Microtubules (MTs) and associated motors play a central role in nuclear migration, which is crucial for diverse biological functions including cell division, polarity, and sexual reproduction. In this paper, we report a dual mechanism underlying nuclear congression during fission yeast karyogamy upon mating of haploid cells. Using microfluidic chambers for long-term imaging, we captured the precise timing of nuclear congression and identified two minus end–directed motors operating in parallel in this process. Kinesin-14 Klp2 associated with MTs may cross-link and slide antiparallel MTs emanating from the two nuclei, whereas dynein accumulating at spindle pole bodies (SPBs) may pull MTs nucleated from the opposite SPB. Klp2-dependent nuclear congression proceeds at constant speed, whereas dynein accumulation results in an increase of nuclear velocity over time. Surprisingly, the light intermediate chain Dli1, but not dynactin, is required for this previously unknown function of dynein. We conclude that efficient nuclear congression depends on the cooperation of two minus end–directed motors.
Introduction
Controlling nuclear positioning within cells is crucial for many cellular processes including cell division, polarity, and motility in unicellular and multicellular organisms. Pronuclear migration, during which male and female pronuclei migrate toward the center of the zygote, is a key step for sexual reproduction and embryogenesis (Reinsch and Gönczy, 1998).
In zygotes of most species, including human, a microtubule (MT) aster is nucleated from the centrosome associated with the male pronucleus, and migration is dependent on the minus end–directed motor dynein (Clift and Schuh, 2013). Dynein accumulates on the female pronuclear envelope mediating translocation toward the aster center. Simultaneously, the male pronucleus is pulled to the cell center in a MT length– and dynein-dependent manner (Kimura and Onami, 2005; Wühr et al., 2010).
In yeast, nuclear congression is observed after fusion of two haploid cells and requires another minus end–directed motor, kinesin-14 Kar3 (Meluh and Rose, 1990). Its localization along MTs proposes that Kar3 pulls nuclei together either by sliding overlapping antiparallel MTs nucleated from spindle pole bodies (SPBs; Meluh and Rose, 1990) or by cross-linking depolymerizing MT plus ends (Molk et al., 2006). A recent study supports a model whereby pulling forces are generated by SPB-anchored Kar3 (Gibeaux et al., 2013).
Distinct MT-dependent mechanisms underlying nuclear movements have been characterized in the fission yeast Schizosaccharomyces pombe. In interphase, first, the nucleus is tightly associated with MT minus ends on multiple sites along the nuclear envelope including the SPB, whereas dynamic MT plus ends grow toward the cell tips, where they exert, upon growth against the cortex, repulsive pushing forces centering the nucleus (Drummond and Cross, 2000; Tran et al., 2001). Organization of interphase MT bundles partly depends on kinesin-14 Klp2, which mediates sliding of MTs of opposite polarity relative to each other and groups MT minus ends at the cell center (Carazo-Salas et al., 2005; Janson et al., 2007). Moreover, Klp2 dissociates from MTs at the end of mitosis to prevent Klp2-dependent clustering of daughter cell nuclei in the cell middle that occurs in telophase, when Klp2 dissociation from MTs is impaired (Mana-Capelli et al., 2012). This demonstrates that Klp2 can directly generate forces to translocate nuclei.
Second, during fission yeast meiosis, the diploid nuclei undergo long-range oscillations known as nuclear horsetail movement ensuring proper efficiency of homologous recombination between paired chromosomes (Chikashige et al., 1994; Yamamoto et al., 1999). These movements are driven by pulling forces exerted on SPB-bound MTs by dynein cortically anchored via Num1 (Yamamoto et al., 2001; Saito et al., 2006; Yamashita and Yamamoto, 2006; Vogel et al., 2009).
Dynein forms a multiprotein complex consisting of two heavy chains and several subunits, intermediate, light intermediate, and light chains, and interacts with additional regulatory protein complexes such as the dynactin complex. These interactions are thought to target dynein to subcellular structures and cargos and to regulate dynein motor activity (Kardon and Vale, 2009; Vallee et al., 2012; Roberts et al., 2013). In fission yeast, the intermediate chain Dic1, the light intermediate chain Dli1 and the dynactin component Ssm4, homologous to Glued, are all absolutely required to generate dynein-dependent pulling forces during horsetail nuclear movement because dynein localization to MTs is disrupted in their absence (Niccoli et al., 2004; Fujita et al., 2010).
Finally, during mating, nuclear congression preceding nuclear fusion and horsetail movement is MT dependent (Yamashita et al., 2013; Polakova et al., 2014). Studies in fixed fission yeast cells revealed that zygotes lacking both, Klp2 and dynein, often contained two nuclei, compared with only one in wild-type zygotes. The two nuclei proceeded to nuclear horsetail movement, suggesting that the process of nuclear congression or fusion had failed (Troxell et al., 2001). However, to date, no study has been performed to investigate the molecular mechanisms underlying nuclear congression in live cells.
Here, we developed a microfluidic-based device for long-term imaging of mating fission yeast cells to capture precise timing of nuclear congression and perform a systematic screen through all MT-dependent motors to test their role in this process. We demonstrate that nuclear congression is mediated by two minus end–directed motors, dynein and Klp2, and provide evidence that these motors work in parallel mechanisms at distinct subcellular structures to promote efficient nuclear congression.
Results
Klp2 and dynein mediate nuclear congression
We fabricated micrometer-scaled channels based on a combination of soft-lithography and microfluidics technology (Velve-Casquillas et al., 2010) permitting long-term imaging of mating cells (Fig. S1, A and B). Both mating partners expressed Cut11-GFP to visualize the nuclear envelope (West et al., 1998), and one partner also expressed GFP-Atb2 to label MTs (Fig. 1 A). Nuclear congression was defined as the period between cell fusion marked by diffusion of cytoplasmic GFP-labeled tubulin into the unmarked partner and nuclear contact, i.e., congression time and was achieved on average within 29 ± 7 min in wild-type zygotes (Figs. 1 A and S1 C).
Klp2 and dynein perform nuclear congression in a parallel manner. (A) Time-lapse images recorded by spinning-disk confocal microscopy of cells expressing Cut11-GFP (nuclear envelope) and unilaterally GFP-Atb2 (MTs) in wild-type cells at 25°C undergoing nuclear congression. Images are presented as maximum projections of 3D stacks. Dotted lines represent cell outlines. Yellow arrowheads highlight the MT bundle formed between two nuclei. Nuclear congression was defined as the duration between cell fusion (t0) and contact between nuclei. (B) Box plot shows the time of nuclear congression in wild type and single motor mutants at 25°C or 36°C. Mean values for strains tested at 25°C: wild type (wt; 29 ± 7 min, n = 104), klp2Δ (48 ± 13 min, P < 10−22, n = 84), klp3Δ (31 ± 10 min, P = 0.06, n = 76), tea2Δ (42 ± 15 min, P < 10−11, n = 94), klp5Δ (28 ± 9 min, P = 0.38, n = 84), klp6Δ (29 ± 8 min, P = 0.63, n = 78), klp8Δ (29 ± 9 min, P = 0.89, n = 60), klp9Δ (27 ± 9 min, P = 0.39, n = 69), pkl1Δ (27 ± 8 min, P = 0.24, n = 75), and dhc1Δ (34 ± 9 min, P < 10−4, n = 87). Mean values for strains tested at 36°C: wild type (26 ± 11 min, n = 54), cut7-24 (24 ± 10 min, P = 0.29, n = 89), and klp2Δ (42 ± 22 min, P < 10−4, n = 44). (C) Box plot shows the time of nuclear congression klp2Δ double motor mutants at 25°C or 36°C. Mean values for strains tested at 25°C (p-value against klp2Δ): klp2Δ klp3Δ (45 ± 13 min, P = 2, n = 75), klp2Δ tea2Δ (54 ± 16 min, P = 0.015, n = 49), klp2Δ klp5Δ (44 ± 10 min, P = 0.07, n = 73), klp2Δ klp6Δ (44 ± 13 min, P = 0.07, n = 69), klp2Δ klp8Δ (50 ± 12 min, P = 0.26, n = 58), klp2Δ klp9Δ (47 ± 13 min, P = 0.68, n = 74), klp2Δ pkl1Δ (46 ± 13 min, P = 0.31, n = 74), klp2Δ dhc1Δ (155 ± 14 min, n = 2), and dhc1Δ tea2Δ (38 ± 14 min, n = 67). Mean values for strains tested at 36°C: klp2Δ cut7-24 (46 ± 14 min, P = 0.33, n = 60). For the box plots, each box encloses 50% of the data with the median values displayed as lines. The top and bottom of each box mark the minimum and maximum values within the dataset that fall within an acceptable range. Any value outside of this range, called an outlier, is displayed as an individual point. (D–F) Time-lapse images of mating cells expressing Cut11-GFP and unilaterally GFP-Atb2 in klp2Δ (D), dhc1Δ (E), or klp2Δ dhc1Δ (F) strains at 25°C. (G) Percentage of zygotes completing nuclear congression in wild type (100%, n = 104), klp2Δ (100%, n = 84), dhc1Δ (100%, n = 87), and klp2Δ dhc1Δ (2%, n = 85). Bars, 5 µm. *, P < 0.01. n were collected from 2–3 independent experiments.
Klp2 and dynein perform nuclear congression in a parallel manner. (A) Time-lapse images recorded by spinning-disk confocal microscopy of cells expressing Cut11-GFP (nuclear envelope) and unilaterally GFP-Atb2 (MTs) in wild-type cells at 25°C undergoing nuclear congression. Images are presented as maximum projections of 3D stacks. Dotted lines represent cell outlines. Yellow arrowheads highlight the MT bundle formed between two nuclei. Nuclear congression was defined as the duration between cell fusion (t0) and contact between nuclei. (B) Box plot shows the time of nuclear congression in wild type and single motor mutants at 25°C or 36°C. Mean values for strains tested at 25°C: wild type (wt; 29 ± 7 min, n = 104), klp2Δ (48 ± 13 min, P < 10−22, n = 84), klp3Δ (31 ± 10 min, P = 0.06, n = 76), tea2Δ (42 ± 15 min, P < 10−11, n = 94), klp5Δ (28 ± 9 min, P = 0.38, n = 84), klp6Δ (29 ± 8 min, P = 0.63, n = 78), klp8Δ (29 ± 9 min, P = 0.89, n = 60), klp9Δ (27 ± 9 min, P = 0.39, n = 69), pkl1Δ (27 ± 8 min, P = 0.24, n = 75), and dhc1Δ (34 ± 9 min, P < 10−4, n = 87). Mean values for strains tested at 36°C: wild type (26 ± 11 min, n = 54), cut7-24 (24 ± 10 min, P = 0.29, n = 89), and klp2Δ (42 ± 22 min, P < 10−4, n = 44). (C) Box plot shows the time of nuclear congression klp2Δ double motor mutants at 25°C or 36°C. Mean values for strains tested at 25°C (p-value against klp2Δ): klp2Δ klp3Δ (45 ± 13 min, P = 2, n = 75), klp2Δ tea2Δ (54 ± 16 min, P = 0.015, n = 49), klp2Δ klp5Δ (44 ± 10 min, P = 0.07, n = 73), klp2Δ klp6Δ (44 ± 13 min, P = 0.07, n = 69), klp2Δ klp8Δ (50 ± 12 min, P = 0.26, n = 58), klp2Δ klp9Δ (47 ± 13 min, P = 0.68, n = 74), klp2Δ pkl1Δ (46 ± 13 min, P = 0.31, n = 74), klp2Δ dhc1Δ (155 ± 14 min, n = 2), and dhc1Δ tea2Δ (38 ± 14 min, n = 67). Mean values for strains tested at 36°C: klp2Δ cut7-24 (46 ± 14 min, P = 0.33, n = 60). For the box plots, each box encloses 50% of the data with the median values displayed as lines. The top and bottom of each box mark the minimum and maximum values within the dataset that fall within an acceptable range. Any value outside of this range, called an outlier, is displayed as an individual point. (D–F) Time-lapse images of mating cells expressing Cut11-GFP and unilaterally GFP-Atb2 in klp2Δ (D), dhc1Δ (E), or klp2Δ dhc1Δ (F) strains at 25°C. (G) Percentage of zygotes completing nuclear congression in wild type (100%, n = 104), klp2Δ (100%, n = 84), dhc1Δ (100%, n = 87), and klp2Δ dhc1Δ (2%, n = 85). Bars, 5 µm. *, P < 0.01. n were collected from 2–3 independent experiments.
To identify the MT-associated motors involved in nuclear congression, we systematically deleted or inactivated the nine kinesins and the single dynein (heavy chain dhc1) of the fission yeast genome. Nuclear congression was significantly delayed in klp2Δ, dhc1Δ, and tea2Δ zygotes compared with wild type (Fig. 1 B). klp2Δ zygotes (Fig. 1 D) exhibited the greatest delay (20 min delay; 48 min congression time), whereas the delay in dhc1Δ zygotes (Fig. 1 E) was the mildest (5 min delay; 34 min congression time), indicating that Klp2 contributes more to nuclear congression than dynein. We noted that dhc1 deletion also slowed down nuclear fusion after nuclear contact, suggesting that dynein might be particularly important during late stages of karyogamy (Fig. S1 D). Despite a delay, nuclear congression was always successfully completed in all single mutants (Figs. 1 G and S1 E), indicating that these motors may operate in parallel to promote nuclear congression. Therefore, we combined klp2 deletion with deletions of other motors (Figs. 1 C and S1 E). Strikingly, klp2Δ dhc1Δ double mutant cells failed almost completely to bring their nuclei together (Fig. 1, F and G), confirming the observations made by Troxell et al. (2001): out of 87 mating events, nuclei were observed to touch each other in two cases only after a long delay (Fig. 1 C). This suggested that these two motors directly contribute to force generation required for nuclear congression in a parallel manner or, alternatively, may be involved in the organization of the MT array specific for nuclear congression.
We tested this by examining MT organization more closely. We observed that, before and immediately after cell fusion, wild-type cells mainly contained linear MT bundles attached to the nuclear envelope at multiple sites as typically seen during interphase (Fig. 1 A, −5 to 10 min). MTs eventually organized into a radial array exclusively nucleated from the SPB (Ding et al., 1998), and a bright bundle between the two nuclei was often observed, created by overlapping antiparallel MTs emanating from the two SPBs (Fig. 1 A, yellow arrowhead). This MT bundle was still established in the absence of one or both motors (Fig. 1, D–F, yellow arrowheads), indicating that the failure in nuclear congression in klp2Δ dhc1Δ zygotes does probably not result from an aberrant MT array but rather from a lack of MT-dependent forces.
Nuclear congression was also delayed upon deletion of the plus end–directed kinesin tea2 (13 min delay; 42 min congression time), but unlike klp2Δ and dhc1Δ, tea2 deletion was not synergistic with klp2 or dhc1 deletion. These results suggest that Tea2 only plays an indirect role, most likely as a result of its function as a plus tip–interacting protein in the control of MT dynamics, possibly by transporting Tip1, the CLIP170 homologue, toward MT plus ends (Busch et al., 2004). Consistently, tip1Δ zygotes characterized by shorter interphase MTs (Brunner and Nurse, 2000) mimicked the tea2Δ phenotype and exhibited a similar delay in nuclear congression (13 min delay; 42 min congression time; Fig. S1 F), suggesting that proper MT dynamics facilitate nuclear congression, but neither Tea2 nor Tip1 is essential for this process.
Distinct roles for Klp2 and dynein in nuclear congression
Next, we examined the kinetics of nuclear congression in wild-type and mutant zygotes by monitoring the migration of the SPB at 1-min intervals before and after cell fusion until SPBs were juxtaposed (hereafter referred to as SPB fusion), using Sfi1-GFP as a marker (Almonacid et al., 2011). Distances between the two SPBs over time were measured from kymographs (Fig. 2, A and B). Strikingly, in most wild-type cells (80%), nuclei usually started moving immediately upon cell fusion (t0), slowly at the beginning, but migration speeded up toward the end of SPB congression (Fig. 2 B, dark green line). The distance over time plot could be fitted with good accuracy by a second degree polynomial function (Fig. 2 B, black line) whose derivative gives the instantaneous velocity over time. This revealed that, the velocity of the two approaching SPBs increased over time (Fig. 2 B, light green line). This behavior observed in individual wild-type zygotes could be confirmed on the median curve derived from a population of 25 zygotes (Figs. 2 C and S2 A) including the increase of velocity over time (Fig. 2 H, green curve; and Fig. S2 B) in the majority of wild-type zygotes. In the remaining 20% of events, SPB congression proceeded at a rather constant speed.
Klp2 constantly generates pulling forces, whereas dynein contribution increases during nuclear congression. (A, top) Overlay of differential interference contrast and fluorescence images of the SPB marker Sfi1-GFP shown for an individual zygote at 10 min before cell fusion (defined by cytoplasmic diffusion of mCherry-Atb2) until completion of nuclear congression at 26 min after cell fusion. Sfi1-GFP images were taken as 3D stacks at 1-min intervals. Dotted lines represent cell outlines. Dotted white line represents the line used to make the kymograph. (bottom) Kymograph representing Sfi1-GFP dynamics from −10 min until SPBs are juxtaposed. Yellow dotted lines indicate moment of cell fusion (t0). (B) SPB distance and velocity over time from individual example shown in A. Velocity obtained from a differential function of a second degree polynomial (black line) of SPB distances. (C) SPB distances over time normalized for each cell to mean SPB distance before cell fusion calculated from first 10 time points. Individual cells are depicted as light green trajectories (n = 25). Median is represented as a thick line, and upper and lower quartiles are shown as dotted lines. (D, left) Kymograph of Sfi1-GFP dynamics in an individual klp2Δ zygote. (right) SPB distances and velocity over time. Black line represents second degree polynomial function as approximation for SPB distance. (E) Sfi1-GFP dynamics in a subset of dhc1Δ zygotes (12/25 cells) depicted as kymograph (top) or as a function over time (bottom). Velocity obtained from a differential function of a linear regression (black line) of SPB distances. (F) Sfi1-GFP dynamics in a second subset of dhc1Δ zygotes (13/25 cells). Initial period without directed nuclear movement is highlighted. Velocity obtained from a differential function of a linear regression (black line) of SPB distances after initial period. (G and H) Median curves of normalized SPB distances (G) or velocities (H) over time in wild-type, klp2Δ, and a subset of dhc1Δ zygotes. Bars, 5 µm. n were collected from two independent experiments.
Klp2 constantly generates pulling forces, whereas dynein contribution increases during nuclear congression. (A, top) Overlay of differential interference contrast and fluorescence images of the SPB marker Sfi1-GFP shown for an individual zygote at 10 min before cell fusion (defined by cytoplasmic diffusion of mCherry-Atb2) until completion of nuclear congression at 26 min after cell fusion. Sfi1-GFP images were taken as 3D stacks at 1-min intervals. Dotted lines represent cell outlines. Dotted white line represents the line used to make the kymograph. (bottom) Kymograph representing Sfi1-GFP dynamics from −10 min until SPBs are juxtaposed. Yellow dotted lines indicate moment of cell fusion (t0). (B) SPB distance and velocity over time from individual example shown in A. Velocity obtained from a differential function of a second degree polynomial (black line) of SPB distances. (C) SPB distances over time normalized for each cell to mean SPB distance before cell fusion calculated from first 10 time points. Individual cells are depicted as light green trajectories (n = 25). Median is represented as a thick line, and upper and lower quartiles are shown as dotted lines. (D, left) Kymograph of Sfi1-GFP dynamics in an individual klp2Δ zygote. (right) SPB distances and velocity over time. Black line represents second degree polynomial function as approximation for SPB distance. (E) Sfi1-GFP dynamics in a subset of dhc1Δ zygotes (12/25 cells) depicted as kymograph (top) or as a function over time (bottom). Velocity obtained from a differential function of a linear regression (black line) of SPB distances. (F) Sfi1-GFP dynamics in a second subset of dhc1Δ zygotes (13/25 cells). Initial period without directed nuclear movement is highlighted. Velocity obtained from a differential function of a linear regression (black line) of SPB distances after initial period. (G and H) Median curves of normalized SPB distances (G) or velocities (H) over time in wild-type, klp2Δ, and a subset of dhc1Δ zygotes. Bars, 5 µm. n were collected from two independent experiments.
We performed the same analysis in klp2Δ zygotes, in which nuclear congression depends mainly on dynein. In 92% of klp2Δ zygotes, despite a delay, the two nuclei migrated toward each other in a similar fashion as in wild-type zygotes, slowly at the beginning but with an increasing speed over time, as shown for an individual cell (Fig. 2 D) or on the median curve for all monitored examples (Fig. 2 G, red curve; and Fig. S2 C). Consistently, velocities clearly increased over time, albeit at a lower rate in the absence of Klp2-generated forces compared with wild type (Fig. 2, D and H; and Fig. S2 D).
The behavior of the approaching nuclei was most strikingly altered in dhc1Δ zygotes, where nuclear congression was mostly driven by Klp2. In about half of the observed events, distances between the two SPBs decreased in a linear fashion upon cell fusion as shown for an individual zygote (Fig. 2 E) or plotted as a median graph for this subset (12 of 25 cells) of dhc1Δ zygotes (Fig. 2 G, orange curve; and Fig. S2 E). This behavior can be best described by a linear regression (Fig. 2 E, black line) instead of a second degree polynomial (Fig. S2 F), with an approximately constant speed of the moving nuclei (Fig. 2 H, orange curve; and Fig. S2 H).
In 10 other dhc1Δ zygotes (36%), an initial phase with no directed nuclear movement preceded the migration of nuclei toward each other (Fig. 2 F). Congression then happened at a constant velocity, like in the first population of dynein-deficient zygotes (Fig. 2 F), although at a higher speed (Fig. S2 H). As the period between cell fusion and initiation of nuclear congression varied between individual cells, a median curve did not accurately represent the behavior of individual cells (Fig. S2 G). Therefore, we depicted the SPB distances and velocities over time only for the first dhc1Δ subset of mating cells representing typical features of dhc1Δ nuclear congression in comparison to wild type and klp2Δ (Fig. 2, G and H). Nuclear congression in the remaining three zygotes was characterized by an increase in velocity.
Collectively, these observations suggest that Klp2 exerts forces on the nuclei in a rather constant fashion, whereas the contribution of dynein appears to increase toward the end of this process. Based on these observations, we propose that these two motors function in distinct, parallel pathways.
Additionally, we confirmed that a double klp2Δ dhc1Δ mutant (n = 10) failed to congress their nuclei and SPBs (Fig. S2, L and M). However, we observed that some nuclei managed to migrate toward each other to some extent in double klp2Δ dhc1Δ zygotes, without completing nuclear congression. We hypothesized that this observation may be a result of pushing forces exerted by growing MTs against the cortex (Tran et al., 2001). Accordingly, nuclear migrations toward each other were slightly dampened in triple klp2Δ dhc1Δ tea2Δ zygotes that exhibit aberrant MT dynamics (Fig. S2, L and N). This indicates that MT pushing forces exerted by growing MTs against the cortex may also contribute to nuclear congression, although they alone are not sufficient to bring nuclei together.
Klp2 localization at MT plus ends and along MTs
To better understand the role of Klp2 and dynein, we defined their localization in mating cells. Consistent with the distinct phenotype of their deletion mutants, Klp2 and dynein showed distinct localization patterns during nuclear congression.
Klp2 located at MT plus ends in a punctuated manner and more weakly along the lattice of MTs during karyogamy (Fig. 3 A), similar to what was observed during interphase in vegetative cells (Carazo-Salas et al., 2005; Janson et al., 2007). Klp2 localization to the MT plus ends was unaffected in mating dhc1Δ cells (Fig. S3 A), confirming that both motors function independently from one another. These observations suggest that Klp2 contributes to nuclear congression by mediating interactions between MTs.
Distinct localization pattern of Klp2 and dynein during nuclear congression. (A) Cellular distribution of Klp2-GFP in mating cells coexpressing Cut11-mCherry and mCherry-Atb2 at 25°C. Images are represented as maximum z projection of 3D stacks. Dotted lines represent outlines of cells in different stages of meiotic prophase. (B) Cellular distribution of dynein (Dhc1-3×GFP) in mating cells coexpressing Cut11-mCherry and mCherry-Atb2 at 25°C. (C) Colocalization of Dhc1-3×GFP and Sfi1-RFP. Insets show magnified regions of the boxed area. (D) Time-lapse images of Dhc1-3×GFP in mating cells coexpressing Cut11-mCherry spanning time point of initial detection of dynein (−35 min) in one cell until SPB fusion (t = 0). (E) SPB intensities of Dhc1-3×GFP in individual cells depicted in D over time until completion of nuclear congression (SPB fusion; t0). Line represents the moving average best fit to the data points. (F) Box plot shows median time of initial detection of dynein in mating cells before SPB fusion in wild-type and klp2Δ zygotes. Mean values for wild type (wt; 31 ± 29 min) and klp2Δ (49 ± 28 min, P < 10−5) for n = 100 SPBs. For the box plots, each box encloses 50% of the data with the median values displayed as lines. The top and bottom of each box mark the minimum and maximum values within the dataset that fall within an acceptable range. Any value outside of this range, called an outlier, is displayed as an individual point. (G) Mean SPB intensities of Dhc1-3×GFP averaged for a cell population (n = 100 SPBs) from 15 min (for wild type) or 35 min (for klp2Δ) before SPB fusion. Error bars: SD. Dotted line highlights similar dynein level between wild-type and klp2Δ zygotes. n were collected from three independent experiments. a.u., arbitrary unit. Bars: (main images) 5 µm; (insets) 1 µm.
Distinct localization pattern of Klp2 and dynein during nuclear congression. (A) Cellular distribution of Klp2-GFP in mating cells coexpressing Cut11-mCherry and mCherry-Atb2 at 25°C. Images are represented as maximum z projection of 3D stacks. Dotted lines represent outlines of cells in different stages of meiotic prophase. (B) Cellular distribution of dynein (Dhc1-3×GFP) in mating cells coexpressing Cut11-mCherry and mCherry-Atb2 at 25°C. (C) Colocalization of Dhc1-3×GFP and Sfi1-RFP. Insets show magnified regions of the boxed area. (D) Time-lapse images of Dhc1-3×GFP in mating cells coexpressing Cut11-mCherry spanning time point of initial detection of dynein (−35 min) in one cell until SPB fusion (t = 0). (E) SPB intensities of Dhc1-3×GFP in individual cells depicted in D over time until completion of nuclear congression (SPB fusion; t0). Line represents the moving average best fit to the data points. (F) Box plot shows median time of initial detection of dynein in mating cells before SPB fusion in wild-type and klp2Δ zygotes. Mean values for wild type (wt; 31 ± 29 min) and klp2Δ (49 ± 28 min, P < 10−5) for n = 100 SPBs. For the box plots, each box encloses 50% of the data with the median values displayed as lines. The top and bottom of each box mark the minimum and maximum values within the dataset that fall within an acceptable range. Any value outside of this range, called an outlier, is displayed as an individual point. (G) Mean SPB intensities of Dhc1-3×GFP averaged for a cell population (n = 100 SPBs) from 15 min (for wild type) or 35 min (for klp2Δ) before SPB fusion. Error bars: SD. Dotted line highlights similar dynein level between wild-type and klp2Δ zygotes. n were collected from three independent experiments. a.u., arbitrary unit. Bars: (main images) 5 µm; (insets) 1 µm.
Because recent work showed that SPB-anchored Kar3 exerts pulling forces on MTs emanating from the second SPB (Gibeaux et al., 2013), we also tested whether Klp2 is recruited to the SPB in fission yeast. In wild-type cells, the complexity of cytoplasmic MT arrays with MT plus ends extending toward the SPB of the mating partner precluded drawing clear conclusions. We therefore imaged Klp2-GFP in cells depleted for the plus tip–interacting protein Mal3 (EB1 homologue) that display short cytoplasmic MTs (Beinhauer et al., 1997) and fail to load Klp2 at MT plus ends (Mana-Capelli et al., 2012). Klp2 was largely absent from these short MTs in this mutant, although a residual staining was nevertheless observed in close proximity to the nucleus and in particular at SPBs in mating cells (Fig. S3 A, yellow arrowheads). Klp2 SPB localization might result from weak Mal3-independent binding to MTs and accumulation at MT minus ends by motility. Alternatively, Klp2 might interact with a SPB component. In any case, this SPB pool was minor compared with the MT-bound one observed in wild-type cells, indicating that to pull nuclei together, Klp2 dominantly generates forces by cross-linking antiparallel MTs.
Dynein recruitment to the SPB during nuclear congression
To study the localization of dynein, we imaged dynein heavy chain tagged with three copies of GFP (Fujita et al., 2010). Dynein expression is up-regulated during meiosis (Miki et al., 2002). Accordingly, no dynein was detected in nonmating cells, but it localized to one or both SPBs in 89% of cells with congressing nuclei as shown in cells coexpressing either the nuclear envelope protein Cut11-mCherry and mCherry-Atb2 (Fig. 3 B) or the SPB marker Sfi1-RFP (Fig. 3 C). This differs sharply from dynein localization during nuclear horsetail movement, where dynein associates with MTs and the cell cortex (Yamamoto et al., 1999). Noteworthy, in some cells (21%), dynein formed multiple dots which may represent the SPB and telocentrosomes (Fig. S3 B, yellow arrowheads; Yoshida et al., 2013). Dynein was also rarely found (7%) along the MT bundle bridging the two nuclei (Fig. S3 B, magenta arrowhead).
A time-lapse study revealed that dynein was recruited to the SPB during cell mating (Fig. 3 D). It was initially detected at SPBs on average 31 ± 29 min before SPB fusion (Fig. 3 F), approximately coinciding with cell fusion, and then gradually increased in intensity until SPB fusion (Fig. 3, E and G). Furthermore, in klp2Δ cells exhibiting a 20-min delay in nuclear congression, dynein was detected 49 min before SPB fusion, 18 min earlier than in wild-type cells (Figs. 3 F and S3, B–D), and its levels at SPBs were on average 1.6-fold higher upon SPB fusion (Fig. 3 G) compared with wild-type zygotes. Consistently, dynein levels 15 min before SPB fusion in wild-type cells were similar to levels at 30–35 min before SPB fusion in klp2Δ cells. This shows that the increase in dynein intensity in klp2Δ cells results from an extension of the period of dynein recruitment (Fig. 3 G). In conclusion, dynein gradually accumulates at the SPB during the course of nuclear congression in wild-type and klp2Δ zygotes and confirms further that the two motors functions independently from one another.
SPB-bound dynein drives nuclear congression and requires Dli1 for proper function
Dynein gradual accumulation at SPBs is consistent with our hypothesis that dynein-dependent forces increase over time and could provide a molecular basis for the speed increase recorded during nuclear congression in most dynein proficient zygotes. If this is true, dynein localization to the SPB is controlling its function during karyogamy. We decided to test this point further and analyzed how dynein targeting to the SPB is controlled.
Previous work showed that Dhc1 N terminus lacking the motor domain (Dhc1(1–1,266)) is sufficient for its recruitment to the SPB (Fig. 4 C; Yoshida et al., 2013), demonstrating that dynein localization to SPBs is not purely caused by an accumulation at MT minus ends by motility. We found that this truncated version in replacement of full-length Dhc1 was not functional in nuclear congression as it delayed nuclear congression similarly to dhc1 deletion in presence and absence of Klp2 (Fig. 4, A and B). Thus, as expected, dynein requires its motor domain to generate forces capable of pulling nuclei together.
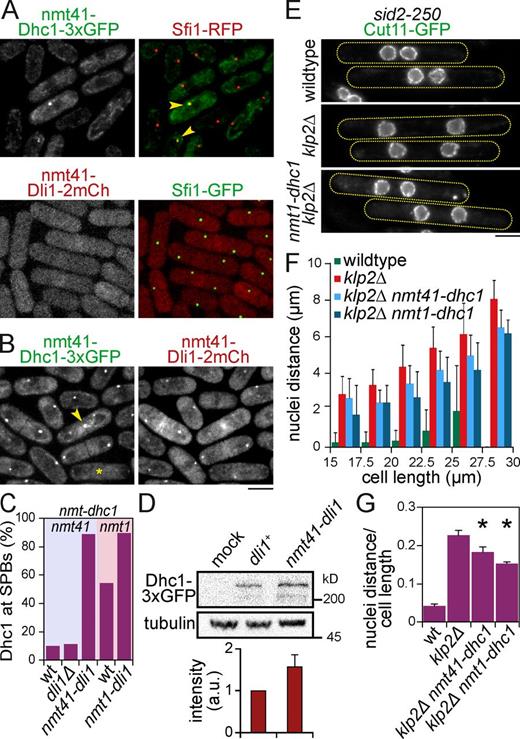
Dynein functions at the SPB during nuclear congression independent of the dynactin complex. (A) Box plot shows time of nuclear congression in wild-type and dynein-related single and klp2Δ double mutants at 25°C. Mean values (p-values against wild type or klp2Δ, respectively): wild type (29 ± 7 min, n = 104), klp2Δ (48 ± 13 min, P < 10−22, n = 84), dhc1Δ (34 ± 9 min, P < 10−4, n = 87), num1Δ (31 ± 9 min, P = 0.09, n = 58), klp2Δ num1Δ (48 ± 16 min, P = 0.85, n = 42), dhc1(1–1,266) (35 ± 10 min, P < 10−4, n = 56), ssm4Δ (29 ± 9 min, P = 0.78, n = 65), klp2Δ ssm4Δ (45 ± 14 min, P = 0.12, n = 96), dli1Δ (33 ± 11 min, P = 0.003, n = 60), klp2Δ dli1Δ (119 ± 65 min, P < 10−6, n = 33/101), dhc1Δ dli1Δ (35 ± 7 min, n = 64), nmt1-dhc1 (28 ± 10 min, P = 0.74, n = 71), and klp2Δ nmt1-dhc1 (40 ± 12 min, P < 10−3, n = 74). (*, P < 0.01, colors indicate strain for comparison.) Note that the highest value of klp2Δ dli1Δ strain is 265 min, beyond the range of the plot. (B) Percentage of zygotes completing nuclear congression in wild type (100%, n = 104), klp2Δ dhc1Δ (2%, n = 85), klp2Δ num1Δ (100%, n = 42), klp2Δ dhc1(1–1266) (2%, n = 56), klp2Δ ssm4Δ (100%, n = 96), and klp2Δ dli1Δ (21%, n = 101). (C) Cellular distribution of N-terminal part of Dhc1-3×GFP in mating dhc1(1–1,266) cells coexpressing Cut11-mCherry and mCherry-Atb2 at 25°C. Images are represented as maximum z projection of 3D stacks. Dotted lines represent outlines of cells in different stages of meiotic prophase. (D) Cellular distribution of Dhc1-3×GFP in mating wild-type, ssm4Δ, dli1Δ, and nmt1-dhc1 cells coexpressing Cut11-mCherry and mCherry-Atb2. (E) Frequency of time points, at which dynein was detected at SPBs relative to SPB fusion in wild-type, ssm4Δ, dli1Δ, and nmt1-dhc1 zygotes (n = 50 cells). Mean values for dynein detection at SPBs relative to SPB fusion: wild type (−36 ± 33 min), ssm4Δ (−32 ± 31 min), dli1Δ (50 ± 26 min, 27/50 events), and nmt1-dhc1 (−62 ± 47 min). (F) Box plot shows SPB intensities of Dhc1-3×GFP at time point of SPB fusion averaged for a cell population of wild-type, ssm4Δ, dli1Δ, and nmt1-dhc1 zygotes (n = 50 SPBs). Mean values for dynein intensities at SPBs at time point of SPB fusion (arbitrary unit [a.u.]): wild type (4,924 ± 2,757), ssm4Δ (3,746 ± 2,160, P = 0.02), dli1Δ (80 ± 277, P < 10−15), and nmt1-dhc1 (13,939 ± 7,979, P < 10−11). (*, P < 0.05; ***, P < 10−4.) Bars, 5 µm. n were collected from 2–3 independent experiments. wt, wild type. For box plots, each box encloses 50% of the data with the median values displayed as lines. The top and bottom of each box mark the minimum and maximum values within the dataset that fall within an acceptable range. Any value outside of this range, called an outlier, is displayed as an individual point.
Dynein functions at the SPB during nuclear congression independent of the dynactin complex. (A) Box plot shows time of nuclear congression in wild-type and dynein-related single and klp2Δ double mutants at 25°C. Mean values (p-values against wild type or klp2Δ, respectively): wild type (29 ± 7 min, n = 104), klp2Δ (48 ± 13 min, P < 10−22, n = 84), dhc1Δ (34 ± 9 min, P < 10−4, n = 87), num1Δ (31 ± 9 min, P = 0.09, n = 58), klp2Δ num1Δ (48 ± 16 min, P = 0.85, n = 42), dhc1(1–1,266) (35 ± 10 min, P < 10−4, n = 56), ssm4Δ (29 ± 9 min, P = 0.78, n = 65), klp2Δ ssm4Δ (45 ± 14 min, P = 0.12, n = 96), dli1Δ (33 ± 11 min, P = 0.003, n = 60), klp2Δ dli1Δ (119 ± 65 min, P < 10−6, n = 33/101), dhc1Δ dli1Δ (35 ± 7 min, n = 64), nmt1-dhc1 (28 ± 10 min, P = 0.74, n = 71), and klp2Δ nmt1-dhc1 (40 ± 12 min, P < 10−3, n = 74). (*, P < 0.01, colors indicate strain for comparison.) Note that the highest value of klp2Δ dli1Δ strain is 265 min, beyond the range of the plot. (B) Percentage of zygotes completing nuclear congression in wild type (100%, n = 104), klp2Δ dhc1Δ (2%, n = 85), klp2Δ num1Δ (100%, n = 42), klp2Δ dhc1(1–1266) (2%, n = 56), klp2Δ ssm4Δ (100%, n = 96), and klp2Δ dli1Δ (21%, n = 101). (C) Cellular distribution of N-terminal part of Dhc1-3×GFP in mating dhc1(1–1,266) cells coexpressing Cut11-mCherry and mCherry-Atb2 at 25°C. Images are represented as maximum z projection of 3D stacks. Dotted lines represent outlines of cells in different stages of meiotic prophase. (D) Cellular distribution of Dhc1-3×GFP in mating wild-type, ssm4Δ, dli1Δ, and nmt1-dhc1 cells coexpressing Cut11-mCherry and mCherry-Atb2. (E) Frequency of time points, at which dynein was detected at SPBs relative to SPB fusion in wild-type, ssm4Δ, dli1Δ, and nmt1-dhc1 zygotes (n = 50 cells). Mean values for dynein detection at SPBs relative to SPB fusion: wild type (−36 ± 33 min), ssm4Δ (−32 ± 31 min), dli1Δ (50 ± 26 min, 27/50 events), and nmt1-dhc1 (−62 ± 47 min). (F) Box plot shows SPB intensities of Dhc1-3×GFP at time point of SPB fusion averaged for a cell population of wild-type, ssm4Δ, dli1Δ, and nmt1-dhc1 zygotes (n = 50 SPBs). Mean values for dynein intensities at SPBs at time point of SPB fusion (arbitrary unit [a.u.]): wild type (4,924 ± 2,757), ssm4Δ (3,746 ± 2,160, P = 0.02), dli1Δ (80 ± 277, P < 10−15), and nmt1-dhc1 (13,939 ± 7,979, P < 10−11). (*, P < 0.05; ***, P < 10−4.) Bars, 5 µm. n were collected from 2–3 independent experiments. wt, wild type. For box plots, each box encloses 50% of the data with the median values displayed as lines. The top and bottom of each box mark the minimum and maximum values within the dataset that fall within an acceptable range. Any value outside of this range, called an outlier, is displayed as an individual point.
It is also known that, during nuclear horsetail movement, dynein localizes along MTs and interacts with the cortex through Num1 to generate pulling forces. In contrast, Num1 was not required for dynein-dependent nuclear congression (Fig. 4, A and B), indicating that dynein interaction with the cortex is not necessary for nuclear congression. We next tested the role of the dynactin complex, by deletion of the component ssm4, homologous to Glued. Surprisingly, in ssm4Δ single as well as in klp2Δ ssm4Δ double mutant zygotes, nuclear congression proceeded like in wild-type cells or klp2Δ single mutants, respectively (Fig. 4, A and B), showing that the dynactin complex is not required for dynein-dependent nuclear congression. Importantly, as previously reported, dynein was absent from MTs throughout meiotic prophase in ssm4Δ cells but was still recruited to SPBs in mating cells (Fig. 4 D; Niccoli et al., 2004). Dynein was detected in the majority of cells (84%) before SPB fusion (Fig. 4 E) and reached almost wild-type levels at the time point of SPB fusion (Fig. 4 F). Similar observations were made in zygotes deficient for dic1 (Fig. S4, A–C), which may link the dynein to the dynactin complex (Vaughan and Vallee, 1995; Fujita et al., 2010). Together, these results show first that dynein localization along MTs is not required for its function in nuclear congression and second, that, even though Dic1 and the dynactin complex are essential for nuclear horsetail movement, they are dispensable for dynein-dependent force generation during nuclear congression.
In contrast, the deletion of the light intermediate chain Dli1 resulted in a delay of 5 min (33 min congression time) compared with wild type, mimicking dhc1 deletion (Fig. 4 A). Furthermore, 79% of a double klp2Δ dli1Δ mutant failed to successfully complete nuclear congression, whereas the remaining 21% exhibited a large delay (Fig. 4, A and B). In contrast, double dhc1Δ dli1Δ mutant behaved like the single mutants (Fig. 4 A), demonstrating that Dli1 functions in the dynein-dependent pathway. Compared with ssm4Δ and dic1Δ zygotes, dynein was not only absent from MTs, but also from SPBs in mating dli1Δ and klp2Δ dli1Δ cells (Figs. 4 D and S4 D), in agreement with a previous study (Fujita et al., 2010). In detail, dynein was never observed at SPBs before SPB fusion. Instead, it was detected at SPBs only after SPB fusion (54%) or never (46%; Fig. 4 E).
In summary, Dli1 appears to play an essential role in dynein-dependent nuclear congression by controlling dynein recruitment to the SPB, whereas the dynactin complex and Dic1 are dispensable for this localization and function. SPB-bound dynein could pull MTs emanating from the nucleus of the mating partner, similar to the proposed role of Kar3 in budding yeast (Gibeaux et al., 2013).
Dli1 is required for proper dynein association with SPBs in vegetative cells
It has been proposed that dynein association with the SPB and more weakly with MTs depends on Dhc1 interaction with Dli1 (Fujita et al., 2010). Potentially, Dli1 might bind to SPBs on its own where it could recruit Dhc1. We tested this in vegetative cells. Dhc1 and Dli1 are meiosis specific and cannot be detected during the vegetative cell cycle, when expressed from their endogenous promoters (Yamamoto et al., 1999; Harigaya et al., 2006). To obtain comparable expression levels throughout the cell cycle, we expressed both genes from the nmt41 promoter in fusion with the three copies of GFP and two copies of mCherry, respectively. When ectopically expressed in vegetative cells, Dli1-2×mCherry did not localize to SPBs, whereas Dhc1-3×GFP was found at SPBs labeled by Sfi1-RFP in 10% of the cells (Fig. 5 A). This is consistent with the observation that Dli1 is absent from SPBs in dhc1Δ zygotes, whereas Dhc1 remains weakly associated to SPBs in dli1Δ zygotes (Fujita et al., 2010). However, when both proteins were coexpressed in vegetative cells, Dli1 could now be found at SPBs and, simultaneously, Dhc1 bound to SPBs at a much higher frequency, in ≤90% cells (Fig. 5, B and C), demonstrating that dynein recruitment to SPBs is increased by the interaction between Dhc1 and Dli1. We noted that Dhc1 was absent from SPBs in mitotic cells (Fig. 5 B, asterisk) and localized to the cell division site during cell separation, presumably to the equatorial MT-organizing center generating the postanaphase MT array (Fig. 5 B, arrowhead). Similarly, when expressed from the nmt1 promoter, Dhc1, but not Dli1, localized to SPBs, but SPB association of both was greatly increased when coexpressed with Dli1 (Figs. 5 C and S5, B and C). Under these conditions, Dhc1 also localized along MTs (Fig. S5 C, arrowhead), indicating that Dli1 also affects Dhc1 association with MTs.
Dynein promotes nuclear clustering in the klp2Δ sid2-250 mutant. (A) Ectopic expression of Dhc1-3×GFP or Dli1-2×mCherry from the nmt41 promoter in vegetative cells coexpressing Sfi1-RFP or -GFP that were exponentially grown 72 h in minimal medium without thiamine. Arrowheads indicate colocalization of Dhc1 with SPB marker Sfi1. (B) Colocalization of Dhc1-3×GFP and Dli1-2×mCherry at SPBs, when coexpressed from the nmt41 promoter. Asterisk labels mitotic cell, in which Dhc1 is absent from SPBs. Arrowhead highlights punctuate localization of Dhc1 at cell division site. (C) Percentage of cells, in which Dhc1-3×GFP expressed from the nmt41 (shaded in blue) or nmt1 promoter (red) is detected at SPBs. Strains with the indicated genotypes were tested. (n ≥ 300 cells, from a single experiment.) (D, top) Western blot of total extracts prepared from dli1Δ or nmt41-dli1-2×mCherry strains expressing nmt41-Dhc1-3×GFP. Western blot was probed with anti-GFP or TAT1 antibodies (tubulin as a loading control). (bottom) Relative amounts of Dhc1 normalized to dli1+. Bars represent means from three independent experiments. a.u., arbitrary unit. (E) sid2-250 and klp2Δ sid2-250 cells expressing Cut11-GFP to visualize nuclei were exponentially grown in minimal medium without thiamine at 25°C and then shifted to 36°C for 4 h to inactive Sid2. Dhc1-3×GFP was expressed from nmt41 or nmt1 promoter (only nmt1 shown) together with nmt41-Dli1-2×mCherry. Dotted lines represent outlines of cells. Cells were observed after fixation with −20°C methanol. Bar graphs show the means and SD of the means of the three replicate experiments with ≥73 cells per replicate. (F) Mean distances between nuclei were plotted against cell length (Fig. S5, D and E) categorized into 2.5-µm intervals (n ≥ 100 cells). (G) Ratio of mean nuclei distance/mean cell length in cells ranging from 20 to 30 µm (n ≥ 73 cells). Mean values for wild type (0.04 ± 0.01, distance = 0.93 ± 0.15 µm, length = 23.02 ± 0.2 µm), klp2Δ (0.23 ± 0.01, distance = 5.64 ± 0.43 µm, length = 24.84 ± 0.43 µm), klp2Δ nmt41-dhc1 nmt41-dli1 (0.18 ± 0.01, distance = 4.65 ± 0.53 µm, length = 25.47 ± 0.9 µm), and klp2Δ nmt1-dhc1 nmt41-dli1 (0.15 ± 0.004, distance = 3.59 ± 0.14 µm, length = 23.6 ± 0.24 µm; *, P < 0.05 to klp2Δ). Bars represent means from three independent experiments. Error bars: SD. wt, wild type. Bars, 5 µm.
Dynein promotes nuclear clustering in the klp2Δ sid2-250 mutant. (A) Ectopic expression of Dhc1-3×GFP or Dli1-2×mCherry from the nmt41 promoter in vegetative cells coexpressing Sfi1-RFP or -GFP that were exponentially grown 72 h in minimal medium without thiamine. Arrowheads indicate colocalization of Dhc1 with SPB marker Sfi1. (B) Colocalization of Dhc1-3×GFP and Dli1-2×mCherry at SPBs, when coexpressed from the nmt41 promoter. Asterisk labels mitotic cell, in which Dhc1 is absent from SPBs. Arrowhead highlights punctuate localization of Dhc1 at cell division site. (C) Percentage of cells, in which Dhc1-3×GFP expressed from the nmt41 (shaded in blue) or nmt1 promoter (red) is detected at SPBs. Strains with the indicated genotypes were tested. (n ≥ 300 cells, from a single experiment.) (D, top) Western blot of total extracts prepared from dli1Δ or nmt41-dli1-2×mCherry strains expressing nmt41-Dhc1-3×GFP. Western blot was probed with anti-GFP or TAT1 antibodies (tubulin as a loading control). (bottom) Relative amounts of Dhc1 normalized to dli1+. Bars represent means from three independent experiments. a.u., arbitrary unit. (E) sid2-250 and klp2Δ sid2-250 cells expressing Cut11-GFP to visualize nuclei were exponentially grown in minimal medium without thiamine at 25°C and then shifted to 36°C for 4 h to inactive Sid2. Dhc1-3×GFP was expressed from nmt41 or nmt1 promoter (only nmt1 shown) together with nmt41-Dli1-2×mCherry. Dotted lines represent outlines of cells. Cells were observed after fixation with −20°C methanol. Bar graphs show the means and SD of the means of the three replicate experiments with ≥73 cells per replicate. (F) Mean distances between nuclei were plotted against cell length (Fig. S5, D and E) categorized into 2.5-µm intervals (n ≥ 100 cells). (G) Ratio of mean nuclei distance/mean cell length in cells ranging from 20 to 30 µm (n ≥ 73 cells). Mean values for wild type (0.04 ± 0.01, distance = 0.93 ± 0.15 µm, length = 23.02 ± 0.2 µm), klp2Δ (0.23 ± 0.01, distance = 5.64 ± 0.43 µm, length = 24.84 ± 0.43 µm), klp2Δ nmt41-dhc1 nmt41-dli1 (0.18 ± 0.01, distance = 4.65 ± 0.53 µm, length = 25.47 ± 0.9 µm), and klp2Δ nmt1-dhc1 nmt41-dli1 (0.15 ± 0.004, distance = 3.59 ± 0.14 µm, length = 23.6 ± 0.24 µm; *, P < 0.05 to klp2Δ). Bars represent means from three independent experiments. Error bars: SD. wt, wild type. Bars, 5 µm.
Because in Drosophila melanogaster, Dli1 is required for the stability of the components of the dynein complex (Mische et al., 2008), Dli1 could regulate dynein localization in fission yeast by promoting the stability of Dhc1 molecules or dimers, core of the dynein complex, resulting in a quantitative increase of the dynein complex globally and driving its accumulation at SPBs and on MTs. Accordingly, Dhc1 levels were enhanced 1.6-fold in vegetative nmt41-Dhc1-3×GFP cells, when Dli1 was expressed from nmt41 promoter instead of its endogenous promoter (Fig. 5 D). This result indicates that Dli1 stabilizes Dhc1 directly or stabilizes the dynein complex with an indirect impact on the stability of Dhc1. Altogether, these data suggest that Dli1 favors Dhc1 association with the SPB and MTs by increasing the Dhc1 cellular pool.
Dynein overexpression speeds up klp2Δ nuclear congression
We next reasoned that if the speed of nuclear congression depends on dynein amounts at the SPB, increasing these levels artificially may speed up the process. To test this, we replaced dhc1 endogenous promoter by the strong nmt1 promoter that is fully active in the absence of thiamine. With this promoter, dynein was expressed at considerably higher levels than with its own promoter in the medium used for cell conjugation that lacks thiamine (Fig. 4 D). Under these conditions, dynein was detected at SPBs ∼30 min earlier than in wild-type zygotes, on average 62 min before SPB fusion (Fig. 4 E), and dynein intensity at SPBs was increased two- to threefold compared with wild-type zygotes (Fig. 4 F), implying that more dynein is present at SPBs during the congression of the two nuclei. Mating nmt1-dhc1 cells took the same time as wild-type cells to bring their nuclei together, but the congression time was reduced from 48 to 40 min in klp2Δ zygotes (Fig. 4 A). This demonstrates that dynein concentration at SPBs controls the speed of nuclear congression in the absence of Klp2 and shows that dynein plays a dose-dependent role in generating forces controlling the migration of the two nuclei.
When we overexpressed Klp2 from the nmt1 promoter, Klp2 levels were increased on average 2.4-fold compared with endogenous Klp2 levels (Fig. S4 E). Curiously, nuclear congression did not proceed faster compared with wild type and was even further delayed in the absence of dynein compared with dhc1Δ zygotes (Fig. S4 F). Currently, we can only speculate why Klp2 overexpression may inhibit nuclear congression under those conditions. Possibly, there might be an optimal cellular kinesin concentration required for efficient nuclear congression and increased levels could lead to motor crowding, which could inhibit the process.
Dynein promotes nuclei clustering in the klp2Δ sid2-250 mutant
Based on this study, we can propose that nuclear congression during fission yeast karyogamy relies on two minus end–directed motors that cooperate to promote efficient and robust congression of nuclei, although each is sufficient for nuclei congression in absence of the other. We thus wondered whether dynein could also replace Klp2 in other nuclear migration processes. In telophase, the segregated nuclei normally remain in the middle of the future daughter cells away from the cell division site. This positioning requires Klp2 inactivation by the septation initiation network, which can otherwise cluster nuclei in the cell middle by bridging MTs from the two nuclei (Mana-Capelli et al., 2012). Accordingly, nuclei are clustered in telophase in the septation initiation network mutant sid2-250 unless Klp2 is deleted, in which case the distance between nuclei increases with cell length (Okazaki and Niwa, 2008). We confirmed these observations in sid2-250 cells expressing Cut11-GFP to visualize nuclei (Fig. 5, E and F; and Fig. S5 D). Moreover, ectopic coexpression of Dhc1 from nmt41 or nmt1 promoters and Dli1 from nmt41 promoter decreased the distance between nuclei in klp2Δ sid2-250 cells compared with cells of similar length that do not express Dhc1 and Dli1, but to a lesser extent than in presence of Klp2 (Fig. 5, E and F; and Fig. S5, D and E). This was shown quantitatively by calculating the ratio between nuclear distance and cell length in a population of cells ranging from 20 to 30 µm in length (Fig. 5 G). Furthermore, this effect was enhanced in presence of higher levels of Dhc1. These data demonstrate that Klp2 and dynein can produce pulling forces in different cellular contexts to drive nuclear migrations.
Discussion
Here, we demonstrate that two minus end–directed motors, the kinesin-14 Klp2 and dynein, are required for efficient nuclear congression in fission yeast. To our knowledge, this is the first report of two minus end–directed motors cooperating to translocate nuclei by distinct mechanisms. Based on our observations, we propose the following model (Fig. 6 A): (1) Klp2 localizes dominantly to MT plus ends via Mal3 and may cross-link and slide MTs emanating from the opposite SPBs in an antiparallel manner; (2) Dynein accumulates at SPBs and pulls on MTs nucleated by the mating partner in a concentration-dependent manner; and (3) MT plus ends can push on the cortex to bring nuclei together. The Klp2-dependent pathway appears dominant, whereas MT pushing only facilitates efficient nuclear congression. Moreover, the delay in nuclear congression in klp2Δ zygotes might be limited by further increase of dynein levels at SPBs that speeds up nuclear migration during the prolongation phase.
Model for nuclear congression in fission yeast. (A) Upon fusion of two haploid cells, MTs dominantly nucleated at SPBs extend into the cytoplasm of the mating partner. (1) Kinesin-14 Klp2 loaded onto MT plus ends via Mal3/EB1 cross-links MTs in an antiparallel fashion. Two models for kinesin-14–dependent generation of pulling forces are proposed: (a) Klp2 slides antiparallel MTs or (b) Klp2 induces depolymerization and cross-links shrinking plus ends. (2) During nuclear congression, dynein accumulates at the SPB resulting in acceleration of nuclear migrations. SPB-bound dynein may exert pulling forces on MTs emanating from the opposite SPB. These parallel mechanisms ensure nuclear congression in fission yeast and illustrate distinct roles for two minus end–directed motor proteins in the same process. (3) Pushing forces generated by growing MTs against the cortex facilitate nuclear congression. (B) Dhc1 molecules or dimers are stabilized by Dli1 resulting in a quantitative increase of dynein at the SPB recruited by an unknown adaptor protein.
Model for nuclear congression in fission yeast. (A) Upon fusion of two haploid cells, MTs dominantly nucleated at SPBs extend into the cytoplasm of the mating partner. (1) Kinesin-14 Klp2 loaded onto MT plus ends via Mal3/EB1 cross-links MTs in an antiparallel fashion. Two models for kinesin-14–dependent generation of pulling forces are proposed: (a) Klp2 slides antiparallel MTs or (b) Klp2 induces depolymerization and cross-links shrinking plus ends. (2) During nuclear congression, dynein accumulates at the SPB resulting in acceleration of nuclear migrations. SPB-bound dynein may exert pulling forces on MTs emanating from the opposite SPB. These parallel mechanisms ensure nuclear congression in fission yeast and illustrate distinct roles for two minus end–directed motor proteins in the same process. (3) Pushing forces generated by growing MTs against the cortex facilitate nuclear congression. (B) Dhc1 molecules or dimers are stabilized by Dli1 resulting in a quantitative increase of dynein at the SPB recruited by an unknown adaptor protein.
Klp2 contains two MT-binding domains, one in the motor domain and an additional one in the tail, allowing it to interact with two MTs at once and to cross-link them (Braun et al., 2009). Kinesin-14s are implicated in MT sliding, and accordingly, Klp2 has been shown to slide newly nucleated MTs toward minus ends (Janson et al., 2007). It is therefore likely that Klp2 generates pulling forces during nuclear congression in a similar manner: by sliding antiparallel MTs relative to each other as suggested previously for budding yeast Kar3 (Meluh and Rose, 1990) and for nuclei clustering at the end of mitosis in fission yeast mutants defective in septation (Mana-Capelli et al., 2012). Alternatively, Klp2 might cross-link shrinking plus ends and potentially induce their depolymerization. Klp2 at kinetochores appears to operate via such a mechanism during mitosis: to shorten kinetochores fibers and facilitate capture of lost chromosomes by spindle poles (Grishchuk and McIntosh, 2006; Gachet et al., 2008). Further support for this model comes from in vitro work showing that Kar3 can depolymerize MTs from plus ends (Sproul et al., 2005).
Sliding activity has been shown to be dependent on kinesin-14 tail and motility (Meluh and Rose, 1990; Braun et al., 2009). Accordingly, a truncated Klp2 protein lacking the tail region (1–186 aa) fails to localize to MTs and causes defects in nuclear congression similar to klp2 deletion (Fig. S4, G–I). A Klp2rigor mutant protein still binds to MTs but also exhibits nuclear congression defects (Fig. S4, G–I), indicating that ATPase and probably sliding activity are required. However, it is currently unclear how kinesin-14 depolymerase activity is regulated, and in vitro data suggest that motility is required to depolymerize MT plus ends (Sproul et al., 2005). Thus, we cannot rule out either model.
It may be also of interest to further investigate the function of Tip1 in this process. Its deletion causes a delay in nuclear congression, which could be simply explained by its role in controlling MT dynamics. Alternatively, Tip1 could function similarly to its budding yeast homologue Bik1, which is proposed to initiate and coordinate MT depolymerization together with Kar3 (Molk et al., 2006).
In our model, the dynein complex anchored at the SPB may interact with MTs emanating from the SPB of the mating partner pulling the two nuclei together, as demonstrated for Kar3 by electron microscopy (Gibeaux et al., 2013). This may be a conserved dynein function. In fertilized oocytes, the female pronucleus is transported toward the male one along MTs emanating from the male pronucleus by dynein accumulating at the nuclear envelope (Reinsch and Gönczy, 1998).
The dynactin complex is dispensable for dynein-dependent nuclear congression in fission yeast. On the other hand, we discovered that the light intermediate chain Dli1 is crucial. We find that dli1 deletion may greatly reduce dynein levels at SPBs and MTs by being required for Dhc1 stability (Fig. 6 B), although we cannot exclude at this stage that other mechanisms may be operating in addition. Dli1 could for instance favor Dhc1 dimerization and enhance as a consequence Dhc1 avidity for the SPB and MTs. Or, Dli1 could also contain low-affinity SPB- and MT-binding sites in the Dli1 molecule not sufficient for Dli1 localization to these structures if dissociated from Dhc1, but increasing the affinity for the SPB and MTs, when both proteins interact.
About 50% of zygotes lacking dynein initiated nuclear congression immediately upon cell fusion, whereas ∼35% exhibited an initial phase without directed nuclear migration. This observation suggests that dynein may favor interactions between antiparallel MTs required for the initiation of nuclear congression. Our preliminary data suggest that this specific role of dynein may become more important in mating cells with more complex geometries adopting S or U shape (Fig. S2, J and K). We hypothesize that in simpler geometries, like straight shapes, MT plus ends might cross each other more easily, favoring their cross-linking by Klp2. In more complex geometries, the presence of a large motor complex at the SPB may help catching MTs from the mating partner, when Klp2 fails to do so.
In mammals, most minus end–directed translocations of organelles, including nuclei and vesicles, are performed by dynein. Cooperation between kinesins, primarily plus end directed, and dynein in nuclear positioning has been reported for several systems and provides bidirectionality of nuclear movements (Fridolfsson and Starr, 2010; Tsai et al., 2010). Our work suggests that the collaboration of dynein with kinesin-14s in parallel pathways may on the other hand increase the efficiency of organelle translocation and provide robustness to these nuclear-positioning events.
In conclusion, the distinct properties as well as specific localization pattern of motor proteins allow differential roles for minus end–directed motors in the same process of nuclear positioning. In the future, it will be interesting to test whether dynein cooperates with kinesin-14 in the fusion of pronuclei and other nuclear migrations in higher eukaryotes.
Materials and methods
Yeast genetics, strains, and plasmids
All S. pombe strains used in this study are isogenic to 972 and listed in Table S1. Standard fission yeast molecular genetics techniques and media were used as previously described (Moreno et al., 1991). Strains from genetic crosses were selected by random spore germination and replica in plates with appropriate supplements or drugs. Transformations were performed using the lithium acetate-DMSO method as previously described, and genomic integrations were achieved through homologous recombination (Bähler et al., 1998).
Production of mutant and tagged strains
Deletion strains and strains expressing Cut11-GFP/mCherry, Sfi1-GFP/mCherry, GFP/mCherry-Atb2, and Klp2-GFP were available in the laboratory strain collection or received as gifts (Dhc1-3×GFP and Dli1-2×mCherry) from M. Yamamoto (University of Tokyo, Tokyo, Japan) and A. Yamamoto (Shizuoka University, Shizuoka, Japan). For those strains, the original study has been indicated in Table S1.
During this study, to produce strains overexpressing dhc1-3×GFP and dli1-2×mCherry genes from their endogenous loci using the thiamine-repressible nmt41 or nmt1 promoter (Maundrell, 1990), NatMX6::nmt1/nmt41, or KanMX6::nmt1/nmt41 were amplified from pFa6A plasmids (Van Driessche et al., 2005) and integrated upstream of the dhc1 or dli1 ORF at the endogenous locus (Bähler et al., 1998). nmt-dhc1-3×GFP and nmt-dli1-2×mCherry strains were grown exponentially in EMM3S without thiamine for 72 h (nmt41) or 24 h (nmt1).
Protein extracts were prepared in extraction buffer (PBS and 2 mM EDTA) containing protease inhibitor cocktail. Western blots were probed with mouse anti-GFP mAb (1:500; Roche) and mouse TAT1 (anti–α-tubulin) mAb (1:500; noncommercial). The TAT1 mAb was a gift from K. Gull (University of Oxford, Oxford, England, UK; Woods et al., 1989). Secondary goat anti–mouse antibody was coupled to peroxidase (Jackson ImmunoResearch Laboratories, Inc.). Signal quantification was performed with Image Lab 4.0.1 (Bio-Rad Laboratories).
Constructs of wild-type, truncated, and rigor Klp2 protein tagged C-terminally to GFP or overexpressing Klp2-GFP (Fig. S4, E–I) were integrated at the leu1 locus under the control of the endogenous klp2 or, in latter case, nmt1 promoter, in a leu1-32 strain carrying a deletion of the endogenous klp2. The plasmids used were created by cloning full-length or truncated (lacking amino acid residues 1–186; Klp2(Δ1–186)) Klp2-GFP between NheI and SalI sites in a pJK148-derived plasmid (gift from K. Gull; Keeney and Boeke, 1994) containing klp2 or nmt1 promoter and nmt1 terminator (pKS92, pKS96, and pKS103). A klp2 rigor mutant with a G568E substitution was produced by site-directed mutagenesis of a pBluescript plasmid carrying the klp2 ORF and cloned into pKS96 containing full-length klp2-GFP under klp2 promoter between AccIII and MluI. Plasmids were integrated at leu1 locus in a leu1-32 strain after digestion with NruI.
Live-cell microscopy of mating cells and data analysis
Under nitrogen starvation and presence of opposite mating types, cells enter the meiotic cycle. Mating occurs only after long incubation in nitrogen-poor medium at random initiation time and at a low frequency. However, with the here described technique, the protocol was optimized to obtain a good number of mating cells during long term imaging.
For long-term imaging of mating cells, we used microfabricated microfluidic flow chambers, based on polydimethylsiloxane (PDMS), previously constructed and described from our laboratory (Terenna et al., 2008). In brief, the design for the flow chamber was drawn using the computer-assisted design software L-Edit (Tanner EDA). The design was then laser etched into a thin layer of chromium on a quartz plate, which served as a photomask (Microtronics). Next, SU8 negative photoresist was spin coated onto a silicon wafer (MicroChem). The features were then transferred from the photomask onto the photoresist layer by exposure and cross-linking with UV light (365 nm) for 20 s. The photoresist was developed with SU8 developer and cleaned with isopropyl alcohol and nitrogen gas. The wafer was then able to serve as a master mold on which repeated replication of molds could be made by casting from PDMS (Dow Corning). Chambers were assembled by peeling off a PDMS replica from a mold, introducing inlet and outlet holes, and bonding the replica to a microscope glass coverslip after surface treatment using a plasma cleaner (Harrick Scientific Products).
Cells of both mating types were separately grown in nitrogen-poor malt extract medium at 25°C overnight. Next, cells were mixed in an equal cell number and were manually injected into a microfluidic chamber (Fig. S1 A). When h90 strains were imaged, for example, to visualize Klp2-GFP and Dhc1-3×GFP, cells were grown in rich YE5S medium overnight, before transferred to malt extract medium and injected into chambers, as these strains initiate mating more quickly. After injection, devices were incubated 6–8 h at 25°C before imaging.
Imaging was usually performed at 25°C and only for cut7-24 strains (Fig. 1, C and D) at 37°C. In this case, cells were shifted to the restrictive temperature 30 min before imaging.
For imaging, we used the spinning-disk confocal head (Yokogawa Electric Corporation) mounted on an inverted microscope (Eclipse Ti; Nikon), equipped with Plan Apochromat 10×/1.4 NA objective lens (Nikon), a PIFOC (perfect image focus) objective stepper, and a charge-coupled device camera (CoolSNAP HQ2; Photometrics) as previously described (Tran et al., 2004). Usually, 3D optical sections were taken. They were analyzed and presented as maximal projections. For imaging purposes, proteins were tagged to GFP or mCherry/RFP.
Images were acquired and processed with MetaMorph 7.7 (Molecular Devices). Data were plotted as box plots generated with KaleidaGraph 4.0 (Synergy). For the box plots, each box encloses 50% of the data, with the median value displayed as a line. The top and bottom of each box mark the minimum and maximum values within the dataset that fall within an acceptable range. Any value outside of this range, called an outlier, is displayed as an individual point. Statistical analyses of data were performed using the Student’s t test for comparison between means in Excel 2010 (Microsoft).
For Sfi1-GFP dynamics, traced lines were drawn along longitudinal axis in zygotes accordingly to their shape to create kymographs. Distances were measured and plotted in diagrams generated by Excel 2010 as a function over time. For each cell, SPB distances were normalized to the mean SPB distance calculated from first 10 time points before cell fusion.
Online supplemental material
Fig. S1 shows the general protocol used in this study to capture and measure nuclear congression. Fig. S2 shows Sfi1-GFP dynamics representing nuclear congression kinetics of wild-type, klp2Δ, dhc1Δ, double mutant klp2Δ dhc1Δ, and triple mutant klp2Δ dhc1Δ tea2Δ zygotes. Fig. S3 shows Klp2-GFP localization in dhc1Δ and mal3Δ zygotes and the SPB localization and accumulation of Dhc1-3×GFP in klp2Δ zygotes. Fig. S4 shows Dhc1-3×GFP localization and nuclear congression characteristics in dic1Δ and klp2Δ dic1Δ zygotes as well as Klp2-GFP localization and nuclear congression characteristics in zygotes expressing klp2-GFP mutants or overexpressing Klp2-GFP. Fig. S5 shows localization of ectopically expressed Dhc1-3×GFP or Dli1-2×mCherry under control of the nmt1 promoter and raw data plots of distances between nuclei over cell length in sid2-250 mutants presented in Fig. 5 F. Table S1 lists the yeast strains used in this study.
Acknowledgments
We thank the laboratory of M. Yamamoto (University of Tokyo), A. Yamamoto (Shizuoka University), and Keith Gull (University of Oxford) for generously providing reagents. The experiments were performed on PICT-IBiSA (Institut Curie, Paris), a member of the France-BioImaging national research infrastructure.
K. Scheffler was supported by a PhD fellowship from Complexité du Vivant–Université Pierre et Marie Curie and Fondation ARC pour la Recherche sur le Cancer. R. Minnes was supported by a post-doctoral fellowship from Human Frontier Science Program. This work was supported by grants from the National Institutes of Health (NIH R01GM102215), Agence Nationale de la Recherche, Institut National du Cancer, and Fondation ARC.
The authors declare no competing financial interests.
Author contributions: K. Scheffler designed and performed experiments, analyzed data, and wrote the paper. R. Minnes initiated the project. V. Fraisier constructed and maintained the microscopes. A. Paoletti and P.T. Tran edited paper.





![Figure 4. Dynein functions at the SPB during nuclear congression independent of the dynactin complex. (A) Box plot shows time of nuclear congression in wild-type and dynein-related single and klp2Δ double mutants at 25°C. Mean values (p-values against wild type or klp2Δ, respectively): wild type (29 ± 7 min, n = 104), klp2Δ (48 ± 13 min, P < 10−22, n = 84), dhc1Δ (34 ± 9 min, P < 10−4, n = 87), num1Δ (31 ± 9 min, P = 0.09, n = 58), klp2Δ num1Δ (48 ± 16 min, P = 0.85, n = 42), dhc1(1–1,266) (35 ± 10 min, P < 10−4, n = 56), ssm4Δ (29 ± 9 min, P = 0.78, n = 65), klp2Δ ssm4Δ (45 ± 14 min, P = 0.12, n = 96), dli1Δ (33 ± 11 min, P = 0.003, n = 60), klp2Δ dli1Δ (119 ± 65 min, P < 10−6, n = 33/101), dhc1Δ dli1Δ (35 ± 7 min, n = 64), nmt1-dhc1 (28 ± 10 min, P = 0.74, n = 71), and klp2Δ nmt1-dhc1 (40 ± 12 min, P < 10−3, n = 74). (*, P < 0.01, colors indicate strain for comparison.) Note that the highest value of klp2Δ dli1Δ strain is 265 min, beyond the range of the plot. (B) Percentage of zygotes completing nuclear congression in wild type (100%, n = 104), klp2Δ dhc1Δ (2%, n = 85), klp2Δ num1Δ (100%, n = 42), klp2Δ dhc1(1–1266) (2%, n = 56), klp2Δ ssm4Δ (100%, n = 96), and klp2Δ dli1Δ (21%, n = 101). (C) Cellular distribution of N-terminal part of Dhc1-3×GFP in mating dhc1(1–1,266) cells coexpressing Cut11-mCherry and mCherry-Atb2 at 25°C. Images are represented as maximum z projection of 3D stacks. Dotted lines represent outlines of cells in different stages of meiotic prophase. (D) Cellular distribution of Dhc1-3×GFP in mating wild-type, ssm4Δ, dli1Δ, and nmt1-dhc1 cells coexpressing Cut11-mCherry and mCherry-Atb2. (E) Frequency of time points, at which dynein was detected at SPBs relative to SPB fusion in wild-type, ssm4Δ, dli1Δ, and nmt1-dhc1 zygotes (n = 50 cells). Mean values for dynein detection at SPBs relative to SPB fusion: wild type (−36 ± 33 min), ssm4Δ (−32 ± 31 min), dli1Δ (50 ± 26 min, 27/50 events), and nmt1-dhc1 (−62 ± 47 min). (F) Box plot shows SPB intensities of Dhc1-3×GFP at time point of SPB fusion averaged for a cell population of wild-type, ssm4Δ, dli1Δ, and nmt1-dhc1 zygotes (n = 50 SPBs). Mean values for dynein intensities at SPBs at time point of SPB fusion (arbitrary unit [a.u.]): wild type (4,924 ± 2,757), ssm4Δ (3,746 ± 2,160, P = 0.02), dli1Δ (80 ± 277, P < 10−15), and nmt1-dhc1 (13,939 ± 7,979, P < 10−11). (*, P < 0.05; ***, P < 10−4.) Bars, 5 µm. n were collected from 2–3 independent experiments. wt, wild type. For box plots, each box encloses 50% of the data with the median values displayed as lines. The top and bottom of each box mark the minimum and maximum values within the dataset that fall within an acceptable range. Any value outside of this range, called an outlier, is displayed as an individual point.](https://cdn.rupress.org/rup/content_public/journal/jcb/209/1/10.1083_jcb.201409087/6/m_jcb_201409087_fig4.jpeg?Expires=1773750950&Signature=YC~Wa6n-0ZSz~mM4ZSydH6vhHg0RC59yg~wZoIaJnu7Ag5TeVXkrLa~vboFRhZJTKx7tX4HpBp~s-245gCsmq94Zk3mA0vJdT4MIvJstSdTS4Qk9keBjAq9-1VirV7bGmD6egcBbSTutfH84z~sWzNt~hs7cLHe5iD6EPEALh~zzYSD2MMDcGIeLOxHnm~bqHlirbtebb3MsbDMzzfausC2Uojg4rWvchHYo7AGEry7zzdBXn8ujrVntD2KlcXCuuPDGh9qrcUVDQZsVFcHuwlU6SG1BivCbkcTi7kgU7lS586NlihSJFWjnDgGkyZp2yzYqaCzZy-NsiOp4xGym~g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)