The kinetochore forms a dynamic interface with microtubules from the mitotic spindle. Live-cell light microscopy–based observations on the dynamic structural changes within the kinetochore suggest that molecular rearrangements within the kinetochore occur upon microtubule interaction. However, the source of these rearrangements is still unclear. In this paper, we analyze vertebrate kinetochore ultrastructure by immunoelectron microscopy (EM) in the presence or absence of tension from spindle microtubules. We found that the inner kinetochore region defined by CENP-A, CENP-C, CENP-R, and the C-terminal domain of CENP-T is deformed in the presence of tension, whereas the outer kinetochore region defined by Ndc80, Mis12, and CENP-E is not stretched even under tension. Importantly, based on EM, fluorescence microscopy, and in vitro analyses, we demonstrated that the N and C termini of CENP-T undergo a tension-dependent separation, suggesting that CENP-T elongation is at least partly responsible for changes in the shape of the inner kinetochore.
Introduction
Faithful chromosome segregation during mitosis is essential for the accurate transmission of genetic material. The kinetochore forms a dynamic interface with microtubules from the mitotic spindle to facilitate chromosome segregation (Cheeseman and Desai, 2008). Importantly, kinetochore–microtubule attachments are capable of generating as much as 700 pN of force (Nicklas, 1988), which functions to both move chromosomes and to generate signals that report on proper attachment status. As might be predicted based on this large amount of force generated at kinetochores, live-cell light microscopy–based observations have suggested that structural rearrangements within the kinetochore occur in the presence of tension (Maresca and Salmon, 2009; Uchida et al., 2009). These works also suggested that these rearrangements are essential for release from a spindle assembly checkpoint–dependent cell cycle arrest. However, the molecular source of these rearrangements is still unclear, as these changes were only evaluated as an increase in intrakinetochore distance (the distance between the inner and outer kinetochore) by light microscopy.
To examine the molecular rearrangements within the kinetochore in detail, it is essential to observe changes in kinetochore ultrastructure using EM. Early EM studies of vertebrate cells revealed that the kinetochore has a trilaminar morphology, including an inner and outer plate (Brinkley and Stubblefield, 1966; Jokelainen, 1967; Comings and Okada, 1971; Rieder, 1982; McEwen et al., 2007). Microtubules attach directly to the outer plate (Brinkley and Stubblefield, 1966; Jokelainen, 1967; Comings and Okada, 1971; McEwen et al., 1998), whereas chromatin-bound proteins, such as CENP-C, localize to the inner plate (Saitoh et al., 1992).
To analyze the structural dynamics of the kinetochore that occur in response to force-dependent changes during mitosis, we combined EM analysis with specific observations of representative kinetochore markers using immuno-EM. We have previously identified multiple kinetochore components and generated a toolkit of antibodies suitable for analyzing the kinetochore ultrastructure (Okada et al., 2006; Hori et al., 2008a; Amano et al., 2009; Ohta et al., 2010). Based on our immuno-EM analyses, we found that the inner kinetochore region is dramatically deformed in the presence of tension from spindle microtubules. In addition, we found that the N and C termini of CENP-T undergo a tension-dependent separation based on EM, fluorescence microscopy, and in vitro analyses, suggesting that CENP-T elongation is responsible for changes in the shape of the inner kinetochore. Thus, this work defines a key force-dependent molecular change within the kinetochore.
Results
High resolution mapping of proteins within the kinetochore by immuno-EM
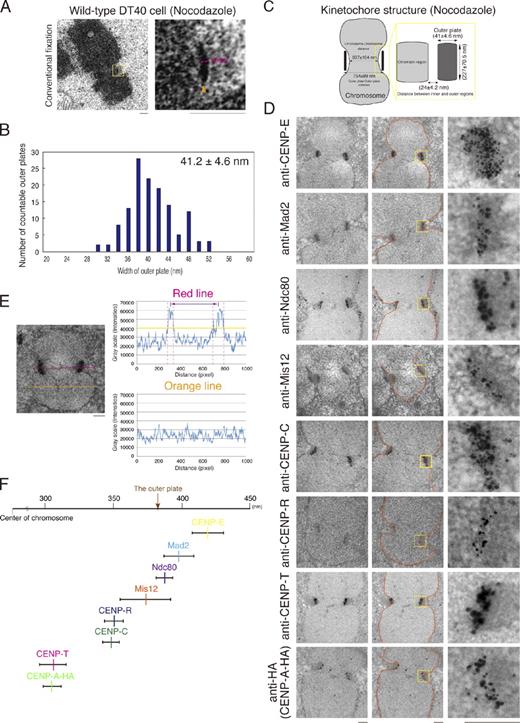
To conduct a comprehensive analysis of the tension-dependent structural changes that occur within the kinetochore, we began by analyzing the kinetochore structure of mitotic chromosomes in chicken DT40 cells in the absence of microtubules (using treatment with the microtubule-depolymerizing drug nocodazole at 500 ng/ml for 3 h). After a conventional fixation with glutaraldehyde and staining with uranyl acetate and lead citrate, we imaged 170-nm-thick serial sections for individual mitotic cells. We observed clear electron-dense kinetochore outer plates in the region of the primary constriction of each chromosome (Fig. 1 A). To define the structure of each kinetochore, we measured the distance between the paired outer kinetochore plates visualized in Fig. 1 A. We also measured the length of the chromatin region at the primary constriction. The mean distance between the paired outer kinetochore plates was 754 nm, the mean length of the chromatin region was 637 nm, and the mean width of the outer plate was 41 nm (Fig. 1, B and C). To conduct an unbiased analysis of this EM data, we used digital images and obtained distribution curves of signal intensity. Using the signal intensity values, we measured the size of the outer plates (Figs. 1, B and C; and S1).
Ultrastructure of the kinetochore in DT40 cells. (A) An image of a chromosome with paired outer plates from DT40 cells observed by EM. The arrowhead shows the outer plate. (B) Distribution of data for the width of the outer plate of 131 kinetochores. The method describing how the size of the outer plate was measured is described in Fig. S1. (C) Summary for the size of the kinetochore region in DT40 cells treated with nocodazole and fixed by glutaraldehyde. We chose chromosomes with the sister kinetochore plates and measured the length and width of the plates by the method described in Fig. S1. The mean of the length and width of the outer region is ∼227 nm (227 ± 70.5 nm) and ∼41 nm (41 ± 4.6 nm), respectively. (D) Immuno-EM images of nocodazole-treated DT40 cells using anti–CENP-E, anti-Mad2, anti-Ndc80, anti-Mis12, anti–CENP-C, anti–CENP-R, anti–CENP-T, and anti-HA (for detection of CENP-A–HA) antibodies. Signals are shown as gold labeling. Orange lines show the distribution of background signals. (E) Measurement of the sister kinetochore distance. Chromosomes with the sister kinetochore plates that were parallel to the plane of one section were chosen. Signal intensities of the red line were measured. The regions between two pink dashed lines show the regions with positive signals (higher than background signals). The yellow line shows the maximum of background signals. Signal intensities of the orange line on the chromosome arm were measured as background signals. The sister kinetochore region is defined by two peaks in the graph for the red line. The distance between two peaks is defined as the distance between the sister kinetochores. (F) Position of each protein in kinetochores. The half-distance between the sister kinetochore signals for each protein was plotted in the graph. CENP-A and CENP-T are located in the most internal region. Mis12, Ndc80, Mad2, and CENP-E are located near the outer plate. Yellow boxes indicate the regions of higher magnification on the right. Bars, 250 nm.
Ultrastructure of the kinetochore in DT40 cells. (A) An image of a chromosome with paired outer plates from DT40 cells observed by EM. The arrowhead shows the outer plate. (B) Distribution of data for the width of the outer plate of 131 kinetochores. The method describing how the size of the outer plate was measured is described in Fig. S1. (C) Summary for the size of the kinetochore region in DT40 cells treated with nocodazole and fixed by glutaraldehyde. We chose chromosomes with the sister kinetochore plates and measured the length and width of the plates by the method described in Fig. S1. The mean of the length and width of the outer region is ∼227 nm (227 ± 70.5 nm) and ∼41 nm (41 ± 4.6 nm), respectively. (D) Immuno-EM images of nocodazole-treated DT40 cells using anti–CENP-E, anti-Mad2, anti-Ndc80, anti-Mis12, anti–CENP-C, anti–CENP-R, anti–CENP-T, and anti-HA (for detection of CENP-A–HA) antibodies. Signals are shown as gold labeling. Orange lines show the distribution of background signals. (E) Measurement of the sister kinetochore distance. Chromosomes with the sister kinetochore plates that were parallel to the plane of one section were chosen. Signal intensities of the red line were measured. The regions between two pink dashed lines show the regions with positive signals (higher than background signals). The yellow line shows the maximum of background signals. Signal intensities of the orange line on the chromosome arm were measured as background signals. The sister kinetochore region is defined by two peaks in the graph for the red line. The distance between two peaks is defined as the distance between the sister kinetochores. (F) Position of each protein in kinetochores. The half-distance between the sister kinetochore signals for each protein was plotted in the graph. CENP-A and CENP-T are located in the most internal region. Mis12, Ndc80, Mad2, and CENP-E are located near the outer plate. Yellow boxes indicate the regions of higher magnification on the right. Bars, 250 nm.
Using these conditions in DT40 cells, it was not possible to visualize clear trilaminar plates, necessitating the use of molecular markers to analyze kinetochore structure. Therefore, we next probed nocodazole-arrested cells with primary antibodies against CENP-C, CENP-E, CENP-R, CENP-T, Mad2, Mis12, or Ndc80 (Fukagawa et al., 1999; Hori et al., 2003, 2008a,b; Kline et al., 2006) and gold-labeled secondary antibodies. Specific information regarding these antibodies, including the antigens used to generate them, is summarized in Materials and methods. In each case, we succeeded in visualizing the labeled proteins with gold particles at DT40 kinetochores (Fig. 1 D). We also stained DT40 cells expressing CENP-A–HA with anti-HA antibodies to visualize CENP-A at kinetochores, as our existing CENP-A antibody was not functional for immuno-EM (Fig. 1 D). We next measured the distance between the two paired foci on sister kinetochores for the eight kinetochore proteins shown in Fig. 1 D. For these measurements, we used a line scan for digital EM images and obtained a signal distribution for the gold particles with two peaks (Fig. 1 E). Then, we measured the distance between the two peaks. The position of each kinetochore protein relative to the center of the chromosome is defined as half of the sister kinetochore distance. These measurements are summarized in Figs. 1 F and S1 E. The mean position of the outer plate was ∼377 ± 50 nm from the center of chromosome, which corresponds to the half-distance of the outer surface of the outer plate. CENP-E, Mad2, Mis12, and Ndc80 were located outside or near the outer plate (Fig. 1 F), which is consistent with previous studies in human cells (Cooke et al., 1997; DeLuca et al., 2005). In contrast, CENP-A, CENP-C, CENP-R, and CENP-T were located internally to the outer plate (Fig. 1 F). CENP-C was previously mapped to the inner kinetochore region using immuno-EM analysis in human cells (Saitoh et al., 1992). We recently demonstrated that CENP-T binds centromere DNA directly (Hori et al., 2008a), and kinetochore single-molecule high resolution colocalization analysis in human cells also suggests that CENP-A, CENP-C, and CENP-T are close to centromeric chromatin (Wan et al., 2009). Thus, our EM analysis, combined with previous studies, demonstrates that CENP-A, CENP-C, CENP-R, and CENP-T are located at the inner kinetochore and that CENP-E, Mad2, Mis12, and Ndc80 are located at the outer kinetochore (Fig. 1 F).
The width of the inner kinetochore region, but not the outer kinetochore, is stretched in the presence of spindle microtubules
The aforementioned studies define a baseline state of kinetochore structure in DT40 cells in the absence of microtubules. To analyze the structural changes that occur in response to tension from microtubule attachments, we next performed EM analysis on cells treated with the proteasome inhibitor MG132, which inhibits the degradation of anaphase-promoting complex/cyclosome substrates, preventing anaphase onset. We confirmed that chromosomes aligned at the metaphase plate with kinetochore-bound microtubules in cells treated with MG132 (unpublished data). We then performed immuno-EM to visualize the localization of diverse inner and outer kinetochore proteins (Figs. 2 and 3). The distance between sister kinetochores (both inner and outer kinetochores) was increased in cells treated with MG132 relative to cells treated with nocodazole (a 180-nm increase for the CENP-T–CENP-T distance; a 160-nm increase for the Ndc80–Ndc80 distance), suggesting that MG132-treated cells generate tension from spindle microtubules. Strikingly, the rectangular-like shape of the inner kinetochore structure occupied by CENP-A, CENP-C, CENP-R, and CENP-T in cells treated with nocodazole was deformed into an ovallike shape in cells treated with MG132 (Fig. 2). Although our antibodies are polyclonal antibodies and it is hard to determine whether there are specific epitopes within each protein, we found that our existing CENP-T antibodies recognized the C-terminal region but not the N terminus (unpublished data). We measured the length and width of the kinetochore region occupied by gold particles. A detailed method for these measurements is described in Fig. S2. The distribution of the inner kinetochore region occupied by the C terminus of CENP-T was significantly altered in the presence of microtubules with a length of 164 ± 40 nm (n = 100) and a width of 86 ± 26 nm (n = 100) in cells treated with MG132 compared with a length of 228 ± 35 nm (n = 100) and a width of 61 ± 18 nm (n = 100) in cells treated with nocodazole (Fig. 2, B and C). This change is statistically significant based on a Student’s t test (Fig. 2). Immuno-EM against CENP-C, CENP-R, and CENP-A–HA revealed similar structural changes in the inner kinetochore in cells treated with MG132 (Fig. 2, D–L). In contrast, the width of the rectangular outer kinetochore region visualized with antibodies against Ndc80, Mis12, and CENP-E is not stretched in cells treated with MG132 or nocodazole (Fig. 3, A–I), whereas the length of the outer kinetochore structure was decreased in cells treated with MG132 (Fig. 2, B, E, and H). The measurements for the length and width of the inner and outer kinetochore in nocodazole- or MG132-treated cells are summarized in Fig. 3 J. In total, these data indicate that the width of the inner kinetochore region, but not the outer kinetochore, is stretched in the presence of spindle microtubules.
The structural deformation of the inner kinetochore when cells are treated with MG132. (A) Immuno-EM images with anti–CENP-T antibodies in DT40 cells treated with nocodazole (Noc) or MG132. (B) Measurement of the length of the inner kinetochore containing CENP-T in cells treated with nocodazole (n = 100) or MG132 (n = 100). (C) Measurement of the width of the inner kinetochore containing CENP-T in cells treated with nocodazole (n = 100) or MG132 (n = 100). (D) Immuno-EM images with anti–CENP-C antibodies in DT40 cells treated with nocodazole or MG132. (E) Measurement of the length of the inner kinetochore containing CENP-C in cells treated with nocodazole (n = 100) or MG132 (n = 100). (F) Measurement of the width of the inner kinetochore containing CENP-C in cells treated with nocodazole (n = 100) or MG132 (n = 100). (G) Immuno-EM images with anti-HA antibodies in CENP-A–HA-expressing DT40 cells treated with nocodazole or MG132. (H) Measurement of the length of the inner kinetochore containing CENP-A–HA in cells treated with nocodazole (n = 70) or MG132 (n = 70). (I) Measurement of the width of the inner kinetochore containing CENP-A–HA in cells treated with nocodazole (n = 70) or MG132 (n = 70). (J) Immuno-EM images with anti–CENP-R antibodies in DT40 cells treated with nocodazole or MG132. (K) Measurement of the length of the inner kinetochore containing CENP-R in cells treated with nocodazole (n = 100) or MG132 (n = 100). (L) Measurement of the width of the inner kinetochore containing CENP-R in cells treated with nocodazole (n = 100) or MG132 (n = 100). Error bars show SDs, and the significance of these measurements was estimated by the Student’s t test (indicated by asterisks). Yellow boxes indicate the regions of higher magnification on the right. Bars, 250 nm.
The structural deformation of the inner kinetochore when cells are treated with MG132. (A) Immuno-EM images with anti–CENP-T antibodies in DT40 cells treated with nocodazole (Noc) or MG132. (B) Measurement of the length of the inner kinetochore containing CENP-T in cells treated with nocodazole (n = 100) or MG132 (n = 100). (C) Measurement of the width of the inner kinetochore containing CENP-T in cells treated with nocodazole (n = 100) or MG132 (n = 100). (D) Immuno-EM images with anti–CENP-C antibodies in DT40 cells treated with nocodazole or MG132. (E) Measurement of the length of the inner kinetochore containing CENP-C in cells treated with nocodazole (n = 100) or MG132 (n = 100). (F) Measurement of the width of the inner kinetochore containing CENP-C in cells treated with nocodazole (n = 100) or MG132 (n = 100). (G) Immuno-EM images with anti-HA antibodies in CENP-A–HA-expressing DT40 cells treated with nocodazole or MG132. (H) Measurement of the length of the inner kinetochore containing CENP-A–HA in cells treated with nocodazole (n = 70) or MG132 (n = 70). (I) Measurement of the width of the inner kinetochore containing CENP-A–HA in cells treated with nocodazole (n = 70) or MG132 (n = 70). (J) Immuno-EM images with anti–CENP-R antibodies in DT40 cells treated with nocodazole or MG132. (K) Measurement of the length of the inner kinetochore containing CENP-R in cells treated with nocodazole (n = 100) or MG132 (n = 100). (L) Measurement of the width of the inner kinetochore containing CENP-R in cells treated with nocodazole (n = 100) or MG132 (n = 100). Error bars show SDs, and the significance of these measurements was estimated by the Student’s t test (indicated by asterisks). Yellow boxes indicate the regions of higher magnification on the right. Bars, 250 nm.
The distribution of outer kinetochore proteins in the absence or presence of spindle microtubules. (A) Immuno-EM images with anti-Ndc80 antibodies in DT40 cells treated with nocodazole (Noc) or MG132. (B) Measurement of the length of the outer kinetochore containing Ndc80 in DT40 cells treated with nocodazole (n = 104) or MG132 (n = 118). (C) Measurement of the width of the outer kinetochore containing Ndc80 in DT40 cells treated with nocodazole (n = 104) or MG132 (n = 118). (D) Immuno-EM images with anti-Mis12 antibodies in DT40 cells treated with nocodazole or MG132. (E) Measurement of the length of the outer kinetochore containing Mis12 in DT40 cells treated with nocodazole (n = 100) or MG132 (n = 100). (F) Measurement of the width of the outer kinetochore containing Mis12 in DT40 cells treated with nocodazole (n = 100) or MG132 (n = 100). (G) Immuno-EM images with anti–CENP-E antibodies in DT40 cells treated with nocodazole or MG132. (H) Measurement of the length of the outer kinetochore containing CENP-E in DT40 cells treated with nocodazole (n = 100) or MG132 (n = 100). (I) Measurement of the width of the outer kinetochore containing CENP-E in DT40 cells treated with nocodazole (n = 100) or MG132 (n = 100). The width is not increased and significantly decreased. (J) The structural change of the inner kinetochore in the presence of microtubules. The inner kinetochore was changed to the ovallike structure from the rectangular structure, and the width of the inner kinetochore (magenta) was significantly stretched. In contrast, the width of the outer kinetochore (blue) was not stretched even in presence of microtubules. Error bars show SDs, and the significance of the length measurements was estimated by the Student’s t test (indicated by asterisks), whereas it was not estimated by the Student’s t test for the width. Yellow boxes indicate the regions of higher magnification on the right. Bars, 250 nm.
The distribution of outer kinetochore proteins in the absence or presence of spindle microtubules. (A) Immuno-EM images with anti-Ndc80 antibodies in DT40 cells treated with nocodazole (Noc) or MG132. (B) Measurement of the length of the outer kinetochore containing Ndc80 in DT40 cells treated with nocodazole (n = 104) or MG132 (n = 118). (C) Measurement of the width of the outer kinetochore containing Ndc80 in DT40 cells treated with nocodazole (n = 104) or MG132 (n = 118). (D) Immuno-EM images with anti-Mis12 antibodies in DT40 cells treated with nocodazole or MG132. (E) Measurement of the length of the outer kinetochore containing Mis12 in DT40 cells treated with nocodazole (n = 100) or MG132 (n = 100). (F) Measurement of the width of the outer kinetochore containing Mis12 in DT40 cells treated with nocodazole (n = 100) or MG132 (n = 100). (G) Immuno-EM images with anti–CENP-E antibodies in DT40 cells treated with nocodazole or MG132. (H) Measurement of the length of the outer kinetochore containing CENP-E in DT40 cells treated with nocodazole (n = 100) or MG132 (n = 100). (I) Measurement of the width of the outer kinetochore containing CENP-E in DT40 cells treated with nocodazole (n = 100) or MG132 (n = 100). The width is not increased and significantly decreased. (J) The structural change of the inner kinetochore in the presence of microtubules. The inner kinetochore was changed to the ovallike structure from the rectangular structure, and the width of the inner kinetochore (magenta) was significantly stretched. In contrast, the width of the outer kinetochore (blue) was not stretched even in presence of microtubules. Error bars show SDs, and the significance of the length measurements was estimated by the Student’s t test (indicated by asterisks), whereas it was not estimated by the Student’s t test for the width. Yellow boxes indicate the regions of higher magnification on the right. Bars, 250 nm.
The stretched distribution of the inner kinetochore depends on tension from spindle microtubules
Our aforementioned analysis demonstrates that inner kinetochore distribution is altered by the presence of spindle microtubules. Although this is likely caused by the force exerted by these microtubules on bioriented kinetochores, we next sought to define the activities responsible for this deformation using specific perturbations that affect kinetochore function. We first tested DT40 cells depleted for the outer kinetochore protein Ndc80. We confirmed that Ndc80 is undetectable by Western blot analysis after the addition of tetracycline to Ndc80 conditional knockout cells (Hori et al., 2003). In addition, Nuf2, another subunit of Ndc80, is degraded in Ndc80-deficient cells, suggesting that complex formation of the Ndc80 complex is compromised (Hori et al., 2003). We previously observed that Ndc80 depletion compromises proper attachment of microtubules to kinetochores and arrests cells at mitosis (Hori et al., 2003). Importantly, depletion of Ndc80 prevented the stretched distribution of the inner kinetochore even in cells blocked in mitosis with MG132 (Fig. 4, A–C). Thus, the lack of proper attachments prevents the deformation of the inner kinetochore.
The proper binding of Ndc80 with the microtubule is essential for the structural deformation within the inner kinetochore. (A) Immuno-EM images with anti–CENP-T antibodies in Ndc80-deficient cells treated with nocodazole (Noc) or MG132. The structural deformation of the inner kinetochore was not observed in Ndc80-deficient cells treated with MG132. (B) Measurement of the length of the inner kinetochore containing CENP-T in Ndc80-deficient cells treated with nocodazole (n = 70) or MG132 (n = 70). (C) Measurement of the width of the inner kinetochore containing CENP-T in Ndc80-deficient cells treated with nocodazole (n = 70) or MG132 (n = 70). (D) Images of the outer plates in wild-type, Ndc80-deficient, and Ndc80 D4 mutant cells. The arrows show the outer plates. (E) The size of the outer plate (means ± SD) in wild-type and Ndc80 D4 mutant cells. (F) Immuno-EM images with anti–CENP-T antibodies in Ndc80 D4 mutant cells treated with nocodazole or MG132. (G) Measurement of the length of the inner kinetochore containing with CENP-T in Ndc80 D4 mutant cells treated with nocodazole (n = 70) or MG132 (n = 70) and untreated cells (n = 70). (H) Measurement of the width of the inner kinetochore containing CENP-T in Ndc80 D4 mutant cells treated with nocodazole (n = 70) or MG132 (n = 70) and untreated cells (n = 70). (I) Immunofluorescence observation using antibodies against CENP-T and α-tubulin in Ndc80 D4 mutant cells treated with MG132. Error bars show SDs, and the significant difference was not detected. Green and magenta rectangles indicate distribution in cells. Yellow boxes indicate the regions of higher magnification on the right. Bars: (A, D, and F) 250 nm; (I) 10 µm.
The proper binding of Ndc80 with the microtubule is essential for the structural deformation within the inner kinetochore. (A) Immuno-EM images with anti–CENP-T antibodies in Ndc80-deficient cells treated with nocodazole (Noc) or MG132. The structural deformation of the inner kinetochore was not observed in Ndc80-deficient cells treated with MG132. (B) Measurement of the length of the inner kinetochore containing CENP-T in Ndc80-deficient cells treated with nocodazole (n = 70) or MG132 (n = 70). (C) Measurement of the width of the inner kinetochore containing CENP-T in Ndc80-deficient cells treated with nocodazole (n = 70) or MG132 (n = 70). (D) Images of the outer plates in wild-type, Ndc80-deficient, and Ndc80 D4 mutant cells. The arrows show the outer plates. (E) The size of the outer plate (means ± SD) in wild-type and Ndc80 D4 mutant cells. (F) Immuno-EM images with anti–CENP-T antibodies in Ndc80 D4 mutant cells treated with nocodazole or MG132. (G) Measurement of the length of the inner kinetochore containing with CENP-T in Ndc80 D4 mutant cells treated with nocodazole (n = 70) or MG132 (n = 70) and untreated cells (n = 70). (H) Measurement of the width of the inner kinetochore containing CENP-T in Ndc80 D4 mutant cells treated with nocodazole (n = 70) or MG132 (n = 70) and untreated cells (n = 70). (I) Immunofluorescence observation using antibodies against CENP-T and α-tubulin in Ndc80 D4 mutant cells treated with MG132. Error bars show SDs, and the significant difference was not detected. Green and magenta rectangles indicate distribution in cells. Yellow boxes indicate the regions of higher magnification on the right. Bars: (A, D, and F) 250 nm; (I) 10 µm.
Although Ndc80 depletion prevents the formation of kinetochore–microtubule attachments, completely eliminating Ndc80 also alters the structure of the kinetochore outer plate (Fig. 4 D; DeLuca et al., 2005). To distinguish between these two functions, we used a conditional replacement mutant of Ndc80 in which four Aurora B phosphorylation sites were replaced with aspartic acid to mimic constitutive phosphorylation (D4 mutant; Welburn et al., 2010). The Ndc80 D4 mutant retains the structural contribution of the Ndc80 complex to the outer kinetochore but reduces interactions with microtubules (Cheeseman et al., 2006; DeLuca et al., 2006; Guimaraes et al., 2008; Miller et al., 2008; Welburn et al., 2010). We confirmed that the outer plate is formed properly in the Ndc80 D4 mutant cells by EM based on its morphology (Fig. 4, D and E). When Ndc80 D4 mutant cells were treated with MG132, we did not detect a deformation of the inner kinetochore compared with cells treated with nocodazole (Fig. 4, F–H). Our results, combined with the previous biochemical data, indicate that proper binding of Ndc80 to microtubules is essential for the structural change of the inner kinetochore. Although the outer plate is formed and microtubules appear to attach to the outer plate in the Ndc80 D4 mutant cells (Fig. 4 I), we found that the distance between sister kinetochores was only slightly increased in the Ndc80 D4 mutant cells treated with MG132 relative to cells treated with nocodazole (a 25-nm increase compared with a 180-nm increase in wild-type cells), suggesting that there is no tension or weak tension in Ndc80 D4 mutant cells.
We next observed the inner kinetochore distribution in conditional mutant cells for the inner kinetochore protein CENP-H. Although the length of the inner kinetochore region was reduced in CENP-H–deficient cells, the width of the rectangular inner kinetochore structure was not stretched in CENP-H–deficient cells treated with MG132 (Fig. 5, A–C), indicating that the structural deformation of the inner kinetochore from the rectangular to the ovallike shape cannot occur in CENP-H–deficient cells even in the presence of microtubules. In CENP-H–deficient cells, the localization of a subset of outer kinetochore proteins is altered (Hori et al., 2003), and the plate structure of outer kinetochores is disorganized (Liu et al., 2006; Hori et al., 2008a), suggesting that proper attachment of microtubules to kinetochores is perturbed similar to Ndc80-deficient cells.
The distribution of inner kinetochore proteins in various knockout cell lines. (A) Immuno-EM images with anti–CENP-T antibodies in CENP-H–deficient cells treated with nocodazole (Noc) or MG132. Although the size of the inner kinetochore in the CENP-H–deficient cells was reduced (length of 120 nm) compared with that of wild-type cells (length of 220 nm; see panel B), the width of the rectangular inner kinetochore structure was not changed in cells treated with MG132, indicating that the structural deformation of the inner kinetochore was not observed in CENP-H–deficient cells treated with MG132. (B) Measurement of the length of the inner kinetochore containing CENP-T in CENP-H–deficient cells treated with nocodazole (n = 70) or MG132 (n = 70). (C) Measurement of the width of the inner kinetochore containing CENP-T in CENP-H–deficient cells treated with nocodazole (n = 70) or MG132 (n = 70). (B and C) The significant difference was not detected. (D) Immuno-EM images with anti–CENP-T antibodies in CENP-C–deficient cells treated with nocodazole or MG132. The size of inner kinetochore of CENP-C–deficient cells was smaller (length of 160 nm). However, the structural deformation of the inner kinetochore was observed in CENP-C–deficient cells treated with MG132. (E) Measurement of the length of the inner kinetochore containing CENP-T in CENP-C–deficient cells treated with nocodazole (n = 70) or MG132 (n = 70). (F) Measurement of the width of the inner kinetochore containing CENP-T in CENP-C–deficient cells treated with nocodazole (n = 70) or MG132 (n = 70). (G) Immuno-EM images with anti–CENP-T antibodies in CENP-S–deficient cells treated with nocodazole or MG132. The structural deformation of the inner kinetochore was observed in CENP-S–deficient cells treated with MG132. (H) Measurement of the length of the inner kinetochore containing CENP-T in CENP-S–deficient cells treated with nocodazole (n = 113) or MG132 (n = 113). (I) Measurement of the width of the inner kinetochore containing CENP-T in CENP-S–deficient cells treated with nocodazole (n = 113) or MG132 (n = 113). (E, F, H, and I) The significance of these measurements was estimated by the Student’s t test (indicated by asterisks). (J) Summary of results for the structural deformation in the inner kinetochore in various cell lines. When tension from spindle microtubules was produced, the structural deformation in the inner kinetochore was observed. When we observed that the distance between the paired sister kinetochores increased >100 nm in cells treated with MG132 relative to cells treated with nocodazole, we judged that tension occurs (+). Therefore, although we observed 4- and 25-nm increases in Ndc80-deficient and Ndc80 D4 cells, respectively, we judged that these values meant that tension does not occur (−). Yellow boxes indicate the regions of higher magnification on the right. Error bars show SDs. Bars, 250 nm.
The distribution of inner kinetochore proteins in various knockout cell lines. (A) Immuno-EM images with anti–CENP-T antibodies in CENP-H–deficient cells treated with nocodazole (Noc) or MG132. Although the size of the inner kinetochore in the CENP-H–deficient cells was reduced (length of 120 nm) compared with that of wild-type cells (length of 220 nm; see panel B), the width of the rectangular inner kinetochore structure was not changed in cells treated with MG132, indicating that the structural deformation of the inner kinetochore was not observed in CENP-H–deficient cells treated with MG132. (B) Measurement of the length of the inner kinetochore containing CENP-T in CENP-H–deficient cells treated with nocodazole (n = 70) or MG132 (n = 70). (C) Measurement of the width of the inner kinetochore containing CENP-T in CENP-H–deficient cells treated with nocodazole (n = 70) or MG132 (n = 70). (B and C) The significant difference was not detected. (D) Immuno-EM images with anti–CENP-T antibodies in CENP-C–deficient cells treated with nocodazole or MG132. The size of inner kinetochore of CENP-C–deficient cells was smaller (length of 160 nm). However, the structural deformation of the inner kinetochore was observed in CENP-C–deficient cells treated with MG132. (E) Measurement of the length of the inner kinetochore containing CENP-T in CENP-C–deficient cells treated with nocodazole (n = 70) or MG132 (n = 70). (F) Measurement of the width of the inner kinetochore containing CENP-T in CENP-C–deficient cells treated with nocodazole (n = 70) or MG132 (n = 70). (G) Immuno-EM images with anti–CENP-T antibodies in CENP-S–deficient cells treated with nocodazole or MG132. The structural deformation of the inner kinetochore was observed in CENP-S–deficient cells treated with MG132. (H) Measurement of the length of the inner kinetochore containing CENP-T in CENP-S–deficient cells treated with nocodazole (n = 113) or MG132 (n = 113). (I) Measurement of the width of the inner kinetochore containing CENP-T in CENP-S–deficient cells treated with nocodazole (n = 113) or MG132 (n = 113). (E, F, H, and I) The significance of these measurements was estimated by the Student’s t test (indicated by asterisks). (J) Summary of results for the structural deformation in the inner kinetochore in various cell lines. When tension from spindle microtubules was produced, the structural deformation in the inner kinetochore was observed. When we observed that the distance between the paired sister kinetochores increased >100 nm in cells treated with MG132 relative to cells treated with nocodazole, we judged that tension occurs (+). Therefore, although we observed 4- and 25-nm increases in Ndc80-deficient and Ndc80 D4 cells, respectively, we judged that these values meant that tension does not occur (−). Yellow boxes indicate the regions of higher magnification on the right. Error bars show SDs. Bars, 250 nm.
Although our analysis suggests that both proper kinetochore structure and the presence of microtubules are necessary for the structural deformation of the inner kinetochore, it is possible that the stretched distribution of the inner kinetochore reflects the physical movement of chromosomes but not the tension-derived force from spindle microtubules. To test this possibility, we used Rad21/Scc1-deficient DT40 cells in which cohesion is disrupted and tension, but not kinetochore–microtubule attachments, is eliminated (Sonoda et al., 2001). Even when Rad21-deficient cells are treated with nocodazole, sister chromatids separate, and the distance between sister kinetochores is increased relative to wild-type cells (Fig. 6, A and B). However, the width of the inner kinetochore is not stretched in Rad21-deficient cells (Fig. 6), suggesting that the physical attachment of kinetochores to microtubules and the movement of the chromosomes are not sufficient for the structural change of the inner kinetochore.
The distribution of inner kinetochore proteins in Rad21-deficient cells. (A) Immuno-EM images with anti–CENP-T antibodies in Rad21-deficient cells treated with nocodazole (Noc). (B) Measurement of the distance between the sister kinetochores stained by anti–CENP-T antibodies in wild-type (n = 51) or Rad21-deficienct DT40 cells (n = 51). The significance of these measurements was estimated by the Student’s t test. (C) Measurement of the width of the inner kinetochore containing CENP-T in wild-type (n = 100) or Rad21-deficient cells (n = 100) treated with nocodazole. The width of the inner kinetochore is significantly decreased. Yellow boxes indicate the regions of higher magnification on the right. Error bars show SDs. Bars, 250 nm.
The distribution of inner kinetochore proteins in Rad21-deficient cells. (A) Immuno-EM images with anti–CENP-T antibodies in Rad21-deficient cells treated with nocodazole (Noc). (B) Measurement of the distance between the sister kinetochores stained by anti–CENP-T antibodies in wild-type (n = 51) or Rad21-deficienct DT40 cells (n = 51). The significance of these measurements was estimated by the Student’s t test. (C) Measurement of the width of the inner kinetochore containing CENP-T in wild-type (n = 100) or Rad21-deficient cells (n = 100) treated with nocodazole. The width of the inner kinetochore is significantly decreased. Yellow boxes indicate the regions of higher magnification on the right. Error bars show SDs. Bars, 250 nm.
The requirements for the structural deformation of the inner kinetochore are summarized in Fig. 5 J. In total, even in the presence of microtubules, the rectangular distribution of the inner kinetochore is not deformed in Ndc80 D4 mutants or Ndc80- and CENP-H–deficient cells, which is consistent with these cells failing to generate tension on the kinetochore. In contrast, CENP-C– and CENP-S–deficient cells, which are able to generate more robust microtubule attachments (Kwon et al., 2007; Hori et al., 2008a; Amano et al., 2009) and produce tension from microtubules (Fig. 5 J), are able to undergo the structural deformation of the inner kinetochore in the presence of microtubules (Fig. 5, D–I). This is consistent with a previous study (Kwon et al., 2007) in which Ndc80 levels at kinetochores are reduced in CENP-H–deficient cells compared with CENP-C–deficient cells. We have shown previously that CENP-C has a distinct function from CENP-T for kinetochore assembly (Hori et al., 2008a). In fact, the stretched distribution of the inner kinetochore (visualized by CENP-C) does not occur in CENP-T–deficient cells treated with MG132 (Fig. S3). These data suggest that CENP-H, CENP-T, and Ndc80 proteins contribute substantially to generating intrakinetochore tension but CENP-C and CENP-S do not (Fig. 5 J). In total, our work demonstrates that a structural deformation of the inner kinetochore occurs when kinetochores form bioriented attachments to microtubules and tension is exerted from the spindle microtubules.
The N-terminal region of CENP-T shows tension-dependent elongation
Although our data suggest that the stretched distribution of the inner kinetochore occurs in the presence of tension from spindle microtubules, it is important to define the molecular basis for this structural deformation. To do this, we sought to identify molecules that are responsible for the deformation of the inner kinetochore. There are two criteria for a candidate protein to contribute to the tension-dependent stretched distribution of the inner kinetochore. First, the candidate protein should be flexible and elongated. Second, the terminal regions of the elongated protein should be associated with the outer kinetochore either directly or indirectly. Structural predications of inner kinetochore proteins using DISOPRED2 or PredictProtein suggested it was possible for CENP-T to form flexible and elongated structures because of a long region (80–500 aa; Fig. S4 A) that does not generate specific secondary structure predictions. In contrast, most other inner kinetochore proteins appear to form secondary structures (unpublished data). Therefore, we chose to focus on CENP-T to determine whether it contributes to the stretched distribution of the inner kinetochore.
If the CENP-T–W complex is responsible for the stretched distribution of the inner kinetochore, the C-terminal DNA-binding region should associate with the inner kinetochore, whereas the N-terminal region should behave more similarly to the outer kinetochore proteins. To distinguish the C terminus and N terminus of CENP-T, we constructed cell lines stably expressing CENP-T–tagged proteins at either the C terminus (monomeric RFP [mRFP] tag) or N terminus (FLAG tag; Fig. 7 A) and performed immuno-EM analyses with anti-mRFP or anti-FLAG antibodies (Fig. 7, B–G). We found that the C-terminal end of CENP-T was stretched in the presence of tension (MG132-treated cells; Fig. 7, B–D), which is consistent with the results of Fig. 2 A in which our anti–CENP-T antibodies recognize the C terminus of CENP-T. In contrast, the N-terminal end of CENP-T detected by anti-FLAG antibodies shows a rectangular distribution even in the presence of tension (Fig. 7, E–G), which is similar to outer kinetochore proteins, such as Ndc80 (Fig. 3 A). Unlike CENP-T, we found that the inner kinetochore protein CENP-N shows a deformation of its distribution within the kinetochore in the presence of tension when detected with either N-terminally or C-terminally tagged antibodies (Fig. 7, H–N), suggesting that not all inner kinetochore proteins contribute to the stretched distribution of the inner kinetochore. These results strongly suggest that the C-terminal region of CENP-T, which associates with chromatin (Hori et al., 2008a), shows an elongated distribution similar to other inner kinetochore proteins, whereas the N-terminal end of CENP-T associates with the outer kinetochore and displays a rectangular distribution even in presence of tension.
The distribution of the N terminus and C terminus of CENP-T or CENP-N in the kinetochore. (A) CENP-T constructs fused with FLAG in the N-terminal end or mRFP in the C-terminal end. (B) Immuno-EM images with anti-mRFP antibodies in CENP-T–deficient cells expressing CENP-T–mRFP (C-terminal fusion) treated with nocodazole (Noc) or MG132. (C) Measurement of the length of the kinetochore region containing CENP-T–mRFP in cells treated with nocodazole (n = 100) or MG132 (n = 100). (D) Measurement of the width of the kinetochore region containing CENP-T–mRFP in cells treated with nocodazole (n = 100) or MG132 (n = 100). (E) Immuno-EM images with anti-FLAG antibodies in CENP-T knockout cells expressing FLAG–CENP-T (N-terminal fusion) treated with nocodazole or MG132. (F) Measurement of the length of the kinetochore region containing FLAG–CENP-T in cells treated with nocodazole (n = 100) or MG132 (n = 100). (G) Measurement of the width of the kinetochore region containing FLAG–CENP-T in cells treated with nocodazole (n = 100) or MG132 (n = 100). (H) CENP-N constructs fused with FLAG in the N-terminal end (FLAG–CENP-N) or in the C-terminal end (CENP-N–FLAG). (I) Immuno-EM images with anti-FLAG antibodies in cells expressing CENP-N–FLAG (C-terminal fusion) treated with nocodazole. (J) Measurement of the length of the inner kinetochore containing CENP-N–FLAG in cells treated with nocodazole (n = 100) or MG132 (n = 100). (K) Measurement of the width of the outer kinetochore containing CENP-N–FLAG in cells treated with nocodazole (n = 100) or MG132 (n = 100). (L) Immuno-EM images with anti-FLAG antibodies in cells expressing FLAG–CENP-N (N-terminal fusion) treated with nocodazole or MG132. (M) Measurement of the length of the inner kinetochore containing FLAG–CENP-N in cells treated with nocodazole (n = 100) or MG132 (n = 100). (N) Measurement of the width of the outer kinetochore containing FLAG–CENP-N in cells treated with nocodazole (n = 100) or MG132 (n = 100). Green and magenta shapes show rectangular and oval distributions. Yellow boxes indicate the regions of higher magnification on the right. Error bars show SDs, and the significance of these measurements was estimated by the Student’s t test (indicated by asterisks) except for G, in which the significant difference was not detected. Bars, 250 nm.
The distribution of the N terminus and C terminus of CENP-T or CENP-N in the kinetochore. (A) CENP-T constructs fused with FLAG in the N-terminal end or mRFP in the C-terminal end. (B) Immuno-EM images with anti-mRFP antibodies in CENP-T–deficient cells expressing CENP-T–mRFP (C-terminal fusion) treated with nocodazole (Noc) or MG132. (C) Measurement of the length of the kinetochore region containing CENP-T–mRFP in cells treated with nocodazole (n = 100) or MG132 (n = 100). (D) Measurement of the width of the kinetochore region containing CENP-T–mRFP in cells treated with nocodazole (n = 100) or MG132 (n = 100). (E) Immuno-EM images with anti-FLAG antibodies in CENP-T knockout cells expressing FLAG–CENP-T (N-terminal fusion) treated with nocodazole or MG132. (F) Measurement of the length of the kinetochore region containing FLAG–CENP-T in cells treated with nocodazole (n = 100) or MG132 (n = 100). (G) Measurement of the width of the kinetochore region containing FLAG–CENP-T in cells treated with nocodazole (n = 100) or MG132 (n = 100). (H) CENP-N constructs fused with FLAG in the N-terminal end (FLAG–CENP-N) or in the C-terminal end (CENP-N–FLAG). (I) Immuno-EM images with anti-FLAG antibodies in cells expressing CENP-N–FLAG (C-terminal fusion) treated with nocodazole. (J) Measurement of the length of the inner kinetochore containing CENP-N–FLAG in cells treated with nocodazole (n = 100) or MG132 (n = 100). (K) Measurement of the width of the outer kinetochore containing CENP-N–FLAG in cells treated with nocodazole (n = 100) or MG132 (n = 100). (L) Immuno-EM images with anti-FLAG antibodies in cells expressing FLAG–CENP-N (N-terminal fusion) treated with nocodazole or MG132. (M) Measurement of the length of the inner kinetochore containing FLAG–CENP-N in cells treated with nocodazole (n = 100) or MG132 (n = 100). (N) Measurement of the width of the outer kinetochore containing FLAG–CENP-N in cells treated with nocodazole (n = 100) or MG132 (n = 100). Green and magenta shapes show rectangular and oval distributions. Yellow boxes indicate the regions of higher magnification on the right. Error bars show SDs, and the significance of these measurements was estimated by the Student’s t test (indicated by asterisks) except for G, in which the significant difference was not detected. Bars, 250 nm.
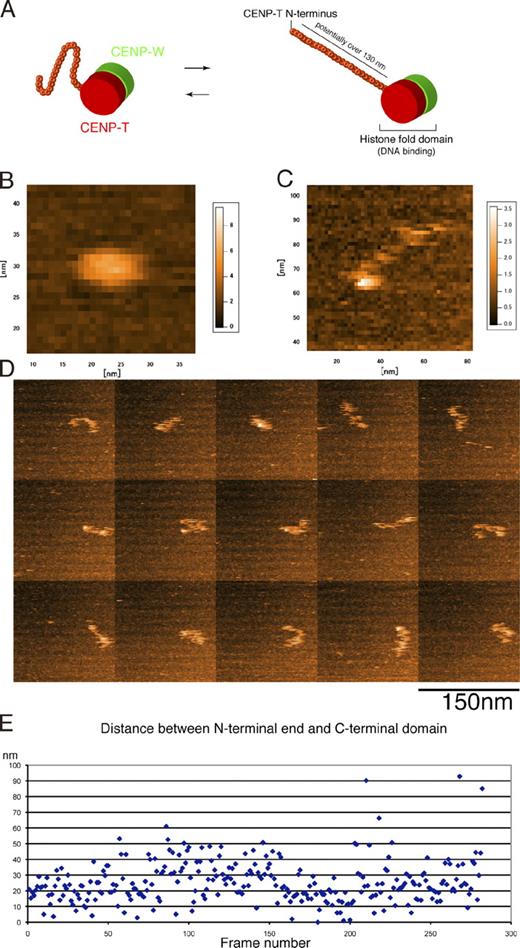
As our EM analyses suggest that the N terminus of CENP-T associates with the outer kinetochore, we next examined whether the long middle region of CENP-T (80–500 aa) has the capacity to form an elongated and flexible structure (Fig. 8 A). To test this, we reconstituted the CENP-T–W complex in vitro (Fig. S4, C and D) and directly visualized the structural dynamics of this complex by high speed atomic force microscopy (AFM; Ando et al., 2001, 2008). Using a complex composed of CENP-W and the histone-fold region of CENP-T, we observed a stable globular structure (Fig. 8 B) similar to a histone molecule. In contrast, a complex with full-length CENP-T and CENP-W showed both a globular region and a flexible elongated region (Fig. 8 C). The AFM images show that the elongated region is quite flexible (Fig. 8 D). We measured the length of the elongated region in each frame and found that its length is variable (25 ± 13 nm, mean ± SD; Fig. 8 E). To confirm this observation, we also used recombinant CENP-T fused to maltose-binding protein (MBP) at its N terminus. As MBP is a globular protein, we could measure the length between the two globular regions (MBP and the histone-fold region; Fig. S5). When we used CENP-W and only the histone-fold region of MBP–CENP-T, the length between MBP and the histone-fold region was constant (Fig. S5, C and E). In contrast, in the complex with MBP fused to full-length CENP-T, the length is variable (Fig. S5, B and D). These results indicate that the long middle region of CENP-T is elongated and flexible in vitro.
The CENP-T–W complex is elongated in vitro. (A) Prediction of the structure of the CENP-T–W complex. The C-terminal region of CENP-T may make a globular-like structure with the entire region of CENP-W because both regions have a histonelike domain. As it is hard to predict a distinct structure of the middle region of CENP-T (80–500 aa; Fig. S4), this region is predicted to be flexible. CENP-T could be potentially elongated to a length of 130 nm as determined by a computer calculation. (B) A high speed AFM image of the complex of CENP-W with the histone-fold region of CENP-T. A stable globular structure is observed. (C) A high speed AFM image of the complex of CENP-W with full-length CENP-T. In addition to a globular structure, an elongated region is observed. (D) Frame images by high speed AFM of the complex of CENP-W with full-length CENP-T. The length of elongated region was flexible. (E) Measurement of the length of the elongated region in each frame image (n = 282).
The CENP-T–W complex is elongated in vitro. (A) Prediction of the structure of the CENP-T–W complex. The C-terminal region of CENP-T may make a globular-like structure with the entire region of CENP-W because both regions have a histonelike domain. As it is hard to predict a distinct structure of the middle region of CENP-T (80–500 aa; Fig. S4), this region is predicted to be flexible. CENP-T could be potentially elongated to a length of 130 nm as determined by a computer calculation. (B) A high speed AFM image of the complex of CENP-W with the histone-fold region of CENP-T. A stable globular structure is observed. (C) A high speed AFM image of the complex of CENP-W with full-length CENP-T. In addition to a globular structure, an elongated region is observed. (D) Frame images by high speed AFM of the complex of CENP-W with full-length CENP-T. The length of elongated region was flexible. (E) Measurement of the length of the elongated region in each frame image (n = 282).
Based on the immuno-EM and AFM analyses, we hypothesized that the conformational change of the CENP-T–W complex may be responsible for the structural deformation of the inner kinetochore. We next sought to directly visualize the CENP-T stretch in the presence of tension by light microscopy. For this experiment, we fused GFP to the N terminus of CENP-T and mRFP to the C terminus and expressed the doubly tagged protein in CENP-T–deficient cells (Fig. 9 A). We confirmed that doubly tagged CENP-T could rescue the CENP-T depletion phenotype, suggesting that this fusion protein is functional (unpublished data). We observed that the GFP and mRFP signals were very close to each other in nocodazole-treated cells (Fig. 9 B). Strikingly, we frequently observed mRFP signals internal to the GFP signals in cells treated with MG132 (Fig. 9 B). We measured the distance between the two paired mRFP signals (L1) or GFP signals (L2) at sister kinetochores and defined (L2 − L1)/2 as CENP-T elongation (Fig. 9 C). When cells were treated with nocodazole, the mean CENP-T elongation was ∼4 nm (4 ± 4 nm, mean ± SEM). However, this distance increased to ∼32 nm (32 ± 5 nm, mean ± SEM) in cells treated with MG132 (Fig. 9 D). This difference is statistically significant based on a Student’s t test (P = 0.0003). These data strongly support the model that CENP-T elongation contributes to the structural deformation of the inner kinetochore in the presence of tension.
The elongation of CENP-T in DT40 cells. (A) Double fusion CENP-T construct fused with mRFP in the C-terminal end and GFP in the N-terminal end. (B) Definition of the value for CENP-T elongation. The distance between paired GFP signals or RFP signals was measured, and the delta was defined as the value. (C) Fluorescence light microscopic images of cells expressing a GFP–CENP-T–mRFP double fusion protein in cells treated with nocodazole (Noc) or MG132. Signal intensities for RFP and GFP were shown in the paired kinetochores from cells treated with nocodazole or MG132. Peaks of signals for GFP and RFP were almost identical from cells treated with nocodazole. In MG132-treated cells, peaks for RFP signals were inside those of GFP signals. (D) Results of the measurement of the values of CENP-T elongation in cells treated with nocodazole or MG132. The mean of the value in cells treated with nocodazole (n = 22) is ∼4 nm (4 ± 4 nm, mean ± SEM), and that in MG132-treated cells (n = 23) is ∼32 nm (32 ± 5 nm, mean ± SEM). The gray bars show means of the distributed dots. The significance of these measurements was estimated by the Student’s t test. (E) A model showing how the structural change of the inner kinetochore occurs. When microtubules (MT) properly attach to the outer kinetochore occupied by KMN network proteins and produce a tension, the structure of the inner kinetochore occupied by constitutive centromere-associated network (CCAN) proteins is deformed to an ovallike structure from the rectangle structure. The width and length of the inner kinetochore is stretched and compressed, respectively, when tension from spindle microtubules occurs. Although the length of outer kinetochore is compressed, the width is not stretched even under tension. The conformational change of the CENP-T–W complex is responsible for the structural deformation. The C terminus of CENP-T makes a complex with CENP-W and associates with chromatin, and the N terminus of CENP-T associates with the outer kinetochore proteins. The long middle region (80–500 aa) of CENP-T is flexible, and the elastic feature is responsible for the inner kinetochore stretch. Other CCAN proteins may contribute to the structural change. There are ∼30 molecules of CCAN per one kinetochore (Johnston et al., 2010), and four to five microtubules attach to the kinetochore plate in DT40 cells (Ribeiro et al., 2009). Therefore, six to seven CCAN units are responsible for one microtubule attachment. Yellow boxes indicate the regions of higher magnification on the right. kMT, kinetochore microtubule. Bars, 1 µm.
The elongation of CENP-T in DT40 cells. (A) Double fusion CENP-T construct fused with mRFP in the C-terminal end and GFP in the N-terminal end. (B) Definition of the value for CENP-T elongation. The distance between paired GFP signals or RFP signals was measured, and the delta was defined as the value. (C) Fluorescence light microscopic images of cells expressing a GFP–CENP-T–mRFP double fusion protein in cells treated with nocodazole (Noc) or MG132. Signal intensities for RFP and GFP were shown in the paired kinetochores from cells treated with nocodazole or MG132. Peaks of signals for GFP and RFP were almost identical from cells treated with nocodazole. In MG132-treated cells, peaks for RFP signals were inside those of GFP signals. (D) Results of the measurement of the values of CENP-T elongation in cells treated with nocodazole or MG132. The mean of the value in cells treated with nocodazole (n = 22) is ∼4 nm (4 ± 4 nm, mean ± SEM), and that in MG132-treated cells (n = 23) is ∼32 nm (32 ± 5 nm, mean ± SEM). The gray bars show means of the distributed dots. The significance of these measurements was estimated by the Student’s t test. (E) A model showing how the structural change of the inner kinetochore occurs. When microtubules (MT) properly attach to the outer kinetochore occupied by KMN network proteins and produce a tension, the structure of the inner kinetochore occupied by constitutive centromere-associated network (CCAN) proteins is deformed to an ovallike structure from the rectangle structure. The width and length of the inner kinetochore is stretched and compressed, respectively, when tension from spindle microtubules occurs. Although the length of outer kinetochore is compressed, the width is not stretched even under tension. The conformational change of the CENP-T–W complex is responsible for the structural deformation. The C terminus of CENP-T makes a complex with CENP-W and associates with chromatin, and the N terminus of CENP-T associates with the outer kinetochore proteins. The long middle region (80–500 aa) of CENP-T is flexible, and the elastic feature is responsible for the inner kinetochore stretch. Other CCAN proteins may contribute to the structural change. There are ∼30 molecules of CCAN per one kinetochore (Johnston et al., 2010), and four to five microtubules attach to the kinetochore plate in DT40 cells (Ribeiro et al., 2009). Therefore, six to seven CCAN units are responsible for one microtubule attachment. Yellow boxes indicate the regions of higher magnification on the right. kMT, kinetochore microtubule. Bars, 1 µm.
Discussion
Structural dynamics of the inner kinetochore
The kinetochore is a macromolecular structure that must both bind to spindle microtubules and couple microtubule dynamics to the production of force and movement. Recent light microscopy (Schittenhelm et al., 2007; Wan et al., 2009) and EM (Liu et al., 2006; Dong et al., 2007) analyses have proposed a model for the kinetochore structure in human cells. Here, we present an analysis of a representative collection of kinetochore components to the DT40 kinetochore that is consistent with this data (Fig. 1). Importantly, our data have also allowed us to define a dynamic structural change that occurs in the inner kinetochore structure during mitosis in response to tension from spindle microtubules (Figs. 2 and 3). The width and length of the inner kinetochore are stretched and compressed, respectively, when tension from spindle microtubules is applied. Although the length of the outer kinetochore is compressed, the width is not stretched even under tension.
Recent light microscopy–based studies suggest that the kinetochore stretch occurs in a tension-dependent manner (Maresca and Salmon, 2009; Uchida et al., 2009). In these studies, the stretch was evaluated as an increase of intrakinetochore distance (the distance between the inner and outer kinetochore) by light microscopy. In this paper, we show that the width of the inner kinetochore region is stretched upon tension from spindle microtubules. Based on our measurements, the inner kinetochore stretch is ∼25 nm (Fig. 3 J), which is close to the value for the increase in intrakinetochore distance defined as “delta” by previous work (Maresca and Salmon, 2009; Uchida et al., 2009) using light microscopy measured by comparing CENP-A (inner kinetochore) and Mis12 or Ndc80 (outer kinetochore). In this case, delta was increased by 15–30 nm in the presence of tension in human cells and by 37 nm in Drosophila melanogaster S2 cells (Maresca and Salmon, 2009; Uchida et al., 2009; Wan et al., 2009). We suggest that both the increased distance between the inner and outer kinetochores and the stretched structure of the inner kinetochore are responsible for the kinetochore stretch observed in the presence of microtubules. Although there may be additional mechanisms that cause the increase in intrakinetochore distance, our immuno-EM analyses suggest that the structural change within the inner kinetochore is primarily responsible for the kinetochore stretch, and this structural change is dependent on tension from spindle microtubules. Although the inner kinetochore is thought to provide a foundation for a “stable” kinetochore structure, our observation suggests that the structure of the inner kinetochore is flexible, and the dynamic deformation of the inner kinetochore can occur during chromosome segregation.
How does the structural deformation of the inner kinetochore occur?
Although we observed a dynamic change within the inner kinetochore structure, it is important to address which molecules are involved in this change. Based on our analyses, we propose that the CENP-T–W complex is a key player in the structural deformation of the inner kinetochore to allow the generation of the kinetochore stretch (Fig. 9 E). CENP-T is a component of inner kinetochore (Fig. 1). Its C-terminal histonelike region forms a complex with CENP-W and shows DNA-binding activity (Hori et al., 2008a). In contrast, the functional role of the N-terminal region was unclear. Expression of CENP-T containing only the N-terminal region does not localize to kinetochores, and expression of CENP-T lacking the N terminus does not complement the CENP-T–deficient phenotype, suggesting that the N-terminal region has a functional role in chromosome segregation (Hori et al., 2008a). Our immuno-EM analyses of CENP-T N-terminally tagged proteins indicate that the N-terminal end still shows a rectangular distribution even in the presence of tension similar to that detected using antibodies against outer kinetochore proteins, such as Ndc80. In our model, the N terminus of CENP-T is associated with the outer kinetochore (Fig. 9 E). In fact, our structural prediction indicates that the N-terminal end (1–80 aa) of CENP-T shows low probability as a disordered region, suggesting this region has a structure that may allow it to associate with outer kinetochore proteins. In contrast, the long middle region of CENP-T (80–500 aa) is flexible, with elastic features that are responsible for the inner kinetochore stretch (Fig. 9 E). Recent analysis of the kinetochore structure with EM suggests an existence of a fibrous structure termed “kinetochore fibrils” (McIntosh et al., 2008). Although it is unclear which molecules are involved in the formation of the structure, an extended CENP-T–W complex may contribute to these observed structures. Although we cannot rule out the possibility that additional proteins contribute to the structural deformation of the inner kinetochore, we propose that CENP-T elongation is a primary factor involved in the dynamic stretching of the inner kinetochore in response to tension.
Materials and methods
DT40 culture
DT40 cells were cultured and transfected as described previously (Fukagawa et al., 1999). All DT40 cells were cultured at 38°C in Dulbecco’s modified medium supplemented with 10% fetal calf serum, 1% chicken serum, 2-mercaptoethanol, penicillin, and streptomycin. To repress the expression of the tetracycline-responsive transgenes, tetracycline (Sigma-Aldrich) was added to the culture medium to a final concentration of 2 µg/ml. CENP-C (Kwon et al., 2007), CENP-H (Fukagawa et al., 2001), CENP-S (Amano et al., 2009), CENP-T (Hori et al., 2008a), and Ndc80 (Hori et al., 2003) conditional knockout cell lines were used in this study.
Observation of the kinetochore outer plate
Observation of the outer plate was performed by a previously described conventional method (Hori et al., 2008a). DT40 cells were treated with 500 ng/ml nocodazole for 3 h and were fixed in 2.5% glutaraldehyde and 0.15% tannic acid in 0.1 M sodium cacodylate buffer for 1 h. Postfixation was performed in 2% OsO4 for 1 h on ice. The cells were dehydrated in ethanol and then infiltrated with Epon 812. Samples were cut with an ultramicrotome (EM UC6 and FC6; Leica) equipped with a diamond knife (Diatome) and examined in a transmission electron microscope (JEM 1010; JEOL).
Immuno-EM
DT40 cells were treated with 500 ng/ml nocodazole for 3 h or 10 µM MG132 for 2 h to enrich mitotic cells. Cells were collected with a cytocentrifuge (Cytospin 3; Shandon) and fixed in 3% paraformaldehyde for 15 min, permeabilized in 0.5% NP-40 in PBS for 15 min, rinsed in 0.5% BSA in PBS, and incubated for 1 h at 37°C with primary antibodies. As primary antibodies, anti–CENP-C (Fukagawa et al., 1999), anti–CENP-E (this study), anti–CENP-T (Hori et al., 2008a), anti–CENP-R (Hori et al., 2008b), anti-Mis12 (Kline et al., 2006), anti-Ndc80 (Hori et al., 2003), anti-Mad2 (this study), and anti-HA (Sigma-Aldrich) antibodies were used. All antibodies against kinetochore proteins are polyclonal antibodies and have generated recombinant proteins as antigens. The following recombinant proteins were used in this study: Gallus gallus CENP-C (ggCENP-C; 1–302 aa), ggCENP-E (1,081–1,501 aa), ggCENP-T (full length), ggCENP-R (full length), ggMis12 (full length), ggNdc80 (465–640 aa), and ggMad2 (full length). As all antibodies are polyclonal, it was hard to determine epitopes. Samples were washed three times in 0.5% BSA in PBS and incubated with 1.4-nm gold-labeled secondary antibodies (Nanoprobes) at a 1:25 dilution for 2 h at 37°C. Samples were washed three times in PBS and fixed with 2.5% glutaraldehyde/3% PFA in 0.1 M cacodylate buffer, pH 7.2, at 4°C for 20 h.
Samples were washed with PBS and silver enhanced using an enhancement kit (HQ Silver; Nanoprobes) according to the manufacturer’s protocol. Postfixation was performed in 0.5% OsO4 on ice. The cells were dehydrated in ethanol and then infiltrated with Epon 812. Polymerization was performed at 60°C for 48 h. Serial sections were cut with an ultramicrotome equipped with a diamond knife (170 nm) and were examined at room temperature with a transmission electron microscope (JEM1010) at 100 kV.
Immunofluorescence and light microscopy imaging
Immunofluorescent staining of whole chicken cells was performed as described previously (Fukagawa et al., 1999). Cells were collected onto slides with a cytocentrifuge, fixed in 3% paraformaldehyde in PBS for 15 min at room temperature, permeabilized in 0.5% NP-40 in PBS for 10 min at room temperature or 100% methanol for 15 min at −20°C, rinsed three times in 0.5% BSA in PBS, and incubated for 1 h at 37°C with a primary antibody. The binding of a primary antibody was then detected with FITC- or Cy3-conjugated goat anti–rabbit IgG (Jackson ImmunoResearch Laboratories, Inc.) diluted to an appropriate concentration in 0.5% BSA in PBS. Chromosomes and nuclei were counterstained with DAPI at 0.2 µg/ml in Vectashield Antifade (Vector Laboratories). All immunofluorescence images were collected at room temperature with a cooled EM charge-coupled device camera (QuantEM; Roper Scientific) mounted on an inverted microscope (IX71; Olympus) with a 100× objective lens together with a filter wheel and a disk-scanning unit confocal unit. Z sections (n = 15–25) were acquired at 0.3-µm steps. All subsequent analysis and processing of images were performed using Metamorph software (Molecular Devices).
Reconstitution and high speed AFM imaging of the CENP-T–W complex
The full-length or histone-fold region of CENP-T and full-length CENP-W were cloned into pET28 (Takara Bio Inc.). For MBP fusion CENP-T constructs, the full-length and histone-fold region of CENP-T were cloned into a pMal-c2x derivative plasmid containing MBP, hexahistidine residues, and three tandem tobacco etch virus protease recognition sites. Full-length CENP-T, the histone-fold region of CENP-T, and CENP-W were expressed in BL21 (DE3) as an inclusion body and purified by Ni-Sepharose under denaturing conditions. CENP-T–W was reconstituted by mixing an equimolar amount of each protein in the presence of 6 M guanidium chloride. The mixture was dialyzed into a 4, 2, 1, 0.5, and 0 M guanidium chloride concentration in a step-wise manner. For the reconstitution of the full-length CENP-T–W complex, 0.4 M arginine hydrochloride was added during the 0.5 M guanidium chloride step to prevent nonspecific aggregation. MBP–CENP-T and CENP-W were coexpressed in BL21 (DE3) with ampicillin and kanamicin selections. The soluble CENP-T–W complex was purified using Ni-Sepharose, an MBPTrap column, and Superdex 200 column (GE Healthcare).
The purified CENP-T–W complex was adsorbed onto a mica surface in PBS solution. The complex was observed using a high speed AFM as described previously (Ando et al., 2001, 2008; Miyagi et al., 2008).
Online supplemental material
Fig. S1 shows the measurement of the size of the outer kinetochore plate observed by EM. Fig. S2 describes the measurement of the size of the inner kinetochore region observed by immuno-EM with anti–CENP-T antibodies. Fig. S3 shows the structure of the inner kinetochore occupied by CENP-C in CENP-T–deficient cells. Fig. S4 shows the prediction of the structure of CENP-T. Fig. S5 shows data of high speed AFM analyses with the complex of MBP-fused CENP-T and CENP-W.
Acknowledgments
The authors are very grateful to I.M. Cheeseman for critical reading of the manuscript and valuable suggestions for several experiments and to T. Ando for AFM imaging. We also thank K. Suzuki, M. Takahashi, and K. Kita for technical assistance and E. Suzuki, H. Kimura, and Y. Hayashi-Takanaka for useful suggestions with EM experiments.
This work was supported by a Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan and the Cabinet Office of the Government of Japan through its Funding Program for Next Generation World-Leading Researchers to T. Fukagawa. T. Hori is supported by Precursory Research for Embryonic Science and Technology of the Japan Science and Technology Agency and by a Grants-in-Aid for Scientific Research from MEXT. T. Nishino is supported by the International Human Frontier Science Program Organization grant and by a Grants-in-Aid for Scientific Research from MEXT.