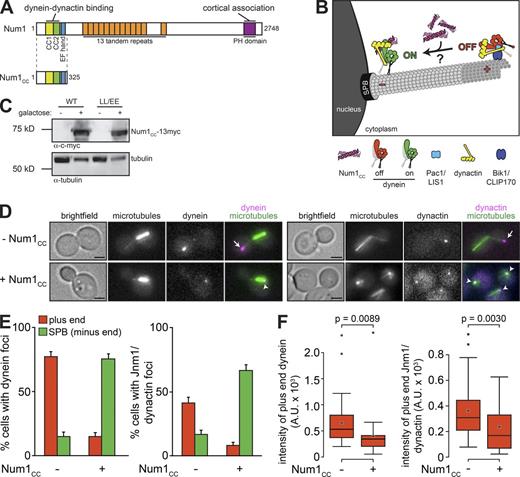
Cortically anchored dynein orients the spindle through interactions with astral microtubules. In budding yeast, dynein is offloaded to Num1 receptors from microtubule plus ends. Rather than walking toward minus ends, dynein remains associated with plus ends due in part to its association with Pac1/LIS1, an inhibitor of dynein motility. The mechanism by which dynein is switched from “off” at the plus ends to “on” at the cell cortex remains unknown. Here, we show that overexpression of the coiled-coil domain of Num1 specifically depletes dynein–dynactin–Pac1/LIS1 complexes from microtubule plus ends and reduces dynein-Pac1/LIS1 colocalization. Depletion of dynein from plus ends requires its microtubule-binding domain, suggesting that motility is required. An enhanced Pac1/LIS1 affinity mutant of dynein or overexpression of Pac1/LIS1 rescues dynein plus end depletion. Live-cell imaging reveals minus end–directed dynein–dynactin motility along microtubules upon overexpression of the coiled-coil domain of Num1, an event that is not observed in wild-type cells. Our findings indicate that dynein activity is directly switched “on” by Num1, which induces Pac1/LIS1 removal.
Introduction
Cytoplasmic dynein is a 1.2-MD multisubunit motor complex that powers movement of various cargoes toward the minus ends of microtubules. This ancient and highly conserved ATPase organizes the intracellular environment throughout the cell cycle; however, its myriad roles become especially apparent during mitosis when it functions in various aspects of spindle morphogenesis and positioning (Rusan et al., 2002; Goshima et al., 2005; Yang et al., 2007; Ferenz et al., 2009; Siller and Doe, 2009). For instance, cortically anchored dynein motors position the spindle at the future site of cytokinesis (Eshel et al., 1993; Li et al., 1993; Carminati and Stearns, 1997), a process that is particularly important during asymmetric cell divisions, when spindle position dictates the plane of cell division, and thus cell fate determination (Pease and Tirnauer, 2011; Williams et al., 2011). The means by which dynein is delivered to the cell cortex and subsequently activated to pull on astral microtubules emanating from spindle poles to move the spindle are unclear.
Recent studies in budding and fission yeast have revealed two distinct mechanisms by which dynein can be targeted to the cell cortex, its site of action in both organisms (Markus and Lee, 2011; Ananthanarayanan et al., 2013). During the meiotic prophase in fission yeast, studies suggest that dynein first binds along astral microtubules that are in close proximity to the cell cortex (Ananthanarayanan et al., 2013). Rather than walking toward the minus end of these microtubules, dynein undergoes one-dimensional diffusion until it encounters Mcp5, its cortical anchor. Once bound to Mcp5, dynein motors switch from diffusive to directed motion and consequently move the spindle appropriately. Thus, in addition to anchoring dynein at the cortex, Mcp5 appears to activate dynein motility by an unknown mechanism.
A similar but somewhat distinct scenario takes place in budding yeast, in which dynein is first targeted to the plus ends of astral microtubules before being offloaded to cortical Num1 (Mcp5 homologue) receptor sites, where it functions to move the mitotic spindle toward the daughter cell (Adames and Cooper, 2000; Lee et al., 2005; Markus and Lee, 2011). It is unknown if dynein motility is activated subsequent to offloading; however, several lines of evidence suggest that its activity is switched from being off at plus ends to being on at the cell cortex. For instance, in spite of its minus end–directed motility, dynein is transported to, and remains associated with, the plus ends of dynamic microtubules (Lee et al., 2003; Carvalho et al., 2004). Plus end targeting requires the dynein motor domain, the +TIP (plus end–tracking protein) Bik1 (CLIP-170 homologue), and the dynein-associated factor Pac1 (homologue of human LIS1; Lee et al., 2003; Sheeman et al., 2003; Markus et al., 2009). Recent studies suggest that Pac1 plays two distinct and important roles in targeting dynein to plus ends. First, Pac1 mediates the interaction between dynein and plus end–bound Bik1 (Sheeman et al., 2003; Roberts et al., 2014). Second, by inhibiting dynein motility (Markus and Lee, 2011; Huang et al., 2012), and/or by prolonging its microtubule attachment (Yamada et al., 2008; McKenney et al., 2010; Huang et al., 2012), Pac1 holds dynein at plus ends by keeping dynein in a nonmotile, or “off” state. Thus, plus end–associated dynein may be poised to walk toward the minus ends of microtubules but is prevented from doing so by Pac1. However, upon offloading to cortical Num1 receptor sites, dynein is active, as apparent by its capacity to pull on astral microtubules and consequently move the spindle. Thus, in budding yeast, as in fission yeast, dynein may undergo a switch in its activity upon attachment to its cortical receptor. However, evidence for such a switch is lacking.
Although factors have been identified that can inhibit dynein motility (e.g., MAP4, She1, and Pac1; Markus and Lee, 2011; Samora et al., 2011; Huang et al., 2012; Markus et al., 2012), it remains unclear whether dynein from organisms other than fission yeast need to be switched on to perform their cellular functions or whether they are constitutively active. Recent studies indicated that purified metazoan dynein is functional for motility in ensemble assays (e.g., microtubule gliding) but requires a stable interaction with the dynactin complex for single-molecule processivity (McKenney et al., 2014; Schlager et al., 2014). Stabilization of the dynein–dynactin interaction by various coiled-coil–containing adaptor proteins (e.g., BicD2, Spindly, Hook3, and Rab11-Fip3) is sufficient to significantly enhance dynein processivity, and thereby “activate” dynein motility. This is in contrast to budding yeast dynein, which is processive in the absence of dynactin or other adaptors or regulators (Reck-Peterson et al., 2006). It is unclear whether such a mechanism generally governs dynein regulation within a cell, given that certain functions of dynein in animal cells, such as centrosome anchoring, pole focusing, and spindle length regulation, have been shown to occur independently of several such adaptors (Raaijmakers et al., 2013). It is also unclear whether dynactin-mediated processivity enhancement is the means by which cortical dynein activity is promoted. Interestingly, plus end association of dynein in higher eukaryotes, which occurs in a dynactin-dependent manner, does not require such adaptor proteins (Splinter et al., 2012; Duellberg et al., 2014). Given the lack of minus end–directed motility of these plus end–associated dynein motors, an interaction between dynein and dynactin is not necessarily sufficient to active dynein motility.
Using budding yeast, we aimed to test the hypothesis that binding of dynein–dynactin to its cortical receptor provides the switch that activates cortical dynein activity. Dynein pathway function is best understood in this genetically tractable organism, in which many of the regulatory components and accessory chains, which are each encoded by only one gene, are highly conserved. We found that overexpression of the dynein–dynactin–interacting coiled-coil domain of Num1 (Num1CC) is sufficient to activate dynein motility, causing a depletion of dynein–dynactin from microtubule plus ends, their accumulation at minus ends, and their apparent minus end–directed motility along astral microtubules. Our data reveal that the mechanism for this activation is likely a Num1CC-mediated release of Pac1, a potent dynein inhibitor, from the dynein motor domain.
Results
Overexpression of Num1CC depletes dynein–dynactin from microtubule plus ends
A recent study revealed that the N-terminal coiled-coil domain of Num1—the dynein cortical receptor—directly interacts with dynein–dynactin complexes (Tang et al., 2012). If this region of Num1 (Num1CC; Fig. 1 A) is sufficient to activate dynein motility, we reasoned that its overexpression would result in (a) the depletion of dynein and dynactin from microtubule plus ends, and (b) an accumulation of dynein–dynactin at spindle pole bodies (SPBs), where microtubule minus ends are anchored (Fig. 1 B). We found this latter phenomenon to be an inherent property of dynein motility, because single-molecule motility experiments demonstrated that upon reaching the end of a microtubule, dynein pauses for several seconds before detaching, much longer than the dwell time of ∼0.1 s per step (Fig. S1 and Video 1). To test our hypothesis, we engineered yeast cells such that the galactose-inducible GAL1 promoter (GAL1p) was immediately upstream of a truncated num1CC allele (Fig. 1 C). Because Num1CC is expressed from the native NUM1 locus, these cells do not possess a full-length Num1. Consequently, dynein and dynactin are excluded from the cell cortex and are unable to properly orient the spindle in these cells.
In vitro motility of single Dyn1-EGFP molecules purified from yeast cells. Time-lapse total internal reflection fluorescence microscope images of single Dyn1-EGFP molecules (green in merge) walking along taxol-stabilized microtubules (blue in merge) and pausing at the minus ends (arrows). Also see Fig. S1. Related to Fig. 1.
In vitro motility of single Dyn1-EGFP molecules purified from yeast cells. Time-lapse total internal reflection fluorescence microscope images of single Dyn1-EGFP molecules (green in merge) walking along taxol-stabilized microtubules (blue in merge) and pausing at the minus ends (arrows). Also see Fig. S1. Related to Fig. 1.
Overexpression of Num1CC depletes dynein and dynactin from microtubule plus ends. (A) Schematic representation of Num1 and Num1CC with domain structure indicated (CC1 and CC2, predicted coiled-coil domains; PH, plecktrin homology domain). (B) Diagram depicting experimental design (see text). (C) Western blot of GAL1p:num1CC-13myc (wild-type or LL/EE mutant) cells grown in the absence or presence of galactose, as indicated, with loading control (anti–α-tubulin). (D) Representative images of GAL1p:num1CC cells expressing mTurquoise2-Tub1 (α-tubulin) and either Dyn1-3mCherry (left) or Jnm1-3mCherry (right) used for quantitation in E and F. Cells were grown to mid-log phase in SD media supplemented with glucose (uninduced; −Num1CC) or galactose plus raffinose (induced; +Num1CC). Each image is a maximum-intensity projection of a 2-µm Z-stack of wide-field images. Arrows indicate plus end foci, and arrowheads indicate SPB foci. Bars, 2 µm. (E) The percentage of cells that exhibit plus end (red) or SPB (green) fluorescent foci is plotted for the strains shown in D. Plus end or SPB foci were identified in two-color movies and scored accordingly (see Materials and methods). Error bars represent the standard error of proportion (n ≥ 114 cells). (F) Box plot of fluorescence intensity values of plus end–associated Dyn1- or Jnm1-3mCherry foci (n ≥ 30 foci). Whiskers define the range of data, boxes encompass the 25th to 75th quartiles, the line depicts the median value, and the “x” depicts the mean value. See also Video 1 and Figs. S1, S2, and S3.
Overexpression of Num1CC depletes dynein and dynactin from microtubule plus ends. (A) Schematic representation of Num1 and Num1CC with domain structure indicated (CC1 and CC2, predicted coiled-coil domains; PH, plecktrin homology domain). (B) Diagram depicting experimental design (see text). (C) Western blot of GAL1p:num1CC-13myc (wild-type or LL/EE mutant) cells grown in the absence or presence of galactose, as indicated, with loading control (anti–α-tubulin). (D) Representative images of GAL1p:num1CC cells expressing mTurquoise2-Tub1 (α-tubulin) and either Dyn1-3mCherry (left) or Jnm1-3mCherry (right) used for quantitation in E and F. Cells were grown to mid-log phase in SD media supplemented with glucose (uninduced; −Num1CC) or galactose plus raffinose (induced; +Num1CC). Each image is a maximum-intensity projection of a 2-µm Z-stack of wide-field images. Arrows indicate plus end foci, and arrowheads indicate SPB foci. Bars, 2 µm. (E) The percentage of cells that exhibit plus end (red) or SPB (green) fluorescent foci is plotted for the strains shown in D. Plus end or SPB foci were identified in two-color movies and scored accordingly (see Materials and methods). Error bars represent the standard error of proportion (n ≥ 114 cells). (F) Box plot of fluorescence intensity values of plus end–associated Dyn1- or Jnm1-3mCherry foci (n ≥ 30 foci). Whiskers define the range of data, boxes encompass the 25th to 75th quartiles, the line depicts the median value, and the “x” depicts the mean value. See also Video 1 and Figs. S1, S2, and S3.
In the absence of Num1CC induction (by growth in glucose-containing media), we observed fluorescent foci of functional Dyn1- (dynein heavy chain) and Jnm1-3mCherry (p50 subunit of dynactin) fusions (Markus et al., 2011) at microtubule plus ends in 77.2% and 41.4% of cells, respectively (Fig. 1, D and E). In contrast, upon induction of Num1CC overexpression (by growth in galactose-containing media), the number of cells exhibiting Dyn1- and Jnm1-3mCherry plus end foci was reduced by approximately fivefold, to 14.8% and 8.1%, respectively. Fluorescence intensity measurements revealed that those plus ends with Dyn1- and Jnm1-3mCherry foci had significantly fewer molecules of each upon Num1CC overexpression (Fig. 1 F). Coincident with the reduction in plus end dynein and dynactin, we also observed a four- to fivefold increase in the number of cells with Dyn1- and Jnm1-3mCherry foci at SPBs upon Num1CC induction (also see Fig. 4 A, bottom). We observed similar results for Pac11- (dynein intermediate chain) and Dyn3-3mCherry (dynein light-intermediate chain; Fig. S2, A–E). In contrast, overexpression of Num1CC had no effect on the extent of plus end localization of Bik1-3mCherry (CLIP170 homologue), a protein that is required for the plus end targeting of dynein (Fig. S2, F and G; Sheeman et al., 2003). To determine whether there is a cell cycle–regulated component to Num1CC-mediated dynein relocalization, we categorized plus end and SPB localization frequencies into the following: G1, preanaphase, and anaphase (as determined by cell and spindle morphology; see Fig. S2 H). Although both G1 and preanaphase cells exhibited an equivalent extent of Num1CC-mediated dynein relocalization, we found that anaphase cells were less susceptible to dynein plus end depletion, in spite of a significant enhancement in the prevalence of SPB localized dynein in these cells (Fig. S2 H). These findings are consistent with the higher frequency of dynein plus end localization noted in wild-type anaphase cells (Sheeman et al., 2003; Markus et al., 2009), and they further suggest that the plus end targeting mechanism is more robust at this point of the cell cycle. Collectively, our data indicate that overexpression of Num1CC specifically depletes dynein–dynactin from microtubule plus ends.
We next wanted to determine whether a Num1-dynein interaction was required for the plus end depletion phenotype. First, we introduced two point mutations within Num1CC (L167E L170E; Num1CCLL/EE; Fig. 2 A) that disrupt its interaction with dynein–dynactin but have no effect on Num1 localization or its mitochondria cortical attachment function (Tang et al., 2012). We found that overexpression of Num1CCLL/EE (Fig. 1 C) had no effect on dynein plus end or SPB localization (Fig. 2, B and C), indicating that an intact dynein–dynactin binding surface within Num1CC is required for plus end depletion. Second, we used two dynein motor domain variants (Dyn1MOTOR and GST-Dyn1MOTOR) that are sufficient for association with microtubule plus ends but lack the N-terminal tail domain that is required for association with Num1 (Fig. 2 D; Markus et al., 2009). We found that Num1CC overexpression had little effect on the frequency by which monomeric (nonmotile) Dyn1MOTOR-3YFP or the dimeric (motile; Reck-Peterson et al., 2006) GST-Dyn1MOTOR-3mCherry fragment was observed at plus ends or SPBs (Fig. 2, E and F). Collectively, these data indicate that Num1CC-mediated plus end depletion of dynein requires an interaction between dynein and Num1CC.
Interaction between dynein and Num1CC is required for plus end depletion. (A) Schematic representation of the Num1CCLL/EE mutant. (B) Representative images of GAL1p:num1CCLL/EE and GAL1p:num1CC cells expressing mTurquoise2-Tub1 and Dyn1-3mCherry used for quantitation in C. (C) The percentage of cells that exhibit plus end (red) or SPB (green) fluorescent foci is plotted for the strains overexpressing wild-type (WT) or mutant (LL/EE) Num1CC. Error bars represent the standard error of proportion (n ≥ 170 cells). (D) Diagram depicting dynein N-terminal (tail) and C-terminal (motor) domains and an artificially dimerized GST-Dyn1MOTOR construct (see text). (E) Representative images of GAL1p:num1CC cells expressing mTurquoise2-Tub1 and Dyn1MOTOR-3YFP used for quantitation in F. (F) The percentage of cells that exhibit plus end (red) or SPB (green) fluorescent Dyn1MOTOR-3YFP or GST-Dyn1MOTOR-3mCherry foci is plotted. Error bars represent the standard error of proportion (n ≥ 87 cells). All images are maximum-intensity projections of a 2-µm Z-stack of wide-field images. Arrows indicate plus end foci, and the arrowhead indicates the SPB focus. Bars, 2 µm.
Interaction between dynein and Num1CC is required for plus end depletion. (A) Schematic representation of the Num1CCLL/EE mutant. (B) Representative images of GAL1p:num1CCLL/EE and GAL1p:num1CC cells expressing mTurquoise2-Tub1 and Dyn1-3mCherry used for quantitation in C. (C) The percentage of cells that exhibit plus end (red) or SPB (green) fluorescent foci is plotted for the strains overexpressing wild-type (WT) or mutant (LL/EE) Num1CC. Error bars represent the standard error of proportion (n ≥ 170 cells). (D) Diagram depicting dynein N-terminal (tail) and C-terminal (motor) domains and an artificially dimerized GST-Dyn1MOTOR construct (see text). (E) Representative images of GAL1p:num1CC cells expressing mTurquoise2-Tub1 and Dyn1MOTOR-3YFP used for quantitation in F. (F) The percentage of cells that exhibit plus end (red) or SPB (green) fluorescent Dyn1MOTOR-3YFP or GST-Dyn1MOTOR-3mCherry foci is plotted. Error bars represent the standard error of proportion (n ≥ 87 cells). All images are maximum-intensity projections of a 2-µm Z-stack of wide-field images. Arrows indicate plus end foci, and the arrowhead indicates the SPB focus. Bars, 2 µm.
Several lines of evidence indicate that an intact dynein–dynactin complex is required for interaction with Num1. Yeast dynactin deletion mutants exhibit a complete loss of cortical dynein and a higher than normal accumulation of dynein at microtubule plus ends (Lee et al., 2003, 2005). This latter observation is also noted in num1Δ mutants (Lee et al., 2003) and is presumed to be attributable to an inability to offload dynein–dynactin to cortical Num1 receptor sites. Finally, a bead-immobilized recombinant Num1CC fragment was sufficient to isolate dynein from cell extracts but was unable to do so from extracts prepared from nip100Δ cells (homologue of human dynactin component p150Glued; Tang et al., 2012). To determine whether Num1CC can deplete plus end dynein in the absence of dynactin, we overexpressed Num1CC in nip100Δ cells. We found that Num1CC overexpression reduced the frequency of dynein plus end localization in nip100Δ cells to a lesser extent than in NIP100 cells (Fig. S3, A and B; 38% reduction in nip100Δ vs. 81% reduction in NIP100 cells; compare with Fig. 1 E); however, fluorescence intensity measurements of plus end dynein revealed no significant difference (Fig. S3 C). These data suggest that the ability of dynein to interact with Num1CC is compromised but not abolished in the absence of dynactin.
We next asked whether plus end targeting of dynein is a requisite for Num1CC-mediated accumulation of dynein at SPBs. To test this, we deleted either Pac1 or Bik1, both of which are required for dynein plus end targeting (Lee et al., 2003; Sheeman et al., 2003). As expected, neither pac1Δ nor bik1Δ cells overexpressing Num1CC exhibited plus end dynein foci; in addition, SPB localization of dynein was apparent in only a small number of cells in both mutants, suggesting that the accumulation of dynein at SPBs requires a pool of dynein–dynactin at plus ends from which to draw (Fig. S3, D–G).
Microtubule binding by dynein is required for plus end depletion
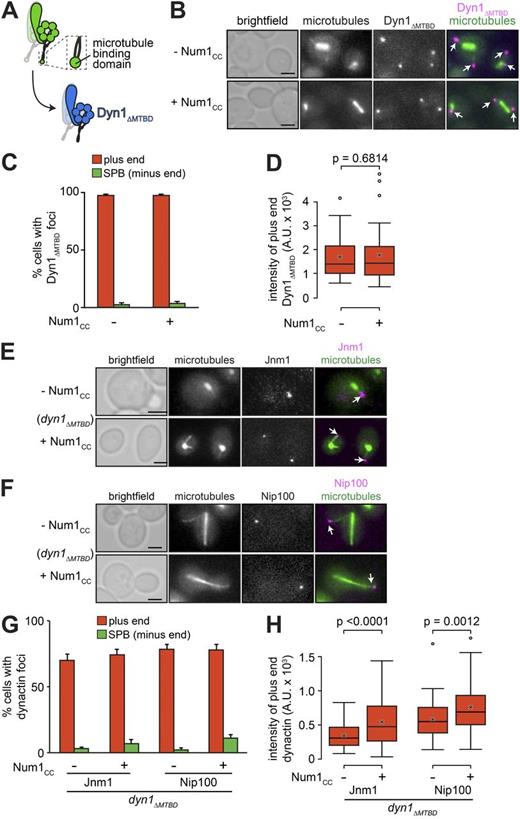
If Num1CC-mediated redistribution of dynein from plus ends to SPBs is a consequence of activated dynein motility, then we reasoned that a motility-incompetent dynein mutant would remain associated with plus ends in the presence of overexpressed Num1CC. To test this, we generated a dynein mutant lacking its microtubule-binding domain (MTBD; Dyn1ΔMTBD; Fig. 3 A). Surprisingly, we found that in the absence of Num1CC induction, Dyn1ΔMTBD was capable of plus end binding (Fig. 3, B and C, top in B), indicating that dynein associates with plus ends independently of its MTBD, and thus likely through its interaction with Pac1 and Bik1. Furthermore, we found that the extent of plus end localization of Dyn1ΔMTBD was greater than wild-type Dyn1 (26% increase in frequency; 148% increase in intensity; P < 0.0001), whereas its SPB localization was lower (Fig. 3, C and D; compare with Fig. 1, E and F). These data suggest that SPB localization of dynein in wild-type cells may be attributable to minus end–directed motility of active dynein motors. Consistent with our hypothesis, upon induction of Num1CC overexpression, Dyn1ΔMTBD localization to plus ends and SPBs remained unchanged (Fig. 3, C and D), suggesting that Num1CC-mediated depletion of dynein from plus ends and accumulation at SPBs both require dynein motility.
The dynein MTBD is dispensable for plus end targeting but is required for Num1CC-mediated plus end depletion. (A) Schematic representation of the Dyn1ΔMTBD mutant. (B) Representative images of GAL1p:num1CC cells expressing mTurquoise2-Tub1 and Dyn1ΔMTBD-3mCherry used for quantitation in C and D. (C) The percentage of cells that exhibit plus end (red) or SPB (green) Dyn1ΔMTBD-3mCherry foci is plotted for cells shown in B. Error bars represent the standard error of proportion (n ≥ 126 cells). (D) Box plot of fluorescence intensity values of plus end–associated Dyn1ΔMTBD-3mCherry (n ≥ 40 foci). (E and F) Representative images of GAL1p:num1CC dyn1ΔMTBD cells expressing mTurquoise2-Tub1 and either Jnm1- (E) or Nip100-3mCherry (F) used for quantitation in G and H. (G) The percentage of dyn1ΔMTBD cells that exhibit plus end or SPB Jnm1- or Nip100-3mCherry foci is plotted for GAL1p:num1CC cells grown in glucose (−Num1CC) or galactose (+Num1CC; n ≥ 100 cells). (H) Box plot of fluorescence intensity values of plus end–associated Jnm1- or Nip100-3mCherry (n ≥ 56 foci). For all box plots, whiskers define the range of data, boxes encompass the 25th to 75th quartiles, the line depicts the median value, and the “x” depicts the mean value. All images are maximum-intensity projections of a 2-µm Z-stack of wide-field images. Arrows indicate plus end foci. Bars, 2 µm. See also Fig. S5.
The dynein MTBD is dispensable for plus end targeting but is required for Num1CC-mediated plus end depletion. (A) Schematic representation of the Dyn1ΔMTBD mutant. (B) Representative images of GAL1p:num1CC cells expressing mTurquoise2-Tub1 and Dyn1ΔMTBD-3mCherry used for quantitation in C and D. (C) The percentage of cells that exhibit plus end (red) or SPB (green) Dyn1ΔMTBD-3mCherry foci is plotted for cells shown in B. Error bars represent the standard error of proportion (n ≥ 126 cells). (D) Box plot of fluorescence intensity values of plus end–associated Dyn1ΔMTBD-3mCherry (n ≥ 40 foci). (E and F) Representative images of GAL1p:num1CC dyn1ΔMTBD cells expressing mTurquoise2-Tub1 and either Jnm1- (E) or Nip100-3mCherry (F) used for quantitation in G and H. (G) The percentage of dyn1ΔMTBD cells that exhibit plus end or SPB Jnm1- or Nip100-3mCherry foci is plotted for GAL1p:num1CC cells grown in glucose (−Num1CC) or galactose (+Num1CC; n ≥ 100 cells). (H) Box plot of fluorescence intensity values of plus end–associated Jnm1- or Nip100-3mCherry (n ≥ 56 foci). For all box plots, whiskers define the range of data, boxes encompass the 25th to 75th quartiles, the line depicts the median value, and the “x” depicts the mean value. All images are maximum-intensity projections of a 2-µm Z-stack of wide-field images. Arrows indicate plus end foci. Bars, 2 µm. See also Fig. S5.
Num1CC has only a modest effect on dynein–dynactin interaction at plus ends
In contrast to yeast dynein, metazoan dynein exhibits mostly nonprocessive, diffusive motility in single-molecule assays (Miura et al., 2010; McKenney et al., 2014; Schlager et al., 2014). A family of coiled-coil–containing adaptor proteins that recruit dynein and dynactin to various cellular sites was recently shown to be sufficient to activate processive single-molecule motility by stably linking dynein to dynactin (McKenney et al., 2014; Schlager et al., 2014). Like these adaptor proteins, Num1 interacts only with intact dynein–dynactin complexes through its coiled-coil domain (Splinter et al., 2012; Tang et al., 2012). Although dynein recruits dynactin to plus ends in a Num1-independent manner, dynactin is present at substoichiometric amounts relative to dynein (approximately three dynein to one dynactin; Markus et al., 2011). Thus, we reasoned that Num1CC may activate dynein motility by enhancing the dynein–dynactin interaction at plus ends. To test this, we assessed the extent by which Num1CC affects dynein-mediated recruitment of dynactin to plus ends. For these experiments, we used dyn1ΔMTBD mutant cells, in which dynein remains associated at plus ends in spite of Num1CC overexpression (see Fig. 3, B–D). We found that Num1CC overexpression had no effect on the frequency of observing plus end–localized dynactin (i.e., Jnm1- or Nip100-3mCherry; Fig. 3, E–G); however, the fluorescence intensities of both Jnm1 and Nip100 at plus ends were modestly, but significantly, increased (Fig. 3 H; 58.9% and 30.9%, respectively; P ≤ 0.0012), suggesting that Num1CC may in fact enhance or stabilize the dynein–dynactin interaction. However, given the small apparent change in dynein–dynactin interaction at plus ends, and the observation that yeast dynein processivity enhancement by dynactin does not require additional factors in vitro (i.e., Num1; Kardon et al., 2009), it is unclear whether stabilization of the dynein–dynactin interaction is the mechanism by which Num1 functions to activate dynein motility. For these reasons, we explored an alternative hypothesis.
Overexpression of Num1CC reduces colocalization of dynein and Pac1
Cells in which Pac1 is deleted exhibit a complete loss of plus end dynein (Lee et al., 2003; Markus et al., 2009), similar to our observations of cells overexpressing Num1CC. We hypothesized that Num1CC may deplete dynein from plus ends by interfering with dynein-Pac1 binding. To test this, we assessed localization of a functional Pac1-3mCherry fusion (Markus et al., 2011) in cells overexpressing Num1CC. As we observed for dynein and dynactin, Pac1-3mCherry foci were depleted from microtubule plus ends upon Num1CC overexpression (Fig. 4, A and B), consistent with their codependence for plus end targeting (Markus et al., 2011). However, in contrast to dynein and dynactin, the fraction of cells exhibiting Pac1-3mCherry foci at SPBs was reduced with respect to cells not expressing Num1CC, suggesting that dynein localizes at SPBs without Pac1 upon Num1CC overexpression. This reduction in localization was not due to a decrease in Pac1 protein expression or stability, as indicated by immunoblotting (Fig. 4 D). Consistent with a Num1CC-mediated reduction in the dynein-Pac1 interaction, we found that the fraction of Dyn1-3YFP and Pac1-3mCherry foci that colocalized (either SPB or plus end) was reduced upon induction of Num1CC (from 59.6% to 26.2%; Fig. 4 E), whereas the fraction of Dyn1-3YFP foci alone (i.e., not colocalized with Pac1) increased (from 28.3% to 62.3%). These data suggest that overexpression of Num1CC may disrupt plus end binding of dynein–dynactin by interfering with the dynein-Pac1 interaction.
Overexpression of Num1CC depletes Pac1 from plus ends and disrupts dynein-Pac1 interaction. (A) Representative images of GAL1p:num1CC cells expressing mTurquoise2-Tub1, Pac1-3mCherry, and Dyn1-3YFP used for quantitation in B, C, and E. The arrow indicates the plus end focus, and arrowheads indicate SPB foci. (B) The percentage of cells that exhibit plus end (red) or SPB (green) fluorescent Pac1-3mCherry foci is plotted for the cells shown in A. Error bars represent the standard error of proportion (n ≥ 122 cells). (C) Box plot of fluorescence intensity values of plus end–associated Pac1-3mCherry (n ≥ 26 foci). (D) Western blot of Pac1-13myc–expressing GAL1p:num1CC or GAL1p:num1CCLL/EE cells (as indicated) grown in galactose-containing media with loading control (anti–α-tubulin). (E) The extent of Pac1-3mCherry and Dyn1-3YFP colocalization is plotted for the indicated cells (n ≥ 61 fluorescent foci). (F) Representative images of GAL1p:PH-DYN1-3mCherry cells expressing Pac1-3YFP and either Num1CC or Num1CCLL/EE. Arrowheads indicate cortical foci. (G and I) The percentage of cortical PH-Dyn1-3mCherry foci that colocalize with either Pac1-3YFP (n ≥ 49 foci; G) or Jnm1-3YFP (n ≥ 55 foci; I) is plotted for cells expressing either Num1CC or Num1CCLL/EE. (H and J) Box plot of fluorescence intensity values for either cortical Pac1-3YFP (n ≥ 25 foci; H) or Jnm1-3YFP foci (n ≥ 44 foci; one outlier was omitted from the plot for display purposes only; J). For all box plots, the whiskers define the range of data, boxes encompass the 25th to 75th quartiles, the line depicts the median value, and the “x” depicts the mean value. All images are maximum-intensity projections of a 2-µm Z-stack of wide-field images. Bars, 2 µm. See also Fig. S4. WT, wild type.
Overexpression of Num1CC depletes Pac1 from plus ends and disrupts dynein-Pac1 interaction. (A) Representative images of GAL1p:num1CC cells expressing mTurquoise2-Tub1, Pac1-3mCherry, and Dyn1-3YFP used for quantitation in B, C, and E. The arrow indicates the plus end focus, and arrowheads indicate SPB foci. (B) The percentage of cells that exhibit plus end (red) or SPB (green) fluorescent Pac1-3mCherry foci is plotted for the cells shown in A. Error bars represent the standard error of proportion (n ≥ 122 cells). (C) Box plot of fluorescence intensity values of plus end–associated Pac1-3mCherry (n ≥ 26 foci). (D) Western blot of Pac1-13myc–expressing GAL1p:num1CC or GAL1p:num1CCLL/EE cells (as indicated) grown in galactose-containing media with loading control (anti–α-tubulin). (E) The extent of Pac1-3mCherry and Dyn1-3YFP colocalization is plotted for the indicated cells (n ≥ 61 fluorescent foci). (F) Representative images of GAL1p:PH-DYN1-3mCherry cells expressing Pac1-3YFP and either Num1CC or Num1CCLL/EE. Arrowheads indicate cortical foci. (G and I) The percentage of cortical PH-Dyn1-3mCherry foci that colocalize with either Pac1-3YFP (n ≥ 49 foci; G) or Jnm1-3YFP (n ≥ 55 foci; I) is plotted for cells expressing either Num1CC or Num1CCLL/EE. (H and J) Box plot of fluorescence intensity values for either cortical Pac1-3YFP (n ≥ 25 foci; H) or Jnm1-3YFP foci (n ≥ 44 foci; one outlier was omitted from the plot for display purposes only; J). For all box plots, the whiskers define the range of data, boxes encompass the 25th to 75th quartiles, the line depicts the median value, and the “x” depicts the mean value. All images are maximum-intensity projections of a 2-µm Z-stack of wide-field images. Bars, 2 µm. See also Fig. S4. WT, wild type.
We next wanted to determine whether the reduction in Pac1-dynein colocalization was a direct consequence of Num1CC-mediated Pac1-dynein dissociation, or whether it was a secondary consequence of dynein plus end depletion. To distinguish between these two possibilities, we ectopically targeted dynein to the plasma membrane using an exogenous pleckstrin homology (PH) domain, and we assessed the degree of colocalization of Pac1 with cortical PH-Dyn1 in the presence of either Num1CCLL/EE or Num1CC. We found that the PH domain was sufficient to target dynein (Fig. 4 F and Fig. S4 A) and dynactin (i.e., Jnm1; Fig. S4 B) to the cell cortex in cells lacking cortical Num1. Interestingly, we found a greater frequency of cortical PH-Dyn1-3mCherry foci in Num1CC-overexpressing cells than in either Num1CCLL/EE-overexpressing cells (45.5% vs. 13.9%, respectively) or NUM1 cells (21.3%; not depicted). In addition, we noted that Num1CC-overexpressing cells exhibited larger cortical patches than either wild-type NUM1 or Num1CCLL/EE-overexpressing cells (Fig. S4 A; data not depicted). These data suggest that PH-Dyn1-Num1CC complexes may be oligomerizing at the cell cortex, which is consistent with the previously described role for the Num1CC domain in the assembly of higher-order cortical patches (Tang et al., 2012). In cells overexpressing Num1CCLL/EE, Pac1-3YFP colocalized with 65.3% of cortical PH-Dyn1-3mCherry foci; however, upon overexpression of Num1CC, only 29.1% of PH-Dyn1-3mCherry foci contained Pac1-3YFP fluorescence (Fig. 4 G). Fluorescence intensity measurements also revealed a significant reduction in the number of Pac1 molecules associated with cortical PH-dynein patches (Fig. 4 H). In contrast, we noted no significant change in either the frequency or intensity of colocalized dynactin (i.e., Jnm1; Fig. 4, I and J; and Fig. S4 B). These data support the notion that Num1CC disrupts the dynein-Pac1 interaction, thereby leading to the plus end depletion phenotype.
An enhanced Pac1 affinity mutant of dynein or Pac1 overexpression reduces the extent of Num1CC-mediated plus end depletion of dynein
If a Num1CC-mediated Pac1 unbinding event is the cause for plus end depletion of dynein–dynactin complexes, we reasoned that we could reduce the extent of Num1CC-mediated dynein depletion from microtubule plus ends by two different means: (a) enhancing the affinity of dynein for Pac1, or (b) overexpression of Pac1. To test the former, we used a yeast strain expressing a well-characterized, motility-competent dynein mutant (Dyn1HL3) that exhibits higher affinity for Pac1 than wild-type dynein (Fig. 5 A; Markus and Lee, 2011). We predicted that Dyn1HL3 and Pac1 would be less susceptible to Num1CC-mediated plus end depletion as a result of their higher affinity. Consistent with our hypothesis, induction of Num1CC overexpression reduced the frequency of observing Dyn1HL3-3YFP and Pac1-3mCherry plus end foci by 40% and 22%, respectively (Fig. 5, B and C), much less than what we observed in wild-type DYN1 cells (respective 81% and 67% reduction; compare with Fig. 1 E and Fig. 4 B). Fluorescence intensity measurements revealed no significant change in the number of Dyn1HL3 or Pac1 molecules at plus ends upon Num1CC overexpression (Fig. 5 D). Thus, both Pac1 and Dyn1HL3 are less susceptible to plus end depletion by Num1CC overexpression in dyn1HL3 cells.
Dyn1HL3 is less susceptible to Num1CC-mediated plus end depletion. (A) Diagram depicting the Dyn1HL3 high Pac1 affinity mutant, in which a helical linker has been inserted between the dynein tail and motor domains (Markus and Lee, 2011). (B) Representative images of GAL1p:num1CC cells expressing mTurquoise2-Tub1 and either Dyn1HL3-3YFP (left) or Dyn1HL3 and Pac1-3mCherry (right) used for quantitation in C and D. Each image is a maximum-intensity projection of a 2-µm Z-stack of wide-field images. Arrows indicate plus end foci, and arrowheads indicate SPB foci. Bars, 2 µm. (C) The percentage of cells that exhibit plus end (red) or SPB (green) fluorescent Dyn1HL3-3YFP (left) or Pac1-3mCherry (right) foci is plotted for the cells shown in B. Error bars represent the standard error of proportion (n ≥ 119 cells). (D) Box plot of fluorescence intensity values of plus end–associated Dyn1HL3-3YFP or Pac1-3mCherry (n ≥ 33 foci). Whiskers define the range of data, boxes encompass the 25th to 75th quartiles, the line depicts the median value, and the “x” depicts the mean value. See also Fig. S5.
Dyn1HL3 is less susceptible to Num1CC-mediated plus end depletion. (A) Diagram depicting the Dyn1HL3 high Pac1 affinity mutant, in which a helical linker has been inserted between the dynein tail and motor domains (Markus and Lee, 2011). (B) Representative images of GAL1p:num1CC cells expressing mTurquoise2-Tub1 and either Dyn1HL3-3YFP (left) or Dyn1HL3 and Pac1-3mCherry (right) used for quantitation in C and D. Each image is a maximum-intensity projection of a 2-µm Z-stack of wide-field images. Arrows indicate plus end foci, and arrowheads indicate SPB foci. Bars, 2 µm. (C) The percentage of cells that exhibit plus end (red) or SPB (green) fluorescent Dyn1HL3-3YFP (left) or Pac1-3mCherry (right) foci is plotted for the cells shown in B. Error bars represent the standard error of proportion (n ≥ 119 cells). (D) Box plot of fluorescence intensity values of plus end–associated Dyn1HL3-3YFP or Pac1-3mCherry (n ≥ 33 foci). Whiskers define the range of data, boxes encompass the 25th to 75th quartiles, the line depicts the median value, and the “x” depicts the mean value. See also Fig. S5.
To test whether overexpression of Pac1 could rescue plus end depletion, we replaced the native PAC1 promoter with the GAL1 promoter, which is sufficient to induce >10-fold higher Pac1 expression levels compared with wild-type cells (Markus et al., 2011). To establish a baseline for Pac1 overexpression–mediated enhancement of dynein plus end targeting, we first assessed dynein localization in cells overexpressing Pac1 and Num1CCLL/EE, the latter of which has no discernible effect on dynein targeting (see Fig. 2, B and C). In these cells, we observed an increase in both frequency and mean fluorescence intensity of plus end dynein by 44% and 170%, respectively (Fig. 6, B and D), compared with cells overexpressing Num1CCLL/EE and expressing native levels of Pac1. In comparison, overexpression of Pac1 and wild-type Num1CC reduced the frequency of plus end dynein by 47% (P < 0.0001; compare GAL1p:PAC1 num1CCLL/EE with GAL1p:PAC1 num1CC), whereas the mean fluorescence intensity was not reduced significantly (30%; P = 0.1636; Fig. 6, B and D). These values are significantly less than the respective 78% and 37% reduction in frequency and intensity we observed when Num1CC alone was overexpressed (compared with Num1CCLL/EE; Fig. 6, C and E). Thus, Pac1 overexpression reduces the extent by which Num1CC depletes dynein from microtubule plus ends.
Overexpression of Pac1 reduces the extent by which Num1CC depletes plus end dynein. (A) Representative images of cells expressing mTurquoise2-Tub1, Dyn1-3mCherry, either Num1CC or Num1CCLL/EE, and either overexpressing Pac1 or Kip2, as indicated. Because of the distorted spindle phenotype in Kip2-overexpressing cells, Spc110-Venus was used to mark SPBs. All cells were grown in galactose-containing media to induce overexpression of Pac1, Kip2, Num1CC, or Num1CCLL/EE, as indicated. Each image is a maximum-intensity projection of a 2-µm Z-stack of wide-field images. For the top row (Pac1 overexpressed), arrows indicate plus end foci, and arrowheads indicate SPB foci. For the bottom row (Kip2 overexpressed), arrows indicate plus ends with or without foci. Bars, 2 µm. (B) The percentage of cells that exhibit plus end (red) or SPB (green) Dyn1-3mCherry foci is plotted for cells shown in A and for cells shown in Figs. 1 D and 2 B. Error bars represent the standard error of proportion (n ≥ 113 cells). (C) Extent by which Num1CC overexpression reduced the frequency of observing dynein plus end foci compared with the respective PAC1 KIP2 isogenic parent strain overexpressing Num1CCLL/EE. Asterisks indicate a statistically significant percent decrease (see B for P values). (D) Box plot of fluorescence intensity values of plus end–associated Dyn1-3mCherry (n ≥ 31 foci). Whiskers define the range of data, boxes encompass the 25th to 75th quartiles, the line depicts the median value, and the “x” depicts the mean value. (E) Extent by which Num1CC overexpression reduced the number of dynein molecules (i.e., fluorescence intensity) at plus ends compared with the isogenic PAC1 KIP2 parent strain overexpressing Num1CCLL/EE. Asterisks indicate a statistically significant percent decrease (see D for P values). See also Fig. S5. WT, wild type.
Overexpression of Pac1 reduces the extent by which Num1CC depletes plus end dynein. (A) Representative images of cells expressing mTurquoise2-Tub1, Dyn1-3mCherry, either Num1CC or Num1CCLL/EE, and either overexpressing Pac1 or Kip2, as indicated. Because of the distorted spindle phenotype in Kip2-overexpressing cells, Spc110-Venus was used to mark SPBs. All cells were grown in galactose-containing media to induce overexpression of Pac1, Kip2, Num1CC, or Num1CCLL/EE, as indicated. Each image is a maximum-intensity projection of a 2-µm Z-stack of wide-field images. For the top row (Pac1 overexpressed), arrows indicate plus end foci, and arrowheads indicate SPB foci. For the bottom row (Kip2 overexpressed), arrows indicate plus ends with or without foci. Bars, 2 µm. (B) The percentage of cells that exhibit plus end (red) or SPB (green) Dyn1-3mCherry foci is plotted for cells shown in A and for cells shown in Figs. 1 D and 2 B. Error bars represent the standard error of proportion (n ≥ 113 cells). (C) Extent by which Num1CC overexpression reduced the frequency of observing dynein plus end foci compared with the respective PAC1 KIP2 isogenic parent strain overexpressing Num1CCLL/EE. Asterisks indicate a statistically significant percent decrease (see B for P values). (D) Box plot of fluorescence intensity values of plus end–associated Dyn1-3mCherry (n ≥ 31 foci). Whiskers define the range of data, boxes encompass the 25th to 75th quartiles, the line depicts the median value, and the “x” depicts the mean value. (E) Extent by which Num1CC overexpression reduced the number of dynein molecules (i.e., fluorescence intensity) at plus ends compared with the isogenic PAC1 KIP2 parent strain overexpressing Num1CCLL/EE. Asterisks indicate a statistically significant percent decrease (see D for P values). See also Fig. S5. WT, wild type.
If Num1CC depletes dynein from plus ends by inducing Pac1 release, then we reasoned that enhancing dynein plus end targeting by a Pac1-independent means would not be able to rescue plus end depletion to the same extent as Pac1 overexpression. To test this, we overexpressed Kip2, a kinesin-7 family member that affects steady-state microtubule length (see Fig. 6 A, bottom) and plays a role in transporting dynein to microtubule plus ends (Carvalho et al., 2004; Markus et al., 2009; Roberts et al., 2014). As with Pac1, we first established a baseline by which Kip2 overexpression enhances dynein plus end localization by imaging Dyn1-3mCherry in GAL1p:KIP2 GAL1p:num1CCLL/EE cells. Consistent with a role for Kip2 in transporting dynein away from minus ends and toward plus ends, overexpression of Kip2 enhanced the frequency of dynein plus end targeting by 19% and reduced the frequency of SPB targeting by 83% (P = 0.0019; Fig. 6 B). Fluorescence intensity measurements revealed a robust 147% increase in the number of dynein molecules per plus end (Fig. 6 D). When both Kip2 and wild-type Num1CC were overexpressed, the frequency and mean fluorescence intensity of plus end dynein was reduced by 59% (P < 0.0001) and 63% (P = 0.0012), respectively, compared with GAL1p:KIP2 GAL1p:num1CCLL/EE cells (Fig. 6, B–E). The extent by which Kip2 overexpression reduces Num1CC-mediated plus end dynein depletion is therefore less than that of Pac1. Collectively, these data are consistent with our hypothesis, and they suggest that Num1CC and Pac1 are competing for binding to dynein–dynactin complexes. It is interesting to note that Pac1 binds to dynein within the C-terminal motor domain (Reck-Peterson et al., 2006; Markus et al., 2009; Huang et al., 2012; Toropova et al., 2014), whereas Num1 associates with dynein via the N-terminal tail domain (Markus et al., 2009). Thus, interference with Dyn1-Pac1 binding by Num1CC likely occurs by an allosteric mechanism.
Num1CC colocalizes with dynein–dynactin
Previous data indicate that Num1CC interacts with dynein at SPBs and is found in the cytoplasm as bright foci (presumably aggregates; see Fig. S5, A and B, open arrows and cytoplasm, respectively; Tang et al., 2012). We found that localization of Num1CC to the SPB is largely dependent on dynein, and more specifically, the MTBD of dynein (Fig. S5, A and B). Furthermore, cells in which dynein plus end targeting is restored—by overexpression of Pac1, deletion of the dynein MTBD, or use of Dyn1HL3—exhibit Num1CC at plus ends, albeit at a low frequency (Fig. S5, A–C). These data indicate that Num1CC can bind plus end dynein (as well as the Dyn1ΔMTBD and Dyn1HL3 mutants) and suggest that Num1 and Pac1 binding to dynein is not entirely mutually exclusive. The latter is consistent with the observation that Dyn1HL3-Pac1 complexes can offload together to Num1 cortical sites (Markus and Lee, 2011).
Direct observation of minus end–directed motion of dynein along astral microtubules
In budding yeast, dynein is targeted to microtubule plus ends by two distinct mechanisms: (a) direct recruitment from the cytoplasm, and (b) Kip2-mediated plus end–directed transport along astral microtubules (Carvalho et al., 2004; Markus et al., 2009). Evidence of the latter is apparent by the movement of fluorescent dynein speckles along astral microtubules toward plus ends (Fig. 7 A and Video 2; Markus et al., 2009). In budding yeast, dynein is never observed moving in the opposite direction—toward the minus ends of astral microtubules. Minus end–directed activity is only apparent when cortically anchored dynein motors move the spindle through interactions with astral microtubules. If depletion of dynein–dynactin from microtubule plus ends is a consequence of these motors being switched “on,” then we reasoned that cells overexpressing Num1CC would exhibit dynein molecules moving in a directed manner toward the minus ends of astral microtubules, as was recently observed in fission yeast expressing a Mcp5ΔPH (Num1 homologue) fragment (Ananthanarayanan et al., 2013).
Plus end–directed motility of dynein molecules in Kip2- and Num1CCLL/EE-overexpressing cells. Time-lapse highly inclined and laminated optical sheet images of Dyn1-3mCherry molecules (inverse fluorescence image in middle panel; red in merge) walking along astral microtubules (inverse fluorescence image in left panel; green in merge) toward the plus ends. Note the long astral microtubule phenotype is attributable to Kip2 overexpression. Related to Fig. 7.
Plus end–directed motility of dynein molecules in Kip2- and Num1CCLL/EE-overexpressing cells. Time-lapse highly inclined and laminated optical sheet images of Dyn1-3mCherry molecules (inverse fluorescence image in middle panel; red in merge) walking along astral microtubules (inverse fluorescence image in left panel; green in merge) toward the plus ends. Note the long astral microtubule phenotype is attributable to Kip2 overexpression. Related to Fig. 7.
Direct observation of Num1CC-mediated minus end motility of dynein and dynactin. (A) Example kymographs of plus end–directed motility of dynein molecules along astral microtubules observed in uninduced GAL1p:num1CC cells (left; –Num1CC) or in cells overexpressing Num1CCLL/EE and Kip2 (right). (B and C) Example kymographs depicting minus end–directed motility of dynein or dynactin (i.e., Jnm1) along astral microtubules in cells overexpressing Num1CC and either Kip2 (B) or Pac1 (C). Kymographs were generated from time-lapse images acquired using highly inclined and laminated optical sheet microscopy (see Materials and methods). Bars: (vertical) 1 min; (horizontal) 1 µm. (D) Velocity values for minus end–directed dynein runs observed in either Pac1- or Kip2-overexpressing cells. See also Fig. S5 and Videos 2 and 3.
Direct observation of Num1CC-mediated minus end motility of dynein and dynactin. (A) Example kymographs of plus end–directed motility of dynein molecules along astral microtubules observed in uninduced GAL1p:num1CC cells (left; –Num1CC) or in cells overexpressing Num1CCLL/EE and Kip2 (right). (B and C) Example kymographs depicting minus end–directed motility of dynein or dynactin (i.e., Jnm1) along astral microtubules in cells overexpressing Num1CC and either Kip2 (B) or Pac1 (C). Kymographs were generated from time-lapse images acquired using highly inclined and laminated optical sheet microscopy (see Materials and methods). Bars: (vertical) 1 min; (horizontal) 1 µm. (D) Velocity values for minus end–directed dynein runs observed in either Pac1- or Kip2-overexpressing cells. See also Fig. S5 and Videos 2 and 3.
In uninduced cells (i.e., not expressing any Num1CC) or in those induced to express Num1CCLL/EE, only plus end–directed motility of dynein (or dyneinΔMTBD) molecules was ever observed (Fig. 7 A and Video 2). We were unable to observe minus end–directed motility of dynein molecules in cells overexpressing Num1CC, likely because of the robust depletion of dynein from microtubule plus ends in these cells. Thus, we chose to focus on cells in which Num1CC-mediated depletion of dynein plus end binding was partially restored: GAL1p:KIP2 and GAL1p:PAC1 cells. Strikingly, we observed numerous instances of minus end–directed motility of dynein in cells overexpressing Kip2 or Pac1 in addition to Num1CC (Fig. 7, B and C; Fig. S5 D; Video 3; and Video 4). The mean velocity of minus end–directed dynein molecules along astral microtubules in GAL1p:KIP2 cells was slightly higher than that in GAL1p:PAC1 cells (55.3 nm/s, n = 22; vs. 41.7 nm/s, n = 9; Fig. 7 D); however, the values from both strains were very similar to the velocity of single molecules of purified dynein (∼70–100 nm/s; Reck-Peterson et al., 2006; Markus and Lee, 2011; Huang et al., 2012) and dynein-mediated spindle movements in this organism (41 nm/s; Markus et al., 2011). Thus, overexpression of Num1CC depletes dynein from microtubule plus ends by activating its minus end–directed motility.
Minus end–directed motility of dynein molecules in Kip2- and Num1CC-overexpressing cells. Time-lapse highly inclined and laminated optical sheet images of Dyn1-3mCherry molecules (inverse fluorescence image in middle panel; red in merge) walking along astral microtubules (inverse fluorescence image in left panel; green in merge) toward the minus ends. Related to Fig. 7.
Minus end–directed motility of dynein molecules in Kip2- and Num1CC-overexpressing cells. Time-lapse highly inclined and laminated optical sheet images of Dyn1-3mCherry molecules (inverse fluorescence image in middle panel; red in merge) walking along astral microtubules (inverse fluorescence image in left panel; green in merge) toward the minus ends. Related to Fig. 7.
Minus end–directed motility of dynein molecules in Kip2- and Num1CC-overexpressing cells. Time-lapse highly inclined and laminated optical sheet images of Dyn1-3mCherry molecules (inverse fluorescence image in middle panel; red in merge) walking along astral microtubules (inverse fluorescence image in left panel; green in merge) toward the minus ends. Related to Fig. 7.
Minus end–directed motility of dynein molecules in Kip2- and Num1CC-overexpressing cells. Time-lapse highly inclined and laminated optical sheet images of Dyn1-3mCherry molecules (inverse fluorescence image in middle panel; red in merge) walking along astral microtubules (inverse fluorescence image in left panel; green in merge) toward the minus ends. Related to Fig. 7.
Although we were able to see Num1CC-mediated minus end motility of dynactin (Jnm1-3YFP) in GAL1p:KIP2 cells (Fig. 7 B, iii), we were unable to observe examples of either Pac1 or Num1CC moving toward the minus ends. The reason for the latter is unclear but may be a result of disengagement of Num1CC from dynein subsequent to activation. Taken together with our other observations (i.e., the lack of Pac1 accumulation at SPBs in Num1CC-overexpressing cells, the reduced colocalization of Dyn1 and Pac1 in DYN1 and PH-DYN1 cells, and the ability of Pac1 overexpression to rescue Dyn1 plus end targeting), the apparent lack of minus end–directed Pac1 molecules further suggests that Num1CC activates plus end dynein by relieving Pac1-mediated inhibition.
Discussion
When purified from various sources, including budding yeast and animal tissue, dynein motors are active as apparent from ATPase and microtubule gliding assays. Yeast dynein requires no additional factors for processive single-molecule motility (Reck-Peterson et al., 2006), whereas dynein isolated from animal tissue requires a combination of dynactin and various adaptor proteins that link dynein to dynactin (McKenney et al., 2014; Schlager et al., 2014). This latter phenomenon helps explain how dynein activity is recruited to different vesicular or organellar compartments in animal cells, and thus how dynein activity is regulated with spatial precision. For instance, one of these adaptors, BicD2, binds to Rab6 on various vesicular cargoes, and thereby recruits dynein–dynactin complexes, which allows for minus end–directed movements of these vesicular cargoes (Matanis et al., 2002). Although mechanistically distinct, we have identified a similar mechanism at play in budding yeast. Specifically, Num1-mediated recruitment of dynein–dynactin complexes to the cell cortex is sufficient to (a) anchor dynein–dynactin complexes at their site of activity, and (b) activate dynein for its spindle orientation function. Unlike animal cells, however, association with dynactin is not sufficient to activate dynein motility in yeast cells. Rather, our studies indicate that a Num1-mediated Pac1 dissociation event is responsible for switching dynein from being off at microtubule plus ends to on at the cell cortex.
Our results revealed a small but significant Num1CC-mediated enhancement in the apparent dynein–dynactin interaction at plus ends (see Fig. 3 H). Although this effect appears to be minor compared with that of Num1CC on the dynein-Pac1 interaction, it is possible that, like the human adaptor proteins (e.g., BicD2 and Spindly), Num1 also plays a role in stabilizing the dynein–dynactin complex. If so, it may be that Num1 may further promote dynactin-mediated processivity enhancement of dynein in a manner reflective of the human adaptor proteins (McKenney et al., 2014; Schlager et al., 2014). Future studies will be needed to directly test this in a reconstituted system.
Although dynactin does not activate yeast dynein motility, per se, it is interesting to note that dynein–dynactin binding is a rate-limiting step during the dynein offloading process. Quantitative fluorescence microscopy revealed a threefold excess of dynein relative to dynein–dynactin complexes at microtubule plus ends (Markus et al., 2011). Given the reliance of dynein on dynactin for offloading to Num1 receptor sites (Lee et al., 2003), the limiting nature of dynactin at plus ends effectively restricts dynein pathway activity by limiting the number of cortical dynein–dynactin complexes. Increasing the number of intact dynein–dynactin complexes at plus ends by deletion of She1, which regulates their interaction in vivo (Woodruff et al., 2009), results in an increased number of cortical dynein–dynactin complexes, as well as enhanced dynein pathway activity (Markus et al., 2011). Thus, through mechanistically distinct processes, dynactin effectively activates dynein-mediated processes both in animal and yeast cells.
It is well established that Pac1 plays a central role in targeting dynein–dynactin to microtubule plus ends in yeast (Lee et al., 2003). Given the ability of Pac1 to reduce dynein motility in vitro (Markus and Lee, 2011; Huang et al., 2012), it has been postulated that Pac1 may hold dynein at plus ends in part by preventing its minus end–directed motility. At some point after or concomitant with dynein offloading, Pac1 dissociates from dynein and is never observed at cortical Num1 sites in wild-type cells (Lee et al., 2003). Given the observations presented here, we hypothesize that Num1 binding to dynein–dynactin triggers the dissociation of Pac1 from dynein. The fact that Dyn1ΔMTBD is insensitive to Num1CC overexpression suggests that microtubule binding by dynein is a requisite for Pac1 dissociation. This suggests a mechanism whereby Num1 binding to the dynein tail domain (Markus et al., 2009) communicates allosteric changes to the motor head that, after microtubule binding, induce Pac1 dissociation from its binding site (at the junction between AAA3 and AAA4; Toropova et al., 2014) and consequently permit minus end–directed motility (see Fig. 8). The requisite microtubule binding by dynein for Num1CC-mediated Pac1 dissociation likely explains the plus end colocalization of Dyn1ΔMTBD with Num1CC (Fig. S5, A–C) and Pac1 (not depicted).
Model for Num1-mediated activation of dynein-mediated spindle positioning. Our data suggest that at the moment of offloading (step 1), contact between dynein–dynactin and cortical Num1 triggers a cascade of events that ultimately leads to Pac1 dissociation (step 3); however, the MTBD (deletion of which interrupts this process) is required to make contact with the microtubule to initiate Pac1 dissociation (step 2).
Model for Num1-mediated activation of dynein-mediated spindle positioning. Our data suggest that at the moment of offloading (step 1), contact between dynein–dynactin and cortical Num1 triggers a cascade of events that ultimately leads to Pac1 dissociation (step 3); however, the MTBD (deletion of which interrupts this process) is required to make contact with the microtubule to initiate Pac1 dissociation (step 2).
It is interesting to note the apparent discrepancies between the requirements for plus end binding of yeast and human dynein. In budding yeast, the dynein motor domain, Pac1, and Bik1 are absolutely essential (Lee et al., 2003; Sheeman et al., 2003; Markus et al., 2009), but dynactin is dispensable for this process (Lee et al., 2003). However, recent in vitro reconstitution experiments with human dynein revealed a distinct plus end binding complex that requires EB1, the p150Glued subunit of dynactin, and the full-length dynein complex (i.e., the motor domain is not sufficient; Duellberg et al., 2014). These latter observations, which did not describe any minus end–directed dynein motility, suggest that dynein and dynactin (or at least p150Glued) can interact at plus ends in the absence of the recently characterized adaptor proteins (e.g., Hook3, Spindly, and BicD2; McKenney et al., 2014; Schlager et al., 2014) and further suggest that their interaction is not sufficient for dynein motility.
Our observation that the MTBD of dynein is dispensable for plus end targeting was surprising and changes our understanding by which dynein recognizes and binds to microtubule plus ends. In light of this observation, we propose that dynein does not directly contact the plus end; rather, dynein associates with plus ends indirectly through its interactions with Pac1 and Bik1 (see Fig. 1 B). Evidence indicates that Pac1 enables dynein tip tracking in part by linking it to the plus end binding protein Bik1 (Sheeman et al., 2003; Roberts et al., 2014). Thus, the ability of Pac1 (and LIS1) to permit prolonged encounters between dynein and microtubules (McKenney et al., 2010; Huang et al., 2012) is likely unrelated to plus end binding by dynein in this organism. However, it is conceivable that by maintaining dynein in an off state at plus ends, Pac1 may prevent minus end motility of dynein motors that are in very close proximity to the microtubule. Upon binding of Num1CC, dynein may pivot (likely in a stochastic manner) such that it may contact the plus end directly through its MTBD, subsequently release Pac1, and then walk toward the minus end. An analogous situation may take place in wild-type cells: after offloading to Num1 receptor sites at the cortex, dynein is well positioned to contact the microtubule to initiate spindle movements, which in turn may trigger Pac1 dissociation (Fig. 8). Recent structural studies support such a possibility: upon microtubule binding, conformational changes within the MTBD affect corresponding changes within the motor ring (Schmidt, 2015; Uchimura et al., 2015). These changes, which are propagated by the antiparallel coiled-coil that lead from the MTBD to the motor ring (via AAA4), could presumably affect Pac1 binding at the AAA3–AAA4 junction (see Fig. 8; Toropova et al., 2014).
Consistent with Num1 affecting the dynein-Pac1 interaction, the dynein mutant with higher than normal affinity for Pac1 (Dyn1HL3) was much less susceptible to Num1CC-mediated plus end depletion. It is unclear why Dyn1HL3 exhibits higher affinity for Pac1. This mutant was engineered such that a helical linker was inserted between the tail and motor domains (Markus and Lee, 2011). In the context of the tertiary structure of the dynein motor, this region lies in close proximity to the Pac1 binding site (between AAA3 and AAA4; see Fig. 5 A). Taken together with an apparent enhanced affinity of a tail-less dynein construct (motor domain only) for Pac1 (Reck-Peterson et al., 2006; Markus et al., 2009), it stands to reason that the tail domain plays a negative regulatory role in affecting Pac1 binding. Thus, whatever allosteric conformational change Num1CC-dynein tail binding induces is likely interrupted by insertion of the helical HL3 linker.
The nature of the Num1CC-dynein–dynactin interaction is currently unknown; however, a recent structural study revealed how human dynein–dynactin interacts with the coiled-coil–containing adaptor protein BicD2 (Urnavicius et al., 2015). Given the importance of the Num1 coiled-coil domain in the dynein–dynactin interaction, and the observation that Num1 (like BicD2) only interacts with intact dynein–dynactin complexes (Splinter et al., 2012), it may be that Num1 exhibits a similar mode of binding (i.e., direct contact with the dynein tail domain, and the Arp1 filament). Future high-resolution structural studies of the dynein tail domain within the context of the Num1CC-dynein–dynactin complex will be necessary to understand the network of interactions that define this enormous protein complex, as well as how the tail domain may possibly affect Pac1-motor domain binding.
Materials and methods
Media and strain construction
All strains are derived from YEF473A (Bi and Pringle, 1996) and are listed in Table S1. We transformed yeast strains using the lithium acetate method (Knop et al., 1999). Strains carrying null mutations or fluorescently tagged components were constructed by PCR product-mediated transformation (Longtine et al., 1998) or by mating followed by tetrad dissection. Strains expressing mTurquoise2-Tub1 were generated as described (Markus et al., 2015). Transformants were clonally purified by streaking to individual colonies on selective media. Proper tagging was confirmed by PCR and, in some cases, sequencing. Yeast synthetic defined (SD) media were obtained from Sunrise Science Products.
To generate a yeast strain with point mutations in Num1CC (L167E L170E; Fig. 2 A), we used the site-specific genomic mutagenesis approach (Gray et al., 2004). In brief, after integration of the URA3 cassette into the num1CC locus (replacing nucleotides 499–510, corresponding to amino acids L167–L170), a PCR product amplified from pSM37 (see Plasmid construction) containing the desired nucleotide substitutions was transformed into the URA3-integrated strain and subsequently selected on 5-fluoroorotic acid–containing plates. 5-Fluoroorotic acid–resistant colonies were selected and confirmed by colony PCR and sequencing of the genomic DNA region. A similar method was used to delete the MTBD (residues 3,102–3,225) from DYN1.
To generate a yeast strain expressing the GAL1p:PH-DYN1 allele, a cassette containing KANR::GAL1p:PH was amplified from pFA6a-kanMX6-pGAL1-PH (see Plasmid construction) and used for integration immediately upstream of the DYN1-3mCherry locus.
Plasmid construction
Using isothermal assembly (Gibson et al., 2009), we generated a plasmid in which the L167E L170E point mutations were engineered into a plasmid encoding Num1CC(95–303)-PCN-S-TEV-ZZ (pBSG02; Tang et al., 2012). In brief, primers were used to separately amplify the N-terminal (nucleotides 283–498, corresponding to amino acids 95–166) and C-terminal (nucleotides 511–909, corresponding to amino acids 171–303) portions of Num1CC, such that the desired nucleotide substitutions were included in the reverse primer for the N-terminal portion and the forward primer for the C-terminal portion. After amplification, the 3′ and 5′ ends of the PCR products corresponding to the N- and C-terminal regions, respectively, contained 20 nucleotides of sequence identity with each other, whereas the 5′ and 3′ ends of the N- and C-terminal regions, respectively, contained 20 nucleotides of sequence identity with the NcoI- and NotI-digested pBSG02 vector. After digesting pBSG02 with NcoI and NotI (to excise Num1CC(95–303) wild type), the gel-purified PCR products and digested vector were assembled in vitro as described (Gibson et al., 2009). Proper assembly was verified by restriction digest and DNA sequencing and resulted in pSM37 (encoding Num1CC(95–303)LL/EE-PCN-S-TEV-ZZ).
To generate a plasmid with which to N-terminally tag Dyn1 with a PH domain, the PH domain of Num1 (amino acids 2,563–2,692) was amplified using a forward primer flanked with an XmaI site and a reverse primer flanked with a SalI site. The PCR product was digested with XmaI and SalI and ligated into pFA6a-kanMX6-PGAL1 (Longtine et al., 1998) digested similarly, yielding pFA6a-kanMX6-PGAL1-PH.
Image acquisition, analysis, and dynein motility assay
Yeast cultures were imaged after growth at 30°C to mid-log phase in synthetic defined media supplemented with either 2% glucose (SD plus glucose) or 2% galactose plus 2% raffinose (SD plus galactose/raffinose; the latter for induction of Num1CC, Pac1, or Kip2, as indicated). To assess the effects of Num1CC on localization of dynein pathway components, GAL1p:num1CC cells were induced in SD plus galactose/raffinose for 6 h before mounting cells for fluorescence microscopy. For wide-field fluorescence microscopy, yeast cells were imaged on an agarose pad containing 50 mM potassium phosphate buffer, pH 7, and images were collected at room temperature using a 1.49 NA 100× objective on a Ti-E inverted microscope equipped with a Ti-S-E motorized stage (Nikon), piezo Z-control (Physik Instrumente), a SOLA SM II LE LED light engine (Lumencor), a motorized filter cube turret, and an iXon X3 DU897 cooled EM-CCD camera (Andor). The microscope system was controlled by NIS-Elements software (Nikon). A step size of 1 µm was used to acquire 2-µm-thick Z-stack images. Sputtered/ET filter cube sets (Chroma Technology) were used for imaging mTurquoise2 (49001), GFP (49002), YFP (49003), and mCherry (49008) fluorescence.
Image analysis was performed using ImageJ software (National Institutes of Health; kymographs in Fig. 7 were generated with the MultipleKymograph plugin). Plus end and SPB foci were identified in two (or three) color movies and scored accordingly. Specifically, plus end molecules were recognized as those foci that localized to the distal tips of dynamic microtubules (identified via mTurquoise2-Tub1 imaging), whereas SPB molecules were recognized as those foci that localized to one of the spindle poles. Cortical molecules (e.g., in PH-Dyn1–expressing cells) were identified as those foci not associated with an astral microtubule plus end that remained stationary at the cell cortex for at least three frames. Two data sets were considered statistically significant if a Student’s t test (assuming unequal variance) returned a p-value < 0.05.
For highly inclined and laminated optical sheet microscopy (Tokunaga et al., 2008), samples were prepared and imaged as above, except 488- and 561-nm lasers were used to excite YFP and mCherry, respectively. The laser illumination angle was adjusted individually for each sample to achieve the maximum signal-to-noise ratio. Emission filters were 525/50 nm for YFP and 600/50 for mCherry.
Purification of TAP (S tag-ZZ)-Dyn1-EGFP and the single-molecule motility assay (Fig. S1 and Video 1) were performed as previously described (Markus and Lee, 2011; Markus et al., 2012).
Cell lysis and immunoblotting
For Western blotting, yeast cultures were grown at 30°C in 3 ml of either SD plus glucose or SD plus galactose/raffinose and harvested. Equal numbers of cells were pelleted and resuspended in 0.2 ml of 0.1 M NaOH and incubated for 5 min at room temperature as described (Kushnirov, 2000). After centrifugation, the resulting cell pellet was resuspended in sample buffer and heated to ∼100°C for 3 min. Lysates were separated on a 10% SDS polyacrylamide gel and electroblotted to PVDF in 25 mM Tris and 192 mM glycine supplemented with 0.05% SDS and 10% methanol for 30 min. Rabbit anti–c-Myc polyclonal (GenScript) or anti–α-tubulin (Applied Biological Materials, Inc.) monoclonal antibodies and HRP-conjugated goat anti–rabbit or anti–mouse antibody (Jackson ImmunoResearch Laboratories) were used at 1:1,000, 1:1,000, 1:3,000, or 1:3,000, respectively. The chemiluminescence signal was acquired on an ImageQuant LAS 500 gel documentation system. Immunoblots were exposed without saturating the camera’s pixels.
Online supplemental material
Fig. S1 depicts the inherent ability of dynein to accumulate at microtubule minus ends in vitro. Fig. S2 shows that Num1CC overexpression depletes dynein light-intermediate (Dyn3) and intermediate (Pac11) chains, but not Bik1 from plus ends, and it also shows the effect of Num1CC overexpression on Dyn1 localization throughout the cell cycle. Fig. S3 shows the role of dynactin (Nip100), Pac1, and Bik1 in the Num1CC-mediated plus end depletion phenotype. Fig. S4 shows additional examples of PH-dynein localization as well as its colocalization with dynactin (Jnm1). Fig. S5 shows the localization of Num1CC-EGFP in various mutant backgrounds and also shows additional examples of the minus end–directed motility of Dyn1 in Num1CC-overexpressing cells. Video 1 shows a representative in vitro dynein single-molecule assay. Video 2 shows plus end–directed molecules of dynein in Kip2- and Num1CCLL/EE-overexpressing cells, whereas Video 3 and Video 4 show minus end–directed molecules of dynein in Kip2- and Num1CC-overexpressing cells. Table S1 shows strains used in this study.
Acknowledgments
We thank Dr. Randa Mahgoub for help with construction of the PH-dynein yeast strains, Dr. Jennifer DeLuca for reagents and critical reading of the manuscript, and members of Dr. DeLuca’s laboratory for valuable discussions throughout this study.
This work was supported by startup funds provided from Colorado State University to S.M. Markus.
The authors declare no competing financial interests.