A key requirement for Rab function in membrane trafficking is site-specific activation by GDP-GTP exchange factors (GEFs), but the majority of the 63 human Rabs have no known GEF. We have performed a systematic characterization of the 17 human DENN domain proteins and demonstrated that they are specific GEFs for 10 Rabs. DENND1A/1B localize to clathrin patches at the plasma membrane and activate Rab35 in an endocytic pathway trafficking Shiga toxin to the trans-Golgi network. DENND2 GEFs target to actin filaments and control Rab9-dependent trafficking of mannose-6-phosphate receptor to lysosomes. DENND4 GEFs target to a tubular membrane compartment adjacent to the Golgi, where they activate Rab10, which suggests a function in basolateral polarized sorting in epithelial cells that compliments the non-DENN GEF Sec2 acting on Rab8 in apical sorting. DENND1C, DENND3, DENND5A/5B, MTMR5/13, and MADD activate Rab13, Rab12, Rab39, Rab28, and Rab27A/27B, respectively. Together, these findings provide a basis for future studies on Rab regulation and function.
Introduction
Rab GTPases are used to encode information about the state of a membrane or membrane domain in order to control specific membrane trafficking events (Zerial and McBride, 2001; Behnia and Munro, 2005). Rabs are activated by specific guanine nucleotide exchange factors (GEFs) promoting the release of GDP and binding of GTP (Pfeffer and Aivazian, 2004). According to the prevailing model, GEFs together with other regulatory factors localize to and act at specific membrane surfaces, and thus provide a means to locally activate their target Rabs (Pfeffer and Aivazian, 2004). This system allows vesicles derived from a particular organelle to be tagged with a specific Rab GTPase, and their movement along the cytoskeleton and tethering to a specified domain on a target membrane to be controlled. Effector protein complexes that are either activated or recruited to the membrane surface by the presence of the GTP-bound Rab mediate these cytoskeletal and membrane tethering functions. GTP hydrolysis triggered either by additional GTPase-activating proteins (GAPs) or spontaneously because of intrinsic activity of the Rab ends the cycle. GEFs and GAPs therefore play a key role in the specific activation and inactivation of Rab GTPases.
The known Rab GEFs and GAPs typically fall into discrete families defined by conserved protein domains (Barr and Lambright, 2010). With the exception of the Rab3GAP1/2 proteins (Fukui et al., 1997; Nagano et al., 1998), Rab GAPs characteristically contain a TBC domain that catalyzes nucleotide hydrolysis by an arginine-glutamine two-finger mechanism (Pan et al., 2006). In humans, the TBC domain family has over 40 members, and it is likely that these regulate all 63 human Rabs, with some TBC domain proteins acting on several closely related Rabs (Haas et al., 2005, 2007; Fuchs et al., 2007). Rab GEFs are more diverse, and several conserved, yet structurally unrelated proteins and protein complexes have been shown to have specific Rab GEF activity (Barr and Lambright, 2010). These are: the TRAPP-I complex activating Ypt1p/Rab1 (Wang et al., 2000; Cai et al., 2008), Vps9 domain proteins activating Rab5/Ypt51p subfamily GTPases (Delprato et al., 2004; Sato et al., 2005; Delprato and Lambright, 2007), Sec2p/Rabin proteins activating Sec4p GTPases (Walch-Solimena et al., 1997; Hattula et al., 2002; Dong et al., 2007; Itzen et al., 2007; Sato et al., 2007b), the Ric1p–Rgp1p complex activating Ypt6p and possibly Rab6 (Siniossoglou et al., 2000), the Mon1p–Ccz1p complex acting on Ypt7p and Rab7 (Nordmann et al., 2010), and the RCC1 domain protein claret, which may act as a GEF for the unique Rab lightoid in Drosophila (Ma et al., 2004). Apart from claret, these GEFs and their target Rab GTPases act in trafficking pathways conserved from mammals to yeasts. However, mammalian cells possess >60 Rabs, compared with the 11 of budding yeast, and therefore require additional GEFs to activate these extra Rabs. At present, most of the 60 mammalian Rabs lack a defined GEF activity, and it is therefore unclear how they would be specifically activated. Additional Rab GEFs are therefore likely to exist.
DENN domain proteins were first implicated as Rab GEFs by the biochemical purification of a Rab3 GEF from bovine brain (Wada et al., 1997). This was subsequently identified as a DENN domain protein, although it remained unclear which domain in the protein was responsible for GEF activity (Coppola et al., 2002). Further studies revealed that the Caenorhabditis elegans MADD homologue AEX-3 was responsible for controlling the activity of Rab3 and Rab27 at the synapse (Iwasaki et al., 1997; Mahoney et al., 2006). More recently, a screen for defective receptor-mediated yolk protein endocytosis in C. elegans identified another DENN domain protein RME-4, and indicated that it acted on Rab35 (Sato et al., 2008). This was confirmed by complementary studies in mammalian cells showing that the RME-4 homologue DENND1A/connecdenn was in fact a Rab35 GEF (Allaire et al., 2010; Marat and McPherson, 2010). These findings support the idea that DENN domain proteins might form a family of Rab GEFs. To investigate this, we have characterized the human DENN domain proteins, and identified their target or substrate Rab GTPases.
Results
Identification of human DENN domain proteins
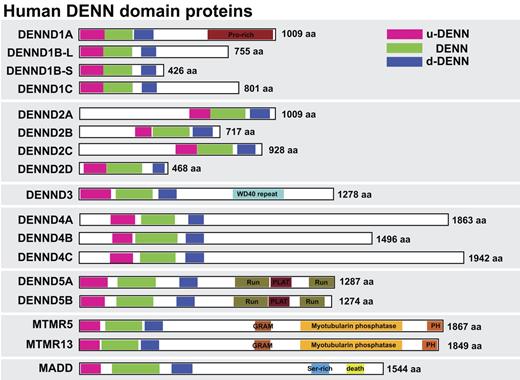
Sequence searches of the human genome using the DENN domain of the Rab3 GEF MADD reveal the presence of 17 proteins sharing this domain (Fig. 1). All these proteins carry a full DENN domain comprising the three upstream (u-DENN), core (DENN), and downstream (d-DENN) subregions (Levivier et al., 2001). Systematic searching of the genome sequence databases reveals that although widely conserved in metazoans and protozoans, these DENN domain proteins are absent from the budding yeast Saccharomyces cerevisiae used as a model for trafficking studies. This suggests that DENN domain proteins may act as GEFs for some of the many additional Rabs found in humans and other metazoans. Further analysis indicates that in addition to these proteins, there is a group of related proteins containing partial DENN homology. Some of these such as Avl9 and its homologues are conserved to budding yeast and other fungi (Harsay and Schekman, 2007). Because Avl9 may be a GEF for the non-Rab Ras family GTPase Gtr2 in late Golgi trafficking (Harsay and Schekman, 2007; Zhang et al., 2010), these are unlikely to be Rab GEFs and they were not pursued further in this study.
DENN proteins form a large family in human cells. A schematic showing the human DENN domain proteins, with the upstream (u-DENN), core DENN, and downstream (d-DENN) regions indicated. Additional domains likely to be of relevance for DENN targeting or regulation are marked and color coded. Sequence alignments of DENNs were done with ClustalX (Chenna et al., 2003) or MUSCLE (Edgar, 2004), and the results were visualized and manipulated with Jalview (Waterhouse et al., 2009). Linear sequence motifs were browsed in the ELM database (Gould et al., 2010). Accession numbers used for this analysis are listed in Table S1.
DENN proteins form a large family in human cells. A schematic showing the human DENN domain proteins, with the upstream (u-DENN), core DENN, and downstream (d-DENN) regions indicated. Additional domains likely to be of relevance for DENN targeting or regulation are marked and color coded. Sequence alignments of DENNs were done with ClustalX (Chenna et al., 2003) or MUSCLE (Edgar, 2004), and the results were visualized and manipulated with Jalview (Waterhouse et al., 2009). Linear sequence motifs were browsed in the ELM database (Gould et al., 2010). Accession numbers used for this analysis are listed in Table S1.
DENND1 regulates Rab35-dependent Shiga toxin trafficking to the TGN
As a first step in the systematic characterization of Rab nucleotide exchange activity, it was important to ensure that a known Rab GEF would give the expected pattern of specificity. The Vps9 domain GEF Rabex-5 was used for this purpose. As expected, Rabex-5 promoted GDP release from Rab5A-C, and displayed some activity toward the Rab5 subfamily GTPases Rab17, Rab21, and Rab22A (Fig. 2 A). Other Rabs fell below the background value set at twice the median. The short form of DENND1B encoding only a DENN domain was then tested for GEF activity using the GDP-releasing assay (Fig. 2 B). This revealed that DENND1B-S promoted GDP release from Rab35 but not the other Rabs tested. For true GEF activities, GTP binding rapidly follows GDP release, and it was therefore important to test this. DENND1B-S specifically promoted GTP binding to Rab35 but not the other Rabs tested (Fig. 2 C). Similar results were obtained for DENND1A and the long form of DENND1B (Fig. 2 D). DENND1C in contrast was most active toward Rab13 and showed no activity to Rab35 (Fig. 2 E). Together, these findings show that DENND1A and DENND1B are Rab35-specific GEFs, and that this activity is caused by the DENN domain.
DENND1A/1B are GEFs for Rab35. (A) Human Rabex-5 was tested against a representative panel of human Rab proteins using the GDP-releasing assay. In brief, 10 µg of each GST-tagged Rab to be tested was incubated in 50 mM Hepes-NaOH, pH 6.8, 0.1 mg/ml BSA, 125 µM EDTA, 10 µM Mg-GDP, and 5 µCi [3H]-GDP (10 mCi/ml; 5,000 Ci/mmol) in a total volume of 200 µl for 15 min at 30°C to load the Rab with the radioactive GDP probe. For standard GDP-releasing GEF assays, 100 µl of the loading reaction was then mixed with 10 µl of 10 mM Mg-GTP and 10 nM His6-tagged Rabex-5 purified from bacteria or a buffer control, then adjusted to 120 µl final volume with assay buffer. The GEF reaction occurred for 20 min at 30°C. After this, 2.5 µl was taken for a specific activity measurement; the remainder was split into two tubes, then incubated with 500 µl of ice-cold assay buffer containing 1 mM MgCl2 and 20 µl of packed glutathione-sepharose for 60 min at 4°C to separate Rab–GDP complexes from free “released” GDP. After washing three times with 500 µl of ice-cold assay buffer, the sepharose was transferred to a vial containing 4 ml of scintillation fluid and counted. The amount of nucleotide exchange was calculated in pmoles of GDP released. (B and C) A representative panel of human Rab proteins was tested against 10 nM of His6-tagged DENND1B-S in the GDP-releasing (B) or GTP-binding assay (C). For GTP-binding assays, the following modifications were made: only unlabeled GDP was used in the loading reaction; in the GEF reaction, 0.5 µl of 10 mM GTP and 1 µCi [35S]-GTPγS (10 mCi/ml; 5000 Ci/mmol) were used. The amount of nucleotide exchange was calculated in pmoles of GTP bound. (D and E) Human DENND1A (D), DENND1B-L (D), and DENND1C (E) were tested against a subset of Rab35-related Rabs using the GTP-binding assay. For these assays, 10 nM of FLAG-tagged DENND1A or DENND1C purified from HeLa cells, or 10 nM of His6-tagged DENND1B-L purified from bacteria were used. Errors bars show the standard error from the mean. The red line marks double the median value taken as a threshold.
DENND1A/1B are GEFs for Rab35. (A) Human Rabex-5 was tested against a representative panel of human Rab proteins using the GDP-releasing assay. In brief, 10 µg of each GST-tagged Rab to be tested was incubated in 50 mM Hepes-NaOH, pH 6.8, 0.1 mg/ml BSA, 125 µM EDTA, 10 µM Mg-GDP, and 5 µCi [3H]-GDP (10 mCi/ml; 5,000 Ci/mmol) in a total volume of 200 µl for 15 min at 30°C to load the Rab with the radioactive GDP probe. For standard GDP-releasing GEF assays, 100 µl of the loading reaction was then mixed with 10 µl of 10 mM Mg-GTP and 10 nM His6-tagged Rabex-5 purified from bacteria or a buffer control, then adjusted to 120 µl final volume with assay buffer. The GEF reaction occurred for 20 min at 30°C. After this, 2.5 µl was taken for a specific activity measurement; the remainder was split into two tubes, then incubated with 500 µl of ice-cold assay buffer containing 1 mM MgCl2 and 20 µl of packed glutathione-sepharose for 60 min at 4°C to separate Rab–GDP complexes from free “released” GDP. After washing three times with 500 µl of ice-cold assay buffer, the sepharose was transferred to a vial containing 4 ml of scintillation fluid and counted. The amount of nucleotide exchange was calculated in pmoles of GDP released. (B and C) A representative panel of human Rab proteins was tested against 10 nM of His6-tagged DENND1B-S in the GDP-releasing (B) or GTP-binding assay (C). For GTP-binding assays, the following modifications were made: only unlabeled GDP was used in the loading reaction; in the GEF reaction, 0.5 µl of 10 mM GTP and 1 µCi [35S]-GTPγS (10 mCi/ml; 5000 Ci/mmol) were used. The amount of nucleotide exchange was calculated in pmoles of GTP bound. (D and E) Human DENND1A (D), DENND1B-L (D), and DENND1C (E) were tested against a subset of Rab35-related Rabs using the GTP-binding assay. For these assays, 10 nM of FLAG-tagged DENND1A or DENND1C purified from HeLa cells, or 10 nM of His6-tagged DENND1B-L purified from bacteria were used. Errors bars show the standard error from the mean. The red line marks double the median value taken as a threshold.
Human Rab35 has previously been identified in a screen for regulators of Shiga toxin trafficking from the plasma membrane to the trans-Golgi network (Fuchs et al., 2007). If DENND1A and DENND1B are specific GEFs for Rab35, then they might be expected to localize to a membrane compartment of this trafficking pathway. DENND1A localization was therefore examined in HeLa cells, where it was found to target to small punctate structures overlapping with clathrin and the plasma membrane AP-2 clathrin adaptor (Fig. 3 A). The AP-1 and AP-3 clathrin adaptors associated with other trafficking steps did not overlap with DENND1A (Fig. 3 A). Analysis of DENND1A complex using mass spectrometry showed that they contained clathrin and components of the AP-2 complex but not other clathrin adaptors (Fig. 3 B). Western blotting confirmed that DENND1A specifically interacts with clathrin and the AP-2 complex but not other adaptors (Fig. 3 C). The other DENND1 family members DENND1B and DENND1C did not interact with clathrin or clathrin adaptors (Fig. 3, B and C). Depletion of clathrin caused loss of the punctate DENND1A plasma membrane staining and overlapping with AP-2, and resulted in a diffuse cytoplasmic and reticular pattern (Fig. 3 D). DENND1A therefore targets to clathrin and the AP-2–positive patches at the plasma membrane, which supports the idea that it functions in some form of endocytic trafficking. To test this idea, HeLa cells were depleted of DENND1A, DENND1B, and clathrin (Fig. 4 A), then tested for receptor-mediated uptake of the growth factor EGF or the transport of Shiga toxin B subunit (STxB) to the trans-Golgi network (Fig. 4, B and C). Cells depleted of DENND1A failed to transport STxB to the trans-Golgi network (Fig. 4, B and D). In the same cells, the uptake of EGF into punctate endosomal structures was not altered (Fig. 4 B). Depletion of clathrin strongly reduced EGF uptake and caused the transferrin receptor to accumulate at the cell surface rather than showing its normal punctate recycling endosome distribution (Fig. 4 C). Although clathrin depletion did not block STxB uptake, uptake efficiency was reduced and it failed to overlap with the trans-Golgi network marker TGN46 after 60 min (Fig. 4, C and D). Together, these findings support the idea that DENND1A is a Rab35 GEF regulating an endocytic trafficking pathway used by the Shiga toxin to reach the trans-Golgi network, but that DENND1A is not essential for the receptor-mediated uptake of EGF.
Localization of DENND1A is clathrin dependent. (A) HeLa cells expressing EGFP-tagged DENND1A (green) were fixed and then stained with antibodies to clathrin heavy chain, and the AP-1, AP-2, and AP-3 clathrin adaptor complexes (red). DNA was stained with DAPI (blue). (B) DENND1A and DENND1B complexes were analyzed by mass spectrometry. The proteins scored highest by the Sequest search algorithm are listed in the table. (C) HeLa cells were transfected with constructs encoding FLAG-tagged DENND1A, DENND1B-L, DENND1B-S, and DENND1C for 48 h. The cells were washed from the dish using PBS with 1 mM EDTA, and the cell pellets were lysed for 20 min on ice in 1 ml cell of lysis buffer (50 mM Tris-HCl, pH 7.4, 1 mM EDTA, 150 mM NaCl, 0.5% Triton X-100, and protease inhibitors cocktails). The FLAG-tagged proteins were immunoprecipitated from the clarified lysate using 20 µl of anti-FLAG M2 affinity gel (Sigma-Aldrich) for 4 h at 4°C. The pellet was washed three times in 1 ml of cell lysis buffer, and bound proteins were eluted with 1 ml of 200 µg/ml FLAG peptide in TBS and then precipitated for 60 min on ice using 10% trichloroacetic acid. The FLAG-tagged DENND1A, DENND1B-L, DENND1B-S, and DENND1C complexes were analyzed by SDS-PAGE on 4–12% gradient gels and Coomassie blue staining, or Western blotted for clathrin heavy chain (CHC) and the AP-1 and AP-2 clathrin adaptors on 10% gels. Asterisks mark proteins that nonspecifically bind to FLAG-agarose and were found in negative control conditions. Molecular mass standards are indicated in kilodaltons. (D) HeLa cells expressing EGFP-tagged DENND1A (green) were transfected for 72 h with siRNA duplexes targeting the clathrin heavy chain, fixed, and then stained with antibodies for clathrin and the AP-2 clathrin adaptor (red). DNA was stained with DAPI (blue). Enlargements are shown to the right to more clearly demonstrate the overlap between DENND1A (green), and clathrin or AP2 (red) in control cells, and the loss of punctate DENND1 staining after clathrin heavy chain depletion. Bars, 10 µm.
Localization of DENND1A is clathrin dependent. (A) HeLa cells expressing EGFP-tagged DENND1A (green) were fixed and then stained with antibodies to clathrin heavy chain, and the AP-1, AP-2, and AP-3 clathrin adaptor complexes (red). DNA was stained with DAPI (blue). (B) DENND1A and DENND1B complexes were analyzed by mass spectrometry. The proteins scored highest by the Sequest search algorithm are listed in the table. (C) HeLa cells were transfected with constructs encoding FLAG-tagged DENND1A, DENND1B-L, DENND1B-S, and DENND1C for 48 h. The cells were washed from the dish using PBS with 1 mM EDTA, and the cell pellets were lysed for 20 min on ice in 1 ml cell of lysis buffer (50 mM Tris-HCl, pH 7.4, 1 mM EDTA, 150 mM NaCl, 0.5% Triton X-100, and protease inhibitors cocktails). The FLAG-tagged proteins were immunoprecipitated from the clarified lysate using 20 µl of anti-FLAG M2 affinity gel (Sigma-Aldrich) for 4 h at 4°C. The pellet was washed three times in 1 ml of cell lysis buffer, and bound proteins were eluted with 1 ml of 200 µg/ml FLAG peptide in TBS and then precipitated for 60 min on ice using 10% trichloroacetic acid. The FLAG-tagged DENND1A, DENND1B-L, DENND1B-S, and DENND1C complexes were analyzed by SDS-PAGE on 4–12% gradient gels and Coomassie blue staining, or Western blotted for clathrin heavy chain (CHC) and the AP-1 and AP-2 clathrin adaptors on 10% gels. Asterisks mark proteins that nonspecifically bind to FLAG-agarose and were found in negative control conditions. Molecular mass standards are indicated in kilodaltons. (D) HeLa cells expressing EGFP-tagged DENND1A (green) were transfected for 72 h with siRNA duplexes targeting the clathrin heavy chain, fixed, and then stained with antibodies for clathrin and the AP-2 clathrin adaptor (red). DNA was stained with DAPI (blue). Enlargements are shown to the right to more clearly demonstrate the overlap between DENND1A (green), and clathrin or AP2 (red) in control cells, and the loss of punctate DENND1 staining after clathrin heavy chain depletion. Bars, 10 µm.
DENND1A is required for Rab35-dependent Shiga toxin trafficking to the trans-Golgi network. (A) HeLa cells expressing EGFP-tagged DENND1A and the long or short forms of DENND1B were transfected with siRNA duplexes to DENND1A, DENND1B, clathrin heavy chain (CHC), or a nonspecific control for 72 h, then Western blotted as indicated. Molecular mass standards are indicated in kilodaltons. The asterisk indicates a nonspecific cross reaction of the clathrin heavy chain antibody. (B) Dual EGF and STxB uptake assays were performed for 60 min as described previously (Fuchs et al., 2007) in cells transfected with control or DENND1A duplexes for 72 h. Cells were fixed and then stained for the Golgi marker golgin-160. (C) Uptake assays were performed as in B using cells transfected with CHC siRNA duplexes. Cells were fixed and then stained for the transferrin receptor (TfR) to mark recycling endosomes or the TGN marker TGN46. Bars, 10 µm. (D) The extent of EGF and Shiga toxin uptake under the various conditions was measured and is plotted in the graphs (n = 3). ImageJ was used to measure colocalization of markers. Error bars indicate standard error of the mean.
DENND1A is required for Rab35-dependent Shiga toxin trafficking to the trans-Golgi network. (A) HeLa cells expressing EGFP-tagged DENND1A and the long or short forms of DENND1B were transfected with siRNA duplexes to DENND1A, DENND1B, clathrin heavy chain (CHC), or a nonspecific control for 72 h, then Western blotted as indicated. Molecular mass standards are indicated in kilodaltons. The asterisk indicates a nonspecific cross reaction of the clathrin heavy chain antibody. (B) Dual EGF and STxB uptake assays were performed for 60 min as described previously (Fuchs et al., 2007) in cells transfected with control or DENND1A duplexes for 72 h. Cells were fixed and then stained for the Golgi marker golgin-160. (C) Uptake assays were performed as in B using cells transfected with CHC siRNA duplexes. Cells were fixed and then stained for the transferrin receptor (TfR) to mark recycling endosomes or the TGN marker TGN46. Bars, 10 µm. (D) The extent of EGF and Shiga toxin uptake under the various conditions was measured and is plotted in the graphs (n = 3). ImageJ was used to measure colocalization of markers. Error bars indicate standard error of the mean.
The DENND2 family regulates Rab9 and lysosome distribution
Having successfully shown that the DENND1 proteins are Rab GEFs, other DENN proteins were then investigated starting with the DENND2 family. The DENND2 family has four members, and one of these, DENND2D, comprises only a DENN domain. This was tested first. DENND2D displayed specific GDP-releasing activity to both Rab9A and Rab9B but not any other Rab tested (Fig. 5 A), which supports the view that the DENN domain alone is responsible for GEF activity. This specificity was confirmed for the three other DENND2 family members (Fig. 5 B). Examination of DENND2 family localization revealed that DENND2A and DENND2B were present on filaments reminiscent of actin, that DENND2C overexpression caused cell shape changes and formed large patches in cell protrusions, and that DENND2D was diffusely located throughout the entire cell (Figs. 5 C and S1). The pronounced actin filament localization of DENND2A was especially intriguing because it was recently shown that the Rho family protein RhoBTB3 is a Rab9 effector protein (Espinosa et al., 2009). Rho family proteins are typically associated with processes controlling the actin cytoskeleton, and this suggests there may be a link between Rab9 function and the actin cytoskeleton. As expected, Rab9 was present on LAMP1-positive lysosomes defined by the marker LAMP1 (Fig. 5 C). Depletion of Rab9 or DENND2A resulted in a similar phenotype, where lysosomes clustered adjacent to the perinuclear region and were lost from the more peripheral regions of the cells (Fig. 5 D). Depletion of other DENND2 family members had no obvious effect in HeLa cells (Fig. 5 D and not depicted). Rab9 has a well-documented function in trafficking of the mannose-6-phosphate receptor (MPR) between the TGN and late endosomes (Lombardi et al., 1993; Díaz et al., 1997), and this function was therefore investigated. Cells depleted of Rab9 or DENND2A showed reduced intensity of MPR staining relative to control cells and a loss of MPR-positive structures in the cell periphery (Fig. 5 E). Fluorescence intensity measurements either integrating the total cell associated signal or taking a transection through the perinuclear region indicate that there is a >60% reduction in MPR staining intensity in DENND2A- and Rab9-depleted cells (Fig. 5 E). Blocking Rab9 function prevents recycling of MPR from late endosomes back to the TGN (Riederer et al., 1994); thus, the MPR may become trapped in late endosomes and may enter the lysosomes, where it will be degraded. Other TGN recycling pathways were not obviously perturbed by Rab9 or DENND2A depletion because the TGN marker TGN46 was not changed by these treatments (Fig. 5 E). Consistent with the biochemical GEF assay data, loss of Rab9 activity either by depleting Rab9 or its GEF regulator DENND2A causes a similar phenotype. These findings support the idea that the DENND2 family members act as GEFs for Rab9 in trafficking between the late endosomes and the TGN. Interestingly, components of the BLOC complex involved in trafficking to lysosome-like organelles have been found to associate with actin filaments (Falcón-Pérez et al., 2002) and to interact with Rab9 (Kloer et al., 2010). The diversity of DENND2 family members suggests that this pathway is regulated differently in different tissues, possibly linking Rab9 regulation at late endosomes and lysosome-related organelles to the status of the actin cytoskeleton.
The DENND2 family regulates Rab9 and lysosomes. (A) A representative panel of human Rab proteins was tested against 10 nM of His6-tagged human DENND2D purified from bacteria using the GDP-releasing assay. (B) Human DENND2A, DENND2B, and DENND2C were expressed as His6-tagged protein in bacteria and then tested against a subset of Rab9-related Rabs. Errors bars show the standard error of the mean. The red line marks double the median value. (C) HeLa cells were transfected with EGFP-tagged DENND2A or Rab9A (green), fixed after 24 h, and stained with the antibodies indicated (red). DNA was stained with DAPI (blue). Inset enlargements are shown to more clearly demonstrate the relationship between Rab9 (green) and LAMP1 (red), which suggests that Rab9 is present on the lysosome membrane. (D) HeLa cells expressing EGFP-tagged Rab9 or DENND2 constructs as indicated were transfected with control, Rab9A, Rab9B, and DENND2A-D siRNA duplexes for 72 h. Western blotting with EGFP antibodies confirmed depletion of the target proteins, whereas tubulin showed that loading was equal for all samples. HeLa cells transfected with control, Rab9A and Rab9B, and DENND2A-D siRNA duplexes for 72 h were fixed, then stained for LAMP1 (red) and DAPI to detect DNA (blue). Molecular mass standards are indicated in kilodaltons. (E) HeLa cells transfected with control, Rab9A and Rab9B, and DENND2A siRNA duplexes for 72 h were fixed, then stained for MPR (green) and TGN46 (red). DNA was stained with DAPI (blue). Bars, 10 µm. (F) Fluorescence intensity for MPR staining from E was measured using ImageJ by drawing a box around the entire cell area and integrating the total signal. An equivalent area with no cell was subtracted for the background. This was performed for 24 cells, and the mean and standard error are plotted on the bar graph. A 20 × 1 µm line measurement was performed across the nuclear region where MPR staining is most clustered. The pixel intensity along the line is plotted in the graph for control, DENND2A, and Rab9a- and Rab9b-depleted cells.
The DENND2 family regulates Rab9 and lysosomes. (A) A representative panel of human Rab proteins was tested against 10 nM of His6-tagged human DENND2D purified from bacteria using the GDP-releasing assay. (B) Human DENND2A, DENND2B, and DENND2C were expressed as His6-tagged protein in bacteria and then tested against a subset of Rab9-related Rabs. Errors bars show the standard error of the mean. The red line marks double the median value. (C) HeLa cells were transfected with EGFP-tagged DENND2A or Rab9A (green), fixed after 24 h, and stained with the antibodies indicated (red). DNA was stained with DAPI (blue). Inset enlargements are shown to more clearly demonstrate the relationship between Rab9 (green) and LAMP1 (red), which suggests that Rab9 is present on the lysosome membrane. (D) HeLa cells expressing EGFP-tagged Rab9 or DENND2 constructs as indicated were transfected with control, Rab9A, Rab9B, and DENND2A-D siRNA duplexes for 72 h. Western blotting with EGFP antibodies confirmed depletion of the target proteins, whereas tubulin showed that loading was equal for all samples. HeLa cells transfected with control, Rab9A and Rab9B, and DENND2A-D siRNA duplexes for 72 h were fixed, then stained for LAMP1 (red) and DAPI to detect DNA (blue). Molecular mass standards are indicated in kilodaltons. (E) HeLa cells transfected with control, Rab9A and Rab9B, and DENND2A siRNA duplexes for 72 h were fixed, then stained for MPR (green) and TGN46 (red). DNA was stained with DAPI (blue). Bars, 10 µm. (F) Fluorescence intensity for MPR staining from E was measured using ImageJ by drawing a box around the entire cell area and integrating the total signal. An equivalent area with no cell was subtracted for the background. This was performed for 24 cells, and the mean and standard error are plotted on the bar graph. A 20 × 1 µm line measurement was performed across the nuclear region where MPR staining is most clustered. The pixel intensity along the line is plotted in the graph for control, DENND2A, and Rab9a- and Rab9b-depleted cells.
DENND4 family proteins are specific GEFs for Rab10 in apical sorting
The role of Rab8 and Rab10 in apical and basolateral sorting is well established. Questions remain, however, about how these two GTPases are independently regulated. Two GEFs carrying the Sec2 domain have been reported to show activity toward Rab8, but the GEF for Rab10 is unknown. Analysis of the DENND4 family proteins showed that these specifically promote GDP release from and GTP binding to Rab10 and have no activity toward Rab8 (Fig. 6, A and B). In contrast, Sec2 domain proteins Rabin3/Rabin8 and Rabin3-like/GRAB are specific GEFs for Rab8A and Rab8B and have no activity toward Rab10 (Fig. 6, C and D). Thus, the Sec2 domain proteins Rabin8 and GRAB, and the DENN domain proteins of the DENND4 family could provide a means to activate Rab8 and Rab10, respectively, and thus independently control polarized trafficking. Although DENND4A and DENND4C showed a diffuse cytoplasmic localization in HeLa cells, DENND4B was present on a tubular membrane compartment emanating from the perinuclear region (Fig. 6 E and S1). Strikingly, DENND4B staining is coincident with that of its target Rab10 (Fig. 6 E). This compartment did not overlap with markers for early endosomes, recycling endosomes, or lysosomes, but did show partial overlap with the Golgi marker GM130 (Fig. 6 E).
DENND4 family proteins are specific GEFs for Rab10. (A) A representative panel of human Rab proteins was tested against 10 nM of human DENND4B expressed as a FLAG-tagged protein in HeLa cells using the GDP-releasing assay. (B) Human DENND4A, DENND4B, and DENND4C were tested against a subset of Rab10-related Rabs using the GTP-binding assay. Again, 10 nM of each FLAG-tagged DENN protein purified from HeLa cells was used for these assays. Error bars show the standard error of the mean. The red line marks double the median value. (C and D) A representative panel of human Rab proteins was tested against 10 nM of His6-tagged human Rabin3/Rabin8 (C) and Rabin3-like/GRAB purified from bacteria using the GDP-releasing assay (D). Error bars indicate standard error of the mean. (E) HeLa cells expressing EGFP-tagged DENND4B were transfected with mCherry-tagged Rab10, or stained for the markers indicated. 4× enlargements of the Rab10-positive tubules are shown in the top panels. Bar, 10 µm.
DENND4 family proteins are specific GEFs for Rab10. (A) A representative panel of human Rab proteins was tested against 10 nM of human DENND4B expressed as a FLAG-tagged protein in HeLa cells using the GDP-releasing assay. (B) Human DENND4A, DENND4B, and DENND4C were tested against a subset of Rab10-related Rabs using the GTP-binding assay. Again, 10 nM of each FLAG-tagged DENN protein purified from HeLa cells was used for these assays. Error bars show the standard error of the mean. The red line marks double the median value. (C and D) A representative panel of human Rab proteins was tested against 10 nM of His6-tagged human Rabin3/Rabin8 (C) and Rabin3-like/GRAB purified from bacteria using the GDP-releasing assay (D). Error bars indicate standard error of the mean. (E) HeLa cells expressing EGFP-tagged DENND4B were transfected with mCherry-tagged Rab10, or stained for the markers indicated. 4× enlargements of the Rab10-positive tubules are shown in the top panels. Bar, 10 µm.
Family-wide assignment of DENN specificity
To complete the family-wide assignment of DENN specificity, the remaining DENN domains proteins DENND3, DENND5A, and DENND5B; the myotubularin-related proteins MTMR5 and MTMR13; and MADD were tested (Fig. 7). This revealed that these proteins also have specific Rab targets. DENND3 is a Rab12 GEF (Fig. 7 A), whereas DENND5A/B act on Rab39 (Fig. 7 B) and MTMR5/13 act on Rab 28 (Fig. 7 C). In agreement with previous reports, MADD showed activity toward Rab27 and Rab27B (Figueiredo et al., 2008), although it did not have activity toward Rab3 family members. This may be caused by the use of bacterially expressed Rab proteins, as it has been reported that C-terminal prenylation may be important for recognition of Rab3 by MADD (Sakisaka and Takai, 2005).
Family-wide assignment of DENN specificity. Human DENND3 (A), DENND5A and DENND5B (B), MTMR5 and MTMR13 (C), and MADD (D) were tested against a representative panel of human Rab proteins using the GDP-releasing assay. All assays used 10 nM of FLAG-tagged DENN protein purified from HeLa cells. Errors bars show the standard error of the mean. The red line marks double the median value.
Family-wide assignment of DENN specificity. Human DENND3 (A), DENND5A and DENND5B (B), MTMR5 and MTMR13 (C), and MADD (D) were tested against a representative panel of human Rab proteins using the GDP-releasing assay. All assays used 10 nM of FLAG-tagged DENN protein purified from HeLa cells. Errors bars show the standard error of the mean. The red line marks double the median value.
In summary, we have assembled a library of full-length human DENN domain proteins and tested their localizations and biochemical specificity (Fig. 8). This approach has revealed that the different DENN proteins targeted to different subcellular compartments (Fig. 8), which is consistent with the idea that they may control Rab activation at unique membrane or cytoskeletal domains. Critically, it also showed that DENN domain proteins have unique and nonoverlapping Rab targets. These findings will therefore be of relevance for many future studies on Rab function in membrane trafficking.
A summary of Rab GEFs indicating their target Rabs. Human (Homo sapiens, hs), fruit fly (Drosophila melanogaster, dm), and nematode (C. elegans, ce) Rabs and budding yeast (Saccharomyces cerevisiae, sc) Ypts were aligned using ClustalX and plotted using NJplot (Larkin et al., 2007). The alignment is annotated to show the known Rab GEFs: TRAPP, Sec2, the Vps9 domain family, Ric1-Rgp1, Mon1-Ccz1, claret, and the DENN domain family. Images to the right indicate the typical localization of the DENN domain family in HeLa cells. The pattern of conservation is summarized in the text to the right. The accession nos. for human, mouse, zebrafish, fruit fly, and nematode DENN domains proteins are listed in Table S1. Bar, 10 µm.
A summary of Rab GEFs indicating their target Rabs. Human (Homo sapiens, hs), fruit fly (Drosophila melanogaster, dm), and nematode (C. elegans, ce) Rabs and budding yeast (Saccharomyces cerevisiae, sc) Ypts were aligned using ClustalX and plotted using NJplot (Larkin et al., 2007). The alignment is annotated to show the known Rab GEFs: TRAPP, Sec2, the Vps9 domain family, Ric1-Rgp1, Mon1-Ccz1, claret, and the DENN domain family. Images to the right indicate the typical localization of the DENN domain family in HeLa cells. The pattern of conservation is summarized in the text to the right. The accession nos. for human, mouse, zebrafish, fruit fly, and nematode DENN domains proteins are listed in Table S1. Bar, 10 µm.
Discussion
DENNs form a conserved family of Rab GEFs
The results presented here provide good evidence that DENN proteins form a family of highly specific Rab GEF regulators controlling specific intracellular transport pathways. DENN proteins are conserved in primitive unicellular eukaryotes such as protozoans of the Naegleria genus (Fritz-Laylin et al., 2010), fission yeast, filamentous fungi, and plants (Levivier et al., 2001); however, they are absent from budding yeast. Consistent with this, none of the DENN target Rabs identified by this study are present in budding yeast. As reported previously, MADD acts on Rab27A, which functions in melanosome transport (Figueiredo et al., 2008). DENND1 and the C. elegans equivalent RME-4 act on Rab35 in endocytic trafficking pathways absent from budding yeast (Sato et al., 2008). Previous investigation of the DENND1/connecdenn proteins in human cells (Allaire et al., 2010; Marat and McPherson, 2010) has shown that Rab35 and DENND1A/connecdenn 1 play a role in recycling of MHC class I at an early endosomal compartment (Allaire et al., 2010). These authors have also shown that connecdenn proteins interact with clathrin (Allaire et al., 2010; Marat and McPherson, 2010), and our findings confirm this for DENND1A/connecdenn 1. We find that clathrin is required for Shiga toxin delivery to the TGN, but not its endocytosis (Fig. 4). This fits with the idea that the STxB traffics through an early endosomal sorting compartment (Fuchs et al., 2007), where it undergoes a clathrin-dependent sorting event (Popoff et al., 2007), before delivery to the TGN. DENND3 and DENND1C, like their targets Rab12 and Rab13, respectively, are present in vertebrates but are absent in invertebrates. In mammalian polarized epithelial cells, Rab13 functions in trafficking between recycling endosomes and the TGN (Nokes et al., 2008). Together with the data presented here, this suggests that the DENND1 family controls endosomal recycling and endosome–TGN trafficking routes involving Rab13 and Rab35.
Putative orthologues of DENND5A/Rab6-interacting protein 1 and its target Rab39 exist in worms, flies, and vertebrates. Interestingly, Rab39 is localized to the Golgi apparatus like its regulators DENND5A/B, and loss-of-function mutations in Rab39B cause X-linked mental retardation (Giannandrea et al., 2010). The underlying defect appears to be caused by altered trafficking required for growth cone and synapse formation, which suggests that further studies of DENND5 should focus on neuronal systems rather than fibroblast-like tissue culture cells. A homologue of the Myotubularin phosphatase domain containing DENN proteins MTMR5 and MTMR13, and their target Rab28, is present in protozoans (Fritz-Laylin et al., 2010), which suggests this has a trafficking function conserved at the cellular level rather than in a tissue-specific pathway. Contradicting this view somewhat, mutations in human MTMR13 and disruption of MMR13 in mice result in an autosomal recessive neuropathy, which suggests a function important for nervous system function (Azzedine et al., 2003; Robinson et al., 2008). Finally, DENND4 family GEFs and Rab10 are also always found together in multicellular organisms with polarized epithelial cell layers.
Rabs of the Rab8 and Rab10 families have been implicated in trafficking to polarized membrane domains at the cell surface (Babbey et al., 2006; Schuck et al., 2007). In budding yeast Sec4p, the Rab8 homologue is required for polarized transport from the late-Golgi into the growing bud (Walch-Solimena et al., 1997). In higher eukaryotes, Rab8 has been found to function in transport to actin-rich membrane protrusions (Peränen et al., 1996), the cilium, and apical surface of polarized epithelial cells (Nachury et al., 2007; Sato et al., 2007a; Yoshimura et al., 2007; Knödler et al., 2010). In contrast, Rab10 is reported to function in basolateral transport (Babbey et al., 2006; Schuck et al., 2007). It seems obvious that Rab8 and Rab10 would require activation by specific GEFs, and this appears to be the case. Sec4p and Rab8 are activated by Sec2 domain GEFs (Walch-Solimena et al., 1997; Hattula et al., 2002; Dong et al., 2007; Itzen et al., 2007; Sato et al., 2007b), whereas DENND4 family GEFs activate Rab10. This is supported by observations that mutations in CRAG, the presumed Drosophila orthologue of human DENND4, result in missorting of cargo such as perlecan and laminin destined for the basolateral surface of cells (Denef et al., 2008). However, the fruit fly poses a problem for this simple idea because it lacks a documented gene encoding a Sec2 domain protein. At present, it is only possible to speculate how Rab8 is activated in fruit flies, but the two most likely possibilities are either that there is a novel Rab8 GEF or that a known GEF family has acquired activity toward Rab8. DENND2 specificity raises similar issues because Rab9 is present in flies, although a readily discernable DENND2 is not. Flies may have some unique features with regard to Rab regulation, and further studies will be necessary to address these issues. Interestingly, DENND2B, also known as suppressor of tumorigenicity 5 (ST5), is mutated in human patients suffering from mental retardation and multiple congenital abnormalities that can result in deafness, cleft palate, and circulatory and kidney function defects (Göhring et al., 2010). Our results suggest that these defects may be caused by defective regulation of Rab9, and hence trafficking between late endosomes and lysosomes. This is reminiscent of lysosomal storage disorders, which are long known to cause mental retardation and other developmental abnormalities.
There appear to be no obvious characteristics distinguishing DENN-modulated Rabs from those controlled by other classes of GEFs and Rabs with as yet unknown GEFs. In a sequence alignment partitioned into these three groups there were no positions that clearly differentiated Rabs under DENN regulation from others (unpublished data). Furthermore, when sequence conservation among DENN-modulated Rabs was mapped onto the surface of the structure of one of them, no additional conserved surface patches—putative DENN-interaction sites—were observed when a comparison was made to Rabs not under DENN control (unpublished data). This suggests that DENNs bind to sites that overlap those seen for other GEFs, perhaps because of shared functional necessities such as switch I displacement (Barr and Lambright, 2010). This and an apparent lack of signature features of DENN-modulated Rabs suggest that only small changes and/or changes at different positions may have been responsible for altering GEF class specificity during Rab evolution. This conclusion is supported by the close relationships between Rabs that are regulated by unrelated GEF classes (Fig. 8).
Are DENN-related proteins Rab GEFs?
As shown here and elsewhere (Allaire et al., 2010; Marat and McPherson, 2010), DENN domain proteins are specific Rab GEFs. It is interesting to note that some of the DENN-related proteins are found throughout the eukaryotic kingdom and are therefore likely to function in conserved cellular processes, including membrane trafficking events. This suggests that the DENN-related proteins might also have Rab GEF activity. However, the only characterized member of this family, budding yeast Avl9p, is possibly a regulator of Gtr2p, a Ras superfamily GTPase outside the Rab subfamily (Harsay and Schekman, 2007; Zhang et al., 2010). Our preliminary evidence also indicates that human Avl9 does not act on any of the human Rabs. Although this supports the idea that DENN and DENN-related domains specify GEF activity toward GTPases of the Ras superfamily, these findings suggest they may not all be Rab GEFs. Further studies will be needed to investigate their specificity and define the cellular processes they act in.
Many conserved Rabs lack GEF regulators
Despite the advances presented here in assigning the specificity of 17 Rab GEFs, several key Rabs involved in membrane trafficking, lipid droplet, and cilium formation are still left without GEF activators. It is intriguing that these often cluster into related groups. For example, see Rab2/4/14 in Fig. 8, which may indicate that these share a family of related but as yet unknown regulators. Here, we argued that because Rabs form a large closely related family, their regulators might do the same and share a common domain. This led us to more closely investigate the DENN domain proteins. Following this line of argument, it may therefore be worthwhile to test conserved domains widely associated with proteins functioning in trafficking for GEF activity. This might include the DENN-related proteins, although there are caveats as discussed, the SNX and BAR domain families (van Weering et al., 2010), and RCC1 domain proteins (Ma et al., 2004). However, it is possible that common domains do not unite the remaining Rab GEFs, and thus other unbiased biochemical and genetic strategies will need to be followed if they are to be identified. Although the work presented here will be useful in informing many future studies on membrane trafficking, we are still some way from defining the full complement of Rab GEF and GAP regulators and effector proteins necessary for a full understanding of Rab function.
Materials and methods
Reagents and antibodies
General laboratory chemicals were obtained from Sigma-Aldrich and Thermo Fisher Scientific. Antibody to EGFP was raised in sheep against the entire coding region of EGFP and affinity purified. Rabbit anti–golgin-160 antibodies were raised and affinity purified against the entire coding region of rat golgin-160 expressed as a His6-tagged protein in bacteria. Mouse anti-clathrin clone X22 was a gift from S. Royle (University of Liverpool, Liverpool, England, UK). Commercially available antibodies were used to α-tubulin (mouse DM1A; Sigma-Aldrich), actin (mouse 2Q1055; Abcam), α-adaptin (clone 8; BD), γ-adaptin (mouse clone 88; BD), δ-adaptin (mouse clone 18; BD), EEA1 (rabbit 2411; Cell Signaling Technology), FLAG antibodies (mouse M2; Sigma-Aldrich), GM130 (mouse clone 35; BD), human LAMP1 (mouse clone 25; BD), TGN46 (sheep AHP500; Serotec), TfR (rabbit CBL47; Millipore), and CI-MPR (mouse 2G11; Abcam). Secondary antibodies raised in donkey to mouse, rabbit, sheep/goat, and human conjugated to HRP, Alexa Fluor 488, Alexa Fluor 555, Alexa Fluor 568, and Alexa Fluor 647 were obtained from Invitrogen and Jackson ImmunoResearch Laboratories.
Molecular biology and protein purification from bacteria and insect cells
Human DENNs were amplified from image clones (Source Bioscience Geneservice) or human fetal cDNA (Marathon ready cDNA; Takara Bio Inc.) using KOD polymerase (EMD). Mutagenesis was performed using the QuikChange method according to the protocol (Agilent Technologies). Duplexes for siRNA were obtained from QIAGEN or Thermo Fisher Scientific. Mammalian expression constructs were made using pcDNA4/TO and pcDNA5/FRT/TO vectors (Invitrogen). Bacterial expression constructs were made using pQE32 (QIAGEN), pMal (New England Biolabs, Inc.), and pFAT2 encoding the His6 tag, His6–maltose-binding protein, and His6–glutathione S-transferase, respectively. His6–glutathione S-transferase–tagged Rab proteins in pFAT2 were expressed in BL21 (DE3) pRIL or BL21 (DE3) pG-KGE8 (Takara Bio Inc.) at 18°C for 12–14 h, then purified using Ni-NTA agarose as described previously (Fuchs et al., 2005). In brief, cell pellets were lysed for 20 min in 10 ml IMAC5 (20 mM Tris-HCl, pH 8.0, 300 mM NaCl, 5 mM imidazole, 0.2% Triton X-100, and protease inhibitor cocktail; Roche) containing 0.5 mg/ml lysozyme, and then sonicated at 70% power four times for 30 s with a 30-s rest period. Lysates were clarified by centrifugation at 14,000 rpm in a JA-17 rotor for 30 min. To purify the tagged protein, 0.5 ml of nickel-charged NTA-agarose (QIAGEN) was added to the clarified lysate and rotated for 2 h. The agarose was washed three times with IMAC20 (IMAC5 with 20 mM imidazole), then the bound proteins were eluted in IMAC200 (IMAC5 with 200 mM imidazole), collecting 0.5 ml fractions. All manipulations were performed on ice or in an 8°C cold room. His6-tagged Rabex5, Rabin3/8, Rabin3-like/GRAB, DENND1B-S, DENND1B-L, and DENND2A-D in pQE32 were expressed in JM109 at 18°C for 12–14 h, then purified using nickel-charged NTA agarose using the same procedure as the Rabs. Rab12 and Rab39 were expressed using the pAcHis-GST vector encoding as His6–glutathione S-transferase tag, which is equivalent to that in pFAT2 in the baculovirus/Sf9 cell expression system (BD). For virus infection, 10 × 15-cm dishes of containing 1.5 × 108 at 5 × 105 cells/ml of Sf9 cells were infected with a virus moiety of infection of 1.0 for 48 h (Neef et al., 2005). The infected cells were harvested, washed once in PBS, and then lysed in IMAC5 for 20 min on ice. Lysates were clarified by centrifugation at 14,000 rpm in a JA-17 rotor (Beckman Coulter) for 30 min. Proteins were purified with the same protocol used for bacterial expression with minor modifications: 100 µl of nickel-charged NTA agarose was used for the purification, and 100-µl fractions of the elution were collected. Purified proteins were dialyzed against TBS (50mM Tris-HCl, pH 7.4, and 150 mM NaCl) and then snap frozen in liquid nitrogen for storage at −80°C. Protein concentration was measured using the Bradford assay.
Recombinant STxB was expressed untagged from pTrc99A in Escherichia coli BL21 (DE3) grown in lysogeny broth for 14 h at 37°C. All subsequent steps were performed at 4°C. The bacterial cell pellet from a 1 liter culture was resuspended in 20 ml of 20 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.1 mg/ml polymyxin B. After mixing for 30 min on a roller, the sample was sonicated twice at 70% power for 10 s. A periplasmic lysate was prepared by removing the cell debris by centrifugation, first at 13,000 g for 15 min then again at 90,000 g for 35 min. This lysate was diluted by the addition of two volumes of 20 mM Tris-HCl, pH 7.5, then loaded on to a 5 ml HiTrapQ column (GE Healthcare). The column was washed with 20 ml of 20 mM Tris-HCl, pH 7.5, then eluted with a 50-ml linear gradient from 0–600 mM NaCl in the same buffer. The flow rate was 2 ml/min throughout. Fractions of 2.5 ml were collected and analyzed by SDS-PAGE, and the peak of STxB pooled to give a 15-ml solution at 1 mg/ml. This was then concentrated to 5 mg/ml using Centricon 10K units (Millipore) centrifuged at 3,000 g. A 500 µl aliquot was then applied at 0.15 ml/min to a Superose 12 gel filtration column equilibrated in PBS, and 1-ml fractions were collected. The peak fractions of pentameric STxB were pooled to give a 1.1 mg/ml solution. A 1-ml aliquot of this solution was labeled by the addition of one vial of monoreactive N-hydroxysuccinimidyl Cy3 (GE Healthcare). After 5 min at room temperature, 100 µl of 1 M Tris-HCl, pH 8.0, was added to stop the reaction. Excess dye was removed by desalting over PD10 columns (GE Healthcare). Fractions of 500 µl were collected, and absorption was measured at 280 and 552 nm. Using molar extinction coefficients of 150,000 M−1cm−1 for Cy3 and 170,000 M−1cm−1 for pentameric STxB, dye labeling was calculated as 1 Cy3 per toxin subunit with a concentration of 0.7 mg/ml.
Cell culture and protein purification from mammalian cells
HeLa and HEK293 cells were cultured in DME containing 10% bovine calf serum (Invitrogen) at 37°C and 5% CO2. For plasmid transfection and siRNA transfection, Mirus LT1 (Mirus Bio LLC), and Oligofectamine (Invitrogen), respectively, were used according to the manufacturers’ instructions. FLAG-tagged forms of DENND1A, DENND1C, DENND3, DENND4A, DENND4B, DENND4C, DENND5A, DENND5B, MTMR5, MTMR13, and MADD in pcDNA5/FRT/TO were transiently expressed in 10 × 15-cm dishes of 70% confluent HeLa cells. After 48 h of growth, the cell pellet was lysed for 20 min on ice in 5 ml of cell lysis buffer (50 mM Tris-HCl, pH 7.4, 1 mM EDTA, 150 mM NaCl, 0.5% Triton X-100, and protease inhibitors cocktails). Cell lysates were split into 1.0 ml aliquots and clarified by centrifugation at 20,000 g in a microcentrifuge (5417R Microfuge; Eppendorf) for 20 min. The FLAG-tagged proteins were immunoprecipitated from the clarified lysate using 100 µl of anti-FLAG M2 affinity gel (Sigma-Aldrich) for 4 h at 4°C. The pellet was washed 10 times in 1 ml of cell lysis buffer, 10 times in 1 ml of high-salt buffer (50 mM Tris-HCl, pH 7.4, and 500 mM NaCl), and 10 times in TBS, and finally the proteins were eluted with 100 µl of 200 µg/ml FLAG-peptide in TBS containing 2 mM dithiothreitol. The eluted proteins were analyzed on 7.5–10% SDS-PAGE gels stained with Coomassie brilliant blue, and concentrations were estimated by comparison to a series of BSA standards in the range of 0.1–1 mg. The peak fractions were snap frozen in liquid nitrogen for storage at −80°C without dialysis.
Nucleotide binding and GEF assays
Nucleotide loading was performed as follows: 10 µg of GST-tagged Rab was incubated in 50 mM Hepes-NaOH, pH 6.8, 0.1 mg/ml BSA, 125 µM EDTA, 10 µM Mg-GDP, and 5 µCi [3H]-GDP (10 mCi/ml; 5,000 Ci/mmol) in a total volume of 200 µl for 15 min at 30°C. For standard GDP-releasing GEF assays, 100 µl of the loading reaction was mixed with 10 µl of 10 mM Mg-GTP and 10–100 nM GEF protein to be tested or a buffer control, and adjusted to 120 µl final volume with assay buffer. The GEF reaction occurred for 20 min at 30°C. After this, 2.5 µl was taken for a specific activity measurement; the remainder was split into two tubes, then incubated with 500 µl of ice-cold assay buffer containing 1 mM MgCl2 and 20 µl of packed glutathione-sepharose for 60 min at 4°C. After washing three times with 500 µl of ice-cold assay buffer, the sepharose was transferred to a vial containing 4 ml of scintillation fluid and counted. The amount of nucleotide exchange was calculated in pmoles of GDP released. For GTP-binding assays the following modifications were made: only unlabeled GDP was used in the loading reaction; in the GEF reaction, 0.5 µl of 10 mM GTP and 1 µCi [35S]-GTPγS (10 mCi/ml; 5,000 Ci/mmol) were used. The amount of nucleotide exchange was calculated in pmoles of GTP bound.
Microscopy
Fixed samples on glass slides were imaged using a 60× 1.35 NA oil immersion objective lens on a standard upright microscope equipped with a CoolSNAP HQ2 camera (Roper Industries) under the control of MetaMorph 7.5 software (MDS Analytical Technologies). Images were cropped in Photoshop CS3 (Adobe) or ImageJ and placed into Illustrator CS3 (Adobe) without performing any other contrast adjustments of image manipulations to produce the figures.
Mass spectrometry
Protein samples for mass spectrometry were separated on 4–12% gradient NuPAGE gels (Invitrogen), then stained using a colloidal Coomassie blue stain. Gel lanes were typically cut into 12 slices, and then digested with trypsin using published methods (Wilm et al., 1996). The resulting tryptic peptide mixtures in 0.05% trifluoroacetic acid were then analyzed by online liquid chromatography tandem mass spectrometry with a nanoAcquity UPLC (Waters) and Orbitrap XL ETD mass-spectrometer (Thermo Fisher Scientific) fitted with a nano-electrospray source (Thermo Fisher Scientific). Peptides were loaded on to a 5 cm × 180 µm BEH-C18 Symmetry trap column (part no. 186003514; Waters) in 0.1% formic acid at 15 µl/minute, and then resolved using a 25 cm × 75 µm BEH-C18 column (part no. 186003815; Waters) in 99–37.5% acetonitrile in 0.1% formic acid at a flow rate of 400 nl/min. The mass spectrometer was set to acquire a mass spectrometry survey scan in the Orbitrap (R = 30,000) and then perform tandem mass spectrometry on the top five ions in the linear quadrupole ion trap after fragmentation using collision ionization (30 ms, 35% energy). A 90-s rolling exclusion list with n = 3 was used to prevent redundant analysis of the same ions. Maxquant and Mascot (Matrix Science) were then used to compile and search the raw data against the human International Protein Index database. Protein group and peptide lists were sorted and analyzed in Excel (Microsoft) and Maxquant (Cox and Mann, 2008). Mass spectrometry and tandem mass spectrometry spectra were manually inspected using Xcalibur Qualbrowser (Thermo Fisher Scientific).
Bioinformatics
Sequence alignments of DENNs and Rabs were done with ClustalX (Chenna et al., 2003) or MUSCLE (Edgar, 2004), and the results were visualized and manipulated with Jalview (Waterhouse et al., 2009). ClustalX was also used to produce a dendrogram illustrative of sequence relationships. Regions of intrinsic disorder were predicted using the metaPrDOS server (Ishida and Kinoshita, 2008), and portions of them with energetic properties appropriate for forming protein–protein interactions were highlighted with ANCHOR (Mészáros et al., 2009). Linear sequence motifs were browsed in the ELM database (Gould et al., 2010).
Online supplemental material
Fig. S1 shows the pattern of localization for all human DENN domain proteins when transfected as EGFP-tagged constructs in Hela cells. Table S1 lists the DENN domain proteins identified in human, mouse, zebrafish, fruit fly, and nematode.
Acknowledgments
We thank Dr. Ricardo Nunes Bastos, Xenia Penate, and Ulrike Gruneberg for advice and discussion.
This work was supported by a Wellcome Trust program grant (08246/Z/07/Z) and a project grant from the Deutsche Forschungsgemeinschaft awarded to F.A. Barr.
References
Author notes
S.-i. Yoshimura’s present address is Dept. of Cell Biology, Graduate School of Medicine, Osaka University, Suita, 565-0871 Osaka, Japan.


![Figure 2. DENND1A/1B are GEFs for Rab35. (A) Human Rabex-5 was tested against a representative panel of human Rab proteins using the GDP-releasing assay. In brief, 10 µg of each GST-tagged Rab to be tested was incubated in 50 mM Hepes-NaOH, pH 6.8, 0.1 mg/ml BSA, 125 µM EDTA, 10 µM Mg-GDP, and 5 µCi [3H]-GDP (10 mCi/ml; 5,000 Ci/mmol) in a total volume of 200 µl for 15 min at 30°C to load the Rab with the radioactive GDP probe. For standard GDP-releasing GEF assays, 100 µl of the loading reaction was then mixed with 10 µl of 10 mM Mg-GTP and 10 nM His6-tagged Rabex-5 purified from bacteria or a buffer control, then adjusted to 120 µl final volume with assay buffer. The GEF reaction occurred for 20 min at 30°C. After this, 2.5 µl was taken for a specific activity measurement; the remainder was split into two tubes, then incubated with 500 µl of ice-cold assay buffer containing 1 mM MgCl2 and 20 µl of packed glutathione-sepharose for 60 min at 4°C to separate Rab–GDP complexes from free “released” GDP. After washing three times with 500 µl of ice-cold assay buffer, the sepharose was transferred to a vial containing 4 ml of scintillation fluid and counted. The amount of nucleotide exchange was calculated in pmoles of GDP released. (B and C) A representative panel of human Rab proteins was tested against 10 nM of His6-tagged DENND1B-S in the GDP-releasing (B) or GTP-binding assay (C). For GTP-binding assays, the following modifications were made: only unlabeled GDP was used in the loading reaction; in the GEF reaction, 0.5 µl of 10 mM GTP and 1 µCi [35S]-GTPγS (10 mCi/ml; 5000 Ci/mmol) were used. The amount of nucleotide exchange was calculated in pmoles of GTP bound. (D and E) Human DENND1A (D), DENND1B-L (D), and DENND1C (E) were tested against a subset of Rab35-related Rabs using the GTP-binding assay. For these assays, 10 nM of FLAG-tagged DENND1A or DENND1C purified from HeLa cells, or 10 nM of His6-tagged DENND1B-L purified from bacteria were used. Errors bars show the standard error from the mean. The red line marks double the median value taken as a threshold.](https://cdn.rupress.org/rup/content_public/journal/jcb/191/2/10.1083_jcb.201008051/9/m_jcb_201008051_rgb_fig2.jpeg?Expires=1773752004&Signature=jNYbj3fNYbefy23IZZ8obaadbr2lfCIZazKy4I37QQsNUU2gsIQQruaTPBzoFmRwnwpb38Yy9oIQXAY4yw-gL1q3ww8l0-YYVTph2ovXUzz1-XkdP8UvObN6bpq81J21wSt~AKMKuS32quHduVH6MvN7VmuVQa0oefbgMVJfYP1TPqbq2ys8xdVT0Le5adYlv6D3gKIbTJnhcNhmGPOFsZcYgRDMe~0EDovGSu-xPB46c02-sFWNer9sBU1qd3OYxUIGjcAQdRfqFIG5~eYo~LHF3wwNFhvxUWBxkwoJoDRwiFrml-NJmwLvhWF8fXdQdERowYlXwD-MM2wz3Shlqw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)








