Physical forces direct the orientation of the cell division axis for cells cultured on rigid, two-dimensional (2D) substrates. The extent to which physical forces regulate cell division in three-dimensional (3D) environments is not known. Here, we combine live-cell imaging with digital volume correlation to map 3D matrix displacements and identify sites at which cells apply contractile force to the matrix as they divide. Dividing cells embedded in fibrous matrices remained anchored to the matrix by long, thin protrusions. During cell rounding, the cells released adhesive contacts near the cell body while applying tensile forces at the tips of the protrusions to direct the orientation of the cell division axis. After cytokinesis, the daughter cells respread into matrix voids and invaded the matrix while maintaining traction forces at the tips of persistent and newly formed protrusions. Mechanical interactions between cells and the extracellular matrix constitute an important mechanism for regulation of cell division in 3D environments.
Introduction
Cell division is an integral part of tissue morphogenesis and homeostasis, and control of the orientation of cell division is essential to proper development and maintenance of tissue architecture (Gillies and Cabernard, 2011; Morin and Bellaïche, 2011). In 2D cultures, when a cell rounds to divide, it maintains numerous short retraction fibers that link the cell body to the substratum. The retraction fibers bear forces that direct the orientation of the mitotic spindle (Théry et al., 2005; Fink et al., 2011) and help the daughter cells to respread and separate (Cramer and Mitchison, 1993; Burton and Taylor, 1997). In vivo, mitotic cells in the developing zebrafish neural tube (Alexandre et al., 2010) or in the nervous system and retina of the mouse embryo (Saito et al., 2003; Kosodo et al., 2008) form daughter cells whose differentiation fates depend on their connections to their extracellular surroundings. It seems likely that physical interaction between cells and the extracellular matrix is crucial for proper regulation of cell division.
Previous studies of cell division in culture have traditionally used glass or plastic dishes. These 2D culture systems have yielded important insights into the mechanism of cell division; however, they present environments that are rigid, uniform, and flat, and thus fail to reflect the character of cell–matrix interactions encountered in vivo. Natural fibrous matrices such as collagen or fibrin mimic more closely the physiological extracellular matrix (Cukierman et al., 2002; Pampaloni et al., 2007; Fraley et al., 2010; Hakkinen et al., 2011). However, the measurement of forces induced by cells fully embedded in 3D matrices is a challenge that requires 3D live-cell imaging and quantitative, minimally invasive tools. Thus, we have a limited understanding of how physical forces regulate cell division in 3D environments. Recent advances have extended measurements of 2D planar stresses to the third dimension by using confocal imaging combined with digital volume correlation (DVC; Maskarinec et al., 2009) or particle tracking algorithms (Legant et al., 2010; Koch et al., 2012) to resolve matrix displacements in all three spatial dimensions.
Previous work has shown that external forces regulate cell division in 2D cultures (Burton and Taylor, 1997; Fink et al., 2011). Here we examine the hypothesis that forces applied by dividing cells against the extracellular matrix (“traction forces”) control the orientation of cell division in three dimensions. We use a physiologically relevant matrix that mimics the essential features of many tissue environments: soft, fibrous, and 3D. By combining 4D (x, y, z, and t) time-lapse imaging with DVC, we mapped full-field matrix displacements to identify sites at which cells apply traction forces. Our measurements resolve highly localized sites of cell–matrix interaction that anchor the mitotic cell to the matrix fibers. We propose that these forces are involved in guiding the orientation of cell division.
Results and discussion
To capture the dynamics of cell division in 3D biomimetic environments, we encapsulated 3T3 fibroblasts in fibrin gels. The gels used in this study support cell adhesion and growth (Lesman et al., 2011), and exhibit fibrillar morphologies and shear moduli characteristic of compliant tissues such as mammary gland and brain (typically ∼100 Pa; Discher et al., 2005; Paszek et al., 2005). We used time-lapse confocal microscopy to collect stacks of images of dividing fibroblasts that expressed an actin-GFP fusion protein (actin-GFP) throughout the cell division cycle (Fig. 1 A, Fig. S1, and Videos 1 and 2).
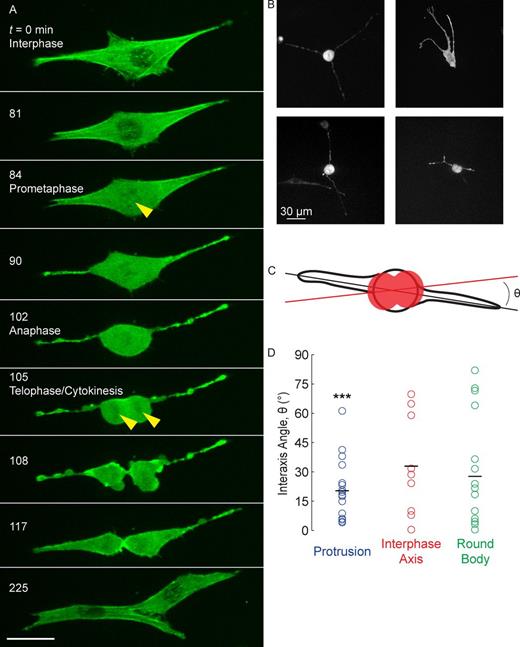
Cells dividing in 3D fibrin matrices extend protrusions that align with the axis of division. (A) A single well-spread actin-GFP fibroblast embedded in a 3D fibrin gel rounding into a sphere before cytokinesis (t = 84–102 min). The nuclear envelope breaks down at t = 84 min, as shown by the influx of GFP into the nuclear volume (yellow arrowheads). After rounding, cytokinesis occurs within minutes (t = 105–108 min). Long, thin protrusions are apparent throughout the division process. After cytokinesis (t = 117–225 min), the daughter cells respread along the long axis of the protrusions. A movie and 3D renderings are shown in Video 1 and Fig. S1, respectively. Bar, 30 µm. (B) The length, number, and morphology of the long, thin protrusions varied from cell to cell. Shown here are representative dividing cells from multiple experiments just before cytokinesis. (C) 3D renderings were used to calculate the angle θ between the cell axis (black line) and the division axis during anaphase (red line). (D) The cell axis is computed from either the protrusion direction during anaphase (“protrusion,” n = 15), the axis of the cell mass during interphase (“interphase axis,” n = 9), or the direction of the elongated mitotic body (“round body,” n = 14). The clustering of the angle θ near zero indicates a correlation between the protrusion and cell division axes (***, the data do not fit a uniform distribution, K-S test, P < 0.001).
Cells dividing in 3D fibrin matrices extend protrusions that align with the axis of division. (A) A single well-spread actin-GFP fibroblast embedded in a 3D fibrin gel rounding into a sphere before cytokinesis (t = 84–102 min). The nuclear envelope breaks down at t = 84 min, as shown by the influx of GFP into the nuclear volume (yellow arrowheads). After rounding, cytokinesis occurs within minutes (t = 105–108 min). Long, thin protrusions are apparent throughout the division process. After cytokinesis (t = 117–225 min), the daughter cells respread along the long axis of the protrusions. A movie and 3D renderings are shown in Video 1 and Fig. S1, respectively. Bar, 30 µm. (B) The length, number, and morphology of the long, thin protrusions varied from cell to cell. Shown here are representative dividing cells from multiple experiments just before cytokinesis. (C) 3D renderings were used to calculate the angle θ between the cell axis (black line) and the division axis during anaphase (red line). (D) The cell axis is computed from either the protrusion direction during anaphase (“protrusion,” n = 15), the axis of the cell mass during interphase (“interphase axis,” n = 9), or the direction of the elongated mitotic body (“round body,” n = 14). The clustering of the angle θ near zero indicates a correlation between the protrusion and cell division axes (***, the data do not fit a uniform distribution, K-S test, P < 0.001).
Monitoring cell division in three dimensions
Before division, the cell has a visible nucleus and is spread with well-developed extensions (Fig. 1 A, t = 0 min). As the cell begins to divide, the characteristic extensions at the poles of the cell body become thinner, the nuclear structure starts to disassemble (Fig. 1 A, t = 84 min), and the cell body adopts a near-spherical shape (Fig. 1 A, t = 102 min). The sphere then starts to split, a cleavage furrow is assembled (Fig. 1 A, t = 105 min), and cytokinesis occurs (Fig. 1 A, t = 105–108 min). Finally, the daughter cells reassemble their nuclei and spread along the long axis of the protrusions (Fig. 1 A, t = 225 min; and Fig. S1). The duration of mitosis in our experiments (∼20 min) is similar to that reported for 2D systems (Fink et al., 2011), which indicates that our imaging protocol has minimal effects on cell viability.
The most striking of these observations is the fact that dividing cells maintain long, thin protrusions (referred to as “mitotic protrusions” hereafter) tethered to the matrix throughout mitosis (Fig. 1 A, Fig. S1, and Videos 1 and 2). The mitotic protrusions showed diverse morphologies, with a mean length of ∼185 µm and a mean width of ∼2 µm (Fig. 1 B, Fig. S1 B, and Video 2). Cells with two protrusions were observed most frequently (65%), but cells with one (18%) or more than two (17%) were also evident (Fig. 1 B). Dividing cells expressing LifeAct-mRuby, which does not interfere with formation of actin filaments, showed similar morphologies, with long, thin protrusions present throughout division (Video 3). To test the hypothesis that the protrusions play a role in guiding the orientation of cell division, we measured the angle between the axis of division (determined at anaphase; Fig. 1 C, red line) and that of the protrusions (determined at cell rounding; Fig. 1 C, black line). We found that 80% of division events were characterized by interaxis angles smaller than 25° and that the angles did not fit a uniform distribution (Fig. 1 D, P < 0.001, Kolmogorov-Smirnov test [K-S test]). We also examined the long axes of the interphase cell and of the elongated mitotic cell body, both of which have been reported to co-orient with the axis of mitosis (Théry and Bornens, 2006; Minc et al., 2011). When we used the two-sample K-S test to compare the distribution of orientations of the protrusion axis to those of the other cellular axes, we found no significant differences (P = 0.31 and 0.55 for comparison with the interphase and mitotic cell axes, respectively). The protrusion axis predicts the orientation of the division axis as reliably as the long axis of either the interphase cell or the mitotic body (Fig. 1 D). These results are consistent with the hypothesis that the protrusions play a role in controlling the orientation of the division axis.
Maximum intensity projections of a 3D image of a 3T3 fibroblast (expressing actin-GFP, shown in Fig. 1) before, during, and after division in a 3D fibrin matrix. Images were acquired with time-lapse confocal microscopy (Swept Field; Nikon). Frames were taken every 3 min for 230 min. The initially spread cell rounds into a sphere while maintaining long, thin protrusions during division. The cell divides and the daughter cells respread along the protrusions.
Maximum intensity projections of a 3D image of a 3T3 fibroblast (expressing actin-GFP, shown in Fig. 1) before, during, and after division in a 3D fibrin matrix. Images were acquired with time-lapse confocal microscopy (Swept Field; Nikon). Frames were taken every 3 min for 230 min. The initially spread cell rounds into a sphere while maintaining long, thin protrusions during division. The cell divides and the daughter cells respread along the protrusions.
Representative maximum intensity projections of dividing cells embedded in 3D fibrin matrices (3T3 actin-GFP fibroblast cells). The movies are synchronized such that each shows 75 min before and after cytokinesis. Images were acquired with time-lapse confocal (Swept Field; Nikon) or two-photon (LSM 710; Carl Zeiss) microscopy. Frames were taken every 5 min.
Representative maximum intensity projections of dividing cells embedded in 3D fibrin matrices (3T3 actin-GFP fibroblast cells). The movies are synchronized such that each shows 75 min before and after cytokinesis. Images were acquired with time-lapse confocal (Swept Field; Nikon) or two-photon (LSM 710; Carl Zeiss) microscopy. Frames were taken every 5 min.
Representative maximum intensity projections of dividing 3T3 fibroblast cells stably expressing LifeAct-mRuby. The movies are synchronized such that each shows 75 min before and after cytokinesis. All of the cells maintain long, thin protrusions throughout the division process. Images were acquired with time-lapse confocal microscopy (LSM 710; Carl Zeiss). Frames were taken every 5 min.
Representative maximum intensity projections of dividing 3T3 fibroblast cells stably expressing LifeAct-mRuby. The movies are synchronized such that each shows 75 min before and after cytokinesis. All of the cells maintain long, thin protrusions throughout the division process. Images were acquired with time-lapse confocal microscopy (LSM 710; Carl Zeiss). Frames were taken every 5 min.
To determine whether long mitotic protrusions are specific to the 3D fibrin environment, we cultured cells on glass dishes or on top of thick fibrin gels. In both kinds of 2D cultures, we observed complete or partial retraction of the protrusions (Fig. 2, A and B); there was no evidence of protrusions for cells entering cytokinesis on glass substrates (uncoated glass; Fig. 2, A, D, and G; and Video 4). In contrast, cells embedded in 3D gels maintained long protrusions throughout the division cycle (Fig. 2, C, F, and G; and Videos 1–3), and the daughter cells always respread along the direction of the protrusions (Fig. 1 A, t = 225 min) even in events where the division and protrusion axes were misaligned (angle > 40°; Fig. S2). The distributions of interaxis angles (as defined in Fig. 1 C) measured for cells on the 2D substrates were not statistically different from a uniform distribution (referred to as “random” hereafter; Fig. 2 H, K-S test, P > 0.19), in contrast to the behavior observed in the 3D fibrin environment (Fig. 2 H, K-S test, P < 0.001). These results indicate that the maintenance of long protrusions during cell division and the co-orientation of the protrusions with the division axis are distinctive characteristics of cells embedded in 3D fibrin gels.
Maximum intensity projections of a 3T3 actin-GFP cell dividing on an uncoated glass substrate (on the left) or on top of a thick (∼200 µm) fibrin gel (on the right), collected with a confocal microscope (Swept Field; Nikon) every 5 min for 150 min. The movie is from the experiments shown in Fig. 2 (A and B). The cell cultured on glass retracts its long protrusions completely during rounding. The cell plated on the fibrin gel partially retracts its long protrusions during rounding.
Maximum intensity projections of a 3T3 actin-GFP cell dividing on an uncoated glass substrate (on the left) or on top of a thick (∼200 µm) fibrin gel (on the right), collected with a confocal microscope (Swept Field; Nikon) every 5 min for 150 min. The movie is from the experiments shown in Fig. 2 (A and B). The cell cultured on glass retracts its long protrusions completely during rounding. The cell plated on the fibrin gel partially retracts its long protrusions during rounding.
Dividing cells maintain intact protrusions during division in 3D matrices. (A–C) Time-lapse images of dividing cells cultured on 2D untreated glass (A), cultured on 200 µm thick fibrin gels (B), and fully embedded in 3D fibrin gels (C). Cells dividing on glass treated with fibrinogen were similar in appearance to those in B. Images shown were recorded just before division (t = 0 min), at the end of cell rounding (t = 20 min), during cytokinesis (t = 25 min), and after spreading of the daughter cells (t = 90 min). Bars, 20 µm. (D–F) Quantification of normalized protrusion length (current length/initial length) over the course of cell rounding demonstrates that dividing cells on untreated glass substrates (D, n = 12) completely and rapidly retract their protrusions, whereas dividing cells on top of 2D fibrin gels (E, n = 11) gradually and partially retract their protrusions. In contrast, cells in 3D fibrin gels (F, n = 17) maintain intact protrusions for the entire division cycle. (G) Normalized protrusion length at the end of cell rounding (5 min before cytokinesis) is plotted for cells dividing on 2D glass (n = 12), on 2D glass coated with fibrinogen (n = 18), on 200-µm-thick fibrin gels (n = 11), and in 3D fibrin matrices (n = 17). Normalized protrusion length is greater for cells in 3D matrices than on any of the 2D substrates (***, significant difference between 3D and 2D data, ANOVA, P < 10−6). (H) The angle θ between the division and protrusion axes is random (K-S test, P > 0.19) for cells on 2D substrates, but correlated in 3D fibrin matrices (K-S test, P < 0.001).
Dividing cells maintain intact protrusions during division in 3D matrices. (A–C) Time-lapse images of dividing cells cultured on 2D untreated glass (A), cultured on 200 µm thick fibrin gels (B), and fully embedded in 3D fibrin gels (C). Cells dividing on glass treated with fibrinogen were similar in appearance to those in B. Images shown were recorded just before division (t = 0 min), at the end of cell rounding (t = 20 min), during cytokinesis (t = 25 min), and after spreading of the daughter cells (t = 90 min). Bars, 20 µm. (D–F) Quantification of normalized protrusion length (current length/initial length) over the course of cell rounding demonstrates that dividing cells on untreated glass substrates (D, n = 12) completely and rapidly retract their protrusions, whereas dividing cells on top of 2D fibrin gels (E, n = 11) gradually and partially retract their protrusions. In contrast, cells in 3D fibrin gels (F, n = 17) maintain intact protrusions for the entire division cycle. (G) Normalized protrusion length at the end of cell rounding (5 min before cytokinesis) is plotted for cells dividing on 2D glass (n = 12), on 2D glass coated with fibrinogen (n = 18), on 200-µm-thick fibrin gels (n = 11), and in 3D fibrin matrices (n = 17). Normalized protrusion length is greater for cells in 3D matrices than on any of the 2D substrates (***, significant difference between 3D and 2D data, ANOVA, P < 10−6). (H) The angle θ between the division and protrusion axes is random (K-S test, P > 0.19) for cells on 2D substrates, but correlated in 3D fibrin matrices (K-S test, P < 0.001).
Previous studies of cell division in 2D culture have described the importance of retraction fibers in the spreading of daughter cells (Cramer and Mitchison, 1993) and in directing the orientation of division (Fink et al., 2011). The protrusions observed in this work in 3D gels are larger, fewer, and more polarized than the retraction fibers observed in 2D systems (Cramer and Mitchison, 1993; Théry et al., 2005; Fink et al., 2011). Studies of cell division in the zebrafish neural tube (Alexandre et al., 2010) or in the nervous system and retina of the mouse embryo (Saito et al., 2003; Kosodo et al., 2008) have described similar extended cellular structures (often referred to as “cellular processes”). The cellular processes attach dividing cells to the apical and basal lamina, extend tens of microns in length (Kosodo et al., 2008), and regulate differentiation of the daughter cells (Alexandre et al., 2010). We propose that long cellular processes may regulate cell division in 3D environments by transmitting mechanical forces in a manner similar to that of retraction fibers observed in two dimensions (Théry et al., 2005; Fink et al., 2011).
Mapping 3D cell-induced matrix displacements during division
To probe the role of cell-induced mechanical forces in regulating the orientation of cell division, we examined the correlation between the protrusion and division axes for cells treated with blebbistatin. Blebbistatin inhibits myosin II contractility but does not disrupt the initiation of mitosis (Straight et al., 2003). After treatment with blebbistatin, the interaxis angles were randomly distributed (Fig. 3 A, K-S test, P > 0.18; and Video 5), in contrast to the behavior observed in the absence of the drug (Fig. 3 A, K-S test, P < 0.001; and Video 2). This result suggests that cell-induced forces play a significant role in determining the orientation of cell division.
Examples of dividing 3T3 fibroblast cells (labeled with actin-GFP) embedded in 3D fibrin matrices in the presence of blebbistatin (50 µM). The movies are synchronized such that each shows 75 min before and after formation of the contractile actin ring. Images were acquired with time-lapse two-photon microscopy (LSM 710; Carl Zeiss). Frames were taken every 5 min.
Examples of dividing 3T3 fibroblast cells (labeled with actin-GFP) embedded in 3D fibrin matrices in the presence of blebbistatin (50 µM). The movies are synchronized such that each shows 75 min before and after formation of the contractile actin ring. Images were acquired with time-lapse two-photon microscopy (LSM 710; Carl Zeiss). Frames were taken every 5 min.
3D traction forces during cell division. (A) For cells treated with blebbistatin, the distribution of angles (θ) between the protrusion and division axes cannot be distinguished from a uniform distribution (K-S test, P > 0.18, n = 17). The mean interaxis angles are significantly different in the presence and absence of blebbistatin (n = 17 and n = 23, respectively; *, P < 0.05, Mann-Whitney-Wilcoxon test). (B) 3D maximum intensity projections of fluorescent particles around a rounding cell (t = 0) and daughter cells (t = 210 min). (C) Quiver plots of 3D matrix displacements show that the cell applies traction to the matrix primarily at the tips of its long protrusions during division (t = 0, arrowheads). Approximately 3 h after division, the daughter cells apply displacements to other regions of the matrix as they respread. See Videos 7–9 for additional time points. (D) During rounding, cells cause larger matrix displacements near the tips of cellular protrusions than near the cell body (**, mean significantly <1; Student’s t test, P < 0.01, n = 7).
3D traction forces during cell division. (A) For cells treated with blebbistatin, the distribution of angles (θ) between the protrusion and division axes cannot be distinguished from a uniform distribution (K-S test, P > 0.18, n = 17). The mean interaxis angles are significantly different in the presence and absence of blebbistatin (n = 17 and n = 23, respectively; *, P < 0.05, Mann-Whitney-Wilcoxon test). (B) 3D maximum intensity projections of fluorescent particles around a rounding cell (t = 0) and daughter cells (t = 210 min). (C) Quiver plots of 3D matrix displacements show that the cell applies traction to the matrix primarily at the tips of its long protrusions during division (t = 0, arrowheads). Approximately 3 h after division, the daughter cells apply displacements to other regions of the matrix as they respread. See Videos 7–9 for additional time points. (D) During rounding, cells cause larger matrix displacements near the tips of cellular protrusions than near the cell body (**, mean significantly <1; Student’s t test, P < 0.01, n = 7).
To quantify the mechanical forces exerted by dividing cells, we applied 3D traction force microscopy. Measurement of cell traction forces has only recently been extended to the third dimension (Hur et al., 2009; Maskarinec et al., 2009; Franck et al., 2011), particularly for cells fully embedded in 3D gels (Legant et al., 2010; Koch et al., 2012). We quantified the full-field matrix displacements induced by dividing cells in 3D fibrous matrices by combining confocal imaging and a DVC algorithm (Franck et al., 2007). In brief, we co-encapsulated cells and 0.5-µm fluorescent particles in fibrin gels (Fig. 3 B and Video 6), acquired confocal volume stacks of the cells and the particles surrounding them, and computed matrix displacements using DVC (Franck et al., 2007). The DVC algorithm was used to correlate all volume stacks to a reference time point acquired after injection of blebbistatin. Treatment with blebbistatin inhibits the generation of contractile forces by the cell (Straight et al., 2003) and allows imaging of a stress-free reference state of the gel. We believe that the experiments described here constitute the first quantitative study of cell-induced matrix displacements during division in three dimensions.
3D maximum intensity projection of fluorescent particles (red; Invitrogen) surrounding a 3T3 dividing fibroblast cell (actin-GFP, green). The movie is from the same experiment as Fig. 3 (B and C). Frames were collected with a confocal microscope (Swept Field; Nikon) every 30 min.
3D maximum intensity projection of fluorescent particles (red; Invitrogen) surrounding a 3T3 dividing fibroblast cell (actin-GFP, green). The movie is from the same experiment as Fig. 3 (B and C). Frames were collected with a confocal microscope (Swept Field; Nikon) every 30 min.
As cells round up, matrix displacements adjacent to the cell body decrease to nearly undetectable values (Fig. 3 C, t = 0; Fig. 3 D; and Videos 7 and 8). These findings are consistent with earlier observations of reduced 2D traction forces near the cell body just before cytokinesis (Burton and Taylor, 1997). Although cells can push on their surroundings during rounding (Stewart et al., 2011), we observed no outward displacement of the 3D fibrin matrix near the rounded cell body (Fig. 3 C, t = 0). In contrast, during mitotic cell rounding, large matrix displacements were observed at the tips of the mitotic protrusions that anchor the cell to the matrix (Fig. 3 C, t = 0, arrowheads; and Video 8). Thus, dividing cells maintain protrusions that not only connect the cell body to the matrix but also support tensile forces. Like thin wires, the protrusions support only uniaxial tension, and therefore only tensile forces propagate along the protrusions and into the cell body during division. Together with a previous finding that cells divide in the direction of externally applied tensile forces (Fink et al., 2011), these observations suggest that tensile forces transmitted through the protrusions act on the cell cortex to direct spindle orientation during mitosis, possibly through polarization of actin and myosin II (Effler et al., 2006; Fink et al., 2011). Finally, as the daughter cells respread after cytokinesis, they pull on the matrix primarily at the tips of persistent and newly developing protrusions (Fig. 3 C, t = 210 min).
Multi-angle view of Fig. 3 C, t = 0, showing vector plots of matrix displacements during mitotic rounding of a 3T3 fibroblast cell labeled with GFP-actin. Displacements were computed using DVC and vector plots, and 3D isosurface renderings of the cell were generated using MATLAB.
Multi-angle view of Fig. 3 C, t = 0, showing vector plots of matrix displacements during mitotic rounding of a 3T3 fibroblast cell labeled with GFP-actin. Displacements were computed using DVC and vector plots, and 3D isosurface renderings of the cell were generated using MATLAB.
Direct imaging of matrix fibers
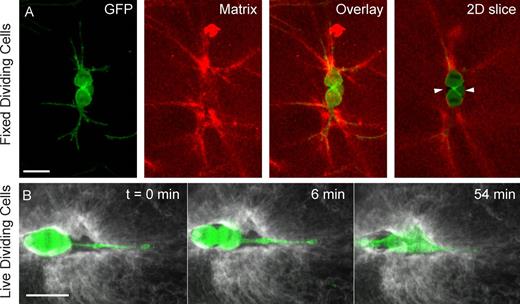
We further examined the interactions of dividing cells with the surrounding matrix through 3D imaging of fibrin gels labeled with fluorescent dyes (Fig. 4 and Video 10). Images of dividing GFP-actin fibroblasts and the surrounding fibrous matrix reveal that, around the mitotic protrusions, the matrix is highly remodeled (Fig. 4 A), which is consistent with the observation that the cell applies localized force at the protrusion tips (Fig. 3 C). Adjacent to the cell body, clear voids separate the cell from the matrix (Fig. 4 A, arrowheads), which is consistent with the observed decline in traction forces in this region (Fig. 3 C). Inspection of the matrix voids revealed that they reflect the shape of the fully spread cell (Fig. 4 B and Fig. S3 A), which suggests that the voids form through proteolysis of the gel as the cell spreads after seeding. The presence of matrix voids may explain our observation that the cell body does not push the matrix outward during rounding (Fig. 3 C, t = 0); the void volume may be large enough to accommodate the rounded cell body. Throughout mitosis, the tips of the cellular protrusions are firmly embedded in the matrix meshwork (Fig. 4 B and Fig. S3, A and B), which is consistent with the large matrix displacements measured at the tips (Fig. 3 C, t = 0, arrowheads). After division, the daughter cells reoccupy the matrix voids (Fig. S3 C).
Time-lapse series of a single confocal plane of a 3T3 dividing fibroblast cell (actin-GFP, green) embedded in a fibrin matrix (labeled with Alexa Fluor 546, gray). Frames were taken every 3 min for 90 min with a confocal microscope (Swept Field; Nikon)
Time-lapse series of a single confocal plane of a 3T3 dividing fibroblast cell (actin-GFP, green) embedded in a fibrin matrix (labeled with Alexa Fluor 546, gray). Frames were taken every 3 min for 90 min with a confocal microscope (Swept Field; Nikon)
Dividing cells embedded in fluorescently labeled fibrin matrices. (A) Fixed gels showing a dividing fibroblast (GFP-actin, green) and the fibrin matrix (red) during cytokinesis. Dividing cells deform the matrix locally around the mitotic protrusions (strong red signal due to dense fibers). Matrix voids are evident between the mitotic cell body and the matrix (“2D slice,” arrowheads). (B) One confocal plane of a live dividing cell (actin-GFP, green) and the labeled fibrin matrix (gray) at cell rounding (t = 0), cytokinesis (t = 6 min), and shortly after division (t = 54 min). See Video 10 and Fig. S3 for the entire sequence of events. Bars, 30 µm.
Dividing cells embedded in fluorescently labeled fibrin matrices. (A) Fixed gels showing a dividing fibroblast (GFP-actin, green) and the fibrin matrix (red) during cytokinesis. Dividing cells deform the matrix locally around the mitotic protrusions (strong red signal due to dense fibers). Matrix voids are evident between the mitotic cell body and the matrix (“2D slice,” arrowheads). (B) One confocal plane of a live dividing cell (actin-GFP, green) and the labeled fibrin matrix (gray) at cell rounding (t = 0), cytokinesis (t = 6 min), and shortly after division (t = 54 min). See Video 10 and Fig. S3 for the entire sequence of events. Bars, 30 µm.
This study has identified important roles for cell–matrix interactions in regulating cell division in 3D environments. The model that emerges is summarized in Fig. 5. During cell rounding, the cell releases adhesive contacts with the matrix around the cell body, but remains anchored by traction forces acting at the tips of polarized long protrusions (Fig. 5, A and B). The dividing cell coaligns its division and protrusion axes as it applies tensile forces against the extracellular matrix. After cytokinesis, the daughter cells assume the shapes of matrix voids that are revealed during cell rounding (Fig. 5 C). Finally, the daughter cells apply traction forces at the protrusion tips as they respread and invade the matrix (Fig. 5 D). Future investigations should reveal the importance of additional physical cues, including matrix stiffness and topography, in regulating cell division in 3D environments.
Illustration of cell division in 3D fibrous matrices. (A) As the cell rounds to divide, its main protrusions thin but stay intact, and a void is created between the cell and matrix (B). The cell divides along the direction of the protrusions while exerting minimal traction around the cell body but applying high traction at the tips of the protrusions. Daughter cells fill the matrix void space (C), and respread into the matrix along the long axis of the protrusions while exerting substantial traction (D).
Illustration of cell division in 3D fibrous matrices. (A) As the cell rounds to divide, its main protrusions thin but stay intact, and a void is created between the cell and matrix (B). The cell divides along the direction of the protrusions while exerting minimal traction around the cell body but applying high traction at the tips of the protrusions. Daughter cells fill the matrix void space (C), and respread into the matrix along the long axis of the protrusions while exerting substantial traction (D).
Materials and methods
Cell culture
Swiss 3T3 fibroblasts (passages 10–20) stably transfected either with GFP-actin or with mRuby-LifeAct (obtained as gifts from S. Fraser, University of Southern California, Los Angeles, CA) were cultured in DMEM medium supplemented with 10% fetal bovine serum and 1× nonessential amino acids in a 37°C humid incubator. Endothelial cells were included to promote fibroblast proliferation (Hirschi et al., 1999). Human umbilical vein endothelial cells (HUVEC) stably transfected with RFP (Angio-Proteomie) were cultured in endothelial growth medium containing 5% serum, growth supplements, and 1× penicillin and streptomycin (passages 4–9; Angio-Proteomie) also in a 37°C humid incubator.
3D fibrin gel preparation
3T3 fibroblasts-GFP-actin (∼3,000 cells) and HUVEC-RFP (∼3,000 cells) were mixed with 20 µl of 5 mg/ml fibrinogen (Omrix Biopharmaceuticals). In a separate vial, red 0.5 µm carboxylated fluorescent particles (Ex 580/Em 605; Invitrogen) were mixed with 20 µl of a 20 U/ml thrombin solution (Omrix Biopharmaceuticals) and vortexed for 1 min to a final particle concentration of 0.05%. The thrombin suspension was placed on a No. 1.5 coverslip in a 35-mm dish (MatTek Corporation), and mixed gently with the fibrinogen suspension. The resulting fibrin gel was placed in the incubator for 15 min to fully polymerize, after which warm medium (50% fibroblast, 50% HUVEC medium) was added to cover the gel. For imaging of cell division without measurement of matrix displacements, fibrin gels were made in a similar way but without including fluorescent particles.
2D culture preparation
2D glass.
3T3 fibroblasts-GFP and HUVEC-RFP were mixed in a 1:1 ratio and plated on a coverslip bottom dish (No. 1.5; MatTek Corporation) uncoated or coated with fibrinogen solution (100 µg/ml, 2 h incubation at room temperature). The 3T3 cells were imaged after overnight incubation while maintaining low confluence.
2D fibrin.
To prepare flat gels for studies of cell division on 2D fibrin substrates, 10 µl of 5 mg/ml fibrinogen was mixed with 10 µl of 20 U/ml thrombin, placed on a No. 1.5 coverslip in a 35-mm dish (MatTek Corporation), and incubated for 15 min. A sterile coverslip was placed on top of the gel to flatten it. After addition of phosphate-buffered saline to fill the well, the coverslip was removed and the cells were cultured on the gels. The thickness of the gel was measured by confocal imaging of gel-embedded fluorescent beads to be ∼200 µm.
Fibrin gel labeling and fixation
Alexa Fluor 546 carboxylic acid, succinimidyl ester (Invitrogen) was mixed with fibrinogen solution in a 7.5:1 molar ratio for 1 h at room temperature and then filtered through a HiTrap desalting column (GE Healthcare) packed with Sephadex G-25 resin to separate the unreacted dye. The labeled fibrinogen was then mixed with thrombin and cells to create labeled, cell-loaded fibrin gels. For the fixation protocols, labeled fibrin gels loaded with 3T3 GFP-actin cells for at least 6 h were immersed in 4% paraformaldehyde for 10 min and subsequently washed with PBS. The nuclear material was labeled with DAPI before imaging.
Fibrin gel characterization
The constitutive mechanical properties of fibrin gels without cells were measured using a stress-controlled AR1000 rheometer equipped with 8-mm-diameter aluminum parallel plates. Frequency sweep and creep tests were performed. The fibrin gel was prepared on the rheometer and placed between sheets of sandpaper to avoid slipping. The bulk material stiffness could not be used to compute traction forces owing to local matrix inhomogeneity at the scale of the cell.
Time-lapse microscopy
After overnight incubation, fibrin gels were imaged with a Swept Field confocal microscope mounted on a Ti stand (Nikon) outfitted with a 40× 1.15 NA Apochromat water immersion objective lens (Nikon) using the microscope’s 30-µm pinhole (for imaging the fluorescent particles), 45-µm pinhole (for imaging the fibrin gel), or 60-µm pinhole (for imaging the cells). Images were captured with a QuantEM:512SC camera (Photometrics) using NIS-Elements Ar software (Nikon). Imaging was performed in a 50/50% mix of fibroblast and HUVEC medium in a custom-built 37°C/5% CO2 incubation chamber after allowing the system to equilibrate for ∼4 h. All experiments on cells embedded in the matrix were conducted on cells positioned at least 100 µm from the bottom surface of the gel.
Confocal z-stacks capturing single fibroblasts were acquired every 15–30 min (for measurement of matrix displacements) or every 2–3 min (for visualizing dividing cells) for ∼6 h. Stacks were collected with a z step size of 0.4 µm (for displacement measurements) to 1 µm (for cell visualization). For experiments involving analysis of matrix displacements, blebbistatin (85 µM; Sigma-Aldrich) was added to the medium at the end of the experiment and stress-free stacks were acquired for an additional 4 h.
Measurement of matrix displacements
To compute matrix displacements, red 0.5 µm fluorescent particles (Invitrogen) were used to create a speckle pattern for a DVC algorithm (Franck et al., 2007) implemented in MATLAB (MathWorks). Before running the DVC algorithm, 3D deconvolution was performed on the volume stacks of the particles in MATLAB using the Lucy-Richardson algorithm as described previously (Franck et al., 2007). The DVC algorithm used a Fourier transform–based correlation to compute the 3D displacements at the center of a subset of 64 × 64 × 64 voxels on a grid of points within the image volume of 512 × 512 × 276 voxels. Real-time displacements were computed by using blebbistatin (final concentration of 85 µM) to inhibit the cell’s myosin II activity, thus allowing the gel to recover to an unstressed state. All stacks collected before injecting blebbistatin were correlated to the unstressed stack acquired after the addition of blebbistatin. To account for minor swelling or shrinking of the fibrin gels during displacement measurements, the mean normal matrix strains were computed. The displacement fields were then corrected by subtracting the displacements associated with mean normal matrix strains.
To quantify the experimental error associated with the displacement measurements, control experiments were performed on fibrin gels without cells. In these experiments, confocal stacks were collected every 30 min for ∼6 h. Then, blebbistatin was injected into the medium to a concentration of 85 µM. Errors in matrix displacements computed with the DVC algorithm were found to be <0.3 µm. Further experiments to analyze the DVC algorithm’s ability to compute matrix displacements and strains were conducted by applying a computational translation or strain to the volume stacks. After correcting for swelling or shrinking of the fibrin gels, displacements were found to be within a standard deviation (∼0.3 µm) of their expected values. A further experiment was performed wherein a fibrin gel was imaged during a compression experiment as described previously (Franck et al., 2007). In this experiment, the fibrin gel was loaded in uniaxial compression using a custom-built loading device designed to fit on the microscope stage (Franck et al., 2007). Displacements of the loaded fibrin gel were computed using DVC and were within a standard deviation of their expected values.
Quantification of protrusion axis, cell axis during interphase, mitotic cell body axis, and division orientation
For cells in 3D matrices, mitotic protrusion directions were determined during cell rounding by fitting a line between the protrusion tip and its intersection with the rounded cell body. Vectors associated with the direction of each protrusion were computed and normalized to unit length. The normalized protrusion vectors were averaged to determine a mean resultant protrusion direction. For cells on flat substrates, protrusions retracted during rounding, and therefore the protrusion directions were measured immediately before cell rounding. Note that these protrusions, measured before rounding in two dimensions, are different from the retraction fibers observed for rounded cells on 2D substrates (Cramer and Mitchison, 1993; Théry et al., 2005; Fink et al., 2011). Resultant protrusion directions for cells on 2D substrates were computed with the same normalization and averaging procedure as for cells in 3D matrices. The cell axis during interphase (a mathematical description of the axis used in the Hertwig rule; Gillies and Cabernard, 2011) was calculated by computing a 3D line of best fit to data points corresponding to the voxels within the cell during interphase. The line of best fit was computed by minimizing the mean square orthogonal distance between each voxel and the line in MATLAB. To calculate the axis of the elongated cell body, deconvolution (Lucy-Richardson algorithm, MATLAB) was applied to volume stacks of the cell to minimize the effect of imaging artifacts in the out-of-plane direction. A line of best fit using the minimum mean square orthogonal distance was then calculated for the voxels within the rounded cell body but not the voxels within the protrusions. The division axis orientation was quantified by fitting spheres to renderings of the two daughter cells collected immediately after cytokinesis, computing the vector connecting the centers of the spheres, and normalizing the computed vector to unit length. The angle between the cell axis and the division axis orientation was computed by taking the dot product of the unit vectors associated with these directions.
Image rendering and plots
Maximum intensity projections were created using ImageJ (National Institutes of Health) or Imaris (version 7.6.4; Bitplane). Matrix displacement vectors were plotted in MATLAB.
Division of blebbistatin-treated cells
3T3 actin-GFP cells encapsulated in fibrin gels for 15–20 h were used in all experiments. Blebbistatin (Sigma-Aldrich) was added to the fibroblast medium at a final concentration of 50 µM. Cells were preincubated for 2–3 h before imaging in the microscope incubator at 37°C to allow for temperature equilibrium and penetration of blebbistatin into the gels. Imaging was performed using ZEN software (Carl Zeiss) on an LSM 710 microscope (Carl Zeiss) using two-photon laser scanning at a wavelength of 900 nm in multiple x-y positions with the use of a photomultiplier tube. Z stacks were captured every 5 min with a z step size of 2–3 µm. The microscope was equipped with a 40× 1.2 NA Apochromat water immersion objective lens (Carl Zeiss).
Statistics
All statistical tests were performed with MATLAB. The K-S test was used to determine whether the interaxis angles fit a uniform distribution. Here, we use the term “random” when the angles are not statistically different from a uniform distribution; we use “correlation” when the angles are statistically different from a uniform distribution. The two-sample K-S test was used to compare distributions between pairs of groups. The Mann-Whitney-Wilcoxon test was used to determine differences in the averages of interaxis angles. The Student’s t test was used to compare the data to a normal random variable with mean of 1. Analysis of variance (ANOVA) was used to compare the means of multiple groups. We use the following convention in the figures to indicate the level of significance: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Online supplemental material
Online supplemental figures show maximum intensity projections and 3D isosurface renderings of dividing cells (Fig. S1 A), protrusion lengths measured during rounding for cells plated on or within fibrin matrices (Fig. S1 B), daughter cells respreading and separating after division (Fig. S2), and images of matrix voids revealed during cell rounding (Fig. S3). Videos 1–3 show projections of cells expressing actin-GFP (Videos 1 and 2) or LifeAct-mRuby (Video 3) while dividing in a 3D matrix. Video 4 shows cells dividing on top of a 2D substrate. Video 5 shows cells in a 3D matrix dividing in the presence of blebbistatin. Video 6 illustrates the method used to compute cell-induced matrix displacements by showing motion of the fluorescent particles surrounding the cell during division. Videos 7–9 show cell-induced matrix displacements at different time points (Video 7) and different angles (Videos 8 and 9). Video 10 shows a time sequence of a cell dividing in a fluorescently labeled fibrous matrix.
Vector plots of matrix displacements and 3D isosurface renderings of a 3T3 fibroblast cell during and after division. Frames were taken every 30 min for 570 min with a confocal microscope (Swept Field; Nikon); displacements were computed using DVC and plotted in MATLAB. The dividing cell shown in Fig. 3 applies inward displacements to the matrix at the tips of its protrusions. After division, the displacements increase at the leading edges of new extensions as the daughter cells begin to spread. Displacements decay to zero after blebbistatin is injected into the medium.
Vector plots of matrix displacements and 3D isosurface renderings of a 3T3 fibroblast cell during and after division. Frames were taken every 30 min for 570 min with a confocal microscope (Swept Field; Nikon); displacements were computed using DVC and plotted in MATLAB. The dividing cell shown in Fig. 3 applies inward displacements to the matrix at the tips of its protrusions. After division, the displacements increase at the leading edges of new extensions as the daughter cells begin to spread. Displacements decay to zero after blebbistatin is injected into the medium.
Multi-angle view of Fig. 3 C, t = 210 min, showing vector plots of matrix displacements around spreading 3T3 fibroblast daughter cells labeled with GFP-actin. Displacements were computed using DVC and vector plots, and 3D isosurface renderings of the cell were generated using MATLAB.
Multi-angle view of Fig. 3 C, t = 210 min, showing vector plots of matrix displacements around spreading 3T3 fibroblast daughter cells labeled with GFP-actin. Displacements were computed using DVC and vector plots, and 3D isosurface renderings of the cell were generated using MATLAB.
Acknowledgments
We thank Scott Fraser for providing the transfected fibroblasts. We thank the Biological Imaging Center at Caltech for use of the two-photon microscope.
This research was supported by grants from the National Science Foundation (Division of Materials Research No. 0520565 and 1206121). J. Notbohm was supported by the Department of Defense through the National Defense Science and Engineering Graduate Fellowship Program. A. Lesman was supported in part by the Rothschild fellowship foundation.
The authors declare no competing financial interests.
References
Author notes
A. Lesman and J. Notbohm contributed equally to this paper.