Meiotic maturation in vertebrate oocytes is an excellent model system for microtubule reorganization during M-phase spindle assembly. Here, we surveyed changes in the pattern of microtubule-interacting proteins upon Xenopus laevis oocyte maturation by quantitative proteomics. We identified the synovial sarcoma X breakpoint protein (SSX2IP) as a novel spindle protein. Using X. laevis egg extracts, we show that SSX2IP accumulated at spindle poles in a Dynein-dependent manner and interacted with the γ-tubulin ring complex (γ-TuRC) and the centriolar satellite protein PCM-1. Immunodepletion of SSX2IP impeded γ-TuRC loading onto centrosomes. This led to reduced microtubule nucleation and spindle assembly failure. In rapidly dividing blastomeres of medaka (Oryzias latipes) and in somatic cells, SSX2IP knockdown caused fragmentation of pericentriolar material and chromosome segregation errors. We characterize SSX2IP as a novel centrosome maturation and maintenance factor that is expressed at the onset of vertebrate development. It preserves centrosome integrity and faithful mitosis during the rapid cleavage division of blastomeres and in somatic cells.
Introduction
The identification and characterization of spindle proteins remains generally important to understand the molecular basis for faithful chromosome segregation. Spindle formation requires, before M phase, the cell cycle–regulated synthesis of microtubule (MT)-associated proteins (MAPs) at M-phase entry. These include MT motor activities, nonmotor proteins to regulate the dynamic behavior of spindle MTs (Manning and Compton, 2008a,b; Walczak and Heald, 2008), a large variety of chromatin proteins (Ohta et al., 2011), and centrosomal proteins to control MT nucleation and organization at mitotic spindle poles (DeLuca, 2007; Sluder and Khodjakov, 2010). Several proteomic approaches identified spindle proteins on the basis of spindle localization or mitotic MT association (Sauer et al., 2005; Nousiainen et al., 2006). However, understanding the cell cycle–dependent changes of MAP patterns at M-phase onset requires a comprehensive comparison of the MT interactomes in late interphase and early M phase.
Here we exploit the Xenopus laevis oocyte model system that enables the preparation of large quantities of naturally arrested, fully synchronous lysates in late interphase (G2/M-arrested oocytes), or M phase (eggs arrested in metaphase II of meiosis), from which MAPs can be purified in preparative scale with unparalleled purity. We apply quantitative differential proteomics using isotope coded protein label (ICPL; Schmidt et al., 2005) and statistical data analysis from repetitive sampling to compare the MAP network before and after entry into M phase. Apart from well-characterized spindle proteins such as TPX2, PRC1, Kif4A, Eg5, and Xkid, we identify synovial sarcoma X breakpoint 2 interacting protein (SSX2IP) as a novel MAP that is expressed at M-phase onset and accumulates at MT minus ends. To test a possible function of SSX2IP in M-phase MT formation, we used X. laevis egg extracts, which faithfully reconstitute spindle formation in a cell-free environment. X. laevis extracts recapitulate centriole-free, meiotic spindle assembly (Heald et al., 1996) and spindle assembly around sperm nuclei. Addition of sperm nuclei reintroduces centrioles, which allows the assembly of centrosome-containing spindles (Sawin and Mitchison, 1991). Strikingly, immunodepletion of SSX2IP leaves centriole-free meiotic spindle formation unaffected but abolishes maturation of mitotic centrosomes and causes severe defects in spindle assembly around sperm nuclei. To confirm the function of SSX2IP in mitotic centrosome function in vivo, we analyzed cell division by time-lapse microscopy in developing medaka (Oryzias latipes) embryos and in human somatic cells. In both systems, SSX2IP down-regulation consistently causes centrosome fragmentation and defects in mitotic progression. We propose that SSX2IP is a novel centrosome maturation factor expressed at meiotic onset, which generally maintains the integrity of the pericentriolar material and proper MT nucleation at mitotic spindle poles.
Results
Identification of SSX2IP as a novel MT-binding protein
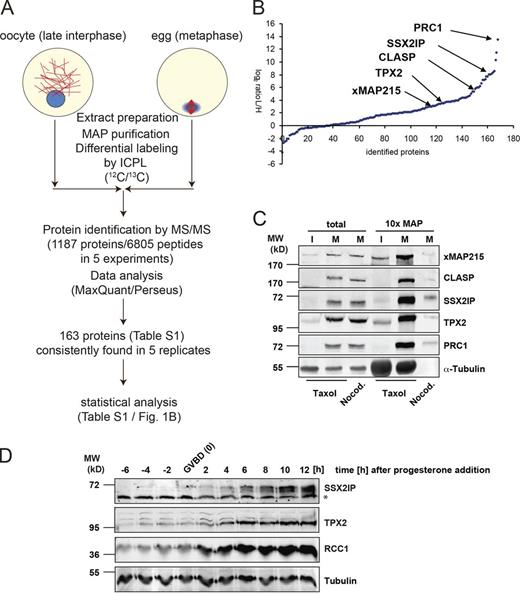
To survey the changes in the MT interactome between interphase and M phase, we purified MAPs derived from G2/M-arrested oocytes, representing late interphase, and from unfertilized X. laevis eggs in metaphase (Fig. 1 A). After purification, MAPs were modified by ICPL (Schmidt et al., 2005), and identified by electrospray ionization (ESI) mass spectrometry in five independent experiments. ICPL uses lysine-modifying reagents containing stable isotopes, which allowed us to assign identified MAPs to either interphase or metaphase, and to quantitatively compare them (Fig. 1, A and B). Mean ratios of proteins found in all five experiments showed an asymmetric distribution. Large differences between the two states were generally caused by increasing amounts in metaphase, which is consistent with a global positive regulation upon M-phase entry (Fig. 1 B). Besides KIF4A, Eg5/KIF11b, XKid, and TPX2 (Table S1), we identified CLASP1/Xorbit and PRC1 as novel targets of M-phase expression regulation (Fig. 1 C). Our screen also revealed MT cosedimentation and maturation-associated expression of X. laevis SSX2IP (XSSX2IP; Fig. 1, B–D; and Fig. S1, A and B), a protein which previously had not been implicated in MT binding. Human SSX2IP had been suggested to be a modulator of the transcriptional repressor SSX2 (de Bruijn et al., 2002), whereas mouse SSX2IP (Afadin DIL domain–interacting protein [ADIP]) had been found to play a role in cell adhesion as an interaction partner of the actin-binding proteins Afadin and α-actinin (Asada et al., 2003). XSSX2IP identified here (Fig. S1 A, 1,800 nt) represents a protein of 65 kD with the highly conserved ADIP motif (see Discussion) and two additional C-terminal coiled-coil motifs. In silico prediction of M phase–dependent expression using characteristic regulatory motifs (Piqué et al., 2008) in the remarkably long 3′ UTR of XSSX2IP suggested translational repression before M phase, and activation upon M phase entry in oocytes (Fig. S1 A).
Identification and validation of SSX2IP as a MAP. (A) Workflow for the comparison of the MT interactome in interphase (stage VI X. laevis oocytes) and metaphase (unfertilized X. laevis eggs) by differential proteomics. (B) Protein abundance ratios (log2 of egg/oocyte ratio) of the 163 proteins found in five of five independent experiments. Median values of the five experiments are shown (see Table S1 for details). Note that most proteins are expressed more highly in metaphase. (C) Validation of the predicted metaphase MT association of identified proteins by immunoblotting. MAPs were purified from oocyte (interphase, I) or egg (metaphase, M) lysates by cosedimentation of taxol-stabilized MTs; nocodazole was used as a negative control. Total lysates were compared with MAP sediments, and tubulin served as a loading control. (D) Progesterone-treated, synchronously maturing X. laevis oocytes were analyzed for expression of SSX2IP, RCC1, and TPX2 (positive controls); tubulin was used as a loading control. Times are indicated with respect to nuclear (i.e., germinal vesicle) breakdown (GVBD). *, note the cross-reacting band at 55 kD in the SSX2IP blot.
Identification and validation of SSX2IP as a MAP. (A) Workflow for the comparison of the MT interactome in interphase (stage VI X. laevis oocytes) and metaphase (unfertilized X. laevis eggs) by differential proteomics. (B) Protein abundance ratios (log2 of egg/oocyte ratio) of the 163 proteins found in five of five independent experiments. Median values of the five experiments are shown (see Table S1 for details). Note that most proteins are expressed more highly in metaphase. (C) Validation of the predicted metaphase MT association of identified proteins by immunoblotting. MAPs were purified from oocyte (interphase, I) or egg (metaphase, M) lysates by cosedimentation of taxol-stabilized MTs; nocodazole was used as a negative control. Total lysates were compared with MAP sediments, and tubulin served as a loading control. (D) Progesterone-treated, synchronously maturing X. laevis oocytes were analyzed for expression of SSX2IP, RCC1, and TPX2 (positive controls); tubulin was used as a loading control. Times are indicated with respect to nuclear (i.e., germinal vesicle) breakdown (GVBD). *, note the cross-reacting band at 55 kD in the SSX2IP blot.
Depletion of SSX2IP leaves centrosome-free spindle assembly unaffected but compromises centrosome maturation
To validate our findings, we first followed expression of XSSX2IP throughout M-phase entry in maturing oocytes and detected de novo expression of XSSX2IP after nuclear (germinal vesicle) breakdown (GVBD; Fig. 1 D), similar to RCC1 (Dumont et al., 2007) and TPX2 (Eliscovich et al., 2008), which we detected as positive controls. Maturation-associated expression and MT binding in egg extracts raised the possibility that the protein functions in meiotic spindle formation around chromatin (Varmark, 2004; Müller-Reichert et al., 2010) or the following mitotic cleavage divisions. We tested both possibilities in living oocytes and in X. laevis egg extracts.
In intact oocytes, we could not detect aberrations in bipolarity, size, or MT density of meiotic spindles after morpholino (MO) XSSX2IP antisense injection, despite inhibition of de novo expression and loss of spindle association of XSSX2IP (Fig. 2 A). When we assayed meiotic spindle formation in X. laevis egg extracts (Heald et al., 1996), spindles were found to assemble normally even after immunodepletion of SSX2IP or direct addition of neutralizing antibodies (Fig. 2, B and C). In contrast, the depletion or inactivation of TPX2 abolished chromatin-driven spindle formation (Gruss et al., 2001; Fig. 2, B and C). Interestingly, XSSX2IP accumulated at spindle poles in both intact oocytes (Fig. 2 A) and chromatin-induced, centrosome-free spindles in extracts (Fig. 2, B and D), which suggested MT minus end binding. Consistent with that, the inhibition of the major minus–end directed MT motor Dynein (Wittmann et al., 1998) abolished XSSX2IP accumulation at spindle poles (Fig. 2 D). Moreover, the addition of RanGTP (Carazo-Salas et al., 1999; Zhang et al., 1999), which mimics the presence of chromatin in egg extracts, induced self-assembly of aster-like structures despite the loss of XSSX2IP (Fig. S2 A). Also here, the inhibition of Dynein motor activity interfered with the accumulation of SSX2IP at focused MT minus ends of Ran-induced structures (Fig. S2 B). To further strengthen this conclusion, we co-pelleted long or short taxol-stabilized MTs (Fig. S2 C), which were incubated in diluted X. laevis egg extracts. Indeed, 1.5-fold more SSX2IP cosedimented with the same mass of MTs when short MTs as opposed to long MTs were pelleted (Fig. S2, C and D). This further argues for preferential minus end binding of SSX2IP. Collectively, we concluded that SSX2IP accumulates at the MT minus end in a Dynein-dependent manner but is not required for chromatin driven MT assembly.
SSX2IP depletion does not interfere with centrosome-free spindle formation. (A) Expression of SSX2IP during in vitro maturation was specifically suppressed by MO oligonucleotides but did not alter meiotic spindle morphology; immunoblotting and indirect immunofluorescence detect SSX2IP and tubulin. (B and D) Chromatin-induced spindle assembly was monitored in X. laevis egg extracts that were mock-depleted (control, ct), immunodepleted of SSX2IP (ΔSSX2IP) or TPX2 (ΔTPX2), or supplemented with control (ctrl.) or anti (α)-SSX2IP antibodies (B). In D, control (ctrl.) antibodies or antibodies against Dynein intermediate chain were used. The blot in B shows depletion efficiencies; images display chromatin bead spindles (red, tubulin; green, SSX2IP; blue, DAPI). Please note that SSX2IP antibody addition efficiently inhibits pole localization of SSX2IP. (C) Quantification of assembled chromatin bead spindles in depleted extracts as indicated (left graph; this experiment was completed once, n = 100), or after addition of specific anti-SSX2IP or anti-TPX2 antibodies (right graph shows mean values ± SD of three independent experiments). The significance was calculated by a Student’s t test (two-tailed) and scored as follows: *, P < 0.05; **, P < 0.01. The magnified panels (magn.) show enlarged views of the boxed regions. Bars: (main panels) 20 µm; (magnified panels) 2 µm.
SSX2IP depletion does not interfere with centrosome-free spindle formation. (A) Expression of SSX2IP during in vitro maturation was specifically suppressed by MO oligonucleotides but did not alter meiotic spindle morphology; immunoblotting and indirect immunofluorescence detect SSX2IP and tubulin. (B and D) Chromatin-induced spindle assembly was monitored in X. laevis egg extracts that were mock-depleted (control, ct), immunodepleted of SSX2IP (ΔSSX2IP) or TPX2 (ΔTPX2), or supplemented with control (ctrl.) or anti (α)-SSX2IP antibodies (B). In D, control (ctrl.) antibodies or antibodies against Dynein intermediate chain were used. The blot in B shows depletion efficiencies; images display chromatin bead spindles (red, tubulin; green, SSX2IP; blue, DAPI). Please note that SSX2IP antibody addition efficiently inhibits pole localization of SSX2IP. (C) Quantification of assembled chromatin bead spindles in depleted extracts as indicated (left graph; this experiment was completed once, n = 100), or after addition of specific anti-SSX2IP or anti-TPX2 antibodies (right graph shows mean values ± SD of three independent experiments). The significance was calculated by a Student’s t test (two-tailed) and scored as follows: *, P < 0.05; **, P < 0.01. The magnified panels (magn.) show enlarged views of the boxed regions. Bars: (main panels) 20 µm; (magnified panels) 2 µm.
As there was no indication that SSX2IP was required for centrosome-free spindle assembly, we tested if SSX2IP may be required for spindle formation around sperm nuclei. Insertion of the sperm basal body into the egg cytoplasm upon fertilization reintroduces centrioles and enables spindle formation in the presence of centriole-based MT-organizing centers (MTOCs; Sawin and Mitchison, 1991). Like in chromatin bead spindles and Ran asters, fluorescently labeled antibodies against XSSX2IP detected accumulation at spindle poles, which was abolished after inhibition of Dynein motor activity (Fig. 3 A). Interestingly, when we immunoprecipitated endogenous XSSX2IP from X. laevis egg extracts, we identified spindle pole and centrosomal components as major XSSX2IP interaction partners including γ-tubulin ring complex (γ-TuRC) proteins (Stearns et al., 1991), X. laevis CCDC52/Spice (Archinti et al., 2010), X. laevis Cep120 (Mahjoub et al., 2010), and X. laevis pericentriolar material protein 1 (PCM-1) by mass spectrometry (Fig. S3 A). Interactions between XSSX2IP, Cep120, γ-tubulin, and PCM-1 were confirmed by immunoblotting (Fig. 3 B). We then assayed spindle assembly in X. laevis egg extracts in the presence of sperm nuclei before and after immunodepletion of XSSX2IP. The translation of an mRNA encoding EGFP-XSSX2IP complemented XSSX2IP depletion (Fig. 3 C). In contrast to control extracts, XSSX2IP-depleted extracts failed in bipolar spindle formation. The majority of structures displayed half-spindle or aster-like morphology (Fig. 3, D and E), which we also saw upon directly neutralizing XSSX2IP by antibody addition (Fig. S2 E). Re-expressed XSSX2IP accumulated at spindle poles and restored bipolar spindle assembly significantly (Fig. 3 E, rescue). Incomplete rescue in this assay was possibly caused by codepletion of the SSX2IP interaction partners γ-TuRC and PCM-1 (Fig. S3 B). We rationalized that impaired spindle assembly in XSSX2IP-depleted extract was either caused by the disturbed activity of centrosomes or the replicated chromatin (Gruss et al., 2002). To differentiate between these alternatives, we inhibited RanGTP production from sperm nuclei by RanT24N to block the Ran-dependent activity of chromatin in spindle formation. Astral arrays of MTs, which still formed under these conditions (Carazo-Salas et al., 2001; Maresca et al., 2009), were significantly smaller in SSX2IP-depleted extracts compared with control or TPX2-depleted extracts (Fig. S2 F). This indicated that defects in centrosome function were causative for spindle assembly defects in SSX2IP-depleted X. laevis egg extracts.
Immunodepletion of SSX2IP interferes with bipolar spindle assembly in centriole-containing spindles. (A) Localization of endogenous SSX2IP in spindles assembled around sperm nuclei in X. laevis egg extracts after addition of control antibodies, or antibodies against Dynein intermediate chain (Dynein IC). Red, tubulin; blue, chromatin; green, α-SSX2IP. Insets show spindle poles (enlarged views from the boxed regions). (B) Proteins identified as SSX2IP interaction partners (PCM-1, γ-tubulin) in X. laevis egg extracts were validated by immunoblotting; TPX2 served as a negative control. Free tubulin does not interact with SSX2IP. (C) Immunodepletion (Δ) of SSX2IP from X. laevis egg extracts was monitored by immunoblotting. A GFP-SSX2IP–encoding mRNA was translated in egg extracts for complementation/rescue. (D and E) Spindles from sperm nuclei were analyzed in control extracts, after depletion (Δ) of TPX2 (only in D) or SSX2IP, or rescue by GFP-SSX2IP. In D: red, Cy3-tubulin; green, SSX2IP; blue, DAPI/chromatin. Magnified panels (magn.) show enlarged views of the boxed regions. (E) Quantification of spindle morphology defects from three independent experiments. The significance was calculated by a Student’s t test (two-tailed) and scored as follows: *, P < 0.05; **, P < 0.01; ****, P < 0.0001. Error bars indicate SD. Bars: (main panels) 20 µm; (insets in A and magnified panels in D) 2 µm.
Immunodepletion of SSX2IP interferes with bipolar spindle assembly in centriole-containing spindles. (A) Localization of endogenous SSX2IP in spindles assembled around sperm nuclei in X. laevis egg extracts after addition of control antibodies, or antibodies against Dynein intermediate chain (Dynein IC). Red, tubulin; blue, chromatin; green, α-SSX2IP. Insets show spindle poles (enlarged views from the boxed regions). (B) Proteins identified as SSX2IP interaction partners (PCM-1, γ-tubulin) in X. laevis egg extracts were validated by immunoblotting; TPX2 served as a negative control. Free tubulin does not interact with SSX2IP. (C) Immunodepletion (Δ) of SSX2IP from X. laevis egg extracts was monitored by immunoblotting. A GFP-SSX2IP–encoding mRNA was translated in egg extracts for complementation/rescue. (D and E) Spindles from sperm nuclei were analyzed in control extracts, after depletion (Δ) of TPX2 (only in D) or SSX2IP, or rescue by GFP-SSX2IP. In D: red, Cy3-tubulin; green, SSX2IP; blue, DAPI/chromatin. Magnified panels (magn.) show enlarged views of the boxed regions. (E) Quantification of spindle morphology defects from three independent experiments. The significance was calculated by a Student’s t test (two-tailed) and scored as follows: *, P < 0.05; **, P < 0.01; ****, P < 0.0001. Error bars indicate SD. Bars: (main panels) 20 µm; (insets in A and magnified panels in D) 2 µm.
To further confirm our hypothesis, we applied an assay for the assembly of functional mitotic centrosomes. X. laevis egg extracts enhance the nucleation capacity of human somatic centrosomes (Yokoyama et al., 2008) in the presence of RanGTP and after TPX2 immunodepletion, which we could clearly confirm (Fig. 4, A and C). More XSSX2IP localized to the stronger nucleating centrosomes in the presence of RanGTP (Fig. 4, A and B). XSSX2IPs centrosomal localization was largely abolished after immunodepletion but could be rescued by GFP-XSSX2IP mRNA addition (please note that SSX2IP is detected by indirect immunofluorescence using antibodies that recognize endogenous and GFP-tagged SSX2IP). The depletion of XSSX2IP resulted in reduced MT nucleation of centrosomes both in the absence and in the presence of RanGTP. Reduced nucleation in SSX2IP-depleted extracts was correlated with reduced recruitment of the γ-TuRC components γ-tubulin, XGrip109/GCP3, and XGrip210/GCP6. Both could be significantly restored after translation of GFP-SSX2IP (Fig. 4, C–F). This confirmed that the activity of centrosomes to assemble MTs was strongly dependent on XSSX2IP and was responsible for the observed spindle assembly defects in spindles around sperm nuclei, possibly because of reduced γ-TuRC activity at mitotic spindle poles. Indeed, directly reducing γ-TuRC levels by 90% in complete X. laevis egg extracts (Fig. 4 G) abolished bipolar spindle formation but still allowed assembly of astral arrays or half spindles (Fig. 4, H and I), as seen after depletion of SSX2IP. Collectively, we concluded that SSX2IP, expressed at the beginning of M phase in X. laevis oocytes, is specifically required for spindle assembly by sperm nuclei in the presence of centriole-based MTOCs.
SSX2IP is required for mitotic centrosome maturation. (A) Localization of SSX2IP on centrosomes. Extracts were incubated with centrosomes and labeled tubulin (red) in the presence or absence of RanGTP. A labeled SSX2IP antibody (green) was added 5 min before squash fixation on coverslips. Note that the RanGTP-dependent centrosome maturation assay was done in TPX2-depleted extracts that do not nucleate self-organized MTs but leave MT nucleation exclusively to centrosomal asters. Insets of the same scale as the images highlight SSX2IP on centrosomes. (B, left) SSX2IP total levels in control, depleted, and reconstituted extracts were quantified by immunoblotting from three independent experiments. (B, right) SSX2IP levels on centrosomes were assayed and quantified as described previously (Yokoyama et al., 2008). (C and E) Samples as in A were fixed after aster assembly, spun down on coverslips, and stained with γ-tubulin antibodies (C, green) or Xgrip109/GCP3 or Xgrip210/GCP6 antibodies (E, green). Insets in C of the same scale as the images highlight γ-tubulin on centrosomes. (D and F) MT intensity and γ-tubulin (D) or Xgrip109 and Xgrip210 (F) levels in centrosomal asters were quantified as described previously (Yokoyama et al., 2008). (G) γ-TuRC levels were reduced in X. laevis egg extracts by immunodepletion using an antibody against γ-tubulin (Zheng et al., 1995); numbers indicate relative γ-tubulin levels. (H) Representative images from bipolar spindles in control (100% γ-TuRC levels) and half-spindles after removing 90% γ-TuRC (10% residual levels). Red, α-tubulin; green, γ-tubulin; blue, DAPI/DNA. Insets show γ-tubulin signals. (I) Mitotic MT assemblies after removing γ-TuRC were analyzed. n > 80 structures counted in all samples; graph shows mean ± SD from three independent experiments. The significance (100 vs. 10% γ-tubulin) was calculated by a Student’s t test (two-tailed) and scored as **, P < 0.01. Bars: (main panels) 20 µm; (magnified regions indicated by white boxes) 2 µm.
SSX2IP is required for mitotic centrosome maturation. (A) Localization of SSX2IP on centrosomes. Extracts were incubated with centrosomes and labeled tubulin (red) in the presence or absence of RanGTP. A labeled SSX2IP antibody (green) was added 5 min before squash fixation on coverslips. Note that the RanGTP-dependent centrosome maturation assay was done in TPX2-depleted extracts that do not nucleate self-organized MTs but leave MT nucleation exclusively to centrosomal asters. Insets of the same scale as the images highlight SSX2IP on centrosomes. (B, left) SSX2IP total levels in control, depleted, and reconstituted extracts were quantified by immunoblotting from three independent experiments. (B, right) SSX2IP levels on centrosomes were assayed and quantified as described previously (Yokoyama et al., 2008). (C and E) Samples as in A were fixed after aster assembly, spun down on coverslips, and stained with γ-tubulin antibodies (C, green) or Xgrip109/GCP3 or Xgrip210/GCP6 antibodies (E, green). Insets in C of the same scale as the images highlight γ-tubulin on centrosomes. (D and F) MT intensity and γ-tubulin (D) or Xgrip109 and Xgrip210 (F) levels in centrosomal asters were quantified as described previously (Yokoyama et al., 2008). (G) γ-TuRC levels were reduced in X. laevis egg extracts by immunodepletion using an antibody against γ-tubulin (Zheng et al., 1995); numbers indicate relative γ-tubulin levels. (H) Representative images from bipolar spindles in control (100% γ-TuRC levels) and half-spindles after removing 90% γ-TuRC (10% residual levels). Red, α-tubulin; green, γ-tubulin; blue, DAPI/DNA. Insets show γ-tubulin signals. (I) Mitotic MT assemblies after removing γ-TuRC were analyzed. n > 80 structures counted in all samples; graph shows mean ± SD from three independent experiments. The significance (100 vs. 10% γ-tubulin) was calculated by a Student’s t test (two-tailed) and scored as **, P < 0.01. Bars: (main panels) 20 µm; (magnified regions indicated by white boxes) 2 µm.
SSX2IP is required for centrosome integrity and genome stability in developing embryos
We initially found SSX2IP as a target of regulation during M-phase resumption before fertilization, obviously to prepare a developing organism for the fast division during early development. To determine the in vivo function of SSX2IP at this stage in a vertebrate model system we took advantage of medaka (O. latipes, Japanese killifish) embryos that are fully transparent during development and, compared with X. laevis, readily applicable for indirect immunofluorescence and high-resolution live cell imaging at early stages. Medaka SSX2IP displayed 51% identity with human and 48% identity with X. laevis SSX2IP. Using two different SSX2IP antibodies, we confirmed centrosomal association of SSX2IP and colocalization with γ-tubulin and PCM-1 in medaka at blastula stage 10–11 (Fig. 5 A and Fig. S4 A). Next, we inhibited the function of medaka SSX2IP concomitant with fertilization using antisense MO. As a readout for mitotic spindle function, we followed chromosome segregation using Histone2B-GFP in live embryos by digital scanned laser light-sheet fluorescence microscopy (Keller et al., 2010). This allowed us to monitor all divisions in embryos at around stage 9 simultaneously for 40–80 min (Fig. 5, see Videos 1 and 2). Visual data inspection and counting chromosome segregation events (Fig. 5 C, see red arrows in Fig. 5 B) indicated a significantly increased frequency of missegregation after MO-mediated down-regulation of SSX2IP (>20% of all divisions in knockdown embryos; in control or untreated embryos, the frequency of chromosome segregation failures was ∼2–4%; Fig. 5, B and C).
Time-lapse imaging of medaka early embryos expressing GFP-Histone2B after control MO injection. Imaging was performed by DSLM in 1-min intervals for 40 min starting 4 h after fertilization.
Time-lapse imaging of medaka early embryos expressing GFP-Histone2B after control MO injection. Imaging was performed by DSLM in 1-min intervals for 40 min starting 4 h after fertilization.
Time-lapse imaging of medaka early embryos expressing GFP-Histone2B after anti-SSX2IP MO injection. Imaging was performed by DSLM in 1-min intervals for 40 min starting 4 h after fertilization.
Time-lapse imaging of medaka early embryos expressing GFP-Histone2B after anti-SSX2IP MO injection. Imaging was performed by DSLM in 1-min intervals for 40 min starting 4 h after fertilization.
Chromosome segregation defects in SSX2IP2 MO–treated eggs. (A) Low concentrations of SSX2IP antibodies (100 ng/µl, injected at one-cell stage eggs of medaka) were used to localize SSX2IP in embryos developed to stage 10–11. Colors of labels indicate colors of respective proteins. Magnified panels (magn.) show enlarged views of the boxed regions. (B) Medaka eggs were coinjected with either control MO (Cont. MO) or SSX2IP2-MO, and Histone H2B-GFP (H2B-GFP) mRNA. Developing embryos were analyzed around stage 8 with DSLM. (top) Representative global images of MO-treated embryos. Red arrowheads in SSX2IP-MO show abnormal cell divisions. (bottom) Still images of dividing blastomeres in Cont.MO and SSX2IP-MO injected eggs. Yellow arrowheads indicate dividing cells; red arrows indicate lagging chromosomes. Each image was taken with an interval of 1 min. (C) Quantification of abnormal chromosomes/lagging chromosomes in control and MO-injected eggs. The significance was calculated by a Student’s t test (two-tailed) and scored ***, P < 0.001. Error bars indicate SD. (D–F) Rescue of SSX2IP-knockdown eggs by EGFP-OlSSX2IP2 (EGFP-SSX2IP) expression. Eggs were coinjected with SSX2IP2-MO and EGFP-SSX2IP. (D) Quantification of γ-tubulin foci in MO-treated and rescued eggs. Numbers of γ-tubulin foci were categorized as shown in representative images of γ-tubulin staining (top panels). The significance was calculated using six or seven independent experiments, in which at least 100 structures were counted. Significance was scored by a Student’s t test (two-tailed) as follows: ***, P < 0.001; ****, P < 0.0001. Error bars indicate SD. (E) Quantification of abnormal spindles in MO-treated and rescued eggs. The significance was calculated by a Student’s t test (two-tailed) and scored as follows: *, P < 0.05; **, P < 0.01. Error bars indicate SD. (F) γ-tubulin and α-tubulin, in MO-treated and rescued eggs at stage 10, was visualized as indicated by the colored legends. Dashed lines crossed by a bar indicate telophase spindles. Magnified panels (magn.) show enlarged views of the boxed regions. Arrowheads indicate abnormal numbers of centrosomes. Arrows indicate localization of EGFP-SSX2IP at the centrosome. Bars: (A) 10 µm; (B, top) 40 µm; (B, bottom) 20 µm; (D) 20 µm; (F) 10 µm.
Chromosome segregation defects in SSX2IP2 MO–treated eggs. (A) Low concentrations of SSX2IP antibodies (100 ng/µl, injected at one-cell stage eggs of medaka) were used to localize SSX2IP in embryos developed to stage 10–11. Colors of labels indicate colors of respective proteins. Magnified panels (magn.) show enlarged views of the boxed regions. (B) Medaka eggs were coinjected with either control MO (Cont. MO) or SSX2IP2-MO, and Histone H2B-GFP (H2B-GFP) mRNA. Developing embryos were analyzed around stage 8 with DSLM. (top) Representative global images of MO-treated embryos. Red arrowheads in SSX2IP-MO show abnormal cell divisions. (bottom) Still images of dividing blastomeres in Cont.MO and SSX2IP-MO injected eggs. Yellow arrowheads indicate dividing cells; red arrows indicate lagging chromosomes. Each image was taken with an interval of 1 min. (C) Quantification of abnormal chromosomes/lagging chromosomes in control and MO-injected eggs. The significance was calculated by a Student’s t test (two-tailed) and scored ***, P < 0.001. Error bars indicate SD. (D–F) Rescue of SSX2IP-knockdown eggs by EGFP-OlSSX2IP2 (EGFP-SSX2IP) expression. Eggs were coinjected with SSX2IP2-MO and EGFP-SSX2IP. (D) Quantification of γ-tubulin foci in MO-treated and rescued eggs. Numbers of γ-tubulin foci were categorized as shown in representative images of γ-tubulin staining (top panels). The significance was calculated using six or seven independent experiments, in which at least 100 structures were counted. Significance was scored by a Student’s t test (two-tailed) as follows: ***, P < 0.001; ****, P < 0.0001. Error bars indicate SD. (E) Quantification of abnormal spindles in MO-treated and rescued eggs. The significance was calculated by a Student’s t test (two-tailed) and scored as follows: *, P < 0.05; **, P < 0.01. Error bars indicate SD. (F) γ-tubulin and α-tubulin, in MO-treated and rescued eggs at stage 10, was visualized as indicated by the colored legends. Dashed lines crossed by a bar indicate telophase spindles. Magnified panels (magn.) show enlarged views of the boxed regions. Arrowheads indicate abnormal numbers of centrosomes. Arrows indicate localization of EGFP-SSX2IP at the centrosome. Bars: (A) 10 µm; (B, top) 40 µm; (B, bottom) 20 µm; (D) 20 µm; (F) 10 µm.
To analyze whether defects in centrosome structure may be responsible for the observed chromosome segregation defects, we visualized centrosomal markers in embryos 6.5 h (corresponding to around stage 10 in controls) after fertilization and MO or antibody injection. A consistently much higher percentage of cells (Fig. 5 D: ∼45%, mean from six embryos) displayed four or more γ-tubulin signals indicative of centrosome fragmentation. This correlated with high frequencies (up to 50%; Fig. 5 E) of abnormal mitotic figures, including multipolar spindles as well as bent and asymmetric central spindles in anaphase and telophase (Fig. 5 F, see bars and dashed lines that indicate spindle symmetry axes). Under these conditions, cell division was slowed down as judged by a reduced number of blastomeres at early stages of development, which also resulted in later developmental defects such as reduced eye size (Fig. S4 B, red arrowheads). These structural defects satisfactorily explain the chromosome segregation defects seen in the previous live experiment. To confirm the specificity of our observation, we coinjected antisense MO–resistant versions of OlSSX2IP mRNAs along with MOs to compensate down-regulation of endogenous SSX2IP. As expected, exogenous, GFP-tagged SSX2IP localized to centrosomes in interphase and M phase as judged by γ-tubulin colocalization (Fig. 5 F). Importantly, its expression significantly reduced the number of cells with supernumerous γ-tubulin signals, almost reaching the number found in control experiments (Fig. 5 E). Likewise, reexpression of SSX2IP rescued the structural spindle defects seen after antisense MO injection (Fig. 5 D).
SSX2IP maintains the integrity of mitotic centrosomes and is required for faithful spindle assembly in somatic cells
To better characterize the subcellular localization of SSX2IP and to analyze the role of SSX2IP in mitotic centrosome function in more detail, we finally applied experiments in human somatic cells. In contrast to mitotic spindle proteins such as TPX2 or KIF11/Eg5, human SSX2IP (59% identity with the X. laevis protein) was equally detectable in interphase and M phase of human somatic cells (Fig. S5 A). To characterize the subcellular localization of SSX2IP, we visualized SSX2IP by indirect immunofluorescence (Fig. 6, A, B, and E) in granules around centrosomes in interphase and mitosis (Fig. 6 A). Previously, the protein PCM-1 was shown to define ∼80-nm large cytoplasmic granules, termed centriolar satellites. Centriolar satellites move along MTs and accumulate at centrosomes (Bärenz et al., 2011). In interphase, SSX2IP largely merged with PCM-1 in centriolar satellites and was found adjacent to and colocalizing with γ-tubulin (Fig. 6 B, see merges with PCM-1 and γ-tubulin). In mitosis, SSX2IP still associated with γ-tubulin at spindle poles, whereas PCM-1 was found more dispersed around the poles, which is consistent with previously published data (Kubo and Tsukita, 2003; Lopes et al., 2011; Fig. 6 B, see merges of SSX2IP and γ-tubulin).
SSX2IP is required for centrosome stability in human somatic cells. (A) Localization of endogenous SSX2IP in human retinal pigment epithelium (RPE) cells in interphase and metaphase as indicated. Green, α-tubulin; blue, chromatin; red, SSX2IP; magnified merges do not display chromatin. Magnified panels (magn.) show enlarged views of the boxed regions. (B) Colocalization of SSX2IP (red), PCM-1 (green), and γ-tubulin (blue) in RPE-1 cells in interphase and mitosis; merges display colocalization between SSX2IP and PCM-1, or SSX2IP and γ-tubulin as indicated. Merges do not display chromatin. Magnified panels (magn.) show enlarged views of the boxed regions. (C) Immunoblots to document siRNA-mediated down-regulation of SSX2IP with two different siRNA oligos (Ol. 1 and 2). α-Tubulin served as a loading control. (D) Mitotic figures after knockdown of SSX2IP in RPE-1 cells. Green, α-tubulin; red, γ-tubulin; blue, DAPI/chromatin. Arrowheads indicate fragmentation of the γ-tubulin signals. (E–G) Analysis of mitotic figures in HEK293T wt cells and cells constitutively expressing low levels (“low”) of an siRNA-resistant, FLAG-tagged version of human SSX2IP and high levels after addition of doxycycline (“high”). (E) Green, γ-tubulin; red, SSX2IP; blue, DAPI/chromatin. (F) Quantification of mitotic fragmentation of γ-tubulin. The graph shows mean ± SD from five (wt) or three (stable expression of FLAG-SSX2IP) independent experiments; the significance was calculated by a Student’s t test (two-tailed) and scored as *, P < 0.05; **, P < 0.01. (G) SSX2IP (left) and γ-tubulin (right) levels at mitotic centrosomes after knockdown of SSX2IP in wt cells or cell lines stably expressing SSX2IP. The data distribution from one representative experiment out of four repetitions is shown; n > 35. Bars, 10 µm.
SSX2IP is required for centrosome stability in human somatic cells. (A) Localization of endogenous SSX2IP in human retinal pigment epithelium (RPE) cells in interphase and metaphase as indicated. Green, α-tubulin; blue, chromatin; red, SSX2IP; magnified merges do not display chromatin. Magnified panels (magn.) show enlarged views of the boxed regions. (B) Colocalization of SSX2IP (red), PCM-1 (green), and γ-tubulin (blue) in RPE-1 cells in interphase and mitosis; merges display colocalization between SSX2IP and PCM-1, or SSX2IP and γ-tubulin as indicated. Merges do not display chromatin. Magnified panels (magn.) show enlarged views of the boxed regions. (C) Immunoblots to document siRNA-mediated down-regulation of SSX2IP with two different siRNA oligos (Ol. 1 and 2). α-Tubulin served as a loading control. (D) Mitotic figures after knockdown of SSX2IP in RPE-1 cells. Green, α-tubulin; red, γ-tubulin; blue, DAPI/chromatin. Arrowheads indicate fragmentation of the γ-tubulin signals. (E–G) Analysis of mitotic figures in HEK293T wt cells and cells constitutively expressing low levels (“low”) of an siRNA-resistant, FLAG-tagged version of human SSX2IP and high levels after addition of doxycycline (“high”). (E) Green, γ-tubulin; red, SSX2IP; blue, DAPI/chromatin. (F) Quantification of mitotic fragmentation of γ-tubulin. The graph shows mean ± SD from five (wt) or three (stable expression of FLAG-SSX2IP) independent experiments; the significance was calculated by a Student’s t test (two-tailed) and scored as *, P < 0.05; **, P < 0.01. (G) SSX2IP (left) and γ-tubulin (right) levels at mitotic centrosomes after knockdown of SSX2IP in wt cells or cell lines stably expressing SSX2IP. The data distribution from one representative experiment out of four repetitions is shown; n > 35. Bars, 10 µm.
To analyze SSX2IP function in somatic cells, we depleted the protein by RNAi (Fig. 6, C, D, and E). In SSX2IP knockdown cells, we detected reduced γ-tubulin levels at mitotic centrosomes (Fig. 6, D, E, and G) as well as frequent (Fig. 6 F and Fig. S4, C and D) aberrations in structure or stability of mitotic MTOCs indicated by ectopic γ-tubulin–positive foci (Fig. 6, D–F; and Fig. S5 B). Ectopic foci contained additional pericentriolar matrix (PCM) proteins including pericentrin and CDK5RAP2 (Figs. S5 E). The fact that centrin (Fig. S5 E) was not found in ectopic foci suggested fragmentation of PCM rather than overduplication of centrioles. To confirm the specificity of our observation, we generated HEK293T cell lines stably expressing an RNAi-resistant version of human SSX2IP. Constitutive, low-level expression of exogenous, FLAG-tagged SSX2IP (Fig. 6, E and G) rendered these cells insensitive to SSX2IP-RNAi knockdown, which consistently caused PCM fragmentation (Fig. 6 F) and reduced γ-tubulin levels at mitotic centrosomes in wild-type (wt) HEK293T cells (Fig. 6, E and G). SSX2IP overexpression led to a strong accumulation of the protein at mitotic centrosomes and rescued RNAi-mediated mitotic centrosome defects (Fig. 6, E–G).
To analyze the functional significance of our observations for cell division, we applied time-lapse imaging of HeLa cells stably expressing fluorescently labeled Histone2B and α-tubulin (see Videos 3–5) that synchronously proceeded through mitosis after SSX2IP down-regulation (Fig. 7 A). Phenotype annotation using the CellCognition framework (Held et al., 2010) suggested normal mitotic entry but delayed mitotic progression after SSX2IP down-regulation (Fig. 7, B and C), which is indicative of a persistently unsatisfied spindle assembly checkpoint. Further quantification of mitosis revealed mitotic retardation (Fig. 7 D), which was specifically caused by prolonged metaphases (Fig. 7 E, time raised from 29 to 47/45 min) after impaired chromosome alignment (Fig. 7 C, see arrowheads that indicate nonaligned chromosomes). Collectively, these data indicate that the function of SSX2IP is required for proper chromosome segregation in human somatic cells. Mitotic defects are correlated with, and most likely caused by, fragmentation of the pericentriolar material after SSX2IP knockdown. Our results confirm the key function of SSX2IP as a mitotic centrosome maturation and maintenance factor.
48-h time-lapse imaging of Thymidine-synchronized human cells stably expressing GFP-Histone2B and mCherry-tubulin under control conditions. Imaging was performed in 20-min time intervals on an inverted microscope (IX81; Olympus) using a 20× objective lens. Bar, 20 µm.
48-h time-lapse imaging of Thymidine-synchronized human cells stably expressing GFP-Histone2B and mCherry-tubulin under control conditions. Imaging was performed in 20-min time intervals on an inverted microscope (IX81; Olympus) using a 20× objective lens. Bar, 20 µm.
48-h time-lapse imaging of Thymidine-synchronized human cells stably expressing GFP-Histone2B and mCherry-tubulin after SSX2IP knockdown (oligo 1). Imaging was performed in 20-min time intervals on an inverted microscope (IX81; Olympus) using a 20× objective lens. Bar, 20 µm.
48-h time-lapse imaging of Thymidine-synchronized human cells stably expressing GFP-Histone2B and mCherry-tubulin after SSX2IP knockdown (oligo 1). Imaging was performed in 20-min time intervals on an inverted microscope (IX81; Olympus) using a 20× objective lens. Bar, 20 µm.
48-h time-lapse imaging of Thymidine-synchronized human cells stably expressing GFP-Histone2B and mCherry-tubulin after SSX2IP knockdown (oligo 2). Imaging was performed in 20-min time intervals on an inverted microscope (IX81; Olympus) using a 20× objective lens. Bar, 20 µm.
48-h time-lapse imaging of Thymidine-synchronized human cells stably expressing GFP-Histone2B and mCherry-tubulin after SSX2IP knockdown (oligo 2). Imaging was performed in 20-min time intervals on an inverted microscope (IX81; Olympus) using a 20× objective lens. Bar, 20 µm.
Knockdown of SSX2IP compromises mitotic MT nucleation and dynamics, and delays mitotic progression. Mitotic progression of HeLa cells stably expressing Histone 2B was analyzed by time-lapse recording. (A) Experimental setup. (B) Analysis using the CellCognition (Held et al., 2010) image recognition software. An early mitotic index (prometaphase, metaphase) was determined via automatic annotation; one representative analysis (out of at least four analyses) for the indicated situations is shown. (C) Histone 2B still images (20-min intervals) in cells progressing through mitosis in control situations, or after knockdown of SSX2IP. Arrowheads indicate nonaligned chromosomes; see also Videos 4 and 5. Bar, 10 µm. (D and E) Quantification of mitotic timing (D: overall duration; E: single mitotic phases; PRO, prophase; PM, prometaphase; M, metaphase; A, early anaphase; TELO, late anaphase and telophase) in control and SSX2IP knockdown cells that were analyzed by manual tracking for all conditions (n ≥ 34, in 4/5 independent experiments). The graphs represent mean values ± SD. The significance was scored by a two-tailed Student’s t test and scored as **, P < 0.01; ***, P < 0.001.
Knockdown of SSX2IP compromises mitotic MT nucleation and dynamics, and delays mitotic progression. Mitotic progression of HeLa cells stably expressing Histone 2B was analyzed by time-lapse recording. (A) Experimental setup. (B) Analysis using the CellCognition (Held et al., 2010) image recognition software. An early mitotic index (prometaphase, metaphase) was determined via automatic annotation; one representative analysis (out of at least four analyses) for the indicated situations is shown. (C) Histone 2B still images (20-min intervals) in cells progressing through mitosis in control situations, or after knockdown of SSX2IP. Arrowheads indicate nonaligned chromosomes; see also Videos 4 and 5. Bar, 10 µm. (D and E) Quantification of mitotic timing (D: overall duration; E: single mitotic phases; PRO, prophase; PM, prometaphase; M, metaphase; A, early anaphase; TELO, late anaphase and telophase) in control and SSX2IP knockdown cells that were analyzed by manual tracking for all conditions (n ≥ 34, in 4/5 independent experiments). The graphs represent mean values ± SD. The significance was scored by a two-tailed Student’s t test and scored as **, P < 0.01; ***, P < 0.001.
Discussion
In the present work, we identify and characterize SSX2IP as a novel centrosome maturation factor. Our data reveal a unique signature of vertebrate SSX2IP: the levels of SSX2IP are tightly regulated in meiotic cell cycles. Mitotic MAPs and motor proteins, such as TPX2, Eg5, or PRC1, are regulated upon meiotic maturation as well as entry into mitosis in somatic cells. In contrast, SSX2IP can be found in interphase and mitosis of somatic cells. SSX2IP may therefore be regulated only once in the life of an organism during oocyte maturation and then be expressed throughout all somatic cell cycles. Remarkably, although SSX2IP governs mitotic cell division, it seems to be dispensable for meiotic spindle assembly although it is expressed and competent for MT binding at this stage. SSX2IP is the first centrosomal component identified so far whose expression rises in meiosis. Recently, Eckerdt et al. (2011) could show that X. laevis oocyte extracts fail to assemble centrioles de novo whereas egg extracts readily produce centrioles in response to Plx4 addition. This suggests a meiotic maturation program for centrosomal proteins, including SSX2IP, but likely also other core centrosomal proteins that will follow the example of SSX2IP. This program prepares the oocyte cytoplasm to assemble and support rapid centriole duplication and centrosome assembly.
Like many centrosomal proteins, vertebrate orthologues of SSX2IP comprise two conserved C-terminal coiled-coil motifs and contain an ∼150 amino acid–long highly conserved N-terminal domain that has been named ADIP domain after the mouse SSX2IP orthologue. This motif is found in the yeast Schizosaccharomyces pombe, several vertebrate and higher plant species. The S. pombe ADIP domain protein Msd1p was previously shown to be required for anchoring minus ends of spindle MTs to the spindle pole body (Toya et al., 2007) and the chicken orthologue of SSX2IP/ADIP (light-inducible and clock-controlled gene, LCG) was suggested to colocalize with γ-tubulin (Hatori et al., 2006) and hence to the centrosomes. Although previous proteomic surveys of centrosomes (Andersen et al., 2003; Jakobsen et al., 2011) did not identify SSX2IP, the human protein atlas clearly supports centrosomal localization (http://www.proteinatlas.org/search/ssx2ip).
We further show that vertebrate SSX2IP accumulates at MT minus ends and maintains high levels of γ-TuRC and other PCM proteins on mitotic centrosomes. Interaction of SSX2IP with several γ-TuRC components is consistent with the novel function of SSX2IP as a mitotic PCM loading and maintenance factor. Reducing the levels of SSX2IP causes fragmentation of γ-tubulin in mitotic spindle poles of human somatic cells and in developing fish. This finally results in aberrant spindles, metaphase alignment defects, and chromosome segregation failure during the rapid cleavage divisions in early embryos.
The most dramatic spindle defects after reducing SSX2IP levels were seen in X. laevis cell-free extracts. Here, the in vitro system allowed acute and near complete depletion of SSX2IP. Under these conditions, centrosomes displayed severely decreased PCM size (seen by reduced levels of γ-TuRC components). They failed to maintain both basic levels of nucleation and chromatin-dependent increase in nucleation from centrosomes. As a consequence, mitotic MTs did not form bipolar spindles but assembled monopolar arrays or half spindles. Although codepletion of γ-TuRC and PCM-1 (Fig. S3 B) may have prevented a full rescue, bipolar spindle formation was significantly restored upon addition of the SSX2IP-encoding mRNA. We speculate that immunodepletion of SSX2IP still enables the assembly of a first, most likely sperm centriole–derived centrosome, whereas a second functional spindle pole does not form at all. This may seem contradictory to the observation that chromatin drives bipolar spindle formation even around sperm nuclei in X. laevis egg extracts (Maresca et al., 2009). However, it is quite conceivable that severely unbalanced MT nucleation after the loss of SSX2IP, or after directly reducing γ-TuRC levels, generates a dominant-negative situation that abolishes bipolar spindle formation at the expense of monopolar structures or half spindles.
Different to the X. laevis cell-free system, SSX2IP knockdown in somatic cells or embryos leads to a gradual, chronic loss of the protein, resulting in PCM fragmentation rather than complete loss of PCM assembly. Residual SSX2IP levels will prevent complete PCM loss and result in centrosome fragmentation. These fragments in somatic cells do not contain centrin, which implies for PCM fragmentation rather than centriole overduplication.
In embryos, the less active checkpoint signaling compared with somatic cells leads to differences in the cellular response to knockdown of spindle proteins. Reduced responses in early amphibian development to DNA damage and spindle assembly errors are well known (Greenwood et al., 2001; Peng et al., 2008). Efficient activation of the spindle assembly checkpoint prevents entry into anaphase and causes a metaphase delay in somatic cells, but not in blastomeres that therefore face frequent chromosome segregation errors.
In intact embryos as well in somatic cells, SSX2IP colocalizes with the satellite marker PCM-1, and the two proteins interact with each other in X. laevis egg extracts. It thus seems very likely that SSX2IP cooperates with PCM-1 and other satellite components, such as Kizuna, Cep72, or Cep90, which have been shown previously to ensure integrity of the pericentriolar material in mitosis (Oshimori et al., 2006, 2009; Kim and Rhee, 2011). Our data therefore suggest that SSX2IP functions in centriolar satellites to deliver PCM proteins to mitotic centrosomes.
Collectively, our data converge into a model in which SSX2IP functions as an assembly and maturation factor for mitotic centrosomes. It recruits and maintains proteins of the PCM including γ-TuRC, pericentrin, and Cdk5rap2. Evidence for a functional link of SSX2IP is particularly strong for γ-TuRC because mass spectrometry data from X. laevis SSX2IP immunoprecipitations suggest a physical interaction between SSX2IP and γ-TuRC. Both in X. laevis cell-free extracts as well as in intact cells, γ-TuRC accumulation on mitotic centrosomes is significantly reduced after SSX2IP immunodepletion or knockdown. Moreover, in X. laevis cell-free extracts devoid of SSX2IP, γ-TuRC loading and activation of the nucleating capacity of centrosomes was prevented. These data suggest that SSX2IP directly interacts with γ-TuRC. Consistent with this, immunodepletion of γ-TuRC phenocopies the defects observed after SSX2IP depletion. We therefore propose that SSX2IP, possibly as part of mitotic centriolar satellites, delivers the γ-TuRC MT nucleating complex and other pericentriolar material proteins in a Dynein-dependent manner to mitotic centrosomes. Loss of SSX2IP interferes with γ-TuRC loading, initially leading to PCM fragmentation and, finally, to the complete loss of functional spindle poles.
Materials and methods
Antibodies
Primary antibodies (dilutions for immunoblot/immunofluorescence): N-terminal fragments comprising amino acids 1–390 of the human or 1–438 of the X. laevis SSX2IP proteins were used for antibody production in rabbits (500/500) or guinea pigs (500/1,000). X. laevis CLASP antibodies (1,000/−) were generated using three synthetic peptides (amino acids 237–255, 561–577, and 774–798 of X. laevis CLASP) in rabbits. Antibodies against a C-terminal fragment of human PCM-1 corresponding to nt 4,993–6,905 (Dammermann and Merdes, 2002; 500/−), and against the X. laevis C-terminal γ-tubulin peptide AATRPDYISWGTQDK (2,000/500) were generated (Zheng et al., 1995) in rabbits. Antibodies against recombinant full-length X. laevis RCC1 (Dumont et al., 2007; 100/−) and full-length recombinant X. laevis PPM1G (Petri et al., 2007; 500/−) were generated in rabbits. Antibodies against PCM-1 were provided by A. Merdes (Université de Toulouse, Toulouse, France), anti-Cdk5rap2 by F. Gergely (University of Cambridge, Cambridge, England, UK), antibodies against Eg5 by T. Surrey (London Research Institute, London, England, UK), antibodies against XKid by I. Vernos (IRB Barcelona, Barcelona, Spain), antibodies against XGrips by Y. Zheng (Carnegie Institution, Washington DC), and anti-GFP by D. Görlich (MPI for Biophysical Chemistry, Göttingen, Germany). Commercial antibodies were as follows: mouse monoclonal α-tubulin (T9026; 2,000/1,000) and mouse monoclonal γ-tubulin (T6557;10,000/500; both from Sigma-Aldrich); and mouse monoclonal Dynein intermediate chain (ab6304, 0.1 µg/µl; Abcam). Secondary antibodies (10,000/500) were: Alexa Fluor 488 goat anti–rabbit and anti–mouse (A11034 and A11001), Alexa Fluor 546 goat anti–mouse (A11030), and Alexa Fluor 633 goat anti–rabbit (A21070) antibodies (all from Life Technologies); Cy3 goat anti–mouse (115-165-062) or anti–guinea pig (706-165-148), and Dylight 549 goat anti–rabbit (111-005-003; all from Jackson ImmunoResearch Laboratories, Inc.); and Cy5 goat anti–rabbit IgG (AP132S; EMD Millipore).
Purification of MAPs from X. laevis egg extract
100 µl of egg or oocyte extracts (precleared for 1 h at 100,000 g) with 1 mM GTP, 1 mM AMPPNP, 10 µM Cytochalasin B, 8% Pentanediol, and then 50 µM Taxol (or 50 µM nocodazole) were diluted in BRB80 buffer with 1 mM GTP, 1 mM AMPPNP, and 50 µM Taxol, and spun over a cushion at 100,000 g for 10 min at 20°C, then washed once and resuspended for ICPL. To generate short MTs used for the experiment in Fig. S2 (C and D), purified taxol-stabilized MTs were sheared using a 0.22-mm needle.
ICPL labeling and mass spectrometry
Isotopic labeling was done with the Serva ICPL kit (Serva GmbH; Schmidt et al., 2005). In brief, proteins of MAP sediments were denatured using 8 M urea and, after reduction and alkylation of cysteines, labeled with the light (C12) or heavy (C13) ICPL reagent (1-[12/13C61H4]-nicotinoyloxy-succinimide). Labeled proteins were mixed, separated by SDS-PAGE, and trypsin digested using a Digest pro MS liquid handling system (Intavis AG; Catrein et al., 2005). Samples were analyzed with a mass spectrometer (ESI LTQ Orbitrap; Thermo Fisher Scientific).
Database searching
Data analysis was done with MaxQuant (version 1.1.1.1.25; see http://www.maxquant.org; Cox and Mann, 2008) and the database search software Andromeda (see http://www.maxquant.org; Cox et al., 2011). The X. laevis (32,351 entries) and Xenopus tropicalis entries (23,786 entries) were extracted from the NCBI nr database (download 18.02.2009; see: http://www.matrixscience.com). Ratios for proteins only found in all five experiments were determined using the Perseus software (see http://www.matrixscience.com) and assigning 200,000 as an artificial value for egg peptides that were missing a detectable counterpart in oocytes.
Identification of sequence motifs in the X. laevis SSX2IP 3′ UTR
Sequence motifs indicative of expression regulation were predicted as described previously (Piqué et al., 2008). In brief, the combinatorial code of CPEC (cytoplasmic polyadenylation element, conserved), HEX (hexanucleotide consensus), and PBE (Pumilio binding element) generated the predictive combinatorial code for expression regulation.
Preparation of X. laevis and medaka oocytes and eggs
X. laevis oocytes were prepared as described previously (Kuge and Richter, 1995). In brief, ovary lobules were taken from female frogs by an operation, and the tissue was washed and treated with collagenase to yield isolated oocytes (Gurdon, 1968). In vitro maturation was done in the presence of Progesterone (Dumont et al., 2007). For in vitro fertilization of X. laevis eggs (Bohnsack et al., 2006), eggs were mixed with male sperm (dispersed male testis) after removal of the egg jelly coat. Treatment of fish embryos and their preparation was done as described previously (Inoue and Wittbrodt, 2011). In brief, the embryos were incubated at 28°C until stage 10–11 of medaka developmental stage (Iwamatsu, 2004).
Egg and oocyte extracts, spindle assembly, and immunodepletions
Oocyte and egg extracts were prepared as described for egg (CSF) extracts (Murray, 1991) by a single 12,000 g centrifugation for 12 min in a rotor (SW60; Beckman Coulter). After centrifugation, the cytosolic phase was harvested by side punctuation using a 1.1-mm needle. Acentrosomal (Heald et al., 1996) and centrosomal (Sawin and Mitchison, 1991) spindles were assembled as described using either DNA-coated Dyna beads, or sperm nuclei, which were incubated in X. laevis egg extracts for an entire cell cycle. For the RanGTP-dependent centrosome maturation (Yokoyama et al., 2008), extracts were depleted of TPX2 and incubated with or without 20 µM RanQ69L-GTP in the presence of somatic centrosomes. Depletions were performed with 2 × 12.5 µg SSX2IP antibodies (rabbit), 3 × 10 µg PCM-1 antibodies or 3 × 25 µg γ-tubulin (rabbit), or 2 × 25 µg TPX2 antibodies (Gruss et al., 2001) precoupled to protein A Dynabeads (Life Technologies) for 100 µl extracts. Dynein inhibition was performed as described previously (Wittmann et al., 1998) by the addition of an antibody against the Dynein intermediate chain (0.1 µg/µl; ab6304; Abcam). For rescue experiments, XSSX2IP mRNA (mMESSAGE mMachine kit; Life Technologies) was added at 20 ng/µl. Spindles were immunostained after fixation with 0.25% glutaraldehyde and centrifugation on 12-mm round coverslips followed by incubation at −20°C in cold methanol (Wittmann et al., 1998) or squash fixed (Heald et al., 1996) by taking a droplet of the assembly reaction, which was overlayed with a 4% paraformaldehyde solution and covered with a coverslip.
Generation of RanT24N and Ran Q69L
Recombinant RanT24N and Ran Q69L were prepared (Klebe et al., 1995) after recombinant expression in Escherichia coli.
Identification of XSSX2IP interaction partners
Depletions of SSX2IP were performed with 2 × 12.5 µg SSX2IP antibodies (rabbit) for 100 µl of extract. Magnetic beads were washed four times with CSF buffer containing 0.05% Triton X-100 and an additional 100 mM KCl, eluted using SDS sample buffer, and analyzed by a mass spectrometer (ESI LTQ Orbitrap).
Visualization of SSX2IP in medaka embryos
To detect OlSSX2IP, 50 ng/µl of anti–rabbit control IgG or anti-XSSX2IP antibody (rabbit) were injected into medaka eggs that were incubated at 28°C until stage 10–11 (Iwamatsu, 2004) and fixed with 4% PFA overnight. Immunostainings were performed as described previously, omitting the heating step (Inoue and Wittbrodt, 2011). Fixed embryos were dechorionated and equilibrated in 1 × PTw (1 × PBS, pH 7.3, and 0.1% Tween), followed by fluorescent whole-mount immunostaining. All fluorescent whole-mount immunostainings with medaka blastomeres were imaged with an inverted confocal microscope (TCS SPE; Leica), with an ACS Apochromat 20×/0.60 NA IMM CORR objective or a ACS Apochromat 40×/1.15 NA oil CS 0.17/E 0.27 objective lenses. Images were acquired with Leica Application Suite Advanced Fluorescence (LAS AF) software (Leica). Images of confocal stacks of 1 airy unit were used to generate maximum projections in Image JA1 1.44a, which were further processed in Photoshop CS3 (Adobe) to adjust output levels in the same way for all images of the same experiment without changing gamma settings. All whole-mount samples were mounted with 1% low-melting agarose gel in dH2O in a glass-bottom culture dish (P35G-1.5-10C; MatTek Corporation). Eggs injected with control IgG did not show any nonspecific staining.
Knockdown of SSX2IP by MO antisense oligos
0.6 mM of antisense MO oligos with 3′ Lissamine against either OlSSX2IP1 (SSX2IP1-MO, control MO) or OlSSX2IP2 (SSX2IP2-MO) were injected into one-cell stage eggs. These were further incubated at 16°C for 6.5 h or 10 h (stage 10 or 11 in controls). The representative equal intensity of Lissamine-labeled eggs were selected and then fixed with 4% PFA at 4°C overnight. Please note that the OlSSX2IP1 corresponding paralogue was only weakly expressed compared with the OlSSX2IP2 as judged by in situ hybridization. Therefore, this MO was used as a negative control. For rescue experiments, 0.6 mM SSX2IP2-MO was mixed with 1 volume of 100 ng/µl EGFP-SSX2IP mRNA, which is OlSSX2IP fused N-terminally with EGFP to be resistant to MO. As a control, SSX2IP1-MO was diluted 1:1 with dH2O. The mixtures were injected into one-cell stage eggs. Abnormalities in mitotic spindles and centrosome numbers were counted manually with Cell Counter in Image JA1 1.44a using maximum intensity projections.
cDNAs, MO oligos, and plasmid constructs
O. latipes (Ol) SSX2IP2 (ENSORLT00000007560) was cloned from a medaka cDNA library into the pCS2+ or pCRII-TOPO vectors (Life Technologies) or the pEGFP-N1 (Takara Bio Inc.) vector. mRNAs were prepared with the mMESSAGE mMACHINE kit (Life technologies). SSX2IP MO oligos with 3′-Lissamine (Gene Tools, LLC) were designed against two paralogues of OlSSX2IP: SSX2IP1 (ENSORLG00000017720) and -2 (ENSORLT00000007560). The sequences of each MO are as follows: SSX2IP1-MO, 5′-TCTCAGTCAACCCCCAGTCTCCCAT-3′; and SSX2IP2-MO, 5′-AAACCAGGGAGGATTCAGGCATCAT-3′.
Morpholino injection into X. laevis oocytes
Collagenase-treated stage VI oocytes were injected with two MOs (1 µM) and further matured in vitro: No. 1, 5′-ATGGATATTATGTCAAACAGCAGGC-3′; and No. 2, 5′-TGTGAAGGTAATACAGTCAAGCAGT-3′.
Cell culture, transfection of plasmids or siRNAs, and generation of stable SSX2IP expressing cell line
HeLa Kyoto and wt and SSX2IP-expressing HEK293T cells were grown in high-glucose DMEM (Life technologies) and RPE-1 cells in DMEM/F12 (50:50, Life technologies) with 10% FCS/2 mM l-glutamine. Transfections were performed with Lipofectamine 2000 (plasmids) or RNAiMAX (siRNAs; Life Technologies: s42131 [5′-GCAUGUCUAAACUUACUAATT-3′] and s42132 [5′-GGGACAAUCUUAUAGUGCATT-3′] used at 20 nM). Cells were synchronized using a Thymidine block (Stein et al., 1994). In brief, cells were arrested at the G1/S boundary by the addition of 2 mM Thymidine for 19 h before release. A second Thymidine block was used for the immunoblot in Fig. S5 A. For cell lines stably expressing siRNA-resistant human SSX2IP, we mutated three residues pairing to the seed region of oligo s97398, fused the mutated cDNA with a FLAG-tag cDNA, and recombined the construct into HEK293T Flp-In T-Rex cells (Life Technologies).
Life imaging
HeLa Kyoto cells stably expressing mCherry-Histone2B and EGFP–α-tubulin were used for time-lapse experiments. Cells were generated using random integration of pH2B-mCherry-IRES-neo3 and pmEGFP-α-tubulin-IRES puro 2b plasmids (Schmitz et al., 2010). Cells were plated on µ-slide 8-well ibiTreat chambers (ibidi GmbH) and imaged for 48 h on a CellR system (Olympus) in 20-min intervals. We used an inverted microscope (IX81; Olympus) with a UPLS-Apochromat 20×/0.75 NA air objective lens and a camera (ORCA-R2; Hamamatsu), and the Olympus xcellence software for image acquisition. Cells were kept in DMEM/FCS at 37°C and 5% CO2 during imaging. For temperature control we used a microscope stage incubation system (Okolab) with a low-temperature thermostat (Lauda). Images were background subtracted and spectral unmixed. Finally, z-stack (2 µm in z, 0.5 µm step size) images were deconvolved using a Wiener filter (xcellence software; Olympus).
Live imaging of medaka embryos during early cleavage divisions was performed on pairs of siblings, each at the zygote stage injected with either MOs or mock control. Both siblings were rolled on sandpaper to smoothen their chorion and mounted together within a cylinder of 1% low-melting-temperature agarose (Biozym). 100 ng/µl Histone H2B (H2B)-GFP mRNA was coinjected with the MOs. Eggs were incubated for 4–5 h at 26°C. SSX2IP knockdown and control eggs with chorion were mounted on the same sample holder for digital scanned laser light sheet microscopy (DSLM). The blastoderm of both embryos was recorded every minute as described previously (Keller et al., 2010). Subsequent image processing and scoring of nuclei phenotypes was performed using Fiji (Schindelin et al., 2012).
Imaging of fixed specimens
HeLa cells were grown on 0.01% poly-l-lysine-coated coverslips and fixed in methanol at –20°C for 5 min. X. laevis eggs were processed for imaging as described previously (Dumont et al., 2007). In vitro assembled spindles in X. laevis egg extract were processed for immunofluorescence as described in the egg and oocyte extract methods (Wittmann et al., 1998). Images of in vitro assembled structures were taken with either an upright microscope (DMRXA; Leica) equipped with a camera (ORCA-ER; Hamamatsu Photonics) using a Plan-Apochromat 40×/1.25–0.75 NA oil objective lens for image acquisition and the Openlab 4.0.2 software, or a confocal system (LSM780; Carl Zeiss). Here, we used an inverted microscope (Axio Observer; Carl Zeiss) with a C-Apochromat 40×/1.2 NA W Korr-UV-VIS-IR objective lens to image spindles in X. laevis oocytes and a Plan-Apochromat 63×/1.4 NA oil objective lens for X. laevis egg extract samples. Images of confocal stacks of 1 airy unit were used to generate maximum projections in Image J64 1.45S. Images were further processed in Photoshop CS5 to adjust output levels in the same way for all images of the same experiment without changing gamma settings.
Statistical data analysis
Figure 2 C, depletion.
This experiment was completed once, n = 100. Antibody addition: chromatin bead spindles from three independent experiments (samples of 10 µl each) were analyzed and means ± SD were calculated.
Figure 3 E.
At least 46 spindles were analyzed for each condition in three independent experiments. Mean values ± SD were calculated.
Figure 4 B (left).
SSX2IP was quantified by immunoblotting in three independent experiments. Mean values ± SD were calculated using SSX2IP in controls for normalization.
Figure 4 I.
At least 89 spindles were analyzed for each experimental condition in three independent experiments. Mean values ± SD were calculated.
Figure 5 C.
Each counting of abnormal dividing cells relative to the total number of dividing cells in blastomeres was performed within 10 time points (three independent embryos at stage 10–11, minimum cell number: 59 cells/embryo).
Figure 5 D.
At least six eggs per sample from two independent experiments were used. Error bars indicate SD.
Figure 5 E.
Cells stained with α-tubulin antibodies in mitosis were analyzed (10 mitotic cells on average per section). Error bars indicate SD.
Figure 6 F.
Mean values ± SD from five (HEK293T wt) or three (stable cell line) independent experiments. n = 40 each.
Figure 6 G.
The data distribution of a single (out of four) representative experiment with n > 35 spindle poles.
Figure 7 (B and E).
Analysis of the Histone 2B signal was performed using the CellCognition software (Held et al., 2010).
Figure 7 (G and H).
30–50 cells from four (controls) or five (each knockdown) experiments were analyzed manually, and medians from individual experiments were determined, averaged, and plotted ± SD.
Figure S5 (C and D).
All quantifications were done using mean values ± SD from three independent experiments analyzing 100 cells in each experiment.
Significances were calculated by Student’s t tests (two-tailed). The α value for all statistical analyses was 0.05. P-values for all tests: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Online supplemental material
Fig. S1 shows the primary structure of X. laevis SSX2IP and highlights a peptide comprising amino acids 183–191 exclusively identified in egg extracts by quantitative mass spectrometry. Fig. S2 shows MT minus end binding and the function of SSX2IP in spindle formation analyzed in X. laevis egg extracts using immunodepletion and the addition of specific antibodies against SSX2IP or the minus end motor Dynein. Fig. S3 shows interaction partners of SSX2IP identified by mass spectrometry and confirmed by partial co-depletion with SSX2IP from X. laevis egg extracts. Fig. S4 shows SSX2IP localization in medaka embryos analyzed by specific antibodies generated in guinea pigs, and SSX2IP function analyzed after MO-mediated knockdown of the protein. Fig. S5 shows cell cycle–dependent expression of SSX2IP in synchronized human cells and its function in maintaining centrosome integrity evaluated by siRNA mediated knockdown. Videos 1 (control) and 2 (SSX2IP knockdown) show time-lapse imaging (1-min intervals) of developing medaka embryos that were injected with a Histone 2B mRNA to track nuclear divisions. Videos 3 (control), 4 (SSX2IP knockdown 1), and 5 (SSX2IP knockdown 2) show time-lapse imaging of human HeLa cells expressing mCherry-tubulin and EGFP-Histone2B performed for 48 h in 20-min intervals. Table S1 shows information on all identified MAPs and their ratios in egg relative to the oocyte state. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201302122.dv.
Acknowledgments
We thank Marie-Helene Verlhac for discussions and preliminary experiments in mouse oocytes and Gislene Pereira for insightful discussions. Antibodies against PCM-1 were kindly provided by Andreas Merdes, anti-Cdk5rap2 by Fanni Gergely, antibodies against Eg5 by Thomas Surrey, antibodies against XKid by Isabelle Vernos, antibodies against XGrips by Yixian Zheng, and anti-GFP by Dirk Görlich. We thank the Interfakultäre Biomedizinische Forschungseinrichtung (IBF) of Heidelberg University and Kresimir Crnokic from the European Molecular Biology Laboratory (EMBL) animal facility. We are grateful to Christian Hoerth of the ZMBH Light Microscopy Facility for excellent technical support.
F. Bärenz was supported by contract research “Proteinbiochemie/Proteomics“ of Baden-Württemberg Stiftung and the Deutsche Forschungsgemeinschaft (DFG 1737/4-3 to O.J. Gruss), and D. Inoue was supported by a Human Frontier Science Program (HFSP) postdoctoral fellowship.
References
- ADIP
Afadin DIL domain–interacting protein
- DSLM
digital scanned laser light sheet microscopy
- ESI
electrospray ionization
- γ-TuRC
γ-tubulin ring complex
- ICPL
isotope coded protein label
- MAP
microtubule-associated protein
- MO
morpholino
- MT
microtubule
- PCM
pericentriolar material
- PCM-1
pericentriolar material protein 1
- SSX2IP
synovial sarcoma X breakpoint 2 interacting protein
- wt
wild type
- XSSX2IP
X. laevis SSX2IP
Author notes
J. Tegha-Dunghu’s present address is Dept. of Biological Sciences, University of Alberta, Edmonton, Alberta T6G 2E9, Canada.
D. Mayilo’s present address is Friedrich Miescher Institute for Biomedical Research, 4058 Basel, Switzerland.
F. Bärenz’s present address is German Cancer Research Center (DKFZ), 69120 Heidelberg, Germany.