The mitotic checkpoint monitors kinetochore–microtubule attachment and prevents anaphase until all kinetochores are stably attached. Checkpoint regulation hinges on the dynamic localization of checkpoint proteins to kinetochores. Unattached, checkpoint-active kinetochores accumulate multiple checkpoint proteins, which are depleted from kinetochores upon stable attachment, allowing checkpoint silencing. Because multiple proteins are recruited simultaneously to unattached kinetochores, it is not known what changes at kinetochores are essential for anaphase promoting complex/cyclosome (APC/C) inhibition. Using chemically induced dimerization to manipulate protein localization with temporal control, we show that recruiting the checkpoint protein Mad1 to metaphase kinetochores is sufficient to reactivate the checkpoint without a concomitant increase in kinetochore levels of Mps1 or BubR1. Furthermore, Mad2 binding is necessary but not sufficient for Mad1 to activate the checkpoint; a conserved C-terminal motif is also required. The results of our checkpoint reactivation assay suggest that Mad1, in addition to converting Mad2 to its active conformation, scaffolds formation of a higher-order mitotic checkpoint complex at kinetochores.
Introduction
The mitotic checkpoint inhibits the anaphase promoting complex/cyclosome (APC/C) in the presence of unattached kinetochores and silences this inhibition once all kinetochores are stably attached to spindle microtubules. Checkpoint activity (i.e., APC/C inhibition) and silencing are correlated with changes in the kinetochore localization of checkpoint proteins, including Mad1, Mad2, Bub1, BubR1, Bub3, Mps1, and Cdc20 (Kops and Shah, 2012). These proteins are enriched at kinetochores until stable MT attachment and are required for checkpoint function. A central challenge in understanding the mitotic checkpoint is to dissect how local changes in checkpoint protein occupancy at kinetochores drive global changes in checkpoint activity.
Preventing the removal of Mad1 or Mps1 from kinetochores via genetic fusion to the stable kinetochore component Mis12 blocks anaphase, which demonstrates that the removal of these proteins is required for checkpoint silencing (Jelluma et al., 2010; Maldonado and Kapoor, 2011). To probe checkpoint activation (i.e., switching the checkpoint from an “off” state to an “on” state), experimental intervention when the checkpoint is silenced, such as at metaphase, is an attractive approach. The checkpoint can be reactivated at metaphase by disrupting kinetochore–microtubule attachments, using either spindle poisons or laser microsurgery (Clute and Pines, 1999; Dick and Gerlich, 2013). It is unknown if the checkpoint can be reactivated after metaphase without compromising kinetochore–microtubule attachment. Metaphase kinetochores are stably attached and depleted of checkpoint proteins, so they provide a context in which to test the effect of increasing the kinetochore concentration of an individual protein in the absence of the full set of signals associated with the unattached state.
We tested whether increasing kinetochore localization of Mad1 at metaphase is sufficient to reactivate the checkpoint. Mad1 and its partner Mad2 are essential checkpoint proteins (Li and Murray, 1991). Mad1 constitutively binds a single copy of Mad2 in the closed conformation, and this bound population of Mad2 serves as the kinetochore receptor for cytosolic, open-conformation Mad2 (Luo et al., 2004; De Antoni et al., 2005; Lara-Gonzalez et al., 2012). Continual recruitment of open Mad2 and its conversion to the closed conformation, concomitant with binding to Cdc20, constitutes the catalytic engine of the spindle assembly checkpoint at kinetochores (Han et al., 2013). We used an improved technique for rapamycin-induced protein dimerization to achieve temporal control over Mad1 kinetochore localization.
Results and discussion
Rapamycin-induced dimerization is a well-established technique to experimentally control the association of two proteins in living cells (Rivera et al., 1996; Putyrski and Schultz, 2012). Rapamycin is a small molecule that induces the dimerization of the proteins FKBP12 (hereafter, FKBP) and mTOR, or mTOR’s minimal rapamycin binding fragment FRB (Chen et al., 1995). To test the feasibility and kinetics of recruiting an unlocalized protein to kinetochores during mitosis, we generated a stable HeLa cell line constitutively expressing Mis12-GFP fused to a tandem trimer of FKBP (Mis12-GFP-FKBP) and inducibly expressing mCherry-FRB (Fig. 1, A and B). The expression level of mCherry-FRB varied between cells, and rapamycin induced-recruitment of mCherry-FRB to kinetochores was only detectable in cells with high mCherry-FRB expression (Fig. S1, B and C). Highly overexpressing Mad1 can compromise the checkpoint (Ryan et al., 2012; Heinrich et al., 2013), so we sought to improve the efficiency of rapamycin-mediated dimerization.
Endogenous FKBP depletion improves efficiency of rapamycin-mediated recruitment. (A) Diagram of a DNA cassette used to constitutively express Mis12-GFP-FKBP and miRNA, and inducibly express mCherry-FRB. The cassette is integrated between Lox acceptor sites downstream of the EF1a promoter (Khandelia et al., 2011; for details see Materials and methods). (B) Schematic representation of rapamycin-mediated recruitment of mCherry-FRB to kinetochore-localized Mis12-GFP-FKBP. (C) HeLa cells expressing Mis12-GFP-FKBP, mCherry-FRB, and either an empty miRNA backbone or miRNA against the 3′ UTR of endogenous FKBP were imaged before and ∼1 min after the addition of 500 nM rapamycin (rap). Images are representative of three independent experiments (quantified in Fig. S1 C). Bar, 5 µm.
Endogenous FKBP depletion improves efficiency of rapamycin-mediated recruitment. (A) Diagram of a DNA cassette used to constitutively express Mis12-GFP-FKBP and miRNA, and inducibly express mCherry-FRB. The cassette is integrated between Lox acceptor sites downstream of the EF1a promoter (Khandelia et al., 2011; for details see Materials and methods). (B) Schematic representation of rapamycin-mediated recruitment of mCherry-FRB to kinetochore-localized Mis12-GFP-FKBP. (C) HeLa cells expressing Mis12-GFP-FKBP, mCherry-FRB, and either an empty miRNA backbone or miRNA against the 3′ UTR of endogenous FKBP were imaged before and ∼1 min after the addition of 500 nM rapamycin (rap). Images are representative of three independent experiments (quantified in Fig. S1 C). Bar, 5 µm.
We reasoned that a fundamental limitation of this system is competition for FRB binding between an exogenous, targeted FKBP construct and endogenous FKBP, a highly abundant cytosolic protein. Depletion of FKBP, which is not essential (Weiwad et al., 2006; Hoeffer et al., 2008; De Angelis et al., 2009; Gerard et al., 2010), should remove this limitation. We added an artificial miRNA targeting the 3′ UTR of endogenous FKBP to the constitutively expressed Mis12-GFP-FKBP transcript. This miRNA effectively depleted endogenous FKBP, but not exogenous Mis12-GFP-FKBP (Fig. S1 A), without any overt effect on cellular morphology or growth. Endogenous FKBP depletion dramatically improved the efficiency of rapamycin-induced dimerization (Fig. 1 C and Fig. S1, B and C). We also found that the kinetics of FRB recruitment to kinetochores are sensitive to applied rapamycin concentration, with maximal recruitment in ∼10 s using 500 nM rapamycin (Fig. S1 D).
We constructed a cell line that combined constitutive expression of FKBP miRNA and Mis12-GFP-FKBP with doxycycline-inducible expression of FRB-mCherry-Mad1. FRB-mCherry-Mad1 localized robustly to misaligned kinetochores (Fig. S2 A) and was removed as kinetochores aligned at the metaphase plate (Fig. 2 A). In the absence of rapamycin, cells expressing FRB-mCherry-Mad1 proceeded through mitosis normally and rarely entered anaphase with misaligned chromosomes. After addition of rapamycin, both FRB-mCherry-Mad1 and endogenous Mad2 were recruited to kinetochores (Fig. 2 B), which did not compromise metaphase plate organization or chromosome alignment. This is consistent with the results of constitutive tethering (Maldonado and Kapoor, 2011). These results indicate that FRB-mCherry-Mad1 is functional and does not exert dominant-negative effects at the expression levels in our cells.
Mad1 recruitment to metaphase kinetochores activates the mitotic checkpoint. Cells expressing FRB-mCherry-Mad1, FKBP miRNA, and either Mis12-GFP-FKBP (A, C, and D) or Mis12-FKBP (B) are shown. (A) Time lapse shows FRB-mCherry-Mad1 removal from the last mCherry-positive kinetochore pair before anaphase (at 22 min). (B) Cells were treated with rapamycin (rap) or vehicle (DMSO) control for 10 min, fixed, and stained for Mad2. Control cells, which have not recruited mCherry, were also stained for Hec1. Images shown are representative of three independent replicates. (C and D) Cells were treated with either rapamycin or vehicle at metaphase (t = 0), then monitored for anaphase onset for 30 min. (C) Images show Mad1 recruitment and metaphase arrest in rapamycin-treated but not control cells. Bars, 5 µm. (D) The graph shows the cumulative percentage of cells entering anaphase over time (n ≥ 24 cells for each condition, pooled from six independent replicates).
Mad1 recruitment to metaphase kinetochores activates the mitotic checkpoint. Cells expressing FRB-mCherry-Mad1, FKBP miRNA, and either Mis12-GFP-FKBP (A, C, and D) or Mis12-FKBP (B) are shown. (A) Time lapse shows FRB-mCherry-Mad1 removal from the last mCherry-positive kinetochore pair before anaphase (at 22 min). (B) Cells were treated with rapamycin (rap) or vehicle (DMSO) control for 10 min, fixed, and stained for Mad2. Control cells, which have not recruited mCherry, were also stained for Hec1. Images shown are representative of three independent replicates. (C and D) Cells were treated with either rapamycin or vehicle at metaphase (t = 0), then monitored for anaphase onset for 30 min. (C) Images show Mad1 recruitment and metaphase arrest in rapamycin-treated but not control cells. Bars, 5 µm. (D) The graph shows the cumulative percentage of cells entering anaphase over time (n ≥ 24 cells for each condition, pooled from six independent replicates).
To test the effect of recruiting Mad1 to kinetochores after metaphase, we identified metaphase cells in an asynchronous population, added either rapamycin or a vehicle (DMSO) control, then monitored for 30 min to determine progress to anaphase (Fig. 2, C and D). We only considered cells with a clearly defined metaphase plate, without misaligned chromosomes identifiable by Mis12-GFP signal, and without mCherry-Mad1–positive kinetochores. 100% of control cells proceeded to anaphase within 15 min of mock treatment. In contrast, after recruiting Mad1 to metaphase kinetochores, only 34% of cells entered anaphase within 30 min (Fig. 2 D). In a related experiment, rapamycin was added at a fixed time (3 min) after removal of Mad1 from the last unattached kinetochore. In this case, very few (1/11) cells entered anaphase, which is consistent with recent results demonstrating that checkpoint reactivation depends on time in metaphase (Dick and Gerlich, 2013). We conclude that increasing Mad1 levels at bioriented, stably attached kinetochores is sufficient to switch them from a checkpoint-silenced to a checkpoint-active state.
We next asked whether recruiting Mad1 to metaphase kinetochores activated the checkpoint through a concomitant increase in two other key checkpoint proteins, Mps1 and BubR1, that are enriched at unattached kinetochores. Mps1 kinase activity is required upstream of Mad1, to recruit Mad1 to unattached kinetochores, and downstream of Mad1, to recruit cytosolic Mad2 to kinetochore-bound Mad1–Mad2 (Hewitt et al., 2010). BubR1 contains a conserved Mad3 homology domain that is the key inhibitor of Cdc20 and a pseudokinase domain that is an important signaling scaffold (Suijkerbuijk et al., 2012; Han et al., 2013). Levels of other checkpoint proteins, including Bub1, CENP-E, P150, ROD, and ZW-10, at metaphase kinetochores are not affected by constitutive tethering of Mad1 to kinetochores (Maldonado and Kapoor, 2011).
We measured levels of endogenous BubR1 and Mps1 at kinetochores using immunofluorescence. As expected, Mps1 and BubR1 were significantly higher at misaligned prometaphase kinetochores than at aligned metaphase kinetochores in vehicle-treated controls. Recruiting Mad1 to metaphase kinetochores produced no detectable increase in levels of kinetochore-bound BubR1 or Mps1 (Fig. 3, A and B). These data indicate that an increase in Mad1 levels at kinetochores is sufficient to activate the checkpoint without triggering an increase in kinetochore levels of Mps1 or BubR1.
Kinetochore levels of Mps1 and BubR1 are not affected by Mad1 recruitment at metaphase. Cells expressing Mis12-FKBP, FRB-mCherry-Mad1, and FKBP miRNA were treated with rapamycin (rap) or vehicle for 10 min, then fixed. (A) Cells were stained for BubR1 together with Hec1 to label kinetochores in control cells, which have not recruited mCherry, and BubR1 levels at kinetochores were quantified. (B) Cells were stained for Mps1, together with CENP-C to label kinetochores in control cells, and Mps1 levels at kinetochores were quantified. Bars, 5 µm. Error bars represent SEM from three independent experiments.
Kinetochore levels of Mps1 and BubR1 are not affected by Mad1 recruitment at metaphase. Cells expressing Mis12-FKBP, FRB-mCherry-Mad1, and FKBP miRNA were treated with rapamycin (rap) or vehicle for 10 min, then fixed. (A) Cells were stained for BubR1 together with Hec1 to label kinetochores in control cells, which have not recruited mCherry, and BubR1 levels at kinetochores were quantified. (B) Cells were stained for Mps1, together with CENP-C to label kinetochores in control cells, and Mps1 levels at kinetochores were quantified. Bars, 5 µm. Error bars represent SEM from three independent experiments.
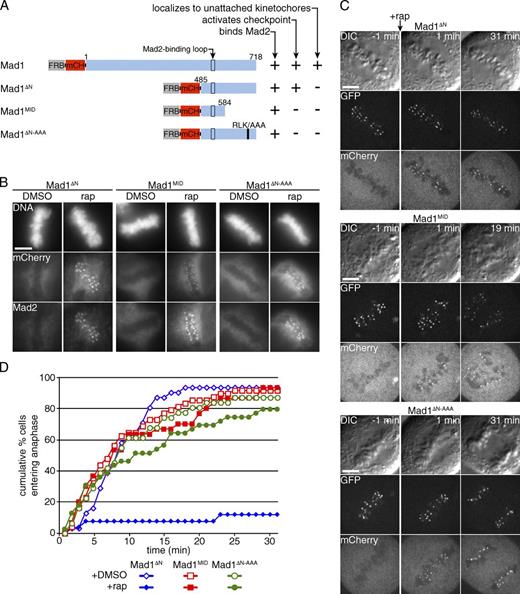
To determine what functional domains of Mad1 are necessary for checkpoint activation, we first tested the role of the N-terminal domain, which is required for kinetochore localization in Xenopus laevis (Chung and Chen, 2002) and mammalian cells (Kim et al., 2012). It is not known whether this domain contributes to checkpoint activation beyond its role in localization. We removed the first 484 residues to generate Mad1ΔN, which matches a fragment used for in vitro studies of Mad2 conformational templating (De Antoni et al., 2005). FRB-mCherry-Mad1ΔN fails to localize to unattached kinetochores in the absence of rapamycin (Fig. S2 A), as expected, but recruits Mad2 to metaphase kinetochores upon treatment with rapamycin (Figs. 4 B and S3 C) and induces a metaphase arrest (Fig. 4, C and D). These results show that the N-terminal domain of Mad1 is not required for checkpoint activation beyond localizing Mad1 to kinetochores.
A conserved RLK motif in the C-terminal domain of Mad1 is required to activate the checkpoint. (A) Schematic representation of Mad1 constructs. The black outline indicates the Mad2-binding region and the black bar indicates an RLK motif mutated to AAA. (B–D) Cells expressing Mis12-GFP-FKBP, FKBP miRNA, and FRB-mCherry-Mad1 fragments as indicated. (B) Cells were treated with rapamycin (rap) or vehicle for 10 min, then fixed and stained for DNA and Mad2. Images are representative of three independent experiments (quantified in Fig. S3 C). (C) Cells were treated with rapamycin at metaphase (t = 0), then monitored for anaphase onset for 30 min. Vehicle controls are shown in Fig. S2. (D) The graph shows the cumulative percentage of cells entering anaphase over time (n ≥ 26 cells for each condition, pooled from at least independent replicates). Bars, 5 µm.
A conserved RLK motif in the C-terminal domain of Mad1 is required to activate the checkpoint. (A) Schematic representation of Mad1 constructs. The black outline indicates the Mad2-binding region and the black bar indicates an RLK motif mutated to AAA. (B–D) Cells expressing Mis12-GFP-FKBP, FKBP miRNA, and FRB-mCherry-Mad1 fragments as indicated. (B) Cells were treated with rapamycin (rap) or vehicle for 10 min, then fixed and stained for DNA and Mad2. Images are representative of three independent experiments (quantified in Fig. S3 C). (C) Cells were treated with rapamycin at metaphase (t = 0), then monitored for anaphase onset for 30 min. Vehicle controls are shown in Fig. S2. (D) The graph shows the cumulative percentage of cells entering anaphase over time (n ≥ 26 cells for each condition, pooled from at least independent replicates). Bars, 5 µm.
Because Mad2 binding is the best-characterized activity of Mad1, we tested whether Mad2 binding is sufficient for Mad1 to activate the checkpoint. We truncated Mad1ΔN at the C terminus to construct Mad1MID, comprising residues 485–584, including the Mad2 interaction loop flanked by α-helical regions, as shown in the crystal structure of the tetrameric Mad1–Mad2 complex (Sironi et al., 2002). FRB-mCherry-Mad1MID recruits Mad2 to metaphase kinetochores upon treatment with rapamycin to similar levels as Mad1 and Mad1ΔN (Figs. 4 B and S3 C) but does not induce a metaphase arrest (Fig. 4, C and D). In contrast, deletion of the Mad2-binding region prevents checkpoint reactivation (Fig. S3), which is consistent with previous results (Maldonado and Kapoor, 2011). These results indicate that localizing Mad2 to kinetochores is necessary but not sufficient to activate the checkpoint, and that the C-terminal domain of Mad1 is also required.
In Saccharomyces cerevisiae, mitotic checkpoint activity can be abolished by mutating three conserved amino acids in the C terminus of S. cerevisiae Mad1 to alanine: R653, L654, and K655, which correspond to residues 617–619 in human Mad1 (Brady and Hardwick, 2000). Mutating either R617 or K619 in human cells impairs kinetochore targeting, presumably by disrupting interactions with other kinetochore proteins (Kim et al., 2012), but the role of the RLK motif in checkpoint activation independent of targeting has not been tested. To determine whether checkpoint activation depends on the C-terminal RLK motif, we mutated these three residues to alanines in the context of Mad1ΔN. Mutating the RLK motif does not reduce Mad2 recruitment (Figs. 4 B and S3 C), but almost completely abolishes the ability of Mad1 to activate the checkpoint when recruited to metaphase kinetochores (Fig. 4, C and D). This result demonstrates that a conserved protein interaction motif in the C terminus of Mad1 is required for full checkpoint activity.
In conclusion, our results show that the spindle assembly checkpoint can be reactivated after silencing by relocalizing Mad1 to metaphase kinetochores. Checkpoint activation does not require the increased levels of Mps1 and BubR1 associated with unattached kinetochores: the basal levels of these proteins present at metaphase kinetochores are sufficient to sustain APC/C inhibition when combined with increased Mad1–Mad2. Both Mps1 and BubR1 exhibit rapid turnover at metaphase kinetochores (Howell et al., 2004), so a large fraction of Mps1 and BubR1 may transiently encounter kinetochore-localized Mad1–Mad2 even though steady-state levels of Mps1 and BubR1 at kinetochores remain low. Additionally, Mps1 and BubR1 play other roles at the kinetochore independent of their checkpoint functions, for example regulating microtubule interactions (Lampson and Kapoor, 2005; Maure et al., 2007; Jelluma et al., 2008; Meyer et al., 2013). It is possible that the increased levels of some checkpoint proteins at unattached kinetochores contribute to functions other than APC/C inhibition.
The results of our Mad1 mutation experiments indicate that the role of Mad1 in APC/C inhibition is not limited to localizing Mad2 to kinetochores. The potent effect of mutating the Mad1 RLK motif strongly suggests that the C terminus of Mad1 mediates interactions with other kinetochore proteins that are essential for checkpoint activity in addition to contributing to Mad1 localization. A growing body of evidence argues that APC/C inhibition is a multistep process in which Cdc20 first binds closed Mad2 (cMad2), which catalyzes the subsequent binding of BubR1 to Cdc20, and that BubR1–Cdc20 is the primary inhibitor of the APC/C (Nilsson et al., 2008; Kulukian et al., 2009; Westhorpe et al., 2011; Han et al., 2013). The C terminus of Mad1 may act as a scaffold to promote either the binding of Cdc20 to cMad2 or the transfer of Cdc20 from Mad2 to BubR1. An important future goal is to determine which proteins interact with the C terminus of Mad1, and what role these interactions play in checkpoint activation.
Finally, our work demonstrates the potential for inducible protein dimerization as a tool to manipulate protein localization at kinetochores on a rapid timescale. Dynamic localization is a hallmark of many mitotic proteins, and much has been learned from disrupting localization by mutating targeting domains or conferring new localization through constitutive tethering, but these techniques are fundamentally limited by a lack of temporal control. Chemically induced dimerization overcomes this limitation, and allows us to study the effects of altering protein localization in real time.
Materials and methods
Cell culture, creation of stable cell lines, and rapamycin treatment
All experiments were performed with stable HeLa cell lines generated by recombinase-mediated cassette exchange (RMCE) using the HILO RMCE system (obtained from E.V. Makeyev, Nanyang Technological University, Singapore; reported in Khandelia et al., 2011). This system allowed us to reproducibly insert transgene cassettes at a single genomic locus. In brief: a monoclonal acceptor cell line with LoxP and Lox2272 recombination sites at a single chromosomal locus was cotransfected with a donor plasmid containing a transgenic cassette flanked by LoxP and Lox2272 sites and a second plasmid expressing Cre recombinase, followed by selection for a marker in the donor cassette. Cells were cultured in growth medium (DMEM with 10% FBS and penicillin/streptomycin) at 37°C in a humidified atmosphere with 5% CO2. Cells at ∼60% confluency in a single well of a six-well plate were transfected with 1 µg of donor plasmid + 10 ng of Cre plasmid pEM784 (see the following section for plasmid details) using Fugene 6 (Promega). 2 d after transfection, 1 µg/ml puromycin was added to the growth medium for selection of stable cell lines. 125 ng/ml doxycycline was added to the growth medium 2 d before the experiment to induce expression of protein from the tetracycline-responsive element (TRE) Tet-responsive promoter. Although dox-induced protein expression was variable within each population of cells, the variation was reproducible and similar between different cell lines produced with this technique. All rapamycin experiments were done at a final working concentration of 500 nM rapamycin, except as noted in Fig. S1 D. Rapamycin was stored as a 500 mM stock in DMSO, then diluted in medium to a 0.75- or 1-mM intermediate dilution in medium and prewarmed before addition to cells.
Plasmids
pEM784 expressing nuclear-localized Cre recombinase and the donor cassette plasmid pEM791 were obtained from E.V. Makeyev (Khandelia et al., 2011). All donor cassettes used for this study were derived from pEM791. pEM791 is designed for inducible expression of miRNA-based shRNA and a reporter gene. pEM791 contains: a Puro resistance gene (Pur) positioned for constitutive transcription from the EF1α promoter upstream of the LoxP site in the acceptor locus, the gene for reverse-tetracycline transactivator 3 (rtTA3) constitutively expressed from a UBC promoter, and a TRE promoter for inducible transcription of an artificial miRNA-based shRNA nested in an intron upstream of a GFP reporter gene (Du et al., 2006; Khandelia et al., 2011).
For this study, we modified pEM791 for constitutive expression of an additional miRNA and protein sequence. Between the LoxP site and Pur, we added: (1) an miRNA-based shRNA against the 3′ UTR of FKBP12, nested within an intron; (2) Mis12-GFP-FKBP or Mis12-FKBP (tandem trimers of FKBP in both cases); and (3) an internal ribosome entry sequence (IRES). These modifications allowed constitutive polycistronic coexpression of FKBP miRNA, Mis12-targeted FKBP, and the Puro resistance gene from the EF1α promoter in the acceptor locus. FKBP miRNA was designed using the miR RNAi function within the Block-iT RNAi Designer (Life Technologies). The following oligos were used for FKBP miRNA: 5′-TGCTGATATGGATTCATGTGCACATGGTTTTGGCCACTGACTGACCATGTGCATGAATCCATAT-3′, 5′-CCTGATATGGATTCATGCACATGGTCAGTCAGTGGCCAAAACCATGTGCACATGAATCCATATC-3′. mCherry-FRB-Mad1 constructs were cloned in place of GFP downstream of TRE. No shRNA sequences were added to the empty miRNA backbone in the inducible transcript for this study. The genes for FRB and FKBP were cloned from plasmids pC4EN-F1, pC4M-F2E, and pC4-RHE (obtained from Ariad Pharmaceuticals, Inc.).
Immunofluorescence
All cell fixation procedures were done at room temperature (∼22°C). For Mad2 staining in Figs. 2, 4, and S3, cells were fixed in PBS + 3.7% formaldehyde with 0.5% Triton X-100 for 10 min. For BubR1staining in Fig. 3 A, cells were preextracted in PBS + 0.5% Triton X-100 for 1 min, then fixed in PBS + 3.7% formaldehyde and 0.5% Triton X-100 for 10 min. For Mps1 staining in Fig. 3 B, cells were prefixed for 5 s in PBS + 3.7% formaldehyde, extracted in PBS + 0.5% Triton X-100 for 1 min, then fixed in PBS + 3.7% formaldehyde with 0.5% Triton X-100 for 10 min. The following primary antibodies were used: rabbit polyclonal anti-Mad2 (1:500; PRB-452C; Covance), mouse monoclonal anti-Hec1 9G3 (1:1,000; ab3613; Abcam), rabbit anti-BubR1 polyclonal (1:1,000; a gift from W. Dai, New York Medical College, Valhalla, NY), rabbit anti–CENP-C polyclonal (1:1,000; a gift from B. Black, University of Pennsylvania, Philadelphia, PA), and mouse anti-Mps1 monoclonal NT (1:100; 05-682; EMD Millipore).
Immunoblotting
Whole cell lysates were prepared from asynchronous populations of cells. Western blot analysis was performed using the following antibodies: rabbit anti-FKBP12 polyclonal (1:2,000; ab2918; Abcam), rabbit anti-GFP polyclonal (1:10,000; a gift from B. Black), and mouse anti-tubulin monoclonal DM1α (1:10,000; Sigma-Aldrich).
Image acquisition and processing
For live imaging, cells were plated on 22 × 22-mm glass coverslips (no. 1.5; Thermo Fisher Scientific) coated with poly-d-lysine (Sigma-Aldrich). Coverslips were mounted in magnetic chambers (Chamlide CM-S22-1, LCI) using L-15 medium without phenol red (Invitrogen) supplemented with 10% FBS and penicillin/streptomycin. Temperature was maintained at ∼35°C using an environmental chamber (Incubator BL; PeCon GmbH).
Except for Figs. 4 B and S3 B, all images were acquired with a spinning disk confocal microscope (DM4000; Leica) with a 100× 1.4 NA objective lens, an XY Piezo-Z stage (Applied Scientific Instrumentation), a spinning disk (Yokogawa Corporation of America), an electron multiplier charge-coupled device camera (ImageEM; Hamamatsu Photonics), and a laser merge module equipped with 488- and 593-nm lasers (LMM5; Spectral Applied Research) controlled by MetaMorph software (Molecular Devices). Images in Figs. 4 B and S3 D, and the images quantified in Fig. S3 C were acquired on a microscope (DM6000; Leica) with a 63× 1.4 NA objective lens and a charge-coupled device camera (ORCA-AG; Hamamatsu Photonics) controlled by Micro-Manager software (Edelstein et al., 2010).
All image processing and analysis was performed using ImageJ (Schneider et al., 2012). For BubR1 and Mps1 quantification (Fig. 3), kinetochores were identified by Hec1 or CenpC costaining. In rapamycin-treated cells, kinetochores were identified via recruited FRB-mCherry-Mad1 fluorescence. Each kinetochore was defined as a circular region 6 pixels (0.72 µm) in diameter, and BubR1 or Mps1 intensity was measured in these regions and background corrected. 20 individual kinetochores were measured in each of five cells for each condition, and the 100 individual kinetochore measurements were averaged to yield a value for each of three independent replicates. The values shown in Fig. 3 are averages of the three independent replicates, normalized to the metaphase control condition. Each bar, therefore, represents an average of three averages of 100 kinetochores (20 each from five cells). Error bars represent the SEM of the three independent replicates.
For mCherry-FRB recruitment quantification in Fig. S1 C, total mCherry-FRB expression was measured for each cell in the pre-rapamycin image as the mean mCherry intensity within a 135 pixel (16.2 µm) diameter circle centered on the cell, then background corrected. Images were cropped to a 200 × 200-pixel region centered on each cell, then kinetochore regions were identified in pre-rapamycin and post-rapamycin GFP images by thresholding using the MaxEntropy algorithm in ImageJ. mCherry intensity within the corresponding kinetochore regions was measured, and the mean pre-rapamycin intensity was subtracted from the post-rap intensity to yield a raw mCherry-FRB recruitment value. Both mCherry-FRB expression and recruitment were thus measured on the same scale of mCherry pixel intensity. These measurements were made on 138 empty-miRNA control cells and 151 FKBP miRNA cells pooled from six independent replicates. Expression and recruitment values were normalized to the cell with the highest mCherry-FRB expression level.
For Mad2 quantification in Fig. S3 C, kinetochore regions in metaphase cells were first defined in Hec1 costaining images with a manually determined intensity threshold. Mean Mad2 intensity within the kinetochore region was measured and background subtracted. Mad2 levels from at least eight cells were averaged for each cell type in three independent replicates. The values from the three replicates were averaged and normalized to full-length Mad1.
Online supplemental material
Fig. S1 shows the effects of endogenous FKBP knockdown on rapamycin-mediated recruitment to kinetochores. Fig. S2 shows a characterization of Mad1 truncation mutants. Fig. S3 shows that Mad2-binding is required for checkpoint activation by Mad1.
Acknowledgments
We thank B. Black for antibodies, helpful discussions, and critical reading of the manuscript.
This work was supported by National Institutes of Health grant GM083988 to M.A. Lampson.
The authors declare no competing financial interests.