Efficient and accurate protein localization is essential to cells and requires protein-targeting machineries to both effectively capture the cargo in the cytosol and productively unload the cargo at the membrane. To understand how these challenges are met, we followed the interaction of translating ribosomes during their targeting by the signal recognition particle (SRP) using a site-specific fluorescent probe in the nascent protein. We show that initial recruitment of SRP receptor (SR) selectively enhances the affinity of SRP for correct cargos, thus committing SRP-dependent substrates to the pathway. Real-time measurement of cargo transfer from the targeting to translocation machinery revealed multiple factors that drive this event, including GTPase rearrangement in the SRP–SR complex, stepwise displacement of SRP from the ribosome and signal sequence by SecYEG, and elongation of the nascent polypeptide. Our results elucidate how active and sequential regulation of the SRP–cargo interaction drives efficient and faithful protein targeting.
Introduction
To ensure proper biogenesis of proteins in the crowded cellular environment, cells have evolved sophisticated molecular machineries that are recruited to the polypeptide exit site of the ribosome early in translation (Kramer et al., 2009). An example is the universally conserved signal recognition particle (SRP), which delivers ∼30% of the proteome to the eukaryotic ER or the bacterial plasma membrane (Keenan et al., 2001). SRP recognizes the N-terminal signal sequence of a protein as it emerges from the translating ribosome (Fig. 1 A, Recognition). The ribosome nascent chain complex (RNC; or cargo) is delivered to the target membrane via interactions of SRP with its receptor (SR; Fig. 1 A, Targeting). At the membrane, RNC is transferred to the Sec61p or SecYEG translocation machinery (Rapoport, 2007), where the nascent protein is either integrated into or translocated across the membrane (Cross et al., 2009). GTP hydrolysis disassembles SRP and SR and recycles them for additional rounds of targeting (Fig. 1 A, last step).
Cotranslational protein targeting by SRP. (A) The targeting of ribosomes carrying SRP signal sequences (magenta) to the membrane requires three sequential steps: cargo recognition by SRP, cargo targeting to the membrane via interaction of SRP with SR, and cargo transfer to the SecYEG translocon. Ffh is in blue, FtsY is in green, and the SRP RNA is in pink. “T” and “D” denote GTP and GDP, respectively. (B) Scheme of the FRET probes used. The nascent chain was labeled with a donor dye (green star) at the signal sequence. SRP was labeled with an acceptor dye (red star) in the M or N domain. (C) The signal sequences of substrates used in this study. The position of the donor dye is denoted by the asterisks. Colored letters indicate the hydrophobic core of the signal sequences.
Cotranslational protein targeting by SRP. (A) The targeting of ribosomes carrying SRP signal sequences (magenta) to the membrane requires three sequential steps: cargo recognition by SRP, cargo targeting to the membrane via interaction of SRP with SR, and cargo transfer to the SecYEG translocon. Ffh is in blue, FtsY is in green, and the SRP RNA is in pink. “T” and “D” denote GTP and GDP, respectively. (B) Scheme of the FRET probes used. The nascent chain was labeled with a donor dye (green star) at the signal sequence. SRP was labeled with an acceptor dye (red star) in the M or N domain. (C) The signal sequences of substrates used in this study. The position of the donor dye is denoted by the asterisks. Colored letters indicate the hydrophobic core of the signal sequences.
Cotranslational protein targeting involves a series of molecular events that present conflicting requirements for the targeting machinery. In the cytosol, SRP must efficiently select its substrates from ∼100-fold excess of translating ribosomes. SRP does so by interacting with RNC via two domains in the universally conserved SRP54 protein (Ffh in bacteria; Poritz et al., 1990). The Ffh M domain binds the signal sequence of the nascent protein (Keenan et al., 1998; Janda et al., 2010; Hainzl et al., 2011), whereas its N domain interacts with L23 and L29 at the polypeptide exit site of the ribosome (Pool et al., 2002; Gu et al., 2003; Halic et al., 2006a; Schaffitzel et al., 2006). How this bidentate interaction enables effective and accurate substrate selection by SRP remains unclear.
Cargo-bound SRP is targeted to the membrane through the binding of its NG domain, composed of the N domain and a GTPase, G domain, to a homologous NG domain in the SRP receptor (SR; FtsY in bacteria; Egea et al., 2004; Focia et al., 2004). SRP and FtsY belong to a novel class of GTPases regulated by GTP-dependent dimerization (Gasper et al., 2009; Shan et al., 2009). Discrete conformational changes, from a transient “early” intermediate upon initial FtsY binding to a GTP-stabilized “closed” complex and finally an “activated” complex, occur in the SRP–SR dimer and culminate in their GTPase activation (Shan et al., 2004; Zhang et al., 2009, 2011). These rearrangements are strongly regulated by the cargo, anionic phospholipids, and the SecYEG translocon, and thus couple the recognition of cargo by SRP to its delivery to the membrane (Braig et al., 2009; Zhang et al., 2009; Lam et al., 2010; Stjepanovic et al., 2011; Akopian et al., 2013). For example, the cargo for SRP strongly stabilizes the otherwise labile early intermediate, and thus accelerates formation of the closed targeting complex (Zhang et al., 2009). Whether and how the SRP/SR GTPases reciprocally regulate the SRP–cargo interaction is poorly understood. This regulation would be particularly important at the target membrane, where SRP must switch from a “cargo-binding” mode to a “cargo-releasing” mode.
Handover of the cargo to the SecYEG translocon remains one of the least understood aspects of the pathway. SecYEG binds the translating ribosome via its cytosolic loops c4 and c5 (Cheng et al., 2005; Ménétret et al., 2007). A lateral gate formed by two transmembrane helices (TM2 and TM7) of SecY binds signal sequence (van den Berg et al., 2004; du Plessis et al., 2009). As both SRP and SecYEG bind RNC via L23 on the ribosome and the signal sequence, the transfer of RNC to SecYEG (Jungnickel and Rapoport, 1995; Song et al., 2000) would require RNC to first detach from SRP. How the loss of cargo during this handover is prevented remains unclear. Recent studies suggest that such abortive events are minimized by formation of a RNC–SRP–SR–SecYEG quaternary intermediate (Shen et al., 2012; Akopian et al., 2013); however, the precise molecular mechanism of cargo transfer is not understood.
A major limitation in addressing these questions has been the lack of quantitative assays that directly and quantitatively report on the interaction of the translating ribosome with SRP and SecYEG. Recently, we developed an efficient method to site-specifically label the nascent protein on a translating ribosome using a fluorescent nonnatural amino acid (Saraogi et al., 2011). Close to 100% incorporation efficiency was achieved, yielding milligram quantities of purified fluorescent RNC (Saraogi et al., 2011). This allowed us, for the first time, to quantitatively measure the dynamic changes in the interaction of the translating ribosome with SRP and SecYEG at discrete stages of the targeting reaction, and generate a detailed molecular picture of the SRP pathway. We found that the targeting and translocation machineries actively regulate the conformation, energetics, and dynamics of the cargo–SRP interaction, thereby ensuring the efficient capture, delivery, and coordinated unloading of the cargo.
Results
General experimental design
To analyze the interaction of SRP with the nascent protein, we used Förster resonance energy transfer (FRET) between a donor fluorophore on the nascent protein (Fig. 1 B, green star) and an acceptor fluorophore on SRP (Fig. 1 B, red star). We generated stalled RNCs of 85 residues with a fluorescent probe, 7-hydroxycoumaryl ethylglycine (Cm), incorporated at the signal sequence (Fig. 1 C, asterisk; Saraogi et al., 2011). BODIPY-FL, a FRET acceptor for Cm, was introduced in the SRP M domain at residue 421 (Fig. 1 B, M domain pair) or the SRP N domain at residue 11 (Fig. 1 B, N domain pair). Structural data (Halic et al., 2006a) suggest that both positions lie within <30 Å of the signal sequence in the RNC–SRP complex, well within the estimated Förster radius of this dye pair (Saraogi et al., 2011).
Using both FRET pairs, we measured and compared the energetics, dynamics, and conformation of SRP–RNC interactions at every stage of the SRP pathway, from the RNC–SRP complex to the early and closed RNC–SRP–SR targeting complexes. Finally, the transfer of cargo from the SRP–SR to SecYEG complex was measured using a combination of the FRET probes (Fig. 1 B) and the increase in Cm fluorescence upon binding SecYEG, which reports on a functional interaction of the signal sequence with the translocon.
As model SRP substrates, we used two engineered signal sequences, 1A9L and 3A7L (Fig. 1 C), shown to direct efficient cotranslational protein targeting (Doud et al., 1993; Zhang et al., 2010). To understand how SRP rejects borderline substrates, we used the alkaline phosphatase (phoA) signal sequence (Doud et al., 1993; Zhang et al., 2010), which is primarily targeted via the SecA/B pathway. A mutant signal sequence, in which two leucine residues in 3A7L are replaced by arginine, serves as a negative control (Fig. 1 C, 3A5L2R).
Ribosomes bearing SRP substrates are selectively retained by SRP
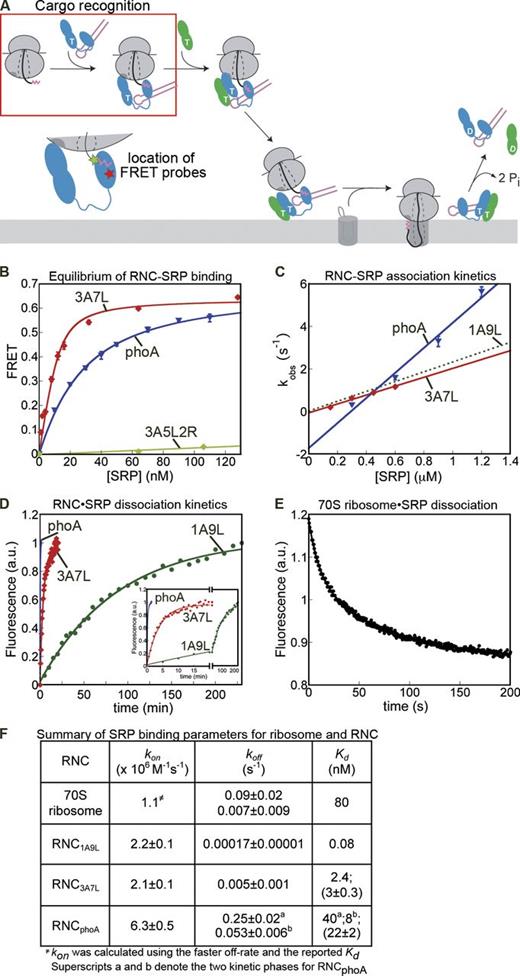
We first asked if SRP could effectively discriminate between signal sequences during cargo recognition (Fig. 2 A, boxed region). Equilibrium titrations based on the M domain FRET pair (Fig. 1 B, left) showed that RNC3A7L bound to SRP with a dissociation constant (Kd) of 3 nM, whereas the binding of RNCPhoA was sevenfold weaker (Fig. 2 B). RNC3A5L2R did not induce a significant FRET signal, which supports the specificity of the assay (Fig. 2 B, green; and Fig. S1 A).
Ribosomes bearing SRP substrates are selectively retained by SRP. (A) Highlight of the cargo recognition step (box) measured in this figure and the FRET probes used. (B) Equilibrium titration for SRP binding to RNC3A7L (red) and RNCPhoA (blue). The data were fit to Eq. 2, and obtained Kd values are summarized in F in parenthesis. The FRET signal with RNC3A5L2R (light green) was too small to be quantified (see also Fig. S1 A). (C) Observed rate constants (kobsd) for RNC–SRP association plotted against SRP concentration. Values of kon were obtained from a fit of the data to Eq. 4 and summarized in F. The data for RNC1A9L (dotted line) is from Saraogi et al. (2011) and shown for comparison. See Fig. S1 (B and C) for representative time courses. (D) Rate constants of SRP dissociation from RNC. The data were fit to Eq. 5a for RNC1A9L and RNC3A7L, and Eq. 5b for RNCPhoA (see Fig. S1 D). The koff values are summarized in F. The inset shows a magnification of the plot. (E) Kinetics of SRP dissociation from the 70S ribosome. These data were fit to Eq. 5b, and koff values are reported in F. (F) Summary of the rate constants obtained in C–E. The Kd values were calculated from measured rate constants using Kd = koff/kon, or from equilibrium titrations (values in parenthesis). For RNCPhoA, the measured Kd was the same, within error, as the weighted mean of the Kd values for the two kinetic phases (indicated by superscripts a and b). See the Materials and methods section and Fig. S1 D for a discussion of the biphasic behavior of RNCPhoA. The kon value for SRP-70S ribosome association was calculated from kon = koff/Kd (Zhang et al., 2010). Error bars in B and C indicate mean ± SEM. Values in F are mean of 2–4 experiments ± SD.
Ribosomes bearing SRP substrates are selectively retained by SRP. (A) Highlight of the cargo recognition step (box) measured in this figure and the FRET probes used. (B) Equilibrium titration for SRP binding to RNC3A7L (red) and RNCPhoA (blue). The data were fit to Eq. 2, and obtained Kd values are summarized in F in parenthesis. The FRET signal with RNC3A5L2R (light green) was too small to be quantified (see also Fig. S1 A). (C) Observed rate constants (kobsd) for RNC–SRP association plotted against SRP concentration. Values of kon were obtained from a fit of the data to Eq. 4 and summarized in F. The data for RNC1A9L (dotted line) is from Saraogi et al. (2011) and shown for comparison. See Fig. S1 (B and C) for representative time courses. (D) Rate constants of SRP dissociation from RNC. The data were fit to Eq. 5a for RNC1A9L and RNC3A7L, and Eq. 5b for RNCPhoA (see Fig. S1 D). The koff values are summarized in F. The inset shows a magnification of the plot. (E) Kinetics of SRP dissociation from the 70S ribosome. These data were fit to Eq. 5b, and koff values are reported in F. (F) Summary of the rate constants obtained in C–E. The Kd values were calculated from measured rate constants using Kd = koff/kon, or from equilibrium titrations (values in parenthesis). For RNCPhoA, the measured Kd was the same, within error, as the weighted mean of the Kd values for the two kinetic phases (indicated by superscripts a and b). See the Materials and methods section and Fig. S1 D for a discussion of the biphasic behavior of RNCPhoA. The kon value for SRP-70S ribosome association was calculated from kon = koff/Kd (Zhang et al., 2010). Error bars in B and C indicate mean ± SEM. Values in F are mean of 2–4 experiments ± SD.
To more accurately determine the binding affinity between RNC and SRP and to gain information into the dynamics of this interaction, we measured the association (kon) and dissociation (koff) rate constants for their binding. SRP bound to RNC3A7L with a kon value of 2.1 ×106 M−1s−1 (Fig. 2, C and F; and Fig. S1 B), similar to that of the more hydrophobic RNC1A9L (Fig. 1 C; Fig. 2 C, dotted line; and Fig. 2 F; Saraogi et al., 2011). RNCPhoA bound to SRP threefold more rapidly (Fig. 2, C and F; and Fig. S1 C). Nevertheless, SRP dissociated from RNCPhoA at a rate comparable to nontranslating ribosomes (Fig. 2, D–F; and Fig. S1 D). In contrast, the dissociation of RNC3A7L and RNC1A9L was 20- and 1,000-fold slower, respectively (Fig. 2, D and F). These findings are consistent with a recent report (Holtkamp et al., 2012; see Fig. S1 E and the Discussion section for a detailed explanation). Thus, SRP binds quickly to ribosomes with or without SRP substrates, but RNCs bearing strong signal sequences form kinetically more stable complexes with SRP and hence persist much longer.
FtsY actively regulates the SRP–cargo interaction
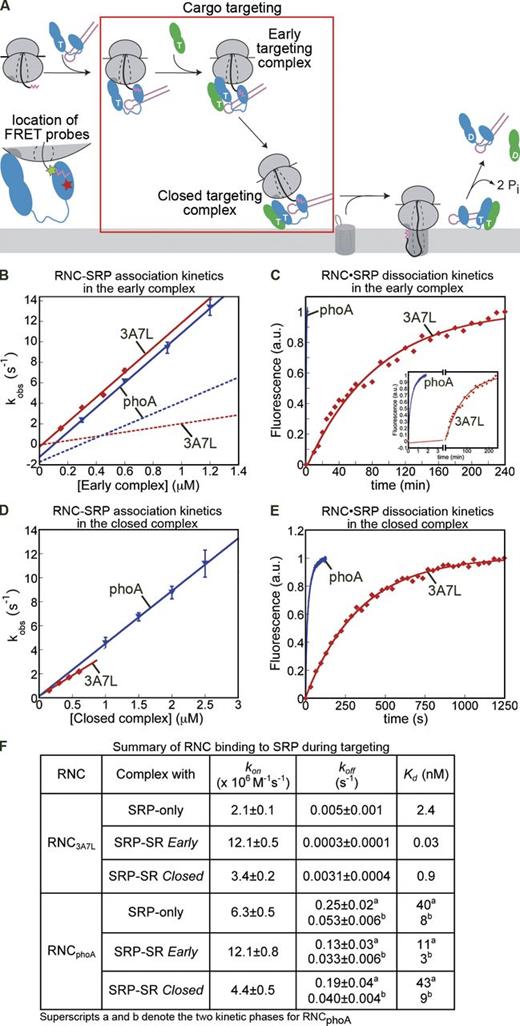
In the next step, RNC is targeted to the membrane via the SRP–SR interaction (Fig. 3 A, boxed area), which proceeds through an early intermediate (Zhang et al., 2008). This intermediate is strongly stabilized by the RNC (Zhang et al., 2009), and its stability directly correlates with the rate of formation of the closed SRP–SR complex (Zhang et al., 2008, 2009, 2010; Shen and Shan, 2010; Shen et al., 2011) and with cotranslational protein targeting (Zhang et al., 2008). Because the early complex can form with or without GTP but its subsequent rearrangements are strictly GTP dependent (Zhang et al., 2008; Zhang et al., 2009), a homogenous early targeting complex can be isolated by incubating RNC, SRP, and SR in the absence of GTP. We tested how the initial recruitment of SR modulates the dynamics and stability of the RNC–SRP interaction. Compared with free SRP, the SRP–SR early complex bound to RNC3A7L sixfold faster (Figs. 3 B and S2) and dissociated from it 10-fold slower (Fig. 3 C). Overall, the interaction of RNC3A7L with SRP was stabilized 80-fold upon formation of the early targeting complex, bringing the Kd value to the picomolar range (Fig. 3 F). In contrast, the corresponding stabilization for RNCPhoA was less than fourfold (Fig. 3, B, C, and F). Thus, the initial recruitment of FtsY increases the specificity of SRP for correct substrates from sevenfold in the RNC–SRP complex to >100-fold in the early targeting complex (Fig. 3 F).
FtsY actively regulates the interaction of SRP with its substrates. (A) Highlight of the targeting step (box) measured in this figure and the FRET probes used. (B and D) Kinetics of RNC binding to the SRP–FtsY early complex (B) or closed complex (D) with RNC3A7L (red) and RNCPhoA (blue). The data were fit to Eq. 4, and the kon values obtained are summarized in Fig. 3 F. See Fig. S2 for representative time courses. The dotted lines indicate the corresponding binding kinetics in the absence of FtsY (from Fig. 2 C) and are shown for comparison. Values are averages of 2–3 experiements ± SD (error bars). (C and E) Kinetics of RNC dissociation from the early (C) or closed (E) targeting complex. The data were fit to Eq. 5a or Eq. 5b, and the obtained koff values are summarized in F. The inset in C shows a magnification of the plot. (F) Summary of the RNC–SRP binding affinity at different stages of the targeting pathway. The values for SRP only are from Fig. 2 F and are shown for comparison. Kd values were calculated from Kd = koff/kon. The two kinetic phases for RNCPhoA are indicated by superscripts a and b. Values in F summarizing data from B–E are averages of 3–4 experiments ± SD.
FtsY actively regulates the interaction of SRP with its substrates. (A) Highlight of the targeting step (box) measured in this figure and the FRET probes used. (B and D) Kinetics of RNC binding to the SRP–FtsY early complex (B) or closed complex (D) with RNC3A7L (red) and RNCPhoA (blue). The data were fit to Eq. 4, and the kon values obtained are summarized in Fig. 3 F. See Fig. S2 for representative time courses. The dotted lines indicate the corresponding binding kinetics in the absence of FtsY (from Fig. 2 C) and are shown for comparison. Values are averages of 2–3 experiements ± SD (error bars). (C and E) Kinetics of RNC dissociation from the early (C) or closed (E) targeting complex. The data were fit to Eq. 5a or Eq. 5b, and the obtained koff values are summarized in F. The inset in C shows a magnification of the plot. (F) Summary of the RNC–SRP binding affinity at different stages of the targeting pathway. The values for SRP only are from Fig. 2 F and are shown for comparison. Kd values were calculated from Kd = koff/kon. The two kinetic phases for RNCPhoA are indicated by superscripts a and b. Values in F summarizing data from B–E are averages of 3–4 experiments ± SD.
Although a strong SRP–RNC interaction is beneficial in the cytosol, it will render cargo release at the target membrane difficult. Notably, anionic phospholipids strongly stabilize the closed SRP–SR complex. Reciprocally, formation of the closed complex exposes a lipid binding helix of FtsY and allows the targeting complex to associate much more stably with the membrane (Lam et al., 2010; Braig et al., 2011; Stjepanovic et al., 2011). We therefore asked whether rearrangement of the RNC–SRP–SR complex from the early to the closed state (Fig. 3 A), which is coupled to stable membrane attachment of the targeting complex, could help the release of cargo. Indeed, in a closed targeting complex assembled using the nonhydrolyzable GTP analogue GppNHp, RNC–SRP association was three to fourfold slower, and their dissociation was 10-fold faster compared with the early targeting complex (Fig. 3, D, E, and F; and Fig. S2). Overall, the affinity of RNC3A7L for SRP was weakened 30-fold in the closed state (Fig. 3 F), which suggests that the early → closed rearrangement primes the cargo for subsequent unloading. This regulation was not observed with RNCPhoA (Fig. 3 F). Collectively, the results in this section show that FtsY assists SRP in sensing distinct stages of targeting and that it does so only for the correct substrates.
Molecular basis for FtsY-regulated SRP–RNC interaction
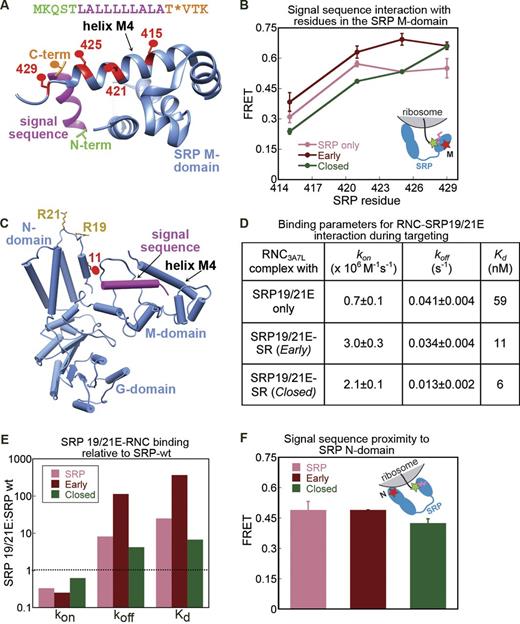
To understand how FtsY alters the energetics of the SRP–RNC interaction, we examined the interaction of SRP with the ribosome and signal sequence. To probe the interaction of the signal sequence with the SRP M domain, we measured maximal FRET efficiencies between Cm at 3A7L signal sequence and BODIPY-FL at residues 415, 421, 425, or 429 along helix M4 that lines the signal sequence-binding groove (Fig. 4 A, red). The cysteine mutations and BODIPY labeling do not affect RNC–SRP binding (Fig. S3 A) or protein targeting (Fig. S3 B). Upon formation of the early targeting complex, the FRET efficiency between Cm at the signal sequence C terminus and residues 425 and 429 in helix M4 increased (Fig. 4 B, pink vs. dark red). A similar trend was observed with Cm at the signal sequence N terminus (Fig. S3 C). The anisotropy of the dyes was low and comparable in all constructs, which indicates that there is no significant contribution of dye orientation to the observed FRET (Table S1), and the position of Cm on the signal sequence did not significantly affect RNC–SRP binding (Fig. S3 D). These results suggest that formation of the early targeting complex allows the signal sequence to pack more tightly against the signal peptide–binding groove.
Molecular basis for FtsY-induced changes in SRP–RNC binding affinity. (A) Crystal structure of a signal sequence (magenta) bound to the Methanocaldococcus jannaschii Ffh M domain (blue; Protein Data Bank accession no. 3NDB; Hainzl et al., 2011). The donor dye at the signal sequence N and C termini are in green and orange, respectively. The acceptor dyes on Ffh helix M4 are in red. Residue numbering is for homologous residues in E. coli Ffh. (B) FRET efficiency between Cm at the signal sequence C terminus and BODIPY-FL at indicated residues in SRP helix M4 in the RNC–SRP complex (pink) and the early (dark red) and closed (green) targeting complexes. The inset shows a diagram of the FRET pair used for this experiment. (C) Crystal structure of Ffh (blue) modeled into the cryo-EM density (Protein Data Bank accession no. 2J28; Halic et al., 2006a) for the SRP–RNC complex. The signal sequence is in magenta. Conserved residues 19 and 21 in the Ffh N domain are shown in gold. Red denotes the FRET acceptor labeled at C11 in the N domain. The ribosome, which sits directly above R19 and R21, is not shown for clarity. (D) A summary of the kinetic parameters for SRP19/21E mutant binding to RNC3A7L at various stages of targeting (see Fig. S4, B–G, for time courses). (E) The defect displayed by mutant SRP19/21E in binding RNC3A7L at different stages of targeting. The ratios of the rate or equilibrium constants for RNC3A7L binding to SRP19/21E (Fig. 4D) relative to wt SRP (Fig. 3 F) are plotted. The dotted line indicates the expected ratio of 1 if the mutant displays no defect (see Fig. 4 D and Fig. S4). (F) Maximal FRET efficiency between Cm-labeled RNC3A7L and BODIPY-labeled SRP C11 (C) during the targeting cycle. The inset shows a diagram of the FRET pair used for this experiment. Molecular graphics were generated using UCSF Chimera (Pettersen et al., 2004). Values in B are means of four experiments ± SEM (error bars). Values in D and F are means of 2–3 experiments ± SD.
Molecular basis for FtsY-induced changes in SRP–RNC binding affinity. (A) Crystal structure of a signal sequence (magenta) bound to the Methanocaldococcus jannaschii Ffh M domain (blue; Protein Data Bank accession no. 3NDB; Hainzl et al., 2011). The donor dye at the signal sequence N and C termini are in green and orange, respectively. The acceptor dyes on Ffh helix M4 are in red. Residue numbering is for homologous residues in E. coli Ffh. (B) FRET efficiency between Cm at the signal sequence C terminus and BODIPY-FL at indicated residues in SRP helix M4 in the RNC–SRP complex (pink) and the early (dark red) and closed (green) targeting complexes. The inset shows a diagram of the FRET pair used for this experiment. (C) Crystal structure of Ffh (blue) modeled into the cryo-EM density (Protein Data Bank accession no. 2J28; Halic et al., 2006a) for the SRP–RNC complex. The signal sequence is in magenta. Conserved residues 19 and 21 in the Ffh N domain are shown in gold. Red denotes the FRET acceptor labeled at C11 in the N domain. The ribosome, which sits directly above R19 and R21, is not shown for clarity. (D) A summary of the kinetic parameters for SRP19/21E mutant binding to RNC3A7L at various stages of targeting (see Fig. S4, B–G, for time courses). (E) The defect displayed by mutant SRP19/21E in binding RNC3A7L at different stages of targeting. The ratios of the rate or equilibrium constants for RNC3A7L binding to SRP19/21E (Fig. 4D) relative to wt SRP (Fig. 3 F) are plotted. The dotted line indicates the expected ratio of 1 if the mutant displays no defect (see Fig. 4 D and Fig. S4). (F) Maximal FRET efficiency between Cm-labeled RNC3A7L and BODIPY-labeled SRP C11 (C) during the targeting cycle. The inset shows a diagram of the FRET pair used for this experiment. Molecular graphics were generated using UCSF Chimera (Pettersen et al., 2004). Values in B are means of four experiments ± SEM (error bars). Values in D and F are means of 2–3 experiments ± SD.
To probe the interaction of the Ffh N domain with the ribosome, we looked for an Ffh mutant defective in ribosome binding. Based on the cryo-EM model of SRP bound to an RNC (Halic et al., 2006a), sequence conservation (Fig. S4 A), previous cross-linking (Gu et al., 2003; Ullers et al., 2003), and the electrostatic nature of the interaction of Ffh with acidic residues in L23 (Schaffitzel et al., 2006; del Alamo et al., 2011), we introduced charge reversal mutations at two highly conserved basic residues of Ffh, R19, and R21 (Fig. 4 C, gold). Mutant SRP19/21E was defective in SRP-dependent targeting (Fig. S4 A, right), which supports the importance of these residues in SRP function.
We measured the extent to which the SRP19/21E mutations weaken the binding between SRP and RNC3A7L (Fig. S4, B and C; and Fig. 4 D). This provides an empirical measure for the energetic contribution of the ribosomal contacts with R19/21 at different stages of targeting. In the RNC–SRP complex, the SRP19/21E mutations weakened SRP–RNC binding 24-fold (Fig. 4 E, pink; and Fig. S4, B and C). In the early complex, these mutations caused a >300-fold defect (Fig. 4 E, dark red; and Fig. S4, D and E), which suggests that the interaction of R19/21 with the ribosome becomes stronger. Together, these results show that tighter RNC–SRP binding in the early targeting complex arises from stronger interactions of both the signal sequence and the ribosome with the M and N domains of SRP, respectively.
We then asked whether these contacts were reorganized in the closed targeting complex, resulting in weaker RNC–SRP interaction (Fig. 3 F). To test if the signal sequence rearranged in the M domain in the closed complex, we measured FRET efficiencies between Cm at the signal sequence and acceptor dyes in helix M4 of Ffh, as described earlier (Fig. 4 A). The FRET efficiencies with residues 415, 421, and 425 were reduced upon the early → closed rearrangement (Fig. 4 B, green), which suggests repositioning of the signal sequence in the M domain.
To test if the contact of SRP N domain with the ribosome is altered in the closed targeting complex, we measured the effect of the SRP19/21E mutations on the RNC–SRP interaction. In the closed complex, these mutations caused only a sixfold weaker binding (Fig. 4 E, green; and Fig. S4, F and G). The actual mutational defect is likely smaller because even in the presence of GppNHp, a significant fraction of the RNC–SRP–SR complex remains in the early state (Zhang et al., 2009), in which these mutations are highly destabilizing. The rescue of the SRP19/21E mutant in the closed targeting complex strongly suggests that the interaction of the SRP N domain with the ribosome is substantially weakened at this stage.
Recent studies show that at late stages of protein targeting, the SRP–SR NG domain complex localizes to the distal end of the SRP RNA (Ataide et al., 2011; Shen et al., 2012). This large-scale movement would vacate L23 at the ribosome exit site for interaction with SecYEG. We therefore asked if this movement had occurred in the closed targeting complex. To probe for the proximity of the N domain to the signal sequence at the ribosome exit site, we used the N domain FRET pair (Figs. 1 B and 4 C). Consistent with structural data (Halic et al., 2006a), we observed efficient FRET (∼0.5) in the RNC–SRP complex (Fig. 4 F, pink). The FRET value, albeit slightly lower, persisted in the closed targeting complex (Fig. 4 F, green). Thus, although previous work (Pool et al., 2002; Halic et al., 2006b) and our mutational analysis suggest that the SRP N domain interacts weakly with the ribosome at this stage, the NG domain complex remains in the vicinity of the nascent polypeptide exit site.
Cm labeled signal sequence reports on RNC–SecYEG interaction
At the membrane, SecYEG must engage RNC. To monitor these late events in targeting, we first tested if the Cm dye can be used to detect the interaction of a signal sequence with SecYEG (Fig. 5 A). Indeed, addition of SecYEG, solubilized in 0.02% DDM (Dalal and Duong, 2010; Akopian et al., 2013), induced an approximately twofold increase in the fluorescence of Cm-labeled RNC3A7L (Fig. 5 B, green). Equilibrium titrations based on this fluorescence change showed that RNC3A7L was bound tightly to wt-SecYEG (Kd = 3 ± 2 nM; Fig. 5 C), in agreement with previous reports (Wu et al., 2012). The following observations demonstrate the specificity of this assay. First, a SecYEG c4/c5 mutant harboring charge reversal mutations (R255/256/357E) in two cytosolic loops critical for ribosome binding (Cheng et al., 2005; Ménétret et al., 2007; Akopian et al., 2013) showed no detectable binding to RNC3A7L (Fig. 5, B and C, gray). Second, introduction of two arginines into the signal sequence (RNC3A5L2R or mt-RNC) weakened RNC–SecYEG binding 46-fold (Fig. 5 C, light green), showing that the interaction is specific to a functional signal sequence. Third, when Cm-labeled RNC3A7L is bound to SecYEG labeled with BODIPY-FL at residue 180, efficient FRET was observed (Fig. S5 A). These results support the specificity of the Cm probe in detecting a functional RNC–SecYEG interaction.
Cm-labeled signal sequence reports on RNC binding to SecYEG. (A) A scheme of RNC binding to SecYEG. The green star denotes the Cm dye. (B) Fluorescence spectra of 20 nM Cm-labeled RNC3A7L in the absence (black) and presence of 300 nM wt (green) or c4/c5 mutant SecYEG (gray). (C) Equilibrium titration of 20 nM RNC3A7L with wt-SecYEG (green) or c4/c5 mutant SecYEG (gray) and of 30 nM RNC3A5L2R (mt-RNC) with wt-SecYEG (light green). The data were fit to Eq. 6 and gave Kd values of 3 ± 2 nM (mean ± SD, with n = 20) and 140 nM with RNC and mt-RNC, respectively. The data in B and C are representative of at least four experiments. The mt-RNC plot in C is representative of two independent experiments.
Cm-labeled signal sequence reports on RNC binding to SecYEG. (A) A scheme of RNC binding to SecYEG. The green star denotes the Cm dye. (B) Fluorescence spectra of 20 nM Cm-labeled RNC3A7L in the absence (black) and presence of 300 nM wt (green) or c4/c5 mutant SecYEG (gray). (C) Equilibrium titration of 20 nM RNC3A7L with wt-SecYEG (green) or c4/c5 mutant SecYEG (gray) and of 30 nM RNC3A5L2R (mt-RNC) with wt-SecYEG (light green). The data were fit to Eq. 6 and gave Kd values of 3 ± 2 nM (mean ± SD, with n = 20) and 140 nM with RNC and mt-RNC, respectively. The data in B and C are representative of at least four experiments. The mt-RNC plot in C is representative of two independent experiments.
SecYEG effectively displaces the SRP–SR complex from RNC
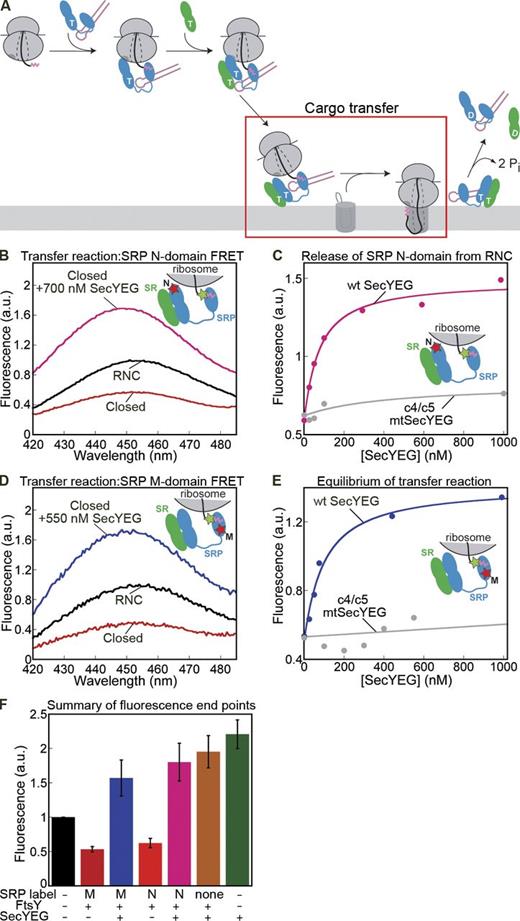
As SecYEG enhances Cm fluorescence upon RNC binding, whereas BODIPY-labeled SRP reduces Cm fluorescence due to FRET, this provides a sensitive assay to monitor the transfer of RNC from the targeting complex to SecYEG (Fig. 6 A, boxed area). To test whether SecYEG can displace the SRP–SR complex from RNC in this minimal system, we added SecYEG to a preformed closed targeting complex containing Cm-labeled signal sequence. The transfer reaction was performed either with the N domain FRET pair (Fig. 1 B) to monitor displacement of the NG domain complex from the ribosome exit site (Fig. 6, B and C) or with the M domain FRET pair to monitor the transfer of the nascent chain from the signal sequence binding site (Fig. 6, D and E). In both cases, addition of SecYEG induced a similar dose-dependent increase in the fluorescence of Cm-labeled RNC (Fig. 6, B and C, magenta; and Fig. 6, D and E, blue; see also Fig. S5 B). The SecYEG-induced increase in Cm fluorescence far exceeded that expected from loss of FRET due to dissociation of the RNC–SRP complex (Fig. 6, B and D; and Fig. 6 F, compare blue or magenta vs. black), which indicates engagement of the signal sequence with SecYEG (see the next paragraph). The effect of SecYEG during the transfer reaction was saturable, with an EC50 value of 39 ± 26 nM (Fig. 6 E), which indicates that SecYEG efficiently displaces SRP from the RNC. The c4/c5 mutations abolished this reaction (Fig. 6, C and E, gray; and Fig. S5 B), which indicates that functional cytosolic loops are required for the observed cargo transfer reaction.
SecYEG effectively unloads cargo from the closed targeting complex. (A) Highlight of the cargo transfer step in the targeting pathway. (B) Fluorescence spectra of Cm-labeled RNC3A7L by itself (black), in the closed targeting complex with SRP labeled with BODIPY-FL at C11 (N domain; red), and upon addition of SecYEG to the closed targeting complex (pink). The inset shows a diagram of the FRET pair used for this experiment. (C) Titration of wt (pink) or mutant (gray) SecYEG into the closed targeting complex formed with BODIPY-labeled SRP (C11; also see Fig. S5 B). (D) As in B, except that the M domain FRET pair (inset and Fig. 1 B) was used. (E) As in C, except that the M domain FRET pair was used. The data were fit to Eq. 6 to give an EC50 for transfer of 39 ± 26 nM. (F) Summary of the changes in fluorescence end points upon addition of saturating SecYEG to the closed targeting complex, formed with unlabeled SRP (brown) or with SRP labeled at the M (blue) or N (magenta) domain. The fluorescence signal upon direct addition of RNC to SecYEG is in green. All experiments were performed with 20 nM Cm-labeled RNC3A7L, 40 nM SRP, 500 nM FtsY, and 100 µM GppNHp. All fluorescence values are normalized relative to those of RNC (black). Data in B–E are representative of three to four experiments. Values in F are means of at least three experiments ± SD (error bars).
SecYEG effectively unloads cargo from the closed targeting complex. (A) Highlight of the cargo transfer step in the targeting pathway. (B) Fluorescence spectra of Cm-labeled RNC3A7L by itself (black), in the closed targeting complex with SRP labeled with BODIPY-FL at C11 (N domain; red), and upon addition of SecYEG to the closed targeting complex (pink). The inset shows a diagram of the FRET pair used for this experiment. (C) Titration of wt (pink) or mutant (gray) SecYEG into the closed targeting complex formed with BODIPY-labeled SRP (C11; also see Fig. S5 B). (D) As in B, except that the M domain FRET pair (inset and Fig. 1 B) was used. (E) As in C, except that the M domain FRET pair was used. The data were fit to Eq. 6 to give an EC50 for transfer of 39 ± 26 nM. (F) Summary of the changes in fluorescence end points upon addition of saturating SecYEG to the closed targeting complex, formed with unlabeled SRP (brown) or with SRP labeled at the M (blue) or N (magenta) domain. The fluorescence signal upon direct addition of RNC to SecYEG is in green. All experiments were performed with 20 nM Cm-labeled RNC3A7L, 40 nM SRP, 500 nM FtsY, and 100 µM GppNHp. All fluorescence values are normalized relative to those of RNC (black). Data in B–E are representative of three to four experiments. Values in F are means of at least three experiments ± SD (error bars).
If complete transfer of RNC to SecYEG occurred, FRET between Cm-labeled signal sequence and SRP should be abolished. Hence, the Cm fluorescence after the transfer reaction would be the same for targeting complexes containing labeled or unlabeled SRP. When we repeated the transfer reaction using unlabeled SRP, this was indeed the case (Fig. 6 F, compare blue and magenta vs. brown). The Cm fluorescence at the end of the transfer reaction is similar to that from direct addition of saturating SecYEG to RNC (Figs. 5 B and 6 F; green), which indicates that close to complete cargo transfer was achieved. These results demonstrate for the first time that detergent solubilized SecYEG can drive the transfer of cargo from SRP.
Step-wise transfer of RNC from SRP to SecYEG
To probe the mechanism of the molecular relay between SRP and SecYEG, we analyzed the kinetics of the transfer reaction monitored using either the M or N domain FRET pair (Fig. 1 B). As the nascent protein continues to elongate during targeting, we examined the transfer reaction using RNCs with different nascent chain length. Several observations suggest that the transfer occurs in a stepwise mechanism. First, the SecYEG-driven release of the SRP N domain from the RNC-85 was threefold faster than that of signal sequence release from the SRP M domain (Fig. 7 A, magenta vs. blue; and Fig. 7 D). Second, when we repeated the transfer reaction with RNC3A7L containing 50 additional residues (RNC-135), the initial release of the SRP N domain was fourfold faster than that of signal sequence from the M domain (Fig. 7, B–D). Significantly, the transfer reaction of RNC-135 monitored with the N domain FRET pair exhibited biphasic kinetics with a pronounced burst, which indicates at least two steps during this transfer (Fig. 7 C).
Kinetics of cargo transfer to SecYEG. (A) Kinetics of transfer of RNC3A7L-85 upon addition of 1 µM SecYEG to the closed targeting complex formed by SRP labeled with BODIPY-FL at the M (blue) or N (pink) domain. The data were fit to Eq. 5a to obtain the observed rate constants listed in D. (B) Kinetics of transfer of RNC3A7L-135 upon addition of SecYEG to the closed targeting complex formed by SRP labeled with BODIPY-FL at the M domain (purple). Data for RNC3A7L-85 (blue) are shown for comparison. (C) As in B, except that the closed targeting complex was formed by SRP labeled with BODIPY-FL at the N domain (magenta). (D) Summary of the observed rate constants for RNC transfer from A–C. (E) Alternative models for transfer of signal sequence from SRP to SecYEG. (F) Simulated kinetic behavior of the transfer reaction according to model (i), monitored using FRET acceptor labeled at the SRP M (blue) or N (pink) domain. (G) Simulated kinetic behavior of the transfer reaction according to model (ii), as the equilibrium to form the transfer complex was progressively less favorable (top to bottom). The value of K4 = k4/k−4 was set to 25, 5, 1, 0.2, and 0.1, respectively. Only results with the SRP N domain are shown for clarity (see Materials and methods for details). Values in A and B are representative of three experiments. Values in D are averages of two to three experiments ± SD.
Kinetics of cargo transfer to SecYEG. (A) Kinetics of transfer of RNC3A7L-85 upon addition of 1 µM SecYEG to the closed targeting complex formed by SRP labeled with BODIPY-FL at the M (blue) or N (pink) domain. The data were fit to Eq. 5a to obtain the observed rate constants listed in D. (B) Kinetics of transfer of RNC3A7L-135 upon addition of SecYEG to the closed targeting complex formed by SRP labeled with BODIPY-FL at the M domain (purple). Data for RNC3A7L-85 (blue) are shown for comparison. (C) As in B, except that the closed targeting complex was formed by SRP labeled with BODIPY-FL at the N domain (magenta). (D) Summary of the observed rate constants for RNC transfer from A–C. (E) Alternative models for transfer of signal sequence from SRP to SecYEG. (F) Simulated kinetic behavior of the transfer reaction according to model (i), monitored using FRET acceptor labeled at the SRP M (blue) or N (pink) domain. (G) Simulated kinetic behavior of the transfer reaction according to model (ii), as the equilibrium to form the transfer complex was progressively less favorable (top to bottom). The value of K4 = k4/k−4 was set to 25, 5, 1, 0.2, and 0.1, respectively. Only results with the SRP N domain are shown for clarity (see Materials and methods for details). Values in A and B are representative of three experiments. Values in D are averages of two to three experiments ± SD.
To explain the experimentally observed kinetics of the transfer reaction and to better understand its underlying mechanism, we performed numerical simulations. In the simplest model (Fig. 7 E, model (i)), the binding of SecYEG first results in the displacement of the SRP N domain from the ribosome. This generates a transfer intermediate, from which the signal sequence is transferred to SecYEG. To validate this model, we simulated the expected rise in Cm fluorescence based on the experimentally observed rate and equilibrium constants of the transfer reaction (for details see Materials and methods). Simulations based on model (i) predict pronounced biphasic increase in Cm fluorescence during the transfer reaction with N domain–labeled SRP (Fig. 7 F, magenta). This is because assembly of SecYEG with the targeting complex to form the transfer complex would occur rapidly at saturating SecYEG concentrations. The resultant loss of FRET between the signal sequence and SRP N domain would far precede the additional increase in Cm fluorescence caused by the slower docking of the signal sequence into SecYEG. Such biphasic kinetics were observed with RNC-135 (Fig. 7 C) but were barely detectable with RNC-85 (Fig. 7 A, magenta), which suggests that model (i) is insufficient to explain all the experimental data.
The absence of a burst expected for formation of the transfer complex indicates that such a complex is transient during the transfer reaction, and cannot be formed simply by bimolecular association between SecYEG and the targeting complex. We therefore considered the possibility that formation of transfer complex with RNC-85 is preceded by and is in unfavorable equilibrium with an initial encounter complex, in which the SRP N domain is not yet removed from the ribosome (Fig. 7 E, model (ii)). Kinetic simulations based on this model showed that, indeed, as the equilibrium to form the transfer complex from the encounter complex (Fig. 7 E, K4 = k4/k−4) was made less favorable (Fig. 7 G, red arrow), the burst associated with release of the N domain–labeled SRP from RNC was progressively removed. At values of K4 ≤ 0.2, the burst expected for removal of the SRP N domain from RNC was barely detectable, and the simulated results closely matched experimental data with RNC-85. A more favorable equilibrium to form the transfer complex (K4 ∼5) generated curves that better matched the results obtained with RNC-135 (Fig. 7 G). These simulations strongly suggest that formation of the productive transfer complex required for efficient cargo transfer is more favorable with RNC-135, whereas the shorter nascent chain in RNC-85 does not provide a strong cue for fast removal of the N domain (Park and Rapoport, 2011). In agreement with this, an overall faster rate of cargo transfer was observed with RNC-135 than RNC-85 (Fig. 7 D). The slow kinetics of RNC-85 transfer to SecYEG could also be explained, in part, by the unfavorable equilibrium to generate the transfer complex.
Discussion
Proper localization of membrane and secretory proteins requires the efficient and accurate execution of a series of molecular events, including capture of the cargo protein, its delivery to the membrane, and its productive handover from the targeting to the translocation machinery (Saraogi and Shan, 2011). Here, we address the molecular mechanisms underlying these events in the SRP pathway, which has served as a paradigm for understanding the molecular basis of protein localization. For the first time, nonnatural amino acid technology has allowed us to study, at an unprecedented resolution, the dynamic interaction of the translating ribosome with SRP as it enters, progresses along, and finally exits the SRP pathway.
How does SRP identify the correct substrates in the crowded cytosol, given a 10–100-fold excess of ribosomes that bind to SRP with substantial affinity (80–100 nM; Flanagan et al., 2003; Bornemann et al., 2008; Zhang et al., 2010)? Our results show that SRP binds quickly to ribosomes with or without an SRP signal sequence (Fig. 8, step 1), but the kinetic stability of the RNC–SRP complex increases significantly with stronger signal sequences. A recent paper (Holtkamp et al., 2012) reached similar conclusions using FRET between L23 and the SRP M domain. Triphasic RNC–SRP binding kinetics were observed by Holtkamp et al. (2012), which could arise from multiple factors. The fluorescent probes placed on L23 likely reported on transient SRP–ribosome interactions before signal sequence docking into the M domain. In contrast, our assay specifically reports on the engagement of the signal sequence with the SRP M domain when a stable RNC–SRP complex is formed. The specificity of the probe allowed us to obtain more straightforward kinetic data in most cases (see Materials and methods).
A model for highly coordinated delivery of RNC from the cytosol to the translocon, driven sequentially by the SRP/SR GTPases and SecYEG at different stages of the targeting cycle. Color notations are the same as in Fig. 1 A. The question mark indicates the unknown nature and structure of the encounter complex.
A model for highly coordinated delivery of RNC from the cytosol to the translocon, driven sequentially by the SRP/SR GTPases and SecYEG at different stages of the targeting cycle. Color notations are the same as in Fig. 1 A. The question mark indicates the unknown nature and structure of the encounter complex.
Holtkamp et al. (2012) further postulate that the SRP rapidly “scans” translating ribosomes for the presence of signal sequences, based on rapid RNC–SRP binding and dissociation rate constants. However, the reported “rapid” binding (k1 = ∼108 M−1s−1) refers to the formation of a highly labile intermediate that forms independently of the signal sequence. Formation of the final RNC–SRP complex, in which the correct and incorrect RNCs are distinguished, requires this intermediate to undergo two additional slow rearrangements and hence proceeds at much slower rates. Holtkamp et al. (2012) also reported rapid “koff” values because, rather than treating SRP dissociation as a sequential process as proposed in the same paper, their koff values were obtained from averaging the rate constants of three steps during dissociation. Hence, the rapid dissociation rate from <10% of highly labile complexes dominated the calculation. This led to overall koff values for a sequential reaction that are much faster than substeps in the reaction sequence, which is physically impossible. A kinetic simulation based solely on the three-step model and individual rate constants reported by Holtkamp et al. (2012) predicts that SRP dissociates from the final stable RNC–SRP complex at a rate constant of 0.0058 s−1 for RNCs bearing a model SRP substrate Lep, and 0.36 s−1 from the 70S ribosome (Fig. S1 E). These values are in good agreement with our measurements (Fig. 2 F). Thus, although it remains possible that SRP rapidly “scans” translating ribosomes, such a mechanism would have to be enabled by additional components in vivo, rather than by the intrinsic property of SRP–RNC interaction.
Indeed, our results indicate that the intrinsic difference in binding between an SRP-dependent substrate, 3A7L, and a SecA/B-dependent substrate, phoA, is fairly modest, only sevenfold. Importantly, the interaction of SRP with RNC3A7L is strongly enhanced upon initial recruitment of FtsY to form the early targeting complex (Fig. 8, step 2), whereas that with RNCphoA is marginally affected. This stabilization was predicted by Zhang et al. (2009) from a thermodynamic analysis of the SRP–SR GTPase cycle. Our data fulfill this prediction and provide direct evidence that the SRP/SR GTPases regulate the cargo–SRP interaction. Further, we show that this stabilization arises from stronger interactions of both the SRP M and N domains with the signal sequence and the ribosome, respectively. This gives the early targeting complex, in which the lipid-binding helix of FtsY is not yet exposed (Lam et al., 2010), an extended time window to search for the membrane. The residence time of SRP on RNCs bearing correct signal sequences is 3,000–5,000 s, much longer than the 3–5-s time window estimated for the SRP to complete targeting (Zhang et al., 2010). Thus, once a correct cargo binds SRP, it likely undergoes subsequent steps in the pathway without dissociating from it. In contrast, the early targeting complex formed by incorrect cargos, in which neither the RNC–SRP interaction (this paper) nor the SRP–FtsY interaction (Zhang et al., 2009) is stabilized, is more likely to disassemble prematurely in the cytosol. These results demonstrate that formation of the RNC–SRP–FtsY early targeting complex is a major commitment step at which the correct substrates selectively enter the SRP pathway.
However, a strong interaction between SRP and RNC poses a challenge to its subsequent release at the membrane. How is this barrier overcome? Notably, anionic phospholipids drive the rearrangement of the GTPases from the early to the closed conformation, which associates with the membrane more stably (Lam et al., 2010; Braig et al., 2011). Here, we demonstrate that this conformational switch (Fig. 8, step 3) also weakens the affinity of SRP for its cargo 30-fold, thus coupling the membrane localization of the targeting complex to the release of cargo. The reduction in binding affinity arises from substantially weaker interaction of the SRP N domain with the ribosome, and possibly a repositioning of the signal sequence in the M domain. These results agree with earlier studies of eukaryotic SRP, which showed loss of electron density of the SRP NG domain (Halic et al., 2006b) and loss of SRP-L23a cross-link (Pool et al., 2002) upon assembly of a stable GTP-dependent complex between RNC, SRP, and SR.
Nevertheless, in the closed targeting complex, the SRP NG domain remains adjacent to the ribosome exit site, and the signal sequence is still bound to the M domain, which indicates that the GTPase rearrangements are insufficient to drive the complete release of cargo. The targeting cycle is completed by SecYEG, which displaces the SRP–SR complex from the RNC to drive cargo unloading. Two recent studies show that SecYEG induces relocalization of the SRP–SR NG domains away from the ribosome exit site, to the distal end on SRP RNA where GTP hydrolysis can be activated (Shen et al., 2012; Akopian et al., 2013). Under these conditions, the RNC remains attached to the SRP–SR complex. In this work, we observed that detachment of the SRP N domain from the ribosome is faster and precedes the release of signal sequence from the SRP M domain and signal sequence docking into SecYEG. Collectively, these data suggest a highly coordinated, step-wise mechanism of cargo transfer (Fig. 8): after the initial encounter with the targeting complex (step 4, encounter complex), SecYEG first displaces the SRP NG domain from the ribosome exit site via interaction of its cytosolic loops with L23 (step 5, transfer complex). This is followed by transfer of the signal sequence from the SRP M domain to SecYEG (step 6). The step-wise transfer prevents abortive loss of RNC from the membrane. During the transfer process, the SRP–SR NG domain complex relocalizes to the distal site of the SRP RNA, where GTP hydrolysis is activated (Ataide et al., 2011; Shen et al., 2012) to reset the targeting cycle (Fig. 8, step 6).
The RNC transfer reactions observed here proceed with a half time (t1/2 = ln2/k) of 70–140 s. This is slower than what is expected physiologically, but is not unexpected for a reaction reconstituted with minimal components and suggests that additional in vivo factors could allosterically regulate SRP, SR, or SecYEG to accelerate transfer. Quantitative analysis of cargo transfer coupled with kinetic simulations in this minimal system helps define the rate-limiting barriers that can be facilitated by these additional factors. A major barrier arises from an unfavorable equilibrium to form the transfer complex (Fig. 8, step 5). This is consistent with observations in single-molecule experiments, in which only ∼30% of the SRP–SR NG domain complex relocalizes to the SRP RNA distal site in the presence of RNC-85 and SecYEG (Shen et al., 2012). Indeed, elongation of the nascent polypeptide facilitates the transfer reaction, and this stimulation appears to arise from more facile release of the NG domain from the ribosome to form the transfer complex. A low probability of signal sequence docking into SecYEG (Fig. 8, step 6), which requires opening of the lateral gate, could present an additional barrier. We speculate that under physiological conditions, both formation of the transfer complex and gate opening in SecYEG could be stimulated by additional factors, including anionic phospholipid membranes and other components of the holo-translocon (Duong and Wickner, 1997).
In summary, we show that the SRP/SR GTPase cycle and the SecYEG translocon actively regulate the conformation, energetics, and dynamics of the SRP–cargo interaction, giving rise to highly coordinated assembly, commitment, and disassembly of the targeting complex. The challenges faced by SRP are general to targeting machineries. Hence, the active regulation of cargo interactions and the stepwise mechanism of cargo transfer observed here could represent general phenomena in protein targeting pathways. The bidentate nature of RNC’s interactions with SRP and SecYEG plays a key role in this regulatory mechanism since the individual contacts can be sequentially formed, dissolved, and exchanged, thus ensuring productive “relay” between upstream and downstream factors in the pathway. In addition to SRP and SecYEG, numerous other cellular machineries contact the ribosome exit site (e.g., chaperones, maturation, and quality control enzymes, etc.), many of which recognize both the ribosome and sequence motifs on the nascent protein. These multidentate interactions may provide an effective handling of the translating ribosome during the biogenesis of nascent proteins. The use of fluorescently labeled RNCs will be a powerful tool for a detailed molecular understanding of the multiplicity of fates that await the nascent protein as it exits the ribosome (Kramer et al., 2009).
Materials and methods
Materials
RNCs labeled with Cm were generated as described previously (Saraogi et al., 2011). In brief, cell-free extract containing engineered Cm-specific tRNA was supplemented with the cognate coumarin-tRNA synthetase, T7 polymerase, and Cm (Sigma-Aldrich) to generate stalled RNCs labeled with Cm during in vitro translation. RNCs were isolated via affinity chromatography using Strep-Tactin Sepharose resin (IBA) and sedimentation over a sucrose cushion.
Ffh and FtsY expression were driven by the T7 promoter on pET3 and pET9 plasmids, respectively (EMD Millipore). Ffh mutants were constructed using the QuikChange mutagenesis procedure (Agilent Technologies). Wild-type (wt) and mutant Ffh were expressed and purified as described previously (Peluso et al., 2001). In brief, Ffh was expressed from BL21(DE3)-pLysE cells with 1 mM IPTG at 37°C, at OD600 = 0.8. Ffh was purified on SP Sepharose Fast Flow resin (GE Healthcare) using a gradient of 150–500 mM NaCl, and then on a gel filtration column (Superose 12; GE Healthcare). Single cysteine mutants of Ffh were labeled with BODIPY-Fluorescein-N-(2-aminoethyl)-maleimide (BODIPY-FL; Invitrogen) as described previously (Zhang et al., 2008, 2009). In brief, ∼50 µM Ffh was treated with 2 mM Tris(2-carboxyethyl)phosphine (TCEP) for 2 h followed by incubation with a fivefold molar excess of the dye at 4°C. Labeled protein was purified using Sephadex G-25 resin (Sigma-Aldrich). Full-length Escherichia coli FtsY was expressed and purified as described previously (Peluso et al., 2001). In brief, FtsY on pET9a (a gift from W. Wintermeyer, Max Planck Institute for Biophysical Chemistry, Gottingen, Germany) was expressed from BL21(DE3) cells with 1 mM IPTG at 37°C, OD600 = 0.6. FtsY was purified using Q Sepharose Fast Flow resin (GE Healthcare) using a gradient of 150– 500 mM NaCl, Ni-NTA agarose resin (QIAGEN), and one or more rounds of anion exchange on a monoQ 10/100 GL column over a gradient of 150–400 mM NaCl (GE Healthcare).
70S ribosomes were purified from MRE600 cells as described previously (Moazed and Noller, 1989; Zhang et al., 2009). In brief, cells at OD600 0.4–0.6 were lysed by a French press at 16,000 psi. Clarified lysate was ultracentrifuged through a 1.1 M sucrose cushion at 100,000 g for 21 h. The ribosome pellet was washed with high ammonium sulfate buffer and further purified over a 10–40% sucrose gradient (SW32, 14 h at 50,000 g). Fractions containing 70S ribosomes were sedimented over a 1.1 M sucrose cushion (100,000 g, 17 h), dissolved, and stored at −80°C.
Cysteineless SecYEG was expressed from pEK20 driven by a trc promoter (a gift from A. Driessen, University of Groningen, Groningen, Netherlands) and purified as described previously (Akopian et al., 2013). For SecYEG labeling, a cysteine was introduced at position 180 and labeled using a published protocol (Kedrov et al., 2011), with modifications. In brief, 400 µl of 40 µM SecYEG purified using Ni-NTA agarose was dialyzed against 50 mM KHepes, pH 7.5, 150 mM NaCl, 10% glycerol, and 0.02% DDM, rebound to 800 µl Ni-NTA agarose, and incubated with a fivefold molar excess of BODIPY-FL in a total of 2 ml for 2 h at 4°C in labeling buffer (50 mM KHepes, pH 7.0, 300 mM NaCl, 20% glycerol, and 0.02% DDM). The beads were washed using dialysis buffer with 20 mM Imidazole, and the protein was eluted using dialysis buffer with 500 mM Imidazole.
Fluorescence measurements
All measurements were performed at 25°C in assay buffer (50 mM KHepes, pH 7.5, 150 mM KOAc, 10 mM Mg(OAc)2, 2 mM DTT, and 10% glycerol, supplemented with 0.02% DDM in assays with SecYEG) on a Fluorolog-3-22 spectrofluorometer (Horiba Jobin Yvon) or a Kintek stopped flow apparatus (for fast reactions). FRET measurements used an excitation wavelength of 360 nm and an emission wavelength of 453 nm, and RNCs labeled two residues downstream of the signal sequence. FRET efficiency (E) was calculated according to Eq. 1:
in which FDA and FD are the fluorescence intensities of the donor with or without the acceptor present, respectively.
SRP–RNC equilibrium binding affinities were measured using 20 nM Cm-labeled RNC and the indicated concentrations of BODIPY-FL–labeled SRP. The data were fit to Eq. 2:
in which Emax is the FRET efficiency at saturating SRP concentration and Kd is the dissociation constant of the RNC–SRP complex.
Observed rate constants for RNC–SRP binding (kobsd) were measured by mixing 40–50 nM Cm-labeled RNC with indicated concentrations of BODIPY-FL–labeled SRP, or an early/closed complex preformed using full-length FtsY and 100 µM GDP or GppNHp. The reaction time course was fit to Eq. 3:
in which Fobsd is the observed fluorescence, F0 is the initial fluorescence, ΔF1 is the amplitude of fluorescence change, and klin represents a very slow linear term included to obtain the best fitting. As the significance of this term was not apparent and its contribution to the fitting was small, we did not pursue this further. The dependence of kobsd on SRP concentration was fit to Eq. 4,
in which kon and koff are the association and dissociation rate constants, respectively, for the RNC–SRP complex. As koff values from this fit are inaccurate, these were measured directly as described in the next paragraph.
To determine the dissociation rate constants (koff) of RNCs from SRP at various stages of targeting, a preformed SRP–RNC early or closed targeting complex using Cm-labeled RNC (20 nM) and BODIPY-FL–labeled SRP (40 nM) was chased with 0.75–1 µM unlabelled SRP. The reaction time course was fit to Eq. 5a or Eq. 5b, in which Fobsd is the observed fluorescence, F0 is the initial fluorescence, ΔF1 and ΔF2 are the amplitudes of fluorescence changes, and k1 and k2 are the dissociation rate constants.
Eq. 5b was needed to analyze the dissociation kinetics with RNCphoA, which exhibited two kinetic phases that differed by approximately fivefold in rate constants (Fig. 2 F and Fig. S1 D). This could arise from different conformations of RNCphoA-SRP complexes or from multistep dissociation of SRP from the RNC that was more apparent with RNCphoA. The two phases responded similarly to FtsY (Fig. 3 F). Therefore, only the rate constants from the faster dissociating phase are used in the main text.
To determine the dissociation rate constant of SRP from 70S ribosomes, a preincubated solution of 100 nM BODIPY-labeled SRP and 500 nM 70S ribosomes was chased with 120–300 nM Cm-labeled RNC1A9L. Under these conditions, RNC1A9L-SRP association is much faster than SRP–ribosome complex dissociation. Thus, observed dissociation is rate-limited by and reflects the dissociation rate of the SRP–ribosome complex. The reaction time course was fit to Eq. 5b.
RNC–SecYEG equilibrium binding affinities (Kd) were measured using 20 nM Cm-labeled RNC and the indicated concentrations of SecYEG. A parallel titration of SecYEG into buffer was used to correct for light scattering from SecYEG. The data were fit to Eq. 6:
in which F is the observed fluorescence, F0 is the fluorescence of RNC in the absence of SecYEG, and ΔF is the amplitude of fluorescence change. For measurements of FRET between RNC and SecYEG, 30 nM of labeled SecYEG was incubated with 40 nM Cm-labeled RNC. Because the addition of SecYEG induces a strong environmental sensitivity in Cm fluorescence, RNC bound to unlabeled SecYEG was used to obtain the donor-only signal and quantify FRET.
To measure the equilibrium of RNC transfer, SecYEG (wt or mutant) was titrated into a preformed closed targeting complex. The data were fit to Eq. 6, in which Kd was replaced by EC50 and denotes the amount of SecYEG needed for 50% transfer. To measure the rate of RNC transfer, a preformed closed targeting complex was mixed with 1 µM SecYEG. The reaction time course was fit to Eqs. 5a or 5b to give the observed rate constant (kobs) of transfer.
Kinetic simulations
Berkeley Madonna was used to simulate reaction time courses according to models (i) and (ii) in Fig. 7 E. For model (i), the following equations were used:
With acceptor in the SRP M domain, the Cm fluorescence was determined by: F(M) = 50[RNC] + 25[RNC–SRP–SR] + 25[RNC–SRP–SR–SecYEG] + 90 [RNC–SecYEG].
With acceptor in the SRP N domain, the Cm fluorescence was determined by: F(N) = 50[RNC] + 30[RNC–SRP–SR] + 60[RNC–SRP–SR–SecYEG] + 90 [RNC–SecYEG].
The coefficients denote the relative fluorescence of the corresponding species, as obtained from Fig. 6 F.
For model (ii), the following reactions were used: (asterisks represent the encounter complex)
With acceptor in the SRP M domain, the Cm fluorescence was determined by: F(M) = 50[RNC] + 25[RNC–SRP–SR] + 25[RNC–SRP–SR–SecYEG*]+ 25 [RNC–SRP–SR–SecYEG] + 90[RNC–SecYEG].
With acceptor in the SRP N domain, the Cm fluorescence was determined by: F(N) = 50[RNC] + 30[RNC–SRP–SR] + 30[RNC–SRP–SR–SecYEG*] + 60 [RNC–SRP–SR–SecYEG] + 90[RNC–SecYEG].
Simulation used the following values. Concentrations: 20 nM RNC, 40 nM SRP–SR, and 1 µM SecYEG. Experimentally determined constants: k5 = 3.4 µM−1s−1; k-5 = 0.003 s−1; k2 = 0.005 s−1; k−1/k1 × k−2/k2 = 0.039 µM (EC50 of transfer; Fig. 6, C and E). Estimated rate constants: k1 = 1.2 µM−1s−1; k−1 = 0.363 s−1; k3 = 1 µM−1s−1; k−3 = 0.08 s−1; k−2 = 0.0006 s−1.
To simulate the dissociation of a stable RNC–SRP or ribosome–SRP complex according to the three-step model of Holtkamp et al. (2012), the following equations (Eqs. 9a–9f) were used. R denotes RNC or ribosome, I1 and I2 denote the intermediates described by Holtkamp et al. (2012), chase refers to unlabelled SRP used in koff measurements, and I1chase and I2chase refer to the corresponding intermediates formed between “R” and chase molecules.
Assuming that our fluorescence assay reports specifically on the final stable complex (R–SRP), the normalized fluorescence is denoted as 1 × [R–SRP]. Values of k1, k−1, k2, k−2, k3, and k−3 were from Holtkamp et al. (2012). The initial concentrations of each species were calculated based on the conditions used (0.05 µM R, 0.1 µM SRP, and 2 µM chase) and the equilibrium constants reported by Holtkamp et al. (2012).
Online supplemental material
Figs. S1 show additional data for RNC binding and dissociation from SRP. Fig. S2 shows the time courses for RNC binding to the SRP–SR early and closed complexes. Fig. S3 shows signal sequence binding to the SRP M domain. Fig. S4 describes the SRP 19/21E mutant. Fig. S5 shows the interaction of the Cm probe with SecYEG. Table S1 lists the anisotropy of the Cm and BODIPY-FL fluorophores.
Acknowledgments
We thank Tom Miller and Connie Wang for discussions and members of the Shan group for comments on the manuscript.
This work was supported by National Institutes of Health grant GM078024 to S.-o. Shan. S.-o. Shan was supported by the Henry Dreyfus teacher-scholar award and the Packard and Lucile Fellowship in science and engineering. This project is funded in part by the Gordon and Betty Moore Foundation through grant GBMF2939 to S.-o. Shan.
The authors declare no competing financial interests.
References
Author notes
I. Saraogi and D. Akopian contributed equally to this paper.
I. Saraogi’s present address is Dept. of Chemistry, Indian Institute of Science Education and Research Bhopal, Bhopal 462066, India.