Identification of the transmitted/founder virus makes possible, for the first time, a genome-wide analysis of host immune responses against the infecting HIV-1 proteome. A complete dissection was made of the primary HIV-1–specific T cell response induced in three acutely infected patients. Cellular assays, together with new algorithms which identify sites of positive selection in the virus genome, showed that primary HIV-1–specific T cells rapidly select escape mutations concurrent with falling virus load in acute infection. Kinetic analysis and mathematical modeling of virus immune escape showed that the contribution of CD8 T cell–mediated killing of productively infected cells was earlier and much greater than previously recognized and that it contributed to the initial decline of plasma virus in acute infection. After virus escape, these first T cell responses often rapidly waned, leaving or being succeeded by T cell responses to epitopes which escaped more slowly or were invariant. These latter responses are likely to be important in maintaining the already established virus set point. In addition to mutations selected by T cells, there were other selected regions that accrued mutations more gradually but were not associated with a T cell response. These included clusters of mutations in envelope that were targeted by NAbs, a few isolated sites that reverted to the consensus sequence, and bystander mutations in linkage with T cell–driven escape.
On sexual transmission, HIV-1 infects CD4+CCR5+ T cells and remains localized in genital/rectal mucosa and draining lymph nodes for ∼10 d (1). Virus then spreads via the blood to other lymphoid tissue, especially that in the gut (2). There, it replicates profusely and the level of free virus in the blood rises exponentially to reach a peak, often millions of virus copies per milliliter of plasma, 21–28 d after infection. Virus level then falls, rapidly at first, until a stable level is reached (3, 4). This set point varies across patients and is partially predictive of the later course of disease in the absence of antiretroviral drugs; patients with low set points progress to AIDS more slowly (5).
It is uncertain how the peak viremia of acute HIV-1 infection is controlled. Some mathematical models (6–8) suggest that the rampant early infection simply destroys so many CD4 T cells in the gut (2, 9) and elsewhere that the cell substrate becomes limiting. However, reduction of peak viremia in rhesus macaques infected with simian immunodeficiency virus (SIV) was dependent on the presence of CD8 cells (10), either T or NK cells, or both. In HIV-1 infection, virus-specific CD8 T cells first appear in the blood just before the viremia peaks and then expand and contract as virus load falls (11–13). HIV-1–specific CD8 T cells are detectable before seroconversion and long before neutralizing antibodies (NAbs) (14). However, the mere presence of such T cells does not prove that they control virus. Indirect evidence for their importance comes from HLA class I allelic associations with low virus set point and with delayed progression to AIDS (15), but these effects, which are probably mediated by HLA class I–restricted T cells, could occur after the initial drop in viremia. Overall, this leaves the relative importance of CD8 T cells and the loss of infectable target cells to the decline of viremia of acute HIV-1 infection unresolved.
Further clues for a role of CD8 T cells in HIV-1 containment comes from studies demonstrating that they select virus escape mutations as early as 30–54 d after the peak viremia of acute infection (16, 17) and continue to select mutations throughout chronic infection (18–26). Nearly all these observations have been made after the steep decline from peak viremia, as the virus is stabilizing at its set point, or later. Few studies have directly measured T cell escape; it is more often argued that changes in virus sequence in regions that correspond to CD8 T cell epitopes matching the HLA type of the patient are selected by T cells (e.g., see reference 28). However, this approach can be confounded by the many overlapping epitopes, that are presented by different HLA types (27). Approaches based on sequence alone also miss T cell responses to invariant epitopes as well as mutants selected by other immune responses.
In this paper, a comprehensive study was made of the ontogeny of the primary T cell response in acute HIV-1 infection. The single genome amplification (SGA) sequencing technique (28, 29) identified the single transmitted/founder virus sequence in each of four preseroconversion patients, before the immune response had made a discernable imprint on the virus quasispecies. The adaptive immune responses to the transmitted virus proteome were then followed using peptides matched to autologous founder virus and to subsequent mutants, to capture responses that might otherwise be missed because of virus variability (30). This approach identified the first T cell responses in HIV-1–infected humans and showed that virus escape mutants were rapidly selected as viremia fell from its peak in acute infection. A mathematical model quantified the downward pressure exerted by these CD8 T cell responses on the virus, showing that they contributed to the concurrent reduction of viremia. These data imply that vaccines that stimulate strong and broad early CD8 T cell responses to HIV-1 should be valuable.
RESULTS
Experimental strategy
A strategy was designed to detect the first T cell responses induced by the single founder/transmitted virus in patients CH40, CH58, and CH77, each identified (31) in Fiebig stage II (3) (Table S1 and Fig. S1 show clinical details). Another untreated patient, SUMA0874, who was previously described (16, 21), was included after sequencing of his transmitted/founder full-length viral genome (29). At the time of screening, there was no evidence of prior immune selection (29). Thereafter, patients were bled regularly, including at time points when virus load was falling from peak. T cell responses were initially measured by ELISpot and/or intracellular cytokine staining (ICS) 6 mo after enrollment using peptides matched to the founder virus sequence. Earlier time points were also tested using peptides reactive at 6 mo and also peptides spanning mutational clusters that were candidates for immune selection. The scale of the experiment and the limited number of PBMC available precluded peptide titrations, but at the 2 µg/ml (∼10−6 M) peptide concentration used in each experiment, there was either no recognition or clearly impaired recognition of the mutant peptide. This relatively high peptide concentration should have allowed the detection of even relatively low avidity T cell responses to the mutated epitope (32), so lack of response was convincing evidence that these were true escape mutations.
Amino acid sequence changes from transmitted/founder virus
Three distinct statistics were used to capture different aspects of positive selection and immune evasion for CH40, CH77, and CH58 (Fig. 1). The first used a sliding window of 45 bases to identify concentrated mutations, either highly mutated single sites or clusters of mutations, within the window. The second test identified clustered mutations in a 27-base sliding window. Finally, a tree-based statistic was used to identify single sites under positive selection (33, 34). When combined, 49 localized regions were identified in the three patients where significant variation emerged during the course of the study. The statistical significance of these regions of interest and a summary of all immunological responses identified are presented in Table S2 and supported by the supplemental text. In the three patients, 22 variable regions were embedded in experimentally confirmed T cell or NAb epitopes. An additional 10 T cell epitopes were identified that were not under detectable selective pressure during the first 12–18 mo of infection.
Identification of putative sites of positive selection using three independent tests. Subjects shown are CH40 (A), CH58 (B), and CH77 (C). The horizontal axis indicates nucleotide positions aligned to HXB2 (nt 750–9750), and the vertical axis indicates significance from each of three tests. Tests for enriched mutations and clustered mutations (black and red bars, respectively) used sliding windows of 45 and 27 nt, respectively. Fixed-effects likelihood (FEL) test results from HYPHY are color coded to reflect the reading frame of the codon (+1, blue; +2, cyan; +3, green). Because overlapping reading frames or recombination might influence FEL results, the FEL test was repeated for aligned fragments without regions where reading frames overlap and partitioned at potential recombination breakpoints (black x). Epitope locations for CTL (gray stripes) and NAb (black stripes) epitopes are juxtaposed. For reference, a map of the HXB2 genome appears below. Methods are detailed in the text. Not shown are 10 (4 in CH40, 4 in CH58, 2 in CH77) regions that were identified as under selection by the first test but were predominantly the consequence of clusters of silent mutations, at least in the standard reading frames for HIV proteins. Some might represent cryptic epitopes, T cell response encoded in unconventional reading frames (63).
Identification of putative sites of positive selection using three independent tests. Subjects shown are CH40 (A), CH58 (B), and CH77 (C). The horizontal axis indicates nucleotide positions aligned to HXB2 (nt 750–9750), and the vertical axis indicates significance from each of three tests. Tests for enriched mutations and clustered mutations (black and red bars, respectively) used sliding windows of 45 and 27 nt, respectively. Fixed-effects likelihood (FEL) test results from HYPHY are color coded to reflect the reading frame of the codon (+1, blue; +2, cyan; +3, green). Because overlapping reading frames or recombination might influence FEL results, the FEL test was repeated for aligned fragments without regions where reading frames overlap and partitioned at potential recombination breakpoints (black x). Epitope locations for CTL (gray stripes) and NAb (black stripes) epitopes are juxtaposed. For reference, a map of the HXB2 genome appears below. Methods are detailed in the text. Not shown are 10 (4 in CH40, 4 in CH58, 2 in CH77) regions that were identified as under selection by the first test but were predominantly the consequence of clusters of silent mutations, at least in the standard reading frames for HIV proteins. Some might represent cryptic epitopes, T cell response encoded in unconventional reading frames (63).
Fig. 2 provides details for these 49 sites of putative positive selection. Validated T cell epitopes are boxed and NAb epitopes are highlighted in blue. All sites are given a lowercase letter to facilitate discussion. Distinct evolutionary trajectories were seen in the 18 proven CD8 T cell epitopes. In 3/18 cases, the first variant in the epitope region simply replaced transmitted virus (Fig. 3 A, CH77.b; and Fig. S2). In a single case, five different variants of the peptide were explored, but the transmitted form remained dominant throughout the study period (Fig. S2, CH40.q). In the remaining 14/18 cases, multiple variants were explored with the final dominant form appearing later in the process (Fig. 3, B and C, CH77.cc, CH77.c; and Fig. S2). These included examples where mutations accrued not only within the defined epitope but proximal to it as well; the nearby mutations might be compensatory or impact processing. NAb-driven escape mutations (CH40.k, CH40.l, CH77.o, and CH77.p) did not begin to accumulate until after 100 d into the infection and also had complex escape patterns (Fig. 2, blue text; and supplemental text). Each was found in a variable loop in gp120 (V1 and V3 in CH40 and V2 in CH77; unpublished data). Overall, these complex patterns are signatures of immune selection for CD8 T cell and NAb selected escapes.
Virus sequence changes emerge both early and rapidly at multiple sites across the HIV-1 proteome over the 12 mo after infection. Full-length SGA sequencing was performed at screening (0 d) for CH40, CH77, and CH58. Sequencing at multiple time points (indicated to the left of each block as days) subsequent to screening used SGA techniques but was derived from separate sequencing of the 5′ (Gag and Pol) and 3′ (Vif, Vpr, Tat, Rev, Vpu, Env, and Nef) regions of the virus genome. The sites in which significant variation was detected by 6 mo after infection are identified in columns including the percentage of SGA sequences that contained nonsynonymous (n.s.) changes at each time point. Some percentages are reported in either red or blue text. Red text indicates early and rapid escapes where n.s. changes occurred in at least 50% of SGA sequences by ∼50 d after screening. Blue text identifies those sites that were experimentally confirmed as containing Nab epitopes. Boxed columns identify T cell epitopes, with dotted boxes (red or black) indicating known or putative CTL motifs according to each patient's HLA type (four digits), whereas solid boxes (red or black) identify those sites found in reactive peptides confirmed in T cell assays. Above solidly boxed columns, the reactive 18-mer peptide, relative to HXB2 standards, is shown and, if confirmed, the optimal CTL peptide. Below each column, n.s. substitutions are shown in parentheses; orange text indicates reversions to consensus clade B. * identifies those n.s. substitutions that were present in >50% of SGA sequences at the final visit. # denotes an insertion. The lower case letter preceding the protein (e.g., a. Gag) is a reference used throughout the paper and supplements to denote regions of positive selection. CH77.l and CH58.f identify regions of change in overlapping reading frames.
Virus sequence changes emerge both early and rapidly at multiple sites across the HIV-1 proteome over the 12 mo after infection. Full-length SGA sequencing was performed at screening (0 d) for CH40, CH77, and CH58. Sequencing at multiple time points (indicated to the left of each block as days) subsequent to screening used SGA techniques but was derived from separate sequencing of the 5′ (Gag and Pol) and 3′ (Vif, Vpr, Tat, Rev, Vpu, Env, and Nef) regions of the virus genome. The sites in which significant variation was detected by 6 mo after infection are identified in columns including the percentage of SGA sequences that contained nonsynonymous (n.s.) changes at each time point. Some percentages are reported in either red or blue text. Red text indicates early and rapid escapes where n.s. changes occurred in at least 50% of SGA sequences by ∼50 d after screening. Blue text identifies those sites that were experimentally confirmed as containing Nab epitopes. Boxed columns identify T cell epitopes, with dotted boxes (red or black) indicating known or putative CTL motifs according to each patient's HLA type (four digits), whereas solid boxes (red or black) identify those sites found in reactive peptides confirmed in T cell assays. Above solidly boxed columns, the reactive 18-mer peptide, relative to HXB2 standards, is shown and, if confirmed, the optimal CTL peptide. Below each column, n.s. substitutions are shown in parentheses; orange text indicates reversions to consensus clade B. * identifies those n.s. substitutions that were present in >50% of SGA sequences at the final visit. # denotes an insertion. The lower case letter preceding the protein (e.g., a. Gag) is a reference used throughout the paper and supplements to denote regions of positive selection. CH77.l and CH58.f identify regions of change in overlapping reading frames.
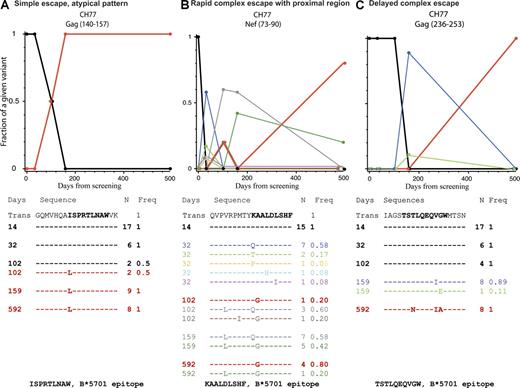
Examples of different patterns of escape. The alignment shows the transmitted form of the peptide that was susceptible to T cells with the variants that were observed over the course of the study underneath. Dashes indicate aa that are identical to the transmitted form and letters indicate aa substitutions. The days from screening (left), the sequence (center), and occurrence of each variant and frequency (right) are shown. A plot of the frequencies of each form over time is shown above the alignment; the colors of the sequences correspond to those of the lines. The transmitted form is shown in black, the most common form at the last time point in red. The pattern shown in A, where a single variant completely replaced the transmitted form, was rare. The pattern in B was typical of rapid escape; the transmitted form was lost very quickly, but many mutated forms were rapidly explored by the virus, and the dominant form at the last time point was not among the earliest escape mutations. The pattern in C was typical of more gradual escape with continuing evolution, but this is a Gag epitope, where escape patterns tended to be of reduced complexity overall. Kinetic patterns of variation for all 19 peptides that stimulated T cell responses are shown in Fig. S2. All alignments and frequencies for all regions of interest are in the supplemental text.
Examples of different patterns of escape. The alignment shows the transmitted form of the peptide that was susceptible to T cells with the variants that were observed over the course of the study underneath. Dashes indicate aa that are identical to the transmitted form and letters indicate aa substitutions. The days from screening (left), the sequence (center), and occurrence of each variant and frequency (right) are shown. A plot of the frequencies of each form over time is shown above the alignment; the colors of the sequences correspond to those of the lines. The transmitted form is shown in black, the most common form at the last time point in red. The pattern shown in A, where a single variant completely replaced the transmitted form, was rare. The pattern in B was typical of rapid escape; the transmitted form was lost very quickly, but many mutated forms were rapidly explored by the virus, and the dominant form at the last time point was not among the earliest escape mutations. The pattern in C was typical of more gradual escape with continuing evolution, but this is a Gag epitope, where escape patterns tended to be of reduced complexity overall. Kinetic patterns of variation for all 19 peptides that stimulated T cell responses are shown in Fig. S2. All alignments and frequencies for all regions of interest are in the supplemental text.
12 regions of positive selection were identified within 2–4 wk of screening (Fig. 2, red text; details about timing of responses and acquisition of variants can be found in Table S2 and the supplemental text). Six sites were selected by detectable T cell responses (Fig. 2, solid red boxes), four of which rapidly declined once the mutant virus became fixed (Figs. 4–7,567, CH40.t, CH77.s, CH77.cc, and CH58.e). At six other rapidly selected sites there were no detectable T cell responses (CH40.a, d, h, and i, CH58.g, and CH77.i). These mutations might be explained by mechanisms other than direct escape from T cells. Two cases are candidates for early reversions, as the dominant early mutation in these regions restored the consensus amino acid (CH77.l and CH40.i). Four (CH40.a, h, and i and CH77.i; Table S2) covaried with other known early T cell escape mutations and could, therefore, be bystanders that were carried along with an escape mutation (discussed further in the next Results section). This left two sites, CH40.d and CH58.g, unexplained. CH40.d may be a processing mutation (discussed further in the next Results section). The site CH58.g was neither a good candidate for a reversion event nor a bystander mutation. There was no known or predicted T cell epitope in this site, but the rapid depletion of the transmitted form, with subsequent complex patterns of variation (detailed in supplemental text), is suggestive of immune escape.
Early and rapid loss of the transmitted form and acquisition of ns mutations within the first weeks of HIV-1 infection result from T cell selection pressure. Within each panel, the HXB2 position sequence of both the wild-type and mutated peptides are given. The percentage of founder virus, which is the proportion of SGA sequences produced which contain founder sequence spanning that relevant 18-mer peptide, is also given. The wild-type and mutated peptides, as well as HLA-matched known or predicted optimal epitopes, were tested in ex vivo IFN-γ ELISpot or ICS. CH40, A and B; CH77, C–E; CH58, F. ELISpot data are expressed as the mean spot forming units (SFU) per million PBMC (SFU/106) ± SEM and ICS data as percentage of CD8 memory T cells secreting IFN-γ, IL-2, MIP-1β, TNF-α, and/or CD107a.
Early and rapid loss of the transmitted form and acquisition of ns mutations within the first weeks of HIV-1 infection result from T cell selection pressure. Within each panel, the HXB2 position sequence of both the wild-type and mutated peptides are given. The percentage of founder virus, which is the proportion of SGA sequences produced which contain founder sequence spanning that relevant 18-mer peptide, is also given. The wild-type and mutated peptides, as well as HLA-matched known or predicted optimal epitopes, were tested in ex vivo IFN-γ ELISpot or ICS. CH40, A and B; CH77, C–E; CH58, F. ELISpot data are expressed as the mean spot forming units (SFU) per million PBMC (SFU/106) ± SEM and ICS data as percentage of CD8 memory T cells secreting IFN-γ, IL-2, MIP-1β, TNF-α, and/or CD107a.
T cell responses induced in acute HIV-1 infection in CH40. Ex vivo IFN-γ ELISpots were performed in CH40 over the first 12 mo of infection. (A) T cell responses that were both >50 SFU/106 and 4× background at one or more time points are shown. Data are expressed as the mean SFU/106 PBMC ± SEM. Line color is used to group T cell responses that peaked at different times during the first year of HIV-1 infection. (B) Mapped T cell responses against the transmitted HIV-1 virus 181 d after screening for CH40. Each panel shows the T cell response to the 18-mer peptide (position relative to HXB2 standards) described in the title. (C–K) Ex vivo IFN-γ ELISpots were performed on PBMC at the days indicated using peptides matching the transmitted virus. Data are graphed against percentage of transmitted virus, which reflects the proportion of SGA sequences produced at each time point that contain the transmitted virus sequence spanning that 18-mer peptide. The purple dotted line in H shows the percentage of transmitted virus in the proximal site CH40.d that may have had an impact on the processing of the Pol 80–97 epitope.
T cell responses induced in acute HIV-1 infection in CH40. Ex vivo IFN-γ ELISpots were performed in CH40 over the first 12 mo of infection. (A) T cell responses that were both >50 SFU/106 and 4× background at one or more time points are shown. Data are expressed as the mean SFU/106 PBMC ± SEM. Line color is used to group T cell responses that peaked at different times during the first year of HIV-1 infection. (B) Mapped T cell responses against the transmitted HIV-1 virus 181 d after screening for CH40. Each panel shows the T cell response to the 18-mer peptide (position relative to HXB2 standards) described in the title. (C–K) Ex vivo IFN-γ ELISpots were performed on PBMC at the days indicated using peptides matching the transmitted virus. Data are graphed against percentage of transmitted virus, which reflects the proportion of SGA sequences produced at each time point that contain the transmitted virus sequence spanning that 18-mer peptide. The purple dotted line in H shows the percentage of transmitted virus in the proximal site CH40.d that may have had an impact on the processing of the Pol 80–97 epitope.
T cell responses induced in acute HIV-1 infection in CH77. (A) The kinetics of the T cell cytokine response to the three epitopes that underwent early rapid escape and the B*5701 TW10 response that became immunodominant at 6 mo were described using ICS measuring total cytokine production in memory CD8 T cells. The dotted gray box highlights the first 50 d after screening when the virus load is declining to set point. Virus escapes that occurred in this period are denoted as both early and rapid. (B) Mapped T cell responses against the transmitted HIV-1 virus 159 d after screening for CH77 with overlapping peptides group with brackets with B*5701 epitopes highlighted in red. (C–F) Ex vivo IFN-γ ELISpots (C) or ICS (D–F) were performed on PBMC from patient CH77 at the days indicated using peptides matching the transmitted virus. Data from both patients are graphed against the percentage of wild-type virus, which reflects the proportion of SGA sequences produced at each time point that contain the transmitted virus sequence spanning that 18-mer peptide. Data are expressed as the mean SFU/106 PBMC. Error bars in C show SEM. ICS data represent the percentage of the background-subtracted IFN-γ, TNF-α, MIP-1β, IL-2, and CD107α production in memory CD8 T cells. Additional ELISpot data are shown as insets in D and E.
T cell responses induced in acute HIV-1 infection in CH77. (A) The kinetics of the T cell cytokine response to the three epitopes that underwent early rapid escape and the B*5701 TW10 response that became immunodominant at 6 mo were described using ICS measuring total cytokine production in memory CD8 T cells. The dotted gray box highlights the first 50 d after screening when the virus load is declining to set point. Virus escapes that occurred in this period are denoted as both early and rapid. (B) Mapped T cell responses against the transmitted HIV-1 virus 159 d after screening for CH77 with overlapping peptides group with brackets with B*5701 epitopes highlighted in red. (C–F) Ex vivo IFN-γ ELISpots (C) or ICS (D–F) were performed on PBMC from patient CH77 at the days indicated using peptides matching the transmitted virus. Data from both patients are graphed against the percentage of wild-type virus, which reflects the proportion of SGA sequences produced at each time point that contain the transmitted virus sequence spanning that 18-mer peptide. Data are expressed as the mean SFU/106 PBMC. Error bars in C show SEM. ICS data represent the percentage of the background-subtracted IFN-γ, TNF-α, MIP-1β, IL-2, and CD107α production in memory CD8 T cells. Additional ELISpot data are shown as insets in D and E.
T cell responses induced in acute HIV-1 infection in CH58. (A) The kinetics of the T cell cytokine response using ICS measuring the total IFN-γ, TNF-α, MIP-1β, IL-2, and CD107α T cell response in memory CD8 T cells. (B) Mapped T cell responses against the transmitted HIV-1 virus 145 d after screening for CH58. In C–G, each panel shows the T cell response, either by ICS or IFN-γ ELISpot, to the 18-mer peptide (position relative to HXB2 standards) described in the title and is graphed against the percentage of wild-type virus, which reflects the proportion of SGA sequences produced at each time point that contain the transmitted virus sequence spanning that 18-mer peptide. The inset in E shows that the T cell response, as measured by ELISpot to a pool of mutant peptides (M), RRQDILDLWVYHTQGYFP, KRQEILDLWVYHTQGYFP, was half that of the response to the transmitted sequence peptide KRQDILDLWYHTQGYFP (T). The purple dotted line in F shows the percentage of transmitted virus in the proximal site CH58.h that may have impacted on processing of the Nef 113–130 epitope. The inset in F shows the ELISpot response at day 70 to the optimal B*5701-restricted epitope HW10. In G, only three SGA sequences were produced at the indicated timepoint (+), so although some variation from the transmitted virus was detected, the changes were not significant in any of the three tests used (i.e., not listed in Fig. 2). Error bars in B, E, and F indicate SEM.
T cell responses induced in acute HIV-1 infection in CH58. (A) The kinetics of the T cell cytokine response using ICS measuring the total IFN-γ, TNF-α, MIP-1β, IL-2, and CD107α T cell response in memory CD8 T cells. (B) Mapped T cell responses against the transmitted HIV-1 virus 145 d after screening for CH58. In C–G, each panel shows the T cell response, either by ICS or IFN-γ ELISpot, to the 18-mer peptide (position relative to HXB2 standards) described in the title and is graphed against the percentage of wild-type virus, which reflects the proportion of SGA sequences produced at each time point that contain the transmitted virus sequence spanning that 18-mer peptide. The inset in E shows that the T cell response, as measured by ELISpot to a pool of mutant peptides (M), RRQDILDLWVYHTQGYFP, KRQEILDLWVYHTQGYFP, was half that of the response to the transmitted sequence peptide KRQDILDLWYHTQGYFP (T). The purple dotted line in F shows the percentage of transmitted virus in the proximal site CH58.h that may have impacted on processing of the Nef 113–130 epitope. The inset in F shows the ELISpot response at day 70 to the optimal B*5701-restricted epitope HW10. In G, only three SGA sequences were produced at the indicated timepoint (+), so although some variation from the transmitted virus was detected, the changes were not significant in any of the three tests used (i.e., not listed in Fig. 2). Error bars in B, E, and F indicate SEM.
There were 23 positively selected regions that arose more slowly (Fig. 2, black text) that were neither embedded in demonstrable T cell epitopes nor targeted by NAb. Most, however, were embedded either in a previously documented CD8 T cell epitope or in a predicted epitope, based on anchor motifs given the patient's HLA types (ELF software, Los Alamos National Laboratory [LANL]), identified as dashed boxes in Fig. 2 (details in Table S2). Eight regions (CH40.j, m, n, and s and CH77.t, u, w, and x) were only under positive selection pressure in the last sample, >1 yr after infection. These immune responses might not have been detectable at 6 mo when samples were screened using autologous peptide sets. 11 selected regions were in Env and might be attributable to undetected NAb escape. Four regions may demonstrate reversion to consensus sequence at later time points (CH40.r, CH58.a, CH77.f, and CH77.g; Table S2). Together with CH40.i and CH77.l, there are six candidates for reversion independent of immune escape; all were simple escapes involving only 1- or 2-aa mutations. Further support for a role for reversion in early selection is that mutations toward consensus were enriched in regions under positive selection that were not in experimentally defined epitopes (21/66), compared with selected sites that were in defined epitopes (8/58; P = 0.014 by a Fisher's exact test). Reversions within epitopes recognized by T cells probably assisted escape (Fig. 2, CH40 Gag 395–403 and CH77 Gag 240–249 and Nef 82–90). A handful of positions among the slower to evolve set were otherwise unexplained but covaried with a site in a selected epitope (Table S2). The mutational patterns within patients were highly interlinked, and some selected mutations may have carried along bystander mutations that would eventually be lost through recombination.
Evolution of T cell responses in early HIV-1 infection
Patient CH40.
In patient CH40, a typical HIV-1–infected patient (set point, 13,183 cc/ml), escape in two epitopes was detected in the acute phase (Fig. 2; and Fig. 4, A and B). Several clustered residues in Nef 185–202 (Fig. 2, CH40.t), predominately an R192H change to the clade B consensus, were found in greater than half of the sequences 14 d after screening. The founder sequence was completely lost 29 d later when R192Q and S188R/N changes dominated. Both the transmitted and mutant forms of the 18-mer peptide (unpublished data) and a predicted HLA-A*3101-restricted nonamer peptide, Nef 188–196 SLAFRHVAR, within it were made and tested for T cell recognition in ELISpot assays. This SLAFRHVAR epitope, which has not been previously described, elicited a response, whereas mutant peptides were not recognized (Fig. 4 A). Another rapid change, R403K (an APOBEC-driven mutation, again toward the clade B consensus) that occurred within the Gag peptide 389–406 (CH40.b) became fixed between 16 and 45 d after screening, as the virus load was falling. At 111 d from screening, another change, I401L (away from consensus), was observed with both changes persisting (though not occurring on the same virus molecule) at 1 yr from screening. The peptide that matched the founder virus sequence, but not its mutant, was recognized by T cells. From within the longer peptide, a predicted new HLA A*3101-restricted epitope, Gag 395–404 CGKEGHIAR, was strongly recognized by T cells, whereas nonamers representing each of the mutants were not (Fig. 4 B).
Four other sites contained early and rapid mutations (Fig. 2, CH40, red text with and without dashed red box, and CH40.a, d, h, and i) but were not detected in T cell assays at or around the time of complete loss of the transmitted virus sequence (not depicted). Between 16 and 45 d after screening, an A120D substitution in Gag 113–130 (CH40.a) moving away from consensus became fixed. An additional V128G substitution was detected from day 111, and by day 412 an insertion was observed at position 128. This region did not correspond to a known HLA-matched CD8+ T cell epitope, but a previous study described a possible CD4 T cell epitope at this site (35). Vpr 74–91 (CH40.i), which corresponded to an HLA-matched T cell motif, became fully mutated by day 45 with complex changes including an initial S84R change to consensus persisting for the first year. Vif 161–178 (CH40.h) exhibited a single amino acid change, which had a high mutual association with nucleotide position 8752 in the Nef epitope region 186–202 (CH40.t) which was selected by T cells (see the third paragraph in the previous Results section), suggesting that this was a mutation carried along by immune selection elsewhere. Lastly, a single Pol K76R change was observed in >50% SGA sequences in Pol (CH40.d) between days 16 and 45, with almost full loss of its transmitted form by day 412 (Fig. 2). This residue did not occur in a known HLA epitope but was proximal to the invariant T cell epitope Pol 80–97, which elicited a response in this patient. The Pol K76R change may therefore represent a mutation that alters processing of Pol 80–97.
Fig. 5 A details the evolving pattern of T cell responses in CH40 over the first 12 mo of infection. The T cell responses to the first two epitopes described in the first paragraph of this section were present in the first PBMC sampled on day 16. The response to the Nef 185–202 (CH40.t) epitope decayed rapidly after the transmitted/founder form was lost (Fig. 5, A–C). The T cell response to the Gag 389–406 (CH40.b) peptide may already have been declining by day 16 but persisted at a low level after the loss of the founder form (Fig. 5, A, B, and D). Three other T cell responses, which expanded after day 16, all selected escape mutants but more slowly: Vif 113–130 (CH40.g), Rev 49–66 (CH40.o), and Env 830–847 (CH40.q). The first two lost the founder form over 200 and 400 d, respectively, and the third never fully replaced the transmitted with escape forms (Fig. 5, E–G). There were also four T cell responses (Gag 481–498, Pol 80–97, Pol 824–841, and Env 765–682), 40% of the whole response, that selected no escape (Fig. 5, H–K); although, as described in the previous paragraph, there is the possibility that the rapidly emerging K76R (CH40.d) mutation may have had an impact on the kinetics of the Pol 80–97 T cell response (Fig. 5 H). Overall, there were three phases of the T cell response in CH40: an early period of falling virus load and rapid escapes within 35 d of screening, then an increasing T cell response dominated by the Rev 49–66 (CH40.o)-specific response, and, lastly, a very strongly dominant response to an invariant Pol 824–841 epitope (Fig. 5 A). The T cell responses were initially focused and then broadened over time.
Patient CH77.
This patient was HLA B*5701 positive and controlled virus better than CH40, with viral load <2,000 cc/ml 6 mo after screening, although a year later virus load increased to 9,165 cc/ml (Fig. S1). There were three early T cell responses that rapidly selected escape mutants in Env 350–368 (CH77.s), Nef 17–34 (CH77.aa), and Nef 73–90 (CH77.cc). In each case, the T cells recognized the founder sequence but did not recognize the mutant peptides or recognized them poorly (Fig. 4, C–E). The T cell response to Nef 17–34 (CH77.aa) epitopes was no longer detectable by the ELISpot assay once the founder virus sequence was lost. Both responses to Env 352–360 (CH77.s) and Nef 73–90 (CH77.cc) declined but remained detectable using ICS at 6 mo (Fig. 6, A–E).
Sequencing of single genomes meant that potential linkage between these escape mutations could be examined (Table I). The Env 352–360 (CH77.s) epitope QFRNKTIVF, which was a new epitope restricted by HLA Cw0401, escaped first between days 0 and 14. Changes were complex and included an APOBEC-driven mutation, R355K, which occurred within the optimal epitope and remained dominant over time. Next, the Nef 17–34 (CH77.aa) epitope mutated between days 0 and 32 and then the Nef epitope 73–90 (CH77.cc) mutated between days 14 and 32. For the Nef 81–90 KAALDLSHF epitope restricted by HLA B*5701, two changes to clade B consensus, A83G and V76L, emerged at day 102 from screening, within and proximal to the epitope, with the A83G dominant at the final visit. Thus, in 32 d, three complete escapes had been selected in rapid succession and accumulated on the same virus molecules.
CH77 3′ SGA direct sequencing spanning CTL epitopes
| Day | SGA clone | ENV 343-363 (CH77.s) | NEF 17-35 (CH77.aa) | NEF 75-94 (CH77.cc) |
| 0 | Consensus | SHVVDKLREQFRNKTIVFNH | RERRRRDEPAAVGVGPASQ | PVRPMTYKAALDLSHFLKEK |
| 14 | CH77_WGA_B3a | ...........K........ | ................... | .................... |
| 14 | CH77_WGA_C7 | ...........K........ | ................... | .................... |
| 14 | CH77_WGA_C9 | ...........K........ | ................... | .................... |
| 14 | CH77_WGA_C4 | ...........K........ | ................... | .................... |
| 14 | CH77_WGA_D5 | ...........K........ | ................... | .................... |
| 14 | CH77_WGA2b | .................... | ................... | .................... |
| 14 | CH77E_L7 | ...........S........ | ......N............ | .................... |
| 14 | CH77E_L8 | ...........K........ | .....K............. | .................... |
| 14 | CH77E_M6 | ..............A..... | ....K.............. | .................... |
| 32 | CH77_WK03_3_B1 | ...........K........ | ....G.............. | .......Q............ |
| 32 | CH77_WK03_3_D1 | ...........K........ | ........S.......... | ............I....... |
| 32 | CH77_WK03_3_D2 | ...........K........ | ......E............ | .......P............ |
| 32 | CH77_WK03_3_D3 | ...........K........ | .....K............. | .......Q............ |
| 32 | CH77_WK03_3_TB1 | ...........K........ | .....K............. | .......Q............ |
| 32 | CH77_WK03_3_TB2 | ...........K........ | ......E............ | .......T............ |
| 32 | CH77_WK03_3_TB4 | ...........K........ | ........S.......... | ..........H......... |
| 32 | CH77_WK03_3_TB5 | ...........K........ | ......N............ | .......Q............ |
| 32 | CH77_WK03_3_TB6 | ...........K........ | ..GA.ERR........... | .......Q............ |
| 32 | CH77_WK03_3_TB7 | ...........K........ | .....K............. | .......Q............ |
| 32 | CH77_WK03_3_TB8 | ...........K........ | ......E............ | .......T............ |
| 32 | CH77_WK03_3_TC1 | ...........K........ | ......N............ | .......Q............ |
| 102 | CH77_WK12_3_E1 | .Y.........K........ | ......N............ | .L.....Q............ |
| 102 | CH77_WK12_3_TA1 | ...........K........ | .....K............. | .L.....Q............ |
| 102 | CH77_WK12_3_TA2 | ...........N........ | .....T............. | .L.....Q............ |
| 102 | CH77_WK12_3_TB1 | ...........K........ | ....G.............. | ........G........... |
| 102 | CH77_WK12_3_TB2 | ...........K........ | .....G............. | ....I...G........... |
| 159 | CH77_WK24_3_TA1 | ...........K........ | K.....E............ | .L......G........#.. |
| 159 | CH77_WK24_3_TA2 | ...N.......K........ | .....G.........T... | .L.....Q........... |
| 159 | CH77_WK24_3_TA3 | ...........K........ | .....G....-------.. | .L.....Q............ |
| 159 | CH77_WK24_3_TA4 | ...........K........ | ......N........S... | .L.....Q............ |
| 159 | CH77_WK24_3_TA6 | ...N.......K........ | ......E............ | .L......G........... |
| 159 | CH77_WK24_3_TB1 | ...N.......K........ | .....G............R | .L.....Q.........#.. |
| 159 | CH77_WK24_3_TB2 | N..........K........ | .....T............. | .L.....Q............ |
| 159 | CH77_WK24_3_TB3 | ...N.......K........ | .....K..........V.. | .L.....Q............ |
| 159 | CH77_WK24_3_TB4 | ...........N........ | .....K..........V.. | .L.....Q............ |
| 159 | CH77_WK24_3_TB5 | G..........N........ | .....T............R | .L......G........... |
| 159 | CH77_WK24_3_TB6 | ...N.......K........ | ......E.........V.. | .L......G........... |
| 159 | CH77_WK24_3_TB7 | ...N.......K........ | .....G............. | .L......G........... |
| 592 | CH77_WK84_J1 | .Y.........K........ | ....GK............R | ........G........... |
| 592 | CH77_WK84_G7 | .Y.........K..A..... | ....GK............R | ........G........... |
| 592 | CH77_WK84_H12 | ...A.......KD....... | ....GK............R | ........G........... |
| 592 | CH77_WK84_C9 | .Y.........K........ | ......E...........R | .L......G........... |
| 592 | CH77_WK84_H3 | .Y.........K........ | .....G............R | ........G........... |
| 592 | CH77_WK84_D10 | .Y.........K........ | ....GK............R | ........G........... |
| Day | SGA clone | ENV 343-363 (CH77.s) | NEF 17-35 (CH77.aa) | NEF 75-94 (CH77.cc) |
| 0 | Consensus | SHVVDKLREQFRNKTIVFNH | RERRRRDEPAAVGVGPASQ | PVRPMTYKAALDLSHFLKEK |
| 14 | CH77_WGA_B3a | ...........K........ | ................... | .................... |
| 14 | CH77_WGA_C7 | ...........K........ | ................... | .................... |
| 14 | CH77_WGA_C9 | ...........K........ | ................... | .................... |
| 14 | CH77_WGA_C4 | ...........K........ | ................... | .................... |
| 14 | CH77_WGA_D5 | ...........K........ | ................... | .................... |
| 14 | CH77_WGA2b | .................... | ................... | .................... |
| 14 | CH77E_L7 | ...........S........ | ......N............ | .................... |
| 14 | CH77E_L8 | ...........K........ | .....K............. | .................... |
| 14 | CH77E_M6 | ..............A..... | ....K.............. | .................... |
| 32 | CH77_WK03_3_B1 | ...........K........ | ....G.............. | .......Q............ |
| 32 | CH77_WK03_3_D1 | ...........K........ | ........S.......... | ............I....... |
| 32 | CH77_WK03_3_D2 | ...........K........ | ......E............ | .......P............ |
| 32 | CH77_WK03_3_D3 | ...........K........ | .....K............. | .......Q............ |
| 32 | CH77_WK03_3_TB1 | ...........K........ | .....K............. | .......Q............ |
| 32 | CH77_WK03_3_TB2 | ...........K........ | ......E............ | .......T............ |
| 32 | CH77_WK03_3_TB4 | ...........K........ | ........S.......... | ..........H......... |
| 32 | CH77_WK03_3_TB5 | ...........K........ | ......N............ | .......Q............ |
| 32 | CH77_WK03_3_TB6 | ...........K........ | ..GA.ERR........... | .......Q............ |
| 32 | CH77_WK03_3_TB7 | ...........K........ | .....K............. | .......Q............ |
| 32 | CH77_WK03_3_TB8 | ...........K........ | ......E............ | .......T............ |
| 32 | CH77_WK03_3_TC1 | ...........K........ | ......N............ | .......Q............ |
| 102 | CH77_WK12_3_E1 | .Y.........K........ | ......N............ | .L.....Q............ |
| 102 | CH77_WK12_3_TA1 | ...........K........ | .....K............. | .L.....Q............ |
| 102 | CH77_WK12_3_TA2 | ...........N........ | .....T............. | .L.....Q............ |
| 102 | CH77_WK12_3_TB1 | ...........K........ | ....G.............. | ........G........... |
| 102 | CH77_WK12_3_TB2 | ...........K........ | .....G............. | ....I...G........... |
| 159 | CH77_WK24_3_TA1 | ...........K........ | K.....E............ | .L......G........#.. |
| 159 | CH77_WK24_3_TA2 | ...N.......K........ | .....G.........T... | .L.....Q........... |
| 159 | CH77_WK24_3_TA3 | ...........K........ | .....G....-------.. | .L.....Q............ |
| 159 | CH77_WK24_3_TA4 | ...........K........ | ......N........S... | .L.....Q............ |
| 159 | CH77_WK24_3_TA6 | ...N.......K........ | ......E............ | .L......G........... |
| 159 | CH77_WK24_3_TB1 | ...N.......K........ | .....G............R | .L.....Q.........#.. |
| 159 | CH77_WK24_3_TB2 | N..........K........ | .....T............. | .L.....Q............ |
| 159 | CH77_WK24_3_TB3 | ...N.......K........ | .....K..........V.. | .L.....Q............ |
| 159 | CH77_WK24_3_TB4 | ...........N........ | .....K..........V.. | .L.....Q............ |
| 159 | CH77_WK24_3_TB5 | G..........N........ | .....T............R | .L......G........... |
| 159 | CH77_WK24_3_TB6 | ...N.......K........ | ......E.........V.. | .L......G........... |
| 159 | CH77_WK24_3_TB7 | ...N.......K........ | .....G............. | .L......G........... |
| 592 | CH77_WK84_J1 | .Y.........K........ | ....GK............R | ........G........... |
| 592 | CH77_WK84_G7 | .Y.........K..A..... | ....GK............R | ........G........... |
| 592 | CH77_WK84_H12 | ...A.......KD....... | ....GK............R | ........G........... |
| 592 | CH77_WK84_C9 | .Y.........K........ | ......E...........R | .L......G........... |
| 592 | CH77_WK84_H3 | .Y.........K........ | .....G............R | ........G........... |
| 592 | CH77_WK84_D10 | .Y.........K........ | ....GK............R | ........G........... |
Italics, nonsynonymous mutation occurring two or more times; underline, confirmed optimal T cell epitope; -, deletion; #, insertion; ., amino acids that are identical to the transmitted form.
Representative of five further SGA clones (unpublished data).
Representative of one further SGA clone (unpublished data).
A fourth very early sequence change was found in Tat in a highly variable region, 55–74 (CH77.l), spanning the nuclear localization signal and the C-terminal end of domain 1 (Fig. 2; and Table S2, CH77.l). Despite testing, both 14 and 21 d from screening, no T cell response was detected, including in the highly sensitive cultured ELISpot assay (unpublished data) (33). This seemed to be the fastest escape mutation in the three patients, and the complex pattern of escape strongly suggested immune escape. However, the T to C change at nucleotide position 5447, resulting in the I64T nonsynonymous change, was a reversion to consensus in Tat. This region of the Tat gene overlaps part of Rev in another reading frame where the nucleotide change at position 5477 is synonymous. The nucleotides encoding the putative Tat epitope actually fall within the region encoding a detected T cell epitope Rev 9–26 (CH77.l), which appeared to escape at a slower rate with amino acid changes in both reading frames. Thus, although an early T cell response against Tat might have been undetected in the earliest assays, an alternative explanation is that rapid reversion in Tat was the first mutation and subsequent immune escape in Rev generated the later changes in Tat.
The pattern of the predominant T cell responses in patient CH77 over the first 200 d is detailed in Fig. 6. After the three early escapes in Nef and Env, all of which occurred when virus load was falling toward set point, a strong immunodominant response developed to the HLA B*5701-restricted Gag 240–249 (CH77.c) TW10 epitope TSTLQEQVGW (Fig. 6, A–F). By 6 mo (159 d) from screening, there were also strong responses to two epitopes in 334–351 (CH77.r) and overlapping peptides Env 597–614 and 605–622 (CH77.v), the latter of which also contained a B*5701-restricted epitope, TTTVPWNVSW. Both epitopes showed evidence of positive selection detectable at days 159 and 592, respectively (Fig. 2). Weaker responses to eight other epitopes (assuming unmapped responses to adjacent overlapping peptides were a single response), which included the HLA B*5701-restricted epitopes Gag 146–155 (CH77.b) IW9 ISPRTLNAW and Rev 9–18 (CH77.l) KTVRLIKRLY, were also detected at 6 mo (Fig. 6 B and Table S2). Although the breadth of the T cell response in CH77 was impressive, 10/15 T cell epitopes were subject to virus escape over the first 18 mo after infection, with escape from the HLA B*5701-restricted TW10 epitope being one of the slowest. There were 10 other sites of positive selection in the virus sequence that occurred at a slower rate, for which no T cell response was found (Fig. 2). Six of these, two of which were in Env, had complex patterns of amino acid change, implying immune escape.
Patient CH58.
This patient, who was also B*5701 positive, achieved very good control of virus load to 263 cc/ml (Fig. S1 and Table S1). As with CH77, there was a very early escape within CH58.e, beginning just 9 d after screening (Fig. 2). The T cell response to the Env 584–592 ERYLKDQQL epitope spanning this site was initially recognized by a high proportion (3.8%) of memory CD8 T cells (Fig. 7 A). Two codons within this epitope underwent selection, one away from consensus and the other involving multiple variants. At 70 d from screening, a response was detected to the transmitted peptide, but not the mutant peptide, in ELISpot (Fig. 4 F). This epitope could be restricted by either B*1402 or Cw*0701 (Table S2). Escape from this T cell response preceded escape from the B*5701-restricted response to TW10 (CH58.c), which was subdominant 9 d after screening but expanded and became immunodominant by 6 mo (Fig. 7, A, B, and D). This TW10 epitope also underwent T242N and G248E changes that were found in most sequences 85 d from screening. Two nonsynonymous changes, K110R and D113E, occurred in Nef 105–122 (CH58.h) at a similar rate to that of escape from the TW10 T cell response. A T cell response was detected against the transmitted 18-mer at 70 d after screening (with 50% loss in magnitude against mutated peptides) but not at day 154 (Fig. 7 E). Another detected response in this patient reacted to the adjacent peptide Nef 113–130 and mapped to the HLA B*5701-restricted epitope 116–125 HTQGYFPDWQ. This T cell response declined rapidly (detectable in ELISpot but undetectable by ICS at day 154; Fig. 7, compare B and G) but no mutation occurred in this epitope. The two fixed nonsynonymous changes K110R and D113E, just described in CH58.h, were located upstream of this epitope and could be close enough to affect antigen presentation (34). It is possible, therefore, that these mutations shown in Fig. 7 F (dotted line) not only served as escape mutations against the Nef 105–122–specific T cell response but could also have impacted on processing of the adjacent HTQGYFPDWQ epitope. Finally, a T cell response was also made to the B*5701-restricted epitope Gag 146–155 IW9, which was still strong at 6 mo and showed signs of mutation at around 1 yr (Fig. 7 G), although the number of SGA sequences at this visit was very low and the mutation was not significant (not a site of positive selection; listed in Fig. 2). There were three additional mutating regions, two of which were in Env. All had complex patterns of escape but none was in an HLA-matched epitope region and no T cell responses were detected (Fig. 2).
Noteworthy in this patient was the very early dominant but transient T cell response to the escaping epitope in Env, which was not HLA B*5701 restricted (with a possible second response to Env CH58.g which also mutated rapidly), followed by strong responses by three HLA B*5701-restricted T cells where escapes were selected in the previously described order, TW10 before IW9 (24). These five T cell responses comprised the whole detectable response in this HLA B*5701 patient who achieved excellent control of virus load.
Quantifying rates of CD8 T cell escape
For modeling, additional data were included from a fourth patient, SUMA0874, after identification of a cluster of mutations in one short region in Rev by SGA sequencing, which was not previously detected by population sequencing because of failure to distinguish multiple variants. These mutations first appeared 20 d after onset of symptoms (DFOSx), near peak viremia, and were consolidated by 34 d with 78% of sequences showing mutations (Fig. S3). There was a strong T cell response to the transmitted/founder sequence, with three of the variant peptides not recognized and weaker responses to the other two, collectively suggesting T cell selection pressure at this site. There was a brief increase of viremia between days 25 and 50, shortly after this escape occurred (Fig. S3). The direction comparison of sequencing strategies in this patient, illustrates the power of the SGA approach in characterizing early virus selection.
Quantification of CD8 T cell escape in the four study subjects summarized in Table II is based on a simple model (see Materials and methods), in which each epitope acts independently and is under pressure by an independent CD8 T cell response. Fig. 8 shows how the model fitted the data from each patient. If there are no fitness costs associated with escape mutations, then the rate of escape is the rate at which cells infected by founder viruses are depleted by CD8 T cells that target the epitope under consideration, as the escape variant is not killed and, hence, its growth advantage is equal to the killing rate. If there are fitness costs associated with escape, then the rate of escape is lower. Hence, the rate of escape as shown is a lower bound on the CD8 T cell killing rate because of recognition of a single epitope. The median rate of escape observed in experimentally confirmed T cell epitopes was 0.14 d−1 and the maximum rate was 0.36 d−1. These estimates are substantially higher than prior estimates. For example, Asquith et al. (35) estimated a median rate of killing of HIV-1–infected cells by a single CD8 T cell response of 0.04 d−1 during primary infection, which is 3.5-fold lower than our estimate but based on previously published virus escapes that were later than those described here. Furthermore, the current estimate of the rate of death of productively infected cells based on the rate of viral decline during potent drug therapy is δ = 1 d−1 (36), suggesting that the CD8 T cell response to a single epitope could be responsible for, on average, ∼15% of this killing and, in some cases, could be responsible for as much as 35%. This implies that these CD8 T cells contribute substantially to reduction of virus load by shortening the life span of HIV-1–infected cells that are replicating virus.
Mathematical modeling fitted to the he measured changes in the frequency of T cell escape mutations suggests that the first T cells induced by HIV-1 infection contribute to the control of acute viremia. Data shown is from patients CH40 (A), CH77 (B), CH58 (C), and SUMA0874 (D). CH77.b and CH58.b mutations occurred within the IW9 Gag epitope. CH77.c and CH58.c mutations occurred within the Gag TW10 epitope.
Mathematical modeling fitted to the he measured changes in the frequency of T cell escape mutations suggests that the first T cells induced by HIV-1 infection contribute to the control of acute viremia. Data shown is from patients CH40 (A), CH77 (B), CH58 (C), and SUMA0874 (D). CH77.b and CH58.b mutations occurred within the IW9 Gag epitope. CH77.c and CH58.c mutations occurred within the Gag TW10 epitope.
Higher escape rates are observed in the first T cells of HIV-1 infection
| Patient | Reference | Mutation/sa | Average rate escape, kb | t50c | CD8 T cell epitoped | Confirmed T cell response (HXB2 position)e |
| day−1 | days | |||||
| CH40 | a. | Gag A120D | 0.17 | 31 | no | no |
| CH40 | b. | Gag I401L, R403K | 0.17 | 31 | yes | yes (Gag389-406) |
| CH40 | d. | Pol K76R | 0.015 | 119 | yes | nof |
| CH40 | h. | Vif T167I | 0.37 | 38 | yes | no |
| CH40 | i. | Vpr S84R | 0.16 | 31 | yes | no |
| CH40 | t. | Nef S188R/N, R196Q | 0.22 | 15 | yes | yes, (Nef185-202) |
| CH77 | l. | Tat I64TS | 0.42 | 8 | yes | nog |
| CH77 | s. | Env R355K/S/N,T358A | 0.36 | 9 | yes | yes, (Env350-368) |
| CH77 | aa. | Nef R21K/G, R22K/G/,TD23E/N, P25S | 0.30 | 19 | yes | yes, (Nef17-34) |
| CH77 | cc. | Nef K82Q/T/P, A83G, L85H, L87I | 0.29 | 24 | yes | yes, (Nef81-98) |
| CH77 | b. | Gag I147L | 0.063 | 101 | yes | yes, (Gag140-157) IW9 |
| CH77 | c. | Gag T242N,V247I, G248E/S | 0.035 | 124 | yes | yes, (Gag236-253) TW10 |
| CH58 | e. | Env Y586H | 0.10 | 21 | yes | yes, (Env576-596) |
| CH58 | g. | Env F829V/S/L, I830M/L/F, A832T, V833I | 0.12 | 27 | no | no |
| CH58 | b. | Gag I147L | 0.007 | 430 | yes | yes, (Gag140-157) IW9 |
| CH58 | c. | Gag T242N, G248E | 0.085 | 60 | yes | yes, (Gag236-253), TW10 |
| SUMA0874 | Rev R48K, Q51H, Q53R, S54L, L55I | 0.32 | 30 | yes | yes, (Rev47-55) | |
| Median | all demonstrated T cell epitopes | 0.17 (max. 0.42) | 0.14 (max. 0.36) |
| Patient | Reference | Mutation/sa | Average rate escape, kb | t50c | CD8 T cell epitoped | Confirmed T cell response (HXB2 position)e |
| day−1 | days | |||||
| CH40 | a. | Gag A120D | 0.17 | 31 | no | no |
| CH40 | b. | Gag I401L, R403K | 0.17 | 31 | yes | yes (Gag389-406) |
| CH40 | d. | Pol K76R | 0.015 | 119 | yes | nof |
| CH40 | h. | Vif T167I | 0.37 | 38 | yes | no |
| CH40 | i. | Vpr S84R | 0.16 | 31 | yes | no |
| CH40 | t. | Nef S188R/N, R196Q | 0.22 | 15 | yes | yes, (Nef185-202) |
| CH77 | l. | Tat I64TS | 0.42 | 8 | yes | nog |
| CH77 | s. | Env R355K/S/N,T358A | 0.36 | 9 | yes | yes, (Env350-368) |
| CH77 | aa. | Nef R21K/G, R22K/G/,TD23E/N, P25S | 0.30 | 19 | yes | yes, (Nef17-34) |
| CH77 | cc. | Nef K82Q/T/P, A83G, L85H, L87I | 0.29 | 24 | yes | yes, (Nef81-98) |
| CH77 | b. | Gag I147L | 0.063 | 101 | yes | yes, (Gag140-157) IW9 |
| CH77 | c. | Gag T242N,V247I, G248E/S | 0.035 | 124 | yes | yes, (Gag236-253) TW10 |
| CH58 | e. | Env Y586H | 0.10 | 21 | yes | yes, (Env576-596) |
| CH58 | g. | Env F829V/S/L, I830M/L/F, A832T, V833I | 0.12 | 27 | no | no |
| CH58 | b. | Gag I147L | 0.007 | 430 | yes | yes, (Gag140-157) IW9 |
| CH58 | c. | Gag T242N, G248E | 0.085 | 60 | yes | yes, (Gag236-253), TW10 |
| SUMA0874 | Rev R48K, Q51H, Q53R, S54L, L55I | 0.32 | 30 | yes | yes, (Rev47-55) | |
| Median | all demonstrated T cell epitopes | 0.17 (max. 0.42) | 0.14 (max. 0.36) |
Summary of characteristics of several mutations that became fixed in the virus population within several weeks after the onset of symptoms.
Note, that in the majority of cases, several different nonsynonymous mutations occurred in a given peptide in the model. All such escape variants are assumed not to be recognized by the corresponding CTL responses. Analysis of the extension of this model assumption will be presented elsewhere.
The rate of escape, k, determines the mean rate at which mutations accumulated in the viral population.
t50 is the predicted time of the mutant to reach 50% frequency in the viral population.
The mutations occur in known CTL epitopes or in sites with CTL anchor binding motifs given the patient's HLA type (ELF software, LANL).
A CD8 T cell response (as measured by ELISpot or ICS) to a peptide corresponding to the transmitted virus was detected at that site.
This mutation is proximal to the Pol 80–97 epitope and may have had an impact on its processing. See also Fig. 5 H.
This region partially overlaps with Rev in another reading frame. See paragraph three of the CH77 section of the Results for further discussion.
DISCUSSION
In this study, five findings were made that advance previous understanding of the role of T cell responses in acute HIV-1 infection. The exact numbers of potential and actual T cell epitopes in the transmitted virus were determined. The kinetics of virus escape, occurring as the virus load was declining from peak viremia, before set point, were shown to be earlier and faster than previously thought, extending previous observations on sequence change in acute infection (37). Some T cell responses to the transmitted form of the virus vanished after selecting early virus escape mutants. A small number of mutated sites were found that were not explained by escape from T cells, NAb, or reversion to consensus sequence; some of these were probably bystander mutations carried along with immune escape mutations elsewhere. Finally, evidence is provided that the earliest CD8 T cell responses are causally linked to decline of virus load in acute infection.
The intensive strategy used in this paper can only be applied to small patient numbers, unlike previous methods which either incompletely sample the T cell response (11, 21) or rely on sequence changes alone (20, 24, 37). The latter approach, though elegant and applicable to large cohorts, makes assumptions as to whether the sequence has changed from consensus during earlier stages of infection and whether mutations are selected by T cells. As found in this paper, mutations can be selected by other immune responses, by mutual association, by selection in another reading frame, or by reversion to the consensus form. Also, when sequencing is performed at later time points, the focus tends to be directed at well described epitopes, presented by more common HLA types, that escape relatively slowly. T cell responses to the new epitopes that were found in this paper in the early immune responses may be missed, particularly when such early T cell responses vanish after the escape. For instance, it has been estimated that the Gag TW10 epitope, presented by HLA B*5701, was the most rapid to escape in a large HIV-1–infected cohort (24), but in CH58 and CH77 there were CD8 T cell responses that fully replaced the transmitted epitope well ahead of any escape in TW10 selected by T cells. There has also been some debate about the extent of amino acid sequence reversion to consensus (assumed to be optimally fit virus) in early HIV-1 infection. Some have argued that such reversions of escaped sequences, originally selected in the patient's sexual partner who has a different HLA type, can account for many of the early sequence changes (23, 26, 38–40). In this study, although six possible examples of reversion were identified, these represented a minority of all regions of mutation in early infection, which is in agreement with Liu et al. (17). Although in the accompanying paper Salazar et al. (29) found only random mutations in founder virus before peak viremia and no evidence of earlier selected mutations, we cannot exclude that some reversion to the most fit virus at the time of transmission occurred to account for the outgrowth of a single virus.
Another difficulty with the sequence-only approach is that it necessarily relies on the assumption that HIV-1 will eventually escape from all T cells and will leave an imprint on the virus. This assumption may be correct, but in the absence of data on the actual T cell responses and sequence information as to how virus has evolved from transmitted/founder virus, the contribution of those T cells that have not yet selected escape mutations on virus load cannot be addressed. In this study, 34% (10/29) of epitopes that stimulated T cell responses remained invariant over the first year after infection. They were mostly in the more conserved virus proteins, with six in Gag, three in Pol, and one in the Nef core. In CH40, there was an invariant Env epitope 765–682 in a coding region of gp41 that is in an overlapping reading frame with Rev, so nonsynonymous changes may be disadvantageous at this locus. The contribution to virus control by T cells specific for epitopes that did not mutate over the first year of infection might be important but is hard to assess. These T cells either could be functionally defective or, more consistent with the observation that invariant epitopes occurred in conserved regions of HIV-1, escape mutations might have too great a fitness cost for the virus (41, 42). In the patients studied in this paper, T cells specific for such epitopes became immunodominant once set point was established (Figs. 5–7,67). If vaccines could bring these responses forward, they might be effective in reducing peak viremia to lower levels than are seen in natural infection.
The early sampling window in this study enabled detection of very early T cell responses that select rapid escapes and then disappear. The rapid decay of several T cell responses once the mutation was fixed was surprising. It suggests, particularly when there is no further selection on the epitope, that these escapes have little fitness cost to the virus because there is no tendency for the mutation to revert and maintain antigenic stimulation. It also implies either complete loss of the epitope, involving alteration in the amino acids which anchor the peptide to the HLA molecule, or failure of T cell clones to recognize the mutant epitopes. This finding also gives information on the half-life of specific CD8 T cells in HIV infection, which is short compared with previous estimates of longevity of HIV-specific memory CD8 T cells in patients given antiretroviral therapy (43) but is consistent with the rapid turnover of all memory CD8 T cells in HIV-1–infected patients measured by McCune et al. (44). It is possible that this is a consequence of impaired CD4 T cell help, which has been shown in mice to be critical for the development of long-term T cell memory (45).
A key question is whether the earliest T cell responses contribute to the early fall in viremia at the same time as they are selecting escape mutations. For complete escape to occur, all virus-infected cells that contribute to the viremia and express the original sequence must be replaced by cells infected by virus with the novel sequence. The new virus has the advantage that it is unlikely to be recognized by the selecting T cells. In CH77, such events were occurring in rapid succession (Table I). Therefore, it is likely that these CD8 T cells contribute to the reduction in virus load. A mathematical model was applied to the data, calculating the rates of virus escape and the contribution of the T cell responses to the fall in virus load. The contribution of experimentally confirmed T cells to killing of virus-infected cells in vivo, calculated from the rates of escape, was 3.5-fold higher than previous estimates made in later stages of infection (35). When compared with the current estimate of the death rate of virus-infected cells, it was found that a CD8 T cell response to a single epitope could account for between 15 and 35% of that killing. This implies that the earliest T cell responses are indeed helping to control the early viremia, even when the virus can escape. T cells specific for different epitopes may cooperate in suppressing virus and these effects should be additive to other causes of the falling viremia, including the loss of target cells for the virus.
The T cells that select rapid escape mutants must therefore be potent. The often observed pattern of multiple clustered mutations within and proximal to an epitope, which is common enough to be considered a signature of immune escapes, implies a real struggle between the virus and antiviral T cells. In this study, the first T cells can change the virus very rapidly, but this occurred at a time when there were very high levels of virus replication, reflecting very rapid turnover of infected cells. Later, as viral set point is reached, the virus turns over more slowly, possibly reflecting reservoirs in different cell populations, the broadened T cell response, and the development of NAbs requiring multiple sequential/simultaneous changes for successful virus escape and replication. Thus, although the suppressive power of the T cell response in the first weeks of infection appears similar to that of a single antiretroviral drug (46), the comparison should best be made when the latter is given at the same stage of infection.
The virus SGA sequencing showed that there were several regions of change, frequently in Env, that were subject to positive selection but with no corresponding T cell response. It is possible that some responses were missed because of the timing of the cellular assays relative to the waning T cell response, that T cells might have been sequestered in sites other than the blood, or that they might not respond to peptide by releasing IFN-γ. However the assays used are sensitive and further attempts to reveal several of these T cell responses by culturing patients' PBMC with peptides and IL-2, which greatly increases sensitivity, failed to detect low-frequency responses (33; not depicted). Nevertheless, some of these mutations were clustered like proven T cell and antibody-selected escapes and spanned regions with known or predicted HLA-appropriate potential epitopes (Table S2), suggesting undefined immune selection pressure.
It is possible that some mutations were selected by immune responses other than CD8 T cells, including CD4 T cells, antibody, and NK cells. CD4 T cells, restricted by class II HLA, also recognize peptides, and HIV escape by mutation in an epitope has been previously described (47). The epitopes recognized by CD4 T cells are less well characterized, but none were detected by ICS using peptides spanning regions of positive selection. Selection by NAb was seen in patients CH40 and CH77 in four sites that occurred in epitopes in the V1 and V3 region. Mutations first appeared in these sites after day 100, which is consistent with previously described NAb kinetics in HIV-1 infection. It is possible that other Env mutations could have been selected by antibody, although in some cases this would imply an earlier antiviral effect of antibody than generally recognized, possibly involving antibody-dependent cell cytotoxicity (48). NK cell recognition can be influenced by the peptides binding to the HLA class I molecules they recognize (49, 50), so this could account for some of the selected mutations, although this type of HIV-1 escape is yet to be demonstrated experimentally. However, an escape mutation in an HLA B27-bound peptide was found to increase binding to the ILT4 receptor found on monocytes (51). Future studies to explain putative immune escapes where no T cell response can be detected will be important and could reveal other forms of immune protection.
Other causes of isolated selected mutations included some that were in mutual association (linkage) with a selected epitope. The mutations could be carried along passively or incomplete mutual associations within the same proteins could represent attempts to compensate for mutations in the epitope. In one example, the mutation occurred on a different virus protein and so cannot compensate for an escape in the same protein but might affect virus RNA structure. Immune pressure that targets codons found in overlapping reading frames may result in mutational clusters in more than one protein (the T to C change at position 5447 in CH77.l) or conversely limit virus escape because of fitness constraints imposed on the other open reading frame (see the Patient CH77 section of the Results for discussion of CH77.1).
Despite the limited numbers investigated, this study offered the opportunity to observe the earliest T cell responses in two patients with HLA B*5701, both of whom initially controlled the virus well. In both cases there were very rapid T cell responses selecting escape mutants while the acute viremia was falling. These preceded the establishment of the classical B*5701-restricted T cell responses to the TW10 and IW9 epitopes, suggesting that these responses play more of a role in maintaining a low set point than in establishing it. The relatively early appearance of these T cell responses to conserved epitopes, where escape has a known virus fitness penalty (41), may be the critical difference between these patients and CH40 and SUMA0874, who did not control virus so well in early infection. Ongoing studies will address this issue, which could be important for vaccine design.
Studies in the SIV challenge model in macaques have reported findings similar to these (52, 53). However, there are differences between this experimental model and HIV-1 infection of humans that made it important to examine human infection in this detail. The infection in rhesus macaques is artificial, and large inoculating virus doses have to be used to ensure infection. The challenge virus is more aggressive and the kinetics of infection and disease progression are faster than in humans (52, 54). Nevertheless, in Mamu A*01-positive animals there was an early Tat-specific CD8 T cell response that selected escape mutations quickly while virus load was falling, and this was followed by a response to the stable Gag CM9 epitope which escapes rarely and slowly (52). The findings were extended to other animals with different MHC types, with similar findings of early epitope escape mutations selected by T cells which recognized antigen with high functional avidity and later responses to more conserved epitopes (53). Thus, this model of infection mimics well the human responses observed in this study. The macaque studies, however, did not explore the full breadth of responses, and other T cells specific for epitopes that did not mutate may have been missed. Nor did these studies address the role of the early escaping T cell responses in controlling early viremia. The rhesus macaque model would provide an experimental opportunity to distinguish the relative roles in early virus control of the Mamu A*01-restricted Tat-specific response versus other immune responses to more conserved epitopes.
Overall, the findings reported in this paper show that the first CD8 T cells, despite limited breadth and very rapid virus escape, suppressed HIV-1 as virus load was declining from its peak. The influence of these T cell responses declined once the virus had escaped, but some control of virus was maintained by further waves of T cell responses, some of which did not select mutations or did so slowly. Many of these later T cells focused on more conserved regions of the virus and, if they were functionally competent, were likely to be important in longer term suppression of the infection. It is probable that the mutations that escaped slowly had a fitness cost for the virus, which could have been beneficial to the patient. Modeling implied that a single T cell response was contributing as much as 15–35% of viral decline with multiple T cell responses. The implication of these observations is that vaccine-induced HIV-1–specific T cells will contribute to control of acute viremia if they are activated early in subsequent HIV-1 infection. However, because of the very rapid escape that occurs within the first few weeks of infection, T cell vaccines will need to stimulate a considerable breadth of T cell responses, clearly greater than the median of three epitopes induced by the Merck vaccine (Corey, L. 2008. AIDS Vaccine 2008 Capetown. http://www.hivvaccineenterprise.org/conference/archive/2008/Presentations/Tuesday/Special%20Session%2001/14h35%20L%20Corey.pdf). Supporting this view is the promising finding that a SIV Gag vaccine using two different adenovirus vectors in a prime-boost combination gave strong CD8 T cell responses to a mean of eight epitopes and gave substantial protection in macaques against SIVmac251 challenge (55). In addition, attempts should be made to focus the earliest T cell responses on conserved epitopes that remain invariant or mutate with a fitness cost.
MATERIALS AND METHODS
Patients and samples.
Patients 700010040 (CH40), 700010058 (CH58), and 700010077 (CH77) gave full informed consent to enroll in to the acute infection arm of the CHAVI 001 cohort. Human experiments were approved by the Prevention Sciences Review Committee Division of Acquired Immunodeficiency Syndrome. In brief, acute infection was defined as positive for HIV-1 viral RNA in plasma but negative or discordant for HIV-1 serology at a screening visit. Patients were bled at multiple time points, as indicated in Figs. 2–6,3456, over 2 yr. Table S1 shows the demographical and clinical data of each patient. Patient SUMA0874 was detected in symptomatic acute HIV-1 infection as described by Jones et al. (21). Patient PBMCs were isolated from blood after centrifugation over ficoll. Cells were washed, counted, and then cryopreserved (∼107 cells/vial). Plasma was obtained after centrifugation of blood collected in EDTA tubes or from ficoll gradients of blood. Viral load set point was determined using an average of viral loads within a set point window (4). The criteria for defining the set point window for a given participant included removing outlying viral loads based on three major categories: values from the initial up- or downswing of viremia, values from the accelerating phase of the disease, and apparent outlying values during the set point interval.
Sequencing and alignments.
RNA extraction, complementary DNA synthesis, and SGA was previously described by Salazar et al. (29). Peptides and substitutions are aligned relative to HXB2 standards (www.hiv.lanl.gov/content/sequence/QUICK_ALIGN/QuickAlign.html).
HLA typing.
HLA typing (Weatherall Institute of Molecular Medicine [WIMM], Oxford, England, UK) was performed using the sequence-specific primer method adapted from Bunce (56), which uses allele-specific primer combination in PCR amplification to provide absolute HLA resolution to two digits and high-probability resolution to four digits. CH40: A*0201/3101, B*4001/4402, and Cw*0302/0501; CH58: A*0101/2301, B*1402/5701, and Cw*0701/0802; and CH77: A*0205, B*5301/5701, and Cw*0401/1801.
Peptides.
18-mer peptides overlapping by 10 aa were synthesized (Sigma-Aldrich; Medical Research Council Human Immunology Unit, WIMM, Oxford UK) to match the transmitted sequence of CH40, CH58, and CH77. Approximately 400 peptides for each patient (including >200 unique to that patient relative to consensus Clade A-D peptide sets) were arranged into pools in a matrix format using the Peptide Portal program (Statistical Center for HIV/AIDS Research and Prevention) adapting code from the Deconvolute this! program (57). Each peptide was repeated four times in the matrix peptide plate. LANL ELF software was used to identify HLA-matched known and predicted CTL epitopes (http://www.hiv.lanl.gov/content/sequence/ELF/epitope_analyzer.html). Epitopes were predicted when the amino acid sequence contained anchor residues that match HLA motifs but occurred in epitopes not found on the LANL database. When such peptides gave a positive T cell response, it was considered likely that this identified both the optimal epitope and its HLA restriction.
Ex vivo ELISpot assays.
Cryopreserved PBMCs were thawed and rested for 2 h before being placed in the ELISpot plates at 1 × 105 cells/well. Antigens in the peptide plates were mixed 1:1 with PBMC in the ELISpot plate to a final concentration of 2 µg/ml and incubated for 20 h at 37°C in 5% CO2. Coating, development, and reading of ELISpot plates has been described previously (33). A low threshold of positivity (30 SFU/106 PBMCs, >3× background) was applied to peptide pools to maximize sensitivity. Data from positive pools were then deconvoluted (57) to identify candidate epitope peptides. Putative positive peptide-specific T cell responses were confirmed in triplicate in a follow up IFN-γ ELISpot with 1 × 105 cells/well and a peptide concentration of 2 µg/ml. A more stringent definition of positive responses was applied: ≥50 SFU/ million, >4× background. IFN-γ ELISpot assays performed at other time points were performed similarly to the deconvolution assays, although, in some cases, when cells were limiting, 0.5 × 104 cells/well were used against 9-mer peptides. For all assays, six negative control wells (media only) and at least 1 positive control well (10 µg/ml PHA [Sigma-Aldrich]) were used.
Flow cytometry.
Cryopreserved PBMCs were thawed and rested overnight. PBMCs were stimulated with 2 µg/ml peptide with 1 mg/ml of anti-CD28 mAb (L293; BD) and 1 mg/ml of anti-CD49d mAb (L25; BD) in the presence of anti-CD107a Alexa Fluor 680 (H4A3; BD), 5 µg/ml brefeldin A (Sigma-Aldrich), and 1 µg/ml of Golgi Stop (BD) for 5.5 h at 37°C in 5% CO2. After washing, the PBMCs were surface stained with anti–CD14–Cascade blue (M5E2), anti–CD19–Cascade blue (HIB 19), anti–CD4-Cy5.5-PE (M-T477), anti–CD8-QD705 (RPA-T8), anti–CD27-Cy5-PE (M-T271), anti–CD57-QD605 (NK-1), and anti–CD45RO-PE–Texas Red (all obtained from BD) for 20 min at room temperature. PBMCs were then fixed and permeabilized with Cytofix/Cytoperm and Perm/Wash buffer (both obtained from BD). PBMCs were stained with anti–CD3-Cy7-APC (SK7), anti–IFN-γ–FITC (B27), anti–IL-2–APC (MQ1-17H12), anti–TNF-α–Cy7-PE (MAb11), and anti–MIP-1b-PE (D21-1351; all obtained from BD) for 1 h at 4°C. After washing and fixation, samples were run on a custom built LSRII (BD). A minimum of 300,000 total events were acquired and data analysis was performed with FlowJo software (Tree Star, Inc.) with Pestle and Spice (provided by M. Roederer, Vaccine Research Center, Bethesda, MD). Memory CD8 T cells are defined as all live lymphocytes that stain positively with CD3 and CD8, excluding any cells that have the CD27 high and CD45RO negative phenotype.
Methods for detecting positive selection.
For each of three cases with longitudinal samples obtained from follow up (CH40, CH58, and CH77), complementary analytic methods were used to identify nucleotide positions enriched for positive selection. Because T cell epitopes often escape by multiple routes, tests were devised that could identify localized regions of selection, as well as selection acting on single sites. The first method used identifies both. This enriched mutation statistic was computed as follows. Using a sliding window of 45 nt, the mutations in that window were counted. Mutations were then randomly shuffled 100×, looping through the number of sequences and, keeping the number of mutations per sequence unchanged, randomly reassigned the positions of mutated sites. The number of mutations per window observed in the randomized set was then counted. After randomization, each count belongs to the null distribution, giving effectively 100 × N randomized replicates, where N is the total number of windows. The p-value for each observed window is the number of randomized windows that contain at least the observed number of mutations, divided by the number of replicate shufflings. The false-discovery rate (denoted as q) was computed, to correct for multiple testing (58), using q < 0.1 to detect windows where significant differences between observed and shuffled mutations occurred. High q-values were used to be inclusive, and so some false positives were expected.
The second method was called the clustered mutation statistic, which used a 27-nt sliding window and computed p-values from a Fisher's exact test on counts in a 2 × 3 contingency table. Rows in the table reflected mutations in the window versus outside the window, and columns represent the number of aligned nucleotide sites with 0, 1, or more sequences that differed from the consensus sequence at that site. Again, the false-discovery rate was used to correct for multiple testing with q < 0.1 to identified windows significantly enriched for mutated positions relative to the frequency of mutations outside the window. The code to perform these window-based statistics for positive selection can be found at ftp://ftp-t10.lanl.gov/pub/WindowSelectionStatistics. It is written in R and Perl.
Third, protein-coding regions for all nine genes in the HIV-1 genome were extracted with GeneCutter and tested for specific codons under positive selection using FEL (59). A neighbor-joining tree and nucleotide substitution model were computed for each aligned gene before FEL analysis (60). Sites with greater nonsynonymous than synonymous substitution rates (dN > dS) and P < 0.2 were taken as significant for positive selection. FEL results were not corrected for multiple testing, according to Pond and Frost (59), and verified robustness of results in the presence of alternative substitution models, discarding sites whose significance could not be repeated using another substitution model. Because the FEL tests can be influenced by recombination, and because synonymous substitution is not properly defined for genes whose reading frames overlap with other genes in different frames, the FEL tests were repeated using subsets of the alignment with overlapping reading frames excised and partitioned, where GARD (61) identified potential recombination breakpoints. The p- and q-values for all tests are shown in Fig. 1 and provided in Table S2I.
Modeling.
To estimate the fitness advantage of the CTL escape variant as compared with the wild type, several previously published approaches were used (35, 62). The changes in the frequency of the escape mutant in the population were fitted by the following model: f(t)= f0/(f0 + (1 − f0)e−kt), where f(t) is the frequency of the escape variant in the population and k is the rate of escape. The models assume that the escape variant is present at time t = 0 at the frequency f0. This equation is equivalent to the equation (2) in Asquith et al. (35). The rate at which an escape variant replaces the wild type is the rate of escape, and it is determined by the balance between the efficiency of the CTL clones evaded in killing the wild-type infected cells and the fitness cost of the mutations (35). The estimated rate of escape was robust to several changes in the model and the fitting procedure, such as including the generation of the escape mutant from the founder by mutation, transforming the frequency data using the arsin(sqrt) transformation, or by fitting the log-transformed ratio of the frequency of the mutant to the founder. Because for the analyzed early escape variants there were rarely two consecutive time points in which both the wild type and the mutant were present, to estimate the escape rates we replace the measured 0 and 100% frequency of the escape mutant with 1/(n + 1) × 100% and n/(n+1) × 100%, respectively, where n is the number of viral sequences analyzed at these time points (35). The time at which the mutant is present at a frequency of 50% is t50 = [ln (1/f0 - 1]/k.
Online supplemental material.
Table S1 provides demographical and clinical data for each study volunteer. Table S2 provides full details of each site of putative positive selection, giving significance for each statistical test applied and listing nonsynonymous changes to and from consensus within each site and whether a putative or validated T cell or antibody epitope corresponded to that region. The supplemental text provides additional detail of alignments and changes over time that support Table S2. Fig. S1 graphs viral load versus time for CH40, CH77, and CH58. Fig. S2 graphs variants versus transmitted founder sequence over time at each mutations cluster for demonstrated T cell epitopes. Fig. S3 shows virus escape from the Rev (47–55) T cell response in acute infection in SUMA0874.
Acknowledgments
We thank CHAVI and DUKE management and support teams for study coordination, A. Williams, C. Margaret, C. deBoer, S. Cheeti, and R. Thomas of Statistical Center for HIV/AIDS Research and Prevention for database support, J. Roberts for administrative support, T. Rostron for HLA typing, K. diGleria and Z. Yu for peptide synthesis, and S.L. Kosakovsky Pond for advice on methods to test for positive selection.
This work was supported by the Center for HIV/AIDS Vaccine Immunology grant A1067854-03. Additional support came from the Medical Research Center Human Immunology Unit, the National Institute for Health Research Oxford Biomedical Research Center, and grant 37874 from the Bill and Melinda Gates Foundation. P. Borrow is supported by a Jenner Fellowship and M.S. Cohen is supported by NIDDK Award DK049381. Portions of this work were done under the auspices of the U. S. Department of Energy under contract DE-AC52-06NA25396, and V.V. Ganusov was supported by their Laboratory Directed Research and Development program.
The authors have no conflicting financial interests.
References
Abbreviations used: FEL, fixed-effects likelihood; ICS, intracellular cytokine staining; NAb, neutralizing antibody; SFU, spot forming unit; SGA, single genome amplification; SIV, simian immunodeficiency virus.
Author notes
N. Goonetilleke, M.K.P. Liu, and J.F. Salazar-Gonzalez contributed equally to this paper.