Secretory granules, such as neuronal dense core vesicles, are specialized for storing cargo at high concentration and releasing it via regulated exocytosis in response to extracellular stimuli. Here, we used expression profiling to identify new components of the machinery for sorting proteins into mucocysts, secretory granule-like vesicles in the ciliate Tetrahymena thermophila. We show that assembly of mucocysts depends on proteins classically associated with lysosome biogenesis. In particular, the delivery of nonaggregated, but not aggregated, cargo proteins requires classical receptors of the sortilin/VPS10 family, which indicates that dual mechanisms are involved in sorting to this secretory compartment. In addition, sortilins are required for delivery of a key protease involved in T. thermophila mucocyst maturation. Our results suggest potential similarities in the formation of regulated secretory organelles between even very distantly related eukaryotes.
Introduction
Across a broad swath of eukaryotic lineages, cells possess organelles that undergo rapid fusion with the plasma membrane in response to extracellular stimuli, termed regulated exocytosis. In some cases, regulated exocytosis involves the retargeting of organelles such as lysosomes or endosomes, but the best-studied regulated exocytic organelles are dedicated secretory reservoirs called secretory granules. Secretory granules, which in animal cells include several classes of dense-core vesicles found in endocrine, neuronal, and other tissues, are critical to both development and behavior, as they underlie extracellular signaling based on the release of peptide hormones like insulin and growth factors like bone-derived neurotrophic factor, as well as neuropeptides (Meldolesi et al., 2004).
A key feature of secretory granules is the presence of a macroscopic core consisting of condensed cargo molecules, which facilitates storage at high concentration. Pioneering work on granule biogenesis in mammalian cells, drawing largely on cell biological and biochemical approaches, established that aggregation also plays a key role in protein sorting in a multistep pathway beginning at the TGN and continuing as a maturation process during which granule cargo is refined, in part by withdrawal of missorted extraneous proteins (Tooze and Huttner, 1990; Chanat and Huttner, 1991; Kuliawat and Arvan, 1992; Arvan et al., 2002; Kim et al., 2006; Morvan and Tooze, 2008). A key implication of these studies was that the positive sorting of granule cargo was independent of classical receptors or of the cytoplasmic coat machinery that are critical to many membrane trafficking pathways.
Recently, a variety of genetic approaches, both in invertebrates and in mammals, have revealed that additional mechanisms may be involved in granule content sorting. One insight, drawn from analysis in Caenorhabditis elegans, was that sorting of aggregated granule core proteins was resilient to mutations that dramatically inhibited the sorting of more soluble dense core vesicle (DCV) cargo, including physiologically important peptides (Edwards et al., 2009). The sufficiency of aggregation-based sorting was directly challenged by an RNAi-based screen in Drosophila melanogaster S2 cells, which revealed that at least two different classes of membrane proteins failed to be efficiently sorted to granules upon knockdown of the AP-3 adaptor complex, a finding that extended to mammalian cells, and which was likely to involve AP-3 function in positive sorting at the level of the TGN (Asensio et al., 2010). Although the AP-3 adaptor had been linked with granule formation in a previous mouse genetics study, neither the mechanism of action nor the AP-3 binding partners have yet been identified for granule formation (Grabner et al., 2006). A key issue, from both mechanistic and evolutionary perspectives, is whether AP-3–based sorting to granules depends on determinants that are identical or homologous to the proteins involved in sorting to lysosome-related organelles, a comparatively well-characterized AP-3–dependent pathway (Braulke and Bonifacino, 2009). Intriguingly, there is some evidence that sorting of bone-derived neurotrophic factor (BDNF) to neuronal DCVs depends on sortilin/VPS10 proteins, a family of receptors that are classically associated with AP-3–dependent trafficking to lysosome-related organelles (Chen et al., 2005). However, whether BDNF sorting also involves AP-3 and other lysosome-related organelle–associated machinery has not been reported. Sortilin-family receptors are found very widely through eukaryotes, although they have been selectively lost in invertebrate lineages and therefore cannot be investigated using the C. elegans or D. melanogaster models (Koumandou et al., 2011).
Whether evolutionarily related secretory granules exist in nonanimal lineages, which constitute the majority of eukaryotic diversity, is currently difficult to assess, given the lack of molecular studies in nonanimal systems. One exception may be ciliates, single-celled protists that, though very distantly related from animals, also possess specialized secretory vesicles that undergo regulated exocytosis (Rosati and Modeo, 2003). These vesicles, which are functionally analogous to secretory granules, have been studied at the molecular level in two species: Tetrahymena thermophila (where the granules are termed mucocysts) and Paramecium tetraurelia (where they are known as trichocysts). The ciliate granules share few identified molecular components with those in animals but share a striking number of biochemical and cell biological features, including extensive processing of proproteins to generate the biologically active cargo peptides (Turkewitz, 2004). Like many granule proteins in animals, the most abundant ciliate granule cargo proteins undergo self-aggregation that is important for their sorting both at the TGN and during maturation; the latter, as in animal granules, depends on proteolytic processing of cargo proproteins (Collins and Wilhelm, 1981; Adoutte, 1988; Turkewitz, 2004). In T. thermophila, proteins in the Grl (Granule lattice) family form obligate hetero-oligomers in the endoplasmic reticulum (Cowan et al., 2005) and then much larger aggregates while en route through the secretory pathway (Rahaman et al., 2009). The aggregates are then reorganized during mucocyst maturation to form a crystalline dense core, a process involving extensive proteolytic processing of core proteins (Verbsky and Turkewitz, 1998; Cowan et al., 2005). Granule proteins in P. tetraurelia, called tmps, as well as in other ciliates that have been studied, show similar behavior (Madeddu et al., 1995), including forming large aggregates during sorting (Garreau de Loubresse, 1993; Peck et al., 1993).
More recently, a second family of T. thermophila mucocyst proteins came to light, and two members of the 13-member gene family, named Grt1p and Igr1p (for Granule tip, and Induced upon granule regeneration, respectively), have been characterized (Haddad et al., 2002; Bowman et al., 2005a,b). Neither undergoes aggregation or proteolytic processing, which suggests that their sorting depends on different mechanisms from those of the Grl proteins. To investigate the mechanisms involved in mucocyst localization of Grt1p, we have now used expression profiling to identify the sorting machinery, taking advantage of both the striking coregulation of mucocyst proteins in T. thermophila and an extensive publicly accessible expression database (Rahaman et al., 2009; Xiong et al., 2011). Our results provide strong evidence that aggregated and nonaggregated proteins depend on distinct mechanisms for delivery to mucocysts, and demonstrate a definitive role for sortilin receptors in this pathway.
Results
Expression profiling reveals co-regulation of mucocyst cargo proteins, sortilin-family receptors, and other proteins predicted to function in protein trafficking
To investigate the sorting of Grt proteins to mucocysts, we followed up on previous observations that genes encoding the lumenal and membrane proteins in T. thermophila mucocysts are coregulated over a wide range of physiological states. These include growth, starvation, and conjugation, as well as during synchronous mucocyst synthesis that can be triggered by stimulating cells to undergo complete exocytosis (Haddad and Turkewitz, 1997; Rahaman et al., 2009). To extend these findings, we used tools available through the Tetrahymena Gene Expression Database (Xiong et al., 2011; subsequently reorganized as the Tetrahymena Functional Genomics Database) to ask whether additional coregulated genes might encode the machinery required for mucocyst synthesis, including proteins involved in the sorting of Grt cargo. Surprisingly, this informatics-based screen advanced the sortilin/Vps10 receptors as candidate actors in mucocyst biogenesis.
Sortilins are Type I transmembrane proteins first characterized in Saccharomyces cerevisiae as the product of the VPS10 gene, which functions as a sorting receptor for vacuolar hydrolases (Marcusson et al., 1994). Although the sortilin gene family has undergone broad expansions throughout the major eukaryotic supergroups, the family has been lost in several lineages that include Arabidopsis thaliana, D. melanogaster, and C. elegans. (Fig. 1 and Fig. S1). Sortilins in vertebrates are receptors for sorting to lysosome-related organelles, in addition to other functions (Hermey, 2009). In T. thermophila, all four of the sortilin genes have similar expression profiles, which are also strikingly similar to those of known mucocyst-associated genes (Fig. 2 A and Fig. S2 A). The four T. thermophila sortilins, called SOR1–SOR4, have diverged significantly from one another as judged by amino acid sequence (∼30% identity within the VPS10 domains; ∼12% identity within the cytosolic tails). The four genes fall into two clades. SOR1 and -3 belong to a clade including members from non-ciliates, whereas SOR2 and -4 belong to a ciliate-restricted clade, and therefore are likely to have arisen via gene duplications that occurred within the ciliate lineage (Fig. 1 and Fig. 2 B).
The T. thermophila sortilins fall into two major groups. A maximum likelihood phylogeny of the ciliate VPS10 domains shown in Fig. 2 B, together with the most highly related homologues (as judged by BLAST scores) present in a variety of organisms from the other major eukaryotic lineages. VPS10 domain–containing genes appear to have been entirely lost in numerous organisms including A. thaliana and D. melanogaster. In some fungi, VPS10 domains are present as tandem repeats, depicted as h1 and h2. Species are abbreviated as follows: Aspergillus nidulans (An), Coccomyxa subellipsoidea (Cs), Daphnia pulex (Dap), Dictyostelium discoideum (Dd), Dictyostelium fasciculatum (Df), Dictyostelium purpureum (Dp), Homo sapiens (Hs), Ichthyophthirius multifiliis (Im), M. pusilla (Mp), Micromonas sp. RCC299 (Mr), Mus musculus (Mm), Naegleria gruberi (Ng), Naumovozyma castellii (Nac), Ostreococcus lucimarinus (Ol), Ostreococcus tauri (Ot), Paracoccidioides brasiliensis (Pab), P. tetraurelia (Pt), Polysphondylium pallidum (Pp), Punctularia strigosozonata (Ps), S. cerevisiae (Sc), T. thermophila (Tt), Trichoplax adhaerens (Tra), Xenopus (Silurana) tropicalis (Xt), Yarrowia lipolytica (Yl). See also Table S1 for a list of accession numbers for the sequences used to assemble this phylogeny.
The T. thermophila sortilins fall into two major groups. A maximum likelihood phylogeny of the ciliate VPS10 domains shown in Fig. 2 B, together with the most highly related homologues (as judged by BLAST scores) present in a variety of organisms from the other major eukaryotic lineages. VPS10 domain–containing genes appear to have been entirely lost in numerous organisms including A. thaliana and D. melanogaster. In some fungi, VPS10 domains are present as tandem repeats, depicted as h1 and h2. Species are abbreviated as follows: Aspergillus nidulans (An), Coccomyxa subellipsoidea (Cs), Daphnia pulex (Dap), Dictyostelium discoideum (Dd), Dictyostelium fasciculatum (Df), Dictyostelium purpureum (Dp), Homo sapiens (Hs), Ichthyophthirius multifiliis (Im), M. pusilla (Mp), Micromonas sp. RCC299 (Mr), Mus musculus (Mm), Naegleria gruberi (Ng), Naumovozyma castellii (Nac), Ostreococcus lucimarinus (Ol), Ostreococcus tauri (Ot), Paracoccidioides brasiliensis (Pab), P. tetraurelia (Pt), Polysphondylium pallidum (Pp), Punctularia strigosozonata (Ps), S. cerevisiae (Sc), T. thermophila (Tt), Trichoplax adhaerens (Tra), Xenopus (Silurana) tropicalis (Xt), Yarrowia lipolytica (Yl). See also Table S1 for a list of accession numbers for the sequences used to assemble this phylogeny.
Expression profiling suggests a role for sortilin-family receptors in mucocyst biogenesis in T. thermophila. (A) Sortilins are coregulated with genes encoding mucocyst contents in T. thermophila. The expression profiles of the four T. thermophila sortilins (SOR1–4; right), are similar to those of genes (GRL1, GRL3, GRT1, and IGR1; left) encoding mucocyst cargo proteins. Expression profiles are derived from the Tetrahymena Functional Genomics Database, with each profile normalized to that gene’s maximum expression level. Points on the x axis correspond to successive time points and represent growing, starved, and mating cultures, including three different culture densities (low [Ll], medium [Lm], and high [Lh]), 7 samples taken during 24 h of starvation, and 10 samples subsequently taken during 18 h after conjugation. (B) Expansion of the sortilin family in ciliates. The maximum likelihood tree illustrates a phylogeny of VPS10 domain–containing receptors (sortilins) in alveolates, the taxonomic group consisting of ciliates, apicomplexans, and dinoflagellates. Two of the T. thermophila sortilins, marked by black circles, cluster with the sortilins from other alveolates. In contrast, T. thermophila SOR2 and SOR4, marked by maroon diamonds, belong to an expansion of sortilins restricted to ciliates. Species are abbreviated as follows: Babesia microti (Bm), Cryptosporidium hominis (Ch), Cryptosporidium muris (Cm), I. multifiliis (Im), Neospora caninum (Nc), P. tetraurelia (Pt), Perkinsus marinus (Pm), Plasmodium berghei (Pb), Plasmodium cynomolgi (Pc), Plasmodium falciparum (Pf), Plasmodium knowlesi (Pk), Plasmodium vivax (Pv), Plasmodium yoelii yoelii (Py), T. thermophila (Tt), Theileria annulata (Ta), Theileria orientalis (To), Theileria parva (Tp), T. gondii (Tg). See Tables S1 and S2 for a list of accession numbers for all of the sequences. (C) Verification of the nonessential sortilin knockouts. cDNA was prepared from wild-type, Δsor1, Δsor2, and Δsor4 cells, and the SOR1, SOR2, and SOR4 sequences were PCR amplified using gene-specific primers. As shown in this 1% ethidium bromide–stained agarose gel, each of the gene knockout lines lacks the amplified product corresponding to the targeted gene, but shows wild-type levels of the other transcripts that serve as loading controls (see also Fig. S2 E). The lanes shown are all part of a single gel, but their order has been rearranged for this figure.
Expression profiling suggests a role for sortilin-family receptors in mucocyst biogenesis in T. thermophila. (A) Sortilins are coregulated with genes encoding mucocyst contents in T. thermophila. The expression profiles of the four T. thermophila sortilins (SOR1–4; right), are similar to those of genes (GRL1, GRL3, GRT1, and IGR1; left) encoding mucocyst cargo proteins. Expression profiles are derived from the Tetrahymena Functional Genomics Database, with each profile normalized to that gene’s maximum expression level. Points on the x axis correspond to successive time points and represent growing, starved, and mating cultures, including three different culture densities (low [Ll], medium [Lm], and high [Lh]), 7 samples taken during 24 h of starvation, and 10 samples subsequently taken during 18 h after conjugation. (B) Expansion of the sortilin family in ciliates. The maximum likelihood tree illustrates a phylogeny of VPS10 domain–containing receptors (sortilins) in alveolates, the taxonomic group consisting of ciliates, apicomplexans, and dinoflagellates. Two of the T. thermophila sortilins, marked by black circles, cluster with the sortilins from other alveolates. In contrast, T. thermophila SOR2 and SOR4, marked by maroon diamonds, belong to an expansion of sortilins restricted to ciliates. Species are abbreviated as follows: Babesia microti (Bm), Cryptosporidium hominis (Ch), Cryptosporidium muris (Cm), I. multifiliis (Im), Neospora caninum (Nc), P. tetraurelia (Pt), Perkinsus marinus (Pm), Plasmodium berghei (Pb), Plasmodium cynomolgi (Pc), Plasmodium falciparum (Pf), Plasmodium knowlesi (Pk), Plasmodium vivax (Pv), Plasmodium yoelii yoelii (Py), T. thermophila (Tt), Theileria annulata (Ta), Theileria orientalis (To), Theileria parva (Tp), T. gondii (Tg). See Tables S1 and S2 for a list of accession numbers for all of the sequences. (C) Verification of the nonessential sortilin knockouts. cDNA was prepared from wild-type, Δsor1, Δsor2, and Δsor4 cells, and the SOR1, SOR2, and SOR4 sequences were PCR amplified using gene-specific primers. As shown in this 1% ethidium bromide–stained agarose gel, each of the gene knockout lines lacks the amplified product corresponding to the targeted gene, but shows wild-type levels of the other transcripts that serve as loading controls (see also Fig. S2 E). The lanes shown are all part of a single gel, but their order has been rearranged for this figure.
Interestingly, in opisthokonts, including S. cerevisiae and mammals, sortilin-dependent trafficking to lysosome-related organelles depends on the heterotetrameric AP-3 adapter (Odorizzi et al., 1998), and we also identified the T. thermophila AP-3 adapter as a top hit with the sortilins for mucocyst-coregulated genes (Fig. S2, A and B). T. thermophila also expresses AP-1, AP-2, and AP-4 adaptor complexes, but none of these is coregulated with mucocyst-associated genes (Fig. S2 C; Elde et al., 2005; Nusblat et al., 2012).
The two ciliate-restricted sortilins, SOR2 and SOR4, are nonessential genes that are required for secretion from mucocysts
To test whether any of the sortilin genes are required for mucocyst function, we targeted each of them by homologous recombination, using standard approaches that result in disruption of all macronuclear (expressed) copies for nonessential genes (Fig. S2 D; Cassidy-Hanley et al., 1997). We obtained complete knockouts for SOR1, -2, and -4 (Fig. 2 C and Fig. S2 E); our inability to obtain complete knockouts of SOR3, and the slow growth of cells after even partial knockdown, suggested that SOR3 is an essential gene. Preliminary analysis of the Δsor1, Δsor2, and Δsor4 knockout lines indicated that SOR2 and SOR4, but not SOR1, were essential for mucocyst formation and/or exocytosis (Fig. S4 and later in this paper). We focused primarily on Δsor4 because it showed stronger deficiencies in secretion. Importantly, Δsor4 cells grew at a similar rate to wild type, which strongly suggests that the absence of SOR4 does not lead to a general disruption of membrane traffic or compromise the function of any essential organelle (Fig. S2 F). Mucocysts themselves are dispensable for normal growth of T. thermophila under laboratory conditions (Orias et al., 1983; Melia et al., 1998).
Sor4p is required for sorting of Grt1p to mucocysts
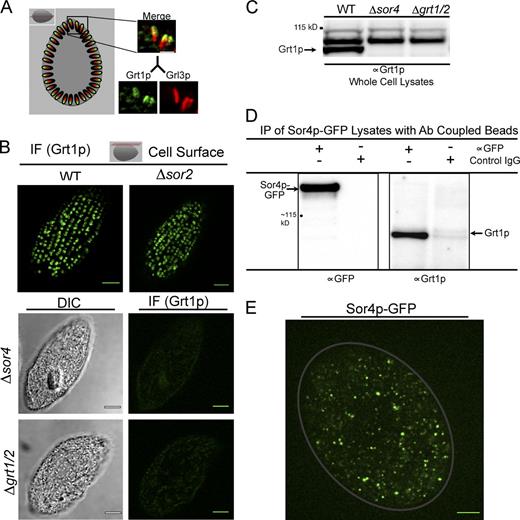
To ask whether the SOR4 gene product, Sor4p, played a role in sorting of mucocyst cargo proteins, we immunolocalized members of the Grt and Grl families (Grt1p and Grl3p, respectively) using previously characterized monoclonal antibodies. In wild-type cells, Grt1p localizes in a polarized fashion to the docked end of mucocysts (Bowman et al., 2005a; Fig. 3 A). Remarkably, we found that Δsor4 cells were completely defective in accumulation of Grt1p in mucocysts, as judged by indirect immunofluorescence (Fig. 3 B), and confirmed by Western blotting of whole cell lysates using a polyclonal antibody (Fig. 3 C and Fig. S3 A). The Δsor4 cells had no defect in Grt1p synthesis per se, as Grt1p was readily detected in cell culture medium. Because sortilins function as ligand-binding receptors, these results suggested that Sor4p acts as a sorting receptor for Grt1p. Consistent with this idea, we could immunoprecipitate Grt1p using an antibody against GFP in cells that were expressing Sor4p-GFP at the endogenous SOR4 locus (Fig. 3 D). Importantly, the GFP fusion protein is functional, as cells expressing Sor4p-GFP in lieu of the wild-type protein were fully exocytosis competent; i.e., did not manifest any SOR4 deficiency (Fig. S3 B). The robust interaction between Sor4p and Grt1p indicated by coprecipitation suggests that the interaction between these proteins is likely to be direct. Moreover, the SOR4-dependent missorting of Grt1p was specific, as disruption of the related T. thermophila paralogue, SOR2, did not produce any apparent defect in the accumulation of Grt1p (Fig. 3 B).
Sortilin 4 is required for the sorting of Grt1p, a member of the Granule tip family of mucocyst cargo proteins. (A) Grt1p localizes to a subdomain of mucocysts, whereas Grl3p is found through the mucocyst core. Grl3p and Grt1p in wild-type cells were simultaneously visualized using mAbs (5E9 and 4D11, respectively) directly conjugated to two different fluorophores. Visualization along the long mucocyst axis (demonstrated best in a cross section of the cell, illustrated by the red plane in the diagram shown at the top for reference) demonstrates that Grt1p is concentrated at the pole where docking occurs (right). Bar, 1 µm. (B) Grt1p is mistargeted in Δsor4 cells. Immunolocalization of Grt1p in wild-type cells (top left) shows that Grt1p accumulates in the expected array of docked mucocysts at the surface (illustrated by the red plane in the diagram at the top), and the same pattern is seen in Δsor2 cells (top right). Grt1p was visualized using mAb 4D11. In contrast, there is only background staining of Grt1p in Δsor4 cells, comparable to the signal in Δgrt1/Δgrt2 cells that lack the mAb target entirely (bottom). Note that docked mucocysts are still present in the Δsor4 cells (Fig. 5, A and B). Images of Δsor4 and Δgrt1/Δgrt2 cells were auto-adjusted to show the cell outlines. Bars, 5 µm. (C) Grt1p is absent from Δsor4 cells. Western blotting of whole cell lysates, using polyclonal anti-Grt1p antiserum, confirms the defect in Grt1p accumulation in Δsor4 relative to wild type. An uncropped version of this Western blot is shown in Fig. S3 A. (D) Biochemical interaction between Sor4p and Grt1p: coprecipitation of Sor4p and Grt1p. Sor4p was immunoprecipitated using anti-GFP antiserum from lysates of cells that express Sor4p-GFP from the endogenous SOR4 locus and that were actively synthesizing new mucocysts. Immunoprecipitated samples were analyzed by Western blotting with anti-GFP antiserum (left), confirming that full-length Sor4p-GFP is expressed, and with anti-Grt1p antiserum (right) to show coprecipitation of Grt1p. (E) Sor4p-GFP localizes to mobile cytoplasmic vesicles but not to docked mucocysts. A frame from Video 1 is shown, in which Sor4p-GFP was tracked in immobilized live cells. Sor4p-GFP is present in mobile cytoplasmic puncta and not present in docked mucocysts. The gray line traces the approximate outline of the cell. Bar, 5 µm.
Sortilin 4 is required for the sorting of Grt1p, a member of the Granule tip family of mucocyst cargo proteins. (A) Grt1p localizes to a subdomain of mucocysts, whereas Grl3p is found through the mucocyst core. Grl3p and Grt1p in wild-type cells were simultaneously visualized using mAbs (5E9 and 4D11, respectively) directly conjugated to two different fluorophores. Visualization along the long mucocyst axis (demonstrated best in a cross section of the cell, illustrated by the red plane in the diagram shown at the top for reference) demonstrates that Grt1p is concentrated at the pole where docking occurs (right). Bar, 1 µm. (B) Grt1p is mistargeted in Δsor4 cells. Immunolocalization of Grt1p in wild-type cells (top left) shows that Grt1p accumulates in the expected array of docked mucocysts at the surface (illustrated by the red plane in the diagram at the top), and the same pattern is seen in Δsor2 cells (top right). Grt1p was visualized using mAb 4D11. In contrast, there is only background staining of Grt1p in Δsor4 cells, comparable to the signal in Δgrt1/Δgrt2 cells that lack the mAb target entirely (bottom). Note that docked mucocysts are still present in the Δsor4 cells (Fig. 5, A and B). Images of Δsor4 and Δgrt1/Δgrt2 cells were auto-adjusted to show the cell outlines. Bars, 5 µm. (C) Grt1p is absent from Δsor4 cells. Western blotting of whole cell lysates, using polyclonal anti-Grt1p antiserum, confirms the defect in Grt1p accumulation in Δsor4 relative to wild type. An uncropped version of this Western blot is shown in Fig. S3 A. (D) Biochemical interaction between Sor4p and Grt1p: coprecipitation of Sor4p and Grt1p. Sor4p was immunoprecipitated using anti-GFP antiserum from lysates of cells that express Sor4p-GFP from the endogenous SOR4 locus and that were actively synthesizing new mucocysts. Immunoprecipitated samples were analyzed by Western blotting with anti-GFP antiserum (left), confirming that full-length Sor4p-GFP is expressed, and with anti-Grt1p antiserum (right) to show coprecipitation of Grt1p. (E) Sor4p-GFP localizes to mobile cytoplasmic vesicles but not to docked mucocysts. A frame from Video 1 is shown, in which Sor4p-GFP was tracked in immobilized live cells. Sor4p-GFP is present in mobile cytoplasmic puncta and not present in docked mucocysts. The gray line traces the approximate outline of the cell. Bar, 5 µm.
Drawing from classical studies of the mannose-6-phosphate receptor in mammalian cells and what is known of VPS10 trafficking in S. cerevisiae, our working model is that Sor4p binds Grt1p at the level of the trans-Golgi and delivers it, potentially via an endosomal intermediate, to immature mucocysts. The interaction between Grt1p and Sor4p is not disrupted at low pH (Fig. S3 C), so ligand binding and dissociation are likely to be controlled by other factors. Consistent with this idea, the immature granules in another ciliate, P. tetraurelia, are not measurably acidic (Lumpert et al., 1992). However, our results could also be explained if Sor4p-Grt1p binding were required to retain Grt1p during mucocyst maturation. However, GFP-tagged Sor4p showed no localization to mucocysts but instead appeared in numerous cytoplasmic puncta (Fig. 3 E). These puncta, many of them highly mobile, are clearly distinguishable from stationary mature mucocysts docked at the plasma membrane (Video 1). The localization also indicates that Sor4p is not primarily associated with the Golgi, which has a distinct cortical distribution in these cells (Kurz and Tiedtke, 1993; Bright et al., 2010).
Sor4p localizes to mobile cytoplasmic puncta and not to mature mucocysts docked at the plasma membrane. Real-time live imaging of immobilized T. thermophila expressing Sor4p-GFP at the endogenous SOR4 locus as a complete gene replacement. Images were acquired with a 100-ms exposure (Marianas Yokogawa type spinning disk inverted confocal microscope; 3i) and displayed in the video at 10 frames/s. There is no detectable Sor4p-GFP in docked mucocysts.
Sor4p localizes to mobile cytoplasmic puncta and not to mature mucocysts docked at the plasma membrane. Real-time live imaging of immobilized T. thermophila expressing Sor4p-GFP at the endogenous SOR4 locus as a complete gene replacement. Images were acquired with a 100-ms exposure (Marianas Yokogawa type spinning disk inverted confocal microscope; 3i) and displayed in the video at 10 frames/s. There is no detectable Sor4p-GFP in docked mucocysts.
Sor4p is required for sorting of other Grt family members
Missorting of Grt1p would not by itself produce a dramatic mucocyst defect, as cells in which GRT1 was deleted together with the closely related GRT2 showed only a mild secretion phenotype (Rahaman et al., 2009). In contrast, Δsor4 cells showed a complete absence of mucocyst discharge on stimulation, as assessed by two different methods. In wild-type T. thermophila, the entire set of docked mucocysts undergo exocytosis after cells are exposed to either the polycation Alcian blue or dibucaine. Alcian blue binds to acidic mucocyst proteins as they exit, entrapping each wild-type cell in a blue-stained capsule (Tiedtke, 1976), but Δsor4 cells showed no trace of capsule formation (Fig. 4 A). The mucocyst contents released from dibucaine-treated wild-type cells form large pelletable aggregates (Satir, 1977), but these were entirely absent in dibucaine-treated Δsor4 cultures, and greatly reduced in Δsor2 cultures (Fig. 4 B). Both the capsule-formation defect and the absence of flocculent are comparable to mutants in mucocyst formation or exocytosis that have previously been characterized in this organism (Orias et al., 1983; Melia et al., 1998; Cowan et al., 2005). The disparity between the strong Δsor4 and mild Δgrt1 phenotypes indicated that Grt1p was unlikely to be the only ligand that depends on Sor4p for sorting to mucocysts. Because Grt1p belongs to a 13-member family of mucocyst content proteins, the other members were obvious candidates for Sor4p ligands.
Δsor4 cells are defective in regulated exocytosis and in sorting of a second Grt family protein. (A) A qualitative assay for mucocyst discharge. Individual T. thermophila cells, fixed and photographed after treatment with the secretagogue Alcian blue. The wild-type cell (left) is surrounded by a translucent capsule made up of the released contents of exocytosed mucocysts. In contrast, Δsor4 cells (right) never form visible capsules after stimulation. Images are differential interference contrast micrographs. The same wild-type control is shown again in Fig. S4 A. Bars, 5 µm. (B) A semiquantitative assay for mucocyst discharge. Identical numbers of wild-type and Δsor4 cells were stimulated with dibucaine, and immediately centrifuged. The wild-type culture produces a two-layer pellet, in which a thick layer of flocculent (below the broken line) resulting from mucocyst discharge sits atop of the packed cells (below the dotted line). Stimulated Δsor4 cultures, in contrast, produce no flocculent layer. Stimulated Δsor2 cultures generate an intermediate amount of the mucocyst-derived flocculent. (C) Δsor4 cells show defective sorting to mucocysts of a second Grt family protein, Igr1p. Igr1p-GFP, expressed from an inducible promoter, accumulates in docked mucocysts in wild-type cells (left), but is absent from the periphery of Δsor4, instead found in small highly mobile cytoplasmic puncta. Images are of GFP autofluorescence in live, immobilized cells. Bars, 5 µm.
Δsor4 cells are defective in regulated exocytosis and in sorting of a second Grt family protein. (A) A qualitative assay for mucocyst discharge. Individual T. thermophila cells, fixed and photographed after treatment with the secretagogue Alcian blue. The wild-type cell (left) is surrounded by a translucent capsule made up of the released contents of exocytosed mucocysts. In contrast, Δsor4 cells (right) never form visible capsules after stimulation. Images are differential interference contrast micrographs. The same wild-type control is shown again in Fig. S4 A. Bars, 5 µm. (B) A semiquantitative assay for mucocyst discharge. Identical numbers of wild-type and Δsor4 cells were stimulated with dibucaine, and immediately centrifuged. The wild-type culture produces a two-layer pellet, in which a thick layer of flocculent (below the broken line) resulting from mucocyst discharge sits atop of the packed cells (below the dotted line). Stimulated Δsor4 cultures, in contrast, produce no flocculent layer. Stimulated Δsor2 cultures generate an intermediate amount of the mucocyst-derived flocculent. (C) Δsor4 cells show defective sorting to mucocysts of a second Grt family protein, Igr1p. Igr1p-GFP, expressed from an inducible promoter, accumulates in docked mucocysts in wild-type cells (left), but is absent from the periphery of Δsor4, instead found in small highly mobile cytoplasmic puncta. Images are of GFP autofluorescence in live, immobilized cells. Bars, 5 µm.
We expressed a GFP-tagged copy of a second family member, Igr1p (Haddad et al., 2002; Bowman et al., 2005b), in wild-type and Δsor4 cells. Strikingly, Igr1p-GFP accumulated in mucocysts in wild-type but not Δsor4 cells (Fig. 4 C). These results indicate that Sor4p acts as a sorting receptor to mucocysts for multiple, and perhaps all, proteins in the Grt family. The mucocyst secretion defect in Δsor4 cells may therefore be related to missorting of multiple Grt-family proteins. In addition, as shown below, SOR4 function is also essential for processing, but not sorting, of the second major family of mucocyst proteins in T. thermophila, and disruption of processing is itself expected to inhibit mucocyst maturation and exocytosis (Verbsky and Turkewitz, 1998).
A GRL family protein is sorted to mucocysts independently of sortilin receptors
Previous work suggested that the major granule core proteins in ciliates are sorted via aggregation, and would consequently not be expected to depend on receptors. We therefore tested the prediction that delivery of Grl proteins to mucocysts in T. thermophila would be independent of SOR4 function. In both wild-type and Δsor4 cells, Grl3p immunofluorescence was concentrated in linear arrays of puncta at the cell surface, representing the cohort of docked mucocysts (Cowan et al., 2005) Similarly, Grl3p accumulated in docked mucocysts in Δsor2 cells (Fig. 5 A). Thus, Grl3p targeting does not depend on either of the sortilin receptors that are associated, based on their knockout phenotypes, with mucocyst function.
Grl protein sorting to mucocysts, though not subsequent proteolytic processing, is independent of SOR4. (A and B) Grl3p localizes to mature mucocysts in the absence of Sor4p. Localization of Grl3p, one of a family of proteins that assembles to form the mucocyst core, in wild-type, Δsor2, and Δsor4 cells. Grl3p was visualized by indirect immunofluorescence (IF) using mAb 5E9. At the cell surface (A; illustrated by the red plane in the diagram shown in the top right and in the differential interference contrast micrographs to the left), Grl3p can be seen localized to mucocysts in both wild-type (top) and Δsor4 (bottom) cells. When seen in cross section (B), the mucocysts of Δsor4 cells appear roughly spherical, in contrast to the elongated wild-type mucocysts (compare the bottom insets of the IF panel). Δsor2 mucocysts show an intermediate morphology. The cross-section image of the wild-type control is shown again in Fig. S4 C. Insets are enlarged from the indicated regions. Bars: (main panels) 5 µm; (inset) 0.5 µm. (C) Ultrastructure of Δsor4 mucocysts. Electron micrographs of docked mucocysts (labeled with asterisks) in wild-type (top) and Δsor2 and Δsor4 cells (bottom). The mucocyst cores in Δsor2 and Δsor4 cells do not contain the visible lattice that is characteristic of wild-type mucocyst cores. The same wild-type control is shown again in Fig. S4 D. Bars, 200 nM. (D) Grl proprotein processing is defective in Δsor4 cells. Whole cell lysates of wild-type and Δsor4 cells were separated by SDS-PAGE and Western blotted with an antibody against Grl1p, which undergoes proteolytic processing during mucocyst maturation. In wild-type cells, Grl1p accumulates primarily in its fully processed form. In Δsor4 cells, most Grl1p remains as an incompletely processed precursor. The unprocessed and processed forms of Grl1p are indicated by arrows.
Grl protein sorting to mucocysts, though not subsequent proteolytic processing, is independent of SOR4. (A and B) Grl3p localizes to mature mucocysts in the absence of Sor4p. Localization of Grl3p, one of a family of proteins that assembles to form the mucocyst core, in wild-type, Δsor2, and Δsor4 cells. Grl3p was visualized by indirect immunofluorescence (IF) using mAb 5E9. At the cell surface (A; illustrated by the red plane in the diagram shown in the top right and in the differential interference contrast micrographs to the left), Grl3p can be seen localized to mucocysts in both wild-type (top) and Δsor4 (bottom) cells. When seen in cross section (B), the mucocysts of Δsor4 cells appear roughly spherical, in contrast to the elongated wild-type mucocysts (compare the bottom insets of the IF panel). Δsor2 mucocysts show an intermediate morphology. The cross-section image of the wild-type control is shown again in Fig. S4 C. Insets are enlarged from the indicated regions. Bars: (main panels) 5 µm; (inset) 0.5 µm. (C) Ultrastructure of Δsor4 mucocysts. Electron micrographs of docked mucocysts (labeled with asterisks) in wild-type (top) and Δsor2 and Δsor4 cells (bottom). The mucocyst cores in Δsor2 and Δsor4 cells do not contain the visible lattice that is characteristic of wild-type mucocyst cores. The same wild-type control is shown again in Fig. S4 D. Bars, 200 nM. (D) Grl proprotein processing is defective in Δsor4 cells. Whole cell lysates of wild-type and Δsor4 cells were separated by SDS-PAGE and Western blotted with an antibody against Grl1p, which undergoes proteolytic processing during mucocyst maturation. In wild-type cells, Grl1p accumulates primarily in its fully processed form. In Δsor4 cells, most Grl1p remains as an incompletely processed precursor. The unprocessed and processed forms of Grl1p are indicated by arrows.
Though not required for sorting of Grl proteins, sortilin function is required for proteolytic maturation of Grl proproteins in mucocysts
The docked mucocysts in Δsor4 cells did not exhibit the elongated form of wild-type mucocysts. Instead, Δsor4 mucocysts seen in profile appeared spherical, as well as smaller than elongated wild-type mucocysts (Fig. 5 B). The aberrant mucocyst morphology was confirmed by electron microscopy, which also revealed that Δsor4 mucocysts lack any discernible crystalline structure within the dense core (Fig. 5 C). In wild-type mucocysts, formation of the dense core is strongly correlated with proteolytic maturation of Grl proproteins (Collins and Wilhelm, 1981; Turkewitz et al., 1991; Cowan et al., 2005), which suggests that the morphological defects in Δsor4 mucocysts might be caused by processing defects. Comparison of the lysates of wild-type and Δsor4 cells, by Western blotting with an antibody against Grl1p, indicated that proteolytic maturation was indeed defective, as much of Grl1p was still present in the unprocessed form (Fig. 5 D). These results raised the possibility that Sor4p, in addition to acting as sorting receptor for Grt family proteins, is also required to deliver one or more factors required for the proteolytic processing of pro-Grl cargo.
The simplest hypothesis was that Sor4p was required to deliver the proteases that process pro-Grl proteins. Those proteases have been studied indirectly but had not yet been identified in any ciliate. However, strong candidates for these enzymes emerged from the same expression screening approach, described earlier, which led us to focus on sortilins. Four of the putative processing enzymes are cathepsins, named CTH1–4, and disruption of the CTH3 gene in particular resulted in a near-complete failure to process Grl proproteins or synthesize mucocysts (unpublished data). To validate the inference that Cth3p functioned directly in mucocyst maturation, we transiently expressed the protein as a CFP fusion. Consistent with a role in pro-Grl processing, Cth3p-CFP showed significant localization to mucocysts (Fig. 6). This is likely due to specific targeting signals, as neither an unrelated cathepsin that has been studied in T. thermophila nor GFP attached to a signal sequence accumulate in mucocysts (Haddad et al., 2002; Jacobs et al., 2006). Importantly, the targeting of Cth3p-CFP to mucocysts, as measured by the colocalization of Cth3p-CFP with Grl3p, was reduced in Δsor4 cells (Fig. 6 and Fig. S5). These results support the hypothesis that Δsor4 cells are deficient in mucocyst delivery of both Grt family proteins and also one or more processing enzymes needed for Grl proprotein processing, with the latter defect sufficient to explain the aberrant mucocyst morphology shown in Fig. 5 C. Sor2p may also be involved in delivery to mucocysts of proteins required for pro-Grl processing, as Δsor2 cells showed comparable, though less severe, defects in Grl-based core formation (Fig. 5, A–C).
Cth3p, an aspartyl protease, is targeted to mucocysts in a Sor4p-dependent manner. Cth3p-CFP was inducibly expressed with 0.75 µg/ml CdCl2 for 2 h in wild-type and Δsor4 cells. Cth3p-CFP was localized in fixed, permeabilized cells using a polyclonal anti-GFP antibody, and endogenous Grl3p was immunolocalized using mAb 5E9. (A) Cth3p-CFP expressed in wild-type cells colocalizes with Grl3p in mucocysts at the cell periphery (top). In contrast, Cth3p-CFP expressed in Δsor4 cells shows reduced colocalization with Grl3p (middle). Bars, 5 µm. (B) Cth3p-CFP shows reduced colocalization with Grl3p in Δsor4 versus wild-type cells. Colocalization was quantified in 15 wild-type and Δsor4 cells, using the Manders’ correlation coefficient M2, and then a mean M2 value for each population was determined from the sample. Reduced colocalization was observed whether measuring all puncta (M2 values for wild type: mean = 0.615, SEM = 0.036; for Δsor4: M = 0.337, SEM = 0.049; P < 0.01 as determined by one-tailed t test) or the subset near the cell periphery, which is enriched in docked mucocysts (wild type: M = 0.731, SEM = 0.036; Δsor4: M = 0.373, SEM = 0.054; P < 0.01; right graph). Details of the image analysis are provided in Materials and methods, and a range of representative images is shown in Fig. S5.
Cth3p, an aspartyl protease, is targeted to mucocysts in a Sor4p-dependent manner. Cth3p-CFP was inducibly expressed with 0.75 µg/ml CdCl2 for 2 h in wild-type and Δsor4 cells. Cth3p-CFP was localized in fixed, permeabilized cells using a polyclonal anti-GFP antibody, and endogenous Grl3p was immunolocalized using mAb 5E9. (A) Cth3p-CFP expressed in wild-type cells colocalizes with Grl3p in mucocysts at the cell periphery (top). In contrast, Cth3p-CFP expressed in Δsor4 cells shows reduced colocalization with Grl3p (middle). Bars, 5 µm. (B) Cth3p-CFP shows reduced colocalization with Grl3p in Δsor4 versus wild-type cells. Colocalization was quantified in 15 wild-type and Δsor4 cells, using the Manders’ correlation coefficient M2, and then a mean M2 value for each population was determined from the sample. Reduced colocalization was observed whether measuring all puncta (M2 values for wild type: mean = 0.615, SEM = 0.036; for Δsor4: M = 0.337, SEM = 0.049; P < 0.01 as determined by one-tailed t test) or the subset near the cell periphery, which is enriched in docked mucocysts (wild type: M = 0.731, SEM = 0.036; Δsor4: M = 0.373, SEM = 0.054; P < 0.01; right graph). Details of the image analysis are provided in Materials and methods, and a range of representative images is shown in Fig. S5.
Discussion
Previous work on granules in ciliates demonstrated that the major core proteins, like those in animal cells, undergo dramatic aggregation that appears important for sorting at multiple steps. In this paper, we show that mucocyst formation in T. thermophila depends on dual sorting mechanisms. In contrast to the core-forming Grl proteins that undergo aggregation, a second group of abundant proteins is instead sorted via a receptor-mediated pathway. The relevant receptors are sortilins, members of a family associated with the biogenesis of lysosome-related organelles in diverse lineages (Marcusson et al., 1994; Braulke and Bonifacino, 2009; Canuel et al., 2009). T. thermophila belongs to the alveolate lineage, which also includes the Apicomplexan parasites in which one sortilin has recently been characterized. The single sortilin gene in Toxoplasma gondii is required for the formation of at least two classes of complex secretory organelles, which appear related to lysosomes, that function during host invasion (Ngô et al., 2004; Sloves et al., 2012). The sortilin family expanded in ciliates, and T. thermophila expresses two ciliate-restricted sortilins, called SOR2 and SOR4, both of which have orthologues in P. tetraurelia. The disruption of either SOR2 or SOR4 produced gross defects in the formation of T. thermophila mucocysts. In contrast, two other T. thermophila sortilins are more highly conserved, as they fall in a clade containing non-ciliate (e.g., apicomplexan) homologues. Secretion from mucocysts was largely unaffected by SOR1 knockout, whereas SOR3 was found to be an essential gene. Because mucocysts are nonessential for T. thermophila viability in the laboratory (Orias et al., 1983), the main role of SOR3 is unlikely to be mucocyst-related, and we hypothesize that both SOR1 and SOR3 maintain an ancestral function in protein trafficking to lysosomes. T. thermophila possesses Rab7-positive lysosomes (Bright et al., 2010), but the pathways involved in their formation have been little investigated in this species (Hünseler et al., 1988). There is also evidence for secretory lysosomes in T. thermophila; importantly, these are clearly distinct from mucocysts (Hünseler et al., 1987; Hünseler and Tiedtke, 1992).
Although both SOR2 and SOR4 are required for mucocyst biogenesis, the knockout of SOR4 produced the more striking defects, and we therefore focused primarily on this gene. Cells lacking Sor4p failed to transport two different members of the Grt family, Grt1p and Igr1p, to mucocysts, and Grt1p was instead found in the culture medium, which is consistent with the idea that constitutive secretion represents a default route for Grt1p in the Δsor4 cells. Δsor4 cells also showed a defect in the sorting of a mucocyst-associated protease, Cth3p. Given the known role of S. cerevisiae Vps10p as a sorting receptor for vacuolar enzymes, our data suggest that T. thermophila Sor4p acts as sorting receptor for lysosome-related proteases that function in mucocyst proprotein processing (Marcusson et al., 1994). Sor4p is likely to act directly as a receptor for Grt1p, as the two proteins could be robustly coimmunoprecipitated. The interaction is specific because Grt1p sorting is, in contrast, independent of Sor2p. Sor2p, which shares 34% amino acid identity with Sor4p in the ligand-binding VPS10 domain, may serve as a receptor for a different set of mucocyst proteins, and our data suggest that this includes proteins required for Grl proprotein processing.
We have not yet identified the Grt1p determinant recognized by Sor4p, but our data provide a strong hint. The two mucocyst proteins Grt1p and Igr1p, which are both mis-sorted in Δsor4 cells, share a predicted C-terminal β/γ crystallin domain (Bowman et al., 2005b). This domain, which we previously found to be sufficient for targeting to mucocysts, may be the direct ligand for Sor4p (Haddad et al., 2002).
Our data provide strong support for the idea that mucocyst biogenesis and lysosome biogenesis rely on shared machinery, particularly because the T. thermophila expression data, from which we identified the sortilins, also implicate AP-3 in mucocyst biogenesis. One possible interpretation of our data is that mucocysts should be considered as a class of lysosome-related organelles. However, the lysosome-related pathway is just one of two trafficking routes contributing to formation of mucocysts, which might therefore be considered as innovative hybrid organelles. In this regard, a key question is whether Grl and Grt proteins, sorted by two different mechanisms, are nonetheless transported in the same vesicular carriers, or whether the formation of mucocysts involves the coalescence of vesicles with distinct origins. Interestingly, the latter scenario is suggested by observations in other ciliates, even though molecular details are lacking. In T. thermophila, Grl proteins localize throughout the mucocyst lumen, whereas Grt1p localizes to the pole where docking and exocytic fusion occur. This sublocalization is strongly reminiscent of a well-defined tip structure on P. tetraurelia granules, called trichocysts (Adoutte, 1988). The trichocyst tip proteins are unknown, but are likely to be Grt homologues that also terminate in a β/γ crystallin domain (see Discussion in Haddad et al., 2002). Most pointedly, there is indirect evidence that the P. tetraurelia tip proteins are delivered in different vesicles from those containing the core proteins, which are homologous to the T. thermophila Grl proteins (Garreau de Loubresse, 1993; Vayssié et al., 2001). Compelling morphological evidence for two populations of pretrichocyst vesicles, but without any molecular data, is also available in another ciliate, Pseudomicrothorax dubius (Peck et al., 1993). If these pathways of granule formation are conserved throughout ciliates, it appears likely that T. thermophila Grt and Grl proteins are transported via separate vesicles. This would also be consistent with the apparent absence of biochemical interactions between Grt1p and Grl proteins (Rahaman et al., 2009).
Heterotypic vesicle coalescence can generate a complex compartment, as recently argued for peroxisome biogenesis (van der Zand et al., 2012). It may also have been important in the evolutionary origin of granules in ciliates, if a critical step was allowing fusion between two distinct, already functional compartments. An alternative evolutionary scenario is that lysosome-related organelles in a ciliate ancestor acquired secretory granule characteristics when they were “invaded” by self-aggregating cargo proteins, a model that may be relevant for certain mammalian organelles like platelet granules (Gunay-Aygun et al., 2004).
Before this work, molecular characterization of secretory granules in ciliates failed to identify homologous components to those in animals, leading to our hypothesis that functionally analogous secretory organelles in ciliates and animals arose independently and that the similarities primarily reflect convergence (Elde et al., 2007). Our current findings may, for the first time, point instead to deep shared ancestry. Secretory granules are complex organelles for which, until recently, a relatively simple mechanism relying on protein self-aggregation appeared sufficient to explain much of the obligatory protein sorting during biogenesis. The self-aggregation mechanism was discovered and largely tested via biochemical analysis of highly abundant proteins that condense to form the granule core in endocrine cells. More recently, genetic approaches in both invertebrates and vertebrates have indicated that alternative sorting mechanisms, as yet poorly understood, may pertain to different sets of granule proteins (Sumakovic et al., 2009; Burgess et al., 2011). Intriguingly, results from both mammalian cells and D. melanogaster provided evidence that proteins classically associated with targeting to lysosomes or lysosome-related organelles are involved in cargo sorting to granules (Chen et al., 2005; Asensio et al., 2010). However, since the precise roles of the lysosome-associated proteins in granule formation have not yet been established, it is not yet possible to conclude that these proteins play a direct role in the process.
Alternatively, our results may represent a further level, in addition to the properties of the cargo proteins themselves, at which mechanisms have converged between distant lineages to create secretory granules with similar properties. In this regard, our finding that a cathepsin protease in T. thermophila, belonging to a family that is classically associated with lysosomes, is targeted to mucocysts in a Sor4p-dependent fashion, may be analogous or homologous to the recruitment of cathepsin L for proprotein processing in mammalian neuropeptide-containing DCVs (Funkelstein et al., 2010). These alternative models should be clarified by further work in both animal and ciliate systems on the role of receptors and AP-3 adaptors in secretory granule formation. Convergent evolution is a well-documented phenomenon at many levels of biology, and is of particular interest because it can cast light on both the selective pressures that have shaped specific features and on the boundary conditions that may limit possible outcomes (Kronforst et al., 2012; Yates and Campbell, 2012; Zakon, 2012). Convergent evolution at the level of organellar structure and function may receive more attention as cell biology encompasses a broader swath of eukaryotic diversity, enhancing the traditional focus on animal and fungal cells, two groups that are relatively closely related (Stechmann and Cavalier-Smith, 2003). One hurdle will be to identify cases in which proteins have taken on new functions, a particular challenge in studies based on comparative genomic approaches. Our study and others suggest that relatively simple tools like expression profiling could facilitate the assignment of gene products, for which vast datasets already exist, to specific pathways and structures (Gurkan et al., 2005; Nusblat et al., 2012). Such approaches, applied to a range of diverse eukaryotes, would help to reveal the full range of solutions to universal cellular challenges.
Materials and methods
Cell culture
All T. thermophila strains were cultured in SPP media (1% proteose peptone, 0.2% dextrose, 0.1% yeast extract, and 0.003% ferric EDTA) at 30°C while shaking at 99 rpm. Experimental cultures were grown to medium density (log phase: 150–300,000 cells/ml) during an overnight incubation (at least 12 h) in a volume of SPP equivalent to one fifth of the total volume of the culture flask. Culture densities were determined with a Z1 Coulter Counter (Beckman Coulter).
pmEGFP-neo4 vector construction
pmEGFP-neo4 is a modification of the vector pEGFP-neo4 (provided by K. Mochizuki, Institute of Molecular Biotechnology, Vienna, Austria), which is designed to GFP-tag genes at their endogenous loci. We site-specifically mutagenized the ORF of GFP in the original vector to create a variant of GFP that is largely monomeric in order to avoid localization artifacts caused by oligomerization. The monomeric mutation of pmEGFP-neo4, encoding an alanine-to-lysine substitution at position 207 of the GFP ORF (Zacharias et al., 2002), was introduced into pEGFP-NEO4 by QuikChange site-directed mutagenesis (Agilent Technologies) with the primer pair 067A and 067B (Table S3).
Expression of Sor4p-GFP
mEGFP was fused at the C terminus of the SOR4 (Tetrahymena Functional Genomics Database accession no. TTHERM_00313130) macronuclear ORF via homologous recombination, using linearized pSOR4-mEGFP-neo4. This construct contains the C-terminal ∼700 bp of the SOR4 genomic locus (minus the stop codon) followed by mEGFP, the BTU1 terminator, a neo4 drug resistance cassette, and ∼600 bp of SOR4 downstream genomic sequence. To create pSOR4-mEGFP-neo4, the C-terminal region of the SOR4 genomic locus lacking the stop codon was amplified with the primer pair 093A and 093B (Table S3). The 5′ region of these primers contains ∼15 bp of homology to pmEGP-neo4 linearized with BamHI for In-Fusion (Takara Bio Inc.)-mediated insertion into the vector. Similarly, the genomic region downstream of the SOR4 locus, amplified with the primer pair 094A and 094B (Table S3), was inserted into the preceding construct linearized with HindIII. Wild-type CU428 cells were then biolistically transformed with the final construct, pSOR4-mEGFP-neo4, which was first linearized with Xho1 and Nhe1. Initial transformants were selected based on paromomycin resistance, and then serially transferred for 3–4 wk in increasing drug concentrations to drive fixation of the GFP-tagged allele. Consistent with the complete replacement of the endogenous locus by the tagged allele, the transformants maintained both Sor4p-GFP expression as well as drug resistance for at least 1 yr after initial selection.
SOR1-4 disruption
The SOR4 macronuclear locus was replaced with a neo4 drug resistance cassette via homologous recombination with the linearized construct pSOR4MACKO-neo4. This construct contains a neo4 construct flanked by <750 bp of the genomic regions immediately upstream and downstream of SOR4. pSOR4MACKO-neo4 was derived from the pSOR4-mEGFP-NEO4 construct described earlier. To complete the KO construct, the genomic region upstream of SOR4 was first amplified with the primer pair 109A and 109B (Table S3). The 5′ region of both of these primers also contains an ∼15-bp 5′ sequence homologous to the ends of pSOR4-mEGFP-neo4 linearized with PstI and Not1 (which removes the 3′ genomic coding region of SOR4 and mEGFP, and the 3′ BTU1 terminator) for In-Fusion–mediated insertion into the vector. The same strategy was used to disrupt the other sortilins (SOR1, Tetrahymena Functional Genomics Database accession no. TTHERM_00420610; SOR2, TTHERM_00410210; and SOR3, TTHERM_00467390), using primers listed in Table S3.
Expression of Igr1p-GFP
The IGR1-eGFP construct (Cowan et al., 2005) was linearized with SfiI and biolistically transformed into CU428 and ΔSOR4. The IGR1 ORF (minus the stop codon) was first cloned into the pVGF.MTT vector, upstream of GFP, via PmeI and XhoI. The sequence encoding Igr1p-GFP was then cloned into the ncvB vector for blasticidin-based selection, using the PmeI and ApaI restriction sites.
Expression of Cth3p-CFP
The CTH3 (TTHERM_00321680) ORF was cloned into the pBSICC Gateway vector, a gift from D. Chalker (Washington University, St. Louis, MO), with primer pair S001 and S002 with Invitrogen’s Gateway cloning system. In brief, the CTH3 ORF was first cloned into the entry vector pENTR/D-TOPO and then inserted into the pBSICC Gateway destination vector with the LR Clonase II recombinase (Life Technologies). The CTH3 ORF is flanked upstream in the destination vector by the cadmium inducible MTT1 promoter (Shang et al., 2002) followed by the 3′ end of the RPL29 locus, modified to contain a mutation that confers cycloheximide resistance (Yao and Yao, 1991), and flanked downstream by the 3′ RPL29 genomic region. After linearization, this construct can integrate at the end of the RPL29 locus for the transient expression of the cloned ORF. The construct was linearized with BaeI and SpeI and biolistically transformed into CU428 and ΔSOR4.
Biolistic transformations
Target cultures were grown to log phase and starved for 18–24 h in 10 mM Tris, pH 7.0. Gold particles (Seashell Technology) were prepared as recommended with 15 µg of total linearized plasmid DNA and then applied to the macrocarrier flying disk for use in a Biolistic PDS-100000/He device (Bio-Rad Laboratories) with the following settings: 27–28 in Hg vacuum, 1/4 in gap distance, 8 mm macrocarrier travel, and a target distance of 9 cm. Cells were concentrated to 1 mL (from 30 ml) and loaded into the apparatus on filter paper. After the shot, the cells were transferred on the filter paper to a prewarmed flask containing 50 ml of SPP. To select for positive transformants, drug was added 4 h after bombardment to cultures shaken at 30°C. Transformants were selected in paromomycin sulfate (120 µg/ml + 1 µg/ml CdCl2), blasticidin (60 µg/ml + 2 µg/ml CdCl2), or cycloheximide (12 µg/ml). Drug-resistant transformants were identified after 3–6 d. Transformants were then serially transferred every 2–3 d in decreasing concentrations of CdCl2 for at least 2 wk before further testing. At least two independent transformants were tested for each line.
RT-PCR assessment of SOR1-4 disruption
RNA was harvested with the RNeasy Mini kit (QIAGEN) from 106 cells grown to 1.5–3.0 × 105 cells/ml, washed and starved for 2 h in 10 mM Tris pH 7.0, and lysed after resuspension in buffer RLT by passage (5–7×) through a 1-ml tuberculin syringe. cDNA was generated from the harvested RNA with the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems). The presence of SOR1–4 transcripts was assayed by PCR amplification of purified cDNA with the same primer pair used to amplify the C-terminal coding region used to construct the GFP fusions (Table S3). Knockouts were confirmed by the persistent absence of the corresponding transcript after 4 wk of growth in the absence of drug selection (3–4 serial transfers/wk).
Dibucaine stimulation
Cultures were grown to stationary phase and then incubated for an additional 24 h before being concentrated, at least 10-fold, into a loose pellet and stimulated with 2.5 mM dibucaine. Stimulated cultures were mixed gently for ∼30 s and restored to their original volume with 10 mM Hepes and 5 mM CaCl2. After gently mixing, the culture was then centrifuged at 1,200 g for 2 min, resulting in the formation of a cell pellet/flocculent bilayer.
Immunofluorescence
Cells were washed and fixed (∼3 ml of culture) in an equal volume of ice-cold 4% paraformaldehyde (in 50 mM Hepes, pH 7.0, for 10 min). After two washes in ice-cold Hepes, cells were then permeabilized in ice-cold 0.1% Triton X-100 in Hepes for 8 min on ice. After two more washes with ice-cold Hepes, cells were resuspended in blocking solution (1% BSA in TBS [10 mM Tris, pH 7.5, and 154 mM NaCl]) and warmed to room temperature while rotating slowly for 30 min. For 1° antibody incubation, fixed cells were resuspended in 100 µL of hybridoma supernatant diluted 1:5 for 4D11 or 1:9 for 5E9 in the 1% BSA blocking solution. After 30 min, with mild agitation every 5 min to prevent the cells from settling, the cells were washed three times with 0.1% BSA in TBS, pelleted, and then resuspended in 100 µl Texas-red-X–coupled goat anti–mouse IgG (Life Technologies) diluted 1:100 in 1% BSA blocking solution. After 30 min, with mild agitation every 5 min, the cells were washed once with 0.1% BSA in TBS and twice with 10 mM Hepes, pH 7.0. Cells were resuspended in a final volume of 150 µl of 10 mM Hepes, pH 7.0, and then mixed with an equal volume of mounting media (30% glycerol and 0.1% trolox) immediately before slide preparation. For the colocalization of Grl3p and Cth3p-CFP, the former was decorated using mAb 5E9 as described earlier, the latter using polyclonal anti-GFP antibody (Life Technologies) diluted 1:400. The 2° antibodies, which were similarly coincubated with samples, were Texas red–coupled goat anti–mouse IgG and 488-coupled donkey anti–rabbit IgG (Life Technologies), diluted 1:250. Cells were imaged with a super-resolution laser scanning confocal microscope (TCS SP5 II STED-CW; Leica) with photomultiplier tube–based detection and a 100×/1.40 NA oil objective lens at room temperature. Images were captured with the LAS_AF confocal software (Leica) for Windows 7. Image data were colored, denoised, and adjusted in brightness/contrast with the program ImageJ. The images of the protease-expressing cells were also colored, but only their brightness/contrast was additionally adjusted. All cells, as shown in Fig. 6, were treated identically and adjusted to the same brightness/contrast values established by a representative wild-type image. The simultaneous localization of Grt1p and Grl3p was performed as described earlier, but the mAbs 4D11 and 5E9 were directly conjugated to Dylight 488 and 649, respectively (Thermo Fisher Scientific), and mixed 1:1 before incubation with samples. These images were then processed with Huygens Professional (Scientific Volume Imaging).
Colocalization measurement
The M2 correlation coefficient was determined with the ImageJ plugin JACoP (Bolte and Cordelières, 2006). For this analysis, a representative wild-type image was selected to establish the threshold values that were then used to determine the M2 value for all other images. To limit the analysis to the cell periphery, each cell outline was traced based on the docked mucocyst signal, and this line was broadened to create a band that encompassed all fluorescent signals at the cell periphery. The broadening factor was 60% larger for wild-type cells than for Δsor4 cells to compensate for the larger size of mucocysts in the former. The signal outside of this selected band was then cleared and the resulting images were processed with JACoP using the same threshold values identified above. A macro for the above, entitled “JACoP band measure macro” was created in ImageJ and is available for download as a supplemental file.
Western blots
Whole cell lysates were prepared from 105 cells starved for 2 h in 10 mM Tris, pH 7.0, washed once and then resuspended in an equal volume, and precipitated with TCA (10% [final]). TCA precipitates were incubated on ice for 30 min, centrifuged (18,000 g, 10 min, 4°C), washed with ice-cold acetone, centrifuged again (18,000 g, 10 min, 4°C), and resuspended in 2× sample buffer. Proteins were resolved with the Novex NuPAGE SDS-PAGE Gel system (4–12% Bis-Tris gels; Life Technologies) and transferred to 0.45 µm PVDF membranes (Thermo Fisher Scientific) at 100 V for 1 h. Protein was then reversibly stained with Ponceau S and the blot was subsequently blocked with 5% dried milk, 50 mM Tris, pH 7.8, 0.02% NP-40, and 2 mM CaCl2 for 1 h. The 1° antibody incubation, diluted in the blocking buffer, was at least 1 h. The blot was then washed four times, for 5 min each, in 50 mM Tris, pH 7.8, 0.02% NP-40, and 2 mM CaCl2. The 2° antibody incubation, diluted in the blocking buffer, was also at least 1 h. After four more washes, the blot was then incubated with the appropriate substrate and imaged. The anti-Grl1p, anti-Grt1p, and monoclonal anti-GFP (Covance) 1° antibodies were diluted 1:5,000, 1:500, and 1:5,000, respectively. Protein was visualized with either ECL horseradish peroxidase–linked rabbit (NA934) or mouse (NA931; GE Healthcare) secondary antibody diluted 1:20,000 and SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific).
Electron microscopy
Cells grown to stationary phase were fixed in 2% glutaraldehyde, 1% sucrose, and 1% osmium at 25°C in 0.1 M sodium cacodylate buffer and section-stained with uranyl acetate and lead citrate after embedding. Thin sections were viewed on a Tecnai G2 F30 Super Twin microscope (FEI).
Alcian blue stimulation/coimmunoprecipitation
Sor4p-GFP was immunoprecipitated with polyclonal anti-GFP antibody coupled to magnetic Dynabeads (Life Technologies), from lysates of cells expressing Sor4p-GFP that were undergoing regranulation after Alcian blue–stimulated degranulation (Haddad and Turkewitz, 1997). In brief, 100-ml cultures were starved for 6 h in 10 mM Tris, pH 7.0, concentrated to 6 mL, transferred to a 500 ml flask, stimulated for 15 s with 2 ml of 0.2% Alcian Blue, rescued with 92 ml of 0.25% proteose peptone + 0.5 mM CaCl2, washed once and suspended in 36 ml of 10 mM Tris, and allowed to recover for 20 min in 50-ml conical tubes. After 20 min, ∼30 ml of swimming cells were withdrawn, pelleted, and resuspended in 9 ml of lysis buffer (25 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40, and 5% glycerol) plus two complete Ultra protease inhibitor tablets per sample (Roche). After 30 min on ice, lysates were cleared by centrifugation at 10,000 g for 20 min, and split into two aliquots. 1.5 mg of anti-GFP–coupled Dynabeads, or rabbit anti–mouse IgG as negative control, were added to each aliquot and mixed for 8 h at 4°C. The beads were then washed three times with lysis buffer (or 25 mM citrate buffer, pH 5.0, 150 mM NaCl, 1 mM EDTA, 1% NP-40, and 5% glycerol) and suspended in 70 µl 100°C SDS-PAGE sample buffer.
Phylogenetic tree construction
Using a protein BLAST (blastp), the T. thermophila SOR4 VPS10 domain was used to identify homologues in ciliates and other alveolates as documented in Table S2, and then homologues in other eukaryotic lineages as documented in Table S1. For tree building, the top hits were selected from each lineage plus the VPS10 domains from sortilins in humans and budding yeast. These VPS10 domains were assembled and aligned with MUSCLE (http://www.ebi.ac.uk/Tools/msa/muscle/), and phylogenetic trees were constructed with MEGA5 (Molecular Evolutionary Genetics Analysis; http://www.megasoftware.net/). The maximum likelihood trees are based on the Whelan and Goldman model. The Neighbor-joining tree uses the JTT matrix-based method to compute evolutionary distances. For both trees, positions containing gaps or missing data were eliminated. Bootstrap consensus trees were inferred from 1,000 replicates.
Live cell imaging
Cultures grown to 150–300,000 cells/ml were starved for 2 h in 10 mM Tris, pH 7.0, before being pelleted and resuspended in 6% polyethylene oxide (Sigma-Aldrich) to immobilize the swimming cells. For cells expressing Igr1p-GFP under the control of the MTT1 promoter, 0.1 µg/ml of CdCl2 was added for 2 h to induce transgene expression. The IGR1-GFP lines were imaged on an spinning disk inverted confocal microscope (DSU; Olympus) with a 100×/1.35 NA oil objective lens at room temperature. Images were captured with a back-thinned charge-coupled device (CCD) camera (Evolve; Photometrics) in SlideBook software (3i). The SOR4-GFP lines were imaged using a Marianas Yokogawa type spinning disk inverted confocal microscope (3i) with a 100×/1.45 NA oil objective lens at room temperature. Images were captured with an Evolve back-thinned air-chilled CCD camera in SlideBook. The brightness/contrast of the images were adjusted in ImageJ. Additionally, background/noise in Video 1 were edited using a variation on the 3 × 3 2D Median Hybrid Filter ImageJ plugin (by C.P. Mauer; available for download as a supplemental file).
Online supplemental material
Fig. S1 shows that the T. thermophila sortilins fall into two major groups. Fig. S2 shows that expression profiling identifies genes associated with mucocysts, including sortilins. Fig. S3 shows that Sor4p-GFP interacts with Grt1p, and is required for the transport of Grt1p to mucocysts. Fig. S4 shows that Δsor1 is not essential for mucocyst formation and secretion. Fig. S5 shows that sorting efficiency of the exogenously expressed mucocyst protease Cth3p-CFP is reduced in Δsor4 cells. Table S1 gives accession numbers from which VPS10 domain sequences were obtained to create the phylogeny in Fig. 1 and Fig. S1. Table S2 gives accession numbers from which VPS10 domain sequences were obtained to create the phylogeny in Fig. 2 B. Table S3 is a master primer list. Video 1 shows that Sor4p localizes to mobile cytoplasmic puncta and not to mature mucocysts docked at the plasma membrane. An ImageJ macro that selects a band at the cell periphery for colocalization analysis with the JACoP plugin using user defined thresholds and a variation on the 3 × 3 2D Median Hybrid Filter ImageJ plugin are available for download. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201305086.dv.
Acknowledgments
Lydia Bright made important early contributions to analyzing mucocyst-associated gene expression in our laboratory. We heartily thank Cassie Kontur, Mahima Joiya, Yusuke Fukuda, and Ania Łuchniak for their continuous and generous support; Christine Labno and Vytas Bindokas of the University of Chicago Light Microscopy Core Facility and Nicholas VanKuren for help with the phylogenetic analysis; Wei Miao (Chinese Academy of Sciences) for discussing expression data; Kazufumi Mochizuki and Doug Chalker for generously providing vectors; Yimei Chen of the Advanced Electron Microscopy Facility; and Ben Glick, Chip Ferguson, Alejandro Nusblat, Sabrice Guerrier, Mohan Gupta, and Nels Elde for many helpful discussions.
J.S. Briguglio was partially supported by National Institutes of Health Training grant T32 GM007191 and by the National Science Foundation (NSF) through a Graduate Research Fellowship. This work was funded by the Chicago Biomedical Consortium with support from The Searle Funds at The Chicago Community Trust, and by NSF grant MCB-1051985 to A.P. Turkewitz. A.P. Turkewitz is on the scientific advisory committee of Tetragenetics, Inc.



![Figure 2. Expression profiling suggests a role for sortilin-family receptors in mucocyst biogenesis in T. thermophila. (A) Sortilins are coregulated with genes encoding mucocyst contents in T. thermophila. The expression profiles of the four T. thermophila sortilins (SOR1–4; right), are similar to those of genes (GRL1, GRL3, GRT1, and IGR1; left) encoding mucocyst cargo proteins. Expression profiles are derived from the Tetrahymena Functional Genomics Database, with each profile normalized to that gene’s maximum expression level. Points on the x axis correspond to successive time points and represent growing, starved, and mating cultures, including three different culture densities (low [Ll], medium [Lm], and high [Lh]), 7 samples taken during 24 h of starvation, and 10 samples subsequently taken during 18 h after conjugation. (B) Expansion of the sortilin family in ciliates. The maximum likelihood tree illustrates a phylogeny of VPS10 domain–containing receptors (sortilins) in alveolates, the taxonomic group consisting of ciliates, apicomplexans, and dinoflagellates. Two of the T. thermophila sortilins, marked by black circles, cluster with the sortilins from other alveolates. In contrast, T. thermophila SOR2 and SOR4, marked by maroon diamonds, belong to an expansion of sortilins restricted to ciliates. Species are abbreviated as follows: Babesia microti (Bm), Cryptosporidium hominis (Ch), Cryptosporidium muris (Cm), I. multifiliis (Im), Neospora caninum (Nc), P. tetraurelia (Pt), Perkinsus marinus (Pm), Plasmodium berghei (Pb), Plasmodium cynomolgi (Pc), Plasmodium falciparum (Pf), Plasmodium knowlesi (Pk), Plasmodium vivax (Pv), Plasmodium yoelii yoelii (Py), T. thermophila (Tt), Theileria annulata (Ta), Theileria orientalis (To), Theileria parva (Tp), T. gondii (Tg). See Tables S1 and S2 for a list of accession numbers for all of the sequences. (C) Verification of the nonessential sortilin knockouts. cDNA was prepared from wild-type, Δsor1, Δsor2, and Δsor4 cells, and the SOR1, SOR2, and SOR4 sequences were PCR amplified using gene-specific primers. As shown in this 1% ethidium bromide–stained agarose gel, each of the gene knockout lines lacks the amplified product corresponding to the targeted gene, but shows wild-type levels of the other transcripts that serve as loading controls (see also Fig. S2 E). The lanes shown are all part of a single gel, but their order has been rearranged for this figure.](https://cdn.rupress.org/rup/content_public/journal/jcb/203/3/10.1083_jcb.201305086/6/m_jcb_201305086_fig2.jpeg?Expires=1772616955&Signature=DIAAj~sw7E4SuzufnOZgbG6qrqap96yXaPDMYTaS0stuloD9IrELXWcSYXQL70rCAaRuApqOLpdcVQ8i-1EUqhSE-87dLoafiNm1VbHzYOTspLl~KJ0uYJeoVz9nS8jiaxNlyT8ZUeBBnwA8du03O9yTq2XP0nPhl72vtQJLOIBAWw8fRkP940WjJUEeC-61J4E4jCGNsrO8B0-4S0xU1ASx~ikfHdJEJydDCdt4gLE4i-HWIlt72nOD~urF1tLT8MF2uwK6bddely1pviz5QApTobpkHr~dzZugMEI74wNCVfc5aDWh9yg7BBVIRGHbqHL~SF4wkMSeisIPa0pRWw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)