During embryonic development and embryonic stem cell (ESC) differentiation, the different cell lineages of the mature heart arise from two types of multipotent cardiovascular progenitors (MCPs), the first and second heart fields. A key question is whether these two MCP populations arise from differentiation of a common progenitor. In this paper, we engineered Mesp1–green fluorescent protein (GFP) ESCs to isolate early MCPs during ESC differentiation. Mesp1-GFP cells are strongly enriched for MCPs, presenting the ability to differentiate into multiple cardiovascular lineages from both heart fields in vitro and in vivo. Transcriptional profiling of Mesp1-GFP cells uncovered cell surface markers expressed by MCPs allowing their prospective isolation. Mesp1 is required for MCP specification and the expression of key cardiovascular transcription factors. Isl1 is expressed in a subset of early Mesp1-expressing cells independently of Mesp1 and acts together with Mesp1 to promote cardiovascular differentiation. Our study identifies the early MCPs residing at the top of the cellular hierarchy of cardiovascular lineages during ESC differentiation.
Introduction
The heart is composed of multiple cell types, including cardiomyocytes (CMs), endothelial cells (ECs), and smooth muscle cells (SMCs; Martin-Puig et al., 2008). The mammalian heart is divided in four chambers: two atria and two ventricles, which are connected to the pulmonary and the general circulation by four vessels (Olson, 2006). During embryonic development, the heart is formed by two sources of multipotent cardiovascular progenitors (MCPs), with an additional contribution of neural crest cells (Buckingham and Desplan, 2010). The first heart field (FHF) MCPs, which form the cardiac crescent around embryonic day 7 during mouse development, give rise to the cells of both atria and to all CMs of the left ventricle. The second heart field (SHF) MCPs, which derive from the pharyngeal mesoderm, give rise to the cells of the right ventricle, some cells in both atria, as well as cells that form the outflow tract. Random labeling of cardiac precursors during embryonic development also revealed the existence of rare clones that contributed to both FHF and SHF lineages and that could represent a common cardiovascular progenitor for both heart fields (Meilhac et al., 2004). Recent studies showed that, during mouse embryonic development, tripotent MCPs that are able to differentiate at the clonal level into CMs, SMCs, and ECs can be marked and isolated based on Brachyury (Bry) and Flk1 (Kattman et al., 2006) or Isl1 and Flk1 expression (Moretti et al., 2006), whereas bipotent MCPs that give rise to CM and SMC lineages can be isolated based on Nkx2-5 and c-Kit expression (Wu et al., 2006). These studies demonstrated that cardiac cells arise from the differentiation of multipotent progenitors, with the ability to differentiate at the clonal level into the different cardiovascular lineages (Kattman et al., 2006; Moretti et al., 2006; Wu et al., 2006).
During the spontaneous differentiation of embryonic stem cells (ESCs), cardiovascular cells are generated through a biological process that recapitulates the cellular and molecular events normally occurring during embryonic development (Kattman et al., 2007; Murry and Keller, 2008). Using the same markers as to isolate the different MCPs during embryonic development, mouse and human bipotent and tripotent MCPs have been isolated during ESC differentiation, giving rise to CMs, SMCs, and ECs similar to their in vivo potential (Kattman et al., 2006; Moretti et al., 2006; Wu et al., 2006; Yang et al., 2008; Bu et al., 2009). The spontaneous appearance of cardiovascular cells during the differentiation of ESCs has created great enthusiasm among developmental biologists for studying, using reductionist in vitro approaches, the complex cellular and molecular mechanisms governing cardiovascular differentiation and cardiovascular diseases as well as providing a means of generating cardiovascular cells for cellular therapy and drug or toxicity screening (Murry and Keller, 2008).
Mesp1 is the earliest marker of cardiovascular development in vivo (Saga et al., 2000; Bondue and Blanpain, 2010). Mesp1 is expressed very transiently during early mesoderm specification in the primitive streak that migrates anterolaterally along with the cardiac mesoderm (Saga et al., 1996, 1999). Mesp1 lineage tracing experiments in mice revealed that almost all cells of the future heart as well as cells of the main vessels derived from cells that had expressed Mesp1 at one point during embryonic development (Saga et al., 1999, 2000). In addition to being the earliest marker of cardiovascular development, Mesp1 also plays a very important role during the earliest step of cardiovascular differentiation. Although genetic mutation of Mesp1 in mice does not lead to the absence of cardiac and vascular cells, possibly because the compensation is mediated by the massive up-regulation of its closest homologue Mesp2 (Saga et al., 1999; Kitajima et al., 2000), the combined deletion of Mesp1 and Mesp2 leads to the absence of mesoderm and cardiac specification (Kitajima et al., 2000). Recently, we and others have shown that Mesp1 overexpression greatly promotes the generation of multiple cardiovascular cell lineages during ESC differentiation, including derivatives of FHF and SHF progenitors (Bondue et al., 2008; David et al., 2008; Lindsley et al., 2008). Transcriptional profiling of Mesp1-expressing cells combined with chromatin immunoprecipitation experiments revealed that Mesp1 directly and rapidly induces the expression of many transcription factors implicated in cardiovascular specification. (Bondue et al., 2008; Lindsley et al., 2008).
Although rapid progress is being made in characterizing MCPs of the FHF and SHF, little is known about their specification. Do these MCPs arise from a common progenitor? If so, do these earliest MCPs represent a homogenous cell population common for both heart fields? What are the cell surface markers expressed by the early MCPs allowing their prospective isolation? What are the transcription factors expressed by the early MCPs that act alone or in combination with Mesp1 to promote MCP specification and cardiovascular lineage differentiation? To address these questions, we generated Mesp1-GFP reporter ESCs that allowed tracking and isolation of the earliest Mesp1-expressing cells during ESC differentiation. We showed that these early Mesp1-expressing cells are enriched for MCPs of both heart fields, which give rise upon differentiation to all cardiovascular cell lineages both in vitro and in vivo. By transcriptionally profiling the early Mesp1-expressing cells, we uncovered cell surface markers allowing their prospective isolation and cellular and molecular characterization. Using gain and loss of Mesp1 function during ESC differentiation, we demonstrated that Mesp1 is required to promote the specification of MCPs and the expression of cardiovascular transcription factors in MCPs. We found that Isl1 is expressed in a subpopulation of Mesp1-expressing cells and stimulates cardiovascular commitment in these early MCPs. Our study provides novel insights into the cellular and transcriptional hierarchy acting during the early steps of cardiovascular differentiation.
Results
Mesp1-GFP–expressing cells represent the earliest source of cardiovascular progenitors during ESC differentiation
To investigate the cellular and molecular characteristics of Mesp1-expressing cells during ESC differentiation, we generated an ESC line expressing Venus-GFP under the control of the 5.6-kb regulatory region upstream of the Mesp1 coding sequence, which faithfully recapitulates endogenous Mesp1 expression in the cardiogenic mesoderm of transgenic mice (Fig. 1 A; Haraguchi et al., 2001). We electroporated this Mesp1-GFP reporter construct into ESCs, isolated neomycin resistant clones, and selected several different Mesp1-GFP ESC clones presenting temporal expression of GFP that closely followed that of Mesp1 mRNA (Fig. 1, B–D). No GFP-positive cells were observed in undifferentiated ESCs, but during ESC differentiation, Mesp1-GFP–positive cells appeared around day 2 (D2), peaked at D3, were maintained at D4, and rapidly decreased thereafter to become undetectable at D6 (Fig. 1 D). This transient expression of Mesp1-GFP during ESC differentiation is consistent with the early and transient expression of Mesp1 in the nascent mesoderm during embryonic development (Saga et al., 1996, 1999). Using RT-PCR analysis, we showed that Mesp1 and GFP transcripts are enriched in Mesp1-GFP–expressing cells isolated at the peak of Mesp1-GFP expression (D3) during ESC differentiation (Fig. 1, D and E), demonstrating that our Mesp1-GFP reporter ESC line recapitulates the temporal endogenous expression of Mesp1.
Engineering ESCs expressing Venus-GFP under the regulatory region of Mesp1. (A) Schematic representation of the Mesp1 reporter transgene. Venus-GFP is cloned under the regulatory sequences of Mesp1 that allowed transgene expression in the cardiogenic mesoderm. (B, right) Detection of GFP in Mesp1-GFP ESCs at D3 of differentiation. (left) Unmodified ESCs at the same day of differentiation are used as a control. Bars, 50 µm. (C and D) Kinetics of Mesp1 mRNA expression measured by RT–quantitative PCR (C), and Mesp1-GFP expression as detected by FACS (D). Results are normalized for Mesp1 expression in undifferentiated ESCs (C) or represent the percentage of Mesp1-GFP–positive cells (D). (E) Relative expression of Mesp1 and GFP transcripts in Mesp1-GFP–expressing cells (GFP positive [pos]) and in Mesp1-GFP–nonexpressing cells (GFP negative [neg]) isolated by FACS at D3. Results are normalized for the expression of the transcripts in all sorted cells (gray bars). Error bars indicate means ± SEM; n = 3. FRT, flippase recognition target. pA, polyadenylation. PGK, phosphoglycerate kinase.
Engineering ESCs expressing Venus-GFP under the regulatory region of Mesp1. (A) Schematic representation of the Mesp1 reporter transgene. Venus-GFP is cloned under the regulatory sequences of Mesp1 that allowed transgene expression in the cardiogenic mesoderm. (B, right) Detection of GFP in Mesp1-GFP ESCs at D3 of differentiation. (left) Unmodified ESCs at the same day of differentiation are used as a control. Bars, 50 µm. (C and D) Kinetics of Mesp1 mRNA expression measured by RT–quantitative PCR (C), and Mesp1-GFP expression as detected by FACS (D). Results are normalized for Mesp1 expression in undifferentiated ESCs (C) or represent the percentage of Mesp1-GFP–positive cells (D). (E) Relative expression of Mesp1 and GFP transcripts in Mesp1-GFP–expressing cells (GFP positive [pos]) and in Mesp1-GFP–nonexpressing cells (GFP negative [neg]) isolated by FACS at D3. Results are normalized for the expression of the transcripts in all sorted cells (gray bars). Error bars indicate means ± SEM; n = 3. FRT, flippase recognition target. pA, polyadenylation. PGK, phosphoglycerate kinase.
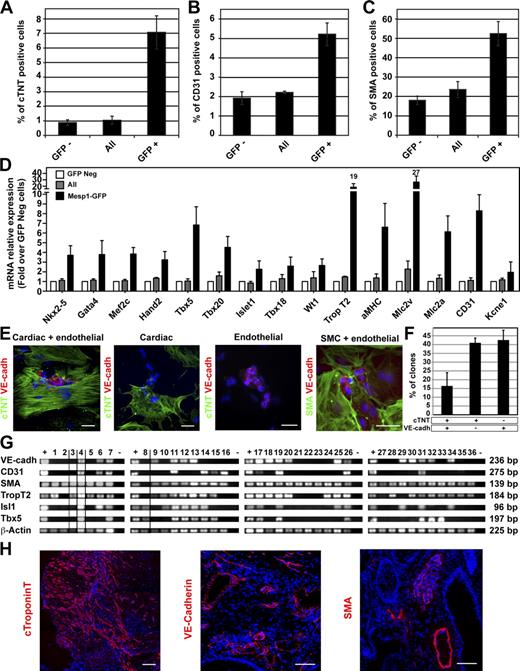
To determine whether Mesp1-expressing cells contained the early cardiovascular progenitors, we isolated Mesp1-GFP–expressing cells at D3 and cultured these cells in a serum-free medium, allowing cardiac terminal differentiation in vitro (Kattman et al., 2006). After 8 d of culture, beating cells were greatly enriched in the Mesp1-GFP–derived cells (Video 1) compared with GFP-negative (Video 2) or all sorted cells (Video 3). We analyzed and quantified the differentiation potential of Mesp1-GFP cells using FACS, immunostaining, and RT-PCR analysis and found that early Mesp1-expressing cells are enriched for progenitors with the potential to differentiate into CMs (marked by cardiac troponin T [cTNT] expression; Fig. 2 A), ECs (CD31; Fig. 2 B), and SMCs (smooth muscle actin [SMA]; Fig. 2 C and Fig. S1 A). Altogether, the three main lineages arising from the differentiation of MCPs represented ∼65% of all cells in Mesp1-GFP isolated cells. RT-PCR showed that Mesp1-GFP–derived cells are enriched in cardiac transcription factors of the FHF and SHF (Nkx2-5, Gata4, Mef2c, Hand2, Tbx5, Tbx20, and Isl1); in pan- (TropT2 and aMHC), atrial (Mlc2a), and ventricular (Mlc2v) cardiac markers; in epicardial markers (Tbx18 and Wt1); and EC markers (CD31; Fig. 2 D). These results demonstrate that Mesp1-GFP–expressing cells are greatly enriched in early MCPs and suggest that early Mesp1-expressing cells give rise to the previously described MCPs of the FHF and the SHF during ESC differentiation (Kattman et al., 2006; Moretti et al., 2006; Wu et al. 2006).
Beating areas in differentiated Mesp1-expressing cells. Time-lapse imaging of Mesp1-GFP–expressing cells after 8 d of differentiation after their isolation and replating at D3 of ESC differentiation. Video displays 15 images/s.
Beating areas in differentiated Mesp1-expressing cells. Time-lapse imaging of Mesp1-GFP–expressing cells after 8 d of differentiation after their isolation and replating at D3 of ESC differentiation. Video displays 15 images/s.
Absence of beating areas in differentiated Mesp1-GFP–negative cells. Time-lapse imaging of Mesp1-GFP–negative cells after 8 d of differentiation after their isolation and replating at D3 of ESC differentiation. Video displays 15 images/s.
Absence of beating areas in differentiated Mesp1-GFP–negative cells. Time-lapse imaging of Mesp1-GFP–negative cells after 8 d of differentiation after their isolation and replating at D3 of ESC differentiation. Video displays 15 images/s.
Beating areas in all sorted cells. Time-lapse imaging of all sorted cells after 8 d of differentiation after their isolation and replating at D3 of ESC differentiation. Video displays 15 images/s.
Beating areas in all sorted cells. Time-lapse imaging of all sorted cells after 8 d of differentiation after their isolation and replating at D3 of ESC differentiation. Video displays 15 images/s.
Isolation and functional characterization of early Mesp1-GFP–expressing cells. (A–C) Expression of cardiovascular markers after 8 d of differentiation of the indicated cell populations isolated at D3. Cardiac and endothelial differentiation were quantified by FACS using a cardiac-specific isoform of the troponin T (cTNT; A) and the endothelial marker CD31 (B). SMC differentiation was assessed by counting the percentage of cells expressing smooth muscle actin (SMA) on cytospin slides (C; also see Fig. S1 A). n = 4. (D) Relative mRNA expression of cardiovascular markers in Mesp1-GFP positive–derived cells (black bars) and in all sorted cells (gray bars) assessed by real-time RT-PCR 8 d after replating. Results are normalized to the expression of the different transcripts in the Mesp1-GFP negative (Neg)–derived cells (white bars). n = 4. (E) Immunostaining for cTNT (CMs), VE-cadherin (VE-cadh; ECs), and SMA (SMCs) in individual colonies obtained after the replating at the clonal density of isolated Mesp1-GFP cells at D3 and cultured for 13 d. Bars, 50 µm. (F) Quantification of colonies expressing cardiovascular (cTNT and VE-cadherin), cardiac (cTNT), and endothelial (VE-cadherin) markers as obtained in E. n = 3. (G) RT-PCR analysis of cardiovascular markers in colonies derived from a single Mesp1-GFP isolated cell in 96 wells after 13 d of differentiation. Only clones positive for β-actin are shown, with dividing lines indicating the removal of intervening lanes from the gels. Samples tested in different experiments are shown as distinct panels with their respective positive (+) and negative (−) control samples. (H) Cardiovascular potential of Mesp1-GFP isolated cells at D3 of ESC differentiation, which were transplanted under the kidney capsule of nonobese diabetic/severe combined immunodeficient mice. Cardiovascular differentiation was assessed after 4 wk by immunostaining for cTNT, VE-cadherin, and SMA. n = 3. Bars, 100 µm. Error bars indicate means ± SEM.
Isolation and functional characterization of early Mesp1-GFP–expressing cells. (A–C) Expression of cardiovascular markers after 8 d of differentiation of the indicated cell populations isolated at D3. Cardiac and endothelial differentiation were quantified by FACS using a cardiac-specific isoform of the troponin T (cTNT; A) and the endothelial marker CD31 (B). SMC differentiation was assessed by counting the percentage of cells expressing smooth muscle actin (SMA) on cytospin slides (C; also see Fig. S1 A). n = 4. (D) Relative mRNA expression of cardiovascular markers in Mesp1-GFP positive–derived cells (black bars) and in all sorted cells (gray bars) assessed by real-time RT-PCR 8 d after replating. Results are normalized to the expression of the different transcripts in the Mesp1-GFP negative (Neg)–derived cells (white bars). n = 4. (E) Immunostaining for cTNT (CMs), VE-cadherin (VE-cadh; ECs), and SMA (SMCs) in individual colonies obtained after the replating at the clonal density of isolated Mesp1-GFP cells at D3 and cultured for 13 d. Bars, 50 µm. (F) Quantification of colonies expressing cardiovascular (cTNT and VE-cadherin), cardiac (cTNT), and endothelial (VE-cadherin) markers as obtained in E. n = 3. (G) RT-PCR analysis of cardiovascular markers in colonies derived from a single Mesp1-GFP isolated cell in 96 wells after 13 d of differentiation. Only clones positive for β-actin are shown, with dividing lines indicating the removal of intervening lanes from the gels. Samples tested in different experiments are shown as distinct panels with their respective positive (+) and negative (−) control samples. (H) Cardiovascular potential of Mesp1-GFP isolated cells at D3 of ESC differentiation, which were transplanted under the kidney capsule of nonobese diabetic/severe combined immunodeficient mice. Cardiovascular differentiation was assessed after 4 wk by immunostaining for cTNT, VE-cadherin, and SMA. n = 3. Bars, 100 µm. Error bars indicate means ± SEM.
To determine whether Mesp1-expressing cells represent common progenitors for both heart fields, we performed clonal analysis of Mesp1-expressing cells isolated at D3. Immunostaining of individual colonies arising from the differentiation of single Mesp1-expressing cells showed that almost all colonies contain SMA-positive cells, ∼15% of the clones presented both cardiac and vascular cells, 40% only expressed cTNT, and 40% only expressed vascular endothelial (VE) cadherin (Fig. 2, E and F), although the proportion of cells expressing these different markers is influenced by the culture conditions (not depicted). To determine whether derivatives of the FHF and SHF are present within the tripotent colonies, we performed RT-PCR on colonies arising from the differentiation of a single Mesp1-expressing cell. Similar to the results obtained by immunostaining, the vast majority of the colonies expressed SMA; among them some colonies also expressed EC or CM markers, some colonies expressed markers of all three lineages, and Tbx5 and Isl1 were both expressed in ∼50% of the tripotent colonies (Fig. 2 G), supporting the notion that a fraction of Mesp1-expressing cells represents common progenitors for both heart fields.
To identify the other cell types into which Mesp1-GFP cells can differentiate, we analyzed the expression of a panel of markers that are representative of different cell lineages from the three germ layers. In addition to differentiating into cardiovascular cells, Mesp1-GFP cells could also differentiate into skeletal muscle and bone cells (Myogenin, Runx2, and Col1a1; Fig. S1 B), which is consistent with the in vivo Mesp1 lineage-tracing experiments that showed that Mesp1-expressing cells give rise to some muscles and bones of the face (McBratney-Owen et al., 2008; Yoshida et al., 2008; Harel et al., 2009). However, not all mesoderm derivatives were increased in Mesp1-GFP cells; e.g., no increase in hematopoietic markers, such as Gata1 and HoxB1, was observed.
To investigate the in vivo differentiation potential of the early Mesp1-GFP–expressing cells, we isolated these cells by FACS at D3 and transplanted them under the kidney capsule of nonobese diabetic/severe combined immunodeficient mice. 4 wk after their transplantation, no teratomas were observed, whereas Mesp1-GFP–negative cells, grafted under the other kidney capsule as a control, generated teratomas (unpublished data). Immunostaining of the grafts demonstrated that Mesp1-GFP cells mainly differentiated into CMs, although expression of EC and SMC markers was also present within the graft (Fig. 2 H). Altogether these data show that Mesp1-expressing cells contain the earliest MCPs specified during ESC differentiation, which give rise upon differentiation to CMs, ECs, and SMCs in vitro and in vivo, and a fraction of Mesp1-expressing cells represent common progenitors for FHF and SHF MCPs.
Transcriptional profiling of early Mesp1-GFP cells during ESC differentiation
To better characterize the early molecular events occurring in Mesp1-expressing cells during MCP specification, we used microarray analysis to define the molecular signature of Mesp1-GFP–expressing cells during ESC differentiation. We determined which genes displayed a change in expression of ≥1.5-fold between Mesp1-GFP–positive and –negative cells at D3 of ESC differentiation in two separate biological replicates. Using these criteria, we found that 1,151 probes out of 45,101 presented a differential expression between Mesp1-positive and Mesp1-negative cells. Among them, 281 probes were found to be up-regulated in Mesp1-expressing cells, corresponding to 212 unique annotated genes (Table I). In addition to the differentially expressed genes found in our duplicate microarray analyses, a certain number of genes were found to be up-regulated in only one of the two replicates, probably because of low level expression, but were confirmed by RT-PCR on different biological replicates.
Microarray analysis of Mesp1-GFP–expressing cells
| Category | Up-regulated genes |
| Transcription factors and chromatin remodeling | Hoxb1 (3.7), Foxc1* (3.5), Lmo1 (3.2), Foxc2* (3.0), Foxf1a (2.9), Pdlim4 (2.9), Isl1 (2.8), Hoxb2 (2.6), Mesp1 (2.6), Etv2 (2.4), Prrx2 (2.4), Tbx3 (2.4), Tbx6 (2.4), Snai1 (2.4), Lef1 (2.3), Msx2 (2.3), Smarcd3 (2.3), Mesp2 (2.2), Tbx2 (2.2), Evx1 (2.1), Hand1 (2.1), Meis2 (2.1), Prdm6 (2.1), Zcchc12 (2.1), Gata4 (2), Hand2 (2), Klhl6 (2), Msx1 (2), Six2 (2), Twist1* (2), Vax1 (2), Zeb2* (2), Bhlhe22 (1.9), Tbx20 (1.9), Zbtb7c (1.9), Ets1 (1.9), Hey2* (1.9), Smad6* (1.9), Zdhhc20* (1.9), Zcchc24* (1.9), Hmga2 (1.8), Hoxd1 (1.8), Pdlim5 (1.8), Tshz1 (1.8), Zfp516 (1.8), Hey1* (1.7), Smad1 (1.7), Cbx4 (1.7), Zfp423 (1.7), Foxh1 (1.6), Nfatc1* (1.6), Twist2* (1.6), Zeb1* (1.6) |
| Signaling pathways (other than receptors) | |
| Notch | Dll1 (2.1), Dll3 (2.0), Hey2 (1.9), Hey1 (1.7) |
| Wnt | Wnt5a (3), Wnt2 (2.8), Wnt5b (2.1), Apcdd1 (1.8), Lef1 (1.8), Wnt3 (1.7) |
| FGF | Fgf3 (2.3), Fgf15 (1.7), Fgf10 (1.6) |
| TGF-b | Gdf10 (2.7), Tgfb2 (2.0), Lefty2 (1.9), Tgfbi (1.9), Tgfb1i1 (1.8), Vasn (1.7) |
| Bmp | Bmper (2.4), Bmp4 (2.0), Bambi (1.7), Bmp6 (1.6), Smad6 (1.9), Smad1 (1.6) |
| Others | Rasgrp3 (2.4), Rgs5 (2.7), Crabp1 (2.7), Htr1d (2.6), Adcyap1r1 (2.5), S1pr5 (2.4), Ptgds (2.3), Dusp9 (2.3), Cap2 (2.0), Dlc1 (1.9), Tnfsf13b (1.9), Adcy3 (1.9), Dok4 (1.9), Efna3 (1.8), Braf (1.7), Prkd1 (1.7), Alox15 (1.6), Pgr (1.6), Vegfc (1.6) |
| Membrane proteins and receptors | Pcdh19 (3.1), Pdgfra (3.1), Ceacam10 (2.9), Cmklr1 (2.5), Gp1bb (2.5), Plac1 (2.5), Cacna1c (2.4), Odz4 (2.3), Nrp2 (2.3), Vldlr (2.3), Cdh4 (2.3), Pcdh18 (2.3), Adrb1 (2.2), Kdr (2.1), Rftn1 (2.1), Unc5c (2.1), Lhfp (2.0), Kcnd3 (2.0), Il13ra1 (2.0), Amhr2 (2.0), Cd160 (1.9), L1cam (1.9), Cxcr4 (1.9), Aplnr (1.9), Pcdh7 (1.9), Cxcr7 (1.9), Slc4a4 (1.9), Gpr177 (1.8), Itga8 (1.8), Prtg (1.8), Ednra (1.8), Kcnc1 (1.8), Cdh11 (1.7), Pcdh7 (1.7), Pdgfrb (1.7), Gfra2 (1.7), Trpc3 (1.7), Nrp1 (1.7), Cdh2 (1.7), Tmem88 (1.6), Il1rap (1.6), Lrp1 (1.6), Ms4a4d (1.6), Kctd15 (1.5) |
| Extracellular matrix | Col6a1 (3.5), Col9a1 (2.6), Emid2 (2.4), Leprel1 (2.3), Fbln2 (2.2), Fbln7 (2.1), Lor (2.0), Col13a1 (2.0), Fn1 (1.9), Has2 (1.8), Vcan (1.6), Flnb (1.6), Mmp2 (1.6) |
| Others | Spp1 (5.8), Fabp4 (5.5), H60a (4.5), Ugt1a1 (4.0), Papss2 (3.8), Agpat9 (3.1), Ccdc109b (2.8), Phlda2 (2.6), Egln3 (2.5), Gna14 (2.5), Pcsk5 (2.4), Atp1a2 (2.4), Dock10 (2.3), Hs3st3b1 (2.3), Morc4 (2.3), Chst2 (2.3), Pmp22 (2.3), Adamts20 (2.2), St6galnac4 (2.2), Exoc3l (2.2), Fam123c (2.1), Myl7 (2.1), Prdm6 (2.1), Susd5 (2.1), Rbm24 (2.1), Siah2 (2.1), Mex3b (2.1), Chst7 (2.0), Nin (2.0), Actc1 (2.0), Kif26b (2.0), Ccnd2 (2.0), Itih5 (1.9), Man1c1 (1.9), Cbln1 (1.9), Mn1 (1.9), Sh3bp1 (1.9), Fam82a1 (1.8), Olfm1 (1.8), Serpinb9 (1.8), Cdkn1c (1.8), Phldb2 (1.8), Pmaip1 (1.8), Gas1 (1.8), Abtb2 (1.8), Adamts3 (1.7), Sgcb (1.7), Sbsn (1.7), Cyp2s1 (1.7), Adam19 (1.7), Brp44 (1.7), Cyp4f15 (1.7), Dclk1 (1.7), Slco3a1 (1.7), Bace2 (1.7), Car3 (1.7), Aard (1.6), Oaf (1.6), Zadh2 (1.6), As3mt (1.6), Grrp1 (1.6), Ablim1 (1.6), Fam122b (1.6), Gne (1.6), Ptprm (1.6), Rps6ka6 (1.6), Lmna (1.5), Man1a (1.5), Pwwp2b (1.5) |
| Category | Up-regulated genes |
| Transcription factors and chromatin remodeling | Hoxb1 (3.7), Foxc1* (3.5), Lmo1 (3.2), Foxc2* (3.0), Foxf1a (2.9), Pdlim4 (2.9), Isl1 (2.8), Hoxb2 (2.6), Mesp1 (2.6), Etv2 (2.4), Prrx2 (2.4), Tbx3 (2.4), Tbx6 (2.4), Snai1 (2.4), Lef1 (2.3), Msx2 (2.3), Smarcd3 (2.3), Mesp2 (2.2), Tbx2 (2.2), Evx1 (2.1), Hand1 (2.1), Meis2 (2.1), Prdm6 (2.1), Zcchc12 (2.1), Gata4 (2), Hand2 (2), Klhl6 (2), Msx1 (2), Six2 (2), Twist1* (2), Vax1 (2), Zeb2* (2), Bhlhe22 (1.9), Tbx20 (1.9), Zbtb7c (1.9), Ets1 (1.9), Hey2* (1.9), Smad6* (1.9), Zdhhc20* (1.9), Zcchc24* (1.9), Hmga2 (1.8), Hoxd1 (1.8), Pdlim5 (1.8), Tshz1 (1.8), Zfp516 (1.8), Hey1* (1.7), Smad1 (1.7), Cbx4 (1.7), Zfp423 (1.7), Foxh1 (1.6), Nfatc1* (1.6), Twist2* (1.6), Zeb1* (1.6) |
| Signaling pathways (other than receptors) | |
| Notch | Dll1 (2.1), Dll3 (2.0), Hey2 (1.9), Hey1 (1.7) |
| Wnt | Wnt5a (3), Wnt2 (2.8), Wnt5b (2.1), Apcdd1 (1.8), Lef1 (1.8), Wnt3 (1.7) |
| FGF | Fgf3 (2.3), Fgf15 (1.7), Fgf10 (1.6) |
| TGF-b | Gdf10 (2.7), Tgfb2 (2.0), Lefty2 (1.9), Tgfbi (1.9), Tgfb1i1 (1.8), Vasn (1.7) |
| Bmp | Bmper (2.4), Bmp4 (2.0), Bambi (1.7), Bmp6 (1.6), Smad6 (1.9), Smad1 (1.6) |
| Others | Rasgrp3 (2.4), Rgs5 (2.7), Crabp1 (2.7), Htr1d (2.6), Adcyap1r1 (2.5), S1pr5 (2.4), Ptgds (2.3), Dusp9 (2.3), Cap2 (2.0), Dlc1 (1.9), Tnfsf13b (1.9), Adcy3 (1.9), Dok4 (1.9), Efna3 (1.8), Braf (1.7), Prkd1 (1.7), Alox15 (1.6), Pgr (1.6), Vegfc (1.6) |
| Membrane proteins and receptors | Pcdh19 (3.1), Pdgfra (3.1), Ceacam10 (2.9), Cmklr1 (2.5), Gp1bb (2.5), Plac1 (2.5), Cacna1c (2.4), Odz4 (2.3), Nrp2 (2.3), Vldlr (2.3), Cdh4 (2.3), Pcdh18 (2.3), Adrb1 (2.2), Kdr (2.1), Rftn1 (2.1), Unc5c (2.1), Lhfp (2.0), Kcnd3 (2.0), Il13ra1 (2.0), Amhr2 (2.0), Cd160 (1.9), L1cam (1.9), Cxcr4 (1.9), Aplnr (1.9), Pcdh7 (1.9), Cxcr7 (1.9), Slc4a4 (1.9), Gpr177 (1.8), Itga8 (1.8), Prtg (1.8), Ednra (1.8), Kcnc1 (1.8), Cdh11 (1.7), Pcdh7 (1.7), Pdgfrb (1.7), Gfra2 (1.7), Trpc3 (1.7), Nrp1 (1.7), Cdh2 (1.7), Tmem88 (1.6), Il1rap (1.6), Lrp1 (1.6), Ms4a4d (1.6), Kctd15 (1.5) |
| Extracellular matrix | Col6a1 (3.5), Col9a1 (2.6), Emid2 (2.4), Leprel1 (2.3), Fbln2 (2.2), Fbln7 (2.1), Lor (2.0), Col13a1 (2.0), Fn1 (1.9), Has2 (1.8), Vcan (1.6), Flnb (1.6), Mmp2 (1.6) |
| Others | Spp1 (5.8), Fabp4 (5.5), H60a (4.5), Ugt1a1 (4.0), Papss2 (3.8), Agpat9 (3.1), Ccdc109b (2.8), Phlda2 (2.6), Egln3 (2.5), Gna14 (2.5), Pcsk5 (2.4), Atp1a2 (2.4), Dock10 (2.3), Hs3st3b1 (2.3), Morc4 (2.3), Chst2 (2.3), Pmp22 (2.3), Adamts20 (2.2), St6galnac4 (2.2), Exoc3l (2.2), Fam123c (2.1), Myl7 (2.1), Prdm6 (2.1), Susd5 (2.1), Rbm24 (2.1), Siah2 (2.1), Mex3b (2.1), Chst7 (2.0), Nin (2.0), Actc1 (2.0), Kif26b (2.0), Ccnd2 (2.0), Itih5 (1.9), Man1c1 (1.9), Cbln1 (1.9), Mn1 (1.9), Sh3bp1 (1.9), Fam82a1 (1.8), Olfm1 (1.8), Serpinb9 (1.8), Cdkn1c (1.8), Phldb2 (1.8), Pmaip1 (1.8), Gas1 (1.8), Abtb2 (1.8), Adamts3 (1.7), Sgcb (1.7), Sbsn (1.7), Cyp2s1 (1.7), Adam19 (1.7), Brp44 (1.7), Cyp4f15 (1.7), Dclk1 (1.7), Slco3a1 (1.7), Bace2 (1.7), Car3 (1.7), Aard (1.6), Oaf (1.6), Zadh2 (1.6), As3mt (1.6), Grrp1 (1.6), Ablim1 (1.6), Fam122b (1.6), Gne (1.6), Ptprm (1.6), Rps6ka6 (1.6), Lmna (1.5), Man1a (1.5), Pwwp2b (1.5) |
Fold changes are indicated in parentheses. Asterisks indicate genes found only in one of the two array replicates and confirmed by RT-PCR on different biological samples. Bold indicates genes found to be also up-regulated after Mesp1 overexpression.
Functional annotation clustering of the 212 probes up-regulated in the duplicate microarray analysis of Mesp1-expressing cells at D3 was performed using the Database for Annotation, Visualization, and Integrated Discovery bioinformatics resources (Huang et al., 2009). The functional annotation chart revealed that the first term retrieved in Mesp1-enriched genes is heart development (10% of the genes) followed by muscle, embryonic, mesoderm, tube, blood vessel, and vasculature development (Table S1).
We have recently demonstrated that Mesp1 overexpression rapidly promotes the expression of many genes implicated in cardiovascular development (Bondue et al., 2008). To determine which of these genes are naturally expressed within Mesp1-expressing cells during ESC differentiation, we compared the list of genes up-regulated upon Mesp1 gain of function with the genes enriched in Mesp1-GFP–expressing cells at D3 and found that ∼35% of the genes up-regulated by Mesp1 overexpression were also gene enriched in Mesp1-GFP–expressing cells (Table I). To validate the significance of this enrichment, we compared the fold change direction of the probes that were significantly up-regulated or down-regulated in Mesp1-GFP cells and after Mesp1 overexpression. The proportion of coherent genes (27%), in which the probe is affected in the same direction in Mesp1-GFP cells and after Mesp1 gain of function, is significantly much higher than the incoherent ones (3%; Fig. S2). These data reinforced the notion that Mesp1 directly or indirectly controls a significant proportion of the cardiac differentiation program during ESC differentiation.
Isolation and functional characterization of Mesp1-expressing cells using a combination of monoclonal antibodies
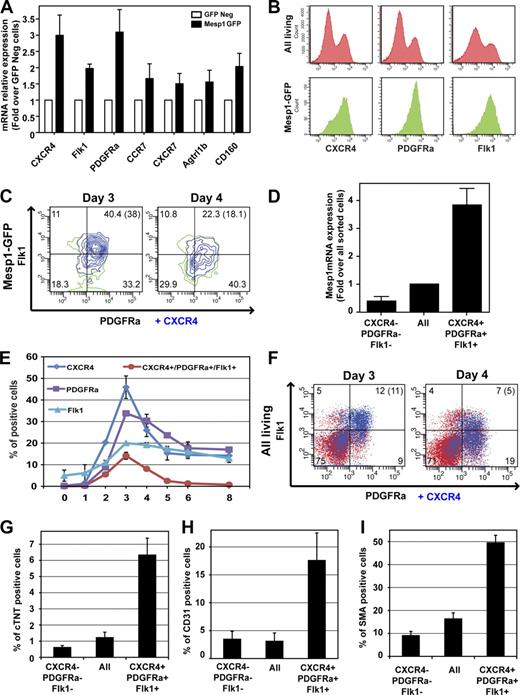
Our microarray and RT-PCR analysis of Mesp1-expressing cells demonstrated that early MCPs preferentially express a variety of cell surface proteins (Fig. 3 A and Table I). Among them, only CXCR4, PDGFRa, or Flk1, which have previously been associated with later stages of cardiovascular progenitors during ESC differentiation (Iida et al., 2005; Moretti et al., 2006; Nelson et al., 2008; Hidaka et al., 2010), was expressed at a high level in almost all Mesp1-GFP–expressing cells at D3 (Fig. 3 B). At this time point, Mesp1-expressing cells consisted of a relatively homogenous population of cells coexpressing a high level of CXCR4, PDGFRa, and Flk1, whereas 24 h later at D4, Mesp1-expressing cells were more heterogeneous with regard to the level of expression of these markers (Fig. 3 C). Cells coexpressing high levels of CXCR4, PDGFRa, and Flk1 at D3 were enriched for Mesp1 mRNA (Fig. 3 D), and these triple-positive (TP) cells presented a temporal appearance (Fig. 3, E and F) similar to Mesp1-GFP–expressing cells (Fig. 1 D), strongly suggesting that this combination of cell surface markers mirrors well the endogenous Mesp1 expression.
Isolation and functional characterization of early MCPs using a combination of monoclonal antibodies. (A) Cell surface marker expression in Mesp1-GFP–expressing cells as measured by real-time RT-PCR in isolated Mesp1-GFP–expressing cells at D3. Results are normalized for the mRNA expression in GFP-negative cells. n = 3. (B) Detection of CXCR4, PDGFRa, and Flk1 by FACS at D3 in all living cells (top) and in the Mesp1-GFP population (bottom). (C) Multicolor FACS analysis gated on Mesp1-GFP cells of CXCR4, PDGFRa, and Flk1 expression at D3 and D4. (D) Enrichment of Mesp1 expression in TP cells at D3 as measured by RT-PCR on FACS-isolated cells. Results are normalized for the relative transcript expression in all sorted cells. n = 3. (E) Temporal expression of CXCR4, PDGFRa, and Flk1 during ESC differentiation as detected by FACS. n = 2. (F) Combined detection of CXCR4, PDGFRa, and Flk1 expression at D3 and D4 in all living cells. (C and F) Percentages of cells in each quadrant are shown, and the percentage of CXCR4/PDGFRa/Flk1 TP cells are shown in parentheses. (G–I) Cardiac (G), endothelial (H), and SMC (I; also see Fig. S1 C) differentiation of TP cells as performed in Fig. 2 (A–C). n = 4. Error bars indicate means ± SEM.
Isolation and functional characterization of early MCPs using a combination of monoclonal antibodies. (A) Cell surface marker expression in Mesp1-GFP–expressing cells as measured by real-time RT-PCR in isolated Mesp1-GFP–expressing cells at D3. Results are normalized for the mRNA expression in GFP-negative cells. n = 3. (B) Detection of CXCR4, PDGFRa, and Flk1 by FACS at D3 in all living cells (top) and in the Mesp1-GFP population (bottom). (C) Multicolor FACS analysis gated on Mesp1-GFP cells of CXCR4, PDGFRa, and Flk1 expression at D3 and D4. (D) Enrichment of Mesp1 expression in TP cells at D3 as measured by RT-PCR on FACS-isolated cells. Results are normalized for the relative transcript expression in all sorted cells. n = 3. (E) Temporal expression of CXCR4, PDGFRa, and Flk1 during ESC differentiation as detected by FACS. n = 2. (F) Combined detection of CXCR4, PDGFRa, and Flk1 expression at D3 and D4 in all living cells. (C and F) Percentages of cells in each quadrant are shown, and the percentage of CXCR4/PDGFRa/Flk1 TP cells are shown in parentheses. (G–I) Cardiac (G), endothelial (H), and SMC (I; also see Fig. S1 C) differentiation of TP cells as performed in Fig. 2 (A–C). n = 4. Error bars indicate means ± SEM.
To determine whether the CXCR4/PDGFRa/Flk1 TP cells are enriched in early MCPs during ESC differentiation, we isolated TP cells by FACS at D3 and cultured them in a serum-free medium for a supplemental 8 d. Similar to what we found for the differentiation of Mesp1-expressing cells, beating cells were preferentially observed in TP cells compared with all sorted cells and triple negative cells. Quantification of cardiac and vascular differentiation revealed that TP cells were similarly enriched in CM (Fig. 3 G), EC (Fig. 3 H), and SMC (Fig. 3 I and Fig. S1 C) differentiation as Mesp1-GFP–expressing cells (Fig. 2, A–C), suggesting that the combination of these three monoclonal antibodies closely tracks with Mesp1 expression at the time of MCP specification and can be used to monitor and isolate early MCPs during ESC differentiation.
To determine how Mesp1-expressing cells are related to the previously described Bry-GFP+/Flk1+ MCPs (Kattman et al., 2007), we analyzed the expression of Mesp1 and CXCR4, PDGFRa, and Flk1 in Bry-GFP/Flk1–expressing cells at different times of ESC differentiation (Fig. S3). At D3, Bry-GFP/Flk1–expressing cells can be separated into two distinct populations, one coexpressing CXCR4, PDGFRa, and Flk1 and the other expressing Flk1/CXCR4 but negative for PDGFRa (Fig. S3 B). Mesp1 was enriched to a similar level in CXCR4/PDGFRa/Flk1 TP cells and in Bry-GFP/Flk1/PDGFRa TP cells, whereas no Mesp1 enrichment was found in Bry-GFP+/Flk1+/PDGFRa–negative cells (Fig. S3 D). In contrast, Scl, a marker of hemangioblast lineage, was strongly enriched in Bry-GFP+/Flk1+/PDGFRa–negative cells but not in CXCR4/PDGFRa/Flk1 TP or in Bry-GFP/Flk1/PDGFRa–positive cells. These data indicated that Mesp1-expressing cells correspond to a subpopulation of the previously described Bry-GFP/Flk1–positive progenitors.
Mesp1 rapidly promotes and is required for MCP specification during ESC differentiation
Using Mesp1 gain of function in ESCs, we and others have previously shown that Mesp1 expression greatly increased and accelerated the differentiation of ESCs into cardiac, vascular, and smooth muscle lineages (Bondue et al., 2008; David et al., 2008; Lindsley et al., 2008). The increase in cells expressing Flk1 and PDGFRa after Mesp1 expression (Lindsley et al., 2008) suggests that Mesp1 expression can promote MCP specification. To determine whether Mesp1 rapidly promotes MCP specification, we assessed the relative frequency of CXCR4/PDGFRa/Flk1 TP cells at different early time points after Mesp1 expression using a doxycyclin (Dox)-inducible Mesp1 ESC line (Fig. 4 A). We added Dox at D2 of ESC differentiation and monitored the expression of TP cells after 24 and 48 h. As early as 24 h after Dox addition, a major increase in the proportion of the TP cell population was observed in Mesp1-overexpressing cells compared with unstimulated Mesp1 ESCs (Fig. 4 B). This effect persisted and increased 48 h after Dox addition (Fig. 4 C), showing that forced expression of Mesp1 during ESC differentiation rapidly promotes MCP specification.
Mesp1 rapidly promotes and is required for MCP specification and cardiac differentiation. (A) Schematic representation of Dox-inducible Mesp1 ESCs. (B) FACS analysis of the expression of CXCR4, PDGFRa, and Flk1 in Mesp1 Dox-inducible ESCs at D3, 24 h after Dox addition. (C) FACS quantification of CXCR4/PDGFRa/Flk1 TP cells in Mesp1 Dox-inducible ESCs 24 (D3) and 48 h (D4) after Dox addition. n = 3. (D and E) FACS quantification of proliferation (BrdU; D) and apoptosis (active caspase-3; E) in PDGFRa+/Flk1+ cells and in all Mesp1-inducible ESCs in the presence and absence of Dox for 24 h (D3). n = 2. (F) Schematic representation of Dox-inducible Engrailed (Engr)-Mesp1 ESCs (EN-Mesp1). (G) FACS analysis of CXCR4, PDGFRa, and Flk1 expression in EN-Mesp1–inducible ESCs at D4, 48 h after Dox addition. (B and G) Percentages of cells in each quadrant are shown, and the percentage of CXCR4/PDGFRa/Flk1 TP cells are shown in parentheses. (H) FACS quantification of TP cells in EN-Mesp1–inducible ESCs 24 (D3) and 48 h (D4) after Dox addition. Results are normalized to unstimulated cells. n = 3. (I) Quantification of beating areas in EN-Mesp1 ESCs in the presence or in the absence of Dox at D8. n = 3. (J and K) FACS quantification of cTNT (J) and CD31 (K) in EN-Mesp1–expressing cells. n = 3. Error bars indicate means ± SEM. TRE, tetracycline-responsive element. EB, embryoid body.
Mesp1 rapidly promotes and is required for MCP specification and cardiac differentiation. (A) Schematic representation of Dox-inducible Mesp1 ESCs. (B) FACS analysis of the expression of CXCR4, PDGFRa, and Flk1 in Mesp1 Dox-inducible ESCs at D3, 24 h after Dox addition. (C) FACS quantification of CXCR4/PDGFRa/Flk1 TP cells in Mesp1 Dox-inducible ESCs 24 (D3) and 48 h (D4) after Dox addition. n = 3. (D and E) FACS quantification of proliferation (BrdU; D) and apoptosis (active caspase-3; E) in PDGFRa+/Flk1+ cells and in all Mesp1-inducible ESCs in the presence and absence of Dox for 24 h (D3). n = 2. (F) Schematic representation of Dox-inducible Engrailed (Engr)-Mesp1 ESCs (EN-Mesp1). (G) FACS analysis of CXCR4, PDGFRa, and Flk1 expression in EN-Mesp1–inducible ESCs at D4, 48 h after Dox addition. (B and G) Percentages of cells in each quadrant are shown, and the percentage of CXCR4/PDGFRa/Flk1 TP cells are shown in parentheses. (H) FACS quantification of TP cells in EN-Mesp1–inducible ESCs 24 (D3) and 48 h (D4) after Dox addition. Results are normalized to unstimulated cells. n = 3. (I) Quantification of beating areas in EN-Mesp1 ESCs in the presence or in the absence of Dox at D8. n = 3. (J and K) FACS quantification of cTNT (J) and CD31 (K) in EN-Mesp1–expressing cells. n = 3. Error bars indicate means ± SEM. TRE, tetracycline-responsive element. EB, embryoid body.
To determine whether Mesp1 promotes MCP specification through a selective mechanism, we used multicolor FACS analysis to directly measure cell proliferation and apoptosis within the PDGFRa+/Flk1+ population after Mesp1 gain of function. Mesp1 expression did not increase cell proliferation (Fig. 4 D) or apoptosis (Fig. 4 E) in PDGFRa+/Flk1+-positive cells, which is consistent with previous observations suggesting that Mesp1 promotes MCP specification through an instructive rather than selective mechanism (Bondue et al., 2008; Lindsley et al., 2008).
To determine whether Mesp1 is required for MCP specification during ESC differentiation, we generated an ESC line allowing inducible expression of a fusion protein of Mesp1 with the repressor domain of Drosophila melanogaster Engrailed (EN; EN-Mesp1; Fig. 4 F; Han and Manley, 1993). Transient expression of EN-Mesp1 at D2 and D3 led to a complete absence of CXCR4/PDGFRa/Flk1 TP cells (Fig. 4, G and H), a complete absence of beating cells (Fig. 4 I), a dramatic reduction in CM differentiation (Fig. 4 J), and a significant reduction in EC differentiation (Fig. 4 K), showing that Mesp1 is required for MCP specification and cardiovascular differentiation during ESC differentiation.
Mesp1 regulates the expression of cardiovascular and epithelial to mesenchymal transition (EMT) transcription factors in MCPs
Our microarray analysis of Mesp1-GFP–expressing cells during ESC differentiation showed that many key transcription factors involved in early cardiovascular development are enriched in Mesp1-GFP–expressing cells (Fig. 5 A and Table I). A large fraction of these transcriptional regulators were previously shown to be up-regulated upon Mesp1 expression, whereas others were not affected (i.e., Isl1) or even down-regulated (i.e., Mesp2) after Mesp1 gain of function (Bondue et al., 2008). In addition to the cardiovascular transcription factors, several transcription factors mediating EMT, such as Snail1, Twist1/2, and Foxc1/2 (Fig. 5 B), were also up-regulated in Mesp1-GFP–expressing cells. Indeed, the vast majority of Mesp1-GFP cells expressed low levels of epithelial (E) cadherin, which is consistent with the notion that Mesp1-GFP cells undergo EMT during MCP specification (Fig. 5 C). RT-PCR analysis performed on FACS-isolated CXCR4/PDGFRa/Flk1 TP cells showed that MCPs isolated using monoclonal antibodies present a similar enrichment for the expression of cardiovascular transcriptional regulators compared with Mesp1-GFP cells (Fig. 5 D), some of which (Hand1, Hand2, Nkx2-5, Gata6, and Tbx20) increased between D3 and D4, suggesting that early specified MCPs undergo a progressive maturation toward cardiovascular differentiation over time.
Cardiovascular and EMT transcription factors regulated by Mesp1 in early MCPs. (A and B) Real-time RT-PCR analysis of mRNA relative expression of cardiovascular (A) and EMT (B) transcription factors in FACS-isolated Mesp1-GFP cells at D3 (black bars). Results are normalized for the transcript expression in Mesp1-GFP–negative (Neg) cells (white bars). (C) E-Cadherin expression in all cells and in Mesp1-expressing cells as measured by FACS. (D) Real-time RT-PCR analysis of the expression of cardiovascular transcription factors in CXCR4/PDGFRa/Flk1 TP cells isolated at D3 (white bars) and D4 (black bars). Results are normalized for the mRNA expression in CXCR4−/PDGFRa−/Flk1− cells. Numbers at the top of the bars indicate the fold change. (E) Real-time RT-PCR analysis of the expression of cardiovascular transcription factors within the TP population in Dox-inducible Mesp1 ESCs isolated at D4 in the presence or in the absence of Dox for 48 h. Results are normalized for transcript expression in unstimulated TP cells. (F) RT-PCR analysis of cardiovascular transcription factor expression in single TP isolated cells from Mesp1-inducible ESCs in the presence or in the absence of Dox for 48 h. Only clones positive for β-actin are shown, with dividing lines indicating the removal of intervening lanes from the gels. Samples tested in different experiments are shown as distinct panels with their respective positive (+) and negative (−) control samples. (G) Expression of cardiovascular transcription factors in Dox-inducible EN-Mesp1 ESCs at D3. Results are normalized for the expression in Dox-untreated cells. Error bars indicate means ± SEM (n = 3).
Cardiovascular and EMT transcription factors regulated by Mesp1 in early MCPs. (A and B) Real-time RT-PCR analysis of mRNA relative expression of cardiovascular (A) and EMT (B) transcription factors in FACS-isolated Mesp1-GFP cells at D3 (black bars). Results are normalized for the transcript expression in Mesp1-GFP–negative (Neg) cells (white bars). (C) E-Cadherin expression in all cells and in Mesp1-expressing cells as measured by FACS. (D) Real-time RT-PCR analysis of the expression of cardiovascular transcription factors in CXCR4/PDGFRa/Flk1 TP cells isolated at D3 (white bars) and D4 (black bars). Results are normalized for the mRNA expression in CXCR4−/PDGFRa−/Flk1− cells. Numbers at the top of the bars indicate the fold change. (E) Real-time RT-PCR analysis of the expression of cardiovascular transcription factors within the TP population in Dox-inducible Mesp1 ESCs isolated at D4 in the presence or in the absence of Dox for 48 h. Results are normalized for transcript expression in unstimulated TP cells. (F) RT-PCR analysis of cardiovascular transcription factor expression in single TP isolated cells from Mesp1-inducible ESCs in the presence or in the absence of Dox for 48 h. Only clones positive for β-actin are shown, with dividing lines indicating the removal of intervening lanes from the gels. Samples tested in different experiments are shown as distinct panels with their respective positive (+) and negative (−) control samples. (G) Expression of cardiovascular transcription factors in Dox-inducible EN-Mesp1 ESCs at D3. Results are normalized for the expression in Dox-untreated cells. Error bars indicate means ± SEM (n = 3).
We have recently demonstrated that Mesp1 rapidly promotes the expression of many transcription factors involved in cardiovascular differentiation during ESC differentiation and have shown that some of these genes are direct Mesp1 target genes (Bondue et al., 2008). To determine to which extent the up-regulation of these transcription factors is regulated by Mesp1, we measured the expression of these cardiovascular transcription factors in CXCR4/PDGFRa/Flk1 TP cells after Mesp1 overexpression. These data showed that Mesp1 overexpression further increased the level of expression of cardiovascular transcription factors, such as Hand2, Myocardin, or Nkx2-5, within the CXCR4/PDGFRa/Flk1 TP population (Fig. 5 E). To determine whether the increase in the expression of these transcription factors was the consequence of a homogenous change in gene expression mediated by Mesp1 in the entire TP cell population or whether Mesp1 only up-regulated the expression of these transcriptions in a fraction of these cells, we performed single-cell RT-PCR on FACS-isolated CXCR4/PDGFRa/Flk1 TP cells after Mesp1 gain of function. In the absence of Mesp1 overexpression, the vast majority of TP cells only expressed one or the other cardiac transcription factors, whereas upon Mesp1 overexpression, a much higher proportion of TP cells expressed multiple cardiac transcription factors at the same time in the same cell (Fig. 5 F). In addition, overexpression of EN-Mesp1 down-regulated the expression of these transcription factors (Fig. 5 G). Altogether, these data strongly suggest that Mesp1 directly or indirectly controls the expression of many key cardiovascular transcription factors in MCPs and increases the probability of cardiac commitment in individual cells.
Isl1 is expressed independently of Mesp1 in a subset of early Mesp1-expressing cells
Isl1 expression has been previously used to mark tripotent MCPs at D5 of ESC differentiation (Moretti et al., 2006). Isl1 is expressed in SHF progenitors and is required for SHF development (Cai et al., 2003), although recent studies reported Isl1 expression in embryonic regions corresponding to the FHF (Brade et al., 2007; Prall et al., 2007). It remains unclear whether Isl1 is also expressed earlier during ESC differentiation at the time of MCP specification. Our microarray and RT-PCR analysis revealed that Mesp1-expressing cells are enriched for the Isl1 transcript as early as D3 of ESC differentiation (Fig. 5 A and Table I). In contrast to direct or indirect Mesp1 target genes, Isl1 is enriched in Mesp1-expressing cells (Fig. 5 A) and in TP cells (Fig. 5 D) but is not up-regulated by Mesp1 overexpression (Fig. 5 E) or down-regulated after EN-Mesp1 expression (Fig. 5 G), strongly suggesting that Isl1 is expressed in early MCPs independently of Mesp1.
To better characterize the relation between Mesp1 and Isl1 expression, we performed immunostaining for Isl1 and GFP expression on cytospin preparations of Mesp1-GFP cells after ESC differentiation. Mesp1-GFP was expressed in 4 and 1.5% of cells at D3 and D4, respectively (Fig. 6 A). Although the level of Isl1 expression was lower than in later stages of differentiation, Isl1 expression was already detected at D3 and D4 in ∼10% of cells (Fig. 6 B). At D3, ∼20% of Mesp1-expressing cells coexpressed Isl1 (Fig. 6, C and E). At D4, the level of Isl1 expression increased, and ∼50% of Mesp1-expressing cells coexpressed Isl1 (Fig. 6, D and E). The Mesp1/Isl1 double-positive cells represent 10 and 6% of Isl1-expressing cells at D3 and D4, respectively (Fig. 6 F). These data show that Isl1 is coexpressed together with Mesp1 in a fraction of early Mesp1-expressing cells.
Isl1 is expressed in a subset of early Mesp1-expressing cells. (A and B) Quantification of Mesp1-GFP (A) and Isl1 (B) expression as measured by immunostaining of GFP and Isl1 on cytospin slides of Mesp1-GFP cells at D3 and D4. n = 3. (C and D) Confocal microscopy analysis of GFP (Mesp1) and Isl1 immunostaining in Mesp1-GFP cells at D3 (C) and D4 (D). (right) Magnification of the insets, and arrows indicate cells that coexpress Mesp1 and Isl1. Bars, 30 µm. (E and F) Quantification of Isl1 expression in Mesp1-GFP–expressing cells (E), and Mesp1 (GFP) expression in Isl1-expressing cells (F) at D3 and D4. More than 300 cells were counted in each condition. n = 3. Error bars indicate means ± SEM.
Isl1 is expressed in a subset of early Mesp1-expressing cells. (A and B) Quantification of Mesp1-GFP (A) and Isl1 (B) expression as measured by immunostaining of GFP and Isl1 on cytospin slides of Mesp1-GFP cells at D3 and D4. n = 3. (C and D) Confocal microscopy analysis of GFP (Mesp1) and Isl1 immunostaining in Mesp1-GFP cells at D3 (C) and D4 (D). (right) Magnification of the insets, and arrows indicate cells that coexpress Mesp1 and Isl1. Bars, 30 µm. (E and F) Quantification of Isl1 expression in Mesp1-GFP–expressing cells (E), and Mesp1 (GFP) expression in Isl1-expressing cells (F) at D3 and D4. More than 300 cells were counted in each condition. n = 3. Error bars indicate means ± SEM.
Isl1 cooperates with Mesp1 to promote endothelial or cardiac cell lineage commitment, depending on the stage of cardiovascular differentiation
To determine the functional consequences of Isl1 expression in Mesp1-expressing cells, we generated an ESC line that allows Dox-inducible expression of Isl1 alone or in combination with Mesp1 (Fig. 7 A). Dox administration in Isl1-inducible ESCs increased transgene expression to a similar level and in the same proportion of cells as in the Mesp1-inducible ESCs (Fig. S4). Isl1 overexpression during the early stage of ESC differentiation (D2 and D3), corresponding to the time of MCP specification, did not increase the proportion of the CXCR4/PDGFRa/Flk1 TP cells at D3 or D4, and the coexpression of Mesp1 and Isl1 had no additive or synergistic effect compared with Mesp1 expression alone (Fig. 7, B and C). Early expression of Isl1 during ESC differentiation only moderately promoted cardiac differentiation (Fig. 7, D and E) but strongly increased endothelial differentiation (Fig. 7, F and G). Combined expression of Mesp1 and Isl1 further increased endothelial differentiation compared with Mesp1 alone (Fig. 7 F). Overexpression of Isl1 during later stages of differentiation (between D5 and D6) did not promote vascular differentiation but increased cardiac differentiation, which was further enhanced by Mesp1 expression (Fig. 7, H and I).
Isl1 and Mesp1 cooperate in promoting cardiovascular differentiation in ESCs. (A) Schematic representation of Mesp1, Isl1, and Mesp1/Isl1 Dox-inducible ESCs. (B) FACS analysis of CXCR4, PDGFRa, and Flk1 expression in Isl1-inducible ESCs at D4, 48 h in the presence or absence of Dox treatment. Percentages of cells in each quadrant are shown, and the percentage of CXCR4/PDGFRa/Flk1 TP cells are shown in parentheses. (C) FACS quantification of CXCR4/PDGFRa/Flk1 TP cells at 24 (D3) and 48 h (D4) in the presence or absence of Dox in Mesp1, Isl1, and Mesp1/Isl1 Dox-inducible ESCs. n = 4. (D) FACS quantification of cTNT expression at D8 in Mesp1, Isl1, and Mesp1/Isl1 Dox-inducible ESCs in the presence or absence of Dox from D2 to D4. n = 4. (E) Immunostaining of cTNT at D8 of differentiation in Dox-inducible Mesp1, Isl1, and Mesp1/Isl1 ESCs in the presence or absence of Dox from D2 to D4. Images shown are mosaïc acquisitions representative of at least four biologically independent experiments. Bars, 500 µm. (F) FACS quantification of CD31 expression at D7 in Mesp1, Isl1, and Mesp1/Isl1 Dox-inducible ESCs in the presence or absence of Dox from D2 to D4. n = 4. (G) Immunostaining for VE-Cadherin expression at D7 in Dox-inducible Mesp1, Isl1, and Mesp1/Isl1 ESCs in the presence or absence of Dox from D2 to D4. Images shown are representative of four biologically independent experiments. Bars, 100 µm. (H and I) FACS quantification of cTNT (H) and CD31 expression (I) in Mesp1, Isl1, and Mesp1/Isl1 Dox-inducible ESCs at D8 and D7 of differentiation, respectively, in the presence or absence of Dox from D2 to D4 or from D5 to D6. n = 4. Error bars indicate means ± SEM. TRE, tetracycline-responsive element.
Isl1 and Mesp1 cooperate in promoting cardiovascular differentiation in ESCs. (A) Schematic representation of Mesp1, Isl1, and Mesp1/Isl1 Dox-inducible ESCs. (B) FACS analysis of CXCR4, PDGFRa, and Flk1 expression in Isl1-inducible ESCs at D4, 48 h in the presence or absence of Dox treatment. Percentages of cells in each quadrant are shown, and the percentage of CXCR4/PDGFRa/Flk1 TP cells are shown in parentheses. (C) FACS quantification of CXCR4/PDGFRa/Flk1 TP cells at 24 (D3) and 48 h (D4) in the presence or absence of Dox in Mesp1, Isl1, and Mesp1/Isl1 Dox-inducible ESCs. n = 4. (D) FACS quantification of cTNT expression at D8 in Mesp1, Isl1, and Mesp1/Isl1 Dox-inducible ESCs in the presence or absence of Dox from D2 to D4. n = 4. (E) Immunostaining of cTNT at D8 of differentiation in Dox-inducible Mesp1, Isl1, and Mesp1/Isl1 ESCs in the presence or absence of Dox from D2 to D4. Images shown are mosaïc acquisitions representative of at least four biologically independent experiments. Bars, 500 µm. (F) FACS quantification of CD31 expression at D7 in Mesp1, Isl1, and Mesp1/Isl1 Dox-inducible ESCs in the presence or absence of Dox from D2 to D4. n = 4. (G) Immunostaining for VE-Cadherin expression at D7 in Dox-inducible Mesp1, Isl1, and Mesp1/Isl1 ESCs in the presence or absence of Dox from D2 to D4. Images shown are representative of four biologically independent experiments. Bars, 100 µm. (H and I) FACS quantification of cTNT (H) and CD31 expression (I) in Mesp1, Isl1, and Mesp1/Isl1 Dox-inducible ESCs at D8 and D7 of differentiation, respectively, in the presence or absence of Dox from D2 to D4 or from D5 to D6. n = 4. Error bars indicate means ± SEM. TRE, tetracycline-responsive element.
Discussion
Our study revealed that, during ESC differentiation, early Mesp1-GFP–expressing cells are greatly enriched for progenitors with the ability to differentiate into the different cardiovascular cell lineages both in vitro and in vivo, similar to the differentiation potential of Mesp1 found in vivo. Clonal analysis revealed that Mesp1-expressing cells differentiate into both FHF and SHF derivatives, indicating that Mesp1-expressing cells represent a common progenitor for the MCPs of both heart fields, which appears several days later (between D5 and D6; Fig. 8; Moretti et al., 2006; Wu et al., 2006). Our data suggest that Mesp1-expressing cells represent a subpopulation of the previously identified Bry-GFP/Flk1 MCPs (Kattman et al., 2006). Bry-GFP/Flk1–expressing cells can be subdivided into two subpopulations: one negative for PDGFRa and representing hemangioblast progenitors and another expressing high levels of PDGFRa, corresponding to the Mesp1-enriched population (Fig. 8).
Model of the cellular hierarchy acting during cardiovascular lineage commitment. During ESC differentiation, Mesp1-expressing cells represent early tripotent cardiovascular progenitors that are able to differentiate at the clonal level into CMs, ECs, and SMCs, representing early common progenitors for all cardiovascular lineages.
Model of the cellular hierarchy acting during cardiovascular lineage commitment. During ESC differentiation, Mesp1-expressing cells represent early tripotent cardiovascular progenitors that are able to differentiate at the clonal level into CMs, ECs, and SMCs, representing early common progenitors for all cardiovascular lineages.
The transcriptional profiling of early Mesp1-expressing cells identified cell surface markers that can be used in combination to enrich for Mesp1-expressing cells during ESC differentiation and represent an ideal method to monitor and isolate the early MCPs generated during ESC differentiation. Interestingly, these markers have been previously reported to be expressed by progenitors of later stages of cardiovascular differentiation (Iida et al., 2005; Moretti et al., 2006; Nelson et al., 2008; Hidaka et al., 2010). PDGFRa and Flk1 are expressed in the cardiac crescent in vivo (Ema et al., 2006; Prall et al., 2007; Takakura et al., 1997), and lineage tracing has shown that Flk1-expressing cells give rise to all cardiovascular lineages (Ema et al., 2006), suggesting that these markers could be used in future studies to isolate the early MCPs during mouse embryonic development.
Isl1 is expressed in the SHF progenitors during embryonic development, and Isl1 expression can be used to isolate tripotent MCPs during mouse and human ESC differentiation (Moretti et al., 2006; Bu et al., 2009). Our study revealed that a fraction of Mesp1-GFP–expressing cells coexpressed Isl1 independently of Mesp1. Interestingly, in Ciona intestinalis, a primitive chordate, a fraction of Mesp1-expressing cells coexpresses Isl1 (Stolfi et al., 2010), suggesting that the expression of Isl1 in a subpopulation of the Mesp1 field has been conserved throughout vertebrate evolution. In vertebrates, several recent studies showed that Isl1 is expressed transiently in the progenitor of the FHF during embryonic development, and some Isl1-derived cells can give rise to both FHF and SHF derivatives (Brade et al., 2007; Prall et al., 2007; Sun et al., 2007; Ma et al., 2008). Isl1 and Mesp1 gain-of-function studies at different times of ESC differentiation revealed that Isl1 cooperates with Mesp1 to promote cardiovascular differentiation. Isl1 promotes endothelial fate at the early step of MCP specification and stimulates cardiac differentiation during latter stages, and these effects were additive with those mediated by Mesp1, suggesting that Mesp1 and Isl1 cooperate to promote cardiovascular lineage commitment and control distinct transcriptional programs at different stages of cardiovascular differentiation. Consistent with the cardiac-promoting effect of late Isl1 overexpression, loss of Isl1 function in differentiating ESC inhibits cardiac differentiation (Kwon et al., 2009).
Our study provides novel insights into the cellular and transcriptional hierarchy that operates during the early step of cardiovascular progenitor specification and provides a means of isolating cardiovascular progenitors during ESC differentiation, increasing the generation of cardiac cells in vitro for cellular therapy or drug screening. Mesp1-GFP ESCs will be a powerful method to screen for new intrinsic and extrinsic regulators of cardiovascular progenitor specification and differentiation.
Materials and methods
Reporter ESC line
A 5.6-kb genomic fragment upstream of the Mesp1 translation start (Haraguchi et al., 2001) was amplified by PCR, sequence verified, and subcloned upstream of the Venus-GFP sequence in a PL451 vector (Liu et al., 2003). The construct was linearized and electroporated in ESCs. Resistant ES cell clones were selected with neomycin and screened for expression of the GFP during ESC differentiation. Bry-GFP ESC line generation and use were previously described elsewhere (provided by G. Keller, McEwen Center for Regenerative Medicine, Toronto, Ontario, Canada; Kattman et al., 2006).
Tetracycline-inducible ES cell lines
Isl1 ORF was amplified by PCR, sequence verified, and cloned in place of Mesp1–3×Flag in the p2LoxMesp1–3×Flag-IRES-EGFP vector (Bondue et al., 2008). Combined expression of Mesp1 and Isl1 in A2lox cells was obtained by generating cell lines containing two tetracycline operators in tandem in the p2Lox backbone (Kyba et al., 2002), introducing the Mesp1–3×Flag sequence after the first tetracycline operator and the Isl1 sequence after the second one. To generate the EN-Mesp1 construct, we performed a fusion protein between the first 298 amino acids of the repressor domain of Drosophila EN and Mesp1 ORF. All these constructs were electroporated in A2Lox cells, and stable cell lines were selected as previously described (Bondue et al., 2008).
Flow cytometry
Staining for cTNT, BrdU, and active caspase-3 was performed as previously described (Bondue et al., 2008). Flk1 (VEGFR2) was stained using a biotinylated antibody at 1:100 (clone Avas12a1; eBioscience) revealed by a streptavidin-phosphatidylethanolamine (PE)-Cy7 secondary antibody at 1:400 (BD). PDGFRa was stained using a PE- or an allophycocyanin-coupled rat monoclonal antibody at 1:75 (clone APA5; eBioscience). CXCR4 was stained using an A647-coupled rat monoclonal antibody at 1:100 (clone 2B11; eBioscience). CD31 expression was detected using a PE-coupled rat monoclonal antibody at 1:100 (clone MEC 13.3; BD). Living cells were gated by propidium iodide dye exclusion. FACS analyses were performed on a FACSCanto or a FACSCalibur device (BD), and isolation of the cells was performed using a cell sorter (FACSAria; BD).
ESC culture and differentiation
ESCs were cultured on irradiated mouse embryonic fibroblasts in DME supplemented with 15% ESC-qualified FBS (Invitrogen), 0.1 mM nonessential amino acids (Invitrogen), 1 mM sodium-pyruvate (Invitrogen), 0.1 mM β-mercaptoethanol (Sigma-Aldrich), 100 U/ml penicillin (Invitrogen), 100 µg/ml streptomycin (Invitrogen), and 1,000 U/ml leukemia inhibitory factor (ESGRO). ESC differentiation was performed in hanging drops of 1,000 cells in 25 µl as previously described (Bondue et al., 2008). To assess the cardiovascular potential of Mesp1-GFP and CXCR4/PDGFRa/Flk1 TP cells, ESCs were cultured for 3 d in hanging drops in differentiation medium consisting of the same medium without leukemia inhibitory factor but containing 15% of ESC-qualified serum (Invitrogen) and 0.5 mM ascorbic acid (Sigma-Aldrich; Bondue et al., 2008). At D3, dissociated cells were stained and sorted in HBSS containing 2% FBS, washed, and replated on gelatin-coated dishes in a serum-free medium based on StemPro-34 (Invitrogen) supplemented with 100 U/ml penicillin, 100 µg/ml streptomycin, 2 mM l-glutamine, 0.5 mM ascorbic acid (Sigma-Aldrich), 10 ng/ml basic FGF, 25 ng/ml FGF10, 5 ng/ml VEGF, 100 ng/ml PDGFRa, and 150 ng/ml hDKK1 (Kattman et al., 2006). All growth factors were purchased from R&D Systems. Medium was replaced on D5, D7, and D9 of differentiation. For low density culture assays, 50 isolated cells were replated in each well of an 8-well Lab-Tek glass chamber slide (Thermo Fisher Scientific) with Y-27632 (EMD) at a final concentration of 10 µM for the first 48 h. Dox-inducible ESC lines were differentiated in DME containing 15% ESC-qualified serum and 0.5 mM ascorbic acid (Sigma-Aldrich). After 4 d in hanging drops, ESCs were replated on gelatin-coated dishes for further differentiation. Dox (Sigma-Aldrich) was added to hanging drops at corresponding days to a final concentration of 1 µg/ml as previously described (Bondue et al., 2008).
Immunofluorescence analysis
Fixation, blocking, and primary and secondary antibodies as well as mounting medium used in this study were previously described (Bondue et al., 2008), except for the anti-GFP staining (rabbit polyclonal; 1:1,500; Invitrogen). Counterstaining of nuclei was performed with Hoechst (1:2,000; Invitrogen). Immunostaining was acquired using a microscope (Axio Observer.Z1), a camera (AxioCam MR3 or MRc5), and the Axiovision software (Carl Zeiss, Inc.). Acquisitions were performed at room temperature using 10 and 20× EC Plan Neofluar objectives (10× = 0.3 numerical aperture and 20× = 0.4 numerical aperture; Carl Zeiss, Inc.). Mosaics were generated by the Axiovision software using a 10% overlap between each single acquisition. Confocal pictures were acquired at room temperature using a multiphoton confocal microscope (LSM510 NLO; Carl Zeiss, Inc.) fitted on an inverted microscope (Axiovert M200; Carl Zeiss, Inc.) equipped with C-Apochromat (40× = 1.2 numerical aperture and 63× = 1.2 numerical aperture) water immersion objectives (Carl Zeiss, Inc.). 0.35-mm-thick, 512 × 512–pixel optical sections were collected sequentially for each fluorochrome. The datasets generated were merged and displayed with the LSM510 software and exported in TIF image format.
Live-sample imaging
8 d after cell isolation at D3 and replating on gelatin-coated dishes, beating areas were imaged by time-lapse bright-field acquisitions using a microscope (Axio Observer.Z1) and the Axiovision software. All acquisitions were performed at room temperature using a 10× EC Plan Neofluar objective (0.3 numerical aperture). Image sequences were compiled with the Axiovision software, and video files display 15 images/s.
RNA isolation, reverse transcription, quantitative PCR, and single-cell PCR
RNA extraction, DNase treatment, and RT-PCR were performed as previously described (Bondue et al., 2008). Quantitative PCR was performed using Brilliant II Fast SYBR qPCR Master Mix (Agilent Technologies) on a real-time PCR system (Mx3005P; Agilent Technologies). All primers were designed using Lasergene 7.2 software (DNAStar, Inc.) and are listed in Table S2. Single-cell PCR, generation of cDNA, and PCR amplification were performed as previously described (Jensen and Watt, 2006). In brief, after cDNA synthesis, two rounds of 35 cycles of amplifications were performed by PCR, and the amplification product was used as a PCR template for the detection of gene expression. For single-cell PCR experiments, CXCR4/PDGFRa/Flk1 TP cells were sorted directly in 96-well plates containing the first strand buffer. The cDNA amplification procedure was used for the expression profiling of colonies obtained after differentiation of single Mesp1-GFP cells sorted in 96 wells.
Microarray analysis
For microarray analysis, Mesp1-GFP cells were sorted at D3 directly in 350 µl lysis buffer of the Absolutely microRNA kit (Agilent Technologies). RNA isolation and microarray analysis were performed in two biologically independent replicates as previously described (Bondue et al., 2008) using mouse genome 430 2.0 arrays (Affymetrix). To compare Mesp1-GFP and Mesp1 overexpression experiments, we plotted the distribution of the fold change of the probes that are significantly differentially expressed in the two experiments (fold change >1.5), representing a total of 1,425 probes. Distributions were compared using χ2 test.
Online supplemental material
Fig. S1 shows the differentiation potential of Mesp1-GFP and Flk1/PDGFRa/CXCR4 TP cells. Fig. S2 compares the fold change of probes affected in Mesp1 gain-of-function and Mesp1-GFP experiments. Fig. S3 shows the expression of Mesp1 in a subpopulation of Bry/Flk1-expressing cells. Fig. S4 characterizes inducible gene expression in Mesp1, Isl1, and Mesp1/Isl1 ESCs. Video 1 shows beating areas in differentiated Mesp1-expressing cells. Video 2 displays beating areas in differentiated Mesp1-negative cells. Video 3 shows beating areas in differentiated all sorted cells. Table S1 displays the functional annotation chart of Mesp1-enriched genes. Table S2 shows primers used for RT-PCR and single-cell PCR.
Acknowledgments
We thank M. Buckingham and members of the Blanpain and Vanderhaeghen laboratories for their suggestions during the realization of this study. We thank Gordon Keller for his comments on the manuscript and providing Bry-GFP ESCs. We thank Kim Jensen for providing technical advices on single-cell PCR.
C. Blanpain and A. Bondue are, respectively, chercheur qualifié and chargé de recherche of the Belgian Fonds National de la Recherche Scientifique. M. Ramialison is funded by a European Molecular Biology Organization (EMBO) long-term fellowship. This work was supported by a career development award of the Human Frontier Science Program Organization, a research grant from the Schlumberger Foundation, the program CIBLES of the Wallonia Region, a research grant from the Fondation Contre le Cancer and the Fond Gaston Ithier, a starting grant of the European Research Council, and the EMBO Young Investigator Program.
References
- Bry
Brachyury
- CM
cardiomyocyte
- cTNT
cardiac troponin T
- Dox
doxycyclin
- EC
endothelial cell
- EMT
epithelial to mesenchymal transition
- EN
Engrailed
- ESC
embryonic stem cell
- FHF
first heart field
- MCP
multipotent cardiovascular progenitor
- PE
phosphatidylethanolamine
- SHF
second heart field
- SMA
smooth muscle actin
- SMC
smooth muscle cell
- TP
triple positive
- VE
vascular endothelial
Author notes
A. Bondue, S. Tännler, and G. Chiapparo contributed equally to this paper.


![Figure 1. Engineering ESCs expressing Venus-GFP under the regulatory region of Mesp1. (A) Schematic representation of the Mesp1 reporter transgene. Venus-GFP is cloned under the regulatory sequences of Mesp1 that allowed transgene expression in the cardiogenic mesoderm. (B, right) Detection of GFP in Mesp1-GFP ESCs at D3 of differentiation. (left) Unmodified ESCs at the same day of differentiation are used as a control. Bars, 50 µm. (C and D) Kinetics of Mesp1 mRNA expression measured by RT–quantitative PCR (C), and Mesp1-GFP expression as detected by FACS (D). Results are normalized for Mesp1 expression in undifferentiated ESCs (C) or represent the percentage of Mesp1-GFP–positive cells (D). (E) Relative expression of Mesp1 and GFP transcripts in Mesp1-GFP–expressing cells (GFP positive [pos]) and in Mesp1-GFP–nonexpressing cells (GFP negative [neg]) isolated by FACS at D3. Results are normalized for the expression of the transcripts in all sorted cells (gray bars). Error bars indicate means ± SEM; n = 3. FRT, flippase recognition target. pA, polyadenylation. PGK, phosphoglycerate kinase.](https://cdn.rupress.org/rup/content_public/journal/jcb/192/5/10.1083_jcb.201007063/6/m_jcb_201007063_rgb_fig1.jpeg?Expires=1773774360&Signature=zllBF-pdEZqjCa6daOsGeiIUwq6NyYH8DUKfbNsmDOLzxcWxkYmH1bD99eyHZdgTKIkkfCDLpQYBSQGToJ~a5Fb2VKJ7ApobtfzXFjPqkC7G1DYKiZmjQn-ol779dksNY9XMqo6DDeAFjalSNfT5cpqitUAsRv2qmnY11LDCaTAO7t4jwLVthqUw4HWAIu1j8uTk5DFB0Nw9pj2-nWFtb4EQwTmiIsWKqGo42SFe27QLAjzwH-sjfSt8BXS47QjV-SbtNuVl-SRI~bCAyHhjVk-ZDjtfPGaKJRdhReeH45iO7x4ilfmFeIRAvSQq254YWUxCrdUeDOUXnXhT2dAwyg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)