Unwanted proteins in the endoplasmic reticulum (ER) are exported into the cytoplasm and degraded by the proteasome through the ER-associated protein degradation pathway (ERAD). Disturbances in ERAD are linked to ER stress, which has been implicated in the pathogenesis of several human diseases. However, the composition and organization of ERAD complexes in human cells is still poorly understood. In this paper, we describe a trimeric complex that we propose functions in ERAD. Knockdown of erasin, a platform for p97/VCP and ubiquilin binding, or knockdown of ubiquilin in human cells slowed degradation of two classical ERAD substrates. In Caenorhabditis elegans, ubiquilin and erasin are ER stress-response genes that are regulated by the ire-1 branch of the unfolded protein response pathway. Loss of ubiquilin or erasin resulted in activation of ER stress, increased accumulation of polyubiquitinated proteins, and shortened lifespan in worms. Our results strongly support a role for this complex in ERAD and in the regulation of ER stress.
Introduction
The ER is a fundamental eukaryotic organelle where proteins are made and where stringent quality-control systems operate to ensure that only correctly folded and properly assembled protein complexes are allowed to exit the organelle for delivery to their intended sites of function (Ellgaard et al., 1999; Kostova and Wolf, 2003; Hebert and Molinari, 2007). The mechanisms by which the ER operates to monitor and dispose of misfolded and misassembled protein complexes are beginning to be understood. One mechanism is ER-associated protein degradation (ERAD), a regulated process by which proteins in the ER are exported to the cytoplasm for destruction by the proteasome (Kanehara et al., 2007; Vembar and Brodsky, 2008; Hirsch et al., 2009). Inefficient clearance of misfolded and misassembled proteins from the ER leads to ER stress, which results in the activation of a signaling cascade called the unfolded protein response (UPR; Schröder and Kaufman, 2005; Malhotra and Kaufman, 2007; Ron and Walter, 2007). In mammals, UPR is regulated by three main sensors called IRE1, PERK, and ATF6, each of which triggers, via distinct mechanisms, a coordinated intracellular response designed to restore protein homeostasis in the ER. If, however, ER stress is not alleviated, a terminal cell death program is then executed.
There is growing evidence that disturbances in ERAD are involved in human disease. For example, several diseases appear to result from mutations in proteins that cause them to misfold in the ER during synthesis, where they are retained and eliminated by ERAD (Schröder and Kaufman, 2005; Zhao and Ackerman, 2006; Brodsky, 2007). Although in this scenario, ERAD is detrimental and leads to disease, under normal circumstances, it presumably functions as a safety mechanism to remove damaged and misassembled proteins before they accumulate to toxic levels. However, inefficient ERAD has also been linked to disease. For example, mutant proteins involved in Huntington's disease and amyotrophic lateral sclerosis were recently shown to induce ER stress by interfering with ERAD (Nishitoh et al., 2008; Duennwald and Lindquist, 2008).
Our knowledge of proteins involved in ERAD is expanding (for reviews see Kanehara et al., 2007; Vembar and Brodsky, 2008; Hirsch et al., 2009). Derlin proteins are thought to form the portal in the ER through which misfolded and misassembled proteins are exported. p97 (valosin-containing protein: p97/VCP) provides the main driving force for extraction of the ERAD substrate (Lilley and Ploegh 2004; Ye et al., 2004). p97/VCP is recruited to the ER for ERAD by binding to ER-localized proteins, candidates of which are gp78, derlin, VIMP, ubx2, Hrd1, erasin, and UBXD8. During extraction, the misfolded protein is tagged with a polyubiquitin chain by ubiquitin ligases (Kostova et al., 2007); the polyubiquitin chain serves as a signal for the recognition and destruction of the protein by the proteasome. At least five ER resident ubiquitin ligases—Hrd1/Der3, gp78, Doa10/Teb4, RMA1, and RFP2—conduct this modification, although others probably exist. The length of the ubiquitin chain that is tagged onto substrates is regulated by Ufd2, which is recruited to the ERAD complex by binding to p97/VCP (Richly et al., 2005). Ufd2, in turn, binds two additional factors, Rad23 and Dsk2, both of which contain ubiquitin-associated (UBA) and ubiquitin-like (UBL) domains, which appear to function in the transfer of the ubiquitinated substrate to the proteasome for degradation (Richly et al., 2005; Elsasser and Finley, 2005). The UBA domains of Rad23 and Dsk2 bind polyubiquitin chains, and their UBL domains bind the proteasome, thus positioning these ubiquitin–proteasome shuttle proteins as the terminal factors that deliver the substrate to the proteasome for degradation (Verma et al., 2004; Raasi and Wolf, 2007).
Many of the ERAD components mentioned were first identified in yeast. Interestingly, mammals contain additional ERAD components for which no counterparts exist in yeast, which suggests that mammals use a more elaborate repertoire of proteins in ERAD (Carvalho et al., 2006; Jentsch and Rumpf, 2007; Kanehara et al., 2007).
In this paper, we demonstrate that human ubiquilin proteins (Wu et al., 1999; Mah et al., 2000; Kleijnen et al., 2000) that are found in the cytoplasm and nucleoplasm (Mah et al., 2000), and which are believed to function as ubiquitin–proteasome shuttle factors, interact with erasin, an ER localized protein (Liang et al., 2006), in a novel complex that functions in ERAD. Using a variety of techniques, including in vitro binding assays, coimmunoprecipitation and biochemical fractionation of proteins from cells, and silencing of the proteins in cells and worms, we obtained evidence in support of this novel complex in ERAD.
Results
In vitro interaction between erasin and ubiquilin
A yeast two-hybrid (Y2H) screen using the central domain of human ubiquilin-1 (UBQLN1ΔUBLΔUBA; Fig. 1 A) netted four clones containing different portions of the N-terminal end of erasin, an ER-localized protein that promotes ERAD (Liang et al., 2006). The longest clone interacted 100-fold stronger with the same ubiquilin bait than with the UBA domain of ubiquilin or with unrelated baits, which indicates strong and specific interaction between erasin and the central domain of ubiquilin (Fig. 1 B).
Erasin interaction with ubiquilin-1, p97/VCP, and hHR23A. (A) Schematic drawing of ubiquilin-1. Ubiquilin-1ΔUBLΔUBA (178–428 aa) was used as the bait in a Y2H screen. (B) Yeast interaction assay of an erasin prey clone with different baits measured in five independent colonies showing the mean and SD (error bars). (C) GST pull-down assays of ubiquilin-1 (UBQ) fusion proteins with 35S-radiolabeled erasin. (C, right) Schematic drawings of the GST constructs and a summary of their binding (+, binding; −, no binding). (C, left) Autoradiogram (top panel) and Coomassie-stained gel (bottom panel) of the pull-down experiment. (D) Coomassie-stained gel of purified proteins used for pull-down assays. The black line indicates that intervening lanes have been spliced out. (E) GST pull-down assays showing that erasin-His binds to GST-ubiquilin but not to GST or GST-hHR23A proteins. The input erasin, the recovered erasin, and GST proteins are shown. (F) GST pull-down assays showing that erasin, p97/VCP, and ubiquilin bind together in a trimeric complex. The input p97/VCP and erasin proteins and the recovered GST complexes (ponceau) are also shown. Arrowheads indicate the position of full-length GST-fusion proteins.
Erasin interaction with ubiquilin-1, p97/VCP, and hHR23A. (A) Schematic drawing of ubiquilin-1. Ubiquilin-1ΔUBLΔUBA (178–428 aa) was used as the bait in a Y2H screen. (B) Yeast interaction assay of an erasin prey clone with different baits measured in five independent colonies showing the mean and SD (error bars). (C) GST pull-down assays of ubiquilin-1 (UBQ) fusion proteins with 35S-radiolabeled erasin. (C, right) Schematic drawings of the GST constructs and a summary of their binding (+, binding; −, no binding). (C, left) Autoradiogram (top panel) and Coomassie-stained gel (bottom panel) of the pull-down experiment. (D) Coomassie-stained gel of purified proteins used for pull-down assays. The black line indicates that intervening lanes have been spliced out. (E) GST pull-down assays showing that erasin-His binds to GST-ubiquilin but not to GST or GST-hHR23A proteins. The input erasin, the recovered erasin, and GST proteins are shown. (F) GST pull-down assays showing that erasin, p97/VCP, and ubiquilin bind together in a trimeric complex. The input p97/VCP and erasin proteins and the recovered GST complexes (ponceau) are also shown. Arrowheads indicate the position of full-length GST-fusion proteins.
To confirm binding, and to map the erasin-binding site in ubiquilin-1, we conducted GST pull-down experiments examining whether GST fusions of different regions of ubiquilin-1 bind radiolabeled erasin protein (Fig. 1 C). Fusion proteins containing the central domain of ubiquilin-1 bound erasin efficiently, whereas those that lacked the domain failed to bind erasin (Fig. 1 C).
By additional pull-down experiments and systematic N- and C-terminal deletion of the erasin polypeptide, we mapped the ubiquilin-1–binding site to the N-terminal end of erasin (between amino acids 1 and 200; see Fig. S1), which is distinct from the p97/VCP-binding site located at the ubiquitin regulatory X (UBX) domain (amino acids 316–395) of the protein (Liang et al., 2006).
Interaction between erasin and other ubiquitin–proteasome shuttle factors
We next investigated, using similar pull-down assays, whether erasin binds hHR23A, another ubiquitin–proteasome shuttle protein, which like ubiquilin contains both UBL and UBA domains (Elsasser and Finley, 2005; Raasi and Wolf, 2007). As shown in Fig. 1 (D and E), erasin-His was efficiently pulled down by GST-ubiquilin but not by GST-hHR23A or GST-His. These data suggest that erasin binds selectively to ubiquilin-1 and not to all ubiquitin–proteasome shuttle factors.
Recombinant erasin, ubiquilin, and p97/VCP proteins bind together in a single complex
Given that erasin binds p97/VCP (Liang et al., 2006; Alexandru et al., 2008), we next examined whether recombinant erasin, ubiquilin, and p97/VCP proteins bind together in one complex. In these experiments, purified GST-ubiquilin or GST-His protein alone, which served as a control for nonspecific binding, were mixed with purified p97/VCP-His, erasin-His, or both p97/VCP-His and erasin-His proteins, and then the GST proteins were affinity purified. The purified complexes were examined by immunoblotting for recovery of the added His-tagged proteins (Fig. 1 F). The blots show that erasin-His bound strongly to GST-ubiquilin but not to GST alone. Also, p97/VCP-His was isolated in a complex with GST-ubiquilin, but only after addition of erasin-His. These results strongly suggest that p97/VCP binds ubiquilin through erasin and that all three proteins coexist in a single complex. Quantification of the stoichiometry of the complex suggests that erasin-His and p97/VCP-His bind GST-ubiquilin in an ∼1:1 ratio (see Fig. S2). Because p97/VCP self-assembles to form a homohexamer, it is likely that in these in vitro binding assays, erasin has saturated each of the six UBX-binding sites of the p97/VCP hexamer (Meyer et al., 2000; Dreveny et al., 2004).
Erasin, ubiquilin, and p97/VCP coimmunoprecipitate with each other in cells
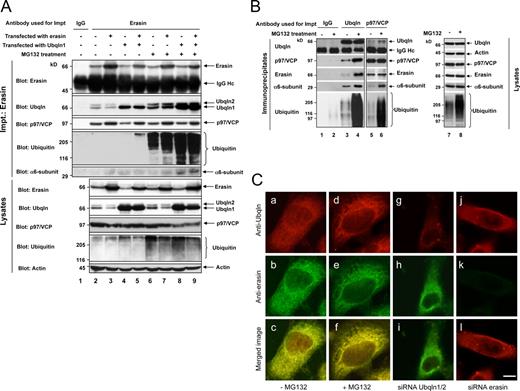
To determine if erasin, ubiquilin, and p97/VCP form a complex in cells, we immunoprecipitated each of the proteins from HEK293 cells and found that the other two proteins were indeed coimmunoprecipitated (Fig. 2 A, lane 2; and Fig. 2 B, lanes 3 and 5). The specificity of these immunoprecipitations is illustrated by the failure to detect any of the proteins in parallel immunoprecipitations conducted with two different control antibodies (Fig. 2 A, lane 1; and Fig. 2 B, lane 1).
Erasin, ubiquilin, and p97/VCP coimmunoprecipitate with each other in cells. (A) HEK293 cells were either left untransfected (lanes 1, 2, and 6), transfected with either erasin (lanes 3 and 7) or ubiquilin-1 cDNA (lane 4 and 8), or cotransfected with both cDNAs (lane 5 and 9). After 20 h, MG132 was added for 4 h to one set of the cultures (lanes 6–9), whereas the other set was left untreated (lanes 1–5). Equivalent amounts of protein lysates were used for immunoprecipitation using either an erasin antibody (lanes 2–9) or control IgG antibody (lane 1). The immunoprecipitates as well as 1/10 the amount of each protein lysate were probed by immunoblotting for the indicated proteins. (B) Similar to A except that cells were not transfected and the antibodies used for immunoprecipitation were a different control antibody (lanes 1 and 2), a ubiquilin antibody (lanes 3 and 4), and a p97/VCP antibody (lanes 5 and 6). (lanes 7 and 8) Blots for proteins in equal amounts of the lysates used in the experiments. (C) Double immunofluorescence localization of ubiquilin and erasin proteins in HEK293 cells that were either not treated (−) or treated (+) with MG132, or transfected with siRNAs designed to knock down ubiquilin-1 and ubiquilin-2 proteins together, or erasin protein alone. Quantification of the colocalization of the proteins is provided in Fig. S3. Bar, 5 µm.
Erasin, ubiquilin, and p97/VCP coimmunoprecipitate with each other in cells. (A) HEK293 cells were either left untransfected (lanes 1, 2, and 6), transfected with either erasin (lanes 3 and 7) or ubiquilin-1 cDNA (lane 4 and 8), or cotransfected with both cDNAs (lane 5 and 9). After 20 h, MG132 was added for 4 h to one set of the cultures (lanes 6–9), whereas the other set was left untreated (lanes 1–5). Equivalent amounts of protein lysates were used for immunoprecipitation using either an erasin antibody (lanes 2–9) or control IgG antibody (lane 1). The immunoprecipitates as well as 1/10 the amount of each protein lysate were probed by immunoblotting for the indicated proteins. (B) Similar to A except that cells were not transfected and the antibodies used for immunoprecipitation were a different control antibody (lanes 1 and 2), a ubiquilin antibody (lanes 3 and 4), and a p97/VCP antibody (lanes 5 and 6). (lanes 7 and 8) Blots for proteins in equal amounts of the lysates used in the experiments. (C) Double immunofluorescence localization of ubiquilin and erasin proteins in HEK293 cells that were either not treated (−) or treated (+) with MG132, or transfected with siRNAs designed to knock down ubiquilin-1 and ubiquilin-2 proteins together, or erasin protein alone. Quantification of the colocalization of the proteins is provided in Fig. S3. Bar, 5 µm.
Because erasin promotes ERAD (Liang et al., 2006), we considered that the erasin–ubiquilin–p97/VCP complex might function for this purpose. We reasoned that erasin might recruit p97/VCP and ubiquilin to the ER during ERAD for separate purposes: p97/VCP to aid in the retrotranslocation of ERAD substrates from the ER, whereas ubiquilin might function to bridge binding of the extracted ubiquitinated substrate and the proteasome through interaction with its UBA and UBL domains, respectively. To test this idea, we made a comparison to see if assembly of the complex was altered in cells treated with or without the proteasome inhibitor MG132. We predicted that proteasome inhibition would lead to a buildup of the complex in cells because of the inability of ubiquilin to deliver the ERAD substrate to the proteasome for degradation. Indeed, more ubiquilin was coimmunoprecipitated with erasin or p97/VCP proteins, or vice versa, in cells cultured in the presence of MG132 than in the absence of the inhibitor (Fig. 2, A and B). Furthermore, more α6 subunit components of the proteasome and ubiquitinated proteins were coimmunoprecipitated with ubiquilin, erasin, and p97/VCP proteins after proteasome inhibition (Fig. 2, A and B). Interestingly, the amount of p97/VCP or erasin proteins that coimmunoprecipitated with each other was not altered after proteasome inhibition (Fig. 2, A and B), which indicates that association of ubiquilin, but not p97/VCP, with erasin is regulated by proteasome activity.
To explore the interaction between ubiquilin and erasin further, we examined whether overexpression of either protein affects assembly of the complex. Overexpression of erasin predominantly increased the amount of p97/VCP, but not ubiquilin, that coimmunoprecipitated with erasin (Fig. 2 A). However, overexpression of ubiquilin-1 increased the amount of ubiquilin-1, but not p97/VCP, that coimmunoprecipitated with erasin (Fig. 2 A). Similar results were found upon repetition of the experiments with cells treated with MG132: binding of ubiquilin to erasin was sensitive to both proteasome inhibition and ubiquilin expression (Fig. 2 A, last four lanes). Interestingly, overexpression of ubiquilin resulted in increased amounts of polyubiquitinated proteins that coimmunoprecipitated with erasin, which was especially evident after proteasome inhibition (Fig. 2 A).
Proteasome inhibition leads to increased colocalization of ubiquilin with erasin
To obtain additional evidence that ubiquilin and erasin interact in cells, we performed double immunofluorescence staining of untransfected HEK293 cells and found that the two endogenous proteins colocalize to some extent (Fig. 2 C, a–c). However, this colocalization increased upon treatment of cells with MG132, which is consistent with our coimmunoprecipitation results (Fig. 2 C, d–f; and Fig. S2). The specificity of the staining is illustrated by loss of colocalization of the proteins after siRNA knockdown of either protein (Fig. 2 C, g–l).
Ubiquilin is recruited to the ER during ERAD
We hypothesized that if ubiquilin interacts with erasin during ERAD, then overexpression of an ERAD substrate should increase the likelihood of observing interaction of the proteins. We monitored ubiquilin binding at the ER by fractionating homogenates of cells in which we either artificially induced, or did not induce, ERAD on density gradients (Fig. 3 A). The ER fractionated mainly across the middle-to-heavy-density portions of the gradients, as revealed by the distribution of calnexin, a classical ER marker (Fig. 3 A). Erasin cofractionated with calnexin, as expected. In untreated HEK293 cells, only a faint amount of ubiquilin cofractionated with erasin, with the vast majority of the protein located in lower density end of the gradient (Fig. 3, A and F). In contrast, MG132 treatment increased migration of ubiquilin toward the ER-containing fractions, which were also enriched for ubiquitinated proteins (Fig. 3, B and F). Similarly, overexpression of the ERAD substrates CD3δ, a membrane-bound protein, or α1-anti-trypsin null Hong Kong mutant (α1ATNHK), a misfolded luminal protein, also promoted migration of ubiquilin to the ER fractions (Fig. 3, C, D, and F). In contrast, overexpression of wild-type α1AT, which is not misfolded and should not be a target for ERAD, did not promote migration of ubiquilin to the ER (Fig. 3, D and F). Treatment of cells with tunicamycin, which causes protein misfolding and increased ERAD, also promoted ubiquilin migration to the ER (Fig. 3, E and F). These results collectively suggest that ubiquilin may be recruited to the ER for ERAD.
Proteasome inhibition or expression of ERAD substrates leads to a redistribution of ubiquilin to the ER. Immunoblot analysis of HEK293 cell homogenates separated on 0–25% iodixanol gradients. (A–E) The panels show the distribution of erasin, calnexin, and ubiquilin in fractions of the gradient (heavy, bottom; light, top) of cells that were either left untreated; treated with MG132 for 4 h; transfected with either CD3δ, α1ATNHK, or wild-type α1AT cDNAs; or treated with tunicamycin for 4 h (+Tun). Ubiquitin immunoreactivity is shown for the −MG132 and +MG132 and treatments. Because its profile did not change significantly, p97/VCP is only shown for the control. (F) The amount of ubiquilin in the ER fractions was estimated by dividing the intensity of the anti-ubiquilin–immunoreactive bands in nine fractions surrounding the calnexin peak (boxed with broken lines in A–E) by the sum of the bands in the whole gradient.
Proteasome inhibition or expression of ERAD substrates leads to a redistribution of ubiquilin to the ER. Immunoblot analysis of HEK293 cell homogenates separated on 0–25% iodixanol gradients. (A–E) The panels show the distribution of erasin, calnexin, and ubiquilin in fractions of the gradient (heavy, bottom; light, top) of cells that were either left untreated; treated with MG132 for 4 h; transfected with either CD3δ, α1ATNHK, or wild-type α1AT cDNAs; or treated with tunicamycin for 4 h (+Tun). Ubiquitin immunoreactivity is shown for the −MG132 and +MG132 and treatments. Because its profile did not change significantly, p97/VCP is only shown for the control. (F) The amount of ubiquilin in the ER fractions was estimated by dividing the intensity of the anti-ubiquilin–immunoreactive bands in nine fractions surrounding the calnexin peak (boxed with broken lines in A–E) by the sum of the bands in the whole gradient.
Ubiquilin promotes degradation of membrane-bound and luminal ERAD substrates
We next investigated whether reduction of ubiquilin in HEK293 cells affects the turnover of the two classical ERAD substrates: CD3δ and α1ATNHK. siRNA-induced knockdown of ubiquilin-1 and -2 proteins in HEK293-CD3δ cells, a HEK293 cell line stably expressing orphan CD3δ protein (Ballar et al., 2007), slowed degradation of CD3δ, whereas overexpression of ubiquilin-1 slightly enhanced CD3δ turnover (Fig. 4 A). Similarly, knockdown of the ubiquilin proteins stalled turnover of α1ATNHK (Fig. 4, B and D). Because of their association with each other, we also examined if loss of erasin expression affects the turnover of the substrates. Like ubiquilin, knockdown of erasin expression reduced degradation of both substrates (Fig. 4, C and D; Liang et al., 2006). These results strongly suggest that both ubiquilin and erasin promote degradation of membrane and luminal ERAD substrates.
Knockdown of ubiquilin or erasin proteins impedes ERAD. (A) HEK293-CD3δ cells were transfected with either a ubiquilin-1 expression plasmid, the empty vector, or with UBQLN1 and UBQLN2 siRNAs. 48 h after transfection, cycloheximide was added to the cultures, and protein lysates were collected at intervals as indicated. Equal amounts of protein lysates were immunoblotted for CD3δ (HA-tag), ubiquilin, and actin. The graph shows the profile of CD3δ turnover determined from three independent experiments showing the mean and the SD (error bars). (B–D) Similar to A, except that turnover of α1ATNHK was analyzed after knockdown of either ubiquilin (B) or erasin (C). In these experiments, HEK293 cells were transfected with an α1ATNHK-HA expression construct 40 h after transfection of the siRNAs, with the turnover studies conducted 20 h later. Equal amounts of protein were immunoblotted for α1ATNHK (HA-tag), tubulin, and erasin or ubiquilin, depending on which of them was knocked down. (D) Quantification of α1ATNHK turnover after knockdown of either ubiquilin or erasin proteins, determined from three separate experiments showing the mean and SD (error bars).
Knockdown of ubiquilin or erasin proteins impedes ERAD. (A) HEK293-CD3δ cells were transfected with either a ubiquilin-1 expression plasmid, the empty vector, or with UBQLN1 and UBQLN2 siRNAs. 48 h after transfection, cycloheximide was added to the cultures, and protein lysates were collected at intervals as indicated. Equal amounts of protein lysates were immunoblotted for CD3δ (HA-tag), ubiquilin, and actin. The graph shows the profile of CD3δ turnover determined from three independent experiments showing the mean and the SD (error bars). (B–D) Similar to A, except that turnover of α1ATNHK was analyzed after knockdown of either ubiquilin (B) or erasin (C). In these experiments, HEK293 cells were transfected with an α1ATNHK-HA expression construct 40 h after transfection of the siRNAs, with the turnover studies conducted 20 h later. Equal amounts of protein were immunoblotted for α1ATNHK (HA-tag), tubulin, and erasin or ubiquilin, depending on which of them was knocked down. (D) Quantification of α1ATNHK turnover after knockdown of either ubiquilin or erasin proteins, determined from three separate experiments showing the mean and SD (error bars).
Expression of dominant-negative erasin mutants inhibits ERAD
To confirm that the function of erasin interaction with ubiquilin is connected with ERAD, we overexpressed erasin cDNAs encoding different portions of the protein, expecting that mutants containing the ubiquilin-binding site would compete for ubiquilin binding but would not create a functional ERAD complex. The erasin mutants selected were chosen based on their in vitro ubiquilin-binding properties (Fig. S1). We selected erasin fragments 1–310 aa and 1–168 aa as putative dominant-negative candidates because they both bound ubiquilin-1 but are not targeted to the ER (Liang et al., 2006), and erasin fragment 414–508 aa as an ineffective mutant because it was devoid of ubiquilin-1 binding. We compared the rate of turnover of CD3δ, stably expressed in HEK293 cells, after overexpression of the mutant constructs. As shown in Fig. 5 (A and B), erasin fragments 1–310 and 1–168 both slowed the turnover of CD3δ, but the 414–508 fragment had little effect. Furthermore, consistent with our idea of how the dominant-negative erasin mutants might function, expression of the 1–310-aa erasin mutant displaced ubiquilin binding from the ER (Fig. 5 C). These studies strongly suggest that proper binding of ubiquilin with erasin at the ER is required for promoting ERAD.
Dominant-negative erasin fragments interfere with CD3δ degradation. (A) HEK293-CD3δ cells were either mock transfected (control) or transfected with erasin deletion constructs encoding portions of the erasin polypeptide from amino acids 1–310, 414–508, or 1–168. Protein turnover was analyzed similar to Fig 4. Both a long and short exposure of the CD3δ signal is shown. Please note that the erasin 414–508 runs aberrantly on SDS-PAGE. (B) Graphs of CD3δ turnover determined from three separate experiments showing the mean and SD (error bars). (C) Immunoblots showing ubiquilin and erasin distribution in fractions of HEK293-CD3δ cell homogenates separated on 0–25% iodixanol gradients.
Dominant-negative erasin fragments interfere with CD3δ degradation. (A) HEK293-CD3δ cells were either mock transfected (control) or transfected with erasin deletion constructs encoding portions of the erasin polypeptide from amino acids 1–310, 414–508, or 1–168. Protein turnover was analyzed similar to Fig 4. Both a long and short exposure of the CD3δ signal is shown. Please note that the erasin 414–508 runs aberrantly on SDS-PAGE. (B) Graphs of CD3δ turnover determined from three separate experiments showing the mean and SD (error bars). (C) Immunoblots showing ubiquilin and erasin distribution in fractions of HEK293-CD3δ cell homogenates separated on 0–25% iodixanol gradients.
Abrogation of MG132-induced accumulation of ubiquilin, p97/VCP, and proteasomes in microsomes after erasin knockdown
The preceding results were in accord with the notion that erasin functions to recruit p97/VCP and ubiquilin to the ER during ERAD. To obtain additional evidence in support of this idea and to examine if ubiquilin, in turn, functions to recruit proteasomes to the ER during ERAD, we compared the protein composition of microsomes isolated from HEK293 cells expressing endogenous levels of erasin and ubiquilin proteins with those from cells silenced for expression of erasin and/or ubiquilin-1 and -2. Microsomes contain an enriched source of ER fragments, which we used to monitor changes in protein composition of the ER.
As shown in Fig. 6 A, four ER proteins—erasin, calnexin, derlin-1, and derlin-2—were detected in the microsome fraction but not the cytosolic fraction, which confirms that our microsome preparations were indeed enriched for ER components. Microsomes that were isolated from cells transfected with erasin siRNAs contained ∼80% less erasin than those isolated from mock-transfected cells, which indicates successful depletion of erasin protein content of microsomes by RNAi (Fig. 6 A).
Microsomes of cells depleted of erasin contain less ubiquilin, p97/VCP, and proteasomes. HEK293 cells were either mock transfected or transfected with siRNAs against erasin. 72 h after transfection, the cells were incubated with MG132 for an additional 4 h, whereas a duplicate set was left untreated. (A) Equal amounts of protein in microsome and supernatant fractions of the cells were separated by SDS-PAGE and probed for the proteins shown. Blots of equal amounts of proteins in total lysates of the cultures are also shown. (B) Similar to A, except that this time erasin or ubiquilin-1 and -2 were knocked down separately, or simultaneously. Additionally, only the microsome fractions were probed for proteasome subunits (α6, α7, and Rpn10), ubiquilin, and calnexin (to ensure equal protein loading). Also shown are the blots of the total lysates of the cultures. (C) Assays of proteasome activity in microsomes and total lysates of HEK293-CD3δ cells that were either mock transfected or transfected with siRNAs against erasin or ubiquilin-1 and -2 of three separate cultures showing the mean and SD (error bars). (D) A repeat of the experiment shown in A, but this time we immunoprecipitated ubiquilin from the cells and examined equal fractions of the precipitates by immunoblotting for the indicated proteins. Blots of the total lysates are also shown. (E) Ubiquilin was immunoprecipitated from HEK293-CD3δ cells that were either left untreated or treated with MG132 for 4 h. Equal portions of the immunoprecipitates were probed for the proteins shown.
Microsomes of cells depleted of erasin contain less ubiquilin, p97/VCP, and proteasomes. HEK293 cells were either mock transfected or transfected with siRNAs against erasin. 72 h after transfection, the cells were incubated with MG132 for an additional 4 h, whereas a duplicate set was left untreated. (A) Equal amounts of protein in microsome and supernatant fractions of the cells were separated by SDS-PAGE and probed for the proteins shown. Blots of equal amounts of proteins in total lysates of the cultures are also shown. (B) Similar to A, except that this time erasin or ubiquilin-1 and -2 were knocked down separately, or simultaneously. Additionally, only the microsome fractions were probed for proteasome subunits (α6, α7, and Rpn10), ubiquilin, and calnexin (to ensure equal protein loading). Also shown are the blots of the total lysates of the cultures. (C) Assays of proteasome activity in microsomes and total lysates of HEK293-CD3δ cells that were either mock transfected or transfected with siRNAs against erasin or ubiquilin-1 and -2 of three separate cultures showing the mean and SD (error bars). (D) A repeat of the experiment shown in A, but this time we immunoprecipitated ubiquilin from the cells and examined equal fractions of the precipitates by immunoblotting for the indicated proteins. Blots of the total lysates are also shown. (E) Ubiquilin was immunoprecipitated from HEK293-CD3δ cells that were either left untreated or treated with MG132 for 4 h. Equal portions of the immunoprecipitates were probed for the proteins shown.
We next examined whether microsomes of cells depleted of erasin contained less ubiquilin, p97/VCP, and proteasomes than those of cells not depleted of erasin. We used antibodies against the α6 subunit and Rpt1, which are contained in the 20S catalytic core and 19S complex of the proteasome (Pickart and Cohen 2004), respectively, as markers for proteasomes. The amount of p97/VCP in microsomes was reduced by ∼50% after erasin knockdown in both MG132-treated and untreated cultures (Fig. 6 A), which suggests that p97/VCP binding to microsomes is somewhat dependent on erasin expression. Similarly, the amounts of ubiquilin, Rpt1, and α6 subunit in microsomes of cells depleted of erasin were all reduced compared with cells in which erasin levels were not altered, but only in cells that were treated with MG132 (Fig. 6 A).
The preceding results were in accord with the idea that erasin located in the ER binds ubiquilin, which, in turn, binds the proteasome. However, because reduction of erasin expression by RNAi resulted in a concomitant reduction in ubiquilin protein in microsomes, we next asked if reduction of ubiquilin protein alone was sufficient to reduce proteasome recruitment to the ER during ERAD. As shown in Fig. 6 B, microsomes of cells transfected with siRNA against ubiquilin-1 and -2, or a combination of siRNAs against both ubiquilins and erasin, failed to manifest the MG132-dependent increase in proteasome binding, as measured using three different proteasome components, that occurred in cells in which erasin or ubiquilin expression was not silenced. Furthermore, consistent with the idea that knockdown of erasin or ubiquilin proteins reduces proteasome recruitment to the ER, we found that proteasome activity in microsomes isolated cells stably expressing CD3δ in which ubiquilin or erasin proteins were knocked down was reduced ∼20% compared with cells in which the proteins were not targeted for RNAi (Fig. 6 C).
Effect of erasin knockdown on coimmunoprecipitation of p97/VCP, ubiquilin, and the proteasome
We next examined whether reduction of erasin in cells affects assembly of the ERAD complex. HEK293 cells transfected with erasin siRNAs, or cells that were mock transfected, were cultured with or without MG132, and ubiquilin proteins were immunoprecipitated from the cells. The precipitates were examined for erasin, p97/VCP, and the proteasome by immunoblotting. As shown in Fig. 6 D, more erasin, p97/VCP, and the α6 subunit of the proteasome coimmunoprecipitated with ubiquilin from cells treated with MG132 than from those not treated with MG132. Cells that were transfected with siRNAs against erasin and treated with MG132 contained lower levels of erasin, p97/VCP, and the α6 subunit than untransfected cells treated with MG132, which suggests that knockdown of erasin expression interferes with formation or stability of the ERAD complex. Immunoblot analysis of total protein lysates confirmed that erasin levels were reduced by ∼80% after the knockdown. Ubiquilin levels were slightly elevated in the erasin knockdown cells compared with the mock-transfected cells, but the levels of p97/VCP and the α6 subunit remained relatively constant. Thus, the decreased coimmunoprecipitation of p97/VCP and the α6 subunit with ubiquilin after erasin knockdown was not caused by a global change in the expression of the proteins. We should point out that the cells depleted of erasin still displayed a slight MG132-dependent increase in the coimmunoprecipitation of p97/VCP and the α6 subunit with ubiquilin. At present, we cannot distinguish whether this is from incomplete silencing of erasin expression or interactions mediated by proteins other than erasin. Nevertheless, these data suggest that erasin may be required for the assembly of ubiquilin, p97/VCP, and the proteasome in a complex.
To demonstrate that ubiquilin functions as a shuttle factor in delivery of ERAD substrates to the proteasome for degradation, we immunoprecipitated ubiquilin from HEK293 cells stably expressing CD3δ and probed them for the presence of CD3δ and the proteasome. As shown in Fig. 6 E, CD3δ as well as 19 and 20S components of the proteasome were found in the ubiquilin immunoprecipitates, and, as expected, more of them were present after MG132 treatment of the cells.
Ubiquilin and erasin expression is induced by ER stress in Caenorhabditis elegans
Because chronic disruption of ERAD causes ER stress, we predicted that loss of ubiquilin or erasin expression would lead to ER stress. We tested this hypothesis in C. elegans. Worms were used for several reasons. First, they are an ideal model organism to study ER stress, with several mutant and reporter lines available for such purposes (Shen et al., 2001, 2005). Second, unlike mammals, worms contain single ubiquilin and erasin genes, making it easier to inactivate the genes without the complication of compensation by redundant family members. And third, because of the efficacy of RNAi in worms.
We first determined whether ubiquilin and erasin are part of the compendium of genes that are induced during the UPR. Accordingly, worms containing an integrated hsp4::gfp transcriptional reporter, widely used to monitor ER stress (Shen et al., 2001; Calfon et al., 2002), were treated with tunicamycin to induce ER stress for different periods of time. As expected, transcripts of the classical ER stress-regulated gene, hsp-4 (the worm homologue of human BiP), were induced by the drug treatment, with peak expression occurring 4 h after drug treatment, but then its expression was attenuated (Fig. 7, A and B). Interestingly, both ubiquilin and erasin transcripts increased with somewhat similar profiles to that of hsp-4, which suggests that they are part of the UPR response (Fig. 7, A–D). Expression of ama-1, which is not part of the UPR, did not change over this period, as expected (Fig. 7 A).
Ubiquilin and erasin expression are induced by ER stress in C. elegans. (A) Mixed-stage zcls4 [hsp-4::GFP] worms were grown on plates with 28 µg/ml tunicamycin for 0, 4, 8, 16, and 48 h, and transcript levels of hsp-4, ubiquilin, erasin, and ama-I were measured by semiquantitative PCR and gel electrophoresis. Quantification of Hsp-4 (B), ubiquilin (C), and erasin (D) expression relative to the 0 time point. Similar trends were observed in two other experiments. (E) Schematic diagram of the hsp-4::GFP reporter construct. (F) Immunoblot analysis of equal amounts of protein lysate (two independent samples for each time point) prepared from worms treated with 28 µg/ml of tunicamycin for the periods indicated. An unknown protein whose expression was insensitive to ER stress was used as a loading control. (G) Schematic diagram of the C. elegans ubiquilin promoter fragment fused to GFP. (H) Immunoblot analysis of equal amounts of protein lysate of worms treated with 0, 2.5, or 28 µg/ml tunicamycin for 24 h. (I) Quantification of GFP expression determined by scanning of immunoblots as described in H showing the mean and SD (error bars) seen in three different experiments. (J) GFP fluorescence of worms containing the integrated ubiquilin::GFP fusion construct treated for 24 h with 0, 2.5, or 28 µg/ml tunicamycin. Bar, 0.2 mm.
Ubiquilin and erasin expression are induced by ER stress in C. elegans. (A) Mixed-stage zcls4 [hsp-4::GFP] worms were grown on plates with 28 µg/ml tunicamycin for 0, 4, 8, 16, and 48 h, and transcript levels of hsp-4, ubiquilin, erasin, and ama-I were measured by semiquantitative PCR and gel electrophoresis. Quantification of Hsp-4 (B), ubiquilin (C), and erasin (D) expression relative to the 0 time point. Similar trends were observed in two other experiments. (E) Schematic diagram of the hsp-4::GFP reporter construct. (F) Immunoblot analysis of equal amounts of protein lysate (two independent samples for each time point) prepared from worms treated with 28 µg/ml of tunicamycin for the periods indicated. An unknown protein whose expression was insensitive to ER stress was used as a loading control. (G) Schematic diagram of the C. elegans ubiquilin promoter fragment fused to GFP. (H) Immunoblot analysis of equal amounts of protein lysate of worms treated with 0, 2.5, or 28 µg/ml tunicamycin for 24 h. (I) Quantification of GFP expression determined by scanning of immunoblots as described in H showing the mean and SD (error bars) seen in three different experiments. (J) GFP fluorescence of worms containing the integrated ubiquilin::GFP fusion construct treated for 24 h with 0, 2.5, or 28 µg/ml tunicamycin. Bar, 0.2 mm.
An examination of the changes in proteins in the tunicamycin-treated worms revealed that the GFP protein expression, under the control of the hsp-4 promoter (Fig. 7 E), increased temporally, with high accumulation observed 16 h after drug treatment (Fig. 7 F). This was accompanied by a parallel increase in ubiquitinated proteins, which is consistent with the idea that tunicamycin treatment causes accumulation of misfolded proteins in the worms. Ubiquilin and erasin protein levels increased fivefold and threefold, respectively, 4 h after tunicamycin treatment, but thereafter their levels remained constant. The difference between the levels of ubiquilin and erasin transcripts and their respective proteins suggests possible posttranscriptional and/or posttranslational regulation of the genes.
To determine if ubiquilin and erasin promoters are direct targets of the UPR, we cloned the GFP open reading frame downstream of a 2-kb fragment containing the putative C. elegans ubiquilin promoter and isolated transgenic worms containing the integrated ubiquilin::gfp promoter fusion plasmid (Fig. 7 G). As shown in Fig. 7 (H–J), ubiquilin::gfp transgenic worms manifested a tunicamycin dose-dependent increase in GFP expression by both immunoblotting and immunofluorescence microscopy. Interestingly, GFP expression was particularly prominent in the pharyngeal muscle, hypodermis, and intestine of the worms, which suggests that the reporter might be unevenly expressed across the worm. Similarly, worms carrying an erasin::GFP reporter were also reported to display a similar tunicamycin dose-dependent increase in GFP expression (Yamauchi et al., 2007). Together, these results suggest that ubiquilin and erasin promoters contain UPR-response elements.
Expression of ubiquilin and erasin during ER stress is regulated by ire-1
We next investigated which arm of the UPR regulates ubiquilin and erasin expression. Like in mammals, the UPR in C. elegans is induced through the action of three main sensors: ire-1, pek-1 (called protein kinase-like ER kinase [PERK] in mammals), and atf-6. We used worms containing inactive versions of the sensors to determine which pathway regulates tunicamycin induction of ubiquilin and erasin expression. In the control wild-type N2 worms, expression of ubiquilin and erasin transcripts increased after 6 h of tunicamycin treatment (Fig. 8). However, the induction of both genes was almost completely attenuated in ire-1 (v33) mutant worms, and partially attenuated in the atf-6 (ok551) and pek-1 (ok275) mutants (Fig. 8). Together, these results indicate that in C. elegans, both ubiquilin and erasin genes are chiefly regulated by ire-1.
The ire-1 branch of the UPR regulates induction of ubiquilin and erasin expression during ER stress. (A) Mixed-stage worms of the indicated mutant strains were grown on plates with 28 µg/ml tunicamycin for 0 or 6 h. Transcript levels of hsp-4 (regulated by ire-1; Shen et al., 2001), ubiquilin, erasin, and CBP were analyzed by PCR and agarose gel electrophoresis. (B–E) Quantification of the relative change in transcript expression determined from the experiment shown in A. Levels were normalized to the 0 h time point in each mutant. Similar results were observed in two other experiments.
The ire-1 branch of the UPR regulates induction of ubiquilin and erasin expression during ER stress. (A) Mixed-stage worms of the indicated mutant strains were grown on plates with 28 µg/ml tunicamycin for 0 or 6 h. Transcript levels of hsp-4 (regulated by ire-1; Shen et al., 2001), ubiquilin, erasin, and CBP were analyzed by PCR and agarose gel electrophoresis. (B–E) Quantification of the relative change in transcript expression determined from the experiment shown in A. Levels were normalized to the 0 h time point in each mutant. Similar results were observed in two other experiments.
RNAi of ubiquilin or erasin results in accumulation of misfolded proteins and induction of ER stress
We next examined whether reduction of ubiquilin or erasin expression in worms causes ER stress. Reduction of either ubiquilin or erasin expression in hsp-4::gfp worms by bacteria-mediated RNAi resulted in a threefold and fivefold increase in GFP expression, respectively (Fig. 9 A). In both cases, GFP expression was lower than that produced by treatment of the worms with tunicamycin, which suggests that loss of ubiquilin or erasin expression induces more subtle ER stress than that produced by tunicamycin treatment. Interestingly, knockdown of ubiquilin expression led to an induction of erasin expression, and vice versa (Fig. 9, A and B). Moreover, RNAi of either ubiquilin or erasin led to an accumulation of ubiquitinated proteins in the worms, which is consistent with the idea that the proteins function in the disposal of misfolded proteins from cells.
Knockdown of ubiquilin or erasin proteins in worms results in the induction of the ER stress reporter hsp-4::GFP, accumulation of misfolded proteins, and a shorter lifespan. (A) Mixed-stage zcls4 [hsp-4::GFP] worms were either fed standard bacteria in the presence or absence of 28 µg/ml tunicamycin or bacteria to induce RNAi of the ubiquilin gene. Immunoblot analysis of equal amounts of protein lysate prepared after 48 h of treatment for GFP, ubiquilin, erasin, ubiquitin, and an unknown protein used as a loading control. (B) Identical to A, except that worms were fed bacteria to specifically knock down erasin. (C) Lifespan curves comparing N2 (wild-type) worms fed bacteria transformed with the empty RNAi vector or the vector containing cDNAs to induce RNAi of either ubiquilin or erasin genes. Results are cumulative from three independent experiments, with ≥60 animals per trial. Log-rank (Mantel-Cox) and Gehan-Breslow-Wilcoxon tests, P < 0.0001 versus control.
Knockdown of ubiquilin or erasin proteins in worms results in the induction of the ER stress reporter hsp-4::GFP, accumulation of misfolded proteins, and a shorter lifespan. (A) Mixed-stage zcls4 [hsp-4::GFP] worms were either fed standard bacteria in the presence or absence of 28 µg/ml tunicamycin or bacteria to induce RNAi of the ubiquilin gene. Immunoblot analysis of equal amounts of protein lysate prepared after 48 h of treatment for GFP, ubiquilin, erasin, ubiquitin, and an unknown protein used as a loading control. (B) Identical to A, except that worms were fed bacteria to specifically knock down erasin. (C) Lifespan curves comparing N2 (wild-type) worms fed bacteria transformed with the empty RNAi vector or the vector containing cDNAs to induce RNAi of either ubiquilin or erasin genes. Results are cumulative from three independent experiments, with ≥60 animals per trial. Log-rank (Mantel-Cox) and Gehan-Breslow-Wilcoxon tests, P < 0.0001 versus control.
Reduction of worm lifespan by RNAi of ubiquilin or erasin
Finally, we investigated whether RNAi of ubiquilin or erasin alters lifespan in the worm. As shown in Fig. 9 C, RNAi of erasin shortened lifespan in worms from a mean survival time of 21 d to 12 d, whereas RNAi of ubiquilin reduced it to 17 d. Together, these data suggest that loss of either ubiquilin or erasin expression shortens the worm lifespan.
Discussion
Here, we describe a novel erasin-containing protein complex involved in ERAD in human cells. Our results suggest that erasin acts as a platform to recruit both p97/VCP and ubiquilin to the complex, and that ubiquilin, in turn, recruits proteasomes. These findings, together with the known functions of the proteins with which erasin coimmunoprecipitates, have enabled us to construct a model of how the complex might function in ERAD. Specifically, we propose that misfolded protein substrates that are extracted from the ER interact sequentially with a series of proteins and are eventually delivered to the terminal degradation apparatus of the cell, the proteasome (Fig. 10). The model we propose predicts that delivery of ER-derived substrates to the proteasome is highly coupled in a manner that prevents premature release of the substrate into the cytosol, where it might accumulate or aggregate into a potentially toxic species.
Model of the mammalian erasin-assembled ERAD complex. A schematic diagram of the putative erasin-containing ERAD complex (see Discussion).
Model of the mammalian erasin-assembled ERAD complex. A schematic diagram of the putative erasin-containing ERAD complex (see Discussion).
Our model focuses on erasin, an ER resident protein that is thought to embed into the membrane via a short hydrophobic sequence close to its C terminus, with the remainder of the protein facing the cytoplasm (Fig. 10 A; Liang et al., 2006). We propose that erasin forms a complex with derlin and gp78/Hrd1 ligases either through direct or indirect interaction of the proteins, which is supported by coprecipitation of the proteins (Liang et al., 2006; Mueller et al., 2008). Erasin recruits p97/VCP to the complex via direct interaction with its UBX domain (Fig. 10 B; Liang et al., 2006). On the basis of previous studies of the mechanism by which ERAD is thought to occur (Ye et al., 2001; Jarosch et al., 2002; Vembar and Brodsky, 2008), we propose that the misfolded substrate in the ER is polyubiquitinated by gp78/Hrd1 and/or related ubiquitin ligases and extracted through the derlin channel by p97/VCP (Fig. 10 B). We speculate that the polyubiquitin chain that is conjugated onto the substrate is then recognized by the UBA domain of ubiquilin, which promotes ubiquilin binding to erasin, or vice versa (Fig. 10 B). Ubiquilin in turn recruits the proteasome to the ERAD complex, probably via interactions with its UBL domain, bringing it in close proximity for the delivery and degradation of the ERAD substrate that is anchored to the UBA domain of ubiquilin (Fig. 10, C and D). After degradation of the substrate, we speculate that the complex most likely disassembles. The model incorporates many of our results with the well-established functions of the proteins mentioned. However, gaps remain, as discussed below.
A key aspect of our model emphasizes the apparent difference in the binding of p97/VCP and ubiquilin proteins with erasin that we found occurs during ERAD. According to our in vitro binding results, recombinant ubiquilin, p97/VCP, and erasin proteins can all bind together in one complex. Yet according to immunoprecipitation studies, we noticed differences in the amount of the proteins that were bound to erasin under different conditions: the amount of p97/VCP that was bound to erasin was dependent on the amount of erasin protein expressed, whereas ubiquilin binding to erasin was stimulated by either overexpression of ubiquilin itself, by proteasome inhibition, or by enhancing ERAD. On the basis of these results, we suggest that binding of p97/VCP to erasin is governed principally by the amount of erasin in cells, whereas binding of ubiquilin to erasin is transitory, and might only occur during delivery of the polyubiquitinated substrate to the proteasome for degradation. According to this model, inhibition of the proteasome would arrest binding of ubiquilin with erasin because of the inability of ubiquilin to deliver its bound substrate to the proteasome for degradation.
The ERAD-enhanced and proteasome inhibition–dependent increase in ubiquilin interaction with erasin provides important clues as to the possible mechanisms by which the complex is assembled. We speculate that during ERAD, the polyubiquitinated chain that is conjugated onto the substrate might first be recognized by the UBA domain of ubiquilin, bringing it in closer proximity to erasin, thereby enhancing their probability of interaction. However, we cannot rule out the possibility that ubiquilin first interacts with erasin, and then with the polyubiquitinated substrate. Both mechanisms are supported by in vitro binding studies. For example, we found that ubiquilin binds erasin with very high affinity (with a KD of 30 nm; Fig. S4). Also, the UBA domain of ubiquilin binds a wide variety of ubiquitin moieties with similar high affinity, including K48-linked tetra-ubiquitin that is principally conjugated onto proteins that are degraded by proteasomes (Ko et al., 2004; Massey et al., 2004; Raasi et al., 2005).
In the next step of our model, we propose that erasin-bound ubiquilin recruits proteasomes to the complex, probably via interaction of its UBL domain with subunits of the proteasome. This proposal is based on previous demonstration that the UBL domain binds the proteasome (Kleijnen et al., 2000; Ko et al., 2004). In support of such a mechanism, we found that silencing of ubiquilin expression abrogated proteasome recruitment to the erasin complex. Furthermore, silencing of erasin expression led to a similar outcome, which is expected because erasin serves to recruit ubiquilin to the complex. Finally, inhibition of the proteasome resulted in increased amounts of CD3δ (the ERAD substrate) and more proteasome subunits that coimmunoprecipitated with ubiquilin, as predicted by the model.
The recruitment of proteasomes to the ER by the ERAD complex we have described is somewhat similar to JNK-associated membrane protein (JAMP), another protein involved in ERAD (Tcherpakov et al., 2008, 2009). We therefore examined whether erasin and JAMP are part of the same complex, but found that JAMP does not coimmunoprecipitate with erasin (Fig. S5), which suggests that they form distinct complexes. Additional coimmunoprecipitation studies indicated that Ufd1, a p97/VCP-interacting factor, is not associated with erasin (Fig. S5), which is similar to a previous report showing that Ufd1 and p97/VCP are not always found bound together in ERAD (Ballar et al., 2006).
Our in vitro binding results indicated that ubiquilin-1, but not hHR23A, binds erasin, which suggests that these two human ubiquitin–proteasome shuttle proteins cannot substitute for each other, at least for binding to the erasin-containing ERAD complex. In a recent study, ubiquilins were found to interact with Herp and regulate degradation of CD3δ (Kim et al., 2008). Herp is another ER-associated protein that shares many properties with erasin. Its topology is similar to that of erasin (Kokame et al., 2000), and like erasin, it associates with derlin-1, p97/VCP, and Hrd1 (Schulze et al., 2005; Okuda-Shimizu and Hendershot, 2007). Also, Herp expression is induced by ER stress (Kokame et al., 2000; Ma and Hendershot, 2004). At present, it is unclear whether ubiquilins discriminate between or bind equally well to erasin and Herp. Interestingly, Herp does not coimmunoprecipitate with erasin (Fig. S5), which suggests they are in distinct complexes.
In accord with their involvement in ERAD, knockdown of either ubiquilin or erasin in C. elegans induced ER stress. The results are consistent with the strong coordination that is known to exist between ERAD and UPR: interference of ERAD induces the UPR, whereas UPR induction increases ERAD capacity (Travers et al., 2000). UPR induction of both erasin and ubiquilin genes in C. elegans appeared to be under the control of ire-1. The ire-1–dependent regulation of erasin and ubiquilin expression is in accord with previous studies indicating that this arm of the UPR is principally involved in regulating expression of ERAD components (Travers et al., 2000).
Our finding, that erasin and ubiquilin are induced by ER stress, might help explain the increased staining of the proteins found in brains of individuals afflicted with Alzheimer's disease (AD; Mah et al., 2000; Liang et al., 2006). The simplest explanation is that ER stress is involved in AD pathogenesis. Obviously, this remains speculative, but it is consistent with the observation that UPR markers are activated AD (Hamos et al., 1991; Hoozemans et al., 2005). In fact, ER stress has now been linked to other neurodegenerative diseases, such as amyotrophic lateral sclerosis and Huntington's disease (Nishitoh et al., 2008; Duennwald and Lindquist, 2008). It is worth noting that in the case of Huntington's disease, overexpression of ubiquilin, which we now show here promotes ERAD, suppresses the toxicity of expanded polyglutamine proteins in cell and animal models of the disease (Wang et al., 2006).
We found that reduction of ubiquilin or erasin expression shortened the life span of worms, which suggests that they are important for normal survival. Similar to worms, loss of ubiquilin in Drosophila shortens life span (Li et al., 2007). To our knowledge, there is only one other report of the consequences of loss of erasin in animals: knockdown by morpholino in zebrafish, which produced highly abnormal embryos (Pickart et al., 2006).
In summary, our results provide strong evidence that ubiquilin and erasin proteins bind together in a novel ERAD complex and that they play important roles in regulating protein degradation and ER stress.
Materials and methods
Y2H experiments
A cDNA fragment encoding the central domain of ubiquilin-1 was cloned in-frame with the LexA coding sequence in the yeast Y2H bait vector, pEG202 (Golemis et al., 2008). The resulting bait was transformed into yeast strain EGY48, which was then used to screen a human fetal brain cDNA library for interactors by selection on plates lacking uracil, histidine, tryptophan, and leucine (Mah et al., 2000). Putative interactors were further characterized by DNA sequencing and by interaction with additional baits as listed in Fig. 1 B. The strength of interaction between the different preys and baits was quantified by β-galactosidase activity assays using the chromogenic substrate o-nitrophenyl-β-d-galactopyranoside (Golemis et al., 2008).
In vitro transcription/translation and GST pull-down assays
[35S]methionine-radiolabeled full-length and partial-length erasin and lamin polypeptides were synthesized in rabbit reticulocyte lysates using a coupled in vitro transcription and translation system (Promega). The 35S-labeled proteins were incubated with 3 µg of purified GST-His or different GST–ubiquilin-1 fusion proteins for 1 h at 4°C under constant rotation. Glutathione-agarose (Sigma-Aldrich) was then added to the mixtures and the incubation was continued for another 1 h. The agarose beads were recovered by centrifugation and washed in standard PBS containing 0.5% NP-40, then equal portions of the samples were separated by SDS-PAGE and stained with Coomassie blue, and an autoradiogram was prepared.
Using a similar strategy, we identified the ubiquilin-binding site in erasin. The deletion constructs and the results of this mapping study are shown in Fig. S1.
In vitro binding of full-length recombinant proteins to one another was analyzed by GST pull-down assays. Recombinant full-length mouse p97/VCP, human erasin, human hHR23A (the expression construct was provided by the late C. Pickart, Johns Hopkins University, Baltimore, MD), and human ubiquilin-1 proteins were expressed in BL21(DE3) bacteria as either His- or GST-tagged fusion proteins. The p97/VCP-His and erasin-His bacterially expressed proteins were purified by nickel–nitrilotriacetic acid–agarose (QIAGEN) affinity chromatography and the GST–ubiquilin-1 and GST-hHR23A proteins by glutathione-agarose affinity chromatography. The purified proteins were dialyzed against 20 mM Tris, pH 8.0, 150 mM NaCl, and 10% glycerol. GST pull-down experiments were conducted as follows: the GST proteins were first incubated with GS-agarose beads in pull-down buffer (20 mM Tris, pH 8.0, 150 mM NaCl, 3 mM MgCl2, 10% glycerol, and 0.1% Triton X-100) for 2 h at 4°C. The beads were recovered, washed, and then incubated with various combinations of erasin-His, p97/VCP-His, or both erasin-His and p97/VCP-His in pull-down buffer for ∼1.5 h at 4°C. Beads were then washed three times with pull-down buffer containing 800 mM NaCl. Proteins that were retained on beads were released with SDS sample buffer and analyzed by immunoblotting.
Cell culture, DNA and siRNA transfection, and immunofluorescence microscopy
HEK293 cells were grown in DME supplemented with 10% FBS. Cells were transiently transfected with plasmid DNA using Lipofectamine 2000 (Invitrogen) or with siRNA SMARTpools (Thermo Fischer Scientific) against erasin, ubiquilin-1, or ubiquilin-2 using the DharmaFECT reagent 1 (Thermo Fisher Scientific) according to the manufacturer's instructions. The turnover of HA-tagged CD3δ and a1ATNHK proteins was measured by immunoblotting equal amounts of protein lysate that were collected at various time intervals, as indicated in the figures, from cultures to which cycloheximide was added to a final concentration 100 µM to block new protein synthesis (Liang et al., 2006; Ballar et al., 2007). Association of erasin with JAMP was analyzed by coimmunoprecipitation assays using a FLAG-tagged JAMP expression construct (provided by Z. Ronai, Burnham Institute for Medical Research, La Jolla, CA). For immunofluorescence microscopy, HEK293 cells grown on coverslips were fixed with 4% paraformaldehyde and stained with rabbit anti-erasin and mouse anti-ubiquilin primary antibodies followed by fluorescein-conjugated donkey anti–rabbit and rhodamine-conjugated donkey anti–mouse secondary antibodies, after which the coverslips were mounted onto glass slides using Aqua Polymount (Polysciences, Inc.). Fluorescence images were captured at room temperature on an LSM 510 confocal microscope using an Axiovert 100M microscope, a 63× oil-immersion lens (NA 1.4) with Carl Zeiss 4.2 software (all from Carl Zeiss, Inc.). Colocalization of the images was performed with iVision-Mac software (BioVision Technologies) without any other manipulation of the images. The extent of colocalization of ubiquilin and erasin is shown as Fig. S3.
Protein preparation, SDS-PAGE, immunoprecipitation, and immunoblotting
For immunoprecipitation (IP) assays, cells were harvested with standard 1× PBS buffer and then collected by centrifugation at 5,000 g for 5 min. The resulting pellets were resuspended in IP buffer (50 mM Tris-Hcl, pH 7.5, 150 mM NaCl, 2 mM EDTA, and 0.1% NP-40), and the cells were thoroughly lysed by passage of the mixture 30 times through a 25-gauge needle. The resulting mixture was centrifuged at 3,000 g for 10 min, after which the supernatant was used for IP. For these assays, 5 µl of an appropriate antibody was added together with ∼100 µl of protein A–Sepharose CL-4B beads (GE Healthcare) and the cell supernatant containing ∼500 µg of protein in a 1.5-ml tube. The contents of the tube were mixed by rotation for 2 h at 4°C, after which the beads were recovered by centrifugation at 5,000 g for 5 min followed by 5 washes with IP buffer. The bound proteins were eluted with standard Laemmli gel sample buffer (Laemmli 1970) and separated by SDS-PAGE. The proteins were then transferred onto 0.4-µm-pore-size Immobilon-P membranes (Millipore), according to the manufacturer's instructions, followed by standard immunoblotting of proteins using the SuperSignal West Pico detection method (Thermo Fisher Scientific). The primary antibodies used in this study were obtained from the following sources. Goat polyclonal anti-actin (Santa Cruz Biotechnology Inc.); rabbit polyclonal antibodies anti-erasin, anti-ubiquilin, anti-p97/VCP, anti-GST (generated by our laboratories), anti–derlin-1 (MBL International), anti–derlin-2 (MBL International), and anti–calnexin-C (StressGen); and mouse monoclonal antibodies anti-ubiquilin (Invitrogen), anti-p97(VCP) (BD), anti-Herp (Abgent), anti-Ufd1 (a gift of H. Meyer, Swiss Federal Institute of Technology, Zurich, Switzerland), anti-His (GE Healthcare), anti-FLAG, anti-HA, anti-tubulin (all from Sigma-Aldrich), anti-ubiquitin (Santa Cruz Biotechnology, Inc.), and α6, α7, Rpt1, and Rpn10 subunits of the proteasome (all from Enzo Life Sciences, Inc.).
Cell fractionation and proteasome activity
HEK293 cells that were either treated with MG132 (50 nM final concentration) or left untreated were homogenized in 1 ml of buffer B (0.25 M sucrose, 1 mM EDTA, and 10 mM Hepes-NaOH, pH 7.4) by 10 up-and-down strokes with the pestle using a Dounce homogenizer, then centrifuged at 3,000 g for 10 min at 4°C, after which the supernatant was collected and layered on top of 10-ml preformed 0–25% iodixanol gradients made with buffer B. The layered gradients were then centrifuged at 200,000 g at 4°C for 2.5 h in a rotor (SW 41Ti; Beckman Coulter). After centrifugation, 0.65-ml fractions were collected from the bottom of the tube. An equal volume of each fraction was immunoblotted for different proteins.
Microsomes were prepared from HEK293 cells that were either mock transfected or transfected with siRNAs (10 nM final concentration) against erasin, or ubiquilin-1 and ubiquilin-2 proteins using hypotonic lysis and the differential centrifugation procedure (Kokame et al., 2000). In brief, after transfection, cells were resuspended in 1 ml of buffer (10 mM Hepes-KOH, pH 7.4, 250 mM sucrose, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, and 1 mM EGTA) and then sheared by passing the suspension 30 times through a 25-gauge needle. A portion of the sample was saved to monitor differences in protein expression between the samples. The remainder of the sample was centrifuged at 1,000 g for 5 min at 4°C, and the resulting supernatant removed and respun at 100,000 g for 1 h at 4°C. The resulting pellet and supernatant fractions were either dissolved in our standard protein lysis buffer containing protease inhibitors for SDS-PAGE analysis or used for proteasome activity assays. Equal amounts of protein in the supernatant and pellet fractions of microsomes were analyzed by immunoblotting for the presence of various proteins as shown in Fig. 6. Corresponding immunoblots were conducted of the samples that were saved before the high-speed centrifugation step.
Proteasome activity was measured using the 20S proteasome activity assay kit (Millipore), which uses N-succinyl-Leu-Leu-Val-Try-AMC (7-amino-4-methycoumarin) as a fluorogenic substrate. In these assays, equal amounts of protein contained in microsomes prepared from HEK293-CD3δ cells that were either mock transfected or transfected with siRNAs against erasin or ubiquilin-1/2 were incubated with the fluorogenic substrate with and without the proteasome inhibitor lactacystin. After 1 h incubation of the reactions at 37°C, the fluorescence (380 nm excitation/460 nm emission) was measured using a fluorescent plate reader (SpectraMax Gemini; MDS Analytical Technologies), and the proteasome activity reported was calculated by subtracting the fluorescence reading of the reaction incubated with lactacystin from that lacking the inhibitor. Proteasome activity after erasin and ubiquilin-1/2 knockdown was normalized to that in the mock-transfected cells. Proteasome activity was also determined in fractions of the total cell lysates that were saved before the microsome preparations. The linear responsiveness of the fluorescence reader for the activity assays was established using increasing amounts of 7-amino-4-methycoumarin (Enzo Life Sciences, Inc.).
C. elegans strains and drug treatments
C. elegans strains were cultivated under standard conditions (Brenner, 1974). The strain N2 (Bristol) was used as the wild-type strain. The reporter strain zcls4 [hsp-4::GFP] and the mutant strains ire-1 (v33), pek-1 (ok275), and atf-6 (ok551) were obtained from the Caenorhabditis Genetics Center, University of Minnesota. For the ER stress tests, nematodes were grown on plates containing 0, 2.5, or 28 µg/ml of tunicamycin (Sigma-Aldrich) for up to 48 h.
PCR analysis of transcripts expressed in worms
Total RNA was isolated from mixed-stage nematodes by using TRIzol reagent (Invitrogen). First-strand cDNA was synthesized using oligo-dT primer (Invitrogen), and 0.5 µg/µl of the cDNA was amplified using primers for ubiquilin (F15C11.2) 5′-ATGGCTACAGAGAGTGCACTCATCAAAGTT-3′ and 5′-CTATGGAGAGTTGAGAAGACGTTCGACAGC-3′; erasin (ZK353.8) 5′-GCTGATCGCGAAGCTGCTCAGAAAAAGTTCGGG-3′ and 5′-ACGAACTTCCGCACTCCTTGGCATCCCGCG-3′; hsp-4 (F43E2.8) 5′-CGTGGCAAACGCGTACTGTGATGAAGGAGC-3′ and 5′-CAGTTCATCATGATCCTCCGATTGCTCCTC-3′; ama-1 (F36A4.7) 5′-CAGTGGCTCATGTCGAGTTTCCAGA-3′ and 5′-CGACCTTCTTTCCATCATTCATCGG-3′; cAMP response element–binding protein (CBP; F40F12.7) 5′-ATGCTTGCACTGCATCAAGA-3′ and 5′-TCAATCATGGAACAACTGTG-3′; and act-3 (T04C12.4.1) 5′-TTTGTTATTGCCCCAAAAGG-3′ and 5′-GTCGGTATGGGACAGAAGGA-3′. The products were separated on a 0.8% agarose gel and visualized by ethidium bromide staining.
RNAi of genes in worms
The entire open reading frame encoding the C. elegans ubiquilin protein was cloned between the BglII and KpnI sites of the L4440 vector (provided by B. Vogel, University of Maryland Biotechnology Institute, Baltimore, MD) that is widely used for inducing RNAi in worms using the bacterial feeding protocol (Timmons and Fire, 1998; Timmons et al., 2001). An erasin cDNA cloned in the same vector was obtained from Thermo Fisher Scientific. The ubiquilin and erasin RNAi plasmids were transformed into Escherichia coli strain HT115 (DE3), and the resulting transformants were cultured overnight at 37°C in Luria broth containing 50 µg/ml ampicillin and 12.5 µg/ml tetracycline. The cultures were diluted into fresh broth and grown to an OD600 of 0.5, after which IPTG was added to a final concentration of 0.4 mM and the incubation continued for another 2 h. The bacteria were then seeded onto nematode growth medium plates containing IPTG, and worms were added to the plates by chunking a piece of agar from a stock plate of the zcls4 strain of worms. The worms were allowed to feed for ∼24–36 h, after which they were collected and protein lysates were prepared and analyzed by immunoblotting (Wang et al., 2006).
Lifespan assay
Nematodes were synchronized by bleaching to obtain hermaphrodites in the fourth larval stage (L4). On day zero, L4 hermaphrodites were placed on individual 60-mm nematode growth medium agar plates containing 2′-deoxy-5-fluorouridine to prevent progeny growth and seeded with OP50 E. coli bacteria or RNAi bacteria. Nematodes were grown for the duration of adult life at 20°C and were scored every 2 d. Animals were classified as dead if they failed to respond to a gentle tap on the head and tail with a platinum wire. Lifespan was defined as day zero to the day the worm was scored as dead. Statistical analysis software was used to compare mean lifespan of each treatment.
Online supplemental material
Fig. S1 shows in vitro binding assays mapping the ubiquilin-binding site in erasin. Fig. S2 shows GST pull-down assays to estimate the stoichiometry of erasin and p97/VCP binding in the trimeric complex. Fig. S3 shows the degree of colocalization of ubiquilin and erasin for Fig. 2 C and the colocalization of calnexin with erasin and ubiquilin. Fig. S4 shows the binding affinity of ubiquilin and erasin determined from a Biacore experiment. Fig. S5 shows that JAMP, Herp, and Ufd1 do not coimmunoprecipitate with erasin.
Acknowledgments
We thank Drs. Bruce Vogel, Hemmo Meyer, Ze'ev Ronai and the late Dr. Cecil Pickart for providing antibodies and expression constructs; the Caenorhabditis Genetics Center for providing worms lines; Chaobo Yin, Angela Weese, and Dr. Robert Bloch for help in these studies; and Dr. Ann Pluta for helpful comments on the manuscript.
This work was supported by National Institutes of Health grants GM066287 and AG016839 to M.J. Monteiro, and GM06696 to S. Fang.
References
Author notes
J. Liang's present address is Dept. of Biochemistry and Molecular Biology, Peking University Health Science Center, Beijing 100191, China.







![Figure 7. Ubiquilin and erasin expression are induced by ER stress in C. elegans. (A) Mixed-stage zcls4 [hsp-4::GFP] worms were grown on plates with 28 µg/ml tunicamycin for 0, 4, 8, 16, and 48 h, and transcript levels of hsp-4, ubiquilin, erasin, and ama-I were measured by semiquantitative PCR and gel electrophoresis. Quantification of Hsp-4 (B), ubiquilin (C), and erasin (D) expression relative to the 0 time point. Similar trends were observed in two other experiments. (E) Schematic diagram of the hsp-4::GFP reporter construct. (F) Immunoblot analysis of equal amounts of protein lysate (two independent samples for each time point) prepared from worms treated with 28 µg/ml of tunicamycin for the periods indicated. An unknown protein whose expression was insensitive to ER stress was used as a loading control. (G) Schematic diagram of the C. elegans ubiquilin promoter fragment fused to GFP. (H) Immunoblot analysis of equal amounts of protein lysate of worms treated with 0, 2.5, or 28 µg/ml tunicamycin for 24 h. (I) Quantification of GFP expression determined by scanning of immunoblots as described in H showing the mean and SD (error bars) seen in three different experiments. (J) GFP fluorescence of worms containing the integrated ubiquilin::GFP fusion construct treated for 24 h with 0, 2.5, or 28 µg/ml tunicamycin. Bar, 0.2 mm.](https://cdn.rupress.org/rup/content_public/journal/jcb/187/2/10.1083_jcb.200903024/6/m_jcb_200903024r_rgb_fig7.jpeg?Expires=1771331275&Signature=L4ZJdLTAjBFqjzd6D3j7hBxnp1tA8SVgJJcMq3KOKN53WJ3wiRKjweZ14nZGffNeUZja-CtMSS143zhWo~wMbG8IWKmoqloLMs9zuZMCjJbcL2HJMShpp8NuDEJipjk709WLU7t9tGVb4icAjC1cJfqW9RD4GUt8e1gQ4IQvqgNYdAAcvK8KSVr2Sw~AwKu8qTWPLXX-mhcNpcJd18oVBPY~F-yNYOmpoE3Or8rUnjNf2HMwXPK8~Nuc6NHzeTNmG7gzpQfcerFlNY9nxQF1oGhL5~8ZdA0wqOZzldoElUjUB0NVJxaFEAXVH49a-2U7nB0OBtobSEDDYthA8-m1zQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 9. Knockdown of ubiquilin or erasin proteins in worms results in the induction of the ER stress reporter hsp-4::GFP, accumulation of misfolded proteins, and a shorter lifespan. (A) Mixed-stage zcls4 [hsp-4::GFP] worms were either fed standard bacteria in the presence or absence of 28 µg/ml tunicamycin or bacteria to induce RNAi of the ubiquilin gene. Immunoblot analysis of equal amounts of protein lysate prepared after 48 h of treatment for GFP, ubiquilin, erasin, ubiquitin, and an unknown protein used as a loading control. (B) Identical to A, except that worms were fed bacteria to specifically knock down erasin. (C) Lifespan curves comparing N2 (wild-type) worms fed bacteria transformed with the empty RNAi vector or the vector containing cDNAs to induce RNAi of either ubiquilin or erasin genes. Results are cumulative from three independent experiments, with ≥60 animals per trial. Log-rank (Mantel-Cox) and Gehan-Breslow-Wilcoxon tests, P < 0.0001 versus control.](https://cdn.rupress.org/rup/content_public/journal/jcb/187/2/10.1083_jcb.200903024/6/m_jcb_200903024_rgb_fig9.jpeg?Expires=1771331275&Signature=4z6xCt6i3Qyqz9h4PHS~eNTuoEsj4D8a6v82q9HdIIOcH7xo-Qa1nrY7FgjYNEVzEeTFpPuI50O9~PWzg-ucV6hvrEC0f0~jIz-~VpXAeO4OwmmQouANiVLvRtaWi58shIk7MB3UFPaZ81c7S2kTHDWfSbQMWyUauXJKzXlBM6iScuFsHa8w1IlzFOBfC2vbQW15FVN-gSUrAV6z7tOxQYKEUMXL52-5dGF0Hd3aQ0WHhOKzJvscdcSUbNmXSI2jJ1zWrtoVmoqCImYKaCK5pGQIR~uprbjJlqyK8y8xYORn79ueEfRgdHag1osL5dRE16Wx7Mnnu98LT3q1zC9lzQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)



