Transcription factors play important roles in lymphopoiesis. We have previously demonstrated that Bcl11a is essential for normal lymphocyte development in the mouse embryo. We report here that, in the adult mouse, Bcl11a is expressed in most hematopoietic cells and is highly enriched in B cells, early T cell progenitors, common lymphoid progenitors (CLPs), and hematopoietic stem cells (HSCs). In the adult mouse, Bcl11a deletion causes apoptosis in early B cells and CLPs and completely abolishes the lymphoid development potential of HSCs to B, T, and NK cells. Myeloid development, in contrast, is not obviously affected by the loss of Bcl11a. Bcl11a regulates expression of Bcl2, Bcl2-xL, and Mdm2, which inhibits p53 activities. Overexpression of Bcl2 and Mdm2, or p53 deficiency, rescues both lethality and proliferative defects in Bcl11a-deficient early B cells and enables the mutant CLPs to differentiate to lymphocytes. Bcl11a is therefore essential for lymphopoiesis and negatively regulates p53 activities. Deletion of Bcl11a may represent a new approach for generating a mouse model that completely lacks an adaptive immune system.
Hematopoietic stem cells (HSCs) can both self-renew and differentiate to all blood cells (Spangrude et al., 1988) and are found in Lin−Sca1+Kit+ (LSK) fraction of BM cells. Generation of lymphocytes from HSCs is a stepwise process through multiple progenitors, including multipotent progenitors (MPPs), lymphoid-primed MPPs (LMPPs; Adolfsson et al., 2005), and common lymphoid progenitors (CLPs; Kondo et al., 1997). In the LSK compartment, MPPs and LMPPs have increased expression of the transmembrane receptor fms-like tyrosine kinase 3 (Flt3; Adolfsson et al., 2005), which is associated with progressive loss of potential to megakaryocyte-erythroid progenitors (MEPs; Adolfsson et al., 2005; Månsson et al., 2007). Two transcription factors, Pu.1 and Ikaros, are important for lymphoid lineage development in LMPPs (Yoshida et al., 2006; Ng et al., 2009; Carotta et al., 2010).
In the adult mouse, lymphocyte development from early progenitors occurs primarily in the BM for B cells or in the thymus for T cells. B cell development requires the interplay of transcription factors and external cues of the microenvironment (Nutt and Kee, 2007). Specification of the B cell lineage program and loss of alternative lineage potential require a network of transcription factors, including E2A (Tcf3), Ebf1, Pax5, and Foxo1 (Lin et al., 2010; Mandel and Grosschedl, 2010). E2A proteins are implicated at multiple stages of B cell development (Bain et al., 1994; Zhuang et al., 1994; Kwon et al., 2008) and have functions in LMPPs, early T cell progenitors (ETPs), and lineage priming (Dias et al., 2008). Deletion of Ebf1, Foxo1, and Pax5 also blocks early B cell development (Urbanek et al., 1994; Lin and Grosschedl, 1995; Dengler et al., 2008). Additional key regulators in B cell development include Myb, Lrf1, Miz1, Foxp1, and Mysm1 (Hu et al., 2006; Maeda et al., 2007; Greig et al., 2010; Kosan et al., 2010; Jiang et al., 2011). Similar to B cells, thymocyte development in the thymus can be divided into several stages, but requires a combination of transcription regulators including Notch1, Gata3, Bcl11b, Tcf1, and Lyl-1 (Liu et al., 2010; Rothenberg et al., 2010; Weber et al., 2011; Zohren et al., 2012).
Bcl11a encodes a C2H2 zinc finger transcription factor that was initially discovered as a retroviral insertion site (Evi9) in myeloid leukemia tumors in the BXH-2 mouse (Nakamura et al., 1996, 2000), and was subsequently found to be overexpressed in some B cell lymphomas caused by chromosomal translocations (Satterwhite et al., 2001). We previously found that a Bcl11a germline-null allele caused neonatal lethality in the homozygous mutant, and identified its essential role in fetal lymphocyte development with a complete absence of B cells in the fetal liver and abnormal T cell development in the fetal thymus (Liu et al., 2003b). Recent studies have implicated that Bcl11a is a potential target of E2A, Ebf1, and Foxo1, which links Bcl11a into the common B cell transcription regulation framework (Doulatov et al., 2010; Lin et al., 2010; Treiber et al., 2010). We thus aimed to investigate whether Bcl11a is required in adult lymphocyte development.
RESULTS
Bcl11a is expressed in both hematopoietic progenitors and differentiated cells
We determined Bcl11a expression at the single-cell level by making and analyzing an eGFP reporter mouse where an IRES-eGFP-FRT-Neo-FRT cassette was targeted to the 3′UTR of the Bcl11a locus (Fig. 1 A). The homozygous Bcl11aeGFP/eGFP mice had normal hematopoiesis and were used for detection of Bcl11a expression (GFP+) by flow cytometry.
Dynamic expression patterns of Bcl11a in hematopoiesis. (A) Schematic diagram of the Bcl11a-eGFP reporter allele. The eGFP reporter cassette flanked by two F3 sites is introduced to the 3′UTR region of Bcl11a, 8 bp after the stop codon TAG. (B) Flow cytometry tracking Bcl11a expression using the Bcl11aeGFP/eGFP reporter mice. Cell surface markers for defining these cells are described in Table S1. (C) Expression of Bcl11a (eGFP+) and Bcl11b (Tdtomato+) in double negative (DN) thymocytes was measured by flow cytometry. DN thymocytes are identified as described in Fig. S1 D. (D) qRT-PCR analysis of Bcl11a expression in sorted hematopoietic populations. Data represent mean values of three independent biological replicates and all values are normalized to the expression of the Gapdh gene. Error bars indicate the SD. Thy, thymus; PB, peripheral blood; M, macrophages; G, granulocytes; Mk, megakaryocytes; B, BM CD19+ B cells; T, spleen CD3+T cells. In all flow cytometry assays, at least four mice were analyzed for each cell type in independent experiments.
Dynamic expression patterns of Bcl11a in hematopoiesis. (A) Schematic diagram of the Bcl11a-eGFP reporter allele. The eGFP reporter cassette flanked by two F3 sites is introduced to the 3′UTR region of Bcl11a, 8 bp after the stop codon TAG. (B) Flow cytometry tracking Bcl11a expression using the Bcl11aeGFP/eGFP reporter mice. Cell surface markers for defining these cells are described in Table S1. (C) Expression of Bcl11a (eGFP+) and Bcl11b (Tdtomato+) in double negative (DN) thymocytes was measured by flow cytometry. DN thymocytes are identified as described in Fig. S1 D. (D) qRT-PCR analysis of Bcl11a expression in sorted hematopoietic populations. Data represent mean values of three independent biological replicates and all values are normalized to the expression of the Gapdh gene. Error bars indicate the SD. Thy, thymus; PB, peripheral blood; M, macrophages; G, granulocytes; Mk, megakaryocytes; B, BM CD19+ B cells; T, spleen CD3+T cells. In all flow cytometry assays, at least four mice were analyzed for each cell type in independent experiments.
GFP+ cells were detected in almost all Lin− BM cells, including HSCs, MPPs, LMPPs, CLPs, common myeloid progenitors (CMPs), granulocyte-monocyte progenitors (GMPs), MEPs, monocyte-dendritic precursors, and common dendritic precursors (CDPs; Fig. 1 B and Fig. S1 A). Bcl11a expression was also detected in erythroid progenitors (EPs), differentiated macrophages, granulocytes and megakaryocytes, and at high levels in plasmacytoid DCs and conventional DCs (Fig. 1 B and Fig. S1 B).
B cell development from CLPs can be divided into several stages based on expression of cell surface markers and intracellular molecules (Hardy et al., 1991; Inlay et al., 2009). CLPs are a heterogeneous population of cells that can be divided into all-lymphoid progenitor and B cell–committed cells based on Ly6d expression (Inlay et al., 2009; Mansson et al., 2010). Bcl11a is expressed in all CLPs (Fig. 1 B), suggesting a possible role for Bcl11a in both B lineage cells and progenitors primed for T and NK cells. Bcl11a expression remains high in pro–B and starts to be down-regulated in pre–B and immature B cells (Fig. 1 B). Spleen B cells, including transitional (Trans), mature (Mat), follicular (FO), and marginal zone (MZ) B cells, also expressed Bcl11a (Fig. 1 B; Fig. S1 C).
T cell development in the thymus can also be divided into several stages. The earliest thymocytes are double-negative (DN) cells that do not express the two TCR co-receptors, CD4 and CD8. Based on expression of c-Kit and/or CD44 and CD25, four DN subpopulations are described (Ciofani and Zúñiga-Pflücker, 2007; Rothenberg et al., 2008): ETPs and DN2–4 (Fig. S1 D). ETPs are thought to be the earliest progenitors to seed in the thymus and still retain non–T cell lineage differentiation potential (Rothenberg et al., 2008). Almost all ETPs expressed Bcl11a (Fig. 1 B). DN2 thymocytes can be further divided into DN2a (c-Kithigh) and DN2b (c-Kitmid; Rothenberg et al., 2008). T cell commitment is suggested to occur between DN2a and DN2b, and is marked by sharp up-regulation of Bcl11b, which is the paralogue of Bcl11a and essential for T cell commitment (Tydell et al., 2007; Ikawa et al., 2010; Li et al., 2010a,b). Although almost all DN2 thymocytes expressed Bcl11a (Fig. 1 B), in DN2b, Bcl11a expression levels (eGFP+) decreased drastically concomitant with the dramatically increased Bcl11b expression (TdTomato+; Li et al., 2010b; Fig. 1 C). Therefore, high levels of Bcl11a expression mark uncommitted early thymocytes whereas high levels of Bcl11b define the committed thymocytes. Bcl11a expression levels further decreased in DN3–4 thymocytes and became undetectable in peripheral T cells (Fig. 1, B and C).
Distinct stages of NK cell development in the BM have been described in recent years (Di Santo, 2006; Carotta et al., 2011; Fathman et al., 2011). Although Bcl11a expression is undetectable in mature BM NK cells (Fig. 1 B), one fraction of the earliest NK progenitors (prepro–NKa), and NK progenitors (CD122+CD3−NK1.1−) clearly expressed Bcl11a (Fig. 1 B; Fig. S1 E). Nevertheless, the other subset of prepro–NK cells, prepro–NKb, did not. Bcl11a expression thus distinguishes the two prepro–NK cell subsets, which appear to be phenotypically and functionally similar (Carotta et al., 2011).
Expression of Bcl11a in hematopoietic lineages was further determined by qRT-PCR analysis, which showed that Bcl11a was highly expressed in the HSC compartment (Lin−Sca1+c-Kit+Flt3−CD48−CD150+) but the levels decreased in LMPPs (Fig.1 D). Both CLPs and B cells were highly enriched for Bcl11a expression, whereas T cells and NK cells had barely detectable Bcl11a expression. Expression of Bcl11a in CMPs, GMPs, and MEPs was low and further down-regulated in differentiated macrophages and granulocytes (Fig. 1 D).
Deletion of Bcl11a in adult mice caused loss of early B cells and depletion of lymphoid progenitors
To investigate Bcl11a in adult hematopoiesis, we produced and analyzed the Bcl11a conditional knockout (cko) mice (Liu et al., 2003a). The cko mice were crossed to Rosa26-CreERT2 mice so that Bcl11a could be deleted in a tamoxifen (Tam)-inducible manner (Hameyer et al., 2007). Activation of Cre recombinase by Tam permits investigation of the immediate consequences of acute loss of Bcl11a. The CreERT2; Bcl11aflox/flox mice (referred to as flox/flox in this work) were grossly normal and fertile.
To delete Bcl11a in vivo, we treated 6–8-wk-old flox/flox and the control mice (CreERT2; Bcl11a+/+ or CreERT2; Bcl11a+/flox) with Tam. The use of control mice would help exclude the potential phenotypes caused by Cre toxicity. The flox/flox mice usually died in 10–14 d upon Tam treatment, likely because of the functions of Bcl11a in nonhematopoeitic tissues (John et al., 2012). We thus analyzed lymphocyte development 1 wk after Tam administration. Genotyping of BM cells showed clean deletion of Bcl11a (Fig. S2 A), which was confirmed by nondetectable Bcl11a protein (Fig. S2 B). We first investigated B cell development in the BM. Flow cytometry analysis revealed severe defects in the B cell compartment with the total BM B cell numbers in the flox/flox mice being reduced to 30% of those in the wild-type control mice (Fig. 2, A and B). Pro–B cells and pre–B cells were affected the most, with few presenting in the flox/flox BM. Immature B cells reduced to approximately one-fifth of the wild-type number. However, within the experiment time window, B cells in the spleen, lymph node, and peripheral blood were not obviously reduced, indicating less dependency of these cells on Bcl11a (unpublished data). In contrast to B cells, other hematopoietic lineages in the BM, including T cells, NK cells, macrophages and granulocytes, and EPs, were not obviously affected (unpublished data).
Profound defects of lymphoid development in Bcl11a-deficient adult mouse. B and T cells at different developmental stages were analyzed in the BM or thymus 1 wk after the mice were injected with Tam (n = 4/genotype, and the experiment was repeated 4 times). (A) BM B cells. Numbers refer to percentages of B cells in total BM-nucleated cells. (B) Cellularity of BM B cells in mice of the indicated genotypes. Cells were collected from two femurs of each mouse. (C) Thymocytes of various development stages. Lin− thymocytes were further analyzed by expression of c-Kit and CD25 for DN thymocytes. (D) Cellularity of DN thymocytes in mice of indicated genotypes. (E) Analysis of LSKs, LMPPs, CLPs, and Ly6d+CLPs. Numbers refer to percentages of progenitors in total BM-nucleated cells. (F) Total cell numbers. +/+, CreERT2; Bcl11a+/+. +/flox, CreERT2; Bcl11a+/flox. flox/flox, CreERT2; Bcl11aflox/flox. *, P < 0.05; **, P < 0.01.
Profound defects of lymphoid development in Bcl11a-deficient adult mouse. B and T cells at different developmental stages were analyzed in the BM or thymus 1 wk after the mice were injected with Tam (n = 4/genotype, and the experiment was repeated 4 times). (A) BM B cells. Numbers refer to percentages of B cells in total BM-nucleated cells. (B) Cellularity of BM B cells in mice of the indicated genotypes. Cells were collected from two femurs of each mouse. (C) Thymocytes of various development stages. Lin− thymocytes were further analyzed by expression of c-Kit and CD25 for DN thymocytes. (D) Cellularity of DN thymocytes in mice of indicated genotypes. (E) Analysis of LSKs, LMPPs, CLPs, and Ly6d+CLPs. Numbers refer to percentages of progenitors in total BM-nucleated cells. (F) Total cell numbers. +/+, CreERT2; Bcl11a+/+. +/flox, CreERT2; Bcl11a+/flox. flox/flox, CreERT2; Bcl11aflox/flox. *, P < 0.05; **, P < 0.01.
In addition to the absence of B cells in the Bcl11a mutant fetal liver, Bcl11a germline-null mutation also resulted in defective fetal thymocyte development (Liu et al., 2003b). 1 wk after Tam treatment, the flox/flox thymus had few ETP cells and DN2 thymocytes (Fig. 2, C and D). However, the numbers of DN3 and DN4 thymocytes, as well as CD4+CD8+ double-positive (DP) and CD4+ or CD8+ single-positive thymocytes, were not substantially changed (Fig. 2, C and D, and not depicted).
Because Bcl11a was also highly expressed in lymphoid progenitors (Fig. 1, B and D), we further investigated the effect of Bcl11a deletion in the progenitor compartments. The flox/flox mice had no Ly6d+ CLPs and a sevenfold reduction of total CLPs (Fig. 2, E and F). Moreover, significantly reduced frequencies and numbers of LSKs and LMPPs were also revealed in the flox/flox mice (Fig. 2, E and F). In contrast to the defects in lymphoid progenitors, CMPs, GMPs, and MEPs were not obviously affected (unpublished data). These results suggest that, in the adult mouse, acute loss of Bcl11a causes lymphoid-specific defects and that Bcl11a has functions in both developing lymphocytes and lymphoid progenitors.
In addition to the severe lymphopoiesis defects caused by acute loss of Bcl11a in the conditional knockout mouse, loss of one copy of Bcl11a in the heterozygous mutant mice (±) that carry the null allele of Bcl11a (Liu et al., 2003b) also affected lymphopoiesis. Compared with the wild-type control, the heterozygous mutants had ∼50% of LMPPs, CLPs, ETPs, and DN2 thymocytes (unpublished data), demonstrating a dose-dependent requirement for Bcl11a in lymphoid progenitors and in lymphocyte development.
Complete loss of lymphoid development caused by Bcl11a deficiency is cell autonomous
We next investigated whether the defects in lymphoid lineages caused by Bcl11a deficiency were cell autonomous, and whether a relatively longer time window (>1 wk) would reveal more profound phenotypes. We intravenously injected Lin− BM cells from flox/flox and control mice into lethally irradiated CD45.1+ C57B6 recipients. The recipients were allowed 6–8 wk for engraftment before Tam administration, and the BM chimeras were subsequently harvested for analysis either 1 or 4 wk later.
Compared with the Tam-treated flox/flox mice described above, the flox/flox BM chimeras exhibited similar lymphoid defects when cells were analyzed 1 wk after Tam treatment (unpublished data). However, 4 wk after Tam administration, more severe lymphoid defects were revealed: no pro–B, pre–B, or immature B cells could be detected, and the total BM B cells decreased to 5% of the wild-type (Fig. 3 A). There were also few ETPs or DN2 thymocytes (Fig. 3 B). The longer time window after Bcl11a deletion also revealed a drastic decrease of DN3 and DN4 thymocytes. The complete depletion of Bcl11a-deficient early B cells and thymocytes indicate that these cells either gradually differentiated or died and that they were not replenished from lymphoid progenitors. Additionally, Bcl11a deletion depleted all NK progenitors, and only few NK cells were present in the BM (Fig. 3, C and D).
Severe defects in B, T, and NK cell precursors and lymphoid progenitor compartments in recipient mice engrafted with Lin− BM cells (CD45.1−). Lethally irradiated recipient mice (CD45.1+) engrafted with BM cells of indicated genotypes were treated with Tam and analyzed 4 wk later (n = 4/group, and the experiment was repeated 4 times). Panels show total cell numbers (bar graphs) and/or flow cytometry dot plots of donor BM B cells (A), donor thymocytes (B), donor BM NK cells (C and D), and donor lymphoid progenitors (E). Numbers in the flow cytometry plots refer to percentages of gated cells in total donor nucleated BM cells or thymocytes. *, P < 0.05; **, P < 0.01.
Severe defects in B, T, and NK cell precursors and lymphoid progenitor compartments in recipient mice engrafted with Lin− BM cells (CD45.1−). Lethally irradiated recipient mice (CD45.1+) engrafted with BM cells of indicated genotypes were treated with Tam and analyzed 4 wk later (n = 4/group, and the experiment was repeated 4 times). Panels show total cell numbers (bar graphs) and/or flow cytometry dot plots of donor BM B cells (A), donor thymocytes (B), donor BM NK cells (C and D), and donor lymphoid progenitors (E). Numbers in the flow cytometry plots refer to percentages of gated cells in total donor nucleated BM cells or thymocytes. *, P < 0.05; **, P < 0.01.
The BM chimeras also allowed us to examine the defects in lymphoid progenitors 4 wk after Bcl11a deletion, and again more severe phenotypes were found: essentially no CLPs could be detected, and both the cell numbers and frequencies of LMPPs and LSKs were significantly reduced in the recipients engrafted with flox/flox BM cells (Fig. 3 E). We subsequently harvested donor CD45.1− BM cells from the Tam-treated primary recipients (1 mo of age), and transplanted them into irradiated secondary recipients. The secondary recipients had myeloid cells, erythrocytes, and megakaryocytes derived from the donor BM cells, but no B cells, T cells, or NK cells if the donor BM cells were of the flox/flox genotype (unpublished data).
After Bcl11a deletion, complete loss of lymphoid progenitors such as CLPs over time suggests that these progenitors had undergone apoptosis or differentiation and/or no new CLPs were derived from the LMPPs, or even HSCs. The substantial reduction of cell numbers in LMPPs or LSKs reflects the possible defective lymphoid development potential in Bcl11a-deficient LMPPs or HSCs.
Bcl11a is required for survival of early B cells
To investigate the cellular mechanism of the lymphopoiesis defects caused by Bcl11a deficiency, we first explored the possibility that Bcl11a deletion might cause apoptosis in developing lymphocytes or progenitors. To this end, we measured apoptosis in BM B cells of the flox/flox mice 4–5 d after Tam administration to capture the immediate consequence of Bcl11a deletion. Dramatically increased apoptosis was observed in pro–B, pre–B, and immature B cells (Fig. 4 A). Bcl11a, thus, has a direct and essential role in committed B cells.
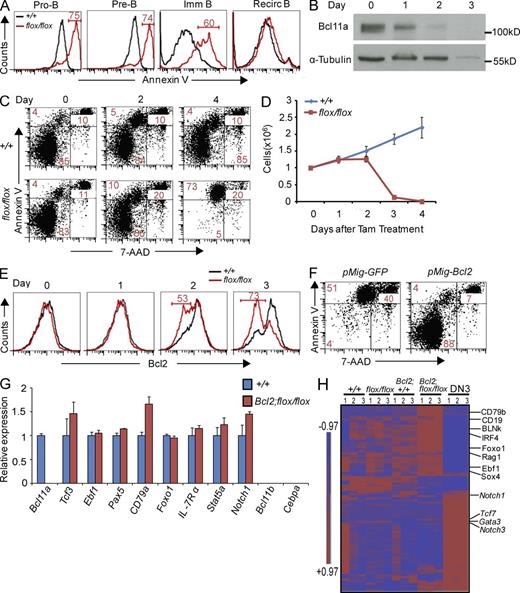
Apoptosis in Bcl11a-deficient B cell precursors could be rescued by exogenous Bcl2. (A) Apoptosis was measured by flow cytometry in Bcl11a-deficient early B cells in the BM. Numbers in histograms are percentages of the indicated gates. (B) Western blot analysis of Bcl11a protein at indicated time points after Tam treatment. (C) Apoptosis in pro–B cells (IMDM supplemented with 50 ng/ml of IL-7, Flt3L, and SCF) after Bcl11a deletion detected by Annexin V and 7-AAD. Numbers in the flow cytometry plots refer to percentages of gated cells. (D) 1 million cultured pro–B cells were treated with Tam for 4 d, and live cells were enumerated at indicated time points. (E) Detection of intracellular Bcl2 in cultured pro–B cells at indicated time points after Tam treatment. (F) Apoptosis in cultured pro–B cells infected with retrovirus-expressing Bcl2. GFP+ cells (infected) were gated for analysis. An empty retroviral vector expressing GFP was used as the control. (G) qRT-PCR analysis of key lymphoid genes in cultured Bcl11a-deficient pro–B cells. Data represent mean values of three independent biological replicates and all values are normalized to Gapdh expression. Error bars indicate the SD. (H) Two-way hierarchical clustering of expression of 100 important lymphoid genes (Table S4) in cultured pro–B cells. Expression of these genes in wild-type DN3 thymocytes was used as an expression control. Scale indicates the log2 value of normalized signal level. For A, C, D, E, F, and G, data represent at least three independent experiments.
Apoptosis in Bcl11a-deficient B cell precursors could be rescued by exogenous Bcl2. (A) Apoptosis was measured by flow cytometry in Bcl11a-deficient early B cells in the BM. Numbers in histograms are percentages of the indicated gates. (B) Western blot analysis of Bcl11a protein at indicated time points after Tam treatment. (C) Apoptosis in pro–B cells (IMDM supplemented with 50 ng/ml of IL-7, Flt3L, and SCF) after Bcl11a deletion detected by Annexin V and 7-AAD. Numbers in the flow cytometry plots refer to percentages of gated cells. (D) 1 million cultured pro–B cells were treated with Tam for 4 d, and live cells were enumerated at indicated time points. (E) Detection of intracellular Bcl2 in cultured pro–B cells at indicated time points after Tam treatment. (F) Apoptosis in cultured pro–B cells infected with retrovirus-expressing Bcl2. GFP+ cells (infected) were gated for analysis. An empty retroviral vector expressing GFP was used as the control. (G) qRT-PCR analysis of key lymphoid genes in cultured Bcl11a-deficient pro–B cells. Data represent mean values of three independent biological replicates and all values are normalized to Gapdh expression. Error bars indicate the SD. (H) Two-way hierarchical clustering of expression of 100 important lymphoid genes (Table S4) in cultured pro–B cells. Expression of these genes in wild-type DN3 thymocytes was used as an expression control. Scale indicates the log2 value of normalized signal level. For A, C, D, E, F, and G, data represent at least three independent experiments.
To experimentally confirm the apoptosis in early B cells in vitro, we cultured BM pro–B cells (CD43+CD19+). Once the cells reached confluence (1 wk), Tam was added to the culture to induce Bcl11a deletion, which led to rapid loss of Bcl11a protein (Fig. 4 B). Concomitant with the decrease of Bcl11a protein, the cultured B cells became apoptotic (Fig. 4 C). Indeed, within 4 d of Tam treatment, no live B cells could be found in the culture (Fig. 4 D), confirming the essential role of Bcl11a in these cells. We subsequently performed an in vitro pre–B cell assay to examine the requirement of Bcl11a in pre–B cells. No pre–B cells colonies were obtained from Bcl11a-deficient BM cells, again revealing the essential role of Bcl11a in these cells (unpublished data).
Apoptosis in Bcl11a-deficient pro–B cells is rescued by exogenous Bcl2
Bcl2 is an important antiapoptotic factor. Once Bcl11a is deleted in cultured B cells, the Bcl2 protein levels were rapidly decreased, as detected in flow cytometry (Fig. 4 E). We next explored whether expressing exogenous Bcl2 or other antiapoptotic factors in Bcl11a-deficient B cells might be able to rescue them from apoptosis. We infected the cultured B cells with retrovirus expressing either Bcl2, Bcl-xL (Bcl2l1), or Mcl1 (all coexpressing GFP). Apoptosis in the GFP+ pro–B cells was blocked by the expression of these antiapoptotic factors (Fig. 4 F and not depicted).
E2A, Ebf1, and Pax5 work in concert to guide B cell development. E2A and Ebf1 are considered to be competence and specification factors, respectively, whereas Pax5 acts as a commitment factor for B cell identity. Indeed, overexpression of Ebf1 can rescue defective B cell development caused by loss of Ikaros, E2A, and Runx1 (Seet et al., 2004; Reynaud et al., 2008; Seo et al., 2012). We thus used retrovirus expressing E2A (both E12 and E47), Ebf1, Pax5, and Stat5CA (constitutively active form). None of these key B cell genes was able to rescue the Bcl11a-deficient pro–B cells, as GFP+ cells were still apoptotic and the culture eventually did not have live cells (unpublished data). The rescue of Bcl11a-deficient B cells by Bcl2, but not by the core B cell genes, demonstrates that Bcl11a functions distinctly and is normally required for cell survival in B cell development. Not surprisingly, deletion of Bcl11a did not significantly change the expression of E2A (Tcf3), Ebf1, or Pax5 in B cells (Fig. 4 G).
We subsequently performed global gene expression of Bcl2-rescued, Bcl11a-deficient cultured pro–B cells and compared it to that of the wild-type control B cells. Neither substantial decrease of key B cell genes nor increase of key genes for T or myeloid lineages such as Bcl11b and Cebpa was detected in the rescued B cells (Fig. 4, G and H; Table S4), indicating that Bcl11a-deficient B cells retained their B cell identity and were not converted to another cell type. Therefore, unlike Bcl11b (in regard to maintaining T cell identity; Li et al., 2010b), Bcl11a is not required to maintain B cell identity in the committed B cells and likely plays a general, permissive role in these cells. This is consistent with the fact that Bcl11a is expressed in most hematopoietic cells.
Coexpression of Mdm2 and Bcl2 rescues both apoptotic and proliferative defects in Bcl11a-deficient pro–B cells
Although overexpression of Bcl2, Bcl-xL, or Mcl1 efficiently rescued Bcl11a-deficient B cells from apoptosis, these B cells appeared to be in senescence because no cells were in S phase (Bcl2; flox/flox in Fig. 5 A), and the cell numbers in the culture did not increase over 6 d (Fig. 5 B). Consistent with a cell cycle defect, expression of the cyclin-dependent kinase inhibitor p21 (also known as Cdkn1a, p21WAF1/Cip1), but not of p27 (Cdkn1b) or p57 (Cdkn1c), was up-regulated in Bcl2; flox/flox B cells (Fig. 5 C). p21 promotes cell cycle arrest in response to many stimuli, and is induced by p53 (Abbas and Dutta, 2009). To further investigate the effect of Bcl11a deletion on the p53 pathway, we performed Western blot analysis of p53 and Mdm2 in Bcl11a-deficient BM B cells. Bcl11a deletion resulted in barely detectable Mdm2 and accumulation of p53 (Fig. S2 C). Mdm2 inhibits p53 transcriptional activity, favors its nuclear export, and stimulates its degradation (Wade et al., 2010; Vucic et al., 2011). Lower levels of Mdm2 cause lymphopoiesis defects (Mendrysa et al., 2003). The apoptosis and cell cycle defect in Bcl11a-deficient B cells might be caused by the aberrant Mdm2–p53–p21 pathway, and therefore, overexpressing Mdm2 together with Bcl2 should relieve the cell cycle block. As shown in Fig. 5 A, 4–5% of Bcl11a-deficient B cells were in S phase when exogenous Mdm2 and Bcl2 were coexpressed (Bcl2/Mdm2; flox/flox). The rescue by Bcl2/Mdm2 was also revealed by the increased cell numbers in the culture (Fig. 5 B).
Genetic interaction of Bcl11a and p53 in lymphoid development. (A) Cell cycle of cultured pro–B cells infected with retrovirus expressing Bcl2 and/or Mdm2 was analyzed by Edu/7-AAD staining. (B) 3 d after Tam treatment, 1 million pro–B cells were cultured and counted daily for viability. (C) qRT-PCR analysis of p21, p27, and p57 in Bcl11a-deficient pro–B cells infected with the Bcl2-expressing retrovirus. (D and E) Presence of pro–B, pre–B, and immature B cells (D), or lymphoid progenitors (E), in BM of Dflox/flox mice 5 d after Tam treatment. Numbers refer to percentages of gated cells in total donor nucleated BM cells. Data in A, B, C, D, and E represent at least three independent experiments. *, P < 0.05; **, P < 0.01.
Genetic interaction of Bcl11a and p53 in lymphoid development. (A) Cell cycle of cultured pro–B cells infected with retrovirus expressing Bcl2 and/or Mdm2 was analyzed by Edu/7-AAD staining. (B) 3 d after Tam treatment, 1 million pro–B cells were cultured and counted daily for viability. (C) qRT-PCR analysis of p21, p27, and p57 in Bcl11a-deficient pro–B cells infected with the Bcl2-expressing retrovirus. (D and E) Presence of pro–B, pre–B, and immature B cells (D), or lymphoid progenitors (E), in BM of Dflox/flox mice 5 d after Tam treatment. Numbers refer to percentages of gated cells in total donor nucleated BM cells. Data in A, B, C, D, and E represent at least three independent experiments. *, P < 0.05; **, P < 0.01.
Genetic interaction of Bcl11a and p53 in lymphopoiesis
Although Bcl2/Mdm2 coexpression was able to rescue cultured pro–B cells from apoptosis and cell cycle defects, compared with the wild-type control cells where 35% of cells were in S phase, the rescued B cells still had only 4–5% of cells in S phase and proliferated slowly (Fig. 5, A and B). To further dissect the genetic interaction between Bcl11a and the p53 pathway, we crossed the flox/flox mice to p53 conditional knockout mice (Jonkers et al., 2001) to produce Rosa26CreERT2; Bcl11aflox/flox/p53flox/flox mice (referred to as Dflox/flox). We first cultured BM cells from the Dflox/flox mice in B cell media to obtain pro–B cells, and then deleted both Bcl11a and p53, which led to the absence of both proteins (Fig. S2 C). Deleting p53 enabled 20–25% of Bcl11a-deficient pro–B cells to be in S phase, which also proliferated robustly (Fig. 5, A and B).
The in vitro rescue of Bcl11a-deficient B cells by p53 deficiency led us to examine the genetic interaction of Bcl11a and p53 in vivo. We treated Dflox/flox mice with Tam and harvested BM cells 5 d later. Efficient deletion of both Bcl11a and p53 was obtained at this time (Fig. S2 D). Flow cytometry and cell number analysis demonstrated a significant increase of pro–B, pre–B, and immature B cells in the Dflox/flox mice compared with those in the flox/flox mice (Fig. 5 D), indicating that p53 deficiency was indeed able to rescue early B cell development.
Over time, Bcl11a deletion caused complete loss of CLPs and reduced cellularity of LMPPs and LSKs (Fig. 3 E). We next examined whether p53 deficiency could also help Bcl11a-deficient lymphoid progenitors. 5 d after Bcl11a deletion, more CLPs and Ly6d+CLPs were found in Dflox/flox mice (Fig. 5 E). However, surprisingly, p53 deficiency did not appear to help LSKs or LMPPs as their numbers were not increased (Fig. 5 E). It is possible that Bcl11a-deficient early lymphoid progenitors such as LSKs and LMPPs require additional rescue mechanisms apart from p53 deficiency.
To further study the genetic interaction of Bcl11a and p53 in lymphoid development, we made BM chimeras from BM cells of Dflox/flox mice. The BM chimeras would allow us to investigate the phenotypes of loss of Bcl11a and p53 over a longer period of time, and to assess cell autonomous phenotypes, specifically in hematopoietic lineages. The recipient mice were allowed 6–8 wk for donor cell engraftment, and were analyzed 4 wk after Tam treatment. Surprisingly, unlike the presence of pro–B, pre–B, and immature B cells in Dflox/flox mice 5 d after Tam treatment, the BM chimeras now had few early B cells, and the total B cell numbers from donor BM cells were also drastically decreased (Fig. 6 A). Strikingly, all CLPs were depleted with decreases of LMPPs and LSKs (Fig. 6 B).
p53 deficiency did not rescue long-term B cell development or lymphoid progenitor defects. Lethally irradiated recipient mice (CD45.1+) were transplanted with Lin− BM cells (CD45.1−) and were treated with Tam to delete Bcl11a. Recipients were subsequently analyzed 4 wk later (n = 4/genotype). (A) Total cell numbers of donor BM B cells of indicated genotypes. (B) Total numbers of donor LSKs, LMPPs, and CLPs. (C) Freshly sorted CLPs (5,000) from mice of indicated genotypes 3 d after Tam treatment were cultured on OP9 stromal cells in the presence of 10 ng/ml of IL-7 and Flt3L for 14 d. A–C represent at least three independent experiments. *, P < 0.05; **, P < 0.01.
p53 deficiency did not rescue long-term B cell development or lymphoid progenitor defects. Lethally irradiated recipient mice (CD45.1+) were transplanted with Lin− BM cells (CD45.1−) and were treated with Tam to delete Bcl11a. Recipients were subsequently analyzed 4 wk later (n = 4/genotype). (A) Total cell numbers of donor BM B cells of indicated genotypes. (B) Total numbers of donor LSKs, LMPPs, and CLPs. (C) Freshly sorted CLPs (5,000) from mice of indicated genotypes 3 d after Tam treatment were cultured on OP9 stromal cells in the presence of 10 ng/ml of IL-7 and Flt3L for 14 d. A–C represent at least three independent experiments. *, P < 0.05; **, P < 0.01.
To test whether the CLPs presented in Dflox/flox mice shortly after Tam treatment were alive and functional, we isolated CLPs 4–5 d after Tam treatment, and cultured them on OP9 stromal cells in the presence of IL7, SCF, and Flt3L. B cells were cultured out from CLPs of Dflox/flox and the wild-type control mice, but not from CLPs of the flox/flox mice (Fig. 6 C). Therefore, p53 deficiency was able to rescue the developing B cells and CLPs. The fact that the rescued early lymphoid precursors or progenitors were eventually depleted over time suggests that no fresh progenitors are produced from LSKs. These results also demonstrate that in developing B cells and CLPs, Bcl11a primarily functions for survival and proliferation through regulating the p53 pathway, and thus likely plays a permissive role similar to IL-7Rα in T cells (Maraskovsky et al., 1997).
Complete loss of lymphoid development potential in Bcl11a-deficient LSKs
To investigate whether Bcl11a-deficient LSKs lost the ability to produce any lymphoid progenitors or lymphocytes and to test the function of LSKs with both Bcl11a and p53 deficiency in lymphoid development, we purified LSKs from the Tam-treated flox/flox, Dflox/flox, or control mice and cultured them on OP9 stromal cells. B cells were cultured out only from the wild-type LSKs, but not from the flox/flox or Dflox/flox LSKs (unpublished data), confirming that LSKs of both flox/flox and Dflox/flox were defective in lymphoid development potential. We next injected purified LSKs (from Tam-treated mice) with CD45.1+ helper BM cells into recipients and examined their in vivo differentiation potential to exclude the possibility that Dflox/flox LSKs may require the BM microenvironment for proper proliferation and/or differentiation.
6–8 wk after engraftment, the recipients were analyzed. As shown in Fig. 7, A and B, LSKs of flox/flox and Dflox/flox mice did not produce any CLPs in the BM or ETPs in the thymus, which led to an absence of B, T, and NK cells in the BM or the thymus (Fig. 7, A and B), whereas myeloid progenitors were not noticeably changed (unpublished data). The absence of mature lymphocytes from donor LSKs also excluded the possibility that the lymphoid progenitors were initially generated, but subsequently differentiated and thus depleted.
Loss of lymphoid development potential in LSKs of flox/flox and Dflox/flox mice. Flow cytometric–sorted LSKs from Tam-treated mice were transplanted to recipients with helper BM cells. Donor cells (CD45.1−) were analyzed 8 wk after transplantation. (A) Total numbers of donor LSK, HSC, LMPP, CLP and ETP of indicated genotypes. (B) Populations of B, T, and NK cells analyzed by flow cytometry. Numbers indicate percentages of lymphocytes in total BM-nucleated cells. (C) Apoptosis in Bcl11a-deficient stem cells or progenitors (red line) analyzed by Annexin V staining. Apoptosis in these cells were rescued by p53 deficiency (green line). BM cells were harvested and stained 4 d after Tam treatment. (D and E) qRT-PCR analysis of cell cycle, lymphoid, and myeloid genes in freshly sorted HSCs. (F) qRT-PCR analysis of lymphoid and myeloid genes in LMPPs. Data represent mean values of three independent biological replicates and all values are normalized to Gapdh expression. Error bars indicate the SD. A–F represent at least three independent experiments. *, P < 0.05; **, P < 0.01.
Loss of lymphoid development potential in LSKs of flox/flox and Dflox/flox mice. Flow cytometric–sorted LSKs from Tam-treated mice were transplanted to recipients with helper BM cells. Donor cells (CD45.1−) were analyzed 8 wk after transplantation. (A) Total numbers of donor LSK, HSC, LMPP, CLP and ETP of indicated genotypes. (B) Populations of B, T, and NK cells analyzed by flow cytometry. Numbers indicate percentages of lymphocytes in total BM-nucleated cells. (C) Apoptosis in Bcl11a-deficient stem cells or progenitors (red line) analyzed by Annexin V staining. Apoptosis in these cells were rescued by p53 deficiency (green line). BM cells were harvested and stained 4 d after Tam treatment. (D and E) qRT-PCR analysis of cell cycle, lymphoid, and myeloid genes in freshly sorted HSCs. (F) qRT-PCR analysis of lymphoid and myeloid genes in LMPPs. Data represent mean values of three independent biological replicates and all values are normalized to Gapdh expression. Error bars indicate the SD. A–F represent at least three independent experiments. *, P < 0.05; **, P < 0.01.
In addition to a complete lack of lymphocytes and CLPs, a significant or substantial reduction of the LSKs, HSCs, and LMPPs was found in the recipients of the flox/flox and Dflox/flox LSK donors (Fig. 7 A). These results thus demonstrate that deletion of Bcl11a specifically abolishes lymphoid development potential in hematopoietic progenitors and that p53 deficiency is not sufficient to rescue this early defect.
We next explored what caused the reduced cell numbers in the LSK compartment and examined apoptosis in these cells. As shown in Fig. 7 C, many LSK, HSC, and LMPP compartment cells went through apoptosis once Bcl11a was deleted (red line in Fig. 7 C). Bcl11a-deficient CLPs also underwent apoptosis. Similar to rescue of apoptosis of early B cells by Bcl2/Mdm2, p53 deficiency essentially rescued apoptosis in LSKs, HSCs, LMPPs, and CLPs (green line in Fig. 7 C). Rescue of Bcl11a-deficient CLPs from apoptosis by p53 deficiency enables CLPs of the Dflox/flox mice to survive and to differentiate to lymphocytes (Fig. 6 C). Nevertheless, LSKs and LMPPs of the Dflox/flox mice were not rescued either in cell number or lymphoid development potential.
Deletion of Bcl11a caused increased p21 expression and cell cycle defects in early B cells (Fig. 5 C). We next investigated whether p21 expression was changed by p53 deficiency in HSCs of Dflox/flox mice. p53 deficiency indeed reduced p21 expression to the wild-type level, whereas p27 was slightly decreased and p57 was slightly increased (Fig. 7 D), suggesting that cell cycle defects were unlikely to cause lymphoid development defects in Bcl11a-deficient LSKs or HSCs and that Bcl11a functions by another mechanism.
Several transcription factors, such as Pbx1, Runx1, Ikzf1 (Ikaros), Sfpi1 (Pu.1), Tcf3 (E2A), and Foxo1, are required for the early stages of lymphoid development. In HSCs, Pbx1 showed significant decrease when Bcl11a was deleted (Fig. 7 E). Pbx1 is reported to regulate self-renewal of long-term HSCs by maintaining their quiescence (Ficara et al., 2008), and is required for generation of CLPs (Sanyal et al., 2007). However, lower Pbx1 is probably not the cause of complete loss of lymphoid potential, as Pbx1−/− stem cells can produce T cells (Sanyal et al., 2007). Besides, Pbx1, Runx1, and Foxo1 were also down-regulated. In contrast, the myeloid gene myeloperoxidase (Mpo) was significantly up-regulated (Fig. 7 E). Gata1 and Klf1 are critical for megakaryocyte/erythroid development (Iwasaki et al., 2003; Siatecka and Bieker, 2011). Klf1 is reported to regulate Bcl11a (Zhou et al., 2010). Once Bcl11a was deleted, expression of both Gata1 and Klf1 was significantly up-regulated in HSCs (Fig. 7 E), which surprisingly did not lead to increased MEPs (unpublished data). Consistent with the inability of p53 deficiency to rescue lymphoid development from LSKs, expression levels of most of the examined genes, including Ikzf1, Sfpi1, Tcf3, and Foxo1, were not altered by p53 deficiency.
We also quantified expression of key lymphoid regulators or markers in LMPPs (Fig. 7 F). Several of them had decreased expression in Bcl11a-deficient LMPPs, including Runx1, which interacts with Ebf1 to promote activation of B cell–specific genes (Lukin et al., 2010; Maier et al., 2004); Flt3, which marks LMPPs (Adolfsson et al., 2005); IL7R which is expressed in CLPs and is required for normal lymphocyte development (Peschon et al., 1994; von Freeden-Jeffry et al., 1995); Foxo1, which has distinct functions at various stages of B cell differentiation (Amin and Schlissel, 2008; Dengler et al., 2008); and Notch1, which is required for normal T cell development. However, expression of genes important in LMPPs, including Ikzf1, Sfpi1, and Tcf3, was not significantly altered. Similarly, expression of Mef2c, which regulates lymphoid versus myeloid fate choice (Stehling-Sun et al., 2009), was not changed (Fig. 7 F). Bcl11a therefore has essential functions in the early hematopoietic progenitors that are distinct from that of Pu.1, Ikaros, E2A, and Mef2c.
Similar to that in HSCs, expression of Mpo, together with Cebpa, was up-regulated in Bcl11a-deficient LMPPs (Fig. 7 F). LSKs of the flox/flox or Dflox/flox mice, however, did not produce more myeloid cells to fill in the space of the depleted lymphoid compartments (unpublished data). The inability of p53 deficiency to restore lymphoid potential in Bcl11a-deficient LMPPs is therefore reflected on the fact that deletion of p53 did not substantially change the expression patterns of the key lymphoid genes in these progenitors.
Genome-wide analysis of Bcl11a binding sites identifying downstream target gene candidates
Bcl11a is a zinc finger transcription factor that binds to its target sites in the genome (Avram et al., 2002; Liu et al., 2006). The aforementioned genetic studies indicate that Bcl11a regulates p53 via Mdm2 or Mdm4 in lymphocyte development. To confirm this regulation, we performed chromatin immunoprecipitation (ChIP) in mouse B cells with an anti-Bcl11a antibody. Bcl11a was reported to have a consensus binding site as GGCCGG (Avram et al., 2002). Computational prediction indicated that there were several putative binding sites in the Mdm2 and Mdm4 loci (Fig. 8 A). Quantitative real-time PCR (qPCR) of ChIP pull-down DNA showed a 4.95-fold enrichment using the Bcl11a antibody compared with the IgG control in the promoter region of Mdm4 locus. A 3.08-fold enrichment of Bcl11a binding was also found at the Mdm2 locus (Fig. 8 A). Bcl11a deletion caused rapid down-regulation of Bcl2 in early B cells. Analysis of ENCODE (The Encyclopedia of DNA Elements) data shows that in human B lymphocytes, there are 15 putative Bcl11a binding sites at the human BCL2 intronic and downstream regions. We analyzed these genomic regions and found that 12 of them were conserved in the mouse (Fig. 8 A). qPCR results showed that a higher than twofold enrichment of Bcl11a binding was found at seven regions, with the highest fold enrichment (fivefold) being identified in an intronic region (Fig. 8 A).
Analysis of Bcl11a binding sites in mouse pro–B cells. (A) qPCR validation of Bcl11a binding at the Mdm2, Mdm4, and Bcl2 loci in ChIP assay. Potential Bcl11a binding sites (red boxes) at the Mdm2 and Mdm4 loci, predicted as GGCCGG-containing sequences. Black bars indicate the qPCR-amplified regions of the validated sites. Potential Bcl11a binding sites (arrows) at the mouse Bcl2 locus, predicted as homologous regions to BCL11A-binding sites at the human BCL2 locus (ENCODE project), are analyzed by ChIP assay. The fold-enrichments of the amplified regions are (from 5′ to 3′) are as follows: 1.31 ± 0.40, 1.69 ± 0.32, 5.32 ± 0.92 (red arrow), 1.87 ± 0.001, 2.37 ± 0.60, 2.80 ± 0.43, 1.32 ± 0.21, 2.12 ± 0.44, 1.88 ± 0.21, 2.29 ± 0.40, 3.00 ± 0.16, and 2.68 ± 0.72. (B) Distribution of Bcl11a-binding sites in the genome from ChIP-Seq analysis. The 10-kb region upstream from transcription start site is defined as Proximal Promoter, and the 10-kb region downstream from transcription stop site is defined as downstream. (C) qPCR validation of Bcl11a binding at the genomic loci of Bcl-xL, p21, Pax5, and E2A. Genomic DNA pull-down using IgG was used as a control. A–C represent three independent experiments. *, P < 0.05; **, P < 0.01.
Analysis of Bcl11a binding sites in mouse pro–B cells. (A) qPCR validation of Bcl11a binding at the Mdm2, Mdm4, and Bcl2 loci in ChIP assay. Potential Bcl11a binding sites (red boxes) at the Mdm2 and Mdm4 loci, predicted as GGCCGG-containing sequences. Black bars indicate the qPCR-amplified regions of the validated sites. Potential Bcl11a binding sites (arrows) at the mouse Bcl2 locus, predicted as homologous regions to BCL11A-binding sites at the human BCL2 locus (ENCODE project), are analyzed by ChIP assay. The fold-enrichments of the amplified regions are (from 5′ to 3′) are as follows: 1.31 ± 0.40, 1.69 ± 0.32, 5.32 ± 0.92 (red arrow), 1.87 ± 0.001, 2.37 ± 0.60, 2.80 ± 0.43, 1.32 ± 0.21, 2.12 ± 0.44, 1.88 ± 0.21, 2.29 ± 0.40, 3.00 ± 0.16, and 2.68 ± 0.72. (B) Distribution of Bcl11a-binding sites in the genome from ChIP-Seq analysis. The 10-kb region upstream from transcription start site is defined as Proximal Promoter, and the 10-kb region downstream from transcription stop site is defined as downstream. (C) qPCR validation of Bcl11a binding at the genomic loci of Bcl-xL, p21, Pax5, and E2A. Genomic DNA pull-down using IgG was used as a control. A–C represent three independent experiments. *, P < 0.05; **, P < 0.01.
To screen for more putative downstream target genes of Bcl11a in lymphocytes, we conducted ChIP followed by next generation DNA sequencing (ChIP-Seq) using mouse pro–B cell nuclear extract. A total of 34.2 million mappable reads were analyzed and 10,865 peaks were identified. The fraction of peaks with different genomic distribution was determined, and majority of the peaks were located in intergenic, intronic, and promoter regions (Fig. 8 B). In total, Bcl11a may bind to 4,517 genomic loci, among which were E2A, Pax5, Bcl11b, and Bcl-xL. Potential binding sites of Bcl11a were also found at the p21 locus but not at p27 or p57. We next repeated the ChIP experiments followed by qPCR to validate several predicted ChIP-Seq peaks, and showed that Bcl11a strongly bound to the intronic region of Bcl-xL locus, which resulted in 95-fold enrichment in Bcl11a antibody pull-down DNA (Fig. 8 C). Other validated loci included p21, Pax5, and E2A loci. However, deletion of Bcl11a in B cells did not affect expression of Pax5 or E2A or other key B cell genes (Fig. 4, G and H). Therefore, although Bcl11a possibly participates in regulating Pax5 or E2A expression, these loci are mainly regulated by other mechanisms and Bcl11a is primarily involved in the survival and proliferation of developing early B cells.
DISCUSSION
Bcl11a is suggested to have multiple functions in normal hematopoiesis and leukemia (Nakamura et al., 2000; Satterwhite et al., 2001; Liu et al., 2003b; Yin et al., 2009). Deletion of Bcl11a causes absence of B cells in the fetal liver (Liu et al., 2003b). Conversely, ectopic Bcl11a expression from chromosomal translocations leads to B cell lymphomas in patients (Satterwhite et al., 2001). This study reveals the essential functions of Bcl11a in CLPs and developing lymphocytes, and uncovers the critical role of Bcl11a for lymphoid developmental potential in early hematopoietic progenitors.
During lymphocyte development, the expression of Bcl11a is found to be highly enriched in CLPs, ETPs, DN2 thymocytes, and B cells. These progenitors and early lymphocytes are completely lost when Bcl11a is deleted in the adult mouse. Interestingly, in thymocyte development, Bcl11a is drastically down-regulated in DN2b cells, the earliest committed T cells, whereas Bcl11b is sharply up-regulated (Liu et al., 2010; Rothenberg et al., 2010). Further investigation is needed to dissect the interaction between these two paralogues in T cell commitment.
Genetic analysis demonstrates that Bcl11a has direct roles in developing lymphocytes. Bcl11a deletion causes apoptosis in early B cells both in vivo and in vitro. Once apoptosis is blocked by expressing exogenous Bcl2 or Bcl-xL, the rescued B cells express high levels of p21 and appear to be in G0/G1 phase thus uncovering the proliferation defect in these cells. Both Mdm2 and Bcl2 are down-regulated in Bcl11a-deficient B cells. ChIP experiments indicate that Bcl11a binds Mdm2 and Bcl2 loci. Coexpressing Mdm2 with Bcl2 rescues both apoptosis and proliferation defects caused by Bcl11a deficiency. Bcl11a deletion also leads to p53 accumulation in lymphocytes, likely caused by the decreased Mdm2/Mdm4. The genetic interaction of Bcl11a and p53 is further revealed by rescuing Bcl11a-deficient B cells and CLPs both in vitro and in vivo by p53 deficiency.
Bcl11a deletion also causes apoptosis in LMPPs and LSKs, which is rescued by p53 deficiency. Surprisingly, lymphoid potential is not restored, proving that Bcl11a has additional functions in these early progenitors besides regulating p53. The HSC compartment consists of heterogeneous population of stem cells. Bcl11a expression in HSCs detected in the Bcl11a-eGFP reporter mouse does not have a bimodal pattern, suggesting that either variations of Bcl11a expression in the lymphoid-primed stem cells, or progenitors might be relatively small, or there exist posttranslational modifications of Bcl11a protein. Recent studies suggest that CD150low HSCs have both lymphoid and myeloid potential, whereas CD150high HSCs primarily produce myeloid cells even though they have higher self-renewal capability (Beerman et al., 2010; Morita et al., 2010). However, deletion of Bcl11a does not appear to cause noticeable difference between CD150high and CD150low HSCs (unpublished data). Several key regulators are known in lymphocyte development. Several of these genes, such as Pbx1, Runx1, Notch1, Flt3, Il7r, and Foxo1, are down-regulated in Bcl11a-deficient HSCs or LMPPs. In contrast, the myeloid-specific gene Mpo, and two key genes in erythrocyte/megakaryocyte development, Gata1 and Klf1, are up-regulated. However, their increased expression does not translate to increased CMPs, GMPs, or MEPs, indicating that the requirement for Bcl11a in either LMPPs or HSCs is not simply repressing the nonlymphoid lineages. It is more possible that deletion of Bcl11a may simply deplete all the lymphoid-primed or –biased stem cells. It will thus be necessary in the future investigation to perform single-cell analysis, combining with genetic studies, to further dissect the lymphoid developmental defects in the HSC compartment. It is worth noting that although we did not observe obvious myeloid development defects when Bcl11a was deleted, a more thorough investigation focusing on myeloid development and functions in the adult mouse is needed.
The p53 pathway is known to be critical for HSC self-renewal and quiescence (TeKippe et al., 2003; Liu et al., 2009), and Bcl11a is also implicated in HSCs (Kustikova et al., 2005). Unlike deleting p53 alone, loss of both p53 and Bcl11a does not lead to an expanded HSC compartment. In contrast, HSCs were substantially decreased, suggesting that Bcl11a is likely epistatic to p53 in HSCs. It was recently suggested in an in vitro system that p53 might negatively regulate Bcl11a (Jabbour et al., 2012). The complex interaction of Bcl11a and p53 in homeostasis of HSCs, including the target genes regulated by Bcl11a and p53, and the signals and factors that regulate these two genes, awaits further investigation. In addition to the p53 pathway in this study, Bcl11a is known to be regulated by Klf1 (Zhou et al., 2010), to interact with repressor complexes (Sankaran et al., 2008), to modulate chromosomal loop formation (Xu et al., 2010) in erythrocytes, and to interact with COUP-TFs (Avram et al., 2000). These studies will eventually help dissect the molecular and cellular mechanisms of Bcl11a’s functions in HSCs or LSKs in lymphoid development.
The regulation of p53 by Mdm2 is also important in developing lymphocytes, as lower levels of Mdm2 induce apoptosis and proliferation defects (Mendrysa et al., 2003). In developing lymphocytes, Bcl11a acts to prevent p53 activities by directly regulating Bcl2, Bcl-xL, Mdm2, and Mdm4. Although ChIP-Seq identifies several key lymphoid genes, such as Pax5 and E2A, to be Bcl11a’s potential downstream targets, their expression is not substantially altered in Bcl11a-deficient B cells or in Bcl2 rescued Bcl11a-deficient B cells, suggesting that their regulation by Bcl11a in B cells is not critical in these cells. On the other hand, the core B cell transcription factors Ebf1, Foxo1, and E2A bind the Bcl11a locus (Doulatov et al., 2010; Lin et al., 2010; Treiber et al., 2010) and possibly regulate its expression for cell survival and proliferation in developing lymphocytes. Therefore, although Bcl11a functions differently from these core B cell development genes, it is still a key component of the transcription factor network in B cell development.
It was reported that human BCL11A occupies discrete binding sites in the β-globin gene cluster in adult erythroid progenitors and executes its function as a transcription repressor to control fetal-to-adult switch of hemoglobin expression (Xu et al., 2010). As revealed by ChIP-seq in B cells, the nearest Bcl11a binding site to the mouse β-globin cluster is 85 kb upstream of Hbb-y and 4 kb upstream of the mouse olfactory receptor gene MOR5′β5. The lack of Bcl11a binding events at the β-globin locus in B cells possibly reflects that Bcl11a occupies differential gene targets in different cell types.
In summary, we have identified Bcl11a as an essential transcription factor in lymphoid development. Bcl11a is required for the survival and proliferation of early B cells and CLPs by suppressing the p53 pathway, and has additional essential functions in LMPPs and HSCs for lymphoid development, which warrants further characterization.
MATERIALS AND METHODS
Mice.
Bcl11a-eGFP reporter allele was generated by introducing a loxP-F3-IRES-eGFP-polyA-F3-FRT-PGK-E7-Neo-polyA-FRT cassette into the 3′UTR of Bcl11a and a loxP site upstream of exon 4 of Bcl11a. Bcl11aeGFP/eGFP mice were generated on C57BL/6 background. The Bcl11aflox/flox mice were crossed to Cre-ERT2 mice to generate Cre-ERT2; Bcl11aflox/flox (flox/flox) mice. flox/flox, flox/+, Cre-ERT2, Bcl11a germline heterozygous mutant (+/−), and the wild-type control mice were of a mixed C57BL/6 and 129S5 background. C57BL/6CD45.1+ mice were used as recipients for transplantation of BM cells or LSKs. All mice were 8–10 wk of age at the time of experiments, and at least 4 mice of one genotype were used in each experiment. The primers used for genotyping were listed in Table S3. All mice used were from colonies maintained at RSF of the Sanger Institute. Housing and breeding of mice and experimental procedures were according to the UK 1986 Animals Scientific Procedure Act and local institute ethics committee regulations.
Antibodies.
All antibodies used are described in Table S2.
Flow cytometry and cell sorting.
Cells from BM (femoral bone), spleen, thymus, and lymph node were mechanically dissociated and the red blood cells were removed using ACK lysis buffer (Lonza). Blood was collected into EDTA tubes (Sarstedt). In vitro–cultured cells were collected and washed with PBS containing 2% FBS before antibody labeling. For all cells, Fc receptors were blocked with anti-CD16 (2.4G2) before antibody labeling. Cells were stained with specific combinations of antibodies (Table S2) on ice for 20 min before washing. Data acquisition was performed using Fortessa (BD) or LSRII (BD) with dead cells excluded based on scatter profile or propidium iodide inclusion. Analysis was performed using FlowJo (Tree Star) software. Sorting was performed using a MoFlo XDP (BD).
Determination of hematopoietic cell population size.
The absolute numbers of viable cells of the BM (two femurs), spleen, thymus, lymph node, and peripheral blood were determined by Trypan blue exclusion. The absolute numbers of hematopoietic cells in an organ were calculated by the percentage of each sub-gate in the viable cell gate and the total cell numbers (propidium iodide–negative).
In vivo and in vitro apoptosis assays.
For the in vivo assay, the flox/flox and control mice were treated with 4.0 mg Tam (Sigma-Aldrich) by intraperitoneal injection over three consecutive days. 5–7 d later, BM cells were collected from these treated mice and immediately stained with Annexin V and 7-AAD (BD) according to the manufacturer’s instructions for detection of apoptotic cells.
BM cells from the flox/flox and control mice were cultured in the presence of 50 ng/ml IL-7, 50 ng/ml Flt3L, and 50 ng/ml SCF for 7 d, followed by Tam administration (final concentration 0.1 µM) in the media to delete Bcl11a. Cells were collected at different time point for staining with Annexin V and 7-AAD (BD) according to the manufacturer’s instructions for detection of apoptotic cells.
BM chimeras.
Single-cell suspensions of BM cells from the flox/flox, the Dflox/flox and the control (+/+ or +/flox) mice were injected intravenously into lethally irradiated (2 × 500 cGy) recipient mice (CD45.1+) for reconstitution for 6–8 wk.
For HSC reconstitution in vivo, the LSK cells were purified from BM of the flox/flox, the Dflox/flox, and the control mice (+/+ or +/flox) treated with Tam for 4–5 d. The sorted LSK cells (2,000 cells) were injected with helper CD45.1+ BM cells (2 × 105 cells) into lethally irradiated (2 × 500 cGy) recipient mice (CD45.1+) via the tail vein.
In vitro culture assay for differentiation of LSKs.
OP9 stromal cells were cultured in α-MEM (Sigma-Aldrich) with 10% FCS, 1% penicillin/streptomycin, and 2 mM l-glutamine (Life Technologies). Cells were passaged every 2–3 d by trypsinization (Invitrogen). A monolayer (70–80% confluent) of OP9 cells was prepared 24 h before co-culture. Sorted LSKs or sorted CLPs from flox/flox, Dflox/flox, and control (+/+) mice treated with Tam 4 d earlier were cultured for 14 d in the presence of for Flt3L (10 ng/ml; PeproTech) and IL-7 (10 ng/ml; PeproTech). After incubation, cells were stained for markers specific for B cells and myeloid cells and analyzed on Fortessa (BD).
Pre–B colony-forming assay.
After ACK lysis (Lonza), single-cell suspension of the BM from flox/flox and control (+/+) mice treated with Tam were plated in 60 mm dishes, (5 × 104 cells/dish) with MethoCult M3630 (Stem Cell Technologies) according to the manufacturer’s instructions. Cells were culture at 37°C with 5% CO2 and colonies were counted and morphologically assessed at different days of culture.
Retroviral transduction of BM cells.
The Phoenix retroviral packaging system was used with the transfection reagent FuGENE 6 (Roche). Retroviral supernatants were centrifuged (2,000 g) at 32°C for 90 min twice on 50 µg/ml RetroNectin precoated nontissue 12-well plates according to the manufacturer’s instructions. BM-derived pro–B cells were transduced by centrifuge at 500 g for 30 min. Bcl2 was obtained from S. Durum (National Cancer Institute, Frederick, MD). E12, E47, and Ebf1 were obtained from B. Kee (University of Chicago, Chicago, IL). Pax5 was obtained from C. Klug (The University of Alabama at Birmingham, Birmingham, AL). Bcl-xL (8790) and Mdm2 (16233) were purchased from Addgene. Stat5aCA was purchased from Cell Biolabs. The cDNA of Mcl1 (BC021638.1) was cloned into pMig empty vector. The vectors were sequence-confirmed before they were used for transfection into Phoenix packaging cells. All retroviral vectors expressed GFP, and an empty retroviral vector carrying GFP only was used as the control.
Microarray analysis and qRT-PCR.
RNA was extracted from cultured pro–B cells. The quality and quantity of RNA samples were tested by Bioanalyzer (Agilent Technologies). Total RNA was amplified using the Illumina Total Prep RNA Amplification kit (Ambion) according to the manufacture’s instructions. The biotinylated cRNA (1.5 g per sample) was applied to Illumina Mouse-6 Expression BeadChips and hybridized overnight at 58°C. Chips were washed, detected, and scanned according to the manufacture’s instruction, and the scanner output was imported into BeadStudio software (Illumina). Data are deposited to Gene Expression Omnibus (GEO) with the accession no. GSE40746.
qRT-PCR was performed with TaqMan Master Mix (ABgene). cDNA input was standardized and PCR was performed for 40 cycles. Primers for qRT-PCR are listed in Table S3.
Statistical analysis.
Data were statistically analyzed and figures were prepared using Microsoft Excel. A two-tailed Student’s t test was used throughout this work to evaluate statistical significance. Significance is indicated as follows: *, P < 0.05; **, P < 0.01.
ChIP-seq analysis.
ChIP was performed as previously described (Forsberg et al., 2000). Chromatin was prepared from 5 × 107 in vitro cultured mouse pro–B cells and sonicated to produce sheared DNA fragments ranging from 200–600 bp. Immunoprecipitation was performed with a polyclonal anti-Bcl11a antibody (Bethyl Laboratories) or IgG (Cell Signaling Technology). ChIP pull-down DNA was analyzed with qPCR in triplicate according to the manufacturer’s instructions, and the primers were listed in Table S3. For ChIP-seq analysis, the DNA library was constructed from anti-Bcl11a ChIP material and sequenced with Illumina Genome Analyzer IIx system. The sequence reads were mapped to mouse genome (version NCBI37) using Bowtie v0.12.1, and the peaks were identified using MACS v2.0.9 software. Motif study was performed with MEME motif-based sequence analysis tools (version 4.8.1). Genome-wide localization of binding peaks was analyzed with reference mouse genome downloaded from National Center for Biotechnology Information (version 37). Data are deposited in the online database ArrayExpress Archive (http://www.ebi.ac.uk/arrayexpress/) with the accession no. E-ERAD-98.
Online supplemental materials.
Fig. S1 contains information on gating strategies of hematopoietic population. Fig. S2 describes genotyping strategies. Table S1 gives definitions of the hematopoietic populations. Table S2 lists the antibodies used in this study. Table S3 has the primer information in this study. Table S4 lists the 100 lymphoid genes chosen for performing clustering analysis.
Acknowledgments
We thank the Sanger Institute RSF (Andrea Kirton, Michael Robinson, Robert Ellis, Sophie Jolley, and Marie Hitcham), flow cytometry core facility (Bee-Ling Ng and William Cheng), microarray facility, and DNA sequencing pipeline for technical assistance. We would also like to thank Drs. Scott Durum, Barbara Kee, and Chris Klug for reagents.
This work is supported by Wellcome Trust (grant number 098051; to P. Liu). Y. Yu is supported by a Fellowship from China Scholarship Council.
The authors declare no competing financial interests.
Author contributions: Y. Yu designed and did most of the experiments, analyzed and interpreted data, and contributed to the writing of the manuscript; J. Wang generated the GFP reporter mouse and performed ChIP experiments; W. Khaled produced Dflox/flox mice; J. Wang, S. Burke, W. Khaled, and P. Li. did experiments, provided intellectual input, and contributed to the writing of the manuscript; X. Chen and W. Yang analyzed and interpreted data; N. Jenkins and N. Copeland provided reagents; S. Zhang provided financial support to Y. Yu; and P. Liu designed the experiments, supervised the research, and wrote the manuscript.
References
- ChIP
chromatin immunoprecipitation
- CLP
common lymphoid progenitor
- CMP
common myeloid progenitor
- DN
double negative
- EP
erythroid progenitor
- ETP
early T cell progenitor
- GMP
granulocyte-monocyte progenitor
- HSC
hematopoietic stem cell
- LMPP
lymphoid-primed MPP
- LSK
Lin−Sca1+c-Kit+
- MEP
megakaryocyte-erythroid progenitor
- MPP
multipotent progenitor
- Tam
Tamoxifen
Author notes
Y. Yu, J. Wang, and W. Khaled contributed equally to this paper.