One component of memory in the antibody system is long-lived memory B cells selected for the expression of somatically mutated, high-affinity antibodies in the T cell–dependent germinal center (GC) reaction. A puzzling observation has been that the memory B cell compartment also contains cells expressing unmutated, low-affinity antibodies. Using conditional Bcl6 ablation, we demonstrate that these cells are generated through proliferative expansion early after immunization in a T cell–dependent but GC-independent manner. They soon become resting and long-lived and display a novel distinct gene expression signature which distinguishes memory B cells from other classes of B cells. GC-independent memory B cells are later joined by somatically mutated GC descendants at roughly equal proportions and these two types of memory cells efficiently generate adoptive secondary antibody responses. Deletion of T follicular helper (Tfh) cells significantly reduces the generation of mutated, but not unmutated, memory cells early on in the response. Thus, B cell memory is generated along two fundamentally distinct cellular differentiation pathways. One pathway is dedicated to the generation of high-affinity somatic antibody mutants, whereas the other preserves germ line antibody specificities and may prepare the organism for rapid responses to antigenic variants of the invading pathogen.
In T cell–dependent B cell responses, activated B cells migrate into the B cell follicles where they proliferate, with a fraction of cells undergoing Ig class switch recombination (CSR; Coffey et al., 2009; Pereira et al., 2010). Although some of the activated cells mediate the primary antibody response through differentiation into plasma cells, others are recruited into the germinal center (GC) reaction (Pereira et al., 2010). This is accompanied by up-regulation of the transcriptional repressor Bcl6, on which GC B cell differentiation depends (Dent et al., 1997; Ye et al., 1997). Bcl6 up-regulation is also required for the differentiation of follicular (FO) T helper (Tfh) cells. These cells are critical for the selection of B cells expressing high-affinity antibodies in the GC environment (Crotty, 2011).
Within the GC, B cells undergo massive proliferation accompanied by CSR and somatic hypermutation (SHM) of their rearranged Ig V region genes, a process in which cells preferentially survive which have acquired mutations that increase antibody affinity for the immunizing antigen (Rajewsky, 1996). This selection process critically depends on antigen presented to the B cells by FO DCs in the GC microenvironment and, in turn, presented by the B cells in the form of antigenic peptides to antigen-specific Tfh cells, resulting in the delivery of survival signals for the B cells involved (Victora et al., 2010). The selected high-affinity GC cells are then believed to differentiate into memory B and long-lived plasma cells, a large fraction of which express somatically mutated Ig V region genes and which persist for long periods of time after termination of the GC response (Rajewsky, 1996; Tarlinton, 2006).
Although the precursor-product relationship of GC and memory B cells seems firmly established, a puzzling observation has been that not all memory B cells carry somatic mutations in their Ig V regions (Takahashi et al., 2001; Blink et al., 2005; Anderson et al., 2007; Zotos et al., 2010). In addition, ICOS blockade early in the immune response caused a reduction in the frequency of mutated memory and GC B cells but did not affect total memory B cell numbers (Inamine et al., 2005). These findings led to the view that some memory cells emerge from the early GC reaction (Good-Jacobson and Shlomchik, 2010) or may even be GC independent, as unmutated memory cells can be generated in irradiated mice reconstituted with Bcl6-deficient BM (Toyama et al., 2002).
However, Bcl6 germline deletion causes multiple immunological dysfunctions, such as arrested Tfh and conventional DC development (Crotty, 2011; Ohtsuka et al., 2011), as well as aberrant macrophage function (Mondal et al., 2010). Furthermore, Bcl6 germline deletion causes a prominent inflammatory disease owing to overexpression of Th2 cytokines (Ye et al., 1997; Dent et al., 1997) and affects the properties of B cells before immunization (Shaffer et al., 2000). Thus, there is no evidence for a GC-independent pathway of memory cell generation under physiological conditions. Moreover, even if such a pathway exists, its timing in the response and impact on B cell memory, and the properties of the participating cells remain elusive.
To obtain a comprehensive understanding of the population dynamics underlying GC-independent and -dependent memory B cell development under physiological conditions, we deleted Bcl6 in the B or T cell lineage through a conditional Bcl6 allele and complemented these experiments by antibody-mediated ablation of the GC response in genetically intact animals. Focusing on antigen-specific IgG1-expressing memory cells, which can be conveniently isolated and distinguished from GC B cells by the level of CD38 expression (Ridderstad and Tarlinton, 1998; Takahashi et al., 2001), we then pursued the fate of these cells in the T cell–dependent immune response and characterized their properties, genetic signature, life span, and functional activity.
Our work not only provides definitive evidence for a GC-independent pathway of memory cell generation under physiological conditions but also a comprehensive view of the strikingly distinct population dynamics underlying GC-independent and -dependent memory B cell development with the help of distinct T cell subsets. The two classes of memory cells establish the memory compartment jointly and at comparable frequencies and attain functional maturation through distinct though related transcriptional programs.
RESULTS
Bcl6 deletion in B cells impairs GC but not memory B cell development
We established mutant mice carrying a loxP-flanked Bcl6 exon 7–9 allele (Bcl6f/f), with these exons encoding the Bcl6 zinc finger (ZF) domains ZF1 to ZF5 (Fig. 1). As the ZF domains are critical for Bcl6 function (Basso and Dalla-Favera, 2010), deletion of exons 7–9 is predicted to cause a complete loss of the capacity of Bcl6 to repress transcription of target genes.
Targeted insertion of loxP sites into the Bcl6 gene. (A). A DNA fragment which contains exons 7–9 of the Bcl6 gene was flanked by loxP sites (solid triangles) with an FRT-flanked pGK-Neo cassette inserted next to it. The pGK-Neo cassette was removed from the mouse germline by breeding heterozygous mice to FLPe-expressing deleter mice. Xb, XbaI site; Hd, HindIII site. (B) Southern blot analysis of XbaI digested genomic DNA from a WT and a targeted ES clone using the probe depicted by the thick bar in A. (C) Removal of the pGK-Neo cassette in a heterozygous floxed mouse was confirmed by genomic PCR. Location of the primer set is indicated by the two arrowheads on the bottom of A. (D–G) Bcl6flox mice were crossed to mb1-cre mice. Conditional Bcl6-deletion in B cells does not affect BM B cell development (D), B cell numbers in the spleen (E), and B cell characteristics before immunization (F and G). BM cells and splenocytes were prepared from Bcl6+/+ (+/+), Bcl6+/f (+/f), and Bcl6f/f (f/f) mice heterozygous for mb1-cre (n = 3). The frequency of B cell subsets in BM (D) and T and B cells in the spleen (E) was analyzed by FACS. Each symbol represents the number of cells in an individual mouse. Bars represent the mean number for each group. (F) Representative FACS plots of splenic B cells with expression of CD44 and CD69 (activated B cell phenotype). Numbers in the plots indicate the percentage of cells in quadrants. (G) The levels of cytokines in culture supernatants produced by splenic B cells upon stimulation with anti-IgM and anti-CD40 mAb or LPS for 3 d (see Materials and methods). Bars represent means for each group. The data are representative of two independent experiments in D–G.
Targeted insertion of loxP sites into the Bcl6 gene. (A). A DNA fragment which contains exons 7–9 of the Bcl6 gene was flanked by loxP sites (solid triangles) with an FRT-flanked pGK-Neo cassette inserted next to it. The pGK-Neo cassette was removed from the mouse germline by breeding heterozygous mice to FLPe-expressing deleter mice. Xb, XbaI site; Hd, HindIII site. (B) Southern blot analysis of XbaI digested genomic DNA from a WT and a targeted ES clone using the probe depicted by the thick bar in A. (C) Removal of the pGK-Neo cassette in a heterozygous floxed mouse was confirmed by genomic PCR. Location of the primer set is indicated by the two arrowheads on the bottom of A. (D–G) Bcl6flox mice were crossed to mb1-cre mice. Conditional Bcl6-deletion in B cells does not affect BM B cell development (D), B cell numbers in the spleen (E), and B cell characteristics before immunization (F and G). BM cells and splenocytes were prepared from Bcl6+/+ (+/+), Bcl6+/f (+/f), and Bcl6f/f (f/f) mice heterozygous for mb1-cre (n = 3). The frequency of B cell subsets in BM (D) and T and B cells in the spleen (E) was analyzed by FACS. Each symbol represents the number of cells in an individual mouse. Bars represent the mean number for each group. (F) Representative FACS plots of splenic B cells with expression of CD44 and CD69 (activated B cell phenotype). Numbers in the plots indicate the percentage of cells in quadrants. (G) The levels of cytokines in culture supernatants produced by splenic B cells upon stimulation with anti-IgM and anti-CD40 mAb or LPS for 3 d (see Materials and methods). Bars represent means for each group. The data are representative of two independent experiments in D–G.
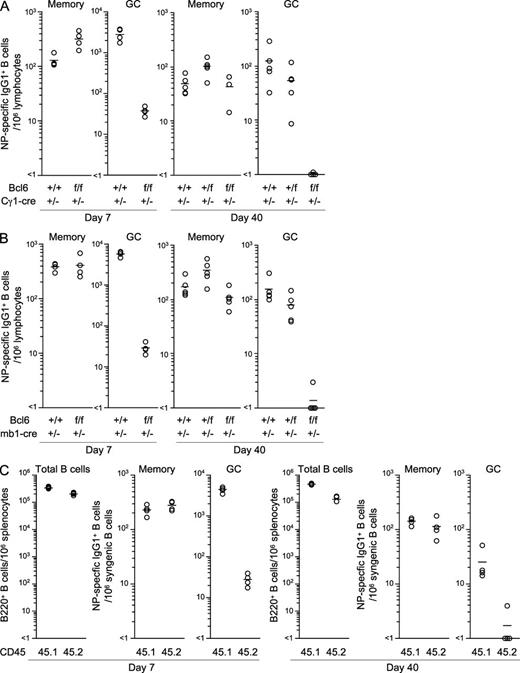
Whereas mice with Bcl6 deletion in the germline exhibit aberrant B cell development in the BM and an activated phenotype in naive splenic B cells (Shaffer et al., 2000; Duy et al., 2010), the conditional deletion of Bcl6 in B cells had no effect on B cell numbers, phenotype, or cytokine production after stimulation through the B cell receptor or Toll-like receptor (Fig. 1). To dissect IgG1 memory B cell development, Bcl6f/f mice were crossed with mice in which a cre cDNA is knocked into the Cγ1 locus (Cγ1-cre; Casola et al., 2006). We immunized the compound mutants and control mice with chicken γ-globulin (CG) coupled to (4-hydroxy-3-nitrophenyl)acetyl (NP-CG) and tracked NP-specific/IgG1+ memory (CD38+/peanut agglutinin [PNA]low) and GC (CD38low/PNA+) B cells in the spleen of immunized mice by multicolor flow cytometry (Fig. S1; Takahashi et al., 2001). GC reactions have been reported to persist for a long time after immunization (Dogan et al., 2009). Indeed, we found that approximately one fourth of the NP-specific/IgG1 B cells consisted of GC B cells 150 d after immunization (Fig. S1).
Bcl6 protein expression is limited to the GC stage of B cell differentiation (Basso and Dalla-Favera, 2010). Bcl6 was brightly stained in the nuclei of PNAhigh/CD38+ and CD38dull B cells at day 7 after immunization, supporting the notion that the transition of GC B cells from a CD38high to a CD38dull phenotype occurs early in the immune response (Shinall et al., 2000). Contrarily, Bcl6 was not detected in the nucleus of WT memory B cells (unpublished data). The conditional deletion of Bcl6 reduced the number of NP-specific/IgG1+ GC B cells in the spleen at days 7 and 40 after immunization to <2% of controls (Fig. 2 A). Early GC B cells were also deleted in the mutant mice (unpublished data). In striking contrast, the number of memory B cells was unaffected. These results suggest that Bcl6 expression in B cells is essential for GC but not memory B cell development.
IgG1+ memory B cells develop independently of the GC reaction. Splenocytes were recovered from Bcl6+/+, Bcl6+/f, and Bcl6f/f mice heterozygous for Cγ1-cre (A) or mb1-cre (B) at day 7 or 40 after immunization with NP-CG in alum and NP-specific/IgG1+ memory and GC B cells were enumerated from 106 cells within the lymphocyte gate (see Materials and methods). Circles represent the number of cells in individual mice (n = 3–5). Bars indicate the mean number in each group. (C) BM cells from Bcl6f/f mice heterozygous for mb1-cre (CD45. 2+) and CD45.1+ WT or heterozygous mb1-cre mice were mixed 1:1 and transferred into CD45.2+Rag-1−/− mice (n = 4). After 8 wk, the recipient mice were immunized with NP-CG. Splenocytes were stained with anti-CD45.1 mAb to distinguish the WT (45.1) and conditional Bcl6 KO (45.2) compartments at days 7 and 40 after immunization. The number of B cells and NP-specific memory and GC B cells in individual spleens was determined as in A. Data are representative of two (A) and three (B and C) independent experiments. See also Fig. S1.
IgG1+ memory B cells develop independently of the GC reaction. Splenocytes were recovered from Bcl6+/+, Bcl6+/f, and Bcl6f/f mice heterozygous for Cγ1-cre (A) or mb1-cre (B) at day 7 or 40 after immunization with NP-CG in alum and NP-specific/IgG1+ memory and GC B cells were enumerated from 106 cells within the lymphocyte gate (see Materials and methods). Circles represent the number of cells in individual mice (n = 3–5). Bars indicate the mean number in each group. (C) BM cells from Bcl6f/f mice heterozygous for mb1-cre (CD45. 2+) and CD45.1+ WT or heterozygous mb1-cre mice were mixed 1:1 and transferred into CD45.2+Rag-1−/− mice (n = 4). After 8 wk, the recipient mice were immunized with NP-CG. Splenocytes were stained with anti-CD45.1 mAb to distinguish the WT (45.1) and conditional Bcl6 KO (45.2) compartments at days 7 and 40 after immunization. The number of B cells and NP-specific memory and GC B cells in individual spleens was determined as in A. Data are representative of two (A) and three (B and C) independent experiments. See also Fig. S1.
To confirm this possibility, we crossed the Bcl6f/f mice with mice expressing Cre recombinase from the mb-1 locus and thus from early on in B cell development (Hobeika et al., 2006). Cre-mediated recombination in WT mice was detected in >98% of BM and splenic B cells, but not in prepro–B and CD4+ T cells (unpublished data). The conditional deletion of Bcl6 through mb1-cre essentially abolished the early development of IgG1+ GC B cells in the spleen on day 5 after immunization (unpublished data). This in turn resulted in an ∼200-fold reduction in the number of GC B cells on day 7 after immunization, compared with the controls (Fig. 2 B). We also generated mixed BM chimeras by transferring BM cells of congenic CD45.1+ WT and CD45.2+ conditional Bcl6-deficient mice into irradiated Rag-1−/− mice and immunized the animals with NP-CG 8 wk after reconstitution. In the chimeric mice, Bcl6-deficient B cells did not generate GC B cells (Fig. 2 C), confirming that Bcl6 expression in B cells is essential for GC B cell development.
In contrast, the number of IgG1+ memory B cells was again comparable in Bcl6-deficient (Bcl6f/f/mb1-cre+/−) and control mice (Bcl6+/+/mb1-cre+/−) at days 7 and 40 after immunization (Fig. 2 B). In BM chimeric mice, memory B cells derived from WT and conditional Bcl6-deficient mice coexisted at almost equal frequency in the spleen (Fig. 2 C), suggesting that GC-dependent and -independent pathways of memory cell generation operate side by side.
Early GC-independent memory B cells are in a resting state and have not undergone SHM
To examine cell cycle progression in NP-specific/IgG1+ memory B cells, B cells were prepared from individual spleens of mutant (Bcl6f/f/mb1-cre+/−) and control (Bcl6+/+/mb1-cre+/−) mice at days 7 and 40 after immunization and stained with the DNA specific dye Hoechst 33342, followed by FACS analysis of their DNA content. Memory B cells in the mutant mice were mostly in a resting state, whereas ∼20% of GC B cells in the controls are in the S/G2/M phases of the cell cycle at these time points (Fig. 3 A and Fig. S2 A). We also performed FACS analysis for NP-specific/IgG1+ memory and GC B cells in individual spleens of FUCCI transgenic mice at days 7 and 40 after immunization (Fig. 3 B). These mice constitutively express monomeric Azami Green fused to partial human geminin protein (Sakaue-Sawano et al., 2008) so that cells in S/G2/M can be easily distinguished by green fluorescence (Fig. S2 B). Fig. 3 B shows that day 7 and day 40 IgG1+ memory B cells in WT mice were mostly in a resting state, in contrast to their GC counterparts. This was confirmed by staining for the Ki67 nuclear protein (unpublished data).
The cell cycle position and VH186.2 gene mutations in NP-specific memory and GC B cells. (A) DNA content in memory (Me) and GC B cells in conditional Bcl6-deficient (Bcl6f/f/mb-1-cre+/−) and control (Bcl6+/+/mb-1-cre+/−) mice at days 7 and 40 after immunization with NP-CG. Each symbol represents the percentage of positive cells in an individual mouse (n = 3). Data are representative of three independent experiments. (B) The cell cycle position of memory B cells (Me) and GC B cells (GC) in FUCCI transgenic mice (n = 3) at indicated times. Each symbol represents the percentage of cells positive for Azami Green in an individual mouse. Data are representative of three (day 7) and two (day 40) independent experiments. (C and D) Single NP-specific/IgG1+ memory and GC B cells were purified from the pooled spleens of Bcl6+/+ (+/+) and Bcl6f/f (f/f) mice heterozygous for mb1-cre (n = 6–13) at day 7 (C) or day 40 (D) after immunization and subjected to RT-PCR to amplify rearranged VH186.2-Cγ1 cDNA for sequencing (see Materials and methods). (E) CD45.1+ (45.1) WT memory and GC B cells and CD45.1− (45.2) Bcl6-deficient memory B cells were purified as single cells from pooled spleens of BM chimeras (n = 9) at day 40 after immunization and their VH genes were sequenced as in C. Number of mutated clones/number of V186.2 genes sequenced are also shown in C–E. Circles represent the number of mutations in individual clones. Closed circles represent W33L clones.
The cell cycle position and VH186.2 gene mutations in NP-specific memory and GC B cells. (A) DNA content in memory (Me) and GC B cells in conditional Bcl6-deficient (Bcl6f/f/mb-1-cre+/−) and control (Bcl6+/+/mb-1-cre+/−) mice at days 7 and 40 after immunization with NP-CG. Each symbol represents the percentage of positive cells in an individual mouse (n = 3). Data are representative of three independent experiments. (B) The cell cycle position of memory B cells (Me) and GC B cells (GC) in FUCCI transgenic mice (n = 3) at indicated times. Each symbol represents the percentage of cells positive for Azami Green in an individual mouse. Data are representative of three (day 7) and two (day 40) independent experiments. (C and D) Single NP-specific/IgG1+ memory and GC B cells were purified from the pooled spleens of Bcl6+/+ (+/+) and Bcl6f/f (f/f) mice heterozygous for mb1-cre (n = 6–13) at day 7 (C) or day 40 (D) after immunization and subjected to RT-PCR to amplify rearranged VH186.2-Cγ1 cDNA for sequencing (see Materials and methods). (E) CD45.1+ (45.1) WT memory and GC B cells and CD45.1− (45.2) Bcl6-deficient memory B cells were purified as single cells from pooled spleens of BM chimeras (n = 9) at day 40 after immunization and their VH genes were sequenced as in C. Number of mutated clones/number of V186.2 genes sequenced are also shown in C–E. Circles represent the number of mutations in individual clones. Closed circles represent W33L clones.
The anti-NP response in C57BL/6 mice is dominated by λ light chain bearing antibodies whose V region of the IgH chain is encoded by a rearranged V186.2 V gene segment (Allen et al., 1988). As shown in Fig. 3 C, two thirds of the V186.2 rearrangements sequenced from IgG1+PNA+CD38+ and CD38dull GC B cells in Bcl6-proficient control mice were somatically mutated at day 7 after immunization, whereas IgG1+ memory B cells from mice with B cell–specific Bcl6 ablation (Bcl6f/f/mb1-cre+/−) were unmutated. This was also true for day 7 memory B cells in Bcl6 proficient controls, except for a few cells carrying rearranged VH genes with a single mutation. The latter may represent early GC B cell progeny. At day 40 after immunization (Fig. 3 D), all rearranged VH genes sequenced from GC B cells had accumulated a large number of mutations in control mice, and about half of the sequences from memory cells of these mice were mutated. The Trp to Leu exchange at aa position 33 of VH186.2 (W33L), which confers a 10-fold increase in affinity for the hapten NP (Allen et al., 1988), was detected in ∼15–45% of day 40 mutated memory and GC B cells (Fig. 3 D) and further enriched in mutated VH186.2 genes joined to DFL16.1 in day 40 memory B cells (27–64%; not depicted), comparable to the observations by Weiss and Rajewsky (1990). In contrast, day 40 memory B cells in Bcl6-deficient mice were still free of somatic mutations (Fig. 3 D). Similarly, in mixed BM chimeric mice (Fig. 2 C), Bcl6-deficient memory B cells did not accumulate mutations (Fig. 3 E), confirming that even in the presence of GC reactions Bcl6 deficiency precludes the acquisition of SHMs by B cells.
GC-independent memory B cells arise from dividing precursors, are long-lived, and enter the memory compartment before GC B cell progeny
To determine the lifespan of unmutated memory B cells, we administered BrdU intraperitoneally into mutant (Bcl6f/f/mb1-cre+/−) and control (Bcl6+/+/mb1-cre+/−) mice on days 4 and 6 after immunization, to mark splenic B cells deriving from precursors dividing during the labeling period. We then isolated NP-specific/IgG1+ memory B cells 7 and 40 d after immunization and analyzed the frequency of BrdU-labeled cells by confocal microscopy (Fig. 4 A; Takahashi et al., 2001; Toyama et al., 2002). More than 95% of day 7 memory B cells from both mutant and control mice had incorporated BrdU, and this frequency declined only slightly in the mutant mice by day 40 after immunization. As IgG1+ memory B cells are in a resting state from day 7 on (Fig. 3), this suggests that most GC-independent memory B cells have a long lifespan. In contrast, in the control mice, the frequency of BrdU-labeled memory B cells was reduced at day 40 to ∼60% (P = 0.005).
GC-independent memory B cells are maintained for a long period and later joined by somatically mutated GC descendents. (A) Bcl6+/+ (+/+), Bcl6+/f (+/f), and Bcl6f/f (f/f) mice heterozygous for mb1-cre (n = 5–25) were injected with BrdU twice at days 4–6 after immunization with NP-CG. Memory (Me) and GC B cells (GC) were then purified at indicated times. BrdU+ B cells were determined as in Materials and methods. Circles represent the percentage of BrdU+ cells in three independent experiments. (B) C57BL/6 mice (n = 5–25) were immunized and injected with BrdU as described in A. On days 6, 8, and 10 after immunization, mice were either not treated (−) or injected with either hamster anti–CD40-ligand mAb (MR1) or normal hamster IgG (Ig) and sacrificed at day 20 or 40 for purification of NP-specific/IgG1+ memory (Me) and GC B cells (GC). BrdU incorporation was determined as in A. Circles represent the percentage of BrdU+ cells in three independent experiments. (C) Accumulation of mutations in VH186.2 genes that were PCR amplified from single NP-specific/IgG1+ memory B cells in immunized WT mice (n = 10–22) at the indicated times. Number of mutated clones/number of V186.2 genes sequenced are also shown. Symbols are as in Fig. 3. The results were evaluated statistically by Mann-Whitney nonparametric test, with P < 0.05 regarded as significant. (D) C57BL/6 mice (n = 5) were immunized with NP-CG in alum and injected with or without (−) anti-CD40 ligand mAb (MR1) or normal Ig (Ig) as described in B. At day 20 or 40, the number of NP-specific/IgG1+ memory and GC B cells was analyzed by FACS. Each symbol represents the number of cells in individual mice. The data are representative of two independent experiments. (E) Immunofluorescence analysis of splenic cryosections from NP-CG–immunized C57BL/6 mice treated with MR1 or normal Ig was performed. Sections were stained with anti-B220 (blue) and PNA (red; see Materials and methods). Representative images of three mice are shown. Bars, 300 µm. (F) Mutational analysis of VH186.2 rearrangements from single day 40 memory B cells of immunized mice (n = 20–40). Mice were either not treated (−) or injected with anti-CD40 ligand mAb (MR1) or normal Ig (Ig) as described in B. Number of mutated clones/number of V186.2 genes sequenced are also shown. Symbols are as in Fig. 3.
GC-independent memory B cells are maintained for a long period and later joined by somatically mutated GC descendents. (A) Bcl6+/+ (+/+), Bcl6+/f (+/f), and Bcl6f/f (f/f) mice heterozygous for mb1-cre (n = 5–25) were injected with BrdU twice at days 4–6 after immunization with NP-CG. Memory (Me) and GC B cells (GC) were then purified at indicated times. BrdU+ B cells were determined as in Materials and methods. Circles represent the percentage of BrdU+ cells in three independent experiments. (B) C57BL/6 mice (n = 5–25) were immunized and injected with BrdU as described in A. On days 6, 8, and 10 after immunization, mice were either not treated (−) or injected with either hamster anti–CD40-ligand mAb (MR1) or normal hamster IgG (Ig) and sacrificed at day 20 or 40 for purification of NP-specific/IgG1+ memory (Me) and GC B cells (GC). BrdU incorporation was determined as in A. Circles represent the percentage of BrdU+ cells in three independent experiments. (C) Accumulation of mutations in VH186.2 genes that were PCR amplified from single NP-specific/IgG1+ memory B cells in immunized WT mice (n = 10–22) at the indicated times. Number of mutated clones/number of V186.2 genes sequenced are also shown. Symbols are as in Fig. 3. The results were evaluated statistically by Mann-Whitney nonparametric test, with P < 0.05 regarded as significant. (D) C57BL/6 mice (n = 5) were immunized with NP-CG in alum and injected with or without (−) anti-CD40 ligand mAb (MR1) or normal Ig (Ig) as described in B. At day 20 or 40, the number of NP-specific/IgG1+ memory and GC B cells was analyzed by FACS. Each symbol represents the number of cells in individual mice. The data are representative of two independent experiments. (E) Immunofluorescence analysis of splenic cryosections from NP-CG–immunized C57BL/6 mice treated with MR1 or normal Ig was performed. Sections were stained with anti-B220 (blue) and PNA (red; see Materials and methods). Representative images of three mice are shown. Bars, 300 µm. (F) Mutational analysis of VH186.2 rearrangements from single day 40 memory B cells of immunized mice (n = 20–40). Mice were either not treated (−) or injected with anti-CD40 ligand mAb (MR1) or normal Ig (Ig) as described in B. Number of mutated clones/number of V186.2 genes sequenced are also shown. Symbols are as in Fig. 3.
In immunized WT mice, BrdU-labeled memory B cells decreased from >95% on day 7 to ∼70% on day 20 (P = 0.023) and ∼60% on day 40 (P ≤ 0.005) after immunization (Fig. 4 B), in parallel with an increase in the frequency of mutated cells from ≤5% at day 7 to 30% at day 20, and 50–66% at day 40 (Fig. 4 C). There was no significant increase in the frequency of mutated cells in the memory B cell population from days 40 to 100 after immunization (Fig. 4 C). The frequency of BrdU+ cells in the GC B cell population of the control mice significantly decreased from >95% on day 7 to <10% on day 40 (Fig. 4, A and B). This suggests that mutated GC progeny cells which have lost their BrdU label during proliferative expansion are gradually recruited into the memory compartment (Fig. 4 A).
To address this issue more directly, immunized WT mice were labeled with BrdU and then treated with the anti-CD40L mAb MR1 or control Ig at days 6 to 10 after immunization (Fig. 4 B). MR1 administration interfered with GC B cell development (Fig. 4, D and E; Takahashi et al., 1998) and the recruitment of mutated cells into the memory compartment (Fig. 4 F). This was accompanied by a strongly diminished loss of BrdU-labeled cells from the memory compartment over time (Fig. 4 B). Together, these results indicate that GC-independent memory B cells that develop within the first week of the response are maintained for a long period and joined by mutated GC B cell progeny as the immune response progresses. In both conditional Bcl6-deficient and control mice, IgG1+ memory B cells localized within the follicles in the spleen at days 7 and 40 after immunization (unpublished data).
Early memory B cells gradually attain functional maturity
To determine the functional activity of memory B cells appearing early in the immune response, we tested their ability to generate an adoptive secondary response (Takahashi et al., 2005). To this end, we purified IgM−/IgD−/B220+ B cells from the pooled spleens of mutant (Bcl6f/f/mb1-cre+/−) and control (Bcl6+/+/mb1-cre+/− and Bcl6+/f/mb1-cre+/−) mice at days 7 and 40 after immunization with NP-CG and determined the frequency of NP-specific/IgG1+ memory B cells in each population by FACS. An equivalent of 1,500 memory B cells was then transferred into Rag-1−/− mice together with CG-primed T cells, followed by stimulation with soluble NP-CG. Fig. 5 A shows that both day 7 and day 40 memory cells from mutant as well as control mice induced an anti-NP IgG1 secondary response, as determined by enumerating splenic antibody-secreting cells (ASCs). However, in all cases the response of the day 40 memory cells was more vigorous, with five to six times as many plasma cells generated. As expected, the antibodies produced by the mutant mice were mostly low affinity, in contrast to the mixture of high- and low-affinity antibodies produced by control mice. These results demonstrate that unmutated GC-independent memory B cells have the capacity to generate a low-affinity secondary response and, like GC-derived memory cells, attain functional maturation as the immune response progresses.
Memory B cells attain functional maturity as the immune response progresses. (A) Splenocytes were recovered from either naive or NP-CG–primed Bcl6+/+ (+/+), Bcl6+/f (+/f), and Bcl6f/f (f/f) mice (n = 4–14) heterozygous for mb1-cre (mb1-cre+/−) at days 7 and 40 after immunization. B cells depleted of plasma cells were enriched by MACS and memory B cell frequency was estimated by FACS. B cell populations containing 1.5 × 103 NP-specific/IgG1+ memory B cells were transferred into Rag-1−/− mice, together with CG-primed CD4+ T cells and naive B cells, followed by immunization with soluble NP-CG. The number of total and high-affinity anti-NP/IgG1+ ASCs in the spleen was measured by ELISPOT at day 10 after immunization. Shown are the total number of anti-NP/IgG1+ ASCs (left) and the ratio of high-affinity ASCs/total number of ASCs (right). (B) On days 6, 8, and 10 after immunization, mice were either not treated (−) or injected with either anti-CD40 ligand mAb (MR1) or normal Ig (Ig) and sacrificed at day 7 (d7, n = 5) or 40 (d40, n = 22–25) for purification of NP-specific/IgG1+ memory B cells. Memory B cells (1 × 103), CG-primed CD4+ T cells (4 × 106), and naive B cells (106), as filler cells, were transferred into Rag-1−/− mice (n = 4), followed by immunization as in A. Anti-NP/IgG1+ ASCs in the spleen were determined as in A. (C) B cells were prepared from mutant (Bcl6f/f/mb-1-cre+/−, n = 4) or control mice (Bcl6+/+/mb-1-cre+/−, n = 4) at day 70 (d70) after immunization with NP-CG. NP-primed B cells or naive B cells (Naive) were transferred into Rag-1−/− mice (n = 5), together with CG-primed T cells, followed by immunization with NP-CG. The number of total and high-affinity anti-NP/IgM ASCs in the spleen was measured by ELISPOT at day 5 after immunization. Data are representative of two (C) and three (A and B) independent experiments. Circles represent cell numbers in individual mice.
Memory B cells attain functional maturity as the immune response progresses. (A) Splenocytes were recovered from either naive or NP-CG–primed Bcl6+/+ (+/+), Bcl6+/f (+/f), and Bcl6f/f (f/f) mice (n = 4–14) heterozygous for mb1-cre (mb1-cre+/−) at days 7 and 40 after immunization. B cells depleted of plasma cells were enriched by MACS and memory B cell frequency was estimated by FACS. B cell populations containing 1.5 × 103 NP-specific/IgG1+ memory B cells were transferred into Rag-1−/− mice, together with CG-primed CD4+ T cells and naive B cells, followed by immunization with soluble NP-CG. The number of total and high-affinity anti-NP/IgG1+ ASCs in the spleen was measured by ELISPOT at day 10 after immunization. Shown are the total number of anti-NP/IgG1+ ASCs (left) and the ratio of high-affinity ASCs/total number of ASCs (right). (B) On days 6, 8, and 10 after immunization, mice were either not treated (−) or injected with either anti-CD40 ligand mAb (MR1) or normal Ig (Ig) and sacrificed at day 7 (d7, n = 5) or 40 (d40, n = 22–25) for purification of NP-specific/IgG1+ memory B cells. Memory B cells (1 × 103), CG-primed CD4+ T cells (4 × 106), and naive B cells (106), as filler cells, were transferred into Rag-1−/− mice (n = 4), followed by immunization as in A. Anti-NP/IgG1+ ASCs in the spleen were determined as in A. (C) B cells were prepared from mutant (Bcl6f/f/mb-1-cre+/−, n = 4) or control mice (Bcl6+/+/mb-1-cre+/−, n = 4) at day 70 (d70) after immunization with NP-CG. NP-primed B cells or naive B cells (Naive) were transferred into Rag-1−/− mice (n = 5), together with CG-primed T cells, followed by immunization with NP-CG. The number of total and high-affinity anti-NP/IgM ASCs in the spleen was measured by ELISPOT at day 5 after immunization. Data are representative of two (C) and three (A and B) independent experiments. Circles represent cell numbers in individual mice.
To confirm that this maturation process also takes place in WT mice, we administered the MR1 mAb or control Ig into immunized WT mice, as described in Fig. 4 B. We then purified day 7 and 40 NP-specific/IgG1+ memory B cells from the two groups of mice and transferred 1,000 such cells together with CG-primed T cells into Rag-1−/− mice to analyze their secondary response. Fig. 5 B shows that day 7 memory B cells respond to boosting antigen, consistent with the results in Fig. 5 A. Significantly, day 40 memory B cells from MR1-treated mice produced larger numbers of ASCs, comparable to those produced by control day 40 memory B cells. However, in contrast to the latter they expressed exclusively low-affinity antibodies.
There is evidence that not only class-switched but also IgM+ memory B cells are generated in both T cell–dependent and –independent responses (Klein et al., 1997; Weller et al., 2001; Obukhanych and Nussenzweig, 2006; Anderson et al., 2007; Dogan et al., 2009: Pape et al., 2011; Taylor et al., 2012). Fig. 5 C shows that NP-primed B cells of conditional Bcl6-deficient and control mice generated a modest IgM adoptive secondary response consisting mostly of low-affinity antibodies. This raises the possibility that IgM+ memory B cells can be generated through a GC-independent pathway (see Discussion).
Early and late memory B cells have closely related gene expression profiles
Gene expression was assessed using Affymetrix GeneChip technology in three replicates of day 7 and 40 NP-specific/IgG1+ memory B cells, GC B cells, and plasma cells in immunized mice and naive marginal zone (MZ) and FO B cells, before and after activation in vitro. Hierarchical cluster analysis for all arrays on the expression of 45037 probes displayed a dendrogram with five major branches (Fig. 6 A). Early and late memory B cells were included in the same group, next to naive B cells and clearly separated from other B cell types.
Gene expression profiles between days 7 and 40 memory B cells are closely related. (A) A hierarchical clustering of triplicate samples. Colors in the heat map depict the Pearson’s correlation coefficient between a pair of samples. A higher value is represented by dark blue as shown in the vertical bar. An AU (approximately unbiased) p-value (percentage) was calculated and placed on a branch of a cluster dendrogram. The values in red color indicate that the microarray pairs are significantly clustered. NP-specific/IgG1+ memory (Me) and GC B cells (GC) and plasma cells (Pc, day 7 only) were purified from WT mice at day 7 (n = 14–20) and day 40 (n = 30–39) after immunization for extraction of total RNA. FO and MZ B cells were purified from unimmunized mice (n = 3). FO B cells were stimulated with anti-IgM and anti-CD40 mAb in vitro for 6 h (FO + stimulation). (B) qRT-PCR analysis for candidate genes identified in the microarray data analysis. NP-specific/IgG1+ memory (Me) and GC B cells and plasma cells (Pc) were purified from WT mice (n = 10) at day 7 after immunization. Day 40 memory B cells were purified from the pooled spleens of immunized mice (n = 18–25), treated with anti-CD40L mAb (MR1), control Igs, or left untreated. cDNA synthesized from total RNA was used for qRT-PCR with the indicated gene-specific primers (see Table S1) and standardized to the relative expression of β-actin. Shown here are gene expression profiles of naive FO (a) and MZ B cells (b), and memory (c), GC (d), and plasma cells (e) at day 7 after immunization. Also shown are day 40 memory B cells that developed in untreated mice (f) or those treated with either control Igs (h) or MR1 (j), together with day 40 GC B cells in untreated mice (g) or those treated with Igs (i). FO B cells stimulated as in A for 6 h (k) and 24 h (l) are also shown. Data are representative of two independent experiments. Error bars represent ±SD. See also Fig. S3.
Gene expression profiles between days 7 and 40 memory B cells are closely related. (A) A hierarchical clustering of triplicate samples. Colors in the heat map depict the Pearson’s correlation coefficient between a pair of samples. A higher value is represented by dark blue as shown in the vertical bar. An AU (approximately unbiased) p-value (percentage) was calculated and placed on a branch of a cluster dendrogram. The values in red color indicate that the microarray pairs are significantly clustered. NP-specific/IgG1+ memory (Me) and GC B cells (GC) and plasma cells (Pc, day 7 only) were purified from WT mice at day 7 (n = 14–20) and day 40 (n = 30–39) after immunization for extraction of total RNA. FO and MZ B cells were purified from unimmunized mice (n = 3). FO B cells were stimulated with anti-IgM and anti-CD40 mAb in vitro for 6 h (FO + stimulation). (B) qRT-PCR analysis for candidate genes identified in the microarray data analysis. NP-specific/IgG1+ memory (Me) and GC B cells and plasma cells (Pc) were purified from WT mice (n = 10) at day 7 after immunization. Day 40 memory B cells were purified from the pooled spleens of immunized mice (n = 18–25), treated with anti-CD40L mAb (MR1), control Igs, or left untreated. cDNA synthesized from total RNA was used for qRT-PCR with the indicated gene-specific primers (see Table S1) and standardized to the relative expression of β-actin. Shown here are gene expression profiles of naive FO (a) and MZ B cells (b), and memory (c), GC (d), and plasma cells (e) at day 7 after immunization. Also shown are day 40 memory B cells that developed in untreated mice (f) or those treated with either control Igs (h) or MR1 (j), together with day 40 GC B cells in untreated mice (g) or those treated with Igs (i). FO B cells stimulated as in A for 6 h (k) and 24 h (l) are also shown. Data are representative of two independent experiments. Error bars represent ±SD. See also Fig. S3.
To identify genes selectively up-regulated in the memory cells, we quantified gene expression by statistical analyses and detected 94 candidate genes (Fig. S3). Assessing the expression of these genes by quantitative RT-PCR (qRT-PCR) in memory B cells of MR1-treated or control animals, we identified 24 genes whose increased expression in either or both day 7 and 40 memory B cells was reproduced in two independent biological replicas (Fig. 6 B). The levels of these transcripts were comparable between day 40 memory B cells from MR1-treated and control animals, suggesting that GC-independent and -dependent memory cells share at least part of their gene expression signature. In addition, the level of transcripts was comparable between day 7 memory B cells in both the conditional Bcl6-deficient and control mice (unpublished data).
The differentially expressed transcripts fall into three groups: transcription factors, which include the AP-1 family of JunB and Fra-2 (van Dam and Castellazzi, 2001) and the NR4A subfamily of nuclear orphan receptors (Maxwell and Muscat, 2006), receptors, such as Tnfrsf1b encoding for TNFR2, and signaling intermediates, including the suppressor of cytokine signaling (SOCS) family, Socs3 (Alexander and Hilton, 2004).
Day 7 and 40 GC-independent memory B cells shared a group of transcripts, including NR4a2, Fra-2, and Socs3, suggesting that these cells sustained the expression of a group of transcripts over time from day 7 onward. The level of certain other transcripts increased or decreased in GC-independent memory B cells over time (Fig. 6 B). An example of this is leukemia inhibitory factor receptor (Lifr), which is preferentially expressed in early memory B cells. In contrast, expression of NR4a1 and NR4a3 was higher in cells harvested at day 40. These results suggest gene expression changes in GC-independent memory cells over time, perhaps in response to environmental cues.
GC-independent and -dependent memory B cells develop with the help of different T cell subsets
IgG1+ memory and GC B cells were generated neither in CD40-deficient mice (in which T cell help cannot be delivered to B cells; Kawabe et al., 1994 and Fig. 7 A) upon NP-CG immunization nor in WT mice immunized with NP-Ficoll, a T cell–independent antigen (not depicted). These results indicate that the development of IgG1+ memory B cells requires T cell help. To address the role of Tfh cells in memory B cell development, we crossed the Bcl6f/f mice with mice carrying a cre transgene under the control of the cd4 enhancer/promoter (CD4-cre). The conditional deletion of Bcl6 through CD4-cre did not affect T and B cell numbers and their phenotype in naive animals (not depicted) but impaired Tfh development in mice immunized with NP-CG (Fig. 7 B). Mutant mice had significantly reduced numbers of splenic NP-specific/IgG1+ GC B cells (Fig. 7 C) at day 7 after immunization, but the frequency of mutations in VH gene rearrangements was comparable in mutant and control mice (Fig. 7 F).
Different subsets of T cells support GC-independent and -dependent memory B cell development. (A) CD40 KO (−/−) and control (+/+) mice were immunized with NP-CG and generation of NP-specific/IgG1+ B cells was analyzed by FACS (left). Circles represent the number of cells in individual mice (n = 3, right). (B–D) Frequencies of Tfh (B), NP-specific/IgG1+ GC (C), and memory B cells (D) in spleens of Bcl6+/+ (+/+) and Bcl6f/f (f/f) mice carrying a CD4-cre transgene at days 7 and day 40 after immunization with NP-CG. Unimmunized mice were used as a control in B. Circles represent the number of cells in individual mice (n = 4–6). (E) The frequency of day 7 NP-specific/IgG1+ memory and GC B cells in the spleens of mutant (Bcl6f/f/mb1-cre+/− and CD4-cre) and control (Bcl6+/+/mb1-cre+/− and CD4-cre) mice. Circles represent the number of cells in individual mice (n = 4). (F and G) Mutational analysis of VH186.2 rearrangements from NP-specific/IgG1+ memory and GC B cells of Bcl6+/+ (+/+) and Bcl6f/f (f/f) mice carrying a CD4-cre transgene at days 7 (F) and 40 (G) after immunization (n = 4–6). Circles represent the number of mutations in individual clones. Number of mutated clones/number of V186.2 genes sequenced are also shown in F and G. Closed circles represent high-affinity clones (W33L). (H) NP-specific/IgG1+ memory B cells were purified from Bcl6+/+ (+/+) and Bcl6f/f (f/f) mice carrying a CD4-cre transgene (n = 10–19) at day 40 after immunization and their secondary response was assessed as in Fig. 5. Response by naive B cells was also assessed as a control. Circles represent the number of anti-NP/IgG1+ ASCs in the spleens of individual mice at day 10 after immunization (n = 4). The data are representative of two independent experiments in A–E and H.
Different subsets of T cells support GC-independent and -dependent memory B cell development. (A) CD40 KO (−/−) and control (+/+) mice were immunized with NP-CG and generation of NP-specific/IgG1+ B cells was analyzed by FACS (left). Circles represent the number of cells in individual mice (n = 3, right). (B–D) Frequencies of Tfh (B), NP-specific/IgG1+ GC (C), and memory B cells (D) in spleens of Bcl6+/+ (+/+) and Bcl6f/f (f/f) mice carrying a CD4-cre transgene at days 7 and day 40 after immunization with NP-CG. Unimmunized mice were used as a control in B. Circles represent the number of cells in individual mice (n = 4–6). (E) The frequency of day 7 NP-specific/IgG1+ memory and GC B cells in the spleens of mutant (Bcl6f/f/mb1-cre+/− and CD4-cre) and control (Bcl6+/+/mb1-cre+/− and CD4-cre) mice. Circles represent the number of cells in individual mice (n = 4). (F and G) Mutational analysis of VH186.2 rearrangements from NP-specific/IgG1+ memory and GC B cells of Bcl6+/+ (+/+) and Bcl6f/f (f/f) mice carrying a CD4-cre transgene at days 7 (F) and 40 (G) after immunization (n = 4–6). Circles represent the number of mutations in individual clones. Number of mutated clones/number of V186.2 genes sequenced are also shown in F and G. Closed circles represent high-affinity clones (W33L). (H) NP-specific/IgG1+ memory B cells were purified from Bcl6+/+ (+/+) and Bcl6f/f (f/f) mice carrying a CD4-cre transgene (n = 10–19) at day 40 after immunization and their secondary response was assessed as in Fig. 5. Response by naive B cells was also assessed as a control. Circles represent the number of anti-NP/IgG1+ ASCs in the spleens of individual mice at day 10 after immunization (n = 4). The data are representative of two independent experiments in A–E and H.
The number of IgG1+ memory B cells was comparable at day 7 after immunization in control and mutant mice with conditional deletion of Bcl6 in T cells (Fig. 7 D) and in both B and T cells (Fig. 7 E), indicating that a Tfh subset of T cells is not required for the generation of GC-independent memory B cells. However, at day 40 after immunization, memory B cell numbers were reduced in the mutant mice by ∼40% (Fig. 7 D) as a result of a significant reduction in the number of mutated cells in the memory B cell population (Fig. 7 G). Consistent with these data, day 40 memory cells from both mutant and control mice initiated an IgG1 secondary response, but the antibodies produced by the mutant cells were mostly of low affinity, in contrast to the mixture of high- and low-affinity antibodies produced by the controls (Fig. 7 H). The results suggest that Tfh depletion impaired the development of mutated memory cells, owing to a failure in expansion and/or maintenance of GC B cells.
Cell surface markers on GC-independent and -dependent memory B cells
PD-L1, PD-L2, CD80, CD73, and CD35 have all been reported to be expressed in memory B cells in the spleen (Anderson et al., 2007; Good-Jacobson et al., 2010). Fig. 8 A shows that the level of PD-L2 and CD80 was increased in day 7 and 40 memory B cells in conditional Bcl6-deficient and control mice compared with naive and GC B cells. Interestingly, both GC-independent and -dependent memory B cells increased their levels of PD-L2 on the surface from day 7 to day 40 after immunization. CD73 expression was increased in GC and a subset of memory B cells in WT mice as the immune response progresses. Fig. 8 B shows that ∼80% of CD73+ memory B cells accumulated somatic mutations compared with ∼30% of CD73− memory cells. Thus, mutated memory B cells are enriched in the CD73+ subset, as suggested by Taylor et al. (2012).
Expression of PD-L2 and CD80 on GC-independent and -dependent memory B cells. (A) Histograms represent the relative intensity of indicated surface markers on naive B cells (darkly shaded), day 7 and day 40 memory B cells (solid line), and GC B cells (dashed line) in mutant (Bcl6f/f/mb-1-cre) and control (Bcl6+/+/mb-1-cre) mice. Representative results of two independent experiments with three mice per group are shown. (B) Accumulation of mutations in VH186.2 genes that were PCR amplified from single CD73+ and CD73− NP-specific/IgG1+ memory B cells and GC B cells in immunized WT mice (n = 5) at day 40 after immunization. Number of mutated clones/number of V186.2 genes sequenced are also shown. Symbols are as in Fig. 3.
Expression of PD-L2 and CD80 on GC-independent and -dependent memory B cells. (A) Histograms represent the relative intensity of indicated surface markers on naive B cells (darkly shaded), day 7 and day 40 memory B cells (solid line), and GC B cells (dashed line) in mutant (Bcl6f/f/mb-1-cre) and control (Bcl6+/+/mb-1-cre) mice. Representative results of two independent experiments with three mice per group are shown. (B) Accumulation of mutations in VH186.2 genes that were PCR amplified from single CD73+ and CD73− NP-specific/IgG1+ memory B cells and GC B cells in immunized WT mice (n = 5) at day 40 after immunization. Number of mutated clones/number of V186.2 genes sequenced are also shown. Symbols are as in Fig. 3.
Cell population dynamics in memory B cell development
To study the generation of GC-independent memory B cells from naive B cell precursors, we used mice (CD45.1+) in which the frequency of NP-specific naive B cells is enhanced (B1-8hi; Shih et al., 2002). Splenic B cells of these mice were transferred into syngeneic CD45.2+ recipients and immunized with NP-CG. This causes the synchronous activation of many NP-specific donor B cells, allowing us to dissect their early response in vivo. Fig. 9 (A and B) shows that a small fraction of NP-binding donor B cells acquired sIgG1 expression and increased in number from day 3 to 4 after immunization, a period in which PNA binding cells were barely detectable. The IgG1+ donor B cells expressed memory surface markers, but not intracellular Bcl6 (Fig. 9 C), and increased expression level of genes, which were detected in day 7 memory B cells in the adoptive recipients and intact immunized mice (Fig. 9 D). Administration of MR1 mAb into the recipients from days 3–6 after immunization blocked Bcl6 expression and reduced the expansion of NP-specific/IgG1+ B cells (Fig. 9, B and C). However, these cells still acquired expression of memory surface markers and genes which we had detected in day 7 memory B cells developing in the absence of MR1 treatment (Fig. 9, C and D). The levels of these transcripts were comparable between memory cells from MR1-treated and control animals (Fig. 9 D). The IgG1+ B cells did not detectably express the GC B cell–specific gene M17 (Christoph et al., 1994) or Blimp-1 associated with plasma cell differentiation (Angelin-Duclos et al., 2000). Together, these data suggest that antigen-activated IgG+ B cells can differentiate toward memory B cells through initial proliferative expansion, in the absence of Bcl6 expression.
Antigen-engaged IgG1+ B cells differentiate into memory cells prior to the appearance of Bcl6 expressing pre-GC B cells. (A) C57BL/6 (CD45.2) mice (n = 3–4) were injected with B220+ B cells from B1-8hi mice (CD45.1) and immunized with NP-CG, followed by FACS analysis. NP-specific/B220+ B cells of donor origin (CD45.1+) in the immunized or unimmunized recipients were gated and their PNA binding and IgG1 expression were analyzed (left). (B) Numbers of NP-specific/IgG1+/PNA− (−) and PNA+ (+) B cells from day 3–4 (left) and NP-specific/IgG1+ memory B cells (CD38+/PNA−, Me) and GC B cells (CD38dull/PNA+, GC) at day 7 (right) after immunization. MR1 was administered from days 3 to 6 after immunization as in Fig. 4 B. (C) Expression of surface PD-L2 and CD80 and intracellular Bcl6 analyzed by FACS in NP-binding B cells from unimmunized recipients (control B cells) and NP-specific/IgG1+ B cells and GC and memory B cells in recipients at different time points after immunization. Memory B cells were purified from recipients treated or not treated with MR1. Representative results of three independent experiments are shown in A–C. (D) Gene expression profiles in NP-specific/IgG1+ donor B cells at day 3 (b) and day 4 (c) after immunization and day 7 NP-specific/IgG1+ memory B cells in MR1-treated (e) or -untreated (d) recipients, together with GC B cells in untreated recipients (f). NP-binding B cells from unimmunized recipients served as controls (a). Total RNA was purified from B cells and cDNAs were synthesized for qRT-PCR with the indicated gene-specific primers. Also shown are gene expression profiles of naive FO B cells (g), MZ B cells (h), day 7 and day 40 memory B cells (i and l), GC B cells (j and m), plasma cells (k), and activated FO B cells (compare Figure 6; FO + Stim). Error bars represent ±SD. Data are representative of two independent experiments and standardized to the expression of β-actin.
Antigen-engaged IgG1+ B cells differentiate into memory cells prior to the appearance of Bcl6 expressing pre-GC B cells. (A) C57BL/6 (CD45.2) mice (n = 3–4) were injected with B220+ B cells from B1-8hi mice (CD45.1) and immunized with NP-CG, followed by FACS analysis. NP-specific/B220+ B cells of donor origin (CD45.1+) in the immunized or unimmunized recipients were gated and their PNA binding and IgG1 expression were analyzed (left). (B) Numbers of NP-specific/IgG1+/PNA− (−) and PNA+ (+) B cells from day 3–4 (left) and NP-specific/IgG1+ memory B cells (CD38+/PNA−, Me) and GC B cells (CD38dull/PNA+, GC) at day 7 (right) after immunization. MR1 was administered from days 3 to 6 after immunization as in Fig. 4 B. (C) Expression of surface PD-L2 and CD80 and intracellular Bcl6 analyzed by FACS in NP-binding B cells from unimmunized recipients (control B cells) and NP-specific/IgG1+ B cells and GC and memory B cells in recipients at different time points after immunization. Memory B cells were purified from recipients treated or not treated with MR1. Representative results of three independent experiments are shown in A–C. (D) Gene expression profiles in NP-specific/IgG1+ donor B cells at day 3 (b) and day 4 (c) after immunization and day 7 NP-specific/IgG1+ memory B cells in MR1-treated (e) or -untreated (d) recipients, together with GC B cells in untreated recipients (f). NP-binding B cells from unimmunized recipients served as controls (a). Total RNA was purified from B cells and cDNAs were synthesized for qRT-PCR with the indicated gene-specific primers. Also shown are gene expression profiles of naive FO B cells (g), MZ B cells (h), day 7 and day 40 memory B cells (i and l), GC B cells (j and m), plasma cells (k), and activated FO B cells (compare Figure 6; FO + Stim). Error bars represent ±SD. Data are representative of two independent experiments and standardized to the expression of β-actin.
DISCUSSION
Memory B cells are long-lived quiescent B cells capable of eliciting more rapid and robust antibody responses upon antigenic stimulation than antigen-inexperienced naive B cells (Tangye and Tarlinton 2009). Based on this generally accepted definition, we pursued the fate of NP-specific/IgG1-expressing memory cells in the T cell–dependent response and characterized their development, properties, genetic signature, and functional activity. As GC reactions persisted over 5 mo after priming after immunization with NP-CG, we used FACS with multicolor parameters throughout to distinguish memory cells from GC B cells (Takahashi et al., 2001; Blink et al., 2005).
A GC-independent pathway of memory B cell generation had been predicted or inferred in previous studies (Weller et al., 2001; Toyama et al., 2002; Blink et al., 2005; Inamine et al., 2005; Chan et al., 2009; Zotos et al., 2010: Taylor et al., 2012). To obtain definitive evidence for this differentiation pathway, we generated a conditional allele of Bcl6, a transcriptional repressor essential for GC formation (Dent et al., 1997; Ye et al., 1997), and deleted it specifically in B cells. We also interfered with GC formation using an antagonistic Ab against CD40L, whose interaction with the CD40 co-receptor on B cells is critical for the GC reaction (Kawabe et al., 1994). In contrast to deletion in the germ line, conditional Bcl6 deletion in the B cell lineage affected neither B cell development and subset distribution in terms of numbers nor the initial expansion of antigen-activated B cells after immunization; however, it precluded the development of GC B cells from day 5 after immunization, the earliest time point at which such cells became apparent in WT mice.
Under these conditions, IgG1+ memory B cells developed within 7 d after immunization like in WT mice, apparently independently of the GC reaction. BrdU incorporation experiments demonstrated that after initial proliferative expansion these cells acquire a resting state already at day 7 after immunization and persist for long periods of time. In immunized WT mice, the memory pool initially contains a very large proportion of nonmutated cells. However, the frequency of nonmutated memory B cells gradually decreases to approximately one half as the immune response progresses. Blocking cell influx from GCs prevented the recruitment of mutated cells into the memory compartment, indicating that these cells represent GC progeny, which access the memory compartment as the GC reaction unfolds. Consistent with this model, blockade of CD40-CD40L interaction by anti-CD40L mAb at days 20–24 after immunization results in the dissolution of established GCs, whereas the number of day 40 memory B cells is largely unaffected (Takahashi et al., 2001). Collectively, these results suggest that nonmutated GC-independent memory B cells develop early in the response of both WT and conditional Bcl6-deficient mice and are maintained for long periods of time.
Dissecting the early T cell–dependent B cell response in vivo through an adoptive transfer system with largely synchronous activation of NP-specific donor B cells (Shih et al., 2002), we obtained evidence that antigen-engaged IgG1+ B cells can differentiate into memory cells before the appearance of Bcl6 expressing pre-GC B cells. Bcl6 up-regulation in the latter cells is thought to promote their interactions with Tfh cells, which are required for GC formation (Crotty 2011; Kitano et al., 2011). In accord with this notion, we observed that the loss of Tfh cells through T cell–specific deletion of Bcl6 significantly reduced the generation of mutated memory B cells in T cell–dependent responses as a consequence of impaired GC development. In contrast, the generation of GC-independent memory cells followed its normal path in the absence of Tfh cells and is therefore driven by a separate T helper cell subset. Thus, different T cell signals, perhaps through differential engagement of CD40 (Taylor et al., 2012), may drive naive B cells into distinct pathways of memory cell generation.
While the present paper was in revision, Taylor et al. (2012) reported that antigen-specific B cells with a memory phenotype (CD38+/GL7−) are generated in GC-independent and -dependent manners in WT and Bcl6−/− BM chimeric or anti-CD40L–treated mice in response to PE. Although the functional properties of these cells were not analyzed, it was satisfying to see cells of similar phenotypes as observed in our study arising in response to yet another T cell–dependent antigen. However, there were important discrepancies between the present study and Taylor et al. (2012) with respect to the persistence of GC-independent memory cells. Taylor et al. (2012) found these cells at a strong disadvantage in competition with GC progeny in mixed BM chimeras. In contrast, our results not only suggest peaceful coexistence of the two populations but also their functional maturation over time, an issue not addressed in Taylor et al. (2012). Among possible reasons for this discrepancy are the use of CD73 as a marker for GC-derived memory B cells (a marker which we find expressed by ∼80% mutated and ∼30% unmutated memory cells), the genetic heterogeneity of the mixed BM chimeras used by Taylor et al. (2012), and the possibility that Freund’s complete adjuvant–induced Th1-dominated helper activity (Billiau and Matthys, 2001; McKee et al., 2007; Taylor et al., 2012) and alum-induced Th2-dominated helper activity differentially drive and sustain GC-independent B cell memory.
Another issue raised by Taylor et al. (2012) relates to IgM expressing memory cells. Although the prominent isotypes expressed in secondary antibody responses require CSR, there is evidence that a subset of memory cells retains IgM expression and may even play a specific role in an antigen-dependent pathway of memory propagation (Anderson et al., 2007; Dogan et al., 2009). IgM-expressing memory B cells may also be generated in T cell–independent antibody responses (Obukhanych and Nussenzweig, 2006). Taylor et al. (2012) report that upon immunization, CD38+/GL7−/IgM+ B cells expand in a GC-independent manner, ∼100-fold above the level of GC-independent isotype-switched Ig+ memory cells. Our results suggest that these former cells do not contribute significantly to the memory response, given the low magnitude of their adoptive secondary response.
Although the global gene expression analysis performed on GC-dependent and -independent IgG1+ memory cells remains descriptive at this stage, it has provided interesting insights into the development and relatedness of these cells. Thus, we identified a distinct gene expression signature distinguishing both types of memory B from other classes of B cells already at day 7 after immunization. This signature was shared by GC-dependent and -independent memory cells and included several transcription factors specifically up-regulated in these cells. Interestingly, most of the genes up-regulated in either or both day 7 and 40 memory B cells discovered in the present experiments were not detected in earlier studies addressing memory B cell–specific gene expression (Bhattacharya et al., 2007; Tomayko et al., 2008), with the exception of Lifr and adenosine receptor A2a (Adora2a; Tomayko et al., 2008) and Nidogen1 (Bhattacharya et al., 2007). These discrepancies may be a result of the fact that in the earlier studies GC B and plasma cells were not depleted from the memory B cell preparations. Alternatively, they may reflect differences between memory B cells expressing different Ig isotypes.
Although the functional significance of the memory B cell specific genes and also of changes of gene expression in these cells over time remain to be determined, it is interesting to speculate about a possible connection between the functional maturation of memory cells over time and the up-regulation of certain surface markers known to be involved in their interaction with T helper and DCs, such as PD-L2, CD80, and CD73 and expressed on subsets of memory B cells (Anderson et al., 2007; Good-Jacobson et al., 2010). We confirmed that PD-L2, CD80, and also costimulatory molecules like MHC class II and CD40 are expressed on both GC-independent and -dependent memory B cells, with an interesting increase of PD-L2 expression from day 7 to day 40 after immunization. As expression of PD-L2 is required for optimal ASC generation (Good-Jacobson et al., 2010), the functional activity of memory B cells could be related to the level of PD-L2 expression.
As a general perspective, our results reveal that the immune system has evolved distinct differentiation pathways to select two classes of antibody binding sites into the memory compartment, namely those that have been subject to evolutionary selection and others selected for high-affinity antigen binding in the GC reaction, through rapid somatic evolution. In the context of immune defense against infection, it seems reasonable to assume that the germ line–encoded antibody repertoire is constantly evolving to provide cross-reacting specificities that are particularly useful as a first line of defense against common pathogens and whose exclusion from the memory compartment would be wasteful. The generation of high-affinity somatic mutants, in contrast, allows the system to focus on a particular pathogen at the expense of loosing useful cross-reactivities. We speculate that responses to variants of invading pathogens may involve the rapid recruitment of cells from the pool of unmutated memory cells into GC reactions. In this picture, the memory compartment consists of two layers of cells, those optimally adapted to the invading pathogen and others providing the substrate for the selection of somatic mutants adapted to antigenic variants arising in the course of infection. It is possible that this second layer of memory cells also contains non–class-switched IgM+ cells, which we have been unable to analyze because of the lack of suitable surface markers. Finally, it is tempting to speculate, that the two classes of human B cell chronic leukemia, which are distinguished by the presence or absence of somatic mutations in their antibody V region genes and whose clinical prognosis differs dramatically (Hamblin et al., 1999), may derive from the two classes of memory cells described here.
MATERIALS AND METHODS
Mice and immunizations.
8–10-wk-old C57BL/6 female mice were purchased from Clea Inc. Mb1-cre mice were provided by Dr. M. Reth (University of Freiburg and Max-Planck Institute for Immunobiology, Freiburg, Germany). Fucci-expressing transgenic mice were provided by Dr. Miyawaki (RIKEN Brain Science Institute, Saitama, Japan). Cγ1-cre mice have been described previously. FLPe-expressing deleter mice were provided by the RIKEN BRC through the National Bio-Resource Project of the MEXT, Japan. Rag-1−/− and CD4-cre mice were obtained from Taconic. Mice were immunized intraperitoneally with 100 µg NP15-CG precipitated in alum (Takahashi et al., 2001) or, in some experiments, with 50 µg NP-Ficoll (Biosearch Technologies) in PBS. All experiments were performed in accordance with guidelines established by the RIKEN Animal Safety Committee.
Generation of Bcl6 conditional knockout mice.
Bcl6f mice were generated by targeted insertion of loxP sites to flank exon 7–9 of the Bcl6 gene (Fig. 1). To construct the targeting vector, we subcloned the genomic DNA fragment encoding exons 5–10 of Bcl6 gene from the C57BL/6 embryonic stem (ES) cell line Bruce4 (Köntgen et al., 1993) by PCR. A loxP site was inserted between exons 6 and 7, and the Gateway rfA cassette (Invitrogen) between exons 9 and 10. The diphtheria toxin A selection cassette from pEZ-Frt-lox-DT was joined at the 3′ adjacent position of exon 10. The inserted Gateway rfA cassette was then replaced with a fragment containing loxP site and combined with an FRT-flanked neo-resistance cassette by a recombination reaction using LR clonase (Invitrogen). The targeting construct was transfected into Bruce4 ES cells by electroporation. G418-resistant colonies were expanded, and the appropriately targeted clones were screened by PCR and Southern blot analysis. Established ES clones were injected into blastocysts from BALB/c mice to produce chimeric mice. The pGK-Neo cassette was removed from the mouse germline by breeding heterozygous mice to FLPe-expressing deleter mice (Kanki et al., 2006). Bcl6f/+ mice were crossed to mb1-cre mice, Cγ1-cre mice, or CD4-cre mice to obtain conditional knockout strains.
Flow cytometric analysis for memory and GC B cells.
This was performed as previously described (Takahashi et al., 2001, 2005). In brief, to prepare single cell suspension, spleens were minced and incubated with 200 U/ml collagenase IV (Sigma-Aldrich) and 20 µg/ml DNase I (Roche) for 30 min at 37°C. After washing, splenocytes were depleted of red blood cells and incubated with anti-FcγRII/III mAb (2.4G2; American Type Culture Collection). For memory and GC B cell analysis, cells were incubated with a mixture of biotinylated mAbs against IgM, IgD, CD3, CD5, CD90, TER119, Gr-1, F4/80, DX5, AA4.1, and NK1.1 (eBioscience), followed by staining with anti-CD38 (CS2; Inamine et al., 2005) conjugated with Alexa Fluor 647 (CD38AlexaFluor647), anti-B220 conjugated with PE-Cy7 (anti-B220PE-Cy7; eBioscience), anti-IgG1 conjugated with Pacific Blue (anti-IgG1PacificBlue; BD), PNA conjugated with FITC (PNAFITC; Vector Laboratories), (4-hydroxy-5-iodo-3-nitrophenyl)acetyl (NIP)-BSA conjugated with PE (NIP-BSAPE), and streptavidin conjugated with PE–Texas Red (streptavidinPE-TexasRed; BD). In C57BL/6 mice (IgHb haplotype), λ-bearing B cells responsive to NP generally produce antibodies that have higher affinities for analogues of NIP than for NP itself (heteroclicity; Reth et al., 1979). Therefore, we used NIP-BSAPE, instead of NP-BSA, for detection of NP-specific B cells in the spleen of immunized and nonimmunized mice. Cells were washed, resuspended in a staining buffer containing 1 µg/ml propidium iodide, and analyzed using a FACSAria (BD). To improve the accuracy of lymphocyte subset analysis, only cells exhibiting forward and large-angle scatter typical of lymphocytes (the lymphocyte gate; eliminating monocytes and granulocytes) were analyzed (Hayakawa et al., 1987; Takahashi et al., 2001). For all experiments reported here, only cells negative for streptavidinPE-TexasRed and PI staining were gated for further analysis. More than one million total events were collected for each file in and then analyzed using FlowJo software (Tree Star).
Analysis of memory and GC B cells of BM chimeric mice.
Splenocytes from NP-CG immunized mice were incubated with a mixture of biotinylated mAbs, as described in the previous section, followed by staining with anti-CD45.1APC (eBioscience), anti-IgG1PacificBlue, anti-B220 conjugated with efluor605 (anti-B220efluor605; eBioscience) or anti-B220 conjugated with V500 (anti-B220V500; BD), anti-CD38 conjugated with PE-Cy7 (anti-CD38PE-Cy7; BioLegend), NIP-BSAPE, PNAFITC, and streptavidinPE-TexasRed at day 7 after immunization. For analysis of day 40 memory and GC B cells, cells were stained with anti-CD45.1APC, anti-IgG1PacificBlue, anti-CD38PE-Cy7, NIP-BSAPE, anti-B220FITC, and streptavidinPE-TexasRed. In single cell sorting for VH sequence analysis, Igκ−IgM−IgD− B cells were selected using a MACS system (Miltenyi Biotec) as described in Takahashi et al., (2001), followed by staining with fluorescent reagents, including anti-CD45.1APC.
Analysis of surface antigens on memory B cells.
Splenocytes were incubated with a mixture of biotinylated mAbs as described in flow cytometric analysis, followed by staining with anti-B220V500, anti-CD38PE-Cy7, NIPAPC, NIP-BSAPE, PNAFITC, or anti-IgG1PacificBlue for immunized mice or anti-IgMPacificBlue for unimmunized mice and streptavidinPE-TexasRed. In addition anti-PD-L2PE, anti-CD73PE, anti-CD80PE, anti-CD35AlexaFluor647, anti-CD40APC, or anti–MHC class IIAPC mAbs were stained.
Analysis of BM B cells.
BM B cells were prepared from Bcl6+/+, Bcl6+/f, and Bcl6f/f mice heterozygous for mb1-cre. BM cells were incubated with biotinylated mAbs against CD3, CD5, CD90, TER119, Gr-1, CD11b, DX5, and NK1.1, followed by staining with anti-B220PE-Cy7, anti-IgMFITC, anti-AA4.1APC, anti-CD24PacificBlue, anti-CD43PE, and streptavidinPE-TexasRed for FACS analysis. BM B220+ cells were separated into AA4.1+ and AA4.1− cells (immature and mature B cell populations, respectively; Allman et al., 2001) by FACS and the frequency of prepro–B cells (CD24lowCD43hi), pro–B cells (CD24intCD43int), pre–B cells (CD24hiCD43lo), and IgM+ cells in immature (AA4.1+B220+) and mature B cell populations (AA4.1−B220+) was analyzed (Nagaoka et al., 2000).
Analysis of FO and MZ B cells.
Splenocytes from unimmunized mice were incubated with biotinylated mAbs against CD95, CD138, CD43, CD3, CD5, CD90, TER119, Gr-1, CD11b, DX5, and NK1.1, followed by staining with B220PE-Cy7, CD21FITC, AA4.1APC, and IgMPacificBlue streptavidinPE-TexasRed. Viable B220+AA4.1− cells were separated into FO (IgMintCD21int) and MZ (IgMhighCD21high) B cells.
Analysis of Tfh cells in mice with conditional Bcl6 deletion through CD4-cre.
Splenocytes from NP-CG immunized mice were stained with anti-CXCR5APC (BD), anti-PD1PE (BioLegend), anti–TCR-βPacificBlue (BioLegend), anti-B220PE-Cy7, and anti-CD4FITC (BD). Viable B220−TCRβ+CD4+ T cells were analyzed for CXCR5 and PD1 expression by FACS.
Cell purification.
To purify memory and GC B cells, splenocytes from NP-CG immunized mice were incubated with a mixture of biotinylated mAbs as described in Flow cytometric analysis for memory and GC B cells, followed by incubation with streptavidin microbeads (Miltenyi Biotec). Thereafter, the cells negatively selected by the MACS system (Miltenyi Biotec) were stained with anti-CD38AlexaFluor647, anti-B220PE-Cy7, anti-IgG1PacificBlue, PNAFITC, NIP-BSAPE, and streptavidinPE-TexasRed. In single cell sorting for VH sequence analysis, biotinylated anti-Igκ (BD) was added into a mixture of biotinylated mAbs or anti-IgλFITC (BD) was used instead of PNAFITC. To purify plasma cells, splenocytes from NP-CG immunized mice were incubated with a biotinylated Ab cocktail and streptavidin microbeads. Thereafter, the cells negatively selected by the MACS system were stained with anti-CD138APC (BD), anti-B220PE-Cy7, anti-IgG1PacificBlue, anti-IgλFITC, NIP-BSAPE, and streptavidinPE-TexasRed. To purify FO and MZ naive B cells, splenocytes from unimmunized mice were incubated with biotinylated mAbs against as described in Analysis of FO and MZ B cells, followed by incubation with streptavidin microbeads. Thereafter, the cells negatively selected by the MACS system were stained with B220PE-Cy7, CD21FITC, CD23PE, AA4.1APC, and streptavidinPE-TexasRed. Viable B220+AA4.1− cells were separated into CD21intCD23+ FO and CD21hiCD23− MZ B cells.
BrdU incorporation assay.
Mice were injected intraperitoneally with 0.6 mg BrdU (Sigma-Aldrich) twice per day from days 4 to 6 after immunization with NP-CG/alum. On days 7, 20, and 40 after immunization, NP-specific/B220+/IgG1+ memory B cells (CD38+/PNAlow) and GC B cells (CD38dull/PNAhigh) were purified under the exclusion of dead cells by FACS from the pooled spleens of immunized mice and then mounted on a glass slide, fixed, and stained with FITC-labeled anti-BrdU mAb or FITC-labeled isotype control mAb (BD) and DAPI, or biotinylated anti-BrdU mAb (Abcam), followed by staining with streptavidinAlexaFluor546 and DAPI. The frequency of stained cells was determined by confocal microscopic inspection of >100 B220+ cells, as previously described (Kimoto et al., 1997; Takahashi et al., 2001; Toyama et al., 2002), and the data were expressed as percentage of B220+ cells that exhibited nuclear BrdU incorporation (SP2AOBS; Leica).
Cell cycle analysis of memory B cells.
Splenocytes were prepared from conditional Bcl6-deficient mice and controls at days 7 and 40 after immunization and incubated with 10 µg/ml Hoechst 33342 solution (Dojindo) for 45 min at 37°C, followed by incubation with a mixture of biotinylated mAbs as described in Flow cytometric analysis for memory and GC B cells. Thereafter, the cells were stained with anti-B220PE-Cy7, anti-CD38AlexaFluor647, anti-IgG1FITC (BD), NIP-BSAPE, and streptavidinPE-TexasRed for FACS analysis. For cell cycle analysis in memory and GC B cells in immunized WT mice, we used Fucci transgenic mice, which constitutively express monomeric Azami Green fused to partial human geminin protein (Sakaue-Sawano et al., 2008). Splenocytes from NP-CG immunized Fucci transgenic mice were incubated with a mixture of biotinylated mAbs and streptavidin microbeads. Thereafter, the cells negatively selected by the MACS system (Miltenyi Biotec) were stained with anti-B220efluor605, PNA conjugated with Alexa Fluor 647 (PNAAlexaFluor647; Invitrogen), anti-CD38PE-Cy7, anti-IgG1PacificBlue, NIP-BSAPE, and streptavidinPE-TexasRed for FACS analysis.
Analysis for early memory B cell development.
To study the generation of GC-independent memory B cells from naive B cell precursors, we used mice in which the frequency of NP-specific naive B cells is enhanced through the knockin of a rearranged, mutant V186.2 gene segment encoding high-affinity binding for the hapten NP into the IgH locus (B1-8hi; Shih et al., 2002). Splenic B cells of these mice, which bear the CD45.1 genetic marker on their lymphocytes were transferred into syngeneic CD45.2 recipients and immunized with NP-CG. This causes synchronous activation of many NP-specific donor B cells, allowing us to dissect their early response in vivo. 3, 4, and 7 d after immunization, Igκ− B cells in the immunized recipients were analyzed or enriched by MACS and subjected to FACS and qPCR analyses. For staining intracellular Bcl6, MACS-enriched cells were first stained with Fixable Viability Dye eFluor780 (eBioscience), followed by staining for surface antigens. Cells were then fixed, permeabilized using the Foxp3 staining buffer set (eBioscience), and stained with anti-Bcl6PE mAb (BD).
Immunohistofluorescence assay.
Immunohistofluorescence assay was performed as described previously (Inamine et al., 2005).
B cell stimulation.
Splenic B cells from individual mice (n = 3) were purified using a MACS system and stimulated in triplicate with 10 µg/ml anti–mouse IgM F(ab’)2 (Jackson ImmunoResearch Laboratories) and 2 µg/ml rat anti–mouse CD40 mAb (eBioscience) or 20 µg/ml LPS (Sigma-Aldrich) for 3 d. The cytokines levels in the culture supernatants were determined by Bio-Plex cytokine multiplex assay (Bio-Rad Laboratories) according to the manufacturer’s instructions.
DNA microarray analysis.
The DNA microarray analysis for memory B cells from wild-type mice was performed with Affymetrix GeneChip Mouse 430 2.0 Arrays. The total RNAs were extracted using TRIzol reagent (Invitrogen). RNA samples were labeled using an Ovation RNA Amplification System V2 and FL-Ovation cDNA Biotin Module V2 kits (Nugen) for memory B cell analysis (Fig. 6). Image files were scanned and processed by GCOS (GeneChip Operating Software) and the microarray data were normalized with GCRMA. A cluster analysis was performed using pvclust (Suzuki and Shimodaira, 2006) as follows: Pearson’s correlation coefficients in each pair of the gene expression profiles were calculated to make a dendrogram for the samples. The AU (approximately unbiased) p-value (percentage) for each branch of the tree was calculated and placed on a branch of a cluster dendrogram. If a branch had a value >99, we considered that cluster of samples as significantly separated from the other samples (Fig. 6 A). Memory B cell–specific genes were identified with two distinct statistical methods (Fig. S3), Tukey’s multiple comparison test and ROKU, a tissue-specific gene identification method (Kadota et al., 2006). If both methods labeled a specific gene as highly expressed in memory B cells, that gene is considered as a memory-specific gene.
qRT-PCR analysis.
Total RNA was purified from B cell subsets and cDNAs were synthesized using a SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen) according to the manufacturer’s instruction. qRT-PCR was performed in triplicate with Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen) using an ABI Prism7500 instrument (Applied Biosystems). Standard curves were created by serial dilution of cDNA synthesized from naive splenocytes. The mRNA level for each gene was normalized to the expression of β-actin mRNA. PCR conditions were as follows: 50°C for 2 min, 95°C for 2 min, 40 cycles of 95°C for 15 s, 60°C for 45 s, and then the dissociation stage to confirm the presence of a unique PCR product. Primer sequences are listed in Table S1.
Sequence analysis of the heavy chain variable region of NP-specific B cells.
Single NP-specific/IgG1+/Igλ+ memory B cells and GC B cells were sorted directly into 10 µl water containing 50 ng of carrier RNA (QIAGEN). 15 µl of the RT-PCR mixture (SuperScript One-Step High Fidelity kit; Invitrogen) was added and subjected to two rounds of nested PCR using primer pairs consisting of V186.2 sense (5′-TTCTTGGCAGCAACAGCTACA-3′) and Cγ1 external antisense (5′-GGATCCAGAGTTCCAGGTCACT-3′) or V186.2 sense and Cμ external antisense (5′-AAATGGTGCTGGGCAGGAAG-3′). 1 µl of the first RT-PCR product was used for the second round of PCR that used primer pairs consisting of V186.2 sense and Cγ1 internal antisense (5′-GGAGTTAGTTTGGGCAGCAG-3′) or V186.2 sense and Cμ internal antisense (5′-AGCCCATGGCCACCAGATT-3′) and Platinum Pfx DNA polymerase (Invitrogen). The PCR products were purified and directly sequenced using Cγ1 internal antisense and/or Cμ internal antisense primers. The PCR cycle conditions and primer sequences used in this study are listed in the Supplemental material. To estimate the Taq-induced misincorporation rate under these single cell RT-PCR conditions, we sequenced 72 VH186.2-Cμ segment transcripts from NP-specific/Igλ+/IgM+ B cells from the pooled spleens of nonimmunized C57BL/6 mice. Two errors were observed in a total of 21,168 VH186.2 nucleotides, which corresponds to an error rate of 9.5 × 10−5. This error rate would correspond to 0.03 mutations per VH186.2 region, indicating that PCR error mutations are negligible in interpreting our results.
Adoptive transfer experiment.
Adoptive transfer experiments were performed as described previously (Takahashi et al., 2001, 2005; Inamine et al., 2005).
ELSIPOT assay.
ELISPOT assays were performed as described previously (Takahashi et al., 2001).
Statistical analysis.
Student’s t test and the Mann-Whitney nonparametric (two-tailed) test were used with KaleidaGraph 4.0 software (Synergy Software). P < 0.05 was considered to indicate a significant difference.
Accession nos.
Complete sequence data are available from the DNA Data Bank of Japan (DDBJ), the EMBL Nucleotide Sequence Database, and GenBank under the following accession nos.: memory and GC B cells from Bcl6 (+/+) or (f/f) mice heterozygous for Cγ1-cre (Fig. 2), AB517255–AB517374; memory and GC B cells from Bcl6 (+/+) or (f/f) mice heterozygous for mb1-cre (Fig. 3), AB517382–AB517552, AB571337–AB571477, and AB589136–AB589232; memory and GC B cells from Bcl6 (+/+) or (f/f) mice carrying CD4-cre (Fig. 7), AB677961–AB678197 and AB678233–AB678346; memory and GC B cells from BM chimeras (Fig. 3), AB574252–AB574328 and AB589233–AB589329; WT memory B cells (day 7–100; Fig. 4 C), AB370693–AB370799, AB370836–AB370868, and AB516477–AB516495; WT GC B cells (day 20–70), AB370432–AB370548; memory and GC B cells from MR1-treated, nIg-treated, or control mice (Fig. 4 F), AB516549–AB516653 and AB588990–AB589135; and CD73− and CD73+ memory B cells and CD73+ GC B cells (Fig. 8 B), AB705493–AB705620. All microarray data have been deposited in the NCBI’s GEO database under the accession no. GSE11961.
Online supplemental material.
Fig. S1 shows the strategy for analysis of NP-specific B cell subpopulations. Fig. S2 shows cell cycle analysis of memory B cells. Fig. S3 shows microarray analysis of memory B cells. Table S1 shows primers for qRT-PCR.
Acknowledgments
We are grateful to Atsushi Miyawaki and Asako Sakaue-Sawano for Fucci-expressing transgenic mice, Michael Reth for mb1-cre mice, Michel Nussenzweig for B1-8hi mice, Garnett Kelsoe for anti-CD40L mAb, and Yuichi Aiba for discussion.
K. Rajewsky is supported by the National Institutes of Health of the USA.
The authors declare no competing financial interests.