Microtubules (MTs) play critical roles in various cellular events, including cell migration. End-binding proteins (EBs) accumulate at the ends of growing MTs and regulate MT end dynamics by recruiting other plus end–tracking proteins (+TIPs). However, how EBs contribute to MT dynamics through +TIPs remains elusive. We focused on tau-tubulin kinase 2 (TTBK2) as an EB1/3-binding kinase and confirmed that TTBK2 acted as a +TIP. We identified MT-depolymerizing kinesin KIF2A as a novel substrate of TTBK2. TTBK2 phosphorylated KIF2A at S135 in intact cells in an EB1/3-dependent fashion and inactivated its MT-depolymerizing activity in vitro. TTBK2 depletion reduced MT lifetime (facilitated shrinkage and suppressed rescue) and impaired HeLa cell migration, and these phenotypes were partially restored by KIF2A co-depletion. Expression of nonphosphorylatable KIF2A, but not wild-type KIF2A, reduced MT lifetime and slowed down the cell migration. These findings indicate that TTBK2 with EB1/3 phosphorylates KIF2A and antagonizes KIF2A-induced depolymerization at MT plus ends for cell migration.
Introduction
The microtubule (MT) cytoskeleton is essential for various physiological phenomena such as directional cell migration, proliferation, and morphogenesis. Because of their intrinsic polarity, MTs contain two distinct ends: a slowly growing minus end and a rapidly growing plus end (Desai and Mitchison, 1997). In cells, MT minus ends are primarily anchored to the MT organizing center and the Golgi apparatus and do not exhibit dynamics (Mitchison and Kirschner, 1984; Efimov et al., 2007), whereas MT plus ends exhibit stochastic oscillation between phases of growth and shrinkage, defined as dynamic instability (Desai and Mitchison, 1997).
Several MT-associated proteins (MAPs) regulate MT integrity and dynamics. Plus end–tracking proteins (+TIPs), a subclass of MAPs, preferentially accumulate at the growing ends of MTs. An interactive and cooperative framework among +TIPs regulates MT dynamics during cellular events, including cell division and cell migration (Howard and Hyman, 2007; Akhmanova and Steinmetz, 2008). The core end-binding proteins (EBs) autonomously accumulate at growing MT plus ends and recruit other +TIPs via interaction with EB-binding domains, the cytoskeleton-associated protein glycine-rich domain, or the SxIP motif (Akhmanova and Steinmetz, 2008; Honnappa et al., 2009). Accumulating evidence has demonstrated the interactive associations between EBs and regulators of MT growth. During MT polymerization, XMAP215 (chTOG) acts as a processive MT polymerase by facilitating the addition of tubulin to the ends, although it can also catalyze depolymerization (Shirasu-Hiza et al., 2003; Brouhard et al., 2008). On the other hand, several classes of kinesin families use their catalytic activities to depolymerize MTs. The most studied of these kinesins is the kinesin-13 family, which is composed of KIF2A, 2B, and 2C (mitotic centromere–associated kinesin [MCAK]; Walczak et al., 2013). These MT depolymerizers diffusely move along the MT lattice and target to the ends to catalyze tubulin removal from the ends using the energy of ATP hydrolysis (Desai et al., 1999; Hunter et al., 2003; Helenius et al., 2006). Among the members, only MCAK has the SxIP motif and is effectively recruited to MT plus ends by EBs (Mennella et al., 2005; Moore et al., 2005; Lee et al., 2008), possibly promoting the rapid switching of MT dynamics (Montenegro Gouveia et al., 2010). Although MT end dynamics are, at least in part, determined by the balance between the activities of the machineries that facilitate MT growth and shrinkage, their interplay is largely not understood.
Tau-tubulin kinase 2 (TTBK2) belongs to the casein kinase I family and was originally identified as a kinase that phosphorylates tau and tubulin (Ikezu and Ikezu, 2014). Mutations in the TTBK2 gene are known to cause a neurodegenerative disorder termed spinocerebellar ataxia type 11 (SCA11), which is characterized by progressive ataxia and atrophy of the cerebellum (Houlden et al., 2007). Although the pathogenic mechanism causing SCA11 is not clear, the inherited mutations in TTBK2 generate premature stop codons, resulting in the truncation of TTBK2 immediately after the kinase domain (SCA11-associated form; Houlden et al., 2007). In addition, it has been reported that TTBK2 acts as an SxIP-containing +TIP (Jiang et al., 2012). However, the role of TTBK2 in MT regulation has yet to be addressed.
In this study, we identified the kinesin-13 family MT-depolymerizing kinesin KIF2A as a novel substrate of TTBK2 via a proteomic approach. TTBK2 phosphorylated and inactivated KIF2A in vitro by inhibiting its association with MTs. TTBK2 phosphorylated KIF2A and removed it from MTs in intact cells in an EB-dependent manner. We propose that TTBK2 phosphorylates KIF2A and antagonizes KIF2A-induced depolymerization at MT plus ends for cell migration.
Results
TTBK2 tracks MT plus ends in an EB-dependent manner
EB1 and EB3 play crucial roles in recruiting other +TIPs to regulate MT dynamics (Akhmanova and Steinmetz, 2008). We sought to isolate the +TIPs that interact with EB1 and EB3 with a pull-down assay using whole rat brain lysates and mass spectrometry. We identified TTBK1 and TTBK2 as EB-binding proteins (Fig. S1), similar to results reported by others (Jiang et al., 2012). Both TTBKs display similar domain organization: a kinase domain at the N terminus and two EB-binding SxIP motifs in the C terminus (Fig. 1 A). Then, we characterized the EB3-binding and end-tracking properties of TTBK2. The formation of complexes between TTBKs and EB3 was confirmed by immunoprecipitation (Fig. 1 B). A pull-down assay using GST-EB3 demonstrated the common binding properties of the SxIP motifs in TTBK2 (Fig. 1 C): EB3 full length (FL) and EB3 Δ3 aa, which lacks the final 3 aa that recognize the cytoskeleton-associated protein glycine-rich domain (Komarova et al., 2005; Mishima et al., 2007), were associated with monomeric GFP (mGFP)–TTBK2, but this association was diminished after further deletion of the C-terminal region of EB3, which is responsible for its binding to the SxIP motif (Honnappa et al., 2009). Furthermore, mutation of both SxIP motifs in TTBK2 almost completely abolished its binding to EB3, although each SxIP motif differentially contributed to EB3 binding (Fig. 1 D). Consistently, the accumulation of mGFP-TTBK2 at MT ends was lost when both SxIP motifs were mutated (Fig. 1 E). The C-terminal region of TTBK2 (TTBK2-C; Fig. 1 A), which harbors the two SxIP motifs, was sufficient for end-tracking (Fig. 1 F). In addition, purified mGFP-TTBK2 required EB3 for its accumulation at the growing ends of MTs in vitro (Fig. 1 G). Collectively, the data indicate that TTBK2 acts as a typical SxIP motif–containing +TIP.
TTBK2 tracks MT plus ends in an EB-dependent manner. (A) Schematic diagram of TTBK1 and TTBK2. Kinase, protein kinase domain; SxIP, EB-binding motif. The red X indicates the mutation site in the SxIP motif. (B) Immunoprecipitation (IP) between TTBK2 and EB3 using COS-7 cells. R IgG indicates rabbit IgG. (C) COS-7 cells expressing mGFP-TTBK2 were lysed and incubated with GST-fused proteins. mGFP-TTBK2 coprecipitated with EB3-FL and EB3 lacking 3 aa from the C terminus but not with the further deletion mutant EB3 1–251 aa. Note that GST–EB3 1–251 aa and GST–EB3 Δ3 aa contain a 13-aa linker after the GST tag (see Protein purification and biochemistry in Materials and methods). IB, immunoblotting. CBB, Coomassie Brilliant blue staining. (D) Mutation of both SxIP motifs diminished the association between TTBK2 and EB3. (E) COS-7 cells expressing TTBK2 were imaged using epifluorescence microscopy. mGFP–TTBK2-WT accumulated at EB3-positive MT plus ends, but the mutant TTBK2-m1/2 did not. Bar, 10 µm. (F) mGFP–TTBK2-C colocalized with EB3 at the MT ends. Bar, 5 µm. (G) In vitro reconstitution of TTBK2 tracking. Purified mGFP–TTBK2-FL tracked the MT plus ends only in the presence of EB3. Protein concentrations are as follows: 15-µM tubulin (containing 3.2% rhodamine-labeled tubulin), 25-nM FLAG–mGFP-TTBK2, and 400-nM EB3. Kymograph images are presented. Horizontal and vertical bars, 5 µm and 50 s, respectively. See Fig. S1 for identification of the TTBKs as EB-binding proteins.
TTBK2 tracks MT plus ends in an EB-dependent manner. (A) Schematic diagram of TTBK1 and TTBK2. Kinase, protein kinase domain; SxIP, EB-binding motif. The red X indicates the mutation site in the SxIP motif. (B) Immunoprecipitation (IP) between TTBK2 and EB3 using COS-7 cells. R IgG indicates rabbit IgG. (C) COS-7 cells expressing mGFP-TTBK2 were lysed and incubated with GST-fused proteins. mGFP-TTBK2 coprecipitated with EB3-FL and EB3 lacking 3 aa from the C terminus but not with the further deletion mutant EB3 1–251 aa. Note that GST–EB3 1–251 aa and GST–EB3 Δ3 aa contain a 13-aa linker after the GST tag (see Protein purification and biochemistry in Materials and methods). IB, immunoblotting. CBB, Coomassie Brilliant blue staining. (D) Mutation of both SxIP motifs diminished the association between TTBK2 and EB3. (E) COS-7 cells expressing TTBK2 were imaged using epifluorescence microscopy. mGFP–TTBK2-WT accumulated at EB3-positive MT plus ends, but the mutant TTBK2-m1/2 did not. Bar, 10 µm. (F) mGFP–TTBK2-C colocalized with EB3 at the MT ends. Bar, 5 µm. (G) In vitro reconstitution of TTBK2 tracking. Purified mGFP–TTBK2-FL tracked the MT plus ends only in the presence of EB3. Protein concentrations are as follows: 15-µM tubulin (containing 3.2% rhodamine-labeled tubulin), 25-nM FLAG–mGFP-TTBK2, and 400-nM EB3. Kymograph images are presented. Horizontal and vertical bars, 5 µm and 50 s, respectively. See Fig. S1 for identification of the TTBKs as EB-binding proteins.
Identification of TTBK2 substrates by a proteomic approach
No functional domains, except for the kinase domain and the SxIP motifs, were found in the TTBK2 amino acid sequence. To gain insight into the functions of TTBK2, we sought to identify TTBK2 substrates. We previously reported the pull-down assay using the affinity beads coated with the kinase domain to isolate substrates (Amano et al., 2010). GST-fused TTBK2-cat (Fig. S2 A) was expressed in bacteria and was purified. Using the purified GST–TTBK2-cat proteins, we isolated the TTBK2-cat–interacting proteins from rat brain lysates. Silver staining revealed several candidate binding partners for GST–TTBK2-cat (Fig. S2 B). The selected SDS-PAGE bands (numbered from 1 to 10 in Fig. S2 B) were excised from the gel and processed for mass spectrometry analysis and identification. The identified proteins are listed in Table S1. Among these potential substrates, we focused on the kinesin-13 family MT depolymerase KIF2A because it is possible that TTBK2 regulates MT dynamics through KIF2A (Desai et al., 1999; Homma et al., 2003).
TTBK2 phosphorylates the MT depolymerase KIF2A
We first confirmed the presence of KIF2A in precipitates of GST–TTBK2-cat. Immunoblot analysis revealed that KIF2A coprecipitated with GST–TTBK2-cat, whereas the closely related protein KIF2C was not detected (Fig. S2 C). When purified KIF2A was incubated with the TTBK2 catalytic domain, TTBK2 efficiently phosphorylated KIF2A under conditions in which RhoGDI was not phosphorylated (Fig. 2 B, left). The stoichiometry of KIF2A phosphorylation calculated from three independent experiments was 2.54 ± 0.13. Among the various fragments of KIF2A examined (Fig. 2 A), KIF2A-N was predominantly phosphorylated by TTBK2 (Fig. 2 B, right), indicating that TTBK2 primarily phosphorylates residues in the N-terminal region of KIF2A. To determine the phosphorylation sites of KIF2A by TTBK2, we performed mass spectrometry analysis of GST–KIF2A-N phosphorylated by TTBK2 in vitro. Several serine or threonine residues were identified as phosphorylation sites (Table S2). Among these residues, S135 exhibited the highest frequency of phosphorylation (Table S2) and is considered to be a main phosphorylation site of KIF2A by TTBK2. S135 is located in the N-terminal region of KIF2A and is conserved among vertebrates (Fig. 2 C). Next, we generated an antibody specifically recognizing KIF2A phosphorylated at S135 (anti–pS135-KIF2A antibody). The specificity of the antibody was confirmed by in vitro phosphorylation analysis. Purified GST–KIF2A-N–wild type (WT) or -S135A, in which S135 was mutated to alanine, was incubated with TTBK2 in the presence or absence of ATP. The anti–pS135-KIF2A antibody only detected KIF2A-N-WT incubated in the presence of ATP, indicating that the antibody specifically recognizes KIF2A phosphorylated at S135 (Fig. 2 D).
TTBK2 phosphorylates the MT depolymerase KIF2A. (A) Schematic diagram of KIF2A. The domain organization of KIF2A and its fragments is indicated. (B) Phosphorylation of KIF2A by TTBK2 in vitro. (Left) KIF2A-FL and control RhoGDI from Sf9 cells were incubated with GST–TTBK2-cat. TTBK2 effectively phosphorylated KIF2A at a stoichiometry of 2.54 ± 0.13. The asterisk indicates the autophosphorylation of TTBK2-cat. (Right) GST-KIF2A fragments from bacteria were incubated with TTBK2-FL. TTBK2 predominantly phosphorylated KIF2A-N. The intact bands of the KIF2A fragments are labeled with red dots. (C) Schematic diagram of KIF2A and multiple sequence alignment of KIF2A from several organisms. A TTBK2-mediated phosphorylation site in KIF2A (S135 in human KIF2A) is located in the N-terminal region of KIF2A. S135 on KIF2A is conserved among vertebrates, and those are highlighted in red. Hs, Homo sapiens; Mm, Mus musculus; Rn, Rattus norvegicus; Gg, Gallus gallus; XI, Xenopus laevis. (D) Purified GST–KIF2A-N-WT or -S135A was incubated with TTBK2 in the presence or absence of ATP followed by silver staining or immunoblot analysis using an anti–pS135-KIF2A antibody. The anti–pS135-KIF2A antibody only detected KIF2A-N-WT incubated in the presence of ATP, indicating that the antibody specifically recognizes KIF2A phosphorylated at S135. The red dot indicates the intact band of GST–KIF2A-N. (E) HeLa cells were transfected with the indicated siRNAs and cultured for 48 h. The phosphorylation of KIF2A at S135 was visualized by immunoblot analysis using an anti–pS135-KIF2A antibody. TTBK2 depletion reduced the phosphorylation of endogenous KIF2A at S135. (F) GST-KIF2A was coexpressed with the indicated HA-fused proteins in COS-7 cells. Coexpression of TTBK2-WT increased the phosphorylation of KIF2A at S135. All of the results are representative of more than four independent experiments. See Fig. S2 for the determination of TTBK2 substrates and Table S2 for identification of the TTBK2-mediated phosphorylation sites of KIF2A. IB, immunoblotting.
TTBK2 phosphorylates the MT depolymerase KIF2A. (A) Schematic diagram of KIF2A. The domain organization of KIF2A and its fragments is indicated. (B) Phosphorylation of KIF2A by TTBK2 in vitro. (Left) KIF2A-FL and control RhoGDI from Sf9 cells were incubated with GST–TTBK2-cat. TTBK2 effectively phosphorylated KIF2A at a stoichiometry of 2.54 ± 0.13. The asterisk indicates the autophosphorylation of TTBK2-cat. (Right) GST-KIF2A fragments from bacteria were incubated with TTBK2-FL. TTBK2 predominantly phosphorylated KIF2A-N. The intact bands of the KIF2A fragments are labeled with red dots. (C) Schematic diagram of KIF2A and multiple sequence alignment of KIF2A from several organisms. A TTBK2-mediated phosphorylation site in KIF2A (S135 in human KIF2A) is located in the N-terminal region of KIF2A. S135 on KIF2A is conserved among vertebrates, and those are highlighted in red. Hs, Homo sapiens; Mm, Mus musculus; Rn, Rattus norvegicus; Gg, Gallus gallus; XI, Xenopus laevis. (D) Purified GST–KIF2A-N-WT or -S135A was incubated with TTBK2 in the presence or absence of ATP followed by silver staining or immunoblot analysis using an anti–pS135-KIF2A antibody. The anti–pS135-KIF2A antibody only detected KIF2A-N-WT incubated in the presence of ATP, indicating that the antibody specifically recognizes KIF2A phosphorylated at S135. The red dot indicates the intact band of GST–KIF2A-N. (E) HeLa cells were transfected with the indicated siRNAs and cultured for 48 h. The phosphorylation of KIF2A at S135 was visualized by immunoblot analysis using an anti–pS135-KIF2A antibody. TTBK2 depletion reduced the phosphorylation of endogenous KIF2A at S135. (F) GST-KIF2A was coexpressed with the indicated HA-fused proteins in COS-7 cells. Coexpression of TTBK2-WT increased the phosphorylation of KIF2A at S135. All of the results are representative of more than four independent experiments. See Fig. S2 for the determination of TTBK2 substrates and Table S2 for identification of the TTBK2-mediated phosphorylation sites of KIF2A. IB, immunoblotting.
Immunoblot analysis revealed that KIF2A was phosphorylated at S135 in HeLa cells (Fig. 2 E). When TTBK2 was depleted, the phosphorylation of KIF2A at S135 was reduced. The expression of TTBK2-WT in COS-7 cells increased the phosphorylation of exogenous or endogenous KIF2A at S135, whereas that of TTBK2–kinase inactive (KN) K50A (Bouskila et al., 2011) did not (Fig. 2 F and see Fig. 5 D). These results indicate that TTBK2 is responsible for the phosphorylation of KIF2A at S135 in intact cells and that KIF2A is a physiological substrate of TTBK2.
TTBK2-mediated phosphorylation inactivates KIF2A by inhibiting its binding to MTs
Next, we examined whether TTBK2-mediated phosphorylation affects the MT-depolymerizing activity of KIF2A. Phosphorylated and nonphosphorylated KIF2A were prepared via incubation with TTBK2 in the presence or absence of ATP, respectively, immediately before the depolymerization experiments. Consistent with a previous study (Desai et al., 1999), purified KIF2A depolymerized guanosine-5'-[(α,β)-methyleno]triphosphate (GMPCPP)–MTs in a dose-dependent manner (Fig. 3 A). Moreover, KIF2A phosphorylation reduced KIF2A activity (∼54.5% and 34.4% reduction at 5-nM and 10-nM KIF2A, respectively). MTs remained detectable in the precipitation fraction of phosphorylated KIF2A (Fig. 3 A). The visualization of remnant GMPCPP-MTs after a short exposure to KIF2A demonstrated that TTBK2-mediated phosphorylation indeed inhibited the MT-depolymerizing activity of KIF2A (Fig. 3 B). To further address the effects of KIF2A phosphorylation, we examined the association between KIF2A and GMPCPP/taxol-stabilized MTs to minimize KIF2A-mediated depolymerization (Hertzer et al., 2006). An assay using a range of MTs demonstrated that KIF2A phosphorylation decreased its binding to MTs (by ∼45.0% and 63.7% at 0.1-µM and 0.5-µM MTs, respectively; Fig. 3 C). Collectively, these data show that TTBK2 inhibits the MT-depolymerizing activity of KIF2A by reducing the interaction between KIF2A and MTs.
TTBK2-mediated phosphorylation inactivates KIF2A by inhibiting its binding to MTs. (A) The ability of nonphosphorylated and phosphorylated KIF2A to depolymerize GMCPP-MTs. Increasing concentrations of KIF2A were incubated with GMPCPP-stabilized MTs. The supernatant (S) and precipitation (P) fractions were visualized by Coomassie Brilliant blue staining. Phosphorylation by TTBK2 decreased the MT-depolymerizing activity of KIF2A. The plot shows the quantification of remnant MTs in the precipitation fraction as the mean ± SD of three independent experiments. *, P < 0.05 versus nonphosphorylated KIF2A (Student’s t test). (B) Visualization of GMPCPP-MTs using an antitubulin antibody after brief incubation with KIF2A. Bar, 10 µm. (C) The association between KIF2A and GMPCPP/taxol-stabilized MTs. Increasing concentrations of MTs were incubated with a fixed concentration of KIF2A, followed by ultracentrifugation. Bound (precipitation fraction [Ppt]) and unbound (supernatant fraction [Sup]) KIF2A were visualized by immunoblot analysis. The plot shows the amount of KIF2A bound to MTs relative to the total amount of KIF2A as the mean ± SD of three independent experiments. **, P < 0.01 versus nonphosphorylated KIF2A (Student’s t test).
TTBK2-mediated phosphorylation inactivates KIF2A by inhibiting its binding to MTs. (A) The ability of nonphosphorylated and phosphorylated KIF2A to depolymerize GMCPP-MTs. Increasing concentrations of KIF2A were incubated with GMPCPP-stabilized MTs. The supernatant (S) and precipitation (P) fractions were visualized by Coomassie Brilliant blue staining. Phosphorylation by TTBK2 decreased the MT-depolymerizing activity of KIF2A. The plot shows the quantification of remnant MTs in the precipitation fraction as the mean ± SD of three independent experiments. *, P < 0.05 versus nonphosphorylated KIF2A (Student’s t test). (B) Visualization of GMPCPP-MTs using an antitubulin antibody after brief incubation with KIF2A. Bar, 10 µm. (C) The association between KIF2A and GMPCPP/taxol-stabilized MTs. Increasing concentrations of MTs were incubated with a fixed concentration of KIF2A, followed by ultracentrifugation. Bound (precipitation fraction [Ppt]) and unbound (supernatant fraction [Sup]) KIF2A were visualized by immunoblot analysis. The plot shows the amount of KIF2A bound to MTs relative to the total amount of KIF2A as the mean ± SD of three independent experiments. **, P < 0.01 versus nonphosphorylated KIF2A (Student’s t test).
EB3 changes the autoinhibitory conformation of TTBK2
In the search for TTBK2 substrates as described in the previous paragraph, TTBK2 itself was also identified as a TTBK2-cat–interacting protein (Fig. S2 A and Table S1). Several kinases are structured in a closed conformation because of the association between their kinase domain and their pseudosubstrate region. Thus, we examined this possibility. The lysates of COS-7 cells expressing mGFP–TTBK2-FL were subjected to a pull-down assay using GST–TTBK2-cat, -N2, or -M fragments. We found that the kinase domain specifically bound to TTBK2-FL (Fig. 4 B). TTBK2-Δcat and -C were associated with GST–TTBK2-cat, but TTBK2-N2, -M, and -ΔC were not (Fig. 4 C). These results suggest an intramolecular association between the kinase domain and the C-terminal region of TTBK2. Because the two SxIP motifs in TTBK2-C are responsible for its end-tracking activity (Fig. 1 E), we hypothesized that the TTBK2 kinase domain inhibits its end-tracking activity. Supporting our hypothesis, the end-tracking efficiency of TTBK2-FL was less than that of the TTBK2-C fragment (Fig. 4 D). Consistently, the expression of mCherry–TTBK2-cat impaired the end-tracking activity of TTBK2-C, but the control mCherry did not (Fig. 4 E). The expression of TTBK2-cat increased the percentage of cells exhibiting diffuse localization of TTBK2-C to ∼40% without affecting the accumulation of EB3 at the MT ends (Fig. 4 E). These results indicate that the TTBK2 kinase domain binds to its C terminus and competes with its binding to EB3. Thus, we next examined whether the association between the TTBK2 kinase domain and its C terminus is interrupted by EB3. We found that purified EB3 specifically inhibited the association between TTBK2-cat and -C in a dose-dependent manner, but the control protein RhoGDI did not (Fig. 4 F). This inhibitory effect was not observed for TTBK2-C-m1/2, which lacks EB3-binding ability (Fig. 1 D and Fig. 4 F). This result suggests that EB3 interacts with TTBK2 and changes its autoinhibitory conformation, thereby liberating the kinase domain.
EB3 changes the autoinhibitory conformation of TTBK2. (A) Schematic diagram of TTBK2. The domain organization of TTBK2 and its fragments is presented with a summary of their ability to bind to TTBK2-cat. (B) COS-7 cells expressing mGFP–TTBK2-FL were lysed and incubated with the indicated GST-fused proteins. mGFP–TTBK2-FL specifically coprecipitated with GST–TTBK2-cat. CBB, Coomassie Brilliant blue staining. (C) The TTBK2 fragments were expressed in COS-7 cells, followed by pull-down. The TTBK2 fragments containing the C-terminal region were coprecipitated with GST–TTBK2-cat. Asterisks indicate the intact bands of the TTBK2 fragments. (D) HeLa cells expressing GFP–TTBK2-FL or -C were fixed and stained with the indicated antibodies. TTBK2-C end-tracked more efficiently than TTBK2-FL. The GFP intensity at the MT plus ends was measured by averaging the peak intensities of >20 GFP-TTBK2 comets for each cell and then plotting against the mean GFP intensity in the whole cell. Each data point represents an individual cell (≥25 cells for each condition). The boxes in the left panels are enlarged in the right panels. Bars, 10 µm. A.U., arbitrary unit. (E) mGFP–TTBK2-C was expressed in COS-7 cells together with control mCherry or mCherry–TTBK2-cat. mGFP–TTBK2-C failed to exhibit localized distribution at MT plus ends in ∼40% of cells expressing mCherry–TTBK2-cat. The graph shows the quantitative results. We counted the numbers of cells exhibiting the localized distribution of the indicated mGFP construct in the presence of mCherry or mCherry–TTBK2-cat. Error bars indicate the SEM. ***, P < 0.001 versus the respective control (Student’s t test). All of the results are representative of more than four independent experiments. The boxes in the left panels are enlarged in the rightmost panels. Bars: (left) 20 µm; (magnified images) 10 µm. (F) TTBK2-C or -C-m1/2 was expressed in COS-7 cells, and the lysates were subjected to pull-down using GST–TTBK2-cat in the presence of the control RhoGDI or EB3. Excess EB3 inhibited the association between TTBK2-cat and -C in a dose-dependent manner. The graph shows the relative amounts of bound TTBK2-C from more than three independent experiments as a box plot. ***, P < 0.001 (one-way analysis of variance [ANOVA], Tukey's test). IB, immunoblotting.
EB3 changes the autoinhibitory conformation of TTBK2. (A) Schematic diagram of TTBK2. The domain organization of TTBK2 and its fragments is presented with a summary of their ability to bind to TTBK2-cat. (B) COS-7 cells expressing mGFP–TTBK2-FL were lysed and incubated with the indicated GST-fused proteins. mGFP–TTBK2-FL specifically coprecipitated with GST–TTBK2-cat. CBB, Coomassie Brilliant blue staining. (C) The TTBK2 fragments were expressed in COS-7 cells, followed by pull-down. The TTBK2 fragments containing the C-terminal region were coprecipitated with GST–TTBK2-cat. Asterisks indicate the intact bands of the TTBK2 fragments. (D) HeLa cells expressing GFP–TTBK2-FL or -C were fixed and stained with the indicated antibodies. TTBK2-C end-tracked more efficiently than TTBK2-FL. The GFP intensity at the MT plus ends was measured by averaging the peak intensities of >20 GFP-TTBK2 comets for each cell and then plotting against the mean GFP intensity in the whole cell. Each data point represents an individual cell (≥25 cells for each condition). The boxes in the left panels are enlarged in the right panels. Bars, 10 µm. A.U., arbitrary unit. (E) mGFP–TTBK2-C was expressed in COS-7 cells together with control mCherry or mCherry–TTBK2-cat. mGFP–TTBK2-C failed to exhibit localized distribution at MT plus ends in ∼40% of cells expressing mCherry–TTBK2-cat. The graph shows the quantitative results. We counted the numbers of cells exhibiting the localized distribution of the indicated mGFP construct in the presence of mCherry or mCherry–TTBK2-cat. Error bars indicate the SEM. ***, P < 0.001 versus the respective control (Student’s t test). All of the results are representative of more than four independent experiments. The boxes in the left panels are enlarged in the rightmost panels. Bars: (left) 20 µm; (magnified images) 10 µm. (F) TTBK2-C or -C-m1/2 was expressed in COS-7 cells, and the lysates were subjected to pull-down using GST–TTBK2-cat in the presence of the control RhoGDI or EB3. Excess EB3 inhibited the association between TTBK2-cat and -C in a dose-dependent manner. The graph shows the relative amounts of bound TTBK2-C from more than three independent experiments as a box plot. ***, P < 0.001 (one-way analysis of variance [ANOVA], Tukey's test). IB, immunoblotting.
TTBK2 phosphorylates KIF2A and removes it from MTs in an EB-dependent manner
We have shown that TTBK2 phosphorylates KIF2A, thereby decreasing its MT-binding activity in vitro. We next investigated how TTBK2 regulates KIF2A localization to MTs in HeLa cells. We used HeLa cells expressing GFP-KIF2A for the analysis because using an anti-GFP antibody enabled us to observe fine changes in KIF2A distribution. The effectiveness of siRNAs targeting TTBK2 was confirmed by immunoblot analysis (Fig. 2 E). In control cells, KIF2A exhibited diffuse distribution in the cytoplasm, where the overlap of KIF2A with MTs was barely observed (Fig. 5 A). When TTBK2 was depleted, KIF2A evidently localized to MTs (Fig. 5 A). Another siRNA targeting TTBK2 yielded essentially similar results (unpublished data). To measure the colocalization of GFP-KIF2A and MTs, we calculated Pearson’s correlation coefficient (Rr) in the peripheral region within 10 µm of the cell edge because we were able to observe clear MT organization in the cell periphery. Theoretical values for this correlation coefficient can range from 1 to −1, where a value of 1 indicates that there is a perfect positive correlation. The calculation showed that TTBK2 depletion significantly increased the correlation of KIF2A with MTs (0.42 in control cells vs. 0.58 in TTBK2-depleted cells; Fig. 5 C), indicating that TTBK2 reduces KIF2A localization to MTs. The expression of siRNA-insensitive TTBK2 (resTTBK2)-WT partially reversed the alterations in KIF2A distribution caused by TTBK2 depletion in that the increase in the correlation of KIF2A with MTs was suppressed (Fig. 5, B and C). Under these conditions, resTTBK2 mutant defective in EB binding (m1/2) or the KN mutant did not rescue the phenotype (Fig. 5, B and C), indicating that the EB-binding activity and kinase activity of TTBK2 are required for the removal of KIF2A from MTs. We then examined whether the EB-binding activity of TTBK2 is required for KIF2A phosphorylation. Immunoblot analysis using anti–pS135-KIF2A antibody revealed that expression of TTBK2-WT increased the phosphorylation of KIF2A, but TTBK2-m1/2 or -KN did not increase it (Fig. 5 D). These results suggest that TTBK2 phosphorylates KIF2A in an EB-dependent manner. To further explore the mechanism by which TTBK2 regulates KIF2A, we monitored KIF2A localization in living HeLa cells. For this experiment, EB3-mRuby was coexpressed with GFP-KIF2A to mark growing MT ends. Cells expressing comparable levels of GFP-KIF2A were selected for analysis. Similar to the results from fixed cells (Fig. 5 A), KIF2A in control cells exhibited diffuse distribution and no apparent colocalization with EB3 (Fig. 5 E and Video 1). A line profile of growing MT ends showed that in control cells, the intensity of KIF2A was reduced at MT ends where EB3 accumulated (Fig. 5 F). In TTBK2-depleted cells, however, KIF2A exhibited filamentous localization that partially overlapped with EB3 (Fig. 5 E and Video 2). The line profile demonstrated that TTBK2 depletion increased KIF2A localization to MTs, particularly at the EB3-containing MT ends (Fig. 5 F). Collectively, these results suggest that TTBK2 phosphorylates KIF2A on MTs and effectively removes KIF2A from MT ends.
HeLa cells were transfected with control scramble siRNA and cultured for 24 h. Then, the cells were transfected with GFP-KIF2A and EB3-mRuby. Images were analyzed by time-lapse confocal microscopy using a laser-scanning confocal microscope (LSM 780; Carl Zeiss). After image acquisition in λ mode, the spectra of GFP and mRuby were unmixed with the linear unmixing function for ZEN software (Carl Zeiss). Frames were recorded every 2.09 s. The frame rate is 15 frames/s. Bar, 5 µm.
HeLa cells were transfected with control scramble siRNA and cultured for 24 h. Then, the cells were transfected with GFP-KIF2A and EB3-mRuby. Images were analyzed by time-lapse confocal microscopy using a laser-scanning confocal microscope (LSM 780; Carl Zeiss). After image acquisition in λ mode, the spectra of GFP and mRuby were unmixed with the linear unmixing function for ZEN software (Carl Zeiss). Frames were recorded every 2.09 s. The frame rate is 15 frames/s. Bar, 5 µm.
HeLa cells were transfected with TTBK2 siRNA and cultured for 24 h. Then, the cells were transfected with GFP-KIF2A and EB3-mRuby. Images were analyzed by time-lapse confocal microscopy using a laser-scanning confocal microscope. After image acquisition in λ mode, the spectra of GFP and mRuby were unmixed with the linear unmixing function for ZEN software. Frames were recorded every 2.09 s. The frame rate is 15 frames/s. Bar, 5 µm.
HeLa cells were transfected with TTBK2 siRNA and cultured for 24 h. Then, the cells were transfected with GFP-KIF2A and EB3-mRuby. Images were analyzed by time-lapse confocal microscopy using a laser-scanning confocal microscope. After image acquisition in λ mode, the spectra of GFP and mRuby were unmixed with the linear unmixing function for ZEN software. Frames were recorded every 2.09 s. The frame rate is 15 frames/s. Bar, 5 µm.
TTBK2 phosphorylates KIF2A and removes it from MTs in an EB-dependent manner. (A) The distribution of KIF2A in HeLa cells transfected with scramble or TTBK2 siRNA. TTBK2 depletion increased KIF2A localization to MTs. (B) Rescue experiments for the distribution of KIF2A using resTTBK2-WT and its mutants. The expression of resTTBK2-WT canceled the extensive KIF2A binding to MTs, but its m1/2 and KN mutants did not. (C) Quantification of the colocalization between GFP-KIF2A and MTs. A correlation coefficient (Rr) was calculated for the region within 10 µm of the cell edge (see Calculation of the correlation coefficient in Materials and methods). TTBK2 depletion significantly increased the correlation of KIF2A with MTs. This increase in the correlation caused by TTBK2 depletion was partially rescued by resTTBK2-WT but not by resTTBK2-m1/2 or -KN mutants. Error bars indicate the SD (≥30 cells were analyzed for each condition). ***, P < 0.001 (one-way ANOVA, Tukey’s honest significant difference test [HSD]). (D) COS-7 cells were transfected with control HA or the indicated HA-TTBK2 mutants. The expression of TTBK2-WT increased the phosphorylation of KIF2A at S135, but that of TTBK2-m1/2 or -KN did not. The graph shows the relative band intensity (pS135-KIF2A/KIF2A) as the mean ± SD of three independent experiments. **, P < 0.01 versus control (one-way ANOVA, Tukey’s HSD). (E) Live imaging of HeLa cells expressing GFP-KIF2A and EB3-mRuby. The boxed regions were magnified and are shown as a time series. TTBK2 depletion altered the distribution of KIF2A. Arrowheads indicate the EB3-containing MT ends. The time is given in seconds. Also see Videos 1 and 2. (F) The intensity profiles of GFP-KIF2A and EB3-mRuby along growing MTs in living cells. Cells with mean GFP intensities of 6.0–8.5 A.U. were selected for analysis under both conditions. TTBK2 depletion increased the binding of KIF2A to MTs, especially at the EB3-containing MT ends. Error bars indicate the SEM (>50 MTs of five cells were analyzed for each condition). The horizontal axis was adjusted to 1.0 at the site of peak EB3 intensity. (G) The subcellular distribution of KIF2A-WT and its nonphosphorylatable mutant S135A. KIF2A-S135A extensively localized to the MTs in HeLa cells. (A, B, and G) The boxes in the left panels are enlarged in the right panels. (H) The effect of the SCA11-associated TTBK2 mutant on the distribution of KIF2A. The expression of the TTBK2 mutant increased the distribution of KIF2A along MTs. A.U., arbitrary unit. Bars: (A, B, and G; left) 10 µm; (A, B, and G magnified images) 5 µm; (E and H) 5 µm.
TTBK2 phosphorylates KIF2A and removes it from MTs in an EB-dependent manner. (A) The distribution of KIF2A in HeLa cells transfected with scramble or TTBK2 siRNA. TTBK2 depletion increased KIF2A localization to MTs. (B) Rescue experiments for the distribution of KIF2A using resTTBK2-WT and its mutants. The expression of resTTBK2-WT canceled the extensive KIF2A binding to MTs, but its m1/2 and KN mutants did not. (C) Quantification of the colocalization between GFP-KIF2A and MTs. A correlation coefficient (Rr) was calculated for the region within 10 µm of the cell edge (see Calculation of the correlation coefficient in Materials and methods). TTBK2 depletion significantly increased the correlation of KIF2A with MTs. This increase in the correlation caused by TTBK2 depletion was partially rescued by resTTBK2-WT but not by resTTBK2-m1/2 or -KN mutants. Error bars indicate the SD (≥30 cells were analyzed for each condition). ***, P < 0.001 (one-way ANOVA, Tukey’s honest significant difference test [HSD]). (D) COS-7 cells were transfected with control HA or the indicated HA-TTBK2 mutants. The expression of TTBK2-WT increased the phosphorylation of KIF2A at S135, but that of TTBK2-m1/2 or -KN did not. The graph shows the relative band intensity (pS135-KIF2A/KIF2A) as the mean ± SD of three independent experiments. **, P < 0.01 versus control (one-way ANOVA, Tukey’s HSD). (E) Live imaging of HeLa cells expressing GFP-KIF2A and EB3-mRuby. The boxed regions were magnified and are shown as a time series. TTBK2 depletion altered the distribution of KIF2A. Arrowheads indicate the EB3-containing MT ends. The time is given in seconds. Also see Videos 1 and 2. (F) The intensity profiles of GFP-KIF2A and EB3-mRuby along growing MTs in living cells. Cells with mean GFP intensities of 6.0–8.5 A.U. were selected for analysis under both conditions. TTBK2 depletion increased the binding of KIF2A to MTs, especially at the EB3-containing MT ends. Error bars indicate the SEM (>50 MTs of five cells were analyzed for each condition). The horizontal axis was adjusted to 1.0 at the site of peak EB3 intensity. (G) The subcellular distribution of KIF2A-WT and its nonphosphorylatable mutant S135A. KIF2A-S135A extensively localized to the MTs in HeLa cells. (A, B, and G) The boxes in the left panels are enlarged in the right panels. (H) The effect of the SCA11-associated TTBK2 mutant on the distribution of KIF2A. The expression of the TTBK2 mutant increased the distribution of KIF2A along MTs. A.U., arbitrary unit. Bars: (A, B, and G; left) 10 µm; (A, B, and G magnified images) 5 µm; (E and H) 5 µm.
Consistent with our hypothesis, a KIF2A mutant that was nonphosphorylatable by TTBK2 (KIF2A-S135A) exhibited preferential localization to MTs in a similar manner to KIF2A-WT in TTBK2-depleted cells (Fig. 5 G). Expression of a SCA11-associated TTBK2 mutant (1284_5delAG or 1329InsA: 1–449 aa or 1–450 aa; Houlden et al., 2007), in which TTBK2 is truncated such that two thirds of the protein from the C terminus is lacking, caused extensive KIF2A accumulation at the MTs (Fig. 5 H and unpublished data), indicating the dominant-negative effects of these SCA11-associated mutants.
TTBK2 regulates MT dynamics via KIF2A
Next, we addressed the roles of TTBK2 in regulating MTs by measuring MT dynamics. For this purpose, we used HeLa cells expressing ensconsin MT-binding domain (EMTB)–2xEGFP, which labels MTs (Faire et al., 1999). In control HeLa cells, MTs at the cell edge exhibited slowed growth and continuous catastrophe/rescue (Fig. 6 A and Video 3). When we plotted the life history of representative MTs, those MTs demonstrated longer lives near the edges, including frequent oscillations between growth and shrinkage (Fig. 6 B). Alternatively, the MTs in TTBK2-depleted cells exhibited short lives (<1.5 min in most cases) after a few rescue events (Fig. 6, A and B; and Video 4). The quantification of MT dynamics revealed that TTBK2 depletion significantly increased the MT shrink rate (13.0 ± 5.2 µm/min in control cells vs. 19.5 ± 6.1 µm/min in TTBK2-depleted cells) and decreased the rescue (transition from shrinkage to growth) frequency (6.12 ± 2.1 min−1 in control cells vs. 4.30 ± 2.4 min−1 in TTBK2-depleted cells; Fig. 6 C). No significant difference was detected in the growth rate or catastrophe (transition from growth to shrinkage) frequency. Conversely, KIF2A depletion increased the rescue frequency (7.88 ± 1.7 min−1; Fig. 6, A–C; and Video 5), suggesting that KIF2A has the ability to suppress rescue in the cell peripheral region. Furthermore, changes in MT dynamics caused by TTBK2 depletion were recovered by additional knockdown of KIF2A in TTBK2-depleted cells (Fig. 6, A–C; and Video 6). Consistent with the effects of TTBK2 depletion, the cells expressing KIF2A-S135A exhibited increased shrink rates and decreased rescue frequencies compared with cells expressing KIF2A-WT (Fig. S3 and Video 7, Video 8, and Video 9). Collectively, with the changes in KIF2A distribution at MTs caused by TTBK2 depletion, these results indicate the antagonistic effects of the EB–TTBK2 complex on the MT-depolymerizing kinesin KIF2A.
HeLa cells expressing EMTB-2xEGFP were transfected with control scramble siRNA. Images were analyzed by time-lapse confocal microscopy using a laser-scanning confocal microscope. Frames were recorded every 2.19 s. The frame rate is 30 frames/s. Bar, 5 µm.
HeLa cells expressing EMTB-2xEGFP were transfected with control scramble siRNA. Images were analyzed by time-lapse confocal microscopy using a laser-scanning confocal microscope. Frames were recorded every 2.19 s. The frame rate is 30 frames/s. Bar, 5 µm.
HeLa cells expressing EMTB-2xEGFP were transfected with TTBK2 siRNA. Images were analyzed by time-lapse confocal microscopy using a laser-scanning confocal microscope. Frames were recorded every 2.19 s. The frame rate is 30 frames/s. Bar, 5 µm.
HeLa cells expressing EMTB-2xEGFP were transfected with TTBK2 siRNA. Images were analyzed by time-lapse confocal microscopy using a laser-scanning confocal microscope. Frames were recorded every 2.19 s. The frame rate is 30 frames/s. Bar, 5 µm.
HeLa cells expressing EMTB-2xEGFP were transfected with KIF2A siRNA. Images were analyzed by time-lapse confocal microscopy using a laser-scanning confocal microscope. Frames were recorded every 2.19 s. The frame rate is 30 frames/s. Bar, 5 µm.
HeLa cells expressing EMTB-2xEGFP were transfected with KIF2A siRNA. Images were analyzed by time-lapse confocal microscopy using a laser-scanning confocal microscope. Frames were recorded every 2.19 s. The frame rate is 30 frames/s. Bar, 5 µm.
HeLa cells expressing EMTB-2xEGFP were transfected with TTBK2 and KIF2A siRNAs. Images were analyzed by time-lapse confocal microscopy using a laser-scanning confocal microscope. Frames were recorded every 2.19 s. The frame rate is 30 frames/s. Bar, 5 µm.
HeLa cells expressing EMTB-2xEGFP were transfected with TTBK2 and KIF2A siRNAs. Images were analyzed by time-lapse confocal microscopy using a laser-scanning confocal microscope. Frames were recorded every 2.19 s. The frame rate is 30 frames/s. Bar, 5 µm.
HeLa cells expressing EMTB-2xEGFP were transfected with mRuby. Images were analyzed by time-lapse confocal microscopy using a laser-scanning confocal microscope. Frames were recorded every 2.19 s. The frame rate is 30 frames/s. Bar, 5 µm.
HeLa cells expressing EMTB-2xEGFP were transfected with mRuby. Images were analyzed by time-lapse confocal microscopy using a laser-scanning confocal microscope. Frames were recorded every 2.19 s. The frame rate is 30 frames/s. Bar, 5 µm.
HeLa cells expressing EMTB-2xEGFP were transfected with mRuby–KIF2A-WT. Images were analyzed by time-lapse confocal microscopy using a laser-scanning confocal microscope. Frames were recorded every 2.19 s. The frame rate is 30 frames/s. Bar, 5 µm.
HeLa cells expressing EMTB-2xEGFP were transfected with mRuby–KIF2A-WT. Images were analyzed by time-lapse confocal microscopy using a laser-scanning confocal microscope. Frames were recorded every 2.19 s. The frame rate is 30 frames/s. Bar, 5 µm.
HeLa cells expressing EMTB-2xEGFP were transfected with mRuby–KIF2A-S135A. Images were analyzed by time-lapse confocal microscopy using a laser-scanning confocal microscope. Frames were recorded every 2.19 s. The frame rate is 30 frames/s. Bar, 5 µm.
HeLa cells expressing EMTB-2xEGFP were transfected with mRuby–KIF2A-S135A. Images were analyzed by time-lapse confocal microscopy using a laser-scanning confocal microscope. Frames were recorded every 2.19 s. The frame rate is 30 frames/s. Bar, 5 µm.
TTBK2 regulates MT dynamics via KIF2A. (A) Time series of MT dynamics in HeLa cells. HeLa cells expressing EMTB-2xEGFP were transfected with the indicated siRNAs, and MT dynamics were monitored. The white and yellow arrowheads indicate static MT ends and MT ends undergoing rapid depolymerization at the cell edge, respectively. The time is given in seconds. Bars, 5 µm. (B) Life history plots of representative MTs in control (blue), TTBK2-depleted (red), KIF2A-depleted (yellow), and TTBK2- and KIF2A-depleted cells (green). TTBK2 depletion shortened the lifetime of the MTs, and these MTs underwent continuous depolymerization. (C) Quantification of the parameters of MT dynamics. Four parameters (growth rate, shrink rate, catastrophe frequency, and rescue frequency) were measured for a total of ≥50 MTs in >10 cells for each condition. TTBK2 depletion significantly increased the shrink rate and decreased the rescue frequency. Error bars indicate the SD. ***, P < 0.001 (one-way ANOVA, Tukey’s HSD). All of the results are representative of more than four independent experiments. Also see Videos 3–6.
TTBK2 regulates MT dynamics via KIF2A. (A) Time series of MT dynamics in HeLa cells. HeLa cells expressing EMTB-2xEGFP were transfected with the indicated siRNAs, and MT dynamics were monitored. The white and yellow arrowheads indicate static MT ends and MT ends undergoing rapid depolymerization at the cell edge, respectively. The time is given in seconds. Bars, 5 µm. (B) Life history plots of representative MTs in control (blue), TTBK2-depleted (red), KIF2A-depleted (yellow), and TTBK2- and KIF2A-depleted cells (green). TTBK2 depletion shortened the lifetime of the MTs, and these MTs underwent continuous depolymerization. (C) Quantification of the parameters of MT dynamics. Four parameters (growth rate, shrink rate, catastrophe frequency, and rescue frequency) were measured for a total of ≥50 MTs in >10 cells for each condition. TTBK2 depletion significantly increased the shrink rate and decreased the rescue frequency. Error bars indicate the SD. ***, P < 0.001 (one-way ANOVA, Tukey’s HSD). All of the results are representative of more than four independent experiments. Also see Videos 3–6.
TTBK2 regulates cell migration via KIF2A
Finally, we examined whether TTBK2 regulates cell migration via KIF2A because appropriate MT dynamics and integrity are required for cell migration (Watanabe et al., 2005; Kuijpers and Hoogenraad, 2011; Etienne-Manneville, 2013). We performed wound healing assays using HeLa cells. The control HeLa cells migrated toward the gap during wound healing, but TTBK2 depletion inhibited this cell migration (Fig. 7 A). Tracking of each cell demonstrated the directional and persistent migration of control cells, whereas TTBK2-depleted cells migrated less efficiently, including a loss of directionality toward the wound (Fig. 7 B). When quantifying migratory properties (Fig. 7 C), control HeLa cells moved at a velocity of 14.1 ± 3.25 µm/h for a total distance of 125 ± 35.8 µm from their original position (n > 150). TTBK2 depletion decreased the velocity to 10.4 ± 3.48 µm/h and the total distance to 77.4 ± 37.4 µm (siTTBK2-1; n > 150). Essentially, similar results were obtained using a different TTBK2 siRNA (unpublished data). The expression of resTTBK2-WT rescued the inhibitory effects of TTBK2 depletion on HeLa cell migration (Fig. 7 C and Fig. S4 A), establishing the essential role of TTBK2 in these migratory properties. We then examined the effects of KIF2A depletion on cell migration and found that KIF2A depletion also inhibited the migration (Fig. 7, B and C; and Fig. S4 A). TTBK2 and KIF2A double depletion, however, partially rescued the migration defects caused by TTBK2 or KIF2A single depletion (Fig. 7, B and C; and Fig. S4 A). Considering that TTBK2 inactivates KIF2A, these results indicate that the proper activity level of KIF2A is required for cell migration, and activities of TTBK2 and KIF2A should be tightly balanced for effective cell migration.
TTBK2 regulates cell migration via KIF2A. (A) Still images during wound healing of HeLa cells transfected with control or TTBK2 siRNA. TTBK2 depletion inhibited the migration of HeLa cells. (B) Migratory tracks of representative control, TTBK2-depleted, KIF2A-depleted, or TTBK2- and KIF2A-depleted cells. (C) Three parameters (migration distance, velocity, and directionality) were measured. For the control, TTBK2 depletion, and TTBK2/KIF2A double depletion, ≥150 cells from more than four independent experiments were analyzed. For KIF2A depletion or rescue with TTBK2-WT, ≥100 cells or ≥60 cells from three independent experiments were analyzed, respectively. ***, P < 0.001 (one-way ANOVA, Tukey’s HSD). (D) MT organization in migrating HeLa cells during wound healing. The boxes in the top panels are enlarged in the bottom panels. MT density at the cell front was measured as the mean intensity in the region indicated. TTBK2 depletion decreased MT intensity at the cell front. The decrease in MT intensity was reversed by co-depletion of KIF2A. Single-plane images focused on the cell periphery were used for the analysis. The fluorescence intensity was quantified in >20 cells for each condition and is shown as a ratio to the control cell value. The data represent the mean ± SD of three independent experiments (total of >60 cells for each condition). ***, P < 0.001 (one-way ANOVA, Tukey’s HSD). (E) The effects of TTBK2 depletion on the migration of cerebellar granule neurons in vivo. Granule neurons expressing control shRNA migrated toward the IGL through the ML, but neurons expressing TTBK2 shRNA exhibited only minimal migration. The graph shows the quantitative results. Error bars indicate the SEM. ***, P < 0.001 (one-way ANOVA). The data used for the statistical analysis were obtained from 12 slices of three brains (>300 cells). Bars: (A and B) 100 µm; (D) 10 µm; (D, magnification) 5 µm; (E) 50 µm.
TTBK2 regulates cell migration via KIF2A. (A) Still images during wound healing of HeLa cells transfected with control or TTBK2 siRNA. TTBK2 depletion inhibited the migration of HeLa cells. (B) Migratory tracks of representative control, TTBK2-depleted, KIF2A-depleted, or TTBK2- and KIF2A-depleted cells. (C) Three parameters (migration distance, velocity, and directionality) were measured. For the control, TTBK2 depletion, and TTBK2/KIF2A double depletion, ≥150 cells from more than four independent experiments were analyzed. For KIF2A depletion or rescue with TTBK2-WT, ≥100 cells or ≥60 cells from three independent experiments were analyzed, respectively. ***, P < 0.001 (one-way ANOVA, Tukey’s HSD). (D) MT organization in migrating HeLa cells during wound healing. The boxes in the top panels are enlarged in the bottom panels. MT density at the cell front was measured as the mean intensity in the region indicated. TTBK2 depletion decreased MT intensity at the cell front. The decrease in MT intensity was reversed by co-depletion of KIF2A. Single-plane images focused on the cell periphery were used for the analysis. The fluorescence intensity was quantified in >20 cells for each condition and is shown as a ratio to the control cell value. The data represent the mean ± SD of three independent experiments (total of >60 cells for each condition). ***, P < 0.001 (one-way ANOVA, Tukey’s HSD). (E) The effects of TTBK2 depletion on the migration of cerebellar granule neurons in vivo. Granule neurons expressing control shRNA migrated toward the IGL through the ML, but neurons expressing TTBK2 shRNA exhibited only minimal migration. The graph shows the quantitative results. Error bars indicate the SEM. ***, P < 0.001 (one-way ANOVA). The data used for the statistical analysis were obtained from 12 slices of three brains (>300 cells). Bars: (A and B) 100 µm; (D) 10 µm; (D, magnification) 5 µm; (E) 50 µm.
We next tested the effects of KIF2A-S135A expression on cell migration. The transient expression of GFP–KIF2A-S135A decreased the migration velocity and distance, but the expression of GFP–KIF2A-WT did not (Fig. S4 B), suggesting that TTBK2-mediated phosphorylation of KIF2A at S135 is required for effective cell migration. Furthermore, we examined whether TTBK2 or KIF2A depletion affects MT organization in migrating HeLa cells. A dense MT network was observed at the front of control cells in the wound edge (Fig. 7 D). TTBK2 depletion appeared to reduce the density of this MT network. To quantify the MT density at the cell front, we measured the fluorescence intensity of MTs in the region indicated in Fig. 7 D. The quantification revealed that the intensity of MTs at the cell front was significantly decreased by TTBK2 depletion, although KIF2A depletion did not affect the intensity. Furthermore, double depletion of TTBK2 and KIF2A recovered the decrease in MT intensity caused by TTBK2 depletion to the levels in control cells (Fig. 7 D). This result supports our hypothesis that TTBK2 regulates cell migration through KIF2A inhibition and control of MT dynamics.
We examined the involvement of TTBK2 in the migration of cerebellar granule neurons because TTBK2 may be involved in normal cerebellar development and tissue homeostasis (Houlden et al., 2007). The cerebellums of mice on postnatal day 5 were electroporated with an shRNA expression vector and mCherry and fixed at postnatal day 7 to monitor the migration of granule neurons from the external granular layer toward the internal granular layer (IGL) through the molecular layer (ML; Fig. 7 E). The proportions of migrating neurons in the ML and IGL were significantly decreased after TTBK2 depletion. Two independent shRNAs targeting TTBK2 resulted in similar inhibitory effects (Fig. 7 E). Collectively, our results indicate that TTBK2 is generally required for cell migration and that the functional depletion of TTBK2 impairs cerebellar development by affecting cell migration.
Discussion
TTBK2 regulates MT dynamics through KIF2A phosphorylation
Based on their crucial interactions with EBs, several +TIPs have been revealed to regulate MT dynamics at the growing MT ends. Our results suggest a new mechanism by which the MT depolymerization is antagonized at MT plus ends. We have shown that a +TIP TTBK2 phosphorylates KIF2A and inhibits the binding of KIF2A to MTs, thereby decreasing the MT-depolymerizing activity of KIF2A in vitro (Fig. 3, A–C). Cells lacking TTBK2 exhibit extensive KIF2A localization to MTs (Fig. 5, A and C). KIF2A phosphorylation and the removal of KIF2A from MTs require the binding of TTBK2 to EBs (Fig. 5, B–D). These results suggest that TTBK2 phosphorylates KIF2A primarily at growing MT ends and counteracts the depolymerization activity of KIF2A. TTBK2 can weakly localize to the MT lattice (Fig. 1 E), where, in addition to the MT ends, EBs can be localized (Sandblad et al., 2006; Maurer et al., 2012). Thus, TTBK2 possibly phosphorylates KIF2A not only at the MT ends but also on the MT lattice, thereby removing KIF2A from the MT lattice.
TTBK2 depletion induces short-lived MTs with an increased shrink rate and a decreased rescue frequency (Fig. 6). Conversely, KIF2A depletion increases rescue frequency (Fig. 6). The effects of TTBK2 depletion on MT dynamics are reversed by the co-depletion of KIF2A (Fig. 6). We also found that TTBK2 phosphorylates KIF2C (MCAK), but its inhibitory effects on MT-depolymerizing activity are not vigorous compared with KIF2A (unpublished data). Thus, the EB–TTBK2 axis appears to antagonize the MT-depolymerizing machinery primarily involving KIF2A. Possible reasons for why only the shrink rate and the rescue frequency were affected by TTBK2 depletion include the following explanations. (a) KIF2A association with MTs accelerates MT shrinkage and suppresses rescue, unlike KIF2C, which increases the catastrophe frequency (Walczak et al., 1996; Kline-Smith et al., 2004). Consistently, KIF2A depletion increased the rescue frequency but did not affect the catastrophe frequency (Fig. 6). Indeed, previous studies demonstrated the functional differences among the members of the kinesin-13 family (Mennella et al., 2005; Ohi et al., 2007; Walczak et al., 2013). (b) The EB3–TTBK2 axis removes KIF2A from MTs at the time of the rescue. (c) TTBK2 is not involved in MT growth and catastrophe, and the machinery facilitating MT growth in cells is sufficiently robust even in the presence of KIF2A binding to MT ends; therefore, in the presence of EBs, KIF2A exhibits no depolymerizing activity at MT ends. It cannot be ruled out, however, that other TTBK2 substrates such as MAP2 also play a role in regulating MT dynamics (Takahashi et al., 1995). Because phosphorylations of MAPs are generally thought to induce their dissociation from MTs, these phosphorylations may not explain the increase in MT shrink rate caused by TTBK2 depletion. Fully understanding these mechanisms requires further investigation via refined reconstitution assays and high-resolution structural analyses.
Regulation of TTBK2 activity via autoinhibitory conformation
In their resting states, several kinases form an inactive closed conformation as a result of the association between their kinase domain and a pseudosubstrate sequence. Here, we found that the N-terminal kinase domain of TTBK2 binds to its C-terminal region, which contains tandem SxIP motifs, and inhibits its binding to EBs (Fig. 4, B, C, and E). Conversely, EB3 interrupts this association (Fig. 4 F). In addition, TTBK2 requires the binding to EBs for its ability to phosphorylate KIF2A and remove it from MTs (Fig. 5, B–D). Collectively, we have hypothesized that TTBK2 is in a closed inactive form in the cytoplasm and that the binding to EBs opens it up, thereby activating TTBK2. This mechanism would enable TTBK2 to act specifically at the growing MT plus ends. Further investigation is required to fully elucidate the activation mechanism of TTBK2.
Materials and methods
Plasmids, siRNAs, chemical reagents, and antibodies
The cDNA used in this study was obtained as follows: partial human TTBK1 KIAA1855 (Kazusa DNA Research Institute and Thermo Fisher Scientific), partial human TTBK2 KIAA0847 (Kazusa DNA Research Institute and Invitrogen), human KIF2A (DNAFORM), mCherry (R. Tsien, University of California, San Diego, La Jolla, CA), and human EB3 (Thermo Fisher Scientific). FL TTBK1 and TTBK2 were generated using standard PCR techniques based on the nucleotide sequences of TTBK1 (RefSeq accession no. NM_032538.1) and TTBK2 (RefSeq accession no. NM_173500.3) in the National Center for Biotechnology Information database. We subcloned the PCR-amplified cDNA fragments into pENTR-D/TOPO (Invitrogen). After sequencing, these fragments were further subcloned into appropriate commercial or homemade destination vectors via Gateway technology (Invitrogen) according to the manufacturer’s instructions. The siRNAs were obtained from Sigma-Aldrich. The following siRNA sequences were used: scramble control, 5′-CAGUCGCGUUUGCGACUGG-3′; siTTBK2-1, 5′-CCAUAUCUCUUCUUUGGAU-3′; siTTBK2-2, 5′-CUAACAACCACCACUACUU-3′; siKIF2A-1, 5′-GAAGCUAUUCUUGAGCAAA-3′; and siKIF2A-2, 5′-CAGAACAUCCGGUCAAACA-3′. The shRNA for TTBK2 was designed by Nagase, and the following sequences were used: control scramble, 5′-CAGTCGCGTTTGCGACTGG-3′; shTTBK2-2, 5′-GGGACAGTTCGTTATGCAT-3′; and shTTBK2-3, 5′-AGACCATGTTTGTAGATTT-3′. These shRNAs were introduced into the pSico vector (Addgene). For the rescue experiments, we used an siRNA-insensitive TTBK2 mutant generated using a site-directed mutagenesis kit (Agilent Technologies) by introducing silent mutations within the siRNA target sequence of TTBK2. The SxIP motifs were mutated into SxNN as described previously (Honnappa et al., 2009). EMTB-2xEGFP was generated based on a previous study (Faire et al., 1999) and was used to monitor MT dynamics in cultured cells. The antibodies used were as follows: anti-TTBK1 and anti-TTBK2 (Sigma-Aldrich), anti-TTBK1 (from T. Ikezu, Boston University, Boston, MA), anti-KIF2A (Novus Biologicals), anti-GFP (Roche or MBL International), antitubulin (DM1A, Sigma-Aldrich; or YOL 1/34, EMD Millipore), and anti-myc (9E10). The anti–pS135-KIF2A rabbit polyclonal antibody was raised against the phosphopeptide GS(-P)VSDISPVQA (Sigma-Aldrich) and was affinity purified against the phosphopeptide as described previously (Goto and Inagaki, 2007). Tubulin and GMPCPP were purchased from Cytoskeleton, Inc. and Jena Bioscience, respectively. The GST-fused TTBK2 catalytic domain was obtained from Carna Biosciences and was used for the phosphorylation assay. Other chemicals were obtained from commercial sources.
Cell culture
COS-7 and HeLa cells were maintained in DMEM (Sigma-Aldrich) supplemented with 10% FBS (Sigma-Aldrich). HeLa cells expressing GFP-KIF2A were provided by R. Uehara (Hokkaido University, Sapporo, Japan) and maintainedas described previously (Uehara et al., 2013). The transfection of HeLa cells was performed using RNAiMax (Invitrogen) for siRNA and Lipofectamine or Lipofectamine LTX (Invitrogen) for transient plasmid transfection. COS-7 cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. For the rescue experiment using the wound healing assay, HeLa cells expressing siRNA-insensitive GFP–TTBK2-FL were generated using the piggyback transposon system (System Biosciences), followed by FACS (Aria; BD) analysis to isolate the GFP-positive cell population. This pooled cell population was used for the rescue experiment.
Protein purification and biochemistry
Recombinant GST-fused proteins were produced in Escherichia coli (XL-1 blue, BL21DE3, or RosettaDE3) using isopropyl-β-d-thiogalactopyranoside and were purified as described previously (Wang et al., 2012; Kakeno et al., 2014). To produce GST, GST-EB1, and GST-EB3, we used pGEX (GE Healthcare). For other GST-fused proteins (EB3 1–251 aa, EB3 Δ3 aa, TTBK2 fragments, and KIF2A fragments), we used pDEST15 (Invitrogen), which contains a 13-aa linker region between GST tag and cloning sites. His-fused KIF2A-FL was expressed in Sf9 cells and was purified based on a previous study (Maney et al., 1998) using HisTrap and an Akta purifier (GE Healthcare). All of the protein purification procedures were performed at 4°C.
In vitro reconstitution of MTs
The end-tracking assay was performed as previously described (Kakeno et al., 2014). Flow chambers (22 mm wide × 1 mm high × 0.15 mm deep) were assembled between a coverslip and a precleaned glass microslide using double-sided tape. The silanized glass coverslip was coated with antibiotin (1% in MRB80; Invitrogen), and the nonspecific surface was blocked with Pluronic F127 (1% in MRB80; Invitrogen). Biotinylated MT seeds (50-µM tubulin solution containing 10% biotinylated tubulin, 10% rhodamine-labeled tubulin, and 1-mM GMPCPP) were specifically attached to the functionalized surface via biotinylated tubulin–antibiotin interactions. After the chamber was washed with MRB80, MT growth was initiated by perfusing 15-µM tubulin (containing 3.2% rhodamine-labeled tubulin) and 25-nM FLAG–mGFP-TTBK2 from mammalian cells with or without 400-nM EB3 into the assay buffer (MRB80, 75-mM KCl, 1-mM GTP, 0.5 mg/ml κ-casein, and 0.1% methylcellulose; Sigma-Aldrich) and an oxygen scavenger system (50-mM glucose, 400 µg/ml glucose oxidase, 200 µg/ml catalase, and 4-mM DTT). The samples were sealed using candle wax. During the experiments, the samples were maintained at 25 ± 1°C. Images were collected every 3 s for 2.5 min via total internal reflection fluorescence microscopy using a microscope (TE2000E; Nikon), a camera (iXonEM+ 897; Andor Technology), a 100× 1.49 NA objective, and a 488/561-nm excitation laser (Melles Griot). The microscopes were controlled using Micro-Manager software (Vale laboratory, University of California, San Francisco). Kymographs were generated for MT seeds using ImageJ software (National Institutes of Health).
Pull-down and immunoprecipitation assays
For the pull-down assay, transfected COS-7 cells were washed with PBS and lysed using buffer A (20-mM Tris/HCl, pH 7.5, 1% NP-40, 50-mM NaCl, 5-mM MgCl2, and protease inhibitors). After removing debris via centrifugation, the lysates were incubated with glutathione beads coated with 200 pmol of the GST-fused protein for 1 h at 4°C. The beads were washed with buffer A and dissolved in SDS sample buffer. For the immunoprecipitation assay, appropriate antibodies and protein A–Sepharose (GE Healthcare) were used instead of the GST-fused protein and the glutathione beads. To identify interacting molecules, rat brain lysates were used. The entire adult rat brain, except for the cerebellum, was lysed using buffer B (50-mM Tris/HCl, pH 7.5, 1% NP-40, 0.1% SDS, 0.1% deoxycholate, 150-mM NaCl, 5-mM MgCl2, and protease inhibitors). The GST-fused proteins were used at 500 pmol.
In vitro phosphorylation assay
The kinase assay was performed in 50 µl kinase buffer C (50-mM Tris-HCl, pH 7.5, 5-mM MgCl2, 1-mM DTT, and 200 µM ATP) containing 6.2-nM recombinant GST-TTBK2 catalytic domain (Carna Biosciences) or 10-nM GFP–TTBK2-FL (homemade), a substrate (80-nM KIF2A-FL or 160-nM KIF2A fragments), and 0.1-mM γ-[32P]ATP. After incubation for 30 min at 30°C, the reaction mixtures were boiled in SDS sample buffer and subjected to SDS-PAGE and silver staining. The radiolabeled bands were visualized using an image analyzer (Typhoon FLA 9000; GE Healthcare). To prepare phosphorylated and nonphosphorylated KIF2A for the biochemical experiments, purified KIF2A was incubated with recombinant GST-TTBK2 catalytic domain for 30 min at 30°C in the presence or absence of ATP, respectively. The buffer conditions and the kinase concentration were the same as described in the kinase assay. After the incubation, the reaction mixtures were immediately subjected to MT depolymerization assays or MT cosedimentation assays.
MT-related assays
GMPCPP-tubulin was prepared by cycling tubulin polymerization in the presence of 1-mM GMPCPP based on a previous study (Stumpff et al., 2007). In brief, 20-µM tubulin was incubated in 1-mM GMPCPP on ice for 10 min and then polymerized at 37°C for 30 min. Polymerized tubulin was pelleted at 150,000 g at 25°C for 10 min. The pellet was resuspended at a concentration of 20 µM. Then, tubulin was further cycled once in the presence of 1-mM GMPCPP. MT depolymerization assays were performed using a previously described method (Hertzer et al., 2006). GMPCPP-MTs at 1 µM in buffer D (80-mM K-Pipes, pH 6.9, 1-mM MgCl2, 1-mM EGTA, 2-mM Mg-ATP, 1-mM DTT, and 50-mM KCl) were incubated in KIF2A at 22°C for 10 min. The reaction mixture was subjected to ultracentrifugation at 350,000 g at 22°C for 10 min. To visualize the MTs, the MT-depolymerizing reaction was stopped after 5 min of incubation by adding glutaraldehyde at a final concentration of 1%. The mixture was sedimented onto coverslips coated with poly-l-lysine, postfixed in methanol, rehydrated, blocked with BSA, and processed for immunofluorescence using an antitubulin antibody.
Mass spectrometry analysis
To identify the EB- or TTBK2-interacting proteins, the obtained eluates were separated by SDS-PAGE and were subjected to silver staining using SilverQuest (Invitrogen). Each band of interest was excised from the gel. These proteins were reduced, S-carboxymethylated, and digested using trypsin. The digested peptide fragments were subjected to nanoelectrospray tandem mass spectrometry analysis using a mass spectrometer (Finnigan LTQ/Orbitrap; Thermo Fisher Scientific) combined with an HPLC system (Paradigm MS4; Michrom BioResources, Inc.).
Identification of phosphorylation sites in KIF2A
To identify the TTBK2-mediated phosphorylation sites on KIF2A, purified GST–KIF2A-N was subjected to a nonradioactive phosphorylation assay using TTBK2 and then precipitated using glutathione beads. The precipitated samples were eluted with guanidine solution (50-mM NH4HCO3 and 7-M guanidine-HCl). The eluates were subjected to reduction, alkylation, demineralization, and peptide enrichment, followed by digestion using trypsin. The phosphopeptides were enriched using the Titansphere Phos-TiO kit (GL Sciences) according to the manufacturer’s instructions. Nanoelectrospray tandem mass spectrometry analysis was performed using a mass spectrometry system (Q Exactive; Thermo Fisher Scientific) combined with an ultra HPLC system (Nano-Advance; Bruker-Michrom, Inc.). The raw data were analyzed using Proteome Discoverer software (Thermo Fisher Scientific) with the Sequest algorithm at a precursor mass accuracy of 15 ppm and a tandem mass spectrometry tolerance of 0.02 D. The peptide search was performed against the UniProtKB Homo sapiens reference proteome dataset (release 2012_10) using a 1% false discovery rate threshold. The most likely phosphorylation site was determined using the PhosphoRS algorithm within the Proteome Discoverer software (Thermo Fisher Scientific).
Immunohistochemistry and imaging
HeLa cells were fixed using methanol for 10 min at −30°C followed by postfixation using 4% formaldehyde in PBS for 10 min at RT. The cells were blocked with 1% BSA (Sigma-Aldrich) for 30 min at RT and incubated in the primary antibody for 1 h at RT. The secondary antibodies included Alexa Fluor 488–, 555–, and 647–conjugated antibodies against mouse, rabbit, or rat IgG (Invitrogen). Confocal images were recorded using a microscope (Axiovert Observer LSM780; Carl Zeiss) equipped with Plan Apochromat 63× 1.40 NA and Plan Apochromat 100× 1.40 NA objectives under the control of Zen software (Carl Zeiss). For live imaging of GFP-KIF2A and EB3-mRuby, cells were observed at 37°C in Hanks’ balanced salt solution supplemented with 10% FBS using an LSM780 microscope equipped with a Plan Apochromat 40× 1.40 NA objective. Images were acquired in λ mode at 2.09-s intervals for 3.5 min and then processed with the linear unmixing function of Zen software to unmix the GFP and mRuby spectra. For dynamic detection of +TIPs in living cells, the cells were observed at 37°C in Hanks’ balanced salt solution supplemented with 10% FBS or CO2-independent medium (Invitrogen) using an inverted microscope (IX-81; Olympus) equipped with a Plan Apo 60× 1.42 NA oil objective and a camera (iXonEM+ DV885; Andor Technology) under the control of MetaMorph software (Molecular Devices). We used filter sets from Semrock (GFP-3035B and TxRed-4040C). The images were processed using LSM Zen (Carl Zeiss), ImageJ, and MetaMorph software.
Calculation of the correlation coefficient
To calculate the correlation coefficient of GFP-KIF2A and MTs at the cell periphery, the region within 10 µm of the cell edge was defined using MetaMorph software as follows. After subtracting the background, the cell outline was manually traced, and the region was shrunk to a 10-µm distance from the cell edge to create the inner region. The area outside the inner region and within the cell outline was used for the analysis. Pearson’s correlation coefficient was calculated using ImageJ.
Analysis of MT dynamics
For the analysis of MT dynamics, cells expressing EMTB-2xEGFP were transfected with the indicated siRNA. At 48 h after transfection, the cells were observed at 37°C in Hanks’ balanced salt solution supplemented with 10% FBS using an LSM780 microscope. The cells were imaged at 2.19-s intervals for 3.65 min. To measure the MT dynamics, life history plots of MTs were generated and analyzed as previously described (Li et al., 2011).
HeLa cell migration during wound healing
At 24 h after siRNA transfection, the HeLa cells were reseeded on the culture inserts (ibidi). During wound healing, the cells were imaged at 10-min intervals for 12 h using an inverted microscope (IX-81) equipped with a UPlan SApo 10× 0.40 NA objective in the presence of L-15 medium (Sigma-Aldrich) supplemented with 10% FBS. Before collecting the images, the cells were stained with Hoechst 33342 at 0.3 µg/ml for 30 min. The movement of each nucleus was determined via object tracking using MetaMorph software. In Fig. 7, the migration distance is expressed as the direct distance from the original position. The directionality was calculated as the migration distance divided by the total track distance in accordance with a previous study (Pegtel et al., 2007). For the observation of MT organization during wound healing, cells were incubated for 6 h after removing the culture insert and then fixed with methanol for 10 min at −30°C, followed by immunostaining with an antitubulin antibody (YOL 1/34). Nuclei were stained with Hoechst 33342. MT density at the cell front was measured as the mean intensity in the region indicated in Fig. 7 D. Single-plane images focused on the cell periphery were used for the analysis.
In vivo electroporation
Animals were handled in accordance with guidelines established by Nagoya University and the National Center of Neurology and Psychiatry. Postnatal day 5 ICRmice were anesthetized on ice and injected with plasmid DNA. 2 µl of plasmid DNA (mixture of 1.0 mg/ml pSico-shRNA and 0.5 mg/ml pCAG-mCherry) in TE buffer (10-mM Tris/HCl, pH 7.5, and 1-mM EDTA) containing Fast green were injected into the interlobular space between lobules V and VI using a glass micropipette (G-1.0; Narishige). After the injection, forceps-type electrodes were placed on the occipital regions, and 50-ms electronic pulses of 80 mV were delivered seven times at intervals of 150 ms using an electroporator (CUY21; Nepa Gene). The pups were revived at 37°C and returned to the litter. The brains were fixed using 4% paraformaldehyde at 48 h after electroporation, cryoprotected in 30% sucrose/PBS, and then sectioned into 40-µm-thick sagittal slices using a cryostat (CM3050S; Leica). Immunofluorescence was performed according to a previous study (Seto et al., 2014). In brief, the sections were blocked with 0.5% BSA in PBS containing 0.2% Triton X-100 at RT for 1 h and subsequently incubated in an anti-RFP antibody (MBL International) in blocking buffer at 4°C for 20 h. The specimens were subsequently rinsed with PBS and incubated in an Alexa Fluor 555–conjugated anti–rabbit IgG antibody and in DAPI at RT for 2 h.
Online supplemental material
Fig. S1 shows the identification of EB-binding proteins. Fig. S2 shows the identification of TTBK2-interacting proteins by a proteomics approach and the presence of KIF2A in GST–TTBK2-cat precipitates. Fig. S3 shows the effects of KIF2A-S135A expression on MT dynamics. Fig. S4 shows representative images of HeLa cells during wound healing under the conditions indicated in Fig. 7 and demonstrates the effects of KIF2A-S135A expression on cell migration. Videos 1 and 2 show the live imaging of GFP-KIF2A in a control cell and the TTBK2-depleted cell, respectively. Videos 3–6 show the MT dynamics in a control cell, a TTBK2-depleted cell, a KIF2A-depleted cell, and a cell co-depleted of TTBK2 and KIF2A, respectively. Videos 7–9 show the MT dynamics in cells expressing control mRuby, mRuby–KIF2A-WT, and mRuby–KIF2A-S135A, respectively. Table S1 shows the list of proteins identified as candidate binding partners of TTBK2-cat. Table S2 shows the results of the mass spectrometry analysis performed to identify TTBK2-mediated phosphorylation sites of KIF2A. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201412075.dv.
Acknowledgments
We thank Dr. R. Tsien for the mCherry cDNA, Dr. M. Dogterom (Foundation for Fundamental Research on Matter Institute for Atomic and Molecular Physics, Amsterdam, Netherlands) and Dr. L. Laan (Harvard University, Cambridge, MA) for developing the in vitro reconstitution method, Dr. R. Uehara for sharing materials and for helpful discussions, Dr. T. Ikezu for sharing the anti-TTBK antibody, Ms. K. Yokoi (Nagoya University, Nagoya, Japan) for initiating this work, all members of the Kaibuchi laboratory for discussion and technical support, and Ms. T. Ishii for secretarial assistance. We also acknowledge the Division for Medical Research Engineering (I. Mizuguchi, Y. Ito, M. Takana, K. Taki, K. Itakura, and Y. Fujita) and the Radioisotope Center at the Medical Branch of the Nagoya University School of Medicine (N. Hamada and Y. Nakamura).
This study was supported by grants from Japan Science and Technology Agency Core Research for Evolutionary Science and Technology to K. Kaibuchi, Grants-in-Aid for Scientific Research to K. Kaibuchi (20227006) and T. Watanabe (20790225), Global Centers of Excellence to K. Kaibuchi, and Special Coordination Funds for Promoting Science and Technology to T. Watanabe.
The authors declare no competing financial interests.
References
- ANOVA
analysis of variance
- EB
end-binding protein
- EMTB
ensconsin MT-binding domain
- FL
full length
- GMPCPP
guanosine-5'-[(α,β)-methyleno]triphosphate
- HSD
honest significant difference
- IGL
internal granular layer
- MAP
MT-associated protein
- MCAK
mitotic centromere–associated kinesin
- mGFP
monomeric GFP
- ML
molecular layer
- MT
microtubule
- SCA11
spinocerebellar ataxia type 11
- +TIP
plus end–tracking protein
- TTBK
tau-tubulin kinase
- WT
wild type
Author notes
T. Watanabe and M. Kakeno contributed equally to this paper.
T. Watanabe’s present address is Dept. of Pharmacology, University of North Carolina at Chapel Hill, School of Medicine, Chapel Hill, NC 27599.


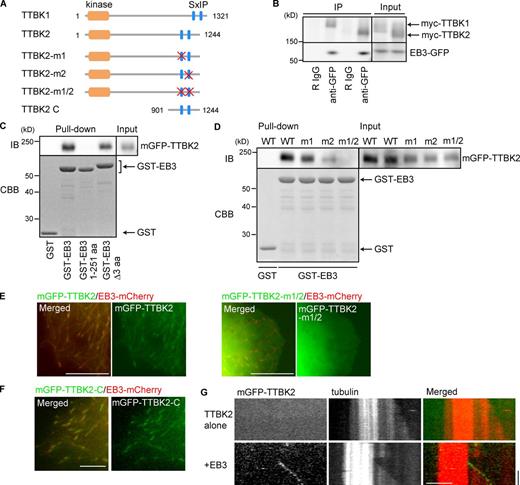
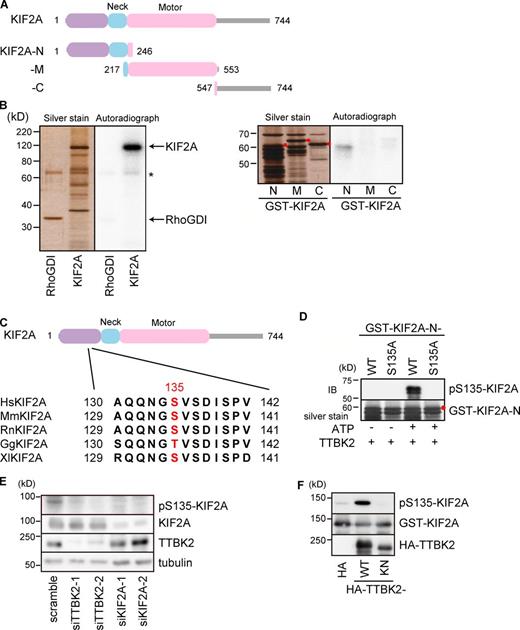
![Figure 3. TTBK2-mediated phosphorylation inactivates KIF2A by inhibiting its binding to MTs. (A) The ability of nonphosphorylated and phosphorylated KIF2A to depolymerize GMCPP-MTs. Increasing concentrations of KIF2A were incubated with GMPCPP-stabilized MTs. The supernatant (S) and precipitation (P) fractions were visualized by Coomassie Brilliant blue staining. Phosphorylation by TTBK2 decreased the MT-depolymerizing activity of KIF2A. The plot shows the quantification of remnant MTs in the precipitation fraction as the mean ± SD of three independent experiments. *, P < 0.05 versus nonphosphorylated KIF2A (Student’s t test). (B) Visualization of GMPCPP-MTs using an antitubulin antibody after brief incubation with KIF2A. Bar, 10 µm. (C) The association between KIF2A and GMPCPP/taxol-stabilized MTs. Increasing concentrations of MTs were incubated with a fixed concentration of KIF2A, followed by ultracentrifugation. Bound (precipitation fraction [Ppt]) and unbound (supernatant fraction [Sup]) KIF2A were visualized by immunoblot analysis. The plot shows the amount of KIF2A bound to MTs relative to the total amount of KIF2A as the mean ± SD of three independent experiments. **, P < 0.01 versus nonphosphorylated KIF2A (Student’s t test).](https://cdn.rupress.org/rup/content_public/journal/jcb/210/5/10.1083_jcb.201412075/7/m_jcb_201412075_fig3.jpeg?Expires=1772603788&Signature=QvlrpLnBmYMj94Z1UIjR9XpsFsWNOKPkGK0NhsDxln8Yg3PAhNPu87YQGFlXe4vMdMKSKb~O-v6fh8BXVobbKzINL9Z-vkCxSv9hRgyQt36XABGkXDiIAE6mdkc-AW1Mce0Yykt6P5fD38BTO0K~f8wxHCkjTjTLHErT3wwcw4CwfvtUv1O91vaOkk8FrEQDD-Mpt0dHdxZWdUVYZbF~zgQEC92CXwzlTuC7EfHaEf70e-NZFczd8numm5EdkfuaNSFO0TJUZdYwU6yrcYSet74txU0cUyk2TQqpfINDQlipTvZsCZvt-RqKHJT6HJcP943g6CX5YveNI4Z9COVZng__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. EB3 changes the autoinhibitory conformation of TTBK2. (A) Schematic diagram of TTBK2. The domain organization of TTBK2 and its fragments is presented with a summary of their ability to bind to TTBK2-cat. (B) COS-7 cells expressing mGFP–TTBK2-FL were lysed and incubated with the indicated GST-fused proteins. mGFP–TTBK2-FL specifically coprecipitated with GST–TTBK2-cat. CBB, Coomassie Brilliant blue staining. (C) The TTBK2 fragments were expressed in COS-7 cells, followed by pull-down. The TTBK2 fragments containing the C-terminal region were coprecipitated with GST–TTBK2-cat. Asterisks indicate the intact bands of the TTBK2 fragments. (D) HeLa cells expressing GFP–TTBK2-FL or -C were fixed and stained with the indicated antibodies. TTBK2-C end-tracked more efficiently than TTBK2-FL. The GFP intensity at the MT plus ends was measured by averaging the peak intensities of >20 GFP-TTBK2 comets for each cell and then plotting against the mean GFP intensity in the whole cell. Each data point represents an individual cell (≥25 cells for each condition). The boxes in the left panels are enlarged in the right panels. Bars, 10 µm. A.U., arbitrary unit. (E) mGFP–TTBK2-C was expressed in COS-7 cells together with control mCherry or mCherry–TTBK2-cat. mGFP–TTBK2-C failed to exhibit localized distribution at MT plus ends in ∼40% of cells expressing mCherry–TTBK2-cat. The graph shows the quantitative results. We counted the numbers of cells exhibiting the localized distribution of the indicated mGFP construct in the presence of mCherry or mCherry–TTBK2-cat. Error bars indicate the SEM. ***, P < 0.001 versus the respective control (Student’s t test). All of the results are representative of more than four independent experiments. The boxes in the left panels are enlarged in the rightmost panels. Bars: (left) 20 µm; (magnified images) 10 µm. (F) TTBK2-C or -C-m1/2 was expressed in COS-7 cells, and the lysates were subjected to pull-down using GST–TTBK2-cat in the presence of the control RhoGDI or EB3. Excess EB3 inhibited the association between TTBK2-cat and -C in a dose-dependent manner. The graph shows the relative amounts of bound TTBK2-C from more than three independent experiments as a box plot. ***, P < 0.001 (one-way analysis of variance [ANOVA], Tukey's test). IB, immunoblotting.](https://cdn.rupress.org/rup/content_public/journal/jcb/210/5/10.1083_jcb.201412075/7/m_jcb_201412075_fig4.jpeg?Expires=1772603788&Signature=qgEG4NkDtxtzA5VT4qShAjzAnMXha0qcE26W2iyzz2Cw9-vmFT3X8-v9eL8~Uv4fOYnJyk-uJa0fVNMQuzyOp8D92BMoiimbj1JtE2mEBq6KTQ3HMN0RtGtKOMu0H3ATAcWvBPgMRsVueqHbY9Jc8bUSVIHcSvCNzaf55NZKwHGo1DKO4Ql0H4y~v3lQRV6vIgyoK22p2F5-z7HSPF-XZyC~OKiYR4OjYfetINUSZCqp~JNU1qvC1NLGJIhRw8XhQO6A53hph9IgcjgfV56Y306yvh~wFplTJQYVG-Ajc-gHxV~ZhZ-m8ENqLkqgGq0tRHqL1P~mJjPPDYICzheodw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 5. TTBK2 phosphorylates KIF2A and removes it from MTs in an EB-dependent manner. (A) The distribution of KIF2A in HeLa cells transfected with scramble or TTBK2 siRNA. TTBK2 depletion increased KIF2A localization to MTs. (B) Rescue experiments for the distribution of KIF2A using resTTBK2-WT and its mutants. The expression of resTTBK2-WT canceled the extensive KIF2A binding to MTs, but its m1/2 and KN mutants did not. (C) Quantification of the colocalization between GFP-KIF2A and MTs. A correlation coefficient (Rr) was calculated for the region within 10 µm of the cell edge (see Calculation of the correlation coefficient in Materials and methods). TTBK2 depletion significantly increased the correlation of KIF2A with MTs. This increase in the correlation caused by TTBK2 depletion was partially rescued by resTTBK2-WT but not by resTTBK2-m1/2 or -KN mutants. Error bars indicate the SD (≥30 cells were analyzed for each condition). ***, P < 0.001 (one-way ANOVA, Tukey’s honest significant difference test [HSD]). (D) COS-7 cells were transfected with control HA or the indicated HA-TTBK2 mutants. The expression of TTBK2-WT increased the phosphorylation of KIF2A at S135, but that of TTBK2-m1/2 or -KN did not. The graph shows the relative band intensity (pS135-KIF2A/KIF2A) as the mean ± SD of three independent experiments. **, P < 0.01 versus control (one-way ANOVA, Tukey’s HSD). (E) Live imaging of HeLa cells expressing GFP-KIF2A and EB3-mRuby. The boxed regions were magnified and are shown as a time series. TTBK2 depletion altered the distribution of KIF2A. Arrowheads indicate the EB3-containing MT ends. The time is given in seconds. Also see Videos 1 and 2. (F) The intensity profiles of GFP-KIF2A and EB3-mRuby along growing MTs in living cells. Cells with mean GFP intensities of 6.0–8.5 A.U. were selected for analysis under both conditions. TTBK2 depletion increased the binding of KIF2A to MTs, especially at the EB3-containing MT ends. Error bars indicate the SEM (>50 MTs of five cells were analyzed for each condition). The horizontal axis was adjusted to 1.0 at the site of peak EB3 intensity. (G) The subcellular distribution of KIF2A-WT and its nonphosphorylatable mutant S135A. KIF2A-S135A extensively localized to the MTs in HeLa cells. (A, B, and G) The boxes in the left panels are enlarged in the right panels. (H) The effect of the SCA11-associated TTBK2 mutant on the distribution of KIF2A. The expression of the TTBK2 mutant increased the distribution of KIF2A along MTs. A.U., arbitrary unit. Bars: (A, B, and G; left) 10 µm; (A, B, and G magnified images) 5 µm; (E and H) 5 µm.](https://cdn.rupress.org/rup/content_public/journal/jcb/210/5/10.1083_jcb.201412075/7/m_jcb_201412075_fig5.jpeg?Expires=1772603788&Signature=5J4PwnjlPwYhZn3Mf3X65dEMFhi5PdV8r9HYD10VLMhukGR5pV2P6g8oblmwd1LSsHhWR0Nu3wze-MWVLcUxBX5korLDrowv8Lx7Q~~H1DaEt1wtYhfhHLjcEXWBB1ABgRjBGpU7~E1ADW-NlH8iTL8sa4-KqOBdRlUZEpRB23tk9AzAue~xwCq9jMbqX~K8LjQm~FPCBhVRJ-pOZNfOVP55nv9Fgkq8T4seSqyWaTJ7GkjHDo6EylGWg41k4EHXJxL0hyOuAdX-bo88-WuX-TLLAXU7DoG3wcCNULfergk2VvvE0mkkxj0WBMQKOQ3ozcQ7mFD5TGTlIotZzDp7YQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)




