Keratins, the largest subgroup of intermediate filament (IF) proteins, form a network of 10-nm filaments built from type I/II heterodimers in epithelial cells. A major function of keratin IFs is to protect epithelial cells from mechanical stress. Like filamentous actin, keratin IFs must be cross-linked in vitro to achieve the high level of mechanical resilience characteristic of live cells. Keratins 5 and 14 (K5 and K14), the main pairing occurring in the basal progenitor layer of epidermis and related epithelia, can readily self-organize into large filament bundles in vitro and in vivo. Here, we show that filament self-organization is mediated by multivalent interactions involving distinct regions in K5 and K14 proteins. Self-organization is determined independently of polymerization into 10-nm filaments, but involves specific type I–type II keratin complementarity. We propose that self-organization is a key determinant of the structural support function of keratin IFs in vivo.
Introduction
Intermediate filaments (IFs) play a major role of structural support in both the cytoplasm and nucleus of a broad variety of cell types (Fuchs and Cleveland, 1998; Kim and Coulombe, 2007). This role is important in tissues routinely subjected to trauma, e.g., surface epithelia and muscle. In such settings, IFs are abundant and fully integrated within a supracellular network responsible for tissue integrity, owing in part to their attachment at cell–cell and cell–matrix adhesion sites. Mutation-based defects in IF structure, organization, or regulation underlie a large number of tissue fragility conditions affecting humans (Omary et al., 2004; Gu and Coulombe, 2007; Szeverenyi et al., 2008).
Similarly to F-actin assemblies (Janmey, 1991), the mechanical resilience of IF networks is primarily influenced by two factors: concentration and length of filaments, and presence of stable linkages between them (Ma et al., 2001; Yamada et al., 2002). Ample evidence shows that both F-actin and IFs must be “cross-linked” to yield the mechanical properties characteristic of living cells in culture (Janmey, 1991; Yamada et al., 2000). (Note, “cross-linking” here means stable filament–filament linkages achieved through noncovalent interactions). A large number of proteins participate in organizing F-actin into various network configurations in vivo (Alberts et al., 2002). Likewise, integration of IFs at sites of cell–cell and cell–matrix adhesions, at the nuclear surface, and with other cytoskeletal networks, is mediated by the plakin family of IF-associated proteins (Green et al., 2005; Wilhelmsen et al., 2005; Sonnenberg and Liem, 2007). However, the mechanisms responsible for IF cross-linking in the general cytoplasm are ill-defined. This is the case for most epithelial cells in surface tissues like epidermis, oral mucosa, and cornea.
Part of the solution to this apparent paradox may lie in the observation that keratin IFs, in particular, can self-organize into large bundles in vitro (e.g., Eichner et al., 1986; Wilson et al., 1992; Ma et al., 2001; Yamada et al., 2002). This property is revealed through slightly modifying the unusual assembly buffer conditions optimal for in vitro polymerization of epidermal keratins (5 mM Tris-HCl and 5 mM β-mercaptoethanol, pH 7.5; Aebi et al., 1983). Thus, raising the ionic strength, adding a modest amount of salt, or acidifying the pH from 7.5 to 7.0 each cause loosely arranged keratin filaments to undergo formation of large bundles (Ma et al., 2001; Yamada et al., 2002). The mechanical resilience of keratin IF networks in vitro is dramatically enhanced (from ∼10 dynes/cm2 to >500 dynes/cm2) by such bundling (Ma et al., 2001; Yamada et al., 2002), and approaches the values measured in the cytoplasm of live epithelial cells in culture (Yamada et al., 2000; Beil et al., 2003). The property of self-organization, which we refer to as the intrinsic pathway of keratin IF cross-linking (Coulombe et al., 2000), is markedly influenced by type I and II keratin complementarity (Yamada et al., 2002).
The type II keratin 5 (K5) and type I keratin 14 (K14) represent the main keratin pairing expressed in the progenitor basal layer of epidermis and related stratified epithelia. Mutations abrogating the ability of K5 or K14 to participate in the formation of a mechanically resilient filament network in basal keratinocytes causes epidermolysis bullosa simplex, a dominantly inherited fragility condition primarily affecting the epidermis (Gu and Coulombe, 2007). Here, we show that the distal half of K14's tail domain and two distinct regions in K5's rod domain interact to mediate the intrinsic pathway of IF cross-linking, independent of 10-nm filament assembly, in vitro and in vivo. Our findings legitimize the property of keratin filament self-organization, and significantly extend our understanding of the structural support function of keratin filaments.
Results
Defining the C-terminal tail domain of K14
We previously reported that the C-terminal portion of K14 binds keratin IFs in vitro and in vivo, and is required for self-driven bundling of K5/K14 filaments. These studies made use of an arbitrarily defined tail domain modified with a short epitope tag (Bousquet et al., 2001). Here, we used a combination of partial proteolysis and mass spectrometry to map the N-terminal boundary of the tail domain of K14 at residue Gly421 (TYRLLEGE, the Gly421 is underlined; Fig. S1, a and b). These findings are in agreement with an x-ray crystallography–based study of vimentin (Strelkov et al., 2002). This newly defined, untagged tail domain of K14, which is 52 residues long, binds K5/K14 filaments, thus reproducing our previous findings (Fig. S1, c and d).
Two distinct regions of K5 interact with the tail domain of K14
To define how the presence of K14's tail domain promotes K5/K14 filament bundling, we sought to identify its binding determinants using far Western blot assays. The latter involves membrane-bound protein monomers as potential targets, a desirable feature given the insolubility and polymerization-prone character of keratin proteins. GFP-T14, in which GFP is fused to the N terminus of K14's tail domain, binds electrophoresed, membrane-immobilized K5, but not K14 (Fig. 1 a). The extent of specific binding observed in such a setting may exceed expectations based on the known affinity between K14's tail domain and K5/K14 filaments in solution (∼2 µM). This likely reflects the effect of concentrating one of the ligands (namely, K5) via nitrocellulose immobilization, along with the chemical cross-linking step applied to stabilize the interaction (see Materials and methods).
The K14 tail domain binds two determinants in K5 and related type II keratins. (a) Recombinant K5 and K14 proteins were resolved SDS-PAGE, followed by blot overlay experiments using either GFP (control) or GFP-fused K14 tail domain (GFP-T14) as a ligand. Binding was detected with rabbit anti-GFP and HRP-conjugated goat anti–rabbit antibodies. (b) Other type II keratins also bind K14's tail domain, as indicated by far Western blotting (see a). K4, K5, and K6 interact with GFP-T14, whereas K8 does not. When added to the ligand mixture, the unfused tail domain of K14 competes for binding to these type II targets in a concentration-dependent fashion (1:1 and 1:5 ratio). (c) Schematic of the K5 deletion mutants tested. (d) Far Western blot results obtained when using GFP-T14 as the ligand. The numbering of constructs on the left in c corresponds to the gel lanes in d. (e) Schematic conveying the secondary structure of wild-type K5 and K8, of the K5H and K8H (single-domain swap) chimeras, and of the K5bd and K8bc (dual domain swap) chimeras. (f) Testing the binding of GFP-T14 to each of K5, K8, K5bd, and K8bc. See main text for further details. MW, molecular weight.
The K14 tail domain binds two determinants in K5 and related type II keratins. (a) Recombinant K5 and K14 proteins were resolved SDS-PAGE, followed by blot overlay experiments using either GFP (control) or GFP-fused K14 tail domain (GFP-T14) as a ligand. Binding was detected with rabbit anti-GFP and HRP-conjugated goat anti–rabbit antibodies. (b) Other type II keratins also bind K14's tail domain, as indicated by far Western blotting (see a). K4, K5, and K6 interact with GFP-T14, whereas K8 does not. When added to the ligand mixture, the unfused tail domain of K14 competes for binding to these type II targets in a concentration-dependent fashion (1:1 and 1:5 ratio). (c) Schematic of the K5 deletion mutants tested. (d) Far Western blot results obtained when using GFP-T14 as the ligand. The numbering of constructs on the left in c corresponds to the gel lanes in d. (e) Schematic conveying the secondary structure of wild-type K5 and K8, of the K5H and K8H (single-domain swap) chimeras, and of the K5bd and K8bc (dual domain swap) chimeras. (f) Testing the binding of GFP-T14 to each of K5, K8, K5bd, and K8bc. See main text for further details. MW, molecular weight.
Extending this analysis to other type II keratins, GFP-T14 also binds K4 and K6, but not K8 (Fig. 1 b). GFP-T14's binding to immobilized K5 is enhanced by salt (Fig. S1 e), which is known to trigger K5/K14 filament cross-linking (Bousquet et al., 2001). To assess binding specificity, the fusion-free K14 tail was added as a competitor to the GFP-T14 ligand mixture in the far Western assay. This decreased the binding of GFP-T14 to each of K4, K5, and K6 in a concentration-dependent manner (Fig. 1 b). Thus, conserved regions in K5 and closely related type II keratins are able to interact specifically with the K14 tail domain.
We next sought to define the regions of K5 bound by K14's tail domain. Experiments involving BNPS-Skatole, which chemically cleaves proteins at Trp residues (Crimmins et al., 1990; Gardner et al., 1994), and a first set of K5/K8 chimeric proteins suggested that there are two such binding sites (not depicted), located upstream and downstream from the L1 linker domain (see Fig. 1 c for diagram of IF domain structure). Deletion mutagenesis was next used to generate a set of nine overlapping K5 fragments, covering the relevant portions of K5. Several of the fragments tested (Fig. 1 c) retained the ability to bind GFP-T14, whereas others did not (Fig. 1 d and not depicted). Collectively, our findings point to two regions in K5—the rod-proximal head domain and L2-2B—as being able to mediate this property. Whether these two regions bind the tail domain of K14 in a sequence-specific or conformation-specific fashion is not resolved by our data. These findings are consistent with the higher sequence identity over these two regions in K5, K4, and K6 relative to K8.
To test the role of these two binding determinants in the context of a full-length protein, we constructed a second set of chimeric K5/K8 cDNAs. The coding sequence corresponding to the relevant portion of K5's head-1A domain (82 amino acid residues) and the L2-2B domain (128 residues long) were swapped between K5 and K8 cDNAs, individually or in combination. The resulting double chimeras were designated K5bd and K8bc (Fig. 1 e), with “bd” and “bc” referring to “bundling-deficient” and “bundling-competent,” respectively (see Fig. 2). In far Western blotting assays, K8bc showed a markedly higher binding affinity for GFP-T14, similar to that of K5 (Fig. 1 f). Conversely, K5bd showed a markedly reduced affinity for GFP-T14 that is similar to K8 (Fig. 1 f). Individual chimeras including K5H and K8H displayed intermediate affinities for GFP-T14 (unpublished data). These findings, and others presented in this paper, further the evidence showing that the rod-proximal head and L2-2B domains in K5 both contribute to K14's tail domain binding.
Assessing the network-forming properties of select type II keratin variants in vitro. The assemblies tested here were produced under cross-linking–promoting buffer conditions. (a) A low-speed sedimentation assay (8,000 g, 30 min) followed by electrophoretic analysis of supernatant (s) and pellet (p) fractions. The type I–type II combination tested is shown at the top. MW, molecular weight. (b) Densitometry-based analysis of the fractions illustrated in frame (a). Data represent a mean from three independent assays (error bars indicate SEM). *, significantly different from wild-type K5/K14; one-way ANOVA, P < 0.01. (c–f). Visualization of keratin assemblies using DIC microscopy. The keratin combination being tested is indicated in the lower right corner of each micrograph. Double and single arrows, respectively, depict large and small keratin filament bundles. Bars, 10 µm. (g) Viscometric analysis of the gelation state of keratin assemblies. Each sample was added to a 1-ml syringe, incubated overnight. A metal ball was then placed at the top of the sample–air interface (arrowheads) when syringes were laid down flat. This picture was taken 1 s after all four syringes were simultaneously oriented upright.
Assessing the network-forming properties of select type II keratin variants in vitro. The assemblies tested here were produced under cross-linking–promoting buffer conditions. (a) A low-speed sedimentation assay (8,000 g, 30 min) followed by electrophoretic analysis of supernatant (s) and pellet (p) fractions. The type I–type II combination tested is shown at the top. MW, molecular weight. (b) Densitometry-based analysis of the fractions illustrated in frame (a). Data represent a mean from three independent assays (error bars indicate SEM). *, significantly different from wild-type K5/K14; one-way ANOVA, P < 0.01. (c–f). Visualization of keratin assemblies using DIC microscopy. The keratin combination being tested is indicated in the lower right corner of each micrograph. Double and single arrows, respectively, depict large and small keratin filament bundles. Bars, 10 µm. (g) Viscometric analysis of the gelation state of keratin assemblies. Each sample was added to a 1-ml syringe, incubated overnight. A metal ball was then placed at the top of the sample–air interface (arrowheads) when syringes were laid down flat. This picture was taken 1 s after all four syringes were simultaneously oriented upright.
The K14 tail domain and two regions in K5 mediate keratin cross-linking in vitro
Distinct assays were used, in vitro and in vivo, to assess the polymerization properties of the wild-type and chimeric keratins generated in this study. First, the ability to form 10-nm filaments was tested using three distinct assays (Bousquet et al., 2001; Ma et al., 2001; Yamada et al., 2002). High-speed sedimentation (150,000 g, which pellets fully formed IFs) coupled with electrophoresis of pellet and supernatant fractions, and negative staining and electron microscopy, were used for in vitro–reconstituted assemblies. Indirect immunofluorescence was performed in transfected fibroblasts and keratinocytes to assess 10-nm filament-forming properties in cells. Second, the ability of filaments to sustain the intrinsic pathway of keratin filament cross-linking was tested using four distinct assays. For polymers reconstituted in vitro, we used a low-speed sedimentation assay (8,000 g, which does not pellet 10-nm filaments) and differential interference contrast (DIC) microscopy, which does not resolve 10-nm IFs (Ma et al., 2001; Yamada et al., 2002); we adapted an existing actin viscometric assay (MacLean-Fletcher and Pollard, 1980; Perng et al., 1999) to test the functionality of cross-linked networks. Quantitative immunofluorescence of digitally processed images was used, finally, to assess the formation of IF bundles in transfected fibroblasts and keratinocytes (technical details are provided in the Materials and methods section).
In vitro, K8, K5bd, K8bc, and, of course, K5, each could efficiently polymerize into 10-nm filaments with K14 as a type I keratin partner, when using standard polymerization conditions, as indicated by high-speed sedimentation as well as electron microscopy (Fig. S2). K8, K5bd, and K8bc also were polymerization-competent when coexpressed with K14 in fibroblasts, or expressed alone in keratinocytes (see Fig. 3). Altogether, this establishes that the swapping of both “head-1A” and L2-2B” regions between K5 and K8 did not significantly alter their ability to form the basic 10-nm filament polymer.
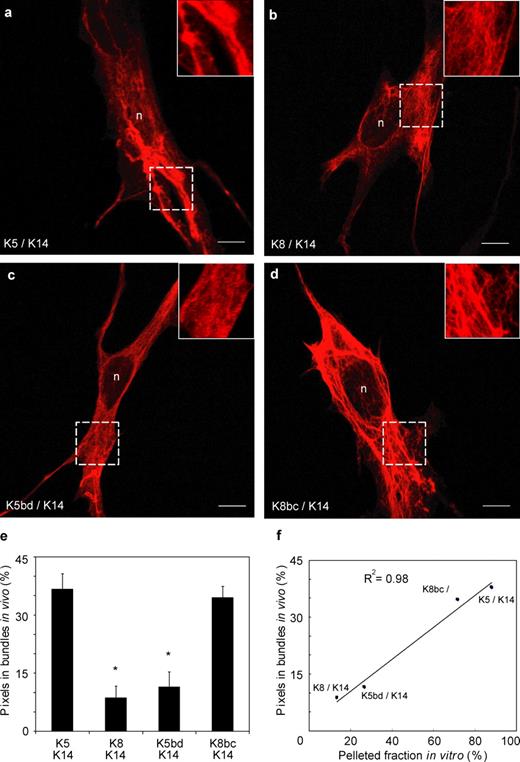
De novo keratin network formation in transfected BHK-21 fibroblasts. Bicistronic expression vectors including a GFP reporter and a type II keratin cDNA (K5, K8, K5bd, or K8bc) were individually cotransfected with a K14 expression vector into keratin-free baby hamster kidney fibroblasts (BHK-21). Cells were fixed and processed for analysis 72 h later. GFP fluorescence was used as an indicator of type II keratin expression. (a–d) Indirect immunofluorescence for K14 using confocal microscopy. The combinations transfected are given in the bottom left corners. The areas in the squares with broken lines are magnified in the upper right insets. n, nucleus. Bars, 10 µm. (e) Quantitation of the fluorescence signal specifically associated with large filament bundles in transfected cells. Image processing is detailed under Materials and methods and illustrated in Fig. S3. Each bar represents the mean and SEM (error bars) from 45 cells from three transfection experiments. *, significantly different from wild-type K5/K14; one-way ANOVA, P < 0.001. (f) Correlation between in vitro (pellet fraction in the low speed sedimentation assay; Fig. 2 B) and in vivo assembly behavior (bundle-associated fluorescence in transfected BHK-21 cells; Fig. 3E) for the K5/K14, K8/K14, K5bd/K14, and K8bc/K14 combinations.
De novo keratin network formation in transfected BHK-21 fibroblasts. Bicistronic expression vectors including a GFP reporter and a type II keratin cDNA (K5, K8, K5bd, or K8bc) were individually cotransfected with a K14 expression vector into keratin-free baby hamster kidney fibroblasts (BHK-21). Cells were fixed and processed for analysis 72 h later. GFP fluorescence was used as an indicator of type II keratin expression. (a–d) Indirect immunofluorescence for K14 using confocal microscopy. The combinations transfected are given in the bottom left corners. The areas in the squares with broken lines are magnified in the upper right insets. n, nucleus. Bars, 10 µm. (e) Quantitation of the fluorescence signal specifically associated with large filament bundles in transfected cells. Image processing is detailed under Materials and methods and illustrated in Fig. S3. Each bar represents the mean and SEM (error bars) from 45 cells from three transfection experiments. *, significantly different from wild-type K5/K14; one-way ANOVA, P < 0.001. (f) Correlation between in vitro (pellet fraction in the low speed sedimentation assay; Fig. 2 B) and in vivo assembly behavior (bundle-associated fluorescence in transfected BHK-21 cells; Fig. 3E) for the K5/K14, K8/K14, K5bd/K14, and K8bc/K14 combinations.
The ability to sustain the intrinsic bundling pathway was next investigated in vitro. After low-speed sedimentation, a significant fraction of K5/K14, but not of K8/K14, is retrieved in the pellet fraction (Fig. 2, a and b). Using DIC microscopy, large arrays of bundled filaments can readily be seen in the K5/K14, but not K8/K14, assemblies (Fig. 2, c and d). In the viscometry assay, the K5/K14 combination, but not the K8/K14 combination, prevented the metal ball from penetrating the sample phase (Fig. 2 g). These findings extend an earlier study showing that K5/K14 but not K8/K14 can sustain the intrinsic pathway of filament bundling in vitro (Yamada et al., 2002).
Whether in the sedimentation, DIC, or viscometry assay, the K5bd/K14 assemblies consistently behave similarly to K8/K14 ones, whereas K8bc/K14 assemblies behave like K5/K14 ones (Fig. 2, a–g). Two distinct behaviors, cross-linking–competent and –noncompetent, could be distinguished. We infer that the ability to sustain the intrinsic pathway segregates with the property of K14 tail binding, and is mediated by the head-1A and L2-2B regions in K5.
The availability of single domain chimeras provided an opportunity to test whether the two relevant regions in K5 function in cis or in trans to mediate filament cross-linking. The K5H and K8H chimeras, in which only the L2-2B subdomains have been swapped, foster a level of self-driven bundling (with K14) that is intermediate between those of K5 and K8 (Fig. S2 g). When they are both present, K5H and K8H (with K14) foster the formation of large bundles that efficiently sediment during low-speed centrifugation (Fig. S2 g). Thus, the head-1A and L2-2B regions of K5 can operate in trans to foster the intrinsic pathway to filament bundling.
Assaying intrinsic pathway determinants in cultured fibroblasts in vivo
We next tested keratin assembly properties in cultured fibroblasts, which are keratin free and thus offer the opportunity to test for keratin assembly on a de novo basis in vivo. BHK-21 cells transfected with various keratin pairings were subjected to indirect immunofluorescence by confocal microscopy, followed by image analysis to quantify the extent of filament bundling. The K5, K8, K5bd, and K8bc cDNAs were subcloned in a bicistronic vector, pIRES-2–GFP. As detailed elsewhere (see Materials and methods and Fig. S3, a and b), the resulting diffuse GFP signal throughout the cytoplasm makes it possible to not only correct for differences in expression level between cells, but also to “subtract” the weaker fluorescence associated with thin keratin filaments. As expected, coexpression of GFP does not interfere with keratin assembly in these cells.
Representative examples of BHK-21 fibroblasts transfected with a combination of K14 and either K5, K8, K5bd, and K8bc are shown in Fig. 3 (a–d). All the combinations tested gave rise to filamentous arrays in transfected cells, extending the in vitro evidence showing that the chimeras can copolymerize with K14. Also, Western blot analyses showed that the four keratin combinations transfected were expressed to similar levels (Fig. S3 c), which makes it possible to compare them directly. Coexpression of either K5/K14 or K8bc/K14 fostered the formation of thick arrays of keratin IFs in the cytoplasm of BHK-21 cells. By comparison, coexpression of the K8/K14 or K5bd/K14 combination gave rise to thinner filamentous structures that were more homogeneously distributed throughout the cytoplasm of transfected cells. This interpretation is fully supported by calculation of a “bundling index,” which corresponds to the fraction of pixels specifically associated with thicker bundles. This quantitative method clearly sorts the four combinations tested into two groupings: “bundle-rich” (K5/K14 and K8bc/K14) and “bundle-poor” (K8/K14 and K5bd/K14) cells (Fig. 3 e). Very similar trends were observed when an experienced observer blinded to the keratin combinations used a qualitative scale to assess bundling (unpublished data). The availability of quantitative indices allows us to relate the transfection findings in vivo to the sedimentation-based data involving pure assemblies in vitro. This reveals a strong correlation in the behavior of K5/K14, K8bc/K14, K8/K14, and K5bd/K14 in these two different settings (Fig. 3 f).
To further validate the image analysis–based findings, thin-section transmission electron microscopy was performed on BHK-21 cells transfected with either GFP (negative control), K5/K14 (positive control), or K8bc/K14, and processed for preembedding immunogold labeling using an antibody against K14 (see Materials and methods). Both keratin combinations gave rise to abundant filaments in the cytoplasm, and to extensive filament bundling. No evidence of protein aggregation was detected in the large number of transfected cells analyzed (Fig. S3, d–g).
K5/K14 filaments self-organize using the same determinants in skin keratinocytes in culture
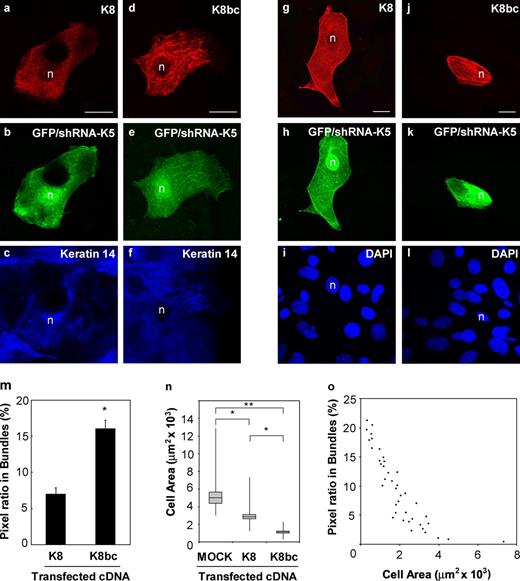
We next set out to test whether this remarkable behavior of keratin filaments, and in particular the newly found role of determinants located within the head and L2-2B subdomains of K5, also apply to skin keratinocytes. This analysis requires a setting where endogenous type II keratin expression is reduced to a minimum. In primary culture, skin keratinocytes express K5, K6a, and K6b as type II keratins. Taking advantage of an existing mouse strain with a double null mutation at the K6a/K6b locus (Wong et al., 2000), we tested several small hairpin RNAs (shRNAs) and found one that was effective at knocking down K5 protein levels in 308 mouse keratinocytes (Fig. S4, a and b) and newborn K6a/K6b-null keratinocytes in primary culture (not depicted). In this latter context, we compared the impact of expressing K8 (the bundling-deficient reference) and K8bc on the organization of K14-immunostained filaments. A clear difference occurred in the frequency with which K8 (Fig. 4, a–c) and K8bc (Fig. 4, d–f) fostered the formation of thick keratin filament bundles in transfected K6a/K6b-null, K5 knockdown keratinocytes (see quantification in Fig. 4 m). These findings extend those obtained in BHK-21 fibroblasts and, moreover, establish that the bundling-promoting determinants in K5 also function in skin keratinocytes.
Testing the assembly properties of wild-type K8 and the K8bc chimera in type II keratin-depleted skin keratinocytes. Skin keratinocytes from newborn K6a/K6b-null mice were seeded in primary culture on glass coverslips (a–f) or collagen IV–coated coverslips (g–l), treated with an shRNA construct (in the context of a bicistronic vector also fostering GFP expression) to knock down K5 expression (see Materials and methods and Fig. S5), and simultaneously transfected with an expression vector for either K8 or K8bc, tested as rescue type II keratins. Cells were fixed 72 h later, and processed for triple immunostaining. GFP fluorescence was used as an indicator of K5 knockdown (Fig. S5). (a–c) Representative cell expressing K8. Some areas in the cytoplasm do not show well in this focal plane. (d–f) Representative cell expressing K8bc. (g–i) Representative cell expressing K8 on the collagen IV–coated surface. (j–l) Representative cell expressing K8bc on the collagen IV–coated surface. The antigen being detected is given in the top right corner. n, nucleus. Bars, 10 µm. (m) Quantitation of the fluorescence signal specifically associated with large filament bundles in transfected cells cultured on glass coverslips. Image processing is detailed under Materials and methods and illustrated in Fig. S3. Each bar represents the mean and SEM (error bars) from 15 cells from three independent transfection experiments. *, significantly different from wild-type K8/K14; t test, P < 0.05. (n) Quantification of cell area in K8-expressing and K8bc-expressing cells cultured on a collagen IV–coated surface. Each box represents the mean and SEM (error bars) from 20 cells from three independent transfection experiments. Solid lines show minima and maxima values for each group. *, significantly different, P < 0.05; **, significantly different, P < 0.01. One-way ANOVA was used. (o) Correlation between the bundle-associated fluorescence signal in transfected cells, and their surface area, for the K8-transfected and K8bc-transfected keratinocytes cultured on a collagen IV–coated surface.
Testing the assembly properties of wild-type K8 and the K8bc chimera in type II keratin-depleted skin keratinocytes. Skin keratinocytes from newborn K6a/K6b-null mice were seeded in primary culture on glass coverslips (a–f) or collagen IV–coated coverslips (g–l), treated with an shRNA construct (in the context of a bicistronic vector also fostering GFP expression) to knock down K5 expression (see Materials and methods and Fig. S5), and simultaneously transfected with an expression vector for either K8 or K8bc, tested as rescue type II keratins. Cells were fixed 72 h later, and processed for triple immunostaining. GFP fluorescence was used as an indicator of K5 knockdown (Fig. S5). (a–c) Representative cell expressing K8. Some areas in the cytoplasm do not show well in this focal plane. (d–f) Representative cell expressing K8bc. (g–i) Representative cell expressing K8 on the collagen IV–coated surface. (j–l) Representative cell expressing K8bc on the collagen IV–coated surface. The antigen being detected is given in the top right corner. n, nucleus. Bars, 10 µm. (m) Quantitation of the fluorescence signal specifically associated with large filament bundles in transfected cells cultured on glass coverslips. Image processing is detailed under Materials and methods and illustrated in Fig. S3. Each bar represents the mean and SEM (error bars) from 15 cells from three independent transfection experiments. *, significantly different from wild-type K8/K14; t test, P < 0.05. (n) Quantification of cell area in K8-expressing and K8bc-expressing cells cultured on a collagen IV–coated surface. Each box represents the mean and SEM (error bars) from 20 cells from three independent transfection experiments. Solid lines show minima and maxima values for each group. *, significantly different, P < 0.05; **, significantly different, P < 0.01. One-way ANOVA was used. (o) Correlation between the bundle-associated fluorescence signal in transfected cells, and their surface area, for the K8-transfected and K8bc-transfected keratinocytes cultured on a collagen IV–coated surface.
Given attachment to cell–cell and/or cell–matrix adhesion sites, the induction of massive cytoskeletal bundling should generate an inward tension that may hamper cell spreading and/or alter cell morphology. There is compelling experimental evidence that this occurs when causing F-actin bundling in various settings (Gross and Kinzy, 2005; Karakozova et al., 2006; Kotadiya et al., 2008). We assessed this possibility by comparing the surface area of K6a/K6b-null, K5 knockdown keratinocytes transfected with either K8/K14 (Fig. 4, g–i) or K8bc/K14 (Fig. 4, j–l; see Materials and methods). Relative to mock-transfected keratinocytes, reintroduction of K8 and K8bc each caused a decrease in keratinocyte surface area, with K8bc exerting a significantly stronger effect, as anticipated (Fig. 4 n). Further, there is a strong correlation between the extent of keratin filament bundling, as determined by our fluorescence-based quantification, and the surface area of transfected keratinocytes expressing either the K8/K14 or K8bc/K14 combinations (Fig. 4 o). These findings provide a glimpse of the physiological consequences associated with variations in the degree of keratin filament bundling in skin keratinocytes.
The tail domain of K14 comprises two distinct functional subdomains
The C-terminal tail domain of K14 is highly conserved among mammals and does not show any significant homology to known structural motifs (unpublished data). Visual inspection reveals a serine residue–rich character over the rod-proximal half in the K14 tail (Fig. 5 a). Alignment of tail domains from K14 (human and mouse), K15, K16, K17, and K18 suggest the existence of two distinct moieties: a serine-rich T1 region (27 residues) proximal to the rod, and a T2 region (25 residues) at the extreme C terminus (Fig. 5 a). T2 comprises a motif, DGKVVS, which, as noted previously (Franke, 1987), is conserved among several type I keratins. Interestingly, K17's tail domain consists primarily of a T2-like motif, whereas K16's tail is missing T2 and resembles a duplicated T1 segment. Accordingly, a series of K14 constructs were generated in which the entire tail domain, the T1 segment, or the T2 segment were deleted, or replaced by the tail domains of K16 and K17 (with no epitope tag). The network-forming properties of the resulting K14 mutants (Fig. 5 b), all of which are competent for 10-nm filament formation with K5 as a partner (Fig. S5), were tested in vitro and in transfected BHK-21 fibroblasts.
Assessing the network-forming properties of type I keratin tail domain variants in vitro. (a) Alignment of the C-terminal tail domain sequence (which begins at the residue indicated by the arrowhead) from several human type I keratins (indicated on the left). The color code reflects the properties of individual amino acids at conserved sites. Two domains, T1 and T2, can be distinguished based on sequence homology. (b) Schematic conveying the secondary structure of wild-type K14 (top), tailless K14 (K14ΔT), and other deletion mutants and tail domain chimeras. See text for further details. (c) Low-speed sedimentation assay (8,000 g, 30 min) followed by electrophoretic analysis of supernatant (s) and pellet (p) fractions for type I–type II combinations (see labels at the top) subjected to assembly under cross-linking–promoting conditions. MW, molecular weight. (d) Densitometry-based analysis of the fractions illustrated in c. Data represents the mean from three independent assays (error bars indicate SEM). *, significantly different from wild-type K5/K14; one-way ANOVA, P < 0.01.
Assessing the network-forming properties of type I keratin tail domain variants in vitro. (a) Alignment of the C-terminal tail domain sequence (which begins at the residue indicated by the arrowhead) from several human type I keratins (indicated on the left). The color code reflects the properties of individual amino acids at conserved sites. Two domains, T1 and T2, can be distinguished based on sequence homology. (b) Schematic conveying the secondary structure of wild-type K14 (top), tailless K14 (K14ΔT), and other deletion mutants and tail domain chimeras. See text for further details. (c) Low-speed sedimentation assay (8,000 g, 30 min) followed by electrophoretic analysis of supernatant (s) and pellet (p) fractions for type I–type II combinations (see labels at the top) subjected to assembly under cross-linking–promoting conditions. MW, molecular weight. (d) Densitometry-based analysis of the fractions illustrated in c. Data represents the mean from three independent assays (error bars indicate SEM). *, significantly different from wild-type K5/K14; one-way ANOVA, P < 0.01.
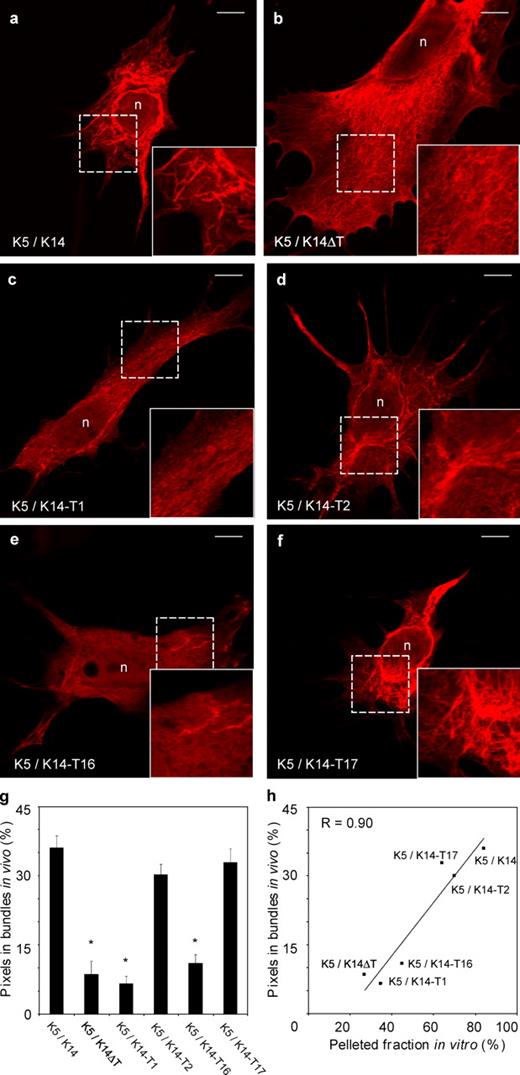
When tested in vitro using low-speed sedimentation (Fig. 5, c and d) or DIC microscopy (not depicted), wild-type and tailless K14 (K14ΔT) show the greatest and lowest potential, respectively, for salt-induced bundle formation with K5 as a partner. K14-T1 behaved largely like K14ΔT, whereas K14-T2 and K14-T17 exhibited properties akin to that of full-length, intact K14 (Fig. 5, c and d; and not depicted). The K14-T16 construct showed a behavior intermediate between K14 and K14ΔT (Fig. 5, c and d). Next, each of these untagged K14 constructs (pIRES-2–GFP–K14 variants) was expressed in combination with K5 (via pCMV-K5) in BHK-21 fibroblasts, and then subjected to indirect immunofluorescence for K5, confocal microscopy, and image processing (as shown in Fig. S3). In this setting, the behavior of the K14 mutants was even more polarized, in that intact K14, K14-T2, and K14-T17 showed a greater and similar ability to promote bundle formation in transfected fibroblasts, whereas K14ΔT, K14-T1, and K14-T16 showed a much weaker potential in this regard (Fig. 6, a–g). Again, here, an excellent correlation is seen between the low-speed sedimentation data, obtained in vitro, and the transfection results. Two distinct assembly behaviors are revealed by integrating these in vitro and in vivo assays: bundle-rich (K14, K14-T2, and K14-T17) versus bundle-poor (K14ΔT, K14-T1, and K14-T16; Fig. 6 h). Thus, the T2 moiety in the K14 tail domain is the main entity promoting the intrinsic pathway of K5/K14 filament cross-linking.
De novo keratin network formation in transfected BHK-21 fibroblasts. Bicistronic expression vectors consisting of a GFP reporter and a type I keratin cDNA (K14, K14ΔT, K14-T1, K14-T2, K14-T16, and K14-T17; see Fig. 5 B) were individually cotransfected with a K5 expression vector into keratin-free baby hamster kidney fibroblasts (BHK-21). Cells were fixed and processed for analysis 72 h later. GFP fluorescence was used as an indicator of type I keratin expression. (a–f) Indirect immunofluorescence for K5 using confocal microscopy. The combinations transfected are given in the lower left corner. The areas included in the squares with broken lines are magnified in the bottom right insets. n, nucleus. Bars, 10 µm. (g) Quantitation of the fluorescence signal specifically associated with large filament bundles in transfected cells. Image processing is detailed under Materials and methods and illustrated in Fig. S3. Bars represent the mean and SEM (error bars) from 45 cells from three transfection experiments. *, significantly different from wild-type K5/K14; one-way ANOVA, P < 0.001. (h) The correlation between in vitro (pellet fraction in the low speed sedimentation assay; Fig. 5 D) and in vivo assembly behavior (bundle-associated fluorescence in transfected BHK-21 cells; Fig. 6 G) for the various type I tail domain mutants tested in this series of experiments.
De novo keratin network formation in transfected BHK-21 fibroblasts. Bicistronic expression vectors consisting of a GFP reporter and a type I keratin cDNA (K14, K14ΔT, K14-T1, K14-T2, K14-T16, and K14-T17; see Fig. 5 B) were individually cotransfected with a K5 expression vector into keratin-free baby hamster kidney fibroblasts (BHK-21). Cells were fixed and processed for analysis 72 h later. GFP fluorescence was used as an indicator of type I keratin expression. (a–f) Indirect immunofluorescence for K5 using confocal microscopy. The combinations transfected are given in the lower left corner. The areas included in the squares with broken lines are magnified in the bottom right insets. n, nucleus. Bars, 10 µm. (g) Quantitation of the fluorescence signal specifically associated with large filament bundles in transfected cells. Image processing is detailed under Materials and methods and illustrated in Fig. S3. Bars represent the mean and SEM (error bars) from 45 cells from three transfection experiments. *, significantly different from wild-type K5/K14; one-way ANOVA, P < 0.001. (h) The correlation between in vitro (pellet fraction in the low speed sedimentation assay; Fig. 5 D) and in vivo assembly behavior (bundle-associated fluorescence in transfected BHK-21 cells; Fig. 6 G) for the various type I tail domain mutants tested in this series of experiments.
Discussion
The wealth of available evidence indicates that keratin filaments must be cross-linked to play a significant role of structural support in vivo (compare with data in the Introduction). We propose that keratin filament cross-linking is mediated by both intrinsic and extrinsic determinants in basal keratinocytes of epidermis (Fig. 7) and related stratified epithelia. Here, we provide conclusive evidence that interactions between three specific protein segments, the newly defined T2 moiety in K14's C-terminal tail domain, and the head-1A and L2-2B regions in K5, play a significant role in the intrinsic pathway of K5/K14 filament bundling, in vitro and in vivo. We show that the property of K5/K14 filament self-organization is determined independently from the 10-nm filament assembly, and can be transferred from K5 to K8 (and vice versa) by swapping the head-1A and L2-2B regions. We show that the assembly character of K16 (bundling-deficient; see Yamada et al., 2002) and K17 (bundling-competent; in this paper) is conferred to K14 upon C-terminal tail domain transfer. Complementarity between specific type I and type II keratins thus plays a key role in defining a potential for polymerization that goes beyond 10-nm filament formation.
Mechanisms of keratin filament bundling in basal keratinocytes of epidermis. Ultrastructural examination shows that keratin filaments are abundant and show a loosely bundled organization in basal layer keratinocytes of epidermis. Although the molecular basis for the attachment of keratin filaments at cell–cell and cell–matrix adhesion sites is well understood, the mechanisms responsible for filament bundling in the general cytoplasm are unknown. We propose that in basal keratinocytes and related cell types, this organization results from contributions from both intrinsic (e.g., self-driven) and extrinsic (e.g., associated proteins such as plectin and epiplakin) determinants. Both determinants are postulated to be required for the key structural support role fulfilled by K5/K14 filaments in basal keratinocytes. Self-organization of K5/K14 filaments into cross-linked networks involves interaction between the short T2 segment at the extreme C terminus of K14, and two distinct regions in K5 (the “head-1A” and “L2-2B” regions) contributed by adjacent filaments within a bundle. Exposure of the K14 tail domain at the filament surface is directly supported by experiments that involved partial proteolysis of assembled K5/K14 filaments. The two regions in K5 rod domain are both required for full expression of the self-organization potential. The “head-1A” and “L2-2B regions are ∼300 Å apart in K5, implying that the small K14's T2 segment (25 residues long) is too short to engage both of them within the same K5 molecule. By virtue of its richness in threonine and especially serine residues, K14's T1 domain is poised to regulate T2 function, and thus the intrinsic pathway of K5/K14 filament organization, via phosphorylation.
Mechanisms of keratin filament bundling in basal keratinocytes of epidermis. Ultrastructural examination shows that keratin filaments are abundant and show a loosely bundled organization in basal layer keratinocytes of epidermis. Although the molecular basis for the attachment of keratin filaments at cell–cell and cell–matrix adhesion sites is well understood, the mechanisms responsible for filament bundling in the general cytoplasm are unknown. We propose that in basal keratinocytes and related cell types, this organization results from contributions from both intrinsic (e.g., self-driven) and extrinsic (e.g., associated proteins such as plectin and epiplakin) determinants. Both determinants are postulated to be required for the key structural support role fulfilled by K5/K14 filaments in basal keratinocytes. Self-organization of K5/K14 filaments into cross-linked networks involves interaction between the short T2 segment at the extreme C terminus of K14, and two distinct regions in K5 (the “head-1A” and “L2-2B” regions) contributed by adjacent filaments within a bundle. Exposure of the K14 tail domain at the filament surface is directly supported by experiments that involved partial proteolysis of assembled K5/K14 filaments. The two regions in K5 rod domain are both required for full expression of the self-organization potential. The “head-1A” and “L2-2B regions are ∼300 Å apart in K5, implying that the small K14's T2 segment (25 residues long) is too short to engage both of them within the same K5 molecule. By virtue of its richness in threonine and especially serine residues, K14's T1 domain is poised to regulate T2 function, and thus the intrinsic pathway of K5/K14 filament organization, via phosphorylation.
The implications of our findings are schematized in Fig. 7. The model presented argues for key contributions of intrinsic determinants, as studied here, as well as extrinsic determinants (e.g., plectin and other IF-associated proteins) in shaping both the form and function of the cross-linked K5/K14 filament network in basal keratinocytes and related cell types. In its representation of the mechanisms underlying the intrinsic pathway, the model asserts that the extreme C terminus of the short K14 tail domain is exposed at the filament surface, as supported by the outcome of limited proteolysis of assembled keratin filaments (e.g., this study). K14's T2 segment reaches out to bind a K5 molecule from a distinct, proximal filament, thus promoting filament–filament linkages. The relatively high density of K5 and K14 molecules in the filament wall (>300 copies each per micrometer of length) allows for individual interactions of intrinsically low affinity (low micromolar range) to mediate stable linkages between adjacent filaments (this paper; Bousquet et al., 2001). The model (Fig. 7) also conveys the notion that the T2 portion of K14's tail domain is likely too small to mediate simultaneous interactions with both the “head-1A” and “L2-2B” segments in a K5 molecule. The latter agrees with our in vitro finding that the “head-1A” and “L2-2B” segments in K5 operate in trans to successfully mediate filament–filament interactions. Proximity of the bulkier type II keratin tail domain, which is twice the size of K14's tail, does not appear to impact the proposed mechanism, as self-organization occurs with both K5/K14 and K8bc/K14. Further insight into these interactions would greatly benefit from an atomic-level understanding of keratin filament structure, particularly its surface features.
Our findings to date point to a significant role for charged residues in mediating filament cross-linking via the intrinsic pathway. Such interactions, if verified, would be ideally suited for regulation via phosphorylation, already shown to play a role in regulating various aspects of IF structure and function in vivo (Izawa and Inagaki, 2006; Omary et al., 2006). The IF-binding region in several known IF-binding proteins tends to be rich in basic amino acid residues (see Coulombe, 2002). The T1 moiety in K14's tail domain, being serine rich and positioned between the rod and T2, seems poised to regulate the intrinsic pathway. Studies still in progress suggest that K14 is phosphorylated on its T1 domain in vivo. The task of defining which residues in the “head-1A” and “L2-2B” segments of K5, and in the T2 segment of K14, mediate filament self-organization should be facilitated by the high homology prevailing among keratin proteins. Residues of interest include those that are charged and also are: (1) conserved in K4, K5, and K6 but dissimilar in K8 over the relevant segments; and (2) conserved in K14 and K17 but dissimilar in K16 in the distal half of their tail domains (Fig. 5). The limited number of residues that meet these criteria are scattered within these three domains (Fig. 5 and not depicted), such that loss of any single one of them, e.g., through mutations, should not result in a marked loss-of-function phenotype. Accordingly, mutations at such sites have not yet been shown to cause EBS disease (unpublished data; Szeverenyi et al., 2008).
Our study also provides a rationale for common and distinct characteristics among three highly homologous type I keratins: K14, K16, and K17. Comparing studies from Paladini and Coulombe (1999) and Kerns et al. (2007), one infers that K17 is likely superior to K16 in its ability compensate for the loss of K14, and the associated keratinocyte fragility, in mouse skin in vivo. K14 and K17 share the T2 conserved motif in their tail domain, a key determinant of self-organization. Of further interest, a newly defined subpopulation of keratinocytes in the outer root sheath of rodent hair follicles express primarily K5 and K17: their keratin filaments are organized into very large, tightly packed bundles (Larouche et al., 2008). These observations provide physiological correlates for the findings described here. In addition, our observation of a tight correlation between keratinocyte surface area (and, potentially, cell shape as well; see Fig. 4) and the degree of keratin filament bundling suggests that the latter may play a role in the profound morphological changes occurring in keratinocytes as they undergo terminal differentiation in epidermis and related epithelia.
Yamada et al. (2002) showed that the K8/K18 pairing is nearly as capable as K5/K14 in undergoing the intrinsic pathway of filament cross-linking in vitro, but that pairing these type I and II keratins in an “unnatural fashion” yields filaments with markedly different self-organization properties. Thus, K8/K14 filaments cannot undergo the intrinsic pathway under the range of conditions tested, whereas, surprisingly, K5/K18 filaments do so to an extent that exceeds that shown by K5/K14 (Yamada et al., 2002). The tail domain of each type I keratin has seemingly evolved to mediate an optimal interaction with its target binding sites on the natural type II partner. When “excessive,” the property of self-organization may be counterproductive, a notion that is supported by the inability of K18 to rescue the K14-null phenotype in mouse (Hutton et al., 1998).
How relevant and important is the property of self-organization to the structural support function of K5/K14 filaments in basal layer keratinocytes? In K8bc and K5bd, we have now generated tools that can help us formally test this notion in vivo, e.g., in basal keratinocytes of K5-null mice. The latter exhibit a dramatic skin fragility phenotype (Peters et al., 2001), providing a convenient readout for testing rescue transgenes in the setting of living skin. Our hypothesis (Fig. 7) predicts that K8bc should fully rescue the K5-null phenotype, whereas the extent of rescue when expressing K5bd in K5-null mice will yield a sense of the relative importance of the intrinsic and extrinsic pathways of keratin IF cross-linking in vivo. As examples of extrinsic factors, known proteins, e.g., epiplakin (Wang et al., 2006; Spazierer et al., 2008) and plectin (Andra et al., 1997; Steinbock et al., 2000; Osmanagic-Myers et al., 2006), are present in basal keratinocytes and are capable of cross-linking keratin filaments (Fig. 7). In particular, studies in plectin-null keratinocytes revealed a profound influence of plectin on keratin IF bundling and network organization (Osmanagic-Myers et al., 2006). Therefore, effectors of the intrinsic and extrinsic pathways likely complement and regulate one another depending on the physical stress circumstances faced by keratinocytes (Fig. 7; Kreplak and Fudge, 2007).
Finally, the conditions that promote the intrinsic pathway of K5/K14 filament bundling remain to be identified in basal keratinocytes. Other than the general ionic strength and (slightly acidic) pH conditions known to prevail in epidermal keratinocytes (Behne et al., 2002), specific ions such as calcium (Hofmann et al., 1991) and/or physical events such as cellular stretching (Fudge et al., 2008) or shear stress (Flitney et al., 2009) could also have a role in fostering interfilament interactions via the intrinsic pathway. Alternatively, specific events, e.g., phosphorylation of the nonhelical head domain of K5 upon stress signaling (Toivola et al., 2002), could alter the conformation or exposure of a key binding site (in this case, the head-1A region) and render it more likely to engage its binding partners. These and many other important aspects of the intrinsic pathway of keratin filament organization require and deserve additional studies.
Materials and methods
Plasmids
Plasmids pET-K14, pET-K5 (Coulombe and Fuchs, 1990), and pET-K8 (Yamada et al., 2002) have been described previously and enable the expression of human keratins in Escherichia coli (Studier et al., 1990). Plasmids pET-K5bd and pET-K8bc were created by subcloning the relevant cDNAs into pET-24a (Yamada et al., 2002). Using a QuickChange site-specific mutagenesis kit (Agilent Technologies), DraIII and XbaI restriction sites were introduced in the K5 and K8 cDNAs to allow for the swapping of coding sequences overlapping a portion of the head domain and the L2-2B domain. Oligonucleotide primers 1–4 were used for this purpose (Table S1).
Because the boundary sequences of the variable regions/head domains of K5 and K8 do not have any available sites for silent mutations to generate restriction enzyme sites, a PCR-generated overlap extension was used (Warrens et al., 1997; Heckman and Pease, 2007) to conjugate the head domain of K5 and variable region of K8 or the head domain of K8 and variable region of K5 using oligonucleotide primers 5–10 (Table S1).
For expression in mammalian cells, we cloned the cDNAs for K5, K5bd, K8, and K8bc into a pIRES-2–GFP vector (BD) using Nhe I and Not I restriction sites. The wild-type K5 and K14 cDNAs were cloned into pBK-CMV vector (Clonetech Laboratories, Inc.) using the same sites.
A tailless mutant of K14 was generated using oligonucleotides 11 and 12 (Table S1), which also created a new NgoMI site without altering codon significance. NgoM I was used to make an adhesive overhang, allowing for the attachment of K16 tail domain or K17 tail domain sequences. NgoM I sites were inserted into K16 or K17 cDNA using oligonucleotides 13 and 14 (Table S1). Plasmids pET-K16 (Paladini et al., 1995) and pET-K17 (Wawersik et al., 1997) contain a natural NgoM I site at the 3′ end of the coding sequence. A K14 chimera with the tail domain of K16 or K17 was built by performing NgoM I treatment of K14 tailless mutant cDNA, and of the K16 and K17 cDNAs, followed by ligation. To make a K14 deletion mutant lacking the T2 domain, we used oligonucleotide primers 11 and 15 (Table S1). To make a K14 deletion mutant lacking the T1 domain, a PCR-based overlap extension method was used using oligonucleotides 11, 16, 17, and 18 (Table S1). All constructs were verified by DNA sequencing. The K14 mutants generated were subcloned into pET-3d or pET-15b for recombinant protein production or a pIRES-2–GFP vector for mammalian expression.
Keratin purification and reconstitution of keratin filament in vitro
pET-K5, pET-K5bd, pET-K8, pET-K8bc, and pET-K14 were transformed into the E. coli strain BL21 (DE3) to produce the corresponding recombinant human proteins as inclusion bodies (Coulombe and Fuchs, 1990). The proteins were purified in multiple steps consisting of preparation of inclusion bodies, urea-based solubilization of keratin proteins, and ion exchange chromatography using Hi-TrapQ and MonoQ columns (GE Healthcare) as described previously (Ma et al., 2001; Yamada et al., 2002). Heterotypic complexes containing type I and type II keratins in an equimolar ratio were prepared and purified using the MonoQ column. Suitable fractions were identified, their concentration was adjusted to 0.5 or 1 mg/ml, and they were used for filament reconstitution by serial dialysis using the following three buffers at room temperature: (a) 9 M urea and 25 mM Tris-HCl, pH 7.4, with 25 mM β-ME for 4 h; (b) 2 M urea and 5 mM Tris-HCl, pH 7.4, with 5 mM β-ME, for 1 h; and (c) 5 mM Tris-HCl, pH 7.5, with 5 mM β-ME overnight. The NaCl concentration of the last buffer was adjusted to 0 mM (standard assembly condition) or 10 mM (to promote cross-linked network formation; Bousquet et al., 2001; Ma et al., 2001; Yamada et al., 2002).
Assessing keratin filament assemblies
The physical states of the polymers and polymerization efficiencies were assessed by high-speed sedimentation (150,000 g; 30 min) as described previously (Ma et al., 2001). Filament ultrastructure was examined by negative staining (1% uranyl acetate) and transmission electron microscopy using Philips BioTwin CM120 (FEI Company) or Hitachi 7600 (Hitachi) instruments. Formation of cross-linked networks was assessed by subjecting the assembled keratin filaments to low-speed sedimentation (8,000 g for 30 min; Pollard and Cooper, 1982), followed by SDS-PAGE, Coomassie blue staining, and measurement of band density using ImageJ (National Institutes of Health). Network morphology was examined by DIC light microscopy using an IX81 instrument (Olympus) equipped with an electron-multiplying charge-coupled device camera (C9100-02; Hamamatsu Photonics). Images were acquired at room temperature using a 100× NA 1.4 oil UIS2 objective lens (Olympus). Optimal contrast and brightness were adjusted using Photoshop software (Adobe). For falling ball viscometry assay, we applied a steel ball (diameter 4 mm; weight 260 mg) into a 1-ml syringe (BD) containing a 0.5 ml-solution of each of the keratin filaments (1 mg/ml) under bundling-promoting assembly condition (10 mM NaCl, 5 mM TrisHCl, pH 7.5, and 5 mM β-ME).
Protease digestion and mass spectrometry
To prepare samples for mass spectrometry analysis, reconstituted K5/K14 filaments were treated with 1:100 (wt/wt) of endoprotease Glu-C (Roche) for 4 h at room temperature. Cleavage was assessed by using monoclonal antibodies directed against the K14 tail domain (LL001; Santa Cruz Biotechniology, Inc.), the rod domains of type I (AE1; or type II keratins [PCK26]; both from Abcam). Digestion products were separated by Centricon (Millipore), with a 3-kD cutoff, to collect small peptides as a flow-through fraction. Larger-sized digestion products were analyzed through 8.5% SDS-PAGE. Enriched small peptide fractions were evaporated under a vacuum, diluted in 10 µl of formic acid 0.5%, and desalted using C18 Zip Tips (Millipore). Peptides were eluted using 5 µl of 80% acetonitrile. Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry was performed on a Voyager DE-STR mass spectrometer (Applied Biosystems). 0.5 µl of the peptide solution was mixed on target with 0.5 µl of matrix (10 µg/µl α-cyano-4-hydroxycinnamic acid [Sigma-Aldrich]) solution in 50% acetonitrile/0.3% trifluoroacetic acid) via the dried droplet method. All MALDI spectra were calibrated with Calibration Mixture 2 (Applied Biosystems). Identification of fragments from MALDI/MS spectra was performed using PeptideCutter (http://ca.expasy.org/tools/peptidecutter/).
Generation of K5 fragments
All K5 deletion mutants for far Western screening were fused to EGFP from a pEGFP-c3 vector (Clontech Laboratories, Inc.), except for the larger K5-2A-L2-2B-T fragment, which was used as an intact, unfused fragment. EGFP was subcloned through a PCR strategy using oligonucleotide primers 19 and 20 (Table S1), then transferred into a pT7-HMT vector, which provides a hexa-histidine tag for one-step purification (Geisbrecht et al., 2006).
The following oligonucleotide primers were used to construct K5 fragments from the wild-type K5 cDNA template (see Table S1): K5 EV (Met 1–Gly 39) using oligonucleotides 21 and 22; K5 VH (Gly 39–Gln 141) using oligonucleotides 23 and 24; K5 H (Gly 138–Ser 181) using oligonucleotides 19 and 20; K5 H-1A (Gly 138–Gly 207) using oligonucleotides 25 and 26; K5 L2 (Ala 350–Gly 371) using oligonucleotides 27 and 28; K5 L2-2B (Ala 350–Glu 479) using oligonucleotides 27 and 29; K14 L2-2B (Ala 295–Ala 424) using oligonucleotides 30 and 31; and K5 Tail (Gly 477–Stop) using oligonucleotides 32 and 33. K5-2A-L2-2B-T was subcloned by subjecting the pET-K5 plasmid to restriction with Nco I (spanning Met 327 in the middle of linker L12) and EcoRI (located at the 3′ end of the K5 coding sequence) and to ligation directly into the pT7-HMT vector without a fusion tag. All K5 deletion mutants were confirmed by sequencing.
K5 deletion fragments were expressed in E. coli, and insoluble fragments (K5 EV, K5 VH, K5 H-1A, and K5 Tail) were recovered in 3M urea. Cell lysates containing HisTag-EGFP fusion proteins were applied to Ni-NTA agarose resin (QIAGEN) to purify the fragment polypeptides using the manufacturer's instructions. To purify the K5 2A-L2-2B-T fragment, cell lysates in TBS buffer were centrifuged and fractionated to isolate inclusion bodies, which were solubilized in 3M urea.
Far Western analysis
Far Western blotting was used as described previously (Edmondson and Roth, 2001) to identify binding regions of the K14 tail domain in K5. In brief, purified fragments were resolved by SDS-PAGE, and transferred to a nitrocellulose membrane. After blocking with phosphate buffer (5 mM sodium phosphate, pH 7.5, and 0.1% Tween-20, with 10 mM NaCl) containing 5% milk, membranes were incubated with purified GFP-fused K14 tail domain (20 µg/ml) for 1 h at room temperature. Membranes were washed with phosphate buffer, and fixed using the cross-linker dithiobis(succinimidyl)propionate (DSP; Thermo Fisher Scientific) dissolved (0.2 µg/ml) in 5 mM sodium phosphate, 0.1% Tween 20, and 10 mM NaCl for 30 min, followed by quenching in 25 mM Tris-HCl, pH 7.5, 0.1% Tween 20, and 5% milk, for 30 min at room temperature. Standard Western blot assays were performed using a rabbit anti-GFP antibody (Invitrogen) diluted 1:2,000 or a rabbit polyclonal anti-K14 antibody (Covance) diluted 1:5,000, followed by a goat anti–rabbit antibody conjugated to horseradish peroxidase (Sigma-Aldrich) diluted 1:10,000. Enhanced chemiluminescence (Thermo Fisher Scientific) was performed according to the manufacturer's instructions.
Fibroblast culture, transfection assays, and fluorescence quantification
The BHK-21 cell line (baby hamster kidney fibroblasts; American Type Culture Collection) was used to analyze the assembly properties of keratin proteins in a keratin-free, in vivo setting (Paladini et al., 1996). Cotransfections of type II keratin and type I keratin were performed using Fugene 6 (Roche) according to the manufacturer's instructions. For assessing type II keratin bundling, one of pIRES-2–GFP–K5, pIRES-2–GFP–K8, pIRES-2–GFP–K5bd, and pIRES-2–GFP–K8bc was cotransfected with pCMV-K14. In the type I tail domain studies, each pIRES-2–GFP–K14 variant was cotransfected with pCMV-K5. At 72 h after transfection, cells were fixed with 3.3% paraformaldehyde for 15 min, extracted with 0.1% Triton X-100 in PBS buffer for 5 min, and processed for indirect immunofluorescence using a rabbit polyclonal antiserum against K14 (AF-64 [Covance]; 1:1,000 dilution) or K5 (AF-138 [Covance]; 1:1,000 dilution), and a rhodamine-conjugated goat anti–rabbit secondary antibody (1:2,000; Kirkegaard and Perry Laboratories, Inc.). Cell images were obtained at room temperature using a confocal microscope (Axiovert 200 microscope with a 510 Meta module; Carl Zeiss, Inc.) fitted with a 63× 1.4 NA Plan-Apochromat oil objective and a 100× 1.4 NA Plan-Apochromat oil objective lenses. Microscope operation and image acquisition was done using the LSM 510 software (Carl Zeiss, Inc.). Pixel information from the confocal images was analyzed using ImageJ software with the following plug-ins: LSM reader, Calculator Plus (http://rsbweb.nih.gov/ij/plugins/index.html) and Just Another Colocalization Plugin (JACoP; Bolte and Cordelieres, 2006). Quantification of pixels in keratin bundles was performed in two steps. First, the rhodamine channel signal (reflecting keratin filaments) was corrected by subtracting the intensity emanating from the GFP channel signal (reflecting the expression level and thus allowing for a between-cell correction) for the whole cell via the Calculator Plus plug-in. After filtering, the remaining rhodamine signals, representing the most intense pixels, are typically associated with thick filament bundles. The final data are reported as the red channel signal intensity after filtering divided by the original signal intensity, prefiltering via JACoP. This image analysis procedure is illustrated in Fig. S3 b.
Electron microscopy of transiently transfected fibroblasts in culture
Transfected BHK-21 cells were fixed with 3.3% paraformaldehyde in PBS buffer for 15 min, then washed with PBS three times. Fixed cells were then permeabilized with 0.5% Triton X-100 in PBS for 5 min, and washed with PBS three times. To identify keratin polymers, the samples were incubated with a rabbit polyclonal K14 antibody for 3 h, washed with PBS, and further incubated with 10 nm of colloidal gold/Alexa Fluor 488–conjugated goat anti–rabbit IgG (Invitrogen), also for 3 h, followed by three PBS washes. Samples were then further fixed with 2% glutaraldehyde in PBS, rinsed in buffer for 30 min, and treated with 1% osmium tetroxide reduced with 0.8% potassium ferrocyanide in 0.1 M sodium cacodylate and 3 mM CaCl2 for 1 h at 4°C in the dark. Samples were rinsed with distilled H2O and en-bloc stained with 2% aqueous uranyl acetate for 1 h in the dark. Plates were dehydrated in a graded series of ethanol and then embedded in Eponate 12 (Ted Pella Inc.) overnight without a catalyst. The next day, dishes were further infiltrated with fresh epon supplemented with 1.5% DMP-30 (catalyst), then cured in a 37°C oven for 3 d. Dishes were then transferred to a 60°C oven for final curing. Upon hardening, the epon discs were separated from the dishes, and 3-mm buttons were removed with a heavy duty hole punch. Samples were mounted cell side up to epon blanks and trimmed for sectioning. 80-nm thin compression free sections were obtained with a diamond knife (Diatome). Sections were picked up on 200 mesh copper grids (Polysciences, Inc.) and further stained with uranyl acetate followed by lead citrate. Grids were examined on a H-7600 transmission electron microscope (Hitachi) operating at 80 Kv. Images were digitally captured with an AMT 1,000 × 1,000 charge-coupled device camera. Image contrast and brightness were enhanced using Photoshop software (Adobe).
Skin keratinocyte culture and keratin knockdown studies
The mouse 308 keratinocyte cell line has been described previously (Strickland et al., 1988). Primary cultures of newborn keratinocytes from K6a/K6b-null mice were established as described previously (Bernot et al., 2005). K5 protein levels were reduced using an shRNA-expressing plasmid, generated according to the pSUPER RNAi system (Oligoengine). A K5 targeting sequence was designed as follows. shRNA K5 (the targeting region in K5′s 3′ untranslated region is underlined): 5′-GATCCGGTCCGTTGTTTGAACAGTACATCTCGAGATGTACTGTTCAAACAACGGATTTTTG-3′.
K6a/K6b-null keratinocytes were transfected with shRNA and/or either K8 (pCMV-K8) or K8bc (pCMV-K8bc) cDNA using GeneJuice (EMD). At 72 h after transfection, cells were fixed and processed for indirect immunofluorescence and image analysis as described in Fig. S3. K14 was detected using the rabbit anti-K14 antibody followed by Pacific Blue goat anti–rabbit IgG (Invitrogen). K8 and K8bc were detected using the chicken polyclonal antibody (ab14053; Abcam) followed by rhodamine-conjugated goat anti–chicken IgY (Abcam). The GFP signal was intensified by using mouse anti-GFP antibody (clone GFP-20; Sigma-Aldrich) with FITC-conjugated goat anti–mouse IgG (Kirkegaard and Perry Laboratories, Inc.).
Measurement of keratinocyte surface area
Keratinocytes expressing the pSuper-shRNA/GFP reporter were imaged at room temperature in the appropriate green channel using a confocal microscope (LSM Meta 510; Carl Zeiss, Inc.), and processed through the ImageJ software to yield binary images. Images were originally acquired using a 12 bit, 1,024 × 1,024 mode, and then converted to 8 bit for further processing by ImageJ. Keratinocyte surface area was computed by counting the number of pixels using the “Analyze Particles” command. DAPI staining was used to identify single cells, thereby excluding multiple cell aggregates from this analysis.
Statistical analyses
Statistical analyses were performed using SPSS software version 13 (SPSS, Inc.). A standard t test was used to compare the cross-linking potential of K8 versus K8bc in transfected keratinocytes. One-way analysis of variance (ANOVA) analysis was used in all instances for which more than three groups were being compared. Differences among groups were assessed by a post-hoc Tukey test. A statistical P-value of <0.05 was considered significant.
Online supplemental material
Fig. S1 shows experiments to determine the rod-tail domain boundary in keratin 14. Fig. S2 shows sedimentation assays and transmission electron micrographs to assess the filament-forming properties of select type II keratin variants in vitro. Fig. S3 shows biochemical and morphological analyses of transfected keratin proteins in cultured BHK-21 fibroblasts. Fig. S4 shows Western blots and fluorescent microscope images for analysis of shRNA-mediated knockdown of K5 protein levels in cultured mouse skin keratinocytes. Fig. S5 shows sedimentation assays and transmission electron micrographs that assess the filament-forming properties of select type I keratin tail variants in vitro. Table S1 shows DNA oligomers for the generation of the mutant constructs.
Acknowledgments
We are most grateful to Hee Jin Bae for her contribution on the cloning of K14 mutants. We thank Robert Cole (mass spectrometry), Mike Delannoy, Carol Cooke (electron microscopy), Douglas Murphy (DIC microscopy), and Susan Craig (high speed ultracentrifugation) for their help and advice, and M. Bishr Omary for providing an expression vector for K8.
These studies were supported by grant AR42047 from the National Institutes of Health to P.A. Coulombe.
References
Abbreviations used in this paper: ANOVA, analysis of variance; DIC, differential interference contrast; IF, intermediate filament; shRNA, small hairpin RNA.