Disassembly of the nuclear lamina is a key step during open mitosis in higher eukaryotes. The activity of several kinases, including CDK1 (cyclin-dependent kinase 1) and protein kinase C (PKC), has been shown to trigger mitotic lamin disassembly, yet their precise contributions are unclear. In this study, we develop a quantitative imaging assay to study mitotic lamin B1 disassembly in living cells. We find that CDK1 and PKC act in concert to mediate phosphorylation-dependent lamin B1 disassembly during mitosis. Using ribonucleic acid interference (RNAi), we showed that diacylglycerol (DAG)-dependent PKCs triggered rate-limiting steps of lamin disassembly. RNAi-mediated depletion or chemical inhibition of lipins, enzymes that produce DAG, delayed lamin disassembly to a similar extent as does PKC inhibition/depletion. Furthermore, the delay of lamin B1 disassembly after lipin depletion could be rescued by the addition of DAG. These findings suggest that lipins activate a PKC-dependent pathway during mitotic lamin disassembly and provide evidence for a lipid-mediated mitotic signaling event.
Introduction
In multicellular eukaryotes, the nuclear envelope (NE) is underpinned by an intermediate filament meshwork of lamin proteins (Hetzer et al., 2005). Lamins interact with inner nuclear membrane proteins and chromatin components and are essential for nuclear structure and function (Parnaik, 2008). Vertebrates have three lamin genes, lamin A, B1, and B2. B-type lamins are essential and ubiquitous, whereas A-type lamins are expressed only in differentiated cells. All lamins contain a central rod that is essential for oligomerization (Stuurman et al., 1998).
NE breakdown (NEBD) precedes the segregation of chromosomes by the metazoan spindle during mitosis. NEBD requires disassembly of all NE components, including lamins (Güttinger et al., 2009). The disassembly of A-type lamins begins in prophase, and they are released into the cytoplasm on NEBD. Farnesylated B-type lamins remain membrane bound during mitosis but disperse throughout the ER (Gerace et al., 1978; Georgatos et al., 1997).
Although lamina breakdown involves microtubule-mediated pulling forces and carboxydemethylation, lamin filament depolymerization is triggered by mitotic phosphorylation (Gerace and Blobel, 1980; Chelsky et al., 1987; Beaudouin et al., 2002; Salina et al., 2002; Mühlhäusser and Kutay, 2007). Conserved CDK1 consensus motifs have been identified in all lamins, and their phosphorylation was shown to be important for mitotic lamin A disassembly in mammalian cells and for CDK1-dependent disassembly of chicken B-type lamins in vitro (Heald and McKeon, 1990; Peter et al., 1990, 1991). PKC-βII has also been reported to phosphorylate B-type lamins during mitosis (Hocevar et al., 1993; Goss et al., 1994). In addition, evidence from various species suggests direct involvement of PKC activity in lamina disassembly (Thompson and Fields, 1996; Collas et al., 1997; Collas, 1999). This suggests that multiple kinases promote efficient mitotic lamin disassembly.
Lipins are conserved enzymes that convert phosphatidate to DAG (Han et al., 2006). Lipins affect membrane proliferation and NE morphology in yeast (Tange et al., 2002; Santos-Rosa et al., 2005), and mammalian lipins transcriptionally activate genes of lipid metabolism (Finck et al., 2006). In rapidly dividing Caenorhabditis elegans embryos, lipin is essential for lamin disassembly during NEBD, but the mechanism by which lipin affects lamin disassembly is unclear (Golden et al., 2009; Gorjánácz and Mattaj, 2009). Because the mammalian lamin kinase PKC-βII is activated by DAG, we tested whether lipins might trigger lamin phosphorylation and disassembly by producing the lipid-signaling molecule DAG to activate PKCs.
Results and discussion
CDK1 and PKC consensus phosphorylation sites on lamin B1 contribute to efficient mitotic disassembly
During mitosis, CDK1 and PKCs phosphorylate lamin B1 at sites flanking its central rod domain (Fig. 1 A; Heald and McKeon, 1990; Peter et al., 1990, 1991; Ward and Kirschner, 1990; Hocevar et al., 1993; Goss et al., 1994). To test the contribution of these to lamin B1 disassembly in living cells, we generated fluorescent lamin B1 reporters with serine (S) to alanine (A) mutations at CDK1, PKC, and both CDK1 and PKC consensus phosphorylation sites (Fig. 1 B). We transiently transfected the reporters into HeLa cells stably expressing the chromatin marker H2B-mCherry and monitored mitotic progression by confocal microscopy. The disassembly of the PKC mutant lamin B1 reporter was indistinguishable from wild type, whereas mutation of the CDK1 phosphorylation sites slightly delayed disassembly and ER translocation of lamin B1. Cytoplasmic fragments of the CDK1 mutant lamin B1 remained visible throughout mitosis (Fig. 1 C). Combined mutation of both CDK1 and PKC phosphorylation motifs resulted in more pronounced disassembly defects (Fig. 1 C), suggesting phosphorylation of both CDK1 and PKC sites contributes to mitotic lamin B1 disassembly. Unlike Heald and McKeon (1990), we did not observe a continuous signal of any mutant lamin B1 reporter surrounding chromatin throughout mitosis. This might be because of biological differences between lamin A and B1 and differences between CHO and HeLa cells or reflect differences in the lamin expression levels between the experiments.
Phosphorylation sites on lamin B1 promote mitotic disassembly. (A) Schematic representation of lamin B1 domain structure and phosphorylation (UniProt). The central rod is flanked by several mitotic phosphorylation sites (indicated by asterisks) that exhibit CDK1 and PKC consensus motifs (Pearson and Kemp, 1991). (B) Overview of lamin B1 reporters containing serine (S) to alanine (A) mutations at CDK1 and/or PKC consensus phosphorylation sites. (C) Confocal time-lapse microscopy images of HeLa cells stably expressing H2B-mCherry and transiently expressing the indicated lamin B1 reporter tagged with GFP (gray). Progression through mitosis was followed, and time relative to chromatin condensation in prophase is shown. Arrowheads indicate cytoplasmic lamin B1 fragments present after prometaphase. Bars, 10 µm.
Phosphorylation sites on lamin B1 promote mitotic disassembly. (A) Schematic representation of lamin B1 domain structure and phosphorylation (UniProt). The central rod is flanked by several mitotic phosphorylation sites (indicated by asterisks) that exhibit CDK1 and PKC consensus motifs (Pearson and Kemp, 1991). (B) Overview of lamin B1 reporters containing serine (S) to alanine (A) mutations at CDK1 and/or PKC consensus phosphorylation sites. (C) Confocal time-lapse microscopy images of HeLa cells stably expressing H2B-mCherry and transiently expressing the indicated lamin B1 reporter tagged with GFP (gray). Progression through mitosis was followed, and time relative to chromatin condensation in prophase is shown. Arrowheads indicate cytoplasmic lamin B1 fragments present after prometaphase. Bars, 10 µm.
A quantitative in vivo lamin B1 disassembly assay
High-throughput wide-field microscopy of HeLa cells stably expressing GFP–lamin B1 and H2B-mCherry enabled us to record several hundred mitotic lamin B1 disassembly events with high time resolution (Fig. 2 A; Neumann et al., 2006). Individual mitotic cells were automatically tracked and classified into mitotic phases using CellCognition (Fig. 2 B; Held et al., 2010; Walter et al., 2010). We monitored lamin B1 disassembly by measuring GFP–lamin B1 fluorescence intensity at chromatin (assembled) and the surrounding cytoplasm (disassembled) throughout mitosis (Fig. 2, C and D). This allowed us to robustly estimate the time required for lamin disassembly (disassembly duration) and the time from prophase to anaphase (mitotic progression) in hundreds of living cells for each experiment.
Quantitative lamin B1 disassembly assay. (A) High-throughput automated time-lapse wide-field microscopy of HeLa cells stably coexpressing H2B-mCherry and GFP–lamin B1. A single cell entering mitosis is shown. The last interphase frame marks time point 0. For each condition, several hundred mitotic events were acquired with a time-lapse of 2 min, and up to eight different experimental conditions were imaged in parallel. (B) H2B-mCherry signals were used for automatic tracking of nuclei and for classification-based annotation of mitotic stages. In addition, error-corrected mitotic classification results were used to determine mitotic progression (sum of pro-, prometa-, and metaphase duration; Held et al., 2010). (C) GFP–lamin B1 intensities were measured at the chromatin and in the cytoplasm throughout mitosis. Segmentation of the H2B-mCherry signal was used to define the chromatin region (in) and by dilation the cytoplasmic region (out). (D) Automated analysis of lamin B1 disassembly. The difference between the GFP–lamin B1 intensity at chromatin (in) and in the cytoplasm (out) was determined during mitosis and normalized to the difference during interphase. Using a mathematical model, disassembly durations were determined as the time in which the intensity difference changed from 95 to 5% (green bars). a.u., arbitrary units. Bars, 10 µm.
Quantitative lamin B1 disassembly assay. (A) High-throughput automated time-lapse wide-field microscopy of HeLa cells stably coexpressing H2B-mCherry and GFP–lamin B1. A single cell entering mitosis is shown. The last interphase frame marks time point 0. For each condition, several hundred mitotic events were acquired with a time-lapse of 2 min, and up to eight different experimental conditions were imaged in parallel. (B) H2B-mCherry signals were used for automatic tracking of nuclei and for classification-based annotation of mitotic stages. In addition, error-corrected mitotic classification results were used to determine mitotic progression (sum of pro-, prometa-, and metaphase duration; Held et al., 2010). (C) GFP–lamin B1 intensities were measured at the chromatin and in the cytoplasm throughout mitosis. Segmentation of the H2B-mCherry signal was used to define the chromatin region (in) and by dilation the cytoplasmic region (out). (D) Automated analysis of lamin B1 disassembly. The difference between the GFP–lamin B1 intensity at chromatin (in) and in the cytoplasm (out) was determined during mitosis and normalized to the difference during interphase. Using a mathematical model, disassembly durations were determined as the time in which the intensity difference changed from 95 to 5% (green bars). a.u., arbitrary units. Bars, 10 µm.
CDK1 and PKC kinase activities boost mitotic progression and lamin B1 disassembly
Applying this assay to our transiently transfected lamin B1 reporters with physiological reporter expression levels (see Materials and methods) confirmed that mutation of the CDK1 sites and combined mutation of CDK1 and PKC consensus motifs significantly delayed the disassembly process (Fig. 3 A). Interestingly, no delay in mitotic progression was observed (Fig. 3 B). To investigate the contribution of CDK1 and PKCs to lamin disassembly more directly, we applied kinase inhibitors to thymidine-synchronized GFP–lamin B1 and H2B-mCherry coexpressing cells ∼30–60 min before mitotic entry. The PKC inhibitor Gö6976 and the CDK1 inhibitor roscovitine caused significant delays in lamin B1 disassembly duration and mitotic progression (Fig. 3, C and D; Martiny-Baron et al., 1993; Meijer et al., 1997). We observed cytoplasmic lamin B1 fragments throughout mitosis after inhibition of either CDK1 or PKC. Although combined inhibition of CDK1 and PKC resulted in an overall decrease of mitotic events in the cell population (Table S2), the cells that entered mitosis exhibited more striking delays in lamin B1 disassembly and mitotic progression compared with cells treated with the individual kinase inhibitors (Fig. 3, C and D). Interestingly, single-cell analysis revealed that lamin B1 disassembly and mitotic progression were only poorly correlated (Fig. S1). This suggests that CDK1 and PKC are essential for efficient phosphorylation-dependent lamin B1 disassembly as well as timely mitotic progression but that the two processes are not directly or causally linked. Because neither the co-inhibition nor the combined mutation of CDK1 and PKC phosphorylation sites completely blocked lamin B1 disassembly, further kinases and/or additional phosphorylation sites may be involved in lamin disassembly. These could include cAMP-dependent protein kinase and S6 kinase II, which also phosphorylate lamin proteins (Ward and Kirschner, 1990; Stuurman, 1997). In addition, carboxydemethylation or microtubule-mediated tearing might compensate for the lack of CDK1- and PKC-mediated disassembly (Chelsky et al., 1987; Beaudouin et al., 2002; Salina et al., 2002; Mühlhäusser and Kutay, 2007).
CDK1 and PKC kinase activities drive mitotic lamin B1 disassembly. (A) HeLa cells stably expressing H2B-mCherry were transiently transfected with mutated GFP–lamin B1 reporters (see also Fig. 1), and mean mitotic lamin B1 disassembly was quantified as described in Fig. 2. (B) Mean mitotic durations of cells quantified in A. Mitotic progression in the transfected H2B-mCherry cells took longer than in the stable GFP–lamin B1– and H2B-mCherry–coexpressing cells in D. (C) HeLa cells stably coexpressing H2B-mCherry and GFP–lamin B1 were synchronized by double thymidine arrest. Approximately 30–60 min before mitotic entry, cells were treated with DMSO (mock), 2 µM Gö6976 (PKC inhibition), 50 µM roscovitine (CDK1 inhibition), or a combination of 2 µM Gö6976 and 50 µM roscovitine, and mean lamin B1 disassembly was determined as in A. (D) Mean mitosis durations of cells quantified in C. The data represent the mitotic events from at least three replicate experiments. n > 100 for each condition with *, P < 0.05 and ***, P < 0.0001 compared with control (U test; Table S1 and Table S2). Dotted lines indicate mean control timing. Error bars indicate 95% confidence intervals.
CDK1 and PKC kinase activities drive mitotic lamin B1 disassembly. (A) HeLa cells stably expressing H2B-mCherry were transiently transfected with mutated GFP–lamin B1 reporters (see also Fig. 1), and mean mitotic lamin B1 disassembly was quantified as described in Fig. 2. (B) Mean mitotic durations of cells quantified in A. Mitotic progression in the transfected H2B-mCherry cells took longer than in the stable GFP–lamin B1– and H2B-mCherry–coexpressing cells in D. (C) HeLa cells stably coexpressing H2B-mCherry and GFP–lamin B1 were synchronized by double thymidine arrest. Approximately 30–60 min before mitotic entry, cells were treated with DMSO (mock), 2 µM Gö6976 (PKC inhibition), 50 µM roscovitine (CDK1 inhibition), or a combination of 2 µM Gö6976 and 50 µM roscovitine, and mean lamin B1 disassembly was determined as in A. (D) Mean mitosis durations of cells quantified in C. The data represent the mitotic events from at least three replicate experiments. n > 100 for each condition with *, P < 0.05 and ***, P < 0.0001 compared with control (U test; Table S1 and Table S2). Dotted lines indicate mean control timing. Error bars indicate 95% confidence intervals.
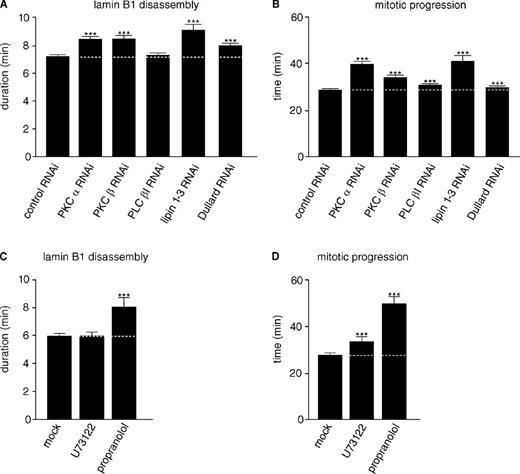
PKC-α and -β can trigger lamin B1 disassembly during mitosis
The function and regulation of PKCs during mitosis is not fully understood. Gö6976 is reported to inhibit classical PKCs (cPKCs; α, β, and γ; Martiny-Baron et al., 1993). Because PKC-γ is specifically expressed in neurons, we wanted to test whether inhibition of PKC-α and -β can account for the mitotic defects we observed upon application of Gö6976 (Liu and Heckman, 1998). We therefore monitored lamin B1 disassembly in unsynchronized cells after PKC-α and -β (isoform I and II) RNAi (Fig. S2 C). Interestingly, RNAi of PKC-α or -β resulted in delayed lamin B1 disassembly and mitotic progression (Fig. 4, A and B). Combined depletion of both PKC-α and -β did not delay these processes further (Table S2). This suggests that PKC-α and -β may work together in boosting mitotic lamin B1 disassembly and mitotic progression. Consistently, PKC-β activity is reported to peak before NEBD, and experiments from fixed mammalian cells suggested that PKC-βII triggers phosphorylation-dependent lamin disassembly (Goss et al., 1994; Thompson and Fields, 1996; Dai et al., 2007).
cPKCs, lipins, and Dullard boost mitotic lamin B1 disassembly. (A) HeLa cells stably coexpressing H2B-mCherry and GFP–lamin B1 were transfected with siRNA oligos against the indicated transcripts or control oligos before quantitation of mean mitotic lamin B1 disassembly as described in Fig. 2. (B) Mean mitosis durations of cells quantified in A. (C) HeLa cells stably coexpressing H2B-mCherry and GFP–lamin B1 were synchronized by double thymidine arrest. Approximately 30–60 min before mitotic entry cells were treated with DMSO (mock), 4 µM U73122 (PLC inhibition), or 200 µM propranolol (lipin inhibition), and mean lamin B1 disassembly was determined as in A. (D) Mean mitosis durations of cells quantified in C. The data represent the mitotic events from two replicate experiments. n > 200 for each condition with ***, P < 0.0001 (U test; Table S2). Dotted lines indicate mean control timing. Error bars indicate 95% confidence intervals.
cPKCs, lipins, and Dullard boost mitotic lamin B1 disassembly. (A) HeLa cells stably coexpressing H2B-mCherry and GFP–lamin B1 were transfected with siRNA oligos against the indicated transcripts or control oligos before quantitation of mean mitotic lamin B1 disassembly as described in Fig. 2. (B) Mean mitosis durations of cells quantified in A. (C) HeLa cells stably coexpressing H2B-mCherry and GFP–lamin B1 were synchronized by double thymidine arrest. Approximately 30–60 min before mitotic entry cells were treated with DMSO (mock), 4 µM U73122 (PLC inhibition), or 200 µM propranolol (lipin inhibition), and mean lamin B1 disassembly was determined as in A. (D) Mean mitosis durations of cells quantified in C. The data represent the mitotic events from two replicate experiments. n > 200 for each condition with ***, P < 0.0001 (U test; Table S2). Dotted lines indicate mean control timing. Error bars indicate 95% confidence intervals.
PLC affects mitotic progression but not lamin B1 disassembly
Because cPKCs require calcium and DAG for activation, regulation of DAG or calcium levels could be involved in their mitotic activation (Mellor and Parker, 1998). Nuclear calcium stimulates early stages of mitosis, and nuclear DAG levels peak at the G2/M-phase transition of the cell cycle (Deacon et al., 2002; Rodrigues et al., 2007). PLC hydrolyses phosphatidylinositol 4,5-bisphosphate, generating DAG and the calcium-mobilizing second messenger inositol 1,4,5-trisphosphate. Previous studies used flow cytometry to show that PLC-βI affects cell cycle progression and speculated that DAG produced by PLC-βI activates cPKCs (Sun et al., 1997; Fiume et al., 2009). Consistently, we found that the general PLC inhibitor U73122 and PLC-βI RNAi slightly but significantly delayed mitotic progression (Fig. S2 C and Fig. 4, B and D). However, no significant effect on lamin B1 disassembly was observed (Fig. 4, A and C). If mitotic lamin disassembly is indeed regulated by DAG, factors other than PLC must be involved.
Lipins trigger mitotic progression and lamin B1 disassembly
The enzyme lipin, which also generates DAG, was recently shown to affect mitotic lamin disassembly and NEBD in C. elegans (Golden et al., 2009; Gorjánácz and Mattaj, 2009). Mammals have three lipin genes, which are all expressed and can be efficiently depleted by RNAi (Fig. S2, A and C). Lipins 1 and 2, but not 3, were detectable by Western blotting in HeLa cells (Fig. S2 B). No reproducible delay in lamin B1 disassembly duration was observed upon down-regulation of individual lipins by RNAi (unpublished data). However, the three mammalian lipins are reported to be functionally redundant (Donkor et al., 2007). Indeed, combined RNAi treatment against all three lipin isoforms resulted in a striking delay of lamin B1 disassembly and mitotic progression (Fig. 4, A and B), whereas the morphology of the ER was unaffected as judged by calnexin immunofluorescence (not depicted). To more directly test potential mitotic functions of lipins, we applied the lipin enzyme inhibitor propranolol to thymidine-synchronized cells before mitotic entry (Havriluk et al., 2008). Propranolol addition phenocopied the defects observed after lipin 1–3 co-RNAi (Fig. 4, C and D). These data indicate that mammalian lipins have redundant functions during lamin disassembly and mitotic progression involving their enzymatic production of DAG, which is further supported by the fact that down-regulation of Dullard (CTDNEP1), the phosphatase that is known to dephosphorylate and thereby activate lipins at the NE (Santos-Rosa et al., 2005; Kim et al., 2007), caused a similar, albeit milder, delay in mitotic progression and lamin B1 disassembly (Fig. 4, A and B). This is in agreement with the finding that Dullard down-regulation affected NEBD to a lesser extent than lipin down-regulation in C. elegans (Han et al., 2012). Phosphorylation of lipins by several kinases, including CDK1, reduced membrane association and enzymatic activity of lipins during mitosis (Grimsey et al., 2008; Choi et al., 2011). It seems likely that Dullard is an activator of lipins at the NE that acts to trigger mitotic lamin disassembly and NEBD, whereas at the same time, a general decrease of lipin activity is mediated by mitotic CDK1 activity (Karanasios et al., 2010).
Lipins and cPKCs also affect mitotic lamin A disassembly
We next performed high-throughput wide-field microscopy of HeLa cells stably expressing GFP–lamin A and H2B-mCherry, to test whether lipins and cPKCs were also required for A-type lamin disassembly. We observed significant disassembly delays on RNAi-mediated codepletion of lipin 1–3 or PKC-α and -β (Fig. S3, A and B). We could also confirm the mitotic delays observed in the GFP–lamin B1 H2B-mCherry cell line (Fig. S3 C). This shows that lipins, as well as cPKCs, affect the disassembly of both A- and B-type lamins.
Localization of lipins and cPKCs at NEBD
To learn more about the localization of lipins and PKC-α and -βII during NEBD, we tracked GFP-tagged versions of the proteins by confocal microscopy. All reporters were mainly cytoplasmic before NEBD. However, they rapidly enter the nuclear volume upon NEBD and could then trigger lamin disassembly (Videos 1, 2, 3, 4, and 5). Therefore, lipin and PKC activity rather than localization might be regulated during mitosis. In addition, DAG can diffuse very rapidly within membranes, and it might not be necessary to produce it locally (Prieto et al., 1994).
Lipins promote mitotic lamin B1 disassembly through DAG
If indeed the reduced DAG levels are the reason for the lamin disassembly defects upon lipin down-regulation, addition of exogenous DAG should rescue these defects. To test this, we combined lipin 1–3 co-RNAi with thymidine synchronization and supplemented lipin-depleted cells with DAG ∼30–60 min before mitosis. Addition of DAG to control RNAi-treated cells had no striking effect on lamin B1 disassembly duration or mitotic progression (Fig. 5, A and B), indicating that exogenous DAG has no adverse effects on the cells during mitosis. However, DAG was able to restore normal lamin B1 disassembly timing and partially rescue the mitotic progression delay in cells depleted of lipins (Fig. 5, A and B). The mitotic delay upon lipin depletion occurs mainly during prometaphase and metaphase when chromosomes are congressing (Table S2), and it will be very interesting to characterize the mechanism that brings about this delay.
Lipid signal mediates mitotic effect of lipins. (A) HeLa cells stably coexpressing H2B-mCherry and GFP–lamin B1 were transfected with siRNA oligos against lipin 1–3 or control oligos and synchronized by double thymidine arrest. Approximately 30–60 min before mitotic entry, cells were supplemented with DMSO (mock) or 200 µM DAG, and mean lamin B1 disassembly was determined as in Fig. 2. (B) Mean mitosis durations of cells quantified in A. The data represent the mitotic events from three replicate experiments. n > 100 for each condition with *, P < 0.05 and ***, P < 0.0001 (U test; Table S2). Dotted lines indicate mean control timing. Error bars indicate 95% confidence intervals. (C) Model of lipid-mediated mitotic lamin disassembly. Dullard dephosphorylates and activates lipins that in turn produce DAG from phosphatidate. DAG activates cPKCs, which, in concert with CDK1, can trigger phosphorylation-dependent mitotic lamin disassembly. NPC, nuclear pore complex; Nups, nucleoporins; INM, inner nuclear membrane; PA, phosphatidate; P, phosphorylation.
Lipid signal mediates mitotic effect of lipins. (A) HeLa cells stably coexpressing H2B-mCherry and GFP–lamin B1 were transfected with siRNA oligos against lipin 1–3 or control oligos and synchronized by double thymidine arrest. Approximately 30–60 min before mitotic entry, cells were supplemented with DMSO (mock) or 200 µM DAG, and mean lamin B1 disassembly was determined as in Fig. 2. (B) Mean mitosis durations of cells quantified in A. The data represent the mitotic events from three replicate experiments. n > 100 for each condition with *, P < 0.05 and ***, P < 0.0001 (U test; Table S2). Dotted lines indicate mean control timing. Error bars indicate 95% confidence intervals. (C) Model of lipid-mediated mitotic lamin disassembly. Dullard dephosphorylates and activates lipins that in turn produce DAG from phosphatidate. DAG activates cPKCs, which, in concert with CDK1, can trigger phosphorylation-dependent mitotic lamin disassembly. NPC, nuclear pore complex; Nups, nucleoporins; INM, inner nuclear membrane; PA, phosphatidate; P, phosphorylation.
Conclusions
To study lamin disassembly in vivo, we developed a high-throughput wide-field microscopy assay to quantify lamin B1 disassembly in large numbers of individual dividing human cells. Exploiting this assay, we have shown that PKC-α and -β trigger rate-limiting steps during mitotic lamin B1 disassembly. This supports the notion that mitotic lamin disassembly is dependent on several kinases (Thompson and Fields, 1996; Collas et al., 1997; Collas, 1999). Multiple kinases also act during nuclear pore complex disassembly; this therefore might represent a common mechanism to ensure robust mitotic entry and NEBD (De Souza et al., 2004; Laurell et al., 2011).
We also show that mammalian lipin proteins, which can enzymatically generate the cPKC activator DAG, affect mitotic lamin B1 disassembly. This is consistent with results from C. elegans embryos and demonstrates that lipins play a role during mitotic lamin disassembly that is conserved from worms to humans (Golden et al., 2009; Gorjánácz and Mattaj, 2009). Finally, we established that DAG transduces the mitotic effect of lipins. This suggests that the lipid signaling pathway from lipins via DAG, PKC activation, and lamin phosphorylation plays an important role in NEBD. Based on our data, we propose a model in which lipins are locally protected from inhibitory phosphorylation by CDK1 through the NE-localized phosphatase Dullard (Grimsey et al., 2008; Karanasios et al., 2010; Choi et al., 2011) and can therefore elevate DAG levels during NEBD, which in turn is essential for complete activation of cPKCs and efficient mitotic disassembly of both A- and B-type lamins (Fig. 5 C). Further work is needed to understand the spatiotemporal activation of lipins during mitosis and to integrate the contribution of calcium to this mitotic signaling pathway.
Materials and methods
Chemicals
Gö6976, propranolol, roscovitine, and U-73122 were purchased from EMD Millipore. DAG was purchased from Avanti Polar Lipids, Inc., and thymidine was purchased from Sigma-Aldrich.
DNA constructs and cell lines
pS65T-C1-lamin A (Broers et al., 1999) and pEGFP-C1-lamin B1 (Daigle et al., 2001) were used to produce stable cell lines and to generate phosphorylation site mutants using the multisite-directed mutagenesis kit (QuikChange; Agilent Technologies). Human lipin 1 cDNA (Grimsey et al., 2008) and human PKC-α cDNA (Reither et al., 2006) were provided by S. Siniossoglou (Cambridge Institute for Medical Research, Cambridge England, UK) and G. Reither (European Molecular Biology Laboratory, Heidelberg, Germany), respectively. Lipin 2 (OCABo5050C0320D), lipin 3 (IRCBp5005A1912Q), and PKC-βII (IRATp970H0335D) cDNA constructs were purchased from ImaGenes. For mammalian expression, full-length lipins and PKCs were cloned into pEGFP-N3 (Takara Bio Inc.) using Bgl II–Sac II and Nhe I–Xho I restriction sites, respectively. For bacterial expression and antibody generation, fragments of lipin 1 (aa 128–377), lipin 2 (aa 259–381), and lipin 3 (aa 133–377) were cloned into pET28a (EMD Millipore) using Nhe I–Xho I restriction sites. A complete list of DNA primers used in this study can be found in Table S3.
H2B-mCherry–expressing HeLa Kyoto cells were a gift of D. Gerlich (Institute of Molecular Biotechnology of the Austrian Academy of Sciences, Vienna, Austria; Schmitz et al., 2010). H2B-mCherry GFP-lamin–coexpressing cells were generated by transient transfection and selection of H2B-mCherry–expressing cells. The levels of GFP-lamin reporters were around (GFP–lamin A, 99%) or below (GFP–lamin B1, 33%) endogenous lamin levels as judged by Western blotting using lamin antibodies detecting both the endogenous and the GFP-lamin reporters. H2B-mCherry–expressing cells were grown in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum, 2 mM glutamine, 1 mM sodium pyruvate, 100 µg/ml penicillin and streptomycin, and 500 ng/ml puromycin (Invitrogen), and coexpressing cells were selected and cultured in the same medium supplemented with 500 µg/ml G418 (Invitrogen).
Recombinant proteins, antibodies, and Western blotting
Recombinant His-tagged lipin fragments were expressed in Escherichia coli Rosetta (DE3; EMD Millipore) for 4 h at room temperature, purified via Ni–nitrilotriacetic acid agarose (QIAGEN), and eluted in lysis buffer (20 mM Tris, pH 7.6, 0.5 M NaCl, 1 mM MgCl2, and 8 mM imidazole) containing 0.4 M imidazole followed by dialysis against PBS supplemented with 0.5 M NaCl. Lipin antibodies were raised by injection of recombinant protein fragments into rabbits and affinity purified using antigen-coupled Affi-Gel 15 (Bio-Rad Laboratories). Lamin antibodies (Schirmer et al., 2001) were provided by E. Schirmer (Wellcome Trust Centre for Cell Biology and Institute of Cell Biology, Edinburgh, Scotland, UK), α–glyceraldehyde 3-phosphate dehydrogenase (GAPDH; sc-32233) was purchased from Santa Cruz Biotechnology, Inc., and secondary horseradish peroxidase antibodies were purchased from Molecular Probes. Western blotting after SDS-PAGE separation was performed on whole-cell protein samples resuspended in SDS sample buffer containing Benzonase (Merck).
Cell transfection and synchronization
Transfections for transient expression experiments and stable cell line generation were performed using FuGENE 6 (Roche). H2B-mCherry–expressing cells were transfected with plasmids (1.5-nM final concentration) for 24 h before live imaging and selection.
Transfections for RNAi experiments were performed using INTERFERin (Polyplus Transfection). Indicated cell lines were transfected with siRNA oligonucleotides (oligos; 30-nM final concentration) for 48 h before live imaging and protein or RNA sample preparation. The following siRNA oligos from Ambion were used: control (s814), 5′-UCGUAAGUAAGCGCAACCCTT-3′; lipin 1 (s23207), 5′-UUCUAUUCAAAGACACUCCTA-3′; lipin 2 (s18592), 5′-UGCAAGCUAAGGAUCAUGGGA-3′; lipin 3 (s35074), 5′-AUUUUCACCACUAGGUUUGGG-3′; Dullard (s23741), 5′-UCUAUUACCACCUUGAGGATG-3′; PKC-α (s11094), 5′-UCCAUGACGAAGUACAGCCGA-3′; PKC-β (s11096), 5′-AAAUCGGUCAGUUUCAUCCGG-3′; and PLC-βI (s23358), 5′-UCUACUUCCACGUAAGUCCCA-3′.
The indicated cell lines were synchronized by double thymidine arrest. Cells were grown in medium containing 2 mM thymidine for 18 and 17 h intersected by 8.5-h release without thymidine. 7 h after the second release (∼30–60 min before mitotic entry), chemicals were applied in imaging medium before live imaging. If chemical treatment was combined with RNAi, cells were supplemented with siRNA transfection mixture just before seeding in medium containing 2 mM thymidine.
Reverse transcription and quantitative real-time PCR
RNA was harvested using the spin cell RNA mini kit (Invisorb; Invitek) and reverse transcribed to cDNA using oligo(dT) primers and the synthesis kit (RETROscript; Ambion). Quantitative real-time PCR was performed from cDNA templates using the SYBR green PCR master mix and a real-time PCR system (7500; Applied Biosystems). Transcript levels were determined after normalization against GAPDH using the relative expression software tool (Pfaffl et al., 2002). A complete list of real-time PCR primers used in this study can be found in Table S3.
Microscopy imaging
Confocal imaging of live samples was performed on laser-scanning confocal microscopes (LSM510 and LSM780; Carl Zeiss) equipped with 63×, NA 1.4 Plan Apochromat oil objectives using the LSM and ZEN software, respectively (Carl Zeiss).
Live imaging for quantitative in vivo lamin disassembly experiments was performed on automated microscopes (IX81; Olympus) equipped with 10×, NA 0.4 Plan Apochromat air objectives using a charge-coupled device camera (C8484-05C; Hamamatsu Photonics) and an image-based autofocus and the scan∧R software (Olympus; Neumann et al., 2010). Up to 24 separate locations were imaged in parallel for 12–24 h at a time lapse of 2 min. All live-cell imaging was performed at 37°C using CO2-independent medium without phenol red (Invitrogen) containing 20% fetal bovine serum, 2 mM l-glutamine, and 100 mg/ml penicillin and streptomycin in 8-well dishes (Lab-Tek; Thermo Fisher Scientific).
Quantitative image analysis
Automated quantitative analysis of dividing H2B-mCherry GFP-lamin–coexpressing cells was used to monitor both mitotic progression and lamin disassembly in single cells. For this, nuclei were detected in the H2B-mCherry channel and classified as previously described (Held et al., 2010; Walter et al., 2010). Nuclei with low expression levels and small nuclei (<100 pixels), which typically correspond to debris, were removed. For classification, we defined 10 morphological classes: interphase, prophase, prometaphase, metaphase, early anaphase (chromosome sets are still together), late anaphase (chromosome sets are separated), telophase, cell death, deformed (grouping polylobed and multiple nuclei as well as artifact nuclei), and MAP (metaphase alignment problems corresponding to partially aligned chromosomes). The training set contained 4,166 manually labeled nuclei, which were detected with an overall accuracy of 90.0% (fivefold cross-validation). Cells were tracked with a constrained nearest-neighbor tracking procedure, and mitotic onset was detected as interphase–prophase or interphase–prometaphase transition. To reduce the effect of classification errors on phase length measurements, classification results were corrected with hidden Markov models (Held et al., 2010).
The segmentation result from the H2B-mCherry channel was used to define the chromatin region (in), and a ring around this region was defined as outer region (out), which was assumed to lie at least partly in the cytoplasm. To compare fluorescence intensity in the inner and outer regions, we calculated the difference between the 70th percentiles in both regions (the 70th percentile was used instead of the mean to be robust with respect to parts of the outer ring being outside the cell). This difference feature was then normalized to its mean value in interphase before entering mitosis. The resulting time series therefore quantified the translocation kinetics of fluorescent proteins from or to the nuclear region. It is of interest that the translocation kinetics from or to the nuclear region mirrored the translocation kinetics from or to the nuclear rim as tested on a control dataset.
To robustly estimate the disassembly duration and time point, we fitted a model (sum of one, two, or three sigmoidals) to the experimental data using the BFGS (Broyden–Fletcher–Goldfarb–Shanno) method (Nocedal and Wright, 2006) with a quadratic error term and a differentiable penalty term that incorporated a priori knowledge about the shape of the model (namely the order of disassembly, reassembly, and decondensation and the sign of the sigmoidals). The number of sigmoidals was selected to best represent the experimental data. Lamin disassembly duration was defined as the time the signal needed to decrease from 5 to 95% of its total drop. Quality control was applied to all extracted tracks. Tracks were removed if the sequence of morphological classes was obviously aberrant (anaphase skipped in completed mitosis, a total mitotic duration of <8 min), if too many dead cells were identified in the track (>3), if the drop was too low (<0.2), or if the fit was of low quality. In addition, tracks were removed if the lamin expression level in interphase was <8 or >100 (in a dynamic range of 0–255). Within this range, the expression levels did not correlate with mitotic progression timings (Spearman correlation = −0.026 and 0.025 with n > 1,000 for GFP–lamin A and B1, respectively) and lamin disassembly durations (Spearman correlation = 0.109 and 0.124 with n > 1,000 for GFP–lamin A and B1, respectively), showing that reporter expression did not affect mitotic progression and lamin disassembly.
The image processing was performed with a modified version of CellCognition (Held et al., 2010). R (R Project) was used for statistical tests, generation of plots (with the geneplotter package for Fig. S1), and numerical optimization for fitting the model. The analysis pipeline was written in Python.
Online supplemental material
Fig. S1 shows that lamin B1 disassembly and mitotic progression are not coupled in single cells. Fig. S2 shows lipin expression in HeLa Kyoto cells and provides validation of RNAi efficiency. Fig. S3 shows that PKC-α and -β as well as lipins 1–3 trigger mitotic lamin A disassembly. Videos 1–5 show the localization of GFP-tagged PKC-α, -βII, and lipins 1, 2, and 3, respectively, during NEBD in living HeLa Kyoto cells. Table S1 shows quantification of lamin B1 disassembly and mitotic progression in cells stably expressing H2B-mCherry transfected with GFP–lamin B1. Table S2 shows quantification of lamin B1 disassembly and mitotic progression in cells stably coexpressing GFP–lamin B1 and H2B-mCherry. Table S3 shows a complete list of DNA primers used in this study.
Localization of PKC-α during NEBD in mammalian cells. HeLa cells stably expressing H2B-mCherry and transiently expressing PKC-α tagged with GFP (green and gray) at the C terminus during mitotic entry. Progression into mitosis was analyzed by time-lapse microscopy using a laser-scanning confocal microscope (LSM 510). Frames were taken every 30 s for 3 min, and time relative to NEBD is shown. Bar, 10 µm.
Localization of PKC-α during NEBD in mammalian cells. HeLa cells stably expressing H2B-mCherry and transiently expressing PKC-α tagged with GFP (green and gray) at the C terminus during mitotic entry. Progression into mitosis was analyzed by time-lapse microscopy using a laser-scanning confocal microscope (LSM 510). Frames were taken every 30 s for 3 min, and time relative to NEBD is shown. Bar, 10 µm.
Localization of PKC-βII during NEBD in mammalian cells. HeLa cells stably expressing H2B-mCherry and transiently expressing PKC-βII tagged with GFP (green and gray) at the C terminus during mitotic entry. Progression into mitosis was analyzed by time-lapse microscopy using a laser-scanning confocal microscope (LSM 510). Frames were taken every 30 s for 3 min, and time relative to NEBD is shown. Bar, 10 µm.
Localization of PKC-βII during NEBD in mammalian cells. HeLa cells stably expressing H2B-mCherry and transiently expressing PKC-βII tagged with GFP (green and gray) at the C terminus during mitotic entry. Progression into mitosis was analyzed by time-lapse microscopy using a laser-scanning confocal microscope (LSM 510). Frames were taken every 30 s for 3 min, and time relative to NEBD is shown. Bar, 10 µm.
Localization of lipin 1 during NEBD in mammalian cells. HeLa cells stably expressing H2B-mCherry and transiently expressing lipin 1 tagged with GFP (green and gray) at the C terminus during mitotic entry. Progression into mitosis was analyzed by time-lapse microscopy using a laser-scanning confocal microscope (LSM 510). Frames were taken every 30 s for 3 min, and time relative to NEBD is shown. Bar, 10 µm.
Localization of lipin 1 during NEBD in mammalian cells. HeLa cells stably expressing H2B-mCherry and transiently expressing lipin 1 tagged with GFP (green and gray) at the C terminus during mitotic entry. Progression into mitosis was analyzed by time-lapse microscopy using a laser-scanning confocal microscope (LSM 510). Frames were taken every 30 s for 3 min, and time relative to NEBD is shown. Bar, 10 µm.
Localization of lipin 2 during NEBD in mammalian cells. HeLa cells stably expressing H2B-mCherry and transiently expressing lipin 2 tagged with GFP (green and gray) at the C terminus during mitotic entry. Progression into mitosis was analyzed by time-lapse microscopy using a laser-scanning confocal microscope (LSM 510). Frames were taken every 30 s for 3 min, and time relative to NEBD is shown. Bar, 10 µm.
Localization of lipin 2 during NEBD in mammalian cells. HeLa cells stably expressing H2B-mCherry and transiently expressing lipin 2 tagged with GFP (green and gray) at the C terminus during mitotic entry. Progression into mitosis was analyzed by time-lapse microscopy using a laser-scanning confocal microscope (LSM 510). Frames were taken every 30 s for 3 min, and time relative to NEBD is shown. Bar, 10 µm.
Localization of lipin 3 during NEBD in mammalian cells. HeLa cells stably expressing H2B-mCherry and transiently expressing lipin 3 tagged with GFP (green and gray) at the C terminus during mitotic entry. Progression into mitosis was analyzed by time-lapse microscopy using a laser-scanning confocal microscope (LSM 510). Frames were taken every 30 s for 3 min, and time relative to NEBD is shown. Bar, 10 µm.
Localization of lipin 3 during NEBD in mammalian cells. HeLa cells stably expressing H2B-mCherry and transiently expressing lipin 3 tagged with GFP (green and gray) at the C terminus during mitotic entry. Progression into mitosis was analyzed by time-lapse microscopy using a laser-scanning confocal microscope (LSM 510). Frames were taken every 30 s for 3 min, and time relative to NEBD is shown. Bar, 10 µm.
Acknowledgments
We thank A. Wünsche, P. Ikrenyi, and J. Thiry for generation of reagents, M. Held for assistance using CellCognition, and members of the Advanced Light Microscopy Facility, Genomics Core Facility, and Laboratory Animal Resources (European Molecular Biology Laboratory, Heidelberg, Germany) for excellent technical assistance. We also thank U. Kutay and E. Laurell for support during in vitro NEBD experiments, D. Gerlich, H. Meyer, and K. Weis for helpful discussions, and members of the Mattaj and Ellenberg groups for helpful discussions and critical reading of the manuscript.
J. Ellenberg acknowledges funding by the European Commission within the MitoSys and SystemsMicroscopy consortia (FP7/2007-2013-241548 and FP7/2007-2013-258068).