NUCKS1 (nuclear ubiquitous casein kinase and cyclin-dependent kinase substrate 1) is a chromatin-associated, vertebrate-specific, and multifunctional protein with a role in DNA damage signaling and repair. Previously, we have shown that NUCKS1 helps maintain homologous recombination (HR) DNA repair in human cells and functions as a tumor suppressor in mice. However, the mechanisms by which NUCKS1 positively impacts these processes had remained unclear. Here, we show that NUCKS1 physically and functionally interacts with the DNA motor protein RAD54. Upon exposure of human cells to DNA-damaging agents, NUCKS1 controls the resolution of RAD54 foci. In unperturbed cells, NUCKS1 prevents RAD54’s inappropriate engagement with RAD51AP1. In vitro, NUCKS1 stimulates the ATPase activity of RAD54 and the RAD51–RAD54-mediated strand invasion step during displacement loop formation. Taken together, our data demonstrate that the NUCKS1 protein is an important new regulator of the spatiotemporal events in HR.
Introduction
Homologous recombination (HR) DNA repair is a complex DNA damage repair pathway that serves to maintain genome stability and suppress cancer. HR is intimately linked to DNA replication, and the involvement of HR in the recovery of stalled and collapsed replication forks ensures the faithful replication and segregation of chromosomes (Lambert et al., 2007). HR entails template-dependent DNA strand invasion and DNA synthesis to repair or tolerate DNA damage, such as DNA double-strand breaks (DSBs), single-stranded DNA (ssDNA) gaps, and interstrand DNA cross-links (Daley et al., 2014; Daley et al., 2013; Li and Heyer, 2008). In the HR reaction, the 5′ strand at the DNA break site undergoes a resection to generate a 3′ ssDNA overhang (Daley et al., 2015; Symington, 2014). This overhang is quickly protected by the ssDNA-binding protein RPA. For the HR reaction to continue, RPA must be replaced by the RAD51 recombinase, a rate-limiting process that is dependent on the action of multiple HR mediators (Dosanjh et al., 1998; Godin et al., 2016; Sung, 1997; Sung et al., 2003; Zelensky et al., 2014; Zhao et al., 2015). Then, the RAD51-ssDNA nucleoprotein filament, also known as the presynaptic filament, captures the duplex DNA and generates a displacement loop (D-loop) upon location of the homologous DNA target sequence. Subsequent DNA synthesis and resolution completes the HR reaction (Daley et al., 2013).
Nuclear ubiquitous casein kinase and cyclin-dependent kinase substrate 1 (NUCKS1) is a nuclear, ubiquitously expressed, chromatin-associated, and highly posttranslationally modified protein (Grundt et al., 2004; Grundt et al., 2007; Grundt et al., 2002; Parplys et al., 2015; Wisniewski et al., 2008). NUCKS1 shares extensive amino acid sequence homology with RAD51-associated protein 1 (RAD51AP1; Parplys et al., 2015), a RAD51-binding protein intimately involved in HR DNA repair (Dray et al., 2011; Dray et al., 2010; Dunlop et al., 2012; Dunlop et al., 2011; Modesti et al., 2007; Wiese et al., 2007). Based on this finding, we previously tested and uncovered a role for NUCKS1 in HR DNA repair and DNA replication (Parplys et al., 2015). In experiments involving gene-specific knockdown of NUCKS1 by siRNA, we showed that the down-regulation of NUCKS1 impairs HR in human cells. We also showed that loss of NUCKS1 function renders S/G2-phase cells hypersensitive to ionizing radiation (IR) and that NUCKS1 loss increases cellular sensitivity to chemotherapeutic agents and the susceptibility to IR-induced cancers in mice (Parplys et al., 2015; Yue et al., 2016). However, the mechanisms by which NUCKS1 influences HR and confers tumor suppression had remained unclear.
The DNA motor protein RAD54 belongs to the SWI2/SNF2 protein family of DNA-dependent ATPases and has multiple functions in HR-mediated DSB repair (Ceballos and Heyer, 2011). One critical function of RAD54 lies after presynaptic filament formation. Upon the successful engagement of a homologous duplex sequence by the RAD51-ssDNA filament, RAD54 promotes the conversion of the synaptic complex into a heteroduplex DNA joint, the D-loop (Jaskelioff et al., 2003; Petukhova et al., 1999; Solinger and Heyer, 2001; Solinger et al., 2001; Van Komen et al., 2002). RAD54 also removes RAD51 from the nascent D-loop, so as to make the 3′ end in the D-loop accessible to DNA polymerases for repair synthesis (Ceballos and Heyer, 2011; Liu et al., 2017; Wright and Heyer, 2014). These functions of RAD54 require its ATPase activity. Independent of its ATPase activity, RAD54 stabilizes the RAD51 nucleoprotein filament and promotes the robustness of DNA damage–induced RAD51 foci in cells (Agarwal et al., 2011; Mazin et al., 2003; Tan et al., 1999; van Veelen et al., 2005).
In this study, we show that the NUCKS1 protein is an important new regulator of the spatiotemporal events in the HR reaction. Upon exposure of human cells to DNA-damaging agents, NUCKS1 is required for the timely resolution of DNA damage–induced RAD54 foci. NUCKS1 is epistatic with RAD54, interacts with RAD54 in vitro and in cells, and functions in conjunction with RAD54 in the RAD51-mediated DNA joint molecule formation as measured by the D-loop assay. NUCKS1 competes with RAD51AP1 to interact with RAD54. As such, NUCKS1 spatially and temporally controls the progression of the HR pathway. Consistent with a role for NUCKS1 in genome stability and tumor suppression, HR impairment in NUCKS1-deficient cells leads to the increased utilization of error-prone nonhomologous end joining (NHEJ) to facilitate DSB repair.
Results
NUCKS1 is required for the timely resolution of DNA damage–induced RAD54 foci
Human NUCKS1 is a 243–amino acid residue protein containing two peptide motifs for nuclear localization (NLS1 and NLS2) and one DNA-binding domain (Fig. 1 A; Grundt et al., 2007; Grundt et al., 2002). Human cells depleted for NUCKS1 (by siRNA) are impaired in HR and sensitive to DNA damage–inducing agents but can form damage-induced RAD51 foci (Parplys et al., 2015).
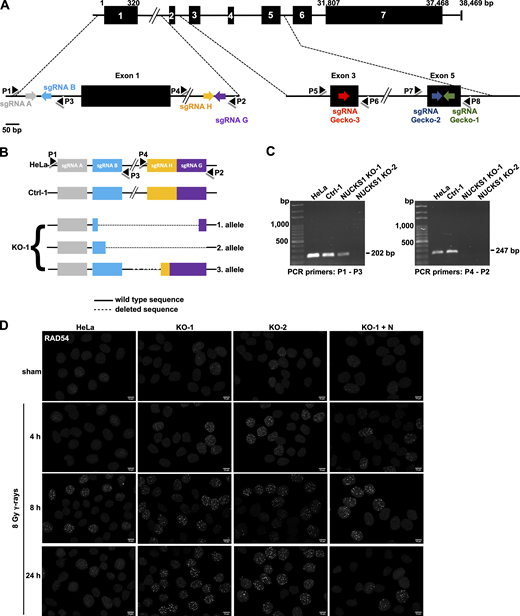
NUCKS1 deficiency leads to an increase in RAD54 foci upon exposure of cells to DNA-damaging agents. (A) Schematic of the primary structure of the 243–amino acid human NUCKS1 protein with two nuclear localization sequences (NLS1 and NLS2; Grundt et al., 2007) and one DNA binding domain (DBD; Grundt et al., 2002). (B) Representative micrographs to show pan-nuclear expression of NUCKS1 in HeLa cells, a clonal isolate of HeLa cells transfected with a nontargeting sgRNA (Ctrl-1), two clonal isolates of HeLa cells that are KO for NUCKS1 (KO-1 and KO-2), and one clonal isolate of KO-1 cells ectopically expressing NUCKS1. Scale bar, 10 µm. (C) Western blots of fractionated nuclear extracts (nuclear soluble and chromatin bound). The signals for PARP1 and Histone H3 serve as loading and fractionation controls, respectively. (D) In response to 8 Gy γ-rays, NUCKS1 KO cells form more RAD54 foci. KO-1+N, KO-1 cells ectopically expressing NUCKS1. Bars represent the mean from two to seven independent experiments (symbols). Error bars, ±1 SEM; ***, P < 0.001; ****, P < 0.0001; ns, not significant; two-way ANOVA analysis. (E) In response to MMC treatment, NUCKS1 KO cells form more RAD54 foci. Bars represent the mean from two independent experiments (symbols). Error bars, ±1 SEM; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant; two-way ANOVA analysis. (F) Results from clonogenic survival assays to show that HeLa cells depleted for RAD54, KO-1 cells, and KO-1 cells depleted for RAD54 display the same sensitivity to the cytotoxic effects of MMC. Data points represent the mean from three independent experiments. Error bars, ± 1SEM. ***, P < 0.001; ****, P < 0.0001; ns, not significant; two-way ANOVA analysis. (G) Representative Western blots from whole cell extracts to show RAD54 knockdown in cells used in F. si-control, nondepleting control siRNA. siRAD54, RAD54-depleting siRNA. PARP1, loading control.
NUCKS1 deficiency leads to an increase in RAD54 foci upon exposure of cells to DNA-damaging agents. (A) Schematic of the primary structure of the 243–amino acid human NUCKS1 protein with two nuclear localization sequences (NLS1 and NLS2; Grundt et al., 2007) and one DNA binding domain (DBD; Grundt et al., 2002). (B) Representative micrographs to show pan-nuclear expression of NUCKS1 in HeLa cells, a clonal isolate of HeLa cells transfected with a nontargeting sgRNA (Ctrl-1), two clonal isolates of HeLa cells that are KO for NUCKS1 (KO-1 and KO-2), and one clonal isolate of KO-1 cells ectopically expressing NUCKS1. Scale bar, 10 µm. (C) Western blots of fractionated nuclear extracts (nuclear soluble and chromatin bound). The signals for PARP1 and Histone H3 serve as loading and fractionation controls, respectively. (D) In response to 8 Gy γ-rays, NUCKS1 KO cells form more RAD54 foci. KO-1+N, KO-1 cells ectopically expressing NUCKS1. Bars represent the mean from two to seven independent experiments (symbols). Error bars, ±1 SEM; ***, P < 0.001; ****, P < 0.0001; ns, not significant; two-way ANOVA analysis. (E) In response to MMC treatment, NUCKS1 KO cells form more RAD54 foci. Bars represent the mean from two independent experiments (symbols). Error bars, ±1 SEM; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant; two-way ANOVA analysis. (F) Results from clonogenic survival assays to show that HeLa cells depleted for RAD54, KO-1 cells, and KO-1 cells depleted for RAD54 display the same sensitivity to the cytotoxic effects of MMC. Data points represent the mean from three independent experiments. Error bars, ± 1SEM. ***, P < 0.001; ****, P < 0.0001; ns, not significant; two-way ANOVA analysis. (G) Representative Western blots from whole cell extracts to show RAD54 knockdown in cells used in F. si-control, nondepleting control siRNA. siRAD54, RAD54-depleting siRNA. PARP1, loading control.
Similar to RAD51, the DNA motor protein RAD54 also accumulates into dynamic protein foci at the sites of DNA damage (Essers et al., 2002; Lisby et al., 2004; Tan et al., 1999), and the formation of RAD54 foci is dependent on RAD51 (Lisby et al., 2004). Hence, we explored if the formation of DNA damage–induced RAD54 foci would be altered in human cells deficient in NUCKS1. We generated several HeLa cell derivatives that are knockout (KO) for NUCKS1 using CRISPR/Cas9. We confirmed that these KO cell lines do not express any NUCKS1 protein by immunocytochemistry (ICC) and Western blot analyses (Fig. 1, B and C). We also sequenced across the Cas9 cleavage sites to verify the disruption of NUCKS1 alleles (Fig. S1, A–C). In the experiments described below, a clonal isolate of HeLa cells transfected with a Cas9 vector expressing nontargeting gRNA (Ctrl-1), parental HeLa cells, and NUCKS1 KO cells ectopically expressing the untagged NUCKS1 ORF (KO-1+NUCKS1; KO-1+N) served as controls.
Data supporting Fig. 1. (A) Schematic of the human NUCKS1 gene with its seven exons, the locations of the targeting sequences of the sgRNAs (see also Table S3) and of PCR primers (P1–P8; see also Table S2) used in this study. Note the NUCKS1 gene is located on the minus strand of the human reference genome. Here, NUCKS1 has been flipped 180° for simplicity. (B) Schematics of the three NUCKS1 KO alleles detected in HeLa NUCKS1 KO-1 cells. PCR products obtained by amplifying genomic DNA from NUCKS1 KO-1 cells with P1-P3 and P4-P2, respectively, were isolated and amplified by topoisomerase-based cloning (Invitrogen) and sequenced. A fourth NUCKS1 KO allele could not be detected, potentially due to absence or due to a large deletion encompassing the binding sequences for P1 and/or P2. (C) Representative agarose gels obtained after amplifying genomic DNA from different cell lines with primers P1-P3 and P4-P2, as indicated. (D) Representative micrographs of nuclear RAD54 foci obtained for the results presented in Fig. 1 D. Scale bars, 10 µm.
Data supporting Fig. 1. (A) Schematic of the human NUCKS1 gene with its seven exons, the locations of the targeting sequences of the sgRNAs (see also Table S3) and of PCR primers (P1–P8; see also Table S2) used in this study. Note the NUCKS1 gene is located on the minus strand of the human reference genome. Here, NUCKS1 has been flipped 180° for simplicity. (B) Schematics of the three NUCKS1 KO alleles detected in HeLa NUCKS1 KO-1 cells. PCR products obtained by amplifying genomic DNA from NUCKS1 KO-1 cells with P1-P3 and P4-P2, respectively, were isolated and amplified by topoisomerase-based cloning (Invitrogen) and sequenced. A fourth NUCKS1 KO allele could not be detected, potentially due to absence or due to a large deletion encompassing the binding sequences for P1 and/or P2. (C) Representative agarose gels obtained after amplifying genomic DNA from different cell lines with primers P1-P3 and P4-P2, as indicated. (D) Representative micrographs of nuclear RAD54 foci obtained for the results presented in Fig. 1 D. Scale bars, 10 µm.
To explore if NUCKS1 status affects RAD54 foci formation, exponentially growing cell cultures were exposed to 8 Gy γ-irradiation and then fixed and stained for RAD54 at 4 h, 8 h, and 24 h after exposure. RAD54 foci were more prevalent in NUCKS1 KO than in control cells at all times after IR exposure (Fig. 1 D and Fig. S1 D). Importantly, for all cell lines investigated, the expression levels of RAD54 protein and cell cycle profiles were similar (Fig. 1 C and Fig. S2, A and B). NUCKS1 KO cells also contained more RAD54 foci upon exposure of cells to the interstrand cross-linking agent mitomycin C (MMC; Fig. 1 E and Fig. S2 E), and, after MMC treatment, the cell cycle profiles for NUCKS1-deficient cells were not different from control cells (Fig. S2 F). Importantly, at 24 h after treatment of cells with either IR or MMC, significantly more nuclei with RAD54 foci remained in NUCKS1 KO cells compared with control cells (Fig. 1, D and E). To exclude cell type–specific effects, we used CRISPR/Cas9 to generate NUCKS1-deficient HT1080 cells (Fig. S2 C). Upon exposure of these cells to γ-irradiation, more nuclei with RAD54 foci were detected in NUCKS1-deficient HT1080 cells compared with control cells (Fig. S2 D). Together, these results show that DNA damage–induced RAD54 foci are more abundant and persist longer in NUCKS1-deficient cells.
Data supporting Fig. 1 . (A) Western blots to show that the amount of nuclear-soluble and chromatin-bound RAD54 is not different between control cells (HeLa and KO-1+N) and NUCKS1 KO cells (KO-1 and KO-2). (B) Results from flow cytometry showing two-color fluorescence of cell cycle profiles of the cells used in Fig. 1 D. Y axis, EdU; x axis, SYTOX. (C) Western blot to show loss of NUCKS1 expression in HT1080 NUCKS1 KO cells. QM, loading control. (D) HT1080 NUCKS1 KO cells form more nuclei with RAD54 foci than HT1080 control cells (Ctrl, clonal isolate of HT1080 cells transfected with a nontargeting sgRNA). Data are from one experiment. (E) Representative micrographs of nuclear RAD54 foci obtained for the results presented in Fig. 1 E. Scale bars, 10 µm. (F) Results from flow cytometry showing two-color fluorescence of cell cycle profiles of the cells used in Fig. 1 E. Y axis, EdU; x axis; SYTOX. (G) Results from clonogenic survival assays to show that NUCKS1 KO HeLa cells (KO-1 and KO-2) are more sensitive to the cytotoxic effects of MMC than Ctrl-1 cells. Data points represent the mean from three independent experiments. Error bars, ±1 SD. *, P < 0.05; ****, P < 0.0001; ns, not significant; two-way ANOVA analysis.
Data supporting Fig. 1 . (A) Western blots to show that the amount of nuclear-soluble and chromatin-bound RAD54 is not different between control cells (HeLa and KO-1+N) and NUCKS1 KO cells (KO-1 and KO-2). (B) Results from flow cytometry showing two-color fluorescence of cell cycle profiles of the cells used in Fig. 1 D. Y axis, EdU; x axis, SYTOX. (C) Western blot to show loss of NUCKS1 expression in HT1080 NUCKS1 KO cells. QM, loading control. (D) HT1080 NUCKS1 KO cells form more nuclei with RAD54 foci than HT1080 control cells (Ctrl, clonal isolate of HT1080 cells transfected with a nontargeting sgRNA). Data are from one experiment. (E) Representative micrographs of nuclear RAD54 foci obtained for the results presented in Fig. 1 E. Scale bars, 10 µm. (F) Results from flow cytometry showing two-color fluorescence of cell cycle profiles of the cells used in Fig. 1 E. Y axis, EdU; x axis; SYTOX. (G) Results from clonogenic survival assays to show that NUCKS1 KO HeLa cells (KO-1 and KO-2) are more sensitive to the cytotoxic effects of MMC than Ctrl-1 cells. Data points represent the mean from three independent experiments. Error bars, ±1 SD. *, P < 0.05; ****, P < 0.0001; ns, not significant; two-way ANOVA analysis.
Next, we tested if knockdown of RAD54 in NUCKS1 KO cells would further increase their sensitivity to MMC. As observed for HeLa cells depleted for NUCKS1 (Parplys et al., 2015), NUCKS1 KO HeLa cells showed increased sensitivity to the cytotoxic effects of MMC (Fig. S2 G). NUCKS1 KO cells transfected with a nondepleting siRNA, and NUCKS1 KO cells and HeLa cells each depleted for RAD54 showed the same sensitivity to the cytotoxic effects of MMC (Fig. 1, F and G). These results suggest epistasis between NUCKS1 and RAD54 in human cells after MMC treatment.
NUCKS1 deficiency delays the formation of DNA damage–induced RAD51 foci
We endeavored to test if RAD54 function was compromised in NUCKS1 KO cells. Since defects in RAD54 were shown to diminish the robustness of RAD51 foci (Agarwal et al., 2011; Tan et al., 1999; van Veelen et al., 2005), we tested if this also was the case in NUCKS1 KO cells. Compared with HeLa cells, less RAD51 foci were detected in NUCKS1 KO cells at early times (2 h and 4 h) after IR exposure (Fig. 2 A and Fig. S3 A). However, at later times (8 h and 24 h) after IR, NUCKS1 KO and control cells displayed more similar levels of RAD51 foci (Fig. 2 A and Fig. S3 A). These results suggest that the formation of IR-induced RAD51 foci is transiently suppressed in NUCKS1 KO cells. The results also show that similar levels of RAD51 foci remain in NUCKS1-deficient and control cells at 24 h after IR exposure.
NUCKS1 deficiency leads to a transient delay in RAD51 foci formation, and NUCKS1 and RAD54 functionally interact. (A) The formation of IR-induced RAD51 foci is delayed in KO-1 and KO-2 cells. Bars represent the mean from two to three independent experiments (symbols). Error bars, ±1 SEM; *, P < 0.05; **, P < 0.01; ****, P < 0.0001; ns, not significant; two-way ANOVA analysis. (B) RAD54 knockdown impairs RAD51 foci formation in NUCKS1-expressing cells (HeLa and KO-1+N), but not in NUCKS1 KO cells (KO-1 and KO-2). Bars represent the mean from two independent experiments (symbols). Error bars, ±1 SEM; **, P < 0.01; ***, P < 0.001; ns, not significant; two-way ANOVA analysis. (C i) Schematic of the D-loop reaction with naked pBluescript II SK(−) plasmid DNA. (C ii) NUCKS1 (60, 120, 600, or 1,200 nM) does not promote the RAD51-mediated D-loop reaction (lanes 2–5). RAD54 (100, 200, or 400 nM) promotes the RAD51-mediated D-loop reaction (lanes 6–8). NUCKS1 (60 or 600 nM) promotes the RAD51–RAD54-mediated D-loop reaction (compare lanes 9 and 10 with lane 6). (C iii) Quantification of the results from three independent experiments. Bars are the means. Error bars, ±1 SD. *, P < 0.05; **, P < 0.01; multiple t test analysis. (Di) Schematic of the D-loop reaction with chromatinized pBluescript II SK(−) plasmid DNA. (Dii) RAD54 (100 or 200 nM) promotes the RAD51-mediated D-loop reaction on chromatinized DNA (lanes 2 and 3). NUCKS1 (600 nM) promotes the RAD51-mediated D-loop reaction in the presence of RAD54 (lane 5), but not in the absence of RAD54 or ATP (lanes 7 and 6, respectively). (D iii) Quantification of the results from three independent experiments. Bars are the means. Error bars, ±1 SD. **, P < 0.01; ns, not significant; multiple t test analysis. (E i) Thin-layer chromatogram to test for the ATPase activity on dsDNA (125 ng) of RAD54 (100 nM), RAD54+NUCKS1 (100 nM + 300 nM), and NUCKS1 alone (300 and 600 nM). (E ii) Quantification of the results. Data are from one experiment only. (F i) Thin-layer chromatogram to show that NUCKS1 (150 and 300 nM) stimulates the ATPase activity of RAD54 on dsDNA. (F ii) Quantification of the results. Data points are the mean from three independent experiments. Error bars, ±1 SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; multiple t test analysis.
NUCKS1 deficiency leads to a transient delay in RAD51 foci formation, and NUCKS1 and RAD54 functionally interact. (A) The formation of IR-induced RAD51 foci is delayed in KO-1 and KO-2 cells. Bars represent the mean from two to three independent experiments (symbols). Error bars, ±1 SEM; *, P < 0.05; **, P < 0.01; ****, P < 0.0001; ns, not significant; two-way ANOVA analysis. (B) RAD54 knockdown impairs RAD51 foci formation in NUCKS1-expressing cells (HeLa and KO-1+N), but not in NUCKS1 KO cells (KO-1 and KO-2). Bars represent the mean from two independent experiments (symbols). Error bars, ±1 SEM; **, P < 0.01; ***, P < 0.001; ns, not significant; two-way ANOVA analysis. (C i) Schematic of the D-loop reaction with naked pBluescript II SK(−) plasmid DNA. (C ii) NUCKS1 (60, 120, 600, or 1,200 nM) does not promote the RAD51-mediated D-loop reaction (lanes 2–5). RAD54 (100, 200, or 400 nM) promotes the RAD51-mediated D-loop reaction (lanes 6–8). NUCKS1 (60 or 600 nM) promotes the RAD51–RAD54-mediated D-loop reaction (compare lanes 9 and 10 with lane 6). (C iii) Quantification of the results from three independent experiments. Bars are the means. Error bars, ±1 SD. *, P < 0.05; **, P < 0.01; multiple t test analysis. (Di) Schematic of the D-loop reaction with chromatinized pBluescript II SK(−) plasmid DNA. (Dii) RAD54 (100 or 200 nM) promotes the RAD51-mediated D-loop reaction on chromatinized DNA (lanes 2 and 3). NUCKS1 (600 nM) promotes the RAD51-mediated D-loop reaction in the presence of RAD54 (lane 5), but not in the absence of RAD54 or ATP (lanes 7 and 6, respectively). (D iii) Quantification of the results from three independent experiments. Bars are the means. Error bars, ±1 SD. **, P < 0.01; ns, not significant; multiple t test analysis. (E i) Thin-layer chromatogram to test for the ATPase activity on dsDNA (125 ng) of RAD54 (100 nM), RAD54+NUCKS1 (100 nM + 300 nM), and NUCKS1 alone (300 and 600 nM). (E ii) Quantification of the results. Data are from one experiment only. (F i) Thin-layer chromatogram to show that NUCKS1 (150 and 300 nM) stimulates the ATPase activity of RAD54 on dsDNA. (F ii) Quantification of the results. Data points are the mean from three independent experiments. Error bars, ±1 SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; multiple t test analysis.
Data supporting Fig. 2 . (A) Representative micrographs of nuclear RAD51 foci obtained for the results presented in Fig. 2 A. Scale bars, 10 µm. (B) Representative Western blots to show RAD54 knockdown in cells used in Fig. 2 B. (C) Representative micrographs of nuclear RAD51 foci obtained for the results presented in Fig. 2 B. Scale bars, 10 µm. (D) SDS-PAGE to show purified human RAD54-FLAG and MBP-RAD51AP1-(His)6. (E) SDS-PAGE to show purified human RAD51. (F) SDS-PAGE to show purified human GST-NUCKS1 and (His)6-RAD51AP1-FLAG. (G) Naked and chromatinized pBluescript II SK(−) DNA after micrococcal nuclease (MNase) digest and deproteinization to show periodicity of nucleosomes and nucleosomal repeat lengths (lanes 5–7). (H) Western blot to show lost expression of RAD54 in RAD54 KO HeLa cells. MW, molecular weight.
Data supporting Fig. 2 . (A) Representative micrographs of nuclear RAD51 foci obtained for the results presented in Fig. 2 A. Scale bars, 10 µm. (B) Representative Western blots to show RAD54 knockdown in cells used in Fig. 2 B. (C) Representative micrographs of nuclear RAD51 foci obtained for the results presented in Fig. 2 B. Scale bars, 10 µm. (D) SDS-PAGE to show purified human RAD54-FLAG and MBP-RAD51AP1-(His)6. (E) SDS-PAGE to show purified human RAD51. (F) SDS-PAGE to show purified human GST-NUCKS1 and (His)6-RAD51AP1-FLAG. (G) Naked and chromatinized pBluescript II SK(−) DNA after micrococcal nuclease (MNase) digest and deproteinization to show periodicity of nucleosomes and nucleosomal repeat lengths (lanes 5–7). (H) Western blot to show lost expression of RAD54 in RAD54 KO HeLa cells. MW, molecular weight.
We asked if RAD51 foci formation in NUCKS1 KO cells would be impacted further by RAD54 loss. To do so, we depleted RAD54 in NUCKS1 KO and control cells (Fig. S3 B), irradiated exponentially growing cell cultures with 8 Gy γ-rays, and quantified RAD51 foci in fixed samples at 2–24 h after exposure. In agreement with prior studies performed in mouse embryonic stem cells (Agarwal et al., 2011; Tan et al., 1999; van Veelen et al., 2005), we found that RAD54 knockdown transiently suppresses the formation of RAD51 foci in HeLa cells and in NUCKS1 KO cells ectopically expressing NUCKS1 (here: KO-1+N; Fig. 2 B and Fig. S3 C). At 2–8 h after IR, in both RAD54-depleted HeLa and RAD54-depleted KO-1+N cells, less nuclei with RAD51 foci were detected than in HeLa and KO-1+N cells transfected with negative control siRNA (Fig. 2, compare A with B). At 24 h, more nuclei with RAD51 foci remained in RAD54-depleted HeLa and KO-1+N cells than in the same cell lines without RAD54 depletion (Fig. 2, compare A with B). These results suggest that RAD54 is important for both the formation and the timely resolution of RAD51 foci in human cells expressing the NUCKS1 protein. Surprisingly, in NUCKS1 KO cells (KO-1 and KO-2), depletion of RAD54 largely alleviated the transient delay in RAD51 foci formation (Fig. 2 B), suggesting that, in the absence of NUCKS1, RAD54 is not required for the formation of a robust presynaptic RAD51 filament. This is in contrast to what is observed in NUCKS1 KO cells that express wild-type levels of RAD54 and in which a transient delay in IR-induced RAD51 foci formation is observed (Fig. 2 A). However, similar to control cells, NUCKS1 KO cells depend on RAD54 to resolve RAD51 foci to wild-type levels at 24 h (Fig. 2 B).
NUCKS1 stimulates the RAD51–RAD54-mediated D-loop reaction
In the HR reaction, the joint DNA molecule, also known as the D-loop, is a central intermediate that forms when the RAD51-ssDNA nucleoprotein filament invades a homologous double-stranded DNA (dsDNA) donor molecule. RAD51-mediated strand invasion and the impact of accessory proteins on this process can be monitored in vitro using the oligonucleotide-based D-loop assay (Raynard and Sung, 2009). RAD54 functionally cooperates with RAD51 in this assay (Petukhova et al., 1998; Petukhova et al., 1999; Sigurdsson et al., 2002). Given our results that suggested a functional relationship between RAD54 and NUCKS1, we asked if NUCKS1 may affect D-loop formation in the presence of RAD54. In accord with our previous study (Parplys et al., 2015), the addition of increasing amounts of NUCKS1 to the D-loop reaction (in the absence of RAD54) did not stimulate RAD51-mediated strand invasion (Fig. 2 C, lanes 2–5; and Fig. S3, D–F). Addition of RAD54 to RAD51-ssDNA nucleoprotein filaments led to ∼2–5% of the input oligonucleotide being converted into D-loops (Fig. 2 C, lanes 6–8; and Fig. S3 D). Notably, the addition of an amount of NUCKS1 sub-stoichiometric to that of RAD51 enhanced RAD51–RAD54-mediated D-loop formation further two- to fourfold (Fig. 2 C, lanes 9 and 10; P < 0.05 and P < 0.01, respectively).
NUCKS1 is a chromatin-associated protein and greatly prefers binding to chromatin over naked dsDNA (Østvold et al., 1985; Parplys et al., 2015). Hence, we decided to also explore the addition of NUCKS1 to a D-loop reaction in which chromatinized plasmid DNA was used as a template (Fig. 2 D and Fig. S3 G). In this reaction, RAD54 overcomes the nucleosome barrier to the homologous pairing reaction catalyzed by RAD51 (Alexeev et al., 2003; Alexiadis and Kadonaga, 2002; Jaskelioff et al., 2003), as also observed by us (Fig. 2 D, lanes 2 and 3). Addition of NUCKS1 to this reaction stimulated D-loop formation four- to sixfold compared with RAD54 alone (Fig. 2 D, lane 5; P < 0.01). These results show that the NUCKS1 protein is able to stimulate the RAD51–RAD54-mediated homologous pairing reaction with either dsDNA or chromatin. Since RAD54’s ATPase activity is required to promote homologous pairing between the RAD51-ssDNA nucleoprotein filament and the dsDNA or chromatin donor (Jaskelioff et al., 2003; Petukhova et al., 1998; Petukhova et al., 1999), we asked if the ATPase activity of RAD54 would be affected in the presence of NUCKS1. In the presence of dsDNA, NUCKS1 itself did not have any ATPase activity (Fig. 2 E, lanes 4 and 5) but greatly stimulated ATP hydrolysis by RAD54 (Fig. 2 E, compare lane 1 with lane 2). Compared with RAD54 and dsDNA alone (Fig. 2 F, lanes 1, 4, 7, and 10), a much higher rate of ATP hydrolysis was observed when NUCKS1 was combined with RAD54 in this assay (Fig. 2 F, lanes 2, 3, 5, 6, 8, 9, 11, and 12). These results suggest that the stimulatory effect of NUCKS1 on homologous strand pairing results from an enhancement of the RAD54 ATPase function.
NUCKS1 and RAD54 interact in vitro and in cells
To determine if NUCKS1 and RAD54 physically interact, a GST pull-down assay with the purified proteins was performed. RAD54 copurified with GST-NUCKS1 on glutathione agarose beads (Fig. 3 A, lane 3). Similarly, NUCKS1 copurified with FLAG-RAD54 on anti-FLAG beads (Fig. 3 B, lane 4). Moreover, RAD51 copurified with RAD54 in the presence of NUCKS1 (Fig. 3 C, lane 3).
NUCKS1 and RAD54 physically interact. (A) SDS-PAGE to show the interaction between GST-NUCKS1 and RAD54 in a GST pull-down assay (lane 3). RAD54 does not precipitate nonspecifically (lane 6). S, supernatant containing unbound proteins; W, wash; E, SDS eluate of glutathione resin. (B) Western blot to show the interaction between FLAG-RAD54 and NUCKS1 precipitated by anti-FLAG M2 affinity resin (lane 4). NUCKS1 does not precipitate nonspecifically (lane 8). S, supernatant containing unbound proteins; W, wash 1 and 2, respectively; E, eluate. (C) Western blot to show the interaction between FLAG-RAD54, NUCKS1, and RAD51 precipitated by anti-FLAG M2 affinity resin (lane 3). RAD51 does not precipitate nonspecifically (lane 4). (D) Western blot to show that endogenous NUCKS1 interacts weakly with full-length RAD54 (lane 6), the RAD54 lobe1+2 fragment (lane 9), and the C-terminal domain of RAD54 (lane 10), strongly with the RAD54 lobe 1 fragment (lane 8), and not with the RAD54 N-terminal domain (lane 7). IP, immunoprecipitation.
NUCKS1 and RAD54 physically interact. (A) SDS-PAGE to show the interaction between GST-NUCKS1 and RAD54 in a GST pull-down assay (lane 3). RAD54 does not precipitate nonspecifically (lane 6). S, supernatant containing unbound proteins; W, wash; E, SDS eluate of glutathione resin. (B) Western blot to show the interaction between FLAG-RAD54 and NUCKS1 precipitated by anti-FLAG M2 affinity resin (lane 4). NUCKS1 does not precipitate nonspecifically (lane 8). S, supernatant containing unbound proteins; W, wash 1 and 2, respectively; E, eluate. (C) Western blot to show the interaction between FLAG-RAD54, NUCKS1, and RAD51 precipitated by anti-FLAG M2 affinity resin (lane 3). RAD51 does not precipitate nonspecifically (lane 4). (D) Western blot to show that endogenous NUCKS1 interacts weakly with full-length RAD54 (lane 6), the RAD54 lobe1+2 fragment (lane 9), and the C-terminal domain of RAD54 (lane 10), strongly with the RAD54 lobe 1 fragment (lane 8), and not with the RAD54 N-terminal domain (lane 7). IP, immunoprecipitation.
To map the NUCKS1 interaction domain on RAD54, we ectopically expressed HA-tagged full-length RAD54 or RAD54 deletions—i.e., the N-terminal domain (residues 1–155), lobe 1 (residues 156–404), lobe 1+2 (residues 156–667), and the C-terminal domain (residues 668–747)—in HeLa RAD54 KO cells (Fig. S3 H) and purified anti-HA protein complexes. NUCKS1 interacts moderately with full-length RAD54, with lobe 1+2, and with the RAD54 C-terminal domain, and more strongly with lobe 1 (Fig. 3 D, lanes 6, 9, 10, and 8, respectively). NUCKS1 does not interact with the N-terminal domain of RAD54 (Fig. 3 D, lane 7), which contains a RAD51 interaction motif (Chi et al., 2006a; Kovalenko et al., 2006; Mazin et al., 2000; Petukhova et al., 1999; Raschle et al., 2004; Sigurdsson et al., 2002).
NUCKS1 governs the timely formation of RAD54–RAD51 and RAD54–RAD51AP1 protein complexes
To further elucidate the consequences of impaired RAD54 activity in NUCKS1 KO cells, we assessed DNA repair synthesis in both HeLa and NUCKS1 KO-1 cells. We exposed cells to 4 or 8 Gy γ-rays and monitored the pulsed incorporation of the nucleotide analogue EdU (as a marker for ongoing DNA repair synthesis) in G2-phase cells 8 h after γ-ray exposure (Fig. S4 A). First, we tested the effects of RAD54 knockdown on DNA repair synthesis in both HeLa and NUCKS1 KO-1 cells in G2 phase (identified by their pan-nuclear CENP-F staining). In agreement with a previous study (Spies et al., 2016), we found that knockdown of RAD54 completely abrogated DNA repair synthesis in HeLa cells (Fig. S4, B and C). Knockdown of RAD54 also completely abrogated DNA repair synthesis in NUCKS1 KO cells (Fig. S4, B and C). In the presence of RAD54, EdU patches arose within 4 h after IR in both HeLa and NUCKS1 KO-1 cells, and, although overall not significantly different from HeLa cells, in individual experiments more nuclei with repair patches were observed in NUCKS1 KO-1 cells at later times (8–48 h) after exposure (Fig. S4 D). These results could suggest that the absence of NUCKS1 may delay HR DNA repair synthesis. However, in contrast to cells with RAD54 knockdown (Spies et al., 2016 and this study), loss of NUCKS1 does not abrogate DNA repair synthesis.
Data supporting Fig. 4 . (A) Representative micrographs to show EdU repair patches (red) in nuclei with RAD54 foci (green) at 8 h after 8 Gy γ-irradiation. Scale bar, 10 µm. (B) DNA repair synthesis after γ-irradiation (as determined by EdU incorporation in G2-phase cells) is abrogated in both HeLa and NUCKS1 KO-1 cells after RAD54 knockdown. (C) Representative micrographs obtained for the data shown in B. Scale bars, 10 µm. S, S-phase cells; G2, G2-phase cells. (D) DNA repair synthesis after γ-irradiation is slightly delayed in NUCKS1 KO-1 cells. Bars represent the means from one to three independent experiments (symbols). Error bars, ±1 SEM; ns, not significant (P > 0.185 for all data points); two-way ANOVA analysis; MW, molecular weight.
Data supporting Fig. 4 . (A) Representative micrographs to show EdU repair patches (red) in nuclei with RAD54 foci (green) at 8 h after 8 Gy γ-irradiation. Scale bar, 10 µm. (B) DNA repair synthesis after γ-irradiation (as determined by EdU incorporation in G2-phase cells) is abrogated in both HeLa and NUCKS1 KO-1 cells after RAD54 knockdown. (C) Representative micrographs obtained for the data shown in B. Scale bars, 10 µm. S, S-phase cells; G2, G2-phase cells. (D) DNA repair synthesis after γ-irradiation is slightly delayed in NUCKS1 KO-1 cells. Bars represent the means from one to three independent experiments (symbols). Error bars, ±1 SEM; ns, not significant (P > 0.185 for all data points); two-way ANOVA analysis; MW, molecular weight.
Given the results described above, we reasoned that NUCKS1 may impact HR at a stage different from the repair synthesis stage. Hence, we used the in situ proximity ligation assay (PLA; for schematic, see Fig. S5 A; Fredriksson et al., 2002; Gullberg et al., 2003), a multistep technique by which close-proximity protein interactions within a range <40 nm are detected (Gauthier et al., 2015), to monitor the associations of RAD51 and RAD51AP1 with RAD54 in cells and upon exposure to IR. First, we ensured that all PLA signals detected were specific using the appropriate negative controls (Fig. S5, B–D). Next, we compared the number of RAD54–RAD51 PLA signals/nucleus between HeLa cells and NUCKS1 KO-1 cells (Fig. 4 A). We found that the formation of RAD54–RAD51 PLA signals was significantly delayed in KO-1 cells at all times after exposure to 8 Gy γ-rays (P < 0.0001). Moreover, sham-irradiated HeLa cells contained significantly more RAD54–RAD51 PLA foci/nucleus than sham-irradiated KO-1 cells (0.52 ± 0.81 vs. 0.10 ± 0.38, respectively; P < 0.0001). Similarly, at all times after IR, the formation of RAD54–RAD51AP1 PLA signals occurred much later in KO-1 cells than in HeLa cells (P < 0.0001). However, sham-irradiated KO-1 cells contained significantly more RAD54–RAD51AP1 PLA foci/nucleus than sham-irradiated HeLa cells (0.38 ± 0.69 vs. 0.07 ± 0.29, respectively; P < 0.0001).
Data supporting Fig. 4 . (A) Schematic of the PLA. (B) Representative micrographs obtained after PLA assay without primary antibodies to show that PLA Plus and Minus probes (red) and goat anti-mouse AlexaFluor-488 secondary antibody (green) do not lead to nonspecific signals in HeLa and KO-1 cells. Scale bar, 10 µm. (C) Top: RAD54–RAD51 PLA signals (red) and RAD54 foci (green) are obtained in HeLa cells transfected with a nondepleting siRNA, but not in HeLa cells that are KO for RAD54 (Fig. S3 H). Knockdown of RAD51 in HeLa cells (si-RAD51) also abrogates RAD54–RAD51 PLA signals (red) and almost all RAD54 foci (green). Bottom: RAD54–RAD51AP1 PLA signals do not form in RAD54 KO and RAD51AP1 KO (here, AP1 KO) HeLa cells. RAD51AP1 KO cells were described earlier (Liang et al., 2019). In contrast, IR-induced RAD54 foci (green) are formed in RAD51AP1 KO, but not in RAD54 KO cells. Scale bar, 10 µm. (D) Top: RAD54–RAD51 PLA signals (red) are detected in sham-irradiated HeLa cells, but not in RAD54 KO cells. Bottom: Few RAD54–RAD51AP1 PLA signals (red) and no RAD54 foci (green) are detected in sham-irradiated HeLa cells. No RAD54–RAD51AP1 PLA signals (red) are detected in RAD54 KO cells. Scale bar, 10 µm. (E) Good overlap (white arrows) between RAD54 foci (green) and the RAD54–RAD51AP1 PLA signals (red) in irradiated HeLa cells. Blue line indicates nuclear periphery filtered on the DAPI signal. Scale bar, 10 µm. (F) Western blots to show the extent of RAD51AP1 knockdown in HeLa and KO-1 cells. RAD54, loading control. *, nonspecific signal. si-AP1, RAD51AP1-depleting siRNA. (G) Western blots to show the extent of XRCC4 knockdown in HeLa and KO-1 cells. PARP1, loading control; si-control, nondepleting negative control siRNA; si-XRCC4, XRCC4-depleting siRNA.
Data supporting Fig. 4 . (A) Schematic of the PLA. (B) Representative micrographs obtained after PLA assay without primary antibodies to show that PLA Plus and Minus probes (red) and goat anti-mouse AlexaFluor-488 secondary antibody (green) do not lead to nonspecific signals in HeLa and KO-1 cells. Scale bar, 10 µm. (C) Top: RAD54–RAD51 PLA signals (red) and RAD54 foci (green) are obtained in HeLa cells transfected with a nondepleting siRNA, but not in HeLa cells that are KO for RAD54 (Fig. S3 H). Knockdown of RAD51 in HeLa cells (si-RAD51) also abrogates RAD54–RAD51 PLA signals (red) and almost all RAD54 foci (green). Bottom: RAD54–RAD51AP1 PLA signals do not form in RAD54 KO and RAD51AP1 KO (here, AP1 KO) HeLa cells. RAD51AP1 KO cells were described earlier (Liang et al., 2019). In contrast, IR-induced RAD54 foci (green) are formed in RAD51AP1 KO, but not in RAD54 KO cells. Scale bar, 10 µm. (D) Top: RAD54–RAD51 PLA signals (red) are detected in sham-irradiated HeLa cells, but not in RAD54 KO cells. Bottom: Few RAD54–RAD51AP1 PLA signals (red) and no RAD54 foci (green) are detected in sham-irradiated HeLa cells. No RAD54–RAD51AP1 PLA signals (red) are detected in RAD54 KO cells. Scale bar, 10 µm. (E) Good overlap (white arrows) between RAD54 foci (green) and the RAD54–RAD51AP1 PLA signals (red) in irradiated HeLa cells. Blue line indicates nuclear periphery filtered on the DAPI signal. Scale bar, 10 µm. (F) Western blots to show the extent of RAD51AP1 knockdown in HeLa and KO-1 cells. RAD54, loading control. *, nonspecific signal. si-AP1, RAD51AP1-depleting siRNA. (G) Western blots to show the extent of XRCC4 knockdown in HeLa and KO-1 cells. PARP1, loading control; si-control, nondepleting negative control siRNA; si-XRCC4, XRCC4-depleting siRNA.
NUCKS1 deficiency negatively impacts HR protein complex formation. (A) RAD54–RAD51 PLA foci in sham-irradiated and irradiated HeLa and NUCKS1 KO-1 cells. Symbols, average values for 250 nuclei from two to three independent experiments each. Bars, overall means. Error bars, ±1 SD. ****, P < 0.0001; one-way ANOVA analysis. (B) RAD54–RAD51AP1 PLA foci in sham-irradiated and irradiated HeLa and NUCKS1 KO-1 cells. Symbols, average values for 250 nuclei from two to three independent experiments each. Bars, overall means. Error bars, ±1 SD. ****, P < 0.0001; one-way ANOVA analysis. (C) Western blots to show that increasing amounts of RAD51AP1 can interfere with the RAD54–RAD51 complex. Interaction between FLAG-RAD54 and RAD51 precipitated by anti-FLAG M2 affinity resin (lane 1). Interaction between FLAG-RAD54 and MBP-RAD51AP1 precipitated by anti-FLAG M2 affinity resin (lane 2). Increasing the amounts of RAD51AP1 competes with the interaction between FLAG-RAD54 and RAD51 (lanes 3 and 4, respectively). RAD51 does not precipitate nonspecifically; excessive MBP-RAD51AP1 (2 µM) does precipitate nonspecifically on anti-FLAG M2 affinity resin (lane 5). Blue, relative signal intensities for RAD51. (D) Western blot to show that purified NUCKS1 and RAD51AP1 interact (lane 3). NUCKS1 does not precipitate nonspecifically on anti-FLAG M2 resin (lane 2). (E) Preincubation of NUCKS1 with RAD51AP1 diminishes the amount of either protein precipitated in anti-FLAG RAD54 complexes (lane 5). Neither NUCKS1 nor RAD51AP1 precipitates nonspecifically on anti-FLAG M2 resin (lane 2). RAD51AP1 interacts with RAD54 (lane 3). NUCKS1 interacts with RAD54 (lane 4). Blue, relative signal intensities for NUCKS1 and RAD51AP1. (F) Knockdown of RAD51AP1 (here, AP1) in NUCKS1 KO-1 cells reduces IR-induced RAD54 foci. Bars represent the means from two to four independent experiments (symbols). Error bars, ±1 SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant; two-way ANOVA analysis. si-control, nondepleting negative control siRNA. si-AP1, RAD51AP1-depleting siRNA (see Fig. S5 F). (G) Results from clonogenic survival assays to show that KO-1 cells with XRCC4 knockdown are more sensitive to the cytotoxic effects of γ-rays than HeLa cells with XRCC4 knockdown. Data points are the mean from three independent experiments. Error bars, ±1 SEM. ***, P < 0.001; ****, P < 0.0001; ns, not significant; two-way ANOVA analysis. si-control, nondepleting negative control siRNA; si-XRCC4, XRCC4-depleting siRNA (see Fig. S5 G).
NUCKS1 deficiency negatively impacts HR protein complex formation. (A) RAD54–RAD51 PLA foci in sham-irradiated and irradiated HeLa and NUCKS1 KO-1 cells. Symbols, average values for 250 nuclei from two to three independent experiments each. Bars, overall means. Error bars, ±1 SD. ****, P < 0.0001; one-way ANOVA analysis. (B) RAD54–RAD51AP1 PLA foci in sham-irradiated and irradiated HeLa and NUCKS1 KO-1 cells. Symbols, average values for 250 nuclei from two to three independent experiments each. Bars, overall means. Error bars, ±1 SD. ****, P < 0.0001; one-way ANOVA analysis. (C) Western blots to show that increasing amounts of RAD51AP1 can interfere with the RAD54–RAD51 complex. Interaction between FLAG-RAD54 and RAD51 precipitated by anti-FLAG M2 affinity resin (lane 1). Interaction between FLAG-RAD54 and MBP-RAD51AP1 precipitated by anti-FLAG M2 affinity resin (lane 2). Increasing the amounts of RAD51AP1 competes with the interaction between FLAG-RAD54 and RAD51 (lanes 3 and 4, respectively). RAD51 does not precipitate nonspecifically; excessive MBP-RAD51AP1 (2 µM) does precipitate nonspecifically on anti-FLAG M2 affinity resin (lane 5). Blue, relative signal intensities for RAD51. (D) Western blot to show that purified NUCKS1 and RAD51AP1 interact (lane 3). NUCKS1 does not precipitate nonspecifically on anti-FLAG M2 resin (lane 2). (E) Preincubation of NUCKS1 with RAD51AP1 diminishes the amount of either protein precipitated in anti-FLAG RAD54 complexes (lane 5). Neither NUCKS1 nor RAD51AP1 precipitates nonspecifically on anti-FLAG M2 resin (lane 2). RAD51AP1 interacts with RAD54 (lane 3). NUCKS1 interacts with RAD54 (lane 4). Blue, relative signal intensities for NUCKS1 and RAD51AP1. (F) Knockdown of RAD51AP1 (here, AP1) in NUCKS1 KO-1 cells reduces IR-induced RAD54 foci. Bars represent the means from two to four independent experiments (symbols). Error bars, ±1 SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant; two-way ANOVA analysis. si-control, nondepleting negative control siRNA. si-AP1, RAD51AP1-depleting siRNA (see Fig. S5 F). (G) Results from clonogenic survival assays to show that KO-1 cells with XRCC4 knockdown are more sensitive to the cytotoxic effects of γ-rays than HeLa cells with XRCC4 knockdown. Data points are the mean from three independent experiments. Error bars, ±1 SEM. ***, P < 0.001; ****, P < 0.0001; ns, not significant; two-way ANOVA analysis. si-control, nondepleting negative control siRNA; si-XRCC4, XRCC4-depleting siRNA (see Fig. S5 G).
As RAD54 is critical for robust DNA damage–induced RAD51 foci (Agarwal et al., 2011; Tan et al., 1999; van Veelen et al., 2005), we reasoned that the increased association between RAD54 and RAD51AP1 in unperturbed NUCKS1 KO-1 cells may interfere with RAD54’s supporting role in RAD51-ssDNA nucleoprotein filament formation. To test if RAD51AP1 would interfere with the RAD54–RAD51 interaction in vitro, we used the purified proteins in a competitive pull-down assay. In accord with prior studies (Clever et al., 1997; Golub et al., 1997), we show that RAD54 and RAD51 physically interact (Fig. 4 C, lane 1). RAD54 also physically interacts with RAD51AP1 (Fig. 4 C, lane 2). Notably, the addition of increasing amounts of RAD51AP1 to the preformed RAD54–RAD51 complex diminishes the interaction between RAD54 and RAD51 (Fig. 4 C, lanes 3 and 4). Together, these results suggest that RAD54–RAD51, RAD54–RAD51AP1, and RAD51AP1–RAD51 are competitive protein complexes that exist both in vitro and in cells.
Due to the lack of an appropriate combination of antibodies, we were unable to monitor NUCKS1-RAD51AP1/RAD54 PLA signals in cells. Hence, we used purified proteins to test for a direct and competitive interaction between NUCKS1, RAD51AP1, and RAD54 in vitro. We show that NUCKS1 copurifies with FLAG-RAD51AP1 on anti-FLAG resin (Fig. 4 D, lane 3). Next, we asked if the presence of RAD51AP1 would affect the interaction between NUCKS1 and RAD54. As expected, both RAD51AP1 and NUCKS1 copurified with FLAG-RAD54 on anti–FLAG-M2 resin (Fig. 4 E, lanes 3 and 4, respectively). However, preincubation of RAD51AP1 with NUCKS1 diminished the amounts of both RAD51AP1 and NUCKS1 precipitated in anti–FLAG-RAD54 complexes (Fig. 4 E, lane 5). These results suggest that both RAD51AP1 and NUCKS1 can exchange with a protein complex involving RAD54 in vitro.
Depletion of RAD51AP1 in NUCKS1 KO cells reduces RAD54 foci
Since, under unperturbed conditions, more RAD54 is associated with RAD51AP1 in NUCKS1 KO cells than in HeLa cells (Fig. 4 B), we asked if knockdown of RAD51AP1 would affect RAD54 foci formation. We show that knockdown of RAD51AP1 in NUCKS1 KO-1 cells diminishes the amount of RAD54 foci to close to the levels detected in HeLa cells (Fig. 4 F and Fig. S5 F). In contrast, knockdown of RAD51AP1 in HeLa cells increases their RAD54 foci in both sham-irradiated cells and after exposure to IR (Fig. 4 F). These results suggest that RAD51AP1 is required to maintain wild-type kinetics of RAD54 foci in HeLa cells. In NUCKS1-deficient cells, however, the presence of RAD51AP1 appears to contribute to the elevated levels of RAD54 foci detected.
NUCKS1 KO cells more prominently engage NHEJ after radiation damage
We hypothesized that the abnormality in protein focus and complex formation of HR proteins in NUCKS1 KO cells (Fig. 1, D and E; Fig. 2, A and B; and Fig. 4, A and B) may force these cells to more frequently use the NHEJ pathway upon treatment with γ-irradiation. To test this, we knocked down the NHEJ protein XRCC4 in both HeLa and NUCKS1 KO-1 cells (Fig. S5 G) and assessed clonogenic survival in these cell populations after increasing doses of γ-rays. Compared with XRCC4-depleted HeLa cells, XRCC4-depleted NUCKS1 KO cells showed increased sensitivity to the cytotoxic effects of γ-rays (Fig. 4 G). These results show that NUCKS1-deficient HeLa cells more heavily rely on the NHEJ repair pathway to mend DSBs induced by IR.
Discussion
In the study we present here, the mechanisms by which NUCKS1 regulates DNA repair by HR are uncovered. We show that NUCKS1 both physically and functionally interacts with the DNA motor protein RAD54. We quantified the formation of heteroduplex DNA in vitro using the D-loop assay. We found that NUCKS1 stimulates this reaction exclusively in the presence of RAD54. We previously showed that NUCKS1 and RAD51 do not interact (Parplys et al., 2015). Hence, we infer that the stimulation of the RAD54–RAD51-mediated D-loop reaction is, at least in part, a consequence of NUCKS1 interacting with RAD54.
RAD54 is known to stabilize the RAD51 filament (Mazin et al., 2003), and RAD54-deficient cells show a transient delay in the formation of DNA damage–induced RAD51 foci (Agarwal et al., 2011; Tan et al., 1999; van Veelen et al., 2005). A transient delay in RAD51 foci formation also is observed in NUCKS1 KO cells, presumably resulting from their inability to timely engage RAD51 with RAD54. Interestingly, RAD51 foci are formed with wild-type kinetics in NUCKS1 KO cells depleted for RAD54. We speculate that, in the absence of RAD54 (but not in the presence of impaired RAD54), a different protein can substitute in stabilizing the RAD51 filament, such as the RAD54 paralog RAD54B or RAD51AP1. In this context, it is worth noting that in unperturbed NUCKS1-deficient cells, the association between RAD54 and RAD51 is diminished. This is not the case in unperturbed HeLa cells, in which the existing RAD54–RAD51 complexes readily may allow the stabilization of the newly formed RAD51-ssDNA nucleoprotein filament after DNA damage has occurred.
RAD54 competitively interacts with both RAD51 and RAD51AP1 (Fig. 4 C). In accord with these findings, RAD51AP1 knockdown in NUCKS1 KO cells reverses RAD54 foci to close to the wild-type level (Fig. 4 F). RAD54 now may be able to engage with RAD51 to stabilize the RAD51 filament and then drive homology search and strand invasion. In contrast, in HeLa cells expressing NUCKS1 (but no RAD51AP1), RAD54 foci are increased due to reduced RAD51 turnover and longer persisting RAD51 foci, as we and others have shown (Modesti et al., 2007; Parplys et al., 2014; Wiese et al., 2007).
NUCKS1 interacts with RAD54 and interferes with the RAD54–RAD51AP1 complex (Fig. 4 E). Combined with the PLA data presented above, these findings suggest that, in the presence of NUCKS1, the formation of the RAD54–RAD51AP1 complex is limited. We suggest that NUCKS1 is a new regulator of spatiotemporal events in the HR reaction, as it (1) controls the early interaction between RAD51 and RAD54 (preceding the D-loop) and (2) promotes the RAD51–RAD54-mediated strand invasion step during D-loop formation (Fig. 5). In the absence of NUCKS1, RAD54’s association with RAD51AP1 is not restricted, and the spatiotemporal pattern of proteins recruited to the RAD51 filament and involved in the formation of heteroduplex DNA is delayed. Given the relatively mild increase in MMC cytotoxicity of NUCKS1-deficient cells, and given their ability to eventually form RAD54–RAD51 and RAD54–RAD51AP1 protein complexes, it appears that a backup pathway may operate in the absence of NUCKS1. Whether this alternative mechanism of RAD54 activation acts in parallel or stems from the continuous absence of NUCKS1 in these KO cells remains to be investigated.
Model to explain how NUCKS1 regulates spatiotemporal events in the HR reaction. In undamaged cells, RAD54 and RAD51AP1 are associated with NUCKS1 to prevent the inappropriate association of RAD54 with RAD51AP1. Upon exposure of cells to IR, RAD54 is able to (1) stabilize the presynaptic RAD51 filament (which does not require RAD54 ATPase activity) and (2) stimulate the RAD54 ATPase for strand invasion and formation of heteroduplex DNA. In the absence of NUCKS1, RAD54 is inappropriately complexed with RAD51AP1. This complex negatively impacts the timely formation of RAD51 foci (see Fig. 2 A), as it delays the interaction between RAD54 and RAD51 that is required for a stable RAD51-ssDNA nucleoprotein filament (Agarwal et al., 2011; Mazin et al., 2003; Tan et al., 1999; van Veelen et al., 2005). RAD51AP1 also promotes strand invasion (Modesti et al., 2007; Wiese et al., 2007).
Model to explain how NUCKS1 regulates spatiotemporal events in the HR reaction. In undamaged cells, RAD54 and RAD51AP1 are associated with NUCKS1 to prevent the inappropriate association of RAD54 with RAD51AP1. Upon exposure of cells to IR, RAD54 is able to (1) stabilize the presynaptic RAD51 filament (which does not require RAD54 ATPase activity) and (2) stimulate the RAD54 ATPase for strand invasion and formation of heteroduplex DNA. In the absence of NUCKS1, RAD54 is inappropriately complexed with RAD51AP1. This complex negatively impacts the timely formation of RAD51 foci (see Fig. 2 A), as it delays the interaction between RAD54 and RAD51 that is required for a stable RAD51-ssDNA nucleoprotein filament (Agarwal et al., 2011; Mazin et al., 2003; Tan et al., 1999; van Veelen et al., 2005). RAD51AP1 also promotes strand invasion (Modesti et al., 2007; Wiese et al., 2007).
Ataxia telangiectasia mutated (ATM)–deficient DT40 cells show similarly altered kinetics of Rad54 and Rad51 foci, findings that were among the first to demonstrate that HR proceeds abnormally in ATM-deficient cells (Morrison et al., 2000). However, unlike in ATM-deficient DT40 cells, in which both Rad51 and Rad54 foci remain elevated (at least up to 8 h after 4 Gy γ-irradiation), RAD51 foci are back to wild-type levels in NUCKS1 KO cells at 24 h after exposure, whereas RAD54 foci, albeit declining, remain higher than in control cells. While this may seem perplexing, we think that many of the RAD54 foci in NUCKS1 KO cells result from unproductive RAD54–RAD51AP1 complexes not associated with RAD51. However, this could also be due to the accumulation of stable heterologous associations and nonproductive recombination intermediates, as recently shown for Saccharomyces cerevisiae Rad54 (Tavares et al., 2019), with potentially less detectable RAD51 protein, or to a technical issue in ICC and/or microscopy.
NUCKS1 is an ATM target and phosphorylated quickly (i.e., within 1 h) upon exposure of human cells to IR (Bennetzen et al., 2010; Bensimon et al., 2010; Matsuoka et al., 2007; Parplys et al., 2015). Similar to the findings reported by Morrison et al. (2000), we show that knockdown of the NHEJ protein XRCC4 further increases the sensitivity of NUCKS1 KO cells to IR. This effect is more pronounced in KO than in wild-type cells, suggesting that NUCKS1 KO cells more heavily rely on NHEJ after radiation damage. Such increased dependence on NHEJ may explain why NUCKS1 plays a role in protecting mice from radiation carcinogenesis (Yue et al., 2016) and human cells from chromosomal aberrations (Parplys et al., 2015). Whether or not phosphorylation of NUCKS1 by ATM is critical for RAD54 activity will be an important question to address in the future.
Taken together, the results from our study show that the NUCKS1 protein is a new regulator of the HR reaction in human cells: (1) NUCKS1 regulates access of RAD54 to RAD51 at the presynaptic stage for RAD51 filament stabilization, and (2) NUCKS1 promotes RAD51–RAD54-mediated DNA strand invasion. Since NUCKS1 is one of the most highly posttranslationally modified proteins in the human proteome (Wiśniewski et al., 2008; Qiu et al., 2015; Grundt et al., 2017), it will be of great interest to investigate if and how HR is affected by NUCKS1 modifications, a trait that further contributes to complicating our efforts directed toward a better understanding of the biology of this multifaceted protein and its role in genome maintenance mechanisms.
Materials and methods
Cell culture, transfection, and siRNAs
HeLa and HT1080 cells were obtained from American Type Culture Collection and were maintained as recommended. siRNAs were obtained from Qiagen. For the nondepleting negative control siRNA, the following target sequence was used: 5′-GATTCGAACGTGTCACGTCAA-3′ (Parplys et al., 2015; Zafar et al., 2010). The target siRNA sequences to knockdown RAD54, XRCC4, and RAD51AP1 were 5′-AAGCATTTATTCGAAGCATTT-3′, 5′-AGCCGCTATTACCGTATCTTA-3′, and 5′-AACCTCATATCTCTAATTGCA-3′ (Modesti et al., 2007; Parplys et al., 2015; Wiese et al., 2007; Zafar et al., 2010), respectively. siRNA forward transfections were performed using lipofectamine RNAiMAX (Invitrogen) on two consecutive days and according to the instructions of the manufacturer (Invitrogen). The concentration of siRNAs in transfections was 20 nM each. Cells were treated with drugs or IR at 96 h after the first transfection, and the extent of protein knockdown or expression was assessed by Western blot analysis.
Generation of NUCKS1 KO, RAD54 KO, and NUCKS1-complemented cells
To generate HeLa cell derivatives deficient for NUCKS1 by CRISPR/Cas9, two different approaches were used. First, single guide RNA (sgRNA) A and sgRNA G (Table S3) were cloned into pSpCas9n(BB)-2A-GFP (PX461; Addgene plasmid #48140), and sgRNA B and sgRNA H (Table S3) were cloned into pSpCas9n(BB)-2A-Puro (PX462; Addgene plasmid #48141). Both PX461 and PX462 were a gift from Feng Zhang (Massachusetts Institute of Technology, Cambridge, MA; Ran et al., 2013). HeLa cells were transfected with all four plasmids simultaneously, as described below, to generate clonal isolates in which the NUCKS1 gene is disrupted (Fig. S1, A–C). HeLa NUCKS1 KO-1 is one derivative generated through this approach. In the second approach, a combination of three NUCKS1 CRISPR/Cas9 KO plasmids, each containing one of three different sgRNAs (i.e., sgRNA Gecko-1, sgRNA Gecko-2, or sgRNA Gecko-3; Table S3) was purchased from Santa Cruz Biotechnology (sc-413018) and used to transfect HeLa and HT1080 cells, as described below. HeLa NUCKS1 KO-2 and HT1080 NUCKS1 KO are derivatives generated through this second approach.
To generate RAD54 KO HeLa cells, a combination of two RAD54 CRISPR/Cas9n(D10A) KO plasmids, each containing one of two different sgRNAs (i.e., sgRNA (54)-A and sgRNA (54)-B; Table S3) was purchased from Santa Cruz Biotechnology (sc-401750-NIC) and used to transfect HeLa cells.
To generate KO clones, cells were plated at 105 cells/well in regular growth medium without antibiotics in 24-well tissue culture plates 24 h before transfection. Cells were transfected using the cocktails of the sgRNA-containing plasmids described above and Lipofectamine 2000 (Thermo Fisher Scientific) according to the instructions by the manufacturer. At 24 h after transfection, the medium was replaced, and successful transfection with CRISPR/Cas9 KO plasmids was confirmed by fluorescent microscopy (GFP expression). Where appropriate, transfected cells were then further selected short term (i.e., for 24 h in 2 µg/ml puromycin) before cells were trypsinized, counted, and reseeded at 0.3 cells/well in 96-well cell culture plates and in regular growth medium without puromycin. Colonies were picked and expanded after 2 wk and tested for loss of NUCKS1 or RAD54 expression by Western blot analysis and ICC. Genomic DNA was isolated from HeLa cells, nonedited control, and edited clones using DNeasy Blood & Tissue Kit (Qiagen). NUCKS1- and RAD54-specific loci were PCR amplified using PCR primers, as listed in Table S1 and Table S2. PCR products were cloned into pCR-Blunt vector and transformed into TOP10 Escherichia coli cells, and plasmid DNA was prepared using ZR Plasmid Miniprep-Classic Kit (Zymo Research) and submitted for Sanger sequencing.
For expression of NUCKS1 in NUCKS1 KO cells, the NUCKS1 cDNA was cloned from NotI to BamHI into pcDNA3.1/hygro, and NUCKS1 KO cells were transfected with this plasmid and Lipofectamine 2000 (Thermo Fisher Scientific) according to the instructions by the manufacturer. For selection of clonal isolates, cells were plated at 10 cells/well in regular growth medium containing 700 µg/ml hygromycin in 96-well tissue culture plates 2 d after transfection. Ectopic expression of NUCKS1 was tested for by Western blot analysis and indirect immunofluorescence.
Generation of human RAD54 deletion constructs
Five constructs were made to express either full-length RAD54 or RAD54 truncations with C-terminal HA-tag in HeLa RAD54 KO cells. The full-length RAD54 cDNA (corresponding to amino acid residues 1–747) and RAD54 fragments (N-terminal domain [amino acid residues 1–155], lobe 1 [amino acid residues 156–404], Lobes 1+2 without the C-terminal domain [amino acid residues 156–667], and the C-terminal domain [amino acid residues 668–747]) were PCR amplified (for primer pairs, see Table S1) from pET32a-RAD54 (Raschle et al., 2004) and cloned from KpnI to NotI into pcDNA3.1/hygro.
Exposure to genotoxic agents and Western blot analysis
Acute exposure of cells to MMC (Sigma) occurred in regular growth medium at 37°C for 2 h and at the concentrations, as indicated. The medium was removed, and monolayers were washed twice in warm PBS before cells were incubated in regular growth medium at 37°C until fixation. For chronic exposure to MMC, cells were plated in regular growth medium containing MMC and kept at 37°C for 12 d, at which time the cells were fixed and stained with crystal violet to visualize colonies. Exposure to γ-rays and Western blot analyses followed our standard protocols (Parplys et al., 2014; Parplys et al., 2015; Wiese et al., 2006; Zhao et al., 2015). The primary antibodies that were used are α-RAD51AP1 (NB100-1129; Novus; 1:5,000; and our own α-RAD51AP1 antibody, as previously described in Dray et al., 2010), α-RAD54 (F-11; sc-374598; Santa Cruz Biotechnology; 1:500); α-NUCKS1 (Østvold et al., 2001), α-RAD51 (Ab-1; EMD Millipore; 1:4,000), α-PARP1 (ab6079; Abcam; 1:5,000), α-H3 (ab1791; Abcam; 1:10,000), α-XRCC4 (AHP387; AbDSerotec; 1:1,000), and α-FLAG (F3165; Sigma; 1:1,000). HRP-conjugated goat anti-rabbit or goat anti-mouse IgG (Jackson ImmunoResearch Laboratories; 1:10,000) were used as secondary antibodies. Western blot signals were quantified using Image Lab software version 5.2.1 (BioRad).
Cell cycle analysis and flow cytometry
Cell cycle analysis was performed according to the manufacturer’s instructions using SYTOX Red (ThermoFisher Scientific). Briefly, 106 cells were pelleted, washed in ice-cold PBS, pelleted again, and resuspended in 50 µl PBS. Cells were fixed by adding 2 ml of ice-cold 70% ethanol/PBS and kept at 4°C for at least 2 d. Fixed cells were pelleted, resuspended in 2 ml PBS, and incubated at room temperature for 60 min to allow rehydration. Cells were then permeabilized in 0.25% Triton X-100/PBS at room temperature for 30 min. The Click-iT EdU AlexaFluor-488 reaction was performed as described by the manufacturer (Thermo Fisher Scientific). First, cells were washed once in 1% BSA/PBS, and then 0.5 ml of a prepared Click-iT reaction cocktail was added to each sample. Samples were incubated at room temperature in the dark for 30 min and then washed once in 0.1% Triton X-100/1% BSA/PBS and once in 1% BSA/PBS. Cells were washed again, once in 0.1% Triton X-100/0.5% BSA/PBS, once in 0.1% Triton X-100/0.25% BSA/PBS, and once in 0.1% Triton X-100/0.1% BSA/PBS and finally pelleted and resuspended in PBS containing 0.5 µl/ml SYTOX Red and 80 µg/ml RNase A. Cells were stored in the dark for at least 2 h or overnight before data acquisition was conducted using a CyAn ADP cell analyzer (Beckman Coulter) with a 488-nm laser with a 530/40-nm band pass filter to detect EdU and a 635-nm laser with a 665/20-nm band pass filter to detect SYTOX. FlowJo version X (Tree Star) was used to fit the data for cell cycle analysis.
Cell fractionation
Cell fractionation was performed using the Subcellular Protein Fractionation Kit (Thermo Fisher Scientific) as described by the manufacturer.
Indirect immunostaining, microscopy, and image analysis
4-well chamber slides were seeded with 20,000 cells per chamber 72 h before treatment. After treatment, cells were washed in PBS once and fixed in 2% PFA/PBS for 15 min. Slides were washed twice in PBS. To detect RAD51 foci, cells were first pre-extracted with cytoskeleton buffer (10-mM Pipes, pH 7.0, 100-mM NaCl, 300-mM sucrose, and 3-mM MgCl2) containing 0.5% Triton X-100 for 3 min at room temperature and then washed with PBS once before fixation with 2% PFA for 15 min. Cells were permeabilized in PBS/0.25% Triton X-100 for 15 min at room temperature, washed with PBS, and blocked with PBS/0.1% Tween-20 (PBS-T) containing 1% BSA. Chamber slides were incubated with primary antibodies in PBS-T/1% BSA overnight and then washed with PBS-T and incubated with the appropriate AlexaFluor-488 or AlexaFluor-594 goat secondary antibodies (Thermo Fisher Scientific; 1:750) in PBS-T/1% BSA for 45 min at room temperature. After several washes in PBS, chamber slides were mounted in ProLong Gold with DAPI (Thermo Fisher Scientific). The working concentrations of the primary antibodies used were as follows: α-RAD51 (H-92; Santa Cruz Biotechnology; 1:1,000), α-RAD54 (F-11; sc-374598; Santa Cruz Biotechnology; 1:1,000), α-RAD51AP1 (NBP2-13197; Novus Biologicals; 1:1,000), and α-NUCKS1 (ab84710; Novus Biologicals; 1:300).
Images were taken using a 63× oil objective and a Zeiss Axio-Imager.Z2 microscope equipped with Zen Blue software (Carl Zeiss Microscopy). Images were obtained as Z-stack sections of 0.2 µm per section containing 18 Z-stacks for each channel. Image processing for foci quantification proceeded by separating the channels and producing a maximum projection file to identify and count both the number of foci and the number of cells. To do so, a combination of both ImageJ (http://imagej.nih.gov/ij/) and Cell Profiler (http://www.cellprofiler.org/) software packages was used with the following voxel settings: minimum voxel size, 10; maximum voxel size, 100; rolling ball for stacks, 150. 100–200 cells were assessed, and nuclei with more than five foci per nucleus were counted as positive.
DNA repair synthesis assay
Cells were exposed to either 4 or 8 Gy γ-rays and pulse labeled in medium containing 10-µM EdU for 20 min before fixation at 4–48 h after IR. Cells were fixed with 2% PFA at room temperature for 15 min and permeabilized in PBS/0.25% Triton X-100 for 15 min. Cells were incubated in mouse α-RAD54 antibody (F-11; sc-374598; Santa Cruz Biotechnology; 1:1,000) in PBS/1% BSA at 4°C overnight. Following two washes in PBS, cells were incubated in secondary goat anti-mouse AlexaFluor-488 antibody (Thermo Fisher Scientific; 1:750) in PBS/1% BSA for 45 min at room temperature and then fixed a second time in 2% PFA for 5 min. Sequentially, mouse α-CENP-F (610768; BD Transduction Laboratories; 1:500) and goat anti-mouse AlexaFluor-647 (Thermo Fisher Scientific; 1:750) antibodies were used to detect cells in G2 phase, and the Click-It reaction was performed with AlexaFluor-555 azide (Thermo Fisher Scientific) as instructed by the manufacturer.
Microscopy was performed as described above. Processing and analysis of micrographs were performed by ImageJ. Z-stacks were separated by channel, and G2 patches were processed and three-dimensionally reconstructed using the following voxel settings: minimum voxel size, 10; maximum voxel size, 100; rolling ball for stacks, 150. Then, individual cells were manually classified for presence of repair patches in G2-phase cells. Colocalization of G2-repair patches with RAD54 foci was determined by ImageJ using the JaCoP plugin.
PLA
Cells were seeded at 13,000 cells/well in 8-well chamber slides 48 h before their treatment with 8 Gy γ-rays. After treatment, cells were washed twice with PBS, fixed with 2% PFA for 15 min, and permeabilized with PBS/0.25% Triton X-100 for 10 min at room temperature. Cells were washed once in PBS, blocked with 1× Duolink blocking solution for 45 min at 37°C, and then incubated with primary antibodies in Duolink antibody diluent at 4°C overnight. On the next day, the slides were incubated with Duolink in situ PLA Plus and Minus probes (Sigma) in Duolink antibody diluent at 37°C for 1 h. Slides were then washed twice in wash buffer A (0.01-M Tris-HCl, 0.15-M NaCl, and 0.05% Tween 20, pH 7.4) for 5 min each. The ligation mix was prepared by diluting Duolink ligation stock (1:5) and ligase (1:40) in high-purity water. Slides were placed in a humidity chamber, and the ligation mix was added onto the slides (40 µl/well) and incubated at 37°C for 30 min. Slides were then washed twice in wash buffer A for 5 min each. The amplification mix was prepared by diluting the Duolink amplification stock (1:5) and rolling circle polymerase (1:80) in high-purity water. Slides were placed into the humidity chamber, and the amplification mix was placed onto the slides (40 µl/well) and incubated at 37°C for 100 min. Slides were washed twice in 60 ml of wash buffer B (0.2-M Tris-HCl and 0.1-M NaCl, pH 7.4) for 10 min each and once in 0.01× wash buffer B for 1 min. When detecting RAD54 foci along with the PLA foci, the slides were washed in PBS for 5 min at room temperature and incubated with goat anti-mouse AlexaFluor-488 antibody (1:500) for 45 min at room temperature. The slides were then washed in PBS twice for 5 min each, excess liquid was tapped off, and Prolong Gold antifade with DAPI (Thermo Fisher Scientific) was added to each well to mount slides with glass coverslips.
Purification of recombinant proteins
GST-NUCKS1, MBP-RAD51AP1, and RAD51 were purified as reported earlier (Parplys et al., 2015; Sigurdsson et al., 2001; Wiese et al., 2007).
For the expression and purification of human RAD54, E. coli Rosetta cells harboring the plasmid that expresses human RAD54 with N-terminal Trx-(His)6-tag and C-terminal FLAG tag were grown in Luria broth at 37°C until the A600 reached 0.8, followed by treatment with 0.4-mM IPTG for 16 h at 16°C to induce protein expression. All the purification steps were performed at 0–4°C. To prepare extract, 10 g of cell paste was suspended in 50 ml of cell breakage buffer A (25-mM Tris-HCl, pH 7.5, 10% glycerol, 0.5-mM EDTA, 150-mM KCl, 0.01% Igepal, and 1-mM DTT) and protease inhibitors (aprotinin, chymostatin, leupeptin, and pepstatin A at 3 µg/ml each and 1-mM PMSF) for sonication. After centrifugation (100,000×g for 90 min), the clarified lysate was incubated with 5 ml Ni2+-NTA agarose (Qiagen) for 1 h. The matrix was poured into a column (2.5 × 10 cm), washed with 50 ml of buffer B (25-mM Tris-HCl, pH 7.5, 10% glycerol, 0.5-mM EDTA, 0.01% Igepal, 1-mM DTT, and 100-mM KCl) and then with 10 ml each of 30- and 50-mM imidazole in buffer A before being eluted with 10 ml of 200-mM imidazole in buffer A. The 200-mM imidazole eluate was incubated with 3 ml anti-FLAG M2 affinity resin (Sigma) for 2 h. The resin was transferred to a column (1.5 × 15 cm) and washed with 50 ml of buffer C (25-mM Tris-HCl, pH 7.5, 300-mM KCl, 10% glycerol, 0.5-mM EDTA, 0.01% Igepal CA-630, 1-mM 2-mercaptoethanol, 5-mM MgCl2, and 2-mM ATP) before the bound proteins were eluted four times with 2 ml of buffer C containing the single FLAG peptide (200 µg/ml). The eluates were combined and mixed with 32 ml of buffer D (25-mM Tris-HCl, pH 7.5, 10% glycerol, 0.5-mM EDTA, 0.01% Igepal CA-630, and 1-mM 2-mercaptoethanol) and further fractionated in a 1-ml Mono S column (Amersham Pharmacia Biotech) using a 15-ml gradient of 150–500-mM KCl in buffer A. RAD54-containing fractions (330–360-mM KCl) were pooled and concentrated in a Centricon-30 concentrator (Amicon). The concentrated protein was divided into small aliquots and stored at −80°C.
For the expression of (His)6-RAD51AP1-FLAG protein in E. coli, RAD51AP1 was amplified from pOK24 (Kovalenko et al., 1997) using the primer pairs listed in Table S1 and cloned from BamHI to SalI into pQE-80L (Qiagen). For purification of this RAD51AP1 protein, E. coli Rosetta cells harboring the plasmid were grown in Luria broth at 37°C until the A600 reached 0.6, followed by treatment with 0.4-mM IPTG for 6 h at 37°C to induce protein expression. All the purification steps were performed at 0–4°C. To prepare extract, 5 g of cell paste was suspended in 25 ml of cell breakage buffer A (50-mM Tris-HCl, pH 7.5, 300-mM KCl, 1-mM EDTA, and 30-mM imidazole [GoldBio]) with protease inhibitors. Clarified lysate was incubated with pre-equilibrated Ni-resin (Invitrogen) for 2 h with gentle rotation. Ni-resin was washed four times with two-column volumes of buffer A containing 50-mM imidazole. Bound protein was eluted with buffer A containing 300-mM imidazole and immediately diluted 1:1 with buffer B (50-mM Tris-HCl, pH 7.5, 300-mM KCl, and 1-mM EDTA). Diluted protein was mixed with 1 ml of pre-equilibrated anti-FLAG M2 affinity resin (Sigma) in buffer B and incubated at 4°C for 2 h with gentle rotation. RAD51AP1 bound to anti-FLAG resin was washed four times with buffer B, and bound protein was eluted in buffer B containing 200 ng/µl 3× FLAG peptide. Eluted protein was dialyzed overnight against buffer C (50-mM Tris-HCl, pH 7.5, 300-mM KCl, 1-mM EDTA, 20% glycerol, and 2-mM DTT). Protein was quantified by SDS-PAGE and BSA standard.
In vitro affinity pull-down assays
GST-tagged NUCKS1 (1.3 µM) was incubated with human RAD54 (1 µM) in 30 µl of buffer B (25-mM Tris-HCl at pH 7.5, 10% glycerol, 0.5-mM EDTA, 0.05% Igepal, 1-mM DTT, and 100-mM KCl) in the presence of Benzonase (100 U; Sigma) on ice for 30 min, and then 10 µl of glutathione resin (GE Healthcare) was added. The GST-tagged protein and associated proteins were captured on the resin by gentle mixing at 4°C for 1 h. The resin was then washed three times with buffer B and treated with 20 µl of 2% SDS to elute the proteins. The supernatant, final wash, and SDS-eluted fractions (10 µl each) were analyzed by 4–15% SDS-PAGE and Coomassie Blue staining.
FLAG pull-downs in Fig. 3 were performed using anti-FLAG M2 affinity resin (Sigma) essentially as described by the manufacturer. Briefly, anti-FLAG resin was equilibrated in binding buffer (50-mM Tris-HCl at pH 7.5, 150-mM NaCl, 0.1% Triton X-100, and 2% BSA [when RAD51 was present in the reaction]). Purified RAD54 or RAD51AP1 (100 nM each) was added to the equilibrated beads and incubated at 4°C for 1 h with gentle agitation. Unbound protein was removed by centrifugation at 3,000 rpm for 3 min. Recombinant GST-NUCKS1 (150 nM) was added in a binding buffer and incubated at 4°C for 2 h with gentle agitation in the presence of DNase I (1 U/µg protein). For the competitive FLAG pull-down assay in Fig. 4 C, recombinant FLAG-RAD54 (180 nM) was incubated with equilibrated anti-FLAG M2 resin at 4°C for 1 h with gentle rotation. Unbound RAD54 was removed from the resin by centrifugation at 3,000 rpm for 5 min. Then RAD51 (180 nM), MBP-RAD51AP1 (250 nM), or RAD51 (180 nM) with increasing amounts of MBP-RAD51AP1 (1.25 µM, 2.5 µM) were incubated with the resin in 30 µl of binding buffer (25-mM Tris-HCl, pH 7.5, 10% glycerol, 0.5-mM EDTA, 150-mM KCl, 2-mM ATP, 2-mM MgCl2, 1-mM DTT, and 0.05% Igepal) at 4°C for 1 h with gentle rotation. The resin was washed twice in 200 µl of binding buffer, and bound protein was eluted in 15 µl of 2× SDS-loading buffer and boiled at 95°C for 5 min. 2 µl of the sample was fractionated by 10% SDS-PAGE, transferred onto a nitrocellulose membrane, and detected by Western blot analysis. For the competitive FLAG pull-down assay in Fig. 4 D, recombinant FLAG-RAD54 (100 nM) was incubated with equilibrated anti-FLAG M2 resin at 4°C for 1 h with gentle rotation. In the meantime, GST-NUCKS1 and MBP-RAD51AP1 (50 nM each) were incubated individually and together in 50 µl of binding buffer on ice for 1 h. Unbound RAD54 was removed from the resin by centrifugation at 3,000 rpm for 3 min. Then RAD51AP1, NUCKS1, or RAD51AP1/NUCKS1 (50 nM each) was added to the resin-bound RAD54 and incubated at 4°C for 2 h with gentle rotation and in the presence of DNase I (1 U/µg protein). The resin was washed four times in 100 µl of binding buffer each, and bound protein was eluted in binding buffer containing 150 ng/µl of 3× FLAG peptide (Sigma). Eluted protein was fractionated by 10% SDS-PAGE, transferred onto a polyvinylidene fluoride membrane, and detected by Western blot analysis.
To monitor formation of the trimeric RAD51–RAD54–NUCKS1 complex, GST-NUCKS1 was incubated with RAD51 first before FLAG-RAD54 was added, and the trimer was purified by anti-FLAG M2 affinity resin as described above for Fig. 4 D.
Anti-HA immunoprecipitations
HA-tagged full-length RAD54 or RAD54 truncations were expressed in RAD54 KO HeLa cells. 24 h after transfection, cells were trypsinized, precipitated, and lysed in lysis buffer (50-mM Tris-HCl, pH 7.5, 300-mM NaCl, and 0.5% NP-40 with protease and phosphatase inhibitors [Thermo Fisher Scientific] as described by the manufacturers). The lysates were cleared by centrifugation at 4°C, adjusted to 150-mM NaCl and 0.1% NP-40, and 1.5 mg of total protein was incubated with 25 µl of settled resin volume anti-HA affinity matrix (Roche) and DNase (0.1 U/µg of protein) on a rotator at 4°C for 2 h. Anti-HA beads were precipitated by centrifugation and washed 4× in 500 µl of binding buffer. SDS-eluted proteins (30 µl each) were detected by 10% SDS-PAGE and Western blot analysis.
D-loop and ATPase assay
The D-loop reaction was performed as described previously (Chi et al., 2006b; Zhao et al., 2014). Briefly, 32P-labeled 90-mer oligonucleotide (2.4-µM nucleotides) was preincubated with RAD51 (1 µM) for 5 min at 37°C in the reaction buffer E (25-mM Tris-HCl, pH 7.5, 60-mM KCl, 1-mM DTT, 1-mM MgCl2, 100-mM CaCl2, 2-mM ATP, and 100 µg/ml BSA). This was followed by the incorporation of RAD54 and/or NUCKS1 and a 5-min incubation at 37°C. The D-loop reaction was initiated by adding naked or chromatinized pBluescript II SK(−) replicative form I DNA (37-µM base pairs) and incubating at 37°C for 7 min. The molar ratio of the 90-mer to pBluescript II SK(−) plasmid in the reactions was 2.1:1. After electrophoresis in a 1% agarose gel, phosphorimaging analysis was performed to visualize the radiolabeled DNA species (Chi et al., 2006b).
RAD54 (50 nM) was incubated with [γ-32P] ATP (100 µM) and 125 ng pBluescript II SK(−) without or with NUCKS1 (150 and 300 nM) at 25°C in 10 µl of reaction buffer (20-mM Tris-HCl, pH 7.4, 25-mM KCl, 1-mM DTT, 4-mM MgCl2, and 100 µg/ml BSA) for the indicated times. The level of ATP hydrolysis was determined by thin-layer chromatography as described previously (Petukhova et al., 1998). For the experiments described in Fig. 2 E, RAD54 (100 nM), RAD54 (100 nM) together with NUCKS1 (300 nM), and NUCKS1 alone (300 and 600 nM) were incubated for 10 min to exclude ATPase activity by NUCKS1 protein alone.
Statistical analysis
The statistical analysis was performed using Prism 8 GraphPad Software on the data from at least two independent experiments, as specified. Statistical significance was assessed by one-way or two-way ANOVA, or multiple t test analyses, as indicated. P ≤ 0.05 was considered significant.
Online supplemental material
Fig. S1 delineates the strategy followed to generate NUCKS1 KO cell lines and shows representative micrographs of nuclear RAD54 foci in HeLa, NUCKS1 KO, and complemented cell lines after IR. Fig. S2 shows representative micrographs of RAD54 foci in HeLa, NUCKS1 KO, and complemented cell lines after exposure to MMC and the cell cycle profiles of these cell lines obtained after IR and MMC treatment. Fig. S3 provides representative micrographs of nuclear RAD51 foci in HeLa, NUCKS1 KO, and complemented cell lines after IR and without or with RAD54 knockdown. This figure also provides information on protein expression in NUCKS1 and RAD54 KO cells, shows representative SDS-PAGE gels of the purified proteins, and the results obtained after micrococcal nuclease digest of naked and chromatinized plasmid DNA. Fig. S4 shows the results obtained after monitoring DNA repair synthesis in NUCKS1 KO cells and in HeLa cells depleted for RAD54. Fig. S5 shows representative micrographs of a compilation of negative and positive control experiments that confirm the specificity of the PLA results obtained. Table S1 provides a list of all PCR primers used. Table S2 shows the lengths of the PCR products analyzed to monitor loss of NUCKS1 sequence in NUCKS1 KO cells and the generation of ectopic RAD54 constructs. Table S3 provides a list of all sgRNAs used.
Acknowledgments
The authors wish to thank Charles McKeon for help with image acquisition and the Colorado State University Flow Cytometry and Cell Sorting Facility for their help in method optimization and sample analysis.
This work was supported by research grants from the National Institutes of Health (ES021454 and ES029206) to C. Wiese and by foundation grants (V Scholar V2019.Q13 from the V Foundation for Cancer Research and a Young Investigator Award from Max and Minnie Tomerlin Voelcker Fund) to W. Zhao.
The authors declare no competing financial interests.
Author contributions: D.G. Maranon, N. Sharma, Y. Huang, P. Selemenakis, M. Wang, N. Altina, W. Zhao, and C. Wiese performed experiments and analyzed data. C. Wiese and W. Zhao designed the experiments and wrote the manuscript with input from all authors. C. Wiese and W. Zhao conducted the acquisition of funding.
References
Author notes
D.G. Maranon, N. Sharma, and Y. Huang contributed equally to this paper.