Vascular endothelial growth factors (VEGFs) and their receptors (VEGFRs) are quintessential for the development and maintenance of blood and lymphatic vessels. However, genetic interactions between the VEGFRs are poorly understood. VEGFR2 is the dominant receptor that is required for the growth and survival of the endothelium, whereas deletion of VEGFR1 or VEGFR3 was reported to induce vasculature overgrowth. Here we show that vascular regression induced by VEGFR2 deletion in postnatal and adult mice is aggravated by additional deletion of VEGFR1 or VEGFR3 in the intestine, kidney, and pancreas, but not in the liver or kidney glomeruli. In the adult mice, hepatic and intestinal vessels regressed within a few days after gene deletion, whereas vessels in skin and retina remained stable for at least four weeks. Our results show changes in endothelial transcriptomes and organ-specific vessel maintenance mechanisms that are dependent on VEGFR signaling pathways and reveal previously unknown functions of VEGFR1 and VEGFR3 in endothelial cells.
Introduction
Blood vessels have specific organotypic features that allow them to carry out vascular bed–specific functions (Augustin and Koh, 2017; Potente and Mäkinen, 2017). Angiogenesis, the formation of blood vessels from preexisting ones, is vital for tissue development and organ function, but also occurs in pathological processes such as cardiovascular diseases, cancer, and inflammation (Carmeliet and Jain, 2011). Vascular endothelial growth factors (VEGFs) are the principal drivers of angiogenesis and lymphangiogenesis via binding to their tyrosine kinase receptors (VEGFRs) to elicit various downstream effects in endothelial cells (ECs).
VEGF signaling in blood vascular ECs is mediated predominantly via activation of VEGFR2. Autocrine signaling of VEGF was shown to be essential for the maintenance of quiescent vasculature in adult mice by mediating EC survival in a cell-autonomous way (Lee et al., 2007). VEGF’s binding affinity to VEGFR1 is ∼10-fold higher than to VEGFR2, but VEGFR1 kinase and signaling activity in ECs is low (Gille et al., 2000; Waltenberger et al., 1994). According to current understanding, VEGFR1 and its soluble isoform sVEGFR1 rather act as decoy receptors for VEGF by sequestering it from interacting with VEGFR2. Embryos with a targeted inactivation of Vegfr1 gene die at embryonic day (E) 8.5 due to abnormal blood vasculature involving an excess of ECs, whereas mice lacking only the VEGFR1 kinase domain develop normal vasculature and survive (Fong et al., 1995; Hiratsuka et al., 1998). Conditional global deletion of Vegfr1 in neonatal or adult mice promotes EC proliferation by increasing both the availability of VEGF to VEGFR2 and the expression of VEGFR2 on ECs (Ho et al., 2012). Interestingly, VEGF administration can also induce hepatocyte growth factor secretion by liver sinusoidal ECs independently of angiogenesis (LeCouter et al., 2003), thus contributing to angiocrine signals for organ growth and maintenance (Rafii et al., 2016).
Despite having an order of magnitude lower affinity for VEGF binding than VEGFR1, VEGFR2 exhibits a much stronger VEGF-induced tyrosine kinase activity than VEGFR1. This is why VEGFR2 is considered to be the major VEGF signaling receptor. VEGFR2 is one of the earliest markers of endothelial progenitors, but it has been shown to be expressed by hematopoietic, cardiomyocyte, and hepatic progenitor cells as well (Chung et al., 2002; Goldman et al., 2013; Kattman et al., 2006; Millauer et al., 1993). VEGFR2 levels are higher in angiogenic vessels than in quiescent vascular beds, for example during development or wound healing (Ehling et al., 2013; Zhang et al., 2004), and VEGFR2 is also expressed in lymphatic endothelium (Secker and Harvey, 2015). VEGFR2-null mice die between E8.5 and E9.5 due to severe defects in the development of endothelial and hematopoietic cells (Shalaby et al., 1995). Conditional somatic deletion of VEGFR2 in neonatal and adult mice caused stunted angiogenesis in pups and vascular regression in the liver, kidney glomeruli, and thyroid in adult mice (Gerber et al., 1998; Jang et al., 2017; Sison et al., 2010). However, even very low amounts of VEGFR2 were enough to sustain angiogenesis in the developing mouse retina (Zarkada et al., 2015).
VEGFR3, which binds VEGF-C and VEGF-D, is the main receptor that drives lymphangiogenesis (Alitalo, 2011). Its expression starts at E8.5 in the cardinal vein and the angioblasts of the head mesenchyme, and is later increased in the developing veins and emerging lymphatic ECs (E11.5–E12.5), after which VEGFR3 is predominantly expressed by the lymphatic ECs and fenestrated and sinusoidal blood vascular ECs (Karaman et al., 2017). VEGFR3-null embryos die at E10.5, before the lymphatic vessels develop, due to defective arterio-venous remodeling of the primary vascular plexus (Dumont et al., 1998; Hamada et al., 2000). Conditional deletion of VEGFR3 in neonatal mice or blockade of VEGF-C/VEGFR3 signaling with a soluble form of the receptor selectively arrests lymphatic vessel growth (Haiko et al., 2008; Karpanen et al., 2006; Mäkinen et al., 2001). Interestingly, VEGFR3 deletion led to hypersprouting of growing blood vessels via disruption of Notch signaling in postnatal mice (Tammela et al., 2011; Tammela et al., 2008) and increased vascular permeability via up-regulation of VEGFR2 in adult mice (Heinolainen et al., 2017).
Although the effects of individual deletions of VEGFRs are already well-documented, little is known about their roles and interplay in promoting the essential functions in ECs. We found that single deletion of each VEGFR or their combinations led to organ-specific phenotypes in which vascular development and vessel maintenance are differently affected. We also found that VEGFR1 and VEGFR3 can support EC survival in the absence of VEGFR2 in a vascular bed–specific manner and that the combined deletion of VEGFR1 and VEGFR3 hyperactivates VEGFR2 signaling, resulting in organotypic differences in developmental angiogenesis.
Results
Endothelial VEGFR1 deletion increases vascular density
We first established mice in which VEGFRs can be deleted selectively in the ECs with high efficiency. To do so, we crossed Vegfr1flox/flox, Vegfr2flox/flox, and Vegfr3flox/flox mice with vascular endothelial cadherin (VE-cadherin)–CreERT2 mice and injected 4-hydroxy-tamoxifen (4-OHT) between postnatal days (P)7 and P9 to generate single, double, and triple compound deletions of VEGFRs in the vasculature. Considering that Cre-mediated gene deletion efficacy may vary between different tissues, we first analyzed the VEGFR levels in various tissues that were harvested at P10 to assess gene function without having secondary effects. After deletion, liver, kidney, pancreas, skin, lung, and heart lysates showed only 1–13% of remaining Vegfr1, Vegfr2, and Vegfr3 mRNAs. Only slightly more Vegfr1 mRNA (19%) remained in the postnatal pancreas, skin, and lungs, which could be due to Vegfr1 expression by macrophages (Hiratsuka et al., 1998; Fig. 1 A).
Endothelial VEGFR2 deletion causes widespread vessel regression in postnatal mice.(A) Quantification of Vegfr transcripts in different tissues after 3 d of deletion in R1R2R3iΔEC pups and littermate controls (n = control [CTRL] = 5, R1R2R3iΔEC = 8 mice pooled from three independent experiments were used for the data shown). Vegfrs are normalized to 36b4 and Cdh5 transcripts. (B) Experimental setup. (C) Pup weights on P10. (D) Spleen and heart weights normalized to body weight. (E) Quantification of Vegfr transcripts normalized to 36b4 and Cdh5 transcripts; and Cdh5 and Pecam1 transcripts normalized to 36b4 transcripts. (F) Western blots and quantifications of VEGFRs normalized to HSC70 and VE-Cadherin (VEC) in lung lysates. (G) Staining and quantification of blood vessels (EMCN, red) in kidneys and glomeruli (insets). Scale bar = 100 µm, inset = 20 µm. (H) Staining and quantification of blood vessels (PODXL, red) in the liver and (I) pancreas; dashed lines mark Langerhans islets. (J) Intestinal whole-mounts stained for blood (CD31, green) and lymphatic (LYVE1, red) vessels and their quantification. (K) EC staining with isolectin B4 (iB4, white) in retinal whole-mounts and their quantification. A, arteries; V, veins. Only the superficial plexus is shown. Scale bar = 500 µm, inset = 200 µm. (L) Tracheal whole-mount staining and quantification of blood and lymphatic vessels (CD31, green; LYVE1, white). EMCN, endomucin; PODXL, podocalyxin. Error bars = mean ± SEM (n = control [CTRL] = 19, R2iΔEC = 14 mice pooled from four independent experiments were used for the data shown in the panels). Statistical significance was determined using Student’s t test (two-tailed, unpaired). *, P < 0.05; **, P < 0.01; ***, P < 0.001. BV, blood vessel; LV, lymphatic vessel. Source data are available for this figure: SourceData F1.
Endothelial VEGFR2 deletion causes widespread vessel regression in postnatal mice.(A) Quantification of Vegfr transcripts in different tissues after 3 d of deletion in R1R2R3iΔEC pups and littermate controls (n = control [CTRL] = 5, R1R2R3iΔEC = 8 mice pooled from three independent experiments were used for the data shown). Vegfrs are normalized to 36b4 and Cdh5 transcripts. (B) Experimental setup. (C) Pup weights on P10. (D) Spleen and heart weights normalized to body weight. (E) Quantification of Vegfr transcripts normalized to 36b4 and Cdh5 transcripts; and Cdh5 and Pecam1 transcripts normalized to 36b4 transcripts. (F) Western blots and quantifications of VEGFRs normalized to HSC70 and VE-Cadherin (VEC) in lung lysates. (G) Staining and quantification of blood vessels (EMCN, red) in kidneys and glomeruli (insets). Scale bar = 100 µm, inset = 20 µm. (H) Staining and quantification of blood vessels (PODXL, red) in the liver and (I) pancreas; dashed lines mark Langerhans islets. (J) Intestinal whole-mounts stained for blood (CD31, green) and lymphatic (LYVE1, red) vessels and their quantification. (K) EC staining with isolectin B4 (iB4, white) in retinal whole-mounts and their quantification. A, arteries; V, veins. Only the superficial plexus is shown. Scale bar = 500 µm, inset = 200 µm. (L) Tracheal whole-mount staining and quantification of blood and lymphatic vessels (CD31, green; LYVE1, white). EMCN, endomucin; PODXL, podocalyxin. Error bars = mean ± SEM (n = control [CTRL] = 19, R2iΔEC = 14 mice pooled from four independent experiments were used for the data shown in the panels). Statistical significance was determined using Student’s t test (two-tailed, unpaired). *, P < 0.05; **, P < 0.01; ***, P < 0.001. BV, blood vessel; LV, lymphatic vessel. Source data are available for this figure: SourceData F1.
Three tamoxifen injections into VE-cadherin–CreERT2;Vegfr1flox/flox mice (R1iΔEC) between P7 and P9 resulted in almost complete deletion of the Vegfr1 mRNA and protein in the lungs when analyzed at P10 (8 and 9% remaining, respectively; Data S1). This did not affect the weight of the pups at P10, but the relative heart weight was increased (Fig. S1, A–C). VEGFR2 and VEGFR3 protein levels in the lungs were significantly elevated, whereas VE-cadherin mRNA and protein levels were unaltered (Fig. S1, D and E). VEGFR1 deletion resulted in increased vascular density in most of the tissues analyzed (Fig. S1, F–K), most prominently in the kidney, pancreas, and intestine (Fig. S1 F, H, and I), whereas the liver vasculature was not affected (Fig. S1 G). Tracheal platelet and EC adhesion molecule 1 (CD31 or PECAM1)+ blood vessel area was significantly increased in the R1iΔEC mice, but lymphatic vessel endothelial hyaluronan receptor 1 (LYVE1)+ lymphatic vessels were not affected (Fig. S1 K). Isolectin-B4 (iB4)+ EC area was increased in the superficial vascular plexus of the retina, but the number of branch points per vessel area was not changed (Fig. S1 J).
Vascular phenotypes in postnatal mice after VEGFR1 or VEGFR3 deletion.(A and L) Experimental setup. (B and M) Pup weights at P10. (C and N) Spleen and heart weights normalized to body weight. (D and O) Quantification of Vegfr transcripts normalized to 36b4 and Cdh5 transcripts; and Cdh5 and Pecam1 transcripts normalized to 36b4 transcripts. (E and P) Western blots and quantification of VEGFR bands normalized to HSC70 and VE-cadherin (VEC) bands in lung lysates. (F and Q) Staining and quantification of blood vessels (EMCN in red) in kidneys and their glomeruli (insets). Scale bar = 100 µm, inset = 20 µm. (G and R) Staining and quantification of blood vessels (PDXL staining in red) in the liver and (H and S) pancreas; the dashed lines mark Langerhans islets (scale bars = 100 µm). (I and T) Intestinal whole-mounts stained for blood (CD31, green) and lymphatic (LYVE1, red) vessels and their quantification (scale bar = 200 µm). (J and U) EC staining with isolectin B4 (iB4, white) in retinal whole-mounts and their quantification. A, arteries; V, veins. Only the superficial plexus is shown (scale bars = 500 µm, insets = 200 µm). (K and V) Tracheal whole-mount staining and quantification of blood and lymphatic vessels (CD31, green; LYVE1, white; scale bar = 200 µm). EMCN: endomucin; PODXL: podocalyxin. Error bars = mean ± SEM. Control (CTRL) = 25, R1iΔEC = 20 mice pooled from six independent experiments were used for the VEGFR1 deletions. CTRL = 18, R3iΔEC = 19 mice pooled from five independent experiments were used for the VEGFR3 deletions. Statistical significance was determined using Student’s t test (two-tailed, unpaired) with Welch’s correction in case of unequal variances. *, P < 0.05; **, P < 0.01; ***, P < 0.001. BV, blood vessel; LV, lymphatic vessel. Source data are available for this figure: SourceData FS1.
Vascular phenotypes in postnatal mice after VEGFR1 or VEGFR3 deletion.(A and L) Experimental setup. (B and M) Pup weights at P10. (C and N) Spleen and heart weights normalized to body weight. (D and O) Quantification of Vegfr transcripts normalized to 36b4 and Cdh5 transcripts; and Cdh5 and Pecam1 transcripts normalized to 36b4 transcripts. (E and P) Western blots and quantification of VEGFR bands normalized to HSC70 and VE-cadherin (VEC) bands in lung lysates. (F and Q) Staining and quantification of blood vessels (EMCN in red) in kidneys and their glomeruli (insets). Scale bar = 100 µm, inset = 20 µm. (G and R) Staining and quantification of blood vessels (PDXL staining in red) in the liver and (H and S) pancreas; the dashed lines mark Langerhans islets (scale bars = 100 µm). (I and T) Intestinal whole-mounts stained for blood (CD31, green) and lymphatic (LYVE1, red) vessels and their quantification (scale bar = 200 µm). (J and U) EC staining with isolectin B4 (iB4, white) in retinal whole-mounts and their quantification. A, arteries; V, veins. Only the superficial plexus is shown (scale bars = 500 µm, insets = 200 µm). (K and V) Tracheal whole-mount staining and quantification of blood and lymphatic vessels (CD31, green; LYVE1, white; scale bar = 200 µm). EMCN: endomucin; PODXL: podocalyxin. Error bars = mean ± SEM. Control (CTRL) = 25, R1iΔEC = 20 mice pooled from six independent experiments were used for the VEGFR1 deletions. CTRL = 18, R3iΔEC = 19 mice pooled from five independent experiments were used for the VEGFR3 deletions. Statistical significance was determined using Student’s t test (two-tailed, unpaired) with Welch’s correction in case of unequal variances. *, P < 0.05; **, P < 0.01; ***, P < 0.001. BV, blood vessel; LV, lymphatic vessel. Source data are available for this figure: SourceData FS1.
Endothelial VEGFR2 deletion results in widespread blood vessel regression
In contrast with VEGFR1 deletion, deletion of VEGFR2 (R2iΔEC) in the pups resulted in reduced body weight, whereas spleen weight was significantly increased (Fig. 1, B–D). VEGFR1 and VEGFR3 mRNA and protein levels were decreased in the lungs (Fig. 1, E and F; and Data S1), concomitantly with a rapid and significant loss of vasculature in all studied tissues at P10. Trachea, kidney glomeruli, liver, retina, pancreas, and its Langerhans islets were the most affected tissues, showing 79%, 69%, 63%, 52%, 50%, and 65%, reduction in blood vessel density, respectively (Fig. 1, G–I, K, and L). Yet the intestine showed only a 24% decrease in vascular density (Fig. 1 J). The intestinal absorptive lymphatic vessels (lacteals) were not affected in the R2iΔEC mice, whereas the tracheal lymphatic vessel area was significantly reduced, indicating organ specificity of VEGFR2 function in the lymphatic vasculature (Fig. 1, J and L).
Endothelial VEGFR3 deletion leads to hypersprouting of retinal blood vessels and regression of lymphatic vessels
Endothelial Vegfr3 deletion (R3iΔEC) had no effect on body weight, but it increased spleen weight significantly (Fig. S1, L–N). The R3iΔEC pups showed less Vegfr1 mRNA and more VEGFR2 protein in lung lysates (Benedito et al., 2012; Tammela et al., 2011), suggesting increased VEGF/VEGFR2 signaling capacity (Fig. S1, O and P). Interestingly, however, the deletion of Vegfr3 between P7 and P9 did not result in significant changes in blood vessel density in the tissues analyzed at P10, except that it induced hypersprouting in the superficial and deep retinal vascular plexuses (Tammela et al., 2011; Fig. S1, Q–V). The R3iΔEC mice also showed a decreased lymphatic vessel area in the intestine and trachea (Fig. S1, T and V). These results confirmed that VEGFR1 and VEGFR3 attenuate angiogenesis mediated by VEGFR2 in the developing control pups.
Additional deletion of VEGFR1 or VEGFR3 aggravates vessel regression in R2iΔEC mice
Next, we investigated the phenotypes of the compound deletions that included VEGFR2. Upon combined endothelial deletion of VEGFR1 and VEGFR2 (R1R2iΔEC), mouse body weight was reduced, spleen weight was increased, and heart weight was not affected (Fig. 2 A–E). These results confirmed that the cardiac hypertrophy induced by endothelial Vegfr1 deletion is, indeed, mediated by Vegfr2 (Kivelä et al., 2019). VEGFR1 has been considered a decoy receptor because deletion of its intracellular part including its tyrosine kinase domain shows no phenotype, whereas complete deletion of VEGFR1 leads to increased vascular density (Fong et al., 1995; Hiratsuka et al., 1998). However, instead of increased vascular density, the R1R2iΔEC mice showed more extensive blood vessel regression than the R2iΔEC mice, especially in the intestine (36% versus 24%), kidney (42% versus 30%), retina (60% versus 52%), and trachea (88% versus 79%; Fig. 2, F–K). Analysis of the remaining VEGFR2 protein levels in total lung lysates indicated that the Vegfr2 deletion efficacy was not responsible for this difference (Data S1).
VEGFR1 deletion aggravates vessel regression induced by VEGFR2 loss.(A) Experimental setup. (B) Pup weights on P10. (C) Spleen and heart weights normalized to body weight. (D) Quantification of Vegfr transcripts normalized to 36b4 and Cdh5 transcripts; and Cdh5 and Pecam1 transcripts normalized to 36b4 transcripts. (E) Western blots and quantifications of VEGFRs normalized to HSC70 and VE-Cadherin (VEC) in lung lysates. (F) Staining and quantification of blood vessels (EMCN, red) in kidneys and glomeruli (insets). Scale bar = 100 µm, inset = 20 µm. (G) Staining and quantification of blood vessels (PDXL, red) in the liver and (H) pancreas; dashed lines mark Langerhans islets. (I) Intestinal whole-mounts stained for blood (CD31, green) and lymphatic (LYVE1, red) vessels and their quantification. (J) EC staining with isolectin B4 (iB4, white) in retinal whole-mounts and their quantification. A, arteries; V, veins. Only the superficial plexus is shown. Scale bar = 500 µm, inset = 200 µm. (K) Tracheal whole-mount staining and quantification of blood and lymphatic vessels (CD31, green; LYVE1, white). EMCN, endomucin; PODXL, podocalyxin. Error bars = mean ± SEM (n = control [CTRL] = 21, R1R2iΔEC = 17 mice pooled from five independent experiments were used for the data shown in the panels). Statistical significance was determined using Student’s t test (two-tailed, unpaired). *, P < 0.05; **, P < 0.01; ***, P < 0.001. BV, blood vessel; LV, lymphatic vessel. Source data are available for this figure: SourceData F2.
VEGFR1 deletion aggravates vessel regression induced by VEGFR2 loss.(A) Experimental setup. (B) Pup weights on P10. (C) Spleen and heart weights normalized to body weight. (D) Quantification of Vegfr transcripts normalized to 36b4 and Cdh5 transcripts; and Cdh5 and Pecam1 transcripts normalized to 36b4 transcripts. (E) Western blots and quantifications of VEGFRs normalized to HSC70 and VE-Cadherin (VEC) in lung lysates. (F) Staining and quantification of blood vessels (EMCN, red) in kidneys and glomeruli (insets). Scale bar = 100 µm, inset = 20 µm. (G) Staining and quantification of blood vessels (PDXL, red) in the liver and (H) pancreas; dashed lines mark Langerhans islets. (I) Intestinal whole-mounts stained for blood (CD31, green) and lymphatic (LYVE1, red) vessels and their quantification. (J) EC staining with isolectin B4 (iB4, white) in retinal whole-mounts and their quantification. A, arteries; V, veins. Only the superficial plexus is shown. Scale bar = 500 µm, inset = 200 µm. (K) Tracheal whole-mount staining and quantification of blood and lymphatic vessels (CD31, green; LYVE1, white). EMCN, endomucin; PODXL, podocalyxin. Error bars = mean ± SEM (n = control [CTRL] = 21, R1R2iΔEC = 17 mice pooled from five independent experiments were used for the data shown in the panels). Statistical significance was determined using Student’s t test (two-tailed, unpaired). *, P < 0.05; **, P < 0.01; ***, P < 0.001. BV, blood vessel; LV, lymphatic vessel. Source data are available for this figure: SourceData F2.
We next analyzed the combined deletion of VEGFR2 and VEGFR3 (R2R3iΔEC; Fig. S2, A–K) or all three VEGFRs (R1R2R3iΔEC; Fig. 3, A–K). Both of these compound deletions resulted in stronger vascular regression than observed in the R2iΔEC mice, for example, in the intestine and the superficial retinal plexus (Fig. S2, A–K; and Fig. 3, A–K). These results indicated that even though VEGFR1 and VEGFR3 attenuate VEGFR2 signaling in blood vessels, both VEGFR1 and VEGFR3 can support EC survival in the absence of VEGFR2.
Vascular phenotypes in postnatal mice after VEGFR2;VEGFR3 deletion or VEGFR1;VEGFR3 deletion. (A and L) Experimental setup. (B and M) Pup weights at P10. (C and N) Spleen and heart weights normalized to body weight. (D and O) Quantification of Vegfr transcripts. (E and P) Western blots and quantifications of VEGFR bands in lung lysates. (F and Q) Staining and quantification of blood vessels (EMCN in red) in kidneys and glomeruli (insets). Scale bar = 100 µm, inset = 20 µm. (G and R) Staining and quantification of blood vessels (PDXL in red) in the liver and (H and S) pancreas; the dashed lines mark Langerhans islets (scale bars = 100 µm). (I and T) Intestinal whole-mounts stained for blood (CD31, green) and lymphatic (LYVE1, red) vessels and their quantification (scale bar = 200 µm). (J and U) EC staining with isolectin B4 (iB4, white) in retinal whole-mounts and their quantification. A, arteries; V, veins. Only the superficial plexus is shown (scale bars = 500 µm, insets = 200 µm). (K and V) Tracheal whole-mount staining and quantification of blood and lymphatic vessels (CD31, green; LYVE1, white, scale bar = 200 µm). EMCN: endomucin; PODXL: podocalyxin. Error bars = mean ± SEM. Control (CTRL) = 16, R2R3iΔEC = 12 mice pooled from four independent experiments were used for the VEGFR2;VEGFR3 deletions. CTRL = 13, R1R3iΔEC = 11 mice pooled from three independent experiments were used for the VEGFR1;VEGFR3 deletions. Statistical significance was determined using Student’s t test (two-tailed, unpaired) with Welch’s correction in case of unequal variances. *, P < 0.05; **, P < 0.01; ***, P < 0.001. BV, blood vessel; LV, lymphatic vessel. Source data are available for this figure: SourceData FS2.
Vascular phenotypes in postnatal mice after VEGFR2;VEGFR3 deletion or VEGFR1;VEGFR3 deletion. (A and L) Experimental setup. (B and M) Pup weights at P10. (C and N) Spleen and heart weights normalized to body weight. (D and O) Quantification of Vegfr transcripts. (E and P) Western blots and quantifications of VEGFR bands in lung lysates. (F and Q) Staining and quantification of blood vessels (EMCN in red) in kidneys and glomeruli (insets). Scale bar = 100 µm, inset = 20 µm. (G and R) Staining and quantification of blood vessels (PDXL in red) in the liver and (H and S) pancreas; the dashed lines mark Langerhans islets (scale bars = 100 µm). (I and T) Intestinal whole-mounts stained for blood (CD31, green) and lymphatic (LYVE1, red) vessels and their quantification (scale bar = 200 µm). (J and U) EC staining with isolectin B4 (iB4, white) in retinal whole-mounts and their quantification. A, arteries; V, veins. Only the superficial plexus is shown (scale bars = 500 µm, insets = 200 µm). (K and V) Tracheal whole-mount staining and quantification of blood and lymphatic vessels (CD31, green; LYVE1, white, scale bar = 200 µm). EMCN: endomucin; PODXL: podocalyxin. Error bars = mean ± SEM. Control (CTRL) = 16, R2R3iΔEC = 12 mice pooled from four independent experiments were used for the VEGFR2;VEGFR3 deletions. CTRL = 13, R1R3iΔEC = 11 mice pooled from three independent experiments were used for the VEGFR1;VEGFR3 deletions. Statistical significance was determined using Student’s t test (two-tailed, unpaired) with Welch’s correction in case of unequal variances. *, P < 0.05; **, P < 0.01; ***, P < 0.001. BV, blood vessel; LV, lymphatic vessel. Source data are available for this figure: SourceData FS2.
VEGFR1/2/3 compound deletion results in the most severe regression of blood and lymphatic vessels in postnatal mice.(A) Experimental setup. (B) Pup weights on P10. (C) Spleen and heart weights normalized to body weight. (D) Quantification of Vegfr transcripts normalized to 36b4 and Cdh5 transcripts; and Cdh5 and Pecam1 transcripts normalized to 36b4 transcripts. (E) Western blots and quantifications of VEGFRs normalized to HSC70 and VE-cadherin (VEC) in lung lysates. (F) Staining and quantification of blood vessels (EMCN, red) in kidneys and glomeruli (insets). Scale bar = 100 µm, inset = 20 µm. (G) Staining and quantification of blood vessels (PDXL, red) in the liver and (H) pancreas; dashed lines mark Langerhans islets. (I) Intestinal whole-mounts stained for blood (CD31, green) and lymphatic (LYVE1, red) vessels and their quantification. (J) EC staining with isolectin B4 (iB4, white) in retinal whole-mounts and their quantification. A = arteries, V = veins. Only the superficial plexus is shown. Scale bar = 500 µm, inset = 200 µm. (K) Tracheal whole-mount staining and quantification of blood and lymphatic vessels (CD31, green; LYVE1, white). EMCN: endomucin; PODXL: podocalyxin. Error bars = mean ± SEM (n = control [CTRL] = 21, R1R2R3iΔEC = 20 mice pooled from seven independent experiments were used for the data shown in the panels). Statistical significance was determined using Student’s t test (two-tailed, unpaired). *, P < 0.05; ***, P < 0.001. BV, blood vessel; LV, lymphatic vessel. Source data are available for this figure: SourceData F3.
VEGFR1/2/3 compound deletion results in the most severe regression of blood and lymphatic vessels in postnatal mice.(A) Experimental setup. (B) Pup weights on P10. (C) Spleen and heart weights normalized to body weight. (D) Quantification of Vegfr transcripts normalized to 36b4 and Cdh5 transcripts; and Cdh5 and Pecam1 transcripts normalized to 36b4 transcripts. (E) Western blots and quantifications of VEGFRs normalized to HSC70 and VE-cadherin (VEC) in lung lysates. (F) Staining and quantification of blood vessels (EMCN, red) in kidneys and glomeruli (insets). Scale bar = 100 µm, inset = 20 µm. (G) Staining and quantification of blood vessels (PDXL, red) in the liver and (H) pancreas; dashed lines mark Langerhans islets. (I) Intestinal whole-mounts stained for blood (CD31, green) and lymphatic (LYVE1, red) vessels and their quantification. (J) EC staining with isolectin B4 (iB4, white) in retinal whole-mounts and their quantification. A = arteries, V = veins. Only the superficial plexus is shown. Scale bar = 500 µm, inset = 200 µm. (K) Tracheal whole-mount staining and quantification of blood and lymphatic vessels (CD31, green; LYVE1, white). EMCN: endomucin; PODXL: podocalyxin. Error bars = mean ± SEM (n = control [CTRL] = 21, R1R2R3iΔEC = 20 mice pooled from seven independent experiments were used for the data shown in the panels). Statistical significance was determined using Student’s t test (two-tailed, unpaired). *, P < 0.05; ***, P < 0.001. BV, blood vessel; LV, lymphatic vessel. Source data are available for this figure: SourceData F3.
The tracheal lymphatic vessel area was reduced by 31% and 23% in the R2iΔEC and R3iΔEC mice, respectively (Fig. 1 L, and Fig. S1 V), but by 62% in the R2R3iΔEC and 63% in R1R2R3iΔEC mice, indicating that both VEGFR2 and VEGFR3 support lymphatic vessel survival as well (Fig. S2 K and Fig. 3 K). On the other hand, the R1R2iΔEC mice showed a 28% decrease in tracheal lymphatic vessel area, suggesting that VEGFR1 deletion does not contribute to the lymphatic phenotype (Fig. 2 K). Unlike the tracheal lymphatic vessels, the lacteal vessels in the gut villi regressed only when VEGFR3 was deleted, yet they were enlarged in the R1R2iΔEC mice (Fig. 2 I).
Compound deletion of Vegfr1 and Vegfr3 induces global angiogenesis
We expected that a combined deletion of VEGFR1 and VEGFR3 (R1R3iΔEC) would result in a dramatic hypersprouting phenotype, as it should boost VEGF concentration in tissue fluids and VEGFR2 protein expression in ECs. Consistent with the results from VEGFR1 deletion, we found increased heart weights in the R1R3iΔEC pups, but no changes in body weight (Fig. S2, L–N). The combined deletion of VEGFR1 and VEGFR3 led to increased VE-cadherin (Cdh5) transcripts and VEGFR2 protein levels in the lungs (Fig. S2, O and P; and Data S1). This correlated with a higher vascular density in mice deleted of both Vegfr1 and Vegfr3 than in mice deleted only of Vegfr1 or Vegfr3, for example, in the kidney glomeruli, liver, Langerhans islets, intestine, and trachea (Fig. S2, Q–T and V). Surprisingly, the retina in the R1R3iΔEC pups showed a dual phenotype. In the retinal angiogenic front, the hypersprouting phenotype seen in R3iΔEC pups was normalized by the additional deletion of VEGFR1 (Fig. S1 U and Fig. S2 U). In contrast, in the more mature vessels proximal to the optic nerve, we found an increase of vessel area and branching (Fig. S2 U). Thus the retina is the only tissue analyzed that had less angiogenesis in R1R3iΔEC mice as compared with the single deletions of VEGFR1 and VEGFR3, suggesting that VEGFR1 and VEGFR3 attenuate VEGFR2-mediated angiogenesis in an additive manner in other tissues.
EC apoptosis contributes to vessel regression
To gain further insight into how vessel regression occurs in mice with Vegfr2 single and compound deletions, we injected 4-OHT into R1R2R3iΔEC and littermate pups between P7 and P9 and analyzed the retinal vasculature at P10 (Fig. 4, A–C). As expected, the R1R2R3iΔEC pups had a significantly reduced iB4+ vessel area, but also a reduced percentage of ETS-related gene (ERG)–stained EC nuclei per vessel area (Fig. 4, B and C). In the control pups, ∼1% of the ERG+ retinal EC nuclei were stained for cleaved caspase 3 (cCasp3), presumably because of the pruning of vasculature at this developmental stage (Fig. 4 B). Instead, the R1R2R3iΔEC pups had a 14-fold increase in cCasp3+/ERG+ nuclei, suggesting that EC apoptosis contributed to the vascular regression (Fig. 4, B and C).
EC apoptosis contributes to vessel regression. (A) Experimental setup. (B and C) iB4/ERG/cCasp3 triple staining and quantification in P10 retinas of the R1R2R3iΔEC pups compared with control littermates. Arrowheads indicate cCasp3/ERG double-positive endothelial nuclei. Error bars = mean ± SEM (data from n = control [CTRL] = 4, R1R2R3iΔEC = 9 mice pooled from three independent experiments). Statistical significance was determined using Student’s t test (two-tailed, unpaired). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
EC apoptosis contributes to vessel regression. (A) Experimental setup. (B and C) iB4/ERG/cCasp3 triple staining and quantification in P10 retinas of the R1R2R3iΔEC pups compared with control littermates. Arrowheads indicate cCasp3/ERG double-positive endothelial nuclei. Error bars = mean ± SEM (data from n = control [CTRL] = 4, R1R2R3iΔEC = 9 mice pooled from three independent experiments). Statistical significance was determined using Student’s t test (two-tailed, unpaired). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Organotypic effects of VEGFR deletions in quiescent adult vasculature
Because we considered that the varying growth rate of vessels in various tissues during postnatal development could affect the organ-specific phenotypes we observed, we next studied the effects of VEGFR deletions in adult mice. We deleted the VEGFRs with three consecutive tamoxifen administrations in 8–12-wk-old mice and analyzed the vasculature 8 d later. After gene deletion, only 2–12% of VEGFR mRNAs remained in the liver, kidney, pancreas, ear, lung, and heart lysates of adult R1R2R3iΔEC mice (Fig. S3 A). An exception was that 19% Vegfr2 mRNA remained in the lung lysates (Fig. S3 A), yet only 1.6% of the Vegfr2 mRNA remained in isolated pulmonary ECs from the same mice (Fig. S3 A).
VEGFR deletion levels in various tissues of adult mice.(A) qPCR analysis of Vegfr1, Vegfr2, and Vegfr3 mRNA levels on day 8 after three doses of tamoxifen in various tissues and their isolated EC populations from R1R2R3iΔEC mice and their control littermates (control [CTRL] = 3, R1R2R3iΔEC = 4 mice pooled from two independent experiments). (B) Schematic of the VEGFR deletion in adult mice. (C) Mouse weights after 8 d of gene deletion. (D) Representative Western blots and corresponding protein densitometry (total of 285 mice shown). (E) quantification of VEGFR levels in lung lysates. (F) qPCR analysis of Vegfr mRNA levels in lung, liver, and kidney lysates from the same cohorts. VEGFR1: CTRL = 21, R1iΔEC = 20 mice pooled from six independent experiments; VEGFR2: CTRL = 21, R2iΔEC = 20 mice pooled from five independent experiments; VEGFR3: CTRL = 16, R3iΔEC = 16 mice pooled from five independent experiments; VEGFR1/VEGFR2: CTRL = 19, R1R2iΔEC = 22 mice pooled from six independent experiments; VEGFR1/VEGFR3: CTRL = 20, R1R3iΔEC = 24 mice pooled from five independent experiments; VEGFR2/VEGFR3: CTRL = 14, R2R3iΔEC = 11 mice pooled from four independent experiments; and VEGFR1/VEGFR2/VEGFR3: CTRL = 31, R1R2R3iΔEC = 30 mice pooled from nine independent experiments. Statistical significance was determined using Student’s t test (two-tailed, unpaired) with Welch’s correction in cases of unequal variance. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Source data are available for this figure: SourceData FS3.
VEGFR deletion levels in various tissues of adult mice.(A) qPCR analysis of Vegfr1, Vegfr2, and Vegfr3 mRNA levels on day 8 after three doses of tamoxifen in various tissues and their isolated EC populations from R1R2R3iΔEC mice and their control littermates (control [CTRL] = 3, R1R2R3iΔEC = 4 mice pooled from two independent experiments). (B) Schematic of the VEGFR deletion in adult mice. (C) Mouse weights after 8 d of gene deletion. (D) Representative Western blots and corresponding protein densitometry (total of 285 mice shown). (E) quantification of VEGFR levels in lung lysates. (F) qPCR analysis of Vegfr mRNA levels in lung, liver, and kidney lysates from the same cohorts. VEGFR1: CTRL = 21, R1iΔEC = 20 mice pooled from six independent experiments; VEGFR2: CTRL = 21, R2iΔEC = 20 mice pooled from five independent experiments; VEGFR3: CTRL = 16, R3iΔEC = 16 mice pooled from five independent experiments; VEGFR1/VEGFR2: CTRL = 19, R1R2iΔEC = 22 mice pooled from six independent experiments; VEGFR1/VEGFR3: CTRL = 20, R1R3iΔEC = 24 mice pooled from five independent experiments; VEGFR2/VEGFR3: CTRL = 14, R2R3iΔEC = 11 mice pooled from four independent experiments; and VEGFR1/VEGFR2/VEGFR3: CTRL = 31, R1R2R3iΔEC = 30 mice pooled from nine independent experiments. Statistical significance was determined using Student’s t test (two-tailed, unpaired) with Welch’s correction in cases of unequal variance. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Source data are available for this figure: SourceData FS3.
Efficient deletion of VEGFR mRNAs and proteins was validated also in adult tissues (Fig. S3, B–F). Vascular phenotyping indicated that the organ-specific effects found in postnatal mice were preserved in the liver, kidney, intestines, and Langerhans islets of the adult mice (Fig. 5 A–F; and Fig. S4, A–H). The heatmap in Fig. 6 provides a comparison of changes in the vascular densities between the VEGFR-deleted postnatal and adult mice and their littermate controls. Overall, these results suggested that the observed organ-specific effects are independent of the vascular maturity in these tissues, and that VEGFR1 and VEGFR3 can support vascular maintenance in the absence of VEGFR2 in adult mice.
Organotypic effects of VEGFR deletions in quiescent adult vasculature. (A–F) Representative confocal micrographs of vascular staining of (A) liver (podocalyxin, red), (B) intestine (podocalyxin, green), (C) kidney (endomucin, red), (D) pancreas (podocalyxin, red), (E) trachea (podocalyxin, green), and (F) retina (isolectin B4 [iB4], white) tissues from adult mice deleted of single, double, or triple combinations of Vegfrs. The corresponding quantifications are shown in Fig. S4. Insets in C show close-up images of kidney glomeruli; Langerhans islets are demarcated by the dashed lines in D. Scale bars = 200 μm (B, E, and F), 100 μm (A, C, and D), and 50 μm (insets in C). Deep p., deep plexus.
Organotypic effects of VEGFR deletions in quiescent adult vasculature. (A–F) Representative confocal micrographs of vascular staining of (A) liver (podocalyxin, red), (B) intestine (podocalyxin, green), (C) kidney (endomucin, red), (D) pancreas (podocalyxin, red), (E) trachea (podocalyxin, green), and (F) retina (isolectin B4 [iB4], white) tissues from adult mice deleted of single, double, or triple combinations of Vegfrs. The corresponding quantifications are shown in Fig. S4. Insets in C show close-up images of kidney glomeruli; Langerhans islets are demarcated by the dashed lines in D. Scale bars = 200 μm (B, E, and F), 100 μm (A, C, and D), and 50 μm (insets in C). Deep p., deep plexus.
Effect of VEGFR deletion on vascular density in various tissues in adult mice. Data in addition to Fig. 5. (A) Representative confocal micrographs of whole-mounted ears stained for CD31 (green, all vessels), and LYVE1 (white, lymphatic vessels). Scale bars = 200 μm. (B) Vessel density quantifications from the ear whole-mounts in A. (C–H) Vascular density quantifications from liver (C), intestine (D), kidney (E, total and glomerular [Glom.]), pancreas (F), trachea (G, blood and lymphatic vessels), and superficial (Sup.) and deep plexus in the retina (H). The heatmap in Fig. 6 provides comparison of the corresponding percent changes in the vascular densities. VEGFR1: control (CTRL) = 14–21, R1iΔEC = 14–20 mice pooled from four to six independent experiments; VEGFR2: CTRL = 10–21, R2iΔEC = 10–20 mice pooled from three to five independent experiments; VEGFR3: CTRL = 9–16, R3iΔEC = 7–16 mice pooled from three to five independent experiments; VEGFR1/VEGFR2: CTRL = 16–19, R1R2iΔEC = 15–22 mice pooled from five or six independent experiments; VEGFR1/VEGFR3: CTRL = 17–20, R1R3iΔEC = 20–24 mice pooled from four or five independent experiments; VEGFR2/VEGFR3: CTRL = 14, R2R3iΔEC = 11 mice pooled from four independent experiments; and VEGFR1/VEGFR2/VEGFR3: CTRL = 12–31, R1R2R3iΔEC = 11–30 mice pooled from three to nine independent experiments. Statistical significance was determined using Student’s t test (two-tailed, unpaired) with Welch’s correction in cases of unequal variance. *, P < 0.05; **, P < 0.01; ***, P < 0.001. BV, blood vessel; LV, lymphatic vessel; EMCN, endomucin; PDXL, podocalyxin.
Effect of VEGFR deletion on vascular density in various tissues in adult mice. Data in addition to Fig. 5. (A) Representative confocal micrographs of whole-mounted ears stained for CD31 (green, all vessels), and LYVE1 (white, lymphatic vessels). Scale bars = 200 μm. (B) Vessel density quantifications from the ear whole-mounts in A. (C–H) Vascular density quantifications from liver (C), intestine (D), kidney (E, total and glomerular [Glom.]), pancreas (F), trachea (G, blood and lymphatic vessels), and superficial (Sup.) and deep plexus in the retina (H). The heatmap in Fig. 6 provides comparison of the corresponding percent changes in the vascular densities. VEGFR1: control (CTRL) = 14–21, R1iΔEC = 14–20 mice pooled from four to six independent experiments; VEGFR2: CTRL = 10–21, R2iΔEC = 10–20 mice pooled from three to five independent experiments; VEGFR3: CTRL = 9–16, R3iΔEC = 7–16 mice pooled from three to five independent experiments; VEGFR1/VEGFR2: CTRL = 16–19, R1R2iΔEC = 15–22 mice pooled from five or six independent experiments; VEGFR1/VEGFR3: CTRL = 17–20, R1R3iΔEC = 20–24 mice pooled from four or five independent experiments; VEGFR2/VEGFR3: CTRL = 14, R2R3iΔEC = 11 mice pooled from four independent experiments; and VEGFR1/VEGFR2/VEGFR3: CTRL = 12–31, R1R2R3iΔEC = 11–30 mice pooled from three to nine independent experiments. Statistical significance was determined using Student’s t test (two-tailed, unpaired) with Welch’s correction in cases of unequal variance. *, P < 0.05; **, P < 0.01; ***, P < 0.001. BV, blood vessel; LV, lymphatic vessel; EMCN, endomucin; PDXL, podocalyxin.
Quantification of changes in vascular density after Vegfr deletions. Heatmap summary of percent changes in blood vascular densities in postnatal (P) and adult (A) mice quantified from single, double, and triple compound deletions of the VEGFRs in liver, intestine, kidney (total and glomerular), pancreas (total and Langerhans islet–specific), trachea, retina (superficial and deep plexus), and heart tissues. Color scale indicates increased (red) or decreased (blue) vascular area; numbers indicate the percent change as compared with littermate controls. VEGFR1: control [CTRL] = 11–21, R1iΔEC = 10–20 mice pooled from four to six independent experiments; VEGFR2: CTRL = 10–21, R2iΔEC = 10–20 mice pooled from three to five independent experiments; VEGFR3: CTRL = 9–16, R3iΔEC = 7–16 mice pooled from three to five independent experiments; VEGFR1/VEGFR2: CTRL = 10–19, R1R2iΔEC = 12–22 mice pooled from three to six independent experiments; VEGFR1/VEGFR3: CTRL = 13–20, R1R3iΔEC = 15–24 mice pooled from three to five independent experiments; VEGFR2/VEGFR3: CTRL = 11–14, R2R3iΔEC = 9–11 mice pooled from three or four independent experiments; and VEGFR1/VEGFR2/VEGFR3: CTRL = 12–31, R1R2R3iΔEC = 11–30 mice pooled from three to nine independent experiments. Asterisks indicate statistical significance (Student’s t test; two-tailed, unpaired). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Quantification of changes in vascular density after Vegfr deletions. Heatmap summary of percent changes in blood vascular densities in postnatal (P) and adult (A) mice quantified from single, double, and triple compound deletions of the VEGFRs in liver, intestine, kidney (total and glomerular), pancreas (total and Langerhans islet–specific), trachea, retina (superficial and deep plexus), and heart tissues. Color scale indicates increased (red) or decreased (blue) vascular area; numbers indicate the percent change as compared with littermate controls. VEGFR1: control [CTRL] = 11–21, R1iΔEC = 10–20 mice pooled from four to six independent experiments; VEGFR2: CTRL = 10–21, R2iΔEC = 10–20 mice pooled from three to five independent experiments; VEGFR3: CTRL = 9–16, R3iΔEC = 7–16 mice pooled from three to five independent experiments; VEGFR1/VEGFR2: CTRL = 10–19, R1R2iΔEC = 12–22 mice pooled from three to six independent experiments; VEGFR1/VEGFR3: CTRL = 13–20, R1R3iΔEC = 15–24 mice pooled from three to five independent experiments; VEGFR2/VEGFR3: CTRL = 11–14, R2R3iΔEC = 9–11 mice pooled from three or four independent experiments; and VEGFR1/VEGFR2/VEGFR3: CTRL = 12–31, R1R2R3iΔEC = 11–30 mice pooled from three to nine independent experiments. Asterisks indicate statistical significance (Student’s t test; two-tailed, unpaired). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Because we observed a notable regression of intestinal and hepatic vasculatures 24–48 h after the gene deletions involving Vegfr2, we chose to characterize their effects 72 h after deletion in postnatal mice and 8 d after deletion in adult mice. We found that 24 h after R1R2R3 deletion induced by a single dose of 4-OHT injected to the pups, the intestinal vasculature had already regressed by 32% (Fig. 7, A and B). In the adult mice, two doses of tamoxifen led to regression of liver vasculature by 39%, 48 h after the first tamoxifen administration (Fig. 7, C and D). Unlike the intestinal and hepatic vasculatures, other vascular beds including those in skin and retina, did not show vascular regression even after 4 wk of gene deletion (Fig. 7, E and F). At this time point, the livers were already cirrhotic, but only occasional mice had died. Thus, the mice survived for at least 4 wk after the triple compound VEGFR deletion, unlike what was reported after the double deletion of the ERK1/2 kinases that act downstream of VEGFRs and several other receptors (Ricard et al., 2019).
Organ-specific temporal responses in VEGFR1/2/3 compound deletion.(A) Representative confocal micrographs and quantifications showing rapid regression of small intestine vasculature 24 h after 4-OHT administration to R1R2R3iΔEC pups (n = 3) compared with littermate controls (n = 3). Scale bar = 200 μm. (B) Western blots and quantification of VEGFRs normalized to HSC70 and VE-cadherin (VEC) in lung lysates at 24 h (control n = 6, R1R2R3iΔECn = 6, two independent experiments). (C) Representative confocal micrographs and quantification showing regression of hepatic vasculature 48 h after tamoxifen (TAM) administration to R1R2R3iΔEC (n = 3) adult mice and controls (n = 3). Scale bar = 100 μm. (D) Western blots and quantifications of VEGFRs in lung lysates at 48 h in R1R2R3iΔEC (n = 2) and control mice (n = 2). (E) Experimental setup, Western blots, and quantification of VEGFRs in lung lysates after 4 wk of Vegfr deletions in R1R2R3iΔEC (n = 4–6) and control mice (n = 6). (F) Representative images and quantification of dermal and retinal vasculature and changes in hepatic gross morphology after 4 wk of deletion. Blood vessels are stained for podocalyxin (PDXL), PECAM1 (CD31), or isolectin B4 (iB4), and lymphatic vessels are stained for LYVE1. Scale bars = 200 μm (ear images) and 100 μm (retina images). Error bars = mean ± SEM. Statistical significance was determined using Student’s t test (two-tailed, unpaired). *, P < 0.05; ***, P < 0.001. Source data are available for this figure: SourceData F7.
Organ-specific temporal responses in VEGFR1/2/3 compound deletion.(A) Representative confocal micrographs and quantifications showing rapid regression of small intestine vasculature 24 h after 4-OHT administration to R1R2R3iΔEC pups (n = 3) compared with littermate controls (n = 3). Scale bar = 200 μm. (B) Western blots and quantification of VEGFRs normalized to HSC70 and VE-cadherin (VEC) in lung lysates at 24 h (control n = 6, R1R2R3iΔECn = 6, two independent experiments). (C) Representative confocal micrographs and quantification showing regression of hepatic vasculature 48 h after tamoxifen (TAM) administration to R1R2R3iΔEC (n = 3) adult mice and controls (n = 3). Scale bar = 100 μm. (D) Western blots and quantifications of VEGFRs in lung lysates at 48 h in R1R2R3iΔEC (n = 2) and control mice (n = 2). (E) Experimental setup, Western blots, and quantification of VEGFRs in lung lysates after 4 wk of Vegfr deletions in R1R2R3iΔEC (n = 4–6) and control mice (n = 6). (F) Representative images and quantification of dermal and retinal vasculature and changes in hepatic gross morphology after 4 wk of deletion. Blood vessels are stained for podocalyxin (PDXL), PECAM1 (CD31), or isolectin B4 (iB4), and lymphatic vessels are stained for LYVE1. Scale bars = 200 μm (ear images) and 100 μm (retina images). Error bars = mean ± SEM. Statistical significance was determined using Student’s t test (two-tailed, unpaired). *, P < 0.05; ***, P < 0.001. Source data are available for this figure: SourceData F7.
Transcriptional alterations in adult endothelia in response to VEGFR deletions
Growth factor signaling leads to short- and long-term readjustment of gene expression in their target cells. To validate that the growth factor–specific signaling pathways were affected even in endothelia that did not show significant histological phenotypes in the organ-specific vasculatures, we employed transcriptomic analysis of single ECs from the tissues. Fig. 8 A shows the workflow of single-cell RNA sequencing (scRNAseq) of the cardiac and pulmonary ECs after 48 h of VEGFR deletions. After filtering the data for mitochondrial and ribosomal gene content (see Materials and methods and Data S2 for details), we performed batch correction and dimensional reduction of the data. Fig. 8, B and C, show Uniform Manifold Approximation and Projection (UMAP) plots of the aggregated clustering of ECs from tamoxifen-injected control mice. We identified 15 transcriptionally distinct cell clusters from 12 samples of cardiac ECs, and 22 clusters from 12 samples of pulmonary ECs. The clusters shown in the figure were identified based on previously published markers; examples are shown as stacked violin plots (Niethamer et al., 2020; Räsänen et al., 2021; Schupp et al., 2021; Fig. 8, B and C; Data S3; Data S4; Data S5; and Data S6). Cell counts and relative frequencies of the cells in clusters are shown in Data S7. Differential gene expression in each cluster after single or compound Vegfr deletions is shown in Data S8, Data S9, Data S10, Data S11, Data S12, and Data S13 for cardiac ECs, and Data S14, Data S15, Data S16, Data S17, Data S18, and Data S19 for pulmonary ECs. Because of the critical dependency of certain endothelia on Vegfr2, we additionally confirmed its deletion using quantitative PCR (qPCR; Fig. S5). The raw data and the raw count matrix files can be obtained from GEO (accession no. GSE185823). These data are also available for browsing in our web-based ShinyApp at http://combodel.it.helsinki.fi.
ScRNAseq analysis of cardiac and pulmonary ECs from mice with single and compound deletion of VEGFRs. (A) Experimental outline. Cardiac and pulmonary ECs of mice were harvested 48 h after Vegfr deletion and subjected to scRNAseq. (B and C) UMAP plots and cluster specific markers from cardiac (B) and pulmonary ECs (C), aggregated from control samples. Cap., capillary; NK, natural killer.
ScRNAseq analysis of cardiac and pulmonary ECs from mice with single and compound deletion of VEGFRs. (A) Experimental outline. Cardiac and pulmonary ECs of mice were harvested 48 h after Vegfr deletion and subjected to scRNAseq. (B and C) UMAP plots and cluster specific markers from cardiac (B) and pulmonary ECs (C), aggregated from control samples. Cap., capillary; NK, natural killer.
Feature scRNAseq heatmaps visualizing the expression of Flt1, Kdr, Flt4, and Cdh5 in cardiac and pulmonary ECs of mice with single and compound deletions of VEGFRs versus their controls. (A and B) Cardiac (A) and pulmonary EC (B) datasets are shown. Gene expression scale: low (blue) to high (red). (C) Violin plots for Kdr expression in the R2iΔEC dataset shown in B, suggesting incomplete deletion of Kdr. qPCR of the deleted exon in ECs isolated from the same mice indicates almost complete deletion of exon 3. (D) Violin plots for Kdr expression in the R1R2R3iΔEC dataset and qPCR of exon 30 versus exon 3 sequences. CTRL, control; DEL, deletion.
Feature scRNAseq heatmaps visualizing the expression of Flt1, Kdr, Flt4, and Cdh5 in cardiac and pulmonary ECs of mice with single and compound deletions of VEGFRs versus their controls. (A and B) Cardiac (A) and pulmonary EC (B) datasets are shown. Gene expression scale: low (blue) to high (red). (C) Violin plots for Kdr expression in the R2iΔEC dataset shown in B, suggesting incomplete deletion of Kdr. qPCR of the deleted exon in ECs isolated from the same mice indicates almost complete deletion of exon 3. (D) Violin plots for Kdr expression in the R1R2R3iΔEC dataset and qPCR of exon 30 versus exon 3 sequences. CTRL, control; DEL, deletion.
Because we were interested in the early transcriptional changes and possible new trajectories that the EC gene expression might take after VEGFR deletions, we next performed RNA velocity analysis using scVelo (Bergen et al., 2020). The top ranked genes for each cluster that drive the velocity vectors are shown in Data S20, Data S21, Data S22, and Data S23. Among several changes observed in the velocity trajectories, one of the most notable was the loss of the strong directionality of velocity vectors from the EC III arterial capillary cluster to the EC IV arterial cluster in the R1R2R3iΔEC cardiac samples (Fig. 9). This finding suggested that VEGFR deletions alter arterial differentiation, which should be explored in further studies. This exemplifies the conclusion that both VEGFR deletion–sensitive and –resistant ECs undergo profound transcriptional changes after the gene deletions. We encourage the readers to explore our datasets as a resource for further discoveries.
RNA velocity analysis of cardiac EC III and EC IV arterial clusters in R1R2R3iΔEC mice. Note the loss of directionality of the velocity vectors in the R1R2R3iΔEC cardiac arterial EC clusters in comparison to the control (circled in blue).
RNA velocity analysis of cardiac EC III and EC IV arterial clusters in R1R2R3iΔEC mice. Note the loss of directionality of the velocity vectors in the R1R2R3iΔEC cardiac arterial EC clusters in comparison to the control (circled in blue).
Discussion
Here we show that VEGFR1 and VEGFR3 support vascular maintenance in the absence of VEGFR2. This means that VEGFR1 is not only a decoy receptor, as thought so far, but can also sustain EC survival, likely by relaying growth factor signals. Similarly, VEGFR3 in blood vessel endothelium not only attenuates VEGFR2 signaling but also promotes EC survival when VEGFR2 signaling is lost. These results indicate genetic robustness in VEGFR signaling, as reported also for some other receptor tyrosine kinases (RTKs; Amit et al., 2007). Thus, VEGFR1 and VEGFR3 can at least partially substitute for VEGFR2 function when angiogenesis is inhibited by targeting only the VEGFR2 pathway (Rafii et al., 2016). These results suggest that blocking all three receptors could be considered when targeting the tumor vasculature.
The vascular regression caused by VEGFR2 deletion was tissue specific. Notably, VEGFR2 deletion resulted in a simultaneous 64% decrease in VEGFR3 protein levels and led to a substantial (∼60%) loss of vessels that have fenestrated endothelium after VEGFR2 deletion in, e.g., the kidney glomeruli, liver, and Langerhans islets, where vascular regression was not aggravated by the additional deletion of VEGFR1 or VEGFR3. A significant regression occurred also in the nonfenestrated tracheal vessels in both postnatal and adult mice and in retinal blood vessels in postnatal mice. Our data thus indicate that VEGFR2 is required for the maintenance of certain vascular beds in both postnatal and adult mice, and that in its absence, VEGFR1 and VEGFR3 can partially support vessel survival. On the other hand, the compound deletion of Vegfr1 and Vegfr3 without Vegfr2 deletion in the pups seemed to hyperactivate VEGFR2 signaling globally, and to increase angiogenesis in the retinal periphery. Yet the hypersprouting induced by VEGFR3 deletion seemed to be attenuated (Fig. S2 U). This is probably because an excess of VEGF signaling during postnatal development of the retina pushes ECs with high VEGFR2 levels toward quiescence (Pontes-Quero et al., 2019).
Although our focus was on blood vessels, we observed that VEGFR2 participates also in lymphatic vessel development independently of VEGFR3 (Dellinger et al., 2013), since the combined deletion of VEGFR2 and VEGFR3 led to additional regression of the tracheal lymphatic vasculature. Deletion of only VEGFR3 was sufficient to cause regression of the intestinal lacteals. These data indicate that the lymphatic vessel response to the loss of VEGFR signaling is also tissue specific.
Previous studies have documented that inhibitors of VEGFR2 signaling, such as small molecule tyrosine kinase inhibitors AG013736 (axitinib; Inai et al., 2004), VEGF-Trap (Inai et al., 2004), soluble VEGFR1 and VEGFR2 (Gerber et al., 1999; Kamba et al., 2006), and VEGFR2-blocking antibodies (Yang et al., 2013), lead to regression of fenestrated capillaries, but not retinal or cardiac vasculature in adult mice (Kamba and McDonald, 2007). However, the use of signaling inhibitors seldom leads to the same phenotypes as deleting their target molecules, as even an inactive RTK protein can provide a docking site for phosphorylation by other kinases (Carmeliet et al., 2001; Saharinen et al., 2005). Our study shows, for the first time, that additional deletions of VEGFR1 and VEGFR3 lead to further reduction of vasculature, as opposed to what was previously described for singular targeting of these receptors.
For therapeutic translation of our results, several questions remain to be addressed. For example, do the organ-specific differences in vascular responses that we describe here contribute to resistance to anti-angiogenic therapy in ocular diseases or in tumors? Should the results encourage the use of combinations of receptor-blocking antibodies or ligand traps (Jeltsch et al., 2013)? Overall, our results highlight the importance of organotypic sensitivity of the ECs to VEGFR signaling and the unexpected robustness in the VEGFR system, warranting further investigations and generation of new ideas concerning the resistance to VEGF-blocking therapies.
Materials and methods
Mice and gene deletions
Mice were maintained in a temperature-controlled, specific pathogen–free facility, with a 12-h light/dark cycle and ad libitum access to regular mouse chow and water. All animal experiments were approved by the Committee for Animal Experiments of the District of Southern Finland, and all procedures were performed in accordance with these protocols.
We interbred the following mouse lines to generate single, double, or triple compound endothelial-specific deletions of the VEGFRs: Cdh5-BAC-CreERT2 (a kind gift of Y. Kubota; Okabe et al., 2014), Vegfr1flox/flox (Ambati et al., 2006), Vegfr2flox/flox (Hooper et al., 2009), and Vegfr3flox/flox (Haiko et al., 2008). All lines were backcrossed for at least eight generations into C57BL/6J before the interbreeding.
To induce gene deletion, mouse pups were injected intraperitoneally with 100 µg of 4-OHT dissolved in corn oil on P7–9. For short-term experiments shown in Fig. 7, A and B, the pups were injected on P7 for P8, P7–8 for P9, and P7–9 for P10 harvesting. For the induction of gene deletion in the adult mice, 8–12-wk-old Vegfr-floxed mice and Cre-negative littermates were administered tamoxifen in corn oil via oral gavage using a feeding needle (2 mg per day, 100 µl volume) for 3 consecutive days, and the tissues were harvested for analysis on day 8. For scRNAseq experiments and short-term deletion experiments shown in Fig. 7 C, and D, female mice 8–10 wk of age were used. The mice were administered two consecutive doses of tamoxifen, and tissues were harvested 48 h after the first tamoxifen administration at the same time of day to avoid changes introduced by circadian regulation. For the long-term deletion experiments shown in Fig. 7, E and F, the mice were given three consecutive tamoxifen administrations in the beginning, and the gene deletions were maintained by additional tamoxifen dosing once a week until the harvesting. For analysis of vessel density and gene deletion levels, both male and female mice were used, and in each independent cohort, mice of the same sex were compared. Routine genotypings were performed using respective protocols that are available on request.
For tissue harvesting, P10 or adult mice were administered a lethal dose of ketamine (Ketaminol; Intervet) and xylazine (Rompun; Bayer) by intraperitoneal injection. For vessel density analyses, 50-mg pieces of lung from the postnatal mice or 50-mg pieces of lung, liver, and kidney from the adult mice were snap-frozen for downstream analyses. The mice were then trans-cardially perfused with ice-cold 1% paraformaldehyde (PFA) in PBS (pH 7.4), and tissues were collected for downstream quantifications. For the measurement of organ-specific Vegfr deletion levels, the mice were administered a lethal dose of ketamine/xylazine by intraperitoneal injection, and pieces of the tissues were immediately snap-frozen for measuring the deletion levels in total lysates. The remaining tissues from adult mice were processed for isolation of ECs for measuring EC-specific deletion levels.
RNA isolation
Tissue lysates
Total RNA from lung, liver, kidney, skin, intestine, pancreas, and heart tissues were isolated via homogenization using Trisure (Bioline) with zirconium oxide bead tubes (Next Advance Inc.; CAT#MB2ZO15). The RNA was isolated from the homogenate using the NucleoSpin RNA Kit (Macherey-Nagel) according to the manufacturer’s instructions.
Isolated ECs
Total RNA from enriched ECs were isolated using RNeasy Micro kit (Qiagen; CAT#74004) following the manufacturer’s instructions.
Quantitative real-time PCR (qRT-PCR)
The quality of the total and EC-enriched RNA samples was determined with Nanodrop ND-1000 (Thermo Fisher Scientific). cDNA was synthetized from 1 µg of total RNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems; CAT#10400745) for total RNA, or 100–200 µg RNA of the EC-enriched RNA. qRT-PCR was performed with primers for 36b4, Vegfr1, Vegfr2 (exon 3), Vegfr2 (exon 30), Vegfr3, Cdh5, and Pecam1 (sequences listed under Oligonucleotides used for PCR) and Fast Start Universal SYBR green Master containing ROX (Roche; CAT# 4913914001). qRT-PCR was performed using a BIO-RAD C1000 Thermal cycler according to the standardized protocol. The data were normalized to the housekeeping gene 36b4 to compensate for experimental variations. Fold-changes were then calculated using the comparative cycle threshold (CT) method (Pfaffl, 2001). The Vegfr gene expression levels in total tissues were additionally normalized to Cdh5 to compensate for possibly altered EC numbers in the gene-deleted mice (except for pancreas tissue), and values were expressed relative to the control mice.
Oligonucleotides used for qPCR
We used the following primers for measuring mRNA levels in mouse tissues: mVegfr2_exon30_F: 5′-TGGTTGGTTTGCTCTCCAGAT-3′; mVegfr2_exon30_R: 5′-ATAAGCACACAGGCAGAAACC-3′; mVegfr1_F: 5′-CAAGCAGAAGCAGGAGGGG-3′; mVegfr1_R: 5′-TGACCATGGTGAGCAAGACG-3′; mVegfr2_exon3_F: 5′-GCGTGATTCTGAGGAAAGGGT-3′; mVegfr2_exon3_R: 5′-CCCTGGGAATGGTGAGTGTT-3′; mVegfr3_F: 5′-CTGGCAAATGGTTACTCCATGA-3′; mVegfr3_R: 5′-ACAACCCGTGTGTCTTCACTG-3′; mCdh5_F: 5′-GGACAAGGCACCCGTACTGA-3′; mCdh5_R: 5′-CTGGGGAAAGTTAGGGCCTG-3′; mPecam1_F: 5′-CTGCTCCGTCTCGGGCACAC-3′; and mPecam1_R: 5′-GGGCTTCGAGAGCATTTCGCACA-3′.
Western blotting
Equal amounts of cleared lysates with 25 µg protein from the lungs were separated in Novex WedgeWell 4–20% TRIS-Glycine gel 1.0 mm × 15 well (Invitrogen; CAT#XP04205BOX). After blotting to polyvinylidene fluoride membranes (Immobilon-FL PVDF; Merck Millipore; CAT#IPFL00010), the proteins were detected using goat anti-mouse VEGFR1 (1:1,000; R&D Systems; CAT#AF471), goat anti-mouse VEGFR2 (1:1,000; R&D Systems; CAT#AF644), goat anti-mouse VEGFR3 (1:1,000; R&D Systems; CAT#AF743), rat anti-mouse CD144 (1:1,000; BD PharMingen; CAT#555289,) and mouse anti-HSC70 (1:5,000; Santa Cruz Biotechnology; CAT#SC-7298) primary antibodies. The blots were then probed with HRP-labeled rabbit anti-goat secondary antibody (1:2,000; Dako; CAT#P0449), and the signal was visualized with SuperSignal West Pico Chemiluminescent Substrate (Pierce; CAT#34079) or SuperSignal West Femto Maximum Sensitivity Substrate (Pierce; CAT#34096). HSC70 was probed using IRDye 680RD donkey anti-Mouse IgG (1:10,000; LI-COR Biosciences; CAT#925-68024) and was detected using LI-COR Odyssey Fc (LI-COR Biosciences). Densitometric analysis of the blots was performed using ImageStudio Lite (Version 5.2.5; LI-COR Biosciences). The values were normalized to HSC70 for protein loading and to VE-cadherin for EC content.
Immunofluorescence
Harvested tissues were fixed overnight in 4% PFA in PBS at 4°C, washed in PBS, and then processed for paraffin embedding. Kidney, liver, heart, and pancreas blocks were cut at 5-µm thickness, deparaffinized, and rehydrated.
Antigen retrieval was done by boiling the sections in high pH antigen retrieval solution (10 mM Tris, 1 mM EDTA, and 0.05% Tween20 in PBS, pH 9.0) in a microwave, 5 min at 70% and 10 min at 30% power. After washing with PBS, the sections were blocked in blocking buffer (0.2% BSA [Biowest, CAT#P6154] and 2.5% normal donkey serum [Biowest; CAT#S2170-500] in PBS) for 1 h and incubated with the following primary antibodies diluted in blocking buffer overnight at 4°C: rat anti-mouse Endomucin (1:500; Santa Cruz; CAT#sc-65495) and goat anti-mouse Podocalyxin (1:250; R&D Systems; CAT#AF1556). The next day, sections were washed 10 min with Tris-NaCl-Tween20 buffer (150 mM NaCl, 100 mM Tris, 0.05% Tween20) and incubated with secondary antibodies (donkey anti-rat Alexa Fluor 594; 1:500; Invitrogen; CAT#A21209 or donkey anti-goat Alexa Fluor 594; 1:500; Invitrogen; CAT#A11058) and Hoechst (1:1,000; Invitrogen; CAT#H3570) diluted in 0.05% PBS-Tween20, for 1 h at room temperature. After washing with Tris-NaCl-Tween20 buffer, the sections were post-fixed with 1% PFA for 5 min, rinsed with PBS for 10 min, and mounted with Prolong Gold antifade reagent (Invitrogen; CAT#P36930).
Whole-mount immunostaining
Harvested tissues were fixed in 4% PFA in PBS at 4°C overnight and washed in PBS.
Retina staining
Retinas were dissected and stained as described with minor modifications (Zarkada et al., 2015). Briefly, for iB4 stainings, the retinas were equilibrated 30 min in PBlec (0.1 mM CaCl2, 0.1 mM MgCl2, 0.1 mM MnCl2, and 1% Triton X-100 in PBS, pH 6.8) and incubated with biotinylated Griffonia simplicifolia lectin I iB4 (1:25; Vector; CAT#B-1205) diluted in PBlec, overnight at 4°C. iB4 was detected by incubation with streptavidin-conjugated Alexa Fluor 488 (1:200; Invitrogen; CAT#S-32354) overnight. For iB4/ERG/cCasp3 combination stainings shown in Fig. 4, following iB4 staining and post-fixation in 1% PFA in PBS for 5 min at room temperature, the retinas were washed with PBS for 30 min, blocked with donkey immunomix (DIM; 5% normal donkey serum [Biowest], 2% BSA [Biowest], and 0.3% Triton X-100 [Sigma-Aldrich] in PBS) for 1 h at room temperature and incubated with rabbit anti-cCasp3 (1:300; Cell Signaling Technology; CAT#9664) diluted in DIM overnight at 4°C, which was detected with Alexa Fluor 594–donkey anti-rat IgG (1:500; Invitrogen; CAT#A-21209). The retinas were then washed all day in PBS and incubated with rabbit-anti-ERG-Alexa 647 conjugated antibody (1:100; Abcam; CAT#ab196149) for 2 d before post-fixation and mounting.
Intestine, trachea, and ear staining
After fixation, the tissues were briefly rinsed with PBS, permeabilized with PBS-T (PBS with 0.3% Triton X-100), and blocked in DIM for 1 h at room temperature, followed by incubation with the following primary antibodies in DIM overnight at 4°C: rat anti-mouse CD31 (1:500; BD PharMingen; CAT#553370), rabbit anti-mouse LYVE1 (1:1,000; Karkkainen et al., 2004), and goat-anti mouse Podocalyxin (1:300; R&D Systems; CAT#AF1556). The following day, the samples were washed all day in PBS and incubated with the secondary antibodies overnight at 4°C diluted in PBS: Alexa Fluor 488-donkey anti-goat IgG (1:500; Invitrogen; CAT#A-32814), Alexa Fluor 488-donkey anti-rat IgG (1:500; Invitrogen; CAT#A-21208), Alexa Fluor 594-donkey anti-rabbit IgG (1:500; Invitrogen; CAT#A-21207), or Alexa Fluor 647-donkey anti-rabbit IgG (1:500; Invitrogen; CAT#A-31573). All tissues were washed with PBS for several hours, post-fixed with 1% PFA for 10 min at room temperature, and flat-mounted with VECTASHIELD Antifade Mounting Medium (Vector; CAT#H-1000).
Microscopy and image processing
Confocal images of intestinal, retinal, dermal, and tracheal whole-mounts were acquired using an LSM 780 confocal microscope (ZEISS) equipped with 10× Plan-Apochromat with numerical aperture (NA) 0.45 and 20× Plan-Apochromat with NA 0.80 air objectives using Zen 2012 software. Three-dimensional maximal projections were digitally constructed from confocal Z-stacks. Kidney and liver sections were imaged using an AxioImager (Carl Zeiss) equipped with 10× EC Plan Neofluar with NA 0.3 and 20× Plan-Apochromat with NA 0.80 air objectives and a 40× C-Apochromat with NA 1.2 (water immersion) objective through Hamamatsu Orca Flash 4.0 LT camera and Zen Pro 2.3 software. Digital scans of pancreas and heart sections were generated with a Pannoramic 250 FLASH II digital slide scanner (3DHISTECH), and snapshots of regions of interest were taken using CaseViewer software (version 2.4, 3DHISTECH). Vessel areas were quantified from the acquired images using ImageJ software (Image J 1.52h; National Institutes of Health), and branch points of retinas were analyzed using AngioTool (version 0.6a; Zudaire et al., 2011). The images were pseudocolored and merged using Photoshop software (version CC 2021; Adobe), and the figures were assembled in Adobe Illustrator software (version CC 2021; Adobe).
Image quantification
For kidney and liver vessel density (Fig. 1, G and H; Fig. 2, F and G; Fig. 3, F and G; Fig. 5, A and C; Fig. 7 C; Fig. S1, F, G, Q, and R; and Fig. S2, F, G, Q, and R), endomucin-positive (kidney) and podocalyxin-positive (liver) areas were quantified as a percentage of area of a region of interest (ROI, 665 µm × 665 µm). Three images of kidney or liver were analyzed per mouse, and the mean value per mouse is represented as a dot.
For kidney glomerular vessel density (shown in insets in Fig. 1 G; Fig. 2 F; Fig. 3 F; Fig. 5 C; Fig. S1, F and Q; and Fig. S2, F and Q), glomerular areas were identified from nuclear staining, and endomucin-positive areas were quantified as a percentage of glomerular area. Three images of kidney were analyzed per mouse, and the mean value per mouse is represented as a dot.
For pancreas vessel density (Fig. 1 I; Fig. 2 H; Fig. 3 H; Fig. 5 D; Fig. S1, H and S; and Fig. S2, H and S), podocalyxin-positive areas were quantified as a percentage of area of an ROI (750 µm × 450 µm). Four images of pancreas were analyzed per mouse, and the mean value per mouse is represented as a dot.
For pancreatic Langerhans islet vessel density (Fig. 1 I, dashed lines; Fig. 2 H; Fig. 3 H; Fig. 5 D; Fig. S1, H and S; and Fig. S2, H and S), Langerhans islets were identified from nuclear staining, and podocalyxin-positive areas were quantified as a percentage of islet area. Five images of pancreas were analyzed per mouse, and the mean value per mouse is represented as a dot.
For intestine blood and lymphatic vessel density Fig. 1 J; Fig. 2 I; Fig. 3 I; Fig. 5 B; Fig. 7 A; Fig. S1, I and T; and Fig. S2, I and T), CD31 (in postnatal mice)– and podocalyxin (in adult mice)–positive blood vessel areas were quantified as a percentage of lamina propria area. LYVE1-positive lymphatic vessel areas were quantified as a percentage of villus area. At least 10 villi from two images of intestines were analyzed per mouse, and the mean value per mouse is plotted as a dot.
For retina vascular density, branches and lacunarity (Fig. 1 K; Fig. 2 J; Fig. 3 J; Fig. 5 F; Fig. 7 F; Fig. S1, J and U; and Fig. S2, J and U), in postnatal mice, tile-scanned images of flat-mounted retinas stained with iB4 were used for analysis. iB4-positive areas of the superficial and deep plexuses were quantified as a percentage of total retinal area. Average values of the two retinas from one mouse are plotted as a dot. Retinal branch point and lacunarity quantifications were performed using standard measurements in AngioTool software (version 0.6a). The number of branch points (junctions) was normalized to iB4-positive area and plotted as a dot. The lacunarity quantified per total retinal area and mean of two retinas per mouse are plotted as a dot. For adult mice, iB4-positive areas of the superficial and deep plexuses were quantified as percentage of ROI (850 µm × 850 µm). Five images from the retinas of one mouse were analyzed, and the mean value per mouse is plotted as a point.
For trachea blood and lymphatic vessel density (Fig. 1 L; Fig. 2 K; Fig. 3 K; Fig. 5 E; Fig. S1, K and V; and Fig. S2, K and V), CD31 (in postnatal mice)– and podocalyxin (in adult mice)–positive blood vessel areas and LYVE1-positive lymphatic vessel areas were quantified as percentage of an ROI (850 µm × 850 µm). Two images per trachea or ear were analyzed per mouse, and the mean value per mouse is plotted as a dot.
Ear blood and lymphatic vessel density (Fig. 7 F and Fig. S4 A)
CD31-positive blood vessel areas and LYVE1-positive lymphatic vessel areas were quantified as percentage of an ROI (850 µm × 850 µm). Two images per ear were analyzed per mouse, and the mean value per mouse is plotted as a dot.
Retina blood vessel, EC nuclei, and cCasp3 quantifications (Fig. 4, B and C)
Maximal projection images of flat-mounted retinas stained with iB4, cCasp3, and ERG were used for analysis. The iB4-positive blood vessel areas were quantified as a percentage of area of ROI. ERG-positive nuclei and ERG/cCasp3 double-positive nuclei were counted manually. Three images of retina were analyzed per pup, and the mean value per pup is represented as a dot.
Enrichment of ECs
Pulmonary ECs
Left lobes of the lungs were finely minced using scissors in a mixture of 0.5 mg/ml Collagenase H (Roche; CAT#11074059001), 0.5 U/ml of Dispase II (Sigma-Aldrich; CAT#D4693), 1 U/ml DNase I (Thermo Fisher Scientific; CAT#EN0521) and 1% (wt/vol) BSA (Biowest; CAT#P6154) in 4 ml of PBS and incubated at 37°C with constant agitation for 20 min. The digestion was stopped by addition of 10 ml of 10% heat-inactivated FBS in FACS buffer (1% heat-inactivated FBS and 2 mM EDTA in PBS). The slurry was then passed through a 70 µm nylon cell strainer (Corning) and pelleted. Erythrocyte lysis was performed using 1 ml Red Blood Cell Lysis buffer (Sigma-Aldrich; CAT#R7757). The following antibodies were used for blocking and staining: rat-anti-mouse CD16/CD32 (Mouse Fc-block; 1:200; BD Biosciences; CAT#553141), rat-anti-mouse CD31-APC (clone: MEC13.3; 1:100; BD Biosciences; CAT#551262). DAPI (at 1 µg/ml final concentration in milli-Q water; Tocris; CAT#5748) was used to determine the dead cells. ECs were enriched by sorting DAPI−CD31+ cells using a BD Influx flow cytometer in 1.0 Drop Single sort mode with a 100-µm nozzle. The cells were used for either scRNAseq or RNA isolation.
Cardiac ECs
ECs from the hearts were isolated exactly as described previously (Hemanthakumar et al., 2021). Briefly, the hearts were rinsed in cooled Dulbecco’s PBS (GIBCO BRL; CAT#14190–094) supplemented with 0.3 mM calcium chloride. The hearts were then minced in 4 ml of prewarmed digestion buffer containing collagenase type I (CAT#17100–017), type II (CAT#17101–015), and type IV (CAT#17104–019), all from Gibco and all at 1 mg/ml in Dulbecco’s PBS with 0.3 mM CaCl2, and incubated in a water bath at 37°C for 25 min, with gentle agitation (vortexing) every 5 min. The slurry was next passed through a T10 serological pipette 20 times. The digestion was stopped with the addition of 10 ml Dulbecco’s modified Eagle’s medium (GIBCO BRL; CAT#31053–028) supplemented with 10% heat-inactivated FCS. The slurry was then filtered through the 70-µm nylon cell strainer (Corning; CAT#352350). The cells were stained with the rat-anti CD31-FITC (eBioscience; CAT#25–1401), rat-anti-PDGFRa/CD140a-PE-cyanine7 (eBioscience; CAT#25–1401), rat-anti-CD45-Pacific Blue (BioLegend; CAT#103125), and rat-anti-Ter119-Pacific Blue (BioLegend; CAT#116231) antibodies for 30 min. DAPI (at 1 µg/ml final concentration in milli-Q water; Tocris; CAT#5748) was used to determine the dead cells. Prior to FACS, the cells were rinsed twice with the staining buffer and filtered through 5-ml cell strainer tubes (Corning; CAT#352235). CD45−Ter119−CD140a−DAPI− and CD31+ live cardiac ECs were sorted using a FACS Aria II (BD Biosciences). The cells were used for either scRNAseq or RNA isolation.
Liver, pancreas, intestine, and ear ECs
ECs of liver, pancreas, intestine, and ear were isolated from adult R1R2R3iΔEC mice after 8 d of Vegfr deletion. For tissue digestion, the organs were finely minced using scissors. For liver and pancreas, a mixture of 0.25 mg/ml Collagenase H (Roche; CAT#11074059001), 0.25 U/ml Dispase II (Sigma-Aldrich; CAT#D4693), 0.5 U/ml DNase I (Thermo Fisher Scientific; CAT#EN0521), and 1% (wt/vol) BSA (Biowest; CAT#P6154) in 4 ml of PBS was used for digestion. For small intestine (20 cm), the digestion mixture contained 2 mg/ml Collagenase IV (GIBCO BRL; CAT#17104-109), 0.05 U/ml DNase I (Roche; CAT#11284932001), 2 mM CaCl2, and 1% (wt/vol) BSA (Biowest; CAT#P6154) in 4 ml of PBS. For ears, the digestion mix contained 5 mg/ml Collagenase IV (GIBCO BRL; CAT#17104-109), 0.2 U/ml DNase I (Roche; CAT#11284932001), and 0.5% (wt/vol) heat-inactivated FBS (GIBCO BRL; CAT#10500-64) in 4 ml of PBS. The samples were incubated at 37°C with constant agitation for 10 (intestine), 15 (liver and pancreas), or 20 min (ear). The digestion was stopped by addition of 10 ml of 10% heat-inactivated FBS in FACS buffer (1% heat-inactivated FBS and 2 mM EDTA in PBS). The slurry was then passed through a 70-µm nylon cell strainer (Falcon; CAT#35–23-50) and pelleted. Erythrocyte lysis was performed for the liver samples with Red Blood Cell Lysis buffer (Sigma-Aldrich; CAT#R7757). After final centrifugation, the pellets were resuspended in 100 µl (liver and ear) or 200 µl (pancreas and intestine) FACS buffer. Liver ECs were isolated with flow cytometry using the same setup as lung ECs, with an additional staining. For liver ECs, CD31/CD146 double-positive cells were sorted (rat-anti CD146-PE-Cy7; BioLegend; CAT#134713). For ear, pancreas, and intestine ECs, Dynabead-mediated magnetic sorting was performed. For negative selection, CD45 and LYVE1 were used, and for positive selection, CD31 was used. Cell suspensions were first incubated with biotinylated rat-anti-CD45 (BioLegend; CAT#103103)– and rabbit anti-mouse LYVE1 (Karkkainen et al., 2004)–bound magnetic beads (DynaBeads sheep-anti-rat; Invitrogen; CAT#11035; and Dynabeads sheep-anti-rabbit; Invitrogen; CAT#11203D). The nonbound cell fraction was then incubated with rat anti-mouse CD31 (BD PharMingen; CAT#553370) bound to the corresponding magnetic beads (DynaBeads sheep-anti-rat; Invitrogen; CAT#11035) following the manufacturer’s instructions. Both selections were performed by first incubating the cell suspension with the coated beads at 2–8°C with constant agitation for 40 min. After incubation, the sample volumes were adjusted to 1 ml using FACS buffer, and sorting was performed using a magnet. The cells were then lysed in RLT buffer (Qiagen) with 1% β-mercaptoethanol, vortexed, and snap-frozen.
scRNAseq
Sorted cardiac and pulmonary ECs were supplemented with 4% BSA in PBS to a final concentration of 0.04% BSA in PBS, and cell counting and single-cell library preparation were performed. Briefly, single-cell gene expression profiles were studied using the 10x Genomics Chromium Single Cell 3′RNaseq platform. The Chromium Single Cell 3′RNaseq run and library preparation were done using the Chromium Single Cell 3′ Reagent 2 chemistry. Data processing and analysis were performed using 10x Genomics Cell Ranger v2.1.1 pipelines. Cell Ranger includes several pipelines, of which the “cellranger mkfastq” was used to produce FASTQ (raw data) files and “cellranger count” to perform alignment, filtering, and unique molecular identifier counting. Mkfastq was run using the Illumina bcl2fastq v2.2.0, and alignment was done against mouse genome mm10.
The raw scRNAseq count matrices were imported to RStudio v1.4 (R v4.0.3) and analyzed using Seurat v4.0 (Hao et al., 2021). Features that appeared in over three cells and cells with >200 features were included in the generated Seurat objects. The following parameters were used to further filter out low-quality cells: 300 < nFeature_RNA < 2,500, pt.mito < 10, and 5 < pt.rRNA < 28 for cardiac ECs, and 500 < nFeature_RNA < 3,500, pt.mito < 8, and 5 < pt.rRNA < 28 for the pulmonary ECs, respectively. Approximately 93% of the raw cell counts was preserved by the filtering. For sample-specific filtered cell counts, see Data S2. All data objects were normalized on a log scale of 10,000, and 2,000 variable features were calculated using default parameters. All deletions and respective controls from cardiac and pulmonary ECs were integrated to merged objects for each tissue, and the cell cycle scores were calculated for each cell using the Regev laboratory table of cell cycle genes (Kowalczyk et al., 2015). The integrated data objects were scaled regressing for RNA counts, mitochondrial gene percentage, and ribosomal RNA percentage and clustered using graph-based clustering with 30 (heart ECs) or 40 (lung ECs) dimensions and with a resolution of 0.4. Since we were interested in VEGFR deletion–induced changes in ECs, including proliferation and apoptosis, cell-cycle regression was not performed. The clustering resulted in 15 clusters for the cardiac ECs and 22 clusters for the pulmonary ECs that were visualized using UMAP plots. The clusters were labeled according to marker genes described in previous publications (Niethamer et al., 2020; Räsänen et al., 2021; Schupp et al., 2021). See Data S3, Data S4, Data S5, and Data S6 for cluster-specific markers. Differential gene expression analysis was performed using the FindMarkers() function and the default Wilcoxon rank-sum test, comparing each deletion to its corresponding control in both tissues (see Data S8, Data S9, Data S10, Data S11, Data S12, Data S13, Data S14, Data S15, Data S16, Data S17, Data S18, and Data S19). Here, a log fold change threshold of 0.25 was used for including only the most significant sets of genes.
RNA velocity analysis
The unspliced/spliced RNA count matrices were generated using Velocyto.py v0.17.16 (La Manno et al., 2018) with the Puhti supercomputer provided by CSC Finland. Here, the genome annotation file was prepackaged with the 10x Genomics Cell Ranger, the masked expressive repetitive elements file provided by the University of California, Santa Cruz genome browser was used as reference, and aligned bam-files were used as input. The generated loom-files were imported to a Python v3.6 environment and merged with counts, UMAP embeddings, and metadata extracted from the previously generated Seurat objects using scVelo v0.2.3 (Bergen et al., 2020). Using the scVelo pipeline, velocity embeddings were estimated using both the stochastic and the dynamical model and projected on two-dimensional UMAP embeddings previously calculated using Seurat v4.0. The top velocity markers were calculated and ranked using the scv.tl.rank_velocity_genes() and scv.tl.rank_dynamical_genes() functions with default parameters, and listed in Data S20, Data S21, Data S22, and Data S23.
Statistical analysis
Statistical details can be found in the figure legends, supplemental datasets, and Materials and methods section. For all datasets, the unpaired two-tailed Student’s t test was used to determine statistical significance between deletion and control mice. In case of unequal variations between groups, the unpaired t test with Welch’s correction was applied, as indicated in figure legends. A P value <0.05 was considered statistically significant. No statistical test was used to predetermine sample size, and the number of mice used represents biological replicates. The graphs were prepared in GraphPad prism v9.0.
Online supplemental material
Fig. S1 shows data from single deletions of VEGFR1 and VEGFR3 in postnatal mice. Fig. S2 shows data from compound deletions of VEGFR3 with VEGFR2 or VEGFR1 in postnatal mice. Fig. S3 shows data from VEGFR deletion levels at the mRNA level in total tissues and isolated ECs from R1R2R3iΔEC adult mice, and Western blots from lung lysates and qPCRs from lung, liver, and kidneys of all VEGFR deletion combinations in adult mice. Fig. S4 shows representative staining from the skin and quantification of vessels in the skin, liver, intestine, kidney, pancreas, trachea, and retina tissues in adult mice with the VEGFR deletion combinations shown in Fig. 5. Fig. S5 shows scRNAseq feature heatmaps for Fms-related tyrosine kinase 1 (Flt1), kinase insert domain receptor (Kdr), Fms-related RTK 4 (Flt4), and Cdh5 expression after 48 h of VEGFR deletion, and confirmation of Vegfr2 deletion by using qPCR. Data S1 shows VEGFR mRNA and protein levels from total lung tissues normalized to 36B4 for mRNA and HSC70 for protein, then normalized to VE-cadherin in single and combination deletions of VEGFRs in postnatal and adult mice. For each litter/cohort, control values are set to 1.0, and Vegfr deletion values are shown relative to control values obtained from tamoxifen-treated and gender-matched littermates. Data S2 shows parameters used for quality control, cell filtering, normalization, and data integration. It shows the number of cells in each dataset before and after filtering and the number of cell clusters calculated for each merged dataset. Data S3 shows significantly up- and downregulated genes in each cluster of the merged heart dataset. Data S4 shows significantly up- and downregulated genes in each cluster of merged heart control datasets. Data S5 shows significantly up- and downregulated genes in each cluster of the merged lung dataset. Data S6 shows significantly up- and downregulated genes in each cluster of merged lung control datasets. Data S7 shows cell numbers and relative cell frequencies in each annotated cluster in cardiac and pulmonary datasets with VEGFR deletions. Data S8 shows differentially expressed (DE) genes calculated with the Wilcoxon rank-sum test. It shows comparison of the R1iΔEC and control datasets of heart tissue. Data S9 shows DE genes calculated with the Wilcoxon rank-sum test. It shows comparison of the R2iΔEC and control datasets of heart tissue. Data S10 shows DE genes calculated with the Wilcoxon rank-sum test. It shows comparison of the R3iΔEC and control datasets of heart tissue. Data S11 shows DE genes calculated with the Wilcoxon rank-sum test. It shows comparison of the R1R2iΔEC and control datasets of heart tissue. Data S12 shows DE genes calculated with the Wilcoxon rank-sum test. It shows comparison of the R2R3iΔEC and control datasets of heart tissue. Data S13 shows DE genes calculated with the Wilcoxon rank-sum test. It shows comparison of the R1R2R3iΔEC and control datasets of heart tissue. Data S14 shows DE genes calculated with the Wilcoxon rank-sum test. It shows comparison of the R1iΔEC and control datasets of lung tissue. Data S15 shows DE genes calculated with the Wilcoxon rank-sum test. It shows comparison of the R2iΔEC and control datasets of lung tissue. Data S16 shows DE genes calculated with the Wilcoxon rank-sum test. It shows comparison of the R3iΔEC and control datasets of lung tissue. Data S17 shows DE genes calculated with the Wilcoxon rank-sum test. It shows comparison of the R1R2iΔEC and control datasets of lung tissue. Data S18 shows DE genes calculated with the Wilcoxon rank-sum test. It shows comparison of the R2R3iΔEC deletion and control datasets of lung tissue. Data S19 shows DE genes calculated with the Wilcoxon rank-sum test. It shows comparison of the R1R2R3iΔEC and control datasets of lung tissue. Data S20 shows rankings of markers of dynamical RNA velocity calculated using the scVelo scv.tl.rank_dynamical_genes() function. This table includes velocity markers for the single and compound deletions of Vegfr1, Vegfr2, and Vegfr3 and their controls of heart tissue. Data S21 shows rankings of markers of stochastic RNA velocity calculated using the scVelo scv.tl.rank_velocity_genes() function. This table includes velocity markers for the single and compound deletions of Vegfr1, Vegfr2, and Vegfr3 and their controls of heart tissue. Data S22 shows rankings of markers of dynamical RNA velocity calculated using the scVelo scv.tl.rank_dynamical_genes() function. This table includes velocity markers for the single and compound deletions of Vegfr1, Vegfr2, and Vegfr3 and their controls of lung tissue. Data S23 shows rankings of markers of stochastic RNA velocity calculated using the scVelo scv.tl.rank_velocity_genes() function. This table includes velocity markers for the single and compound deletions of Vegfr1, Vegfr2, and Vegfr3 and their controls of lung tissue.
Data availability
Processed and raw scRNAseq data are deposited in GEO under accession no. GSE185823. The web application framework R Shiny v1.6.0 was used to create an application for easy exploration of cluster-specific gene expression across conditions in the scRNAseq datasets (Chang et al., 2021). The application is employed via a virtual server maintained by University of Helsinki IT services and is available at http://combodel.it.helsinki.fi.
Acknowledgments
We would like to thank the Biomedicum Molecular Imaging Unit, Genome Biology Units for microscopy and slide scanning services, Institute for Molecular Medicine Finland (FIMM) Single Cell Analytics (SCA) unit for the scRNAseq services (all supported by Biocenter Finland, Helsinki, Finland), HiLife Flow Cytometry Unit at Biomedicum for assistance with FACS experiments, and University of Helsinki IT services. We gratefully acknowledge Mari Jokinen, Katja Salo, Yelin Subasi, Alicia Prapotnich, Akseli Bonsdorff, Tapio Tainola, and the personnel of the Laboratory Animal Center of the University of Helsinki for excellent technical assistance. The graphical abstract was created using BioRender.
This work was supported by the Jenny and Antti Wihuri Foundation, the Academy of Finland (grants 307366, 314498, and 335721), the Finnish Brain Foundation, the European Research Council under the European Union’s Horizon 2020 research and innovation program (grant agreements 743155 and 874708), the Novo Nordisk Foundation (NNF16OC0023554), the Sigrid Jusélius Foundation, and the Cancer Foundation Finland (all to K. Alitalo); the Orion Research Foundation, the Finnish Foundation for Cardiovascular Research, and the University of Helsinki (to S. Karaman); and the Mary and Georg C. Ehrnrooth Foundation (to S. Paavonsalo). S. Karaman was supported by grants from the Swiss National Science Foundation (Advanced Postdoc.Mobility grant P300PB_164732), the Maud Kuistila Memorial Foundation, and the Academy of Finland (grant 330053). S. Paavonsalo was supported by the the Instrumentarium Foundation, the Mary and Georg C. Ehrnrooth Foundation, the Onni and Hilja Tuovinen Foundation, the Emil Aaltonen Foundation, and the Waldemar von Frenckell Foundation. K. Heinolainen was supported by the Ida Montini Foundation, the Biomedicum Helsinki Foundation, the K. Albin Johansson Foundation, and the Maud Kuistila Memorial Foundation. M.H. Lackman was supported by the Nylands Nation, the University of Helsinki, and the Swedish Student Foundation.
Author contributions: S. Karaman designed, performed, supervised, and analyzed experiments, prepared figures, provided funding, and wrote the manuscript; S. Paavonsalo performed and analyzed experiments, prepared figures, and wrote the Materials and methods; K. Heinolainen designed, performed, and analyzed experiments; M.H. Lackman analyzed scRNAseq data, prepared figures, generated the online data exploration tool, and wrote the Materials and methods; A. Ranta performed and analyzed experiments; K.A. Hemanthakumar contributed to the design of experiments and isolated cardiac ECs; Y. Kubota provided mice and advice; and K. Alitalo designed and supervised the work, provided funding, and wrote the manuscript.
References
Competing Interests
Disclosures: The authors declare no competing interests exist.
Author notes
S. Paavonsalo and K. Heinolainen contributed equally to this paper.
K. Heinolainen’s present address is Novartis Finland Oy, Espoo, Finland.



![Endothelial VEGFR2 deletion causes widespread vessel regression in postnatal mice.(A) Quantification of Vegfr transcripts in different tissues after 3 d of deletion in R1R2R3iΔEC pups and littermate controls (n = control [CTRL] = 5, R1R2R3iΔEC = 8 mice pooled from three independent experiments were used for the data shown). Vegfrs are normalized to 36b4 and Cdh5 transcripts. (B) Experimental setup. (C) Pup weights on P10. (D) Spleen and heart weights normalized to body weight. (E) Quantification of Vegfr transcripts normalized to 36b4 and Cdh5 transcripts; and Cdh5 and Pecam1 transcripts normalized to 36b4 transcripts. (F) Western blots and quantifications of VEGFRs normalized to HSC70 and VE-Cadherin (VEC) in lung lysates. (G) Staining and quantification of blood vessels (EMCN, red) in kidneys and glomeruli (insets). Scale bar = 100 µm, inset = 20 µm. (H) Staining and quantification of blood vessels (PODXL, red) in the liver and (I) pancreas; dashed lines mark Langerhans islets. (J) Intestinal whole-mounts stained for blood (CD31, green) and lymphatic (LYVE1, red) vessels and their quantification. (K) EC staining with isolectin B4 (iB4, white) in retinal whole-mounts and their quantification. A, arteries; V, veins. Only the superficial plexus is shown. Scale bar = 500 µm, inset = 200 µm. (L) Tracheal whole-mount staining and quantification of blood and lymphatic vessels (CD31, green; LYVE1, white). EMCN, endomucin; PODXL, podocalyxin. Error bars = mean ± SEM (n = control [CTRL] = 19, R2iΔEC = 14 mice pooled from four independent experiments were used for the data shown in the panels). Statistical significance was determined using Student’s t test (two-tailed, unpaired). *, P < 0.05; **, P < 0.01; ***, P < 0.001. BV, blood vessel; LV, lymphatic vessel. Source data are available for this figure: SourceData F1.](https://cdn.rupress.org/rup/content_public/journal/jem/219/3/10.1084_jem.20210565/2/m_jem_20210565_fig1.png?Expires=1773732510&Signature=WL-TU0GSW14OgCXM~wcCB4SBVrqerZ4TnL0El9g4gFeJ5uDQcwa3LOYimr8OSJcViwJuqbiYTEI5b9nGUlZhU46O8j06SoTkxKT~YkR5Ec8PjpYu6t5EieJzuKZPldjcTAgtXGlnV4MHPiY39fm95b2~S~UR~y3LGqbPrBmEDhcmDiw1Ax5MSZ0fwH4pLpfumnkcNDTogpcgZaB5sB0JmUcUU1URAgktspsc3sKbEtaGxJeqpNXhqNplGzSlpbKtqw7RtK~p86kezlX0Nbi1h6alQWkd3zY1lddWI3rZ4xFxPPgbquN2IBh8mRN5HXqw~yubWw3WdexDehnjLLJYGQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![VEGFR1 deletion aggravates vessel regression induced by VEGFR2 loss.(A) Experimental setup. (B) Pup weights on P10. (C) Spleen and heart weights normalized to body weight. (D) Quantification of Vegfr transcripts normalized to 36b4 and Cdh5 transcripts; and Cdh5 and Pecam1 transcripts normalized to 36b4 transcripts. (E) Western blots and quantifications of VEGFRs normalized to HSC70 and VE-Cadherin (VEC) in lung lysates. (F) Staining and quantification of blood vessels (EMCN, red) in kidneys and glomeruli (insets). Scale bar = 100 µm, inset = 20 µm. (G) Staining and quantification of blood vessels (PDXL, red) in the liver and (H) pancreas; dashed lines mark Langerhans islets. (I) Intestinal whole-mounts stained for blood (CD31, green) and lymphatic (LYVE1, red) vessels and their quantification. (J) EC staining with isolectin B4 (iB4, white) in retinal whole-mounts and their quantification. A, arteries; V, veins. Only the superficial plexus is shown. Scale bar = 500 µm, inset = 200 µm. (K) Tracheal whole-mount staining and quantification of blood and lymphatic vessels (CD31, green; LYVE1, white). EMCN, endomucin; PODXL, podocalyxin. Error bars = mean ± SEM (n = control [CTRL] = 21, R1R2iΔEC = 17 mice pooled from five independent experiments were used for the data shown in the panels). Statistical significance was determined using Student’s t test (two-tailed, unpaired). *, P < 0.05; **, P < 0.01; ***, P < 0.001. BV, blood vessel; LV, lymphatic vessel. Source data are available for this figure: SourceData F2.](https://cdn.rupress.org/rup/content_public/journal/jem/219/3/10.1084_jem.20210565/2/m_jem_20210565_fig2.png?Expires=1773732510&Signature=Pkd893rzds0ysLtpuGbEFAwr~kew4hXwxhCJPMMnlPbyfxmpNflWlt6PdeJ65RA2T-V~gKEhd7kakPOBmiSOlglYX3Khi7DZJ7uRYwzeAfkpq-BqWXD8P2vGo3A5ihFRtQnHJl4s0YT7-N8xlwCDLqUt5wGOIwFRCAqrTy0J8fYSp8KOADW-JsSlhTT4Da-dqC4VJQRnc8uBVG6msdBSHEsCvyi8EaWTJEoUxmO2LphG5z1OsGidZ0lFCjfl4DntOiJuPLQG~Ep8K72zd87o3GDMgCDYZuA2gmxskj9Zlga~4kvF1MN6BBpelmhHAGC-bDpLisP81b0JMnZ-cGmIsQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
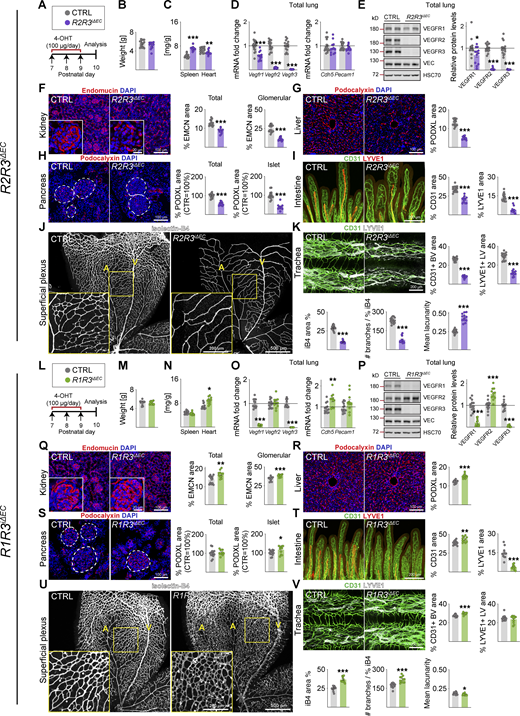
![VEGFR1/2/3 compound deletion results in the most severe regression of blood and lymphatic vessels in postnatal mice.(A) Experimental setup. (B) Pup weights on P10. (C) Spleen and heart weights normalized to body weight. (D) Quantification of Vegfr transcripts normalized to 36b4 and Cdh5 transcripts; and Cdh5 and Pecam1 transcripts normalized to 36b4 transcripts. (E) Western blots and quantifications of VEGFRs normalized to HSC70 and VE-cadherin (VEC) in lung lysates. (F) Staining and quantification of blood vessels (EMCN, red) in kidneys and glomeruli (insets). Scale bar = 100 µm, inset = 20 µm. (G) Staining and quantification of blood vessels (PDXL, red) in the liver and (H) pancreas; dashed lines mark Langerhans islets. (I) Intestinal whole-mounts stained for blood (CD31, green) and lymphatic (LYVE1, red) vessels and their quantification. (J) EC staining with isolectin B4 (iB4, white) in retinal whole-mounts and their quantification. A = arteries, V = veins. Only the superficial plexus is shown. Scale bar = 500 µm, inset = 200 µm. (K) Tracheal whole-mount staining and quantification of blood and lymphatic vessels (CD31, green; LYVE1, white). EMCN: endomucin; PODXL: podocalyxin. Error bars = mean ± SEM (n = control [CTRL] = 21, R1R2R3iΔEC = 20 mice pooled from seven independent experiments were used for the data shown in the panels). Statistical significance was determined using Student’s t test (two-tailed, unpaired). *, P < 0.05; ***, P < 0.001. BV, blood vessel; LV, lymphatic vessel. Source data are available for this figure: SourceData F3.](https://cdn.rupress.org/rup/content_public/journal/jem/219/3/10.1084_jem.20210565/2/m_jem_20210565_fig3.png?Expires=1773732510&Signature=xA1kItPhMnpw468d-d~8hApTY1oM811VZpcbV9Wb5a6ZcREPWreCUBHU-5bmVYebX3DEtcGHatEg7bkv~IUcLf3huhetr37QK6ifew3xp5~T50HBzDG17J5u6P0dGB-lLxnI9bChCt9wO4pIYeVNzvu1DfgVq3GfXJ6u8hFv5bxmcms2j7Ule~7teF3uWCoE465cb0HGUbAIQhBDMY-9Tr6R92Ks6jDuxU~3j~27Q2chZO1kMOz5dtbO7NEncPXhuF508Ujfmne55G5zbTzylvNdjvS3VcZUAimxZs5T5IQ7AyyKvGvvDDgFVWUBzUGym0T~ZbRkitS7ZzDY75OgvA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![EC apoptosis contributes to vessel regression.(A) Experimental setup. (B and C) iB4/ERG/cCasp3 triple staining and quantification in P10 retinas of the R1R2R3iΔEC pups compared with control littermates. Arrowheads indicate cCasp3/ERG double-positive endothelial nuclei. Error bars = mean ± SEM (data from n = control [CTRL] = 4, R1R2R3iΔEC = 9 mice pooled from three independent experiments). Statistical significance was determined using Student’s t test (two-tailed, unpaired). *, P < 0.05; **, P < 0.01; ***, P < 0.001.](https://cdn.rupress.org/rup/content_public/journal/jem/219/3/10.1084_jem.20210565/2/m_jem_20210565_fig4.png?Expires=1773732510&Signature=u4uoHJMDpyNyIyp6zg5p505Nn5eQz6QPU1cLNc81v3BNo8C6jh1qDsEt5nL0B-w8Da0TwbxG3XLT1iZJ8kes-XsR1xIhLohJrM7hMvMLO1v1sfyx~xxZmyvuMUBXxSBYwG7JXg3Ek0cYfJrIlgO7x8fX~GLOyqkr5ZUhd5cJaEYfv5JkVVMUhUfjIA2MHuRuU9KmkHZiOSCqIEhxB2Tq9kuM1Vil8BgbdtSyBt5mL5eoesJE1K-0odiWts~WXKu~hguiTWd3jr4kqYJ1WZ85i2vx~HH3InA-evi7xe8-kcjCk7SN64nJCPx6A8kFobB3sbvcE8uPzhzgCOe7bWPNoA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![VEGFR deletion levels in various tissues of adult mice.(A) qPCR analysis of Vegfr1, Vegfr2, and Vegfr3 mRNA levels on day 8 after three doses of tamoxifen in various tissues and their isolated EC populations from R1R2R3iΔEC mice and their control littermates (control [CTRL] = 3, R1R2R3iΔEC = 4 mice pooled from two independent experiments). (B) Schematic of the VEGFR deletion in adult mice. (C) Mouse weights after 8 d of gene deletion. (D) Representative Western blots and corresponding protein densitometry (total of 285 mice shown). (E) quantification of VEGFR levels in lung lysates. (F) qPCR analysis of Vegfr mRNA levels in lung, liver, and kidney lysates from the same cohorts. VEGFR1: CTRL = 21, R1iΔEC = 20 mice pooled from six independent experiments; VEGFR2: CTRL = 21, R2iΔEC = 20 mice pooled from five independent experiments; VEGFR3: CTRL = 16, R3iΔEC = 16 mice pooled from five independent experiments; VEGFR1/VEGFR2: CTRL = 19, R1R2iΔEC = 22 mice pooled from six independent experiments; VEGFR1/VEGFR3: CTRL = 20, R1R3iΔEC = 24 mice pooled from five independent experiments; VEGFR2/VEGFR3: CTRL = 14, R2R3iΔEC = 11 mice pooled from four independent experiments; and VEGFR1/VEGFR2/VEGFR3: CTRL = 31, R1R2R3iΔEC = 30 mice pooled from nine independent experiments. Statistical significance was determined using Student’s t test (two-tailed, unpaired) with Welch’s correction in cases of unequal variance. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Source data are available for this figure: SourceData FS3.](https://cdn.rupress.org/rup/content_public/journal/jem/219/3/10.1084_jem.20210565/2/m_jem_20210565_figs3.png?Expires=1773732510&Signature=FNHpSMBKtUV~G-QL12fn9NLHAnSv5Sxs1kFXJlmOaOP~1fscIqt~Pysq7biW3TfvBpxkB~MY1hnHoRLJtzcJncjHwyTfB7vOiP1mLZ0JTL-BSpy~STPT~AYAxsClNomLlNBnWE4GpZqxihTI1jyNHgOwdaffN5wi8DqG4NtK4nyLU4AOn~Io2-FyiBeViDmV1y0iA9s5ABjn41KJSkWoosmADcR-mKoEI-z5-ee--LXl4kZgMvIV5n4jPM1g4KCHa1M6La4k65opXB8kLKlgoTvEzTEzJ~wTICVEXaF9ueZkEqBbVp-gyKZ6DUw8vxOZqRga9~N7clvRyRc4TRqyXQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Organotypic effects of VEGFR deletions in quiescent adult vasculature.(A–F) Representative confocal micrographs of vascular staining of (A) liver (podocalyxin, red), (B) intestine (podocalyxin, green), (C) kidney (endomucin, red), (D) pancreas (podocalyxin, red), (E) trachea (podocalyxin, green), and (F) retina (isolectin B4 [iB4], white) tissues from adult mice deleted of single, double, or triple combinations of Vegfrs. The corresponding quantifications are shown in Fig. S4. Insets in C show close-up images of kidney glomeruli; Langerhans islets are demarcated by the dashed lines in D. Scale bars = 200 μm (B, E, and F), 100 μm (A, C, and D), and 50 μm (insets in C). Deep p., deep plexus.](https://cdn.rupress.org/rup/content_public/journal/jem/219/3/10.1084_jem.20210565/2/m_jem_20210565_fig5.png?Expires=1773732510&Signature=oYPxSPSDblGir~KdIMj-dXGW1pwLupw7TmDogJiVNSNvvI3irj1MYxGXiqRU49QajAXbhSXTDlTaHfySO8L2cgdQ7DzlSFHC0146txu4W9a2lsd7dcgHl2TYdUDekSU5gg9J7p1rag-CxNUahA8K0Gm3lB593FI7~03UtBOtFVUO7Ur0Ji0Z24B2psTCkLMGPaliftPK-52DeIDeFMZwSlbvD06rjUxFRuRaPC6nxLf1yZGxVj4L4txhq0XRDWJ1eGLeayAOlCR2C42NKucNoAkl~fsmAos5jrXEdudgCVxvspu3hSa1KuKu9iELSLkhhDj12zJ~KEUed2Sra8ciPw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Effect of VEGFR deletion on vascular density in various tissues in adult mice. Data in addition to Fig. 5. (A) Representative confocal micrographs of whole-mounted ears stained for CD31 (green, all vessels), and LYVE1 (white, lymphatic vessels). Scale bars = 200 μm. (B) Vessel density quantifications from the ear whole-mounts in A. (C–H) Vascular density quantifications from liver (C), intestine (D), kidney (E, total and glomerular [Glom.]), pancreas (F), trachea (G, blood and lymphatic vessels), and superficial (Sup.) and deep plexus in the retina (H). The heatmap in Fig. 6 provides comparison of the corresponding percent changes in the vascular densities. VEGFR1: control (CTRL) = 14–21, R1iΔEC = 14–20 mice pooled from four to six independent experiments; VEGFR2: CTRL = 10–21, R2iΔEC = 10–20 mice pooled from three to five independent experiments; VEGFR3: CTRL = 9–16, R3iΔEC = 7–16 mice pooled from three to five independent experiments; VEGFR1/VEGFR2: CTRL = 16–19, R1R2iΔEC = 15–22 mice pooled from five or six independent experiments; VEGFR1/VEGFR3: CTRL = 17–20, R1R3iΔEC = 20–24 mice pooled from four or five independent experiments; VEGFR2/VEGFR3: CTRL = 14, R2R3iΔEC = 11 mice pooled from four independent experiments; and VEGFR1/VEGFR2/VEGFR3: CTRL = 12–31, R1R2R3iΔEC = 11–30 mice pooled from three to nine independent experiments. Statistical significance was determined using Student’s t test (two-tailed, unpaired) with Welch’s correction in cases of unequal variance. *, P < 0.05; **, P < 0.01; ***, P < 0.001. BV, blood vessel; LV, lymphatic vessel; EMCN, endomucin; PDXL, podocalyxin.](https://cdn.rupress.org/rup/content_public/journal/jem/219/3/10.1084_jem.20210565/2/m_jem_20210565_figs4.png?Expires=1773732510&Signature=MRzml7Dqx69mu9qsDxMACYEm0sSsNlQgMhSYuzHHPWN5bY-Htqc6fiSh1onHMngXjCdFJdafuhqLGUl1VggRKDT9IwAaKLtO2irD8karXeP1EmGu3eHgA71tTjfkJS3tRw5r3JcqZNzCwUosrYd-YCVOqZxQILT0A-539-XbD025iobxln6pa0RBYvWHz2NOWMIVqPNXIH8s2O9YZfGXW8C20vilyvZhg0CE0reFuXl5QDtlVJwbFFHxeeS7r3c8DwvQVf8a0KCwvcgNIds6mNwxNSM9DfucWV0jqlzTpr0cTPnFHPcLxGGimrZVpot4hMr3OTkk-fHmRSzjJF9PEg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Quantification of changes in vascular density after Vegfr deletions. Heatmap summary of percent changes in blood vascular densities in postnatal (P) and adult (A) mice quantified from single, double, and triple compound deletions of the VEGFRs in liver, intestine, kidney (total and glomerular), pancreas (total and Langerhans islet–specific), trachea, retina (superficial and deep plexus), and heart tissues. Color scale indicates increased (red) or decreased (blue) vascular area; numbers indicate the percent change as compared with littermate controls. VEGFR1: control [CTRL] = 11–21, R1iΔEC = 10–20 mice pooled from four to six independent experiments; VEGFR2: CTRL = 10–21, R2iΔEC = 10–20 mice pooled from three to five independent experiments; VEGFR3: CTRL = 9–16, R3iΔEC = 7–16 mice pooled from three to five independent experiments; VEGFR1/VEGFR2: CTRL = 10–19, R1R2iΔEC = 12–22 mice pooled from three to six independent experiments; VEGFR1/VEGFR3: CTRL = 13–20, R1R3iΔEC = 15–24 mice pooled from three to five independent experiments; VEGFR2/VEGFR3: CTRL = 11–14, R2R3iΔEC = 9–11 mice pooled from three or four independent experiments; and VEGFR1/VEGFR2/VEGFR3: CTRL = 12–31, R1R2R3iΔEC = 11–30 mice pooled from three to nine independent experiments. Asterisks indicate statistical significance (Student’s t test; two-tailed, unpaired). *, P < 0.05; **, P < 0.01; ***, P < 0.001.](https://cdn.rupress.org/rup/content_public/journal/jem/219/3/10.1084_jem.20210565/2/m_jem_20210565_fig6.png?Expires=1773732510&Signature=fkMlC3tWW~9STqvWrEq8HZ18UXndeN1GfRyv51kML0G9TsM78lpfhJhpm87IgBRvWk6yrditsZ6Rc2OYIc2F-7YQ20nPx0ltJIIkfO~xIiVsyqLBqvT66Wcg2~RAmOwDAtg76nAVvL5v0nrqlR4CUfM8Z15RTL-cz~uwx2TaJ5QyuoWXb6Z9GLkxhtc1ezeV7mW2gU2z4zp7l120Tz2uNEpuXjQiZwv5Y6WtHORk-gn1X2VbMzixmd45QXx0P2xjexoXUEsj4Mroi85c30AH8whkmuZwoRB6YIR9aStacvbWDaVsv8Az~PILp6yfVl5sQyazvFu0Jk4jwsevDAKZ6A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)






