Mutant huntingtin, which causes Huntington’s disease (HD), is ubiquitously expressed but induces preferential loss of striatal neurons by unclear mechanisms. Rab11 dysfunction mediates homeostatic disturbance of HD neurons. Here, we report that Rab11 dysfunction also underscores the striatal vulnerability in HD. We profiled the proteome of Rab11-positive endosomes of HD-vulnerable striatal cells to look for protein(s) linking Rab11 dysfunction to striatal vulnerability in HD and found XK, which triggers the selective death of striatal neurons in McLeod syndrome. XK was trafficked together with Rab11 and was diminished on the surface of immortalized HD striatal cells and striatal neurons in HD mouse brains. We found that XK participated in transporting manganese, an essential trace metal depleted in HD brains. Introducing dominantly active Rab11 into HD striatal cells improved XK dynamics and increased manganese accumulation in an XK-dependent manner. Our study suggests that impaired Rab11-based recycling of XK onto cell surfaces for importing manganese is a driver of striatal dysfunction in Huntington’s disease.
Introduction
Selective neuronal vulnerability is a fundamental feature shared by neurodegenerative disorders for which no effective treatment exists (Fu et al., 2018; Muddapu et al., 2020; Paß et al., 2021; Subramaniam, 2019). Understanding the molecular mechanisms underlying the preferential loss of certain populations of neurons is highly important for developing targeted disease-modifying treatments. However, studies on biological factors that make particular populations of neurons preferentially prone to degeneration whereas other neuronal populations relatively resilient have been challenging because most of the disease-causing or relevant proteins are not specific for the vulnerable neurons or brain regions, but ubiquitously expressed in many other cell types and tissues.
Huntington’s disease (HD) is a slowly progressive neurodegenerative disease caused by an elongation of the polyglutamine tract near the NH2 terminus of huntingtin (HTT), a cytoplasmic protein with a function in multiple cellular pathways (MacDonald et al., 1993; Bates et al., 2015; Saudou and Humbert, 2016). Individuals with HD suffer a preferential loss of medium spiny neurons in the striatum in the early stages of the disease and severe atrophy of the whole brain in advanced stages (Tabrizi et al., 2020; Vonsattel and DiFiglia, 1998). While great progress has been made in discovering cellular pathways dysfunctional in HD, mechanisms that underlie the early loss of striatal neurons in HD are still poorly understood (Bates et al., 2015; Saudou and Humbert, 2016; Tabrizi et al., 2020). Previous studies have shown that some proteins that are differentially expressed in the striatum contribute to such vulnerability of striatal neurons in HD. For example, Rhes, a SUMO E3 ligase, is highly enriched in the striatum, and HAP1, an interactor for HTT, is expressed at low levels in the striatum (Liu et al., 2020; Subramaniam et al., 2009). These studies also suggest that preferential vulnerability of striatal neurons in HD is a multifarious process.
We have shown that the HD mutation diminishes the activity of Rab11 by compromising a factor for activating Rab11, and that Rab11 dysfunction mediates the homeostatic disturbance of HD neurons (Li et al., 2009a; Li et al., 2012; Li et al., 2010; McClory et al., 2014). Recently, we identified this Rab11 activator as a transport protein particle II (TRAPPII) and suggested that Kalirin linked HTT to TRAPPII (Ke et al., 2020; McClory et al., 2018; Wang et al., 2020). While these studies strengthen our idea that Rab11 dysfunction is a key mediator of disease progression in HD, a gap in knowledge exists between Rab11 dysfunction and the preferentially early loss of striatal neurons in HD because Rab11 is not specific for striatal neurons but ubiquitously expressed in all types of brain cells and other tissues. Considering that Rab11 regulates endosomal recycling of receptor and transporter proteins back onto cell surfaces for exerting functions (Welz et al., 2014), we speculate that the striatal vulnerability in HD is a consequence of deficient Rab11-based recycling of protein(s) necessary for the function and survival of striatal neurons but relatively dispensable to other cell populations.
XK is a multipass membrane protein covalently linked to the glycoprotein Kell on the surface of red blood cells; its deficiency induces early loss of striatal neurons in McLeod syndrome, an X-linked form of neuroacanthocytosis (Ho et al., 1994; Russo et al., 1998). Unlike red blood cells, which express both XK and Kell, neurons in the brain express only XK, which is mainly intracellular (Claperon et al., 2007). The function of XK is not known. Studies from XK null mice suggest that XK is engaged in cellular homeostasis of divalent cations (Rivera et al., 2013). In observance that McLeod syndrome and HD exhibit a preferentially early loss of striatal neurons, Danek and colleagues proposed that XK and HTT belong to a common pathway that renders striatal neurons preferentially vulnerable to degeneration (Danek et al., 2001). However, this pathway still remains elusive.
STHdhQ111/Q111 cells are neuronal progenitors established from the embryonic striatum of mice having HTT exon-1 replaced by human HTT exon-1 bearing 111 CAG repeats, whereas the wild-type (WT) counterparts (STHdhQ7/Q7) are from embryonic striata of WT littermates (Trettel et al., 2000). These cells present defects seen in cells of HD patients, e.g., energy metabolism disturbance, impaired autophagy, oxidative stress, defective endosomal trafficking, and Rab11-regulated recycling (Gines et al., 2003; Martinez-Vicente et al., 2010; McClory et al., 2014; Milakovic and Johnson, 2005; Pal et al., 2006). Therefore, we utilized these cells to find protein(s) linking Rab11 dysfunction to the preferential vulnerability of striatal neurons in HD. We focused on the proteome of Rab11-positive endosomes and found XK. Our live cell imaging and biochemical and histological studies showed that XK trafficking was regulated by Rab11 and was impaired in HD striatal cells. We further found that XK was involved in transporting manganese (Mn), an essential trace metal highly enriched in the striatum and reduced in HD brains. Expression of dominantly active Rab11 in STHdhQ111/Q111 cells improved XK dynamics in endosomes and increased Mn accumulation in an XK-dependent manner. Collectively, our study suggests that the early loss of striatal neurons in HD involves defective Rab11-regulated recycling of XK onto neuronal surfaces for importing Mn, thus sharing with McLeod syndrome a common pathway for the degeneration of striatal neurons.
Results
Proteomic analysis detects XK in Rab11-positive endosomes of striatal cells
We proposed Rab11 dysfunction to be a key mediator of disease progression in HD (Li et al., 2009a; Li et al., 2009b; Li et al., 2012; Li et al., 2010). We speculate that the contribution of Rab11 dysfunction to the selective vulnerability of spiny neurons in HD is mediated by a protein, which relies on Rab11 for recycling back onto neuronal surfaces; such a mediator protein would be necessary for the survival of striatal neurons but relatively dispensable to other populations of cells. We focused on the proteome analysis of Rab11-positive endosomes in HD vulnerable cells that are in the process of recycling back onto cell surfaces as a way to identify such a mediator protein.
To isolate Rab11-positive endosomes, we developed a two-step method, which required the expression of 6xhistidine-tagged Rab11 (His-Rab11) to label endosomes in cells. In the first step, cytosolic and membrane-bound (endosomes) His-Rab11 were enriched with Nickle-based affinity chromatography, whereas in the second step, membrane-bound and cytosolic His-Rab11 were separated over a sucrose gradient (Fig. 1 A). Prior to endosome isolation, cells were treated with β-cycloheximide to minimize the detection of newly synthesized proteins. Isolated endosomes from STHdhQ7/Q7 cells by Western blot analysis showed His-Rab11 was enriched in eluates (step 1) and centrifugation pellets (step 2); cytosolic EGFP was not detectable in eluates or endosomes (Fig. 1 B). GRASP55, which identifies the Golgi apparatus, was undetected, but Calnexin, which marks the endoplasmic reticulum (ER), was weakly present in prepared endosomes (Fig. 1 B). Electron microscopic analysis of membrane-bound His-Rab11 revealed clusters of tubulovesicular profiles (Fig. 1 C), reminiscent of the morphology of Rab11-positive recycling endosomes (Prekeris et al., 2000; Ullrich et al., 1996). Proteomics analysis of prepared His-Rab11 endosomes led us to 947 proteins, which included those functioning as receptors, transporters, and adhesion molecules (Table S1). As expected, proteins known to have a role in or travel through Rab11-positive endosomes were found in our analyses (Fig. 1 D). Interestingly, McLeod syndrome causal protein XK was included in the proteins identified in our proteomics analyses. Western blot analysis using a commercial XK antibody, which showed reactivity to XK as well as other proteins (Fig. S1, A–C), verified the presence of XK in the prepared Rab11 endosomes (Fig. 1 B). In contrast to XK, Calnexin was not enriched in prepared endosomes. In addition, prior to endosome isolation, STHdhQ7/Q7 cells were treated with β-cycloheximide to allow newly synthesized XK to move out of the ER. Hence, we concluded that XK detected in prepared endosomes did not stem from the contaminated ER membranes.
Proteomic analysis of Rab11 endosomes uncovers XK. (A) Schematic representation of two-step isolation of Rab11 endosomes. STHdhQ7/Q7 cells were transfected with pcDNA3.1-6xHis-Rab11 to label endosomes. Homogenates were prepared using a ball-bearing homogenizer and centrifuged to remove nuclei and then incubated with Nickel resins. Eluates from Nickel resins were overlaid on a sucrose gradient and centrifuged to obtain membrane-bound His-Rab11 in pellets (endosomes). (B) Western blot analysis of eluates from nickel resins and ultracentrifuge pellets (membrane-bound His-Rab11) with indicated antibodies. Shown are blot analyses from one of the four individual endosomal preparations. The open arrowhead indicates a protein likely to be an isoform of XK, as signals for this protein were very low at the basal state and appeared to increase when different amounts of XK-expressing plasmids were transfected (Fig. S1). Star symbols indicate signals for exogenous and endogenous Rab11, which were still persistent during the incubation of the blots with anti-EGFP and secondary antibodies. (C) Membrane-bound His-Rab11 in ultracentrifugation pellets were resuspended in glutaraldehyde and dropped onto grids for electron microscopic analysis. Images in both panels showed profiles of tubulovesicular clusters, which are reminiscent of recycling endosomes in cells. Shown are electron microscopic images from one of four individual endosomal preparations.(D) Examples of known Rab11 interactors and/or known cargo proteins identified in the proteomic analysis of isolated Rab11 endosomes are shown. Shown are the proteins identified in all four endosomal preparations. Source data are available for this figure: SourceData F1.
Proteomic analysis of Rab11 endosomes uncovers XK. (A) Schematic representation of two-step isolation of Rab11 endosomes. STHdhQ7/Q7 cells were transfected with pcDNA3.1-6xHis-Rab11 to label endosomes. Homogenates were prepared using a ball-bearing homogenizer and centrifuged to remove nuclei and then incubated with Nickel resins. Eluates from Nickel resins were overlaid on a sucrose gradient and centrifuged to obtain membrane-bound His-Rab11 in pellets (endosomes). (B) Western blot analysis of eluates from nickel resins and ultracentrifuge pellets (membrane-bound His-Rab11) with indicated antibodies. Shown are blot analyses from one of the four individual endosomal preparations. The open arrowhead indicates a protein likely to be an isoform of XK, as signals for this protein were very low at the basal state and appeared to increase when different amounts of XK-expressing plasmids were transfected (Fig. S1). Star symbols indicate signals for exogenous and endogenous Rab11, which were still persistent during the incubation of the blots with anti-EGFP and secondary antibodies. (C) Membrane-bound His-Rab11 in ultracentrifugation pellets were resuspended in glutaraldehyde and dropped onto grids for electron microscopic analysis. Images in both panels showed profiles of tubulovesicular clusters, which are reminiscent of recycling endosomes in cells. Shown are electron microscopic images from one of four individual endosomal preparations.(D) Examples of known Rab11 interactors and/or known cargo proteins identified in the proteomic analysis of isolated Rab11 endosomes are shown. Shown are the proteins identified in all four endosomal preparations. Source data are available for this figure: SourceData F1.
Characterization of anti-XK antibodies and the XK-EGFP reporter. (A) Western blot analysis of lysates of cells transfected with the indicated amounts of plasmids expressing WT XK. (B) Densitometry of blot analyses in A showed that signals for the protein band of ∼43 kD identified with an arrow increased in a plasmid dose-dependent manner. Signals for the protein band of ∼52 kD indicated by an open arrowhead were very weak at basal state, but also appeared to increase in a plasmid dose-dependent manner. Signals for the protein bands of 90 and 100 kD marked by a star symbol remained constant regardless of the amount of plasmids transfected, suggesting that they are crossreactive. (C) Western blot analysis of cytosol and total membranes prepared from STHdhQ7/Q7 cells. The protein bands of 43 and 52 kD, respectively, were present in total membranes, whereas the protein bands of 90 and 100 kD were cytosolic, further supporting their crossreactivity to XK antibodies. (D) Schematic representation of the XK-EGFP reporter. (E) STHdhQ7/Q7 cells were transfected with pcDNA3.1-XK for 16 h and further cultured in the presence or absence of β-cycloheximide (CYX) for 5 h. Cells were collected for preparing homogenates by passing through a 25-gauge needle. Postnuclear supernatants were overlaid on a discontinuous Nycodenz gradient and centrifuged as in Materials and methods. The same volume of each fraction was analyzed by Western blot. Protein bands identified by fixed arrowheads were detected by other antibodies left over in the XK antibody solutions which were reused. XK-EGFP was expressed at the expected size. The open arrowhead indicated the 52 kD isoform of XK. Source data are available for this figure: SourceData FS1.
Characterization of anti-XK antibodies and the XK-EGFP reporter. (A) Western blot analysis of lysates of cells transfected with the indicated amounts of plasmids expressing WT XK. (B) Densitometry of blot analyses in A showed that signals for the protein band of ∼43 kD identified with an arrow increased in a plasmid dose-dependent manner. Signals for the protein band of ∼52 kD indicated by an open arrowhead were very weak at basal state, but also appeared to increase in a plasmid dose-dependent manner. Signals for the protein bands of 90 and 100 kD marked by a star symbol remained constant regardless of the amount of plasmids transfected, suggesting that they are crossreactive. (C) Western blot analysis of cytosol and total membranes prepared from STHdhQ7/Q7 cells. The protein bands of 43 and 52 kD, respectively, were present in total membranes, whereas the protein bands of 90 and 100 kD were cytosolic, further supporting their crossreactivity to XK antibodies. (D) Schematic representation of the XK-EGFP reporter. (E) STHdhQ7/Q7 cells were transfected with pcDNA3.1-XK for 16 h and further cultured in the presence or absence of β-cycloheximide (CYX) for 5 h. Cells were collected for preparing homogenates by passing through a 25-gauge needle. Postnuclear supernatants were overlaid on a discontinuous Nycodenz gradient and centrifuged as in Materials and methods. The same volume of each fraction was analyzed by Western blot. Protein bands identified by fixed arrowheads were detected by other antibodies left over in the XK antibody solutions which were reused. XK-EGFP was expressed at the expected size. The open arrowhead indicated the 52 kD isoform of XK. Source data are available for this figure: SourceData FS1.
XK localizes to Rab11-positive endosomes in striatal cells
Due to the crossreactivity of the XK antibody, we generated a reporter expressing EGFP-fused XK to facilitate the study of the subcellular localization of XK. We inserted EGFP into the second extracellular loop of XK to minimize disturbing the targeting signal(s) in XK (Fig. S1 D). Chimeric XK-EGFP was expressed at an expected size and distributed in density-gradient centrifugation fractions with a pattern overlapping that of endogenous XK (Fig. S1 E), suggesting that the XK-EGFP reporter mimics endogenous XK in subcellular localization. To determine the localization of XK in the endocytic pathway, STHdhQ7/Q7 cells were transfected with XK-EGFP-expressing plasmids along with plasmids expressing mCherry-Rab4, mCherry-Rab5, mCherry-Rab11, or dsRed-Vps35. Prior to being processed for fluorescent microscopy, cells were treated with β-cycloheximide for 5 h. Under these conditions, XK-EGFP was colocalized with mCherry-Rab11 at both small and large punctate structures and was rarely detected at structures labeled with mCherry-Rab4, mCherry-Rab5, or dsRed-Vps35 (Fig. 2, A and B). These data indicate the association of XK with Rab11-positive recycling endosomes in cells.
Rab11 regulates XK trafficking. (A) Colocalization of XK-EGFP with mCherry-Rab11 in striatal cells. SThdhQ7/Q7 cells were transfected with plasmids expressing XK-EGFP and with plasmids expressing dsRed-Rab4, dsRed-Rab5, dsRed-Vps35, or mCherry-Rab11. After treatment with β-cycloheximide, cells were processed for fluorescence microscopy. Shown are representative confocal images, which were captured individually for each channel and merged. Yellow structures indicate where the colocalization of XK-EGFP with mCherry-fused endosomal marker proteins took place. Scale bars: 10 μm (merge) and 2 μm (inset). (B) Pearson’s coefficient of colocalization. Digital images were analyzed with the JACoP plugin of the NIH ImageJ Fiji. Each symbol represents one cell. Data are mean ± SD. (C) Western blot of biotinylated proteins and postnuclear supernatants of SThdhQ7/Q7 cells transfected with plasmids expressing dNrab11 or dArab11 or empty vectors. Shown are blot analyses from one of the three individual experiments. (D) Densitometry for blot analyses in C. Data are mean ± SD. One-way ANOVA and post hoc Tukey’s analysis: F(2,6) = 44.26, P < 0.001; Tukey’s test * P < 0.01, # P < 0.001. (E) Effects of dominant active and inactive mutants of Rab11 on the subcellular distribution of XK-EGFP. STHdhQ7/Q7 cells were transfected with plasmids expressing XK-EGFP along with plasmids expressing mCherry-fused dArab11 or dNrab11. Confocal images showed that XK-EGFP and mCherry-dArab11 colocalized at small vesicular structures distributed throughout the cytoplasm and clustered at perinuclear regions, whereas the colocalization of XK-EGFP and mCherry-dNrab11 occurred at large tubulovesicular structures. Scale bars: 20 μm (merge) and 3 μm (inset). Source data are available for this figure: SourceData F2.
Rab11 regulates XK trafficking. (A) Colocalization of XK-EGFP with mCherry-Rab11 in striatal cells. SThdhQ7/Q7 cells were transfected with plasmids expressing XK-EGFP and with plasmids expressing dsRed-Rab4, dsRed-Rab5, dsRed-Vps35, or mCherry-Rab11. After treatment with β-cycloheximide, cells were processed for fluorescence microscopy. Shown are representative confocal images, which were captured individually for each channel and merged. Yellow structures indicate where the colocalization of XK-EGFP with mCherry-fused endosomal marker proteins took place. Scale bars: 10 μm (merge) and 2 μm (inset). (B) Pearson’s coefficient of colocalization. Digital images were analyzed with the JACoP plugin of the NIH ImageJ Fiji. Each symbol represents one cell. Data are mean ± SD. (C) Western blot of biotinylated proteins and postnuclear supernatants of SThdhQ7/Q7 cells transfected with plasmids expressing dNrab11 or dArab11 or empty vectors. Shown are blot analyses from one of the three individual experiments. (D) Densitometry for blot analyses in C. Data are mean ± SD. One-way ANOVA and post hoc Tukey’s analysis: F(2,6) = 44.26, P < 0.001; Tukey’s test * P < 0.01, # P < 0.001. (E) Effects of dominant active and inactive mutants of Rab11 on the subcellular distribution of XK-EGFP. STHdhQ7/Q7 cells were transfected with plasmids expressing XK-EGFP along with plasmids expressing mCherry-fused dArab11 or dNrab11. Confocal images showed that XK-EGFP and mCherry-dArab11 colocalized at small vesicular structures distributed throughout the cytoplasm and clustered at perinuclear regions, whereas the colocalization of XK-EGFP and mCherry-dNrab11 occurred at large tubulovesicular structures. Scale bars: 20 μm (merge) and 3 μm (inset). Source data are available for this figure: SourceData F2.
Rab11 regulates XK trafficking
Having verified a connection of XK with Rab11-positive endosomes, we next examined whether XK was a trafficking cargo of Rab11. We manipulated Rab11 activity in STHdhQ7/Q7 cells by expressing dominant negative Rab11 (dNrab11) and dominant active Rab11 (dArab11) mutants, which are widely used for investigating the effects of Rab11 on cargo trafficking (Prekeris et al., 2000; Ullrich et al., 1996). In previous studies, we characterized glucose transporter 3 as a Rab11-dependent trafficking cargo using a biotinylation assay, in which cell surface proteins are labeled with biotin and separated from nonlabeled cytoplasmic proteins (McClory et al., 2014). Our Western blot analysis showed that ectopic expression of dNrab11 or dArab11 did not affect overall levels of XK in STHdhQ7/Q7 cells (Fig. 2, C and D). However, the levels of biotinylated cell surface XK were increased in cells expressing dArab11 and decreased in cells expressing dNrab11 (Fig. 2, C and D). Consistent with a regulatory role for Rab11 in XK trafficking, XK-EGFP and mCherry-dArab11 were colocalized at vesicular structures distributed throughout the cytoplasm and clustered at perinuclear regions, whereas XK-EGFP and mCherry-dNrab11 were codistributed at large tubulovesicular structures (Fig. 2 E). Taken together, these data suggest that Rab11 regulates XK trafficking to cell surfaces.
XK traffics together with Rab11 and exhibits poor dynamics in HD striatal cells
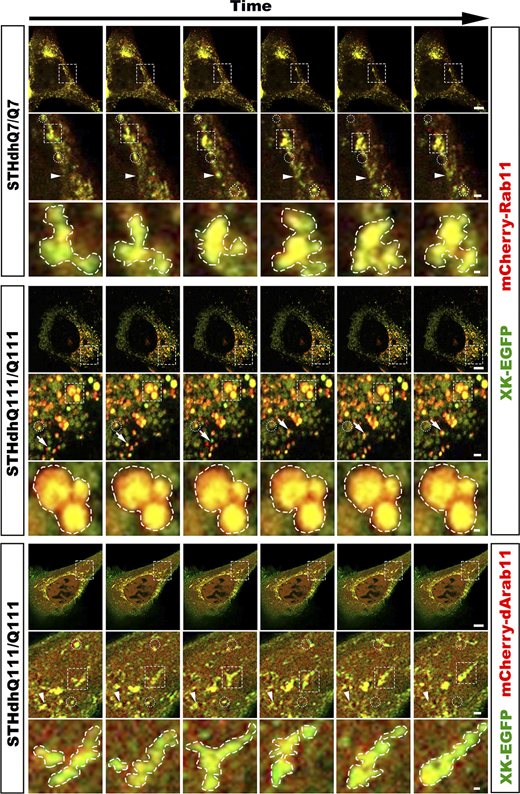
To further demonstrate that Rab11 regulates XK trafficking, we performed real-time live cell imaging with STHdhQ7/Q7 cells coexpressing XK-EGFP and mCherry-Rab11. Consistent with the above findings in static cell imaging studies, XK-EGFP and mCherry-Rab11 trafficked together on small motile vesicles and were colocalized at large tubulovesicular structures; small motile vesicles containing XK-EGFP and mCherry-Rab11 were frequently seen in STHdhQ7/Q7 cells (Fig. 3, Fig. S2; and Videos 1 and 2). The morphology of large tubulovesicular structures underwent frequent changes with motile vesicles joining in and coming out, indicating their highly dynamic nature in WT cells. In STHdhQ111/Q111 cells, small motile vesicles containing both XK-EGFP and mCherry-Rab11 were barely seen, and large tubulovesicular structures were far less dynamic than those in WT cells (N = 5 WT and 6 HD cells, mean ± SD motile vesicles per cell, WT vs. HD: 208.2 ± 104.9 vs. 30.8 ± 12.2; two-tailed Student’s t test: P < 0.005; Fig. 3, Fig. S2; and Videos 3 and 4). Collectively, these data suggest that XK-EGFP trafficking is impaired in HD striatal cells.
Dynamics of Rab11 endosomes in HD striatal cells declines and is enhanced upon expression of dominantly active Rab11. Time-lapse cell imaging of SThdhQ7/Q7 cells expressing XK-EGFP and mCherry-Rab11, SThdhQ111/Q111 expressing XK-EGFP and mCherry-Rab11, and SThdhQ111/Q111 expressing XK-EGFP and mCherry-dArab11. Prior to imaging and during the whole period of imaging, cells were treated with β-cycloheximide to ascertain the detection of XK-EGFP in the endocytic pathway. A series of six consecutive frames were chosen from Videos 1, 3, and 5, respectively, to highlight cotrafficking of XK-EGFP with mCherry-Rab11 or with mCherry-dArab11 in motile vesicles and the dynamics of large tubulovesicular structures labeled with XK-EGFP and mCherry-Rab11 or with XK-EGFP and mCherry-dArab11. Boxed regions were enlarged and shown below the corresponding frame. Arrows in insets trace motile structures containing both XK-EGFP and mCherry-Rab11/dArab11, whereas arrowheads point to structures changing in their size. Dashed circles indicate motile structures containing both XK-EGFP and mCherry-Rab11/dArab11 disappearing in the following images, and dashed polygons identify those appearing in the following images. Enlarged dashed contours indicate dynamic changes in the morphology of large vesiculotubular structures, likely reflecting events of vesicle fusion and budding. Scale bars in the last frame of each of SThdhQ7/Q7, SThdhQ111/Q111, and SThdhQ111/Q111 + dArab11: 10 μm (upper), 2 μm (middle), and 0.5 μm (lower). Source data are available for this figure: SourceData F3.
Dynamics of Rab11 endosomes in HD striatal cells declines and is enhanced upon expression of dominantly active Rab11. Time-lapse cell imaging of SThdhQ7/Q7 cells expressing XK-EGFP and mCherry-Rab11, SThdhQ111/Q111 expressing XK-EGFP and mCherry-Rab11, and SThdhQ111/Q111 expressing XK-EGFP and mCherry-dArab11. Prior to imaging and during the whole period of imaging, cells were treated with β-cycloheximide to ascertain the detection of XK-EGFP in the endocytic pathway. A series of six consecutive frames were chosen from Videos 1, 3, and 5, respectively, to highlight cotrafficking of XK-EGFP with mCherry-Rab11 or with mCherry-dArab11 in motile vesicles and the dynamics of large tubulovesicular structures labeled with XK-EGFP and mCherry-Rab11 or with XK-EGFP and mCherry-dArab11. Boxed regions were enlarged and shown below the corresponding frame. Arrows in insets trace motile structures containing both XK-EGFP and mCherry-Rab11/dArab11, whereas arrowheads point to structures changing in their size. Dashed circles indicate motile structures containing both XK-EGFP and mCherry-Rab11/dArab11 disappearing in the following images, and dashed polygons identify those appearing in the following images. Enlarged dashed contours indicate dynamic changes in the morphology of large vesiculotubular structures, likely reflecting events of vesicle fusion and budding. Scale bars in the last frame of each of SThdhQ7/Q7, SThdhQ111/Q111, and SThdhQ111/Q111 + dArab11: 10 μm (upper), 2 μm (middle), and 0.5 μm (lower). Source data are available for this figure: SourceData F3.
Another set of cells to illustrate that the dynamics of Rab11 endosomes in HD striatal cells declines and is enhanced upon the expression of dominantly active Rab11, as in Fig. 3 . Boxed regions were enlarged and shown below the corresponding frame. Arrows in Insets trace motile structures containing both XK-EGFP and mCherry-Rab11/dArab11, whereas arrowheads point to structures changing in their size. Dashed circles indicate motile structures containing both XK-EGFP and mCherry-Rab11/dArab11 disappearing in the following images, and dashed polygons identify those appearing in the following images. Enlarged dashed contours indicate dynamic changes in the morphology of large tubulovesicular structures, likely reflecting events of vesicle fusion and budding. Scale bars in the last frame of each of SThdhQ7/Q7, SThdhQ111/Q111, and SThdhQ111/Q111 + dArab11: 10 μm (upper), 2 μm (middle), and 0.5 μm (lower).
Another set of cells to illustrate that the dynamics of Rab11 endosomes in HD striatal cells declines and is enhanced upon the expression of dominantly active Rab11, as in Fig. 3 . Boxed regions were enlarged and shown below the corresponding frame. Arrows in Insets trace motile structures containing both XK-EGFP and mCherry-Rab11/dArab11, whereas arrowheads point to structures changing in their size. Dashed circles indicate motile structures containing both XK-EGFP and mCherry-Rab11/dArab11 disappearing in the following images, and dashed polygons identify those appearing in the following images. Enlarged dashed contours indicate dynamic changes in the morphology of large tubulovesicular structures, likely reflecting events of vesicle fusion and budding. Scale bars in the last frame of each of SThdhQ7/Q7, SThdhQ111/Q111, and SThdhQ111/Q111 + dArab11: 10 μm (upper), 2 μm (middle), and 0.5 μm (lower).
Dynamics of XK-EGFP and mCherry-Rab11 in WT striatal cells. STHdhQ7/Q7 cells were cotransfected with plasmids expressing XK-EGFP and plasmids expressing mCherry-Rab11 for 16 h, treated with β-cycloheximide at 37°C for 5 h, and imaged size through a 63×/1.40 Oil CorrM27 objective at 33°C with a 1-s interval at a pixel resolution of 0.10 μm (fast-AS mode) in XY and 0.2 μm interval in Z step-size using the Zeiss LSM 900 confocal laser scanning microscopy system. Frame rate is 1 s.
Dynamics of XK-EGFP and mCherry-Rab11 in WT striatal cells. STHdhQ7/Q7 cells were cotransfected with plasmids expressing XK-EGFP and plasmids expressing mCherry-Rab11 for 16 h, treated with β-cycloheximide at 37°C for 5 h, and imaged size through a 63×/1.40 Oil CorrM27 objective at 33°C with a 1-s interval at a pixel resolution of 0.10 μm (fast-AS mode) in XY and 0.2 μm interval in Z step-size using the Zeiss LSM 900 confocal laser scanning microscopy system. Frame rate is 1 s.
Dynamics of XK-EGFP and mCherry-Rab11 in WT striatal cells. STHdhQ7/Q7 cells were cotransfected with plasmids expressing XK-EGFP and plasmids expressing mCherry-Rab11 for 16 h, treated with β-cycloheximide at 37°C for 5 h, and imaged size through a 63×/1.40 Oil CorrM27 objective at 33°C with a 1-s interval at a pixel resolution of 0.10 μm (fast-AS mode) in XY and 0.2 μm interval in Z step-size using the Zeiss LSM 900 confocal laser scanning microscopy system. Frame rate is 1 s.
Dynamics of XK-EGFP and mCherry-Rab11 in WT striatal cells. STHdhQ7/Q7 cells were cotransfected with plasmids expressing XK-EGFP and plasmids expressing mCherry-Rab11 for 16 h, treated with β-cycloheximide at 37°C for 5 h, and imaged size through a 63×/1.40 Oil CorrM27 objective at 33°C with a 1-s interval at a pixel resolution of 0.10 μm (fast-AS mode) in XY and 0.2 μm interval in Z step-size using the Zeiss LSM 900 confocal laser scanning microscopy system. Frame rate is 1 s.
Dynamics of XK-EGFP and mCherry-Rab11 in HD striatal cells. STHdhQ111/Q111 cells were cotransfected with plasmids expressing XK-EGFP and plasmids expressing mCherry-Rab11 for 16 h, treated with β-cycloheximide at 37°C for 5 h, and imaged through a 63×/1.40 Oil CorrM27 objective at 33°C with a 1-s interval at a pixel resolution of 0.10 μm (fast-AS mode) in XY and 0.2 μm interval in Z step-size using the Zeiss LSM 900 confocal laser scanning microscopy system. Frame rate is 1 s.
Dynamics of XK-EGFP and mCherry-Rab11 in HD striatal cells. STHdhQ111/Q111 cells were cotransfected with plasmids expressing XK-EGFP and plasmids expressing mCherry-Rab11 for 16 h, treated with β-cycloheximide at 37°C for 5 h, and imaged through a 63×/1.40 Oil CorrM27 objective at 33°C with a 1-s interval at a pixel resolution of 0.10 μm (fast-AS mode) in XY and 0.2 μm interval in Z step-size using the Zeiss LSM 900 confocal laser scanning microscopy system. Frame rate is 1 s.
Dynamics of XK-EGFP and mCherry-Rab11 in HD striatal cells. STHdhQ111/Q111 cells were cotransfected with plasmids expressing XK-EGFP and plasmids expressing mCherry-Rab11 for 16 h, treated with β-cycloheximide at 37°C for 5 h, and imaged through a 63×/1.40 Oil CorrM27 objective at 33°C with a 1-s interval at a pixel resolution of 0.10 μm (fast-AS mode) in XY and 0.2 μm interval in Z step-size using the Zeiss LSM 900 confocal laser scanning microscopy system. Frame rate is 1 s.
Dynamics of XK-EGFP and mCherry-Rab11 in HD striatal cells. STHdhQ111/Q111 cells were cotransfected with plasmids expressing XK-EGFP and plasmids expressing mCherry-Rab11 for 16 h, treated with β-cycloheximide at 37°C for 5 h, and imaged through a 63×/1.40 Oil CorrM27 objective at 33°C with a 1-s interval at a pixel resolution of 0.10 μm (fast-AS mode) in XY and 0.2 μm interval in Z step-size using the Zeiss LSM 900 confocal laser scanning microscopy system. Frame rate is 1 s.
As Rab11 activation is compromised in HD cells, we wondered whether XK-EGFP trafficking could be improved by enhancing the activity of Rab11. To this end, STHdhQ111/Q111 cells were cotransfected with plasmids expressing mCherry-dArab11 and XK-EGFP, respectively, and imaged in real time with the same imaging parameters as above. Upon expression of dArab11, the number of small motile vesicles produced and the dynamics of large tubulovesicular structures were vastly increased; events of vesicle joining in and coming out from large tubulovesicular structures were frequently seen (N = 4 HD-dArab11 cells, mean ± SD motile vesicles per cell, two-tailed Student’s t test, HD-dArab11 vs. WT: 306 ± 199.9 vs. 208.2 ± 104.9, P = 0.37; HD-dArab11 vs. HD: 306 ± 199.9 vs. 30.8 ± 12.2, P < 0.01; Fig. 3, Fig. S2; and Videos 5 and 6). These data suggest that the impaired trafficking of XK-EGFP in HD striatal cells is a result of the deficient activity of Rab11.
Dynamics of XK-EGFP and mCherry-dArab11 in HD striatal cells. STHdhQ111/Q111 cells were transfected with plasmids expressing XK-EGFP along with plasmids expressing mCherry-tagged dominant active Rab11 mutant for 16 h, treated with β-cycloheximide at 37°C for 5 h, and imaged through a 63×/1.40 Oil CorrM27 objective at 33°C with a 1-s interval at a pixel resolution of 0.10 μm (fast-AS mode) in XY and 0.2 μm interval in Z step-size using the Zeiss LSM 900 confocal laser scanning microscopy system. Frame rate is 1 s.
Dynamics of XK-EGFP and mCherry-dArab11 in HD striatal cells. STHdhQ111/Q111 cells were transfected with plasmids expressing XK-EGFP along with plasmids expressing mCherry-tagged dominant active Rab11 mutant for 16 h, treated with β-cycloheximide at 37°C for 5 h, and imaged through a 63×/1.40 Oil CorrM27 objective at 33°C with a 1-s interval at a pixel resolution of 0.10 μm (fast-AS mode) in XY and 0.2 μm interval in Z step-size using the Zeiss LSM 900 confocal laser scanning microscopy system. Frame rate is 1 s.
Dynamics of XK-EGFP and mCherry-dArab11 in HD striatal cells. STHdhQ111/Q111 cells were transfected with plasmids expressing XK-EGFP along with plasmids expressing mCherry-tagged dominant active Rab11 mutant for 16 h, treated with β-cycloheximide at 37°C for 5 h, and imaged through a 63×/1.40 Oil CorrM27 objective at 33°C with a 1-s interval at a pixel resolution of 0.10 μm (fast-AS mode) in XY and 0.2 μm interval in Z step-size using the Zeiss LSM 900 confocal laser scanning microscopy system. Frame rate is 1 s.
Dynamics of XK-EGFP and mCherry-dArab11 in HD striatal cells. STHdhQ111/Q111 cells were transfected with plasmids expressing XK-EGFP along with plasmids expressing mCherry-tagged dominant active Rab11 mutant for 16 h, treated with β-cycloheximide at 37°C for 5 h, and imaged through a 63×/1.40 Oil CorrM27 objective at 33°C with a 1-s interval at a pixel resolution of 0.10 μm (fast-AS mode) in XY and 0.2 μm interval in Z step-size using the Zeiss LSM 900 confocal laser scanning microscopy system. Frame rate is 1 s.
Endogenous XK deficiently traffics to cell surfaces in HD striatal cells
Our above studies showed an impairment of the dynamics of XK inside HD striatal cells. To know whether the decline in XK dynamics changes its expression level on the cell surface in HD striatal cells, we replaced the EGFP moiety in XK-EGFP with the pH-sensitive GFP, pHluorin, which is fluorescent only in environments with a neutral pH, e.g., extracellular spaces (Miesenbock et al., 1998). Under the same transfection conditions, the signal intensities of pHluorin on the cell surface of STHdhQ111/Q111 were lower than those of STHdhQ7/Q7 cells (Fig. 4, A and B). To determine whether the trafficking of endogenous XK onto cell surfaces was also impaired in HD striatal cells, we first compared overall as well as cell surface levels of endogenous XK between STHdhQ7/Q7 and STHdhQ111/Q111 cells. Compared with STHdhQ7/Q7 cells, STHdhQ111/Q111 cells expressed comparable levels of total XK protein, but exhibited a decrease in levels of biotinylated XK on cell surfaces (Fig. 4, C–E), a sign of defective trafficking of endogenous XK to the cell surface in STHdhQ111/Q111 cells. We then took advantage of the commercial XK antibody, which recognizes an extracellular epitope and thus is suitable for selectively labeling cell surface located XK molecules. Although other proteins were crossreactive to the antibody, their cytosolic localization would not interfere with the accuracy in detecting cell surface XK in brain sections (Fig. S1). To ascertain the integrity of plasma membranes being maintained, detergent was omitted in all solutions used for immunolabeling and washing brain sections as previously described (Ke et al., 2020). Under these conditions, the levels of XK on and/or in close proximity to the surface of striatal neurons in brain sections of HD140Q/140Q mice were reduced relative to those of WT mice (Fig. 4 F). The reduction of XK on cell surfaces was not a consequence of cell shrinkage or altered expression of XK in the brain of HD140Q/140Q mice (Fig. 4, G and H; and Fig. S3, A and B). Similar results were obtained with the diaminobenzidine-based immunohistochemistry method to detect cell surface XK in brain sections (Fig. S3, C–E). These results suggest that endogenous XK is deficiently delivered to neuronal surfaces in the brain of HD mice.
Defective trafficking of XK onto cell surfaces in HD cells. (A) Confocal images of SThdhQ7/Q7 and SThdhQ111/Q111 cells transiently transfected with plasmids expressing XK-pHluorin showed that pHluorin signals occurred at cell surfaces and intracellular compartments. Arrows point to cells with XK-pHluorin signals concentrated in the perinuclear regions. (B) Densitometry of pHluorin signals on the sharp edges of each of SThdhQ7/Q7 and SThdhQ111/Q111 cells showed a decline in the expression of XK-pHluorin on the surface of HD striatal cells. (C) Western blot analysis of biotinylated proteins and postnuclear supernatants of SThdhQ7/Q7 and SThdhQ111/Q111 cells with indicated antibodies. Shown are blot analyses from one of the three experiments. (D and E) Densitometry of signals of XK in postnuclear supernatants (total, D) and biotinylated XK (cell surfaces, E) for blot analyses in (C). (F) Images of XK staining in a brain section of WT and HD140Q/140Q mice. A series of consecutive coronal brain sections cut through the striatum were processed for labeling with antibodies for XK, with detergents omitted in all solutions to ensure the integrity of the plasma membrane. Images were captured from the nucleus accumubens of each brain section with the same settings. (G and H) Digital images from three brain sections for each of 3 WT and 3 HD mice were analyzed with the NIH ImageJ Fiji software to measure the cross-sectional area (G) and signal intensity (H) of striatal neurons immunoreactive to the XK antibody. Arrowheads in F indicate examples of neurons applied for densitometry and for measuring their cross-section areas. Scale bars: 20 μm (A), 100 μm (F), and 5 μm (inset in F). Data are mean ± SD. Each symbol in B, G, and H represents one cell and in D and E represents one experiment. Two-tailed Student’s t test. Source data are available for this figure: SourceData F4.
Defective trafficking of XK onto cell surfaces in HD cells. (A) Confocal images of SThdhQ7/Q7 and SThdhQ111/Q111 cells transiently transfected with plasmids expressing XK-pHluorin showed that pHluorin signals occurred at cell surfaces and intracellular compartments. Arrows point to cells with XK-pHluorin signals concentrated in the perinuclear regions. (B) Densitometry of pHluorin signals on the sharp edges of each of SThdhQ7/Q7 and SThdhQ111/Q111 cells showed a decline in the expression of XK-pHluorin on the surface of HD striatal cells. (C) Western blot analysis of biotinylated proteins and postnuclear supernatants of SThdhQ7/Q7 and SThdhQ111/Q111 cells with indicated antibodies. Shown are blot analyses from one of the three experiments. (D and E) Densitometry of signals of XK in postnuclear supernatants (total, D) and biotinylated XK (cell surfaces, E) for blot analyses in (C). (F) Images of XK staining in a brain section of WT and HD140Q/140Q mice. A series of consecutive coronal brain sections cut through the striatum were processed for labeling with antibodies for XK, with detergents omitted in all solutions to ensure the integrity of the plasma membrane. Images were captured from the nucleus accumubens of each brain section with the same settings. (G and H) Digital images from three brain sections for each of 3 WT and 3 HD mice were analyzed with the NIH ImageJ Fiji software to measure the cross-sectional area (G) and signal intensity (H) of striatal neurons immunoreactive to the XK antibody. Arrowheads in F indicate examples of neurons applied for densitometry and for measuring their cross-section areas. Scale bars: 20 μm (A), 100 μm (F), and 5 μm (inset in F). Data are mean ± SD. Each symbol in B, G, and H represents one cell and in D and E represents one experiment. Two-tailed Student’s t test. Source data are available for this figure: SourceData F4.
XK is expressed normally in the striatum but is diminished on the cell surface of striatal neurons in CAG140 knock-in mice. (A and B) Postnuclear striatal and cortical supernatants of WT (N = 7) and HD (N = 6) mice were analyzed by Western blot (A) followed by densitometry (B) to examine expression levels of XK. The age of the mice was 10 mo. (C–E) A series of three consecutive coronal brain sections cut through the striatum of WT and HD140Q/140Q mice were processed for labeling with antibodies for XK with the same procedures as in Fig. 4. Immunolabeled XK molecules were detected by the avidin–biotin peroxidase method. (C) Images of immunolabeled XK in one brain section of one animal for WT and HD. Boxed regions were enlarged and shown below the corresponding photograph. (D and E) Digital images captured from the striatum of four brain sections for each genotype were analyzed with the NIH ImageJ/Fiji software to measure the cross-sectional areas (D) and signal intensity (E) of striatal neurons immunoreactive to the XK antibody. Scale bars: 150 μm. Intensities of XK, as well as GAPDH (A), immunoreactive signals were measured with the NIH ImageJ/Fiji software. Data are mean ± SD. Each symbol represents one animal (A) and one cell (D and E), respectively. Two-tailed Student’s t test was done for comparison. Source data are available for this figure: SourceData FS3.
XK is expressed normally in the striatum but is diminished on the cell surface of striatal neurons in CAG140 knock-in mice. (A and B) Postnuclear striatal and cortical supernatants of WT (N = 7) and HD (N = 6) mice were analyzed by Western blot (A) followed by densitometry (B) to examine expression levels of XK. The age of the mice was 10 mo. (C–E) A series of three consecutive coronal brain sections cut through the striatum of WT and HD140Q/140Q mice were processed for labeling with antibodies for XK with the same procedures as in Fig. 4. Immunolabeled XK molecules were detected by the avidin–biotin peroxidase method. (C) Images of immunolabeled XK in one brain section of one animal for WT and HD. Boxed regions were enlarged and shown below the corresponding photograph. (D and E) Digital images captured from the striatum of four brain sections for each genotype were analyzed with the NIH ImageJ/Fiji software to measure the cross-sectional areas (D) and signal intensity (E) of striatal neurons immunoreactive to the XK antibody. Scale bars: 150 μm. Intensities of XK, as well as GAPDH (A), immunoreactive signals were measured with the NIH ImageJ/Fiji software. Data are mean ± SD. Each symbol represents one animal (A) and one cell (D and E), respectively. Two-tailed Student’s t test was done for comparison. Source data are available for this figure: SourceData FS3.
XK is involved in importing manganese
The results above suggest that striatal neurons in HD are deficient in XK function. However, how the deficient function of XK leads to preferential degeneration of striatal neurons remains elusive. Previous studies suggest that XK is involved in the cellular homeostasis of divalent cations in red blood cells (Rivera et al., 2013). Manganese (Mn) is an essential trace metal that is broadly distributed in the brain but has its highest levels in the striatum and is reduced in postmortem brains of HD patients (Horning et al., 2015; Rosas et al., 2012). Defective Mn enrichment has been reported in STHdhQ111/Q111 cells and YAC128 HD mice as well as in human iPSC-derived striatal neuroprogenitor cells (Joshi et al., 2019; Pfalzer et al., 2020; Williams et al., 2010). These lines of evidence led us to the idea that XK maintains the homeostasis of cellular Mn.
To test this idea, we first conducted bioinformatics analyses to determine whether XK contained a Mn-binding site. Metal ion interaction analysis predicted 15 sites where a Mn2+ ion could bind to XK, among which the two sites formed by 230T-232V-233L and 221T-223V-224I, respectively, were exposed to external environments or faced the predicted pore (Fig. S4 A). Molecular dynamic simulation studies revealed that upon the binding of Mn2+ ion at the 230T-232V-233L site, the predicted pore underwent a transition from a compact and closed conformation to an open basket shape, which might facilitate Mn2+ transport (Fig. S4 B and Video 7). This molecular simulation analysis indicates the potential of XK to be a Mn transporter.
Analysis of the feasibility for XK to act as a manganese transporter. (A) Metal ion interaction analysis predicted 15 sites to which a Mn2+ ion could bind in XK. Arrows point to two predicted sites exposed to external environments or the pore, and their corresponding residues were also displayed. (B) The simulated trajectory of the Mn2+ movement through the pore formed by the helices of XK. At the beginning of the simulation, the Mn2+ ions were clustered in the same place. At 10 ns, Mn2+ binding to the predicted site (230t-232V-233L) triggered the pore to change from a compact closed conformation to an open basket shape, which facilitated the inward movement of the Mn2+ ion. Once the channel was in the open state, the Mn2+ inward movement might not be driven by the electrostatic interaction with the binding sites but be driven by a concentration gradient. The ribbon structure was rotated at 30, 40, and 50 ns, respectively, to illustrate that the Mn2+ ion moved through the pore. Arrows in the ribbon structures point to the Mn2+ ion located inside the pore during the simulation.
Analysis of the feasibility for XK to act as a manganese transporter. (A) Metal ion interaction analysis predicted 15 sites to which a Mn2+ ion could bind in XK. Arrows point to two predicted sites exposed to external environments or the pore, and their corresponding residues were also displayed. (B) The simulated trajectory of the Mn2+ movement through the pore formed by the helices of XK. At the beginning of the simulation, the Mn2+ ions were clustered in the same place. At 10 ns, Mn2+ binding to the predicted site (230t-232V-233L) triggered the pore to change from a compact closed conformation to an open basket shape, which facilitated the inward movement of the Mn2+ ion. Once the channel was in the open state, the Mn2+ inward movement might not be driven by the electrostatic interaction with the binding sites but be driven by a concentration gradient. The ribbon structure was rotated at 30, 40, and 50 ns, respectively, to illustrate that the Mn2+ ion moved through the pore. Arrows in the ribbon structures point to the Mn2+ ion located inside the pore during the simulation.
Simulation of manganese movement through the predicted pore of the XK protein. The simulation was carried out in a physiological salt solution system using an NVT ensemble for 100 ps followed by a short run equilibration and minimization using an NPT ensemble for 12 ps. The long-range electrostatic interactions were calculated with the radius for the coulomb interactions fixed at 9 Å and the bonded forces for each trajectory were calculated with a time step of 2 fs. Frame rate is 100 ps.
Simulation of manganese movement through the predicted pore of the XK protein. The simulation was carried out in a physiological salt solution system using an NVT ensemble for 100 ps followed by a short run equilibration and minimization using an NPT ensemble for 12 ps. The long-range electrostatic interactions were calculated with the radius for the coulomb interactions fixed at 9 Å and the bonded forces for each trajectory were calculated with a time step of 2 fs. Frame rate is 100 ps.
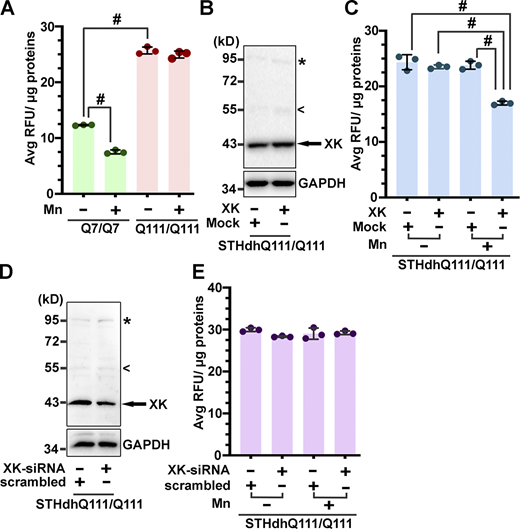
We then examined whether the levels of cellular Mn were correspondingly altered upon manipulating the expression of XK. To this end, we took advantage of the Fura-2 fluorescence quenching assay for measuring cellular Mn described by Kwakye and colleagues (Kwakye et al., 2011). In this assay, the higher the readout of fura-2 fluorescence signals, the lower the concentration of cellular Mn. Consistent with previous findings (Williams et al., 2010), STHdhQ111/Q111 cells contained significantly lesser Mn than STHdhQ7/Q7 cells (Fig. S5 A). Using this assay, we found that the ectopic expression of XK did not influence levels of cellular Mn at the basal state, but enhanced Mn-stimulated enrichment of Mn (Fig. S5, B and C; and Fig. 5 A and B). On the other hand, siRNA-mediated knockdown of XK mitigated Mn-triggered accumulation of Mn (Fig. 5, C and D). These data suggest that XK is involved in transporting Mn.
Effects of altered levels of XK on manganese accumulation in HD striatal cells. (A) Deficiency of Mn in STHdhQ111/Q111 cells. At basal and Mn-triggered states, STHdhQ111/Q111 cells enriched less Mn than STHdhQ7/Q7 cells, suggesting that HD striatal cells have a deficit in taking up Mn. (B and C) Ectopic expression of XK significantly improved STHdhQ111/Q111 cells to accumulate Mn. (D and E) However, XK-siRNA treatment had little effect on Mn accumulation in STHdhQ111/Q111 cells, though XK was efficiently lowered down upon transfection with XK-specific siRNA. Blot analyses in B and D were from one of three experiments. Data in C and E are mean ± SD. One-way ANOVA and post hoc Tukey’s analysis: F(3,8) = 1,083, P < 0.0001 (A); F(3,8) = 55.8, P < 0.001 (C); F(3,8) = 2.495, P = 0.134 (E); Tukey’s test: # P < 0.0001. Source data are available for this figure: SourceData FS5.
Effects of altered levels of XK on manganese accumulation in HD striatal cells. (A) Deficiency of Mn in STHdhQ111/Q111 cells. At basal and Mn-triggered states, STHdhQ111/Q111 cells enriched less Mn than STHdhQ7/Q7 cells, suggesting that HD striatal cells have a deficit in taking up Mn. (B and C) Ectopic expression of XK significantly improved STHdhQ111/Q111 cells to accumulate Mn. (D and E) However, XK-siRNA treatment had little effect on Mn accumulation in STHdhQ111/Q111 cells, though XK was efficiently lowered down upon transfection with XK-specific siRNA. Blot analyses in B and D were from one of three experiments. Data in C and E are mean ± SD. One-way ANOVA and post hoc Tukey’s analysis: F(3,8) = 1,083, P < 0.0001 (A); F(3,8) = 55.8, P < 0.001 (C); F(3,8) = 2.495, P = 0.134 (E); Tukey’s test: # P < 0.0001. Source data are available for this figure: SourceData FS5.
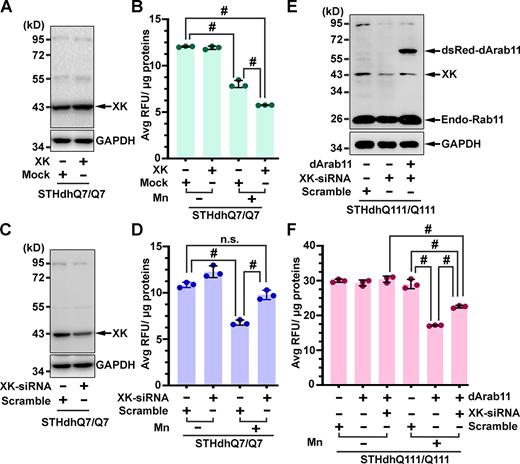
XK is involved in transporting manganese. (A–D) Mn accumulation in SThdhQ7/Q7 cells was elevated and reduced upon ectopic expression (A and B) and knockdown (C and D) of XK, respectively. SThdhQ7/Q7 cells were transfected with pcDNA3.1-XK or empty vector (Mock) or XK-specific or Scramble siRNA. After exposure to Mn, cells were harvested for verifying changes in expression levels of XK by Western blot analysis (A and C) and for measuring the contents of cellular Mn (B and D). Blot analyses shown in A and C were obtained from one of the three experiments. (E and F) Expression of dArab11 in SThdhQ111/Q111 cells promotes Mn accumulation in an XK-dependent manner. SThdhQ111/Q111 cells were transfected with plasmids expressing dsRed-dArab11 along with XK-specific or Scramble siRNA. After Mn exposure, cells were collected for Western blot analysis (E) and measurement of cellular Mn (F). Data in B, D, and F are mean ± SD. One-way ANOVA and post hoc Tukey’s analysis: F(3,8) = 619.5, P < 0.0001 (B), F(3,8) = 81.47, P < 0.0001 (D), F(5,12) = 144, P < 0.0001 (F); Tukey’s test: # P < 0.0001. Source data are available for this figure: SourceData F5.
XK is involved in transporting manganese. (A–D) Mn accumulation in SThdhQ7/Q7 cells was elevated and reduced upon ectopic expression (A and B) and knockdown (C and D) of XK, respectively. SThdhQ7/Q7 cells were transfected with pcDNA3.1-XK or empty vector (Mock) or XK-specific or Scramble siRNA. After exposure to Mn, cells were harvested for verifying changes in expression levels of XK by Western blot analysis (A and C) and for measuring the contents of cellular Mn (B and D). Blot analyses shown in A and C were obtained from one of the three experiments. (E and F) Expression of dArab11 in SThdhQ111/Q111 cells promotes Mn accumulation in an XK-dependent manner. SThdhQ111/Q111 cells were transfected with plasmids expressing dsRed-dArab11 along with XK-specific or Scramble siRNA. After Mn exposure, cells were collected for Western blot analysis (E) and measurement of cellular Mn (F). Data in B, D, and F are mean ± SD. One-way ANOVA and post hoc Tukey’s analysis: F(3,8) = 619.5, P < 0.0001 (B), F(3,8) = 81.47, P < 0.0001 (D), F(5,12) = 144, P < 0.0001 (F); Tukey’s test: # P < 0.0001. Source data are available for this figure: SourceData F5.
Expression of dominantly active Rab11 improves manganese enrichment in HD striatal cells
The above studies show Rab11-dependent trafficking of XK and establish a role for XK in Mn uptake. To determine if the defect of Mn accumulation in STHdhQ111/Q111 cells could be mitigated by enhancing the activity of Rab11, STHdhQ111/Q111 cells were transfected with plasmids expressing dArab11. The expression of dArab11 improved STHdhQ111/Q111 cells to enrich Mn upon exposure to Mn (Fig. 5 E and F). To know whether the beneficial effect of dArab11 on Mn accumulation was dependent on XK, STHdhQ111/Q111 cells were transfected with dArab11-expressing plasmids along with XK-specific or scramble siRNA. Transfection of XK-siRNA along with or without dArab11-expressing plasmids efficiently reduced the expression level of XK (Fig. 5 E). Under these conditions, STHdhQ111/Q111 cells transfected with dArab11-expressing plasmids along with XK-siRNA were less enriched in Mn than STHdhQ111/Q111 cells transfected with dArab11-expressing plasmids together with a scramble siRNA (Fig. 5 F), suggesting that XK mediates the beneficial effect of dArab11 on the enrichment of Mn. Taking all the above lines of evidence together, we predicted a model that defective Rab11-regulated recycling of XK onto neuronal surfaces for importing manganese is a primary driver of dysfunction and early death of striatal neurons in HD (Fig. 6).
A model of selective degeneration of striatal neurons in HD. The idea that Rab11 activation is achieved by a protein complex composed of HTT, Kalirin, and TRAPPII was based on our recent studies (Ke et al., 2020; McClory et al., 2018; Wang et al., 2020). Mutant HTT compromises the Kalirin–TRAPPII complex in activating Rab11 on endosomal membranes (1), thereby impeding XK recycling back onto neuronal surfaces (2) and reducing XK-mediated import of Mn into neurons (3). Constant deficiency of Mn along with the chronic decline of functions of other Rab11-regulated trafficking proteins render the dysfunction of striatal neurons (4) and eventually leads to degeneration of striatal neurons and atrophy of the striatum (5).
A model of selective degeneration of striatal neurons in HD. The idea that Rab11 activation is achieved by a protein complex composed of HTT, Kalirin, and TRAPPII was based on our recent studies (Ke et al., 2020; McClory et al., 2018; Wang et al., 2020). Mutant HTT compromises the Kalirin–TRAPPII complex in activating Rab11 on endosomal membranes (1), thereby impeding XK recycling back onto neuronal surfaces (2) and reducing XK-mediated import of Mn into neurons (3). Constant deficiency of Mn along with the chronic decline of functions of other Rab11-regulated trafficking proteins render the dysfunction of striatal neurons (4) and eventually leads to degeneration of striatal neurons and atrophy of the striatum (5).
Discussion
In this study, we report four novel findings. First, we developed a method for isolating Rab11-positive endosomes from cultured cells. The expression of His-Rab11 can be mediated with lentivirus or adeno-associated virus to enable the isolation of Rab11-positive endosomes from animal tissues. Second, we identified a set of proteins functioning at or trafficking through Rab11-positive endosomes in striatal cells and verified XK as a new trafficking cargo protein of Rab11. Third, we demonstrated that the expression of dominantly active Rab11 in HD striatal cells improved endosomal dynamics by increasing the fusion and fission of cargo-laden vesicles at endosomes. This finding not only strengthens our idea that mutant HTT impairs vesicle formation from Rab11-positive endosomes, but also provides a mechanism for the regulatory role of dominantly active Rab11 in improving endosomal recycling. Lastly, we clarified a function for XK and thus a pathogenic mechanism for McLeod syndrome. XK has been predicted to be a membrane transporter and to maintain the homeostasis of cellular divalent cations (Rivera et al., 2013). However, the substrate cation is unknown. Our study revealed that the levels of cellular Mn were increased in striatal cells ectopically expressing XK and decreased in cells treated with XK-specific siRNA, suggesting that XK is involved in transporting Mn into cells.
HD and McLeod syndromes are inherited neurodegenerative disorders presenting a similar pattern of selective neurodegeneration in the striatum, which leads to the idea that HTT and XK belong to a common pathway (Danek et al., 2001). This pathway has not been clarified. Here, we suggest that Rab11-dependent recycling is such a pathway. HTT is a cytoplasmic protein associated with intracellular membrane-bound structures including endosomes. Several lines of evidence suggest that HTT is involved in the process of conversion from Rab5- to Rab11-controlled endosomal transport (Ke et al., 2020; Li et al., 2008; Li et al., 2009b; McClory et al., 2018; Pal et al., 2006; Wang et al., 2020). In this scenario, Rab5 through HAP40 recruits HTT onto endosomal membranes, where HTT regulates the activation of Rab11 by association with a protein complex composed of Kalirin and TRAPPII. Mutant HTT interferes with the Kalirin–TRAPPII complex in activating Rab11, thereby leading to the dysfunction of Rab11 and slowing endosomal recycling of a host of proteins including those involved in transporting nutrients. XK is a transmembrane protein covalently linked to Kell on the surface of red blood cells. In the brain, XK and Kell are independently expressed with XK in neurons and Kell restricted to blood cells in vessels (Claperon et al., 2007). In neurons, XK is mainly intracellular, hence resembling the subcellular localization of Rab11-regulated trafficking cargo proteins (Claperon et al., 2007; Li et al., 2010; McClory et al., 2014). These lines of evidence suggest that XK is likely to recycle through Rab11-positive endosomes. Indeed, we found that XK was physically associated with Rab11-labeled endosomes and trafficked together with Rab11 in motile transporting vesicles. Consistent with impaired recycling, XK was expressed normally but diminished on cell surfaces of immortalized STHdhQ111/Q111 cells and of striatal neurons in brain sections of HD140Q/140Q mice. Our study suggests that the deficient function of XK is a common pathogenic mechanism shared by McLeod syndrome and HD. In McLeod syndrome, the deficient function of XK arises from genetic mutations rendering the absence of XK, whereas in HD, XK functional deficiency originates from impaired recycling back onto cell surfaces because of Rab11 dysfunction.
How the deficient function of XK triggers preferential degeneration of striatal neurons is not clear. Based on the domain structure, XK has been suggested to be a membrane transporter (Ho et al., 1994), but its substrate remains elusive. Consistently, red blood cells of XK null mice exhibit homeostatic disturbance of divalent cations and have a reduction in the activity of sodium-dependent magnesium transport, which coincides with an increase in the levels of cellular magnesium (Rivera et al., 2013). However, the same study showed that red blood cells of mice null for both XK and Kell manifest an increase in the activity of sodium-dependent magnesium transport, arguing against the idea that XK is a magnesium transporter. We found that ectopic expression of XK promoted striatal cells to enrich Mn, whereas siRNA-mediated knockdown of XK reduced Mn accumulation, suggesting that XK is a Mn transporter. Mn is a trace metal essential for brain physiology and function by serving as a cofactor for various enzymes, e.g., Mn-dependent superoxide dismutase 2, an enzyme converting the superoxide byproducts produced in mitochondria to hydrogen peroxide and molecular oxygen to protect cells from oxidative stress (Chen et al., 2018; Horning et al., 2015; Smith et al., 2017). Chronic excessive exposure to Mn can lead to manganism, an irreversible condition that resembles Parkinson’s disease (Guilarte and Gonzales, 2015; Kwakye et al., 2015). Despite its vital role in all cells, Mn is not evenly distributed in the brain but exhibits the highest levels in the striatum (Bonilla et al., 1982). Such a distribution pattern of Mn in the brain may be a reason for the preferential vulnerability of striatal neurons to the deficient function of XK.
Mn deficiency is seen in immortalized STHdhQ111/Q111 cells and in human striatal neuroprogenitor cells derived from induced pluripotent stem cells, as well as in postmortem brains of HD patients (Joshi et al., 2019; Rosas et al., 2012; Williams et al., 2010). In YAC128 transgenic mice, which express human mutant HTT and manifest selective loss of striatal neurons characteristic of HD (Slow et al., 2003), deficient accumulation of Mn is restricted to the striatum (Williams et al., 2010). On the other hand, excessive Mn induces toxicity specifically to striatal neurons (Guilarte and Gonzales, 2015; Kwakye et al., 2015). Therefore, Mn deficiency is likely to dictate the selectivity of death of striatal neurons in McLeod syndrome as well as in HD. Consistent with Mn deficiency, activities of Mn-dependent enzymes, e.g., superoxide dismutase 2 and arginase 2, are reduced in postmortem brains of HD patients; the decline of their activities may relate to oxidative stress and impaired urea cycle in HD brains (Browne et al., 1997; Butterworth, 1986; Handley et al., 2017; Patassini et al., 2015). Very recently, Wilcox and colleagues reported that YAC128 mice were protected from a Mn-dependent decline in motor performance upon chronic administration of a low dose of Mn (Wilcox et al., 2021). These lines of evidence highlight that Mn deficiency contributes to the pathogenesis of HD and also suggest that strategies that mitigate Mn deficiency may be beneficial in treating HD. However, the mechanism of Mn deficiency in HD is unknown. We showed here that XK was a Mn transporter and deficiently recycled in immortalized HD striatal cells and striatal neurons in HD mouse brains. Our data suggest that Mn deficiency in HD results from defective Rab11-regulated recycling of XK onto neuronal surfaces for importing Mn. Consistent with this idea, an expression of dArab11 in STHdhQ111/Q111 cells improved XK dynamics in endosomes and led to an increase in Mn enrichment, which was attenuated upon suppressing the expression of XK. However, it is not clear whether the function of XK in transporting Mn is coupled with other cations or substrates, e.g., lipids, as XK was recently shown to be a phospholipid scramblase (Adlakha et al., 2022; Ryoden et al., 2022). Mn deficiency in HD may also involve other transporter proteins, e.g., transferrin receptor, which can transport Mn and deficiently recycle in HD cells (Horning et al., 2015; Li et al., 2009a; Li et al., 2009b). ATP13A1, one of the five P5-type ATPase transporters displaying Mn-transporting activities (Cohen et al., 2013), was identified in our proteomics analysis. It is necessary for future studies to examine whether ATP13A1 is a trafficking cargo of Rab11 and/or contributes to Mn deficiency in HD.
How XK imports Mn from outside the neurons in vivo remains speculative. Bioinformatics analyses predicted two potential sites capable of binding to extracellular Mn, demonstrating the feasibility of XK as a Mn transporter. Molecular dynamics simulation studies revealed that upon the binding of Mn at the predicted binding site, the predicted pore formed by the helices of XK underwent a transition from a closed compact conformation to an open basket shape to facilitate ion influx. Our data showed that XK resembled a transferrin receptor in the subcellular distribution, mainly intracellular, and Rab11-regulated endocytic recycling. Transferrin receptor is well known to rely on endocytosis to transport iron into cells. An alternative way by which XK transports Mn into cells may be the endocytosis of the XK-Mn complexes.
Notably, vacuolar protein sorting-associated protein 13A (Vps13A), which causes selective death of striatal neurons in autosomal recessive neuro-acanthocytosis (Rampoldi et al., 2001; Ueno et al., 2001), was also included in the proteins identified in our proteomics analysis. How mutations of Vps13A render selective death of striatal neurons has not been defined. Vps13A locates at mitochondrion–ER, mitochondrion–endosome, and ER–lipid droplet contact sites and plays a key role in transporting lipids between these intracellular organelles (Dziurdzik and Conibear, 2021; Kumar et al., 2018; Munoz-Braceras et al., 2019; Yeshaw et al., 2019). Very recently, Vps13A was found to colocalize and interact with XK; a disease-causative mutation in Vps13A hindered Vps13A to colocalize with XK (Park and Neiman, 2020; Urata et al., 2019), providing an explanation for their similar pathogenicity in triggering preferential degeneration of striatal neurons. It will be interesting to know whether Vps13A has a role in Rab11-positive endosomes and whether Vps13A participates in HD onset.
Rab11 is emerging as a central node involved in the pathogenesis of a group of neuropsychiatric disorders. Genetic mutations of Rab11 cause severe intellectual disability with brain structural malformations (Lamers et al., 2017). We have shown that impaired activation of Rab11 contributes to HD pathogenesis and demonstrated that Rab11 activation in mammals is achieved by a protein complex composed of TRAPPII, Kalirin, and HTT (Ke et al., 2020; McClory et al., 2018; Wang et al., 2020). Genetic studies have linked several subunits of TRAPPII to brain developmental disorders, whereas a coding variant of Kalirin is associated with schizophrenia (Bogershausen et al., 2013; Harripaul et al., 2018; Marin-Valencia et al., 2018; Milev et al., 2017; Mir et al., 2009; Mochida et al., 2009; Mohamoud et al., 2018; Philippe et al., 2009; Russell et al., 2018; Van Bergen et al., 2020). Like mutant HTT, genetic variants of TRAPPII subunits and Kalirin may disable Rab11 activation and therefore impinge on the function of Rab11. However, Rab11-dependent endosomal traffic has been an underexplored area of research in neuroscience. Our development of a method for purifying Rab11-positive endosomes, which can be adapted for use in brain tissues, should aid future studies to discover molecular factors that mediate the pathogenesis of the above brain developmental disorders.
In summary, we develop a method for isolating Rab11-positive endosomes and profile the proteome of Rab11-positive endosomes of mouse striatal cells. We characterize XK as a new cargo of Rab11 and unveil a function for XK in transporting manganese. Our study provides a common mechanism for the preferentially early loss of striatal neurons in McLeod syndrome and Huntington’s disease.
Materials and methods
Animals
HD140Q/140Q and WT mice were housed under a regular 12-h light/dark cycle and a constant temperature with food and water ad libitum. Experimental procedures regarding intracardiac perfusion and collection of brain tissues were performed according to the institutional and U.S. National Institute of Health guidelines and approved by the Massachusetts General Hospital Subcommittee on Research Animal Care.
Plasmids construction
Mouse XK was amplified from cDNA using forward primers 5′-ATGAAATTCCCGGCCTCGGTG-3′ and reverse primers 5′-TTAAGCAGAGCACAGATCAACAGCAGT-3′, purified, and cloned into the Xho I and Xba I sites of pcDNA3.1. The XK-EGFP reporter was designed by inserting an enhanced green fluorescent protein (EGFP) moiety into the second extracellular loop of XK. The N-terminal and C-terminal portions of XK were amplified with primers (5′-ATGAAATTCCCGGCCTCGGTG-3′ and 5′-GCGGTCGGACTGACAGTAGATACA-3′ for XK-N) and primers (5′-CCAGAATGAAGAACCTTATGTGAGC-3′ and 5′-TTAAGCAGAGCACAGATCAACAGCAGT-3′ for XK-C), respectively. PCR products were purified and digested with Age I and Not I for XK-N and with Xho I and Asc I for XK-C. XK-N and XK-C along with Not I and Xho I treated EGFP open-reading frame were cloned into the Age I and Asc I sites of pET41a, from which the DNA fragment coding the XK-EGFP chimera was released and subcloned into pcDNA3.1. Constructs expressing dsRed-fused Rab4, Rab5, Rab11, and Vps35 as well as plasmids expressing mCherry-Rab11 were made as described previously (Ding et al., 2021). The pcDNA3.1-mCherry-dArab11 plasmid was constructed by replacing pcDNA3.1-mCherry-Rab11 with the dArab11 coding fragment. All plasmids were verified by DNA sequencing before being used for studies.
Cell culture and transfection
STHdhQ7/Q7 and STHdhQ111/Q111 cells were maintained in Dulbecco's modified eagle medium (DMEM) supplemented with 10% fetal bovine serum, l-glutamine, and penicillin/streptomycin at 33°C in a humidified CO2 incubator (Thermo Fisher Scientific). DMEM medium, fetal bovine serum, and supplements were purchased from Thermo Fisher Scientific. The day before transfection, STHdhQ7/Q7 and/or STHdhQ111/Q111 cells were seeded onto glass coverslips in a 24-well plate, or in 35 mm glass-bottomed dishes (Eppendorf), or in 10 cm dishes at 5 × 105 cells ml−1. Transfection was conducted with Lipo8000 (Beyotime Biotechnology) according to the manufacturer’s protocol. We used 2 μl Lipo8000 per 1 μg of plasmids for each transfection and equal molar concentration of DNAs, where different plasmids were used for transfection and comparisons. For siRNA transfections, 20 nM of annealed siRNA oligos (Genewiz, XK sense: 5′-UCAUCGUCCUGGUCCUCUUUATT-3′ and antisense: 5′-UAAAGAGGACCAGGACGAUGATT-3′; Genewiz Scramble) were mixed with 0.2 μg of empty pcDNA3.1 vector. The mixture of liposomes and nucleic acids were incubated at room temperature for 25 min with gentle mixing every 5 min and then applied evenly onto cells. Cells were cultured for indicated times.
Purification of Rab11-positive endosome
STHdhQ7/Q7 cells transfected with plasmids bicistronically expressing His-Rab11 and EGFP were harvested 24 h after transfection, washed in cold PBS, and resuspended in 1 ml of cold homogenization buffer (0.25 M sucrose buffer, 10 mM HEPES, 5 mM MgCl2, 1 mM PMSF, and protease inhibitor cocktail). Homogenates were prepared by passing cells through an isobiotec Balch homogenizer (Heidelberg) with a 12-μm ball clearance for 25 times. Homogenates were centrifuged at 12,000 rpm for 15 min (4°C). The postnuclear supernatant fraction was incubated with Ni-NTA resins for 1 h at 4°C in a rotating shaker. After centrifugation for 2 min at 4°C and 2,000 rpm, the resins were washed with five resin-bed volumes (2 × 500 µL) of washing buffer (0.25 M sucrose buffer, 10 mM HEPES, and 5 mM MgCl2). Proteins on Ni-NTA resins were eluted twice using one resin-bed volume of elution buffer (0.25 M sucrose buffer, 10 mM HEPES, 5 mM MgCl2, and 250 mM imidazole). Eluates were combined and overlaid with 0.3 M sucrose (10 mM HEPES, 5 mM MgCl2) in 5 ml Beckman Coulter Open-Top Thinwall Ultra-Clear Tube (Beckman Coulter). After centrifugation at 4°C and 41,000 rpm for 1 h, the aqueous fraction was discarded and the pellet was resuspended into 100 μl of resuspension buffer (0.25 M sucrose buffer, 10 mM HEPES, and 5 mM MgCl2) or in 2.5% glutaraldehyde for analyses. For Western blot and mass spectrometry analyses, the concentration of proteins in endosome samples was determined with the bicinchoninic acid (BCA) assay (Thermo Fisher Scientific).
Subcellular fractionation
To examine whether the XK-EGFP reporter mimicked endogenous XK in subcellular distribution, STHdhQ7/Q7 cells in 10-cm dishes were transfected with pcDNA3.1-XK-EGFP for 16 h and cultured further in the presence or absence of 50 μg/ml of β-cycloheximide for 5 h. Cells were then scraped into culture media and collected by centrifugation at 4°C 1,000 rpm for 5 min. After sequential washes with cold PBS and cold homogenization buffer (25 mM Tris/Cl pH 7.5 and 130 mM KCl), cells were resuspended in homogenization buffer containing protease inhibitors and homogenized by passing through a 25-gauge needle 20 times on ice. Lysates were cleared by centrifugation at 4°C and 2,000 rpm for 5 min. Postnuclear supernatants were overlaid on a discontinuous Nycodenz gradient premade in an SW41 tube (Beckman Coulter) as 0.66 ml of 40% and 5 ml each of 25 and 5% Nycodenz (Axis-Shield/Alere Technologies AS) and centrifuged at 4°C and 30,000 rpm for 1 h. Twelve fractions of 1 ml each were collected from top to bottom of each gradient. Equal volumes of each fraction were used for precipitating the proteins with three volumes of methanol/chloroform (2:1, v/v). After being dried at 37°C for 1 h, precipitated proteins in each fraction were resuspended in 1XSDS sample buffer and analyzed by SDS-PAGE and Western blot.
Transmission electron microscopy
Purified endosome samples were fixed with 2.5% glutaraldehyde solution at 4°C for 30 min and overlaid on a ultrathin carbon support film or microgrid for 5 min. Then microgrids were washed three times with ddH2O for 5 min each. After washing, the microgrids were stained with phosphotungstic acid hydrate (pH 6.5; Sigma-Aldrich) for 5 min at room temperature and then washed three times with ddH2O for 5 min each. After being air-dried at room temperature, samples on microgrids were examined under a 120-kV transmission electron microscope (Talos L120C G2). The dot resolution of 0.37 nm and the line resolution of 0.23 nm were used.
Liquid chromatography/mass spectrometry (LC/MS) and data analysis
His-Rab11-labeled endosomes, 100 μg of proteins, were reduced with 10 mM DTT for 30 min at 56°C, followed by alkylation with 20 mM iodoacetamide for 30 min in the dark at room temperature. Then, the sample was transferred to a 10 kDa ultrafiltration tube and washed with 25 mM NH4HCO3 three times. The samples were treated with trypsin (sequencing-grade; Promega) at a final protease-to-protein ratio of 1:100 (w/w) overnight at 37°C. The digestive reaction was stopped by adding formic acid, and the peptides were desalted using Pierce C18 Spin Tips (Thermo Fisher Scientific) then stored at −20°C for later LC-MS analysis at Shanghai Jiao Tong University Instrumental Analysis Center.
Trypsin-treated samples were analyzed using an EASY-nLC 1,200 system coupled online to an Orbitrap Q Exactive Plus mass spectrometer (Thermo Fisher Scientific). Reverse-phase separation was accomplished using analytical columns (15 cm long, 5 mm inner diameter) ranging from 2 to 100% buffer B at a flow rate of 300 nl/min. The total analysis time was 60 min. The mobile phase consisted of 0.1% formic acid solution (A) and 0.1% formic acid in 80% acetonitrile solution (B). MS data were acquired using a data-dependent top-20 method, with a mass-to-charge ratio (m/z) range between 350 and 1,500 most abundant precursor ions in positive ionization mode. The resolution was set to 700,000 at m/z 200 for MS scans, followed by 17,500 for MS/MS scans of the top 20 ion candidates per cycle. The automatic gain control (AGC) was set to 5e4 ions for MS and 1e5 for MS/MS. MS was put on a dynamic exclusion list for 30 s.
Protein identification was performed using the Proteome Discoverer software 2.4 with Sequest HT search engine. Search parameters were set as follows: (i) species, Mus musculus; (ii) protein database, UniProtKB/Swiss-Prot (including 16,996 sequences); (iii) digestion, trypsin (full), allowing up to two missed cleavage; (iv) static modifications, carbamidomethyl; (v) dynamic modifications, methionine oxidation/+15.995 Da (M); acetyl/+42.011 Da (N-Terminus); Met-loss/−131.040 Da (M); Met-loss+acetyl/−89.030 Da (M; vi) precursor mass tolerance = 10 ppm, fragment mass tolerance = 0.02 Da and (vii) FDR targets: 0.01.
SDS-PAGE and Western blot analysis
SDS-PAGE and Western blot analysis were conducted as standard procedures. Primary antibodies used for Western blot analysis included rabbit anti-XK (1:2,000, ARP33809_P050; Aviva System Biology), rabbit anti-Calnexin (1:2,000, ab10286; Abcam), mouse anti-Rab11A (1:1,000, AB_397984; BD Bioscience, BD Transduction Laboratories), mouse anti-Transferrin receptor (1:2,000; AB_2533029; Thermo Fisher Scientific), mouse anti-EGFP (1:2,000; AT0028; Engibody Biotechnology), mouse anti-GRASP55 (1:1,000; ab211531; Abcam), and mouse anti-GAPDH (1:5,000; 60004-1-Ig; ProteinTech). All secondary antibodies used for Western blot analysis were diluted at 1:10,000. Peroxidase AffiniPure goat anti-mouse (115035003) and anti-rabbit (111035003) IgG (H+L) were purchased from Jackson ImmunoResearch. Blots were developed using enhanced PierceTM enhanced chemiluminescence (Thermo Fisher Scientific) and imaged with the ChemiDocTM MP Imaging System (Bio-Rad). Densitometry was conducted using the NIH ImageJ/Fiji software 1.48v. Background signals were removed. Signal intensities for each relevant protein were normalized with the signal of GAPDH or actin in the corresponding sample.
Immunohistochemistry
Mice at age of 3–4 mo (3 WT and 3 HD) were deeply anesthetized and processed for intracardiac perfusion with 50 ml phosphate saline followed by 50 ml 4% (w/v) paraformaldehyde (Sigma-Aldrich) in 0.1 M in phosphate buffer (pH 7.4). Fixed brains of WT and HD140Q/140Q mice were cut into 30-μm-thick coronal sections on a cryostat. For each animal, a series of three consecutive brain sections cut through the striatum were used for immunohistochemistry. Immunolabeling was performed with free-floating brain sections, with Triton X-100 being omitted in all buffers to preserve the integrity of the plasma membrane. Immunolabeled XK in brain sections was detected on incubation with BODIPY FL conjugated goat anti-rabbit IgG (H+L; Invitrogen) or with the avidin–biotin peroxidase method (Vectastain Elite ABC kit; Vector Laboratories). Brain sections were mounted onto glass slides with ProLong Gold or Cytoseal (Thermo Fisher Scientific). Cells in brain sections were identified by Hoechst 34580 staining of nuclei. Digital images were acquired through a Zeiss LSM 880 confocal microscope (Carl Zeiss) using the Zeiss Zen Black software with the same settings, including pinhole, laser strength, signal gain, and offset. For brain sections detected with the peroxidase method, digital images were collected using a SPOT camera.
Images were analyzed with NIH ImageJ/Fiji by examiners blinded to genotypes. From each image, 10–15 cells with a clear edge were chosen for analysis. The outer and inner edges of the soma of a chosen neuron were tracked using the freehand selection tool to generate two contours. Signal intensities within the outer and inner contours for each neuron were measured separately. The difference in the signal intensities within the two contours was determined. Cross-sectional areas of the outer contour of each chosen neuron were also measured and used as a measure of cell shrinkage in HD140/140Q mouse brains.
Confocal microscopy
After transfection with corresponding plasmids for 16 h, STHdhQ7/Q7 cells on glass coverslips were treated with 50 μg/ml β-cycloheximide, fixed in 4% paraformaldehyde, and processed for confocal microscopy. Cells were identified by staining the nuclei with Hoechst 33258 (1:2,000, H3569; Thermo Fisher Scientific). All images were taken with a Zeiss LSM 900 confocal microscope (Carl Zeiss) with a 63× oil immersion objective (1.4 numerical aperture) using the Zeiss Zen Black software. Optical slices within the z-stack were taken at 1.00or 2.00 μm intervals. High-resolution images were acquired using the Zeiss LSM 900 with Airyscan under the 63× oil immersion objective in a super-resolution mode. Optical slices within the z-stack were 0.13 μm with a frame size of 2,210 × 2,210 pixels. Images were merged and analyzed using the NIH ImageJ/Fiji software by at least two examiners who were blinded to experimental conditions. The JACoP plugin was applied to determine Pearson’s correlation coefficient as a measure of the frequency of protein colocalization in cells. Images used for colocalization analysis were obtained from at least three independent transfections or experiments. Statistical analyses were conducted using GraphPad Prism 7 software. All data were represented as mean ± SD.
Live cell imaging
STHdhQ7/Q7 and STHdhQ111/Q111 cells for time-lapse imaging were seeded in 35-mm glass-bottomed dishes (Eppendorf) the day before transfection. After transfection with pcDNA3.1-XK-EGFP along with pcDNA3.1-mCherry-Rab11 or with mCherry-dArab11 for 16 h, cells were treated with β-cycloheximide and imaged in phenol red–free DMEM media using a Zeiss LSM 900 confocal laser scanning microscope with the Zen Blue software 2.1 (Carl Zeiss AG), which was equipped with an Airyscan with live cell capabilities and fitted with a fast-AS module. The NA oil immersion alpha Plan-Apochromat 63×/1.40 Oil CorrM27 objective (Zeiss) was used with Carl Zeiss Immersol Immersion Oil 518 F (ne = 1.518 [23 _C]; Carl Zeiss). We used the 488-nm excitation (green) and 561-nm excitation (red) for imaging the XK-EGFP and mCherry-Rab11/dArab11, respectively. Time-resolved images were acquired with a 1-s interval at a pixel resolution of 0.10 µm (fast-AS mode) in XY and 0.2 µm interval in Z step-size using the piezo drive. The microscope was equipped with an environmental chamber that maintained the environment at 33°C with humidified 5% CO2 gas during the entire imaging. All cells were imaged with the same settings. Image analysis was conducted as previously described (Ding et al., 2021). We determined the dynamics of the structures containing XK-EGFP and mCherry-Rab11/dARab11 in two ways. First, we counted the number of small motile structures, which were defined as those present in a previous image but absent in the following one(s), and vice versa, and/or those moving from one place to another with a distance of at least three times the diameter of the motile vesicle. Second, we tracked the change in their size and/or morphology.
Cellular fura-2 manganese extraction assay
Measurement of cellular Mn was conducted with the cellular fura-2 Mn extraction assay (CFMEA) as previously described (Kwakye et al., 2011). STHdhQ7/Q7 and/or STHdhQ111/Q111 cells transfected with corresponding plasmids with or without XK-specific or scramble siRNAs for 16 h were further cultured in the presence and absence of 100 μM of Mn dichloride tetrahydrate (MnCl2·4H2O) for 6 h. After Mn exposure, cells were quickly washed three times with 0.5 ml PBS. The cells were extracted at 33°C for 1 h in 0.5 ml PBS containing 0.1% Triton X-100 and 0.5 μM of fura-2. After extraction, 100 μl of cell lysates were transferred into a 96-well plate for measuring signals of fura-2 fluorescence on a plate reader at Ex360 (filter bandwidth ±35 nm) and Em535 (filter bandwidth ±25 nm) with an integration time of 200 ms using the multimode analysis software (version 3.2.0.6) and top-read settings. The graph was plotted after normalizing average raw fluorescence signal values (RFU) with the total protein concentration for individual readings. One-way ANOVA followed by Tukey’s test was conducted to determine statistical significance. The remaining cell lysates were used for Western blot analysis to determine levels of proteins.
Molecular dynamics simulation of manganese transport
The initial coordinate of XK for simulations was obtained from the AlphaFold Protein Structure Database (https://alphafold.ebi.ac.uk/entry/Q9QXY7). The binding sites of Mn2+ ion in XK were predicted using the metal ion-binding site prediction and docking server, with the protein energy minimized using 1,000 steps of conjugate gradient algorithm (https://www.cgl.ucsf.edu/chimera/). The simulations were carried out in a physiological salt solution system using the Desmond 2020.1 from Schrödinger. The system was initially equilibrated using an NVT (constant number of particles, volume, and temperature) ensemble for 100 ps to retrain the XK and Mn2+ complex, followed by a short run equilibration and minimization using an NPT (constant number of particles, pressure, and temperature) ensemble for 12 ps. The NPT ensemble was set up using the Nose-Hoover chain coupling scheme with a temperature of 27°C, a relaxation time of 1.0 ps, and a pressure of 1 bar. The time step was 2 fs. The Martyna–Tuckerman–Klein chain coupling scheme barostat method was used for pressure control with a relaxation time of 2 ps. The particle mesh Ewald method was used for calculating long-range electrostatic interactions, with the radius for the coulomb interactions fixed at 9 Å. The RESPA integrator was applied to calculate the bonded forces for each trajectory with a time step of 2 fs. The root mean square deviation, radius of gyration, root mean square fluctuation, and radius of gyration were calculated to monitor the stability of the molecular dynamics simulation.
Online supplemental material
Fig. S1 shows the characterization of the antibody for XK and of the XK-EGFP reporter. Fig. S2 shows supporting data for Fig. 3. Fig. S3 shows supporting data for Fig. 4. Fig. S4 shows the predicted Mn-binding sites in XK and the simulated movement of Mn through XK. Fig. S5 shows the effects of altered XK expression on Mn accumulation in STHdhQ111/Q111 cells. Videos 1 and 2 show the dynamics of XK-EGFP and mCherry-Rab11 in STHdhQ7/Q7 cells. Videos 3 and 4 show the dynamics of XK-EGFP and mCherry-Rab11 in STHdhQ111/Q111 cells. Videos 5 and 6 show the dynamics of XK-EGFP and mCherry-dArab11 in STHdhQ111/Q111 cells. Video 7 shows the simulation of Mn ion movement through the predicted pore of XK. Table S1 shows the proteins identified by mass spectrometry analysis of the proteome of His-Rab11 endosomes isolated from STHdhQ7/Q7 cells.
Acknowledgments
We thank Drs. Aaron B Bowman, Michael Aschner, and Gunnar Kwakye for their suggestions on the Fura-2 fluorescence quenching assay for measuring cellular manganese.
This work was supported by Endowments from the Pao Foundation (Anna Peiqing Sohmen) to X. Li, who is also supported by the Hereditary Disease Foundation, the Dake Family Fund, and the Infectious Diseases Society of America. G. Chhetri was sponsored by the Chinese scholarship council, and M. DiFiglia was supported by the Dake Family Fund and the CHDI foundation.
The authors declare no competing financial interests.
Author contributions: G. Chhetri, Y. Ke, P. Wang, Y. Li, E. Sapp, A. Ghosh, A. Boudi and X. Wang designed, performed, and analyzed experiments. M. Usman, M.A. Islam and Y. Li conducted data analysis. G. Chhetri, M. DiFiglia, and X. Li wrote the manuscript. X. Li. conceived, designed, analyzed, and supervised the overall execution of the study.
References
Author notes
G. Chhetri and Y. Ke contributed equally to this paper.
Yuting Ke’s present address is Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge, MA.