Primary cilia play critical roles in development and disease. Their assembly and disassembly are tightly coupled to cell cycle progression. Here, we present data identifying KIF14 as a regulator of cilia formation and Hedgehog (HH) signaling. We show that RNAi depletion of KIF14 specifically leads to defects in ciliogenesis and basal body (BB) biogenesis, as its absence hampers the efficiency of primary cilium formation and the dynamics of primary cilium elongation, and disrupts the localization of the distal appendage proteins SCLT1 and FBF1 and components of the IFT-B complex. We identify deregulated Aurora A activity as a mechanism contributing to the primary cilium and BB formation defects seen after KIF14 depletion. In addition, we show that primary cilia in KIF14-depleted cells are defective in response to HH pathway activation, independently of the effects of Aurora A. In sum, our data point to KIF14 as a critical node connecting cell cycle machinery, effective ciliogenesis, and HH signaling.
Introduction
The primary cilium is an antenna-like structure typically present on the surface of nondividing cells. While it was originally described as a vestigial organelle, and hence largely neglected, recent years have indisputably proven its status as a seminal structure for sensing various extracellular stimuli (Anvarian et al., 2019; Bangs and Anderson, 2017). In vertebrates, primary cilia govern many important aspects of embryonic development as well as tissue homeostasis in adulthood (Gerdes et al., 2009; Goetz and Anderson, 2010; Reiter and Leroux, 2017). Consequently, deregulation of primary cilia assembly, maintenance, or function is linked to numerous human diseases, collectively termed “ciliopathies” (Badano et al., 2006; Baker and Beales, 2009; Braun and Hildebrandt, 2017; Mitchison and Valente, 2017).
Primary cilium assembly, as well as disassembly, is intimately connected to cell cycle progression and in turn to the centrosome duplication cycle. In cultured cells, primary cilia formation generally occurs after exiting mitosis, in G0, and conversely, resorption of primary cilia starts upon entry into the new cell cycle (Sánchez and Dynlacht, 2016). Cells in the G0/G1 phase typically contain one centrosome with two centrioles. A hallmark of the older, fully mature, so-called mother centriole is a set of distal and subdistal appendages, which decorate its distal end (Bowler et al., 2019; Yang et al., 2018). As the immature, daughter centriole lacks those structures, only a mother centriole is capable of serving as the basal body (BB) to allow ciliogenesis (Fırat-Karalar and Stearns, 2014; Kobayashi and Dynlacht, 2011; Nigg and Holland, 2018). However, the picture of the mutual interactions between primary cilia and the cell cycle still remains far from complete (Seeley and Nachury, 2010).
The fully grown primary cilium is composed of the BB, a mother centriole anchored to the plasma membrane via its distal appendages (Anderson, 1972; Kobayashi and Dynlacht, 2011); the transition zone, a specialized domain at the ciliary base involved in targeting and sorting of proteins to and from the ciliary compartment (Reiter et al., 2012); and the axoneme, a microtubule-based structure protruding into the extracellular space and enclosed within the ciliary membrane (Garcia-Gonzalo and Reiter, 2017; Satir et al., 2010).
The molecular aspects of primary cilium formation have started to emerge only recently, yet many questions remain to be answered. Ciliogenesis seems to be initiated by the docking of small vesicles to the distal appendages of a mother centriole and their subsequent fusion into one large ciliary vesicle (Lu et al., 2015; Sorokin, 1962; Wu et al., 2018). Correct assembly and composition of the distal appendages is essential here, as the absence of many individual distal appendage components (CEP164, CEP83, CEP89, and SCLT1) prevents vesicle docking (Schmidt et al., 2012; Sillibourne et al., 2013; Tanos et al., 2013). A further step in ciliogenesis and mother centriole to BB transition resides in the removal of CP110 and CEP97 from the distal end of the mother centriole (Huang et al., 2018; Spektor et al., 2007). This process requires the activity of TTBK2, recruited to the BB by CEP164 (Čajánek and Nigg, 2014; Goetz et al., 2012; Oda et al., 2014). TTBK2 and the distal appendage protein FBF1 are also involved in the recruitment of components of the intraflagellar transport (IFT) system (Čajánek and Nigg, 2014; Goetz et al., 2012; Wei et al., 2013). In turn, IFT particles are responsible for moving cargos to the tip (anterograde IFT-B complex) of the axoneme and back (retrograde IFT-A complex). IFT-based transport is crucial for not only ciliogenesis but also primary cilium maintenance and sensory function (Lechtreck, 2015; Rosenbaum and Witman, 2002; Scholey, 2003).
Primary cilia seem to be implicated in the modulation of several pathways in vertebrates (Anvarian et al., 2019; Bryja et al., 2017), but it is indisputably Hedgehog (HH) signaling where the importance of primary cilia is best characterized (Bangs and Anderson, 2017; Mukhopadhyay and Rohatgi, 2014). In the absence of a ligand, the Patched receptor localizes to the primary cilium and is thought to inhibit the entry of the Smoothened (SMO) receptor into the primary cilium. Following ligand binding to Patched, SMO moves to the ciliary membrane and promotes the accumulation and activation of GLI proteins at the tip of the primary cilia. Activated GLIs are then transported out of the primary cilia by IFT complexes, and, in turn, regulate proliferation, migration, and differentiation via the induction of target gene expression in the cell nucleus (Bangs and Anderson, 2017; Briscoe and Thérond, 2013).
As we have already mentioned, cell cycle machinery is thought to be connected to primary cilium formation, maintenance, and disassembly (Izawa et al., 2015). The exact mechanisms underlying this relationship are only beginning to emerge. It is generally accepted that cells initiate primary cilium resorption upon cell cycle reentry (G1/S) and that primary cilia are fully resorbed before the onset of mitosis (late G2). Thus, primary cilium formation can be readily triggered by serum starvation of various cultured cell lines, and conversely, primary cilium resorption can be induced by the addition of serum. Molecules implicated in controlling primary cilium disassembly seem to include many well-established regulators of mitotic progression (Plotnikova et al., 2012; Sánchez and Dynlacht, 2016; Seeley and Nachury, 2010). Among these, mitotic kinase Aurora A (AURA) has turned out to be a key regulatory element of primary cilium disassembly (Inoko et al., 2012; Pan et al., 2004; Plotnikova et al., 2012; Pugacheva et al., 2007). Activation of AURA in the context of primary cilium resorption is at least in part mediated by the adaptor protein HEF1/NEDD9 (Gradilone et al., 2013; Pugacheva et al., 2007) and in turn leads to the promotion of primary cilium disassembly by a mechanism expected to involve the deacetylase HDAC6 (Gradilone et al., 2013; Pugacheva et al., 2007). Additional levels of mutual crosstalk between the cell cycle tool kit and primary cilium assembly/disassembly are likely to exist. Indeed, recent evidence has implied the possibility of a connection at the level of regulation and/or regulators of cytokinesis (Bernabé-Rubio et al., 2016; Smith et al., 2011).
Here, we report on a new role of kinesin KIF14 in primary cilium formation. Kinesins are molecular motors capable of binding microtubules through their motor domain, and they are involved in various aspects of intracellular transport, including ciliogenesis (Hirokawa et al., 2009; Reilly and Benmerah, 2019; Silverman and Leroux, 2009). They may serve different functions in primary cilia. While kinesin II complex is important for driving anterograde transport of IFT particles from the base of the cilium to its tip (Cole et al., 1998; Marszalek et al., 1999; Morris and Scholey, 1997), KIF7 seems to organize the HH signaling platform at the tip of the primary cilium (Endoh-Yamagami et al., 2009; He et al., 2014), and it has been proposed that KIF24 negatively regulates the early steps of primary cilium formation (Kobayashi et al., 2011) and mediates cilium disassembly (Kim et al., 2015). KIF14, a member of the kinesin-3 subfamily, has been previously implicated in the regulation of chromosome segregation (Zhu et al., 2005) and cytokinesis and midbody formation (Bassi et al., 2013; Carleton et al., 2006; Gruneberg et al., 2006; Moawia et al., 2017; Ohkura et al., 1997). Interestingly, recent reports have suggested a possible link between KIF14 mutations and ciliopathies (Filges et al., 2014; Makrythanasis et al., 2018; Reilly et al., 2019), yet no evidence that KIF14 is involved in ciliogenesis has been provided. In this work, we demonstrate through various means that KIF14 depletion is associated with primary cilium defects and hence KIF14 is important for primary cilium formation and primary cilium–related signaling. Moreover, we identify deregulated AURA as a mediator of at least some of the KIF14 depletion phenotypes.
Results
To find novel regulators of primary cilium formation, we used proteomics and literature data mining. In our proteomics approaches, we focused on the identification of CEP164 interactome. We have combined two independent strategies to obtain our candidates: coimmunoprecipitation (coIP) of endogenous CEP164 in hTERT RPE-1 (retinal pigment epithelium cells immortalized with human telomerase reverse transcriptase) cells and proximity-dependent biotinylation by inducibly expressed CEP164-BirA* in HEK293 (human embryonal kidney) cells. By combining the results from both experiments (Table S1 and Table S2) with literature data mining, we selected five candidates to focus on: CCDC92, LUZP1, SCYL2, KIF7, and KIF14 (Fig. 1 A).
KIF14 knockdown causes ciliogenesis defects. (A) Graphical summary illustrating rational behind the selection of candidates. (B–H) hTERT RPE-1 cells were transfected with indicated siRNA and 24 h serum starved. (B) Quantification of ARL13B+ primary cilia formation. CEP164 siRNA was used as a positive control. For SCYL2 and CEP164 silencing, we used single siRNA oligos; for LUZP1, CCDC92, KIF7, and KIF14, we used a 50 nM mix of oligos 1 and 2 (Table S3). (C) Representative images of immunofluorescent staining of ARL13B (green), γ-tubulin (red), and DNA (blue); scale bar, 10 µm (crops 1 and 2 show examples of “long” primary cilia; crop 3 shows “no” cilium; and crop 4 shows a “short” primary cilium; scale bar, 2 µm). Silencing of KIF3A was used as a positive control. (D) Graph of cilia length (only cells with ciliary axoneme are included). (E–G) Test of KIF14 knockdown efficiency using different siRNA oligos. (E) Western blot analysis of protein expression in total hTERT RPE-1 cell lysates. (F) Representative images of IF staining of ARL13B (green), γ-tubulin (red), and DNA (blue); scale bar, 10 µm. (G) Quantification of ARL13B+ primary cilia formation after KIF14 knockdown using different siRNA oligos. KIF3A siRNA was used as a positive control. (H) IF staining of ciliogenesis defects rescue, caused by KIF14 siRNA #2–mediated depletion, by expression of GFP-KIF14si2res upon DOX induction (GFP-KIF14, green; Ac-tub, red; and DNA, blue); scale bar, 10 µm. Green arrows point to long primary cilia (>3.3 µm), and red arrows point to short primary cilia (<3.3 µm). (I) Quantification of ciliogenesis rescue. Note that ciliated cells here refer to a sum of short-cilium and long-cilium cells. Asterisks indicate statistical significance determined using an unpaired t test (D and I; ciliated cells = short + long) or the Holm–Sidak method (B, G, and I; categories).
KIF14 knockdown causes ciliogenesis defects. (A) Graphical summary illustrating rational behind the selection of candidates. (B–H) hTERT RPE-1 cells were transfected with indicated siRNA and 24 h serum starved. (B) Quantification of ARL13B+ primary cilia formation. CEP164 siRNA was used as a positive control. For SCYL2 and CEP164 silencing, we used single siRNA oligos; for LUZP1, CCDC92, KIF7, and KIF14, we used a 50 nM mix of oligos 1 and 2 (Table S3). (C) Representative images of immunofluorescent staining of ARL13B (green), γ-tubulin (red), and DNA (blue); scale bar, 10 µm (crops 1 and 2 show examples of “long” primary cilia; crop 3 shows “no” cilium; and crop 4 shows a “short” primary cilium; scale bar, 2 µm). Silencing of KIF3A was used as a positive control. (D) Graph of cilia length (only cells with ciliary axoneme are included). (E–G) Test of KIF14 knockdown efficiency using different siRNA oligos. (E) Western blot analysis of protein expression in total hTERT RPE-1 cell lysates. (F) Representative images of IF staining of ARL13B (green), γ-tubulin (red), and DNA (blue); scale bar, 10 µm. (G) Quantification of ARL13B+ primary cilia formation after KIF14 knockdown using different siRNA oligos. KIF3A siRNA was used as a positive control. (H) IF staining of ciliogenesis defects rescue, caused by KIF14 siRNA #2–mediated depletion, by expression of GFP-KIF14si2res upon DOX induction (GFP-KIF14, green; Ac-tub, red; and DNA, blue); scale bar, 10 µm. Green arrows point to long primary cilia (>3.3 µm), and red arrows point to short primary cilia (<3.3 µm). (I) Quantification of ciliogenesis rescue. Note that ciliated cells here refer to a sum of short-cilium and long-cilium cells. Asterisks indicate statistical significance determined using an unpaired t test (D and I; ciliated cells = short + long) or the Holm–Sidak method (B, G, and I; categories).
KIF14 knockdown causes ciliogenesis defects
To assess the potential role of our candidates in ciliogenesis, we used siRNA to knock down their expression in the hTERT RPE-1 cell line (Fig. S1 A). Having confirmed efficient depletion of individual mRNAs, we next examined the possible impact of RNAi on the formation of ARL13B+ primary cilium (Fig. 1 B). Interestingly, while knockdown of SCYL2, LUZP1, CCDC92, or KIF7 showed no apparent effect on primary cilium formation, KIF14 depletion led to prominent ciliogenesis defects (Fig. 1, B and C). Specifically, the proportion of cells forming fully grown primary cilia (“long”; >3.3 µm) after KIF14 knockdown was reduced compared with the control, while populations of cells with short (≤3.3 µm) or no primary cilia were increased (Fig. 1, B and C). We subsequently examined the effect on primary cilium length more closely and found that primary cilia were on average ∼50% shorter in KIF14-depleted cells than in controls (Fig. 1, C and D). To further corroborate these findings, obtained using a mix of two siRNAs, we tested the effects of KIF14 siRNA on ciliogenesis in hTERT RPE-1 cells using three individual KIF14 siRNA oligos. KIF14 siRNA #1 and #2 were used in our original experiments; KIF14 siRNA #3 had previously been shown to efficiently deplete KIF14 levels (Carleton et al., 2006). We tested their KD efficiency on protein (Fig. 1 E and,Fig. S1 B) and mRNA (Fig. S1 C) levels and in turn examined their effects on the formation of ARL13B+ primary cilia. Importantly, all tested KIF14 siRNAs led to a similar defect in the formation of ARL13B+ primary cilia (Fig. 1, F and G; and Fig. S1, D–F). Furthermore, we confirmed the defect in primary cilium formation following KIF14 depletion using another marker of primary cilia, acetylated tubulin (Ac-tub) in hTERT RPE-1 (Fig. S1, G–I). Finally, to fully prove the specificity of ciliogenesis defects detected in the KIF14 siRNA condition, we established a stable hTERT RPE-1 cell line inducibly expressing a GFP-KIF14 mutant resistant to KIF14 siRNA #2 (GFP-KIF14siRNA2res; Fig. S1 J). Importantly, the expression of GFP-KIF14siRNA2res, following the depletion of endogenous KIF14, was able to fully restore the formation of primary cilia to levels comparable to controls (Fig. 1, H and I).
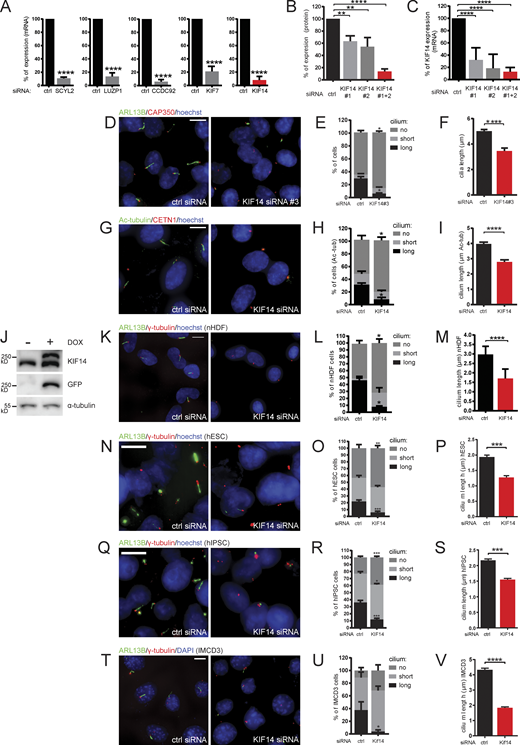
KIF14 knockdown causes ciliogenesis defects in different cell types. (A) Quantitative RT-PCR mRNA expression analysis for validation of siRNA knockdown efficiency (related to Fig. 1 B). For SCYL2 silencing, we used a single siRNA oligo; for LUZP1, CCDC92, KIF7, and KIF14, we used a 50 nM mix of oligos 1 and 2 (Table S3). (B and C) Test of KIF14 knockdown efficiency using different siRNA oligos in hTERT RPE-1 cells. (B) Quantification of Fig. 1 E. Western blot analysis of protein expression in total cell lysates using two different siRNA KIF14 oligos. (C) Quantitative RT-PCR expression analysis of KIF14 mRNA. (D–F) Test of the effect of KIF14 siRNA oligo #3 (Carleton et al., 2006; Table S3) on hTERT RPE-1 cells. (D) Representative image of IF staining of ARL13B (green), CAP350 (red), and DNA (blue); scale bar, 10 µm. (E and F) Graphs of primary cilia formation ability after KIF14 siRNA #3 knockdown (E) and the effect on ARL13B+ cilia length (F). (G–I) Validation of the effect of KIF14 knockdown on ciliogenesis in hTERT RPE-1 cells using Ac-tub as a ciliary marker. (G) Representative images of IF staining of Ac-tub (green), CETN1 (red),and DNA (blue); scale bar, 10 µm. Quantification of the experiment is shown as a percentage of ciliated cells (H) and by Ac-tub+ cilia length (I). (J) Validation of Flp-in T-REx RPE-1 cell line by Western blot analysis upon DOX induction expressing GFP-KIF14 mutant resistant to KIF14 siRNA 2 (GFP-KIF14siRNA2res; related to Fig. 1, H and I). (K–V) Examination of phenotypes after KIF14 depletion in nHDFs (K–M), hESCs (N–P), hiPSCs (Q–S), and IMCD3s (T–V). (K, N, Q, and T) Representative images of experiments detecting IF staining of ARL13B (green), γ-tubulin (red), DNA (blue); scale bars, 10 µm. Quantifications of experiments (by percentage of ciliated cells) are shown in L, O, R, and U, and graphs showing ARL13+ ciliary lengths are shown in M, P, S, and V. Asterisks indicate statistical significance using an unpaired t test.
KIF14 knockdown causes ciliogenesis defects in different cell types. (A) Quantitative RT-PCR mRNA expression analysis for validation of siRNA knockdown efficiency (related to Fig. 1 B). For SCYL2 silencing, we used a single siRNA oligo; for LUZP1, CCDC92, KIF7, and KIF14, we used a 50 nM mix of oligos 1 and 2 (Table S3). (B and C) Test of KIF14 knockdown efficiency using different siRNA oligos in hTERT RPE-1 cells. (B) Quantification of Fig. 1 E. Western blot analysis of protein expression in total cell lysates using two different siRNA KIF14 oligos. (C) Quantitative RT-PCR expression analysis of KIF14 mRNA. (D–F) Test of the effect of KIF14 siRNA oligo #3 (Carleton et al., 2006; Table S3) on hTERT RPE-1 cells. (D) Representative image of IF staining of ARL13B (green), CAP350 (red), and DNA (blue); scale bar, 10 µm. (E and F) Graphs of primary cilia formation ability after KIF14 siRNA #3 knockdown (E) and the effect on ARL13B+ cilia length (F). (G–I) Validation of the effect of KIF14 knockdown on ciliogenesis in hTERT RPE-1 cells using Ac-tub as a ciliary marker. (G) Representative images of IF staining of Ac-tub (green), CETN1 (red),and DNA (blue); scale bar, 10 µm. Quantification of the experiment is shown as a percentage of ciliated cells (H) and by Ac-tub+ cilia length (I). (J) Validation of Flp-in T-REx RPE-1 cell line by Western blot analysis upon DOX induction expressing GFP-KIF14 mutant resistant to KIF14 siRNA 2 (GFP-KIF14siRNA2res; related to Fig. 1, H and I). (K–V) Examination of phenotypes after KIF14 depletion in nHDFs (K–M), hESCs (N–P), hiPSCs (Q–S), and IMCD3s (T–V). (K, N, Q, and T) Representative images of experiments detecting IF staining of ARL13B (green), γ-tubulin (red), DNA (blue); scale bars, 10 µm. Quantifications of experiments (by percentage of ciliated cells) are shown in L, O, R, and U, and graphs showing ARL13+ ciliary lengths are shown in M, P, S, and V. Asterisks indicate statistical significance using an unpaired t test.
Having established that KIF14 is needed for efficient formation of primary cilia in hTERT RPE-1, we next examined its implication in ciliogenesis in several other cell lines. Importantly, we observed similar phenotypes following the depletion of KIF14 in neonatal human dermal fibroblasts (nHDFs; Fig. S1, K–M), human embryonic stem cells (hESCs; Fig. S1, N–P), human induced pluripotent stem cells (hiPSCs; Fig. S1, Q–S), and IMCD3s (a mouse inner medullary collecting duct cell line; Fig. S1, T–V). We conclude that the function of KIF14 in ciliogenesis is not restricted to one particular cell type.
KIF14 localizes to primary cilia in interphase cells
KIF14 was reported to localize to midbody during cytokinesis (Carleton et al., 2006; Gruneberg et al., 2006), which we also observed in our models (not shown). Given the effects of KIF14 depletion we found, we examined KIF14 localization in cells in interphase, when the cells can form primary cilia. First, we transiently expressed GFP-KIF14 and untagged KIF14, respectively, in hTERT RPE-1 and found that both proteins could localize to primary cilia (Fig. 2, A and B). To corroborate this observation, we examined the localization of endogenous KIF14. Importantly, while we detected the signal of the KIF14 antibody in primary cilia in hTERT RPE-1, the signal was diminished in cells transfected with KIF14 siRNA, thereby demonstrating the specificity of the KIF14 staining (Fig. 2 C).
KIF14 localizes to primary cilia in interphase cells. (A) Representative image of IF staining detecting GFP-KIF14 (green) localization to the ciliary axoneme after overexpression in hTERT RPE-1, ARL13B (red), or centrin (blue); scale bar, 10 µm. (B) Representative image of IF staining detecting untagged KIF14 (red) localization to the ciliary axoneme after overexpression in hTERT RPE-1, GFP (green), or DNA (blue); scale bar, 10 µm. (C) Representative images of high-resolution microscopy analyses of endogenous KIF14 (green) localization to the ciliary axoneme and Ac-tub (red) in control vs. KIF14-depleted hTERT RPE-1 cells; scale bar, 5 µm. (D) Scheme of KIF14 mutants (Reilly et al., 2018) transfected into hTERT RPE-1 cells. PRC1, Protein Regulator of Cytokinesis 1 - binding domain; CIT, Citron Kinase - binding domain; FHA, forkhead-associated domain. (E) Representative images of IF microscopy analysis of KIF14 truncated mutant localization, ARL13B (red), GFP-KIF14 (green), and centrin (blue); scale bar, 20 µm. Note the accumulation of GFP-KIF14 in the ciliary tip upon transfection of C-terminally truncated mutants. (F) Graphical overview of GFP-KIF14 mutant localization. (G) Quantification of the GFP-KIF14 ciliary tip accumulation for different KIF14 mutants (relative fluorescent signal, arbitrary units). Asterisks indicate statistical significance determined using an unpaired t test, n = 2, N ≥ 55. AU, arbitrary units.
KIF14 localizes to primary cilia in interphase cells. (A) Representative image of IF staining detecting GFP-KIF14 (green) localization to the ciliary axoneme after overexpression in hTERT RPE-1, ARL13B (red), or centrin (blue); scale bar, 10 µm. (B) Representative image of IF staining detecting untagged KIF14 (red) localization to the ciliary axoneme after overexpression in hTERT RPE-1, GFP (green), or DNA (blue); scale bar, 10 µm. (C) Representative images of high-resolution microscopy analyses of endogenous KIF14 (green) localization to the ciliary axoneme and Ac-tub (red) in control vs. KIF14-depleted hTERT RPE-1 cells; scale bar, 5 µm. (D) Scheme of KIF14 mutants (Reilly et al., 2018) transfected into hTERT RPE-1 cells. PRC1, Protein Regulator of Cytokinesis 1 - binding domain; CIT, Citron Kinase - binding domain; FHA, forkhead-associated domain. (E) Representative images of IF microscopy analysis of KIF14 truncated mutant localization, ARL13B (red), GFP-KIF14 (green), and centrin (blue); scale bar, 20 µm. Note the accumulation of GFP-KIF14 in the ciliary tip upon transfection of C-terminally truncated mutants. (F) Graphical overview of GFP-KIF14 mutant localization. (G) Quantification of the GFP-KIF14 ciliary tip accumulation for different KIF14 mutants (relative fluorescent signal, arbitrary units). Asterisks indicate statistical significance determined using an unpaired t test, n = 2, N ≥ 55. AU, arbitrary units.
Mutations in KIF14 have been implicated in the etiology of microcephaly and kidney development defects (Filges et al., 2014; Makrythanasis et al., 2018; Moawia et al., 2017; Reilly et al., 2019). Given the observation of ciliary localization of KIF14, we in turn examined KIF14 mutants described by Reilly and colleagues (Reilly et al., 2019; Fig. 2 D), in terms of their ability to localize to BB and axoneme, respectively. We found that following transient transfection in hTERT RPE-1, all tested variants could be readily detected at centrin+ centrioles (Fig. 2, E and F). Intriguingly, KIF14 mutations leading to truncated protein moieties (Q1380x, Q1304x, and R1189x) showed a strong enrichment in the ciliary tip compartment (Fig. 2, E–G), in contrast to N-terminal motor domain mutants (R364C and T456M; previously demonstrated as being defective in the motor domain function (Reilly et al., 2019), which showed diminished signal intensity in the ciliary tip (Fig. 2, E–G). These results suggest that both N-terminal and C-terminal parts of KIF14 are implicated in the correct transport of KIF14 molecules in primary cilium axoneme.
KIF14 affects dynamics of cilia growth
Given the phenotypes we found, we next examined the dynamics of ciliogenesis defects following KIF14 depletion by tracking primary cilium growth using time-lapse microscopy (Fig. 3 A). First, we confirmed the consistency of the primary cilium formation defects after KIF14 depletion in the hTERT RPE-1 cell line expressing GFP-ARL13B upon doxycycline (DOX) induction (Fig. S2, A and B). Next, we tracked GFP-ARL13B+ cilia growth over time. We examined changes in the length of primary cilia within a period of 9 h post primary cilium induction and found that primary cilia after KIF14 knockdown were notably shorter than controls, with the differences becoming more pronounced at the later time points of our time-lapse experiment (Fig. 3 B). In agreement with this observation, our measurements further revealed that the initial speed of primary cilium elongation was comparable between controls and KIF14-depleted cells, but the growth in the KIF14 siRNA condition dramatically dropped at later time points and subsequently completely stopped (please note the effects on average and current speed of cilia growth, respectively (Fig. 3, C and D). Together, these results have established that the depletion of KIF14 hampers the dynamics of primary cilium growth, mainly at the later stage of axoneme elongation.
KIF14 depletion affects dynamics of primary cilia growth. (A–D) Time-lapse imaging microscopy analyses of ciliogenesis in stable GFP-ARL13B hTERT RPE-1 during 9 h after serum starvation. (A) Representative stills of control or KIF14 siRNA-transfected cells; scale bars, 20 µm (ctrl siRNA) and 10 µm (KIF14 siRNA). Green arrows point to long (control) primary cilia, and red arrows point to short primary cilia. T indicates time in hours. (B) Quantification of cilia length during time-lapse microscopy of GFP-ARL13B+ primary cilia. Note the increasing difference between control and KIF14-depleted cells at later time points. (C and D) Measurements of the speed of ciliary axoneme elongation. The average primary cilia growth speed (µm/h) reflects actual PC length reached at the indicated time (C), and the current primary cilia speed of growth (µm/h) represents an increment of length per hour (D). Asterisks indicate statistical significance determined using an unpaired t test.
KIF14 depletion affects dynamics of primary cilia growth. (A–D) Time-lapse imaging microscopy analyses of ciliogenesis in stable GFP-ARL13B hTERT RPE-1 during 9 h after serum starvation. (A) Representative stills of control or KIF14 siRNA-transfected cells; scale bars, 20 µm (ctrl siRNA) and 10 µm (KIF14 siRNA). Green arrows point to long (control) primary cilia, and red arrows point to short primary cilia. T indicates time in hours. (B) Quantification of cilia length during time-lapse microscopy of GFP-ARL13B+ primary cilia. Note the increasing difference between control and KIF14-depleted cells at later time points. (C and D) Measurements of the speed of ciliary axoneme elongation. The average primary cilia growth speed (µm/h) reflects actual PC length reached at the indicated time (C), and the current primary cilia speed of growth (µm/h) represents an increment of length per hour (D). Asterisks indicate statistical significance determined using an unpaired t test.
KIF14 knockdown affects localization of BB components and IFT-B machinery. (A) Validation of KIF14 siRNA knockdown efficiency by Western blot protein expression analysis in Flp-in T-REx RPE-1 cell line (referring to time-lapse imaging experiments in Fig. 3) upon doxycyclin (DOX) induction expression GFP-ARL13B. (B) Quantification of ciliogenesis ability in GFP-ARL13B inducible RPE-1 cells after KIF14 siRNA knockdown. hTERT RPE-1 cells were transfected with the indicated siRNA, serum starved for 24 h, and analyzed by IF microscopy (related to Fig. 4, C–Q). γ-tubulin or CAP350 staining were used to visualize centrosomes, Ac-tubulin or ARL13B staining detected primary cilia. (C–F) Examination of protein levels of BB and ciliary markers used for normalization; levels are not affected by KIF14 siRNA. Relative CAP350 (C) and γ-tubulin (D) BB intensities and relative ARL13B (E) and Ac-tubulin (F) ciliary intensity are quantified in arbitrary units (AU) after background subtraction. (G) KIF14 depletion does not affect process of CP110 removal from the distal end of mother centriole as quantified by number of CP110 dots. TTBK2 knockdown was used as positive control. (H) Representative images of IF staining detects CEP164 (green), γ-tubulin (red), and DNA (blue); scale bar, 2 µm. Quantification of CEP164 intensity on BB (normalized to γ-tubulin) in I (CEP164 siRNA was used as positive control). (J) Representative images of IF staining of IFT88 (green), CAP350 (yellow), and Ac-tub (red); scale bar, 3 µm. (K and L) Quantification of IFT88 intensity (normalized to CAP350) at BBs (K) and a decrease of IFT88 in the ciliary tip (L) are shown. (M) Representative images of IF staining of IFT140 (green), CAP350 (yellow), and Ac-tub (red); scale bar, 3 µm. (N and O) IFT140 intensity (normalized to CAP350) at BBs (N) or in the ciliary tip (O) was not affected by KIF14 depletion. (P–Q) Histograms demonstrate distribution of CAP350 (red), Ac-tub (gray), and IFT (green) proteins along the ciliary axoneme (N = 20). Quantification was done for typical control long cilia and short defective cilia after KIF14 siRNA and for additional control also within a subset of short primary cilia in controls. Asterisks or ns indicates statistical significance determined by an unpaired t test.
KIF14 knockdown affects localization of BB components and IFT-B machinery. (A) Validation of KIF14 siRNA knockdown efficiency by Western blot protein expression analysis in Flp-in T-REx RPE-1 cell line (referring to time-lapse imaging experiments in Fig. 3) upon doxycyclin (DOX) induction expression GFP-ARL13B. (B) Quantification of ciliogenesis ability in GFP-ARL13B inducible RPE-1 cells after KIF14 siRNA knockdown. hTERT RPE-1 cells were transfected with the indicated siRNA, serum starved for 24 h, and analyzed by IF microscopy (related to Fig. 4, C–Q). γ-tubulin or CAP350 staining were used to visualize centrosomes, Ac-tubulin or ARL13B staining detected primary cilia. (C–F) Examination of protein levels of BB and ciliary markers used for normalization; levels are not affected by KIF14 siRNA. Relative CAP350 (C) and γ-tubulin (D) BB intensities and relative ARL13B (E) and Ac-tubulin (F) ciliary intensity are quantified in arbitrary units (AU) after background subtraction. (G) KIF14 depletion does not affect process of CP110 removal from the distal end of mother centriole as quantified by number of CP110 dots. TTBK2 knockdown was used as positive control. (H) Representative images of IF staining detects CEP164 (green), γ-tubulin (red), and DNA (blue); scale bar, 2 µm. Quantification of CEP164 intensity on BB (normalized to γ-tubulin) in I (CEP164 siRNA was used as positive control). (J) Representative images of IF staining of IFT88 (green), CAP350 (yellow), and Ac-tub (red); scale bar, 3 µm. (K and L) Quantification of IFT88 intensity (normalized to CAP350) at BBs (K) and a decrease of IFT88 in the ciliary tip (L) are shown. (M) Representative images of IF staining of IFT140 (green), CAP350 (yellow), and Ac-tub (red); scale bar, 3 µm. (N and O) IFT140 intensity (normalized to CAP350) at BBs (N) or in the ciliary tip (O) was not affected by KIF14 depletion. (P–Q) Histograms demonstrate distribution of CAP350 (red), Ac-tub (gray), and IFT (green) proteins along the ciliary axoneme (N = 20). Quantification was done for typical control long cilia and short defective cilia after KIF14 siRNA and for additional control also within a subset of short primary cilia in controls. Asterisks or ns indicates statistical significance determined by an unpaired t test.
KIF14 knockdown affects BB components and IFT-B anterograde transport
To better understand the function of KIF14 in primary cilium formation, we performed a series of IF microscopy analyses of BB and/or primary cilium components, previously implicated in key steps of ciliogenesis. First, we confirmed that tested markers of centriole or primary cilium (CAP350 [Yan et al., 2006], γ-tubulin, ARL13B, and Ac-tub) were not affected in terms of either localization or intensity by KIF14 KD (Fig. S2, C–F), and hence can be used for subsequent normalization to accurately determine the relative abundance of any further tested component in hTERT RPE-1. Next, we examined the BB components TTBK2 (Fig. 4, A and B), CP110 (Fig. S2 G), and CEP164 (Fig. S2, H and I), implicated in the early steps of primary cilium formation (Čajánek and Nigg, 2014; Goetz et al., 2012; Graser et al., 2007; Schmidt et al., 2012; Spektor et al., 2007), and found their levels at BB not affected by KIF14 knockdown (Fig. 4, A and B; and Fig. S2, G–I). Interestingly, FBF1, a protein localizing between distal appendage sheets (Wei et al., 2013; Yang et al., 2018), and SCLT1, localizing to distal appendage sheets together with CEP164 (Tanos et al., 2013; Yang et al., 2018), were present at BB in significantly lower levels in KIF14-depleted cells than in controls (Fig. 4, C–F). In addition, levels of ODF2, a component of subdistal appendages (Ishikawa et al., 2005), were also modestly decreased after KIF14 depletion (Fig. 4, G and H). Additional proteins tested (CEP290, INVS, and CEP97) were not affected by KIF14 siRNA knockdown (data not shown).
KIF14 knockdown affects BB components and IFT-B anterograde transport. hTERT RPE-1 cells were transfected with the indicated siRNA, serum starved for 24 h, and analyzed by IF microscopy. γ-tubulin or CAP350 staining was used to visualize centrosomes, and Ac-tub staining detected primary cilia. (A) TTBK2 localization (green) is not affected by KIF14 depletion (TTBK2 siRNA knockdown was used as a positive control); γ-tubulin, red; scale bar, 2 µm. (B) Quantification of TTBK2 signal (normalized to γ-tubulin). (C) FBF1 protein intensity (green) is significantly affected by KIF14 depletion; γ-tubulin, red; scale bar, 2 µm. (D) Quantification of FBF1 signal (normalized to γ-tubulin). (E) SCLT1 protein intensity (green) is significantly affected by KIF14 depletion; CAP350, red; scale bar, 2 µm. (F) Quantification of SCLT1 signal (normalized to CAP350). (G) ODF2 protein intensity (green) is significantly affected by KIF14 depletion; γ-tubulin, red; scale bar, 2 µm. (H) Quantification of ODF2 signal (normalized to γ-tubulin). (I) Localization of IFT57 (green) is significantly different after KIF14 depletion (CAP350, yellow; Ac-tubulin, red); scale bar, 3 µm. (J and K) Quantification of IFT57 signal at BBs (J; "norm." means normalized to CAP350) and in the ciliary tip (K; "normal." means normalized to CAP350). (L) Histograms showing distribution of IFT57 (green), CAP350 (red), and Ac-tub (gray) and its signal intensities along primary cilia. Quantification was done for typical "control long" cilia and "short" defective cilia after KIF14 siRNA (N = 10) and for additional control also within a subset of short primary cilia in controls (N = 5). Asterisks or “ns” indicates statistical significance determined by unpaired t test (ns, not significant).
KIF14 knockdown affects BB components and IFT-B anterograde transport. hTERT RPE-1 cells were transfected with the indicated siRNA, serum starved for 24 h, and analyzed by IF microscopy. γ-tubulin or CAP350 staining was used to visualize centrosomes, and Ac-tub staining detected primary cilia. (A) TTBK2 localization (green) is not affected by KIF14 depletion (TTBK2 siRNA knockdown was used as a positive control); γ-tubulin, red; scale bar, 2 µm. (B) Quantification of TTBK2 signal (normalized to γ-tubulin). (C) FBF1 protein intensity (green) is significantly affected by KIF14 depletion; γ-tubulin, red; scale bar, 2 µm. (D) Quantification of FBF1 signal (normalized to γ-tubulin). (E) SCLT1 protein intensity (green) is significantly affected by KIF14 depletion; CAP350, red; scale bar, 2 µm. (F) Quantification of SCLT1 signal (normalized to CAP350). (G) ODF2 protein intensity (green) is significantly affected by KIF14 depletion; γ-tubulin, red; scale bar, 2 µm. (H) Quantification of ODF2 signal (normalized to γ-tubulin). (I) Localization of IFT57 (green) is significantly different after KIF14 depletion (CAP350, yellow; Ac-tubulin, red); scale bar, 3 µm. (J and K) Quantification of IFT57 signal at BBs (J; "norm." means normalized to CAP350) and in the ciliary tip (K; "normal." means normalized to CAP350). (L) Histograms showing distribution of IFT57 (green), CAP350 (red), and Ac-tub (gray) and its signal intensities along primary cilia. Quantification was done for typical "control long" cilia and "short" defective cilia after KIF14 siRNA (N = 10) and for additional control also within a subset of short primary cilia in controls (N = 5). Asterisks or “ns” indicates statistical significance determined by unpaired t test (ns, not significant).
The primary cilium defects we observed following KIF14 depletion (observation of short cilia with altered axoneme elongation) implied possible perturbation of IFT machinery. Indeed, microscopy analysis revealed that IFT57 normalized signal intensities at the base of primary cilia (Fig. 4, I and J) and in the ciliary tip (Fig. 4, I and K) were both significantly decreased upon KIF14 depletion. Importantly, this seems to be specifically related to KIF14 deficiency rather than the length of primary cilia, as we observed similar behavior when comparing primary cilia of KIF14 depleted cells to control primary cilia of comparable length (Fig. 4 L). Further, IFT88, another component of IFT-B complex, also showed reduced levels after KIF14 depletion, both at the BB (Fig. S2, J, K, and P) and the tip (Fig. S2, J, L, and P), while the levels of IFT140, a member of IFT-A complex, were found to be unchanged by the lack of KIF14 (Fig. S2, M–O and Q).
Together, these data suggest that KIF14 knockdown leads to changes in BB and IFT-B within the primary cilia.
KIF14 and CIT knockdown leads to minor effects on cell cycle in RPE-1
Given that KIF14 was described as playing a role in mitosis, particularly during cytokinesis (Carleton et al., 2006; Gruneberg et al., 2006; Reilly et al., 2019), we examined possible links between phenotypes we found and cell cycle/cytokinesis. Surprisingly, we found no evidence of defective cytokinesis in serum-starved hTERT RPE-1, as tested by immunofluorescence microscopy analyses of binucleated cells (Fig. 5 A). Next, we examined the cell cycle distribution after KIF14 knockdown by FACS or phospho-histone H3 (ph-H3; marker of G2/M cells) immunofluorescence staining. The results from nonstarved cells, depleted of KIF14 by siRNA, revealed a slight increase in S- and G2/M-cells, respectively (Fig. S3, A–D), in line with the reported mitotic role of KIF14 (Carleton et al., 2006; Gruneberg et al., 2006; Reilly et al., 2019). Importantly, we did not observe any alterations of cell cycle distribution after KIF14 knockdown at the endpoint phase of our experimental setup (cells after 24 h serum starvation), as examined by FACS (Figs. 5 B and,S3 E) or ph-H3 immunofluorescence staining (Fig. 5, C and D). To corroborate these data further, we used siRNA targeting Citron Rho-interacting kinase (CIT; Fig. S3, F–H), an additional regulator of cytokinesis and KIF14 interactor (Bassi et al., 2011; Gruneberg et al., 2006). Importantly, we found that CIT siRNA had similar effects to the depletion of KIF14 in RPE-1 (Fig. S3, I–O).
KIF14 function in primary cilia formation is related to interphase. (A–D) hTERT RPE-1 cells were transfected with the indicated siRNA, serum starved (SFM) for 24 h, and analyzed. (A) Quantification of proportion of binucleated cells by microscopy analysis. Treatment with 0.5 µM cytochalasin D (Cyto D) was used as a positive control. (B) Cell cycle distribution analysis of control or KIF14 siRNA-transfected cells with a comparable proportion of cells in G2/M phase. (C) Representative images of detection of ph-H3+ cells (green and pointed out with arrows; DNA, blue); scale bar, 50 µm. (D) Quantification of the percentage of ph-H3+ cells, where a total of 40,000 nuclei were counted. (E and F) Examination of the effects of prolonged mitosis, induced by 10 µM monastrol for 2–4 h, on primary cilia formation in hTERT RPE-1 cells (please see the scheme of the experiment in E). (F) Monastrol-mediated mitotic arrest and subsequent release into 10 µM (R)-roscovitine (a cyclin-dependent kinase inhibitor) had only a modest effect on primary cilia formation (please compare black columns). Depletion of KIF14 leads to a significantly lower percentage of ciliated cells, both in controls (“leftover”) and cells that experienced temporal mitotic arrest from monastrol treatment. (G–J) hTERT RPE-1 cells were synchronized in G0 by serum starvation (24 h before siRNA transfection), transfected by the indicated siRNA, and serum starved for an additional 48 h before fixation (please see the scheme of the experiment in G). (H) Representative images (ARL13B in green, γ-tubulin in red, and DNA in blue; scale bar, 20 µm) show a significant effect of KIF14 depletion on ciliogenesis independent of mitosis, quantified as a decreased percentage of ciliated cells (I) and cilia length (J). (K–N) Same experimental setup as in G–J, with an additional 20 µM (R)-roscovitine to halt any proliferation before the transfection with siRNA (please see the scheme of the experiment in K, representative images in L; scale bar, 20 µm, and quantifications in M and N). Asterisks or "ns" indicates statistical significance determined using an unpaired t test (A, B, D, F, J, and N) or the Holm–Sidak method (I and M).
KIF14 function in primary cilia formation is related to interphase. (A–D) hTERT RPE-1 cells were transfected with the indicated siRNA, serum starved (SFM) for 24 h, and analyzed. (A) Quantification of proportion of binucleated cells by microscopy analysis. Treatment with 0.5 µM cytochalasin D (Cyto D) was used as a positive control. (B) Cell cycle distribution analysis of control or KIF14 siRNA-transfected cells with a comparable proportion of cells in G2/M phase. (C) Representative images of detection of ph-H3+ cells (green and pointed out with arrows; DNA, blue); scale bar, 50 µm. (D) Quantification of the percentage of ph-H3+ cells, where a total of 40,000 nuclei were counted. (E and F) Examination of the effects of prolonged mitosis, induced by 10 µM monastrol for 2–4 h, on primary cilia formation in hTERT RPE-1 cells (please see the scheme of the experiment in E). (F) Monastrol-mediated mitotic arrest and subsequent release into 10 µM (R)-roscovitine (a cyclin-dependent kinase inhibitor) had only a modest effect on primary cilia formation (please compare black columns). Depletion of KIF14 leads to a significantly lower percentage of ciliated cells, both in controls (“leftover”) and cells that experienced temporal mitotic arrest from monastrol treatment. (G–J) hTERT RPE-1 cells were synchronized in G0 by serum starvation (24 h before siRNA transfection), transfected by the indicated siRNA, and serum starved for an additional 48 h before fixation (please see the scheme of the experiment in G). (H) Representative images (ARL13B in green, γ-tubulin in red, and DNA in blue; scale bar, 20 µm) show a significant effect of KIF14 depletion on ciliogenesis independent of mitosis, quantified as a decreased percentage of ciliated cells (I) and cilia length (J). (K–N) Same experimental setup as in G–J, with an additional 20 µM (R)-roscovitine to halt any proliferation before the transfection with siRNA (please see the scheme of the experiment in K, representative images in L; scale bar, 20 µm, and quantifications in M and N). Asterisks or "ns" indicates statistical significance determined using an unpaired t test (A, B, D, F, J, and N) or the Holm–Sidak method (I and M).
KIF14 and CIT knockdown leads to minor effects on cell cycle in RPE-1, related to Fig. 5, A–E . Cell cycle analyses demonstrating distribution of hTERT RPE-1 cells 48 h transfected with control or KIF14 siRNA and 24 h proliferating in normal 10% FBS medium (A–D) or serum starved (E). (A and B) Graph (A) and histograms (B) of flow cytometry analysis of cycling cells show minor increase of G2/M cells (quantification indicates increased ploidy = cells with >G2/M DNA content). (C and D) Representative images (C; phospho-histone H3 in green and DNA in blue; scale bar, 100 µm) and graph of quantification (D) show minor increment of phospho-histon H3 positive (ph-H3+) proliferating cells. (E) Histograms of flow cytometry analyses quantified in Fig. 5 B graph. (F–H) Validation of CIT siRNA knockdown efficiency in hTERT RPE-1 cells by Western blot analysis on protein level (F) and quantitative RT-PCR analysis of CIT mRNA depletion (G). (H) Representative images of IF microscopy detecting CIT (green), CETN1 (red), and DNA (blue); scale bar, 5 µm. (I–O) Cell cycle analyses demonstrating distribution of hTERT RPE-1 cells 48 h transfected with control or CIT siRNA and proliferating for 24 h in normal 10% FBS medium (J, K, N, and O) or serum starved (I and L–M). (I) Quantification of percentage of binucleated cells (cytochalasin D treatment was used as positive control). (J–M) Flow cytometry cell cycle analyses demonstrating minor increase in proportion of G2/M phase in 10% FBS and unaffected G2/M proportion in serum-starved conditions (quantification indicates increased ploidy = cells with >G2/M DNA content). (N and O) Representative images of ph-H3 (green) and DNA (blue) IF staining show minor increase of proliferating cells after CIT depletion in 10% FBS condition; scale bar, 100 µm (quantified in O). (P) P53 Western blot analysis of whole hTERT RPE-1 cell lysates transfected for 48 h with control or KIF14 siRNA (150 nM centrinone + 10 µM MG132 treatment was used as positive control). (Q) Graph showing ARL13B+ cilia length (related to Fig. 5, E and F; please see scheme of the experiment in Fig. 5 E). (R and S) Analysis of effectivity of serum starvation to synchronize hTERT RPE-1 cells in G0 phase (related to Fig. 5, G–J). (R) Representative images show IF staining detecting the proliferation marker Ki-67 (green) and DNA (red); scale bar, 20 µm. (S) Quantification of proportion of cells synchronized in G0 (Ki-67+). Asterisks or "ns" indicate statistical significance determined using an unpaired t test.
KIF14 and CIT knockdown leads to minor effects on cell cycle in RPE-1, related to Fig. 5, A–E . Cell cycle analyses demonstrating distribution of hTERT RPE-1 cells 48 h transfected with control or KIF14 siRNA and 24 h proliferating in normal 10% FBS medium (A–D) or serum starved (E). (A and B) Graph (A) and histograms (B) of flow cytometry analysis of cycling cells show minor increase of G2/M cells (quantification indicates increased ploidy = cells with >G2/M DNA content). (C and D) Representative images (C; phospho-histone H3 in green and DNA in blue; scale bar, 100 µm) and graph of quantification (D) show minor increment of phospho-histon H3 positive (ph-H3+) proliferating cells. (E) Histograms of flow cytometry analyses quantified in Fig. 5 B graph. (F–H) Validation of CIT siRNA knockdown efficiency in hTERT RPE-1 cells by Western blot analysis on protein level (F) and quantitative RT-PCR analysis of CIT mRNA depletion (G). (H) Representative images of IF microscopy detecting CIT (green), CETN1 (red), and DNA (blue); scale bar, 5 µm. (I–O) Cell cycle analyses demonstrating distribution of hTERT RPE-1 cells 48 h transfected with control or CIT siRNA and proliferating for 24 h in normal 10% FBS medium (J, K, N, and O) or serum starved (I and L–M). (I) Quantification of percentage of binucleated cells (cytochalasin D treatment was used as positive control). (J–M) Flow cytometry cell cycle analyses demonstrating minor increase in proportion of G2/M phase in 10% FBS and unaffected G2/M proportion in serum-starved conditions (quantification indicates increased ploidy = cells with >G2/M DNA content). (N and O) Representative images of ph-H3 (green) and DNA (blue) IF staining show minor increase of proliferating cells after CIT depletion in 10% FBS condition; scale bar, 100 µm (quantified in O). (P) P53 Western blot analysis of whole hTERT RPE-1 cell lysates transfected for 48 h with control or KIF14 siRNA (150 nM centrinone + 10 µM MG132 treatment was used as positive control). (Q) Graph showing ARL13B+ cilia length (related to Fig. 5, E and F; please see scheme of the experiment in Fig. 5 E). (R and S) Analysis of effectivity of serum starvation to synchronize hTERT RPE-1 cells in G0 phase (related to Fig. 5, G–J). (R) Representative images show IF staining detecting the proliferation marker Ki-67 (green) and DNA (red); scale bar, 20 µm. (S) Quantification of proportion of cells synchronized in G0 (Ki-67+). Asterisks or "ns" indicate statistical significance determined using an unpaired t test.
Role of KIF14 in primary cilium formation seems independent of its role in mitosis
As the lack of binucleated cells following KIF14/CIT depletion is evidence, but not proof, of mitosis/cytokinesis occurring correctly, we sought other means to corroborate our results.
Prolonged mitosis typically leads to up-regulation of p53, which in turn triggers apoptosis, differentiation, or cell cycle arrest (Bazzi and Anderson, 2014; Orth et al., 2012; Renzova et al., 2018). To probe for any possible contribution of mitotic stress to the effect of KIF14 depletion on primary cilium formation, we first tested whether KIF14 siRNA could possibly elevate p53 levels. However, we found no evidence of such an effect (Fig. S3 P). Next, we examined the effects of prolonged mitosis, induced by 10 µM monastrol for 2‒4 h, on primary cilium formation in hTERT RPE-1 (please see the scheme of the experiment in Fig. 5 E). We found that monastrol-mediated mitotic arrest and subsequent release had only a modest effect on primary cilium formation (please compare the first column in the “leftover” condition to the first column in the “M-arrest” condition; Fig. 5 F), and showed no effect on ciliary length (Fig. S3 Q). Importantly, however, the depletion of KIF14 led to a significantly lower percentage of ciliated cells, both in controls (“leftover”) and cells that experienced temporal mitotic arrest from monastrol treatment (Fig. 5 F). Based on these results, we conclude that any stress generated during mitotic arrest is unlikely to mediate the effects of KIF14 depletion on primary cilium formation.
Further, we aimed to test whether going through mitosis is required for the primary cilium phenotypes related to KIF14 knockdown. First, we depleted KIF14 by siRNA in hTERT RPE-1 cells that were serum starved for 24 h before transfection (Fig. 5 G). As expected, 24 h serum starvation showed a profound negative effect on the number of KI-67+ (maker of active cell cycle) cells (Fig. S3, R and S). Importantly, however, the cells transfected with KIF14 siRNA following the serum starvation still showed impaired primary cilium formation (Fig. 5, H–J) in a manner comparable to our initial observations (Fig. 1). To strengthen this observation further, we used the cyclin-dependent kinases inhibitor (R)-roscovitine (Meijer et al., 1997) to halt any remaining proliferation before the transfection with siRNA (Fig. 5 K). As shown in Fig. 5 L and quantified in Fig. 5, M and N, KIF14 depletion in hTERT RPE-1, cultured in serum-free medium + 20 µM roscovitine, again demonstrated negative effects on the proportion of cells with primary cilium (in Fig. 5 M) and the primary cilium length (Fig. 5 N). From these experiments, we conclude that impaired ciliogenesis following KIF14 depletion is not a simple reflection of mitotic delay or long-term effects on cell cycle; rather, KIF14 has a novel function in primary cilium formation, which seems to be independent of its role during mitosis/cytokinesis. This prompted us to examine alternative explanations for KIF14 action toward primary cilium formation.
KIF14 depletion deregulates AURA
AURA is a kinase known for its function in centrosome maturation and mitotic progression. Intriguingly, recent studies have also implicated AURA in the resorption of primary cilia (Inaba et al., 2016; Inoko et al., 2012; Plotnikova et al., 2012; Pugacheva et al., 2007). In light of our data, we hypothesized that abrogated AURA activity might explain some of the defects observed in KIF14-depleted cells.
To this end, we examined levels of AURA phosphorylated at Thr288 within its kinase domain T-loop (phospho-AURA [pAURA]), which typically indicates increased activity of the kinase (Littlepage et al., 2002; Walter et al., 2000). Our Western blot analysis (Fig. S4 A) suggested an up-regulation of pAURA and hence its increased activity in the KIF14 siRNA condition. We confirmed this intriguing observation through immunofluorescence microscopy analyses of pAURA levels (Fig. 6, A and B; and Fig. S4 B). As these data were not able to resolve whether there is indeed an increase of pAURA at BB, we turned to high-resolution confocal microscopy to specifically examine pAURA levels in this compartment. Importantly, as shown in Fig. 6, C and D, we detected a significant elevation of activated pAURA at BB after depletion of KIF14.
KIF14 depletion deregulates AURA. (A) Western blot analysis of whole hTERT RPE-1 cell lysates to detect an increase in pAURA protein levels after KIF14 depletion (100 ng/ml nocodazole treatment was used to enrich for mitotic cells). (B) Quantification of pAURA fluorescence intensity (normalized to Hoechst signal intensity) in nonstarved hTERT RPE-1 cells after control or KIF14 siRNA transfection. (C) Graph showing cilia lengths after MYC-AURA overexpression in hTERT RPE-1 cells (related to Fig. 6, E and F). (D–G) Related to Fig. 6, G–L. (D) Representative images of IFT88 (green), Ac-tub (red), and CAP350 (yellow) IF staining of hTERT RPE-1 cells transfected with mock or MYC-AURA (scale bar, 2 µm), where the proportion of ciliated cells is decreased, as quantified in E. (F) Representative images of ODF2 (green), CAP350 (red), and MYC (yellow) IF staining of hTERT RPE-1 cells transfected with mock or MYC-AURA (scale bar, 1 µm), where the proportion of ciliated cells is not changed (as quantified in G). Asterisks or "ns" indicates statistical significance compared with control determined using an unpaired t test.
KIF14 depletion deregulates AURA. (A) Western blot analysis of whole hTERT RPE-1 cell lysates to detect an increase in pAURA protein levels after KIF14 depletion (100 ng/ml nocodazole treatment was used to enrich for mitotic cells). (B) Quantification of pAURA fluorescence intensity (normalized to Hoechst signal intensity) in nonstarved hTERT RPE-1 cells after control or KIF14 siRNA transfection. (C) Graph showing cilia lengths after MYC-AURA overexpression in hTERT RPE-1 cells (related to Fig. 6, E and F). (D–G) Related to Fig. 6, G–L. (D) Representative images of IFT88 (green), Ac-tub (red), and CAP350 (yellow) IF staining of hTERT RPE-1 cells transfected with mock or MYC-AURA (scale bar, 2 µm), where the proportion of ciliated cells is decreased, as quantified in E. (F) Representative images of ODF2 (green), CAP350 (red), and MYC (yellow) IF staining of hTERT RPE-1 cells transfected with mock or MYC-AURA (scale bar, 1 µm), where the proportion of ciliated cells is not changed (as quantified in G). Asterisks or "ns" indicates statistical significance compared with control determined using an unpaired t test.
KIF14 depletion deregulates AURA. (A) Representative images of IF detection of pAURA (green), Ac-tub (red), CAP350 (yellow), and DNA (blue) in serum-starved hTERT RPE-1 cells; scale bar, 5 µm. (B) Quantification of pAURA fluorescent intensity (normalized to Hoechst signal intensity) in serum-starved whole cells. (C) Detection of a BB-specific pool of pAURA (green) in nonstarved cells using high-resolution microscopy (CAP350, blue; Ac-tub, red); scale bar, 0,4 µm. (D) Quantification of pAURA signal intensity on BB (normalized to CAP350 signal intensity). (E–L) Effect of MYC-AURA overexpression on 24 h serum-starved hTERT RPE-1 cells. (E) Representative images of IF detection of MYC-tagged (green), Ac-tub+ primary cilia (red; CAP350, yellow; DNA, blue) in mock- or MYC-AURA–transfected cells; scale bar, 10 µm. (F) Graph quantifying statistically significant decrease of ciliated cells percentage after MYC-AURA overexpression. (G and H) Examination of FBF1 (green) localization and decreased intensity after MYC-AURA overexpression. Representative images (CAP350 in red, MYC in yellow) are shown in G (scale bar, 2 µm) and intensity quantification (normalized to CAP350) in H. (I and J) Examination of SCLT1 (green) localization and decreased intensity after MYC-AURA overexpression. Representative images (CAP350 in red, MYC in yellow) are shown in I (scale bar, 1 µm) and intensity quantification (normalized to CAP350) in J. (K and L) Examination of IFT57 (green) localization and decreased intensity at BBs after MYC-AURA overexpression. Representative images (Ac-tub in red, CAP350 in yellow) are shown in K (scale bar, 2 µm) and BB intensity quantification (normalized to CAP350) in L. Asterisks indicate statistical significance determined using an unpaired t test.
KIF14 depletion deregulates AURA. (A) Representative images of IF detection of pAURA (green), Ac-tub (red), CAP350 (yellow), and DNA (blue) in serum-starved hTERT RPE-1 cells; scale bar, 5 µm. (B) Quantification of pAURA fluorescent intensity (normalized to Hoechst signal intensity) in serum-starved whole cells. (C) Detection of a BB-specific pool of pAURA (green) in nonstarved cells using high-resolution microscopy (CAP350, blue; Ac-tub, red); scale bar, 0,4 µm. (D) Quantification of pAURA signal intensity on BB (normalized to CAP350 signal intensity). (E–L) Effect of MYC-AURA overexpression on 24 h serum-starved hTERT RPE-1 cells. (E) Representative images of IF detection of MYC-tagged (green), Ac-tub+ primary cilia (red; CAP350, yellow; DNA, blue) in mock- or MYC-AURA–transfected cells; scale bar, 10 µm. (F) Graph quantifying statistically significant decrease of ciliated cells percentage after MYC-AURA overexpression. (G and H) Examination of FBF1 (green) localization and decreased intensity after MYC-AURA overexpression. Representative images (CAP350 in red, MYC in yellow) are shown in G (scale bar, 2 µm) and intensity quantification (normalized to CAP350) in H. (I and J) Examination of SCLT1 (green) localization and decreased intensity after MYC-AURA overexpression. Representative images (CAP350 in red, MYC in yellow) are shown in I (scale bar, 1 µm) and intensity quantification (normalized to CAP350) in J. (K and L) Examination of IFT57 (green) localization and decreased intensity at BBs after MYC-AURA overexpression. Representative images (Ac-tub in red, CAP350 in yellow) are shown in K (scale bar, 2 µm) and BB intensity quantification (normalized to CAP350) in L. Asterisks indicate statistical significance determined using an unpaired t test.
Having established a link between KIF14 depletion and AURA activation, we analyzed the effects of increased pAURA levels on primary cilium formation in our model (Fig. 6 E). In agreement with previous reports (Inoko et al., 2012; Plotnikova et al., 2012; Pugacheva et al., 2007), we found that overexpression of MYC-AURA decreased the proportion of hTERT RPE-1 cells able to build primary cilia (Fig. 6, E and F) and that those primary cilia formed were shorter than controls (Fig. 6 E and,Fig. S4 C). Interestingly, we further found that MYC-AURA expression led to reduced BB levels of FBF1 (Fig. 6, G and H), SCLT1 (Fig. 6, I and J), IFT57 (Fig. 6, K and L), and IFT88 (Fig. S4, D and E), but not ODF2 (Fig. S4, F and G). These data not only indicated that AURA may play a role in recruitment of the examined BB components but importantly also suggested that there is not only a phenotypic correlation but probably also a causality between KIF14 depletion, AURA activation, and defective ciliogenesis.
AURA activity mediates effects of KIF14 depletion on primary cilia formation
To fully prove this hypothesis, we treated control or KIF14 siRNA-transfected hTERT RPE-1 cells with 100 nM AURA inhibitor TCS7010 (Fig. 7 A). First, we examined the effect of the KIF14 depletion in combination with AURA inhibition on the recruitment of FBF1 and found that while its levels at BB decreased in KIF14-depleted cells, treatment with AURA inhibitor restored its levels to that of controls (Fig. 7, B and C). Interestingly, we observed a similar rescue effect on KIF14 depletion for BB levels of SCTL1 (Fig. 7, D and E). Next, we examined possible rescue effects of KIF14 siRNA by AURA inhibitor on levels of IFT-B proteins. In line with earlier observations, KIF14 siRNA led to lower levels of IFT57 at the BB and ciliary tip (Fig. 7, F and G). Importantly, the addition of AURA inhibitor restored IFT57 ciliary levels in KIF14-depleted cells (Fig. 7, F and G), although we noted that distribution of the IFT57 signal along the axoneme was still somewhat different than in controls. A similar rescue effect was observed in the case of IFT88 (Fig. S5 A). Remarkably, the use of TCS7010 inhibitor also led to the rescue of two key phenotypes associated with KIF14 depletion in serum-starved cells: the reduction of the proportion of cells forming primary cilia (Fig. 7, H and I) and the defective cilia length (Fig. 7 J).
AURA activity mediates effects of KIF14 depletion on primary cilia formation. (A) Experimental design of KIF14 siRNA effects rescue using 100 nM TCS7010 (AURA inhibitor). (B–J) hTERT RPE-1 cells were transfected with indicated siRNA 48 h before fixation and last 24 h serum starved and AURA inhibited by TCS7010. (B and C) Examination of FBF1 (green) localization and intensity. Representative images (CAP350 in red; scale bar, 1 µm) are shown in B and intensity quantification (normalized to CAP350) in C. (D and E) Examination of SCLT1 (green) localization and intensity. Representative images (CAP350 in red; scale bar, 1 µm) are shown in D and intensity quantification (normalized to CAP350) in E. (F and G) Examination of IFT57 (green) localization and intensity. Representative images (CAP350 in yellow, Ac-tub in red; scale bar, 2 µm) are shown in F and intensity quantification histograms (normalized to CAP350; N = 5) in G ("norm." means normalized to CAP350). (H) Representative images of AURA inhibition rescue experiment of ciliogenesis defect caused by KIF14 knockdown. Detection of ARL13B+ primary cilia (green; γ-tubulin, red; DNA, blue); scale bar, 10 µm. (I) Quantification of ARL13B+ primary cilia formation. (J) Effects on ARL13B+ primary cilia length. Asterisks or "ns" indicates statistical significance determined using Tukey's multiple comparisons test (C, E, and J), an unpaired t test (I; ciliated cells = short + long), or the Holm–Sidak method (I; categories).
AURA activity mediates effects of KIF14 depletion on primary cilia formation. (A) Experimental design of KIF14 siRNA effects rescue using 100 nM TCS7010 (AURA inhibitor). (B–J) hTERT RPE-1 cells were transfected with indicated siRNA 48 h before fixation and last 24 h serum starved and AURA inhibited by TCS7010. (B and C) Examination of FBF1 (green) localization and intensity. Representative images (CAP350 in red; scale bar, 1 µm) are shown in B and intensity quantification (normalized to CAP350) in C. (D and E) Examination of SCLT1 (green) localization and intensity. Representative images (CAP350 in red; scale bar, 1 µm) are shown in D and intensity quantification (normalized to CAP350) in E. (F and G) Examination of IFT57 (green) localization and intensity. Representative images (CAP350 in yellow, Ac-tub in red; scale bar, 2 µm) are shown in F and intensity quantification histograms (normalized to CAP350; N = 5) in G ("norm." means normalized to CAP350). (H) Representative images of AURA inhibition rescue experiment of ciliogenesis defect caused by KIF14 knockdown. Detection of ARL13B+ primary cilia (green; γ-tubulin, red; DNA, blue); scale bar, 10 µm. (I) Quantification of ARL13B+ primary cilia formation. (J) Effects on ARL13B+ primary cilia length. Asterisks or "ns" indicates statistical significance determined using Tukey's multiple comparisons test (C, E, and J), an unpaired t test (I; ciliated cells = short + long), or the Holm–Sidak method (I; categories).
AURA activity mediates the effects of KIF14 depletion on primary cilia formation. (A) Histograms show localization of CAP350 (red), Ac-tub (gray), and distribution of IFT88 (green) along the primary cilium (N = 5, "norm." means normalized to CAP350) rescued by AURA inhibition (related to to Fig. 7). (B and C) hTERT RPE-1 cells were 48 h transfected with control or KIF14 siRNA, together with either additional siRNA targeting the individual AURA activator or in combination with small-molecule calmidazolium chloride treatment (4 µM) to inhibit calmodulin (CaM) and 24 h serum starved before fixation. (B) Graph showing percentage of ciliated cells and pinpointing NEDD9 and PIFO as modest rescuers of the ciliogenesis defect caused by KIF14 depletion. (C) Graph showing partial cilia length rescues of defects caused by KIF14 depletion by all used AURA activators. (D) Graph quantifying the percentage of cells with SMO localized to the ciliary axoneme after control or KIF14 siRNA transfection, 24 h serum starvation, and 2 µM SAG treatment to HH pathway activation (related to Fig. 8 E). Asterisks indicate statistical significance determined using Tukey's multiple comparisons test.
AURA activity mediates the effects of KIF14 depletion on primary cilia formation. (A) Histograms show localization of CAP350 (red), Ac-tub (gray), and distribution of IFT88 (green) along the primary cilium (N = 5, "norm." means normalized to CAP350) rescued by AURA inhibition (related to to Fig. 7). (B and C) hTERT RPE-1 cells were 48 h transfected with control or KIF14 siRNA, together with either additional siRNA targeting the individual AURA activator or in combination with small-molecule calmidazolium chloride treatment (4 µM) to inhibit calmodulin (CaM) and 24 h serum starved before fixation. (B) Graph showing percentage of ciliated cells and pinpointing NEDD9 and PIFO as modest rescuers of the ciliogenesis defect caused by KIF14 depletion. (C) Graph showing partial cilia length rescues of defects caused by KIF14 depletion by all used AURA activators. (D) Graph quantifying the percentage of cells with SMO localized to the ciliary axoneme after control or KIF14 siRNA transfection, 24 h serum starvation, and 2 µM SAG treatment to HH pathway activation (related to Fig. 8 E). Asterisks indicate statistical significance determined using Tukey's multiple comparisons test.
Activation of AURA is typically tightly controlled, with a number of proteins proposed to act as activators of AURA activity (Hirota et al., 2003; Hutterer et al., 2006; Inaba et al., 2016; Kinzel et al., 2010; Kozyreva et al., 2014; Kufer et al., 2002; Plotnikova et al., 2012; Ran et al., 2015). Having established elevated AURA activity as a mediator of several primary cilium and BB defects in KIF14-depleted cells, we in turn asked to what extent individual AURA activators contributed to the KIF14 depletion defects we observed. To this end, we transfected hTERT RPE-1 with KIF14 siRNA, together with either additional siRNA targeting the individual AURA activator or in combination with small-molecule calmidazolium chloride treatment (4 µM) to inhibit calmodulin. Interestingly, we found that targeting individual candidate activators had only modest rescue effects on the KIF14 depletion phenotypes; NEDD9 and PIFO siRNA conditions partially rescued the percentage of ciliated cells (Fig. S5 B), as well as the cilium length (Fig. S5 C). Depletion of the remaining candidates showed modest rescue of the primary cilium length defect (Fig. S5 C) but failed to produce statistically significant effects for the percentage of ciliated cells (Fig. S5 B). This implies that the mechanism of AURA activation in KIF14-depleted cells is rather complex and that in this case, the participation of several regulators of its activity seems more plausible than a scenario with one “master regulator.”
Together, these data have demonstrated a connection between KIF14 depletion and the up-regulation of the resorption pathway caused by elevated activation of AURA, which results in a short cilia phenotype and defects in ciliogenesis.
KIF14 regulates HH signaling independently of AURA activity
To examine any functional consequences of the observed defects in primary cilium formation, we tested the ability of KIF14-depleted cells to respond to HH pathway activation. As expected, when KIF14 was depleted, nHDF cells failed to induce expression of GLI1, an HH pathway target gene (Lee et al., 1997), in response to a treatment with the agonist of the SMO receptor, smoothened agonist (SAG; Fig. 8 A). Furthermore, we examined the translocation of SMO into the primary cilia, which is a prerequisite for the activation of the expression of the target gene GLI1. Interestingly, while in control nHDF cells SMO readily translocated into primary cilia upon SAG treatment, the SMO translocation was impaired in KIF14-depleted nHDF cells able to form primary cilia (Fig. 8, B and C; and Fig. S5 D). These data suggest that a deficiency of KIF14 protein leads not only to a somewhat expected defect in response to HH pathway stimulation on the cell population level (in line with the observed reduction in the number of cells forming primary cilia) but intriguingly also to HH activation response abnormalities in short primary cilia formed upon KIF14 depletion.
KIF14 regulates HH signaling independently of AURA activity. (A–C) Functional analyses of HH pathway activation following control or KIF14 silencing and mock or 2 µM SAG treatment in nHDFs. (A) Quantitative RT-PCR quantification of the effect of GLI1 on mRNA levels.(B and C) Analysis of the effect of KIF14 depletion on SMO translocation to the ciliary axoneme and its intensity. (B) Representative images of SMO (green) and ARL13B (red) IF detection; scale bar, 2 µm. (C) Quantification of changes in relative SMO intensity. (D–F) Examination of effect of TCS7010 AURA inhibition on response of nHDF cells transfected with the indicated siRNA to HH pathway activation. Experimental setup was the same as in Fig. 7 A, but with an additional 0.5 µm SAG treatment for the last 24 h. (D) Representative images of SMO (green) and ARL13B (red) IF detection in nHDF cells transfected with ctrl or KIF14 siRNA and treated with mock or AURA inhibitor; scale bar = 2 µm. Quantification of changes in SMO intensity (normalized to ARL13B) is shown in E and quantitative RT-PCR quantification of effect on mRNA level of GLI1 in F. Asterisks or “ns” indicate statistical significance determined using Tukey's multiple comparisons test.
KIF14 regulates HH signaling independently of AURA activity. (A–C) Functional analyses of HH pathway activation following control or KIF14 silencing and mock or 2 µM SAG treatment in nHDFs. (A) Quantitative RT-PCR quantification of the effect of GLI1 on mRNA levels.(B and C) Analysis of the effect of KIF14 depletion on SMO translocation to the ciliary axoneme and its intensity. (B) Representative images of SMO (green) and ARL13B (red) IF detection; scale bar, 2 µm. (C) Quantification of changes in relative SMO intensity. (D–F) Examination of effect of TCS7010 AURA inhibition on response of nHDF cells transfected with the indicated siRNA to HH pathway activation. Experimental setup was the same as in Fig. 7 A, but with an additional 0.5 µm SAG treatment for the last 24 h. (D) Representative images of SMO (green) and ARL13B (red) IF detection in nHDF cells transfected with ctrl or KIF14 siRNA and treated with mock or AURA inhibitor; scale bar = 2 µm. Quantification of changes in SMO intensity (normalized to ARL13B) is shown in E and quantitative RT-PCR quantification of effect on mRNA level of GLI1 in F. Asterisks or “ns” indicate statistical significance determined using Tukey's multiple comparisons test.
Next, we asked whether the defect in SMO localization we found is also mediated by elevated AURA, as we observed for the ciliogenesis phenotypes. Remarkably, however, the accumulation of SMO in primary cilia in KIF14-depleted cells treated with AURA inhibitor was still impaired, even though the AURA inhibition showed prominent rescue effects on the percentage of ciliated cells and the primary cilium length (Fig. 8, D and E; and Fig. 7, H–J). In agreement with this result, we found that KIF14-depleted cells treated with AURA inhibitor were not able to respond to SAG agonist treatment through the induction of GLI1 mRNA (Fig. 8 F).
Together, those data propose that KIF14 has a dual role in relation to primary cilia formation and function, with the elevated AURA activity responsible for the ciliogenesis defects, but not the HH signaling abnormalities found.
Discussion
Cell cycle regulation and ciliogenesis are intimately interconnected via a regulatory network, the extent of which is only starting to become apparent. Here, we have explored this connection by examining the role of KIF14, a protein hinted at during our initial proteomics experiments, in primary cilium formation. We have found that KIF14 is required for correct BB biogenesis and primary cilium formation in several cell lines, as well as for proper response of primary cilia to HH pathway activation. Further, we have pinpointed deregulated AURA activity as a downstream mediator of KIF14 deficiency responsible for BB and primary cilium defects (Fig. 9).
Final scheme. Graphic summary of prominent phenotypes observed after KIF14 depletion and effects AURA inhibitionDAP, distal appendage proteins; KD, siRNA knockdown; sDA, subdistal appendage. KIF14 localizes to the ciliary axoneme. KIF14-depleted cells have increased pAURA levels on BBs, decreased intensities of distal appendage proteins FBF1 and SCLT1, and changed distribution of IFT57 and IFT88. This in turn leads to lower proportion of ciliated cells, reduced ciliary length, and failure to respond to HH pathway activation (reduced SMO ciliary localization and diminished expression of target gene Gli1). Treatment of KIF14-depleted cells by AURA inhibitor rescues the localization defects of FBF1, SCLT1, IFT57, and IFT88; defective cilia formation; and reduced ciliary length but fails to restore competence of KIF14-depleted primary cilia to respond to HH pathway activation.
Final scheme. Graphic summary of prominent phenotypes observed after KIF14 depletion and effects AURA inhibitionDAP, distal appendage proteins; KD, siRNA knockdown; sDA, subdistal appendage. KIF14 localizes to the ciliary axoneme. KIF14-depleted cells have increased pAURA levels on BBs, decreased intensities of distal appendage proteins FBF1 and SCLT1, and changed distribution of IFT57 and IFT88. This in turn leads to lower proportion of ciliated cells, reduced ciliary length, and failure to respond to HH pathway activation (reduced SMO ciliary localization and diminished expression of target gene Gli1). Treatment of KIF14-depleted cells by AURA inhibitor rescues the localization defects of FBF1, SCLT1, IFT57, and IFT88; defective cilia formation; and reduced ciliary length but fails to restore competence of KIF14-depleted primary cilia to respond to HH pathway activation.
Our testing of five putative regulators of primary cilium formation has revealed that depletion of KIF14 leads to a penetrant decrease in the proportion of cells with primary cilia and reduced ciliary length, in contrast to depletion of SCYL2, LUZP1, CCDC92, or KIF7, which shows no apparent effect on ciliogenesis. In a similar vein, and in line with reports on connections between KIF7 and ciliopathies (Ali et al., 2012; Dafinger et al., 2011), the absence of KIF7 does not seem to lead to major ciliogenesis defects related to IFT or trafficking of HH pathway proteins into primary cilia; instead, KIF7 has been demonstrated to reside in ciliary tips, where it is responsible for fine-tuning the response to HH pathway activation (Endoh-Yamagami et al., 2009; He et al., 2014). It should be noted that the absence of KIF7 led to increased primary cilium length in mouse embryonic fibroblasts (He et al., 2014), which we did not observe in hTERT RPE-1 cells following KIF7 depletion. SCYL2, LUZP1, and CCDC92 have previously been found to be associated with centrioles/BBs (Chaki et al., 2012; Gupta et al., 2015), but as the absence of obvious primary cilium formation phenotypes implies, if they somehow function in primary cilium formation, their role is likely modulatory and/or redundant.
Previous studies established KIF14 as a regulator of cytokinesis (Carleton et al., 2006; Gruneberg et al., 2006,Makrythanasis et al., 2018; Moawia et al., 2017; Ohkura et al., 1997; Reilly et al., 2019) and, given the reported developmental kidney and brain defects linked to KIF14 mutations, also suggested its possible participation in the pathophysiology of ciliopathies (Filges et al., 2014; Heidet et al., 2017; Reilly et al., 2019). Reilly and colleagues showed that a lower incidence of primary cilia following KIF14 ablation in zebrafish is a consequence of the accumulation of cells blocked in mitosis, which in turn prevents primary cilium formation, as mitotic cells are not able to make a primary cilium. However, our data suggest that the relationship between KIF14 and primary cilium formation is more complex. First, we (this study) and others have detected KIF14 associated with BB components (Gupta et al., 2015; Jakobsen et al., 2011) or localized to primary cilium axoneme. Second, in agreement with the reports on the mitotic role of KIF14, we have also detected alterations in the cell cycle progression of KIF14-depleted cells. However, as those changes are restricted to serum-cultured cells and are somewhat modest, we argue that they do not explain the prominent defects in ciliogenesis and HH pathway activation we detected following serum starvation of those cells. Third, the presence of binucleated cells, a hallmark of dysfunctional cytokinesis, showed very low to no phenotypic penetrance in our models, in line with a report of a very modest incidence of binucleation in nontransformed cell lines over cancerous cell lines (McKenzie and D’Avino, 2016). With all that said, our data clearly indicate the requirement of KIF14 for effective ciliogenesis in several cell types we tested, encompassing human cells of embryonic origin (hPSCs), hTERT RPE-1, nHDF-1, and the murine cell line IMCD3.
There might be several reasons as to why the primary cilium formation phenotypes related to KIF14 deficiency were not detected in previous studies. Even though we consistently observed the KIF14 depletion–linked primary cilium defects in several cell lines, we cannot formally exclude it plays a cell-type–specific role in vivo. In fact, this possibility has support in earlier reports describing cell-type–restricted requirement of KIF14 or CIT for efficient cytokinesis in Drosophila (Ohkura et al., 1997), mouse embryos (Di Cunto et al., 2000), and humans (Filges et al., 2014; Reilly et al., 2019). In addition, the role of KIF14 in primary cilium formation may be redundant with a yet-unknown factor. In turn, its deficit may not be sufficiently penetrant in a model of constitutive knockout (Reilly et al., 2019) due to genetic compensation (El-Brolosy and Stainier, 2017). In fact, striking phenotypic differences between the acute depletion and the long-term loss have been reported for several regulators of primary cilium formation (Hall et al., 2013; Yadav et al., 2016).
Both cytokinesis and ciliogenesis rely on strict coordination of microtubule organization and directed membrane trafficking; both have been intensively studied, yet evidence of a “shared” regulatory toolbox is surprisingly sparse. The first indication that such crosstalk might indeed be plausible came from a study showing the localization of a set of central spindle/midbody proteins (PRC1, MKLP-1, INCENP, and centriolin) to BBs in vertebrate ciliated epithelial cells and in turn reporting primary cilium phenotypes for the aforementioned mutants in Caenorhabditis elegans (Smith et al., 2011). Conversely, it was also shown that IFT88 depletion leads to mitotic defects in HeLa cells, mouse kidney cells, and zebrafish embryos (Delaval et al., 2011). Furthermore, Bernabé-Rubio and colleagues (Bernabé-Rubio et al., 2016, 2019,ib13Preprint) showed that the midbody is required for primary cilia formation. In addition, CIT mutations in rats were reported to cause alterations in primary cilium length (Anastas et al., 2011), and CIT protein was found to be associated with primary cilia in mice (Mick et al., 2015). Intriguingly, both CIT and KIF14 siRNA were observed to have an effect on primary cilium formation in recent large-scale screens (Kuhns et al., 2013; Wheway et al., 2015), and the proteins were found to be associated with centrosomes (Jakobsen et al., 2011). Our experiments have demonstrated that KIF14 depletion impairs primary cilium formation even if the cells have been starved and chemically arrested before siRNA transfection. This indicates that primary cilium phenotypes linked to the depletion of KIF14 are not a mere consequence of impaired mitosis but rather that KIF14 has a novel role in interphase cells. Given that, it should be rewarding to explore in detail the extent of cooperation between regulators of cytokinesis and ciliogenesis, respectively.
Additional evidence of a specific, interphase-related role for KIF14 relates to its ability to localize to primary cilia. In this regard, our data suggest that both the N-terminal motor domain and the C-terminal part of KIF14 are required for its efficient transport in primary cilium axoneme. The fact that the motor domain mutants R364C and T456M were to some extent able to get inside the primary cilia, albeit with reduced efficiency, suggests a possible participation of a different motor protein. One point to bear in mind is that these experiments were performed in the KIF14 WT background, so it is plausible that the presence of WT KIF14 is somewhat responsible for this observation. Alternatively, although the mutations R364C and T456M lead to several impairments of KIF14 motor function (Reilly et al., 2019), we cannot exclude the possibility that some residual motor activity is still retained.
C-terminal KIF14 mutations (Q1380*, Q1304*, and R1189*) used in our study give rise to truncated protein moieties, with functional motor domain. Nonetheless, their ability to participate in either intermolecular or intramolecular interactions is expected to be hampered (Reilly et al., 2019; Verhey et al., 2011). In agreement with this prediction, all these mutants showed prominent accumulation in the tip of primary cilium (at microtubule ends), which we suspect relates to their constitutively active character in terms of microtubule binding (see also Reilly et al., 2019) and in turn the inability to reload on retrograde transport complexes to be transported back to ciliary base.
Activation of AURA is typically linked to resorption of primary cilia (Inaba et al., 2016; Inoko et al., 2012; Plotnikova et al., 2012; Pugacheva et al., 2007). Primary cilium phenotypes related to KIF14 depletion in our models include hampered speed of primary cilium elongation, hindered responsiveness of primary cilium to HH pathway stimulation, and defective recruitment and/or distribution of a subset of components of BB appendages (FBF1, SCLT1, and ODF2) or IFTs (IFT88 and IFT57). Our data imply that aberrant AURA activity is responsible for the defects in BB biogenesis and dynamics of primary cilium formation. Indeed, altered distribution of IFT88 along the axoneme, reminiscent of what we see following KIF14 depletion, was observed upon inhibition of AURA (Pugacheva et al., 2007). In addition, we speculate that FBF1, which is involved in the assembly of IFT complexes (Wei et al., 2013) and that we found regulated by KIF14 and AURA, might also be implicated in the observed alterations in the distribution of IFT components. Furthermore, SCLT1 and FBF1 are incorporated into distal appendages during centriole maturation in the G2 phase of the cell cycle (Tanos et al., 2013), and FBF1 undergoes temporal removal from distal appendages during mitosis (Bowler et al., 2019), when AURA activity peaks. Importantly, our data represent the first evidence implicating AURA in the control of distal appendage components in human cells, in line with the recently reported role of AURA in the regulation of mitotic removal of CEP164 from distal appendages in the murine IMCD3 cell line (Bowler et al., 2019). Of note, previous work by Tanos and colleagues (Tanos et al., 2013) showed that CEP164 requires SCLT1 for its localization to distal appendage. As our data demonstrate that elevated AURA activity affects SCLT1, but not CEP164, we speculate the residual SCTL1 present in distal appendage suffices for the correct localization of CEP164.
Our data suggest that the role of KIF14 in primary cilium formation is related to the activation of AURA in interphase by a currently unknown mechanism. AURA activity is typically regulated by interactions with adaptor proteins, with TPX2, AJUBA, and BORA playing the most prominent roles in the activation of AURA during mitosis (Carmena et al., 2009). Recent studies hinted that nonmitotic activation of AURA in the context of primary cilium resorption is at least in part mediated by the adaptor protein HEF1/NEDD9 (Korobeynikov et al., 2017; Pugacheva et al., 2007). Additional proteins implicated in AURA activation in interphase include PIFO (Kinzel et al., 2010), Trichoplein (Inoko et al., 2012), and CPAP (Gabriel et al., 2016). We have attempted to pinpoint a key factor responsible for AURA activation in KIF14-depleted cells. Nonetheless, our data argue against a model with one “master” activator mediating AURA activation in our models and rather suggest the participation of several regulators, which we speculate might act in a synergistic fashion. A detailed examination of the mechanism of how KIF14 regulates AURA should not only bring the necessary mechanistic insight but could also provide a means for possible future therapy to mitigate defects related to KIF14 mutations. Importantly, not all KIF14 depletion phenotypes seem to be linked to elevated AURA activity. Our data clearly show that inhibition of AURA in KIF14-depleted cells is able to almost completely rescue the defective localization of SCLT1, FBF1, IFT88, and IFT57, and, in turn, the proportion of cells able to assemble a fully grown primary cilium. However, although the primary cilia form in numbers and lengths similar to controls, these primary cilia fail to respond accordingly to the stimulation of the HH pathway with SAG. Thus, these data imply that KIF14 also plays a specific role in the activation of the HH signaling pathway, which is distinct from the effects on primary cilium formation mediated by the elevated AURA activity.
In sum, our study demonstrates a novel function for KIF14 in the regulation of primary cilium formation dynamics and responsiveness to HH signaling in human cells. It identifies AURA activity as a downstream mediator of at least some of the KIF14 depletion–related primary cilium defects and points toward KIF14 as an important element connecting cell cycle machinery, effective ciliogenesis, and HH signaling.
Materials and methods
Cell culture, transfections, and treatments
All cell lines were cultured at 5% CO2 at 37°C. hTERT RPE-1 (Čajánek and Nigg, 2014) and Flp-In T-REx RPE-1 (both kind gifts from E. Nigg, Biozentrum, University of Basel, Basel, Switzerland) and mouse innermedullary collecting duct cells (IMCD-3; kind gift from G. Jansen, Erasmus University Medical Center, Rotterdam, Netherlands) were cultured in DMEM/F12 supplemented with 10% FBS, 1% L-Glutamine (all from ThermoFisher), and 1× penicillin/streptomycin (Biosera). HEK293T and Flp-In T-REx 293 cells (#R78007; Invitrogen) were cultured in GlutaMax DMEM supplemented with 10% FBS (both from ThermoFisher) and 1× penicillin/streptomycin (Biosera). Inducible cell lines were cultivated in standard medium, with tetracycline-free FBS, for induction and media supplemented with 1 µg/ml DOX. nHDFs (#C0045C; Gibco) were cultivated in knockout DMEM supplemented with 10% heat-inactivated FBS, 1% L-Glutamine (all from ThermoFisher), 1× nonessential amino acids, 1× penicillin/streptomycin (both from Biosera), and 1% β-mercaptoethanol (Sigma-Aldrich). hESCs (CCTL14; https://hpscreg.eu/cell-line/MUNIe007-A) were cultured in feeder-free conditions on Matrigel-coated cell culture plastic in daily exchanged mouse embryonic fibroblast–conditioned hESC medium (DMEM/F12, 15% knockout serum replacement; both from ThermoFisher), 2 mM L-glutamine, 1× nonessential amino acids, 0.5% Pen/Strep (all from Biosera) + 0.5× ZellShield (Minerva Biolabs), 100 µmol/Lβ-mercaptoethanol (Sigma-Aldrich), and 10 ng/ml hFGF2 (ThermoFisher). hiPSCs (Neo1, derived as described previously; Barta et al., 2016) were cultured in feeder-free conditions on Matrigel-coated plastic in daily exchanged mTeSR medium (StemCell Technologies).
Cells were transfected 24 h after seeding at 60% confluency (both human stem cell lines were seeded in high density to reach full confluency at the end of the experiment). DNA transfections of RPE-1 cells were done with Lipofectamine 3000 (ThermoFisher) or FuGENE HD Transfection (Promega) 24 h before analysis, and siRNAs were transfected with Lipofectamine RNAiMAX (ThermoFisher) 48 h before analysis. HEK293 cells were transfected using polyethylenimine (Sigma-Aldrich) 24 h before analysis. For knockdowns of KIF14, CIT, KIF7, SCYL2, LUZP1, CCDC92, AJUBA, BORA, NEDD9, PIFO, TCHP, or TPX2, the cells were transfected using different siRNA oligos (25 nM #1, 25 nM #2, or 25 nM #3 separately, where indicated) or 50 nM mix (50 nM mix of 25 nM #1 and 25 nM #2, where indicated). For a list of all siRNA sequences used, see Table S3.
For cilia analysis, cells were starved in serum-free medium for 24 h before analysis. To induce HH pathway activity, nHDF cells were starved in serum-free media and treated with 0.5–2 µM SAG (SMO agonist; Sigma-Aldrich) for 24 h before analysis (Fig. 8, A–C; and Fig. S5 D). Where indicated, cells were treated for indicated time period with either vehicle (DMSO) or the following compounds at the indicated dose: 100 nM TCS7010 (AURA inhibitor; Tocris Bioscience; Fig. 7, A–J;,Fig. S5 A; and Fig. 8, D–F), 10–20 µM (R)-roscovitine (Bukanov et al., 2006; Fig. 5, E, F, and K–N), or 10 µM monastrol (Fig. 5, E and F).
DNA constructs and stable cell lines
To generate destination vector with N-terminal MYC-BirA* tag, MYC-BirA* was amplified by PCR from pcDNA3.1 MYC-BirA* (43920; Addgene) using forward primer NO°1 and reverse primer NO°2 (Table S4) to create AflII and BstBI site–containing overhangs and cloned into TOPO entry vector (using TOPO TA Cloning Kit, K450001; ThermoFisher) sites. Next, following digest with AflII and BstBI (enzymes from NEB) the MYC-BirA* fragment was ligated (K1422; ThermoFisher) into AflII and BstBI sites of pgLAP2 vector (19703; Addgene). MYC-BirA*-CEP164 construct was generated from pDONOR-CEP164 (Čajánek and Nigg, 2014) and MYC-BirA* destination vector described above by LR Gateway cloning reaction (ThermoFisher), and subsequently validated for correct expression and localization by Western blot and immunofluorescence microscopy analyses following transfection of HEK293T cells.
MYC-AURA was kind gift from E. Nigg (Meraldi et al., 2002). pcDNA3.1 (Invitrogen) empty vector was used as transfection control for AURA overexpression experiments.
KIF14 construct resistant to KIF14 siRNA #2 sequence: 5′-TTTAATAGTCAACATTGCT-3′ was generated by site-directed mutagenesis of pENTR KIF14 vector (generated by Gateway cloning from KIF14 plasmid kindly provided by F. Barr, University of Oxford, Oxford, UK; Gruneberg et al., 2006) in seven nucleotides (t2536a, a2539g, a2542c, c2545a, c2548t, t2551c, and t2554c) using PCR with mutagenesis forward NO°3 and reverse NO°4 primers (Table S4). Expression vector for KIF14siRNA2res was generated using LR Gateway cloning (ThermoFisher) of pENTR KIF14siRNA2res into pgLAP1_Neo destination vector. KIF14siRNA2res-pgLAP1_Neo (GFP-KIF14siRNA2res) was subsequently used for cotransfection with pOG44 Flp-recombinase expression vector (V600520; ThermoFisher) at a 1:30 ratio (total amount of DNA 2 μg/10-cm-diameter dish) by Lipofectamine 3000 (ThermoFisher) to create stable inducible RPE-1 Flp-in T-REx (kind gift from E. Nigg; Čajánek and Nigg, 2014). Cells were selected with 700 µg/ml G418 and 5 µg/ml of blasticidin S (both from Sigma-Aldrich).
To generate pgLAP1_Neo vector, Neomycin ORF was amplified by PCR from pcDNA3.1 (ThermoFisher) and cloned into TOPO entry vector (using TOPO TA Cloning Kit K450001; ThermoFisher) with SacII and NheI site–containing overhangs using primers NO°5 and NO°6 (Table S4). Next, following digest with SacII and NheI (NEB) the Neomycin ORF fragment was cloned using Rapid DNA Ligation Kit (K1422; ThermoFisher) into SacII and NheI sites of modified pgLAP1 vector; the NheI site was generated by site-directed mutagenesis of pgLAP1 (19702; Addgene) using primers NO°7 and NO°8 (Table S4).
Plasmids encoding WT, 1–356, 1–718, and 800–1648 human KIF14 GFP-fusions were kind gifts from F. Barr and U. Gruneberg (University of Oxford, Oxford, UK; Gruneberg et al., 2006). Mutant forms (R364C, T456M, Q1380*, Q1304*, and R1189*) corresponding to mutations identified in fetuses with syndromic microcephaly were described previously (Reilly et al., 2019). The lentiviral vector, pReceiver-Lv225, encoding for untagged human KIF14, EX-T0642-Lv225, was obtained from GeneCopoeia.
The ARL13B-GFP Flp-in RPE-1 cell line was kind gift from V. Varga (Institute of Molecular Genetics of the Czech Academy of Sciences, Prague, Czech Republic).
Stable inducible Flp-In T-REx 293 cell lines were selected by 500 µg/ml hygromycin and 5 µg/ml blasticidin S (both from Sigma-Aldrich) selection of cells cotransfected with BioID construct (MYC-BirA*-CEP164 or MYC-BirA* empty vector) and pOG44 Flp-recombinase expression vector (V600520; ThermoFisher) at a 1:9 ratio (total amount of DNA 12 µg/10-cm-diameter dish) using polyethylenimine.
All stable cell lines were validated by Western blot analysis and immunofluorescence staining.
Rescue experiments
A hTERT RPE-1 cell line inducibly (1 µg/ml DOX for 5 d before analysis) expressing GFP-KIF14 resistant to siRNA 2 (GFP-KIF14si2res) was transfected with 50 nM KIF14 siRNA #2 or control siRNA 48 h prior to analysis (Fig. 1, H and I). For rescue experiments with AURA inhibition, hTERT RPE-1 cells were seeded to 50% confluency, transfected 72 h prior to analysis with control or KIF14 siRNA, starved with serum-free medium 48 h prior analysis, and treated with 100 nM TCS7010 AURA inhibitor for 24 h (Fig. 7, A–J; Fig. S5 A; and Fig. 8, D–F).
Cells were then fixed with ice-cold MeOH and analyzed by immunofluorescence staining.
Antibodies
For complete list of antibodies used, see Table S5. MYC antibody was described previously (Evan et al., 1985), as was CAP350 antibody (Yan et al., 2006).
Immunofluorescence staining
Cells were washed with PBS and fixed by either ice-cold MeOH or 3.7% PFA (Fig. 1, C and D; and Fig. S3, C, D, N, and O) followed by permeabilization by PBS + 0.1% TRITON X-100, blocked in blocking buffer (1% BSA in PBS), and subjected to incubation with primary and secondary antibodies, respectively. Cell nuclei were stained with 1 µg/ml Hoechst (Life Technologies) or DAPI (Sigma-Aldrich), and coverslips were mounted using Glycergel (Agilent) or Mowiol (Sigma-Aldrich).
Microscopes, image acquisition, measurements, quantifications, and time-lapse microscopy analyses
Microscopy analyses of fixed cells were done in 25°C on microscopes DeltaVision Elite (GE Healthcare) with a 100×/Zeiss Plan-ApoChromat 1.4 objective and DeltaVision softWoRx acquisition software (here, the image stacks were taken with a Z distance of 0.2 µm, deconvolved with three to five cycles of conservative ratio deconvolution algorithm, and directly projected as maximal intensity images), Zeiss Axio Observer.Z1 with confocal unit LSM 800 with 63×/Zeiss Plan-ApoChromat 1.4 objective, and Zeiss AxioImager.Z2 with Hamamatsu ORCA Flash 4.0 camera and 63×/Plan-ApoChromat 1.4 objective (both with ZEN Blue acquisition software). For acquisition, a Zeiss Axio Observer.Z1 and AxioCam 503 charge-coupled device camera in widefield mode was used (ZEN Blue software). For higher resolution microscopy (Fig. 2 C; Fig. 6, C and D; and Fig. S4 B), a laser scanning confocal microscope was used (Zeiss LSM 880 with Airyscan Fast module and 100×/M27 oil differential interference contrast Plan-ApoChromat 1.46 objective and ZEN Black acquisition software). Time-lapse microscopy analyses were done in 37°C/5% CO2/FluorBright DMEM media (ThermoFisher) on Zeiss Axio Observer.Z1 with 10×/0.3 EC Plan-Neofluar, 25×/LCI Plan-Neofluar 0.8, and 40×/Plan-ApoChromat 1.3 objectives using ZEN Blue software for acquisition.
All images within given experiment were acquired with the same laser intensities and exposition times and projected to maximum Z intensity (ranging from 0.1- to 0.25-µm interval and total Z ranging from 2 to 50 slices) before analysis in ImageJ (National Institutes of Health). Where appropriate, contrast and/or brightness adjustment and cropping of final images were done using ZEN Black software (Zeiss) or ImageJ, with identical settings applied to all images from the given experiment.
For Fig. 2, A, B, and E–G, stained cells were imaged using an epi-illumination microscope (DMR; Leica) with a cooled charge-coupled device camera (DFC3000G; Leica). Images were acquired with LAS (V4.6; Leica) and processed with ImageJ and Photoshop CS2 (Adobe Systems).
Cilia lengths were measured using straight- or segmented-line tool for selection of fluorescent signal of ciliary marker in maximum Z intensity projected images in ImageJ. Cilia were categorized as follows: no cilia (no ARL13B/Ac-tub signal); hTERT RPE-1: short cilia, ≤3.3 µm; long cilia, ≥3.3 µm; nHDF, hESC CCTL14, and hiPSC: short cilia, ≤2.2 µm; long cilia, ≥ 2.2 µm; IMCD-3: short cilia, ≤4 µm; long cilia, ≥4 µm.
SDS-PAGE and Western blotting
Cells were washed in PBS and lysed in SDS lysis buffer (50 mM Tris-HCl, pH 6.8, 10% glycerol, and 1% SDS). Protein concentration was measured using the DC Protein Assay Kit (Bio-Rad). Samples were adjusted to equal concentration, mixed with bromphenol blue (0.01%) and β-mercaptoethanol (2.5%), boiled, and sonicated. Proteins were separated by SDS-PAGE, transferred onto polyvinylidene fluoride membranes (Millipore), and blocked with 5% milk in TBS-Tween buffer (10 mM Tris-HCl, pH 7.4, 100 mM NaCl, and 0.05% Tween). Membranes were incubated with primary and secondary antibodies, respectively (Table S5). Chemiluminescent signal was revealed by ECL Prime (GE Healthcare) and film (Agfa Healthcare) or Image Reader LAS-4000 (GE Healthcare). Subsequent crops and quantification analysis were done using ImageJ software.
BioID
For affinity capturing of biotinylated proteins, we adopted protocol published by Roux et al. (2012). Briefly, cells were incubated for 24 h in complete media supplemented with 1 µg/ml DOX and 50 µM biotin. After three PBS washes (all following steps on ice), cells (4 × 107) were washed with wash buffer (50 mM Tris-HCl, pH 7.4), lysed in 1 ml lysis buffer (50 mM Tris, pH 7.4, 250 mM NaCl, 0.4% SDS, 1 mM DTT, 2% Triton X-100, and 1× Complete protease inhibitor [4693132001; Roche]) and sonicated. Then, an equal volume of ice-cold wash buffer was added, and samples were centrifuged at 16,000 relative centrifugal force. Supernatants were incubated with 40 µl Streptactin Sepharose High Performance beads (#28935599; GE Healthcare) overnight at 4°C. Beads were collected and washed three times with 25 mM Tris-HCl (pH 7.4), three times with wash buffer, and twice with 50 mM NH4HCO3. Samples were analyzed by mass spectrometry.
Immunoprecipitation
hTERT RPE-1 cells (3 × 15-cm plates per condition) were grown to confluency, washed in PBS, scraped, pelleted by centrifugation (400 g/5 min/4°C), resuspended in immunoprecipitation (IP) lysis buffer (20 mM Tris-Cl, pH 7.4, 150 mM NaCl, 0.5% NP-40, 0.5% Triton-X-100, 0.1 mM DTT [43815; Merck], and 1× Complete proteasome inhibitors [4693132001; Roche]) and lysed (15 min on ice). Following centrifugation (15,000 g for 10 min at 4°C), cleared extracts were incubated 2–6 h at 4°C in an orbital shaker with either anti-CEP164 antibody (Graser et al., 2007) or control IgG antibody (sc-66931; Santa Cruz), coupled to protein G Sepharose beads (17–0618-01; GE Healthcare). Bound protein complexes were washed four times with IP lysis buffer and four times with wash buffer (20 mM Tris-Cl, pH 7.4, and 150 mM NaCl).
Protein digest and liquid chromatography with tandem mass spectrometry (LC-MS/MS) analysis of peptides
On-beads digestion
Following IP washes, bead-bound protein complexes were digested directly on beads by addition of 0.75 µg (1 µg/µl) trypsin (sequencing grade; Promega) in 50 mM NaHCO3 buffer. Beads were gently tapped to ensure even suspension of trypsin solution and incubated at 37°C with mild agitation for 2 h. Beads were then vortexed to ensure the complex falls of the beads and the partially digested complex was transferred to the clean tubes to separate it from the beads and incubated at 37°C for 16 h without agitation. Resulting peptides were extracted into LC-MS vials by 2.5% formic acid (FA) in 50% acetonitrile (ACN) and 100% ACN with addition of polyethylene glycol (20,000; final concentration 0.001%; Stejskal et al., 2013) and concentrated in a SpeedVac concentrator (ThermoFisher).
LC-MS/MS analysis of peptides
LC-MS/MS analyses of peptide mixture were done using the RSLCnano system connected to an Orbitrap Elite hybrid spectrometer (ThermoFisher). Prior to LC separation, tryptic digests were online concentrated and desalted using trapping column (100 µm × 30 mm; flow rate of 4 μl/min) filled with 3.5-µm X-Bridge BEH 130 C18 sorbent (Waters). After washing of trapping column with 0.1% FA, the peptides were eluted (flow 300 nl/min) from the trapping column onto an analytical column (Acclaim Pepmap100 C18, 3 µm particles, 75 µm × 500 mm; ThermoFisher) by a 100-min nonlinear gradient program (1–56% of mobile phase B; mobile phase A: 0.1% FA in water; mobile phase B: 0.1% FA in 80% ACN). Equilibration of the trapping column and the column was done before sample injection to sample loop. The analytical column outlet was directly connected to the Digital PicoView 550 (New Objective) ion source with PicoTip emitter SilicaTip (FS360-20-15-N-20-C12; New Objective). ABIRD (active background ion reduction device) was installed.
MS data were acquired in a data-dependent strategy selecting up to top 10 precursors based on precursor abundance in the survey scan (350–2,000 m/z). The resolution of the survey scan was 60,000 (400 m/z) with a target value of 1 × 106 ions, one microscan and maximum injection time of 200 ms. HCD MS/MS spectra were acquired with a target value of 50,000 and resolution of 15,000 (400 m/z). The maximum injection time for MS/MS was 500 ms. Dynamic exclusion was enabled for 45 s after one MS/MS spectra acquisition and early expiration was disabled. The isolation window for MS/MS fragmentation was set to 2 m/z.
The analysis of the mass spectrometric RAW data files was performed using the Proteome Discoverer software (version 1.4; ThermoFisher) with in-house Mascot (version 2.6; Matrixscience) and Sequest search engines utilization. MS/MS ion searches were done at first against modified cRAP database (based on http://www.thegpm.org/crap/; 111 sequences in total) containing protein contaminants like keratin and trypsin. MS/MS spectra assigned by the Mascot search engine to any cRAP protein peptide with a Mascot ion score >30 were excluded from the next database searches. Remaining filtered MS/MS ions were further searched against UniProtKB proteome database for Homo sapiens (taxonomy ID 9606), downloaded on January 17, 2017 (number of proteins, 21,031). Mass tolerance for peptides and MS/MS fragments were 10 ppm and 0.05 D, respectively. Oxidation of methionine, deamidation (N, Q), and acetylation (protein N terminus) as optional modification and two-enzyme missed cleavages were set for all searches. Percolator was used for post-processing of the search results. Only peptides with q-value <0.01, rank 1, and at least six amino acids were considered. Proteins matching the same set of peptides were reported as protein groups. Protein groups/proteins were reported only if they had at least one unique peptide. Label-free quantification using protein area calculation in Proteome Discoverer was used (top three protein quantification; Silva et al., 2006).
Obtained lists of candidate binding partners were manually curated to remove false-positive hits and nonspecific binders. For coIP experiments, hits commonly detected in both mock (IP/IgG) and IP/CEP164 were deleted, and hits with a CRAPome ratio (number of entries/total entries × 100%; https://www.crapome.org/) >10% were also deleted. Only candidates detected at least in two biological replicates were considered (Table S1). For BioID experiments, only candidates with an identified peptide ratio in CEP164/control ≥2 in two consecutive experiments and with ≥10 identified peptides in CEP164 sample in at least one experiment at the same time were considered (Table S2).
Quantitative RT-PCR
1 µg RNA, isolated using the RNeasy Mini Kit (Qiagen), was used for cDNA synthesis using the Transcriptor First Strand cDNA Synthesis Kit (Roche). Reactions were done in triplicates with LightCycler 480 SYBR Green I Master according to the manufacturer’s protocol and monitored in real time using LightCycler 480 Instrument II (Roche). Relative gene expression was calculated using the 2−ΔΔCT method; GAPDH was used as reference gene (for primer sequences, see Table S4).
Cell cycle distribution and flow cytometry
Cells were siRNA transfected for 48 h (0.5 µM Cytochalasin D; 24-h treatment was used as positive control to enrich the proportion of G2/M cells) and serum starved for 24 h where indicated. Cells were fixed (30 min, ice-cold 70% ethanol), washed with PBS + 0.5% BSA, and treated with RNase A (0.016 mg/ml, 30 min, 37°C). DNA was stained by propidium iodide (20 µg/ml; both from Sigma-Aldrich). The cell cycle profile was determined using FACS CANTO II (BD Biosciences) and FlowJo software (www.flowjo.com); at least 40,000 events per sample were analyzed. For graph creation, doublets were excluded. Quantification in histograms indicates increased ploidy (cells with >G2/M DNA content).
Mitotic shake-off
To increase the proportion of mitotic cells, 24 h after seeding, cells were transfected by KIF14 or control siRNA; 2 h after siRNA transfection, cells were treated for additional 2–4 h with 10 µM monastrol (Sigma-Aldrich). Following treatment, mitotic cells were shaken off in PBS. Leftover nonmitotic cells were also passaged by trypsin. All conditions were counted and seeded in the same density. Cells were further maintained in 1% serum medium and, where indicated, supplemented with 10–20 µM (R)-roscovitine (an inhibitor of cyclin-dependent kinases; Bukanov et al., 2006) for 2 d to induce ciliogenesis and block the cell cycle progression. Cells were then fixed and analyzed by immunofluorescence (see illustrative scheme in Fig. 5 E).
Statistics
All statistical analyses were done and graphically visualized in GraphPad Prism Software v. 6.0 (GraphPad Software; www.graphpad.com). All data are presented as mean ± SEM from at least three independent experiments (n ≥ 3), unless otherwise stated in the figure legends. In all experiments, at least 100 cells/cilia were counted or measured per condition (N ≥ 100), unless otherwise stated in the figure legends. P values < 0.05 were considered significant (*, P < 0.05; **, P < 0.005; ***, P < 0.001; ****, P < 0.0001), and asterisks indicate statistical significance when compared with controls, unless otherwise stated. To determine statistical significance between two categories (simple data groups), an unpaired t test was used. To determine statistical significance between more than two groups, Tukey's multiple comparisons test was used (comparing the mean of each column with the mean of every other column). For group data with more than one category in columns, statistical significance was determined using the Holm–Sidak method (α = 5%), and each category was analyzed individually, without assuming a consistent SD.
Online supplemental material
Fig. S1 shows that KIF14 knockdown causes ciliogenesis defects in different cell types. Fig. S2 shows that KIF14 knockdown affects localization of BB components and IFT-B machinery. Fig. S3 shows that KIF14 and CIT knockdown leads to minor effects on cell cycle in RPE-1. Fig. S4 shows that KIF14 depletion deregulates AURA. Fig. S5 shows that AURA activity mediates effects of KIF14 depletion on primary cilia formation. Table S1 lists candidate binding partners for CEP164 coIP. Table S2 lists the candidate binding partners from CEP164 BioID experiments, showing proximity-dependent biotinylation by CEP164-BirA*. Table S3 lists all siRNA sequences used in this study. Table S4 lists the primers used in this study. Table S5 lists the antibodies used in this study.
Acknowledgments
We thank Erich Nigg (Biozentrum, University of Basel, Basel, Switzerland), Vladimir Varga (Institute of Molecular Genetics of the Czech Academy of Sciences, Prague, Czech Republic), Stjepan Uldrijan (Masaryk University, Brno, Czech Republic), Francis Barr (University of Oxford, Oxford, UK), Ulrike Gruneberg (University of Oxford, Oxford, UK), Lumir Krejci (Masaryk University, Brno, Czech Republic), Pavel Krejci (Masaryk University, Brno, Czech Republic), Gert Jansen (Erasmus University Medical Center, Rotterdam, Netherlands), Peter Jackson (Stanford University School of Medicine, Stanford, CA), Kyle Roux (Stanford University School of Medicine, Stanford, CA), and Vladimir Rotrekl (Masaryk University, Brno, Czech Republic) for sharing reagents or instruments.
This work was supported by the Czech Science Foundation (grants 16-03269Y and 19-05244S to L. Cajanek), the Swiss National Science Foundation (grant IZ11Z0_166533 to L. Cajanek), and the Fondation pour la Recherche Médicale (grant DEQ20130326532 to A. Benmerah). O. Bernatik was supported by Faculty of Medicine MU junior researcher funds. Z. Zdrahal was supported by the Ministry of Education, Youth and Sports of the Czech Republic (MEYS CR; project CEITEC 2020, LQ1601). M.L. Reilly was supported by a Paris Diderot University fellowship. We acknowledge the core facility CELLIM of CEITEC supported by the Czech-BioImaging large RI project (MEYS CR grant LM2018129) for their support with obtaining scientific data presented in this paper. CIISB research infrastructure project (MEYS CR grant LM2018127) is gratefully acknowledged for the financial support of the LC-MS/MS measurements at the Proteomics Core Facility.
The authors declare no competing financial interests.
Author contributions: P. Pejskova performed and analyzed most of the experiments and assembled figures. M.L. Reilly performed and analyzed experiments in Fig. 2, A, B, and D–G. L. Bino performed and analyzed experiments in Fig. S1, N–V and assembled Fig. 9. O. Bernatik constructed cell line for experiment in Fig. 1, H and I, and Fig. S1 J. L. Dolanska performed and analyzed experiments in Fig. 8, A and C and Fig. S5 D. R.S. Ganji and Z. Zdrahal performed the proteomics analysis. A. Benmerah conceptualized and supervised work related to Fig. 2, A, B, and D–G. L. Cajanek conceptualized and supervised the study and, together with P. Pejskova, Z. Zdrahal, and A. Benmerah, wrote the paper. All authors approved the final version of the manuscript.