The primary method for probing DNA replication dynamics is DNA fiber analysis, which utilizes thymidine analog incorporation into nascent DNA, followed by immunofluorescent microscopy of DNA fibers. Besides being time-consuming and prone to experimenter bias, it is not suitable for studying DNA replication dynamics in mitochondria or bacteria, nor is it adaptable for higher-throughput analysis. Here, we present mass spectrometry–based analysis of nascent DNA (MS-BAND) as a rapid, unbiased, quantitative alternative to DNA fiber analysis. In this method, incorporation of thymidine analogs is quantified from DNA using triple quadrupole tandem mass spectrometry. MS-BAND accurately detects DNA replication alterations in both the nucleus and mitochondria of human cells, as well as bacteria. The high-throughput capability of MS-BAND captured replication alterations in an E. coli DNA damage-inducing gene library. Therefore, MS-BAND may serve as an alternative to the DNA fiber technique, with potential for high-throughput analysis of replication dynamics in diverse model systems.
Introduction
Accurate duplication of the genome is essential for the faithful transmission of genetic information to daughter cells. Complete genome duplication demands the unimpeded progression of replication forks. To this end, the replisome must work in tandem with a variety of cellular processes, such as transcription, DNA repair, and chromatin assembly. DNA lesions, alterations in deoxynucleotide pools, and conflicts with transcription are some of the potential threats to the completion of replication. Cells have evolved several repair pathways to safeguard genome integrity and to ensure timely completion of DNA replication (Ashour and Mosammaparast, 2021; Berti et al., 2020; Cortez, 2019). Understanding the cellular responses to replication stress is emerging as a central theme in cell survival, genome stability, and human disease. Indeed, genetic mutations or changes in the expression levels of replication stress response factors are associated with greater cancer risk. At the same time, cancer cells rely on replication stress response mechanisms to survive, making these potential targets for cancer therapy (Ngoi et al., 2021; Thomas et al., 2021).
Currently, the gold standard for analyzing replication dynamics is the DNA fiber technique, which quantifies alterations in replication dynamics genome-wide at single DNA molecule resolution. This technique depends on the labeling of nascent DNA strands with distinct halogenated nucleosides, typically 5-Iodo-2′-deoxyuridine (IdU) and 5-Chloro-2′-deoxyuridine (CldU), followed by isolation and stretching of DNA fibers on a microscope slide. Subsequently, the labeled DNA tracts are stained with antibodies directed against these non-native thymidine analogs, which are then visualized by fluorescence microscopy. This labeling scheme allows monitoring the changes in replication fork elongation, termination events, fork symmetry, origin firing, and can also be adapted to quantify DNA gaps (Quinet et al., 2017; Vindigni and Lopes, 2017).
While being a major advance for studying DNA replication, DNA fiber analysis has a number of limitations. First, DNA fibers have a stretching range of 2–3 kb per micrometer of DNA, limiting the range of fibers analyzed to less than 300 kb fragments (Pham et al., 2013). Second, the technique is not easily adaptable to study DNA replication dynamics in yeast and bacteria. This is due to the low uptake and incorporation rate of exogenous thymidine analogues in the genome of these organisms (Breier et al., 2005; Pham et al., 2013). Moreover, the average rate of DNA synthesis in Escherichia coli is exceptionally fast, nearing 1 kb per second. This requires the pulse-labeling with the analogues to be limited to a few minutes. Longer labeling time will lead to longer DNA tracks that would be potentially out of the microscopic field of view (Breier et al., 2005; Pham et al., 2013). Specific strains have been developed to increase the incorporation rate of labeled thymidine in bacteria. However, these genetic manipulations to increase thymidine analog incorporation induce replication stress responses (Pham et al., 2013).
Beyond these limitations, the standard DNA fiber technique is difficult to adapt for studying mitochondrial DNA (mtDNA) replication. Human mtDNA consists of a 16.6-kb circular genome and encodes 37 genes that function primarily in oxidative phosphorylation (Anderson et al., 1981). mtDNA replication is accomplished with a distinct set of enzymes from the nuclear replisome, utilizing polymerase γ (PolG), Twinkle helicase, and the mitochondrial single-stranded DNA binding protein as minimal factors for the process (Korhonen et al., 2004). Like nuclear replication, mtDNA replication arrests at sites of template damage (Chiang et al., 2017; Dahal et al., 2018; Fontana and Gahlon, 2020; Huang et al., 2018). Attempts to adapt the DNA fiber technique to measure mtDNA replication have led to the development of mitochondrial single-molecule analysis of replicating DNA (Tigano et al., 2020). Mitochondrial single-molecule analysis of replicating DNA depends on super-resolution microscopy to delineate the boundaries between CldU and IdU, and the technique also requires FISH probes to mark the mtDNA molecules. These technical requirements, combined with the potential subjectivity and time necessary to quantify DNA fiber samples, call for the development of an alternative approach with wider utility.
Here, we develop mass spectrometry–based analysis of nascent DNA (MS-BAND) as a highly sensitive and quantitative technique to study DNA replication dynamics. In this assay, replication dynamics are still assessed through the incorporation of IdU or CldU in replicating cells; however, these labels are instead quantified in minutes using rapid liquid chromatography–mass spectrometry (LC-MS) rather than fluorescent microscopy (Fig. 1 a). We have validated the application of MS-BAND to study DNA replication dynamics in eukaryotic cells, bacteria, and mitochondria. The versatility of the technique permits higher-throughput screens for factors or compounds that disrupt DNA replication in various model organisms, which we demonstrate using an established library of DNA damage–inducing factors in bacteria.
MS-BAND detects IdU- and CldU-labeled nascent DNA. (a) Overview of the MS-BAND method to detect and quantify ldU and CldU incorporation in biological samples. (b) DNA labeling scheme. Increased labeling time would be detected by DNA fiber analysis as an increase in IdU/CldU track lengths, whereas it can be detected by MS-BAND as an increase in the IdU/dG and CldU/dG ion intensity ratios. (c) Size distribution of IdU-labeled (left) and CldU-labeled (right) DNA track length in human 293T cells. At least 200 tracks were measured per sample. Bars and colored numbers designate median length. Statistical differences in DNA fiber tract lengths were determined by the Mann–Whitney test (****P < 0.0001). (d) IdU/dG (left) and CldU (right) ion intensity ratios of labeled DNA from cells in c measured by MS-BAND. Data represent the mean of five replicates ±SD. Data were analyzed using one-way ANOVA (****P < 0.0001). (e) Chromatogram of unlabeled DNA is shown on the left, with analogous chromatogram of IdU/CldU-labeled DNA is shown in comparison on the right.
MS-BAND detects IdU- and CldU-labeled nascent DNA. (a) Overview of the MS-BAND method to detect and quantify ldU and CldU incorporation in biological samples. (b) DNA labeling scheme. Increased labeling time would be detected by DNA fiber analysis as an increase in IdU/CldU track lengths, whereas it can be detected by MS-BAND as an increase in the IdU/dG and CldU/dG ion intensity ratios. (c) Size distribution of IdU-labeled (left) and CldU-labeled (right) DNA track length in human 293T cells. At least 200 tracks were measured per sample. Bars and colored numbers designate median length. Statistical differences in DNA fiber tract lengths were determined by the Mann–Whitney test (****P < 0.0001). (d) IdU/dG (left) and CldU (right) ion intensity ratios of labeled DNA from cells in c measured by MS-BAND. Data represent the mean of five replicates ±SD. Data were analyzed using one-way ANOVA (****P < 0.0001). (e) Chromatogram of unlabeled DNA is shown on the left, with analogous chromatogram of IdU/CldU-labeled DNA is shown in comparison on the right.
Results
Triple quadrupole MS to quantify IdU and CldU
We first developed LC-MS/MS methods for the detection and quantification of CldU and IdU, as well as 2′-deoxyguanosine (dG), which was used for normalization. Nucleosides were separated and eluted off a reverse-phase Zorbax C18 column and negative parent ions for each compound were detected by a triple quadrupole mass spectrometer in selected ion monitoring (SIM) mode (Fig. 1 and Fig. S1 a). A standard curve ranging from 5 to 1,000 ng/ml was generated for each nucleoside. All R2 values were >0.99 and relative SD values were less than 3.2% (Fig. S1, b–e). Next, we sought to determine whether our optimized LC-MS/MS method could accurately quantify IdU- and CldU-labeled DNA from biological samples. We incubated HEK-293T cells with IdU followed by CldU for increasing amounts of time and measured analog incorporation through both traditional DNA fiber analysis and MS-BAND (Fig. 1, b–d). Labeled DNA was extracted from cells and digested down to the nucleoside level prior to LC-MS/MS analysis. As expected, the lengths of both IdU- and CldU-labeled fibers increased with increased incubation times (Fig. 1 c). Similarly, the IdU/dG and CldU/dG ion intensity ratios measured by MS-BAND also increased with longer labeling times, demonstrating that MS-BAND can detect nascent DNA in biological samples (Fig. 1 d). Increasing the labeling of time for the second analog (CldU) while keeping the first analog (IdU) incubation time constant resulted in a proportionally higher CldU/dG ratio but did not significantly alter the IdU/dG ratio, as expected (Fig. S1, f and g). Importantly, background signal for IdU and CldU was undetectable in non-labeled DNA samples (Fig. 1, d and e), indicating that our LC-MS/MS method is specific for quantifying these non-native nucleosides.
LC-MS/MS quantitation of dG, CldU, and IdU in negative SIM mode. (a) Chromatogram of dG, CldU, and IdU standards. (b) Retention times, R2 values, and relative standard deviations for dG, CldU, and IdU standard curves. (c–e) Standard curves for each nucleoside, as labeled (n = 5 replicates for each nucleoside). (f) Detection of nascent DNA with the indicated labeling scheme. HeLa cells were labeled with IdU for 15 min, followed by CldU for 15, 30, or 45 min. (g) MS-BAND was then used to measure the ion intensity of IdU/dG and CldU/dG. Data represent the mean of three replicates ±SD of the mean. Data were analyzed using one-way ANOVA (****P < 0.0001).
LC-MS/MS quantitation of dG, CldU, and IdU in negative SIM mode. (a) Chromatogram of dG, CldU, and IdU standards. (b) Retention times, R2 values, and relative standard deviations for dG, CldU, and IdU standard curves. (c–e) Standard curves for each nucleoside, as labeled (n = 5 replicates for each nucleoside). (f) Detection of nascent DNA with the indicated labeling scheme. HeLa cells were labeled with IdU for 15 min, followed by CldU for 15, 30, or 45 min. (g) MS-BAND was then used to measure the ion intensity of IdU/dG and CldU/dG. Data represent the mean of three replicates ±SD of the mean. Data were analyzed using one-way ANOVA (****P < 0.0001).
MS-BAND accurately quantifies slowing of nascent DNA synthesis in response to replication stress
We next compared the MS-based assay to the DNA fiber technique in measuring replication fork stalling (Fig. 2 a). Cells were pulse-labeled for 30 min with IdU, followed by 30 min with CldU and increasing doses of hydroxyurea (HU). While IdU-labeled fiber lengths were equal for all samples, CldU-labeled fibers were shortened in an HU dose-dependent manner, indicating replication fork stalling (Fig. 2 b). The same result was detected using MS-BAND, as the IdU/dG ratios remained constant in HU-treated samples, while the CldU/dG ratio decreased with increasing HU concentration (Fig. 2 c). Comparison of the MS-BAND results and DNA fiber lengths in these experiments produced a correlation coefficient >0.97, demonstrating the accuracy of MS-BAND compared to the traditional assay (Fig. 2 d). These results were confirmed using two other known DNA replication inhibitors; specifically, the addition of camptothecin (CPT), a topoisomerase I inhibitor, as well as doxorubicin (DOXO), a topoisomerase II inhibitor. Both of these inhibitors resulted in reductions in replication fork progression as detected by either the DNA fiber (Fig. 2 e) or MS-BAND approaches (Fig. 2 f). Moreover, MS-BAND detected fork slowing by aphidicolin, an established DNA polymerase inhibitor (Fig. S2).
MS-BAND detects of DNA replication inhibition in human cells. (a) Schematic of experiments using MS-BAND or traditional DNA fiber analysis to detect decrease in fork speed in the presence of replication inhibitor or DNA damaging agent. This phenotype would be detected by DNA fiber analysis as decrease in CldU track length, where it can be detected by MS-BAND as a decrease in the CldU/dG ion intensity ratio. (b) Size distribution of IdU- (left) and CldU- (right) labeled DNA track lengths in 293T cells. At least 200 tracks were scored for each sample. Colored bars and numbers indicate median track length. Statistical differences in DNA fiber tract lengths were determined by the Mann-Whitney test (****P < 0.0001). (c) MS-BAND ion intensity ratios for IdU/dG (left) and CldU/dG (right). Data represent the mean of three biological replicates ±SD. Data were analyzed using one-way ANOVA (****P < 0.0001). (d) Linear correlation between median DNA fiber length and CldU/dG ion intensity ratio. Plotted values are from experiments in Fig. 1, c and d; and Fig. 2, b and c. (e and f) The indicated labeling/inhibitor scheme was used either for DNA fiber experiments (performed in biological duplicate; e) or MS-BAND experiments (f) in HeLa cells. Statistical analysis of the data was determined as in b and c. Statistical significance was defined as *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
MS-BAND detects of DNA replication inhibition in human cells. (a) Schematic of experiments using MS-BAND or traditional DNA fiber analysis to detect decrease in fork speed in the presence of replication inhibitor or DNA damaging agent. This phenotype would be detected by DNA fiber analysis as decrease in CldU track length, where it can be detected by MS-BAND as a decrease in the CldU/dG ion intensity ratio. (b) Size distribution of IdU- (left) and CldU- (right) labeled DNA track lengths in 293T cells. At least 200 tracks were scored for each sample. Colored bars and numbers indicate median track length. Statistical differences in DNA fiber tract lengths were determined by the Mann-Whitney test (****P < 0.0001). (c) MS-BAND ion intensity ratios for IdU/dG (left) and CldU/dG (right). Data represent the mean of three biological replicates ±SD. Data were analyzed using one-way ANOVA (****P < 0.0001). (d) Linear correlation between median DNA fiber length and CldU/dG ion intensity ratio. Plotted values are from experiments in Fig. 1, c and d; and Fig. 2, b and c. (e and f) The indicated labeling/inhibitor scheme was used either for DNA fiber experiments (performed in biological duplicate; e) or MS-BAND experiments (f) in HeLa cells. Statistical analysis of the data was determined as in b and c. Statistical significance was defined as *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
MS-BAND detects inhibition of DNA replication in cells. MS-BAND ion intensity ratios for CldU/dG for control versus aphidicolin treated samples. Data represent the mean of three biological replicates ±SD of the mean. Data were analyzed using one-way ANOVA (****P < 0.0001).
MS-BAND detects inhibition of DNA replication in cells. MS-BAND ion intensity ratios for CldU/dG for control versus aphidicolin treated samples. Data represent the mean of three biological replicates ±SD of the mean. Data were analyzed using one-way ANOVA (****P < 0.0001).
Using MS-BAND to detect nascent DNA fork degradation
Nascent DNA fork protection has been revealed to be a major function of certain factors involved in DNA replication and repair, most notably the BRCA1 and BRCA2 tumor suppressor proteins (Byrum et al., 2019; Lemaçon et al., 2017; Ray Chaudhuri et al., 2016; Schlacher et al., 2011). We therefore wished to test whether MS-BAND could detect degradation of stalled DNA forks in the setting of BRCA1 deficiency. To this end, we used sequential labeling with IdU and CldU, followed by replication inhibition with HU, to test the degree of fork protection in an established BRCA1-deficient (UWB1.289) cell line, versus the isogenic BRCA1-complemented controls (Fig. 3, a–c). We noticed a significant reduction in the CldU/IdU ratio in the BRCA1-deficient cell line, as determined by traditional DNA fiber analysis, and a much smaller but statistically insignificant fork degradation in the BRCA1-complemented cells (Fig. 3 b). MS-BAND also detected a similar degree of nascent DNA loss in the BRCA1-deficient cell line upon HU stress, as measured by normalized CldU/IdU ratios (Fig. 3 c). A statistically significant but smaller degree of fork degradation was seen in the BRCA1 proficient cells, as determined by MS-BAND. Thus, it appears that MS-BAND is also capable of detecting a loss of DNA fork protection.
MS-BAND can detect nascent DNA fork degradation in BRCA1-deficient cells. (a) Schematic of experiments using MS-BAND or traditional DNA fiber analysis to detect degradation of stalled DNA forks in the presence of the replication inhibitor, HU. The results of UWB1.289 (BRCA1 deficient) cells were compared to BRCA1 complemented cells. This phenotype would be detected by DNA fiber analysis as a decrease in CldU/IdU track length, whereas it can be detected by MS-BAND as a decrease in the CldU/IdU ion intensity ratio. (b) Size distribution of CldU/IdU-labeled DNA track lengths. At least 100 tracks were scored for each biological replicate (n = 2 for each genotype). Statistical differences in DNA fiber tract lengths were determined by the Mann–Whitney test (****P < 0.0001). (c) MS-BAND ion intensity ratios for CldU/IdU. Data represent the mean of five biological replicates ± SD of the mean. Data were analyzed using one-way ANOVA (*P < 0.05 and ****P < 0.0001).
MS-BAND can detect nascent DNA fork degradation in BRCA1-deficient cells. (a) Schematic of experiments using MS-BAND or traditional DNA fiber analysis to detect degradation of stalled DNA forks in the presence of the replication inhibitor, HU. The results of UWB1.289 (BRCA1 deficient) cells were compared to BRCA1 complemented cells. This phenotype would be detected by DNA fiber analysis as a decrease in CldU/IdU track length, whereas it can be detected by MS-BAND as a decrease in the CldU/IdU ion intensity ratio. (b) Size distribution of CldU/IdU-labeled DNA track lengths. At least 100 tracks were scored for each biological replicate (n = 2 for each genotype). Statistical differences in DNA fiber tract lengths were determined by the Mann–Whitney test (****P < 0.0001). (c) MS-BAND ion intensity ratios for CldU/IdU. Data represent the mean of five biological replicates ± SD of the mean. Data were analyzed using one-way ANOVA (*P < 0.05 and ****P < 0.0001).
Assessing replication dynamics in mitochondria using MS-BAND (mito-MS-BAND)
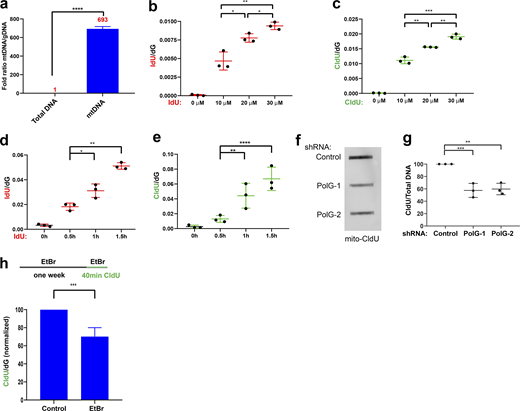
Due to its sensitivity in detecting thymidine analog incorporation, we reasoned that we could apply MS-BAND to study DNA replication in mitochondria. We first established a protocol to isolate highly pure mtDNA (see Materials and methods) that consistently produced significant enrichment in mtDNA (>500-fold enrichment; Fig. S3 a). We then pulse-labeled HeLa cells with CIdU and IdU at different concentrations and lengths of time. We again observed an increase in both the IdU/dG and CIdU/dG signals in a time- and concentration-dependent manner (Fig. S3, b–e). To validate that the assay measures mtDNA replication, we depleted DNA PolG using two independent shRNAs, both of which significantly reduced PolG protein expression (Fig. 4 a). Mitochondrial MS-BAND (mito-MS-BAND) analysis demonstrated a significant reduction in the rate of CldU incorporation in PolG knockdown cells compared to controls (Fig. 4 b). We also confirmed this result in PolG-depleted cells by measuring the incorporation of CldU into mtDNA using slot-blot analysis (Fig. S3 f). It is established that ethidium bromide (EtBr) reduces the mtDNA copy number and reduces mtDNA replication (King and Attardi, 1989). Consistently, mito-MS-BAND demonstrated a reduction in the replication rate of mtDNA after prolonged incubation with EtBr (Fig. S3 g). We then tested the effect of different inhibitors on mtDNA replication. HU, CPT, and etoposide all reduced the rate of DNA replication in mtDNA as determined by our assay. On the other hand, neither Poly(ADP-Ribose) Polymerase nor Ataxia Telangiectasia and Rad3-related protein (ATR) inhibitors had a significant effect as determined by mito-MS-BAND (Fig. 4, c and d). As these results are consistent with the effect of these inhibitors on mitochondrial replication (Chiang et al., 2017; Huang et al., 2018), our results further confirm the ability of MS-BAND to measure replication dynamics in mitochondria. Finally, evidence suggests a crosstalk between mtDNA damage and nuclear DNA damage (Fang et al., 2016; Saki and Prakash, 2017; Tigano et al., 2021; Wiese and Bannister, 2020). We therefore used the inducible AsiSI restriction enzyme system to induce nuclear DNA double-strand breaks (DSBs; Aymard et al., 2014; Harding et al., 2017). As expected, induction of DSBs in the nucleus with this system led to a reduction in the rate of nuclear DNA synthesis as determined by MS-BAND. Interestingly, the DSBs in the nucleus also resulted in a reduction in the mtDNA synthesis (Fig. 4 f). Further investigation will be required to identify the factor(s) that link nuclear DNA damage and mtDNA replication.
Mitochondrial MS-BAND. (a) Assessment of the purity of mtDNA by quantitative PCR, using an mtDNA target normalized to GAPDH as control. Data represent the mean of three biological replicates ± SD. Data were analyzed using Student’s t test (****P < 0.0001). (b and c) Mito-MS-BAND results of cells labeled with different concentrations of ldU or CldU and grow at 37°C for 30 min. (d and e) Mito-MS-BAND results of cells labeled with ldU or CldU for increasing periods of time. Data in b–e represent the mean of three biological replicates ± SD. Data were analyzed using one-way ANOVA. Statistical significance was defined as *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. (f) Dot blot results using material from Fig. 3 a. Cells were labeled with CldU for 1 h, followed by mtDNA purification. Dot blot was performed using an anti-CldU antibody using equal amounts of DNA per sample. (g) Quantification of f. Data represent the mean of three biological replicates ± SD. Data were analyzed using one-way ANOVA, and statistical significance was defined as **P <0.01 and ***P <0.001. (h) Schematic of experiment using mito-MS-BAND to determine the effect of long-term EtBr on mtDNA replication. Data represent the mean of three biological replicates ±SD. Data were analyzed using Student’s t test (***P < 0.001). Source data are available for this figure: SourceData FS3.
Mitochondrial MS-BAND. (a) Assessment of the purity of mtDNA by quantitative PCR, using an mtDNA target normalized to GAPDH as control. Data represent the mean of three biological replicates ± SD. Data were analyzed using Student’s t test (****P < 0.0001). (b and c) Mito-MS-BAND results of cells labeled with different concentrations of ldU or CldU and grow at 37°C for 30 min. (d and e) Mito-MS-BAND results of cells labeled with ldU or CldU for increasing periods of time. Data in b–e represent the mean of three biological replicates ± SD. Data were analyzed using one-way ANOVA. Statistical significance was defined as *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. (f) Dot blot results using material from Fig. 3 a. Cells were labeled with CldU for 1 h, followed by mtDNA purification. Dot blot was performed using an anti-CldU antibody using equal amounts of DNA per sample. (g) Quantification of f. Data represent the mean of three biological replicates ± SD. Data were analyzed using one-way ANOVA, and statistical significance was defined as **P <0.01 and ***P <0.001. (h) Schematic of experiment using mito-MS-BAND to determine the effect of long-term EtBr on mtDNA replication. Data represent the mean of three biological replicates ±SD. Data were analyzed using Student’s t test (***P < 0.001). Source data are available for this figure: SourceData FS3.
Replication dynamics in mitochondria as determined by MS-BAND. (a) HeLa cells were transduced with the indicated shRNAs. After selection, Western blots were performed from whole cell lysates using the indicated antibodies. Molecular weight (Mw) marker positions (in kD) are shown. (b) Mito-MS-BAND results using cells from a; cells were labeled with CldU for 1 h. The CldU/dG ion intensity for each mitochondrial sample was normalized to the control. Data represent the mean of three biological replicates ± SD. Data were analyzed using one-way ANOVA (*P < 0.05, **P < 0.01). (c) Mito-MS-BAND results of untreated, CPT, or HU treated samples. Cells were labeled with CldU for 1 h in the presence of each inhibitor at the indicated concentration. The CldU/dG ion intensity for each mitochondrial sample was normalized to the untreated control. Data represent the mean of three biological replicates ±SD. Data were analyzed using one-way ANOVA (****P < 0.0001). (d) Mito-MS-BAND results of cells treated with etoposide (10 μM), olaparib (10 μM), or AZD6738 (ATR inhibitor; 5 μM), compared to untreated controls. Cells were labeled with CldU for 1 h. The CldU/dG ion intensity for each sample was normalized to control. Data represent the mean of three biological replicates ± SD and were analyzed as in c. (e) U2OS cells expressing a nuclear targeted endonuclease (AsiSI) were induced using a combination of Shield1 ligand and 4-hydrotamoxifen (4-OHT). Western blots were performed from whole cell lysates using the indicated antibodies. (f) Nuclear versus mito-MS-BAND results of cells from e using the schematic shown. The CldU/dG ion intensity for each sample was normalized to control. Data represents the mean of three biological replicates ± SD and were analyzed using Student’s t test. Statistical significance was defined as **P < 0.01, ****P < 0.0001. Source data are available for this figure: SourceData F4.
Replication dynamics in mitochondria as determined by MS-BAND. (a) HeLa cells were transduced with the indicated shRNAs. After selection, Western blots were performed from whole cell lysates using the indicated antibodies. Molecular weight (Mw) marker positions (in kD) are shown. (b) Mito-MS-BAND results using cells from a; cells were labeled with CldU for 1 h. The CldU/dG ion intensity for each mitochondrial sample was normalized to the control. Data represent the mean of three biological replicates ± SD. Data were analyzed using one-way ANOVA (*P < 0.05, **P < 0.01). (c) Mito-MS-BAND results of untreated, CPT, or HU treated samples. Cells were labeled with CldU for 1 h in the presence of each inhibitor at the indicated concentration. The CldU/dG ion intensity for each mitochondrial sample was normalized to the untreated control. Data represent the mean of three biological replicates ±SD. Data were analyzed using one-way ANOVA (****P < 0.0001). (d) Mito-MS-BAND results of cells treated with etoposide (10 μM), olaparib (10 μM), or AZD6738 (ATR inhibitor; 5 μM), compared to untreated controls. Cells were labeled with CldU for 1 h. The CldU/dG ion intensity for each sample was normalized to control. Data represent the mean of three biological replicates ± SD and were analyzed as in c. (e) U2OS cells expressing a nuclear targeted endonuclease (AsiSI) were induced using a combination of Shield1 ligand and 4-hydrotamoxifen (4-OHT). Western blots were performed from whole cell lysates using the indicated antibodies. (f) Nuclear versus mito-MS-BAND results of cells from e using the schematic shown. The CldU/dG ion intensity for each sample was normalized to control. Data represents the mean of three biological replicates ± SD and were analyzed using Student’s t test. Statistical significance was defined as **P < 0.01, ****P < 0.0001. Source data are available for this figure: SourceData F4.
MS-BAND can detect replication alterations in bacteria
E. coli has served as a leading model system for clarifying the molecular mechanisms underlying DNA replication. The use of the DNA fiber assay to study DNA replication in bacteria is limited, due to the low incorporation rate of CIdU and IdU in the bacterial genome (Pham et al., 2013). Moreover, the restriction in the microscopic field of view mentioned above requires very short labeling time, making it a less practical method for bacteria (Breier et al., 2005; Pham et al., 2013). Since our LC-MS/MS method can readily quantify concentrations of CIdU and IdU to at least as low as 5 ng/ml (Fig. S1), we reasoned that MS-BAND could quantify small amounts of CldU and IdU incorporation in the genome of wildtype bacterial strains. We grew E. coli DH10β in Luria Bertani Herskowitz medium, then diluted the cultures to equal optical density while simultaneously pulse-labeling the cultures with increasing concentrations of CldU or IdU. The cells were collected, and genomic DNA was extracted and analyzed by MS-BAND. As anticipated, MS-BAND could detect a concentration-dependent incorporation of IdU and CldU into bacterial genomic DNA (Fig. S4, a and b). Moreover, an increase in labeling time resulted in an increase in the IdU/dG and CldU/dG ion intensity ratios (Fig. S4, c and d). In order to confirm that the detected IdU and CIdU signal was due to bona fide incorporation into the bacterial genome, we quantified dG, IdU, and CldU nucleosides in nuclease digested and undigested DNA samples. The undigested samples showed minimal signal for all three nucleosides, whereas the digested, unlabeled sample only produced a signal for dG (Fig. S4, e–g). Only the labeled and digested samples could produce signals for IdU and CldU, confirming that the MS-BAND signal is due to the incorporation of these thymidine analogs into the bacterial genome (Fig. S4, f and g).
Measuring replication dynamics in bacteria by MS-BAND. (a and b) MS-BAND results of E. coli DH10β bacteria labeled with different concentrations of ldU or CldU and grown at 37°C for 30 min. (c and d) MS-BAND results of E. coli DH10β labeled with ldU or CldU for increasing periods of time. Data in a–d represent the mean of three biological replicates ± SD. Data were analyzed using one-way ANOVA, and statistical significance was defined as *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. (e–g) MS-BAND results of bacterial strains unlabeled, labeled with ldU, or labeled with CldU. Subsequently, samples were left undigested by nucleases or digested overnight to single nucleosides for LC-MS analysis. (h) Normalized OD600 of the indicated bacterial strains before (left) and after (right) heat shock at 42°C for 30 min, corresponding to experiment in Fig. 4 a.
Measuring replication dynamics in bacteria by MS-BAND. (a and b) MS-BAND results of E. coli DH10β bacteria labeled with different concentrations of ldU or CldU and grown at 37°C for 30 min. (c and d) MS-BAND results of E. coli DH10β labeled with ldU or CldU for increasing periods of time. Data in a–d represent the mean of three biological replicates ± SD. Data were analyzed using one-way ANOVA, and statistical significance was defined as *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. (e–g) MS-BAND results of bacterial strains unlabeled, labeled with ldU, or labeled with CldU. Subsequently, samples were left undigested by nucleases or digested overnight to single nucleosides for LC-MS analysis. (h) Normalized OD600 of the indicated bacterial strains before (left) and after (right) heat shock at 42°C for 30 min, corresponding to experiment in Fig. 4 a.
Next, we tested the ability of MS-BAND to detect replication slowing in bacterial strains with established temperature sensitive defects in the replication machinery. We determined the replication speed in a wildtype strain in comparison with temperature sensitive mutants for replication initiation factor (dnaA), the catalytic subunit of DNA polymerase III (dnaE), and replicative helicase (dnaB) under permissive and non-permissive conditions. Strikingly, the MS-BAND signal demonstrated a rapid fourfold induction of thymidine analogue incorporation in control cells after shifting to 42°C. On the other hand, there was a reduction in thymidine analogue incorporation after shifting from 30 to 42°C in all the mutant strains compared to control, although this effect was more prominent with the dnaA and dnaB mutants (Fig. 5 a). The relative OD600 of the three mutant strains did not change significantly upon shifting of the temperature conditions (Fig. S4 h). This validates the ability of MS-BAND to quantify the slowing of nascent DNA synthesis in response to genetic alteration of canonical replication factors.
Replication dynamics in bacteria as determined by MS-BAND. (a) MS-BAND results of the four indicated E. coli strains. All strains were initially grown to early log phase at 30°C, a portion was then labeled with CldU and shifted immediately to 42°C for 30 min. In parallel, each strain was also labeled with CldU and continued to grow at 30°C. The CldU/dG ion intensity at 42°C was normalized to the 30°C result for each individual strain. Data represent the mean of three biological replicates ±SD. Data were analyzed using Student’s t test. Statistical significance was defined as *P < 0.05 and ****P < 0.0001. (b–d) OD600 and MS-BAND results of E. coli DH10β bacteria labeled with CldU in the presence or absence of each antibiotic: ampicillin (b), kanamycin (c), or tetracycline (d). Cells were grown at 37°C for 1.5 h. The results were normalized to untreated samples. Data represent the mean of three biological replicates ±SD. Data were analyzed using one-way ANOVA. Statistical significance was defined as *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Replication dynamics in bacteria as determined by MS-BAND. (a) MS-BAND results of the four indicated E. coli strains. All strains were initially grown to early log phase at 30°C, a portion was then labeled with CldU and shifted immediately to 42°C for 30 min. In parallel, each strain was also labeled with CldU and continued to grow at 30°C. The CldU/dG ion intensity at 42°C was normalized to the 30°C result for each individual strain. Data represent the mean of three biological replicates ±SD. Data were analyzed using Student’s t test. Statistical significance was defined as *P < 0.05 and ****P < 0.0001. (b–d) OD600 and MS-BAND results of E. coli DH10β bacteria labeled with CldU in the presence or absence of each antibiotic: ampicillin (b), kanamycin (c), or tetracycline (d). Cells were grown at 37°C for 1.5 h. The results were normalized to untreated samples. Data represent the mean of three biological replicates ±SD. Data were analyzed using one-way ANOVA. Statistical significance was defined as *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
We next tested the ability of MS-BAND to detect changes in replication dynamics in bacteria in response to antibiotics, as recent results have suggested that antibiotic stress may increase replication rate as part of the filamentation stress response, which induces re-replication (Slager et al., 2014). We treated E. coli with increasing concentrations of ampicillin, tetracycline, or kanamycin. Interestingly, incubation with all three antibiotics, but most notably the latter two agents, demonstrated increased thymidine analog incorporation compared to control conditions (Fig. 5, b–d). This is despite the fact that the OD600 was the same or moderately reduced under such conditions. Our result is consistent with a recent report suggesting that certain classes of antibiotics increase the replication by re-initiation near the replication origin (Slager et al., 2014).
MS-BAND can be used as screening tool for replication defects
Because our LC-MS/MS method is relatively fast (∼5 min per sample after DNA digestion), we reasoned that MS-BAND could be used as a screening tool for identifying conditions of DNA replication alteration in a higher throughput capacity. To test MS-BAND for such an application, we screened a bacterial library of 208 factors previously shown to encode inducers of DNA damage (also known as DNA damage proteins, or DDPs; Xia et al., 2019). The genes that are part of this overexpression library are known to induce the bacterial SOS response, and over half of them appear to promote replication fork reversal (Xia et al., 2019). The overexpression of each DDP in the library was induced with IPTG for 6 h before the addition of CldU (Fig. 6 a). The majority of these genes (104; 50%) reduced the DNA synthesis rate by at least 20%, while 28 genes (13.5%) increased the rate of replication by at least 20% (Fig. 6 a and Table S1). We validated these results using 28 initial hits from the library, 21 of which reduced DNA synthesis, and 7 of which increased DNA synthesis (Fig. 6, b and c). The initial MS-BAND screen results were confirmed in 28/28 of these cases. A reduced CldU incubation time of 7 or 14 min gave similar results in five of these hits, further validating the approach (Fig. S5, a and b). Among the confirmed DDPs that led to slowing of DNA synthesis, dinB (DNA pol IV), which encodes a poorly processive, error-prone translesion DNA polymerase (Bunting et al., 2003; Tang et al., 2000; Wagner et al., 1999), gave the strongest phenotype. It is likely that DNA pol IV outcompetes or inhibits the normal replication machinery, leading to slower replication, consistent with a previous explanation (Uchida et al., 2008). Another was diaA, which encodes a DnaA associating factor crucial to ensure the timely initiation of chromosomal replication (Ishida et al., 2004; Keyamura et al., 2007). Furthermore, both hda and seqA are established negative regulators of replication re-initiation, and both reduced the MS-BAND signal (Fig. 6 b; Kato and Katayama, 2001; Lu et al., 1994; Slater et al., 1995). MS-BAND also demonstrated that induction of ftsX and ftsK, which coordinate cell division and chromosome segregation (de Leeuw et al., 1999; Wang and Lutkenhaus, 1998; Yu et al., 1998), increase the apparent rate of DNA synthesis in our assay (Fig. 6 c). Amongst DDPs that increased thymidine analog incorporation, dnaA, which is responsible for initiating DNA replication (Boye et al., 1996; Erzberger et al., 2002; Murray and Errington, 2008), was the top hit. A number of other DDPs known to participate in metabolism (guaD, yedQ, yeaW, nudC, panE, and prpD) and metabolite transport (rhmT, manZ, and aroP) also gave strong phenotypes in reducing the MS-BAND signal. To our knowledge, the exact roles for these proteins in replication are unclear. However, it has been suggested that the increased transporter activity and perturbation in metabolic pathways can provoke high ROS levels that can induce DNA damage (Xia et al., 2019), which may lead to replication slowing.
Screening of bacterial inducible library for replication defects by MS-BAND. (a) Library of 208 E.coli DDPs was analyzed by MS-BAND using the schematic as indicated. The results were normalized to the vector control sample. The Z-score of the results for each strain is shown. (b) Confirmation of the results of 21 genes that lead to an apparent reduction in the replication rate. (c) Confirmation of the results of seven genes that lead to an apparent increase in the replication rate. Data in b and c represent the mean of three biological replicates ±SD. Data were analyzed using one-way ANOVA. Statistical significance was defined as *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. (d) Lower MS-BAND signal (slower replication progression) in the DDPs correlates with higher ROS levels, anucleate cells, and sensitivities to phleomycin (r < or >0.2, P < 0.001). The Z values for the ROS, anucleate cells, and phleomycin sensitivities were described previously (Xia et al., 2019). Each point represents a single E. coli DDP, with the red dot indicating the vector-only control. Pearson correlation coefficient r and P values were calculated for each of the three correlations. Blue line: linear regression line. Red lines: 95% confidence intervals. (e) Working model indicating how E. coli DDPs may cause replication alterations through different mechanisms. (1) A DDP may act in a dominant-negative manner to perturb replisome stoichiometry. (2) A DDP may generate DNA damage directly to affect replication dynamics, or (3) induce ROS, which could slow replication progression. These can lead to chromosome segregation defects that further exacerbate DNA damage.
Screening of bacterial inducible library for replication defects by MS-BAND. (a) Library of 208 E.coli DDPs was analyzed by MS-BAND using the schematic as indicated. The results were normalized to the vector control sample. The Z-score of the results for each strain is shown. (b) Confirmation of the results of 21 genes that lead to an apparent reduction in the replication rate. (c) Confirmation of the results of seven genes that lead to an apparent increase in the replication rate. Data in b and c represent the mean of three biological replicates ±SD. Data were analyzed using one-way ANOVA. Statistical significance was defined as *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. (d) Lower MS-BAND signal (slower replication progression) in the DDPs correlates with higher ROS levels, anucleate cells, and sensitivities to phleomycin (r < or >0.2, P < 0.001). The Z values for the ROS, anucleate cells, and phleomycin sensitivities were described previously (Xia et al., 2019). Each point represents a single E. coli DDP, with the red dot indicating the vector-only control. Pearson correlation coefficient r and P values were calculated for each of the three correlations. Blue line: linear regression line. Red lines: 95% confidence intervals. (e) Working model indicating how E. coli DDPs may cause replication alterations through different mechanisms. (1) A DDP may act in a dominant-negative manner to perturb replisome stoichiometry. (2) A DDP may generate DNA damage directly to affect replication dynamics, or (3) induce ROS, which could slow replication progression. These can lead to chromosome segregation defects that further exacerbate DNA damage.
Applications and correlations of bacterial MS-BAND. (a) Schematic of experiments using bacterial MS-BAND with shorter CldU pulse labeling. (b) MS-BAND (using 7 or 14 min of CldU, as indicated) was used to measure the ion intensity of CldU/dG of three bacterial clones that resulted in reduction in analogue incorporation, and two clones that lead to an apparent decrease in analogue incorporation. Data represent the mean of three biological replicates ± SD. Data were analyzed using one-way ANOVA. Statistical significance was defined as ****P < 0.0001. (c) GamGFP (DSB reporter), reversed forks, and mitomycin C and H2O2 sensitivity in E. coli DDPs do not correlate with slower forks as measured by MS-BAND. The Z values for GamGFP, reversed forks, and mitomycin C and H2O2 sensitivity were described previously (Xia et al., 2019). Each point represents a single E. coli DDP. Pearson correlation coefficient r and P values were calculated for each of the four correlations. Blue line: linear regression line. Red lines: 95% confidence intervals.
Applications and correlations of bacterial MS-BAND. (a) Schematic of experiments using bacterial MS-BAND with shorter CldU pulse labeling. (b) MS-BAND (using 7 or 14 min of CldU, as indicated) was used to measure the ion intensity of CldU/dG of three bacterial clones that resulted in reduction in analogue incorporation, and two clones that lead to an apparent decrease in analogue incorporation. Data represent the mean of three biological replicates ± SD. Data were analyzed using one-way ANOVA. Statistical significance was defined as ****P < 0.0001. (c) GamGFP (DSB reporter), reversed forks, and mitomycin C and H2O2 sensitivity in E. coli DDPs do not correlate with slower forks as measured by MS-BAND. The Z values for GamGFP, reversed forks, and mitomycin C and H2O2 sensitivity were described previously (Xia et al., 2019). Each point represents a single E. coli DDP. Pearson correlation coefficient r and P values were calculated for each of the four correlations. Blue line: linear regression line. Red lines: 95% confidence intervals.
We performed correlation analysis with other functional assays of the 208 E. coli DDP clones. This revealed a moderate correlation (defined as r < or >0.2) between lower MS-BAND signal and ROS, anucleate cells, and phleomycin sensitivity (Fig. 6 d). These are characteristics of a specific cluster of genes from a previous study (Xia et al., 2019; cluster 2), where DNA replication and repair proteins are overrepresented, consistent with our findings here that most clones with a strong phenotype in MS-BAND play key roles in DNA replication and repair. However, MS-BAND did not meet this criterion of correlation with four other functional assays related to DNA damage, specifically a DNA DSB reporter, formation of reversed forks, or sensitivity to mitomycin and hydrogen peroxide (Fig. S5 c). This suggests that while MS-BAND can detect a broad range of alterations that directly or indirectly impact DNA replication (Fig. 6 e), not all DNA damage–inducing factors will have a significant change in thymidine analogue incorporation. Taken together, our results suggest that MS-BAND can be used as a tool for targeted screening of alterations in replication.
Discussion
While the reconstitution of the eukaryotic replisome with purified proteins has provided many insights about replisome assembly and function (Yeeles et al., 2015; Yeeles et al., 2017), this system requires several dozen factors, which makes this in vitro approach challenging for studying the replication stress response. Moreover, assaying replisome function in living cells under a variety of genetic and environmental conditions requires a cell-based approach. While the preferred method for measuring replication progression is DNA fiber analysis or DNA combing, this technique is laborious and cannot readily be used to study the replication of bacteria or mitochondria. Here, we describe a simple, straightforward MS-based approach to measure global replication dynamics in different biological model systems. The high sensitivity and adaptability of the technique enable the investigation of replication dynamics in particular cell types or cellular compartments where the standard DNA fiber assay is not readily applicable.
We show that MS-BAND can detect fork slowing in human cell lines in a manner comparable to the DNA fiber assay (Fig. 2). This suggests that MS-BAND is capable of quantifying the alteration in nascent DNA synthesis under a variety of replication stress conditions. Our data suggest that in certain circumstances, MS-BAND gives stronger phenotypes compared to the traditional fiber assay for specific agents such as CPT and DOXO; since we are measuring bulk replication, new origin firing is likely suppressed, which likely exacerbates the reduction of apparent replication relative to the DNA fiber technique. While flow cytometry analysis for the quantitation of newly synthesized DNA may be a more feasible approach for most labs who may not have ready access to quantitative MS, the MS approach is far more sensitive relative to flow cytometry; this sensitivity is likely necessary for analyzing more subtle phenotypes, such as nascent fork degradation (Fig. 3). There are additional advantages of using MS to quantify nascent DNA; for example, there is no requirement for a specific antibody, and therefore any nucleoside analogue can be quantified. Therefore, it would allow more complex labeling schemes to be used, such as ones that use more than two different sequential thymidine analogs. For example, a third analog (e.g., BrdU) can be used to analyze replication dynamics after two different replication inhibiting drugs are used sequentially, or at the same drug used at different concentrations. In addition, this may also allow MS-BAND to be used for looking at incorporation of damaged nucleosides during replication. This sensitivity of MS also allowed us to explore the potential use of MS-BAND for studying replication dynamics in biological models that cannot be readily investigated using the DNA fiber assay, demonstrating the use of the MS-BAND technique to measure replication dynamics in mitochondria. MS-BAND was able to measure replication slowing in mtDNA in response to PolG knockdown or in the presence of certain replication inhibitors, such as etoposide and HU (Fig. 4). Our data also suggests that nuclear DNA damage may affect mtDNA replication, consistent with reports of damage-induced crosstalk between these two cellular compartments (Fang et al., 2016; Saki and Prakash, 2017; Tigano et al., 2021; Wiese and Bannister, 2020). While we cannot rule out the possibility that induction of nuclear AsiSI causes indirect DNA damage to mitochondria, mtDNA does not contain any AsiSI sites, and therefore this possibility is less likely. Overall, our work indicates a need for caution when interpreting the impact of agents or factors that alter both nuclear and mtDNA replication; that is, reduction in mitochondrial replication may be due to effects secondary to nuclear DNA damage.
The DNA fiber assay is limited in measuring the replication dynamics in bacteria, due to the low incorporation rate of thymidine analogs in the bacterial genome, and also due to the restriction in the microscopic field of view. In order to test the ability of MS-BAND to detect replication slowing in bacteria, we used temperature-sensitive strains for genes that are established to have a role in DNA replication. MS-BAND can quantify the reduction in nascent DNA replication after a rapid shift to the non-permissive temperature. These results suggest that MS-BAND could readily measure DNA replication in bacteria. Interestingly, our use of MS-BAND to determine the response to commonly used antibiotics demonstrates the ability of the assay to detect a relative increase in nascent DNA synthesis under certain stress conditions. These results are consistent with previous reports showing that, in response to antibiotics, bacteria initially increase the copy number of antibiotic resistant genes near the replication origin as part of the filamentation response (Slager et al., 2014).
As far as we are aware, there are no known techniques that can perform high-throughput screening for replication defects. We decided to take advantage of the unbiased, quantitative, and the relatively fast readout of MS-BAND to screen an established bacterial library of genes previously shown to induce the bacterial SOS response (Xia et al., 2019). Indeed, more than half of the genes reduced the replication rate by at least 20%, and several of these also increased the apparent replication rate. In addition to validating known regulators for replication among these hits, we have also identified many metabolic and transport regulators with unexpected roles in DNA replication. Comparing MS-BAND results with other readouts of DNA damage sensitivity or replication demonstrated correlations with some of these functional outcomes but not others (Fig. 6 d and Fig. S5). Most notably, MS-BAND did not seem to correlate with induction of reversed fork structures (Fig. S5). It is not clear why MS-BAND does not correlate with a marker of replication fork reversal in bacteria. There are certain possible limitations of using the regressed fork marker as a readout for replication alterations in bacteria. First, the fork reversal marker would not identify an increase in overall replication rate. Second, the fork regression marker can only detect slowing in replication if it leads to fork reversal, but replication fork slowing or initiation inhibition may not always produce a reversed fork intermediate in bacteria. Further work is necessary to understand these complex phenotypic relationships.
While MS-BAND has several advantages to the DNA fiber, it also has a number of limitations. One disadvantage of the MS-BAND technique is the loss of information gleaned from a single-molecule assay. For example, events such as fork asymmetry and frequency of origin firing necessitate single-molecule DNA fiber analysis, and we cannot envision a modification of MS-BAND that would make this possible. MS-BAND is also not ideal for analyzing fork restart, as this typically requires analysis of single molecules that have incorporated two analogs sequentially on the same strand of DNA as an indication of a restarted fork. With our global analysis of nascent DNA synthesis using MS-BAND, in a fork restart replication scheme, we cannot rule out the possibility that some of the second analog signal is the result of new origin firing, as opposed to replication restart of a previously replicating DNA. Modification of MS-BAND may be able to overcome this issue. Specifically, the cells can be labeled with EdU followed by the addition of desired replication inhibiting drug, and then finally a pulse of IdU or CldU is added. Isolation of this EdU-labeled DNA and performing MS-BAND for the IdU or CldU would give a more specific readout of restart, while depletion of this EdU-labeled material and doing MS-BAND on the unbound material would give a readout of new origin firing. Further work is required to determine whether such an approach is feasible. Another disadvantage is the high up-front cost of the LC-MS/MS, although a triple quadrupole machine is relatively inexpensive compared to other quantitative MS technologies. From our E. coli DDP screen, defects that only indirectly impact replication rates will also be detected, requiring further workup to determine the precise role or phenotype in DNA replication.
Besides what we have presented here, we envision that MS-BAND may have other potential applications. Technically, there is no reason that MS-BAND cannot be readily applied to yeast or other model organisms. While we chose IdU and CldU because they are the most commonly used thymidine analogs for fiber analysis, any nucleoside can be detected and quantified by this type of mass spectrometer. In another potential application, it should be possible to perform chromatin immunoprecipitation of a telomere-associated protein after an IdU or CldU pulse to determine replication at telomeric regions. As suggested earlier, ribonucleoside incorporation or incorporation of damaged nucleosides can also be coupled to the MS-BAND approach. Indeed, we have shown this type of MS can be used to demonstrate association of an RNA-binding protein with alkylated ribonucleosides upon methyl methanesulfonate–mediated damage (Tsao et al., 2021; Tsao et al., 2022). Further development of MS-BAND will be necessary to demonstrate its utility, which we reason may be quite broad.
Materials and methods
Cell lines and cell culture
HeLa and HEK293T cells were cultured in DMEM media supplemented with 10% FBS and 100 U/ml penicillin and 100 μg/ml streptomycin (1%) at 37°C with 5% CO2. AsiSI-Ind MCF10A cells (a gift from Roger Greenberg, University of Pennsylvania, Philadelphia, PA, USA) were cultured in DMEM-F12 HEPES (Gibco) supplemented with 5% horse serum (Gibco), 20 ng/ml hEGF (Sino Biologicals), 0.5 mg/ml hydrocortisone (Sigma-Aldrich), 100 ng/ml cholera toxin (Sigma-Aldrich), 10 μg/ml insulin (BOC Sciences), 2 mM l-glutamine (Gibco), 100 U/ml penicillin–streptomycin (Gibco), 0.1 mM MEM non-essential amino acids (Gibco), and 200 μM uridine (Sigma-Aldrich). AsiSI expression was induced with 2.0 μM 4-OHT (Sigma-Aldrich) and 3.0 μM Shield-1 (Takara Bio) for the indicated length of time. The BRCA1-null ovarian cancer cells UWB1.289 and its BRCA1 complemented derivative, UWB1.289 + BRCA1 (kindly provided by Dr. Lee Zou, Harvard Medical School, Boston, MA, USA; Lemaçon et al., 2017), were cultivated in 50% MEGM BulletKit (Lonza CC-3150), 50% RPMI media, supplemented with 5% FBS, 100 U/ml penicillin, and 100 mg/ml streptomycin at 37°C with 5% CO2. The culture media for UW + BRCA1 cells was supplemented with 400 mg/ml of G418 (Millipore Sigma).
The E. coli cell strains used in this study were as follows: The temperature-sensitive replication mutants were FC40 derivatives (McKenzie et al., 2000), including FC40 dnaEts486 zae∷Tn10d-Cam (Hastings et al., 2010), FC40 dnaBts22 malB::Tn9 (Tarkowski et al., 2002), and FC40 dnaA46(ts) ∆tnaA::FRTKanFRT (Xia et al., 2016).
Lentiviral delivery
The bacterial glycerol stocks harboring sequence-verified shRNA lentiviral plasmid vectors for human genes were selected from MISSION shRNA Library (Sigma). Specific clones used for depletion of PolG were TRCN0000296704 (shRNA sequence: 5′-TTCCTTTGACCGAGCTCATAT-3′) and TRCN0000290536 (shRNA sequence: 5′-GCGCTTACTAATGCAGTTTAA-3′). Replication incompetent viral particles were produced in packaging cells (HEK293T) by co-transfection with compatible packaging plasmids. Subsequently, target cell lines were transduced using supernatant from transfected HEK 293T cells, in the presence of 8 μg/ml polybrene (Sigma-Aldrich). Cells were selected with 1 μg/ml puromycin for 5 d and recovered 1 additional day before analysis by Western blot. Scrambled shRNA vector was used as a negative control (SHC002 from Sigma-Aldrich; shRNA sequence: 5′-CAACAAGATGAAGAGCACCAA-3′).
MS-BAND and DNA fiber reagents
dG, IdU, and CldU were all purchased from Sigma-Aldrich. Nucleoside standards were dissolved in tissue culture-grade H2O and filtered with a 0.22-µm filter for LC-MS analysis. For DNA labeling, IdU and CldU stocks were dissolved in tissue culture-grade H2O and filtered with a 0.45-µm filter prior to use. All nucleosides were quantified using an Agilent 1290 Infinity II UHPLC coupled to an Agilent 6470 ESI triple-quadrupole mass spectrometer operated under SIM mode. The UHPLC was fitted with a Zorbax Eclipse Plus C18 reverse phase column (2.1 × 50 mm, 1.8 µm; Agilent) operating at 35°C. The injection volume was 10 µl. The gradient used contained solvent A (water, 0.1% formic acid) and B (acetonitrile, 0.1% formic acid) starting at 2% B for 8 min, ramping to 10% over 2 min, then to 20% in 1 min, to 50% in 1 min, ramping to 90% in 1 min, and holding for an additional 1 min at 90% B. The column was then re-equilibrated to 2% B for 20 s. The flow rate was held at 0.5 ml/min. MS detection was performed using negative electrospray ionization, monitoring the parent ions for each compound (dG, 266; CldU, 261; IdU, 353).
DNA fiber analysis
Unless otherwise indicated, cells were pulse-labeled with 20 µM IdU for the indicated time, washed twice with PBS, then pulse-labeled with 200 µM CldU for the indicated time, followed by two washes with PBS. In the case of fork slowing experiments (using HU, CPT, or DOXO), CldU was added concomitantly with the indicated doses of each inhibitor. In the case of fork degradation experiments, cells were pulse-labeled with 20 µM IdU for 30 min, washed twice with PBS, then pulse-labeled with 200 µM CldU for 40 min, followed by two washes with PBS. The cells were then incubated for 4 h with 4 mM HU.
2 µl of cells were mixed with 6 µl of lysis buffer (200 mM Tris-HCl, pH 7.5, 50 mM EDTA, 0.5% SDS in water) on top of a positively charged glass slide. After 5 min incubation at room temperature, slides were tilted at a 20–45° angle to spread the fibers at a constant, low speed. After air drying for 10–15 min at room temperature, DNA was fixed onto the slides with a freshly prepared solution of methanol:glacial acetic acid at 3:1 for 5 min, then stored at 4°C overnight. For immuno-staining of DNA fibers, DNA was washed with PBS, then denatured with 2.5 M HCl for 1 h at room temperature. Slides were then washed with PBS three times and blocked with 5% BSA at 37°C for 1 h. DNA fibers were immuno-stained with mouse-anti-BrdU (347580; 1:20, BD Biosciences) and rat anti-BrdU (Ab6326; 1:100, Abcam) for 1.5 h at room temperature, and washed three times with PBS-0.1%Tween-20 for 5 min. The slides were then incubated with secondary antibodies; anti-rat Alexa Fluor 488 and anti-mouse Alexa Fluor 568 (1:100 each; A21470 and A21123; Thermo Fisher Scientific, respectively) for 1 h at room temperature, and then washed with PBS-0.1%Tween-20 three times. Slides were put in PBS before mounting with Prolong Gold Antifade Reagent (P36930; Thermo Fisher Scientific; Quinet et al., 2020). Images were acquired with either LAS AF software using a Leica DMi8 confocal microscope with a 40×/1.15 oil immersion objective, or cellSens Dimension software using an Olympus BX-53 fluorescence microscope with a UPlanS-Apo 100×/1.4 oil immersion objective. At least 10 images were taken across the whole slide using only one channel to select the regions for the images in order to avoid potential bias. At least 100–200 individual tracts were scored for each dataset. Only DNA fiber tracts where the beginning and end of each labeling was unambiguously defined were analyzed. IdU and CldU tracts were only measured on forks characterized by contiguous IdU–CldU signals (i.e., progressing replication forks). The length of each tract was measured manually using ImageJ software. The pixel values were converted into micrometers using the microscope software scale bar. Size distribution of tract lengths was plotted as scatter dot plot with the line representing the median. Data were pooled from independent experiments, and statistical analysis was performed as indicated in the figure legends. For the fork degradation experiments, nascent DNA degradation was assessed by plotting the CldU/IdU ratio for each individual fiber. Decrease in the median of CldU/IdU distribution reflects degradation of the CldU tracts that were incorporated prior to HU treatment.
Nuclear MS-BAND sample preparation
Cells were cultured in 10-cm plates to 70–90% confluency and labeled with the two thymidine analogs: 20 μM IdU, followed by 200 μM CldU for the indicated times according to the labeling and treatment scheme used. Cells were washed 2–3 times with either pre-warmed 1X PBS or regular media between labeling and/or drug treatment incubations. After labeling, the cells were harvested and resuspended in 10-ml cold 1X PBS. Where required a small aliquot of each sample was used to perform DNA fiber analysis. Cells for MS-BAND analysis were then pelleted and DNA was purified using the DNeasy blood and tissue kit (Qiagen) according to the manufacturer’s protocol. DNA (5 μg) was then digested with DNA Degradase Plus (Zymo research) or nucleoside digestion mix (NEB) overnight at 37°C according to the manufacturer’s protocol. Digested samples were filtered through a 0.22-µm filter (Millipore) and analyzed for IdU and CldU ion intensity by LC-MS/MS. Samples were diluted 1:100 for LC-MS measurement of dG content, which was used to normalize analog ion intensities.
Mito-MS-BAND sample preparation
Human tissue culture cells were labeled with IdU or CldU at 100 μM for the indicated times according to the labeling and treatment scheme used. 1 15-cm plate of cells at 70–90% confluency was used per sample. After labeling, the cells were harvested and resuspended in 10 ml cold 1X PBS. Mitochondrial isolation was performed using the Qproteome mitochondria isolation kit (Qiagen) according to manufacturer’s instructions. The mitochondria were then resuspended in 55 Kunitz units of DNase (Qiagen) for 20 min, and then washed twice by PBS. mtDNA was then extracted using DNeasy blood and tissue kit (Qiagen). The purity of the mtDNA was assessed by quantitative PCR (SYBR Green method) comparing mtDNA to nuclear encoded GAPDH. For mtDNA, the forward and reverse primer sequences were: 5′-TCACCCCGCTAAATCCCCTA-3′ and 5′-TGACGTGAAGTCCGTGGAAG-3′, respectively; for GAPDH, the forward and reverse primer sequences were: 5′-TCAGTTGCAGCCATGCCTTA-3′ and GCGCCCAATACGACCAAATC. Similar to nuclear MS-BAND, DNA was then digested overnight and analyzed for IdU and CldU ion intensity by LC-MS.
Western blotting
The cells were collected and lysed in lysis buffer (40 mM Tris-HCL, pH 7.5, 1% Triton X-100, 100 mM NaCl, 1 mM DTT, and protease inhibitors). Protein concentration was determined using the Bradford assay. Equal amounts of total protein were boiled with SDS-PAGE sample buffer at 95°C for 5 min and 50 µg of protein was loaded for separation by 12% SDS-PAGE. Electrophoresed proteins were then transferred onto polyvinylidene difluoride membranes (Millipore). The membranes were blocking in 5% non-fat milk for 1 h. Then, the membranes were incubated with primary antibodies; rabbit anti-PolG (PA5-21314; Thermo fisher Scientific), mouse anti-DD tag (631073; Takara), and rabbit anti-GAPDH (ab9485; Abcam) for 1 h at room temperature. After three washes in PBS with 0.05% Tween-20 (PBS-T), membranes were then incubated with anti-rabbit or anti-mouse HRP-linked secondary antibodies (7074 and 7076; Cell Signaling, respectively) for 1 h at room temperature. After three washes in PBS-T, visualization was performed by ECL detection reagent (1705061; Bio-Rad). Imaging was performed using the ChemiDoc system (Bio-Rad).
mtDNA slot blot analysis
DNA was labeled with CldU for 1 h, then the cells were pelleted and mtDNA was purified as described above. DNA concentration was measured by Qubit dsDNA HS assay kit (Q32854; Thermo fisher Scientific) and Qubit Flex Fluorometer apparatus (Thermo Fisher Scientific). Equal amount of DNA from each sample was denatured in buffer (0.4 M NaOH and 10 mM EDTA) for 10 min at 100°C. The solution was rapidly neutralized by equal volume of 2 M ammonium acetate, pH 7. Then, the ssDNA was then applied onto nitrocellulose membrane using a slot blot apparatus (Bio-Rad). The membrane was washed by Tris-EDTA buffer (10 mM Tris-HCl, and 1 mM EDTA, pH 8), then blocked with 5% milk. The membrane was then incubated with rat anti-BrdU (Ab6326; Abcam) for 1 h. After three washes with Tween 20-PBS, the membrane was incubated with anti-rabbit HRP-linked secondary antibody (7074; Cell Signaling) for 1 h at room temperature. After three washes in PBS-T, visualization was performed by ECL detection reagent (1705061; Bio-Rad). Imaging was performed using the ChemiDoc system (Bio-Rad).
Bacterial MS-BAND sample preparation
E. coli DH10β, derivative of K12, was grown in Luria Bertani Herskowitz rich medium. Unless otherwise indicated, the liquid cultures were treated with IdU or CldU at 100 μM for the indicated times according to the described labeling and treatment scheme. The cells were then pelleted and processed for genomic DNA purification using DNeasy blood and tissue kit (Qiagen). Similar to nuclear MS-BAND, DNA was then digested overnight and analyzed for IdU and CldU ion intensity by LC-MS.
Screening of E. coli DDP library
The E. coli gene library has been previously described (Xia et al., 2019). Each strain was grown on M9-glucose media (1× M9 salt [Sigma-Aldrich], 0.1% glucose, 0.4 mM CaCl2, 1 mM MgSO4, 10 μg/ml Vitamin B1, and 20 μg/ml carbenicillin) plates and incubated at 37°C overnight. A single colony was picked from each plate and used for culturing a 5 ml M9-glucose media in Falcon tubes and incubated at 37°C overnight. The cultures were diluted to 1:100 in 5 ml M9-glycerol media (1× M9 salt [Sigma-Aldrich], 0.1% glycerol, 0.4 mM CaCl2, 1 mM MgSO4, 10 μg/ml Vitamin B1, and 20 μg/ml carbenicillin) in Falcon tubes and incubated at 37°C for 10 additional hours. IPTG was added for 6 h at a final concentration of 100 μM to induce target gene. The cultures were further diluted to 1:5 in M9-glycerol media. CldU was added to a final concentration of 100 μM and incubated for 30–40 min at 37°C. The liquid cultures were collected by centrifugation followed by DNA extraction and degradation, and then applied to LC-MS/MS. Validation experiments were performed using the same method in biological triplicate.
Statistical methods
Statistical significances were measured by one-way ANOVA, Student’s t test, or Mann–Whitney U-test as indicated in the figure legends. When t-tests were used, data distribution was assumed to be normal, but this was not formally tested. Correlation analysis using the bacterial library was performed using the Z-values of seven functional assays of the clones, as determined in a previous publication (Xia et al., 2019). Pearson correlation coefficient r and P values were calculated for each of the seven functional assays with MS-BAND measurements. r > 0.2 and P < 0.001 are used as a cutoff for correlations.
Online supplemental material
Fig. S1 shows material related to Fig. 1 (LC-MS/MS chromatograms of nucleoside standards, LC and MS characteristics of nucleosides, quantitative standard curves, and detection of nascent DNA using MS-BAND). Fig. S2 shows material related to Fig. 2 (MS-BAND based quantitation of thymidine analogue incorporation after DNA polymerase inhibition with aphidicolin). Fig. S3 shows material related to Fig. 4 (mtDNA purification, IdU and CldU incorporation into mtDNA as determined by MS-BAND, validation of PolG phenotype using slot blotting, and mito-MS-BAND phenotype of cells treated with EtBr). Fig. S4 is related to Fig. 5 (IdU and CldU incorporation into bacterial genomic DNA as determined by MS-BAND, and optical densities of control bacteria and temperature-sensitive mutant strains). Fig. S5 is related to Fig. 6 (validation of MS-BAND results in bacterial strains using a shorter thymidine analog incubation time and lack of correlation between MS-BAND and specific markers of DNA damage responses in bacteria). Table S1 is related to Fig. 6 (Z-scores of bacterial MS-BAND screen).
Acknowledgments
We thank Peter Burgers, Ben Garcia, Hani Zaher, John Tainer, and members of the Mosammaparast and Vindigni labs for discussion of this work. We thank Roger Greenberg for the inducible AsiSI MCF10A cell line.
This work was supported by a fellowship from the American Italian Cancer Foundation (to A. Meroni), the Alvin J. Siteman Cancer Center Siteman Investment Program (supported by The Foundation for Barnes-Jewish Hospital, Cancer Frontier Fund, to A. Vindigni and N. Mosammaparast), the U.S. Department of Defense Breast Cancer Research Program Expansion Award (BC191374 to A. Vindigni), the State of Nebraska (LB595 and LB692 to J. Xia) the National Institutes of Health (K99 ES033259 to J. Xia; R01 CA250905 and DP1 AG072751 to S.M. Rosenberg; R35 GM139508 to R. Galletto; R01 CA237263 and R01 CA248526 to A. Vindigni; R01 CA193318 and P01 CA092584 to N. Mosammaparast), an American Cancer Society Research Scholar Award (RSG-18-156-01-DMC to N. Mosammaparast), the Centene Corporation (to N. Mosammaparast), and the Barnard Foundation (to A. Vindigni and N. Mosammaparast).
Author contributions: M.E. Ashour, A. K. Byrum, and N. Mosammaparast conceived the project. A. K. Byrum initiated MS-BAND optimization and feasibility experiments in human cells. M.E. Ashour performed MS-BAND experiments. M.E. Ashour, A. K. Byrum, and A. Meroni performed DNA fiber experiments. J. Xia, S. Singh, and R. Galletto contributed key reagents. M.E. Ashour, A. Meroni, and J. Xia analyzed the data with guidance from S. M. Rosenberg, A. Vindigni, and N. Mosammaparast. S. M. Rosenberg supervised J. Xia. A. Vindigni supervised A. Meroni. R. Galletto supervised S. Singh. N. Mosammaparast supervised M.E. Ashour and A. K. Byrum. N. Mosammaparast and M.E. Ashour wrote the manuscript with input from all other authors.
References
Author notes
Disclosures: The authors declare no competing interests exist.