Mitochondria are highly dynamic double membrane–bound organelles that maintain their shape in part through fission and fusion. Mitochondrial fission is performed by a dynamin-related protein, Dnm1 (Drp1 in humans), that constricts and divides the mitochondria in a GTP hydrolysis–dependent manner. However, it is unclear whether factors inside mitochondria help coordinate the process and if Dnm1/Drp1 activity is sufficient to complete the fission of both mitochondrial membranes. Here, we identify an intermembrane space protein required for mitochondrial fission in yeast, which we propose to name Mdi1 (also named Atg44). Loss of Mdi1 causes mitochondrial hyperfusion due to defects in fission, but not the lack of Dnm1 recruitment to mitochondria. Mdi1 is conserved in fungal species, and its homologs contain an amphipathic α-helix, mutations of which disrupt mitochondrial morphology. One model is that Mdi1 distorts mitochondrial membranes to enable Dnm1 to robustly complete fission. Our work reveals that Dnm1 cannot efficiently divide mitochondria without the coordinated function of Mdi1 inside mitochondria.
Introduction
Mitochondria are multifunctional double membrane–bound organelles distributed throughout cells as part of a dynamic network of tubules (Friedman and Nunnari, 2014). Mitochondrial subcellular localization is in part controlled by the ability of mitochondria to fuse together or undergo fission. These opposing processes are in balance at steady state, and disruption of either fusion or fission leads to altered distribution of mitochondria (Quintana-Cabrera and Scorrano, 2023). Mitochondrial fusion and fission are mediated by conserved members of a family of large GTPases, the dynamin-related proteins (DRPs; Gao and Hu, 2021; Kraus et al., 2021). Fusion requires the coordinated sequential action of DRPs on the outer mitochondrial membrane (OMM) and inner mitochondrial membrane (IMM). During fission, a DRP (Dnm1 in yeast, Drp1 in humans) is recruited to the OMM by receptor proteins, assembles in an oligomeric structure that circumscribes mitochondria, and constricts and divides the membranes via a confirmational change driven by GTP hydrolysis (Kraus et al., 2021).
Several factors contribute to the spatial regulation of mitochondrial division. Inter-organelle contacts, primarily between the ER and mitochondria, mark sites of membrane preconstriction that precedes fission in both yeast and humans (Abrisch et al., 2020; Friedman et al., 2011; Guo et al., 2018; Kleele et al., 2021). Additionally, sites of replication of mitochondrial DNA (mtDNA), which occurs in the matrix compartment interior to the IMM, are linked to division sites in human cells (Lewis et al., 2016). However, it is unknown how mitochondrial division is spatially coordinated across both membranes and whether factors inside mitochondria play a direct role. Additionally, it has remained an open question whether the activity of the Dnm1/Drp1 division GTPase is sufficient to fully drive the fission of both mitochondrial membranes, whether a separate IMM fission machinery exists, or if other factors assist in completing membrane scission (Anand et al., 2014; Chakrabarti et al., 2018; Fonseca et al., 2019; Kamerkar et al., 2018; Klecker et al., 2015; Lee et al., 2016). For example, the yeast IMM protein Mdm33 has been implicated as an auxiliary mediator of mitochondrial division, though its absence does not lead to mitochondrial morphology defects that phenocopy loss of other known mitochondrial fission proteins (Klecker et al., 2015; Messerschmitt et al., 2003).
Here, we characterize the mitochondrial intermembrane space (IMS) yeast protein mitochondria class one 8 kD (Mco8; named for its molecular weight and its high confidence localization to mitochondria [Morgenstern et al., 2017]). We show that Mco8 can be spatially linked to sites of Dnm1-dependent mitochondrial fission and that its loss leads to a disruption of mitochondrial morphology due to the inability of Dnm1 to complete the scission of the organelle. While this work was in revision, a study revealed a requirement for Mco8 for normal mitochondrial morphology and fission associated with mitophagy, leading the authors to name the protein Atg44 (Fukuda et al., 2023). However, based on its role in Dnm1-dependent mitochondrial fission, we propose the more general name Mitochondrial Division IMS 1 (Mdi1). Mdi1 is characterized by a structurally conserved putative amphipathic α-helix, and mutations in this motif prevent its function in maintaining normal mitochondrial morphology. These data are consistent with a model that Mdi1 locally binds and/or distorts mitochondrial membranes from the inside of the organelle, enabling Dnm1 to complete fission. Thus, we have identified the first mitochondrial IMS-localized factor directly involved in the process of mitochondrial division.
Results and discussion
Mitochondrial division is perturbed in the absence of Mdi1
To explore a functional role for the IMS protein Mdi1, we examined the mitochondrial morphology of ∆mdi1 yeast cells expressing the matrix marker mito-dsRed by fluorescence microscopy. In contrast to the tubular morphology observed in wild-type cells, mitochondria in ∆mdi1 cells had an aberrant net-like appearance, phenocopying cells deficient for the mitochondrial division DRP, Dnm1 (∆dnm1), or its OMM receptor, Fis1 (∆fis1; Fig. 1, A and B). Tubular mitochondrial networks are formed by a balance between opposing fission and fusion processes (Bleazard et al., 1999; Nunnari et al., 1997; Sesaki and Jensen, 1999), and we reasoned that the net-like morphology caused by the loss of Mdi1 could be due either to a deficiency in mitochondrial fission or excessive mitochondrial fusion. To address this, we utilized a temperature-sensitive allele of the gene encoding the mitochondrial OMM fusion DRP Fzo1 (fzo1-1; Hermann et al., 1998). Consistent with published observations, the growth of fzo1-1 cells at a non-permissive temperature (37°C) caused a deficiency in Fzo1-dependent mitochondrial fusion, and within 20 min, mitochondria appeared fragmented and/or aggregated (Fig. 1, C and D). We then asked how the loss of Dnm1 or Mdi1 impacted mitochondrial morphology in the fzo1-1 yeast strain. As expected, mitochondria in ∆dnm1 fzo1-1 cells appeared tubular and/or net-like, even at elevated temperatures (Fig. 1, C and D; Mozdy et al., 2000). Remarkably, ∆mdi1 fzo1-1 cells similarly maintained interconnected mitochondria in nearly all cells (Fig. 1, C and D). These data suggest that Mdi1 plays a positive role in mitochondrial division.
Mitochondrial division is perturbed in the absence of Mdi1. (A) Representative single-plane deconvolved epifluorescence microscopy images of the indicated budding yeast strains expressing the mitochondrial matrix marker mito-dsRed. (B) A graph of the categorization of mitochondrial morphology from cells as in A. Data shown represent at least 75 cells per strain in each of the three independent experiments, and bars indicate SEM. Asterisks (***P < 0.001) represent unpaired two-tailed t tests. (C) Maximum intensity projections of epifluorescence microscopy images of the indicated strains expressing mito-dsRed that were grown to log phase at room temperature and imaged before (top) and after (bottom) growth at 37°C for 20 min. (D) A graph depicting the categorization of mitochondrial morphology as in C. Data shown represent at least 70 cells per strain in each of the three independent experiments and bars indicate SEM. Asterisks (***P < 0.001) represent unpaired two-tailed t tests. (E) Serial dilutions of the indicated yeast cells were plated on media containing the non-fermentable carbon source ethanol/glycerol (YPEG) and grown at the indicated temperatures. Cell boundaries are indicated with dotted lines. Scale bars = 2 µm (A), 3 µm (C). See also Fig. S1.
Mitochondrial division is perturbed in the absence of Mdi1. (A) Representative single-plane deconvolved epifluorescence microscopy images of the indicated budding yeast strains expressing the mitochondrial matrix marker mito-dsRed. (B) A graph of the categorization of mitochondrial morphology from cells as in A. Data shown represent at least 75 cells per strain in each of the three independent experiments, and bars indicate SEM. Asterisks (***P < 0.001) represent unpaired two-tailed t tests. (C) Maximum intensity projections of epifluorescence microscopy images of the indicated strains expressing mito-dsRed that were grown to log phase at room temperature and imaged before (top) and after (bottom) growth at 37°C for 20 min. (D) A graph depicting the categorization of mitochondrial morphology as in C. Data shown represent at least 70 cells per strain in each of the three independent experiments and bars indicate SEM. Asterisks (***P < 0.001) represent unpaired two-tailed t tests. (E) Serial dilutions of the indicated yeast cells were plated on media containing the non-fermentable carbon source ethanol/glycerol (YPEG) and grown at the indicated temperatures. Cell boundaries are indicated with dotted lines. Scale bars = 2 µm (A), 3 µm (C). See also Fig. S1.
Mitochondrial fusion is required for the maintenance of mtDNA in yeast and is therefore required for mitochondrial respiration (Hermann et al., 1998). Thus, fzo1-1 cells are inviable at elevated temperatures when grown on media containing a carbon source that requires respiration (ethanol/glycerol; Fig. 1 E; Hermann et al., 1998). However, simultaneous loss of both division and fusion machinery prevents mitochondrial fragmentation and allows for genome maintenance, albeit with an increased rate of mtDNA mutation and loss (Bleazard et al., 1999; Mozdy et al., 2000; Osman et al., 2015; Tieu and Nunnari, 2000). We therefore asked whether ∆mdi1 fzo1-1 cells were viable on respiration-requiring media at a non-permissive temperature. We found that, unlike ∆dnm1 fzo1-1 cells, ∆mdi1 fzo1-1 cells were inviable at 37°C (Fig. 1 E). However, loss of Mdi1 promoted growth to fzo1-1 cells at an intermediate temperature of 32°C. These data suggest that in cells lacking Mdi1, mitochondrial fission is deficient, though may not be completely absent.
Loss of mitochondrial membrane potential has previously been established to induce rapid Dnm1-dependent mitochondrial fission in yeast (Hughes et al., 2016; Klecker et al., 2015). As a complementary approach, we asked whether loss of Mdi1 prevented mitochondrial fragmentation induced by treatment with the uncoupler CCCP. As expected, mitochondria in the majority of wild-type cells appeared fragmented after CCCP treatment (25 µM, 45 min; Fig. S1). In contrast, loss of Dnm1 prevented mitochondrial fragmentation in most cells. Notably, mitochondria in ∆dnm1 cells treated with CCCP frequently appeared hyperconstricted with a beads-on-a-string appearance, indicative of Dnm1-independent mitochondrial constriction (Legesse-Miller et al., 2003; Fig. S1, see arrows). As in ∆dnm1 cells, loss of Mdi1 appeared to largely prevent mitochondrial fragmentation, though not hyperconstriction, upon CCCP treatment (Fig. S1, see arrows). Altogether, these data indicate that loss of Mdi1 leads to hyperfused mitochondria due to deficiency in mitochondrial fission.
Mdi1 is required for CCCP-induced mitochondrial division. (A) Maximum intensity projections of deconvolved images of the indicated yeast strains expressing mito-dsRed before (top) or after (bottom) treatment with 25 µM CCCP for 45 min. Arrows mark mitochondrial hyper-constrictions that occur after CCCP treatment. (B) A graph depicting the categorization of mitochondrial morphology of cells as in A. Data shown represent at least 75 cells per strain in each of the three independent experiments and bars indicate SEM. Asterisks (***P < 0.001) represent unpaired two-tailed t tests. Cell boundaries are indicated with dotted lines. Scale bar = 3 µm.
Mdi1 is required for CCCP-induced mitochondrial division. (A) Maximum intensity projections of deconvolved images of the indicated yeast strains expressing mito-dsRed before (top) or after (bottom) treatment with 25 µM CCCP for 45 min. Arrows mark mitochondrial hyper-constrictions that occur after CCCP treatment. (B) A graph depicting the categorization of mitochondrial morphology of cells as in A. Data shown represent at least 75 cells per strain in each of the three independent experiments and bars indicate SEM. Asterisks (***P < 0.001) represent unpaired two-tailed t tests. Cell boundaries are indicated with dotted lines. Scale bar = 3 µm.
Mdi1 is a soluble IMS protein
Previous work determined Mdi1 submitochondrial localization using a protease protection assay that indicated that the protein resides in the IMS (Morgenstern et al., 2017). As Mdi1 is not suggested to have transmembrane domains according to hydrophobicity/transmembrane prediction programs such as TMHMM and Phobius (Käll et al., 2004; Krogh et al., 2001), Mdi1 likely influences Dnm1-dependent mitochondrial fission from inside the organelle. To our knowledge, depletion of no other internal mitochondrial factor leads to a net-like mitochondrial morphology that phenocopies loss of Dnm1, which motivated us to confirm its localization.
Several different chromosomally integrated C-terminal fusions to Mdi1 led to mitochondrial morphology defects similar to ∆mdi1 cells, indicating that these tags interfere with its function. Therefore, to confirm the localization of Mdi1, we generated an in-frame 2xFLAG tag fusion (Mdi1*-2xFLAG) in an unstructured region of the protein based on AlphaFold2 predictions (Fig. S2 A; Jumper et al., 2021; Varadi et al., 2022). This fusion, when chromosomally integrated and expressed using the native MDI1 promoter, was capable of fully rescuing the mitochondrial morphology defect of ∆mdi1 cells (Fig. S2 B). Treatment of mitochondria isolated from Mdi1*-2xFLAG expressing cells with an alkaline solution of sodium carbonate indicates that Mdi1 could readily be extracted from membranes, suggesting it is not a membrane-integral protein (Fig. S2 C). Further, consistent with published work, a protease protection analysis suggests that Mdi1*-2xFLAG is protected from proteolytic cleavage unless the OMM is disrupted, similar to the soluble IMS protein Mia40 (Fig. S2 D; Chacinska et al., 2004). Thus, these data indicate that Mdi1 is a soluble, IMS-localized protein.
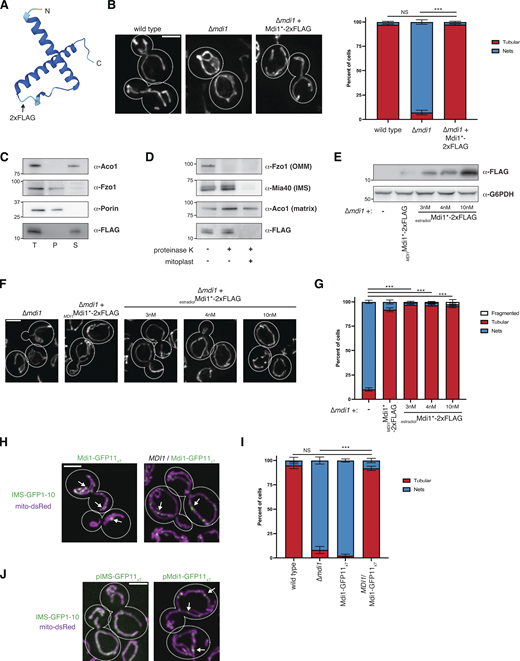
Mdi1 is a soluble intermembrane space protein that concentrates at discrete submitochondrial foci. (A) AlphaFold2 predicted structure of Mdi1 (Jumper et al., 2021; Varadi et al., 2022). Arrow marks the site of insertion of the internal 2xFLAG tag. (B) Left: Representative deconvolved maximum intensity projections of the indicated yeast strains expressing mito-DsRed. Right: A graph depicting categorization of mitochondrial morphology of cells as shown on left. Data shown represent at least 75 cells per strain in each of three independent experiments and bars indicate SEM. Asterisks (***P < 0.001) represent unpaired two-tailed t test. NS indicates not statistically significant. (C) Western analysis with the indicated antibodies of mitochondria isolated from a ∆mdi1 strain expressing Mdi1*-2xFLAG and subjected to alkaline extraction. Crude membranes were incubated with 0.1 M Na2CO3 pH11.5, and total (T), pellet (P), and supernatant (S) fractions were collected after centrifugation. (D) Western analysis with the indicated antibodies of mitochondria isolated from a ∆mdi1 strain expressing Mdi1*-2xFLAG and subjected to protease protection analysis. Mitochondria were treated where indicated with proteinase K. Mitoplast sample indicates selective disruption of the OMM by a combination of osmotic swelling and mechanical disruption. (E) Western analysis of whole-cell lysates of the indicated yeast strains grown in SCD media and expressing Mdi1*-2xFLAG driven by the indicated promoter and treated as indicated with β-estradiol. (F) Maximum intensity projections of deconvolved images of cells expressing mito-dsRed and grown as in E. (G) A graph depicting categorization of mitochondrial morphology from the indicated strains as in E and F. Data shown represent at least ∼75 cells per strain in each of three independent experiments and bars indicate SEM. Asterisks (***P < 0.001) represent unpaired two-tailed t tests. (H) Maximum intensity projections of confocal images of chromosomally tagged Mdi1-GFP11x7 haploid cells (left) or heterozygous diploid cells (right) cells coexpressing IMS-targeted GFP1-10 (green) and mito-dsRed (magenta). Arrows mark focal assemblies of Mdi1-GFP11x7. (I) A graph depicting the categorization of mitochondrial morphology from the indicated strains as in H. Data shown represent at least 75 cells per strain in each of three independent experiments and bars indicate SEM. Asterisks (***P < 0.001) represent unpaired two-tailed t test. NS indicates not statistically significant. (J) Maximum intensity projections of confocal images of wild-type cells co-expressing mito-dsRed, IMS-targeted GFP1-10, and plasmid-borne IMS-GFP11x7 (left) or chromosomally integrated Mdi1-GFP11x7 (right). Arrows mark focal accumulations of Mdi1-GFP11x7. Cell boundaries are indicated with dotted lines. Scale bar = 3 µm. Source data are available for this figure: SourceData FS2.
Mdi1 is a soluble intermembrane space protein that concentrates at discrete submitochondrial foci. (A) AlphaFold2 predicted structure of Mdi1 (Jumper et al., 2021; Varadi et al., 2022). Arrow marks the site of insertion of the internal 2xFLAG tag. (B) Left: Representative deconvolved maximum intensity projections of the indicated yeast strains expressing mito-DsRed. Right: A graph depicting categorization of mitochondrial morphology of cells as shown on left. Data shown represent at least 75 cells per strain in each of three independent experiments and bars indicate SEM. Asterisks (***P < 0.001) represent unpaired two-tailed t test. NS indicates not statistically significant. (C) Western analysis with the indicated antibodies of mitochondria isolated from a ∆mdi1 strain expressing Mdi1*-2xFLAG and subjected to alkaline extraction. Crude membranes were incubated with 0.1 M Na2CO3 pH11.5, and total (T), pellet (P), and supernatant (S) fractions were collected after centrifugation. (D) Western analysis with the indicated antibodies of mitochondria isolated from a ∆mdi1 strain expressing Mdi1*-2xFLAG and subjected to protease protection analysis. Mitochondria were treated where indicated with proteinase K. Mitoplast sample indicates selective disruption of the OMM by a combination of osmotic swelling and mechanical disruption. (E) Western analysis of whole-cell lysates of the indicated yeast strains grown in SCD media and expressing Mdi1*-2xFLAG driven by the indicated promoter and treated as indicated with β-estradiol. (F) Maximum intensity projections of deconvolved images of cells expressing mito-dsRed and grown as in E. (G) A graph depicting categorization of mitochondrial morphology from the indicated strains as in E and F. Data shown represent at least ∼75 cells per strain in each of three independent experiments and bars indicate SEM. Asterisks (***P < 0.001) represent unpaired two-tailed t tests. (H) Maximum intensity projections of confocal images of chromosomally tagged Mdi1-GFP11x7 haploid cells (left) or heterozygous diploid cells (right) cells coexpressing IMS-targeted GFP1-10 (green) and mito-dsRed (magenta). Arrows mark focal assemblies of Mdi1-GFP11x7. (I) A graph depicting the categorization of mitochondrial morphology from the indicated strains as in H. Data shown represent at least 75 cells per strain in each of three independent experiments and bars indicate SEM. Asterisks (***P < 0.001) represent unpaired two-tailed t test. NS indicates not statistically significant. (J) Maximum intensity projections of confocal images of wild-type cells co-expressing mito-dsRed, IMS-targeted GFP1-10, and plasmid-borne IMS-GFP11x7 (left) or chromosomally integrated Mdi1-GFP11x7 (right). Arrows mark focal accumulations of Mdi1-GFP11x7. Cell boundaries are indicated with dotted lines. Scale bar = 3 µm. Source data are available for this figure: SourceData FS2.
Mdi1 localizes at discrete focal structures that can be spatially linked to sites of Dnm1-marked mitochondrial division
Given the genetic involvement of Mdi1 in mitochondrial fission, we next wanted to ascertain Mdi1 sublocalization within mitochondria and its spatial relationship to division sites. We therefore visualized Mdi1*-2xFLAG by immunofluorescence microscopy. Due to its low expression, Mdi1 could not be robustly detected with this approach when expressed using the native promoter. However, immunolabeled Mdi1*-2xFLAG appeared focal compared with mito-dsRed when mildly overexpressed to levels that did not alter mitochondrial morphology using an estradiol-driven promoter (Fig. 2 A and Fig. S2, E–G).
Mdi1 localizes at discrete focal structures that can be spatially linked to sites of Dnm1-marked mitochondrial division. (A) Maximum intensity projections of confocal images of ∆mdi1 cells co-expressing mito-dsRed (magenta) and Mdi1*-2xFLAG driven by an estradiol-controlled promoter. Cells were grown exponentially in SCD media supplemented with 4 nM β-estradiol, fixed, and immunolabeled with FLAG antibody (green). Arrows mark focal concentrations of Mdi1. (B) A schematic depicting the tandem tag split GFP approach utilized to visualize Mdi1. (C) Maximum intensity projections of confocal fluorescence microscopy images of ∆mdi1 (left) or wild-type (right) cells coexpressing IMS-targeted GFP1-10 and chromosomally integrated Mdi1-GFP11x7 driven by its native promoter (green) and mito-dsRed (magenta). Arrows mark focal accumulations of Mdi1-GFP11x7. (D) A graph depicting the categorization of mitochondrial morphology from the indicated strains as in C. Data shown represent at least 75 cells per strain in each of three independent experiments and bars indicate SEM. Asterisks (***P < 0.001) represent unpaired two-tailed t test. NS indicates not statistically significant. (E) Timelapse microscopy images of wild-type cells expressing Mdi1-GFP11x7 as in C. Images shown are a maximum intensity projection of three confocal images with 0.4 µm z-steps captured at the indicated time intervals. Arrows mark sites of Mdi1-GFP11x7-marked mitochondrial division. (F) Categorization of the frequency that mitochondrial division events captured as in E were spatially linked to Mdi1-GFP11x7 foci. (G) Maximum intensity projection confocal images of MDI1/Mdi1-GFP11x7 heterozygous diploid cells co-expressing IMS-targeted GFP1-10 (green), Dnm1-mCherry (magenta), and mito-TagBFP (blue). White arrows mark Mdi1 focal assemblies that colocalize with Dnm1. Dnm1 and Mdi1 assemblies that do not colocalize are marked with pink and yellow arrows, respectively. (H) Time-lapse microscopy images are shown for cells as in G. Images were captured and displayed as in E. (I) Images as in C of the indicated strains. Cell boundaries are indicated with dotted lines. Scale bars = 2 µm (A, E, G, and H), 3 µm (C–I). See also Fig. S2; and Videos 1, 2, 3, 4, and 5.
Mdi1 localizes at discrete focal structures that can be spatially linked to sites of Dnm1-marked mitochondrial division. (A) Maximum intensity projections of confocal images of ∆mdi1 cells co-expressing mito-dsRed (magenta) and Mdi1*-2xFLAG driven by an estradiol-controlled promoter. Cells were grown exponentially in SCD media supplemented with 4 nM β-estradiol, fixed, and immunolabeled with FLAG antibody (green). Arrows mark focal concentrations of Mdi1. (B) A schematic depicting the tandem tag split GFP approach utilized to visualize Mdi1. (C) Maximum intensity projections of confocal fluorescence microscopy images of ∆mdi1 (left) or wild-type (right) cells coexpressing IMS-targeted GFP1-10 and chromosomally integrated Mdi1-GFP11x7 driven by its native promoter (green) and mito-dsRed (magenta). Arrows mark focal accumulations of Mdi1-GFP11x7. (D) A graph depicting the categorization of mitochondrial morphology from the indicated strains as in C. Data shown represent at least 75 cells per strain in each of three independent experiments and bars indicate SEM. Asterisks (***P < 0.001) represent unpaired two-tailed t test. NS indicates not statistically significant. (E) Timelapse microscopy images of wild-type cells expressing Mdi1-GFP11x7 as in C. Images shown are a maximum intensity projection of three confocal images with 0.4 µm z-steps captured at the indicated time intervals. Arrows mark sites of Mdi1-GFP11x7-marked mitochondrial division. (F) Categorization of the frequency that mitochondrial division events captured as in E were spatially linked to Mdi1-GFP11x7 foci. (G) Maximum intensity projection confocal images of MDI1/Mdi1-GFP11x7 heterozygous diploid cells co-expressing IMS-targeted GFP1-10 (green), Dnm1-mCherry (magenta), and mito-TagBFP (blue). White arrows mark Mdi1 focal assemblies that colocalize with Dnm1. Dnm1 and Mdi1 assemblies that do not colocalize are marked with pink and yellow arrows, respectively. (H) Time-lapse microscopy images are shown for cells as in G. Images were captured and displayed as in E. (I) Images as in C of the indicated strains. Cell boundaries are indicated with dotted lines. Scale bars = 2 µm (A, E, G, and H), 3 µm (C–I). See also Fig. S2; and Videos 1, 2, 3, 4, and 5.
To visualize Mdi1 localization in live cells, we utilized a C-terminal 7x tandem GFP11 tag inserted either at the endogenous chromosomal locus or using an exogenous genome-integrating plasmid. This tag could be visualized by artificially targeting the complementary GFP1-10 to the IMS via the N-terminal sequence of Cyb2 (Fig. 2 B; Beasley et al., 1993; Kamiyama et al., 2016). While Mdi1-GFP11x7 cannot rescue the mitochondrial morphology defect of ∆mdi1 cells, the tagged protein does not adversely affect mitochondrial morphology in the presence of a wild-type copy of the MDI1 gene (Fig. 2, C and D; and Fig. S2, H and I). In cells expressing both wild-type Mdi1 and Mdi1-GFP11x7, each expressed using the native MDI1 promoter, Mdi1-GFP11x7 concentrated in discrete focal structures that were distributed throughout the tubular mitochondrial network (Fig. 2 C and Fig. S2 H, arrows). To further validate that Mdi1 focal structures were not an artifact of a non-functional fusion, we compared Mdi1-GFP11x7 localization with that of GFP11x7 targeted to the IMS by fusion to the N-terminus of Cyb2 (IMS-GFP11x7). When complemented with GFP1-10, IMS-GFP11x7 uniformly labeled the mitochondrial as compared with Mdi1-GFP11x7 (Fig. S2 J). Collectively, the immunofluorescence and live cell imaging data suggest Mdi1 localizes into foci within mitochondria.
To examine the dynamic behavior of Mdi1-GFP11x7 in cells co-expressing wild-type Mdi1, we performed time-lapse confocal microscopy. While Mdi1-GFP11x7 is susceptible to photobleaching, we could regularly observe examples of discrete Mdi1 foci marking sites of mitochondrial division (Fig. 2, E and F, arrows; Videos 1, 2, and 3; 77% of mitochondrial division events had Mdi1 foci spatially linked at or adjacent to the division site). Importantly, Mdi1 localized to numerous foci distributed throughout the mitochondrial network, and during a 2-min period, only a minor subset of foci marked an active mitochondrial division event.
Example of a Mdi1-marked mitochondrial fission event. Timelapse microscopy images of a wild-type cell co-expressing Mdi1-GFP11x7 and IMS-GFP1-10 (green) and mito-dsRed (magenta). Images shown are maximum intensity projections of three confocal images with 0.4 µm z-steps captured at the indicated time intervals. Time is indicated in mm:ss. Scale bar = 2 µm. Still images of Video 1 are shown in Fig. 2 E.
Example of a Mdi1-marked mitochondrial fission event. Timelapse microscopy images of a wild-type cell co-expressing Mdi1-GFP11x7 and IMS-GFP1-10 (green) and mito-dsRed (magenta). Images shown are maximum intensity projections of three confocal images with 0.4 µm z-steps captured at the indicated time intervals. Time is indicated in mm:ss. Scale bar = 2 µm. Still images of Video 1 are shown in Fig. 2 E.
Example of a Mdi1-marked mitochondrial fission event. Timelapse microscopy images of a wild-type cell co-expressing Mdi1-GFP11x7 and IMS-GFP1-10 (green) and mito-dsRed (magenta). Images shown are maximum intensity projections of three confocal images with 0.4 µm z-steps captured at the indicated time intervals. Time is indicated in mm:ss. Scale bar = 2 µm. Still images of Video 2 are shown in Fig. 2 E.
Example of a Mdi1-marked mitochondrial fission event. Timelapse microscopy images of a wild-type cell co-expressing Mdi1-GFP11x7 and IMS-GFP1-10 (green) and mito-dsRed (magenta). Images shown are maximum intensity projections of three confocal images with 0.4 µm z-steps captured at the indicated time intervals. Time is indicated in mm:ss. Scale bar = 2 µm. Still images of Video 2 are shown in Fig. 2 E.
Example of a Mdi1-marked mitochondrial fission event. Time-lapse microscopy images of a wild-type cell co-expressing Mdi1-GFP11x7 and IMS-GFP1-10 (green) and mito-dsRed (magenta). Images shown are maximum intensity projections of three confocal images with 0.4 µm z-steps captured at the indicated time intervals. Time is indicated in mm:ss. Scale bar = 2 µm. Still images of Video 3 are shown in Fig. 2 E.
Example of a Mdi1-marked mitochondrial fission event. Time-lapse microscopy images of a wild-type cell co-expressing Mdi1-GFP11x7 and IMS-GFP1-10 (green) and mito-dsRed (magenta). Images shown are maximum intensity projections of three confocal images with 0.4 µm z-steps captured at the indicated time intervals. Time is indicated in mm:ss. Scale bar = 2 µm. Still images of Video 3 are shown in Fig. 2 E.
We next wanted to determine the spatial relationship between Mdi1 and the Dnm1 division machinery. We exogenously expressed Dnm1-mCherry in heterozygous diploid cells co-expressing both wild-type Mdi1 and Mdi1-GFP11x7, IMS-GFP1-10, and mito-TagBFP, and imaged by confocal microscopy. Consistent with previous results, Dnm1 localized to numerous foci associated with the mitochondrial network and only a subset of these occasionally marked a mitochondrial fission event (Lackner et al., 2013; Legesse-Miller et al., 2003; Mozdy et al., 2000; Sesaki and Jensen, 1999). Dnm1 labeled ∼10–30 mitochondrial-associated foci per cell, approximately half of which co-localized with Mdi1 (n = 163 of 340 foci from 25 cells; Fig. 2 G, compare white and pink arrows). Mdi1 also concentrated in ∼10–30 focal structures within mitochondria per cell and 47% colocalized with Dnm1 (n = 164 of 346 foci; Fig. 2 G, compare white and yellow arrows). However, consistent with our analysis of Mdi1 relative to division sites, in time-lapse movies, Mdi1 foci localized at or adjacent to 72% of Dnm1-mCherry-marked mitochondrial division events (n = 65 Dnm1-mCherry marked division events; Fig. 2 H and Videos 4 and 5). Due to photobleaching issues, it was not possible to draw conclusions regarding the temporal relationship of Dnm1 and Mdi1 at mitochondrial division sites. However, with the caveat that the tagged form may not accurately reflect the behavior of the endogenous protein, our data suggest a subset of Mdi1 foci dynamically localize in proximity to Dnm1 during mitochondrial division events.
Example of Mdi1 focal localization at a Dnm1-marked mitochondrial fission event. A montage of synchronized merged timelapse microscopy images of a MDI1/Mdi1-GFP11x7 heterozygous diploid cell co-expressing IMS-GFP1-10 (green), Dnm1-mCherry (magenta), and mito-TagBFP (blue). Images shown are maximum intensity projections of three confocal images with 0.4 µm z-steps captured at the indicated time intervals. Time is indicated in mm:ss. Scale bar = 2 µm. Still images of Video 4 are shown in Fig. 2 H.
Example of Mdi1 focal localization at a Dnm1-marked mitochondrial fission event. A montage of synchronized merged timelapse microscopy images of a MDI1/Mdi1-GFP11x7 heterozygous diploid cell co-expressing IMS-GFP1-10 (green), Dnm1-mCherry (magenta), and mito-TagBFP (blue). Images shown are maximum intensity projections of three confocal images with 0.4 µm z-steps captured at the indicated time intervals. Time is indicated in mm:ss. Scale bar = 2 µm. Still images of Video 4 are shown in Fig. 2 H.
Example of Mdi1 focal localization at a Dnm1-marked mitochondrial fission event. A montage of synchronized merged timelapse microscopy images of a MDI1/Mdi1-GFP11x7 heterozygous diploid cell co-expressing IMS-GFP1-10 (green), Dnm1-mCherry (magenta), and mito-TagBFP (blue). Images shown are maximum intensity projections of three confocal images with 0.4 µm z-steps captured at the indicated time intervals. Time is indicated in mm:ss. Scale bar = 2 µm. Still images of Video 5 are shown in Fig. 2 H.
Example of Mdi1 focal localization at a Dnm1-marked mitochondrial fission event. A montage of synchronized merged timelapse microscopy images of a MDI1/Mdi1-GFP11x7 heterozygous diploid cell co-expressing IMS-GFP1-10 (green), Dnm1-mCherry (magenta), and mito-TagBFP (blue). Images shown are maximum intensity projections of three confocal images with 0.4 µm z-steps captured at the indicated time intervals. Time is indicated in mm:ss. Scale bar = 2 µm. Still images of Video 5 are shown in Fig. 2 H.
We next asked how the loss of Dnm1 or Fis1 impacted Mdi1-GFP11x7 localization within mitochondria. In both ∆dnm1 or ∆fis1 cells, even though the mitochondria appeared net-like, Mdi1 retained its focal distribution within the mitochondrial network (Fig. 2 I, arrows). Thus, the propensity of Mdi1-GFP11x7 to concentrate at discrete focal structures within mitochondria is Fis1 and Dnm1-independent, suggesting Mdi1 distribution is influenced by other determinants.
Dnm1 fails to complete mitochondrial division in the absence of Mdi1
Mitochondrial division requires the coordinated scission of both the OMM and the IMM of the organelle. However, several groups have reported evidence of the IMM constricting upstream and independently of Dnm1/Drp1 during mitochondria division in yeast, worms, mice, and human cells (Chakrabarti et al., 2018; Cho et al., 2017; Friedman et al., 2011; Ishihara et al., 2009; Labrousse et al., 1999; Legesse-Miller et al., 2003). Indeed, we observed apparent constrictions during CCCP-induced division, even in the absence of Dnm1 (Fig. S1 A and Fig. 3 A). Given Mdi1 localization to the IMS and its ability to concentrate at discrete Dnm1-independent focal assemblies, we hypothesized that the protein could be responsible for the constriction of the organelle upstream of Dnm1 recruitment. However, as in ∆dnm1 cells, mitochondria in ∆mdi1 cells also appear constricted after treatment with CCCP (Fig. S1 A and Fig. 3 A, arrows). We considered the possibility that in the absence of Mdi1, Dnm1 is sufficient to cause mitochondria constrictions, and therefore examined morphology in ∆mdi1 ∆dnm1 cells. However, even in the absence of both Dnm1 and Mdi1, mitochondria in CCCP-treated cells appear hyperconstricted (Fig. 3 A, arrows). Together, these data indicate that Dnm1-independent mitochondrial constrictions do not require Mdi1.
Dnm1 fails to complete mitochondrial division in the absence of Mdi1. (A) Maximum intensity projections of deconvolved images of the indicated yeast strains expressing mito-dsRed grown to log phase and either directly imaged (top) or treated for 40 min with 25 µM CCCP (bottom) prior to imaging. Arrows mark sites of mitochondrial constriction. (B) Maximum intensity projections of deconvolved images of the indicated yeast strains expressing chromosomally integrated EGFP-Fis1 (green) and mito-DsRed (magenta). Arrows mark sites of focal accumulation of Fis1. (C) As in B for cells expressing an endogenous chromosomal Dnm1-EGFP tag (green). Arrows mark sites of Dnm1 association with mitochondria. (D) As in C for cells treated for 30 min with 25 µM CCCP prior to imaging. Dashed yellow line corresponds to the fluorescence intensity linescan shown below. (E) Single plane (left) and maximum intensity projection (right) confocal fluorescence microscopy images are shown of the indicated yeast strains expressing Tom20-mCherry (OMM, magenta), mito-TagBFP (matrix, blue), and Dnm1-EGFP (green) and treated for 40 min with 25 µM CCCP prior to imaging. Arrows mark sites of Dnm1-marked hyperconstriction where the OMM label appears continuous. Dashed yellow lines correspond to the fluorescence intensity linescans shown at right. (F) Maximum intensity projections of confocal microscopy images of the indicated yeast strains expressing chromosomally tagged Tom20-mCherry (OMM, magenta) and Pam17-EGFP (IMM, green). (G) As in F for the indicated yeast strains treated for 45 min with 25 µM CCCP. Dashed yellow lines correspond to the fluorescence intensity linescans shown at right. Quantification shown in the figure represents the percentage of Tom20-positive mitochondrial hyperconstrictions that appeared positive for Pam17-EGFP. Cell boundaries are indicated with dotted lines. Scale bars = 2 µm (E), 3 µm (A–D and F–G).
Dnm1 fails to complete mitochondrial division in the absence of Mdi1. (A) Maximum intensity projections of deconvolved images of the indicated yeast strains expressing mito-dsRed grown to log phase and either directly imaged (top) or treated for 40 min with 25 µM CCCP (bottom) prior to imaging. Arrows mark sites of mitochondrial constriction. (B) Maximum intensity projections of deconvolved images of the indicated yeast strains expressing chromosomally integrated EGFP-Fis1 (green) and mito-DsRed (magenta). Arrows mark sites of focal accumulation of Fis1. (C) As in B for cells expressing an endogenous chromosomal Dnm1-EGFP tag (green). Arrows mark sites of Dnm1 association with mitochondria. (D) As in C for cells treated for 30 min with 25 µM CCCP prior to imaging. Dashed yellow line corresponds to the fluorescence intensity linescan shown below. (E) Single plane (left) and maximum intensity projection (right) confocal fluorescence microscopy images are shown of the indicated yeast strains expressing Tom20-mCherry (OMM, magenta), mito-TagBFP (matrix, blue), and Dnm1-EGFP (green) and treated for 40 min with 25 µM CCCP prior to imaging. Arrows mark sites of Dnm1-marked hyperconstriction where the OMM label appears continuous. Dashed yellow lines correspond to the fluorescence intensity linescans shown at right. (F) Maximum intensity projections of confocal microscopy images of the indicated yeast strains expressing chromosomally tagged Tom20-mCherry (OMM, magenta) and Pam17-EGFP (IMM, green). (G) As in F for the indicated yeast strains treated for 45 min with 25 µM CCCP. Dashed yellow lines correspond to the fluorescence intensity linescans shown at right. Quantification shown in the figure represents the percentage of Tom20-positive mitochondrial hyperconstrictions that appeared positive for Pam17-EGFP. Cell boundaries are indicated with dotted lines. Scale bars = 2 µm (E), 3 µm (A–D and F–G).
We next examined how the loss of Mdi1 impacted the mitochondrial fission machinery. We expressed chromosomally integrated EGFP-tagged Fis1, the OMM receptor for Dnm1, under the control of its native promoter. Fis1 appeared distributed throughout the mitochondria but also enriched in discrete foci (Fig. 3 B, arrows). Loss of Dnm1 caused Fis1 to redistribute and more uniformly decorate mitochondria, which is consistent with the notion that Fis1 focal localization is promoted by Dnm1 recruitment to the OMM (Fig. 3 B). However, in contrast, Fis1 remained focal along the hyperfused mitochondria in ∆mdi1 cells, suggesting Dnm1 recruitment may not be affected (Fig. 3 B, arrows).
We then examined Dnm1 localization with an endogenous chromosomal C-terminal EGFP fusion. In wild-type cells, Dnm1 appeared in focal structures that were predominantly associated with mitochondria (Fig. 3 C, arrows). As expected, in ∆fis1 cells, Dnm1 did not localize to mitochondria and instead appeared in discrete cytosolic foci (Fig. 3 C; Mozdy et al., 2000). In contrast, in ∆mdi1 cells, Dnm1 associated with the hyperfused mitochondrial membrane, which is consistent with our observation that Fis1 localized to discrete foci in ∆mdi1 cells (Fig. 3 C, arrows). Thus, the defect in mitochondrial fission in the absence of Mdi1 is not due to the loss of Dnm1 recruitment to mitochondria.
Given that Dnm1 can be recruited to mitochondria in the absence of Mdi1, we asked how Dnm1 localization was affected during acutely induced mitochondrial division. We therefore treated cells expressing Dnm1-EGFP with CCCP and imaged them by fluorescence microscopy. In wild-type cells, fragments of mitochondria were often associated with Dnm1 foci (Fig. 3 D, arrows). In contrast, in ∆fis1 cells, Dnm1 failed to be recruited to mitochondria and often concentrated in bright focal structures in the cytosol (Fig. 3 D). However, consistent with our observations in ∆dnm1 cells (Fig. 3 A), mitochondrial hyperconstriction upon CCCP treatment still occurred in the absence of Fis1. Remarkably, Dnm1 was able to assemble in focal structures on the hyperconstricted mitochondria only in ∆mdi1 cells (Fig. 3 D). Indeed, the mitochondria in these cells appeared nearly fragmented, with Dnm1 commonly interspersed between each “bead” of mitochondrial signal at sites where the matrix marker signal appeared diminished (see Fig. 3 D, linescan). To verify that mitochondrial division was not completed after CCCP treatment in ∆mdi1 cells when mitochondria appeared hyperconstricted, we co-expressed Dnm1-EGFP, mito-TagBFP, and Tom20-mCherry (OMM; Hughes et al., 2016), and imaged cells by confocal microscopy (Fig. 3 E). Even at sites where Dnm1 was recruited to mitochondria and the matrix marker appeared to be discontinuous, the OMM appeared to remain connected (Fig. 3 E, arrows and linescan). Together, these data indicate that Dnm1 recruitment to sites of constriction is insufficient to drive mitochondrial division to completion in the absence of Mdi1.
We considered that the role of Mdi1 could be to divide the IMM independently of Dnm1. To test this possibility, we coexpressed chromosomally tagged markers for the OMM (Tom20-mCherry) and the IMM (Pam17-EGFP; Wurm and Jakobs, 2006) in wild-type cells or in cells lacking Dnm1 or Mdi1. The expression of the tagged mitochondrial proteins did not negatively impact mitochondrial morphology, which appeared tubular as expected in wild-type cells and net-like in the absence of either Dnm1 or Mdi1 (Fig. 3 F). We then treated with CCCP to induce mitochondrial hyperconstrictions in cells lacking either Dnm1 or Mdi1, which we could readily detect with Tom20-mCherry as thin threads between adjacent sections of mitochondria (Fig. 3 G). We then asked whether the IMM marker, Pam17-EGFP, could be identified at these hyperconstrictions, finding it was present in 100% of constrictions, both in the absence of Mdi1 (n = 96) and in ∆dnm1 cells, where Mdi1 was still present (n = 118; Fig. 3 G, see linescan). With the limitation that mitochondrial membrane integrity cannot be directly visualized using this approach to confirm that scission has not occurred, these data suggest that Mdi1 does not mediate fission of the IMM independent of Dnm1.
Mdi1 plays a functionally conserved role in mitochondrial division
Evolutionary analysis using CLIME (Li et al., 2014) and InterPro (Paysan-Lafosse et al., 2023) indicates that sequence homologs of Mdi1 are widely found within fungal species, though they are notably absent from metazoans (Fig. 4 A). To determine if Mdi1 plays a similar role in mitochondrial division in other species, we asked whether the Mdi1 sequence homolog from the fission yeast Schizosaccharomyces pombe could functionally complement S. cerevisiae ∆mdi1 cells. We expressed an intronless version of S. pombe Mdi1 under the control of the S. cerevisiae MDI1 promoter and found that mitochondrial morphology defects associated with loss of Mdi1 could be fully rescued by expression of the S. pombe Mdi1 homolog (Fig. 4, B and C).
Mdi1 plays a functionally conserved role in mitochondrial division. (A) CLIME analysis (Li et al., 2014) of Mdi1 identifies conservation among fungal, but not plant or metazoan, species. (B) Maximum intensity projections of deconvolved fluorescence microscopy images of the indicated S. cerevisiae strains expressing mito-dsRed and, where indicated, chromosomally integrated S. pombe Mdi1 (SpMdi1) driven by the S. cerevisiae MDI1 promoter. (C) A graph depicting the categorization of mitochondrial morphology of cells as in B. Data shown represent ∼100 cells per strain in each of the three independent experiments and bars indicate SEM. Asterisks (***P < 0.001) represents unpaired two-tailed t test. (D) Single-plane deconvolved fluorescence microscopy images of the indicated S. pombe strains expressing the matrix marker mito-mCherry. (E) A graph depicting the categorization of mitochondrial morphology of cells as in D. Data shown represent ∼100 cells per strain in each of three independent experiments and bars indicate SEM. Asterisks (***P < 0.001) represent unpaired two-tailed t tests. (F) Single plane (left) and maximum intensity projection (right) deconvolved fluorescence microscopy images of wild-type (top) and ∆mdi1 (bottom) fission yeast cells expressing chromosomally tagged Tom20-mCherry (magenta) and Dnm1-EGFP (green). Arrows mark sites of Dnm1 recruitment to mitochondria. (G) As in F for cells treated for 30 min with 25 µM CCCP. Cell boundaries are indicated with dotted lines. Scale bars = 3 µm (B), 4 µm (D), 5 µm (F and G).
Mdi1 plays a functionally conserved role in mitochondrial division. (A) CLIME analysis (Li et al., 2014) of Mdi1 identifies conservation among fungal, but not plant or metazoan, species. (B) Maximum intensity projections of deconvolved fluorescence microscopy images of the indicated S. cerevisiae strains expressing mito-dsRed and, where indicated, chromosomally integrated S. pombe Mdi1 (SpMdi1) driven by the S. cerevisiae MDI1 promoter. (C) A graph depicting the categorization of mitochondrial morphology of cells as in B. Data shown represent ∼100 cells per strain in each of the three independent experiments and bars indicate SEM. Asterisks (***P < 0.001) represents unpaired two-tailed t test. (D) Single-plane deconvolved fluorescence microscopy images of the indicated S. pombe strains expressing the matrix marker mito-mCherry. (E) A graph depicting the categorization of mitochondrial morphology of cells as in D. Data shown represent ∼100 cells per strain in each of three independent experiments and bars indicate SEM. Asterisks (***P < 0.001) represent unpaired two-tailed t tests. (F) Single plane (left) and maximum intensity projection (right) deconvolved fluorescence microscopy images of wild-type (top) and ∆mdi1 (bottom) fission yeast cells expressing chromosomally tagged Tom20-mCherry (magenta) and Dnm1-EGFP (green). Arrows mark sites of Dnm1 recruitment to mitochondria. (G) As in F for cells treated for 30 min with 25 µM CCCP. Cell boundaries are indicated with dotted lines. Scale bars = 3 µm (B), 4 µm (D), 5 µm (F and G).
We next sought to determine the effect of loss of Mdi1 in the fission yeast S. pombe. As in budding yeast, fission yeast mitochondria labeled with mito-mCherry appear as part of a semicontinuous tubular network (Fig. 4, D and E). In wild-type cells, mitochondria occasionally appeared mildly hyperfused and contained smaller nets in ∼20% of cells. However, in the absence of mitochondrial fission (∆dnm1), consistent with published observations (Dong et al., 2022), mitochondria formed extensive hyperfused nets in the majority of cells (Fig. 4, D and E). Occasionally, the mitochondria in fission yeast ∆dnm1 cells appeared aggregated or hyperconstricted, similar to budding yeast ∆dnm1 cells that were treated with CCCP. In the absence of Mdi1, mitochondria regularly formed hyperfused nets as in ∆dnm1 cells, though they also had abundant hyperconstricted, but interconnected, mitochondria (Fig. 4, D and E).
We then asked whether Dnm1 remains recruited to mitochondria in the absence of Mdi1 in fission yeast as it does in budding yeast. We generated endogenous chromosomally integrated fusions of Tom20-mCherry and Dnm1-EGFP in wild-type and ∆mdi1 cells. As in mito-mCherry expressing cells, Tom20-mCherry labeled mitochondria appeared net-like in the absence of Mdi1 (Fig. 4 F). Additionally, Dnm1-EGFP localized to discrete foci along the mitochondrial network in wild-type cells, consistent with published observations (Fig. 4 F, arrows; Jourdain et al., 2009). Comparable with our results in budding yeast, the loss of Mdi1 did not impact Dnm1 recruitment to mitochondria and Dnm1 foci appeared associated with the hyperfused net structures (Fig. 4 F, arrows). Next, we induced fragmentation of the mitochondrial network with CCCP. Strikingly, in ∆mdi1 cells, after CCCP treatment, Tom20-mCherry-labeled mitochondria frequently had a hyperconstricted beads-on-a-string appearance and Dnm1 could be observed at discrete foci at each constriction site (Fig. 4 G, arrows). Thus, our data indicate that Mdi1 plays a conserved role as a profission factor that is not required for Dnm1 recruitment but is required to facilitate the completion of mitochondrial division.
A putative amphipathic alpha-helix is required for Mdi1 function
Analysis using InterPro indicates that Mdi1 is defined by a domain of unknown function (DUF1748; Paysan-Lafosse et al., 2023). AlphaFold2 predictions of fungal DUF1748-containing proteins suggest that they all contain two lengthy alpha-helices, although, notably, our data suggest that Mdi1 is not an integral membrane protein (Fig. S2 C). We therefore examined the structural characteristics of Mdi1 homologs to identify conserved motifs and found that the second predicted α-helix appears amphipathic in helical wheel projection plots (Fig. 5 A).
A putative amphipathic alpha-helix is required for Mdi1 function. (A) Helical wheel projection plots generated by HeliQuest (Gautier et al., 2008) of the second α-helical region of the indicated fungal orthologues of Mdi1. Valine 56 of S. cerevisiae Mdi1 is circled in black. (B) Maximum intensity projections of deconvolved images of the indicated yeast strains expressing mito-dsRed. Mdi1*-2xFLAG is expressed with the MDI1 promoter. Mdi1*(V56A)-2xFLAG and Mdi1*(V56E)-2xFLAG are driven by an estradiol-controlled GalL promoter and were grown in the presence of 3 nM β-estradiol. (C) A graph depicting the categorization of mitochondrial morphology of cells as in B. Data shown represent at least 75 cells per strain in each of three independent experiments and bars indicate SEM. Asterisks (***P < 0.001) represent unpaired two-tailed t tests. (D) A model for a potential role of Mdi1 in facilitating mitochondrial fission. Cell boundaries are indicated with dotted lines. Scale bar = 3 µm. See also Fig. S3.
A putative amphipathic alpha-helix is required for Mdi1 function. (A) Helical wheel projection plots generated by HeliQuest (Gautier et al., 2008) of the second α-helical region of the indicated fungal orthologues of Mdi1. Valine 56 of S. cerevisiae Mdi1 is circled in black. (B) Maximum intensity projections of deconvolved images of the indicated yeast strains expressing mito-dsRed. Mdi1*-2xFLAG is expressed with the MDI1 promoter. Mdi1*(V56A)-2xFLAG and Mdi1*(V56E)-2xFLAG are driven by an estradiol-controlled GalL promoter and were grown in the presence of 3 nM β-estradiol. (C) A graph depicting the categorization of mitochondrial morphology of cells as in B. Data shown represent at least 75 cells per strain in each of three independent experiments and bars indicate SEM. Asterisks (***P < 0.001) represent unpaired two-tailed t tests. (D) A model for a potential role of Mdi1 in facilitating mitochondrial fission. Cell boundaries are indicated with dotted lines. Scale bar = 3 µm. See also Fig. S3.
Amphipathic α-helices are common structural motifs that can promote membrane targeting of proteins. The hydrophobic side of the alpha helix can embed between hydrocarbon phospholipid tails, often causing lipid packing defects that have the capacity to bend or locally deform the membrane (Giménez-Andrés et al., 2018). To determine if the amphipathic character of the Mdi1 α-helix contributes to Mdi1 function, we generated mutations in budding yeast Mdi1, changing a valine residue on the hydrophobic side of the α-helix to a less bulky alanine (V56A) and to a charged glutamate (V56E; Fig. 5 A, circled in black). The mutant Mdi1*-2xFLAG constructs failed to express stably, and to counter this, we expressed them conditionally in the presence of estradiol to exceed the expression of wild-type Mdi1*-2xFLAG controlled by its native promoter (Fig. S3 A). Remarkably, these Mdi1 mutations led to defects in protein function as assayed by their ability to promote normal mitochondrial morphology. Unlike strains that express Mdi1*-2xFLAG, which appear to have normal mitochondrial morphology, cells expressing Mdi1*(V56A)-2xFLAG had net-like mitochondria in nearly half of cells, and mitochondria in Mdi1*(V56E)-2xFLAG expressing cells appeared similar to those of ∆mdi1 cells (Fig. 5, B and C; Fig. S2 E, and Fig. S3 G). Notably, these mutations did not affect the ability of Mdi1-GFP11x7 to localize to focal structures within the mitochondrial network (Fig. S3 B). Because the Mdi1*(V56E)-2xFLAG mutant failed to rescue mitochondrial morphology defects of ∆mdi1 cells, we performed a protease protection assay and confirmed that the protein was correctly targeted to the IMS (Fig. S3 C). These data suggest that although the mutant protein localizes to the correct mitochondrial compartment, mitochondrial fission remains defective in these cells. In summary, our findings indicate that the amphipathic nature of a predicted Mdi1 α-helix is critical for Mdi1 to promote mitochondrial division.
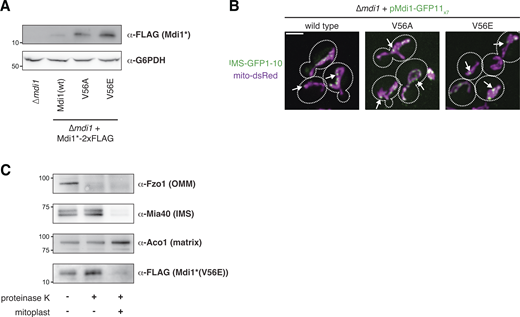
The V56E mutation does not prevent Mdi1 focal assembly or targeting to the IMS. (A) Western analysis with the indicated antibodies of whole cell lysates of the indicated yeast strains. Mdi1*-2xFLAG is expressed with the MDI1 promoter. Mdi1*(V56A)-2xFLAG and Mdi1*(V56E)-2xFLAG are expressed by an estradiol-controlled GalL promoter and grown in the presence of 3 nM β-estradiol. (B) Maximum intensity projections of confocal microscopy images of ∆mdi1 cells expressing the indicated allele of chromosomally integrated Mdi1-GFP11x7 controlled by the native promoter and co-expressing IMS-targeted GFP1-10 (green) and mito-dsRed (magenta). (C) Western analysis with the indicated antibodies of mitochondria isolated from an ∆mdi1 strain expressing Mdi1*(V56E)-2xFLAG and subjected to protease protection analysis. Mitochondria were treated where indicated with proteinase K. Mitoplast sample indicates selective disruption of the OMM by a combination of osmotic swelling and mechanical disruption. Cell boundaries are indicated with dotted lines. Scale bar = 3 µm. Source data are available for this figure: SourceData FS3.
The V56E mutation does not prevent Mdi1 focal assembly or targeting to the IMS. (A) Western analysis with the indicated antibodies of whole cell lysates of the indicated yeast strains. Mdi1*-2xFLAG is expressed with the MDI1 promoter. Mdi1*(V56A)-2xFLAG and Mdi1*(V56E)-2xFLAG are expressed by an estradiol-controlled GalL promoter and grown in the presence of 3 nM β-estradiol. (B) Maximum intensity projections of confocal microscopy images of ∆mdi1 cells expressing the indicated allele of chromosomally integrated Mdi1-GFP11x7 controlled by the native promoter and co-expressing IMS-targeted GFP1-10 (green) and mito-dsRed (magenta). (C) Western analysis with the indicated antibodies of mitochondria isolated from an ∆mdi1 strain expressing Mdi1*(V56E)-2xFLAG and subjected to protease protection analysis. Mitochondria were treated where indicated with proteinase K. Mitoplast sample indicates selective disruption of the OMM by a combination of osmotic swelling and mechanical disruption. Cell boundaries are indicated with dotted lines. Scale bar = 3 µm. Source data are available for this figure: SourceData FS3.
Conclusions
While extensive work has been done to characterize Dnm1/Drp1 behavior in vitro and demonstrate its ability to utilize GTP hydrolysis to constrict membranes (Basu et al., 2017; Francy et al., 2015; Ingerman et al., 2005; Kalia et al., 2018; Kamerkar et al., 2018; Mears et al., 2011), it has remained unclear whether it performs unassisted scission of both mitochondrial membranes. Our data indicate that the soluble, IMS-localized protein Mdi1 plays a conserved role in facilitating Dnm1-dependent mitochondrial division in fungal species. Loss of Mdi1 in either budding yeast or fission yeast leads to mitochondrial hyperfusion caused by an inability of Dnm1 to complete fission of the organelle, even despite its recruitment to hyperconstriction sites during induced division.
Mdi1 localizes to discrete focal structures within the IMS that can be spatially linked to sites of Dnm1-marked mitochondrial division, though it can concentrate at focal structures in the absence of Dnm1. Indeed, Mdi1 localizes to many foci within mitochondria that do not appear to colocalize with Dnm1 and vice versa. However, Mdi1 is neither required for Dnm1-independent mitochondrial preconstriction nor does it appear to promote IMM division independently of Dnm1, even when overexpressed. Mdi1 orthologs have a structurally conserved amphipathic α-helix, mutations of which prevent Mdi1 function in mitochondrial division, suggesting it may work in part by associating with and/or remodeling membranes. One model for a role for Mdi1 is that its amphipathic α-helix may allow it to bind and locally distort regions of the IMM and/or OMM, allowing Dnm1 to more easily constrict and simultaneously divide both the OMM and the IMM when Dnm1 is recruited to Mdi1-enriched sites (Fig. 5 D). An alternative, non-mutually exclusive, model is that Mdi1 may serve as a bridging factor between the OMM and IMM during mitochondrial division.
While Dnm1 is recruited to mitochondria via the OMM tail-anchored receptor Fis1, Fis1 has no functional domains exposed to the IMS, raising the question of how Mdi1 assemblies are positioned and associated with mitochondrial division sites. One possibility is that Mdi1 senses and assembles with specific phospholipids that enrich at discrete sites along the OMM or IMM that stochastically are in proximity with a subset of Dnm1 assemblies on the OMM. Alternatively, Mdi1 may have binding partners that are integral to either mitochondrial membrane that help position it near productive division sites. While our working model is that Mdi1 performs a direct role in division, it remains possible that Mdi1 may work indirectly via yet-to-be-determined factors. Additionally, while our data suggest that Mdi1 cannot independently divide the IMM, we cannot rule out the possibility that it participates in a step of IMM scission that requires Dnm1-mediated hyperconstriction.
Our findings also beg the question of whether an IMS-localized factor works analogously to Mdi1 outside of fungal species. Some primitive eukaryotes such as the red algae Cyanidioschyzon merolae utilize the bacterial division machinery FtsZ to help facilitate mitochondrial division by constricting the IMM from within the matrix during Dnm1-mediated fission (Nishida et al., 2003; Osteryoung and Nunnari, 2003). Meanwhile, work from the Voeltz lab has suggested that in human cells, Drp1 cannot complete the fission of mitochondria without the subsequent activity of Dynamin 2 at division sites (Lee et al., 2016). Thus, it is possible that primitive eukaryotes, yeast, and humans each evolved to solve this issue through different mechanisms. However, Dynamin 2 is not universally required for mitochondrial division (Fonseca et al., 2019; Kamerkar et al., 2018), and Drp1 is capable of severing membrane tubules in vitro (Kamerkar et al., 2018). That said, it is not clear whether Drp1 is sufficient to completely divide two membrane bilayers in vivo. Thus, while Mdi1 is not conserved in human cells, structurally or functionally analogous proteins could conceivably work from inside mitochondria to cooperate in Drp1-mediated division in metazoans.
Materials and methods
Yeast growth
Saccharomyces cerevisiae strains were constructed in the W303 genetic background (ade2-1; leu2-3; his3-11, 15; trp1-1; ura3-1; can1-100). The fzo1-1 and fzo1-1 ∆dnm1 yeast strains were a kind gift from Laura Lackner (Northwestern, Evanston, IL). Routine cell growth was performed in YPD (1% yeast extract, 2% peptone, 2% glucose), YPEG (1% yeast extract, 2% peptone, 3% ethanol, 3% glycerol), or synthetic complete dextrose (SCD; 2% glucose, 0.7% yeast nitrogen base, and amino acids). Wild-type haploid S. pombe was a kind gift from Mike Henne (UT Southwestern, Dallas, TX). All S. pombe strains were routinely grown in YES (0.5% yeast extract, 3% glucose, 225 mg/l adenine, 225 mg/l leucine, 225 mg/l histidine, 225 mg/l uracil, 225 mg/l lysine) or as indicated in EMM (Sunrise Science; supplemented with 225 mg/l adenine, 225 mg/l leucine, 225 mg/l histidine, 225 mg/l uracil, 225 mg/l lysine, 20 mg/l thiamine).
Plasmids and yeast strain construction
All deletions in S. cerevisiae or S. pombe were made using PCR-based homologous recombination replacing the entire ORF with the HIS, NatMX6, or HphMX6 cassettes from pFA6a-series plasmids using lithium acetate transformation (Longtine et al., 1998). C-terminal protein fusions were integrated at the indicated endogenous chromosomal locus and driven by the native promoter, except where indicated for Mdi1, and were generated using pFA6a-link-yEGFP-SpHIS5, pFA6a-link-yEGFP-Kan, pFA6a-link-yEGFP-NatMX, pFA6a-mCherry-HphMX6 (see below), or pFA6a-mCherry-Kan (Lackner et al., 2013; Sheff and Thorn, 2004; Tirrell et al., 2020). pFA6a-mCherry-HphMX6 was generated by cloning the HphMX6 cassette into the BglII/EcoRV sites of pFA6a-mCherry-Kan, replacing the KanMX cassette.
The following plasmids were used to visualize the mitochondrial matrix in S. cerevisiae: pYX142 mito-dsRed (Friedman et al., 2011), pRS304 mito-DsRed, and pRS304 mito-TagBFP. pRS304 mito-DsRed was generated by cloning the mito-DsRed cassette from pRS305 mito-DsRed (pLL19; Abrisch et al., 2020) into the NotI/SacI sites of pRS304 (Sikorski and Hieter, 1989). pRS304 mito-TagBFP was generated by PCR amplifying mito-TagBFP from pVT100u-mt-TagBFP (Friedman et al., 2015) and cloning into the NcoI/AscI sites of pRS304 mito-DsRed, replacing mito-DsRed.
An expression plasmid of internally tagged Mdi1*-2xFLAG (pRS306 Mdi1(Leu37)-2xFLAG) was generated by isothermal assembly into the XhoI/NotI sites of pRS306 with DNA fragments consisting of the Mdi1 promoter, the Mdi1 coding sequence, an internal 2xFLAG tag after amino acid Leu37 flanked on each side by an additional glycine and serine, and the Mdi1 terminator.
To overexpress Mdi1*-2xFLAG and mutant alleles, the endogenous Mdi1 promoter of pRS306-Mdi1(Leu37)-2xFLAG was replaced with the GalL promoter that was PCR-amplified from pYM-N29 (Janke et al., 2004), generating pRS306 GalLprom-Mdi1(Leu37)-2xFLAG. V56A and V56E mutations of Mdi1 were subsequently generated by site-directed mutagenesis. Mdi1 expression plasmids were linearized and integrated into the yeast genome at the ura3 locus. To control expression of Mdi1 constructs driven by the GalL promoter with β-estradiol, pAGL (Veatch et al., 2009) or pAGL-KanMX6 (Gok et al., 2022) were used in concert with these expression plasmids and were linearized and replaced the leu2-3 locus.
To visualize Mdi1 dynamics in live cells, Mdi1-GFP11x7 was tagged at the endogenous MDI1 chromosomal locus or exogenously expressed from a plasmid as follows. First, pFA6a-GFP11x7-Kan was generated by replacing the yEGFP cassette of pKT127 (Sheff and Thorn, 2004) with GFP11x7 amplified from pHRm-NLS-dCas9-GFP11x7-NLS (70224; Addgene; Kamiyama et al., 2016). Then, Mdi1 was C-terminally chromosomally tagged with GFP11x7 preceded by a 24-amino acid linker by PCR-based homologous recombination as described above. Where indicated, this strain was mated to wild-type S. cerevisiae to generate MDI1/Mdi1-GFP11x7 heterozygous diploids. To generate pRS306 Mdi1-GFP11x7 (also referred to as pMdi1-GFP11x7), genomic DNA was isolated from the above strain and used to amplify the Mdi1 promoter, coding sequence, linker, GFP11x7, and terminator into the XhoI/NotI sites of pRS306 by isothermal assembly. V56A and V56E mutations of pRS306 Mdi1-GFP11x7 were subsequently generated by site-directed mutagenesis. Mdi1 expression plasmids were linearized and integrated into the yeast genome at the ura3 locus.
A GFP1-10 expression plasmid that coexpresses mito-DsRed, which enabled visualization of Mdi1-GFP11x7 (pRS304 Cyb2-GFP1-10 mito-DsRed), was generated by isothermal assembly of (1) the GPD promoter from pYM-N17 (Janke et al., 2004), (2) sequence coding for amino acids 1–167 of Cyb2, and (3) GFP1-10 from pFA6a-link-yGFP1-10-CaURA3MX (86419; Addgene; Smoyer et al., 2016), and subsequent cloning into the XhoI/NotI sites of pRS304 mito-DsRed. pRS304 Cyb2-GFP1-10 mito-TagBFP was generated by subsequently subcloning the Cyb2-GFP1-10 insert into the XhoI/NotI sites of pRS304 mito-TagBFP. These plasmids were linearized and integrated at the trp1 locus.
An EGFP-Fis1 expression plasmid (pRS306; EGFP-Fis1) was generated by PCR amplifying (1) the Fis1 promoter, (2) the yEGFP coding sequence, and (3) the Fis1 coding sequence and terminator and cloning into the XhoI/NotI sites of pRS306 by isothermal assembly. The Fis1 expression plasmid was linearized and integrated into the yeast genome at the ura3 locus. To visualize Dnm1, an EGFP tag was chromosomally integrated at the endogenous locus, as described above, or pHS20 Dnm1-mCherry (Lackner et al., 2009) was used.
To visualize GFP11x7 targeted to the intermembrane space, pRS316 Cyb2-GFP11x7 was generated by PCR amplifying (1) the MIC60 promoter, (2) sequence coding for amino acids 1–167 of Cyb2, (3) sequence coding for GFP11x7, and (4) the ADH terminator and cloning into the KpnI/NotI sites of pRS316 by isothermal assembly.
To exogenously express S. pombe Mdi1 in S. cerevisiae, pRS306 SpMdi1 was generated by PCR amplifying (1) the S. cerevisiae MDI1 promoter, (2) the SpMdi1 coding sequence from cDNA isolated from wild type S. pombe, and (3) the ADH terminator and cloning into the XhoI/NotI sites of pRS306 by isothermal assembly.
Mitochondrial matrix-targeted fluorescent proteins in budding yeast were either expressed from low-copy plasmids maintained by auxotrophic selection or linearized and integrated into the trp1 locus. Combinations of multiple tags and/or deletions were generated by strain crossing and tetrad dissection and/or by serial PCR-based homologous recombination. To visualize the mitochondrial matrix in S. pombe, mito-mCherry (mitoRED::Hyg; Kraft and Lackner, 2019) was linearized with NotI and introduced into the leu1 locus.
Fluorescence microscopy and analysis
All epifluorescence microscopy was performed with a Nikon Eclipse Ti inverted epifluorescence microscope equipped with a Hamamatsu Orca-Fusion sCMOS camera and a Nikon 100 × 1.45-NA objective and acquired with Nikon Elements. Z-series images were acquired with a 0.2 µm step size. All images were deconvolved using AutoQuant X3 (10 iterations, blind deconvolution, and low noise).
All confocal fluorescence microscopy, where noted, was performed on a Nikon Spinning Disk Confocal microscope with Yokogawa CSU-W1 SoRa and equipped with a Hamamatsu Orca-Fusion sCMOS camera and a Nikon 100 × 1.45 NA objective and acquired with Nikon Elements. Z-series images were acquired with a 0.2-µm step size (except for time-lapse microscopy as noted below) and the standard resolution disk with 50-µm pinholes.
Linear adjustments to images were made with ImageJ/Fiji. Linescan analysis was performed with slight modifications to the RGB Profiles Tool macro (https://imagej.nih.gov/ij/macros/tools/RGBProfilesTool.txt).
Strains were grown to exponential phase in SCD media with the appropriate auxotrophic selection (fission yeast were grown in EMM media), concentrated, and immobilized on a 3% agarose bed of growth media on cavity microscope slides. Multiple fields of view were imaged per strain or condition in each of at least three independent experiments. Except where indicated below, cells were grown at 30°C and imaged at room temperature. For experiments involving fzo1-1 strains, cells were grown to exponential phase at room temperature and imaged at room temperature or shifted to 37°C for 20 min and imaged at 37°C. For CCCP treatment, cells were grown to the exponential phase at 30°C, media was supplemented with CCCP (25 µM; C2759; Sigma-Aldrich), and cells were allowed to continue growth at 30°C for the indicated times prior to imaging at room temperature. For estradiol-driven expression of Mdi1*-2xFLAG and point mutants, where indicated, standard imaging analysis was performed for cells grown in liquid media constitutively in the presence of the indicated concentrations of β-estradiol (3301; Calbiochem).
To assess mitochondrial morphology, images were blinded prior to analysis, cells were manually categorized as indicated in figures, and the number of cells analyzed per experiment is indicated in the associated figure legend. Statistical comparison was performed between each of the indicated samples by unpaired two-tailed t test with Welch’s correction of the tubular or interconnected mitochondrial morphology category.
To characterize Mdi1 localization to mitochondrial division sites, three images centered on the midplane of cells were captured in 0.4-µm steps at 10-s intervals for 2 min. Maximum intensity projections were generated in ImageJ, and mitochondrial division sites were identified with mito-dsRed blind to Mdi1-GFP11x7/IMS-GFP1-10 signal. Mdi1 focal localization relative to the division site was then manually assessed.
To characterize Mdi1 localization relative to Dnm1, z-series images in 0.2-µm steps were captured throughout the volume of yeast cells. Dnm1 and Mdi1 foci were identified relative to mito-TagBFP signal blind to the presence of the other, and foci were manually scored as positive or negative for colocalization. To characterize Mdi1 localization relative to Dnm1 at mitochondrial division sites, three images centered on the midplane of cells were captured in 0.4-µm steps at 10-s intervals for 1 min. Maximum intensity projections were generated in ImageJ and Dnm1-marked mitochondrial division sites were identified with mito-dsRed blind to Mdi1-GFP11x7/IMS-GFP1-10 signal. Mdi1 focal localization relative to the division site was then manually assessed.
To assess IMM connectivity at hyperconstricted mitochondria, maximum intensity projections of images of CCCP-treated cells coexpressing Pam17-EGFP (IMM) and Tom20-mCherry (OMM) were generated using ImageJ. Tom20-marked mitochondrial hyperconstrictions were identified blind to the Pam17-EGFP signal and then manually assessed for continuity of Pam17 fluorescence at the constriction site.
Indirect immunofluorescence
Immunofluorescence of yeast was performed as described (English et al., 2020) with minor modifications. Cells expressing mito-DsRed and Mdi1*-2xFLAG from an estradiol-driven GalL promoter were grown exponentially in SCD media treated with 4 nM β-estradiol for 16 h to ∼0.5 OD600/ml. Cells were fixed in growth media with paraformaldehyde (4%; EMS) for 1 h at 30°C. Cells were then washed twice (100 mM Tris pH 8, 1.2 M sorbitol), treated twice with DTT buffer (10 mM DTT, 0.1 M Tris pH 9.4) for 10 min each at room temperature, and spheroplasts were generated by incubation in zymolyase buffer (0.1 M KPi pH 6.5, 1.2 M sorbitol, 0.25 mg/ml zymolyase) for 30 min at 30°C. Spheroplasts were pelleted and resuspended in wash buffer, diluted in 1× PBS, and allowed to adhere to glass-bottom cover dishes coated with poly-D-lysine (MatTek) for 30 min at room temperature. Unbound cells were washed with PBS and adherent spheroplasts were permeabilized with cold 0.1% Triton X-100 in 1× PBS (10 min, 4°C). Samples were incubated in blocking buffer (1% BSA, 0.1% Triton X-100 in 1× PBS) for 30 min at room temperature. Samples were then incubated in primary antibody (mouse α-FLAG [F1804; Sigma-Aldrich] diluted 1:500 in blocking buffer) overnight at 4°C, washed 3× for 5 min each (0.1% Triton X-100 in 1× PBS), and incubated in secondary antibody (donkey anti-mouse conjugated to Alexa Fluor 488 [A-21202; Thermo Fisher Scientific] diluted 1:400 in blocking buffer) for 1 h at room temperature. Cells were washed 3× for 5 min each (0.1% Triton X-100 in 1× PBS) followed by 3× washes with 1× PBS before imaging by confocal fluorescence microscopy as described above.
Cell growth analysis
For analysis of the growth of yeast cells on ethanol/glycerol media, assays were performed by growing cells to exponential phase in YPD at room temperature, pelleting, and resuspending cells in water at a concentration of 0.5 OD600/ml. 5 μl of 10-fold serial dilutions of cells were plated on YPEG plates and incubated at the indicated temperatures.
Mitochondria isolation, alkaline extraction, and protease protection analysis
Mitochondria were isolated by differential centrifugation, as previously described (Meeusen et al., 2004), with the following modifications. Briefly, 500–1,000 ODs of yeast cells were grown to exponential phase in YPEG media (Mdi1*(V56E)-2xFLAG was grown in the presence of 1 nM estradiol). After harvesting cells and washing them with water, spheroplasts were generated by digesting cell walls with 4 mg/ml zymolyase 20T (Sunrise Science) in 1.2 M sorbitol. Spheroplasts were washed and resuspended in a minimal volume of cold MIB (0.6 M sorbitol, 50 mM KCl, 20 mM HEPES pH7.4) and homogenized with a tight-fitting dounce. Unlysed cells and large debris were removed by low-speed centrifugation (2,400 g, 5 min, 4°C). Crude mitochondria were enriched by centrifugation of the supernatant (13,000 g, 15 min, 4°C). The mitochondrial pellet was resuspended in a minimal volume of cold MIB to a final concentration of 5–10 mg/ml. Protein concentration was measured by a Bradford assay (Bio-Rad), and 200 µg aliquots were flash-frozen in liquid nitrogen and stored at −80°C.
Alkaline extraction to determine the solubility of Mdi1 was performed as previously described, with minor modifications (Hoppins et al., 2011). Equivalent amounts of crude mitochondria were washed, and pellets were either directly resuspended in Laemmli sample buffer or resuspended in 0.1 M Na2CO3 pH 11.5 and incubated for 30 min on ice. Na2CO3-treated mitochondria were subjected to centrifugation (60 min, 100,000 g, 4°C) and the pellet was resuspended in an equivalent amount of Laemmli sample buffer. Supernatant proteins were precipitated with TCA (12.5%, 30 min on ice), pelleted by centrifugation (10 min, 16,000 g, 4°C), washed with cold acetone, and resuspended in Laemmli sample buffer. Samples were analyzed by SDS-PAGE and Western blot as described below.
Protease protection analysis of Mdi1 was performed as described previously with minor modifications (Hoppins et al., 2011). Crude mitochondria were washed with MIB, distributed into three equivalent tubes, pelleted, and resuspended in equivalent volumes as follows: two tubes were resuspended in MIB buffer and one tube was resuspended in mitoplast buffer (20 mM HEPES pH 7.4). Mitoplast samples were incubated on ice for 15 min and then mechanically disrupted by pipetting up and down 15 times. MIB-resuspended mitochondria were mock-treated or treated with proteinase K (100 µg/ml), mitoplast samples were treated with proteinase K, and all samples were incubated on ice for an additional 15 min. 2 mM PMSF was added to all samples to stop the protease reaction and incubated for 5 min on ice. Samples were subjected to centrifugation (10,400 g, 15 min, 4°C), pellets were resuspended in MIB with 1× protease inhibitor cocktail (539131; MilliporeSigma), and precipitated with TCA as above. Samples were resuspended in Laemmli sample buffer and analyzed by SDS-PAGE/Western blotting as described below.
Whole-cell extracts and Western analysis
For whole-cell extracts, cells were grown to exponential phase in SCD. 0.25 OD600 cells were pelleted, washed with dH20, and extracts were prepared by alkaline extraction (0.255 M NaOH, 1% 2-mercaptoethanol) followed by precipitation in 9% trichloroacetic acid. Precipitates were washed with acetone, dried, and resuspended in 50 μl Laemmli protein sample buffer prior to Western analysis.
Whole-cell lysates or crude mitochondria extracts were freshly prepared and incubated at 70°C (to enable optimal detection of Mdi1*-2xFLAG) or 95°C for 10 min prior to SDS-PAGE, transferred to 0.2-µm pore size PVDF membranes, and immunoblotted with the following primary antibodies: mouse α-FLAG (F1804; 1:1,000; Sigma-Aldrich), rabbit α-G6PDH (A9521; 1:2,000, Sigma-Aldrich), mouse α-Porin (459500; 1:2,000; Thermo Fisher Scientific), rabbit α-Fzo1 (1:1000; a kind gift of Jodi Nunnari, Altos Labs), α-Aco1 (1:10,000, a kind gift of Anju Sreelatha, UT Southwestern), or α-Mia40 (1:10,000, a kind gift of Anju Sreelatha, UT Southwestern). To detect Mdi1*-2xFLAG, goat anti-mouse HRP (A4416; Sigma-Aldrich) was used (1:10,000) and the signal was visualized with SuperSignal West Femto Substrate (Thermo Fisher Scientific). All other proteins were detected with secondary antibodies conjugated to DyLight800 (SA5-35521 and SA5-35571, 1:10,000; Thermo Fisher Scientific). Images were acquired with a ChemiDoc MP Imaging System (Bio-Rad). Linear adjustments to images were made with Photoshop (Adobe).
Online supplemental material
Fig. S1 shows that Mdi1 is required for CCCP-induced mitochondrial division. Fig. S2 shows that Mdi1 is a soluble IMS protein that is enriched at discrete sites within mitochondria. Fig. S3 shows the analysis of Mdi1 mutant alleles. Videos 1, 2, and 3 show examples of Mdi1-marked mitochondrial division events. Videos 4 and 5 show examples of Mdi1 localization relative to Dnm1 during mitochondrial division events.
Data availability
All the data underlying this study are available in the published article and its online supplemental material.
Acknowledgments
We thank Laura Lackner (Northwestern University) for her critical reading of the manuscript, helpful discussions, and kindly providing yeast strains and plasmids. We thank Mike Henne (UT Southwestern) and Maya Schuldiner (Weizmann Institute of Science) for their helpful discussions. We thank Jodi Nunnari (Altos Labs) and Anju Sreelatha (UT Southwestern) for kindly providing antibodies.
The UT Southwestern Quantitative Light Microscopy Facility, which is supported in part by National Institutes of Health P30CA142543, provided access to the Nikon SoRa microscope (purchased with NIH 1S10OD028630-01 to Kate Luby-Phelps) and deconvolution software. This work was supported by a grant from the NIH to J.R. Friedman (R35GM137894).
Author contributions: S.K. Matta: Investigation and writing—review and editing, J.R. Friedman: Conceptualization, formal analysis, funding acquisition, investigation, supervision, and writing (original draft, review, and editing).
References
Author notes
Disclosures: The authors declare no competing interests exist.