Talin is a large scaffolding molecule that plays a major role in integrin-dependent cell–matrix adhesion. A role for talin in cell–cell attachment through cadherin has never been demonstrated, however. Here, we identify a novel calpain-dependent proteolytic cleavage of talin that results in the release of a 70-kD C-terminal fragment, which serves as a substrate of posttranslational arginylation. The intracellular levels of this fragment closely correlated with the formation of cell–cell adhesions, and this fragment localized to cadherin-containing cell–cell contacts. Moreover, reintroduction of this fragment rescued the cell–cell adhesion defects in arginyltransferase (Ate1) knockout cells, which normally have a very low level of this fragment. Arginylation of this fragment further enhanced its ability to rescue cell–cell adhesion formation. In addition, arginylation facilitated its turnover, suggesting a dual role of arginylation in its intracellular regulation. Thus, our work identifies a novel proteolytic product of talin that is regulated by arginylation and a new role of talin in cadherin-dependent cell–cell adhesion.
Introduction
Cell adhesion is a key process that regulates such critical biological events as embryogenesis, cell motility, and cancer, and is responsible for maintaining tissue integrity and architecture (Thiery, 2003). Two major types of adhesions in cells are generated by cell attachment to extracellular matrix through a transmembrane protein integrin, and cell attachment to each other through another type of transmembrane protein, cadherin (Wegener and Campbell, 2008; Watanabe et al., 2009). Both types of adhesions depend on the actin cytoskeleton; however, they are believed to be mediated by different protein complexes. Integrin adhesions (including focal adhesions) are mediated by talin, a large protein that scaffolds the key components of the actin cytoskeleton to the integrin complex (Arnaout et al., 2007; Campbell, 2008). The composition of the cadherin complex is less clear (Yamada et al., 2005). Although genetic evidence suggests that talin and cadherin can interact in situ (Kostin et al., 2000), the functional involvement of talin in cadherin-mediated adhesions was never directly demonstrated.
Talin is a rod-like molecule containing multiple sites for binding of vinculin, actin, and integrin (Nayal et al., 2004; Critchley, 2005). Its function in focal adhesions is regulated by calpain-dependent proteolysis between amino acids 433 and 434, with the release of an N-terminal regulatory domain during focal adhesion maturation and turnover (Fox et al., 1985; Franco et al., 2004b). It has been shown that other types of proteases can generate different talin fragments in vitro (Critchley, 2004); however, no other functionally significant talin proteolysis in vivo has been observed. Our recent analysis of proteins arginylated in vivo revealed that talin undergoes arginylation (Wong et al., 2007) and that arginylation-deficient cells with the knockout (KO) of arginyltransferase (Ate1) have adhesion defects (Kurosaka et al., 2010). Here we analyzed the arginylation-dependent talin function and found that talin arginylation is coupled to novel proteolytic processing with the release of a functional C-terminal 70-kD fragment. Because this fragment contains vinculin- and actin-binding sites, and a dimerizing domain, we termed it VAD fragment. Remarkably, we found that this talin fragment plays a major role in the formation of cadherin-mediated cell–cell adhesions, and that arginylation appears to be a novel mechanism that regulates both the function and the half-life of this fragment in vivo.
Results
Talin’s 70-kD VAD fragment is generated in vivo
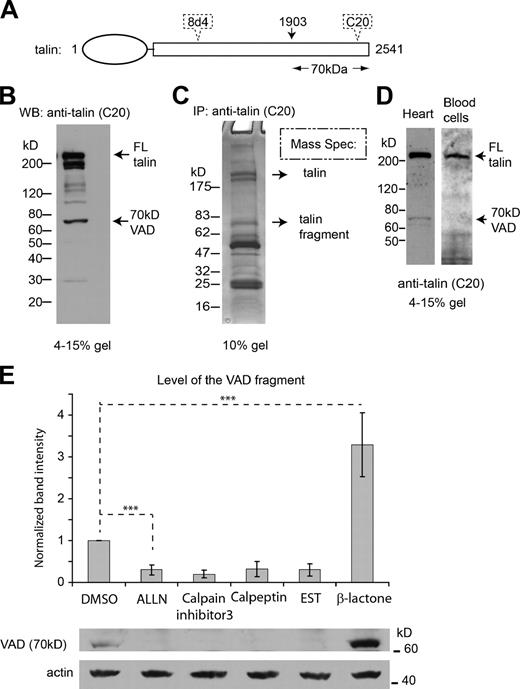
During our global analysis of proteins arginylated in vivo (Wong et al., 2007), we found that talin was arginylated on Ala1903, located in the last ∼1/4 of the molecule. Because arginylation on Ala should require an N-terminally exposed α amino group that could serve as an Arg acceptor site, it must be preceded by proteolysis and is predicted to generate a C-terminal proteolytic product of talin with the molecular weight of ∼70 kD (Fig. 1 A and Fig. S1 A). Because much of the data about protein arginylation were based on analysis of protein samples from immortalized mouse embryonic fibroblasts (MEFs), we went back to test if such a fragment is indeed generated in these cells. To do that, we probed whole-cell lysates from confluent MEFs with the commercially available antibody raised specifically against talin’s C terminus (talin C20 antibody from Santa Cruz Biotechnology, Inc.; see Fig. 1 A and Fig. S1 A for the map of the antibody binding site) and found that, in addition to the full-length talin and its previously characterized 180-kD proteolytic fragment (containing the C-terminal 180-kD portion of talin with the release of the N-terminal FERM domain), such cell lysates contained a prominent 70-kD band, immunoreactive with the talin C20 antibody (Fig. 1 B; see Fig. S1 B for other representative immunoblots showing the variation between preparations). Immuno-pulldowns using talin C20 antibody confirmed that a 70-kD band was consistently present in such preparations in the amounts comparable to the full-length talin band. Mass spectrometry identified talin-derived peptides as the major peptides in both regions excised from the gels (Fig. 1 C). Because disruption of either arginylation or talin function leads to prominent abnormalities of the adhesion structures in tissues and organs, especially the heart (Kostin et al., 2000; Kwon et al., 2002; Rai et al., 2008), we tested whether this VAD fragment also exist in the heart tissue. Indeed, we found a prominent 70-kD talin-derived band in the heart. As a comparison, such a band was absent in circulating blood cells (Fig. 1 D). Thus, a 70-kD talin fragment is generated in vivo, with variations between preparations of cultured cells and different tissue types, suggesting that additional factors are involved in regulating the generation of this fragment in vivo.
A novel 70-kD C-terminal talin fragment is generated in vivo. (A) A map of mouse talin 1, showing the putative cleavage site before Ala 1903, expected to generate a 70-kD fragment. The approximate epitope positions of the two antibodies against the talin protein used in this study, C20 and 8d4, are indicated by dotted outlines. See Fig. S1 A for a more detailed depiction. (B) A representative immunoblot probed with anti-talin C-20 that recognizes the C terminus of talin, showing the cleavage pattern of talin in confluent cultured MEFs. The positions of the full-length (FL) talin and the 70-kD VAD fragment are indicated. See also Fig. S1 B for other blots showing the variation between preparations. (C) A representative SDS-PAGE showing the product from immuno-pulldown with anti-talin C20. The top band and the 70-kD band shown by arrows were analyzed by mass spectrometry to confirm the dominant presence of talin in these two bands. Peptides from mouse talin isoforms 1 and 2 were identified in both bands. (D) Representative immunoblots of the homogenates of mouse heart muscle and circulating blood cells, probed with anti-talin C-20. The 70-kD band is present in the heart but not in the blood. (E) Top: quantification of the level of the 70-kD VAD fragment in cells treated with calpain inhibitors ALLN (50 µM), calpain inhibitor 3 (50 µM), calpeptin (50 µM), and EST (50 µM), and with the proteasome inhibitor β-lactone (3.3 µM) that has no reported crossreactivity with calpain; DMSO was used as a control treatment. Numbers represent the level of the 70-kD band detected by Western blots as shown in the representative image in the bottom panel, quantified against the actin loading control and normalized to that in the DMSO-treated cells. Error bars represent SEM (n = 3). ***, P < 0.01.
A novel 70-kD C-terminal talin fragment is generated in vivo. (A) A map of mouse talin 1, showing the putative cleavage site before Ala 1903, expected to generate a 70-kD fragment. The approximate epitope positions of the two antibodies against the talin protein used in this study, C20 and 8d4, are indicated by dotted outlines. See Fig. S1 A for a more detailed depiction. (B) A representative immunoblot probed with anti-talin C-20 that recognizes the C terminus of talin, showing the cleavage pattern of talin in confluent cultured MEFs. The positions of the full-length (FL) talin and the 70-kD VAD fragment are indicated. See also Fig. S1 B for other blots showing the variation between preparations. (C) A representative SDS-PAGE showing the product from immuno-pulldown with anti-talin C20. The top band and the 70-kD band shown by arrows were analyzed by mass spectrometry to confirm the dominant presence of talin in these two bands. Peptides from mouse talin isoforms 1 and 2 were identified in both bands. (D) Representative immunoblots of the homogenates of mouse heart muscle and circulating blood cells, probed with anti-talin C-20. The 70-kD band is present in the heart but not in the blood. (E) Top: quantification of the level of the 70-kD VAD fragment in cells treated with calpain inhibitors ALLN (50 µM), calpain inhibitor 3 (50 µM), calpeptin (50 µM), and EST (50 µM), and with the proteasome inhibitor β-lactone (3.3 µM) that has no reported crossreactivity with calpain; DMSO was used as a control treatment. Numbers represent the level of the 70-kD band detected by Western blots as shown in the representative image in the bottom panel, quantified against the actin loading control and normalized to that in the DMSO-treated cells. Error bars represent SEM (n = 3). ***, P < 0.01.
It has been previously found that the regulatory proteolysis between residues 433 and 434 of talin during focal adhesion retraction is mediated by calpain (Franco et al., 2004b). To test if calpain is also responsible for the generation of the 70-kD VAD fragment, we used a panel of calpain inhibitors and tested their effect on the intracellular level of the VAD fragment. We found that all the tested inhibitors considerably reduced the level of the fragment, making it virtually undetectable (Fig. 1 E). Because some of these inhibitors have also been suggested to have an effect on the proteasome, we treated a control cell culture with the proteasome inhibitor β-lactone, which has no known effect on calpain. Unlike calpain inhibitors, this treatment did not inhibit the endogenous VAD fragment generation—in fact, the levels of this fragment were increased, suggesting that the proteasome activity may be responsible for the turnover of this fragment. Consistent results were observed in a calpain-4–deficient cell line (calpain-4 [CPN4] KO MEF), which lacks the small regulatory subunit for the two major calpain proteases (calpain-1 and calpain-2; Tan et al., 2006). In these cells the level of the 70-kD VAD fragment was significantly reduced, and this effect was at least partially rescued by reintroduction of calpain-4 (Fig. S1 C). Thus, the regulatory proteolysis responsible for releasing the VAD fragment from the full-length talin is likely mediated by calpain. Out of the two major calpain proteases (calpain-1 and calpain-2) in mammalian cells, calpain-2 is often seen located at the cell periphery and is considered responsible for talin’s proteolytic cleavage at residue 434 (Franco et al., 2004a). To test whether calpain-2 also plays a role in the cleavage at residue 1903 of talin, we knocked down the endogenous calpain-1 and calpain-2 in MEFs by stable transfection with specific small interfering RNAs (siRNAs). We found that the knockdown of calpain-2, but not calpain-1, leads to a decrease in the VAD fragment to levels below those detectable by Western blots (Fig. S1 D), suggesting that calpain-2 is likely mediating the cleavage at Ala 1903 of talin.
Generation of talin’s 70-kD fragment depends on cell density and cell–cell contact area
To test if the generation of the 70-kD talin fragment depends on the cell-physiological state, we tested its presence in confluent and scarce cultures of MEFs by Western blotting and found that although this fragment is prominently present in confluent cells, it is absent, or nearly absent, from scarce cultures plated at single-cell density (Fig. 2 A), suggesting that some characteristics of confluent, and not scarce cells, favor this type of talin proteolysis.
The level of the 70-kD VAD fragment is dependent on cell–cell contact area. (A) The 70-kD band is prominent in confluent cell cultures, but is undetectable in scarce cultures where cells are largely isolated from each other. Actin was used as loading control. (B) The levels of the VAD fragment in confluent cultures plated at different densities. Left, a representative immunoblot probed with anti-talin C-20 is shown for samples prepared from overnight confluent cultures seeded at different densities (1x: 3 million cells; 1.5x: 4.5 million cells; 2x: 6 million cells in 10-cm tissue culture dishes). Right, top: a diagram depicting the increase of the cell–cell contact area with increasing cell density; middle, representative phase-contrast images showing overnight cultures plated at different seeding densities (imaged by 10x lens); bottom, quantification of the level of the VAD fragment in overnight cell cultures with different densities. Numbers represent the intensities of the 70-kD fragment detected on Western blots at different seeding densities, normalized to that of 1x seeding density. Error bars represent SEM (n = 5). *, P < 0.1. See also Fig. S2 for the level of the VAD fragment in cell cultures with different densities cultured for a shorter duration (5 h). (C) Change of the level of the VAD fragment in cells after their adhesions have been disrupted by trypsin. Top: representative images showing the morphologies of MEF before and after a brief treatment with trypsin (which is then neutralized by 10% FBS medium) imaged by 20x lens. At 200 min after the treatment, cells either partially reestablished cell–cell adhesions (semi-confluent) or remained in scarce culture due to an intentional dilution after the trypsin treatment (control: scarce). Middle left and bottom left: representative immunoblots showing the changes of the levels of the VAD fragment during these treatments. The VAD fragment promptly decreased after the cell adhesions were disrupted, and only recovered when the cells were allowed to reestablish cell–cell adhesions, but not when cells were kept in scarce culture at the time point of 200 min. β-Tubulin and actin were used as loading controls. Bottom right: quantifications of the level of the VAD fragment before and after trypsinization. Numbers represent the intensity of the 70-kD fragment detected on Western blots, adjusted to the loading controls and normalized to the values of the samples at 0 min in each group. Error bars represent SEM (n = 4 for cells allowed to enter semi-confluent state at 200 min, n = 3 for cells kept in scarce state at 200 min).
The level of the 70-kD VAD fragment is dependent on cell–cell contact area. (A) The 70-kD band is prominent in confluent cell cultures, but is undetectable in scarce cultures where cells are largely isolated from each other. Actin was used as loading control. (B) The levels of the VAD fragment in confluent cultures plated at different densities. Left, a representative immunoblot probed with anti-talin C-20 is shown for samples prepared from overnight confluent cultures seeded at different densities (1x: 3 million cells; 1.5x: 4.5 million cells; 2x: 6 million cells in 10-cm tissue culture dishes). Right, top: a diagram depicting the increase of the cell–cell contact area with increasing cell density; middle, representative phase-contrast images showing overnight cultures plated at different seeding densities (imaged by 10x lens); bottom, quantification of the level of the VAD fragment in overnight cell cultures with different densities. Numbers represent the intensities of the 70-kD fragment detected on Western blots at different seeding densities, normalized to that of 1x seeding density. Error bars represent SEM (n = 5). *, P < 0.1. See also Fig. S2 for the level of the VAD fragment in cell cultures with different densities cultured for a shorter duration (5 h). (C) Change of the level of the VAD fragment in cells after their adhesions have been disrupted by trypsin. Top: representative images showing the morphologies of MEF before and after a brief treatment with trypsin (which is then neutralized by 10% FBS medium) imaged by 20x lens. At 200 min after the treatment, cells either partially reestablished cell–cell adhesions (semi-confluent) or remained in scarce culture due to an intentional dilution after the trypsin treatment (control: scarce). Middle left and bottom left: representative immunoblots showing the changes of the levels of the VAD fragment during these treatments. The VAD fragment promptly decreased after the cell adhesions were disrupted, and only recovered when the cells were allowed to reestablish cell–cell adhesions, but not when cells were kept in scarce culture at the time point of 200 min. β-Tubulin and actin were used as loading controls. Bottom right: quantifications of the level of the VAD fragment before and after trypsinization. Numbers represent the intensity of the 70-kD fragment detected on Western blots, adjusted to the loading controls and normalized to the values of the samples at 0 min in each group. Error bars represent SEM (n = 4 for cells allowed to enter semi-confluent state at 200 min, n = 3 for cells kept in scarce state at 200 min).
To address the question of which properties of the confluent cells are responsible for this differential talin processing, we considered the following differences between the confluent and scarce cultures: (i) lack of persistent directional motility in confluent monolayers; (ii) inhibition of cell proliferation in confluent cells; and (iii) a considerable decrease of cell–cell contact area in scarce cells. To address the first two possibilities, we plated cells at a low density and transferred them to serum-free medium—a treatment that inhibits both cell proliferation and cell motility. Comparison of such scarce serum-starved cells to the control scarce cultures grown at normal serum concentrations showed that serum starvation did not result in an increase in the talin 70-kD fragment levels (Fig. S2), suggesting that the generation of this fragment is not a direct consequence of reduced cell motility or proliferation.
To test whether the generation of the 70-kD VAD fragment correlates with the cell–cell contact area, we plated MEFs at increasing densities, thus forcibly increasing the cell–cell contact area in the monolayer (see the diagram in Fig. 2 B). We found that such increase in cell density resulted in a dose-dependent increase in the level of the 70-kD VAD fragment (Fig. 2 B and Fig. S3). These data suggest that the generation of this fragment is directly proportional to cell density and likely depends on cell–cell contact area. To test this further, we probed the level of the VAD fragment in a confluent cell culture recovering after trypsinization, where the cell contacts are first forcibly broken by trypsin digestion, and then reestablish after cell attachment and spreading. In this experiment, the VAD fragment, initially present at high levels, gradually disappeared after trypsinization and reattachment, and then came back again after the cells reached a semi-confluent state and reestablished cell–cell contacts (Fig. 2 C). If the cells after respreading were plated scarcely, so that the cell–cell contacts could not reestablish, the levels of the VAD fragment remained low (Fig. 2 C). Thus, the VAD fragment generation indeed depends on the establishment of cell–cell contacts, and not on cell adhesion to the substrate.
Talin’s 70-kD VAD fragment is localized to cadherin-containing cell–cell adhesion sites
To test directly whether the VAD fragment associates with cadherin-dependent cell–cell adhesions, we probed the intracellular localization of the recombinant GFP-fused VAD fragment and found that it localized to the prominent plaques mainly at the cell periphery but not in the central regions of the cell (Fig. 3 A, top center and top right). Many spots with the VAD-GFP signal at least partially colocalized with the established focal adhesion marker anti-talin 8d4 (raised against an epitope in the middle of talin and recognizing both the full-length and the conventional calpain cleavage product of talin, but not the VAD fragment; see Fig. 1 A and Fig. S1 A for the map of the antibody-binding site). However, a prominent subset of VAD-containing spots localized to distinct sites at the borders between cells, from which anti-talin 8d4 signal was prominently absent (Fig. 3 A, bottom panels). Co-staining of integrin β1, a marker for integrin-mediated adhesions, generated similar results: the VAD-GFP localized to the cell borders where the integrin staining was absent, suggesting that those sites were not the conventional focal adhesions (Fig. 3 B). To test whether such sites represent true cell–cell adhesions, we compared the VAD-GFP distribution with anti-cadherin staining (Fig. 3 B) and found that VAD-GFP prominently colocalizes with cadherin at the sites of cell–cell interface (Fig. 4 A, arrows), confirming that the VAD fragment at least in some cases is associated with cadherin cell–cell adhesion sites.
The VAD fragment, unlike full-length talin, partially localizes at integrin-independent cell–cell contact areas. (A) Comparison of the localization of VAD-GFP fusion (green) and anti-talin 8d4 that recognizes full-length talin but not the VAD fragment (red). Top panels: VAD-GFP only colocalizes with anti-talin 8d4 at the cell periphery (arrowheads) but not in the central regions (in which the anti-talin staining may represent fibrillar adhesions, a mature form of focal adhesions); VAD-GFP also localizes to the borders between cells, devoid of anti-talin (8d4) staining (arrows). Bottom panels: representative photos showing the VAD-GFP incorporated into dots at the cell–cell contact regions (arrowheads), from which the staining by anti-talin (8d4) is prominently absent, suggesting that those are not conventional focal adhesion sites. (B) Comparison of the localization of VAD-GFP (green) and anti-integrin β1 (localized at the sites of cell adhesion to the extracellular matrix, red). White arrows point to a line of zipper-like structures along the cell–cell border that contain VAD-GFP but not integrin. All the fluorescence images here were acquired with a 40x dry lens.
The VAD fragment, unlike full-length talin, partially localizes at integrin-independent cell–cell contact areas. (A) Comparison of the localization of VAD-GFP fusion (green) and anti-talin 8d4 that recognizes full-length talin but not the VAD fragment (red). Top panels: VAD-GFP only colocalizes with anti-talin 8d4 at the cell periphery (arrowheads) but not in the central regions (in which the anti-talin staining may represent fibrillar adhesions, a mature form of focal adhesions); VAD-GFP also localizes to the borders between cells, devoid of anti-talin (8d4) staining (arrows). Bottom panels: representative photos showing the VAD-GFP incorporated into dots at the cell–cell contact regions (arrowheads), from which the staining by anti-talin (8d4) is prominently absent, suggesting that those are not conventional focal adhesion sites. (B) Comparison of the localization of VAD-GFP (green) and anti-integrin β1 (localized at the sites of cell adhesion to the extracellular matrix, red). White arrows point to a line of zipper-like structures along the cell–cell border that contain VAD-GFP but not integrin. All the fluorescence images here were acquired with a 40x dry lens.
The VAD fragment, unlike full-length talin, colocalizes with cadherin-containing cell–cell contact sites. (A) Comparison of the localization of VAD-GFP (green) and cadherin (red) at the region around cell–cell borders. White arrows point to a few locations where VAD-GFP and cadherin staining overlaps. (B) Anti-talin C20, which recognizes both the full-length endogenous talin and its VAD fragment (green), partially colocalizes with cadherin (red) in cell–cell contact areas (indicated by white arrows). (C) Full-length talin (green, represented by recombinant full-length talin protein fused with GFP on its N terminus) does not show prominent colocalization with cadherin (red). Asterisks were used to mark the positions for comparison. All the fluorescent images here were acquired with a 40x dry lens.
The VAD fragment, unlike full-length talin, colocalizes with cadherin-containing cell–cell contact sites. (A) Comparison of the localization of VAD-GFP (green) and cadherin (red) at the region around cell–cell borders. White arrows point to a few locations where VAD-GFP and cadherin staining overlaps. (B) Anti-talin C20, which recognizes both the full-length endogenous talin and its VAD fragment (green), partially colocalizes with cadherin (red) in cell–cell contact areas (indicated by white arrows). (C) Full-length talin (green, represented by recombinant full-length talin protein fused with GFP on its N terminus) does not show prominent colocalization with cadherin (red). Asterisks were used to mark the positions for comparison. All the fluorescent images here were acquired with a 40x dry lens.
To further test this conclusion, we compared the intracellular distribution of talin stained by C20 antibody (which reacts with the talin’s C terminus and therefore stains both the full-length talin and its VAD fragment) and cadherin (Fig. 4 B). We found a number of prominent sites where these two types of staining overlapped, suggesting that a portion of the endogenous talin (likely its VAD fragment) indeed localizes to the cadherin cell–cell adhesion sites. However, when talin with GFP fused to its N terminus was used to visualize the localization of the full-length talin, it didn’t show any prominent colocalization with cadherin (Fig. 4 C), suggesting that localization to cell–cell adhesions is specific to the proteolytic product of talin.
Talin’s 70-kD VAD fragment is essential for cadherin-mediated cell–cell adhesions
We have previously reported that knockout of arginyltransferase Ate1 results in a reduction in cell spreading and the size and the number of focal adhesions (Karakozova et al., 2006; Kurosaka et al., 2010). To test whether Ate1 KO cells also have a reduction in cell–cell contacts, we compared cell contact area and morphology in confluent wild-type (WT) and Ate1 KO MEFs, using cadherin as a marker. Although the WT MEF formed prominent cadherin-containing cell–cell adhesions in confluent culture, Ate1 KO cells formed fewer cell–cell adhesions and the size and number of cadherin-containing plaques in these cells was noticeably reduced (Fig. 5, A and B), despite the fact that these cells had similar levels of cadherin, vinculin, β-catenin, and talin compared with the control (Fig. S4; Kurosaka et al., 2010). To test whether the reduced cell adhesion in these cells also correlates with the reduced levels of the VAD fragment, we compared the intracellular level of this fragment in WT MEFs to those in Ate1 KO cells in confluent and scarce cultures. Surprisingly, we found that the level of the VAD fragment in Ate1 KO cells was negligibly low or absent regardless of the cell density (Fig. 5 A), suggesting that arginylation is essential for maintaining the high intracellular level of this fragment in confluent cells. Treatment of confluent Ate1 KO cells with the proteasome inhibitor β-lactone did not increase the level of the endogenous VAD fragment (Fig. S4 B), suggesting the lack of this fragment in these cells is not due to its preferential degradation in the absence of ATE1. Because the VAD fragment is likely generated by calpain cleavage (Fig. 1 E), we tested the hypothesis that Ate1 knockout inhibits the calpain-dependent proteolysis. To do this, we checked the activity of calpain-dependent proteolysis by the spectrin cleavage assay, by probing extracts from these cells with an antibody specific to the calpain-dependent proteolytic product of spectrin that does not recognize the full-length protein (Roberts-Lewis et al., 1994; Simpkins et al., 2003), and found that calpain-dependent spectrin proteolysis (seen by the abundance of spectrin’s proteolytic product) was significantly lower in Ate1 KO cells compared with the control (Fig. S4 C). At the same time, the intracellular levels of the major calpain isoforms were unchanged in Ate1 KO compared with WT (Fig. S4 D). Thus, lack of arginylation inhibits the activity of calpain, and the absence of the VAD fragment in Ate1 KO cells is likely due to this inhibition.
The VAD fragment at least partially rescues the cell–cell adhesion defects in Ate1 knockout cells. (A) Left: a representative immunoblot probed with anti-talin C-20, showing the cleavage pattern of talin in wild-type (WT) and arginyltransferase1 (Ate1) knockout (KO) cells. The position of the full-length talin and the VAD fragment are indicated by arrows. The VAD fragment is notably absent in Ate1 KO cells regardless of culture densities. β-Tubulin was used as loading control. Right: representative fluorescence images showing the cell–cell adhesions stained by anti-cadherin in confluent cultures of WT and Ate1 KO cells imaged with a 40x dry lens. (B) Left: representative fluorescence images showing cadherin-mediated cell–cell adhesions in WT and Ate1 KO MEFs stably transfected with GFP or VAD-GFP as indicated, imaged with a 40x dry lens. White arrows point to the locations of a few cadherin-containing plaques in each cell. Right: quantification of the sizes of individual cadherin-containing plaques in cells transfected with GFP or VAD-GFP. In each group, 15 fluorescent images containing at least 1,000 properly focused cadherin-positive cell–cell contact spots were analyzed, and average size of the cadherin plaques calculated from each image were pooled together. Numbers in the graph represent the average size of cadherin plaques from all images. Error bars represent SEM (n = 15). ***, P < 0.01. (C) Left: representative time-lapse fluorescence images of MEFs, stably transfected with N-cadherin–mCherryFP and GFP for wild-type and Ate1 KO cells (WT:GFP and KO:GFP, respectively) and Ate1 KO cells stably transfected with VAD-GFP (KO:VAD-GFP) imaged with a 60x water immersion lens with 2 × 2 binning in acquisition. The dynamics of the cadherin-containing punctae in those cells was tested by FRAP. Right: the graph shows the times of half-recovery of the original intensity after bleaching (t1/2), used as an indicator of cadherin dynamics. WT:GFP and KO:VAD-GFP had larger t1/2 value than the KO:GFP, indicating that their cadherin puncta were more stable. Error bars represent SEM (n = 11 for WT:GFP, n = 7 for Ate1 KO:GFP, n = 6 for Ate1:VAD-GFP). *, P < 0.1, **, P < 0.05.
The VAD fragment at least partially rescues the cell–cell adhesion defects in Ate1 knockout cells. (A) Left: a representative immunoblot probed with anti-talin C-20, showing the cleavage pattern of talin in wild-type (WT) and arginyltransferase1 (Ate1) knockout (KO) cells. The position of the full-length talin and the VAD fragment are indicated by arrows. The VAD fragment is notably absent in Ate1 KO cells regardless of culture densities. β-Tubulin was used as loading control. Right: representative fluorescence images showing the cell–cell adhesions stained by anti-cadherin in confluent cultures of WT and Ate1 KO cells imaged with a 40x dry lens. (B) Left: representative fluorescence images showing cadherin-mediated cell–cell adhesions in WT and Ate1 KO MEFs stably transfected with GFP or VAD-GFP as indicated, imaged with a 40x dry lens. White arrows point to the locations of a few cadherin-containing plaques in each cell. Right: quantification of the sizes of individual cadherin-containing plaques in cells transfected with GFP or VAD-GFP. In each group, 15 fluorescent images containing at least 1,000 properly focused cadherin-positive cell–cell contact spots were analyzed, and average size of the cadherin plaques calculated from each image were pooled together. Numbers in the graph represent the average size of cadherin plaques from all images. Error bars represent SEM (n = 15). ***, P < 0.01. (C) Left: representative time-lapse fluorescence images of MEFs, stably transfected with N-cadherin–mCherryFP and GFP for wild-type and Ate1 KO cells (WT:GFP and KO:GFP, respectively) and Ate1 KO cells stably transfected with VAD-GFP (KO:VAD-GFP) imaged with a 60x water immersion lens with 2 × 2 binning in acquisition. The dynamics of the cadherin-containing punctae in those cells was tested by FRAP. Right: the graph shows the times of half-recovery of the original intensity after bleaching (t1/2), used as an indicator of cadherin dynamics. WT:GFP and KO:VAD-GFP had larger t1/2 value than the KO:GFP, indicating that their cadherin puncta were more stable. Error bars represent SEM (n = 11 for WT:GFP, n = 7 for Ate1 KO:GFP, n = 6 for Ate1:VAD-GFP). *, P < 0.1, **, P < 0.05.
To test whether the lack of the VAD fragment in Ate1 KO cells is at least partially responsible for the defects in cadherin-mediated cell–cell adhesions (and/or focal adhesions) in these cells, we investigated whether reintroduction of the VAD fragment into Ate1 KO cells results in a rescue of their cell–cell or focal adhesions, by transfecting Ate1 KO cells with either GFP alone, or GFP-fused VAD fragment. Remarkably, although such transfections had no apparent effect on focal adhesions (Fig. S5), reintroduction of the VAD fragment into Ate1 KO cells resulted in a significant increase of the size of cadherin-containing plaques (Fig. 5 B). A similar rescue effect was observed when we probed the dynamics of the cell–cell contacts by monitoring the recovery of the RFP (mCherrryFP)-fused N-cadherin (the major cadherin isoform in fibroblasts) in cell–cell contact area after photobleaching. We found that the cadherin punctae in Ate1 KO cells had shorter recovery half-time after bleaching than those in WT cells, and such a difference was abolished after reintroduction of the VAD fragment into the KO cells (Fig. 5 C), suggesting that stable association of cadherin with cell–cell adhesion sites is at least partially dependent on the VAD fragment. Thus, the VAD fragment is an important player in cadherin-dependent cell–cell adhesion.
It is known that actin polymerization is essential for the stabilization of the adhesion complex mediated by cadherin (Cavey et al., 2008). The VAD fragment contains several functional domains that can directly or indirectly promote actin polymerization. Thus, one of the possible mechanisms of the VAD-dependent cadherin adhesion rescue is in inducing localized actin polymerization at the cadherin adhesion sites, which would then stabilize such sites and mediate the recruitment of essential cell–cell adhesion components. To test if this mechanism is likely responsible for the VAD-dependent rescue effect, we looked at the actin distribution at cell–cell adhesion sites and found that the size of the actin patches adjacent to the cadherin plaques in the cell–cell contact sites in Ate1 KO cells was significantly reduced compared with the WT cells. This defect was at least partially rescued after reintroduction of the VAD fragment (Fig. 6 A). Treatment with proteasome inhibitor MG132 significantly increased the cellular levels of the GFP-VAD fragment and such increase resulted in a proportional increase of F-actin in the cell, suggesting that the VAD fragment indeed enhances actin polymerization in vivo (Fig. 6 B). Thus, the VAD-dependent rescue effect on cadherin cell–cell adhesions is likely mediated, at least in part, by increased actin polymerization after its localization to these sites.
The VAD fragment likely stabilizes the cadherin-mediated adhesions by enhancing actin polymerization in situ. (A) Left: representative fluorescence images showing the actin patch at the cadherin-containing cell–cell contact site in WT MEFs stably transfected with GFP (left) and Ate1 KO MEFs stably transfected with GFP (middle) or VAD-GFP (right) imaged with a 40x dry lens. Right: quantification of the average size of the actin patches in the cadherin-containing cell–cell contacts in WT cells stably transfected with GFP, Ate1 KO cells stably transfected with GFP or VAD-GFP. The measurement was performed in more than 20 cells from 9 images for each group. *, P < 0.1; ***, P < 0.001. (B) Left: immunoblots showing the levels of the GFP and VAD-GFP in Ate1 KO cells treated with MG132 for different durations. The level of the VAD fragment, but not GFP, was specifically increased by this treatment. Right: immunoblots with actin antibody showing the level of total actin protein in cell lysates (input) and high speed pellets representing actin polymer in cells. Increase of the VAD-GFP leads to a proportional increase in polymerized actin.
The VAD fragment likely stabilizes the cadherin-mediated adhesions by enhancing actin polymerization in situ. (A) Left: representative fluorescence images showing the actin patch at the cadherin-containing cell–cell contact site in WT MEFs stably transfected with GFP (left) and Ate1 KO MEFs stably transfected with GFP (middle) or VAD-GFP (right) imaged with a 40x dry lens. Right: quantification of the average size of the actin patches in the cadherin-containing cell–cell contacts in WT cells stably transfected with GFP, Ate1 KO cells stably transfected with GFP or VAD-GFP. The measurement was performed in more than 20 cells from 9 images for each group. *, P < 0.1; ***, P < 0.001. (B) Left: immunoblots showing the levels of the GFP and VAD-GFP in Ate1 KO cells treated with MG132 for different durations. The level of the VAD fragment, but not GFP, was specifically increased by this treatment. Right: immunoblots with actin antibody showing the level of total actin protein in cell lysates (input) and high speed pellets representing actin polymer in cells. Increase of the VAD-GFP leads to a proportional increase in polymerized actin.
Arginylation of the VAD fragment regulates its physiological functions as well as turnover
In our previous screen for arginylated proteins, we found the VAD fragment arginylated on its N terminus in vivo (Wong et al., 2007), suggesting that in addition to the generation of the VAD fragment, arginylation also regulates its properties in the cell, either by changing the function, localization, or the metabolic fate of this fragment, as seen with other arginylated proteins (Varshavsky, 1997; Davydov and Varshavsky, 2000; Karakozova et al., 2006; Carpio et al., 2010). To test this hypothesis, we tested the effects of expression of the arginylated VAD fragment compared with the nonarginylated one by expressing GFP fusion constructs in cultured cells (Fig. 7 A). During such expression, arginylated VAD (R-VAD) proved to be much less abundant than nonarginylated VAD at stationary levels, an effect that could be countered by treatment with the proteasome inhibitor MG132 (Fig. 7 B), suggesting that this fragment in vivo is preferentially degraded by the proteasome. We have recently shown (Zhang et al., 2010) that differences in the stationary levels between the arginylated and nonarginylated proteins could be due either to the differences in their cotranslational processing (an effect, which is irrelevant to the R-VAD fragment function in vivo because the endogenous talin VAD fragment is not translated de novo, but is generated through posttranslational proteolysis), or to differences in their posttranslational degradation rates. To test if the posttranslational degradation rates are different for these two fragments, we tested changes in their levels over time in cells treated with cycloheximide to inhibit de novo protein synthesis. We found that under such conditions the decrease of the arginylated VAD fragment is only moderately faster than the nonarginylated species with a half-life for both fragments estimated to be more than 5 h, which should allow sufficient time for exerting their physiological functions (Fig. 7 C). The intracellular distribution of R-VAD-GFP proved to be very similar to VAD-GFP, with similar extent of localization to cadherin-positive cell–cell contacts, suggesting that arginylation does not change the intracellular targeting of the VAD fragment to the cell adhesion structures (Fig. 7 D).
Arginylation regulates VAD fragment turnover but does not affect its intracellular localization. (A) Diagram describing the recombinant proteins used to mimic arginylated and nonarginylated VAD fragment in cells. The N-terminal ubiquitin moiety is cotranslationally removed, exposing the N-terminal amino acid residues indicated in the diagram. (B) Representative immunoblots showing the stationary levels of the GFP-fused nonarginylated and arginylated VAD fragments (VAD-GFP and R-VAD-GFP, respectively) in transiently transfected MEFs treated with proteasome inhibitor MG132 or DSMO (as control) for 5 h. Actin was used as a loading control. Both proteins accumulate in the presence of MG132, suggesting that they both undergo proteasome degradation; however, the differences in their stationary levels in control cells and the degree of their accumulation in the presence of the proteasome inhibitor suggest that R-VAD is much more unstable in vivo, due to cotranslational or posttranslational degradation. (C) Representative immunoblots showing the levels of VAD and R-VAD-GFP in transiently transfected HEK 293T cells during treatment with 100 µg/ml cycloheximide to inhibit de novo protein expression and detect posttranslational degradation. Actin was used as a loading control. Right, a graph showing the posttranslational degradation dynamics of VAD-GFP and R-VAD-GFP as the change of their levels in the presence of cycloheximide over time. Error bar represents SEM (n = 4). (D) Localization of the VIA-GFP and R-VIA-GFP in stably transfected WT MEFs after treatment with MG132 for 6 h to induce accumulation of the transfected proteins and facilitate their detection. Top panels: fluorescence images showing the GFP signal of the VIA-GFP and R-VIA-GFP. Both proteins are mainly localized to the periphery of the cells as well as the borders between cells, as indicated by arrows. Bottom panel: fluorescence images showing the cell–cell adhesion sites stained with anti-cadherin (left, red), R-VIA-GFP detected by staining with anti-GFP (middle, green), and the merged images showing the partial colocalization of these two markers. All images were acquired with a 40x dry lens. White arrows indicate selected spots of colocalization.
Arginylation regulates VAD fragment turnover but does not affect its intracellular localization. (A) Diagram describing the recombinant proteins used to mimic arginylated and nonarginylated VAD fragment in cells. The N-terminal ubiquitin moiety is cotranslationally removed, exposing the N-terminal amino acid residues indicated in the diagram. (B) Representative immunoblots showing the stationary levels of the GFP-fused nonarginylated and arginylated VAD fragments (VAD-GFP and R-VAD-GFP, respectively) in transiently transfected MEFs treated with proteasome inhibitor MG132 or DSMO (as control) for 5 h. Actin was used as a loading control. Both proteins accumulate in the presence of MG132, suggesting that they both undergo proteasome degradation; however, the differences in their stationary levels in control cells and the degree of their accumulation in the presence of the proteasome inhibitor suggest that R-VAD is much more unstable in vivo, due to cotranslational or posttranslational degradation. (C) Representative immunoblots showing the levels of VAD and R-VAD-GFP in transiently transfected HEK 293T cells during treatment with 100 µg/ml cycloheximide to inhibit de novo protein expression and detect posttranslational degradation. Actin was used as a loading control. Right, a graph showing the posttranslational degradation dynamics of VAD-GFP and R-VAD-GFP as the change of their levels in the presence of cycloheximide over time. Error bar represents SEM (n = 4). (D) Localization of the VIA-GFP and R-VIA-GFP in stably transfected WT MEFs after treatment with MG132 for 6 h to induce accumulation of the transfected proteins and facilitate their detection. Top panels: fluorescence images showing the GFP signal of the VIA-GFP and R-VIA-GFP. Both proteins are mainly localized to the periphery of the cells as well as the borders between cells, as indicated by arrows. Bottom panel: fluorescence images showing the cell–cell adhesion sites stained with anti-cadherin (left, red), R-VIA-GFP detected by staining with anti-GFP (middle, green), and the merged images showing the partial colocalization of these two markers. All images were acquired with a 40x dry lens. White arrows indicate selected spots of colocalization.
To test the effect of arginylation on the physiological function of the VAD fragment, we compared the phenotype rescue effects by the VAD and R-VAD fragments on cell–cell and focal adhesions in Ate1 KO cells. To counter the effect produced by differences in their intracellular levels and to test any dose-dependence of their action in the cells, we used proteasome inhibitor MG132 to induce accumulation of the transfected VAD and R-VAD fragments for different amounts of time, from 0 to 7 h (Fig. 8 A). Under these conditions, neither fragment had any significant effect in rescuing the focal adhesion defects in Ate1 KO cells (Fig. S5). However, both the VAD and R-VAD fragments were able to increase the size of the cadherin-containing cell–cell adhesions in a similar dose-dependent manner when compared with the GFP control group (Fig. 8, B and C). Moreover, increasing levels of the R-VAD fragment caused a significant increase of the density of cadherin plaques, which was not seen with the VAD fragment (Fig. 8, B and C). This improved rescue in the presence of arginylated VAD fragment resulted in an improvement in the overall morphology of the confluent Ate1 KO cell monolayer, making it appear more similar to the WT culture (Fig. 8 B, bottom right). Thus, arginylation facilitates the function of the talin VAD fragment in establishing and/or maintaining cell–cell adhesions.
Arginylation of the VAD fragment regulates its physiological function. (A) Accumulation of VAD-GFP and R-VAD-GFP in stably transfected Ate1 KO cells treated with MG132 for different time intervals. In “0 h” of MG132, 5 h treatment with DMSO was used. The levels of the GFP fusion proteins were detected using anti-GFP. Actin was used as a loading control. (B) Representative fluorescence images showing the morphology of the cadherin-stained cell–cell adhesions in WT and Ate1 KO cells stably transfected with GFP, VAD-GFP, or R-VAD-GFP as indicated, and either treated with DMSO for 5 h (MG132: 0 h) or with MG132 for 5 h and 7 h as indicated, and then allowed to recover overnight for 16 h, imaged by a 40x dry lens. White arrows point to a few cadherin plaques along the cell–cell borders in each picture. (C). Quantification of the average size of the cadherin plaques in Ate1 KO cells transfected with VAD-GFP, R-VAD-GFP, or GFP control and treated with MG132 for the indicated time intervals as described in B. The measurement was performed in more than 30 cells from 15 images for each group. (D) Quantification of the cadherin plaque density in cells stably transfected with different protein constructs (VAD-GFP, R-VAD-GFP, and GFP alone) and treated with MG132 for different time intervals as described in B. The numbers represent average coverage percentage on lines drawn across the cell borders along the cadherin plaques with the intensity and size above the threshold. The measurement was performed in more than 30 cells from 15 images for each group. **, P < 0.05; ***, P < 0.01.
Arginylation of the VAD fragment regulates its physiological function. (A) Accumulation of VAD-GFP and R-VAD-GFP in stably transfected Ate1 KO cells treated with MG132 for different time intervals. In “0 h” of MG132, 5 h treatment with DMSO was used. The levels of the GFP fusion proteins were detected using anti-GFP. Actin was used as a loading control. (B) Representative fluorescence images showing the morphology of the cadherin-stained cell–cell adhesions in WT and Ate1 KO cells stably transfected with GFP, VAD-GFP, or R-VAD-GFP as indicated, and either treated with DMSO for 5 h (MG132: 0 h) or with MG132 for 5 h and 7 h as indicated, and then allowed to recover overnight for 16 h, imaged by a 40x dry lens. White arrows point to a few cadherin plaques along the cell–cell borders in each picture. (C). Quantification of the average size of the cadherin plaques in Ate1 KO cells transfected with VAD-GFP, R-VAD-GFP, or GFP control and treated with MG132 for the indicated time intervals as described in B. The measurement was performed in more than 30 cells from 15 images for each group. (D) Quantification of the cadherin plaque density in cells stably transfected with different protein constructs (VAD-GFP, R-VAD-GFP, and GFP alone) and treated with MG132 for different time intervals as described in B. The numbers represent average coverage percentage on lines drawn across the cell borders along the cadherin plaques with the intensity and size above the threshold. The measurement was performed in more than 30 cells from 15 images for each group. **, P < 0.05; ***, P < 0.01.
Discussion
Our study identifies a new type of posttranslational processing of talin and a novel role of this processing in cadherin-mediated cell–cell adhesion. Talin has been conventionally considered exclusively associated with the integrin-dependent cell–matrix adhesions as the major scaffolding and regulatory protein (Nayal et al., 2004). Our study demonstrates for the first time that the C-terminal VAD fragment of talin plays a major role in cadherin-dependent cell–cell adhesion, revealing a previously unknown function of talin in cell adhesion. This is also the first demonstration of a functional proteolytic product of a protein regulated by arginylation (Fig. 9 A).
Talin’s novel VAD fragment regulated by an arginylation-dependent mechanism stabilizes cadherin-mediated cell–cell adhesions by enhancing actin polymerization in situ. (A) Multiple levels of talin and the VAD fragment regulation by Ate1, associated with the formation of the cadherin-mediated adherens junctions. The VAD fragment, a newly discovered talin cleavage product, is generated by calpain, and is involved in the formation of the cadherin-mediated cell–cell adhesions (adherens junctions). Ate1 regulates this fragment at three different levels: (1) its generation (likely through its previously demonstrated modification of calmodulin; Wong et al., 2007), (2) its physiological function, and (3) its turnover, and appears to play a central role in this hierarchical order. Additional pathways, represented by dotted lines, may be involved in Ate1-dependent regulation of cell adhesion, but have not been directly demonstrated. (B) The generation of the VAD fragment is likely coupled to the force-induced conformation change of the talin molecule that exposes the hidden calpain-sensitive sites. The talin molecule contains a strong integrin binding site on its N-terminal FERM domain and two strong F-actin binding sites (marked with “A”) in the middle of the rod domain and in the C terminus, respectively. The scheme illustrates a possible scenario in which the three anchorage sites are bound to different components in the cell. The competition between the nascent integrin and cadherin-mediated adhesions near the cell–cell contact region exerts stress force on the talin molecule to expose the protease-sensitive sites at aa 434 (which will generate classic calpain cleavage site that are required for turnover of focal adhesions) and at aa1903 (which will generate the VAD fragment as observed in this study that has an essential role in stabilizing the cadherin-mediated cell–cell adhesions).
Talin’s novel VAD fragment regulated by an arginylation-dependent mechanism stabilizes cadherin-mediated cell–cell adhesions by enhancing actin polymerization in situ. (A) Multiple levels of talin and the VAD fragment regulation by Ate1, associated with the formation of the cadherin-mediated adherens junctions. The VAD fragment, a newly discovered talin cleavage product, is generated by calpain, and is involved in the formation of the cadherin-mediated cell–cell adhesions (adherens junctions). Ate1 regulates this fragment at three different levels: (1) its generation (likely through its previously demonstrated modification of calmodulin; Wong et al., 2007), (2) its physiological function, and (3) its turnover, and appears to play a central role in this hierarchical order. Additional pathways, represented by dotted lines, may be involved in Ate1-dependent regulation of cell adhesion, but have not been directly demonstrated. (B) The generation of the VAD fragment is likely coupled to the force-induced conformation change of the talin molecule that exposes the hidden calpain-sensitive sites. The talin molecule contains a strong integrin binding site on its N-terminal FERM domain and two strong F-actin binding sites (marked with “A”) in the middle of the rod domain and in the C terminus, respectively. The scheme illustrates a possible scenario in which the three anchorage sites are bound to different components in the cell. The competition between the nascent integrin and cadherin-mediated adhesions near the cell–cell contact region exerts stress force on the talin molecule to expose the protease-sensitive sites at aa 434 (which will generate classic calpain cleavage site that are required for turnover of focal adhesions) and at aa1903 (which will generate the VAD fragment as observed in this study that has an essential role in stabilizing the cadherin-mediated cell–cell adhesions).
Consistent with the newly discovered role of talin in cell–cell adhesions, we find a high level of the VAD fragment in heart muscle but not in circulating blood cells that form no stable connections to each other and exist in a suspended state. It appears likely that the generation of this fragment occurs only in those tissues where cell–cell adhesion plays a prominent role. Perturbing talin’s function in heart cells is associated with abnormalities in cell–cell attachment and heart muscle integrity (Belkin et al., 1986; Kostin et al., 2000). In addition, talin (or a talin fragment) was detected in the intercalated discs (cadherin-dominant adhesions between cardiomyocytes) when a polyclonal antisera raised against full-length talin protein was used (Belkin et al., 1986). However, the interaction between talin and the cadherin-mediated junctions in heart tissue remained unclear (Zuppinger et al., 2000). Our study opens up a novel possibility that talin’s function may also be mediated by the formation of its C-terminal VAD fragment that plays an important role in maintaining the cardiac structure.
We have demonstrated that the VAD fragment is incorporated into the cadherin-mediated cell–cell adhesion complex. However, direct binding between talin, or any of its fragments, with cadherin was not shown (or ever been looked into), and this possibility deserves further investigation. At the same time, the presence of the vinculin-binding site in the VAD fragment suggests that this fragment can interact with other components of the cadherin complex that have been shown to bind vinculin (e.g., β-catenin), and this interaction may indirectly target the VAD fragment to cell–cell adhesions. These possibilities constitute exciting future research directions.
The VAD fragment also contains a reported binding site for integrin, albeit one with low affinity, leading to a speculation on whether this fragment is relevant to integrin-mediated focal adhesions. However, studies of the spatial composition of the focal adhesion complex show that only the N-terminal region (that contains a high affinity site for integrin), but not the C-terminal region of talin, directly binds integrin (Kanchanawong et al., 2010). In our study, the endogenous VAD fragment was nearly absent from scarce cells, despite the dominance of integrin-mediated cell–matrix adhesions in these cells. Furthermore, the recombinant VAD fragment did not rescue the defects of focal adhesions in Ate1 KO cells (where the endogenous VAD fragment is absent), suggesting either that the VAD fragment is not a major player in focal adhesions or that additional factors affected by arginylation are also required in mediating its focal adhesion–dependent functions. Further studies may be needed to elucidate these possibilities. Interestingly, evidence from protein domain evolution suggests that the C-terminal domains of talin evolved much earlier than either integrin or cadherin, because in yeast a protein carrying a similar domain already exists and is essential for the formation of the actin patches near the endocytic sites (Baggett et al., 2003). Therefore, the C terminus of talin released by limited proteolysis as found in our study may represent an ancient mechanism of talin to enhance actin polymerization and is not necessarily restricted by integrin, which mainly interacts with the N terminus of talin.
Our data suggest that arginylation regulates the talin VAD fragment at two levels: upstream, through affecting the calpain-dependent generation of this fragment, and downstream, by modifying the fragment directly to facilitate its function (Fig. 8 D) as well as its turnover (Fig. 7 C). These multiple levels of regulation by arginylation of the VAD fragment resemble the case of another important type of posttranslational modification, phosphorylation, which can simultaneously modulate the physiological function and promote the degradation of a target protein (Wu et al., 2000; Biggs et al., 2005; Cho and Tsichlis, 2005). Understanding the detailed molecular mechanisms and the hierarchical relationships within this multistep regulation by arginylation constitutes an exciting direction of further studies.
Evidence generated in vitro and in structural studies suggest that the talin molecule can undergo force-dependent conformational changes, which would expose the otherwise hidden functional domains and the largely hydrophobic linker sequences in between that are likely sensitive to proteases (Hytönen and Vogel, 2008; del Rio et al., 2009). Notably, the size distribution of the talin fragments detected by the C20 antibody in our tests appears to closely match the prediction from those putative cleavage sites, suggesting that the generation of the talin fragments may be a downstream event or a consequence of the conformational change induced by “stretch” force applied on the talin molecule during cell motility and adhesion (Fig. 8 B; Rauzi et al., 2010), which in turn likely regulates the migration and adhesion events (Papusheva and Heisenberg, 2010; Stricker et al., 2011). Although the cleavage pattern of talin appears to be more complicated than we expected, the 70-kD fragment is the most prominent fragment that is different between the confluent and scarce MEF in our tests. Other fragments are either not associated with the cell density (such as the 180-kD fragment and a series of fragments smaller than 70 kD), or are less abundant than the 70-kD fragment, and in many cases are present only in occasional preparations. Similarly, we found the 70-kD fragment as the prominent talin fragment in the heart muscle. It is likely that Ala 1903 is the preferential site for cleavage in fibroblasts and the cardiomyocytes, which may reflect their physical properties (such as stiffness or physical stress), or may be relevant to the major cadherin isoform expressed in these cells (N-cadherin). Consistent with these speculations, we were able to identify a prominent level of the 70-kD VAD fragment in another fibroblast cell line (NIH 3T3; Fig. 10 A). A lower level of the VAD fragment with a slightly different cleavage pattern of talin (which still fits with the predicted force-induced cleavage positions on the molecule) was observed in a mouse endothelial cell line CLS1, whose adhesion mainly relies on VE-cadherin (Fig. 10 B). In these cells, like in fibroblasts, the recombinant VAD-GFP also colocalized with cadherin (Fig. 10 C). However, the 70-kD VAD fragment was not observed in epithelial cells (tested in the mouse mammary epithelial cell, NMuMG, which mainly expresses E-cadherin). Further investigation may be needed to test the dependency of talin cleavage on the cells’ physical properties and/or cadherin isoforms.
The role of the VAD fragment in other cell lines. (A) The cleavage pattern of talin in confluent NIH 3T3 mouse fibroblasts and 50% confluent mouse endothelial cell CLS1 (which forms clustered cell islands) visualized by Western blots with anti-talin C20. The positions of the 70-kD VAD fragment in each cell line are indicated by arrows. (B) Representative fluorescence images showing the locations of the cadherin (stained by anti-PAN cadherin antibody) and the VAD-GFP (stained by anti-GFP) and the merged image. The images were acquired by a 40x dry lens.
The role of the VAD fragment in other cell lines. (A) The cleavage pattern of talin in confluent NIH 3T3 mouse fibroblasts and 50% confluent mouse endothelial cell CLS1 (which forms clustered cell islands) visualized by Western blots with anti-talin C20. The positions of the 70-kD VAD fragment in each cell line are indicated by arrows. (B) Representative fluorescence images showing the locations of the cadherin (stained by anti-PAN cadherin antibody) and the VAD-GFP (stained by anti-GFP) and the merged image. The images were acquired by a 40x dry lens.
Regulatory proteolysis of many transmembrane proteins that are relevant to cell–cell adhesion, including cadherin and EpCAM, was shown to be regulated by the formation of the cell–cell contacts and proposed to be part of the mechanisms regulating the contact inhibition, a poorly understood phenomenon that distinguishes normal and cancer cells (Parisiadou et al., 2004; Denzel et al., 2009; McCusker and Alfandari, 2009). Interestingly, genetic evidence also suggests that talin regulates the expression of cadherin (Bécam et al., 2005). Our finding of the cell contact–dependent cleavage of talin and its participation in cadherin-mediated adhesions provides a new angle in studying the poorly understood molecular mechanism of contact inhibition.
Materials and methods
Plasmids and constructs
The VAD region of mouse talin isoform 1 (from Ala1903 to the C terminus) was cloned from a plasmid containing the coding sequence of the full-length talin (Franco et al., 2004a). To generate the constructs representing the nonarginylated or arginylated talin VAD fragments, the VAD domain with or without a preceding Arg residue (coded by the nucleotide triplet AGG) was inserted into a sandwich containing N-terminal ubiquitin and C-terminal eGFP by restriction sites SacII and BamHI, as described in Karakozova et al. (2006). To maximize the chance for the translation to start from the correct position a Kozak translation initiation sequence (Kozak, 1987) was placed before the N-terminal ubiquitin domain. To minimize the interference of possible internal translation starting sites, the sequence CCACCATCAATG positioned at nucleotides 5890–5991 in the talin 1 coding region was replaced with the sequence CAACAATAAATG, without changing the corresponding amino acid sequence. The constructs were then inserted into the plasmid pEGFP-N2 (Takara Bio Inc.) using restriction sites of XhoI and NotI for transient expression, or into plasmid pQC-XIP (Takara Bio Inc.) using restriction sites of NotI and EcoRI for retroviral infection and stable expression.
Full-length N-cadherin fused with eGFP was a gift from Dr. Cécile Gauthier-Rouvière (CRBM-CNRS, Montpellier, France; Taulet et al., 2009). The coding sequence of N-cadherin together with its native upstream flanking RNA sequence was amplified with the following primers: upstream 5′-AATTACGCGTCCACTGCCCGGAGCCGACTCC-3′, and downstream 5′-ATATTGATCACGTCGTCACCACCGCCGTACATGTC-3′. The PCR product is then digested by MluI and BclI and inserted into modified pBabe-puro vector carrying a C-terminal fusion of mCherryFP (without ATG start codon) that was digested by MluI and BamHI. The final linker sequence between the N-cadherin and mCherryFP is 5′-GTGATCCACCGGCCTGCTTCTGGCTCT-3′.
Retroviral vectors pSUPER.retro.puro carrying siRNA specific for mouse calpain-1 and calpain-2 were gifts from Dr. Anna Huttenlocher (University of Wisconsin-Madison, Madison, WI; Franco et al., 2004a).
Cell culture
Wild-type and arginyltransferase 1 (Ate1) KO MEF cell lines were obtained by immortalization of primary cells as described previously (Kwon et al., 2002; Karakozova et al., 2006). Human embryonic kidney (HEK) 293T cells (clone #17) were purchased from American Type Culture Collection. Floxed wild-type (PZ/PZ), calpain-4 knockout, and rescued MEF cell lines (stably expresses recombinant calpain-4) were gifts from Dr. Peter Greer (Queen’s University, Ontario, Canada). Mouse endothelial cell line CLS1 was obtained from the University of Pennsylvania Cell Center. NIH 3T3 mouse fibroblasts were a gift from Dr. Narayan Avadhani (Department of Animal Biology, University of Pennsylvania, Philadelphia, PA). Mouse mammary epithelial cell line NMuMG was a gift from Dr. Dmitry A. Goncharov (Department of Medicine, University of Pennsylvania, Philadelphia, PA). All cells were maintained in a 1:1 mixture of DME (high glucose with glutamax) and F10 nutrient mix (Invitrogen) supplemented with 10% fetal bovine serum and antibiotics (Antibiotic-Antimycotic solution; Invitrogen). Cells were passaged at the ratio of 1:4 every 3–4 d.
Cell transfection
All plasmids for transfection were purified by the Endo-free Maxi Prep kit (QIAGEN). For transient transfection, Lipofectamine reagent (Invitrogen) alone (for HEK 293T) or with the Plus Reagent (for MEFs) was used. For stable transfection of the VAD constructs and the GFP control, retrovirus was produced using plasmids derived from pQC-XIP (Takara Bio Inc.) in combination with the standard GAG-Pol and VsvG vectors (Takara Bio Inc.) following the manufacturer’s protocol. The target cells were infected with the virus three times in the presence of polybrene (Sigma-Aldrich). Cells expressing GFP-fused proteins were enriched by puromycin selection and fluorescence sorting. In the case of cells stably transfected with R-VAD-GFP, the fluorescence signal was boosted by treatment of the cells with MG132 for 5 h to inhibit the degradation of the arginylated protein before sorting. For stable transfection of the N-cadherin–mCherryFP construct into MEF, plasmids derived from pBabe-puro vector carrying the desired construct were used together with GAG-Pol and VsvG to produce the retrovirus for infection of the cells using the protocol similar to that described above. Cells with the red fluorescence were enriched by fluorescence sorting and allowed to grow and proliferate for at least three passages to recover before any further treatment or test. For stable knockdown of mouse calpain-1 or calpain-2 in MEFs, retroviral vectors carrying specific siRNA were used with GAG-Pol and VsvG to produce virus to infect the cells using a protocol similar as above. Infected cells were enriched by selection with puromycin.
Preparation of cultured cell, tissue, and blood cell lysate samples
For cultured cell lysates, treated cells were harvested by scraping and centrifugation at 3,000 rpm in 50-ml conical tubes in a medical centrifuge for 3 min at room temperature. The cell pellet was promptly resuspended in dPBS, transferred to a microtube, and spun down twice to thoroughly remove the supernatant. This procedure was generally finished within 10 min after the cells were harvested. The pellet was then weighed and lysed in 10 vol of 1x SDS loading buffer by boiling for 5 min. For heart tissue lysates, pieces of ventricle were harvested from freshly sacrificed mice (C57BL/6J mice line) and minced into small pieces by a knife, then washed by dPBS twice and frozen by liquid nitrogen. Frozen tissue was ground into powder using a porcelain mortar and pestle and then lysed in 2x SDS loading buffer by boiling for 10 min. For the lysates of blood cells, freshly harvested mouse blood was washed with ice-cold dPBS five times and then the blood cells were spun down, collected for lysis in 2x SDS loading buffer, and boiled for 10 min.
Testing the density-dependent properties of cultured cells
Cells were maintained in culture for at least 1 wk by repeated splitting into a new dish at the ratio of 1:4 in every 3 d to keep them in a constant growth phase. On the day of the test, cells were washed with dPBS and resuspended by brief treatment with 0.25% trypsin. The suspended cells were filtered through cell strainers with 40 µm pore size (from Thermo Fisher Scientific) and then counted for numbers using a hemocytometer. Desired numbers of cells were added to culture dishes and incubated as indicated in the figure legends. For 10-cm tissue culture dishes used in our test, a newly confluent culture of WT MEF monolayer contained ∼4 million cells. For 6-cm dishes, a confluent monolayer contained 1.2–1.4 million cells.
Metabolic assays and inhibitor treatment
To measure posttranslational protein degradation dynamics, the change of protein level over time in the presence of cycloheximide was monitored. Cells in each experiment were transfected for 6–8 h and then equally split into individual wells of the 6-well plates for overnight cultures. After ∼20 h, culture media containing 100 µg/ml cycloheximide (EMD) was added to each well and the cells were harvested at time points as indicated for analysis of the protein levels. For inhibition of calpain activity, WT MEFs (5 million cells for a 10-cm dish) were resuspended by trypsinization and allowed to resettle for 3 h to reach confluent state, followed by a treatment with calpain inhibitors (EMD) including ALLN, calpectin, calpain inhibitor 3, and EST, for additional 2 h. Proteasome-specific inhibitor β-lactone (Sigma-Aldrich), which has minimum crossreactivity to calpain, was used as a control for a possible effect of the calpain inhibitors to the proteasome. To induce accumulation of the exogenously expressed recombinant proteins, transfected cells were treated with 20 µM MG132 (EMD) for durations as indicated. Treatment with 0.1% DMSO was used as control for all reagent treatment unless otherwise indicated.
Western blotting
Cell lysate samples were separated by electrophoresis in SDS-PAGE of 4–15% or 10% as indicated and then transferred to nitrocellulose or PVDF membranes. The endogenous talin bands were detected by polyclonal goat anti-talin (C20) antibody that reacts with sequences on the C termini of mouse talin 1 and 2 (catalog no. sc-7534; Santa Cruz Biotechnology, Inc.), unless otherwise indicated. The recombinant eGFP fusion proteins were detected by mouse anti-GFP (catalog no. 11814460001; Roche). β-Catenin was detected by rabbit antibody mAb (catalog no. 1247; Epitomics). Cadherin was detected by rabbit anti-PAN cadherin, which recognizes most mouse cadherin isoforms (catalog no. C3678; Sigma-Aldrich). The levels of calpain proteases were detected by rabbit antisera targeting the highly conserved N-terminal sequence of calpain-1 and the peptide sequences CDSYKKWKLTK of calpain-2, respectively (generous gifts from Dr. Marek Ma and Robert W. Neumar, University of Pennsylvania; similarly described in Bevers et al. [2009]). The cellular level of full-length α-spectrin was detected by a mouse monoclonal antibody (clone #AA6, catalog no. MAB1622; Millipore). The level of the cleaved form of α-spectrin was detected by a specific antibody that only reacts to the cleaved form but not the full-length protein (anti-α spectrin AB38, a generous gift from Dr. David Lynch, University of Pennsylvania; described in Roberts-Lewis et al. [1994]; Simpkins et al. [2003]). The endogenous actin or tubulin was used as a loading control (detected with rabbit polyclonal anti-actin [catalog no. AAN01; Cytoskeleton] or mouse monoclonal anti–β-tubulin [catalog no. T4026; Sigma-Aldrich]). Chemifluorescence visualization was performed with secondary antibodies conjugated with HRP and reagents provided in the BM Chemifluorescence Western Blotting kit Mouse/Rabbit (Roche) or the SuperSignal West Femto Chemiluminescence kit (Thermo Fisher Scientific). In most cases at least three different exposures with time ranging from 5 s to 1 min on x-ray films were performed and the films showing moderate intensity of chemifluorescence signals were chosen for quantification. The corresponding films were scanned by an Epson 4490 Perfection scanner into grayscale digital files with 1,200 dpi. The densitometry of the protein bands was analyzed with ImageQuantTL 1D gel analysis software pack, version 7.0 (GE Healthcare) according to the manufacturer’s manual. In brief, regions containing the expected protein bands were chosen, and the background was removed by using the automatic “minimal profile” function in ImageQuant software. The “net volume” of each protein band calculated as the integration of pixel area and pixel intensity was obtained as the level of protein in each band.
Immunoprecipitation
Cells were washed by dPBS twice, then lysed in cold lysis buffer containing 50 mM Tris-Cl, pH 7.9, 150 mM NaCl, 1% NP-40, mammalian protease inhibitor cocktail (Sigma-Aldrich), and 1 mM PMSF. The mixture was rotated at 4°C for 30 min with occasional vortexing. The lysate was precleaned with resin beads conjugated with Protein A (catalog no. 15918–014; Invitrogen) and Protein G (catalog no. 15920–010; Invitrogen). The endogenous talin and talin fragments were immunoprecipitated using a polyclonal goat anti-talin (C20) antibody that reacts with the C-terminal region of mouse talin 1 and 2 (catalog no. sc-7534; Santa Cruz Biotechnology, Inc.). The antibody–protein complexes were pulled down using protein A and G beads and washed six times with lysis buffer. The beads were resuspended in 2x SDS-loading buffer and boiled for 10 min. The resulting protein samples were analyzed by SDS-PAGE.
Immunofluorescence
Cells cultured on coverslips were fixed by treatment with chilled methanol (−20°C) for 20 min followed by chilled acetone (−20°C) for 5 min. The treated cells were rehydrated in dPBS for 5 min and then blocked with 2% BSA in PBS supplemented with 0.1% Tween 20. For detection of integrin-mediated focal adhesions, rat anti–mouse integrin β-1 (catalog no. 553715; BD) was used. For detection of cadherin-mediated adherens junctions, polyclonal rabbit anti-PAN cadherin (catalog no. C3678; Sigma-Aldrich) was used. For detection of talin in focal adhesions, mouse anti talin (8d4, catalog no. T3287; Sigma-Aldrich) was used. For detection of β-actin, monoclonal mouse anti–β-actin, clone AC15 (catalog no. A1978; Sigma-Aldrich) was used. For detection of talin and the C-terminal fragments of talin, polyclonal goat anti-talin (C20, catalog no. sc-7534; Santa Cruz Biotechnology, Inc.) was used. GFP was either detected by its own fluorescence signal or by enhancement with mouse anti-GFP (catalog no. 11814460001; Roche), as indicated. For visualization with fluorescent chromophores, conjugated secondary antibodies with minimum species crossreactivity were used, including donkey anti–mouse Alexa 594 (catalog no. A21203; Invitrogen), goat anti–mouse Alexa 488 (catalog no. A11001; Invitrogen), donkey anti–goat Alexa 488 (catalog no. A11055; Invitrogen), and donkey anti–rabbit Alexa 488 (catalog no. A21206; Invitrogen). All the pseudo-red color fluorescent images reflect the signal from Alexa 594, and all the pseudo-green color fluorescent images reflect the signal from Alexa 488. The stained cells were mounted in Aqua Poly/Mount solution (Polysciences) and sealed with nail polish.
Fluorescence recovery after photobleaching (FRAP)
Cells stably transfected with N-cadherin–mCherryFP cultured in phenol-red free DME with 10% FBS and antibiotics were visualized using a Nikon CFI Plan Apochromat violet corrected (VC) 60x water immersion lens (NA 1.27) in a Nikon Ti-E inverted fluorescent microscope (with PFS) equipped with a Photometrics Evolve 512 camera and controlled by the Nikon NIS-Elements AR software (version 4.00, 64 bit). The image was binned by 2 × 2 to increase acquisition speed. The photobleaching of the red fluorescent color mCherryFP was performed by a 10-s exposure under a 10-mw, 561-nm wavelength laser source facilitated by the built-in module in the NIS-Element software. To measure the recovery dynamics of the bleached region, the time needed to recover to half of the intensity before the bleaching event was calculated with the built-in function in the software. The background and the reference region that were not exposed to the laser were used to normalize the dynamic curve of the bleached region.
Separation of actin polymers
Cells were harvested and lysed in F-actin stabilization buffer containing 50 mM Pipes, pH 6.9, 50 mM NaCl, 5 mM MgCl2, 5 mM EGTA, 5% glycerol, 0.1% NP-40, 0.1% Triton X-100, 0.1% Tween 20, 0.1% 2-mercaptoethanol, 1 mM ATP, and protease inhibitor cocktail (catalog no. P8340; Sigma-Aldrich. The resulting lysates were sedimented by a series of low speed centrifugations at 37°C: 200 g for 5 min; 1,500 g for 15 min; and 16,000 g for 15 min. The supernatant from the last step was used as total actin input and was spun down at 66,000 g for 60 min at 37°C. The pellet (containing the actin polymers) was dissolved in 8 M urea in PBS of a half-volume of the input, and then further lysed by 2x SDS-PAGE loading buffer of another half-volume of the input. Protein concentrations in all the fractions were quantified by Western blots using antibodies to total actin (catalog no. AAN01; Cytoskeleton).
Microscopy
For most epifluorescence imaging, the slides were examined using an inverted fluorescence microscope (TE 300; Nikon) equipped with a series of objective lenses (Plan 10x, NA 0.25; Plan 20x, NA 0.40; Plan Fluor 40x, NA 0.75; all dry lenses), Orca AG camera (Hamamatsu Photonics), and computerized image acquisition software (MetaMorph; Molecular Devices). Acquisition parameters were controlled so that the acquired signal did not reach saturation point (maximal gray level 4095 a.u.). For total internal reflection fluorescence (TIRF) microscopy, an inverted optical microscope (Eclipse TI; Nikon) was equipped with a Nikon oil immersion TIRF objective (60x, NA 1.45) and a 300-mW, 532-nm laser was used. For FRAP, a Nikon TI-E microscope was used (see more details in the methods section for FRAP). For live imaging, the cells were cultured in phenol-red free DME with 10% FBS and antibiotics overnight and imaged with the same medium. All imaging (including live cell imaging) was performed at room temperature and in open air unless otherwise indicated.
Image analysis
The acquired images were analyzed by MetaMorph imaging software. In analysis of confluent cells stained with anti-cadherin, to reduce nonspecific background and out-of-focal plane signal, the background of fluorescence images were filtered by the built-in background flatten function in “background and shading correction” with the object size set at 2 pixels in the MetaMorph software. To minimize any arbitrary bias in selection of the objects for quantification, the thresholds of the images were first set by using the automatic function of “autothreshold for light objects” in the software, and then subjected to minor manual adjustment usually within 10 units of gray value to closely fit the edge of the thresholded object to the fluorescence signal. The sizes of the cadherin plaques, reflected by the total pixel area of each thresholded object were measured using the “integrated morphometry analysis” function of the MetaMorph software. To further minimize the impact of nonspecific antibody staining and background noise, only cadherin-stained objects with pixel area of at least 5 pixels under a 40× objective lens (equals 0.126 µm2) were included in the final statistics unless otherwise indicated. Similarly, when the size of the actin “patch” near the cadherin plaque was measured, only the objects with pixel area of at least 4 pixels and less than 500 pixels with a 40x objective lens were included in the final statistical analysis. The density of the cadherin plaques along the border between cells was reflected by the percentage of the area that is covered by the cadherin fluorescence on the curvy line chased through the border between the cells, which was measured by the built-in function in “region statistics” in MetaMorph. In the analysis of scarce cells stained with anti-talin 8d4, the thresholds of the images were set by using the built-in function of “autothreshold for light objects” in the software. The area sizes of the focal adhesions, reflected by the total pixel area of thresholded objects with minimum value of 5 pixels (equals 0.126 µm2), were measured using the “integrated morphometry analysis” function of the MetaMorph software. In the analysis of confluent cells stained with anti-talin 8d4, the focal adhesions close to the basal surface of the cells were visualized by TIRF microscopy. To further remove the out-of-focal plane fluorescence signals, the images were subjected to background filtering by the built-in background flatten function of “background and shading correction” with the object size set at 2 pixels in the MetaMorph software.
Image processing
The representative images shown in the paper were processed with Photoshop (Adobe) to obtain displayed portion and to alter picture size. The contract and brightness of the images were subjected to linear adjustment by changing the display levels. All linear adjustments were applied equally to the whole image.
Statistics
Calculation of standard deviation (SD) and standard error of means (SEM) was based on Student’s t-distribution. The statistical significance (P-value) between two groups of samples was calculated based on Student’s t test with the assumption of unpaired, two tailed, and equal variances.
Online supplemental material
Fig. S1 shows diagrams and Western blots demonstrating the structure of talin, the generation of the VAD fragment, and its dependence on calpain. Fig. S2 shows that the VAD fragment is absent in scarce cultures regardless of their motile and proliferative state modulated by serum starvation. Fig. S3 shows that the level of the VAD fragment increases with the increase in cell density during a 5-h incubation to avoid the effects of change of cell numbers by cell division. Fig. S4 shows that lack of the VAD fragment in Ate1 knockout cells correlates with the decreased calpain activity in these cells (evidenced by reduced spectrin cleavage) and is not related to proteasome-dependent degradation or altered calpain protein levels. Fig. S5 shows that reintroduction of the VAD fragment into Ate1 KO cells does not rescue the focal adhesion defects in scarce or confluent cultures.
Acknowledgments
We thank Dr. Chao-Xing Yuan from the University of Pennsylvania Proteomics Core Facility for the analysis of talin-derived bands in C20 antibody precipitates and members of the Kashina Laboratory for helpful discussions. We thank Dr. Taniya Das and Dr. Yale Goldman from the University of Pennsylvania, Department of Physiology for helping with the TIRF imaging. We also thank Joel Silfies and John Zentmeyer from Nikon for their technical assistance with the FRAP experiments.
This work was supported by National Institutes of Health grants RO1HL084419 and R01HL084419-03S1 to A. Kashina.
References
- AJ
adherens junction
- ALLN
N-Acetyl-Leu-Leu-Nle-CHO
- Con
confluent
- CPN
calpain
- EST
(2S,3S)-trans-epoxysuccinyl-l-leucylamido-3-methylbutane ethyl ester
- FA
focal adhesion
- FL
full length
- KO
knockout
- mCHFP
monomeric cherry fluorescent protein
- MEF
mouse embryonic fibroblast
- MG132
carbobenzoxy-l-leucyl-l-leucyl-l-leucinal, Z-LLL-CHO
- SC
scarce
- TIRF
total internal reflection fluorescence
- WT
wild type
Author notes
F. Zhang’s present address is Department of Molecular and Cellular Pharmacology, University of Miami, Miller School of Medicine, Miami, FL 33136.
S. Saha’s present address is Department of Molecular Biology and Biotechnology, Tezpur University, Napaam 784028, India.