribution (head, neck, base) of syndapin I at membranes of thin (G), stubby (H), and mushroom spines (I). (J) Simplified summary of anti–syndapin I cluster analyses at dendritic membranes and at membranes of spines (full data, Fig. S5 H). (K) Quantitation of the density of clustered anti–syndapin I labeling in different spine regions and at dendritic membranes. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Data represent mean ± SEM (error bars).
Visualization of membrane-associated syndapin I via immunolabeling of freeze-fractured replica of hippocampal neurons. (A) Electron micrograph of an anti–syndapin I immunogold labeled (arrowheads) replica of a dendrite section of rapid-frozen, freeze-fractured primary rat hippocampal neurons (DIV 16). Examples of intramembrane particles (arrows) identifying the P-face. Right, inverted image (light = C/Pt shadowing direction). (B) Quantitation of anti–syndapin I labeling densities in spine, dendrite, and control areas. (C–E) Electron micrographs of anti–syndapin I–labeled (arrowheads) freeze-fracture replica showing thin (C), stubby (D), and mushroom spines (E). Bars, 200 nm. (F) Labeling densities in the different spines. (G–I) Intra-spine distribution (head, neck, base) of syndapin I at membranes of thin (G), stubby (H), and mushroom spines (I). (J) Simplified summary of anti–syndapin I cluster analyses at dendritic membranes and at membranes of spines (full data, Fig. S5 H). (K) Quantitation of the density of clustered anti–syndapin I labeling in different spine regions and at dendritic membranes. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Data represent mean ± SEM (error bars).
Incubation of freeze-fracture replica with anti–syndapin I antibodies showed that spine membrane surfaces of hippocampal cultures are accessible for immunoelectron microscopic examinations when anti–syndapin I labeling was achieved (Fig. 8 A). In line with its lack of membrane-binding domains, ProSAP1 was not preserved at membranes of freeze-fractured neurons (unpublished data).
Quantitative analysis of syndapin I labeling densities demonstrated a specific anti–syndapin I labeling at P-faces of dendrites and spines. Unspecific labeling at control surfaces was very low (0.9 particles/µm2; Fig. 8 B). Further experiments proved that labeling in secondary antibody controls was almost undetectable (0.1 particles/µm2) and that labeling at syndapin I KO material was also low (2.6 particles/µm2; Fig. S5 F).
Interestingly, syndapin I was detected at the PM of spines more than twice as often as at dendritic membranes (16.6 particles/µm2 vs. 7.3 particles/µm2; Fig. 8 B). Further analyses showed that the labeling densities at the membranes of the different spine classes (thin, stubby, and mushroom) ranged from 14 to 22 particles/µm2 (Fig. 8, C–F). Thus, membrane-associated syndapin I is enriched in dendritic spines.
Syndapin I nanoclusters accumulate at the PM of head regions of dendritic spines
Anti–syndapin I immunolabeling at spine membranes did not appear to be uniform (Fig. 8, C–E). We therefore determined the labeling densities in the spine subareas head, neck, and base separately for all types of spines. The density of anti–syndapin I immunogold labeling was significantly increased in the head areas of all types of spines analyzed (Fig. 8, G–I). The values for the heads were ∼2–3 times as high as for neck regions (Fig. 8, G and I). Data for neck regions were comparable to the moderate labeling density observed at dendritic membranes (Fig. 8, G and I). PM-attached syndapin I thus has a preference for spine head regions.
Biochemical studies suggested that syndapin I oligomerization may lead to the formation of multimeric syndapin lattices (Kessels and Qualmann, 2006). To address whether such syndapin I clusters do indeed occur at neuronal membranes, we next systematically analyzed the distribution of all individual syndapin I molecules detected. TEM analysis of immunolabeled freeze-fracture replica provided direct visual evidence for the proposed formation of syndapin I lattices at membranes in vivo (Fig. 8). About half of the detected syndapin I was found in clusters of at least three at membranes of both heads and bases of spines. About 15% of the detected syndapin I occurred in the form of clusters of an even higher order (Fig. 8 J). The size of the syndapin I nanoclusters ranged from 40 to 100 nm. Interestingly, mushroom, thin, and stubby spines all contained elevated levels of clustered syndapin I. Up to eight anti–syndapin I labels per membrane nanodomain were observed (Fig. S5, G and H).
These clusters were not immunolabeling artifacts, as higher order clusters preferentially occurred in the heads and at the bases of spines but were rare in dendrites and absent from control surfaces as well as from spine neck membrane regions (P = 0.0131; Fig. 8 J and Fig. S5, G and H). The densities of highly clustered anti–syndapin I labeling were increased particularly in the head of spines, and this increase was highly statistically significant (Fig. 8 K).
Additional control experiments demonstrated that anti–syndapin I–labeled clusters remained unaltered when the antibody concentration was increased (Fig. S5 I). Syndapin I clustering is thus a specific feature of syndapin I enriched at the membranes of head and base regions of dendritic spines.
Postsynaptic syndapin I loss-of-function leads to reduced frequencies of miniature excitatory postsynaptic currents (mEPSCs), and its physiological consequence thereby resembles that of ProSAP1 KO
If syndapin I indeed provides interconnection, membrane linkage, and spatial cues for ProSAP1 in postsynapses, this may lead to defects in synaptic transmission resembling those of ProSAP1 loss-of-function. Whole-cell patch clamp recordings from rat hippocampal neurons transfected with syndapin I RNAi revealed that mEPSCs showed a significant reduction in frequency (−40%) when compared with both pRNAT and scrambled RNAi-expressing control cells (Fig. 9, A, B, and D). The mEPSC amplitudes were not altered (Fig. 9, A, C, and E). Our whole-cell patch clamp recordings from syndapin I RNAi cells revealed that the mEPSC phenotype is of postsynaptic origin. Similarly, a decrease in mEPSC frequency by ∼30% and unchanged amplitudes were also observed upon ProSAP1/Shank2 KO (Schmeisser et al., 2012).
Syndapin I depletion reduces the frequencies of mEPSCs. (A and F) Sample traces of whole-cell patch clamp recordings of mEPSCs from individual primary rat hippocampal neurons transfected at DIV 12 and analyzed 48 h later. (B) The frequency of mEPSCs was reduced in syndapin I RNAi neurons when compared with pRNAT and scrambled RNAi, respectively (B and D), whereas the mEPSC amplitudes did not differ (C and E). (F–J) Syndapin I RNAi rescue experiments with coexpression of mCherry–syndapin I and mutants thereof showing that both SH3 domain protein interactions and F-BAR domain–mediated membrane interactions are crucial for syndapin I functions in postsynaptic neurotransmission. *, P < 0.05. Data represent mean ± SEM (error bars).
Syndapin I depletion reduces the frequencies of mEPSCs. (A and F) Sample traces of whole-cell patch clamp recordings of mEPSCs from individual primary rat hippocampal neurons transfected at DIV 12 and analyzed 48 h later. (B) The frequency of mEPSCs was reduced in syndapin I RNAi neurons when compared with pRNAT and scrambled RNAi, respectively (B and D), whereas the mEPSC amplitudes did not differ (C and E). (F–J) Syndapin I RNAi rescue experiments with coexpression of mCherry–syndapin I and mutants thereof showing that both SH3 domain protein interactions and F-BAR domain–mediated membrane interactions are crucial for syndapin I functions in postsynaptic neurotransmission. *, P < 0.05. Data represent mean ± SEM (error bars).
Rescue experiments affirmed that the impairments in mEPSCs are specifically caused by loss of syndapin I (Fig. 9, F–J). Reexpression of WT syndapin I protein completely suppressed the syndapin I RNAi effects and restored WT mEPSC frequencies. Interestingly, further analyses with mutant syndapin I proteins revealed that both SH3 domain association and lipid binding are critical, as neither syndapin IΔSH3 nor syndapin II122E/M123E were able to restore normal mEPSC frequencies, and are thus important aspects for proper function of synapses (Fig. 9, F–J).
Syndapin I acts as a spatial organizer for ProSAP1/Shank2 in postsynapses
The syndapin I enrichment at spine head membrane areas, its preferential occurrence in nanoclusters in spine heads (Fig. 8), the requirement of membrane interactions for syndapin I functions (Fig. 7), and the similarities of syndapin I and ProSAP1 loss-of-function in spine and synapse formation (Fig. 6), as well as in synaptic function (Fig. 9), suggested that syndapin I may help to organize ProSAP1 at particular spine membrane areas. This was revealed by quantitative image analyses of anti–PSD-95– and anti-ProSAP1–labeled postsynapses of syndapin I KO neurons in comparison to WT neurons. Although neither the area of PSD-95–positive puncta, their mean anti–PSD-95 immunolabeling intensity, nor the area of colocalization with ProSAP1 were altered, the area of ProSAP1-positive puncta was significantly enlarged upon syndapin I KO (Fig. 10, A–E).
Syndapin I acts as a spatial organizer for ProSAP1/Shank2 in postsynapses. (A) Maximal intensity projections (top) of anti–PSD-95 and anti-ProSAP1 coimmunolabeling of WT (+/+) and syndapin I KO (−/−) neurons (hippocampal cultures, DIV 14) subjected to surface reconstruction using Imaris software (bottom). Bars, 2.5 µm. (B–G) Quantitative analyses of indicated immunolabeling parameters of PSD-95– and ProSAP1-labeled postsynapses. Note that identical amounts of ProSAP1 (Σ ProSAP1 intensity of pixels per puncta; G) are distributed over significantly larger postsynaptic areas upon syndapin I KO (E), as marked by arrows in A. **, P < 0.01. Data represent mean ± SEM (error bars). (H and I) Schematic representation (H) and models (I) depicting syndapin I loss-of-function effects on ProSAP1 organization (H and I) and syndapin I distribution during synapse formation in nanodomains at the base and the head regions of dendritic spines (I).
Syndapin I acts as a spatial organizer for ProSAP1/Shank2 in postsynapses. (A) Maximal intensity projections (top) of anti–PSD-95 and anti-ProSAP1 coimmunolabeling of WT (+/+) and syndapin I KO (−/−) neurons (hippocampal cultures, DIV 14) subjected to surface reconstruction using Imaris software (bottom). Bars, 2.5 µm. (B–G) Quantitative analyses of indicated immunolabeling parameters of PSD-95– and ProSAP1-labeled postsynapses. Note that identical amounts of ProSAP1 (Σ ProSAP1 intensity of pixels per puncta; G) are distributed over significantly larger postsynaptic areas upon syndapin I KO (E), as marked by arrows in A. **, P < 0.01. Data represent mean ± SEM (error bars). (H and I) Schematic representation (H) and models (I) depicting syndapin I loss-of-function effects on ProSAP1 organization (H and I) and syndapin I distribution during synapse formation in nanodomains at the base and the head regions of dendritic spines (I).
The total amount of ProSAP1 (sum of ProSAP1 intensity) was unchanged, and the averaged anti-ProSAP1 immunoreactivity per synaptic site appeared to be slightly decreased (Fig. 10, F and G). The enlarged areas of ProSAP1 localization thus did not reflect an increased abundance of ProSAP1 in postsynapses but a broader distribution of anti-ProSAP1 signals within spine heads (Fig. 10 H). These data are in line with syndapin I acting as a spatial organizer of ProSAP1 functions at membranes of postsynaptic specializations (Fig. 10 I).
Discussion
The formation of morphologically distinct, compartmentalized synapses is a prerequisite for information processing in the brain. The formation of dendritic spines as postsynaptic compartments is thought to rely on neurotransmitter receptor clustering by scaffold proteins and on the remodeling of the cortical actin cytoskeleton. Here we reveal that syndapin I–mediated functions represent further, thus far unrecognized molecular mechanisms crucial for synapse formation.
Our data reveal that the brain-enriched F-BAR domain protein syndapin I is a key player in postsynapse formation. The density of dendritic spines was significantly reduced in CA1 neurons of syndapin I KO mice. Evaluation of individual neurons depleted for syndapin I by RNAi led to a similar phenotype: densities of spines as well as of pre- and postsynapses were significantly reduced. Syndapin I RNAi also proved that the impaired formation of dendritic spines is specifically caused by loss of syndapin I in the postsynaptic cell. Consistently, syndapin I overexpression (Schael et al., 2013; this study) had the opposite effects and increased synapse density.
Expression of an RNAi-insensitive syndapin IΔSH3 mutant failed to rescue syndapin I loss-of-function phenotypes in synapse and spine formation. Thus, syndapin I’s role in synaptogenesis is strictly dependent on SH3 domain interactions. Importantly, with ProSAP1, we were able to identify a binding partner for the syndapin I SH3 domain that is specifically localized to postsynapses. Syndapin I associated with ProSAP1 in vitro and in vivo. Both endogenous and heterologously expressed proteins are coprecipitated specifically by their respective binding partner. Syndapin I–ProSAP1 complex formation depended on the syndapin I SH3 domain as well as on a motif in the N-terminal part of ProSAP1 closely fitting to a recently identified consensus sequence for syndapin SH3 domain binding (Schwintzer et al., 2011). The interaction could be reconstituted in vitro with purified proteins as well as in intact cells. Thus, the syndapin I interaction with ProSAP1 revealed in coimmunoprecipitation analyses is direct and is not a postsolubilization artifact. In line with an in vivo relevance of complex formation between syndapin I and ProSAP1, both proteins were enriched in the hippocampus and colocalized in dendritic spines.
Syndapin I had thus far been largely characterized as an axonally and presynaptically acting component (Anggono et al., 2006; Andersson et al., 2008; Dharmalingam et al., 2009; Koch et al., 2011). However, our analyses revealed a colocalization with pre- and postsynaptic markers as well as with dendrite markers, and furthermore unveiled specific anti–syndapin I labeling at membranes of dendritic spines. These findings are in line with some postembedding immunogold labeling reported at chemically fixed hippocampal CA1 synapses (Pérez-Otaño et al., 2006).
Functionally, ProSAP1-mediated effects seemed to critically rely on syndapin I association. This is underscored by the fact that three different manipulations interfering with such complexes (quench via the syndapin I SH3 domain, mutation of the syndapin binding site in ProSAP1, and syndapin I RNAi) all consistently abolished ProSAP1-mediated spine head enlargements.
In line with a close functional relationship of syndapin I and ProSAP1, the syndapin I loss-of-function defects we observed qualitatively and quantitatively phenocopied the effects of ProSAP1 loss-of-function. ProSAP1 RNAi led to an ∼20% reduction in synapse density. Our ProSAP1 data are in agreement with recent synapse density analyses in dissociated neurons depleted for ProSAP1 by RNAi (Grabrucker et al., 2011) and in CA1 neurons of ProSAP1 KO mice (Schmeisser et al., 2012). Another study failed to reproduce the ProSAP1 loss-of-function effects on synapse density (Won et al., 2012).
Our electrophysiological analyses revealed that the close functional relationship of syndapin I and ProSAP1, as well as the syndapin I loss-of-function phenotypes we unraveled in spine and synapse formation and in spatial organization of ProSAP1 in synapses, are also reflected in studies on synapse function. The frequencies of mEPSCs were significantly reduced upon syndapin I depletion. The reduced spontaneous synaptic activity observed in electrophysiological recordings was therefore clearly of postsynaptic origin, as recordings were from syndapin I RNAi cells, and may reflect the reductions of synapse numbers observed upon syndapin I deficiency. The reduced mEPSC frequencies upon syndapin I RNAi furthermore are in line with a recent study on the physiological consequences of ProSAP1 KO. ProSAP1 KO also led to a reduction in mEPSC frequency (Schmeisser et al., 2012). Further supporting our conclusion that syndapin I and ProSAP1 work together closely in postsynapse formation, the increased postsynapse density observed upon syndapin I overexpression was suppressed by ProSAP1 RNAi.
In contrast to ProSAP1/Shank2, Shank1 did not interact with syndapin I (this study), appears late in synapse formation (Grabrucker et al., 2011), and may be rather important for integrity of existing postsynapses (Hung et al., 2008).
Our analyses of the molecular mechanisms underlying the important role of syndapin I in synapse formation showed that syndapin I is able to associate with and to interconnect ProSAP1 to membranes, that membrane-bound syndapin I is specifically enriched in spine heads, that most membrane-associated syndapin I at the bases of spines and in spine heads is clustered, and that syndapin I’s membrane association is required for synapse formation and for proper synaptic function, as revealed by electrophysiological studies. Consistently, syndapin I roles in synapse formation and function were not rescued by restoring SH3 domain interactions and self-association; they also required F-BAR–mediated membrane binding. Together, these findings suggest that membrane-associated syndapin I lattices help to ensure cortical localization and interconnection of binding partners, and that syndapin I–enriched nanodomains at the spine membrane provide important spatial cues for postsynaptic scaffold components during synapse formation (Fig. 10).
Mechanistically, ignition of local actin nucleation would have to occur at certain dendritic membrane micro- or nanodomains to give rise to spines and at sites slightly set back from spine tips to efficiently generate the forces required for head formation (Fig. 10). Branched filament networks containing the actin nucleator Arp2/3 complex were observed at the bases of spines and in spine heads (Korobova and Svitkina, 2010). In these steps of synapse formation, syndapin I may cooperate with further ProSAP1 binding partners, as the ProSAP1 binding partner and F-actin–associating protein Abp1 is also critical in spine and synapse formation. It is, however, also possible that ProSAP1–Abp1 complexes work downstream of ProSAP1–syndapin I complexes and play an important role in promoting head formation processes after syndapin I ignited such processes. In line with such an early role of syndapin I in synapse formation, our high-resolution analyses revealed a selective occurrence of syndapin I clusters at membranes at both the base and the head of spines, providing high local concentrations of docking sites for syndapin I binding partners. Such syndapin I lattices are ideally placed for efficient assembly of multicomponent superstructures. Our analyses of the ProSAP1 organization in syndapin I–depleted postsynapses revealed a lack of spatial confinement of ProSAP1 to PSD-95–marked postsynaptic areas.
In line with morphological measurements of PSD width in syndapin I KO sections (Koch et al., 2011), we did not observe any effects on the PSD-95 scaffold itself but found that the spatial organization of the newly identified syndapin I interaction partner ProSAP1 was specifically impaired in syndapin I–depleted postsynapses. The spatial cues for this organizing role of syndapin I and for syndapin I cluster formation are likely to result from signaling cascades converging on syndapin I (Schael et al., 2013), from syndapin I’s affinity for certain lipids (Itoh et al., 2005; Dharmalingam et al., 2009) and from its preference for membrane topologies with positive curvatures and saddle points fitting to the banana- and tilde-shaped geometry, and to the different lipid binding interfaces of syndapin I dimers (Qualmann et al., 2011).
In contrast, WRP/srGAP3, another F-BAR domain protein recently implicated in spine formation, has a convex lipid-binding surface and was proposed to be selective for strongly negatively curved membranes. Overexpressed fusion proteins of the WRP/srGAP3 F-BAR domain consistently accumulated in filopodia (Carlson et al., 2011). Different F-BAR domain proteins may therefore set distinct spatial cues during synapse formation.
Based on our functional, mechanistic, and high-resolution studies of syndapin I in developing neuronal networks, we propose that membrane association and topology modulation as well as local organization of receptor scaffolding components are important mechanistic requirements for the formation of excitatory synapses in dendritic spines.
Materials and methods
Animals
All experimental procedures were performed according to the German Tierschutzgesetz (license 02-011/10). Syndapin I KO (−/−) mice were generated via excision of exon 1 of the syndapin I gene using the Cre/loxP system (ES 129/SV) and backcrossed on C57/BL6J as described previously (Koch et al., 2011).
DNA constructs
Plasmids encoding for GFP (pEGFP-C1; Takara Bio Inc.)-, GST (pGEX-2T; GE Healthcare)-, Xpress (pcDNA 3.1/HisC; Invitrogen)-, and FLAG-tagged (pCMV-Tag2b; Agilent Technologies) full-length syndapin I as well as for GST–syndapin I SH3 domain (aa 376–441, pGEX-2T; aa 378–441, pGEX-5X-1; GE Healthcare), GST–syndapin I SH3P434L (GST-Sdp I SH3*; aa 376–441, pGEX-2T), and GST–syndapin IΔSH3 (aa 1–382, pGEX-2T) were described in Qualmann et al. (1999), Qualmann and Kelly (2000), and Kessels and Qualmann (2006), respectively.
A vector expressing FLAG-mCherry (FLAGc) was generated by replacing GFP with mCherry in a pCMV-based FLAG-GFP expressing vector (Ahuja et al., 2007). FLAGc-tagged syndapin I SH3 domain (FLAGc-Sdp I SH3) was generated by subcloning. Plasmids encoding for mCherry-tagged syndapin I fusion proteins targeted to the outer mitochondrial membrane (Mito-Sdp I) were generated by subcloning of syndapin I into Mito-FLAGc-pCMV-Tag2 vector (generated by replacing GFP of Mito-FLAG-GFP-pCMV [Kessels and Qualmann, 2002] with mCherry).
Plasmids encoding for GST-tagged full-length syndapin II (aa 1–488, pGEX-2T) and full-length GST–syndapin III (pGEX-5X-1) were described in Qualmann and Kelly (2000) and Braun et al. (2005), respectively. A plasmid encoding for a His-thioredoxin (Trx)-tagged full-length syndapin I (HisTrx-Sdp I) was generated by subcloning into pET32 (EMD Millipore). The plasmid encoding for a GST-tagged SH3 domain of Abp1 (aa 371–433, pGAT2 vector) was generated as described previously in Kessels et al. (2001).
Generation of an RNAi-insensitive full-length syndapin I mutant (pCMV-Tag2b) was described in Dharmalingam et al. (2009). A FLAGc-tagged version thereof was generated by subcloning into FLAGc. RNAi-insensitive syndapin IΔSH3 (aa 1–382) deletion mutants in pCMV-Tag2 or FLAGc were generated by subcloning thereof.
Rat syndapin I wedge loop mutation (syndapin II122E/M123E) was designed according to Wang et al. (2009) and introduced by PCR (primer sequence below) and cloning into pBluescript SK+ (GenBank).
Syndapin I forward primer, 5′-GCCTACCACAAGCAGGAGGAGGGCGGCTTCAAGGAG-3′; syndapin I reverse primer, 5′-CTCCTTGAAGCCGCCCTCCTCCTGCTTGTGGTAGGC-3′.
Plasmids encoding GFP-, FLAG-, and GST-tagged syndapin II122E/M123E were generated by subcloning in pEGFP-C1, pCMV-Tag2b, and pGEX-2T, respectively.
RNAi-insensitive syndapin II122E/M123E was generated by introducing the mutation rendering syndapin I RNAi insensitive by PCR into syndapin II122E/M123E (primer sequences below) and cloning into pCMV-Tag2 and FLAGc.
PCR1 forward primer, T3 primer; PCR1 reverse primer, 5′-CGGAATTCTTCACCTCCTGGTGCAG-3′; PCR2 forward primer, 5′-GCGAATTCATTGTTAAATGAAGACCTGGAGAAAGTC-3′; PCR2 reverse primer, 5′-GTCCTCGAGTCTATATAGCCTCAAC-3′.
The PM-mCherry–expressing vector (a derivative of the pEGFP-F vector; Takara Bio Inc.) was a gift of M. Korte (Technische Universität Braunschweig, Braunschweig, Germany).
Plasmids encoding for GFP-ProSAP1 (pEGFP-C1) and GFP-ProSAP2 (pEGFP-C2) were provided by T. Boeckers (University of Ulm, Ulm, Germany). GFP-Shank1 (pEGFP-C1) was provided by C. Sala (Consiglio Nazionale delle Ricerche Institute of Neuroscience, Milan, Italy).
Generation of deletion constructs of ProSAP1 (NCBI Protein database accession no. NP_001004133) was described in Haeckel et al. (2008). A syndapin I binding-deficient full-length ProSAP1 mutant (GFP-ProSAP*) was generated by PCR with primers encoding for GAGAAAAAAG instead of the syndapin I–binding motif RKKAPPPPKR and cloned into pEGFP-C. GFP-ProSAP1 1–235 was generated by subcloning into pEGFP-C using an internal HindIII site. Constructs coding for GST fusion proteins were generated by subcloning in pGEX-4T.
Fusion proteins of the second KRRAPPPP motif of Cobl (aa 318–332) have been described previously (Schwintzer et al., 2011). Constructs encoding for +++APPPP motifs of ProSAP1 (aa 139–153) and ProSAP2 (aa 671–685; accession no. NP_067708) were generated by annealing primers (sequence below) and cloning into pEGFP-C2 and pGEX-4T.
ProSAP1 forward primer, 5′-AATTCACAGCCAGAAAGAAAGCTCCCCCACCTCCAAAGCGGGCTCCGACCG-3′; ProSAP1 reverse primer, 5′-TCGACGGTCGGAGCCCGCTTTGGAGGTGGGGGAGCTTTCTTTCTGGCTGTG-3′; ProSAP2 forward primer, 5′-AATTCAGTGCTCGGCGCAGAGCCCCACCACCTCCCAAGAGGGCCCCCAGCG-3′; ProSAP2 reverse primer, 5′-TCGACGCTGGGGGCCCTCTTGGGAGGTGGTGGGGCTCTGCGCCGAGCACTG-3′.
For syndapin I RNAi, sequence No. 2 was used as described and characterized in Dharmalingam et al. (2009). Syndapin I RNAi sequence, 5′-GATCCGCTCGTTCAGTAGGCTGTTCTTGAT-ATCCGGAACAGCCTACTGAACGAGTTTTTTA-3′ and 5′-AGCTTAAAAAACTCGTTC-AGTAGGCTGTTCCGGATATCAAGAACCAGCCTACTGAACGAGG-3′.
A nonsilencing, scrambled RNAi was generated by annealing primers (sequence below) and cloning in pRNAT-H1.1.
Scrambled RNAi-forward primer, 5′-GATCCCACAGCGTCTGAACGTAGTTTCAAGAGAACTACGTTCAGACGCTGTGTTTTT-3′; scrambled RNAi reverse primer, 5′-AGCTTAAAAACACAGCGTCTGAACGTAGTTCTCTTGAAACTACGTTCAGACGCTGTG-3′.
RNAi plasmids directed against ProSAP1 were generated with annealing primers according to shRNAs sequence No. 2 (Berkel et al., 2012).
Derivatives coexpressing PM-mCherry were generated by replacing the GFP reporter encoded by pRNAT-H1.1-GFP (GenScript) by PM-mCherry using the NheI and SmaI sites of pRNAT-H1.1.
Correct cloning by PCR and/or primer annealing was verified by sequencing in all cases.
Antibodies, recombinant proteins, and dyes
Rabbit anti–syndapin I and anti-GST antibodies were raised against GST–syndapin IΔSH3 and affinity-purified using immobilized MBP–syndapin IΔSH3 and GST protein, respectively, as described previously (Qualmann et al., 1999). Guinea pig anti–syndapin I and anti-GST antibodies were raised against GST–syndapin IΔSH3 and affinity-purified using MBP–syndapin IΔSH3 and GST protein, respectively, as described previously (Braun et al., 2005). Guinea pig anti-HisTrx antibodies were raised against HisTrx-Cobl aa 750–1,005 and affinity-purified using HisTrx protein as described in Haag et al. (2012).
Monoclonal anti–ProSAP1 (N23B/6) antibodies were from NeuroMab. Aliquots of rabbit and guinea pig anti-ProSAP1 antibodies were provided by T. Boeckers. Antibodies were raised against GST-ProSAP1 aa 355–509/aa 826–1,259 and affinity-purified as described in Boeckers et al. (1999a). Polyclonal rabbit anti-GFP and anti-MAP2 antibodies as well as monoclonal anti–PSD-95 (6G6-1C9) antibodies were from Abcam. Monoclonal mouse anti-GFP antibodies (B34, JL-8) were from Covance and Takara Bio Inc., respectively. Monoclonal mouse anti-synapsin 1 (46.1) antibodies were from Synaptic Systems. Polyclonal rabbit and monoclonal mouse anti-FLAG (M2) and anti-MAP2 (HM-2) antibodies were from Sigma-Aldrich. Rabbit anti-Bassoon (sap7f; tom Dieck et al., 1998) antibodies were provided by E.D. Gundelfinger (Leibniz Institute for Neurobiology, Magdeburg, Germany). Monoclonal mouse anti-Xpress antibodies were from Invitrogen.
Secondary antibodies used included Alexa Fluor 488– and 568–labeled goat anti–guinea pig antibodies; Alexa Fluor 488– and 568–labeled donkey anti–mouse as well as Alexa Fluor 647–labeled goat anti–mouse antibodies; Alexa Fluor 488–labeled donkey anti–rabbit and Alexa Fluor 568– and 647–labeled goat anti–rabbit antibodies; Alexa Fluor 680–labeled goat anti–rabbit and anti–mouse antibodies (Molecular Probes); goat anti–rabbit, anti–guinea pig, and anti–mouse peroxidase antibodies (Dianova); and DyLight800-conjugated goat anti–rabbit and anti–mouse antibodies (Thermo Fisher Scientific) and donkey anti–guinea pig antibodies coupled to IRDye680 and IRDye800, respectively (LI-COR Biosciences). For immunoelectron microscopy, goat anti–rabbit conjugated to 10 nm gold (British Biocell International) served as secondary antibodies.
GST fusion proteins as well as HisTrx-tagged proteins were expressed in Escherichia coli (BL21) at 37°C for 4 h according to standard methods, and purified from cell lysates with high-affinity GST matrix (GenScript) in PBS or talon superflow metal affinity matrix (Takara Bio Inc.) in 50 mM sodium phosphate and 300 mM sodium chloride, pH 7.0, respectively, in the presence of protease inhibitor Complete EDTA-free (Roche), 1% (vol/vol) Triton X-100, DNase I, and 10 mM magnesium chloride. Elution was done with 20 mM glutathione, 50 mM Tris/HCl, 120 mM sodium chloride, pH 8.0 (for GST fusion proteins), and 50 mM sodium phosphate, 300 mM sodium chloride, and 150 mM imidazole, pH 7.0 (for HisTrx fusion proteins).
Preparation of HEK293 cell lysates
After 24–48 h of transfection, HEK293 cells were washed with PBS containing 0.5 mM MgCl2 and 0.5 mM CaCl2, harvested, spun down gently, and subjected to sonification for 10 s. Cell lysis was followed by an incubation in 10 mM Hepes, pH 7.4, 1 mM EGTA, 0.1 mM MgCl2, and 1% (vol/vol) Triton X-100 (lysis buffer) supplemented with 150 mM NaCl and protease inhibitor Complete EDTA-free (Roche) for 1 h at 4°C. Clarification of cell lysates was done by centrifugation for 20 min at 16,000 g at 4°C.
Coprecipitation of proteins from HEK293 cell lysates
For coprecipitation experiments, extracts from an HEK293 cell expressing different GFP fusion proteins were incubated for 3 h at 4°C with purified GST fusion protein immobilized on glutathione Sepharose beads (GenScript). GST fusion protein binding, lysate incubation, and washing were performed in lysis buffer with 150 mM NaCl. Bound protein complexes were eluted with 20 mM reduced glutathione, 120 mM NaCl, and 50 mM Tris/HCl, pH 8.0, and analyzed by anti-GFP and anti-GST immunoblotting. Every experiment included samples with only GST instead of GST fusion proteins attached to the matrix to control for specificity and was conducted several times to reproduce data.
Brain lysates and subcellular fractionations for coprecipitation of endogenous proteins
For coprecipitation of endogenous proteins from rat brain lysates, freshly frozen brains were dissected, homogenized in lysis buffer containing 150 mM NaCl, and subjected to ultracentrifugation according to Qualmann et al. (2004). Obtained brain lysates were used to precipitate endogenous protein complexes as described for HEK293 cell lysates, 10 mg S3 was used for precipitation of endogenous syndapin I. Eluates from the beads were analyzed by immunoblotting with anti-GST and anti–syndapin I antibodies.
In additional experiments, endogenous ProSAP1 was precipitated with syndapin I SH3 domain using the following procedure: two mouse cortices were lysed in buffer A (5 mM Hepes, pH 7.4, 0.32 M sucrose, protease inhibitor Complete EDTA-free), and mouse brain membranes (P2) were prepared according to Boeckers et al. (1999b). A soluble protein fraction was obtained after lysis in buffer B (150 mM NaCl, 2 mM EGTA, 1% [vol/vol] deoxycholate, 0.1% [vol/vol] Triton X-100, and 20 mM Tris/HCl, pH 8.1, for 1 h at 4°C) and subsequent centrifugation for 20 min at 12,000 g.
Equal amounts of supernatant (1.25 mg total protein) were transferred to beads with immobilized GST–syndapin I SH3 domain (40 µg) or GST alone (30 µg) as a control. After extensive washing with wash buffer (150 mM NaCl, 2 mM EGTA, 1% [vol/vol] Triton X-100, and 20 mM Tris/HCl, pH 8.1), bound proteins were eluted and tested for the presence of ProSAP1 by immunoblotting.
All data were reproduced by several independent experiments.
Coimmunoprecipitation analyses
Lysates from HEK293 cells transfected with different GFP- and FLAG-tagged fusion proteins were incubated for 3 h at 4°C with 6 µg of monoclonal anti-FLAG antibody, 3 µg of rabbit anti-FLAG antibody, mouse anti-GFP antibody, and nonimmune mouse and rabbit IgG (Santa Cruz Biotechnology, Inc.) prebound to protein A/G–agarose (5 h, 4°C; Santa Cruz Biotechnology, Inc.). Lysate preparation, matrix coating, binding, and washing were performed in lysis buffer containing 75 mM NaCl (for syndapin I–ProSAP1 coimmunoprecipitation) and 100 mM NaCl (for syndapin I/syndapin I coimmunoprecipitation). Coimmunoprecipitated proteins were analyzed by immunoblotting using rabbit polyclonal anti-FLAG antibodies, rabbit polyclonal and mouse monoclonal anti-GFP antibodies, and rabbit anti–syndapin I antibodies.
Several independent experiments were conducted to reproduce data.
Analyses of direct protein–protein interactions
Direct protein–protein interactions were demonstrated by coprecipitations with recombinant proteins purified from E. coli. Immobilized WT and mutant GST-ProSAP1 1–235 were incubated with HisTrx–syndapin I (HisTrx-Sdp I) in 10 mM Hepes, pH 7.4, 1 mM EGTA, 0.1 mM MgCl2, and 1% (vol/vol) Triton X-100 (lysis buffer) containing 300 mM NaCl. Eluted proteins were analyzed by SDS-PAGE and subsequent anti-HisTrx and anti-GST immunoblotting. Several independent experiments were conducted to reproduce data.
Cross-link experiments
Cross-link studies (n = 2 experimental sets) with N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC; Sigma-Aldrich), FLAG-tagged syndapin I, and a mutant thereof were performed as described previously (Kessels and Qualmann, 2006). 20-µl HEK293 cell extracts were incubated with different concentrations of EDC (0 mM, 2 mM, 5 mM, 15 mM, and 20 mM) in 10 mM Hepes, 1 mM EGTA, 0.1 mM MgCl2, 100 mM NaCl, 1% (vol/vol) Triton X-100, and protease inhibitor Complete EDTA-free for 20 min at RT. Afterward, the reaction mixtures were boiled in SDS-PAGE sample buffer and subjected to immunoblotting.
Liposome-binding assays
Liposomes were prepared with Folch fraction type I lipids (Sigma-Aldrich) using 4 mg of lipids in 0.56 ml chloroform-methanol (53:1) as described previously (Koch et al., 2012b). Dried and subsequently water-saturated lipids were incubated overnight at 37°C in 30 ml cytosol buffer (CB; 25 mM Hepes, 25 mM KCl, 2.5 mM magnesium acetate, and 100 mM potassium glutamate), collected at 28°C for 1 h at 72,000 g, and resuspended in 500 µl of the supernatant.
For floatation of protein complexes (n = 2 experimental sets), 5 µg of fusion protein and 30 µl of protein complex–containing eluates from coprecipitation experiments, respectively, were mixed with 30 µl liposomes in 100 µl CB containing 6.16% (wt/vol) sucrose and incubated at 37°C (15 min at 800 rpm). The samples were then mixed with 150 µl 75% (wt/vol) sucrose in CB, overlaid with 200 µl 35% (wt/vol) sucrose in CB and 200 µl CB, and subsequently centrifuged at 200,000 g for 30 min at 28°C. Density gradient fractions of 100 µl were collected, ethanol-precipitated, and subjected to immunoblotting.
Cell culture, transfection, and immunostaining
HEK293 and COS-7 cells were maintained in 10 ml DMEM containing 2 mM l-glutamine, 10% (vol/vol) fetal bovine serum, and penicillin/streptomycin (Invitrogen) at 37°C and 5% CO2.
HEK293 and COS-7 cells were transfected using TurboFect (Thermo Fisher Scientific). Mitochondria were stained with 0.2 µM MitoTracker Deep Red FM (Molecular Probes) in medium at 37°C and fixed after 1 h with 4% PFA for 7 min. Mitochondrial targeting experiments were conducted at least twice for each condition tested.
Primary rat hippocampal neurons were prepared from embryonic day 18 hippocampi as described previously (Qualmann et al., 2004). In brief, hippocampi were dissected in ice-cold HBSS (Invitrogen), rinsed in HBSS, and trypsinized with 0.05% trypsin/EDTA (Invitrogen) for 15 min at 37°C. The supernatant was exchanged for Neurobasal medium (Invitrogen) supplemented with 0.5 mM l-glutamine, 0.025 mM l-glutamate, and B-27 supplement (Invitrogen), and the tissue was dissociated into single cells by trituration with 1-ml Pasteur pipettes. Neurons were plated on a poly-d-lysine–coated coverslip (60,000 cells per 12-mm coverslip) and maintained at 37°C and 5% CO2.
Calcium phosphate transfections of rat hippocampal neurons at 12 d in vitro (DIV) were performed using 1.3 µg DNA, 2.5 µl CaCl2, and 40 µl 1× HBS, pH 7.12, per well (24-well plate). Cells were fixed after 48 h (DIV 14) in 4% (wt/vol) PFA in PBS, pH 7.4, at RT for 4–6 min.
For primary mouse hippocampal neurons, postnatal day 0–4 hippocampi were prepared, rinsed, and trypsinized similar to rat hippocampi. After rinsing in HBSS, cells were dissociated by trituration with 1 ml Pasteur pipettes in MEM (Invitrogen) supplemented with 10% (vol/vol) horse serum, 0.6% (wt/vol) glucose, and 4 µg/µl DNase I. Cells were plated on poly-l-lysine–coated coverslips (18 mm). After 45–60 min of incubation, cells were further maintained in Neurobasal medium supplemented with 2 mM GlutaMAX supplement (Invitrogen), 1% (vol/vol) horse serum, B-27 supplement, 5 µM β-mercaptoethanol, and 28% (vol/vol) conditioned primary rat hippocampal culture medium.
Immunolabeling was done after quenching with 25 mM glycine in PBS for 30 min. Cells were permeabilized and blocked with 10% (vol/vol) horse serum and 5% (wt/vol) BSA in PBS (blocking solution) with 0.2% (vol/vol) Triton X-100. Primary and secondary antibody incubations were done according to Qualmann et al. (2004), i.e., in blocking solution for 1 h at RT with PBS washing steps. Coverslips were dipped in distilled water and mounted on glass slides using Mowiol 4-88.
Immunolabeling and Golgi staining of mouse brain sections
Brain sections were prepared of PFA-perfused, adult (7 wk) male mice, as described previously (Haag et al., 2012). In brief, mice were anesthetized by intraperitoneal injections (100 µl per 10 g mouse) with a mixture of 1.2% (wt/vol) ketamine hydrochloride, 0.17% (vol/vol) Rompun, and 0.9% (wt/vol) sodium chloride. Perfusion was performed transcardially with PBS and subsequently with 4% (wt/vol) PFA. Removed brains were postfixed with 4% (wt/vol) PFA for 16 h at 4°C, rinsed with water, and placed first in 10% (wt/vol) and then in 30% (wt/vol) sucrose in PBS at 4°C for 24 h, respectively.
Brains were sagittally sectioned in 55-µm-thick slices on dry ice using a sliding microtome (SM2000R; Leica). The obtained slices were rocked very slowly twice in PB (77.4 mM Na2HPO4 and 22.6 mM NaH2PO4), permeabilized, and blocked for 1 h at RT in blocking solution (5% [vol/vol] goat serum, 0.25% [vol/vol] Triton X-100 in PB). Primary antibody labeling was done in blocking solution for 48 h at 4°C. Slices were washed three times and then incubated with secondary antibody conjugates (1:1,000 in PB) for 24 h at 4°C. Staining of nuclei was done with DAPI diluted 1:1,000 in PB for 30 min at RT. Slices were transferred into PBS and mounted on glass slides with Fluoromount G (SouthernBiotech) on HistoBond slides (Marienfeld-Superior).
Golgi staining of brain sections from male, adult (25–27 wk) mice (3 male mice/genotype) was performed with an FD Rapid GolgiStain kit (FD NeuroTechnologies, Inc.). Mice were deeply anesthetized with halothane (Sigma-Aldrich) and sacrificed by decapitation. Brains were removed and incubated for 21 d in impregnation solution at RT and then transferred to solution C for 6 d of incubation at 4°C. For sectioning, brains were dipped in dry ice-cold isopentane for 2 min, embedded in 2% agarose solution on dry ice, and coronally sectioned in 25-µm-thick slices at −23°C using a cryostat (CM3050S; Leica). Slices were mounted on gelatin-coated (0.3% [wt/vol] gelatin and 0.05% [wt/vol] chromium potassium sulfate dodecahydrate) slides and air dried at RT. Slices were then rinsed twice with water and stained for 30 s in staining solution. After rinsing twice with water, sections were dehydrated in an ascending ethanol series (50%, 75%, 95%, 4× 99%, all vol/vol), then cleared three times in xylene and mounted onto coverslips with Roti-Histokit II (Roth).
Light microscopy
Confocal images were recorded using a TCS SP5 microscope (Leica; equipped with HCX Apochromat U-V-I 40×/0.75 NA dry UV and HCX Plan-Apochromat lambda blue 63×/1.4 NA oil UV objective lenses; LAS AF software) or an AxioObserver.Z1 microscope (Carl Zeiss) equipped with an ApoTome. Brightfield microscopy was performed using an AxioObserver.Z1. Both Carl Zeiss microscopes were equipped with Plan-Apochromat 63×/1.4 NA oil, 40×/1.3 NA oil, and 20×/0.5 NA dry objective lenses and a charge-coupled device (CCD) camera (AxioCam MRm; Carl Zeiss).
Digital images from Carl Zeiss microscopes were recorded with AxioVision Software (Vs40 4.8.2.0). Images for quantitative analyses of dendritic spines and synapses were acquired as z series (0.2–0.3-µm intervals). Nonquantitative image processing was done with Photoshop (Adobe). Quantitative image processing was done by ImageJ 1.46r or Imaris 7.6 software (Bitplane), respectively.
Quantitative analyses of dendritic spine morphology and synapses in neuronal cultures
Spine analyses of cultured neurons were performed with ≥3 independent neuronal preparations on 2–6 independent coverslips each. All experiments included the full set of controls. Neurons were selected at random.
Densities of spines and puncta of anti–synapsin 1 (presynaptic marker) and anti–PSD-95 (postsynaptic marker) immunolabeling were determined by using clearly evaluable areas of ≥50 µm of primary dendrites (n = 10–20 for spine densities; n = 50–100 for synapse densities). All individual spiny protrusions present on the entire dendrite section were included. All immunolabeled synapses that showed some spatial overlap with the area of the transfected cells (outlined by PM-mCherry) were included irrespective of whether they appeared to be located at spines or dendrites. Quantification of spine and synaptic puncta densities as well as head width measurements were performed using ImageJ 1.46r on maximal intensity projections. Synapse densities were normalized to control cells of the same neuronal preparation processed for immunofluorescence in parallel. Data are means and SEM.
The n numbers of synapse density data shown (Figs. 2, 6, 7, S2, and S3) are as follows. Fig. 2 F: pRNAT, n = 196; Sdp I RNAi, n = 210; Sdp I RNAi+Sdp Ires, n = 164; Sdp I RNAi+Sdp IΔSH3, n = 71. Fig. 2 G: pRNAT, n = 186; Sdp I RNAi, n = 219; Sdp I RNAi+Sdp Ires, n = 181; Sdp I RNAi+Sdp IΔSH3, n = 84 dendrite sections. Fig. 2 (F and G): one-way analysis of variance (ANOVA) with Tukey’s post test. Corresponding data in absolute numbers are as follows. Fig. S2 A: pRNAT, n = 196; Sdp I RNAi, n = 210; Sdp I RNAi+Sdp Ires, n = 164. Fig. S2 B: pRNAT, n = 186; Sdp I RNAi, n = 219; Sdp I RNAi+Sdp Ires, n = 181; one-way ANOVA with Tukey’s post test. Fig. S2 M: control, n = 196; Sdp I, n = 80. Fig. S2 N: control, n = 186; Sdp I, n = 67 dendrite sections. Fig. S2 O: pRNAT, n = 71; Sdp I RNAi+Sdp IΔSH3, n = 71. Fig. S2 P: pRNAT, n = 57; Sdp I RNAi+Sdp IΔSH3, n = 84 dendrite sections. Fig. S2 (M, N, O, and P): unpaired, two-tailed Student’s t-test. Fig. 6 B: pRNAT, n = 196; ProSAP1 RNAi, n = 86; ProSAP1 RNAi+Sdp I, n = 77 dendrite sections. Fig. 6 C: pRNAT, n = 186; ProSAP1 RNAi, n = 69; ProSAP1 RNAi+Sdp I, n = 85 dendrite sections. Fig. 6 (B and C) one-way ANOVA with Tukey’s post test. Fig. S3 A: pRNAT, n = 69; ProSAP1 RNAi, n = 86; ProSAP1 RNAi+Sdp I, n = 77 dendrite sections. Fig. S3 B: pRNAT, n = 64; ProSAP1 RNAi, n = 69; ProSAP1 RNAi+Sdp I, n = 85 dendrite sections. Fig. S3 C: control, n = 196; ProSAP1, n = 45 dendrite sections. Fig. S3 D: control, n = 54; ProSAP1, n = 45 dendrite sections. Fig. S3 (C and D), unpaired, two-tailed Student’s t test (both not significantly different). Fig. 7 F: pRNAT, n = 196; Sdp I RNAi, n = 210; Sdp I RNAi+Sdp II122E/M123E, n = 59 dendrite sections. Fig. 7 G: pRNAT, n = 186; Sdp I RNAi, n = 219; Sdp I RNAi+Sdp II122E/M123E, n = 70 dendrite sections; one-way ANOVA with Tukey’s post test.
Spine morphologies were categorized as thin and filopodia-like, stubby, mushroom, or branched. Definitions used were: stubby spines, 0.75 µm maximal length; thin and filopodia-like spines, 5 µm maximal length and head width indistinguishable from neck or <0.3 µm; mushroom spines, length ≤5 µm and head width ≥0.3 µm. Coexpression of PM-mCherry was used to visualize cell morphology.
For densities of spine morphology groups, each spiny protrusion was measured and assigned to the morphological groups. In most cases 500–1,000 spines (from 10–20 dendrite sections; 10–16 neurons) were analyzed.
The n numbers of spine morphology data shown (Figs. 2, 7, and S3) are as follows. Fig. 2 (B–D): pRNAT, n = 23; scrambled RNAi, n = 14; Sdp I RNAi, n = 22; Sdp I RNAi+Sdp Ires, n = 18 dendrite sections; one-way ANOVA with Tukey’s post test. Fig. 2 (I–K): pRNAT, n = 10; Sdp I RNAi+Sdp IΔSH3, n = 14 dendrite sections; unpaired, two-tailed Student’s t test. Fig. S2 (H–K): control, n = 19; Sdp I, n = 16 dendrite sections; unpaired, two-tailed Student’s t test. Fig. S3 E: GFP, n = 20; ProSAP1, n = 23 dendrite sections. Fig. S3 F: GFP, n = 20; GFP+FLAGc, n = 11; ProSAP1, n = 23; ProSAP1+Sdp I SH3, n = 16; GFP+Sdp I SH3, n = 11 dendrite sections. Fig. S3 G: GFP, n = 20; ProSAP1, n = 23; ProSAP1*, n = 14 dendrite sections. Fig. S3 H: pRNAT, n = 23; GFP+pRNAT, n = 8; ProSAP1+pRNAT, n = 15; ProSAP1+Sdp I RNAi, n = 19; Sdp I RNAi, n = 22 dendrite sections. Fig. S3 (E–H), one-way ANOVA with Tukey’s post test. Fig. 7 (C–E): pRNAT, n = 10, Sdp I RNAi+SdpII122E/M123E, n = 14 dendrite sections; unpaired, two-tailed Student’s t test.
Spine morphology measurements also were based on coexpressed PM-mCherry highlighting the cell morphology of transfected neurons. 182–663 spines were analyzed. The distance from the base of the neck to the furthest point of the spine head was taken as the spine length. The head width was the maximal width of the spine head perpendicular to spine neck axis.
The n numbers of spine morphology data shown (Fig. 5) are as follows. Fig. 5 B: GFP, n = 471; GFP+FLAGc, n = 269; (GFP-)ProSAP1, n = 663; ProSAP1+(FLAGc-)Sdp I SH3, n = 242; GFP+(FLAGc-)Sdp I SH3, n = 235 spines. Fig. 5 E: GFP, n = 471; (GFP-)ProSAP1, n = 663; (GFP-)ProSAP1*, n = 205 spines. Fig. 5 G: pRNAT, n = 332; GFP+pRNAT, n = 182; (GFP-)ProSAP1+pRNAT, n = 308; (GFP-)ProSAP1+Sdp I RNAi, n = 246; Sdp I RNAi, n = 222 spines. Fig. 5 H: control (PM-mCherry), n = 277; (Xpress-)Sdp I (+PM-mCherry), n = 310 mushroom spines; unpaired, two-tailed Student’s t test. Fig. 5 I: pRNAT, n = 332; scrambled RNAi, n = 273; Sdp I RNAi, n = 222; Sdp I RNAi+Sdp Ires, n = 256 mushroom spines. Fig. 5 J: ProSAP1 RNAi causes a decrease in head width not seen upon syndapin I RNAi and not rescued by syndapin I coexpression. pRNAT, n = 360; ProSAP1 RNAi, n = 303; ProSAP1 RNAi+Sdp I, n = 376 spines. Fig. 5 (B, E, G, I, and J), one-way ANOVA with Tukey’s post test. Morphological analyses are based on coexpressed PM-mCherry.
Quantitative analyses of dendritic spine morphology in brain sections
Quantification of spine density in Golgi-stained mouse brain sections was done using three mice per genotype. Analyses were performed with Imaris 7.6 software (Bitplane) on maximal intensity projections from inverted brightfield images of recorded dendritic branches from CA1 pyramidal neurons in the stratum radiatum of the hippocampus. Dendrite and 3D spine reconstruction was done using the Imaris Filament Tracer set with the shortest distance algorithm, spine head ≥0.2 µm and spine length <5 µm.
Classification into spine morphology groups was performed using the Matlab spine classification tool in the Imaris software with settings as follows: stubby spines, length(spine) < 0.75; mushroom spines, length(spine) < 5 and max_width(head) > mean_width(neck)*1.5; long, thin spines, max_width(head) >= mean_width(neck); filopodia, true; head length, 45%; ground length, 25%.
N numbers of evaluated dendrite sections in Fig. 1 (B–F): +/+, n = 109; −/−, n = 93 dendrite sections from 3 male mice/genotype; unpaired, two-tailed Student’s t test.
Surface reconstruction and quantitative analysis of anti-ProSAP1 labeling
Areas of anti–PSD-95– and anti-ProSAP1–immunolabeled puncta in primary neuronal cultures from WT and syndapin I KO mice were reconstructed using the Imaris 7.6 software Surface tool on single channels or colocalization channel built via the Imaris Coloc tool on maximal intensity projections. Surfaces were built using a surface area detail level of 0.1 µm, thresholding by absolute intensity and taking all voxel >1.0 into account. The area sizes of individual anti–PSD-95 and anti-ProSAP1 immunostained puncta, the sizes of colocalization areas, the mean intensity of the puncta, and the sum of all intensities within individual immunostained puncta were analyzed and the mean values calculated. 40 cells per each genotype (mean values 200–2,000 synapses per cell) were evaluated and compared (unpaired, two-tailed Student’s t test).
Preparation and freeze-fracturing of primary neuronal cultures
Hippocampal neurons (several independent preparations) were grown for 16 d on poly-l-lysine–coated sapphire discs (4 mm diameter) in 24-well plates. Cells were washed in PBS, covered with 20% (wt/vol) BSA and a copper head sandwich profile, and plunged immediately into liquid nitrogen–cooled ethane/propane (1:1). Cooling rates were measured using a thermocouple element sandwiched between two copper profiles and a 50 MHz digital storage oscilloscope (99 spectrometer Series II; Fluke) according to Ryan et al. (1990) and Koch et al. (2012a). Cooling rates reached values of >4,000 K/s.
Sandwiches were then put into a detachable cold table cooled by liquid nitrogen, transferred into the recipient of a BAF400T freeze-fracture unit (Leica), and freeze-fractured at 10−6 mbar and a temperature of −140°C. Fractured membranes were evaporated first with 15–20 nm carbon and then with 2 nm platinum/carbon (35° angle). This order of coating resulted in improved immunolabeling. The thickness of the carbon layer was controlled visually. Platinum/carbon coating was followed by a thin layer quartz crystal monitor. Freeze-fracture replicas were then transferred to 2.5% (wt/vol) SDS and 30 mM sucrose in 10 mM Tris/HCl, pH 8.4, and incubated with gentle shaking overnight (Fujimoto, 1995).
Freeze-fracturing of brain slices
For preparation of freeze-fracture replica directly from brain, WT and syndapin I KO mice (two each) were anesthetized with halothane, the brains were removed, and the hippocampi were prepared and cut sagittally in 300-µm-thick slices using a McIlwain Tissue Chopper (Ted Pella, Inc.). The sections were transferred into PBS and the CA1 region was frozen immediately in a droplet of 20% (wt/vol) BSA between a copper head sandwich profile using a 1:1, liquid nitrogen–cooled ethane/propane mixture.
The copper sandwiches were freeze-fractured at 5 × 10−7 mbar and a temperature of −140°C, shadowed with carbon and platinum/carbon, and incubated in 5% (wt/vol) SDS and 30 mM sucrose in 10 mM Tris/HCl, pH 8.4, overnight. The n number evaluated (Fig. S5 F) was 20 spines each.
Immunolabeling and TEM of freeze-fractured neuronal membranes
Replica from several independent freeze-fracturing experiments were washed (three times with PBS); blocked with 1% (wt/vol) BSA, 0.5% (wt/vol) gelatin, and 0.0005% (vol/vol) Tween 20 in PBS; and incubated with rabbit anti–syndapin I antibodies (1:50; 4°C overnight) and 10 nm gold-conjugated secondary antibodies (2 h, RT). Experiments controlling for a putative dependence of clustered anti–syndapin I immunolabeling on antibody concentration were performed with a 1:20 dilution of anti–syndapin I antibodies (Fig. S5 I; 1:20 dilution of antibody [Ab 1:20], n = 32 spines; Ab 1:50, n = 134 spines; two-way ANOVA, not significantly different). Further controls addressing the specificity of anti–syndapin I labeling of freeze-fracture replica included evaluations of labeling at the E-face (almost no labeling) and quantitative analyses of syndapin I labeling densities at control surfaces not representing cellular membranes (very low immunogold labeling; 0.9 particles/µm2; n = 54 areas [∑96.3 µm2]), secondary antibody control incubations (almost undetectable labeling; 0.1 particles/µm2; n = 20 spine membrane areas), and labeling trials with syndapin I KO material (also low labeling; 2.6 particles/µm2; n = 20 spine membrane areas). Subsequently, replica were washed three times with PBS and fixed with 0.5% (vol/vol) glutaraldehyde, collected on uncoated copper grids (300 mesh), and viewed with an electron microscope (EM902A; Carl Zeiss) run at 80 kV to explore grids systematically. Images were taken with a CCD camera (1,000 FastScan) and software (Tietz Video and Image Processing Systems), and processed with Photoshop.
Quantitative EM analyses
All immunolabeled spines with recognizable head structures (thin, stubby, mushroom) were analyzed by counting gold particles per spine, per spine subcompartment, and per square micrometer, respectively. Areas were calculated using ImageJ. Filopodia-like protrusions (no head, pointed tip, length ≤5 µm) were relatively rare and therefore not evaluated quantitatively.
Spine morphology definitions: stubby, spines with a head but without an elongated neck region; thin, head-carrying with the head width ≤1.5× neck width; mushroom, head width >1.5× neck width. Spine subcompartments: base, extrapolated dendrite border ±150 nm perpendicular; neck, PM running parallel; and head. Data are based on several independent neuronal preparations, sapphire discs, and freeze-fracture experiments.
The data shown in Figs. 8 and S5 are based on the following n numbers. Spines, n = 134 (∑50.72 µm2); dendrite, n = 54 (∑28.42 µm2); control, n = 54 (∑96.3 µm2); one-way ANOVA with Tukey’s post test. The n number of spines was separated into the different spine classes as follows: thin, n = 30; stubby, n = 48; mushroom, n = 56; one-way ANOVA with Tukey’s post test.
Cluster analyses were done with 100-nm diameter regions of interest, and statistical significance calculations were performed using two-way ANOVA.
Patch clamp recordings
Primary hippocampal neuron cultures (transfected at DIV 12/13) were placed in a submerged recording chamber mounted on an upright fluorescence microscope (BX51WI; Olympus) 48 h later. The extracellular solution was composed of 120 mM NaCl, 3.5 mM KCl, 1.3 mM MgSO4, 1.25 mM NaH2PO4, 2.5 mM CaCl2, 10 mM d-glucose, and 10 mM Hepes, pH 7.3. Cultures were superfused continuously with a flow of 2–3 ml/min at 32°C. mEPSCs were isolated by adding tetrodotoxin (0.5 µm; Tocris Bioscience) and bicuculline methiodide (20 µm; Enzo Life Sciences) to block action potential–induced glutamate release and GABAA receptor-mediated mIPSCs, respectively. Whole-cell recordings of mEPSCs were performed and analyzed as described previously (Sinning et al., 2011). The n numbers of recordings were as follows. pRNAT = pRNATGFP+mCherry, n = 19; and Sdp I RNAi = Sdp I RNAiGFP+mCherry, n = 21 from 6 experiments each; Sdp I RNAi+Sdp Ires, n = 15 from 4 experiments; Sdp I RNAi+Sdp IΔSH3, n = 12; and Sdp I RNAi+SdpII122E/M123E, n = 12 from 2 experiments each; unpaired, two-tailed Student’s t test (Fig. 9, F–J). Data in Fig. 9 (A–E) are n = 12 from four experiments each; one-way ANOVA with a Newman-Keuls post test.
Statistical analyses
Statistical analysis was performed using Prism 5.03 (GraphPad Software). Statistical significances of spine and synapse analyses were tested using the unpaired, two-tailed Student’s t test and one-way ANOVA with Tukey’s post test, respectively. Statistical analyses of clustered syndapin I localization at membranes of dendritic spines was performed using two-way ANOVA. Electrophysiological differences (mEPSC frequency and amplitude) were tested for statistical significance using an unpaired, two-tailed Student’s t test and one-way ANOVA with Newman-Keuls post test. Data from constructions of sub-spine ProSAP1 and PSD-95 labeling surfaces of WT and syndapin I KO neurons were tested for statistical significance using an unpaired, two-tailed Student’s t test.
Online supplemental material
Fig. S1 shows that endogenous as well as recombinant syndapin I is present in both the axonal/presynaptic as well as the dendritic/postsynaptic compartment of cultured primary rat hippocampal neurons. Fig. S2 displays the absolute values for the normalized data shown in Fig. 2 (F and G), contains further details on the synapse and spine morphology analyses in syndapin I RNAi neurons (compare with Fig. 2, E–G), and presents a comprehensive analysis of spine morphology and synapse density changes upon syndapin I overexpression (G–N). Fig. S3 shows the absolute values for the normalized data shown in Fig. 6 (B and C) and the synapse and mushroom spine densities upon ProSAP1 overexpression neurons (compare with Fig. 5, B, E, and G). Fig. S4 shows cross-link and heterologous coimmunoprecipitation experiments of the syndapin I wedge loop mutant demonstrating that F-BAR domain–mediated self-association remains intact upon I122E/M123E mutation. Fig. S5 shows a characterization of primary rat hippocampal neurons cultured on sapphire discs, highlights the different spiny structures preserved upon freeze-fracturing, demonstrates the specificity of the anti–syndapin I labeling, shows detailed data on the syndapin I cluster analysis (compare with Fig. 8, J and K), and confirms the saturation of the anti–syndapin I immunolabeling. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201307088.dv.
Acknowledgments
We thank M. Korte, T.M. Boeckers, E.D. Gundelfinger, and C. Sala for plasmids and antibodies, respectively. We thank A. Haeckel for initial observations and discussions, A. Kreusch, B. Schade, C. Scharf, and D. Wolf for technical help, and L. McMillan for proofreading.
This work was supported by grants from the Schram Foundation to B. Qualmann (T287/16245/2006) and from the Deutsche Forschungsgemeinschaft to C.A. Hübner (HU800/6-1), M.M. Kessels (KE685/2-3), and B. Qualmann (Qu116/5-1 and 5-2 and QU116/6-1).
The authors declare no competing financial interests.
References
Author notes
R. Ahuja’s present address is Institute of Biotechnology, University of Helsinki, 00014 Helsinki, Finland.