Faithful chromosome segregation depends on the precise timing of chromatid separation, which is enforced by checkpoint signals generated at kinetochores. Here, we provide evidence that the C-terminal domain (CTD) of DNA topoisomerase IIα (Topo II) provides a novel function at inner centromeres of kinetochores in mitosis. We find that the yeast CTD is required for recruitment of the tension checkpoint kinase Ipl1/Aurora B to inner centromeres in metaphase but is not required in interphase. Conserved CTD SUMOylation sites are required for Ipl1 recruitment. This inner-centromere CTD function is distinct from the catalytic activity of Topo II. Genetic and biochemical evidence suggests that Topo II recruits Ipl1 via the Haspin–histone H3 threonine 3 phosphorylation pathway. Finally, Topo II and Sgo1 are equally important for Ipl1 recruitment to inner centromeres. This indicates H3 T3-Phos/H2A T120-Phos is a universal epigenetic signature that defines the eukaryotic inner centromere and provides the binding site for Ipl1/Aurora B.
Introduction
For chromosomes to segregate equally during mitotic cell division, they must be bioriented on the spindle. This requires that the duplicated sister chromatids remain paired until anaphase. One mechanism of chromatid pairing is provided by the entanglement of the newly replicated sister DNA molecules (Holm et al., 1985; Cook, 1991). All of these entanglements (catenations), between every pair of sister chromatids, must then be removed to allow chromosome segregation in anaphase. DNA topoisomerase IIα (Topo II) catalyzes this resolving activity and is thus essential for mitosis in all eukaryotes (Nitiss, 2009a). To achieve this, the Topo II enzyme performs a unique catalytic cycle known as the strand passage reaction (SPR), where a transient double-strand break in one double helix is made, a second helix is passed through the break, and then the first helix is religated (Wang, 2002). Extensive biochemical and structural studies of the catalytic core of Topo II have revealed the mechanism of the SPR but, intriguingly, have also revealed that the C-terminal domain (CTD) of Topo II is dispensable for the catalytic cycle (Jensen et al., 1996; Dickey and Osheroff, 2005). This is of interest because, in vivo, the CTD of Topo II is nevertheless required for faithful chromosome segregation from yeast to humans. The crucial function of the Topo II CTD remains to be elucidated (Bachant et al., 2002; Dickey and Osheroff, 2005; Lane et al., 2013).
The majority of DNA catenations are removed by Topo II during DNA replication and in the G2 phase of the cell cycle (Downes et al., 1994; Larsen et al., 1996). Congruent with this bulk activity, Topo II binds to sites throughout the genome during interphase (Fachinetti et al., 2010; Dykhuizen et al., 2013). In mitosis, however, Topo II redistributes to become most abundant at the centromere region of chromosomes, although no centromere or kinetochore function has been ascribed to the enzyme (Porter and Farr, 2004). Given that it is important for accurate chromosome segregation, one possible function of the Topo II CTD, independent of the catalytic cycle, is therefore at the centromere/kinetochore in mitosis. The unanswered questions are: What is the function of the Topo II CTD in chromosome segregation? What is the putative function at centromeres/kinetochores during mitosis?
As well as the physical linkages between sister chromatids that permit biorientation and are resolved by Topo II, successful mitosis depends on checkpoint controls that monitor biorientation. The spindle assembly checkpoint (SAC) monitors kinetochore–microtubule attachment (often termed kinetochore “occupancy”), whereas the tension checkpoint is thought to directly assess the tension on the kinetochore mediated by microtubules (Jia et al., 2013). Microtubules and kinesin 5 motors that exert force on the kinetochores are opposed by the physical pairing of the sister chromatids, and therefore, this architecture imparts a precise degree of mechanical tension on bioriented sister kinetochores (e.g., 4–6 pN tension in yeast; Chacón et al., 2014). The force is sensed by the tension checkpoint via Aurora B kinase, and this mechanism requires that Aurora B resides at the inner-centromere region of kinetochores in mitosis (Cheeseman et al., 2006; Cimini et al., 2006; Pinsky et al., 2006; Welburn et al., 2010; Jia et al., 2013). However, the mechanism of Aurora B recruitment to the inner centromere is only partly understood.
Aurora B is a component of the chromosomal passenger complex (CPC), also consisting of inner centromere protein (INCENP), Survivin, and Borealin (Klein et al., 2006). In higher eukaryotes, Aurora B–CPC redistributes in prometaphase to almost exclusively occupy binding sites at the inner centromere of chromosomes (Ainsztein et al., 1998; Adams et al., 2000; Kaitna et al., 2000; Gassmann et al., 2004; Nozawa et al., 2010). These precise binding sites are established epigenetically by a pair of inner-centromere–specific nucleosome modifications, phosphorylation of histone H3 threonine 3 (H3 T3-Phos), and histone H2A threonine 120 (H2A T120-Phos), both of which are required for Aurora B–CPC recruitment to inner centromeres (Kelly et al., 2010; Jeyaprakash et al., 2011; Carmena et al., 2012; Sawicka and Seiser, 2014). First, the BIR domain of Survivin interacts with the ART motif at the very N terminus of H3 specifically when T3 is phosphorylated (Kelly et al., 2010; Jeyaprakash et al., 2011). Second, Shugoshin binds to H2A T120-Phos and then directly binds to Borealin (Kawashima et al., 2007; Tsukahara et al., 2010; Yamagishi et al., 2010). The histone marks themselves therefore coexist at, and in part establish the epigenetic identity of, the inner centromere (Yamagishi et al., 2010), and it is the intersection of these nucleosomes that form the Aurora B–CPC binding sites where Aurora B functions in tension sensing. H2A T120-Phos is generated by Bub1 kinase, which has a well-understood mechanism of inner-centromere association (Sawicka and Seiser, 2014). In contrast, H3 T3-Phos is generated by Haspin kinase (Dai and Higgins, 2005; Wang et al., 2010), but the mechanism of Haspin recruitment to inner centromeres is not known, and therefore, our understanding of the molecular requirements for tension checkpoint activation is incomplete. There is evidence that Topo II may play some role in Aurora B activation, suggesting a possible function of Topo II at the inner centromere of kinetochores, but there is no mechanistic insight into this possible link (Coelho et al., 2008; Warsi et al., 2008).
Here, we determine a novel inner-centromere function of the Topo II CTD. In budding yeast mitosis, we find that both the CTD and the lysine residues that become SUMOylated are essential for the recruitment of Aurora B/Ipl1 to inner centromeres. This mechanism also requires Haspin kinases (Alk1 and Alk2) and histone H3 threonine 3 phosphorylation. This Topo II–dependent pathway and the Sgo1-dependent pathway are each important for Ipl1 recruitment to inner centromeres in yeast mitosis. Therefore, both H2A and H3 tails work in concert to form the inner-centromere–specific nucleosomal binding site for Ipl1. Moreover, the CTD of Topo II has a noncatalytic inner-centromere function in mitosis, which can explain the importance of the CTD for faithful chromosome segregation, distinct from the catalytic activity of Topo II.
Results
Yeast kinetochores assemble properly in top2 mutants
In mitosis, Topo II is most abundant at kinetochores, but no function within this complex has been revealed (Porter and Farr, 2004). A previous study of yeast Topo II (top2) mutants analyzed the attachment of a single chromosome to the mitotic spindle in prometaphase and found no gross defect in biorientation (Andrews et al., 2006). To extend this analysis, we asked if kinetochores assemble correctly in loss-of-function top2-4 mutants (Holm et al., 1985). We reasoned that proper localization of an outer-kinetochore protein, Nuf2-GFP, serves to indicate assembly of the underlying inner-centromere chromatin and central regions of the kinetochore. To identify mitotic cells, we simultaneously observed Spc110-mCherry, a component of spindle pole bodies. In both wild type and top2-4, grown at the nonpermissive temperature for top2-4, Nuf2-GFP localized to the clustered kinetochores in almost all metaphase cells, each cluster of sister kinetochores being in line with the spindle axis (Fig. 1 A). Thus, there is no gross defect in kinetochore structure, and thereby the inner centromere, in the absence of Topo II catalytic activity.
Structural and functional mitotic kinetochore integrity in top2 mutants. (A) Representative images and quantification of Nuf2-GFP localization to kinetochores in prometaphase/metaphase (0.5–2.0-µm spindles) wild type and top2-4 (grown at 30°C for 1 h; the nonpermissive temperature for top2-4). Spc110-mCherry indicates spindle poles. (B) Representative images and quantification of Mad2-GFP localization to nuclear pores (left) and kinetochores (right) in mitotic wild type and top2-4 (grown at 30°C for 1 h) with (right) and without (left) nocodazole. Spc110-mCherry indicates spindle poles. Bars, 1 µm. n = wild type, 0 h (902 cells) and 1 h (1,072 cells); top2-4, 0 h (902 cells) and 1 h (1,022 cells; from three experimental repeats). Error bars, standard deviation.
Structural and functional mitotic kinetochore integrity in top2 mutants. (A) Representative images and quantification of Nuf2-GFP localization to kinetochores in prometaphase/metaphase (0.5–2.0-µm spindles) wild type and top2-4 (grown at 30°C for 1 h; the nonpermissive temperature for top2-4). Spc110-mCherry indicates spindle poles. (B) Representative images and quantification of Mad2-GFP localization to nuclear pores (left) and kinetochores (right) in mitotic wild type and top2-4 (grown at 30°C for 1 h) with (right) and without (left) nocodazole. Spc110-mCherry indicates spindle poles. Bars, 1 µm. n = wild type, 0 h (902 cells) and 1 h (1,072 cells); top2-4, 0 h (902 cells) and 1 h (1,022 cells; from three experimental repeats). Error bars, standard deviation.
Second, we assessed a functional aspect of kinetochore integrity by asking if the SAC protein Mad2 can be recruited to kinetochores in top2-4 cells. In untreated top2-4 metaphase cells, we observed the expected localization of Mad2-GFP at nuclear pore complexes, decorating the periphery of the nucleus (Fig. 1 B, left; Iouk et al., 2002). In the presence of nocodazole, which disrupts microtubules and therefore activates the SAC, we observed efficient relocalization of Mad2-GFP to the clustered kinetochores (Fig. 1 B), as previously reported for wild-type cells (Iouk et al., 2002). Therefore, the underlying structures and molecular cues required for Mad2-GFP recruitment to kinetochores are intact in the absence of Topo II catalytic activity.
Top2 is required for inner-centromere localization of Ipl1 in mitosis
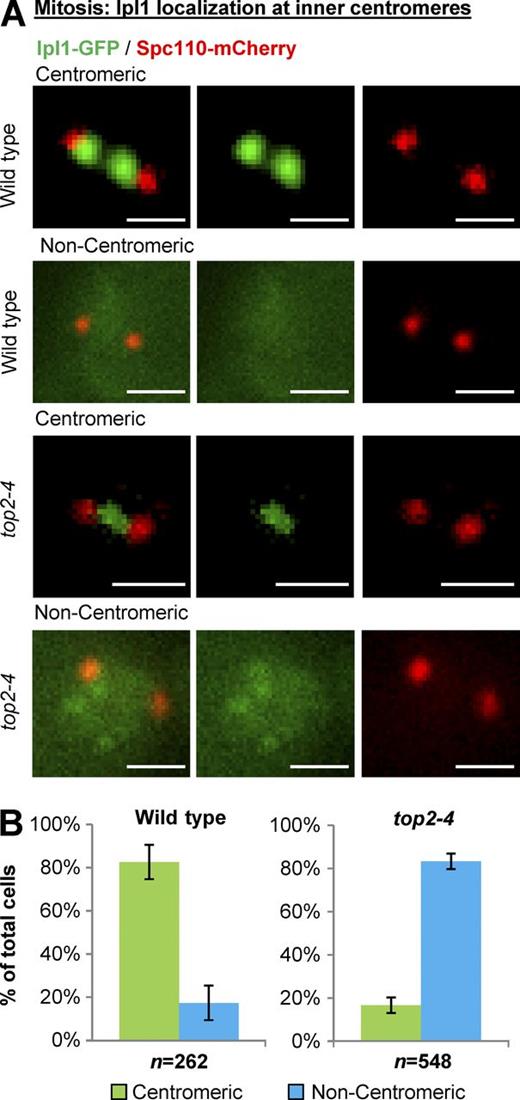
We then focused on Ipl1 (yeast Aurora B kinase), which localizes to the inner-centromere region underlying the kinetochores throughout the cell cycle in yeast and is required for the kinetochores to sense the mechanical tension that is imparted by spindle microtubules and motor proteins (Biggins and Murray, 2001; Pinsky et al., 2003, 2006). In interphase cells with a single spindle pole body, Ipl1-GFP localized to inner centromeres in almost all wild-type and top2-4 cells (Fig. 2 and Fig. S1). Strikingly, however, Ipl1-GFP was drastically delocalized from inner centromeres in metaphase top2-4 cells (Fig. 3). The phenotype was readily apparent based on classification of cells into categories of inner centromere, nucleoplasmic, partially diffuse, or diffuse Ipl1 (Fig. S2), the same criteria used in a previous study (Peplowska et al., 2014). For simplicity, however, and given that the phenotype in top2-4 cells was highly penetrant and resulted in dramatic loss of Ipl1 from inner centromeres, we will refer to Ipl1 localization as either centromeric or noncentromeric.
Yeast Top2 is not required for Ipl1 recruitment to inner centromeres during interphase. Representative images (A) and quantification (B) of Ipl1-GFP localization to inner centromeres in interphase yeast strains (grown at 30°C for 1 h; the nonpermissive temperature for top2-4). Interphase was defined as cells with a single spindle pole body, corresponding to G1 and early S phase. Bars, 1 µm. n is total number of cells scored from three experimental repeats. Error bars, standard deviation. Analysis of other strains used in this study is presented in Fig. S1.
Yeast Top2 is not required for Ipl1 recruitment to inner centromeres during interphase. Representative images (A) and quantification (B) of Ipl1-GFP localization to inner centromeres in interphase yeast strains (grown at 30°C for 1 h; the nonpermissive temperature for top2-4). Interphase was defined as cells with a single spindle pole body, corresponding to G1 and early S phase. Bars, 1 µm. n is total number of cells scored from three experimental repeats. Error bars, standard deviation. Analysis of other strains used in this study is presented in Fig. S1.
Yeast Top2 is required for Ipl1 recruitment to inner centromeres in mitosis. Representative images (A) and quantification (B) of Ipl1-GFP localization to inner centromeres in prometaphase/metaphase (0.5–2.0-µm spindles) wild type and top2-4 (grown at 30°C for 1 h; the nonpermissive temperature for top2-4). Spc110-mCherry indicates spindle poles. Bars, 1 µm. n is the total number of cells scored from three experimental repeats. Error bars, standard deviation. P-value for top2-4 versus wild type (P = 0.0002), Student's t test. Computational analysis and analysis of cells binned by spindle length is shown in Fig. S3.
Yeast Top2 is required for Ipl1 recruitment to inner centromeres in mitosis. Representative images (A) and quantification (B) of Ipl1-GFP localization to inner centromeres in prometaphase/metaphase (0.5–2.0-µm spindles) wild type and top2-4 (grown at 30°C for 1 h; the nonpermissive temperature for top2-4). Spc110-mCherry indicates spindle poles. Bars, 1 µm. n is the total number of cells scored from three experimental repeats. Error bars, standard deviation. P-value for top2-4 versus wild type (P = 0.0002), Student's t test. Computational analysis and analysis of cells binned by spindle length is shown in Fig. S3.
We also used a quantitative approach to measure whether the Ipl1 fluorescence was localized (Fig. 4 and Fig. S3; see Materials and methods; Chacón and Gardner, 2013). A strong localization pattern, with Ipl1 at centromeres, but not at poles or in the central region of the spindle, will yield two bright foci of GFP pixels surrounded by dim pixels. Together, these pixels will have a high standard deviation in pixel intensity. In contrast, a diffuse localization pattern will have similar pixel intensities in the pixels from pole to pole and therefore a low standard deviation in pixel intensity. We measured this standard deviation, and it recapitulated the classification approach (Fig. S2). It revealed strong localization of Ipl1 in wild-type cells but a striking dispersal of Ipl1 in mitotic top2-4 cells (Fig. 4). As a control, to quantitatively compare this diffuse localization in top2-4 cells with that previously seen in a CPC mutant (Shimogawa et al., 2009), we analyzed bir1-107 cells at the nonpermissive temperature, which again revealed a diffuse localization pattern similar to top2-4 (Fig. 4).
Analysis of Ipl1-GFP distribution between mitotic spindle poles. Computational analysis of Ipl1-GFP distribution between spindle poles in prometaphase/metaphase (1.25–2.0 µm spindles) wild type, top2-4, and bir1-107 strains. Images from the dataset in Fig. 3 were analyzed as described previously (Chacón and Gardner, 2013) to determine the standard deviation of normalized Ipl1-GFP pixel intensities along line scans between the spindle poles. Higher standard deviations indicate localized signals caused by dim regions near poles and bright regions near centromeres (see Materials and methods). From left to right: Merged, example merged image (Ipl1-GFP, Spc110-mCherry); Poles finder, algorithm to locate poles; Ipl1 line scans, image of Ipl1-GFP between poles with positions of line scans overlaid; Ipl1 intensity vector, plot of Ipl1-GFP intensity between poles; and deviation in signal, mean standard deviation of Ipl1-GFP pixel intensities between spindle poles. Bars, 0.5 µm. Analysis of the other strains used in this study is shown in Fig. S3. P-values for each mutant versus wild type are 0.00063 (top2-4) and 0.00009 (bir1-107), Student’s t test. a.u., arbitrary units; st. dev., standard deviation.
Analysis of Ipl1-GFP distribution between mitotic spindle poles. Computational analysis of Ipl1-GFP distribution between spindle poles in prometaphase/metaphase (1.25–2.0 µm spindles) wild type, top2-4, and bir1-107 strains. Images from the dataset in Fig. 3 were analyzed as described previously (Chacón and Gardner, 2013) to determine the standard deviation of normalized Ipl1-GFP pixel intensities along line scans between the spindle poles. Higher standard deviations indicate localized signals caused by dim regions near poles and bright regions near centromeres (see Materials and methods). From left to right: Merged, example merged image (Ipl1-GFP, Spc110-mCherry); Poles finder, algorithm to locate poles; Ipl1 line scans, image of Ipl1-GFP between poles with positions of line scans overlaid; Ipl1 intensity vector, plot of Ipl1-GFP intensity between poles; and deviation in signal, mean standard deviation of Ipl1-GFP pixel intensities between spindle poles. Bars, 0.5 µm. Analysis of the other strains used in this study is shown in Fig. S3. P-values for each mutant versus wild type are 0.00063 (top2-4) and 0.00009 (bir1-107), Student’s t test. a.u., arbitrary units; st. dev., standard deviation.
To examine the top2-4 phenotype further in terms of mitotic stage, we then binned centromeric and noncentromeric Ipl1 cells according to spindle length, based on the distance between separated spindle pole bodies (Figs. 5 A and S3 B). This revealed that more cells with very short spindles had Ipl1 localized at inner centromeres compared with metaphase cells with slightly longer spindles. When mitotic spindles were 0.5–1.0 µm long, 37% of top2-4 cells had Ipl1 at inner centromeres. When mitotic spindles were 1.25–2.0 µm long, only 5% of top2-4 cells had Ipl1 at inner centromeres. Ipl1 was present at inner centromeres in almost all wild-type cells at all spindle lengths. Thus, the need for Topo II to localize Ipl1 to inner centromeres increases throughout early mitosis to a maximum at full preanaphase spindle length, corresponding with prometaphase/metaphase in other eukaryotes. This is the first indication that Topo II has an important function at kinetochores in mitosis to recruit the Ipl1 kinase to the inner centromere in prometaphase/metaphase.
Ipl1 is recruited to mitotic inner centromeres via a noncatalytic function of Top2. (A) Quantification of Ipl1-GFP localization to inner centromeres in cells binned according to spindle length (determined by measuring the distance between the centers of each spindle pole). Spindles 0.5–1.25 µm long correspond to stages from spindle pole separation to spindles in the process of assembly. Spindles 1.25–2.0 µm long correspond to prometaphase/metaphase. Yeast strains were grown at 30°C for 1 h (the nonpermissive temperature for top2-4) before imaging. Error bars, SEM. Representative images (B) and quantification (C) of Ipl1-GFP localization to inner centromeres in prometaphase/metaphase (0.5–2.0 µm spindles) cells of a top2-4 strain carrying an additional allele of catalytically dead top2-Y782F (grown at 30°C for 1 h; the nonpermissive temperature for top2-4). Bars, 1 µm. n is the total number of cells scored from three experimental repeats. Error bars, standard deviation. P-value for top2-4 versus top2-4 top2-YF is 0.04, Student's t test.
Ipl1 is recruited to mitotic inner centromeres via a noncatalytic function of Top2. (A) Quantification of Ipl1-GFP localization to inner centromeres in cells binned according to spindle length (determined by measuring the distance between the centers of each spindle pole). Spindles 0.5–1.25 µm long correspond to stages from spindle pole separation to spindles in the process of assembly. Spindles 1.25–2.0 µm long correspond to prometaphase/metaphase. Yeast strains were grown at 30°C for 1 h (the nonpermissive temperature for top2-4) before imaging. Error bars, SEM. Representative images (B) and quantification (C) of Ipl1-GFP localization to inner centromeres in prometaphase/metaphase (0.5–2.0 µm spindles) cells of a top2-4 strain carrying an additional allele of catalytically dead top2-Y782F (grown at 30°C for 1 h; the nonpermissive temperature for top2-4). Bars, 1 µm. n is the total number of cells scored from three experimental repeats. Error bars, standard deviation. P-value for top2-4 versus top2-4 top2-YF is 0.04, Student's t test.
Top2 localizes Ipl1 to mitotic inner centromeres independently of the SPR
top2-4 is a loss-of-function allele; at the nonpermissive temperature, the Top2-4 enzyme is catalytically dead (Holm et al., 1985). To determine if the SPR enzyme cycle of Topo II is required for Ipl1 recruitment to inner centromeres, we asked if the phenotype in top2-4 could be rescued by expression of top2-Y782F, which carries a point mutation in the enzyme active site (Liu and Wang, 1998). Top2-Y782F can bind to DNA and undergo conformational changes associated with the enzyme cycle (e.g., opening and closure of the N-terminal gate of the enzyme) but is unable to break the helix bound at the catalytic core (Liu and Wang, 1998). Surprisingly, these properties of Top2-Y782F were sufficient to partially restore Ipl1 inner-centromere recruitment in top2-4 cells (Fig. 5, B and C). Therefore, Top2 may only need to be bound to DNA at inner centromeres, and not engaged in the catalytic cycle, in order to recruit Ipl1.
The conserved SUMOylation sites in the CTD of Top2 are required for mitotic inner-centromere localization of Ipl1
The ability of catalytically dead top2-Y782F to rescue Ipl1 inner-centromere localization indicated that this is a function independent of the SPR. To explore this further, we first examined strains where the CTD of Topo II had been deleted (residues 1,243 to the C terminus; top2ΔCTD). The CTD is dispensable for the SPR but is required for faithful chromosome segregation (Bachant et al., 2002; Dickey and Osheroff, 2005; Lane et al., 2013), indicating that it has an unknown function distinct from the catalytic cycle. Strikingly, top2ΔCTD had the same phenotype as top2-4: Ipl1 recruitment to inner centromeres was abolished, and specifically in prometaphase/metaphase (Fig. 6 A and Figs. S1–S3). Thus, the CTD of Topo II is required for Ipl1 recruitment to inner centromeres in mitosis. Further, expression of top2-Y782F was able to partially rescue Ipl1 inner-centromere localization in top2ΔCTD cells (Fig. 6 B). Therefore, a Top2 protein that possesses a CTD but is catalytically inactive is sufficient for Ipl1 recruitment. The data indicate that Top2 has a noncatalytic function in mitosis to recruit Ipl1 to inner centromeres.
SUMOylation sites in the CTD of Top2 are required for Ipl1 recruitment to mitotic inner centromeres. Representative images and quantification of Ipl1-GFP localization to inner centromeres in prometaphase/metaphase (0.5–2.0-µm spindles). (A) top2ΔCTD strain lacking the CTD. (B) top2ΔCTD carrying an additional allele of catalytically dead top2-Y782F. (C) top2-SNM strain, lacking the major CTD SUMOylation sites. All strains were shifted to 30°C for 1 h before imaging to maintain consistent imaging conditions between all strains. Spc110-mCherry indicates spindle poles. Bars, 1 µm. n is the total number of cells scored from three experimental repeats. Error bars, standard deviation. P-values for top2ΔCTD versus wild type and top2ΔCTD versus top2ΔCTD top2-YF are 0.00004 and 0.002, respectively, Student's t test. Computational analysis and analysis of cells binned by spindle length is shown in Fig. S3.
SUMOylation sites in the CTD of Top2 are required for Ipl1 recruitment to mitotic inner centromeres. Representative images and quantification of Ipl1-GFP localization to inner centromeres in prometaphase/metaphase (0.5–2.0-µm spindles). (A) top2ΔCTD strain lacking the CTD. (B) top2ΔCTD carrying an additional allele of catalytically dead top2-Y782F. (C) top2-SNM strain, lacking the major CTD SUMOylation sites. All strains were shifted to 30°C for 1 h before imaging to maintain consistent imaging conditions between all strains. Spc110-mCherry indicates spindle poles. Bars, 1 µm. n is the total number of cells scored from three experimental repeats. Error bars, standard deviation. P-values for top2ΔCTD versus wild type and top2ΔCTD versus top2ΔCTD top2-YF are 0.00004 and 0.002, respectively, Student's t test. Computational analysis and analysis of cells binned by spindle length is shown in Fig. S3.
The CTD of Topo II is not required for the SPR, but it is subjected to multiple posttranslational modifications (Cardenas and Gasser, 1993; Isaacs et al., 1998; Bachant et al., 2002). Although the biological consequences of these modifications are largely unexplored, mutation of five conserved SUMOylation sites from lysine to arginine (the top2-SNM mutant) renders yeast cells with reduced fidelity of chromosome segregation (Bachant et al., 2002). To ask if SUMO modification plays a role in Ipl1 targeting, we examined this top2-SNM mutant, revealing that most mitotic cells failed to properly localize Ipl1 to the inner centromeres (Fig. 6 C and Figs. S2 and S3). As with top2-4 and top2ΔCTD cells, the phenotype was specific to prometaphase/metaphase cells and there was no defect in interphase (Figs. S1–S3). Together, the data suggest a noncatalytic function of the Top2 CTD in mitotic Ipl1 localization, in particular mediated by SUMOylation of the Top2 CTD.
Top2 CTD SUMOylation is dispensable for chromatin association
One explanation for the CTD SUMOylation-dependent recruitment of Ipl1 to the inner centromeres in mitosis could be that these posttranslational modifications play a role in chromatin association of Topo II. To test this, we tagged the endogenously produced Top2, Top2ΔCTD, and Top2-SNM proteins with GFP to directly observe and quantify their abundance on chromatin. We prepared chromatin in situ by making spheroplasts and extracting with detergent. As a control, we observed a soluble GFP-tagged nuclear protein (TetR-GFP) that does not bind to chromatin and that was readily dispersed upon extraction (Fig. 7). In contrast, Top2 was robustly retained on chromatin after extraction, as was Top2-SNM (Fig. 7). Therefore, the SUMOylation sites that are important for Ipl1 recruitment to inner centromeres are not needed for the association of Top2 with chromatin. These data indicate Top2 that is associated with chromatin, but not necessarily engaged in the catalytic cycle, becomes SUMOylated to stimulate Ipl1 recruitment. Examination of Top2ΔCTD revealed a reduction in abundance on chromatin after detergent extraction (Fig. 7). Therefore, it is possible that CTD motifs other than the SUMOylation sites function in the association of Top2 with chromatin or in the residence time of Top2 on chromatin. This could potentially influence the capacity of Top2 to recruit Ipl1 to inner centromeres. However, SUMOylation of CTD lysines is likely to be the predominant Ipl1 recruitment signal.
The Top2 CTD, but not Top2 SUMO modification, is required for robust Top2 association with chromatin. (A) Representative images of DAPI staining and GFP signal in spheroplasted yeast with and without detergent extraction. Logarithmic phase TOP2-GFP, top2-SNM-GFP, top2ΔCTD-GFP, and TetR-GFP strains, along with untagged controls (strain CRY1), were arrested in nocodazole for 2 h. To assess Top2 association with chromatin, cells were spheroplasted to partially remove the cell wall, and permeabilized with 1% Triton X-100. Bars, 4 µm. (B) Distribution of chromatin association with and without detergent extraction. To quantify chromatin association, regions of GFP signal from spheroplasted and extracted cells were assigned and thresholded to obtain values for the intensity of the GFP signal. With the exception of the TetR-GFP strain (n = 25), at least 50 cells were scored per sample. In the case of the TetR-GFP experiment, the active region was defined using the corresponding DAPI signal. P-values, two-tailed Student’s t test.
The Top2 CTD, but not Top2 SUMO modification, is required for robust Top2 association with chromatin. (A) Representative images of DAPI staining and GFP signal in spheroplasted yeast with and without detergent extraction. Logarithmic phase TOP2-GFP, top2-SNM-GFP, top2ΔCTD-GFP, and TetR-GFP strains, along with untagged controls (strain CRY1), were arrested in nocodazole for 2 h. To assess Top2 association with chromatin, cells were spheroplasted to partially remove the cell wall, and permeabilized with 1% Triton X-100. Bars, 4 µm. (B) Distribution of chromatin association with and without detergent extraction. To quantify chromatin association, regions of GFP signal from spheroplasted and extracted cells were assigned and thresholded to obtain values for the intensity of the GFP signal. With the exception of the TetR-GFP strain (n = 25), at least 50 cells were scored per sample. In the case of the TetR-GFP experiment, the active region was defined using the corresponding DAPI signal. P-values, two-tailed Student’s t test.
Top2 is not required for Sgo1 localization to inner centromeres in mitosis
Two pathways are implicated in Aurora B recruitment to inner centromeres in eukaryotes: the Sgo1–histone H2A T120-Phos pathway and the Haspin kinase–histone H3 T3-Phos pathway (Jia et al., 2013). In yeast, the Sgo1-dependent mechanism that relies on Bub1 phosphorylation of H2A was found to be important for Ipl1 recruitment to mitotic inner centromeres (Peplowska et al., 2014). We confirmed these data using sgo1Δ strains (Fig. 8 A and Figs. S1–S3). Importantly, these data also revealed that the phenotypes of sgo1Δ, top2-4, top2ΔCTD, and top2-SNM are similarly penetrant in terms of the Ipl1 localization defect at metaphase. In interphase, neither Top2 nor Sgo1 was required for Ipl1 localization. This suggests that Topo II and Sgo1 are equally important for Ipl1 localization to inner centromeres in mitosis. Top2 functions either in the Sgo1 pathway or in a parallel pathway that is equally crucial for Ipl1 localization.
Top2 is not required for localization of Sgo1 to inner centromeres in mitosis. Representative images and quantification of Ipl1-GFP (A) and Sgo1-GFP (B) localization to inner centromeres in prometaphase/metaphase (0.5–2.0-µm spindles) wild type, top2-4, and top2ΔCTD (grown at 30°C for 1 h, the nonpermissive temperature for top2-4). Spc110-mCherry indicates spindle poles. Bars, 1 µm. n is the total number of cells scored from three experimental repeats. Error bars, standard deviation. P-value for sgo1Δ versus wild type is 0.00004, Student's t test. Computational analysis of Ipl1-GFP localization and analysis of cells binned by spindle length is shown in Fig. S3.
Top2 is not required for localization of Sgo1 to inner centromeres in mitosis. Representative images and quantification of Ipl1-GFP (A) and Sgo1-GFP (B) localization to inner centromeres in prometaphase/metaphase (0.5–2.0-µm spindles) wild type, top2-4, and top2ΔCTD (grown at 30°C for 1 h, the nonpermissive temperature for top2-4). Spc110-mCherry indicates spindle poles. Bars, 1 µm. n is the total number of cells scored from three experimental repeats. Error bars, standard deviation. P-value for sgo1Δ versus wild type is 0.00004, Student's t test. Computational analysis of Ipl1-GFP localization and analysis of cells binned by spindle length is shown in Fig. S3.
A simple explanation for Ipl1 mislocalization during mitosis in top2 mutants could be that Sgo1 fails to localize to inner centromeres. This would place Top2 upstream of Sgo1 and indicate that Top2 functions via the H2A T120-Phos pathway. To test this directly, we tagged endogenous Sgo1 with a three GFP fusion (3xGFP) at the C terminus of Sgo1. As seen in previous studies examining Sgo1-GFP (Peplowska et al., 2014), we observed that Sgo1-3xGFP was present within the foci of clustered centromeres in the majority (60–70%) of metaphase cells (Fig. 8 B). Robust localization to centromeres was also observed in top2-4 mutants grown at the nonpermissive temperature and top2ΔCTD mutants (Fig. 8 B), under conditions identical to those where Ipl1-GFP was dramatically mislocalized. Therefore, Top2 is not needed for Sgo1 localization to the inner centromeres in mitosis. Top2 cannot act via Sgo1 recruitment in order to fulfill its function in Ipl1 recruitment.
Yeast Haspin kinase and histone H3 threonine 3 are required for mitotic inner-centromere localization of Ipl1
The mitotic role of Sgo1 in Ipl1 recruitment to the inner centromere has been largely described, and the data in Fig. 8 B demonstrated that Top2 does not contribute to Sgo1 inner-centromere localization. In contrast, the mechanism of Haspin-mediated H3 T3 phosphorylation and its contribution to Ipl1 recruitment to inner centromeres are less well understood. In Saccharomyces cerevisiae in particular, there is no evidence that Haspin kinase or H3 T3-Phos are involved in Ipl1 recruitment to inner centromeres. Interestingly, however, Xenopus laevis Haspin kinase coprecipitated with the Topo II CTD from Xenopus egg extract, and the Topo II CTD was required for recruitment of Haspin to centromeres (see Yoshida et al. in this issue). To determine if yeast Topo II might function via Haspin kinase and H3 T3-Phos, we first examined Haspin mutants. In strains lacking both Haspin orthologues (alk1Δ and alk2Δ), Ipl1 inner-centromere localization in mitosis was severely disrupted (Fig. 9 A), and once more, this phenotype was specific to prometaphase/metaphase cells (Figs. S1–S3). Importantly, the same phenotype was observed in yeast carrying a T3A substitution of the histone H3 gene, where phosphorylation of threonine 3 could not occur (h3-T3A; Fig. 9 A and Figs. S1–S3). Therefore, yeast Haspin kinases, and presumably phosphorylation of yeast H3 threonine 3 at inner centromeres, is required for Ipl1 recruitment in mitosis.
Haspin kinases and histone H3 threonine 3 phosphorylation are required for mitotic Ipl1 inner-centromere localization. (A and C) Representative images and quantification of Ipl1-GFP localization to inner centromeres in prometaphase/metaphase (0.5–2.0-µm spindles) alk1Δ alk2Δ, h3-T3A, top2-4 alk1Δ alk2Δ, and top2ΔCTD alk1Δ alk2Δ (grown at 30°C for 1 h, the nonpermissive temperature for top2-4). Spc110-mCherry indicates spindle poles. Bars, 1 µm. n is the total number of cells scored from three experimental repeats. Error bars, standard deviation. Computational analysis of Ipl1-GFP localization and analysis of cells binned by spindle length is shown in Fig. S3. P-values for alk1Δ alk2Δ versus top2-4 alk1Δ alk2Δ and alk1Δ alk2Δ versus top2ΔCTD alk1Δ alk2Δ are 0.248726748 and 0.282585966, respectively, Student's t test. (B) Western blotting of histone H3 phosphorylated at threonine 3 in cells arrested in mitosis after inactivation of Mtw1. The indicated strains were incubated for 2 h at the nonpermissive temperature (37°C) for the mtw1-1 allele (present in all of the strains) to achieve arrest. H3 phosphorylated at threonine 3 was quantified relative to loading control (α-tubulin). Error bars show standard deviation from at least three experimental repeats for each strain.
Haspin kinases and histone H3 threonine 3 phosphorylation are required for mitotic Ipl1 inner-centromere localization. (A and C) Representative images and quantification of Ipl1-GFP localization to inner centromeres in prometaphase/metaphase (0.5–2.0-µm spindles) alk1Δ alk2Δ, h3-T3A, top2-4 alk1Δ alk2Δ, and top2ΔCTD alk1Δ alk2Δ (grown at 30°C for 1 h, the nonpermissive temperature for top2-4). Spc110-mCherry indicates spindle poles. Bars, 1 µm. n is the total number of cells scored from three experimental repeats. Error bars, standard deviation. Computational analysis of Ipl1-GFP localization and analysis of cells binned by spindle length is shown in Fig. S3. P-values for alk1Δ alk2Δ versus top2-4 alk1Δ alk2Δ and alk1Δ alk2Δ versus top2ΔCTD alk1Δ alk2Δ are 0.248726748 and 0.282585966, respectively, Student's t test. (B) Western blotting of histone H3 phosphorylated at threonine 3 in cells arrested in mitosis after inactivation of Mtw1. The indicated strains were incubated for 2 h at the nonpermissive temperature (37°C) for the mtw1-1 allele (present in all of the strains) to achieve arrest. H3 phosphorylated at threonine 3 was quantified relative to loading control (α-tubulin). Error bars show standard deviation from at least three experimental repeats for each strain.
Mitotic histone H3 threonine 3 phosphorylation is reduced in alk1Δ alk2Δ and top2ΔCTD mutants
Histone H3 threonine 3 phosphorylation has been observed in many eukaryotes but has to our knowledge not been detected before in S. cerevisiae (Panigada et al., 2013). Indeed, in extracts made from log-phase cultures of yeast, we were unable to detect this modification using anti-H3 T3-Phos antisera (unpublished data). To investigate this further, we reasoned that cells may need to be synchronized in mitosis under conditions where Ipl1 is known to be active and localized at inner centromeres. This can be achieved using the mtw1-1 allele, with a temperature-sensitive mutation in a component of the MIND (Mtw1–Nnf1–Nsl1–Dsn1) kinetochore complex. To provide a negative control, we also constructed an mtw1-1 h3-T3A mutant strain, in which histone H3 cannot be phosphorylated at threonine 3. Cultures were harvested after growth at the mtw1-1 nonpermissive temperature for 2 h, which was sufficient for mitotic arrest, and then cell extracts were subjected to SDS-PAGE and Western blotting. Under these conditions, H3 T3 phosphorylation was consistently observed in mtw1-1 extracts but was substantially reduced in mtw1-1 h3-T3A extracts (Fig. 9 B). Because there was a residual signal even when threonine 3 could not be phosphorylated, the antibody must have a base level of affinity for H3 lacking the modification at T3. This control provided an important baseline of signal for subsequent analyses.
Having characterized the specificity of the antibody, we then asked if H3 T3 phosphorylation requires Haspin kinases in S. cerevisiae by analyzing extracts from arrested mtw1-1 alk1Δ alk2Δ cells. This yielded a signal similar to that seen in the h3-T3A mutant (Fig. 9 B), revealing that yeast Haspin kinases are indeed required for H3 T3 phosphorylation in mitosis.
If Top2 functions upstream of Haspin kinases for Ipl1 recruitment to inner centromeres in mitosis, then H3 T3 phosphorylation is predicted to require Top2. To test this possibility, we examined extracts from mtw1-1 top2ΔCTD cells. Importantly, this revealed a similar reduction in the H3 T3-Phos signal as observed in the Haspin mutant and the h3-T3A mutant.
These biochemical studies are consistent with the genetic analysis of the requirements for Ipl1 inner-centromere localization in mitosis and place Top2 in the Haspin kinase–H3 T3-Phos pathway. If Top2 and Haspin kinases do function in a common pathway, then from a genetic point of view, we would predict that the lack of Haspin kinases ought not to enhance the Ipl1 inner-centromere localization defect seen in top2 mutants. Corroborating the biochemical analysis, we observed that top2-4 alk1Δ alk2Δ and top2ΔCTD alk1Δ alk2Δ mutants were no more defective in Ipl1 localization than the single top2 mutants, respectively (Fig. 9 C). Together, the data indicate that Top2 acts with Haspin to mediate H3 T3-phosphorylation and thereby facilitates Ipl1 recruitment to the inner centromeres in mitosis.
A phosphomimetic histone H3 threonine 3 bypasses the requirement for Top2 and Haspin for Ipl1 inner-centromere recruitment in mitosis
If the role of Topo II in Ipl1 inner-centromere recruitment is to promote H3 T3 phosphorylation by Haspin, then a phosphomimetic H3 T3 residue would be predicted to bypass the requirement for Topo II in this mechanism. To test this, we first asked if expression of wild-type H3 can restore Ipl1 inner-centromere localization in the h3-T3A mutant to provide a baseline measurement of rescue efficiency. The temporal parameters for this experiment were important to establish because expression of wild-type H3 would allow only a gradual displacement of mutant h3-T3A histone from the genome and from inner centromeres (∼50% replacement per replication cycle). In time course experiments, after induction of wild-type H3 (HHT2) expression from the GAL10 promoter (Fig. S4), a peak of rescued Ipl1 inner-centromere localization at metaphase was observed after 2 h (Fig. 10 A). Given that this is approximately the length of one cell cycle, during which not more than half of the mutant H3 is expected to be replaced as a consequence of DNA replication, the rescue of Ipl1 localization to ∼40% of cells is striking. At 4 h after induction (and at later time points), we observed that the cells had large vacuoles, consistent with previous studies that overexpression of H3 is detrimental to growth (Singh et al., 2010). These data therefore establish the window of time that is optimal for assessing rescue of Ipl1 inner-centromere localization in metaphase.
Phosphomimetic histone h3-T3E bypasses the requirements for Top2 and Haspin kinases for mitotic Ipl1 inner-centromere localization. (A–C) Quantification of Ipl1-GFP localization to inner centromeres in prometaphase/metaphase cells (0.5–2.0-µm spindles) after induction of wild-type H3 in the h3-T3A strain or induction of phosphomimetic H3-T3E mutant in the top2ΔCTD and alk1Δ alk2Δ strains. Bars, 1 µm. n is the total number of cells scored from three experimental repeats. Error bars, standard deviation. P-values for t = 0 versus t = 2 h time points are 0.005, wild-type H3 (GAL10-HHT2) in h3-T3A (hht2-T3A) strain; 0.009, h3-T3E (GAL10-hht2-T3E) in top2ΔCTD strain; and 0.00882, h3-T3E (GAL10-hht2-T3E) in alk1Δ alk2Δ strain, Student's t test. (D) Proposed pathways contributing to Aurora B/Ipl1 recruitment to mitotic inner centromeres. Bub1 kinase phosphorylates the H2A C-terminal tail, providing a binding site for Sgo1, which itself binds to the CPC. The Top2 CTD mediates Haspin-dependent phosphorylation of the H3 N-terminal tail, providing a binding site for the CPC. Key shows vertebrate and S. cerevisiae orthologues of the proteins depicted.
Phosphomimetic histone h3-T3E bypasses the requirements for Top2 and Haspin kinases for mitotic Ipl1 inner-centromere localization. (A–C) Quantification of Ipl1-GFP localization to inner centromeres in prometaphase/metaphase cells (0.5–2.0-µm spindles) after induction of wild-type H3 in the h3-T3A strain or induction of phosphomimetic H3-T3E mutant in the top2ΔCTD and alk1Δ alk2Δ strains. Bars, 1 µm. n is the total number of cells scored from three experimental repeats. Error bars, standard deviation. P-values for t = 0 versus t = 2 h time points are 0.005, wild-type H3 (GAL10-HHT2) in h3-T3A (hht2-T3A) strain; 0.009, h3-T3E (GAL10-hht2-T3E) in top2ΔCTD strain; and 0.00882, h3-T3E (GAL10-hht2-T3E) in alk1Δ alk2Δ strain, Student's t test. (D) Proposed pathways contributing to Aurora B/Ipl1 recruitment to mitotic inner centromeres. Bub1 kinase phosphorylates the H2A C-terminal tail, providing a binding site for Sgo1, which itself binds to the CPC. The Top2 CTD mediates Haspin-dependent phosphorylation of the H3 N-terminal tail, providing a binding site for the CPC. Key shows vertebrate and S. cerevisiae orthologues of the proteins depicted.
We next asked if expression of a phosphomimetic h3-T3E histone is able to alleviate the Haspin kinase and Topo II requirements for Ipl1 inner-centromere recruitment in mitosis. As predicted, if Haspin kinases are required for H3 T3 phosphorylation in S. cerevisiae, Ipl1 inner-centromere localization in mitosis was partly restored by h3-T3E histone in alk1Δ alk2Δ mutant cells (Fig. 10 B and Fig. S4). Importantly, when expressed in the top2ΔCTD mutant (Fig. S4), bypass of the requirement for the CTD was also observed, with Ipl1 localizing correctly in ∼47% of cells 2 h after induction of the h3-T3E mutant (Fig. 10 C). These data provide evidence directly linking the yeast Topo II CTD to Haspin-mediated H3 T3 phosphorylation, required for Ipl1 recruitment to inner centromeres in metaphase. We conclude that the centromere function of the Topo II CTD is to recruit Ipl1 via the Haspin–H3 T3-Phos pathway.
Discussion
The SPR of Topo II has been studied extensively using biochemical and structural approaches because it is the target of several important classes of antitumor drugs (Nitiss, 2009b). However, binding of Topo II to DNA occurs even in the absence of the catalytic activity of the enzyme, and it has remained unknown whether Topo II plays additional roles in cells that are not associated with the DNA topology changes induced by the SPR. In particular, an important region of the enzyme is the CTD, which is dispensable for the SPR but is nevertheless required for faithful chromosome segregation from yeast to human cells (Jensen et al., 1996; Bachant et al., 2002; Dickey and Osheroff, 2005; Lane et al., 2013). The CTD of Topo II is heavily modified by posttranslational modifications. Of particular note are lysine residues prominently modified by SUMOylation. When five of these lysines are mutated in yeast cells (i.e., the top2-SNM mutant), one consequence is reduced chromosome segregation fidelity (Bachant et al., 2002; Takahashi et al., 2006). At the molecular level, however, there is no clear understanding of the role of these SUMOylated lysines or even of the CTD as a whole.
Here, we set out to ask if Topo II may have an important function in chromosome segregation that is independent of the SPR. During mitosis, Topo II is enriched at the centromeres of chromosomes, and so we began by asking if we could detect any defects in kinetochore assembly. Based on the localization of outer kinetochore component Nuf2 and the recruitment of Mad2 to the kinetochore when microtubules were depolymerized, we did not observe obvious defects in kinetochore structure or function. However, the inner-centromere kinase Ipl1 (Aurora B) was drastically delocalized in top2 mutants, including top2ΔCTD, which has normal catalytic activity, and top2-SNM, which lacks the major CTD lysines that are modified by SUMOylation.
Analysis of chromatin in situ demonstrated that Top2-SNM protein robustly associates with the chromatin. Therefore, if Top2 functions in the recruitment of Ipl1 to centromeres, the role of the CTD is not solely that of an anchor that tethers Top2 to chromosomes. These data are consistent with substantial evidence that SUMOylation is essential for accurate chromosome segregation (Tanaka et al., 1999; Biggins et al., 2001; Bachant et al., 2002; Azuma et al., 2003; Díaz-Martínez et al., 2006). Other evidence has revealed that vertebrate Topo II is primarily SUMOylated at the centromere region in mitosis (Azuma et al., 2005; Ryu et al., 2010a,b). Together, the data suggest that Topo II is a critical target of SUMO ligases that ensure mitotic fidelity. It is important to note that top2ΔCTD retains one SUMOylated lysine that is mutated in top2-SNM (K1220). It is therefore possible that K1220 is not a major SUMO acceptor site or that it is not sufficient for Ipl1 recruitment to inner centromeres.
In eukaryotes other than S. cerevisiae, determinants of inner-centromere targeting of the Ipl1 orthologue, Aurora B, have been described (Kelly et al., 2010; Jeyaprakash et al., 2011; Carmena et al., 2012; Sawicka and Seiser, 2014). The data indicate that two pathways contribute inner-centromere–specific epigenetic marks to provide the binding site for the CPC, of which Aurora B is a component (Fig. 10 D). Bub1 kinase phosphorylates histone H2A at threonine 120, facilitating interaction with Sgo1, which then binds directly to the CPC protein Borealin. In addition, Haspin kinase phosphorylates histone H3 at threonine 3, providing an interaction surface for the CPC protein Survivin. Consistent with these pathways being conserved in yeast, previous work demonstrated that sgo1Δ yeast mutants are defective in Ipl1 centromere targeting (Peplowska et al., 2014), and we observed similar defects in a Haspin kinase-deficient mutant (alk1Δ alk2Δ) and also an H3 mutant where threonine 3 could not be phosphorylated (h3-T3A).
The yeast top2, h3-T3A, and Haspin mutants were all specifically defective in mitotic Ipl1 inner-centromere localization: normal inner-centromere localization was observed in interphase cells with a single spindle pole body (Fig. S1). This is consistent with the restriction of H3 T3 phosphorylation to mitosis in other eukaryotes (Dai et al., 2005; Wang et al., 2010; Ghenoiu et al., 2013). Moreover, the data show that both Sgo1 and Topo II pathways are important for Ipl1 recruitment to inner centromeres in mitosis. Therefore, consistent with studies in higher eukaryotes (Yamagishi et al., 2010; Sawicka and Seiser, 2014), the H3 T3-Phos and H2A T120-Phos marks likely provide a conserved epigenetic feature to specify mitotic inner centromeres. This conserved molecular mechanism of Ipl1 recruitment must also have arisen independently of whether a kinetochore is built upon a point centromere, in S. cerevisiae, or the greatly different regional centromeres found in most other eukaryotes (Steiner and Henikoff, 2015).
In Xenopus egg extract, the Topo II CTD promotes the recruitment of Haspin kinase to centromeres (Yoshida et al., 2016). We attempted to directly observe yeast Haspin kinases at centromeres by tagging Alk1 and Alk2 with 3xGFP. This revealed that both kinases are broadly distributed throughout the nucleus and cytoplasm (Fig. S4). These localization patterns indicate that Haspin kinases may have multiple substrates, consistent with their genetic and physical interaction maps (Stark et al., 2006; Gilmore et al., 2012). We did not observe any obvious changes in Alk1-3xGFP or Alk2-3xGFP localization in top2 mutants (Fig. S4), but typical of kinases, this could be explained if their association with centromeres is transient.
Several lines of genetic and biochemical evidence have placed Top2 in the Haspin-mediated pathway and not the Sgo1 pathway (Fig. 10 D). First, we determined that top2 mutants are not defective in Sgo1 recruitment to centromeres in mitotic yeast cells. Therefore, the function of Top2 in Ipl1 recruitment does not occur upstream of Sgo1 centromere targeting. Because Sgo1 bridges one of the interactions between Ipl1 and the inner centromere by binding directly to H2A T120-Phos and Borealin of the CPC, these data make it unlikely that Top2 influences Ipl1 localization via Sgo1. We attempted to perform epistasis analysis between top2 and sgo1Δ mutants, but after tetrad dissection, top2 sgo1Δ double mutants were viable at a very low frequency and the surviving isolates had a synthetic sick phenotype, making analysis of Ipl1 localization problematic (we have not been able to derive an Ipl1-3xGFP strain combined with top2 sgo1Δ). These synthetic genetic interactions are, however, consistent with Top2 and Sgo1 having an overlap in function. The relevant function could perhaps be in Ipl1 recruitment to the inner centromeres in mitosis.
Consistent with Top2 acting in the Haspin pathway, we observed no additive defect in Ipl1 recruitment in a top2-4 alk1Δ alk2Δ mutant. An additive defect would be expected if Top2 and Haspin function by separate, partially redundant, mechanisms. Perhaps more strikingly, we observed that a phosphomimetic H3 threonine 3 (h3-T3E) mutant was able to bypass both the requirements for Top2 and Alk1,2 in Ipl1 recruitment. These data indicate that both Top2 and Haspin kinases act upstream of histone H3. Together, the data provide genetic evidence that Top2 and Haspin are required to establish the H3 T3-Phos component of the CPC binding site, and this was corroborated by biochemical evidence that Top2 and Haspin are required for mitotic histone H3 threonine 3 phosphorylation.
In the accompanying manuscript, Yoshida et al. provide evidence that SUMOylation of the Topo II CTD promotes the formation of Topo II–Haspin complexes at centromeres to facilitate Aurora B recruitment. Because these studies were performed using Xenopus egg extracts, the data indicate a conserved mechanism in yeast and vertebrates where Topo II CTD SUMOylation establishes part of the binding surface at the inner centromere for Ipl1/Aurora B. The genetic and biochemical analyses presented here provide further evidence that this mechanism acts independently of the histone H2A–Sgo1 binding interface. Therefore, the CPC most likely has a two-component binding surface where it associates both with the phosphorylated N-terminal tail of histone H3 (T3-Phos) and the phosphorylated C-terminal tail of histone H2A (T120-Phos).
Materials and methods
Yeast strains and plasmids
Yeast strains used in this study are derivatives of W303 as described in Table S1. Strains MMWY72-22D (Nuf2-GFP) and PWY334-1B (Ipl1-3xGFP) were provided by T. Davis (University of Washington, Seattle, WA). Strain SBY2668 (mtw1-1) was provided by S. Biggins (Fred Hutchinson Cancer Research Center, Seattle, WA). Strains used here to analyze Mad2-3xGFP localization were derived from a strain provided by T. Tanaka (University of Dundee, Dundee, Scotland, UK). Yeast were modified by standard genetic approaches (Burke, 2000). Plasmid p646 (a gift from J. Berman, Tel Aviv University, Tel Aviv, Israel), which expresses HHT2 under control of GAL10, was mutated by amplifying the plasmid using primers 5′-GGCCAGAGAAAAACAAACAGCTAGAAAATCC-3′ and 5′-GCTGTTTGTTTTTCTCTGGCCATTGTGGAGTG-3′. The overlapping PCR ends were joined using Gibson Assembly (New England Biolabs, Inc.) to create an intact plasmid. 3X-GFP C-terminal–tagged proteins were generated by cloning a C-terminal fragment of ALK1, ALK2, and SGO1 into either p682(His) or p683(Ura), which were gifts from J. Berman. These constructs were then cut in the C-terminal fragment and used to transform yeast by a homologous recombination based protocol at the endogenous loci. Strains harboring TOP2-GFP, top2-SNM-GFP, and top2ΔCTD-GFP, replacing TOP2 at the endogenous locus, were constructed using pGS001, pGS002, and pJBN340, respectively. The top2ΔCTD-GFP allele encoded in pJBN340 is truncated after amino acid 1,235. Yeast were grown in rich medium with yeast extract, peptone, and 2% glucose at 30°C, except where stated.
Analysis of Ipl1 localization
Yeast were grown at 26°C overnight in rich medium supplemented with additional adenine, diluted to an OD of 0.15–0.3 and cultured for 4 h, then shifted to 30°C for 1 h. Yeast were then washed and cultured in complete synthetic medium in a microfluidic chamber, as follows. Cells were immobilized on a coverslip following a method described previously (Chacón et al., 2014). Briefly, flow chambers were prepared with a washed (1 M NaOH-treated) 22 × 22-mm imaging coverslip, overlaid with thin strips of Parafilm, which were melted to attach an untreated 18 × 18-mm coverslip to form the top of the chamber. 0.5 mg/ml concanavalin A was flowed in and allowed to sit in the flow chamber for 20 min at room temperature, which bound to the imaging coverslip. Excess concanavalin A was then washed out of the chamber with water. Cells were pipetted into the chamber and adhered to the concanavalin A–coated imaging coverslip. Excess cells that did not adhere to the coverslip were removed with vacuum.
Unless otherwise stated, yeast cells were imaged using total internal reflection fluorescence (TIRF) microscopy with an Eclipse Ti microscope (Nikon) using 488- and 561-nm Sapphire lasers (Coherent) to visualize GFP and mCherry. A rapid switching FireWire setup allowed for near-simultaneous imaging between the red and green lasers. An electron-multiplying charge-coupled iXon3 camera (Andor Technology) fitted with a 2.5× projection lens was used to capture images with a 64-nm pixel size in the field of view. Cells were imaged with a CFI Apochromat 100×, 1.49-NA oil objective (Nikon). NIS Elements software (Nikon) was used for image acquisition, and all images were contrast enhanced using ImageJ (Fiji). All imaging was performed at 30°C except for the analysis of bir1-107, which was imaged at 37°C. Imaging medium was complete synthetic medium unless otherwise described.
Two independent methods were used to analyze Ipl1 localization: (1) subjective categorization and (2) quantification. For subjective categorization, images were binned into the following categories (see Fig. S1 B): (a) centromeric (inner centromere), where the GFP signal was restricted to two compact foci clustered within the spindle axis; (b) nuclear, where the GFP signal was homogenous within the nucleoplasm; (c) partially diffuse, where the GFP signal was homogenous within the nucleoplasm but with some regions of higher intensity; and (d) diffuse, where the GFP signal was homogenous but not completely restricted to the nucleoplasm. For quantification, we used a simple way to quantify whether Ipl1-GFP was strongly localized or diffuse that uses the standard deviation in Ipl1-GFP pixel intensity between the spindle poles (Chacón and Gardner, 2013). Unlocalized Ipl1-GFP will lack bright foci, and therefore, the pixel intensities between the poles will be similar. In contrast, localized Ipl1-GFP will have very dim regions (near the poles) and very bright regions (near the centromeres). Therefore, the standard deviation in pixel intensity within a line scan between the poles will be higher for localized Ipl1-GFP than for unlocalized.
The method was applied as follows: Poles were first identified using 2D Gaussian fitting. We then measured the Ipl1-GFP pixel intensity in a 15-pixel band from pole to pole, with a 15-pixel width chosen because it captures the width of the spindle. The pixel values in the columns of this band, where the columns correspond to the direction perpendicular to the pole to pole direction, were summed. This gave us a single vector of data containing the Ipl1-GFP signal from pole to pole. Next, the vector was rebinned into 50 bins to reduce confounds from different spindle lengths. Finally, we normalized the pixel intensities in the vector by dividing each bin by the vector’s sum and took the standard deviation of these 50 normalized intensities, which are plotted in Fig. 2 C.
Determination of spindle length to bin cells with a mitotic spindle
ImageJ (Fiji) and MATLAB (MathWorks, Inc.) were used to subtract background, identify spindle pole bodies, and calculate the distance between poles. The poles were identified by thresholding-based image segmentation, and the MATLAB centroid function was used to identify the center of the poles in the segmented image. This automation sorted images by spindle length, allowing subsequent rapid analysis of Ipl1 localization according to cells binned by spindle length.
Analysis of Ipl1 localization after histone H3 allele induction
Strains carrying a centromeric plasmid harboring histone H3 or the mutant T3E derivative, both under control of the GAL10 promoter, were grown in synthetic medium with 4% raffinose and lacking tryptophan or uracil at 26°C. The histone H3 genes were induced with 2% galactose for the indicated time points. Cells were shifted to 30°C an hour before collecting cells for imaging to assay Ipl1 localization.
Analysis of Mad2-GFP localization in live cells
Strains expressing Mad2-3xGFP were grown overnight in synthetic complete medium with 2% dextrose then grown to log phase after dilution. Cells were then visualized after growth for 1 h at 30°C with or without 15 µg/ml nocodazole. Cells were categorized into kinetochore or nuclear pore localization based on criteria used previously (Iouk et al., 2002). For kinetochore localization, the Mad2-GFP was tightly clustered within a single focus within the interior of the nucleus. For nuclear pore localization, the Mad2-GFP was distributed into numerous smaller foci that defined the circumference of the nucleus. Representative images were acquired by TIRF microscopy as described in the Analysis of Ipl1 localization section with or without nocodazole in synthetic complete culture medium. At least 900 cells were categorized per sample across three experimental repeats to calculate a mean and standard deviation.
Analysis of Top2 on chromatin in situ
To analyze Top2 chromatin association, OD600 0.6–0.8 cultures in YPD media supplemented with 20 µg/ml adenine were arrested in 50 µg/ml nocodazole for 2 h at 30°C. Cells from 1 ml of the culture were recovered by brief centrifugation and washed into 100 µl YPD containing 1.2M sorbitol, 40 mM DTT, 15 mM BME and protease inhibitors (1 µg/ml leupeptin, 2 µg/ml aprotinin, 1.6 µg/ml benzamidine, and 1 µg/ml phenylmethylsulfonyl fluoride). 1 µl of a 20-mg/ml solution of Zymolyase (100T; ICN) was added to initiate spheroplasting, and the reaction was incubated for 10 min at 30°C. The optimal time for spheroplasting varied for different preparations of Zymolyase and is a crucial variable. After spheroplasting, 1 ml PBS (150 mM NaCl and 50 mM KPO4 7.5) containing 0.5 M sorbitol and protease inhibitors (PBS-SI) was added to the reaction, and the spheroplasts were recovered by centrifugation for 3 min at 3,000 rpm. The pellet was washed with an additional 1 ml PBS-SI and resuspended in 100 µl PBS-SI. For detergent extraction, 5 µl of 10% Triton X-100 was added to 45 µl of the resuspended spheroplasts. Cell permeabilization was performed at ambient temperature for 5 min, and 1 ml of PBS-SI was added immediately to the sample, which was then centrifuged at 3,000 rpm for 3 min. The resulting pellet was gently rinsed without resuspension using 1 ml PBS-SI and gently resuspended in 20 µl PBS-SI. For microscopy, 5 µl of each sample was spotted onto a 0.1% polylysine-coated slide, along with 5 µl of mounting medium supplemented with DAPI (Vecta-shield; Vector Laboratories). Slide preparations remained suitable for microscopy for at least 3 d when the slides were stored at 4°C.
Statistical analysis
To determine the statistical significance of differences in Ipl1 inner-centromere localization between strains, t tests were performed, yielding the following values: Fig. 3, top2-4 versus wild type (P = 0.0002); Fig. 4, wild type versus top2-4 (P = 0.00063), wild type versus bir1-107 (P = 0.00009), Fig. 5, top2-4 versus top2-4 top2-YF (P = 0.04), Fig. 6, top2ΔCTD versus wild type (P = 0.00004), top2ΔCTD vs. top2ΔCTD top2-YF (P = 0.002); Fig. 8, sgo1Δ vs. wild type (P = 0.00004); Fig. 9, alk1Δ alk2Δ versus wild type (P = 0.0005), hht2-T3A versus wild type (P = 0.0001); Fig. 10, hht2-T3A plus GAL10-HHT2 (0 h vs. 1 h induction, P = 0.2; 2 h induction, P = 0.005; 3 h induction, P = 0.009; 4 h induction, P = 0.2), alk1Δ alk2Δ plus GAL10-hht2-T3E (0 vs. 2 h induction, P = 0.00882), and top2ΔCTD plus GAL10-hht2-T3E (0 h vs. 2 h induction, P = 0.009).
Western blotting of histone H3
Yeast cells were lysed in lysis buffer (1% deoxycholic acid, 1% Triton X-100, 0.1% SDS, 50 mM Tris-HCl, pH 7.5, 1 mM sodium pyrophosphate, and 250 mM NaCl) supplemented with Complete protease inhibitor cocktail (one tablet per 50 ml, #11697498001; Roche), PhosStop phosphatase inhibitor cocktail (one tablet per 10 ml, #04906837001; Roche), and phosphatase inhibitor cocktail 3 (100 µl in 10 ml, #P0044; Sigma-Aldrich). Protein concentration was quantified using the Bio-Rad Protein Assay (#500-00006), and an equal amount of protein from each sample was loaded onto a 12% SDS-PAGE and run for 1.5 h at 150 V on a Bio-Rad Mini Protein II apparatus. The proteins were transferred onto a PVDF membrane using CAPSO transfer buffer (10 mM CAPSO salts, pH 10, and 20% methanol) on a Bio-Rad Mini Protein II transfer apparatus at 150 V for 1 h. Blots were blocked with 3% BSA-TBST for 1 h. Anti-H3 T3 phos antibody (#ab78531, 1:1,000; Abcam), anti-H3 antibody (#ab1791, 1:10,000; Abcam), or anti-tubulin antibody (#ab80779, 1:10,000; Abcam) were diluted in 3% BSA-TBST and used for protein detection. A goat anti–rabbit secondary antibody (HRP conjugated, #31463, 1:15,000; Thermo Fisher Scientific) was used to detect the H3 and H3T3-Phos primary antibodies. A goat anti–mouse secondary antibody (HRP conjugated, #031430, 1:15,000; Pierce) was used to detect the tubulin primary antibody. The secondary antibodies were detected using the SuperSignal West Femto Maximum Sensitivity Substrate (#34095; Thermo Fisher Scientific). ImageJ was used to quantify signals on exposed film.
Online supplemental material
This material includes analysis of Ipl1 inner-centromere localization in interphase cells, categorization and computation of mitotic Ipl1 localization defects, mitotic Ipl1 localization in cells binned by spindle length, analysis of GAL10-HHT2 and GAL10-hht2-T3E induction, and visualization of Alk1-3xGFP and Alk2-3xGFP in live yeast cells.
Acknowledgments
We thank the Clarke, Gardner, Titus, and Luxton laboratories at the University of Minnesota, the Azuma laboratory at the University of Kansas, and the Stark laboratory at the University of Dundee for continued discussions during the course of this work. We are especially grateful to the Azuma laboratory for delaying publication of their study while we completed revisions to our manuscript. We thank S. Biggins, T. Tanaka, T. Davis, J. Berman, M. Stark, C. Moore, G. Zeng, and Z. Storchova for providing strains and reagents.
Funding was provided by National Institutes of Health grant GM112793. J. Bachant acknowledges funding from the California Cancer Research Coordinating Committee that contributed to this work.
The authors declare no competing financial interests.