The cadherin extracellular region produces intercellular adhesion clusters through trans- and cis-intercadherin bonds, and the intracellular region connects these clusters to the cytoskeleton. To elucidate the interdependence of these binding events, cadherin adhesion was reconstructed from the minimal number of structural elements. F-actin–uncoupled adhesive clusters displayed high instability and random motion. Their assembly required a cadherin cis-binding interface. Coupling these clusters with F-actin through an α-catenin actin-binding domain (αABD) dramatically extended cluster lifetime and conferred direction to cluster motility. In addition, αABD partially lifted the requirement for the cis-interface for cluster assembly. Even more dramatic enhancement of cadherin clustering was observed if αABD was joined with cadherin through a flexible linker or if it was replaced with an actin-binding domain of utrophin. These data present direct evidence that binding to F-actin stabilizes cadherin clusters and cooperates with the cis-interface in cadherin clustering. Such cooperation apparently synchronizes extracellular and intracellular binding events in the process of adherens junction assembly.
Introduction
The transmembrane adhesive receptor classical cadherin forms several types of cell–cell adhesive structures generally known as adherens junctions. Despite significant morphological and structural differences, all types of adherens junctions share a similar molecular organization: they consist of cadherin clusters in which their ectodomain produces intercellular adhesive contacts, and its intracellular domain anchors the clusters to the cytoskeleton. The mechanism of cadherin clustering and its regulation are the key aspects of cadherin adhesion because the adhesive activity of individual cadherin molecules is negligible. The dependency of cadherin adhesion on the level of its clustering is the conceptual framework in the biology of cell–cell contacts (Yap et al., 1997; Gumbiner, 2005; Nelson, 2008; Niessen et al., 2011). But despite its importance, the mechanisms of cadherin cluster formation are not known.
Adherens junctions are very dynamic; they continuously gain and lose all of their structural components (de Beco et al., 2009; Hong et al., 2010). The continuous renewal of adherens junctions is based on the balance of two opposite reactions—the assembly and disassembly of cadherin clusters. Recent biophysical, structural, and cell biology studies shed light on the initial events leading to the assembly of adhesive clusters (Brasch et al., 2012; Troyanovsky, 2012). First, the membrane-distant domain of the cadherin extracellular region establishes an adhesive bond with the cadherin molecule residing on the opposite cell through strand-swap trans-dimerization. This bond stabilizes the cadherin cis-binding interface that, in turn, triggers lateral interactions of trans-dimers (Wu et al., 2010; Harrison et al., 2011). Eventually, these two extracellular binding reactions—the formation of trans-dimers and their cis-arrangement—produce a cadherin adhesive cluster without any assistance from intracellular proteins. What happens next? In particular, how is the assembly of these clusters coordinated with their anchorage to the cytoskeleton? How does this anchorage change the adhesive clusters? All of these crucial questions remain unanswered.
Numerous observations have shown that abnormalities in the actin cytoskeleton affect adherens junction formation. For example, disruption of microfilaments dramatically perturbs both cell–cell adhesion (Angres et al., 1996; Chu et al., 2004; Ivanov et al., 2005) and adhesion of cadherin-coated beads to the cells (Lambert et al., 2002; Baumgartner et al., 2003). Assembly and maintenance of adherens junctions are also significantly perturbed by the inactivation of the major cadherin–F-actin adaptor protein α-catenin (Torres et al., 1997; Vasioukhin et al., 2000; Pappas and Rimm, 2006; Kwiatkowski et al., 2010). Vice versa, the augmentation of F-actin in cell–cell junctions by Dia/RhoA overexpression enhances adherens junction formation (Carramusa et al., 2007). Different models were proposed to explain the actin dependency of adherens junctions, the simplest of which is that actin filaments form an intracellular scaffold that anchors the cadherin cluster molecules, thereby reinforcing the adhesive contact (Adams et al., 1998; Mège et al., 2006; Lambert et al., 2007). However, direct evidence for this hypothesis is missing because experimental distortions in the actin cytoskeleton or cadherin–actin interactions can produce multiple secondary effects affecting adhesion by alternative mechanisms. Furthermore, the stabilizing role of actin is difficult to reconcile with the active dynamics of the actin cytoskeleton proximal to the junctions (Nelson, 2008), which may, in fact, add instability to the cadherin clusters assembled through extracellular interactions. The role of F-actin in the biology of the transmembrane receptors is of general interest: integrin clusters in focal adhesions (Vicente-Manzanares et al., 2009) and clusters of acetylcholine receptors in neuromuscular junctions (Dai et al., 2000) have been also suggested to be maintained by F-actin.
In addition to the proposed role of the actin cytoskeleton in stability of the clusters, live-imaging observations have shown that spotlike adherens junctions move in the apical direction by the actin-dependent mechanism (Kametani and Takeichi, 2007; Hong et al., 2010). Whether the junctions are simply “dragged” by the actin cytoskeleton along the lateral plasma membrane or their movement is based on more sophisticated mechanisms remains to be studied.
To directly analyze the role of F-actin in the formation and maintenance of cadherin clusters, we reconstructed cadherin adhesion clusters that are engaged or disengaged from the actin cytoskeleton using a minimal set of structural components. This system unambiguously shows that binding to F-actin stabilizes cadherin clusters and sets up their directional movement. Furthermore, it demonstrates that binding to F-actin cooperates with the cadherin cis-interface, thereby promoting the cadherin clustering process. Finally, cluster dynamics and, eventually, the behavior of the resulting adhesive structures depend on the number of the cluster molecules interacting with actin.
Results
Stability of trans-dimers determines the tailless cadherin cluster dynamics
To elucidate cadherin cluster formation, we first analyzed the adhesive clusters produced in cadherin-deficient A431D cells by the tailless E-cadherin mutant EcDnΔ (Fig. S1 a). In agreement with our previous data (Hong et al., 2010), these clusters were observed as distinct dots of different intensities along the cell–cell contacts (Fig. 1 a, Δ). Individual clusters could be resolved only in the cell–cell contact areas exhibiting low levels of the mutant. The clusters were randomly localized with no particular pattern. Our initial attempts to visualize the clusters in live-cell imaging experiments using 1-s-long acquisition time (the standard microscope setting) were unsuccessful: only general recruitment of the mutant into the cell–cell contacts was detected (Fig. S2 a). However, the clusters could be satisfactorily monitored using the fastest available speed of the microscope, a stream mode with 100 ms of image acquisition time (Fig. 1 b and Video 1). In this mode, some of the clusters could be tracked through 1 s or more of the 2-s-long live-imaging experiments (Fig. 1 f). To verify that these clusters are based on the cis-arrangement of the strand-swapped trans-dimers, we studied A431D cells expressing the EcDnΔ mutant bearing in addition either W2A or V81D/V175D point mutations inactivating trans- or cis-interactions, correspondingly (Harrison et al., 2010, 2011; Hong et al., 2011). As expected, these mutations completely abolished the formation of discernible clusters (Fig. 1 a, Cis-Δ; and Fig. S1 c, W-Δ). Therefore, trans- and cis-intercadherin interactions are the two binding reactions that drive the formation of the “tailless” cadherin clusters.
The tailless cadherin clusters are unstable and move randomly. A431D cells expressing cadherin tailless mutant EcDnΔ were analyzed by time-lapse wild-field microscopy. Images were acquired in a stream mode at 100 ms/frame for 2 s. The central area of the video was used for Fig. 1 b.
The tailless cadherin clusters are unstable and move randomly. A431D cells expressing cadherin tailless mutant EcDnΔ were analyzed by time-lapse wild-field microscopy. Images were acquired in a stream mode at 100 ms/frame for 2 s. The central area of the video was used for Fig. 1 b.
Properties of the “tailless” cadherin clusters. (a) Immunofluorescence microscopy of A431D cells expressing EcDnΔ (Δ) or its cis-interface-defective version, cis-EcDnΔ (Cis-Δ). The cells were stained with anti-Dendra antibody. Higher magnifications of the selected regions (indicated by dashed boxes) are shown in the insets. Note that cell–cell contacts exhibit numerous EcDnΔ clusters of different intensities. The individual clusters are not resolved at high level of mutant expression (arrow). The clusters became undetectable upon mutation of the cis-interface. Bars: (main images) 15 µm; (insets) 5 µm. (b) Time-lapse analyses of EcDnΔ clusters present in an area boxed in the left image. The sequence is assembled from Video 1. Some clusters, two of which are marked with blue and yellow circles, persist over the entire 1.5-s-long video. Others (red circles) are much more transient. Bars: (left) 10 µm; (right) 3 µm. (c) Time-lapse analyses of D1A-EcDnΔ clusters (from Video 2). Point mutation D1A increases stability of the cadherin trans-dimers. Note that this mutation significantly decreases cluster dynamics. Bar, 3 µm. (d) 1-s-long trajectories of the six fastest clusters in cells expressing EcDnΔ (Δ), D1A-EcDnΔ (D1A-Δ), and EcDnΔ-αABD (α). Two independent videos for each cell line were analyzed. Note that the EcDnΔ clusters (in Δ), although not moving in particular directions, covered distances of ∼0.2 µm. The D1A mutation or αABD notably restricted cluster motility. (e) Dendra photoconversion assay of different EcDnΔ mutants: EcDnΔ (Δ), D1A-EcDnΔ (D1A-Δ), EcDnΔ-αABD (Δ-αABD), and D1A-EcDnΔ-αABD (D1A-Δ-αABD). The graph shows changes in intensity of the red fluorescence over time (in seconds) in the entire laser-irradiated area (diameter = 0.25 µm; see Fig. S3). The error bars represent SDs (n = 30). (f) The lifetime of the arbitrary EcDnΔ (Δ) and D1A-EcDnΔ (D1A-Δ) clusters from two independent videos. Note that D1A-EcDnΔ clusters, in contrast to the parental EcDnΔ clusters, could typically be followed over the entire 2-s-long periods.
Properties of the “tailless” cadherin clusters. (a) Immunofluorescence microscopy of A431D cells expressing EcDnΔ (Δ) or its cis-interface-defective version, cis-EcDnΔ (Cis-Δ). The cells were stained with anti-Dendra antibody. Higher magnifications of the selected regions (indicated by dashed boxes) are shown in the insets. Note that cell–cell contacts exhibit numerous EcDnΔ clusters of different intensities. The individual clusters are not resolved at high level of mutant expression (arrow). The clusters became undetectable upon mutation of the cis-interface. Bars: (main images) 15 µm; (insets) 5 µm. (b) Time-lapse analyses of EcDnΔ clusters present in an area boxed in the left image. The sequence is assembled from Video 1. Some clusters, two of which are marked with blue and yellow circles, persist over the entire 1.5-s-long video. Others (red circles) are much more transient. Bars: (left) 10 µm; (right) 3 µm. (c) Time-lapse analyses of D1A-EcDnΔ clusters (from Video 2). Point mutation D1A increases stability of the cadherin trans-dimers. Note that this mutation significantly decreases cluster dynamics. Bar, 3 µm. (d) 1-s-long trajectories of the six fastest clusters in cells expressing EcDnΔ (Δ), D1A-EcDnΔ (D1A-Δ), and EcDnΔ-αABD (α). Two independent videos for each cell line were analyzed. Note that the EcDnΔ clusters (in Δ), although not moving in particular directions, covered distances of ∼0.2 µm. The D1A mutation or αABD notably restricted cluster motility. (e) Dendra photoconversion assay of different EcDnΔ mutants: EcDnΔ (Δ), D1A-EcDnΔ (D1A-Δ), EcDnΔ-αABD (Δ-αABD), and D1A-EcDnΔ-αABD (D1A-Δ-αABD). The graph shows changes in intensity of the red fluorescence over time (in seconds) in the entire laser-irradiated area (diameter = 0.25 µm; see Fig. S3). The error bars represent SDs (n = 30). (f) The lifetime of the arbitrary EcDnΔ (Δ) and D1A-EcDnΔ (D1A-Δ) clusters from two independent videos. Note that D1A-EcDnΔ clusters, in contrast to the parental EcDnΔ clusters, could typically be followed over the entire 2-s-long periods.
One of the most remarkable features of the EcDnΔ clusters was their motility. Fig. 1 d (Δ) shows tracks of several of the most motile clusters (among them are those circled in Fig. 1 b), which they traversed over 1-s intervals. These clusters, although not moving in particular directions, covered the distance of ∼0.2 µm. Their largest displacement for 1 s was close to 0.2 µm, corresponding to their diffusion coefficient D = 0.01 µm2/s (note that the D value for the mean cluster was not studied). Such a diffusion rate is similar to that found for the F-actin–disengaged cadherin clusters (D = 0.04 µm2/s), measured by video microscopy of cadherin-coated beads (Lambert et al., 2002). This fast but random motion could be explained by two principal mechanisms. First, the adhesive cluster could move as an entire junctional unit as a result of Brownian motion. Alternatively, it could include the constant reassembly process. For example, the imbalanced molecular turnover at the cluster periphery resulted in the recruitment of new molecules more on one side than the other, leading to the random motion of the clusters.
Continuous reassembly of adhesive bonds is a prerequisite for fast cluster movement
To assess the role of cadherin turnover in cluster motility, we introduced into the EcDnΔ mutant a point mutation, D1A, that had been shown to increase stability of cadherin trans-dimerization (Laur et al., 2002; Troyanovsky et al., 2007). If a cluster moved as a whole unit, its motion would be independent from the strength of its trans-dimerization. In contrast, the experiments with this mutant showed that the D1A mutation notably restricted motility of individual clusters (Fig. 1, c and d; and Video 2) and significantly increased their lifetime (Fig. 1 f). Using a Dendra photoconversion diffusion assay, we determined the diffusion rates of the tailless mutants in the cell–cell contacts. This assay followed the expansion of the photoconverted (2.5 µm in diameter) areas within the cell–cell contacts (Fig. 2). Importantly, this assay determined the diffusion of cadherin molecules in the cell–cell contact but not that of the particular clusters. It showed that D1A mutation dropped the diffusion coefficient (D) of the tailless mutant from 0.012 to 0.003 µm2/s. The mutation of the cis-interface, in contrast, increased the diffusion coefficient of the tailless mutant to 0.05 µm2/s.
D1A mutation stabilizes the clusters. The clusters formed in A431D cells by the tailless cadherin mutant D1A-EcDnΔ were analyzed by time-lapse wild-field microscopy as indicated in Video 1. The central area of the video was used for Fig. 1 c. Note that the clusters are much more stable and immobile than in Video 1.
D1A mutation stabilizes the clusters. The clusters formed in A431D cells by the tailless cadherin mutant D1A-EcDnΔ were analyzed by time-lapse wild-field microscopy as indicated in Video 1. The central area of the video was used for Fig. 1 c. Note that the clusters are much more stable and immobile than in Video 1.
Dendra diffusion assay with cells expressing EcDnΔ, D1A-EcDnΔ, and Cis-EcDnΔ. (a) The circular areas (diameter = 2.5 µm) in the selected cell–cell contact of EcDnΔ cells, which express relatively high level of the mutants (−3), was photoconverted from green to red fluorescence (the photoconverted area is circled in the red channel image). The region was imaged at 3 and 23 s (0 and 20) after photoconversion in both green and red channels. Image acquisition time was 1 s. Bar, 10 µm. (b) The experiment shown in panel a was repeated 10 times with each of the indicated cell lines. Error bars indicate SDs. The graph shows the expansion of the red fluorescence area.
Dendra diffusion assay with cells expressing EcDnΔ, D1A-EcDnΔ, and Cis-EcDnΔ. (a) The circular areas (diameter = 2.5 µm) in the selected cell–cell contact of EcDnΔ cells, which express relatively high level of the mutants (−3), was photoconverted from green to red fluorescence (the photoconverted area is circled in the red channel image). The region was imaged at 3 and 23 s (0 and 20) after photoconversion in both green and red channels. Image acquisition time was 1 s. Bar, 10 µm. (b) The experiment shown in panel a was repeated 10 times with each of the indicated cell lines. Error bars indicate SDs. The graph shows the expansion of the red fluorescence area.
To confirm that strengthening of trans-dimerization did stabilize the individual clusters, we examined the turnover of cluster molecules using a Dendra photoconversion assay. In this experiment, fluorescent intensities of the individual cluster groups inside the 2.5-µm photoconverted contact areas were traced over time. These experiments showed that the adhesive clusters of the parental tailless mutant were in fast dynamic monomer/cluster equilibrium. The D1A mutation significantly reduced the molecular exchange rate in the clusters (Fig. 1 e and Fig. S3). Collectively, these results clearly showed that cadherin molecules in the clusters continuously rearrange their adhesive bonds. The bond instability is required for motility of both the individual clusters and the entire pool of junctional cadherin molecules.
α-Catenin actin-binding domain (αABD) stabilizes cadherin clusters and guides their movement
The aforementioned experiments showed that the cadherin tailless mutant formed adhesive clusters. Rapid turnover of this mutant within the clusters led to high instability of the clusters and their fast motion along the contacts. We next examined how binding to F-actin changes the dynamic properties of these “tailless” clusters. To couple the clusters to microfilaments, we added the αABD, which had been mapped between the residues 671 and 906 at the α-catenin C terminus (Pokutta et al., 2002; Pappas and Rimm, 2006; Rangarajan and Izard, 2013), to the C terminus of EcDnΔ (Fig. S1 a).
A431D cells expressing the EcDnΔ-αABD chimera exhibited dramatic changes in the morphology of their cell–cell contacts: their subapical regions produced numerous long filopodia that radially protruded to the adjacent cells. Such protrusions had been never observed in EcDnΔ-expressing cells. These filopodia and the surrounding subapical area of the cell–cell contacts recruited the majority of cadherin clusters (Fig. 3 a, arrowheads). Some individual clusters were also present along the lateral cell–cell contact areas (Fig. 3 a, arrow). Staining for F-actin showed that most of the clusters, including those on the lateral surfaces, were aligned with actin filament bundles (Fig. 3 a′).
Distribution and dynamics of the EcDnΔ-αABD clusters. (a–b′′) Double-label immunofluorescence microscopy of A431D cells expressing EcDnΔ-αABD (a, α) or D1A-EcDnΔ-αABD (b, D1Aα) chimeras. Cells were stained for chimeras using the anticadherin antibody (a and b) and for F-actin using Alexa Fluor 555 phalloidin (a′ and b′, actin). The corresponding images are merged in a′′ and b′′. Note that most of the cadherin clusters (two of the subapical and one of the basal clusters are indicated by arrowheads and an arrow in the insets) associated with actin bundles. Bars: (main images) 40 µm; (insets) 5 µm. (c) Time-lapse series (right) of EcDnΔ-αABD clusters (frames are indicated according to Video 3) present in the boxed region of the left image. Arrows of the same color indicate the same clusters in consequent frames. Note that the clusters produced by this chimera had a much longer lifetime than the tailless clusters and exhibited directional movements. Bars: (left) 10 µm; (right) 5 µm.
Distribution and dynamics of the EcDnΔ-αABD clusters. (a–b′′) Double-label immunofluorescence microscopy of A431D cells expressing EcDnΔ-αABD (a, α) or D1A-EcDnΔ-αABD (b, D1Aα) chimeras. Cells were stained for chimeras using the anticadherin antibody (a and b) and for F-actin using Alexa Fluor 555 phalloidin (a′ and b′, actin). The corresponding images are merged in a′′ and b′′. Note that most of the cadherin clusters (two of the subapical and one of the basal clusters are indicated by arrowheads and an arrow in the insets) associated with actin bundles. Bars: (main images) 40 µm; (insets) 5 µm. (c) Time-lapse series (right) of EcDnΔ-αABD clusters (frames are indicated according to Video 3) present in the boxed region of the left image. Arrows of the same color indicate the same clusters in consequent frames. Note that the clusters produced by this chimera had a much longer lifetime than the tailless clusters and exhibited directional movements. Bars: (left) 10 µm; (right) 5 µm.
Live-cell imaging of these clusters revealed remarkable behavior. In contrast to their tailless counterparts, the clusters containing αABD were completely immobile during the 2-s-long observations (Fig. 1 d, α). Imaging with a slower mode showed that these clusters had a much longer lifetime than the tailless clusters. Most remarkable was the behavior of the lateral clusters. They continuously assembled at the base of the cell–cell contacts and moved apically (Fig. 3 c and Video 3). Taking in consideration the approximate height of A431D cells (1.6 µm), the velocity of the clusters was ∼0.4 µm/min—much slower than the random displacement of the tailless clusters. Strikingly, several subsequently assembled clusters used the same or very similar tracks in their movements (Fig. 3 c and Video 3). By these features, the lateral clusters of the EcDnΔ-αABD mutant were similar to the spotlike adherens junctions of wild-type A431 cells, which also form at the base of cell–cell contacts and then climb to the subapical region (Kametani and Takeichi, 2007; Hong et al., 2010).
αABD provides directionality to cluster movement. The clusters formed in A431D cells by the cadherin–α-catenin chimera EcDnΔ-αABD were analyzed by time-lapse wild-field microscopy. Frames were acquired at 30-s intervals for 10 min. Image acquisition time was 1 s. Selected frames from this video were used in Fig. 3 c. Note that the clusters form at the edge of the lamella contacting with another cell and then move in an apical direction.
αABD provides directionality to cluster movement. The clusters formed in A431D cells by the cadherin–α-catenin chimera EcDnΔ-αABD were analyzed by time-lapse wild-field microscopy. Frames were acquired at 30-s intervals for 10 min. Image acquisition time was 1 s. Selected frames from this video were used in Fig. 3 c. Note that the clusters form at the edge of the lamella contacting with another cell and then move in an apical direction.
The relatively slow motility of the αABD-containing clusters and their longer persistence on the cell surface suggested that αABD stabilized cadherin molecules in the clusters. Accordingly, the Dendra photoconversion assay showed that αABD significantly slowed down the turnover of the tailless cadherin mutant in the clusters (Fig. 1 e).
Cadherin turnover in the EcDnΔ-αABD clusters is required for their movement
We showed that the random motion of the tailless cadherin clusters requires their continuous reassembly. Therefore, we tested whether the directional and much slower movement of the EcDnΔ-αABD clusters also depended on the same process. To this end, we inserted the D1A mutation into the EcDnΔ-αABD chimera. This insertion resulted in a complete disappearance of the extrajunctional cadherin fluorescence (Fig. 3 b) and reduced the exchange rate of the chimera in the clusters assessed by the photoconversion assay (Fig. 1 e). Both these effects confirmed that the D1A mutation stabilized the mutant within the clusters. In addition, the D1A mutation notably increased the amount of apical junctions, concomitantly reducing the pool of lateral cadherin clusters. The only occasional formation of lateral clusters in these cells prevented us from evaluating the role of cadherin turnover in their motility.
To enrich D1A-EcDnΔ-αABD–expressing cells with motile lateral clusters, we set calcium-switch experiments in which we expected to follow the clusters starting from the moment of their formation. The cells expressing the parental EcDnΔ-αABD mutant produced numerous clusters as fast as 1 min after the low to high calcium switch (Fig. 4 a and Video 4). Importantly, the absolute majority of the clusters appeared at the basal edge of the cell–cell contacts and then persistently moved in the apical direction. Many of these clusters proceeded the entire way to the apical surface (Fig. 4 a, arrows). The D1A mutation did not affect the initial assembly of the clusters at the edge of the contacts (Fig. 4 b and Video 5). However, the basal to apical movement of the D1A-EcDnΔ-αABD clusters was significantly impeded. Most of the clusters were immobile (Fig. 4 b, arrows). In some cases, they could move for about a minute or so, passing no more than halfway to the apical surface (Fig. 4 c). These data clearly showed that the movement of αABD-containing cadherin clusters, just as the random movement of the tailless clusters, required the continuous turnover of the cluster molecules.
Clustering of the EcDnΔ-αABD chimera in the calcium-switch assay. Cells were preincubated with low calcium for 3 h. High calcium media were added at time 0. Images were acquired at 30-s intervals for 10 min (1 s/frame). This video was used in Fig. 4 a. Note that most clusters were assembled at the basal edge of cell–cell contacts immediately after calcium addition; once assembled, most of the clusters moved in an apical direction.
Clustering of the EcDnΔ-αABD chimera in the calcium-switch assay. Cells were preincubated with low calcium for 3 h. High calcium media were added at time 0. Images were acquired at 30-s intervals for 10 min (1 s/frame). This video was used in Fig. 4 a. Note that most clusters were assembled at the basal edge of cell–cell contacts immediately after calcium addition; once assembled, most of the clusters moved in an apical direction.
Clustering of the D1A-EcDnΔ-αABD chimera in the calcium-switch assay. Experiment was performed exactly as Video 4. This video was used in Fig. 4 b. Note that the majority of the assembled clusters do not show directional movement.
Clustering of the D1A-EcDnΔ-αABD chimera in the calcium-switch assay. Experiment was performed exactly as Video 4. This video was used in Fig. 4 b. Note that the majority of the assembled clusters do not show directional movement.
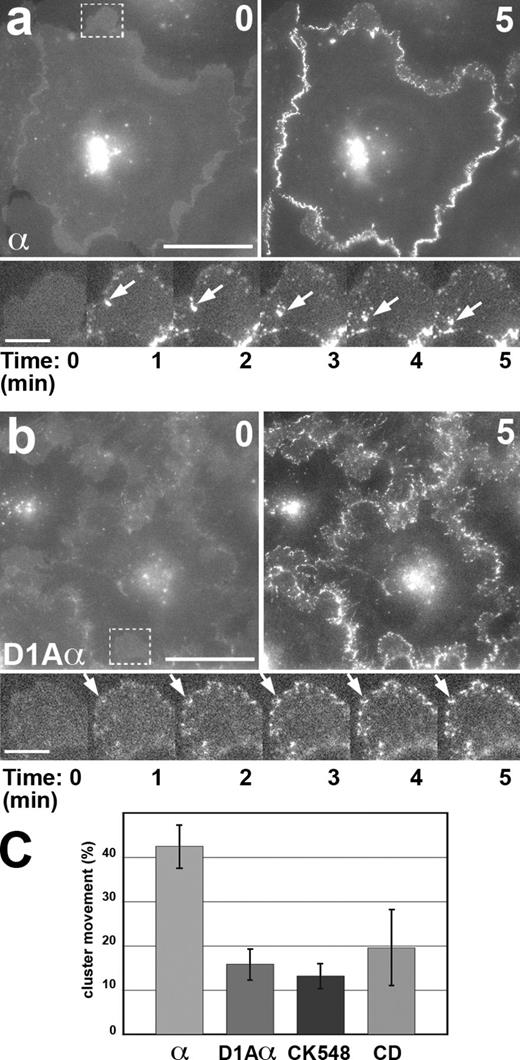
The D1A mutation impeded the basal to apical movement of the EcDnΔ-αABD clusters. (a and b) The calcium-switch assay with the cells expressing EcDnΔ-αABD (a, α) and D1A-EcDnΔ-αABD (b, D1Aα). The cells immediately before the switch (0) and 5 min after the switch (5) are presented. Time-lapse images (numbering according to Videos 4 and 5) of the boxed regions in a and b are shown at the bottom. Note that the D1A mutation did not affect the initial assembly of the clusters but significantly reduced their movement. Arrows show a large motile (in a) and stationary (in b) clusters. Bars: (top) 20 µm; (bottom) 5 µm. (c) The path of the mean cluster (presented as a fraction of the basal to apical distance, assuming the entire distance for 100%; error bars are SDs) over 5 min after the calcium switch in cells expressing EcDnΔ-αABD (α), D1A-EcDnΔ-αABD (D1Aα), or in cells expressing EcDnΔ-αABD in the presence of 100 µM CK548 (CK548) or the low dose (25 nM) of cytochalasin D (CD), inhibitors of actin polymerization (see also Fig. 5).
The D1A mutation impeded the basal to apical movement of the EcDnΔ-αABD clusters. (a and b) The calcium-switch assay with the cells expressing EcDnΔ-αABD (a, α) and D1A-EcDnΔ-αABD (b, D1Aα). The cells immediately before the switch (0) and 5 min after the switch (5) are presented. Time-lapse images (numbering according to Videos 4 and 5) of the boxed regions in a and b are shown at the bottom. Note that the D1A mutation did not affect the initial assembly of the clusters but significantly reduced their movement. Arrows show a large motile (in a) and stationary (in b) clusters. Bars: (top) 20 µm; (bottom) 5 µm. (c) The path of the mean cluster (presented as a fraction of the basal to apical distance, assuming the entire distance for 100%; error bars are SDs) over 5 min after the calcium switch in cells expressing EcDnΔ-αABD (α), D1A-EcDnΔ-αABD (D1Aα), or in cells expressing EcDnΔ-αABD in the presence of 100 µM CK548 (CK548) or the low dose (25 nM) of cytochalasin D (CD), inhibitors of actin polymerization (see also Fig. 5).
αABD-mediated stability of the clusters and their directional movement are based on αABD–F-actin interactions
Did the direct binding of αABD to F-actin, and not other possible αABD activities, mediate the directional motility and extra stability of EcDnΔ-αABD clusters? To address this question, we first tested these clusters for the presence of ZO1 and Eplin, two proteins that had been shown to interact with αABD (Imamura et al., 1999; Abe and Takeichi, 2008). However, most of the clusters were negative for these two proteins (Fig. S4).
Then, we inserted a short, 42-aa-long terminal deletion into the αABD. This mutation had been shown to inactivate αABD binding to F-actin without detectable effects on αABD folding (Pokutta et al., 2002; Pappas and Rimm, 2006). This deletion completely abolished the formation of the stable cadherin clusters typical for EcDnΔ-αABD, resulting, instead, in a phenotype indistinguishable from that of the tailless EcDnΔ mutant (Fig. 5 a, α-Δ42). The clusters of this mutant could not be captured by the 1-s-long “snapshot” of live cells (Fig. S2 c).
Cluster stability depends on the actin cytoskeleton. (a) Immunolocalization of the EcDnΔ-αABD-Δ42 chimera (Δ42). Note that the 42-aa-long terminal deletion of the αABD that inactivates binding of this domain to F-actin abolished apical concentration of the clusters and resulted in formation of numerous small clusters typical for EcDnΔ. Bar, 10 µm. (b–g) Effect of actin depolymerization on cadherin clustering during low to high calcium switch: Cells expressing EcDnΔ-αABD chimera (b and c; α), tailless EcDnΔ chimera (d and e; Δ), and full-size Dendra-tagged E-cadherin (f and g; EcD) were cultured at low calcium and then moved to high calcium media in control conditions (b, d, and f; DMSO) or in presence of 8 µM of latrunculin A (c, e, and g; LatA). Cells were fixed 5 min after the calcium switch and immunostained for cadherin and the actin cytoskeleton (actin, shown only in c′). The boxed areas are enlarged in the insets. Magnifications are the same in all images. Note that depolymerization of F-actin by latrunculin A prevented concentration of the EcDnΔ-αABD clusters along the apical area of cell–cell contacts and significantly inhibited formation of the EcDn junctions but has little effect on clustering of the tailless EcDnΔ cadherin. Bars: (main images) 10 µm; (insets) 5 µm.
Cluster stability depends on the actin cytoskeleton. (a) Immunolocalization of the EcDnΔ-αABD-Δ42 chimera (Δ42). Note that the 42-aa-long terminal deletion of the αABD that inactivates binding of this domain to F-actin abolished apical concentration of the clusters and resulted in formation of numerous small clusters typical for EcDnΔ. Bar, 10 µm. (b–g) Effect of actin depolymerization on cadherin clustering during low to high calcium switch: Cells expressing EcDnΔ-αABD chimera (b and c; α), tailless EcDnΔ chimera (d and e; Δ), and full-size Dendra-tagged E-cadherin (f and g; EcD) were cultured at low calcium and then moved to high calcium media in control conditions (b, d, and f; DMSO) or in presence of 8 µM of latrunculin A (c, e, and g; LatA). Cells were fixed 5 min after the calcium switch and immunostained for cadherin and the actin cytoskeleton (actin, shown only in c′). The boxed areas are enlarged in the insets. Magnifications are the same in all images. Note that depolymerization of F-actin by latrunculin A prevented concentration of the EcDnΔ-αABD clusters along the apical area of cell–cell contacts and significantly inhibited formation of the EcDn junctions but has little effect on clustering of the tailless EcDnΔ cadherin. Bars: (main images) 10 µm; (insets) 5 µm.
We next studied whether complete depolymerization of the actin cytoskeleton by a high dose of latrunculin A (8 µM) inhibited the assembly of the stable EcDnΔ-αABD cadherin clusters after the calcium switch. Indeed, such treatment destabilized the clusters (Fig. S2 d) and prevented their recruitment into the apical area of cell–cell contacts (Fig. 5, b and c). As a result, general distribution of EcDnΔ-αABD clusters in cells treated with latrunculin A was indistinguishable from that of tailless clusters formed by EcDnΔ chimera (Fig. 5). Formation of these clusters and their features was completely latrunculin A independent (Fig. 5, d and e). Interestingly, Dendra-tagged full-size E-cadherin (EcDn) in the latrunculin A–treated A431D cells also was completely unable to form stable junctions: only sparse EcDnΔ-like clusters could be detected in the cell–cell contacts of such cells 5 min after a calcium switch (Fig. 5, f and g). Clearly, for both proteins, EcDnΔ-αABD chimera, and full-size cadherin, the intact actin cytoskeleton was essential for junction formation.
In contrast to latrunculin A, inhibitors of actin polymerization, such as ARP2/3 inhibitors, CK-548, or CK-636, did not attenuate stable cluster formation (Fig. 6 a). However, these inhibitors immobilized the clusters. The same effects were observed at a low dose of cytochalasin D (25 nM; Fig. 6 b), which is known to block actin polymerization by capping the free barbed ends of actin filaments (Cooper, 1987). In both of these cases, the clusters were associated with the remaining actin cytoskeleton (Fig. 6, c–c′′). Collectively, these observations confirmed that EcDnΔ-αABD cluster stability and their directional movement were mediated by direct αABD interaction with F-actin.
Directional motility of cadherin clusters depends on the actin polymerization. (a and b) The calcium-switch assay with EcDnΔ-αABD–expressing cells in the presence of Arp2/3 inhibitor CK548 (a, αCK548) and a low dose (25 nM) of cytochalasin D (b, αCD). The cells immediately before the switch (0) and 5 min after the switch (5) are presented. Time-lapse images of the boxed regions are shown at the bottom of each image pairs (numbers indicate minutes after the switch). Note that both inhibitors did not stop cluster formation but immobilized the clusters. Bars: (main images) 10 µm; (insets) 5 µm. (c) The calcium-switch assay with EcDnΔ-αABD–expressing cells in the presence of a low dose (25 nM) of cytochalasin D. Cells were fixed 5 min after addition of calcium and stained for the chimera (c, α) and actin (c′, actin). Note that clusters (three of them are indicated by arrows) appear in proximity to the remaining F-actin structures. Bars: (main images) 5 µm; (insets) 3 µm.
Directional motility of cadherin clusters depends on the actin polymerization. (a and b) The calcium-switch assay with EcDnΔ-αABD–expressing cells in the presence of Arp2/3 inhibitor CK548 (a, αCK548) and a low dose (25 nM) of cytochalasin D (b, αCD). The cells immediately before the switch (0) and 5 min after the switch (5) are presented. Time-lapse images of the boxed regions are shown at the bottom of each image pairs (numbers indicate minutes after the switch). Note that both inhibitors did not stop cluster formation but immobilized the clusters. Bars: (main images) 10 µm; (insets) 5 µm. (c) The calcium-switch assay with EcDnΔ-αABD–expressing cells in the presence of a low dose (25 nM) of cytochalasin D. Cells were fixed 5 min after addition of calcium and stained for the chimera (c, α) and actin (c′, actin). Note that clusters (three of them are indicated by arrows) appear in proximity to the remaining F-actin structures. Bars: (main images) 5 µm; (insets) 3 µm.
αABD cooperates with the cadherin cis-interface for cluster formation
The aforementioned data suggested that the cortical F-actin establishes a multivalent platform holding EcDnΔ-αABD molecules in the clusters. Such a multivalent platform could play, in theory, another important function—it can facilitate the cadherin-clustering process. By this scenario, cluster stabilization by F-actin enhances the entrapment of trans-dimers in the clusters, thereby contributing to the clustering reaction. If so, binding to F-actin could play a function similar to that of the cis-cadherin interface, which also mediates cadherin clustering through cluster stabilization (Wu et al., 2010) Therefore, we studied whether αABD could rescue the clustering of the cis-interface EcDnΔ mutant.
Fig. 7 a shows that in contrast to the cis-EcDnΔ (see Fig. 1 a, cis-Δ), which was completely unable to form clusters, the cis-EcDnΔ-αABD chimera produced numerous clusters in the cell–cell contacts, and these clusters were associated with the actin filament bundles. Live-cell imaging experiments showed that these clusters were much more transient than the parental EcDnΔ-αABD clusters: they could be monitored for a period no longer than 3–4 min (Video 6). However, these clusters could be captured by the 1-s-long frames in live-cell imaging (Fig. S2 e), and many of them exhibited directional albeit short-distance basal to apical movement (Fig. 7 b). The Dendra photoconversion assay confirmed that these clusters were much more unstable than their intact counterparts (Fig. 7 c). Interestingly, reinforcement of these clusters by the additional D1A mutation slowed down cluster dynamics to the level of the parental EcDnΔ-αABD (Fig. 7 c). Together, these observations suggested that αABD cooperates with the cis-interface in a process of cadherin cluster assembly.
The cis-EcDnΔ-αABD mutant forms short-lived clusters. A431D cells expressing the cis-EcDnΔ-αABD chimera were analyzed by time-lapse microscopy. Images were acquired at 1-min intervals for 20 min (1 s/frame). Fragments of this video were used in Fig. 7 b. Note that similar to the cis-interface–intact version of this mutant, EcDnΔ-αABD, the clusters mainly form at the basal edge of cell–cell contacts. They are much more transient, whereas some of them move in an apical direction.
The cis-EcDnΔ-αABD mutant forms short-lived clusters. A431D cells expressing the cis-EcDnΔ-αABD chimera were analyzed by time-lapse microscopy. Images were acquired at 1-min intervals for 20 min (1 s/frame). Fragments of this video were used in Fig. 7 b. Note that similar to the cis-interface–intact version of this mutant, EcDnΔ-αABD, the clusters mainly form at the basal edge of cell–cell contacts. They are much more transient, whereas some of them move in an apical direction.
αABD cooperates with the cadherin cis-interface. (a–a′′) Immunostaining for the cis-EcDnΔ-αABD chimera (a) and for actin filaments (a′). Note that many cadherin clusters were still associated with the actin filaments (some are indicated by the arrows) and concentrated along the apical region of the cell–cell contacts. Bars: (main images) 20 µm; (insets) 2.5 µm. (b) Time-lapse imaging of the cis-EcDnΔ-αABD clusters (frames are indicated according to Video 6). The movement of one of the clusters is traced by the arrows. Note that the clusters are stable and exhibit basal to apical movement. Bar, 5 µm. (c) Dendra photoconversion assay of the clusters in cells expressing EcDnΔ-αABD (Δ-αABD), cis-EcDnΔ-αABD (Cis-Δ-αABD), and D1A/Cis-EcDnΔ-αABD (D1A/Cis-Δ-αABD). The graph shows changes in intensity of the red fluorescence in the individual junctions over time (in minutes) after the Dendra2 green to red photoconversion. The error bars represent SD (n = 30).
αABD cooperates with the cadherin cis-interface. (a–a′′) Immunostaining for the cis-EcDnΔ-αABD chimera (a) and for actin filaments (a′). Note that many cadherin clusters were still associated with the actin filaments (some are indicated by the arrows) and concentrated along the apical region of the cell–cell contacts. Bars: (main images) 20 µm; (insets) 2.5 µm. (b) Time-lapse imaging of the cis-EcDnΔ-αABD clusters (frames are indicated according to Video 6). The movement of one of the clusters is traced by the arrows. Note that the clusters are stable and exhibit basal to apical movement. Bar, 5 µm. (c) Dendra photoconversion assay of the clusters in cells expressing EcDnΔ-αABD (Δ-αABD), cis-EcDnΔ-αABD (Cis-Δ-αABD), and D1A/Cis-EcDnΔ-αABD (D1A/Cis-Δ-αABD). The graph shows changes in intensity of the red fluorescence in the individual junctions over time (in minutes) after the Dendra2 green to red photoconversion. The error bars represent SD (n = 30).
The utrophin actin-binding domain (UtrABD) induces cadherin clustering in the cis-interface–independent mode
To obtain additional evidence that cadherin binding to F-actin drives cadherin clustering, we replaced αABD in our chimera with the F-actin–binding domain of human utrophin (UtrABD). This domain (261 aa), of a size similar to αABD (235 aa), binds to F-actin as a monomer with slightly lower affinity than αABD (10 vs. 1 µM; Keep, 2000; Pappas and Rimm, 2006; Lin et al., 2011). We reasoned that if αABD drove cadherin clustering by binding to F-actin, its replacement with UtrABD should also result in the same effect. Indeed, this chimera was efficiently recruited into cell–cell junctions, forming giant, elongated cadherin clusters extending along the contacts (Fig. 8 a). These clusters were associated with the actin bundles. In contrast to EcDnΔ-αABD and wild-type lateral adherens junctions, UtrABD-based junctions were remarkably static, exhibiting a very slow molecular turnover rate based on our photoconversion assay (Fig. 8 c). Such features of the clusters possessing UtrABD suggested that this domain is even more efficient than αABD in cadherin clustering. Remarkably, the inactivation of the cis-interface in this chimera changed neither the morphology (Fig. 8 b) nor the dynamics of the resulting clusters (Fig. 8 c). Therefore, combining with UtrABD, cis-interface makes a negligible contribution to both cadherin cluster assembly and the maintenance of the assembled clusters.
Clusters of the EcDnΔ-UtrABD chimera are cis-interface independent. (a–b′) Immunostaining for the EcDnΔ-UtrABD (a, Utr) or its cis mutant, cis-EcDnΔ-UtrABD (b, cis-Utr), and actin filaments (a′ and b′, actin). The area indicated by the arrow is enlarged in the insets. Bars: (main images) 10 µm; (insets) 2.5 µm. (c) Dendra photoconversion assay of the clusters made of EcDnΔ-UtrABD (Δ-Utr) and cis-EcDnΔ-UtrABD (Cis-Δ-Utr) in comparison with those made of EcDnΔ-αABD (Δ-αABD). See Fig. 7 c for details. Note that UtrABD mediated formation of the giant cadherin clusters extending along the actin bundles. Inactivation of the cis-interface had little effect on the morphology of the clusters or their internal dynamics. The error bars represent SD (n = 30).
Clusters of the EcDnΔ-UtrABD chimera are cis-interface independent. (a–b′) Immunostaining for the EcDnΔ-UtrABD (a, Utr) or its cis mutant, cis-EcDnΔ-UtrABD (b, cis-Utr), and actin filaments (a′ and b′, actin). The area indicated by the arrow is enlarged in the insets. Bars: (main images) 10 µm; (insets) 2.5 µm. (c) Dendra photoconversion assay of the clusters made of EcDnΔ-UtrABD (Δ-Utr) and cis-EcDnΔ-UtrABD (Cis-Δ-Utr) in comparison with those made of EcDnΔ-αABD (Δ-αABD). See Fig. 7 c for details. Note that UtrABD mediated formation of the giant cadherin clusters extending along the actin bundles. Inactivation of the cis-interface had little effect on the morphology of the clusters or their internal dynamics. The error bars represent SD (n = 30).
Cluster dynamics depend on the hinge joining cadherin with ABD
Why were the adhesive structures mediated by UtrABD and αABD so different? They were immobile in the case of UtrABD and dynamic in the case of αABD. This remarkable divergence was unlikely based on the higher affinity of UtrABD to F-actin; on the contrary, UtrABD was shown to bind F-actin more weakly (Keep, 2000; Pappas and Rimm, 2006). Inspecting the available crystal structures of the utrophin and α-catenin relative vinculin (Keep, 2000; Bakolitsa et al., 2004; Borgon et al., 2004), we noticed that the UtrABD module used in our work contained an unstructured N-terminal region (26 aa). The αABD module in EcDnΔ-αABD, in contrast, had no disordered region at all. This difference can be crucial: the flexible joint between the Dendra tag and the actin-binding unit could reinforce cadherin clusters because it allows connecting nearly every cadherin molecule of the cluster to the underlying F-actin scaffold (Fig. 9 a). The absence of such a joint in EcDnΔ-αABD not only may impede such robust association but could even produce a destabilizing effect caused by a structural clash between cadherin organization in the clusters and actin molecules in the microfilaments.
A hinge between Dendra and αABD changes properties of the clusters. (a) The models of cadherin clusters made from EcDnΔ-αABD (EcDnΔ-αABD), from hinge-containing EcDnΔ-HαABD (EcDnΔ-HαABD), and from a combination of EcDnΔ-HαABD and the tailless mutant EcChΔ (EcDnΔ-HαABD + EcChΔ). Note that the hinge (blue) connecting the Dendra2 tag (Dn) and αABD (yellow) allows connection of each cadherin molecule in the cluster with F-actin. Such clusters could be very stable. In contrast, the lack of the hinge hampers interactions of the majority of cluster molecules with F-actin, resulting in cluster instability. ABD, actin-binding domain; TM, transmembrane domain; mCh, mCherry. (b and b′) Immunostaining of EcDnΔ-HαABD–expressing cells for the chimera (b, HαABD) and for actin filaments (b′, actin). Note that the apical cell–cell contact areas exhibit no filopodia-like protrusions, and the lateral areas (arrowhead) have no lateral clusters. (c) Dendra photoconversion assay of the clusters in cells expressing EcDnΔ-αABD (Δ-αABD) and EcDnΔ-HαABD (Δ-HαABD). See Fig. 7 c for other details. Note that the hinge significantly stabilized the clusters. The error bars represent SD (n = 30). (d–d′′) A431D cells coexpressing EcDnΔ-HαABD (d, green, HαABD) and EcChΔ (d′, red, Δ). Both images are merged in d′′. Note that, upon coexpression with the tailless cadherin mutant, the hinge-containing EcDnΔ-HαABD produced many lateral clusters (arrowhead) and resulted in formation of numerous filopodia on the subapical area of cell–cell contact regions. Bars: (main images) 10 µm; (insets) 3 µm.
A hinge between Dendra and αABD changes properties of the clusters. (a) The models of cadherin clusters made from EcDnΔ-αABD (EcDnΔ-αABD), from hinge-containing EcDnΔ-HαABD (EcDnΔ-HαABD), and from a combination of EcDnΔ-HαABD and the tailless mutant EcChΔ (EcDnΔ-HαABD + EcChΔ). Note that the hinge (blue) connecting the Dendra2 tag (Dn) and αABD (yellow) allows connection of each cadherin molecule in the cluster with F-actin. Such clusters could be very stable. In contrast, the lack of the hinge hampers interactions of the majority of cluster molecules with F-actin, resulting in cluster instability. ABD, actin-binding domain; TM, transmembrane domain; mCh, mCherry. (b and b′) Immunostaining of EcDnΔ-HαABD–expressing cells for the chimera (b, HαABD) and for actin filaments (b′, actin). Note that the apical cell–cell contact areas exhibit no filopodia-like protrusions, and the lateral areas (arrowhead) have no lateral clusters. (c) Dendra photoconversion assay of the clusters in cells expressing EcDnΔ-αABD (Δ-αABD) and EcDnΔ-HαABD (Δ-HαABD). See Fig. 7 c for other details. Note that the hinge significantly stabilized the clusters. The error bars represent SD (n = 30). (d–d′′) A431D cells coexpressing EcDnΔ-HαABD (d, green, HαABD) and EcChΔ (d′, red, Δ). Both images are merged in d′′. Note that, upon coexpression with the tailless cadherin mutant, the hinge-containing EcDnΔ-HαABD produced many lateral clusters (arrowhead) and resulted in formation of numerous filopodia on the subapical area of cell–cell contact regions. Bars: (main images) 10 µm; (insets) 3 µm.
To test this model, we designed EcDnΔ-HαABD chimera. Its α-catenin–derived module, HαABD (residues 631–906), was 40 aa longer than that in the original EcDnΔ-αABD version. These 40 aa apparently constitute a hinge connecting the central domain of α-catenin to its actin-binding domain. Such a role had been suggested by vinculin structure and completely supported by a recent α-catenin structure analysis (Rangarajan and Izard, 2013). Indeed, in agreement with our model, the EcDnΔ-HαABD chimera produced intercellular junctions remarkably similar to those produced by EcDnΔ-UtrABD: their major features were the complete absence of both lateral mobile clusters and filopodia-like protrusions (Fig. 9 b) as well as a very low turnover rate of cluster molecules (Fig. 9 c). In contrast, expression of the EcDnD-Δ26UtrABD chimera, which lacked the 26-aa-long unstructural region of UtrABD, resulted in formation of the EcDnΔ-αABD–like junctions featuring numerous lateral clusters and filopodia (Fig. S5).
These experiments suggested that the overall dynamics of the adhesive structures depend on the ratio between free and actin-bound molecules in the clusters. To directly demonstrate this hypothesis, we coexpressed EcDnΔ-HαABD chimera with the tailless cadherin mutant tagged with a red fluorescent protein, mCherry (EcChΔ). We expected that incorporating the tailless mutant into EcDnΔ-HαABD clusters would decrease their actin-bound cadherin fraction, converting the clusters from the static to the dynamic phenotype (Fig. 9 a). Indeed, the adhesive structures in the cotransfected cells were similar to those produced by the hinge-deficient EcDnΔ-αABD chimera: they featured numerous apical protrusions and multiple spotlike lateral clusters exhibiting basal to apical movement (Fig. 9 d).
Discussion
The intracellular face of adherens junctions intimately interacts with actin filaments. Although the structural aspects of this interaction are yet to be clarified, this interaction has traditionally been assigned to the key role in stabilizing cadherin clusters (Adams et al., 1998; Kusumi et al., 1999; Mège et al., 2006; Lambert et al., 2007; Cavey et al., 2008). However, this role has been never formally proven because of the high complexity of the junctions and because the actin cytoskeleton is indispensable to a variety of crucial cellular processes. To eventually elucidate the contribution of F-actin to cadherin cluster stability, we analyzed the clusters formed by a tailless cadherin mutant lacking any known intracellular binding sites and therefore unable to interact with F-actin. Then, we examined how tethering these “tailless” clusters to F-actin changes their properties. This study revealed dramatic effects of cadherin–actin interactions: it does not just merely increase cadherin cluster stability, but it also drives the clustering process itself.
The formation of cadherin adhesive clusters exclusively through the cadherin ectodomain has been recently shown (Hong et al., 2010; Harrison et al., 2011). Here, we determine that the lifetime of such ectodomain-built tailless clusters is in the range of seconds. The clusters are unstable not only in time but also structurally: they are at fast dynamic equilibrium with monomers. The clusters become undetectable upon targeted inactivation of the cadherin cis- or trans-binding interfaces. In contrast, the strengthening of the cadherin trans-bonds stabilizes the clusters. Collectively, our results confirm that the two binding reactions—formation of trans-homodimers and subsequent reinforcement of these trans-dimers by their cis-interactions—assemble adhesive clusters. However, these clusters are highly unstable, and to maintain adhesion, they have to be reinforced. Scaffolding of the cadherin clusters by the actin cytoskeleton could be one of the reinforcement mechanisms.
To test this possibility, we added to the C terminus of the cadherin tailless mutant the αABD. This 235-aa-long domain was the only active element in the intracellular portion of the resulting EcDnΔ-αABD chimera. This simple design allows for straightforward data interpretation that was not possible with previously designed chimeras, which contained a p120 binding site along with α-catenin–derived modules (Nagafuchi et al., 1994; Ozawa, 1998; Imamura et al., 1999).
The αABD binds to F-actin as a monomer with the dissociation constant of ∼1 µM (Pokutta et al., 2002; Pappas and Rimm, 2006). Its addition to EcDnΔ significantly changed the properties of cadherin clusters: their lifetime increased from seconds to minutes, the rate of their molecular turnover decreased, and they acquired directional basal to apical movement. Control experiments confirmed that cluster stabilization is maintained by direct αABD to F-actin binding. First, the clusters were deficient for ZO1 and Eplin, two proteins that have been also shown to interact with αABD (Imamura et al., 1999; Abe and Takeichi, 2008). Second, both the short C-terminal deletion of αABD that uncouples this domain from F-actin (Pokutta et al., 2002; Pappas and Rimm, 2006) and F-actin depolymerization by latrunculin A completely abolished cluster stability. Finally, alternative coupling of the tailless cadherin to F-actin, through UtrABD, also stabilized cadherin clusters. This latter domain also interacts as a monomer with F-actin with a binding affinity of ∼10 µM (Keep, 2000; Lin et al., 2011). Together, these data convincingly show that just binding to F-actin is sufficient to achieve cadherin cluster stabilization.
One of the most remarkable effects of cadherin binding to actin was that this interaction cooperates with the cadherin cis-interface in a cadherin-clustering process. This interface is known to organize the trans-dimers into the ordered structure (Wu et al., 2010; Harrison et al., 2011) and is indispensable for the tailless cadherin clustering. We found, however, that the cis-interface is less important for clustering of the αABD-containing counterpart. The UtrABD made this process completely cis-interface independent. Strong compensation of the cis-interface mutation was also observed in the D1A/cis-cadherin-αABD chimera. These observations showed that three parameters—cis- and trans-intercadherin interactions as well as cadherin binding to F-actin—determine cadherin monomer–cluster equilibrium. The requirement for the cis-interface for clustering is decreased once the strength of trans-interactions or interactions with F-actin is increased. Several mechanisms can be proposed to explain the clustering potential of actin-binding domains. First, by stabilizing the clusters, this domain decreases their off rate, thereby promoting recruitment of cadherin into the clusters. Second, considering that the cis-interface mutant cannot form an organized structure, binding to actin may enhance cadherin clustering just by limiting trans-dimer diffusion in the junctions. Indeed, it was shown that binding to F-actin, although not preventing cadherin surface diffusion, can significantly slow it down (Sako et al., 1998). Our analyses demonstrate that trans-dimerization also limits diffusion of cadherin molecules (Fig. 2). Therefore, in cooperation, trans-dimerization and binding to actin might be sufficient to immobilize cadherin in the sites of cell–cell contact even without cis-interface contribution. Molecular organization of these two types of clusters, built with and without cis-interface participation, could be, however, very different.
Another important observation we made in our work is that the D1A mutation significantly impedes cluster motility. Because this mutation reduces turnover of cadherin molecules, this observation shows that the movement of the adhesive clusters requires their continuous reassembly. Whether this is true for wild-type adherens junctions remains to be studied. It may be the case because continuous reassembly is the normal state of adherens junctions (Troyanovsky et al., 2006; de Beco et al., 2009). The mechanism of such reassembly, however, is much more complex than just dynamic monomer–cluster equilibrium: it is based on the ATP-dependent disassembly of junctions (Troyanovsky et al., 2006; Hong et al., 2010, 2011). An attractive possibility is that the regulated cadherin–F-actin disengagement in the adherens junctions contributes to this mechanism. In this case, such disengagement may control both the general rate and exact topography of cadherin turnover in the junctions. More experiments are required to fully understand these complex processes and their physiological significance.
Our analysis, although performed using the simple model system, is indispensable for understanding several important aspects of adherens junction biology. First, it reveals two modes of the cadherin-clustering process. In a sharp difference from the EcDnΔ-αABD chimera, clustering of the intact cadherin–catenin complex is strictly cis-interface dependent (Harrison et al., 2011). Therefore, by this feature, the intact cadherin–catenin complex is similar to the tailless cadherin mutant. Such striking similarity might be explained by the fact that the intact complex, just as the tailless mutant, does not interact with F-actin (Nelson, 2008). The lack of interactions between the intact complex and actin suggests that only cis-interface–driven clustering can form an adherens junction, at least at the initial step of its assembly. This initial clustering might then activate the αABD (or other cadherin-associated actin-binding proteins), which, in turn, completes the clustering process using a much more efficient actin-dependent mechanism. The requirement for this second step explains why junctions with strength sufficient to mediate cell–cell adhesion form only in association with the actin cytoskeleton. The actin-dependent mode of cadherin clustering can be especially prominent in regions of high F-actin concentration, such as lamellipodia and filopodia (Vasioukhin et al., 2000; Mège et al., 2006).
F-actin can also regulate the maintenance of the junctions in a steady state. Such possibility is suggested by observations showing that adherens junctions are the sites of actin polymerization (Vasioukhin et al., 2000; Zhang et al., 2005). The exact role of this process, however, is not very well understood. Our data suggest an attractive possibility that adherens junctions themselves produce the actin scaffold, and the rate of the junction-associated actin polymerization regulates the size and stability of the junctions.
Finally, our results demonstrate that the density, size, and flexibility of the links between cadherin clusters and the actin scaffold contribute to the dynamics and mechanics of the junctions. Just by increasing the size of the linker connecting αABD with the rest of the cadherin molecule, we converted the relatively unstable, motile junctions into remarkably robust, nondynamic adhesive structures. The combination of actin-binding proteins in the junction, their activities, and total abundance could, therefore, determine the basic properties of the junction. It might explain the complexity and large variety of cadherin-associated actin-binding proteins (Nelson, 2008; Niessen et al., 2011; Yonemura, 2011). Numerous examples of specific actin-binding proteins in particular sets of adherens junctions are well known (Watabe-Uchida et al., 1998; Nola et al., 2011; Taguchi et al., 2011).
In conclusion, we show that cooperation between extracellular intercadherin interactions and intracellular cadherin interaction with F-actin is one of the key processes in the formation of adherens junctions. This cooperation not only powers cadherin clustering but apparently plays a key role in the synchronization of extracellular and intracellular assembly events. The stabilizing role of F-actin for cadherin clusters makes it possible to regulate the strength and organization of adherens junctions dynamically and reversibly. This regulation can be achieved by changes in α-catenin–F-actin binding affinities, by changes in the number, complexity, and activities of other actin-associated proteins within the cluster, and by changes in the dynamics of the junction-associated actin cytoskeleton.
Materials and methods
Plasmids
The plasmids, which all were based on the vector pRc/cytomegalovirus (Invitrogen), encoding EcDnΔ (in previous works, EcDendra-Δ748-KL), its W2A, cis, and D1A mutants, and full-size E-cadherin (EcDn) were previously described (Hong et al., 2010). The EcDnΔ plasmid was used for constructing the plasmids encoding chimeric proteins (see Fig. S1 a) with αABD, HαABD (aa 671–906 or 631–906 of human α-catenin, correspondingly), UtrABD, Δ26UtrABD (aa 1–261 or 27–161 of human utrophin), and also EcChΔ, the version of EcDnΔ but with an mCherry tag. In all cases, the inserts were placed between Hind III–Not I sites of pRc/cytomegalovirus. The point mutations changing trans- and cis-cadherin interfaces (Fig. S1 a) were reported (Laur et al., 2002; Harrison et al., 2011; Hong et al., 2011). All plasmid inserts were completely sequenced before use.
Cell culture and transfection
Transfection and growth of A431D cells (provided by J.K. Wahl, University of Nebraska Medical Center, Omaha, NE) were performed as previously described (Hong et al., 2010). In brief, cells were grown in DMEM supplemented with 10% FBS. Cells were transfected using Lipofectamine 2000 (Invitrogen) according to the company protocol. After selection of the Geneticin-resistant cells (0.5 mg/ml), the cells were sorted for transgene expression by FACS, and only moderate-expressing cells were used. All cell sublines expressed similar levels of the recombinant proteins (Fig. S1 b).
Immunofluorescence microscopy
The following antibodies were used: mouse anti–E-cadherin, clone HECD1 (Invitrogen); rabbit anti-Dendra2 (Evrogen); mouse anti-ZO1 (BD); and mouse anti-Eplin (sc-136399; Santa Cruz Biotechnology, Inc.). Alexa Fluor 555 phalloidin was purchased from Invitrogen. For the calcium-switch assay, cells were cultivated in a low calcium media (20 µM Ca2+) overnight. The inhibitors of actin polymerization (all obtained from Sigma-Aldrich) were added into the media 15 min before the switch. The same inhibitors were also present in the high calcium media. The inhibitor’s concentrations are indicated in the figure legends.
For immunofluorescence, cells grown for 2 d on glass coverslips were fixed and permeabilized either with methanol-acetone or with 3% formaldehyde–1% Triton X-100 as previously described (Troyanovsky et al., 2006). Both fixation protocols produced the same results in respect to the subcellular distribution of the cadherin chimeras. The actin cytoskeleton was studied using a formaldehyde-based protocol. No variations in the staining patterns for the same cell line were observed. Wide-field images were taken using microscope (Plan Apochromat 100×/1.40 NA objective lens; Eclipse 80i; Nikon) and a digital camera (CoolSNAP EZ; Photometrics). The images were then processed using NIS-Elements software (Nikon).
Live-cell imaging and data processing
These experiments were performed essentially as described earlier (Hong et al., 2010). In brief, cell suspension (∼105 cells) was plated into a homemade chamber built on cover glass. The next day, the culture media were replaced with imaging media (L-15 plus 10% FBS), and the chamber was imaged by microscope (Eclipse Ti-E; Nikon) at 37°C controlled with NIS-Elements software. The microscope was equipped with an incubator chamber, a camera (CoolSNAP HQ2), Plan Apochromat 60×/1.40 NA and Plan Apochromat VC 100×/1.40 NA lenses, and halogen and mercury light sources. Time-lapse images were taken in both FITC and TRITC filter sets using halogen light that minimized phototoxicity and photobleaching. To analyze cadherin junctional turnover, we used a junctional Dendra photoconversion assay (Hong et al., 2010) in which the point of interest (ϕ = 2.5 µm) was photoconverted by a 100-ms-long exposure to the 402-nm wavelength laser. Time-lapse images were then taken in red channel in a stream mode with 100 ms (Fig. 1 e) or in 20-s intervals with 1 s (Fig. 7, Fig. 8, and Fig. 9) of image acquisition time.
All images were saved as TIFF files and processed using ImageJ software (National Institutes of Health). In the Dendra photoconversion assay, the red fluorescent intensity (I) was normalized in such a way that 0 and 1 corresponded to the background and the initial (I0; immediately after activation) values. The background value was obtained from the image taken right before the photoconversion. The time course of the intensity change was produced from 10 sets of independent experiments. Mean values were calculated for each time point. Error bars indicate SD.
Online supplemental material
Fig. S1 provides a detailed characterization of the cadherin mutants and chimeric proteins used in the work. Fig. S2 shows snapshots from the standard videos in which the acquisition time of each frame was 1 s. Fig. S3 shows an example of the Dendra photoconversion assay. Fig. S4 shows double staining of EcDnΔ-αABD–expressing cells for Dendra2 tag and ZO1 or Eplin. Fig. S5 compares cadherin clusters formed by the EcDnΔ-UtrABD and its shorter version lacking a hinge between the Dendra tag and UtrABD. Videos 1, 2, 3, and 6 show the behavior of cadherin clusters formed from EcDnΔ, its D1A mutant, EcDnΔ-αABD, and cis-EcDnΔ-αABD chimeras, respectively. Videos 4 and 5 demonstrate calcium-switch experiments with EcDnΔ-αABD and its D1A mutant, respectively.
Acknowledgments
We are grateful to Dr. I. Indra (Northwestern University, Chicago, IL) and Drs. L. Shapiro, B. Honig, and O. Harrison (Columbia University, New York, NY) for helpful discussion. We are also thankful to Dr. J.K. Wahl for providing A431D cells. Sequencing and flow cytometry were performed at the Northwestern University Genetic and Flow Cytometry Core Facilities.
The work was supported by grants AR44016 and AR057992 from the National Institutes of Health.