Secretory protein folding is monitored by endoplasmic reticulum (ER) quality control mechanisms. Misfolded proteins are retained and targeted to ER-associated degradation (ERAD) pathways. At their core are E3 ubiquitin ligases, which organize factors that recognize, ubiquitinate, and translocate substrates. Of these, we report that the Hrd1 complex manages three distinct substrate classes. A core complex is required for all classes and is sufficient for some membrane proteins. The accessory factors Usa1p and Der1p adapt the complex to process luminal substrates. Their integration is sufficient to process molecules bearing glycan-independent degradation signals. The presence of Yos9p extends the substrate range by mediating the recognition of glycan-based degradation signals. This modular organization enables the Hrd1 complex to recognize topologically diverse substrates. The Hrd1 system does not directly evaluate the folding state of polypeptides. Instead, it does so indirectly, by recognizing specific embedded signals displayed upon misfolding.
Introduction
Cellular quality control pathways monitor protein folding and target flawed products for turnover. In the secretory pathway, ER quality control (ERQC) mechanisms must also regulate trafficking to prevent the premature export of misfolded proteins. Proteins deemed misfolded are routed to ER-associated degradation (ERAD) pathways. These pathways are defined by specialized E3 ubiquitin ligases that organize factors to receive and extract polypeptides from the ER membrane. On the cytosolic face, substrates are ubiquitinated and degraded by the 26S proteasome (Sifers, 2003; Sitia and Braakman, 2003; Römisch, 2005; Vembar and Brodsky, 2008).
In budding yeast, the Hrd1p and Doa10p ubiquitin ligases represent two distinct pathways (Huyer et al., 2004; Vashist and Ng, 2004; Carvalho et al., 2006). Metazoans have homologues of these E3s and have also evolved an expanded repertoire of ERAD-processing centers (for review see Nakatsukasa and Brodsky, 2008). Of the yeast ERAD-processing sites, the Doa10 complex is the simplest. At Doa10 sites, substrate ubiquitination is mediated by the E2 ubiquitin–conjugating enzyme Ubc7p, which is attached to Doa10p via Cue1p. The Cdc48p (p97 in mammals)–Npl4p–Ufd1p subcomplex is linked via Ubx2p and extracts substrates from the membrane (Ye et al., 2001; Rabinovich et al., 2002; Lilley and Ploegh, 2004; Schuberth and Buchberger, 2005; Carvalho et al., 2006; Gauss et al., 2006b). Doa10p clients include folded cytosolic proteins and membrane proteins bearing misfolded cytosolic domains (ERAD-C; Swanson et al., 2001; Huyer et al., 2004; Vashist and Ng, 2004; Ravid et al., 2006; Metzger et al., 2008). The Hrd1 complex contains all known partners of Doa10p and several more. Directly bound to Hrd1p is Hrd3p, a tetratricopeptide repeat–containing protein involved in substrate binding (Gardner et al., 2000; Denic et al., 2006; Gauss et al., 2006b). Associated with Hrd3p (SEL1L in mammals) is the lectin-like factor Yos9p (OS-9 and XTP3-B in mammals; Carvalho et al., 2006; Denic et al., 2006; Mueller et al., 2006; Christianson et al., 2008; Hosokawa et al., 2008). Yos9p is the receptor for glycans containing a terminal α1,6-linked mannose generated by Htm1p (also known as Mnl1p; Nakatsukasa et al., 2001; Olivari et al., 2006; Hosokawa et al., 2007; Quan et al., 2008; Clerc et al., 2009). Htm1p is the final enzyme of a glycan-trimming cascade that forms the basis of a de facto folding timer. Unfolded proteins modified by Htm1p display an ERAD signal comprised of the glycan and adjacent unfolded peptide segment (Xie et al., 2009). This pathway has been termed ERAD-L because it detects misfolded luminal domains (Vashist and Ng, 2004). Two additional factors of the Hrd1 complex, Usa1p (HERP in mammals) and Der1p (Derlin-1 in mammals), are required for ERAD-L (Knop et al., 1996a; Lilley and Ploegh, 2004; Ye et al., 2004; Carvalho et al., 2006; Okuda-Shimizu and Hendershot, 2007). The Hrd1p complex also detects proteins with misfolded membrane segments through a mode termed ERAD-M (ERAD membrane; Carvalho et al., 2006).
Together, these systems appear to account for the quality control mechanisms for nascent polypeptides that traffic through the ER and provide a unifying model for ERAD, at least in budding yeast (Carvalho et al., 2006). However, one substrate class, nonglycosylated soluble proteins, seems to fall outside of this framework. A nonglycosylated variant of the mating pheromone precursor, ΔgppαF, is retained in the ER and degraded by the proteasome (McCracken and Brodsky, 1996). Unlike other known ERAD substrates, degradation is independent of ubiquitination and all known components of the E3 complexes (Brodsky and McCracken, 1999). Cholera toxin and the ricin A chain may hijack a similar ubiquitin-independent mechanism for translocation from the ER to the cytosol (Simpson et al., 1999; Rodighiero et al., 2002). In contrast, the mammalian Hrd1 pathway degrades unassembled immunoglobulin κ light chain and the NHK-QQQ variant of α1-antitrypsin, both of which are nonglycosylated soluble proteins (Okuda-Shimizu and Hendershot, 2007; Hosokawa et al., 2008). Required components for their degradation include Hrd1, p97, Derlin-1, Herp, and XTP3-B. In a mammalian cell-free system, ΔgppαF retrotranslocation does not require the activity of p97 but requires Derlin-1, a member of the Hrd1 complex (Wahlman et al., 2007). Do these findings indicate an evolutionary divergence for how this substrate class is degraded in mammals?
In this study, we report that the yeast Hrd1 complex has the capacity to process three substrate classes. The pattern of substrate specialization among components reveals functional modularity in its organization. At the most basic level is a core complex whose factors are required for all substrates. These are comprised of factors common to both Doa10 and Hrd1 complexes. The integration of accessory factors Usa1p and Der1p expands the client pool to include luminal proteins. The addition of another factor, Yos9p, adapts the complex to recognize misfolded glycoproteins vetted through a glycan timer mechanism. Importantly, we demonstrate that utilization of luminal pathways is specified by signals embedded in proteins that are displayed only when misfolded. Together, these data show that the Hrd1 E3 complex manages distinct substrate classes by assimilating factors that diversify signal recognition.
Results
Novel PrA variants reveal a third substrate class of the yeast Hrd1 complex
A folding defective mutant of the vacuolar proteinase A (PrA) called PrA*-Ab is a glycan-dependent substrate of the ERAD-L pathway (Fig. 1 A; Finger et al., 1993; Spear and Ng, 2005). Deletion analysis identified two variants, PrA*-AbΔ147–183 and PrA*-AbΔ295–331, degraded rapidly with a reduced dependence on Htm1p, suggesting a glycan-independent mode of ERAD-L (Fig. S1; Xie et al., 2009). To confirm the assertion, a nonglycosylated PrA (ngPrA) variant called ngPrA*Δ295–331 (Fig. 1 A) was tested. Its rapid degradation in control cells and stability in Δhrd1 cells demonstrate the existence of the pathway in budding yeast (Fig. 1 B). As expected, cells lacking HTM1 failed to retard its degradation and even slightly accelerated it (Fig. 1 B). For this pathway, the Δ295–331 lesion alone is sufficient to cause glycan-independent degradation of PrA. The variant ngPrAΔ295–331 (Fig. 1 A) that lacks the “star” deletion (amino acids 55–91) behaved identically (Fig. 1 C). Substrates of ERQC are typically retained in the ER even if degradation is blocked (Gething et al., 1986; de Silva et al., 1990; Loayza et al., 1998). Using indirect immunofluorescence, ngPrAΔ295–331 colocalized with the ER marker Kar2p in Δhrd1 cells, confirming it as a bona fide substrate of ERQC (Fig. 1 D).
ngPrA variants are substrates of the Hrd1p-dependent ERAD pathway. (A) Schematic representation of PrA variants with positions of deletions under each diagram. N-linked glycans are represented by branched symbols, signal sequences are shown in dark gray, prodomains are colored light gray, and HA epitope tags are indicated. PrA* differs from PrA by lacking sequences Leu55 through Tyr91. (B) Wild-type (WT), Δhrd1, and Δhtm1 cells expressing ngPrA*Δ295–331 were pulse labeled for 10 min with [35S]methionine/cysteine at 30°C followed by a cold chase for the times indicated. Immunoprecipitation of ngPrA*Δ295–331 was performed using anti-HA mAb and resolved by SDS-PAGE. Decay kinetics were quantified by phosphorimager analysis. The data reflect three independent experiments with the SEM indicated by error bars. Representative phosphor scan images are shown. (C) Wild-type, Δhrd1, and Δhtm1 cells expressing ngPrAΔ295–331 were analyzed by pulse-chase analysis as described in B. (D) Intracellular localization of ngPrAΔ295–331 in Δhrd1 cells was analyzed by indirect immunofluorescence. The substrate was detected using anti-HA antibodies (green), the ER was visualized using anti-Kar2p antisera (red), and the positions of nuclei were stained by DAPI. Bar, 2 µM.
ngPrA variants are substrates of the Hrd1p-dependent ERAD pathway. (A) Schematic representation of PrA variants with positions of deletions under each diagram. N-linked glycans are represented by branched symbols, signal sequences are shown in dark gray, prodomains are colored light gray, and HA epitope tags are indicated. PrA* differs from PrA by lacking sequences Leu55 through Tyr91. (B) Wild-type (WT), Δhrd1, and Δhtm1 cells expressing ngPrA*Δ295–331 were pulse labeled for 10 min with [35S]methionine/cysteine at 30°C followed by a cold chase for the times indicated. Immunoprecipitation of ngPrA*Δ295–331 was performed using anti-HA mAb and resolved by SDS-PAGE. Decay kinetics were quantified by phosphorimager analysis. The data reflect three independent experiments with the SEM indicated by error bars. Representative phosphor scan images are shown. (C) Wild-type, Δhrd1, and Δhtm1 cells expressing ngPrAΔ295–331 were analyzed by pulse-chase analysis as described in B. (D) Intracellular localization of ngPrAΔ295–331 in Δhrd1 cells was analyzed by indirect immunofluorescence. The substrate was detected using anti-HA antibodies (green), the ER was visualized using anti-Kar2p antisera (red), and the positions of nuclei were stained by DAPI. Bar, 2 µM.
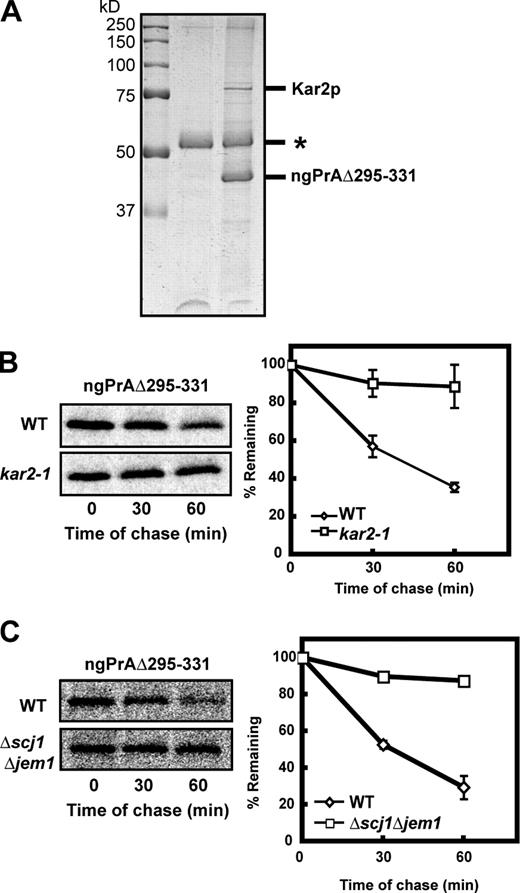
To identify direct interactors of ngPrAΔ295–331, the substrate was purified from a microsomal fraction under nondenaturing conditions. A major species migrating near the 75-kD marker was identified as the ER chaperone Kar2p (BiP) by mass spectrometry (Fig. 2 A and Table S1). To test the requirement of the chaperone in the glycan-independent mode of the Hrd1 pathway, ngPrAΔ295–331 turnover was analyzed in the kar2-1 and Δscj1Δjem1 mutants. The kar2-1 allele disrupts ERAD without affecting Kar2p’s essential housekeeping functions (Kabani et al., 2003). Scj1p and Jem1p are ER DnaJ family proteins that functionally interact with Kar2p (Silberstein et al., 1998; Nishikawa et al., 2001). Likely because of redundancy, SCJ1 and JEM1 single mutants display weak and no ERAD phenotypes, respectively, whereas the double mutant is strongly defective (Nishikawa et al., 2001). ngPrAΔ295–331 is strongly stabilized in both strains, demonstrating a critical role of the chaperone system in glycan-independent ERAD (Fig. 2, B and C).
The Kar2 chaperone system is required for glycan-independent ERAD-L. (A) ngPrAΔ295–331 protein complexes purified from microsomes were resolved by SDS-PAGE and visualized by Coomassie brilliant blue. The band labeled Kar2p was excised and identified by mass spectrometry (Table S1). The asterisk indicates the position of IgG heavy chain. (B and C) Wild-type (WT), kar2-1, and Δscj1Δjem1 cells expressing ngPrAΔ295–331 were analyzed by metabolic pulse-chase analysis as described in Fig. 1 B. The plotted data reflect three independent experiments with the SEM indicated by error bars.
The Kar2 chaperone system is required for glycan-independent ERAD-L. (A) ngPrAΔ295–331 protein complexes purified from microsomes were resolved by SDS-PAGE and visualized by Coomassie brilliant blue. The band labeled Kar2p was excised and identified by mass spectrometry (Table S1). The asterisk indicates the position of IgG heavy chain. (B and C) Wild-type (WT), kar2-1, and Δscj1Δjem1 cells expressing ngPrAΔ295–331 were analyzed by metabolic pulse-chase analysis as described in Fig. 1 B. The plotted data reflect three independent experiments with the SEM indicated by error bars.
The glycan-independent luminal pathway requires most but not all factors of the Hrd1 ERAD system
We next determined the factors of the Hrd1 complex required to recognize and degrade ngPrAΔ295–331. This analysis extends the important mammalian findings by providing a more comprehensive accounting of relevant factors (Okuda-Shimizu and Hendershot, 2007; Hosokawa et al., 2008). First, functions in common with the Doa10 complex were tested. ngPrAΔ295–331 is strongly stabilized in cdc48-1 and Δcue1 strains, indicating that its degradation follows a classical ERAD mechanism (Fig. 3, A and B).
ngPrAΔ295–331 degradation requires multiple components of the Hrd1 complex. Metabolic pulse-chase experiments were performed and presented as in Fig. 1 B. Strains and substrates are indicated. (A) Wild-type (WT) and cdc48-1 cells expressing ngPrAΔ295–331 were grown to log phase at 25°C and shifted to 30°C for 30 min before the experiment. (B) Wild-type and Δcue1 cells expressing ngPrAΔ295–331. (C) Wild-type, Δder1, and Δusa1 cells expressing ngPrAΔ295–331. (D) Wild-type and Δyos9 cells expressing ngPrAΔ295–331. (E) Wild-type, Δyos9, and yos9-R200A cells expressing PrA*-Ab. (F) Wild-type, Δyos9, and yos9-R200A cells expressing ngPrAΔ295–331. Error bars indicate SEM.
ngPrAΔ295–331 degradation requires multiple components of the Hrd1 complex. Metabolic pulse-chase experiments were performed and presented as in Fig. 1 B. Strains and substrates are indicated. (A) Wild-type (WT) and cdc48-1 cells expressing ngPrAΔ295–331 were grown to log phase at 25°C and shifted to 30°C for 30 min before the experiment. (B) Wild-type and Δcue1 cells expressing ngPrAΔ295–331. (C) Wild-type, Δder1, and Δusa1 cells expressing ngPrAΔ295–331. (D) Wild-type and Δyos9 cells expressing ngPrAΔ295–331. (E) Wild-type, Δyos9, and yos9-R200A cells expressing PrA*-Ab. (F) Wild-type, Δyos9, and yos9-R200A cells expressing ngPrAΔ295–331. Error bars indicate SEM.
Next, genes encoding components exclusive to the Hrd1 complex were examined. Pulse-chase analysis shows that DER1 and USA1 are also essential for ngPrAΔ295–331 degradation (Fig. 3 C). The analysis was not performed for Hrd3p because its presence is required for Hrd1p stability (Gardner et al., 2000). In contrast, degradation of ngPrAΔ295–331 is efficient in a strain deleted of YOS9 (Fig. 3 D). This differs from glycan-dependent substrates, which depend heavily on Yos9p for degradation (Buschhorn et al., 2004; Bhamidipati et al., 2005; Kim et al., 2005; Szathmary et al., 2005). Nevertheless, we did observe slight stabilization over wild type. We wondered whether this result reflects a possible role in the glycan-independent pathway. In mammals, reduction of the Yos9 homologue hXTP3-B (long form) stabilizes the nonglycosylated NHK-QQQ substrate (Hosokawa et al., 2008). Alternatively, the effect we observed could be indirect, caused structurally by physically eliminating a member of the complex. To confirm or rule out the latter possibility, we analyzed degradation in the yos9-R200A variant (Bhamidipati et al., 2005). In this strain, a conserved residue required for glycan recognition is mutated. By pulse-chase analysis, the degradation defect of PrA*-Ab (glycan-dependent substrate) in the yos9-R200A strain is equal to Δyos9, demonstrating the efficacy of the point mutation (Fig. 3 E). In contrast, ngPrAΔ295–331 turnover in the yos9-R200A was as efficient as wild type (Fig. 3 F). Collectively, the profile of ngPrAΔ295–331 degradation mirrors that of misfolded glycoproteins, except Htm1p and Yos9p are dispensable (Table I). Although they are established mediators of glycan-based ERAD, these data provide new evidence of their functional specificity.
Genetic requirements for the degradation of Hrd1-dependent substrates
| Substrate | HRD1 | CUE1 | CDC48 | USA1 | DER1 | YOS9 | HTM1 |
| CPY*, PrA* | + | + | + | + | + | + | + |
| ngPrAΔ295–331 | + | + | + | + | + | − | − |
| Hmg2p | + | + | + | − | − | − | − |
| Substrate | HRD1 | CUE1 | CDC48 | USA1 | DER1 | YOS9 | HTM1 |
| CPY*, PrA* | + | + | + | + | + | + | + |
| ngPrAΔ295–331 | + | + | + | + | + | − | − |
| Hmg2p | + | + | + | − | − | − | − |
ngPrAΔ295–331 competes with the glycan-dependent substrate CPY* for degradation
A previous study provided evidence of Hrd1 subcomplexes, differing only by the occupancy of Yos9p (Gauss et al., 2006a). Combined with the results presented in this study, the data raise the intriguing possibility of distinct receptor sites devoted to glycosylated and nonglycosylated substrates. We applied an in vivo substrate competition assay to test the possibility. CPY* regulated by the inducible GAL1 promoter was selected as the glycan-dependent competitor (Johnston and Davis, 1984; Knop et al., 1996b). Test proteins were constitutively expressed from their native promoters, and turnover was monitored by metabolic pulse-chase analysis. As expected, CPY* competes with PrA*-Ab, the glycan-dependent control (Fig. 4 A). When the assay was applied to ngPrAΔ295–331, a similar reduction was observed, indicating that the two substrate classes compete in ERAD (Fig. 4 B). To determine whether competition is specific to the Hrd1p complex or includes general factors of ERAD, the assay was repeated with the Doa10p-dependent substrate Ste6-166p. In this study, Ste6-166p degradation was as efficient as the negative control after a slight delay at the first chase point (Fig. 4 C). These data show that the two luminal substrate classes can compete for the same Hrd1 complexes.
Glycan-independent and glycan-dependent substrates of ERAD-L are competitors for degradation. (A) Wild-type cells carrying GAL1-regulated CPY* (or nonexpressing control vector) and PrA*-Ab regulated by its native promoter were shifted to galactose media to induce CPY* expression. Turnover of PrA*-Ab was analyzed as described in Fig. 1 B. (B) The experiment was performed as described for A with ngPrAΔ295–331 as the test substrate. (C) The experiment was performed as described for A with Ste6-166p as the test substrate. OE, overexpression. Error bars indicate SEM.
Glycan-independent and glycan-dependent substrates of ERAD-L are competitors for degradation. (A) Wild-type cells carrying GAL1-regulated CPY* (or nonexpressing control vector) and PrA*-Ab regulated by its native promoter were shifted to galactose media to induce CPY* expression. Turnover of PrA*-Ab was analyzed as described in Fig. 1 B. (B) The experiment was performed as described for A with ngPrAΔ295–331 as the test substrate. (C) The experiment was performed as described for A with Ste6-166p as the test substrate. OE, overexpression. Error bars indicate SEM.
The glycan-independent mode of ERAD-L recognizes distinct degradation signals
The classical ERAD substrates CPY* and PrA* contain bipartite degradation signals composed of specific glycans and their adjacent unfolded peptide segments (Quan et al., 2008; Clerc et al., 2009; Xie et al., 2009). Without these glycans, the molecules are stable, suggesting that the remaining segments are not recognized by a glycan-independent mode (Kostova and Wolf, 2005; Spear and Ng, 2005). To identify a glycan-independent determinant, we started with our shortest variant, PrA*-AbΔ147–183/295–331 (Fig. S1 E), and eliminated its remaining glycan to generate PrA*-GI (glycan independent; Fig. 5 A). As shown in Fig. 5 B, its degradation profile shows that it is a substrate of ERAD. A systematic deletion series was generated for PrA*-GI to identify essential ERAD determinants if they exist (Fig. 5 A). Deleting any of its three N-proximal segments had little effect on degradation (Fig. 5 C). This includes PrA*-GIΔ2, which eliminated the peptide portion of the glycan-dependent determinant (Xie et al., 2009). Deletion of the C-terminal segment, however, strongly stabilized the molecule, indicating that an important element was altered (Fig. 5 C, PrA*-GIΔ4). However, PrA*-GIΔ4 is less stable than PrA*-GI in ERAD mutants (Fig. 5, compare C with B), suggesting the possible presence of a second, less-important determinant elsewhere in PrA.
PrA contains a distinct determinant for glycan-independent ERAD. (A) Schematic representation of PrA*-GI variants. N-linked glycans are represented by branched symbols, signal sequences are shaded dark gray, prodomains are colored light gray, and positions of HA epitope tags are indicated. Dashed lines mark deleted regions. (B, C, and E–G) Substrate decay rates were measured and presented as described in Fig. 1 B. Strains and substrates are indicated. (B) PrA*-GI decay rates in wild-type (WT), Δhtm1, and Δhrd1 cells. (C) Degradation profiles for PrA*-GIΔ1, PrA*-GIΔ2, PrA*-GIΔ3, or PrA*-GIΔ4 in wild-type cells. (D) Intracellular localization of PrA*-GIΔ4 in wild-type cells was analyzed by confocal microscopy. The substrate was detected using anti-HA antibodies (green), the ER was visualized using anti-Kar2p antisera (red), and nuclei were stained by DAPI. Bar, 5 µM. (E) Wild-type and Δhtm1 cells expressing PrA*-GIΔ4CHO. (F) Wild-type, Δhrd1, Δusa1, and Δder1 cells expressing PrA-CTD. (G) Wild-type, Δyos9, and Δhtm1 cells expressing PrA-CTD. Error bars indicate SEM.
PrA contains a distinct determinant for glycan-independent ERAD. (A) Schematic representation of PrA*-GI variants. N-linked glycans are represented by branched symbols, signal sequences are shaded dark gray, prodomains are colored light gray, and positions of HA epitope tags are indicated. Dashed lines mark deleted regions. (B, C, and E–G) Substrate decay rates were measured and presented as described in Fig. 1 B. Strains and substrates are indicated. (B) PrA*-GI decay rates in wild-type (WT), Δhtm1, and Δhrd1 cells. (C) Degradation profiles for PrA*-GIΔ1, PrA*-GIΔ2, PrA*-GIΔ3, or PrA*-GIΔ4 in wild-type cells. (D) Intracellular localization of PrA*-GIΔ4 in wild-type cells was analyzed by confocal microscopy. The substrate was detected using anti-HA antibodies (green), the ER was visualized using anti-Kar2p antisera (red), and nuclei were stained by DAPI. Bar, 5 µM. (E) Wild-type and Δhtm1 cells expressing PrA*-GIΔ4CHO. (F) Wild-type, Δhrd1, Δusa1, and Δder1 cells expressing PrA-CTD. (G) Wild-type, Δyos9, and Δhtm1 cells expressing PrA-CTD. Error bars indicate SEM.
By analogy to glycan-dependent signals, if the deletion specifically removed a degradation signal, PrA*-GIΔ4 is expected to be retained stably in the ER as a misfolded protein. Alternatively, mislocalization or the formation of a structure incompatible with ERAD could explain the phenotype. To address these possibilities, two experiments were performed. Indirect immunofluorescence shows that PrA*-GIΔ4 colocalizes with the ER marker Kar2p (Fig. 5 D). This result shows that the molecule is retained by ERQC but incompetent for ERAD. To assess whether PrA*-GIΔ4 generated a molecule generally incapable of entering ERAD, the A glycosylation site was restored to reconstitute the original glycan-dependent signal. The resulting molecule, PrA*-GIΔ4CHO (Fig. 5 A), degraded rapidly in an HTM1-dependent fashion (Fig. 5 E). This supports the conclusion that the Δ4 lesion eliminated a critical degradation signal. To determine whether the element is sufficient to signal glycan-independent degradation, the C-terminal domain (CTD) containing this domain was expressed behind the PrA signal sequence (Fig. 5 A, PrA-CTD). The ngPrA-CTD is degraded rapidly in wild-type cells and strongly stabilized in Δhrd1, Δusa1, and Δder1 strains, confirming that it uses the Hrd1 pathway (Fig. 5 F). As expected, PrA-CTD degradation is efficient in cells lacking HTM1 or YOS9 (Fig. 5 G). This result confirms that the Hrd1 complex can efficiently process nonglycosylated luminal substrates in the absence of Yos9p. These data show that determinants recognized by the glycan-dependent and -independent modes of the Hrd1p complex can be nonoverlapping and distinct within the same molecule.
Discussion
The Hrd1 complex is functionally modular in its management of three substrate classes
This study extends the known Hrd1 client range in budding yeast and demonstrates that the surveillance of glycosylated and nonglycosylated luminal substrates by the Hrd1 complex is conserved. Based on these results, we questioned how a single complex could manage such a diverse range of substrates. An interesting pattern emerges from comparison of data from previous studies and this study (Table I; Bays et al., 2001; Carvalho et al., 2006; Sato et al., 2009). ERAD-M substrates like Sec61-2p and Hmg2p have the simplest requirements for substrate recognition and degradation. These include the Hrd1/Hrd3 E3 dimer, the E2 dimer (Cue1p/Ubc7p), and the Cdc48p subcomplex (Cdc48p–Npl4p–Ufd1p–Ubx2p; Fig. S2). These components, which are interchangeable between the Hrd1 and Doa10 complexes, are required for all identified ubiquitin-dependent ERAD substrates thus far. Adding Der1p and Usa1p to the complex expands the Hrd1 range to handle luminal substrates bearing specific peptide signals. The additional presence of Yos9p endows the capacity to recognize glycan signals marking substrates whose time windows for folding have expired (Quan et al., 2008; Clerc et al., 2009).
The primary functions of ERAD E3 complexes include substrate sorting, ubiquitination, and membrane extraction. By extension, the Hrd1 core complex might be sufficient to carry out these activities for ERAD-M. Recently, a systematic study revealed that Hrd1p transmembrane segments detect structural deviations in substrate transmembrane domains (Sato et al., 2009). This suggests that Hrd1p itself recognizes degradation signals for this class. With the source of ubiquitination and substrate extraction activities already known, this study explains why components outside the core can be dispensable for some substrates of this class. The accessory factors Der1p and Usa1p are required for all substrates of the Hrd1 pathway bearing luminal lesions (Fig. 3 C; Knop et al., 1996a; Carvalho et al., 2006). Their inclusion is sufficient for the Hrd1 complex to process luminal substrates bearing glycan-independent determinants. An important function of Usa1p is to link Der1p to the Hrd1 complex (Carvalho et al., 2006; Horn et al., 2009). Although Sec61-2p and Hmg2p (single Myc tagged) are efficiently degraded in the absence of Usa1p, recent studies have shown that it is not dispensable for all membrane substrates (Horn et al., 2009; Carroll and Hampton, 2010). Their data suggest that Usa1p may also play a broader role in maintaining the integrity of the Hrd1 complex. Although the precise role of Der1p remains unclear, the prevailing evidence suggests that it facilitates the movement of luminal domains to the cytosol. The mammalian Der1p homologue Derlin-1 is in proximity to MHC class I molecules during retrotranslocation across ER membranes (Lilley and Ploegh, 2004; Ye et al., 2004). This makes Derlin-1 a candidate component of the ERAD translocation pore. Although a role in signal recognition isn’t ruled out for Der1p and Usa1p, Hrd3p more likely plays this role in ERAD-L (in addition to Yos9p). Hrd3p binds misfolded CPY* with or without its glycans (Gauss et al., 2006a). Thus, Hrd3p is the best candidate as the substrate receptor for the glycan-independent luminal pathway. The further addition of Yos9p adapts the complex to recognize glycan-dependent substrates (Buschhorn et al., 2004; Bhamidipati et al., 2005; Kim et al., 2005; Szathmary et al., 2005). The requirement for Der1p and Usa1p indicates that both luminal substrate classes share the same processing mechanism after recognition. Therefore, it is not surprising that the pathogenic protein cholera toxin has coopted this pathway for its cytosolic entry from the ER (Bernardi et al., 2008). Collectively, the Hrd1 core complex exhibits remarkable functional plasticity. The addition of three accessory factors, Der1p, Usa1p, and Yos9p, is sufficient to expand its client range from a one class to three. Indeed, recent studies have shown that the inclusion of Usa1p alone extends the range of ERAD-M substrates processed by the Hrd1p complex (Horn et al., 2009; Carroll and Hampton, 2010). This theme is expanded in the mammalian system. The Yos9p homologues OS-9, XTP3-B, and their variants display substrate specificities that may be used to further diversify the substrate range (Christianson et al., 2008; Hosokawa et al., 2008).
Substrates display specific signals recognized by ERAD
One of the longstanding questions of ERQC revolves around the molecular cues used to differentiate folded, actively folding, and terminally misfolded proteins. It is now well established that glycoprotein substrates can display highly specific signals for degradation. Normally, N-linked glycans of nascent polypeptides are progressively trimmed in the ER by glucosidase I, glucosidase II, and α-mannosidase I to the GlcNAc2Man8 structure. Should a molecule fail to fold at this point, the Htm1p mannosidase cleaves a single residue to expose a terminal α1,6-linked mannose, the ligand for the Yos9p receptor (Quan et al., 2008; Clerc et al., 2009). This combines with adjacent disordered peptide segments to form a bipartite ERAD determinant (Xie et al., 2009). PrA may form a class of substrates that harbor determinants for both modes of the Hrd1 complex. Our analyses show that the PrA glycan-independent determinant is located in a region distinct from its glycan-dependent determinant (Fig. 5). These experiments show that protein misfolding is not inherently sufficient to trigger ERAD. Instead, ERAD responds to specific cues in substrates that coevolved with the system. This might explain why yeast and mammalian ERAD pathways are sometimes unable to recognize misfolded proteins from other organisms (Hong et al., 1996; Mancini et al., 2003; Coughlan et al., 2004). Although much of the general mechanisms are conserved, the specific nature of some signals may have diverged.
The novel PrA variants demonstrate the conservation of the ubiquitin–proteasome system in the quality of control of soluble nonglycosylated proteins. Our experiments are consistent with the current understanding of how this class is processed by the mammalian Hrd1 pathway (Okuda-Shimizu and Hendershot, 2007; Hosokawa et al., 2008). Therefore, it is likely that the advances represented by this study are also applicable to mammals. The discovery of the glycan-independent mode of ERAD-L also shows that budding yeast have at least two distinct mechanisms for soluble, nonglycosylated substrates, one ubiquitin dependent and another ubiquitin independent (for review see Nakatsukasa and Brodsky, 2008). Although not fully characterized, there are indications of a similar arrangement in mammals. Evidence of a mammalian ubiquitin-independent mechanism comes from detailed analysis using an elegant cell-free system. In this assay, the yeast ΔgppαF substrate undergoes retrotranslocation across ER membranes independently of ubiquitination and p97 (the Cdc48p orthologue; Wahlman et al., 2007). This result indicates that the structural cues used to sort this substrate in yeast are faithfully recognized in mammals and strengthens the view that ERQC mechanisms use specific determinants embedded in substrates to signal degradation if misfolded.
Concluding remarks
The ERAD client portfolio is perhaps the most varied among quality control systems. To tackle the challenge, substrate receptor sites have evolved to handle the topological diversity of substrates. It is now clear that the clearance of defective molecules is caused by signals embedded within the substrates themselves. Molecules not bearing features recognized by ERAD but are nevertheless misfolded may be handled by alternative pathways. Some use ubiquitin-independent mechanisms of ERAD and others are sent to the vacuole (the yeast lysosome) through autophagy or via the classical secretory pathway (Chang and Fink, 1995; Hong et al., 1996; Jenness et al., 1997; VanSlyke et al., 2000; Ravikumar et al., 2002; Kruse et al., 2006; Sarkar et al., 2007). Understanding how these molecules are differentiated from folded proteins remains a major target for future studies.
Materials and methods
Strains and antibodies
Saccharomyces cerevisiae strains used in this study are listed in Tables S3 and S4. Anti-HA (HA.11) antibody, HA probe, and anti-Myc antibody were purchased from Covance. Anti-hexokinase antibody was purchased from United States Biological Inc. Anti-Kar2p rabbit polyclonal antiserum was provided by P. Walter (University of California, San Francisco, San Francisco, CA).
Plasmids used in this study
Plasmids and primers used in this study are listed in Table S2. Plasmids were constructed using standard cloning protocols. All coding sequences of constructs used in this study were sequenced in their entirety. Unless indicated, all substrate proteins described in this study contain a single C-terminal HA epitope tag used for detection (Spear and Ng, 2005).
PrA expression vectors and variants
Plasmids pKK129, pKK159, pKK163, and pKK261 carry PrA*-Ab, PrA*-AbΔ147–183, PrA*-AbΔ295–331, and ngPrAΔ295–331 driven by their endogenous promoter, respectively, were described previously (Xie et al., 2009). Plasmid pKK249, carrying ngPrA*Δ295–331, was created as follows: a 1.6-kb fragment encoding PrA*-Ab and its promoter was released from plasmid pKK150 by digesting with ClaI. This fragment was ligated into pKK163 digested with ClaI to create pKK249.
Yos9 and Yos9R200A expression vectors
Plasmid pKK278, carrying Yos9 driven by its endogenous promoter, was created as follows: HA epitope–tagged Yos9p driven by the endogenous YOS9 promoter was described previously (Kim et al., 2005). HA epitope tag was deleted from it by site-directed mutagenesis using KKN303 primer to create the plasmid pKK278. Plasmid pKK284, carrying Yos9R200A driven by the endogenous YOS9 promoter, was created by site-directed mutagenesis to make a point mutation R200A using KKN306 primer. Plasmid pKK278 was used as template for site-directed mutagenesis.
CPY* expression vector
Plasmid pKK286, carrying CPY* driven by a galactose-inducible promoter, was created by site-directed mutagenesis to delete HA epitope tag sequence from plasmid pES67 using KKN307 primer (Spear and Ng, 2003).
Hmg2p expression vector
Plasmid pRH244 was provided by R. Hampton (University of California, San Diego, La Jolla, CA). The following plasmids were generated using site-directed mutagenesis of ERAD substrate clones (Sawano and Miyawaki, 2000). Substrate variant names are indicated in parentheses. Primer sequences are listed in Table S4. All substrate genes are controlled by the endogenous promoter and encode the HA epitope tag at their C termini.
pKK214 (PrA*-AbD147–183/295–331).
Using the KKN136 primer, a segment encoding Thr295 through Arg331 was deleted from pKK159.
pKK232 (PrA*-GI).
Using the KKN228 primer, the N-linked glycosylation site was mutagenized on pKK214.
pWX41 (PrA*-GID1).
Using the WXN41 primer, a segment encoding Lys23 through Leu71 was deleted from pKK232.
pWX29 (PrA*-GID2).
Using the WXN29 primer, a segment encoding Asp72 through Ala142 was deleted from pKK232.
pWX30 (PrA*-GID3).
Using the WXN30 primer, a segment encoding Glu143 through Glu213 was deleted from pKK232.
pWX31 (PrA*-GID4).
Using the WXN31 primer, a segment encoding Ser214 through Ile284 was deleted from pKK232.
pWX51 (PrA*-GID4CHO).
Using the WXN31 primer, a segment encoding Ser214 through Ile284 was deleted from pKK214.
pKK252 (PrA-CTD).
Using the KKN279 primer, a segment encoding Lys26 through Ile284 was deleted from pKK150.
Indirect immunofluorescence
Strains were grown to early log phase in synthetic complete media lacking the appropriate component for plasmid selection. Formaldehyde (analytical grade; Merck) was added directly to 3–9 ml medium to 3.7% at 30°C for 90 min. After fixation, cells were collected by centrifugation and washed with 5 ml ice-cold 0.1 M potassium phosphate buffer, pH 7.5, for 5 min. Cells were resuspended and incubated in 90 µl 0.1 M potassium phosphate buffer, pH 7.5, containing 1.2 M sorbitol and 1 mg/ml zymolyase 20T (United States Biological Inc.) for 10–20 min at room temperature to digest the cell wall.
To terminate digestion, cells were washed twice in 0.1 M potassium phosphate buffer, pH 7.5, containing 1.2 M sorbitol. Cells were resuspended in 30 µl 0.1 M potassium phosphate buffer, pH 7.5, containing 1.2 M sorbitol and applied to a clean glass slide (precoated with 0.1% poly L-lysine) for 10 min at room temperature. Slides were washed once with ice-cold TBS buffer (50 mM Tris–HCl and 150 mM NaCl, pH 7.4) for 3 min, soaked in methanol (−20°C) for 6 min followed by acetone for 30 s, rinsed in ice-cold TBS buffer for 3 min, and air dried. Subsequent steps were performed at room temperature. 30 µl of blocking buffer (TBS buffer with 5% nonfat milk and 0.05% Tween 20) was added to each well and incubated for 30 min followed by a TBS buffer wash for 10 min. Primary antibodies HA.11 mAb (Covance) and polyclonal rabbit α-Kar2p were applied at 1:500 and 1:1,000 dilutions, respectively, in 30 µl of blocking buffer for 90 min. Wells were washed twice for 10 min with TBS buffer. 30 µl of blocking buffer containing Alexa Fluor 488 goat anti–mouse and Alexa Fluor 594 goat anti–rabbit (Invitrogen) as secondary antibodies was added to each well and incubated for 90 min in the dark. Slides were washed for 10 min twice with TBS buffer. Each well was applied with 15 µl of mounting medium (PBS buffer, pH 9.0, 90% glycerol, and 0.05 µg/ml DAPI) and a glass coverslip. Slides were sealed with clear nail polish and stored at −20°C in a dry and dark container.
Cells were visualized using an inverted microscope (LSM 510 META; Carl Zeiss, Inc.) with a Plan Apochromat 100× 1.4 NA Ph3 objective (Carl Zeiss, Inc.) in immersion oil (Immersol 518F; Carl Zeiss, Inc.) at room temperature. Image acquisition was performed using standard photomultiplier tube with LSM 510. Images were archived using LSM 5 Image Examiner (Carl Zeiss, Inc.) and Photoshop (version 7.0; Adobe), and no additional software adjustments were performed on images after acquisition other than cropping.
Cell labeling and immunoprecipitation (IP)
3.0 A600 OD units of cells grown to log phase were harvested by centrifugation and resuspended in 0.9 ml of synthetic complete media lacking methionine and cysteine. After a 30-min incubation at the appropriate temperature, 150 µCi [35S]methionine/cysteine (Pro-mix; GE Healthcare) was added to cells for 10 min. A chase was initiated by adding cold methionine/cysteine to a final concentration of 2 mM. Ice-cold TCA was added to a final concentration of 10% to terminate each time point. Cells were homogenized by the addition of 0.4 ml of 0.5-mm zirconium beads followed with agitation in a mini-bead beater cell disrupter two times for 30 s (Biospec Products). The homogenate was transferred to a fresh tube, pooled with a subsequent 10% TCA bead wash, and centrifuged at 18,000 g for 15 min. The pellet was resuspended in 120 µl of TCA suspension buffer (100 mM Tri-HCl, 3% SDS, and 3 mM DTT) and heated to 100°C for 5 min. Insoluble debris was pelleted, and 40 µl detergent lysate was added to 560 µl IPS II (13.3 mM Tris–HCl, 150 mM NaCl, 1% Triton X-100, 0.02% NaN3, 1 mM PMSF, and 1 µl protease inhibitor cocktail; Sigma-Aldrich) and the appropriate antiserum. After a 2-h incubation at 4°C, the sample was centrifuged for 15 min at 18,000 g, and the supernatant transferred to a fresh tube containing protein A–Sepharose beads. The tube was rotated for 1 h and washed three times with IPS I (0.2% SDS, 1% Triton X-100, 20 mM Tris–HCl, pH 7.4, 150 mM NaCl, and 0.02% NaN3) and once with TBS (20 mM Tris–HCl, pH 7.4, and 150 mM NaCl). Immunoprecipitated proteins were eluted with SDS gel sample buffer and resolved by gel electrophoresis. SDS-PAGE gels were exposed to phosphor screens for 24–48 h. Exposed screens were scanned using a phosphorimager (Typhoon; GE Healthcare) and quantified using ImageQuant TL software (GE Healthcare). All data plots reflect three independent experiments with SEM indicated with error bars.
Cycloheximide chase analysis
Cells were grown logarithmically at 30°C in synthetic media. Cessation of protein synthesis was initiated with the addition of cycloheximide to 200 µg/ml. 1.08 A600 OD U cells were collected at each time point by low speed centrifugation and resuspended in 1 ml 10% TCA. Cells were homogenized by agitation in a mini-bead beater cell disrupter with 0.4 ml 0.5-mm zirconium beads (Biospec Products). The homogenate was transferred to a fresh tube and pooled with a subsequent 10% TCA bead wash. After precipitation in 10% TCA, proteins were resuspended in urea SDS sample buffer (8 M urea, 5% SDS, 200 mM Tris–HCl, pH 6.8, 0.1 mM EDTA, and 100 mM DTT) and incubated for 30 min at room temperature. Detergent lysates were clarified by centrifugation (18,000 g for 10 min). Before loading on gels, proteins were incubated for 10 min at 37°C in sample buffer. Proteins were resolved by SDS-PAGE and transferred to nitrocellulose membranes. Membranes were incubated in blocking buffer provided by the manufacturer (LI-COR Biosciences). After incubation with anti-Myc and anti-hexokinase antibodies, membranes were washed in PBS containing 0.1% Tween 20. Secondary antibodies conjugated to IRDye 680 (infrared dye) or 800 were diluted 15,000-fold in blocking buffer containing 0.1% Tween 20. After washing, membranes were scanned for direct infrared fluorescence using an infrared imaging system (Odyssey) and quantified using the manufacturer’s software (LI-COR Biosciences).
Induction of CPY* expression by galactose
Wild-type strains expressing CPY* using the GAL1 promoter (or empty vector as control) were preincubated overnight in synthetic complete media with 3% raffinose as the sole carbon source. Cells were harvested by low speed centrifugation (3,000 g for 5 min), inoculated in synthetic complete media with 2% galactose as carbon source, and grown to log phase. 9.0 OD600 equivalents of cells were harvested by low speed centrifugation (3,000 g for 5 min) and resuspended in 3.0 ml synthetic complete media with 2% galactose followed by metabolic pulse-chase analysis.
Purification of proteins for mass spectrometry
Strains were grown to late log phase in synthetic complete media lacking uracil. 4,000 OD600 equivalents of cell culture were harvested by low speed centrifugation (3,000 g for 10 min) and washed once in ice-cold water. The cells were collected by centrifugation (3,000 g for 10 min), and the pellet was resuspended in 40 ml ice-cold 50 mM Tris–HCl, pH 7.4. The cells were collected by centrifugation (3,000 g for 10 min) and transferred in a mortar filled with liquid nitrogen. The cells were ground by a pestle to a fine powder in liquid nitrogen. The disrupted cells were resuspended in 20 ml Tris-–IP buffer (50 mM Tris–HCl, pH 7.4, and 150 mM NaCl) containing 1.0 mM PMSF. After a low speed spin (800 g for 5 min) to pellet debris, the lysate was subjected to ultracentrifugation (30,000 g for 30 min). The pellet was resuspended in 20 ml Tris–IP buffer containing 0.5% Triton X-100 and 1 mM PMSF on ice for 1 h. The solubilized microsome fraction was collected by ultracentrifugation (30,000 g for 10 min). 50 µl agarose-immobilized anti-HA antibody (Santa Cruz Biotechnology, Inc.) was added to the supernatant and incubated at 4°C with rocking for 2 h. The beads were washed gently three times in ice-cold Tris–IP buffer with 0.5% Triton X-100 and once in ice-cold Tris–IP buffer. Proteins were eluted from the beads by boiling in SDS loading buffer. Proteins were boiled for 5 min, resolved by SDS-PAGE, and visualized by Coomassie brilliant blue staining. Protein bands were excised for in-gel trypsinization and identified by matrix-assisted laser desorption/ionization mass spectrometry/mass spectrometry (National University of Singapore Mass Spectrometry Laboratory). Analysis of ngPrAΔ295–331-binding proteins is shown in Table S1.
Online supplemental material
Fig. S1 shows PrA variants with reduced dependence on the glycan-dependent ERAD factor Htm1p for degradation. Fig. S2 shows the degradation profile of the ERAD-M substrate Hmg2p (single Myc tagged). Table S1 lists the peptide mass data of the ngPrAΔ295–331-binding protein. Tables S2–S4 list the strains, plasmids, and oligonucleotide primers used in this study.
Acknowledgments
We thank members of the Ng Laboratory for discussion and comments. We also thank the Temasek Life Sciences Laboratory and National University of Singapore core facilities for providing excellent technical support. We thank Drs. Randy Hampton and Peter Walter for gifts of plasmids and antibodies.
This work was supported by funds from the Temasek Trust and by a grant from the Japan Society for the Promotion of Science to K. Kanehara (Postdoctoral Fellowship for Research Abroad).


![Figure 1. ngPrA variants are substrates of the Hrd1p-dependent ERAD pathway. (A) Schematic representation of PrA variants with positions of deletions under each diagram. N-linked glycans are represented by branched symbols, signal sequences are shown in dark gray, prodomains are colored light gray, and HA epitope tags are indicated. PrA* differs from PrA by lacking sequences Leu55 through Tyr91. (B) Wild-type (WT), Δhrd1, and Δhtm1 cells expressing ngPrA*Δ295–331 were pulse labeled for 10 min with [35S]methionine/cysteine at 30°C followed by a cold chase for the times indicated. Immunoprecipitation of ngPrA*Δ295–331 was performed using anti-HA mAb and resolved by SDS-PAGE. Decay kinetics were quantified by phosphorimager analysis. The data reflect three independent experiments with the SEM indicated by error bars. Representative phosphor scan images are shown. (C) Wild-type, Δhrd1, and Δhtm1 cells expressing ngPrAΔ295–331 were analyzed by pulse-chase analysis as described in B. (D) Intracellular localization of ngPrAΔ295–331 in Δhrd1 cells was analyzed by indirect immunofluorescence. The substrate was detected using anti-HA antibodies (green), the ER was visualized using anti-Kar2p antisera (red), and the positions of nuclei were stained by DAPI. Bar, 2 µM.](https://cdn.rupress.org/rup/content_public/journal/jcb/188/5/10.1083_jcb.200907055/6/m_jcb_200907055_rgb_fig1.jpeg?Expires=1773614255&Signature=N3c2gnwvFQB264ZN8qZXIW2gesaqoe08WRjJTZRlJvLmT9ajbzoINtAKUfqkUHfpBNGFCY7aNP~fcmc3VTZFOagJZ~rK5KkP5z6YVdaXiugsjtMzxCqaXRdmLqFGU5ypHU5pWTBbkRz9BDobdJ02-GULKzTZ5ddy1-yzOi8O4Mh6tynQDtfmpOotoYR8-zcJmWHFYFXIQXclxFRiAptR74v-aBL-pQ4ULNnQttBa4kyiqYecFX9sOi5SELWzkZ5xlywu3v8osNfXcdYdh3IrNMy6Q6SvqzstUul2-lEgMqQnDyvZbKTI8kHFOwyEPx7uWFBStZXW~1YecLAC1Ul2Lw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)