The bacterial ATPase SecA and protein channel complex SecYEG form the core of an essential protein translocation machinery. The nature of the conformational changes induced by each stage of the hydrolytic cycle of ATP and how they are coupled to protein translocation are not well understood. The structure of the SecA–SecYEG complex revealed a 2-helix-finger (2HF) of SecA in an ideal position to contact the substrate protein and push it through the membrane. Surprisingly, immobilization of this finger at the edge of the protein channel had no effect on translocation, whereas its imposition inside the channel blocked transport. This analysis resolves the stoichiometry of the active complex, demonstrating that after the initiation process translocation requires only one copy each of SecA and SecYEG. The results also have important implications on the mechanism of energy transduction and the power stroke driving transport. Evidently, the 2HF is not a highly mobile transducing element of polypeptide translocation.
Introduction
Most secretory proteins are transported outside cells via the Sec complex. In bacteria, the process begins by preprotein recognition and engagement at the plasma membrane by the motor ATPase SecA, peripherally associated with the SecYEG protein channel complex (Brundage et al., 1990; Lill et al., 1990). Its initiation is thought to involve the dissociation of solution-state SecA dimers (Or et al., 2002; Woodbury et al., 2002), followed by the docking of the monomeric form onto SecYEG. The translocation of preprotein then proceeds through a channel enclosed by a single copy of SecYEG (Van den Berg et al., 2004; Cannon et al., 2005). A back-to-back dimeric arrangement of SecYEG has been observed in the membrane (Breyton et al., 2002), which is thought to be required for the binding and activation of SecA (Osborne and Rapoport, 2007; Deville et al., 2011; Dalal et al., 2012). However, the second passive nontranslocating copy of SecYEG is not necessarily fixed or essential for transport (Park and Rapoport, 2012).
The protein channel, formed in the center of SecY between the pseudo-symmetric halves of trans-membrane segments (TMS) 1–5 and 6–10, is kept closed by a central ring of hydrophobic amino acid side chains and a short helix, or plug inserted from the periplasmic side (Van den Berg et al., 2004; Park and Rapoport, 2011). The structure of membrane-bound SecYEG determined with a shortened preprotein shows that the signal sequence acts on a single copy of the dimeric complex, at the lateral gate formed between the two halves of SecY (Hizlan et al., 2012). The resultant conformational change of the adjacent TMS 2b and 7 serve to displace the plug and thereby unlock the complex (Tam et al., 2005; Robson et al., 2009a). Channel opening is presumably then achieved upon separation of the two halves, like a crab’s claw (Van den Berg et al., 2004; Egea and Stroud, 2010), allowing the passage of polypeptide either through or into the membrane.
SecA alone has been crystallized in monomeric and dimeric states (Hunt et al., 2002; Osborne et al., 2004). In the SecA–SecYEG complex, there is one copy of each (Fig. 1 A; Zimmer et al., 2008); the nontranslocating passive copy having been removed by exposure to detergents during the purification procedure (Deville et al., 2011). The structure of SecA reveals a binding site for ATP at the interface between two nucleotide-binding domains (NBD1 and NBD2; Hunt et al., 2002) and one for signal sequence, between the preprotein cross-linking domain (PPXD) and the helical wing domain (HWD; Fig. 1 A; Gelis et al., 2007).
Interaction of the 2HF of SecA with SecYEG. (A) Structure of T. maritima SecA bound to SecYEG viewed from the side facing the lateral gate with the α-helices shown as cylinders (Zimmer et al., 2008); SecY (white), SecE (gray), SecG (dark gray) and SecA: 2HF (yellow), PPXD (green), HWD (red), NBD1 (pale blue), and NBD2 (pink). Key residues (corresponding to E. coli numbering) have also been highlighted: SecYK268 (purple space fill), SecYI183 (cyan space fill), SecAY794 (red space fill), and SecAA795 (orange space fill). Residues shown to be in close proximity to the translocating preprotein are shown for SecA (blue sticks; Bauer and Rapoport, 2009) and SecY (magenta sticks; Cannon et al., 2005). (B) Color coding and numbering as in A. Close-up showing the cross-linking site between SecYK268C (Y-268) and both SecAY794C (A-794) and SecAA795C (A-795). The alternative cross-linking site in SecY is also shown (Y-183). The pathway for the preprotein is shown by a dashed line, on the basis of known cross-links to SecA and SecY. (C) Color coding and numbering as in A. Equivalent view as B of a molecular model based on the same SecA–SecYEG structure (Zimmer et al., 2008) constrained by a cross-link between the SecYI183C (cyan space fill) and SecAA795C (orange space fill). The blocked pathway for the preprotein is shown by the dashed line.
Interaction of the 2HF of SecA with SecYEG. (A) Structure of T. maritima SecA bound to SecYEG viewed from the side facing the lateral gate with the α-helices shown as cylinders (Zimmer et al., 2008); SecY (white), SecE (gray), SecG (dark gray) and SecA: 2HF (yellow), PPXD (green), HWD (red), NBD1 (pale blue), and NBD2 (pink). Key residues (corresponding to E. coli numbering) have also been highlighted: SecYK268 (purple space fill), SecYI183 (cyan space fill), SecAY794 (red space fill), and SecAA795 (orange space fill). Residues shown to be in close proximity to the translocating preprotein are shown for SecA (blue sticks; Bauer and Rapoport, 2009) and SecY (magenta sticks; Cannon et al., 2005). (B) Color coding and numbering as in A. Close-up showing the cross-linking site between SecYK268C (Y-268) and both SecAY794C (A-794) and SecAA795C (A-795). The alternative cross-linking site in SecY is also shown (Y-183). The pathway for the preprotein is shown by a dashed line, on the basis of known cross-links to SecA and SecY. (C) Color coding and numbering as in A. Equivalent view as B of a molecular model based on the same SecA–SecYEG structure (Zimmer et al., 2008) constrained by a cross-link between the SecYI183C (cyan space fill) and SecAA795C (orange space fill). The blocked pathway for the preprotein is shown by the dashed line.
The pathway for translocation has been defined by cross-linking the preprotein to SecA (Bauer and Rapoport, 2009) and SecYEG (Fig. 1 A; Cannon et al., 2005); several interesting features of the SecA–SecYEG structure relate to the substrate-binding site. The PPXD swings away from the HWD toward NBD2, a conformational change that would effectively release the bound signal sequence and form a “clamp” for translocating polypeptide, thus creating a continuous channel together with the pore through SecYEG (Gelis et al., 2007; Zimmer et al., 2008; Zimmer and Rapoport, 2009; Fig. 1 A, blue residues in SecA and magenta in SecY).
Where the two channels meet, the 2HF of the SecA helical scaffold domain (HSD) pokes into the opening in SecYEG, ideally placed to make contact with the translocating preprotein. Cross-linking experiments show that it does indeed come into close proximity with the substrate in transit (Erlandson et al., 2008a; Bauer and Rapoport, 2009). It has been proposed that translocation is achieved by coordinated ATP-dependent conformational changes of the clamp and the 2HF, whereby the polypeptide is pushed through the membrane by a direct contact with the conserved tyrosine-794 (Escherichia coli numbering) at the fingertip (Erlandson et al., 2008a; Zimmer et al., 2008).
We set out to investigate the nature and timing of the conformational changes at the interface between SecA and SecYEG, as well as by the PPXD and clamp of SecA in an associated study (Gold et al., 2012). Here, the dynamic role of the 2HF of SecA was monitored by a fluorescent reporter at the entrance of the protein channel (Robson et al., 2007), as well as by the analysis of the consequences of its immobilization at this site (Robson et al., 2009b). The immobilization of the 2HF at the native binding site at the channel entrance of SecY had very little effect on the ability of SecA to drive translocation through the membrane. The results also resolve an ongoing debate on the stoichiometry of the active translocon.
Results
The binding of ATP to SecA promotes the insertion of the 2HF into the protein channel
Initial experiments were established to monitor the interaction of the 2HF with the SecYEG complex, and to determine the effects of the incorporation of cysteine mutants at the tip (E. coli sites Y794 and A795). The labeling of SecYEG with a fluorescein at the entrance to the channel at a unique cysteine at position 268 (designated SecY268FlEG; Fig. 1, purple residue) preserves the activity and monitors a long-range conformational change in the SecA–SecYEG complex (Robson et al., 2007). The binding of a nonhydrolyzable analogue of ATP results in a conformational change reported by a quench in fluorescence 45 Å away (Fig. 2 A). The effect is due to the juxtaposition of the 2HF of SecA against the cytoplasmic loop between TMS 6 and 7 of SecY. In the structure of the ATP-bound configuration of SecA–SecYEG, SecAY794 directly contacts SecYK268 (Fig. 1, A and B, red and purple residues, respectively; Zimmer et al., 2008).
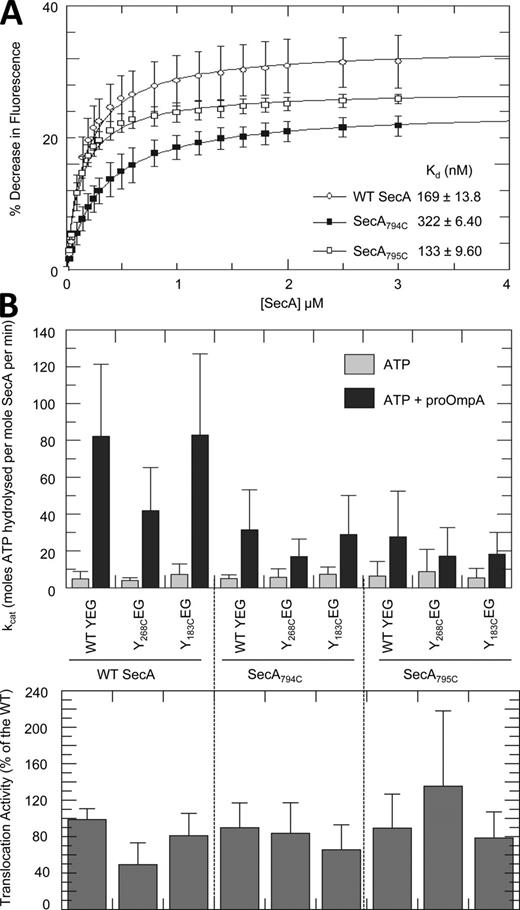
Activity of the wild-type and mutant forms of SecA. (A) Equilibrium titration of wild-type and mutant SecA into 29 nM SecY268FlEG in the presence of 1 mM AMPPNP. Data were fitted to a binding equation (Eq. 1) giving the Kd values shown. The results are the average of three independent experiments. (B) (Top) steady-state ATPase activity of 0.3 µM wild-type and mutant SecA in the presence of 0.3 µM wild-type or mutant SecYEG proteoliposomes, measured with and without 1 µM proOmpA. (Bottom) relative levels of translocation activity achieved in each case analyzed by anti-proOmpA immunoblot and quantification of translocation activity using ImageJ software; n = 3. See Fig. S1 for a representative blot and time dependency of this experiment.
Activity of the wild-type and mutant forms of SecA. (A) Equilibrium titration of wild-type and mutant SecA into 29 nM SecY268FlEG in the presence of 1 mM AMPPNP. Data were fitted to a binding equation (Eq. 1) giving the Kd values shown. The results are the average of three independent experiments. (B) (Top) steady-state ATPase activity of 0.3 µM wild-type and mutant SecA in the presence of 0.3 µM wild-type or mutant SecYEG proteoliposomes, measured with and without 1 µM proOmpA. (Bottom) relative levels of translocation activity achieved in each case analyzed by anti-proOmpA immunoblot and quantification of translocation activity using ImageJ software; n = 3. See Fig. S1 for a representative blot and time dependency of this experiment.
The fluorescence change of SecY268FlEG was measured as a function of SecA concentration to determine an affinity for SecYEG, with the finger in the “inserted” conformation, of ∼170 nM (Fig. 2 A). The apparent affinity of SecYEG for SecA in the ADP-bound state is appreciably less (∼500 nM; Robson et al., 2009b), indicative of a loosening of the SecA–SecYEG complex at that stage of the hydrolysis cycle.
Next, the consequences of single cysteine mutants at the tip of the 2HF were assessed upon the interaction with SecYEG. The affinity of the ATP-bound configuration of SecAA795C for SecYEG, determined by the extrinsic fluorescent probe, was unchanged, whereas substitution of the tyrosine-794 resulted in a modest (twofold) reduction in affinity compared with the wild type (Fig. 2 A).
To examine the consequence of the weakened SecA–SecYEG interaction, the mutants were challenged in ATP-driven protein translocation. The addition of preprotein (proOmpA) to SecA usually stimulates the ATPase activity required to drive translocation into the interior of phospholipid vesicles reconstituted with SecYEG (Fig. 2 B; Robson et al., 2009b). The level of translocation-stimulated ATPase activity was about the same for SecAY794C and SecAA795C, slightly less than 50% of the wild type (Fig. 2 B). This was true for wild-type SecYEG, and for all the SecYEG mutant complexes used in the study, with single cysteine residues at either the entrance to the channel or deeper within it (respectively, SecYK268CEG or SecYI183CEG, shown in Fig. 1 B). In all cases, the hydrolysis of ATP was successfully coupled to protein transport (Fig. 2 B). Translocation time-course experiments also show that the rates attained by the SecAY794C and SecAA795C mutants were comparable to wild-type SecA (Fig. S1 A). Therefore, tyrosine-794 contributes to the binding site for SecYEG and is required for a high-affinity contact, but not for effective protein translocation.
Generation and purification of specific cross-linked single-copy complexes of SecA and SecYEG
To test the hypothesis that the 2HF contacts and actively pushes the preprotein across the membrane (Erlandson et al., 2008b; Zimmer et al., 2008), we decided to engineer forms of the SecA–YEG complex with the finger immobilized by a disulphide bond to the SecY complex. On the basis of the close proximity of the 2HF tip to the protein channel entrance (Fig. 1, A and B), either SecAY794C or SecAA795C was selected for cross-linking to SecYK268C. Inner membranes from E. coli overexpressing SecYK268C were incubated in turn with the SecA single-cysteine mutants and AMPPNP (an ATP analogue that promotes 2HF insertion into SecY) before oxidation with copper phenanthroline. The membranes were subsequently re-isolated and solubilized with detergent and the resultant extract was subject to nickel chelating and ion exchange and gel filtration chromatography. In both cases cross-linked complexes (hereafter referred to as SecA794/795–SecY268EG) could be isolated in sufficient quantities and purity for further characterization (Fig. 3, A and C).
Purification of cross-linked SecA–SecYEG complexes. Samples of purified SecYEG, SecA, and their cross-linked products were analyzed by (A and B) SDS-PAGE: A794–Y268EG (SecAY794C cross-linked to SecYK268CEG), and similarly, A795–Y268EG, A794–Y183EG, and A795–Y183EG. The SecYEG and SecA samples were loaded purely as markers and the quantities loaded have no relation to the amounts used in the cross-linking experiment (see Materials and methods for further details). Molecular weight standards are shown on the left side of each panel. (C) Size exclusion chromatography: SecY183CEG (YEG), SecA794C (A) and A795–Y268EG (A-YEG), and a dimer of SecYEG produced by a genetic fusion of two SecY subunits (Y-Y; Duong, 2003). See Fig. S2 for analysis of the eluting A795-Y268EG fractions by SDS-PAGE. Again, the SecYEG, SecA, and SecY-Y samples were loaded in order to calibrate the column and their quantities have no relation to the cross-linked samples. Note that the mutant forms of SecA and SecYEG used elute from the column like their wild-type counterparts.
Purification of cross-linked SecA–SecYEG complexes. Samples of purified SecYEG, SecA, and their cross-linked products were analyzed by (A and B) SDS-PAGE: A794–Y268EG (SecAY794C cross-linked to SecYK268CEG), and similarly, A795–Y268EG, A794–Y183EG, and A795–Y183EG. The SecYEG and SecA samples were loaded purely as markers and the quantities loaded have no relation to the amounts used in the cross-linking experiment (see Materials and methods for further details). Molecular weight standards are shown on the left side of each panel. (C) Size exclusion chromatography: SecY183CEG (YEG), SecA794C (A) and A795–Y268EG (A-YEG), and a dimer of SecYEG produced by a genetic fusion of two SecY subunits (Y-Y; Duong, 2003). See Fig. S2 for analysis of the eluting A795-Y268EG fractions by SDS-PAGE. Again, the SecYEG, SecA, and SecY-Y samples were loaded in order to calibrate the column and their quantities have no relation to the cross-linked samples. Note that the mutant forms of SecA and SecYEG used elute from the column like their wild-type counterparts.
The SecA795–SecY268EG preparation was cleaner than the SecA794–SecY268EG sample, which contained detectable quantities of the uncross-linked components (Fig. 3 A, see also Fig. 6 A). The uncross-linked SecA and SecYEG in the latter most likely results from their natural tendency to homodimerize, and hence associate with the respective component of the SecA–SecYEG complex. The initial encounter presumably occurs between SecA monomers and SecYEG dimers (Deville et al., 2011), wherein the cross-link forms between a single SecA and SecY subunit. The passive copy of SecYEG is then entirely (SecA795–SecY268EG) or partially (SecA794–SecY268E) removed upon exposure to detergent and the depletion of lipids such as cardiolipin (Bessonneau et al., 2002; Gold et al., 2010).
Similar experiments were performed in order to fix the 2HF deeper inside the channel (SecYI183C; Fig. 1 C). The SecA794–Y183EG and SecA795–Y183EG complexes could also be cross-linked. However, the efficiency was reduced and the preparations were not quite as pure (Fig. 3 B), most likely due to the greater distances between the sites (Fig. 1; Zimmer et al., 2008).
The aggregation seen at the void volume of the cross-linked samples (shown for SecA795–SecY268EG in Fig. 3 C) probably results from the exposure of the membrane lipids and proteins to the oxidizing agent. However, these aggregates were removed by the gel filtration step and did not interfere with the subsequent functional analysis.
The basal ATPase activity of SecA is dramatically stimulated when the 2HF is permanently fixed within SecY
The ATPase activity of SecA was measured in order to monitor a functional interaction with the SecY complex. The low basal ATPase activity of SecA (kcat ∼1 min−1; Fig. 4) is stimulated by lipids, SecYEG, and especially during protein translocation (Lill et al., 1990; Robson et al., 2009b). This basal activity remained low for SecAY794C (kcat ∼0.9 min−1), and was increased somewhat in SecAA795C (kcat ∼4.7 min−1). The stimulation of this activity by SecYEG in detergent solution is dependent on the presence of lipids, preferably cardiolipin (Gold et al., 2010). Therefore, in this case when SecYEG is stripped of lipids by detergent extraction the stimulation of the SecA ATPase is marginal. These results were reproduced here for the wild type (1.7-fold increase), SecAY794C (1.3-fold) and SecAA795C (1.3-fold; Fig. 4). We were therefore surprised that the activity of the SecA794–Y268EG and SecA795–Y268EG cross-linked complex in detergent were both activated in the absence of lipids by a factor of ∼20 (Fig. 4).
Analysis of ATPase activity of SecA–SecYEG cross-linked complexes in detergent solution. Steady-state ATPase activities were measured in TKM buffer (plus 0.03% C12E9) containing 0.3 µM SecA in the absence or presence of saturating (1 µM) SecYK268CEG. The cross-linked SecA–SecYEG complexes (A794–Y268EG or A795–Y268EG) were analyzed on their own. The results were averaged from three independent experiments. The inset zooms in on the much lower level uncross-linked enzyme.
Analysis of ATPase activity of SecA–SecYEG cross-linked complexes in detergent solution. Steady-state ATPase activities were measured in TKM buffer (plus 0.03% C12E9) containing 0.3 µM SecA in the absence or presence of saturating (1 µM) SecYK268CEG. The cross-linked SecA–SecYEG complexes (A794–Y268EG or A795–Y268EG) were analyzed on their own. The results were averaged from three independent experiments. The inset zooms in on the much lower level uncross-linked enzyme.
Consequences of the immobilization of the 2HF on protein translocation
Experiments were conducted in order to assess the transport capability of the cross-linked complexes. The SecA–SecYEG complexes with the 2HF cross-linked at SecY268 at the edge of the protein channel (Figs. 1 A and 3 A) were reconstituted into proteoliposomes and tested for their ability to translocate preprotein. Remarkably, both SecA794–Y268EG and SecA795–Y268EG were fully capable of translocation-associated ATPase activity and transport of proOmpA (Fig. 5 A). The ATPase activity was diminished when SecAY794C was used, but the translocation efficiency was unaffected. The SecA795–Y268EG was more prone to higher levels of uncoupled ATPase activity, as was the case in detergent solution (Fig. 4).
Analysis of ATP-driven translocation activity of reconstituted cross-linked SecA–SecYEG complexes. (A) (Top) steady-state ATPase activity of 0.3 µM SecA, SecAY794C, or SecAA795C in the presence of saturating (1 µM) membrane reconstituted SecYK268CEG, compared with proteoliposomes containing only the cross-linked complexes (A794–Y268EG and A795–Y268EG); in each case with or without 0.7 µM proOmpA. The results are the average of three independent experiments. (Bottom) corresponding levels of translocation activity analyzed by anti-proOmpA immunoblot and compared with wild-type levels of translocation. The results were quantified using ImageJ software; n = 3 (bottom). See Fig. S2, A and B for representative blots and time courses of the translocation assays. (B) As in A but using SecY183. See Fig. S2 C for a representative blot. The ATPase results shown are the average of five independent experiments.
Analysis of ATP-driven translocation activity of reconstituted cross-linked SecA–SecYEG complexes. (A) (Top) steady-state ATPase activity of 0.3 µM SecA, SecAY794C, or SecAA795C in the presence of saturating (1 µM) membrane reconstituted SecYK268CEG, compared with proteoliposomes containing only the cross-linked complexes (A794–Y268EG and A795–Y268EG); in each case with or without 0.7 µM proOmpA. The results are the average of three independent experiments. (Bottom) corresponding levels of translocation activity analyzed by anti-proOmpA immunoblot and compared with wild-type levels of translocation. The results were quantified using ImageJ software; n = 3 (bottom). See Fig. S2, A and B for representative blots and time courses of the translocation assays. (B) As in A but using SecY183. See Fig. S2 C for a representative blot. The ATPase results shown are the average of five independent experiments.
The same experiments were conducted with the finger covalently linked to a position deep inside the channel (Figs. 1 C and 3 B). The ATPase activities of the cross-linked SecA794–Y183EG and SecA795–Y183EG complexes without transport substrate were unaffected; however, the response of ATP turnover to preprotein and ability to transport was severely compromised (Fig. 5 B).
Verification of the cross-link stability and the immobilized state of the 2HF
The mobility (or immobility) of the 2HF of SecA has important implications for the molecular mechanism of protein translocation. Therefore, it was important to verify that the cross-link remained intact throughout the translocation reaction, and that its presence brought about the expected immobilization of the 2HF.
Exposure of the cross-linked complex to reducing agent (DTT) resulted in the release of SecA from SecYEG (Fig. 6 A). The uncross-linked material contained trace amounts of SecA (<10% of the form cross-linked to SecY) and no detectable amounts SecY (Fig. 6 A). Obviously, the conditions used in the translocation assay were nonreducing in order to preserve the cross-link. Analysis of the assay mixture containing the SecA795–Y268EG complex after translocation, by Western blotting, demonstrated that this was indeed the case (Fig. 6 B). Comparison with the standard translocation mixture containing uncross-linked SecYEG confirms the levels of free SecYEG were undetectable. Therefore, translocation must have been driven through the complex with the 2HF cross-linked to the cytosolic surface of SecY.
Stability of the disulphide bond formed within the SecA795–Y268EG complex. (A) Before translocation, the purified SecA795–Y268EG complex (2 µg), untreated or reduced with 50 mM DTT, was analyzed by SDS-PAGE and stained with Coomassie brilliant blue dye. SecYEG is used as a marker on the left, the quantities of which have no relation to those used in the cross-linking experiment. (B) Equivalent quantities of spent translocation reactions (before proteinase K treatment) containing the cross-linked (SecA795–Y268EG; 1 µg) or uncross-linked (wild-type SecYEG; 0.43 µg) complexes were analyzed by SDS-PAGE and Western blotting for SecY. The lower molecular weight form of SecY (SecY′) in the latter sample is a result of a well known C-terminal cleavage product (Brundage et al., 1990).
Stability of the disulphide bond formed within the SecA795–Y268EG complex. (A) Before translocation, the purified SecA795–Y268EG complex (2 µg), untreated or reduced with 50 mM DTT, was analyzed by SDS-PAGE and stained with Coomassie brilliant blue dye. SecYEG is used as a marker on the left, the quantities of which have no relation to those used in the cross-linking experiment. (B) Equivalent quantities of spent translocation reactions (before proteinase K treatment) containing the cross-linked (SecA795–Y268EG; 1 µg) or uncross-linked (wild-type SecYEG; 0.43 µg) complexes were analyzed by SDS-PAGE and Western blotting for SecY. The lower molecular weight form of SecY (SecY′) in the latter sample is a result of a well known C-terminal cleavage product (Brundage et al., 1990).
To test the assumption that a disulphide bond at this position would be expected to immobilize or severely restrict the mobility of the 2HF within the complex, molecular dynamics (MD) simulations were run of a model of the E. coli translocon (Deville et al., 2011) embedded in a POPC membrane, with and without a disulfide at position SecA795–Y268EG. The simulations were analyzed by measuring the distance between the centers of mass of the two groups of atoms comprising the eight residues around positions SecA795 and Y268EG, respectively. Fig. 7 shows how this distance lengthens from ∼6.5 Å to 7.5 Å at 10 ns in the disulphide bridged complex and then remains between 7 and 8 Å for the rest of the 0.1 µs. In contrast, this distance in the wild type undergoes repeated fluctuations between 7 and 9 Å during the simulation. These results suggest that the inherent mobility of the 2HF is indeed reduced by the cross-link and there is no indication that the disulphide would destabilize this area or promote the mobility of the 2HF. Given also that the cross-link is buried deep in the interior of the SecA–YEG structure, it is difficult to imagine under these circumstances how the 2HF could move very far.
Molecular dynamics simulations of the wild-type SecA–SecYEG and cross-linked SecA795–Y268EG complexes. Distances are between the centers of mass of the atoms comprising the groups of residues 791–799 (SecA) and 264–272 (SecY), respectively. Red is the wild-type (average: 7.81 ± 0.56 Å) and blue is SecA795–Y268EG cross-linked (average: 7.51 ± 0.28 Å). These statistical data were obtained from the single simulations shown here, which each represent an accumulation of force calculations for ∼570,000 atoms (50 million times for each trajectory).
Molecular dynamics simulations of the wild-type SecA–SecYEG and cross-linked SecA795–Y268EG complexes. Distances are between the centers of mass of the atoms comprising the groups of residues 791–799 (SecA) and 264–272 (SecY), respectively. Red is the wild-type (average: 7.81 ± 0.56 Å) and blue is SecA795–Y268EG cross-linked (average: 7.51 ± 0.28 Å). These statistical data were obtained from the single simulations shown here, which each represent an accumulation of force calculations for ∼570,000 atoms (50 million times for each trajectory).
Protein translocation requires only one copy each of SecA and SecYEG
Next, the oligomeric state of the cross-linked SecA–SecYEG complex was confirmed in order to resolve the stoichiometry of the active complex. The SecA795–SecY268EG complex eluted from a size exclusion column with, as expected, an average apparent molecular weight higher than both SecYEG and SecA alone, but smaller than the dimer of SecYEG produced by a genetic fusion of two SecYs (Fig. 3 C; Duong, 2003). Therefore, the complex, at least in solution, most likely only contains one copy of SecYEG. The complex can only contain one SecA, as a second uncross-linked copy would be have been identified by SDS-PAGE (Figs. 3 A and 6 A). A complex of one SecA and one SecYEG is consistent with the x-ray structure of the complex (Zimmer et al., 2008), where the second copy of SecYEG has been lost from the complex during exposure to detergent during the purification (Deville et al., 2011).
To prove that this single-copy complex was the active species we needed to show that the complex did not form higher order oligomers upon membrane incorporation; for example, by formation of SecA795–SecY268EG dimers about the back-to-back membrane interface of SecYEG (Fig. S3; Breyton et al., 2002; Deville et al., 2011). Therefore, the reconstituted vesicles were subject to nonspecific cross-linking. Previously, photo-induced cross-linking of unmodified proteins (PICUP) has proved useful in the verification of the dimeric membrane-bound state of SecYEG (Deville et al., 2011).
Treatment of proteoliposomes containing uncross-linked SecYEG, followed by analysis by SDS-PAGE and Western blotting revealed the expected presence of dimers (SecY-Y cross-links) in addition to higher aggregated states (Fig. 8, left-hand side). In the case of vesicles containing the disulphide-linked SecA–SecYEG complex no dimers could be detected (Fig. 8, right-hand side). The PICUP cross-linking reaction was quenched with DTT, which broke the disulphide linking SecA to SecY; the released SecY was entirely monomeric. The time-dependent increase in DTT resistant SecA-Y was presumably the result of PICUP cross-links between SecA and SecY, preserving the connection even after breaking the disulphide bond between SecA795 and SecY268. Therefore, in contrast to the SecYEG alone, the disulphide-linked SecA795–SecY268EG form does not form dimers in the membrane. This demonstration defines the single-copy stoichiometry of SecA and SecYEG in the translocating complex.
Nonspecific cross-linking of membrane-bound wild-type SecYEG and SecA795–Y268EG. Proteoliposomes containing SecYEG or the cross-linked complex (A795–Y268EG) were cross-linked using Tris-bipyridylruthenium and exposure to light, as indicated in seconds (see Materials and methods), followed by treatment with DTT to quench the reaction and break the disulphide bond. Proteins were separated by SDS-PAGE and visualized by anti-SecY immunoblot. u/c refers to protein controls where no cross-linker was added and SecY-Y to a genetically fused SecY dimer (Duong, 2003). Bands corresponding to uncross-linked SecY, to two conjoined or cross-linked SecY subunits (Y-Y), and specifically/nonspecifically cross-linked SecA-SecY (A-Y) are shown.
Nonspecific cross-linking of membrane-bound wild-type SecYEG and SecA795–Y268EG. Proteoliposomes containing SecYEG or the cross-linked complex (A795–Y268EG) were cross-linked using Tris-bipyridylruthenium and exposure to light, as indicated in seconds (see Materials and methods), followed by treatment with DTT to quench the reaction and break the disulphide bond. Proteins were separated by SDS-PAGE and visualized by anti-SecY immunoblot. u/c refers to protein controls where no cross-linker was added and SecY-Y to a genetically fused SecY dimer (Duong, 2003). Bands corresponding to uncross-linked SecY, to two conjoined or cross-linked SecY subunits (Y-Y), and specifically/nonspecifically cross-linked SecA-SecY (A-Y) are shown.
Discussion
The experiments presented here address the dynamic mechanism of protein translocation through the SecYEG complex. It has been proposed that both the 2HF and the PPXD cooperate to push preproteins through the membrane (Erlandson et al., 2008a; Zimmer et al., 2008). We have addressed this hypothesis here and elsewhere by using intra- and intermolecular disulphide bonds to immobilize the 2HF (this paper) and the PPXD (Gold et al., 2012) and assess their dynamic roles in protein transport.
Here, the experiments have relied on single cysteine mutants in SecY and SecA. Replacement of the tyrosine-794 at the end of the 2HF of SecA causes a reduction in the binding affinity of SecYEG, but retains activity. This decrease in affinity may explain the loss of activity of mutants at this position, and elsewhere on the fingertip, observed previously (Erlandson et al., 2008a). The addition of higher concentrations of SecA in our transport assay (300 nM) compared with the previous reports (200 nM; Erlandson et al., 2008a), together with other differences in conditions (such as temperature) may be the cause of this apparent discrepancy. We found it necessary to include 10 mM DTT to prevent covalent dimerization of SecAY794C, which may also account for the inactivation seen previously at 1 mM DTT (Erlandson et al., 2008a).
The demonstrated ability of cross-linked single copies each of SecA and SecYEG to drive protein translocation has several important implications for the mechanism. The stoichiometry of the components that make up the active translocon continues to be highly contentious. Therefore, the results provide a timely and definitive resolution of this problem. The likelihood that the cross-linked SecA–SecYEG complex forms higher-order oligomeric (e.g. dimeric) assemblies in the membrane is fairly implausible (Fig. S3), and was verified not to occur experimentally (Fig. 6). Therefore, protein translocation requires only one copy of each, and the second respective copies do not have a direct role in the transmission of protein across the membrane. SecA dimers are required to maintain a low basal rate of ATP turnover in the resting state (Gold et al., 2007), and its dissociation is critical for the activation of the enzyme and preprotein binding (Or et al., 2002; Duong, 2003; Gold et al., 2012).
The single copy cross-linked complex is activated (increased ATPase activity) due to the transposition of the PPXD toward the NBDs upon contact of SecA with SecYEG (Zimmer et al., 2008; Gold et al., 2012), and thereby fully capable of translocation. However, in the normal situation, in the absence of the activating cross-link, the initiation process probably requires the assistance of second copies of both components. SecA, for the transfer of preprotein to the channel (Gold et al., 2012) and SecYEG, for a high affinity association and activation of SecA (Deville et al., 2011; Dalal et al., 2012). Once translocation is underway only single copies of each are required. An in vivo analysis of the SecYEG shows that the dimeric interface is somewhat plastic (Park and Rapoport, 2012). Therefore, the passive second complex may be highly dynamic and is not absolutely essential for secretion. It may even be replaced by alternative facilitators of secretion or membrane protein insertion, such as SecDF and YidC.
The 2HF of SecA could be cross-linked to alternative positions in the protein channel. When fixed at the usual position at the edge of the channel (Zimmer et al., 2008), there were no adverse effects on activity. MD simulations, as expected, show that the disulphide bond restricts the mobility in this region. Given the imposition of a covalent tether at the tip of the 2HF, it seems unlikely that the 2HF could move sufficiently to push stretches of polypeptide across the membrane. Immobilizing the finger to an alternative location deeper inside the channel, in contrast, results in loss of function, presumably by blocking the polypeptide path (Fig. 1 C).
On the basis of its proximity with the preprotein and location near the translocation channel, the 2HF may have an alternative role in transport; perhaps it helps to hold the channel in a secretion-compatible conformation. The insertion of the finger may prize open the channel and prime it for translocation; for example, the opening of a “window” in the lateral gate (Zimmer et al., 2008) or the partitioning of the two halves of SecY (Egea and Stroud, 2010) may be stabilized by this incursion. Alternatively, the finger may act to prevent backsliding of the translocating polypeptide in the ATP-bound configuration (Erlandson et al., 2008b).
If the 2HF is not the direct driver of protein translocation, then what is? The PPXD is also in the vicinity of the translocating substrate, and on this basis has been implicated with a mobile role required for the activation of the enzyme (Ding et al., 2003; Cannon et al., 2005; Karamanou et al., 2007; Zimmer et al., 2008; Bauer and Rapoport, 2009; Zimmer and Rapoport, 2009). However, a related study shows that the major movements of this domain are only required for the initiation of translocation (Gold et al., 2012). Therefore, the translocation of preprotein must be driven by very subtle movements of the PPXD or 2HF, or by some other as yet undisclosed factor. Alternatively, there may be an indirect mechanism at play.
Materials and methods
Chemicals and biochemicals
All lipids were purchased from Avanti Polar Lipids, Inc., detergents were supplied by Glycon, BioBeads from Bio-Rad Laboratories, NuPAGE precast gels from Invitrogen, all chromatographic material from GE Healthcare, the QuikChange kit from Agilent Technologies, and all other reagents from Sigma-Aldrich. The monoclonal antibody to SecY (10C11) was produced by Cocalico Biologicals, Inc. The proOmpA anitobody was raised in sheep against the purified protein and the serum was used at 1:5,000 dilutions for Western blotting.
Molecular modeling of the cross-linked SecA795–Y183EG and SecA795–Y268EG complexes
The crystal structure of the Thermatoga maritima SecA–SecYEG complex (PDB code 3DIN) was used as the basis for the cross-linked complex. The residues in T. maritima (SecA 782 and SecY 183) corresponding to 795 and 183 in E. coli were changed to cysteine to form the appropriate mutant pair. The loop region and adjacent helical turns of the SecA 2HF were remodeled to bring position 795 adjacent to position 183 in SecY, and cystine formed to generate the cross-linked model. The cross-linked loop in the model was relaxed by energy minimization. The remodeled region had a similar geometric quality to the original crystal structure, as evaluated by Procheck. Modeling was performed using InsightII-2005 and energy calculations using Discover 2.98 (Accelrys).
Cloning, expression, and purification of the translocation components
Cloning, expression, and purification of the translocation components and specific mutants thereof were conducted exactly as previously described (Robson et al., 2009b; Deville et al., 2011). In brief, the proteins were overexpressed in E. coli. SecA (pET21), proOmpA Δ176-296 (pTrc99A), and SecYEG (pBAD22) were produced by conventional overexpression protocols, in LB (SecA) or 2× TY (proOmpA and SecYEG) liquid media at 37°C. SecA (and mutants) was purified from the soluble cell extracts by Ni-chelating, anion exchange, and size exclusion chromatography. The proOmpA preprotein substrate formed inclusion bodies, which were solubilized in 6M urea and purified by anion exchange chromatography. SecYEG and various mutants were extracted using the detergent n-dodecyl-β-maltoside (DDM) from total membranes prepared after overexpression. The complex was then purified sequentially by Ni-chelating and size exclusion and anion exchange chromatography.
Interaction between SecA and SecY268FlEG
Fluorescence quenching of SecY268FlEG was monitored in a Jobin Yvon Fluorolog (Horiba Scientific) using an excitation wavelength of 495 nm, and an emission wavelength of 515 nm. 29 nM SecY268FlEG and 1 mM AMPPNP was added to 1 ml of SecYEG buffer (20 mM Tris-HCl, pH 8, 130 mM NaCl, 10% glycerol, 0.02% DDM, 2 mM MgCl2, and 10 mM DTT). Baseline fluorescence was recorded. Wild-type SecA, SecA794C, or SecA795C was titrated in over a 0–3-µM concentration range, and fluorescence readings taken 200 s after each SecA addition.
Data were analyzed using Grafit (Erithacus). Results were presented as percentage of fluorescence quench relative to baseline, and fitted according to a one site weak binding equation:
where [L] is the total concentration of ligand, F is the fluorescence signal change, Fmax is the maximum signal change, and Kd is the dissociation constant.
Purification of cross-linked versions of SecA–SecYEG
Total membranes extracted from 12 liters worth of E. coli overexpressing SecYK268CEG or SecYI183CEG were resuspended in 100 ml of TSGM buffer (20 mM Tris-HCl, pH 8, 130 mM NaCl, 10% glycerol, and 2 mM MgCl2). In each case either 100 nmol of SecAY794C or SecAA795C was added together with 25 µM AMPPNP, before oxidation with 0.6 mM copper phenanthroline for 45 min at 4°C. The oxidizing agent was then removed by dialysis before the membranes were re-isolated by centrifugation. Membranes containing oxidized SecYI183CEG were washed twice in 100 ml of TSG to help remove the uncross-linked excess SecA. The membranes were then solubilized in TSGM buffer including 2% (wt/vol) DDM for 1 h at 4°C, and the insoluble material was removed by centrifugation. The cross-linked complexes were then purified by successive Ni-chelating and Q-Sepharose high performance and size-exclusion chromatography in a manner similar to that used previously for the wild-type and mutant SecYEG complexes (Robson et al., 2009b). As before, the uncross-linked SecYEG complexes washed through the Q-column (XK 16/10, ∼20-ml bed volume; GE Healthcare) in TSGM with 130 mM NaCl and 0.1% C12E9 (wt/vol). The bound cross-linked SecA–SecYEG and free SecA were eluted separately by a NaCl gradient (130–1,000 mM in 100 ml) in TSGM with 0.1% C12E9. The appropriate fractions were pooled and further purified by gel filtration using a Superdex 200HR XK16/60 in TSGM (130 mM NaCl) with 0.1% C12E9 (Fig. 3 B); see Fig. S2 for SDS-PAGE analysis of the eluting fractions from the corresponding gel filtration column.
Steady-state ATPase and in vitro protein translocation assays
Protein translocation assays and steady-state SecA ATPase measurements were performed as previously described (Robson et al., 2009b). In brief, ATPase activities were assayed in TKM buffer (50 mM triethanolamine, 50 mM KCl, and 2 mM MgCl2, pH 7.5) containing 1 U lactate dehydrogenase, 1.4 U pyruvate kinase, 0.2 mM NADH, and 2 mM phosphoenol pyruvate in 100 µl cuvettes at 25°C. 10 mM DTT was added to reaction buffer for uncross-linked SecA. Detergent solution experiments used 0.03% C12E9. Other reaction components were at concentrations stated in the text. Reactions were initiated by addition of 1 mM ATP, and the change in absorbance at 340 nm monitored for 10 min. 0.7 µM proOmpA was added, and the absorbance monitored for a further 20 min.
To measure in vitro translocation activity, 25-µl end-point samples from the ATPase reactions were incubated on ice with 0.2 mg/ml proteinase K for 1 h. For translocation time courses, multiple reaction volumes were incubated in a single tube at 25°C, and samples of 25 µl removed to 0.2 mg/ml proteinase K on ice at time points indicated in the figures. t = 0 samples represent translocation after initial mixing of assay components, which took ∼10 s. Protease-protected proOmpA was detected by immunoblot using an antibody raised against proOmpA as described elsewhere (Deville et al., 2011). Translocation activity was quantified from three immunoblots using ImageJ software (National Institutes of Health).
Before the proteinase K treatment the translocation reactions containing the wild-type and cross-linked (SecA795–Y268EG) complexes were analyzed for SecY by SDS-PAGE (4–12% Bis-Tris gels; Invitrogen) and immunoblotting with a monoclonal antibody to SecY.
Molecular dynamics simulations of the wild-type and cross-linked (SecA795–Y268EG) SecA–YEG complex
Molecular dynamics simulations were performed on the model of the E. coli wild-type membrane-bound SecA(YEG)2 translocon previously described (Deville et al., 2011). Hydrogen atoms were added consistent with pH 7.0 and the protein was parameterized with the OPLS/AA force field and MgATP parameterized as described previously (Piggot et al., 2012b). A POPC bilayer comprising 1,152 lipids was constructed with the utility genconf and the set of coordinates for an equilibrated POPC bilayer available on the Tieleman web site as the protein databank format file (http://moose.bio.ucalgary.ca/files/popc128a.pdb) and parameterized with a modified Berger-lipid force field (Berger et al., 1997; Piggot et al., 2012a). The PBC box containing the lipid was extended to 16.227 nm in Z (i.e., the axis normal to the plane of the bilayer). The box was filled with SPC water and Na+ Cl− ions to an ionic strength of 130 mM. The translocon model was placed at an appropriate depth in the bilayer near the center of the box by inspection, using VMD 1.9. The utility g_membed (Wolf et al., 2010) was used to embed the protein into the membrane and remove overlapping lipid water and ions. An in-house utility was used to remove water molecules within the hydrophobic core of the bilayer and Na+ Cl− ions swapped or deleted to ensure electrical neutrality of the simulation box. Molecular dynamics simulations were performed at 310 K and 1 bar as NPT ensembles, applying Nose-Hover temperature coupling and Parrinello-Rahman pressure coupling, under periodic boundary conditions. Electrostatic interactions were treated by the PME method with a real-space cut-off of 1.0 nm and a dispersion correction for long-range van der Waals interactions. Version 4.5.5 of GROMACS was used throughout.
An initial 1-ns simulation was performed on the wild-type while the protein atoms were restrained to their initial positions, which were subsequently removed and the simulation continued for a further 30 ns. This simulation was analyzed to measure the distance between the Cα–Cα and Cβ–Cβ atoms of residues 795 and 268 to identify the structure with the most appropriate geometry to be converted to the SecA795–Y268EG disulphide mutant. The structure at 14.4 ns was selected (Cα–Cα 7.9 Å; Cβ–Cβ 9.0 Å) and the cysteine created and energy minimized to generate the starting structure for the SecA795–Y268EG cross-linked simulation. The unmodified structure at 14.4 ns was also energy minimized to provide the starting structure for the comparison wild-type simulation. Both complexes were simulated for 100 ns and the resulting trajectories analyzed with g_bond.
Nonspecific cross-linking by PICUP
PICUP was performed in a total reaction volume of 10 µl. In the absence of light, 2 mM ammonium persulphate and 0.1 mM Tris-bipyridylruthenium were added to 0.85 µg of WT SecYEG or A795–Y268EG proteoliposomes in 20 mM Tris-Cl, pH 8, 130 mM NaCl, and 10% (wt/vol) glycerol. Proteins were cross-linked by irradiating with a 250 W slide projector for 2, 10, or 30 s, and reactions quenched by addition of 100 mM DTT. Cross-linked protein complexes were separated by SDS-PAGE on 4–12% Bis-Tris gels (Invitrogen), and detected by anti-SecY immunoblot (as above).
Online supplemental material
Fig. S1 depicts representative blots of single time points and a time course of protein translocation by wild-type and mutant SecA and SecYEG. Fig. S2 depicts representative blots of single time points and time courses of protein translocation assays of the purified mutant and cross-linked complexes. Fig. S3 depicts ribbon representations of various hypothetical oligomeric arrangements of SecA-SecYEG.
Acknowledgments
We thank Sir John Walker for providing the E. coli C43(DE3) strain, Tom Rapoport for donating the plasmids for the overexpression of SecAY794C and SecAA795C, and Franck Duong for the pTrcEHisYYG expression construct. We thank the University of Bristol Advanced Computing Research Centre and the e-Infrastructure South Consortium (UK) for provision of high performance computing. We gratefully acknowledge our technical support team, in particular Ms. Kalpana Austin for keeping the laboratory in order.
This work was supported by the BBSRC (A doctoral training grant PhD studentship to S. Whitehouse and Project Grants BB/I008675/1) and the Wellcome Trust (Equipment Grant 082140 for the bioreactor).
References
Author notes
V.A.M. Gold’s present address is Max Planck Institute of Biophysics, Department of Structural Biology, Max-von-Laue-Straβe 3, D-60438 Frankfurt am Main, Germany.