To prevent accumulation of mutations, cells respond to DNA lesions by blocking cell cycle progression and initiating DNA repair. Homology-directed repair of DNA breaks requires CtIP-dependent resection of the DNA ends, which is thought to play a key role in activation of ATR (ataxia telangiectasia mutated and Rad3 related) and CHK1 kinases to induce the cell cycle checkpoint. In this paper, we show that CHK1 was rapidly and robustly activated before detectable end resection. Moreover, we show that the key resection factor CtIP was dispensable for initial ATR–CHK1 activation after DNA damage by camptothecin and ionizing radiation. In contrast, we find that DNA end resection was critically required for sustained ATR–CHK1 checkpoint signaling and for maintaining both the intra–S- and G2-phase checkpoints. Consequently, resection-deficient cells entered mitosis with persistent DNA damage. In conclusion, we have uncovered a temporal program of checkpoint activation, where CtIP-dependent DNA end resection is required for sustained checkpoint signaling.
Introduction
DNA double-strand breaks (DSBs) are dangerous lesions that threaten genomic integrity, and it is critically important for cells to detect and properly repair them to maintain genome stability (Jackson and Bartek, 2009). Cells are equipped with a DNA damage response (DDR) to detect and correct these lesions (Ciccia and Elledge, 2010). This response includes the cell cycle checkpoints that induce cell cycle arrest in G1 and G2 phases as well as slowing down of DNA synthesis (Iliakis et al., 2003). Checkpoints allow time for DSB repair, which is mediated by either homologous recombination (HR) or nonhomologous end joining (NHEJ; Iliakis et al., 2003; Jackson and Bartek, 2009).
There are major differences in the two DSB repair pathways, which are manifested already at the initial DNA end processing stage. Whereas NHEJ only requires little end processing, HR requires the removal of up to several hundred bases of the 5′ strand to generate a long 3′ single-stranded DNA (ssDNA) tail through the process of DNA end resection (Huertas, 2010; Mimitou and Symington, 2011). This tail is subsequently covered with RAD51, forming the structure that searches for the homologous sequence in the sister chromatid (Moynahan and Jasin, 2010). The mechanism and regulation of end resection of DSBs are not fully understood, although it requires the activity of several nucleases and helicases (Gravel et al., 2008; Huertas, 2010; Mimitou and Symington, 2011). The MRN (MRE11–RAD50–NBS1) complex may carry out the initial stage of DNA end processing by removing a few nucleotides of the 3′ strand DNA from a DSB end (Huertas, 2010; Mimitou and Symington, 2011). The CtIP protein also plays an important role in HR repair through regulation of DNA end resection (Sartori et al., 2007). CtIP is recruited to sites of DNA damage, where it is thought to cooperate with the MRN complex (Sartori et al., 2007). The exonuclease activity of MRE11 has 3′–5′ directionality, whereas generation of the resected overhang requires 5′–3′ resection, which suggests the action of other nucleases such as EXO1 and DNA2 exonucleases, which are capable of generating 3′ ssDNA overhangs (Eid et al., 2010; Huertas, 2010; Mimitou and Symington, 2011). The ssDNA generated from the resection process is immediately coated by replication protein A (RPA), which promotes both checkpoint activation and HR repair (Cimprich and Cortez, 2008; Huertas, 2010).
A key feature of the DDR is the activation of the PI3-kinase–related protein kinases ataxia telangiectasia mutated (ATM), ATR (ATM and Rad3 related), and DNA–protein kinase, which target a large number of substrates to regulate the cellular responses to DNA damage, including checkpoint activation (Ciccia and Elledge, 2010). A critical target of the checkpoint signaling cascade is CHK1 kinase, which is required for the S- and G2-phase checkpoints in mammalian cells (Zhao et al., 2002; Sørensen et al., 2003). CHK1 restrains the activity of the CDC25A phosphatase, an activator of the Cdks, through phosphorylation-dependent ubiquitylation and degradation (Busino et al., 2004). CHK1 activity is regulated by ATR phosphorylation on S317 and S345 (Cimprich and Cortez, 2008). ATR is constitutively associated with ATR-interacting protein, which binds directly to RPA-coated ssDNA (Cortez et al., 2001), which can be generated by nuclease- and helicase-dependent processing of DNA (Cimprich and Cortez, 2008). The junctions between ssDNA and double-stranded DNA are also important for ATR activation, likely by recruiting the ATR-activating factor TopBP1 (Kumagai et al., 2006; Cimprich and Cortez, 2008). Importantly, end resection during HR generates both ssDNA and junctions between single and double strands, which has led to the model that resection is critical for ATR/CHK1 activation (You and Bailis, 2010).
Here, we have investigated the interplay between DNA end resection and checkpoint responses. We now show that the CHK1-mediated checkpoint response is activated rapidly after DSB induction in a manner independent of resection by CtIP, EXO1, and DNA2. Importantly, resection is crucial for checkpoint maintenance, thereby linking DNA processing with prolonged cell cycle arrest to allow sufficient time for DNA repair.
Results and discussion
Checkpoint activation and DNA end resection are temporally separated
The interplay between DNA repair and checkpoint signaling is not fully understood. To address this, we treated U2OS cells with the DNA topoisomerase I poison camptothecin (CPT), which generates DSBs during DNA replication. Activation of checkpoint pathways measured by CHK1 and CHK2 phosphorylation was relatively rapid and detectable within minutes after CPT addition (Fig. 1 A). The CHK1 target CDC25A was also found to be rapidly degraded, indicative of checkpoint activation. In contrast, we found that RPA2 phosphorylation, which has been used as a readout for DNA end resection (Bunting et al., 2010), was delayed and not detectable until 40 min after CPT addition (Fig. 1 A). To investigate this further, we monitored the timing of DNA end resection by observing the occurrence of ssDNA after CPT treatment (Fig. 1 B). The formation of ssDNA was apparent 30–40 min after drug addition, which supported the finding that DNA end resection occurs considerably later than checkpoint activation. Interestingly, this indicated that DNA end resection, which is critical for the initiation of HR-mediated repair, might play a limited role in the initial activation of the ATR–CHK1 pathway. Moreover, the temporal separation of ATR–CHK1 activation and RPA phosphorylation was confirmed in a primary cell line (Fig. S1 A). We also noted a significant increase in the level of CHK1 phosphorylation at later time points in CPT-treated cells that coincided with the initiation of DNA end resection, indicating a functional relation (Fig. 1 A).
Checkpoint initiation and CtIP-dependent DNA end resection are temporally separated. (A) U2OS cells were either left untreated or treated with 1 µM CPT for the indicated time. Cells were harvested and processed for immunoblotting with the indicated antibodies. (B) U2OS cells were seeded on coverslips and incubated with BrdU for 24 h. Cells were treated with 1 µM CPT for the indicated time, fixed, and stained with BrdU and 53BP1 antibodies under native conditions. Pictures were acquired using epifluorescence microscopy. Bar, 10 µm. (C) U2OS cells were transfected for 2 d with the indicated siRNAs. Cells were treated with 1 µM CPT for the indicated time and then processed as in A. Numbers represent quantification of phosphorylated CHK1. siUNC, Universal Negative Control siRNA. (D) U2OS cells were transfected for 2 d with the indicated siRNAs. The cells were treated with 0.1 µM or 1 µM CPT and then processed as in A. Numbers are as in C.
Checkpoint initiation and CtIP-dependent DNA end resection are temporally separated. (A) U2OS cells were either left untreated or treated with 1 µM CPT for the indicated time. Cells were harvested and processed for immunoblotting with the indicated antibodies. (B) U2OS cells were seeded on coverslips and incubated with BrdU for 24 h. Cells were treated with 1 µM CPT for the indicated time, fixed, and stained with BrdU and 53BP1 antibodies under native conditions. Pictures were acquired using epifluorescence microscopy. Bar, 10 µm. (C) U2OS cells were transfected for 2 d with the indicated siRNAs. Cells were treated with 1 µM CPT for the indicated time and then processed as in A. Numbers represent quantification of phosphorylated CHK1. siUNC, Universal Negative Control siRNA. (D) U2OS cells were transfected for 2 d with the indicated siRNAs. The cells were treated with 0.1 µM or 1 µM CPT and then processed as in A. Numbers are as in C.
Initial ATR–CHK1 activation occurs independently of DNA end resection
CtIP is a critical factor in DNA end resection after DNA damage (Sartori et al., 2007). Given the temporal separation of ATR–CHK1 activation and extensive end resection, we addressed whether abrogation of resection would have an impact on activation of ATR/CHK1. We found that the ATR–CHK1 pathway was activated rapidly after CPT treatment in CtIP-depleted cells. This was apparent at the earliest time point, as measured by increased CHK1 phosphorylation and CDC25A decrease (Fig. 1 C). Notably, when looking at time points after the onset of extensive DNA end resection, we found that the level of phosphorylated CHK1 was substantially reduced in CtIP-depleted cells compared with mock-depleted cells (Figs. 1 C and S1 B). To test whether the initial CHK1 phosphorylation was dependent on ATR activity, we depleted cells for the key ATR activator TopBP1. In response to CPT treatment, we found that depletion of TopBP1 led to a complete abrogation of CHK1 but not CHK2 phosphorylation (Fig. 1 C). We subsequently addressed whether the observed increase in CHK1 phosphorylation at later time points was indeed dependent on resected DNA ends. To investigate this, we used a low concentration of CPT (0.1 µM) that leads to little DNA breakage (Ray Chaudhuri et al., 2012) and consequently limited resection, as measured by RPA hyperphosphorylation. ATR–CHK1 activation was unaffected by CtIP depletion at low-dose CPT (Fig. 1 D), supporting the notion that the increase in ATR–CHK1 activity was caused by extensive DNA resection.
CPT induces DNA DSB damage specifically at the replication fork. To corroborate our findings, we treated cells with ionizing radiation (IR), which results in stochastic replication-independent DNA damage (DSBs). As was the case for CPT-treated cells, initial activation of ATR–CHK1 and CDC25A degradation appeared largely independent of CtIP upon IR (Figs. 2 A and S1 C). However, CtIP was required to support CHK1 phosphorylation at later time points. In line with this, CDC25A reappeared prematurely in CtIP-depleted cells, indicative of insufficient ATR–CHK1 activity and defective checkpoint maintenance. Furthermore, the initial CHK1 phosphorylation was abolished by TopBP1 depletion, indicating that CHK1 is targeted by ATR-mediated phosphorylation after IR (Fig. 2 A). We found that DNA end resection was apparent at ∼45 min after irradiation, as observed by both RPA foci formation (Fig. 2 B) and by immunostaining for ssDNA (Fig. S1 D). A large part of CHK1 signaling in asynchronously growing cells is originating from S-phase cells. To understand whether G2-phase cells share the same regulation, we measured IR-induced checkpoint responses in synchronized cells (Fig. S1 E). We found that CtIP is dispensable for initial CHK1 activation but is required for sustained CHK1 phosphorylation (Fig. 2 C). These findings are in agreement with our findings for CPT-treated cells.
CtIP-dependent DNA end resection is required for sustained checkpoint signaling. (A) U2OS cells were transfected for 2 d using the indicated siRNAs. Cells were irradiated (10 Gy), collected at the indicated time points, and processed as in Fig. 1 A. Numbers are as in Fig. 1 C. siUNC, Universal Negative Control siRNA. (B) U2OS cells were seeded on coverslips and siRNA transfected for 2 d. After IR, cells were fixed at the indicated times and stained for RPA and γH2AX. Pictures were acquired using confocal microscopy. Bar, 10 µm. (C) U2OS cells were transfected with control or CtIP siRNA and synchronized in G2 phase by double thymidine treatment. Cells were irradiated (15 Gy), collected at the indicated time points, and then processed for immunoblotting with the indicated antibodies. Numbers are as in Fig. 1 C. AS, asynchronized growing cells.
CtIP-dependent DNA end resection is required for sustained checkpoint signaling. (A) U2OS cells were transfected for 2 d using the indicated siRNAs. Cells were irradiated (10 Gy), collected at the indicated time points, and processed as in Fig. 1 A. Numbers are as in Fig. 1 C. siUNC, Universal Negative Control siRNA. (B) U2OS cells were seeded on coverslips and siRNA transfected for 2 d. After IR, cells were fixed at the indicated times and stained for RPA and γH2AX. Pictures were acquired using confocal microscopy. Bar, 10 µm. (C) U2OS cells were transfected with control or CtIP siRNA and synchronized in G2 phase by double thymidine treatment. Cells were irradiated (15 Gy), collected at the indicated time points, and then processed for immunoblotting with the indicated antibodies. Numbers are as in Fig. 1 C. AS, asynchronized growing cells.
Maintenance of the G2/M- and S-phase checkpoints is dependent on CtIP
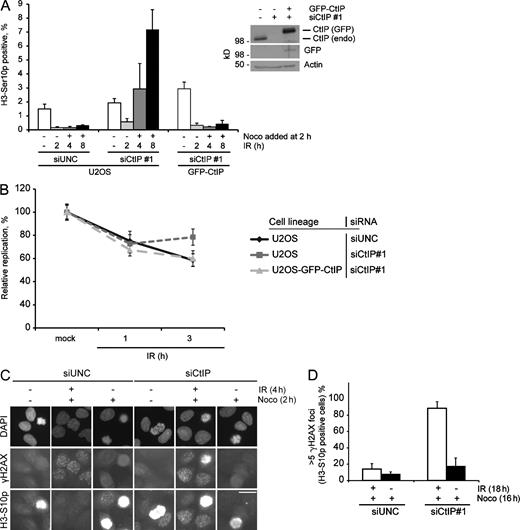
Having established that CtIP has a temporally distinct impact on CHK1 activity, we next investigated the role of CtIP in cell cycle checkpoints. In response to irradiation of control siRNA–depleted cells, the fraction of mitotic cells was significantly reduced both at relatively early and late time points (Fig. 3 A). Interestingly, CtIP-depleted cells did initially activate the checkpoint, as indicated by a significant decrease in mitotic cells, comparable with that of the control-depleted cells. However, at later time points, there was a dramatic increase in the percentage of mitotic cells in spite of irradiation, indicating a critical role for CtIP in checkpoint maintenance (Fig. 3 A). Similar data were observed when G2-phase–synchronized cells were analyzed (Fig. S2 B). As expected, CHK1 was required for the early and late phases of the G2/M checkpoint (Fig. S2 C). Similar temporal dependency on CtIP was observed for the intra–S-phase checkpoint. Initially, DNA replication was reduced in CtIP-depleted cells to a level indistinguishable from control-depleted cells. However, at later time points, DNA replication was substantially elevated in CtIP-depleted cells relative to the control cells (Fig. 3 B). To rule out off-target effects of the siRNA depletions, we performed reconstitution experiments using an U2OS cell line stably expressing an siRNA-insensitive GFP-CtIP allele. Importantly, both the G2/M- and intra–S-phase DNA damage checkpoint defects caused by CtIP down-regulation were rescued using this cell line (Fig. 3, A and B). In addition, CtIP depletion had only little effect on cell cycle progression, ruling out cell cycle defects as an explanation for the phenotypes (Fig. S2 A).
CtIP is critical for IR-induced checkpoint maintenance. (A) U2OS cells or U2OS cells stably expressing siRNA-resistant GFP-CtIP were transfected with the indicated siRNA for 2 d, and samples were fixed at the indicated time points after IR. Nocodazole (Noco) was added 2 h after IR. The bar chart shows the percentage of Histone H3-Ser10p–positive cells measured by flow cytometry. Data are the means of three independent experiments. Error bars represent SDs. The inset blot shows expression levels of GFP-CtIP and endogenous (endo) CtIP in the used cells lines. siUNC, Universal Negative Control siRNA. (B) RDS assay was performed using U2OS cells or U2OS cells stably expressing siRNA-resistant GFP-CtIP. Cells were siRNA transfected for 2 d with the indicated siRNAs and subjected to RDS assay. Data are the means of three independent experiments. Error bars represent SDs. (C) U2OS cells were seeded on coverslips and transfected for 2 d with the indicated siRNAs. The samples were treated with IR and nocodazole, as indicated, followed by fixation. Cells were stained for Histone H3-S10p and γH2AX, and images were acquired using epifluorescence microscopy. Bar, 10 µm. (D) U2OS cells were treated as in Fig. S2 D. The bar chart shows the percentage of Histone H3-S10p–positive cells with more than five γH2AX foci for a given treatment. Data are the means of three independent experiments. For each sample, at least 50 Histone H3-S10p–positive cells were counted. Error bars represent SDs.
CtIP is critical for IR-induced checkpoint maintenance. (A) U2OS cells or U2OS cells stably expressing siRNA-resistant GFP-CtIP were transfected with the indicated siRNA for 2 d, and samples were fixed at the indicated time points after IR. Nocodazole (Noco) was added 2 h after IR. The bar chart shows the percentage of Histone H3-Ser10p–positive cells measured by flow cytometry. Data are the means of three independent experiments. Error bars represent SDs. The inset blot shows expression levels of GFP-CtIP and endogenous (endo) CtIP in the used cells lines. siUNC, Universal Negative Control siRNA. (B) RDS assay was performed using U2OS cells or U2OS cells stably expressing siRNA-resistant GFP-CtIP. Cells were siRNA transfected for 2 d with the indicated siRNAs and subjected to RDS assay. Data are the means of three independent experiments. Error bars represent SDs. (C) U2OS cells were seeded on coverslips and transfected for 2 d with the indicated siRNAs. The samples were treated with IR and nocodazole, as indicated, followed by fixation. Cells were stained for Histone H3-S10p and γH2AX, and images were acquired using epifluorescence microscopy. Bar, 10 µm. (D) U2OS cells were treated as in Fig. S2 D. The bar chart shows the percentage of Histone H3-S10p–positive cells with more than five γH2AX foci for a given treatment. Data are the means of three independent experiments. For each sample, at least 50 Histone H3-S10p–positive cells were counted. Error bars represent SDs.
It was previously shown that the activity of CtIP restricts repair by NHEJ (Yun and Hiom, 2009). Thus, a potential explanation of the observed defects in checkpoint maintenance could be that the faster NHEJ pathway substitutes for the slower HR pathway in CtIP-depleted cells. In theory, this could result in faster repair kinetics and therefore mimic premature termination of the checkpoint response (Shibata et al., 2011). To test this, we investigated whether all DNA damage had indeed been repaired before mitotic entry in CtIP-depleted cells. We found that irradiated cells devoid of CtIP showed a marked presence of γH2AX foci in mitosis (Fig. 3 C). Because of the fact that control-depleted cells do not enter mitosis before all damage has been repaired, we quantified the amount of mitotic cells with γH2AX foci 18 h after irradiation. Even at this late time point, CtIP-depleted mitotic cells had a significant amount of persisting γH2AX foci, whereas mock-depleted cells had almost no γH2AX foci (Figs. 3 D and S2 D). These findings clearly indicate that CtIP-depleted cells enter mitosis with unrepaired lesions after IR treatment.
Extensive DNA end resection is critical for checkpoint maintenance
CtIP is functionally involved in additional cellular processes, such as transcriptional control, that could affect DDRs (Wu and Lee, 2006). To further address the role of CtIP in checkpoint maintenance, we made use of cell lines stably expressing a T847A mutant and a C-terminal truncated version of RNAi-resistant CtIP (Fig. 4 B). These mutants are resection deficient (Sartori et al., 2007; Huertas and Jackson, 2009), and the corresponding cell lines were unable to resect DNA (Fig. S3 A and not depicted). Whereas wild-type CtIP restored checkpoint function, the two mutant versions were unable to do so (Fig. 4 A), indicating that the resection activity of CtIP is responsible for checkpoint maintenance. ATM kinase has previously been shown to be required for resection of DSBs as well as regulation of the cell cycle checkpoint (Iliakis et al., 2003; Jazayeri et al., 2006). To test the temporal role of ATM in the G2/M checkpoint, we used a specific inhibitor (Hickson et al., 2004). As expected, we found that inhibition of ATM completely abolished ATM autophosphorylation, CtIP phosphorylation, and resection after IR (Fig. S3 B). Also, inhibition of ATM abolished the G2/M checkpoint to a similar extent as depletion of CtIP. Interestingly, we found that ATM was dispensable for checkpoint maintenance when inhibition occurred after initiation of resection. In contrast to this, inhibition of CHK1 both before and after initiation of resection almost completely abrogated the G2/M checkpoint (Fig. 4 C). This indicates that ATM mainly contributes to G2/M checkpoint maintenance through resection-dependent activation of ATR/CHK1.
Checkpoint maintenance is dependent on DNA end resection. (A) U2OS cells or U2OS cells stably expressing siRNA-resistant GFP-CtIP were transfected with the indicated siRNA for 2 d. Samples were fixed 8 h after IR and 6 h after nocodazole addition and then processed as in Fig. 3 A. For each sample, the H3Ser10p value was divided by the corresponding nocodazole (Noco) control to normalize for mitotic entry in the absence of IR. Data are the means of three independent experiments. Error bars represent SDs. WT, wild type. (B) U2OS cells or U2OS cells stably expressing siRNA-resistant GFP-CtIP were transfected for 2 d using the indicated siRNAs. The cells were harvested and processed for immunoblotting using the indicated antibodies. endo, endogenous. (C) U2OS cells were treated with DMSO or the ATM inhibitor (ATMi) 30 min before or 1 h after IR (5 Gy), as indicated. Nocodazole was applied 1.5 h after IR, and samples were fixed 6 h after IR. The bar chart shows the percentage of Histone H3-S10p–positive cells measured by flow cytometry and calculated as in A. Data are the means of three independent experiments. Error bars represent SDs. (D) U2OS cells were transfected with the indicated siRNA for 2 d, and samples were fixed at the indicated time points after IR and then processed as in A. Data are the means of three independent experiments. Error bars represent SDs. siUNC, Universal Negative Control siRNA. (E) Illustration of resection-independent checkpoint activation followed by resection-dependent maintenance of the ATR–CHK1-dependent S- and G2-phase checkpoints. Red circles indicate RPA. P, phosphorylation.
Checkpoint maintenance is dependent on DNA end resection. (A) U2OS cells or U2OS cells stably expressing siRNA-resistant GFP-CtIP were transfected with the indicated siRNA for 2 d. Samples were fixed 8 h after IR and 6 h after nocodazole addition and then processed as in Fig. 3 A. For each sample, the H3Ser10p value was divided by the corresponding nocodazole (Noco) control to normalize for mitotic entry in the absence of IR. Data are the means of three independent experiments. Error bars represent SDs. WT, wild type. (B) U2OS cells or U2OS cells stably expressing siRNA-resistant GFP-CtIP were transfected for 2 d using the indicated siRNAs. The cells were harvested and processed for immunoblotting using the indicated antibodies. endo, endogenous. (C) U2OS cells were treated with DMSO or the ATM inhibitor (ATMi) 30 min before or 1 h after IR (5 Gy), as indicated. Nocodazole was applied 1.5 h after IR, and samples were fixed 6 h after IR. The bar chart shows the percentage of Histone H3-S10p–positive cells measured by flow cytometry and calculated as in A. Data are the means of three independent experiments. Error bars represent SDs. (D) U2OS cells were transfected with the indicated siRNA for 2 d, and samples were fixed at the indicated time points after IR and then processed as in A. Data are the means of three independent experiments. Error bars represent SDs. siUNC, Universal Negative Control siRNA. (E) Illustration of resection-independent checkpoint activation followed by resection-dependent maintenance of the ATR–CHK1-dependent S- and G2-phase checkpoints. Red circles indicate RPA. P, phosphorylation.
CtIP has been shown to promote resection by supporting the activation of DNA nucleases (Mimitou and Symington, 2011). We reasoned that such nucleases operating downstream of CtIP could also participate in regulating G2/M checkpoint maintenance. To address this, we depleted cells of EXO1 and DNA2, two nucleases that are important exonucleases in the resection process (Nimonkar et al., 2011). Both nucleases were dispensable for checkpoint activation but were required for efficient checkpoint maintenance (Fig. 4 D). However, the role of the nucleases was less pronounced than CtIP. Depletion of DNA2 and EXO1 did not affect cell cycle progression, as cells progressed well into mitosis in the absence of IR (Fig. S3 C). Both nucleases were efficiently depleted, and their individual depletion led to some decrease in resection, as measured by RPA2 hyperphosphorylation, although, again, not as marked as CtIP depletion (Fig. S3 D). Codepletion experiments targeting both DNA2 and EXO1 were not possible as a result of marked cell cycle progression issues and occurrence of spontaneous DNA damage (unpublished data).
It has previously been demonstrated that the initial checkpoint response in S and G2 phase is dependent on CHK1 kinase, which inhibits CDC25 phosphatases (Zhao et al., 2002; Sørensen et al., 2003). Here, we show that this pathway operates before extensive DNA end resection, thus facilitating a rapid inhibition of DNA replication and mitotic entry of cells with damaged DNA (Fig. 4 E). Once HR repair is initiated, the CtIP-dependent checkpoint pathway then ensures sufficient time for repair by restraining mitotic entry until the repair process is complete. The rapid CHK1 activation could be through MRN complex–dependent loading of TopBP1 at the break site (Yoo et al., 2009). This pathway is strongly stimulated by ATM-mediated phosphorylation of TopBP1, and it can explain the role of ATM, TopBP1, and ATR in a resection-independent checkpoint pathway. It is also possible that short ssDNA junctions generated at the newly formed DSBs could activate the ATR pathway (MacDougall et al., 2007). These junctions may provide a loading site for the RAD9–RAD1–HUS1 complex during S and G2 phases, thereby recruiting TopBP1 to activate ATR (Kumagai et al., 2006; Cimprich and Cortez, 2008).
CtIP has previously been implicated in regulation of the G2/M checkpoint; however, the authors suggested a role for CtIP in early but not late stages of the checkpoint (Yu and Chen, 2004). The observed discrepancy may relate to the different experimental setups, as the authors relied on immunofluorescence to score checkpoint status. In addition to its role in G2/M checkpoint maintenance, we discovered that CtIP-deficient cells are defective in late stages of the S-phase checkpoint. A previous study found no role of CtIP in this checkpoint (Greenberg et al., 2006). However, the authors investigated one time point 30 min after IR, which is at an early stage that precedes DNA end resection. Premature initiation of DNA replication using a damaged template could lead to loss of genomic integrity. In this regard, it is noteworthy that CtIP is a haploinsufficient tumor suppressor in the mouse (Chen et al., 2005).
Materials and methods
Cell culture, chemicals, and siRNA
Human U2OS osteosarcoma and Tig3 TERT cell lines were grown in DME medium with 10% FBS. U20S cell lines stably expressing siRNA-resistant N-terminally EGFP-tagged CtIP pEGFP-C1 vector were selected with G418 (0.5 mg/ml) and tested for expression (by immunofluorescence and immunoblotting). The ΔC truncation of CtIP is lacking amino acids 790–897. ATM inhibitor (KU55933) was used at 10 µM (Tocris Bioscience). UCN-01, nocodazole, and CPT were used at 300 nM, 100 ng/µL, and 1 µM, respectively (Sigma-Aldrich). IR was given using an x-ray apparatus (Faxitron Bioptics LLC) calibrated to give 1 Gy γ-irradiation per 2 min. The following siRNA sequences were used in this study: CtIP1#1, 5′-GCUAAAACAGGAACGAAUC-3′ (Sartori et al., 2007); CtIP#2, 5′-UCCACAACAUAAUCCUAAU-3′ (Sartori et al., 2007); TopBP1, 5′-AGACCUUAAUGUAUCAGUA-3′; Exo1#1, 5′-GCCUGAGAAUAAUAUGUCU-3′; DNA2#2, 5′-GGAAUGGAUAACCGGUACC-3′; and Universal Negative Control siRNA. All siRNA duplexes (Sigma-Aldrich) were transfected at a final concentration of 50 nM using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s instructions.
Cell synchronization
U2OS cells were synchronized using a double thymidine block by treating cells with 2 mM thymidine (Sigma-Aldrich) for 20 h. Cells were released into fresh media supplemented with 24 µM 2-deoxycytidine (Sigma-Aldrich) for 10 h followed by a second thymidine block for 15 h. To obtain a G2-enriched population, cells were released in fresh media supplemented with 2-deoxycytidine for 8.5 h.
Microscopy and immunofluorescence
Immunofluorescence was essentially performed as previously described (Jørgensen et al., 2007). In brief, cells were grown on coverslips and treated as indicated in the figure legends. To visualize generation of ssDNA upon CPT or IR treatment, cells were incubated with BrdU for 24 h and subjected to BrdU immunofluorescence under nondenaturing conditions. To remove soluble proteins before immunofluorescence, cells grown on coverslips were preextracted for 2 min on ice using ice-cold CSK buffer (0.5% Triton X-100, 10 mM Pipes, pH 6.8, 300 mM sucrose, 100 mM NaCl, and 1.5 mM MgCl2) before fixation with 4% formaldehyde. Cells from all immunofluorescence experiments were fixed as previously described, except the experiments with Histone H3-S10p and γH2AX staining. Here, the coverslips were washed briefly in PBS followed by fixation using 1:1 methanol/acetone (ice cold), incubated for 10 min at room temperature, left to air dry, rehydrated with PBS, and stored at 4°C. Fluorescence-conjugated anti–mouse and –rabbit IgG were purchased from Invitrogen (Alexa Fluor 488 and 594). After antibody staining, cells were mounted using VECTASHIELD mounting medium with or without DAPI (Vector Laboratories). Grayscale images were acquired on a microscope (Leitz DMRXE; Leica) using an HCX PL FLUOTAR 40×/0.75 in air or PL APO 63×/1.32–0.6 objective and type F immersion liquid (Leica). Pictures were obtained at room temperature using a DFC340 FX camera (Leica) and FireCam software (v3.1; Leica) for Mac software. Color pictures were acquired using confocal microscopy (LSM510 Axiovert 200M [Carl Zeiss], using a 63× C-Apochromat objective with an NA of 1.2 in H2O). Pictures were analyzed using LSM 510 META software and LSM image examiner software (Carl Zeiss). All pictures were exported in preparation for printing using Photoshop (Adobe).
Immunoblotting and antibodies
Cells were lysed on ice in EBC buffer (50 mM Tris, pH 7.4, 120 mM NaCl, 0.5% NP-40, and 1 mM EDTA) followed by sonication using a digital sonifier (102C CE Converter; Branson). Proteins were separated on SDS-PAGE gel and transferred to a nitrocellulose membrane. The membranes were incubated with primary antibody diluted in 5% milk followed by incubation with secondary antibody (HRP-conjugated anti–mouse or –rabbit IgG; Vector Laboratories). Antibody to CHK1 (DCS310) has been previously described (Sørensen et al., 2005). Commercially available antibodies from Cell Signaling Technology were phospho–CHK1 S317 (#23445), phospho–CHK1 S345 (#2348), and phospho–CHK2 T68 (#2661). Antibodies from Millipore were phosphorylated γH2AX (#05-636) and phospho–Histone H3-S10 (#06-570). Antibodies from Bethyl Laboratories, Inc. were CtIP (#BL1914) and phospho–RPA32 S4/8 (#A300-245). Antibodies from Santa Cruz Biotechnology, Inc. were CDC25A (#F-6), CHK2 (#sc-56296), 53BP1 (#sc-22760), and GFP (#sc-8334). Antibodies from Abcam were TopBP1 (#2402) and DNA2 (ab96488). Other commercial antibodies were actin (#AB1501; Millipore), BrdU (#RPN20AB; GE Healthcare), Exo1 Ab-4 clone 266 (Thermo Fisher Scientific), RPA2 (#NA19L; EMD), and vinculin (#V9131; Sigma-Aldrich). CHK1 phosphorylation relative to loading control (vinculin for Figs. 1 [C and D] and 2 C and RPA2 for Fig. 2 A) was quantified using ImageJ software (National Institutes of Health).
Flow cytometry
Cells were prepared for flow cytometry as previously described (Jørgensen et al., 2007). In brief, cells were fixed 48 h after transfection in 70% ethanol and stained with antibodies toward phospho–Histone H3-S10 (1:200; Millipore) and γ-H2AX (1:1,000; Millipore) for 1 h followed by a 1-h incubation with Alexa Fluor 488 and 647 secondary antibodies (1:1,000; Invitrogen). DNA was stained using 0.1 mg/ml propidium iodide containing RNase for 30 min at 37°C. Flow cytometry analysis was performed on FACSCalibur using CellQuest Pro software (BD). Data were analyzed using FlowJo software (v7.2.2; Tree Star).
Radioresistant DNA synthesis (RDS) assay
The RDS assay was performed as previously described (Sørensen et al., 2003). In brief, U2OS cells or U2OS cells stably expressing siRNA-resistant GFP-CtIP were transfected with siRNAs. After washing off the transfection, cells were incubated for 24 h with 20 nCi/ml [14C]thymidine (NEC156010UC; PerkinElmer) followed by another 24-h incubation in nonradioactive medium. Cells were either left untreated or irradiated (10 Gy), and [3H]thymidine (NET027A250UC; PerkinElmer) at a final concentration of 2.5 µCi/ml was added 15 min before fixing the cells in 70% methanol. The cells were washed in PBS, and radioactivity was measured in a liquid scintillation counter. The ratios of [3H]/[14C] were calculated, and DNA synthesis after irradiation was expressed as a percentage of nonirradiated values.
Online supplemental material
Fig. S1 shows that the temporal separation of CHK1 phosphorylation and DNA end resection is observed in Tig3 fibroblast cells. We also detected very little DNA end resection in CtIP-depleted cells, both after IR and CPT treatment. CHK1 phosphorylation is not affected by CtIP depletion shortly after IR was applied. Finally, we show cell cycle profiles of double thymidine–synchronized cells. Fig. S2 shows that G2-synchronized, CtIP-depleted cells are not entering mitosis within the first hour after IR treatment. In the absence of IR, CtIP-depleted cells and cells stably expressing GFP-CtIP are progressing into mitosis as mock-treated cells. Moreover, CHK1 is required for the G2/M checkpoint at all time points after IR treatment. Representative microscope pictures of damage (γH2AX) in mitotic cells show that the damage is persisting in CtIP-depleted cells that enter mitosis after IR. Fig. S3 shows that DNA end resection after either IR or CPT is strongly suppressed in ATM-inhibited cells, CtIP-depleted cells, and cells expressing ΔC-CtIP. Also, DNA end resection is partly suppressed in DNA2 and Exo1-depleted cells after CPT. Finally, the siRNAs efficiently depleted DNA2 and Exo1 and had no observable impact on cell cycle progression.
Acknowledgments
This work was supported by The Novo Nordisk Foundation, The Danish Cancer Society, The Lundbeck Foundation, The Danish Medical Research Council, and the Vontobel Foundation (to A.A. Sartori).