Homotypic membrane fusion catalyzed by the atlastin (ATL) GTPase sustains the branched endoplasmic reticulum (ER) network in metazoans. Our recent discovery that two of the three human ATL paralogs (ATL1/2) are C-terminally autoinhibited implied that relief of autoinhibition would be integral to the ATL fusion mechanism. An alternative hypothesis is that the third paralog ATL3 promotes constitutive ER fusion with relief of ATL1/2 autoinhibition used conditionally. However, published studies suggest ATL3 is a weak fusogen at best. Contrary to expectations, we demonstrate here that purified human ATL3 catalyzes efficient membrane fusion in vitro and is sufficient to sustain the ER network in triple knockout cells. Strikingly, ATL3 lacks any detectable C-terminal autoinhibition, like the invertebrate Drosophila ATL ortholog. Phylogenetic analysis of ATL C-termini indicates that C-terminal autoinhibition is a recent evolutionary innovation. We suggest that ATL3 is a constitutive ER fusion catalyst and that ATL1/2 autoinhibition likely evolved in vertebrates as a means of upregulating ER fusion activity on demand.
Introduction
The structure and continuity of the ER membrane network in metazoans are generated and maintained through iterative cycles of membrane tubule extension and homotypic membrane fusion that is seemingly constitutive (Friedman et al., 2010; Lee and Chen 1988; Waterman-Storer and Salmon, 1998; Westrate et al., 2015). The atlastin (ATL) GTPases mediate this fusion reaction and are critical for ER network maintenance (Hu and Rapoport 2016; McNew et al., 2013; Orso et al., 2009). The Drosophila ATL ortholog (DATL) purified from E. coli was shown over a decade ago to be sufficient to catalyze membrane fusion in vitro (Orso et al., 2009), with the absence of ATL causing disruptions to ER structure in both insect and mammalian cells (Hu et al., 2009; Hu et al., 2015; Morin-Leisk et al., 2011; Orso et al., 2009; Zhao et al., 2016). Notably, while Drosophila and invertebrates in general appear to have a single ATL, humans have three paralogs (ATL1-3) that are broadly important for human health, with mutations in ATL3 causing hereditary sensory neuropathy (HSN; Fischer et al., 2014; Kornak et al., 2014) and ATL1 mutations causing both hereditary spastic paraplegia (HSP; Blackstone 2012; Zhao et al., 2001) and HSN (Leonardis et al., 2012).
Demonstration that DATL is sufficient to drive in vitro membrane fusion not only established the ATL GTPases as ER fusion catalysts but enabled mechanistic understanding of the ATL fusion mechanism (Hu and Rapoport 2016; McNew et al., 2013). In brief, GTP-binding induced GTPase (G) domain dimerization in trans, accompanied by a crossover rotation of a 3-helical bundle (3HB) attached to the G domain through a short linker, leads to formation of a crossed-over dimer in which the 3HBs of ATLs on opposing membranes are closely aligned and contact the G domain of the other subunit (Bian et al., 2011; Byrnes et al., 2013; Byrnes and Sondermann, 2011). Crossover formation together with insertion of a juxta-membrane amphipathic helix (AH) into the bilayer triggers lipids to mix for fusion (Faust et al., 2015; Liu et al., 2012; Winsor et al. 2017). After fusion, GTP hydrolysis within the G domains drives disassembly of the crossover dimer and resets the fusion machinery for subsequent reactions (Crosby and Lee, 2022; Winsor et al., 2018).
Despite substantial advances using DATL, reconstitution of fusion activity by the human ATLs had proved refractory until recently, when our lab demonstrated that ATL1/2 expressed and purified from HEK cells, rather than E. coli, has robust fusion activity (Crosby et al., 2022). Unexpectedly, fusion reconstitution also revealed that each contains a C-terminal autoinhibitory domain located just downstream of the required AH (Crosby et al., 2022). For ATL2, the presence of the C-terminal extension inhibits fusion to the extent that the full-length purified protein lacks nearly all detectable activity, and removal of the extension increases the initial fusion rate by 500-fold. For ATL1, the presence of the C-terminal extension is inhibitory, but does not completely suppress activity, leaving a partially active fusion protein whose initial rate is further increased (by fivefold) upon removal of the C-terminus. Mapping of the C-terminal inhibitory elements in ATL1/2 revealed distinct requirements (Crosby et al., 2022), suggesting that the autoinhibitory mechanism could differ between paralogs.
The mechanisms of ATL1/2 autoinhibition remain to be worked out, but their existence implies a potential requirement for relief of autoinhibition during each iterative fusion reaction cycle. Arguably, the ER network in neuronal cells, which predominantly express ATL1 (Rismanchi et al., 2008), could be sustained by the partial activity of ATL1. This, however, would not be the case in nonneuronal cells, which typically express only low levels of ATL1 (Rismanchi et al., 2008). As the ATL2 C-terminus suppresses fusion activity almost entirely (Crosby et al., 2022), each fusion reaction cycle would presumably require relief of autoinhibition. On the other hand, it is also plausible that the third human ATL3 paralog, which is broadly expressed together with ATL2 across diverse cell and tissue types (Rismanchi et al., 2008), serves a basal, constitutive fusion function to sustain the ER network under normal circumstances and that ATL2 is activated on demand when cell physiology dictates a need to further increase fusion capacity.
Notably, ATL3 was not investigated in our earlier study, though the three human paralogs share a high degree of sequence and structural similarity within the protein core (Bian et al., 2011; Byrnes and Sondermann, 2011; Kelly et al., 2021; O’Donnell et al., 2017), and so ATL3 might have been expected to share the fusion function of ATL1/2. Consistent with this, early RNAi studies supported a requirement for depletion of both ATL2 and ATL3 to render an ER structural defect in cultured cells (Hu et al., 2009; Hu et al., 2015; Morin-Leisk et al., 2011). Further, overexpression of ATL3 was shown to increase the frequency of ER fusion events (Krols et al., 2018). However, a flurry of recent studies has raised doubts regarding ATL3’s fusion capabilities. First, there are reports of ATL2 but not ATL3 depletion being sufficient to perturb ER structure, implying a lesser role, if any, for ATL3 in ER structural maintenance (Chen et al., 2019; Pawar et al., 2017). Second, ATL3 was shown incapable of rescuing ER morphological defects in ATL2/3 RNAi knockdown cells (Hu et al., 2015). Another study reported successful rescue by both ATL1 and ATL3 in ATL1/2/3 triple KO cells, but only the ATL1 data were shown (Zhao et al., 2016). Further, there are conflicting reports on the ATL3 GTPase rate relative to ATL1/2, with similar maximal rates reported early on for the soluble domains of ATL1/2/3 (Hu et al., 2015), but a fourfold lower kcat for ATL3 than for ATL1 reported more recently (Kelly et al., 2021; O’Donnell et al., 2017). Lastly, it was recently reported that ATL3 purified from E. coli is indeed capable of catalyzing the fusion of liposomes with an ER-like lipid composition (Jang et al., 2023). However, Jang et al. (2023) propose a model wherein ATL3 on its own does not function as a fusion protein in cells, but rather, serves to support fusion driven by the autoinhibited ATL2 paralog. Taken together, the recent studies cast doubt on the fusion capability of ATL3.
To add to the uncertainty regarding its role, ATL3 has been implicated in diverse other processes. It has been identified as a possible ER-autophagy receptor that interacts with GABARAP proteins, with HSN disease mutations disrupting this interaction (Chen et al., 2019). Additionally, ATL2/3 depletion impairs protein targeting to the inner nuclear membrane (Pawar et al., 2017), protein export from the ER (Niu et al., 2019), as well as both selective (Liang et al., 2018) and nonselective autophagy (Liu et al., 2021), the latter through loss of ATL2/3 interactions with ULK1 and ATG13. Lastly, ATL3 has been linked to flavivirus replication, with demonstrations of interactions between ATL3 and both nonstructural and structural viral proteins (Monel et al., 2019; Neufeldt et al., 2019). Some of these diverse roles could stem from a primary role for ATL3 in ER structural maintenance, but they could also indicate one or more ATL3 functions outside of membrane fusion.
Here we investigate ATL3’s fusion capability both in vitro and in cells. We report that ATL3 is fully capable of lipid bilayer fusion in vitro and of sustaining ER network structure in ATL triple knockout cells. While ATL3’s G domain dimer affinity is measurably lower than that of ATL2, its kcat is comparable, and the lesser dimer affinity of ATL3 is offset by its higher relative abundance in cells. Importantly, and consistent with a constitutive fusion function, we find that ATL3 altogether lacks the C-terminal autoinhibition observed for ATL1/2. Finally, phylogenetic analysis indicates that ATL1/2 C-terminal autoinhibition is a recent vertebrate innovation, and this is supported by the fly protein DATL also lacking C-terminal autoinhibition. Together, this work more firmly establishes an ER membrane fusion function for ATL3 that, unlike ATL1/2, is most likely constitutive. It lays the foundation for future investigation of the interplay between the constitutive and regulated ATL paralogs in maintaining ER network structure in diverse tissues under divergent physiological conditions.
Results
ATL3 catalyzes membrane fusion
We purified His-tagged human ATL3 from a transiently transfected HEK293-derived suspension cell line (Fang et al., 2017) as previously described for ATL1/2 (Crosby et al., 2022). Purified protein (Fig. S1 A) was incorporated into synthetic liposomes (Fig. S1 B) and assessed for fusion activity via an established lipid mixing assay that monitors dequenching of fluorescently labeled lipids upon mixing and dilution with unlabeled lipids (Orso et al., 2009). ATL3 incorporated at a 1:1,000 M protein/lipid ratio catalyzed robust lipid mixing. The initial rate (0.02 s−1) mimicked the previously characterized full-length, partially active ATL1 at the same concentration, though the final extent of lipid mixing was higher (Fig. 1 A). Notably, both ATL3 and ATL1, when purified from HEK293-derived cells, are fusion competent even when the liposomes lack cholesterol and PE. This contrasts with the strict requirement for cholesterol and PE when the same proteins are purified from E. coli (Jang et al., 2023).
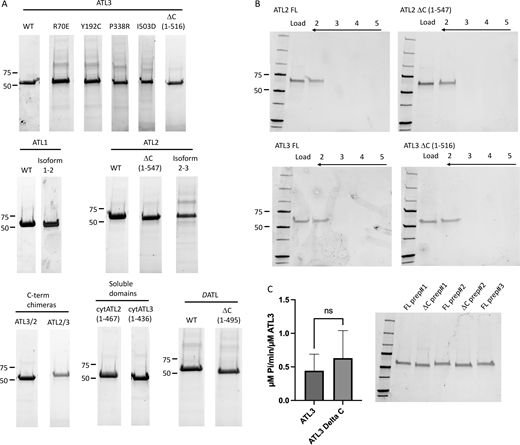
Proteins, flotation gradients, and GTPase assays. (A) Peak Ni+2-NTA fractions of all His-tagged ATL1, ATL2, ATL3, and DATL proteins and variants in this study are shown after resolving on SDS-PAGE. (B) Flotation of ATL2 FL, ATL2DC, ATL3 FL, and ATL3DC proteo-liposomes in a Nycodenz step gradient demonstrates similarly efficient incorporation for each protein. Fraction 5 is the bottom of each gradient, and fraction 2 is the 0/45% interface. For comparison, an equivalent fraction of the load is also shown. (C) GTPase assays of FL ATL3 and ATL3ΔC proteo-liposomes demonstrate similar GTPase rates (+/− SD) at the concentration typically used in the fusion assay (1:1,000 protein/lipid). The data represent 2–3 independent measurements from two to three independent proteo-liposome preps, with three technical replicates for each measurement. An unpaired t test reveals no statistically significant difference between the two sets of proteo-liposomes (P = 0.56). Source data are available for this figure: SourceData FS1.
Proteins, flotation gradients, and GTPase assays. (A) Peak Ni+2-NTA fractions of all His-tagged ATL1, ATL2, ATL3, and DATL proteins and variants in this study are shown after resolving on SDS-PAGE. (B) Flotation of ATL2 FL, ATL2DC, ATL3 FL, and ATL3DC proteo-liposomes in a Nycodenz step gradient demonstrates similarly efficient incorporation for each protein. Fraction 5 is the bottom of each gradient, and fraction 2 is the 0/45% interface. For comparison, an equivalent fraction of the load is also shown. (C) GTPase assays of FL ATL3 and ATL3ΔC proteo-liposomes demonstrate similar GTPase rates (+/− SD) at the concentration typically used in the fusion assay (1:1,000 protein/lipid). The data represent 2–3 independent measurements from two to three independent proteo-liposome preps, with three technical replicates for each measurement. An unpaired t test reveals no statistically significant difference between the two sets of proteo-liposomes (P = 0.56). Source data are available for this figure: SourceData FS1.
Human ATL3 catalyzes GTP-dependent membrane fusion in vitro. (A) Purified HA-6His-tagged ATL3 or ATL1 was reconstituted into donor and acceptor vesicles, and fusion was monitored as the dequenching of NBD-labeled lipid present in donor vesicles over time at 37°C after addition of GTP. (B and C) Inner leaflet mixing by ATL3 was monitored by quenching outer leaflet NBD with membrane-impermeable sodium dithionite at the times indicated prior to GTP addition (3 mM dithionite final concentration). The raw fluorescence trace is shown in B and the trace after normalization to the maximal signal in detergent in C. For reference, the data in C are plotted relative to a parallel lipid mixing reaction without dithionite pretreatment. (D) Fusion by the R70E, I503D, Y192C, and P388R variants of ATL3. All lipid mixing was initiated with 2 mM GTP (final concentration) at a 1:1,000 protein/lipid ratio and a 1:2 donor/acceptor vesicle ratio. The traces are the average of two technical replicates and representative of two biological replicates.
Human ATL3 catalyzes GTP-dependent membrane fusion in vitro. (A) Purified HA-6His-tagged ATL3 or ATL1 was reconstituted into donor and acceptor vesicles, and fusion was monitored as the dequenching of NBD-labeled lipid present in donor vesicles over time at 37°C after addition of GTP. (B and C) Inner leaflet mixing by ATL3 was monitored by quenching outer leaflet NBD with membrane-impermeable sodium dithionite at the times indicated prior to GTP addition (3 mM dithionite final concentration). The raw fluorescence trace is shown in B and the trace after normalization to the maximal signal in detergent in C. For reference, the data in C are plotted relative to a parallel lipid mixing reaction without dithionite pretreatment. (D) Fusion by the R70E, I503D, Y192C, and P388R variants of ATL3. All lipid mixing was initiated with 2 mM GTP (final concentration) at a 1:1,000 protein/lipid ratio and a 1:2 donor/acceptor vesicle ratio. The traces are the average of two technical replicates and representative of two biological replicates.
To confirm that ATL3 catalyzes fusion of both leaflets of the bilayer, we used the membrane impermeant compound sodium dithionite to selectively reduce outer leaflet NBD-labeled lipids to the nonfluorescent ABD (McIntyre and Sleight, 1991; Meers et al., 2000). Following two rounds of dithionite addition to ensure complete reduction of the outer leaflet, GTP addition induced inner leaflet lipid mixing (Fig. 1 B) with the expected delay compared to the total lipid mixing kinetic observed without dithionite (Fig. 1 C).
To assess whether the fusion activity observed for ATL3 is mechanistically similar to that of ATL1/2, we tested R70E, a mutation equivalent to R77E in ATL1, that blocks both GTP binding and trans G domain dimerization (Bian et al., 2011; Byrnes and Sondermann, 2011). This mutation abolished all lipid mixings (Fig. 1 D). Fusion by ATL3 was also disrupted by a I503D mutation, similar to the block by an equivalent I507D mutation in ATL1 (Crosby et al., 2022) and L482D in DATL (Liu et al., 2012), the latter shown to disrupt the nonpolar face of the AH thereby preventing the membrane insertion of the AH necessary for fusion (Faust et al., 2015; Liu et al., 2012). Additionally, the HSN disease mutations Y192C and P338R (Fischer et al., 2014; Kornak et al., 2014), the former being equivalent to the Y196C HSP disease mutation in ATL1 (Byrnes and Sondermann, 2011), and both shown to weaken the ATL3 soluble domain crossover dimer (Krols et al., 2018), were inhibitory (Fig. 1 D), in agreement with recent data on ATL3 Y192C purified from E. coli (Jang et al., 2023). These observations indicate that the three human ATL paralogs catalyze membrane fusion using a shared mechanism.
ATL3 is sufficient to restore ER network structure in ATL1/2/3 KO cells
From our in vitro reconstitution demonstrating the sufficiency of ATL3 in fusion catalysis, it seemed that ATL3 should, like ATL1/ATL2 (Hu et al., 2015), be capable of maintaining the tubular ER network in cells. As noted above, however, there have been conflicting results on whether endogenous ATL3 is sufficient to maintain ER structure in the absence of ATL1/2 (Chen et al., 2019; Hu et al., 2015; Pawar et al., 2017). Similarly, studies differ on whether exogenous expression of ATL3 alone can maintain the ER network (Hu et al., 2015; Zhao et al., 2016). To address this discrepancy, we used the well-characterized E5 NIH-3T3 cell line where all three ATLs have been knocked out by CRISPR (Zhao et al., 2016) to ask if exogenous ATL3 expression alone could restore and maintain the tubular ER network.
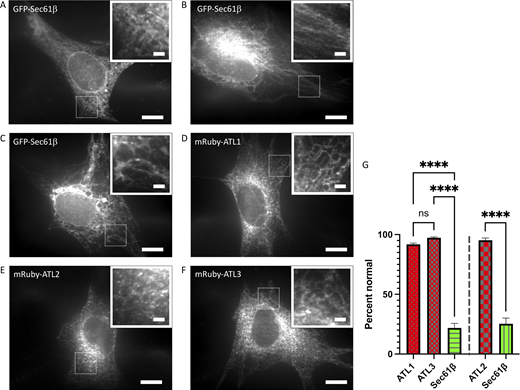
As anticipated, whereas the parental 3T3 cells expressing the ER marker protein GFP-Sec61β showed a normal branched ER network morphology (Fig. 2 A), 3T3 E5 triple ATL knockout cells expressing GFP-Sec61β only infrequently displayed normal ER morphology (Zhao et al., 2016), with the ER typically exhibiting a stringy/unbranched morphology (Fig. 2, B and C, quantified in G). In contrast, either mRuby-ATL1 (Fig. 2 D), mRuby-ATL2 (Fig. 2 E), or mRuby-ATL3 (Fig. 2 F) expression rescued these defects (Fig. 2 G). Notably, ATL3 restored a branched ER network morphology in ∼97% of expressing cells. Similarly, ATL1 and ATL2 restored ER morphology in ∼92% and ∼94% of expressing cells, respectively. There was no significant difference in rescue between ATL3 and ATL1. Thus, when introduced exogenously into ATL1/2/3 KO cells as the sole source of ATL, ATL3 can restore and maintain a normal ER network.
Human ATL3 can maintain the ER network in ATL1/2/3 KO cells. (A–F) NIH 3T3 parental (A) or triple ATL1/2/3 KO E5 cells (B–F) were transfected with GFP-Sec61β (A–C), mRuby-ATL1 (D), mRuby-ATL2 (E), or mRuby-ATL3 (F). 48 h later, cells were fixed and visualized by confocal microscopy; scale bar, 10 µm. Also shown are magnified insets of the boxed area within each image; scale bar, 2 µm. (G) The percent cells expressing the indicated proteins that had a normal branched ER network morphology ± SD, n = 3 independent experiments, an average of 44 cells per experiment. Quantification for analysis of ATL1 and ATL3 with respect to Sec61β (left) was blinded, and quantification of analysis of ATL2 with respect to Sec61β (right) was done separately without blinding. ns, P > 0.05; ****, P < 0.0001 as determined by one-way ANOVA analysis (ATL1/ATL3/Sec61β) or unpaired t test (ATL2/Sec61β). Source data are available for this figure: SourceData F2.
Human ATL3 can maintain the ER network in ATL1/2/3 KO cells. (A–F) NIH 3T3 parental (A) or triple ATL1/2/3 KO E5 cells (B–F) were transfected with GFP-Sec61β (A–C), mRuby-ATL1 (D), mRuby-ATL2 (E), or mRuby-ATL3 (F). 48 h later, cells were fixed and visualized by confocal microscopy; scale bar, 10 µm. Also shown are magnified insets of the boxed area within each image; scale bar, 2 µm. (G) The percent cells expressing the indicated proteins that had a normal branched ER network morphology ± SD, n = 3 independent experiments, an average of 44 cells per experiment. Quantification for analysis of ATL1 and ATL3 with respect to Sec61β (left) was blinded, and quantification of analysis of ATL2 with respect to Sec61β (right) was done separately without blinding. ns, P > 0.05; ****, P < 0.0001 as determined by one-way ANOVA analysis (ATL1/ATL3/Sec61β) or unpaired t test (ATL2/Sec61β). Source data are available for this figure: SourceData F2.
ATL3 lacks autoinhibition by the C-terminus
The recently discovered C-terminal autoinhibitory domain in ATL1/2 is located just downstream of the required and well-conserved AH (Crosby et al., 2022). For canonical ATL2 (ATL2-1), inhibition maps to two short discrete regions within a broad stretch of nearly 40 amino acids (548–583) that, unless removed, blocks in vitro fusion altogether. For canonical ATL1 (ATL1-1), an analogous element mapping to a single short stretch within the C-terminus (525–528) suppresses fusion activity, albeit only partially (as seen in the ATL1 trace in Fig. 1 A). Interestingly, the AH and the C-terminal extensions are separated by an intron, and alternative splicing of ATL1 and ATL2 generates multiple isoforms of each that differ solely in the C-terminal extension (Crosby et al., 2022; see below). In contrast to ATL1/2, there appear to be no documented C-terminal variants of ATL3. To test for autoinhibition in ATL3, residues beyond the AH (Fig. 3 A) were deleted to generate an ATL3ΔC construct (1–516). This truncation was at a position equivalent to that previously used to generate disinhibited and maximally active ATL2ΔC (1–547) and ATL1ΔC (1–520; Crosby et al., 2022), and was incorporated into liposomes as efficiently as full-length ATL3 (Fig. S1 B). Strikingly, ATL3 truncation had no effect on the fusion rate (Fig. 3 B) and no significant effect on GTPase activity when measured under these fusion conditions (Fig. S1 C). This lack of effect of C-terminal deletion on the ATL3 fusion rate contrasted sharply with the substantial stimulation seen when the C-terminus of ATL1 is deleted (Fig. 3 C; Crosby et al., 2022). Further, ATL3ΔC retained the ability to catalyze mixing of both inner and outer leaflets of the bilayer (Fig. 3 D).
Human ATL3 lacks C-terminal autoinhibition. (A) Alignment of ATL1/2/3 C-terminal extensions with a black bar indicating the residues deleted to produce the respective ΔC constructs. ATL1/2 C-terminal sequences are from the canonical splice isoforms ATL1-1 and ATL2-1. (B) Lipid mixing by full-length (FL) ATL3 or ATL3 (1-516) lacking its C-terminus (ΔC). (C) Comparison of the effect of C-terminal deletion on ATL3 fusion kinetics (replotted from B) to that of C-terminal deletion on ATL1 fusion kinetics. (D) Inner leaflet mixing kinetics by ATL3ΔC after dithionite treatment compared to total lipid mixing of the same without dithionite. (E) Lipid mixing by full-length ATL2 or an ATL2 chimera with the ATL3 C-terminus. (F) Lipid mixing by full-length ATL3 or an ATL3 chimera with the ATL2 C-terminus. (G) Lipid mixing by ATL2 in donor vesicles (lab) and ATL3 in acceptor vesicles (unlab) and vice versa. Lipid mixing by ATL2 only and ATL3 only are shown for reference. All traces are the average of two technical replicates and representative of two biological replicates.
Human ATL3 lacks C-terminal autoinhibition. (A) Alignment of ATL1/2/3 C-terminal extensions with a black bar indicating the residues deleted to produce the respective ΔC constructs. ATL1/2 C-terminal sequences are from the canonical splice isoforms ATL1-1 and ATL2-1. (B) Lipid mixing by full-length (FL) ATL3 or ATL3 (1-516) lacking its C-terminus (ΔC). (C) Comparison of the effect of C-terminal deletion on ATL3 fusion kinetics (replotted from B) to that of C-terminal deletion on ATL1 fusion kinetics. (D) Inner leaflet mixing kinetics by ATL3ΔC after dithionite treatment compared to total lipid mixing of the same without dithionite. (E) Lipid mixing by full-length ATL2 or an ATL2 chimera with the ATL3 C-terminus. (F) Lipid mixing by full-length ATL3 or an ATL3 chimera with the ATL2 C-terminus. (G) Lipid mixing by ATL2 in donor vesicles (lab) and ATL3 in acceptor vesicles (unlab) and vice versa. Lipid mixing by ATL2 only and ATL3 only are shown for reference. All traces are the average of two technical replicates and representative of two biological replicates.
To further demonstrate that the ATL3 C-terminal extension lacks an autoinhibitory activity, an ATL2/3 chimera was constructed. As anticipated, replacing the C-terminal extension of ATL2 with that of ATL3 resulted in an ATL2/3 chimera that largely lacks autoinhibition (Fig. 3 E). The inability of the ATL3 C-term to autoinhibit ATL2 was unlikely simply due to incompatibility between paralog domains because the reciprocal chimera consisting of ATL2’s C-terminal extension on ATL3 had strongly reduced fusion activity compared to full-length ATL3 (Fig. 3 F). On the other hand, the ATL3/2 chimera (Fig. S2) restored the ER network to a similar extent as either ATL3 (Fig. 2 F, quantified in G) or ATL2 (Fig. 2 E, quantified in G) when introduced into E5 cells, indicating that the reduced activity of the purified ATL3/2 chimera was unlikely due to protein misfolding. Presumably, the autoinhibition observed for purified ATL3/2 is relieved in the context of E5 cells just as observed for full-length ATL2. Interestingly, expression of the ATL2/3 chimera, which mimics the ATL2 protein lacking C-terminal autoinhibition in our fusion assay (Fig. 3 E), led largely to the “collapsed ER” morphology (Fig. S2) previously observed for ATL2 variants that lack autoinhibition (Crosby et al., 2022). Together, these data indicate that ATL3 is unique among the canonical human ATL paralogs in its lack of a C-terminal autoinhibitory domain.
ATL3_2 and ATL2_3 chimera are functional in ATL1/2/3 KO E5 cells. (A–F) National Institutes of Health 3T3 triple ATL1/2/3 KO E5 cells were transfected with GFP-Sec61β (A), mRuby-ATL3_2 (B), HA-ATL3_2 (C), or HA-ATL2_3 (D–F). 48 h later, cells were fixed and visualized directly by confocal microscopy (A and B) or visualized after immunostaining with anti-HA antibodies (C–F); scale bar, 10 µm. (G) The percent cells expressing the indicated proteins that had a normal branched ER network or collapsed ER morphology ± SD, n = 3 independent experiments, an average of 20 cells per experiment. Ruby-ATL3_2 and Sec61β (left) was performed separately from the HA-ATL3_2 and HA-ATL2_3 (right) experiments and analyzed separately. All analysis was performed without blinding. ****, P < 0.0001; **, P = 0.0030 as determined by unpaired t tests (Ruby-ATL3_2/Sec61β; HA-ATL3_2/HA-ATL2_3) comparing percent cells with normal ER morphology. Source data are available for this figure: SourceData FS2.
ATL3_2 and ATL2_3 chimera are functional in ATL1/2/3 KO E5 cells. (A–F) National Institutes of Health 3T3 triple ATL1/2/3 KO E5 cells were transfected with GFP-Sec61β (A), mRuby-ATL3_2 (B), HA-ATL3_2 (C), or HA-ATL2_3 (D–F). 48 h later, cells were fixed and visualized directly by confocal microscopy (A and B) or visualized after immunostaining with anti-HA antibodies (C–F); scale bar, 10 µm. (G) The percent cells expressing the indicated proteins that had a normal branched ER network or collapsed ER morphology ± SD, n = 3 independent experiments, an average of 20 cells per experiment. Ruby-ATL3_2 and Sec61β (left) was performed separately from the HA-ATL3_2 and HA-ATL2_3 (right) experiments and analyzed separately. All analysis was performed without blinding. ****, P < 0.0001; **, P = 0.0030 as determined by unpaired t tests (Ruby-ATL3_2/Sec61β; HA-ATL3_2/HA-ATL2_3) comparing percent cells with normal ER morphology. Source data are available for this figure: SourceData FS2.
ATL3 does not synergize with full-length autoinhibited ATL2
Fusion catalysis by full-length ATL3 was robust, as indicated by its final extent of fusion being near the maximal extent of fusion possible under our assay conditions at this protein concentration (Crosby et al., 2022). However, its initial fusion rate (0.02 s−1) was lower than the disinhibited and maximally active ATL1ΔC and ATL2ΔC (0.1–0.3 s−1) reported previously at the same concentrations (Crosby et al., 2022). Instead, the ATL3 initial fusion rate resembled the partially inhibited full-length ATL1 as noted above (Fig. 1 A). Importantly, ATL3 is expressed together with ATL2 in many cell and tissue types, and the two proteins co-precipitate, at least under overexpression conditions (Rismanchi et al., 2008). Additionally, recent work showed that ATL2 fusion activity is enhanced in cis by a sub-fusogenic concentration of ATL3 (Jang et al., 2023). Therefore, we wondered if ATL3 and full-length, autoinhibited ATL2 might complement and mutually activate one another in trans. To test for this, a lipid mixing assay was performed where either the donor or acceptor proteo-liposomes contained full-length autoinhibited ATL2, while the other contained full-length ATL3. Notably, both proteins were incorporated at the same 1:1,000 protein/lipid ratio. In either case, only a very low fusion rate (0.004 s−1) was observed (Fig. 3 G), demonstrating that although the two proteins are capable of heterodimerizing in trans and may complement one another in cis (Jang et al., 2023), they do not complement one another in trans.
ATL3 fusion is accelerated at higher concentrations
Since exogenously expressed ATL3 was clearly sufficient to sustain the ER network (Fig. 2 G), we wondered whether the lower initial fusion rate of ATL3 in vitro might be countered simply by increasing its concentration. When ATL3 was incorporated at either two- or approximately threefold higher protein/lipid ratios of 1:500 or 1:300, respectively, it catalyzed fusion at dramatically higher initial rates (Fig. 4 A). Doubling the ATL3 concentration was sufficient to increase its initial fusion rate by fivefold (Fig. 4 A, compare 1:1,000 to 1:500). When a similar titration was performed using the disinhibited form of ATL2ΔC (1–547), a trend emerged: When present at approximately twice the molar concentration, ATL3 catalyzed fusion with a similar rate to that of ATL2ΔC (Fig. 4 B). Notably, all proteins (ATL3, ATL2ΔC, ATL3ΔC, and ATL2) incorporated into liposomes with similar efficiency (Fig. S1 B).
ATL3 has a lower G domain dimer affinity than ATL2 but is more abundant in cells. (A) Lipid mixing by ATL3 at varying protein/lipid ratios. (B) Lipid mixing by ATL2ΔC at varying protein/lipid ratios. ATL3 lipid mixing traces (dotted lines) are replotted from (A) for ease of comparison. (C and D) Steady state GTPase assays on purified truncated soluble domain cyt ATL3 (C) and cyt ATL2 (D) as a function of ATL concentration measured at saturating 500 µM GTP. The data were fit to a dimerization equation (red lines) to extract the indicated KD and kobs values. Measurements were averaged from technical triplicates across two biological replicates. (E and F) ATL3 is more abundant than ATL2 in cells. (E) Equal amounts of total lysate prepared from the indicated cell lines were probed with antibodies against ATL3, ATL2 or calnexin. The relative abundance of ATL3 and ATL2 (ng) were determined using a standard curve generated with HA-6His-tagged purified versions of each protein. (F) The ratio of ATL3/ATL2 in each cell line. Quantifications are the average of two independent experiments. Source data are available for this figure: SourceData F4.
ATL3 has a lower G domain dimer affinity than ATL2 but is more abundant in cells. (A) Lipid mixing by ATL3 at varying protein/lipid ratios. (B) Lipid mixing by ATL2ΔC at varying protein/lipid ratios. ATL3 lipid mixing traces (dotted lines) are replotted from (A) for ease of comparison. (C and D) Steady state GTPase assays on purified truncated soluble domain cyt ATL3 (C) and cyt ATL2 (D) as a function of ATL concentration measured at saturating 500 µM GTP. The data were fit to a dimerization equation (red lines) to extract the indicated KD and kobs values. Measurements were averaged from technical triplicates across two biological replicates. (E and F) ATL3 is more abundant than ATL2 in cells. (E) Equal amounts of total lysate prepared from the indicated cell lines were probed with antibodies against ATL3, ATL2 or calnexin. The relative abundance of ATL3 and ATL2 (ng) were determined using a standard curve generated with HA-6His-tagged purified versions of each protein. (F) The ratio of ATL3/ATL2 in each cell line. Quantifications are the average of two independent experiments. Source data are available for this figure: SourceData F4.
The above trend suggested that the lower ATL3 fusion rate at any given protein/lipid ratio might be explained by a correspondingly lower G domain dimer affinity. To test this, we carried out steady-state GTPase assays on the truncated soluble domains of ATL3 and ATL2 as a function of ATL concentration (Fig. 4, C, and D). As anticipated, a fit of the observed data to a dimerization equation revealed dimerization constants of ∼2.1 µM for ATL3 (Fig. 4 C) and ∼0.9 µM for ATL2 (Fig. 4 D), respectively, indicating an approximately twofold lower G domain dimer affinity for ATL3 compared to ATL2. Thus, the lower fusion rate for ATL3 relative to ATL2ΔC observed under our standard assay conditions is readily explained by its G domain dimer affinity. Notably, at the 1:1,000 protein/lipid ratio typically used in our fusion assay, the protein concentration is 0.6 µM, in which case ATL2ΔC present near its estimated KD, whereas ATL3 is significantly below it. Lastly, the fit of the data revealed an intrinsic GTP hydrolysis rate for ATL3 (kobs ∼4.3 µM/min) that was at least as high, if not higher, than ATL2 (kobs ∼3.4 µM/min).
To gain insight into how ATL3’s lower dimer affinity might impact its effectiveness as an ER fusion catalyst in cells, we measured the relative abundance of ATL3/ATL2 in commonly used cultured cell lines using antibodies specific to each paralog (Fig. 4 E). Potential differences in antibody binding efficiency were accounted for by simultaneously probing a series of known quantities of purified His-tagged full-length ATL2 and ATL3 to generate a standard curve for each antibody. While there was some variability in the levels of ATL3 across cell types, particularly in HEK cells, which also express some ATL1 (Rismanchi et al., 2008), quantification of the immunoblots revealed the abundance of ATL3 to be several-fold higher than ATL2 in all cell lines assessed (Fig. 4 F). Thus, the lower G domain dimer affinity of ATL3 relative to ATL2 appears to be sufficiently offset by its consistently higher relative abundance in cells.
Invertebrate DATL also lacks C-terminal autoinhibition
The above findings established a role for ATL3 in mammalian ER structural maintenance. However, the complete lack of C-terminal autoinhibition in ATL3 was somewhat surprising and led us to wonder the extent to which autoinhibition is conserved across metazoans. Previously, it was observed that removal of residues C-terminal to the AH in the single Drosophila ortholog DATL did not dramatically alter fusion activity (Moss et al., 2011). However, fusion catalyzed by the truncated DATL was also noticeably slower than the full length. To clarify the role of the C-terminus in DATL, we compared the fusion kinetic of full-length DATL to that truncated at the same position used to generate ATL3ΔC (Fig. 5 A). Moreover, the DATL proteins were expressed and purified in a manner identical to that used for the human proteins (Crosby et al., 2022). Strikingly, the initial fusion rate of DATLΔC (1-495) was indistinguishable from the full length (Fig. 5 B), clearly indicating that DATL, like ATL3, altogether lacks C-terminal autoinhibition.
Model for emergence of autoinhibition based on the lack of autoinhibition in ATL3/DATL and the ATL core gene tree. (A) Alignment of ATL3 and DATL C-terminal extensions with a black bar indicating the residues deleted from DATL to produce the respective ΔC construct. (B) Lipid mixing by full-length (FL) DATL or DATLΔC (1-495). Lipid mixing for both was performed at a 1:1,000 protein/lipid ratio and the traces are the average of two technical replicates. (C) ATL gene tree constructed using maximum likelihood estimation from amino acid sequences comprising the G, 3HB, TM, and AH domains showing that vertebrate ATLs arose through two duplications in rapid succession prior to the divergence of bony vertebrates and cartilaginous fish. Longer branches of the ATL3 clade (red) indicates rapid sequence divergence in this paralog relative to ATL1 (green) and ATL2 (blue). Branches with support values >70% shown in bold and branch lengths in substitutions per site. (D) Evolutionary hypotheses, as described in the text, for the distribution of C-terminal autoinhibitory function in present-day ATLs. Green + indicates gain of autoinhibition, while red x indicates loss of autoinhibition.
Model for emergence of autoinhibition based on the lack of autoinhibition in ATL3/DATL and the ATL core gene tree. (A) Alignment of ATL3 and DATL C-terminal extensions with a black bar indicating the residues deleted from DATL to produce the respective ΔC construct. (B) Lipid mixing by full-length (FL) DATL or DATLΔC (1-495). Lipid mixing for both was performed at a 1:1,000 protein/lipid ratio and the traces are the average of two technical replicates. (C) ATL gene tree constructed using maximum likelihood estimation from amino acid sequences comprising the G, 3HB, TM, and AH domains showing that vertebrate ATLs arose through two duplications in rapid succession prior to the divergence of bony vertebrates and cartilaginous fish. Longer branches of the ATL3 clade (red) indicates rapid sequence divergence in this paralog relative to ATL1 (green) and ATL2 (blue). Branches with support values >70% shown in bold and branch lengths in substitutions per site. (D) Evolutionary hypotheses, as described in the text, for the distribution of C-terminal autoinhibitory function in present-day ATLs. Green + indicates gain of autoinhibition, while red x indicates loss of autoinhibition.
The ATL gene tree indicates ATL1 and ATL2 autoinhibition could have arisen independently
The absence of autoinhibition in both ATL3 and the single Drosophila ortholog suggested constitutive, rather than regulated fusion activity, to be the default mode. This suggested that the C-terminal autoinhibition seen in ATL1/2 (Crosby et al., 2022) might represent a vertebrate specialization rather than a core-conserved feature of the ATL fusion machinery. Deciphering the evolutionary history of C-terminal autoinhibition would shed light on this hypothesis but would require an understanding of the phylogeny of the ATLs and their C-termini. Notably, ATL phylogeny had been reported previously, but there was some disagreement, with one analysis showing a closer relationship between ATL1 and ATL2 than either to ATL3 (Rismanchi et al., 2008); and a more recent analysis showing a closer relationship between ATL2 and ATL3 (Neufeldt et al., 2019). Additionally, no prior analysis had explicitly addressed the evolution of the C-terminal extensions. Therefore, we undertook an independent analysis herein.
We assembled a set of 18 diverse vertebrate species with high-quality genomes (Table S1 and Fig. S3 A). Blast searches recovered ATL1 and ATL2 homologs from all 18 genomes. Curiously, we found ATL3 to be absent from birds and cartilaginous fish. Next, we searched for ATL homologs in a set of 15 insect species comparable in taxonomic breadth to the vertebrate data set (Fig. S3 A) and found that all insect genomes harbor a single ATL-related gene, which we termed ATLi. In all, we retrieved a total of 63 genes that fall into four ATL subfamilies: ATLi, ATL1, ATL2, and ATL3 (Table S2). This data set was used to reconstruct the ATL phylogeny based on the conserved core region comprising of the G, 3HB, TM, and AH domains. The resulting gene tree (Figs. 5 C and S4 A) indicates that ATL1/2/3 arose through two duplications at the base of the vertebrate lineage, with ATL1 branching first and a subsequent duplication leading to ATL2 and ATL3, in line with the more recently reported phylogeny (Neufeldt et al., 2019). Significantly, ATL3, which altogether lacks autoinhibition, and ATL2, the most strongly autoinhibited paralog (Crosby et al., 2022), are sister taxa, while ATL1 and ATL2, the two paralogs that exhibit autoinhibition, are not (Figs. 5 C and S4 A); thus, the presence or absence of autoinhibition does not appear to correlate with the overall evolutionary relationship between paralogs. Based on the gene tree, the presence and absence of autoinhibition in the canonical ATL paralogs could be most parsimoniously explained by three distinct models (Fig. 5 D): (model 1) ancestral autoinhibition, followed by independent losses of autoinhibition in ATL3 and in ATLi; (model 2) lack of autoinhibition in the ancestor with recent independent gains of autoinhibition in ATL1 and ATL2; or (model 3) lack of ancestral autoinhibition, gain of autoinhibition in the vertebrate ancestor, and a later loss in ATL3.
Phylogeny of selected species and ATL C-terminus alignment. (A) Phylogeny for the 18 vertebrate and 15 insect genera represented in our data set downloaded from Timetree (http://timetree.org/). Branch lengths represent time in millions of years. (B) Multiple alignment of the sequences encoded by the final exons in ATL2-2, ATL1-1, ATL2-1, and ATLi. Sequence features (black boxes) previously shown to be required for autoinhibition (Crosby et al., 2022) in ATL1-1 and ATL2-1 are conserved throughout the vertebrates. Note that the ATL1-1 feature differs in composition and does not overlap with the features in ATL2-1, indicating likely independent origin of autoinhibition in these paralogs.
Phylogeny of selected species and ATL C-terminus alignment. (A) Phylogeny for the 18 vertebrate and 15 insect genera represented in our data set downloaded from Timetree (http://timetree.org/). Branch lengths represent time in millions of years. (B) Multiple alignment of the sequences encoded by the final exons in ATL2-2, ATL1-1, ATL2-1, and ATLi. Sequence features (black boxes) previously shown to be required for autoinhibition (Crosby et al., 2022) in ATL1-1 and ATL2-1 are conserved throughout the vertebrates. Note that the ATL1-1 feature differs in composition and does not overlap with the features in ATL2-1, indicating likely independent origin of autoinhibition in these paralogs.
ATLi/1/2 C-termini are the descendants of the C-terminus of a common ATL ancestor
To distinguish among the above models and to further probe the evolutionary history of ATL autoinhibition, we asked whether the C-termini in the various ATL subfamilies share common ancestry. As vertebrate ATLs undergo C-terminal alternative splicing, which we previously showed to be functionally significant for ATL2 variants (Crosby et al., 2022), we first examined the exon structure of all ATLs in our data set (Fig. 6 A and Table S3). This analysis revealed the exon structure of the C-termini to be highly conserved within, but not across, paralogs. For instance, all ATL3 homologs in our data set harbor a single C-terminal exon, whereas ATL1 homologs generally harbor two C-terminal exons: a short 5 amino acid (AA) encoding exon followed by a longer 30–40 AA exon. On the other hand, most ATL2 homologs harbor three C-terminal exons: a short 5 AA exon followed by two longer 30–40 AA exons, with only one of the longer exons being expressed in any single ATL2 variant. Finally, like ATL3, the majority of ATLi homologs in our data set harbor a single C-terminal exon. Interestingly, in vertebrates, the paralogs that encode C-terminal autoinhibition also have more complex exon structure within the C-terminus, allowing for a diversity of splice variants, with there being at least two distinct expressed C-terminal splice variants of ATL1 (ATL1-1, 1-2) and three distinct C-terminal variants of ATL2 (ATL2-1, 2-2, 2-3; Fig. 6 A). It should be noted that alternative splicing of N-terminal exons further increases the diversity of ATL1/2 paralogs, but the N-termini were not studied herein as we previously showed that deletion of the N-terminal extension of ATL1/2 did not affect fusion activity (Crosby et al., 2022).
C-terminal exon structure of ATL1/2/3/i and evolutionary relationships between C-terminal exons with and without autoinhibitory function. (A) Schematic of C-terminal exon structure of ATL1/2/3/i. The lengths of exons are to scale and colored dark green when the exon contains autoinhibitory function and light gray when the exon does not. The exons comprising the C-terminus of each splice variant are also indicated to the right. (B and C) Multisequence alignment (B) and phylogeny of the C-terminus (C) reveal the evolutionary relationships between the final exons of ATL2-2 (exon 2B), ATL1-1 (exon 1B), ATL2-1 (exon 2C), and ATLi (exon iA) isoforms. Exons 1B and 2B are most closely related and share marked sequence similarity, while exon 2C shares sequence features with the insect C-terminus. Sequence features previously shown to be required for autoinhibition (Crosby et al., 2022) in ATL1-1 and ATL2-1 (black boxes) differ in amino acid composition and are positioned in different columns, suggesting that those features do no share common ancestry. In B, branches with support values of >70% are shown in bold and branch lengths in substitutions per site.
C-terminal exon structure of ATL1/2/3/i and evolutionary relationships between C-terminal exons with and without autoinhibitory function. (A) Schematic of C-terminal exon structure of ATL1/2/3/i. The lengths of exons are to scale and colored dark green when the exon contains autoinhibitory function and light gray when the exon does not. The exons comprising the C-terminus of each splice variant are also indicated to the right. (B and C) Multisequence alignment (B) and phylogeny of the C-terminus (C) reveal the evolutionary relationships between the final exons of ATL2-2 (exon 2B), ATL1-1 (exon 1B), ATL2-1 (exon 2C), and ATLi (exon iA) isoforms. Exons 1B and 2B are most closely related and share marked sequence similarity, while exon 2C shares sequence features with the insect C-terminus. Sequence features previously shown to be required for autoinhibition (Crosby et al., 2022) in ATL1-1 and ATL2-1 (black boxes) differ in amino acid composition and are positioned in different columns, suggesting that those features do no share common ancestry. In B, branches with support values of >70% are shown in bold and branch lengths in substitutions per site.
The conservation of exon structure within each ATL subfamily, as well as the similarity in C-terminal exon lengths across the ATL1/2/i subfamilies, is suggestive of common ancestry. To further assess this, we constructed a multiple sequence alignment of the amino acid sequences encoded by the final exons in ATLi, ATL1, ATL2-1, and ATL2-2 (Fig. 6 B and Fig. S3 B). From the alignment, the similarity of the vertebrate and invertebrate C-terminal sequences is readily apparent, supporting the hypothesis that the C-terminus of the ancestral ATL was the progenitor of the C-termini of all present-day ATL1/2/i. We also observed that the amino acid sequences of the short exons in ATL1 and ATL2 C-termini bear little similarity (GSTNE versus RSPRK) and are likely independent acquisitions. Strikingly, both major C-terminal exons of ATL2 (Fig. 6 A, exons 2B and 2C) are easily aligned with ATL1/i and with each other, suggesting that the two adjacent exons are paralogous. To investigate how this doubling of exons could have occurred, we reconstructed the phylogeny of the ATL C-termini (Fig. 6 C and Fig. S4 B) and applied Notung-DM software (Stolzer et al., 2015) to the resulting tree to infer the most parsimonious history of events in the C-terminal region (Fig. S5). Interestingly, although the two major exons in ATL2 are adjacent, they are not closest relatives in the C-terminal gene tree (Fig. 6 C), suggesting that the C-terminal expansion is not the product of a recent tandem duplication as their spatial arrangement would suggest. Rather, formal comparison of the C-terminal tree and the core gene tree with Notung-DM (Fig. S5) indicates that a copy of the ATL1 C-terminal exon was inserted into the C-terminus of an ancestral ATL2 gene, giving rise to the first of the major C-terminal exons (Fig. 6 A, exon 2B) in present day ATL2 homologs. In stark contrast to the ATL1/2 C-termini, the ATL3 C-terminus is encoded by a single exon, is notably shorter (20–29 AA) and cannot be unambiguously aligned with them. This is likely due to sequence divergence and streamlining of a C-terminus that lacks autoinhibition activity.
C-terminus phylogeny reconstructed using Neighbor Joining from the multiple sequence alignment shown in Fig. S3 . The final exon in ATL2-2 (exon 2B) is more closely related to the final exon in ATL1 (exon 1B) than it is to the exon adjacent to it (exon 2C). Branches with support values >70% shown in bold and branch lengths in substitutions per site.
C-terminus phylogeny reconstructed using Neighbor Joining from the multiple sequence alignment shown in Fig. S3 . The final exon in ATL2-2 (exon 2B) is more closely related to the final exon in ATL1 (exon 1B) than it is to the exon adjacent to it (exon 2C). Branches with support values >70% shown in bold and branch lengths in substitutions per site.
Phylogenetic reconstruction reveals the evolutionary history of the ATLs and their C-termini. (A) Phylogenetic reconciliation of the ATL core tree in Fig. 5 C with the species tree in Fig. S2 using Notung (Stolzer et al., 2015) shows that ATL1/2/3 arose through two duplications after the separation of insects and vertebrates and prior to the divergence of cartilaginous fish and bony vertebrates. ATL3 was subsequently lost in birds and cartilaginous fish. Tree reconstructed from ATL core sequence from a sample of 8 insect and 10 vertebrate species. (B) Phylogenetic reconciliation of the C-terminal tree (Fig. 6 C) with the reconciled core tree (A) using Notung-DM (Stolzer et al., 2015) reveals the most parsimonious history of gains and losses that explain the branching order of the C-terminal tree. The gene duplication that gave rise to ATL1 and the ancestor of ATL2/3 indicated by blue box in red border. Subsequently, a copy of the final exon of ATL1 was inserted into the C-terminus of ATL2 (purple arrow), giving rise to a more complex ATL2 C-terminus that harbors two adjacent major exons.
Phylogenetic reconstruction reveals the evolutionary history of the ATLs and their C-termini. (A) Phylogenetic reconciliation of the ATL core tree in Fig. 5 C with the species tree in Fig. S2 using Notung (Stolzer et al., 2015) shows that ATL1/2/3 arose through two duplications after the separation of insects and vertebrates and prior to the divergence of cartilaginous fish and bony vertebrates. ATL3 was subsequently lost in birds and cartilaginous fish. Tree reconstructed from ATL core sequence from a sample of 8 insect and 10 vertebrate species. (B) Phylogenetic reconciliation of the C-terminal tree (Fig. 6 C) with the reconciled core tree (A) using Notung-DM (Stolzer et al., 2015) reveals the most parsimonious history of gains and losses that explain the branching order of the C-terminal tree. The gene duplication that gave rise to ATL1 and the ancestor of ATL2/3 indicated by blue box in red border. Subsequently, a copy of the final exon of ATL1 was inserted into the C-terminus of ATL2 (purple arrow), giving rise to a more complex ATL2 C-terminus that harbors two adjacent major exons.
Sequence features associated with autoinhibition arose independently in ATL1 and ATL2
Finally, to gain insight into the possible evolutionary history of C-terminal autoinhibition, we considered how sequence features previously shown to confer autoinhibition in ATL1 and ATL2 (Crosby et al., 2022) are positioned in the alignment (Fig. 6 B and Fig. S3 B). As anticipated, within each subfamily, these features are conserved from mammals to cartilaginous fish. For instance, the ME(E/D)N and LT(E/D)Q sequences, which mediate autoinhibition in ATL2-1, are present in all ATL2-1 homologs examined. Similarly, the (Y/F; K/R)LY associated with ATL1 autoinhibition is also present in all ATL1 homologs. But strikingly, the sequence features in ATL1 and ATL2-1 differ in both composition and location within the alignment (Fig. 6 B and Fig. S3 B). Thus, the C-terminal extension shares ancestry across vertebrates and insects, but the sequence features that mediate autoinhibition most likely arose after the divergence of vertebrate ATL paralogs. Taken together, these analyses strongly favor the second of the three models proposed above (Fig. 5 D). That is, ATL1/2 C-terminal autoinhibition arose independently in the vertebrate lineage; therefore, it is unlikely to represent a core conserved feature of the ATL fusion mechanism.
Short 5 AA exons in ATL1/2 further diversify the extent of C-terminal autoinhibition
From our analysis of C-terminal exon structure above, we noticed that the 5 AA exon encoding the residues GSTNE is perfectly conserved in all ATL1-1 orthologs examined but that the ATL1-2 variant lacks it (Fig. 7 A). Similarly, the 5 AA exon encoding the residues RSPRK is present in ATL2-3 but not ATL2-2 (Fig. 7 A), the latter which we previously determined to confer only very weak autoinhibition as compared to ATL2-1 (Crosby et al., 2022). To test the functional significance of inclusion/exclusion of these short exons, we directly compared variants with or without these additions. Intriguingly, C-terminal autoinhibition in ATL1-2 was weakened by absence of the GSTNE motif found in ATL1-1 (Fig. 7 B); and similarly, autoinhibition in ATL2-3 was further weakened by the presence of the RSPRK motif (Fig. 7 C). Thus, C-terminal autoinhibition may be additionally fine-tuned by the inclusion or exclusion of an additional short exon, supporting our hypothesis that this form of regulation is a vertebrate innovation that serves to provide fine control of ER fusion activity. In stark contrast, the lack of complexity in the C-terminus of either ATL3 or ATLi is consistent with a role for these homologs in basal and constitutive ER fusion.
Short 5 AA acids in ATL1/2 further modulate autoinhibition by the C-terminus. (A) Depiction of additional five AAs encoded by the short exons (boxed in orange) and their positions relative to the autoinhibitory elements (yellow boxes) in the splice variants ATL1-1 and ATL2-3. (B) Lipid mixing by the splice variant ATL1-2 that differs from the canonical ATL1-1 solely by the exclusion of the short exon encoding GSTNE. For reference, the maximally disinhibited ATL1ΔC is also shown. (C) Lipid mixing by the splice variant ATL2-3 that differs from ATL2-2 solely by the inclusion of the exon encoding RSPRK. For reference, the canonical ATL2-1 and the maximally disinhibited ATL2ΔC are also shown. The ATL2-2 trace is replotted from Crosby et al. (2022). All lipid mixing was performed at a 1:1,000 protein/lipid ratio, and the traces are the average of two technical replicates.
Short 5 AA acids in ATL1/2 further modulate autoinhibition by the C-terminus. (A) Depiction of additional five AAs encoded by the short exons (boxed in orange) and their positions relative to the autoinhibitory elements (yellow boxes) in the splice variants ATL1-1 and ATL2-3. (B) Lipid mixing by the splice variant ATL1-2 that differs from the canonical ATL1-1 solely by the exclusion of the short exon encoding GSTNE. For reference, the maximally disinhibited ATL1ΔC is also shown. (C) Lipid mixing by the splice variant ATL2-3 that differs from ATL2-2 solely by the inclusion of the exon encoding RSPRK. For reference, the canonical ATL2-1 and the maximally disinhibited ATL2ΔC are also shown. The ATL2-2 trace is replotted from Crosby et al. (2022). All lipid mixing was performed at a 1:1,000 protein/lipid ratio, and the traces are the average of two technical replicates.
Discussion
Our findings herein support a model in which C-terminal autoinhibition of human ATL1/2 fusion activity is not a core conserved feature of the ATL mechanism but more likely a regulatory adaptation. First, we find that the previously understudied human ATL paralog ATL3 is a robust fusion catalyst that unlike ATL1/2, altogether lacks autoinhibition. Second, an in-depth phylogenetic analysis reveals that C-terminal autoinhibition in ATL1 and ATL2 are recent innovations that likely arose independently during the diversification of vertebrates.
The precise mechanism of ATL2 (and ATL1) autoinhibition has yet to be uncovered and therefore how and under what conditions ATL1/2 undergoes relief of autoinhibition remains unknown. Nevertheless, we can speculate that there may be conditions, such as during cell differentiation or during ER stress, when there is a need for rapid ER network expansion (Brewer and Jackowski, 2012; Schuck et al., 2009). In those situations, the existing pool of active ATL3 may be insufficient, particularly if its concentration drops below its dimer affinity within an expanding ER. A reserve pool of autoinhibited ATL2, particularly with its higher dimer affinity, may then provide a means of quickly and reversibly increasing ER fusion capacity on demand. We further speculate that this reserve pool of ATL2 may also be called to action in cells that experience loss of ATL3 due to experimental manipulation, either through CRISPR deletion or RNAi knock down. Such a scenario would help to explain why ATL3 depletion by itself is insufficient to render ER network morphological defects. Finally, if relief of autoinhibition were mediated by a post-translational modification, then ATL2 could be activated quickly and transiently and subsequently returned to its autoinhibited state once the need for increased ER fusion capacity has passed. In support of this possibility, ATL2 has been observed to undergo extensive post-translational modifications (Dephoure et al., 2008; Guo et al., 2014; Wagner et al., 2011).
The lack of autoinhibition in ATL3 and a corresponding lack of autoinhibition in the single Drosophila ortholog leads us to speculate that having at least one constitutive form of ATL might be beneficial to cells and to organisms. Notable exceptions though are that ATL3 may be expressed at only low levels in the brain in mammals (Rismanchi et al., 2008), and it was not found in birds and cartilaginous fish (Table S2). On the other hand, we previously showed that a splice variant of ATL2 (ATL2-2), whose C-terminus is only weakly autoinhibitory, is expressed specifically in the brain (Crosby et al., 2022). Though the precise pattern of ATL2-2 expression within the brain remains to be determined, it is tempting to speculate that a lack of constitutively active ATL3 may be compensated in a tissue specific manner by a more active splice variant of ATL1/2. In the case of birds and cartilaginous fish, more widespread expression of a constitutively active ATL1/2 splice variant would be needed to offset the apparent lack of ATL3 in the genome.
It should be noted that the lack of C-terminal autoinhibition in ATL3 does not rule out the possibility of ATL3 regulation by other means. Indeed, ATL3 interacting proteins (Chen et al., 2019; Liu et al., 2021) and ATL3 post-translational modifications have been reported (Choudhary et al., 2009; Deeb et al., 2014; Kelly et al., 2021; Zhou et al., 2013), though these have yet to be explored for possible effects on fusion activity. However, if ATL3 regulation were to occur, it may not involve the C-terminus, as the ATL3 C-terminus is substantially shorter and less well conserved across vertebrate homologs as compared to the C-termini of ATL1/2.
Finally, we note that our data on ATL3 disagrees with some but not all previous work. Our enzyme kinetic analysis reveals similar intrinsic GTPase rates for ATL3 and ATL2 soluble domains when assessed at protein concentrations at which G domain dimerization is not rate-limiting. However, due to the lower dimer affinity of ATL3 relative to that of ATL2, the GTPase rates diverge when compared at lower protein concentrations. This is largely consistent with an earlier study in which the GTP hydrolysis rates of the three human paralogs were found to differ at lower ATL concentrations (0.2 µM), with ATL3 having the lowest rate, but converging to a similar rate at higher enzyme concentrations (2.5 µM; Hu et al., 2015). In contrast, a more recent study reported the soluble domains of ATL3 and ATL1 to have similar G domain dimer affinities but markedly different catalytic rates, with the kcat of ATL3 being fourfold lower than that of ATL1 (Kelly et al., 2021). The basis for the discrepancy is unclear and may at least in part be attributed to differences in proteins purified from E. coli. However, another recent report also observed no difference in GTPase activity between full-length human ATLs, though the protein concentrations used in that study were not reported (Jang et al., 2023).
Like our assessment of the catalytic capability of ATL3 provided herein, our observation that ATL3 is sufficient to sustain normal ER network morphology in cells is consistent with some, but not all, previously published work. Early on, we and others showed that RNAi depletion of ATL2 is insufficient to cause an observable network defect in HeLa and COS7 cells (Hu et al., 2009; Hu et al., 2015; Morin-Leisk et al., 2011). More recently however, ATL2 RNAi alone in HeLa (Pawar et al., 2017) and ATL2 KO alone in COS7 (Chen et al., 2019) was reported to cause an observable ER morphological defect, implying that ATL3 is insufficient to maintain the ER network. Similarly, exogenous ATL3 was shown insufficient to restore normal network morphology to COS7 cells depleted of ATL2/3 by RNAi (Hu et al., 2015). While there is no clear unifying explanation for the discrepancies, it is clear from our results herein that ATL3 is capable, at least under certain cell type and culture conditions, to sustain a morphologically normal ER. In cases where ATL3 was observed as insufficient to rescue defects, one possibility is that exogenous expression produced an ATL3 concentration lower than its KD. Future investigation of the precise interplay between ATL paralogs and their splice variants under diverse physiological conditions may shed light on some of the experimental discrepancies.
Materials and methods
Reagents and constructs
All ATL constructs used for protein purification were in the pGW1-CMV vector (gifted in the form of HA-ATL2 by C. Blackstone, Massachusetts General Hospital, Boston, MA, USA) and had a 6-His tag inserted between the HA tag and the ATL N-term. All mutations were made with the standard QuikChange protocol using PfuTurbo polymerase (Agilent Technologies), and all mutagenic oligos were from IDT. All constructs were fully sequenced (GENEWIZ) before use. HA-6His-ATL3 was made by amplifying ATL3 (gifted by J. McNew, Rice University, Houston, TX, USA) with forward primer 5′-GCTCTAGAATGTTGTCCCCTCAGCGAGTGGC-3′ and reverse primer 5′-GCCAATTGTCATTGAGCTTTTTTATCCATGGATGGTCTTCC-3′ and cloning the fragment after digestion with XbaI/MfeI into HA-His-ATL1/pGW1-CMV cut with XbaI and MfeI. The XbaI site follows the HA-6His tags and precedes the ATL1 coding sequence. The ATL2/3 chimera ([1-536] ATL2/[506-541] ATL3) was made by amplifying the C-term of ATL3 (506-541) with forward primer 5′-GCGTGATCAGGGTGCCGCATATGTGTTG-3′ and reverse primer 5′-CGCTGATCACGCGAGCTCGGTACGGATCCAGACATG-3′ and cloning the fragment after digestion with BclI into HA-6His-ATL2/pGW1-CMV cut at a BclI site internal to ATL2 and in the vector. The ATL3/2 chimera (ATL3 [1-502]/[534-583] ATL2) was made by amplifying the N-term of ATL3 (1–502) with forward primer 5′-GGAAGACTTAAGGCAGCGGCAGAAGAAGATGC-3′ and reverse primer 5′-GCAATCTGATCAATAGCTCCGCCCAGCTCACG-3′ and cloning the fragment after digestion with AflII/BclI into HA-6His-ATL2/pGW1-CMV cut at an AflII site in the vector and BclI internal to ATL2 (after removing the BclI site in pGW1-CMV). The ATL2-3 splice variant was constructed using gBlocks Gene Fragments (IDT) and cloning into ATL2-1/pGW1-CMV cut with BclI. mRuby-ATL3 was made by amplifying ATL3 with forward primer 5′-TAAGCAGGATCCGGTGGAAGTGGTGGAATGTTGTCCCCTCAGCGAGTG-3′ and reverse primer 5′-TGCGCATCTAGATCATTGAGCTTTTTTATCCATGG-3′ and cloning the fragment after digestion with BamHI and XbaI into an mRuby2/pcDNA3 plasmid cut with the same. This created an mRuby-ATL3 construct with a flexible linker (GGSGG) between mRuby2 and ATL3. mRuby-ATL1, mRuby-ATL2, and mRuby-ATL3/2 chimera (ATL3 [1-502]/[534-583] ATL2) cloning used the same procedure. GFP-Sec61β was from E. Snapp (Albert Einstein College of Medicine, New York, NY, USA). All restriction enzymes were from New England Biolabs. GTP (Sigma-Aldrich) was reconstituted in a 100 mM stock in TE buffer (10 mM Tris, pH 8.0, and 1 mM EDTA), aliquoted into single-use aliquots and stored at −80°C. All lipids were purchased from Avanti Polar Lipids.
Protein expression and purification
HA-6His-ATL/pGW1-CMV plasmids were transiently transfected into 200 ml of Expi293 cells cultured at 37°C/8% CO according to the manufacturer’s protocol (Thermo Fisher Scientific). The optimum time of harvest after transfection varied from 30 to 54 h depending on the construct. Cells were harvested by centrifugation (5,000 × g, 15 min), washed once with 15 ml of cold PBS, centrifuged again (5,000 × g, 10 min) and then the pellet flash frozen. Proteins were purified with chilled buffers on ice or at 4°C. Frozen pellets were thawed on ice and resuspended in 20 ml of Lysis 0% buffer (50 mM Tris, pH 8.0, 5 mM MgCl2, 300 mM NaCl, 10 mM imidazole, 10% glycerol, 0.5 mM PMSF, 1 µg/ml pepstatin, 1 µM leupeptin, and 2 mM 2-mercaptoethanol [2-ME]). 5 ml of Lysis 10% buffer (50 mM Tris, pH 8.0, 5 mM MgCl2, 300 mM NaCl, 10 mM imidazole, 10% glycerol, 10% Triton X-100, 0.5 mM PMSF, 1 µg/ml pepstatin, 1 µM leupeptin, and 2 mM 2-mercaptoethanol [2-ME]) was then added and mixed. The suspension was diluted 1:1 with addition of 25 ml of Lysis 0% buffer and rotated for 30 min at 4°C. Samples were then centrifuged at 12,000 rpm for 30 min at 4°C (F20-12 × 50 LEX rotor; Thermo Fisher Scientific). The supernatant was transferred to pre-chilled tubes and centrifuged at 50,000 rpm for 1 h (Ti70 rotor; Beckman Coulter). The final supernatant was filtered through 0.45-µm filters (Millex) and mixed with 0.25 ml Ni+2-NTA agarose (Qiagen) overnight at 4°C for ∼20 h. Ni+2-NTA beads were then poured into a column support and washed with 30 ml of Wash 1 (50 mM Tris, pH 8.0, 5 mM MgCl2, 100 mM NaCl, 20 mM imidazole, 10% glycerol, 1% Triton X-100, and 2 mM 2-ME), followed by 30 ml of Wash 2 (50 mM Tris, pH 8.0, 5 mM MgCl2, 100 mM NaCl, 20 mM imidazole, 10% glycerol, 0.1% Anapoe X-100, and 2 mM 2-ME). Protein was eluted into 0.25 ml fractions using Elution buffer (50 mM Tris, pH 8.0, 5 mM MgCl2, 100 mM NaCl, 250 mM imidazole, 10% glycerol, 0.1% Anapoe X-100, and 2 mM 2-ME). Fractions were resolved by SDS-PAGE to verify purity, flash-frozen, and stored at −80°C. The soluble forms of ATL2 (1-467) and ATL3 (1-436) were purified as described above; however, Wash 1 contained 0.1% Triton X-100 instead of 1%. Wash 2 and Elution both had no detergent.
Liposome preparation
Lipids in chloroform were dried by rotary evaporation and then resuspended in A100 buffer (25 mM HEPES, pH 7.4, 100 mM KCl, 10% glycerol, 1 mM EDTA, 2 mM 2-ME) for a final lipid concentration of 10 mM. Lipids were then subjected to 11 freeze-thaw cycles in liquid nitrogen and a 45°C water bath. Liposomes were formed by extruding using the LipoFast LF-50 extruder (Avestin) 15 times through 100-nm polycarbonate filters. Liposome size was verified using dynamic light scattering (Zen3600; Malvern Panalytical). Unlabeled liposomes were 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (PC) and 1,2-dioleoyl-sn-glycero-3-phospho-l-serine (PS) at an 85:15 M ratio and labeled liposomes were PC:PS:1,3-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2-1,3-benzoxadiazol-4-yl; NBD):1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) at an 82:15:1.5:1.5 M ratio.
Protein incorporation and lipid-mixing assay
Purified protein was incorporated into unlabeled and labeled liposomes at a detergent/lipid ratio of ∼0.7. The liposomes and proteins were incubated together at 4°C for 1 h while rotating, and then subsequently four 1-h detergent removal incubations were performed at 4°C for 1 h while rotating. For detergent removal samples were incubated with SM-2 Bio-Beads (Bio-Rad) at 1 g beads per 70 mg Anapoe X-100. Incorporations were then desalted over a 2.4 ml Sephadex G-25 column into A100 after which the peak fractions (1 ml total) were pooled. Samples were stored on ice and used that day, or flash-frozen and stored at −80°C. All incorporations were performed at a 1:1,000 protein/lipid molar ratio unless otherwise specified. In instances where the protein/lipid molar ratio was modified, the lipid concentration was held constant, and therefore only the protein concentration modified.
Fusion activity was assessed using an established lipid mixing assay (Orso et al., 2009). Assays were performed with 0.6 mM total lipids at a 1:2 labeled/unlabeled ratio. For all assays, proteoliposomes were mixed with A100 buffer containing 5 mM MgCl2, and then pre-incubated at 37°C for 5 min in a 96-well plate. Following pre-incubation, the plate was placed in a Spark plate reader (Tecan), two baseline readings were taken, and then 2 mM GTP or A100 buffer of equal volume was added with a multichannel pipette. For 60 min NBD dequenching was monitored at 10 s intervals (excitation at 460 nm, dequenching at 538 nm). Maximum possible dequenching was determined after 60 min via the addition of 0.5% Anapoe X-100, with two more cycles read on the plate reader after Anapoe addition. Fmax was calculated by taking the average of these two post-Anapoe cycles. Fusion calculations were performed using the function . In instances where initial kinetics were extremely rapid and the first reading post-GTP addition was not representative of time zero, the average of two readings taken immediately before GTP addition was used to calculate initial fluorescence observed. This difference in calculation of initial fluorescence was used for ATL3 at 1:300 and ATL2 1-547 at 1:500.
To assess inner leaflet lipid mixing, the assay was performed as described above, however after placement into the plate reader 2 mM sodium dithionite (final concentration) was added via multichannel pipette and fluorescence subsequently monitored, until a flat baseline was reached (∼10 min). Following the observation of the baseline, 1 mM more of sodium dithionite was added and fluorescence monitored (∼5 min) to ensure that the outer NBD was fully reduced. In total this reduction took ∼15 min and was performed at 37°C. Following the second dequenching, GTP was added, and the assay was performed as described above.
GTPase assay
GTPase activity was measured using the EnzCheck Phosphate Assay Kit (Molecular Probes; Thermo Fisher Scientific). Purified soluble proteins HA-6His-ATL2 (1-467) and HA-6His-ATL3 (1-436) were desalted into SEC buffer (25 mM Tris, pH 7.5, 100 mM NaCl, 5 mM MgCl2, 2 mM EGTA, 1 mM imidazole), and addition of the desalted proteins at the specified final concentrations were used to start the reactions. GTPase assays of membrane-inserted proteins were performed as described above; however, proteoliposomes incorporated at 1:1,000 and desalted into A100 were used to start reactions at a final concentration of 0.6 µM protein. Standard reactions were prepared by mixing 1 U/mol purine nucleoside phosphorylase (PNP), 0.2 mM 2-amino-6-mercapto-7-methylpurine riboside (MESG), and 500 mM GTP in a total volume of 200 µl and absorbance at 360 nm monitored at 37°C in a Spark plate reader (Tecan). A phosphate standard curve was used to convert the increase in absorbance to phosphate produced over time, after which the rate of phosphate production per ATL at the specified ATL concentrations was calculated. Fitting of the data to a simple dimerization equation and extraction of KD and kcat was performed using Microsoft Excel.
Immunofluorescence
NIH-3T3 E5 cells (Zhao et al., 2016; gifted from C. Blackstone) were cultured in high glucose DMEM + 10% FBS + 1% penicillin-streptomycin. The parent NIH-3T3 cell line (Zhao et al., 2016; gifted from C. Blackstone) was cultured as described above, however DMEM was used instead of high glucose DMEM. For transfection cells were cultured on 12-mm glass coverslips in a 24-well plate with antibiotic-free media. E5 cells were transfected with Ruby-ATL3 (200 ng), Ruby-ATL2 (100 ng), Ruby-ATL1 (200 ng), Ruby-ATL3/2 (200 ng), HA-His-ATL3/2 (200 ng), HA-His-ATL2/3 (200 ng) or GFP-Sec61β (200 ng) and WT 3T3 cells were transfected with GFP-Sec61β (200 ng) using 1.5 µl of Lipofectamine 3000 (Thermo Fisher Scientific) following manufacturer’s instructions. 1 d after transfection media was replaced with antibiotic containing media, and 2 d after transfection cells were fixed in 3% paraformaldehyde (PFA) and mounted for viewing. For HA transfections, cells were fixed as described above and then stained with mouse anti-HA antibody (ab18181; 1:500; Abcam) in blocking solution (PBS containing 0.2 M glycine, 2.5% calf serum, and 0.1% Triton X-100), followed by Alexa Fluor 568-conjugated goat anti-mouse (1:400; Thermo Fisher Scientific). Images (Fig. 2, A–F and Fig. S2, A–F) were obtained using a spinning disk confocal scanhead (Yokagawa; PerkinElmer) mounted on an Axiovert 200 microscope (Zeiss) with a 100× 1.4-NA objective (Zeiss) and acquired with a 12-bit CCD camera (Hamamatsu Photonics). Maximal projections of 2-4 sections at 0.2 µm spacing were acquired with Micromanager open-source software (University of California, San Francisco).
ER rescue analysis
For quantification of ER morphological rescue, PFA-fixed cells expressing similarly moderate levels of the specified constructs were imaged using an Axioplan microscope (Zeiss) with a 40× 1.4-NA objective (Zeiss) equipped with a 12-bit CCD camera (Hamamatsu) and acquired with HCImage software (Hamamatsu). Imaged cells for the comparison of mRuby-ATL3, mRuby-ATL1 and GFP-Sec61β were randomized and blinded prior to analysis. The comparisons of mRuby-ATL2 to GFP-Sec61β, mRuby-ATL3/2 to GFP-Sec61β, and HA-His-ATL3/2 to HA-His-ATL2/3, were carried out separately and not blinded. Qualitative analysis was performed to categorize cells as displaying normal or abnormal ER morphology. For analysis of HA-His-ATL2/3 a third phenotype of a collapsed ER reminiscent of the ATL2 delta C phenotype previously observed (Crosby et al., 2022) was also included in the categorization. Each construct was assessed in n = 3 independent experiments, with a mean of 44 cells (range of 22–70) per experiment for the experiments shown in Fig. 2, and a mean of 20 cells per experiment (range 12–28) for the experiments shown in Fig. S2. In instances where cells displayed ambiguous morphological phenotypes, occurring in <10% of imaged cells, they were not scored. The percentage of cells displaying normal morphology (%normal) was calculated for all conditions, as well as the percentage of cells displaying the collapsed ER phenotype for the HA-His-ATL2/3 condition.
Statistical analysis
For the ER rescue analysis in Fig. 2 G, an ordinary one-way ANOVA with multiple comparisons was performed in Prism 8 (GraphPad) to analyze % cells with normal ER across conditions for the ATL1/ATL3/Sec61β set of experiments. For the analysis of ATL2/Sec61β in Fig. 2 G, as well as for the analysis of Ruby-ATL3-2/Sec61β and HA-His-ATL3-2/HA-His-ATL2/3 in Fig. S2 G, an unpaired two-tailed Student’s t test was performed in Prism 8. Data distribution for all rescue analyses were assumed to be normal but were not formally tested. For the comparison of GTPase activities of FL ATL3 and ATL3ΔC in Fig. S1 C, an unpaired two-tailed Student’s t test was performed in Prism 8. Data distribution was assumed to be normal but was not formally tested.
Immunoblot analysis for ATL2/3 quantification
Total lysates from NIH 3T3, COS-7, HeLa, HEK 293, and U2OS cells were prepared from near confluent 10 cm dishes. After washing in PBS, cells were harvested in 0.5 ml RSB, boiled, and passed through a 25-gauge needle before resolving by SDS-PAGE. All SDS-PAGE loadings contained 0.1 ml of total lysate and identical samples were used for ATL2, ATL3, and calnexin immunoblots. Immunoblots were performed using a standard protocol. The following antibodies were used: rabbit anti-ATL2 (16688-1-AP; 1:1,500; Proteintech), rabbit anti-ATL3 (ab117819; 1:5,000; Abcam), rabbit anti-Calnexin (ab22595; 1:1,000; Abcam), goat anti-rabbit-HRP (1706515; 1:2,500; Biorad). Standard curves for quantifying ATL2 and ATL3 were generated using known amounts of purified full-length HA-6His-ATL2 and full-length HA-6His-ATL3. Signal detection used ECL (Pierce; Thermo Fisher Scientific), images acquired on a Biorad ChemiDoc imager, and quantified in ImageJ (National Institutes of Health).
Liposome floatation
Proteoliposomes incorporated at 1:1,000 were floated via (80/45/0%) Nycodenz (Axis-Shield, Dundee, Scotland) step gradient in A100 without glycerol. Gradients were centrifuged at 40,000 rpm overnight for ∼17 h (SW50.1 rotor; Beckman Coulter). Gradient fractions were then resolved via SDS-PAGE.
Species and genome selection
We compiled representative samples of vertebrate species with high quality genomes that are broadly distributed taxonomically to minimize long branch attraction artifacts. For comparison, a set of insect species representing major taxonomic lineages within the insecta was selected to be roughly comparable to the vertebrate set with respect to the number of species, evolutionary time span, and genome quality. Species divergence times were estimated using TimeTree (Kumar et al., 2017). Genome quality was assessed using the BUSCO (Manni et al., 2021), assembly status, and N50 statistics reported for National Center for Biotechnology Information refseq genomes. This procedure resulted in a set of 18 vertebrate and 15 insect genomes (Table S1).
Sequence retrieval
Atlastin-like sequences were retrieved from these 33 genomes (Table S1), using the tblastn search function embedded in the NCBI genome browser to permit inspection of the splice variants retrieved. For each genome, three tblastn searches were carried out with default parameters using the three human paralogs (ATL1, NP_056999.2; ATL2, NP_001129145.1; and ATL3, XP_047282681.1) as query sequences. The resulting fasta files were concatenated and redundant sequences were removed. In vertebrates, the retrieved sequences were identified as ATL1, ATL2, or ATL3 homologs based on the human paralog that provided the best match. This procedure retrieved 3 ATL-like sequences in all vertebrate genomes searched, except for the bird and cartilaginous fish genomes, which lacked an ATL3-like sequence. Additional tblastn searches against all Aves (resp. Chondrichthyes) in the NCBI nr database found no ATL3-like sequences, consistent with independent losses in the bird and cartilaginous fish ancestors. In 13 insect genomes, tblastn searches retrieved a single ATL-like sequence. Two ATL-like sequences, each, were recovered from the silkworm and tsetse fly genomes. In both genomes, one of the two sequences retrieved displayed the length and domain composition characteristic of insect ATL sequences and was retained. The other sequence, in both cases, deviated in length by more than 100 residues and notably lacked the typical ATL transmembrane region and amphipathic helix. These were deemed to be domain-based matches, not ATL homologs, and were excluded from further analysis. This procedure resulted in a set of 63 ATL homologs (ATL1: 18; ATL2: 18; ATL3: 12; ATLi: 15) in 33 species (Table S2), which formed the basis for the reconstruction of the ATL core phylogeny and the analysis of the C-terminal region.
Atlastin core phylogeny
We compiled a set of isoforms, one per homolog, that cover the full extent of the ATL core, comprising the G, 3HB, TM, and AH domains as annotated by InterPro (Blum et al., 2021). Following alignment with MUSCLE (Edgar, 2004) from the JalView dashboard (Waterhouse et al., 2009; Troshin et al., 2011) the core sequence set was obtained by removing N-terminal sequences upstream from the initial (D/Q)HF motif in the G domain and C-terminal sequences downstream from the final W(E/D; Q/N) motif in the AH domain. Prior to phylogenetic analysis, columns with more than 50% gaps were removed from the alignment.
The Atlastin core phylogeny was reconstructed using maximum likelihood estimation (MLE) implemented in IQ-TREE 2 (Minh et al., 2020) with default settings, including built-in automated model selection by ModelFinder (Kalyaanamoorthy et al., 2017). Branch support values were estimated using UFBoot2 (Hoang et al., 2018). The resulting tree was rooted using mid-point rooting in Figtree (http://tree.bio.ed.ac.uk/software/Figtree/), which produces the same result as outgroup rooting with the insect clade (Fig. S4 A).
Phylogenetic analysis of the ATL C-terminus
For this analysis, only the C-termini of ATL1-2, ATL2-1, ATL2-2, and ATLi were considered, where the C-terminus was defined to be the sequence downstream of the conserved W(E/D; Q/N) motif that terminates the AH domain. The ATL3 sequences were eliminated from consideration because an unambiguous alignment to other ATL C-termini could not be determined. The two 5 AA sequences encoded by the short exons at the beginning of the ATL1 and ATL2 C-termini, respectively, were also eliminated from consideration. The ATL1 5-mer and ATL2 5-mer have no amino acids in common; within each group, the sequences are too conserved to be phylogenetically informative. The resulting set of 67 C-terminal sequences was aligned with MUSCLE (Edgar, 2004) from the JalView dashboard (Troshin et al., 2011; Waterhouse et al., 2009). Following alignment, 7 sequences that were 100% identical to another sequence in the data set were removed, resulting in a final set of 60 C-terminal sequences.
A C-terminus phylogeny was constructed from this alignment using the BioNJ (Gascuel, 1997) implementation of Neighbor Joining (NJ) in the Seaview (v. 5) package (Gouy et al., 2021), with “Observed” distances and 100 bootstrap replicates. NJ was used because the alignment was deemed too short to be appropriate for MLE. Prior to analysis with Seaview, columns with more than 50% gaps were removed from the alignment. Within Seaview, the “Ignore all gap sites” feature was suppressed. The resulting unrooted tree was rooted using the insect clade as the outgroup.
ATL2-1 and ATL2-2 do not group together in the resulting C-terminus tree. This branching pattern suggests a history of exon gain and/or losses. To infer this history, we compared the C-terminus tree with the core ATL tree and the species phylogeny using Notung-DM phylogenetic reconciliation software (Stolzer et al., 2015). This analysis reveals the most parsimonious explanation for the two major terminal exons in ATL2: following the divergence of ATL1 and ATL2, a copy of the final exon in ATL1 was inserted into ATL2, upstream from the existing terminal exon.
Online supplemental material
Fig. S1 shows all the ATL3/2/1 and DATL proteins used in this study resolved by SDS-PAGE; the efficiency of proteo-liposome incorporation of ATL3, ATL3ΔC, ATL2 and ATL2ΔC; and the comparable GTPase activities of ATL3 and ATL3ΔC. Fig. S2 shows E5 cell morphological rescue of the ER network by the ATL3/2 chimera. Fig. S3 A shows the species tree for the 18 vertebrates and 15 insects used in this study; and the full multiple sequence alignment of the ATL1/2/i C-terminal extensions (associated with Fig. 6 B). Fig. S4 A shows the full ATL core phylogeny (associated with Fig. 5 C), and Fig. S4 B shows the full ATL1/2/i C-terminus phylogeny (associated with Fig. 6 C). Fig. S5 shows the Notung reconciliation of the core and C-terminal extension trees that reveal the evolutionary history of the ATLs and their C-termini. Table S1 shows the full set of species used in this study. Table S2 shows the full set of ATL genes used in this study. Table S3 shows the full set of ATL exons.
Data availability
The source data consisting of all purification gels, flotation gels, and immunoblots underlying Fig. 4 and Fig. S1, as well as representative immunofluorescence images shown in Fig. 2 and Fig. S2, are available in the published article and its online supplemental material. All other raw experimental and computational data generated and/or analyzed during the current study are available at https://doi.org/doi:10.5061/dryad.j6q573nk8 and https://doi.org/doi:10.5061/dryad.zpc866td7, respectively.
Acknowledgments
The authors thank C. Blackstone for his gift of E5 cells, E. Snapp for the gift of GFP-Sec61β, and J. McNew for the gift of the ATL3 construct. A. Linstedt contributed helpful comments throughout.
This work was supported by a grant from the National Institutes of Health T32 training grant GM133353 to S. Bryce, and a grant to T.H. Lee from the National Institutes of Health/National Institutes for General Medical Sciences R01GM107285. D. Durand, R. Yang, and M. Stolzer were supported in part by National Science Foundation grants DBI-1838344 and DBI-1759943. D. Durand wishes to thank M. Hiller, Senckenberg Society for Nature Research, Goethe University, Frankfurt, for sharing comparative genomics expertise and resources developed in his laboratory.
Author contributions. S. Bryce and T.H. Lee conceived, designed, and analyzed the experiments. S. Bryce carried out the experiments with assistance from D. Crosby. The evolutionary analysis was designed by M. Stolzer and D. Durand and carried out by M. Stolzer, R. Yang, and D. Durand. The manuscript was written by S. Bryce, M. Stolzer, D. Durand, and T.H. Lee.
References
Author notes
Disclosures: The authors declare no competing interests exist.