Cytoplasmic dynein transports membranous cargoes along microtubules, but the mechanism of dynein–cargo interaction is unclear. From a genetic screen, we identified a homologue of human Hook proteins, HookA, as a factor required for dynein-mediated early endosome movement in the filamentous fungus Aspergillus nidulans. HookA contains a putative N-terminal microtubule-binding domain followed by coiled-coil domains and a C-terminal cargo-binding domain, an organization reminiscent of cytoplasmic linker proteins. HookA–early endosome interaction occurs independently of dynein–early endosome interaction and requires the C-terminal domain. Importantly, HookA interacts with dynein and dynactin independently of HookA–early endosome interaction but dependent on the N-terminal part of HookA. Both dynein and the p25 subunit of dynactin are required for the interaction between HookA and dynein–dynactin, and loss of HookA significantly weakens dynein–early endosome interaction, causing a virtually complete absence of early endosome movement. Thus, HookA is a novel linker important for dynein–early endosome interaction in vivo.
Introduction
Microtubule-based intracellular transport of membranous organelles depends on motor proteins, such as cytoplasmic dynein and kinesins, whose proper functions are essential for brain development and survival of neurons (Vale, 2003; Hirokawa et al., 2010; Perlson et al., 2010; Ori-McKenney et al., 2011). For a motor to carry its cargo in vivo, it must be physically connected to the cargo, and thus, the mechanism of motor–cargo interaction is a topic of great interest (Akhmanova and Hammer, 2010; Stephens, 2012). For the minus end–directed cytoplasmic dynein, the dynactin complex is implicated in linking dynein to membranous cargoes (Holleran et al., 1998; Schroer, 2004; Akhmanova and Hammer, 2010). This function of dynactin has recently been specifically demonstrated in dynein-mediated transport of the early endosome (Zhang et al., 2011; Yeh et al., 2012). However, mechanistically, how dynactin connects dynein to early endosomes is unclear, and one outstanding question is whether additional proteins are required for linking dynactin–dynein to early endosomes.
For in vivo transport, additional factors may also be required for enhancing or regulating motor–cargo–track interactions. Proteins of the cytoplasmic linker proteins family are thought to facilitate intracellular transport by weakly linking membranous organelles to microtubules (Pierre et al., 1992; Rickard and Kreis, 1996; Schroer, 2000). The prototypic cytoplasmic linker protein is the mammalian CLIP-170 originally identified as an endosome-binding protein that contains a microtubule-binding domain (Pierre et al., 1992). CLIP-170 has emerged to be a prototypic microtubule plus end–tracking protein that regulates microtubule dynamics and recruits dynactin to the microtubule plus end (Perez et al., 1999; Schuyler and Pellman, 2001; Lansbergen et al., 2004; Miller et al., 2006; Akhmanova and Steinmetz, 2008). These functions of CLIP-170 are important for the initiation of vesicle transport in melanocytes and neurons (Lomakin et al., 2009, 2011; Moughamian et al., 2013) but not critical for vesicle distribution in other types of cultured cells (Lansbergen et al., 2004; Akhmanova et al., 2005; Watson and Stephens, 2006). In addition, the CLIP-170 homologue in the fungus Ustilago maydis is not required for dynein-mediated early endosome transport (Lenz et al., 2006).
Proteins of the Hook family are also considered cytoplasmic linker proteins, as they have a domain structure similar to that of CLIP-170 (Simpson et al., 2005). They contain an N-terminal microtubule-binding domain (Walenta et al., 2001), an extended central coiled-coil domain implicated in homodimerization (Xu et al., 2008), and a divergent C-terminal domain implicated in organelle association (Walenta et al., 2001). The founding member of this family is the Drosophila melanogaster Hook protein, which is required for the proper formation or stabilization of multivesicular bodies in the endocytic pathway (Krämer and Phistry, 1996, 1999; Sunio et al., 1999). There are three Hook proteins in mammalian cells. Although Hook3 is associated with Golgi (Walenta et al., 2001), Hook2 is involved in centrosome function, aggresome formation, and primary cilium morphogenesis (Szebenyi et al., 2007a,b; Baron Gaillard et al., 2011). Hook1 is involved in sperm head morphogenesis (Mendoza-Lujambio et al., 2002), just like CLIP-170 (Akhmanova et al., 2005), and it is required for endosome sorting of nonclathrin cargo in a microtubule-dependent fashion (Maldonado-Báez et al., 2013). A novel Hook-related protein (HkRP) family has been identified (Simpson et al., 2005). The HkRPs have a domain organization similar to the Hook proteins, but they are larger in size (Simpson et al., 2005). Overexpression of the C-terminal domain of HkRP1 affects distribution of the early endosome marker sorting nexin 1 but not the EEA1 (early endosome antigen-1), suggesting that HkRP1 may only affect the tubulation of early endosome subdomains (Simpson et al., 2005). Thus, it remains unknown whether any member of this family is required for dynein-mediated early endosome transport.
The filamentous fungus Aspergillus nidulans is a well-established genetic system for discovering novel factors regulating cytoplasmic dynein function, as previous work in this system first linked the function of LIS1 and NudE/Nudel to dynein function (Xiang et al., 1995a; Efimov and Morris, 2000; Kardon and Vale, 2009; Ori-McKenney et al., 2011; Egan et al., 2012a; Peñalva et al., 2012). In A. nidulans, cytoplasmic dynein accumulates at the plus ends of microtubules at the hyphal tip in a kinesin-1– and dynactin-dependent fashion (Xiang et al., 2000; Han et al., 2001; Zhang et al., 2003, 2008; Egan et al., 2012b; Yao et al., 2012). Although plus-end dynein in budding yeast is exclusively used for spindle orientation (Lee et al., 2003; Sheeman et al., 2003; Winey and Bloom, 2012), in filamentous fungi, plus-end dynein drives early endosome transport in a LIS1- and dynactin-dependent fashion (Lenz et al., 2006; Abenza et al., 2009, 2010; Zekert and Fischer, 2009; Zhang et al., 2010, 2011; Schuster et al., 2011; Egan et al., 2012b). In particular, the p25 protein of the dynactin complex is important for dynein–early endosome interaction in A. nidulans (Zhang et al., 2011). Based on this finding, we performed a classical genetic screen to identify additional proteins involved in the dynein–early endosome interaction. This genome-wide search has led to the discovery of HookA, the A. nidulans Hook orthologue, as a new factor essential for dynein-mediated early endosome transport. Furthermore, results from imaging and biochemical analyses indicate that HookA serves as a novel linker between early endosomes and dynein–dynactin to allow early endosome transport.
Results
HookA is required for dynein-mediated early endosome motility in A. nidulans
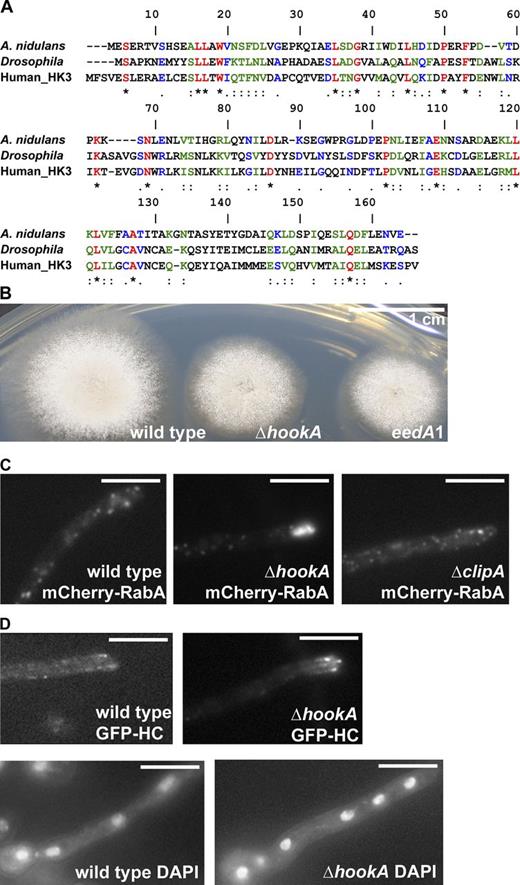
We used a classical genetic approach to screen for eed (early endosome distribution) mutants in A. nidulans. Because we are particularly interested in identifying novel factors involved in the dynein–early endosome interaction, we looked for mutants that resemble the Δp25 mutant (Fig. 1, A and B; Zhang et al., 2011). From ∼20,000 survivors of a UV mutagenesis, we selected several hundred colonies that were slightly more compact than wild-type colonies and then used three criteria for a microscopy-based screen: First, the mutants exhibit an abnormal buildup of early endosomes at the hyphal tip. Second, the mutants exhibit normal accumulation of dynein at microtubule plus ends, as indicated by the presence of the plus-end dynein comets (Han et al., 2001). Third, the mutants exhibit normal nuclear distribution. The A. nidulans strain constructed for this genetic experiment contains GFP–dynein heavy chain (HC; Zhuang et al., 2007) and mCherry-RabA, an early endosome marker in A. nidulans (Abenza et al., 2009, 2010), which allowed us to monitor the localization of dynein and early endosomes in living cells. The eedA1 mutant was isolated from the initial screen, and it fitted perfectly with our screening criteria (Fig. 1, C–E).
Phenotype of the eedA1 mutant and rescue of the mutant phenotype by the gene encoding HookA. (A) A schematic diagram depicting the phenotype of the Δp25 mutant in comparison to the ΔnudA (dynein HC) mutant. Note that early endosomes abnormally accumulate at the hyphal tip in both the ΔnudA and Δp25 mutants but a nuclear distribution phenotype is shown only in the ΔnudA mutant. Red, early endosomes. Dark blue, nuclei. Black lines, microtubules. (B) A brief outline of the mutant-screening procedure. (C) Colony phenotypes of the eedA1 mutant and a wild-type strain. (D) Microscopic images showing the distributions of mCherry-RabA–labeled early endosomes (mCherry-RabA) and GFP-labeled dynein HC (GFP-HC). The same cells are shown for both the mCherry-RabA and GFP-HC images. Although bidirectional movements of mCherry-RabA–labeled early endosomes are not completely abolished, ∼83% of eedA1 hyphal tips show obvious accumulation of mCherry-RabA signals (n = 140), whereas none of the wild-type hyphal tips show this accumulation (n = 100). Dynein comets are present in all wild-type and mutant cells. (E) Images of nuclei stained by a DNA dye, DAPI, in wild type and the eedA1 mutant. The pattern of nuclear distribution in the eedA1 mutant is normal, as none of the mutant cells show any cluster of four or more nuclei when grown under the same conditions that allow us to see the hyphal tip mCherry-RabA accumulation (n > 100 for wild type, and n > 100 for the mutant). (F) Rescue of the eedA1 mutant phenotype with the HookA-encoding DNA. (left) Colony phenotypes of the eedA1 mutant, eedA1 mutant transformed with the HookA-encoding gene, and a wild-type strain. (right) Distribution of mCherry-RabA–labeled early endosomes in an eedA1 mutant transformed with the HookA-encoding gene. None of the hyphal tips show accumulation of mCherry-RabA signals (n = 30). Bars, 5 µm.
Phenotype of the eedA1 mutant and rescue of the mutant phenotype by the gene encoding HookA. (A) A schematic diagram depicting the phenotype of the Δp25 mutant in comparison to the ΔnudA (dynein HC) mutant. Note that early endosomes abnormally accumulate at the hyphal tip in both the ΔnudA and Δp25 mutants but a nuclear distribution phenotype is shown only in the ΔnudA mutant. Red, early endosomes. Dark blue, nuclei. Black lines, microtubules. (B) A brief outline of the mutant-screening procedure. (C) Colony phenotypes of the eedA1 mutant and a wild-type strain. (D) Microscopic images showing the distributions of mCherry-RabA–labeled early endosomes (mCherry-RabA) and GFP-labeled dynein HC (GFP-HC). The same cells are shown for both the mCherry-RabA and GFP-HC images. Although bidirectional movements of mCherry-RabA–labeled early endosomes are not completely abolished, ∼83% of eedA1 hyphal tips show obvious accumulation of mCherry-RabA signals (n = 140), whereas none of the wild-type hyphal tips show this accumulation (n = 100). Dynein comets are present in all wild-type and mutant cells. (E) Images of nuclei stained by a DNA dye, DAPI, in wild type and the eedA1 mutant. The pattern of nuclear distribution in the eedA1 mutant is normal, as none of the mutant cells show any cluster of four or more nuclei when grown under the same conditions that allow us to see the hyphal tip mCherry-RabA accumulation (n > 100 for wild type, and n > 100 for the mutant). (F) Rescue of the eedA1 mutant phenotype with the HookA-encoding DNA. (left) Colony phenotypes of the eedA1 mutant, eedA1 mutant transformed with the HookA-encoding gene, and a wild-type strain. (right) Distribution of mCherry-RabA–labeled early endosomes in an eedA1 mutant transformed with the HookA-encoding gene. None of the hyphal tips show accumulation of mCherry-RabA signals (n = 30). Bars, 5 µm.
We first mapped the eedA1 mutation to chromosome V using parasexual genetics (Pontecorvo et al., 1953; McCully and Forbes, 1965). Subsequently, by combining classical genetic mapping with whole genome sequencing (Otogenetics Corp.), we identified two mutations in the AN5126 gene. These two mutations are close to each other, causing the replacement of residues Leu150 and Glu151 by Pro and Lys, respectively. We confirmed that AN5126 is eedA by using a genomic DNA fragment containing the AN5126 gene to rescue the mutant phenotype (Fig. 1 F).
AN5126 encodes a homologue of the Hook proteins (Fig. 2 A and Fig. S1), which contain an N-terminal putative microtubule-binding domain followed by coiled-coil domains and a C-terminal cargo-binding domain (Walenta et al., 2001). The size of the A. nidulans Hook homologue is 638 aa, similar to that of the Hook proteins but not to the HkRPs. We named the translation product of AN5126 HookA (standing for Hook in A. nidulans) and further studied its function by constructing a ΔhookA deletion mutant. We initially suspected that ΔhookA might be lethal because Drosophila Hook is implicated in endosome maturation (Krämer and Phistry, 1996, 1999; Sunio et al., 1999), and endosome maturation is essential for A. nidulans (Abenza et al., 2010, 2012). However, the ΔhookA mutant is not lethal but exhibits a colony phenotype and defect in early endosome distribution similar to that of the eedA1 mutant (Fig. 2, B and C; and Videos 1 and 2). This is in contrast to the normal distribution of early endosomes in ΔclipA (Fig. 2 C), the null mutant of the CLIP-170 orthologue ClipA (Efimov et al., 2006). Importantly, nuclear distribution and the accumulation of GFP–dynein HC to the dynamic microtubule plus end are normal in the ΔhookA mutant (Fig. 2 D), indicating that HookA is not required for the overall function and localization of dynein.
Sequence and functional analyses of HookA. (A) A sequence alignment of the N-terminal putative microtubule-binding domain of HookA (A. nidulans), Hook (Drosophila), and Hook3 (human_HK3). The alignment was performed using CLUSTALW (Pôle BioInformatique Lyonnais Network Protein Sequence Analysis). Residues that are identical (asterisks), strongly similar (double dots), or weakly similar (single dots) are shown as red, green, and blue characters, respectively. Also see Fig. S1. (B) Colony phenotype of the ΔhookA mutant. (C) Distributions of mCherry-RabA–labeled early endosomes in the ΔhookA mutant (also see Videos 1 and 2) and the ΔclipA mutant. An obvious accumulation of mCherry-RabA signals is found at ∼80% of the hyphal tips in the ΔhookA mutant (n = 326), whereas none of the hyphal tips in wild type (n = 240) or the ΔclipA mutant (n = 73) show the same accumulation. (D, top) GFP–dynein HC (GFP-HC) signals in wild type and the ΔhookA mutant. Maximal signal intensities (arbitrary units) of the plus-end GFP-HC comets in wild-type cells and in ΔhookA cells are 276 ± 262 (n = 25) and 268 ± 225 (n = 30), respectively, and there is no statistical difference between the values at P = 0.05. (Bottom) Images of nuclei stained by DAPI in wild type and the ΔhookA mutant. The pattern of nuclear distribution in the ΔhookA mutant is normal, as none of the mutant cells shows any cluster of four or more nuclei when grown under the same conditions that allow us to see the hyphal tip mCherry-RabA accumulation (n > 100 for wild type, and n > 100 for the mutant). Bars, 5 µm.
Sequence and functional analyses of HookA. (A) A sequence alignment of the N-terminal putative microtubule-binding domain of HookA (A. nidulans), Hook (Drosophila), and Hook3 (human_HK3). The alignment was performed using CLUSTALW (Pôle BioInformatique Lyonnais Network Protein Sequence Analysis). Residues that are identical (asterisks), strongly similar (double dots), or weakly similar (single dots) are shown as red, green, and blue characters, respectively. Also see Fig. S1. (B) Colony phenotype of the ΔhookA mutant. (C) Distributions of mCherry-RabA–labeled early endosomes in the ΔhookA mutant (also see Videos 1 and 2) and the ΔclipA mutant. An obvious accumulation of mCherry-RabA signals is found at ∼80% of the hyphal tips in the ΔhookA mutant (n = 326), whereas none of the hyphal tips in wild type (n = 240) or the ΔclipA mutant (n = 73) show the same accumulation. (D, top) GFP–dynein HC (GFP-HC) signals in wild type and the ΔhookA mutant. Maximal signal intensities (arbitrary units) of the plus-end GFP-HC comets in wild-type cells and in ΔhookA cells are 276 ± 262 (n = 25) and 268 ± 225 (n = 30), respectively, and there is no statistical difference between the values at P = 0.05. (Bottom) Images of nuclei stained by DAPI in wild type and the ΔhookA mutant. The pattern of nuclear distribution in the ΔhookA mutant is normal, as none of the mutant cells shows any cluster of four or more nuclei when grown under the same conditions that allow us to see the hyphal tip mCherry-RabA accumulation (n > 100 for wild type, and n > 100 for the mutant). Bars, 5 µm.
HookA associates with early endosomes independently of dynein–early endosome interaction
To visualize the HookA protein in hyphae, we constructed a HookA-GFP fusion with GFP fused to the C terminus of HookA. This fusion was used to replace the endogenous HookA, and its expression is driven by the endogenous hookA promoter. The HookA-GFP fusion is functional, as indicated by the normal early endosome distribution in a strain containing both HookA-GFP and mCherry-RabA (Fig. 3 A). HookA-GFP signals moving along microtubule-like structures were observed, and some HookA-GFP signals appeared to colocalize with mCherry-RabA signals. As shown by the kymographs obtained by dual-view fluorescent imaging, some of the HookA-GFP motile dots clearly colocalized with moving early endosomes (Fig. 3 B and Fig. S2). To confirm that HookA-GFP is associated with early endosomes and to address whether the association of HookA-GFP with early endosomes depends on dynein/dynactin on the same early endosomes, we examined HookA-GFP in the ΔkinA and Δp25 mutants. KinA (kinesin-1 in A. nidulans) is required for dynein accumulation at the microtubule plus ends (Zhang et al., 2003, 2010; Egan et al., 2012b), which is important for dynein-mediated early endosome transport away from the hyphal tip (Lenz et al., 2006; Abenza et al., 2009; Zekert and Fischer, 2009; Zhang et al., 2010; Egan et al., 2012b). In the ΔkinA mutant, dynein and dynactin signals are localized along microtubules and clearly are not seen at the hyphal tip, where early endosomes accumulate (Zhang et al., 2010; Yao et al., 2012). However, a dramatic accumulation of HookA-GFP signals was seen at almost every hyphal tip, largely overlapping with mCherry-RabA signals (Fig. 3 C). Similarly, in the Δp25 mutant in which dynein–early endosome interaction is defective (Zhang et al., 2011), a dramatic accumulation of HookA-GFP signals at the hyphal tip was seen, which largely overlapped with early endosome signals (Fig. 3 D). These data further indicate that HookA is associated with early endosomes, and this association can occur independently of association of dynein–dynactin with the same early endosomes.
Colocalization of HookA-GFP signals with mCherry-RabA–labeled early endosomes. (A) Images of HookA-GFP and mCherry-RabA in the same cell. (B) Kymographs of the GFP and mCherry signals obtained via duel-view imaging. Arrows are shown to indicate that some HookA-GFP signals are associated with motile early endosomes. (C) HookA-GFP and mCherry-RabA in the ΔkinA mutant. HookA-GFP signals were concentrated at every hyphal tip where early endosomes accumulate (n = 50). (D) HookA-GFP and mCherry-RabA in the Δp25 mutant. HookA-GFP signals were concentrated at every hyphal tip where early endosomes accumulate (n = 50). The same minimal medium containing 1% glycerol as a carbon source was used for cells shown in A, C, and D. For the medium used for growing the cells viewed by the dual-view imaging (B), 0.1% fructose instead of 1% glycerol was used as a carbon source to reduce the intensity of the mCherry signals. Bars, 5 µm.
Colocalization of HookA-GFP signals with mCherry-RabA–labeled early endosomes. (A) Images of HookA-GFP and mCherry-RabA in the same cell. (B) Kymographs of the GFP and mCherry signals obtained via duel-view imaging. Arrows are shown to indicate that some HookA-GFP signals are associated with motile early endosomes. (C) HookA-GFP and mCherry-RabA in the ΔkinA mutant. HookA-GFP signals were concentrated at every hyphal tip where early endosomes accumulate (n = 50). (D) HookA-GFP and mCherry-RabA in the Δp25 mutant. HookA-GFP signals were concentrated at every hyphal tip where early endosomes accumulate (n = 50). The same minimal medium containing 1% glycerol as a carbon source was used for cells shown in A, C, and D. For the medium used for growing the cells viewed by the dual-view imaging (B), 0.1% fructose instead of 1% glycerol was used as a carbon source to reduce the intensity of the mCherry signals. Bars, 5 µm.
The C terminus of HookA is critical for HookA–early endosome interaction
The C-terminal sequences of Hook proteins are quite divergent, but importantly, the C-terminal 130 aa of hHK3 (human Hook3) have been shown to be sufficient for targeting Hook to Golgi (Walenta et al., 2001). SMART (Simple Modular Architecture Research Tool) protein sequence analysis suggests that within the 638 aa of HookA, there are three coiled-coil domains, aa 143–395, aa 454–493, and aa 532–589, and the final coiled-coil domain is followed by 49 aa residues. To determine the function of the C-terminal region, we made two separate deletion mutants. The first deletion mutant (ΔC) was made by deleting 37 aa within the last 49 aa at the C terminus of HookA after the last coiled-coiled domain, and the second deletion mutant (ΔC1) was made by deleting only 13 aa at the end of the last coiled-coil domain (Fig. 4 A). Using homologous recombination, we replaced the endogenous HookA gene with alleles encoding ΔC-HookA–GFP or ΔC1-HookA–GFP. Interestingly, both the ΔC-HookA mutant and the ΔC1-HookA mutant showed the same phenotypes as the ΔhookA mutant. Similar to the ΔhookA mutant, these two new mutants both formed colonies that were slightly more compact than wild type (Fig. 4 B). In addition, they both showed an obvious accumulation of mCherry-RabA–labeled early endosomes at the hyphal tip just like the ΔhookA mutant (Fig. 4 C). Furthermore, we were unable to find any mCherry-RabA signals moving away from the hyphal tip in both the ΔhookA and ΔC-HookA mutants (zero movements in 160 s in the mutants vs. 30 movements in 160 s in wild type; Fig. 4 D and Videos 2 and 3).
The C-terminal deletion mutants of HookA exhibit a defect in the HookA–early endosome interaction. (A) A diagram showing the wild-type HookA protein and the two C-terminal deletion mutants, ΔC-HookA and ΔC1-HookA, in which different amino acids are deleted. The red box indicates the putative microtubule-binding domain, the blue boxes indicate the three predicted coiled-coil domains, and the brown box indicates the C-terminal cargo-binding domain. (B) The ΔC-HookA and ΔC1-HookA mutants exhibit the same colony phenotype as that exhibited by the ΔhookA mutant. (C) The ΔC-HookA and ΔC1-HookA mutants show an obvious accumulation of mCherry-RabA–labeled early endosomes at the hyphal tip. The accumulation can be seen in ∼75% of the hyphal tips of both the ΔC-HookA and the ΔC1-HookA mutants (n = 98 for the ΔC-HookA mutant, and n = 102 for ΔC1-HookA mutant). See Video 3 for the phenotype of the ΔC-HookA mutant. (D) Kymographs showing an obvious accumulation of mCherry-RabA–labeled early endosomes at the hyphal tip in the ΔC-HookA and ΔhookA mutants and nonmotile early endosomes along the hyphae. (E) ΔC-HookA–GFP or ΔC1-HookA–GFP do not colocalize with the hyphal tip–accumulated early endosomes (100%, n = 50 for each mutant). (F) Western blots are shown to demonstrate that the ΔC-HookA–GFP or ΔC1-HookA–GFP proteins are expressed and stable. By measuring protein signal intensity on the Western blots in relation to protein loading as indicated by Ponceau S staining, we found that the level of ΔC-HookA–GFP relative to HookA-GFP is 1.17 ± 0.28 (mean ± SD; n = 3) and that of ΔC1-HookA–GFP relative to HookA-GFP is 0.99 ± 0.18 (mean ± SD; n = 3). There is no significant difference between the value of either mutant and that of the wild type at P = 0.05. Bars, 5 µm.
The C-terminal deletion mutants of HookA exhibit a defect in the HookA–early endosome interaction. (A) A diagram showing the wild-type HookA protein and the two C-terminal deletion mutants, ΔC-HookA and ΔC1-HookA, in which different amino acids are deleted. The red box indicates the putative microtubule-binding domain, the blue boxes indicate the three predicted coiled-coil domains, and the brown box indicates the C-terminal cargo-binding domain. (B) The ΔC-HookA and ΔC1-HookA mutants exhibit the same colony phenotype as that exhibited by the ΔhookA mutant. (C) The ΔC-HookA and ΔC1-HookA mutants show an obvious accumulation of mCherry-RabA–labeled early endosomes at the hyphal tip. The accumulation can be seen in ∼75% of the hyphal tips of both the ΔC-HookA and the ΔC1-HookA mutants (n = 98 for the ΔC-HookA mutant, and n = 102 for ΔC1-HookA mutant). See Video 3 for the phenotype of the ΔC-HookA mutant. (D) Kymographs showing an obvious accumulation of mCherry-RabA–labeled early endosomes at the hyphal tip in the ΔC-HookA and ΔhookA mutants and nonmotile early endosomes along the hyphae. (E) ΔC-HookA–GFP or ΔC1-HookA–GFP do not colocalize with the hyphal tip–accumulated early endosomes (100%, n = 50 for each mutant). (F) Western blots are shown to demonstrate that the ΔC-HookA–GFP or ΔC1-HookA–GFP proteins are expressed and stable. By measuring protein signal intensity on the Western blots in relation to protein loading as indicated by Ponceau S staining, we found that the level of ΔC-HookA–GFP relative to HookA-GFP is 1.17 ± 0.28 (mean ± SD; n = 3) and that of ΔC1-HookA–GFP relative to HookA-GFP is 0.99 ± 0.18 (mean ± SD; n = 3). There is no significant difference between the value of either mutant and that of the wild type at P = 0.05. Bars, 5 µm.
In sharp contrast to wild-type HookA-GFP, which colocalizes with early endosomes accumulated at the hyphal tip (Fig. 3, C and D), ΔC-HookA–GFP and ΔC1-HookA–GFP signals appear to be largely diffuse in the cytoplasm (Fig. 4 E), although some faint punctuates can be seen to move along microtubule-like structures (Videos 4 and 5; Video 4 is presented to show HookA-GFP as a wild-type control, and Video 5 shows the ΔC-HookA–GFP images from the same cell shown in Fig. 4 E). The signals clearly do not colocalize with accumulated early endosomes at the hyphal tip. The mutant proteins are stably expressed (Fig. 4 F), indicating that the defect in endosomal localization is not caused by protein instability. Thus, the C terminus of HookA is indeed necessary for HookA–early endosome interaction, which itself is essential for dynein-mediated transport of early endosomes.
HookA-GFP signals in a wild-type strain. Bidirectional movements of the signals can be observed, and many of these signals are associated with early endosomes (Fig. S2). The DNA encoding the HookA-GFP fusion protein was transformed into a wild-type strain carrying the mCherry-RabA marker although only GFP signals are shown in this video. An inverted fluorescence microscope (IX70; Olympus) was used for capturing images. 12 frames were taken with a 0.1-s exposure time and a 1-s interval between frames. Binning is 2 × 2. The video speed has been increased fivefold.
HookA-GFP signals in a wild-type strain. Bidirectional movements of the signals can be observed, and many of these signals are associated with early endosomes (Fig. S2). The DNA encoding the HookA-GFP fusion protein was transformed into a wild-type strain carrying the mCherry-RabA marker although only GFP signals are shown in this video. An inverted fluorescence microscope (IX70; Olympus) was used for capturing images. 12 frames were taken with a 0.1-s exposure time and a 1-s interval between frames. Binning is 2 × 2. The video speed has been increased fivefold.
ΔC-HookA–GFP signals. The signals are largely diffuse, but some puncta can be observed to move along microtubule-like structures. The DNA encoding the ΔC-HookA–GFP fusion protein was transformed into a wild-type strain carrying the mCherry-RabA marker although only GFP signals are shown in this video. An inverted fluorescence microscope (IX70; Olympus) was used for capturing images. 12 frames were taken with a 0.1-s exposure time and a 1-s interval between frames. Binning is 2×2. The video speed has been increased fivefold.
ΔC-HookA–GFP signals. The signals are largely diffuse, but some puncta can be observed to move along microtubule-like structures. The DNA encoding the ΔC-HookA–GFP fusion protein was transformed into a wild-type strain carrying the mCherry-RabA marker although only GFP signals are shown in this video. An inverted fluorescence microscope (IX70; Olympus) was used for capturing images. 12 frames were taken with a 0.1-s exposure time and a 1-s interval between frames. Binning is 2×2. The video speed has been increased fivefold.
To test directly whether the C-terminal domain mediates HookA–early endosome interaction, we constructed a strain in which the endogenous hookA allele is replaced by the C-HookA–GFP allele containing the coding sequence of 64 aa at the C terminus of HookA linked with GFP (all upstream untranslated region and the coding sequence for the first 7 aa of HookA were retained to ensure normal expression of the fusion protein). Remarkably, the C-HookA–GFP fusion largely colocalized with hyphal tip–accumulated early endosomes (Fig. 5 A), suggesting that the C terminus of HookA is capable of interacting with early endosomes, either directly or indirectly. To confirm this possibility, we performed pull-down experiments using extracts from the strains containing the HookA-GFP and C-HookA–GFP fusions. We found that early endosomes marked with mCherry-RabA were pulled down with either HookA-GFP or C-HookA–GFP by the anti-GFP antibody, and the amount of mCherry-RabA pulled down with C-HookA–GFP was even higher than that with HookA-GFP (Fig. S3). Thus, the C terminus indeed is the region within HookA that mediates HookA–early endosome interaction.
The C-terminal domain of HookA is capable of interacting with early endosomes. (A) Two examples showing that the C-HookA–GFP signals are concentrated at the hyphal tip where mCherry-RabA–marked early endosomes accumulate and the GFP and mCherry signals largely overlap (100% hyphal tips that show the concentrated GFP signals show the mCherry-RabA accumulation; n = 50). (B) A Western blot showing that the protein level of C-HookA–GFP expressed under the gpdA promoter (gpdA-C-HookA–GFP) is much higher than that expressed under the endogenous hookA promoter (C-HookA–GFP). The proteins were pulled down by the anti-GFP antibody, and the Western blot was probed by the anti-GFP antibody. Quantitation of the Western blots suggests that the level of gpdA-C-HookA–GFP is significantly higher than that of C-HookA–GFP (P < 0.005, n = 3). If we set the values of C-HookA–GFP as 1, the mean ± SD value of gpdA-C-HookA–GFP is 3.1 ± 0.4. This is likely to be an underestimate of the gpdA-C-HookA–GFP protein level as there seems to be a lot of degradation products, which are hard to include in the measurements. (C) Phenotypic analysis of the diploids showing that overexpression of C-HookA (gpdA-C-HookA–GFP) in the wild-type background produced a dominant-negative phenotype in early endosome distribution. Bars, 5 µm.
The C-terminal domain of HookA is capable of interacting with early endosomes. (A) Two examples showing that the C-HookA–GFP signals are concentrated at the hyphal tip where mCherry-RabA–marked early endosomes accumulate and the GFP and mCherry signals largely overlap (100% hyphal tips that show the concentrated GFP signals show the mCherry-RabA accumulation; n = 50). (B) A Western blot showing that the protein level of C-HookA–GFP expressed under the gpdA promoter (gpdA-C-HookA–GFP) is much higher than that expressed under the endogenous hookA promoter (C-HookA–GFP). The proteins were pulled down by the anti-GFP antibody, and the Western blot was probed by the anti-GFP antibody. Quantitation of the Western blots suggests that the level of gpdA-C-HookA–GFP is significantly higher than that of C-HookA–GFP (P < 0.005, n = 3). If we set the values of C-HookA–GFP as 1, the mean ± SD value of gpdA-C-HookA–GFP is 3.1 ± 0.4. This is likely to be an underestimate of the gpdA-C-HookA–GFP protein level as there seems to be a lot of degradation products, which are hard to include in the measurements. (C) Phenotypic analysis of the diploids showing that overexpression of C-HookA (gpdA-C-HookA–GFP) in the wild-type background produced a dominant-negative phenotype in early endosome distribution. Bars, 5 µm.
It has previously been shown in mammalian cells that expression of the C-terminal region of Hook1 causes a dominant-negative phenotype in endosome sorting (Maldonado-Báez et al., 2013). To test whether expression of the HookA C terminus also causes a dominant-negative phenotype, we constructed a strain in which the endogenous hookA allele is replaced by the gpdA-C-HookA–GFP allele, thereby allowing overexpression of the C-HookA–GFP fusion driven by the gpdA promoter (Pantazopoulou and Peñalva, 2009). The protein level of the C-HookA–GFP fusion is significantly higher in this strain compared with the strain in which the fusion is driven by the endogenous promoter of hookA (Fig. 5 B). We made a heterozygous diploid containing the gpdA-C-HookA–GFP allele and the wild-type hookA allele. Remarkably, an obvious accumulation of early endosomes was observed in ∼85% of hyphal tips in this diploid (n = 73; Fig. 5 C). In contrast, a heterozygous diploid containing a wild-type hookA allele and the C-HookA–GFP allele under its own promoter shows largely a wild-type phenotype (Fig. 5 C), with only ∼6% (n = 48) of hyphal tips containing an obvious accumulation of early endosomes. Similarly, a heterozygous diploid containing a wild-type hookA allele and the original hookAL150P,E151K mutant allele also shows largely a wild-type phenotype (Fig. 5 C), with only ∼10% (n = 62) of hyphal tips containing an obvious accumulation of early endosomes. Together, these results demonstrate that the C terminus of HookA mediates HookA–early endosome interaction, and its overexpression leads to a dominant-negative phenotype, most likely by saturating the HookA binding sites on early endosomes.
The N terminus of HookA is important for the interaction between HookA and dynein–dynactin
The N terminus of HookA is homologous to the microtubule-binding domain of human Hook3 (Fig. 2 A) and contains a calponin homology fold as recognized by PHYRE, a protein homology recognition engine. To study its function, we made a ΔN-HookA mutant by homologously replacing the endogenous HookA gene with a ΔN-HookA–GFP allele missing the codons for 127 aa of the putative microtubule-binding domain (Fig. 6 A). The ΔN-HookA mutant has essentially the same colony phenotype as the ΔhookA mutant (Fig. 6 B), and an obvious buildup of mCherry-RabA–labeled early endosomes at the hyphal tip was observed (Fig. 6, C and D). The ΔN-HookA–GFP fusion concentrated at the hyphal tip and colocalized with the hyphal tip–accumulated early endosomes (Fig. 6 E), indicating that the N-terminal domain is not required for the interaction between HookA and early endosomes.
The N-terminal domain of HookA is critical for the HookA–dynein–dynactin interaction. (A) A diagram showing the wild-type HookA protein and the N-terminal deletion mutant protein, ΔN-HookA, in which the putative microtubule-binding domain is deleted. (B) The ΔN-HookA mutant exhibits the same colony phenotype as that exhibited by ΔhookA. (C) The ΔN-HookA mutant showed an obvious accumulation of mCherry-RabA–labeled early endosomes at the hyphal tip (∼71% of the hyphal tips show this accumulation, n = 112. Also see Video 6). (D) Kymographs showing an obvious accumulation of mCherry-RabA–labeled early endosomes at the hyphal tip in the ΔN-HookA and ΔhookA mutants. An arrowhead indicates one early endosome that moved away from the hyphal tip in the ΔN-HookA mutant. (E) The ΔN-HookA–GFP signals were concentrated at the hyphal tip where mCherry-RabA–marked early endosomes accumulate, and the GFP and mCherry signals largely overlap (100% hyphal tips that show the concentrated GFP signals show the mCherry-RabA accumulation; n = 50). (F) The dynein HC, the p150 subunit of dynactin, and NudF/LIS1 can be pulled down with HookA-GFP, ΔC-HookA–GFP, and ΔC1-HookA–GFP, but the amounts of these proteins pulled down with ΔN-HookA–GFP were obviously decreased. (G) A quantitative analysis of the Western results shown in F. The ratio of pulled down dynein HC, dynactin p150, or NudF/LIS1 to HookA-GFP was calculated. Values of all the mutants are relative to the wild-type values, which are set at 1. Mean and SD values were calculated from multiple independent pull-down experiments, and the number of experiments is indicated as n. For the ratio of dynein to HookA (dynein/HookA), the mean ± SD value for ΔN is 0.08 ± 0.11 (n = 4, P < 0.001), and the values for ΔC and ΔC1 are 1.77 ± 0.96 (n = 4) and 1.26 ± 0.71 (n = 3), respectively. Note that a p-value is provided only when the values are statistically different from the wild-type value, and the values of ΔC and ΔC1 are not different from the wild-type value at P = 0.05. For the ratio of dynactin to HookA (dynactin/HookA), the mean ± SD value for ΔN is 0.22 ± 0.18 (n = 4, P < 0.001), and the values for ΔC and ΔC1 are 1.5 ± 0.28 (n = 4, P < 0.05) and 1.6 ± 0.87 (n = 3), respectively. For the ratio of NudF/LIS1 to HookA (LIS1/HookA), the mean ± SD value for ΔN is 0.15 ± 0.1 (n = 4, P < 0.001), and the values for ΔC and ΔC1 are 1.37 ± 0.21 (n = 4, P < 0.05) and 2.0 ± 1.1 (n = 3), respectively. (H) A diagram showing the wild-type, ΔC-HookA, ΔN-ΔC-HookA, and ΔN1-ΔC-HookA mutant proteins. (I) Dynein HC and dynactin p150 could be pulled down with ΔC-HookA–GFP, but the amounts of these proteins were obviously diminished when the pull-down was performed with ΔN-ΔC-HookA–GFP and were nearly undetectable when the pull-down was performed with ΔN1-ΔC-HookA–GFP. (J) A quantitative analysis of the Western results shown in I. Values of ΔN-ΔC-HookA–GFP and ΔN1-ΔC-HookA–GFP are relative to the ΔC-HookA–GFP values, which are set at 1. Values of ΔN-ΔC-HookA–GFP and ΔN1-ΔC-HookA–GFP are significantly lower than that of ΔC-HookA–GFP. For the ratio of dynein to HookA (dynein/HookA), the mean ± SD values for ΔN-ΔC and ΔN1-ΔC are 0.2 ± 0.3 (n = 3, P < 0.05) and 0 ± 0 (n = 3, P < 0.001), respectively. For the ratio of dynactin to HookA (dynactin/HookA), the mean ± SD values for ΔN-ΔC and ΔN1-ΔC are 0.16 ± 0.2 (n = 3, P < 0.005) and 0.02 ± 0.03 (n = 3, P < 0.001), respectively. However, the values for ΔN-ΔC and ΔN1-ΔC are not significantly different from each other at P = 0.05. Bars, 5 µm.
The N-terminal domain of HookA is critical for the HookA–dynein–dynactin interaction. (A) A diagram showing the wild-type HookA protein and the N-terminal deletion mutant protein, ΔN-HookA, in which the putative microtubule-binding domain is deleted. (B) The ΔN-HookA mutant exhibits the same colony phenotype as that exhibited by ΔhookA. (C) The ΔN-HookA mutant showed an obvious accumulation of mCherry-RabA–labeled early endosomes at the hyphal tip (∼71% of the hyphal tips show this accumulation, n = 112. Also see Video 6). (D) Kymographs showing an obvious accumulation of mCherry-RabA–labeled early endosomes at the hyphal tip in the ΔN-HookA and ΔhookA mutants. An arrowhead indicates one early endosome that moved away from the hyphal tip in the ΔN-HookA mutant. (E) The ΔN-HookA–GFP signals were concentrated at the hyphal tip where mCherry-RabA–marked early endosomes accumulate, and the GFP and mCherry signals largely overlap (100% hyphal tips that show the concentrated GFP signals show the mCherry-RabA accumulation; n = 50). (F) The dynein HC, the p150 subunit of dynactin, and NudF/LIS1 can be pulled down with HookA-GFP, ΔC-HookA–GFP, and ΔC1-HookA–GFP, but the amounts of these proteins pulled down with ΔN-HookA–GFP were obviously decreased. (G) A quantitative analysis of the Western results shown in F. The ratio of pulled down dynein HC, dynactin p150, or NudF/LIS1 to HookA-GFP was calculated. Values of all the mutants are relative to the wild-type values, which are set at 1. Mean and SD values were calculated from multiple independent pull-down experiments, and the number of experiments is indicated as n. For the ratio of dynein to HookA (dynein/HookA), the mean ± SD value for ΔN is 0.08 ± 0.11 (n = 4, P < 0.001), and the values for ΔC and ΔC1 are 1.77 ± 0.96 (n = 4) and 1.26 ± 0.71 (n = 3), respectively. Note that a p-value is provided only when the values are statistically different from the wild-type value, and the values of ΔC and ΔC1 are not different from the wild-type value at P = 0.05. For the ratio of dynactin to HookA (dynactin/HookA), the mean ± SD value for ΔN is 0.22 ± 0.18 (n = 4, P < 0.001), and the values for ΔC and ΔC1 are 1.5 ± 0.28 (n = 4, P < 0.05) and 1.6 ± 0.87 (n = 3), respectively. For the ratio of NudF/LIS1 to HookA (LIS1/HookA), the mean ± SD value for ΔN is 0.15 ± 0.1 (n = 4, P < 0.001), and the values for ΔC and ΔC1 are 1.37 ± 0.21 (n = 4, P < 0.05) and 2.0 ± 1.1 (n = 3), respectively. (H) A diagram showing the wild-type, ΔC-HookA, ΔN-ΔC-HookA, and ΔN1-ΔC-HookA mutant proteins. (I) Dynein HC and dynactin p150 could be pulled down with ΔC-HookA–GFP, but the amounts of these proteins were obviously diminished when the pull-down was performed with ΔN-ΔC-HookA–GFP and were nearly undetectable when the pull-down was performed with ΔN1-ΔC-HookA–GFP. (J) A quantitative analysis of the Western results shown in I. Values of ΔN-ΔC-HookA–GFP and ΔN1-ΔC-HookA–GFP are relative to the ΔC-HookA–GFP values, which are set at 1. Values of ΔN-ΔC-HookA–GFP and ΔN1-ΔC-HookA–GFP are significantly lower than that of ΔC-HookA–GFP. For the ratio of dynein to HookA (dynein/HookA), the mean ± SD values for ΔN-ΔC and ΔN1-ΔC are 0.2 ± 0.3 (n = 3, P < 0.05) and 0 ± 0 (n = 3, P < 0.001), respectively. For the ratio of dynactin to HookA (dynactin/HookA), the mean ± SD values for ΔN-ΔC and ΔN1-ΔC are 0.16 ± 0.2 (n = 3, P < 0.005) and 0.02 ± 0.03 (n = 3, P < 0.001), respectively. However, the values for ΔN-ΔC and ΔN1-ΔC are not significantly different from each other at P = 0.05. Bars, 5 µm.
Interestingly, although the hyphal tip accumulation of mCherry-RabA is conspicuous in the ΔN-HookA mutant, at a lower frequency, some early endosomes were still seen to move away from the hyphal tip with a normal speed (mean ± SD: 1.65 ± 0.40 µm/s [n = 16] for wild type and 1.65 ± 0.56 µm/s [n = 20] for ΔN-HookA; Fig. 6 D, arrows). This is in contrast to the absence of any early endosome movement in the ΔC-HookA and ΔhookA mutant hyphae grown under the same conditions (Videos 2, 3, and 6), indicating that the putative microtubule-binding domain is not as critical as the C-terminal cargo-binding domain for HookA function.
Intriguingly, the N-terminal putative microtubule-binding domain of the Caenorhabditis elegans Hook homologue Zyg-12, a protein involved in nucleus–centrosome coupling, has been proposed to bind dynein because a dynein light intermediate chain was identified as one of the interacting proteins in a yeast two-hybrid assay (Malone et al., 2003). This observation has never been followed up in any experimental systems by biochemical analysis, and thus, Hook proteins are not generally considered as dynein-interacting proteins or cargo–dynein linkers for vesicle transport. Therefore, we tested whether the N-terminal putative microtubule-binding domain of A. nidulans HookA is responsible for linking HookA to dynein. In pull-down experiments using cell extracts containing HookA-GFP-fusions, both dynactin p150 and dynein HC were pulled down with HookA-GFP by the anti-GFP antibody (Fig. 6 F). NudF/LIS1 was pulled down, also as expected, because it associates with dynein (Sasaki et al., 2000; McKenney et al., 2010; Huang et al., 2012). Importantly, the ΔC1-HookA–GFP and ΔC-HookA–GFP proteins, which fail to associate with early endosomes, were still able to pull down dynein HC, dynactin p150, and NudF/LIS1, and the amounts of these proteins pulled down with ΔC1-HookA–GFP or ΔC-HookA–GFP are definitely not lower than that pulled down with HookA-GFP. This result indicates that the HookA–dynein–dynactin interaction does not depend on the association between HookA and early endosomes. The amount of dynein HC pulled down with ΔC-HookA–GFP, however, is significantly lower than that pulled down with dynactin p150-GFP (Fig. S4), indicating that HookA might only interact with a small portion of dynein in the cell.
In contrast to ΔC1-HookA–GFP and ΔC-HookA–GFP, ΔN-HookA–GFP pulled down significantly lower amounts of dynactin, dynein, and NudF/LIS1 (Fig. 6, F and G). Because ΔN-HookA–GFP signals are largely associated with that of hyphal tip–accumulated early endosomes, we were concerned about the possibility that the weakened interaction between ΔN-HookA–GFP and dynein–dynactin might be caused indirectly by the unavailability of HookA protein in the soluble pool to interact with dynein–dynactin. To address this concern, we made a ΔN-ΔC-HookA–GFP fusion with both the C-terminal and N-terminal deletions (Fig. 6 H). Because the ΔC mutation prevented HookA from associating with early endosomes, there should be sufficient ΔN-ΔC-HookA–GFP in the soluble pool to interact with dynein–dynactin. However, the amount of pulled down dynein–dynactin with ΔN-ΔC-HookA–GFP is still significantly reduced compared with that pulled down with ΔC-HookA–GFP (Fig. 6 I). These results strongly indicate that the N-terminal putative microtubule-binding domain of HookA is indeed important for HookA–dynein–dynactin interaction.
In these experiments, we noticed that residual amounts of dynein HC can still be pulled down by the ΔN-ΔC-HookA–GFP fusion (Fig. 6 I), suggesting that a region other than the N-terminal microtubule-binding region might also contribute to HookA–dynein–dynactin interaction. Therefore, we deleted a larger region of the N terminus to remove both the microtubule binding site and the first half of the first coiled coil, and we named this deletion ΔN1 (Fig. 6 H). When the ΔN1-ΔC-HookA–GFP fusion was used in the same pull-down experiment, the mean values of the amounts of pulled down dynein and dynactin are further reduced (Fig. 6, I and J). However, the differences are not statistically significant at P = 0.05. Thus, although our results show convincingly that the N-terminal part of HookA is important for the HookA–dynein–dynactin interaction, the exact binding sites would need to be mapped more precisely in the future.
Dynein HC and dynactin p25 are codependent for HookA–dynein–dynactin interaction
We next sought to determine whether it is the dynein complex or the dynactin complex that mediates the interaction between HookA and dynein–dynactin. In the dynein complex, dynein HC is the major component, and its loss destabilizes dynein intermediate chain (Zhang et al., 2002; Caviston et al., 2007; Levy and Holzbaur, 2008), a component that binds to the light chains, the dynactin complex, and NudE/Nudel (Karki and Holzbaur, 1995; Vaughan and Vallee, 1995; Susalka et al., 2002; McKenney et al., 2011; Wang and Zheng, 2011; Wang et al., 2013). In the dynactin complex, Arp1 forms a minifilament (Fig. 7 A), and its loss results in the disruption of the complex (Schafer et al., 1994; Minke et al., 1999; Schroer, 2004; Haghnia et al., 2007; Zhang et al., 2008). To test the requirement for dynein and dynactin in the HookA–dynein–dynactin interaction, we introduced the HookA-GFP fusion into a dynein HC conditional null mutant alcA-nudAHC and an Arp1 conditional null mutant alcA-nudKArp1 and performed pull-down experiments using the anti-GFP antibody. Expression of the nudA dynein HC gene or the nudK Arp1 gene driven by the alcA promoter is prevented in rich medium containing glucose (Xiang et al., 1995b; Zhang et al., 2008), causing disruption of the dynein or the dynactin complex, respectively. Interestingly, in both mutants, pulled down dynein or dynactin is diminished (Fig. 7, B and C).
Dynein and p25 of dynactin are codependent for the HookA–dynein–dynactin interaction. (A) A diagram showing the dynactin complex (Schroer, 2004) and the heavy chain (HC) and intermediate chain (IC) of dynein. (B) Western blots showing that HookA–dynein–dynactin interaction is defective in a dynein HC conditional null mutant, alcA-nudAHC, and an Arp1 conditional null mutant, alcA-nudKArp1. Cells were grown in rich medium YG that contains glucose to prevent expression of the nudA dynein HC gene and the nudK Arp1 gene. (C) A quantitative analysis of the Western results shown in B. The ratio of pulled down dynein HC or dynactin to HookA-GFP was calculated. Values of all the mutants are relative to the wild-type values, which are set at 1. Mean and SD values were calculated from multiple independent pull-down experiments, and the number of experiments is indicated as n. For the ratio of dynein to HookA (dynein/HookA), the mean ± SD values for alcA-nudAHC and alcA-nudKArp1 are 0.1 ± 0.17 (n = 3, P < 0.001) and 0.08 ± 0.13 (n = 3, P < 0.001), respectively. For the ratio of dynactin to HookA (dynactin/HookA), values for alcA-nudAHC and alcA-nudKArp1 are 0.04 ± 0.07 (n = 3, P < 0.001) and 0.01 ± 0.01 (n = 3, P < 0.001), respectively. (D) Western blots showing that the HookA–dynein–dynactin interaction is defective in the alcA-nudAHC and the Δp25 mutant. (E) A quantitative analysis of the Western results shown in D. Values of all the mutants are relative to the values of ΔC1-HookA in the wild-type background, which are set at 1. For the ratio of dynein to ΔC1-HookA (dynein/ΔC1-HookA), the mean ± SD values for alcA-nudAHC and Δp25 are 0.04 ± 0.08 (n = 3, P < 0.001) and 0.01 ± 0.03 (n = 4, P < 0.001), respectively. For the ratio of dynactin to ΔC1-HookA (dynactin/ΔC1-HookA), values for alcA-nudAHC and Δp25 are 0.1 ± 0.09 (n = 3, P < 0.001) and 0.06 ± 0.12 (n = 4, P < 0.001), respectively. (F) Western blots showing that the HookA–dynein–dynactin interaction is defective in the Δp25 mutant. For the pull-down experiments presented in F, a supernatant of the 100,000 g high-speed centrifugation was used. (G) A quantitative analysis of the Western results shown in F. Values of the mutants are relative to the values of ΔC1-HookA in the wild-type background, which are set at 1. For the ratio of dynein to ΔC1-HookA (dynein/ΔC1-HookA), the mean ± SD value for Δp25 is 0.12 ± 0.2 (n = 3, P < 0.001). For the ratio of dynactin to ΔC1-HookA (dynactin/ΔC1-HookA), the mean ± SD value for Δp25 is 0.02 ± 0.03 (n = 3, P < 0.001).
Dynein and p25 of dynactin are codependent for the HookA–dynein–dynactin interaction. (A) A diagram showing the dynactin complex (Schroer, 2004) and the heavy chain (HC) and intermediate chain (IC) of dynein. (B) Western blots showing that HookA–dynein–dynactin interaction is defective in a dynein HC conditional null mutant, alcA-nudAHC, and an Arp1 conditional null mutant, alcA-nudKArp1. Cells were grown in rich medium YG that contains glucose to prevent expression of the nudA dynein HC gene and the nudK Arp1 gene. (C) A quantitative analysis of the Western results shown in B. The ratio of pulled down dynein HC or dynactin to HookA-GFP was calculated. Values of all the mutants are relative to the wild-type values, which are set at 1. Mean and SD values were calculated from multiple independent pull-down experiments, and the number of experiments is indicated as n. For the ratio of dynein to HookA (dynein/HookA), the mean ± SD values for alcA-nudAHC and alcA-nudKArp1 are 0.1 ± 0.17 (n = 3, P < 0.001) and 0.08 ± 0.13 (n = 3, P < 0.001), respectively. For the ratio of dynactin to HookA (dynactin/HookA), values for alcA-nudAHC and alcA-nudKArp1 are 0.04 ± 0.07 (n = 3, P < 0.001) and 0.01 ± 0.01 (n = 3, P < 0.001), respectively. (D) Western blots showing that the HookA–dynein–dynactin interaction is defective in the alcA-nudAHC and the Δp25 mutant. (E) A quantitative analysis of the Western results shown in D. Values of all the mutants are relative to the values of ΔC1-HookA in the wild-type background, which are set at 1. For the ratio of dynein to ΔC1-HookA (dynein/ΔC1-HookA), the mean ± SD values for alcA-nudAHC and Δp25 are 0.04 ± 0.08 (n = 3, P < 0.001) and 0.01 ± 0.03 (n = 4, P < 0.001), respectively. For the ratio of dynactin to ΔC1-HookA (dynactin/ΔC1-HookA), values for alcA-nudAHC and Δp25 are 0.1 ± 0.09 (n = 3, P < 0.001) and 0.06 ± 0.12 (n = 4, P < 0.001), respectively. (F) Western blots showing that the HookA–dynein–dynactin interaction is defective in the Δp25 mutant. For the pull-down experiments presented in F, a supernatant of the 100,000 g high-speed centrifugation was used. (G) A quantitative analysis of the Western results shown in F. Values of the mutants are relative to the values of ΔC1-HookA in the wild-type background, which are set at 1. For the ratio of dynein to ΔC1-HookA (dynein/ΔC1-HookA), the mean ± SD value for Δp25 is 0.12 ± 0.2 (n = 3, P < 0.001). For the ratio of dynactin to ΔC1-HookA (dynactin/ΔC1-HookA), the mean ± SD value for Δp25 is 0.02 ± 0.03 (n = 3, P < 0.001).
We next tested the involvement of the p25 protein, which is part of the pointed end complex, and similar to p27, its loss does not significantly affect the integrity/function of the core dynactin complex (Eckley et al., 1999; Lee et al., 2001; Zhang et al., 2011; Yeh et al., 2012, 2013). Because the ΔC1-HookA–GFP fusion is not associated with early endosomes but is able to interact with dynein–dynactin, we used it rather than the full-length HookA-GFP for this analysis. Specifically, we introduced the ΔC1-HookA–GFP fusion into the alcA-nudAHC and Δp25 mutants and performed pull-down experiments. Interestingly, in both mutants, neither dynein nor dynactin could be effectively pulled down (Fig. 7, D and E). To confirm that the effect is independent of early endosomes, we centrifuged the lysate at 100,000 g to remove membranes and used the supernatant for the pull-down assay (Fig. 7 F). Although ΔC1-HookA–GFP was able to pull down dynein and dynactin from the wild-type supernatant, the amounts of dynein and dynactin pulled down from the Δp25 supernatant were significantly diminished (Fig. 7, F and G). Together, our results suggest that both the dynein complex and p25 dynactin are required for the physical interaction between HookA and dynein–dynactin.
HookA enhances dynein–early endosome interaction
To test whether HookA is involved in the physical interaction between the dynein motor and the early endosome cargo, we performed pull-down experiments using strains containing dynein HC–GFP (GFP-HC) and mCherry-RabA. In the absence of detergent, the anti-GFP antibody pulled down mCherry-RabA–labeled early endosomes from the wild-type cell extract (Fig. 8 A). Importantly, the amount of mCherry-RabA pulled down from the ΔhookA mutant extract under the same conditions was significantly reduced (P < 0.001; Fig. 8, A and B), indicating that HookA enhances dynein–early endosome interaction.
Loss of HookA or the HookA–dynein–dynactin interaction significantly weakens the interaction between dynein and early endosomes. (A) Western blots showing that the amount of mCherry-RabA–labeled early endosomes pulled down with GFP–dynein HC from the ΔhookA mutant extract is significantly lower than that from the wild type. The mCherry-RabA signals in the extracts used for the pull-down experiments are also shown. (B) A quantitative analysis of the Western results (shown in A). The ratio of mCherry-RabA to GFP–dynein HC (RabA/dynein) was calculated. Values are relative to the wild-type value, which is set at 1. The mean ± SD value for the ΔhookA mutant is 0.19 ± 0.22 (n = 4, P < 0.001). (C) The HookAL150P,E151K-GFP signals were concentrated at the hyphal tip where mCherry-RabA–marked early endosomes accumulate, and the GFP and mCherry signals largely overlap. Bars, 5 µm. (D) The dynein HC and the p150 subunit of dynactin could be pulled down with HookA-GFP, but the amounts of these proteins pulled down with HookAL150P,E151K-GFP were obviously decreased. For the ratio of dynein to HookA, if we set the wild-type mean values to 1, the mean ± SD value for HookAL150P,E151K was 0.05 ± 0.09 (n = 3, P < 0.001). For the ratio of dynactin to HookA, if we set the wild-type mean values to 1, values for HookAL150P,E151K were 0.09 ± 0.16 (n = 3, P < 0.001). In contrast, the amount of mCherry-RabA–labeled early endosomes pulled down was apparently not decreased. The mCherry-RabA signals in the extracts used for the pull-down experiments are also shown. (E) Western blots showing that the amount of mCherry-RabA–labeled early endosomes pulled down with dynein HC-GFP from the HookAL150P,E151K mutant extract is significantly lower than that from the wild type. The mCherry-RabA signals in the extracts used for the pull-down experiments are also shown. (F) A quantitative analysis of the Western results (shown in E). The ratio of mCherry-RabA to GFP–dynein HC (RabA/dynein) was calculated. Values are relative to the wild-type value, which is set at 1. The mean ± SD value for the ΔhookA mutant is 0.25 ± 0.15 (n = 4, P < 0.001). (G) A working model showing that HookA on an early endosome links dynein–dynactin to the cargo for its movement along the microtubule track. Several dimers of HookA are depicted. A possibility not excluded is that HookA also facilitates cargo–track interaction, which is likely to be dynamic rather than static. For simplicity, HookA is depicted as the only protein linking dynein–dynactin to the early endosome, but it is likely that additional proteins are required for bridging the HookA–dynein–dynactin and HookA–early endosome interactions.
Loss of HookA or the HookA–dynein–dynactin interaction significantly weakens the interaction between dynein and early endosomes. (A) Western blots showing that the amount of mCherry-RabA–labeled early endosomes pulled down with GFP–dynein HC from the ΔhookA mutant extract is significantly lower than that from the wild type. The mCherry-RabA signals in the extracts used for the pull-down experiments are also shown. (B) A quantitative analysis of the Western results (shown in A). The ratio of mCherry-RabA to GFP–dynein HC (RabA/dynein) was calculated. Values are relative to the wild-type value, which is set at 1. The mean ± SD value for the ΔhookA mutant is 0.19 ± 0.22 (n = 4, P < 0.001). (C) The HookAL150P,E151K-GFP signals were concentrated at the hyphal tip where mCherry-RabA–marked early endosomes accumulate, and the GFP and mCherry signals largely overlap. Bars, 5 µm. (D) The dynein HC and the p150 subunit of dynactin could be pulled down with HookA-GFP, but the amounts of these proteins pulled down with HookAL150P,E151K-GFP were obviously decreased. For the ratio of dynein to HookA, if we set the wild-type mean values to 1, the mean ± SD value for HookAL150P,E151K was 0.05 ± 0.09 (n = 3, P < 0.001). For the ratio of dynactin to HookA, if we set the wild-type mean values to 1, values for HookAL150P,E151K were 0.09 ± 0.16 (n = 3, P < 0.001). In contrast, the amount of mCherry-RabA–labeled early endosomes pulled down was apparently not decreased. The mCherry-RabA signals in the extracts used for the pull-down experiments are also shown. (E) Western blots showing that the amount of mCherry-RabA–labeled early endosomes pulled down with dynein HC-GFP from the HookAL150P,E151K mutant extract is significantly lower than that from the wild type. The mCherry-RabA signals in the extracts used for the pull-down experiments are also shown. (F) A quantitative analysis of the Western results (shown in E). The ratio of mCherry-RabA to GFP–dynein HC (RabA/dynein) was calculated. Values are relative to the wild-type value, which is set at 1. The mean ± SD value for the ΔhookA mutant is 0.25 ± 0.15 (n = 4, P < 0.001). (G) A working model showing that HookA on an early endosome links dynein–dynactin to the cargo for its movement along the microtubule track. Several dimers of HookA are depicted. A possibility not excluded is that HookA also facilitates cargo–track interaction, which is likely to be dynamic rather than static. For simplicity, HookA is depicted as the only protein linking dynein–dynactin to the early endosome, but it is likely that additional proteins are required for bridging the HookA–dynein–dynactin and HookA–early endosome interactions.
To confirm that HookA is important for dynein–early endosome interaction, we further analyzed the HookAL150P,E151K (eedA1) mutant (Fig. 1). Just like ΔN-HookA–GFP, HookAL150P,E151K-GFP signals were concentrated at the hyphal tip where mCherry-RabA–labeled early endosomes accumulate (Fig. 8 C). In pull-down assays, HookAL150P,E151K-GFP pulled down much lower amounts of dynein HC and the p150 subunit of dynactin but apparently higher amounts of mCherry-RabA–labeled early endosomes than HookA-GFP (Fig. 8 D). Thus, the HookAL150P,E151K mutant is defective in the HookA–dynein–dynactin interaction but not in HookA–early endosome interaction. When dynein HC-GFP was used for the pull-down assay, the amount of mCherry-RabA–labeled early endosomes pulled down with dynein HC-GFP from the HookAL150P,E151K mutant extract is significantly lower than that from the wild type (P < 0.001; Fig. 8, E and F). Thus, the ability of HookA to bind dynein–dynactin is important for the dynein–early endosome interaction.
So far, it is unclear whether HookA is involved in enhancing early endosome–microtubule interaction. In our current study, we have not been able to detect a direct interaction between HookA and microtubules in a microtubule-pelleting assay (Fig. S5). In addition, the N-HookA–GFP fusion containing only the N-terminal putative microtubule-binding domain of HookA does not obviously decorate microtubules in live cells (Fig. S5).
HookA function is involved in the proper distribution of peroxisomes
Although we have discovered the function of HookA in dynein-mediated early endosome transport, it is unclear whether a similar function of HookA is involved in the transport of other dynein cargoes. In A. nidulans, peroxisomes are another known dynein cargo (Egan et al., 2012b). In a dynein-null mutant, peroxisomes accumulate at the hyphal tip area (Egan et al., 2012b). Intriguingly, a hyphal tip accumulation of peroxisomes was also observed in the ΔuncA kinesin-3–null mutant (Egan et al., 2012b), and the mechanism behind this interesting phenomenon is currently not understood. To this end, we tested the requirement for HookA in peroxisome transport by introducing a peroxisome marker, GFP-labeled PexK (Pex11) protein (Hynes et al., 2008; Egan et al., 2012b), into a ΔhookA mutant via genetic crossing. In most wild-type cells, peroxisomes distribute along the hyphae as previously described (Egan et al., 2012b). Interestingly, the majority of ΔhookA cells (71%, n = 107) showed an accumulation of PexK-GFP within ∼5 µm of the hyphal apex (Fig. 9). In images in which mCherry-RabA and PexK-GFP were observed in the same cells, PexK-GFP accumulation appeared slightly behind the mCherry-RabA signals (Fig. 9). Although a minor fraction of wild-type cells (13%, n = 88) also showed accumulation of PexK-GFP at a similar position, the majority of wild-type cells (87%, n = 88) did not show such an accumulation. Based on these observations, we conclude that Hook function is involved in peroxisome distribution, either directly or indirectly. However, as we have not yet obtained any evidence for the physical interaction between HookA and peroxisomes, mechanistically, how HookA affects peroxisome distribution will be an interesting question to address in the future.
Distribution of PexK-GFP, a peroxisomal marker, is abnormal in the ΔhookA mutant. A wild-type control and several ΔhookA cells are shown. For the ΔhookA cell presented on the top right, PexK-GFP, mCherry-RabA, and their merger are all shown. For the ΔhookA cells presented on the bottom, PexK-GFP and the merger of PexK-GFP and mCherry-RabA are shown. Arrows indicate the abnormal accumulation of PexK-GFP signals. Bars, 5 µm.
Distribution of PexK-GFP, a peroxisomal marker, is abnormal in the ΔhookA mutant. A wild-type control and several ΔhookA cells are shown. For the ΔhookA cell presented on the top right, PexK-GFP, mCherry-RabA, and their merger are all shown. For the ΔhookA cells presented on the bottom, PexK-GFP and the merger of PexK-GFP and mCherry-RabA are shown. Arrows indicate the abnormal accumulation of PexK-GFP signals. Bars, 5 µm.
Discussion
By using a classical genetic approach combined with whole genome sequencing of an A. nidulans mutant, we identified HookA as a new factor essential for dynein-mediated early endosome transport in vivo. HookA is a homologue of the human Hook proteins with a putative microtubule-binding domain at its N terminus and a cargo-binding domain at its C terminus (Walenta et al., 2001). Although the microtubule-binding domain of human Hook3 binds directly to microtubules as judged by a microtubule-pelleting assay (Walenta et al., 2001), microtubule binding of HookA is not detectable (Fig. S5). Thus, the affinity of HookA for microtubules is low or the binding is dynamic. Alternatively, HookA might have a low affinity for microtubules made from mammalian tubulins, and it will be worthwhile in the future to perform microtubule-binding assays using A. nidulans tubulins (Widlund et al., 2012). Our biochemical analyses, however, clearly demonstrate that the putative microtubule-binding domain of HookA is important, albeit not essential, for HookA to interact with dynein–dynactin and that HookA links dynein to early endosomes for long-distance early endosome movement.
The Hook–dynein interaction has been previously proposed for the C. elegans Hook homologue, Zyg-12, based on the yeast two-hybrid data showing that the putative microtubule-binding domain binds to a dynein light intermediate chain (Malone et al., 2003). This observation has never been confirmed by biochemical analysis in any experimental system, and thus, Hook has never been considered a motor adapter for vesicle transport. In this study, we found that both dynein and the p25 protein of the dynactin complex are critical for HookA–dynein–dynactin interaction (Fig. 7). The HookA–dynein–dynactin interaction is specific rather than nonspecifically mediated by early endosomes, as it can be detected in the nonmembrane fraction, and the two C-terminal truncation mutants of HookA unable to bind early endosomes are fully capable of interacting with dynein–dynactin. Together, our results suggest that HookA is more likely to bind to the dynein–dynactin supercomplex rather than to the individual dynein or dynactin complex and that p25 is necessary for mediating the interaction. This is consistent with the fact that HookA and p25 are both required for dynein–early endosome interaction (Fig. 8; Zhang et al., 2011) and that they both associate with motile early endosomes driven by dynein (Fig. 3; Egan et al., 2012b). However, the HookA–p25 interaction may not be direct, as this newly identified linkage system may contain additional bridging proteins.
In this study, we provide strong evidence to indicate that the C terminus of HookA mediates HookA–early endosome interaction. However, it is likely that the HookA–early endosome interaction requires additional proteins, and how HookA interacts with early endosomes awaits further studies. In higher eukaryotic cells, dynein uses a variety of mechanisms to bind to vesicles/organelles (Akhmanova and Hammer, 2010; Tan et al., 2011; Splinter et al., 2012; Yadav et al., 2012; Zhou et al., 2012; Fu and Holzbaur, 2013), and whether and how Hook proteins are involved in dynein-mediated transport of various cargoes will need to be determined. It should be noted that a similar function of Hook in early endosome transport has recently been found in another filamentous fungus U. maydis (Steinberg, G., personal communication). Interestingly, the budding yeast Hook homologue is a v-SNARE–binding protein containing the C-terminal part of Hook but misses the putative microtubule-binding domain (Kama et al., 2007), which we show here to be important for interacting with dynein–dynactin for early endosome transport. This may be related to the notion that budding yeast dynein has evolved to be required exclusively for spindle orientation (Winey and Bloom, 2012) and that budding yeast dynactin lacks the p25 subunit required for HookA–dynein–dynactin interaction (Eckley et al., 1999; Moore et al., 2008).
In summary, we have discovered HookA as a novel motor–cargo linker essential for dynein-mediated early endosome movement in vivo. Although a genome-wide RNAi-based screen in higher eukaryotes has recently been used to find novel regulators of organelle/vesicle transport (Winter et al., 2012), a classical genetic screen specifically designed for discovering motor–cargo linkers has never been reported. HookA represents the first product from such a screen, which is far from being saturated, and it is likely that additional proteins may participate in the HookA-dependent dynein–early endosome linkage. Moreover, it is unclear whether any HookA-independent linkage exists between dynein–dynactin and early endosomes in A. nidulans. Because our pull-down results show a significant decrease but not abolition of dynein–early endosome interaction in the absence of HookA, it is possible that dynein can still interact with early endosomes via mechanisms independent of HookA. However, in the absence of HookA, any interaction between dynein and early endosomes is not sufficiently strong for supporting transport in vivo, as indicated by a virtually complete absence of early endosome transport in the absence of HookA. As all dynein regulators found in A. nidulans are evolutionarily conserved (Kardon and Vale, 2009), the identification of HookA will undoubtedly open new avenues for understanding dynein–early endosome interactions in a variety of experimental systems.
Materials and methods
A. nidulans strains, media, and mutagenesis
A. nidulans strains used in this study are listed in Table S1. For biochemical experiments, YG (yeast extract plus glucose) + UU (or YUU) liquid medium was used. UV mutagenesis on spores of A. nidulans strains was performed as previously described (Willins et al., 1995; Xiang et al., 1999). For DAPI staining of nuclei, cells were incubated in YUU liquid medium for 8 h at 37°C or in liquid minimal medium containing 1% glycerol plus supplements overnight at 32°C. For live-cell imaging experiments, liquid minimal medium containing 1% glycerol plus supplements was used, and cells were cultured at 32°C overnight and observed at room temperature. For dual-view imaging of HookA-GFP and mCherry-RabA, 0.1% fructose instead of 1% glycerol was used. The PexK-GFP strains were obtained from M. Hynes (The University of Melbourne, Melbourne, Australia), S. Reck-Peterson, and K. Tan (Harvard Medical School, Boston, MA; Hynes et al., 2008; Egan et al., 2012b). The original strain carrying the DkinA::pyr4 allele was obtained from R. Fischer (Karlsruhe Institute of Technology, Karlsruhe, Germany; Requena et al., 2001). The original strain carrying the DnkuA::argB allele was obtained from B. Oakley (The University of Kansas, Lawrence, KS; Nayak et al., 2006).
Live-cell imaging and analyses
Fluorescence microscopy of live A. nidulans hyphae was as previously described (Zhang et al., 2011). Cells were grown at 32°C overnight using the chambered cover glass system (Lab-Tek; Thermo Fisher Scientific), and liquid minimal medium containing 1% glycerol plus supplements was used for culturing the cells.
All images except for those presented in Fig. 3 and Fig. S2 were captured at room temperature using an inverted fluorescence microscope (IX70; Olympus) linked to a cooled charge-coupled device camera (SensiCAM QE; PCO/Cooke Corporation). A U Plan Apochromat 100× objective lens (oil) with a 1.35 numerical aperture was used. A filter wheel system with GFP/mCherry-ET Sputtered Series with high transmission (BioVision Technologies) was used. The IPLab software (BioVision Technologies) was used for image acquisition and analysis. For the PexK-GFP images shown in Fig. 9, the bright-field images were taken simultaneously with the GFP images, which allowed us to see the shapes of the hyphae in these images. All images were transferred to Photoshop (Adobe) for annotation and saved as TIFF files.
For measuring the signal intensity of the individual GFP-HC comets, an area containing the whole comet was selected as a region of interest (ROI), and the Max/Min tool of the IPLab program was used to measure the maximal intensity within the ROI. Then, the ROI box was dragged outside of the cell to take the background value, which was then subtracted from the value of the comet. Because comets that have arrived at the hyphal tip show the highest signal intensity, as we have described previously, we only selected frames within a sequence in which the comets are seen at the hyphal tip region, and only those comets that have arrived at the hyphal tip were measured.
For the simultaneous imaging of mCherry-RabA and HookA-GFP (presented in Fig. 3 and Fig. S2) in the red and green channels, respectively, liquid minimal medium containing 0.1% fructose plus supplements was used. For these experiments, we used an inverted microscope (DMI6000 B; Leica) and a stage-coupled incubation chamber set at 28°C. The microscope, driven by MetaMorph (Molecular Devices) software, was equipped with a 63× HCX Plan Aprochromat objective (1.4 numerical aperture; Leica) and an external light source (EL6000; Leica) for epifluorescence excitation. Images were collected with a beam splitter (Dual-View; Photometrics), set for the GFP and mCherry channels, and a camera (ER-II; Hamamatsu Photonics; Abenza et al., 2012). To capture fast-moving early endosomes, we used the streaming function of MetaMorph. Image analysis was performed with MetaMorph standard functions: The time stacks corresponding to the red and green channels were used to derive kymographs, which were aligned and combined using the color align menu of the MetaMorph software. The color kymographs were saved as 8-bit RGB images also using MetaMorph, whereas the kymographs from the individual channels were converted to grayscale to facilitate visualization. All images were transferred to Photoshop for annotation and saved as TIFF files.
Complementation of the eedA1 mutant
Complementation of the eedA1 (HookAL150P,E151K) mutant was performed by using the genomic DNA fragment amplified using the following two oligonucleotides (oligos): 41U, 5′-CATGCTTGCTTCCTCTTGC-3′, and 41D, 5′-GCTCGTACCAGGCTGAACTC-3′. The DNA fragment was used to transform the JZ542 strain.
DNA constructs for generating HookA mutants and for making GFP-tagged HookA
For making all the following mutants, the strain used for transformation carries ΔnkuA, which facilitates the selection of transformants in which the DNA fragments integrate into the genome via homologous recombination (Nayak et al., 2006; Szewczyk et al., 2007). Homologous integrations were confirmed by Southern blots, PCR, and sequencing analyses and/or Western analyses.
For constructing the ΔhookA mutant, the following six oligos were used to make the ΔhookA construct with the selective marker pyrG from Aspergillus fumigatus, AfpyrG, in the middle of the linear construct (Szewczyk et al., 2007): HKd5, 5′-TACACCGGACTCTGTGATAG-3′; HKNN3, 5′-GGTACGCTCCGACTCCAT-3′; HKPG5, 5′-ATGGAGTCGGAGCGTACCTGCTCTTCACCCTCTTCGCG-3′; HKPG3, 5′-GGAGCTATCATGGACCAGCCTGTCTGAGAGGAGGCACTG-3′; HKDD5, 5′-GCTGGTCCATGATAGCTCC-3′; and HKd3, 5′-CCTGACAGTGGTCGTACTAA-3′. The deletion construct was obtained via fusion PCRs and used to transform the RQ54 strain containing mCherry-RabA, ΔnkuA, and pyrG89.
For constructing the HookA-GFP fusion, we used the following six oligos to amplify genomic DNA and the GFP-AfpyrG fusion from the plasmid pFNO3 (deposited in the Fungal Genetics Stock Center by S. Osmani, The Ohio State University, Columbus, OH; Yang et al., 2004; McCluskey et al., 2010): HKORFF, 5′-AAACGACGAGCAGCAGCTG-3′; HKORFR, 5′-TGGACCAGCAACGGCACTTC-3′; HKFusF, 5′-AAGAAGTGCCGTTGCTGGTCCAGGAGCTGGTGCAGGCGCTGGAG-3′; HKFusR, 5′-AGGGGCTGCAGGAGCTATCACTGTCTGAGAGGAGGCACTGATG-3′; HKUTRF, 5′-TGATAGCTCCTGCAGCCCC-3′; and HKUTRR, 5′-TAACTGTTGAAGGAGATCC-3′. The fusion PCRs generated the HookA-GFP-AfpyrG fragment that we used to transform into RQ54.
For constructing the ΔC-HookA–GFP fusion that lacks 37 aa of the C terminus, oligos HKORFF, HKUTRR, HKFusF, and ΔC37R (5′-TGGACCAGCAACGGCACTTCTTTGTGAACTCATGAGGGCGAG-3′) were used for fusion PCR, and the product was transformed into RQ54, which resulted in the deletion of 37 aa at the C terminus of HookA (AWYELQSKLHSTNNVPTSRYRHGSAGLVDAQKSWLAR). For constructing the ΔC1-HookA–GFP fusion that lacks the 13 aa near the end of the last coiled-coil domain near the C terminus, oligos HKORFF, HKUTRR, HKmbf (5′-GTGTGCTAACAAATACGTCTTCCAGCGCGCCCTCATGAGTTCAGCCTGGTACGAGC-3′), and HKmbr (5′-GCGCTGGAAGACGTATTTGTTAGCACAC-3′) were used for fusion PCR using the Hook-GFP-AfpyrG DNA as a template. This fragment was transformed into RQ54, which resulted in the deletion of 13 aa close to the C terminus of HookA (ALEKQLDALTREL).
For constructing the ΔN-HookA–GFP fusion that lacks the microtubule binding site, oligos 41U, 41D, HKdMTf (5′-ATGGAGTCGGAGCGTACCGTCATTCAGAAACTTGATAGTCCG-3′), and HKdMTr (5′-CTGAATGACGGTACGCTCCGACTCC-3′) were used for fusion PCR to obtain the N-terminal HookA fragment missing the sequence encoding 127 aa at the N terminus of HookA, from aa 8 to aa 134, SHSEALLAWVNSFDLVGEPKQIAELSDGRIIWDILHDIDPERFPDVTDPKKSNLENLVTIHGRLQYNILDLRKSEGWPRGLDPEPNLIEFAENNSARDAEKLLKLVFFAATITAKGNTASYETYGDA. This fragment was cotransformed with the HooKA-GFP-AfpyrG, the ΔC-HookA–GFP-AfpyrG, or the ΔC1-HookA–GFP-AfpyrG fragment into RQ54.
For constructing the ΔN1-ΔC-HookA–GFP fusion that lacks the microtubule-binding domain and the first half of the first coiled-coil domain in addition to the ΔC deletion, oligos 41U, 41D, HKdMTCf (5′-ATGGAGTCGGAGCGTACCGTCGACCTCGACCAGAAGACCGAG-3′), and HKdMTCr (5′-GGTCGACGGTACGCTCC-3′) were used for fusion PCR to obtain the N-terminal HookA fragment missing the sequence encoding 226 aa at the N terminus of HookA, from aa 8 to aa 233, SHSEALLAWVNSFDLVGEPKQIAELSDGRIIWDILHDIDPERFPDVTDPKKSNLENLVTIHGRLQYNILDLRKSEGWPRGLDPEPNLIEFAENNSARDAEKLLKLVFFAATITAKGNTASYETYGDAIQKLDSPIQESLQDFLENVEEGQYELDDLARESRESQLVKTIEELKQENTVLREKYVKTEQRVLELEYAEENYKSELEFMKERIEVLKSGKGEFGFSKR. This fragment was cotransformed with the ΔC-HookA–GFP-AfpyrG fragment into RQ54.
For constructing the C-HookA–GFP fusion that contains only 64 aa of the C terminus, oligos 41U, HKCF (5′-ATGGAGTCGGAGCGTACCGTCCGCGCCCTCGAGAAACAGCTCGAT-3′), HKCR (5′-CGCGGACGGTACGCTCCGACTCC-3′), and HKUTRR were used for fusion PCR using the genomic DNA from the strain containing Hook-GFP-AfpyrG as a template. This fusion product also contains all the necessary sequence for HookA protein translation together with the coding sequence for the first 7 aa of HookA followed by the coding sequence for the 64 aa at the HookA C terminus, RALEKQLDALTRELALMSSAWYELQSKLHSTNNVPTSRYRHGSAGLVDAQKSWLARQRSAVAGP. This fragment was transformed into RQ54.
For constructing the strain containing the gpdA promoter–driven C-HookA–GFP (gpdA-C-HookA–GFP), oligos 41U, HKpr2 (5′-GGATATGTCCAAGTAATCGCTG-3′), gpdAF2 (5′-CAGCGATTACTTGGACATATCCGACTCGAGTACCATTTAATTCTAT-3′), gpdAR2 (5′-ACGGTACGCTCCGACTCCATTGTGATGTCTGCTCAAGC-3′), HKCF2 (5′-AGTCGGAGCGTACCGTCCGCGCCCTCGAGAAACA-3′), and HKUTRR were used for fusion PCR using the genomic DNA from the strain containing Hook-GFP-AfpyrG as a template. In this fragment, the ∼1.2-kb upstream sequence of the gpdA gene (before the start codon ATG) is inserted after the 1-kb upstream sequence of hookA, which allows homologous integration at the hookA locus. For constructing the strain containing the N-HookA–GFP, oligos 41U, HKUTRR, HK134F (5′-GAGTTACGAGACATACGGCGATGCCGGAGCTGGTGCAGGCGCTGGAG-3′), and HK134R (5′-GGCATCGCCGTATGTCTCGTAACTC-3′) were used for fusion PCR using the genomic DNA from the strain containing Hook-GFP-AfpyrG as a template.
Analyses of protein–protein interactions and interaction of dynein with early endosomes
The µMACS GFP-tagged protein isolation kit (Miltenyi Biotec) was used to determine whether GFP-tagged HookA pulls down dynein–dynactin. This was performed as described in Qiu et al. (2013). Strains were grown overnight in liquid rich medium YG or minimal medium containing 0.4% (wt/vol) fructose. About 0.4 g hyphal mass was harvested from overnight culture for each sample, and cell extracts were prepared using a lysis buffer containing 50 mM Tris-HCl, pH 8.0, and 10 µg/ml of a protease inhibitor cocktail (Sigma-Aldrich). Cell extract was centrifuged at 16,000 g for 30 min at 4°C, and the supernatant was used for the pull-down experiment. In some experiments, the supernatant was further centrifuged at 100,000 g for 40 min, and the supernatant was used for further experiments. To pull down GFP-tagged protein, 25 µl anti-GFP MicroBeads was added into the cell extracts for each sample and incubated at 4°C for 30 min. The MicroBeads/cell extracts mixture was then applied to the µColumn followed by gentle wash with the lysis buffer and the wash buffer 2 provided in the kit (Miltenyi Biotec). Preheated (95°C) SDS-PAGE sample buffer was used as elution buffer.
For the biochemical analyses of dynein–early endosome or HookA–early endosome interactions, we used the strains containing GFP-labeled dynein HC or HookA and mCherry-RabA–labeled early endosomes. The protocol was the same as described in Qiu et al. (2013) except that the strains were grown overnight in liquid minimal medium containing 0.4% (wt/vol) fructose. The cell extract was centrifuged at 16,000 g for 30 min at 4°C, and the supernatant was used for the pull-down experiment. The rabbit anti-mCherry antibody used on the Western blots to detect mCherry-RabA was purchased from BioVision Research Products. Western analyses were performed using the AP system, and blots were developed using the AP color development reagents obtained from Bio-Rad Laboratories, Inc. Quantitation of the protein band intensity was performed using the IPLab software as described previously (Qiu et al., 2013). Specifically, an area containing the whole band was selected as a ROI, and the intensity sum within the ROI was measured. Then, the ROI box was dragged to the equivalent region of the negative control lane to take the background value, which was then subtracted from the intensity sum. The intensity ratio of the pulled down mCherry-RabA to GFP-HC or HookA-GFP was calculated. The ratios calculated from the wild-type samples were set as 1, and the relative ratios of the mutant were calculated and presented. For the analyses of HookA–dynein–dynactin interactions, we used the strains containing GFP-labeled wild-type or mutant HookA proteins for pull-down experiments. Rabbit polyclonal antibodies against A. nidulans dynein HC, intermediate chain, dynactin p150, and NudF/LIS1 were described previously (Xiang et al., 1995a,b; Zhang et al., 2008). The antigens for the anti-HC, –intermediate chain, -p150, and -NudF/LIS1 antibodies were a portion of the HC protein fused with LacZ (Xiang et al., 1995b), a portion of the intermediate chain protein fused with 6× histidine (Zhang et al., 2008), a portion of p150 protein fused with 6× histidine (Zhang et al., 2008), and the NudF protein fused with LacZ (Xiang et al., 1995a), respectively. A rabbit anti-GFP antibody from Takara Bio Inc. (polyclonal) was also used for Western analyses. The intensity ratio of the pulled down dynein HC, dynactin p150, or NudF/LIS1 to GFP-labeled HookA proteins was calculated.
Online supplemental material
Fig. S1 contains a sequence alignment of A. nidulans HookA with Drosophila Hook and the three human Hook proteins. Fig. S2 contains kymographs showing colocalization of HookA-GFP signals with mCherry-RabA–labeled early endosomes in a hyphal segment. Fig. S3 contains data of pull-down assays to show that the C terminus of HookA physically interacts with early endosomes. Fig. S4 shows that the amount of dynein pulled down with the ΔC-HookA–GFP is significantly lower than that pulled down with dynactin p150-GFP. Fig. S5 shows that the HookA–microtubule interaction is not readily detectable. Table S1 includes A. nidulans strains used in this study. Video 1 shows movements of early endosomes in a wild-type strain of A. nidulans. Video 2 shows early endosomes in the ΔhookA strain. Video 3 shows early endosomes in the ΔC-HookA strain. Video 4 shows HookA-GFP signals in a wild-type strain. Video 5 shows ΔC-HookA–GFP signals. Video 6 shows early endosomes in the ΔN-HookA strain.
Movements of early endosomes in a wild-type strain of A. nidulans. Early endosomes are labeled by the mCherry-RabA fusion protein that was initially introduced to A. nidulans by transformation of the DNA encoding the fusion. The mCherry-RabA signals undergo bidirectional movements in wild-type hyphae. An inverted fluorescence microscope (IX70; Olympus) was used for capturing images. 30 frames were taken with a 0.1-s exposure time and a 0.3-s interval between frames. Binning is 1 × 1. The video speed has been increased fivefold.
Movements of early endosomes in a wild-type strain of A. nidulans. Early endosomes are labeled by the mCherry-RabA fusion protein that was initially introduced to A. nidulans by transformation of the DNA encoding the fusion. The mCherry-RabA signals undergo bidirectional movements in wild-type hyphae. An inverted fluorescence microscope (IX70; Olympus) was used for capturing images. 30 frames were taken with a 0.1-s exposure time and a 0.3-s interval between frames. Binning is 1 × 1. The video speed has been increased fivefold.
Early endosomes in the ΔhookA strain. Early endosomes labeled by mCherry-RabA are largely accumulated at the hyphal tip. Directed movements of early endosomes away from the hyphal tip are not observed. The ΔhookA construct was transformed into a wild-type strain carrying the mCherry-RabA marker. An inverted fluorescence microscope (IX70; Olympus) was used for capturing images. 30 frames were taken with a 0.1-s exposure time and a 0.3-s interval between frames. Binning is 1 × 1. The video speed has been increased fivefold.
Early endosomes in the ΔhookA strain. Early endosomes labeled by mCherry-RabA are largely accumulated at the hyphal tip. Directed movements of early endosomes away from the hyphal tip are not observed. The ΔhookA construct was transformed into a wild-type strain carrying the mCherry-RabA marker. An inverted fluorescence microscope (IX70; Olympus) was used for capturing images. 30 frames were taken with a 0.1-s exposure time and a 0.3-s interval between frames. Binning is 1 × 1. The video speed has been increased fivefold.
Early endosomes in the ΔC-HookA strain. Early endosomes labeled by mCherry-RabA are largely accumulated at the hyphal tip. Directed movements of early endosomes away from the hyphal tip are not observed. The construct containing the ΔC-HookA mutation was transformed into a wild-type strain carrying the mCherry-RabA marker. An inverted fluorescence microscope (IX70; Olympus) was used for capturing images. 30 frames were taken with a 0.1-s exposure time and a 0.3-s interval between frames. Binning is 1 × 1. The video speed has been increased fivefold.
Early endosomes in the ΔC-HookA strain. Early endosomes labeled by mCherry-RabA are largely accumulated at the hyphal tip. Directed movements of early endosomes away from the hyphal tip are not observed. The construct containing the ΔC-HookA mutation was transformed into a wild-type strain carrying the mCherry-RabA marker. An inverted fluorescence microscope (IX70; Olympus) was used for capturing images. 30 frames were taken with a 0.1-s exposure time and a 0.3-s interval between frames. Binning is 1 × 1. The video speed has been increased fivefold.
Early endosomes in the ΔN-HookA strain. Early endosomes labeled by mCherry-RabA are largely accumulated at the hyphal tip, but a few can be seen to move away from the hyphal tip. The construct containing the ΔN-HookA mutation was transformed into a wild-type strain carrying the mCherry-RabA marker. An inverted fluorescence microscope (IX70; Olympus) was used for capturing images. 30 frames were taken with a 0.1-s exposure time and a 0.3-s interval between frames. Binning is 1 × 1. The video speed has been increased fivefold.
Early endosomes in the ΔN-HookA strain. Early endosomes labeled by mCherry-RabA are largely accumulated at the hyphal tip, but a few can be seen to move away from the hyphal tip. The construct containing the ΔN-HookA mutation was transformed into a wild-type strain carrying the mCherry-RabA marker. An inverted fluorescence microscope (IX70; Olympus) was used for capturing images. 30 frames were taken with a 0.1-s exposure time and a 0.3-s interval between frames. Binning is 1 × 1. The video speed has been increased fivefold.
Acknowledgments
We thank Drs. Xuanli Yao, Rachel Cox, Bo Liu, Xiangfeng Wang, and Tian Jin for helpful discussions and Henry Zhou for technical help at the beginning of this work. We thank Drs. Samara L. Reck-Peterson, Kaeling Tan, Michael Hynes, Berl Oakley, and Reinhard Fischer for A. nidulans strains, the Fungal Genetic Stock Center for the pFNO3 plasmid, and Steve Osmani for depositing it. We also thank Gero Steinberg for communicating unpublished results on the U. maydis Hook homologue. Services for primer synthesis and DNA sequencing were provided by the Biomedical Instrumentation Center of the Uniformed Services University.
This work was funded by the National Institutes of Health grant RO1 GM097580 (to X. Xiang), a Uniformed Services University intramural grant (to X. Xiang), the Biotechnology and Biological Sciences Research Council grant BB/F01189X/1 (to H.N. Arst and Elaine Bignell), the Wellcome Trust grant 084660/Z/08/Z (to H.N. Arst and Joan Tilburn), the Spanish Government grant BIO2012-30695 (to M.A. Peñalva), and the Comunidad de Madrid grant S2012/BMD2414 (to M.A. Peñalva).
The authors declare no competing financial interests.
References
Author notes
J. Zhang and R. Qiu contributed equally to this paper.