Two to three years after infection, a fraction of HIV-1–infected individuals develop serologic activity that neutralizes most viral isolates. Broadly neutralizing antibodies that recognize the HIV-1 envelope protein have been isolated from these patients by single-cell sorting and by neutralization screens. Here, we report a new method for anti–HIV-1 antibody isolation based on capturing single B cells that recognize the HIV-1 envelope protein expressed on the surface of transfected cells. Although far less efficient than soluble protein baits, the cell-based capture method identified antibodies that bind to a new broadly neutralizing epitope in the vicinity of the V3 loop and the CD4-induced site (CD4i). The new epitope is expressed on the cell surface form of the HIV-1 spike, but not on soluble forms of the same envelope protein. Moreover, the new antibodies complement the neutralization spectrum of potent broadly neutralizing anti-CD4 binding site (CD4bs) antibodies obtained from the same individual. Thus, combinations of potent broadly neutralizing antibodies with complementary activity can account for the breadth and potency of naturally arising anti–HIV-1 serologic activity. Therefore, vaccines aimed at eliciting anti–HIV-1 serologic breadth and potency should not be limited to single epitopes.
A significant number of HIV-1–infected individuals develop serum antibodies that neutralize large numbers of viral variants at low concentrations (Sather et al., 2009; Simek et al., 2009; Stamatatos et al., 2009; Walker et al., 2010; Doria-Rose et al., 2010; Gray et al., 2011; Mikell et al., 2011). In the few instances where these antibody responses have been characterized at a monoclonal level, the serologic activity seems to result from the combination of different antibodies (Scheid et al., 2009a; Bonsignori et al., 2012) or from a single broad neutralizing antibody clone that targets either the CD4 binding site (CD4bs), the variable loops, the membrane-proximal external region (MPER) or a carbohydrate-dependent epitope (Walker et al., 2009, 2011; Wu et al., 2010; Bonsignori et al., 2011; Moir et al., 2011; Morris et al., 2011; Scheid et al., 2011; Overbaugh and Morris, 2012).
Whereas many broadly neutralizing anti-CD4bs antibodies were obtained by single-cell sorting methods, which directly identified HIV-1–reactive B cells (Scheid et al., 2009a,b, 2011; Wu et al., 2010), broad neutralizing antibodies to carbohydrate-dependent epitopes were identified and cloned by screening directly for neutralizing activity (Walker et al., 2009, 2011). Among the latter, the antibodies PG9 and PG16 stand out because although they target the gp160 HIV-1 spike, they preferentially bind to this glycoprotein when it is expressed on a cell or viral membrane (Walker et al., 2009). Therefore, it has been proposed that PG9 and PG16 target an epitope found preferentially on the HIV-1 envelope protein expressed on cell membranes (Walker et al., 2009). Potent neutralizing antibodies to such epitopes would be difficult to obtain by single B cell sorting with soluble protein baits and to date have only been obtained by functional screens (Walker et al., 2009, 2011; Bonsignori et al., 2011). Nevertheless, serologic studies indicate that antibodies targeting quaternary epitopes may comprise a significant proportion of serum-neutralizing activity (Walker et al., 2010).
To facilitate the cloning of anti–HIV-1 antibodies directed at epitopes expressed on the cell surface form of the HIV-1 envelope spike, we developed a single B cell sorting method that uses gp160ΔcBaL-expressing cells as bait. Here, we report on the results of the cloning experiments, which revealed a broad neutralizing antibody to a novel conformational epitope that complements the activity of a potent anti-CD4bs antibody identified in the same individual (Scheid et al., 2011).
Results
Using cell surface–expressed gp160ΔcBaL to identify HIV-1–neutralizing antibodies
To determine whether HIV-1–neutralizing activity correlates with antibody binding to the HIV-1 spike expressed on cell surfaces, we used 293T cells expressing GFP and the HIV-1BaL envelope protein gp160 lacking the cytoplasmic tail (Δc; pMX-gp160ΔcBaL-IRES-GFP referred to as GFP-293TBaL; Pietzsch et al., 2010). We measured the binding of 51 monoclonal antibodies (Scheid et al., 2009a; Mouquet et al., 2011) and 81 polyclonal IgG samples purified from serum of HIV-1–infected volunteers (Fig. S1 A) to GFP-293TBaL by flow cytometry. There was a significant correlation between the reactivity of monoclonal antibodies with GFP-293TBaL and their neutralizing activity against the BaL.26 virus (rho = −0.458; P = 0.0074; Fig. 1 A). Similarly, polyclonal IgG binding to GFP-293TBaL was correlated with the amount of serologic neutralizing activity in the patients (rho = −0.505; P = 0.0004; Fig. 1 B and Fig. S1 A).
Binding and adsorption of HIV-1–reactive antibodies and purified IgGs to GFP-293TBaL cells. (A) Dot plots show mean fluorescence intensity (MFI) of staining by 51 HIV-1–reactive mAbs (y axis) tested on gp160ΔcBaL-expressing 293T cells versus IC50 measured in TZM-bl assay using the BaL.26 pseudovirus. On the left, antibodies are grouped into those with no or low neutralizing activity (IC50 > 50 µg/ml, black), and those that neutralize (IC50 <50 µg/ml, red). On the right, the IC50 values for neutralizing antibodies (IC50 < 50 µg/ml) are shown. P-values were determined using the Mann-Whitney U test (left); correlation (right) was analyzed by Spearman correlation coefficient (rho). (B) As in A using IgG purified from the serum of 81 HIV-1–infected patients. (C) Graphs show the neutralization activity against BaL.26 of IgG from 4 patients (3B, 7A, 8A, and C69) after adsorption with 293T cells transfected with plasmids encoding gp160ΔcBaL (blue) or no-insert (green). Antibody binding to gp160ΔcBaL-expressing 293T cells (A and B) was measured in at least two independent experiments and one representative dataset is shown. Neutralization activity of adsorbed patients’ IgGs was analyzed in duplicate.
Binding and adsorption of HIV-1–reactive antibodies and purified IgGs to GFP-293TBaL cells. (A) Dot plots show mean fluorescence intensity (MFI) of staining by 51 HIV-1–reactive mAbs (y axis) tested on gp160ΔcBaL-expressing 293T cells versus IC50 measured in TZM-bl assay using the BaL.26 pseudovirus. On the left, antibodies are grouped into those with no or low neutralizing activity (IC50 > 50 µg/ml, black), and those that neutralize (IC50 <50 µg/ml, red). On the right, the IC50 values for neutralizing antibodies (IC50 < 50 µg/ml) are shown. P-values were determined using the Mann-Whitney U test (left); correlation (right) was analyzed by Spearman correlation coefficient (rho). (B) As in A using IgG purified from the serum of 81 HIV-1–infected patients. (C) Graphs show the neutralization activity against BaL.26 of IgG from 4 patients (3B, 7A, 8A, and C69) after adsorption with 293T cells transfected with plasmids encoding gp160ΔcBaL (blue) or no-insert (green). Antibody binding to gp160ΔcBaL-expressing 293T cells (A and B) was measured in at least two independent experiments and one representative dataset is shown. Neutralization activity of adsorbed patients’ IgGs was analyzed in duplicate.
To determine whether the neutralizing activity could be depleted by cell surface–expressed gp160ΔcBaL, we adsorbed total IgG purified from the serum of four selected patients with high titers of broadly neutralizing serum activity on GFP-293TBaL cells (Fig. 1 C; and Fig. S1, B and C). To this end, purified IgGs were incubated for 20 min with GFP-293TBaL or GFP-293Tempty (negative control) cells followed by centrifugation and removing and testing the supernatant. In all four patients, neutralizing activity against BaL.26 was nearly fully adsorbed with GFP-293TBaL but not GFP-293Tempty (Fig. 1 C). We conclude that GFP-293TBaL cells express an HIV-1 spike protein that is recognized by broadly neutralizing serum antibodies.
Isolation and analysis of single HIV-1–reactive B cells captured by GFP-293TBaL cells
To determine whether GFP-293TBaL cells can be used to clone anti–HIV-1 antibodies, we performed sorting and cloning experiments of B cells captured on gp160ΔcBaL-expressing 293T cells (Fig. 2 A). PBMCs from HIV-1–infected individuals were mixed with GFP-293TBaL cells and incubated at 4°C for 40 min before staining for CD20 and IgG to detect memory B cells. The mixture was sorted for live cells (DAPI negative) expressing GFP, CD20, and IgG, and antibody cloning was performed as described for protein baits (Fig. 2, A; Scheid et al., 2009a,b, 2011). Visualization by ImageStreamX identified B cell/GFP-293TBaL doublets (Fig. 2 A, right).
Antibodies cloned by single-cell sorting using cell surface–expressed gp160ΔcBaL. (A) Cell sorting strategy. Dot plot (left) shows percentage of doublets composed of GFP-293TBaL cells and CD20+ B cells from one HIV-1–infected donor (patient 3B). IgG-expressing B cell/GFP-293TBaL doublets (middle) were sorted into single wells and subjected to antibody cloning procedure. (right) ImageStreamX visualization of a PE-stained B cell (orange) attached or in close proximity to a larger GFP-positive gp160ΔcBaL-transfected 293T cell. (B) Pie charts depicting expansion of clonally related antibodies (colored) cloned from four HIV-1–infected individuals (3B, 7A, 8A, and C69). The numbers within the inner circles (top row) indicate the total number of IgH sequences analyzed. Percentages of clonally related sequences are shown in the middle row. Total numbers of antibody clones are displayed within the inner circle (bottom row) and expansion of clones are proportionally displayed in pie charts (bottom row). (C) mAbs were tested by FACS (y axis shows MFI) for binding to 293T cells transfected with GFP-harboring plasmids encoding gp160ΔcBaL (black bar), gp160ΔcYU2 (gray bar), or no-insert (white bar). All 15 antibodies that bound to gp160ΔcBaL are shown. PG16, b12 (positive controls), and mGO53 (negative control) were included. (D) The same antibody panel as in C was tested for binding to soluble HIV-1 proteins (gp140BaL and gp140YU2) by ELISA. Graphs show OD405nm (y axis) and antibody concentration in µg/ml (x axis). Binding analysis of generated antibodies to soluble protein (D) and cell surface–expressed gp160ΔcBaL/gp160ΔcYU2 (C) was at least performed in duplicates.
Antibodies cloned by single-cell sorting using cell surface–expressed gp160ΔcBaL. (A) Cell sorting strategy. Dot plot (left) shows percentage of doublets composed of GFP-293TBaL cells and CD20+ B cells from one HIV-1–infected donor (patient 3B). IgG-expressing B cell/GFP-293TBaL doublets (middle) were sorted into single wells and subjected to antibody cloning procedure. (right) ImageStreamX visualization of a PE-stained B cell (orange) attached or in close proximity to a larger GFP-positive gp160ΔcBaL-transfected 293T cell. (B) Pie charts depicting expansion of clonally related antibodies (colored) cloned from four HIV-1–infected individuals (3B, 7A, 8A, and C69). The numbers within the inner circles (top row) indicate the total number of IgH sequences analyzed. Percentages of clonally related sequences are shown in the middle row. Total numbers of antibody clones are displayed within the inner circle (bottom row) and expansion of clones are proportionally displayed in pie charts (bottom row). (C) mAbs were tested by FACS (y axis shows MFI) for binding to 293T cells transfected with GFP-harboring plasmids encoding gp160ΔcBaL (black bar), gp160ΔcYU2 (gray bar), or no-insert (white bar). All 15 antibodies that bound to gp160ΔcBaL are shown. PG16, b12 (positive controls), and mGO53 (negative control) were included. (D) The same antibody panel as in C was tested for binding to soluble HIV-1 proteins (gp140BaL and gp140YU2) by ELISA. Graphs show OD405nm (y axis) and antibody concentration in µg/ml (x axis). Binding analysis of generated antibodies to soluble protein (D) and cell surface–expressed gp160ΔcBaL/gp160ΔcYU2 (C) was at least performed in duplicates.
Whereas the efficiency of obtaining large groups of clonally related antibodies was high with protein baits (Scheid et al., 2009a, 2011; Mouquet et al., 2011), many fewer clones were found using the cell-based anti–HIV-1 B cell capture method (Fig. 2 B). In the 4 patients studied, we found that an average of 14% (1–26%) of all antibodies was clonally related from a total of 734 heavy chain sequences (Fig. 2 B). When tested for binding to GFP-293TBaL cells 15 antibodies (13 out of 25 from clones and 2 out of 12 from the single antibodies tested) showed specific reactivity, and 10 of those also bound to GFP-293TYU2 cells, but not to GFP-293Tempty (Fig. 2 C). Like the antibodies cloned using soluble protein baits, the majority of the antibodies obtained by cell-based capture were more mutated than average human IgG (38 mutations on average per IGVH gene segment; Table S1; Mouquet et al., 2011; Scheid et al., 2009a, 2011) and the majority were also polyreactive (9/16; Fig. S2; Mouquet et al., 2010). We conclude that cell-based anti–HIV-1 B cell capture can be used for antibody cloning, but at low efficiency. To determine whether the antibodies cloned by cell-based capture recognized soluble forms of the HIV-1 spike, we tested their binding to soluble proteins corresponding to those expressed on GFP-293TBaL and GFP-293TYU2 (BaL and YU2 gp140 and gp120) by ELISA and surface plasmon resonance (SPR). Only 4 of the 15 antibodies with reactivity to GFP-293TBaL or GFP-293TYU2 bound to soluble forms of BaL and or YU2, but these corresponded to high affinity interactions as measured by SPR (Fig. 2 D and Table S3). We conclude that most of the antibodies obtained by cell-based capture do not recognize the soluble form of the envelope proteins tested.
3BC176 and 3BC315 complement HIV-1–neutralizing activity in a patient carrying a highly potent anti-CD4bs antibody clone
To determine the HIV-1–neutralizing activity of the antibodies that showed binding to GFP-293TBaL, we performed TZM-bl neutralization experiments using a panel of 10 viral envelopes representing Tier 1–3 viruses from clades A, B, and C (Table S2; Seaman et al., 2010). Only 6 of the 15 antibodies tested showed neutralizing activity, and 3 of these bound to soluble protein (Table S2, Fig. 2 D, and Table S3). Of the antibodies with no measurable binding activity against the tested soluble proteins, 3BC176 and 3BC315 showed the greatest breadth and potency. These antibodies were tested on an extended panel of pseudoviruses representing most clades, but with an emphasis on viruses resistant to broadly neutralizing anti-CD4bs antibodies identified in the same patient (i.e., 3BNC117, 3BNC55; Fig. 3; Scheid et al., 2011). 3BC176 and 3BC315 showed significant potency and breadth on the expanded viral panel, neutralizing 25 of the 39 viruses tested (Fig. 3). However, the most impressive observation is that the spectrum of viruses neutralized complemented the viruses neutralized by the broadly neutralizing anti-CD4bs antibodies that had been obtained from the same HIV-1–infected individual using soluble protein bait (Scheid et al., 2011). 3BNC117 shows similar breadth, but greater potency than VRC01, neutralizing 90% of the viruses in a large panel of different HIV-1 isolates (Wu et al., 2010; Scheid et al., 2011). 10 of the 13 viruses in the selected panel of 36 that were not neutralized by 3BNC117 and its clonal relative 3BNC55 were neutralized by 3BC176 or 3BC315 (Fig. 3). Moreover, the combined neutralizing activity of the broadly neutralizing anti-CD4bs antibodies (3BNC117, 3BNC55) and the new antibodies reconstitutes the neutralizing activity of the total IgG obtained from patient 3B, with the exception of 1 out of the 36 viruses (Fig. 3). We conclude that patient 3B produced at least two separate clones of broad neutralizing antibodies that have a complementary spectrum of activity against different HIV-1 isolates.
Heat map showing neutralization activity of mAbs and total IgG isolated from patient 3B. The IC50 values for 3BC176, 3BC315, and total IgG on a panel of 39 viral strains comprising multiple clades compared with published data on the neutralization of the same viruses by 3BNC117 and 3BNC55 (Scheid et al., 2011). Top group displays viruses neutralized by both sets of antibodies (clone 1 and clone 2). Second and third group lists viruses that are neutralized only by 3BNC117 and 3BNC55 or only by 3BC176 and 3BC315, respectively. Three virus strains (bottom) were not neutralized by any of these antibodies. IC50 values are color-coded: IC50 < 0.1, red; IC50 between 0.1 and 1 µg/ml, dark orange; IC50 between 1 and 10 µg/ml, light orange; IC50 between 10 and 100 µg/ml, dark yellow; and IC50 > 100 µg/ml, yellow. IC50 values above the measured concentration, as well as viruses that were not determined (nd), are not highlighted. All neutralization assays were performed in duplicate. Additionally, 3BC176 and 3BC315 were reproduced and retested against a panel of 10 different virus strains, confirming the neutralizing activity.
Heat map showing neutralization activity of mAbs and total IgG isolated from patient 3B. The IC50 values for 3BC176, 3BC315, and total IgG on a panel of 39 viral strains comprising multiple clades compared with published data on the neutralization of the same viruses by 3BNC117 and 3BNC55 (Scheid et al., 2011). Top group displays viruses neutralized by both sets of antibodies (clone 1 and clone 2). Second and third group lists viruses that are neutralized only by 3BNC117 and 3BNC55 or only by 3BC176 and 3BC315, respectively. Three virus strains (bottom) were not neutralized by any of these antibodies. IC50 values are color-coded: IC50 < 0.1, red; IC50 between 0.1 and 1 µg/ml, dark orange; IC50 between 1 and 10 µg/ml, light orange; IC50 between 10 and 100 µg/ml, dark yellow; and IC50 > 100 µg/ml, yellow. IC50 values above the measured concentration, as well as viruses that were not determined (nd), are not highlighted. All neutralization assays were performed in duplicate. Additionally, 3BC176 and 3BC315 were reproduced and retested against a panel of 10 different virus strains, confirming the neutralizing activity.
Characteristics of the epitope targeted by 3BC176 and 3BC315
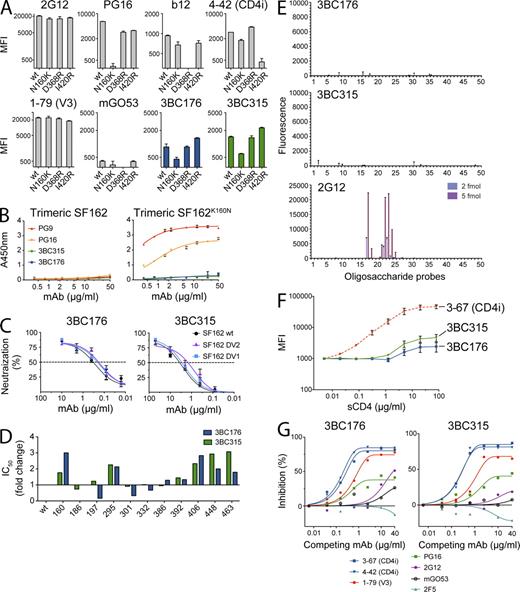
To examine the region of the HIV-1 spike targeted by 3BC176 and 3BC315, we initially tested the antibodies in a peptide ELISA with an overlapping set of peptides that cover all of gp120 (Mouquet et al., 2011). Moreover, we assayed binding by ELISA to gp41 and a series of additional monomeric (Q461.e2, Q259.d2, Q769.h5) and trimeric (Q461.e2, Q259.d2, Q769.h5, 349C, Q168.a2) HIV-1 envelope proteins. Neither 3BC176 nor 3BC315 showed significant binding to gp41, the peptides, or the HIV-1 envelope proteins that were partly reactive to PG9 and PG16 (Davenport et al., 2011; unpublished data). Because 3BC176 and 3BC315 did not bind to the tested soluble proteins, we tested both antibodies against mutants of surface-expressed gp160ΔcBaL carrying single mutations in the CD4bs (D368R), CD4i site (I420R) or in the glycosylation site in the V2 loop that alters the binding of PG9 and PG16-like antibodies (N160K). Whereas GFP-293TBaL-D368R, GFP-293TBaL-I420R, and GFP-293TBaL-N160K abolished binding of b12 (Burton et al., 1994), 4–42 (a CD4i antibody; Scheid et al., 2009a), and PG16 (Walker et al., 2009), respectively, these mutations had either no (D368R, I420R) or less pronounced (N160K) effects on 3BC176 and 3BC315 (Fig. 4 A). Although there is an effect of GFP-293TBaL-N160K on 3BC176 and 3BC315, the epitope recognized by these antibodies differs from PG16 in that they, unlike PG9 and PG16, fail to bind to soluble SF162K160N (Davenport et al., 2011; Fig. 4 B). Furthermore, 3BC176, 3BC315, and controls were assayed for neutralization of SF162 variants. Deletion of variable loops V1 and V2 from SF162 did not alter the neutralizing activity of 3BC176 or 3BC315 and therefore neither of these loops seems to play an essential role forming the actual epitope recognized by these antibodies (Fig. 4 C). We conclude that the epitope recognized by 3BC176 and 3BC315 differs from the epitopes that are recognized by traditional anti-V3 loop, anti-CD4bs, and anti-CD4i antibodies as well as PG9- and PG16-like antibodies. Moreover, 3BC176 and 3BC315 do not require the V1/V2 loops for neutralizing the HIV-1 strain SF162.
Characterization of the epitope recognized by 3BC176 and 3BC315. (A) MFIs of the antibodies 3BC176, 3BC315, and controls measured at 20 µg/ml on GFP-293TBaL wt and the indicated mutants (N160K, D368R, and I420R). (B) ELISA for binding to SF162 wt (left) and to SF162K160N mutant (right) by PG9, PG16, 3BC176, and 3BC315. Graphs show OD405 nm (y axis) and antibody concentration in µg/ml (x axis). (C) Pseudovirus neutralization measured in TZM-bl assay. Graphs show percent neutralization (y axis) by increasing concentrations of 3BC176 or 3BC315 (x axis) of wt SF162 and mutants lacking the V1 or the V2 loop (SF162 DV1 and SF162 DV2, respectively). (D) IC50 of 3BC176 and 3BC315 neutralization of SF162 wt and 11 SF162 pseudoviruses carrying single mutations at different glycosylation sites or the K160N mutatio.n Position of mutated glycosylation site (x axis) according to HXBc2. For both antibodies, the fold increase or decrease of the IC50 values are visualized. (E) Microarray analyses of 3BC176, 3BC315, and 2G12 binding using a set of 50 oligosaccharide probes as neoglycolipids. Table S4 gives designations of the oligosaccharide probes. The binding signals (fluorescence intensities) shown are the mean values of duplicate spots, printed at 2 and 5 fmol per spot (the error bars represent half of the difference between the two values). (F) Graph shows enhancement of Alexa Fluor 647–labeled antibody binding to GFP-293TBaL in the presence of sCD4. Staining intensity is measured by MFI (y axis), and the starting value normalized to MFI 1,000. 3–67 is a CD4-induced site (CD4i) antibody and was used as control (Scheid et al., 2011). (G) Inhibition of 3BC176 and 3BC315 binding to gp160 ΔcBaL-transfected 293T cells in the presence of sCD4 (10 µg/ml). Graphs show inhibition (in percentage) of Alexa Fluor 647–labeled 3BC176 or 3BC315 (y axis) in the presence of increasing concentrations of the indicated antibodies (x axis). All experiments were at least performed in duplicate, and results of representative experiments (D and G) are shown. Standard errors are shown for A–C and F.
Characterization of the epitope recognized by 3BC176 and 3BC315. (A) MFIs of the antibodies 3BC176, 3BC315, and controls measured at 20 µg/ml on GFP-293TBaL wt and the indicated mutants (N160K, D368R, and I420R). (B) ELISA for binding to SF162 wt (left) and to SF162K160N mutant (right) by PG9, PG16, 3BC176, and 3BC315. Graphs show OD405 nm (y axis) and antibody concentration in µg/ml (x axis). (C) Pseudovirus neutralization measured in TZM-bl assay. Graphs show percent neutralization (y axis) by increasing concentrations of 3BC176 or 3BC315 (x axis) of wt SF162 and mutants lacking the V1 or the V2 loop (SF162 DV1 and SF162 DV2, respectively). (D) IC50 of 3BC176 and 3BC315 neutralization of SF162 wt and 11 SF162 pseudoviruses carrying single mutations at different glycosylation sites or the K160N mutatio.n Position of mutated glycosylation site (x axis) according to HXBc2. For both antibodies, the fold increase or decrease of the IC50 values are visualized. (E) Microarray analyses of 3BC176, 3BC315, and 2G12 binding using a set of 50 oligosaccharide probes as neoglycolipids. Table S4 gives designations of the oligosaccharide probes. The binding signals (fluorescence intensities) shown are the mean values of duplicate spots, printed at 2 and 5 fmol per spot (the error bars represent half of the difference between the two values). (F) Graph shows enhancement of Alexa Fluor 647–labeled antibody binding to GFP-293TBaL in the presence of sCD4. Staining intensity is measured by MFI (y axis), and the starting value normalized to MFI 1,000. 3–67 is a CD4-induced site (CD4i) antibody and was used as control (Scheid et al., 2011). (G) Inhibition of 3BC176 and 3BC315 binding to gp160 ΔcBaL-transfected 293T cells in the presence of sCD4 (10 µg/ml). Graphs show inhibition (in percentage) of Alexa Fluor 647–labeled 3BC176 or 3BC315 (y axis) in the presence of increasing concentrations of the indicated antibodies (x axis). All experiments were at least performed in duplicate, and results of representative experiments (D and G) are shown. Standard errors are shown for A–C and F.
To investigate glycan dependency, we measured the neutralizing activity on variants of SF162 that carry mutations in glycosylation sites and the K160N mutation (Fig. 4 D). The mutations had only limited effects, indicating a minor role for the glycosylation sites tested in SF162 (Fig. 4 D). Finally, neither of the antibodies showed significant binding to any of the fifty oligosaccharide probes in an N-glycan microarray that were used to characterize glycan-dependent anti–HIV-1 antibodies (Pejchal et al., 2011), in contrast to 2G12, which gave strong binding to Man7(D1), Man8, and Man9 N-glycans (Fig. 4 E). We conclude that 3BC176 and 3BC315 are unlikely to recognize a glycan-dependent epitope.
To examine the potential dependence of 3BC176 and 3BC315 on CD4 binding, we measured their binding to GFP-293TBaL (FACS) or gp140BaL (ELISA) in the presence or absence of soluble CD4 (sCD4). Although less pronounced than for the CD4i antibody 3–67 (Scheid et al., 2009a), we found that sCD4 enhances the binding of both of the antibodies to GFP-293TBaL, but not to soluble gp140BaL (Fig. 4 F; ELISA not depicted). Like CD4, some anti-CD4bs antibodies are able to enhance binding of anti-CD4i antibodies to the HIV-1 envelope. However, the broad and potent anti-CD4bs antibodies cloned from patient 3B did not enhance binding of 3BC176 or 3BC315 to GFP-293TBaL (unpublished data). We conclude that the two antibodies recognize an epitope expressed on the cell surface form of gp160ΔcBaL, which is revealed in part or stabilized by CD4 binding (Fig. 4 F).
To uncover the region of the HIV-1 spike recognized by 3BC176 and 3BC315, we performed competition experiments with antibodies to the V3 loop (1–79), the CD4i site (3–67 and 4–42; Scheid et al., 2009a), the MPER (2F5; Purtscher et al., 1994), and glycan-dependent epitopes using PG16 (Walker et al., 2009) and 2G12 (Trkola et al., 1996). The antibody mGO53 does not react to HIV-1 envelope proteins and was used as a control (Wardemann et al., 2003). mGO53, 2G12, and 2F5 (Muster et al., 1993) did not have significant effects on 3BC176 or 3BC315 binding to GFP-293TBaL in the presence of sCD4. Limited effects were detected by competition with PG16 (Fig. 4 G), but the most potent blocking effects were seen with the two anti-CD4i antibodies (3–67 and 4–42), followed by the anti-V3 loop antibody (1–79; Fig. 4 G). This indicates that 3BC176 and 3BC315 recognize an HIV-1 spike epitope that is exposed in part by CD4 binding, and is in the vicinity of the V3 loop and the CD4i site, without being susceptible to the I420R mutation.
DISCUSSION
A significant fraction of the individuals infected with HIV-1 develop broadly neutralizing antibodies, but only after a prolonged period of infection (Sather et al., 2009; Simek et al., 2009; Stamatatos et al., 2009; Doria-Rose et al., 2010; Walker et al., 2010; Gray et al., 2011; Mikell et al., 2011). These antibodies do not resolve the infection, but exert selective pressure on the virus as indicated by the emergence of HIV-1 variants that are antibody resistant (Albert et al., 1990; Bunnik et al., 2008; Richman et al., 2003; Wei et al., 2003). Despite the viruses’ ability to avert the antibody response, broad HIV-1–neutralizing antibodies are of interest because they can prevent infection in nonhuman primates and have been shown to delay viral rebound (Mascola et al., 1999; Shibata et al., 1999; Mascola et al., 2000; Trkola et al., 2005; Hessell et al., 2009a,b). Additionally, the presence of envelope-reactive IgGs correlates with protection in the vaccination trial RV144 (Haynes et al., 2012).
Notwithstanding their potential importance, little was known about the molecular composition of the HIV-1–neutralizing response until recently (Stamatatos et al., 2009; Mascola and Montefiori, 2010; McMichael et al., 2010; Moir et al., 2011). The introduction of HIV-1–specific single-cell antibody cloning methods (Scheid et al., 2009a,b; Tiller et al., 2008) resulted in the cloning of dozens of antibodies, and today HIV-1–neutralizing antibodies with far greater potency and breadth are available (Walker et al., 2009, 2011; Corti et al., 2010; Wu et al., 2010; Bonsignori et al., 2011; Diskin et al., 2011; Scheid et al., 2011). Here, we report on a new antibody cloning method based on the use of cell surface–expressed HIV-1 envelope protein gp160ΔcBaL as bait. The cellular bait is far less efficient than the soluble protein bait; nevertheless, it captured unique antibodies that target an epitope that has not previously been associated with broad neutralization.
Although the recent HIV-1 antibody cloning experiments have uncovered broad and potent antibodies, they have not revealed new antibody target regions on the HIV-1 spike. For example, the recently identified broadly neutralizing anti-CD4bs antibodies, which are the most potent and broadest of the new antibodies to date (Wu et al., 2010; Diskin et al., 2011; Scheid et al., 2011), recognize the CD4bs, a well-known target of previously characterized, less potent antibodies (Stamatatos et al., 2009; Mascola and Montefiori, 2010; McMichael et al., 2010). In addition, PG9 and PG16 target epitopes on the variable loops (Walker et al., 2009; Davenport et al., 2011; McLellan et al., 2011) that are also seen by the strain-specific antibody 2909 (Gorny et al., 2005) and the antibodies CH1-CH4 (Bonsignori et al., 2011). The PGT antibodies (Walker et al., 2011) recognize the V3 loop, which has been studied extensively as a target for broadly neutralizing antibodies (Hioe et al., 2010). Similarly, although we have not pinpointed the precise target of 3BC176 and 3BC315, the epitope is likely to be in close proximity to the V3 loop and the CD4i site (Stamatatos et al., 2009; Mascola and Montefiori, 2010; McMichael et al., 2010). However, the epitope recognized by 3BC176 and 3BC315 differs from previously described epitopes. Unlike PG9, PG16, 2909, and CH01-CH04, 3BC176 and 3BC315 are able to neutralize HIV-1 strains that do or do not carry a glycosylation site at position 160 of the gp120 envelope molecule (HXBc2-numbering). Furthermore, 3BC176 and 3BC315 failed to recognize any of the panel of soluble envelope monomers or trimers tested. Further analysis may reveal soluble envelope proteins that react with 3BC176 and 3BC315, and these envelopes would be promising candidates to elicit 3BC176/3BC315-type antibodies by immunization.
Only a small number of patients have been studied in depth by single-cell antibody cloning methods, but the results reveal that there is significant heterogeneity in the antibodies responsible for anti–HIV-1 serologic activity (Scheid et al., 2009a, 2011; Walker et al., 2009, 2010, 2011; Corti et al., 2010; Wu et al., 2010, 2011; Bonsignori et al., 2011, 2012; Morris et al., 2011). The serologic activity in patient 3B appears to result from at least two different types of broadly neutralizing antibody clones to two different sites on the HIV-1 spike: one that targets the CD4bs and a second that targets a conformational epitope likely to be in the vicinity of the CD4i site. The anti-CD4bs antibodies from this patient, typified by 3BNC117 and 3BNC55, were obtained by single-cell sorting using soluble protein baits (Scheid et al., 2011).
Although patient 3B’s broadly neutralizing anti-CD4bs antibodies cover nearly 90% of all viruses initially tested, there are significant holes in the repertoire, many of which are common to other known broadly neutralizing anti-CD4bs antibodies (Scheid et al., 2011; Wu et al., 2011). 3BC176 and 3BC315 complement the spectrum of viruses neutralized by the broadly neutralizing anti-CD4bs antibodies. As a result 10 out of 13 of the anti-CD4bs antibody resistant viruses in the 39 virus panel tested were sensitive to 3BC176 and/or 3BC315.
Development of broad HIV-1–neutralizing antibodies is likely to involve iterative cycles of germinal center formation, antibody gene somatic mutation, and viral escape. This is evident from the appearance of viral escape mutants (Albert et al., 1990; Richman et al., 2003; Wei et al., 2003; Bunnik et al., 2008) and the exceedingly high levels of somatic hypermutation found on HIV-1–neutralizing antibodies (Corti et al., 2010; Wu et al., 2010, 2011; Scheid et al., 2011). In addition to the emergence of unique clones of broadly neutralizing antibodies to a single epitope, HIV-1 infection can also elicit broadly neutralizing antibodies to different epitopes with complementary breadth (Bonsignori et al., 2012). We would like to propose that vaccine strategies aimed at producing such antibodies should include the maximum possible number of epitopes recognized by such antibodies to optimize the opportunities for development of anti–HIV-1 antibody breadth and potency.
MATERIALS AND METHODS
Patient samples.
Serum samples (Fig. 1, A and B) were collected from HIV-1–infected individuals after signed informed consent and in accordance to the Institutional Review Board (IRB; protocol 09–281, University of Cologne, Germany). Based on broad serum-neutralizing activity (Fig. S1 C), patients 3, 7, and 8 were recruited from the Elite Controller Study of the Partners Aids Research Center and patient C69 from the University of Cologne, Germany. Participants were enrolled in the IRB-reviewed protocol MNU-0628 at The Rockefeller University to collect PBMCs for further analysis.
Antibodies and IgG.
51 HIV-1–reactive antibodies recognizing different envelope epitopes (CD4bs, CD4i, V3-loop, gp41) were selected from previous studies (Mouquet et al., 2011; Scheid et al., 2009a) based on available neutralization data for BaL.26. In addition, total IgG was purified from 81 serum samples of HIV-1–infected patients and tested for neutralization activity for BaL.26 (Fig. 1 B and Fig. S1).
Binding of total IgG or mAbs to GFP-293TBaL/YU2.
HEK 293T/17 (American Type Culture Collection; CRL11268) cells were grown to 70% confluency and transfected with pMX-gp160ΔcBaL-IRES-GFP (Pietzsch et al., 2010), pMX-IRES-GFP, and pMX-gp160ΔcBaL-IRES-GFP mutants (N160K, D368R, I420R; Fig. 4 A) or pMX-gp160ΔcYU2-IRES-GFP (Fig. 2 C; Scheid et al., 2011) using FuGENE 6 (Roche) at a 1:2 plasmid/FuGENE ratio. 36–48 h later, cells were washed with 1×PBS and detached with trypsin-free cell dissociation buffer (Invitrogen) and resuspended in 1×PBS containing 0.5% BSA and 2 mM EDTA. The transfected cells were stained with total IgGs or mAbs at 20 µg/ml for 25 min that were visualized using APC mouse anti–human IgG (BD) and analyzed using a BD FACSCalibur or LSR II. Mann-Whitney U test was used for comparison of neutralizing with non- or weakly neutralizing mAbs (IC50 > 50 µg/ml) or IgGs (IC50 >200 µg/ml). Correlation (rho) was calculated using nonparametric correlation (Spearman).
Adsorption on GFP-293TBaL cells.
HEK 293T/17 cells were transfected with pMX-gp160ΔcBaL-IRES-GFP or pMX-IRES-GFP as described in the previous paragraph. After 36 h, 5 × 107 GFP-293TBaL or GFP-293Tempty cells were collected in trypsin-free cell dissociation buffer (Invitrogen) and incubated with 500 µl of 1 mg/ml purified IgG (patients 3, 7, 8, and C69). Samples were incubated on ice for 20 min before centrifugation for 8 min at 1,500 rpm. Supernatants were removed and transferred to a second tube containing the same number of cells. This procedure was repeated 5 times followed by filtration through Amicon Ultra-0.5 ml Centifugal Filters (Millipore) and an IgG purification step using Protein G–Sepharose 4 Fast Flow (GE Healthcare). Recovered IgGs from adsorption on GFP-293TBaL and GFP-293Tempty were analyzed for gp120BaL contamination by SDS gel electrophoresis and silver staining, and neutralizing activity was measured in TZM-bl assay.
Sorting single B cells captured by GFP-293TBaL.
HEK 293T/17 cells transfected with pMX-gp160ΔcBaL-IRES-GFP were resuspended in 1×PBS containing 0.5% BSA and 10 mM EDTA and mixed with purified B cells (20–40 × 106) at a ratio of 1:1 (sample 3, 8, and 7) or 1:1.5 (patient C69). The mixture was subjected to a dual centrifugation step starting with 10 min at 1,250 rpm, followed by 30 min at 625 rpm. After spinning, the mix was resuspended gently and stained on ice with APC-H7 Mouse Anti–Human CD20 (BD), PE Mouse Anti–Human IgG (BD), and DAPI (Invitrogen). CD20+, IgG+, GFP+, and DAPI− doublets were sorted into 96-well PCR plates (Eppendorf) containing 4 µl lysis buffer (Tiller et al., 2008) on a FACSAria III cell sorter (BD).
mAb cloning, expression and sequence analysis.
Amplification of immunoglobulin gene segments, as well as data analysis, cloning and production of antibodies, was performed as previously described (Tiller et al., 2008; Scheid et al., 2009a,b, 2011; Mouquet et al., 2011).
ELISA.
Monoclonal antibodies were tested for binding to gp140BaL, gp140YU2 (plasmid provided by R. Wyatt, The Scripps Research Institute, La Jolla, CA), gp120YU2 (plasmid provided by J.R. Mascola, National Institutes of Health, Bethesda, MD), and gp41IIIB (ProSpec) as previously described (Mouquet et al., 2011). In brief, 96-well ELISA plates (Costar) were coated overnight at room temperature with 125 ng/well of purified protein in 1×PBS. Plates were blocked with PBST (0.1% Tween-20) and 2% BSA for 1 h, and antibodies were applied in a 1:3 serial dilution with a starting concentration of 8 µg/ml. Subsequently, plates were incubated with HRP-conjugated goat anti–human IgG (Jackson ImmunoResearch Laboratories) for 1 h. ABTS single solution (Invitrogen) was used for development, and optical density was measured at 405 nm. Between all steps, plates were washed 3× with distilled water.
To analyze binding of 3BC176 and 3BC315 to gp140BaL in the presence of sCD4, plates were coated with gp140BaL (125 ng/well) and sCD4 (Progenics) at a 1:1 molar ratio. ELISAs measuring binding of 3BC176 and 3BC315 to soluble monomeric and trimeric gp140 proteins derived from HIV-1 strains SF162 (Fig. 4 B), 349C, Q168.a2, Q259.d2, Q461.e2, and Q769.h5 were performed as previously described (Srivastava et al., 2003; Davenport et al., 2011). To determine polyreactivity, antibodies were tested for binding to ssDNA, dsDNA, insulin, and LPS as described previously (Wardemann et al., 2003; Mouquet et al., 2011). Antibodies were considered polyreactive when binding was detected for at least two of the four assayed antigens. Peptide ELISA was performed as previously described (Mouquet et al., 2006) using a gp120YU2 peptide library consisting of 94 overlapping peptides (Mouquet et al., 2011).
Competition experiment.
GFP-293TBaL cells were resuspended in FACS buffer (1×PBS, 0.5% BSA, 2 mM EDTA) at a concentration of 106 cells/ml. After 10 min, the antibodies 2G12, PG16, 2F5, 3–67, 4–42, and 1–79 or the control antibody mGO53 were added in eight 1:4 serial dilutions (starting concentration 40 µg/ml). After an incubation period of 15 min, cells were washed and stained for 25 min with Alexa Fluor 647–labeled 3BC176 and 3BC315. Cells were analyzed using an LSRFortessa cell analyzer (BD). All staining and washing steps were performed at 4°C.
Surface plasmon resonance.
Experiments were performed with a Biacore T100 (Biacore, Inc.) as previously described (Mouquet et al., 2010). In brief, gp140BaL and gp140YU2 were immobilized on CM5 chips (Biacore, Inc.) at immobilization levels of 100 RUs. IgGs and Fabs were injected through flow cells at 700 nM and 1,400 nM, respectively and 4 successive 1:2 dilutions in HBS-EP+ running buffer (Biacore, Inc.). Off-rate (kd (s−1)), on-rate (ka (M−1 s−1)), and binding constants KD (M) or KA(M−1) were calculated using the Biacore T100 Evaluation software (Table S3).
Neutralization assay.
Total IgG purified from serum of HIV-1–infected volunteers, as well as generated antibodies were tested for neutralizing activity as described earlier (Li et al., 2005; Montefiori, 2005; Saunders et al., 2005; Kraft et al., 2008). In brief, HIV-1-Env-pseudoviruses were mixed with serial dilutions of total IgG or antibodies and applied to TZM-bl cells for a single round of infection. Neutralization activity was determined by measuring reduction in luciferase reporter gene expression resulting in reduced relative luminescence units (RLUs) compared with the controls (Fig. 3, Fig. 4 C, Fig. 4 D, Fig. S1, and Table S2).
Carbohydrate microarray analysis.
Microarrays were generated by robotically printing glycan probes as neoglycolipids onto nitrocellulose-coated glass slides as previously described (Palma et al., 2006; Liu et al., 2012) at two levels (2 and 5 fmol/spot) in duplicate. The microarrays (designated N-glycan-related Array Set 1) containing 50 neoglycolipid probes were used for the analyses (Table S4); the sequences of the probes are as in Table S6 of Pejchal et al. (2011). The binding assays were performed as previously described (Pejchal et al., 2011). In brief, 3BC315 and 3BC176, and, as a control, 2G12 (Trkola et al., 1996; from Polymun Scientific) were precomplexed with biotinylated anti–human IgG (Vector Laboratories) at a 1:3 ratio, wt/wt, before applying onto the slides at a final concentration of 10 µg/ml, followed by Alexa Fluor 647–labeled streptavidin (Invitrogen). The analyses of 3BC176 and 3BC315 were performed in two separate experiments, and the results showed good reproducibility; those of one of the experiments in which antibody 2G12 was also analyzed are shown in Fig. 4 E.
Online supplemental material.
Fig. S1 shows the clinical information of patients whose serum or PBMC samples were used in experiments. Fig. S2 shows polyreactivity measured by ELISA for the 15 antibodies that demonstrated binding activity to GFP-293TBaL cells. Fig. S3 shows the amino acid sequences for 3BC176 and 3BC315 heavy and light chains. Immunoglobulin features and neutralization activity of generated antibodies that bind to surface-expressed gp160ΔcBaL are shown in Tables S1 and S2, respectively. The same set of antibodies was tested for binding to gp140BaL and gp140YU2 by using SPR (Table S3). Table S4 shows the 50 neoglycolipid probes that were used in the N-glycan-related Array Set 1.
Acknowledgments
We thank all HIV-infected volunteers who participated in this study. We also thank Klara Velinzon and Yelena Shatalina for cell sorting, and members of the Glycosciences Laboratory for their collaboration in the establishment of the neoglycolipid-based microarray system. We thank Mila Jankovic, Dr. Pamela Bjorkman, Dr. Ron Diskin, and the members of the Nussenzweig laboratory for helpful discussions; Florencia Pereyra and Gisela Kremer for patient coordination; and Christopher Bare from the RU Flow Cytometry Resource Center for helping to use ImageStreamX.
This research was supported by The Rockefeller University, the UK Research Council’s Basic Technology Initiative ‘Glycoarrays’ (GRS/79268), EPSRC Translational Grant (EP/G037604/1), the National Cancer Institute Alliance of Glycobiologists (U01CA128416), and the National Institutes of Health grant to M.C. Nussenzweig (NIH 1 PO1 AI081677). F. Klein was supported by the German Research Foundation (DFG, KL 2389/1-1). C. Gaebler was supported by The German National Academic Foundation. C. Lehmann and G. Fätkenheuer are supported by the German Federal Ministry of Education and Research (BMBF 01 KI 0771). M.S. Seaman and L. Morris were supported by the Bill and Melinda Gates Foundation’s Comprehensive Antibody Vaccine Immune Monitoring Consortium, grant number 1032144. M.C. Nussenzweig is a Howard Hughes Medical Institute investigator. M.C. Nussenzweig and F. Klein have a pending patent application for the antibodies 3BC176 and 3BC315 with the United States Patent and Trademark Office. These reagents are available with a Material Transfer Agreement.
The authors declare no conflicting financial interests.