In budding yeast, the phosphatase Cdc14 orchestrates progress through anaphase and mitotic exit, thereby resetting the cell cycle for a new round of cell division. Two consecutive pathways, Cdc fourteen early anaphase release (FEAR) and mitotic exit network (MEN), contribute to the progressive activation of Cdc14 by regulating its release from the nucleolus, where it is kept inactive by Cfi1. In this study, we show that Cdc14 activation requires the polo-like kinase Cdc5 together with either Clb–cyclin-dependent kinase (Cdk) or the MEN kinase Dbf2. Once active, Cdc14 triggers a negative feedback loop that, in the presence of stable levels of mitotic cyclins, generates periodic cycles of Cdc14 release and sequestration. Similar phenotypes have been described for yeast bud formation and centrosome duplication. A common theme emerges where events that must happen only once per cycle, although intrinsically capable of oscillations, are limited to one occurrence by the cyclin–Cdk cell cycle engine.
Introduction
The eukaryotic cell cycle is driven by a series of complexes comprising cyclins and Cdks. In budding yeast, cell cycle progression is controlled by a single Cdk, Cdc28, whose specificity is dictated by its cyclin regulatory subunit. When Cdk forms complexes with mitotic (M) cyclins (Clb1-4, with Clb2 being the prevalent species), the cells enter mitosis. For cells to exit from mitosis, S phase and M cyclins (collectively called Clbs) must be inactivated (Morgan, 2007). In budding yeast, inactivation of Clb–Cdk is achieved by two redundant mechanisms (Donovan et al., 1994; Schwab et al., 1997; Visintin et al., 1997): (1) accumulation of the Clb–Cdk kinase inhibitor Sic1 (Mendenhall, 1993; Schwob et al., 1994) and (2) degradation of the Clb cyclins by a ubiquitin-dependent proteolysis machinery (Schwab et al., 1997; Visintin et al., 1997; Shirayama et al., 1998). The latter occurs in two steps. At anaphase onset, a specialized ubiquitin ligase known as the anaphase-promoting complex/cyclosome (APC/C), bound to its cofactor Cdc20 (APC/CCdc20), targets S phase and a subset of M cyclins for degradation by the proteosome. During late anaphase and the next G1, the APC/C in complex with Cdh1 (APC/CCdh1) contributes to the full removal of M cyclins (for review see Sullivan and Morgan, 2007). Besides Clb–Cdk inactivation, exit from mitosis requires the reversal of the phosphorylation events mediated by these kinases (for review see Sullivan and Morgan, 2007). The conserved protein phosphatase Cdc14 plays a fundamental role in this process by both inactivating and reversing Clb–Cdk activity (Jaspersen et al., 1998; Visintin et al., 1998; Zachariae et al., 1998; Jin et al., 2008). Cdc14 activity is negatively regulated by Cfi1 (also known as Net1), which sequesters Cdc14 in the nucleolus from G1 up to metaphase (Shou et al., 1999; Visintin et al., 1999). The release of Cdc14 into the nucleus first and later into the cytoplasm responds to different cell cycle cues and is orchestrated in space and time by the interplay of two signal transduction pathways known as Cdc fourteen early anaphase release (FEAR) and mitotic exit network (MEN; for review see Stegmeier and Amon, 2004). This sequential release of Cdc14 dictates the order of dephosphorylation of the multiple Clb–Cdk substrates (Jin et al., 2008), thereby establishing the correct execution of exit from mitosis (for reviews see Stegmeier and Amon, 2004; Rock and Amon, 2009).
At anaphase entry, the FEAR network initiates the release of Cdc14 from the nucleolus (Pereira et al., 2002; Stegmeier et al., 2002; Yoshida and Toh-e, 2002). The activation of this signaling cascade is triggered by the APC/CCdc20 that targets for the degradation of securin, thereby activating separase, an element of FEAR (for review see Stegmeier and Amon, 2004). Via a series of reactions that still need to be fully understood, separase activation eventually leads to the Clb2–Cdk-dependent phosphorylation of Cfi1, which results in the dissociation of Cdc14 from its inhibitor and movement of the phosphatase into the nucleus (Azzam et al., 2004; Queralt et al., 2006). Although FEAR initiates Cdc14 release, it is the MEN pathway that contributes to the full activation of Cdc14 by maintaining it released and allowing its accumulation into the cytoplasm (Shou et al., 1999; Visintin et al., 1999; Mohl et al., 2009). To date, the most downstream component of the MEN is the protein kinase Dbf2 (for review see Stegmeier and Amon, 2004) and is therefore considered the MEN effector. As soon as exit from mitosis is completed, Cdc14 is quickly resequestered into the nucleolus (Shou et al., 1999; Visintin et al., 1999).
Central to the regulation of Cdc14 is the polo-like kinase Cdc5. Cdc5 belongs to both the FEAR network (Pereira et al., 2002; Stegmeier et al., 2002; Visintin et al., 2008) and MEN (Hu et al., 2001; Hu and Elledge, 2002; Geymonat et al., 2003). It is also the only protein among the FEAR and MEN network components that, when overexpressed, can promote the release of Cdc14 from the nucleolus in stages other than anaphase (Visintin et al., 2003). The importance of Cdc5 in releasing Cdc14 is also underscored by the observation that its depletion is necessary and sufficient for the return of the phosphatase into the nucleolus (Visintin et al., 2008).
A common theme of the networks controlling Cdc14 localization is the role played by kinases. The polo-like kinase Cdc5, Clb2–Cdk, and likely the MEN kinase Dbf2 all converge to promote Cdc14 release (Hu et al., 2001; Hu and Elledge, 2002; Pereira et al., 2002; Stegmeier et al., 2002; Yoshida and Toh-e, 2002; Geymonat et al., 2003; Visintin et al., 2003, 2008; Azzam et al., 2004; Queralt et al., 2006; Mohl et al., 2009). How the three kinases interact to induce a timely and complete release of Cdc14 is unknown. In this study, we addressed this question by modulating the activity of these kinases either alone or in mutual combination at various cell cycle stages. We found that the combination of Cdc5 with either Clb–Cdk or Dbf2 is necessary and sufficient to promote Cdc14 release from the nucleolus. Our results provide an explanation of how the interplay of the aforementioned kinases results in the release of Cdc14 at the right time of the cell cycle. Once fully released, Cdc14 triggers a negative feedback loop that, in the presence of stable levels of M cyclins, gives rise to periodic cycles of Cdc14 release and sequestration. Similar oscillatory phenotypes have been described for yeast bud formation and centrosome duplication, suggesting a paradigm whereby events capable of repeating themselves multiple times are restrained to occur once per cycle by their coupling to the cyclin–Cdk engine.
Results
Cdc5 and either Clb–Cdk or the MEN are required for Cdc14 release
Three kinases control Cdc14 release from the nucleolus, Cdc5, Clb2–Cdk, and the MEN kinase Dbf2 (Queralt and Uhlmann, 2008; Mohl et al., 2009). We aimed to evaluate what is the minimal kinase requirement capable of promoting the release of Cdc14. Because these kinases are all active during mitosis, we decided to minimize the side effects caused by their ectopic manipulation by analyzing them in G1 and S phase. In these phases of the cell cycle, both Clb2 and Cdc5 are absent or just start to accumulate (Morgan, 2007), MEN is inactive (for review see Stegmeier and Amon, 2004), and thus, Cdc14 is sequestered in the nucleolus (Shou et al., 1999; Visintin et al., 1999).
When cells are blocked in G1, neither overexpression of an allele of Cdc5 resistant to degradation (GAL-CDC5dBΔ; Shirayama et al., 1998) nor overexpression of a hyperactive allele of Cdc15 (GAL-CDC15(1–750)), known to ectopically activate Dbf2 (Bardin et al., 2003; Visintin et al., 2003), was able to induce the release of Cdc14. However, the combined expression of these alleles of Cdc5 and Cdc15 led to Cdc14 release (Fig. 1 a). To overexpress genes of interest, their open reading frame was placed under the control of the inducible GAL1-10 promoter. Because high levels of Clb2 quickly drive cells blocked in G1 into S phase (Morgan, 2007), the contribution of overexpressing Clb2 was assessed directly in cells blocked in S phase. In this phase of the cell cycle, neither the overexpression of a nondegradable allele of Clb2 (GAL-CLB2dBΔ; Fig. 1 b; Surana et al., 1993), of a hyperactive allele of Cdc15, nor the combined overexpression of these alleles of Clb2 and Cdc15 induced Cdc14 release (Fig. 1 c). Different than in G1, ectopic expression of Cdc5 in S phase–arrested cells was able to trigger Cdc14 release (Fig. 1, b and c; Visintin et al., 2003), but it did so in a Cdk-dependent manner, as shown by the lack of release when the cdc28-as1 ATP analogue–sensitive allele of Cdc28 (Bishop et al., 2001) was inhibited (Fig. 1 b). As for cells arrested in G1, the concomitant activation of Cdc5 and MEN rendered the presence of Clb–Cdk dispensable (Fig. 1 c). Our results show that Cdc14 is released from the nucleolus via a two-hit mechanism, which requires the combined activity of two different kinases, Cdc5 and either MEN kinase Dbf2 or Clb–Cdk.
Cdc5 requires a partner kinase for promoting Cdc14 release. (a) GAL-CDC5dBΔ (Ry1358; GAL-CDC5ΔN70), GAL-CDC15(1–750) (Ry995), and GAL-CDC5dBΔ GAL-CDC15(1–750) (Ry1345) cells were arrested in G1 by α-factor in YEPR. When arrest was complete, the media were supplemented with 2% galactose to induce the expression of CDC5dBΔ and CDC15(1–750). The percentage of cells with Cdc14 released from the nucleolus was determined at the indicated times. (b) GAL-CDC5dBΔ (Ry1358), GAL-CLB2dBΔ (Ry448), and GAL-CDC5dBΔ cdc28-as1 (Ry1356) cells were arrested in G1 with α-factor in YEPR and released in YEPR fresh media supplemented with HU to induce a synchronous arrest in S phase. Once cells were arrested, the media were supplemented with 2% galactose to induce the expression of CDC5dBΔ and CLB2dBΔ together with the Cdc28-as1 inhibitor (1NM-PP1 analogue 9; Bishop et al., 2001). Cell samples were analyzed as in a. (c) GAL-CDC5dBΔ (Ry1358), GAL-CDC15(1–750) (Ry995), GAL-CDC5dBΔ GAL-CDC15(1–750) (Ry1345), GAL-CDC5dBΔ GAL-CDC15(1–750) cdc28-as1 (Ry1353), and GAL-CLB2dBΔ GAL-CDC15(1–750) (Ry1547) cells were treated and analyzed as described in b.
Cdc5 requires a partner kinase for promoting Cdc14 release. (a) GAL-CDC5dBΔ (Ry1358; GAL-CDC5ΔN70), GAL-CDC15(1–750) (Ry995), and GAL-CDC5dBΔ GAL-CDC15(1–750) (Ry1345) cells were arrested in G1 by α-factor in YEPR. When arrest was complete, the media were supplemented with 2% galactose to induce the expression of CDC5dBΔ and CDC15(1–750). The percentage of cells with Cdc14 released from the nucleolus was determined at the indicated times. (b) GAL-CDC5dBΔ (Ry1358), GAL-CLB2dBΔ (Ry448), and GAL-CDC5dBΔ cdc28-as1 (Ry1356) cells were arrested in G1 with α-factor in YEPR and released in YEPR fresh media supplemented with HU to induce a synchronous arrest in S phase. Once cells were arrested, the media were supplemented with 2% galactose to induce the expression of CDC5dBΔ and CLB2dBΔ together with the Cdc28-as1 inhibitor (1NM-PP1 analogue 9; Bishop et al., 2001). Cell samples were analyzed as in a. (c) GAL-CDC5dBΔ (Ry1358), GAL-CDC15(1–750) (Ry995), GAL-CDC5dBΔ GAL-CDC15(1–750) (Ry1345), GAL-CDC5dBΔ GAL-CDC15(1–750) cdc28-as1 (Ry1353), and GAL-CLB2dBΔ GAL-CDC15(1–750) (Ry1547) cells were treated and analyzed as described in b.
Nondegradable Clb2 is not sufficient to maintain Cdc14 released in the absence of MEN
The observation that Cdc5 requires Clb–Cdk or MEN activity to release Cdc14 in phases other than anaphase suggests an appealing interpretation to explain how the correct timing of Cdc14 activation is achieved during a normal cell cycle. At anaphase onset, Cdc5 and Clb2–Cdk would be responsible to initiate Cdc14 release. At this stage, although Cdc5 and Clb2–Cdk are active, MEN is not (for review see Stegmeier and Amon, 2004). During mid- to late anaphase, MEN becomes activated (for review see Stegmeier and Amon, 2004) concomitantly with the reduction of Clb2 levels triggered by the APC/CCdc20 (Morgan, 2007). Thus, it can maintain Cdc14 in its released state by taking over the role of Clb2–Cdk when the activity of the latter goes below a critical threshold. This mechanism would also explain Cdc14 dynamics in mutants lacking MEN, which is in agreement with a previously proposed model (Queralt et al., 2006). FEAR and MEN are not redundant; in MEN mutants, the release of Cdc14 brought about by the FEAR network is transient and limited to the nucleus and is thus insufficient to complete exit from mitosis (Fig. 2, a and c; Pereira et al., 2002; Stegmeier et al., 2002; Yoshida and Toh-e, 2002). In this study, we will refer to this phenotype as the FEAR phenotype. Only by the additional contribution of MEN can the release of Cdc14 be prolonged in time, resulting in Cdc14 spreading into the cytoplasm as well (Fig. 2, b and d; Mohl et al., 2009; for review see Stegmeier and Amon, 2004). According to this model, the decrease of Clb2–Cdk activity is not compensated in MEN mutants and might be the reason why the FEAR-mediated Cdc14 release is transient.
Cdc14 localization and Cdc5 degradation in wild-type cells and MEN mutants. (a–f) cdc15-as1 (Ry1132; a, c, and e) and wild-type (Ry278; b, d, and f) cells were arrested in G1 with α-factor in YEPD at 23°C and released in fresh YEPD media supplemented with the Cdc15-as1 inhibitor (1NM-PP1 analogue 9; Bishop et al., 2001). (a and b) At the indicated time points, samples were taken to determine the percentages of metaphase spindles (closed squares), anaphase spindles (closed circles), and cells with Cdc14 released from the nucleolus (open circles). Examples of Cdc14 localization for the 105-min time point are shown. (c and d) Cdc14 is shown in red, tubulin in green, and DAPI in blue. Bars, 3 µm. (e and f) For the entire time course, we analyzed Clb5, Clb2, Cdc5, and Pgk1 protein levels. Pgk1 protein was used as an internal loading control in immunoblots. Black lines indicate that intervening lanes have been spliced out. Size markers on the sides of the gel blots indicate kilodaltons.
Cdc14 localization and Cdc5 degradation in wild-type cells and MEN mutants. (a–f) cdc15-as1 (Ry1132; a, c, and e) and wild-type (Ry278; b, d, and f) cells were arrested in G1 with α-factor in YEPD at 23°C and released in fresh YEPD media supplemented with the Cdc15-as1 inhibitor (1NM-PP1 analogue 9; Bishop et al., 2001). (a and b) At the indicated time points, samples were taken to determine the percentages of metaphase spindles (closed squares), anaphase spindles (closed circles), and cells with Cdc14 released from the nucleolus (open circles). Examples of Cdc14 localization for the 105-min time point are shown. (c and d) Cdc14 is shown in red, tubulin in green, and DAPI in blue. Bars, 3 µm. (e and f) For the entire time course, we analyzed Clb5, Clb2, Cdc5, and Pgk1 protein levels. Pgk1 protein was used as an internal loading control in immunoblots. Black lines indicate that intervening lanes have been spliced out. Size markers on the sides of the gel blots indicate kilodaltons.
If this hypothesis is correct, it should be possible to make Cdc14 release permanent even in the absence of MEN by stabilizing Clb2. To test this hypothesis, we tracked the kinetic of Cdc14 localization in a synchronous population of cells overexpressing a nondegradable form of Clb2 (GAL-CLB2dBΔ) while being impaired in MEN because of the ATP analogue–sensitive allele of CDC15 (cdc15-as1; Bishop et al., 2001). We expressed Clb2dBΔ at low and high doses to cover a wide range of Clb2 concentrations. As for the low doses, we first determined the minimal galactose concentration necessary to arrest the cells in mitosis. GAL-CLB2dBΔ cells exit from mitosis when grown in media with 0.025% galactose (unpublished data) but are blocked in the presence of 0.05% galactose. Thus, we grew GAL-CLB2dBΔ cdc15-as1 cells in 0.05% galactose to express what we call low doses of nondegradable Clb2. For the high doses, we used 1% galactose. Nucleolar integrity was assessed by imaging the nucleolar protein Nop1 (Fig. S1 a; Tollervey et al., 1991).
In the presence of nondegradable Clb2 expressed at high doses, more cdc15-as1 mutant cells released Cdc14 as compared with the cdc15-as1 single mutant (compare the release at 135 min in Fig. 3 a with the release at 105 min in Fig. 2 a). Interestingly, we noticed that the spatial pattern of Cdc14 release changed from a nuclear FEAR type to a nuclear and cytoplasmic release, resembling the one mediated by MEN (compare Fig. 3 b [135 min] with Fig. 2, c and d). However, this shift toward a MEN-type release was incomplete, suggesting that Clb2–Cdk expressed at high doses and MEN have overlapping but not identical roles. This conclusion is consistent with recent results showing that Cdc14 accumulation in the cytoplasm is mediated by phosphorylation of Cdc14 by Dbf2 (Mohl et al., 2009). When nondegradable Clb2 was expressed at low doses, Cdc14 localization dynamics remained similar to that observed in an MEN single mutant for both the small fraction of anaphase cells with Cdc14 released (Fig. 3 c) and the nuclear localization (unpublished data).
Nondegradable Clb2 promotes periodic cycles of Cdc14 release and sequestration. (a and b) GAL-CLB2dBΔ cdc15-as1 (Ry1394) cells were arrested with α-factor in YEPR and released into medium supplemented with 1% galactose and 5 µM 1NM-PP1 analogue 9 (Bishop et al., 2001) to induce the expression of Clb2 and inactivate the cdc15-as1 allele, respectively. (a) The percentages of cells with metaphase spindles (closed squares), anaphase spindles (closed circles), and cells with Cdc14 released from the nucleolus (open circles) were determined at the indicated times. (b) The localization of Cdc14 and the morphology of the mitotic spindle and nuclei for representative fields of cells are shown for various time points (Videos 1 and 2). Bar, 3 µm. (c) GAL-CLB2dBΔ cdc15-as1 (Ry1394) cells were arrested with α-factor in YEPR and released into fresh medium supplemented with 0.05% galactose and 5 µM Cdc15-as1 inhibitor (1NM-PP1 analogue 9; Bishop et al., 2001). Cell samples were analyzed as in a.
Nondegradable Clb2 promotes periodic cycles of Cdc14 release and sequestration. (a and b) GAL-CLB2dBΔ cdc15-as1 (Ry1394) cells were arrested with α-factor in YEPR and released into medium supplemented with 1% galactose and 5 µM 1NM-PP1 analogue 9 (Bishop et al., 2001) to induce the expression of Clb2 and inactivate the cdc15-as1 allele, respectively. (a) The percentages of cells with metaphase spindles (closed squares), anaphase spindles (closed circles), and cells with Cdc14 released from the nucleolus (open circles) were determined at the indicated times. (b) The localization of Cdc14 and the morphology of the mitotic spindle and nuclei for representative fields of cells are shown for various time points (Videos 1 and 2). Bar, 3 µm. (c) GAL-CLB2dBΔ cdc15-as1 (Ry1394) cells were arrested with α-factor in YEPR and released into fresh medium supplemented with 0.05% galactose and 5 µM Cdc15-as1 inhibitor (1NM-PP1 analogue 9; Bishop et al., 2001). Cell samples were analyzed as in a.
Regardless of the presence of nondegradable Clb2, Cdc14 eventually reentered the nucleolus in both cases. Although with low doses it remained there, after some time with high doses, Cdc14 was surprisingly released again. Noticeably, the alternation of sequestration and release of Cdc14 was not an isolated event but occurred three to four times during the 5 h of observation (Fig. 3 a), and during each cycle, Cdc14 was always fully released into the cytoplasm (Fig. 3 b; and Videos 1 and 2). In conclusion, our data suggest that the stabilization of Clb2 is not sufficient to maintain Cdc14 released in MEN mutants.
Clb–Cdk stabilization maintains Cdc14 released in the absence of MEN
If in MEN mutants, Cdc14 is sequestered even in the presence of stable Clb2, what other mechanisms contribute to Cdc14 sequestration? We aimed to verify whether other Clbs besides Clb2 could play a role in this process. This hypothesis is consistent with the results of our experiments in cells arrested in S phase, where Cdc5 requires Clb–Cdk activity to release Cdc14 (Fig. 1 b), as by inactivating Cdc28 directly, we inhibited all Clb–Cdk complexes at once. To test whether stabilization of Clbs other than Clb2 can maintain Cdc14 released in MEN mutants, we stabilized all Clbs at physiological levels by depleting Cdc20 in a strain lacking PDS1 (Cdc20 depletion was achieved by repressing transcription of CDC20 from the methionine-repressible MET3 promoter MET-CDC20 pds1Δ; Fig. S2 a; Mountain and Korch, 1991). These cells arrest in telophase with Cdc14 released (Fig. 4 a; Shirayama et al., 1999). Strikingly, the same phenotype was observed when we inactivated MEN via the cdc15-as1 allele (Fig. 4, compare b with e; and Fig. S2 b). These results indicate that degradation of other Clbs besides Clb2 is the reason why Cdc14 is resequestered in MEN mutants.
Stabilization of S and M phase cyclins leads to constant Cdc14 release. (a–e) MET-CDC20 pds1Δ cdc15-as1 (Ry1558; a and b), MET-CDC20 pds1Δ cdc15-as1 clb5Δ (Ry1538; c and d), and cdc15-as1 (Ry1132; e) cells were arrested in G1 with α-factor in a synthetic complete medium lacking methionine. When arrest was complete, cells were released into YEPD media lacking the pheromone but with an added 8 mM methionine to repress the expression of Cdc20 in the presence (b, c, and e) or absence (a and d) of 5 µM Cdc15-as1 inhibitor (1NM-PP1 analogue 9; Bishop et al., 2001). The percentages of cells with metaphase spindles (closed squares), anaphase spindles (closed circles), and cells with Cdc14 released from the nucleolus (open circles) were determined at the indicated times. The presence or absence of the inhibitor is indicated as + or −, respectively.
Stabilization of S and M phase cyclins leads to constant Cdc14 release. (a–e) MET-CDC20 pds1Δ cdc15-as1 (Ry1558; a and b), MET-CDC20 pds1Δ cdc15-as1 clb5Δ (Ry1538; c and d), and cdc15-as1 (Ry1132; e) cells were arrested in G1 with α-factor in a synthetic complete medium lacking methionine. When arrest was complete, cells were released into YEPD media lacking the pheromone but with an added 8 mM methionine to repress the expression of Cdc20 in the presence (b, c, and e) or absence (a and d) of 5 µM Cdc15-as1 inhibitor (1NM-PP1 analogue 9; Bishop et al., 2001). The percentages of cells with metaphase spindles (closed squares), anaphase spindles (closed circles), and cells with Cdc14 released from the nucleolus (open circles) were determined at the indicated times. The presence or absence of the inhibitor is indicated as + or −, respectively.
As Clb5 is the predominant species among S phase cyclins and it is degraded at the metaphase to anaphase transition (Fig. 2, e and f), we set to verify its contribution to Cdc14 release in the absence of MEN. To this aim, we analyzed the consequences of deleting CLB5 in a MET-CDC20 pds1Δ cdc15-as1 mutant. To our surprise, in this mutant, we found that Cdc14 was never released from the nucleolus (Fig. 4 c). This result points to Clb5 as a novel member of the FEAR pathway. Indeed, the pattern of Cdc14 release of the MET-CDC20 pds1Δ cdc15-as1 clb5Δ mutant in the absence of the Cdc15 inhibitor, and thus with active MEN, closely resembles the typical phenotype of FEAR mutants (Fig. 4 d) in which Cdc14 release is delayed during early anaphase. We conclude that the stabilization of both S phase and M cyclins is sufficient to render full and stable the release of Cdc14. This result is consistent with the hypothesis that S phase cyclin degradation contributes to Cdc14 sequestration.
Oscillations are impaired in Cdc5 mutants and in the absence of both MEN- and Clb2–Cdk-mediated phosphorylation of Cfi1
Although Cdc14 was resequestered in the presence of both high and low doses of nondegradable Clb2, oscillations arose only in cells blocked in anaphase by high doses. In this setting, it is also possible that Clb5 is required for Cdc14 release and, accordingly, that the first sequestration event is related with its degradation. However, at the metaphase to anaphase transition, the APC/CCdc20 is activated, and thus, it is unlikely that Clb5 is present during the ensuing cycles of Cdc14 release and sequestration. In this setting, we can hypothesize that in MEN mutants, high doses of nondegradable Clb2 can make up for the lack of Clb5 and trigger an additional mechanism that leads to multiple cycles of Cdc14 release and sequestration. Therefore, what is the molecular circuit underlying the oscillatory behavior? First, we asked whether the aforementioned kinase requirement for Cdc14 release (i.e., the two-hit hypothesis whereby Cdc5 and either MEN or Clb–Cdk are necessary for Cdc14 release) was satisfied during the oscillations. According to this model, oscillations (a) should persist in the presence of at least one between MEN and Clb2–Cdk, (b) should be abolished by the simultaneous removal of both MEN and Clb2–Cdk, (c) should occur when both are present, and (d) should be abolished by the inactivation of Cdc5.
Thus, we moved to test the predictions. We have already shown that high doses of Clb2–Cdk can sustain oscillations in the absence of MEN (Fig. 3 a). To test whether MEN alone can drive the oscillations, we interfered with Clb2–Cdk ability to release Cdc14 by using an allele of Cfi1 (cfi1-6Cdk) lacking six Clb2–Cdk phosphorylation sites. This mutant releases Cdc14 with the kinetic typical of FEAR mutants (Azzam et al., 2004). In agreement with our prediction, we found that in the presence of high doses of stable Clb2, in the cfi1-6Cdk mutant cells, although Cdc14 was released with a delay, oscillations nevertheless arose (Fig. 5 a). The simultaneous removal of both MEN and Clb2–Cdk can arrest the oscillatory behavior, as observed when cdc15-as1 was inhibited with the analogue-sensitive inhibitor in combination with the cfi1-6Cdk allele both when the inhibitor was added before or after the initial release (Fig. 5, b and c). Finally, we confirmed that oscillations arise when both MEN and Clb2–Cdk are present (Fig. 5 d). So far, we have observed oscillations only in the presence of high levels of stabilized Clb2. Given that both Clb2–Cdk and MEN alone can sustain the oscillations, we asked whether the presence of MEN would allow low levels of stable Clb2–Cdk to undergo periodical cycles of Cdc14 release and resequestration, and we found this to be the case (Fig. 5 e). Because the release of Cdc14 in the simultaneous presence of Clb2–Cdk and MEN resembles that observed in wild-type cells (Fig. S1, b and c), we decided to investigate the molecular nature of the oscillations in this setting, using both high and low doses of nondegradable Clb2.
Cdc14 oscillations fulfill the kinase requirement of the two-hit model. (a) GAL-CLB2dBΔ cfi1-6Cdk (Ry1545) cells were arrested with α-factor in YEPR and released into medium supplemented with 1% galactose to induce the expression of Clb2. The percentages of cells with metaphase spindles (closed squares), anaphase spindles (closed circles), and cells with Cdc14 released from the nucleolus (open circles) were determined at the indicated times. (b and c) GAL-CLB2dBΔ cfi1-6Cdk cdc15-as1 (Ry1715) cells were arrested with α-factor in YEPR and released into medium supplemented with 1% galactose. 195 min into the release (b) or at the moment of the release (c), 5 µM 1NM-PP1 analogue 9 inhibitor (Bishop et al., 2001) was added to inactivate the cdc15-as1 allele. Cell samples were analyzed as in a. (d and e) GAL-CLB2dBΔ (Ry430) cells were arrested in G1 with α-factor in YEPR and released into fresh medium supplemented with 1 (d) or 0.05% (e) galactose. Cell samples were analyzed as in a. (f and g) GAL-CLB2dBΔ cdc5-1 (Ry1393) cells were treated as in d and e, respectively. Cells were shifted at the restrictive temperature 165 min after the release. Cell samples were analyzed as in a. Arrows indicate the time of inhibitor addition in b (+In) and the shift to the restrictive temperature in f and g.
Cdc14 oscillations fulfill the kinase requirement of the two-hit model. (a) GAL-CLB2dBΔ cfi1-6Cdk (Ry1545) cells were arrested with α-factor in YEPR and released into medium supplemented with 1% galactose to induce the expression of Clb2. The percentages of cells with metaphase spindles (closed squares), anaphase spindles (closed circles), and cells with Cdc14 released from the nucleolus (open circles) were determined at the indicated times. (b and c) GAL-CLB2dBΔ cfi1-6Cdk cdc15-as1 (Ry1715) cells were arrested with α-factor in YEPR and released into medium supplemented with 1% galactose. 195 min into the release (b) or at the moment of the release (c), 5 µM 1NM-PP1 analogue 9 inhibitor (Bishop et al., 2001) was added to inactivate the cdc15-as1 allele. Cell samples were analyzed as in a. (d and e) GAL-CLB2dBΔ (Ry430) cells were arrested in G1 with α-factor in YEPR and released into fresh medium supplemented with 1 (d) or 0.05% (e) galactose. Cell samples were analyzed as in a. (f and g) GAL-CLB2dBΔ cdc5-1 (Ry1393) cells were treated as in d and e, respectively. Cells were shifted at the restrictive temperature 165 min after the release. Cell samples were analyzed as in a. Arrows indicate the time of inhibitor addition in b (+In) and the shift to the restrictive temperature in f and g.
The final prediction of the two-hit model is that Cdc5 is required to generate oscillations. If this is the case, then a loss of function allele of CDC5 should sequester Cdc14 without further release. Because Cdc5 is required for the release of Cdc14, the allele was inactivated after a large fraction of the population had reached anaphase (as assessed by DAPI staining). The prediction was confirmed experimentally by using the temperature-sensitive mutant cdc5-1 with no distinction between high and low doses (Fig. 5, f and g). The resequestration of Cdc14 was not a mere consequence of the temperature shift because the GAL-CLB2dBΔ single-mutant strain shows oscillations at 37°C as well (Fig. S2 c). In conclusion, our data suggest that the periodical release and sequestration of Cdc14 is in agreement with the kinase requirement as proposed by the two-hit model.
Cdc14 oscillations require Cdc5, Cdc14, and Cdh1
The simultaneous inactivation of either MEN and Clb2–Cdk or Cdc5 interfered with the oscillatory phenotype. Thus, we set out to understand which of these two activities is periodically regulated during the oscillations. The simultaneous and coordinated activation and inactivation of Clb2–Cdk and MEN is difficult to envision. A much easier option is that oscillations are driven by regulation of the polo-like kinase Cdc5. At the end of mitosis, Cdc5 is negatively regulated by a negative feedback loop composed of Cdc5, Cdc14, and APC/CCdh1 (Visintin et al., 2008). In this setting, Cdc14 induces the activation of APC/CCdh1, which leads to Cdc5 degradation and, thus, to Cdc14 resequestration. Noticeably, theoretical studies proved that the minimal circuit capable of oscillations is a negative feedback loop composed by three elements (Novák and Tyson, 2008). Thus, we set to verify whether this is the circuit underlying the oscillatory behavior.
If Cdc14 belongs to the loop and is required for its own resequestration, as recently proposed (Visintin et al., 2008; Tomson et al., 2009), the prediction is that in a Cdc14 loss of function mutant, the loop cannot be closed, and Cdc14 should always be released. We tested this hypothesis using the temperature-sensitive cdc14-1 mutant and found the prediction to be true for both high and low doses (Fig. 6, a and b). A possible caveat to this experiment is that the mutant protein at the restrictive temperature could be misfolded and thus unable to bind its inhibitor. In this case, we would observe Cdc14 constantly released independently from Cdc5 stabilization. We tackled this problem in two ways: (1) we assessed whether the cdc14-1 arrest was reversible once cells were shifted back to the permissive temperature with the de novo protein synthesis inhibited by cycloheximide, and (2) we asked whether in the presence of a wild-type copy of CDC14 the Cdc14-1–6HA mutant protein could bind Cfi1 at the restrictive temperature. We found that the cdc14-1 arrest is reversible (Fig. S3 a), and the mutant Cdc14-1–6HA protein in a heterozygous diploid CDC14/cdc14-1–6HA strain undergoes a wild-type kinetic of release and sequestration at the restrictive temperature after release from an S phase arrest (Fig. S3, b and c). We concluded that in our experimental setup, Cdc14-1 stays released because it is enzymatically inactive.
Mutants that interfere with the oscillatory behavior. (a and b) GAL-CLB2dBΔ cdc14-1 (1575) cells were arrested with α-factor in YEPR and released into medium supplemented with 1 (a) or 0.05% (b) galactose to induce the expression of Clb2. (a) The percentages of cells with metaphase spindles (closed squares), anaphase spindles (closed circles), and cells with Cdc14 released from the nucleolus (open circles) were determined at the indicated times. Arrows indicate the time of the temperature shift (180 min). (c and d) GAL-CLB2dBΔ CDC5dBΔ (Ry1392) cells were treated as in a and b, respectively. (e and f) GAL-CLB2dBΔ cdh1Δ (Ry1466) cells were treated as in a and b, respectively. (c–f) Cell samples were analyzed as in a.
Mutants that interfere with the oscillatory behavior. (a and b) GAL-CLB2dBΔ cdc14-1 (1575) cells were arrested with α-factor in YEPR and released into medium supplemented with 1 (a) or 0.05% (b) galactose to induce the expression of Clb2. (a) The percentages of cells with metaphase spindles (closed squares), anaphase spindles (closed circles), and cells with Cdc14 released from the nucleolus (open circles) were determined at the indicated times. Arrows indicate the time of the temperature shift (180 min). (c and d) GAL-CLB2dBΔ CDC5dBΔ (Ry1392) cells were treated as in a and b, respectively. (e and f) GAL-CLB2dBΔ cdh1Δ (Ry1466) cells were treated as in a and b, respectively. (c–f) Cell samples were analyzed as in a.
In the negative feedback loop, Cdc14 induces its own sequestration by indirectly destabilizing Cdc5. If the loop works as expected, we predict that stabilizing Cdc5 should abolish the oscillations. We tested this possibility using a nondegradable mutant of Cdc5 (Visintin et al., 2008), and indeed, in this setting, Cdc14 was always in a released state for both high and low doses (Fig. 6, c and d). Although it is possible that other mechanisms such as the effect of opposing phosphatases might also contribute to modulate Cdc5 activity, the result is in agreement with the notion that the negative regulation occurs via Cdc5 degradation. This is not obvious, as in the presence of high levels of Clb2, APC/CCdh1 should be inhibited (for review see Peters, 2006). This possibility, however, allows a clear prediction: in a cdh1Δ strain, oscillations should be lost and Cdc14 fully released. We tested this prediction and confirmed that oscillations were indeed lost, which is a result confirmed for both high and low doses (Fig. 6, e and f).
APC/CCdh1 is not the only form of APC/C capable of degrading Cdc5
The oscillatory phenotype observed in the cdh1Δ mutant only partially confirmed the prediction; indeed, Cdc14 localization in anaphase cells was intermediate in nature. In cells with high doses of nondegradable Clb2, approximately half of the cells showed Cdc14 released, whereas the other half showed Cdc14 sequestered (Fig. 6 e). Cells with low doses of Clb2 also had an intermediate phenotype, but in these conditions, >50% of anaphase cells had Cdc14 released (Fig. 6 f). The reasons for this discrepancy are at the moment unclear. Because cdh1Δ cells are difficult to arrest in G1 (Schwab et al., 1997), we reasoned that such an intermediate phenotype could be a consequence of poor synchronization and, thus, the result of an averaging of the oscillations observed at the population level. However, this possibility is unlikely because wild-type cells released from a metaphase arrest induced by nocodazole, although poorly synchronous, show oscillations, whereas the cdh1Δ mutant confirmed the intermediate phenotype (Fig. S2, e and f). Thus, we decided to further assess the contribution of degradation to the oscillatory behavior by analyzing the pattern of Cdc14 localization in cells carrying a temperature-sensitive allele of CDC23 (cdc23-1), a core component of the APC/C (for review see Peters, 2006). Because APC/C is required for the metaphase to anaphase transition, the allele was inactivated after a large fraction of the population had reached anaphase. In these cells, both at high and low doses of Clb2dBΔ, Cdc14 does not oscillate, but, contrarily to cdh1Δ, it is released in the entire anaphase population (Fig. 7, a and b), which is in agreement with the possibility that APC/C-dependent degradation of Cdc5 promoted by Cdc14 is required for the oscillatory behavior. We confirmed the role of protein synthesis and degradation in the generation of oscillations by blocking protein synthesis with cycloheximide 180 min after having overexpressed nondegradable Clb2. In these conditions, Cdc14 was sequestered and never released (Fig. S2 d). Moreover, we observed a reduction of Cdc5 levels in the presence of high levels of stabilized Clb2 with live cell imaging (Videos 3 and 4). These results prompted us to verify whether the periodic release of Cdc14 was affected by Cdc20, the other cofactor of the APC/C (for review see Peters, 2006). When CLB2dBΔ was overexpressed in a cdc20-3 mutant, we observed periodic sequestration and release of Cdc14 regardless of the expression levels of nondegradable Clb2 (Fig. 7, c and d), thus showing that Cdc20 is not implicated in the oscillatory loop. This result, together with what we observed in the cdc23-1 and cdh1Δ mutants, strongly points to a role for Cdh1 in the generation of oscillations. This finding raises the possibility that the spatial localization of the components of this circuit allows Cdc14 to activate a subset of Cdh1 even in the presence of high levels of Clb2–Cdk.
APC/C-dependent degradation closes the loop. (a–d) GAL-CLB2dBΔ cdc23-1 (Ry1452; a and b) and GAL-CLB2dBΔ cdc20-3 (Ry1457; c and d) cells were arrested with α-factor in YEPR and released into fresh medium supplemented with 1 (a and c) or 0.05% (b and d) galactose to induce the expression of Clb2. Cells were shifted at the restrictive temperature 180 min after the release (arrows). The percentages of cells with metaphase spindles (closed squares), anaphase spindles (closed circles), and cells with Cdc14 released from the nucleolus (open circles) were determined at the indicated times.
APC/C-dependent degradation closes the loop. (a–d) GAL-CLB2dBΔ cdc23-1 (Ry1452; a and b) and GAL-CLB2dBΔ cdc20-3 (Ry1457; c and d) cells were arrested with α-factor in YEPR and released into fresh medium supplemented with 1 (a and c) or 0.05% (b and d) galactose to induce the expression of Clb2. Cells were shifted at the restrictive temperature 180 min after the release (arrows). The percentages of cells with metaphase spindles (closed squares), anaphase spindles (closed circles), and cells with Cdc14 released from the nucleolus (open circles) were determined at the indicated times.
As for the distribution of Cdc14 in cdh1Δ cells, we hypothesize that although in this strain Cdc5 is degraded in an APC-dependent manner, the negative feedback loop is destroyed, and thus, oscillations are lost. In this context, Cdc5 synthesis (Clb2–Cdk dependent; Darieva et al., 2003) and Cdc5 degradation (APC dependent; for review see Peters, 2002) find an equilibrium. This intermediate value of Cdc5 could result in some cells having Cdc14 sequestered and others released. As for the APC-dependent and Cdh1-independent mechanism that drives Cdc5 degradation, our results hint to a possible role for Cdc20. In cdc15-as1 cells, APC/CCdh1 is not activated because Cdc14 is only transiently released from the nucleolus early in anaphase, and thus, Cdc20, a substrate of APC/CCdh1, is stabilized. In this setting, Cdc5 levels decrease with the same kinetics as Clb2 (Fig. 2, e and f), suggesting that similarly to Clb2, Cdc5 could also be partially degraded in an APC/CCdc20-dependent manner.
In conclusion, the oscillations in Cdc14 localization are driven by the negative feedback loop, which includes Cdc14, Cdh1, and Cdc5 and controls Cdc14 relocalization at the end of mitosis (Visintin et al., 2008). The novel oscillatory phenotype and the underlying molecular circuitry described in this study were also observed in the laboratory of F. Cross using live cell imaging (Lu and Cross, 2010).
Simulations point to an additional role for Cdc14
Our results show that cdc23-1 (Fig. 7, a and b) and cdc14-1 (Fig. 6, a and b) mutants share the same phenotype, having Cdc14 permanently released, contrarily to cdh1Δ (Fig. 6, e and f), where Cdc14 is only partially released. The fact that cdh1Δ and cdc14-1 show different phenotypes is not consistent with the network we have analyzed so far (Cdc14 → Cdh1⊣ Cdc5 → Cdc14), in which the only role played by Cdc14 is to activate Cdh1. This discrepancy suggests that Cdc14 performs additional roles to induce its own sequestration. Given that both the inactivation of Cdc14 and APC/C (i.e., cdc23-1) cause the complete release of Cdc14, one possibility is that the cdc14-1 mutant stabilizes Cdc5 as efficiently as the cdc23-1 mutant. We can envision one scenario (the Cdc5 stabilization hypothesis) whereby Cdc5 can be stabilized by phosphorylation (possibly by Clb2–Cdk), whereas its dephosphorylation, catalyzed by Cdc14, makes the kinase a good substrate for both APC/CCdc20 and APC/CCdh1. Alternatively, the different phenotypes of cdh1Δ and cdc14-1 could be explained if Cdc14 would induce its own sequestration, besides triggering Cdc5 degradation, via dephosphorylating other substrates, a likely candidate being Cfi1 (the Cfi1 dephosphorylation hypothesis). A detailed description of the Cfi1 dephosphorylation hypothesis is given in Fig. S4 a, whereas the molecular details of the Cdc5 stabilization hypothesis are shown in Fig. S5 a.
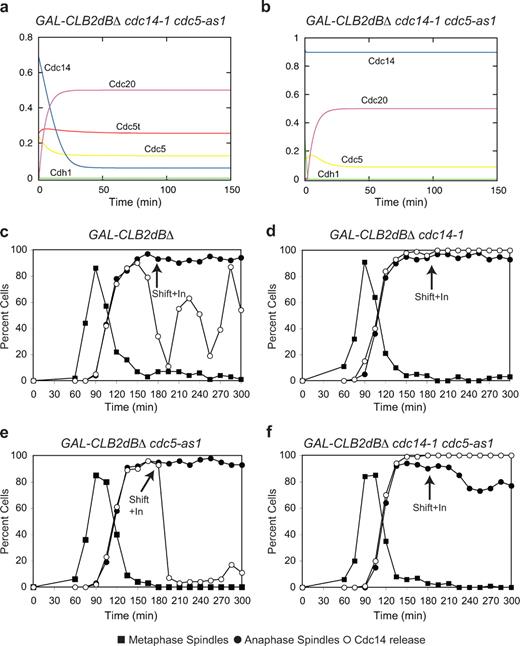
We then set out to verify whether any of these two hypotheses can account for all the results presented so far. Given the complexity of the aforementioned proposed circuits, we preferred to analyze them with the help of mathematical analysis. Leaning on previous work (Queralt et al., 2006), we translated the two networks into mathematical models (Tables S1 and S2; and see supplemental equations). Simulations confirmed that both networks are consistent with the experimental results (Figs. S4 and S5) and showed that the two models predict quite a different outcome if both Cdc5 and Cdc14 are inactivated after the initial release of Cdc14. According to the Cdc5 stabilization hypothesis, Cdc14 should be resequestered and never released again in the double mutant because the stabilization of Cdc5 resulting from the lack of Cdc14 would be overcome by the inactivation of Cdc5 itself (Fig. 8 a). Contrarily, the Cfi1 dephosphorylation model predicts that Cdc14 would not be sequestered in a cdc5 cdc14 double mutant because if Cfi1 phosphorylation could not be increased, neither could Cfi1 dephosphorylation (Fig. 8 b). In cells blocked in mitosis by high doses of Clb2, the single mutants, as expected, blocked the oscillations, with Cdc14 being constantly released in the cdc14-1 strain and resequestered in the cdc5-as1 mutant (Fig. 8, c–e; Zhang et al., 2005). In agreement with the Cfi1 dephosphorylation hypothesis, when we performed the experiment in a strain carrying both of the cdc14-1 and cdc5-as1 alleles, we found that Cdc14 was not resequestered (Fig. 8 f). Interestingly, experimental data show that Cdc14 in the cdc14-1 cdc5-as1 double mutant is constitutively released as much as in a simple cdc14-1 mutant, whereas simulations show a larger release in the latter case. The discrepancy might point to additional ways by which Cdc14 induces its own resequestration. We conclude that Cdc14 induces its own resequestration via additional pathways besides the destabilization of Cdc5 mediated by APC/CCdh1.
Cdc14 resequestration requires both Cdc5 inactivation and Cdc14 phosphatase activity. (a) The Cdc5 stabilization model predicts that Cdc14 is resequestered in the double mutant cdc14-1 cdc5-as1. Parameters and equations in the supplemental equations are shown in the text and Table S2, except for kph_cfi = 0, kdeph_cdh = 0, and kdeph_c5 = 0. (b) The Cfi1 dephosphorylation model predicts that when both Cdc5 and Cdc14 are not active, the phosphatase is not resequestered. Parameters and equations are shown in the supplemental equations paragraph and Table S1, except for kph_cfi = 0, kdeph_cdh = 0, and kdeph_cfi = 0. (c–f) GAL-CLB2dBΔ (Ry1387; c), GAL-CLB2dBΔ cdc14-1 (Ry1575; d), GAL-CLB2dBΔ cdc5-as1 (cdc5L158G; Ry1607; e; Zhang et al., 2005; Snead et al., 2007), and GAL-CLB2dBΔ cdc14-1cdc5-as1 (cdc5L158G; Ry1606; f) cells were arrested with 5 µg/ml α-factor. After 3 h, cells were released into medium lacking pheromone supplemented with 1% galactose to induce the expression of Clb2. 180 min after the release, cells were shifted at 37°C and supplemented with 10 µM cdc5-as1 inhibitor (CMK; arrows; Snead et al., 2007). The percentages of cells with metaphase spindles (closed squares), anaphase spindles (closed circles), and cells with Cdc14 released from the nucleolus (open circles) were determined at the indicated times.
Cdc14 resequestration requires both Cdc5 inactivation and Cdc14 phosphatase activity. (a) The Cdc5 stabilization model predicts that Cdc14 is resequestered in the double mutant cdc14-1 cdc5-as1. Parameters and equations in the supplemental equations are shown in the text and Table S2, except for kph_cfi = 0, kdeph_cdh = 0, and kdeph_c5 = 0. (b) The Cfi1 dephosphorylation model predicts that when both Cdc5 and Cdc14 are not active, the phosphatase is not resequestered. Parameters and equations are shown in the supplemental equations paragraph and Table S1, except for kph_cfi = 0, kdeph_cdh = 0, and kdeph_cfi = 0. (c–f) GAL-CLB2dBΔ (Ry1387; c), GAL-CLB2dBΔ cdc14-1 (Ry1575; d), GAL-CLB2dBΔ cdc5-as1 (cdc5L158G; Ry1607; e; Zhang et al., 2005; Snead et al., 2007), and GAL-CLB2dBΔ cdc14-1cdc5-as1 (cdc5L158G; Ry1606; f) cells were arrested with 5 µg/ml α-factor. After 3 h, cells were released into medium lacking pheromone supplemented with 1% galactose to induce the expression of Clb2. 180 min after the release, cells were shifted at 37°C and supplemented with 10 µM cdc5-as1 inhibitor (CMK; arrows; Snead et al., 2007). The percentages of cells with metaphase spindles (closed squares), anaphase spindles (closed circles), and cells with Cdc14 released from the nucleolus (open circles) were determined at the indicated times.
Discussion
A two-hit model for Cdc14 release
Our results indicate that Clb–Cdk, Cdc5, and MEN, likely via kinase Dbf2, are important for Cdc14 activation, albeit to a different extent. Although Cdc5 is always required for an effective release of Cdc14, MEN and Clb–Cdk have partially overlapping roles. Besides Clb2–Cdk, our data suggest that the S phase cyclin Clb5 contributes to Cdc14 release during early anaphase. The role of Clb5 in displacing and thus activating Cdc14, although at odds with its well-known role of inhibitor of anaphase entry and progression (Shirayama et al., 1999; Jin et al., 2009), could underlie the mechanism whereby cells coordinate DNA replication with the completion of mitosis.
Based on the two-hit model, we can envision that during a normal cell cycle, Cdc5 cooperates with Clb–Cdk to induce the first release of Cdc14. At the metaphase to anaphase transition, Clb5 is degraded, shortly followed by a partial degradation of Clb2. In the meantime, MEN becomes active and compensates for the inactivation of the two Clbs, making sure that either Clb–Cdk or MEN is always present throughout mitosis to contribute to Cdc14 release. As Cdh1 is activated, Cdc5 degradation causes the resequestration of Cdc14. If MEN is missing, the overall decrease in Clb2–Cdk and Clb5–Cdk activity may account for the shortened timing of Cdc14 release in MEN mutants.
What are the molecular implications of the two-hit model? The observation that the activity of Cdc5 is always required for Cdc14 release offers an appealing answer to this question. Polo-like kinases, of which Cdc5 is the only yeast member, contain in their C terminus a conserved sequence motif called the polo box domain (PBD; Lee et al., 1998, 1999; Elia et al., 2003a). PBD mediates the interaction between the kinase and its substrates after they have undergone phosphopriming by another kinase (Elia et al., 2003b). Therefore, we speculate that priming by Clb–Cdk and Dbf2 serves to build up Cdc5 activity. Cdc5 phosphorylation of Cfi1 and/or of Cdc14 could then promote the release of Cdc14 from Cfi1. The validation of this hypothesis awaits the identification of residues in Cdc14 and/or Cfi1 whose phosphorylation status is cell cycle regulated. Of particular interest will be the identification of those residues that are specifically phosphorylated during anaphase, the cell cycle stage during which the Cdc14/Cfi1 interaction is disrupted. In support of this idea, six Clb2–Cdk-dependent phosphoresidues have been identified in Cfi1. Cells carrying a mutant allele of Cfi1 in which these amino acids have been mutated into alanine (cfi1-6Cdk), mimicking lack of phosphorylation, are delayed in the release of Cdc14. Notably, one of these residues (threonine 212) is part of an optimal phosphobinding motif recognized by PBD (Elia et al., 2003a). Recently, we identified another residue in Cfi1, specifically phosphorylated during anaphase that lies in a putative Dbf2 recognition sequence (Mah et al., 2005) and is also part of a minimal PBD-phosphobinding motif (Elia et al., 2003a; unpublished data). The consequences of mutating the latter residue into alanine will be a matter of further studies. As for the role of Clb5, the observation that recombinant Cfi1 is phosphorylated in vitro by Clb2–Cdk but not by Clb5–Cdk (Azzam et al., 2004) argues against the two Clbs sharing the same function. Understanding how Clb5 impinges on the FEAR network will be an important quest for the future.
A model for Cdc14 dynamics at the exit from mitosis
We challenged the two-hit model by analyzing Cdc14 localization in the presence of nondegradable Clb2, which is expressed both at high and low levels. In the presence of high doses, Clb2–Cdk has a double role: it both prevents cells from exiting mitosis and induces the release of Cdc14 (together with MEN, which is dispensable in this setting). This latter role is shared with Cdc5, with both of them being required for the release of Cdc14. When Cdc5 is degraded as a consequence of Cdc14 release and the ensuing APC/CCdh1 activation, Clb2–Cdk cannot promote Cdc14 release alone, and thus the phosphatase goes back into the nucleolus. Nevertheless, Clb2–Cdk manages to block cells in mitosis, allowing Cdc5 to be synthesized again and to subsequently induce a new round of Cdc14 release and sequestration. In this context, Clb2–Cdk likely plays an additional third role, promoting the transcription of Cdc5, that together with CLB2 is a member of the CLB2 cluster genes, a set of ∼30 genes whose expression depends on Clb2 (Darieva et al., 2003).
When the doses of nondegradable Clb2 are lowered, the presence of MEN becomes essential for full Cdc14 release similarly to a normal cell cycle. As Cdh1 is activated, Cdc5 degradation causes the resequestration of Cdc14 followed by MEN-driven cycles of Cdc14 release and sequestration. In both high and low doses of Clb2, the role of Clb5 is likely limited to the initial release of Cdc14, after which it is degraded. Its presence is dispensable at high levels of stable Clb2, whereas it becomes essential when Clb2 is stabilized at low doses in the absence of MEN.
We were surprised to observe that APC/CCdh1 was required for the oscillations in the presence of nondegradable Clb2. This result contrasts with the notion that Clb2–Cdk inhibits APC/CCdh1 (Nasmyth, 1996). Similarly, Clb2–Cdk has been reported to inhibit MEN (König et al., 2010; for review see Stegmeier and Amon, 2004). In our system, however, MEN seems to be active even in the presence of high levels of stable Clb2. When we used the cfi1-6Cdk mutant, we found that oscillation raised in an MEN-dependent manner in the presence of active Clb2–Cdk-arresting cells in mitosis. At present, we cannot give an explanation for these contrasting results. They possibly point to the fact that subcellular localization of the kinases and phosphatases, whose delicate balance controls Cdc14 localization, cannot be neglected if we aim to understand the network controlling exit from mitosis.
Why oscillations?
Our results raise the problem of the physiological significance of the oscillations of Cdc14 localization. We believe that what we report contributes to the understanding of the principles underlying the coordination between the cell cycle engine (i.e., cyclin–Cdk activities) and the events triggered by the engine itself.
The oscillatory behavior reported in this study is not an isolated example but resembles other cases of periodical phenotypes that emerge when the cell cycle engine is blocked. In budding yeast, the deletion of Clbs results in periodical budding in G1 (Haase and Reed, 1999). In both budding yeast and Drosophila melanogaster, the deletion or knockdown of M cyclins arrest cells before mitosis, but centrosomes nevertheless undergo periodic rounds of divisions (Haase et al., 2001; McCleland and O’Farrell, 2008). Mutants for three major control mechanisms preventing DNA rereplication can be blocked in G2/M with high levels of Clb2–Cdk and undergo multiple cycles of rereplication (Nguyen et al., 2001). These seemingly different systems share some basic properties among themselves and with our system: they are all triggered by the cell cycle engine and normally occur once per cell cycle. This last property is caused by their coupling with the cell cycle engine because, as we said, they show periodic behavior when the engine is blocked.
Do these systems also share properties at the molecular level? The molecular details of the circuits we have mentioned are not as well known as the circuit we analyzed in this study. However, their capability to oscillate implies that they all include at least one negative feedback loop. This fits very well with the need for these circuits to reset to their initial state after one cycle, ready for the forthcoming cell cycle. However, their capability to oscillate implies much stronger requirements than the simple negative feedback loop: they must have enough nonlinearity, sufficient time delays, and proper balancing of the rate constants, all conditions required for oscillations (Novák and Tyson, 2008). All of these conditions are not obvious, and indeed, the oscillatory behavior in the presence of constant high levels of mitotic kinase came as a surprise.
Thus, we believe that an important question to be asked is why events coupled to the cell cycle engine that take place only once per cycle have evolved to an oscillatory regime, normally hidden by their coupling to the cell cycle machinery. The answer to this question is unknown but surely worth further investigations. In this study, we propose that an oscillatory dynamic guarantees a resetting of the molecular circuit to its initial conditions less dependent on the Cdk input than a simple negative feedback loop, thus avoiding intermediate outcomes of the circuit. In the case of Cdc14, that would be a state with only a fraction of Cdc14 resequestered in the nucleolus at the end of mitosis, APC/CCdh1 only partially active, and intermediate levels of Clb2. Such a condition would leave cells in a state of unclear definition, partially mitosis and partially G1. The oscillatory dynamics, even if limited to one cycle, make sure that this fate is avoided.
Materials and methods
Yeast strains
All strains were derivatives of strain W303 (K699). In Table S3, the relevant genotypes of the strains used in this study are indicated.
Growth conditions
Cell cycle arrest and synchronization experiments were performed as described previously (Amon, 2002).
Cell cycle arrest experiments pertinent to Fig. 1.
Cells were grown at 23°C in yeast extract peptone (YEP) supplemented with 2% raffinose (YEPR) and arrested in G1 by adding 5 µg/ml of the α-factor pheromone. For the G1 block experiments, when the arrest was complete (>90% unbudded cells), 2% galactose was added to drive the overexpression of the genes of interest. To maintain the G1 block, 1 h after galactose addition, 2.5 µg/ml α-factor was readded to the media. For the S phase block experiments, when >90% of cells reached G1, cells were released into fresh YEPR supplemented with 10 mg/ml HU to synchronously arrest them in the next S phase. When >90% of cells were arrested (cells have small- to medium-size buds), 2% galactose and 5 µM of the 1NM-PP1 analogue 9 were added, respectively, to induce the overexpression of genes of interest and to inhibit ATP analogue–sensitive alleles when present.
Synchronization experiments pertinent to Fig. 2.
Cells were grown at 23°C in YEP supplemented with 2% glucose (YEPD) and arrested in G1 by adding 5 µg/ml of the α-factor pheromone. When arrest was complete, cells were released in fresh YEPD media lacking the pheromone supplemented with 5 µM of the 1NM-PP1 analogue 9 to inhibit the cdc15-as1 allele of CDC15.
Synchronization experiments pertinent to Fig. 4.
MET3-CDC20 cells were grown in synthetic medium lacking methionine and arrested in G1 by adding 5 µg/ml of the α-factor pheromone. When arrest was complete, cells were released in YEPD medium supplemented with 8 mM methionine to shut off the MET3 promoter in the presence or absence of 5 µM of the 1NM-PP1 analogue 9.
Oscillation experiments
Cells were grown at 23°C in YEPR and arrested in G1 by adding 5 µg/ml of the α-factor pheromone. When arrest was complete, cells were released in fresh YEPR media lacking the pheromone but with an added 1% galactose (YEPR + galactose high doses) or 0.05% galactose (YEPR + galactose low doses) to induce the expression of GAL-CLB2dBΔ. Strains carrying temperature-sensitive alleles were shifted at the restrictive temperature (37°C) after the majority of the population had undergone the metaphase to anaphase transition, as assessed by DAPI staining. The time of the shift for each experiment is indicated in Figs. 5–8 by an arrow. cdc15-as1 cells were inactivated by adding 5 µM Cdc15-as1 inhibitor (1NM-PP1 analogue 9) already at the time of release or as indicated in the legend of Fig. 8. cdc5-as1 cells were inactivated by adding 5 µM Cdc5-as1 inhibitor (CMK; Snead et al., 2007) after the majority of the population had reached anaphase, as assessed by DAPI staining.
Immunoblot analysis
Cells were lysed in 50 mM Tris, pH 7.5, 1 mM EDTA, 1 mM PNP, 50 mM DTT, 1 mM PMSF, and 2 µg/ml pepstatin with glass beads for 1 min and boiled in 1× sample buffer. Immunoblot analysis of the total amount of Clb2 and Pgk1 was performed as described previously (Cohen-Fix et al., 1996) using α-Clb2 (provided by A. Amon, Massachusetts Institute of Technology, Cambridge, MA) and α-Pgk1 (Invitrogen). Immunoblot analysis of the total amount of endogenous Cdc5 and Clb5 was performed according to the manufacturer’s guidelines using α-Cdc5 and α-Clb5 antibodies (Santa Cruz Biotechnology, Inc.) as described previously (Visintin et al., 2008).
Immunostaining
Indirect in situ immunofluorescence was performed as described previously (Visintin et al., 1999). Spindle formation and elongation was detected by α-tubulin immunostaining with the YOL34 monoclonal antibody (AbD Serotec) followed by indirect immunofluorescence using FITC-conjugated anti–rat antibody (Jackson ImmunoResearch Laboratories, Inc.). Cdc14 immunostaining was performed using polyclonal Cdc14 antibodies (Santa Cruz Biotechnology, Inc.) followed by indirect immunofluorescence using CY3-conjugated anti–goat antibody (Santa Cruz Biotechnology, Inc.). Nucleolar integrity was detected by α-Nop1 (EnCor Biotechnology) immunostaining followed by indirect immunofluorescence using FITC-conjugated anti–mouse antibodies (Jackson ImmunoResearch Laboratories, Inc.).
Scoring of indirect immunofluorescence samples
Cell cycle progression was scored by analysis of spindle morphology. Spindles were divided into three categories: (1) interphase microtubules, which include cells in the G1, S, and G2 phases of the cell cycle, (2) metaphase spindles, characteristic of cells in metaphase, and (3) anaphase spindles, which include cells in anaphase and telophase. Cdc14 was scored as sequestered when the large majority of the protein was inside the nucleolus, whereas it was considered released when the majority of the protein was outside the nucleolus. We did not distinguish between nuclear and cytoplasmic release; as soon as Cdc14 had left the nucleolus, we considered it released. This category also included cells containing a small amount of the phosphatase in the nucleolus. Examples of the aforementioned categories can be found in Visintin et al. (2008).
Images acquisition and analysis
Images of immunofluorescence were taken using either a microscope (IX81; Olympus) with a 60× 1.4 NA Plan Apo oil objective (Olympus) with an additional enlargement of 1.6 (Fig. 2), a 100× 1.35 NA UPlan Apo oil objective (Olympus) with an additional enlargement of 1.6 (Fig. 3 and Fig. S3), or a microscope (BX51; Olympus) with a 60× 1.4 NA Plan Apo oil objective (Olympus) with an additional enlargement of 1.25 (Fig. S1). All images were acquired with a 12-bit camera (CoolSNAP; Photometrics). Analysis of the data was performed using MetaMorph software (version 7.5.6.0; MDS Analytical Technologies). No manipulations were performed other than adjustments in brightness and contrast.
Online supplemental material
Fig. S1 shows that the first release of Cdc14 is similar in wild-type cells and in cells expressing nondegradable Clb2. Fig. S2 contains controls pertinent to different experiments: Clb2 and Clb5 are stabilized in a MET-CDC20 pds1Δ block, oscillations persist at 37°C, oscillations are lost when protein synthesis is inhibited, and oscillations persist in wild-type cells released from a nocodazole block but are abolished in a cdh1Δ mutant. Fig. S3 shows that the mutant Cdc14-1 protein can bind Cfi1 at the restrictive temperature. Fig. S4 shows simulations of the Cfi1 dephosphorylation model. Fig. S5 shows simulations of the Cdc5 stabilization model. The Cfi1 dephosphorylation model equation paragraph includes all of the algebraic differential equations for the Cfi1 dephosphorylation model, whose parameters can be found in Table S1. The Cdc5 stabilization model equation paragraph includes all of the algebraic differential equations for the Cdc5 stabilization model, whose parameters can be found in Table S2. Table S3 contains a list of yeast strains used in this study. Videos 1 and 2 show the periodical localization of Cdc14 in cells blocked in mitosis by high doses of nondegradable Clb2. Videos 3 and 4 show that Cdc5 levels are reduced periodically when cells are blocked in mitosis by nondegradable Clb2.
Periodic release and resequestration of Cdc14-GFP in a GAL-CLB2dB Δ strain.GAL-CLB2dB Δ CDC14-GFP cells (Ry1470) were grown at 23°C in synthetic complete medium with 2% sucrose and arrested in G1 by adding 10 µm/ml α-factor pheromone. When arrest was complete, cells were loaded in a microfluidic chamber (Y2 plates; Cellasic) according to the manufacturer’s instructions and released in synthetic complete medium with 2% sucrose and 2% galactose to induce the expression of GAL-CLB2dB Δ. The pressure applied to control the medium flow and loading of the cells in the chamber was 2.5 psi. Time-lapse imaging was performed at 30°C using an epifluorescence microscope equipped with an oil objective and a camera. Frames were taken every 15 min. Images were deconvolved using standard deconvolution (enhanced ratio [aggressive]) with 10 iterations and a medium noise filtering. Bar, 5 µm.
Periodic release and resequestration of Cdc14-GFP in a GAL-CLB2dB Δ strain.GAL-CLB2dB Δ CDC14-GFP cells (Ry1470) were grown at 23°C in synthetic complete medium with 2% sucrose and arrested in G1 by adding 10 µm/ml α-factor pheromone. When arrest was complete, cells were loaded in a microfluidic chamber (Y2 plates; Cellasic) according to the manufacturer’s instructions and released in synthetic complete medium with 2% sucrose and 2% galactose to induce the expression of GAL-CLB2dB Δ. The pressure applied to control the medium flow and loading of the cells in the chamber was 2.5 psi. Time-lapse imaging was performed at 30°C using an epifluorescence microscope equipped with an oil objective and a camera. Frames were taken every 15 min. Images were deconvolved using standard deconvolution (enhanced ratio [aggressive]) with 10 iterations and a medium noise filtering. Bar, 5 µm.
Periodic release and resequestration of Cdc14-GFP in a GAL-CLB2dB Δ strain.GAL-CLB2dB Δ CDC14-GFP cells (Ry1470) were grown at 23°C in synthetic complete medium with 2% sucrose and arrested in G1 by adding 10 µm/ml α-factor pheromone. When arrest was complete, cells were loaded in a microfluidic chamber (Y2 plates) according to the manufacturer’s instructions and released in synthetic complete medium with 2% sucrose and 2% galactose to induce the expression of GAL-CLB2dB Δ. The pressure applied to control the medium flow and loading of the cells in the chamber was 2.5 psi. Time-lapse imaging was performed at 30°C using an epifluorescence microscope equipped with an oil objective and a camera. Frames were taken every 15 min. Images were deconvolved using standard deconvolution (enhanced ratio [aggressive]) with 10 iterations and a medium noise filtering. Bar, 5 µm.
Periodic release and resequestration of Cdc14-GFP in a GAL-CLB2dB Δ strain.GAL-CLB2dB Δ CDC14-GFP cells (Ry1470) were grown at 23°C in synthetic complete medium with 2% sucrose and arrested in G1 by adding 10 µm/ml α-factor pheromone. When arrest was complete, cells were loaded in a microfluidic chamber (Y2 plates) according to the manufacturer’s instructions and released in synthetic complete medium with 2% sucrose and 2% galactose to induce the expression of GAL-CLB2dB Δ. The pressure applied to control the medium flow and loading of the cells in the chamber was 2.5 psi. Time-lapse imaging was performed at 30°C using an epifluorescence microscope equipped with an oil objective and a camera. Frames were taken every 15 min. Images were deconvolved using standard deconvolution (enhanced ratio [aggressive]) with 10 iterations and a medium noise filtering. Bar, 5 µm.
Live cell imaging of Cdc5-Venus in a GAL-CLB2dB Δ strain.GAL-CLB2dB Δ CDC5-Venus cells (Ry1687) were grown at 23°C in synthetic complete medium with 2% sucrose and imaged in synthetic complete medium with 2% sucrose and 2% galactose to induce the expression of GAL-CLB2dB Δ. Cells were loaded in a microfluidic chamber (Y2 plates) according to the manufacturer’s instructions. The pressure applied to control the medium flow and loading of the cells in the chamber was 2.5 psi. Time-lapse imaging was performed at 30°C using an epifluorescence microscope equipped with an oil objective and a camera. Frames were taken every 20 min. Images were deconvolved using standard deconvolution (enhanced ratio [aggressive]) with 10 iterations and a medium noise filtering. Bar, 5 µm.
Live cell imaging of Cdc5-Venus in a GAL-CLB2dB Δ strain.GAL-CLB2dB Δ CDC5-Venus cells (Ry1687) were grown at 23°C in synthetic complete medium with 2% sucrose and imaged in synthetic complete medium with 2% sucrose and 2% galactose to induce the expression of GAL-CLB2dB Δ. Cells were loaded in a microfluidic chamber (Y2 plates) according to the manufacturer’s instructions. The pressure applied to control the medium flow and loading of the cells in the chamber was 2.5 psi. Time-lapse imaging was performed at 30°C using an epifluorescence microscope equipped with an oil objective and a camera. Frames were taken every 20 min. Images were deconvolved using standard deconvolution (enhanced ratio [aggressive]) with 10 iterations and a medium noise filtering. Bar, 5 µm.
Live cell imaging of Cdc5-Venus in a GAL-CLB2dB Δ strain.GAL-CLB2dB Δ CDC5-Venus cells (Ry1687) were grown at 23°C in synthetic complete medium with 2% sucrose and imaged in synthetic complete medium with 2% sucrose and 2% galactose to induce the expression of GAL-CLB2dB Δ. Cells were loaded in a microfluidic chamber (Y2 plates) according to the manufacturer’s instructions. The pressure applied to control the medium flow and loading of the cells in the chamber was 2.5 psi. Time-lapse imaging was performed at 30°C using an epifluorescence microscope equipped with an oil objective and a camera. Frames were taken every 20 min. Images were deconvolved using standard deconvolution (enhanced ratio [aggressive]) with 10 iterations and a medium noise filtering. Bar, 5 µm.
Live cell imaging of Cdc5-Venus in a GAL-CLB2dB Δ strain.GAL-CLB2dB Δ CDC5-Venus cells (Ry1687) were grown at 23°C in synthetic complete medium with 2% sucrose and imaged in synthetic complete medium with 2% sucrose and 2% galactose to induce the expression of GAL-CLB2dB Δ. Cells were loaded in a microfluidic chamber (Y2 plates) according to the manufacturer’s instructions. The pressure applied to control the medium flow and loading of the cells in the chamber was 2.5 psi. Time-lapse imaging was performed at 30°C using an epifluorescence microscope equipped with an oil objective and a camera. Frames were taken every 20 min. Images were deconvolved using standard deconvolution (enhanced ratio [aggressive]) with 10 iterations and a medium noise filtering. Bar, 5 µm.
Acknowledgments
We are grateful to Angelika Amon, Ray Deshaies, and Yves Barral for generous gifts of strains and antibodies, to Fred Cross for communicating results before publication, and to Yves Barral for helping us to perform the live cell experiments in his laboratory. We thank Angelika Amon, Kathy Chen, Andrea Musacchio, and Simonetta Piatti for critical reading of the manuscript and Bela Novak and Angelika Amon for discussions.
This work was supported by an Armenise-Harvard foundation career development program grant and a grant from the Associazione Italiana Ricerca sul Cancro (AIRC; to R. Visintin), an AIRC “Enrico Ghezzi” fellowship (to F. Montani), and a National Institutes of Health grant (R01-GM079207 to A. Ciliberto).
References
Author notes
R. Manzoni and F. Montani contributed equally to this paper.