Cilia formation is a multi-step process that starts with the docking of a vesicle at the distal part of the mother centriole. This step marks the conversion of the mother centriole into the basal body, from which axonemal microtubules extend to form the ciliary compartment. How vesicles are stably attached to the mother centriole to initiate ciliary membrane biogenesis is unknown. Here, we investigate the molecular role of the mother centriolar component Cep164 in ciliogenesis. We show that Cep164 was indispensable for the docking of vesicles at the mother centriole. Using biochemical and functional assays, we identified the components of the vesicular transport machinery, the GEF Rabin8 and the GTPase Rab8, as interacting partners of Cep164. We propose that Cep164 is targeted to the apical domain of the mother centriole to provide the molecular link between the mother centriole and the membrane biogenesis machinery that initiates cilia formation.
Introduction
Primary cilia are evolutionarily conserved organelles that play an essential role in embryonic development and tissue homeostasis in adulthood (D’Angelo and Franco, 2009; Tasouri and Tucker, 2011). The primary cilium is composed of a basal body and a microtubule based axoneme that is enclosed within a ciliary membrane. The basal body is formed from the centriole that is shared between the cilium and the centrosome. Each centrosome comprises a mother and daughter centriole, surrounded by the proteinaceous pericentriolar material (PCM) from which astral microtubules are organized. The two centrioles are structurally and functionally distinct. The mother centriole (M-centriole) contains electron-dense materials at the subdistal and distal ends (the so-called appendages), whereas the daughter centriole lacks these substructures. Only the M-centriole is capable of forming a cilium (Nigg and Raff, 2009). This property can be attributed to the role of centriolar appendages, as cilia formation is impaired in their absence (Ishikawa et al., 2005; Mikule et al., 2007). The molecular mechanisms by which appendage proteins contribute to cilia formation are not yet understood.
Ciliogenesis also relies upon a specialized transport system formed by motor proteins in complex with components of the intraflagellar transport (IFT) machinery. The IFT machinery promotes cilia formation and extension via anterograde and retrograde transport of cargo along the ciliary microtubules (Pedersen and Rosenbaum, 2008). Cilia membrane biogenesis and the delivery of membrane proteins to the cilium are coordinated by polarized vesicle trafficking under the control of the conserved GTPases of the Rab and Arf families (Li and Hu, 2011). Rab8, Rab11, Rab17, and Rab23 all execute a prominent role during ciliogenesis (Nachury et al., 2007; Yoshimura et al., 2007; Boehlke et al., 2010; Knödler et al., 2010). Although the function of Rab17 and Rab23 is less clear, a Rab cascade involving Rab11, Rab8, and the Rab8 guanine nucleotide exchange factor (GEF), Rabin8, contribute to polarized membrane trafficking to the centrosome during the initial stages of ciliogenesis (Nachury et al., 2007; Yoshimura et al., 2007; Knödler et al., 2010). Recently, the coat assembly complex, named the BBSome, and the transport protein particle II complex, TRAPPII, were also shown to cooperate with the Rab11–Rab8 system in vesicular transport to the centrosome (Nachury et al., 2007; Jin et al., 2010; Knödler et al., 2010; Westlake et al., 2011).
Formation of the primary cilium, which starts at the G1/G0 phase of the cell cycle, is a multi-step process that has been characterized by detailed ultrastructural analysis of ciliated cells (Sorokin, 1962; Pedersen et al., 2008). Ciliogenesis can be subdivided into two physiologically relevant pathways referred to as intra- and extracellular pathways (Sorokin, 1968; Molla-Herman et al., 2010; Ghossoub et al., 2011). In the extracellular pathway, the M-centriole first docks to the plasma membrane after which axonemal microtubules extend from this centriolar template to form the ciliary shaft. In the intracellular pathway, the extension of the cilium starts in the cytoplasm upon association of the M-centriole with vesicles, named the ciliary vesicles (CV), which most likely derive from the Golgi apparatus (Huber et al., 1993). Although the morphology of this process was described in the early sixties by electron microscopy, the mechanism by which vesicles stably attach at the nonmembraneous M-centriole to initiate cilia membrane biogenesis remains to be elucidated.
In this study, we establish links between the centrosomal protein Cep164 and ciliation with detailed characterization of Cep164 function during ciliogenesis. We show that Cep164 is essential for the initial establishment of the ciliary membrane at the distal end of the M-centriole. We identified the components of the vesicular machinery, Rabin8 and Rab8, as Cep164 interactors and established that Cep164 is required for Rab8 centrosome binding. We therefore propose that Cep164 provides the molecular link connecting the M-centriole to components of the machinery that initiate ciliary membrane biogenesis.
Results
Cep164 localizes at the centrosome in a cell cycle–dependent manner
To investigate Cep164 function, we monitored Cep164 localization in human telomerase immortalized retinal pigment epithelial (RPE1) cells using specific antibodies raised against the N-terminal (anti-Cep164-N) and middle domains of Cep164 (anti-Cep164-M; Fig. S1 A). The specific recognition of Cep164 by these antibodies was demonstrated by the loss of the signal upon Cep164 depletion by RNAi (Fig. S1, B–G). In addition, Cep164 labeled by either antibody completely colocalized with distal but not subdistal appendage components (Fig. S1, H–J), confirming that our Cep164 antibodies decorate the distal end of the M-centriole (Graser et al., 2007). As previously reported for U2OS cells (Graser et al., 2007), Cep164 associated with the M-centriole during interphase; however, we noted a drastic reduction in Cep164 levels at the M-centriole during mitosis (Fig. 1, A and B). Similar results were obtained using anti–Cep164-M (Fig. S2, A and B), an established Cep164 antibody (Fig. S2, C and D; Graser et al., 2007), and RPE1 cell lines stably expressing LAP-Cep164 (Cep164 fused to the localization and affinity purification tag; Cheeseman and Desai, 2005; Fig. S2, E and F). The protein levels of Cep164 did not drastically decrease in mitosis (Fig. 1 C, lanes 4, 5, and 7), excluding the possibility that the diminished centrosomal recruitment is due to Cep164 degradation. The levels of Cep164 found at the basal body in ciliated cells were comparable to those found at centrosomes during interphase (unpublished data). Moreover, although faster migrating forms of Cep164 gradually accumulated upon serum starvation, we did not observe an increase in Cep164 protein levels upon induction of ciliogenesis (Fig. 1 D), implying that expression of Cep164 is not up-regulated in response to ciliation.
Cep164 centrosomal localization is cell cycle dependent independently of microtubules. (A) RPE1 cells were stained for Cep164 (anti-Cep164-N), γ-tubulin, and DNA. Centrosomes of representative cells in G1 (arrowheads) and mitosis (arrows) are indicated. (B) Quantification of A. A.U., arbitrary units. (C and D) Western blot analysis of Cep164 protein levels during the cell cycle. (C) RPE1 cells were released from double thymidine block (lanes 1–6) or from double thymidine block followed by nocodazole arrest (lanes 7–9) and harvested at the indicated time points. In D, cycling cells (t = 0) were serum starved as indicated. Plk1 is up-regulated during mitosis and served as mitotic marker. Actin served as a loading control. Low and high exposures are shown in D. The percentage of ciliated cells in D was based on polyglutamylated tubulin staining. Data are means ± SD. (E) Microtubule regrowth assay in RPE1 cells. Staining for α-tubulin in the left panels indicates depolymerization and repolymerization of microtubules. Representative cells are shown for the indicated time points. Regions inside the white boxes are shown at higher magnification in the top right corner of the corresponding panels.
Cep164 centrosomal localization is cell cycle dependent independently of microtubules. (A) RPE1 cells were stained for Cep164 (anti-Cep164-N), γ-tubulin, and DNA. Centrosomes of representative cells in G1 (arrowheads) and mitosis (arrows) are indicated. (B) Quantification of A. A.U., arbitrary units. (C and D) Western blot analysis of Cep164 protein levels during the cell cycle. (C) RPE1 cells were released from double thymidine block (lanes 1–6) or from double thymidine block followed by nocodazole arrest (lanes 7–9) and harvested at the indicated time points. In D, cycling cells (t = 0) were serum starved as indicated. Plk1 is up-regulated during mitosis and served as mitotic marker. Actin served as a loading control. Low and high exposures are shown in D. The percentage of ciliated cells in D was based on polyglutamylated tubulin staining. Data are means ± SD. (E) Microtubule regrowth assay in RPE1 cells. Staining for α-tubulin in the left panels indicates depolymerization and repolymerization of microtubules. Representative cells are shown for the indicated time points. Regions inside the white boxes are shown at higher magnification in the top right corner of the corresponding panels.
We next asked whether Cep164 mimics centrosomal-associated components including PCM1, a component of the centriolar satellites (Kubo et al., 1999), in requiring microtubules to associate with centrosomes. In contrast to PCM1, the centrosomal localization of Cep164 was not decreased after treatment of cells with the microtubule-depolymerizing drug nocodazole or upon overexpression of dynamitin, which acts as a dominant-negative mutant of the dynactin complex and so blocks microtubule-based traffic of PCM1 to the centrosome (Fig. 1 E and unpublished data; Vaughan and Vallee, 1995; Burkhardt et al., 1997). Our data thus indicate that in RPE1 cells Cep164 associates with the M-centriole in a cell cycle–dependent manner, independently of microtubules.
Localization of Cep164 at the M-centriole is mediated by its C terminus
Cep164 contains a predicted tryptophan/tryptophan (WW) domain at its N terminus followed by three coiled coil regions and a C-terminal region that does not resemble known protein domains (Fig. 2 A). WW domains facilitate protein–protein interactions (Macias et al., 2002). To understand how the different regions of Cep164 contribute to ciliogenesis, we assessed which of these domains are required to target Cep164 to the M-centriole and basal body. A series of GFP-fusion constructs were transiently overexpressed in RPE1 cells (Fig. 2 A). All constructs generated proteins of the predicted molecular weights (Fig. 2 B). Only the two truncations containing the C-terminal amino acids 1200–1460 of Cep164 (Cep164-ΔN and Cep164-C) colocalized with the centrosomal marker γ-tubulin, which decorates the pericentriolar region of the two centrioles (Fig. 2 C; Stearns et al., 1991). Similarly, comparisons with the distribution of the daughter centriole–specific protein centrobin (Zou et al., 2005) revealed that GFP-Cep164 and GFP-Cep164-C associated only with the M-centriole (Fig. 2 D). Depletion of endogenous Cep164 by RNAi did not affect the recruitment of GFP-Cep164-C to the M-centriole (Fig. 2 E), implying that the C-terminal region of Cep164 is sufficient to mediate Cep164 centrosomal localization. To confirm this hypothesis, we fused the C-terminal region of Cep164 to the nuclear protein CTNNBL1 (Ganesh et al., 2011). Whereas GFP-CTNNBL1 was mainly nuclear, the GFP-CTNNBL1–Cep164-C chimera was recruited to the M-centrioles in 67% of the transfected cells (Fig. 2, F and G). Therefore, the C-terminal domain of Cep164 is necessary and sufficient to target Cep164 to the M-centriole.
Localization of Cep164 at the centrosome depends on its C terminus. (A) Schematic representation of Cep164 constructs used in this study. Numbers indicate amino acid positions. (B) Western blot analysis of the indicated GFP-Cep164 truncations. Specific protein bands are indicated with red asterisks. (C and D) RPE1 cells were transiently transfected with the indicated GFP-Cep164 constructs for 24 h and stained for γ-tubulin and DNA in C or for centrobin and γ-tubulin in D. (E) RPE1 cells expressing low levels of GFP-Cep164-C were control depleted or depleted for Cep164 for 24 h with serum followed by 48 h without serum. Cells were fixed and stained for Cep164, γ-tubulin, and acetylated tubulin. (F) RPE1 cells were transiently transfected with the indicated GFP constructs as in C and stained for centrobin and γ-tubulin. (G) Quantification for F. The left panels of C–F show merged images. Regions within the white boxes are shown at higher magnification to the right.
Localization of Cep164 at the centrosome depends on its C terminus. (A) Schematic representation of Cep164 constructs used in this study. Numbers indicate amino acid positions. (B) Western blot analysis of the indicated GFP-Cep164 truncations. Specific protein bands are indicated with red asterisks. (C and D) RPE1 cells were transiently transfected with the indicated GFP-Cep164 constructs for 24 h and stained for γ-tubulin and DNA in C or for centrobin and γ-tubulin in D. (E) RPE1 cells expressing low levels of GFP-Cep164-C were control depleted or depleted for Cep164 for 24 h with serum followed by 48 h without serum. Cells were fixed and stained for Cep164, γ-tubulin, and acetylated tubulin. (F) RPE1 cells were transiently transfected with the indicated GFP constructs as in C and stained for centrobin and γ-tubulin. (G) Quantification for F. The left panels of C–F show merged images. Regions within the white boxes are shown at higher magnification to the right.
Overexpression of the N and C terminus of Cep164 has a dominant-negative effect on ciliogenesis
To identify domains that are required for ciliogenesis, we assessed the ability of cells expressing truncated forms of GFP-Cep164 to form cilia. Overproduced GFP-Cep164-C (codons 1200–1460) localized at the M-centriole in serum-starved cells, yet blocked the ability of these cells to form cilia (Fig. 3, A and B). A dominant-negative impact upon ciliogenesis was also observed after overexpression of full-length GFP-Cep164 and GFP-Cep164-N (codons 1–298), but not of the coiled coil–containing Cep164-ΔN/C construct (codons 298–1250; Fig. 3, A and B). Importantly, GFP-Cep164 overexpression did not stop cells from arresting at the G1/G0 phase of the cell cycle upon serum starvation (Fig. 3 C). To investigate whether endogenous Cep164 still localized at the centrosome upon overexpression of the truncated GFP-tagged constructs, we stained endogenous Cep164 with antibodies against the N-terminal or middle domains of Cep164 (anti–Cep164-N or anti–Cep164-M; Fig. S1). These antibodies detected endogenous Cep164 but not the respective Cep164 truncations (Fig. 3, D and E). Interestingly, GFP-Cep164-C, but neither GFP-Cep164-N nor GFP-Cep164-ΔN/C, displaced endogenous Cep164 from the centrosome when overproduced (Fig. 3, D and E). Importantly, expression of low levels of GFP-tagged Cep164 or Cep164-C had no impact upon the generation of cilia (unpublished data). The dominant-negative effect of Cep164-N was abolished by inactivation of the WW domain by substitution of the conserved residues tryptophan 62 and 84 (W62/84) or tyrosine 73 and 74 (Y73/74) to alanine (Fig. 3, F and G; Macias et al., 2002). Taken together, these results indicate that full-length and both the N and C terminus of Cep164 are critical for ciliogenesis and can act as dominant-negative mutants once overproduced. Overexpression of Cep164-C inhibits ciliogenesis presumably by displacing endogenous Cep164, whereas Cep164-N and high levels of Cep164 full length impair ciliogenesis most likely in a WW domain–dependent manner, presumably by titrating out key components away from the centrosome.
Overexpression of Cep164 truncations acts dominantly negative on ciliogenesis. (A) GFP-Cep164 truncations were transiently overexpressed in RPE1 cells for 24 h, serum starved for another 24 h, and stained for γ-tubulin and acetylated tubulin and DNA. (B) Quantification of A. Data are means ± SD. (C) RPE1 cells with the indicated constructs were treated as in A. Cells in G0 or G1 were determined by absent or nucleolar Ki67 staining. n > 100 from a single experiment. (D) GFP-Cep164 truncations were overexpressed in RPE1 cells for 24 h and stained for γ-tubulin. Endogenous Cep164 was visualized with anti–Cep164-M (top two panels) or anti–Cep164-N (bottom three panels). The top panels on the right (a) show untransfected cells, whereas the bottom panels (b) show transfected cells. (E) Quantification of D. Data are means ± SD. (F) GFP-Cep164-N fusion proteins were overexpressed in RPE1 cells as described in A. Cells were stained for γ-tubulin and acetylated tubulin and DNA. WT, wild type. (G) Quantification of F. Data are means ± SD. Merged images are shown in the left panels of A and D. Regions within the white boxes in A, D, and F are shown at higher magnification to the right in A and D and in the top right corner in F. Please note that overexpression in A and D was high, whereas overexpression in Fig. 2 E was low.
Overexpression of Cep164 truncations acts dominantly negative on ciliogenesis. (A) GFP-Cep164 truncations were transiently overexpressed in RPE1 cells for 24 h, serum starved for another 24 h, and stained for γ-tubulin and acetylated tubulin and DNA. (B) Quantification of A. Data are means ± SD. (C) RPE1 cells with the indicated constructs were treated as in A. Cells in G0 or G1 were determined by absent or nucleolar Ki67 staining. n > 100 from a single experiment. (D) GFP-Cep164 truncations were overexpressed in RPE1 cells for 24 h and stained for γ-tubulin. Endogenous Cep164 was visualized with anti–Cep164-M (top two panels) or anti–Cep164-N (bottom three panels). The top panels on the right (a) show untransfected cells, whereas the bottom panels (b) show transfected cells. (E) Quantification of D. Data are means ± SD. (F) GFP-Cep164-N fusion proteins were overexpressed in RPE1 cells as described in A. Cells were stained for γ-tubulin and acetylated tubulin and DNA. WT, wild type. (G) Quantification of F. Data are means ± SD. Merged images are shown in the left panels of A and D. Regions within the white boxes in A, D, and F are shown at higher magnification to the right in A and D and in the top right corner in F. Please note that overexpression in A and D was high, whereas overexpression in Fig. 2 E was low.
Cep164 is required for the establishment of vesicular docking sites at the M-centriole
Cilia formation was drastically reduced upon Cep164 knockdown using four independent siRNAs (Fig. 4, A and B; Graser et al., 2007). This phenotype was partly rescued by expressing a siRNA-resistant Cep164 construct (FLAG-Cep164-R1), as Cep164-depleted cells that were transfected with FLAG-Cep164-R1 showed a three- to fourfold increase in the percentage of ciliated cells compared with GFP-expressing cells (Fig. 4, C–E). To get more insights into Cep164 function, we next asked which step of ciliogenesis required Cep164. One of the first steps of ciliogenesis is the docking of Golgi-derived vesicles at the distal part of the M-centriole (Sorokin, 1962). Subsequent steps comprise the formation of the ciliary shaft and extension of axonemal microtubules (Fig. 4 F). We thus analyzed the phenotype related to Cep164 depletion by transmission electron microscopy (TEM). For this, RPE1 cells were treated with nontargeting or Cep164 siRNA for 48 or 72 h and serum starved for the last 24 or 48 h before analysis by TEM. The successful depletion of Cep164 was confirmed by immunofluorescence staining with anti-Cep164 antibodies (unpublished data). We also established by DNA-content analysis that Cep164 depletion abolished cilia formation without interfering with the G1/G0 cell cycle arrest that was induced by serum withdrawal (unpublished data). The incubation of control cells in serum-free medium for a short period of time (24 h) triggered early steps of ciliogenesis (Fig. 4 F, steps 1–3). These included M-centrioles that were associated with vesicles or ciliary pockets with an extended axoneme (Fig. 4, G, H, and L). In Cep164-depleted cells, however, the number of M-centrioles with docked vesicles was drastically reduced, although vesicles accumulated around the centrosomes upon cilia induction (Fig. 4, I–K; arrowheads indicating vesicles [V]). Extending the period of incubation in serum-free medium did not compensate for this defect (Fig. 4 L). Thus, Cep164 plays a crucial role in the initial steps of ciliogenesis leading to vesicle docking at the M-centriole.
Vesicular docking to the M-centriole is impaired in Cep164-depleted cells. (A) RPE1 cells were treated with the indicated siRNAs for 48 h and serum starved for another 24 h. Ciliated cells were counted based on polyglutamylated tubulin staining. Data are means ± SD. (B) Western blot analysis of cells treated as in A. (C) RPE1 cells transfected with the indicated constructs for 30 h were subsequently treated with control or Cep164_1 siRNA for 24 h before serum starvation for another 24 h. Cells were stained for FLAG, Cep164, γ-tubulin, and acetylated tubulin. (D) Quantification of C. Data are means ± SD. (E) Western Blot of cells treated as in C. (F) Scheme illustrating the different stages of ciliogenesis (Molla-Herman et al., 2010; Ghossoub et al., 2011). The M-centriole with distal (DA) and subdistal appendages (SDA) is shown (0). During early stages, primary ciliary vesicles (PCVs) dock at the M-centriole (1), while nearby secondary vesicles (V) are subjected to later fusion with the PCVs. Accumulation of electron-dense material at the distal end of the M-centriole builds the so-called ciliary bud (CB), which invaginates the PCV (2). The axonemal shaft (AS) grows out and the ciliary pocket (CiPo) forms (3). Last, the cilia membrane fuses with the apical plasma membrane (PM) (4). M-centriole and axonemal shaft are illustrated in black, and membrane structures are shown in red. (G–K) Electron micrographs showing serial sections of RPE1 cells serum starved after control (G and H) or Cep164 depletion (I–K). CP, coated pit; MT, microtubules. (L) Quantifications for G–K. KD, knockdown.
Vesicular docking to the M-centriole is impaired in Cep164-depleted cells. (A) RPE1 cells were treated with the indicated siRNAs for 48 h and serum starved for another 24 h. Ciliated cells were counted based on polyglutamylated tubulin staining. Data are means ± SD. (B) Western blot analysis of cells treated as in A. (C) RPE1 cells transfected with the indicated constructs for 30 h were subsequently treated with control or Cep164_1 siRNA for 24 h before serum starvation for another 24 h. Cells were stained for FLAG, Cep164, γ-tubulin, and acetylated tubulin. (D) Quantification of C. Data are means ± SD. (E) Western Blot of cells treated as in C. (F) Scheme illustrating the different stages of ciliogenesis (Molla-Herman et al., 2010; Ghossoub et al., 2011). The M-centriole with distal (DA) and subdistal appendages (SDA) is shown (0). During early stages, primary ciliary vesicles (PCVs) dock at the M-centriole (1), while nearby secondary vesicles (V) are subjected to later fusion with the PCVs. Accumulation of electron-dense material at the distal end of the M-centriole builds the so-called ciliary bud (CB), which invaginates the PCV (2). The axonemal shaft (AS) grows out and the ciliary pocket (CiPo) forms (3). Last, the cilia membrane fuses with the apical plasma membrane (PM) (4). M-centriole and axonemal shaft are illustrated in black, and membrane structures are shown in red. (G–K) Electron micrographs showing serial sections of RPE1 cells serum starved after control (G and H) or Cep164 depletion (I–K). CP, coated pit; MT, microtubules. (L) Quantifications for G–K. KD, knockdown.
Cep164 contributes to distal but not subdistal appendage formation
We next asked whether Cep164 influenced the centrosomal levels of known appendage components because depletion of subdistal appendage proteins, such as ODF2, centriolin, and ninein, was shown to impair ciliogenesis (Ishikawa et al., 2005; Graser et al., 2007; Mikule et al., 2007). We did not observe a major difference in the morphology of the subdistal appendages of the control versus Cep164-depleted cells in longitudinal TEM sections (Fig. 4, G–K; arrowheads indicating subdistal appendages [SDA]). In addition, the levels of the subdistal appendage proteins ODF2, centriolin, and ninein did not differ between serum-starved control and Cep164 siRNA-treated cells, as determined by quantitative fluorescence microscopy (Fig. S3, A–F; unpublished data). Given that subdistal appendages are involved in anchoring microtubules at the centrosome (Mogensen et al., 2000; Azimzadeh and Bornens, 2007), we also asked whether this function was maintained in the absence of Cep164. Importantly, we observed microtubules attached to subdistal appendages independently of cells being control or Cep164 depleted (Fig. 4, G, J, and K; arrowheads indicating microtubules [MT]). We therefore consider it unlikely that Cep164 influences subdistal appendage integrity or function.
We next asked whether the formation of distal appendages was influenced by Cep164. The integrity of the distal appendages was difficult to evaluate by TEM because of the low electron density of these substructures in our preparations. We therefore asked whether Cep164 influenced the localization of the distal appendage proteins nephrocystin-1 (NPHP1), IFT88, and Cep123. Like Cep164, NPHP1, IFT88, and Cep123 decorate the distal appendages and the transition zone of the cilium (Fig. 5, A and B; Fig. S3 G; Winkelbauer et al., 2005; Fliegauf et al., 2006; Sillibourne et al., 2011). In addition, IFT88 accumulates along the ciliary axoneme and tip (Fig. 5 B; Jurczyk et al., 2004). NPHP1-specific antibodies (Fig. S3, I–M) revealed a reduction in NPHP1 levels at the centrosome upon Cep164 depletion using four independent siRNAs (Fig. 5 A, C, E, and F). Similarly, the levels of centrosomal associated IFT88 strongly decreased upon Cep164 depletion (Fig. 5, B and D–F). Importantly, the overall protein levels of NPHP1 or IFT88 was not influenced by Cep164 depletion (Fig. 5 F). The defective localization of NPHP1 and IFT88 was rescued by expression of a siRNA-resistant form of Cep164 (Fig. 5, G and H). These data imply that association of NPHP1 and IFT88 largely depended on Cep164. Interestingly, the centrosomal levels of Cep123 were not significantly affected by Cep164 knockdown (Fig. S3, G and H). Similar results were obtained by an independent group (Bornens, M., and J. Sillibourne, personal communication). The fact that Cep164 was dispensable for Cep123 centrosomal localization but needed to target NPHP1 and IFT88 to the M-centriole led us to the conclusion that Cep164 partly contributes to distal appendage integrity.
Cep164 is involved in recruiting the distal appendage proteins NPHP1 and IFT88 to the M-centriole. (A) RPE1 cells stably expressing LAP-CSAP were treated with control or Cep164 siRNA for 24 h and serum starved for 48 h. Cells were stained for NPHP1 and Cep164. LAP-CSAP localizes to both centrioles and the axoneme and served as a centriolar and ciliary marker (Backer et al., 2012). (B) RPE1 cells, treated as in A, were stained for IFT88, Cep164, and polyglutamylated tubulin. In control depleted cells, IFT88 decorates the M-centriole of nonciliated cells and additionally the axoneme of ciliated cells. (C–E) Box-and-whisker plots showing NPHP1 (C), IFT88 (D), and Cep164 (E) signal intensity at the centrosome in RPE1 cells treated with the indicated siRNAs for 48 h and serum starved for another 24 h. (F) Western blot analysis of the experiment shown in C–E. Please note that the top and bottom panels are identical to Fig. 4 B, as these experiments were performed together. (G and H) Box-and-whisker plots show the relative signal intensity of NPHP1 (G) and IFT88 (H) at centrosomes in control and Cep164-depleted RPE1 cells expressing GFP or FLAG-Cep164-R1. Cells were treated as in Fig. 4 C. Knockdown efficiency was confirmed by Western blot analysis (Fig. 4 E). The left panels of A and B show merged images. Regions within the white boxes are shown at higher magnification to the right. A.U., arbitrary units.
Cep164 is involved in recruiting the distal appendage proteins NPHP1 and IFT88 to the M-centriole. (A) RPE1 cells stably expressing LAP-CSAP were treated with control or Cep164 siRNA for 24 h and serum starved for 48 h. Cells were stained for NPHP1 and Cep164. LAP-CSAP localizes to both centrioles and the axoneme and served as a centriolar and ciliary marker (Backer et al., 2012). (B) RPE1 cells, treated as in A, were stained for IFT88, Cep164, and polyglutamylated tubulin. In control depleted cells, IFT88 decorates the M-centriole of nonciliated cells and additionally the axoneme of ciliated cells. (C–E) Box-and-whisker plots showing NPHP1 (C), IFT88 (D), and Cep164 (E) signal intensity at the centrosome in RPE1 cells treated with the indicated siRNAs for 48 h and serum starved for another 24 h. (F) Western blot analysis of the experiment shown in C–E. Please note that the top and bottom panels are identical to Fig. 4 B, as these experiments were performed together. (G and H) Box-and-whisker plots show the relative signal intensity of NPHP1 (G) and IFT88 (H) at centrosomes in control and Cep164-depleted RPE1 cells expressing GFP or FLAG-Cep164-R1. Cells were treated as in Fig. 4 C. Knockdown efficiency was confirmed by Western blot analysis (Fig. 4 E). The left panels of A and B show merged images. Regions within the white boxes are shown at higher magnification to the right. A.U., arbitrary units.
Cep164 interacts with Rabin8 in vivo and in vitro
To get more insight on how Cep164 contributes to vesicle docking on a molecular level, we performed a yeast two-hybrid screen with Cep164-N and Cep164-C as bait to identify Cep164-interacting proteins. Cep164-N constructs were strongly self-activating, whereas Cep164-C localized in the yeast nucleus without detectable self-activation (unpublished data; Fig. 6 A). Using Cep164-C as bait, we screened a human testis cDNA library for putative Cep164 interactors. Among the most prominent Cep164-C binding partners, we found a plasmid encoding the C terminus of the Rab8 GEF, Rabin8 (Fig. 6 A; Hattula and Peränen, 2005). This finding was particularly interesting because Rabin8 has been reported to activate Rab8 to initiate membrane formation during early ciliogenesis (Yoshimura et al., 2007; Westlake et al., 2011). The interaction between Rabin8 and Cep164-C could be further validated by coimmunoprecipitation using GFP-Rabin8 and FLAG-Cep164 constructs. GFP-Rabin8 interacted strongly with full-length FLAG-Cep164 and FLAG-Cep164-C but not with FLAG-Cep164-N or anti-FLAG beads (Fig. 6 B). Thus, the C terminus of Cep164 interacts with Rabin8.
The C terminus of Cep164 interacts with Rabin8 and Rab8. (A) Yeast strains expressing the indicated gene fusions were grown on selective plates lacking either leucine and tryptophan (−LW) or leucine, tryptophan, histidine, and adenine (−LWHA). Growth on −LWHA plates indicates interaction. Duplicates are shown. Negative (Lamin C and T7-T-antigen) and positive (p53 and T7-T-antigen) controls were included. Colonies from noninteractors appear red on −LW plates because cells deficient for ADE2 expression accumulate a red pigment. (B–E) HEK293T cells were transiently cotransfected with the indicated constructs. Immunoprecipitations (IP) were performed using anti-FLAG agarose and interacting proteins were detected by Western blot. Arl6-GFP in D, a small GTPase with a role in ciliogenesis (Wiens et al., 2010), served as negative control.
The C terminus of Cep164 interacts with Rabin8 and Rab8. (A) Yeast strains expressing the indicated gene fusions were grown on selective plates lacking either leucine and tryptophan (−LW) or leucine, tryptophan, histidine, and adenine (−LWHA). Growth on −LWHA plates indicates interaction. Duplicates are shown. Negative (Lamin C and T7-T-antigen) and positive (p53 and T7-T-antigen) controls were included. Colonies from noninteractors appear red on −LW plates because cells deficient for ADE2 expression accumulate a red pigment. (B–E) HEK293T cells were transiently cotransfected with the indicated constructs. Immunoprecipitations (IP) were performed using anti-FLAG agarose and interacting proteins were detected by Western blot. Arl6-GFP in D, a small GTPase with a role in ciliogenesis (Wiens et al., 2010), served as negative control.
The interaction between Cep164 and components of the Rab pathway was not limited to Rabin8, as Cep164 also coimmunoprecipitated with Rab8 (Fig. 6 C, lane 1). Importantly, Cep164 interacted preferentially with Rab8a compared with Rab8b (Fig. 6 C, lanes 1 and 2), although both isoforms are involved in ciliogenesis (Yoshimura et al., 2007; Westlake et al., 2011). The GTPase Rab11, which is involved in trafficking and activation of Rabin8 (Knödler et al., 2010; Westlake et al., 2011), did not coimmunoprecipitate with Cep164 (Fig. 6 C, lanes 3 and 4), highlighting the specificity of interaction between Cep164 and Rab8a.
To test whether perturbations in the GDP/GTP cycle of Rab8a would affect its association with Cep164, we made use of the established inactive GDP-locked Rab8a-T22N and constitutively active GTP-locked Rab8a-Q67L mutants (Peränen et al., 1996). Cep164 bound to both Rab8a-T22N and Rab8a-Q67L (Fig. 6 D, lanes 2 and 3). As observed for Rabin8, the C-terminal region of Cep164 contributed to its binding to Rab8a wild-type or mutant forms (Fig. 6 E; unpublished data). Collectively, these data suggest that Cep164 binds to the GEF Rabin8 and the GTPase Rab8 independently of its nucleotide status.
We next asked whether Cep164 directly interacted with Rabin8 and Rab8. For this, we performed in vitro binding assays using bacterially purified maltose-binding protein (MBP)-Cep164-C, 6His-Rabin8, or NusA-Rab8a-T22N-6His (Fig. 7, A–D). Interestingly, 6His-Rabin8 bound to MBP-Cep164-C but not to MBP alone (Fig. 7 A). The interaction of MBP-Cep164-C was specific to Rabin8, as NusA-Rab11-6His was capable to bind to MBP-Rabin8, but no binding was observed between MBP-Cep164-C and NusA-Rab11-6His (Fig. 7 B). In contrast to Rabin8, NusA-Rab8a-T22N-6His failed to bind to Cep164-C in vitro (Fig. 7 C). In addition, we could not observe any interaction between Cep164-C and Rab8 or its mutants in yeast two-hybrid analysis (unpublished data), suggesting that Rabin8, but not Rab8, directly interacts with the C-terminal domain of Cep164 in vitro.
Cep164-C interacts with Rabin8, but not with Rab8, in vitro. (A–C) In vitro binding assays using bacterially purified proteins. (A) MBP or MBP-Cep164-C beads were incubated with purified 6His-Rabin8. (B) MBP, MBP-Rabin8, or MBP-Cep164-C beads were incubated with NusA-Rab11a-6His. (C) MBP-fusion proteins (immobilized on beads) were incubated with purified NusA-Rab8a-T22N-6His. Proteins were visualized by Western blot. (D) Coomassie blue–stained gel of proteins used in A–C. The asterisks indicate unspecific protein bands.
Cep164-C interacts with Rabin8, but not with Rab8, in vitro. (A–C) In vitro binding assays using bacterially purified proteins. (A) MBP or MBP-Cep164-C beads were incubated with purified 6His-Rabin8. (B) MBP, MBP-Rabin8, or MBP-Cep164-C beads were incubated with NusA-Rab11a-6His. (C) MBP-fusion proteins (immobilized on beads) were incubated with purified NusA-Rab8a-T22N-6His. Proteins were visualized by Western blot. (D) Coomassie blue–stained gel of proteins used in A–C. The asterisks indicate unspecific protein bands.
Cep164 is required for the centrosomal recruitment of Rab8
We next asked whether recruitment of Rabin8 and Rab8 to centrosomes requires Cep164 function. All attempts to detect Rabin8 at centrosomes using commercially available antibodies or GFP-tagged Rabin8 constructs failed, most likely due to its very transient recruitment to the centrosome after serum withdrawal (Nachury et al., 2007; Yoshimura et al., 2007; Westlake et al., 2011). We therefore focused upon an analysis of the dependency between Rab8 recruitment and Cep164, as Rab8 centrosomal recruitment was shown to depend upon Rabin8 (Nachury et al., 2007; Westlake et al., 2011). Rab8 accumulates at the centrosome during early ciliogenesis and decorates the ciliary membrane during cilium growth, but disappears from the fully elongated cilium (Nachury et al., 2007; Yoshimura et al., 2007; Westlake et al., 2011). To investigate whether Cep164 influenced Rab8 localization, we generated RPE1 cells stably expressing GFP-Rab8a and analyzed Rab8 localization in control and Cep164 siRNA–treated cells after serum withdrawal. In ∼30% of the control depleted cells GFP-Rab8a was not observed at centrosomes or matured cilia (Fig. 8 A, a; unpublished data; Fig. 8 B). In the remaining 70% of cells, GFP-Rab8a was either only partially colocalized with Cep164 at the centrosome (not attached to a cilium, Fig. 8 A, b) or decorated the ciliary membrane, irrespectively of whether tubulin acetylation was detected or not (Fig. 8 A, c and d; Fig. 8 B). The presence of GFP-Rab8a at cilia with nonmodified tubulin has been reported previously (Nachury et al., 2007; Westlake et al., 2011). In cells depleted for Cep164 using four independent siRNAs, GFP-Rab8a showed a cytoplasmic and Golgi-like localization but was no longer observed at centrioles in >99% of cells (Fig. 8 A, e; Fig. 8 B). Thus, Cep164 does not prevent Rab8a binding to locations other than at the centrosome. The decreased centrosomal localization of Rab8a upon Cep164 siRNA treatment could be rescued by a siRNA-resistant form of Cep164 (Fig. 8, C and D).
Rab8 centrosomal/ciliary localization requires Cep164. (A) RPE1 cells stably overexpressing GFP-Rab8a were treated with control or Cep164 siRNA for 48 h, serum starved for 24 h, and stained for Cep164, γ-tubulin, and acetylated tubulin. Representative fluorescence micrographs are shown for the localization patterns of GFP-Rab8a (a–e). The left panels show merged images. Regions within the white boxes are shown at higher magnification to the right. (B) Quantification of cells treated with the indicated siRNAs as described in A. Only cells with complete loss of Cep164 were considered. (C) Centrosomal/ciliary localization of Cherry-Rab8a in serum-starved RPE1 cells expressing GFP or FLAG-Cep164-R1 for 30 h. Cells were then transfected with the indicated siRNAs for 30 h and serum starved for 18 h. Data are means ± SD. (D) Box-and-whisker plots showing endogenous Cep164 signal intensity at the centrosome of cells treated as in C, confirming efficient depletion of Cep164. Please note that the efficiency of Cep164 knockdown in RPE1 cells co-overexpressing Cherry-Rab8a and FLAG-Cep164-R1 was estimated based on the quantification of endogenous Cep164 centrosomal signal from cells not expressing FLAG-Cep164-R1 within the same population (FLAG-negative cells). A.U., arbitrary units.
Rab8 centrosomal/ciliary localization requires Cep164. (A) RPE1 cells stably overexpressing GFP-Rab8a were treated with control or Cep164 siRNA for 48 h, serum starved for 24 h, and stained for Cep164, γ-tubulin, and acetylated tubulin. Representative fluorescence micrographs are shown for the localization patterns of GFP-Rab8a (a–e). The left panels show merged images. Regions within the white boxes are shown at higher magnification to the right. (B) Quantification of cells treated with the indicated siRNAs as described in A. Only cells with complete loss of Cep164 were considered. (C) Centrosomal/ciliary localization of Cherry-Rab8a in serum-starved RPE1 cells expressing GFP or FLAG-Cep164-R1 for 30 h. Cells were then transfected with the indicated siRNAs for 30 h and serum starved for 18 h. Data are means ± SD. (D) Box-and-whisker plots showing endogenous Cep164 signal intensity at the centrosome of cells treated as in C, confirming efficient depletion of Cep164. Please note that the efficiency of Cep164 knockdown in RPE1 cells co-overexpressing Cherry-Rab8a and FLAG-Cep164-R1 was estimated based on the quantification of endogenous Cep164 centrosomal signal from cells not expressing FLAG-Cep164-R1 within the same population (FLAG-negative cells). A.U., arbitrary units.
The Golgi-like staining observed for Rab8 implied that Golgi structure was not disrupted. However, because an intact Golgi apparatus was reported to be important for post-Golgi membrane traffic from the trans-Golgi network to the centrosome to initiate cilia formation and receptor delivery to the ciliary membrane (Deretic and Papermaster, 1991; Haller and Fabry, 1998; Stephens, 2001), we analyzed Golgi integrity in more detail. For this, we labeled GM130, Golgin97, and TGN46, which served as markers for the cis- and trans-Golgi network, respectively (Fig. S4 A; Nakamura et al., 1995; Banting and Ponnambalam, 1997; Yoshino et al., 2003). We did not observe any major difference in respect to Golgi structure when comparing control and Cep164-depleted cells (Fig. S4, A, C, and D), thereby excluding the possibility that Cep164 is required for Golgi integrity. Finally, given that the microtubule cytoskeleton is involved in Rab trafficking to the cilium (Westlake et al., 2011), we asked whether microtubule-based transport to the centrosome was impaired in Cep164-depleted cells. We therefore analyzed the behavior of the pericentriolar matrix proteins PCM1 and CEP290, as they depend on microtubules for centrosomal targeting (Kubo et al., 1999; Kim et al., 2008). PCM1 and CEP290 accumulated around centrosomes irrespective of the presence or absence of Cep164 (Fig. S4, B–D; unpublished data), suggesting that Cep164 depletion did not perturb microtubule-dependent transport. Collectively, our data show that Cep164 is required for the centrosomal binding of Rab8.
Rab8 overexpression compensates for reduced Cep164 levels
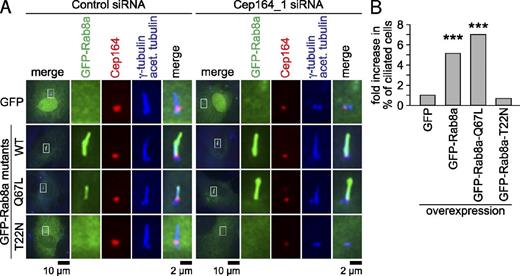
We hypothesized that the mislocalization of Rab8 could be responsible for the defect in cilia formation in Cep164-depleted cells. To test this idea, we asked whether ectopically overproduced wild-type GFP-Rab8a or the GTP-locked Rab8a-Q67L mutant could restore ciliogenesis in case of Cep164 depletion (Fig. 9 A). The inactive GDP-locked Rab8a-T22N construct and GFP alone were used as negative controls. GFP, GFP-Rab8a, GFP-Rab8a-Q67L, and GFP-Rab8a-T22N were transiently overexpressed in RPE1 cells for 24 h before subjecting the cells to treatment with control or Cep164 siRNA and subsequent serum withdrawal. The degree of centrosomal Cep164 depletion was determined by Western blot (Fig. S5 A) and quantitative fluorescence microscopy using Cep164-specific antibodies (Fig. S5, B and C). The residual levels of Cep164 upon depletion were correlated with the presence or absence of a cilium (based on tubulin acetylation staining) in individual cells. These quantifications were particularly important to establish the threshold for the amount of Cep164 left at the centrosome still able to support cilia formation. Cilia formation was strongly suppressed by a 40–85% reduction of the initial Cep164 centrosome-associated signal (Fig. S5, B and C). A reduction to levels below 85% of normal Cep164 levels completely abolished ciliogenesis (Fig. S5 C). We analyzed the ability of Rab8 constructs to rescue ciliogenesis in cells in which centrosomal Cep164 was either strongly depleted (0–10% of Cep164 left at the centriole; “strong depletion”) or substantially depleted (10–60% of Cep164 left at the centriole; “medium depletion”). Strong depletion of Cep164 did not support cilia formation irrespective of whether GFP, GFP-Rab8a, GFP-Rab8a-Q67L, or GFP-Rab8a-T22N was overexpressed (Fig. S5 C). Importantly, in Cep164 medium-depleted cells, ciliogenesis was significantly increased (five- to sevenfold) upon overexpression of GFP-Rab8a or the GTP-locked form of Rab8a (GFP-Rab8a-Q67L; Fig. 9, A and B; Fig. S5 C). This effect was specific to active Rab8a, as ciliogenesis was not rescued in Cep164 medium-depleted cells carrying inactive Rab8a (GFP-Rab8a-T22N) or GFP alone (Fig. 9, A and B; Fig. S5 C). This implies that Cep164 function during ciliogenesis can be compensated, but not completely replaced, by overproduction of active Rab8. Collectively, the data indicate that Cep164 is required for Rab8 centrosomal recruitment and function to promote ciliogenesis.
Overproduction of Rab8a rescues Cep164-dependent ciliogenesis defects. (A) The indicated GFP constructs were transiently overexpressed in RPE1 cells for 24 h. Cells were subjected to control or Cep164 depletion for 48 h and serum starved for 24 h. Cep164, γ-tubulin, and acetylated tubulin staining are shown. Equal knockdown efficiencies were confirmed by measuring Cep164 signal intensity (Student’s t test analysis). The left panels show merged images. Regions within the white boxes are shown at higher magnification to the right. (B) Graph shows fold increase in the percentage of ciliated cells processed as in A. Only medium depleted cells, in which 10–60% of initial Cep164 staining remained at the centrosome, were considered (see Fig. S5 for details). A single representative experiment out of three repeats is shown. For the experiment shown, 40–45 cells were observed per cell line.
Overproduction of Rab8a rescues Cep164-dependent ciliogenesis defects. (A) The indicated GFP constructs were transiently overexpressed in RPE1 cells for 24 h. Cells were subjected to control or Cep164 depletion for 48 h and serum starved for 24 h. Cep164, γ-tubulin, and acetylated tubulin staining are shown. Equal knockdown efficiencies were confirmed by measuring Cep164 signal intensity (Student’s t test analysis). The left panels show merged images. Regions within the white boxes are shown at higher magnification to the right. (B) Graph shows fold increase in the percentage of ciliated cells processed as in A. Only medium depleted cells, in which 10–60% of initial Cep164 staining remained at the centrosome, were considered (see Fig. S5 for details). A single representative experiment out of three repeats is shown. For the experiment shown, 40–45 cells were observed per cell line.
Discussion
In contrast to several membrane biogenesis events, which involve the fusion of two membrane compartments, the formation of the ciliary membrane at the centriole is unique in the sense that it initially requires the stable attachment of vesicles to the nonmembranous mother centriolar structure. This step, which is defined as the primary ciliary vesicle formation, was described at an ultrastructural level more than four decades ago (Sorokin, 1962), yet the molecular mechanisms have remained elusive. Our data now establish that the distal appendage protein Cep164 is indispensable to promote this vesicular docking that initiates cilia membrane biogenesis (Fig. 10).
A model of Cep164 function during ciliogenesis. At the end of mitosis, the amount of Cep164 that associates with the M-centriole increases, reaching a maximum in interphase. Upon induction of ciliogenesis, centrosomal Cep164 is needed for the recruitment of Rab8 and docking of vesicles at the M-centriole to initiate the formation of the cilium (see Discussion for details).
A model of Cep164 function during ciliogenesis. At the end of mitosis, the amount of Cep164 that associates with the M-centriole increases, reaching a maximum in interphase. Upon induction of ciliogenesis, centrosomal Cep164 is needed for the recruitment of Rab8 and docking of vesicles at the M-centriole to initiate the formation of the cilium (see Discussion for details).
Cell cycle association of Cep164 with the M-centriole
Cep164 localizes at the distal appendages of the M-centriole and the transition zone of ciliated cells (Graser et al., 2007). We show that the C-terminal region of Cep164 is required and sufficient to target Cep164 to the M-centriole. In fact, Cep164-C was sufficient to relocate the nuclear protein CTNNBL1 to the M-centriole, demonstrating that Cep164-C contains all the information for this recruitment step. Recently it was reported that a mutation in the stop codon of Cep164 leads to an extended version of Cep164 and can cause a form of nephronophthisis-related ciliopathy (Chaki et al., 2012). Together with our data, this implies that an intact C terminus is critical for the localization and function of Cep164.
In contrast to a previous report (Graser et al., 2007), we found that Cep164 associated with the centrosome in a cell cycle–dependent manner, as Cep164 protein levels at the centrosome decrease during mitosis to peak at this location in interphase of cycling or serum-starved cells. The decrease of Cep164 at the M-centriole during mitosis observed here is unlikely to arise from protein degradation or from a change in the accessibility of the protein to antibodies, as Cep164 levels were not decreased in metaphase-arrested cells and we obtained similar results with LAP-tagged Cep164. Reduction in centrosomal association during mitosis has been previously reported for other centrosomal proteins, including Cep170, Nlp, ninein, and Cep55 (Casenghi et al., 2003; Chen et al., 2003; Fabbro et al., 2005; Guarguaglini et al., 2005). The enrichment of Cep164 at the M-centriole during G1/G0 phases fits perfectly with the essential function of Cep164 during ciliogenesis at this point of the cell cycle and may be a key regulatory step in the promotion of ciliogenesis. Therefore, it would be important to define the cell cycle determinants of Cep164 centrosomal localization. We observed that Cep164 and the centrosomal targeting region of the C-terminal domain of Cep164 showed several forms with a decreased mobility in SDS-PAGE due to phosphorylation (unpublished data). It is therefore possible that cell cycle–dependent phosphorylation might influence Cep164 centrosomal binding. Alternatively, the Cep164 binding partner could equally well be regulated in a cell cycle–dependent manner. The protein OFD1, which is mutated in the orofaciodigital syndrome 1, was shown to be required for distal appendage formation and Cep164 centrosomal localization (Singla et al., 2010). However, OFD1 and Cep164 were not found in common complexes, suggesting that OFD1 might not bind Cep164 directly.
A role of Cep164 for centriole–ciliary membrane association
Our data suggest that Rab8-positive vesicles are stabilized at centrosomes in a Cep164-dependent manner (Fig. 10). Rab8 was recently shown to be under control of a Rab cascade involving Rabin8 and Rab11. In this cascade, the GTPase Rab11 promotes the transport of Rabin8 to the centrosome and the GEF activity of Rabin8 toward Rab8 (Knödler et al., 2010; Westlake et al., 2011). The generation of active Rab8 seems to be critical for its centrosomal localization and ciliary function, as in the absence of Rabin8 Rab8 does not bind to centrosomes and ciliogenesis is impaired (Nachury et al., 2007; Westlake et al., 2011). Cep164 interacted with Rabin8 both in vivo and in vitro, suggesting that Cep164 may be directly involved in recruiting Rabin8 to promote the local activation of Rab8 by Rabin8 at the centrosome. The fact that Rab8 failed to localize at the centrosome in the absence of Cep164 and overproduction of Rab8 or a constitutively active GTP-locked Rab8 mutant partially rescued the ciliogenesis defect arising from the reduction in Cep164 levels at the centrosome are consistent with the hypothesis that Cep164 promotes accumulation of GTP-Rab8 at the centrosome. Furthermore, a local activation of Rab8 at the centrosome has been proposed previously (Nachury et al., 2007; Westlake et al., 2011). Importantly, Cep164 depletion neither interfered with the centrosomal accumulation of the proteins PCM1 or CEP290, which have been recently reported to be required for Rab8 targeting to the primary cilium (Kim et al., 2008), nor with an intact Golgi structure, which is also required for appropriate membrane trafficking to the centrosome during ciliogenesis (Deretic and Papermaster, 1991; Haller and Fabry, 1998; Stephens, 2001). Interestingly, the centrosomal and Golgi-resident protein Hook2 was recently shown to interact with PCM1 and Rab8 and to be essential for PCM1 centrosomal association and ciliary vesicle docking (Baron Gaillard et al., 2011). Although Hook2 coordinates vesicular docking of Rab8-positive vesicles at the M-centriole most likely by regulating microtubule-dependent traffic of cargos to the centrosome via PCM1 (Baron Gaillard et al., 2011), the distal appendage protein Cep164 may function as an acceptor molecule at the M-centriole allowing vesicular docking to the distal appendages most likely by facilitating the local activation of Rab8. Our data, however, do not exclude the possibility that Cep164 might additionally contribute to the initial docking of Rab8-positive vesicles at the distal appendages of the M-centriole by an alternative mechanism, i.e., by bringing together components that promote and facilitate vesicle tethering and fusion processes (Fig. 10). Most likely, the conserved N-terminal WW domain of Cep164 might play a crucial role in this process, as the overexpression of this domain has a dominant-negative effect upon ciliogenesis that was lost after its inactivation by the introduction of site-specific mutations (Fig. 3).
Remarkably, centrosomes of fission and budding yeast (referred to as spindle pole bodies; SPBs), serve as membrane nucleators during the meiotic cycle (Moreno-Borchart and Knop, 2003; Shimoda, 2004). The initiation of membrane biogenesis at SPBs involves a developmentally controlled modification of the SPB cytoplasmic plaque, which acquires a proteinaceous complex involved in docking of post-Golgi–derived vesicles. This docking event seems to be a prerequisite for fusion/membrane extension and it requires the yeast Rab8 homologue, Sec4, in addition to the exocyst complex (Moreno-Borchart and Knop, 2003; Shimoda, 2004). Interestingly, the subdistal appendage component centriolin, which is in close proximity to Cep164, physically interacts with components of the exocyst and SNARE fusion machineries during cytokinesis (Gromley et al., 2005; Hehnly et al., 2012). In addition, the exocyst component Sec10 and the SNAREs, SNAP25 and syntaxin 3, have been reported to localize to the cilium (Low et al., 1998; Rogers et al., 2004; Mazelova et al., 2009). Collectively, these observations raise the exciting possibility that Cep164 might contribute to the formation of a multi-functional platform that promotes the initial docking of Rab8-positive vesicles at the distal appendages of the M-centriole to facilitate further tethering and fusion events (Fig. 10).
Defects in transition zone proteins have been reported to be linked to heritable ciliopathies, such as Meckel and Joubert syndromes (Hildebrandt et al., 2009). Considering the importance of Cep164 in converting the M-centriole into the basal body in RPE1 cells, it will be appealing to investigate how Cep164 mutations are associated with the etiology of ciliopathies (Chaki et al., 2012).
Materials and methods
Plasmids
cDNA for Cep164 was kindly provided by Ingrid Hoffmann (DKFZ, Heidelberg, Germany) and the ORF was cloned by PCR into pCMV-3Tag-1b or pEGFP-C1 to fuse with N-terminal FLAG or GFP. cDNA for CTNNBL1 was a kind gift of Oliver Gruss (ZMBH, University of Heidelberg, Germany) and cloned by PCR into pEGFP-C1 to fuse with N-terminal GFP. Cep164 and ODF2 were cloned by PCR into pIC113 (kind gift of Iain Cheeseman, Whitehead Institute, Cambridge, MA) to fuse the ORF with the N-terminal LAP-tag (Cheeseman and Desai, 2005). LAP-Cep164 was subcloned in pENTR1A and recombination was performed in pLenti6.2/V5-Dest (Invitrogen) for stable expression via genomic integration. The retroviral ODF2 expression vector, which contained the TET-on inducible promoter, was generated by subcloning of LAP-ODF2 into pMOWSIN-TREl (Pfeifer et al., 2010). For generation of stable NIH 3T3 cells, retroviral expression vectors were cotransduced into NIH 3T3 cells together with the cDNA for the transactivator protein contained in pMOWS-rtTAM2 (Pfeifer et al., 2010). Other plasmids were generated by PCR amplification from total cDNA obtained from RPE1 cells. RNA was isolated using Absolutely RNA Microprep kit (Agilent Technologies) and reverse transcribed in cDNA using RevertAid First Strand cDNA Synthesis kit (Thermo Fisher Scientific). FLAG, GFP, Cherry, 6His, MBP, and NusA/6His in-frame fusions were generated using standard vectors (pCMV-3Tag-1 [Agilent Technologies]; pEGFP-C1, pEGFP-N1, pmCherry-C1 [Takara Bio Inc.]; pET28 [EMD Millipore]; pMAL-c2X [New England Biolabs, Inc.]; and pQE80-NusA-Tev-6His [a kind gift of Elmar Schiebel, ZMBH, University of Heidelberg, Germany]). Truncated versions of Cep164 and the CTNNBL1-Cep164-C fusion protein were generated using a PCR-based strategy. Point mutations in Cep164 and Rab8a were generated by site-directed mutagenesis. FLAG-Cep164-R1 is resistant to Cep164_1 siRNA and was generated as a rescue construct by introducing six point mutations without altering the amino acid sequence. All constructs were confirmed by DNA sequencing.
Generation of Cep164 polyclonal antibodies
Rabbit polyclonal antibodies were generated against amino acids 1–298 (anti-Cep164-N) and 665–873 (anti-Cep164-M) of human Cep164 (PSL GmbH, Heidelberg, Germany). Antigens were expressed as 6xHis-fusion proteins in Rosetta 2(DE3) and purified under native conditions. Antibodies were purified from preabsorbed serum by affinity purification using the immobilized antigen.
Antibodies
Primary antibodies used for indirect immunofluorescence were rabbit anti-Cep164 (residues 1–298; 1:1,000; Graser et al., 2007), rabbit anti-Cep164-N (1:500–1,000; this study), rabbit anti–Cep164-M (1:1,000; this study), rabbit anti-PCM1 (residues 665–2024; 1:150; a kind gift of Andreas Merdes, CNRS, Toulouse, France; Dammermann and Merdes, 2002), mouse GTU88 and rabbit anti–γ-tubulin (1:500; Sigma-Aldrich), mouse anti-acetylated tubulin C3B9 (1:100; Sigma-Aldrich), mouse anti-polyglutamylated tubulin GT335 (1:1,000; synthetic peptide mimicking the structure of polyglutamylated sites of α-tubulin was used as antigen; a kind gift of Carsten Janke, Institute Curie, Orsay/Paris, France; Wolff et al., 1992), rabbit anti-centriolin (residues 938–2711; 1:50; a kind gift of Elmar Schiebel, ZMBH, University of Heidelberg, Germany), mouse anti-NPHP1 (residues 12–205; 1:2; a kind gift of Hanswalter Zentgraf, DKFZ, Heidelberg; Liebau et al., 2011), rabbit anti-Odf2 (residues 41–266 of mouse Odf2; 1:100; a kind gift of Sachito Tsukita, Osaka University, Osaka, Japan; Ishikawa et al., 2005), rabbit anti-centrobin (1:500; Sigma-Aldrich), rabbit anti-ninein (1:200; Abcam), rat anti–α-tubulin (1:50; clone YL1/2; Sera Laboratory), rabbit anti-Ki67 (1:1,000; Novocastra Laboratories), goat anti-IFT88 (generated against the peptides EDDLYSGFNDYNPAY and DDFADEELGDDLLSE of mouse IFT88; 1:100; a kind gift of Joseph C. Besharse, Medical College of Wisconsin, Milwaukee, WI; Baker et al., 2003), rabbit anti-Cep123 (residues 1–230; 1:1,000, a kind gift of Michel Bornens; Sillibourne et al., 2011), mouse anti-FLAG M2 (1:1,000; Sigma-Aldrich), rabbit anti-FLAG (1:500; Sigma-Aldrich), rabbit anti-TGN46 (1:100; Novus Biologicals), mouse anti-GM130 (1:500; BD), and mouse anti-Golgin97 (1:800; Invitrogen). For Western blot analysis, rabbit anti-Cep164-N (1:400), rabbit anti-Cep164-M (1:400), rabbit anti-Cep164 (1:1,000; Graser et al., 2007), mouse anti-Plk1 (1:500; Santa Cruz Biotechnology, Inc.), mouse anti-FLAG M2 antibody (1:10,000; Sigma-Aldrich), mouse anti-NPHP1 (1:10), rabbit anti-Rab8a (1:250; Sigma-Aldrich), mouse anti-Actin (1:1,000; EMD Millipore), rabbit anti-GFP antibody (1:250; a kind gift of Elmar Schiebel), mouse anti-MBP antibody (1:10,000; New England Biolabs, Inc.), and mouse anti-His antibody (1:1,000; GE Healthcare) were used. Secondary antibodies for immunofluorescence analysis were coupled to Alexa Fluor 350, 488, or 546 dyes (Molecular Probes). Secondary antibodies for Western blot analysis were coupled to horseradish peroxidase (Dianova).
Cell culture and transfection
hTERT-RPE1 (RPE1) cells (a kind gift of Stephen Doxsey, University of Massachusetts, Worcester, MA) and RPE1 cells stably expressing LAP-CSAP (a kind gift of Iain Cheeseman, Whitehead Institute, Cambridge, MA; stable integration of LAP-CSAP after transduction with Moloney murine leukemia retrovirus using the plasmid pBABEblast; Backer et al., 2012) were grown in DMEM/Ham’s F12 medium supplemented with 10% FCS, 2 mM l-glutamine, and 0.348% sodium bicarbonate. HEK293T cells (a kind gift of Thomas Benzing, University of Cologne, Cologne, Germany) and Phoenix Eco cells were cultured in DMEM supplemented with 10% FCS. NIH 3T3 cells were grown in DMEM supplemented with 10% newborn calf serum. Cell lines were grown at 37°C under 5% CO2. HEK293T and Phoenix Eco cells were transiently transfected with plasmid DNA using the calcium phosphate precipitation method. HEK293T cells were harvested 32 h after transfection, after serum starvation for the last 5–8 h. RPE1 cells were transiently transfected with plasmid DNA using Fugene 6 (Roche) and fixed 24–96 h after transfection. Cilia formation in RPE1 cells was induced by serum withdrawal for 48 h. To observe early stages of ciliogenesis, cells were serum starved for a period of 18–24 h. Stable RPE1 cell lines were selected with 800 µg/ml G418 (Invitrogen) for 2 wk. Lentiviral stocks were prepared according to standard procedures. Transduced RPE1 cells were selected with 20 µg/ml blasticidin S (MP Biomedicals) for 2 wk and single clones were obtained by dilution.
Cell cycle–related analysis
Mitotic cells were identified by immunofluorescence microscopy based on their chromosome morphology (condensed chromosomes). Cells with noncondensed chromosomes and splitted centrioles were classified as in the S–G2 phase of the cell cycle.
To synchronize RPE1 cells, cells were incubated in 3 mM thymidine (Sigma-Aldrich) for 19 h to enrich for mitotic cells. Thymidine was washed out and cells were released in fresh medium. After 8 h, cells were again treated with 3 mM thymidine for 17 h. After release in fresh medium for 6 h, cells were arrested in 100 nM nocodazole (Sigma-Aldrich) for 4 h. Cells were harvested at the time points indicated after release from double thymidine block or double thymidine block followed by nocodazole treatment.
Microtubule regrowth assay
RPE1 cells were treated with 8 µM nocodazole (Sigma-Aldrich) at 37°C for 60–90 min before the drug was washed out to allow microtubules to regrow. Microtubule de- and repolymerization were visualized by immunofluorescence microscopy using antibodies against α-tubulin.
RNA interference
RPE1 cells were reverse transfected with 50 nM siRNA oligonucleotides using DharmaFECT3 (Thermo Fisher Scientific). Cells were fixed 48–72 h after transfection. The siRNA used in this study (purchased from Ambion) targeted the following sequences: human Cep164 siRNA1, 5′-CAGGTGACATTTACTATTTCA-3′ (Graser et al., 2007); Cep164 siRNA2, 5′-AAGAAGATACAGGAAGCTCAA-3′ (Sivasubramaniam et al., 2008); Cep164 siRNA3, 5′-ACCACTGGGAATAGAAGACAA-3′; and Cep164 siRNA4, 5′-GACCTTGAAACCAGAGCTAAA-3′. Cep164 siRNA3 and siRNA4 were kindly provided by Alwin Krämer (DKFZ, Heidelberg; Germany). Human NPHP-1 siRNA1, 5′-GGGAATCAATTTCGAGCAA-3′ and NPHP-1 siRNA2, 5′-CTGATGGTTGGTGGATAGCTA-3′. As negative controls, siRNA targeting Luciferase (5′-AACGTACGCGGAATACTTCGA-3′; Knödler et al., 2010), or nontargeting siRNA (5′-AATTCTCCGAACGTGTCACGT-3′) were used (Spektor et al., 2007).
Rescue experiments
For rescue experiments, 1–3 µg of a plasmid bearing FLAG-Cep164-R1 were transiently transfected in RPE1 cells. After 30 h, cells were transfected with Cep164 siRNA_1, which allows the knockdown of endogenous Cep164 but not FLAG-Cep164-R1. After 24 h (Fig. 4, C–E; Fig. 5, C and D) or 30 h (Fig. 8, C and D), cells were serum starved for the next 24 h (Fig. 4, C–E; Fig. 5, C and D) or 18 h (Fig. 8, C and D). Cells were subsequently fixed and processed for immunofluorescence analysis. Efficient depletion of endogenous Cep164 was confirmed by Western blot (Fig. 4 E) or signal intensity measurements (Fig. 8 D).
Immunofluorescence staining and microscopy
Cells were grown on coverslips, which were precoated with collagen A (Biochrom AG) for NIH 3T3 cells, and either fixed in ice-cold methanol for 3 min and permeabilized in ice-cold acetone for 20 s or fixed in 3% PFA for 20 min. Cilia staining with anti-acetylated tubulin included a previous cold treatment for 10 min (methanol fixation) or 25 min (PFA fixation) to depolymerize cytoplasmic microtubules. PFA fixation was followed by a 10-min quenching step in 50 mM NH4Cl and a 5-min permeabilization step in 0.1% Triton X-100. Cells were blocked with 0.4% BSA in 0.1% Triton X-100/PBS for 30 min at room temperature and incubated with primary antibodies at 37°C for 1 h. Primary antibodies were detected with secondary antibodies for 30–60 min at room temperature. DAPI (Sigma-Aldrich) was included with the secondary antibodies for DNA staining except when a triple staining was performed. All antibodies were diluted in blocking solution. Coverslips were mounted on glass slides in Mowiol (EMD Millipore). Images were acquired as z-stacks (0.3-µm steps spacing 3 µm) using a microscope (Axiophot; Carl Zeiss) equipped with a 63x NA 1.4 or 100x 1.45 NA Plan-Fluor oil immersion objective lens (Carl Zeiss), a charge-coupled device camera (Cascade 1K; Photometrics), and MetaMorph software (Universal Imaging Corp.). All images were taken at room temperature. GFP fusion proteins were visualized by the fluorescence signal. Color balance was adjusted to the same level in MetaMorph and image brightness and contrast were adjusted equally in Adobe Photoshop CS3. The z-plane of focus was selected for images. Figures were assembled in Illustrator CS3 (Adobe).
Fluorescence intensity measurement
Quantification of fluorescence intensity was performed using ImageJ software (National Institutes of Health, Bethesda, MD). The intensity of centrosomal signals was determined using the summed z-planes. An area of 36 square pixels was defined around the centrosome identified by γ-tubulin costaining. Background signals were measured in the near proximity of each centrosome and subtracted from the centrosomal measurement. Images from one dataset were acquired at the same day and equal exposure times were set between different samples. As mitotic cells contain two centrosomes, the signal intensities for both were measured. Only the measurement of the centrosome with the higher signal intensity was included in the final graph. The highest value of each experiment was set to one.
Cell lysis, immunoprecipitation, and Western blot
RPE1 cells were lysed in RIPA buffer (50 mM Tris-HCl, pH 8, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS, 1 mM DTT, 2 mM Na3VO4, 1 mM PMSF, and complete EDTA-free protease inhibitor cocktail [Roche]). Total extracts were incubated rotating at 4°C for 20 min and centrifuged at 21,000 g for 15 min. Protein concentrations were determined with Bradford reagent (Sigma-Aldrich). Immunoprecipitations were performed as described previously (Nachury, 2008). In brief, HEK293T cells were lysed in 1.4 ml lysis buffer containing 50 mM Hepes, pH 7.4, 300 mM KCl, 5 mM EDTA, 10% glycerol, 0.5 mM DTT, 2 mM Na3VO4, 1 mM PMSF, and complete EDTA-free protease inhibitor cocktail (Roche). Cell lysates were incubated rotating at 4°C with 0.32% NP-40 and 10 µg/ml cytochalasin D (Applichem) for 20 min. Total extracts were centrifuged at 21,000 g for 15 min. The supernatant was diluted in 700 µl LAP0 buffer containing 50 mM Hepes, pH 7.4, 5 mM EDTA, 10% glycerol, 0.05% NP-40, 2 mM Na3VO4, 1 mM PMSF, and complete EDTA-free protease inhibitor cocktail and further clarified by centrifugation at 27,000 g for 20 min. Protein concentrations were measured using Advanced Protein Assay 01 (ADV01) reagent (Cytoskeleton) and adjusted. FLAG-tagged proteins were immunoprecipitated using anti-FLAG M2 Agarose (Sigma-Aldrich) for 1–2 h. The agarose beads were washed three times in LAP200 buffer containing 50 mM Hepes, pH 7.4, 200 mM KCl, 5 mM EDTA, 10% glycerol, and 0.35–0.4% NP-40. Bound proteins were eluted in SDS sample buffer and resolved by SDS-PAGE. After transfer to a nitrocellulose or PVDF membrane, immunoreactive proteins were detected with the respective antibodies and visualized with ECL.
Recombinant proteins and in vitro binding assay
6His-Rabin8, NusA-Rab8a-6His (wild-type and mutant forms), and NusA-Rab11a-6His were expressed at 23°C for 4 h with 0.4 mM IPTG and purified with Ni-NTA-agarose (QIAGEN). MBP, MBP-Rabin8, and MBP-Cep164-C were expressed under the same conditions and purified with Amylose resin (New England Biolabs, Inc.). MBP fusion proteins (immobilized on beads) were incubated with the appropriate purified His fusion protein in binding buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 10 mM MgCl2, 0.5% TritonX-100, 1 mM DTT, and 0.5 mg BSA/ml) for 1.5–2 h at 4°C. After extensive washing in binding buffer lacking BSA, bound proteins were eluted with SDS sample buffer and analyzed by Western blot analysis.
Yeast two-hybrid screen
The yeast two-hybrid screening was performed using the Matchmaker two-hybrid system 3 and a human testis cDNA library (Takara Bio Inc.). The amino acids 1200–1460 of Cep164 (Cep164-C) were used as bait and fused to the GAL4 DNA-binding domain in vector pGBKT7 (Takara Bio Inc.). Yeast growth conditions and strain manipulations were performed as described previously (Sherman, 1991). Self-activation of Cep164-C was determined by cotransformation of pGBKT7-Cep164-C with pACT2 (Takara Bio Inc.). Saccharomyces cerevisiae strain AH109 (Takara Bio Inc.) carrying pGBKT7-Cep164-C as bait was transformed with the human testis cDNA library and selected on plates lacking leucine, tryptophan, and histidine. Growing colonies were restreaked on selective plates lacking leucine, tryptophan, histidine, and adenine and further analyzed for β-galactosidase activity. Plasmids of positive clones were isolated from yeast, bacterially amplified, retransformed in AH109, and retested for interaction with Cep164-C or pACT2 (self-activation). The identity of putative interactors was determined by BLAST sequence analysis after plasmid sequencing.
Transmission electron microscopy
RPE1 cells were grown on coverslips and fixed with 2.5% glutaraldehyde for 20 min starting at 30°C with a temperature decrease to 4°C. Glutaraldehyde was washed out with 50 mM cacodylate buffer. Cells were post-fixed in 2% OsO4 for 1–2 h at 4°C and incubated in 0.5% aqueous uranyl acetate overnight. Cells were dehydrated at 4°C using ascending ethanol concentrations and propylene oxide. The samples were incubated in a 1:1 mixture of Epon/propylene oxide for 2 h, infiltrated with Epon for 2 h, and polymerized at 60°C for 48 h. Serial ultrathin sections (70 nm) were stained with uranyl acetate and lead citrate and examined under a microscope (Philips EM 410; FEI Eindhofen) equipped with a digital camera (BioScan 792; Gatan) and DigitalMicrograph software (Gatan). Image brightness and contrast were adjusted in Adobe Photoshop CS3.
FACS analysis
Standard protocols were applied for staining of cells with propidium iodide (Sigma-Aldrich). Cells were subjected to flow cytometry by using a FACScan (BD) flow cytometer.
Statistical analysis and sampling size
Significance was determined using the χ2-test (Fig. 8 B; Fig. S5, B and C) or the two-tailed Student t test (all other experiments). P < 0.05 was considered statistically significant. ***, P < 0.001; **, P < 0.01; and *, P < 0.05. Box-and-whisker plots: boxes show the upper and lower quartiles (25–75%) with a line at the median and whiskers extend from the minimum to the maximum of all data.
The sampling size for quantifications shown is as follows: Fig. 1 B, 100 cells were scored per cell cycle phase. Fig. 1 D, >200 cells were scored per time point. Fig. 3 B, 150–200 cells were scored for each condition from two independent experiments. Fig. 3 C, >100 cells were scored for each condition. Fig. 3 E, 190–200 cells were scored for each condition from two independent experiments. Fig. 3 G, >300 cells for each condition from three independent experiments. Fig. 4 A, >240 cells were scored for each condition from two independent experiments. Fig. 4 D, 90–250 cells were scored for each condition from three independent experiments. Fig. 5, C–E, >50 cells were scored for each condition. Figs. 5, G and H, >60 cells were scored for each condition. Fig. 8 B, >100 cells from one representative experiment out of two were scored. Fig. 8 C, >170 cells were scored for each condition from two independent experiments. Fig. 8 D, >30 cells were quantified. Fig. 9: One representative experiment out of three is shown, 100–150 cells were scored per sample.
Online supplemental material
Fig. S1 shows the characterization of anti-Cep164 antibodies. Fig. S2 shows the localization of Cep164 during the cell cycle. Fig. S3 shows that Cep164 is dispensable for the centrosomal localization of ODF2, centriolin, and Cep123 (A–H) and confirms the specificity of the NPHP1 antibody (I–M). Fig. S4 shows that Golgi integrity and PCM1 localization are not perturbed in Cep164-depleted cells. Fig. S5 provides the statistical analysis for the experiment shown in Fig. 9.
Acknowledgments
This paper is dedicated to the memory of Hanswalter Zentgraf.
We thank Alwin Krämer, Ingrid Hoffmann, Oliver Gruss, Iain Cheeseman, Elmar Schiebel, Andreas Merdes, Sachiko Tsukita, Carsten Janke, Stephen Doxsey, Michel Bornens, Joseph C. Besharse, and Thomas Benzing for reagents; Astrid Hofmann and members of the Pereira laboratory for excellent technical assistance; Marc Arnold and Ingrid Hoffmann for support with the yeast two-hybrid system; Karsten Richter for support with electron microscopy; Simon Anderhub, Francesca Fabretti, and Bernhard Schermer for technical advice; Franz Meitinger for help with yeast manipulations; Ayse Koca-Caydasi for help with statistics; Ana Martin-Villalba for help with stable cell lines; and Elmar Schiebel, Iain Hagan, Anne Spang, Kerry Tucker, Alwin Krämer, and members of the Pereira laboratory for critical comments on the manuscript.
K.N. Schmidt and S. Kuhns were funded by the Helmholtz Association grant HZ-NG-111 and are members of the Helmholtz International Graduate School for Cancer Research (HIGS). This work was funded by grant HZ-NG-111 (to G. Pereira).