Skip Nav Destination
Close Modal
Update search
- Title
- Author
- Author Affiliations
- Full Text
- Abstract
- Keyword
- DOI
- ISBN
- EISBN
- ISSN
- EISSN
- Issue
- Volume
- References
- Title
- Author
- Author Affiliations
- Full Text
- Abstract
- Keyword
- DOI
- ISBN
- EISBN
- ISSN
- EISSN
- Issue
- Volume
- References
- Title
- Author
- Author Affiliations
- Full Text
- Abstract
- Keyword
- DOI
- ISBN
- EISBN
- ISSN
- EISSN
- Issue
- Volume
- References
- Title
- Author
- Author Affiliations
- Full Text
- Abstract
- Keyword
- DOI
- ISBN
- EISBN
- ISSN
- EISSN
- Issue
- Volume
- References
- Title
- Author
- Author Affiliations
- Full Text
- Abstract
- Keyword
- DOI
- ISBN
- EISBN
- ISSN
- EISSN
- Issue
- Volume
- References
- Title
- Author
- Author Affiliations
- Full Text
- Abstract
- Keyword
- DOI
- ISBN
- EISBN
- ISSN
- EISSN
- Issue
- Volume
- References
NARROW
Date
1-20 of 81643
Follow your search
Access your saved searches in your account
Would you like to receive an alert when new items match your search?
1
Sort by
Journal Articles
Journal:
Journal of Experimental Medicine
J Exp Med (2026) 223 (4): e20252509.
Published: 20 February 2026
Journal Articles
Kathrynne A. Warrick, Anne Katrine Z. Johansen, Mengchi Jiao, Megan E. Linnemann, Irene Saha, Suh-Chin J. Lin, Charles N. Vallez, Thomas Hagan, Jeffery D. Molkentin, Chandrashekhar Pasare
Journal:
Journal of Experimental Medicine
J Exp Med (2026) 223 (4): e20251717.
Published: 20 February 2026
Includes: Supplementary data
Journal Articles
Dominic P. Golec, Pedro H. Gazzinelli-Guimaraes, Daniel Chauss, Kang Yu, Hiroyuki Nagashima, Anthony C. Cruz, Tom Hill, Sundar Ganesan, Jennifer L. Cannons, Jillian K. Perry, Luis Nivelo, Ilin Joshi, Nicolas Pereira, Fabrício Marcus Silva Oliveira, Yufan Zheng, Makheni Jean Pierre, Kirk M. Druey, Justin B. Lack, Eric V. Dang, Thomas B. Nutman, Alejandro V. Villarino, John J. O’Shea, Behdad Afzali, Pamela L. Schwartzberg
Journal:
Journal of Experimental Medicine
J Exp Med (2026) 223 (4): e20252154.
Published: 20 February 2026
Includes: Supplementary data
Images
in Immune checkpoint inhibitor–induced myocarditis is dependent on CD8 T cell–derived TNF and TNFR2 signaling
> Journal of Experimental Medicine
Published: 20 February 2026
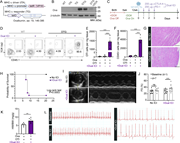
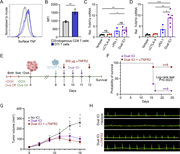
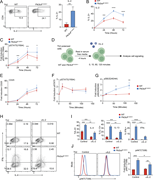
Figure 1. Immune checkpoint inhibition drives myocarditis in a novel mouse model. (A) Schematic diagram illustrating the DTG tetracycline–repressor system, driven by a myosin heavy chain, α isoform (MHC-α) promoter, for inducible overexpression More about this image found in Immune checkpoint inhibition drives myocarditis in a novel mouse model. (A)...
Images
in Immune checkpoint inhibitor–induced myocarditis is dependent on CD8 T cell–derived TNF and TNFR2 signaling
> Journal of Experimental Medicine
Published: 20 February 2026
Figure 2. PD-1 blockade induces de novo priming and differentiation of antigen-specific CD8 T cells. (A and B) Representative flow cytometry contour plots and cell numbers showing the kinetics of OTI T cell expansion in the MedLN at day 4.5, 6, More about this image found in PD-1 blockade induces de novo priming and differentiation ...
Images
in Immune checkpoint inhibitor–induced myocarditis is dependent on CD8 T cell–derived TNF and TNFR2 signaling
> Journal of Experimental Medicine
Published: 20 February 2026
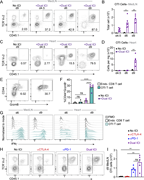
Figure 3. Cardiac-specific CD8 T cells drive innate inflammation and myocardial damage. (A) Quantification of cardiac-infiltrating myeloid cells (CD45.2+CD11b+ and CD45.2+CD11c+) and total T cells (endogenous and OTI T cells, CD45.2+CD90.2+) More about this image found in Cardiac-specific CD8 T cells drive innate inflammation and myocardial damag...
Images
in Immune checkpoint inhibitor–induced myocarditis is dependent on CD8 T cell–derived TNF and TNFR2 signaling
> Journal of Experimental Medicine
Published: 20 February 2026
Figure 4. CD8 T cell – derived TNF is required for ICI-induced myocarditis pathogenesis and mortality. (A) qRT-PCR analysis of Tnf mRNA expression from bulk cardiac tissue (n = 3–6). (B) Infiltration of leukocytes (represented as cell number More about this image found in CD8 T cell – derived TNF is required for ICI-induced myocarditis pathogenes...
Images
in Immune checkpoint inhibitor–induced myocarditis is dependent on CD8 T cell–derived TNF and TNFR2 signaling
> Journal of Experimental Medicine
Published: 20 February 2026
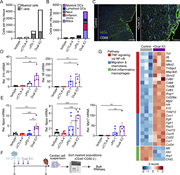
Figure 5. TNFR2 blockade protects against myocarditis and preserves antitumor efficacy of ICI. (A) Flow cytometry analysis of surface TNF staining in expanded OTI or endogenous CD8 T cell populations in the presence of ICI. Cells were purified More about this image found in TNFR2 blockade protects against myocarditis and preserves antitumor efficac...
Images
in A PI3Kδ-Foxo1-FasL signaling amplification loop rewires CD4+ T cell signaling and differentiation
> Journal of Experimental Medicine
Published: 20 February 2026
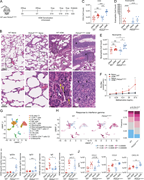
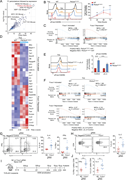
Figure 1. Activated PI3Kδ reshapes the HDM-induced immune response. (A–J) WT and Pik3cdE1020K/+ animals were sensitized intranasally with HDM extracts (200 μg on days 0 and 7; 50 μg on days 14, 16, and 18) and lungs examined on day 20. (A) More about this image found in Activated PI3Kδ reshapes the HDM-induced immune response. (A–J) WT and ...
Images
in A PI3Kδ-Foxo1-FasL signaling amplification loop rewires CD4+ T cell signaling and differentiation
> Journal of Experimental Medicine
Published: 20 February 2026
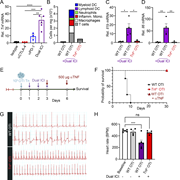
Figure 2. Aberrant Th1 responses at the expense of Th2 immunity in Pik3cd E1020K/+ lungs following HDM sensitization. (A) Scatter plot comparing gene expression in CD4 T cells from WT and Pik3cdE1020K/+ HDM-treated mice (cluster 1 from Fig. 1 More about this image found in Aberrant Th1 responses at the expense of Th2 immunity in Pik3cd...
Images
in A PI3Kδ-Foxo1-FasL signaling amplification loop rewires CD4+ T cell signaling and differentiation
> Journal of Experimental Medicine
Published: 20 February 2026
Figure 3. Hyperactivated PI3Kδ disrupts Th2 lineage restriction. (A–E) Naïve CD4 T cells were activated with αCD3 + αCD28 in the presence of WT T-depleted APCs under Th2-polarizing conditions (IL-4 + αIL-12) for 72 h. (A) Left: Representative More about this image found in Hyperactivated PI3Kδ disrupts Th2 lineage restriction. (A–E) Naïve CD4 T c...
Images
in A PI3Kδ-Foxo1-FasL signaling amplification loop rewires CD4+ T cell signaling and differentiation
> Journal of Experimental Medicine
Published: 20 February 2026
Figure 4. Dysregulated IL-2 signaling rewires Th2 differentiation of Pik3cd E1020K/+ CD4 T cells. (A) Left: Representative flow cytometry plots showing IL-2 and CD4 expression in Th2-polarized cells from the indicated mice. Right: Percentages More about this image found in Dysregulated IL-2 signaling rewires Th2 differentiation of Pik3cd...
Images
in A PI3Kδ-Foxo1-FasL signaling amplification loop rewires CD4+ T cell signaling and differentiation
> Journal of Experimental Medicine
Published: 20 February 2026
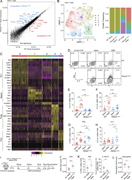
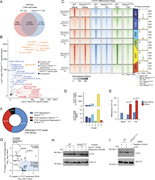
Figure 5. Inactivation of Foxo1 in Pik3cd E1020K/+ CD4 + T cells impairs Th2 lineage restriction. (A) Pathway enrichment of TF perturbations followed by expression gene sets performed using Enrichr ( Xie et al., 2021 ): significantly More about this image found in Inactivation of Foxo1 in Pik3cd E1020K/+...
Images
in A PI3Kδ-Foxo1-FasL signaling amplification loop rewires CD4+ T cell signaling and differentiation
> Journal of Experimental Medicine
Published: 20 February 2026
Figure 6. Pik3cd E1020K reshapes the epigenetic landscape of CD4 + T cells. (A and B) Naïve CD4 T cells from WT and Pik3cdE1020K/+ mice were polarized under Th2 conditions and evaluated by ATACseq (n = 3). A total of 71,040 peaks were More about this image found in Pik3cd E1020K reshapes the e...
Images
in A PI3Kδ-Foxo1-FasL signaling amplification loop rewires CD4+ T cell signaling and differentiation
> Journal of Experimental Medicine
Published: 20 February 2026
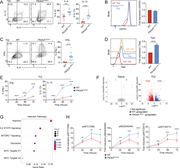
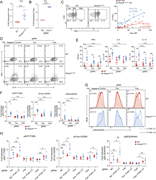
Figure 7. Fas-FasL signaling potentiates T cell activation, exacerbating CD4 + T cell dysregulation in the presence of activated PI3Kδ. (A) Fold surface FasL expression (MFI normalized to WT) on Th2-polarized live CD4+ T cells, measured by More about this image found in Fas-FasL signaling potentiates T cell activation, exacerbating CD4 + ...
Images
in A PI3Kδ-Foxo1-FasL signaling amplification loop rewires CD4+ T cell signaling and differentiation
> Journal of Experimental Medicine
Published: 20 February 2026
Figure 8. Fas-induced T cell activation occurs in the absence of FADD. (A) Naïve CD4 T cells from WT and Pik3cdE1020K/+ mice were nucleofected with Cas9-gRNA complexes containing NC or Fadd-targeting gRNAs and polarized under Th2 conditions in More about this image found in Fas-induced T cell activation occurs in the absence of FADD. (A) Naïve CD4...
Images
in A PI3Kδ-Foxo1-FasL signaling amplification loop rewires CD4+ T cell signaling and differentiation
> Journal of Experimental Medicine
Published: 20 February 2026
Figure 9. Fas interacts with the TCR complex and costimulates TCR signaling. (A) Confocal imaging of CD3ε and Fas in CD4 T cells. Naïve and stimulated (αCD3+αCD28 or αCD3+αCD28+FasL-LZ) CD4 T cells from the indicated mice were stained with More about this image found in Fas interacts with the TCR complex and costimulates TCR signaling. (A) Con...
Images
in A PI3Kδ-Foxo1-FasL signaling amplification loop rewires CD4+ T cell signaling and differentiation
> Journal of Experimental Medicine
Published: 20 February 2026
Figure 10. PI3Kδ regulates CD4 + T cell differentiation through integration of TCR, IL-2 receptor, and Fas signaling, driving Foxo1 inactivation and transcriptional reprogramming. More about this image found in PI3Kδ regulates CD4 + T cell differentiation through integrati...
Journal Articles
Journal:
Journal of Experimental Medicine
J Exp Med (2026) 223 (3): e20241266.
Published: 19 February 2026
Images
in Next-generation CRISPR screens enable causal systems immunology
> Journal of Experimental Medicine
Published: 19 February 2026
Figure 1. Biological applications of CRISPR perturbations and expanded modalities of bulk CRISPR screens in T cells. (A) Biological applications of CRISPR perturbations include the discovery of master regulator genes, identification of More about this image found in Biological applications of CRISPR perturbations and expanded modalities of ...
1