Somatic mutations occurring on key enzymes are extensively studied and targeted therapies are developed with clinical promises. However, context-dependent enzyme function through distinct substrates complicated targeting a given enzyme. Here, we develop an algorithm to elucidate a new class of somatic mutations occurring on enzyme-recognizing motifs that cancer may hijack to facilitate tumorigenesis. We validate BUD13-R156C and -R230Q mutations evading RSK3-mediated phosphorylation with enhanced oncogenicity in promoting colon cancer growth. Further mechanistic studies reveal BUD13 as an endogenous Fbw7 inhibitor that stabilizes Fbw7 oncogenic substrates, while cancerous BUD13-R156C or -R230Q interferes with Fbw7Cul1 complex formation. We also find this BUD13 regulation plays a critical role in responding to mTOR inhibition, which can be used to guide therapy selections. We hope our studies reveal the landscape of enzyme-recognizing motif mutations with a publicly available resource and provide novel insights for somatic mutations cancer hijacks to promote tumorigenesis with the potential for patient stratification and cancer treatment.
Introduction
Cancer leverages various signaling deregulations to facilitate tumor growth, metastasis, and drug resistance (Ellisen, 2017) largely through genetic changes. Point mutations that alter enzyme protein sequence exert gene dose-independent function by modifying downstream targets to directly modulate cellular function. Advances in deep sequencing have enabled extensive cataloging of somatic single-nucleotide variants in large cohorts of cancer patients. Through these studies, hyperactivating mutations in oncogenes encoding enzymes, such as mTOR (Grabiner et al., 2014), Ras (Muñoz-Maldonado et al., 2019), PIK3CA (Samuels and Waldman, 2010), and BRAF (Holderfield et al., 2014), as well as inactivating mutations in tumor suppressor genes, including PTEN (Yin and Shen, 2008), TSC2/TSC1 (Papadopoulou et al., 2018), and NF1 (Philpott et al., 2017) have been commonly detected in a broad spectrum of human cancers. Agents specifically targeting an oncogenic variant, such as the Ras-G12V mutation, have been developed and have shown clinical promise (Shin et al., 2020). Investigation of the pathophysiological function of somatic mutations has traditionally relied on cell-based assays and murine cancer models (Koren and Bentires-Alj, 2013). The latter approach typically is restricted to analysis of a limited number of mutations (usually one or two), which is also time-consuming and costly. Notably, many enzymes exert context-dependent function, which complicates therapeutic attempts at targeting these enzymes, such as the kinase PKA (Zhang et al., 2020) and the E3 ligase β-TRCP (Wang et al., 2014). Extensive efforts have been devoted to identifying and deciphering frequent point mutations in oncoprotein enzymes due to their druggable nature. However, these mutated enzymes rely on their downstream substrates to impact cell function and drive cancer, and the effects of mutations in the target enzyme substrates have been little explored.
A recent effort connects protein-coding variants with human diseases (Sun et al., 2022), and similarly cancer-driven mutations in non-protein coding regions were mapped in a pan-cancer study (Rheinbay et al., 2020). Synonymous mutations were also reported to function as driver mutations in facilitating tumorigenesis (Supek et al., 2014). Here, we develop algorithms to systematically identify substrate motif mutations for key cancer-related enzymes in a pan-cancer landscape and validate the importance of enzyme-substrate mutations in fueling tumorigenesis to fill this knowledge gap.
Results
AGC kinase motif mutations are mostly frequently observed in The Cancer Genome Atlas (TCGA) database using a newly developed algorithm
We developed an algorithm that can be used to search for gained or lost substrate motifs for a given enzyme in any public or customized sequencing databases by locating the enzyme-substrate motifs in all proteins first, followed by determining if any identified motifs are mutated in cancer (Fig. 1 A). We applied this algorithm to the TCGA database, searching for altered motifs for well-characterized enzymes, including E3 ubiquitin ligases [Fbw7 (TPPLSP [Hao et al., 2007]), Keap1 (ETGE [McMahon et al., 2003]), etc.] and protein kinases [AMPK (LxRxxS/T [Gwinn et al., 2008]), LATS1 (HxRxxS/T), AGC kinases (R/KxR/KxxS/T), etc.; Fig. S1, A and B]. We found that in all enzyme-recognizing motifs we focused on, AGC kinase motifs, including both gain and loss of AGC kinase motifs (Fig. 1 B), showed the highest number of mutations (2,192 mutations identified in all cancer types, Fig. 1 C). AGC (named after PKA, PKG and PKC) is a superfamily of serine/threonine kinases with ∼ 60 members, who share a similar kinase structure composition and regulation (Pearce et al., 2010). AGC kinases phosphorylate and regulate a broad set of substrates, playing critical roles in numerous pivotal cellular processes including proliferation, metastasis, drug resistance, and development (Pearce et al., 2010). Among all cancer types with AGC kinase motif alternations, melanoma evinces the greatest number of substrate mutations (Fig. 1 D). In addition, frequently mutated genes may bear mutations in more than one kinase/enzyme-recognizing motif (Fig. 1 E), suggesting mutations on distinct kinase motifs may cooperate to exert synergistic or redundant oncogenic effects. We have built a publicly available website (https://xianming-tan.shinyapps.io/motif/) where readers can search for up-to-date mutation profiles for any given enzyme substrate motifs in pan-cancer.
Systematic identification of enzyme-recognizing motif mutations in pan-cancer by bioinformatic approaches. (A) A cartoon illustration of the bioinformatic analyses pipeline to systematically mine any given enzyme-recognizing motif mutations in cancer databases. A given enzyme-recognizing motif sequence is first identified in all protein sequences in uniport. Captured motif sequences in a given protein are further searched in TCGA database for comparison, and mutated motifs are recorded. Mutations occurring on essential motif residues likely to influence motif function are further grouped. This illustration is generated by Biorender.com. (B) An illustration for both gain and loss of AGC kinase motif. (C) A pie chart showing the total number of enzyme-recognizing motif mutations identified grouped by the enzymes. (D) A pie chart illustrating distribution of gained AGC kinase motif mutations in indicated cancer types. The acronym for indicated cancer types can be found at https://gdc.cancer.gov/resources-tcga-users/tcga-code-tables/tcga-study-abbreviations. (E) An illustration of mutated enzyme-recognizing motifs in these listed proteins. (F) Normalized number of mutated motifs and motifs affecting AGC phosphorylation sites in indicated molecules. (G) An illustration of two BUD13 mutations in cancer patients that disable AGC kinase motifs from cBioportal. (H) Sequence conservation alignment of BUD13 R156 and R230 across species.
Systematic identification of enzyme-recognizing motif mutations in pan-cancer by bioinformatic approaches. (A) A cartoon illustration of the bioinformatic analyses pipeline to systematically mine any given enzyme-recognizing motif mutations in cancer databases. A given enzyme-recognizing motif sequence is first identified in all protein sequences in uniport. Captured motif sequences in a given protein are further searched in TCGA database for comparison, and mutated motifs are recorded. Mutations occurring on essential motif residues likely to influence motif function are further grouped. This illustration is generated by Biorender.com. (B) An illustration for both gain and loss of AGC kinase motif. (C) A pie chart showing the total number of enzyme-recognizing motif mutations identified grouped by the enzymes. (D) A pie chart illustrating distribution of gained AGC kinase motif mutations in indicated cancer types. The acronym for indicated cancer types can be found at https://gdc.cancer.gov/resources-tcga-users/tcga-code-tables/tcga-study-abbreviations. (E) An illustration of mutated enzyme-recognizing motifs in these listed proteins. (F) Normalized number of mutated motifs and motifs affecting AGC phosphorylation sites in indicated molecules. (G) An illustration of two BUD13 mutations in cancer patients that disable AGC kinase motifs from cBioportal. (H) Sequence conservation alignment of BUD13 R156 and R230 across species.
Identification of enzyme motif mutations in TCGA by bioinformatic approaches. (A) A list of characterized kinase motifs. (B) A list of characterized E3 ligase binding motifs. (C) An illustration showing both BUD13-R156 and R230 residues are flexibly accessible from AlphaFold predicted BUD13 structure. (D) DepMap Portal (https://depmap.org/portal/) indicates CRISPR-mediated depletion of BUD13 reduces proliferation in most cancer types. (E) IB analyses of WCL and GST-pulldown from HEK293T cells transfected with indicated GST-BUD13 constructs. (F) IB analyses of WCL and GST-pulldown from HeLa cells transfected with indicated GST-BUD13 constructs. Where indicated, cells were starved in FBS-free media overnight and stimulated with insulin (100 ng/ml for 30 min) or EGF (100 ng/ml for 10 min) before cell collection. (G) IB analyses of WCL and GST-pulldown from HeLa cells transfected with indicated GST-BUD13 constructs. Where indicated, cells were starved in FBS-free media overnight and stimulated with 100 ng/ml EGF for 10 min before cell collection. (H) A list of putative AGC kinases with tumor suppressive roles in cancer. (I) IB analyses of WCL from DLD1 cells infected with indicated sgRNAs by lentiviral infection. Cells were selected with 1 µg/ml puromycin for 72 h to eliminate non-infected cells before cell collection. (J) IB analyses of WCL and HA-IP from HEK293T cells transfected with indicated DNA constructs. All the data shown are representative of three independent experiments. Source data are available for this figure: SourceData FS1.
Identification of enzyme motif mutations in TCGA by bioinformatic approaches. (A) A list of characterized kinase motifs. (B) A list of characterized E3 ligase binding motifs. (C) An illustration showing both BUD13-R156 and R230 residues are flexibly accessible from AlphaFold predicted BUD13 structure. (D) DepMap Portal (https://depmap.org/portal/) indicates CRISPR-mediated depletion of BUD13 reduces proliferation in most cancer types. (E) IB analyses of WCL and GST-pulldown from HEK293T cells transfected with indicated GST-BUD13 constructs. (F) IB analyses of WCL and GST-pulldown from HeLa cells transfected with indicated GST-BUD13 constructs. Where indicated, cells were starved in FBS-free media overnight and stimulated with insulin (100 ng/ml for 30 min) or EGF (100 ng/ml for 10 min) before cell collection. (G) IB analyses of WCL and GST-pulldown from HeLa cells transfected with indicated GST-BUD13 constructs. Where indicated, cells were starved in FBS-free media overnight and stimulated with 100 ng/ml EGF for 10 min before cell collection. (H) A list of putative AGC kinases with tumor suppressive roles in cancer. (I) IB analyses of WCL from DLD1 cells infected with indicated sgRNAs by lentiviral infection. Cells were selected with 1 µg/ml puromycin for 72 h to eliminate non-infected cells before cell collection. (J) IB analyses of WCL and HA-IP from HEK293T cells transfected with indicated DNA constructs. All the data shown are representative of three independent experiments. Source data are available for this figure: SourceData FS1.
We summarized all identified gain-of-AGC kinase motif mutations (Table S1) and loss-of-AGC kinase motif mutations (Table S2) at a pan-cancer level. To examine if these cancerous enzyme-recognizing motif mutations increase the oncogenicity potential, we found that BUD13 AGC kinase motif mutations were observed in nine types of cancers and seven identified AGC kinase motif mutations hit on five potential AGC kinase phosphorylation sites (Fig. 1 F). In addition, two mutations are particularly enriched in cancer including R156C/H/P, which may disrupt T159 phosphorylation, and R230Q, which may interfere with S235 phosphorylation (Fig. 1 G and Fig. S1 C). The motif containing R156 is conserved through evolution, while the R230-containing motif is unique to human (Fig. 1 H). Importantly, phosphorylation of both T159 and S235 has been confirmed by mass spectrometry analyses in multiple mammalian cell lines at endogenous levels (Bouhaddou et al., 2020; Mertins et al., 2016). BUD13 is a component of the activated spliceosome that controls pre-mRNA splicing (Zhang et al., 2018), and recently BUD13 was reported to be essential to antagonize intron retention (Frankiw et al., 2019). In addition, BUD13 polymorphisms have been observed and associated with metabolic syndrome (Kim et al., 2019; Zhang et al., 2017). Query of the DepMap portal suggests that BUD13 is likely a common essential gene in CRISPR-mediated loss-of-function screens (Fig. S1 D; http://DepMap.org). Thus, we chose BUD13-R156C and -R230Q mutations for a proof-of-principle study to evaluate if mutations in BUD13 AGC kinase motifs affect tumorigenesis.
BUD13-R156C and -R230Q mutations are deficient in RSK3-mediated phosphorylation
First, we tested if BUD13-R156C or -R230Q mutations are deficient in phosphorylation on AGC kinase motifs. To this end, we found ectopically expressed BUD13-R156C/P or -R230Q mutants were deficient in BUD13 phosphorylation at AGC kinase motifs detected by an AGC motif RxRxxpS/pT antibody in cells (Fig. 2, A and B; and Fig. S1 E). Next, we aimed to determine the potential AGC kinase(s) that phosphorylates BUD13 at T159 and S235 sites. Given we observed a relatively high level of basal BUD13 phosphorylation on AGC motifs in cells (Fig. 2 A), we applied a panel of kinase inhibitors but found none of these suppressed BUD13 phosphorylation (Fig. 2 C). In addition, considering well-characterized AGC kinases such as Akt, S6K, and PKC are responsive to growth factor signaling, we performed serum starvation followed by insulin or EGF stimulation but found BUD13 AGC kinase motif phosphorylation was largely non-responsive to growth factor deprivation/stimulation (Fig. S1, F and G), suggesting these defined AGC kinases functioning in growth signaling such as Akt, S6K, and PKC are unlikely to be kinase(s) phosphorylating BUD13 in cells. Previously, AGC kinase(s) has been shown to interact with substrates for phosphorylation in addition to the “kiss-and-run” mechanism (Gao et al., 2009). We hypothesized that BUD13 phosphorylating AGC kinase(s) would bind BUD13 and found that seven AGC kinases we have examined interacted with BUD13 in cells (Fig. 2, D and E), including PRKACB, RSK3, PKC1, PKC2, RPS6KA4, GRK6, and PKN3, all of which are more likely tumor suppressive kinases (Fig. S1 H). To further determine which of these seven kinases phosphorylate(s) BUD13, we depleted each of these kinases by at least three independent single guide RNAs (sgRNAs) and found depletion of RSK3 (Fig. S1 I), PKC1, GRK6, and PKN3, but not PKC2, PRKACB, and RPS6KA4, led to reduced BUD13 phosphorylation in cells (Fig. 2, F and G). Moreover, ectopic expression of only RSK3, but not PKC1, GRK6, or PKN3, increased BUD13 phosphorylation in cells (Fig. 2, H–J). Notably, RSK3-induced BUD13 phosphorylation increases were largely abolished in RSK3-K112R (a kinase-dead version of RSK3 [Woo et al., 2004]) expressing cells (Fig. 2 H), suggesting the RSK3 kinase activity is indispensable for BUD13 phosphorylation. In addition, inhibiting RSK3 by a small molecule pan-RSK1/2/3 inhibitor BI-D1870 reduced BUD13 phosphorylation in a BI-D1870 dose-dependent manner in cells (Fig. 2 K). RSK3 ectopic expression largely promoted WT- but not R156C-BUD13 phosphorylation in cells on AGC kinase motifs (Fig. S1 J). More importantly, we proved that T159 and S235 sites of BUD13 are the major phosphorylated sites of RSK3 kinase in vitro (Fig. 2 L). Moreover, BUD13 interaction with RSK3 was confirmed at endogenous levels in DLD1 cells (Fig. 2 M). Together, these data suggest RSK3 as a possible AGC kinase governing BUD13 phosphorylation on both T159 and S235.
RSK3 is a putative kinase phosphorylating BUD13 on R156 and R230 motifs. (A) IB) analyses of WCL and HA-immunoprecipitates (IP) derived from HEK293T cells transfected with indicated HA-BUD13 constructs. (B and C) IB analyses of WCL and GST-pulldowns derived from HEK293T cells transfected with indicated GST-BUD13 constructs. (D) IB analyses of WCL and GST-pulldowns derived from HEK293T cells transfected with indicated Flag-AGC constructs with GST-BUD13. (E) IB analyses of WCL and GST-pulldowns derived from HEK293T cells transfected with indicated HA-AGC constructs with GST-BUD13. (F and G) IB analyses of WCL and HA-IPs derived from DLD1 cells stably expressing HA-BUD13 by lentiviral infection and depleted of indicated endogenous genes by CRISPR. Cells were selected in 1 µg/ml puromycin for 72 h to eliminate non-infected cells. (H) IB analyses of WCL and HA-IPs derived from HEK293T cells transfected with indicated Flag-RSK3 constructs with HA-BUD13. (I) IB analyses of WCL and HA-IPs derived from HEK293T cells transfected with indicated Flag-AGC constructs with HA-BUD13. (J) IB analyses of WCL and GST-pulldowns derived from HEK293T cells transfected with indicated HA-AGC constructs with GST-BUD13. (K) IB analyses of WCL and GST-pulldowns derived from HEK293T cells transfected with GST-BUD13 and treated with indicated doses of BI-D1870 for 18 h before cell collection. (L) In vitro kinase assay between His-tag-BUD13 protein and active RSK3. (M) IB analysis of input and endogenous BUD13-IP from DLD1 cells. All the data are representative of three independent experiments. Source data are available for this figure: SourceData F2.
RSK3 is a putative kinase phosphorylating BUD13 on R156 and R230 motifs. (A) IB) analyses of WCL and HA-immunoprecipitates (IP) derived from HEK293T cells transfected with indicated HA-BUD13 constructs. (B and C) IB analyses of WCL and GST-pulldowns derived from HEK293T cells transfected with indicated GST-BUD13 constructs. (D) IB analyses of WCL and GST-pulldowns derived from HEK293T cells transfected with indicated Flag-AGC constructs with GST-BUD13. (E) IB analyses of WCL and GST-pulldowns derived from HEK293T cells transfected with indicated HA-AGC constructs with GST-BUD13. (F and G) IB analyses of WCL and HA-IPs derived from DLD1 cells stably expressing HA-BUD13 by lentiviral infection and depleted of indicated endogenous genes by CRISPR. Cells were selected in 1 µg/ml puromycin for 72 h to eliminate non-infected cells. (H) IB analyses of WCL and HA-IPs derived from HEK293T cells transfected with indicated Flag-RSK3 constructs with HA-BUD13. (I) IB analyses of WCL and HA-IPs derived from HEK293T cells transfected with indicated Flag-AGC constructs with HA-BUD13. (J) IB analyses of WCL and GST-pulldowns derived from HEK293T cells transfected with indicated HA-AGC constructs with GST-BUD13. (K) IB analyses of WCL and GST-pulldowns derived from HEK293T cells transfected with GST-BUD13 and treated with indicated doses of BI-D1870 for 18 h before cell collection. (L) In vitro kinase assay between His-tag-BUD13 protein and active RSK3. (M) IB analysis of input and endogenous BUD13-IP from DLD1 cells. All the data are representative of three independent experiments. Source data are available for this figure: SourceData F2.
Cancer patient–derived BUD13-R156C and -R230Q mutants facilitate colon cancer cell growth in vitro and in mice
Next, we went on to examine if these BUD13 cancerous mutants deficient in RSK3-mediated phosphorylation benefit tumorigenesis. To this end, given both BUD13-R156C and R230Q mutants were observed in colorectal adenocarcinoma patients (TCGA), we chose colon cancer as a model to test effects of BUD13-R156C expression that is conserved through evolution or BUD13-R230Q conserved only in human (Fig. 1 H). Depletion of endogenous BUD13 reduced growth of multiple colorectal cancer cell growth in 2D colony formation assays and 3D anchorage-independent growth assays including both colon cancer cells with mutated Ras/Raf such as HCT116 (Fig. 3, A–E), DLD1 (Fig. S2, A–E), and HT29 (Fig. S2, F–J), and colon cancer cells with WT-Ras/Raf such as Colo320 (Fig. S2, K to O) and Caco2 (Fig. S2, P–T). We further obtained isogenic single clones from CRISPR-mediated BUD13-KO in DLD1 cells (Fig. S2 U) and confirmed that BUD13 knockout also reduced DLD1 cell growth (Fig. S2, V and W). Notably, re-expressing WT-BUD13 in isogenic BUD13-KO cells largely rescued growth retardation caused by BUD13 KO (Fig. S2, X–Z1), alleviating off-target concerns in these BUD13-KO clones. We then reintroduced either WT-, R156C-, or R230Q-BUD13 into BUD13-KO DLD1 cells (Fig. 3 F). Compared with WT-BUD13 expressing DLD1 cells, either R156C- or R230Q-BUD13 expression increased DLD1 cell growth in 2D colony formation (Fig. 3, G and H) and 3D soft agar growth (Fig. 3, I and J) assays, as well as in a mouse xenograft assay (Fig. 3, K–M). These data support the notion that cancerous R156C- and R230Q-BUD13 expression facilitate DLD1 cell growth, which might explain why colon cancer prefers to retain this mutation. To reinforce that R156C- or R230Q-BUD13 expression induced colon cancer growth increase is not biased to Ras/Raf mutation status, we reconstituted WT-, R156C-, or R230Q-BUD13 expression in additional Ras/Raf mutated colon cancer cells including HCT116 and HT29, where we found R156C- or R230Q-BUD13 expressing cells exert enhanced growth ability in vitro (Fig. S2, Z2–Z7) and in xenograft models (Fig. S2, Z8–Z10), as well as WT-Ras/Raf expressing Colo320 and Caco2 cells, where similarly R156C- or R230Q-BUD13 expression enhanced cell growth in vitro (Fig. S2, Z11–Z15) and in xenograft models (Fig. 3, N–Q). To further evaluate the contribution of T159 phosphorylation in regulating R156C-BUD13–induced growth increase, we generated the R156C/T159E-BUD13 mutant to mimic T159 phosphorylation and found this mutant significantly suppressed R156C-induced growth increase (Fig. 3, R and S). These data suggest R156C largely promotes colon cancer cell growth by evading T159 phosphorylation.
The cancerous R156C- or R230Q-BUD13 mutant promotes colon cancer cell growth. (A) IB analyses of WCL derived from HCT116 cells depleted of BUD13 by indicated sgRNAs via lentiviral infection. Cells were selected in 1 μg/ml puromycin for 72 h to eliminate non-infected cells. (B and C) Representative images for 2D colony formation assays using cells from A and quantified in C. Scale bar represents 5 mm. (D and E) Representative images for 3D soft agar growth assays using cells from A and quantified in E. Scale bar represents 5 mm. (F) IB analyses of WCL derived from DLD1 cells depleted of BUD13 and re-expressing indicated BUD13-WT or mutants via lentiviral infection. Cells were selected with 2 mg/ml blasticidin for 72 h to eliminate non-infected cells. (G and H) Representative images for 2D colony formation assays using cells from F and quantified in H. Scale bar represents 5 mm. (I and J) Representative images for 3D soft agar growth assays using cells from F and quantified in J. Scale bar represents 5 mm. (K–M) Mouse xenograft experiments (n = 7) were performed with indicated DLD1 cells. The tumor volume changes in indicated days. 25 d after injection, mice were sacrificed, and tumors were dissected (L) and weighed (M). (N–Q) Mouse xenograft experiments (n = 6) were performed with indicated Caco2 cells (N). O indicates the tumor volume changes at indicated days. 22 d after injection, mice were sacrificed, and tumors were dissected (P) and weighed (Q). (R and S) Representative images for 2D colony formation assays using indicated DLD1 cells and quantified in S. Scale bar represents 5 mm. Statistical analysis was performed using one-way ANOVA. Asterisks show the significance between indicated groups. *, P < 0.05. All the data are representative of three independent experiments (error bars, mean ± SD). Source data are available for this figure: SourceData F3.
The cancerous R156C- or R230Q-BUD13 mutant promotes colon cancer cell growth. (A) IB analyses of WCL derived from HCT116 cells depleted of BUD13 by indicated sgRNAs via lentiviral infection. Cells were selected in 1 μg/ml puromycin for 72 h to eliminate non-infected cells. (B and C) Representative images for 2D colony formation assays using cells from A and quantified in C. Scale bar represents 5 mm. (D and E) Representative images for 3D soft agar growth assays using cells from A and quantified in E. Scale bar represents 5 mm. (F) IB analyses of WCL derived from DLD1 cells depleted of BUD13 and re-expressing indicated BUD13-WT or mutants via lentiviral infection. Cells were selected with 2 mg/ml blasticidin for 72 h to eliminate non-infected cells. (G and H) Representative images for 2D colony formation assays using cells from F and quantified in H. Scale bar represents 5 mm. (I and J) Representative images for 3D soft agar growth assays using cells from F and quantified in J. Scale bar represents 5 mm. (K–M) Mouse xenograft experiments (n = 7) were performed with indicated DLD1 cells. The tumor volume changes in indicated days. 25 d after injection, mice were sacrificed, and tumors were dissected (L) and weighed (M). (N–Q) Mouse xenograft experiments (n = 6) were performed with indicated Caco2 cells (N). O indicates the tumor volume changes at indicated days. 22 d after injection, mice were sacrificed, and tumors were dissected (P) and weighed (Q). (R and S) Representative images for 2D colony formation assays using indicated DLD1 cells and quantified in S. Scale bar represents 5 mm. Statistical analysis was performed using one-way ANOVA. Asterisks show the significance between indicated groups. *, P < 0.05. All the data are representative of three independent experiments (error bars, mean ± SD). Source data are available for this figure: SourceData F3.
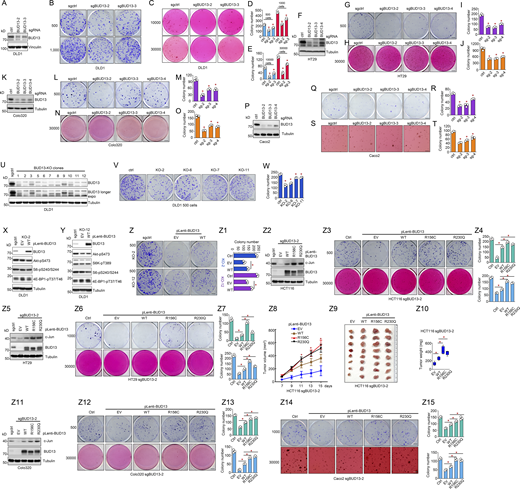
BUD13 AGC kinase motif mutations promote colon cancer growth. (A, F, K, and P) IB analyses of WCL from DLD1 (A), HT29 (F), Colo320 (K), or Caco2 (P) cells infected with indicated sgRNAs by lentiviral infection. Cells were selected with 1 mg/ml puromycin for 72 h to eliminate non-infected cells before cell collection. (B, D, G, I, L, M, Q, and R) Representative images for 2D colony formation assays using indicated number of cells for 10–21 d (B, G, L, and Q) and quantified in D, I, M, and R. Scale bar represents 5 mm. (C, E, H, J, N, O, S, and T) Representative images for 3D soft agar growth assays using indicated number of cells for 2–3 wk (C, H, N, and S) and quantified in E, J, O, and T. Scale bar represents 5 mm. (U) IB analyses of WCL from single clones of DLD1 cells infected with BUD13 sgRNAs by lentiviral infection. (V and W) Representative images for 2D colony formation assays using indicated DLD1 cells for 10 d and quantified in W. Scale bar represents 5 mm. (X and Y) IB analyses of WCL from indicated DLD1-BUD13 KO cells infected with indicated BUD13 viruses. Cells were selected with 2.5 mg/ml blasticidin for 72 h to eliminate non-infected cells before cell collection. (Z and Z1) Representative images for 2D colony formation assays using indicated DLD1 cells for 10 d and quantified in Z1. Scale bar represents 5 mm. (Z2) IB analyses of WCL from indicated HCT116-sgBUD13-2 cells infected with indicated BUD13 viruses. Cells were selected with 2.5 mg/ml blasticidin for 72 h to eliminate non-infected cells before cell collection. (Z3 and Z4) Representative images for 2D and 3D colony formation assays using indicated HCT116 cells and quantified in Z4. Scale bar represents 5 mm. (Z5) IB analyses of WCL from indicated HT29-sgBUD13-2 cells infected with indicated BUD13 viruses. Cells were selected with 2.5 mg/ml blasticidin for 72 h to eliminate non-infected cells before cell collection. (Z6 and Z7) Representative images for 2D and 3D colony formation assays using indicated HT29 cells for 21 d and quantified in Z7. Scale bar represents 5 mm. (Z8–Z10) Mouse xenograft experiments were performed with indicated HCT116 cells (n = 8). 15 d after injection, mice were sacrificed, and tumors were dissected (Z9) and weighed (Z10). (Z11) IB analyses of WCL from indicated Colo-320 sgBUD13-2 cells infected with indicated BUD13 viruses. Cells were selected with 2.5 mg/ml blasticidin for 72 h to eliminate non-infected cells before cell collection. (Z12 and Z13) Representative images for 2D and 3D colony formation assays using indicated Colo-320 cells for 21 d and quantified in Z13. Scale bar represents 5 mm. (Z14 and Z15) Representative images for 2D and 3D colony formation assays using indicated Caco2 cells for 21 d and quantified in Z15. Scale bar represents 5 mm. Data in A, D, K, P, U, X, Y, Z2, Z5, and Z11 are representative data from at least biologically duplicated experiments. Data in B, C, G, H, L, N, Q, S, V, Z, Z3, Z6, Z12, and Z14 are representative data from biologically triplicated experiments. Statistical analysis was performed using one-way ANOVA. Asterisks show the significance between indicated groups. *, P < 0.05. All the data are representative of three independent experiments (error bars, mean ± SD). Source data are available for this figure: SourceData FS2.
BUD13 AGC kinase motif mutations promote colon cancer growth. (A, F, K, and P) IB analyses of WCL from DLD1 (A), HT29 (F), Colo320 (K), or Caco2 (P) cells infected with indicated sgRNAs by lentiviral infection. Cells were selected with 1 mg/ml puromycin for 72 h to eliminate non-infected cells before cell collection. (B, D, G, I, L, M, Q, and R) Representative images for 2D colony formation assays using indicated number of cells for 10–21 d (B, G, L, and Q) and quantified in D, I, M, and R. Scale bar represents 5 mm. (C, E, H, J, N, O, S, and T) Representative images for 3D soft agar growth assays using indicated number of cells for 2–3 wk (C, H, N, and S) and quantified in E, J, O, and T. Scale bar represents 5 mm. (U) IB analyses of WCL from single clones of DLD1 cells infected with BUD13 sgRNAs by lentiviral infection. (V and W) Representative images for 2D colony formation assays using indicated DLD1 cells for 10 d and quantified in W. Scale bar represents 5 mm. (X and Y) IB analyses of WCL from indicated DLD1-BUD13 KO cells infected with indicated BUD13 viruses. Cells were selected with 2.5 mg/ml blasticidin for 72 h to eliminate non-infected cells before cell collection. (Z and Z1) Representative images for 2D colony formation assays using indicated DLD1 cells for 10 d and quantified in Z1. Scale bar represents 5 mm. (Z2) IB analyses of WCL from indicated HCT116-sgBUD13-2 cells infected with indicated BUD13 viruses. Cells were selected with 2.5 mg/ml blasticidin for 72 h to eliminate non-infected cells before cell collection. (Z3 and Z4) Representative images for 2D and 3D colony formation assays using indicated HCT116 cells and quantified in Z4. Scale bar represents 5 mm. (Z5) IB analyses of WCL from indicated HT29-sgBUD13-2 cells infected with indicated BUD13 viruses. Cells were selected with 2.5 mg/ml blasticidin for 72 h to eliminate non-infected cells before cell collection. (Z6 and Z7) Representative images for 2D and 3D colony formation assays using indicated HT29 cells for 21 d and quantified in Z7. Scale bar represents 5 mm. (Z8–Z10) Mouse xenograft experiments were performed with indicated HCT116 cells (n = 8). 15 d after injection, mice were sacrificed, and tumors were dissected (Z9) and weighed (Z10). (Z11) IB analyses of WCL from indicated Colo-320 sgBUD13-2 cells infected with indicated BUD13 viruses. Cells were selected with 2.5 mg/ml blasticidin for 72 h to eliminate non-infected cells before cell collection. (Z12 and Z13) Representative images for 2D and 3D colony formation assays using indicated Colo-320 cells for 21 d and quantified in Z13. Scale bar represents 5 mm. (Z14 and Z15) Representative images for 2D and 3D colony formation assays using indicated Caco2 cells for 21 d and quantified in Z15. Scale bar represents 5 mm. Data in A, D, K, P, U, X, Y, Z2, Z5, and Z11 are representative data from at least biologically duplicated experiments. Data in B, C, G, H, L, N, Q, S, V, Z, Z3, Z6, Z12, and Z14 are representative data from biologically triplicated experiments. Statistical analysis was performed using one-way ANOVA. Asterisks show the significance between indicated groups. *, P < 0.05. All the data are representative of three independent experiments (error bars, mean ± SD). Source data are available for this figure: SourceData FS2.
BUD13 is an endogenous inhibitor for the E3 ligase Fbw7
We next investigated the molecular mechanism(s) underlying R156C- or R230Q-BUD13 mutation in promoting colon cancer cell proliferation. BUD13 polymorphisms have been reported to associate with metabolic syndrome (Zhang et al., 2017), dyslipidemia (Bai et al., 2019), stroke risk (Zhou et al., 2015), and other human disorders for unknown reasons. BUD13 is part of the activated spliceosome (Haselbach et al., 2018; Zhang et al., 2018) as a potential RNA-binding protein. BUD13 regulates IRF7 intron retention to promote type I interferon signaling (Frankiw et al., 2019). BUD13 deletion in yeast resulted in abnormal bipolar budding due to deficiency in pre-mRNA splicing (Tuo et al., 2012). BUD13 was also found to suppress mir-210 splicing to control microRNA levels (Nussbacher and Yeo, 2018) and recruited by circSERPINA3 to stabilize SERPINA3 mRNA in prostate cancer (Xing et al., 2021). Non-RNA splicing-related BUD13 function has not been reported. We found that BUD13-R156C/P and -R230Q mutants displayed comparably binding ability to a BUD13 binding single strand RNA (Frankiw et al., 2019), suggesting increased cell growth in R156C- and R230Q-BUD13 expressing DLD1 cells is unlikely due to BUD13 changes in binding RNA (Fig. S3, A and B). Moreover, BUD13 KO didn’t significantly affect DLD1 cell responses to poly I:C or ISD90 stimulation that mimics RNA or DNA viral infection, and no IRF7 protein abundance changes were found to be associated with BUD13 loss or mutations (Fig. S3, C and D). Furthermore, comparable levels of innate immune activation evidenced by IRF3-pS396 and TBK1-pS172 signals were observed in R156C- or R230Q-BUD13 expressing DLD1 cell upon poly I:C or ISD90 treatment with WT-BUD13 cells with minimal effects on IRF7 expression (Fig. S3, E–H). Together, these data suggest that BUD13-R156C and -R230Q mutants promote DLD1 cell growth independent of reported RNA binding or IRF7-innate immune regulations. Moreover, BUD13-mutant-expressing DLD1 cells showed a similar migration ability in vitro as BUD13-WT cells (Fig. S3, I and J).
BUD13-R156C mutation is not deficient in RNA binding and innate immunity. (A and B) IB analysis of biotin-RNA pulldowns and WCL derived from indicated DLD1 cells. (C–H) IB analyses of WCL derived from indicated DLD1 cells transfected with either 5 mg/ml ISD90 or 5 mg/ml poly I:C for indicated time periods before cell collection. (I and J) Representative images for transwell assays using 1 × 105 DLD1 cells for 24 h and quantified in J. Scale bar represents 5 µm. Statistical analysis was performed using one-way ANOVA. n.s. shows no significance between indicated groups. All the data are representative of three independent experiments (error bars, mean ± SD). Source data are available for this figure: SourceData FS3.
BUD13-R156C mutation is not deficient in RNA binding and innate immunity. (A and B) IB analysis of biotin-RNA pulldowns and WCL derived from indicated DLD1 cells. (C–H) IB analyses of WCL derived from indicated DLD1 cells transfected with either 5 mg/ml ISD90 or 5 mg/ml poly I:C for indicated time periods before cell collection. (I and J) Representative images for transwell assays using 1 × 105 DLD1 cells for 24 h and quantified in J. Scale bar represents 5 µm. Statistical analysis was performed using one-way ANOVA. n.s. shows no significance between indicated groups. All the data are representative of three independent experiments (error bars, mean ± SD). Source data are available for this figure: SourceData FS3.
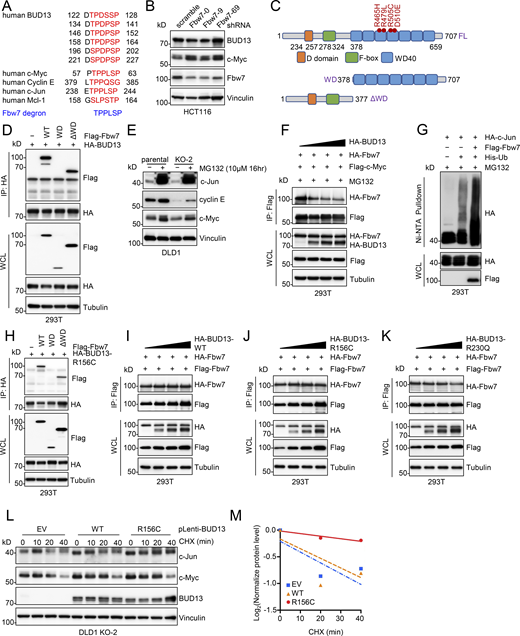
We then examined changes in various signaling pathways that have been reported to be critical for colorectal cancer development, including Wnt (Schatoff et al., 2017), mTOR (Francipane and Lagasse, 2014), and Fbw7 (Wood et al., 2007) signaling. Depletion of BUD13 minimally affected expression of β-catenin and activation of mTOR (evidenced by both Akt-pS473 and S6K-pT389; Fig. 4 A), while significantly downregulating the expression of c-Jun and c-Myc that are well-characterized substrates for the E3 ligase Fbw7 (Davis et al., 2014; Fig. 4 A). Moreover, re-expressing WT-BUD13 in BUD13-KO DLD1 cells partially rescued the expression of c-Jun and c-Myc with minimal effects on β-catenin expression and mTOR activation (Fig. 4 B). These data suggest that BUD13 may interfere with Fbw7-mediated degradation of its substrates including c-Jun. In addition, compared with WT-BUD13, R156C- or R230Q-BUD13 expression led to increased levels of c-Jun (Fig. 4 B), and depletion of c-Jun in R156C-BUD13-expressing DLD1 cells reduced growth (Fig. 4, C–E), supporting that c-Jun partially mediates R156C-BUD13-controlled DLD1 cell growth increase. To examine if BUD13 regulates c-Jun expression functioning through Fbw7-mediated c-Jun protein stability control, we observed BUD13 bound to Fbw7 in cells (Fig. 4 F). Interestingly, a closer examination of human BUD13 protein sequence identified six putative Fbw7 degron motifs including T159 that R156C regulates (Fig. S4 A). Depletion of endogenous Fbw7 didn’t significantly affect BUD13 protein abundance in both DLD1 (Fig. 4 G) and HCT116 (Fig. S4 B) cells. In addition, ectopic expression of Fbw7 failed to degrade BUD13 in cells (Fig. 4 H). These data suggest that although BUD13 interacts with Fbw7, BUD13 is more likely an endogenous Fbw7 inhibitor or a pseudo-substrate. Unlike canonical substrates binding to the Fbw7-WD40 repeat domains, BUD13 largely interacted with Fbw7-ΔWD40 domain (Fig. S4, C and D). Given the F-box domain has been shown to create non-specific-binding, we further confirmed that BUD13 interacted with F-box–deleted Fbw7 in cells (Fig. 4 I), further supporting an interaction between Fbw7 and BUD13. Moreover, treatment with the proteasomal inhibitor MG132 largely rescued BUD13-depletion-induced downregulation of Fbw7 substrates c-Jun, cyclin E, and c-Myc (Fig. S4 E), supporting that BUD13 suppresses Fbw7 function at posttranslational levels. As a result, BUD13 binding to Fbw7 released c-Jun (Fig. 4 J) or c-Myc (Fig. S4 F) from binding to Fbw7. As a potential competitor for Fbw7 substrates in binding Fbw7, ectopic expression of BUD13 stabilized Fbw7 substrates such as c-Jun and c-Myc in DLD1 cells (Fig. 4 K). Mechanistically, WT-BUD13 expression suppressed Fbw7-governed c-Jun ubiquitination in cells, and both R156C- and R230Q-BUD13 exerted an enhanced ability in further suppressing Fbw7-mediated c-Jun ubiquitination (Fig. 4 L and Fig. S4 G). Together, these data suggest that BUD13 may serve as an endogenous Fbw7 inhibitor to stabilize Fbw7 substrates through a competitively binding mechanism (Fig. 4 M).
BUD13 binds and interferes with Fbw7 binding to substrates. (A) IB analyses of WCL derived from DLD1 cells deleted of BUD13 by indicated sgRNAs via lentiviral infection and single clones were selected and validated by Western blotting. (B) IB analyses of WCL derived from indicated DLD1 cells. (C) IB analyses of WCL derived from indicated DLD1 cells depleted of c-Jun by lentiviral infection. Cells were selected in 1 μg/ml puromycin for 72 h to eliminate non-infected cells. (D and E) Representative images for 2D colony formation assays using cells from C and quantified in E. Scale bar represents 5 mm. (F) IB analyses of endogenous BUD13-IP derived from DLD1 cells. (G) IB analyses of WCL derived from DLD1 cells depleted of Fbw7 by indicated shRNAs via lentiviral infection. Cells were selected in 1 μg/ml puromycin for 72 h to eliminate non-infected cells. (H) IB analyses of WCL derived from HEK293T cells transfected with indicated DNA constructs. (I) IB analyses of WCL and GST-pulldowns derived from HEK293T cells transfected with indicated DNA constructs. (J) IB analyses of WCL and Flag-IP derived from HEK293T cells transfected with indicated DNA constructs and treated with 10 mM MG132 overnight before cell collection. (K) IB analyses of WCL derived from DLD1 cells transfected with indicated DNA constructs. (L) IB analyses of Ni-NTA pulldown and WCL derived from HEK293T cells transfected with indicated DNA constructs. (M) A cartoon illustration for a model for BUD13 competing with c-Jun to bind Fbw7 via binding to distinct Fbw7 domains. Statistical analysis was performed using one-way ANOVA. Asterisks show the significance between indicated groups. *, P < 0.05. All the data are representative of three independent experiments (error bars, mean ± SD). Source data are available for this figure: SourceData F4.
BUD13 binds and interferes with Fbw7 binding to substrates. (A) IB analyses of WCL derived from DLD1 cells deleted of BUD13 by indicated sgRNAs via lentiviral infection and single clones were selected and validated by Western blotting. (B) IB analyses of WCL derived from indicated DLD1 cells. (C) IB analyses of WCL derived from indicated DLD1 cells depleted of c-Jun by lentiviral infection. Cells were selected in 1 μg/ml puromycin for 72 h to eliminate non-infected cells. (D and E) Representative images for 2D colony formation assays using cells from C and quantified in E. Scale bar represents 5 mm. (F) IB analyses of endogenous BUD13-IP derived from DLD1 cells. (G) IB analyses of WCL derived from DLD1 cells depleted of Fbw7 by indicated shRNAs via lentiviral infection. Cells were selected in 1 μg/ml puromycin for 72 h to eliminate non-infected cells. (H) IB analyses of WCL derived from HEK293T cells transfected with indicated DNA constructs. (I) IB analyses of WCL and GST-pulldowns derived from HEK293T cells transfected with indicated DNA constructs. (J) IB analyses of WCL and Flag-IP derived from HEK293T cells transfected with indicated DNA constructs and treated with 10 mM MG132 overnight before cell collection. (K) IB analyses of WCL derived from DLD1 cells transfected with indicated DNA constructs. (L) IB analyses of Ni-NTA pulldown and WCL derived from HEK293T cells transfected with indicated DNA constructs. (M) A cartoon illustration for a model for BUD13 competing with c-Jun to bind Fbw7 via binding to distinct Fbw7 domains. Statistical analysis was performed using one-way ANOVA. Asterisks show the significance between indicated groups. *, P < 0.05. All the data are representative of three independent experiments (error bars, mean ± SD). Source data are available for this figure: SourceData F4.
BUD13 is an endogounous inhibitor for Fbw7 and competes with c-Jun to bind Fbw7. (A) Illustration of putative Fbw7 degrons in BUD13 protein sequence. (B) IB analyses of WCL derived from HCT116 cells depleted of Fbw7 by indicated shRNAs via lentiviral infection. Cells were selected in 1 mg/ml puromycin for 72 h to eliminate non-infected cells. (C) A cartoon illustration of Fbw7 domain structures. (D) IB analysis of HA-IP and WCLs derived from HEK293T cells transfected with indicated DNA constructs. (E) IB analysis of WCL derived from indicated DLD1 cells treated with 10 mM MG132 for 16 h before cell collection. (F) IB analyses of WCL and Flag-IP from HEK293T cells transfected with indicated DNA constructs. Cells were treated with 10 mM MG132 for 16 h before cell collection. (G) IB analyses of Ni-NTA pulldown and WCL from HEK293T cells transfected with indicated DNA constructs. Cells were treated with 10 mM MG132 for 16 h before cell collection. (H) IB analyses of WCL and HA-IP from HEK293T cells transfected with indicated DNA constructs. Cells were treated with 10 mM MG132 for 16 h before cell collection. (I–K) IB analyses of WCL and Flag-IP from HEK293T cells transfected with indicated BUD13 and Fbw7 constructs. (L and M) IB analysis of WCL derived from indicated DLD1 cells treated with 200 mg/ml cycloheximide (CHX) for indicated time periods and quantified in M. All the data are representative of three independent experiments. Source data are available for this figure: SourceData FS4.
BUD13 is an endogounous inhibitor for Fbw7 and competes with c-Jun to bind Fbw7. (A) Illustration of putative Fbw7 degrons in BUD13 protein sequence. (B) IB analyses of WCL derived from HCT116 cells depleted of Fbw7 by indicated shRNAs via lentiviral infection. Cells were selected in 1 mg/ml puromycin for 72 h to eliminate non-infected cells. (C) A cartoon illustration of Fbw7 domain structures. (D) IB analysis of HA-IP and WCLs derived from HEK293T cells transfected with indicated DNA constructs. (E) IB analysis of WCL derived from indicated DLD1 cells treated with 10 mM MG132 for 16 h before cell collection. (F) IB analyses of WCL and Flag-IP from HEK293T cells transfected with indicated DNA constructs. Cells were treated with 10 mM MG132 for 16 h before cell collection. (G) IB analyses of Ni-NTA pulldown and WCL from HEK293T cells transfected with indicated DNA constructs. Cells were treated with 10 mM MG132 for 16 h before cell collection. (H) IB analyses of WCL and HA-IP from HEK293T cells transfected with indicated DNA constructs. Cells were treated with 10 mM MG132 for 16 h before cell collection. (I–K) IB analyses of WCL and Flag-IP from HEK293T cells transfected with indicated BUD13 and Fbw7 constructs. (L and M) IB analysis of WCL derived from indicated DLD1 cells treated with 200 mg/ml cycloheximide (CHX) for indicated time periods and quantified in M. All the data are representative of three independent experiments. Source data are available for this figure: SourceData FS4.
The BUD13-R156C or -R230Q mutation disrupts Fbw7Cul1 E3 ligase complex formation and function
We observed that both R156C- and R230Q-BUD13 mutants displayed a similar cellular localization as WT-BUD13 in DLD1 cells (Fig. 5 A). Although R156C- or R230Q-BUD13 expression further stabilized c-Jun compared with WT-BUD13 (Fig. 4 B) and both displayed an enhanced binding with Fbw7 (Fig. 5 B); however, unlike WT-BUD13, R156C-BUD13 didn’t compete with c-Jun in binding Fbw7 (Fig. 5 C), but rather enhanced c-Jun/Fbw7 interactions (Fig. 5 D). Thus, we thought R156C-BUD13 may function through a non-Fbw7 substrate competitor-dependent manner in stabilizing c-Jun. To determine the underlying mechanism(s), we first found that R156C-BUD13 still bound to Fbw7-ΔWD40 domain as WT-BUD13 (Fig. S4 H). Moreover, neither WT-, R156C-, nor R230Q-BUD13 affected Fbw7 dimerization that is necessary for efficient Fbw7 E3 ligase activity (Fig. S4, I–K). Interestingly, we found that WT-BUD13 didn’t affect Fbw7 binding to cullin 1 (Fig. 5 E), while either R156C-BUD13 (Fig. 5 F) or R230Q-BUD13 (Fig. 5 G) expression reduced cullin 1 binding to Fbw7. This observation further confirmed that compared with WT-BUD13, R156C- or R230Q-BUD13 colocalized with Fbw7 (Fig. 5 H) but not cullin 1 (Fig. 5 I) in DLD1 cells. Given Fbw7 complexes with Skp1 and cullin 1 to form a functional E3 ligase complex (Wang et al., 2014), deficiency of R156C- or R230Q-BUD13 in maintaining an active E3 ligase may explain why these cancerous BUD13 mutants fail to promote Fbw7-mediated c-Jun destruction thus leading to c-Jun stabilization (Fig. 5 J). Consistent with this notion, compared with WT-BUD13, extended c-Jun protein half-life was observed in R156C-BUD13 expressing DLD1 cells (Fig. S4, L and M).
BUD13-R156C interferes with Fbw7 E3 ligase complex formation and function. (A) Representative immunofluorescent (IF) images derived from indicated DLD1 cells stained with indicated antibodies. Scale bar represents 50 µm. (B) IB analyses of WCL and Flag-IP derived from HEK293T cells transfected with indicated DNA constructs. (C–E) IB analyses of WCL and Flag-IP derived from HEK293T cells transfected with indicated DNA constructs and treated with 10 mM MG132 overnight before cell collection. (F and G) IB analyses of WCL and Myc-IP derived from HEK293T cells transfected with indicated DNA constructs. (H and I) Representative IF images derived from indicated DLD1 cells stained with indicated antibodies. Scale bar represents 50 µm. (J) A cartoon illustration indicating BUD13-R156C mutant binds to Fbw7 to interfere with cullin 1 binding instead of c-Jun binding. All the data are representative of three independent experiments. Source data are available for this figure: SourceData F5.
BUD13-R156C interferes with Fbw7 E3 ligase complex formation and function. (A) Representative immunofluorescent (IF) images derived from indicated DLD1 cells stained with indicated antibodies. Scale bar represents 50 µm. (B) IB analyses of WCL and Flag-IP derived from HEK293T cells transfected with indicated DNA constructs. (C–E) IB analyses of WCL and Flag-IP derived from HEK293T cells transfected with indicated DNA constructs and treated with 10 mM MG132 overnight before cell collection. (F and G) IB analyses of WCL and Myc-IP derived from HEK293T cells transfected with indicated DNA constructs. (H and I) Representative IF images derived from indicated DLD1 cells stained with indicated antibodies. Scale bar represents 50 µm. (J) A cartoon illustration indicating BUD13-R156C mutant binds to Fbw7 to interfere with cullin 1 binding instead of c-Jun binding. All the data are representative of three independent experiments. Source data are available for this figure: SourceData F5.
RSK3-mediated BUD13 phosphorylation regulates Fbw7 E3 ligase activity
In echoing RSK3 as a major kinase phosphorylating BUD13, we found RSK3 expression reduced BUD13 binding to Fbw7 in cells (Fig. 6 A) presumably through promoting BUD13 phosphorylation (Fig. 6 B). In addition, inhibiting RSK3 kinase activation by BI-D1870 significantly increased BUD13 binding to Fbw7 (Fig. 6 C) and disrupted Fbw7 binding with Cul1 (Fig. 6 D). Therefore, BI-D1870 treatment stabilized c-Jun proteins in DLD1 cells in both a time- (Fig. 6, E and F) and dose- (Fig. 6 G) dependent manner. In further supporting RSK3-mediated BUD13-T159 phosphorylation in disrupting function of BUD13 in binding and stabilizing Fbw7 substrates, consistent with our observation that R156C/T159E expressing cells inhibited R156C-BUD13 expression induced growth increase (Fig. 3, R and S), we found compared with R156C, R156C/T159E expression also reduced expression of Fbw7 substrates including c-Jun and cMyc (Fig. 6 H). Together, these data reveal that the cancerous R156C-BUD13 behaves similarly as RSK3 inactivation in enhanced Fbw7 association to disrupt cullin 1 binding, leading to inactivation of the functional and active Fbw7Cul1 E3 ligase complexes.
RSK3-mediated BUD13 phosphorylation regulates Fbw7 E3 ligase function. (A) IB analyses of WCL from HEK293T cells transfected with indicated DNA constructs. (B) A cartoon illustration revealing RSK3-mediated BUD13 phosphorylation dissociates BUD13 from binding Fbw7. (C and D) IB analyses of WCL and Flag-IP derived from HEK293T cells transfected with indicated DNA constructs and treated with indicated doses of BI-D1870 for 18 h before cell collection. (E) IB analyses of WCL from DLD1 cells treated with 5 mM BI-D1870 for indicated time periods before cell collection. (F) IB analyses of WCL from indicated DLD1 cells. Cells were treated with 5 mM BI-D1870 for indicated time periods before cell collection. (G) IB analyses of WCL from DLD1 cells treated with indicated doses of BI-D1870 for 24 h before cell collection. (H) IB analysis of WCL derived from indicated DLD1 cells. All the data are representative of three independent experiments. Source data are available for this figure: SourceData F6.
RSK3-mediated BUD13 phosphorylation regulates Fbw7 E3 ligase function. (A) IB analyses of WCL from HEK293T cells transfected with indicated DNA constructs. (B) A cartoon illustration revealing RSK3-mediated BUD13 phosphorylation dissociates BUD13 from binding Fbw7. (C and D) IB analyses of WCL and Flag-IP derived from HEK293T cells transfected with indicated DNA constructs and treated with indicated doses of BI-D1870 for 18 h before cell collection. (E) IB analyses of WCL from DLD1 cells treated with 5 mM BI-D1870 for indicated time periods before cell collection. (F) IB analyses of WCL from indicated DLD1 cells. Cells were treated with 5 mM BI-D1870 for indicated time periods before cell collection. (G) IB analyses of WCL from DLD1 cells treated with indicated doses of BI-D1870 for 24 h before cell collection. (H) IB analysis of WCL derived from indicated DLD1 cells. All the data are representative of three independent experiments. Source data are available for this figure: SourceData F6.
BUD13 expression contributes to Torin 2 treatment resistance
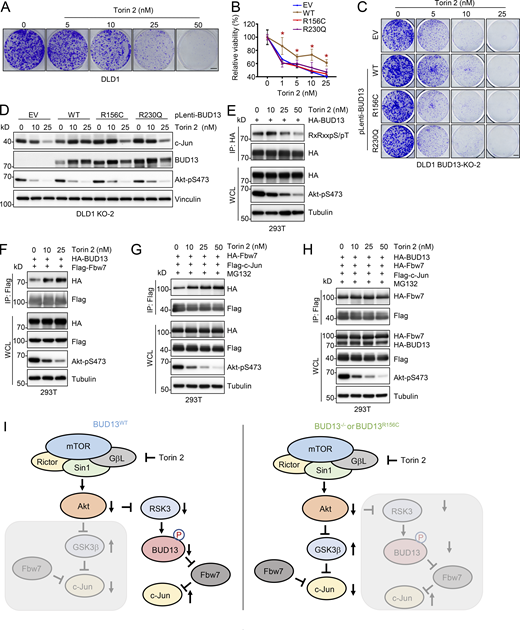
We next tried to find a potential therapeutic direction to treat colon cancer cells expressing these cancerous RSK3 phosphorylation deficient mutations. To this end, we found that either RSK3 mediates mTOR inhibition resistance in breast cancer (Serra et al., 2013) or mTOR contributes to RSK3 inhibition in small-cell lung cancer (Kumari et al., 2021,Preprint). Thus, we examined effects of mTOR inhibitors in regulating growth of DLD1 cells expressing WT-, R156C-, or R230Q-BUD13. Notably, similar levels of growth suppression in all of these cells were observed in treatments by mTORC1-specific inhibitors including rapamycin (Fig. S5 A), Temsirolimus (Fig. S5 B), and Everolimus (Fig. S5 C) or an S6K inhibitor (Fig. S5 D); however, compared with BUD13-deleted DLD1 cells or DLD1 cells expressing R156C- or R230Q-BUD13, WT-BUD13 expressing DLD1 cells displayed significant resistance to treatment by Torin 2 (Fig. 7, A–C), a pan-mTOR inhibitor suppressing both mTORC1 and mTORC2. Notably, rapamycin treatment didn’t significantly change c-Jun protein abundance (Fig. S5 E). On the other hand, Torin 2 treatment significantly reduced c-Jun levels in DLD1 cells expressing R156C- or R230Q-BUD13, but failed to do so in DLD1 cells expressing WT-BUD13 (Fig. 7 D)—this failure in degrading oncogenic c-Jun may explain why WT-BUD13 expressing cells exerted resistance to Torin 2 treatment (Fig. 7, B and C). Consistent with this notion, Torin 2 treatment reduced BUD13 phosphorylation in cells (Fig. 7 E), leading to enhanced BUD13 binding to Fbw7 (Fig. 7 F) that competed with c-Jun for Fbw7 binding. As a result, Torin 2-treatment-induced binding of c-Jun with Fbw7 (presumably through activating GSK3β to promote c-Jun phosphorylation and recognition by Fbw7 [Wei et al., 2005], Fig. 7 G) was largely abolished by BUD13 (Fig. 7 H). To further evaluate a unique role of mTORC2 but not mTORC1 in this regulation, we depleted Rictor, an essential mTORC2 complex component (Fig. S5 F). We found Rictor depletion significantly reduced Akt-pS473 signals (Fig. S5 F), and compared with WT-BUD13 expressing DLD1 cells, Rictor depletion caused more severe growth reduction in R156C- or R230Q-BUD13 expressing DLD1 cells (Fig. S5, G and H). Echoing for mTORC2 activation in negatively regulating BUD13 interactions with Fbw7 (Fig. 7 F), Rictor depletion similarly enhanced BUD13 binding to Fbw7 (Fig. S5 I). Together, these data suggest that in addition to the canonical GSK3-β induced c-Jun recognition and degradation by Fbw7 (Wei et al., 2005), BUD13 may function to protect c-Jun from being degraded by Fbw7 in responding to Torin 2 treatment, leading to cellular resistance to mTOR inhibition. This process depends on BUD13 expression and/or BUD13 phosphorylation such that cancerous R156C- or R230Q-BUD13 evading phosphorylation maintains sensitivity to Torin 2 inhibition (Fig. 7 I).
BUD13 expression contributes to DLD1 cellular resistance to Torin 2 treatment. (A–D) MTT cell viability assays measuring indicated DLD1 cell responses to indicated doses of compounds for 72 h. (E) IB analyses of WCL from DLD1 cells treated with indicated doses of rapamycin for 24 h before cell collection. (F) IB analyses of WCL derived from indicated DLD1 expressing indicated BUD13 mutants further depleted of endogenous Rictor by lentiviral infection. (G and H) Representative images for 2D colony formation assays using 500 indicated DLD1 cells for 10 d and quantified in H. Scale bar represents 5 mm. (I) IB analyses of BUD13-IP and WCL from indicated DLD1 cells. (J–M) IB analyses of WCL from DLD1 (J and L) or Caco2 (K and M) cells treated with indicated doses of JR-AB2-011 (J and K) or Torin 2 (L and M) overnight before cell collection. Statistical analysis was performed using one-way ANOVA. Asterisks show the significance between indicated groups. *, P < 0.05. All the data are representative of three independent experiments (error bars, mean ± SD). Source data are available for this SourceData FS5.
BUD13 expression contributes to DLD1 cellular resistance to Torin 2 treatment. (A–D) MTT cell viability assays measuring indicated DLD1 cell responses to indicated doses of compounds for 72 h. (E) IB analyses of WCL from DLD1 cells treated with indicated doses of rapamycin for 24 h before cell collection. (F) IB analyses of WCL derived from indicated DLD1 expressing indicated BUD13 mutants further depleted of endogenous Rictor by lentiviral infection. (G and H) Representative images for 2D colony formation assays using 500 indicated DLD1 cells for 10 d and quantified in H. Scale bar represents 5 mm. (I) IB analyses of BUD13-IP and WCL from indicated DLD1 cells. (J–M) IB analyses of WCL from DLD1 (J and L) or Caco2 (K and M) cells treated with indicated doses of JR-AB2-011 (J and K) or Torin 2 (L and M) overnight before cell collection. Statistical analysis was performed using one-way ANOVA. Asterisks show the significance between indicated groups. *, P < 0.05. All the data are representative of three independent experiments (error bars, mean ± SD). Source data are available for this SourceData FS5.
BUD13 mutations sensitize colon cancer cells to Torin 2 treatment. (A) Representative images for 2D colony formation assays using 3,000 DLD1 cells treated with indicated doses of Torin 2 for 2 wk. Scale bar represents 5 mm. (B) MTT cell viability assays measuring indicated BUD13 expressing DLD1 cells responses to indicated doses of Torin 2. (C) Representative images for 2D colony formation assays using 3,000 indicated DLD1 cells treated with indicated doses of Torin 2 since day 1 for 2 wk. Scale bar represents 5 mm. (D) IB analysis of WCL derived from indicated DLD1 cells treated with indicated doses of Torin 2 for 72 h before cell collection. (E) IB analysis of HA-IP and WCL derived from HEK293T cells transfected with indicated DNA constructs. (F–H) IB analysis of Flag-IP and WCL derived from HEK293T cells transfected with indicated DNA constructs. Where indicated, cells were treated with indicated doses of Torin 2 overnight or with 10 mM MG132 for 16 h before cell collection. (I) A proposed model for the involvement of BUD13 in cellular responses to Torin 2 treatment. Specifically, in WT-BUD13 cells, Torin 2 treatment inhibits mTORC2/Akt signaling that indirectly suppresses BUD13 phosphorylation, allowing for enhanced BUD13 interaction with Fbw7 to protect c-Jun from Fbw7-mediated ubiquitination and degradation—a process that overrides GSK3β-induced c-Jun degradation by Fbw7. On the other hand, in low BUD13 expressing cells or cells expressing BUD13-R156C phosphorylation-deficient mutants (such as R156C or R230Q), given BUD13 fails to bind Fbw7 to protect c-Jun, Torin 2 treatment induced GSK3β activation primes c-Jun for Fbw7 mediated degradation. Statistical analysis was performed using one-way ANOVA. Asterisks show the significance between indicated groups. *, P < 0.05. All the data are representative of three independent experiments (error bars, mean ± SD). Source data are available for this figure: SourceData F7.
BUD13 mutations sensitize colon cancer cells to Torin 2 treatment. (A) Representative images for 2D colony formation assays using 3,000 DLD1 cells treated with indicated doses of Torin 2 for 2 wk. Scale bar represents 5 mm. (B) MTT cell viability assays measuring indicated BUD13 expressing DLD1 cells responses to indicated doses of Torin 2. (C) Representative images for 2D colony formation assays using 3,000 indicated DLD1 cells treated with indicated doses of Torin 2 since day 1 for 2 wk. Scale bar represents 5 mm. (D) IB analysis of WCL derived from indicated DLD1 cells treated with indicated doses of Torin 2 for 72 h before cell collection. (E) IB analysis of HA-IP and WCL derived from HEK293T cells transfected with indicated DNA constructs. (F–H) IB analysis of Flag-IP and WCL derived from HEK293T cells transfected with indicated DNA constructs. Where indicated, cells were treated with indicated doses of Torin 2 overnight or with 10 mM MG132 for 16 h before cell collection. (I) A proposed model for the involvement of BUD13 in cellular responses to Torin 2 treatment. Specifically, in WT-BUD13 cells, Torin 2 treatment inhibits mTORC2/Akt signaling that indirectly suppresses BUD13 phosphorylation, allowing for enhanced BUD13 interaction with Fbw7 to protect c-Jun from Fbw7-mediated ubiquitination and degradation—a process that overrides GSK3β-induced c-Jun degradation by Fbw7. On the other hand, in low BUD13 expressing cells or cells expressing BUD13-R156C phosphorylation-deficient mutants (such as R156C or R230Q), given BUD13 fails to bind Fbw7 to protect c-Jun, Torin 2 treatment induced GSK3β activation primes c-Jun for Fbw7 mediated degradation. Statistical analysis was performed using one-way ANOVA. Asterisks show the significance between indicated groups. *, P < 0.05. All the data are representative of three independent experiments (error bars, mean ± SD). Source data are available for this figure: SourceData F7.
Discussion
In this study, we developed an algorithm and provided a publicly available resource (https://xianming-tan.shinyapps.io/motif/) that can be used to search for protein motif mutations in TCGA database. Using this tool, we characterize gained oncogenicity of the cancerous BUD13 AGC kinase motif mutations in promoting colon cancer growth. We further identify RSK3 as a major kinase responsible for BUD13 phosphorylation. Our mechanistic studies reveal a new BUD13 physiological function as an Fbw7 E3 ligase binding partner and regulator by competing with Fbw7 substrates binding to Fbw7 in stabilizing Fbw7 oncogenic substrates to exert an oncogenic function. RSK3-mediated BUD13 phosphorylation disrupts Fbw7 binding, leading to enhanced binding of Fbw7 with its substrates and subsequent degradation of these oncogenic substrates such as c-Jun and c-Myc. Thus, cancer tries to evade RSK3-mediated phosphorylation by maintaining RSK3-phosphorylation deficient BUD13 mutations including BUD13-R156C and -R230Q. Interestingly, the BUD13-R156C or -R230Q mutant utilizes a distinct mechanism than BUD13-WT to stabilize Fbw7 oncogenic substrates, and this mutation enhances BUD13 binding with Fbw7 to disrupt cullin 1 binding and formation of an active Fbw7Cul1 E3 ligase complex. This also leads to stabilization of Fbw7 oncogenic substrates, including c-Jun that fuels tumor growth. We further find that specifically inhibiting mTORC2 but not mTORC1 might serve as a potential therapeutic direction for treating colon cancer with no/low BUD13 expression or with RSK3 phosphorylation deficient BUD13 mutants. Interestingly, a previously developed mTORC2 inhibitor JR-AB2-011 (Benavides-Serrato et al., 2017) that interferes with mTOR binding to Rictor failed to inhibit mTORC2/Akt signaling in DLD1 and Caco2 cells (Fig. S5, J and K) where Torin 2 could do so (Fig. S5, L and M). This may urge to search for mTORC2-specific inhibitors more suitable for colon cancer.
Previous work reveals various endogenous inhibitors or pseudo-substrates for E3 ubiquitin ligases, such as Emi1 in suppressing APC/C (Reimann et al., 2001), hnRNP-U in inhibiting β-TRCP (Davis et al., 2002), and p105 NF-kB as a pseudo-substrate for FBXO7 (Udasin et al., 2021). In addition, an N-terminal pseudo-substrate motif in MDM2 has been shown to facilitate interactions of its N-terminal hydrophobic pocket with central acidic domain for MDM2 activation (Worrall et al., 2009). Moreover, LSD1 has also been reported as an Fbw7 pseudo-substrate that promotes Fbw7 self-ubiquitination and degradation by binding Fbw7 (Lan et al., 2019). Rictor has also been reported to be associated with Fbw7 to govern cyclin E and c-Myc protein ubiquitination (Guo et al., 2012). Given inhibiting certain E3 ligases (by blocking protein-protein interactions) shows promise in cancer treatment, including Skp2 inhibitors in blocking cell cycle progression (Wu et al., 2012) and restricting cancer stem cell traits (Chan et al., 2013), CRL4 inhibitors in triggering cell apoptosis (Wu et al., 2021), Mdm2/MdmX inhibitors in stabilizing p53 to cause cell death (Herman et al., 2011), and others, contributions of endogenous E3 ligase modulators including inhibitors/pseudo-substrates would need to be taken into consideration in responding to pharmacological E3 ligase manipulations.
As the growth of hallmarks of cancer in past two decades (Hanahan, 2022; Hanahan and Weinberg, 2011), in addition to hot-spot enzyme mutations that affect a large scale of downstream effectors, we hope our study reveals the existence of a group of enzyme-recognizing motif mutations that cancer hijacks to promote tumorigenesis, and targeting this type of mutations may provide new therapeutic directions for cancer treatments. Of note, a recent study comprehensively defining substrate sequence specificity for more than 300 human serine/threonine kinases (Johnson et al., 2023) provides a further guide to advance our understanding of additional kinase substrate motif mutations in human diseases including cancer.
Materials and methods
Materials
MG132 (S2619), BI-D1870 (S2843), and cycloheximide (S6611) were purchased from Selleck. Puromycin (P8833), Blasticidin (15205), iodonitrotetrazolium chloride (I10406), MTT (475989), anti-Flag agarose beads (A-2220), anti-HA agarose beads (A-2095), and glutathione agarose beads (G4510) were purchased from Sigma-Aldrich. Rapamycin (13346), Torin2 (14185), MK2206 (11593), GSK2110 (17988), S6K1-I (15018), Pazopanib (12097), Crizotinib (12087), Everolimus (11597), and Temsirolimus (11590) were purchased from Cayman Chemical. Active RSK3 protein (50-199-8827) was purchased from Thermo Fisher Scientific. JR-AB2-011 (HY-122022) was purchased from MedChemExpress.
Antibodies
All antibodies were used at a 1:1,000 dilution in TBS with 0.1% Tween 20 detergent buffer with 5% non-fat milk for Western blotting. Anti-Myc-Tag antibody (2278), anti-HA antibody (3724), anti-Phospho-Akt Substrate (RXRXXpS*/pT*) antibody (10001), anti-p-AKT (Ser473; 4060), anti- anti-p-p70 S6 Kinase (Thr389; 9234), anti-p-S6 Ribosomal Protein (Ser240/244; 5364), anti-p-4EBP1 (Thr37/46; 2855), pIRF3 antibody (Ser386; 37829), anti-IRF3 antibody (4302), anti-IRF7 antibody (4920), anti- anti-p-TBK1 (Ser172; 5483), anti-TBK1 antibody (51872), anti-STING antibody (13647), anti-c-Myc antibody (18583), anti-RSK1/2/3 antibody (9347), anti-Rictor antibody (9476), anti-Rbx1 (11922), anti-Skp1 antibody (12248), anti-rabbit IgG, HRP-linked antibody (7074), and anti-mouse IgG, HRP-linked antibody (7076) were obtained from Cell Signaling Technology. Anti-cyclin E antibody (sc-198), anti-β-catenin antibody (sc-59737), anti-c-Jun antibody (sc-45), anti-GST antibody (sc-459), anti-Cul1 (sc-11384), and anti-vinculin antibody (sc-25336) were obtained from Santa Cruz Biotechnology. Polyclonal anti-Flag antibody (F-7425), monoclonal anti-Flag antibody (F-3165, clone M2), and anti-α-tubulin antibody (T-5168) were obtained from Sigma-Aldrich. Anti-BUD13 antibody (20163-1-AP) and anti-Fbw7 antibody (28424-1-AP) were obtained from Proteintech.
Cell culture and transfection
Human colorectal cancer cell lines DLD1, HCT116, HT29, Caco2, Colo320, human immortalized kidney cell lines HEK293T, and human cervical adenocarcinoma cell line HeLa were cultured in DMEM medium supplemented with 10% FBS, 100 U penicillin, and 100 mg/ml streptomycin in 37°C incubator with 5% CO2. Cell transfection was performed using polyethylenimine, as described previously (Jiang et al., 2019; Su et al., 2021). Packaging of lentiviral small hairpin RNA (shRNA) or cDNA expressing viruses, as well as subsequent infection of various cell lines, were performed according to the protocols described previously (Jiang et al., 2019; Liu et al., 2014).
Plasmids
The pRP[Exp]-Bsd-CMV-hBUD13-HA vector was synthesized by Vector Builder. GST-BUD13 was cloned into pCMV-GST vector using XhoI and NotI enzyme sites. HA-BUD13-WT were cloned into pLenti-GFP-Blasticidin vector using XbaI and XhoI sites. R156C-BUD13 and R230Q-BUD13 related constructs were obtained using Site-Directed Mutagenesis Kits (200523) from Agilent. pET28a-BUD13 was cloned into pET28a vector using EcoRI and XhoI enzyme sites. HA-c-Jun and Flag-c-Jun were cloned into pCDNA3-HA and pCDNA3-Flag vectors using BamHI and SalI enzyme sites. Flag-AGC and HA-AGC constructs were either obtained from hORFeome V5.1 library owned by UNC Lineberger Tissue Culture Facility or subcloned into pCDNA3-HA vector. HA-Fbw7 and various truncations constructs were described in Inuzuka et al. (2011). Flag-Fbw7 was cloned into pCDNA3-Flag using BglII and SalI sites. Flag-c-Myc was cloned into pCDNA3-Flag vector using BamHI and SalI vector. Primers used for vector cloning are listed in Table 1. Myc-Culin1 was as described (Inuzuka et al., 2011).
Primer sequences (5′ to 3′) used for vector construction
| . | . |
|---|---|
| BUD13-XhoI-F | 5′-GCATCTCGAGCGGCGGCAGCTCCGCCG-3′ |
| BUD13-EcoRI-F | 5′-GCATGAATTCCGGCGGCAGCTCCGCCG-3′ |
| BUD13-NotI-R | 5′-GCATGCGGCCGCTTACATATCCTCAACACTCCATTTGTAGGC-3′ |
| BUD13-XbaI-F | 5′-GCATGCGGCCGCTTACATATCCTCAACACTCCATTTGTAGGC-3′ |
| BUD13-HA-XbaI-F | 5′-GCATTCTAGAGCCACCATGTACCCATACGATGTTCCAGATTACGCTGCGGCAGCTCCGC-3′ |
| BUD13-R230Q-F | 5′-CCCTCTCCTCCTAGGCAAGCCCGTCATGGTTCC-3′ |
| BUD13-R230Q-R | 5-GGAACCATGACGGGCTTGCCTAGGAGGAGAGGG-3′ |
| BUD13-R156P-F | 5′-CCTAGGAGGGCCCCTCATGACACCCCGGATCC-3′ |
| BUD13-R156P-R | 5′-GGATCCGGGGTGTCATGAGGGGCCCTCCTAGG-3′ |
| BUD13-R156C-F | 5′-CCTAGGAGGGCCTGTCATGACACCCCGGATCC-3′ |
| BUD13-R156C-R | 5′-GGATCCGGGGTGTCATGACAGGCCCTCCTAGG-3′ |
| BUD13-R156CT159E-F | 5′-CCTAGGAGGGCCTGTCATGACGAGCCGGATCC-3′ |
| BUD13-R156CT159E-R | 5′-GGATCCGGCTCGTCATGACAGGCCCTCCTAGG-3′ |
| Biotin-BUD13-RNA-F | biotin-5′-AUCAGUACCAGGUGAGTCUAUCAUG-3′ |
| Biotin-BUD13-RNA-R | 5′-CATGATAGACTCACCTG GTACTGAT-3′ |
| hcJun-BamHI-F | 5′-GCATGGATCCACTGCAAAGATGGAAACGACCTTC-3′ |
| hcJun-SalI-R | 5′-GCATGTCGACTCAAAATGTTTGCAACTGCTGCG-3′ |
| hcMyc-BamHI-F | 5′-GCATGGATCCCCCCTCAACGTTAGCTTCAC-3′ |
| hcMyc-SalI-R | 5′-GCATGTCGACTTACGCACAAGAGTTCCGTAGC-3′ |
| Fbw7-BglII-F | 5′-GCATAGATCTAATCAGGAACTGCTCTCTGTGG-3′ |
| Fbw7-SalI-R | 5′-GCATGTCGACTCACTTCATGTCCACATCAAAGTCC-3′ |
| . | . |
|---|---|
| BUD13-XhoI-F | 5′-GCATCTCGAGCGGCGGCAGCTCCGCCG-3′ |
| BUD13-EcoRI-F | 5′-GCATGAATTCCGGCGGCAGCTCCGCCG-3′ |
| BUD13-NotI-R | 5′-GCATGCGGCCGCTTACATATCCTCAACACTCCATTTGTAGGC-3′ |
| BUD13-XbaI-F | 5′-GCATGCGGCCGCTTACATATCCTCAACACTCCATTTGTAGGC-3′ |
| BUD13-HA-XbaI-F | 5′-GCATTCTAGAGCCACCATGTACCCATACGATGTTCCAGATTACGCTGCGGCAGCTCCGC-3′ |
| BUD13-R230Q-F | 5′-CCCTCTCCTCCTAGGCAAGCCCGTCATGGTTCC-3′ |
| BUD13-R230Q-R | 5-GGAACCATGACGGGCTTGCCTAGGAGGAGAGGG-3′ |
| BUD13-R156P-F | 5′-CCTAGGAGGGCCCCTCATGACACCCCGGATCC-3′ |
| BUD13-R156P-R | 5′-GGATCCGGGGTGTCATGAGGGGCCCTCCTAGG-3′ |
| BUD13-R156C-F | 5′-CCTAGGAGGGCCTGTCATGACACCCCGGATCC-3′ |
| BUD13-R156C-R | 5′-GGATCCGGGGTGTCATGACAGGCCCTCCTAGG-3′ |
| BUD13-R156CT159E-F | 5′-CCTAGGAGGGCCTGTCATGACGAGCCGGATCC-3′ |
| BUD13-R156CT159E-R | 5′-GGATCCGGCTCGTCATGACAGGCCCTCCTAGG-3′ |
| Biotin-BUD13-RNA-F | biotin-5′-AUCAGUACCAGGUGAGTCUAUCAUG-3′ |
| Biotin-BUD13-RNA-R | 5′-CATGATAGACTCACCTG GTACTGAT-3′ |
| hcJun-BamHI-F | 5′-GCATGGATCCACTGCAAAGATGGAAACGACCTTC-3′ |
| hcJun-SalI-R | 5′-GCATGTCGACTCAAAATGTTTGCAACTGCTGCG-3′ |
| hcMyc-BamHI-F | 5′-GCATGGATCCCCCCTCAACGTTAGCTTCAC-3′ |
| hcMyc-SalI-R | 5′-GCATGTCGACTTACGCACAAGAGTTCCGTAGC-3′ |
| Fbw7-BglII-F | 5′-GCATAGATCTAATCAGGAACTGCTCTCTGTGG-3′ |
| Fbw7-SalI-R | 5′-GCATGTCGACTCACTTCATGTCCACATCAAAGTCC-3′ |
shRNA constructs against human Fbw7 are as previously described (Inuzuka et al., 2011). sgRNA plasmids were constructed by inserting synthesized sgRNAs into lentiCRISPRv2-puro vector. Primers used for sgRNA plasmid construction are listed in Table 2.
Primer sequences (5′ to 3′) used for sgRNAs
| . | . |
|---|---|
| BUD13-sg2-F: | 5′-CACCGTCTGAAGCGTTACTTGTCCG-3′ |
| BUD13-sg2-R: | 5′-AAACCGGACAAGTAACGCTTCAGAC-3′ |
| BUD13-sg3-F: | 5′-CACCGCACGGAATAGACCTAGACAC-3′ |
| BUD13-sg3-R: | 5′-AAACGTGTCTAGGTCTATTCCGTGC-3′ |
| BUD13-sg4-F: | 5′-CACCGTCATGACGGGCCCTCCTTGG-3′ |
| BUD13-sg4-R: | 5′-AAACCCAAGGAGGGCCCGTCATGAC-3′ |
| PRKACB-sg1-F: | 5′-CACCGTACTCCAGTCGAACAAGGAA-3′ |
| PRKACB-sg1-R: | 5′-AAACTTCCTTGTTCGACTGGAGTAC-3′ |
| PRKACB-sg2-F: | 5′-CACCGTGGTTATGGAATATGTCCCT-3′ |
| PRKACB-sg2-R: | 5′-AAACAGGGACATATTCCATAACCAC-3′ |
| PRKACB-sg3-F: | 5′-CACCGACCCTTGGAACAGGTTCATT-3′ |
| PRKACB-sg3-R: | 5′-AAACAATGAACCTGTTCCAAGGGTC-3′ |
| PKC1-sg1-F: | 5′-CACCGGCTGTTTTGTGGTCCACAAG-3′ |
| PKC1-sg1-R: | 5′-AAACCTTGTGGACCACAAAACAGCC-3′ |
| PKC1-sg2-F: | 5′-CACCGTCTATGGACTTATCCATCAA-3′ |
| PKC1-sg2-R: | 5′-AAACTTGATGGATAAGTCCATAGAC-3′ |
| PKC1-sg3-F: | 5′-CACCGGAAGAACGTGCACGAGGTGA-3′ |
| PKC1-sg3-R: | 5′-AAACTCACCTCGTGCACGTTCTTCC-3′ |
| PKC2-sg1-F: | 5′-CACCGGCTGTATGGACTCATCCACC-3′ |
| PKC2-sg1-R: | 5′-AAACGGTGGATGAGTCCATACAGCC-3′ |
| PKC2-sg2-F: | 5′-CACCGGCTGCTTTGTGGTGCACAAG-3′ |
| PKC2-sg2-R: | 5'-AAACCTTGTGCACCACAAAGCAGCC-3' |
| PKC2-sg3-F: | 5′-CACCGCCACGTTTTGTGACCACTGT-3′ |
| PKC2-sg3-R: | 5′-AAACACAGTGGTCACAAAACGTGGC-3′ |
| GRK6-sg1-F: | 5′-CACCGGTGCTGACACTGATGAACGG-3′ |
| GRK6-sg1-R: | 5′-AAACCCGTTCATCAGTGTCAGCACC-3′ |
| GRK6-sg2-F: | 5′-CACCGAAGTGACCCCGGATGACAAG-3′ |
| GRK6-sg2-R: | 5′-AAACCTTGTCATCCGGGGTCACTTC-3′ |
| GRK6-sg3-F: | 5′-CACCGGGTGCTGACACTGATGAACG-3′ |
| GRK6-sg3-R: | 5′-AAACCGTTCATCAGTGTCAGCACCC-3′ |
| PKN3-sg1-F: | 5′-CACCGAAGCAGTTTCACCACGTTCT-3′ |
| PKN3-sg1-R: | 5′-AAACAGAACGTGGTGAAACTGCTTC-3′ |
| PKN3-sg2-F: | 5′-CACCGAACACAGGACCGCAAGGCAC-3′ |
| PKN3-sg2-R: | 5′-AAACGTGCCTTGCGGTCCTGTGTTC-3' |
| PKN3-sg3-F: | 5′-CACCGATGACCCACACGTGCGCCAG-3′ |
| PKN3-sg3-R: | 5′-AAACCTGGCGCACGTGTGGGTCATC-3′ |
| RSK3-sg1-F: | 5′-CACCGGTTTTAGGACAAGGATCCTA-3′ |
| RSK3-sg1-R: | 5′-AAACTAGGATCCTTGTCCTAAAACC-3′ |
| RSK3-sg2-F: | 5′-CACCGGGCTTTCTTAAGGACCTTCA-3′ |
| RSK3-sg2-R: | 5′-AAACTGAAGGTCCTTAAGAAAGCCC-3′ |
| RSK3-sg3-F: | 5′-CACCGCTGCTGAAGGTTTTAGGACA-3′ |
| RSK3-sg3-R: | 5′-AAACTGTCCTAAAACCTTCAGCAGC-3′ |
| RSK3-sg4-F: | 5′-CACCGTTGGACACGACGTCCTGCCA-3′ |
| RSK3-sg4-R: | 5′-AAACTGGCAGGACGTCGTGTCCAAC-3′ |
| RPS6KA4-sg1-F: | 5′-CACCGAAATCATCCGTAGCAAGACG-3′ |
| RPS6KA4-sg1-R: | 5′-AAACCGTCTTGCTACGGATGATTTC-3′ |
| RPS6KA4-sg2-F: | 5′-CACCGGAAATCATCCGTAGCAAGAC-3′ |
| RPS6KA4-sg2-R: | 5′-AAACGTCTTGCTACGGATGATTTCC-3′ |
| RPS6KA4-sg3-F: | 5′-CACCGAATGTGCTGCTGGACTCCGA-3′ |
| RPS6KA4-sg3-R: | 5′-AAACTCGGAGTCCAGCAGCACATTC-3′ |
| c-Jun-sg1-F: | 5′-CACCGTGCTCTGTTTCAGGATCTTG-3′ |
| c-Jun-sg1-R: | 5′-AAACCAAGATCCTGAAACAGAGCAC-3′ |
| c-Jun-sg3-F: | 5′-CACCGCAAGCTGGCGTCGCCCGAGC-3′ |
| c-Jun-sg3-R: | 5′-AAACGCTCGGGCGACGCCAGCTTGC-3′ |
| Rictor-sg-F: | 5′-CACCGAAGTGGACTATTTAATAGCT-3′ |
| Rictor-sg-R: | 5′-AAACTAGTAAAGTTATTCAGATGGC-3′ |
| . | . |
|---|---|
| BUD13-sg2-F: | 5′-CACCGTCTGAAGCGTTACTTGTCCG-3′ |
| BUD13-sg2-R: | 5′-AAACCGGACAAGTAACGCTTCAGAC-3′ |
| BUD13-sg3-F: | 5′-CACCGCACGGAATAGACCTAGACAC-3′ |
| BUD13-sg3-R: | 5′-AAACGTGTCTAGGTCTATTCCGTGC-3′ |
| BUD13-sg4-F: | 5′-CACCGTCATGACGGGCCCTCCTTGG-3′ |
| BUD13-sg4-R: | 5′-AAACCCAAGGAGGGCCCGTCATGAC-3′ |
| PRKACB-sg1-F: | 5′-CACCGTACTCCAGTCGAACAAGGAA-3′ |
| PRKACB-sg1-R: | 5′-AAACTTCCTTGTTCGACTGGAGTAC-3′ |
| PRKACB-sg2-F: | 5′-CACCGTGGTTATGGAATATGTCCCT-3′ |
| PRKACB-sg2-R: | 5′-AAACAGGGACATATTCCATAACCAC-3′ |
| PRKACB-sg3-F: | 5′-CACCGACCCTTGGAACAGGTTCATT-3′ |
| PRKACB-sg3-R: | 5′-AAACAATGAACCTGTTCCAAGGGTC-3′ |
| PKC1-sg1-F: | 5′-CACCGGCTGTTTTGTGGTCCACAAG-3′ |
| PKC1-sg1-R: | 5′-AAACCTTGTGGACCACAAAACAGCC-3′ |
| PKC1-sg2-F: | 5′-CACCGTCTATGGACTTATCCATCAA-3′ |
| PKC1-sg2-R: | 5′-AAACTTGATGGATAAGTCCATAGAC-3′ |
| PKC1-sg3-F: | 5′-CACCGGAAGAACGTGCACGAGGTGA-3′ |
| PKC1-sg3-R: | 5′-AAACTCACCTCGTGCACGTTCTTCC-3′ |
| PKC2-sg1-F: | 5′-CACCGGCTGTATGGACTCATCCACC-3′ |
| PKC2-sg1-R: | 5′-AAACGGTGGATGAGTCCATACAGCC-3′ |
| PKC2-sg2-F: | 5′-CACCGGCTGCTTTGTGGTGCACAAG-3′ |
| PKC2-sg2-R: | 5'-AAACCTTGTGCACCACAAAGCAGCC-3' |
| PKC2-sg3-F: | 5′-CACCGCCACGTTTTGTGACCACTGT-3′ |
| PKC2-sg3-R: | 5′-AAACACAGTGGTCACAAAACGTGGC-3′ |
| GRK6-sg1-F: | 5′-CACCGGTGCTGACACTGATGAACGG-3′ |
| GRK6-sg1-R: | 5′-AAACCCGTTCATCAGTGTCAGCACC-3′ |
| GRK6-sg2-F: | 5′-CACCGAAGTGACCCCGGATGACAAG-3′ |
| GRK6-sg2-R: | 5′-AAACCTTGTCATCCGGGGTCACTTC-3′ |
| GRK6-sg3-F: | 5′-CACCGGGTGCTGACACTGATGAACG-3′ |
| GRK6-sg3-R: | 5′-AAACCGTTCATCAGTGTCAGCACCC-3′ |
| PKN3-sg1-F: | 5′-CACCGAAGCAGTTTCACCACGTTCT-3′ |
| PKN3-sg1-R: | 5′-AAACAGAACGTGGTGAAACTGCTTC-3′ |
| PKN3-sg2-F: | 5′-CACCGAACACAGGACCGCAAGGCAC-3′ |
| PKN3-sg2-R: | 5′-AAACGTGCCTTGCGGTCCTGTGTTC-3' |
| PKN3-sg3-F: | 5′-CACCGATGACCCACACGTGCGCCAG-3′ |
| PKN3-sg3-R: | 5′-AAACCTGGCGCACGTGTGGGTCATC-3′ |
| RSK3-sg1-F: | 5′-CACCGGTTTTAGGACAAGGATCCTA-3′ |
| RSK3-sg1-R: | 5′-AAACTAGGATCCTTGTCCTAAAACC-3′ |
| RSK3-sg2-F: | 5′-CACCGGGCTTTCTTAAGGACCTTCA-3′ |
| RSK3-sg2-R: | 5′-AAACTGAAGGTCCTTAAGAAAGCCC-3′ |
| RSK3-sg3-F: | 5′-CACCGCTGCTGAAGGTTTTAGGACA-3′ |
| RSK3-sg3-R: | 5′-AAACTGTCCTAAAACCTTCAGCAGC-3′ |
| RSK3-sg4-F: | 5′-CACCGTTGGACACGACGTCCTGCCA-3′ |
| RSK3-sg4-R: | 5′-AAACTGGCAGGACGTCGTGTCCAAC-3′ |
| RPS6KA4-sg1-F: | 5′-CACCGAAATCATCCGTAGCAAGACG-3′ |
| RPS6KA4-sg1-R: | 5′-AAACCGTCTTGCTACGGATGATTTC-3′ |
| RPS6KA4-sg2-F: | 5′-CACCGGAAATCATCCGTAGCAAGAC-3′ |
| RPS6KA4-sg2-R: | 5′-AAACGTCTTGCTACGGATGATTTCC-3′ |
| RPS6KA4-sg3-F: | 5′-CACCGAATGTGCTGCTGGACTCCGA-3′ |
| RPS6KA4-sg3-R: | 5′-AAACTCGGAGTCCAGCAGCACATTC-3′ |
| c-Jun-sg1-F: | 5′-CACCGTGCTCTGTTTCAGGATCTTG-3′ |
| c-Jun-sg1-R: | 5′-AAACCAAGATCCTGAAACAGAGCAC-3′ |
| c-Jun-sg3-F: | 5′-CACCGCAAGCTGGCGTCGCCCGAGC-3′ |
| c-Jun-sg3-R: | 5′-AAACGCTCGGGCGACGCCAGCTTGC-3′ |
| Rictor-sg-F: | 5′-CACCGAAGTGGACTATTTAATAGCT-3′ |
| Rictor-sg-R: | 5′-AAACTAGTAAAGTTATTCAGATGGC-3′ |
Immunoblot (IB) and immunoprecipitations analyses
Cells were lysed in EBC buffer (50 mM Tris, pH 7.5, 120 mM NaCl, 0.5% NP-40) supplemented with protease inhibitor cocktail (EDTA-free, mini-tablet; Bimake) and phosphatase inhibitor cocktail (Bimake). The protein concentrations of whole-cell lysates (WCL) were measured by NanoDrop OneC using the Bio-Rad protein assay reagent as described previously (Jiang et al., 2019; Su et al., 2021). Equal amounts of WCL were loaded by SDS-PAGE and immunoblotted with indicated antibodies. For GST pulldown and immunoprecipitations analysis, 1 mg of total lysates were incubated with the indicated beads for 3–4 h at 4°C. The recovered immuno-complexes were washed three times with NETN buffer (20 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA, and 0.5% NP-40) before being resolved by SDS-PAGE and immunoblotted with indicated antibodies.
Colony formation assays
Indicated cells were seeded into 6-well plates (500 or 1,000 cells/well) and cultured in 37°C incubator with 5% CO2 for 10–15 d (as indicated in figure legends) until formation of visible colonies. Colonies were washed with distilled water, fixed with fixation buffer (10% acetic acid and 10% methanol) for 1 h, and stained with 1% crystal violet overnight. Colonies were then washed with distilled water and air-dried. Colony numbers were manually counted. Three independent experiments were performed to generate the error bars.
Soft agar assays
The anchorage-independent cell growth assays were performed as described previously (Jiang et al., 2019; Su et al., 2021). Briefly, the assays were preformed using 6-well plates where the solid medium consists of two layers. The bottom layer contains 0.8% noble agar and the top layer contains 0.4% agar suspended with 3 × 104 or indicated number of cells. 1 ml complete DMEM medium with 10% FBS was added every 7 d. About 2–5 wk later the cells were stained with 1 mg/ml iodonitrotetrazolium chloride overnight for colony visualization, imaging, and counting. At least two replicates were performed to generate the error bar.
Cell viability assays
Indicated number of cells were seeded in each well of 96-well plates to monitor cell viability at indicated time periods upon indicated treatments using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) according to manufacturer’s instruction. Briefly, at indicated time points post-cell seeding, MTT solution was added into each well at a final concentration of 0.5 mg/ml and incubated in the culture incubator (37°C with 5% CO2) for 3–4 h until crystal was observed. After thorough mixing, absorbance at 590 nm was measured using the BioTek Cytation 5 Cell Imaging reader.
Transwell assays
1 × 105 cells were plated in an 8.0-mm, 24-well plate chamber insert (catalog no. 3422; Corning Life Sciences) with serum-free DMEM medium at the top of the insert and the same medium containing 20% FBS at the bottom of the insert. Cells were incubated for 24 h and fixed with 4% paraformaldehyde for 15 min. After washing with PBS, cells at the top of the insert were scraped with a cotton swab. Cells adherent to the bottom were stained with 0.5% crystal violet blue for 60 min and then washed with double-distilled H2O. The positively stained cells were examined under the microscope.
Mouse xenograft assays
All mouse work has been reviewed and approved by University of North Carolina Institutional Animal Care and Use Committee under IACUC#19-031. Mouse xenograft assays were performed as described previously (Jiang et al., 2019; Su et al., 2021). Briefly, for mouse xenograft experiments, 2 × 106 DLD1 cells, 2 × 106 HCT116 cells, and 5 × 106 Caco2 cells were suspended in PBS and injected into the flank of indicated female nude mice (NCRNU-M-M from University of North Carolina Animal Facility, 4 wk old). Tumor size was monitored every 2–4 d with a digital caliper, and the tumor volume was determined with the formula L × W2 × 0.52, where L is the longest diameter and W is the shortest diameter. After 3–5 wk, mice were sacrificed, and tumors were dissected and weighed.
In vitro kinase assays
His-BUD13 protein was purified from Escherichia coli strain BL21 induced by 0.3 mM IPTG at 16°C overnight. 1 mg His-BUD13 protein and 250 ng active RSK3 protein were incubated in kinase buffer (pH = 7.5 50 mM Tris, 10 mM MgCl2, 2 mM DTT, 0.5 mM EDTA, 0.5 mM EGTA, 0.2 mM ATP) at 37°C for 1 h. Reaction was terminated by adding SDS loading buffer, and results were detected using relative antibody.
Bioinformatics analyses and data mining
We downloaded the mutation data from all TCGA projects (all cancer types) using the function GDCquery_Maf in the R package TCGAbiolinks. We downloaded the human proteome from the website (https://www.uniprot.org/proteomes/UP000005640). Each mutation record in the TCGA mutation data contains information including the mutation position, WT sequence, and mutated amino acid (AA). Combining this mutation information with the human proteome data, we obtained both the WT and mutated AA sequences around (e.g., ±6 AAs) this mutation position. We call the WT AA sequence “raw sequence,” and the mutated sequence the “mutation sequence.” Under the “supervised search” mode, for a given enzyme-recognizing motif, e.g., (AMPK [LxRxxS/T]), we decide, for each mutation record, if the raw sequence contains this protein kinase and whether the mutation sequence contains this protein kinase. We then decide if, in this mutation record, there was a “gain” of this enzyme-recognizing motif, i.e., the raw sequence does not contain this motif, but the mutation sequence contains this motif, or if there was a “loss” of this enzyme-recognizing motif, i.e., the raw sequence contains this enzyme-recognizing motif, but the mutation sequence does not contain this enzyme-recognizing motif. The TCGA mutation data also include information like case ID, cancer type, etc.; we could thus extract all cases that contain a gain or loss of a protein enzyme-recognizing motif sequence. This way, we obtained summary information like how many unique cases were involved, and what cancer types were involved in the gain or loss of each enzyme-recognizing motif.
Statistical analysis
Statistical analyses were performed using the SPSS 11.5 Statistical Software. P ≤ 0.05 was considered statistically significant. The results are shown as means + SD from at least two or three independent experiments as indicated in figure legends. Differences between control and experimental conditions were evaluated by one-way ANOVA.
Online supplemental materials
Fig. S1 shows a list of enzyme-recognizing motif sequences we searched in TCGA and BUD13 cancerous mutants are deficient in RSK3-mediated phosphorylation (related to Figs. 1 and 2). Fig. S2 shows that depletion of BUD13 reduces colon cancer growth and BUD13 AGC kinase motif mutations promote colon cancer growth (related to Fig. 3). Fig. S3 shows that BUD13-R156C mutation is not deficient in RNA binding and innate immunity (related to Fig. 4). Fig. S4 shows that BUD13 is a pseudo-substrate for Fbw7 and competes with c-Jun to bind Fbw7 (related to Figs. 4, 5, and 6). Fig. S5 shows that BUD13 expression contributes to DLD1 cellular resistance to Torin 2 treatment (related to Fig. 7). Table S1 shows a list of AGC kinase motif gain-of-function mutations in TCGA (related to Fig. 1). Table S2 shows a list of AGC kinase motif loss-of-function mutations in TCGA (related to Fig. 1).
Data availability
All data, code, and materials used in the analysis are available from the corresponding author upon reasonable request.
Acknowledgments
We thank Liu and Tan lab members for critical reading of the manuscript and helpful discussions. We also thank Alton Charles Gayton for help with the cloning processes.
This work is supported by National Institutes of Health grant R21CA270967 (P. Liu), Gabrielle’s Angel Foundation Medical Research Award (P. Liu), Department of Defense Congressionally Directed Medical Research Programs Kidney Cancer Research Program Idea Development Award W81XWH2110419 (P. Liu), North Carolina Biotechnology Center Flash Grant (P. Liu, X. Tan), and University of North Carolina at Chapel Hill University Cancer Research Fund (P. Liu).
Author contributions: Conceptualization: X. Tan and P. Liu; Methodology: J. Chen, X. Zhang, X. Tan, and P. Liu; Investigation: J. Chen and X. Zhang; Visualization: J. Chen and X. Zhang; Funding acquisition: P. Liu; Project administration: P. Liu; Supervision: X. Tan and P. Liu; Writing—original draft: J. Chen and P. Liu; Writing—review & editing: J. Chen, X. Zhang, X. Tan, and P. Liu.
References
Author notes
Disclosures: The authors declare no competing interests exist.