DNA replication depends on a preceding licensing event by Cdt1 and Cdc6. In animal cells, relicensing after S phase but before mitosis is prevented by the Cdt1 inhibitor geminin and mitotic cyclin activity. Here, we show that geminin, like cyclin B1 and securin, is a bona fide target of the spindle checkpoint and APC/CCdc20. Cyclin B1 and geminin are degraded simultaneously during metaphase, which directs Cdt1 accumulation on segregating sister chromatids. Subsequent activation of APC/CCdh1 leads to degradation of Cdc6 well before Cdt1 becomes unstable in a replication-coupled manner. In mitosis, the spindle checkpoint supports Cdt1 accumulation, which promotes S phase onset. We conclude that the spindle checkpoint, APC/CCdc20, and APC/CCdh1 act successively to ensure that the disappearance of licensing inhibitors coincides exactly with a peak of Cdt1 and Cdc6. Whereas cell cycle entry from quiescence requires Cdc6 resynthesis, our results indicate that proliferating cells use a window of time in mitosis, before Cdc6 is degraded, as an earlier opportunity to direct S phase.
Introduction
In each cell cycle, initiation of a new round of DNA replication should be restricted until after completion of the previous nuclear division (Mailand and Diffley, 2005; Arias and Walter, 2007). To prepare for S phase, DNA replication is licensed by the ATP-dependent loading of the MCM2-7 helicase to chromosome-bound ORC1-6 complexes. This process begins after mitosis and is controlled by two licensing factors, the pre-replication complex (preRC) components Cdt1 and Cdc6. Loaded MCM2-7 hexamers are activated toward the end of G1 phase when they unwind DNA to enforce polymerase recruitment and allow progression of the replication fork.
Cyclin–Cdk1 complexes that accumulate between S phase and mitosis form a principle DNA replication inhibitory activity, in part by preventing effective use of Cdc6 (Piatti et al., 1996; Honey and Futcher, 2007). Furthermore, the E3 ligase Cul4–DDB1–Cdt2 eliminates Cdt1 at the onset of DNA replication when it is recruited by chromatin-bound PCNA (Senga et al., 2006).
In animal cells, geminin, a Cdt1 binder and inhibitor that accumulates with similar kinetics in the cell cycle as cyclin B1, safeguards against unscheduled replication, too. However, it is unclear exactly when in the cell cycle mammalian geminin is degraded. Several studies suggested that in re-replicating or endo-reduplicating cells, geminin degradation relies on Cdh1 (Diffley, 2004; Li and Blow, 2004; Di Fiore and Pines, 2007; Narbonne-Reveau et al., 2008; Zielke et al., 2008). Also in proliferating somatic cells, geminin degradation had been attributed to the APC/C activator Cdh1, variably timed to coincide with either sister chromatid disjunction or G1 phase (Diffley, 2004; Li and Blow, 2004; Pines, 2006; Di Fiore and Pines, 2007; Narbonne-Reveau et al., 2008; Sakaue-Sawano et al., 2008; Skaar and Pagano, 2008; Zielke et al., 2008; Colombo et al., 2010; Emanuele et al., 2011). In such a model, degradation of cyclin B1, which inactivates Cdk1 and leads to activation of APC/CCdh1, could initiate degradation of geminin. Alternatively, somatic geminin may be targeted by the mitotic APC/C activator Cdc20, similar to the situation in Xenopus egg extracts (McGarry and Kirschner, 1998). Nevertheless, Cdc20 dependency in itself cannot reveal when geminin is degraded because we and others found that different pools of Cdc20 operate at different times in mammalian mitosis. These contribute to the order of APC/C substrate degradation. For example, proposed APC/CCdc20 substrates Nek2A, p21, cyclin A, and Mcl1 are targeted right after nuclear envelope breakdown (NEB), during prometaphase (Hames et al., 2001; Amador et al., 2007; Wolthuis et al., 2008; Harley et al., 2010), while two other important substrates, cyclin B1 and securin, are stabilized by the spindle checkpoint until sister chromatid bi-orientation on the mitotic spindle is complete (Pines, 2006). Furthermore, several other APC/CCdc20 substrates, including CENP-F and Plk1, are not processed until after sister chromatid disjunction, suggesting a role for Cdc20 activity in anaphase (Floyd et al., 2008; Gurden et al., 2010). Because geminin and cyclin B1–Cdk1 are both potent inhibitors of DNA replication (Diffley, 2004; Hochegger et al., 2007), their inactivation should be coordinated to make licensing decisive, but how this takes place is unknown.
Another question regarding APC/C-dependent timing mechanisms for replication licensing is why, paradoxically, the licensing inhibitor geminin and the MCM loader Cdc6 both become APC/C substrates upon mitotic exit. Furthermore, it is unclear how a reported positive role for geminin in replication licensing could be separated from its well-documented licensing inhibitory role in interphase (Ballabeni et al., 2004). To shed light on these matters, here we investigated in detail how the protein destruction events that connect mitosis to S phase are scheduled in proliferating cells.
Results
Geminin is degraded at the same time as cyclin B1
To compare the destruction of the licensing inhibitor geminin to that of other APC/C substrates, we first immunoblotted synchronized cells collected at different time points after mitosis. This showed that, like cyclin B1, cellular geminin disappeared completely after release from a mitotic block (Fig. 1 A and Fig. S1 A). Geminin roughly remained absent during the first half of G1 phase, even in rapidly cycling U2OS cells. These findings are different from the situation in Xenopus egg extracts that degrade only 50% of geminin (Li and Blow, 2004). Interestingly, degradation of Cdc6, like that of Aurora A, was delayed in comparison to degradation of geminin and cyclin B1. Cdc6 reached its lowest levels ∼2 h after nocodazole release (Fig. 1 A, three independent experiments quantified in Fig. 1 B). To determine when degradation of these different APC/C substrates started in relation to anaphase and cytokinesis, we filmed the levels of their fluorescent versions while cells proceeded through mitosis. Degradation of geminin-Cherry began when all the chromosomes aligned at the metaphase plate, exactly when both cyclin B1–Cherry and securin-Venus started disappearing (Fig. 1 C, panels 1, 2, and 3, respectively; Fig. S1 B). Geminin was lost progressively during metaphase and early anaphase (Fig. 1 C, quantified in Fig. 1 D). The fluorescently tagged version of geminin appeared to be functional because DNA re-replication induced by geminin shRNA was complemented by coexpression of an shRNA-resistant geminin-Venus cDNA (Fig. S1 C). Endogenous and tagged APC/C substrates were degraded with comparable timing and efficiencies (Fig. 1, B and D), regardless of the cell line investigated (Fig. S1, E and F). Summarized, these experiments show that geminin degradation starts at metaphase. In contrast, however, Aurora A–ECFP and Cdc6-Venus fluorescence levels only declined after anaphase onset (Fig. 1 C, panel 4 and 5; Fig. S1 G shows U2OS and RPE-1-TERT cells expressing Cdc6-Venus). We conclude that cyclin B1 and geminin are targeted for destruction at the same time, when Cdc6 and Aurora A are still stable.
Geminin is degraded at the same time as cyclin B1. (A) Endogenous geminin is fully degraded at mitotic exit (also see Fig. S1 A). U2OS cells arrested in mitosis by nocodazole were collected by mitotic shake-off and released for the indicated hours. APC3 is highly phosphorylated in mitosis. Actin is used as a loading control. The data shown are a representative from at least three repeated experiments. (B) Relative protein levels of geminin, cyclin B1, Cdc6, and Aurora A are calculated and corrected for actin loading (data from three independent experiments; mean ± SEM). (C) Live U2OS cells expressing geminin-Cherry (panel 1), cyclin B1-Cherry (panel 2), securin-Venus (panel 3), Cdc6-Venus (panel 4), and Aurora A–ECFP (panel 5) were imaged by differential interference contrast (DIC, bottom) and fluorescence (top) microscopy. Still images of the indicated phases in mitosis are shown. Arrows point to the metaphase plate. Bars, 10 µm. (D) The intensity of fluorescence in U2OS cells expressing the different constructs was plotted against time after anaphase (geminin-Cherry and cyclin B1-Cherry, n = 4; securin-Venus and Aurora A–ECFP, n = 5; Cdc6-Venus, n = 11; mean ± SEM).
Geminin is degraded at the same time as cyclin B1. (A) Endogenous geminin is fully degraded at mitotic exit (also see Fig. S1 A). U2OS cells arrested in mitosis by nocodazole were collected by mitotic shake-off and released for the indicated hours. APC3 is highly phosphorylated in mitosis. Actin is used as a loading control. The data shown are a representative from at least three repeated experiments. (B) Relative protein levels of geminin, cyclin B1, Cdc6, and Aurora A are calculated and corrected for actin loading (data from three independent experiments; mean ± SEM). (C) Live U2OS cells expressing geminin-Cherry (panel 1), cyclin B1-Cherry (panel 2), securin-Venus (panel 3), Cdc6-Venus (panel 4), and Aurora A–ECFP (panel 5) were imaged by differential interference contrast (DIC, bottom) and fluorescence (top) microscopy. Still images of the indicated phases in mitosis are shown. Arrows point to the metaphase plate. Bars, 10 µm. (D) The intensity of fluorescence in U2OS cells expressing the different constructs was plotted against time after anaphase (geminin-Cherry and cyclin B1-Cherry, n = 4; securin-Venus and Aurora A–ECFP, n = 5; Cdc6-Venus, n = 11; mean ± SEM).
Destruction of geminin and Cdc6 requires different APC/C activators
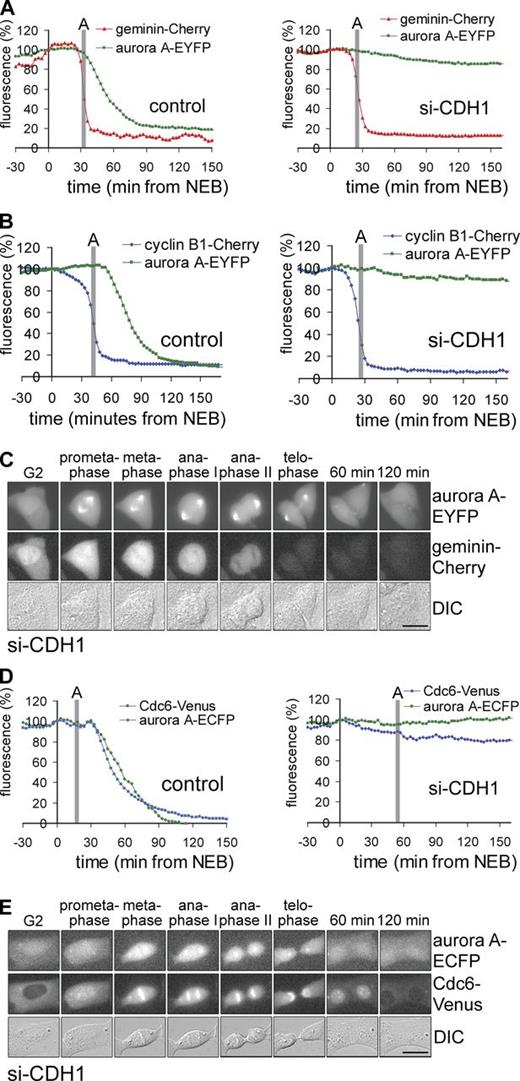
Subsequently, we determined the roles of APC/C activators Cdc20 and Cdh1, and the spindle checkpoint, in the observed degradation events. Cyclin B1 is an APC/CCdc20 substrate but several reports mentioned that somatic geminin is a target of APC/CCdh1. To resolve this issue, we compared live-cell degradation kinetics of cyclin B1 and geminin to that of the established APC/CCdh1 target Aurora A (Floyd et al., 2008; García-Higuera et al., 2008). In control cells, coexpressed Aurora A–EYFP started to be degraded after geminin-Cherry or cyclin B1–Cherry had disappeared (Fig. 2, A and B, left). However, in cells depleted of Cdh1 by siRNA, Aurora A–EYFP was fully stabilized during and after mitosis (Fig. 2 A and 2 B, right) while degradation of geminin-Cherry or cyclin B1–Cherry progressed completely normally and cells exited mitosis without obvious delay (Fig. 2 C; Fig. 2, A and B, right; Fig. S2 A shows the effect of an independent Cdh1 shRNA targeting construct). Importantly, Cdc6-Venus was degraded at the exact same time as Aurora A in control cells, starting right after sister chromatid separation (Fig. 2 D, left). When cells were depleted of Cdh1 by siRNA, Cdc6-Venus, like Aurora A, remained stable even as cells divided and entered G1 (Fig. 2 E; Fig. 2 D, right; Fig. S2 B shows an independent Cdh1 shRNA). In contrast, geminin remained absent independently of APC/CCdh1 at least during the first three hours of G1 phase (Fig. 2 A, right; Fig. S2 A). Altogether, these live-cell destruction assays show that geminin and Cdc6 are degraded at different times and via different APC/C activators.
Destruction of geminin and Cdc6 requires different APC/C activators. (A and B) Live U2OS cells expressing Aurora A–EYFP and geminin-Cherry (A) or cyclin B1–Cherry (B) were imaged by DIC and fluorescence microscopy. Left, control; right, cotransfected with si-CDH1. The intensity of fluorescence was plotted against time after NEB. The start of anaphase (A) was indicated as detected by DIC microscopy. Data are from individual cells representative of three independent experiments. Control of single shRNA is shown in Fig. S2 A. (C) Still images of the cell in A cotransfected with si-CDH1. The different phases of mitosis are indicated. Minutes refer to time passed after anaphase onset. (D) Live U2OS cells expressing Cdc6-Venus and Aurora A–ECFP were imaged by DIC and fluorescence microscopy. Left, control; right, cotransfected with si-CDH1. The intensity of fluorescence was plotted against time after NEB. The start of anaphase (A) was indicated as observed by DIC microscopy. Data are from individual cells representative of three independent experiments. Control of single shRNA is in Fig. S2 B. (E) Still images of the cell in D cotransfected with si-CDH1. Different phases of mitosis are indicated, or minutes after anaphase onset. Nuclear Cdc6 levels start to decline at the last time point by an APC/C-independent pathway (not depicted). Bars, 10 µm.
Destruction of geminin and Cdc6 requires different APC/C activators. (A and B) Live U2OS cells expressing Aurora A–EYFP and geminin-Cherry (A) or cyclin B1–Cherry (B) were imaged by DIC and fluorescence microscopy. Left, control; right, cotransfected with si-CDH1. The intensity of fluorescence was plotted against time after NEB. The start of anaphase (A) was indicated as detected by DIC microscopy. Data are from individual cells representative of three independent experiments. Control of single shRNA is shown in Fig. S2 A. (C) Still images of the cell in A cotransfected with si-CDH1. The different phases of mitosis are indicated. Minutes refer to time passed after anaphase onset. (D) Live U2OS cells expressing Cdc6-Venus and Aurora A–ECFP were imaged by DIC and fluorescence microscopy. Left, control; right, cotransfected with si-CDH1. The intensity of fluorescence was plotted against time after NEB. The start of anaphase (A) was indicated as observed by DIC microscopy. Data are from individual cells representative of three independent experiments. Control of single shRNA is in Fig. S2 B. (E) Still images of the cell in D cotransfected with si-CDH1. Different phases of mitosis are indicated, or minutes after anaphase onset. Nuclear Cdc6 levels start to decline at the last time point by an APC/C-independent pathway (not depicted). Bars, 10 µm.
The spindle checkpoint stabilizes geminin
In prometaphase, the spindle checkpoint critically protects cyclin B1 and securin from the APC/C until all chromosomes reach bi-orientation. Release of the checkpoint at metaphase liberates a fraction of Cdc20 that targets securin, causing Separase activation, and cyclin B1, driving cytokinesis by concomitant inactivation of Cdk1. Does the spindle checkpoint support geminin’s lifetime in mitosis, too? Fluorescence levels of geminin-Cherry were followed in cells transfected with an shRNA vector targeting the checkpoint protein Mad2 (pS-Mad2) or treated with 50 nM of the checkpoint kinase Mps1 inhibitor reversine shortly before mitosis (Fig. 3, A–C). Like cyclin B1, in the absence of an active spindle checkpoint geminin-Cherry disappeared exactly at NEB, which is the time when APC/CCdc20 becomes first activated (Wolthuis et al., 2008) and reached 50% of NEB levels within 13 ± 0.5 min (pS-Mad2, n = 13) or 14 ± 0.4 min (reversine, n = 7). In untreated control cells, geminin-Cherry was not degraded until metaphase and reached ∼50% of its early mitotic levels by anaphase, 32 ± 1.1 min (n = 11) after NEB (Fig. 3 B). Conversely, when cells were kept in a spindle checkpoint–dependent mitotic arrest by adding 1 µM of taxol, geminin was stabilized ∼10-fold, with levels only gradually slipping during the course of the mitotic arrest, similar to cyclin B1 (Fig. 3, D and E; Brito and Rieder, 2006). So, like cyclin B1, but unlike early APC/CCdc20 substrates such as cyclin A and Nek2A, geminin requires spindle checkpoint activity to counteract its premature loss during prometaphase. We conclude that geminin is a bona fide target of the mitotic spindle checkpoint.
The spindle checkpoint stabilizes geminin. (A) U2OS cells stably expressing geminin-Cherry were transfected with pS-MAD2, treated with Mps1 inhibitor reversine or left untreated and imaged. The intensity of fluorescence was plotted against time after NEB. Data are from individual cells representative of three independent experiments. (B) The time from NEB until 50% of geminin was degraded was measured (control, n = 11; pS-MAD2, n = 13; reversine, n = 7; mean ± SEM). (C) U2OS cells stably expressing geminin-Cherry were imaged by DIC and fluorescence microscopy after 50 nM reversine treatment. Images of the indicated phases in mitosis are shown. Note that there is no metaphase plate and that cells divide in two. Bar, 10 µm. (D) Live U2OS cells stably expressing geminin-Cherry were imaged. The mean intensity of fluorescence in taxol-treated U2OS cells expressing geminin-Cherry was plotted against time after NEB (n = 10; mean ± SEM). The graph indicates the quantification until 240 min after NEB, when some cells started to slip out of mitosis. (E) The time from NEB until 30% of geminin was degraded was measured in taxol-treated and control cells (n = 10; mean ± SEM).
The spindle checkpoint stabilizes geminin. (A) U2OS cells stably expressing geminin-Cherry were transfected with pS-MAD2, treated with Mps1 inhibitor reversine or left untreated and imaged. The intensity of fluorescence was plotted against time after NEB. Data are from individual cells representative of three independent experiments. (B) The time from NEB until 50% of geminin was degraded was measured (control, n = 11; pS-MAD2, n = 13; reversine, n = 7; mean ± SEM). (C) U2OS cells stably expressing geminin-Cherry were imaged by DIC and fluorescence microscopy after 50 nM reversine treatment. Images of the indicated phases in mitosis are shown. Note that there is no metaphase plate and that cells divide in two. Bar, 10 µm. (D) Live U2OS cells stably expressing geminin-Cherry were imaged. The mean intensity of fluorescence in taxol-treated U2OS cells expressing geminin-Cherry was plotted against time after NEB (n = 10; mean ± SEM). The graph indicates the quantification until 240 min after NEB, when some cells started to slip out of mitosis. (E) The time from NEB until 30% of geminin was degraded was measured in taxol-treated and control cells (n = 10; mean ± SEM).
Increased licensing opportunity during mitotic exit
We then asked whether geminin degradation depends on mitotic exit or exclusively on APC/CCdc20. When cells coexpressing geminin-Venus and cyclin B1–Cherry were filmed upon depletion of Cdc20, geminin and cyclin B1 were stabilized equally (Fig. 4 A; Fig. S2 C shows the effect of an independent Cdc20 shRNA-targeting construct). Endogenous securin, cyclin B1, and geminin remained stable after forcing siCDC20-arrested cells to exit mitosis by treating them with the Cdk1 inhibitor roscovitine (Fig. 4 B, compare lanes 2 and 4). Furthermore, we followed geminin degradation in live cells expressing nondegradable cyclin B1 (R42A–cyclin B1–Cerulean; Clute and Pines, 1999). This mutant of cyclin B1 maintains mitotic Cdk1 activity, arresting cells in mitosis and repressing APC/CCdh1, while the spindle checkpoint can be satisfied leading to degradation of Cdc20 targets (Fig. 4 C). Endogenous geminin was degraded together with endogenous cyclin B1 and securin but the APC/CCdh1 substrates Cdc6 and Aurora A remained stable in R42A–cyclin B1 expressing metaphase cells (Fig. 4 C). In our time-lapse assay, geminin-Cherry was lost with normal kinetics in mitotic R42A–cyclin B1 cells (Fig. 4, D and E). We conclude that geminin degradation is not an effect of cell cycle progression or reducing Cdk1 activity but depends exclusively on APC/CCdc20 like cyclin B1 and securin. When geminin-Cherry and Cdc6-Venus were coexpressed in the same cells, satisfaction of the spindle checkpoint initiated geminin degradation while Cdc6 started to disappear only after geminin had been degraded (Fig. 4 F). This suggests that mitotic exit creates a peak in the opportunity to license DNA during the narrow window of time that opens by APC/CCdc20 (degradation of cyclin B1 and geminin) and closes by APC/CCdh1 (degradation of the preRC factor Cdc6; Fig. 4 G).
Increased licensing opportunity during mitotic exit. (A) The fluorescence intensity in live U2OS cells coexpressing cyclin B1–Cherry and geminin-Venus, cotransfected with si-CDC20 was plotted against time after NEB. Note that cells arrest in metaphase (n = 7; mean ± SEM). Controls are shown in Fig. S1 B and Fig. S2 C. (B) U2OS cells were treated with si-CDC20 or control RNAi. Mitotic cells were collected by mitotic shake-off and left untreated or treated for 2 h with Cdk1 inhibitor roscovitine. APC4 is used as loading control. The data shown are from a single representative experiment out of three repeats. (C) U2OS cells were transfected with R42A–cyclin B1–Cerulean, thymidine synchronized, and released for 14 h. After mitotic shake-off, cells were replated in fresh medium for indicated hours. APC/C–Cdc20 substrates, and not APC/C–Cdh1 substrates, were degraded in the nondegradable cyclin B1 arrest. Actin is used as a loading control. The data shown are from a single representative experiment out of three repeats. (D and E) The fluorescence intensity of geminin-Cherry and R42A–cyclin B1–Cerulean was plotted against time after NEB. Data are from an individual cell representative of three independent experiments (D). Images of the indicated phases in mitosis are shown E. Note that the cells did not exit mitosis but stayed arrested. Bar, 10 µm. (F) The intensity of fluorescence in live U2OS cells coexpressing geminin-Cherry and Cdc6-Venus was plotted against time from the beginning of anaphase. The fluorescence levels are relative to levels at NEB (n = 11, mean ± SEM). (G) A window of opportunity for DNA replication licensing is created by geminin destruction and closed again by Cdc6 destruction, before S phase.
Increased licensing opportunity during mitotic exit. (A) The fluorescence intensity in live U2OS cells coexpressing cyclin B1–Cherry and geminin-Venus, cotransfected with si-CDC20 was plotted against time after NEB. Note that cells arrest in metaphase (n = 7; mean ± SEM). Controls are shown in Fig. S1 B and Fig. S2 C. (B) U2OS cells were treated with si-CDC20 or control RNAi. Mitotic cells were collected by mitotic shake-off and left untreated or treated for 2 h with Cdk1 inhibitor roscovitine. APC4 is used as loading control. The data shown are from a single representative experiment out of three repeats. (C) U2OS cells were transfected with R42A–cyclin B1–Cerulean, thymidine synchronized, and released for 14 h. After mitotic shake-off, cells were replated in fresh medium for indicated hours. APC/C–Cdc20 substrates, and not APC/C–Cdh1 substrates, were degraded in the nondegradable cyclin B1 arrest. Actin is used as a loading control. The data shown are from a single representative experiment out of three repeats. (D and E) The fluorescence intensity of geminin-Cherry and R42A–cyclin B1–Cerulean was plotted against time after NEB. Data are from an individual cell representative of three independent experiments (D). Images of the indicated phases in mitosis are shown E. Note that the cells did not exit mitosis but stayed arrested. Bar, 10 µm. (F) The intensity of fluorescence in live U2OS cells coexpressing geminin-Cherry and Cdc6-Venus was plotted against time from the beginning of anaphase. The fluorescence levels are relative to levels at NEB (n = 11, mean ± SEM). (G) A window of opportunity for DNA replication licensing is created by geminin destruction and closed again by Cdc6 destruction, before S phase.
Concurrent degradation of geminin and cyclin B1 drives Cdt1 translocation
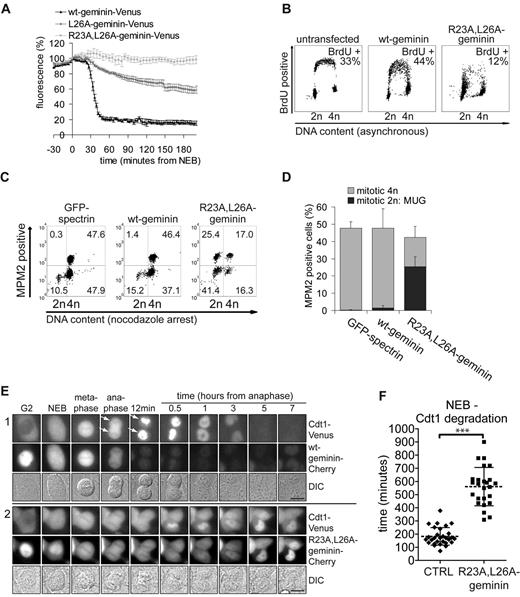
We compared the roles of cyclin B1 degradation and geminin degradation in cell cycle progression and the dynamics of the Cdt1 licensing factor. Xenopus geminin has one conserved, N-terminal destruction box (D-box, RxxLxxI) that is recognized by the APC/C–activator complex (McGarry and Kirschner, 1998). Mutating Leu26 within the homologous D-box of geminin-Venus to Ala was only partially stabilizing, but combining R23A and L26A rendered the protein completely refractory to degradation in mitosis and G1 (Fig. 5 A). Cells expressing this nondegradable geminin completed mitosis without observable defects in cytokinesis or sister chromatid separation (Fig. S3 A). They also progressed through G1 normally but then largely failed to synthesize DNA, detected as a strongly reduced incorporation of BrdU, which correlated with geminin stability (12% of cells incorporated BrdU in R23A,L26A-geminin versus 44% in wt-geminin transfected cells, Fig. 5 B). Typically, cells that expressed high levels of nondegradable geminin retained a 2n DNA content (Fig. 5 C) even when they continued to progress through the cell cycle and entered mitosis, a phenotype described earlier as mitosis with unreplicated genome (MUG; Brinkley et al., 1988; Fig. 5 C, see top-left quadrants, showing anti–MPM2-positive cells with a single genome copy; quantification in Fig. 5 D). In conclusion, whereas a failure to degrade the spindle checkpoint effectors cyclin B1 or securin results in abortion of cytokinesis or a cut-phenotype, respectively (Hagting et al., 2002), stabilizing geminin represses S phase but not initial cell cycle progression. Eventually, this leads to an aberrant mitosis after S phase fails.
Concurrent degradation of geminin and cyclin B1 drives Cdt1 translocation. (A) U2OS cells were transiently transfected with the indicated plasmids. Each curve in the graph shows the average fluorescence intensity of multiple cells (wt-geminin-Venus, n = 4; L26A-geminin-Venus, n = 5; R23A,L26A-geminin-Venus, n = 4; mean ± SEM). (B) Asynchronous cells transfected with the indicated plasmids were, 48 h later, BrdU pulsed and processed for FACS analysis. Percentages of cells incorporating BrdU are indicated. The data shown are from a single representative experiment out of three repeats. (C) U2OS cells were transfected with the indicated plasmids and after overnight nocodazole treatment, processed for FACS analysis. Cells were stained for MPM2 (y-axis) and propidium iodide (x-axis). The numbers in the quadrants indicate the percentages of 4n-mitotic cells (top-right quadrant), G2 phase cells (bottom-right quadrant), G1 plus S phase cells (bottom-left quadrant), and 2n-mitotic cells (top-left quadrant). Values indicate mean percentages from two independent experiments. (D) The percentages of mitotic cells from C being either 4n or 2n, indicated as MUG, are represented in a bar graph (n = 2; mean ± SEM). (E) Live U2OS cells stably expressing both geminin-Cherry and Cdt1-Venus were imaged by DIC and fluorescence microscopy. Cdt1-Venus localization was used as a marker for preRC formation. Still images of control cells (panel 1) and cells wherein R23A,L26A-geminin-Cherry was expressed after RNAi-complementation (panel 2) are shown (for control see Fig. S3 C). The different phases of mitosis as observed by DIC microscopy are indicated. The arrows reveal the onset of Cdt1 chromosome localization in anaphase II as detected by reduced negative staining. Bars, 10 µm. (F) The start of Cdt1 degradation marks the start of DNA replication (see also Fig. S3 C). The time from NEB till Cdt1 degradation start is measured in live U2OS cells expressing Cdt1-Venus and wt-geminin-Cherry or R23A,L26A-geminin (control, n = 29; R23A,L26A-geminin, n = 25; mean ± SD; Mann-Whitney test; ***, P < 0.0001).
Concurrent degradation of geminin and cyclin B1 drives Cdt1 translocation. (A) U2OS cells were transiently transfected with the indicated plasmids. Each curve in the graph shows the average fluorescence intensity of multiple cells (wt-geminin-Venus, n = 4; L26A-geminin-Venus, n = 5; R23A,L26A-geminin-Venus, n = 4; mean ± SEM). (B) Asynchronous cells transfected with the indicated plasmids were, 48 h later, BrdU pulsed and processed for FACS analysis. Percentages of cells incorporating BrdU are indicated. The data shown are from a single representative experiment out of three repeats. (C) U2OS cells were transfected with the indicated plasmids and after overnight nocodazole treatment, processed for FACS analysis. Cells were stained for MPM2 (y-axis) and propidium iodide (x-axis). The numbers in the quadrants indicate the percentages of 4n-mitotic cells (top-right quadrant), G2 phase cells (bottom-right quadrant), G1 plus S phase cells (bottom-left quadrant), and 2n-mitotic cells (top-left quadrant). Values indicate mean percentages from two independent experiments. (D) The percentages of mitotic cells from C being either 4n or 2n, indicated as MUG, are represented in a bar graph (n = 2; mean ± SEM). (E) Live U2OS cells stably expressing both geminin-Cherry and Cdt1-Venus were imaged by DIC and fluorescence microscopy. Cdt1-Venus localization was used as a marker for preRC formation. Still images of control cells (panel 1) and cells wherein R23A,L26A-geminin-Cherry was expressed after RNAi-complementation (panel 2) are shown (for control see Fig. S3 C). The different phases of mitosis as observed by DIC microscopy are indicated. The arrows reveal the onset of Cdt1 chromosome localization in anaphase II as detected by reduced negative staining. Bars, 10 µm. (F) The start of Cdt1 degradation marks the start of DNA replication (see also Fig. S3 C). The time from NEB till Cdt1 degradation start is measured in live U2OS cells expressing Cdt1-Venus and wt-geminin-Cherry or R23A,L26A-geminin (control, n = 29; R23A,L26A-geminin, n = 25; mean ± SD; Mann-Whitney test; ***, P < 0.0001).
It is known that licensing depends on the loading of Cdc6 and Cdt1 to chromosomal replication origins. Our live-cell analyses showed that fluorescent Cdc6 translocates to mitotic chromosomes at NEB, before cyclin B1 and geminin are degraded (Fig. 1 C, panel 4). In marked contrast, in live cells that coexpress full-length Cdt1-Venus and geminin-Cherry, Cdt1 distinctively bound late anaphase and telophase chromatids, in line with observations in fixed cells (Fig. 5 E, panel 1, arrows; Xouri et al., 2007). However, nondegradable geminin-Cherry (expressed by RNAi complementation to prevent geminin overexpression; Fig. S3 B and Fig. 5 E, panel 2) largely prevented the translocation of Cdt1-Venus to separated chromatids and newly formed G1 nuclei. The onset of Cdt1 degradation after mitosis, typically coupled to the start of active DNA replication and coinciding with the appearance of PCNA replication foci (Fig. S3 C; Nishitani et al., 2001; Arias and Walter, 2005), was strongly delayed in nondegradable geminin-expressing cells (Fig. 5 F; 561 ± 145 min after NEB in R23A,L26A-geminin, mean ± SD, versus 183 ± 67 min after NEB in control cells, mean ± SD). Interestingly, Cdt1-Venus also failed to accumulate on segregated chromatids when cytokinesis was inhibited by expressing R42A–cyclin B1, even though geminin and securin were degraded normally in these cells (Fig. S3 D; Hochegger et al., 2007). Altogether, these data indicate that geminin and cyclin B1 regulate Cdt1 independently and may have to be degraded at the same time to help translocation of Cdt1 to chromatids.
Geminin promotes Cdt1 accumulation in prometaphase
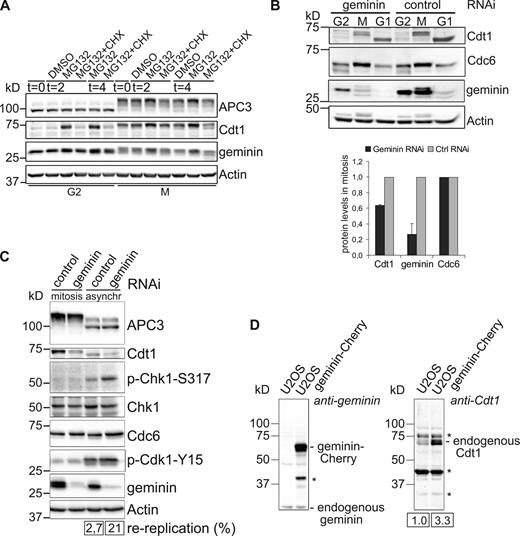
Paradoxically, apart from inhibiting Cdt1, a positive role for geminin in preRC control has been reported too, i.e., to stabilize Cdt1 in mitosis (Ballabeni et al., 2004). We found that Cdt1 is highly unstable during G2 phase and, indeed, first accumulates when cells reach mitosis (Fig. 6 A). When prometaphase-arrested cells were collected and treated with the proteasome inhibitor MG132, geminin and Cdt1 levels increased further. This was prevented by cycloheximide cotreatment. This shows that geminin and Cdt1 are rapidly produced during mitosis (Fig. 6 A, lanes 7–13). When mitotic cells were separated from interphase cells shortly after treatment with geminin RNAi, but before re-replication had occurred, Cdc6 accumulated normally yet Cdt1 increased only to 40% of its normal mitotic levels (Fig. 6 B, blots and bar graph; see Materials and methods for detailed synchronization procedure). This effect is different from the disappearance of both Cdt1 and Cdc6 that is induced by severe DNA damage after re-replication (Hall et al., 2008), because, in our case, mitotic cells were captured before re-replication started. Furthermore, the mitotic cells were negative for the DNA damage marker phospho-Chk1 (Fig. 6 C). In mitosis, Cdc6, but not Cdt1, accumulated normally after partial depletion of geminin (Fig. 6, B and C). In contrast, Chk1 phosphorylation was obvious in a cycling population of geminin RNAi cells of which 21% were re-replicating (Fig. 6 C; Fig. S4 A shows flow cytometry analysis of geminin-depleted cells). Cdc6 levels were not reduced in these cells either, indicating that the level of DNA damage after approximately one round of re-replication is too modest to affect Cdc6 (Hall et al., 2008). In summary, we therefore conclude that geminin is indeed a rate-limiting factor for the accumulation of Cdt1 during mitosis, as first proposed by Ballabeni et al. (2004). This Cdt1-stabilizing role of geminin is further illustrated by increased accumulation of Cdt1 in mitosis after overexpressing geminin-Cherry (Fig. 6 D and Fig. S4 B). Because by immunoprecipitating geminin we could deplete Cdt1 from extracts, all mitotic Cdt1 is in stable complex with geminin (unpublished data), in line with a dependency of Cdt1 stability on complex formation. Indeed, while full-length Cdt1 appears as cells enter mitosis, a mutant of Cdt1 lacking the geminin interaction domain did not accumulate before G1 (Fig. S4 C), at the time when Cdt1 is part of DNA-bound preRC complexes. Slow loss of geminin in spindle checkpoint–arrested cells, by mitotic slippage, related to a similar gradual decrease in Cdt1 during the arrest (unpublished data). Together, this means that a functional spindle checkpoint supports accumulative geminin–Cdt1 complex formation. When geminin is degraded from metaphase onwards, Cdt1 starts to translocate to chromatin where it remains stable until the start of S phase.
Geminin promotes Cdt1 accumulation in prometaphase. (A) Cdt1 is synthesized during prometaphase. Cdt1 levels in cells collected by gentle mitotic shake-off after nocodazole treatment were compared with those in G2 cells. Further protein synthesis of Cdt1 in mitosis is revealed after MG132 treatment of the nocodazole-blocked cells. Actin is used as loading control. The data shown are from a single representative experiment out of two repeats. (B) Top: U2OS cells were during thymidine release transfected with the indicated siRNAs, again thymidine synchronized, and arrested in mitosis by nocodazole treatment. Mitotic cells were collected by mitotic shake-off and released for 2 h after nocodazole wash-out or left untreated. Actin was used as loading control. Bottom: relative Cdt1, geminin, and Cdc6 protein levels in mitosis were corrected for loading and plotted. The graph indicates data from two independent experiments (mean ± SEM). (C) Mitotic cells with reduced geminin levels show no DNA damage signaling. U2OS cells were synchronized in mitosis as in B and compared with asynchronous geminin-depleted cells. Actin is used as loading control. Numbers indicate percentages of re-replicating cells (flow cytometry analysis of parallel samples is in Fig. S4 A). Note that a slight reduction of the Cdt1 signal in lane 4 correlates to the increased population of replicating cells. Replicating cells do not express Cdt1. (D) Geminin is limiting for Cdt1 accumulation. U2OS cells were stably transfected with geminin-Cherry and blotted for Cdt1. Numbers indicate relative Cdt1 levels. Asterisks indicate background bands.
Geminin promotes Cdt1 accumulation in prometaphase. (A) Cdt1 is synthesized during prometaphase. Cdt1 levels in cells collected by gentle mitotic shake-off after nocodazole treatment were compared with those in G2 cells. Further protein synthesis of Cdt1 in mitosis is revealed after MG132 treatment of the nocodazole-blocked cells. Actin is used as loading control. The data shown are from a single representative experiment out of two repeats. (B) Top: U2OS cells were during thymidine release transfected with the indicated siRNAs, again thymidine synchronized, and arrested in mitosis by nocodazole treatment. Mitotic cells were collected by mitotic shake-off and released for 2 h after nocodazole wash-out or left untreated. Actin was used as loading control. Bottom: relative Cdt1, geminin, and Cdc6 protein levels in mitosis were corrected for loading and plotted. The graph indicates data from two independent experiments (mean ± SEM). (C) Mitotic cells with reduced geminin levels show no DNA damage signaling. U2OS cells were synchronized in mitosis as in B and compared with asynchronous geminin-depleted cells. Actin is used as loading control. Numbers indicate percentages of re-replicating cells (flow cytometry analysis of parallel samples is in Fig. S4 A). Note that a slight reduction of the Cdt1 signal in lane 4 correlates to the increased population of replicating cells. Replicating cells do not express Cdt1. (D) Geminin is limiting for Cdt1 accumulation. U2OS cells were stably transfected with geminin-Cherry and blotted for Cdt1. Numbers indicate relative Cdt1 levels. Asterisks indicate background bands.
The spindle checkpoint controls replication
Our observations raise the question of whether the spindle checkpoint, by controlling licensing factors in mitosis, supports a sharply controlled G1-to-S phase transition. Although ablation of the spindle checkpoint shortened mitosis significantly (Fig. 3, A and B; note the different timing of anaphase in relation to NEB), this did not affect the time point when APC/CCdh1 became active in relation to APC/CCdc20 activation. In other words, the window of time separating geminin degradation from Cdc6 degradation was not influenced by the spindle checkpoint (Fig. S5, A and B). Next, the relative increase of Cdt1-Venus was measured from NEB until the start of geminin-Cherry destruction in control and spindle checkpoint ablated cells (reversine). Accumulation of Cdt1-Venus during mitosis was reduced in these cells because prometaphase was shorter (Fig. 7 A). Cdt1 is a limiting factor for DNA replication (Rialland et al., 2002; Maiorano et al., 2005), so our observation suggests that the integrity of the spindle checkpoint could influence the timing of DNA replication. To test this, we measured BrdU incorporation in cells depleted of MCM4, as a positive control, in relation to the effect of depleting the spindle checkpoint component Mad2. MCM4 knockdown reduces functionality of the MCM2-7 helicase, a target of Cdt1, and this indeed delays G1-to-S phase progression (Fig. 7 B, top-right plot; quantification in Fig. 7 C; Ibarra et al., 2008). Interestingly, in Mad2 RNAi cells, S over G1 ratios similarly decreased, while codepletion of MCM4 and Mad2 did not further dampen the G1-to-S transition (Fig. 7 B, bottom plots; Fig. 7 C). We did not observe significant aneuploidy or cytokinesis failure in these Mad2 RNAi cells, arguing against a role for chromosome mis-segregation or DNA damage in delaying S phase onset. In line with this idea, Mad2 depletion did not aggravate the effect of MCM4 depletion. Live-cell analyses, using Cdt1 degradation as a distinct marker, confirmed that S phase onset was equally delayed in either MAD2- or MCM4-depleted cells compared with controls (313 ± 134 min in MAD2 RNAi and 343 ± 181 min in MCM4 RNAi compared with 163 ± 66 min in control RNAi, mean ± SD, Fig. 7 D). In conclusion, although formally we cannot fully rule out a role for DNA damage in the effect of Mad2 on S phase timing, we favor a model in which normal mitosis and a functional spindle checkpoint support Cdt1 function and thereby direct the timing of DNA replication.
The spindle checkpoint controls replication. (A) Live U2OS cells stably expressing both geminin-Cherry and Cdt1-Venus were imaged by DIC and fluorescence microscopy. Cells were treated with reversine or left untreated. Accumulation of Cdt1-Venus was measured from NEB until the start of geminin-Cherry degradation (control, n = 47; reversine, n = 38; mean ± SD; Mann-Whitney test; ***, P < 0.0001). (B) Asynchronous cells transfected with the indicated siRNAs were, 48 h later, pulsed with BrdU and processed for FACS analysis (left) and Western blot (right). Left: BrdU incorporation and propidium iodide staining were analyzed using flow cytometry. Values indicate percentages of cells in the different cell cycle phases. Right: actin was used as loading control. The data shown are from a single representative experiment out of two repeats. (C) G1-to-S phase transition was measured as the ratio S phase over G1 phase cells from two independent experiments (mean ± SEM). (D) Live U2OS cells expressing Cdt1-Venus were treated with the indicated siRNAs and filmed. The time from anaphase until the start of Cdt1 degradation was plotted (control, n = 29; siMAD2, n = 58; siMCM4, n = 51; mean ± SD; Mann-Whitney test; ***, P < 0.0001).
The spindle checkpoint controls replication. (A) Live U2OS cells stably expressing both geminin-Cherry and Cdt1-Venus were imaged by DIC and fluorescence microscopy. Cells were treated with reversine or left untreated. Accumulation of Cdt1-Venus was measured from NEB until the start of geminin-Cherry degradation (control, n = 47; reversine, n = 38; mean ± SD; Mann-Whitney test; ***, P < 0.0001). (B) Asynchronous cells transfected with the indicated siRNAs were, 48 h later, pulsed with BrdU and processed for FACS analysis (left) and Western blot (right). Left: BrdU incorporation and propidium iodide staining were analyzed using flow cytometry. Values indicate percentages of cells in the different cell cycle phases. Right: actin was used as loading control. The data shown are from a single representative experiment out of two repeats. (C) G1-to-S phase transition was measured as the ratio S phase over G1 phase cells from two independent experiments (mean ± SEM). (D) Live U2OS cells expressing Cdt1-Venus were treated with the indicated siRNAs and filmed. The time from anaphase until the start of Cdt1 degradation was plotted (control, n = 29; siMAD2, n = 58; siMCM4, n = 51; mean ± SD; Mann-Whitney test; ***, P < 0.0001).
Discussion
Scheduling licensing regulatory proteins
Here we showed that, apart from cyclin B1 and securin, the spindle checkpoint governs the stability of the DNA licensing inhibitor geminin until all sister chromatids reach bi-orientation on the mitotic spindle. The spindle checkpoint and APC/CCdc20 schedule the degradation of geminin before that of the APC/CCdh1 substrate and critical preRC component, Cdc6 (Petersen et al., 2000). This suggests that high APC/CCdc20 activity is permissive for licensing in cycling cells, whereas high APC/CCdh1 activity reduces licensing competence right after mitosis. The spindle checkpoint, by protecting against geminin disappearance, helps S phase by directing the accumulation of Cdt1, a rate-limiting licensing factor for DNA synthesis that is largely absent from S and G2 phase cells. Our data support a reported dual role for geminin in the control of replication that is in part dependent on the phase in the cell cycle (Ballabeni et al., 2004). Whereas in mitosis geminin protects Cdt1 from degradation and ensures its accumulation, together with mitotic cyclin–Cdk1 activity it also keeps Cdt1 in check. In S phase, in the absence of high mitotic cyclin activity, geminin functions as the predominant inhibitor of any Cdt1 molecules that escape proteolysis. On Western blots or by time-lapse analyses of its fluorescent version, we hardly detect Cdt1 in S and G2 phase, whereas it is clearly present in mitotic cells. This means that Cdt1 accumulates between late G2 phase and metaphase. Unfortunately, attempts to stain Cdt1 by immunofluorescence were inconclusive thus far, so we cannot exclude that Cdt1 starts to accumulate in prophase already. Nevertheless, our live-cell assay clearly shows that ectopic Cdt1 fails to reach its maximum levels when prometaphase is shortened by spindle checkpoint ablation (Fig. 7 A). Upon checkpoint inactivation, coordinated destruction of both geminin and cyclin B1 in metaphase allows Cdt1 to be released from inhibition as it translocates to segregating sister chromatids. The other licensing factor, Cdc6, is completely cytoplasmic in G2 but already binds mitotic chromosomes from prometaphase onward. By telophase, both licensing factors localize to sister chromatids.
Remarkably, in S phase and G2 phase geminin is present yet it does not support Cdt1 accumulation until mitosis begins. This could either be explained by assuming that Cdt1 synthesis is restricted to mitosis or by an inability of geminin to form stabilizing complexes with Cdt1 in interphase. Because we found rapid Cdt1 accumulation after treating G2 cells with proteasome inhibitors (Fig. 6 A), we disfavor the first possibility. In line with the second hypothesis, we were puzzled to see that geminin was nuclear in G2 phase, whereas in cells overexpressing Cdt1-Venus, the weak fluorescent signal detected was strictly cytoplasmic (see Fig. 5 E). We consider it likely that NEB also facilitates efficient complex formation between geminin and Cdt1, contributing to Cdt1 stabilization during mitosis.
Different licensing windows in proliferating cells and cells that start cycling?
In cycling cells, we observed that the degradation of licensing inhibitors coincides with a peak of licensing factors in a narrow window of time around anaphase. During G1 phase, Cdc6 remained absent. This program is likely to be specifically relevant for rapidly proliferating cells. In resting cells that enter the cell cycle, Cdc6 must first accumulate before preRC formation can occur in late G1 phase (Mailand and Diffley, 2005). Cdc6 only appears in quiescent cells when it becomes phosphorylated on its APC/C recognition motif by cyclin E–Cdk2. Cyclin E–dependent phosphorylation ensures that Cdc6 escapes detection by APC/CCdh1, so it stays stable and allows cells to accumulate de novo synthesized Cdc6 (Duursma and Agami, 2005; Mailand and Diffley, 2005). On the basis of our findings we propose that proliferating cells can use an earlier window of time, after geminin and cyclin B1 disappeared but while Cdt1 peaks and Cdc6 is still stable, as the first opportunity to direct S phase. A model in which licensing control is scheduled differently in ongoing cell cycles as compared with starting cell cycles fits with the observation that cyclin E is completely redundant in rapidly dividing cells while quiescent cells require cyclin E to be able to initiate S phase (Geng et al., 2003).
Could APC/CCdh1 protect against relicensing at G1/S?
Cdc6 is degraded upon mitotic exit but Cdt1 remains stable until its Cul4–DDB1–Cdt2-dependent degradation on PCNA begins at the start of S phase (∼200 min after mitosis in U2OS cells, Fig. 5 F; Senga et al., 2006; Arias and Walter, 2007). Cdc6 is a critical licensing factor, in line with observations that Cdc6 depletion by RNAi completely blocks re-replication by geminin RNAi (Melixetian et al., 2004; unpublished data). The drastic drop in Cdc6 levels after mitosis could thus help to safeguard against the risk of relicensing after the first replication fork has started firing but before all Cdt1 is cleared. When the APC/C is inhibited again in S phase, Cdc6 may reappear, but at this time it does not catalyze DNA relicensing because Cdt1 is absent. In such a model, inability to activate APC/CCdh1 and remove Cdc6 before S phase would cause refiring of the first replication forks. Speculatively, this might relate to the occurrence of moderate DNA damage that slowly builds up in Cdh1-depleted cells (Engelbert et al., 2008; García-Higuera et al., 2008) and re-replicating cells (Melixetian et al., 2004). Cdc6 is either redundant during S and G2 or executes a function unrelated to replication licensing. For example, the fission yeast Cdc6 orthologue Cdc18 governs a DNA damage checkpoint in S phase, independently of Cdt1 (Rialland et al., 2002; Hermand and Nurse, 2007). In conclusion, exactly scheduling the levels of licensing control factors to their right time in the cell cycle is likely to support the fidelity of DNA replication and the maintenance of genomic integrity in multiple ways.
Materials and methods
Cell culture and synchronization
The human osteosarcoma cell line U2OS, telomerase-expressing human retinal pigment epithelial RPE-1 TERT cells, and ecotropic-Phoenix cells were cultured in DMEM containing 8% FCS and antibiotics. Cells were plated on 35-mm glass-bottom dishes (Willco Wells) or 9-cm Falcon dishes 24–48 h before synchronization. Cells were synchronized in mitosis by a 24-h thymidine block (2.5 mM final concentration; Sigma-Aldrich) and then released from thymidine in the presence of nocodazole (830 nM final concentration; Sigma-Aldrich) or taxol (1 µM final concentration; Sigma-Aldrich) for 12–16 h. To obtain mitotic cells after geminin RNAi treatment, but before re-replication started, cells were synchronized by a double thymidine protocol while RNAi was added during the first round of thymidine release. Mitotic cells were collected by mitotic shake-off (Wolthuis et al., 2008; van Zon et al., 2010). G2 cells were collected 8 h after release from a thymidine block, after washing away any early mitotic cells. To obtain G1 cells, mitotic cells were collected in nocodazole and either released for the indicated hours or treated with a Cdk1 inhibitor for 2 h (roscovitine #10009569, 50 µM final concentration [Cayman Chemicals]; or RO-3306 #217699, 10 µM final concentration [Calbiochem]). Other drugs, used as indicated: Mps1 inhibitor reversine (#10004412, 50 nM final concentration; Cayman Chemicals); proteasome inhibitor MG132 (#13697, 5 µM final concentration; Cayman Chemicals); translation inhibitor cycloheximide (#C6255, 10 µM final concentration; Sigma-Aldrich).
Plasmids and siRNA
Homo sapiens Geminin cDNA was PCR amplified from pcDNA3-geminin-EGFP (a kind gift from Valeria de Marco and Anastassis Perrakis, Netherlands Cancer Institute [NKI], Amsterdam, Netherlands) and cloned into pVenus-N1 (Takara Bio Inc.) using BglII–HindIII sites. Geminin-Cherry was made by replacing Venus by Cherry in geminin-Venus. The L26A-geminin mutant was made by site-directed mutagenesis using geminin-Venus as a template. The nondegradable R23A,L26A-geminin mutant was made by PCR using a mutant reverse primer (5′-GGCTGAATCATCTTCGCAGTTCTTGCTGGGACAGAACTATTC-3’). After this the product was used as forward primer in the second PCR. Silent mutations to render geminin resistant to the used shRNA were introduced by site-directed mutagenesis of the RNAi target sequence starting at codon 143 by oligo 5′-AGAAGTGGCAGAACACGTA-3’. The mutated nucleotides are underlined. The CDT1-Venus construct was constructed by PCR amplification of CDT1 from pcDNA3-CDT1-EGFP (a kind gift from Valeria de Marco and Anastassis Perrakis, NKI, Amsterdam, Netherlands) and cloning into pVenus-N1 using NheI–SacI restriction sites. The Cdc6-Venus construct was constructed by PCR amplification of Cdc6 from Cdc6-GFP-N1 (Duursma and Agami, 2005) and cloning into pVenus-N1 using NheI–XhoI restriction sites. The following Takara Bio Inc. expression plasmids have all been described previously (Floyd et al., 2008; van Leuken et al., 2008; Wolthuis et al., 2008; van Zon et al., 2010) and have been cloned using restriction sites as indicated: GFP-spectrin (EcoRI–SalI), Aurora A–EYFP (KpnI–BamHI), Aurora A–ECFP (KpnI–BamHI), securin-Venus (KpnI–BamHI), cyclin B1–Cherry (HindIII–BamHI), and R42A–cyclin B1–Cerulean and the pSuper plasmids pS-empty (no target sequence present) and pS-MAD2 (target-sequence 5′-GGAAGAGTCGGGACCACAG-3’). The pSuper plasmid pS-geminin has the target-sequence 5′-AGAAGTAGCAGAACATGTA-3’; pS-Cdh1 has the target-sequence 5′-GGGAAGAAGCTGTCCATGT-3’; and pS-Cdc20 has the target-sequence 5′-GGTGGCTGAACTCAAAGGT-3’. The viral plasmid pLIB–CDT1–Venus was constructed by PCR amplification of CDT1-Venus from pVenus-CDT1-Venus and cloning into pLIB-N1 (from Johan Kuiken, NKI, Amsterdam, Netherlands) using XhoI–NotI restriction sites. The viral plasmids pLIB–geminin–Cherry and pLIB–geminin–Venus were constructed by cloning geminin-Cherry or geminin-Venus from the expression plasmids into pLIB-N1 using BglII–NotI restriction sites. pLIB–cyclin B1–Cherry was constructed by cloning cyclin B1–Cherry from Cherry-N1 into pLIB-N1 using BglII–NotI restriction sites. The viral plasmid pLIB–Cdc6–Venus was constructed by PCR amplification of Cdc6 from Cdc6-Venus and cloning into pLIB-Venus-N1 using XhoI–BamHI restriction sites. All plasmids were sequence verified. The siRNAs to target Cdc20 (CDC20), Cdh1 (FZR), geminin (GMNN), MCM4, and MAD2 were purchased from Thermo Fisher Scientific as set of four individual ON-TARGET-plus oligos.
Transfection
Cells were transfected with expression plasmids and pSuper plasmids using a standard calcium phosphate transfection protocol. For transfection of siRNA pools, we used Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. For time-lapse microscopy, cells plated on Willco Wells dishes were transfected with 40 nM siRNA (ON-TARGET-plus; Thermo Fisher Scientific) or with expression plasmids as indicated in the following amounts: 0.05 µg geminin-Venus, 0.05 µg geminin-Cherry, 0.075 µg securin-Venus, 0.29 µg R42A–cyclin B1–Cerulean, 0.15 µg Cdc6-Venus, 0.15 µg Aurora A–ECFP, 0.15 Aurora A–EYFP, 2 µg pS-MAD2, 2 µg pS-Cdc20, 2 µg pS-Cdh1, or 2 µg pS-geminin. Transfection with expression plasmids is 24 h after siRNA oligo transfection as indicated. U2OS cells were cotransfected on 9-cm Falcon culture dishes, using 2.9 µg R42A–cyclin B1–Cerulean, 1 µg wt-geminin-Cherry, or 1 µg R23A,L26A-geminin-Cherry together with 1 µg of pBABE-Puro and selected with puromycin for 24 h (in the presence of thymidine as indicated). Cells in 9-cm Falcon dishes for MPM2 positivity analysis were cotransfected with GFP-spectrin as indicated. Cells in 9-cm Falcon dishes were transfected with 40 nM pooled siRNA oligos targeting the human protein as indicated; CTRL refers to 1× siRNA buffer.
Ecotropic Phoenix cells were transfected with 20 µg of pLIB-CDT1-Venus, 20 µg pLIB-Cdc6-Venus, 10 µg pLIB–cyclin B1–Cherry, or 10 µg pLIB–geminin–Cherry plasmid (or combinations for coexpressing cell lines) using standard calcium phosphate transfection. Viral supernatant was collected twice, 40 and 48 h after transfection. The supernatant was cleared through a 0.45-µm filter (EMD Millipore). U2OS cells expressing the ecotropic receptor (from Johan Kuiken, NKI, Amsterdam, Netherlands) were infected twice with the virus.
Western blots
Cells were lysed in ELB+ (150 mM NaCl, 50 mM Hepes, pH 7.5, 5 mM EDTA, 0.3% NP-40, 10 mM β-glycerophosphate, 6% glycerol, 5 mM NaF, 1 mM Na2VO3, and protease inhibitor cocktail [Roche]) for 30 min. Lysates were cleared by centrifugation (13,000 g for 8 min at 4°C). Protein levels were measured and equalized using standard Bradford assay (Bio-Rad Laboratories). Protein was separated on SDS-PAGE and blotted on nitrocellulose membranes. For membrane blocking and antibody incubation 4% ELK was used, except for rabbit anti-Cdt1, rabbit anti–p-Chk1-S317, and rabbit anti–p-Cdk1-Y15, for which 4% BSA was used. Quantification of Western blots was done with Image Laboratory (Bio-Rad Laboratories) software.
Antibodies
For detection of proteins the following antibodies were used at the indicated dilution: mouse anti-CDC27 (#610455, 1:1,000; BD), rabbit anti-geminin (FL-209, 1:1,000; Santa Cruz Biotechnology, Inc.), rabbit anti-p55CDC (H-175, sc-8358, 1:1,000; Santa Cruz Biotechnology, Inc.), mouse anti-Cdh1 (Ab-1, DH01, 1:1,000; NeoMarkers), goat anti-APC4 (C-18, sc-21414, 1:1,000; Santa Cruz Biotechnology, Inc.), mouse anti-cyclin B1 (GNS1, sc-245, 1:1,000; Santa Cruz Biotechnology, Inc.), mouse anti-securin (DCS-280, ab3305, 1:500; Abcam), rabbit anti-Aurora A/AIK (#3092, 1:1,000; Cell Signaling Technology), rabbit anti-MCM4 (#559544, 1:2,000; BD), mouse anti-Mad2 (K0167-3, 1:1,000; MBL), goat anti-actin (I-19, sc-1616, 1:1,000; Santa Cruz Biotechnology, Inc.), mouse anti-Cdc6 (180.2, sc-9964, 1:1,000; Santa Cruz Biotechnology, Inc.), goat anti-Cdk4 (C-22, sc-260, 1:1,000; Santa Cruz Biotechnology, Inc.) and mouse anti-BrdU (M0744, clone Bu20a, 1:40; Dako), rabbit anti-Cdt1 (ab70829, 1:1,000, Abcam), rabbit anti–phospho-Cdk1-Y15 (#9111s, 1:1,000, Cell Signaling Technology), rabbit anti–phospho-Chk1-S317 (#2344, 1:1,000, Cell Signaling Technology), and rabbit anti-Chk1 (SC7898, 1:500, Santa Cruz Biotechnology, Inc.). Secondary PO-conjugated antibodies were obtained from Dako.
Flow cytometry
For analysis of DNA replication, cells were pulsed with BrdU (1 µM final concentration) for 20 min, ethanol fixed, followed by 30 min HCl/Triton treatment, borate neutralization, and staining with anti-BrdU antibodies. For analysis of cell cycle distribution, cells were, after fixation in 70% ethanol, stained with mouse anti-MPM2 (#05-368, 1:250; EMD Millipore). For analysis by flow cytometry, cells were counterstained with propidium iodide diluted in RNase-containing PBS. MPM-2 positivity of GFP-positive cells and BrdU positivity was analyzed using CellQuest (BD) and FCS Express 2 (De Novo Software).
Time-lapse fluorescence microscopy
U2OS or RPE1-TERT cells transfected with siRNA and indicated plasmids were followed by fluorescence time-lapse microscopy. Acquisition of DIC and fluorescence images started 46 h after transfection on a microscope (Axio Observer Z1; Carl Zeiss) in a heated culture chamber (5% CO2 at 37°C) using DMEM with 8% FCS and antibiotics. The microscope was equipped with an LD 0.55 condenser and 40× NA 1.40 Plan Apochromat oil DIC objective and CFP/YFP and GFP/HcRed filter blocks (Carl Zeiss) to select specific fluorescence. Images were taken using AxioVision Rel. 4.8.1 software (Carl Zeiss) with a charge-coupled device camera (ORCA R2 Black and White CCD [Hamamatsu Photonics] or Roper HQ [Roper Scientific]) at 100-ms exposure times. For quantitative analysis of degradation, MetaMorph software (Universal Imaging) and Excel (Microsoft) were used. Captured images were processed using Photoshop and Illustrator software.
Online supplemental material
Fig. S1 shows that like cyclin B1, geminin degradation starts at metaphase and is complete by mitotic exit. Further, it shows the timing of degradation of geminin-Cherry and Cdc6-Venus in U2OS and RPE-1 cells. Fig. S2 shows that degradation of Aurora A and Cdc6, but not that of geminin, is prevented in Cdh1-depleted cells. Fig. S3 shows that cells expressing nondegradable R23A,L26A-geminin exit mitosis without defects and that the start of Cdt1-Venus degradation coincides with Cherry-PCNA foci formation. Further, it shows Cdt1-localization is prevented in R42A–cyclin B1–Cerulean arrested cells. In Fig. S4 it is shown that geminin stabilizes Cdt1. Finally, a Cdt1 mutant lacking geminin interaction accumulates only after Cdt1 translocates to chromatin after mitosis. Fig. S5 reveals that spindle checkpoint ablation does not affect the total window of time when Cdc6 is present while licensing inhibitors are absent.
Acknowledgments
We thank Helma van Riel and María-José Villalobos Quesada for directly contributing to this project; and Wouter van Zon, Erik Voets, and our other colleagues for ideas, discussions, and critically reviewing the manuscript. We are grateful to Jon Pines, Cath Lindon, Valeria de Marco, Anastassis Perrakis, Johan Kuiken, Anja Duursma, and Reuven Agami for reagents.
This work was supported by a grant from the Netherlands Cancer Society (KWF 2007-3789 to L. Clijsters and J. Ogink), a Vidi Grant from the Netherlands Organization for Scientific Research (NOW to R. Wolthuis), and an HFSP Project Grant (to R. Wolthuis).