FLT3-ITD–mediated leukemogenesis is associated with increased expression of oncogenic PIM serine/threonine kinases. To dissect their role in FLT3-ITD–mediated transformation, we performed bone marrow reconstitution assays. Unexpectedly, FLT3-ITD cells deficient for PIM1 failed to reconstitute lethally irradiated recipients, whereas lack of PIM2 induction did not interfere with FLT3-ITD–induced disease. PIM1-deficient bone marrow showed defects in homing and migration and displayed decreased surface CXCR4 expression and impaired CXCL12–CXCR4 signaling. Through small interfering RNA–mediated knockdown, chemical inhibition, expression of a dominant-negative mutant, and/or reexpression in knockout cells, we found PIM1 activity to be essential for proper CXCR4 surface expression and migration of cells toward a CXCL12 gradient. Purified PIM1 led to the phosphorylation of serine 339 in the CXCR4 intracellular domain in vitro, a site known to be essential for normal receptor recycling. In primary leukemic blasts, high levels of surface CXCR4 were associated with increased PIM1 expression, and this could be significantly reduced by a small molecule PIM inhibitor in some patients. Our data suggest that PIM1 activity is important for homing and migration of hematopoietic cells through modification of CXCR4. Because CXCR4 also regulates homing and maintenance of cancer stem cells, PIM1 inhibitors may exert their antitumor effects in part by interfering with interactions with the microenvironment.
Genetic alterations that lead to uncontrolled protein tyrosine kinase (PTK) activity are a hallmark of human malignant myeloproliferative disorders. Fusion genes involving ABL or PDGFR are the molecular correlate of chronic myeloproliferative disorders, whereas activating mutations of FLT3 are recurrently found in human acute myeloid leukemia (AML; Chalandon and Schwaller, 2005). The success of small molecules that block oncogenic tyrosine kinase activity, such as imatinib-mesylate (Gleevec; Novartis), provided a proof of principle for targeted antileukemic therapy (Giles et al., 2005). However, the successful clinical use of such compounds has been challenged by the development of drug resistance and a limited clinical efficacy in patients with acute leukemia (von Bubnoff et al., 2003). To overcome these limitations, identification of critical signaling mediators downstream of an oncogenic tyrosine kinase is essential to identify new targets that would allow the development of an efficient combined therapeutic approach. There is strong evidence that most oncogenic tyrosine kinases mediate malignant transformation through parallel activation of several signaling pathways such as JAK–STAT, PI3K–AKT, RAS–RAF–MAPK, or NF-κB (Chalandon and Schwaller, 2005).
Retroviral gene tagging in c-myc–induced lymphomas in mice has led to the identification of the PIM family of serine/threonine kinases that encompasses three members: PIM1, PIM2, and PIM3 (Mikkers et al., 2002). Transgenic mouse models have demonstrated that PIM kinases exert their oncogenic potential mainly in cooperation with various oncogenes. Targeted inactivation of one, two, or all three PIM genes resulted in mice with some growth retardation and mild defects in steady-state hematopoiesis, but these mice were fertile with a normal lifespan (Mikkers et al., 2004). PIM1 has been characterized as one of the first target genes of STAT5 (Mui et al., 1996). In vitro and in vivo experimental work has demonstrated that constitutive STAT5 activation is a hallmark of a wide spectrum of hematological malignancies and that STAT5 is an essential mediator for transformation by oncogenic tyrosine kinases (Schwaller et al., 2000). Interestingly, both PIM1 and PIM2 are frequently overexpressed in leukemia and lymphoma as well as in some solid tumors (Amson et al., 1989; Mizuki et al., 2003; Hammerman et al., 2005). As PIM kinases are constitutively active and their expression is regulated predominantly by transcription and/or proteasomal degradation, this suggests that PIM kinases play an important role in hematological malignancies. Further in vitro functional studies have provided evidence that activation of PIM1 and/or PIM2 is important for malignant transformation by oncogenic tyrosine kinases such as FLT3-ITD, expressed in ∼25% of human AML cases (Kim et al., 2005; Adam et al., 2006; Agrawal et al. 2008). These studies indicated that small molecules inhibiting PIM kinases could provide a promising therapeutic avenue for hematological malignancies. However, it remains unclear whether one or both of the hematopoietically expressed PIMs (PIM1 and PIM2) are essential for mediating disease manifestations induced by oncogenic PTKs.
CXCL12, which is also known as SDF1 (stromal cell–derived factor 1), mediates signals through the chemokine receptor 4 (CXCR4) that are essential for homing and maintenance of hematopoietic stem cells (HSCs) in the bone marrow niche (Lapidot et al., 2005; Ratajczak et al., 2006). There is increasing evidence that CXCR4 is a key regulator of homing and retention of leukemic stem cells within the marrow niche that allows these cells to escape spontaneous and chemotherapy-induced cell death as well as promoting metastasis (Juarez and Bendall, 2004; Burger and Kipps, 2006). These findings are supported by the negative prognostic impact of high CXCR4 expression levels in patients with AML (Rombouts et al., 2004; Spoo et al., 2007). Consequently, targeting leukemic stem cells within the bone marrow by disruption of the CXCL12–CXCR4 interaction by small molecule inhibitors has been recently proposed (Zeng et al., 2006; Juarez et al., 2007). There is strong evidence that the CXCL12–CXCR4-mediated growth promotion and migration is not limited to leukemia but is also essential for the spreading of tumor-initiating cells of many solid tumors (Croker and Allan, 2008).
Using bone marrow cells from PIM knockout mice, we show that in vivo malignant transformation by the FLT3-ITD mutant is independent of PIM2. However, our experiments unexpectedly unravel that PIM1 is important for bone marrow reconstitution in lethally irradiated hosts. Further functional analysis suggests that PIM1 regulates surface CXCR4 expression in hematopoietic cells lines and in AML blasts. Collectively, our work reveals a previously unknown function of the oncogenic PIM1 serine/threonine kinase as regulator of homing and migration of bone marrow cells involving functional interaction with the CXCL12–CXCR4 signaling axis.
RESULTS
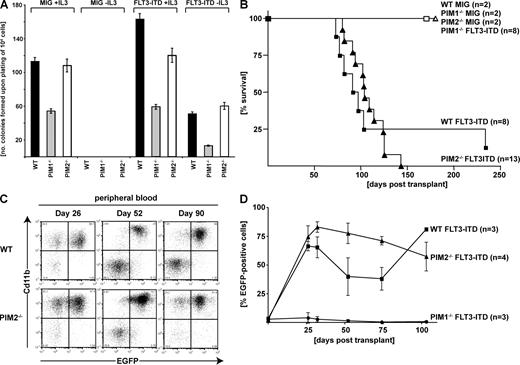
To investigate the role of PIM kinases in hematological diseases induced by oncogenic PTKs, we expressed the constitutively active FLT3-ITD mutant in primary mouse bone marrow cells. Expression of FLT3-ITD in WT cell induced growth factor-independent growth in liquid culture as well as in methylcellulose. Compared with WT bone marrow, expression of FLT3-ITD in PIM1−/− cells resulted in a significantly lower number of colonies growing with and without IL-3 (Fig. 1 A). No differences in colony formation or growth properties were observed when FLT3-ITD was expressed in bone marrow cells from mice lacking the PIM2 gene (PIM2−/−) compared with WT mice when grown in presence or in absence of IL-3 (Fig. 1 A). Liquid culture of the cells over 5 d revealed no difference in viability but a general reduced growth rate of bone marrow cells originating from PIM1−/− animals compared with PIM2−/− or WT mice (Fig. S1, B–D). These in vitro results suggest that PIM1, and not PIM2, is involved in signaling events that are essential for IL-3 and/or FLT3-ITD–regulated proliferation. To further study the role of PIMs in FLT3-ITD–mediated leukemogenesis in vivo, we performed bone marrow transduction/transplantation experiments using WT or PIM1−/− or PIM2−/− donor mice. Mice that were transplanted with cells lacking PIM1 showed no hematological disorder during 1-yr posttransplant observation (Fig. 1 B). Histological analysis of the normal sized spleens of animals that had been transplanted with PIM1−/− FLT3-ITD–expressing cells did not show any significant pathological changes 6 mo after transplant (Fig. S2, top). In contrast, all animals that received a transplant of FLT3-ITD–transduced bone marrow cells from WT or PIM2−/− mice developed a myeloproliferative disorder characterized by a massive expansion of EGFP-positive myeloid (CD11b+) cells in the peripheral blood assessed by flow cytometry 26 d after transplantation (Fig. 1 C). There was no significant difference in survival between recipients of WT versus PIM2−/− bone marrow transduced with FLT3-ITD, with both mice groups succumbing to a fatal disease within a median survival time of 108 and 138 d, respectively (Fig. 1 B). Histopathological examination of the spleens of sick mice revealed extensive infiltration with neoplastic myeloid cells in mice that received a transplant of FLT3-ITD–infected WT as well as PIM2−/− bone marrow cells (Fig. S2, middle and bottom). All mice receiving a transplant of FLT3-ITD–transduced bone marrow developed exactly the same mixed myeloid/T lymphoid phenotype, irrespective of the genetic background of the transplanted bone marrow (Fig. S3). Observation of the EGFP+ reconstituted cells revealed significant expansion of FLT3-ITD–expressing WT or PIM2−/− cells within the first month after transplant. In contrast, although detectable over several weeks, FLT3-ITD–expressing PIM1−/− underwent no significant expansion (Fig. 1 D).
In vitro and in vivo FLT3-ITD–mediated transformation in the presence and absence of PIM serine/threonine kinases. (A) Comparative analysis of clonogenic growth of FLT3-ITD–expressing bone marrow cells from FVB/N (WT) and PIM1−/− or PIM2−/− mice. 104 cells were sorted (EGFP+) 48 h after infection and plated in methylcellulose in presence or absence of IL-3. The figure represents one out of three independent experiments performed in triplicates. (B) Kaplan Meier survival plot. WT, PIM1−/−, or PIM2−/− bone marrow was transduced with FLT3-ITD or empty vector (MigRI) as indicated and transplanted into sublethally irradiated (750 rad) recipient mice. All mice that underwent transplantation with FLT3-ITD–transduced WT or PIM2−/− bone marrow died or were sacrificed because of disease conditions. One mouse transplanted with FLT3-ITD–transduced WT bone marrow showed extended survival and no signs of disease. n, number of mice transplanted and analyzed. Three independent transplantations were performed (see Fig. S3 E for details). (C) Flow cytometric analysis of peripheral blood samples in the course of the disease (26, 52, and 90 d after transplantation). Two-parameter dot plots show expression of myeloid lineage-specific antigen CD11b versus EGFP. The figure represents one out of three (WT) and four (PIM2−/−) analyzed mice, respectively. (D) Percentage of EGFP-positive leukocytes in the peripheral blood of transplanted mice determined at the indicated time points. Data represents the mean ± SD of three to four analyzed animals as indicated.
In vitro and in vivo FLT3-ITD–mediated transformation in the presence and absence of PIM serine/threonine kinases. (A) Comparative analysis of clonogenic growth of FLT3-ITD–expressing bone marrow cells from FVB/N (WT) and PIM1−/− or PIM2−/− mice. 104 cells were sorted (EGFP+) 48 h after infection and plated in methylcellulose in presence or absence of IL-3. The figure represents one out of three independent experiments performed in triplicates. (B) Kaplan Meier survival plot. WT, PIM1−/−, or PIM2−/− bone marrow was transduced with FLT3-ITD or empty vector (MigRI) as indicated and transplanted into sublethally irradiated (750 rad) recipient mice. All mice that underwent transplantation with FLT3-ITD–transduced WT or PIM2−/− bone marrow died or were sacrificed because of disease conditions. One mouse transplanted with FLT3-ITD–transduced WT bone marrow showed extended survival and no signs of disease. n, number of mice transplanted and analyzed. Three independent transplantations were performed (see Fig. S3 E for details). (C) Flow cytometric analysis of peripheral blood samples in the course of the disease (26, 52, and 90 d after transplantation). Two-parameter dot plots show expression of myeloid lineage-specific antigen CD11b versus EGFP. The figure represents one out of three (WT) and four (PIM2−/−) analyzed mice, respectively. (D) Percentage of EGFP-positive leukocytes in the peripheral blood of transplanted mice determined at the indicated time points. Data represents the mean ± SD of three to four analyzed animals as indicated.
As bone marrow harvest, as well as the retroviral infection rate of WT, PIM1−/−, or PIM2−/− bone marrow cells, did not differ (unpublished data), the absence of any FLT3-ITD–mediated disease might be explained by the inability of PIM1−/− to reconstitute irradiated animals. During the first series of experiments, we applied a nonmyeloablative irradiation dose to FVB/N mice (750 rad). To further exclude the possibility of a homing failure in PIM1−/− cells, we increased the irradiation to a lethal dose of 900 rad and repeated the experiments. Again, animals receiving FLT3-ITD–expressing cells from WT and PIM2−/− donors developed an identical myelolymphoproliferative disorder to that described in the previous paragraph (unpublished data). In contrast, all recipients receiving PIM1−/− bone marrow expressing the control vector (Mig-IRES-EGFP) alone or FLT3-ITD died rapidly (within 22 d) because of the lack of bone marrow reconstitution (Fig. 2 A). Careful examination of the hematologic parameters showed that in contrast to animals transplanted with PIM2−/− and WT cells, in recipients receiving PIM1−/− cells the white cell, red cell, and platelet counts declined rapidly in the first 2 wk after transfer (Fig. S4). This unexpected observation suggests that PIM1 might play an essential role in early bone marrow reconstitution. Although previous work has demonstrated that PIM1−/− bone marrow cells show some growth defects (Domen et al., 1993), as shown in colony-forming assays (Fig. 1 A), the number of HSCs, as assessed by the determination of the number of lineage-negative (Lin− Sca1+ Kit+) cells, was not reduced in PIM1−/− when compared with WT animals (FVB/N; not depicted).
The rapid declines in hematopoietic cells of all lineages, leading to death of animals within the first month after transplant, suggest that PIM1−/− HSCs have not only a proliferation defect but also impaired migration and/or homing capacities. To understand the role of PIM1 in HSC homing, we performed a series of experiments in which we labeled bone marrow cells from WT or PIM1−/− mice with a vital dye (CFSE), transplanted equal numbers of positively labeled and viable cells into lethally irradiated recipients, and determined the amount of cells in spleen and bone marrow 4 and 20 h after transplant. As shown in Fig. 2 B, a significant time-independent decrease in CSFE-labeled cells was observed in the bone marrow and spleen after transferring PIM1−/− cells compared with cells from WT donor animals. No significant difference in viability of CFSE-labeled cells was observed between the genotypes (Fig. 2 C). This finding suggests that PIM1−/− bone marrow cells are impaired in their capacity to home to hematopoietic organs.
PIM1−/− bone marrow cells do not reconstitute lethally irradiated recipient animals and have an impaired homing capacity. (A) Overall survival (Kaplan-Meier plot) of lethally irradiated (900 rad) recipients receiving WT, PIM1−/−, or PIM2−/− bone marrow cells. n, number of mice transplanted and analyzed. Two sets of independent transplantations were performed. (B) Homing of CFSE-labeled bone marrow cells from PIM1−/− and WT FVB/N mice to the bone marrow (top) and the spleen (bottom) of WT recipients. Data represent the mean ± SD of three experiments performed in triplicates. (C) Viability of bone marrow cells from PIM1−/− and WT FVB/N mice 4 and 20 h after CFSE stain. Data represent the mean ± SD of three experiments performed in duplicates.
PIM1−/− bone marrow cells do not reconstitute lethally irradiated recipient animals and have an impaired homing capacity. (A) Overall survival (Kaplan-Meier plot) of lethally irradiated (900 rad) recipients receiving WT, PIM1−/−, or PIM2−/− bone marrow cells. n, number of mice transplanted and analyzed. Two sets of independent transplantations were performed. (B) Homing of CFSE-labeled bone marrow cells from PIM1−/− and WT FVB/N mice to the bone marrow (top) and the spleen (bottom) of WT recipients. Data represent the mean ± SD of three experiments performed in triplicates. (C) Viability of bone marrow cells from PIM1−/− and WT FVB/N mice 4 and 20 h after CFSE stain. Data represent the mean ± SD of three experiments performed in duplicates.
The CXCL12–CXCR4 signaling axis has been proposed to be essential for homing and migration of hematopoietic cells (Juarez and Bendall, 2004; Lapidot et al., 2005; Burger and Kipps, 2006; Ratajczak et al., 2006). To explore the role of these signaling mediators, we compared WT, PIM1−/−, and PIM2−/− cells for their capacity to migrate over a CXCL12 gradient. As shown in Fig. 3 A, bone marrow cells lacking PIM1 are significantly impaired in migration. In contrast, PIM2−/− bone marrow cells were not defective in migration toward CXCL12. Interestingly, we found that bone marrow cells from PIM1−/− mice, but not PIM2−/− mice, expressed lower levels of surface CXCR4 when analyzed by flow cytometry (Fig. 3 B) but did not show any differences in the expression of other surface molecules involved in homing such as the integrins α4 and α5 (Fig. S5). In addition, CXCL12-induced Ca2+ flux was significantly reduced in bone marrow cells lacking PIM1 when compared with WT cells (Fig. 3 C). Furthermore, we observed a modest but reproducible impairment of CXCL12-induced phosphorylation of ERK in the lineage-negative subpopulation of PIM1−/− bone marrow cells analyzed by flow cytometry and immunoblotting (Fig. S6 and not depicted). These observations suggested that the serine/threonine kinase PIM1 could functionally regulate CXCR4.
Decreased steady-state CXCR4 surface expression of PIM1−/− bone marrow cells is associated with impaired migration and CXCL12-induced signaling. (A) Bone marrow cells from PIM1−/−, PIM2−/−, and WT FVB/N mice were allowed to migrate toward a 300-ng/ml CXCL12 gradient along with background migration as indicated. The migration index was calculated as a percentage of input cells. Data represent the mean ± SD of three experiments performed in triplicates. (B) Cell surface expression of CXCR4 of bone marrow cells from PIM1−/−, PIM2−/−, and WT mice was analyzed by staining with PE-conjugated anti–mouse CD184 antibody (open histogram). The level of nonspecific binding was assessed using a PE-conjugated rat IgG2b as isotype control (gray histogram). Data represent one out of three experiments. (C) Calcium flux was monitored in freshly isolated WT and PIM−/− bone marrow cells after stimulation with 300 ng/ml CXCL12. Data represent one out of three experiments.
Decreased steady-state CXCR4 surface expression of PIM1−/− bone marrow cells is associated with impaired migration and CXCL12-induced signaling. (A) Bone marrow cells from PIM1−/−, PIM2−/−, and WT FVB/N mice were allowed to migrate toward a 300-ng/ml CXCL12 gradient along with background migration as indicated. The migration index was calculated as a percentage of input cells. Data represent the mean ± SD of three experiments performed in triplicates. (B) Cell surface expression of CXCR4 of bone marrow cells from PIM1−/−, PIM2−/−, and WT mice was analyzed by staining with PE-conjugated anti–mouse CD184 antibody (open histogram). The level of nonspecific binding was assessed using a PE-conjugated rat IgG2b as isotype control (gray histogram). Data represent one out of three experiments. (C) Calcium flux was monitored in freshly isolated WT and PIM−/− bone marrow cells after stimulation with 300 ng/ml CXCL12. Data represent one out of three experiments.
To further experimentally address this possibility, PIM1 expression was knocked down by retrovirally expressed small interfering RNAs (siRNAs) in Ba/F3 cells and resulted in a significant decrease of CXCR4 surface expression (Fig. 4 A). Interestingly, Western blot analysis revealed no change in total CXCR4 protein expression (Fig. 4 B). Furthermore, expression of a dominant-negative PIM1-KD mutant also down-regulated cell surface CXCR4 expression, suggesting that PIM1 kinase activity is necessary (Fig. S7 A). These data suggested that PIM1 does not influence the total protein level of CXCR4, but may control the surface presentation, and that PIM1 activity might be necessary. We therefore tested whether the reexpression of PIM1 in PIM1−/− bone marrow cells could rescue surface CXCR4 expression and migration toward a CXCL12 gradient. As shown in Fig. 4 C, retroviral expression of PIM1 in bone marrow cells from PIM1−/− background increased surface CXCR4 expression to the level of WT littermates. In addition, expression of PIM1 restored the capability of cells to efficiently migrate toward CXCL12 (Fig. 4 D). These results strongly suggested that PIM1 regulates CXCR4 surface expression and function in hematopoietic cells. To further test the hypothesis that PIM1 kinase activity is involved in regulation of CXCR4 surface expression, we used a recently identified potent small molecule inhibitor of human PIM1 (K00486; Pogacic et al., 2007). Human JURKAT acute lymphoblastic leukemia cells that express high levels of PIM1 and surface CXCR4 were used in this experiment. Interestingly, as shown in Fig. 4 E, short-term nontoxic treatment (2 h, 10 µM) with the PIM inhibitor K00486 led to a transient but significant decrease of CXCR4 surface expression and reduced migration of the cells toward a CXCL12 gradient without affecting cellular viability (Fig. 4 F). Treatment of the cells with the PIM inhibitor also impaired CXCL12-induced, but not PMA-induced, ERK phosphorylation (Fig. S7 B). These results strongly suggested that PIM1 might act as functional regulator of the CXCL12–CXCR4 signaling axis.
PIM1 activity regulates CXCR4 surface expression in hematopoietic cells. (A) Down-regulation of PIM1, but not PIM2, resulted in reduced surface expression of CXCR4 in Ba/F3 cells. Ba/F3 cells were retrovirally transduced with pLMP (∅), pLMP-PIM1, or pLMP-PIM2 (expressing siRNA). Cell surface expression of CXCR4 was analyzed by staining with PE-conjugated anti–mouse CD184 antibody (open histogram). The level of nonspecific binding was assessed as described in 3B (gray histogram). Data represent one out of three experiments. (B) Down-regulation of PIM1 and PIM2 protein expression was confirmed by Western blot analysis. No change in total CXCR4 expression was observed. The figure represents one out of three independent experiments. (C) WT and PIM1−/− bone marrow was transduced with pMSCV mPIM1-YFP or empty vector (MYFP) as indicated. Cell surface expression of CXCR4 was analyzed by staining with PE-conjugated anti–mouse CD184 antibody. Data represent the mean of two independent experiments. (D) Bone marrow cells from WT and PIM1−/− FVB/N mice, transduced with pMSCV mPIM1-YFP or empty vector (MYFP) as indicated, were allowed to migrate toward a 300-ng/ml CXCL12 gradient along with background migration as indicated. The migration index was calculated as a percentage of input cells. Data represent the mean ± SD of three independent experiments performed in triplicates (one-way ANOVA: *, P ≤ 0.05). (E) Treatment of human JURKAT leukemia cells with a small-molecule PIM1 inhibitor (K00486, 10 µM) leads to a transient but significant reduction of surface CXCR4 expression after 2 h (dotted line) and 24 h (gray line). Viability of the cells was not significantly changed within the time of the experiment determined by 7-AAD staining (not depicted). Data represent one of three experiments. (F) JURKAT cells were allowed to migrate toward a 100-ng/ml CXCL12 gradient with or without pretreatment with 10 µM of the K00486 PIM inhibitor for 2 h. Data represent the mean ± SD of three experiments.
PIM1 activity regulates CXCR4 surface expression in hematopoietic cells. (A) Down-regulation of PIM1, but not PIM2, resulted in reduced surface expression of CXCR4 in Ba/F3 cells. Ba/F3 cells were retrovirally transduced with pLMP (∅), pLMP-PIM1, or pLMP-PIM2 (expressing siRNA). Cell surface expression of CXCR4 was analyzed by staining with PE-conjugated anti–mouse CD184 antibody (open histogram). The level of nonspecific binding was assessed as described in 3B (gray histogram). Data represent one out of three experiments. (B) Down-regulation of PIM1 and PIM2 protein expression was confirmed by Western blot analysis. No change in total CXCR4 expression was observed. The figure represents one out of three independent experiments. (C) WT and PIM1−/− bone marrow was transduced with pMSCV mPIM1-YFP or empty vector (MYFP) as indicated. Cell surface expression of CXCR4 was analyzed by staining with PE-conjugated anti–mouse CD184 antibody. Data represent the mean of two independent experiments. (D) Bone marrow cells from WT and PIM1−/− FVB/N mice, transduced with pMSCV mPIM1-YFP or empty vector (MYFP) as indicated, were allowed to migrate toward a 300-ng/ml CXCL12 gradient along with background migration as indicated. The migration index was calculated as a percentage of input cells. Data represent the mean ± SD of three independent experiments performed in triplicates (one-way ANOVA: *, P ≤ 0.05). (E) Treatment of human JURKAT leukemia cells with a small-molecule PIM1 inhibitor (K00486, 10 µM) leads to a transient but significant reduction of surface CXCR4 expression after 2 h (dotted line) and 24 h (gray line). Viability of the cells was not significantly changed within the time of the experiment determined by 7-AAD staining (not depicted). Data represent one of three experiments. (F) JURKAT cells were allowed to migrate toward a 100-ng/ml CXCL12 gradient with or without pretreatment with 10 µM of the K00486 PIM inhibitor for 2 h. Data represent the mean ± SD of three experiments.
Elevated surface CXCR4 expression has been demonstrated to be an adverse prognostic marker in patients with AML (Rombouts et al., 2004; Spoo et al., 2007). Because our results suggest that PIM1 is a regulator of surface CXCR4 expression, we compared pim1 expression levels in leukemic samples that have been previously analyzed for surface CXCR4 expression (Spoo et al., 2007). A tendency for higher pim1 expression in AML samples with high CXCR4 surface expression was observed (P < 0.05; Fig. 5 A, left). In contrast, we found no correlation between surface CXCR4 and cxcr4 messenger RNA (mRNA) levels (Fig. 5 A, right). These results suggest that PIM1 signaling is necessary for increased CXCR4 surface expression. When freshly isolated leukemic blasts from six patients with newly diagnosed AML expressing high surface CXCR4 levels received short-term treatment with the PIM inhibitor (K00486), a significant decrease in steady-state surface CXCR4 expression was observed in four out of six samples without significantly impaired viability (Fig. 5 B). These observations suggest that PIM1 is an important regulator of surface CXCR4 expression in primary human cancer cells. To determine if elevated PIM1 levels that are often found in human cancers might affect CXCR4 function, we evaluated migration of Ba/F3 cells stably overexpressing human PIM1 toward a CXCL12 gradient (Pogacic et al., 2007). As shown in Fig. 5 C, transmigration toward a gradient of 10 nM CXCL12 was significantly enhanced for PIM1-overexpressing cells and was significantly impaired in the presence of the PIM inhibitor.
Expression and regulation of PIM1 and CXCR4 in primary AML blasts. (A) Expression of PIM1 and CXCR4 mRNA in leukemic cells from AML patients with high versus low surface CXCR4 expression (as described in Spoo et al. [2007]) by quantitative real-time PCR analysis. The values are normalized to GAPDH levels and represent each AML patient (diamond) and median values (bars) of two independent experiments performed in triplicate (one-way ANOVA: *, P ≤ 0.05). (B) Regulation of surface CXCR4 expression in primary blasts from four out of six AML patients upon treatment with the K00486 PIM inhibitor. Data represents one of two independent experiments. (C) BaF/3 cells stably overexpressing human PIM1, as well as WT BaF/3 cells, were allowed to migrate toward a 10-nM CXCL12 gradient in the presence or absence of 10 µM of the K00486 PIM inhibitor for 2 h. The migration index was calculated as a percentage of input cells. Data represent the mean ± SD of three experiments (one-way ANOVA: **, P ≤ 0.01).
Expression and regulation of PIM1 and CXCR4 in primary AML blasts. (A) Expression of PIM1 and CXCR4 mRNA in leukemic cells from AML patients with high versus low surface CXCR4 expression (as described in Spoo et al. [2007]) by quantitative real-time PCR analysis. The values are normalized to GAPDH levels and represent each AML patient (diamond) and median values (bars) of two independent experiments performed in triplicate (one-way ANOVA: *, P ≤ 0.05). (B) Regulation of surface CXCR4 expression in primary blasts from four out of six AML patients upon treatment with the K00486 PIM inhibitor. Data represents one of two independent experiments. (C) BaF/3 cells stably overexpressing human PIM1, as well as WT BaF/3 cells, were allowed to migrate toward a 10-nM CXCL12 gradient in the presence or absence of 10 µM of the K00486 PIM inhibitor for 2 h. The migration index was calculated as a percentage of input cells. Data represent the mean ± SD of three experiments (one-way ANOVA: **, P ≤ 0.01).
CXCR4 expression is regulated by ligand-induced internalization and surface reexpression of a significant fraction (Busillo and Benovic, 2007). To address a functional connection of CXCR4 and PIM1, we followed CXCR4 surface expression in JURKAT cells upon stimulation with CXCL12 in presence or absence of the PIM inhibitor (Fig. 6 A). In analogy to previous studies, 1.5 h after exposure of JURKAT cells to 10 nM CXCL12, the majority of surface CXCR4 has been internalized (Ding et al., 2003). Washing out of the CXCL12 after 30 min resulted in rapid reexposure of surface CXCR4 to ∼80% of the starting level. Pretreatment of the cells with 10 µM of the PIM inhibitor for 1 h before CXCL12 addition increased the fraction of internalized CXCR4 to >90% after 1.5 h. Interestingly, surface CXCR4 reappearance after washing out was significantly impaired, reading only 40% of the starting level. To visualize the observed effects, we analyzed CXCR4 expression by immunofluorescent staining at the end of the experiment (3.5 h). As shown in Fig. 6 B, CXCL12 treatment resulted in internalization of a significant fraction of CXCR4 and washing out almost completely restored surface CXCR4. In presence of the PIM inhibitor, CXCL12 treatment also led to CXCR4 internalization; however, after washing out, an important fraction of the receptor remained internalized.
Functional inhibition of PIM1 impairs surface expression or reexpression of CXCR4. (A) Delayed ligand-induced surface expression or reexpression of CXCR4 in the presence of PIM1 inhibitor shown by flow cytometric evaluation in JURKAT cells. 0.5-h treatment with 10 nM CXCL12 before wash allows induction of recycling of CXCR4 on the cell surface evaluated 1, 2, and 3 h after wash compared with nonwashed cells. Recycling is delayed when cells are pretreated for 1 h with 10 µM of the PIM inhibitor (K00486). Data represent the mean of three independent experiments. (B) Confocal immunofluorescence analysis of distribution of CXCR4 in JURKAT cells after 3.5 h of treatment as mentioned in A. The left column shows CXCR4 distribution without PIM inhibitor, the right column shows CXCR4 distribution with treatment of 10 µM of the PIM inhibitor (K00486). The figure illustrates one out of three independent experiments. Bar, 10 µm. (C) JURKAT E cells were transduced with pLMP hPIM1-siRNA or control vector. After 1 h of treatment with CXCL12, the cells were washed. Surface expression of CXCR4 was analyzed before and after CXCL12 stimulation and 1, 2, and 3 h after wash. Data represent the mean ± SD of two independent experiments performed in duplicates.
Functional inhibition of PIM1 impairs surface expression or reexpression of CXCR4. (A) Delayed ligand-induced surface expression or reexpression of CXCR4 in the presence of PIM1 inhibitor shown by flow cytometric evaluation in JURKAT cells. 0.5-h treatment with 10 nM CXCL12 before wash allows induction of recycling of CXCR4 on the cell surface evaluated 1, 2, and 3 h after wash compared with nonwashed cells. Recycling is delayed when cells are pretreated for 1 h with 10 µM of the PIM inhibitor (K00486). Data represent the mean of three independent experiments. (B) Confocal immunofluorescence analysis of distribution of CXCR4 in JURKAT cells after 3.5 h of treatment as mentioned in A. The left column shows CXCR4 distribution without PIM inhibitor, the right column shows CXCR4 distribution with treatment of 10 µM of the PIM inhibitor (K00486). The figure illustrates one out of three independent experiments. Bar, 10 µm. (C) JURKAT E cells were transduced with pLMP hPIM1-siRNA or control vector. After 1 h of treatment with CXCL12, the cells were washed. Surface expression of CXCR4 was analyzed before and after CXCL12 stimulation and 1, 2, and 3 h after wash. Data represent the mean ± SD of two independent experiments performed in duplicates.
We next followed surface CXCR4 expression in JURKAT cells that have been transduced with a retrovirus expressing human PIM1-specific siRNA, leading to significant down-regulation of PIM1 protein expression (Fig. S8 A). The cells were stimulated with 10 nM CXCL12 for 30 min before washing out the ligand. Similar to treatment with PIM inhibitor, siRNA-mediated PIM1 knockdown in JURKAT cells significantly impaired CXCR4 surface reexpression (measured 1, 2, and 3 h after washout) to ∼50% of the WT control cells (Fig. 6 C). Collectively, these experiments suggest that PIM1 might play an important role for proper surface reexpression of CXCR4.
Functional regulation of CXCR4 by PIM1 activity raised the question of whether PIM1 kinase directly modifies the CXCR4 receptor. In support of a direct association, immunolocalization studies in JURKAT cells showed partially overlapping signals for CXCR4 and PIM1 predominantly in the cytoplasm (Fig. S8 B). CXCR4 undergoes ligand-dependent and ligand-independent endocytosis and surface reexpression depending on the integrity of the intracellular C-terminal domain rich in serine and threonine residues (Busillo and Benovic, 2007). We identified three putative PIM1 consensus sites in this region (S325, S330, and S339) containing either the preferred -5 or -3 arginine (position relative to the target serine; Fig. 7 A). To assess their potential as a PIM1 phosphorylation site, the C-terminal 46 residues of CXCR4 were expressed as a GST fusion protein (GST-C46) and treated in vitro with purified PIM1 protein. A single PIM1-dependent phosphorylation was detected by a shift of +80 Da in the intact protein mass as determined by electrospray ionization liquid chromatography/mass spectrometry, whereas no phosphorylation was detected on GST alone. To further define the phosphorylation site, a series of C-terminal deletion constructs were prepared, treated with PIM1, and analyzed by mass spectrometry. Phosphorylation was observed for short deletions, including the construct GST-C46Δ13, but was lost upon further truncation, locating the likely substrate site to the C-terminal residue S339 (Fig. 7 A). To confirm this result, the phosphorylation reaction was repeated with an 11-mer peptide derived from this site and a similar peptide containing the mutation S339A (Fig. 7 B). Importantly, MALDI TOF experiments revealed PIM1-dependent phosphorylation of the WT peptide but not the S339A mutant peptide. This data suggests that PIM1 regulation of CXCR4 results from direct phosphorylation of the C-terminal domain. Generation of a phosphospecific antibody has previously allowed the demonstration that phosphorylation of CXCR4-S339 occurs upon stimulation of brain cancer cells by CXCL12 or by phorbol-ester or EGFR (Woerner et al., 2005). We used this antibody in a series of in vitro kinase assays to show that PIM1-dependent phosphorylation of CXCR4 was lost specifically in the S339A mutant but not in the mutants S325A or S330A, which correspond to the other putative PIM1 recognition sites (Fig. 7 C). Treatment of JURKAT cells with the PIM1 inhibitor K00486 resulted in reduced CXCL12 and PMA-induced phosphorylation of CXCR4-S339 (Fig. 7 D, top). Similarly, reduced CXCL12-induced phosphorylation of CXCR4-S339 was observed upon siRNA-mediated knockdown of PIM1 (Fig. 7 D, bottom). In vitro kinase assays furthermore demonstrated that PIM1, but not PIM2, is able to efficiently phosphorylate CXCR4-S339, whereas BAD is phosphorylated by both kinases to a similar extent (Fig. 7 E). These observations suggest that kinase active PIM1, but not PIM2, can regulate CXCR4 phosphorylation.
PIM1 phosphorylates the CXCR4 C terminus at serine 339. (A) Deletion series defining the CXCR4 C-terminal region phosphorylated by PIM1. Schematic representation of human CXCR4 showing the C-terminal 46 aa expressed as a GST fusion (CXCR4 region boxed in blue). Three putative PIM1 recognition sites are shown in red. A series of C-terminal truncations map PIM1 phosphorylation to a peptide region including S339 (indicated by shaded area; ESI-LC/MS spectra, not shown). (B) Identification of Ser339 as a PIM1 phosphorylation site. MALDI TOF spectra of an 11-mer CXCR4 peptide (shaded sequence from A) show PIM1 phosphorylation of the WT sequence but not of a mutant S339A peptide. Inset box shows detail of the WT phosphorylation with or without PIM1 treatment. Spectra of the phosphorylated peptide are offset for clarity. The figure represents one out of two independent analyses. (C) In vitro kinase assay performed with different CXCR4 C-terminal mutants confirms that CXCR4 is phosphorylated by PIM1 at serine 339. Protein is detected by anti-PIM1 or anti–phospho-CXCR4 (Ser339) antibody. The blot represents one out of three independent experiments. (D, top) Determination of the CXCR4-Ser339 phosphorylation state in JURKAT cells incubated for 0.5 h with 10 nM CXCL12 or 40 nM PMA with or without a 1-h pretreatment with 10 µM of PIM inhibitor (K00486) before a 2-h wash. CXCR4-Ser339 phosphorylation is inhibited in the presence of PIM inhibitor. Values are the mean of two independent experiments. (D, bottom) Determination of CXCR4-Ser339 phosphorylation state in JURKAT E cells, transduced with pLMP hPIM1-siRNA or control vector and incubated for 0.5 h with 10 nM CXCL12, followed by a 2-h wash. The blot represents one out of two independent experiments. (E) In vitro kinase assay performed with CXCR4 C-terminal mutant C46 shows that CXCR4 is phosphorylated by PIM1 at serine 339 but not by PIM2. BAD phosphorylation was used as positive control to confirm enzyme activity of PIM1 and PIM2. The blot represents one out of two independent experiments.
PIM1 phosphorylates the CXCR4 C terminus at serine 339. (A) Deletion series defining the CXCR4 C-terminal region phosphorylated by PIM1. Schematic representation of human CXCR4 showing the C-terminal 46 aa expressed as a GST fusion (CXCR4 region boxed in blue). Three putative PIM1 recognition sites are shown in red. A series of C-terminal truncations map PIM1 phosphorylation to a peptide region including S339 (indicated by shaded area; ESI-LC/MS spectra, not shown). (B) Identification of Ser339 as a PIM1 phosphorylation site. MALDI TOF spectra of an 11-mer CXCR4 peptide (shaded sequence from A) show PIM1 phosphorylation of the WT sequence but not of a mutant S339A peptide. Inset box shows detail of the WT phosphorylation with or without PIM1 treatment. Spectra of the phosphorylated peptide are offset for clarity. The figure represents one out of two independent analyses. (C) In vitro kinase assay performed with different CXCR4 C-terminal mutants confirms that CXCR4 is phosphorylated by PIM1 at serine 339. Protein is detected by anti-PIM1 or anti–phospho-CXCR4 (Ser339) antibody. The blot represents one out of three independent experiments. (D, top) Determination of the CXCR4-Ser339 phosphorylation state in JURKAT cells incubated for 0.5 h with 10 nM CXCL12 or 40 nM PMA with or without a 1-h pretreatment with 10 µM of PIM inhibitor (K00486) before a 2-h wash. CXCR4-Ser339 phosphorylation is inhibited in the presence of PIM inhibitor. Values are the mean of two independent experiments. (D, bottom) Determination of CXCR4-Ser339 phosphorylation state in JURKAT E cells, transduced with pLMP hPIM1-siRNA or control vector and incubated for 0.5 h with 10 nM CXCL12, followed by a 2-h wash. The blot represents one out of two independent experiments. (E) In vitro kinase assay performed with CXCR4 C-terminal mutant C46 shows that CXCR4 is phosphorylated by PIM1 at serine 339 but not by PIM2. BAD phosphorylation was used as positive control to confirm enzyme activity of PIM1 and PIM2. The blot represents one out of two independent experiments.
DISCUSSION
Several lines of evidence suggest that the family of PIM serine/threonine kinases plays an important role in the pathogenesis of hematological malignancies. PIM kinases have been reported as being deregulated by oncogenic PTK fusions, such as the FLT3-ITD mutant found in up to a third of human AMLs. These studies have produced partly conflicting results regarding the role of PIM1 or PIM2 (which are both expressed in the hematopoietic system) in the pathogenesis of PTK-mediated leukemogenesis (Mizuki et al., 2003; Kim et al., 2005; Adam et al., 2006; Agrawal et al., 2008). Our study addressed the role of PIM kinases in primary cells, and the results showed that transformation of hematopoietic cells by FLT3-ITD in vitro and in vivo is independent of PIM2 (Fig. 1). This finding is unexpected, as previous work demonstrated that targeting of PIM2 by siRNA or by expressing dominant-negative kinase dead mutants (PIM-KD) significantly impaired growth and survival or FLT3-ITD–expressing hematopoietic cells (Adam et al., 2006; Agrawal et al., 2008). However, the previous experiments were performed in immortalized cell lines that stably express FLT3-ITD, whereas in the present work primary mouse bone marrow cells were used.
The PIM kinases have been identified through serial retroviral gene tagging in c-myc–induced lymphomas in WT mice, as well as in PIM knockout mice, suggesting that the loss of one PIM can be compensated for by deregulation of the others (Mikkers et al., 2004). However, there are studies suggesting that in some situations, the loss of a particular PIM kinase may not be compensated for by the presence of other PIMs. For example, the growth of bone marrow–derived mast cells from PIM1 knockout animals is significantly reduced despite induction of PIM2 by IL-3 (Domen et al., 1993). Our present work clearly demonstrates that in primary mouse bone marrow cells expressing FLT3-ITD, the lack of PIM1 cannot be compensated for by the presence of PIM2. Using bone marrow reconstitution assays, we show that FLT3-ITD induces a lethal myelo- and lymphoproliferative disorder in vivo in the absence of PIM2. Interestingly, PIM1−/− cells expressing an empty retrovirus (MigRI) or FLT3-ITD transplanted in a sublethally irradiated host were detectable but gradually declined over several weeks after transplant (unpublished data). Transplantation of an identical number of PIM1−/− cells into lethally irradiated hosts resulted in early death (2–4 wk) of all recipients, suggesting that cells lacking PIM1 have an inherent homing defect, which was further confirmed in a series of experiments measuring homing to the bone marrow and the spleen 4 and 20 h after transplant (Fig. 2). However, expression of FLT3-ITD in bone marrow cells from PIM1−/− mice resulted in generally impaired growth in vitro (Fig. 1 and Fig. S1), indicating that these cells do not only display a homing defect but also have a growth disadvantage compared with WT or PIM2−/− bone marrow cells.
Homing of HSCs to the bone marrow niche is a complex and still poorly understood process involving several signaling pathways. There is strong evidence that interaction of the chemokine CXCL12, which is expressed on stroma cells, with its receptor CXCR4, which is expressed on HSCs, plays an essential role in this process (Juarez and Bendall, 2004; Lapidot et al., 2005; Burger and Kipps, 2006; Ratajczak et al., 2006). Proper functioning of the CXCL12–CXCR4 axis seems to be crucial for directing homing/engraftment of normal, as well as leukemic, HSCs into bone marrow after transplantation (Peled et al., 1999; Tavor et al., 2004). Our results provide strong evidence that PIM1 can functionally regulate CXCR4. Absence of PIM1 is associated with impaired CXCL12-induced Ca2+ flux and activation of downstream effectors (Fig. 3). In addition, functional down-regulation by siRNAs, expression of a dominant-negative–acting kinase-dead mutant, or treatment with a small molecule PIM kinase inhibitor leads to down-regulation of CXCR4 expression at the surface of hematopoietic cells (Figs. 4 and 5). In addition, colocalization studies, as assessed by immunofluorescence, and in vitro kinase assays suggest that PIM1 may directly modify the intracellular C-terminal domain of CXCR4 (Fig. 7 and Fig. S8 B). However, we cannot rule out that kinase-active PIM1 indirectly regulates the phosphorylation of CXCR4 through another yet unidentified kinase.
Our results show that PIM1 regulates surface expression of CXCR4. PIM1−/− hematopoietic cells have a subtle but measurable defect in cytokine-mediated proliferation (Domen et al., 1993; Fig. 1). Therefore, it is very likely that impaired in vivo homing of PIM1−/− marrow cells might be the result of inappropriate function of several PIM targets including CXCR4. Interestingly, PIM1−/− marrow cells do not completely lack CXCR4. In fact, they express less surface CXCR4, most probably as a consequence of inappropriate reexpression. As PIM1−/− mice are viable and have a normal lifespan, a lower amount of surface CXCR4 seems to be sufficient to regulate steady-state hematopoiesis.
G protein–coupled receptors like CXCR4 are regulated by desensitization, internalization, and degradation (Busillo and Benovic, 2007). CXCR4 seems to undergo ligand-dependent and -independent internalization and surface reexposure in hematopoietic cells. In both internalization and reexposure of CXCR4, the serine/threonine-rich intracellular C terminus seems to play a key role (Orsini et al., 1999; Pelchen-Matthews et al., 1999). Natural mutants in patients with WHIM syndrome, as well as artificial truncation and alanine scanning mutagenesis, have suggested multiple regions in the CXCR4 C-terminal domain as potential phosphoacceptor sites (Hernandez et al., 2003). However, although multiple protein kinases have been proposed (PKC, GRK3, and GRK6), no specific kinase has yet to be clearly implicated (Busillo and Benovic, 2007). In this study, we observed that functional interference of PIM1 leads to a decrease in CXCR4 surface expression, whereas the total amount of the CXCR4 protein remains unchanged (Fig. 3). In addition, our work suggests that PIM1 regulates surface reexpression, whereas internalization of the receptor seems to be not affected (Fig. 6). A similar phenotype has been observed in T cells deficient in SYT3 (synaptotagmin 3), although the underlying mechanism remained unclear (Masztalerz et al., 2007). Our experiments using mass spectrometry and in vitro kinase assays show that PIM1 is able to directly phosphorylate Ser339 in the CXCR4 C-terminal domain in vitro (Fig. 7). It is worth noting that this site is in a motif that closely resembles the consensus PIM recognition motif with −5 arginine and −2 histidine although the preferred −3 arginine is absent (Bullock et al., 2005a). The CXCR4 Ser339 is known being phosphorylated in brain cancer cells upon stimulation with CXCL12, phorbol-ester (PMA), or EGF (Woerner et al., 2005). Interestingly, both CXCL12–CXCR4 and EGF–EGFR lead to activation of the JAK–STAT signaling pathway, which regulates PIM1 expression (Soriano et al., 2003), whereas PMA treatment is also known to produce a rapid induction of PIM1 expression (Wingett et al., 1996). As some of the PIM1 target sites, like the apoptosis regulator BAD (Ser112) or the cell cycle regulator p21(WAF1) (Thr145/Ser146), are also phosphorylated by other protein kinases, such as RAF1, PAK5, RSK2/5 for the former and PKC-δ or AKT for the latter (Aho et al., 2004; Jin et al., 2005; Kim et al., 2006, Oh et al., 2006; Zhang et al., 2007), it is unlikely that PIM1 is the only kinase that phosphorylates CXCR4-Ser339. Although the exact mechanisms remain to be elucidated, based on our results, PIM1-regulated phosphorylation of CXCR4-S339 could provide an interaction platform for proteins that regulate receptor surface reexpression. Elevated PIM1 levels frequently observed in leukemia and solid tumors would facilitate CXCL12-mediated signaling by increasing surface reexpression of the receptor. This hypothesis is supported by our observation of significantly increased migration toward CXCL12 in cells that overexpress PIM1 (Fig. 5 C).
Interaction of CXCL12 with CXCR4 does not only provide signals for efficient migration and homing, but seems also to support survival of hematopoietic progenitor cells (Lataillade et al., 2002). There is increasing evidence that CXCR4 is a key regulator of homing and retention of leukemic stem cells within the marrow niche, allowing these cells to escape spontaneous and chemotherapy-induced cell death (Juarez and Bendall, 2004; Sipkins et al., 2005; Burger and Kipps, 2006). These findings are supported by the negative prognostic impact of high CXCR4 expression levels in patients with AML (Rombouts et al., 2004; Spoo et al., 2007). Consequently, targeting leukemic stem cells within the bone marrow by disruption of the CXCL12–CXCR4 interaction by small molecule inhibitors has been recently proposed (Zeng et al., 2006; Juarez et al., 2007).
The association of elevated PIM1 expression with expression of high levels of surface CXCR4 in leukemic blasts from AML patients (Fig. 5 A) suggests that targeting aberrant PIM activity by small molecules would be rather promising by its effects on interfering not only with self-renewal but also with migration and homing of leukemic cells. Indeed, structural analysis of PIM1 has allowed us to identify a group of selective small molecule inhibitors with potent antileukemic activity in vitro (Pogacic et al., 2007). Short-term treatment of fresh leukemic blasts from six AML patients with the PIM inhibitor (K00486) resulted in a significant decrease of steady-state surface CXCR4 expression in four samples (Fig. 5 B). Ongoing experiments aim to understand why some AML blasts are resistant to CXCR4 regulation after treatment with the PIM inhibitors.
Collectively, we have dissected the role of PIM serine/threonine kinases for FLT3-ITD leukemogenesis in vitro and in vivo. Our work demonstrated that PIM2 is dispensable for transformation by FLT3-ITD and cells lacking PIM1 are impaired for growth and survival that cannot be overridden by FLT3-ITD. Most importantly, our work demonstrates that PIM1 activity directs cellular homing and migration by regulation of surface CXCR4 expression, suggesting that increased PIM1 expression supports CXCL12–CXCR4-mediated homing of leukemic stem cells and could also be implicated in metastasis formation in human cancer.
MATERIALS AND METHODS
DNA constructs and growth factors.
The complementary DNA (cDNA) of mouse FLT3-ITD was provided by H. Serve (University of Frankfurt, Frankfurt, Germany). The retroviral vector was constructed by cloning the FLT3-ITD cDNA into the MigRI retroviral vector coexpressing the green fluorescent protein (EGFP; Grundler et al., 2005). Mouse PIM1 in MigRI has been described previously (Pogacic et al., 2007). Retroviral constructs (pLMP) expressing mouse and human PIM-specific siRNAs have been obtained through Open Biosystems (mPIM1, VMM_46214; mPIM2, VMM_194110; hPIM1, V2HS_170016). A cDNA fragment coding for the 46 C-terminal CXCR4 residues (pcDNA3-hCXCR4fusin was provided by V. Piguet, Geneva University Hospital, Geneva, Switzerland) and various deletion and point mutants were generated by PCR and cloned into pGEX-2T. pcDNA3.1-HA-hCXCR4 was provided by M. Biard-Piechaczyk (Institut de Biologie, Montpelier, France). Recombinant IL-3, IL-6, and mouse stem cell factor were purchased from R&D Systems.
Cell culture, retrovirus preparation, and helper virus assay.
Mouse Ba/F3 cells were grown in RPMI-1640 containing 1 ng/ml IL-3 and 10% FCS. Human JURKAT T cells were obtained through the DSMZ (Braunschweig, Germany) and kept in RPMI-1640 containing 10% FCS. Retrovirus preparation was performed as described previously (Grundler et al., 2005).
Transduction and transplantation of mouse bone marrow.
Syngenic bone marrow transplantations were performed as previously described (Grundler et al., 2005). Bone marrow was harvested from FVB/N (WT), FVB/N PIM1−/−, and FVB/N PIM2−/− donor mice (provided by A. Berns and M. Nawijn, The Netherlands Cancer Institute, Amsterdam, Netherlands) and incubated overnight in IMDM/20% FCS supplemented with growth factors (10 ng/ml mIL-3, 10 ng/ml mIL-6, and 50 ng/ml mouse stem cell factor). Cells were transduced in four rounds of spin infection (1,200 g, 32°C, 90 min) with 4 µg/ml polybrene (Sigma-Aldrich). Subsequently, cells were resuspended in HBSS (Sigma-Aldrich) and injected into the tail vein of sublethally (750 rad) or lethally (900 rad) irradiated FVB/N recipient mice. All procedures were performed in compliance with the German animal welfare law, reviewed by the supervisory animal care committee of the Technical University Munich, and approved by the government of Upper Bavaria.
Flow cytometric immunophenotyping (FACS analysis).
Single-cell suspensions of indicated tissue samples were prepared by passing tissue through 100-µm nylon mesh strainers (Falcon; BD). Red blood cells of peripheral blood were lysed before analysis. Cells were preincubated with Fc block and CD45-PECy5 and subsequently stained with PE-conjugated anti-CD11b (Mac-1), CD45R/B220, CD90.1 (Thy1.1), CD4, or CD8-α antibodies as indicated. Dead cells were excluded by propidium iodide (PI) or 7-amino-actinomycin D (7-AAD) staining as mentioned in the text. Cell surface expression of CXCR4 was analyzed by staining with PE-conjugated anti–mouse CD184 (2B1) or APC-conjugated anti–human CD184 (12G5) antibodies (BD). Unspecific binding was assessed by a PE- or APC-conjugated rat IgG2b as an isotype control. Fc block and all antibodies were purchased from BD.
Bone marrow cell homing.
Female recipient FVB/N mice were lethally irradiated (900 rad) 24 h before transplantation. Donor cells of PIM1−/− or WT FVB/N mice were stained with 1 µM CFSE (Invitrogen) according to the manufacturer’s instructions. Comparable staining and viability of labeled cells was determined by flow cytometry (FACS). Subsequently, cells were resuspended in HBSS and irradiated mice received transplants of 106 CFSE-stained and viable bone marrow cells via tail vein injection. Mice were killed 4 or 20 h after transplantation as indicated. Single cell suspensions were incubated with Fc-blocking reagent and subsequently stained with PE-CY5–conjugated anti-CD45 antibody. Dead cells were excluded by PI staining. Donor cells in bone marrow and spleen were detected by flow cytometry. The CFSE-stained cells were concurrently cultured in IMDM/20% FCS supplemented with growth factors, and viability was determined by PI staining after 4 and 20 h to exclude differences in viability during the homing process.
Cellular migration analysis.
5 × 105 bone marrow cells from PIM1−/−, PIM2−/−, and WT FVB/N mice were allowed to migrate 4 h toward a 300-ng/ml CXCL12 gradient using 5.0-µm pore size transwell chambers (Costar). The migration index was calculated as a percentage of input cells. For Ba/F3 and JURKAT cells, migration toward a 10-nM CXCL12 gradient was allowed for 4 h using transwell chambers, when indicated cells were pretreated for 2 h with 10 µM K00486.
Phosphorylation of GST-CXCR4.
PIM1 was expressed and purified as described previously (Bullock et al., 2005b). GST-CXCR4 fusions were expressed in BL21 (DE3) cells with 0.5 mM IPTG induction for 6 h at 18°C. Lysates were purified by glutathione sepharose chromatography. Phosphorylation reactions were performed for 90 min at room temperature in 125-µl samples containing 1 µM PIM1 and 10 µM GST-CXCR4 buffered in 50 mM Hepes, pH 7.5, 100 mM NaCl, 1 mM DTT, 1 mM ATP, 5 mM MgCl2, and 1 mM MnCl2. Control samples were treated similarly in the absence of PIM1. Reactions were stopped by the addition of 0.1% formic acid and protein masses determined by electrospray ionization liquid chromatography/mass spectrometry.
In vitro kinase assay.
5 µg of recombinant PIM1 or PIM2 was incubated at 30°C for 5 min in kinase buffer (40 mM Tris-HCl, pH 7.4, 20 mM MgCl2, and 0.1 mg/ml bovine serum albumin) containing 500 µM ATP with or without 2 µM PIM inhibitor (K00486; Pogacic et al., 2007). 8 µg of the mentioned GST-CXCR4 construct or BAD-purified protein (Millipore) were then added for a further 10 min. The reactions were terminated by the addition of Laemmli sample buffer. Phosphorylation of CXCR4 was analyzed by Western blotting with anti–phospho-CXCR4 antibody (provided by J. Rubin, Washington University School of Medicine, St. Louis, MO; Woerner et al., 2005). Phosphorylation of BAD was determined by Western blotting with anti–phospho-BAD (Cell Signaling Technology). Equal loading of substrate was evaluated by using anti-GST (GE Healthcare) and anti-BAD (Cell Signaling Technology).
CXCR4 expression after PIM inhibition in AML patient samples.
Mononuclear cells from peripheral blood from AML patients with >90% of leukemic cells were freshly purified using Ficoll gradient. Cells were washed in PBS and seeded in 24-well plates with 106 cells/ml of media containing RPMI and 10% FCS. Cells were treated with DMSO and 5 or 10 µM K00486. After 30 min of treatment, cells were fixed in 3.75% PFA, and CXCR4 surface expression was measured using APC-conjugated anti–human CD184 (12G5) antibody. The work with clinical samples was approved by the ethics board of the Albert-Ludwigs-University Freiburg, Germany (Project 163/03).
Regulation of CXCR4 surface expression.
JURKAT or JURKAT-E (transduced with pLMP of pLMP-hPIM1) cells were plated in RPMI complete at a concentration of 105/ml and pretreated for 1 h with 10 µM K00486 or DMSO at 37°C, as indicated. After this treatment, cells were incubated for 30 or 60 min with 10 nM CXCL12 at 37°C. Half of the cells were washed three times in PBS to remove CXCL12 and K00486 and resuspended in new medium, and the rest were left in the original medium. Samples were collected right after the CXCL12 treatment and 1, 2, and 3 h after washing. Cells were fixed in 4% PFA. Surface CXCR4 was analyzed by staining with APC-conjugated anti–human CD184 (12G5) antibody. Nonspecific binding was determined using the APC-conjugated rat IgG2b as isotype control (BD).
Determination of intracellular Ca2+ flux.
The increase in the intracellular calcium concentration ([Ca2+]i) induced by CXCL12 was monitored by fluorescence spectrophotometry. Freshly isolated BM cells were loaded with 1 µM Fura-2-AM (Invitrogen) in calcium buffer (137 mM NaCl, 2.7 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 5.6 mM glucose, and 0.1% BSA buffered with 20 mM Hepes, pH 7.4). After incubation at 37°C for 30 min, cells were washed twice and suspended in calcium buffer to a final concentration of 2 × 106 cells/ml. Fura-2 fluorescence was monitored in an LS50B luminescence spectrophotometer (PerkinElmer). The excitation wavelengths used were 340 and 380 nm; emission was measured at 510 nm. [Ca2+]i was calculated from the Grynkiewicz equation. The Kd used was 224 nM. Rmax was obtained after lysis of the cells with 0.1% Triton X-100, and Rmin was determined by addition of 10 mM EGTA.
Confocal microscopy analysis.
JURKAT cells were flattened on a glass slide, fixed with acetone for 3 s, and dried overnight. Cells were treated briefly with 0.1 M glycin in PBS followed by 0.3% Triton X-100 in phosphate buffer, incubated overnight at 4°C with anti-CXCR4 (Abcam) and anti-PIM1 (12H8) antibodies (Santa Cruz Biotechnology, Inc.), and washed and incubated for 1 h at room temperature with appropriate secondary antibodies (Invitrogen). TOTO-3 iodide (642/660) was used for DNA staining. Confocal microscopy was performed on a laser-scanning confocal microscope (Radiance 2100; Bio-Rad Laboratories) equipped with a krypton/argon laser and a red laser diode. To reduce bleed-through, confocal images were acquired sequentially. Noise reduction is achieved by Kalman filtering during acquisition. Cells harvested at the last time point of the recycling assay were cytospinned and immunolocalization was performed. Cells were permeabilized with 0.5% Triton X-100 in PBS, treated with RNase, and blocking was done in 0.1% Tween-20/PBS supplemented with 1% BSA. The CXCR4 primary antibody (Abcam) was added for 1 h at room temperature, and slides were washed and incubated in the dark for 1 h with the Alexa Fluor 488 goat anti–rabbit antibody (Invitrogen). PI was used for nuclei staining. Slides were washed in mounted with FluorSafe reagent (EMD). Confocal microscopy is carried on a laser-scanning microscope (LSM 510; Carl Zeiss, Inc.).
MALDI TOF analysis of CXCR4 peptides.
CXCR4 11-mer peptides (WT, KRGGHSSVSTE; and S339A, KRGGHSAVSTE) were phosphorylated for 2 h at room temperature in 300-µl samples containing 3 µM PIM1 and 30 µM CXCR4 peptide buffered in 25 mM Hepes, pH 7.5, 150 mM NaCl, 1 mM DTT, 1 mM ATP, 5 mM MgCl2, and 1 mM MnCl2. Control samples were treated similarly in the absence of PIM1. Samples were desalted using a ZipTipC18 (Millipore) equilibrated with 2% acetonitrile and 0.5% TFA, and bound peptide was washed repeatedly with the same buffer. Peptides were eluted with 2–5 µl of a saturated solution of α-cyano-4-hydroxycinnamic acid in 40% ACN and 0.1% TFA and then deposited directly onto a stainless steel target plate and allowed to air dry. The spots were subsequently analyzed using an ultraflex LIFT II mass spectrometer equipped with a 337-nm N2 laser (Bruker Daltonics) and the peak lists were extracted using Flex Analysis software (Bruker Daltonics).
Immunoblotting.
Cellular expression of PIM1, PIM2, and CXCR4 was analyzed by Western blot analysis using standardized protocols with the following antibodies: α-PIM1 (clone 12H8, 1:1,000), α-PIM2 (clone 1D12, 1:1,000; Santa Cruz Biotechnology); α-CXCR4 (ab1670, 1:1,000; Abcam); and β-actin (clone 8AC-15, 1:5,000; Sigma-Aldrich).
Quantitative real-time PCR analysis.
mRNA expression was determined by quantitative real-time PCR using SYBR Green. 1 µg of total mRNA (isolated with the RNeasy kit; QIAGEN) was used for cDNA synthesis (High Capacity cDNA Reverse Transcription kit; Applied Biosystems), and 50 ng cDNA was applied for real-time PCR using the Power SYBR Green PCR MasterMix (Applied Biosystems) on an ABI Prism 7700 sequence detection system (Applied Biosystems). Expression levels were normalized to GAPDH using the ΔCt method. The following oligonucleotides were used as PCR primers: hPIM1 forward, 5′-CGAGCATGACGAAGAGATCAT-3′, and hPIM1 reverse, 5′-TCGAAGGTTGGCCTATCTGA-3′ (119-bp amplicon); hCXCR4 forward, 5′-ATGAAGGAACCCTGTTCCCGT-3′, and hCXCR4 reverse, 5′-AGATGATGGAGTAGATGGTGGG-3′ (76-bp amplicon); and hGAPDH forward, 5′-GTGGTCTCCCTGACTTTCAACAGC-3′, and hGAPDH reverse, 5′-ATGAGGTCCACCACCTGCTTGCTG-3′ (149-bp amplicon). Statistical significance was assessed by one-way ANOVA.
In vitro colony-forming assays.
Bone marrow was transduced according to the method described in Transduction and transplantation of mouse bone marrow. 10–20 × 103 EGFP-positive bone marrow cells per well were plated in triplicate in methylcellulose media containing 10 ng/ml of recombinant IL3 or no cytokines (StemCell Technologies, Inc.).
Online supplemental material.
Fig. S1 shows that the expression of FLT3-ITD induces growth factor–independent growth in primary mouse bone marrow cells independent of PIM2. Fig. S2 shows the induction of a myeloproliferative disease in mice transplanted with FLT3-ITD expressing WT or PIM2−/− but not PIM1−/− bone marrow. Fig. S3 shows that the absence of PIM2 does not affect the FLT3-ITD–induced phenotype in vivo and summarizes all transplant experiments. Fig. S4 shows the hematopoietic reconstitution of mice transplanted with WT, PIM1−/−, and PIM2−/−. Fig. S5 shows that PIM1 does not influence integrin-α4/α5 expression. Fig. S6 shows ERK phosphorylation in lineage-negative PIM1−/− cells. Fig. S7 shows that the expression of a dominant-negative PIM1 mutant decreases surface CXCR4 expression and that the inhibition of PIM affects CXCL12-induced ERK phosphorylation. Fig. S8 shows the knockdown efficacy of PIM1 and the immunolocalization of PIM1 and CXCR4 in JURKAT cells.
Acknowledgments
We would like to thank M. Wymann, D. Jankovic, V. Stavropoulou, T. Liu, B. Erne, Pamela della Mina, V. Jaeggin, and U. Schibler for their inputs and help and R. Skoda for critically reading the manuscript.
This work has been supported by Oncogene networks in AML to R. Grundler and J. Duyster, funded by the Mildred Scheel Stiftung, and by a grant from the José Carreras Stiftung to J. Duyster. This work has also been supported by grants from the Swiss National Science Foundation (SNF-3100AO-116587/1), ONOCOSUISSE (OCS-01830-02-2006), the Susy-Rueckert Foundation and the Krebsliga beider Basel (Basel, Switzerland), and the Wilhelm Sander Foundation (2008.048.1, Munich, Germany) to J. Schwaller. J. Schwaller is a research professor supported by the Gertrude von Meissner Foundation. The Structural Genomics Consortium is a registered charity organization (number 1097737).
The authors have no conflicting financial interests.
References
Abbreviations used: AML, acute myeloid leukemia; HSC, hematopoietic stem cell; mRNA, messenger RNA; PTK, protein tyrosine kinase; siRNA, small interfering RNA.
Author notes
J. Duyster and J. Schwaller contributed equally to this paper.





![Figure 5. Expression and regulation of PIM1 and CXCR4 in primary AML blasts. (A) Expression of PIM1 and CXCR4 mRNA in leukemic cells from AML patients with high versus low surface CXCR4 expression (as described in Spoo et al. [2007]) by quantitative real-time PCR analysis. The values are normalized to GAPDH levels and represent each AML patient (diamond) and median values (bars) of two independent experiments performed in triplicate (one-way ANOVA: *, P ≤ 0.05). (B) Regulation of surface CXCR4 expression in primary blasts from four out of six AML patients upon treatment with the K00486 PIM inhibitor. Data represents one of two independent experiments. (C) BaF/3 cells stably overexpressing human PIM1, as well as WT BaF/3 cells, were allowed to migrate toward a 10-nM CXCL12 gradient in the presence or absence of 10 µM of the K00486 PIM inhibitor for 2 h. The migration index was calculated as a percentage of input cells. Data represent the mean ± SD of three experiments (one-way ANOVA: **, P ≤ 0.01).](https://cdn.rupress.org/rup/content_public/journal/jem/206/9/10.1084_jem.20082074/7/m_jem_20082074_gs_fig5.jpeg?Expires=1773827663&Signature=lnaIkPYgJRZdVaeSEftgILbWdNnrGULGCl5kqAjrGaX2ooYLgtF5iPXHxtiwGQ30LUpA4Gs9JamiMyLx-t7CDlpIcpCm2Amwx-kgfQGOWaOOeV5UCwmN8txivXM73YB~RInMc1WSZcj8WQM6OtpFVCpKmX865K8SjVPBIad3BlU66ucoIcDSya5PAg6LHGjdrVHNJVJm7yiO~HXb6SEokw6Glwur3C7fH3ugr5WGi7P18IqK2J3ggsK9HhftvqmooXa0gwU~4gno~lvSTSzuWr4am5X8bur1u0S-wpTuaJfnw9xXouKNR8atx1ZcDmb7JNwpDU3hhi7jqqC2gUprWw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)




