Hematopoietic differentiation is controlled by both genetic and epigenetic regulators. Long noncoding RNAs (lncRNAs) have been demonstrated to be important for normal hematopoiesis, but their function in erythropoiesis needs to be further explored. We profiled the transcriptomes of 16 murine hematopoietic cell populations by deep RNA sequencing and identified a novel lncRNA, Gm15915, that was highly expressed in erythroid-related progenitors and erythrocytes. For this reason, we named it lncEry. We also identified a novel lncEry isoform, which was the principal transcript that has not been reported before. lncEry depletion impaired erythropoiesis, indicating the important role of the lncRNA in regulating erythroid differentiation and maturation. Mechanistically, we found that lncEry interacted with WD repeat–containing protein 82 (WDR82) to promote the transcription of Klf1 and globin genes and thus control the early and late stages of erythropoiesis, respectively. These findings identified lncEry as an important player in the transcriptional regulation of erythropoiesis.
Introduction
Hematopoietic stem cells (HSCs) are multipotent precursors with the capacity to self-renew and differentiate into all mature blood cell types (Busch et al., 2015; Luo et al., 2015; Nestorowa et al., 2016; Wilson et al., 2008). During hematopoietic differentiation, long-term HSCs (LT-HSCs) differentiate into multiple blood cellular components (Orkin and Zon, 2008), including short-term HSCs (ST-HSCs), multipotent progenitor cells (MPPs), committed progenitor cells, and mature blood cells (Nakamura et al., 2010; Nilsson et al., 2000). Hematopoiesis is tightly regulated by various regulatory elements, including noncoding RNAs (ncRNAs), to maintain normal biological processes (Delas et al., 2019; Li et al., 2018).
In mammals, approximately two thirds of genomic DNA is pervasively transcribed (Ulitsky and Bartel, 2013), while <2% can be translated to proteins. Thus, genomic DNA transcribed into ncRNAs is better correlated with organismal complexity among species, suggesting that RNA-based regulatory mechanisms might be involved in the complex developmental processes of eukaryotes (Djebali et al., 2012; Fatica and Bozzoni, 2014). These ncRNAs can be divided into two main types: small ncRNAs and long ncRNAs (lncRNAs). lncRNAs are defined as transcripts of >200 nucleotides with no apparent open reading frames (Clark et al., 2015; Li et al., 2018; Mattick and Makunin, 2006). The function of lncRNAs is associated with their cellular localization: nuclear lncRNAs always perform their function through transcriptional regulation, chromatin interactions, and RNA processing, whereas cytoplasmic lncRNAs may regulate mRNA stability or translation and influence cellular signaling cascades (Batista and Chang, 2013; Morlando et al., 2015; Schmitt and Chang, 2016). Numerous functional lncRNAs have been discovered, including those that have a vital role in mediating hematopoiesis. For example, the oncofetal lncRNA gene H19 controls the balance between HSC quiescence and activation by regulating Igf2–Igf1r pathway activation (Venkatraman et al., 2013), promoting pre-HSC and HSC specification via the demethylation of hematopoietic transcription factors such as Runx1 and Spi1 (Zhou et al., 2019), or by participating in tumorigenesis (Raveh et al., 2015). lncHSC-1 and lncHSC-2 regulate the differentiation of myeloid and T cells, respectively (Luo et al., 2015), while lnc-DC regulates monocyte-derived dendritic cell differentiation through STAT3 binding (Wang et al., 2014). Erythropoiesis is also regulated by lncRNAs (An et al., 2014; Arriaga-Canon et al., 2014; Kulczynska and Siatecka, 2016; Paralkar et al., 2014; Shi et al., 2014; Xu and Shi, 2019); for example, lncRNA EPS (Hu et al., 2011) regulates the terminal differentiation of erythroid cells by promoting erythroid progenitor survival; lncRNA UCA1 controls erythropoiesis at the proerythroblast stage through the regulation of heme metabolism (Liu et al., 2018); and lnc-EC1 (Alvarez-Dominguez et al., 2014) and lnc-EC6 (Wang et al., 2015) regulate erythroblast enucleation. Yet, despite these advances in our knowledge of lncRNA identification, the function of most lncRNAs in erythropoiesis regulation remains largely unknown.
Erythroid Krüppel-like factor (EKLF; KLF1; Miller and Bieker, 1993) is a zinc-finger hematopoietic transcription factor that plays a global role in regulating the activation of genes in different stages of erythropoiesis (Gnanapragasam and Bieker, 2017; Mukherjee et al., 2021). The selective expression of Klf1 promotes erythropoiesis and represses megakaryopoiesis (Frontelo et al., 2007; Siatecka and Bieker, 2011). During maturation, nucleated erythrocytes (NuEs) shed their nucleus and progressively gain erythroid characteristics as well as synthesize hemoglobin, changing from NuEs to reticulocytes (Retic-Es) and ultimately mature blood cells. The dysfunction of hemoglobin can induce hemoglobinopathies such as β-thalassemia or sickle cell anemia (Stamatoyannopoulos, 2005). In differentiated erythroid cells, the remote regulatory sequences of the α-globin gene recruit polymerase II and the preinitiation complex, and then bind to transcription factors located in the promoter region to activate α-globin transcription (Vernimmen et al., 2007). Despite the depth of our understanding, we still need to ascertain the complexities of the transcriptional mechanisms regulating Klf1 and globin expression.
Although many functional lncRNAs are recognized as hematopoiesis mediators, the lncRNAs that regulate erythroid differentiation need clarification. Thus, we aimed to search for previously unidentified functional lncRNAs that play a role in erythroid differentiation by using high-throughput RNA sequencing (RNA-seq) approaches. We further investigated the mechanisms of IncRNA in the regulation of erythropoiesis by promoting the transcriptional activation of Klf1 and globin genes at different stages of erythropoiesis.
Results
Gm15915 is highly expressed in an erythroid lineage
Previous studies have confirmed the involvement of lncRNAs in many biological processes, including lineage differentiation (Guttman et al., 2011; Hung et al., 2011; Klattenhoff et al., 2013; Luo et al., 2015). To identify novel lncRNAs with biological relevance, we isolated 16 hematopoietic cell subsets from the bone marrow (BM) of C57BL/6 mice by FACS. These cell types included LT-HSCs, ST-HSCs, MPPs, common lymphoid progenitors (CLPs), common myeloid progenitors (CMPs), granulocyte-macrophage progenitors (GMPs), and megakaryocyte-erythroid progenitors (MEPs). We also isolated 10 mature lineage cell subsets: natural killer (NK) cells, B cells, CD4 T cells, CD8 T cells, monocytes, macrophages, granulocytes, megakaryocytes, and NuEs. Through RNA-seq, we identified 2,250 lncRNAs and 13,168 protein-coding genes. Thus, just a handful of highly expressed lncRNAs constituted the hematopoietic cell landscape (Fig. 1 A and Table S1). Consistent with previous studies (Venkatraman et al., 2013), we found that the classic lncRNA H19 was highly expressed in LT-HSCs (Fig. 1 A), illustrating the validity of our approach.
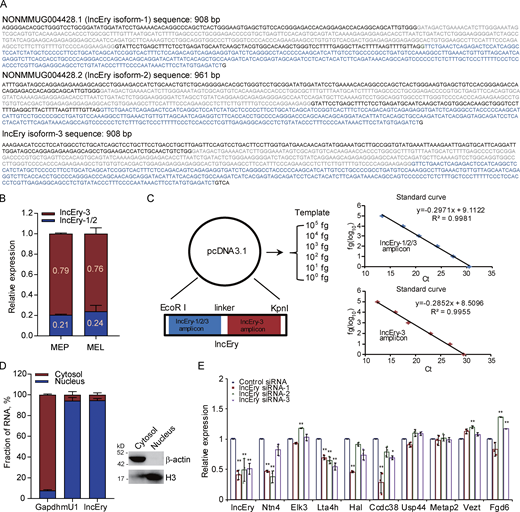
lncRNA Gm15915 is highly expressed in erythroid lineage. (A) Heatmap of lncRNA expression across 16 hematopoietic cell populations. Representative lncRNAs in different cell types are shown on the right. The top 10 lncRNAs are shown in Table S1. (B) RNA-seq analysis of lncEry expression in the 16 hematopoietic cell populations (upper panel). Correlation analysis of lncEry and erythroid lineage–related gene expression in the 16 hematopoietic cell populations (lower panel). Data are represented as mean ± SD (two samples). FPKM, fragments per kilobase of exon per million mapped fragments. (C) Schematic representation of the hematopoietic hierarchy showing the cell types used in this study; the intensity of the red color indicates the level of lncEry expression. MONO, monocytes; MR, macrophages; GR, granulocytes; MK, megakaryocytes. (D) GSEA showing −log10 of the uncorrected P values on the x axis; darker shading corresponds to a greater amount of enriched genes in each term. (E) 5′ and 3′ RACE assays and gel electrophoresis analysis to detect lncEry transcripts in of MEP cells. Three independent experiments. (F) RNA-seq tracks at lncEry loci with different read numbers linked to the different exons of the lncEry isoforms. The shared exons of the different isoforms are marked by the red frame. lncEry isoform-1, NONMMOUG004428.1; lncEry isoform-2, NONMMUG004428.2. (G) Western blot showing that there was no expression of Flag-tagged lncEry isoform-3 in all three reading frames in MEL cells. Flag-tagged KLF4 was used as a positive control. Full-length lncEry isoform-3 was cloned into the eukaryotic expression vector pcDNA3.1(−) with a 5′ terminal start codon ATG and a 3′ terminal Flag tag in all three reading frames; these plasmids were transfected into MEL cells separately and analyzed by Western blotting. Three independent experiments. (H) RNAscope and confocal microscopy analysis of lncEry subcellular localization in MEL cells. Ppib and DapB probes were used as positive and negative controls, respectively. The percentage of different lncEry subcellular localization points in >50 cells was calculated. Scale bar, 10 μm. Four independent experiments. Source data are available for this figure: SourceDataF1.
lncRNA Gm15915 is highly expressed in erythroid lineage. (A) Heatmap of lncRNA expression across 16 hematopoietic cell populations. Representative lncRNAs in different cell types are shown on the right. The top 10 lncRNAs are shown in Table S1. (B) RNA-seq analysis of lncEry expression in the 16 hematopoietic cell populations (upper panel). Correlation analysis of lncEry and erythroid lineage–related gene expression in the 16 hematopoietic cell populations (lower panel). Data are represented as mean ± SD (two samples). FPKM, fragments per kilobase of exon per million mapped fragments. (C) Schematic representation of the hematopoietic hierarchy showing the cell types used in this study; the intensity of the red color indicates the level of lncEry expression. MONO, monocytes; MR, macrophages; GR, granulocytes; MK, megakaryocytes. (D) GSEA showing −log10 of the uncorrected P values on the x axis; darker shading corresponds to a greater amount of enriched genes in each term. (E) 5′ and 3′ RACE assays and gel electrophoresis analysis to detect lncEry transcripts in of MEP cells. Three independent experiments. (F) RNA-seq tracks at lncEry loci with different read numbers linked to the different exons of the lncEry isoforms. The shared exons of the different isoforms are marked by the red frame. lncEry isoform-1, NONMMOUG004428.1; lncEry isoform-2, NONMMUG004428.2. (G) Western blot showing that there was no expression of Flag-tagged lncEry isoform-3 in all three reading frames in MEL cells. Flag-tagged KLF4 was used as a positive control. Full-length lncEry isoform-3 was cloned into the eukaryotic expression vector pcDNA3.1(−) with a 5′ terminal start codon ATG and a 3′ terminal Flag tag in all three reading frames; these plasmids were transfected into MEL cells separately and analyzed by Western blotting. Three independent experiments. (H) RNAscope and confocal microscopy analysis of lncEry subcellular localization in MEL cells. Ppib and DapB probes were used as positive and negative controls, respectively. The percentage of different lncEry subcellular localization points in >50 cells was calculated. Scale bar, 10 μm. Four independent experiments. Source data are available for this figure: SourceDataF1.
To gain insights into the expression of lncRNAs in erythropoiesis, we decided to focus on the lncRNAs specifically expressed in MEPs, of which Gm15915 was the most highly expressed (Fig. 1 A). Gm15915 was also highly expressed in LT-HSCs, CMPs, and NuEs (Fig. 1 B, upper panel). Gm15915 levels positively correlated with the expression of genes associated with erythroid-lineage development, such as Gata1, Klf1, Tal1, and Car1 (Fig. 1 B, lower panel), suggesting that Gm15915 is an erythroid lineage–specific lncRNA (Fig. 1 C). Hence, Gm15915 was named lncEry. To investigate lncEry further, we reanalyzed its expression in hematopoietic cells using a dataset from a previous study (Qian et al., 2016). lncEry was highly expressed in LT-HSCs, ST-HSCs, CMPs, MEPs, and NuEs (Fig. S1 A), confirming our findings. In addition, by analyzing single-cell RNA-seq data from a previous study (Nestorowa et al., 2016), we found lncEry to be highly expressed in the MEP population (Fig. S1 B). Next, we compared the expressions of lncEry in CMPs, GMPs, and MEPs to determine any correlation with cell fate and found that lncEry expression increased as CMPs differentiated into MEPs but not GMPs (Fig. S1 C).
Gm15915 is highly expressed in erythroid lineage. (A) Heatmap of differentially expressed lncRNAs in 17 hematopoietic cell populations and representative lncRNAs in different cell types. Histogram shows the expression of lncEry in each cell population. FPKM, fragments per kilobase of exon per million mapped fragments. (B) Diffusion map of all cells colored according to the expression of lncEry genes. Diffusion of all cells and MEP cells is shown in purple. The color corresponds to a log2 scale of expression ranging from 0 to the maximum value for the gene. Diffusion components 1, 2, and 3 are shown. (C) Volcano plots comparing the DEGs (dots) between MEPs and CMPs, MEPs and GMPs, or GMPs and CMPs. Red or green dots with black circles show representative upregulated and downregulated genes, respectively. Gm15915 is indicated by larger dots. (D) Heatmap and unsupervised hierarchical clustering of DEGs in 16 hematopoietic cell populations. 10 cluster types were grouped according to the transcriptome profiles. (E) Line chart showing cluster 9 gene expression in the 16 hematopoietic cell populations. MONO, monocytes; MR, macrophages; GR, granulocytes; MK, megakaryocytes.
Gm15915 is highly expressed in erythroid lineage. (A) Heatmap of differentially expressed lncRNAs in 17 hematopoietic cell populations and representative lncRNAs in different cell types. Histogram shows the expression of lncEry in each cell population. FPKM, fragments per kilobase of exon per million mapped fragments. (B) Diffusion map of all cells colored according to the expression of lncEry genes. Diffusion of all cells and MEP cells is shown in purple. The color corresponds to a log2 scale of expression ranging from 0 to the maximum value for the gene. Diffusion components 1, 2, and 3 are shown. (C) Volcano plots comparing the DEGs (dots) between MEPs and CMPs, MEPs and GMPs, or GMPs and CMPs. Red or green dots with black circles show representative upregulated and downregulated genes, respectively. Gm15915 is indicated by larger dots. (D) Heatmap and unsupervised hierarchical clustering of DEGs in 16 hematopoietic cell populations. 10 cluster types were grouped according to the transcriptome profiles. (E) Line chart showing cluster 9 gene expression in the 16 hematopoietic cell populations. MONO, monocytes; MR, macrophages; GR, granulocytes; MK, megakaryocytes.
To further investigate lncEry function, we performed unsupervised hierarchical clustering on the expression levels of protein-coding and lncRNA genes. We defined 10 clusters and hypothesized that the genes expressed within the same cluster might have similar functions (Fig. S1 D). The genes in cluster 9 were highly expressed in MEPs and NuEs (Fig. S1 E). We found known erythropoiesis-associated genes, such as Gata1, Klf1, Tal1, and Car1, and lncEry in cluster 9. Finally, gene ontology (GO) enrichment analysis showed that the coding genes in cluster 9 were significantly enriched for erythrocyte differentiation, supporting the functional role of lncEry in erythroid differentiation (Fig. 1 D). Taken together, our data suggest that lncEry is potentially involved in regulating erythroid differentiation.
lncEry is a bona fide lncRNA
To gain insights into the molecular characteristics of lncEry, we performed 5′ and 3′ rapid amplification of cDNA ends (RACE) PCR of MEP clones, followed by Sanger sequencing, to identify lncEry transcript isoforms (Fig. 1 E). Two isoforms (isoform-1, NONMMUG004428.1, and isoform-2, NONMMUG0048.2) are annotated in the NONCODE database, and we discovered a third isoform, isoform-3, that shares four incomplete exons (exons 2, 3, 4, and 6 of isoform-1) with the other two (Fig. 1 F).
After analyzing the detailed sequence information (Fig. S2 A) and the RNA-seq read coverage of the lncEry gene in MEPs, we found lncEry isoform-3 to be the principal transcript. Specifically, ∼79.5 of lncEry was expressed as isoform-3 and ∼20.5% as isoform-1/2 (Fig. S2, B and C). Similarly, 76% of lncEry in the mouse erythroleukemia (MEL) cell line was expressed as isoform-3 (Fig. S2, B and C). Next, we examined the coding capacity of isoform-3. Full-length isoform-3 was inserted into the eukaryotic expression vector pcDNA3.1 with 5′ terminal start codon ATG and 3′ terminal Flag tag in all three reading frames, and the results confirmed the non–protein-coding characteristics of isoform-3 (Fig. 1 G).
lncEry is a bona fide lncRNA. (A) Nucleotide sequences of lncEry isoforms. (B) Expression of different lncEry isoforms in MEP and MEL cells analyzed by qPCR. (C) Schematic of approach used to quantify the relative expression of lncEry isoform-1/2/3 and isoform-3. Both lncEry isoform-1/2/3 and isoform-3 amplicons were cloned in tandem into the same plasmid. Five different dilutions were made for the qPCR to generate the standard curve. The amount of lncEry isoform-1/2/3 and isoform-3 was calculated by fitting the cycle threshold value to the respective standard curve. The amount of lncEry isoform-1/2 was calculated by subtracting the value of lncEry isoform-3 from the value of lncEry isoform-1/2/3 (left). Representative standard curves for lncEry isoform-1/2/3 and isoform-3 (right). (D) The lncEry fraction in the cytosol and nucleus. Gapdh and mU1 were used as markers of the cytosolic and nuclear RNA fractions, respectively. The separation efficiency of each cell component was determined by Western blotting (right). (E) MEL cells were transfected with control or lncEry siRNA, and the expression levels of indicated genes were analyzed by qPCR (n = 3 samples). Three independent experiments for B–E. Data are represented as mean ± SD. *, P < 0.05; **, P < 0.01; one-way ANOVA. Source data are available for this figure: SourceDataFS2.
lncEry is a bona fide lncRNA. (A) Nucleotide sequences of lncEry isoforms. (B) Expression of different lncEry isoforms in MEP and MEL cells analyzed by qPCR. (C) Schematic of approach used to quantify the relative expression of lncEry isoform-1/2/3 and isoform-3. Both lncEry isoform-1/2/3 and isoform-3 amplicons were cloned in tandem into the same plasmid. Five different dilutions were made for the qPCR to generate the standard curve. The amount of lncEry isoform-1/2/3 and isoform-3 was calculated by fitting the cycle threshold value to the respective standard curve. The amount of lncEry isoform-1/2 was calculated by subtracting the value of lncEry isoform-3 from the value of lncEry isoform-1/2/3 (left). Representative standard curves for lncEry isoform-1/2/3 and isoform-3 (right). (D) The lncEry fraction in the cytosol and nucleus. Gapdh and mU1 were used as markers of the cytosolic and nuclear RNA fractions, respectively. The separation efficiency of each cell component was determined by Western blotting (right). (E) MEL cells were transfected with control or lncEry siRNA, and the expression levels of indicated genes were analyzed by qPCR (n = 3 samples). Three independent experiments for B–E. Data are represented as mean ± SD. *, P < 0.05; **, P < 0.01; one-way ANOVA. Source data are available for this figure: SourceDataFS2.
The functions of most lncRNAs are restricted to their subcellular localization (Batista and Chang, 2013; Chen, 2016; Chen and Carmichael, 2010). We thus performed RNAscope assays to identify the location of lncEry in MEL cells. Nearly 90% of lncEry molecules localized to the nucleus (Fig. 1 H), which was confirmed by subcellular fractionation assay followed by quantitative RT-PCR (qRT-PCR; Fig. S2 D). We presumed that the nuclear location of this lncRNA indicated its involvement in transcriptional regulation (Carlevaro-Fita and Johnson, 2019). To prove this hypothesis, we transfected MEL cells with siRNAs targeting lncEry and analyzed the expression of the genes found within 1 Mbp upstream and downstream of the lncEry locus on chromosome 10. lncEry downregulation not only affected the expression levels of adjacent genes, such as Ntn4 and Ccdc38, but also influenced (to some extent) the expression of genes located >100 kbps distant, e.g., Hal and Lta4h (Fig. S2 E). We thus speculated that lncEry has transcription regulatory capacities.
Erythroid differentiation is impaired upon lncEry knockdown in hematopoietic stem and progenitor cells (HSPCs)
To determine the functional impact of lncEry on erythroid differentiation, we transduced cKit+ cells with a GFP-expressing lentivirus carrying lncEry shRNA, and then performed in vitro CFU and in vivo transplantation assays (Fig. S3 A). First, we confirmed that the two shRNA constructs exhibited high-knockdown efficiencies at the mRNA level (Fig. S3 B). The CFU assays showed that lncEry knockdown decreased the number of CFU-GM colonies by 32% on average and more potently decreased the number of BFU-E (burst-forming unit erythroid)forming unit erythroid) and CFU-GEMM (colony-forming unit − granulocyte, erythroid, macrophage, megakaryocyte) colonies (74 and 66% on average, respectively; Fig. S3 C). In addition, colony sizes were significantly decreased upon lncEry knockdown (Fig. S3 D).
Erythroid differentiation is impaired by lncEry knockdown in HSPCs. (A) Schematic of the experimental procedure of gene knockdown. Briefly, GFP-fused control or lncEry shRNA lentiviruses were transduced into donor (CD45.1+) murine cKit+ HSPCs, which were injected into lethally irradiated (9.5 Gy) recipient (CD45.2+) mice with the indicated GFP+ percentage, or GFP+ cells were sorted and analyzed by colony assay. (B) cKit+ cells stably expressing different sets of lncEry shRNAs were collected for qPCR analysis (n = 4). (C) GFP+ cells were cultured for 10–14 d for CFU assays in complete methylcellulose-based medium, and colonies were counted (n = 4). (D) Colony assays of GFP+ cells transfected with control or lncEry shRNAs. Representative images from triplicate experiments are shown. Scale bar, 200 μm. (E) Percentage of GFP+ cells before transplantation into recipient mice (n = 5–7 mice per group). (F) Absolute numbers or concentrations of indicated items in PB 21 d after transplantation (n = 3–7 mice per group). (G and H) Percentage of GFP+ cells in recipient mice PB or BM 21 d after transplantation (n = 3–7 mice per group). (I) Percentage of Pro-Es, basophilic erythroblasts (Baso-Es), polychromatic erythroblasts (Poly-Es), orthochromatic erythroblasts (Ortho-Es), Retic-Es, or RBCs in GFP+ cells of recipient mice BM (n = 3–7 mice per group). Two independent experiments for B–I. Data are represented as mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; one-way ANOVA.
Erythroid differentiation is impaired by lncEry knockdown in HSPCs. (A) Schematic of the experimental procedure of gene knockdown. Briefly, GFP-fused control or lncEry shRNA lentiviruses were transduced into donor (CD45.1+) murine cKit+ HSPCs, which were injected into lethally irradiated (9.5 Gy) recipient (CD45.2+) mice with the indicated GFP+ percentage, or GFP+ cells were sorted and analyzed by colony assay. (B) cKit+ cells stably expressing different sets of lncEry shRNAs were collected for qPCR analysis (n = 4). (C) GFP+ cells were cultured for 10–14 d for CFU assays in complete methylcellulose-based medium, and colonies were counted (n = 4). (D) Colony assays of GFP+ cells transfected with control or lncEry shRNAs. Representative images from triplicate experiments are shown. Scale bar, 200 μm. (E) Percentage of GFP+ cells before transplantation into recipient mice (n = 5–7 mice per group). (F) Absolute numbers or concentrations of indicated items in PB 21 d after transplantation (n = 3–7 mice per group). (G and H) Percentage of GFP+ cells in recipient mice PB or BM 21 d after transplantation (n = 3–7 mice per group). (I) Percentage of Pro-Es, basophilic erythroblasts (Baso-Es), polychromatic erythroblasts (Poly-Es), orthochromatic erythroblasts (Ortho-Es), Retic-Es, or RBCs in GFP+ cells of recipient mice BM (n = 3–7 mice per group). Two independent experiments for B–I. Data are represented as mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; one-way ANOVA.
Next, we infected donor cells with lncEry-shRNA–carrying lentiviruses, and after 48 h of culture, achieved transduction efficiencies of ∼94, 84, and 40% for control, shRNA-1, and shRNA-2, respectively (Fig. S3 E). We then transplanted the transduced cells (without sorting) into lethally irradiated mouse recipients (Fig. S3 A). 21 d after transplantation, two recipients from the lncEry shRNA-1 group died, and the remaining three mice in the lncEry shRNA-1 group showed pale paws and were moribund, indicating severe anemia. The recipients of lncEry shRNA-1 and lncEry shRNA-2 had decreased numbers of white blood cells (WBCs) and RBCs (Fig. S3 F), as well as very low hemoglobin levels compared with the controls (Fig. S3 F). Finally, the lncEry knockdown animals showed a decrease in the percentage of GFP+ cells in the peripheral blood (PB) and BM (Fig. S3, G and H) and in the percentage of Retic-Es and RBCs in the GFP+ cells within the BM (Fig. S3 I). We thus considered that lncEry is involved in erythroid differentiation from HSPCs.
Erythroid differentiation is impaired in lncEry knockout mice
To further study the function of lncEry in erythroid differentiation, we generated lncEryfl/fl (flox/flox) mice (Fig. S4 A). We then generated Mx1-Cre; lncEryfl/fl (Δ/Δ) mice, in which the lncEry deletion could be induced by poly(I:C) administration (Aliprantis et al., 2008; Ruocco et al., 2005). We analyzed the expression of lncEry isoforms in the BM cells of both lncEryfl/fl and Δ/Δ mice: excision of exons 1/2 of lncEry isoform-3 strongly decreased the expression of isoform-3 as well as the other two lncEry isoforms (Fig. 2 A).
Erythroid differentiation is impaired in lncEry Δ/Δ mouse. (A) Schematic of the loxP sequence integration into exons 1–2 of lncEry isoform-3 using CRISPR/Cas9 technology. (B) Cell numbers in flox/flox and Δ/Δ mouse BM (n = 8–9 mice per group). (C) Gating strategies for FACS analysis of T, B, and myeloid cells. MAC, magnetic activated cell sorting; SSC, side scatter. (D) Plot of CD44 vs. FSC of the Ter119-positive cells with gating for Pro-E, Baso-E, Poly-E, Ortho-E, Retic-E, or RBC populations. FSC, forward scatter. (E) Percentage of indicated cell populations in Ter119+ cells from control and lncEry cKO mice (n = 4 mice per group). (F) Gating strategies for CD71/Ter119 staining (R1, Pro-E; R2, Baso-E; R3, Poly-E; R4, Ortho-E, Retic-E, RBC). (G) Percentage of indicated cell populations in the BM of flox/flox or Δ/Δ mice (n = 4 mice per group). (H) Percentage of indicated cell populations in BM of flox/flox or Δ/Δ mice treated with PHZ (100 μg/g) for 72 h (n = 4–5 mice per group). (I) Experimental design for noncompetitive transplantation. Mice were treated with 25 μg/g poly(I:C) three times every other day before transplantation. (J) Absolute numbers or concentrations of the indicated items in the PB 2 mo after transplantation. (K) Percentage of indicated cell populations in the BM of recipient mice 2 mo after transplantation (n = 5–7 mice per group). (L) Gating strategies for FACS of the indicated cells. (M and N) Percentage of indicated cell populations in BM cells from control and lncEry cKO mice (M) and related gating strategies (N; n = 4–5 mice per group). (O–Q) Percentage of indicated cell populations in the spleens of flox/flox or Δ/Δ mice (n = 4–5 mice per group). (R and S) Percentage of indicated cell populations in the fetal livers of Vav-Cre;lncEryfl/fl or lncEryfl/fl mice at E13.5 (n = 6–8 mice per group). (T) BFU-E colony assays of 500 control or lncEry cKO BM cells cultured in methylcellulose-based medium with EPO cytokine stimulation for 10–14 d. Scale bar, 100 μm (n = 5 wells). Three independent experiments for B, E, G, M, and O–T. Two independent experiments for H and J + K. Data are represented as mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; unpaired two-tailed Student’s t test.
Erythroid differentiation is impaired in lncEry Δ/Δ mouse. (A) Schematic of the loxP sequence integration into exons 1–2 of lncEry isoform-3 using CRISPR/Cas9 technology. (B) Cell numbers in flox/flox and Δ/Δ mouse BM (n = 8–9 mice per group). (C) Gating strategies for FACS analysis of T, B, and myeloid cells. MAC, magnetic activated cell sorting; SSC, side scatter. (D) Plot of CD44 vs. FSC of the Ter119-positive cells with gating for Pro-E, Baso-E, Poly-E, Ortho-E, Retic-E, or RBC populations. FSC, forward scatter. (E) Percentage of indicated cell populations in Ter119+ cells from control and lncEry cKO mice (n = 4 mice per group). (F) Gating strategies for CD71/Ter119 staining (R1, Pro-E; R2, Baso-E; R3, Poly-E; R4, Ortho-E, Retic-E, RBC). (G) Percentage of indicated cell populations in the BM of flox/flox or Δ/Δ mice (n = 4 mice per group). (H) Percentage of indicated cell populations in BM of flox/flox or Δ/Δ mice treated with PHZ (100 μg/g) for 72 h (n = 4–5 mice per group). (I) Experimental design for noncompetitive transplantation. Mice were treated with 25 μg/g poly(I:C) three times every other day before transplantation. (J) Absolute numbers or concentrations of the indicated items in the PB 2 mo after transplantation. (K) Percentage of indicated cell populations in the BM of recipient mice 2 mo after transplantation (n = 5–7 mice per group). (L) Gating strategies for FACS of the indicated cells. (M and N) Percentage of indicated cell populations in BM cells from control and lncEry cKO mice (M) and related gating strategies (N; n = 4–5 mice per group). (O–Q) Percentage of indicated cell populations in the spleens of flox/flox or Δ/Δ mice (n = 4–5 mice per group). (R and S) Percentage of indicated cell populations in the fetal livers of Vav-Cre;lncEryfl/fl or lncEryfl/fl mice at E13.5 (n = 6–8 mice per group). (T) BFU-E colony assays of 500 control or lncEry cKO BM cells cultured in methylcellulose-based medium with EPO cytokine stimulation for 10–14 d. Scale bar, 100 μm (n = 5 wells). Three independent experiments for B, E, G, M, and O–T. Two independent experiments for H and J + K. Data are represented as mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; unpaired two-tailed Student’s t test.
Erythroid differentiation is impaired in lncEry Δ/Δ mouse. (A) qRT-PCR analysis of lncEry isoform expression in flox/flox or Δ/Δ BM cells (n = 3 samples). (B and C) Percentage of indicated cell populations in flox/flox or Δ/Δ mice BM (n = 4–5 mice per group). (D) Absolute numbers or concentrations of the indicated items in the PB of flox/flox or Δ/Δ mice (n = 3–4 mice per group). (E and F) Percentage of indicated cell populations in the BM of flox/flox or Δ/Δ mice (n = 4–6 mice per group). (G) 1 × 104 BM cells from flox/flox or Δ/Δ mice were cultured for 10–14 d in assays in complete methylcellulose-based medium, and colony numbers were counted (n = 5 for flox/flox, n = 6 for Δ/Δ). (H) BFU-E CFU assays of 1 × 104flox/flox or Δ/Δ mice BM cells cultured in methylcellulose-based medium with EPO cytokine supplementation for 10–14 d. Representative images from triplicate experiments are shown (n = 6 wells). (I) CFU-E colony assays of 500 flox/flox or Δ/Δ mice MEP cells cultured in methylcellulose-based medium with EPO cytokine for 48 h. Representative images from triplicate experiments are shown. Scale bar, 50 μm (n = 5 wells). (J) Experimental flow chart of competitive transplantation; mice were treated with poly I:C 25 μg/g three times every other day before transplantation. (K) Percentage of CD45.2+ cells in the PB of recipient (CD45.1+) mice (n = 4–7 mice per group). (L) Percentage of CD45.2+ cells in indicated populations 4 mo after transplantation (n = 7 mice per group). (M) Repopulating units (RU) of donor cells calculated after 4-mo reconstitution (n = 7 mice per group). (N) Experimental design for competitive transplantation. Mice were treated with 25 μg/g poly I:C three times every other day before transplantation. 300 LT-HSCs sorted from flox/flox or Δ/Δ mice were transplanted into irradiated recipients together with 3 × 105 competitor cells. (O) Percentage of CD45.2+ cells in the PB of recipient (CD45.1+) mice (n = 4–7 mice). Three to four independent experiments for A–I; two independent experiments for J–O. Data are represented as mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; unpaired two-tailed Student’s t test.
Erythroid differentiation is impaired in lncEry Δ/Δ mouse. (A) qRT-PCR analysis of lncEry isoform expression in flox/flox or Δ/Δ BM cells (n = 3 samples). (B and C) Percentage of indicated cell populations in flox/flox or Δ/Δ mice BM (n = 4–5 mice per group). (D) Absolute numbers or concentrations of the indicated items in the PB of flox/flox or Δ/Δ mice (n = 3–4 mice per group). (E and F) Percentage of indicated cell populations in the BM of flox/flox or Δ/Δ mice (n = 4–6 mice per group). (G) 1 × 104 BM cells from flox/flox or Δ/Δ mice were cultured for 10–14 d in assays in complete methylcellulose-based medium, and colony numbers were counted (n = 5 for flox/flox, n = 6 for Δ/Δ). (H) BFU-E CFU assays of 1 × 104flox/flox or Δ/Δ mice BM cells cultured in methylcellulose-based medium with EPO cytokine supplementation for 10–14 d. Representative images from triplicate experiments are shown (n = 6 wells). (I) CFU-E colony assays of 500 flox/flox or Δ/Δ mice MEP cells cultured in methylcellulose-based medium with EPO cytokine for 48 h. Representative images from triplicate experiments are shown. Scale bar, 50 μm (n = 5 wells). (J) Experimental flow chart of competitive transplantation; mice were treated with poly I:C 25 μg/g three times every other day before transplantation. (K) Percentage of CD45.2+ cells in the PB of recipient (CD45.1+) mice (n = 4–7 mice per group). (L) Percentage of CD45.2+ cells in indicated populations 4 mo after transplantation (n = 7 mice per group). (M) Repopulating units (RU) of donor cells calculated after 4-mo reconstitution (n = 7 mice per group). (N) Experimental design for competitive transplantation. Mice were treated with 25 μg/g poly I:C three times every other day before transplantation. 300 LT-HSCs sorted from flox/flox or Δ/Δ mice were transplanted into irradiated recipients together with 3 × 105 competitor cells. (O) Percentage of CD45.2+ cells in the PB of recipient (CD45.1+) mice (n = 4–7 mice). Three to four independent experiments for A–I; two independent experiments for J–O. Data are represented as mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; unpaired two-tailed Student’s t test.
To monitor the effects of lncEry on the hematopoietic system, we analyzed BM cells from flox/flox and Δ/Δ mice by flow cytometry. We found that the Δ/Δ mice had normal BM cellularity, with unbiased lymphoid and myeloid differentiation (Figs. 2 B and S4, B and C) and a decreased percentage of terminally differentiated erythrocytes in BM (Chen et al., 2009; Liu et al., 2013; Fig. 2 C and Fig. S4, D–G). These findings are consistent with those of our knockdown assay (Fig. S3 I). In this case, however, it seemed that complete loss of lncEry might have impaired the terminal differentiation of erythropoiesis. To investigate how lncEry is involved in terminal erythroid differentiation, we analyzed several parameters in the BM of flox/flox and Δ/Δ mice (Fig. 2 C). We found that knocking out lncEry reduced the number of RBCs and Retic-Es as well as the concentration of hemoglobin (Fig. 2 D) but had no effect on WBCs in Δ/Δ mice. We also detected stress erythropoiesis by using phenylhydrazine (PHZ) treatment and a noncompetitive transplantation. The results showed that stress induced a more severe phenotype compared with steady state (Fig. 2 C and Fig. S4, H–K).
Next, we examined the effects of knocking out lncEry on HSPCs. The percentages of most HSPC subsets (including LT-HSCs, ST-HSCs, MPP2, MPP3/4, CMPs, MEPs, and MKPs) were decreased in the BM of Δ/Δ mice compared with their littermate controls (Fig. 2, E and F; and S4, L–N). Furthermore, the percentages of preMegE, PreCFU-E, and CFU-E/proerythroblast (Pro-E) were significantly reduced in the BM of Δ/Δ mice (Fig. 2 F), whereas CLPs and GMPs were minimally affected (Fig. 2 F and Fig. S4, M and N). Interestingly, lncEry deletion had a mild effect on HSPCs and terminal erythroid cell populations in spleen and fetal liver (Fig. S4, O–S). Therefore, loss of lncEry apparently impairs the differentiation of erythroid lineage cells in adult BM.
To examine the function of lncEry in erythropoiesis, we compared the colony-forming ability of BM cells from flox/flox and Δ/Δ mice during in vitro culture in complete methylcellulose-based medium. We found that the colony numbers of BFU-E, CFU-G, and CFU-GEMM from Δ/Δ mice were lower than those from their littermate controls (Fig. 2 G). We then cultured BM or MEP cells in methylcellulose-based medium with erythropoietin (EPO) and established a BFU-E colony assay; the number and size of BFU-E colonies again decreased in Δ/Δ compared with flox/flox mice (Figs. 2 H and S4 T). Finally, we cultured MEP cells sorted from flox/flox or Δ/Δ mice in methylcellulose-based medium with EPO to support the optimal growth of CFU-E colonies. The colony number decreased for Δ/Δ MEP cells (Fig. 2 I), indicating that loss of lncEry isoform-3 not only affects terminal differentiation during erythropoiesis but also reduces the growth of erythroid progenitor cells.
To directly assess the effect of lncEry on the regenerative function of HSCs in vivo, we transplanted BM cells from flox/flox and Δ/Δ mice (CD45.2) accompanied with competitor cells into irradiated recipients (CD45.1; Fig. 2 J). The BM cells from Δ/Δ mice had a lower reconstitution capacity than cells from control mice (71.1 vs. 85.4%; Fig. 2 K), but donor cells from the two groups gave rise to the same level of myeloid (Mac-1+) and lymphoid (CD3+ and B200+) lineages (Fig. 2 L). The frequency of LT-HSCs in Δ/Δ mice was approximately two- to threefold lower than in the control mice (Figs. 2 E and S4 M), and BM cells from Δ/Δ mice showed a 2.3-fold reduction in donor-cell engraftment 16 wk after transplantation (Fig. 2 M). These data might explain the decreased level of engraftment observed in recipients when unseparated BM cells were transplanted. We also performed another competitive transplantation using purified LT-HSCs from flox/flox and Δ/Δ mice, and the results showed that the reconstitution of LT-HSCs from both mice was equal (Fig. 2, N and O). Together, these results demonstrated that loss of lncEry decreased the growth of MEPs, which ultimately led to suppressed erythroid differentiation and decreased RBC production. Interestingly, although lncEry deletion decreased the number of HSCs, the function of individual HSCs was unaffected.
We generated an additional lncEryfl/fl mouse (flox/flox-2) model by inserting loxP sites around exons 2–6 of lncEry isoform-1 using CRISPR/Cas9 technology (Fig. S5 A). We then generated Mx1-Cre;lncEryfl/fl mice (Δ/Δ-2). After poly(I:C) induction, the knockout efficiency of lncEry was confirmed by qRT-PCR (Fig. S5 B). The deficits of erythroid differentiation and maturation were also observed in the new Δ/Δ-2 mice (Fig. S5, C and D). These results further confirmed the function of lncEry in regulating erythropoiesis.
lncEry deletion impairs erythroid differentiation. (A) Schematic of loxP sequences integrated into the lncEry locus (left). Schematic of cKO mice induced with MX1-Cre for 2 mo before flow cytometry analysis. (B) qPCR analysis of lncEry isoform expression in flox/flox-2 or Δ/Δ-2 BM cells (n = 3 samples). (C) Percentage of indicated populations in the BM of flox/flox-2 or Δ/Δ-2 mice (n = 4–6 mice per group). (D) Absolute numbers or concentrations of indicated items in the PB of flox/flox-2 or Δ/Δ-2 mice (n = 5–6 mice per group). (E) qRT-PCR analysis of indicated genes in lncEry-depleted MEL cells (n = 3–4 samples). (F) GSEA enrichment plot of Klf1 target gene set for DEGs between control and lncEry siRNAs in MEL cells. Three independent experiments for B–E. Data are represented as mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; unpaired two-tailed Student’s t test for B–D; one-way ANOVA for E.
lncEry deletion impairs erythroid differentiation. (A) Schematic of loxP sequences integrated into the lncEry locus (left). Schematic of cKO mice induced with MX1-Cre for 2 mo before flow cytometry analysis. (B) qPCR analysis of lncEry isoform expression in flox/flox-2 or Δ/Δ-2 BM cells (n = 3 samples). (C) Percentage of indicated populations in the BM of flox/flox-2 or Δ/Δ-2 mice (n = 4–6 mice per group). (D) Absolute numbers or concentrations of indicated items in the PB of flox/flox-2 or Δ/Δ-2 mice (n = 5–6 mice per group). (E) qRT-PCR analysis of indicated genes in lncEry-depleted MEL cells (n = 3–4 samples). (F) GSEA enrichment plot of Klf1 target gene set for DEGs between control and lncEry siRNAs in MEL cells. Three independent experiments for B–E. Data are represented as mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; unpaired two-tailed Student’s t test for B–D; one-way ANOVA for E.
lncEry deletion decreases Klf1 expression in MEP cells
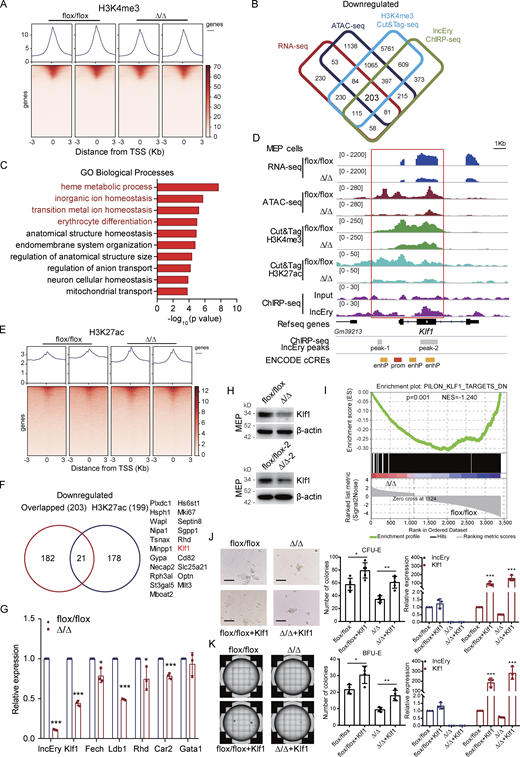
To gain mechanistic insights into the function of lncEry in erythropoiesis, we isolated MEP cells from flox/flox and Δ/Δ mice and performed RNA-seq analysis (Fig. 3 A). Compared with the flox/flox group, 3,256 genes were differentially expressed genes (DEGs), 1,054 of which were downregulated. To explore the changes in chromatin accessibility, we performed transposase-accessible chromatin (ATAC)-seq and observed 7,926 differential peaks (Fig. 3 B), and 4,599 peaks of accessibility were decreased upon deletion of lncEry in MEP cells. Notably, an integrative analysis with RNA-seq and ATAC-seq data revealed a significant correlation between downregulated genes and decreased accessibility, and we identified 421 overlapping genes (Fig. 3 C). GO enrichment analysis showed that the overlapping downregulated genes were enriched for erythrocyte differentiation and some metabolism-related terms (Fig. 3 D). We then examined ROS levels, mitochondrial membrane potential, and glucose uptake of MEP cells (Fig. 3 E), and the results suggest that the function of lncEry in erythropoiesis regulation does not depend on metabolic changes.
lncEry deletion affects erythroid differentiation of MEP cells. (A) Heatmap of DEGs from RNA-seq of MEP cells sorted from BM of flox/flox or Δ/Δ mice. (B) Heatmap showing replication of samples from ATAC-seq of MEP cells sorted from BM of flox/flox or Δ/Δ mice. (C) Experimental flow chart for sorting of MEP cells from BM of flox/flox or Δ/Δ mice and RNA-seq, ATAC-seq, and Cut&Tag (left). The number of overlapping downregulated DEGs and downregulated peak genes in lncEry-depleted MEP cells according to RNA-seq and ATAC-seq, respectively (right). (D) GSEA showing −log10 of the uncorrected P value on the x axis; darker shading corresponds to a greater number of enriched genes in each term. (E) Determination of low cytometric analysis of ROS levels by DCF-DA (left), mitochondrial membrane potential by TMRE (middle), and glucose uptake 2-NBDG (left) in MEP cells from BM of flox/flox or Δ/Δ mice. n = 3 mice per group; three independent experiments. Unpaired two-tailed Student’s t test. (F) qRT-PCR confirming that ChIRP retrieved 17% of cellular lncEry RNA and 0.1% Gapdh RNA. Lac Z probes were used as negative controls. n = 3 samples; three independent experiments. ***, P < 0.001; unpaired two-tailed Student’s t test. (G) Distribution of lncEry-binding sites across the indicated intergenic or intragenic regions in MEL cells, as shown by ChIRP-seq. (H) TSS profile and heatmap showing binding of lncEry from ChIRP-seq in relation to promoter regions. Data are represented as mean ± SD.
lncEry deletion affects erythroid differentiation of MEP cells. (A) Heatmap of DEGs from RNA-seq of MEP cells sorted from BM of flox/flox or Δ/Δ mice. (B) Heatmap showing replication of samples from ATAC-seq of MEP cells sorted from BM of flox/flox or Δ/Δ mice. (C) Experimental flow chart for sorting of MEP cells from BM of flox/flox or Δ/Δ mice and RNA-seq, ATAC-seq, and Cut&Tag (left). The number of overlapping downregulated DEGs and downregulated peak genes in lncEry-depleted MEP cells according to RNA-seq and ATAC-seq, respectively (right). (D) GSEA showing −log10 of the uncorrected P value on the x axis; darker shading corresponds to a greater number of enriched genes in each term. (E) Determination of low cytometric analysis of ROS levels by DCF-DA (left), mitochondrial membrane potential by TMRE (middle), and glucose uptake 2-NBDG (left) in MEP cells from BM of flox/flox or Δ/Δ mice. n = 3 mice per group; three independent experiments. Unpaired two-tailed Student’s t test. (F) qRT-PCR confirming that ChIRP retrieved 17% of cellular lncEry RNA and 0.1% Gapdh RNA. Lac Z probes were used as negative controls. n = 3 samples; three independent experiments. ***, P < 0.001; unpaired two-tailed Student’s t test. (G) Distribution of lncEry-binding sites across the indicated intergenic or intragenic regions in MEL cells, as shown by ChIRP-seq. (H) TSS profile and heatmap showing binding of lncEry from ChIRP-seq in relation to promoter regions. Data are represented as mean ± SD.
The nuclear location of an lncRNA might be suggestive of its function in transcriptional regulation (Chu et al., 2011). To understand the mechanisms of lncEry transcriptional regulation, we sought to determine the binding sites for lncEry in the genome through chromatin isolation by RNA purification sequencing (ChIRP-seq; Chu et al., 2011; Engreitz et al., 2013; Luo et al., 2015). We performed the ChIRP assay on MEL cells and confirmed the identities of the isolated RNAs by qRT-PCR, and ∼17% of lncEry RNA was pulled down (Fig. 3 F). In the sequencing analysis, we identified 1,786 lncEry binding sites compared with input. When we analyzed the locations of these binding sites in the genome, ∼85% were located in promoter regions (Fig. 3 G) and were mainly concentrated within a region 1 kbp from the transcriptional start sites (TSS; Fig. 3 H). Some of these results support our theory of a role of lncEry in transcriptional regulation.
To further explore the transcriptional regulatory function of lncEry in MEP cells, we performed cleavage under targets and tagmentation (Cut&Tag) assays (Kaya-Okur et al., 2019) on lncEry-deficient MEP cells using an antibody against the histone H3K4me3 (which is associated with transcriptional activation; Ptashne and Gann, 1997), followed by sequencing (Fig. 4 A). We then performed integrative analysis of the downregulated genes from RNA-seq, ATAC-seq, ChIRP-seq, and H3K4me3 Cut&Tag data to ascertain the directly regulated target genes, and 203 overlapping genes were identified (Fig. 4 B and Table S2). GO enrichment analysis showed that these target genes were enriched in erythrocyte homeostasis- and differentiation-related terms (Fig. 4 C). In addition, the peaks of H3K4me3 and H3K27ac at the cis-regulate region (CRR) of the Klf1 gene were decreased upon lncEry deletion (Fig. 4, D–F). Importantly, lncEry could bind to the CRR of Klf1, and deletion of lncEry decreased Klf1 expression (Fig. 4, G and H), chromatin accessibility, and the transcriptional active stage of Klf1 CRR (Fig. 4 D), suggesting that lncEry participates in the transcriptional regulation of Klf1 in MEP cells. lncEry deletion also significantly decreased the expression of other erythrocyte differentiation–related genes: Fech, Ldb1, Rhd, and Car2 (Fig. 4 G). In addition, gene set enrichment analysis (GSEA) revealed that deletion of lncEry in MEP cells reduced the enrichment of KLF1-target genes (Fig. 4 I). To confirm the function of lncEry in regulating the early stage of erythropoiesis through Klf1, MEP cells from the BM of flox/flox or Δ/Δ mice were sorted and transduced with lentiviruses carrying FLAG-Klf1-GFP or FLAG-GFP (control). After transduction, we performed in vitro colony assays. The results showed that the decreased numbers of CFU-E and BFU-E colonies caused by lncEry deletion were rescued by overexpression of Klf1 (Fig. 4, J and K). Together, these results indicate that lncEry regulates the transcription of Klf1 to affect the early stage of erythropoiesis.
lncEry deletion decreases Klf1 expression in MEP cells. (A) TSS profile and heatmap showing binding of lncEry from H3K4me3 Cut&Tag in relation to promoter regions. (B) Cut&Tag sequence analysis of H3K4me3 in MEP cells sorted from BM of flox/flox or Δ/Δ mice; the decreased peak genes under lncEry deletion overlapped with the downregulated genes detected by RNA-seq, downregulated peak genes from ATAC-seq, and lncEry-binding peak genes from ChIRP-seq. (C) GO term analysis of the downregulated genes in D, −log10 of the uncorrected P value on the x axis. (D) Visualization of RNA-seq, ATAC-seq, Cut&Tag sequence of H3K4me3 and H3K27ac, and ChIRP-seq peaks and predicted cis-regulate elements in Klf1 regions with IGV software. The sites of enhP and prom were from ENCODE database. (E) TSS profile and heatmap showing binding of lncEry from ChIRP-seq, H3K4me3, and H3K27ac from Cut&Tag in relation to promoter regions. (F) Number of overlapping genes from B and downregulated peak genes from H3K27ac Cut&Tag sequencing of MEP cells. (G) qRT-PCR analysis of the indicated genes in MEP cells sorted from BM of flox/flox or Δ/Δ mice (n = 3 samples). (H) Western blot analysis of the expression of indicated proteins in MEP cells sorted from BM of flox/flox or Δ/Δ mice. (I) GSEA enrichment plot of Klf1 target gene set for DEGs between flox/flox and Δ/Δ MEP cells. DN, downregulated genes. (J and K) The lentiviruses carrying pLVX-FLAG-Klf1-P2A-GFP-IRES-Puro or pLVX-FLAG-GFP-IRES-Puro (control) were transduced into MEP cells sorted from BM of flox/flox or Δ/Δ mice. CFU-E colony assays of 1,000 transduced MEP cells cultured in methylcellulose-based medium with EPO cytokine and puromycin (10 μg/ml) for 48 h (J). Representative images (left), CFU-E numbers (middle, n = 5 wells), and expression levels of lncEry and Klf1 (right, n = 3 samples) are shown. Scale bar, 100 μm. BFU-E colony assays of 1,000 transduced MEP cells cultured in methylcellulose-based medium with EPO cytokine and puromycin (10 μg/ml) for 48 h (K). Representative images (left), BFU-E numbers (middle, n = 4 wells), and expression levels of lncEry and Klf1 (right, n = 3 samples) are shown. Three independent experiments for G + H and J + K. Data are represented as mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; unpaired two-tailed Student’s t test. Source data are available for this figure: SourceDataF4.
lncEry deletion decreases Klf1 expression in MEP cells. (A) TSS profile and heatmap showing binding of lncEry from H3K4me3 Cut&Tag in relation to promoter regions. (B) Cut&Tag sequence analysis of H3K4me3 in MEP cells sorted from BM of flox/flox or Δ/Δ mice; the decreased peak genes under lncEry deletion overlapped with the downregulated genes detected by RNA-seq, downregulated peak genes from ATAC-seq, and lncEry-binding peak genes from ChIRP-seq. (C) GO term analysis of the downregulated genes in D, −log10 of the uncorrected P value on the x axis. (D) Visualization of RNA-seq, ATAC-seq, Cut&Tag sequence of H3K4me3 and H3K27ac, and ChIRP-seq peaks and predicted cis-regulate elements in Klf1 regions with IGV software. The sites of enhP and prom were from ENCODE database. (E) TSS profile and heatmap showing binding of lncEry from ChIRP-seq, H3K4me3, and H3K27ac from Cut&Tag in relation to promoter regions. (F) Number of overlapping genes from B and downregulated peak genes from H3K27ac Cut&Tag sequencing of MEP cells. (G) qRT-PCR analysis of the indicated genes in MEP cells sorted from BM of flox/flox or Δ/Δ mice (n = 3 samples). (H) Western blot analysis of the expression of indicated proteins in MEP cells sorted from BM of flox/flox or Δ/Δ mice. (I) GSEA enrichment plot of Klf1 target gene set for DEGs between flox/flox and Δ/Δ MEP cells. DN, downregulated genes. (J and K) The lentiviruses carrying pLVX-FLAG-Klf1-P2A-GFP-IRES-Puro or pLVX-FLAG-GFP-IRES-Puro (control) were transduced into MEP cells sorted from BM of flox/flox or Δ/Δ mice. CFU-E colony assays of 1,000 transduced MEP cells cultured in methylcellulose-based medium with EPO cytokine and puromycin (10 μg/ml) for 48 h (J). Representative images (left), CFU-E numbers (middle, n = 5 wells), and expression levels of lncEry and Klf1 (right, n = 3 samples) are shown. Scale bar, 100 μm. BFU-E colony assays of 1,000 transduced MEP cells cultured in methylcellulose-based medium with EPO cytokine and puromycin (10 μg/ml) for 48 h (K). Representative images (left), BFU-E numbers (middle, n = 4 wells), and expression levels of lncEry and Klf1 (right, n = 3 samples) are shown. Three independent experiments for G + H and J + K. Data are represented as mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; unpaired two-tailed Student’s t test. Source data are available for this figure: SourceDataF4.
lncEry regulates late-stage erythropoiesis by promoting globin gene expression
To further explore the function of lncEry in the late stage of erythropoiesis, we transfected MEL cells with siRNAs targeting lncEry and performed RNA-seq analysis. Compared with the control group, there were 117 and 134 DEGs after lncEry knockdown with siRNA-1 and siRNA-2, respectively (Fig. 5 A). Of these DEGs, 75 overlapping genes were downregulated (Fig. 5 B and Table S3). When we conducted enrichment analysis of these 75 genes with the Metascape online tool, strikingly, the most enriched term was erythrocyte homeostasis (Fig. 5 C).
lncEry regulates the late stage of erythropoiesis by promoting globin gene expression. (A) Heatmap of DEGs in lncEry-siRNA-treated and control (CT) MEL cells according to RNA-seq. (B) The numbers of overlapping, downregulated DEGs in lncEry-depleted and CT MEL cells using two different targeting siRNAs. (C) GSEA of downregulated genes with −log10 plot of the uncorrected P value on the x axis; darker shading corresponds to a greater level of enriched genes in each term. (D) MEL cells were transfected with control or lncEry siRNA, and cellular extracts were prepared and analyzed by Western blotting. The intensity of each band was quantified by densitometry with ImageJ software and normalized to β-actin (n = 3 samples). (E) qRT-PCR of indicated genes in NuE cells sorted from BM of flox/flox or Δ/Δ mice (n = 3 samples). (F) Cellular extracts of NuE cells sorted from BM of flox/flox, Δ/Δ, flox/flox-2, or Δ/Δ-2 mice were prepared and analyzed by Western blotting. (G) MEL cells were transfected with control siRNA or lncEry siRNA. Soluble chromatin was collected for ChIP-seq analysis using antibodies against Pol II–S5p or H3K4me3; the decreased peak genes under lncEry knockdown overlapped with the downregulated genes detected by RNA-seq. (H) Heatmap showing binding of H3K4me3 and RNA polymerase II S5p in relation to promoter regions. (I) ChIP-seq trace showing Pol II–S5p and H3K4me3 binding of control or lncEry knockdown cells in relation to the indicated gene CRRs, visualized with IGV software. (J) qChIP of Pol II–S5p (left) or H3K4me3 (right) with primers covering the promoters of the indicated genes (n = 3 samples). Three independent experiments for D–F and J. Data are represented as mean ± SD. **, P < 0.01; ***, P < 0.001; unpaired two-tailed Student’s t test for E–J; one-way ANOVA for D. Source data are available for this figure: SourceDataF5.
lncEry regulates the late stage of erythropoiesis by promoting globin gene expression. (A) Heatmap of DEGs in lncEry-siRNA-treated and control (CT) MEL cells according to RNA-seq. (B) The numbers of overlapping, downregulated DEGs in lncEry-depleted and CT MEL cells using two different targeting siRNAs. (C) GSEA of downregulated genes with −log10 plot of the uncorrected P value on the x axis; darker shading corresponds to a greater level of enriched genes in each term. (D) MEL cells were transfected with control or lncEry siRNA, and cellular extracts were prepared and analyzed by Western blotting. The intensity of each band was quantified by densitometry with ImageJ software and normalized to β-actin (n = 3 samples). (E) qRT-PCR of indicated genes in NuE cells sorted from BM of flox/flox or Δ/Δ mice (n = 3 samples). (F) Cellular extracts of NuE cells sorted from BM of flox/flox, Δ/Δ, flox/flox-2, or Δ/Δ-2 mice were prepared and analyzed by Western blotting. (G) MEL cells were transfected with control siRNA or lncEry siRNA. Soluble chromatin was collected for ChIP-seq analysis using antibodies against Pol II–S5p or H3K4me3; the decreased peak genes under lncEry knockdown overlapped with the downregulated genes detected by RNA-seq. (H) Heatmap showing binding of H3K4me3 and RNA polymerase II S5p in relation to promoter regions. (I) ChIP-seq trace showing Pol II–S5p and H3K4me3 binding of control or lncEry knockdown cells in relation to the indicated gene CRRs, visualized with IGV software. (J) qChIP of Pol II–S5p (left) or H3K4me3 (right) with primers covering the promoters of the indicated genes (n = 3 samples). Three independent experiments for D–F and J. Data are represented as mean ± SD. **, P < 0.01; ***, P < 0.001; unpaired two-tailed Student’s t test for E–J; one-way ANOVA for D. Source data are available for this figure: SourceDataF5.
We then performed qRT-PCR to analyze the expression of several DEGs: lncEry knockdown significantly decreased the expression of erythrocyte homeostasis– and differentiation-related genes, such as Hba-a1, Hba-a2 (downregulated in siEry2 parts; two variants of α-globin), Hbb-b1, Hbb-b2 (two variants of β-globin), Alas2, and Rhag in MEL cells (Fig. S5 E). Furthermore, lncEry knockdown significantly reduced the protein levels of α- and β-globin (Fig. 5 D). We also examined DEGs in NuEs sorted from the BM of flox/flox and Δ/Δ mice, and lncEry knockout decreased mRNA expression and protein levels of α- and β-globin (Fig. 5, E and F). Unlike in MEPs, lncEry knockout did not reduce the expression of Klf1 in NuE cells (Fig. 5, E and F), and the DEGs in MEL cells were also not enriched in Klf1-target genes (Fig. S5 F). These results indicate that lncEry participates in erythropoiesis regulation at different stages and using different mechanisms.
To further explore the role of lncEry in regulating DEGs at the transcriptional level, we performed chromatin immunoprecipitation (ChIP) assays in lncEry-deficient cells using antibodies against Ser5-phosphorylated RNA polymerase II (Pol II–S5p) and histone H3K4me3, followed by sequencing. We then compared the peaks enriched by Pol II–S5p and histone H3K4me3 of the gene promoters in control and lncEry-depleted cells. Interestingly, we found four downregulated DEGs in our RNA-seq dataset (Hbb-b1, Hba-a1, Hbb-b2, and Btg2) that overlapped in the Pol II–S5p and H3K4me3 ChIP-seq datasets (Fig. 5, G and H). Consistently, the Pol II–S5p and H3K4me3 enrichment peaks of globin gene cis-regulated regions, such as Hba-a1, Hba-a2, Hbb-b1, and Hbb-b2, sharply declined upon lncEry knockdown (Fig. 5 I), and the results of the quantitative ChIP (qChIP) assays verified these findings (Fig. 5 J). We thus concluded that lncEry depletion affects the transcription of globin genes in the late stage of erythropoiesis.
lncEry is physically associated with Wdr82
lncRNAs are usually associated with numerous cellular functions, most of which require interactions with one or more RNA-binding proteins (Cao et al., 2019; Ferre et al., 2016). To determine whether lncEry acts alone or in concert with other proteins in the different stages of erythropoiesis, we performed RNA-pulldown assays (Fig. 6 A) followed by silver staining and mass spectrometry to identify lncEry isoform-3 interaction partners in MEPs sorted from the BM of wild-type mice and MEL cells. We then performed integrative analysis and found 11 common interacting partners in the two types of cells (Fig. 6, B and D; and Tables S4 and S5). Interestingly, we identified two transcription regulators, WD repeat-containing protein 82 (WDR82) and DEAD-box helicase 5 (DDX5), as likely lncEry binding partners, which may explain some of the mechanisms involved in regulating the DEGs of lncEry-depleted MEP and NuE cells at the transcriptional level. Western blot analyses confirmed the interactions between lncEry and its binding partners (Fig. 6 E).
Identifying lncEry-interacting proteins. (A) Experimental design for identifying lncEry-interacting proteins. In vitro–transcribed lncEry-3 was used as the bait, and anti-lncEry-3 was used as the control. (B and C) RNA pulldown and mass spectrometry analyses of lncEry-interacting proteins. Whole-cell extracts from MEP (B) or MEL (C) were prepared and subjected to RNA pulldown using in vitro–transcribed lncEry-3 or anti-lncEry-3 as the bait. After extensive washing, the bound proteins were eluted and visualized by silver staining on SDS-PAGE. The protein bands on the gel were recovered by trypsinization and analyzed by mass spectrometry. Detailed results from the mass spectrometric analysis are provided in Tables S4 and S5. (D) Overlapping lncEry interaction partners from MEP and MEL cells, as analyzed by mass spectrometry. (E) RNA-pulldown–purified proteins retrieved using the indicated baits were analyzed by Western blotting with antibodies against the indicated proteins. (F and G) CFU colony assays of 2,000 cKit+ cells transfected with control, Wdr82, or Ddx5 shRNAs cultured for 10–14 d in complete methylcellulose-based medium. The colony numbers are provided in G (n = 3–4 wells). Representative images from triplicate experiments are shown in F. (H) Cellular extracts of cKit+ cells transfected with control, Wdr82, or Ddx5 shRNAs were prepared and analyzed by Western blotting with indicated antibodies. (I) BFU-E colony assays of 2,000 cKit+ cells transfected with control or Wdr82 shRNAs cultured for 10–14 d in complete methylcellulose-based medium with EPO cytokine stimulation. The colony numbers were counted (right). Representative images from triplicate experiments are shown (left; n = 3 wells). (J) Flow analysis percentage of CD44+ in Ter119+ cKit+ cells transfected with control or Wdr82 shRNAs cultured in serum-free expansion medium with growth factors SCF, IL-3, and EPO for 7–10 d. Flow analysis of CD44 expression in Ter119+ cells (n = 3 samples). FSC, forward scatter. Three independent experiments for E–J. Data are represented as mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; one-way ANOVA. Source data are available for this figure: SourceDataF6.
Identifying lncEry-interacting proteins. (A) Experimental design for identifying lncEry-interacting proteins. In vitro–transcribed lncEry-3 was used as the bait, and anti-lncEry-3 was used as the control. (B and C) RNA pulldown and mass spectrometry analyses of lncEry-interacting proteins. Whole-cell extracts from MEP (B) or MEL (C) were prepared and subjected to RNA pulldown using in vitro–transcribed lncEry-3 or anti-lncEry-3 as the bait. After extensive washing, the bound proteins were eluted and visualized by silver staining on SDS-PAGE. The protein bands on the gel were recovered by trypsinization and analyzed by mass spectrometry. Detailed results from the mass spectrometric analysis are provided in Tables S4 and S5. (D) Overlapping lncEry interaction partners from MEP and MEL cells, as analyzed by mass spectrometry. (E) RNA-pulldown–purified proteins retrieved using the indicated baits were analyzed by Western blotting with antibodies against the indicated proteins. (F and G) CFU colony assays of 2,000 cKit+ cells transfected with control, Wdr82, or Ddx5 shRNAs cultured for 10–14 d in complete methylcellulose-based medium. The colony numbers are provided in G (n = 3–4 wells). Representative images from triplicate experiments are shown in F. (H) Cellular extracts of cKit+ cells transfected with control, Wdr82, or Ddx5 shRNAs were prepared and analyzed by Western blotting with indicated antibodies. (I) BFU-E colony assays of 2,000 cKit+ cells transfected with control or Wdr82 shRNAs cultured for 10–14 d in complete methylcellulose-based medium with EPO cytokine stimulation. The colony numbers were counted (right). Representative images from triplicate experiments are shown (left; n = 3 wells). (J) Flow analysis percentage of CD44+ in Ter119+ cKit+ cells transfected with control or Wdr82 shRNAs cultured in serum-free expansion medium with growth factors SCF, IL-3, and EPO for 7–10 d. Flow analysis of CD44 expression in Ter119+ cells (n = 3 samples). FSC, forward scatter. Three independent experiments for E–J. Data are represented as mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; one-way ANOVA. Source data are available for this figure: SourceDataF6.
To explore the functional impact of Wdr82 and Ddx5 on HSPCs, we transduced cKit+ cells with a GFP-expressing lentivirus carrying Wdr82 or Ddx5 shRNAs and performed in vitro colony assays (Fig. 6, F and G). The results showed that Wdr82 knockdown decreased the numbers of CFU-G, CFU-M, and BFU-E, whereas Ddx5 knockdown did not decrease BFU-E numbers (Fig. 6, F and G). The knockdown efficiency was confirmed by Western blotting (Fig. 6 H). We then cultured Wdr82 knockdown cKit+ cells in M3436 methylcellulose-based medium with EPO and established a BFU-E colony assay. The colony number and size of the BFU-E colonies decreased in Wdr82-knockdown cells (Fig. 6 I). However, when we cultured the cKit+ cells in vitro for ∼1 wk after they were transduced with Wdr82 shRNA, Wdr82 knockdown was seen to decrease the percentage of late-stage erythropoiesis cells (Ter119+CD44−) and arrest the progress of erythropoiesis (Fig. 6 J). These results suggest that Wdr82, with similar phenotypes to lncEry, was more likely to participate in erythropoiesis with lncEry than Ddx5. Following this, we were interested in exploring the function of lncEry interaction with Wdr82 in regulating the early and late stages of erythropoiesis. We found that both isoform-1 and isoform-3 can interact with WDR82 with similar intensity (Fig. 7 A). Consistent with these findings, RNA immunoprecipitation (RIP) assays further confirmed this interaction between all lncEry isoforms and Wdr82 (Fig. 7 B).
lncEry is physically associated with Wdr82. (A) RNA-pulldown–purified proteins retrieved using the indicated baits were analyzed by Western blotting with antibodies against the indicated proteins. (B) Whole-cell lysates from MEL cells were immunoprecipitated with Wdr82 antibodies; purified RNA was analyzed by qRT-PCR and agarose gel electrophoresis. (C) Schematic of MFE structure and the lncEry isoform-3 truncation mutant. RNA-pulldown–purified proteins retrieved by lncEry truncation mutant baits were analyzed by Western blotting. (D) Schematic of the MFE structure and the lncEry truncation mutants. RNA pull-down purified proteins by lncEry truncation mutant baits were analyzed by Western blotting with antibodies against the indicated proteins. (E) MEL cells with doxycycline-inducible expression of stably integrated Flag-Wdr82 were collected. Cellular extracts were prepared and subjected to affinity purification using an anti-FLAG affinity column. After extensive washing in high salt solution, the purified Wdr82 protein was stained with Coomassie blue. (F) Sensorgrams of lncEry-P5 binding to Wdr82, as measured by SPR technology on a Biacore 3000 instrument. Representative sensorgrams were obtained by injecting various concentrations (0, 36.7, 73.4, 147, and 294 nM) of lncEry-P5 over Wdr82 immobilized on a CM5 sensor chip. (G and H) RNAscope and immunofluorescence assays using lncEry probes and Wdr82 antibodies, respectively, of MEP (G) or MEL (H) cells and analysis by confocal microscopy. Scale bar, 10 μm. Three to four independent experiments for A–H. Data are represented as mean ± SD. ***, P < 0.001; unpaired two-tailed Student’s t test. Source data are available for this figure: SourceDataF7.
lncEry is physically associated with Wdr82. (A) RNA-pulldown–purified proteins retrieved using the indicated baits were analyzed by Western blotting with antibodies against the indicated proteins. (B) Whole-cell lysates from MEL cells were immunoprecipitated with Wdr82 antibodies; purified RNA was analyzed by qRT-PCR and agarose gel electrophoresis. (C) Schematic of MFE structure and the lncEry isoform-3 truncation mutant. RNA-pulldown–purified proteins retrieved by lncEry truncation mutant baits were analyzed by Western blotting. (D) Schematic of the MFE structure and the lncEry truncation mutants. RNA pull-down purified proteins by lncEry truncation mutant baits were analyzed by Western blotting with antibodies against the indicated proteins. (E) MEL cells with doxycycline-inducible expression of stably integrated Flag-Wdr82 were collected. Cellular extracts were prepared and subjected to affinity purification using an anti-FLAG affinity column. After extensive washing in high salt solution, the purified Wdr82 protein was stained with Coomassie blue. (F) Sensorgrams of lncEry-P5 binding to Wdr82, as measured by SPR technology on a Biacore 3000 instrument. Representative sensorgrams were obtained by injecting various concentrations (0, 36.7, 73.4, 147, and 294 nM) of lncEry-P5 over Wdr82 immobilized on a CM5 sensor chip. (G and H) RNAscope and immunofluorescence assays using lncEry probes and Wdr82 antibodies, respectively, of MEP (G) or MEL (H) cells and analysis by confocal microscopy. Scale bar, 10 μm. Three to four independent experiments for A–H. Data are represented as mean ± SD. ***, P < 0.001; unpaired two-tailed Student’s t test. Source data are available for this figure: SourceDataF7.
We were then intrigued to identify the binding sites underlying the lncEry–Wdr82 interaction. To do so, we generated lncEry isoform-3 truncation mutants according to the isoform’s main minimum free energy (MFE) stem-loop regions, which we predicted using RNAfold WebServer tools (http://rna.tbi.univie.ac.at; Fig. 7 C, upper panel). In vitro RNA-pulldown assays showed that the interaction was primarily dependent on the lncEry 3′ terminal loop regions (Fig. 7 C). We also generated two lncEry fragments that are shared by all three isoforms: exons 2, 3, 4 and 6 of lncEry isoform-1. The results of the binding assay revealed that lncEry mainly interacted with Wdr82 through the last exon transcript (exon 6; Fig. 7 D). Next, we conducted surface plasmon resonance (SPR) assays using the GE Healthcare Biacore 3000 platform to examine the binding kinetics of lncEry and Wdr82. Indeed, the last exon region lncEry-P5 (exon 6) shared by each of the three lncEry isoforms directly interacted with Wdr82 that was purified from MEL cell lines (Fig. 7 E) with a Kd value of 38.2 nM (Fig. 7 F). Finally, we performed colocalization assays combining RNAscope with immunofluorescent staining followed by fluorescent confocal microscopy. We observed that lncEry mainly colocalized with Wdr82 in the nuclei of MEP and MEL cells (Fig. 7, G and H). All lncEry isoforms physically associated with Wdr82 in the nuclei of MEP and MEL cells. We thus proposed a hypothesis that the lncEry–Wdr82 complex serves to regulate transcription in these cells.
lncEry–Wdr82 regulates the transcriptional activation of Klf1 in MEP cells
As lncEry was found to be associated with Wdr82, we further explored the molecular mechanisms of the role of lncEry and Wdr82 in MEP cells by performing Cut&Tag assays on lncEry-deficient MEP cells using an antibody against Wdr82. The results showed that lncEry deletion decreased chromatin accessibility and the binding of Wdr82 at CRR around the Klf1 gene body as well as the binding of Wdr82 in the whole genome region (Fig. 8, A and B). Therefore, we speculated that lncEry can physically interact and functionally coordinate with Wdr82 to regulate the transcription of Klf1. To test this, we established two pGL3-luciferase reporters containing CRRs, as shown in Fig. 8 C, then performed reporter assays with pGL3-luciferase reporters containing Klf1 CRRs or mutant CRRs (without the main lncEry binding site) cotransfected into 293T cells together with lncEry, Wdr82, or both, as well as the Renilla luciferase vector for normalization (Fig. 8, C and D). The assays showed that overexpression of lncEry or coexpression of lncEry and Wdr82 enhanced the activity of the Klf1-CRR reporter, but not the Klf1-CRRs-mt reporter (Fig. 8 C). These results suggest that lncEry coordinates with Wdr82 to regulate the transcription of Klf1 in early erythroid differentiation.
lncEry-Wdr82 regulates transcriptional activation of Klf1 and globin genes. (A) Visualization of ATAC-seq, Cut&Tag for H3K4me3 and Wdr82, ChIRP-seq lncEry peaks, predict cis-regulate elements, and luciferase reporter clone CRRs in Klf1 gene regions with IGV software. (B) Peak center profile and heatmap showing binding of Wdr82 at whole genome of MEP cells. (C) Schematic diagrams of the pGL3-CRRs-luciferase reporter constructs; relative luciferase activity was determined by sequential normalization to Renilla and pGL3-vector activity (n = 3 samples). (D) qRT-PCR of lncEry expression and Western blotting of the expression of indicated proteins in the reporter assays shown in C (n = 3 samples). (E) ChIRP-seq trace showing lncEry binding in relation to the indicated gene regions. Red boxes indicate the CRRs of globin genes qPCR and agarose gel electrophoresis analysis of isolated chromatin sequences of indicated gene promoter regions (n = 3 samples). (F) ChIRP-seq trace showing lncEry binding in relation to the indicated gene regions (upper panel). qPCR and agarose gel electrophoresis of chromatin-isolated sequence of the indicated gene regions (n = 3 samples). (G and H) Schematic diagrams of pGL3-CRRs-luciferase reporter constructs (upper panel). For reporter assays, MEL cells were cotransfected with pcDNA3.1-lncEry or Wdr82 or pcDNA3.1-lncEry and Wdr82 together with Renilla and globin gene CRRs luciferase. The relative luciferase activity was determined by sequential normalization to Renilla and pGL3-vector activity (n = 3–4 samples). Western blots of the expression of indicated proteins in the reporter assays are shown (H). Three to four independent experiments for C–H. Data are represented as mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; unpaired two-tailed Student’s t test for D–F; one-way ANOVA for C and G. Source data are available for this figure: SourceDataF8.
lncEry-Wdr82 regulates transcriptional activation of Klf1 and globin genes. (A) Visualization of ATAC-seq, Cut&Tag for H3K4me3 and Wdr82, ChIRP-seq lncEry peaks, predict cis-regulate elements, and luciferase reporter clone CRRs in Klf1 gene regions with IGV software. (B) Peak center profile and heatmap showing binding of Wdr82 at whole genome of MEP cells. (C) Schematic diagrams of the pGL3-CRRs-luciferase reporter constructs; relative luciferase activity was determined by sequential normalization to Renilla and pGL3-vector activity (n = 3 samples). (D) qRT-PCR of lncEry expression and Western blotting of the expression of indicated proteins in the reporter assays shown in C (n = 3 samples). (E) ChIRP-seq trace showing lncEry binding in relation to the indicated gene regions. Red boxes indicate the CRRs of globin genes qPCR and agarose gel electrophoresis analysis of isolated chromatin sequences of indicated gene promoter regions (n = 3 samples). (F) ChIRP-seq trace showing lncEry binding in relation to the indicated gene regions (upper panel). qPCR and agarose gel electrophoresis of chromatin-isolated sequence of the indicated gene regions (n = 3 samples). (G and H) Schematic diagrams of pGL3-CRRs-luciferase reporter constructs (upper panel). For reporter assays, MEL cells were cotransfected with pcDNA3.1-lncEry or Wdr82 or pcDNA3.1-lncEry and Wdr82 together with Renilla and globin gene CRRs luciferase. The relative luciferase activity was determined by sequential normalization to Renilla and pGL3-vector activity (n = 3–4 samples). Western blots of the expression of indicated proteins in the reporter assays are shown (H). Three to four independent experiments for C–H. Data are represented as mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; unpaired two-tailed Student’s t test for D–F; one-way ANOVA for C and G. Source data are available for this figure: SourceDataF8.
lncEry–Wdr82 regulates the transcriptional activation of globin genes through CRRs
In the late stage of erythropoiesis, lncEry depletion decreased the transcription of globin genes. We found that lncEry was located at the CRRs of Hba-a1 and Hba-a2 (Fig. 8 E) but not the genomic regions of Hbb-b1 and Hbb-b2 (Fig. 8 F). Furthermore, using quantitative PCR (qPCR), we detected lncEry binding on the CRRs of globin genes (Fig. 8, E and F). However, we were unable to detect lncEry binding on Hbb-b1 and Hbb-b2 CRRs, possibly because it has lower affinity for these promoters or because cofactors are required for lncEry binding.
Because lncEry associated with Wdr82 and directly bound to the CRRs of globin genes in MEL cells, we speculated that lncEry physically interacts and functionally coordinates with Wdr82 to regulate globin gene transcription in late erythropoiesis. To test this, MEL cells were cotransfected with pGL3-luciferase reporters containing Hba-a1, Hba-a2, Hbb-b1, or Hbb-b2 promoters (Fig. 8 G) along with lncEry, Wdr82, or both, or the Renilla luciferase vector. Reporter assays showed that overexpression of either lncEry or Wdr82 resulted in a significant increase in Hba-a1, Hba-a2, Hbb-b1, and Hbb-b2 reporter activity. Coexpression of lncEry and Wdr82 enhanced Hba-a1 and Hba-a2 (but not Hbb-b1 or Hbb-b2) reporter activity further, which was perhaps because reporter activity was saturated (Fig. 8 G). Flag-Wdr82 expression was confirmed by Western blotting (Fig. 8 H).
To investigate whether the effects of lncEry on the transcriptional activation of globin genes were associated with Wdr82, we transfected Flag-Wdr82 plasmids into lncEry-deficient MEL cells. Subsequent luciferase reporter assays showed that lncEry depletion inhibited Hba-a1, Hba-a2, Hbb-b1, and Hbb-b2 reporter activities, but this inhibition was rescued, at least in part, upon Flag-Wdr82 overexpression (Fig. 9, A–E). Similarly, Wdr82 depletion inhibited the four globin gene reporter activities, and this inhibition was rescued by lncEry overexpression (Fig. 9, A–E).
lncEry-Wdr82 regulates transcriptional activation of globin genes through CRRs. (A–D) MEL cells were cotransfected with the indicated siRNAs or plasmids together with Renilla and the globin gene CRRs luciferase. The relative luciferase activity was determined by sequential normalization to Renilla and pGL3-vector activity (n = 3 samples). (E) Western blots of the expression of indicated proteins in the reporter assays shown in A–D. (F) MEL cells were cotransfected with the indicated siRNAs or plasmids, and the cellular extracts were analyzed by Western blotting with antibodies against the indicated proteins. (G) qRT-PCR of lncEry expression in each sample shown in F (n = 3 samples). (H and I) MEL cells were transfected with control or lncEry siRNAs; the soluble chromatin was immunoprecipitated with antibodies against Wdr82, Set1A (H), or RbBP5, Ash2l (I), and analyzed by qPCR with the indicated primers. The relative fold enrichment was determined by sequential normalization with the cycle threshold values of input and the control siRNA samples (n = 3 samples). Three to four independent experiments for A–I. Data are represented as mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; one-way ANOVA for A–D; unpaired two-tailed Student’s t test for G–I. Source data are available for this figure: SourceDataF9.
lncEry-Wdr82 regulates transcriptional activation of globin genes through CRRs. (A–D) MEL cells were cotransfected with the indicated siRNAs or plasmids together with Renilla and the globin gene CRRs luciferase. The relative luciferase activity was determined by sequential normalization to Renilla and pGL3-vector activity (n = 3 samples). (E) Western blots of the expression of indicated proteins in the reporter assays shown in A–D. (F) MEL cells were cotransfected with the indicated siRNAs or plasmids, and the cellular extracts were analyzed by Western blotting with antibodies against the indicated proteins. (G) qRT-PCR of lncEry expression in each sample shown in F (n = 3 samples). (H and I) MEL cells were transfected with control or lncEry siRNAs; the soluble chromatin was immunoprecipitated with antibodies against Wdr82, Set1A (H), or RbBP5, Ash2l (I), and analyzed by qPCR with the indicated primers. The relative fold enrichment was determined by sequential normalization with the cycle threshold values of input and the control siRNA samples (n = 3 samples). Three to four independent experiments for A–I. Data are represented as mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; one-way ANOVA for A–D; unpaired two-tailed Student’s t test for G–I. Source data are available for this figure: SourceDataF9.
We confirmed the effects of lncEry and Wdr82 on globin gene transcription at the protein level, and again, lncEry depletion decreased the protein expression of α-globin and β-globin, and Wdr82 overexpression partially rescued the effect (Fig. 9 F, left panel) and vice versa (Fig. 9 F, right panel). Interestingly, lncEry depletion decreased the level of H3K4me3, which could be rescued by overexpression of Wdr82, and vice versa (Fig. 9, F and G). These results led us to propose that lncEry might participate in the Set1A/Wdr82 complex to affect H3K4me3 levels and, as a result, regulate the transcriptional activation of globin genes.
To strengthen the above hypothesis, we examined the effects of lncEry on Wdr82 and Set1A recruitment to globin gene CRRs using qChIP assay. To this end, we immunoprecipitated soluble chromatin from control or lncEry-depleted MEL cells using antibodies against Wdr82 or Set1A, and then performed qPCR analysis to identify the precipitated DNA. lncEry depletion specifically decreased Wdr82 and Set1A enrichment on the CRRs of globin genes, but not the two other components of the Set1A/Wdr82 complex, Ash2l and RbBP5 (Wu et al., 2008; Fig. 9, H and I). These findings might be due to the low affinity of Ash2l and RbBp5 antibodies for ChIP samples (Fig. 9, H and I). Collectively, these results support the hypothesis that lncEry recruits Wdr82 and stabilizes the Set1A/Wdr82 complex located on the CRRs of globin genes.
lncEry–Wdr82 regulates the transcriptional activation of globin genes through locus control regions (LCRs)
Because the transcription of globin genes also can be regulated by distal regulatory elements (Cao and Moi, 2002; Krivega et al., 2015), we then investigated whether lncEry/Wdr82 could regulate globin genes through these elements. We generated luciferase reporters for different LCRs, containing several DNase I hypersensitive sites (HS; Hu et al., 2007; Sawado et al., 2001). 14 HSs were individually subcloned into the enhancer insertion site of pGL3-Hba-a1-CRRs-luciferase or pGL3-Hbb-b1-CRRs-luciferase vectors (Fig. 10, A and B). The results of luciferase reporter assays showed that knockdown of either lncEry or Wdr82 resulted in a significant decrease in Hba-a1-CRRs-HS26 and Hbb-b1-CRRs-HS2 reporter activity compared with Hba-a1-CRRs and Hbb-b1-CRRs reporter respectively (Fig. 10, C and D). In addition, coexpression of lncEry and Wdr82 also enhanced Hba-a1-CRRs-HS26 and Hbb-b1-CRRs-HS2 reporter activity compared with Hba-a1-CRRs and Hbb-b1-CRRs reporter respectively (Fig. 10, E and F). When testing the distal regulatory elements of other globin variants with similar strategies, we further confirmed the importance of lncEry/Wdr82 in regulating HS26 and HS2 locus on globin a2 and b2, respectively (Fig. 10, G–I). Next, to further investigate the effect of lncEry on Wdr82 and Set1A recruitment to LCRs of globin genes, the NuE cells from BM of flox/flox or Δ/Δ mice were sorted and qChIP assays were performed. The results showed that lncEry depletion specifically decreased the level of H3K4me3 and the enrichment of Wdr82 and Set1A on the HS26 and HS2 locus in globin gene bodies (Fig. 10 J). All the results indicate that lncEry/Wdr82 could regulate the transcription of globin genes through CRRs but also distal regulatory elements.
lncEry-Wdr82 regulates transcriptional activation of globin genes through LCR. (A and B) Schematic representation of different LCRs related to gene body of α-globin or β-globin variants and pGL3-CRRs-HSs luciferase reporter constructs. (C and D) MEL cells were transfected with control, lncEry or Wdr82 siRNAs, and the knockdown efficiency was examined and shown in C. The knockdown cells were then transfected with the indicated CRRs-HSs and Renilla luciferase reporters. The relative luciferase activity was determined by sequential normalization to the activity of Renilla and pGL3-Hba-a1-CRRs or pGL3-Hbb-b1-CRRs vector and shown in D (n = 3 samples). (E and F) MEL cells were cotransfected with lncEry together with Wdr82 or pcDNA3.1 plasmids, and the overexpression efficiency was examined and shown in E. The overexpressed cells were then transfected with the indicated plasmids of CRRs-HSs and Renilla luciferase reporter. The relative luciferase activity was determined by sequential normalization to the activity of Renilla and each corresponding control cells expressing pcDNA3.1 and shown in F (n = 3 samples). (G) Schematic representation of pGL3-CRRs-HSs luciferase reporter constructs. (H) Reporter assays analogous to D with cells expressing Hba-a2-CRRs-HSs or Hbb-b2-CRRs-HSs luciferase constructs and the indicated siRNAs or genes (n = 3 samples). (I) Reporter assays analogous to F with cells expressing CRRs or CRRs-HSs luciferase constructs and the indicated genes (n = 3 samples). (J) NuE cells were sorted from BM of flox/flox or Δ/Δ mice. Soluble chromatin was immunoprecipitated with antibodies against Wdr82, Set1A, or H3K4me3 and analyzed by qPCR with the indicated primers. The relative fold enrichment was determined by sequential normalization with the cycle threshold values of input and the flox/flox samples (n = 3 samples). (K) Mechanistic model of how lncEry combines with Wdr82 to participate in erythropoiesis regulation. Generally, the novel annotated lncRNA, lncEry, interacts with Wdr82 to control the location of the Set1A/Wdr82 complex and facilitate H3K4me3 binding to CRRs of target genes. In MEP cells, lncEry bind to the CRRs of Klf1 to recruit Wdr82 and elevate H3K4me3 levels at CRRs to promote Klf1 expression and coordinate early erythroid differentiation. In NuE, lncEry promotes Set1A/Wdr82 complex to bind to CRRs as well as LCRs of globin genes to promote transcription of Hba-a1, Hba-a2, Hbb-b1, and Hbb-b2, which promotes terminal erythropoiesis. In brief, lncEry combines with Wdr82 to promote Klf1 and globin gene expression to regulate the early and late stages of erythropoiesis, respectively. Three independent experiments for C–F and H–J. Data are represented as mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; one-way ANOVA for C–D and H; unpaired two-tailed Student’s t test for E + F and I + J.
lncEry-Wdr82 regulates transcriptional activation of globin genes through LCR. (A and B) Schematic representation of different LCRs related to gene body of α-globin or β-globin variants and pGL3-CRRs-HSs luciferase reporter constructs. (C and D) MEL cells were transfected with control, lncEry or Wdr82 siRNAs, and the knockdown efficiency was examined and shown in C. The knockdown cells were then transfected with the indicated CRRs-HSs and Renilla luciferase reporters. The relative luciferase activity was determined by sequential normalization to the activity of Renilla and pGL3-Hba-a1-CRRs or pGL3-Hbb-b1-CRRs vector and shown in D (n = 3 samples). (E and F) MEL cells were cotransfected with lncEry together with Wdr82 or pcDNA3.1 plasmids, and the overexpression efficiency was examined and shown in E. The overexpressed cells were then transfected with the indicated plasmids of CRRs-HSs and Renilla luciferase reporter. The relative luciferase activity was determined by sequential normalization to the activity of Renilla and each corresponding control cells expressing pcDNA3.1 and shown in F (n = 3 samples). (G) Schematic representation of pGL3-CRRs-HSs luciferase reporter constructs. (H) Reporter assays analogous to D with cells expressing Hba-a2-CRRs-HSs or Hbb-b2-CRRs-HSs luciferase constructs and the indicated siRNAs or genes (n = 3 samples). (I) Reporter assays analogous to F with cells expressing CRRs or CRRs-HSs luciferase constructs and the indicated genes (n = 3 samples). (J) NuE cells were sorted from BM of flox/flox or Δ/Δ mice. Soluble chromatin was immunoprecipitated with antibodies against Wdr82, Set1A, or H3K4me3 and analyzed by qPCR with the indicated primers. The relative fold enrichment was determined by sequential normalization with the cycle threshold values of input and the flox/flox samples (n = 3 samples). (K) Mechanistic model of how lncEry combines with Wdr82 to participate in erythropoiesis regulation. Generally, the novel annotated lncRNA, lncEry, interacts with Wdr82 to control the location of the Set1A/Wdr82 complex and facilitate H3K4me3 binding to CRRs of target genes. In MEP cells, lncEry bind to the CRRs of Klf1 to recruit Wdr82 and elevate H3K4me3 levels at CRRs to promote Klf1 expression and coordinate early erythroid differentiation. In NuE, lncEry promotes Set1A/Wdr82 complex to bind to CRRs as well as LCRs of globin genes to promote transcription of Hba-a1, Hba-a2, Hbb-b1, and Hbb-b2, which promotes terminal erythropoiesis. In brief, lncEry combines with Wdr82 to promote Klf1 and globin gene expression to regulate the early and late stages of erythropoiesis, respectively. Three independent experiments for C–F and H–J. Data are represented as mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; one-way ANOVA for C–D and H; unpaired two-tailed Student’s t test for E + F and I + J.
Discussion
Because a number of lncRNAs have been shown to be important for hematopoiesis, in this study, we aimed to investigate the importance of these regulatory factors in erythropoiesis, about which much less is known. We found a novel functional lncRNA, Gm15915 (lncEry), that is specifically and highly expressed in MEPs and NuEs, and we annotated a new and highly expressed lncEry isoform localized in the nucleus. lncEry depletion decreased Klf1 and globin gene expression in erythroid progenitors and mature cells, respectively, and therefore impaired murine erythroid cell differentiation and maturation. Mechanistically, lncEry together with Wdr82 facilitates the binding of the histone H3K4me3 to the CRRs of Klf1 as well as the CRRs and LCRs of globin genes, which in turn regulates the early and late stages of erythropoiesis (Fig. 10 K).
Erythroid differentiation is regulated at multiple levels to ensure the proper generation of mature cells under multiple physiological conditions (Alvarez-Dominguez et al., 2014). Numerous functional lncRNAs that regulate cellular processes, such as cell development, differentiation, division, survival, and death, have been identified in recent years (Gallagher, 2014; Sabin et al., 2013). The nuclear localization of lncEry suggests that it might participate in regulating gene expression by modulating certain nuclear events such as epigenetic modifications, transcription, or mRNA splicing (Sun et al., 2018). Wdr82 is a unique subunit of the Set1A (KMT2F) histone H3-Lys4 methyltransferase complex and can recruit the Set1A complex to the transcription start sites of target genes and bind to Ser 5 phosphorylated RNA polymerase II to promote transcriptional activation (Deng et al., 2013; Lee and Skalnik, 2005; Rao and Dou, 2015). Recruitment of the KMT2 complex to specific chromatin regions by lncRNAs has been reported (Guo et al., 2020), which supports our hypothesis that lncEry interacts with Wdr82 and participates in transcriptional activation at CRRs as well as LCRs. Furthermore, Wdr82 depletion results in dysfunction of the Set1A/Set1B complex, affecting H3K4me3 status and inhibiting the transcriptional activation of Pou5f1, thereby preventing early embryonic development (Bi et al., 2011). Whether lncEry also participates in embryonic development with Wdr82 needs further exploration.
For transcriptional regulation, recruitment of the mixed-lineage leukemia (MLL) complex to specific chromatin regions depends on the presence not only of plant homeodomains but also lncRNAs; for example, a long intergenic noncoding RNA, HOTTIP, guides WDR5 to chromatin to recruit the MLL complex (Wang et al., 2011; Yang et al., 2014). As a member of the Set1A complex, Wdr82 has affinity for the lncEry 3′ terminus, which is a common region of all lncEry isoforms. lncEry can generally target CRRs of Klf1 and globin genes; thus we speculated that binding of the Wdr82/Set1A complex to the genome is likely to partially depend on lncEry. According to previous reports, Wdr82 promotes Set1A-dependent H3K4 trimethylation (Wu et al., 2008) to participate in transcriptional activation. Even so, lncEry depletion decreased enrichment of Wdr82 and the level of H3K4me3 in the CRRs of Klf1 and globin genes, which supports our hypothesis that lncEry directs the Set1A/Wdr82 complex to the CRRs of Klf1 and globin genes in MEP and MEL cells and stabilizes its location. We thus believe we have identified a new and important regulator associated with the Wdr82/Set1A complex that promotes the transcriptional activation of Klf1 and globin genes in early- and late-stage erythropoiesis, respectively.
Most lncRNAs show little conservation between species and exhibit a rapid evolutionary turnover. Indeed, we did not find lncEry in human cells, yet there are two annotated lncRNAs (ENST00000552603.1 and ENST0000055074.1) similar to lncEry located anti-sense of Ntn4. However, we did not manage to show these lncRNAs to have the same function or expression pattern as lncEry (data not shown). Despite this, we cannot exclude the possibility that other functional lncRNAs bind to the Wdr82/Set1A complex to participate in related biological processes.
In conclusion, we have identified a potential component of the Wdr82/Set1A complex that participates in the transcriptional regulation of Klf1 and globin genes and, by extension, the early- and late-stage of erythroid differentiation to regulate erythropoiesis. Whether lncRNAs together with Wdr82 are viable targets for manipulating other processes and their clinical application need to be addressed in additional research.
Materials and methods
Mice
C57BL/6J and B6.SJL mice were purchased from the animal facility of the State Key Laboratory of Experimental Hematology (SKLEH, Tianjin, China). lncEryflox/+ mice were generated by Beijing Biocytogen, Co. All animal procedures were performed in compliance with the animal care guidelines approved by the Institutional Animal Care and Use Committees of the SKLEH and the Institute of Hematology.
Antibodies and reagents
The following antibodies were used in this study: FLAG (F1804; 1:2,000 for Western blot [WB]) and β-actin (A1978; 1:10,000 for WB) from Sigma-Aldrich; histone H3 (ab1791; 1:10,000 for WB), RNA polymerase II (phospho S5; ab5408; 1:200 for ChIP), H3K4me3 (ab8580; 1:200 for ChIP and Cut&Tag), and H3K27ac (ab6002; 1:200 for Cut&Tag) from Abcam; Wdr82 (99715; 1:1,000 for WB; 1:100 for Cut&Tag), DDX5 (9877T; 1:1,000 for WB), and RBBP5 (13171S; 1:200 for ChIP) from Cell Signaling Technology; PCBP2 (NBP2-19715; 1:1,000 for WB) from Novusbio; PTBP1 (32-4800; 1:1,000 for WB) from Invitrogen; α-globin (14537-1-AP; 1:2,000 for WB) from Proteintech; EKLF (OM184222; 1:1,000 for WB) and β-globin (OM256195; 1:1,000 for WB) from OmnimAbs; and ASH2L (A11278; 1:200 for ChIP) from Abclonal. Anti-FLAG M2 affinity gel (A2220), 3× FLAG peptide (F4799), doxycycline (D9891), PHZ (P26252) and 2′,7′-dichlorofluorescin diacetate (DCF-DA; 35845) were purchased from Sigma-Aldrich. Guinea pig anti-rabbit IgG (heavy & light chain) antibody (ABIN101961; 1:100 for Cut&Tag) was from Antibodies Online. Tetramethylrhodamine methyl ester (TMRE; I34361) and 2-NDBG (2-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)-2-deoxyglucose; N13195) were purchased from Invitrogen. Recombinant murine SCF (250-03), murine IL-3 (213-13), and recombinant human EPO (100-64) were obtained from PeproTech.
ROS levels, mitochondrial membrane potential, and glucose uptake analysis
MEP cells from flox/flox or Δ/Δ mice were labeled with surface markers, and after 30 min of incubation at 4°C, the cells were washed with 1 ml staining buffer. The cells were stained with DCF-DA, TMRE, or 2-NDBG for 20 min at 37°C with shaking and washed with 2 ml cold staining buffer. The cells were immediately analyzed by flow cytometry.
Plasmids and viral production
The FLAG-tagged lncEry isoforms or truncation mutants were expressed using a pcDNA3.1 vector. FLAG-tagged Wdr82 was inserted into a pLenti-Tight-Puro vector. Klf1 was inserted into a pLVX-FLAG-GFP-IRES-Puro vector. Klf1, Hba-a1, Hba-a2, Hbb-b1, and Hbb-b2 CRRs or CRRs-HSs and luciferase were ligated into pGL3-luciferase vectors. The lncEry, Wdr82, and Ddx5 LV-shRNA-GFP lentiviruses were produced by GeneChem. For lentiviral production, the target plasmid was transfected together with pSPAX2 and pMD2G into 293T cell lines using Lipofectamine 2000. The supernatant was harvested after 48 and 72 h of culture and concentrated using an Amicon filter (100K NMWL; Millipore).
Cell culture
MEL and HEK293T cells were purchased from American Type Culture Collection and cultured in RPMI 1640 (A10491-01; Gibco) or DMEM (SH30243.01; Hyclone) with 10% FBS, respectively. BM cells were obtained from C57BL/6 mice and cultured in serum-free expansion medium (09650; STEM CELL). Cells that allow protein expression under doxycycline treatment were created using two steps. First, cells were infected with a lentivirus carrying rtTA and selected using neomycin. The established rtTA cells were subsequently infected with a virus expressing pLenti-Tight-Puro vector that encodes Wdr82 and selected using puromycin. All of the cells integrated with rtTA were cultured in Tet-Approved FBS and RPMI 1640. All cells were authenticated by examination of their morphology and growth characteristics and were confirmed to be mycoplasma-free.
Western blotting
Western blotting was performed as described previously (Yang et al., 2018). In brief, cellular extracts were harvested from cells and resuspended in 5× SDS-PAGE loading buffer. The boiled protein samples were then subjected to SDS-PAGE followed by immunoblotting with the appropriate primary antibodies and secondary antibodies.
RACE assays
5′ and 3′ RACE reactions were performed to isolate full-length lncEry from the total RNA of MEP cells using the 5′- and 3′-Full RACE Kits (TaKaRa) in accordance with the manufacturer’s instructions. Primers used for RACE are listed in Table S6.
Flow cytometry
For cell sorting experiments using mouse MEP cells, cKit+ cells were enriched before flow cytometry using cKit magnetic beads (Miltenyi Biotec). The cells were subsequently stained with a lineage cocktail and cKit (eBioscience; 17-1171-82; 1:200), Sca-1 (eBioscience; 25-5981-82; 1:200), CD34 (eBioscience; 13-0341-82; 1:100), and CD16/CD32 (eBioscience; 45-0161-82; 1:200) antibodies. The lineage cocktail included Gr-1 (BioLegend; 108424; 1:400), Mac-1 (BioLegend; 101226; 1:400), B220 (BioLegend; 103224; 1:400), CD4 (BioLegend; 100414; 1:400), CD8 (BioLegend; 100714; 1:400), CD3 (BioLegend; 100330; 1:400), CD71 (BioLegend; 113806; 1:400), and Ter-119 (BioLegend; 116223; 1:400). DAPI (D9542; 1 mg/ml; Sigma-Aldrich) was used to exclude dead cells. For HSPC analysis, nucleated BM cells were stained with lineage-specific antibodies against Sca-1, cKit, CD34, CD16/CD32, and CD127 (BioLegend; 135040; 1:400). For PreMegE and MPP analyses, nucleated BM cells were stained with lineage cocktail, Sca-1, cKit, CD16/CD32, CD41 (eBioscience; 46-0411-82; 1:400), CD150 (BioLegend; 115904; 1:400), CD48 (BioLegend; 103432; 1:400), and CD105 (BioLegend; 120409; 1:400). The lineage cocktail included CD3, CD4, CD8, B220, Gr-1, Ter119, and Mac-1. For Pro-E analysis, nucleated BM cells were stained with CD3, B220, Mac-1, Ter119, and CD44 (eBioscience; 45-0441-82; 1:200). A modified LSR II flow cytometer with four lasers (355, 488, 561, and 633 nm) was used for the analysis, and an Aria III flow cytometer with four lasers (375, 488, 561, and 633 nm) was used for sorting. The analyses were performed using FACS Diva and FlowJo (TreeStar) software.
Immunophenotypes are as follows: LT-HSC, Lin−Sca-1+cKit+Flk2−CD34− or Lin−Sca-1+cKit+CD48−CD150+; ST-HSC, Lin−Sca-1+cKit+Flk2−CD34+; MPP, Lin−Sca-1+cKit+Flk2+CD34+; MPP1, Lin−Sca-1+cKit+CD48−CD150−; MPP2, Lin−Sca-1+cKit+CD48+CD150+; MPP3/4, Lin−Sca-1+cKit+CD48+CD150−; MKP, Lin−Sca-1−cKit+CD41+CD150+; PreMegE, Lin−Sca-1−cKit+CD41−CD16/CD32−CD150+CD105−; PreCFU-E, Lin−Sca-1−cKit+CD41−CD16/CD32−CD150+CD105+; CFU-E + ProE, Lin−Sca-1−cKit+CD41−CD16/CD32−CD150−CD105+; PreGM, Lin−Sca-1−cKit+CD41−CD16/CD32−CD150−CD105−; CMP, Lin−Sca-1−cKit+Flk2+CD34+CD16/CD32−; CLP, Lin−Sca-1lowcKitlowIL7Rα+Flk2+; GMP, Lin−Sca1−cKit+CD34+CD16/CD32+ or Lin−Sca-1−cKit+CD41−CD150−CD16/CD32+; MEP, Lin−Sca-1−cKit+CD34−CD16/CD32−; MK, CD41+SSChigh; NuE, Ter119+CD71+; monocyte, CD3−B220−NK1.1−F4/80−CD115+SSClow; macrophage, CD3−B220−NK1.1−F4/80+CD115−SSClow; granulocyte, CD3−B220−NK1.1−Mac1+Gr1+SSChigh; CD4 T cell, CD3+B220−CD4+CD8−; CD8 T cell, CD3+B220−CD4−CD8+; B cell, CD3−B220+CD19+; NK, CD4−CD8−B220−NK1.1+; and myeloid, CD3−B220−CD11b+.
Colony-forming assays
Murine BM cells were cultured in M3434 complete methylcellulose-based medium (03434; StemCell Technologies) for 10–14 d to generate BFU-E, CFU-G, CFU-M, CFU-GM, and CFU-GEMM colonies. Murine BM or sorted MEP cells were cultured in M3436 medium (03436; StemCell Technologies) for 48 h to generate CFU-E colonies. Murine sorted MEP cells were cultured in M3334 medium (03334; StemCell Technologies) for 10–14 d to generate BFU-E colonies.
Transplantation
For the lncEry knockdown assay, GFP-expressing lentiviruses carrying control or lncEry shRNA were transduced into B6.SJL (CD45.1+) mouse cKit+ HSPCs, and 4 × 105 transduced cells (CD45.1+GFP+) were transplanted together with 1.5 × 105 CD45.2+ BM cells into lethally irradiated C57BL/6J (CD45.2) recipients; repopulation was measured monthly. For competitive BM transplantation, 5 × 105 BM cells or 300 LT-HSCs from flox/flox or Δ/Δ mice were transplanted into lethally irradiated (9.5 Gy) B6.SJL recipient mice in competition with 2.5 × 105 or 3 × 105 CD45.1+ BM cells. For noncompetitive BM transplantation, 2 × 106 total BM cells from flox/flox or Δ/Δ mice were transplanted into lethally irradiated (9.5 Gy) B6.SJL recipient mice.
RNA pulldown and mass spectrometry analyses
Substrate RNAs were synthesized by in vitro transcription using a T7 RNA production system (P1300; Promega) in accordance with the manufacturer’s instructions. The 3′-end desthiobiotin-labeled RNA probes used in the RNA pulldown were generated using an RNA 3′ End Desthiobiotinylation Kit (20163; Thermo Fisher Scientific) following the manufacturer’s instructions. RNA pulldown was performed using a Magnetic RNA-Protein Pull-Down Kit (20164; Thermo Fisher Scientific). In brief, 3′-end desthiobiotin-labeled RNA probes were captured by streptavidin magnetic beads and mixed with MEP or MEL cell extracts (containing 1 mg of protein) in immunoprecipitation lysis buffer (87787; Thermo Fisher Scientific) in protein-RNA-binding buffer and incubated for 30–60 min at 4°C with agitation or rotation. After general washing, the retrieved proteins were detected by Western blotting or subjected to silver staining and LC-MS/MS analysis (Tables S4 and S5).
Nano-high-performance liquid chromatography–tandem mass spectrometry (HPLC-MS/MS) analysis of lncEry-binding proteins
To identify proteins associated with lncEry, LC-MS/MS analysis was performed on an Orbitrap Q Exactive mass spectrometer. Tryptic peptides were separated by reverse-phase liquid chromatography on an easy-nLC 1000 system (Thermo Fisher Scientific) and directly sprayed into a Q Exactive Plus mass spectrometer (Thermo Fisher Scientific). Mass spectrometry analysis was carried out in data-dependent mode with an automatic switch between a full MS and an MS/MS scan on the Orbitrap. For the full MS scan, the automatic gain control target was 1e6, and the scan ranged from 300 to 1,800 with a resolution of 70,000. The 10 most intense peaks with a charge state ≥2 were selected for fragmentation by high-energy collision dissociation with a normalized collision energy of 27%. The MS2 spectra were acquired with 17,500 resolutions. All MS/MS spectra were searched against the Uniport-Human protein sequence database using Mascot 2.2. Peptide sequences were searched using trypsin specificity while allowing for a maximum of two missed cleavages. Cys carbamidomethylation was specified as a fixed modification, and oxidation of methionine was fixed as a variable modification. The peptide mass tolerance was set at ±20 ppm, and the fragment mass tolerance was set at ±0.1 dalton.
RIP
RIP assays were performed using a Magna Nuclear RIP Kit from Merck Millipore (17-10520; Millipore) with 5 µg IgG or anti-Wdr82 antibody, in accordance with the manufacturer’s instructions. The eluted RNAs were purified using TRIzol LS regent (Life Technology). We then performed qRT-PCR analysis with the indicated primers (Table S6).
qRT-PCR
Total cellular RNA was isolated with TRIzol reagent (Invitrogen) and used for first-strand cDNA synthesis via the Reverse Transcription System (Roche). Quantitation of all gene transcripts was made by qRT-PCR using a Power SYBR Green PCR Master Mix (Roche) and Thermo Quant Studio 5 sequence detection system (Thermo) with the expression of ACTB (β-actin) as the internal control. The primers used are listed in Table S6.
RNAscope and immunofluorescence assays
50,000 cells were centrifuged onto slides at 800 g for 5 min. Cells were fixed with 4% paraformaldehyde for 15 min and dehydrated in 50, 70, and 100% ethanol for 2 min each. After incubating with hydrogen peroxide and proteinase IV, the samples were treated using the RNAscope multiplex fluorescent V2 assay kit (323100; ACD) and incubated with the specific probes (Mm-Gm15915-O2: 555551; Negative Control DapB: 310043 and Positive Control Mm-Ppib: 313911 from ACD) and Cy3 dye (designed by ACD) in accordance with the manufacturer’s instructions. Before staining with DAPI, the samples were incubated with the appropriate primary and secondary antibodies coupled to Alexa Fluor 488 (Invitrogen) following the instructions provided by ACD. Confocal images were captured using a Spinning Disk Confocal Microscope system (UltraVIEW VOX) with a ×100 oil objective. To avoid bleed-through effects in the double-staining experiments, each dye was scanned independently in multitracking mode.
Recombinant protein purification
Lysates from MEL cells stably expressing FLAG-Wdr82 were prepared by incubating the cells in lysis buffer (300 mM NaCl, 1% NP-40, 0.1% SDS, 0.5% sodium-deoxycholate, and 50 mM Tris-HCl, pH 8.0) containing a protease inhibitor cocktail (Roche). Anti-FLAG immunoaffinity columns were prepared using anti-FLAG M2 affinity gel (Sigma-Aldrich) following the manufacturer’s instructions. Cell lysates were applied to an equilibrated FLAG column of 200-μl bed volume to allow for protein adsorption to the column resin. After binding, the column was extensively washed with high-salt solution for 5 min five times each (300 mM NaCl, 150 mM KCl, 1% NP-40, 0.1% SDS, 0.5% sodium-deoxycholate, and 50 mM Tris-HCl, pH 8.0). A FLAG peptide (Sigma-Aldrich) was applied to the column to elute the FLAG protein complex, as recommended by the manufacturer. The eluents were collected and visualized by SDS-PAGE followed by Coomassie blue staining. The purified protein was used in subsequent molecular interaction assays.
SPR
Sensorgrams of lncEry-P5 binding to Wdr82 were measured by SPR technology using a Biacore 3000 instrument (GE Healthcare). The flowing-phase RNA lncEry-P5 was synthesized by in vitro transcription using a T7 RNA production system (P1300; Promega) in accordance with the manufacturer’s instructions. The substrate-phase Wdr82 proteins were purified from MEL cells and immobilized on a CM5 sensor chip (BIAcore) with the following procedure. All experiments were carried out using HBS-EP (10 mM Hepes, pH 7.4, 150 mM NaCl, 3.4 mM EDTA, and 0.005% surfactant P20) as the running buffer, at a constant flow rate of 30 μl/min at 25°C. The Wdr82 protein was diluted in 10 mM sodium acetate buffer (pH 4.0) to a final concentration of 2.5 μM and covalently immobilized on the hydrophilic carboxymethylated dextran matrix of the CM5 sensor chip using a standard primary amine coupling procedure. lncEry-P5 was dissolved in the running buffer to concentrations ranging from 36.7 to 294 nM. All data were analyzed by BIA evaluation software, and the sensorgrams were processed by automatic correction for nonspecific bulk refractive index effects. Kinetic analyses of lncEry-P5/Wdr82 binding were performed based on the 1:1 Langmuir binding fit model in accordance with the procedures in the software manual.
RNA interference
All siRNA transfections were performed using Lipofectamine RNAiMAX (Invitrogen) following the manufacturer’s recommendations. The final concentration of the siRNA molecules was 10 nM, and the cells were harvested 72 or 96 h after transfection, depending on the experiment. Control, lncEry, and Wdr82 siRNAs were chemically synthesized by Sigma-Aldrich. The shRNAs against lncEry, Wdr82, or Ddx5 in lentivirus U6-MSX-IRES vectors were purchased from GeneChem. The siRNA and shRNA sequences are provided in Table S6.
RNA-seq and analysis
RNA-Seq included three parts of experiment samples. First, a total of 16 hematopoietic cell populations were sorted from the BM of C57BL/6 mice. Second, MEP cells were sorted from the BM of flox/flox or Δ/Δ mice. Third, MEL cells were transfected with control or lncEry siRNAs. Total RNA was isolated using the RNeasy (Qiagen) purification kit. Libraries were prepared using an Illumina RNA library preparation TruSeq PE kit. High-throughput RNA-Seq was performed on an Illumina Xten sequencer (paired-end 150-bp sequencing). Clean reads were filtered by removing reads including adapters, reads including poly-N, and low-quality reads from raw data. All the following analyses were based on clean data. Clean data were first aligned to the mouse genome (GRCm38) with GENCODE M16 gene annotation using HISAT2 (v2.1.0; Kim et al., 2015). DEG analyses were conducted using Cufflinks and Cuffdiff (v2.2.1) software (Trapnell et al., 2010). Unsupervised hierarchical clustering was conducted using pheatmap R package. Genes with a q value <0.05 in the Cuffdiff results were considered to be significantly different genes.
ATAC-seq
ATAC-seq was performed as described previously (Wang et al., 2019), In brief, 50,000 MEP cells sorted from BM of flox/flox or Δ/Δ mouse cells were lysed in lysis buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% [vol/vol] IGPAL CA-630) for 10 min on ice. After centrifugation at 500 g for 5 min, the obtained nuclei were added to 50 μl transposition reaction buffer (offered by Vazyme TD501-01) followed with incubation at 37°C for 30 min. After tagmentation, VAHTS DNA Clean Beads were used to stop the reaction, and DNA was purified for final library construction (TruePrep DNA Library Prep Kit V2 for Illumina) before paired-end high-throughput sequencing using HiSeq XTe. Clean reads were aligned to the mouse genome (GRCm38) using BWA package, and peaks were called using MACS2 package (v2.2.5; Zhang et al., 2008) with a false discovery rate (FDR) cutoff of 0.05.
Cleavage under targets and tagmentation
Cut&Tag experiments were performed as described previously (Kaya-Okur et al., 2019) together with the Hyperactive TM In-Situ ChIP Library Prep Kit for Illumina from Vazyme (TD902-01). MEP cells sorted from the BM of flox/flox or Δ/Δ mice cells were captured by ConA beads and incubated with primary and secondary antibodies in antibody buffer and dig-wash buffer, respectively, for the time indicated in the manufacturer’s instructions. The cells were then incubated with Hyperactive pA-Tn5 Transposon and fragmented in Tagmentation Buffer at 37°C for 1 h. The extracted DNA fragments were analyzed by high-throughput sequencing. Clean reads were aligned to the mouse genome (GRCm38) using the Bowtie2 package (Langmead and Salzberg, 2012), and peaks were detected using MACS2 callpeak (v2.2.5; Zhang et al., 2008) with an FDR cutoff of 0.05.
ChIP-seq
ChIP-seq was performed as described previously (Shang et al., 2019). In brief, ∼10 million cells were crosslinked with 1% formaldehyde for 10 min at room temperature and quenched by adding glycine to a final concentration of 125 mM for 5 min. The fixed cells were resuspended in SDS lysis buffer (1% SDS, 5 mM EDTA, and 50 mM Tris-HCl, pH 8.1) in the presence of protease inhibitors and subjected to sonication (Bioruptor; Diagenode) to generate chromatin fragments of ∼300 bp in length. For immunoprecipitation, after dilution, the chromatin was incubated with control or specific antibodies (3 μg) overnight at 4°C with constant rotation; 50 μl of 50% (vol/vol) protein G magnetic beads was then added, and the incubation was continued for an additional 2 h. The beads were washed with the following buffers: TSE I (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, and 150 mM NaCl), TSE II (0.1% SDS, 1% Triton X-100, 2 mM EDTA, and 20 mM Tris-HCl), buffer III (0.25 M LiCl, 1% Nonidet P-40, 1% sodium deoxycholate, 1 mM EDTA, and 10 mM Tris-HCl), and Tris-EDTA buffer as described previously (Shang et al., 2019). The cross-links were removed from the pulled-down chromatin complex together with the input sample by incubation at 65°C for 2 h in elution buffer. The eluted DNA was purified using a PCR purification kit (Qiagen) and analyzed by qPCR using the primers detailed in Table S6 or by high-throughput sequencing. Clean reads were aligned to the mouse genome (GRCm38) using the Bowtie2 package (Langmead and Salzberg, 2012). ChIP-seq peaks were detected using MACS2 callpeak (v2.2.5; Zhang et al., 2008) with a minimum FDR cutoff of 0.05.
ChIRP-seq
ChIRP-seq was performed as described previously (Chu et al., 2011). Eluted DNA was purified using a PCR purification kit (Qiagen) and analyzed by qPCR using the qChIP primers described in Table S6 or by high-throughput sequencing. The processing of sequencing data was the same as that for the ChIP-seq data described above. Peaks were annotated using the ChIPseeker R package (v1.24.0; Yu et al., 2015).
Luciferase reporter assay
The modulation of globin gene promoter activity by lncEry was analyzed by luciferase assay. A pGL3 construct carrying the globin gene CRRs sequence was obtained by PCR enrichment using the primers listed in Table S6, and the Klf1 CRRs sequence was synthesized. Luciferase reporter activity was measured using the Dual Luciferase Assay System (E1910; ProMega). Relative luciferase activity was normalized to Renilla luciferase and control vector luciferase reporter activity.
Cell type specifically expressed lncRNA analysis
Statistical analyses
Data from biological triplicate experiments are presented, and data represent means ± SD. An unpaired two-tailed Student’s t test was used to compare two groups of data. ANOVA with Bonferroni’s correction was used to compare multiple groups of data. A P value <0.05 was considered statistically significant. All statistical analyses were performed in SPSS v20.0 software, and graphs were generated by GraphPad Prism v8.0 software.
Data availability
RNA-seq datasets of 12 hematopoietic cell populations were deposited in the NCBI GEO (GSE142216). Other sequencing data have been deposited in the NCBI SRA (PRJNA647682). The uncropped gels and Western blots have been provided in Source Data files.
Online supplemental material
Fig. S1 is related to Fig. 1 and shows the analyses of bulk or single-cell RNA-seq data from previous studies as well as ours. Fig. S2 is related to Fig. 1 and shows the sequence information and expression characteristics of lncEry. Fig. S3 shows erythroid differentiation of HSPCs after lncEry knockdown by shRNAs. Fig. S4 is related to Fig. 2 and shows (i) flow cytometric analysis of hematopoietic populations in BM, SP and fetal liver; (ii) stress erythropoiesis by using PHZ treatment or a noncompetitive transplantation; and (iii) a BFU-E colony of MEP cells from flox/flox and Δ/Δ mice. Fig. S5 is related to Figs. 2 and 5, and shows (i) the results from the second lncEry conditional knockout (cKO) mouse model; and (ii) qRT-PCR and GSEA analyses of RNA-seq data derived from MEL cells. Table S1 lists the top 10 highly expressed lncRNAs in each population. Table S2 lists overlapping downregulated genes in MEP cells (203). Table S3 lists overlapping downregulated genes in MEL cells (75). Table S4 shows mass spectrometry analysis of lncEry-interacting proteins in MEP cells. Table S5 shows mass spectrometry analysis of lncEry-interacting proteins in MEL cells. Table S6 lists 5′ and 3′ RACE primers, qRT-PCR primers, qChIP primers, siRNA sequences, shRNA sequences, ChIP probe sequences, ChIRP probe sequences, and luciferase reporter primers.
Acknowledgments
We thank all lab members for their assistance with the experiments. We thank Dr. Yiran Ren for providing help on SPR assays.
This work was supported by grants from the Ministry of Science and Technology of China (2021YFA1100900, 2020YFE0203000, 2017YFA0103400), the National Natural Science Foundation of China (81922002, 81730006, 81861148029, 81870086, 81900113, 81890993, 81900117, 81920108004, 82070112, 82000115, 32000803, 82000116, 81872289), Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences (2021-I2M-1-019), the Chinese Academy of Medical Sciences Fundamental Research Funds for Central Research Institutes (2019RC310003, 3332021093), and Distinguished Young Scholars of Tianjin (19JCJQJC63400).
Author contributions: S.D. Yang, G.H. Sun, and C. Chen designed and performed the experiments, analyzed the data, and wrote the paper. P. Wu performed bioinformatic analyses. Y.J. Kuang, L. Liu, Z.F. Zheng, Y.C. He, Q. Gu, F.J. Wang, F.L. Gou, Z.N. Yang, X. Lv, X.N. Zhao, and S.H. Lu helped with mouse and biochemistry experiments. T. Lu, C.Y. Zhu, and L. Yang helped with bioinformatic analyses. Y.P. Li helped with experimental design. Y.N. Ma, J. Yu, S.R. Yuan, F. Dong, L.H. Shi, and L.G. Ng assisted with the manuscript. J. Liu, L. Shi, T. Cheng, and H. Cheng proposed the study, designed the experiments, interpreted the results, wrote the paper, and oversaw the research project.
References
Author notes
Disclosures: The authors declare no competing interests exist.
S. Yang, G. Sun, P. Wu, and C. Chen contributed equally to this paper.