Histone H2B monoubiquitination by Rad6/Bre1 is required for the trimethylation of both histone H3K4 and H3K79 by COMPASS and Dot1 methyltransferases, respectively. The dependency of methylation at H3K4 and H3K79 on the monoubiquitination of H2BK123 was recently challenged, and extragenic mutations in the strain background used for previous studies or epitope-tagged proteins were suggested to be the sources of this discrepancy. In this study, we show that H3K4 and H3K79 methylation is solely dependent on H2B monoubiquitination regardless of any additional alteration to the H2B sequence or genome. Furthermore, we report that Y131, one of the yeast histone H2A/H2B shuffle strains widely used for the last decade in the field of chromatin and transcription biology, carries a wild-type copy of each of the HTA2 and HTB2 genes under the GAL1/10 promoter on chromosome II. Therefore, we generated the entire histone H2A and H2B alanine-scanning mutant strains in another background, which does not express wild-type histones.
Introduction
A nucleosome contains 146 bp of DNA wrapped twice around an octamer composed of two copies of each histone protein: H2A, H2B, H3, and H4 (Kornberg, 1974; Kornberg and Lorch, 1999). Nucleosomes are observed as a series of “beads on a string” via electron microscopy with the “beads” being the individual nucleosomes connected by the linker DNA, the “string.” Structural studies have demonstrated that histone N-terminal tails protrude outward from the nucleosome and can be posttranslationally modified by different enzymes (Luger et al., 1997; Shilatifard, 2006). Such posttranslational modifications of histone tails include phosphorylation, acetylation, sumoylation, ADP ribosylation, ubiquitination, and methylation.
The first H3K4 (histone H3 lysine 4) methylase, Set1/COMPASS, was isolated from Saccharomyces cerevisiae and was demonstrated to be capable of mono-, di-, and trimethylating H3K4 (Miller et al., 2001; Roguev et al., 2001; Krogan et al., 2002). This posttranslational modification of H3K4 by COMPASS requires prior H2BK123 (histone H2B lysine 123) monoubiquitination in yeast and H2BK120 in vertebrates, which is a process known as histone cross talk (Dover et al., 2002; Sun and Allis, 2002; Wood et al., 2005; Shilatifard, 2006). It has also been demonstrated that histone H3K79 methylation by Dot1 also requires H2BK123 monoubiquitination (Briggs et al., 2002; Wood et al., 2003a). Monoubiquitination of histone H2BK123 is mediated by the macromolecular complex containing the E2-conjugating enzyme Rad6 and the E3 ligase Bre1 in S. cerevisiae (Robzyk et al., 2000; Hwang et al., 2003; Wood et al., 2003a). This modification has been linked to transcriptional activation and elongation (Henry et al., 2003; Kao et al., 2004; Xiao et al., 2005; Pavri et al., 2006; Shilatifard, 2006; Tanny et al., 2007; Kim et al., 2009). Studies in other eukaryotic organisms also have confirmed that this mode of regulation is well conserved from yeast to human (Pavri et al., 2006; Smith and Shilatifard, 2009). However, a recent study performed by Foster and Downs (2009) argued that monoubiquitination of H2BK123 in yeast is not the sole determinant for the methylation of H3K4 and H3K79. In their study, Foster and Downs (2009) observed the presence of both H3K4 and H3K79 trimethylation in the H2BK123R mutant strain derived from an FY406 parental strain, which is contradictory to the previous findings, whereas both modifications were lost in an H2BK123R mutant derived in the Y131 background. Surprisingly, a triple KSS mutant (H2BK123R-S125A-S126A) in the FY406 background resulted in the loss of trimethylation of both H3K4 and H3K79. Additionally, Foster and Downs (2009) demonstrated that Flag-tagged H2B containing the K123R mutation (Flag-H2BK123R) abolished the trimethylation of H3K4 and H3K79, whereas untagged H2BK123R still possessed normal levels of trimethylation of both of the lysine residues in strain FY406. These observations led them to conclude that monoubiquitination of histone H2B alone is not required for the trimethylation of either H3K4 or H3K79, but that an additional “unknown” alteration to H2B or the mutation in the genome in combination with the K123 mutation caused the loss of histone H3K4 and H3K79 trimethylation. In this study, we have addressed a possible role for the Flag tag on histone H2B in the regulation of H3K4 and H3K79 methylation. Our collective experiments have demonstrated that histone H3K4 and H3K79 methylation is solely dependent on H2B monoubiquitination and is independent of any other unknown factors or genetic backgrounds.
Results and discussion
To further investigate the role of H2BK123 monoubiquitination in H3K4 and H3K79 trimethylation and a possible role for the Flag epitope of H2B in this process, we set out to analyze the role of Flag-less H2B mutated in K123 for H3K4 and H3K79 methylation in several different strain backgrounds. We first wanted to be able to detect the presence of monoubiquitinated H2B in a Flag-less H2B background because the observation made by Foster and Downs (2009) that H3K4 trimethylation could be detected in a Flag-less H2BK123R strain could be because of the inadvertent presence of a wild-type copy of H2B. To address this, we generated polyclonal antibodies specific to monoubiquitinated H2B (Fig. 1 A) and then shuffled a plasmid expressing H2B lacking an N-terminal Flag tag (Fig. 1 B) into several different backgrounds, including Y131, FY406, DY20D, and JHY205. We analyzed the trimethylation levels of both H3K4 and H3K79 as well as the monoubiquitination of H2B in these backgrounds (Fig. 2, A and B). Our antibody generated in this study is specific for monoubiquitinated H2B in yeast and is capable of detecting this modification in the presence or absence of a Flag tag (Figs. 1 A and 2 A; and see Figs. 3 B and 5 B). When this antibody was used, Flag-less wild-type H2B showed a band migrating slightly faster than that of Flag-tagged wild-type H2B as a result of the deletion of the Flag tag (Fig. 2 A). However, no monoubiquitinated form of H2B was detected from K123A mutants in both Y131 and FY406 backgrounds and K123R mutants in DY20D and JHY205, regardless of the attachment of a Flag tag to H2B (Fig. 2, A and B). Furthermore, we tested these strains for the methylation of histone H3K4 and H3K79 (Fig. 2). Regardless of the presence or absence of a Flag tag on H2B, H3K4 methylation was detected in strains bearing wild-type H2B but not in strains in which H2B carries a point mutation at the K123 residue. Similar strains were generated independently in all of our laboratories, and we have each confirmed that there is no difference between Flag-tagged H2B and untagged H2B and that all of the histone modifications are consistent among the different strain backgrounds, including the FY406 background used in the study performed by Foster and Downs (2009; Fig. 2). Therefore, we conclude that H3K4 and H3K79 trimethylation is indeed solely dependent on H2B monoubiquitination regardless of an additional alteration to the H2B sequence or strain backgrounds.
Generation of antibodies specific to K123-monoubiquitinated H2B. (A) Development of polyclonal antibodies specific to monoubiquitinated H2B. Ubiquitinated H2B–specific antibodies generated in rabbit were affinity purified and characterized by Western blot analysis of whole cell extracts prepared from wild type (WT), H2BK123A, ubp8Δ, and rad6Δ. Both wild-type and ubp8Δ strains show the presence of monoubiquitinated H2B (24 kD and 23 kD, respectively); however, an increased amount of the ubiquitinated form of H2B was observed in the ubp8Δ strain as expected. No bands were detected in either H2BK123A or the rad6Δ strains, showing that our H2Bub antibody is capable of specifically recognizing ubiquitinated H2B in yeast. (B) Schematics of the plasmids used in this study. pSN888 was generated from pZS145 by deletion of the N-terminal Flag tag (Fg) from the HTB1 gene. Similarly, the Flag on H2B was removed in pRS315-HHT2-HHF2-HTA1-FlagHTB1 to generate H2B Flag-less strains.
Generation of antibodies specific to K123-monoubiquitinated H2B. (A) Development of polyclonal antibodies specific to monoubiquitinated H2B. Ubiquitinated H2B–specific antibodies generated in rabbit were affinity purified and characterized by Western blot analysis of whole cell extracts prepared from wild type (WT), H2BK123A, ubp8Δ, and rad6Δ. Both wild-type and ubp8Δ strains show the presence of monoubiquitinated H2B (24 kD and 23 kD, respectively); however, an increased amount of the ubiquitinated form of H2B was observed in the ubp8Δ strain as expected. No bands were detected in either H2BK123A or the rad6Δ strains, showing that our H2Bub antibody is capable of specifically recognizing ubiquitinated H2B in yeast. (B) Schematics of the plasmids used in this study. pSN888 was generated from pZS145 by deletion of the N-terminal Flag tag (Fg) from the HTB1 gene. Similarly, the Flag on H2B was removed in pRS315-HHT2-HHF2-HTA1-FlagHTB1 to generate H2B Flag-less strains.
Di- and trimethylation of histone H3K4 and trimethylation of H3K79 are dependent solely on monoubiquitination of H2BK123. (A) Western blotting of whole cell extracts from strains transformed with a plasmid carrying wild-type (WT) H2B or mutant H2BK123 (K123A; K123R) either with or without an N-terminal Flag tag. Cell extracts were prepared from wild-type H2B or H2BK123 mutants in three different strain backgrounds and were subjected to SDS-PAGE and analyzed by Western blot analysis with antibodies to dimethyl H3K4, trimethyl H3K4, trimethyl H3K79 (αH3K4me2, αH3K4me3, and αH3K79me3), monoubiquitinated H2BK123 (αH2Bub), or Flag (αFLAG). An antibody against H3 (αH3) was used as a loading control. The calculated molecular masses of H2B, ubiquitinated H2B, and H3 are 14 kD, 23 kD, and 15 kD, respectively. The calculated molecular masses of Flag-tagged H2B and ubiquitinated, Flag-tagged H2B are 15 kD and 24 kD, respectively. White lines indicate that intervening lanes have been spliced out. (B) Western blotting of whole cell extracts from strains transformed with plasmid carrying wild-type or mutant H2BK123R in FY406 background.
Di- and trimethylation of histone H3K4 and trimethylation of H3K79 are dependent solely on monoubiquitination of H2BK123. (A) Western blotting of whole cell extracts from strains transformed with a plasmid carrying wild-type (WT) H2B or mutant H2BK123 (K123A; K123R) either with or without an N-terminal Flag tag. Cell extracts were prepared from wild-type H2B or H2BK123 mutants in three different strain backgrounds and were subjected to SDS-PAGE and analyzed by Western blot analysis with antibodies to dimethyl H3K4, trimethyl H3K4, trimethyl H3K79 (αH3K4me2, αH3K4me3, and αH3K79me3), monoubiquitinated H2BK123 (αH2Bub), or Flag (αFLAG). An antibody against H3 (αH3) was used as a loading control. The calculated molecular masses of H2B, ubiquitinated H2B, and H3 are 14 kD, 23 kD, and 15 kD, respectively. The calculated molecular masses of Flag-tagged H2B and ubiquitinated, Flag-tagged H2B are 15 kD and 24 kD, respectively. White lines indicate that intervening lanes have been spliced out. (B) Western blotting of whole cell extracts from strains transformed with plasmid carrying wild-type or mutant H2BK123R in FY406 background.
The Y131 strain was originally generated for plasmid shuffling of H2A-H2B genes, as a simultaneous deletion of the HTA1-HTB1 and HTA2-HTB2 loci is lethal (Robzyk et al., 2000). The genotype of Y131 describes that hta1-htb1 was replaced with LEU2, whereas hta2-htb2 was replaced with URA3 in the presence of a HIS3 HTA2-HTB2 plasmid. After selection for tight 5-FOA resistance, a URA3 plasmid carrying HTA1-HTB1 was substituted for the HIS3-HTA2-HTB2 plasmid, thus creating the shuffle strain that was used to introduce a HIS3-marked plasmid containing HTA1 and Flag-HTB1. The Y131 strain expressing Flag-tagged wild-type H2B under either glucose or galactose media has normal levels of H2B monoubiquitination and methylation of H3K4 and K79 (Fig. 3 A, lanes 3–6). To our surprise, when Flag-H2BK123R in the Y131 background was grown continuously in a galactose-containing media, the methylation of both of the H3 residues was present at levels close to those in the wild-type cells (Fig. 3 A, lanes 7–10). Because the N terminus of H2BK123R was tagged with Flag, we tested for the presence of monoubiquitination using a Flag antibody. There was no slower-migrating band in the histone H2BK123R bearing strain, indicating that there is no monoubiquitinated form of Flag-H2B (Fig. 3 A, lanes 3–10). The H3K4 and H3K79 methylation patterns appear to be normal.
The H2A/H2B shuffle strain Y131 contains a galactose-regulated copy of HTA2-HTB2 genes on chromosome II. (A) Western blot analysis of whole cell extracts prepared from Flag-tagged wild-type H2B and Flag-H2BK123R grown under either glucose or galactose media. Cell extracts were subjected to SDS-PAGE and Western analysis with antibodies specific to H3K4me2, H3K4me3, H3K79me3, and Flag. An antibody against H3 (αH3) was used as a loading control. Triangles describe the increasing amounts of proteins loaded onto the gel. Letters D and G denote glucose and galactose, respectively. (B) Western analysis of whole cell extracts prepared from wild-type (WT) BY4742 (FM392) and its derivative rad6Δ, Flag-H2B (Y131 background), and Flag-H2BK123R (Y131 background). Cell extracts were subjected to SDS-PAGE and Western analysis with antibodies to trimethyl H3K4 (αH3K4me3), monoubiquitinated H2BK123 (αH2Bub), or Flag (αFLAG). An antibody against H3 was used as a loading control. The H2B monoubiquitination–specific antibody detected a faster migrating band, which is indicated by red arrows. This band represents an untagged version of H2B, which is only seen in Y131 when cells are grown in galactose-containing media. Blue arrows indicate the slower migrating Flag-tagged, monoubiquitinated H2B seen under both dextrose and galactose growth conditions only in wild-type cells and not H2BK123R. Black lines indicate that intervening lanes have been spliced out.
The H2A/H2B shuffle strain Y131 contains a galactose-regulated copy of HTA2-HTB2 genes on chromosome II. (A) Western blot analysis of whole cell extracts prepared from Flag-tagged wild-type H2B and Flag-H2BK123R grown under either glucose or galactose media. Cell extracts were subjected to SDS-PAGE and Western analysis with antibodies specific to H3K4me2, H3K4me3, H3K79me3, and Flag. An antibody against H3 (αH3) was used as a loading control. Triangles describe the increasing amounts of proteins loaded onto the gel. Letters D and G denote glucose and galactose, respectively. (B) Western analysis of whole cell extracts prepared from wild-type (WT) BY4742 (FM392) and its derivative rad6Δ, Flag-H2B (Y131 background), and Flag-H2BK123R (Y131 background). Cell extracts were subjected to SDS-PAGE and Western analysis with antibodies to trimethyl H3K4 (αH3K4me3), monoubiquitinated H2BK123 (αH2Bub), or Flag (αFLAG). An antibody against H3 was used as a loading control. The H2B monoubiquitination–specific antibody detected a faster migrating band, which is indicated by red arrows. This band represents an untagged version of H2B, which is only seen in Y131 when cells are grown in galactose-containing media. Blue arrows indicate the slower migrating Flag-tagged, monoubiquitinated H2B seen under both dextrose and galactose growth conditions only in wild-type cells and not H2BK123R. Black lines indicate that intervening lanes have been spliced out.
Because H2B monoubiquitination is required for the methylation of these lysine residues, we suspected that one of the two genomic HTB genes was not deleted in Y131 and that the genomic wild-type H2B was expressed in the K123R mutant strain in a galactose-dependent manner. We tested this possibility by using our antibody specific to monoubiquitinated histone H2B (Fig. 3 B). In wild-type Y131 under glucose media, the Flag-tagged wild-type H2B produced only an upper band (Flag-tagged, monoubiquitinated H2B; Fig. 3 B, lane 5, blue arrow). However, in wild-type Y131 under galactose media there are two bands (Fig. 3 B, lane 6). The upper band is Flag-tagged, monoubiquitinated histone H2B, and the lower band is Flag-less, monoubiquitinated histone H2B (Fig. 3 B, lane 6). Similarly, the Flag-H2BK123R strain shows no H2B monoubiquitination under the glucose media (Fig. 3 B, lane 7). However, the Flag-tagged H2BK123R strain grown under galactose media shows the band corresponding to Flag-less, monoubiquitinated histone H2B (Fig. 3 B, lane 8). We have also detected the expression of wild-type H2B under the galactose condition when using H2B-specific antibodies (unpublished data). This suggests that Y131 contains a galactose-inducible version of wild-type H2B somewhere in the genome.
To understand the basis for these results between the two growth conditions, we sequenced the genomic regions around the deleted hta1-htb1 (chromosome IV) and hta2-htb2 (chromosome II) genes in strain Y131. We found that intact HTA2 and HTB2 genes were still present on chromosome II in the Y131 strains, and surprisingly, the bidirectional GAL1/10 promoter was inserted between the two genes to exactly replace the bidirectional HTA2-HTB2 promoter (Fig. 4). The GAL10 promoter drives the expression of HTA2, and GAL1 drives the expression of HTB2. The presence of the GAL1/10 promoter explains why wild-type H2B was expressed only under the galactose media and not under the glucose media. It remains somewhat of a mystery how GAL1/10 was inserted exactly between the two genes in the first place, although the HIS3 plasmid in the original Y131 strain carried a GAL1/10-regulated HTA2-HTB2 locus that may have been incorporated into the genome by a rare recombination event. Because Y131 is a widely used strain, mutants that affect GAL1/10 transcription may not appropriately repress wild-type HTA2-HTB2 in glucose in this strain, and data obtained from its usage under prolonged growth in galactose conditions must be interpreted carefully. Studies using this strain to analyze effects of the H2B-K123R mutation on GAL1 transcription were performed for short periods of galactose induction (2 h) when the expression of the genomic HTA2-HTB2 genes was not detected, and the transcription results have been recapitulated in other strain backgrounds (Xiao et al., 2005; unpublished data).
In the Y131 strain, the GAL1/10 promoter is inserted between HTA2 and HTB2 on chromosome II. Schematic of the regions containing HTA2-HTB2 genes on chromosome II. Although this region was deleted and replaced with URA3 in the Y131 strain, a wild-type copy of HTA2-HTB2 controlled by the GAL1/10 promoter is present in this region.
In the Y131 strain, the GAL1/10 promoter is inserted between HTA2 and HTB2 on chromosome II. Schematic of the regions containing HTA2-HTB2 genes on chromosome II. Although this region was deleted and replaced with URA3 in the Y131 strain, a wild-type copy of HTA2-HTB2 controlled by the GAL1/10 promoter is present in this region.
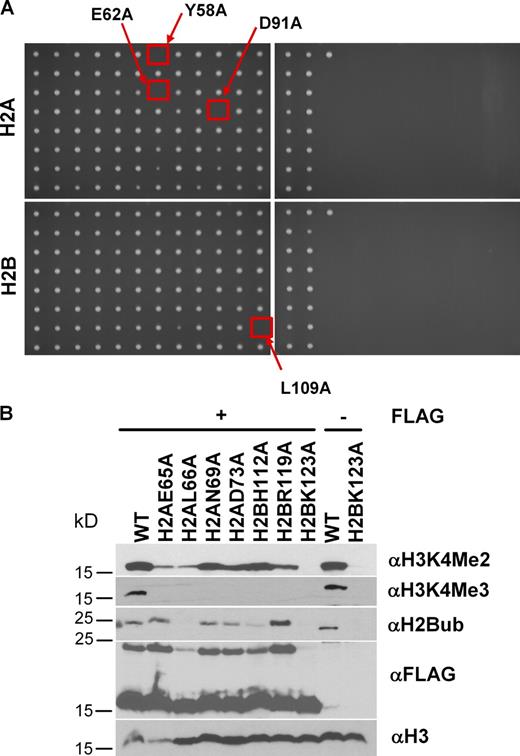
Finally, we have now generated a histone H2A and H2B alanine mutant library in the FY406 background (Fig. 5 A) in addition to the library we reported earlier in Y131 (Nakanishi et al., 2008), as one may wish to use this collection under galactose conditions for different genetic and biochemical screens. From our new library in FY406, we found three H2A (Y58, E62, and D91) residues and one H2B (L109) residue that are essential for viability. These data are in agreement with our previous published results in Y131 (Nakanishi et al., 2008). Furthermore, we tested our new library for H3K4 methylation and H2B monoubiquitination and identified the same residues in Y131, specifically those that regulate normal levels of H3K4 methylation in FY406, as well (Fig. 5 B; Nakanishi et al., 2008). These residues include four residues within H2A (Glu65, Leu66, Asn69, and Asp73) and three residues within H2B (His 112, Arg119, and Lys123). This observation is consistent with our previous findings. Thus, strain differences did not cause any discordant results when grown under glucose media.
Generation of the entire H2A/H2B alanine-scanning collection in an FY406 background. (A) The H2A/H2B alanine-scanning library was generated as described in our previous study (Nakanishi et al., 2008). The complete removal of wild-type (WT) H2B was ensured by multiple rounds of 5-FOA selection and verified by sequencing. Each colony represents a strain expressing histones containing a single alanine substitution mutation of each of the residues of H2A and H2B. Red squares indicate the location of strains that are inviable in SD media containing 5-FOA (lethal mutants). For the key to the corresponding mutant strains within each plate, see Tables S2 and S3. (B) Western blot analysis of mutant strains in the FY406 background identified as defective for proper methylation of H3K4. Cell extracts prepared from each mutant strain were subjected to SDS-PAGE and analyzed by Western analysis with antibodies to dimethyl H3K4, trimethyl H3K4 (αH3K4me2 and αH3K4me3, respectively), monoubiquitinated H2BK123 (αH2Bub), or Flag (αFLAG). An antibody against H3 (αH3) was used as a loading control. White lines indicate that intervening lanes have been spliced out.
Generation of the entire H2A/H2B alanine-scanning collection in an FY406 background. (A) The H2A/H2B alanine-scanning library was generated as described in our previous study (Nakanishi et al., 2008). The complete removal of wild-type (WT) H2B was ensured by multiple rounds of 5-FOA selection and verified by sequencing. Each colony represents a strain expressing histones containing a single alanine substitution mutation of each of the residues of H2A and H2B. Red squares indicate the location of strains that are inviable in SD media containing 5-FOA (lethal mutants). For the key to the corresponding mutant strains within each plate, see Tables S2 and S3. (B) Western blot analysis of mutant strains in the FY406 background identified as defective for proper methylation of H3K4. Cell extracts prepared from each mutant strain were subjected to SDS-PAGE and analyzed by Western analysis with antibodies to dimethyl H3K4, trimethyl H3K4 (αH3K4me2 and αH3K4me3, respectively), monoubiquitinated H2BK123 (αH2Bub), or Flag (αFLAG). An antibody against H3 (αH3) was used as a loading control. White lines indicate that intervening lanes have been spliced out.
In summary, our data clearly demonstrate that monoubiquitination of histone H2B on lysine 123 and the machinery required for its implementation are the sole requirements for the regulation of the trimethylation of H3K4 and H3K79 in yeast S. cerevisiae. Our collective experiments indicate that there are no “unknown” mutations as proposed by Foster and Downs (2009) that function with H2BK123R in the regulation of H3K4 and H3K79 trimethylation. Furthermore, we have discovered that the widely used Y131 strain background expresses a previously undetected copy of the HTA2-HTB2 genes when this strain is grown for many generations in the presence of galactose. Therefore, we generated the entire h2a/h2b mutant collection in a background that can be readily used under galactose conditions and have now made this collection available to the entire scientific community.
Materials and methods
Generation of histone mutants
Strains used in this study are listed in Table S1. We used a plasmid containing HTA1 and HTB1 genes and a plasmid containing all four histones, HTA1, HTB1, HHT2, and HHF2 genes, as shown in Fig. 1 B. Plasmids bearing an alanine or arginine mutation in the Flag-tagged HTB1 gene and Flag-less HTB1 gene were generated by site-directed mutagenesis (QuickChange II kit; Agilent Technologies). Products were transformed into E. cloni 10G ELITE electrocompetent cells (Lucigen). Mutated targets were confirmed by sequencing using the primers (HTBseqF) 5′-GGCAAATACTACCTTGGTTGG-3′ and (HTBseqR) 5′-TTTCGAGAACACAATTTTACAACCGA-3′. Each plasmid was transformed manually into yeast shuffle strains Y131, FY406, DY20D, and JHY205 using a standard yeast transformation protocol, and strains were grown on a synthetic dropout (SD) medium lacking histidine, SD-His (for Y131 and FY406 strains), or a medium lacking leucine, SD-Leu (for DY20D and JHY205 mutants). After 2 d of incubation, transformants were replica plated onto plates containing either SD-His plus 5-FOA or SD-Leu plus 5-FOA to select single-colony cells that had lost the plasmid containing the wild-type histones. Each colony was inoculated into YPD medium (1% yeast extract, 2% peptone, and 2% dextrose) plus 5-FOA.
Western blot analyses
Cells are grown in YPD or YPGal (1% yeast extract, 2% peptone, and 2% galactose) to mid-log phase. Whole cell extracts were prepared from the wild-type and histone mutant strains as previously described (Wood et al., 2003b) with some minor modifications. In brief, cell pellets were washed and resuspended in 400 µl NIB (0.25 M sucrose, 60 mM KCl, 14 mM NaCl, 5 mM MgCl2, 1 mM CaCl2, and 0.8% Triton X-100). After the addition of 250 µl of 0.5-mm glass beads to the tubes containing the suspension, the tubes were vortexed for 20 min at 4°C. Cell lysates were recovered by puncturing the bottom of the tube and centrifuging the contents at 3,000 rpm. After removal of the supernatant, the pellet was suspended in 150 µl of sterile water and 75 µl of 4× loading buffer and heated at 95°C for 5 min. Extracts were subjected to 18% SDS-PAGE electrophoresis, transferred to nitrocellulose membrane, and probed with either anti-Flag or H2B ubiquitin–specific antibodies, as well as antibodies specific for H3K4 di- and trimethylation and H3K79 trimethylation, followed by detection of the bound antibody with horseradish peroxidase–conjugated secondary antibodies. An antibody to histone H3 was used as a loading control.
Generation of H2A/H2B histone mutant library
The h2a/h2b histone mutant library was generated in a FY406 background as described previously (Nakanishi et al., 2008). In brief, plasmids bearing alanine mutations in the HTA1 and HTB1 genes were systematically generated by site-directed mutagenesis in 96-well format. After transformation into E. cloni 10G ELITE electrocompetent cells using a 96-well electroporator, plasmids were prepared with a BioMekFX (Beckman Coulter) using the CosMCPrep kit (Agencourt). Each mutation was confirmed by sequencing using the aforementioned primers, and each plasmid was transformed into FY406 using a standard yeast transformation protocol. Transformants were selected on an SD-His medium followed by the second selection on a plate containing SD-His plus 5-FOA. To ensure the complete removal of the plasmid containing the wild-type histones, each single colony was inoculated into YPD medium plus 5-FOA in 96-well plates. Finally, histone mutant strains were confirmed by sequencing, and the glycerol stocks of the library were generated.
Online supplemental material
Table S1 lists the strains used for this study. Tables S2 and S3 show the key for the histone H2A/H2B library in FY406 background that corresponds to the mutant strains within each plate shown in Fig. 5 A.
Acknowledgments
We thank K. Weaver, B. Miller, and K. Delventhal for technical assistance, L. Shilatifard for editorial assistance, and Dr. E. Smith for conversation and suggestions throughout this study.
This work was supported by the National Institutes of Health (grants CA109355 to Z.W. Sun, GM068088 to B.D. Strahl, and GM069905 to A. Shilatifard) and the March of Dimes Basil O'Connor Award to S.L. Jaspersen. S. Nakanishi is a Fellow of the Leukemia and Lymphoma Society.
References
Abbreviation used in this paper: SD, synthetic dropout.