Cell migration through 3D tissue depends on a physicochemical balance between cell deformability and physical tissue constraints. Migration rates are further governed by the capacity to degrade ECM by proteolytic enzymes, particularly matrix metalloproteinases (MMPs), and integrin- and actomyosin-mediated mechanocoupling. Yet, how these parameters cooperate when space is confined remains unclear. Using MMP-degradable collagen lattices or nondegradable substrates of varying porosity, we quantitatively identify the limits of cell migration by physical arrest. MMP-independent migration declined as linear function of pore size and with deformation of the nucleus, with arrest reached at 10% of the nuclear cross section (tumor cells, 7 µm2; T cells, 4 µm2; neutrophils, 2 µm2). Residual migration under space restriction strongly depended upon MMP-dependent ECM cleavage by enlarging matrix pore diameters, and integrin- and actomyosin-dependent force generation, which jointly propelled the nucleus. The limits of interstitial cell migration thus depend upon scaffold porosity and deformation of the nucleus, with pericellular collagenolysis and mechanocoupling as modulators.
Introduction
Cell migration along and through 3D extracellular matrix (ECM) is fundamental to tissue formation and regeneration, immune cell trafficking, and disease, including cancer invasion and metastasis. Interstitial migration is a cyclic multi-step process consisting of (1) actin polymerization-dependent pseudopod protrusion at the leading edge; (2) integrin-mediated adhesion to ECM; (3) contact-dependent ECM cleavage by cell surface proteases; (4) actomyosin-mediated contraction of the cell body increasing longitudinal tension; and (5) rear retraction and translocation of the cell body (Ridley et al., 2003; Friedl and Wolf, 2009; Friedl and Alexander, 2011). This program is constitutively active in mesenchymal cells, including fibroblasts and solid tumor cells (Wolf et al., 2007; Sanz-Moreno et al., 2008; Sabeh et al., 2009; Grinnell and Petroll, 2010), which display prominent protrusions and spindle-shaped morphology, strong adhesion to ECM, and proteolytic tissue remodeling. In contrast, interstitial leukocyte movement is characterized by an ellipsoid, rapidly deforming morphology with small protrusions, weak adhesion, and lack of proteolysis (Wolf et al., 2003b; Sabeh et al., 2009). Consequently, each step is considered adaptive in response to cell-intrinsic and extracellular chemical or mechanical signals, including regulators of adhesion, cytoskeletal dynamics, proteolysis, deformation of the cell body, and/or ECM geometry (Berton et al., 2009; Lautenschläger et al., 2009; Friedl and Wolf, 2010; Friedl et al., 2011; Tong et al., 2012).
Interstitial invasion of mesenchymal cells, including fibroblasts and tumor cells into collagen-rich ECM is controlled by MMPs (matrix metalloproteinases), particularly membrane-tethered (MT)1-MMP/MMP-14 as the key enzyme degrading intact fibrillar collagen (Sabeh et al., 2004; Wolf et al., 2007; Rowe and Weiss, 2009). Active MT1-MMP focalizes at contacts to collagen and cleaves fibrils that act as barriers to migration, particularly at pseudopod branches and along the cell body, and inhibition of MT1-MMP abrogates collagen cleavage and ECM remodeling (Sabeh et al., 2004; Wolf et al., 2007). As a consequence, nonproteolytic migration is either maintained by amoeboid cell deformation (Wolf et al., 2003a) or is abrogated (Sabeh et al., 2004), dependent on the type of collagen scaffold used as migration substrate (Packard et al., 2009; Sodek et al., 2008; Sabeh et al., 2009). Scaffolds reconstituted from different collagen sources vary in physicochemical properties, including porosity and stiffness (Zaman et al., 2006; Sabeh et al., 2009; Wolf et al., 2009; Yang and Kaufman, 2009; Miron-Mendoza et al., 2010; Yang et al., 2010). However, an integrative concept as to how ECM properties either allow or restrict migration as a function of MMP activity is lacking.
Here, we address the rate-limiting substrate conditions that enable or preclude the migration of different cell types in 3D extracellular matrices. Using live-cell microscopy, we first monitored migration rates and the associated deformation of both the cell body and nucleus in 3D matrices that range from low to high density. After mapping the subtotal and absolute migration limits, we then addressed important molecular modulators of migration efficacy in confined space. By multi-parameter analyses, we identify the ratio between ECM density and cell deformation as the key parameters controlling cell migration in dense tissue environments, with MMP activity, actomyosin-based contractility, and integrin-mediated mechanocoupling as modulators of invasion efficacy.
Results
In vitro reconstitution of collagen matrices
3D hydrogels were reconstituted from either telopeptide-intact covalently cross-linked collagen after acid extraction of rat tail tendon or telopeptide- and crosslink-reduced bovine dermal collagen after acid and pepsin treatment (Wolf et al., 2003a; Sabeh et al., 2004; Sodek et al., 2007; Packard et al., 2009). Equal collagen concentrations (1.7 mg/ml) were compared for assembly speed, fibril architecture, porosity, and matrix stiffness. For controlled culture and imaging conditions, the matrices were anchored using a custom glass chamber (Fig. S1 A). Time-to-polymerization monitored by confocal backscatter microscopy at 37°C yielded 16-fold faster assembly of rat tail (half-maximum polymerization after 30 s) relative to bovine dermal collagen (8 min; Fig. 1 A). The different assembly speeds are consistent with the divergent telopeptide contents of the collagen preparations (Helseth and Veis, 1981; Sabeh et al., 2009). The fibrillar matrix architecture, as detected by confocal reflection microscopy, was verified by collagen type-I immunofluorescence, hence confirming a negligible detection error from backscatter-negative fibrils in vertical orientation (below 3% of signal-containing pixels; Fig. S1, B–D; Jawerth et al., 2010). Whereas rat tendon–derived collagen formed thin fibrils with a 20-nm diameter and a narrow pore size range of 2–5 µm2 (1–2 µm pore diameters), bovine dermis–derived collagen matrices were comprised of fibrils with a diameter of 60 nm and pore cross sections ranging from 6–30 µm2 (2–6 µm pore diameters; Fig. 1 B, D, E; Fig. S2 A, B, D, and E). The fractal box count, measured as fibril-containing fields of decreasing side length, was two- to threefold higher for rat tail collagen compared with concentration-matched bovine collagen (Fig. S2, C and F). To control for fibril density–dependent changes in collagen lattice stiffness, atomic force microscopy (AFM) was used with a 10-µm bead as probe to approximate the size of a cell. The surface of the lattice was repeatedly probed by the cantilever (Fig. S1 A) and both bead penetration and force were co-registered (Fig. 1 C, left). Consistent with previous reports, AFM revealed a twofold lower elastic modulus for bovine dermal compared with rat tail collagen at 1.7 mg/ml (28 vs. 51 Pa; Fig. 1 C, right; Stein et al., 2008; Yang and Kaufman, 2009). Thus, although reconstituted at equivalent concentrations, the collagen preparations substantially diverge in terms of fibril diameter and interfibrillar space, and moderately in network stiffness.
3D collagen lattices of varying origin and cross-link status: Assembly speed, stiffness, and pore sizes at different polymerization conditions. (A) Assembly speed and (B) organization of acid-extracted pepsinized bovine dermal compared with acid-extracted rat tail collagen matrices after in vitro reconstitution (1.7 mg/ml). (A) Real-time confocal reflection microscopy of fibril formation, represented as kymograph (top panel; B, bovine dermal collagen; R, rat tail collagen) and time-dependent signal intensity curves normalized to the end-point of complete polymerization (bottom panel, horizontal solid line; n = 3 ± SD, shaded areas). Dashed lines, half-maximal polymerization after 8 min (bovine) and 30 s (rat tail, also see inset). (B) Collagen fibril diameter and network geometry, detected by transmission and scanning electron microscopy (TEM, SEM) of fixed dehydrated lattices and confocal reflection microscopy (horizontal, xy; vertical, xz) of native hydrated samples. Marked areas in xz represent signal-free regions referred to as pores. Brackets (TEM) indicate representative fibril calibers. (C) Determination of elastic modulus of bovine and rat tail collagen lattices by AFM. Left, force curves while probing bovine dermal or rat tail collagen lattices (1.7 mg/ml). Dotted curves, medians of all values fitted by the Hertz formula. Oscillatory example curves represent each one of 5–35 repeats. Vertical lines, deformation depth at 1,500 pN applied force. Right, stiffness for different collagen types and concentrations. Unless stated otherwise, polymerization temperature was 37°C. ***, P < 0.0001; **, P < 0.01; *, P < 0.05; n.s., not significant; data points represent 5–35 measurements from different locations as average of 2–20 repetitive tappings/position. Gray horizontal lines, median. (D–F) Pore cross sections quantified by confocal reflection xz sections of bovine dermal or rat tail collagen lattices of different polymerization conditions. Horizontal lines, medians. Data show 1–2 representative experiments from n = 3–4; 21 data points each. ***, P < 0.001; **, P < 0.01; *, P < 0.05; n.s., nonsignificant. (F) Right column, SEM from rat tail collagen lattices assembled at different temperatures. Bars: (A; B, confocal reflection) 10 µm; (B, F, SEM) 1 µm; (B, TEM) 0.1 µm.
3D collagen lattices of varying origin and cross-link status: Assembly speed, stiffness, and pore sizes at different polymerization conditions. (A) Assembly speed and (B) organization of acid-extracted pepsinized bovine dermal compared with acid-extracted rat tail collagen matrices after in vitro reconstitution (1.7 mg/ml). (A) Real-time confocal reflection microscopy of fibril formation, represented as kymograph (top panel; B, bovine dermal collagen; R, rat tail collagen) and time-dependent signal intensity curves normalized to the end-point of complete polymerization (bottom panel, horizontal solid line; n = 3 ± SD, shaded areas). Dashed lines, half-maximal polymerization after 8 min (bovine) and 30 s (rat tail, also see inset). (B) Collagen fibril diameter and network geometry, detected by transmission and scanning electron microscopy (TEM, SEM) of fixed dehydrated lattices and confocal reflection microscopy (horizontal, xy; vertical, xz) of native hydrated samples. Marked areas in xz represent signal-free regions referred to as pores. Brackets (TEM) indicate representative fibril calibers. (C) Determination of elastic modulus of bovine and rat tail collagen lattices by AFM. Left, force curves while probing bovine dermal or rat tail collagen lattices (1.7 mg/ml). Dotted curves, medians of all values fitted by the Hertz formula. Oscillatory example curves represent each one of 5–35 repeats. Vertical lines, deformation depth at 1,500 pN applied force. Right, stiffness for different collagen types and concentrations. Unless stated otherwise, polymerization temperature was 37°C. ***, P < 0.0001; **, P < 0.01; *, P < 0.05; n.s., not significant; data points represent 5–35 measurements from different locations as average of 2–20 repetitive tappings/position. Gray horizontal lines, median. (D–F) Pore cross sections quantified by confocal reflection xz sections of bovine dermal or rat tail collagen lattices of different polymerization conditions. Horizontal lines, medians. Data show 1–2 representative experiments from n = 3–4; 21 data points each. ***, P < 0.001; **, P < 0.01; *, P < 0.05; n.s., nonsignificant. (F) Right column, SEM from rat tail collagen lattices assembled at different temperatures. Bars: (A; B, confocal reflection) 10 µm; (B, F, SEM) 1 µm; (B, TEM) 0.1 µm.
Pore size dependence of proteolytic and nonproteolytic migration of HT1080 cells
To recapitulate differences in MMP-independent cell migration rates between the collagen preparations, MT1-MMP–transduced HT1080 (HT/MT1) cells were used as model for efficient collagenolytic invasion converting to collagenolysis-independent migration after broad-spectrum MMP inhibition with GM6001 or MT1-MMP silencing (Wolf et al., 2007; Sabeh et al., 2009). Baseline migration of HT/MT1 cells reached 0.7 µm/min in bovine dermal and 0.3 µm/min in rat tail collagen lattices at 1.7 mg/ml collagen content (Fig. 2, A and C; Video 1 A), suggesting that MMPs enable migration in collagen scaffolds of diverging pore size. Conversely, broad-spectrum MMP inhibition by GM6001 prompted cell immobilization in rat tail but not bovine dermal collagen in three different migration models (Fig. S1 A), including individual cell migration quantified by cell tracking (Fig. 2, A and C; Video 1 B), cell emigration from multicellular spheroids (Fig. S3 A and B [top]; and C [top]; Video 3), and vertical invasion after cell seeding atop 3D collagen lattices (Fig. S3 D). Equivalent inhibition of migration was obtained by transient knockdown of MT1-MMP expression, confirming MT1-MMP as the invasion-promoting collagenase (Fig. S3 B, bottom; Sabeh et al., 2009).
Spontaneous migration of HT1080/MT1 cells, and migration arrest after inhibition of MMP activity by GM6001 in 3D rat tail but not bovine dermal collagen. Imaging by bright-field inverse microscopy (Leica) at 37°C. (A, first sequence) Overview movies of spontaneous HT1080/MT1 cell movement in bovine dermal (left) and rat tail collagen of 1.7 mg/ml (right); representative still images are shown in Fig. 2, A and C. Second clip: as first, plus 20 µM GM6001. Cell tracks indicate the migratory capacity of each condition. Time (h:min:s) as indicated (24-min intervals over 18 h of observation).
Spontaneous migration of HT1080/MT1 cells, and migration arrest after inhibition of MMP activity by GM6001 in 3D rat tail but not bovine dermal collagen. Imaging by bright-field inverse microscopy (Leica) at 37°C. (A, first sequence) Overview movies of spontaneous HT1080/MT1 cell movement in bovine dermal (left) and rat tail collagen of 1.7 mg/ml (right); representative still images are shown in Fig. 2, A and C. Second clip: as first, plus 20 µM GM6001. Cell tracks indicate the migratory capacity of each condition. Time (h:min:s) as indicated (24-min intervals over 18 h of observation).
Emigration of HT1080/MT1 cells from multicellular spheroids into bovine (left) or rat (right) collagen (1.7 mg/ml) in the absence (A, first sequence) or presence (B, second sequence) of MMP inhibitor GM6001 with paths representing the position change of cell centers. Representative still images are shown in Fig. S3 A. Bright-field inverse microscopy (Leica) at 37°C; time (h:min:s) as indicated (16-min intervals over 8.5 h of observation).
Emigration of HT1080/MT1 cells from multicellular spheroids into bovine (left) or rat (right) collagen (1.7 mg/ml) in the absence (A, first sequence) or presence (B, second sequence) of MMP inhibitor GM6001 with paths representing the position change of cell centers. Representative still images are shown in Fig. S3 A. Bright-field inverse microscopy (Leica) at 37°C; time (h:min:s) as indicated (16-min intervals over 8.5 h of observation).
Collagen pore size determines physical migration limits of HT/MT1 cells in 3D collagen lattices. (A, C, and E) Single-cell migration rates in bovine dermal or rat tail collagen lattices of varying concentration or polymerization temperature (1.7 mg/ml) using anchored 3D matrices in migration chambers (shown in Fig. S1 A), monitored by time-lapse microscopy and analyzed by single-cell cell tracking. Migration paths (A and C, left) and averaged single-cell speeds (A and C, right; E) after 24 h of observation in the absence or presence of MMP inhibitor GM6001. Horizontal lines and boxes and whiskers show the medians, 25th/75th, and 5th/95th percentile (50–180 cells, 19–24 h of migration analyzed, 3–6 independent experiments). Dashed lines indicate references for 100% MMP-independent movement and 90% inhibition. ***, P < 0.0001; n.s., not significant. (B, D, and F) Correlation between median pore cross section (from Fig. 1, D–F) and proteolytic and MMP-independent cell speed (A, C, and E) for each collagen condition. Symbols show medians, and whiskers the 5th/95th percentiles. R2 (describing how well the regression line approximates real data points), slopes, and significance between slopes from untreated and GM6001-treated populations were as follows: (B) bovine dermal collagen, untreated control 0.998/0.044; GM6001-treated population 0.991/0.040/n.s.; (D) rat tail collagen, untreated control 0.851/0.010, GM6001-treated cells 0.999/0.017/*, P < 0.05 (P = 0.045); (F) rat tail collagen with varied polymerization temperature, untreated control 0.960/0.007, GM6001-treated cells 0.972/0.017/*, P < 0.05 (P = 0.01). Dashed lines, 90% inhibition of migration. Bars: (A and C) 100 µm.
Collagen pore size determines physical migration limits of HT/MT1 cells in 3D collagen lattices. (A, C, and E) Single-cell migration rates in bovine dermal or rat tail collagen lattices of varying concentration or polymerization temperature (1.7 mg/ml) using anchored 3D matrices in migration chambers (shown in Fig. S1 A), monitored by time-lapse microscopy and analyzed by single-cell cell tracking. Migration paths (A and C, left) and averaged single-cell speeds (A and C, right; E) after 24 h of observation in the absence or presence of MMP inhibitor GM6001. Horizontal lines and boxes and whiskers show the medians, 25th/75th, and 5th/95th percentile (50–180 cells, 19–24 h of migration analyzed, 3–6 independent experiments). Dashed lines indicate references for 100% MMP-independent movement and 90% inhibition. ***, P < 0.0001; n.s., not significant. (B, D, and F) Correlation between median pore cross section (from Fig. 1, D–F) and proteolytic and MMP-independent cell speed (A, C, and E) for each collagen condition. Symbols show medians, and whiskers the 5th/95th percentiles. R2 (describing how well the regression line approximates real data points), slopes, and significance between slopes from untreated and GM6001-treated populations were as follows: (B) bovine dermal collagen, untreated control 0.998/0.044; GM6001-treated population 0.991/0.040/n.s.; (D) rat tail collagen, untreated control 0.851/0.010, GM6001-treated cells 0.999/0.017/*, P < 0.05 (P = 0.045); (F) rat tail collagen with varied polymerization temperature, untreated control 0.960/0.007, GM6001-treated cells 0.972/0.017/*, P < 0.05 (P = 0.01). Dashed lines, 90% inhibition of migration. Bars: (A and C) 100 µm.
To test whether divergence of MMP-independent migration arises as a function of fibril density, matrix porosity was normalized by varying the collagen content. Increasing the concentration of bovine dermal collagen from 1.7 to 15 mg/ml resulted in significantly smaller pore cross sections (median 5 µm2; Fig. 1 D; Fig. S2, A–C), higher stiffness (shown for a bovine lattice of 6.6 mg/ml; Fig. 1 C), and a dose-dependent migration delay, ultimately resulting in the immobilization of HT/MT1 cells in the presence of GM6001 (Fig. 2 A). To categorize this graded response, we operationally defined subtotal and absolute migration limits as 90 and 99% speed delay, respectively. Impeded migration in bovine dermis collagen was a linear function of the pore size for both MMP-mediated and MMP-independent migration with similar slope, but higher offset with MMPs available (Fig. 2 B). Conversely, decreasing the concentration of rat tail collagen to a minimum of 0.3 mg/ml, which lowers scaffold stiffness (Fig. 1 C) but provides larger pore cross sections of 20–30 µm2 (Fig. 1 E; Fig. S2, D–F), fully rescued MMP-independent migration, eventually reaching the speed distribution of control cells (Fig. 2 C). Again, migration was a linear function of pore size, with a significantly steeper slope when MMP activity was inhibited (Fig. 2 D).
To avoid varying collagen concentrations, and to minimize indirect effects unrelated to porosity (e.g., altered density of adhesive ligands [Zaman et al., 2006] or uncontrolled collagen cross-link density in bovine dermal collagen), the porosity of rat tail collagen lattices was altered by reducing the polymerization temperature (Raub et al., 2007) while keeping collagen concentration constant (1.7 mg/ml). At 9°C, fibrillogenesis was delayed (Fig. S2 H, top) and, consequently, pore diameters and related cross sections increased (median diameter 8 µm and median cross section 30 µm2) and fractal box count decreased, while fibril diameter (80 nm) and stiffness increased (Fig. 1, C and F; Fig. S2, H [bottom] and L). Again, as pore size increased, MMP-independent migration in rat tail collagen was rescued with single-cell movement reaching peak speeds equal to proteolytic migration rates (Fig. 2 E; Video 4). This rescue was achieved as well for cell emigration from multicellular spheroids (Fig. S3 C) or vertical invasion cultures (Fig. S3 D). As before, the speed of MMP-independent migration was a linear function of pore size with a significantly steeper slope observed relative to protease-dependent migration rates (Fig. 2 F). Hence, irrespective of collagen preparation, the strategy used to alter pore size, or migration assay used, cell speed was a linear function of pore size. Notably, the availability of MMP activity was sufficient to maintain migration even when cells were confronted with small pores that otherwise impeded or arrested migration (Fig. 2 F).
Rescue of MMP-independent migration of HT1080/MT1 cells in rat tail collagen (1.7 mg/ml) in the presence of GM6001 after reconstitution at different polymerization temperatures indicated in the movies. Bright-field inverse microscopy (Leica) at 37°C. Time (h:min:s) as indicated (32-min intervals over 22 h of observation).
Rescue of MMP-independent migration of HT1080/MT1 cells in rat tail collagen (1.7 mg/ml) in the presence of GM6001 after reconstitution at different polymerization temperatures indicated in the movies. Bright-field inverse microscopy (Leica) at 37°C. Time (h:min:s) as indicated (32-min intervals over 22 h of observation).
MMP-independent migration in dense ECM depends upon deformation of the nucleus
To identify the subcellular compartments that modulate migration in dense 3D environments, the morphokinetic changes of cell body, leading edge, and nucleus were assessed as a function of collagen density. HT/MT1 cells embedded in bovine dermal collagen in the presence of GM6001 moved both their leading edge and cell bodies at approximately equal velocities. In contrast, after migration arrest in rat tail collagen, cells displayed an immobile spherical central body with one or more dynamic, dendrite-like extension (Fig. 3 A; Video 1 B) with zones of collagen deformation at the front pole (Video 2). In addition, pseudopods occasionally broke away from cell bodies and moved with snake-like morphology (Video 3 B, right). The arrested portion of the cell body consisted of cytoplasm and nucleus forming occasional small protrusions (prolapse) pointing toward the leading cell extension (Fig. 3, B and C; Video 2, arrow; Video 5). In arrested cells, nuclear prolapses measured 1–3 µm in diameter (1–7 µm2 cross section), whereas nuclei of cells during MMP-independent migration adopted hourglass deformations that were 3–7 µm in diameter (7–40 µm2 cross section; Fig. 3, B–D). These morphological signatures were distinct from the ellipsoid nuclear shapes (8–11 µm in diameter; 50–100 µm2 in cross section) observed in MMP-competent HT1080 cells that generated proteolytic tracks whose widths match those of the cell diameter (Fig. 3, C and D). Accordingly, irrespective of the collagen preparation or scaffold porosity, the nuclear diameters of MT1-MMP–expressing control cells retained average cross sections of 40–90 µm2 (Fig. 3 E). Conversely, GM6001 caused a gradual decrease of nuclear diameters until conditions of arrest were reached (Fig. 3 E), with either a singular small nuclear prolapse (Fig. 3 D;,Fig. 3 E, pink dotted circles) or a nondeformed spherical shape (Fig. 3 D; Fig. 3 E, black dotted circles). These population data confirm time-lapse imaging data that show immobilized nuclei cycling between prolapse and rounding (Video 2). In summary, hourglass-shaped deformations of the nucleus are sufficient to maintain MMP-independent migration of HT/MT1 cells through 3D collagen lattices with pore diameters above 7 µm2, whereas smaller pore sizes defeat nuclear deformability and prompt physical arrest despite persistent leading edge kinetics and frustraneous force generation. Hence, MMP-independent migration in dense ECM depends upon deformation of the nucleus.
Confocal time sequence of migration arrest. Single HT1080/dual-color cell expressing cytoplasmic DsRed2 (red) and nuclear histone-2B (H2B)–coupled EGFP (green) in rat tail collagen (1.7 mg/ml), in the presence of GM6001. White arrow highlights short-lived repeated prolapses of the nucleus in direction of the leading pseudopod. Imaging at 37°C by laser scanning confocal microscopy (510; Carl Zeiss) and maximum intensity projection of confocal z-stacks with 2.5-min time interval for 100 min.
Confocal time sequence of migration arrest. Single HT1080/dual-color cell expressing cytoplasmic DsRed2 (red) and nuclear histone-2B (H2B)–coupled EGFP (green) in rat tail collagen (1.7 mg/ml), in the presence of GM6001. White arrow highlights short-lived repeated prolapses of the nucleus in direction of the leading pseudopod. Imaging at 37°C by laser scanning confocal microscopy (510; Carl Zeiss) and maximum intensity projection of confocal z-stacks with 2.5-min time interval for 100 min.
Nuclear deformation together with ongoing or abrogated migration of HT1080 cells transfected with histone H2B-GFP and cytoplasmic DsRed2 in the presence of protease inhibitors. Deformation of the nucleus in spheroid culture of HT1080 dual-color cells expressing cytoplasmic DsRed2 (red) and nuclear histone-2B (H2B)–coupled EGFP (green) during migration (cells in green boxes) or immobilization (red boxes) in bovine collagen (6.6 mg/ml) in the presence of MMP and other protease inhibitors (see Materials and methods). Imaging at 37°C by confocal microscopy (SP2; Leica); maximum intensity projection of confocal z-stacks; 5-min frame rate, 85-min observation time.
Nuclear deformation together with ongoing or abrogated migration of HT1080 cells transfected with histone H2B-GFP and cytoplasmic DsRed2 in the presence of protease inhibitors. Deformation of the nucleus in spheroid culture of HT1080 dual-color cells expressing cytoplasmic DsRed2 (red) and nuclear histone-2B (H2B)–coupled EGFP (green) during migration (cells in green boxes) or immobilization (red boxes) in bovine collagen (6.6 mg/ml) in the presence of MMP and other protease inhibitors (see Materials and methods). Imaging at 37°C by confocal microscopy (SP2; Leica); maximum intensity projection of confocal z-stacks; 5-min frame rate, 85-min observation time.
Requirement for deformation of the nucleus during MMP-independent migration at rate-limiting pore conditions. (A) Time-lapse sequence (left) and population speed (right) of leading edge and main cell body in bovine dermal and rat tail collagen (1.7 mg/ml) in the presence of GM6001. Tracking of the leading extension (white dotted lines) and body (green dotted lines) from starting (*) to end (triangle) position after 5 h. Speed is depicted as median with box blot and whiskers (5th/95th percentile; 45 cells, 3 independent experiments). ***, P < 0.0001; n.s., nonsignificant. (B) Nuclear deformation in moving or migration-arrested nuclei in HT1080 cells expressing H2B/EGFP and cytoplasmic DsRed2 in 6.6 mg/ml bovine dermal collagen in the presence of protease inhibitor cocktail (PI). Overview image from movie sequence (top; Video 5) highlighting moving (green arrowheads) and immobilized nuclei (red arrowheads). Middle: time-lapse example sequences of moving and immobilized nuclei from cells marked in the top image (numbers, time in min). Bottom: diameters of deformed nuclei from independent cells during migration or immobilization. 10 cells from one representative experiment out of n = 2; *, P < 0.05. (C) Morphology of cell body and nucleus during migration in 3D bovine dermal or rat tail collagen matrix (numbers, collagen concentration in mg/ml) in the absence or presence of GM6001. Cultures were fixed 10–16 h after polymerization and stained as indicated, including collagen cleavage neoepitope detection (cyan signal). Nuclear deformation imposed by collagen structures in moving cells in the presence of GM6001 (diameters 4–5 µm; empty arrowheads); conditions of immobilization induce nuclear rounding with occasional thin prolapse (white arrowheads). Arrows, direction of migration and protrusion, respectively. (D) Classification of nuclear morphologies under conditions of cell migration (middle) and immobilization (right). Left: representative xz cross section of a nondeformed HT/MT1 cell (50 µm2). Arrows, nuclear diameters used for quantification in B and E. Numbers show arbitrary ranges for diameters and corresponding cross sections for each phenotype. Red symbols (top-left corner) represent population phenotypes marked in E. (E) Nuclear diameters from cells cultured in collagen matrices of varying pore sizes in the absence or presence of GM6001. After 16 h, samples were fixed, stained with DAPI and phalloidin, and cells with polarized morphologies were analyzed by confocal microscopy for nuclear diameters (compare C). Red dashed ovals, subset of deformed nuclei with hourglass shape associated with movement (middle gray zone) or local prolapse in largely arrested cells (dark gray zone). Black dashed ovals highlight round nuclei of arrested phenotypes (light gray zone). Horizontal lines represent the medians (of each 21 nuclei per group; 2–3 independent experiments). Tables indicate the frequency of hourglass and prolapse phenotypes for each condition. Bars: (A) 50 µm; (B–D) 10 µm.
Requirement for deformation of the nucleus during MMP-independent migration at rate-limiting pore conditions. (A) Time-lapse sequence (left) and population speed (right) of leading edge and main cell body in bovine dermal and rat tail collagen (1.7 mg/ml) in the presence of GM6001. Tracking of the leading extension (white dotted lines) and body (green dotted lines) from starting (*) to end (triangle) position after 5 h. Speed is depicted as median with box blot and whiskers (5th/95th percentile; 45 cells, 3 independent experiments). ***, P < 0.0001; n.s., nonsignificant. (B) Nuclear deformation in moving or migration-arrested nuclei in HT1080 cells expressing H2B/EGFP and cytoplasmic DsRed2 in 6.6 mg/ml bovine dermal collagen in the presence of protease inhibitor cocktail (PI). Overview image from movie sequence (top; Video 5) highlighting moving (green arrowheads) and immobilized nuclei (red arrowheads). Middle: time-lapse example sequences of moving and immobilized nuclei from cells marked in the top image (numbers, time in min). Bottom: diameters of deformed nuclei from independent cells during migration or immobilization. 10 cells from one representative experiment out of n = 2; *, P < 0.05. (C) Morphology of cell body and nucleus during migration in 3D bovine dermal or rat tail collagen matrix (numbers, collagen concentration in mg/ml) in the absence or presence of GM6001. Cultures were fixed 10–16 h after polymerization and stained as indicated, including collagen cleavage neoepitope detection (cyan signal). Nuclear deformation imposed by collagen structures in moving cells in the presence of GM6001 (diameters 4–5 µm; empty arrowheads); conditions of immobilization induce nuclear rounding with occasional thin prolapse (white arrowheads). Arrows, direction of migration and protrusion, respectively. (D) Classification of nuclear morphologies under conditions of cell migration (middle) and immobilization (right). Left: representative xz cross section of a nondeformed HT/MT1 cell (50 µm2). Arrows, nuclear diameters used for quantification in B and E. Numbers show arbitrary ranges for diameters and corresponding cross sections for each phenotype. Red symbols (top-left corner) represent population phenotypes marked in E. (E) Nuclear diameters from cells cultured in collagen matrices of varying pore sizes in the absence or presence of GM6001. After 16 h, samples were fixed, stained with DAPI and phalloidin, and cells with polarized morphologies were analyzed by confocal microscopy for nuclear diameters (compare C). Red dashed ovals, subset of deformed nuclei with hourglass shape associated with movement (middle gray zone) or local prolapse in largely arrested cells (dark gray zone). Black dashed ovals highlight round nuclei of arrested phenotypes (light gray zone). Horizontal lines represent the medians (of each 21 nuclei per group; 2–3 independent experiments). Tables indicate the frequency of hourglass and prolapse phenotypes for each condition. Bars: (A) 50 µm; (B–D) 10 µm.
Modulation of migration in confined space by cell actomyosin contractility and integrin-mediated mechanocoupling
The combination of ongoing cytoskeletal activity and impaired migration suggests an active role for mechanocoupling in confined space. We therefore tested both integrin-mediated leading edge traction on substrate using β1 integrin–perturbing mAb 4B4 (Wolf et al., 2003a) and actomyosin-dependent contractility using ROCK inhibitor Y-27632 that inhibits myosin light chain (MLC) phosphorylation and contraction of the cell rear (Fig. 4 A; Ren et al., 2004; Lämmermann et al., 2008). Both approaches gradually impaired HT/MT1 cell-mediated contraction of collagen lattices (Fig. 4 B). Using collagen matrices with 20 µm2 pore area, representing a moderate physical challenge to nuclear deformability (compare Fig. 1 D with Fig. 3 D), both mAb 4B4 and Y-27632 significantly delayed migration rates in a dose-dependent manner, with greater negative impact by GM6001 relative to control cells (Fig. 4 C). Mechanically perturbed force generation by mAb 4B4 was apparent from compromised cell elongation (Fig. 4 D). When mAb 4B4 was used at a concentration that inhibited collagen contraction by ∼50% (1 µg/ml; see Fig. 4 B, left), MMP-independent migration was almost completely blocked, consistent with an impaired ability to generate sufficient adhesion and traction force to translocate the nucleus (Fig. 4 C and F; Video 6 A, left). Consequently, reducing space constraints by enlarging pore diameters to ∼55 µm2 (Fig. 4 E) led to a near-complete rescue of migration (Fig. 4 F; Video 6 A, middle and right). Consistent with the speed delay in constraining matrix (1.7 mg/ml bovine collagen in the presence of GM6001), increased nuclear deformation was observed that, again, could be reversed when matrix porosity was increased or MMPs were engaged (Fig. 4 G). With the integrin system intact, the importance of actomyosin-mediated cell contraction in propelling the nucleus through 3D matrix was underlined by delayed rear retraction and increased cell length in the presence of Y27632 (Fig. 4 H; Video 6 B, left). A concentration causing half-maximum collagen contraction (Fig. 4 B, right), Y27632 (2 µM) effected a partial migration arrest and, concomitantly, strong nucleus deformation in the presence of GM6001 (Fig. 4, C, I, and J). Both of these inhibitors’ effects were effectively reversed by increasing pore size cross sections (Fig. 4, I and J; Video 6 B, middle and right). Thus, both integrin-mediated traction force and actomyosin contractility are required to propel the nucleus forward when dense ECM is transmigrated in cooperation with MMP-mediated pore generation.
Abrogation and physical rescue of nonproteolytic HT1080/MT1 cell migration after interference with cell–matrix adhesion and contractility in bovine dermal collagen lattices of increasing pore sizes. Imaging at 37°C by bright-field inverse microscopy (Leica). Cells migrating in the presence of GM6001 were additionally treated with 4B4 mAb (1 µg/ml; A, first video sequence) or Y-27632 (2 µM; B, second sequence). (A and B) Left, collagen concentration 1.7 mg/ml (high magnification); middle, 1.7 mg/ml; right, 0.8 mg/ml. Time intervals as indicated.
Abrogation and physical rescue of nonproteolytic HT1080/MT1 cell migration after interference with cell–matrix adhesion and contractility in bovine dermal collagen lattices of increasing pore sizes. Imaging at 37°C by bright-field inverse microscopy (Leica). Cells migrating in the presence of GM6001 were additionally treated with 4B4 mAb (1 µg/ml; A, first video sequence) or Y-27632 (2 µM; B, second sequence). (A and B) Left, collagen concentration 1.7 mg/ml (high magnification); middle, 1.7 mg/ml; right, 0.8 mg/ml. Time intervals as indicated.
Modulation of migration efficiency and nuclear deformation in dense collagen lattices by integrin- and actomyosin-mediated force generation. (A) Dose-dependent reduction of pMLCT18S19 content in the presence of ROCK inhibitor Y-27632 in HT/MT1 cells. Signal intensity was calculated by densitometry and normalized to the total MLC signal. (B) Dose-dependent inhibition of collagen contraction by anti–β1 integrin mAb 4B4 (left) or Y-27632 (right). Cell-free and cell-containing collagen lattices treated with GM6001 (20 µM), and mAb 4B4, control IgG1 (3 µg/ml), or Y-27632 at indicated concentration. Matrix contraction was measured from gel areas (top images) after 48 h (left) or 24 h (right) as triplicates (means and SD from one representative experiment). Dashed horizontal lines mark 0, 50, and 100% gel contraction. (C) Mean population speed from single-cell analysis after 19–24 h of cell tracking in bovine collagen (1.7 mg/ml) in the absence or presence of MMP inhibitor GM6001 (20 µM) and mAb 4B4 or Y-27632. Medians and boxes and whiskers from 60–150 cells (n = 2–3 for Y-27632; n = 3–5 for 4B4). Dashed lines indicate 100% reference for MMP-independent movement and 90% inhibition. ***, P < 0.0001; **, P < 0.01; n.s., not significant. (D and H) Median elongation (cell body length divided by width) after 10 h of MMP-independent migration (50–90 cells from n = 2; ***, P < 0.0001; n.s., not significant). Horizontal line, median. (E) xy and xz image cross sections of confocal reflectance from bovine collagen fibrils (top) and quantification of pore cross sections (1.7 vs. 0.8 mg/ml concentration). (F and I) Population speed from single-cell analysis in bovine dermal collagen in the presence of 1 µg/ml mAb 4B4 (F) or 2 µM Y-27632 (I) and, where indicated, 20 µM GM6001. 60–108 cells from 2–3 independent experiments; ***, P < 0.0001; **, P < 0.01; *, P < 0.05; n.s., nonsignificant. (G and J) Nuclear diameter analysis as described in the legend of Fig. 3 E. Red dashed ovals, subset of deformed nuclei with hourglass (“h”) shape associated with movement (middle gray zone) or local prolapse (“p”) in largely arrested cells (dark gray zone). r/e, round or ellipsoid nuclei. Arrowheads, deformation of nuclei; arrows, migration direction. Cells migrated in bovine collagen in the presence of either 1 µg/ml 4B4 antibody or 2 µM Y-27632 and, when indicated, 20 µM GM6001. 11–27 cells each; n = 2. Bars: (B) 5 mm; (D) 25 µm; (E, G, and J) 10 µm.
Modulation of migration efficiency and nuclear deformation in dense collagen lattices by integrin- and actomyosin-mediated force generation. (A) Dose-dependent reduction of pMLCT18S19 content in the presence of ROCK inhibitor Y-27632 in HT/MT1 cells. Signal intensity was calculated by densitometry and normalized to the total MLC signal. (B) Dose-dependent inhibition of collagen contraction by anti–β1 integrin mAb 4B4 (left) or Y-27632 (right). Cell-free and cell-containing collagen lattices treated with GM6001 (20 µM), and mAb 4B4, control IgG1 (3 µg/ml), or Y-27632 at indicated concentration. Matrix contraction was measured from gel areas (top images) after 48 h (left) or 24 h (right) as triplicates (means and SD from one representative experiment). Dashed horizontal lines mark 0, 50, and 100% gel contraction. (C) Mean population speed from single-cell analysis after 19–24 h of cell tracking in bovine collagen (1.7 mg/ml) in the absence or presence of MMP inhibitor GM6001 (20 µM) and mAb 4B4 or Y-27632. Medians and boxes and whiskers from 60–150 cells (n = 2–3 for Y-27632; n = 3–5 for 4B4). Dashed lines indicate 100% reference for MMP-independent movement and 90% inhibition. ***, P < 0.0001; **, P < 0.01; n.s., not significant. (D and H) Median elongation (cell body length divided by width) after 10 h of MMP-independent migration (50–90 cells from n = 2; ***, P < 0.0001; n.s., not significant). Horizontal line, median. (E) xy and xz image cross sections of confocal reflectance from bovine collagen fibrils (top) and quantification of pore cross sections (1.7 vs. 0.8 mg/ml concentration). (F and I) Population speed from single-cell analysis in bovine dermal collagen in the presence of 1 µg/ml mAb 4B4 (F) or 2 µM Y-27632 (I) and, where indicated, 20 µM GM6001. 60–108 cells from 2–3 independent experiments; ***, P < 0.0001; **, P < 0.01; *, P < 0.05; n.s., nonsignificant. (G and J) Nuclear diameter analysis as described in the legend of Fig. 3 E. Red dashed ovals, subset of deformed nuclei with hourglass (“h”) shape associated with movement (middle gray zone) or local prolapse (“p”) in largely arrested cells (dark gray zone). r/e, round or ellipsoid nuclei. Arrowheads, deformation of nuclei; arrows, migration direction. Cells migrated in bovine collagen in the presence of either 1 µg/ml 4B4 antibody or 2 µM Y-27632 and, when indicated, 20 µM GM6001. 11–27 cells each; n = 2. Bars: (B) 5 mm; (D) 25 µm; (E, G, and J) 10 µm.
Divergent kinetics and rate-limits in mononuclear and polymorphonuclear cells
To obtain insight into how different nuclear shape types enable or limit migration in restricted space, we examined the nuclear mechanics of additional cell types with mononuclear or polymorphonuclear organization. Lamin A/C, a central nuclear protein required for nuclear membrane organization and stability (Goldberg et al., 2008), is expressed in tumor cells, including HT/MT1, HT/wt, and MDA-MB-231/MT1 (MDA/MT1) breast cancer cells, but not in human CD4+ T-blasts or polymorphonuclear neutrophils (PMNs; Fig. 5 A). Irrespective of cell type, migration of mononuclear cells through low- to intermediate-density collagen lattices remained unperturbed by GM6001 (Fig. 5, B and D; Fig. S4 A). Ongoing migration was accompanied by deformation of the nucleus with cross section distributions matching the available pore size range of the matrix (Fig. 5, C and E; Fig. S4 B). In HT/wt cells and T-blasts moving through dense matrices, hourglass nuclear shapes predominated, whereas MDA/MT1 cells displayed a spectrum from hourglass to cigar-like shapes. For pore dimensions ranging from 2 to 5 µm2, GM6001 caused migration arrest and nuclear prolapse formation in tumor cells, whereas T-blast migration persisted, albeit at slower speeds (Fig. 5, B–D; Fig. S4, A–C; Video 7). Compared with tumor cells, T-blasts are distinguished by a two- to fourfold smaller nucleus (Fig. 3 C; Fig. 5 E) and the inability to proteolytically degrade fibrillar collagen, thus constitutively depending upon shape changes alone to bypass matrix barriers (Wolf et al., 2003b). Ultimately, however, all T-blasts were immobilized at pore cross sections of 1–2 µm2 (Fig. S2 G; Fig. 5 F) that no longer accommodated nuclear deformations (Fig. 5 E; Fig. S4 D). Thus, MMP-independent mononuclear cell migration uniformly depends upon nuclear deformation in response to lateral compression by tissue structures.
Migration of CD4+ T-blasts and PMNs in rat tail collagen at low and high concentrations indicated in the movie in the presence of MMP-inhibitor GM6001. Imaged at 37°C by bright-field inverse microscopy (Leica); time (h:min:s) as indicated (2-min frame interval; 2 h of observation).
Migration of CD4+ T-blasts and PMNs in rat tail collagen at low and high concentrations indicated in the movie in the presence of MMP-inhibitor GM6001. Imaged at 37°C by bright-field inverse microscopy (Leica); time (h:min:s) as indicated (2-min frame interval; 2 h of observation).
Pore size–dependent migration and deformation of the nucleus in mononuclear breast cancer cells, T-blasts, and PMN. (A) Lamin A/C content in distinct cell types. Lanes were loaded with whole-cell lysates normalized to GAPDH content and immunoblotted for lamin A/C. (B, D, and F) Migration efficiencies of the indicated cell types in rat tail collagen of different density in the absence or presence of GM6001. Steady-state speeds of single cells monitored over 24 h (MDA/MT1) or 2 h (T-blasts, PMNs). Dashed horizontal lines, subtotal (90% inhibition) and absolute (99% inhibition) migration limit (dashed top lane indicating 100% of MMP-independent migration). Box and whiskers show the medians, 25th/75th and 5th/95th percentiles (50–200 cells, 2–6 independent experiments). ***, P < 0.0001; n.s., not significant. (C, E, and G) Moving and immobilized phenotypes of MDA/MT1 cells, T-blasts, and PMN in the presence of GM6001 in rat tail collagen of different pore sizes (numbers, collagen concentration in mg/ml). Cultures were fixed at the end-point (16 h, MDA/MT1; 2 h, leukocytes) and stained as indicated. Insets, DAPI. Arrows, direction of migration and protrusion, respectively. Tables (C and E), ranges of nuclear cross sections and diameters and their frequencies at different collagen density (numbers in percent, full dataset shown in Fig. S4, C and D) associated with intact or abrogated migration (17–30 independent cells). Insets (G), schematics of different nuclear shapes, including rounded (immobilized), fully or partially unfolded. (H) Change of nuclear morphology in migrating PMN (rat tail collagen, 1.7 mg/ml) over 600-s time period (full sequence shown in Fig. S4 E, example 1; and Video 8). Insets, DAPI signal (left) and schematics of nuclear shape (right). Arrowhead, saltatory migration phase. (I) Nuclear elongation index (top graph) and distance between nucleus and leading extension (pseudopod–nucleus distance) plotted over time. Measurement as indicated by black arrows. Graphs show one representative example of a moving cell (red lines; compare to Fig. S4 E, example 2) and immobilized cell (black lines; compare to Fig. S4 F, example 1) out of n = 3. Bars: (C) 10 µm; (E, G, and H) 5 µm.
Pore size–dependent migration and deformation of the nucleus in mononuclear breast cancer cells, T-blasts, and PMN. (A) Lamin A/C content in distinct cell types. Lanes were loaded with whole-cell lysates normalized to GAPDH content and immunoblotted for lamin A/C. (B, D, and F) Migration efficiencies of the indicated cell types in rat tail collagen of different density in the absence or presence of GM6001. Steady-state speeds of single cells monitored over 24 h (MDA/MT1) or 2 h (T-blasts, PMNs). Dashed horizontal lines, subtotal (90% inhibition) and absolute (99% inhibition) migration limit (dashed top lane indicating 100% of MMP-independent migration). Box and whiskers show the medians, 25th/75th and 5th/95th percentiles (50–200 cells, 2–6 independent experiments). ***, P < 0.0001; n.s., not significant. (C, E, and G) Moving and immobilized phenotypes of MDA/MT1 cells, T-blasts, and PMN in the presence of GM6001 in rat tail collagen of different pore sizes (numbers, collagen concentration in mg/ml). Cultures were fixed at the end-point (16 h, MDA/MT1; 2 h, leukocytes) and stained as indicated. Insets, DAPI. Arrows, direction of migration and protrusion, respectively. Tables (C and E), ranges of nuclear cross sections and diameters and their frequencies at different collagen density (numbers in percent, full dataset shown in Fig. S4, C and D) associated with intact or abrogated migration (17–30 independent cells). Insets (G), schematics of different nuclear shapes, including rounded (immobilized), fully or partially unfolded. (H) Change of nuclear morphology in migrating PMN (rat tail collagen, 1.7 mg/ml) over 600-s time period (full sequence shown in Fig. S4 E, example 1; and Video 8). Insets, DAPI signal (left) and schematics of nuclear shape (right). Arrowhead, saltatory migration phase. (I) Nuclear elongation index (top graph) and distance between nucleus and leading extension (pseudopod–nucleus distance) plotted over time. Measurement as indicated by black arrows. Graphs show one representative example of a moving cell (red lines; compare to Fig. S4 E, example 2) and immobilized cell (black lines; compare to Fig. S4 F, example 1) out of n = 3. Bars: (C) 10 µm; (E, G, and H) 5 µm.
Similarly, PMN migration remained unaffected by the absence or presence of GM6001 in low- to intermediate-density collagen matrices (Fig. 5 F). In contrast to “in-whole” deformation of mononuclear nuclei, the segmented nucleus of PMNs displayed interconvertible folding states, including compact configuration or pearl chain–like complete or partial unfolding (Fig. 5 G). These folding states reflected speed changes, alternating between compaction during phases of immobility and unfolding during migration (Fig. 5 H; Fig. S4 E, example 1; Video 8). In high-density rat-tail collagen (3.3 mg/ml, pore cross section 2–3 µm2), PMN migration remained intact irrespective of MMP activity, albeit at reduced speed, and was arrested at higher collagen density (6.6 mg/ml) when pore cross sections decreased to 0.5–1.5 µm2 (Fig. 5 F; Fig. S2 G; Video 7). Not unlike tumor cells, immobilized PMNs exhibited collapsed, spherical nuclei with occasional single-segment prolapse toward the leading oscillating pseudopod (Fig. 5, G and I; Fig. S4, F and G). In summary, stereotypic shape change patterns, as exemplified by hourglass-like compression of mononuclear nuclei or unfolding of polymorphonuclear nuclei, are sufficient to maintain cell navigation through interstitial matrices. In high-density fibrillar collagen matrix with collagenolysis absent, nuclear deformability alone determines migration rates as a function of pore size, reaching near absolute limits at 7 µm2 in tumor cells and 2–3 µm2 in in PMNs and T-blasts (Fig. 6 A). Thus, impaired migration is a linear function of pore size and independent of scaffold stiffness (Fig. 6 B).
Pearl chain configuration of segmented nucleus during neutrophil migration in rat tail collagen (1.7 mg/ml) in the presence of IL-8 (100 ng/ml). Maximum intensity projection of red DNA label (Hoechst 33342 dye) with transmission image obtained by time-lapse confocal microscopy (LSM510; Carl Zeiss). Frame interval, 15 s; duration, 10 min.
Pearl chain configuration of segmented nucleus during neutrophil migration in rat tail collagen (1.7 mg/ml) in the presence of IL-8 (100 ng/ml). Maximum intensity projection of red DNA label (Hoechst 33342 dye) with transmission image obtained by time-lapse confocal microscopy (LSM510; Carl Zeiss). Frame interval, 15 s; duration, 10 min.
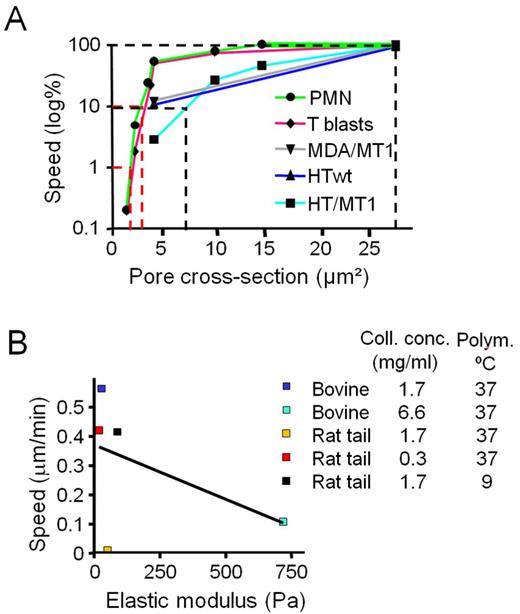
Correlation of migration rates with pore cross section, but not scaffold stiffness. (A) Migration rates as a function of pore cross section in mononuclear tumor cells, T-blasts, and PMN in rat tail collagen of different density in the presence of GM6001. Speed was normalized to the maximum speed in low-density matrix (0.3 mg/ml) as 100% reference, and the subtotal and absolute migration limits are indicated (dotted lines). Data points depict medians from Figs. 1 E, 2 D, 5 (B, D, and F); S2 G, and S4 A. For T-blasts and PMN, data points were included for additional pore size conditions, n = 2–4. Subtotal limits were reached at median pore cross sections of 4–7 µm2 for tumor cells and 3–4 µm2 for leukocytes, and absolute limits at 2 µm2 for leukocytes. (B) Association of migration rates and elastic modulus of 3D collagen lattices. Speed medians were used from Fig. 2 (A, C, and E) obtained for cells migrating in the presence of GM6001 in lattice conditions for which the elastic modulus was measured (compare Fig. 1 C). R2 for linear regression (black line) was 0.2339 (nonsignificant).
Correlation of migration rates with pore cross section, but not scaffold stiffness. (A) Migration rates as a function of pore cross section in mononuclear tumor cells, T-blasts, and PMN in rat tail collagen of different density in the presence of GM6001. Speed was normalized to the maximum speed in low-density matrix (0.3 mg/ml) as 100% reference, and the subtotal and absolute migration limits are indicated (dotted lines). Data points depict medians from Figs. 1 E, 2 D, 5 (B, D, and F); S2 G, and S4 A. For T-blasts and PMN, data points were included for additional pore size conditions, n = 2–4. Subtotal limits were reached at median pore cross sections of 4–7 µm2 for tumor cells and 3–4 µm2 for leukocytes, and absolute limits at 2 µm2 for leukocytes. (B) Association of migration rates and elastic modulus of 3D collagen lattices. Speed medians were used from Fig. 2 (A, C, and E) obtained for cells migrating in the presence of GM6001 in lattice conditions for which the elastic modulus was measured (compare Fig. 1 C). R2 for linear regression (black line) was 0.2339 (nonsignificant).
Physical limits of cell migration through nondegradable substrate
Migration rates through dense 3D collagen gels and broad-spectrum MMP inhibitor treatment might be affected by additional parameters not yet addressed, including residual low-level MMP-independent collagenolysis executed by other classes of proteases; guidance by occasional gaps and fissures present in randomly assembled bio-scaffolds; and/or mechanical rupture of very small or incompletely polymerized collagen fibrils by migrating cells. To exclude such interfering parameters, we used a polycarbonate filter model for cell trafficking through nondegradable and nondeformable barriers. In this system, subtotal inhibition (90%) of migration was reached at pore cross sections of 7–10 µm2 for all mononuclear cell types and 4 µm2 for PMN with complete inhibition (>99%) obtained at pore dimensions below 5–6 µm2 for mononuclear cells and 1 µm2 for PMNs (Fig. 7 A). Similar to 3D collagen matrices, effective transmigration through polycarbonate filter pores resulted from cytoplasmic protrusion followed by nuclear deformation within a pore cross-sectional range of 7–50 µm2 (Fig. 7, B and C). Likewise, the morphology of cells immobilized above small pores (0.8 µm2 cross section) consisted of a long cytoplasmic protrusion extending into and the nucleus atop the pore (Fig. 7, B and C), thereby recapitulating the arrested phenotypes observed in high-density fibrillar collagen. Thus, cell migration through nondegradable pores exhibits physical thresholds that are dependent upon pore size and nuclear deformability, confirming the limiting thresholds obtained for 3D fibrillar collagen matrix.
Physical migration limits of mono- and polymorphonuclear cell types in transwell filter assay. (A) Transmigration of different cell types through polycarbonate membranes of 5-, 3-, 1-, and 0.45-µm pore diameter after 20 h (HT/MT1, HT/wt, and MDA/MT1 cells), 5 h (T-blasts) or 3 h (PMN). The cell number in the bottom compartment was normalized to transmigration through 5-µm pores as 100% reference. Solid and dashed vertical lines represent the pore cross section at 90 and 99% inhibition, respectively, of migration for each cell type (black lines, mononuclear cells; gray area between black lines depicts a range of subtotal migration limits; red lines, PMN). Subtotal limits were reached at 7–10 µm2 for tumor cells and T-blasts, and 4 µm2 for PMNs; and absolute limits at 5 µm2 for tumor cells and T-blasts and 1–2 µm2 for PMNs (n = 3–5; means, SD). (B) Polycarbonate membranes with pore sizes ranging from 8 to 1 µm in diameter (top row). Representative images of nuclear deformation in transmigrating dual-color HT1080 cells and (C) PMNs during passage through pores of decreasing size. Filter membranes were fixed and, in C, stained as indicated (n = 2). Arrows, direction of transmigration. Bars: (B) 10 µm, (C) 5 µm.
Physical migration limits of mono- and polymorphonuclear cell types in transwell filter assay. (A) Transmigration of different cell types through polycarbonate membranes of 5-, 3-, 1-, and 0.45-µm pore diameter after 20 h (HT/MT1, HT/wt, and MDA/MT1 cells), 5 h (T-blasts) or 3 h (PMN). The cell number in the bottom compartment was normalized to transmigration through 5-µm pores as 100% reference. Solid and dashed vertical lines represent the pore cross section at 90 and 99% inhibition, respectively, of migration for each cell type (black lines, mononuclear cells; gray area between black lines depicts a range of subtotal migration limits; red lines, PMN). Subtotal limits were reached at 7–10 µm2 for tumor cells and T-blasts, and 4 µm2 for PMNs; and absolute limits at 5 µm2 for tumor cells and T-blasts and 1–2 µm2 for PMNs (n = 3–5; means, SD). (B) Polycarbonate membranes with pore sizes ranging from 8 to 1 µm in diameter (top row). Representative images of nuclear deformation in transmigrating dual-color HT1080 cells and (C) PMNs during passage through pores of decreasing size. Filter membranes were fixed and, in C, stained as indicated (n = 2). Arrows, direction of transmigration. Bars: (B) 10 µm, (C) 5 µm.
Discussion
By challenging MMP-dependent and -independent cell movement with decreasing scaffold porosity, and by quantitatively co-registering migration speed with the shapes of cell body and nucleus, we have identified the rate-limiting physicochemical determinants of cell migration. First-order determinants are (1) the available spaces between ECM fibrils or within the filter pore that can accommodate the moving cell body and (2) the deformability of the nucleus in response to space constraints. With decreasing cross section, interfibrillar pores mechanically impede cell movement and impose deformation in a cell type–specific manner until net deformability is exceeded and the nucleus becomes mechanically trapped. This physical balance of translocation or arrest is modulated by “second-order” processes that control pore size or nucleus deformation. Here addressed effectors include (1) fibril remodeling by MT1-MMP to create “neo-space” and (2) mechanocoupling by integrins and actomyosin contractility to propel the nucleus forward through constraining spaces.
Variability of ECM porosity in vivo and in vitro
In vivo, fibrillar collagen is organized as fibrils and fibers of diverse caliber, orientation, and interfibrillar spacing (Starborg et al., 2008; Wolf et al., 2009). To reflect the variability of interfibrillar space present in loose versus dense interstitial tissues in vivo with ranges between 2 and 30 µm (Stoitzner et al., 2002; Wolf et al., 2009; Weigelin et al., 2012), we similarly varied the median porosity of 3D collagen matrices. This space range serves to both guide and mechanically challenge moving cells that commonly display cross sections of 30–100 µm2 in a cell type–dependent manner.
The biophysics of subtotal and absolute limits of cell migration
Our data show that migration speed diminishes as a linear function with decreasing pore size in a cell type–specific manner (Fig. 8). The decline in response to increasing mechanical confinement is not a simple “go or stop” decision, but is a gradual process that largely depends upon cell deformation, a coping strategy intrinsic to migrating cells (Lautenschläger et al., 2009; Wolf and Friedl, 2011; Tong et al., 2012). Maximum speed is reached at pore sizes that approximate the cell body where the substrate guides without physically impeding migration (Fig. 8 A; Jacobelli et al., 2010). With decreasing porosity, migration delay is reciprocal with increasing need for cell deformation, resulting in slower, but ongoing migration (Fig. 8 B, up to subtotal limit). Eventually, migration is abrogated when a cell type–specific maximum of deformation is reached (Fig. 8 C, absolute limit). As cell shape is highly adaptable, pores of defined cross sections, but distinct geometry, are equally well accommodated, including discontinuous polygonal-shaped pores in 3D fibrillar collagen, flat and broad cleft-like spaces, evenly shaped cylindrical pores in polycarbonate membranes, or elongated continuous channels in microdevices (Lautenschläger et al., 2009; Jacobelli et al., 2010; Rolli et al., 2010; Ilina et al., 2011; Balzer et al., 2012; Tong et al., 2012). A cross section of 20–30 µm2 identified herein for near-optimal migration of HT1080 and MDA-MB-231 cells through complex-shaped spaces in collagen lattices and monomorphic transwell filter pores matches the 30 µm2 cross-sectional areas of engineered rectangular microchannels that enable high migration speed (Tong et al., 2012). As an extreme example of shape adaptation, keratinocytes are able to traverse gaps of 500 nm in height but unrestricted in width (Brunner et al., 2006), confirming that the cross section of the transmigrated space, rather than its diameter, defines the efficacy of cell migration under confinement. Thus, the limits of cell migration are a two-parameter function of substrate porosity and cell deformability.
Tissue porosity and nuclear deformation jointly determine subtotal and absolute limits of cell migration. Cell migration efficacy is a joint function of substrate porosity and nuclear deformability, with MMP activity and mechanocoupling as modulators. (A) Minor deformation of nuclei and optimal velocity during migration through tissue of sufficient to high porosity, with mononuclear shapes in ellipsoid and polymorphonuclear nuclei in partly unfolded state. (B) Significant deformation of the nucleus and migration delay in cells moving through dense ECM with pore cross sections much below the nondeformed nucleus size. Mononuclear nuclei are compressed in their entirety and adopt hourglass-like or cigar-like shapes, whereas polymorphonuclear deformation consists of unfolding with transient pearl chain–like configuration as maximum. Maximal deformation is reached when the nucleus deforms to ∼10% of its original cross section, reaching a subtotal migration limit. (C) Migration arrest during confrontation with pore cross sections that exhaust the deformation capability of the nucleus. Both mono- and polymorphonuclear nuclei of migration-arrested cells retain a roundish, collapsed morphology with nonproductive transient prolapse, the cross section of which matches the geometry of the pore (∼5–10% of the original nuclear cross section). Modulators of migration rates near the physical limit include (1) pericellular proteolysis by MMPs that widens pore cross sections and (2) mechano-coupling toward ECM determining the force with which the nucleus is transported. CS, cross section; CR, compression ratio (degree of nuclear deformation versus nondeformed state); Tu, tumor cells; T, T blast; PMN, polymorphonuclear neutrophil. White arrows, direction of migration.
Tissue porosity and nuclear deformation jointly determine subtotal and absolute limits of cell migration. Cell migration efficacy is a joint function of substrate porosity and nuclear deformability, with MMP activity and mechanocoupling as modulators. (A) Minor deformation of nuclei and optimal velocity during migration through tissue of sufficient to high porosity, with mononuclear shapes in ellipsoid and polymorphonuclear nuclei in partly unfolded state. (B) Significant deformation of the nucleus and migration delay in cells moving through dense ECM with pore cross sections much below the nondeformed nucleus size. Mononuclear nuclei are compressed in their entirety and adopt hourglass-like or cigar-like shapes, whereas polymorphonuclear deformation consists of unfolding with transient pearl chain–like configuration as maximum. Maximal deformation is reached when the nucleus deforms to ∼10% of its original cross section, reaching a subtotal migration limit. (C) Migration arrest during confrontation with pore cross sections that exhaust the deformation capability of the nucleus. Both mono- and polymorphonuclear nuclei of migration-arrested cells retain a roundish, collapsed morphology with nonproductive transient prolapse, the cross section of which matches the geometry of the pore (∼5–10% of the original nuclear cross section). Modulators of migration rates near the physical limit include (1) pericellular proteolysis by MMPs that widens pore cross sections and (2) mechano-coupling toward ECM determining the force with which the nucleus is transported. CS, cross section; CR, compression ratio (degree of nuclear deformation versus nondeformed state); Tu, tumor cells; T, T blast; PMN, polymorphonuclear neutrophil. White arrows, direction of migration.
The nucleus as a rate-limiting cell compartment
The nucleus is the largest and most rigid cell organelle, based on chromatin content and the stabilizing nuclear lamina, in particular lamin A/C (Dahl et al., 2004; Gerlitz and Bustin, 2011; Chow et al., 2012). Kinetic time-lapse imaging of migrating cells reveals two mechanically distinct forms of nuclear deformation: (1) the global deformation of mononuclear nuclei resulting in transient hourglass-like or cigar-shaped elongated morphologies, and (2) the unfolding of polymorphonuclear nuclei to adopt pearl chain–like configurations in the absence of major deformations in individual nuclear segments (Fig. 8). Albeit transient, these migration-associated nuclear morphologies are frequently detected in vivo during cancer cell dissemination (Yamauchi et al., 2005, 2006; Alexander et al., 2008; Beadle et al., 2008; Friedl et al., 2011) as well as transendothelial migration (Feng et al., 1998; Voisin et al., 2009). Whereas the deformability of the nucleus in dense space is finite, the cytoplasm is able to accommodate nearly any pore size used here, including 1-µm2 gaps in collagen gels and 0.8-µm2 pores of polycarbonate membranes (Schoumacher et al., 2010; Shankar et al., 2010).
Cell movement through dense ECM is impacted by at least three properties of the nucleus, including size, rigidity, and shape, which together control an adaptation range of factor 2–5 when mononuclear tumor cells and PMN are compared directly (Fig. 8). For all tested cell types, we noted a compellingly consistent ratio of minimum nuclear cross sections relative to undeformed state of ≈1/10 (Fig. 8, right column), suggesting that maximum compressibility is an absolute value irrespective of cell type, constitutive nuclear shape, or nuclear rigidity. The nuclear lamina may control nuclear shape and/or deformability rather than the absolute compression limit, which hints toward a noncompressible intranuclear component (e.g., chromatin) in defining the compression maximum and, arguably, the physical limits of cell migration.
Modulation of ECM porosity by pericellular proteolysis
By degrading fibrillar collagen at the cell–matrix interface, MT1-MMP–dependent proteolysis widens collagen pore cross sections to accommodate the cell body and facilitates migration through degradable substrates (Wolf et al., 2007; Fisher et al., 2009; Sabeh et al., 2009). MT1-MMP thus accelerates migration at confining porosity above critical limits and, importantly, secures slow but nonetheless persistent migration even in very dense matrix. Conversely, with porosity high enough to accommodate the deforming cell body, pericellular proteolysis is dispensable and migration persists despite pharmacological or molecular targeting of MMP activity in either tumor cell or leukocyte populations (Fig. 8). MMPs thus act to modulate pore dimensions and thereby impact cell speed, deformation, and the limits of cell migration in biodegradable scaffolds. By establishing the preferential role of MMPs in pore size control, our approach using different collagen preparations side by side resolves controversies on the role of MMPs for cell invasion through 3D collagen scaffolds of different origin and porosity (Wolf et al., 2003a; Sabeh et al., 2004, 2009).
Modulation by mechanocoupling
Forces generated between cell and 3D scaffold impact cell deformation and migration through confined space (Fig. 8). β1 integrins that serve as both main adhesion receptors to fibrillar collagen and mechanotransducers in migrating cells (Huttenlocher et al., 1995; Puklin-Faucher and Sheetz, 2009) mediate the tractional forces needed to propel the nucleus through narrow pores. Likewise, Rho kinase–mediated actomyosin contractility reinforces integrin-mediated cell adhesion and controls retraction of the cell rear (Vicente-Manzanares et al., 2008) that together supports translocation of the nucleus through small spaces (Lämmermann et al., 2008). Thus, by using complementary mechanisms, MMP-dependent ECM degradation and integrin- and Rho-mediated force transmission jointly secure migration, particularly when space is restricted.
Parameters that affect the limits of cell migration indirectly
Additional physicochemical substrate properties likely impact migration rates, such as ECM telopeptide status and stiffness (Sabeh et al., 2009; Miron-Mendoza et al., 2010; Ehrbar et al., 2011). The high telopeptide content of collagen extracted from rat tails accelerates fibril polymerization and increases mechanical strength, but decreases fibril caliber and network porosity (Helseth and Veis, 1981; Elbjeirami et al., 2003; Sabeh et al., 2009; Wolf et al., 2009), which compromises nonproteolytic cell migration. Conversely, by temperature-dependently delaying collagen fibrillogenesis, larger pore sizes were generated that better matched nuclear size and deformability, and enabled MMP-independent cell movement. Thus, irrespective of the telopeptide content of the collagen preparation used, small pores exhaust nuclear deformability and preclude MMP-independent migration. Our data further show no effect of substrate stiffness variation on cell immobilization by space constraints. The stiffness range used here includes 20–700 Pa for reconstituted fibrillar collagen and >106 Pa for polycarbonate membranes, thus encompassing elastic modules of in vivo tissues, ranging from 200 to 1,000 Pa for mammary interstitium and from 1,000 to 10,000 Pa for adipocytes and myofibers (Stein et al., 2008; Butcher et al., 2009; Levental et al., 2009; Buxboim et al., 2010). Thus, when normalized to pore size, both soft and rigid substrates account for near-identical subtotal and absolute migration limits, excluding substrate stiffness above 20 Pa as an independent mechanism for controlling cell deformation or physical migration arrest.
Outlook
This multi-scale analysis of cell–matrix geometries and migration kinetics may serve as biophysical inventory and reference for addressing further modulators of cell migration in confined spaces. These likely include elasticity of the nucleus by regulation of lamin expression, the nucleus–cytoskeleton connection, intracellular and intranuclear pressure and hydration, chromatin organization, and physical “memory” as consequence of repetitive or long-lasting mechanical stress. Likely, the herein defined deformability of the cell body and nucleus matches the 2D and 3D tracks and gaps constitutively present in healthy connective tissues (Wolf et al., 2009; Weigelin et al., 2012). More complex invasion programs, however, may be required for penetration of tumor-associated collagen-rich desmoplastic tissue, scarred stroma, and, likely, basement membranes (Provenzano et al., 2006; Rowe and Weiss, 2009; Tanaka et al., 2010; Salmon et al., 2012). To this end, multi-scale wet-laboratory and in silico analyses of cell migration will jointly delineate the strategies used by cells to traverse ECM barriers encountered in vitro and in vivo.
Materials and methods
Cell culture, cell purification, and reagents
The following cells were used: human HT1080-wild type (HT1080/wt or HT/wt) fibrosarcoma cells (ACC315; DSMZ Braunschweig); HT1080 cells transfected with MT1-MMP/MMP14 (HT1080/MT1 or HT/MT1; Deryugina et al., 1997); HT1080 dual-color cells expressing cytoplasmic DsRed2 and nuclear histone-2B (H2B)–coupled EGFP (Yamamoto et al., 2004); and human MDA-MB-231 breast cancer cells transfected with MT1-MMP (MDA-MB-231/MT1 or MDA/MT1; Moss et al., 2009). For generation of stable cell lines, the following constructs were generated: wild-type MT1-MMP protein in the eukaryotic expression vectors pcDNA3 and pCR3.1-Uni (Invitrogen) for HT1080 and MDA-MB-231 cell lines, respectively; wild-type RFP cDNA and H2B-GFP fusion gene in the retroviral pLNCX2 and pLHCX vectors (Takara Bio Inc.), all under the control of the cytomegalovirus promoter. Retroviral vectors were transfected into the packaging cell line PT67 prior to transduction of HT1080 cells. Cell lines were cultured (37°C at 5% CO2 humidified atmosphere) in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen) supplemented with 10% heat-inactivated fetal calf serum (FCS; Sigma-Aldrich), 100 U/ml penicillin and 100 µg/ml streptomycin (PAA), 2 mM l-glutamine, and 1 mM sodium pyruvate (Invitrogen). Spheroids were generated by 24-h culture in low-adhesive culture dishes (Thermo Fisher Scientific) before incorporation into 3D collagen.
Human CD4+ T-blasts (T-blasts, purity >95%, measured by flow cytometry) were obtained from healthy donors using the mononuclear cell fraction followed by stimulation with 1.25 µg/ml concanavalin A (Oncogene) and 100 U/ml IL-2 (Strathman Biotech) in T-cell medium (TCM; RPMI 1640 medium [Gibco] supplemented as DMEM, except l-glutamine, and additionally with 10 mM Hepes [Gibco], 500 mM 2-mercaptopethanol, and 0.1 mM nonessential amino acids [Gibco]); and after 48 h were purified by positive immunomagnetic separation (Dynabeads) using paramagnetic beads coated with an anti-CD4+ mAb and magnetic separation from unbound cells. Human PMNs (purity >90%) were obtained by density gradient centrifugation (van Spriel et al., 2001). In brief, blood was added on top of two-layered histopaque and ficoll (Lymphoprep), centrifuged, and the PMN fraction isolated for immediate use in cell migration assays.
The following antibodies and reagents were used: polyclonal anti-collagen type I antibody (Rockland); polyclonal rabbit anti-COL23/4Cshort antiserum (IBEX Pharmaceuticals) or affinity purified rabbit antibody (ImmunoGlobe) directed against the C-terminal cleavage neo-epitope of collagens type I and type II (Wolf et al., 2007); function-blocking mouse monoclonal anti–β1 integrin IgG1 (4B4; Coulter-Immunotech); isotypic IgG1 control (NCG01; Thermo Fisher Scientific); monoclonal mouse anti–lamin A/C IgG1 (131C3; Acris); monoclonal rabbit anti-GAPDH antibody (14C10; Cell Signaling Technology); polyclonal rabbit myosin light chain 2 and phospho-myosin light chain 2 (Thr18/Ser19) antibodies (Cell Signaling Technology); secondary goat anti–rabbit antibody conjugated to horseradish peroxidase (Thermo Fisher Scientific) or a fluorophore specific to the Odyssee system (IRDye 680RD; LI-COR Biosciences); secondary Alexa Fluor 647–conjugated preabsorbed goat anti–rabbit IgG (Invitrogen); Alexa Fluor 488–conjugated phalloidin (Invitrogen); DAPI (Roche); Hoechst 33342 (Invitrogen); GM6001 (ilomastat; EMD Millipore); protease inhibitor cocktail consisting of marimastat (50 µM; British Biotech), leupeptin (2 µM; Molecular Probes/Invitrogen), E-64 (50 µM; Sigma-Aldrich), aprotinin (0.7 µM; Sigma-Aldrich) and pepstatin A (50 µM; Sigma-Aldrich; Wolf et al., 2007); LPA (Sigma-Aldrich); Y-27632 (Sigma-Aldrich); and interleukin-8 (IL-8; R&D Systems). Knockdown of MT1-MMP by smart-pool siRNA (Thermo Fisher Scientific) was performed according to the manufacturer’s protocol and as described previously (Wolf et al., 2007).
3D collagen matrix assays
Bovine dermal collagen was obtained as acidified, pepsinized solution (stock concentration 3.1 mg/ml; PureCol, Advanced BioMatrix); non-pepsinized (telopeptide intact) rat tail collagen was purified by acid extraction, stored in acidified lyophilized form (Hotary et al., 2000), and re-solubilized before use in 0.2% acetic acid. Fibrillar collagen matrices were reconstituted in vitro by raising the pH to 7.4 using either final concentrations of 0.345 N NaOH and buffering by 25 µM Hepes (Sigma-Aldrich) for rat tail matrices, or 0.75% Na-bicarbonate solution (Gibco) for bovine dermal lattices, together with mimimum essential Eagle’s medium (Sigma-Aldrich), and polymerized at 37°C, unless indicated otherwise (Hotary et al., 2000; Wolf et al., 2007). High-density matrices of bovine dermal collagen were reconstituted after concentration of the collagen solution (20 mg/ml; speed vac concentrator; Savant). Delayed polymerization speed of rat tail collagen was achieved by lowering the temperature during polymerization to 20, 14, or 9°C. Cell-free collagen lattices were used for bright-field microscopy (vertical invasion studies; Fig. S1 A), 3D confocal and electron microscopy, and atomic force microscopy. For cell migration assays, cells (20,000/100 µl) after detachment with EDTA (2 mM) or multicellular spheroids (2–7 spheroids/100 µl) were suspended in neutralized collagen solution before polymerization and incorporated into a self-constructed chamber (see Fig. S1 A). After addition of medium (containing 10% FCS) spontaneous migration was monitored, except for PMNs which were stimulated by 100 ng/ml IL-8. Inhibition experiments were performed using 20 µM GM6001, a five-component protease inhibitor cocktail, function-perturbing anti-β1 mAb 4B4, or Y-27632. Inhibitors were added to both the cell–collagen mixture and supernatant. For collagen contraction assays, cells (8 or 16 × 104, for incubation with Y-27632 or 4B4, respectively) were incorporated into non-anchored bovine collagen lattices (300 µl) in 48-well plates.
Collagen stiffness measurement
Fibrillar 3D matrices of rat tail or bovine dermal collagen were reconstituted and probed by atomic force microscopy in PBS on a Catalyst BioScope (Bruker Corporation) coupled to a confocal microscope (TCS SP5 II; Leica). A polystyrene microsphere (10-µm diameter; Polysciences, Inc.) was glued to a cantilever (NP-S type D nominal spring constant of 0.06 N/m; Bruker Corporation) by a two-component polyurethane glue (Bison International) and dried overnight. Individual spring constants were calibrated in PBS on glass by acquiring both deflection sensitivity and thermal noise data subsequently fitted and corrected for a V-shaped cantilever (te Riet et al., 2011). Matrix stiffness was measured by repeatedly (2–20 times) bringing the bead-functionalized cantilever into contact with the surface of the lattice at an identical position (contact force ≤2 nN; approach–retraction distance 10 µm; approach velocity 10 µm/s) with a closed z-loop. Force–distance curves from each measurement were converted into force–indentation (F-δ) curves and subsequently fitted over the 0–1,500 pN force range with the Hertz model (Lin et al., 2007) using a custom algorithm (IgorPro 6; Wavemetrics) to calculate the stiffness:
with F, force in Newtons [N]; Rc, bead radius in meters [m]; E, elastic modulus or stiffness in Pascals [Pa]; ν, Poisson ratio of 0.5; and δ, penetration/indentation in meters [m].
Time-lapse microscopy and quantification of cell migration
Cells were monitored by digital time-lapse, bright-field inverse microscopy (air objectives, 10×, NA 0.20; 20×, NA 0.30; 40×, NA 0.50; Leica) at 37°C using CCD cameras (Sentech) and the 16-channel frame grabber software (Vistek) for 24 h with 4-min frame intervals (tumor cells) or for 2 h with 30-s intervals (leukocytes; Friedl and Bröcker, 2004). Migration speed was quantified by computer-assisted cell tracking (Autozell 1.0 software; Center for Computing and Communication Technologies [TZI], University of Bremen, Bremen, Germany) of xy paths with 12-min step intervals (tumor cells) or 30-s intervals (leukocytes). The average speed per cell was calculated from the length of the path divided by time, including “go” and “stop” phases.
Confocal fluorescence and reflection microscopy and image analysis
Confocal microscopy (usually use of a 40×/NA 1.3 oil objective; LSM 510; Carl Zeiss) for 3D reconstruction of cell- and fixative-free hydrated matrices was performed with a minimum of 10 µm (typically 10–40 µm) distance to the cover glass. The pore dimensions were obtained from xy scans of cell-free native hydrated collagen lattices (1-µm slice interval; 30-µm z-depth) and orthogonal reconstruction as xz images. The pore cross sections were measured as areas free of dot-like reflection signal from collagen fibers, and pore diameters as smallest linear distance between two fibrils, as described previously (Friedl et al., 1997). For fixed-sample microscopy, cells migrating in 3D collagen matrix (drop matrix cultures; Fig. S1 A) were fixed (4% buffered PFA), washed, and stained with the indicated antibodies and dyes. Dynamic imaging of HT1080 dual-color cells or PMNs was performed as sequential z-stacks in drop collagen matrices within 12 h (HT1080) or 2 h (PMNs) after collagen polymerization using a temperature- and CO2-controlled stage (37°C, 5%). Neutrophil nuclei were labeled with live-DNA marker Hoechst 33342 (10 µg/ml). Time-lapse sequences were reconstructed as maximum intensity projections from usually each 2-3 fluorescence z-scans and 2 reflection scans (ImageJ; 1.40v; National Institutes of Health). Kymography of collagen fibril assembly was obtained from a maximum projection of three z-slices scanned continuously (37°C or 9°C; up to 120 min) and the polymerization speed was obtained from the mean signal intensity/frame. Images were processed by cropping, rotation, and manual adjustment of contrast and brightness and displayed in virtual colors. Elongation index (length/width) of moving cells was calculated from still images after 10 h of incubation. Non-moving, apoptotic or mitotic cells, as identified morphologically from the movie sequence, were excluded from elongation analysis. For cells treated with Y-27632 inhibitor, elongation was calculated only from cells that showed dendrite formation, measuring the main dendrite and, if present, a second dendrite that extended from the cell body by an angle more than 90° away from the main dendrite.
Scanning/ transmission electron microscopy
For scanning and transmission electron microscopy (SEM, TEM), polymerized collagen lattices were fixed with 2% glutaraldehyde and dehydrated step-wise with ethanol of increasing concentration (30–100%). For SEM, samples were dehydrated by critical point drying using CO2, spattered with gold and scanned (microscope JSM-6340F; JEOL). For TEM, samples were embedded in Epon (Serva), sliced, and imaged (transmission electron microscope 1010; JEOL).
Western blotting
Lamin A/C content was determined by Western blot analysis from whole-cell lysates using anti-GAPDH mAb as loading control, followed by chemiluminescence detection (ECL detection kit; GE Healthcare). pMLCT18S19 content was determined with MLC Ab as loading control, followed by fluorescence detection (Odyssee; LI-COR Biosciences) and densitometric analysis.
Filter transmigration assay
Transwell chambers with polycarbonate filters containing 5-, 3-, 1-, and 0.45-µm diameter pore sizes (BD and Costar) were overlaid with cells and the transmigration conditions were as follows: tumor cells (104; DMEM/2% FCS) migrated from the noncoated filter membrane into the lower chamber containing DMEM/10% FCS and pro-migratory lysophosphatidic acid (LPA; 10 µM); leukocytes (3 × 105; TCM/10% FCS) migrated from the filter membrane toward the lower compartment containing TCM/10% FCS, with precoating of type-I collagen on the lower membrane (40 µg/ml; for CD4+ T-blasts) or addition of IL-8 (100 nM; for PMNs). Transmigration rates were counted as numbers of cells attached to the lower membrane side (tumor cells) or the total cell number in suspension in the lower compartment (CD4+ T-blasts, PMNs).
Statistics
Statistical analysis was performed by the two-tailed nonpaired Mann-Whitney test for samples with non-Gaussian distribution.
Online supplemental material
Fig. S1 introduces the 3D cell–collagen chamber systems used for stable collagen anchorage and confirms confocal reflection microscopy as the method of choice for structural imaging of collagen fibers. Fig. S2 visualizes and quantifies the porosity of collagen structures in vitro. Fig. S3 confirms maintenance or abrogation of cell migration in the presence of MMP inhibitor GM6001 or transient MT1-MMP knockdown for spheroid and vertical cell invasion. Fig. S4 presents speed and nuclear adaptation data from different cell types. Video 1 shows migration of single HT1080/MT1-MMP cells in bovine dermal and rat tail collagen in the absence or presence of MMP inhibitor GM6001. Video 2 shows a high-resolution confocal sequence of an arrested cell in rat tail collagen in the presence of GM6001. Video 3 is as Video 1, but for spheroids. Video 4 shows gradually increasing HT1080/MT1-MMP cell migration in rat tail collagen assembled at decreasing temperatures in the presence of MMP inhibitor GM6001. Video 5 shows nuclear deformation together with ongoing or abrogated migration of HT1080 cells transfected with histone H2B-GFP and cytoplasmic DsRed2 in the presence of protease inhibitors. Video 6 shows cells in the presence of MMP inhibitor GM6001 and cell traction modulators β1 integrin 4B4 mAb or Y-27632 in collagen gels of different pore sizes. Video 7 shows migration of CD4+ T blasts and PMNs (in the presence of chemokine IL-8) in the presence of MMP inhibitor GM6001 in rat tail collagen of varying concentration. Video 8 shows IL-8–enhanced migration of a neutrophil stained with nuclear marker Hoechst 33342 in rat tail collagen. Additional data are available in the JCB Data-Viewer at http://dx.doi.org/10.1083/jcb.201210152.dv.
Acknowledgments
We gratefully acknowledge M. Ott, F. Ogboro, and M. Zegers for technical support and advice; the Microscopical Imaging Centre core support by M. van Dommelen, M. Wijers, H. Croes, G.J. Bakker, and J. Fransen for assistance with imaging or data analysis; and M. Sharon Stack and E. Deryugina for providing MDA-MB-231 and HT1080 cells transfected with MT1-MMP.
This project was supported by the Netherlands Science Organization (NWO-VENI 680-47-421 to J. te Riet; NWO-VIDI 917.10.364 to K. Wolf; NWO-VICI 918.11.626 to P. Friedl), the European Union FP7 Tissue Transmigration Training Network (T3Net 237946 to P. Friedl); and the National Institutes of Health (R01 CA88308 and R01 CA71699 to S.J. Weiss).