Skip Nav Destination
Close Modal




Update search
- Title
- Author
- Author Affiliations
- Full Text
- Abstract
- Keyword
- DOI
- ISBN
- EISBN
- ISSN
- EISSN
- Issue
- Volume
- References
- Title
- Author
- Author Affiliations
- Full Text
- Abstract
- Keyword
- DOI
- ISBN
- EISBN
- ISSN
- EISSN
- Issue
- Volume
- References
- Title
- Author
- Author Affiliations
- Full Text
- Abstract
- Keyword
- DOI
- ISBN
- EISBN
- ISSN
- EISSN
- Issue
- Volume
- References
- Title
- Author
- Author Affiliations
- Full Text
- Abstract
- Keyword
- DOI
- ISBN
- EISBN
- ISSN
- EISSN
- Issue
- Volume
- References
- Title
- Author
- Author Affiliations
- Full Text
- Abstract
- Keyword
- DOI
- ISBN
- EISBN
- ISSN
- EISSN
- Issue
- Volume
- References
- Title
- Author
- Author Affiliations
- Full Text
- Abstract
- Keyword
- DOI
- ISBN
- EISBN
- ISSN
- EISSN
- Issue
- Volume
- References
NARROW
Date
1-20 of 219430
Follow your search
Access your saved searches in your account
Would you like to receive an alert when new items match your search?
1
Sort by
Journal

Journal of Cell Biology (JCB) publishes advances in any area of basic cell biology as well as applied cellular advances in fields such as immunology, neurobiology, metabolism, microbiology, developmental biology, and plant biology. Est. 1955
Journal

Journal of Experimental Medicine (JEM) publishes papers providing novel conceptual insight into immunology, neuroscience, cancer biology, vascular biology, microbial pathogenesis, and stem cell biology. Est. 1896
Journal

Journal of General Physiology (JGP) publishes mechanistic and quantitative cellular and molecular physiology of the highest quality. Est. 1918
Journal
Journal
Journals Gateway
Journal

Journal of Human Immunity (JHI) publishes papers that provide novel insights into the physiology and pathology of human immunity through the study of genetic defects and their phenocopies, including the study of leukocytes and other cells.
Journal Articles
Sachi Inagaki, Housei Wada, Takeshi Itabashi, Yuki Itakura, Reiko Nakagawa, Lin Chen, Kazuyoshi Murata, Atsuko H. Iwane, Shigeo Hayashi
Journal:
Journal of Cell Biology
J Cell Biol (2025) 225 (2): e202503127.
Published: 29 December 2025
Includes: Supplementary data
Journal Articles
Journal:
Journal of Cell Biology
J Cell Biol (2025) 225 (2): e202511183.
Published: 29 December 2025
Journal Articles
Journal:
Journal of Cell Biology
J Cell Biol (2025) 225 (4): e202504003.
Published: 29 December 2025
Images
Published: 29 December 2025
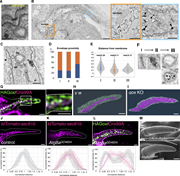
Figure 1. ER-localized Gox/Osi23 controls ER-phagy. (A–C) TEM of the olf hair cell at 44 h APF. (A) An enlarged view of the curved cuticular envelop and the underlying plasma membrane. The lowest point of the curved envelope is often More about this image found in ER-localized Gox/Osi23 controls ER-phagy. (A–C) TEM of the olf hair cell a...
Images
Published: 29 December 2025
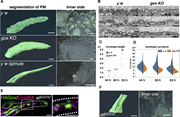
Figure 2. Plasma membrane landscape of the olf hair cell. (A) Plasma membrane structures of the olf and spinule obtained by FIB-SEM. Outer view (left) and inner view (right). (B) Longitudinal TEM views of the plasma membrane and envelope at More about this image found in Plasma membrane landscape of the olf hair cell. (A) Plasma membrane struct...
Published: 29 December 2025
Images
Published: 29 December 2025
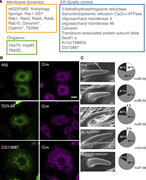
Figure 3. Gox/Osi23-interacting proteins involved in nanopore formation. (A) Mass spectrometric identification of Gox-associated proteins in S2 cells expressing Gox–Flag. An asterisk (*) indicates proteins identified from cells co-expressing More about this image found in Gox/Osi23-interacting proteins involved in nanopore formation. (A) Mass sp...
Images
Published: 29 December 2025
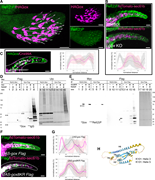
Figure 4. The role of Ref(2)P in Gox localization and ubiquitination. (A) Ref(2)P is specifically enriched in the shaft region of olf hair cells, which is labeled by HA–Gox. Maxillary palp 42 h APF. (B) Subcortical localization of Ref(2) in More about this image found in The role of Ref(2)P in Gox localization and ubiquitination. (A) Ref(2)P is...
Images
Published: 29 December 2025
Figure 5. Dynamin–Gox interaction. (A) Temperature-shift protocol for inactivating dynamin in shits2 mutants. (B) PMI in control and shits2 mutants olf hair cells reconstructed by FIB-SEM. (C) PMI neck width measured from three independently More about this image found in Dynamin–Gox interaction. (A) Temperature-shift protocol for inactivating d...
Images
Published: 29 December 2025
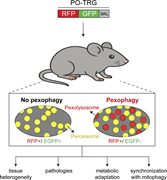
Figure 1. A Tandem Red-Green reporter linked to a targeting signal for the peroxisomal lumen (PO-TRG) has been expressed in mice. Material that has been delivered to lysosomes via selective autophagy shows as red punctae and provides an index of More about this image found in A Tandem Red-Green reporter linked to a targeting signal for the peroxisoma...
Images
in Cell type–specific spatiotemporal control of GFP-tagged protein degradation in mice
> Journal of Cell Biology
Published: 29 December 2025
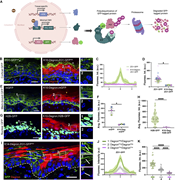
Figure 1. Degron GFP degrades GFP-tagged proteins located in different cell compartments. (A) Diagram of the DegronGFP system. To the left, alleles required for the system: rtTA under a tissue-specific promoter, TRE-VHL-aGFP, and a gene of More about this image found in Degron GFP degrades GFP-tagged proteins located in different c...
Images
in Cell type–specific spatiotemporal control of GFP-tagged protein degradation in mice
> Journal of Cell Biology
Published: 29 December 2025
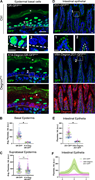
Figure 2. Degron GFP degrades GFP-tagged proteins in different cell populations. (A) Immunofluorescence staining of GFP (green) and degron (red) in E18.5 back skin epidermis of K14-Degron;GR-GFP and control GR-GFP littermate. Insets show only More about this image found in Degron GFP degrades GFP-tagged proteins in different cell popu...
Images
in Cell type–specific spatiotemporal control of GFP-tagged protein degradation in mice
> Journal of Cell Biology
Published: 29 December 2025
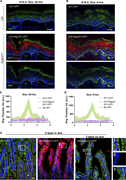
Figure 3. Kinetics of GFP degradation using Degron GFP . (A and B) Immunofluorescence staining of GFP (green) and degron (red) in E18.5 back skin epidermis of K10-Degron;ZO1-GFP and control ZO1-GFP embryos 24 h (A) or 6 h (B) after dox More about this image found in Kinetics of GFP degradation using Degron GFP . (A and B) Immu...
Images
in Cell type–specific spatiotemporal control of GFP-tagged protein degradation in mice
> Journal of Cell Biology
Published: 29 December 2025
Figure 4. Degron GFP efficiently depletes GR-GFP and reproduces knockout phenotypes. (A–C) Immunofluorescence staining of GFP (green) and EdU (magenta) in E18.5 back skin epidermis of GR-GFP (A), K14-Degron;GR-GFP (B), and K10-Degron;GR-GFP More about this image found in Degron GFP efficiently depletes GR-GFP and reproduces knockout...
1
Advertisement