The nucleus is a unique organelle that contains essential genetic materials in chromosome territories. The interchromatin space is composed of nuclear subcompartments, which are defined by several distinctive nuclear bodies believed to be factories of DNA or RNA processing and sites of transcriptional and/or posttranscriptional regulation. In this paper, we performed a genome-wide microscopy-based screening for proteins that form nuclear foci and characterized their localizations using markers of known nuclear bodies. In total, we identified 325 proteins localized to distinct nuclear bodies, including nucleoli (148), promyelocytic leukemia nuclear bodies (38), nuclear speckles (27), paraspeckles (24), Cajal bodies (17), Sam68 nuclear bodies (5), Polycomb bodies (2), and uncharacterized nuclear bodies (64). Functional validation revealed several proteins potentially involved in the assembly of Cajal bodies and paraspeckles. Together, these data establish the first atlas of human proteins in different nuclear bodies and provide key information for research on nuclear bodies.
Complete screen data
Introduction
The nucleus is enclosed by a double-membrane structure termed the nuclear envelope, which serves as a physical barrier to separate nuclear contents from the cytoplasm. Numerous nuclear pores exist as large protein complexes across the nuclear envelope, which allow the transport of water-soluble molecules. Interphase chromosomes occupy distinct subnuclear territories. The interchromatin space is also well organized and harbors multiple nuclear bodies that can be visualized as distinct nuclear foci at the microscopic level. To date, nuclear bodies that have been studied extensively are nucleoli, promyelocytic leukemia (PML) bodies, nuclear speckles, Cajal bodies, paraspeckles, and Polycomb bodies (Spector, 2006).
Tremendous effort has been made and allowed us to understand the distinct functions of several nuclear bodies: (a) Nucleoli are sites of ribosomal DNA transcription, preribosomal RNA processing, and preribosomal assembly. (b) Nuclear speckles may serve as storage and/or modification sites for splicing factors and sites for pre-mRNA splicing. In fact, nuclear speckles are often in close proximity to many active genes, suggesting that transcription and RNA splicing are coupled in the cell. (c) Cajal bodies are involved in the assembly and maturation of small nuclear RNPs (snRNPs; Spector, 2006). Recently, telomerase RNA and telomerase reverse transcription were also shown to localize to Cajal bodies (Zhu et al., 2004; Tomlinson et al., 2008). (d) PML bodies engage in a multitude of cellular events, including apoptosis, DNA repair, and transcription control, by sequestering, modifying, and degrading many partner proteins (Lallemand-Breitenbach and de Thé, 2010). (e) Paraspeckles are involved in nuclear retention of some A-to-I hyperedited mRNAs, and such retention is altered upon environmental stress, which provides a control mechanism for gene expression (Prasanth et al., 2005). (f) Two classes of complexes designated as PRC1 and PRC2 (Polycomb repressive complexes 1 and 2) have been found in Polycomb bodies, which are believed to collaborate to repress gene transcription through epigenetic silencing (Spector, 2006). However, despite the importance of these nuclear bodies, their compositions and regulations are still largely unknown.
There are previous attempts in identifying mammalian proteins localized to nuclear subcompartments (Sutherland et al., 2001), which also include proteomic analysis of the nucleolus (Andersen et al., 2002; Scherl et al., 2002) as well as nuclear speckles (Saitoh et al., 2004). However, an ORFeome-scale systematic approach has yet to be conducted. This is especially important for the studies of nuclear bodies because these nuclear bodies have no membrane and are difficult to isolate using traditional biochemical methods. In this study, we took advantage of the available 15,483 ORFs in the Human ORFeome Library and performed whole-genome screening for proteins localized to distinct nuclear bodies. This study allowed us to expand the inventory of components in various nuclear bodies and to construct the first nuclear body landscape.
Results
Description and validation of the nuclear foci screen
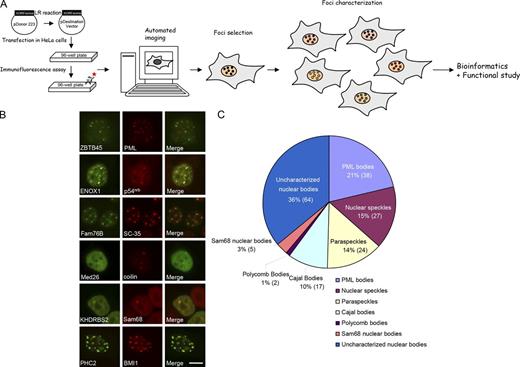
To generate a proteome of nuclear subcompartments, we subcloned the Human ORFeome v5.1 Library into a Gateway-compatible destination vector. Individual plasmid DNA was transfected into HeLa cells in a 96-well format followed by immunofluorescence staining of the tagged proteins. Fluorescent images were captured by an automated fluorescence microscope, subcellular localization of each ORF was reviewed with use of MetaXpress software (Molecular Devices), and proteins forming nuclear foci were selected for further characterization (Fig. 1 A).
Identification and characterization of proteins in various nuclear subcompartments. (A) Overall schematic flow of this protein localization screen. (B) Representative images of proteins that show colocalization with various nuclear body markers. Bar, 10 µm. (C) The pie chart shows the distribution of proteins in various nuclear subcompartments (148 nucleolar proteins were not included). A total of 325 proteins that formed nuclear foci were identified in this screen, including 148 in the nucleolus, 38 in PML bodies, 27 in nuclear speckles, 24 in paraspeckles, 17 in Cajal bodies, 5 in Sam68 nuclear bodies, 2 in Polycomb bodies, and 64 proteins in uncharacterized nuclear subcompartments (please also see Table S1).
Identification and characterization of proteins in various nuclear subcompartments. (A) Overall schematic flow of this protein localization screen. (B) Representative images of proteins that show colocalization with various nuclear body markers. Bar, 10 µm. (C) The pie chart shows the distribution of proteins in various nuclear subcompartments (148 nucleolar proteins were not included). A total of 325 proteins that formed nuclear foci were identified in this screen, including 148 in the nucleolus, 38 in PML bodies, 27 in nuclear speckles, 24 in paraspeckles, 17 in Cajal bodies, 5 in Sam68 nuclear bodies, 2 in Polycomb bodies, and 64 proteins in uncharacterized nuclear subcompartments (please also see Table S1).
To estimate the accuracy of our study, we randomly selected 36 proteins in the ORFeome library for which the antibodies recognizing endogenous proteins are available. By comparing the fluorescence intensities in the transfected and untransfected cells, we estimated that the mean level of overexpression is ∼2.35-fold of that of endogenous protein (Fig. S1). Moreover, 34/36 proteins displayed subcellular localization identical to that of endogenous protein (Fig. S1). These results suggest that the tagged proteins are only moderately expressed, and most of them exhibit proper localization as their endogenous counterparts.
To validate our screening results, we characterized the localization of these proteins that display nuclear foci by costaining with various nuclear foci or nuclear body markers (Fig. 1 B) or based on the distinct nucleolus morphology. In summary, we identified a total of 325 proteins in various nuclear bodies, which include 148 nucleolar proteins, 38 proteins in PML bodies, 27 proteins in nuclear speckles, 24 proteins in paraspeckles, 17 proteins in Cajal bodies, 5 proteins in Sam68 nuclear bodies, 2 proteins in Polycomb bodies, and 64 proteins in uncharacterized nuclear subcompartments (Fig. 1 C, Table S1, and Table S2). We also identified an additional 48 proteins that are targeted to nuclear envelope.
Next, we compared our list of nuclear body proteins to available datasets of various nuclear bodies. For nucleolar proteins, we took advantage of an available Nucleolar Proteome Database (NOPdb; version 3.0). We found that 37.2% (55 out of 148) nucleolar proteins identified in our screen overlapped with those in NOPdb. Interestingly, 29.1% (43/148) nucleolar proteins were exclusively identified in our study but not in NOPdb. More importantly, these 43 proteins have already been verified by other peer-reviewed articles (Fig. 2 and Table S2). This comparison suggests that our screening complements previous biochemical isolation of the nucleolus (Leung et al., 2006; Ahmad et al., 2009) and allows us to identify novel nucleolar proteins. For nuclear speckles, 29.6% (8/27) proteins on our list overlapped with those in the database, whereas 18.5% (5/27) nonoverlapping nuclear speckle proteins were reported elsewhere to be a nuclear speckle component (Fig. 2 and Table S3). Similarly, 13.2% (5/38) PML body proteins we identified are present in other datasets, whereas 7.9% (3/38) of the remaining PML body proteins were reported by others as components of PML bodies (Fig. 2 and Table S3). Overall, ∼40% (134/325) of the proteins on our list are known to be present and/or function in various nuclear compartments.
Comparison of our study with datasets on nuclear subcompartments. Datasets were created for each nuclear subcompartment based on online databases or recent review articles. NOPdb (Ahmad et al., 2009) was used to represent nucleolar proteins. A proteomic analysis of interchromatin granule clusters (Saitoh et al., 2004) was used to represent the nuclear speckles dataset. A PML body interactome analysis (Van Damme et al., 2010) was used to represent proteins in PML bodies. A list of Cajal body proteins from a recent review paper (Machyna et al., 2013) was used to represent proteins in Cajal bodies. A list of paraspeckle proteins from a review (Bond and Fox, 2009) was used to represent proteins in paraspeckles. Venn graphs were used to show the extent of overlapping between an available dataset (green) and our study (blue). A group of proteins that are uniquely identified in our study and have been reported by others in the literature were presented in dark blue. The other groups of proteins that are confirmed by our shRNA screen to be involved in the assembly of Cajal bodies or paraspeckles were presented in yellow. Please also see Table S2 and Table S3.
Comparison of our study with datasets on nuclear subcompartments. Datasets were created for each nuclear subcompartment based on online databases or recent review articles. NOPdb (Ahmad et al., 2009) was used to represent nucleolar proteins. A proteomic analysis of interchromatin granule clusters (Saitoh et al., 2004) was used to represent the nuclear speckles dataset. A PML body interactome analysis (Van Damme et al., 2010) was used to represent proteins in PML bodies. A list of Cajal body proteins from a recent review paper (Machyna et al., 2013) was used to represent proteins in Cajal bodies. A list of paraspeckle proteins from a review (Bond and Fox, 2009) was used to represent proteins in paraspeckles. Venn graphs were used to show the extent of overlapping between an available dataset (green) and our study (blue). A group of proteins that are uniquely identified in our study and have been reported by others in the literature were presented in dark blue. The other groups of proteins that are confirmed by our shRNA screen to be involved in the assembly of Cajal bodies or paraspeckles were presented in yellow. Please also see Table S2 and Table S3.
We also experimentally confirmed that four novel components in Cajal bodies and six in paraspeckles are, respectively, required for the assembly of Cajal bodies and paraspeckles (Fig. 2 and Table S3; please also see Fig. 4, Fig. 5, Fig. 6, and Fig. 7 for details), which indicate that many nuclear bodies are understudied and contain numerous previously unknown components. Of note, we also identified 64 proteins with uncharacterized nuclear subcompartments. Most of them (46/64) form nuclear foci of <2 µM in diameter and have more than three foci per cell (Table S4). The functional significance of these nuclear foci remains to be determined.
Bioinformatics analysis of nuclear foci proteome
We conducted a bioinformatics analysis of the nuclear foci proteome (does not include nucleolar proteins) and found that 62% (110/177) of these proteins had been categorized as nucleus-localized proteins in the Gene Ontology (GO) database (Fig. S2 A and Table S1). Many of these proteins acquire the GO functions in ENSEMBL as protein binding, DNA binding, RNA binding, and chromatin binding properties, all of which are highly relevant to close proximity of these proteins to nuclei acids (Fig. S2 B and Table S1). Our survey of GO processes revealed that about one third of the proteins were associated with regulation of the transcription process, whereas the remaining proteins were associated with RNA splicing as well as mRNA processing and splicing, indicating that many DNA and RNA processing proteins are enriched in these nuclear bodies (Fig. S2 C and Table S1). A literature search further confirmed that 20% (11/52) of proteins annotated with DNA/chromatin binding and 46% (15/33) of proteins annotated with RNA binding were experimentally validated (Table S1). In addition, top protein motifs in each nuclear subcompartment revealed by using the InterProScan database (see the Bioinformatics analysis section) were also presented (Fig. S2 D, Table S5, and Table S6).
Proteomic analysis of nuclear foci proteome
We took advantage of tandem affinity purification to isolate protein complexes that contain a randomly selected protein from the list of each nuclear subcompartment. The rationale is that if the selected protein can interact with relevant proteins at the same nuclear subcompartment, it would give us a high confidence that this is a genuine player in that subcompartment. Moreover, such proteomic analysis may also help us to identify additional components in these subcompartments, which could be missed in our initial screening because of various reasons (e.g., not present in the ORFeome library, mislocalization caused by overexpression, or limited binding partners).
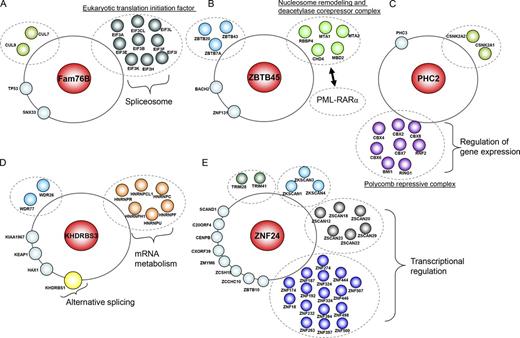
Our initial proteomic profiling revealed that the nuclear speckle–targeting protein Fam76B interacted with several eukaryotic translation initiation factors (Fig. 3 A), which were also reported in the proteomic profiling of human spliceosome (Makarov et al., 2002; Bessonov et al., 2010; Agafonov et al., 2011). However, we found that Fam76B could not interact with several pre-mRNA splicing factors, such as SRSF1, SRSF3, and Sc-35, in coimmunoprecipitation (IP; co-IP) experiments (Fig. S3 A). Therefore, Fam76B is likely not a spliceosomal component.
Interactome analysis of nuclear foci proteome. (A–E) A random selected protein from each nuclear subcompartment was used as the bait for tandem affinity purification and mass spectrometry analysis. The protein–protein interaction networks characterized for Fam76B (nuclear speckles; A), ZBTB45 (PML bodies; B), PHC2 (Polycomb bodies; C), KHDRBS3 (Sam68 nuclear bodies; D), or ZNF24 (paraspeckles; E) are presented in the cartoon (please also see Table S7).
Interactome analysis of nuclear foci proteome. (A–E) A random selected protein from each nuclear subcompartment was used as the bait for tandem affinity purification and mass spectrometry analysis. The protein–protein interaction networks characterized for Fam76B (nuclear speckles; A), ZBTB45 (PML bodies; B), PHC2 (Polycomb bodies; C), KHDRBS3 (Sam68 nuclear bodies; D), or ZNF24 (paraspeckles; E) are presented in the cartoon (please also see Table S7).
The PML body–targeting protein ZBTB45 interacted with nucleosome remodeling and deacetylase corepressor complex components (Fig. 3 B), which were known to be recruited by oncogenic PML–RAR-α to suppress target gene repression (Morey et al., 2008). Polycomb body–associated PHC2 was found to interact with the Polycomb repressive complex (Fig. 3 C), which is relevant to the function of polycomb bodies in transcription regulation. The Sam68 nuclear body–associated protein KHDRBS3 bound to the core component KHDRBS1/Sam68 of this nuclear body, which mediates alternative splicing in response to extracellular signal (Matter et al., 2002). KHDRBS3 also associated with several heterogenous nuclear RNPs (Fig. 3 D), which are required for mRNA metabolism and relevant to the function of sam68 nuclear bodies in mRNA splicing. The paraspeckle-targeting protein ZNF24 interacted with lots of zinc finger–containing proteins (Fig. 3 E), which may bind RNA. The core components of paraspeckles, including pspc1, NONO, and p54nrb, all contain RNA recognition motifs that are required for their localization and function to retain A-to-I hyperedited RNA at paraspeckles (Matter et al., 2002). We showed that ZNF24 interacted with core paraspeckle components PSPC1 and PSF in co-IP experiments (Fig. S3 B), indicating that ZNF24 may act as a peripheral paraspeckle component and only associate with core paraspeckle components in a transient or regulated manner, which was difficult to identify using our tandem affinity purification–mass spectrometry method. In summary, this proteomic analysis not only validates our screen but also provides lists of proteins that could be useful starting points for the expansion of the protein–protein interaction network in each of these nuclear subcompartments.
Functional validation of Cajal body–localized proteins reveals the role of TOE1 in Cajal body biogenesis
We sought to demonstrate that the proteins in our nuclear foci proteome actually play a role in their corresponding nuclear bodies in vivo. To this end, we focused on the proteins localized to Cajal bodies. Cajal bodies are sites where snRNP biogenesis takes place (Kiss, 2004). We first subjected 10 Cajal body proteins to proteomic analysis (Fig. 4 A and Fig. S3, C and D). Next, seven proteins showing prominent Cajal body signals were subjected to shRNA-mediated gene silencing, and coilin foci formation was used as a readout for Cajal body biogenesis (Fig. 4 A and Fig. S3, C and D). Given that TOE1 interacts with several proteins involved in Cajal body function and that its gene silencing affects coilin foci formation, it was picked for further characterization.
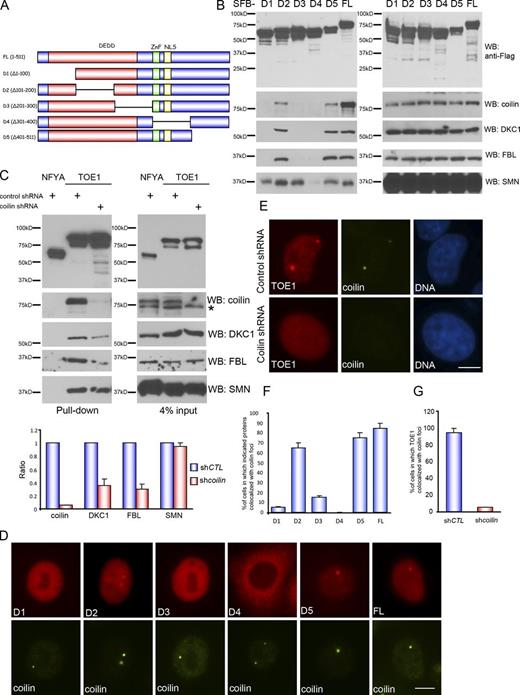
Identification of TOE1 function in regulating Cajal body homeostasis. (A) Schematic workflow showing how the function of proteins localized to Cajal bodies was studied (please also see Fig. S3, C and D). (B) TOE1 is an integral component of Cajal bodies. The localization of TOE1 in HeLa cells was determined by coimmunostaining using anti-TOE1 and anti-coilin (top) or anti-SMN (bottom). Bars, 10 µm. (C) Proteomic analysis of TOE1-containing protein complexes. A cartoon (top part) or a list (bottom part) was presented. (D) SFB-tagged NUAK2 (negative control), TOE1, and TCAB1 were ectopically expressed in HEK293T cells. Pull-down experiments were conducted using streptavidin beads, and immunoblotting was performed with anti-Flag and the indicated antibodies. The asterisk indicates a nonspecific band. (E) Association of endogenous TOE1 and coilin was confirmed by co-IP experiments. Immunoprecipitation (IP) was conducted using the anti-coilin antibody or normal rabbit IgG. WB, Western blot.
Identification of TOE1 function in regulating Cajal body homeostasis. (A) Schematic workflow showing how the function of proteins localized to Cajal bodies was studied (please also see Fig. S3, C and D). (B) TOE1 is an integral component of Cajal bodies. The localization of TOE1 in HeLa cells was determined by coimmunostaining using anti-TOE1 and anti-coilin (top) or anti-SMN (bottom). Bars, 10 µm. (C) Proteomic analysis of TOE1-containing protein complexes. A cartoon (top part) or a list (bottom part) was presented. (D) SFB-tagged NUAK2 (negative control), TOE1, and TCAB1 were ectopically expressed in HEK293T cells. Pull-down experiments were conducted using streptavidin beads, and immunoblotting was performed with anti-Flag and the indicated antibodies. The asterisk indicates a nonspecific band. (E) Association of endogenous TOE1 and coilin was confirmed by co-IP experiments. Immunoprecipitation (IP) was conducted using the anti-coilin antibody or normal rabbit IgG. WB, Western blot.
TOE1 is conserved from Caenorhabditis elegans to mammals. Just like tagged TOE1, endogenous TOE1 colocalized with Cajal body components coilin and survival of motor neuron (SMN; Fig. 4 B). TOE1 copurified with the Cajal body core component coilin, all seven members in the Sm core, box H/ACA RNPs, box C/D RNPs, U5 snRNP/triangular RNP, U4/6 snRNP/triangular RNP, proteins catalyzing U4/6 snRNP recycling, and several serine-rich proteins that localize to nuclear speckles (Fig. 4 C and Fig. S3 E). TOE1 also coimmunoprecipitated with coilin, box H/ACA RNP component DKC1, box C/D RNP component fibrillarin (FBL), Sm-D1/snRNP-D1 protein, and SMN (Fig. 4 D). The affinity of TOE1 to these proteins is comparable to that of TCAB1/WRAP53, a coilin-binding protein essential for Cajal body formation and telomerase trafficking to Cajal bodies (Venteicher and Artandi, 2009; Mahmoudi et al., 2010). Moreover, endogenous TOE1 associated with endogenous coilin (Fig. 4 E). Collectively, these data suggest that TOE1 is an integral component of Cajal bodies.
TOE1 targets to Cajal bodies in a coilin-dependent manner
We constructed a series of internal deletion mutants of TOE1 (Fig. 5 A) and found that only the fragments (D2 and D5) containing the highly conserved N terminus as well as the middle region harboring zinc finger and NLS signals could pull down coilin, DKC1, and FBL (Fig. 5 B and Fig. S4 A). The middle region of TOE1, but not its N terminus, is required for binding to SMN (Fig. 5 B). Moreover, we found that although the binding of TOE1 to dyskerin or FBL requires coilin, its binding to SMN can occur in a coilin-independent manner (Fig. 5 C).
TOE1 is recruited to Cajal bodies in a coilin-dependent manner. (A) Schematic representation of TOE1 mutants used in this study. ZnF, zinc finger domain; DEDD, deadenylation; FL, full length. (B) Mapping coilin-binding domain in TOE1. 293T cells were transfected with constructs encoding full-length or mutant TOE1. Pull-down experiments were performed using streptavidin beads and blotted with anti-Flag (for TOE1 mutants), anti-coilin, anti-DKC1, anti-FBL, or anti-SMN antibodies (please also see Fig. S4 A). (C) Coilin binding is a prerequisite for the loading of DKC1 and FBL, but not SMN, into TOE1-containing complexes. Constructs encoding TOE1 or a negative control NFYA were transfected into HeLa cells stably expressing control shRNA or coilin shRNA. Pull-down experiments were performed using streptavidin beads and blotted with anti-Flag (for negative control NYFA and TOE1), anticoilin, anti-DKC1, anti-FBL, or anti-SMN antibodies. Quantitative results showed the ratio (±SD) of the indicated proteins pulled down by TOE1 in coilin knockdown cells relative to those in control cells (n = 3 independent experiments). The asterisk indicates a nonspecific band. (D) TOE1 localizes to Cajal bodies in a coilin-dependent manner. Indicated mutants of TOE1 were transiently expressed in HeLa cells and then subjected to immunostaining using anti-Flag and anti-coilin antibodies. (E) Cajal body localization of TOE1 was abolished in coilin-depleted cells. Localization of TOE1 was detected in HeLa cells stably expressing control shRNA (top) or coilin shRNA (bottom) by immunostaining using anti-TOE1 and anti-coilin antibodies. (F) Quantitative results showed the percentage of cells (±SD) in which indicated proteins colocalized with coilin (n = 3 independent experiments). (G) Quantitative results showed the percentage of cells (±SD) expressing control or coilin shRNA in which TOE1 colocalized with coilin (n = 3 independent experiments). shCTL, control shRNA; WB, Western blot. Bars, 10 µm.
TOE1 is recruited to Cajal bodies in a coilin-dependent manner. (A) Schematic representation of TOE1 mutants used in this study. ZnF, zinc finger domain; DEDD, deadenylation; FL, full length. (B) Mapping coilin-binding domain in TOE1. 293T cells were transfected with constructs encoding full-length or mutant TOE1. Pull-down experiments were performed using streptavidin beads and blotted with anti-Flag (for TOE1 mutants), anti-coilin, anti-DKC1, anti-FBL, or anti-SMN antibodies (please also see Fig. S4 A). (C) Coilin binding is a prerequisite for the loading of DKC1 and FBL, but not SMN, into TOE1-containing complexes. Constructs encoding TOE1 or a negative control NFYA were transfected into HeLa cells stably expressing control shRNA or coilin shRNA. Pull-down experiments were performed using streptavidin beads and blotted with anti-Flag (for negative control NYFA and TOE1), anticoilin, anti-DKC1, anti-FBL, or anti-SMN antibodies. Quantitative results showed the ratio (±SD) of the indicated proteins pulled down by TOE1 in coilin knockdown cells relative to those in control cells (n = 3 independent experiments). The asterisk indicates a nonspecific band. (D) TOE1 localizes to Cajal bodies in a coilin-dependent manner. Indicated mutants of TOE1 were transiently expressed in HeLa cells and then subjected to immunostaining using anti-Flag and anti-coilin antibodies. (E) Cajal body localization of TOE1 was abolished in coilin-depleted cells. Localization of TOE1 was detected in HeLa cells stably expressing control shRNA (top) or coilin shRNA (bottom) by immunostaining using anti-TOE1 and anti-coilin antibodies. (F) Quantitative results showed the percentage of cells (±SD) in which indicated proteins colocalized with coilin (n = 3 independent experiments). (G) Quantitative results showed the percentage of cells (±SD) expressing control or coilin shRNA in which TOE1 colocalized with coilin (n = 3 independent experiments). shCTL, control shRNA; WB, Western blot. Bars, 10 µm.
Instead of forming nuclear foci–like wild-type TOE1, TOE1 mutants (D1 and D3) defective in coilin binding mainly localized to nucleoplasm, whereas D4, lacking zinc finger and NLS, showed a diffuse pattern in the cytoplasm (Fig. 5, D and F). Moreover, we found that TOE1 failed to localize to nuclear foci in the absence of coilin (Fig. 5, E and G). Together, these data suggest that the interaction between TOE1 and coilin is required for TOE1 localization to Cajal bodies.
TOE1 is required for Cajal body integrity and function
We used siRNAs to knock down the endogenous TOE1 level to <10%, whereas the coilin protein level did not change (Fig. 6 A). However, although coilin usually forms one to four foci per nucleus in control cells, the number of coilin foci increased substantially, and Cajal bodies became dispersed in the nucleoplasm in TOE1 knockdown cells (Fig. 6, B and C). This phenotype was fully rescued by the expression of exogenous TOE1 (Fig. 6, B and C).
TOE1 is required for maintaining Cajal body integrity and efficient splicing. (A) TOE1 was down regulated in HeLa cells transfected by siRNA against TOE1. (B) Knockdown of TOE1 affected the number and homogeneity of coilin foci. A control and two different siRNAs against TOE1 were used to knock down endogenous TOE1 expression in HeLa cells. Localization of TOE1 and coilin was detected by immunostaining using anti-TOE1 and anti-coilin. To recover the expression of TOE1, a construct encoding an siRNA-resistant form of TOE1 was cotransfected with siTOE1-A. The exogenous protein was detected by the anti-Flag antibody. (C) The bar graph shows the percentage of cells (±SD) containing more than four coilin foci after the indicated treatment. WT, wild type. (D and E) Down-regulation of TOE1 disrupted localization of SMN complex and newly synthesized Sm-D1. Control siRNA (siCTL)– or siTOE1-A–treated cells were subjected to coimmunostaining using anti-coilin and anti-SMN antibodies (D) or anti-coilin and anti-Flag (for HA-Flag–tagged Sm-D1) antibodies (E). (F) Quantitative results showed the percentage of cells (±SD) in which the indicated proteins colocalized with coilin (n = 3 independent experiments). (G) TOE1 is required for efficient splicing. A splicing reporter was introduced into HeLa cells or WI-38 cells with the indicated treatment. 24 h later, both spliced and unspliced RNAs were amplified from cDNA using the indicated primer sets. (H) The intensity of unspliced and spliced products was quantified by Quantity One software. The ratios of spliced to unspliced RNAs (±SD) were normalized by controls and presented as a bar graph for the indicated groups (n = 3 independent experiments). (I) TOE1 down-regulation suppresses cell growth. Cells were harvested and counted at day 1–5 after siRNA transfection. The cell numbers (±SD) were plotted against the days after siRNA treatment (n = 3 independent experiments). Bars, 10 µm.
TOE1 is required for maintaining Cajal body integrity and efficient splicing. (A) TOE1 was down regulated in HeLa cells transfected by siRNA against TOE1. (B) Knockdown of TOE1 affected the number and homogeneity of coilin foci. A control and two different siRNAs against TOE1 were used to knock down endogenous TOE1 expression in HeLa cells. Localization of TOE1 and coilin was detected by immunostaining using anti-TOE1 and anti-coilin. To recover the expression of TOE1, a construct encoding an siRNA-resistant form of TOE1 was cotransfected with siTOE1-A. The exogenous protein was detected by the anti-Flag antibody. (C) The bar graph shows the percentage of cells (±SD) containing more than four coilin foci after the indicated treatment. WT, wild type. (D and E) Down-regulation of TOE1 disrupted localization of SMN complex and newly synthesized Sm-D1. Control siRNA (siCTL)– or siTOE1-A–treated cells were subjected to coimmunostaining using anti-coilin and anti-SMN antibodies (D) or anti-coilin and anti-Flag (for HA-Flag–tagged Sm-D1) antibodies (E). (F) Quantitative results showed the percentage of cells (±SD) in which the indicated proteins colocalized with coilin (n = 3 independent experiments). (G) TOE1 is required for efficient splicing. A splicing reporter was introduced into HeLa cells or WI-38 cells with the indicated treatment. 24 h later, both spliced and unspliced RNAs were amplified from cDNA using the indicated primer sets. (H) The intensity of unspliced and spliced products was quantified by Quantity One software. The ratios of spliced to unspliced RNAs (±SD) were normalized by controls and presented as a bar graph for the indicated groups (n = 3 independent experiments). (I) TOE1 down-regulation suppresses cell growth. Cells were harvested and counted at day 1–5 after siRNA transfection. The cell numbers (±SD) were plotted against the days after siRNA treatment (n = 3 independent experiments). Bars, 10 µm.
Because coilin is essential for the assembly of multiple components inside the Cajal bodies, we examined whether other Cajal body protein components would be recruited to residual Cajal bodies after TOE1 down-regulation. We observed that SMN formed cytoplasmic foci instead of nuclear foci (Fig. 6, D and F), suggesting that the SMN complex failed to be recruited to Cajal bodies in the absence of TOE1. Moreover, the number Sm-D1 foci, which normally colocalize with coilin, were also reduced (Fig. 6, E and F). The absence of Sm-D1 in Cajal bodies could also be caused by a failure of Sm proteins to bind to the cytosolic SMN complex, which mediates snRNP assembly (Coady and Lorson, 2011). We therefore tested and found that TOE1 is not required for the association of cytosolic SMN with Sm-D1 (Fig. S4 B), indicating that TOE1 is not involved in snRNP assembly. Collectively, we speculate that TOE1 is likely to function in the maintenance of Cajal body integrity and thereby is required for the docking of SMN and snRNPs to Cajal bodies.
Because the primary role of Cajal bodies is for snRNP maturation and biogenesis, which is needed for efficient RNA splicing (Whittom et al., 2008; Strzelecka et al., 2010b), we attempted to demonstrate the functional relevance of TOE1, especially its potential functions in RNA splicing and cell proliferation. We used an artificial splicing substrate and found that efficient splicing requires coilin as previously reported (Whittom et al., 2008) and TOE1 (Fig. 6, G and H). Double knockdown of TOE1 and coilin did not show any additive defect in splicing (Fig. 6, G and H). Reconstitution of TOE1-depleted cells with siRNA-resistant wild-type TOE1, but not a coilin binding–deficient mutant of TOE1 (TOE1-D3), rescued the splicing defect (Fig. 6, G and H). As a control, we introduced the splicing reporter into WI-38 primary cells that lack Cajal bodies (Fig. S4 C). The splicing efficiency in WI-38 cells was lower than that in HeLa cells. Moreover, knockdown of TOE1 in WI-38 cells did not alter splicing activity (Fig. 6, G and H). Furthermore, we checked the abundance of the spliced mRNA for three endogenous genes (DPP8, NOSIP, and DDX20) and found that silencing TOE1 or coilin reduced the levels of spliced mRNA by 25–70% in HeLa cells but not in WI-38 cells (Fig. S4 D). TOE1 knockdown cells grew slower than mock siRNA-treated cells (Fig. 6 I), a phenotype that was also observed in cells lacking SMN or coilin (Lemm et al., 2006). Introducing wild-type TOE1, but not a TOE1-D3 mutant, into siTOE1-treated cells restored normal cell proliferation (Fig. 6 I). Together, these data suggest that TOE1 is important for Cajal body integrity, which contributes to its roles in splicing as well as cell proliferation.
Identification of proteins involved in paraspeckle formation by shRNA screen
Paraspeckle is a less-characterized nuclear subdomain involved in the control of gene expression via retention of RNA in the nucleus (Bond and Fox, 2009). We first confirmed the localization of newly identified paraspeckle proteins by demonstrating their colocalization with paraspeckle marker protein p54nrb (Fig. 7 A) as well as with NEAT1 long noncoding RNA (Fig. 7 B), which serves as a core structural component for paraspeckle integration (Chen and Carmichael, 2009; Clemson et al., 2009; Sasaki et al., 2009; Sunwoo et al., 2009). Second, we performed an shRNA screen to examine whether any of these newly identified paraspeckle proteins would be required for paraspeckle integrity, which were scored using p54nrb staining or NEAT1 RNA FISH (Fig. 7 C). In addition, NEAT1 expression was also analyzed by quantitative RT-PCR (qRT-PCR; Fig. 7 C). A protein was only considered to be involved in paraspeckle formation if knockdown of such protein leads to ≥30% loss/gain in the number of paraspeckles in the cell, and the phenotype has to be reproducible by at least two independent shRNAs. When compared with RBM14 and NONO, two known components in paraspeckles, we found that knockdown of five other components (HECTD3, FAM53B, ZNF24, XIAP, and ENOX1) also reduced paraspeckle-containing cells, whereas knockdown of another novel component, SH2B1, led to increased paraspeckles in the cell (Fig. 7, D and F), indicating that these proteins are positively or negatively involved in paraspeckle formation. Consistently, down-regulation of five out of eight proteins (HECTD3, RBM14, ZNF24, NONO, and XIAP) required for paraspeckle formation also negatively affect NEAT1 expression (Fig. 7 E).
Identify proteins required for paraspeckle formation using shRNA screen. (A and B) Representative images showed colocalization of newly identified paraspeckle proteins with paraspeckles marker p54nrb (A) or NEAT1 long noncoding RNA (B). (C–E) Phenotypic screen for proteins affecting paraspeckles assembly. (C) A schematic flow for shRNA screen is presented. (D) Bar graph showed the percentage (±SD) of paraspeckles-containing cells determined by p54nrb or NEAT1 staining after the indicated shRNA treatment relative to control mock shRNA-treated cells (n = 3 independent experiments). (E) Bar graph showed relative NEAT1 expression (±SD) to control mock shRNA-treated cells and normalized to GAPDH (n = 3 independent experiments). Dotted lines display the level of control mock shRNA-treated cells for comparison. (F) Representative images showed phenotypes after shRNA transduction. The localization of paraspeckles foci was detected with the use of anti-p54nrb antibodies (top) or FITC-RNA probes against NEAT1 (bottom). Arrows show the paraspeckles foci labeled by anti-p54nrb antibodies (top) or RNA probes against NEAT1 (bottom). CTL, control. Bars, 10 µm.
Identify proteins required for paraspeckle formation using shRNA screen. (A and B) Representative images showed colocalization of newly identified paraspeckle proteins with paraspeckles marker p54nrb (A) or NEAT1 long noncoding RNA (B). (C–E) Phenotypic screen for proteins affecting paraspeckles assembly. (C) A schematic flow for shRNA screen is presented. (D) Bar graph showed the percentage (±SD) of paraspeckles-containing cells determined by p54nrb or NEAT1 staining after the indicated shRNA treatment relative to control mock shRNA-treated cells (n = 3 independent experiments). (E) Bar graph showed relative NEAT1 expression (±SD) to control mock shRNA-treated cells and normalized to GAPDH (n = 3 independent experiments). Dotted lines display the level of control mock shRNA-treated cells for comparison. (F) Representative images showed phenotypes after shRNA transduction. The localization of paraspeckles foci was detected with the use of anti-p54nrb antibodies (top) or FITC-RNA probes against NEAT1 (bottom). Arrows show the paraspeckles foci labeled by anti-p54nrb antibodies (top) or RNA probes against NEAT1 (bottom). CTL, control. Bars, 10 µm.
Paraspeckle proteins are known to accumulate within perinucleolar cap structures when RNA polymerase II transcription is inhibited (Bond and Fox, 2009). Interestingly, the 15 paraspeckles components we identified relocalized to NONO/p54nrb-containing structures after actinomycin D treatment (Fig. S5 A), suggesting that all of these proteins are likely bona fide components of paraspeckles.
Discussion
In this study, we used high throughput microscopic screening to identify hundreds of proteins that form nuclear bodies and therefore put together an atlas of proteins in nuclear domains or nuclear bodies, which is the first step to understanding the dynamic regulations and functions ongoing at these nuclear subcompartments. Nuclear bodies generally represent sites of protein enrichment inside the nucleus. These are likely sites of protein–DNA or –RNA interactions and may be factories for transcriptional and posttranscriptional controls and/or other cellular functions. It is of great interest to identify novel members at various nuclear bodies to gain further understanding of the dynamic regulation of these nuclear bodies. The advantage of our microscopic screen is that it can readily detect nuclear body formation using a straightforward, nonbiased strategy, which does not depend on the availability of high quality antibodies. Moreover, after the discovery of new members at each nuclear body, we could take advantage of the powerful tandem affinity purification approach to further expand the protein–protein interaction network within each nuclear subcompartment.
Of course, there are also shortcomings of this approach. We constructed our ORFeome library based on the existing Human ORFeome V5.1 collection, which sometimes contains truncated genes. We also did not confirm that every ORF was successfully transferred to a destination vector. As a result, our nuclear foci proteome may represent a lion’s share, but not all, of the proteome. Our quality control experiments indicate that the majority of the proteins we tested (34/36) displayed localization identical to that of endogenous protein (Fig. S1). This is likely because the size of the HA-Flag tag is small (<20 amino acids) and the expression level of the tagged protein is moderate (∼2.35-fold of that of endogenous protein). As for further improving our screening, one issue is that the position of epitope tag may influence protein localization. We can subclone our library in a vector with C-terminal HA-Flag fusion and compare the results with that of N-terminal HA-Flag tag fusion proteins used in this study. We can also further reduce the expression of exogenous protein using retrovirus-based vectors. Of course, knocking in an epitope tag at endogenous locus will permit the examination of this gene product at a physiological level, but currently, it is challenging to generate such huge number of knockin cells.
Concerning the validity and accuracy of our screening, we found that one fourth (79/325) of our inventory could be found in various datasets. An additional 57 proteins were previously reported in the literature (Table S2 [blue region] and Table S3 [blue region]). Moreover, we experimentally validated that 10 new proteins (four from Cajal bodies and six from paraspeckles) are required for the assembly of their corresponding nuclear subdomains. Together, ≤45% (146/325) of proteins on our list have been verified either by peer-reviewed articles or in this study.
When compared with NOPdb of the nucleolus, which contains 725 human proteins mainly from two high quality proteomics studies (Andersen et al., 2002; Scherl et al., 2002), the number of nucleolar proteins identified by our screen (148) appears to be quite small. However, there are several differences between our studies. First, we have different selection criteria. We report a protein as a nucleolar protein only when ≥30% of the given protein localizes to the nucleolus. This strict criterion may significantly reduce the number of nucleolar proteins reported in this study, but it ensures that the nucleolar proteins we identified mainly localize in the nucleolus and therefore likely perform major functions in the nucleolus. As a matter of fact, many of them, such as NOLC/NOPP140 and NOP56, are known to play physiological roles in preribosomal RNA processing in the nucleolus (Chen et al., 1999; Hayano et al., 2003; Thiry et al., 2009). On the contrary, a mass spectrometry–based proteomic screen allows the identification of many candidates, only a small fraction of which may primarily reside in the nucleolus. For example, ≥20 chaperone proteins, 16 cytoskeleton proteins, and 21 mitochondria proteins were deposited in NOPdb. The functional significance of these proteins in the nucleolus remains to be verified. Second, although we validated all of our 148 candidates using a secondary screen, the early studies only experimentally confirmed a small fraction of their putative nucleolar proteins. For instance, only 18/271 (∼7%) nucleolar candidates in one of their proteomic experiments were validated by YFP-tagged fusion proteins (Andersen et al., 2002). Third, we found that 66/148 (∼45%) of the nucleolar proteins we identified were already reported in the literature (Table S2). This data confirms the accuracy of our screen. Because only ∼37% of nucleolar proteins in our screen overlapped with those in NOPdb, we believe that the proteomic studies and our cell-based study complement each other, and both of them provide important information for further functional analysis.
An earlier study used the gene trap technology to visualize the localization of fused endogenous proteins and searched for proteins that localize to different nuclear subcompartments (Sutherland et al., 2001). This study has the advantage of protein expression under native promoters. However, the throughput of such a screen is limited (703 clones were analyzed), and it is difficult to expand the screen to genome wide. We found that the efficiency of our screen (2.1% or 325/15,483) is lower but comparable to theirs (4.2% or 29/703). One possible solution to increase the coverage of our screen is to combine various commercially available cDNA libraries, which will allow us to screen more full-length cDNAs.
We also compared our study with a recent review paper (Machyna et al., 2013), which extensively summarized known protein components of Cajal bodies (Fig. 2 and Table S3). There are several discrepancies between our inventory and the published list. First, we listed some small nucleolar RNP maturation factors, such as Nopp140, FBL, NHP2, dyskerin, and Nop56, as nucleolar proteins rather than Cajal body components. This is because these proteins predominantly localize to the nucleolus with only a small fraction localizing to Cajal bodies. We also defined SUMO-1 and PIASy in the same way as they mainly localize in PML nuclear bodies instead of Cajal bodies. Nonetheless, both studies agree on major Cajal body components. In addition to seven well-known components of Cajal bodies, such as Coilin and WRAP53/TCAB1, recovered by our microscopic screen, 13 other already characterized Cajal body components (SMN, TGS1, SART3, FBL, Gar1, Nop10, NHP2, dyskerin, Nop56, Nop58, ELL, LSM10, and LSM11) could also be recovered by the interactome analysis of Cajal bodies as described in our study.
We observed that several proteins (PJA1, CSPP1, ANKRD54, FOSL2, FAM53B, ZNF24, CHMP6, and CSPP1) co-occupy paraspeckles and PML nuclear bodies. One possibility is that a fraction of these proteins originally in paraspeckles may become SUMOylated and thus retained in PML bodies. Another possibility is that there is a functional interaction between paraspeckles and PML bodies, which remains to be elucidated.
The use of the ORFeome library provides an alternative approach that has a better chance to identify proteins directly involved in a cellular process. In this study, we showed that TOE1 plays a critical role in maintaining Cajal body integrity. As for the Cajal body, coilin is believed to be the crucial factor for de novo assembly of Cajal bodies (Kaiser et al., 2008). Coilin could directly recruit spliceosomal Sm protein through protein–protein interactions (Xu et al., 2005; Toyota et al., 2010), a prerequisite for snRNP maturation. The interaction between TCAB1/WRAP53 and coilin was reported to be important for Cajal body formation and for targeting the SMN complex to Cajal bodies (Mahmoudi et al., 2010). More recently, a new SUMO isopeptidase, USPL1, was identified as a novel component of Cajal bodies and required for the integrity of Cajal bodies (Schulz et al., 2012). Mouse embryonic fibroblast cells lacking the 85% C-terminal region of coilin retain residual foci with morphological features similar to those Cajal bodies. However, these foci failed to recruit spliceosomal snRNPs or the SMN complex (Tucker et al., 2001). Similarly, only small nucleolar RNP components, but not U snRNPs, formed detectable foci in coilin-depleted HeLa cells (Lemm et al., 2006). These findings confirm the role of coilin to maintain functional Cajal bodies, which is important for snRNP biogenesis and maturation.
TOE1 was originally discovered to be a target of the EGR1 and responsible for maintaining the cellular level of p21, an inhibitor of cell proliferation (De Belle et al., 2003). However, in this study, we showed that TOE1 localizes in Cajal bodies and interacts with coilin and SMN, indicating that TOE1 may regulate both coilin and SMN. Indeed, coilin was dispersed into numerous heterogenous nuclear foci in TOE1-depleted cells, which is reminiscent of depletion of TGS1, SMN, and PHAX—key players involved in the snRNP biogenesis pathway (Girard et al., 2006; Lemm et al., 2006). snRNP biogenesis involves assembly of the Sm core complex to small nuclear RNAs in cytoplasm. During this process, the SMN complex binds the methylated Sm core complex, allowing specific recruitment of small nuclear RNAs and then guiding the Sm complex onto the Sm binding site on small nuclear RNAs (Coady and Lorson, 2011). However, our result indicated that TOE1 is not required for SMN binding to Sm-D1 (a subunit in the Sm core complex; Fig. S4 B), suggesting that snRNP assembly may not require TOE1. Nevertheless, in the absence of TOE1, SMN foci resided in cytoplasm and failed to be recruited to tiny residual Cajal bodies, which indicates that TOE1 may be required for recruiting the SMN complex to Cajal bodies. Consistent with defective SMN-dependent nuclear import of snRNPs (Narayanan et al., 2004), concentration of newly synthesized Sm-D1 protein at residual coilin foci was also significantly reduced in cells lacking TOE1. Failure of retention of snRNPs in Cajal bodies would lead to incomplete snRNP maturation, which should result in compromised splicing and reduced cell proliferation. Indeed, TOE1-depleted cells showed reduced splicing and proliferation capacity, which phenocopies coilin deficiency. Moreover, the coilin binding–deficient mutant of TOE1 was not able to rescue the splicing activity and cell proliferation in TOE1-depleted cells, suggesting that TOE1 acts with coilin to maintain Cajal body integrity and function. In addition, TOE1 knockdown does not alter splicing efficiency in Cajal body–deficient cells, suggesting that TOE1 functions in pre-mRNA splicing via its role in maintaining Cajal body homeostasis. We speculate that coilin may initiate the nucleation of “nascent” Cajal bodies, whereas assembling of several other factors such as TOE1 would allow Cajal bodies to “grow up.” Eventually, such “mature” Cajal bodies can integrate several small Cajal body–specific RNPs, SMN, and snRNPs to complete snRNPs’ biogenesis, which is important for efficient splicing and cell survival (Fig. S5 B).
The number of Cajal bodies varies with transcriptional and cellular activities, for example, cells have more Cajal bodies to accommodate increasing levels of RNA processing during zebrafish embryogenesis (Strzelecka et al., 2010a). Also, Cajal bodies frequently increase when cells undergo transformation or immortalization (Spector et al., 1992). These findings raise the possibility that cells are capable of forming more Cajal bodies with increased demand for snRNP production. Only a fraction of TOE1 associates with coilin during normal cell proliferation. One possibility is that TOE1 only needs to interact with coilin transiently to carry out its function. Another nonexclusive explanation is that the TOE1–coilin interaction may be regulated and enhanced when the demand for snRNPs increases under certain circumstance, which warrants further investigation.
As another part of validation for our screening, we evaluated paraspeckle formation and NEAT1 expression using shRNAs. We showed that besides the established paraspeckle components such as RBM14 and NONO (Bond and Fox, 2009), down-regulation of three proteins (HECTD3, ZNF24, and XIAP) reduced paraspeckle foci formation as well as NEAT1 expression, which implicates that they may regulate paraspeckles through controlling NEAT1 stability. However, knockdown of other three proteins (SH2B1, FAM5B, and ENOX1) only affected paraspeckle formation without altering NEAT1 expression (Fig. 7, D–F). The underlying mechanisms of how these proteins regulate paraspeckles warrant further investigation.
In summary, our ORFeome screen offers an alternative approach for the identification of proteins involved in various biological functions at distinct nuclear bodies or subnuclear compartments. Expansion of this screen, together with follow up functional analyses, will uncover the roles of these cellular processes in different physiological and pathological conditions.
Materials and methods
Construction of ORFeome library and large-scale screening
A total of 15,483 human ORFs (Human ORFeome v5.1) already in pDONR223 vectors were first transferred into a Gateway-compatible destination vector containing the HA-Flag tag by LR reaction according to the manufacturer’s protocol (Invitrogen). The products were transformed into DH5-α, and the transformants were positively selected with Luria broth medium containing 100 µg/ml ampicillin. The plasmid DNAs were purified using a high quality 96-plasmid DNA purification kit (PureLink; Invitrogen).
A day before transfection, 6 × 103 HeLa cells were seeded on 96-well optical bottom plates (Thermo Fisher Scientific). Plasmid transfection was performed with the use of Lipofectamine 2000 (Invitrogen). 24 h after transfection, cells were subjected to ionizing radiation (IR; 10 Gy) and fixed with 3% paraformaldehyde 6 h later. Next, the cells were permeabilized with a 0.5% Triton X-100 solution and blocked with 3% BSA. Cells were then subjected to incubation with anti-Flag antibodies (1:5,000 dilution) for 2 h, after which they were washed extensively with PBS and incubated with rhodamine-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.) at room temperature for 1 h. Nuclei were counterstained with DAPI. Finally, cells were subjected to automated imaging with the use of ImageXpress Micro (Molecular Devices) equipped with a 20× air objective lens (NA 0.75; Nikon) and a megapixel cooled charge-coupled device camera (CoolSNAP HQ 1.4; Photometrics). The fluorescence images were captured and analyzed using MetaXpress software.
After capturing and analyzing all the images, we selected proteins forming nuclear foci from those that do not form nuclear foci for further characterization. The secondary screen of proteins forming nuclear foci was conducted in untreated or IR-treated cells. Because 325 proteins constitutively form nuclear foci in untreated or IR-treated cells, the validation of 325 proteins with nuclear foci localization was conducted manually using various markers of nuclear bodies or distinct nucleolus morphology in untreated cells. Considering that subcellular localizations of the various nuclear body markers we used are largely distinct from each other, we have not assessed colocalization of each gene with all six marker proteins sequentially. Instead, we scored a protein as positive for a particular nuclear body component in the case of it showing >70% overlapping with any marker protein. During the course of the analysis, we were aware that some proteins colocalize with both PML and paraspeckles, and therefore, we examined whether the identified PML proteins also localize to paraspeckles or vice versa. Eventually, eight proteins were found to localize in both nuclear subcompartments.
To estimate the level of overexpression in our experimental setup, we randomly selected 36 proteins in the ORFeome library for which the antibodies recognizing endogenous proteins are available. We presented the estimation of overexpression of ATRIP as an example. ATRIP is the ATR (ataxia telangiectasia and Rad3 related)-interacting protein, it is in a HA-Flag–tagged expression construct, and it is one of the ORFs in our library. To this aim, we first transfected the HA-Flag ATRIP plasmid into the cells. After paraformaldehyde fixation, the cells were subjected to immunofluorescence staining using anti-Flag (only to indicate which cells express exogenous HA-Flag ATRIP) and using anti-ATRIP antibodies (can stain cells expressing HA-Flag–tagged ATRIP or untransfected cells only expressing endogenous ATRIP). After that, we measured fluorescence intensity from the area of the transfected cells expressing HA-Flag–tagged ATRIP or untransfected cells only expressing endogenous ATRIP (both from the anti-ATRIP channel). Level of overexpression = (Fluorescence intensity transfected cell/Area − Fluorescence intensity background/Area)/(Fluorescence intensity untransfected cell/Area − Fluorescence intensity background/Area). We estimated the level of overexpression for the remaining 35 ORFs using the same strategy as we showed for ATRIP in Fig. S1.
DNA constructs
DNA constructs used in this study were obtained from the human ORFeome v5.1 collection as the pDONR223 entry clone and subsequently transferred to a Gateway-compatible destination vector for protein expression. The SFB tag is a triple-epitope tag (S protein, Flag, and streptavidin binding peptide), which allows efficient detection and purification of exogenously expressed proteins. Internal deletion mutants or point mutations of TOE1 were constructed by using the site-directed mutagenesis kit (QuikChange; Agilent Technologies) and verified by sequencing.
Antibodies
Mouse monoclonal anti–α-tubulin, anti–β-actin, anti-HA, anti-Flag (M2), and anti–sc-35 antibodies were obtained from Sigma-Aldrich; rabbit polyclonal anti-coilin (H-300), anti-PML (H-238); mouse monoclonal anti-sam68 (7–1), anti–Sm-D1 (A-9), and anti-Myc (9E10) antibodies were obtained from Santa Cruz Biotechnology, Inc.; mouse monoclonal anti-SMN, anticoilin, and anti-p54nrb antibodies were obtained from BD; rabbit polyclonal anti-TOE1 and anti-DKC1 antibodies were purchased from Bethyl Laboratories, Inc.; rabbit polyclonal anti-FBL, SFRS1, and SFRS3 antibodies were purchased from Abcam; and the rabbit monoclonal DLC1 antibody was obtained from GeneTex, Inc.
Cell culture and transfection
HeLa, HEK293T (ATCC), and WI-38 cells (obtained from J. Kuang, The University of Texas MD Anderson Cancer Center, Houston, TX) were maintained in DMEM supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. Plasmid transfection was performed using polyethylenimine reagent. To generate a stable cell line expressing SFB-tagged proteins, HEK293T cells were selected with 2 mg/ml puromycin 24 h after transfection. Resistant clones were picked, and expression of the tagged proteins was confirmed by Western blotting and immunofluorescence microscopy. To assess the effect of coilin or TOE1 depletion on the cell growth, cells were harvested and counted by trypan blue exclusion method at 1–5 d after siRNA transfection. To study the effect of actinomycin D on the localization of paraspeckle proteins, 0.5 µg/ml actinomycin D was used to treat HeLa cells for 4 h at 37°C before fixation.
RNAi
siRNA duplexes against TOE1 and Coilin were synthesized (Invitrogen). The sequences of siTOE1-A, 5′-GGGATAGCATCAAGCCTGAAGAAAC-3′; siTOE1-B, 5′-CCTTACCCTGGAGTTCTGCAACTAT-3′; and siCoilin, 5′-AGCAUUGGAAGAGUCGAGAGAACAA-3′ were used. RNAi Negative Control (Medium GC Duplex) was also purchased from Invitrogen. The siRNA duplexes were delivered into cells by transfection using Oligofectamine (Invitrogen).
shRNAs were used to down-regulate components in Cajal bodies and paraspeckles. shRNAs in the pLKO.1 vector were purchased from Sigma-Aldrich, and GIPZ shRNA clones (Thermo Fisher Scientific) were obtained from the Cell Based Assay Screening Service core facility (Baylor College of Medicine). Lentiviral supernatant was generated by transient transfection of 293T cells with the helper plasmids pSPAX2 and pMD2G and harvested 48 h after transfection. Supernatants were passed through a 0.45-µm filter used to infect HeLa cells followed by selection with 2 mg/ml puromycin for 2–3 d.
The sequences of shRNAs obtained from Sigma-Aldrich were as follows: Coilin shRNA-1 (TRCN0000312465), 5′-CCGGGCATTGGAAGAGTCGAGAGAACTCGAGTTCTCTCGACTCTTCCAATGCTTTTTG-3′; SPOPL shRNA-1 (TRCN0000141108), 5′-CCGGCGACAACTTGGGTGTAAAGATCTCGAGATCTTTACACCCAAGTTGTCGTTTTTTG-3′; SPOPL shRNA-4 (TRCN0000140307), 5′-CCGGCAGTTTGGCATTCCACGCAAACTCGAGTTTGCGTGGAATGCCAAACTGTTTTTTG-3′; MED26 shRNA-2 (TRCN0000022009), 5′-CCGGGCACTTGAGGAAACACGACTTCTCGAGAAGTCGTGTTTCCTCAAGTGCTTTTT-3′; TCAB1/WRAP53 shRNA-5 (TRCN0000000312), 5′-CCGGGTTCCTGCATCTTGACCAATACTCGAGTATTGGTCAAGATGCAGGAACTTTTT-3′; EAF2 shRNA-2 (TRCN0000005293), 5′-CCGGGCTATGACTTCAAACCTGCTTCTCGAGAAGCAGGTTTGAAGTCATAGCTTTTT-3′; EAF12 shRNA-12 (TRCN0000005291), 5′-CCGGGCAAATCCTCTACTTCTGATACTCGAGTATCAGAAGTAGAGGATTTGCTTTTT-3′; TOE1 shRNA-7 (TRCN0000151849), 5′-CCGGCCTTATCATTGACACTGATGACTCGAGTCATCAGTGTCAATGATAAGGTTTTTTG-3′; ZGPAT shRNA-9 (TRCN0000162675), 5′-CCGGCCACAAGAAGATGACTGAGTTCTCGAGAACTCAGTCATCTTCTTGTGGTTTTTTG-3′; and control shRNA, 5′-TCTCGCTTGGGCGAGAGTAAG-3′. The clone IDs for each GIPZ shRNA are as follows: CHMP6 (V2LHS_136493, V3LHS_311202, V3LHS_311201, and V3LHS_311200), CPSF6 (V2LHS_149714, V3LHS_640886, V3LHS_640888, and V3LHS_367240), CYBA (V2LHS_257604, V2LHS_84227, V3LHS_358352, and V3LHS_358350), ENOX1 (V2LHS_174882, V2LHS_220987, V3LHS_392270, and V3LHS_392266), FAM53A (V2LHS_259927, V3LHS_330169, and V3LHS_330166), FAM53B (V2LHS_79311, V3LHS_309627, V3LHS_309631, and V3LHS_309629), GATA1 (V2LHS_114063, V3LHS_348340, V3LHS_348337, and V3LHS_348339), HECTD3 (V2LHS_254879, V2LHS_156785, V2LHS_156788, and V3LHS_302340), KLF4 (V2LHS_28276, V2LHS_28277, V2LHS_28349, and V3LHS_376638), LMNB2 (V2LHS_177319, V3LHS_306247, V3LHS_306250, and V3LHS_306248), NONO (V3LHS_644243, V3LHS_644241, V3LHS_644239, and V3LHS_646457), PSPC1 (V2LHS_156677, V3LHS_638976, V3LHS_638975, and V3LHS_348420), RBM14 (V2LHS_178055, V2LHS_275527, V2LHS_178053, and V2LHS_178054), RBM4B (V3LHS_404299, V3LHS_331471, and V3LHS_404298), SCYL1 (V2LHS_247649, V2LHS_57900, V3LHS_638849, and V3LHS_347641), SH2B1 (V2LHS_96745, V2LHS_270857, V3LHS_307685, and V3LHS_400799), XIAP (V2LHS_94577, V2LHS_94576, V2LHS_94574, and V3LHS_302106), ZC3H8 (V2LHS_159014 and V2LHS_159011), ZNF24 (V2LHS_232833, V2LHS_95031, V3LHS_341312, and V3LHS_341309), ZNF444 (V2LHS_175080, V3LHS_392796, V3LHS_392797, and V3LHS_392798), SRSF11 (V3LHS_352519, V3LHS_639450, V3LHS_639446, and V3LHS_639445), and KIAA1683 (V3LHS_328224 and V3LHS_328226).
Immunofluorescence staining
Cells grown on coverslips were fixed either in methanol (−20°C for 10 min) or in 4% paraformaldehyde in PBS at room temperature for 15 min. After fixation, cells were subjected to immunostaining using the same protocol for the large-scale screening. Images were captured with use of a fluorescence microscope (Eclipse E800; Nikon) equipped with a Plan Fluor 40× oil objective lens (NA 1.30; Nikon) and a camera (SPOT; Diagnostic Instruments, Inc.). Images were captured using NIS-Elements basic research imaging software (Nikon) and analyzed using Photoshop CS4 (Adobe).
Tandem affinity purification of SFB-tagged protein complexes
293T cells were transfected with plasmids encoding the protein of interest. Cell lines stably expressing the protein of interest were selected in a cell culture medium containing 2 mg/ml puromycin and were verified by immunostaining and Western blotting. For tandem affinity purification, 293T cells were lysed in NETN (100 mM NaCl, 20 mM Tris-Cl, pH 8.0, 1 mM EDTA, and 0.5% [vol/vol] NP-40) buffer containing protease inhibitors for 20 min at 4°C. Crude lysates were subjected to centrifugation at 14,000 rpm for 30 min. Supernatants were then incubated with streptavidin-conjugated beads (GE Healthcare) for 4 h at 4°C. The beads were washed three times with NETN buffer, and bounded proteins were eluted with NETN buffer containing 2 mg/ml biotin (Sigma-Aldrich) for 1 h twice at 4°C. The elutes were incubated with S-protein beads (EMD Millipore) overnight at 4°C. The beads were eluted with SDS sample buffer and subjected to SDS-PAGE. Protein bands were excised and subjected to mass spectrometry analysis.
Mass spectrometry data analysis
Mass spectrometry analysis was performed by the Taplin Mass Spectrometry Facility at Harvard Medical School. General contaminant proteins, such as heat shock proteins and ribosomal proteins, were discarded after comparison with results from control purifications. The protein of interact was manually sorted based on a literature search by the particular complex they form and/or any common domain they contain. After that, the protein–protein interaction networks were drawn and presented as cartoons in Fig. 3 and Fig. 4.
Bioinformatics analysis
GO analysis was performed with the UniProt-GO Annotation Database. In brief, symbols of the proteins were entered into the database. The annotated GO components, GO process, and GO function for each input would be displayed and then manually recorded in Excel (Microsoft). Finally, the data in the spreadsheet were sorted, and top hits of annotated GO process and GO function among the spreadsheet were presented as bar graphs. Also, a pie chart was used to show the percentage of proteins in the lists annotated with the GO component nucleus. To analyze the protein motif belonging to the listed proteins, we used the InterProScan tool at the European Bioinformatics Institute website (Protein Function Analysis), and this tool consists of a cocktail of databases for protein motif prediction. The protein sequence was first entered into InterProScan. Next, the motifs found for each input were recorded. Top hits of motifs were shown as a bar graph. Nucleolar proteins found in this screen were compared with those deposited in the NOPdb (Nucleolar Proteome Database). Any overlapping nucleolar protein was marked, and the overall results were shown in a bar graph.
Splicing reporter assay
72 h after siRNA transfection, the pSI splicing reporter (obtained from M.D. Hebert, The University of Mississippi Medical Center, Jackson, MS) was introduced into the cells by Lipofectamine 2000. 24 h later, cells were harvested, and total RNAs were extracted by TRIZOL (Invitrogen). The resultant RNAs were subsequently digested by DNase I (Sigma-Aldrich) followed by RT-PCR reaction using primer RP1. Next, primers FP1 and RP1 were used to amplify both spliced and unspliced RNAs with different product sizes. Primers FP1 and RP2 were used to only amplify the intron-containing fragment present in unspliced RNAs. Primers FP2 and RP1 were used to amplify a common fragment in both spliced and unspliced RNAs as the internal loading control among different samples. The PCR products were run on 2% DNA agarose gel. The resulting gel image was exported as TIFF format. Quantity One software (Bio-Rad Laboratories) was used to quantify the intensity of gel bands. The primer sequences are as follows: FP1, 5′-AGGCTTTTGCAAAAAGCTTGATTCTTCTGACACAACAG-3′; FP2, 5′-GTGTCCACTCCCAGTTCAATTACAGCTCTTAAG-3′; RP1, 5′-CTCATCAATGTATCTTATCATGTCTGCTCGAAGCG-3′; and RP2, 5′-GTGGAGAGAAAGGCAAAGTGG-3′.
RNA immunofluorescence FISH
FISH was performed as described previously (Sasaki et al., 2009). In brief, HeLa cells were transduced with mock or shRNA against various paraspeckle proteins and selected with puromycin for 2 d. Then, cells on the coverslips were fixed with 4% paraformaldehyde in PBS at room temperature for 15 min. After dehydration by 70, 95, and 100% ethanol for 5 min each, the coverslips were incubated with prehybridization buffer (2× SSC, Denhardt’s solution, 50% formamide, 10 mM EDTA, 100 µg/ml Escherichia coli tRNA, and 0.01% Tween 20) at 55°C for 2 h. RNA probes against NEAT1 noncoding RNA were prepared with use of a FITC RNA labeling kit (Roche). Prehybridized coverslips were incubated with hybridization buffer (5% dextran sulfate in the prehybridization buffer containing the FITC-labeled RNA probe) at 55°C for 16–18 h and sealed with rubber cement. The plasmid encoding the Neat1 RNA probe was obtained from T. Hirose (Biomedicinal Information Research Center, National Institute of Advanced Industrial Science and Technology, Koto, Tokyo, Japan). After probe incubation, the coverslips were washed twice with wash buffer A (2× SSC, 50% formamide, and 0.01% Tween 20) at 55°C for 20 min and washed once with wash buffer B (2× SSC and 0.01% Tween 20) at 55°C for 20 min and twice with wash buffer C (0.1× SSC and 0.01% Tween 20) at 55°C for 20 min. To detect the probe, the coverslips were first blocked with blocking buffer (1% blocking reagent [Roche] in TBST [TBS with Tween 20]) at room temperature for 1 h and then incubated with anti-FITC antibodies against the RNA probes and/or antibodies against paraspeckle proteins diluted with blocking buffer for 1 h. The coverslips were then washed three times in TBST for 15 min, incubated with the secondary antibodies at room temperature for 1 h, stained with DAPI to visualize the DNA, and mounted onto the glass slides.
qRT-PCR
Total RNAs from siRNA- or shRNA-treated cells were extracted by TRIZOL (Invitrogen). Next, 1 µg/ml RNA was reverse transcribed with use of Moloney murine leukemia virus Taq RT-PCR kit (ProtoScript; New England Biolabs, Inc.). cDNAs were subjected to real-time PCR with use of Power SYBR Green PCR Master Mix (Applied Biosystems) according to the manufacturer’s protocol. The primer sequences are used as follows: NEAT1 forward primer 1, 5′-CAATTACTGTCGTTGGGATTTAGAGTG-3′; NEAT1 reverse primer 1, 5′-TTCTTACCATACAGAGCAACATACCAG-3′; NEAT1 forward primer 2, 5′-TGTGTGTGTAAAAGAGAGAAGTTGTGG-3′; NEAT1 reverse primer 2, 5′-AGAGGCTCAGAGAGGACTGTAACCTG-3′; GAPDH forward primer, 5′-ACAACTTTGGTATCGTGGAAGG-3′; GAPDH reverse primer, 5′-GCCATCACGCCACAGTTTC-3′; DPP8 forward, 5′-TCTATTACCTTGCCATGTCTGGTG-3′; DPP8 reverse, 5′-AATACATTCCATAGTCCAGTGTTG-3′; NOSIP forward, 5′-CTGGAGAAGCCGTCCCGCACGGTG-3′; NOSIP reverse, 5′-CACGGCACACACGTAGCGCTCGCT-3′; DDX20 forward, 5′-TTAAGTACCCAGATTTTGATCTTG-3′; and DDX20 reverse, 5′-AAGTCTGGTTTTGTCTTGTGATAA-3′.
Online supplemental material
Fig. S1 examines the level of overexpression of ORFeome library in our study. Fig. S2 shows a bioinformatics analysis of the nuclear foci proteome. Fig. S3 shows a proteomic analysis of various nuclear subcompartments. Fig. S4 shows that TOE1 is required for endogenous mRNA splicing. Fig. S5 shows the validation of identified paraspeckle proteins. Table S1 is an inventory of nuclear foci proteome with GO analysis. Table S2 shows a comparison with NOPdb. Table S3 shows a comparison with different datasets. Table S4 shows the classification of unknown nuclear foci. Table S5 shows the InterProScan analysis. Table S6 shows the top hit protein motif among various nuclear bodies. Table S7 is a list of interacting proteins from mass spectrometry analysis. Additional data are available in the JCB DataViewer at https://doi.org/10.1083/jcb.201303145.dv.
Acknowledgments
We thank our colleagues in Dr. Junjie Chen’s laboratory for their insightful discussions and technical assistance. We thank Dr. Micheal D. Hebert (The University of Mississippi Medical Center) for providing the pSI splicing reporter plasmid. We thank Dr. Tetsuro Hirose (Functional RNomics Team, Biomedicinal Information Research Center, Japan) for providing the plasmid encoding NEAT1 RNA probes. We thank Jian Kuang (The University of Texas MD Anderson Cancer Center) for providing WI-38 primary cells.
K.-w. Fong is a recipient of The Kimberly Patterson Fellowship in Leukemia Research. This work was supported in part by grants from the National Institutes of Health to J. Chen (CA089239, CA092312, and CA113381) and a start-up fund provided by the MD Anderson Cancer Center; J. Chen is a member of the MD Anderson Cancer Center (CA016672). This study was also supported by the National Basic Research Program (973 Program; 2010CB945401) and the National Natural Science Foundation (31000611, 91019020, and 31171397). We would also like to acknowledge the support of the National Institute of General Medical Sciences (GM095599), the Genome-wide RNAi Screens Cores Shared Resource at the Dan L. Duncan Cancer Center (grant P30CA125123), and the Baylor College of Medicine Intellectual and Developmental Disabilities Research Center (grant 5P30HD024064) from the Eunice Kennedy Shriver National Institute of Child Health and Human Development.