The ubiquitin protein ligase anaphase-promoting complex or cyclosome (APC/C) controls mitosis by promoting ordered degradation of securin, cyclins, and other proteins. The mechanisms underlying the timing of APC/C substrate degradation are poorly understood. We explored these mechanisms using quantitative fluorescence microscopy of GFP-tagged APC/CCdc20 substrates in living budding yeast cells. Degradation of the S cyclin, Clb5, begins early in mitosis, followed 6 min later by the degradation of securin and Dbf4. Anaphase begins when less than half of securin is degraded. The spindle assembly checkpoint delays the onset of Clb5 degradation but does not influence securin degradation. Early Clb5 degradation depends on its interaction with the Cdk1–Cks1 complex and the presence of a Cdc20-binding “ABBA motif” in its N-terminal region. The degradation of securin and Dbf4 is delayed by Cdk1-dependent phosphorylation near their Cdc20-binding sites. Thus, a remarkably diverse array of mechanisms generates robust ordering of APC/CCdc20 substrate destruction.
Introduction
Cell division is a fundamental biological process governed by a complex network of regulatory molecules, and the key to its success lies in having the right molecules become active (or inactive) at the right time. The regulatory network controlling cell division is hierarchical: a few master regulators, primarily the Cdks and the anaphase-promoting complex or cyclosome (APC/C), orchestrate the activities of hundreds of downstream proteins and processes (Morgan, 2007). As the activities of the master regulators rise and fall, they also drive changes in the activities of downstream players. One interesting feature of this regulatory system is that downstream components, even when regulated by the same master regulator, can become active or inactive in an ordered fashion, rather than simultaneously (Pines, 2006; Sullivan and Morgan, 2007). Deciphering how the master regulators discriminate between their substrates and achieve this ordering is crucial to our understanding of the orchestration of the cell cycle and other complex processes.
The APC/C is a ubiquitin protein ligase or E3 that governs mitotic events by promoting timely degradation of key mitotic proteins (Peters, 2006; Barford, 2011; Pines, 2011; Primorac and Musacchio, 2013). Together with its early mitotic activator subunit Cdc20, APC/C promotes the degradation of securin, an inhibitor of separase. Separase then cleaves the cohesins that link the sister chromatid pairs, triggering sister chromatid separation (Fig. 1 A; Nasmyth and Haering, 2009). APC/CCdc20 also promotes the degradation of S and M cyclins, which lowers Cdk activity. In budding yeast, APC/CCdc20-dependent separase activation also leads to the activation of Cdc14, a phosphatase that dephosphorylates numerous Cdk substrates (Stegmeier and Amon, 2004; Queralt et al., 2006; Queralt and Uhlmann, 2008). Among these Cdk substrates is the alternative APC/C activator Cdh1, which together with APC/C promotes the degradation of late-mitotic substrates and drives the completion of mitosis, cytokinesis, and entry into G1 (Fig. 1 A; Sullivan and Morgan, 2007).
Metaphase–anaphase transition in cells carrying GFP-tagged APC/C substrates. (A) Network diagram of the metaphase–anaphase transition in budding yeast. (B) GFP-tagged APC/CCdc20 substrates and mCherry-tagged SPBs in cycling cells, at 3-min intervals. Broken circles indicate cells at the onset of SPB separation, and solid circles mark cells at the onset of spindle elongation.
Metaphase–anaphase transition in cells carrying GFP-tagged APC/C substrates. (A) Network diagram of the metaphase–anaphase transition in budding yeast. (B) GFP-tagged APC/CCdc20 substrates and mCherry-tagged SPBs in cycling cells, at 3-min intervals. Broken circles indicate cells at the onset of SPB separation, and solid circles mark cells at the onset of spindle elongation.
APC/CCdc20 and APC/CCdh1 each have multiple substrates, which are degraded at distinct times in the cell cycle (Pines, 2006; Sullivan and Morgan, 2007). In the case of mammalian APC/CCdc20, the substrates Nek2A and cyclin A are degraded in prometaphase, immediately after nuclear envelope breakdown, whereas securin and cyclin B are degraded in metaphase (den Elzen and Pines, 2001; Geley et al., 2001; Hames et al., 2001; Hagting et al., 2002). Ordered degradation is equally prevalent among APC/CCdh1 substrates in anaphase and G1 (Pines, 2006; Sullivan and Morgan, 2007). It is not clear how the same APC/C complex robustly distinguishes among its substrates and promotes their degradation at different times in the cell cycle.
The timing of APC/CCdc20 substrate degradation in vertebrate cells is influenced by the spindle assembly checkpoint (SAC), which is activated by unattached kinetochores and inhibits APC/CCdc20 activity toward different substrates to varying degrees. Upon SAC activation, kinetochore-localized SAC components stimulate the formation of Mad2–Cdc20 complexes, leading to the formation of the mitotic checkpoint complex (MCC) consisting of Cdc20, Mad2, and Mad3 (in yeast) or BubR1 (in vertebrates), and Bub3 (Musacchio and Salmon, 2007; Lara-Gonzalez et al., 2012). The MCC is the major effector of the SAC. It binds to APC/C and strongly inhibits its activity toward securin and cyclin B, whereas cyclin A and Nek2A can still be degraded in an active checkpoint due to less efficient inhibition by the MCC (den Elzen and Pines, 2001; Geley et al., 2001; Hames et al., 2001; Hagting et al., 2002; Collin et al., 2013; Dick and Gerlich, 2013). When all kinetochores are properly attached to the spindle, the SAC is turned off and the MCC is disassembled to allow APC/CCdc20-dependent degradation of securin and anaphase onset. The protein components and mechanisms of the SAC are highly conserved across species. However, even though the SAC plays an essential role in mammalian cell division (Michel et al., 2001, 2004; Meraldi et al., 2004), disabling the SAC in yeast has very little impact on the cell cycle under normal conditions, and the SAC becomes essential only in the presence of spindle defects (Hoyt et al., 1991; Li and Murray, 1991).
The APC/C recognizes its targets through short sequence motifs called the D box or KEN box, which are often found in unstructured N-terminal regions of APC/C substrates (Glotzer et al., 1991; Pfleger and Kirschner, 2000). Both Nek2A and cyclin A are thought to possess extra binding sites for the APC/C, allowing them to bypass or overcome inhibition by SAC proteins. Nek2A employs a C-terminal motif that resembles Cdc20 and Cdh1 C termini to bind to the APC/C core directly without the need of an activator (Hames et al., 2001; Hayes et al., 2006; Sedgwick et al., 2013). Cyclin A gains additional affinity for the APC/CCdc20 by forming a complex with Cdk and the accessory subunit Cks1 (Wolthuis et al., 2008; Di Fiore and Pines, 2010). Cks1 binds to Cdk and contributes to recognition of Cdk substrates carrying specific phosphothreonines (Brizuela et al., 1987; Hadwiger et al., 1989; Richardson et al., 1990; Tang and Reed, 1993; Kõivomägi et al., 2013; McGrath et al., 2013). There is also evidence that Cks1 binds APC/C directly to promote its phosphorylation by Cdk (Patra and Dunphy, 1998; Shteinberg and Hershko, 1999; Rudner and Murray, 2000). Thus, cyclin A interacts directly with APC/CCdc20 through its D box and also indirectly through Cdk-Cks1.
Modifications of APC/C substrates also influence their ubiquitination by the APC/C (Holt et al., 2008; Singh et al., 2014). Budding yeast securin has two Cdk1 sites near its D box and KEN box, and phosphorylation of these sites inhibits its ubiquitination in vitro (Holt et al., 2008; Holt et al., 2009). Cdc14 dephosphorylates these sites in vitro. Given that securin degradation leads to Cdc14 activation indirectly through separase, these results suggested the existence of positive feedback in securin degradation. Although this phosphoregulation of securin by Cdk1 improves the fidelity of sister chromatid segregation (Holt et al., 2008), it remains unclear how this regulation influences the securin degradation rate and timing. Cdk1 sites are also found inside or near the D box of other budding yeast APC/C substrates (Holt et al., 2009), including Dbf4, the activating subunit for Cdc7 (also known as the Dbf4-dependent kinase [DDK]). DDK collaborates with S cyclin–Cdk1 to initiate DNA replication (Bell and Dutta, 2002). Dbf4 is an APC/CCdc20 substrate (Oshiro et al., 1999; Ferreira et al., 2000; Sullivan et al., 2008), but it is not clear whether Cdk phosphorylation contributes to its degradation timing or dynamics.
Here we explore how the interplay among the SAC, Cdk1, APC/CCdc20, and its substrates lays out the path toward the metaphase–anaphase transition in budding yeast, and we dissect the mechanisms responsible for ordered APC/CCdc20 substrate degradation. We used single cell analyses of fluorescently tagged proteins to show that APC/C substrates are degraded in a specific order, with early degradation of the S cyclin Clb5 followed by degradation of securin, Dbf4, and then finally the M cyclin Clb2. We also show that the SAC is largely turned off before the degradation of Clb5 and thus does not contribute to the degradation timing of later substrates. Instead, we find that Cdk-dependent phosphorylation of securin and Dbf4 delays their degradation, and we present evidence that Cks1 and a previously undiscovered sequence motif in Clb5 promote early Clb5 degradation. Together our results provide a temporal and mechanistic view of the key regulatory steps leading to the metaphase–anaphase transition.
Results
APC/C substrates are degraded in a defined order
We used fluorescence microscopy and in silico synchronization (Clute and Pines, 1999) to analyze the timing and dynamics of APC/C substrate degradation in living yeast cells. We constructed a series of yeast strains in which a single APC/C substrate (Clb5, securin/Pds1, Dbf4, or Clb2) was tagged at its endogenous locus with a C-terminal GFP. In these strains, the spindle pole body (SPB) component Spc42 was also tagged at its endogenous locus with C-terminal mCherry. After their duplication in early S phase, SPBs display two distinctive behaviors that serve as useful indices of mitotic timing: first, at the beginning of mitosis, the two SPBs separate from each other and form a short spindle; and second, at anaphase onset, the two SPBs move quickly away from each other as the spindle begins to elongate, which coincides with separase activation and the onset of sister chromatid separation (Fig. 1 B; Straight et al., 1997; Pearson et al., 2001; Yaakov et al., 2012).
Using spinning-disk confocal microscopy with a 30-s time resolution, we analyzed these fluorescent markers in individual cells in unperturbed, asynchronously proliferating cultures (Fig. 1 B). We first measured the time from SPB separation to spindle elongation in single cells as an estimate of the time from mitotic entry to anaphase onset. This time was highly variable from cell to cell, ranging from 15 to 45 min, with a median of 21 min. Thus, after SPB separation, cells display remarkable variability in the timing of APC/CCdc20 activation and anaphase onset. This timing and variability did not change significantly in any of the strains carrying GFP-tagged APC/C substrates (Fig. 2 A, one-way ANOVA, P = 0.47), which is consistent with the fact that GFP tagging had no effect on the doubling times of all strains (not depicted). We also confirmed that fluorescence imaging had little impact on mitotic duration (Fig. S1 A; see Materials and methods for optimization of imaging conditions).
Timing and dynamics of APC/CCdc20 substrate degradation. (A) Time from SPB separation to spindle elongation in individual cells with GFP tags on APC/C substrates. Each dot represents a single cell. Starting from the left, sample sizes are: n = 49, 90, 121, 82, and 77 cells. For each strain, the middle bar indicates the median value and error bars indicate the 25th and 75th percentiles. (B) GFP intensity of representative individual cells with tagged APC/CCdc20 substrates (from the cell populations analyzed in A). Underlying black lines show the original data, and the colored lines are smoothed traces. The timing of SPB separation and spindle elongation are marked with broken and solid lines, respectively. (C and D) Comparison of different GFP-tagged substrates using SPB separation (C, broken lines) or spindle elongation (D, solid lines) as the timing reference (from the cell populations analyzed in A). In top panels, each line is a smoothed trace of a single cell. A random subset of representative cells is shown for clarity of viewing. Bottom panels show the averaged traces, where unsmoothed traces from all cells were first aligned to the same time reference point, averaged at each time point and then normalized to maximum intensity.
Timing and dynamics of APC/CCdc20 substrate degradation. (A) Time from SPB separation to spindle elongation in individual cells with GFP tags on APC/C substrates. Each dot represents a single cell. Starting from the left, sample sizes are: n = 49, 90, 121, 82, and 77 cells. For each strain, the middle bar indicates the median value and error bars indicate the 25th and 75th percentiles. (B) GFP intensity of representative individual cells with tagged APC/CCdc20 substrates (from the cell populations analyzed in A). Underlying black lines show the original data, and the colored lines are smoothed traces. The timing of SPB separation and spindle elongation are marked with broken and solid lines, respectively. (C and D) Comparison of different GFP-tagged substrates using SPB separation (C, broken lines) or spindle elongation (D, solid lines) as the timing reference (from the cell populations analyzed in A). In top panels, each line is a smoothed trace of a single cell. A random subset of representative cells is shown for clarity of viewing. Bottom panels show the averaged traces, where unsmoothed traces from all cells were first aligned to the same time reference point, averaged at each time point and then normalized to maximum intensity.
Next, in each single cell progressing through mitosis, we monitored the degradation of the GFP-labeled substrate relative to the two SPB events (Figs. 1 B and 2 B). With this information, we could compare different cells by referencing the same SPB event, allowing us to compare cells with different GFP-tagged substrates, as well as cells from the same GFP-tagged strain (Fig. 2, C and D).
Our results revealed sequential degradation of APC/C substrates during mitosis. At 30°C, Clb5 degradation began an average of 10 min after SPB separation and was almost complete when the spindle started to elongate. Degradation of securin and Dbf4 began ∼16 min after SPB separation and was less than half complete when the spindle started to elongate. A small fraction of Clb2 was degraded at the time of anaphase onset, but the majority was degraded later in anaphase (Figs. 1 B and 2, B–D). The substrate ordering we observed is consistent with previous results from population measurements (Ferreira et al., 2000; Lianga et al., 2013). The two phases of Clb2 degradation we observed also support previous evidence that Clb2 degradation is initiated by APC/CCdc20 and later completed by APC/CCdh1 (Bäumer et al., 2000; Yeong et al., 2000; Wäsch and Cross, 2002).
With single cell measurements, we were also able to observe variations in the population. When cells were synchronized in silico with SPB separation, degradation timing of the same substrate was highly variable in different cells, similar to the variability in mitotic timing (Fig. 2 C, top; see also Fig. 3 A, bottom left). This accounts for the fact that the first phase of Clb2 degradation was obscured when we averaged the GFP traces over the population (Fig. 2 C as compared with Fig. 2 D, bottom). Thus, the timing of APC/C activation, and thus its substrate degradation, is not closely correlated with the timing of mitotic onset (SPB separation).
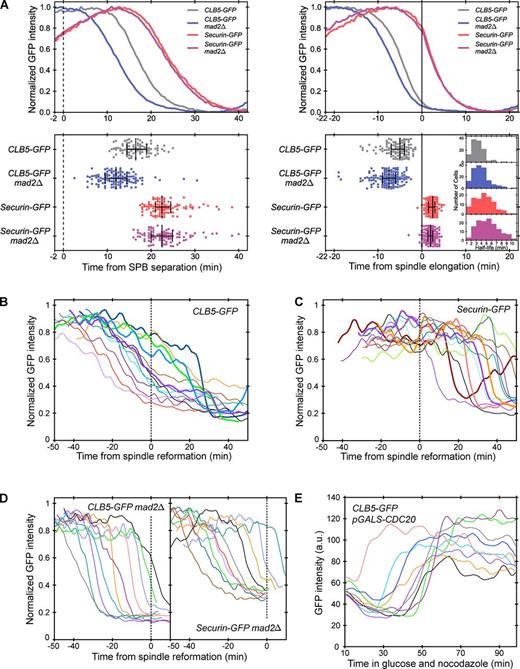
Role of the SAC in APC/CCdc20 substrate degradation. (A) Clb5 and securin degradation profiles in wild-type and mad2Δ cells. Cells are aligned using either SPB separation (left, broken line) or spindle elongation (right, solid line). Top panels show averaged and normalized traces as in Fig. 2, C and D. Bottom panels show the time of 50% substrate degradation in individual cells (Fig. S1 C). Each dot represents a single cell, and n > 90 cells per strain. For each strain, the middle bar indicates the median value, and error bars indicate the 25th and 75th percentiles. The inset in the bottom right panel shows a histogram of protein half-lives in different strains, calculated from single cell traces; n > 100 cells per strain. (B–D) Clb5 and securin degradation in nocodazole-treated cells; n > 20 cells per strain. Asynchronous cells were plated on agarose pads with 15 µg/ml nocodazole 10 min before imaging began. Representative traces from individual cells are normalized and aligned to spindle reformation (broken lines). The traces shown here were selected on the basis of two criteria: minimum overlap among traces for clarity of viewing, and inclusion of only mitotic cells, as judged by bud size. Wild-type (B and C) and mad2Δ cells (D) are shown. In B and C, representative cells with fast substrate degradation after recovery from the SAC arrest are shown in bold lines. (E) Clb5 degradation in nocodazole with CDC20 shut off; n > 20 cells. Asynchronous cells were grown in 2% galactose and plated on an agarose pad with 2% glucose and 15 µg/ml nocodazole. Representative traces began 10 min after cells were plated on the agarose pad and were selected randomly.
Role of the SAC in APC/CCdc20 substrate degradation. (A) Clb5 and securin degradation profiles in wild-type and mad2Δ cells. Cells are aligned using either SPB separation (left, broken line) or spindle elongation (right, solid line). Top panels show averaged and normalized traces as in Fig. 2, C and D. Bottom panels show the time of 50% substrate degradation in individual cells (Fig. S1 C). Each dot represents a single cell, and n > 90 cells per strain. For each strain, the middle bar indicates the median value, and error bars indicate the 25th and 75th percentiles. The inset in the bottom right panel shows a histogram of protein half-lives in different strains, calculated from single cell traces; n > 100 cells per strain. (B–D) Clb5 and securin degradation in nocodazole-treated cells; n > 20 cells per strain. Asynchronous cells were plated on agarose pads with 15 µg/ml nocodazole 10 min before imaging began. Representative traces from individual cells are normalized and aligned to spindle reformation (broken lines). The traces shown here were selected on the basis of two criteria: minimum overlap among traces for clarity of viewing, and inclusion of only mitotic cells, as judged by bud size. Wild-type (B and C) and mad2Δ cells (D) are shown. In B and C, representative cells with fast substrate degradation after recovery from the SAC arrest are shown in bold lines. (E) Clb5 degradation in nocodazole with CDC20 shut off; n > 20 cells. Asynchronous cells were grown in 2% galactose and plated on an agarose pad with 2% glucose and 15 µg/ml nocodazole. Representative traces began 10 min after cells were plated on the agarose pad and were selected randomly.
However, when cells were aligned with the onset of spindle elongation, substrate degradation timing was much less variable (Fig. 2 D, top; see also Fig. 3 A, bottom right), which is consistent with the causal relationship between APC/CCdc20 activation and anaphase onset. Compared with Clb5, the timing of securin degradation had a particularly strong correlation with spindle elongation, in agreement with the fact that securin degradation directly leads to sister chromatid separation and spindle elongation. Interestingly, Dbf4 was not only degraded at the same time as securin, but its degradation timing also strongly correlated with spindle elongation (Fig. 2 D, top; see also Fig. S3 C), which suggests some link in the regulation of their degradation. The first phase of Clb2 degradation also occurred immediately before spindle elongation, similar to the timing of securin and Dbf4 degradation, which is consistent with it being APC/CCdc20 dependent (Fig. 2 D).
The SAC determines the degradation timing of Clb5 but not that of securin
We next pursued the mechanisms underlying the sequential degradation of APC/CCdc20 substrates. To quantify and compare the timing of substrate degradation in each cell, we determined the time point when 50% of the substrate was degraded, and calculated its timing relative to the reference SPB events (Fig. S1, B and C; and Fig. 3 A, bottom; see also Materials and methods). Note that substrate degradation dynamics depend on two factors: the timing of degradation onset and the rate of degradation. Our quantification of the time of 50% degradation provides a combinatorial indication of both factors, and is also more robust than measuring the timing of degradation onset given the noise in our GFP signals (Fig. S1 C, unsmoothed traces). In most cases, we also calculated the rate constant of degradation by fitting each single-cell GFP trace to a single exponential decay, and we present these rates in terms of protein half-life (Fig. 3 A, inset; and Fig. S1 D; see also Materials and methods).
In mammalian cells, the SAC is known to influence the timing of APC/CCdc20 substrate degradation. To assess the contribution of the SAC in our system, we disabled the SAC by deleting MAD2 (Fig. 3 A) or MAD1 (Fig. S2 A), either of which is sufficient to abolish SAC activity (Li and Murray, 1991). To our surprise, disabling the SAC caused Clb5 degradation to occur several minutes earlier than in wild-type cells (Student’s t test, P < 0.001) but left securin degradation timing largely unchanged (Figs. 3 A and S2 A). These data suggest that the SAC normally inhibits Clb5 degradation, and that the timing of Clb5 degradation onset in the absence of the SAC likely indicates the time when APC/CCdc20 becomes active, possibly as a result of APC/C phosphorylation by Cdk1 (Rudner and Murray, 2000; Kraft et al., 2003). In addition, the rate of Clb5 degradation in wild-type cells was very similar to that in mad2Δ cells, if not slightly faster (Fig. 3 A, inset). This suggests that in wild-type cells the SAC is removed abruptly and that APC/CCdc20 is fully activated before Clb5 degradation begins.
The yeast SAC is known to be dispensable for growth in normal conditions but becomes essential under spindle stress. One possibility is that the SAC is activated only in cells that need it, and therefore in normal growth conditions the SAC is activated only in a small subset of cells with kinetochore attachment defects. If this were the case, then there would be a subpopulation of cells with delayed Clb5 degradation due to SAC activation. Disabling the SAC would eliminate this subpopulation and reduce the variability in the timing of Clb5 degradation in the whole population. However, our observations were inconsistent with this possibility. Disabling the SAC led to earlier Clb5 degradation in the entire population without a decrease in variability (Fig. 3 A and Fig. S2 A), which supports the idea that in yeast cells, as in mammalian cells, the SAC operates in most cells as an integral feature of cell cycle control.
Securin degradation was largely unaffected by deletion of SAC components (Figs. 3 A and S2 A), and the timing of anaphase onset was also unchanged (Fig. S2 B). These results are consistent with our evidence from Clb5 timing that the SAC is shut off and APC/CCdc20 is activated several minutes before the onset of securin degradation.
Clb5 can be degraded during an active SAC arrest
Given that transient SAC activation in a normal cell cycle delays Clb5 degradation, we wondered whether a prolonged SAC activation would fully stabilize it. We thus plated cells on media containing the microtubule poison nocodazole, which prevents spindle formation and thereby produces a sustained SAC signal. We observed collapsed spindles immediately after nocodazole treatment, which indicates an active SAC. Interestingly, we also observed that after 1–2 h in the arrest, cells began to assemble a spindle and progress into anaphase, perhaps because nocodazole was inactivated under our experimental conditions. We thus used spindle reformation as a single-cell timing marker, before which the cells should have an active SAC and after which cells are recovering from the SAC arrest.
Consistent with previous observations made on a population level, an active SAC inhibited Clb5 degradation but did not fully stabilize the protein (Keyes et al., 2008). Clb5 was degraded slowly in a nearly linear fashion (Fig. 3 B, before spindle reformation), even though securin was fully stabilized (Fig. 3 C, before spindle reformation). Disabling the SAC by deletion of MAD2 allowed degradation of Clb5 and securin in nocodazole at a normal rate in the absence of a spindle (Fig. 3 D). When we shut off CDC20 expression from a galactose-inducible promoter (Mumberg et al., 1994), Clb5 was fully stabilized in the presence or absence of nocodazole (Fig. 3 E; Keyes et al., 2008), which indicates that the slow degradation in nocodazole depended on APC/CCdc20. We suspect that this slow degradation of Clb5 also occurs in a normal cell cycle, during the brief 4-min time window after APC/CCdc20 becomes active toward Clb5 (indicated by the onset of Clb5 degradation in mad2Δ cells in Fig. 3 A) and before the SAC is turned off (indicated by the onset of fast Clb5 degradation in wild-type cells in Fig. 3 A). However, because this time window is so short, and Clb5 degradation during an active SAC is so slow, this partial Clb5 degradation is not noticeable in wild-type cells.
All nocodazole-treated cells eventually assembled a spindle and entered anaphase after 1–2 h on the nocodazole plate. The reassembly of spindles in these cells suggested that escape from the arrest was due to proper bi-orientation of sister chromatids, and thus inactivation of the SAC, rather than checkpoint adaptation (Vernieri et al., 2013). This fortuitous escape from the checkpoint allowed us to make interesting additional observations. Soon after reformation of the spindle, both Clb5 and securin underwent rapid degradation with a rate very similar to that in an unperturbed cell cycle (Fig. 3, B and C, after spindle reformation), which indicates abrupt activation of APC/CCdc20 upon SAC inactivation, as observed in unperturbed wild-type cells.
Phosphorylation by Cdk1 delays securin and Dbf4 degradation
To further address the mechanisms that determine the differences in the timing of Clb5 and securin degradation, we studied the influence of Cdk1-dependent phosphorylation on securin degradation. Phosphorylation near its KEN and D boxes (Thr27 and Ser71) was shown previously to inhibit securin ubiquitination by APC/CCdc20 in vitro, but it was unclear how this phosphorylation influences the rate or timing of securin degradation in vivo (Holt et al., 2008). To determine the effects of this phosphorylation, we replaced the endogenous copy of the securin gene with a mutant encoding securin-2A (T27A and S71A). The securin-2A mutant was degraded 2 min earlier than the wild-type protein (Fig. 4 A; P < 0.001), revealing that phosphorylation normally delays securin degradation. Interestingly, a larger fraction of securin-2A was degraded at the onset of spindle elongation compared with wild-type securin (Fig. 4 B; P < 0.001). This delay between securin-2A degradation and spindle elongation compensated for the earlier degradation of securin-2A to result in only a small but reproducible decrease in the time between SPB separation and spindle elongation (Fig. S3 A). In addition, securin-2A was degraded at a slightly greater rate than the wild-type protein (Fig. 4 A, inset).
Role of phosphorylation by Cdk1 in APC/CCdc20 substrate degradation. (A) Degradation profiles of GFP-tagged securin-2A, wild-type securin, and Clb5, as in Fig. 3 A. n > 70 cells per strain, and in the inset, n > 160 cells per strain. (B) Fraction of securin or securin-2A remaining when spindle elongation occurs. Single-cell traces of GFP were smoothed, and the fraction remaining was calculated as the GFP intensity at spindle elongation divided by maximum GFP intensity. Each dot represents a single cell (n > 100 cells per strain). For each strain, the middle bar indicates the median value and error bars indicate the 25th and 75th percentiles. (C) Dbf4 ubiquitination by APC/CCdc20 in vitro. Radiolabeled Dbf4 N-terminal fragment (residues 1–236) was produced by in vitro translation and incubated with buffer, purified Clb2-Cdk1, or both Clb2-Cdk1 and Cdc14, before the addition of purified APC/C, Cdc20, and other ubiquitination components for the indicated times. Reaction products were separated by SDS-PAGE and analyzed by autoradiography. (D) Dbf4 and Dbf4-A ubiquitination by APC/CCdc20 or APC/CCdh1 in vitro, as in C. (E) Degradation profiles of Dbf4-A-GFP and wild-type Dbf4-GFP, as in Fig. 3 A; n > 70 cells per strain.
Role of phosphorylation by Cdk1 in APC/CCdc20 substrate degradation. (A) Degradation profiles of GFP-tagged securin-2A, wild-type securin, and Clb5, as in Fig. 3 A. n > 70 cells per strain, and in the inset, n > 160 cells per strain. (B) Fraction of securin or securin-2A remaining when spindle elongation occurs. Single-cell traces of GFP were smoothed, and the fraction remaining was calculated as the GFP intensity at spindle elongation divided by maximum GFP intensity. Each dot represents a single cell (n > 100 cells per strain). For each strain, the middle bar indicates the median value and error bars indicate the 25th and 75th percentiles. (C) Dbf4 ubiquitination by APC/CCdc20 in vitro. Radiolabeled Dbf4 N-terminal fragment (residues 1–236) was produced by in vitro translation and incubated with buffer, purified Clb2-Cdk1, or both Clb2-Cdk1 and Cdc14, before the addition of purified APC/C, Cdc20, and other ubiquitination components for the indicated times. Reaction products were separated by SDS-PAGE and analyzed by autoradiography. (D) Dbf4 and Dbf4-A ubiquitination by APC/CCdc20 or APC/CCdh1 in vitro, as in C. (E) Degradation profiles of Dbf4-A-GFP and wild-type Dbf4-GFP, as in Fig. 3 A; n > 70 cells per strain.
Similar results were obtained with Dbf4. We found that Cdk1 inhibited Dbf4 ubiquitination by the APC/C in vitro, and the effects of Cdk1 were reversed by the phosphatase Cdc14 (Fig. 4 C). Dbf4 has two putative D boxes starting at Arg10 and Arg62. It was previously shown that mutating Arg62 and Leu65 to alanines stabilized Dbf4 in vivo (Ferreira et al., 2000), but we found that mutating Arg10 and Leu13 had a more dramatic inhibitory effect on the ubiquitination of the Dbf4 N-terminal fragment by the APC/C in vitro (Fig. S3 B). Furthermore, Dbf4 is phosphorylated by Cdk1 at Ser11 in vivo (Holt et al., 2009), prompting us to make a Dbf4-A mutant in which Ser11 is mutated to alanine. The ubiquitination of Dbf4-A by APC/C was not inhibited by Cdk1 in vitro (Fig. 4 D). Like securin-2A, Dbf4-A was degraded slightly earlier than the wild-type protein (Fig. 4 E). Although the difference was small, it was consistent whether we synchronized cells to SPB separation (Fig. 4 E; P = 0.035) or to spindle elongation (Fig. S3 C; P < 0.001). Thus, Dbf4 and securin are governed by similar Cdk1-dependent regulatory mechanisms, perhaps explaining why they are degraded simultaneously and why Dbf4 degradation is strongly correlated with spindle elongation.
DNA damage is also thought to inhibit securin degradation through Chk1-dependent phosphorylation of securin at several non-Cdk sites (Wang et al., 2001; Agarwal et al., 2003). We deleted CHK1 in the securin-2A strain and did not observe any effect on the timing of securin-2A degradation (Fig. S3 D), which suggests that this branch of the DNA damage response does not have a significant impact on mitotic timing in an unperturbed cell cycle.
Cks1 binding promotes early degradation of Clb5
Our results indicate that securin phosphorylation accounts for only a part of the difference in the timing of Clb5 and securin degradation. We therefore considered the possibility that there is some feature of Clb5 that promotes its early degradation, perhaps by making it a better APC/CCdc20 substrate. First, we replaced the N-terminal 95 residues of Clb5 with the N-terminal 110 residues of securin-2A. These N-terminal regions contain all of the known APC/CCdc20 binding motifs. This Clb5 chimera was degraded only slightly later than wild-type Clb5 (Fig. S4 A). We therefore hypothesized that early Clb5 degradation depends primarily on features within the C-terminal region of Clb5, starting from residue 96.
The Clb5 C-terminal region contains the globular domain that binds and activates Cdk1 (Jeffrey et al., 1995). Interestingly, the early SAC-resistant degradation of mammalian cyclin A depends in part on its binding to the Cdk1–Cks1 complex (Wolthuis et al., 2008; Di Fiore and Pines, 2010). Yeast APC/CCdc20 can bind directly to Cks1, and this interaction promotes APC/CCdc20 phosphorylation by Cdk1 (Rudner and Murray, 2000). These results motivated us to test the contribution of Cdk1 and Cks1 to Clb5 degradation. Given their essential functions in cell cycle progression, we reasoned that any perturbation in Cdk1 or Cks1 would be likely to have ubiquitous effects on multiple cell cycle processes, in which case it would be difficult to pinpoint the direct role of these proteins in Clb5 degradation. Instead, we analyzed the degradation of a Clb5 mutant that cannot bind Cdk1. Based on structural homology and conservation in the cyclin family (see Materials and methods and Fig. S4 B), we identified four hydrophobic residues (Ile166, Phe169, Phe254, and Phe291) at the predicted Clb5–Cdk1 interface (Fig. S4 C) and mutated a combination of them to aspartate or arginine. We then assessed their interaction with Cdk1 in vivo. Ectopic expression of a stabilized Clb5 protein lacking its N-terminal region (Clb5-ΔN, with residues 2–95 deleted) under control of the CLB5 promoter is known to be lethal because of excess Clb5–Cdk1 activity (Sullivan et al., 2008). If our mutations disrupted Clb5–Cdk1 binding, then introduction of these mutants into Clb5-ΔN should prevent its dominant lethal effects. Indeed, when these mutants were expressed under the control of the CLB5 promoter (582 bp upstream of the CLB5 ORF) in an integration plasmid, we observed improved growth as we increased the number of mutations in Clb5-ΔN (Fig. S4 D), even though all mutants had a similar expression level in the cell (Fig. S4 E). When we combined three mutations (F169D, F254D, and F291D; henceforth the Clb5-3D mutant), the cells grew with a doubling time (85.4 ± 0.2 min) very similar to that of wild-type cells (84.1 ± 0.5 min); adding a fourth mutation (I166D, F169A, F254D, and F291D) did not further improve growth (85.6 ± 0.1 min). Furthermore, the Clb5-3D mutant almost completely failed to associate with Cdk1 in cell lysates (Fig. 5 A). We therefore used the Clb5-3D mutant for the following experiments.
Contribution of Cdk1–Cks1 to Clb5 early degradation. (A) Binding of Cdk1 by Clb5-GFP, Clb5-2D-GFP (I166D and F291D), and Clb5-3D-GFP (F169D, F254D, and F291D). Lysates of cells expressing wild-type or mutant Clb5-GFP from the endogenous CLB5 promoter were incubated with GFP-binding protein coupled to Sepharose beads. After washing, associated proteins were analyzed by SDS-PAGE and Western blotting with anti-GFP and anti-Cdk1 antibodies simultaneously. (B) Degradation profiles of wild-type Clb5-GFP, Clb5-3D-GFP, and Clb5-3D-GFP-Cks1, as in Fig. 3 A; n > 70 cells per strain. (C) Degradation profiles of securin-2A-Cks1-GFP, securin-2A-GFP, and Clb5-GFP in the securin-2A-Cks1 or securin-2A background, as in Fig. 3 A; n > 50 cells in the Clb5-GFP strains and n > 90 cells in the securin-GFP strains. In the inset, n > 100 cells per strain. (D) Fraction of securin-2A or securin-2A-Cks1 remaining when spindle elongation occurs, as in Fig. 4 B; n > 100 cells per strain.
Contribution of Cdk1–Cks1 to Clb5 early degradation. (A) Binding of Cdk1 by Clb5-GFP, Clb5-2D-GFP (I166D and F291D), and Clb5-3D-GFP (F169D, F254D, and F291D). Lysates of cells expressing wild-type or mutant Clb5-GFP from the endogenous CLB5 promoter were incubated with GFP-binding protein coupled to Sepharose beads. After washing, associated proteins were analyzed by SDS-PAGE and Western blotting with anti-GFP and anti-Cdk1 antibodies simultaneously. (B) Degradation profiles of wild-type Clb5-GFP, Clb5-3D-GFP, and Clb5-3D-GFP-Cks1, as in Fig. 3 A; n > 70 cells per strain. (C) Degradation profiles of securin-2A-Cks1-GFP, securin-2A-GFP, and Clb5-GFP in the securin-2A-Cks1 or securin-2A background, as in Fig. 3 A; n > 50 cells in the Clb5-GFP strains and n > 90 cells in the securin-GFP strains. In the inset, n > 100 cells per strain. (D) Fraction of securin-2A or securin-2A-Cks1 remaining when spindle elongation occurs, as in Fig. 4 B; n > 100 cells per strain.
We expressed Clb5-GFP or Clb5-3D-GFP under the control of the CLB5 promoter, using an integration plasmid. Both strains retained the endogenous copy of CLB5 to maintain a normal cell cycle. We found that Clb5-3D displayed two phases of degradation: a slow phase and a fast phase (Fig. 5 B). The slow phase displayed a nearly linear rate and was not dependent on Cdc20 (Fig. S5 A), and so it likely reflected nonspecific degradation of Clb5-3D due to the destabilizing effects of the mutations. The fast phase, however, disappeared if we shut off Cdc20 expression and thus reflected APC/CCdc20-dependent degradation (Fig. S5 A). This fast phase of Clb5-3D degradation was significantly delayed relative to the degradation of wild-type Clb5 (Fig. 5 B; P < 0.001), which suggests that Cdk1 binding contributes to early Clb5 degradation.
These results are consistent with a role for Cdk1 binding in Clb5 degradation, and this is most likely mediated through Cks1, which binds to both Cdk1 and APC/C (Patra and Dunphy, 1998; Shteinberg and Hershko, 1999; Rudner and Murray, 2000; Wolthuis et al., 2008; Di Fiore and Pines, 2010; van Zon et al., 2010). To directly test the role of Cks1, we fused Cks1 to the C terminus of Clb5-3D-GFP. The Cks1 fusion rescued the delay in Clb5-3D degradation (Fig. 5 B; P < 0.001), which indicates that Cks1 facilitates early degradation of Clb5.
We also fused Cks1-GFP to the C terminus of securin-2A and compared degradation of the fusion protein with that of securin-2A-GFP. In both cases, the endogenous copy of securin was replaced to ensure that the cells expressed only one securin variant, the degradation of which would drive sister chromatid separation. Securin-2A-Cks1 was degraded significantly earlier than securin-2A (Fig. 5 C; P < 0.001) and at a slightly faster rate. Accordingly, anaphase onset also occurred significantly earlier (Fig. S5 B; P < 0.001) and sooner relative to Clb5 degradation (Fig. 5 C, right). Interestingly, as in our earlier observations with securin-2A, more securin-2A-Cks1 was degraded than securin-2A when spindle elongation occurred (Fig. 5 D; P < 0.001), which suggests that securin degradation is not the sole determinant of anaphase onset.
However, Cdk1–Cks1 binding did not fully explain early Clb5 degradation relative to securin-2A: Clb5-3D was still degraded earlier than securin-2A (Fig. S5 C; P < 0.005). We suspected that additional mechanisms exist to promote early degradation of Clb5.
The Cdc20-binding “ABBA motif” contributes to Clb5 degradation in the presence or absence of an activated SAC
A short amino acid sequence motif in the yeast protein Acm1 interacts with the WD40 domain of Cdh1 at a site distinct from the binding sites for the D box and KEN box (Enquist-Newman et al., 2008; Burton et al., 2011; He et al., 2013). Recent studies suggest that a related motif exists in cyclin A and facilitates its early mitotic degradation via APC/CCdc20 (J. Pines, personal communication). Originally called the “A motif,” this motif was renamed the “ABBA” motif to reflect its conserved presence in Acm1, Bub1, BubR1, and cyclin A. We tested the possibility that a similar motif exists in Clb5 and helps promote early Clb5 degradation. We performed a motif search in Clb5 homologues from closely related yeast species of the Saccharomyces clade (Davey et al., 2012), and we found a putative ABBA motif at residues 99–105 in Clb5, within a highly conserved region (Fig. 6 A).
Contribution of the “ABBA motif” to Clb5 degradation. (A) Sequence alignment of Clb5 orthologues from closely related yeast species, showing the putative ABBA motif in Clb5 in alignment with other known ABBA motifs. Different colors represent chemical properties of the residues. (B) Degradation profiles of Clb5-2A-GFP and wild-type Clb5-GFP, as in Fig. 3 A; n > 70 cells per strain. In the inset, n > 170 cells per strain. (C) Degradation profiles of Clb5-2A3D-GFP and securin-2A-GFP, as in Fig. 3 A; n > 60 cells per strain. (D) Sequence alignment of budding yeast Cdc20 and Cdh1; purple dots mark the potential ABBA motif interacting residues that are different between Cdc20 and Cdh1. Below is the degradation profile of Clb5-2A-GFP or wild-type Clb5-GFP in a CDC20-GAG background, compared with the wild-type CDC20 background; n > 57 cells per strain. In the inset, n > 75 cells per strain. (E) Analysis of Clb5 and Clb5-2A ubiquitination by APC/CCdh1 or APC/CCdc20-GAG in vitro, as described in the Materials and methods. Reactions with securin were included as controls. (F) Degradation of Clb5-GFP and Clb5-2A-GFP in nocodazole-treated cells. Asynchronous cells were plated on an agarose pad with 15 µg/ml nocodazole 10 min before the start of imaging. Clb5-GFP dynamics before spindle reformation (SAC inactivation) were analyzed. Representative traces were selected to minimize overlap and omit cells that were not in mitosis. The right panel shows the rates of degradation calculated by fitting single-cell GFP traces to a linear decay; n > 55 cells per strain.
Contribution of the “ABBA motif” to Clb5 degradation. (A) Sequence alignment of Clb5 orthologues from closely related yeast species, showing the putative ABBA motif in Clb5 in alignment with other known ABBA motifs. Different colors represent chemical properties of the residues. (B) Degradation profiles of Clb5-2A-GFP and wild-type Clb5-GFP, as in Fig. 3 A; n > 70 cells per strain. In the inset, n > 170 cells per strain. (C) Degradation profiles of Clb5-2A3D-GFP and securin-2A-GFP, as in Fig. 3 A; n > 60 cells per strain. (D) Sequence alignment of budding yeast Cdc20 and Cdh1; purple dots mark the potential ABBA motif interacting residues that are different between Cdc20 and Cdh1. Below is the degradation profile of Clb5-2A-GFP or wild-type Clb5-GFP in a CDC20-GAG background, compared with the wild-type CDC20 background; n > 57 cells per strain. In the inset, n > 75 cells per strain. (E) Analysis of Clb5 and Clb5-2A ubiquitination by APC/CCdh1 or APC/CCdc20-GAG in vitro, as described in the Materials and methods. Reactions with securin were included as controls. (F) Degradation of Clb5-GFP and Clb5-2A-GFP in nocodazole-treated cells. Asynchronous cells were plated on an agarose pad with 15 µg/ml nocodazole 10 min before the start of imaging. Clb5-GFP dynamics before spindle reformation (SAC inactivation) were analyzed. Representative traces were selected to minimize overlap and omit cells that were not in mitosis. The right panel shows the rates of degradation calculated by fitting single-cell GFP traces to a linear decay; n > 55 cells per strain.
To test the function of the putative ABBA motif in Clb5, we replaced the key residues Ile102 and Tyr103 with alanines to generate the Clb5-2A mutant. Clb5-2A was degraded significantly later than Clb5 (Fig. 6 B; P < 0.001), but at a very similar rate (Fig. 6 B, inset). We also analyzed a Clb5 mutant (Clb5-2A3D) in which both Cdk1 binding and the ABBA motif were disrupted. The rapid phase of degradation of this mutant now occurred slightly later than the degradation of securin-2A (Fig. 6 C).
The ABBA motifs in Clb5 homologues differ from those in Acm1 and cyclin A by having conserved basic residues upstream of the core Ile102 and Tyr103 (Fig. 6 A). We wondered whether this was accompanied by differences in the ABBA motif binding site on Cdc20. Based on homology modeling of the Acm1-Cdh1 structure (He et al., 2013), we identified residues on Cdc20 that potentially interact with the ABBA motif, and we compared them to those on Cdh1. One striking difference was a cluster of acidic residues (Asp311, Asp312, and Asp313) in Cdc20 that are absent in Cdh1 (Fig. 6 D). If these residues are important for binding to the basic residues in the Clb5 ABBA motif, then replacing the endogenous CDC20 with a CDC20-GAG (D311G, D312A, and D313G) mutant should delay wild-type Clb5 degradation but have little impact on Clb5-2A degradation. Indeed, this is what we observed (Fig. 6 D; P < 0.001 and P = 0.8775, respectively).
We confirmed these findings with APC/C-dependent ubiquitination reactions in vitro. In reactions with wild-type Cdc20 as the activator, the ubiquitination of Clb5-2A was less efficient than that of wild-type Clb5. Consistent with this result, wild-type Clb5 was less efficiently modified in reactions with the Cdc20-GAG mutant, and ubiquitination of Clb5-2A was similar in reactions with wild-type and mutant Cdc20 (Fig. 6 E).
We conclude that the early degradation of Clb5 in a normal cell cycle depends mostly, if not entirely, on Cdk1–Cks1 and the ABBA motif, both of which provide additional binding sites for APC/CCdc20. We also tested the effect of ABBA motif disruption on Clb5 degradation in nocodazole. Compared with wild-type Clb5, the Clb5-2A mutant was still degraded in a linear fashion but at a significantly slower rate (Fig. 6 F). Thus, the ABBA motif also contributes to Clb5 degradation in an active SAC.
Discussion
Our results, together with those from previous single-cell studies, provide a detailed temporal picture of how yeast cells progress toward the metaphase–anaphase transition (Fig. 7). The process begins with inactivation of the SAC, which inhibits APC/CCdc20 activity until all sister chromatids are properly attached to the spindle. Activated APC/CCdc20 first degrades the S cyclin Clb5 with a mean half-life of 3.4 min. About 6 min later, securin is degraded with a mean half-life of 4.7 min. Soon after securin degradation begins, separase is abruptly activated, and only 1 min is required for separase to cleave enough cohesin to promote sister chromatid separation (Yaakov et al., 2012). By the time of sister chromatid separation, Clb5 is fully degraded but more than half of securin remains.
APC/C substrate degradation timing in yeast and mammalian cells. (A) Summary of the regulatory events leading to the metaphase–anaphase transition in yeast and mammalian cells, and the mechanisms that determine the timing and order of APC/CCdc20 substrate degradation in wild-type cells. (B) In yeast, defects in SAC function cause earlier Clb5 destruction but do not affect the timing of securin destruction. In contrast, in mammalian cells, SAC defects result in earlier cyclin A and securin destruction and anaphase onset. (C) In a prolonged SAC arrest, securin is stable, whereas Cks1 and the ABBA motif promote slow degradation of Clb5 and cyclin A. The diagrams in this figure are based on the current work and many previous studies (J. Pines, personal communication; Clute and Pines, 1999; den Elzen and Pines, 2001; Geley et al., 2001; Hagting et al., 2002; Wolthuis et al., 2008; Di Fiore and Pines, 2010; Shindo et al., 2012; Yaakov et al., 2012; Collin et al., 2013; Dick and Gerlich, 2013).
APC/C substrate degradation timing in yeast and mammalian cells. (A) Summary of the regulatory events leading to the metaphase–anaphase transition in yeast and mammalian cells, and the mechanisms that determine the timing and order of APC/CCdc20 substrate degradation in wild-type cells. (B) In yeast, defects in SAC function cause earlier Clb5 destruction but do not affect the timing of securin destruction. In contrast, in mammalian cells, SAC defects result in earlier cyclin A and securin destruction and anaphase onset. (C) In a prolonged SAC arrest, securin is stable, whereas Cks1 and the ABBA motif promote slow degradation of Clb5 and cyclin A. The diagrams in this figure are based on the current work and many previous studies (J. Pines, personal communication; Clute and Pines, 1999; den Elzen and Pines, 2001; Geley et al., 2001; Hagting et al., 2002; Wolthuis et al., 2008; Di Fiore and Pines, 2010; Shindo et al., 2012; Yaakov et al., 2012; Collin et al., 2013; Dick and Gerlich, 2013).
The SAC is not essential for yeast viability under normal growth conditions (Hoyt et al., 1991; Li and Murray, 1991), and thus it has not been clear what role, if any, the SAC plays in the normal timing of yeast mitotic regulatory events. Our experiments now reveal that the SAC is activated in most yeast cells as an integral part of progression through mitosis. Compared with the SAC in mammalian cells, however, the yeast checkpoint appears to be inactivated relatively early in mitosis and determines the timing of S cyclin degradation and not that of securin. Our results are consistent with the observation that, in yeast, bi-orientation of sister chromatids on the spindle begins immediately after spindle assembly and is possibly complete many minutes before anaphase onset (Goshima and Yanagida, 2000; He et al., 2000; Tanaka et al., 2000; Pearson et al., 2001). Thus, the nonessential nature of the SAC may be due, at least in part, to the waiting period between the proper attachment of sister chromatids (and SAC inactivation) and their separation. Even without the surveillance provided by the SAC, the sister chromatid pairs would normally achieve proper attachment to the spindle minutes before securin degradation triggers their separation.
With the SAC turned off or disabled, we found that the ordered degradation of Clb5, securin, and Dbf4 is established primarily through differences in their interaction with APC/CCdc20. Cdk1-dependent phosphorylation near KEN and D boxes in securin and Dbf4 can delay their degradation, and this similarity in regulation results in almost simultaneous degradation of these substrates. We also found that the early degradation of Clb5 depends on two factors that provide additional binding sites for APC/CCdc20: the interaction of Clb5 with the Cdk1–Cks1 complex and the presence of the ABBA motif in the Clb5 N-terminal region. We still lack a complete mechanistic understanding of how these factors influence the interaction between substrates and APC/CCdc20 inside the cell. They could simply improve substrate-binding affinity for APC/CCdc20, or they might help orient substrates (or lysines in those substrates) for efficient ubiquitin transfer. They might also provide selectivity for specific subpopulations of APC/CCdc20. For example, Cks1 is thought to interact with the phosphorylated subpopulation of the APC, which has higher binding affinity for Cdc20 (Rudner and Murray, 2000).
We can only speculate about how differences in binding interactions with APC/CCdc20 are converted to a robust ordering of substrate degradation. One can imagine two possible scenarios that are not mutually exclusive. In a “threshold” model, APC/CCdc20 activity continues to rise after its initial activation, and efficient ubiquitination of securin or Dbf4 might require a higher level of APC/CCdc20 activity than that of Clb5. Such thresholds could be established by changes in docking interactions between substrates and APC/CCdc20, as well as by phosphoregulation of APC/CCdc20. In an alternative “competition” model, the amount of active APC/CCdc20 is limiting, and substrates compete with each other for APC/CCdc20 binding. In this case, APC/CCdc20 is initially occupied by higher-affinity substrates such as Clb5, and only after destruction of these substrates can efficient securin and Dbf4 ubiquitination begin. Our results seem to argue against this model and are more consistent with a threshold model. For example, when we measured the timing of Clb5-2A degradation, we deleted the endogenous copy of Clb5. If Clb5 competitively inhibited the degradation of lower-affinity substrates, then the prediction would be that destruction of the Clb5-2A mutant would not be delayed because its Clb5 competitor was absent. A complete understanding of substrate ordering will require more detailed quantitative analyses of APC/CCdc20 activation, phosphoregulation, and localization in the cell.
A major difference between mammalian and yeast cells is that cyclin A is thought to be degraded in the presence of an active SAC and needs to compete with SAC proteins for APC/CCdc20 binding (den Elzen and Pines, 2001; Geley et al., 2001), whereas Clb5 degradation appears to occur just after the SAC is turned off. Interestingly, despite these very different circumstances, the same two mechanisms—Cks1 and the ABBA motif—allow cyclin A and Clb5 to be degraded earlier than other substrates (J. Pines, personal communication; Wolthuis et al., 2008; Di Fiore and Pines, 2010). It was shown recently that the degradation of cyclin A and securin seems to remain sequential in mammalian cells without a functional SAC (Collin et al., 2013). We suspect that in this scenario, the same mechanisms promote cyclin A degradation earlier than that of other substrates.
We found that Clb5, like cyclin A, is degraded in cells with an active SAC, but the rate of degradation was much slower than that in the absence of the SAC (Keyes et al., 2008). This slow degradation depends on Cdc20 and on the ABBA motif, which suggests that this motif is capable of driving some interaction with APCCdc20 even in the presence of an active SAC. We also suspect that Clb5 degradation in the presence of the SAC depends on Cdk1–Cks1 binding, but we could not test this possibility due to the intrinsic instability of our Clb5-3D mutant.
Securin degradation leads to sister chromatid separation, but the timing of sister separation also seems to depend on other factors. Among the different variants of securin we tested, including the wild-type protein, securin-2A, and securin-2A-Cks1, earlier degradation correlated with an increase in the amount of securin that was degraded before anaphase onset. This could suggest another branch of regulation in the timing of sister chromatid separation. Indeed, the cohesin subunit Scc1 is phosphorylated by Polo kinase, which increases the rate of cleavage by separase by several-fold (Alexandru et al., 2001; Yaakov et al., 2012). One can imagine that when securin is degraded early, and separase is released early, Scc1 is not yet fully phosphorylated and cohesin cleavage will take longer to complete. Consistent with this idea, Scc1 is cleaved more slowly in securin-2A cells than in wild-type cells (Yaakov et al., 2012).
Our results suggest that there is a 9-min delay between the completion of sister chromatid bi-orientation (SAC satisfaction) and the initiation of sister chromatid separation via securin degradation. Does this time delay serve a purpose? One possibility is that the delay allows time for complete Clb5 degradation before anaphase begins. Clb5–Cdk1 phosphorylates numerous specific substrates that have functions in anaphase (Loog and Morgan, 2005), and these functions are inhibited by Cdk1-dependent phosphorylation. These substrates include the spindle stabilizer Fin1 (Woodbury and Morgan, 2007), the spindle midzone organizer Ase1 (Juang et al., 1997; Khmelinskii et al., 2009), the SPB component Spc110 (Kilmartin et al., 1993; Lianga et al., 2013), the late mitotic APC/C activator Cdh1 (Visintin et al., 1997; Jaspersen et al., 1999), and the kinetochore component Cnn1 (Bock et al., 2012; Schleiffer et al., 2012). Several of these proteins are dephosphorylated by Cdc14 (Jaspersen et al., 1999; Woodbury and Morgan, 2007; Khmelinskii et al., 2009). The early degradation of Clb5, which is completed by the onset of anaphase and coincides with activation of Cdc14, may allow earlier and more abrupt activation of these Clb5 substrates and lead to a more efficient and coherent anaphase. Indeed, removing securin phosphorylation, which disturbs the coordination between Clb5 degradation and anaphase onset, was shown to impede spindle elongation and increase chromosome loss (Holt et al., 2008). Stabilized Clb5 has also been shown to slow down spindle elongation (Lianga et al., 2013) and delay ribosomal DNA (rDNA) segregation (Sullivan et al., 2008). There is also recent evidence in mammalian cells that cyclin A destruction before anaphase is important for the stabilization of kinetochore–microtubule attachments (Kabeche and Compton, 2013). Thus, differences in the relative timing of cyclin and securin degradation are likely to make important contributions to the overall orchestration of mitosis.
Materials and methods
Yeast strain construction
All yeast strains were haploid derivatives of the W303 strain (Table S1). Fluorescent protein tagging, gene replacement, and deletion of genes at their endogenous loci were performed using standard PCR-based homologous recombination (Baudin et al., 1993; Longtine et al., 1998; Goldstein and McCusker, 1999; Jansen et al., 2005), while preserving the endogenous promoters. Addition of genes to the genome was done using an integration plasmid at the genomic TRP1 locus (Sikorski and Hieter, 1989), with promoters as indicated in Table S1, and selected for single-copy integration by PCR and fluorescence intensity.
Fluorescence microscopy
All images were taken with a spinning-disk confocal microscope at the UCSF Nikon Imaging Center with a 60×/1.4 NA oil immersion objective lens, under the control of µManager (Edelstein et al., 2010). The microscope was an inverted microscope (Ti-E; Nikon) equipped with a scanner unit (CSU-22; Yokogawa) and a camera (Evolve EMCCD; Photometrics). Illumination was provided by a 50-mW, 491-nm laser and a 50-mW, 561-nm laser. Imaging sessions were generally 1 h long, or 1.5 h for nocodazole experiments, with 30- or 45-s time intervals. Z stacks were taken across 4 µm of distance with 0.5 µm steps for each time point and each channel. Exposure times for mCherry and GFP channels were <40 ms for each z slice.
All yeast cultures were grown and imaged at 30°C. Before imaging, yeast cells were grown in synthetic complete media with 2% glucose (SD) for 24 h with serial dilution to maintain OD <0.4. For imaging, cells were mounted on a 1.5% agarose pad made with SD, and allowed to continue proliferating on the slide for 40–60 min in a 30°C incubator before imaging. For nocodazole experiments, cells were plated onto an agarose pad containing 15 µg/ml nocodazole for 5–10 min before imaging. For experiments involving the shutting off of galactose-induced promoters, cells were grown in 2% galactose for 24 h, and plated onto an agarose pad containing 2% glucose for 5–10 min before imaging.
All single-cell data in the same plot were obtained within the same week. For every strain, data represent two or three repeats with different transformants. Differences between transformants were negligible, and figures show combined results from all repeats.
Optimization of imaging conditions
To minimize phototoxicity while retaining sufficient temporal resolution and dynamic range of the fluorescent signal, we optimized our imaging conditions in several ways. First, the specific setup of the spinning-disk confocal microscope at the UCSF Nikon Imaging Center allowed much shorter exposure times and more frequent time points than with other microscopes we tested. Second, the yeast nitrogen base in our SD was a significant source of background fluorescence, and we found that fresh yeast nitrogen base from Sigma-Aldrich had less autofluorescence than others. Autoclaving SD ingredients also raised the autofluorescence level, so all media was filtered instead. Third, during the imaging process, we took one frame every 30 or 45 s, which was just enough to capture the features of the dynamics of substrate degradation. For fitting of the degradation rate, a 30-s time interval was the minimum required to obtain enough data points during one degradation event for a good fit. We also took short movies of ∼1 h, which covers only one round of mitosis. Last, we used the minimum level of laser intensity and exposure time to generate fluorescent signals that were minimally sufficient for quantification.
Image processing
To quantify GFP intensity at each time point, we first used ImageJ (Schneider et al., 2012) and its plugin Image5D (http://rsb.info.nih.gov/ij/plugins/image5d.html) to average across each z stack and flatten it to 2D. GFP intensity was then quantified using MATLAB (The MathWorks, Inc.) code previously developed in the Tang laboratory (Yang et al., 2013). Because all of the APC/C substrates we studied were localized to the nucleus, we took the brightest square of 5 × 5 pixels in the cell as an estimate of the protein level (Fig. S1 B). Timing of SPB events was determined based on the temporal 3D positions of the SPB using the mCherry images (Supplemental code S1). SPB separation was defined as the time point when one SPB split into two, and spindle elongation was defined as the time point when two SPBs began to move rapidly away from each other.
Data processing
The time point of 50% substrate degradation in each cell was defined as when GFP intensity was halfway between the maximum and the minimum intensity on a smoothed trace of a degradation event (Fig. S1 C). For Clb5-3D strains in which slow degradation occurred before the fast degradation by APC/CCdc20, the maximum intensity was replaced by the intensity right before the fast degradation began, so that the resulting 50% degradation point corresponds to the midpoint of the fast degradation. Determination of the timing of the 50% drop of GFP signal, or the level of GFP at a certain time point, was performed with newly developed MATLAB code (Supplemental code S2; Crocker and Grier, 1996). The rate of substrate degradation in single cells was calculated by fitting the fast decreasing section of each GFP trace, spanning at least seven time points for a robust fit, to a single exponential decay (Fig. S1 D) with MATLAB code previously developed in the Tang laboratory (Yang et al., 2013). Statistical analysis and plotting were performed in MATLAB and Python (Hunter, 2007; Oliphant, 2007).
Ubiquitination assays in vitro
For analysis of APC/C activity with phosphorylated Dbf4 (Fig. 4, C and D; and Fig. S3 B), wild-type and mutant Dbf4 substrates, carrying a C-terminal ZZ tag and tobacco etch virus (TEV) cleavage site, were translated in vitro with TnT Quick Coupled Transcription/Translation Systems (Promega) in the presence of [35S]methionine. Substrates were immobilized on IgG beads and incubated at 23°C for 60 min with purified Clb2-Cdk1 in kinase buffer (25 mM Hepes, pH 7.4, 150 mM NaCl, 10 mM MgCl2, 1 mM ATP, and 5% glycerol). Beads were washed with QAH buffer (20 mM Hepes, pH 7.4, 150 mM NaCl, 1 mM MgCl2, and 10% glycerol) and cleaved with TEV protease to produce soluble radiolabeled Dbf4 substrates. APC/C was purified from lysates of TAP-CDC16 cdh1Δ W303 strains by affinity chromatography with IgG beads, followed by elution with TEV protease (Carroll and Morgan, 2002). E1 (Uba1-6His) was expressed in yeast and purified by metal-affinity chromatography (Carroll and Morgan, 2002). E2 (Ubc4-6His) was expressed in bacteria and purified by metal-affinity chromatography (Rodrigo-Brenni and Morgan, 2007). Activator was tagged with an N-terminal ZZ tag and TEV cleavage site, and produced with TnT Quick Coupled Transcription/Translation Systems (Promega), followed by purification on IgG beads and TEV cleavage (Foster and Morgan, 2012). E2 charging was performed in the presence of E1 (Uba1, 300 nM), E2 (Ubc4, 50 µM), methyl-ubiquitin (150 µM; Boston Biochem), and ATP (1 mM) at 23°C for 20 min. APC/C reactions were initiated by mixing APC/C, activator, substrate, and charged E2. Reactions were performed at 23°C for 60 min (Dbf4). Reaction products were separated by SDS-PAGE and visualized with a PhosphorImager (Molecular Dynamics).
For analysis of APC/C activity with securin and Clb5 (Fig. 6 E), lysate was prepared from an ∼300 mg pellet of TAP-CDC16 cdh1Δ W303 cells, and APC/C was immobilized on IgG-coupled Dynabeads (Invitrogen) as described previously (Matyskiela and Morgan, 2009). The final concentration of APC/C in each reaction was ∼1 nM. ZZ-tagged substrates were generated in vitro with TnT Quick Coupled Transcription/Translation Systems (Promega) in the presence of [35S]methionine, purified with IgG-coupled Dynabeads, and cleaved using TEV protease. E2 charging and ZZ-tagged activator purification were performed as described in the previous paragraph. APC/C reactions were initiated by the addition of activator, substrate, and charged E2 to immobilized APC/C, and incubated at 23°C for 30 min. Reaction products were separated by SDS-PAGE and visualized with a PhosphorImager.
Design of Clb5 mutant with a Cdk-binding defect
We first used HOMCO (Fukuhara and Kawabata, 2008) to build homology models of the Clb5–Cdk1 complex based on several available cyclin–Cdk structures, including cyclin B–Cdk2 (2jgz; Brown et al., 2007), cyclin A–Cdk2 (1jst; Russo et al., 1996), and cyclin E–Cdk2 (1w98; Honda et al., 2005). Our initial pool of candidate residues was selected from residues predicted to be at the cyclin–Cdk interface. We then selected the residues for mutation using the following criteria: they should be present at the Clb5–Cdk interface in all of the three homologous structures; they should not participate in intramolecular interactions, so that mutations in those residues are less likely to destabilize Clb5; and they should be conserved in all budding yeast cyclins, which all bind to the same Cdk1 (Fig. S4 B). This narrowed the candidates down to four residues (I166, F169, F254, and F291), as shown in Fig. S4 C (Pettersen et al., 2004). We then made several Clb5-ΔN mutants (with residues 2–95 deleted; Sullivan et al., 2008) containing a combination of these residues mutated to arginine or aspartate and expressed them under control of the CLB5 promoter (582 bp upstream of the CLB5 ORF). Cells expressing either wild-type Clb5-ΔN or Clb5-ΔN with a single mutation of F254D were inviable. Clb5-ΔN with double mutations inhibited cell growth more than triple mutations (Fig. S4 D). Cells with Clb5-ΔN triple mutations and quadruple mutations all grew at a rate similar to the parent strain without Clb5-ΔN.
To analyze Cdk1 binding to Clb5 mutants, log phase cells with full-length wild-type or mutant Clb5-GFP expressed under the endogenous CLB5 promoter were lysed by bead-beating in lysis buffer (50 mM Hepes, pH 8.0, 150 mM NaCl, 1% NP-40, 50 mM β-glycerophosphate, 50 mM NaF, 1 mM DTT, 1 µg/ml leupeptin, 1 µg/ml pepstatin, 1 µg/ml aprotinin, 1 mM PMSF, 10% glycerol, 0.63 mg/ml benzamidine, and 5 mM EDTA). Lysates were incubated with GFP-binding protein (Rothbauer et al., 2008) covalently coupled to Sepharose beads (NHS-Activated Sepharose 4 Fast Flow; GE Healthcare) at 4°C for 30 min. The beads were then washed with lysis buffer, and associated proteins were analyzed by Western blotting with a mixture of anti-Cdk1 (Cdc2 p34 [PSTAIRE], sc-53; Santa Cruz Biotechnology, Inc.) and anti-GFP (GFP-FL, sc-8334; Santa Cruz Biotechnology, Inc.).
Online supplemental material
Fig. S1 shows a control experiment and illustrates the data processing methods used in image analysis. Fig. S2 provides additional evidence for the role of the SAC in early Clb5 destruction. Fig. S3 provides additional results to support a role for substrate phosphorylation in the control of APC/C substrate degradation. Fig. S4 describes the design and analysis of Clb5 mutants with Cdk1-binding defects. Fig. S5 provides additional evidence for the role of Cks1 in substrate degradation. Table S1 lists the yeast strains used in this study. Code S1 provides the MATLAB codes used to track SPB positions. Code S2 provides the MATLAB codes used to determine the timing of the 50% drop of GFP intensity.
Acknowledgments
We thank Hana El-Samad, Jaline Gerardin, Wendell A. Lim, Xili Liu, Geoff C. Rollins, Jacob Stewart-Ornstein, Orion D. Weiner, Xiaojing Yang, and members of the Morgan and Tang laboratories for inspiring discussions and comments on the manuscript; Kurt Thorn, Alice Thwin, and DeLaine Larsen at the UCSF Nikon Imaging Center for their critical help with microscopy; Heather Eshleman, Matilde Galli, and Arda Mizrak for assistance; and Jon Pines and Barbara Di Fiore for sharing results before publication.
This work was supported by a fellowship (to D. Lu) from the UC Cancer Research Coordinating Committee (CRCC) and by funding from the National Institute of General Medical Sciences (R01-GM097115 to C. Tang and R37-GM053270 to D.O. Morgan).
The authors declare no competing financial interests.