Asymmetry of cell fate is one fundamental property of stem cells, in which one daughter cell self-renews, whereas the other differentiates. Evidence of nonrandom template segregation (NRTS) of chromosomes during asymmetric cell divisions in phylogenetically divergent organisms, such as plants, fungi, and mammals, has already been shown. However, before this current work, asymmetric inheritance of chromatids has never been demonstrated in differentiating embryonic stem cells (ESCs), and its molecular mechanism has remained unknown. Our results unambiguously demonstrate NRTS in asymmetrically dividing, differentiating human and mouse ESCs. Moreover, we show that NRTS is dependent on DNA methylation and on Dnmt3 (DNA methyltransferase-3), indicating a molecular mechanism that regulates this phenomenon. Furthermore, our data support the hypothesis that retention of chromatids with the “old” template DNA preserves the epigenetic memory of cell fate, whereas localization of “new” DNA strands and de novo DNA methyltransferase to the lineage-destined daughter cell facilitates epigenetic adaptation to a new cell fate.
Introduction
One defining characteristic of stem cells is their ability to divide asymmetrically, such that one daughter cell self-renews to remain stem, whereas the other daughter cell commits to lineage-specific differentiation (Knoblich, 2008). This often coincides with asymmetric inheritance of macromolecules to the daughter cells, for example, misfolded proteins (Rujano et al., 2006), centrioles (Yamashita et al., 2007), and the younger versus older replicated chromatids in different organisms, such as bacteria (Lark, 1966), plants (Lark, 1967), filamentous fungi (Rosenberger and Kessel, 1968), or mammals. In mammals, it has been described in a variety of cell types: epithelium (Potten et al., 1978), intestine (Potten et al., 2002; Falconer et al., 2010; Quyn et al., 2010), mammary (Smith, 2005), neural (Karpowicz et al., 2005), and muscle (Shinin et al., 2006; Conboy et al., 2007; Rocheteau et al., 2012) cells. The earliest observations led to the immortal DNA strand hypothesis, postulating that stem cells avoid accumulating mutations arising from DNA replication by consecutively and infinitely segregating old DNA strands in the stem daughter cell (Cairns, 1975). Aspects of this hypothesis and the underlying phenomenon have been debated (Lansdorp, 2007; Rando, 2007; Steinhauser et al., 2012) because of the lack of evidence supporting the infinite ability of stem cells to sort their DNA, conflicting studies in similar tissues (Potten et al., 2002; Falconer et al., 2010; Quyn et al., 2010; Escobar et al., 2011; Schepers et al., 2011), and the reported inability of some other tissue-specific stem cells to segregate DNA strands nonrandomly, such as blood (Kiel et al., 2007), hair (Waghmare et al., 2008), and skin (Sotiropoulou et al., 2008). Nevertheless, a growing body of evidence supports DNA strand nonrandom template segregation (NRTS) in a variety of asymmetrically dividing stem cells. Asymmetric segregation of epigenetically unequal sister chromatids might be required to affect gene expression and consequently cell fate in asymmetric division. Moreover, such distinct epigenetic marks between sister chromatids might be necessary to sort older versus younger DNA strands during mitosis (Klar, 1994; Lansdorp, 2007). However, before this current work, these notions remained undemonstrated, and the identification of epigenetic marks had been poorly—if at all—documented (Huh and Sherley, 2011), perhaps because of the lack of an in vitro cellular model exhibiting robust NRTS.
Considering that embryonic stem cells (ESCs) do not exhibit NRTS when cultured in self-renewing conditions (Karpowicz et al., 2005; Falconer et al., 2010) and the lack of data on NRTS in these pluripotent stem cells during multilineage differentiation—when a high rate of asymmetric cell divisions is predicted—we decided to investigate NRTS in human ESCs (hESCs) and mouse ESCs (mESCs) that are induced to differentiate into the three germ layers as embryoid bodies (EBs). Our results are the first to unambiguously show that NRTS occurs at a high frequency in differentiating EBs, through the use of conventional microscopy as well as time-lapse imaging. Moreover, this work establishes that NRTS is dependent on DNA methylation and on the activity of de novo DNA methyltransferases (Dnmts) Dnmt3a and Dnmt3b enzymes but not on Dnmt1 or histone deacetylation.
Results
High NRTS occurrence in differentiating human and mouse EBs
By the semiconservative mechanism of DNA replication, each single-stranded DNA of a chromatid serves as a template for synthesizing a new complementary strand (Meselson and Stahl, 1958). By following templates and synthesis over more than one cell division, it can be demonstrated that the replicated sister chromatids are not exact copies: one sister chromatid will have an older template strand than the other one (Fig. 1). All studies of NRTS have been based on variations of one experimental principle: a pulsed incorporation of a thymidine analogue to label synthesized DNA strands and a chase time for those strands to become templates followed by a quantification of mitotic cell pairs that distribute those labeled template strands asymmetrically versus symmetrically to daughter cells.
Single- and double-labeling methods to assess nonrandom template DNA strand segregation. (a and b) Two pairs of chromosomes are represented for clarity. Newly synthesized DNA strands are shown in dashed, colored lines. After one round of cell division, all cells are labeled with BrdU for the single-labeling method (a) or CldU for the double-labeling method (b). It is only after the second division that NRTS can be observed. If cells randomly distribute their template DNA, the two resulting daughter cells will inherit BrdU (a)- or CldU (b)-labeled strands. However, if all template DNA chromosomes are segregated to one daughter cell, the other daughter cell inherits the BrdU (a)- or CldU (b)-labeled newly synthesized DNA strands (after the second round of division, all daughter cells are labeled with IdU in the double-labeling method).
Single- and double-labeling methods to assess nonrandom template DNA strand segregation. (a and b) Two pairs of chromosomes are represented for clarity. Newly synthesized DNA strands are shown in dashed, colored lines. After one round of cell division, all cells are labeled with BrdU for the single-labeling method (a) or CldU for the double-labeling method (b). It is only after the second division that NRTS can be observed. If cells randomly distribute their template DNA, the two resulting daughter cells will inherit BrdU (a)- or CldU (b)-labeled strands. However, if all template DNA chromosomes are segregated to one daughter cell, the other daughter cell inherits the BrdU (a)- or CldU (b)-labeled newly synthesized DNA strands (after the second round of division, all daughter cells are labeled with IdU in the double-labeling method).
Timing is crucial to optimally detect NRTS, mainly because more apparent symmetric events will be observed if the traced cell undergoes more (or less) than two consecutive cell cycles (Fig. 1). Therefore, we determined the population doubling time and performed cell cycle analysis of each cell line used in these studies, for both self-renewing ESCs and differentiating EBs (Fig. S1, b and c). These data are summarized in Table S1 and were used to adequately adjust the timing and duration of the thymidine analogue pulse and chase periods.
To assess the frequency of NRTS, we performed paired cell assays in combination with a BrdU single-labeling method (Fig. 1 a) during differentiation of hESCs. Multilineage differentiation of H9 hESCs was induced by mechanical disaggregation of ESC colonies into large clumps to form EBs grown in suspension on nonadherent Petri dishes in the presence of 10% FBS for 1–7 d. EBs at day 1, 2, 3, 5, or 7 were pulsed with BrdU for less than one round of DNA replication, resulting in BrdU incorporation exclusively into neosynthesized DNA strands (Figs. 1 a and 2 a). EBs were then chased of BrdU during the completion of this first round of division. During the second round of cell division (in the absence of BrdU) EBs were dissociated into single cells by trypsin digestion, plated at very low density onto Matrigel-coated chamber slides, and allowed to complete the second division to form paired daughter cells that were subsequently analyzed for NRTS (Figs. 1 a and 2 a). In cells that randomly distributed chromatids of different template DNA age, the BrdU label is distributed fairly evenly between the two daughter cells (Fig. 2, b and c, top). However, in cells that segregated the template DNA chromosomes by age, only one of the daughter cells inherited almost all the BrdU label (Fig. 2, b and c, bottom). We observed a surprisingly high occurrence of NRTS 1 d after differentiation, with >40% of the cells showing asymmetric segregation of their DNA (Fig. 2 d). This frequency dropped to 28% on day 2 and subsequently increased, with differentiation reaching ∼50% on day 7 (Fig. 2 d). It is not clear why the frequency of NRTS is relatively high 1 d after differentiation, but it might suggest that NRTS is critical for the transition between the pluripotent state and the specification of the three germ layers. It is also possible that a significant portion of the cells in large hESC colonies are poised to differentiate toward one lineage (King et al., 2009; Pal et al., 2009).
High occurrence of NRTS during hESC and mESC differentiation. (a) Experimental design scheme representing how NRTS was assessed in EBs 7 d after differentiation for H9 hESCs that have a population doubling time of 24 h. The first label (BrdU for single labeling or CldU for double labeling) was added to EBs for 8 h between days 6 and 7. This was followed by a 16-h chase without any label. 24 h after addition of the first label, when cells have completed one division and started a second one, EBs were dissociated into single cells, plated at very low density, and allowed to complete the second division (in the presence of IdU for the double-labeling method). Paired cells were then fixed and stained for BrdU or CldU and IdU. (b) Theoretical predicted outcomes of the experiment described in a. (c) H9 hESC pairs that are symmetric (top) or asymmetric (bottom) for BrdU in the single-labeling method. Paired-cell assays were performed on EBs 7 d after differentiation. Representative photographs of cell pairs immunostained for BrdU are shown alongside bright-field (BF) and DAPI nuclear counterstaining images. (d) Quantification of asymmetric pairs during H9 hESC differentiation. Paired-cell assays were performed at different days after EB formation as indicated. Cell pairs were immunostained for BrdU. More than 200 pairs were analyzed per time point and the percentage of pairs showing asymmetric BrdU (i.e., NRTS) was quantified. Data represent means ± SEM (n = 3 independent experiments). (e and f) H9 hESC (e) and R1 mESC (f) pairs that are symmetric or asymmetric for CldU in the double-labeling method. Paired-cell assays were performed on EBs 7 d after differentiation. Cell pairs were immunostained for CldU and IdU. Bright-field images and DAPI nuclear counterstaining are shown. Representative photographs of symmetric (top), asymmetric (middle), and IgG control (bottom) pairs are displayed for each cell line. (g and h) CldU (g) and IdU (h) fluorescence intensity distribution among daughter cells from R1-derived EB pairs experiment using the CldU/IdU double-labeling method. CldU intensity was normalized to IdU intensity for each cell, and IgG background was subtracted from the CldU and IdU signals. The scatter graphs show the quantification of immunofluorescence intensity of visually symmetric (with a distribution from equal to 60:40, as indicated) and visually asymmetric (with a distribution of 90:10 or greater, as indicated) pairs of cells (n = 58 representative pairs from three to four independent experiments). (i) Quantification of the occurrence of NRTS in H9- and R1-derived EBs 7 d after differentiation and IMR-90 human fetal fibroblasts using the CldU/IdU double-labeling method. NRTS frequency was also quantified in R1-derived EBs after the first division in which the CldU label is expected to be segregated to both daughter cells. NRTS occurrence in undifferentiated R1 mESCs was assessed using the BrdU single-labeling method and quantified among Oct4 double-positive pairs. More than 100 pairs were analyzed for each cell line per experiment, and the percentage of cells showing asymmetric CldU or BrdU (i.e., NRTS) was quantified (all cells had symmetric IdU in the double-labeling method). Data represent means ± SEM (n = 3–6 independent experiments).
High occurrence of NRTS during hESC and mESC differentiation. (a) Experimental design scheme representing how NRTS was assessed in EBs 7 d after differentiation for H9 hESCs that have a population doubling time of 24 h. The first label (BrdU for single labeling or CldU for double labeling) was added to EBs for 8 h between days 6 and 7. This was followed by a 16-h chase without any label. 24 h after addition of the first label, when cells have completed one division and started a second one, EBs were dissociated into single cells, plated at very low density, and allowed to complete the second division (in the presence of IdU for the double-labeling method). Paired cells were then fixed and stained for BrdU or CldU and IdU. (b) Theoretical predicted outcomes of the experiment described in a. (c) H9 hESC pairs that are symmetric (top) or asymmetric (bottom) for BrdU in the single-labeling method. Paired-cell assays were performed on EBs 7 d after differentiation. Representative photographs of cell pairs immunostained for BrdU are shown alongside bright-field (BF) and DAPI nuclear counterstaining images. (d) Quantification of asymmetric pairs during H9 hESC differentiation. Paired-cell assays were performed at different days after EB formation as indicated. Cell pairs were immunostained for BrdU. More than 200 pairs were analyzed per time point and the percentage of pairs showing asymmetric BrdU (i.e., NRTS) was quantified. Data represent means ± SEM (n = 3 independent experiments). (e and f) H9 hESC (e) and R1 mESC (f) pairs that are symmetric or asymmetric for CldU in the double-labeling method. Paired-cell assays were performed on EBs 7 d after differentiation. Cell pairs were immunostained for CldU and IdU. Bright-field images and DAPI nuclear counterstaining are shown. Representative photographs of symmetric (top), asymmetric (middle), and IgG control (bottom) pairs are displayed for each cell line. (g and h) CldU (g) and IdU (h) fluorescence intensity distribution among daughter cells from R1-derived EB pairs experiment using the CldU/IdU double-labeling method. CldU intensity was normalized to IdU intensity for each cell, and IgG background was subtracted from the CldU and IdU signals. The scatter graphs show the quantification of immunofluorescence intensity of visually symmetric (with a distribution from equal to 60:40, as indicated) and visually asymmetric (with a distribution of 90:10 or greater, as indicated) pairs of cells (n = 58 representative pairs from three to four independent experiments). (i) Quantification of the occurrence of NRTS in H9- and R1-derived EBs 7 d after differentiation and IMR-90 human fetal fibroblasts using the CldU/IdU double-labeling method. NRTS frequency was also quantified in R1-derived EBs after the first division in which the CldU label is expected to be segregated to both daughter cells. NRTS occurrence in undifferentiated R1 mESCs was assessed using the BrdU single-labeling method and quantified among Oct4 double-positive pairs. More than 100 pairs were analyzed for each cell line per experiment, and the percentage of cells showing asymmetric CldU or BrdU (i.e., NRTS) was quantified (all cells had symmetric IdU in the double-labeling method). Data represent means ± SEM (n = 3–6 independent experiments).
Even with an accurate determination of the cell cycle length, one cannot exclude the possibility that a subset of the population divides slower after differentiation is induced during EB cultures. If this happened to be the case, these cells would appear as “symmetric” cell pairs if they were ending only their first mitosis after pulse labeling at the time the experimenter is expecting the second mitosis. To exclude slower-dividing cells, we discriminated between each strand of DNA: younger template, older template, and newly synthesized complementary (Conboy et al., 2007; Kiel et al., 2007). The experimental procedure is similar to the one described here for the BrdU single-labeling method except that two sequential rounds of DNA synthesis were labeled (Fig. 1 b), allowing tracking of both the neotemplate and the immediately synthesized complementary strands: 5-chloro-2′-deoxyuridine (CldU; Fig. 1 b, green) labeled the DNA synthesized in the first cell cycle (the equivalent of BrdU during the aforementioned single-labeling method) and 5-iodo-2′-deoxyuridine (IdU; Fig. 1 b, red) labeled the DNA synthesized in the second cell cycle (Fig. 2 a). Of the IdU-labeled cell pairs, some showed almost all the CldU label in one daughter cell (Figs. 1 b and 2, b and e, middle), indicating that chromatids containing the “younger” template DNA segregated to that cell and that unlabeled chromatids containing the “older” template DNA segregated to the other cell, i.e., NRTS. Other cell pairs showed a distribution of CldU label to both daughter cells (Fig. 1 b and 2, b and e, top), indicating that “younger” and “older” chromatids segregated randomly between the daughter cells. These results represent the first evidence that NRTS occurs during multilineage differentiation of human EBs. With ∼50% of the cells undergoing nonrandom segregation of the majority of their chromosomes (Fig. 2 i), differentiating human EBs robustly demonstrate NRTS and serve as a useful model to study the phenomenon.
We also evaluated the occurrence of NRTS in differentiating R1 mESCs to confirm the fundamental phenomenon and to take advantage of available mESC mutants to study molecular mechanisms that may regulate NRTS. 7 d after R1 mESC differentiation as EBs, we performed paired-cell assays in combination with CldU/IdU double labeling (Fig. 2 f). The relative fluorescence intensity of quantified CldU signal in the visually asymmetric daughter cells ranged from 90:10 to 100:0, whereas daughter cells with visually symmetric CldU were within the 60:40 range, as expected (Fig. 2 g). CldU intensities were normalized to IdU intensities for each cell to compensate for any potential unequal staining artifacts (antibody–antigen accessibility, chromatin condensation, and plane of focus) between daughter cells (Fig. 2 g). Validating the accuracy of this approach, the IdU intensities were typically within the 60:40 range (Fig. 2 h). Similarly to the results obtained with H9 cells, NRTS occurred at a very high frequency in differentiating R1 cells, with 36% of R1 paired cells showing asymmetric segregation of their chromosomes. NRTS was observed only at 2.9% when cells were assessed after the first mitosis, in which the CldU label is expected to be segregated to both daughter cells, demonstrating the very low frequency of experimental artifacts (Fig. 2 i). In accordance with previous studies (Karpowicz et al., 2005; Falconer et al., 2010), when costained for BrdU and Oct4, undifferentiated R1 mESCs showed only 4.5% NRTS among Oct4 double-positive pairs using the BrdU single-labeling method (Fig. 2 i). NRTS was also observed at only 4.7% in the IMR-90 human fetal fibroblast line used as a non–stem cell control (Fig. 2 i). Additionally, to evaluate the efficiency of EB dissociation into single cells, we quantified the occurrence of paired-cell events quickly (1–2 h) after plating and determined that it happened only 4.2 ± 0.8% of the time. Of note, given the 1–2 h required for cells to attach, we cannot exclude that some of these 4.2% pairs are valid pairs resulting from the cellular division of parent cells in the G2/M phase of the cell cycle at the time of EB dissociation. Hence, the experimental artifacts by such an evaluation might be even lower than 4.2%. In addition, the number of IdU asymmetric cells observed during paired-cell assays—another measure of system artifacts—was very low (4.4 ± 1.6%), providing more support for our observations of significant NRTS.
hESCs and differentiating cells in EBs exhibit notoriously poor survival after dissociation, and substantial apoptosis and necrosis are part of the normal differentiation process in EBs, particularly in the process of cavitation (the EB analogue to blastocoel; Boyd et al., 1984; Choi et al., 2005). To better characterize our experimental system, we performed apoptosis analysis of both H9- and R1-derived EBs 7 d after differentiation. As shown in Fig. S2, 40 and 55% of H9 and R1 EB-dissociated cells in our assay, respectively, are alive at the time of plating to accomplish their second round of cell division during the paired-cell assay. Summarily, the high occurrence of NRTS in EBs generated from both mESCs and hESCs represents a robust, evolutionarily conserved phenomenon that is corroborated by the low levels of NRTS in negative control cells and the low percentage of experimental artifacts.
Time-lapse to directly assess NRTS
The paired-cell assay in combination with pulse incorporation of thymidine analogues (with different permutations of this technique) has been the most commonly used method to assess NRTS (Potten et al., 1978; Conboy et al., 2007; Falconer et al., 2010; Rocheteau et al., 2012). This is primarily caused by the lack of traceable molecules to follow DNA synthesis in live cells. Even though one can control the experimental procedure, such as a precise cell cycle length determination, appropriate cell density, and use of the double-labeling method to minimize the risks of data misinterpretation, this method is based on the assumption that the two observed cells or daughter cells in a pair are the product of the cellular division of a common parent cell. Therefore, one cannot exclude that two nondaughter cells might have been plated next to each other or have migrated toward each other to show an apparent, but artificial, paired-cell pattern. To address this potential caveat, we performed time-lapse microscopy during the second cell cycle of differentiating R1 EBs cultured for the paired-cell assay and labeled for CldU and IdU as described for Fig. 2. CldU incorporation was performed before time-lapse acquisition and later used to confirm that the first cell cycle occurred as well as potential NRTS. IdU was incorporated during time-lapse acquisition and used to confirm that the second cell cycle was completed, which was also confirmed by the direct observation of cell division during time-lapse videos.
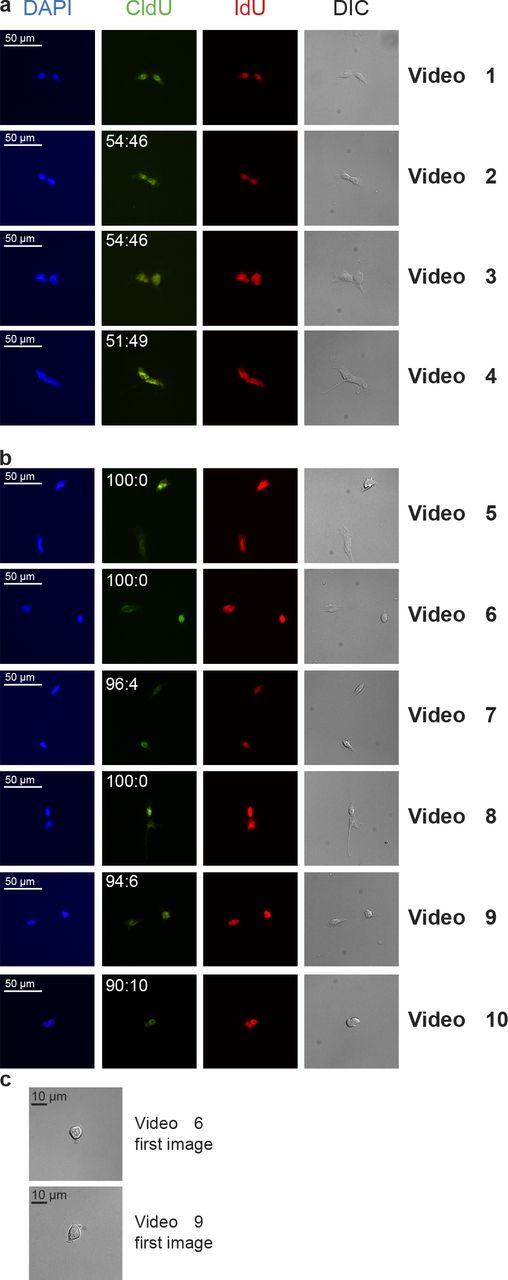
Immediately after the time-lapse imaging, cells were fixed and subsequently analyzed for NRTS by immunofluorescence. This allowed us to track with absolute certainty the daughter cells of each mitotic division and to unambiguously establish whether cells have truly divided or merely happened to be next to each other as a result of the plating and/or migration. In 10 independent experiments, a total of 826 single cells was recorded, and of these, only 99 (28 + 71) were counted as symmetric or asymmetric, for which 10 examples are shown in Fig. 3 and Videos 1, 2, 3, 4, 5, 6, 7, 8, 9, and 10. The 727 other cells did not meet the criterion to be included in the count for multiple reasons: (a) cell did not divide during the time-lapse, (b) cell died during time lapse, (c) cell went out of field during time lapse, (d) cells were CldU or IdU negative after staining, (e) cell was washed out of the slide during the staining process, or (f) it was not possible to clearly identify a nucleus before cell division (Fig. 3 c), unambiguously demonstrating that a single cell was plated and studied for NRTS by this time-lapse approach. Of the IdU-labeled daughter cells meeting the aforementioned criteria, 28 showed the majority of CldU-labeled strands in one daughter cell, i.e., NRTS (Fig. 3 b and Videos 5, 6, 7, 8, 9, and 10), whereas 71 daughter cells randomly distributed CldU-labeled strands (Fig. 3 a and Videos 1, 2, 3, and 4). With 28.3% of daughter cells segregating template DNA strands nonrandomly, these results unequivocally demonstrate the high frequency of NRTS in differentiating EBs.
Single cell division displaying random template strand segregation. This video is related to Fig. 3 a. Time-lapse of the second cell division of a paired-cell assay in combination with the CldU/IdU double-labeling method for a single R1 EB-derived cell 7 d after differentiation. The resulting paired cells display symmetric template strand segregation. DIC pictures were captured using a 20× objective every 5 min for a total of 8–9 h in a humidified chamber at 37°C and 5% CO2 (imaging microscope station [TE2000; Nikon]).
Single cell division displaying random template strand segregation. This video is related to Fig. 3 a. Time-lapse of the second cell division of a paired-cell assay in combination with the CldU/IdU double-labeling method for a single R1 EB-derived cell 7 d after differentiation. The resulting paired cells display symmetric template strand segregation. DIC pictures were captured using a 20× objective every 5 min for a total of 8–9 h in a humidified chamber at 37°C and 5% CO2 (imaging microscope station [TE2000; Nikon]).
Single cell division displaying random template strand segregation. This video is related to Fig. 3 a. Time-lapse of the second cell division of a paired-cell assay in combination with the CldU/IdU double-labeling method for a single R1 EB-derived cell 7 d after differentiation. The resulting paired cells display symmetric template strand segregation. DIC pictures were captured using a 20× objective every 5 min for a total of 8–9 h in a humidified chamber at 37°C and 5% CO2 (imaging microscope station [TE2000; Nikon]).
Single cell division displaying random template strand segregation. This video is related to Fig. 3 a. Time-lapse of the second cell division of a paired-cell assay in combination with the CldU/IdU double-labeling method for a single R1 EB-derived cell 7 d after differentiation. The resulting paired cells display symmetric template strand segregation. DIC pictures were captured using a 20× objective every 5 min for a total of 8–9 h in a humidified chamber at 37°C and 5% CO2 (imaging microscope station [TE2000; Nikon]).
Single cell division displaying random template strand segregation. This video is related to Fig. 3 a. Time-lapse of the second cell division of a paired-cell assay in combination with the CldU/IdU double-labeling method for a single R1 EB-derived cell 7 d after differentiation. The resulting paired cells display symmetric template strand segregation. DIC pictures were captured using a 20× objective every 5 min for a total of 8–9 h in a humidified chamber at 37°C and 5% CO2 (imaging microscope station [TE2000; Nikon]).
Single cell division displaying random template strand segregation. This video is related to Fig. 3 a. Time-lapse of the second cell division of a paired-cell assay in combination with the CldU/IdU double-labeling method for a single R1 EB-derived cell 7 d after differentiation. The resulting paired cells display symmetric template strand segregation. DIC pictures were captured using a 20× objective every 5 min for a total of 8–9 h in a humidified chamber at 37°C and 5% CO2 (imaging microscope station [TE2000; Nikon]).
Single cell division displaying random template strand segregation. This video is related to Fig. 3 a. Time-lapse of the second cell division of a paired-cell assay in combination with the CldU/IdU double-labeling method for a single R1 EB-derived cell 7 d after differentiation. The resulting paired cells display symmetric template strand segregation. DIC pictures were captured using a 20× objective every 5 min for a total of 8–9 h in a humidified chamber at 37°C and 5% CO2 (imaging microscope station [TE2000; Nikon]).
Single cell division displaying random template strand segregation. This video is related to Fig. 3 a. Time-lapse of the second cell division of a paired-cell assay in combination with the CldU/IdU double-labeling method for a single R1 EB-derived cell 7 d after differentiation. The resulting paired cells display symmetric template strand segregation. DIC pictures were captured using a 20× objective every 5 min for a total of 8–9 h in a humidified chamber at 37°C and 5% CO2 (imaging microscope station [TE2000; Nikon]).
Single cell division displaying nonrandom template strand segregation. This video is related to Fig. 3 b. Time-lapse of the second cell division of a paired-cell assay in combination with the CldU/IdU double-labeling method for a single R1 EB-derived cell 7 d after differentiation. The resulting paired cells display nonrandom template strand segregation. DIC pictures were captured using a 20× objective every 5 min for a total of 8–9 h in a humidified chamber at 37°C and 5% CO2 (imaging microscope station [TE2000; Nikon]).
Single cell division displaying nonrandom template strand segregation. This video is related to Fig. 3 b. Time-lapse of the second cell division of a paired-cell assay in combination with the CldU/IdU double-labeling method for a single R1 EB-derived cell 7 d after differentiation. The resulting paired cells display nonrandom template strand segregation. DIC pictures were captured using a 20× objective every 5 min for a total of 8–9 h in a humidified chamber at 37°C and 5% CO2 (imaging microscope station [TE2000; Nikon]).
Single cell division displaying nonrandom template strand segregation. This video is related to Fig. 3 b. Time-lapse of the second cell division of a paired-cell assay in combination with the CldU/IdU double-labeling method for a single R1 EB-derived cell 7 d after differentiation. The resulting paired cells display nonrandom template strand segregation. DIC pictures were captured using a 20× objective every 5 min for a total of 8–9 h in a humidified chamber at 37°C and 5% CO2 (imaging microscope station [TE2000; Nikon]).
Single cell division displaying nonrandom template strand segregation. This video is related to Fig. 3 b. Time-lapse of the second cell division of a paired-cell assay in combination with the CldU/IdU double-labeling method for a single R1 EB-derived cell 7 d after differentiation. The resulting paired cells display nonrandom template strand segregation. DIC pictures were captured using a 20× objective every 5 min for a total of 8–9 h in a humidified chamber at 37°C and 5% CO2 (imaging microscope station [TE2000; Nikon]).
Single cell division displaying nonrandom template strand segregation. This video is related to Fig. 3 b. Time-lapse of the second cell division of a paired-cell assay in combination with the CldU/IdU double-labeling method for a single R1 EB-derived cell 7 d after differentiation. The resulting paired cells display nonrandom template strand segregation. DIC pictures were captured using a 20× objective every 5 min for a total of 8–9 h in a humidified chamber at 37°C and 5% CO2 (imaging microscope station [TE2000; Nikon]).
Single cell division displaying nonrandom template strand segregation. This video is related to Fig. 3 b. Time-lapse of the second cell division of a paired-cell assay in combination with the CldU/IdU double-labeling method for a single R1 EB-derived cell 7 d after differentiation. The resulting paired cells display nonrandom template strand segregation. DIC pictures were captured using a 20× objective every 5 min for a total of 8–9 h in a humidified chamber at 37°C and 5% CO2 (imaging microscope station [TE2000; Nikon]).
Single cell division displaying nonrandom template strand segregation. This video is related to Fig. 3 b. Time-lapse of the second cell division of a paired-cell assay in combination with the CldU/IdU double-labeling method for a single R1 EB-derived cell 7 d after differentiation. The resulting paired cells display nonrandom template strand segregation. DIC pictures were captured using a 20× objective every 5 min for a total of 8–9 h in a humidified chamber at 37°C and 5% CO2 (imaging microscope station [TE2000; Nikon]).
Single cell division displaying nonrandom template strand segregation. This video is related to Fig. 3 b. Time-lapse of the second cell division of a paired-cell assay in combination with the CldU/IdU double-labeling method for a single R1 EB-derived cell 7 d after differentiation. The resulting paired cells display nonrandom template strand segregation. DIC pictures were captured using a 20× objective every 5 min for a total of 8–9 h in a humidified chamber at 37°C and 5% CO2 (imaging microscope station [TE2000; Nikon]).
Single cell division displaying nonrandom template strand segregation. This video is related to Fig. 3 b. Time-lapse of the second cell division of a paired-cell assay in combination with the CldU/IdU double-labeling method for a single R1 EB-derived cell 7 d after differentiation. The resulting paired cells display nonrandom template strand segregation. DIC pictures were captured using a 20× objective every 5 min for a total of 8–9 h in a humidified chamber at 37°C and 5% CO2 (imaging microscope station [TE2000; Nikon])
Single cell division displaying nonrandom template strand segregation. This video is related to Fig. 3 b. Time-lapse of the second cell division of a paired-cell assay in combination with the CldU/IdU double-labeling method for a single R1 EB-derived cell 7 d after differentiation. The resulting paired cells display nonrandom template strand segregation. DIC pictures were captured using a 20× objective every 5 min for a total of 8–9 h in a humidified chamber at 37°C and 5% CO2 (imaging microscope station [TE2000; Nikon])
Single cell division displaying nonrandom template strand segregation. This video is related to Fig. 3 b. Time-lapse of the second cell division of a paired-cell assay in combination with the CldU/IdU double-labeling method for a single R1 EB-derived cell 7 d after differentiation. The resulting paired cells display nonrandom template strand segregation. DIC pictures were captured using a 20× objective every 5 min for a total of 8–9 h in a humidified chamber at 37°C and 5% CO2 (imaging microscope station [TE2000; Nikon])
Single cell division displaying nonrandom template strand segregation. This video is related to Fig. 3 b. Time-lapse of the second cell division of a paired-cell assay in combination with the CldU/IdU double-labeling method for a single R1 EB-derived cell 7 d after differentiation. The resulting paired cells display nonrandom template strand segregation. DIC pictures were captured using a 20× objective every 5 min for a total of 8–9 h in a humidified chamber at 37°C and 5% CO2 (imaging microscope station [TE2000; Nikon])
Time-lapse to assess NRTS. Paired-cell assays were performed on R1-derived EBs 7 d after differentiation using the CldU/IdU double-labeling method. Cells were plated at clonal density, and the second division was followed by time-lapse microscopy. Cells were immediately fixed and immunostained for CldU and IdU. Differential interference contrast (DIC) images and DAPI nuclear counterstaining are shown. (a and b) Representative photographs of symmetric (a) and asymmetric (b) daughter cells are displayed, and CldU fluorescence intensity (normalized to IdU intensity) distribution among daughter cells is shown. (c) Photographs of the first time-lapse image acquired for Videos 6 and 9 unambiguously showed that a single cell was plated. Time-lapse videos corresponding to each set of photographs presented in a and b are specified in the right column.
Time-lapse to assess NRTS. Paired-cell assays were performed on R1-derived EBs 7 d after differentiation using the CldU/IdU double-labeling method. Cells were plated at clonal density, and the second division was followed by time-lapse microscopy. Cells were immediately fixed and immunostained for CldU and IdU. Differential interference contrast (DIC) images and DAPI nuclear counterstaining are shown. (a and b) Representative photographs of symmetric (a) and asymmetric (b) daughter cells are displayed, and CldU fluorescence intensity (normalized to IdU intensity) distribution among daughter cells is shown. (c) Photographs of the first time-lapse image acquired for Videos 6 and 9 unambiguously showed that a single cell was plated. Time-lapse videos corresponding to each set of photographs presented in a and b are specified in the right column.
Moreover, using time-lapse microscopy, we provided yet another evaluation of the artifacts of our experimental system, which were found to be low (and similar to the aforementioned levels). Namely, R1 cells were plated at a similar density to the one used in Fig. 2 i, and out of the cells displaying a paired-cell pattern, only 5% showed NRTS artificially and were nondaughter cells. Subtracting this 5% error, we found a good correlation between the frequency of NRTS obtained using conventional and time-lapse microscopy experiments: 36 ± 5% of R1 cells displayed NRTS by conventional microscopy (Fig. 2 i) compared with 28.3% of cells directly observed using time-lapse videos. These data are the first to directly quantify the experimental error of the paired-cell assay, to validate this experimental setup for studying NRTS phenomena, and to directly demonstrate robust NRTS in differentiating EBs.
NRTS is associated with asymmetric cell fate
NRTS has been associated with cell fate in yeast (Klar, 1987, 1990), and in nearly all previous studies on NRTS in stem cells, the self-renewing daughter cell inherits the “old” template DNA, whereas the differentiating daughter cell inherits the neotemplate strands (Potten et al., 1978; Shinin et al., 2006; Conboy et al., 2007; Karpowicz et al., 2009). During multilineage differentiation, ESCs give rise to differentiated progenitors of the three primary embryonic germ layers: ectoderm, mesoderm, and endoderm. To address whether NRTS is associated with cell fate in this experimental system, we examined H9 EB-differentiated paired cells exhibiting both random (symmetric BrdU) and nonrandom (asymmetric BrdU) segregation of their DNA for the expression of the mesoderm marker Brachyury (Bry) or the primitive endoderm marker GATA4. After 7 d of differentiation, these cells have lost expression of pluripotency markers such as Oct4 (unpublished data).
Consistent with the percentages of NRTS described in Fig. 2 (d and i), we observed that 62% (in the Bry costaining experiments) and 54% (in the GATA4 costaining experiments) of the pairs showed asymmetric BrdU 7 d after differentiation. Of the pairs showing asymmetric inheritance of both BrdU and the differentiation marker, Bry was observed in the BrdU-positive cell in 82 ± 5% and GATA4 in 93 ± 7% of the pairs (Fig. 4, a [bottom] and c). Similar results were seen with the endodermal marker HNF4 (unpublished data). These results clearly demonstrate a strong correlation between asymmetric inheritance of the neotemplate DNA and asymmetry of the daughter cell phenotype. Interestingly, a significant fraction of pairs, 34% (Bry) and 35% (GATA4), were asymmetric for the BrdU-labeled DNA and symmetric for these markers (Fig. 4 c), suggesting that as observed during muscle (Conboy et al., 2007) or neural (Karpowicz et al., 2005) differentiation, DNA template age asymmetry correlates well with marker asymmetry but not necessarily the other way around. Of the pairs exhibiting symmetric BrdU, the majority (over 80%) also showed symmetric expression of Bry or GATA4 (Fig. 4, a [top] and b), suggesting that these cells symmetrically expand as progenitors of mesodermal and endodermal lineages. Overall and consistent with previous studies, we found a direct high correlation between asymmetric inheritance of the neotemplate DNA strand and the differentiated phenotype.
Asymmetric template strand segregation is associated with asymmetric cell fate. (a) Paired-cell assays were performed on H9-derived EBs 7 d after differentiation using the BrdU single-labeling method. Cell pairs were immunostained for BrdU and the mesoderm marker Brachyury (Bry) or the endoderm marker Gata4. Representative photographs of paired cells symmetric (top) or asymmetric (bottom) for BrdU and expressing Bry or Gata4 are shown. (b and c) Data from experiments as in a were quantified. (b) Paired cells symmetric for BrdU were distinguished based on Bry (top) or Gata4 (bottom) expression pattern: symmetric and coincident with BrdU or asymmetric. (c) Paired cells asymmetric for BrdU were distinguished based on Bry (top) or Gata4 (bottom) expression pattern: symmetric, asymmetric, and coincident with BrdU or asymmetric and mutually exclusive with BrdU. More than 100 pairs were quantified for Bry and GATA4 for each experiment. Data represent means ± SEM (n = 3 independent experiments). BF, bright field. **, P < 0.01; ***, P < 0.001.
Asymmetric template strand segregation is associated with asymmetric cell fate. (a) Paired-cell assays were performed on H9-derived EBs 7 d after differentiation using the BrdU single-labeling method. Cell pairs were immunostained for BrdU and the mesoderm marker Brachyury (Bry) or the endoderm marker Gata4. Representative photographs of paired cells symmetric (top) or asymmetric (bottom) for BrdU and expressing Bry or Gata4 are shown. (b and c) Data from experiments as in a were quantified. (b) Paired cells symmetric for BrdU were distinguished based on Bry (top) or Gata4 (bottom) expression pattern: symmetric and coincident with BrdU or asymmetric. (c) Paired cells asymmetric for BrdU were distinguished based on Bry (top) or Gata4 (bottom) expression pattern: symmetric, asymmetric, and coincident with BrdU or asymmetric and mutually exclusive with BrdU. More than 100 pairs were quantified for Bry and GATA4 for each experiment. Data represent means ± SEM (n = 3 independent experiments). BF, bright field. **, P < 0.01; ***, P < 0.001.
NRTS is dependent on DNA methylation and de novo Dnmts
The molecular mechanisms by which cells recognize and differentiate sister chromatids to direct NRTS during mitosis remain unknown. To test the dependence of NRTS on epigenetic differences between sister chromatids, we screened inhibitors of chromatin modification enzymes for ones that alter the occurrence of NRTS in human and mouse EBs. Dnmt inhibitors 5-azacytidine (AZA) and RG108 interfere with DNA methylation through different mechanisms and erase the epigenetic DNA markings (Brueckner et al., 2005; Lyko and Brown, 2005). These were used throughout both ESC and EB culture at doses that maintained cell viability (Fig. S2, a and b) and did not perturb EB formation (Fig. S1 a) or the cell cycle progression (Fig. S1, b and c). Both inhibitors robustly (by 33 and 40%, respectively) decreased the occurrence of NRTS in H9-derived EBs 7 d after differentiation induction as compared with control (DMSO treated) cells (Fig. 5 a). Interestingly, the histone deacetylase inhibitor trichostatin A (TSA) had no effect on NRTS frequency. Similar results were obtained using R1-derived EBs with a significant reduction of NRTS frequency by 32 and 31% after treatment with AZA or RG108, respectively, and no significant attenuation of NRTS by TSA (Fig. 5 a). These results are the first to demonstrate that NRTS is regulated epigenetically and is directly dependent on DNA methylation and independent of histone acetylation.
Epigenetic regulation of NRTS by DNA methylation and de novo Dnmts. (a) H9 hESCs and R1 mESCs were exposed to 1 nM AZA, 50 µM RG108, or 10 nM TSA during proliferation and differentiation. Paired-cell assays were performed on EBs 7 d after differentiation, and cell pairs were immunostained for CldU and IdU and analyzed using the double-labeling method as described in Fig. 2. (b) Quantification of the occurrence of NRTS in Dnmt-null mESCs using the CldU/IdU double-labeling method. Paired-cell assays were performed on EBs 7 d after differentiation using J1 (WT), Dnmt1-null (Dnmt1−/−), Dnmt3a-null (Dnmt3a−/−), Dnmt3b-null (Dnmt3b−/−), and Dnmt3a-Dnmt3b double-null (Dnmt3a-3b−/−) mESCs. A minimum of 100 pairs were analyzed per condition per experiment, and the percentage of cells showing asymmetric CldU (i.e., NRTS) was quantified (all cells had symmetric IdU). Data represent means ± SEM (n = 3 independent experiments). *, P < 0.05; **, P < 0.01; ***, P < 0.001 versus DMSO control (a) or versus WT mESCs (b). (c) Paired-cell assays were performed on H9-derived EBs 7 d after differentiation using the BrdU single-labeling method. Cell pairs were immunostained for BrdU and Dnmt1 (top), Dnmt3a (middle), or Dnmt3b (bottom). Representative photographs of paired cells symmetric or asymmetric for BrdU and expressing Dnmt1, Dnmt3a, or Dnmt3b are shown. (d and e) Data from experiments shown in c were quantified. (d) Paired cells symmetric for BrdU were distinguished based on Dnmt1 (top), Dnmt3a (middle), or Dnmt3b (bottom) expression pattern: symmetric and coincident with BrdU or asymmetric. (e) Paired cells asymmetric for BrdU were distinguished based on Dnmt1 (top), Dnmt3a (middle), or Dnmt3b (bottom) expression pattern: symmetric, asymmetric, and coincident with BrdU or asymmetric and mutually exclusive with BrdU. More than 100 pairs were quantified for Dnmt1, Dnmt3a, and Dnmt3b for each experiment. Data represent means ± SEM (n = 3 independent experiments). ***, P < 0.001. (f) Dnmt1, 3a, and 3b fluorescence intensity distribution among daughter cells from H9-derived EB pairs experiment. IgG background immunofluorescence was subtracted from the Dnmts immunofluorescence, and the quantification of immunofluorescence intensity of representative visually symmetric (with a distribution from equal to 60:40, as indicated) and visually asymmetric pairs of cells are shown in the scatter graph (n = 60 pairs for Dnmt1, n = 34 pairs for Dnmt3a, and n = 54 pairs for Dnmt3b representative of three independent experiments). BF, bright field.
Epigenetic regulation of NRTS by DNA methylation and de novo Dnmts. (a) H9 hESCs and R1 mESCs were exposed to 1 nM AZA, 50 µM RG108, or 10 nM TSA during proliferation and differentiation. Paired-cell assays were performed on EBs 7 d after differentiation, and cell pairs were immunostained for CldU and IdU and analyzed using the double-labeling method as described in Fig. 2. (b) Quantification of the occurrence of NRTS in Dnmt-null mESCs using the CldU/IdU double-labeling method. Paired-cell assays were performed on EBs 7 d after differentiation using J1 (WT), Dnmt1-null (Dnmt1−/−), Dnmt3a-null (Dnmt3a−/−), Dnmt3b-null (Dnmt3b−/−), and Dnmt3a-Dnmt3b double-null (Dnmt3a-3b−/−) mESCs. A minimum of 100 pairs were analyzed per condition per experiment, and the percentage of cells showing asymmetric CldU (i.e., NRTS) was quantified (all cells had symmetric IdU). Data represent means ± SEM (n = 3 independent experiments). *, P < 0.05; **, P < 0.01; ***, P < 0.001 versus DMSO control (a) or versus WT mESCs (b). (c) Paired-cell assays were performed on H9-derived EBs 7 d after differentiation using the BrdU single-labeling method. Cell pairs were immunostained for BrdU and Dnmt1 (top), Dnmt3a (middle), or Dnmt3b (bottom). Representative photographs of paired cells symmetric or asymmetric for BrdU and expressing Dnmt1, Dnmt3a, or Dnmt3b are shown. (d and e) Data from experiments shown in c were quantified. (d) Paired cells symmetric for BrdU were distinguished based on Dnmt1 (top), Dnmt3a (middle), or Dnmt3b (bottom) expression pattern: symmetric and coincident with BrdU or asymmetric. (e) Paired cells asymmetric for BrdU were distinguished based on Dnmt1 (top), Dnmt3a (middle), or Dnmt3b (bottom) expression pattern: symmetric, asymmetric, and coincident with BrdU or asymmetric and mutually exclusive with BrdU. More than 100 pairs were quantified for Dnmt1, Dnmt3a, and Dnmt3b for each experiment. Data represent means ± SEM (n = 3 independent experiments). ***, P < 0.001. (f) Dnmt1, 3a, and 3b fluorescence intensity distribution among daughter cells from H9-derived EB pairs experiment. IgG background immunofluorescence was subtracted from the Dnmts immunofluorescence, and the quantification of immunofluorescence intensity of representative visually symmetric (with a distribution from equal to 60:40, as indicated) and visually asymmetric pairs of cells are shown in the scatter graph (n = 60 pairs for Dnmt1, n = 34 pairs for Dnmt3a, and n = 54 pairs for Dnmt3b representative of three independent experiments). BF, bright field.
Three Dnmts are responsible for the global DNA methylation of mammalian cells: one maintenance Dnmt (Dnmt1) and two de novo Dnmts (Dnmt3a and Dnmt3b; Li et al., 1992; Okano et al., 1999). To address which Dnmt enzymes are involved in the regulation of NRTS, we used Dnmt-null mESCs (Okano et al., 1999). Paired-cell assays were performed using CldU/IdU double labeling 7 d after differentiation as EBs for J1 (wild type [WT]), Dnmt1-null (Dnmt1−/−), Dnmt3a-null (Dnmt3a−/−), Dnmt3b-null (Dnmt3b−/−), and Dnmt3a-Dnmt3b double-null (Dnmt3a-3b−/−) mESCs (Fig. S3). Cell cycle analysis was performed 7 d after differentiation as EBs and no significant differences were observed between WT and mutant cells, ensuring proper labeling of DNA templates (Fig. S4 a and Table S1). The occurrence of NRTS in WT J1-derived EBs was similar to that observed using R1-derived EBs with 38% of the cells nonrandomly segregating almost all of their chromosomes (Figs. 2 i and 5 b). No significant difference in the frequency of NRTS was observed in single-null mutant mESCs, Dnmt1−/−, Dnmt3a−/−, or Dnmt3b−/−, as compared with WT mESCs, although a slight reduction was noticed for each. Conversely, Dnmt3a-3b−/− mESCs showed a dramatic decrease in the occurrence of NRTS: only 15% of paired cells segregated their chromosomes nonrandomly, a 2.5-fold decrease as compared with WT cells (Fig. 5 b). These results suggest that the regulation of NRTS is mediated by de novo Dnmts and likely by de novo methylation. These data are consistent with the known redundancy in function of Dnmt3a and Dnmt3b (Okano et al., 1999) and substantiate our data on the decline in NRTS upon AZA treatment, as the effect of AZA is primarily mediated by de novo Dnmts in mESCs (Oka et al., 2005). In directed differentiation assays, Dnmt mutant cells display increased apoptosis and defective differentiation abilities (Lei et al., 1996; Panning and Jaenisch, 1996; Jackson et al., 2004; Sakaue et al., 2010). However, Dnmt mutant ESCs effectively form EBs, and no difference in apoptosis was observed between WT and mutant cells 7 d after EB differentiation (Figs. S4 b and S2 c). With respect to differentiation, early mesodermal lineage was not affected in Dnmt-null mutants as shown by the mRNA expression of Bry and Wilms tumor gene 1, whereas the expression of Nanog, a pluripotent cell marker, was similarly down-regulated in Dnmt-null mutants. Interestingly, we observed a declined expression of primitive endodermal markers Gata4 and Sox17 in all four Dnmt mutants when compared with WT and an increased expression of primitive ectodermal lineage markers Pax6 and Sox1 in Dnmt3b−/− and Dnmt3a-3b−/− ESC-derived EBs (Fig. S4 c). Because NRTS is altered in Dnmt3a-3b−/− cells, these data further support that NRTS is linked to differentiation.
To extrapolate these data and to further investigate the role of Dnmt enzymes in NRTS, we examined paired cells exhibiting both random (symmetric BrdU) and nonrandom (asymmetric BrdU) segregation of their DNA for the expression of Dnmt1, Dnmt3a, or Dnmt3b. As described for the CldU, the visually asymmetric immunofluorescent signals observed for Dnmts in representative images was 95:5 to 70:30 for Dnmt1 or Dnmt3b. Interestingly, the Dnmt3b asymmetric pairs clustered in two groups: those for which fluorescence intensity ratios between the two daughter cells are within 70:30 to 80:20 (i.e., both daughter cells inherit a significant amount of Dnmt3b, even though asymmetry is observable) and a second cluster for which the fluorescence intensity ratios are greater than or equal to 95:5 between the two daughter cells and virtually all Dnmt3b is localized into one daughter cell (Fig. 5 f). In contrast, Dnmt3a was prominently asymmetric with fluorescence intensity ratios greater than 85:15. These data suggest that asymmetry of Dnmt3a is more prominent than that of Dnmt3b, and based on our data, a knockout of both Dnmt3a and 3b is required for the reduction of NRTS (Fig. 5 b). Immunofluorescent signals corresponding to symmetric events were in the 60:40 range (Fig. 5 f). Because every cell relies on de novo DNA methylation and also needs to maintain an existing epigenome, a 100:0 distribution of Dnmts in asymmetrically dividing cells may not be expected.
Among pairs exhibiting symmetric BrdU, the majority showed symmetric expression of Dnmt1 (84%), Dnmt3a (82%), or Dnmt3b (93%), demonstrating maintenance and de novo methylation of DNA in cells that divide with random segregation of DNA strands (Fig. 5, c and d). Consistent with the idea that de novo DNA methylation is required for the acquisition of a new cell fate, of the pairs asymmetric for both BrdU and Dnmt3, Dnmt3a was observed in the BrdU+ cell in 82 ± 6% of the pairs and Dnmt3b in 92 ± 2% of them (Fig. 5, c and e). The majority (67%) of pairs asymmetric for BrdU were symmetric for Dnmt1 (Fig. 5, c and e). In addition, among pairs both asymmetric for BrdU and Dnmt1, 49 ± 11% expressed Dnmt1 in the BrdU+ daughter cell (Fig. 5, c and e). These results demonstrate the lack of correlation between Dnmt1 and BrdU-labeled strand (younger chromatid) segregation and are consistent with the results obtained using Dnmt1-null mESCs (Fig. 5 b). Of note, among pairs with asymmetric BrdU, significant fractions were symmetric for Dnmt3a or Dnmt3b (41 and 26%, respectively; Fig. 5 e). Collectively, these results are the first to establish that the de novo Dnmts preferentially colocalize with neotemplate DNA strands during asymmetric cell divisions.
Discussion
The results of this work establish the robust evolutionarily conserved phenomenon of NRTS in both hESCs and mESCs undergoing multilineage differentiation into the three germ layers. The time-lapse microscopy provides a direct observation of NRTS in differentiating EBs, which in combination with the high paired-cell numbers quantified throughout the study for both species (≈6,000 for hESCs and 3,000 for mESCs) and the consistent data obtained between the mesodermal and endodermal cell fate determination substantiate the existence of the NRTS process.
Some recent studies have shown a phenomenon, perhaps related to NRTS, in which only part of the chromatids are sorted nonrandomly (Armakolas and Klar, 2006; Falconer et al., 2010). However, as shown by quantifications of fluorescence intensities (Fig. 2 g), in one third of dividing ESCs undergoing multilineage differentiation as EBs, most if not all of the chromatids in these cells must segregate asymmetrically. When NRTS of apparently all thymidine analogue–labeled chromosomes has been demonstrated and subsequently reanalyzed at the single-chromosome resolution using chromosome orientation FISH, both methods of analysis have shown complete agreement (Falconer et al., 2010; Rocheteau et al., 2012). The detection of thymidine analogues shown herein confidently predicts that a similar outcome would be achieved by single-chromatid resolution study and offers an additional option of dual labeling (with CldU and IdU), which provides the assurance that two consecutive cell cycles were analyzed.
In an effort to track nuclei during time-lapse microscopy, we added live Hoechst (Hoechst 33342) during the acquisition of videos at concentrations ranging from 2 µM to 100 nM. Unfortunately, it resulted in a complete abolition of cell division even at the lowest concentration (unpublished data). These data reveal that ESCs are poorly tolerant to alteration of their chromatin structure. A similar observation was made when Dnmt inhibitors and TSA concentrations were tested. ESCs showed much higher sensitivity to these drugs when compared with other cells, which resulted in cell death.
Although the number of artificial daughter cells was low in our sparsely plated cultures of the paired-cell assays, we have also observed that with higher plating density, the doublet artifacts increase because of enhanced cell mobility. Therefore, in addition to the precisely measured cell cycle timing, the correct plating density of examined cells is critically important for detecting and studying NRTS. Interestingly, the 5% NRTS artifact revealed by time-lapse experiments agrees very well with the 4.5 and 4.7% NRTS observed in undifferentiated R1 mESCs and in the non–stem cell control IMR-90 cells, respectively (Fig. 2 i).
DNA repair pathways involving chromatid exchange such as homologous recombination might introduce a bias in the interpretation of the data on NRTS. However, recent studies on eukaryotic cell Holliday junctions, four-way intermediate DNA structures formed before recombination, indicate that they represent a minor pathway of double-strand break repair in mitotic cells as opposed to the major contributor of meiotic recombination. Moreover, when Holliday junctions form in mitotic cells they are often dissolved to prevent crossover outcomes (Bzymek et al., 2010; Liberi and Foiani, 2010). At any rate, recombination would serve to randomize the distribution of thymidine analogue–labeled strands, which is in contrast to the nonrandom phenomenon that we observe. Therefore, it is unlikely that chromatid exchange explains the observed detection of thymidine analogues between asymmetric daughter cells in our experimental system.
With respect to the molecular mechanism, both pharmacological and genetic inhibition of DNA methylation, which disturbed the WT epigenetic DNA profile, negated the NRTS phenomenon. This confirmed the observation of NRTS in the first place and strongly suggested a dependence of this asymmetry on the epigenetic DNA modifications that were produced by the redundant activities of Dnmt3a and Dnmt3b. These data correlate very well with the known functions of Dnmt3a and Dnmt3b that are responsible for the wave of de novo methylation during early embryogenesis, which establishes the somatic methylation pattern (Okano et al., 1999). The dependence of NRTS on both Dnmt3a and 3b, collectively with the differential degrees of asymmetry of these de novo Dnmts in dividing EB-derived cells, summarily suggest that the molecular process of NRTS is closely linked to Dnmt3a asymmetry, and the redundant function of Dnmt3b may even be to maintain NRTS when Dnmt3a becomes lacking. In the future, it would be interesting to examine whether the asymmetry of Dnmt3b becomes more pronounced in the Dnmt3a mutant ESCs that undergo multilineage differentiation as EBs and display NRTS. In alignment with our findings, a recent elegant study showed that sister chromatid segregation of X and Y chromosomes during Drosophila melanogaster male germline stem cell asymmetric division was randomized in Dnmt2 mutants (Yadlapalli and Yamashita, 2013), therefore suggesting a conserved requirement of Dnmts in the NRTS phenomenon across species. Interestingly, Yadlapalli and Yamashita (2013) also had results showing NRTS of autosomes (Sauer et al., 2013; Yadlapalli and Yamashita, 2013).
Injection of early passage Dnmt3a-3b–null ESCs, which display mild genomic demethylation, results in smaller teratomas than WT cells, and injection of late passage Dnmt3a-3b–null ESCs, which display severe genomic demethylation, failed to form teratomas in nude mice (Chen et al., 2003). These data are in complete agreement with our conclusions that deficiency in NRTS that is caused by defective DNA methylation in Dnmt3a-3b–null cells (as we show) affects cell-fate determination and teratoma formation (Chen et al., 2003).
Although previous studies suggested that Dnmt mutant ESCs display defective differentiation potential and enhanced apoptosis upon differentiation (Lei et al., 1996; Panning and Jaenisch, 1996; Jackson et al., 2004; Sakaue et al., 2010), these phenotypes were severe only after prolonged in vitro culture (passage 30 or more) concomitant with the progressive diminution of methylation pattern occurring with passage (Chen et al., 2003; Jackson et al., 2004). In the present study, the Dnmt mutant cells were used for paired cell assays, apoptosis assays, and cell cycle analysis between passages 13 and 30. In our differentiation conditions, we observed no difference in apoptosis between WT cells and Dnmt mutant cells 7 d after differentiation. This discrepancy could be explained either by the difference in differentiation protocols used or by the timing after differentiation at which apoptosis was assessed. It appears that in most studies, enhanced apoptosis was observed later on during differentiation in vitro or in vivo after teratoma formation (Panning and Jaenisch, 1996; Chen et al., 2003; Sakaue et al., 2010). With respect to differentiation, our data are in alignment with previous findings because differentiation of Dnmt mutant cells was partly altered. The concomitant decline of NRTS occurrence and altered differentiation potential of Dnmt mutants reinforce the idea that NRTS strongly correlates with differentiation. However, the dramatic decline in primitive endodermal lineage marker expression and the increase of primitive ectodermal lineage marker expression in the Dnmt3a-3b−/− mutant suggest that in EBs at this time, NRTS is linked to endodermal differentiation.
Proliferation rate determination is crucial for NRTS assessment. Dnmt inhibitors are known to be cytotoxic and to perturb the cell cycle (Lyko and Brown, 2005; Gravina et al., 2010). However, such effects were observed at much higher concentrations than the ones used in this study, explaining why apoptosis or cell cycle progression was not affected versus control-treated cells. Similarly, as shown in Table S1 and Fig. S4 a, the cell cycle was not altered in Dnmt mutant ESCs either in self-renewing conditions or in differentiation conditions. Previous studies suggested that Dnmt mutant cells display no difference in proliferation when self-renewing but showed a declined proliferation rate during differentiation (Lei et al., 1996; Chen et al., 2003; Jackson et al., 2004; Tsumura et al., 2006). However, cell cycle was never precisely assessed during differentiation in these studies. These claims were based on data of teratoma size, proliferation curves, or growth competition assays when co-cultured with WT cells. These assays do not allow for the distinction between lack of proliferation and apoptosis; therefore, the enhanced apoptosis reported at advanced differentiation stages is likely to be accountable for the observed phenotype. The change in NRTS we observe with chemical or genetic perturbation of methylation is a change in the ratio of cells that choose asymmetric to symmetric fate upon a cell division and not a decrease or increase in the absolute number of dividing cells. Although the total number of cell division is not significantly affected in AZA-treated or Dnmt mutant cells, the proportion (or percentage) of asymmetric division is significantly reduced with the loss of methylation.
In addressing the purpose of nonrandom DNA segregation, these data suggest that the main function of NRTS in differentiating human and mouse EBs is to deposit the new, unmethylated DNA strands and de novo methylation enzymes into one daughter cell (facilitating epigenetic change to a new cell fate), while preferentially retaining the already methylated DNA template strands in the other daughter cell, thus maintaining its epigenetic memory. Future advancements upon these findings may provide new strategies for harnessing the regenerative potential of ESCs through the deliberate control of cell fate determination.
Materials and methods
Cell culture
H9 hESCs were cultured in serum-free, feeder-independent mTeSR 1 medium (STEMCELL Technologies; Ludwig et al., 2006) and differentiated as previously described (Itskovitz-Eldor et al., 2000). In summary, cells were fed daily and were passaged as small clumps using 1 mg/ml dispase (STEMCELL Technologies) followed by mechanical disruption of stem cell colonies. Clumps were then plated on Matrigel-coated dishes (30 µg/cm2 hESC-qualified matrix; BD). H9 cells were differentiated as EBs using undirected differentiation medium: DMEM/F-12 (STEMCELL Technologies) supplemented with 10% FBS (Cellgro). EBs were formed from bigger clumps of ESCs and grown in suspension on Petri dishes. EBs were formed overnight in mTeSR 1 medium and shifted the next morning to differentiation medium. All experiments were performed between passages 40 and 70.
R1 mESCs were cultured and differentiated as previously described (Nagy et al., 1993; Wobus et al., 2002) with slight differences. In brief, R1 cells were maintained on mitomycin C (Sigma-Aldrich) inactivated STO (CRL-1503; ATCC) feeder cells and fed daily with DMEM high-glucose medium supplemented with 15% KnockOut serum replacement, 2 mM GlutaMAX-I supplement, 1 mM MEM sodium pyruvate, 0.1 mM MEM nonessential amino acids (all obtained from Invitrogen), 55 µM 2-mercaptoethanol, and 1,000 U/ml ESGRO leukemia inhibitory factor (LIF; EMD Millipore). At each passage or before EB formation, R1 cells were preplated for 1 h on noncoated tissue-culture dishes to remove STO cells. R1 cells were differentiated as EBs using the hanging drop technique in maintenance medium without LIF. EBs were transferred to Petri dishes 2.5 d after formation and fed every other day.
IMR-90 human fetal fibroblasts (ATCC) were cultured following ATCC recommendations, and all experiments were performed between passages 7 and 15.
J1 WT and Dnmt-null mESCs were provided by E. Li (Novartis, Cambridge, MA) and cultured as previously described (Okano et al., 1999) except that cells were grown on gelatin-coated dishes (no feeder cells were used). In brief, undifferentiated ESC were grown in DMEM high-glucose medium supplemented with 15% ESC Qualified FBS, 2 mM GlutaMAX-I supplement, 1 mM MEM sodium pyruvate, 0.1 mM MEM nonessential amino acids (all obtained from Invitrogen), 55 µM 2-mercaptoethanol, and 1,000 U/ml ESGRO LIF. Cells were differentiated as EBs using the hanging drop technique in growth medium without LIF. EBs were transferred to Petri dishes 2.5 d after formation and fed every other day. All experiments were performed between passages 13 and 30.
AZA, RG-108, and TSA were purchased from Sigma-Aldrich and used at final concentrations of 1 nM, 50 µM, and 10 nM, respectively. Stock solutions (dissolved in DMSO) were kept as aliquots at −80°C and freshly diluted into maintenance or differentiation medium upon cell feeding.
Thymidine analogues and labeling
BrdU, CldU, and IdU were purchased from Sigma-Aldrich and used as described in Table S1 to perform single-BrdU or double-CldU/IdU labeling. Labeling duration was optimized for each cell type according to their specific population doubling time. Thymidine analogue and antibody final concentrations were chosen to minimize cross-reactivity of anti-CldU and anti-IdU primary antibodies (Table S1).
Paired-cell assay to assess DNA strand NRTS
At different days after differentiation induction, ESC-derived EBs were labeled with thymidine analogues (BrdU for single- or CldU for double-labeling methods) as described in Table S1. During the second round of cell division (corresponding to IdU labeling for the double-labeling method), EBs were dissociated as single cells by trypsin digestion. Single cells were then plated at very low density (≈10–20 cells/mm2) onto Lab-Tek II 4-well chamber slides coated with Matrigel (30 µg/cm2 hESC-qualified matrix) to accomplish their second round of cell division and form paired cells. Because hESCs exhibit poor survival after cellular dissociation (Watanabe et al., 2007), they were plated at higher cell density (≈100–200 cells/mm2). Paired cells were fixed with 4% PFA, immunostained, and scored as paired cells if they were within one cell diameter from each other and 50 cell diameters away from other cells. The number of cell pairs scored per experiment and the number of replicates are described in the figure legends.
Time-lapse microscopy
Paired-cell assays were performed on R1 mESC-derived EBs using a CldU/IdU double-labeling method. 7 d after differentiation, EBs were dissociated as single cells and plated at clonal density (100 cells/cm2) in differentiation medium onto Lab-Tek II 2-well chamber slides coated with Matrigel in the presence of IdU to accomplish their second round of cell division. Cells were allowed to attach for 2 h. Nonadherent and dead cells were washed away, and cells were staged for live-cell imaging in a humidified chamber at 37°C and 5% CO2 connected to a imaging microscope station (TE2000; Nikon). 20× differential interference contrast (DIC) images were captured every 5 min for a total of 8–9 h, and videos were made using the NIS elements advanced software (Nikon). Cells typically ball up before cytokinesis. A slight change in cell shape caused by the fixation procedure for immunofluorescence is at times noticeable between the last DIC image of the time lapse and its fluorescent image after staining.
Population doubling time determination
For each cell line used in the study, proliferation curves of undifferentiated (self-renewing) cells were performed. Cells were plated at 18,000 cells/cm2 in growth medium for R1, J1, Dnmt mutant, and IMR-90 cells. For H9 hESCs, small clumps were plated. Cell numbers were determined at the indicated times after initial plating, and population doubling times were determined using an online doubling time calculator (http://www.doubling-time.com/compute.php).
Cell cycle analysis
7 d after differentiation, EBs were pulsed with BrdU for 2 h. BrdU was diluted out of EB culture by performing three PBS washes before the addition of fresh differentiation medium. At the indicated time after BrdU removal, EBs were dissociated into single cells fixed with 70% ethanol at 4°C for 30 min and stored at −20°C overnight. Cells were stained with an anti–BrdU-FITC antibody (BD) and 10 µg/ml propidium iodide. Data were acquired on a flow cytometer (Guava easyCyte; EMD Millipore) using Guava InCyte software (EMD Millipore). The percentage of BrdU-positive cells in G0/G1, S, or G2/M phase of the cell cycle was determined using FlowJo (Tree Star) software. Cell cycle length was ascertained as the time required for BrdU-positive cells to complete the next G1 phase.
RNA isolation and quantitative RT-PCR
RNA isolation was performed using an RNeasy kit (QIAGEN) according to manufacturer’s recommendations. 2 µg RNA was reverse transcribed in 20 µl using the RT2 First Strand kit (QIAGEN) according to the manufacturer’s recommendations. Real-time quantitative PCR was performed using 10 µl cDNA diluted 25 times for a 25-µl reaction and RT2 SYBR Green Quantitative PCR Mastermix (QIAGEN). Quantitative PCR was run on a cycler (iQ5; Bio-Rad Laboratories), and data were analyzed by iQ5 optical system software. Values were normalized against the internal control GAPDH and were plotted using the ΔΔcycle threshold method.
Apoptosis analysis
7 d after differentiation, EBs were dissociated into single cells as described in the Paired-cell assay to assess DNA strand NRTS section. Cellular apoptosis and death were measured using a Dead Cell Apoptosis kit with Annexin-V Alexa Fluor 488 and Propidium Iodide for Flow Cytometry (Invitrogen) according to the manufacturer’s recommendations. Data were acquired on a flow cytometer (Epics XL; Beckman Coulter) and analyzed using FlowJo software.
Fluorescence quantification
Fluorescence quantification of daughter cells was performed using ImageJ (National Institutes of Health). All images were converted to 8-bit grayscale before quantification. DAPI images were autothresholded using the dark background default method to identify nuclei, and ImageJ’s watershed algorithm was run to separate nuclei in contact with one another. Particle analysis was then performed, with intensity measurement redirected to the channel of interest. The intensity fraction of each cell was then computed and graphed against the intensity fraction of the other cell in the pair. IgG control background fluorescence intensity was subtracted from CldU and IdU fluorescence intensities. CldU measurements were then normalized to IdU by dividing each CldU intensity measurement by the IdU intensity measurement of the cell. Some images with nuclei that were too close or too faint to be distinguished by the autothresholding algorithm were thresholded manually. All other image processing steps performed on these images were identical to those performed on the autothresholded images.
Antibodies and immunostaining
Mouse antibody clone B44 recognizing IdU (and also BrdU) was obtained from BD and used at 0.5 µg/ml; rat antibody clone BU1/75 (ICR1) recognizing CldU (and also BrdU) was obtained from Novus Biologicals and used at 1 µg/ml for CldU staining and at 5 µg/ml for BrdU staining. Rabbit polyclonal antibodies to Bry (ab20680) and GATA4 (ab61767) were obtained from Abcam and used at 2 µg/ml and 5 µg/ml, respectively. Rabbit polyclonal antibodies to Dnmt1 (ab19905) and Dnmt3a (ab4897) were purchased from Abcam and used at 5 µg/ml. Mouse monoclonal antibody to Dnmt3b (ab13604) was obtained from Abcam and used at 2.5 µg/ml. All primary antibodies were used overnight at 4°C. Secondary antibody staining was performed at room temperature for 2 h. Images were acquired using a microscope (Axio Imager A1; Carl Zeiss), a 20×/0.5 NA EC Plan Neofluar objective lens (Carl Zeiss), a camera (AxioCam MRm; Carl Zeiss), and the AxioVision software (Carl Zeiss).
BrdU staining for flow cytometry analysis was performed using FITC-conjugated mouse anti-BrdU antibody (clone B44) obtained from BD at 1.25 µg/ml. FITC-conjugated mouse IgG1, κ isotype control for flow cytometry was purchased from BD.
Statistics
Quantified data are expressed as means ± SEM. A minimum of three replicates was performed for each described experimental condition. Significant differences between values obtained in different conditions were determined using the two-tailed unpaired Student’s t test. P-values of <0.05 were assigned significant.
Online supplemental material
Fig. S1 documents EB morphology and cell cycle analysis of human and mouse EB-derived cells cultured in the presence of Dnmt inhibitors. Fig. S2 shows apoptosis analysis of human and mouse EB-derived cells exposed to Dnmt inhibitor and Dnmt-null EB-derived cells. Fig. S3 shows examples of Dnmt-null paired cells displaying random or NRTS. Fig. S4 document EB morphology, cell cycle analysis, and differentiation ability of Dnmt-null ESCs. Table S1 summarizes the timing and duration of the thymidine analogues for each cell line used to assess NRTS. Videos 1–10 show single-cell division time-lapse videos of cells displaying random template segregation (Videos 1–4) or NRTS (Videos 5–10).
Acknowledgments
We thank Dr. En Li for providing Dnmt-null ESCs. We thank Andrea Tham, Sunny Kung, and Steven Hendler for their technical help, Eric Jabart for careful reading of the manuscript, and Mary West and the California Institute for Regenerative Medicine (CIRM)/QB3 shared Stem Cell Facility at the University of California, Berkeley, for use of equipment.
This work was supported by the National Institutes of Health National Institute on Aging AG-027252 and CIRM RN1-00532 grants to I.M. Conboy and a Postdoctoral Training Fellowship from CIRM to C. Elabd.
Authors contributions: C. Elabd designed and performed all the experiments: Figs. 1, 2, 3, 4, 5, S1, S2, S3, and S4, analyzed the data, and wrote the manuscript. W. Cousin contributed to Figs. 2 (f and h), 5 (a and b), and S4 c and analyzed the data. R.Y. Chen contributed to Figs. 3, 5 (a and b), and S1 (b and c). M.S. Chooljian contributed to Figs. 2 g, 3, and 5 f. J.T. Pham contributed to Fig. 2 (c and d). I.M. Conboy designed and directed the work, performed pilot studies, interpreted the data, and wrote the manuscript. M.J. Conboy designed and directed the work, performed key preliminary experiments for Figs. 2 and 4, contributed to Figs. S1 (b and c), S2, and S4 a, interpreted the data, and wrote the manuscript.