Skip Nav Destination
Close Modal
Update search
- Title
- Author
- Author Affiliations
- Full Text
- Abstract
- Keyword
- DOI
- ISBN
- EISBN
- ISSN
- EISSN
- Issue
- Volume
- References
- Title
- Author
- Author Affiliations
- Full Text
- Abstract
- Keyword
- DOI
- ISBN
- EISBN
- ISSN
- EISSN
- Issue
- Volume
- References
- Title
- Author
- Author Affiliations
- Full Text
- Abstract
- Keyword
- DOI
- ISBN
- EISBN
- ISSN
- EISSN
- Issue
- Volume
- References
- Title
- Author
- Author Affiliations
- Full Text
- Abstract
- Keyword
- DOI
- ISBN
- EISBN
- ISSN
- EISSN
- Issue
- Volume
- References
- Title
- Author
- Author Affiliations
- Full Text
- Abstract
- Keyword
- DOI
- ISBN
- EISBN
- ISSN
- EISSN
- Issue
- Volume
- References
- Title
- Author
- Author Affiliations
- Full Text
- Abstract
- Keyword
- DOI
- ISBN
- EISBN
- ISSN
- EISSN
- Issue
- Volume
- References
NARROW
Format
Subjects
Journal
Article Type
Date
1-20 of 133
Follow your search
Access your saved searches in your account
Would you like to receive an alert when new items match your search?
1
Sort by
Journal Articles
In Special Collection:
Cancer Cell Biology 2021
, Immune Cell Biology 2021
, The Year in Cell Biology: 2020
Isaac S. Chan, Hildur Knútsdóttir, Gayathri Ramakrishnan, Veena Padmanaban, Manisha Warrier, Juan Carlos Ramirez, Matthew Dunworth, Hao Zhang, Elizabeth M. Jaffee, Joel S. Bader, Andrew Josef Ewald
Journal:
Journal of Cell Biology
J Cell Biol (2020) 219 (9): e202001134.
Published: 09 July 2020
Includes: Supplementary data
Images
in Cancer cells educate natural killer cells to a metastasis-promoting cell state
> Journal of Cell Biology
Published: 09 July 2020
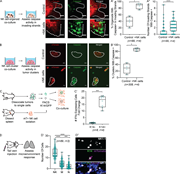
Figure 1. NK cells limit early stages of metastasis in ex vivo models of breast cancer. (A) Dot plot of GFP+ K14+ and K14− tumor cells stained for MHC class I expression. (B) Schema of hNK cell-tumor organoid coculture. Tumor organoids were More about this image found in NK cells limit early stages of metastasis in ex vivo models of breast cance...
Images
in Cancer cells educate natural killer cells to a metastasis-promoting cell state
> Journal of Cell Biology
Published: 09 July 2020
Figure S1. hNK cells limit invasion, growth, and colony formation in the C3(1)-Tag mouse model of breast cancer. (A) Representative DIC images of tumor organoids alone (top) or in coculture with hNK cells (bottom) at 0 and 24 h. Scale bar, 50 More about this image found in hNK cells limit invasion, growth, and colony formation in the C3(1)-Tag mou...
Images
in Cancer cells educate natural killer cells to a metastasis-promoting cell state
> Journal of Cell Biology
Published: 09 July 2020
Figure 2. hNK cells induce apoptosis in K14+ invasive breast cancer cells. (A) Representative confocal images of the invading strands of tumor organoids (mTomato+) and caspase activity (green) cultured alone (top) or in coculture with hNK cells More about this image found in hNK cells induce apoptosis in K14+ invasive breast cancer cells....
Images
in Cancer cells educate natural killer cells to a metastasis-promoting cell state
> Journal of Cell Biology
Published: 09 July 2020
Figure S2. hNK cells induce caspase activity in K14+ invasive cells, and hNK cell cytotoxicity can be increased by using a CD44 antibody specific to K14+ cells. (A and B) Representative confocal images of tumor organoids (A) and tumor clusters More about this image found in hNK cells induce caspase activity in K14+ invasive cells, and hN...
Images
in Cancer cells educate natural killer cells to a metastasis-promoting cell state
> Journal of Cell Biology
Published: 09 July 2020
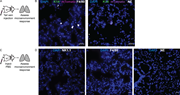
Figure S3. Quantification of macrophage and neutrophil response to early metastatic seeds in the lungs. (A) Schema for assessment of the innate immune response to an initial metastatic seed. Tumor clusters from the mammary tumors of More about this image found in Quantification of macrophage and neutrophil response to early metastatic se...
Images
in Cancer cells educate natural killer cells to a metastasis-promoting cell state
> Journal of Cell Biology
Published: 09 July 2020
Figure S4. Breast cancer organoids are able to overcome hNK cell cytotoxicity over time in 3D culture. (A and A′) Representative tumor organoids isolated from MMTV-PyMT (A) and C3(1)-Tag (A′) mice placed in 3D collagen I alone (top) or in More about this image found in Breast cancer organoids are able to overcome hNK cell cytotoxicity over tim...
Images
in Cancer cells educate natural killer cells to a metastasis-promoting cell state
> Journal of Cell Biology
Published: 09 July 2020
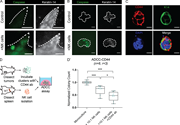
Figure 3. teNK cells promote colony formation. (A) Schema for teNK cell-tumor organoid coculture. (A′) Boxplot of tumor organoid invasion strands of tumor organoids cultured alone or in coculture with teNK cells. Error bars represent 5th to More about this image found in teNK cells promote colony formation. (A) Schema for teNK cell-tumor organ...
Images
in Cancer cells educate natural killer cells to a metastasis-promoting cell state
> Journal of Cell Biology
Published: 09 July 2020
Figure S5. RNA-seq analysis of hNK cells and teNK cells reveals differences in identity and biological processes. Receptor–ligand analysis of hNK cells and K14+ or K14− cells reveals interactions between NK cells and cancer cells. Treatment with More about this image found in RNA-seq analysis of hNK cells and teNK cells reveals differences in identit...
Images
in Cancer cells educate natural killer cells to a metastasis-promoting cell state
> Journal of Cell Biology
Published: 09 July 2020
Figure 4. The teNK cell phenotype can be reversed. (A) Heat map of z-scores of gene expression by hNK cells and teNK cells of genes related to NK cell inhibitory signaling. Hierarchical clustering was used to order the genes. (B) Relationship More about this image found in The teNK cell phenotype can be reversed. (A) Heat map of z-scores of gene...
Images
in Cancer cells educate natural killer cells to a metastasis-promoting cell state
> Journal of Cell Biology
Published: 09 July 2020
Figure 5. Pretreatment of teNK cells with FDA-approved DNMT inhibitors neutralizes the teNK cell phenotype. (A) Heat map of z-scores of gene expression by hNK cells and teNK cells of genes related to DNMTs. Hierarchical clustering was used to More about this image found in Pretreatment of teNK cells with FDA-approved DNMT inhibitors neutralizes th...
Journal Articles
Interferon-stimulated gene 15 accelerates replication fork progression inducing chromosomal breakage
In Special Collection:
The Year in Cell Biology: 2020
Maria Chiara Raso, Nikola Djoric, Franziska Walser, Sandra Hess, Fabian Marc Schmid, Sibylle Burger, Klaus-Peter Knobeloch, Lorenza Penengo
Journal:
Journal of Cell Biology
J Cell Biol (2020) 219 (8): e202002175.
Published: 29 June 2020
Includes: Supplementary data
Images
in Interferon-stimulated gene 15 accelerates replication fork progression inducing chromosomal breakage
> Journal of Cell Biology
Published: 29 June 2020
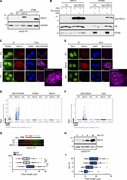
Figure 1. ISG15 localizes at the DNA replication forks and accelerates replication fork progression. (A) ISG15 immunoblot on protein extracts of U2OS FIT cells bearing EV or FLAG-ISG15, induced with doxycycline (dox; 1 µg/ml) for 48 h, and in More about this image found in ISG15 localizes at the DNA replication forks and accelerates replication fo...
Images
in Interferon-stimulated gene 15 accelerates replication fork progression inducing chromosomal breakage
> Journal of Cell Biology
Published: 29 June 2020
Figure S1. ISG15 localizes at the DNA replication forks and accelerates replication fork progression. (A) Schematic representation of the pipeline followed for the generation of CRISPR/Cas9-mediated ISG15 KO cell lines. (B) Analysis of ISG15 More about this image found in ISG15 localizes at the DNA replication forks and accelerates replication fo...
Images
in Interferon-stimulated gene 15 accelerates replication fork progression inducing chromosomal breakage
> Journal of Cell Biology
Published: 29 June 2020
Figure 2. ISG15 expression levels impact on replication fork progression in different systems. (A) Time course of ISG15 expression in U2OS treated with IFN-β (30 U/ml, 2 h) and chased for the indicated time points before lysis. (B) Analysis More about this image found in ISG15 expression levels impact on replication fork progres...
1