Platinum-based chemotherapy drugs can lead to the development of anorexia, a detrimental effect on the overall health of cancer patients. However, managing chemotherapy-induced anorexia and subsequent weight loss remains challenging due to limited effective therapeutic strategies. Growth differentiation factor 15 (GDF15) has recently gained significant attention in the context of chemotherapy-induced anorexia. Here, we report that hepatic GDF15 plays a crucial role in regulating body weight in response to chemo drugs cisplatin and doxorubicin. Cisplatin and doxorubicin treatments induce hepatic Gdf15 expression and elevate circulating GDF15 levels, leading to hunger suppression and subsequent weight loss. Mechanistically, selective activation by chemotherapy of hepatic IRE1α-XBP1 pathway of the unfolded protein response (UPR) upregulates Gdf15 expression. Genetic and pharmacological inactivation of IRE1α is sufficient to ameliorate chemotherapy-induced anorexia and body weight loss. These results identify hepatic IRE1α as a molecular driver of GDF15-mediated anorexia and suggest that blocking IRE1α RNase activity offers a therapeutic strategy to alleviate the adverse anorexia effects in chemotherapy.
Introduction
Platinum-based drugs such as cisplatin (Cis), carboplatin, and oxaliplatin are widely used in cancer therapy, but their optimal use is limited by toxicities, including nausea, vomiting, anorexia, muscle wasting, and weight loss, that compromise quality of life and treatment adherence (Kelland, 2007; Ruggiero et al., 2013). Although antiemetic regimens including 5-HT3R and NK-1 receptor antagonists, dexamethasone, and olanzapine have reduced treatment-related nausea and vomiting, breakthrough and delayed emesis remain common in many patients (Hesketh et al., 2017; Aapro et al., 2018). Mechanistic understanding of platinum-induced weight loss remains incomplete, though preclinical studies implicate NF-κB and ActRII signaling as well as ghrelin receptor activation as potential modulators (Barreto et al., 2017; Chen et al., 2017; Peterson and Guttridge, 2008). Therefore, identification of new causal mechanism(s) of chemotherapy-induced body weight loss represents a critical step to inform novel strategies for optimizing platinum treatment and improving cancer care.
Growth differentiation factor 15 (GDF15) is a stress-responsive cytokine and a distant member of the transforming growth factor β superfamily (Bootcov et al., 1997; Breit et al., 2021; Tan et al., 2000; Tsai et al., 2018). GDF15 has been associated with numerous human diseases including cancer, cardiovascular disease, and inflammatory diseases (Breit et al., 2021; Tsai et al., 2018; Wang et al., 2021). Recently, a growing body of literature has documented the role of GDF15 in anorexia, weight loss, and cachexia in rodents and primates (Borner et al., 2020a, 2020b; Breen et al., 2020; Hsu et al., 2017; Wang et al., 2023). Evidence from rodents and shrews supports that GDF15 triggers anorexia through nausea and emesis, resulting in weight loss (Borner et al., 2020a). Notably, GDF15 levels are increased following administration of chemotherapy drugs (e.g., Cis, bleomycin, and doxorubicin [DOX]) (Breen et al., 2020). Increased GDF15 levels activate the glial cell–derived neurotrophic factor receptor α-like (GFRAL) whose expression is limited to the area postrema (AP) and the nucleus tractus solitarius (NTS) located in the hindbrain (Emmerson et al., 2017; Hsu et al., 2017; Mullican et al., 2017; Suriben et al., 2020). GDF15 administration increased c-Fos activation in AP/NTS, an area associated with cancer anorexia-cachexia syndrome in tumor-bearing animals in which plasma GDF15 levels are increased (Borner et al., 2020a; Hsu et al., 2017). Furthermore, antibody-mediated GDF15 neutralization alleviates chemotherapy-induced anorexia and weight loss in preclinical models (Breen et al., 2020), which implies a therapeutic potential in counteracting chemotherapy-associated side effects. Although the elevated circulating GDF15 and activation of GFRAL-expressing neurons localized in AP/NTS of the brainstem are accountable for chemotherapy-induced anorexia, it remains elusive that peripheral tissues or organs serve as the primary source of GDF15 and contribute to these adverse effects (Borner et al., 2020a; Breen et al., 2020; Hsu et al., 2017).
Originally characterized as a macrophage-secreted factor (Bootcov et al., 1997), the physiological expression of GDF15 is detected in liver, lung, kidney, and distal colon (Breen et al., 2020; Coll et al., 2020). Nevertheless, it appears that GDF15 is a general stress-induced cytokine in a wide variety of cell types (Appierto et al., 2009; Chung et al., 2017; Hsiao et al., 2000; Park et al., 2012; Patel et al., 2019; Yang et al., 2010), and regulation of Gdf15 expression represents a critical mechanism by which serum GDF15 levels are controlled (Breen et al., 2020; Luan et al., 2019; Xie et al., 2022). However, due to its ubiquitous induction in response to various stimulations, regulatory mechanisms controlling Gdf15 expression are highly diversified and context dependent (Breit et al., 2021; Tsai et al., 2018). For instance, the Gdf15 transcription is actively regulated in injured and inflamed tissues and cells, and transcription factors including p53 (Tan et al., 2000) and Egr-1 (Baek et al., 2004) appear to be responsive for Gdf15 induction in these settings (Wang et al., 2021). In a different context, transcriptional factors related to integrated stress response emerge as critical regulators of Gdf15 expression, which is induced in stress conditions or by drug treatments (Patel et al., 2019; Wang et al., 2021). Hence, as regulatory mechanisms of GDF15 production are heterogeneous, the full array of context-dependent transcriptional regulators has yet to be defined in the control of Gdf15 expression.
Herein, we report that chemotherapy drugs acutely stimulate liver GDF15 production via selective activation of the hepatic ER stress sensor IRE1α, an ER-resident transmembrane protein kinase/RNase that conveys a key signaling branch of the unfolded protein response (UPR) (Walter and Ron, 2011), thereby controlling the circulating GDF15 level. Genetic ablation of hepatic IRE1α leads to reduced circulating GDF15, alleviating anorexia and body weight loss following chemotherapy drug treatments in tumor-bearing mice. Mechanistically, chemotherapy drugs activate hepatic IRE1α RNase activity to produce the spliced active form of transcription factor XBP1, consequently promoting Gdf15 expression in hepatocytes. In addition, pharmacological IRE1α RNase inhibitor effectively suppresses liver Gdf15 expression and circulating GDF15 levels, which is associated with improvements in chemotherapy-induced anorexia and body weight loss. Our findings thus unveil a stress-responsive mechanism underlying a liver–brain crosstalk that is pharmacologically targetable for alleviating the anorexic side effects associated with chemotherapy-induced body weight loss.
Results and discussion
Activation of hepatic UPR accompanies body weight loss upon chemotherapy
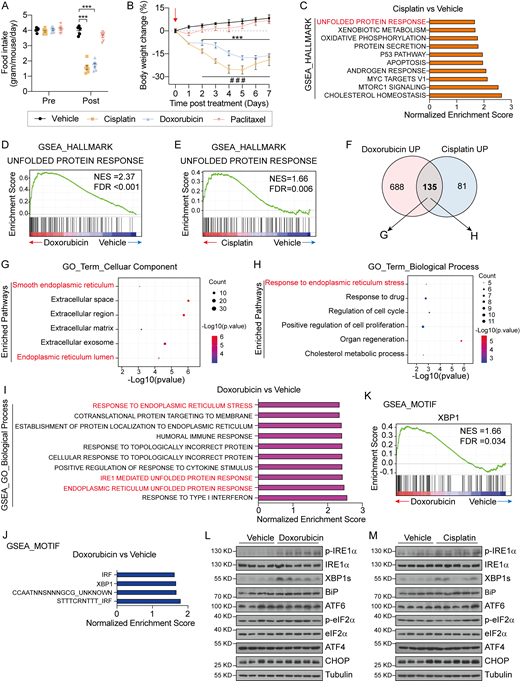
Documented studies have demonstrated that platinum-based chemotherapy (i.e., Cis) could cause body weight loss and anorexia in animals and subjects with cancers (Borner et al., 2020a; Breen et al., 2020; Hsu et al., 2017). To explore whether these side effects of chemotherapy are universal for other chemotherapy drugs, we administered 8-wk-old wild-type healthy mice with one dose of three main types of chemotherapeutic agents, Cis (platinum-based agent), DOX (anthracycline agent), and paclitaxel (PTX; taxane agent). In contrast to PTX, Cis and DOX treatment significantly repressed food intake at 1 day after injection and subsequently resulted in substantial body weight loss of the animals (Fig. S1, A and B), which is in agreement with previous studies (Breen et al., 2020). As liver is the central hub for drug metabolism in systemic administration of chemotherapeutic agents (Ramadori and Cameron, 2010; Tao et al., 2020), we reasoned that DOX and Cis treatment may exert profound impact on the liver. To this end, we employed bulk RNA sequencing (RNA-seq) to analyze the liver samples collected from mice at 1 day after DOX and Cis chemotherapy (Fig. 1 A). The sequencing data revealed extensive alterations in hepatic gene expression profiles from treated animals (Fig. 1, B and C). Whereas treatment by DOX and Cis elicited obviously different patterns of enriched pathways, the UPR appeared as a commonly upregulated pathway among the top 10 enriched pathways by Gene Set Enrichment Analysis (GSEA) of the two treatment groups (Fig. 1 D and Fig. S1, C–E). Notably, the analysis of significantly upregulated genes of the two treatment groups (823 in DOX group and 216 genes in Cis group) revealed that the overlapping 135 transcripts are highly associated with the ER and involved in the response to ER stress according to Gene Ontology (GO) analysis (Fig. S1, F and H). These data indicate a link between hepatic UPR pathways and chemotherapy-associated anorexia and body weight loss.
Chemotherapy with Cis and DOX causes anorexia and selective activation of the IRE1α-XBP1 pathway in the liver. Related to Fig. 1. (A and B) Food intake (A) and body weight changes (B) of C57BL/6 mice treated with one dose of Vehicle (i.p.), DOX (10 mg per kg body weight, i.p.), Cis (10 mg per kg body weight, i.p.) or PTX (10 mg per kg body weight, i.p.) at 1 day. Vehicle, n = 5; DOX, n = 7; Cis, n = 7; PTX, n = 7. (C) Top 10 enriched HALLMARK pathways by the GSEA that are differentially regulated between liver samples from Vehicle- and Cis-treated mice. HALLMARK pathways are defined using the FPKM values of all the detected genes in livers from Cis-treated mice, and ranked according to the NES. (D) GSEA showing DOX-induced enrichment of the “unfolded protein response” gene signature in the liver (NES = 2.37 and FDR q-value < 0.001). The solid bars represent individual genes in the “unfolded protein response” gene set. (E) GSEA showing Cis-induced enrichment of the “unfolded protein response” gene signature in the liver (NES = 1.66 and FDR q-value = 0.006). The solid bars represent individual genes in “unfolded protein response” gene set. (F) Venn diagram showing the upregulated genes (compared to Vehicle treatment) that overlapped in liver samples from DOX- and Cis-treated mice. (G and H) GO analysis of the 135 overlapping genes in F. (G) Bubble plot of the top six enriched pathways by GO Term Biological Process analysis. (H) Bubble plot of the top six enriched pathways by GO Term Cellular Component analysis. The diameter of the circle is proportional to the number of DEGs enriched in the indicated pathways. The color of the circle represents the value of −Log10(P value). (I) The top 10 enriched pathways from GO Biological Process analysis by GSEA based on differentially regulated genes in the liver between Vehicle- and DOX-treated mice. The enriched pathways are defined using the FPKM values of all the detected genes in livers from DOX-treated mice, and ranked according to the NES. (J) The top four enriched motifs according to the MOTIF analysis by GSEA of the liver from Vehicle- and DOX-treated mice. The enriched motifs are defined using the FPKM values of all the detected genes in liver samples from DOX-treated mice and ranked according to the NES. (K) GSEA showing the enrichment of “XBP1” target gene signatures in liver samples from DOX-treated mice (NES = 1.66 and FDR q-value = 0.034). The solid bars represent individual genes in the “XBP1” gene set. (L) Western blot analysis of the indicated proteins in liver lysates from 8-wk-old mice treated with a single dose of DOX (10 mg per kg body weight, i.p.). Liver samples were collected 1 day after injection. Each sample represents an individual animal. (M) Western blot analysis of the indicated proteins in liver lysates from 8-wk-old mice treated with a single dose of Cis (10 mg per kg body weight, i.p.). Liver samples were collected 1 day after injection. Each sample represents an individual animal. Data are representative of two independent experiments and presented as mean ± SEM. ***P < 0.001 (DOX versus Vehicle), ###P < 0.001 (Cis versus Vehicle) by unpaired two-tailed Student’s t test. Source data are available for this figure: SourceData FS1.
Chemotherapy with Cis and DOX causes anorexia and selective activation of the IRE1α-XBP1 pathway in the liver. Related to Fig. 1. (A and B) Food intake (A) and body weight changes (B) of C57BL/6 mice treated with one dose of Vehicle (i.p.), DOX (10 mg per kg body weight, i.p.), Cis (10 mg per kg body weight, i.p.) or PTX (10 mg per kg body weight, i.p.) at 1 day. Vehicle, n = 5; DOX, n = 7; Cis, n = 7; PTX, n = 7. (C) Top 10 enriched HALLMARK pathways by the GSEA that are differentially regulated between liver samples from Vehicle- and Cis-treated mice. HALLMARK pathways are defined using the FPKM values of all the detected genes in livers from Cis-treated mice, and ranked according to the NES. (D) GSEA showing DOX-induced enrichment of the “unfolded protein response” gene signature in the liver (NES = 2.37 and FDR q-value < 0.001). The solid bars represent individual genes in the “unfolded protein response” gene set. (E) GSEA showing Cis-induced enrichment of the “unfolded protein response” gene signature in the liver (NES = 1.66 and FDR q-value = 0.006). The solid bars represent individual genes in “unfolded protein response” gene set. (F) Venn diagram showing the upregulated genes (compared to Vehicle treatment) that overlapped in liver samples from DOX- and Cis-treated mice. (G and H) GO analysis of the 135 overlapping genes in F. (G) Bubble plot of the top six enriched pathways by GO Term Biological Process analysis. (H) Bubble plot of the top six enriched pathways by GO Term Cellular Component analysis. The diameter of the circle is proportional to the number of DEGs enriched in the indicated pathways. The color of the circle represents the value of −Log10(P value). (I) The top 10 enriched pathways from GO Biological Process analysis by GSEA based on differentially regulated genes in the liver between Vehicle- and DOX-treated mice. The enriched pathways are defined using the FPKM values of all the detected genes in livers from DOX-treated mice, and ranked according to the NES. (J) The top four enriched motifs according to the MOTIF analysis by GSEA of the liver from Vehicle- and DOX-treated mice. The enriched motifs are defined using the FPKM values of all the detected genes in liver samples from DOX-treated mice and ranked according to the NES. (K) GSEA showing the enrichment of “XBP1” target gene signatures in liver samples from DOX-treated mice (NES = 1.66 and FDR q-value = 0.034). The solid bars represent individual genes in the “XBP1” gene set. (L) Western blot analysis of the indicated proteins in liver lysates from 8-wk-old mice treated with a single dose of DOX (10 mg per kg body weight, i.p.). Liver samples were collected 1 day after injection. Each sample represents an individual animal. (M) Western blot analysis of the indicated proteins in liver lysates from 8-wk-old mice treated with a single dose of Cis (10 mg per kg body weight, i.p.). Liver samples were collected 1 day after injection. Each sample represents an individual animal. Data are representative of two independent experiments and presented as mean ± SEM. ***P < 0.001 (DOX versus Vehicle), ###P < 0.001 (Cis versus Vehicle) by unpaired two-tailed Student’s t test. Source data are available for this figure: SourceData FS1.
Hepatic IRE1α-XBP1 branch of the UPR is selectively activated by chemotherapy. (A) Schematic illustration of the experimental design. 8-wk-old C57BL/6 male mice were treated with Vehicle (n = 5), a single dose of DOX (10 mg per kg body weight, i.p., n = 5), or Cis (10 mg per kg body weight, i.p., n = 5) for 1 day before liver samples were collected for bulk RNA-seq. (B) Principal component analysis of transcriptomic data obtained by bulk RNA-seq of liver tissue from mice treated with Vehicle, DOX, or Cis for 1 day. (C) Heatmap visualization of the expression patterns of differentially expressed genes (DEGs) following chemotherapeutic drug administration. The cutoff values are |Log2(Foldchange)| > 0.5, and P value < 0.05. For each group, the expression value is shown for five mice. (D) The top 10 enriched HALLMARK pathways by GSEA, which are differentially regulated in livers from Vehicle- and DOX-treated mice. HALLMARK pathways are defined using FPKM (fragments per kilobase million) values of all the detected genes in livers from DOX-treated mice and ranked according to NES. (E) GSEA plot indicating the enrichment (NES = 2.43 and FDR q-value < 0.001) of “IRE1-mediated unfolded protein response” gene signature in DOX-treated mice. The solid bars represent individual genes in this gene set. (F and G) 8-wk-old C57BL/6 male mice were treated with a single dose of (F) DOX (10 mg per kg body weight, i.p.) or (G) Cis (10 mg per kg body weight, i.p.). Liver samples were collected at indicated time (0, 1, 3, and 7 days) after injection. Western blot analysis showing the levels of the indicated proteins in liver lysates from the indicated mice. Each sample represents an individual animal. Tubulin was used as a loading control. (H–J)Xbp1 mRNA splicing (H), mRNA levels of the indicated XBP1s target genes (I), and ER stress markers (J) in livers from mice treated with DOX (10 mg per kg body weight, i.p.), Cis (10 mg per kg body weight, i.p.), or PTX (10 mg per kg body weight, i.p.) for 0 (n = 4), 1 (n = 6), 3 (n = 6), and 7 days (n = 6). Data are representative of two independent experiments and presented as mean ± SEM. *P < 0.05, **P < 0.01, or ***P < 0.001 by one-way ANOVA (H–J). Source data are available for this figure: SourceData F1.
Hepatic IRE1α-XBP1 branch of the UPR is selectively activated by chemotherapy. (A) Schematic illustration of the experimental design. 8-wk-old C57BL/6 male mice were treated with Vehicle (n = 5), a single dose of DOX (10 mg per kg body weight, i.p., n = 5), or Cis (10 mg per kg body weight, i.p., n = 5) for 1 day before liver samples were collected for bulk RNA-seq. (B) Principal component analysis of transcriptomic data obtained by bulk RNA-seq of liver tissue from mice treated with Vehicle, DOX, or Cis for 1 day. (C) Heatmap visualization of the expression patterns of differentially expressed genes (DEGs) following chemotherapeutic drug administration. The cutoff values are |Log2(Foldchange)| > 0.5, and P value < 0.05. For each group, the expression value is shown for five mice. (D) The top 10 enriched HALLMARK pathways by GSEA, which are differentially regulated in livers from Vehicle- and DOX-treated mice. HALLMARK pathways are defined using FPKM (fragments per kilobase million) values of all the detected genes in livers from DOX-treated mice and ranked according to NES. (E) GSEA plot indicating the enrichment (NES = 2.43 and FDR q-value < 0.001) of “IRE1-mediated unfolded protein response” gene signature in DOX-treated mice. The solid bars represent individual genes in this gene set. (F and G) 8-wk-old C57BL/6 male mice were treated with a single dose of (F) DOX (10 mg per kg body weight, i.p.) or (G) Cis (10 mg per kg body weight, i.p.). Liver samples were collected at indicated time (0, 1, 3, and 7 days) after injection. Western blot analysis showing the levels of the indicated proteins in liver lysates from the indicated mice. Each sample represents an individual animal. Tubulin was used as a loading control. (H–J)Xbp1 mRNA splicing (H), mRNA levels of the indicated XBP1s target genes (I), and ER stress markers (J) in livers from mice treated with DOX (10 mg per kg body weight, i.p.), Cis (10 mg per kg body weight, i.p.), or PTX (10 mg per kg body weight, i.p.) for 0 (n = 4), 1 (n = 6), 3 (n = 6), and 7 days (n = 6). Data are representative of two independent experiments and presented as mean ± SEM. *P < 0.05, **P < 0.01, or ***P < 0.001 by one-way ANOVA (H–J). Source data are available for this figure: SourceData F1.
The hepatic IRE1α-XBP1 signaling pathway is selectively activated in mice upon chemotherapy
Given the diverse functions of the three UPR signaling branches in various physiological or pathological contexts (Hetz et al., 2020; Walter and Ron, 2011; Huang et al., 2019), we sought to determine which UPR branch(s) is/are activated in the liver upon chemotherapy. According to the GO Biological Process enrichment analysis, the “IRE1-mediated unfolded protein response” pathway is ranked (normalized enrichment score [NES] = 2.43 and false discovery rate [FDR] q-value < 0.001) in the top 10 pathways enriched in DOX-treated livers (Fig. 1 E and Fig. S1 I). Furthermore, MOTIF enrichment analysis revealed the activation of the IRE1α-XBP1 branch, as evidenced by higher transcriptional activity of XBP1, a versatile transcription factor generated by IRE1α-mediated unconventional mRNA splicing (Walter and Ron, 2011), in DOX-treated livers (Fig. S1, J and K). To further confirm this, we evaluated the activation of the three UPR pathways in the liver at different time intervals following DOX and Cis treatment. In line with the bioinformatics analysis, DOX- and Cis-treated mice displayed notably increased hepatic levels of phosphorylated IRE1α and the spliced form of XBP1 (XBP1s) (Fig. 1, F and G; and Fig. S1, L and M) relative to their vehicle control counterparts. Additionally, enhanced Xbp1 mRNA splicing as well as upregulated expression of XBP1s-target genes (Erdj4, Sec61a1) were also observed in the livers after 1-day chemotherapy (Fig. 1, H and I). However, neither the PERK-eIF2α nor the ATF6 branches were apparently affected by chemotherapy, as evidenced by little changes in eIF2α phosphorylation or the protein/mRNA abundance of BiP, CHOP, ATF4, and ATF6 (Fig. 1, F, G, and J; and Fig. S1, L and M). These results demonstrate that the hepatic IRE1α-XBP1 signaling pathway is selectively activated in response to the treatment by chemotherapeutic drugs.
Loss of hepatic IRE1α alleviates chemotherapy-induced anorexia and body weight loss
Notably, DOX and Cis could acutely exert its activating impact upon the IRE1α-XBP1 pathway that peaked around 1 day after injection (Fig. 1, F and G), coinciding with the instant body mass decline upon chemotherapy (Fig. S1, A and B). These observations prompted us to hypothesize that hepatic IRE1α-XBP1 signaling may be directly involved in the development of chemotherapy-induced anorexia and body weight loss. To test this idea, we intercrossed Ern1flox/flox mice, which harbor two loxP sites flanking the exon 2 of the Ern1 gene (encoding IRE1α protein), with Albumin-Cre transgenic mice (Alb-Cre) to generate hepatocyte-specific Ern1 knockout mouse model (Ern1flox/flox;Alb-Cre; denoted LKO) (Shao et al., 2014). Hepatic IRE1α inactivation abolished the effects of DOX on inducing XBP1s protein accumulation, without affecting the other two UPR pathways in LKO livers (Fig. 2 A). Compared to their control counterparts, LKO mice showed remarkable improvements in food intake reduction and body weight loss induced by single or multiple doses of DOX treatments (Fig. 2, B and C). Moreover, hepatic IRE1α ablation led to similar protection in the setting of Cis treatment (Fig. 2, D–F). These results demonstrate the critical role of hepatic IRE1α in regulating anorexia and body weight loss in animals during chemotherapy.
Hepatic IRE1α ablation alleviates chemotherapy-induced anorexia and body weight loss. (A) Western blot analysis of the indicated proteins in liver lysates from flox/flox (Ern1loxP/loxP, littermate controls) and LKO (Ern1loxP/loxP; Albumin-Cre) mice treated with a single dose of Vehicle or DOX (5 mg per kg body weight, i.p.). Liver samples were collected 1 day after injection. Each sample represents an individual animal. Tubulin was used as a loading control. (B and C) Daily food intake (B) and body weight changes (C) of the indicated groups following Vehicle (Veh.) or DOX injection. flox/flox+Veh., n = 6; LKO+Veh., n = 6; flox/flox+DOX, n = 6; LKO+DOX, n = 7. Yellow arrows indicate the time points of DOX injection (5 mg per kg body weight, i.p.). (D) Western blot analysis of the indicated proteins in liver lysates from flox/flox and LKO mice treated with a single dose of Vehicle or Cis (5 mg per kg body weight, i.p.). Liver samples were collected 1 day after injection. Each sample represents an individual animal. Tubulin was used as a loading control. (E and F) Daily food intake (E) and body weight changes (F) of the indicated groups following Vehicle (Veh.) or Cis injection. flox/flox+Veh., n = 6; LKO+Veh., n = 6; flox/flox+Cis, n = 8; LKO+Cis, n = 7. Brown arrows indicate the time points of Cis injection (5 mg per kg body weight, i.p.). (G) Schematic illustration of the experimental design. 8-wk-old male flox/flox and LKO mice were subcutaneously inoculated with Hepa1-6 cells to induce tumor formation. DOX (5 mg per kg body weight) or Vehicle was i.p. administered, and visible tumors were removed by surgery at the indicated time. (H) Body weight changes in tumor-bearing and -resected flox/flox and LKO mice treated with Vehicle or DOX. flox/flox+Veh., n = 10; LKO+Veh., n = 8; flox/flox+DOX, n = 12; LKO+DOX, n = 10. Yellow arrow indicates the time point of tumor removal. Black arrows indicate the time points of DOX or Vehicle treatment. Data are representative of two independent experiments and presented as mean ± SEM. **P < 0.01, or ***P < 0.001 by unpaired two-tailed Student’s t test (B, C, E, F, and H). Source data are available for this figure: SourceData F2.
Hepatic IRE1α ablation alleviates chemotherapy-induced anorexia and body weight loss. (A) Western blot analysis of the indicated proteins in liver lysates from flox/flox (Ern1loxP/loxP, littermate controls) and LKO (Ern1loxP/loxP; Albumin-Cre) mice treated with a single dose of Vehicle or DOX (5 mg per kg body weight, i.p.). Liver samples were collected 1 day after injection. Each sample represents an individual animal. Tubulin was used as a loading control. (B and C) Daily food intake (B) and body weight changes (C) of the indicated groups following Vehicle (Veh.) or DOX injection. flox/flox+Veh., n = 6; LKO+Veh., n = 6; flox/flox+DOX, n = 6; LKO+DOX, n = 7. Yellow arrows indicate the time points of DOX injection (5 mg per kg body weight, i.p.). (D) Western blot analysis of the indicated proteins in liver lysates from flox/flox and LKO mice treated with a single dose of Vehicle or Cis (5 mg per kg body weight, i.p.). Liver samples were collected 1 day after injection. Each sample represents an individual animal. Tubulin was used as a loading control. (E and F) Daily food intake (E) and body weight changes (F) of the indicated groups following Vehicle (Veh.) or Cis injection. flox/flox+Veh., n = 6; LKO+Veh., n = 6; flox/flox+Cis, n = 8; LKO+Cis, n = 7. Brown arrows indicate the time points of Cis injection (5 mg per kg body weight, i.p.). (G) Schematic illustration of the experimental design. 8-wk-old male flox/flox and LKO mice were subcutaneously inoculated with Hepa1-6 cells to induce tumor formation. DOX (5 mg per kg body weight) or Vehicle was i.p. administered, and visible tumors were removed by surgery at the indicated time. (H) Body weight changes in tumor-bearing and -resected flox/flox and LKO mice treated with Vehicle or DOX. flox/flox+Veh., n = 10; LKO+Veh., n = 8; flox/flox+DOX, n = 12; LKO+DOX, n = 10. Yellow arrow indicates the time point of tumor removal. Black arrows indicate the time points of DOX or Vehicle treatment. Data are representative of two independent experiments and presented as mean ± SEM. **P < 0.01, or ***P < 0.001 by unpaired two-tailed Student’s t test (B, C, E, F, and H). Source data are available for this figure: SourceData F2.
To further determine whether hepatic IRE1α exerts the same action in the development of chemotherapy-associated side effects under pathological conditions, we generated a tumor-bearing mouse model by subcutaneously implanting Hepa1-6 liver cancer cells in LKO and flox/flox control mice. Tumors were surgically removed from animals prior to DOX treatment, a process to mimic clinical chemotherapy received by cancer patients (Fig. 2 G). After the removal of visible tumors, the first dose of DOX injection caused the loss of body weight in both flox/flox and LKO mice (Fig. 2 H). However, the body weight of LKO mice recovered steadily and became indistinguishable from those of vehicle-treated control animals after two DOX injections, whereas the flox/flox group exhibited sustained body weight loss in response to the same treatment of DOX (Fig. 2 H). This affirms that hepatic IRE1α exerts crucial actions in eliciting chemotherapy-induced body weight changes.
Liver-derived GDF15 mediates chemotherapy-induced anorexia and body weight loss
To explore how hepatic IRE1α signaling pathway regulates anorexia and body weight loss during chemotherapy, we first analyzed genes whose expression was altered by chemotherapy drugs (Fig. 3 A). Among the 135 genes whose expression was commonly altered by Cis and DOX treatment, Gdf15 exhibited a most prominent upregulation, and it top-ranked among the 26 genes encoding putative secretory factors, according to the RNA-seq datasets (Fig. 3 B). In line with its anorexic effects, treatment by both DOX and Cis, but not by PTX, led to robust elevation of hepatic Gdf15 mRNA levels and circulating GDF15 protein abundance in mice (Fig. 3, C and D). In human patients, remarkably, circulating levels of GDF15 were also dramatically elevated in colon cancer and breast cancer individuals after receiving the first dose of chemotherapy (Fig. 3 E). To confirm the necessity and sufficiency of GDF15 for chemo-induced anorexia, we employed whole-body Gdf15 gene knockout mouse model (Gdf15KO). Compared to their control counterparts, Gdf15KO mice showed marked improvements in food intake reduction and body weight loss upon DOX or Cis treatment (Fig. S2, A–H). Emerging evidence has shown that GDF15 is a stress-induced hepatokine and acts as a pivotal mediator in regulating chemotherapy-induced anorexia and body weight loss via its obligate receptor GFRAL expressed in neurons localized in AP/NTS of the brainstem (Borner et al., 2020a; Breen et al., 2020; Hsu et al., 2017; Luan et al., 2019). Therefore, we next asked whether liver-derived GDF15 serves as a major contributor to its elevated circulating levels upon chemotherapy. We first assessed Gdf15 mRNA levels in various tissues/organs and found that the liver displayed the most robust expression and upregulation of Gdf15 mRNA in response to both DOX and Cis treatments (Fig. 3 F). Then, we employed adenovirus-associated virus to deliver Gdf15-targeting shRNA (AAV-shGdf15) to knock down the expression of Gdf15 specifically in the liver (Cunningham and Alexander, 2019) (Fig. S2, I and J). AAV-shGdf15 significantly reduced hepatic Gdf15 mRNA induction and subsequently the elevation of circulating GDF15 levels in mice following Cis administration (Fig. 3, G and H). Consistently, mice injected with AAV-shGdf15 exhibited significant improvements in anorexia and body weight loss relative to animals injected with control viruses after the same dose of Cis treatment (Fig. 3, I and J). These results indicate that in the context of chemotherapy, liver-derived GDF15 is a key factor that drives the development of anorexia and body weight loss.
Liver-derived GDF15 mediates chemotherapy-induced anorexia and body weight loss. (A) Volcano plot showing DEGs (FDR < 0.05 and |Log2(Foldchange)| > 2) between the liver samples from Vehicle- and Cis-treated mice. Significantly changed genes are colored in red (upregulated, Up) and blue (downregulated, Down), while non-significantly changed genes (Not sig) are in gray. (B) Heatmap of 26 genes encoding secreted factors among 135 overlapping upregulated genes (compared to Vehicle treatment) in liver samples from both DOX- and Cis-treated mice. (C and D) mRNA levels of Gdf15 in livers (C) and circulating GDF15 protein levels (D) from mice treated with the indicated chemotherapy drugs (DOX, 10 mg per kg body weight, i.p.; Cis, 10 mg per kg body weight, i.p.; or PTX, 10 mg per kg body weight, i.p.) for 0 (n = 4), 1 (n = 6), 3 (n = 6), and 7 days (n = 6). (E) Circulating GDF15 protein levels in healthy volunteers (non-tumor) and in individuals with colon or breast cancer before and after receiving chemotherapy for the first time (healthy volunteer, n = 15; colon cancer patients, n = 12; breast cancer patients, n = 10). (F) mRNA levels of Gdf15 in various tissues isolated from mice treated with Vehicle (n = 6), DOX (n = 6), or Cis (n = 6). Tissues were collected 1 day after injection. gWAT, gonadal white adipose tissue; iWAT, inguinal white adipose tissue; TA, tibialis anterior; Gas, gastrocnemius. (G–J) C57BL/6 male mice were injected with AAV-shNC (n = 12) or AAV-shGdf15-1 (n = 12) for 4 wk before i.p. injection of Cis (10 mg per kg body weight) or Vehicle. (G and H) Liver Gdf15 mRNA abundance (G) and circulating GDF15 protein levels (H) were quantified in liver samples collected 1 day after injection and serum samples harvested at the indicated time points following Cis treatment, respectively. AAV-shNC+Veh., n = 6; AAV-shGdf15+Veh., n = 6; AAV-shNC+Cis, n = 6; AAV-shGdf15+Cis, n = 6. (I and J) Daily food intake (I) and body weight changes (J) of AAV-shNC- and AAV-shGdf15-injected mice following Vehicle or Cis treatment. The arrow indicates the time point of Cis injection. Data are representative of two independent (except A, B, and E) experiments and presented as mean ± SEM. *P < 0.05, **P < 0.01, or ***P < 0.001 by one-way ANOVA (C, D, and F), two-way ANOVA (G), unpaired two-tailed Student’s t test (H–J), or paired Student’s t test (E).
Liver-derived GDF15 mediates chemotherapy-induced anorexia and body weight loss. (A) Volcano plot showing DEGs (FDR < 0.05 and |Log2(Foldchange)| > 2) between the liver samples from Vehicle- and Cis-treated mice. Significantly changed genes are colored in red (upregulated, Up) and blue (downregulated, Down), while non-significantly changed genes (Not sig) are in gray. (B) Heatmap of 26 genes encoding secreted factors among 135 overlapping upregulated genes (compared to Vehicle treatment) in liver samples from both DOX- and Cis-treated mice. (C and D) mRNA levels of Gdf15 in livers (C) and circulating GDF15 protein levels (D) from mice treated with the indicated chemotherapy drugs (DOX, 10 mg per kg body weight, i.p.; Cis, 10 mg per kg body weight, i.p.; or PTX, 10 mg per kg body weight, i.p.) for 0 (n = 4), 1 (n = 6), 3 (n = 6), and 7 days (n = 6). (E) Circulating GDF15 protein levels in healthy volunteers (non-tumor) and in individuals with colon or breast cancer before and after receiving chemotherapy for the first time (healthy volunteer, n = 15; colon cancer patients, n = 12; breast cancer patients, n = 10). (F) mRNA levels of Gdf15 in various tissues isolated from mice treated with Vehicle (n = 6), DOX (n = 6), or Cis (n = 6). Tissues were collected 1 day after injection. gWAT, gonadal white adipose tissue; iWAT, inguinal white adipose tissue; TA, tibialis anterior; Gas, gastrocnemius. (G–J) C57BL/6 male mice were injected with AAV-shNC (n = 12) or AAV-shGdf15-1 (n = 12) for 4 wk before i.p. injection of Cis (10 mg per kg body weight) or Vehicle. (G and H) Liver Gdf15 mRNA abundance (G) and circulating GDF15 protein levels (H) were quantified in liver samples collected 1 day after injection and serum samples harvested at the indicated time points following Cis treatment, respectively. AAV-shNC+Veh., n = 6; AAV-shGdf15+Veh., n = 6; AAV-shNC+Cis, n = 6; AAV-shGdf15+Cis, n = 6. (I and J) Daily food intake (I) and body weight changes (J) of AAV-shNC- and AAV-shGdf15-injected mice following Vehicle or Cis treatment. The arrow indicates the time point of Cis injection. Data are representative of two independent (except A, B, and E) experiments and presented as mean ± SEM. *P < 0.05, **P < 0.01, or ***P < 0.001 by one-way ANOVA (C, D, and F), two-way ANOVA (G), unpaired two-tailed Student’s t test (H–J), or paired Student’s t test (E).
Global Gdf15 deficiency alleviates the chemotherapy-induced anorexia and body weight loss. Related to Figs. 3 and 4. (A–D)Gdf15KO (n = 5) and their littermates (Control, n = 5) were administered with one dose of Vehicle or DOX (5 mg per kg body weight, i.p.). (A) Gdf15 mRNA levels in livers; (B) circulating GDF15 protein levels; (C) daily food intake; (D) body weight change. (E–H)Gdf15KO (n = 5) and their littermates (Control, n = 5) were administered with one dose of Vehicle or Cis (5 mg per kg body weight, i.p.). (E) Gdf15 mRNA levels in livers; (F) circulating GDF15 protein levels; (G) daily food intake; (H) body weight change. (I) mRNA levels of Gdf15 in Hepa1-6 cells infected with AAV-shNC, AAV-shGdf15-1, or AAV-shGdf15-2 for 48 h (n = 4 per group). (J) mRNA levels of Gdf15 in the indicated mouse tissues 4 wk after the injection of AAV-shNC (n = 3) or AAV-shGdf15 (n = 3). (K)Xbp1 mRNA splicing in various tissues isolated from mice treated with Vehicle (n = 6), DOX (n = 6), or Cis (n = 6). Tissues were collected 1 day after injection. gWAT, gonadal white adipose tissue; iWAT, inguinal white adipose tissue; TA, tibialis anterior; Gas, gastrocnemius. (L)XBP1 mRNA splicing in Huh7 cells transfected with control siRNA (si-NC) or siRNA against ERN1 (si-ERN1) prior to treatment with Vehicle or DOX (0.01 μg/ml) for 48 h. n = 3 per group. (M)XBP1 mRNA splicing in Huh7 cells transfected with control siRNA (si-NC) or siRNA against ERN1 (si-ERN1) prior to treatment with Vehicle or Cis (10 μM) for 48 h. n = 3 per group. Data are representative of two independent experiments (A–H and G) or three independent experiments (I–K) and presented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 by unpaired two-tailed Student’s t test (A–H) one-way ANOVA (I–K) and two-way ANOVA (L and M).
Global Gdf15 deficiency alleviates the chemotherapy-induced anorexia and body weight loss. Related to Figs. 3 and 4. (A–D)Gdf15KO (n = 5) and their littermates (Control, n = 5) were administered with one dose of Vehicle or DOX (5 mg per kg body weight, i.p.). (A) Gdf15 mRNA levels in livers; (B) circulating GDF15 protein levels; (C) daily food intake; (D) body weight change. (E–H)Gdf15KO (n = 5) and their littermates (Control, n = 5) were administered with one dose of Vehicle or Cis (5 mg per kg body weight, i.p.). (E) Gdf15 mRNA levels in livers; (F) circulating GDF15 protein levels; (G) daily food intake; (H) body weight change. (I) mRNA levels of Gdf15 in Hepa1-6 cells infected with AAV-shNC, AAV-shGdf15-1, or AAV-shGdf15-2 for 48 h (n = 4 per group). (J) mRNA levels of Gdf15 in the indicated mouse tissues 4 wk after the injection of AAV-shNC (n = 3) or AAV-shGdf15 (n = 3). (K)Xbp1 mRNA splicing in various tissues isolated from mice treated with Vehicle (n = 6), DOX (n = 6), or Cis (n = 6). Tissues were collected 1 day after injection. gWAT, gonadal white adipose tissue; iWAT, inguinal white adipose tissue; TA, tibialis anterior; Gas, gastrocnemius. (L)XBP1 mRNA splicing in Huh7 cells transfected with control siRNA (si-NC) or siRNA against ERN1 (si-ERN1) prior to treatment with Vehicle or DOX (0.01 μg/ml) for 48 h. n = 3 per group. (M)XBP1 mRNA splicing in Huh7 cells transfected with control siRNA (si-NC) or siRNA against ERN1 (si-ERN1) prior to treatment with Vehicle or Cis (10 μM) for 48 h. n = 3 per group. Data are representative of two independent experiments (A–H and G) or three independent experiments (I–K) and presented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 by unpaired two-tailed Student’s t test (A–H) one-way ANOVA (I–K) and two-way ANOVA (L and M).
Hepatic IRE1α-XBP1s pathway controls chemotherapy-induced GDF15 expression
Because the induction of hepatic Gdf15 expression (Fig. 3, C and F) concurs with the activation of the IRE1α-XBP1s signaling pathway during chemotherapy (Fig. 1, F–I; and Fig. S2 K), we asked whether IRE1α-XBP1s signaling is involved in mediating chemotherapy-induced Gdf15 expression in the liver. To test this, we overexpressed IRE1α or XBP1s in human hepatoma Huh7 cells, which led to a robust increase in GDF15 expression (Fig. 4, A and B). Conversely, silencing the expression of ERN1 (the gene encoding human IRE1α) by si-ERN1 efficiently blunted the inducing effects of chemotherapy drugs on GDF15 expression (Fig. 4 C and Fig. S2, L and M). We then tested whether Gdf15 is a direct transcriptional target of the IRE1α-XBP1 pathway. To this end, we engineered a reporter construct in which the expression of luciferase is driven by the promoter portion at 1,028 bp upstream of murine Gdf15 gene transcription start site. Notably, a putative ER stress–response element (ERSE) was identified within this region (Fig. 4 D) (Yamamoto et al., 2004). Luciferase reporter assays showed that XBP1s directly stimulated the Gdf15 promoter activity, which was abolished by mutation of the putative ERSE (Fig. 4 D). Furthermore, we performed chromatin immunoprecipitation (ChIP) assays, using nuclear extracts from primary hepatocytes overexpressing GFP or XBP1s. Indeed, XBP1s could bind to the putative ERSE-containing region of the Gdf15 promoter (Fig. 4 E). Moreover, imaging analyses of mice with engineered Gdf15 promoter-Luc reporter in their livers showed that hepatocyte-specific deletion of IRE1α attenuated chemotherapy-induced activation of the Gdf15 promoter in vivo (Fig. 4 F). Thus, these data demonstrate that XBP1s directly binds to and activates Gdf15 promoter, suggesting that chemotherapy-activated liver IRE1α-XBP1 pathway acts through a transcriptional regulatory mechanism for the associated increase of GDF15 levels and the consequential anorexia.
The IRE1α-XBP1 pathway mediates chemotherapy-induced hepatic GDF15 upregulation. (A and B) Western blot analysis of the indicated proteins (left) and RT-qPCR analysis of GDF15 mRNA abundance (right) in cell lysates of Huh7 cells transfected with control vector, or vector expressing IRE1α (OE-IRE1α) (A) or XBP1s (OE-XBP1s) (B). Tubulin was used as a loading control. (C)GDF15 mRNA levels in Huh7 cells transfected with control siRNA (si-NC) or ERN1 knockdown siRNA (si-ERN1) prior to treatment with DOX (0.01 μg/ml) or Cis (10 μM) for 48 h. n = 3 per group. (D) Schematic of the luciferase constructs of Gdf15 promoter with the ERSE-like sequence indicated. Luciferase reporter assays were performed by co-transfection of HEK293T cells with the control vector or pCMV-XBP1s plasmid together with Luc constructs under the control of the mouse Gdf15 promoter (WT) or those with mutant ERSE sequences (Mut1, Mut2, Mut3). n = 3 per group. (E) ChIP-qPCR analysis of XBP1s binding site on Gdf15 promoter was performed by overexpression of XBP1s in primary hepatocytes (n = 4). ChIP-qPCR of the segments containing XBP1s-binding site (−713 to −515) or no XBP1s-binding site (−1,459 to −1,327) within the Gdf15 promoter. (F)flox/flox (littermates) and LKO mice were engineered to express the empty control construct or the Gdf15-WT Luc reporter in livers. Luciferase activities were monitored in vivo by the imaging system under the indicated treatments. Mice were i.p. administered with DOX (5 mg per kg body weight) or Cis (5 mg per kg body weight) 24 h prior to luciferase assays. (G and H)Xbp1 mRNA splicing (G) and mRNA levels of Gdf15 (H) in livers from flox/flox and LKO mice following DOX treatment (5 mg per kg body weight, i.p.) for 1 day. flox/flox+Veh., n = 4; LKO+Veh., n = 5; flox/flox+DOX, n = 5; LKO+DOX, n = 7. (I) Circulating GDF15 protein levels in flox/flox (n = 7) and LKO (n = 7) mice at the indicated time points following DOX treatment (5 mg per kg body weight, i.p.). (J) Serum GDF15 protein levels in non-tumor-bearing (Non T.B.) flox/flox (n = 12) and LKO (n = 12) mice, and in tumor-bearing (T.B.) flox/flox and LKO mice treated with Vehicle (Veh.; n = 12 for flox/flox, n = 10 for LKO) or DOX (5 mg per kg body weight, i.p., n = 12 for each group) for 1 day. (K) Representative images of immunofluorescence staining against c-Fos and GFRAL at the AP and NTS of the murine brainstem (left). GFRAL+ c-Fos+ cells (indicated by arrowheads, left) in AP or NTS per high power field (HPF) were quantified (right). The frozen brainstem slides were from flox/flox and LKO mice at 1 day after DOX treatment (5 mg per kg body weight, i.p.). Scale bar, 100 μm. (L and M)Xbp1 mRNA splicing (L) and mRNA levels of Gdf15 (M) in liver samples from flox/flox and LKO mice following Cis treatment (5 mg per kg body weight, i.p.) for 1 day. flox/flox+Veh., n = 4; LKO+Veh., n = 5; flox/flox+Cis, n = 5; LKO+Cis, n = 5. (N) Circulating GDF15 protein levels in flox/flox (n = 10) and LKO (n = 10) mice at the indicated time points following Cis treatment (5 mg per kg body weight, i.p.). (O) Representative images of immunofluorescence staining against c-Fos and GFRAL at the area AP and NTS of the murine brainstem (left). GFRAL+ c-Fos+ cells (indicated by arrowheads, left) per HPF were quantified (right). The frozen brainstem slides were from flox/flox and LKO mice at 1 day after Cis treatment (5 mg per kg body weight, i.p.). Scale bar, 100 μm. Data are representative of three independent experiments (A–G) or two independent experiments (H–P) and presented as mean ± SEM. *P < 0.05, **P < 0.01, or ***P < 0.001 by unpaired two-tailed Student’s t test (A, B, I, K, and O) or two-way ANOVA (C–H, J, L–N). Source data are available for this figure: SourceData F4.
The IRE1α-XBP1 pathway mediates chemotherapy-induced hepatic GDF15 upregulation. (A and B) Western blot analysis of the indicated proteins (left) and RT-qPCR analysis of GDF15 mRNA abundance (right) in cell lysates of Huh7 cells transfected with control vector, or vector expressing IRE1α (OE-IRE1α) (A) or XBP1s (OE-XBP1s) (B). Tubulin was used as a loading control. (C)GDF15 mRNA levels in Huh7 cells transfected with control siRNA (si-NC) or ERN1 knockdown siRNA (si-ERN1) prior to treatment with DOX (0.01 μg/ml) or Cis (10 μM) for 48 h. n = 3 per group. (D) Schematic of the luciferase constructs of Gdf15 promoter with the ERSE-like sequence indicated. Luciferase reporter assays were performed by co-transfection of HEK293T cells with the control vector or pCMV-XBP1s plasmid together with Luc constructs under the control of the mouse Gdf15 promoter (WT) or those with mutant ERSE sequences (Mut1, Mut2, Mut3). n = 3 per group. (E) ChIP-qPCR analysis of XBP1s binding site on Gdf15 promoter was performed by overexpression of XBP1s in primary hepatocytes (n = 4). ChIP-qPCR of the segments containing XBP1s-binding site (−713 to −515) or no XBP1s-binding site (−1,459 to −1,327) within the Gdf15 promoter. (F)flox/flox (littermates) and LKO mice were engineered to express the empty control construct or the Gdf15-WT Luc reporter in livers. Luciferase activities were monitored in vivo by the imaging system under the indicated treatments. Mice were i.p. administered with DOX (5 mg per kg body weight) or Cis (5 mg per kg body weight) 24 h prior to luciferase assays. (G and H)Xbp1 mRNA splicing (G) and mRNA levels of Gdf15 (H) in livers from flox/flox and LKO mice following DOX treatment (5 mg per kg body weight, i.p.) for 1 day. flox/flox+Veh., n = 4; LKO+Veh., n = 5; flox/flox+DOX, n = 5; LKO+DOX, n = 7. (I) Circulating GDF15 protein levels in flox/flox (n = 7) and LKO (n = 7) mice at the indicated time points following DOX treatment (5 mg per kg body weight, i.p.). (J) Serum GDF15 protein levels in non-tumor-bearing (Non T.B.) flox/flox (n = 12) and LKO (n = 12) mice, and in tumor-bearing (T.B.) flox/flox and LKO mice treated with Vehicle (Veh.; n = 12 for flox/flox, n = 10 for LKO) or DOX (5 mg per kg body weight, i.p., n = 12 for each group) for 1 day. (K) Representative images of immunofluorescence staining against c-Fos and GFRAL at the AP and NTS of the murine brainstem (left). GFRAL+ c-Fos+ cells (indicated by arrowheads, left) in AP or NTS per high power field (HPF) were quantified (right). The frozen brainstem slides were from flox/flox and LKO mice at 1 day after DOX treatment (5 mg per kg body weight, i.p.). Scale bar, 100 μm. (L and M)Xbp1 mRNA splicing (L) and mRNA levels of Gdf15 (M) in liver samples from flox/flox and LKO mice following Cis treatment (5 mg per kg body weight, i.p.) for 1 day. flox/flox+Veh., n = 4; LKO+Veh., n = 5; flox/flox+Cis, n = 5; LKO+Cis, n = 5. (N) Circulating GDF15 protein levels in flox/flox (n = 10) and LKO (n = 10) mice at the indicated time points following Cis treatment (5 mg per kg body weight, i.p.). (O) Representative images of immunofluorescence staining against c-Fos and GFRAL at the area AP and NTS of the murine brainstem (left). GFRAL+ c-Fos+ cells (indicated by arrowheads, left) per HPF were quantified (right). The frozen brainstem slides were from flox/flox and LKO mice at 1 day after Cis treatment (5 mg per kg body weight, i.p.). Scale bar, 100 μm. Data are representative of three independent experiments (A–G) or two independent experiments (H–P) and presented as mean ± SEM. *P < 0.05, **P < 0.01, or ***P < 0.001 by unpaired two-tailed Student’s t test (A, B, I, K, and O) or two-way ANOVA (C–H, J, L–N). Source data are available for this figure: SourceData F4.
Then, we evaluated the effect of hepatic IRE1α deficiency upon Gdf15 expression and circulating GDF15 levels in LKO mice in vivo. In parallel with the suppression of Xbp1 mRNA splicing (Fig. 4 G), genetic ablation of liver IRE1α markedly attenuated DOX-induced upregulation of hepatic Gdf15 mRNA expression (Fig. 4 H) and serum GDF15 protein level (Fig. 4 I) relative to that in control animals. Moreover, serum GDF15 levels in tumor-bearing LKO mice were significantly reduced to ∼55% of those in control animals after DOX administration (Fig. 4 J). Reported studies have demonstrated that anorexia in chemotherapy can be ascribed to elevated circulating levels of GDF15, which activates c-Fos in GFRAL-expressing neurons localized to AP/NTS of the brainstem (Borner et al., 2020a; Breen et al., 2020; Hsu et al., 2017). In agreement, relative to control animals, the decline of circulating GDF15 protein levels in DOX-treated LKO mice was paralleled with lower abundance of GFRAL+c-Fos+ neurons in AP (Fig. 4 K). Similar effects were also observed in Cis-treated LKO mice (Fig. 4, L–O). Notably, deficiency of hepatic IRE1α did not exert detectable effects on liver homeostatsis or chemo-induced hepatic damages (Fig. S3, A–F). Together, these results support a key role of hepatic IRE1α-XBP1s-GDF15 axis in promoting the development of anorexia during chemotherapy.
Neither hepatic IRE1α ablation nor pharmacological blocking of IRE1α activity has impacts upon the homeostasis or chemo-induced damage of the liver. (A–C) flox/flox (littermates) and LKO mice were i.p. injected with three doses of Vehicle (Veh.) and DOX (5 mg per kg body weight) at the time points shown in Fig. 2 B. (A) Serum aspartate aminotransferase (AST) and alanine aminotransferase (ALT) of the mice. flox/flox+Veh., n = 6; LKO+Veh., n = 6; flox/flox+DOX, n = 6; LKO+DOX, n = 6. (B) Representative H&E (scale bar, 100 μm) and TUNEL (scale bar, 50 μm) staining of liver sections. (C) mRNA levels of Il6, Il1β, and Tnfα genes in the liver samples. flox/flox+Veh., n = 3; LKO+Veh., n = 3; flox/flox+DOX, n = 3; LKO+DOX, n = 3. (D–F)flox/flox (littermates) and LKO mice were i.p. injected with three doses of Vehicle and Cis (5 mg per kg body weight) at the time points shown in Fig. 2 E. (D) Serum AST and ALT of the mice. flox/flox+Veh., n = 6; LKO+Veh., n = 6; flox/flox+ Cis, n = 6; LKO+ Cis, n = 6. (E) Representative H&E (scale bar, 100 μm) and TUNEL (scale bar, 50 μm) staining of liver sections. (F) mRNA levels of Il6, Il1b, and Tnfa in the liver samples. flox/flox+Veh., n = 3; LKO+Veh., n = 3; flox/flox+DOX, n = 3; LKO+DOX, n = 3. (G–I) C57BL/6 male mice were i.p. injected with Vehicle, 4μ8C (3.3 mg per kg body weight, i.p.), DOX (5 mg per kg body weight, i.p.), or the combination of DOX and 4μ8C for three doses at the time points as indicated in Fig. 5 G. (G) Serum AST and ALT of the mice. Veh., n = 6; 4μ8C, n = 6; DOX, n = 6; DOX+4μ8C, n = 6. (H) Representative H&E (scale bar, 100 μm) and TUNEL (scale bar, 50 μm) staining of the liver sections. (I) mRNA levels of Il6, Il1b, and Tnfa in the liver samples. Veh., n = 3; 4μ8C, n = 3; DOX, n = 3; DOX+4μ8C, n = 3. (J–L) C57BL/6 male mice were i.p. injected with Vehicle, 4μ8C (3.3 mg per kg body weight), Cis (5 mg per kg body weight), or the combination of Cis and 4μ8C for three doses at the time points as indicated in Fig. 5 M. (J) Serum AST and ALT of the mice. Veh., n = 6; 4μ8C, n = 6; Cis, n = 6; Cis +4μ8C, n = 6. (K) Representative H&E (scale bar, 100 μm) and TUNEL (scale bar, 50 μm) staining of the liver sections. (L) mRNA levels of Il6, Il1b, and Tnfa in the liver samples. Veh., n = 3; 4μ8C, n = 3; Cis, n = 3; Cis+4μ8C, n = 3. Data are representative of two independent experiments and presented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 by two-way ANOVA (A, C, D, F, G, I, J, and L).
Neither hepatic IRE1α ablation nor pharmacological blocking of IRE1α activity has impacts upon the homeostasis or chemo-induced damage of the liver. (A–C) flox/flox (littermates) and LKO mice were i.p. injected with three doses of Vehicle (Veh.) and DOX (5 mg per kg body weight) at the time points shown in Fig. 2 B. (A) Serum aspartate aminotransferase (AST) and alanine aminotransferase (ALT) of the mice. flox/flox+Veh., n = 6; LKO+Veh., n = 6; flox/flox+DOX, n = 6; LKO+DOX, n = 6. (B) Representative H&E (scale bar, 100 μm) and TUNEL (scale bar, 50 μm) staining of liver sections. (C) mRNA levels of Il6, Il1β, and Tnfα genes in the liver samples. flox/flox+Veh., n = 3; LKO+Veh., n = 3; flox/flox+DOX, n = 3; LKO+DOX, n = 3. (D–F)flox/flox (littermates) and LKO mice were i.p. injected with three doses of Vehicle and Cis (5 mg per kg body weight) at the time points shown in Fig. 2 E. (D) Serum AST and ALT of the mice. flox/flox+Veh., n = 6; LKO+Veh., n = 6; flox/flox+ Cis, n = 6; LKO+ Cis, n = 6. (E) Representative H&E (scale bar, 100 μm) and TUNEL (scale bar, 50 μm) staining of liver sections. (F) mRNA levels of Il6, Il1b, and Tnfa in the liver samples. flox/flox+Veh., n = 3; LKO+Veh., n = 3; flox/flox+DOX, n = 3; LKO+DOX, n = 3. (G–I) C57BL/6 male mice were i.p. injected with Vehicle, 4μ8C (3.3 mg per kg body weight, i.p.), DOX (5 mg per kg body weight, i.p.), or the combination of DOX and 4μ8C for three doses at the time points as indicated in Fig. 5 G. (G) Serum AST and ALT of the mice. Veh., n = 6; 4μ8C, n = 6; DOX, n = 6; DOX+4μ8C, n = 6. (H) Representative H&E (scale bar, 100 μm) and TUNEL (scale bar, 50 μm) staining of the liver sections. (I) mRNA levels of Il6, Il1b, and Tnfa in the liver samples. Veh., n = 3; 4μ8C, n = 3; DOX, n = 3; DOX+4μ8C, n = 3. (J–L) C57BL/6 male mice were i.p. injected with Vehicle, 4μ8C (3.3 mg per kg body weight), Cis (5 mg per kg body weight), or the combination of Cis and 4μ8C for three doses at the time points as indicated in Fig. 5 M. (J) Serum AST and ALT of the mice. Veh., n = 6; 4μ8C, n = 6; Cis, n = 6; Cis +4μ8C, n = 6. (K) Representative H&E (scale bar, 100 μm) and TUNEL (scale bar, 50 μm) staining of the liver sections. (L) mRNA levels of Il6, Il1b, and Tnfa in the liver samples. Veh., n = 3; 4μ8C, n = 3; Cis, n = 3; Cis+4μ8C, n = 3. Data are representative of two independent experiments and presented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 by two-way ANOVA (A, C, D, F, G, I, J, and L).
Pharmacologic blocking of IRE1α RNase alleviates chemotherapy-induced anorexia and body weight loss
Considering the in vitro and in vivo evidence showing the regulation by hepatic IRE1α-XBP1 pathway of chemotherapy-induced GDF15 production in body weight loss, we further examined whether pharmacologic inhibition of IRE1α could effectively alleviate these side effects. Indeed, treatment with 4μ8C, a chemical inhibitor of IRE1α RNase activity, substantially suppressed the induction of GDF15 in both DOX- and Cis-treated Huh7 cells (Fig. 5, A and B). Then, we determined whether chemotherapy-induced anorexia and body weight loss could be corrected through such pharmacologic blocking of IRE1α RNase activity in vivo. To this end, we administered mice with 4μ8C in combination with DOX and observed apparent suppression of Xbp1 mRNA splicing in the liver, indicating that hepatic IRE1α RNAse activity could be efficiently inhibited by 4μ8C treatment (Fig. 5 C). Consistently, 4μ8C treatment robustly decreased hepatic Gdf15 mRNA expression and lowered serum GDF15 levels in DOX-treated mice (Fig. 5, D and E). Notably, a decreased number of GFRAL+c-Fos+ neurons in AP was also observed in mice treated with DOX plus 4μ8C (Fig. 5 F). As expected, 4μ8C treatment markedly dampened the development of anorexia and body weight loss upon chemotherapy in animals (Fig. 5, G and H). In addition, administration of 4μ8C in Cis-treated mice resulted in similar phenotypic effects, i.e., reduced GDF15 induction and alleviated body weight loss (Fig. 5, I–N). Notably, 4μ8C treatment did not exert noticeable effects upon liver homeostatsis or chemo-induced hepatic damages (Fig. S3, G–L). Together, these data demonstrate that pharmacologic inhibition of IRE1α RNase activity can effectively alleviate chemotherapy-associated anorexia and body weight loss.
Pharmacologic blocking of IRE1α activity alleviates chemotherapy-induced anorexia and body weight loss via suppression of hepatic GDF15. (A and B) XBP1 mRNA splicing and GDF15 mRNA levels in Huh7 cells treated with (A) Vehicle, 4μ8C (1 μM), DOX (0.01 μg/ml), or DOX+4μ8C or (B) Vehicle, 4μ8C (1 μM), Cis (10 μM), or Cis+4u8C for 24 h. n = 3 per group. (C–H) Male C57BL/6 mice were treated with Vehicle, 4μ8C (3.3 mg per kg body weight, i.p.), DOX (5 mg per kg body weight, i.p.), or the combination of DOX and 4μ8C at the indicated time points (black arrows) for three doses of treatment. (C and D)Xbp1 mRNA splicing (C) and Gdf15 mRNA levels (D) in liver samples from mice after the treatments. Veh., n = 5; 4μ8C, n = 5; DOX, n = 5; DOX+4μ8C, n = 5. (E) Circulating GDF15 protein levels in mice after the treatments. Veh., n = 6; 4μ8C, n = 6; DOX, n = 6; DOX+4μ8C, n = 6. (F) Representative images of immunofluorescence staining against c-Fos and GFRAL at the area AP and NTS of the murine brainstem (left). GFRAL+ c-Fos+ cells (indicated by arrowheads, left) per HPF were quantified (right). The frozen brainstem slides were from mice at 1 day after the final treatments. Scale bar, 100 μm. (G and H) Daily food intake (G, Veh., n = 4; 4μ8C, n = 4; DOX, n = 4; DOX+4μ8C, n = 4) and body weight changes (H, Veh., n = 6; 4μ8C, n = 6; DOX, n = 6; DOX+4μ8C, n = 6) of mice following the indicated treatments. (I–N) C57BL/6 male mice were treated with three doses of Vehicle, 4μ8C (3.3 mg per kg body weight, i.p.), Cis (5 mg per kg body weight, i.p.), or the combination of Cis and 4μ8C at the indicated time points (black arrows). (I and J)Xbp1 mRNA splicing (I, Veh., n = 7; 4μ8C, n = 8; Cis, n = 8; Cis+4μ8C, n = 8) and Gdf15 mRNA levels (J, Veh., n = 6; 4μ8C, n = 6; Cis, n = 6; Cis+4μ8C, n = 6) in liver samples from mice after the treatments. (K) Circulating GDF15 protein levels in mice after the treatments. Veh., n = 6; 4μ8C, n = 6; Cis, n = 6; Cis+4μ8C, n = 6. (L) Representative images of immunofluorescence staining against c-Fos and GFRAL at the area AP and NTS of the murine brainstem (left). GFRAL+ c-Fos+ cells (indicated by arrowheads, left) per HPF were quantified (right). The frozen brainstem slides were from mice at 1 day after the final treatments. Scale bar, 100 μm. (M and N) Daily food intake (M, Veh., n = 3; 4μ8C, n = 3; Cis, n = 4; Cis+4μ8C, n = 4) and body weight changes (N, Veh., n = 6; 4μ8C, n = 6; Cis, n = 6; Cis+4μ8C, n = 6) of mice following the treatments. (O) Proposed model: Hepatic IRE1α-XBP1 signaling activated by chemo drugs regulates hepatic GDF15 expression and promotes chemotherapy-induced anorexia and body weight Loss. Data are representative of three independent experiments (A and B) or two independent experiments (C–M) and presented as mean ± SEM. *P < 0.05, **P < 0.01, or ***P < 0.001 by two-way ANOVA (A–E and I–K) or unpaired two-tailed Student’s t test (F–H and L–N).
Pharmacologic blocking of IRE1α activity alleviates chemotherapy-induced anorexia and body weight loss via suppression of hepatic GDF15. (A and B) XBP1 mRNA splicing and GDF15 mRNA levels in Huh7 cells treated with (A) Vehicle, 4μ8C (1 μM), DOX (0.01 μg/ml), or DOX+4μ8C or (B) Vehicle, 4μ8C (1 μM), Cis (10 μM), or Cis+4u8C for 24 h. n = 3 per group. (C–H) Male C57BL/6 mice were treated with Vehicle, 4μ8C (3.3 mg per kg body weight, i.p.), DOX (5 mg per kg body weight, i.p.), or the combination of DOX and 4μ8C at the indicated time points (black arrows) for three doses of treatment. (C and D)Xbp1 mRNA splicing (C) and Gdf15 mRNA levels (D) in liver samples from mice after the treatments. Veh., n = 5; 4μ8C, n = 5; DOX, n = 5; DOX+4μ8C, n = 5. (E) Circulating GDF15 protein levels in mice after the treatments. Veh., n = 6; 4μ8C, n = 6; DOX, n = 6; DOX+4μ8C, n = 6. (F) Representative images of immunofluorescence staining against c-Fos and GFRAL at the area AP and NTS of the murine brainstem (left). GFRAL+ c-Fos+ cells (indicated by arrowheads, left) per HPF were quantified (right). The frozen brainstem slides were from mice at 1 day after the final treatments. Scale bar, 100 μm. (G and H) Daily food intake (G, Veh., n = 4; 4μ8C, n = 4; DOX, n = 4; DOX+4μ8C, n = 4) and body weight changes (H, Veh., n = 6; 4μ8C, n = 6; DOX, n = 6; DOX+4μ8C, n = 6) of mice following the indicated treatments. (I–N) C57BL/6 male mice were treated with three doses of Vehicle, 4μ8C (3.3 mg per kg body weight, i.p.), Cis (5 mg per kg body weight, i.p.), or the combination of Cis and 4μ8C at the indicated time points (black arrows). (I and J)Xbp1 mRNA splicing (I, Veh., n = 7; 4μ8C, n = 8; Cis, n = 8; Cis+4μ8C, n = 8) and Gdf15 mRNA levels (J, Veh., n = 6; 4μ8C, n = 6; Cis, n = 6; Cis+4μ8C, n = 6) in liver samples from mice after the treatments. (K) Circulating GDF15 protein levels in mice after the treatments. Veh., n = 6; 4μ8C, n = 6; Cis, n = 6; Cis+4μ8C, n = 6. (L) Representative images of immunofluorescence staining against c-Fos and GFRAL at the area AP and NTS of the murine brainstem (left). GFRAL+ c-Fos+ cells (indicated by arrowheads, left) per HPF were quantified (right). The frozen brainstem slides were from mice at 1 day after the final treatments. Scale bar, 100 μm. (M and N) Daily food intake (M, Veh., n = 3; 4μ8C, n = 3; Cis, n = 4; Cis+4μ8C, n = 4) and body weight changes (N, Veh., n = 6; 4μ8C, n = 6; Cis, n = 6; Cis+4μ8C, n = 6) of mice following the treatments. (O) Proposed model: Hepatic IRE1α-XBP1 signaling activated by chemo drugs regulates hepatic GDF15 expression and promotes chemotherapy-induced anorexia and body weight Loss. Data are representative of three independent experiments (A and B) or two independent experiments (C–M) and presented as mean ± SEM. *P < 0.05, **P < 0.01, or ***P < 0.001 by two-way ANOVA (A–E and I–K) or unpaired two-tailed Student’s t test (F–H and L–N).
Concluding remarks
As the main chemotherapy drugs used in clinical treatment of solid tumors, platinum-based chemotherapeutic agents such as Cis as well as DOX can cause nausea, vomiting, and anorexia, resulting in body weight loss and poor life quality in the patients. Approximately half of the patients undergoing chemotherapy suffer from chemotherapy-induced anorexia (Hong et al., 2006; Winton et al., 2005). Owing to the largely elusive mechanisms underlying these adverse effects, there are limited therapeutic strategies for effective treatment. Documented studies have revealed that Cis treatment leads to excessive serotonin (5-HT) release from the gastrointestinal tract into circulation followed by abnormal dynamics of an hunger-stimulating hormone ghrelin, which may directly stimulate 5-HT receptors in the postrema area of the central nervous system (Minami et al., 2003). Although application of antagonists for 5-HT3R and NK-1 receptor has been shown to improve chemotherapy-induced nausea, breakthrough or delayed emesis could still be observed in a significant percentage of individuals with cancers (Aapro et al., 2018; Einhorn et al., 2017; McCullough, 2017). Recently, a critical role of the GDF15-GFRAL axis has been uncovered in regulating the occurrence of chemotherapy-induced nausea, anorexia, and weight loss in rodents, shrews, and primates (Borner et al., 2020a, 2020b; Breen et al., 2020; Hsu et al., 2017). Elevation of circulating GDF15 levels and activation of the GFRAL neurons in the postrema area of hindbrain were observed in animals treated with platinum-based drugs. Importantly, inhibition of the GDF15-GFRAL axis by neutralizing circulating GDF15 protein with monoclonal antibodies or by ablating Gfral gene profoundly alleviated anorexia and weight loss during platinum-based chemotherapy (Breen et al., 2020; Hsu et al., 2017). Here, our results demonstrate that treatment with DOX, another chemotherapy agent with high clinical emetic scores, can also robustly elevate circulating GDF15 levels along with anorexia and body weight loss in both healthy and tumor-bearing animals, in contrast to PTX, which has low clinical emetic scores. This is in line with previous studies regarding platinum-based drugs (Breen et al., 2020). Importantly, we observed elevated levels of plasma GDF15 not only in animal studies, but also in individuals with colon or breast cancers receiving the first dose of platinum-based or DOX-based chemo drugs (Fig. 3 E). These effects are much more dramatic than the results observed in a previous study involving patients who underwent multiple rounds of undefined chemotherapy. This suggests a significant and immediate response of GDF15 expression to chemotherapy treatment.
Our results from animal models showed that liver-derived GDF15 critically mediates chemotherapy-induced anorexia and body weight loss, supporting its role as an endocrine factor in the liver–brain communication via neural circuitry (Matsubara et al., 2022). A number of hepatocyte-derived secreted factors, namely “hepatokines,” have been documented as essential information transmitters that sense liver metabolic status and regulate systemic metabolism through target organs such as the brain, such as FGF21 and GDF15 (Jensen-Cody and Potthoff, 2021). In response to starvation and stress conditions, hepatocyte-derived FGF21 acts as an endocrine hormone to regulate a variety of physiological aspects including glucose/lipid metabolism, energy expenditure, and insulin/leptin sensitivity (Hill et al., 2019; Maida et al., 2016; Potthoff et al., 2009). Recent studies have also highlighted FGF21 signaling in the control of carbohydrate intake and taste preference through glutamatergic neurons in mice (von Holstein-Rathlou et al., 2016). Likewise, GDF15 acts through its obligate receptor GFRAL, whose expression is strictly limited to hindbrain, a critical area for the physiological control of emesis, nausea, and vomiting (Borner et al., 2020a). Notably, GDF15 is also a ubiquitous stress-responsive endocrine factor, whose expression has been shown to be regulated by integrated stress response transcription factors ATF4 and CHOP (Day et al., 2019; Miyake et al., 2021; Xie et al., 2022). With a lower hepatic expression under basal conditions, liver-secreted GDF15 exerts its metabolic actions in the settings where its expression is induced by stress, injury, or drug administration (Patel et al., 2022). While GDF15’s cellular origin is highly diversified (Patel et al., 2019), the physiological and pathological significance of hepatic GDF15 production has remained obscure in various contexts. An intriguing observation in our study is that mice in some cohorts displayed rapid normalization of food intake, but with relatively slow regaining of body weight during the recovery phase after chemotherapy challenge, similar to patterns as previously documented (Borner et al., 2020a). This delayed regaining of body weight may stem from several factors, such as delayed systemic storage of assimilated energy and nutrients, other chemotherapy-induced, metabolically adverse effects, or multiple metabolic actions of GDF15, e.g., promoting adipose tissue lipolysis for lipid utilization and energy production (Suriben et al., 2020; Hsu et al., 2017; Wang et al., 2023).
In the current study, we have found that the IRE1α-XBP1 branch of the UPR is selectively activated, which subsequently upregulates the expression of Gdf15 gene in the liver upon DOX or Cis treatments. This unveils an ER stress–mediated mechanism that links elevated liver GDF15 production to chemotherapy drug-induced adverse effects. IRE1α has been established as a key metabolic stress sensor and regulator of liver metabolism in response to nutritional states and various stimuli, including fasting/refeeding conditions and many endocrine factors (Huang et al., 2019). Here, our results reveal that chemotherapy drugs can also selectively activate the IRE1α-XBP1 pathway in hepatocytes, but not the PERK or ATF6 branches of UPR (Fig. 1). With respect to their mechanisms of action, Cis is known to form DNA crosslinks, blocking DNA, and RNA synthesis (Dasari and Tchounwou, 2014), and DOX can intercalate DNA, inhibiting topoisomerase II and RNA polymerase II activity and inducing DNA damage (Minotti et al., 2004), whereas PTX may prevent microtubule depolymerization, blocking the G2/M phase of cell division (Jordan and Wilson, 2004). Thus, it remains to be deciphered how the DNA-damaging chemotherapeutic agents can selectively trigger the activation of the IRE1α-XBP1 pathway in the liver.
It is particularly worth noting that hepatic Gdf15 expression is under the control of XBP1s, suggesting a potential therapeutic opportunity to target liver IRE1α RNase activity for ameliorating chemotherapy-induced anorexic effects. Targeting the signaling molecules of the UPR to combat various diseases has gained an increasing attraction in recent years (Marciniak et al., 2022). An array of compounds or drug-like molecules targeting the UPR sensors such as IRE1α has become available and is being tested in clinical trials (Marciniak et al., 2022). For instance, two clinical trials for the chemical compound ORIN1001, a specific inhibitor of IRE1α RNase activity, have been under way in patients with advanced solid tumors (NCT05154201, NCT03950570). Given that IRE1α also serves as a therapeutic target in tumor therapy, our results may point to an additional therapeutic benefit since selective blocking of hepatic IRE1α RNase activity is effective in reducing chemotherapy-induced GDF15 production to ameliorate GDF15-dependent anorexic side effects. It is also noteworthy that both DOX and Cis could still induce slight reductions of food intake and body weight in Gdf15KO mice (Fig. S2, A–H). Besides GDF15, our results also revealed ∼25 secreted factors whose expression increased in response to both chemo drugs (Fig. 3 B). Therefore, it warrants further investigations to explore additional mechanisms by which hepatic IRE1α-XBP1s signaling pathway mediates the adverse effects of chemotherapy beyond its regulation of GDF15 production in the context of not only anorexia, but also nausea and malaise as well as other possible behavioral changes. It would be also of great translational significance to dissect whether targeting the IRE1α pathway can be more efficacious than modulating the GDF15-GFRAL axis in these settings.
In summary, our study demonstrates that IRE1α-dependent upregulation of hepatic Gdf15 expression serves as a key mechanism driving chemotherapy-associated anorexia and body weight loss. Upon chemotherapy, selective activation of hepatic IRE1α-XBP1 pathway leads to upregulation of Gdf15 expression, and consequently, elevation of circulating GDF15 levels results in activation of GFRAL-expressing neurons. This liver–brain crosstalk in turn triggers anorexia and body weight loss. Moreover, pharmacologic inhibition of hepatic IRE1α RNase activity offers a potential therapeutic approach for effectively alleviating these side effects of chemotherapy (Fig. 5 O). Hence, our findings have important translational implications not only for chemotherapy-associated adverse effects, but also for other GDF15-related wasting conditions.
Materials and methods
Animals
Adult wild-type male mice (C57BL/6J; 8 wk old) were obtained from Hangzhou Ziyuan Laboratory Animal Technology Co., Ltd. Liver-specific IRE1α knockout (LKO, Albumin-Cre; Ern1flox/flox) mice on the C57BL/6 background were generated by intercrossing the ER to nucleus signaling 1 (Ern1) floxed (flox/flox, Ern1flox/flox) mice, in which the exon 2 of the Ern1 allele was flanked by loxP sites, with the Albumin-Cre mice as described (Shao et al., 2014). The mice were generated at Hangzhou Ziyuan Laboratory Animal Technology Co., Ltd. and maintained at 23 ± 3°C with a humidity of 35 ± 5% under a 12-h dark–light cycle, with free access to water and food. Whole-body Gdf15 gene knockout mice (Gdf15KO, strain no. T011862) were obtained from Gem-Pharmatech Co., Ltd. All animal experiments were performed according to protocols approved by the Institutional Animal Care and Use Committee at Zhejiang Academy of Traditional Chinese Medicine, Tongde Hospital of Zhejiang Province.
Mouse studies
For the evaluation of chemotherapy effects in healthy mice, 8-wk-old C57BL/6J male mice were randomized into indicated groups before the treatments, respectively. Chemotherapy agents were administered as described in the section Chemotherapy drugs.
For the inhibition of RNase activity of IRE1α in vivo, 8-wk-old male C57BL/6J mice were randomized into the indicated groups and received the i.p. injection of IRE1α inhibitor 4μ8C (3.3 mg/kg body weight, #S7272; Selleck) or DMSO (in 16% vol/vol Cremophor EL; Sigma-Aldrich) in combination with chemotherapeutic drugs. These mice were administered for two additional doses of chemo drugs and 4μ8C at the indicated time. Additional solvents (double distilled water/saline+DMSO+Cremophor EL) in equal doses to the groups of DOX/Cis+4μ8C were i.p. injected into mice of control groups.
For silencing the expression of hepatic GDF15, 6-wk-old C57BL/6J male mice were tail-vein injected with AAV, which ectopic overexpress shRNA targeting at murine Gdf15 mRNA (AAV-shGdf15-1) or control virus (AAV-shNC). 4 wk after the injection, animals were sacrificed for the evaluation of knockdown efficiency or administered with indicated chemotherapy agents for further analysis.
For the study of tumor-bearing mouse model, 8-wk-old male flox/flox or LKO mice received 5 × 105 Hepa1-6 cells by subcutaneous injection, respectively. The mice received the administration of DOX (5 mg per kg body weight) or vehicle at day 9 and 15 after the inoculation, respectively. Tumor growth was monitored daily following inoculation via measuring the tumor size three times per week using digital calipers. At day 7 after the inoculation, the palpable tumors were removed by surgery to mimic the clinical situation. For all the animal studies, body weight and food intake were determined around 09:00 in a day using a digital scale.
Chemotherapy drugs
DOX (#S1208) and PTX (#S1150) were purchased from Selleck. Cis was purchased from Sigma-Aldrich (#C2210000). The chemotherapy agents were delivered via intraperitoneal (i.p.) administration and dosed once: DOX (2 mg/ml in sterile water; 10 mg/kg), Cis (1 mg/ml in 0.9% saline; 10 mg/kg), and PTX (20 mg/ml in 66.6% Cremophor EL: ethanol in 0.9% saline; 10 mg/kg). For multiple chemotherapy dosing experiments, DOX (2 mg/ml in sterile water; 5 mg/kg) was administrated on day 0, day 11, and day 22, and Cis (1 mg/ml in 0.9% saline; 5 mg/kg) was administrated on day 0, day 9, and day 18, respectively. Doses for the chemotherapy agents were chosen in an attempt to induce ∼10–30% weight loss (Breen et al., 2020) with minor modifications according to the experimental results.
AAV
pAAV-U6-shNC-EGFP, pAAV-U6-shGdf15-1-EGFP, and pAAV-U6-shGdf15-2-EGFP plasmids were constructed at GenePharma Co. Ltd. The plasmids were packaged into AAV vector serotype DJ (AAV-DJ) by GenePharma utilizing standard plasmid transfection protocols. The silencing efficiency of pAAV-U6-shGdf15-1-EGFP and pAAV-U6-shGdf15-2-EGFP was evaluated in Hep1-6 cells and AAV-shGdf15-1 was chosen for the subsequent in vivo assays. AAVDJ-U6-shNC-EGFP (titer: 2.77 × 1012 vector genome [V.G]/ml) and AAVDJ-U6-shGdf15-1-EGFP (titer: 9.37 × 1012 V.G/ml) were diluted in PBS and administered at a dose of 1.0 × 1011 V.G/mouse via tail-vein injection (100 μl in total volume). The target sequences for shRNAs against Gdf15 mRNA are shown in Table S6.
RNA-seq analysis
Total RNA was isolated from freshly frozen liver tissues from the mice treated with chemotherapy agents (n = 5 individual mice per group) by TRIzol reagent (#T9424; Invitrogen). RNA library was prepared and sequenced by Berry Genomics. In brief, mRNA was purified from total RNA using polyT and then fragmented into 300–350 bp fragments, the first-strand cDNA was reverse-transcribed using fragmented RNA and dNTPs (dATP, dTTP, dCTP, and dGTP) and second-strand cDNA synthesis was subsequently performed. Remaining overhangs of double-strand cDNA were converted into blunt ends via exonuclease/polymerase activities. After adenylation of 3′ ends of DNA fragments, sequencing adaptors were ligated to the cDNA and the library fragments were purified. The template was enriched by PCR, and the PCR product was purified to obtain the final library. After library preparation and pooling of different samples, the samples were subjected for Illumina sequencing.
The analysis of RNA-seq data was performed as previously described (Shan et al., 2021). Reads with phred quality scores <20 and <35 bp after trimming were removed from further analysis using trimgalore version 0.4.1. Quantity-filtered reads were then aligned to the mouse reference genome GRCm38 (mm10) using the HISAT (v 2.0.1) (Kim et al., 2015) aligner using default settings and marked duplicates using Sembamba version 0.6.6 (Tarasov et al., 2015). Aligned reads were quantified using “featurecount” (v1.4.6) (Liao et al., 2014) per gene ID against mouse Gencode version 20 (Frankish et al., 2019). Generation of normalized counts and analysis of differential gene expression was done using the R package EdgeR. The resulting P values were adjusted using the Benjamini and Hochberg approach for controlling the FDR. All RNA-seq data have been deposited to GEO (GSE235695).
Immunoblotting and antibodies
Immunoblotting assays were performed as previously described (Shan et al., 2017). In brief, cells or tissues were lysed by homogenization in radioimmunoprecipitation assay lysis buffer (150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 50 mM Tris-HCl, pH 7.4). Protein extracts were separated by SDS–polyacrylamide gel electrophoresis and then transferred onto a polyvinylidene difluoride membrane. Following overnight incubation with the indicated primary antibodies at 4°C, membranes were developed with Thermo Fisher Scientific’s SuperSignal West Pico Chemiluminescent substrate or Millipore’s Immunobilon Western Chemiluminescent HRP substrate. The primary antibodies and diluted ratio include: anti-phospho-IRE1α 1:1,000 (Ser724; #NB100-2323; Novus Biologicals); anti-IRE1α 1:1,000 (#3294; Cell Signaling Technology); anti-XBP1 1:1,000 (#ab220783; Abcam); anti-α-tubulin 1:1,000 (#3873; Cell Signaling Technology); anti-BiP 1:1,000 (#3182; Cell Signaling Technology); anti-ATF6 1:1,000 (#65880; Cell Signaling Technology); anti-phospho-eIF2α 1:1,000 (#9721; Cell Signaling Technology); anti-eIF2α 1:1,000 (#9722; Cell Signaling Technology); anti-ATF4 1:1,000 (#11815; Cell Signaling Technology); and anti-CHOP 1:1,000 (#2895; Cell Signaling Technology). Uncropped images of western blots presented in this study are in source data figures.
Immunofluorescence staining
Brain samples were collected from mice perfused and fixed with 4% PFA for 1 h. Tissues were then transferred to 30% sucrose in PBS at 4°C overnight. Fixed samples were O.C.T. embedded and cut into 5-μm sections for staining. Sections were blocked with 3% normal horse serum in PBS at room temperature for 1 h and then incubated with anti-c-Fos (dilution: 1:300; #2250; Cell Signaling Technology) and anti-GFRAL (dilution: 1:100; #PA5-47769; Thermo Fisher Scientific) antibodies at 4°C overnight, followed by incubation with anti-rabbit-lgG-Alexa Fluor 555 (dilution: 1:500; #A32732; Thermo Fisher Scientific) and anti-sheep-IgG-Alexa Fluor 488 (dilution: 1:500; #ab150177; Abcam) antibodies for 2 h. Slides were then mounted with DAPI-containing antifade mounting medium (#BL739A; Biosharp), visualized under the LSM800 confocal laser scanning microscope, and analyzed with ZEN software (Zeiss).
Reverse transcription and quantitative PCR (RT-qPCR)
For RT-qPCR analysis, total RNA was isolated from liver tissues or cells by TRIzol reagent (#T9424; Invitrogen), and cDNA was synthesized using random hexamer primers (Thermo Fisher Scientific; #N8080127) and moloney murine leukemia virus reverse transcriptase (Thermo Fisher Scientific; #28025013) according to the manufacturer’s instructions. Real-time PCR was performed using the SYBR Green PCR system (#4309155; Applied Biosystems). Gapdh was utilized as an internal control for calculation using the ΔΔ-Ct method. All primers sequences used in this study are listed in Table S6.
Human specimens
For the evaluation of human circulating GDF15, plasma samples were obtained from healthy volunteers or individuals with cancers before and after receiving chemotherapy for the first time. Within the 37 individuals, 15 healthy volunteers were from our lab, 12 colon cancer patients were from the Hangzhou Third People’s Hospital, and 10 breast cancer patients were from the First Affiliated Hospital, College of Medicine, Zhejiang University. The collection of peripheral blood samples from donors was approved by Medical Ethics Committee of Tongde Hospital of Zhejiang Province. All the individuals in this study were recruited with written informed consents. The clinical characteristics of all the patients are shown in Table S9.
GDF15 protein measurement
Circulating levels of GDF15 protein were respectively measured using the Mouse/Rat GDF15 ELISA kit (#DY6385; R&D) for murine serum, or human GDF15 ELISA kit (#DGD150; R&D) for human plasma according to the manufacturer’s instructions.
Cell culture
Huh7 cells and Hepa1-6 cells were generously provided by Dr. Daqian Xu (Zhejiang University). HEK293T cells (#CL-0005) were obtained from Procell Life Science & Technology Co., Ltd. Cells were cultured in Dulbecco’s Modified Eagle’s Medium (#C11995500BT; Gibco) supplemented with 10% fetal bovine serum (#164210; Procell) with the supplement of 1% penicillin/streptomycin (#15140122; Gibco). All cells were maintained at 37°C in a humidified atmosphere of 5% CO2. Huh7 cells were seeded in 12-well plates and transfected with control vector or XBP1s-expressing vector using Lipofectamine 2000 (#11668027; Invitrogen) according to the manufacturer’s instruction. 48 h after transfection, the cells were treated with chemotherapy agents and subsequently processed for further analysis.
Isolation of primary murine hepatocytes
Hepatocytes were isolated from mice at 8–12 wk of age as previously described (Liu et al., 2015). In brief, anesthetized mice were subjected to collagenase perfusion through the portal vein with 50 ml of perfusion buffer (Krebs Ringer buffer containing 3.6 mg/ml glucose, 1 M CaCl2, and 5,000 U of collagenase I [Worthington]) at 37°C. The liver was aseptically removed to a sterile 10-cm cell culture dish with 20 ml of ice-cold perfusion buffer without collagenase. The excised liver was cut and hepatocytes were dispersed through aspiration using a large-bore pipette. After filtration through a 70-µm cell strainer (Thermo Fisher Scientific) into a 50-ml centrifuge tube and centrifugation at 50 × g for 2 min at 4°C, cells were then washed with cold Hepatocyte Wash Medium (Gibco) for three times and re-suspended in 15 ml of cold HepatoZYME-SFM (Gibco) medium supplemented with 2 mM L-glutamine, 10 units/ml penicillin, and 10 g/ml streptomycin. Cell viability was then determined by Trypan Blue staining, and hepatocytes were plated at 6 × 105 cells/well in 6-well culture dishes or at 3 × 105 cells/well in 12-well dishes that were pre-coated with collagen. Hepatocytes were cultured for 8 h before transfection with adenoviruses.
ChIP
ChIP was performed as previously described (Shan et al., 2020). Primary murine hepatocytes were prepared and transfected with adenoviruses expressing XBP1s-FLAG (Ad-XBP1s) or GFP (Ad-GFP). Cells were then cross-linked with 1% formaldehyde in PBS for 10 min at 37°C and quenched in 125 mM glycine in PBS for 5 min at 4°C. Cells were then lysed in Farnham lysis buffer (5 mM PIPES, pH 8.0, 85 mM KCl, 0.5% NP-40, 1 mM dithiothreitol [DTT], and protease inhibitor cocktail [#P8340; Sigma-Aldrich]). Crude nuclear pellets were collected by centrifugation before incubation in lysis buffer containing 5 mM Tris-HCl, pH 7.9, 1% SDS, 10 mM EDTA, 1 mM DTT, and protease inhibitor cocktail (#P8340; Sigma-Aldrich). Chromatin fragmentation (∼200–500 bp in length) was performed at 4°C by Bioruptor 300 using the setting of 10 cycles of 30 sec on and 60 sec off. Soluble chromatin was then diluted 1:10 with dilution buffer (20 mM Tris-HCl, pH 7.9, 0.5% Triton X-100, 2 mM EDTA, 150 mM NaCl, 1 mM DTT, and protease inhibitor cocktail [#P8340; Sigma-Aldrich]) and precleared using Protein G Sepharose 4 Fast Flow (#17-0618-01; GE Healthcare Biosciences) for 1 h at 4°C. Precleared samples were incubated with anti-XBP1 antibody (1:100 in dilution, #ab220783; Abcam) overnight at 4°C. Antibody-protein-DNA complexes were captured by incubation with Protein G Sepharose 4 Fast Flow (#17-0618-01; GE Healthcare Biosciences) at 4°C for 2 h. Immunoprecipitated material was consecutively washed with low-salt wash buffer (20 mM Tris-HCl, pH 7.9, 2 mM EDTA, 125 mM NaCl, 0.05% SDS, 1% Triton X-100, and protease inhibitor cocktail [#P8340; Sigma-Aldrich]), high-salt wash buffer (20 mM Tris-HCl, pH 7.9, 2 mM EDTA, 500 mM NaCl, 0.05% SDS, 1% Triton X-100, and protease inhibitor cocktail [#P8340; Sigma-Aldrich]), LiCl wash buffer (10 mM Tris-HCl, pH 7.9, 1 mM EDTA, 250 mM LiCl, 1% NP-40, 1% sodium deoxycholate, and protease inhibitor cocktail [#P8340; Sigma-Aldrich]), and 1× Tris-EDTA. After elution (100 mM NaHCO3, 1% SDS), the immunoprecipitated material was digested with RNase (#11119915001; Roche) and proteinase K (#EO0491; Thermo Fisher Scientific) prior to purification and concentration of the immunoprecipitated genomic DNA by ChIP DNA Clean & Concentrator Kit (#D5201; Zymo Research). ChIP-isolated DNA was subjected to qPCR (ChIP-qPCR). Sequences of the primers used in the assay are listed in Table S6.
Luciferase reporter assay
The Gdf15 promoter-luciferase reporter plasmid and its mutant versions were constructed through a PCR-based cloning strategy. For luciferase activity analysis, HEK293T cells were co-transfected with the reporter plasmid, pCMV-XBP1s, and the pRL-TK-Renilla-luciferase plasmid. Luciferase activity was measured with Dual Luciferase Reporter Gene Assay Kit (#RG027; Beyotime Biotechnology) following the manufacturer’s instructions, and Renilla luciferase activity was used for normalization.
For in vivo luciferase reporter assays, the luciferase reporter plasmids for mouse Gdf15 promoter spanning the region from −1,028 to +1 were constructed in pGL3 (Promega) via a PCR-based cloning strategy. Reporter DNA constructs were introduced into livers of mice through hydrodynamic injection as described (Bell et al., 2007). Briefly, a DNA solution (12.5 mg/ml in sterile saline) was injected at 10% of volume/body weight through the tail vein within 10 sec, and mice were then administered with DOX or Cis 24 h later. Mice were imaged at 24 h after chemotherapeutic drug administration. For in vivo imaging, anesthetized mice were injected i.p. with 150 mg per kg D-Luciferin firefly-potassium salt (#40902ES03; Yeasen Biotechnology), and images were captured after 15 min by the IVIS Spectrum Imaging System and analyzed with Living Image software (PerkinElmer).
Statistical analysis
Statistical analysis was carried out as indicated in the figure legends. All data are presented as the mean ± SEM unless otherwise indicated in the figure legends. Data variance was examined by F test or Bartlett’s test. The data meet the assumptions of the indicated statistical analysis. All tests were performed as two sided. A P value <0.05 was considered statistically significant. All statistical analyses were performed using Microsoft Excel or GraphPad Prism 8.0 (GraphPad Software). All statistical information, including P values, samples sizes, and repetitions, is provided in Table S7.
Online supplemental material
Fig. S1 contains supporting data for chemo-induced anorexia and selective activation of the IRE1α-XBP1 pathway in the liver. Fig. S2 shows the necessity and sufficiency of GDF15 for chemo-induced anorexia. Fig. S3 shows no impacts on liver homeostasis or chemo-induced liver damage upon hepatic IRE1α ablation or pharmacological blocking of IRE1α activity. Table S1 lists GSEA HALLMARK pathways enriched in DOX group. Table S2 lists GSEA HALLMARK pathways enriched in Cis group. Table S3 lists GSEA GO Biological Process enriched in DOX group. Table S4 lists GSEA MOTIF enriched in DOX group. Table S5 lists the overlapping upregulated 135 genes. Table S6 lists oligonucleotide primer sequences. Table S7 lists statistical data. Table S8 lists information of reagents and materials. Table S9 lists clinical characteristics.
Data availability
RNA-seq data have been deposited to Gene Expression Omnibus (accession GSE235695). All other data from this study have been shown in figures and online supplemental material.
Acknowledgments
We thank Daqian Xu (Zhejiang University, Hangzhou, China) for providing the Huh7 and Hepa1-6 cell lines. We thank Chao Bi from the Core Facilities, Zhejiang University School of Medicine for their technical support.
This work was supported by the grants from the National Key Research and Development Program of China (2022YFA1104102) to B. Shan, Leading Innovative and Entrepreneur Team Introduction Program of Zhejiang (2022R01002) to B. Shan, the National Natural Science Foundation of China (32371235, 81972674, 31900543, 82303369, 81972693, 82170891, 82270926, and 32241008) to Y. Wu, Y. Tang, W. Chen, M. Shao, and B. Shan, the Fundamental Research Funds for the Central Universities (226-2023-00061 and 226-2024-00024) to B. Shan, the Natural Science Funding of Zhejiang Province (LQ23H160003, LR20H160001) to Y. Tang and W. Chen, Zhejiang Provincial Program for the Cultivation of High-level Innovative Health Talents to Y. Wu and W. Chen, Shanghai Municipal Science and Technology Project (22140903200) to M. Shao, State Administration of Traditional Chinese Medicine Science and Technology Department-Zhejiang Provincial Administration of Traditional Chinese Medicine Co-construction of Key Laboratory (no. GZY-ZJ-SY-2402), and Institute Specifical Foundation for Zhejiang Provincial Department of Science and Technology (YSZX2401).
Author contributions: Y. Tang, T. Yao, B. Shan, and Y. Wu designed the study and performed most of the experiments and analyzed the data. X. Tian, X. Xia, X. Huang, Z. Qin, L. Zhao, Y. Zhao, B. Diao, Y. Ping, X. Zheng, and T. Qian participated in collection and analysis of the data. Z. Shen collected and provided the human samples used in this study. Y. Xu and H. Chen assisted some of the animal experiments. T. Ma, B. Zhou, S. Xu, Q. Zhou, Y. Liu, M. Shao, and W. Chen contributed reagents, technical support, and provided scientific insight and discussion. M. Shao, B. Shan, and Y. Wu conceptualized the study, interpreted the experiments, and wrote the manuscript. Y. Wu supervised the work. All authors contributed to the manuscript and approved the submitted version.
References
Author notes
Y. Tang and T. Yao contributed equally to this paper.
Disclosures: The authors declare no competing interests exist.
Supplementary data
shows gene sets enriched in the liver samples of DOX group relative to Vehicle group.
shows gene sets enriched in the liver samples of DOX group relative to Vehicle group.