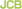Text and Interview by Nicole LeBrasseur
Curing society's ills—social and biological—is Alan Aderem's inspiration. While his Institute for Systems Biology focuses on the biological, Aderem remembers the importance of scientists' role in society.
The matter of immunology often conjures images of the minute: tiny cells on guard against tiny invaders. Many immunologists, not unreasonably, think in microscopic terms—molecular events in disease and resistance. Alan Aderem has likewise studied the infinitesimal. His early work dissected the molecular signaling pathways that are activated within macrophages when these white blood cells sense bacterial intruders (1, 2).
Later, Aderem's view expanded somewhat. He studied the events that lead macrophages to engulf pathogens (3). He also became interested in a large family of immune cell receptors, known as the Toll family, that allows the host to distinguish between various types of pathogens (4, 5).
Now, as cofounder and director of the Institute for Systems Biology (ISB) in Seattle, WA, Aderem deals with the immense. His researchers collect vast amounts of data on a daily basis. He now spends his days organizing and analyzing this enormous load.
MELDING WORLDS
I've heard you had a history of political involvement as a young man in South Africa. I was very active in the anti-Apartheid movement and the African National Congress. I had an active role in the trade unions and community movements and edited a township newspaper. I ended up under house arrest for five years and in prison.
Wow. So how did you go from activist to immunologist? When I left South Africa in 1980, I tried to pull together my two lives: science and the politics of the developing world. The only way I could see doing that was to work on infectious diseases that mattered to people in resource-poor countries, like malaria and TB.
So I went to the Rockefeller University and worked with Zan Cohn, who was the macrophage maven. He convinced me that infections come and go, but the host remains. He said I should work on the host response to infections rather than focus on any one pathogen. That was the best advice I ever had. For example, when HIV came along later, I was well positioned for that.
Zan was a remarkable person and an incredible mentor. At the time, it was a fantastic lab: Ralph Steinman, Gilla Kaplan, Sam Silverstein, Ira Mellman, Carl Nathan, Dan Portnoy, and others.
How did you become involved in systems biology? In '94, I started wondering what was going to happen when the genome was sequenced and how that information could be best used. At that time, I ran into Lee Hood, who was having similar thoughts. We came to the same conclusion—that the way to go would be to combine biology, technology, and computation. I was eager then to move to Seattle, where Lee had also just moved, so we could set up an institute that might do this. Together with Ruedi Aebersold, we cofounded the ISB in 2000.
What was so appealing about Seattle? I was initially moving to the immunology department at the University of Washington, and they had some really good immunologists there that I wanted to interact with, including Sasha Rudensky, Mike Bevan, and Roger Perlmutter.
Seattle was also great because of its computational infrastructures. You had Microsoft and lots of young people around. They'd changed the world once, and they believed they could change it again.
Finally, I'm a serious sailor. I come from Capetown, where you have mountains and sea, and I wanted to get back to that at some level. They say when sailors die and go to heaven, they go to the Pacific Northwest.
So you're already dead? Yes, but rigor mortis hasn't set in.
Your original plan was to establish the institute at the University of Washington? Yes, but for structural reasons we couldn't do it there. We needed a lot of freedom to move, without the walls of departments. To do proper systems biology, you need teamwork. You need to have people with complementary skills working together under one roof: from hardcore mathematicians, physicists, chemists, biologists, physicians, epidemiologists, ecologists, etc. And teamwork's basically the kiss of death for an academic career. They'll always say, “What did she do?” Not, “What did she do with five colleagues?”
We also needed global technologies—robots and very big, very expensive machines, the kind of stuff that you can normally get in a company but not in a university. Within particular departments, you might have one mass spectrometer. But we need more than that.
And we needed to establish relationships with companies. For example, our engineers might build a prototype of an instrument, but to actually build a robust instrument, you need to interface with companies. Those sorts of things are extremely difficult to do in a university.
A SYSTEMS APPROACH
How would you define “systems biology”? My definition is a comprehensive, quantitative analysis of the manner in which all the components of a biological system interact functionally and over time.
The underlying principle is that the question comes from biology. And if it can't be answered by the technology that's available at the time, then the technology is developed. That technology can then revolutionize biology in general. For example, Lee's motivation for developing the DNA sequencer was to answer a question regarding antibody structure. But once it was built, it enabled the genome to be sequenced. Biology drives technology, and then technology revolutionizes biology.
The most important aspect of systems-level analysis is that you have a shot at understanding emergence. Complex systems are not demonstrated by the individual parts, so they can't be predicted even if you completely understand the parts alone. Life is like that. It's not inherent in DNA and RNA and protein and carbohydrate and lipid. You can take all of those things and mix them in a bowl, and you don't have life. Life's a consequence of their actions and interactions.
To identify the emergent properties of a biological system, you've got to capture an enormous amount of information about it, and you need to be able to infer its emergent properties computationally.
Is immunology particularly well-suited for systems analysis? I think so, yes. First, it is a system. And you can isolate the cells in a functional state. This allows you to perturb and interrogate them. Once you have the right tools in hand you can extend your investigation in vivo. You can't easily do that with organs. It is very hard to tease the brain apart and get different types of cells that will communicate with each other in a dish.
What are the institute's long-term goals?
A macrophage extends filopodia to capture bacteria (pink).
Cure is more likely for most diseases if they are identified early. The predictive component comes from two ends. One is your genes: polymorphisms and mutations that predispose you to various diseases. The other is biomarkers that define your particular health status at a given time. Biomarkers must be multi-parameter, for example, the proteome of the blood or the transcriptional profile of a circulating cell.
The preventive side would be intervening by changing your diet or your habits, or taking a drug or vaccine.
The personalized aspect would be tailoring the therapy to the needs of the individual. If you integrate someone's genetic information with their health status, you can individualize the therapy.
And we've thrown in one additional component, which we call the fourth P, for participatory. If you had access to medical information and all your data, you could very much be involved in shaping your treatment and your health.
Do you fear any ethical issues associated with access to that sort of information? Yes, in the sense that insurance companies would love to get a hold of that information, for instance. But those things can be handled. People deal with their banks through the web; safeguards exist when you buy something on the internet; we can protect the information.
There are much more difficult questions. For example, if you have an incurable disease is it better to know? Is it worth leading a constrained life, because you know that at some point in the future, you're going to develop X disease?
LEADING THE CHARGE
What are some of the challenges you've encountered as institute director? One difficulty has been to get scientific integration among the various disciplines. I had initially thought that the physicists would understand biology rather easily, because they'd all splashed around in rock pools and things like that as kids, but that the biologists would have a terrible time with the mathematics of physics.
But it turned out to be exactly the opposite. Because biologists are trained quantitatively, they can sit down and figure out how to program a computer or do the quantitative sciences. But the physicists strive for simplicity, one equation that describes it all. The complication of biology, they hate it. The reason for that, of course, is evolution. Evolution is whatever works; there's often no logic in the design. You can't easily reverse-engineer something that's evolved.
Having been politically involved yourself, do you recommend the same for other scientists?
Absolutely! Serious assaults are being made on science. More than half of the country doesn't believe in evolution. Scientists need to get out there and educate people; for people to practice democracy, they need to understand the issues. You can't vote on stem cells if you don't know what a stem cell is. It's all very well to sit in our ivory towers and say, “It's not our business.” But actually it very much is our business.