Cernunnos is involved in the nonhomologous end-joining (NHEJ) process during DNA double-strand break (DSB) repair. Here, we studied immunoglobulin (Ig) class switch recombination (CSR), a physiological process which relies on proper repair of the DSBs, in B cells from Cernunnos-deficient patients. The pattern of in vivo generated CSR junctions is altered in these cells, with unusually long microhomologies and a lack of direct end-joining. The CSR junctions from Cernunnos-deficient patients largely resemble those from patients lacking DNA ligase IV, Artemis, or ATM, suggesting that these factors are involved in the same end-joining pathway during CSR. By screening 269 mature B cell lymphoma biopsies, we also identified a somatic missense Cernunnos mutation in a diffuse large B cell lymphoma sample. This mutation has a dominant-negative effect on joining of a subset of DNA ends in an in vitro NHEJ assay. Translocations involving both Ig heavy chain loci and clonal-like, dynamic IgA switching activities were observed in this tumor. Collectively, our results suggest a link between defects in the Cernunnos-dependent NHEJ pathway and aberrant CSR or switch translocations during the development of B cell malignancies.
Mammalian B cells require two forms of DNA recombination to produce functional antibody-encoding genes. The first, V(D)J recombination, takes place during early B cell development and mediates assembly of coding regions of the variable (V) domains of antibodies. By combining different V, diversity (D), and joining (J) gene segments, a vast repertoire of antibody specificities can be generated. The second, class switch recombination (CSR), occurs in mature B cells and allows a previously rearranged Ig heavy chain (IGH or Igh when referring to the human and the mouse loci, respectively) V domain to be expressed in association with a different constant (C) domain. CSR does not affect the antibody specificity but leads to the production of different antibody classes (IgG, IgA, or IgE) with improved biological properties.
V(D)J recombination, which is initiated by the lymphocyte-specific proteins RAG1 and RAG2, is a site-specific process as it proceeds through precise DNA cleavage at conserved signal sequences (Jung et al., 2006). CSR, which is initiated by the B cell–specific factor activation-induced cytidine deaminase (AID; Muramatsu et al., 2000), is rather a region-specific process, involving cleavage and recombination of tandemly repeated DNA sequences referred to as switch (S) regions, located upstream of the C regions (Stavnezer, 1996; Pan-Hammarström et al., 2007). However, there are also similarities between the two types of recombination. Both processes are regulated by transcription. Furthermore, DNA double-strand breaks (DSBs) are intermediates for both V(D)J recombination and CSR (Wuerffel et al., 1997), and the nonhomologous end-joining (NHEJ) machinery has been implicated in resolution of the DSBs in both processes (Chaudhuri and Alt, 2004; Jung et al., 2006; Kotnis et al., 2009).
NHEJ is the principle mechanism for DSB repair in vertebrate cells and requires a set of proteins (Lieber et al., 2003; Lieber, 2008). It has been proposed that NHEJ starts with Ku70/80 binding to the DNA ends at the DSBs. Subsequently, Ku recruits the catalytic subunit of the DNA-dependent protein kinase (DNA-PKcs), forming the active DNA-PK holoenzyme, which regulates/facilitates the recruitment of additional factors such as Artemis (Moshous et al., 2001), a nuclease which is thought to be involved in DNA end processing, and XRCC4–DNA ligase IV (Lig4), the ligase complex.
Cernunnos (XLF or NHEJ1) is the latest addition to the NHEJ machinery (Ahnesorg et al., 2006; Buck et al., 2006a). In humans, mutations in the gene encoding Cernunnos result in a rare, autosomal recessive disorder characterized by microcephaly, radiosensitivity, and combined immunodeficiency (Buck et al., 2006a). Defective V(D)J recombination probably accounts for the profound T and B cell lymphocytopenia observed in these patients (Buck et al., 2006a). In addition, a possible CSR defect has been suggested (Buck et al., 2006a). In vitro biochemical studies have further shown that the role of Cernunnos in NHEJ relies on its ability to stimulate incompatible DNA end ligation by the XRCC4–Lig4 complex (Gu et al., 2007b; Lu et al., 2007b; Tsai et al., 2007). The importance of Cernunnos in NHEJ-mediated DSB repair, including the V(D)J recombination process, has been confirmed in Cernunnos-deficient murine embryonic stem cells (Zha et al., 2007). Surprisingly, in Cernunnos-deficient mice, only a modestly decreased number of mature lymphocytes has been observed, and Cernunnos-deficient pro-B cell lines can support nearly normal levels of V(D)J recombination (Li et al., 2008a). A recent study suggested that the redundant functional properties of ATM and Cernunnos in joining DNA breaks might explain this modest defect of lymphocyte development (Zha et al., 2011). If this indeed reflects a lymphocyte-specific compensation for Cernunnos deficiency in V(D)J recombination, it is unclear why such a mechanism would not rescue the development of T and B lymphocytes in humans.
Mature B cells from Cernunnos-deficient mice have been reported to be modestly defective in CSR (Li et al., 2008a). It is unclear whether this again reflects a lymphocyte-specific compensatory mechanism or whether it indicates a nonessential role of Cernunnos in CSR. To address this question, we studied the in vivo pattern of CSR junctions in Cernunnos-deficient human B cells.
Knocking out one of the NHEJ factors (Lig4, XRCC4, Ku70, Ku80, DNA-PKcs, or Artemis) in mice on a p53 deficiency background leads to the development of pro-B cell lymphomas associated with complex Igh translocations (Difilippantonio et al., 2002; Zhu et al., 2002; Gladdy et al., 2003; Rooney et al., 2004). These translocations are RAG dependent and represent an inefficient repair of DSBs on the Igh loci during V(D)J recombination. Inactivation of XRCC4 in p53-deficient mature B cells leads to the development of surface Ig–negative B cell lymphomas, which often harbor CSR-related reciprocal chromosomal Igh/Myc translocations (Wang et al., 2008). These translocations share some features with the AID-dependent Igh translocations observed in IL6-induced murine plasmacytomas (Ramiro et al., 2004) and are likely to be associated with aberrant repair of DSBs during CSR. Cernunnos and p53 double-deficient mice rarely develop pro-B cell lymphomas and instead succumb to thymic lymphomas (Li et al., 2008a). This again could reflect the mild defect of V(D)J recombination in pro-B cells from these mice (Li et al., 2008a). To address the question of whether Cernunnos deficiency and aberrant CSR events are associated with development of malignancies in human B cells, we screened for somatic mutations in the Cernunnos gene in a large cohort of mature B cell lymphomas.
RESULTS
Altered pattern of Sμ-Sα recombination junctions in Cernunnos-deficient cells
This study encompasses seven patients, including four of the five patients described in the original study on Cernunnos deficiency (P1, P2, P4, and P5; Buck et al., 2006a). Patients P6 and P7 were recently diagnosed, and, similar to the previously described patients, they both suffer from microcephaly, developmental delay, and combined immunodeficiency with substantially reduced numbers of T and B cells (Table 1). Patient P8 presented with microcephaly, agammaglobulinemia, and more striking physical anomalies, including microphthalmia, blepharophimosis, posterior cleft palate, and acral anomalies (Verloes et al., 2001). In contrast to the point mutations identified in the other six patients, this patient had a large homozygous deletion in the Cernunnos gene, encompassing exons 2–5 (Fig. 1), which may explain the more severe clinical manifestation observed. Markedly reduced serum levels of IgA and IgG were observed in all patients, whereas the levels of IgM were quite variable; low in P2, P7, and P8, normal in P4 and P5, and high in P1 and P6, when compared with age-matched controls (Table 1). The location of mutations identified in the Cernunnos gene for all patients is depicted in Fig. 1.
Characterization of Cernunnos-deficient patients
| Patient ID | Age at sampling | T cell counts | B cell counts | Serum Ig | References | ||
| IgM | IgG | IgA | |||||
| yr | cells/µl | cells/µl | g/liter | g/liter | g/liter | ||
| P1 | 14 | 630↓ | 0↓ | 6.8↑ | 2.4↓ | <0.06↓ | P1 in Buck et al. (2006a) |
| P2 | 2 | 591↓ | 154↓ | 0.27↓ | 2.37↓ | <0.05↓ | P4 in Buck et al. (2006a) |
| P4 | 2 | 730↓ | 54↓ | 1.39 | <0.33↓ | <0.06↓ | P2 in Buck et al. (2006a) |
| P5 | 7 | 693↓ | 83↓ | 1.05 | 0.6↓ | 0.06↓ | P5 in Buck et al. (2006a) |
| P6 | 6 | 600↓ | 81↓ | 9.0↑ | <1.2↓ | <0.06↓ | This study |
| P7 | 2 | 1,260↓ | 38↓ | 0.53↓ | 1.4↓ | 0.13↓ | This study |
| P8 | 11 | 182↓ | 0↓ | 0.16↓ | 0.1↓ | <0.06↓ | This study |
| Patient ID | Age at sampling | T cell counts | B cell counts | Serum Ig | References | ||
| IgM | IgG | IgA | |||||
| yr | cells/µl | cells/µl | g/liter | g/liter | g/liter | ||
| P1 | 14 | 630↓ | 0↓ | 6.8↑ | 2.4↓ | <0.06↓ | P1 in Buck et al. (2006a) |
| P2 | 2 | 591↓ | 154↓ | 0.27↓ | 2.37↓ | <0.05↓ | P4 in Buck et al. (2006a) |
| P4 | 2 | 730↓ | 54↓ | 1.39 | <0.33↓ | <0.06↓ | P2 in Buck et al. (2006a) |
| P5 | 7 | 693↓ | 83↓ | 1.05 | 0.6↓ | 0.06↓ | P5 in Buck et al. (2006a) |
| P6 | 6 | 600↓ | 81↓ | 9.0↑ | <1.2↓ | <0.06↓ | This study |
| P7 | 2 | 1,260↓ | 38↓ | 0.53↓ | 1.4↓ | 0.13↓ | This study |
| P8 | 11 | 182↓ | 0↓ | 0.16↓ | 0.1↓ | <0.06↓ | This study |
Normal ranges for T and B cell counts and serum Ig levels have been described previously (Buck et al., 2006a). Values that are above or below the normal ranges are indicated by arrows. T cell counts are as follows: 2,300–5,400, 2 yr; 1,900–3,700, 6–7 yr; 1,400–3,300, 13–14 yr. B cell counts are as follows: 390–1,400, 2 yr; 110–570, 6–7 yr; 110–570, 13–14 yr. IgM levels are as follows: 0.72–1.6 g/liter, 2 yr; 0.6–1.75 g/liter, 6–7 yr; 0.4–1.8 g/liter, 13–14 yr. IgG levels are as follows: 5.2–10.8 g/liter, 2 yr; 5.7–13.2 g/liter, 6–7 yr; 6.4–14.2 g/liter, 13–14 yr. IgA levels are as follows: 0.36–1.65 g/liter, 2 yr; 0.65–2.4 g/liter, 6–7 yr; 0.52–2.2 g/liter, 13–14 yr (Buck et al., 2006a).
Schematic of the structure of the human Cernunnos gene. Numbered boxes represent exon sequences, and the filled gray boxes indicate the coding sequences. The approximate positions of mutations identified in Cernunnos-deficient patients (P1, P2, P4, P5, P6, P7, and P8 denote individual patients) and the lymphoma sample (DL8; highlighted in red) are indicated by arrows. All the point mutations are located in the coding regions. He, heterozygous; Ho, homozygous.
Schematic of the structure of the human Cernunnos gene. Numbered boxes represent exon sequences, and the filled gray boxes indicate the coding sequences. The approximate positions of mutations identified in Cernunnos-deficient patients (P1, P2, P4, P5, P6, P7, and P8 denote individual patients) and the lymphoma sample (DL8; highlighted in red) are indicated by arrows. All the point mutations are located in the coding regions. He, heterozygous; Ho, homozygous.
Genomic DNA samples purified from peripheral blood cells were available from all seven patients. To determine whether CSR was affected by a lack of functional Cernunnos, individual Sμ-Sα junctions were amplified using our previously described nested-PCR assay (Pan et al., 2001, 2002). Notably, Sμ-Sα fragments could be amplified from DNA samples from all Cernunnos-deficient patients, even in patients with extremely low levels of serum IgA (P1, P2, P4, P5, P6, and P8) and peripheral B cells (P1 and P8; undetectable by routine flow cytometry analyses). However, the number and intensity of the amplified bands were much lower in patients as compared with controls (Fig. 2 A). The amplified Sμ-Sα fragments, including the very weak bands amplified from the Cernunnos-deficient patients, were subsequently cloned and sequenced and indeed displayed bona fide CSR junctions. Residual IgA switching thus appears to occur in all patients, irrespective of the type of Cernunnos mutations (missense mutations in P1 and P5, truncated or frame-shift mutations in P2, P4, P6, and P7, and large deletions in P8; Fig. 1).
Characterization of Sμ-Sα junctions in Cernunnos-deficient patients and healthy controls. (A) PCR amplification of Sμ-Sα fragments. 10 PCR reactions were run in parallel using DNA from each individual. M, molecular weight marker (1-kb ladder from Invitrogen or 100 plus DNA ladder from Fermentas). (B) Selected sequences of Sμ-Sα junctions from Cernunnos-deficient patients (X7-37a and X8-3) and controls (C5-216 and SC4-20). The recombination junctional sequences are aligned with Sμ (above) and Sα1 or Sα2 (below) reference sequences. Microhomology (perfectly matched sequence homology) is indicated by a box (solid line). Imperfect repeat was determined by identifying the longest overlap region at the switch junction by allowing one mismatch on either side of the breakpoints (the extra nucleotide identified beyond the perfectly matched sequence identity is boxed by a dashed line). The length of microhomology and imperfect repeat (in parenthesis) for each S junction is indicated at the bottom right. Insertion was defined as a nucleotide at the breakpoints that was not identical to either of the switch regions. The Sμ and Sα breakpoints for each switch fragment are indicated by ▾ and ▴, respectively, and their positions in the reference sequences are indicated above or below the arrowheads. OL, overlap. (C) Pie charts demonstrating the microhomology usage at Sμ-Sα junctions in patients and controls. The proportion of switch junctions with a given size of perfectly matched short homology is indicated by the size of the slices. The numbers of switch junctions from each patient are as follows: P1, n = 11; P2, n = 6; P4, n = 8; P5, n = 6; P6, n = 4; P7, n = 31; and P8, n = 18. At least two independent experiments were performed on each patient, and a representative gel picture from P1, P4, and P8 is shown in A, whereas B and C are a highlight and summary of sequencing results obtained from all experiments.
Characterization of Sμ-Sα junctions in Cernunnos-deficient patients and healthy controls. (A) PCR amplification of Sμ-Sα fragments. 10 PCR reactions were run in parallel using DNA from each individual. M, molecular weight marker (1-kb ladder from Invitrogen or 100 plus DNA ladder from Fermentas). (B) Selected sequences of Sμ-Sα junctions from Cernunnos-deficient patients (X7-37a and X8-3) and controls (C5-216 and SC4-20). The recombination junctional sequences are aligned with Sμ (above) and Sα1 or Sα2 (below) reference sequences. Microhomology (perfectly matched sequence homology) is indicated by a box (solid line). Imperfect repeat was determined by identifying the longest overlap region at the switch junction by allowing one mismatch on either side of the breakpoints (the extra nucleotide identified beyond the perfectly matched sequence identity is boxed by a dashed line). The length of microhomology and imperfect repeat (in parenthesis) for each S junction is indicated at the bottom right. Insertion was defined as a nucleotide at the breakpoints that was not identical to either of the switch regions. The Sμ and Sα breakpoints for each switch fragment are indicated by ▾ and ▴, respectively, and their positions in the reference sequences are indicated above or below the arrowheads. OL, overlap. (C) Pie charts demonstrating the microhomology usage at Sμ-Sα junctions in patients and controls. The proportion of switch junctions with a given size of perfectly matched short homology is indicated by the size of the slices. The numbers of switch junctions from each patient are as follows: P1, n = 11; P2, n = 6; P4, n = 8; P5, n = 6; P6, n = 4; P7, n = 31; and P8, n = 18. At least two independent experiments were performed on each patient, and a representative gel picture from P1, P4, and P8 is shown in A, whereas B and C are a highlight and summary of sequencing results obtained from all experiments.
Altogether, 84 unique Sμ-Sα sequences from the Cernunnos-deficient patients were analyzed (Fig. S1), and these were compared with our previously described 137 Sμ-Sα junctions from a group of healthy young individuals (1–6 yr; Du et al., 2008). The majority of CSR junctions represented direct switching from IgM to IgA1 or IgA2 (Sμ-Sα1 or Sμ-Sα2). Sequential switching, involving Sμ and two Sα regions (Sμ-Sα1-Sα2), was occasionally observed in both patients and controls (1% of total CSR junctions analyzed). However, another type of sequential switching, involving recombination of the Sγ regions (Sμ-Sγ-Sα1 or Sμ-Sγ-Sα2), was not observed in patients, whereas this type of CSR junctions could be identified in controls (3%).
The Sμ-Sα junctions from Cernunnos-deficient B cells were characterized by a strong preference of microhomology (perfectly matched sequence homology, on average 8.5 ± 6.9 vs. 3.9 ± 5.1 nt in controls; Student’s t test, P = 4.3 × 10−7). More than 40% of the junctions from patients exhibited a microhomology of ≥10 bp (42 vs. 16% in controls; χ2 test, P = 2.4 × 10−5), with the longest being 32 bp (Fig. 2, B and C; and Table 2). The latter observation is striking, as when we performed a dot plot analysis of the human Sμ (4.5 kb) and Sα1 (3.3 kb) sequences, taking into account the constraints of the PCR method used (i.e., <2.5 kb can be amplified), this is the only position where such a long overlap is theoretically possible (i.e., perfect match, 32 bp, or with one mismatch, 41/42 bp; X7-37a in Fig. 2 B and not depicted). We have not observed such a breakpoint in >700 control Sμ-Sα fragments analyzed (311 from previously published [Pan et al., 2002; Pan-Hammarström et al., 2005; Du et al., 2008] and 419 from unpublished control sequences). Thus, the formation of CSR junctions with long microhomologies at the CSR junctions is probably not an accidental joining of two DNA ends with overlapping overhangs. Rather, active mechanisms of searching for, and employment of, long sequence homologies seem to be required for the alternative end-joining (A-EJ) process when Cernunnos is defective.
Characterization of Sμ-Sα and Sμ-Sγ junctions
| Patients | Perfectly matched short homology | Total number of S fragments | |||||
| 0 bp | 1–3 bp | 4–6 bp | 7–9 bp | ≥10 bp | |||
| 1-bp insertions | No insertions | ||||||
| Sμ-Sα | |||||||
| Cernunnos−/− | 16 (19%) | 2 (2%)*** | 7 (8%)* | 12 (14%) | 12 (14%) | 35 (42%)*** | 84 |
| Artemis−/− | 6 (11%)* | 0 (0%)** | 10 (19%) | 8 (15%) | 9 (17%) | 21 (39%)*** | 54 |
| Controls (1–6 yr) | 34 (25%) | 24 (18%) | 25 (18%) | 21 (15%) | 11 (8%) | 22 (16%) | 137 |
| Lig4−/− | 1 (3%)** | 0 (0%)*** | 7 (23%) | 4 (13%) | 4 (13%) | 14 (47%)*** | 30 |
| ATM−/− | 1 (2%)*** | 1 (2%)** | 15 (34%) | 9 (20%) | 5 (11%) | 13 (30%)*** | 44 |
| Controls (adults) | 39 (25%) | 28 (18%) | 56 (36%) | 15 (10%) | 11 (7%) | 5 (3%) | 154 |
| Sμ-Sγ | |||||||
| Cernunnos−/− | 2 (33%)a | 0 (0%) | 2(33%) | 2 (33%) | 0 (0%) | 0 (0%) | 6 |
| Artemis−/− | 4 (17%) | 5 (21%) | 14 (58%) | 1 (4%) | 0 (0%) | 0 (0%) | 24 |
| Controls (1–6 yr) | 9 (16%) | 13 (22%) | 26 (45%) | 10 (17%) | 0 (0%) | 0 (0%) | 58 |
| Lig4−/− | 11 (32%)* | 4 (12%) | 15 (44%) | 4 (12%) | 0 (0%) | 0 (0%) | 34 |
| ATM−/− | 3 (8%) | 3 (8%) | 23 (61%) | 7 (18%)* | 2 (5%) | 0 (0%) | 38 |
| Controls (adults) | 7 (12%) | 12 (20%) | 37 (63%) | 3 (5%) | 0 (0%) | 0 (0%) | 59 |
| Patients | Perfectly matched short homology | Total number of S fragments | |||||
| 0 bp | 1–3 bp | 4–6 bp | 7–9 bp | ≥10 bp | |||
| 1-bp insertions | No insertions | ||||||
| Sμ-Sα | |||||||
| Cernunnos−/− | 16 (19%) | 2 (2%)*** | 7 (8%)* | 12 (14%) | 12 (14%) | 35 (42%)*** | 84 |
| Artemis−/− | 6 (11%)* | 0 (0%)** | 10 (19%) | 8 (15%) | 9 (17%) | 21 (39%)*** | 54 |
| Controls (1–6 yr) | 34 (25%) | 24 (18%) | 25 (18%) | 21 (15%) | 11 (8%) | 22 (16%) | 137 |
| Lig4−/− | 1 (3%)** | 0 (0%)*** | 7 (23%) | 4 (13%) | 4 (13%) | 14 (47%)*** | 30 |
| ATM−/− | 1 (2%)*** | 1 (2%)** | 15 (34%) | 9 (20%) | 5 (11%) | 13 (30%)*** | 44 |
| Controls (adults) | 39 (25%) | 28 (18%) | 56 (36%) | 15 (10%) | 11 (7%) | 5 (3%) | 154 |
| Sμ-Sγ | |||||||
| Cernunnos−/− | 2 (33%)a | 0 (0%) | 2(33%) | 2 (33%) | 0 (0%) | 0 (0%) | 6 |
| Artemis−/− | 4 (17%) | 5 (21%) | 14 (58%) | 1 (4%) | 0 (0%) | 0 (0%) | 24 |
| Controls (1–6 yr) | 9 (16%) | 13 (22%) | 26 (45%) | 10 (17%) | 0 (0%) | 0 (0%) | 58 |
| Lig4−/− | 11 (32%)* | 4 (12%) | 15 (44%) | 4 (12%) | 0 (0%) | 0 (0%) | 34 |
| ATM−/− | 3 (8%) | 3 (8%) | 23 (61%) | 7 (18%)* | 2 (5%) | 0 (0%) | 38 |
| Controls (adults) | 7 (12%) | 12 (20%) | 37 (63%) | 3 (5%) | 0 (0%) | 0 (0%) | 59 |
The switch junctions from Cernunnos- or Artemis-deficient patients were compared with those from control children (1–6 yr of age), whereas the switch junctions from Lig4- or ATM-deficient patients were compared with adult controls. Statistical analysis was performed using the χ2 test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). The data from Artemis-, Lig4-, and ATM-deficient patients and controls have been described previously (Pan et al., 2002; Pan-Hammarström et al., 2005; Du et al., 2008).
One Sμ-Sγ junction from Cernunnos-deficient cells showed a 3-bp insertion at the junction.
Approximately 20% of the Sμ-Sα junctions from Cernunnos-deficient patients and 25% from controls were classified as having 1-bp insertions (Table 2). However, most of the junctions derived from patients (15 of 16; 93%) were flanked by ≥3/4-bp imperfect repeats, whereas in controls, about one third were not associated with any imperfect repeats (exemplified by X8-3 for patients and SC4-20 for controls in Fig. 2 B). Thus, although all junctions could be classified as 1-bp insertions, the junctions from patients were more likely to be joined by a mechanism that is dependent on imperfect repeats.
The Sμ-Sα junctions obtained from Cernunnos-deficient B cells were also characterized by a markedly reduced number of junctions caused by direct end-joining (no microhomologies, no insertions; 2 vs. 18%; χ2 test, P = 0.0007; Table 2). This feature, defective direct joining, along with the dramatic increased length of microhomology, has previously been observed in B cells deficient in Lig4 (Pan-Hammarström et al., 2005), XRCC4 (Yan et al., 2007), and Artemis (Du et al., 2008; Rivera-Munoz et al., 2009), three important components in the NHEJ machinery, and furthermore in B cells deficient in ATM (Pan et al., 2002), an additional factor which is implicated in regulation of the NHEJ pathway.
The frequencies of mutations around the Sμ-Sα junctions (±15 bp) as well as away from the recombination breakpoints (starting 15 bp upstream) were both significantly reduced in Cernunnos-deficient patients (Table 3). Furthermore, there was a significant shift in mutation targeting, with more mutations occurring at A/T residues (Table 3). The general pattern of these mutations was very similar, but not identical, to that observed in Artemis-deficient patients (Table 3).
Mutations in recombined Sμ-Sα fragments
| Patients | Number of mutations | Total number of mutations | Number of bp sequenced | Frequency (per 1,000 bp) | ||
| AT vs. GC mutations | Transitions | In RGYW/WRCY motifs | ||||
| Close to the junction (±15 bp) | ||||||
| Cernunnos−/− | 8 vs. 6 (57 vs. 43%)* | 6 (43%) | 9 (64%) | 14 | 2,520 | 5.6** |
| Artemis−/− | 8 vs. 1 (89 vs. 11%)*** | 5 (56%) | 5 (56%) | 9 | 1,620 | 5.6* |
| Controls (1–6 yr) | 13 vs. 38 (25 vs. 75%) | 27 (53%) | 28 (55%) | 51 | 4,110 | 12.4 |
| Lig4−/− | 1 vs. 5 (17 vs. 83%) | 2 (33%) | 5 (83%) | 6 | 900 | 6.7 |
| ATM−/− | 1 vs. 5 (17 vs. 83%) | 1 (17%) | 4 (67%) | 6 | 1,320 | 4.5** |
| Controls (adults) | 11 vs. 56 (16 vs. 84%) | 30 (45%) | 53 (79%) | 67 | 4,590 | 14.6 |
| Within Sμ (>15 bp upstream of the junction) | ||||||
| Cernunnos−/− | 36 vs. 28 (56 vs. 44%)*** | 52 (81%)* | 30 (47%)*** | 64 | 23,364 | 2.7*** |
| Artemis−/− | 38 vs. 28 (58 vs. 42%)*** | 43 (65%) | 37 (56%)* | 66 | 15,508 | 4.3 |
| Controls (1–6 yr) | 42 vs. 59 (30 vs. 70%) | 95 (67%) | 101 (72%) | 141 | 26,569 | 5.3 |
| Lig4−/− | 10 vs. 8 (56 vs. 44%) | 14 (78%) | 13 (72%) | 18 | 9,749 | 1.8*** |
| ATM−/− | 20 vs. 16 (56 vs. 44%)* | 31 (86%)** | 24 (67%) | 36 | 11,993 | 3.0** |
| Controls (adults) | 62 vs. 105 (37 vs. 63%) | 99 (59%) | 117 (70%) | 167 | 31,502 | 5.3 |
| Patients | Number of mutations | Total number of mutations | Number of bp sequenced | Frequency (per 1,000 bp) | ||
| AT vs. GC mutations | Transitions | In RGYW/WRCY motifs | ||||
| Close to the junction (±15 bp) | ||||||
| Cernunnos−/− | 8 vs. 6 (57 vs. 43%)* | 6 (43%) | 9 (64%) | 14 | 2,520 | 5.6** |
| Artemis−/− | 8 vs. 1 (89 vs. 11%)*** | 5 (56%) | 5 (56%) | 9 | 1,620 | 5.6* |
| Controls (1–6 yr) | 13 vs. 38 (25 vs. 75%) | 27 (53%) | 28 (55%) | 51 | 4,110 | 12.4 |
| Lig4−/− | 1 vs. 5 (17 vs. 83%) | 2 (33%) | 5 (83%) | 6 | 900 | 6.7 |
| ATM−/− | 1 vs. 5 (17 vs. 83%) | 1 (17%) | 4 (67%) | 6 | 1,320 | 4.5** |
| Controls (adults) | 11 vs. 56 (16 vs. 84%) | 30 (45%) | 53 (79%) | 67 | 4,590 | 14.6 |
| Within Sμ (>15 bp upstream of the junction) | ||||||
| Cernunnos−/− | 36 vs. 28 (56 vs. 44%)*** | 52 (81%)* | 30 (47%)*** | 64 | 23,364 | 2.7*** |
| Artemis−/− | 38 vs. 28 (58 vs. 42%)*** | 43 (65%) | 37 (56%)* | 66 | 15,508 | 4.3 |
| Controls (1–6 yr) | 42 vs. 59 (30 vs. 70%) | 95 (67%) | 101 (72%) | 141 | 26,569 | 5.3 |
| Lig4−/− | 10 vs. 8 (56 vs. 44%) | 14 (78%) | 13 (72%) | 18 | 9,749 | 1.8*** |
| ATM−/− | 20 vs. 16 (56 vs. 44%)* | 31 (86%)** | 24 (67%) | 36 | 11,993 | 3.0** |
| Controls (adults) | 62 vs. 105 (37 vs. 63%) | 99 (59%) | 117 (70%) | 167 | 31,502 | 5.3 |
The mutation patterns from Cernunnos- or Artemis-deficient patients were compared with those from control children (1–6 yr of age), whereas the mutation patterns from Lig4- or ATM-deficient patients were compared with adult controls (Pan-Hammarström et al., 2003). Statistical analysis was performed using the χ2 test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
After performing multiple experiments on DNA samples from each patient, we could only amplify six in vivo generated Sμ-Sγ fragments from Cernunnos-deficient B cells (Fig. S1). No direct end-joining was found at these junctions (Table 2). However, the small sample size does not allow a proper comparison with controls or other patient groups.
Collectively, although some differences do exist, such as the frequency of insertions at CSR junctions and the nature of mutations in the recombined S regions, the pattern of in vivo generated Sμ-Sα CSR junctions derived from Cernunnos-deficient B cells largely resembles those from Lig4-, Artemis-, and ATM-deficient cells. Cernunnos is thus likely to be involved in the Lig4-dependent NHEJ pathway during CSR.
Sequencing the Cernunnos-encoding gene in B cell lymphomas
Given the potential function of Cernunnos in DNA DSB repair and DNA end-joining during CSR, we hypothesized that Cernunnos deficiency and associated aberrant CSR activity might contribute to lymphomagenesis. To test this hypothesis, we performed direct sequencing of the Cernunnos gene in a set of mature B cell lymphoma samples where the postulated normal counterparts of tumor cells are B cells of germinal center or post–germinal center (activated B cells) origin or referred to as antigen-experienced B cells. These include diffuse large B cell lymphoma (DLBCL), follicular lymphoma (FL), Burkitt lymphoma (BL), and chronic lymphocytic leukemia (CLL). We specifically focused on DLBCL samples, as illegitimate CSR events have previously been shown to be associated with this subset of B cell lymphomas (Lenz et al., 2007).
The 5′ untranslated region (exon 1), the coding regions (exons 2–8), and the exon–intron borders of the Cernunnos gene were analyzed in altogether 184 DLBCL samples. A novel missense mutation in exon 6 (c.680A>G; p.Q227R) was identified in one sample (DL8; Figs. 1 and 3 A). In silico analysis using bioinformatics tools at PolyPhen (Ramensky et al., 2002) predicted that this mutation is possibly damaging. 12 known single nucleotide polymorphisms and 4 novel variants (c.1-2297T>C, c.1-2289G>A, c.530-77T>A, and c.588+55G>T) located in the noncoding regions of Cernunnos were also identified with various frequencies (not depicted). We subsequently analyzed 85 additional mature B cell lymphoma samples, obtained from 20 FL, 7 BL, and 58 CLL patients. Only a silent mutation in exon 4 (c.441G>A; p.Q147Q) was identified in two samples (FL3 and BL7), and two novel variants in introns (c.390+13C>T and c.707-57C>G) were found in one BL and one CLL sample, respectively.
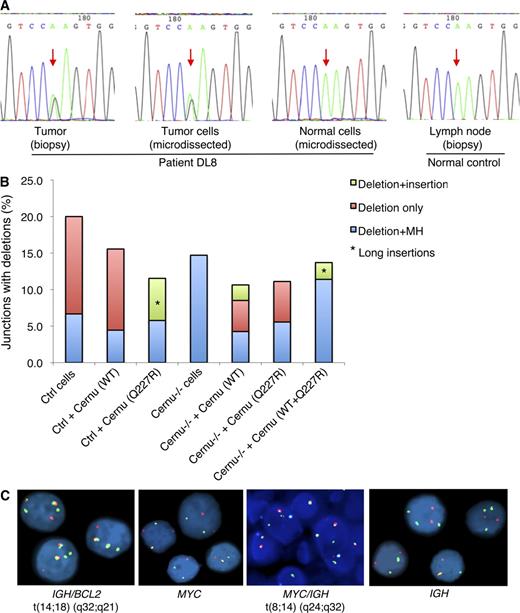
Novel mutation identified in the Cernunnos gene in a DLBCL sample (DL8). (A) Somatically acquired mutation in Cernunnos in tumor cells. The missense mutation (c.680A>G, p.Q227R; indicated by red arrows) was found in a heterozygous form in DNA prepared from the tumor biopsy or microdissected tumor cells but not in microdissected normal cells, nor in controls. The microdissection experiment was performed once, whereas the sequencing was performed on both directions, and the result from the forward direction is shown in the figure. (B) Functional characterization of the p.Q227R mutant using a plasmid-based NHEJ assay. The proportion (%) of different types of end-joining junctions associated with deletions, summarized from three independent experiments, is plotted. A summary of all types of junctions is shown in Table S1. Ctrl, control; Cernu, Cernunnos; MH, microhomology. (C) Detection of chromosomal translocations in tumor cells by FISH assays. IGH/BCL2 (first panel) and MYC/IGH rearrangements (third panel) were identified as orange/green (yellow) fusion signals. MYC (second panel) and IGH (fourth panel) rearrangements were identified as red and green splitting signals. The first panel is based on two independent experiments, whereas the remaining three are based on one experiment. Within each experiment, at least 100 intact, nonoverlapping nuclei were scored for each probe, and representative results are depicted.
Novel mutation identified in the Cernunnos gene in a DLBCL sample (DL8). (A) Somatically acquired mutation in Cernunnos in tumor cells. The missense mutation (c.680A>G, p.Q227R; indicated by red arrows) was found in a heterozygous form in DNA prepared from the tumor biopsy or microdissected tumor cells but not in microdissected normal cells, nor in controls. The microdissection experiment was performed once, whereas the sequencing was performed on both directions, and the result from the forward direction is shown in the figure. (B) Functional characterization of the p.Q227R mutant using a plasmid-based NHEJ assay. The proportion (%) of different types of end-joining junctions associated with deletions, summarized from three independent experiments, is plotted. A summary of all types of junctions is shown in Table S1. Ctrl, control; Cernu, Cernunnos; MH, microhomology. (C) Detection of chromosomal translocations in tumor cells by FISH assays. IGH/BCL2 (first panel) and MYC/IGH rearrangements (third panel) were identified as orange/green (yellow) fusion signals. MYC (second panel) and IGH (fourth panel) rearrangements were identified as red and green splitting signals. The first panel is based on two independent experiments, whereas the remaining three are based on one experiment. Within each experiment, at least 100 intact, nonoverlapping nuclei were scored for each probe, and representative results are depicted.
The missense mutation p.Q227R was found in a heterozygous form in the tumor biopsy (DL8) from a Chinese DLBCL patient (germinal center B cell like, according to Hans’ algorithm [Hans et al., 2004]). The patient was 70 yr old at diagnosis and, as described in Materials and methods, had a poor clinical course. By analyzing the Cernunnos transcript, we observed that the mutant allele was expressed in the tumor sample. By genotyping 429 ethnically matched healthy individuals, we showed that this mutation is not a rare polymorphism in the population. Furthermore, by analyzing the microdissected tumor and nontumor tissues, we could confirm that this mutation was somatically acquired in the tumor cells (Fig. 3 A).
To evaluate whether the p.Q227R mutation affects Cernunnos protein function, we used a previously described plasmid-based NHEJ assay (Verkaik et al., 2002), where a linearized plasmid with defined blunt DNA ends (both with a 6-bp direct repeat; ATCAGC) was transiently transfected into fibroblast cell lines derived from one Cernunnos-deficient patient (P4) and an age-matched healthy individual. In the control cells, the joining of the DNA ends was mainly caused by direct joining (73.3%), where the two blunt DNA ends were joined precisely without nucleotide addition or loss (Table S1). Other types of end-joining, involving the 6-bp direct repeat (microhomology), deletions or insertions, were also observed in the control cells with various frequencies (Table S1). In the Cernunnos-deficient cells, the proportion of junctions formed by direct joining was significantly reduced (4.4 vs. 73.3% in controls; P = 6.7 × 10−19), whereas the proportion of 6-bp direct repeat–mediated end-joining was significantly increased (80.9 vs. 5.7% in controls; χ2 test, P = 5 × 10−24). Furthermore, another type of junction, referred to as “deletion only” (not associated with any microhomologies or insertions), representing 13.3% of junctions from control cells, was totally absent in the Cernunnos-deficient cells (Fig. 3 B and Table S1). By transfecting a construct encoding the WT of the protein into the Cernunnos-deficient cells, direct joining was restored, and the general pattern of end-joining was indistinguishable from that of controls (Table S1). A similar pattern was observed when a construct expressing the mutant form of Cernunnos (p.Q227R) was used, suggesting that this mutant, in a homozygous form, had no effect on end-joining in this assay. However, when this mutant was overexpressed in control cells, the pattern of the recovered junctions was altered: first, like in the Cernunnos-deficient cells, no junctions with “deletion only” were observed (Fig. 3 B and Table S1; 0 vs. 13.3% in controls; χ2 test, P = 0.006); second, an unusual type of joining, characterized by deletions plus the addition of nontemplate nucleotides was noted (referred to as “deletion + insertion”; Fig. 3 B and Table S1; 5.8 vs. 0% in controls; χ2 test, P = 0.01). This type of joining was indeed not observed in any of the recovered junctions from untransfected control cells (105 joints) or control cells transfected with WT Cernunnos (45 joints). One junction derived from Cernunnos-deficient cells (transfected with WT Cernunnos) could also be characterized as “deletion + insertion.” However, only a 1-bp insertion was present, whereas in the mutant-expressing control cells, the insertion could be as long as 12 bp.
To further test the possibility that this mutant, in a heterozygous form, may have a dominant-negative effect on DNA end-joining, Cernunnos-deficient cells were cotransfected with the WT and mutant constructs at a 1:1 ratio. The recovered junctions showed a very similar pattern as those from control cells transfected with the mutant Cernunnos, i.e., the absence of “deletion only” joints and, furthermore, the presence of an unusual type of joining, i.e., “deletion + insertion” (exemplified by a joint with a 64-bp deletion plus a 10-bp insertion; Fig. 3 B and Table S1). Thus, in this plasmid-based NHEJ assay, the p.Q227R mutation has a dominant-negative effect on the end-joining of a subset of DNA ends, where resections are involved, reflected by deletions observed at the junctions. These ends can only be joined when a few base pairs of microhomologies are available and, strikingly, stretches of nontemplate nucleotides can occasionally be added.
Characterization of translocations and aberrant CSR activities in lymphoma cells carrying the Cernunnos mutation
We subsequently performed fluorescence in situ hybridization (FISH) analysis on the paraffin-embedded tumor tissue from DL8 to determine the presence of IGH-associated translocations (Fig. 3 C). Rearrangement of the BCL6 locus was excluded (not depicted). However, using a dual-color, dual-fusion probe, we found the IGH/BCL2 rearrangement corresponding to the t(14;18)(q32;q21) in 85% of the cells. In addition, we found an MYC rearrangement using a break-apart probe in 55% of the cells, which corresponds to the presence of a t(8;14)(q24;q32) as demonstrated by a specific dual-color, dual-fusion probe. Using an IGH break-apart probe, we further confirmed that one or both IGH were rearranged in 39% and 50% of the cells, respectively. Collectively, our data indicate that there is a subclone with only the IGH/BCL2 rearrangement and another with both the IGH/BCL2 and an IGH/MYC rearrangement, the latter probably resulting from clonal evolution of the former. Furthermore, a subset of the tumor cells presented additional isolated signals with the IGH and MYC break-apart probes, which is compatible with the presence of an additional copy of the der(8)t(8;14).
We subsequently performed a series of long-distance PCR assays (Basso et al., 1999; Busch et al., 2004) to map the MYC/IGH translocation breakpoints. Although we could amplify two translocations with MYC, involving the Sμ region in a Ramos cell line or the Sγ region in a BL sample (BL6) included in this study, we could not amplify any translocated fragments from patient DL8, using an MYC primer together with C region primers or a JH primer (not depicted). Possibly, the translocation breakpoint in the MYC gene in this tumor sample is not located in a region where most of the breakpoints from BL occur (exon 1 and intron 1; Basso et al., 1999; Busch et al., 2004). Alternatively, more complex translocations had occurred in this tumor, and thus, a standard long-distance PCR assay might not detect such a translocation. An indirect approach was subsequently applied, where a long-distance PCR assay was used to amplify the germline Sμ region (Pan-Hammarström et al., 2003). In contrast to the Ramos cells or control B cells, where one or two copies of the full-length Sμ region could be amplified, the tumor sample (DL8) had lost the germline Sμ region on both alleles (Fig. 4 A). A clonal-like IgA switching event could be identified, in which a trans-switching event involving both alleles of Sμ was observed (Fig. 4, B and C). Notably, recombination still seems to be ongoing in the tumor cells, as small deletions (affecting the Sμ-Sμ or Sμ-Sα1 junctions), insertions of a small piece of Sα1 (from the other allele), or sequential switching to Sα2 in the proposed primordial Sμ-Sμ-Sα1 fragment was observed (Fig. 4 C). The germline Sγ regions were subsequently amplified, and both copies of the Sγ4 region were undetectable in DL8 (Fig. 4 D). During legitimate CSR, the Sγ4 region can only be disrupted by the switching to Sγ4 or entirely deleted if there is a recombination to Sε or Sα2, either directly from Sμ or from Sμ through another S region located upstream of Sγ4. We have excluded the first possibility as no clonal or clonal-like Sμ-Sγ4 CSR events could be amplified in the tumor sample (Fig. 4 E). Furthermore, because of the structure of the human Ig locus, 5′-Sμ-Sγ3-Sγ1-Sα1-Sγ2-Sγ4-Sε-Sα2-3′, these “legitimate” CSR events would result in deletion of the other Sγ regions located upstream of Sγ4. As Sγ3 and Sγ2 regions could still be detected in DL8, the possibility that both alleles have switched to Sε or Sα2, thus resulting in deletion of both copies of Sγ4 regions, can be excluded. Collectively, we propose that one copy of Sμ in DL8 is recombined to the Sα1 and subsequently the Sα2 region and the other copy is used in a trans-switching process. One copy of Sγ4 is likely to be deleted during legitimate CSR to the Sα2 region, and the other copy is probably disrupted by translocation or other aberrant CSR activities such as intra Sγ4 region recombination.
Aberrant CSR activities in DL8 tumor cells. DL8, a DLBCL sample carrying the Cernunnos mutation and DH translocations (IGH/BCL2 and MYC/IGH); BL6, a BL sample carrying an MYC/IGH(Sγ) translocation; C1/Control 1, a healthy individual as a control; Ramos, a human BL cell line carrying an MYC/IGH(Sμ) translocation; M, molecular marker; N, negative control (H2O as template). All the gel pictures in this figure represent one experiment. (A) Amplification of the full-length germline Sμ region (4.5 kb). (B) Amplification of Sμ-Sα fragments. Five PCR reactions were run in parallel using DNA from DL8 or the control sample. (C) Clonal-like and ongoing IgA switching in DL8. The amplified Sμ-Sα fragments from B were sequenced, and the schematic structure of selected CSR junctions is presented. The proposed original Sμ-Sμ-Sα recombination event, shown on the left, involves trans-switching of two Sμ regions. Small deletions around the Sμ-Sα junction, insertions caused by trans-switching from the other Sα1 allele and sequential switching to Sα2, can be observed (right). (D) Amplification of full-length germline Sγ regions. The white bars indicate the positions of the Sγ4 regions. No Sγ4 could be amplified from DL8. (E) Amplification of Sμ-Sγ4 fragments. No clonal or clonal-like legitimate Sμ-Sγ4 events could be identified in DL8.
Aberrant CSR activities in DL8 tumor cells. DL8, a DLBCL sample carrying the Cernunnos mutation and DH translocations (IGH/BCL2 and MYC/IGH); BL6, a BL sample carrying an MYC/IGH(Sγ) translocation; C1/Control 1, a healthy individual as a control; Ramos, a human BL cell line carrying an MYC/IGH(Sμ) translocation; M, molecular marker; N, negative control (H2O as template). All the gel pictures in this figure represent one experiment. (A) Amplification of the full-length germline Sμ region (4.5 kb). (B) Amplification of Sμ-Sα fragments. Five PCR reactions were run in parallel using DNA from DL8 or the control sample. (C) Clonal-like and ongoing IgA switching in DL8. The amplified Sμ-Sα fragments from B were sequenced, and the schematic structure of selected CSR junctions is presented. The proposed original Sμ-Sμ-Sα recombination event, shown on the left, involves trans-switching of two Sμ regions. Small deletions around the Sμ-Sα junction, insertions caused by trans-switching from the other Sα1 allele and sequential switching to Sα2, can be observed (right). (D) Amplification of full-length germline Sγ regions. The white bars indicate the positions of the Sγ4 regions. No Sγ4 could be amplified from DL8. (E) Amplification of Sμ-Sγ4 fragments. No clonal or clonal-like legitimate Sμ-Sγ4 events could be identified in DL8.
DISCUSSION
Cernunnos/XLF, the latest addition to the NHEJ machinery, was first identified through characterization of a group of patients with combined immunodeficiency, growth retardation, and microcephaly (Buck et al., 2006a) and by searching for XRCC4-interacting factors (Ahnesorg et al., 2006). Similar to defects in six other NHEJ components, Cernunnos deficiency is associated with a defective V(D)J recombination (Buck et al., 2006a; Zha et al., 2007). However, the defect in knockout murine B cells is considerably less severe as compared with Cernunnos-deficient patients, thus raising the possibility that the lymphocyte-specific compensatory mechanism proposed by Li et al. (2008a) is restricted to mouse B cells. To address this question, it is important to study the second recombination process requiring NHEJ in lymphocytes, CSR, in both human and mouse B cells deficient in Cernunnos. In mice, CSR in cultured Cernunnos-deficient B cells is reduced by 50%, and the frequency of Sμ-Sγ direct joints is reduced, along with a small increase in the average length of junctional microhomology (Li et al., 2008a). However, it is notable that compared with the XRCC4- and Lig4-deficient cells, the changes at the CSR junctions are modest (Yan et al., 2007). From the present study, there is nevertheless clear evidence in support of a nonredundant role of Cernunnos in CSR in human lymphocytes. First, although all seven Cernunnos-deficient patients included in this study had remarkably low levels of IgG and IgA in their sera, four of the patients showed normal or even extremely high levels of IgM (Table 1), resembling patients with a hyper IgM syndrome caused by CSR defects (CD40L, CD40, AID, or UNG deficient; Kracker et al., 2010a). Second, the altered pattern of Sμ-Sα junctions derived from patients is not identical, but very similar, to those from ATM-, Artemis-, or Lig4-deficient patients, with long microhomologies and a markedly reduced proportion of direct end-joining. Thus, like in V(D)J recombination, the CSR defect observed in Cernunnos-deficient human B cells is more severe than that in Cernunnos-deficient mouse B cells. We therefore propose that the lymphocyte-specific compensatory mechanism involving ATM (Li et al., 2008a; Zha et al., 2011) is less active in human B cells.
The shift toward the usage of A-EJ or microhomology-based end-joining to repair the DSBs at CSR junctions in Cernunnos-deficient cells supports the proposed function of Cernunnos in NHEJ, i.e., interaction with the XRCC4–Lig4 complex (Ahnesorg et al., 2006; Callebaut et al., 2006), promoting XRCC4–Lig4 complex readenylation (Riballo et al., 2009) and stimulating joining of incompatible DNA ends that lack microhomology (Gu et al., 2007a,b; Lu et al., 2007a; Akopiants et al., 2009). Its role is probably to provide end bridging or stability when terminal microhomology is absent (Gu et al., 2007b) and to facilitate alignment-based gap filling for partially complementary ends (Akopiants et al., 2009). Cernunnos is also required for efficient joining of short cohesive 3′ or 5′ ends in a plasmid-based assay (Akopiants et al., 2009). Interestingly, the proportion of junctions with 1–3 bp of short microhomologies was significantly reduced in Cernunnos- but not in Lig4-deficient cells (Table 1). It is possible that at least some junctions with few base pair microhomologies result from ligation of cohesive ends during CSR and that this process is dependent on Cernunnos but not Lig4. Recently, Cernunnos has also been suggested to have additional roles in the early stages of NHEJ, as it quickly responds to DSB induction, can bind to DNA (Lu et al., 2007a), and interacts with Ku in a DNA-dependent manner (Yano and Chen, 2008; Yano et al., 2008, 2009). In this respect, it is worth noting that a dramatic shift in microhomology usage could also be observed at Sμ-Sα junctions in cells deficient for some of the factors proposed to sense the DSBs and to regulate the early phase of NHEJ, such as ATM (Pan et al., 2002) and RNF168 (Stewart et al., 2007). Thus, there are several possibilities, not mutually exclusive, for the function of Cernunnos in CSR, both in the early and late steps of Lig4-dependent NHEJ and in some of the Lig4-independent reactions, such as simple religation of cohesive ends.
Mutations near the Sμ-Sα junctions (±15 bp) and in the Sμ core (>15 bp upstream of the junctions; referred to somatic hypermutation–like mutations) are frequently observed in normal B cells and are considered to be a consequence of CSR-associated events (Petersen et al., 2001; Pan-Hammarström et al., 2003; Stavnezer et al., 2010). These mutations are normally GC biased (Table 3). The shift to AT mutations observed in Cernunnos-deficient patients was previously observed in Artemis-deficient cells (Du et al., 2008) and, to a lesser extent, in cells from patients carrying heterozygous deletions in the C-terminal part of AID (Kracker et al., 2010b). Interestingly, there is also a significant trend toward the usage of long microhomologies at the CSR junctions in the latter group of patients (Kracker et al., 2010b). As the C-terminal part of AID has been proposed to provide CSR-specific targeting and stabilization or to recruit repair factors (Barreto et al., 2003; Ta et al., 2003; Geisberger et al., 2009), the remarkable similarity of the pattern of CSR junctions and the associated mutation spectra in the aforementioned patient groups suggests that the C-terminal domain of AID may directly recruit or interact with some of the NHEJ components, such as Cernunnos and Artemis. Alternatively, it interacts with an as of yet unknown cofactor that is crucial for the choice between the classical NHEJ and microhomology-based A-EJ.
Mouse studies have provided ample evidence suggesting that NHEJ defects are associated with the occurrence of Igh translocations and the development of lymphoid malignancies (Zhang et al., 2010; Gostissa et al., 2011). In humans, early development of B cell malignancies has been observed in a few patients with Lig4 (Riballo et al., 2001; Buck et al., 2006b) or Artemis deficiency (Moshous et al., 2003), further suggesting a link between NHEJ deficiency and B cell lymphomagenesis. Population-based case-control studies also suggest that common variants in the XRCC4 and Lig4 genes may alter the risk for developing non-Hodgkin lymphoma (Hill et al., 2006; Shen et al., 2010). The mutation (p.Q227R) we observed in Cernunnos in the DLBCL tumor sample (DL8), to our knowledge, represents the first somatically occurring mutation identified in a gene encoding an NHEJ factor in human lymphomas.
In one of the published partial Cernunnos structures (aa 1–233; Li et al., 2008b), the Q227 residue is located near its C terminus. It is exposed to the surface and does not form hydrogen bonds with nearby structures. Thus, mutations in this residue may not directly influence the structure of the protein. However, it may still affect the as of yet unknown structure of the C-terminal region, which has been shown to be required for DNA binding and mismatched end ligation (Andres et al., 2007). In addition, this residue is evolutionarily conserved among higher vertebrates, including human, mouse, rat, dog, and cow (Andres et al., 2007), and the Q to R change is predicted to be possibly damaging by in silico analysis. Moreover, we could show that in the plasmid-based NHEJ assay, the mutation has a dominant-negative effect on the joining of a subset of DNA ends, when resections/deletions are involved. As the functional form of Cernunnos is a homodimer (Li et al., 2008b), the heterozygous mutation may theoretically have an effect on the stabilization of the complex. However, when the mutant and the WT were coexpressed in the Cernunnos-deficient cells, an efficient Cernunnos dimer formation was observed, thus not supporting this hypothesis (unpublished data). Alternatively, the mutant might affect the DNA binding activity of the Cernunnos:XRCC4 filament (Hammel et al., 2010), and lack of this DNA holder activity would require microhomology to stabilize the resected DNA ends.
Although the Cernunnos gene is mutated in only ∼0.5% of the DLBCLs screened (1 of 184 DLBCLs), it should be noted that the tumor carrying the Cernunnos mutation showed an unusual combination of translocations involving the IGH loci (IGH/BCL2 and MYC/IGH) and can thus be referred to as a “double-hit” (DH) lymphoma. DH lymphomas frequently appear in the form of DLBCLs, with a germinal center phenotype, and are particularly aggressive, conferring a poor clinical prognosis. They are sometimes classified as “B cell lymphoma unclassifiable with features intermediate between DLBCL and BL” (Aukema et al., 2011). The incidence of DH lymphomas (defined as carrying a chromosomal breakpoint affecting the MYC/8q24 locus in combination with another recurrent breakpoint) in DLBCLs has been reported to range from 0–14% (Aukema et al., 2011). The incidence of DH lymphomas similar to the one in our patient (DL8) is estimated to be 0–3% of all DLBCL cases (Aukema et al., 2011). If taking into account that the incidence of IGH/BCL2 translocations in Mongoloids is much lower than in Western populations (Biagi and Seymour, 2002; Yoon et al., 2008), this number is probably even lower in Asian populations. Thus, as discussed below, if Cernunnos deficiency is indeed associated with the second hit (MYC translocation) in DH lymphomas, more mutations would be expected if such lymphomas were enriched in the studied materials.
Translocation of the BCL2 gene with IGH, i.e., t(14;18), is a hallmark of FL, and the composition of breakpoints suggests an origin from aberrant Rag1/2-mediated V(D)J recombination during early B cell development (Tsujimoto et al., 1985; Küppers and Dalla-Favera, 2001). The MYC breakpoints in DH lymphomas are likely to represent a secondary event, or second hit (Aukema et al., 2011), and may result from AID-mediated aberrant somatic hypermutation and CSR activity in mature B cells (Ramiro et al., 2004; Robbiani et al., 2008). In DL8, we observed a trans-switching event involving both Sμ alleles, a clonal diversification of the proposed original Sμ-Sμ-Sα1 sequences (with deletions around the CSR junctions, trans-switching involving the other Sα1 allele, and sequential switching to Sα2), and disruption of the Sγ4 region either by MYC/IGH translocations or intra-S region recombination, all pointing to aberrant and dynamic CSR activities in the lymphoma cells. It is tempting to speculate that the mutation we identified in the Cernunnos gene has contributed to the initial illegitimate CSR activities, the subsequent MYC/IGH translocations and the ongoing aberrant CSR activities. Unlike the Nhej1/p53 double mutant mouse models, no TP53 mutations were identified in DL8. However, the preexisting activation of BCL2 caused by the t(14;18) translocation could provide the antiapoptosis activity required, instead of TP53 mutations, and together with the high level of expression of MYC, lead to the transformation to a more aggressive lymphoma in this patient.
In summary, the altered pattern of CSR junctions in Cernunnos-deficient patients suggests that Cernunnos is required in the NHEJ pathway during CSR. Furthermore, the somatically occurring mutation identified in the Cernunnos gene in a DH lymphoma suggests a link between Cernunnos deficiency and aberrant CSR or switch translocations during the development of B cell malignancies.
MATERIALS AND METHODS
Cernunnos-deficient patients.
The study included seven Cernunnos-deficient patients from independent families. The clinical and genetic characterization of patients P1, P2, P4, and P5 has been described previously (Buck et al., 2006a). Patient P6 was from a nonconsanguineous Polish family and suffered from microcephaly, growth retardation, and severe infections (recurrent pneumonia). Patient P7 was the first child to consanguineous Pakistani parents. He had a low birth weight and suffered from developmental delay with microcephaly. He suffered from weight loss with episodic diarrhea, vomiting, and cough at 17 mo of age. Patient P8 was the third child to consanguineous Turkish parents. He had microphthalmia, craniofacial defects, acral anomalies, and severe microcephaly (Verloes et al., 2001). The patient suffered from recurrent lower respiratory tract infections during the first months of life, and a diagnosis of agammaglobulinemia was made at the age of 9 mo. The striking developmental defects observed in this patient could be caused by the large Cernunnos gene deletion, although, as this patient was born to consanguineous parents, other potentially unknown genes might also contribute to the clinical presentation. Table 1 summarizes the mutations in the gene encoding Cernunnos, age of sampling, T and B cell counts, and Ig levels of these patients. 14 healthy children (1–6 yr old) were included in the study as controls. The institutional review board at the Karolinska Institutet approved the study.
Characterization of switch recombination junctions.
Genomic DNA was purified from peripheral blood cells from patients and healthy blood donors using standard methods. Amplification of Sμ-Sα (Pan et al., 2001, 2002), Sμ-Sγ(1, 2, 3) (Pan et al., 1997; Pan-Hammarström et al., 2005), and Sμ-Sγ4 (Pan et al., 1998; Pan and Hammarström, 1999) from in vivo switched cells was performed using a previously described nested PCR assay. A modified version of Taq polymerase (Go Taq; Promega) was used in the PCR reactions. The PCR-amplified switch fragments were gel purified (QIAGEN), cloned into a pGEM-T vector, and sequenced by an automated fluorescent sequencer in Macrogen.
The CSR junctions were determined by aligning the switch fragment sequences with the Sμ (Mills et al., 1990) and Sα1/Sα2 (Islam et al., 1994; Pan et al., 2001) or Sγ1/Sγ2/Sγ3/Sγ4 (Mills et al., 1995; Pan et al., 1998) sequences. Analysis of microhomology usage and mutation pattern at the CSR junctions was performed as described previously (Pan et al., 2002; Pan-Hammarström et al., 2003; Stavnezer et al., 2010).
Lymphoma samples.
Fresh or frozen tumor biopsies were available from 100 Chinese patients (DLBCL = 64, FL = 20, BL = 7, and CLL = 9) and 121 Swedish patients (DLBCL = 120 and CLL = 1). These patients were recruited from the Sun Yat-Sen University Cancer Center and Uppsala University Hospital, respectively. Whenever possible, fresh tumor biopsies were stored in RNAlater solution (Invitrogen/Applied Biosystems) at 4°C overnight and subsequently at −70°C before processing. DNA was extracted using a DNeasy blood and tissue kit (QIAGEN), and total RNA was extracted with TRIZOL (Invitrogen) according to the manufacturer’s instructions. Lymphoma classification was performed by experienced pathologists at the Sun Yat-Sen University Cancer Center or Uppsala University Hospital using the World Health Organization classification scheme. Clinical data, including age, gender, tumor size, sites of involvement, sites of extranodal involvement, performance status, stage, B symptoms, “bulky” disease, and serum LDH, and information on treatment were available on 213 patients. 48 previously characterized Iranian patients (all with CLL) were also included in the study (Hojjat-Farsangi et al., 2009). The institutional review boards at the Sun Yat-Sen University Cancer Center, Uppsala University, Tehran University, and Karolinska Institutet approved the study.
The patient who carried the Q227R mutation in the Cernunnos gene was 70 yr old at diagnosis. The disease involved multiple lymph nodes, including bilateral cervical nodes, mediastinal nodes, paraaortic nodes, and bilateral inguinal nodes. He had no B symptoms (fever, weight loss >10% within 6 mo, or drenching night sweats). The clinical staging revealed a stage IIIA lymphoma, and the international prognostic index was determined as 4 (high risk group). The patient had concomitant tuberculosis, did not receive any treatment, and died 6 mo after diagnosis. No family history of cancer was found.
Sequencing the Cernunnos gene in lymphoma samples.
The exons of the Cernunnos gene were PCR amplified using genomic DNA samples purified from tumor samples. The PCR primer sequences and PCR conditions are shown in Table S2. The PCR products were purified in a 96-well format and subsequently sequenced by an automated fluorescent sequencer in Macrogen. When possible, the forward primers were tagged with universal primer sequence tags M13 or T7 promoter to facilitate automated sequencing.
The g.82751 A>G (c.680A>G; p.Q227R) mutation was genotyped in DNA samples from 429 Chinese healthy controls using matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF; Sequenom Inc.) as previously described (Jurinke et al., 2002). The MassARRAY Typer 4.0 software (Sequenom Inc.) was used to analyze the resulting mass spectra for peak identification. Two independent scorers confirmed all genotypes.
For analysis of Cernunnos transcripts, cDNA was synthesized from total RNA using a first strand cDNA synthesis kit according to the manufacturer’s instruction (GE Healthcare). The PCR primer sequences and RT-PCR conditions are shown in Table S2.
Laser microdissection of tumor cells and nontumor cells.
Paraffin-embedded tissues were fixed on Membrane Slides (Leica) and regular glass slides. Routine hematoxylin and eosin staining and immunohistochemical staining using anti-CD20 were performed to differentiate tumor cells and nontumor cells. The paraffin-embedded tissue sections were treated in xylene three times for 5 min each and air-dried. Laser microdissections of tumor and nontumor cells were subsequently performed using Leica LMD7000 (Leica). Genomic DNA from pooled microdissected cells was extracted using the DNeasy blood and tissue mini kit and further amplified with a GenomePlex complete whole genome amplification kit (Sigma-Aldrich).
Plasmid construction, mutagenesis, and expression of the Cernunnos mutant.
The WT Cernunnos cDNA sequence was cloned in frame with a V5-His tag in the pcDNA3.1D/V5-His-TOPO vector (Invitrogen; Buck et al., 2006a). The Q227R mutation was introduced into the construct by an overlap extension PCR approach, using the following primers: XLF-BamHI-F, 5′-TCCGGATCCGACGATGGAAGAACTGGAGC-3′; XLF-EcoRV-R, 5′-TAGGATATCACTGAAGAGACCCCTTGGCT-3′; XLF-Q227R-F, 5′-GTCACCACACAAGAGGTCCGAGTGGGACAGAAGCATCAA-3′; and XLF-Q227R-R, 5′-TTGATGCTTCTGTCCCACTCGGACCTCTTGTGTGGTGAC-3′. Constructs were subsequently checked for correctness by sequencing.
The WT and mutant-expressing constructs were transiently transfected into human fibroblast cell lines (obtained from a healthy control and a Cernunnos-deficient patient) using Turbofect according to the manufacturer’s instructions (Fermentas). An EGFP construct (BD) was used as a control for transfection efficiency. 48 h after transfection, cells were harvested and subjected to protein, RNA, or DNA isolation. Western blot and real-time quantitative PCR (qPCR) assays were used to monitor the expression of Cernunnos protein and messenger RNA after the transfection. Expression of the Cernunnos–V5-His fusion proteins was tested by immunoblotting using a horseradish peroxidase–coupled monoclonal mouse anti-His antibody (Invitrogen). Expression of Cernunnos messenger RNA was determined by qPCR using the qPCR Core kit for SYBR Green I (Eurogentec) with the primers 5′-AATTCCTTCTTGGAACAATTTATGAT-3′ and 5′-CCTGCAGATTCATGACAAAGG-3′. Relative expression of Cernunnos was normalized to the expression of the housekeeping gene β-actin (Wen et al., 2004). qPCRs were performed on a StepOne Real-Time PCR System (Applied Biosystems).
In vitro NHEJ plasmid assay.
1 µg pDVG94 construct (provided by D.C. van Gent, Erasmus University Rotterdam, Rotterdam, Netherlands) was digested by EcoRV and EcoR47III (Promega), resulting in defined blunt DNA ends with 6-bp direct repeats (ATCAGC; Verkaik et al., 2002). The linearized plasmids were transiently transfected into fibroblast cell lines with or without the Cernunnos WT and mutant-expressing constructs. 48 h after transfection, plasmid DNA was recovered from the cell lines using the DNeasy blood and tissue kit. Recombination junctions were PCR amplified using the primers FM30 and DAR5 (Verkaik et al., 2002). The resulting PCR products were purified using QIAquick gel extraction kit (QIAGEN), cloned into pGEM-T vectors, and sequenced by an automated fluorescent sequencer (Macrogen).
Detection of the chromosomal translocations by FISH and long-distance PCR.
Vysis LSI IGH/BCL2 dual-color, dual-fusion translocation probe, Vysis LSI BCL6 dual-color, break-apart rearrangement probe, Vysis LSI MYC/IGH, CEP 8 tricolor, dual-fusion translocation probe, Vysis LSI MYC dual-color, break-apart rearrangement probe, and Vysis LSI IGH break-apart rearrangement probe were purchased from Abbott Molecular Inc. Sample processing and hybridization were performed according to the manufacturer’s instruction and as described previously (Ribeiro et al., 2006). Slides were counterstained with DAPI (Vector Laboratories), and fluorescent images were sequentially captured with a Cohu 4900 charge-coupled device camera coupled to an Axioplan fluorescence microscope (Carl Zeiss).
For detection of MYC/IGH translocations, the c-myc/M6 or c-myc/M2 primer (Busch et al., 2004) was used together with one of the four primers from the IGH loci, Cμ/03, Cγ/02, Cα/01, or JH, in a previously described long-distance PCR assay (Basso et al., 1999). The combination of c-myc/M6 or c-myc/M2 with the c-myc up primer was used to ensure that the quality of the genomic DNA was sufficient for long-distance PCR amplification (Busch et al., 2004). Amplification of germline Sμ (Pan-Hammarström et al., 2003) and Sγ (Pan et al., 1997, 1998) regions was performed as described previously.
Online supplemental material.
Fig. S1 shows the sequences of the Sμ-Sα and Sμ-Sγ junctions from Cernunnos-deficient patients. Table S1 shows the characterization of the end-joining junctions from the plasmid-based NHEJ assay. Table S2 shows the primer sequences and PCR conditions for amplification of coding regions and transcripts of Cernunnos.
Acknowledgments
We thank Prof. T. Blundell for insightful discussion on structure of Cernunnos, Dr. K. Duvefelt for help with genotyping, Dr. D.C. van Gent for providing the pDVG94 construct, Dr. S. Lisboa for technical support for FISH analysis, and Prof. J. Stavnezer for helpful comments on the manuscript.
This work was supported by the Swedish Cancer Society, the Swedish Research Council, and the European Research Council (242551-ImmunoSwitch).
The authors have no conflicting financial interests.
References
- A-EJ
alternative end-joining
- AID
activation-induced cytidine deaminase
- BL
Burkitt lymphoma
- CLL
chronic lymphocytic leukemia
- CSR
class switch recombination
- DH
double hit
- DLBCL
diffuse large B cell lymphoma
- DSB
double-strand break
- FISH
fluorescence in situ hybridization
- FL
follicular lymphoma
- NHEJ
nonhomologous end-joining
- qPCR
quantitative PCR
Author notes
L. Du and R. Peng contributed equally to this paper.